Abstract
Strigolactones (SLs) are endogenous hormones and exuded signaling molecules in plant responses to low levels of mineral nutrients. Key mediators of the SL signaling pathway in rice include the α/β-fold hydrolase DWARF 14 (D14) and the F-box component DWARF 3 (D3) of the ubiquitin ligase SCFD3 that mediate ligand-dependent degradation of downstream signaling repressors. One perplexing feature is that D14 not only functions as the SL receptor but is also an active enzyme that slowly hydrolyzes diverse natural and synthetic SLs including GR24, preventing the crystallization of a binary complex of D14 with an intact SL as well as the ternary D14/SL/D3 complex. Here we overcome these barriers to derive a structural model of D14 bound to intact GR24 and identify the interface that is required for GR24-mediated D14-D3 interaction. The mode of GR24-mediated signaling, including ligand recognition, hydrolysis by D14, and ligand-mediated D14-D3 interaction, is conserved in structurally diverse SLs. More importantly, D14 is destabilized upon the binding of ligands and D3, thus revealing an unusual mechanism of SL recognition and signaling, in which the hormone, the receptor, and the downstream effectors are systematically destabilized during the signal transduction process.
Keywords: Strigolactones, D14, D3, GR24, receptor/co-receptor
Introduction
Strigolactones (SLs) are endogenous hormones that in response to poor soil nutrients inhibit lateral shoot branching. SLs also function as signal molecules secreted into the soil to promote root colonization by arbuscular mycorrhizal fungi and to stimulate symbiosis with nitrogen-fixing bacteria1,2,3,4,5. The secreted SLs are also recognized and exploited as germination signals by parasitic weeds that cause devastating crop losses in developing countries6,7.
Extensive genetic studies in Arabidopsis, pea, and rice using SL-deficient and -insensitive mutants have identified components of a conserved SL signaling pathway8,9,10,11,12,13, including rice dwarf 14 (d14) and dwarf 3 (d3). Due to the homology between D3 and the auxin and jasmonate receptors, TIR1 and COI1, and between D14 and the gibberellin receptor GID1 and the putative karrikin receptor KAI2, it had been speculated that both D3 and D14 could be candidates for the SL receptor14. In particular, gibberellin signaling involves direct hormone binding to its receptor, the α/β hydrolase-fold protein GID1, which promotes the formation of a complex consisting of gibberellins, GID1, and DELLA transcriptional repressors. This complex is then recognized by the SCFSLY1/GID2 complex, which ubiquitinates DELLA proteins for proteasomal degradation15,16. As GID1 and D14 are members of the α/β-hydrolase family while GID2 and D3 are members of the F-box family, D14 and D3 are assumed to function in SL signaling in a manner similar to that of GID1 and GID2 in gibberellin signaling (reviewed in17).
Perception of and signaling by karrikins, compounds originally identified in the smoke of forest fires, are also associated with D3 and KAI2, an α/β hydrolase that is highly homologous to D1418. Recent studies have identified the putative transcriptional repressor D5319,20 as well as the transcription factor BES1 as SCFD3 substrate targets in response to SL exposure and the D53 paralog SMAX1 in response to karrikin exposure. SLs induce association of D14 with D3/D53 and D3/BES1 complexes and promote SCFD3-mediated degradation of D5319,20, BES121, and D14 itself22.
However, while GID1 is catalytically inactive and KAI2 had been proposed to be incapable of hydrolyzing karrikins23, D14 is catalytically active toward SLs. It can hydrolyze the synthetic SL GR24, albeit with an extremely low turnover rate of only about one molecule GR24 for every one molecule of D14 every three minutes24 or less25, and this hydrolysis has been proposed to be required for SL signaling3,26. Several groups reported high-resolution crystal structures of apo-D14 and its orthologs24,25,26,27 and of KAI2 in apo-23,24,27,28 and karrikin-bound23 states. In contrast, attempts to co-crystallize D14 in complex with GR24 only yielded structures of D14 covalently bound to a GR24 hydrolysis intermediate24 or weakly bound to the butenolide ring (D ring) hydrolysis product26. Similarly, all attempts to crystallize a D14/SL/D3 complex have failed. It therefore remains unknown how D14 recognizes natural and synthetic SLs and how SLs mediate D14-D3 interaction. Here we have used X-ray crystallography, computational modeling, hydrogen-deuterium exchange mass spectrometry (HDX), molecular dynamics (MD), and extensive mutational analyses to determine a detailed model of GR24 binding to the D14 ligand-binding pocket (LBP) and to map binding of D3 to a contiguous surface surrounding the D14 pocket entrance. These results reveal an unexpected mechanism by which key signaling components including the hormone, the receptor, and the downstream effectors are systematically destabilized during the signal transduction process, highlighting the transient nature of SL signaling.
Results
Recombinant D14 binds GR24 or the GR24 D-ring with μM affinity
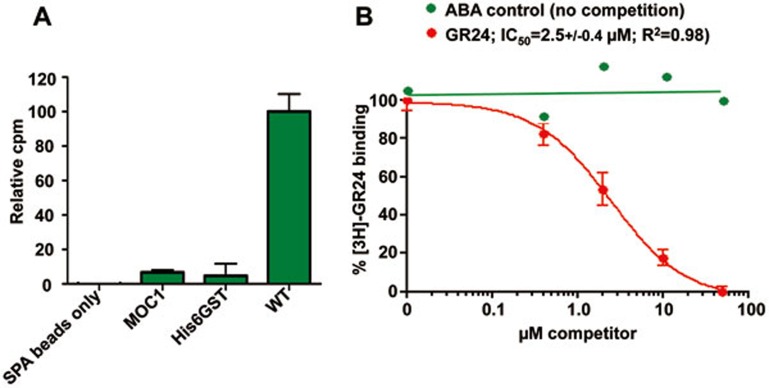
We demonstrated and quantitated the direct binding of GR24 to D14 in a scintillation proximity assay (SPA) using commercially available [3H]-GR24 (Figure 1A). While the free D-ring of GR24 is biologically inactive and does not bind to D14, GR24 hydrolysis proceeds through formation of a covalently bound D-ring-derived reaction intermediate24. Since the radiolabel is localized to the D-ring, the SPA signal could be due to binding of either intact GR24 or the hydrolysis intermediate or product. This binding is specific and reversible as it was readily competed by unlabeled GR24, but not by the unrelated plant hormone ABA, with an IC50 of 2.5 μM (Figure 1B), in agreement with ITC measurements of the Arabidopsis ortholog of D1427.
Figure 1.
Direct binding of GR24 to D14. (A) SPA radio-ligand binding assay of [3H]-GR24 to D14. MOC1 and His6GST tag were included as specificity controls. All binding data were normalized to the SPA bead background signal in the absence of any bound protein; n = 3, error bars = SEM. (B) Competition of the D14- [3H]-GR24 binding by unlabeled GR24 (red) or ABA (green); n = 3, error bars = SEM (n = 1 for the ABA control).
Crystal structure of the complex formed between D14 and GR24
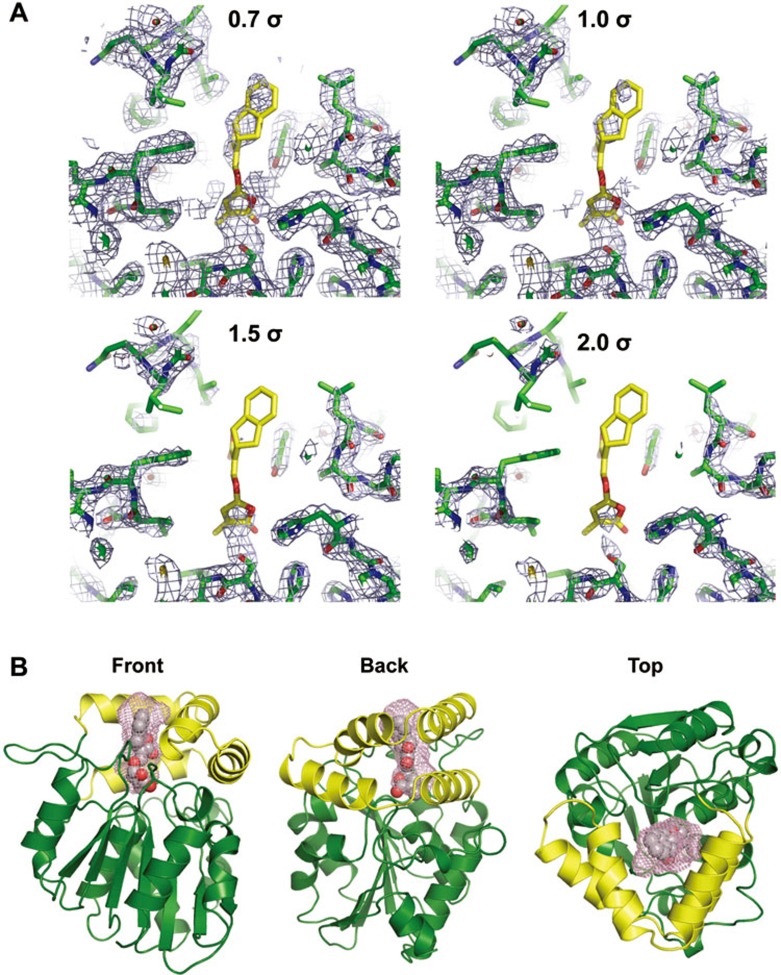
We and others have previously solved the crystal structures of apo-D14 (51-318)24,25,26,27 and the complex of D14 covalently bound to the GR24 hydrolysis intermediate24 or bound to the free, hydrolyzed D-ring26. In order to trap the complex of D14 with intact GR24, we co-crystallized the complex at a high molar ratio of GR24 to D14 and extensively screened crystallization conditions. In addition, initial crystals were further soaked with fresh GR24 to replenish hydrolyzed GR24, which allowed us to isolate well-diffracting crystals and to solve the D14+GR24 structure at a resolution of 2.4 Å (Supplementary information, Table S1). However, while the D14 protein was well resolved, GR24 was poorly resolved, indicating its incomplete occupancy. While we could readily assign density to the GR24 D-ring, in simulated annealing composite omit maps we only detected trace density for the remainder of GR24 (Figure 2A and Supplementary information, Figure S1). The model of the ligand based on the density is most consistent with the structure containing a mixture of intact GR24 and the GR24 D-ring hydrolysis product. We therefore used a combination of chemical constraints and electron density map to derive the final model as shown in Figure 2B. Similar to apo D14, D14 in the complex shows the characteristic α/β-hydrolase fold with an open LBP that is surrounded by the separate open lid domain consisting of two parallel layers of V-shaped helices and an extended loop. In this model, GR24 resides in the LBP with its D-ring facing the catalytic triad at the bottom of the pocket (Figure 2), whereas the A-ring partially protrudes out of the pocket and is directly exposed to the solvent (Figure 3A).
Figure 2.
Structure of the D14-GR24 complex. (A) Simulated annealing 2Fo-Fc electron density omit maps surrounding GR24 and the ligand-binding pocket, contoured at increasing σ levels. (B) The overall structure of the D14-GR24 complex in three different orientations, showing an open pocket that is surrounded by an open lid domain (yellow) consisting of double layer V-shaped helices. The pocket surface is shown as purple mesh and the modeled GR24 as van der Waals spheres.
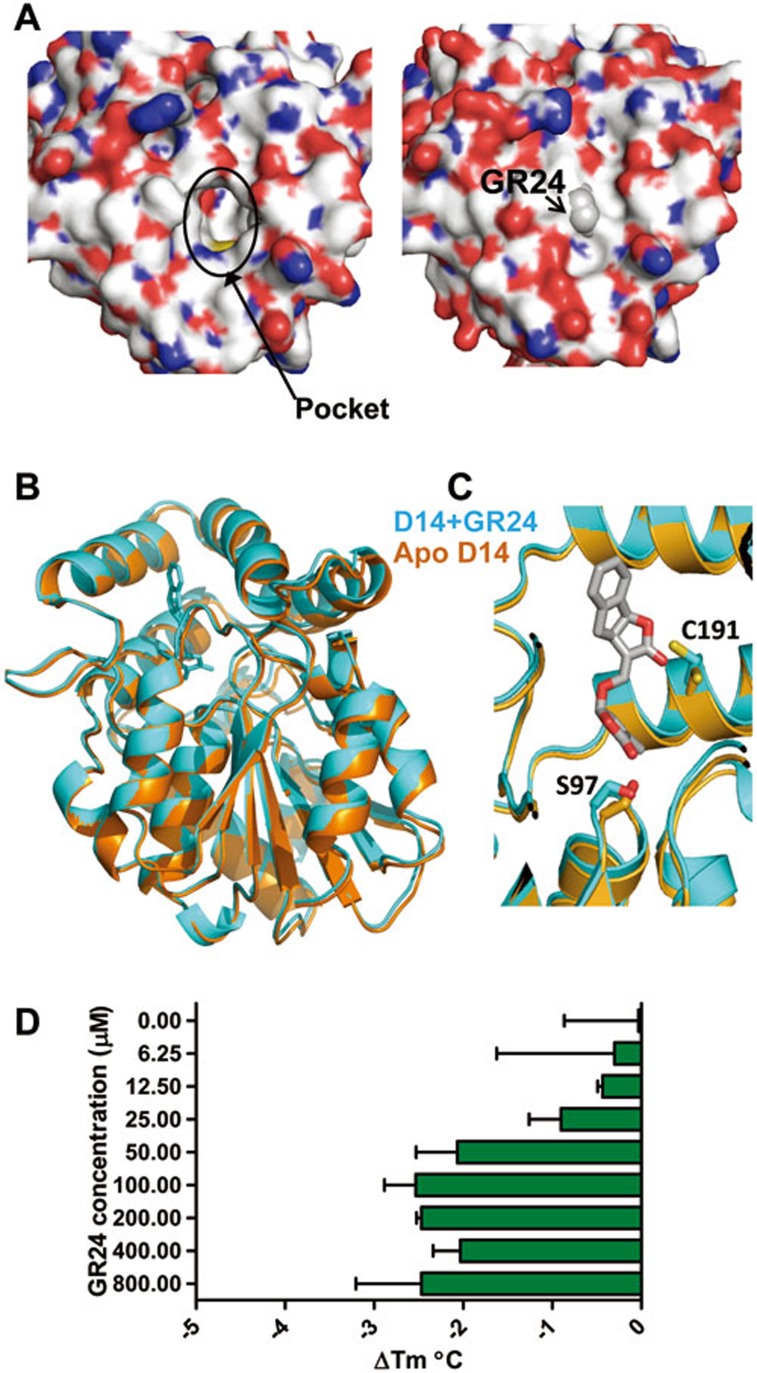
Figure 3.
GR24 binding induces only very minor conformational changes in D14. (A) Surface topology difference between apo- (left panel) and ligand bound- (right panel) D14 structures. (B) Structure overlay of apo D14 (brown) and GR24-bound D14 (cyan). (C) Close-up view of the side chains of S97 and C191, which are rearranged by GR24 binding. (D) The effect of increasing concentrations of GR24 on the thermal stability of D14 as determined by thermal stability shift assay (TSA). The bars represent the change in Tm (midpoint temperature between native and unfolded protein) relative to apo D14 (n = 3, error bars = SD).
Structural comparison between apo-D14 and the D14/GR24 complex revealed that their conformations showed little difference (Figure 3A, 3B), except for (i) flexible charged surface residues whose conformations also differ among the apo-D14 structures (Supplementary information, Figure S2), and (ii) C191 (amino acid numbers refer to the conserved hydrolase fold) and the catalytic residue S9729 deep inside of the LBP, which have become constrained by GR24 binding (Figure 3B and 3C). Similarly, comparison of the D14 apo-structure with the structure of D14 bound to the GR24 hydrolysis intermediate or the D-ring hydrolysis product showed almost identical surface conformations (Supplementary information, Figure S3A). We also analyzed D14 and D14+GR24 by HDX (Supplementary information, Figure S4). HDX measures the accessibility of the backbone amide hydrogen atoms to deuterium exchange in solution, which is closely linked to protein structure and dynamics30. In spite of excellent peptide coverage, addition of 100 μM GR24 failed to cause any significant changes in HDX of D14, consistent with the observed absence of major surface conformational changes.
The overall surface topology thus appears to be the same among the four different states of D14. Yet, it was reported that GR24 binding to the D14 homolog from petunia25, DAD2, induces a marked decrease (9 °C) in DAD2 melting temperature, suggestive of a GR24-mediated conformational destabilization25. We therefore also determined the thermostability of D14 and detected a very small, but reproducible, effect of GR24 on the D14 melting temperature (2.3 °C Tm decrease at [GR24] ≥ 50 μM; Figure 3D). To test whether the large differences in the effect of GR24 on thermostability are due to the difference between D14 and DAD2 or difference in the methods used, we determined hydrolytic activity and thermostability of DAD2. As shown in Supplementary information, Figure S5, our data confirm GR24 hydrolysis by DAD2 as well as a decrease in DAD2 melting temperature (9 °C Tm decrease at [GR24] ≥ 200 μM). Collectively, these results indicate that GR24 binding can markedly destabilize DAD2, but only has a mild effect on D14 conformation.
Molecular recognition of GR24 by D14
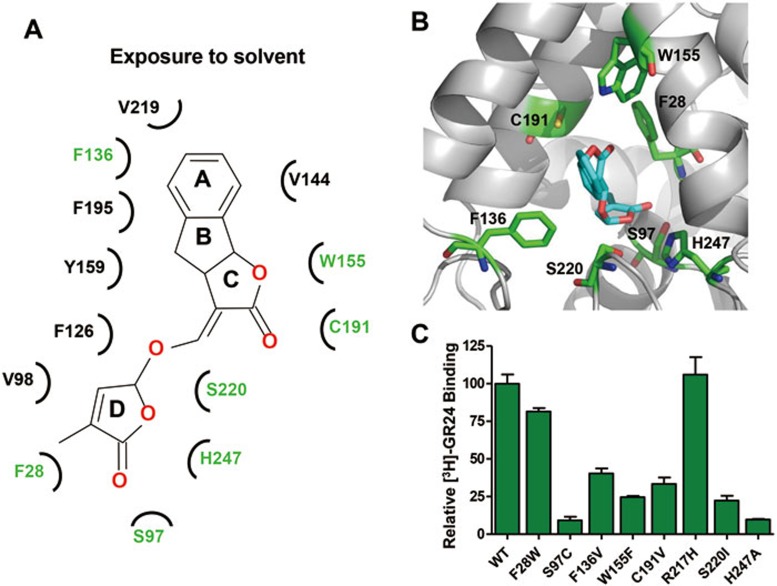
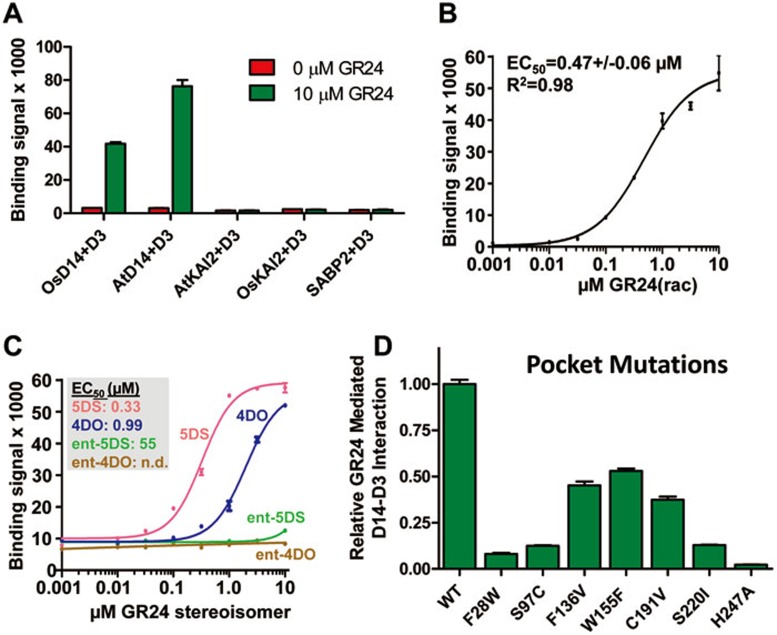
The bound GR24 molecule in the model is nested in the pocket and engaged in specific hydrogen bonds and hydrophobic interactions with pocket residues of D14 (Figure 4A-4B and Supplementary information, Figure S6). We designed a series of mutations for each pocket residue to validate the structural basis of ligand recognition by D14. Mutant D14 proteins that maintained sufficient stability and could be purified as monodisperse species by size exclusion chromatography (indicating that they were not grossly misfolded) were purified to homogeneity for SPAs to measure their ability to bind the ligand (Figure 4C). Most of these mutations reduced or even abolished the binding of D14 to GR24 (or its subsequent hydrolysis product), consistent with an important role of these residues in mediating direct ligand binding, while mutation R217H, which serves as control for a residue outside of the binding pocket, did not alter the binding ability of D14 with GR24. In particular, mutations of S220 and the catalytic triad residues S97 and H247 at the bottom of the binding pocket (Figure 4 and Supplementary information, Figure S6), which are essential for hydrolase activity, nearly abolished the binding ability (Figure 4C). Overall, the data of mutation analyses are in excellent agreement with the binding model of GR24 in the D14 pocket.
Figure 4.
Molecular recognition of GR24 by D14. (A) Main molecular interactions between GR24 and D14 in a cartoon diagram. (B) Location of the pocket residues (green) whose mutations compromised GR24 binding to D14. (C) [3H]-GR24 binding to D14 mutant proteins with replacements in LBP residues.
Binding of GR24 inside of the D14 LBP is required for the interaction between D14 and D3
GR24 can induce an interaction between D14 and D3, which has been shown in cells by yeast 2-hybrid analysis25 and in extracts by pull-down assays19,20. To test whether GR24 directly stimulates an interaction between D14 and D3 in vitro, and to quantitatively analyze such an interaction, we used highly purified recombinant proteins in an AlphaScreen luminescence proximity assay. As shown in Figure 5A, GR24 induced the interaction between D3/ASK7 (D3 requires the SCFD3 complex component ASK7 for stability) and both rice and Arabidopsis D14. The interaction is specific as GR24 did not promote an interaction between D3/ASK7 and D14 paralogs from rice and Arabidopsis, OsKAI2 and AtKAI2, or an unrelated α/β-hydrolase, the salicylic acid-binding protein SABP2. We analyzed the stimulatory effect of GR24 by a dose-response curve, which demonstrated that the ligand can greatly enhance the interaction with an EC50 of 0.47 μM, further supporting the ligand-dependent interaction between D14 and D3 (Figure 5B). Moreover, the stimulatory effect was stereoisomer-specific. We purified small amounts of the four different GR24 stereoisomers and tested them separately for their ability to induce a D14-D3 interaction. Two of the four possible stereoisomers, designated GR245DS and GR244DO, have the same 2′R-configuration of the D-ring as found in all physiological SLs. These two isomers, and particularly GR245DS, are more active in D14-dependent bioassays than the other two isomers31. The same two isomers were selectively capable of stimulating the interaction between D14 and D3, with GR245DS being more potent than GR244DO (Figure 5C), thus closely mimicking the effects of GR24 isomers in plants31. The activity to stimulate the D14-D3 interaction further correlates with the energetically most favorable binding conformation as judged by computational docking (Supplementary information, Figure S7A), and the docking position of the 5DS isomer is most consistent with the GR24 model (Supplementary information, Figure S7B). Comparison of the relative stimulating activity of the isolated stereoisomers with that of the racemic (rac) GR24 mixture (EC50 = 0.47 μM) further indicates that most of the racGR24 activity is due to the 5DS isomer (EC50 = 0.33 μM).
Figure 5.
GR24 induces a direct interaction between D14 and D3. (A) AlphaScreen assay measuring the interactions between His8MBP-D3 and biotinylated OsD14, AtD14, AtKAI2, OsKAI2, and SABP2 in the presence or absence of 10 μM GR24 (n = 3, error bars = SD). (B, C) AlphaScreen data showing the ability of the GR24 stereoisomer mixture (GR24 rac) (B) or of pure GR24 stereoisomers (C) to promote a D14-D3 interaction in a concentration-dependent manner (n = 3, error bars = SD). (D) Proteins with mutations in the D14 LBP residues that compromise GR24 binding are also defective in GR24-mediated D14-D3 interaction. In the AlphaScreen assay, the interaction of wild-type D14 with D3 is normalized to 1.0 (n = 3, error bars = SD).
Next we tested whether the binding of GR24 to LBP is required for the effect of GR24 to stimulate the interaction of D14 with D3. We therefore analyzed the same pocket mutant proteins as shown in Figure 4C for their ability to interact with the D3/ASK7 subcomplex of SCFD3 in the presence of GR24, and found that indeed all pocket mutations reduced GR24-mediated interaction between D14 and D3/ASK7 (Figure 5D). The ability to design loss-of-function mutations of D14 further highlights the quality of our ligand-binding model and confirms that the interaction between D14 and D3/ASK7 requires binding of GR24 inside of the D14 LBP.
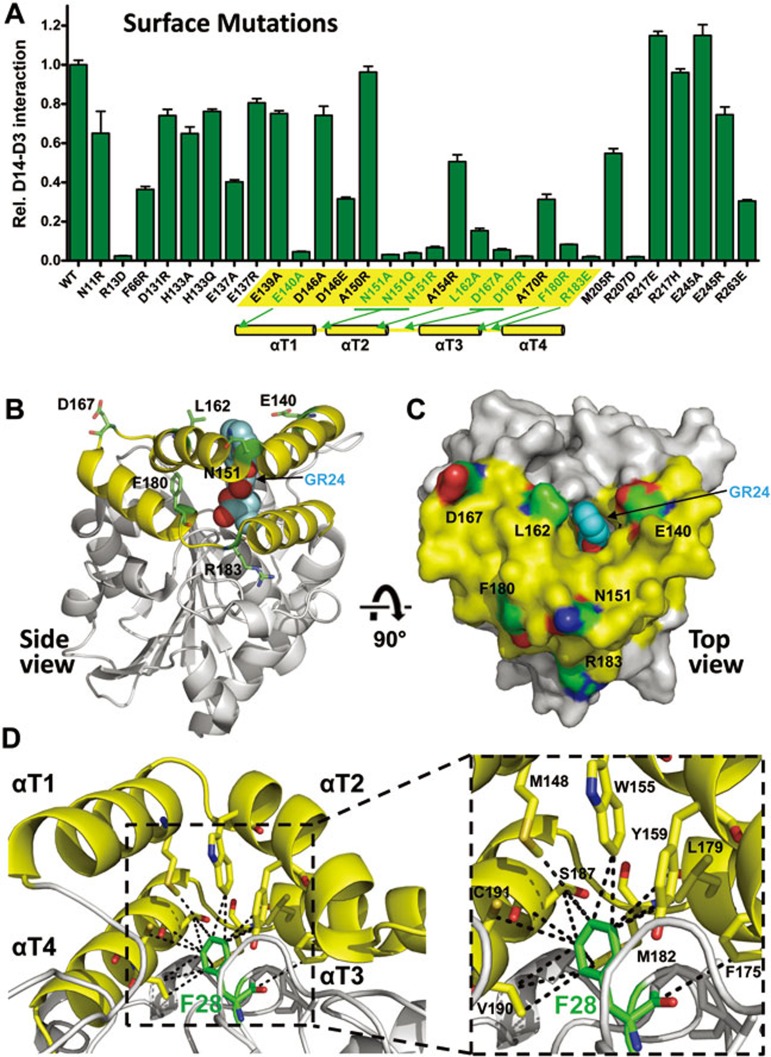
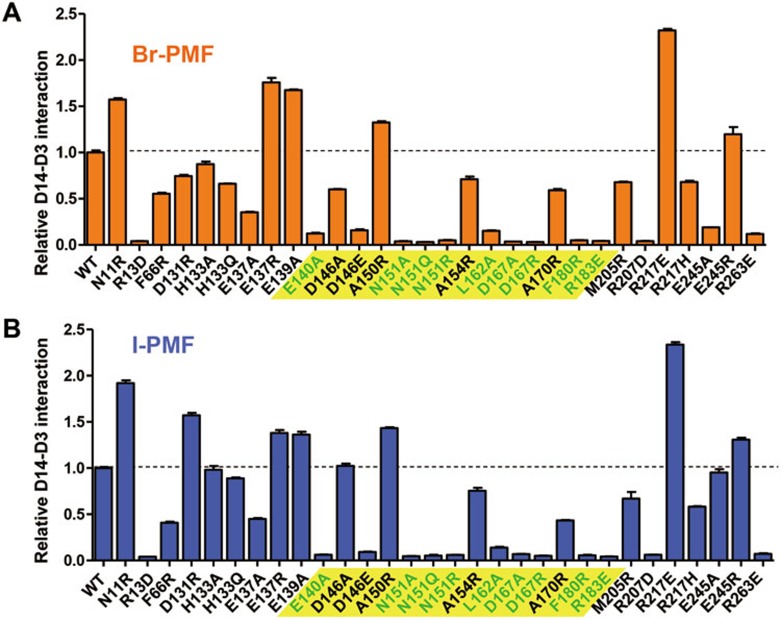
Mutational analysis suggests that D3 binds to and closes the entrance of the D14 LBP
To experimentally determine the interaction interface of D14 with D3/ASK7, we designed a set of 30 surface mutations of D14 and purified the corresponding mutant proteins. Surface mutations that robustly reduced the ability of D14 to interact with D3/ASK7 in the presence of GR24 involve residues surrounding the open ligand-binding pocket and clustering at the junctions between the helices of the lid domain and their connecting loops (Figure 6A-6C), indicating that GR24 binding induces interaction of D3 with the surface of the lid domain of D14. In this position D3 would block the entrance of D14 LBP and could directly interact with the ligand or its hydrolysis products. In addition to the above surface mutations, replacement of the pocket residue F28 with W also strongly reduced D3 interaction (Figure 5C) while only slightly affecting GR24 binding (Figure 4C). The bulky F28 centrally inserts into the lid domain and forms multiple hydrophobic interactions spanning all four lid helices (M148, W155, Y159, F175, L179, M182, S187, V190, and C191; Figure 6D), suggesting that F28 is critical for positioning of the lid helices.
Figure 6.
D14 surface residues that are required for D14-D3 interaction are clustered at the open lid domain surrounding the entrance of the LBP. (A) AlphaScreen interaction assay measuring the ability of D14 surface mutant proteins to interact with D3 in the presence of 10 μM GR24. The lid domain region is highlighted in yellow, with mutations that strongly compromised D14-D3 binding shown in green. The position of the four lid domain helices (αT1-αT4) is shown below the bar graph. (B) Side view ribbon presentation and (C) top view surface presentation of D14 lid domain residues whose mutations strongly compromised D14-D3 interaction. (D) F28 (green stick model) inserts into the lid domain and forms hydrophobic interactions with all four lid helices. Interacting amino acids are shown in stick presentation and are labeled in the close-up view.
Charge-reversal mutations in the lid domain loop cause constitutive D14-D3 interaction
In addition to these loss-of-function mutations, we also explored whether any of the D14 surface mutations might have a gain-of-function phenotype. When we measured D3 interaction in the absence of GR24, charge-reversal mutations of two acidic residues in the long flexible loop connecting the D14 lid domain to the core α/β-hydrolase fold allowed D3 interaction in the absence of SL (Supplementary information, Figure S8). While mutation of D14 E137 at the junction between the lid domain and the loop resulted in partially constitutive D14-D3 interaction, D131R exhibited an even higher D3 binding signal in the absence of ligand than in its presence. Together, these results suggest that the mutant loops of D14 can likely engage in a charge interaction with a negatively charged surface of lid domain-bound D3. Sequence alignment indicates that D14 residues that are important for D3 interaction are conserved across species (Supplementary information, Figure S9).
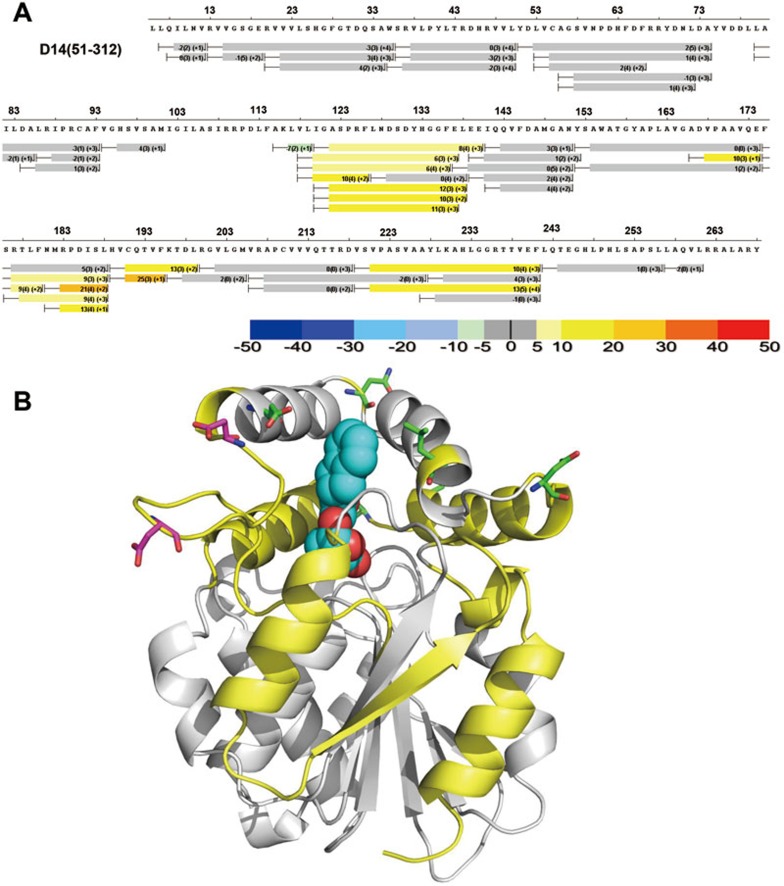
GR24-induced D3 binding destabilizes D14
To gain further insight into the D14-D3 interaction, we performed HDX on the complex consisting of D14, GR24, and D3-ASK7 and compared the profile with that of apo-D14. To our surprise, D3/ASK7 binding markedly increased deuterium exchange in D14 (Figure 7). This is in contrast to the fact that protein binding normally restrains the interaction interface, resulting in a decrease in deuterium exchange. This indicates that GR24-induced D3 binding substantially destabilizes D14, reminiscent of the subtle and strong thermo-destabilizations of D14 and DAD2, respectively, induced by GR24 binding (Figure 3D and Supplementary information, Figure S5B).
Figure 7.
GR24-induced D3 binding destabilizes D14. (A) HDX perturbation heat map comparing deuterium exchange in the D14/GR24+D3/ASK7 complex to exchange of apo D14. The bars below the sequence represent the peptide fragments resolved by mass spectrometry and the color of the bars represents the % change in HDX (positive numbers indicate increased HDX/destabilization). (B) Structural elements that become destabilized (increased HDX) upon GR24-induced D3 binding are colored in yellow. Lid surface residues whose mutation strongly reduces D3 binding are shown as green stick models. The two acidic loop residues whose charge-reversal mutations result in constitutive D14-D3 interaction are shown as magenta stick models.
A structurally distinct class of SL analogs possesses similar ability to stimulate interaction of D3 with various D14 mutants as GR24
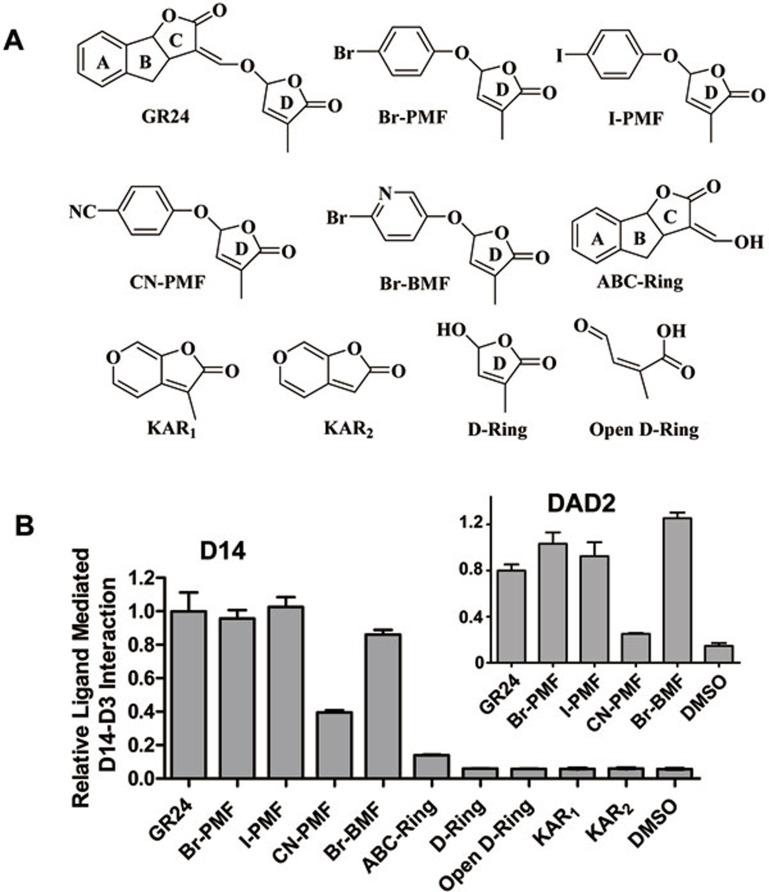
Natural SLs have been isolated from root exudates of various plants, especially those that are parasitized by the noxious weeds Striga and Orobanche spp. As a consequence, isolation and identification of natural SLs are extremely difficult and a series of new SL analogues have been derived from simple and cheap starting materials. The first successful series of analogues were the GR compounds, as is exemplified by GR24. Because of GR24's chemical instability and cost of production, new SL analogs that can be easily synthesized would have important agricultural applications. We show here that several of the new compounds that are active in SL signaling32 can be hydrolyzed by D14 (Supplementary information, Figure S10), can promote D14-D3 (Fig 8A-8B) and DAD2-D3 interactions (Figure 8B inlet), and destabilize D14 and DAD2 (Supplementary information, Figure S11). In contrast, karrikins and the ABC-ring and D-ring of GR24, which are inactive in SL signaling25,33,34, failed to promote D14-D3 interaction (Figure 8B).
Figure 8.
Structure and activity of GR24-like ligands that promote D3-D14 interaction. (A) Chemical structure of GR24, Br-PMF, I-PMF, CN-PMF, Br-BMF, ABC-ring, D-ring and karrikins (KAR1 and KAR2). All these compounds contain a conserved D-ring structure, which is required for SL function. PMF: 5-phenoxy-3-methylfuran-2(5H)-one; BMF: 3-methyl-5-(pyridin-3-yloxy)furan-2(5H)-one. (B) AlphaScreen assay showing interaction between D14 (inlet: DAD2) and D3 promoted by GR24-like ligands. The GR24-induced interaction between D14/DAD2 and D3 is normalized to 1.0 (n = 3, error bars = SD).
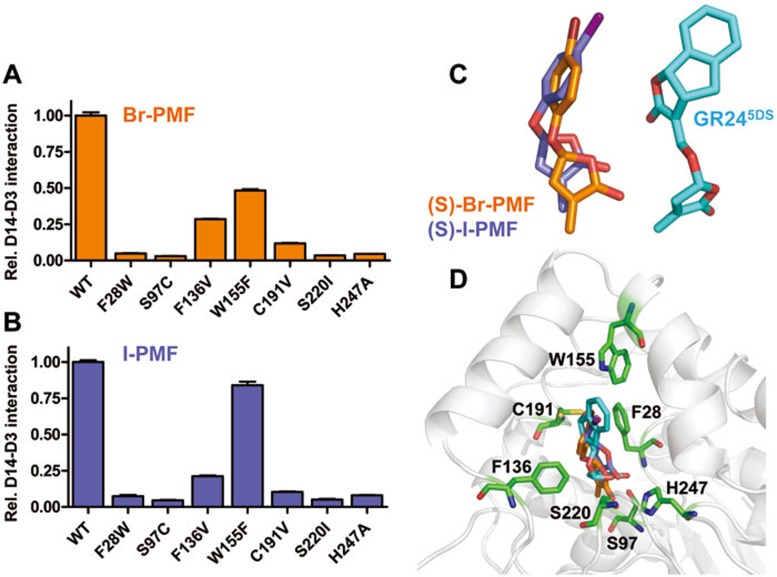
Given the substantially different scaffolds of these analogs, we reasoned that measuring their abilities to stimulate the interaction between D3 and our extended set of D14 mutant proteins might provide insight into the SL signaling mechanism. Upon stimulation with Br-PMF or I-PMF, the two more potent SL analogs, the mutant D14 proteins with either LBP mutations (compare Figures 9A and 9B with Figure 5D) or surface mutations (compare Figures 10A and 10B with Figure 6A) showed remarkably similar response patterns to those induced by GR24. Consistent with the importance of the interaction between D14 and intact SLs, all pocket residues whose mutations reduced D14-D3 interactions are in bonding distance with GR24 and with Br-PMF and I-PMF modeled in the binding pocket (Figure 9D).
Figure 9.
Effect of D14 pocket residue mutations on induction of the interaction with D3/ASK7 by Br-PMF and I-PMF. AlphaScreen interaction assay measuring the ability of D14 LBP mutant proteins to interact with D3 in the presence of 10 μM Br-PMF (A) or I-PMF (B). The interaction of wild-type D14 has been normalized to 1.0 (n = 3, error bars = SD). (C) Docking models of the biologically active (S)-Br-PMF and (S)-I-PMF stereoisomers next to the structure model for GR245DS in the LBP. (D) (S)-Br-PMF, (S)-I-PMF, and GR245DS models overlaid in the D14 LBP. LBP residues analyzed by mutagenesis are shown as stick models.
Figure 10.
Effect of D14 surface residue mutations on induction of the interaction with D3/ASK7 by Br-PMF and I-PMF. AlphaScreen interaction assay measuring the ability of D14 surface mutant proteins to interact with D3 in the presence of 10 μM Br-PMF (A) or I-PMF (B). The interaction of wild-type D14 has been normalized to 1.0 (n = 3, error bars = SD).
Br-PMF and I-PMF only share the butenolide D-ring and an ether linkage to an electron-rich group (aliphatic double bond or aromatic ring) with GR24. Unlike GR24, they would not protrude out of the pocket (Figure 9D) and would expose a more polar head group to solvent than GR24. Yet D3 interaction with all D14 surface mutant proteins was affected almost identically by each of the three ligands, strongly arguing that the three ligands induce D14-D3 interaction via the same mechanism in which the common D-ring and the cleavable ether linkage are key elements to stimulate this interaction.
Discussion
The ability of D14 to bind SLs and to interact with D3 in an SL-dependent manner illustrates a signaling mechanism similar to what was found in several other plant hormone signaling systems. However, the ability of D14 to hydrolyze the bound hormone is unique and represents a barrier to gaining structural insight into SL recognition and signaling. Using a combination of co-crystallization and ligand soaking, we solved the structure of D14 partially occupied by GR24 and the immediate GR24 hydrolysis products. In this structure model, the D-ring is localized at the bottom of the pocket in close proximity to the catalytic triad residues and the ABC-ring is partially retained in the pocket. This model is in full agreement with our mutational analysis of GR24-binding residues (Figure 4). We note that the position of the GR24 D-ring in our structure is in contrast to that in the recent structure of D14 bound to the final D-ring hydrolysis product, in which the D-ring was bound to the opening of LBP26, a position that is not stable based on our MD simulations (Supplementary information, Figure S12).
Our extensive mutational analysis of exposed D14 residues has allowed us to map the likely D3-binding surface on D14. Specifically, we demonstrate that the lid surface surrounding the LBP entrance contains residues whose mutations disrupt the interaction between D14 and D3, suggesting that D3 binds to this surface and blocks the pocket entrance. In further support, mutation of the non-exposed D14 residue F28 (F28W) almost completely abolishes the interaction of D14 with D3, but has only a minor effect on GR24 binding. The bulky F28 side chain inserts into the lid domain and makes interactions with all four lid helices and the lid hinge, consistent with the conformation of the lid being critical for D3 binding. Finally, gain-of-function mutations in the linker between the lid and the α/β-hydrolase domain allow a GR24-indenpendent interaction between D14 and D3.
How does GR24 binding induce a D14-D3 interaction? GR24 binding and the initial phase of GR24 hydrolysis do not induce noticeable conformational changes as the D14 structures in four different states (apo-D14 and D14 bound to GR24, the GR24 covalently bound hydrolysis intermediate, and the D-ring hydrolysis product) show almost identical surface conformations (Supplementary information, Figure S3A-S3B). In further agreement, GR24 binding does not induce changes in HDX protection (Supplementary information, Figure S4). While the intact GR24 ligand itself appears to provide a surface difference by partially exposing its hydrophobic A-ring, our analysis of other SL analogs suggests that exposure of a hydrophobic group in non-hydrolyzed SL is not required for inducing D14-D3 interaction (Figures 8 and 9) or for SL biological activity32. Our results suggest that ligand hydrolysis is required for the ability of GR24 to induce D14-D3 interaction. An important hint came from the observation that GR24 induces a subtle D14 destabilization in a thermostability shift assay and that subsequent formation of a complex with D3 induces a marked destabilization of the top and front of D14 as determined by HDX. Together with the requirement of a hydrolyzable ether bond and suitable leaving group, we speculate that SL hydrolysis may induce a subtle D14 destabilization to allow a transient D3 interaction, which in a multi-step process may cause further D14 destabilization and increased D3 binding. In an alternative model, the hydrolyzed D-ring moves to the opening of the LBP, where it can directly engage in an interaction with D3 as proposed26. However, we do not favor this model as (i) the D-ring itself is inactive in D14 binding25,33,34 and cannot induce a D14-D3 interaction in vitro (Figure 8B), and (ii) our crystal structure (Figure 2) and MD simulations (Supplementary information, Figure S12) suggest that the D-ring at the LBP opening position is not stable.
In contrast to D14, the highly homologous KAI2 appears to be unable to hydrolyze karrikin. While the catalytic serine of KAI2 is required for karrikin signaling35, in the KAI2/karrikin complex structure, karrikin stably binds to the pocket at a position distal from the catalytic triad23. In further contrast to D14/GR24, karrikin binding appears to induce significant conformational changes in ten residues of the lid domain23 that are poorly conserved in D14 (Supplementary information, Figure S9). Though karrikin is not an endogenous KAI2 agonist, D14 and KAI2 may have evolved two different mechanisms to stabilize interaction of D3 with their lid domains.
In addition, despite the little structural similarity between the SL analogs (Br-PMF and I-PMF, Figures 8 and 9) and GR24 other than the butenolide ring and the ether linkage, D14 can hydrolyze all three compounds and mutations of D14 LBP and surface residues affected the ability of all three ligands to induce a D14-D3 interaction in a very similar manner. Consistently, when modeled into D14 LBP, all three ligands could make surprisingly similar interactions with all pocket residues that we have analyzed by mutations. These interactions involve the invariant butenolide ring and a hydrogen bond acceptor in a position comparable to that of the C-ring oxygens. Ligand-mediated D14-D3 interaction also depends on the same set of D14 surface residues, revealing an emerging unified mechanism for SL recognition and signaling.
Furthermore, our results reveal one surprising feature of SL signaling ― the systematic destabilization of key signaling components including the hormone, the receptor and the effectors such as D5319,20. This is clearly distinct from the signaling paradigm for other major plant hormones. While the hormone-dependent degradation of transcriptional repressors through the ubiquitin-proteasome system is similar between SL and other plant hormones, the enzymatic degradation of the hormone by the receptor itself as well as the conformational destabilization of the receptor by the hormone and effector is totally unexpected. This built-in instability of the SL signaling system highlights the transient nature of SL signaling and may be highly relevant to its biological functions in determining the plant branching architecture phenotype. It can be envisioned that once the cell is committed to the branching fate through the activation of SL pathways, the plants want to shut down the branching signals as soon as possible. This could be achieved by destabilization of the receptor D14 upon ligand binding and effector coupling as reported here, followed by ubiquitination-mediated degradation as D14 is subject to proteolysis in cells in the presence of GR2422. While many gaps remain in our understanding of this unusual signaling mechanism, the structural and mutational analyses of the D14/GR24 and D14/GR24/D3 complexes reported in this paper establish the GR24 binding mode in the D14 pocket and identify the D3-binding interface of D14. In addition, given the conserved roles of SLs in symbiosis of mycorrhiza formation and parasite weed germination, the structure of the D14-GR24 model provides a rational template to design better chemical ligands to fence off aggression of parasite weeds and to enhance mycorrhizal and rhizobium symbiosis, which are of great significance in agriculture and plant sciences.
Materials and Methods
Chemical structures of GR-24, Br-PMF, I-PMF, CN-PMF, Br-BMF, ABC-ring, D-ring, and karrikins
GR24 for crystallography and GR24-rac for biochemical assays were purchased from Chiralix (product number: CX23880). ABC-ring, D-ring and other SL mimics were chemically synthesized based on previous published literature32,36,37,38. In brief, for the synthesis of Br-PMF, I-PMF, CN-PMF, and Br-BMF32, the corresponding phenol was coupled with 5-bromo-3-ethylfuran- 2(5H)-one under basic conditions to give the desired products; for the synthesis of the ABC-ring37, first, the intermediate indanylacetic acid was prepared by using 1-indanone as the starting material. Then, it was reduced in the presence of NaBH4 and CeCl3•7H2O, followed by intermolecular esterification to deliver the desired product; the D-ring38 was synthesized through a Kharasch addition catalyzed by (PCy3)2Cl2Ru = CHPh and subsequent hydrolysis in 6 M HCl solution; for the synthesis of KAR1 and KAR236, first, commercially available 3-hydroxy-4H-pyran-4-one was treated with phosphorus pentasulphide. Then, the products were esterified with the corresponding 2-chloroalkyl chloride in the presence of triethylamine, followed by intermolecular cyclization though the classical Friedel-Crafts reaction in acetic anhydride with triphenyl phosphine and sodium acetate to yield the final products.
Protein preparation
Rice D14 (residues 52-318) was expressed as a 6× His-sumo (H6SUMO) fusion protein from the expression vector pSUMO (LifeSensors). The modified fusion protein contains a H6-tag (MKKGHHHHHHG) at the N terminus and SUMO protease site between SUMO and D14. BL21 (DE3) cells transformed with the expression plasmid were grown in LB broth at 16 °C to an OD600 of ∼1.0 and induced with 0.1 mM IPTG for 16 h. Cells were harvested, resuspended in 100 ml extract buffer (20 mM Tris, pH 8.0, 200 mM NaCl, and 10% glycerol) per 6 L of cells, and passed three times through a French press with pressure set at 1 000 Pa. The lysate was centrifuged at 16 000 rpm in a Sorvall SS34 rotor for 30 min, and the supernatant was loaded on a 50 ml Nickel HP column. The column was washed with 600 ml 10% buffer B (20 mM Tris, pH 8.0, 200 mM NaCl, 500 mM imidazole, and 10% glycerol) and eluted in two steps with 200 ml 50% buffer B, and then 100 ml 100% buffer B. The eluted H6SUMO-D14 was dialyzed against extract buffer and cleaved overnight with SUMO protease at a protease/protein ratio of 1:1 000 in the cold room. The cleaved H6SUMO tag was removed by passing through a Nickel HP column, and the protein was further purified by chromatography through a HiLoad 26/60 Superdex 200 gel filtration column in 20 mM Tris, pH 8.0, 200 mM ammonium acetate, 1 mM dithiotreitol and 1 mM EDTA. Apo-D14 was eluted as a sharp single peak with an estimated molecular weight of 30 kD, suggesting that apo-D14 is a monomer in solution (predicted MW of 29.2 kD).
The full-length rice D3 and ASK7 were co-expressed as a 8× His-Maltose-Binding Protein (H8MBP) fusion protein and as untagged protein, respectively, in Sf9 insect cells. The modified D3 fusion protein contains a H8-MBP tag at the N terminus and TEV protease site between the MBP tag and D3. Sf9 insect cells were grown at 28 °C to 2 million cells/ml and then co-infected with D3 and ASK7 viruses. Cells were harvested after 48 h, and resuspended and homogenized in 100 ml (per 2 L cell culture) homogenization buffer (2 mM MgCl2, 20 mM Tris, pH 8.0, 500 mM NaCl, 10% glycerol and 1 mM PMSF). An equal volume of dilution buffer (2 mM MgCl2, 20 mM Tris, pH 8.0, 10% glycerol and 1 mM PMSF) was added after homogenization. The H8MBP-D3/ASK7 complex was isolated from the soluble cell lysate by amylose affinity chromatography, followed by gel filtration chromatography. The H8MBP-D3/ASK7 complex was eluted as a sharp single peak from the HiLoad 26/60 Superdex 200 gelfiltration column with an estimated molecular weight of 140 kD, suggesting that the complex is a monomer in solution (predicted molecular weight of 138 kD).
Small scale purifications of H6-thioredoxin-3C-avitag-tagged D14 of wild-type and mutant proteins were performed by chromatography through a 5 ml Nickel HP column (GE Healthcare). To generate biotinylated proteins for luminescence proximity assays (AlphaScreen), H6-thioredoxin-3C-avitag-tagged D14 were expressed in E. coli BL21(DE3) cells from a pETDuet (Novagen) derivative vector. The first T7 polymerase-driven expression unit of this vector contains the D14 open reading frame as H6-thioredoxin-3C protease cleavage site-avitag fusion, the second site the E. coli biotin-ligase gene BirA. The 14 amino acid avitag functions as a defined in vivo biotinylation site in E. coli. Cells grown in the presence of 40 μM biotin were lysed and fusion protein purified through a Nickel HiTrap column as above. Following 3C protease release of the H6-thioredoxin tag, D14 proteins with biotinylated avitags were purified by 5 ml Nickel HP column chromatography followed by gel filtration chromatography. Residual biotin was removed by extensive dialysis prior to their use in luminescence proximity assays.
Crystallization
The D14-GR24 complex was prepared with 20-30-fold excess of GR24 to D14, and then concentrated to 4-5 mg/ml for crystallization. The crystals were grown at room temperature in hanging drops containing 1.0 μl of the above protein-ligand solutions and 1.0 μl of well solution containing 0.1 M HEPES pH 7.5, 8% v/v ethylene glycol, 10% w/v polyethylene glycol 8 000. Crystals of about 40 μm in length appeared the following day. Crystals were soaked with additional freshly prepared GR24 solution in well buffer and 30% sorbitol for cryo protection prior to flash freezing in liquid nitrogen.
Data collection and structure determination
The X-ray diffraction data sets for apo-D14 and D14-GR24 complex crystals were collected at SSRF (Shanghai Synchrotron Radiation Facility) beam line BL17U. The observed reflections were reduced, merged, and scaled with DENZO and SCALEPACK in the HKL2000 package39. Crystals of apo-D14 and D14-GR24 diffracted to resolutions of 1.5 Å and 2.0 Å, respectively.
Molecular replacement was performed by using the Collaborative Computational Project 4 (CCP4) program Phaser40 using the RbsQ structure as the initial model. Programs O41 and Coot42 were used to manually fit the protein model. Model refinement was performed with CNS43 and the CCP4 program Refmac544. The volumes of the LBP were calculated with the program Voidoo45 by using program default parameters and a probe with a radius of 1.4 Å. All structure figures were prepared by using PyMOL (DeLano Scientific). The statistics of data collection and the model refinement are summarized in Supplementary information, Table S1. While the D-ring of GR24 could be reliably placed into the electron density, the remainder of GR24 was modeled into the structure based on chemical constraints, which matched the trace density in the structure.
Computational docking
GR24 and SL analog stereoisomers were docked into the D14 LBP using the standard flexible-ligand sampling algorithm46 implemented in DOCK 6.747. Ligand placement was prioritized by D-Ring orientation relative to the crystal structure. The combined grid Van-der-Waals and electrostatic (vdw+es) scores and internal energy scores were then used to select energetically favorable docking positions for each ligand. Prior to docking, partial charges from the all-atom AMBER ff14SB348 were assigned to the receptor and semi-empirical AM1-BCC charges were derived for each SL analog.
MD simulation
The initial model for MD simulations was constructed from the 2.1 Å resolution X-ray structure of DWARF14 in complex with the hydroxy D-ring (D-OH) of (±)-GR24 (PDB ID: 3WIO)26. Only chain A and the D-OH product were used for all simulations; crystallographic waters and chain B were removed.
Setup and simulation was performed as follows for all systems using pmemd.cuda in the AMBER14 (update.10)49 suite with the AMBER14SB and GAFF50 force fields. Atomic point changes for 5S/R D-OH ligands were derived using the AM1-BCC charge model in antechamber. Optimal hydrogen bonding and protonation states for titratable residues were determined at pH 7.0 using the PROPKA4 program51 as included in Schrödinger Release 2015–1. Each system was solvated in a 8 Å pad of TIP3P waters and neutralized. Additional NaCl ions were randomly added for a final concentration of 50 mM to mimic experimental conditions noted above. The simulation protocol, previously described by de Waal et al.52, was adopted for the current study with minor alterations. Briefly, the system was maintained at 310.0 K to increase sampling of the D-OH free energy landscape, cutoff of long-range electrostatic interactions was set to 10 Å, and all production simulations were performed in the isothermal-isobaric NPT ensemble using the Monte-Carlo barostat and the Langevin thermostat with a collision frequency of 5.0 ps-1. All simulations were performed in triplicate.
Hydrogen-deuterium exchange (HDX)
HDX of D14 in the absence and presence of 100 μM GR24 in 20 mM Tris pH 8.0, 200 mM NaCl was performed at 4 °C using an automated system described previously53. Briefly, protein was incubated in a D2O buffer for a range of exchange times from 10 s to 1 h before quenching the deuterium exchange reaction with an acidic quench solution (pH 2.4) containing 3 M Urea and 1% TFA. All mixing and digestions were carried out on a LEAP Technologies Twin HTS PAL liquid handling robot housed inside a temperature controlled fridge. Protein digestion was performed in-line with chromatography using an immobilized pepsin column. Mass spectra were acquired on a Q Exactive hybrid quadrupole-Orbitrap mass spectrometer (ThermoFisher Scientific). Three replicates were performed for each HDX time point. Percent deuterium exchange values for peptide isotopic envelopes at each time point were calculated and processed using the Workbench Software54.
AlphaScreen luminescence proximity assay
AlphaScreen assays for determining D14-D3 interaction were performed using an Envision reader (PerkinElmer). Biotinylated D14 was attached to streptavidin-coated donor beads, and the H8MBP-D3/ASK7 complex to nickel-chelated accepted beads. The donor and acceptor beads were brought into proximity by the interactions between D14 and D3/ASK7 complex, which were measured with and without GR24 at the indicated concentrations. When excited by a laser beam of 680 nm, the donor beads emit singlet oxygen that activates thioxene derivatives in the acceptor beads, which then release photons of 520-620 nm as the binding signal. The experiments were conducted with 100 nM of D14 and D3/ASK7 complex proteins in the presence of 5 μg/ml donor and acceptor beads in a buffer of 50 mM HEPES, pH 7.0, 50 mM NaF, 50 mM CHAPS, and 0.1 mg/ml bovine serum albumin. The results were based on an average of three experiments with standard errors typically < 10% of the measurements.
Radio-ligand SPA
35 μM H6Sumo-D14 were incubated with 250 μg of Ytrium silicate copper-chelating SPA beads (GE Healthcare) in a buffer of 50 mM MOPS, pH 7.3, 50 mM NaF, 50 mM CHAPS, and 0.1 mg/ml bovine serum albumin for 70 min shaking on ice. H6Sumo-D14 bound to SPA beads was separated from free H6Sumo-D14 by centrifugation at 5 200× g for 30 s. Bead pellets were washed with 1 ml of the same buffer, then resuspended in 120 μl of the buffer supplemented with 15 nM 3H-labelled GR24 (American Radiolabeled Chemicals), in the presence of indicated concentrations of unlabeled GR24 and ABA. The reactions were incubated shaking for 1 h at room temperature. 3H-labelled GR24-D14 binding brings the radioactive GR24 into the immediate proximity of the scintillant embedded in the SPA beads, resulting in the generation of light, which was quantified in a liquid scintillation counter.
Mutagenesis and purification of d14- mutant proteins
The open reading frame for wild-type D14 with N-terminal Biotin-MBP tag was introduced into a pET-Duet vector containing the BirA biotin ligase gene in the second expression cassette. Mutations were introduced by the QuikChange method (Agilent). All mutant constructs were confirmed by DNA sequencing.
Acknowledgments
This work was supported by the Van Andel Research Institute, Amway (China), the National Natural Science Foundation of China (NSFC 31300245 and NSFC 91217311), the Ministry of Science and Technology of China (2012ZX09301001, 2012CB910403, 2013CB910600, XDB08020303, and 2013ZX09507001), and the National Institute of Health (USA; DK071662 to HEX, and GM102545 and GM104212 to KM). We thank staff members of the Life Science Collaborative Access Team of the Advanced Photon Source (APS) for assistance in data collection at the beam lines of sector 21, which is in part funded by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (085P1000817). Use of APS was supported by the Office of Science of the US Department of Energy, under contract no DE-AC02-06CH11357.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Fo-Fc electron density maps calculated from the GR24 deleted model, contoured at 2 σ (top) or 3 σ (bottom).
A group of flexible charged surface residues adopts different conformations in six different D14 crystal structures.
D14 adopts almost identical surface conformations in its four different states.
HDX perturbation map comparing deuterium exchange of apo D14 to exchange in the D14+GR24 complex.
Binding of GR24 to petunia DAD2 induces GR24 hydrolysis and DAD2 destabilization.
Details of charge (A) and hydrophobic (B) interactions between GR24 and D14 ligand-binding pocket residues.
Unbiased computational docking of GR24 stereoisomers in the D14 LBP.
Charge reverse mutations of two acidic D14 residues cause SL-independent D3 interaction.
D14 residues that are important for D3 interaction are conserved.
Thermal destabilization of D14 and DAD2 by SL analogs.
Simulations of 3WIO D-OH stereoisomer products.
Statistics of D14/GR24 complex data sets and structure refinement
References
- 1Xie X, Yoneyama K. The strigolactone story. Annu Rev Phytopathol 2010; 48:93–117. [DOI] [PubMed] [Google Scholar]
- 2Waldie T, McCulloch H, Leyser O. Strigolactones and the control of plant development: lessons from shoot branching. Plant J 2014; 79:607–622. [DOI] [PubMed] [Google Scholar]
- 3de Saint Germain A, Bonhomme S, Boyer FD, Rameau C. Novel insights into strigolactone distribution and signalling. Curr Opin Plant Biol 2013; 16:583–589. [DOI] [PubMed] [Google Scholar]
- 4Brewer PB, Koltai H, Beveridge CA. Diverse roles of strigolactones in plant development. Mol Plant 2013; 6:18–28. [DOI] [PubMed] [Google Scholar]
- 5Ruyter-Spira C, Al-Babili S, van der Krol S, Bouwmeester H. The biology of strigolactones. Trends Plant Sci 2013; 18:72–83. [DOI] [PubMed] [Google Scholar]
- 6Tsuchiya Y, McCourt P. Strigolactones as small molecule communicators. Mol Biosyst 2012; 8:464–469. [DOI] [PubMed] [Google Scholar]
- 7Lopez-Raez JA, Matusova R, Cardoso C, et al. Strigolactones: ecological significance and use as a target for parasitic plant control. Pest Manag Sci 2009; 65:471–477. [DOI] [PubMed] [Google Scholar]
- 8Arite T, Umehara M, Ishikawa S, et al. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol 2009; 50:1416–1424. [DOI] [PubMed] [Google Scholar]
- 9Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J. Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol 2005; 46:79–86. [DOI] [PubMed] [Google Scholar]
- 10Stirnberg P, van De Sande K, Leyser HM. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 2002; 129:1131–1141. [DOI] [PubMed] [Google Scholar]
- 11Beveridge CA, Ross JJ, Murfet IC. Branching in Pea (Action of Genes Rms3 and Rms4). Plant Physiol 1996; 110:859–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Gomez-Roldan V, Fermas S, Brewer PB, et al. Strigolactone inhibition of shoot branching. Nature 2008; 455:189–194. [DOI] [PubMed] [Google Scholar]
- 13Umehara M, Hanada A, Yoshida S, et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature 2008; 455:195–200. [DOI] [PubMed] [Google Scholar]
- 14Beveridge CA, Kyozuka J. New genes in the strigolactone-related shoot branching pathway. Curr Opin Plant Biol 2010; 13:34–39. [DOI] [PubMed] [Google Scholar]
- 15Murase K, Hirano Y, Sun TP, Hakoshima T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 2008; 456:459–463. [DOI] [PubMed] [Google Scholar]
- 16Shimada A, Ueguchi-Tanaka M, Nakatsu T, et al. Structural basis for gibberellin recognition by its receptor GID1. Nature 2008; 456:520–523. [DOI] [PubMed] [Google Scholar]
- 17Bennett T, Leyser O. Strigolactone signalling: standing on the shoulders of DWARFs. Curr Opin Plant Biol 2014; 22:7–13. [DOI] [PubMed] [Google Scholar]
- 18Nelson DC, Scaffidi A, Dun EA, et al. F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA 2011; 108:8897–8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Jiang L, Liu X, Xiong G, et al. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 2013; 504:401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Zhou F, Lin Q, Zhu L, et al. D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 2013; 504:406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Wang Y, Sun S, Zhu W, Jia K, Yang H, Wang X. Strigolactone/MAX2-induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching. Dev Cell 2013; 27:681–688. [DOI] [PubMed] [Google Scholar]
- 22Chevalier F, Nieminen K, Sanchez-Ferrero JC, et al. Strigolactone promotes degradation of DWARF14, an alpha/beta hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 2014; 26:1134–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Guo Y, Zheng Z, La Clair JJ, Chory J, Noel JP. Smoke-derived karrikin perception by the alpha/beta-hydrolase KAI2 from Arabidopsis. Proc Natl Acad Sci USA 2013; 110:8284–8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Zhao LH, Zhou XE, Wu ZS, et al. Crystal structures of two phytohormone signal-transducing alpha/beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Res 2013; 23:436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Hamiaux C, Drummond RS, Janssen BJ, et al. DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, Strigolactone. Curr Biol 2012; 22:2032–2036. [DOI] [PubMed] [Google Scholar]
- 26Nakamura H, Xue YL, Miyakawa T, et al. Molecular mechanism of strigolactone perception by DWARF14. Nat Commun 2013; 4:2613. [DOI] [PubMed] [Google Scholar]
- 27Kagiyama M, Hirano Y, Mori T, et al. Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes Cells 2013; 18:147–160. [DOI] [PubMed] [Google Scholar]
- 28Bythell-Douglas R, Waters MT, Scaffidi A, Flematti GR, Smith SM, Bond CS. The structure of the karrikin-insensitive protein (KAI2) in Arabidopsis thaliana. PLoS One 2013; 8:e54758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29Derewenda ZS, Vekilov PG. Entropy and surface engineering in protein crystallization. Acta Crystallogr D Biol Crystallogr 2006; 62:116–124. [DOI] [PubMed] [Google Scholar]
- 30Krishna MM, Hoang L, Lin Y, Englander SW. Hydrogen exchange methods to study protein folding. Methods 2004; 34:51–64. [DOI] [PubMed] [Google Scholar]
- 31Scaffidi A, Waters MT, Sun YK, et al. Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol 2014; 165:1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32Fukui K, Ito S, Ueno K, Yamaguchi S, Kyozuka J, Asami T. New branching inhibitors and their potential as strigolactone mimics in rice. Bioorg Med Chem Lett 2011; 21:4905–4908. [DOI] [PubMed] [Google Scholar]
- 33Fukui K, Ito S, Asami T. Selective mimics of strigolactone actions and their potential use for controlling damage caused by root parasitic weeds. Mol Plant 2013; 6:88–99. [DOI] [PubMed] [Google Scholar]
- 34Boyer FD, de Saint Germain A, Pillot JP, et al. Structure-activity relationship studies of strigolactone-related molecules for branching inhibition in garden pea: molecule design for shoot branching. Plant Physiol 2012; 159:1524–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35Waters MT, Scaffidi A, Moulin SL, Sun YK, Flematti GR, Smith SM. A Selaginella moellendorffii ortholog of KARRIKIN INSENSITIVE2 functions in Arabidopsis development but cannot mediate responses to Karrikins or Strigolactones. Plant Cell 2015; 27:1925–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36Flematti GR, Goddard-Borger ED, Merritt DJ, Ghisalberti EL, Dixon KW, Trengove RD. Preparation of 2H-furo[2,3-c]pyran-2-one derivatives and evaluation of their germination-promoting activity. J Agric Food Chem 2007; 55:2189–2194. [DOI] [PubMed] [Google Scholar]
- 37Wang X, Yan Y, Gong B, et al. Efficient synthesis of functionalized 1-oxo-1-phenyl-2-acetic acids through Ru(II)-catalyzed transfer hydrogenation. Bull Korean Chem Soc 2013; 34:3143–3146. [Google Scholar]
- 38Lee B, Schrader T, Martín-Matute B, Kauffman C, Zhang P, Snapper M. (PCy3)2Cl2Ru=CHPh Catalyzed Kharasch additions. Application in a formal olefin carbonylation. Tetrahedron 2004; 60:7391–7396. [Google Scholar]
- 39Otwinowski Z, Borek D, Majewski W, Minor W. Multiparametric scaling of diffraction intensities. Acta Crystallogr A 2003; 59:228–234. [DOI] [PubMed] [Google Scholar]
- 40McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41Kleywegt GJ, Jones TA. Efficient rebuilding of protein structures. Acta Crystallogr D Biol Crystallogr 1996; 52:829–832. [DOI] [PubMed] [Google Scholar]
- 42Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 43Brunger AT, Adams PD, Clore GM, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 1998; 54:905–921. [DOI] [PubMed] [Google Scholar]
- 44Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallogr 1999; 55:247–255. [DOI] [PubMed] [Google Scholar]
- 45Kleywegt GJ, Jones TA. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr D Biol Crystallogr 1994; 50:178–185. [DOI] [PubMed] [Google Scholar]
- 46Mukherjee S, Balius TE, Rizzo RC. Docking validation resources: protein family and ligand flexibility experiments. J Chem Inf Model 2010; 50:1986–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47Allen WJ, Balius TE, Mukherjee S, et al. DOCK 6: Impact of new features and current docking performance. J Comput Chem 2015; 36:1132–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser K, Simmerling C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput 2015. [DOI] [PMC free article] [PubMed]
- 49Case D, Babin J, Berryman J, et al. AMBER 14. University of California, San Francisco 2014.
- 50Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem 2004; 25:1157–1174. [DOI] [PubMed] [Google Scholar]
- 51Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins 2005; 61:704–721. [DOI] [PubMed] [Google Scholar]
- 52de Waal PW, Sunden KF, Furge LL. Molecular dynamics of CYP2D6 polymorphisms in the absence and presence of a mechanism-based inactivator reveals changes in local flexibility and dominant substrate access channels. PLoS One 2014; 9:e108607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53Chalmers MJ, Busby SA, Pascal BD, et al. Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal Chem 2006; 78:1005–1014. [DOI] [PubMed] [Google Scholar]
- 54Pascal BD, Willis S, Lauer JL, et al. HDX workbench: software for the analysis of H/D exchange MS data. J Am Soc Mass Spectrom 2012; 23:1512–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fo-Fc electron density maps calculated from the GR24 deleted model, contoured at 2 σ (top) or 3 σ (bottom).
A group of flexible charged surface residues adopts different conformations in six different D14 crystal structures.
D14 adopts almost identical surface conformations in its four different states.
HDX perturbation map comparing deuterium exchange of apo D14 to exchange in the D14+GR24 complex.
Binding of GR24 to petunia DAD2 induces GR24 hydrolysis and DAD2 destabilization.
Details of charge (A) and hydrophobic (B) interactions between GR24 and D14 ligand-binding pocket residues.
Unbiased computational docking of GR24 stereoisomers in the D14 LBP.
Charge reverse mutations of two acidic D14 residues cause SL-independent D3 interaction.
D14 residues that are important for D3 interaction are conserved.
Thermal destabilization of D14 and DAD2 by SL analogs.
Simulations of 3WIO D-OH stereoisomer products.
Statistics of D14/GR24 complex data sets and structure refinement