Abstract
In eudicot plants, the miR482/miR2118 superfamily regulates and instigates the production of phased secondary small interfering RNAs (siRNAs) from NB-LRR (nucleotide binding leucine-rich repeat) genes that encode disease resistance proteins. In grasses, this miRNA family triggers siRNA production specifically in reproductive tissues from long noncoding RNAs. To understand this functional divergence, we examined the small RNA population in the ancient gymnosperm Norway spruce (Picea abies). As many as 41 miRNA families in spruce were found to trigger phasiRNA (phased, secondary siRNAs) production from diverse PHAS loci, with a remarkable 19 miRNA families capable of targeting over 750 NB-LRR genes to generate phasiRNAs. miR482/miR2118, encoded in spruce by at least 24 precursor loci, targets not only NB-LRR genes to trigger phasiRNA production (as in eudicots) but also noncoding PHAS loci, generating phasiRNAs preferentially in male or female cones, reminiscent of its role in the grasses. These data suggest a dual function of miR482/miR2118 present in gymnosperms that was selectively yet divergently retained in flowering plants. A few MIR482/MIR2118 precursors possess an extremely long stem-loop structure, one arm of which shows significant sequence similarity to spruce NB-LRR genes, suggestive of an evolutionary origin from NB-LRR genes through gene duplication. We also characterized an expanded miR390-TAS3 (TRANS-ACTING SIRNA GENE 3)-ARF (AUXIN RESPONSIVE FACTOR) pathway, comprising 18 TAS3 genes of diverse features. Finally, we annotated spruce miRNAs and their targets. Taken together, these data expand our understanding of phasiRNA network in plants and the evolution of plant miRNAs, particularly miR482/miR2118 and its functional diversification.
Keywords: microRNA, Norway spruce, phasiRNA, tasiRNA, miR482/miR2118, NB-LRR, TAS3
Introduction
In plants, microRNAs (miRNAs) comprise a class of predominantly 20–22 nt small RNAs (sRNAs), which play critical roles in gene regulation in plant development and stress responses (Chen 2009; Voinnet 2009; Sunkar et al. 2012). miRNA biogenesis relies on the formation of a stem-loop structure of a precursor mRNA that is subsequently cleaved by DICER-LIKE 1 (DCL1). This results in the release of a miRNA/miRNA* duplex with a 3′ 2-nt overhang (Jones-Rhoades et al. 2006; Voinnet 2009). The functional strand of the duplex (the miRNA) is loaded into an Argonaute (AGO) protein, the key component of the RNA-induced silencing complex, directing interactions with target genes and resulting in either mRNA cleavage or translation inhibition (Jones-Rhoades et al. 2006; Voinnet 2009). The cleavage of target genes by some miRNAs can trigger the biogenesis of another class of sRNAs; an RNA-dependent RNA polymerase is recruited to convert an upstream or downstream cleaved fragment into double-strand RNA, subsequently chopped by DICER-LIKE 4 into 21-nt sRNAs (Peragine et al. 2004; Vazquez et al. 2004; Allen et al. 2005; Yoshikawa et al. 2005; Axtell et al. 2006). These sRNAs are “in phase” with the miRNA cleavage site, hence named phased, secondary small interfering RNAs (phasiRNAs), some of which function in trans (trans-acting siRNAs, tasiRNA) or in cis (Zhai et al. 2011; Fei et al. 2013). The tasiRNAs from the (TRANS-ACTING SIRNA GENE 3) transcripts, targeting AUXIN RESPONSE FACTOR (ARF)-encoding mRNAs and triggered by miR390, are among the most well-characterized tasiRNAs (Allen et al. 2005; Axtell et al. 2006). TAS3 is also perhaps the earliest-evolved tasiRNA known in plants, a pathway that expanded in flowering plants to include many phasiRNA-generating loci (Krasnikova et al. 2009).
PhasiRNAs are produced from both protein-coding and noncoding genes. In many eudicots, three large gene families generate the majority of phasiRNAs, including those encoding nucleotide binding leucine-rich repeat proteins (NB-LRR genes), pentatricopeptide repeat proteins (PPR genes), and MYB transcription factors (MYB genes) (Fei et al. 2013). Monocot genomes have thus far yielded a contrasting picture in which phasiRNAs are produced predominantly from noncoding transcripts. In rice, >1,000 noncoding loci distributed in all the chromosomes give rise to the production of appreciable 21-nt phasiRNAs (Johnson et al. 2009; Song et al. 2012). In addition, monocots produce a class of 24-nt phasiRNAs, apparently absent in eudicots (Johnson et al. 2009; Vogel et al. 2010; Song et al. 2012; Jeong et al. 2013).
NB-LRR genes, one of the first lines of defense against pathogen infection (Dangl and Jones 2001; Meyers et al. 2005) comprises the largest class of genes producing phasiRNAs (PHAS genes) in eudicots (Fei et al. 2013). A significant trigger of the NB-LRR phasiRNAs is the miR482/miR2118 superfamily, which serves as the master regulator of NB-LRR genes in legumes and Solanaceaous plants (and likely many others) by targeting the region coding for the critical P-loop motif in the highly conserved NBS (nucleotide-binding site) domain (Zhai et al. 2011; Li et al. 2012; Shivaprasad et al. 2012). This miRNA-mediated regulation spawns secondary phasiRNAs, a layer of control with potential roles in defense or symbiosis (Zhai et al. 2011; Shivaprasad et al. 2012). In monocots, in contrast, miR482/miR2118 targets noncoding transcripts, exemplified in rice, maize, and Brachypodium, and instigating secondary phasiRNA production mainly in reproductive tissues (Johnson et al. 2009; Vogel et al. 2010; Song et al. 2012; Jeong et al. 2013). This dramatic functional divergence of an miRNA is rare in plants, perhaps unique to this miRNA family.
Gymnosperms are a group of land plants that emerged more than 300 Ma, well before the angiosperm lineage separated into the two main lineages of modern plants, eudicots and monocots, approximately 150 Ma. Although the first sequenced gymnosperm genome greatly facilitates studies of development, adaptation, and plant evolution (Nystedt et al. 2013), profiling and analyses of sRNAs in gymnosperms, especially phasiRNAs, have been minimal, unlike the ancient bryophytes (mainly the mosses) in which sRNAs are better characterized. In this study, we set out to narrow this gap by genome-wide characterization of miRNAs, their target genes, and PHAS pathways in Norway spruce (Pinus abies). We also assessed the function and evolutionary history of miR482/miR2118, and found that miR482/miR2118 has dual functions in Norway spruce which perhaps were selectively retained in eudicots and monocots. Our results demonstrate the presence of large miRNA population and a greatly expanded miRNA-PHAS-phasiRNA network in Norway spruce, enhancing our understanding of plant miRNAs and their evolution.
Results
miRNAs and Their Target Genes Identified in Norway Spruce
Norway spruce (P. abies) is the first gymnosperm species for which a whole-genome sequence is available (Nystedt et al. 2013). In that 2013 study, sRNAs were sequenced from 22 tissue samples (44 libraries; supplementary table S1, Supplementary Material online), generating a list of spruce miRNAs (Nystedt et al. 2013). However, their miRNA annotation was not exhaustive and it lacked characterization of the sRNA abundance, processing accuracy, and strand specificity. By combining the public sRNA data and genome sequence with our well-developed miRNA annotation pipeline (Jeong et al. 2011; Zhai et al. 2011; Xia et al. 2012), we performed a thorough characterization of spruce miRNAs using a set of stringent criteria (Meyers et al. 2008).
The miRNA annotation was conducted through the workflow shown in supplementary figure S1, Supplementary Material online. Apart from screening for properties of the stem-loop structure, two additional, important filters were applied. First, a strand-bias filter which requires that ≥90% of the reads mapped to a miRNA locus (±300 bp) are from the same strand, as MIRNA precursor mRNA transcripts fold into a structure (stem-loop or hairpin) which by itself gives rise to a mature miRNA and miRNA*. The second filter considers the accuracy of miRNA processing, which requires that the read abundance of the top two sRNA sequences accounts for ≥70% (for new miRNAs) and ≥40% (for known miRNAs) of the total reads from a given MIRNA locus. In addition, for a novel miRNA, the workflow requires that the miRNA* is present in the same library from which the miRNA sequence was identified.
In total, we identified 585 MIRNA genes producing 426 unique miRNA sequences (fig. 1A and supplementary fig. S1, Supplementary Material online). Among these MIRNA genes, 313 (generating 185 unique miRNA sequences) were from 52 miRNA families with homologs in miRBase (version 21), and therefore we labeled these as “known miRNAs” (supplementary table S2, Supplementary Material online). Twenty-one out of the 22 deeply conserved miRNA families in plants (Cuperus et al. 2011) were found in spruce, with the exception of miR827. In addition, we found in spruce another seven miRNA families widely present in plants but absent in some lineages such as monocots; these included miR403, miR482/miR2118, miR529, miR535, miR845, miR828, miR858 and miR4376, and they are considered “less-conserved.” The other 272 MIRNA genes encoding 241 unique miRNA sequences belonged to as many as 181 miRNA families, yet had no homologs in miRBase, and thus we called these “novel” miRNAs (supplementary table S3, Supplementary Material online). As in other plants, the majority (66.2%) of the identified spruce miRNAs was 21 nt in length, and the 22-nt and 20-nt classes accounted for 27.9% (119) and 5.8% (25), respectively (fig. 1A). More than half of the miRNAs in each class contained an initial uridine (U), especially in the 22-nt class for which the U-initiated miRNAs constituted approximately 80%. To get a sense of how many of these novel miRNAs are conserved in gymnosperms, we sequenced sRNAs from Ginkgo biloba (common name, ginkgo), a gymnosperm distantly related to and predating the conifers. Due to the lack of a sequenced genome for ginkgo, we were unable to perform de novo annotation of ginkgo miRNAs. However comparison of homologous sequences (≤4 mismatches) enabled us to identify potential miRNA homologs for 51 of these novel miRNAs, indicating a considerable proportion of miRNAs (∼28%) identified as novel are potentially conserved within the gymnosperms (supplementary table S4, Supplementary Material online).
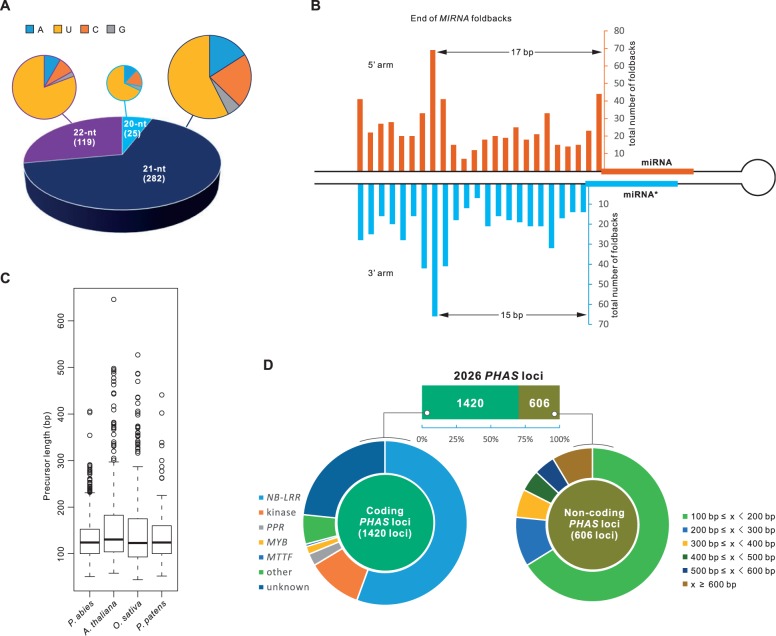
Fig. 1.
miRNA and PHAS loci in Norway spruce. (A) Distribution of miRNA sizes and the initial nucleotide of miRNAs in spruce. (B) Distribution of large loops (≥4 nucleotides on one side) or ends of helices for foldback structures of spruce miRNAs. An enrichment is shown at the site 17/15 nucleotides from the miRNA/miRNA* region. (C) Distribution of MIRNA precursor (stem-loop structure) length of spruce miRNAs compared with three other well-studied plant species. (D) PHAS loci characterized in spruce. PHAS loci were classified into coding and noncoding genes; these were subsequently grouped according to their annotated function and sequence length, respectively.
In flowering plants, the initial processing event by DCL1 depends on a stem length of approximately 15 bp below the miRNA/miRNA* duplex (Mateos et al. 2010; Song et al. 2010; Werner et al. 2010). To check whether this feature is also conserved in gymnosperms, we calculated the stem length between the miRNA/miRNA* duplex and the unpaired region (>4 unpaired mismatches), and found that the stem length is predominantly approximately 15 bp (fig. 1B), suggesting that the approximately 15-bp rule of the initial DCL1 cleavage is true also for gymnosperms. We also compared the stem-loop length of the spruce miRNA precursor genes with three other representative plants and found no apparent differences (fig. 1C). The spruce miRNAs have a slightly but not significantly (one-way ANOVA [analysis of variance] test) shorter stem-loop structure compared with the three other representative species we analyzed (fig. 1C).
To investigate the targets of the spruce miRNAs, we constructed and sequenced four PARE (parallel analysis of RNA end) libraries, which were used for characterization of miRNA target genes (German et al. 2008). Seventeen out of the 21 conserved miRNAs displayed a conserved target relationship; for instance, miR156 targeted genes encoding SQUAMOSA PROMOTER BINDING-LIKE proteins (SPL genes), miR160 and miR167 ARF genes, and miR390 TAS3 (supplementary table S5, Supplementary Material online). Our analyses failed to identify conserved target genes for the other four conserved miRNAs (miR162, miR164, miR168, and miR403), perhaps due to the incomplete annotation of spruce genes or, less likely, that these miRNAs have not evolved to act on their conserved target genes in spruce. Among the less-conserved miRNAs, the eudicot-prevalent miRNA-target interactions of 1) miR482/miR2118 and transcripts encoding nucleotide-binding leucine-rich repeat (NB-LRR) proteins and 2) MYB-targeting of miR828/858 were also found in spruce. Interestingly, in addition to miR482/miR2118, eight known miRNAs also targeted NB-LRR genes; these included miR946a, miR950, miR951, miR1311, miR1312, miR3701, miR3709, and miR3710, all of which have only been reported in the Pinaceae. We also identified target genes for greater than 75% (138 families) of the novel miRNAs, among which some target NB-LRRs (described in more detail below). Compared with targets identified for known/conserved miRNAs, fewer target genes of the novel miRNAs were of high-confidence (supplementary table S6, Supplementary Material online). Transcripts encoding receptor-like protein kinases, NB-LRRs, both the pentatricopeptide repeat (PPR) and tetratricopeptide repeat protein superfamilies, MYBs, and mitochondrial transcription termination factors (MTTFs) are the major groups cleaved by spruce miRNAs (supplementary table S6, Supplementary Material online).
Genome-Wide Characterization of PHAS Loci and Their Triggers
PhasiRNAs are a major class of sRNAs in plants (Fei et al. 2013). They are produced from not only diverse protein-coding genes, including large gene families such as NB-LRRs, PPRs, and MYBs, small gene families, and even single-copy genes, but also noncoding transcripts, especially in reproductive tissues of monocots (Johnson et al. 2009; Zhai et al. 2011; Xia et al. 2012, 2013; Fei et al. 2013). To explore whether phasiRNAs are prevalent and any phasiRNA pathways are conserved in gymnosperms, we performed genome-wide identification of PHAS loci in spruce.
For this analysis, an approach based on P values was used, as described in Xia et al. (2013), with additional stringent filters. Apart from a P-value threshold of 0.001, two other criteria were applied, to ensure that we captured only highly confident PHAS loci: 1) The length of phasiRNA-producing region exceeded 100 bp, and 2) over 30% of siRNAs produced from a given PHAS loci were derived from the same phase (i.e., are “in phase”). From this, we identified the surprisingly high count of 2,026 PHAS loci, named PHAS1–PHAS2026 after ranking according to their phasiRNA abundance in the set of 22 tissues (supplementary table S7, Supplementary Material online). These loci were subjected to further analyses.
Of the 2,026 loci, only 431 overlapped with annotated genes (supplementary table S7, Supplementary Material online); we interpreted this as a low proportion, suggesting that either they are largely noncoding loci, or the spruce genome may be poorly annotated. Therefore, we performed de novo gene annotation and classified the PHAS loci as coding or noncoding loci by evaluation of their sequence similarity to proteins present in public repositories, plus their protein-coding potential (calculated by Coding Potential Calculator; supplementary fig. S2, Supplementary Material online). PHAS loci were considered as noncoding if their longest coded peptide (without stop codons) had less than 100 amino acids and they were 1) of low similarity to known proteins (1e-4, BLASTX), and 2) of protein-coding potential less than 1 (supplementary fig. S2, Supplementary Material online). The 606 loci that passed these criteria were annotated as “noncoding,” with the remaining 1,420 loci considered as “coding.” Among the coding loci, the majority showed sequence similarity to NB-LRR genes (789 or 56%), with additionally large proportions represented by genes coding for kinase, PPR and MYB proteins (fig. 1D). Similarly, many other coding genes were also found to generate phasiRNAs as well, including MTTF, bHLH (encoding BASIC HELIX-LOOP-HELIX proteins), TIR1/AFB (encoding TRANSPORT INHIBITOR RESPONSE 1/AUXIN-RELATED F-BOX proteins), and ACA10 (encoding AUTOINHIBITED CA2+-ATPase proteins); the latter two were reported to produce phasiRNAs in other plants (Si-Ammour et al. 2011; Wang et al. 2011). The noncoding PHAS loci were classified according to the length of the region producing phasiRNAs. Although most noncoding PHAS loci were relatively short (<500 bp), a substantial portion (12.8%) produced phasiRNAs from a region longer than 500 bp, suggesting that noncoding PHAS loci, like coding genes, can generate long transcripts.
We next determined which miRNAs triggered phasiRNA production from these PHAS loci. Integrating PARE data, the miRNAs, and the PHAS loci, we identified miRNAs which serve as triggers of phasiRNA production. As many as 41 families of miRNAs were found to function as phasiRNA triggers (table 1), almost all were 22 nt in length with an initial “U,” consistent with canonical features of miRNA triggers of phasiRNAs (Chen et al. 2010; Cuperus et al. 2010). Several interactions between miRNA triggers and PHAS loci were conserved in many other plants, including miR393-TIR/AFB, miR4376-ACA10, miR828-MYB, miR482/miR2118-NB-LRR, and miR390-TAS3; another 16 miRNAs cataloged in miRBase were identified as triggers of phasiRNA production. Among the novel miRNAs characterized first in this study, 20 instigate phasiRNA production from their target genes (table 1). Interestingly, among these triggers, there were two 21-nt long miRNAs, miR2111 and miR3710, for which the triggering mechanism of phasiRNA production is unclear. Moreover, six miRNAs other than miR390 (three known miRNAs and three novel miRNAs; table 1) triggered phasiRNA production solely from noncoding loci, indicating that diverse TAS-like genes are present in spruce.
Table 1.
miRNA Triggers of PHAS Genes or Loci Identified in Spruce.
| miRNA | Sequence | Length | Target PHAS Genes/Loci |
|---|---|---|---|
| miR393 | UCCAAAGGGAUUGCAUUGAUUC | 22 | Transport inhibitor response 1/ auxin-binding F-box gene (TIR1/AFB) |
| miR4376 | UGCGCAGGGGAGAUGACACUGU | 22 | Autoinhibited Ca(2+)-ATPase 10 (ACA10) |
| miR2111 | UAAUCUGCAUCCUGGGGUUUG | 21 | Galactose oxidase/kelch repeat superfamily |
| miR3699 | UGACAGAAGAUAGACUUUGGUC | 22 | Integrase-type DNA-binding superfamily protein |
| miR11425 | UUUCACAGCUAUAUCUUUUCCU | 22 | Leucine-rich repeat protein kinase family |
| miR11551 | UGUUUUGUUUUCCCUCCGCAAU | 22 | Leucine-rich repeat protein kinase family |
| miR11540 | UGCCGAAGCCUGGAGGAAUAUC | 22 | Leucine-rich repeat protein kinase family |
| miR11562 | UUCUGAUAAUGCUUCACCCUCA | 22 | MTTF family protein |
| miR828 | UCUUGCUCAAAUGAGUGUUCCA | 22 | MYB |
| miR482/miR2118 | UCUUUCCACUUCUACCCAUUUC | 22 | NB-LRR/noncoding |
| miR482_2 | UUGAGAAACUGUGAGCCAAAUC | 22 | NB-LRR |
| miR951 | UGUUCUUGACGUCUGGACCACG | 22 | NB-LRR/noncoding |
| miR946 | CAGCCCUUCUGCUAUCCACAAC | 22 | NB-LRR |
| miR950 | UCUGGGCCCCGGUGGUUUAUGA | 22 | NB-LRR |
| miR1311 | UCAGAGUUUUGCCAGUUCCGCC | 22 | NB-LRR |
| miR1312 | UUCGGAGAGAAAAUGGCGACAU | 22 | NB-LRR |
| miR3697 | UAGCCCCUGACUUCAACAUGAG | 22 | NB-LRR |
| miR3701 | UAAACAGUGCCCACCCUUCAUC | 22 | NB-LRR |
| miR3701_2 | UGGCAAUAGCCUCUAUGUUCUU | 22 | NB-LRR |
| miR3709 | UUCAGAUGCUUUAAAUUCCCGA | 22 | NB-LRR |
| miR3710 | UUGGGAACCUGACGGGUCUCC | 21 | NB-LRR |
| miR11476 | UAAGCAGCCCUUCUCCGAUCCA | 22 | NB-LRR |
| miR11482 | UAACCAGUCCUUCUGCAAUCCA | 22 | NB-LRR |
| miR11506 | UCACAAAACACCGGAAUAAUCU | 22 | NB-LRR |
| miR11511 | UCCAACGAAGAUCAGAAGGCUU | 22 | NB-LRR |
| miR11519 | UCGUAAAACACAGGAAUGAUGG | 22 | NB-LRR |
| miR11523 | UCUCAGCAGAUUUAAUCCCCGA | 22 | NB-LRR |
| miR11528 | UCUGGCAACAUCCUCUAUUUCA | 22 | NB-LRR |
| miR11532 | UGACAUUGUAAAAGACGGGAAU | 22 | NB-LRR |
| miR11546 | UGGUACGGGUUGAAGUAGCACC | 22 | NB-LRR |
| miR947 | UAUCGGAAUCUGUUACUGUUUC | 22 | Unknown/noncoding |
| miR3698 | UAAGCCAAGGCAGAGUUGCAAG | 22 | Unknown |
| miR11462 | UCUGCCAGAGUGUGGCCGCGGA | 22 | Unknown |
| miR3627 | UGGCGCAGAAGAAAUGACACUG | 22 | Unknown |
| miR11570 | UUGCUUGAGUAACUUGAACACA | 22 | Unknown |
| miR11452 | UGGGAGCGAUCGAUGAGGUGUU | 22 | Unknown |
| miR390 | AAGCUCAGGAGGGAUAGCGCC | 21 | TAS3 |
| miR949 | UCCCGGGAAUCCAAUGGGCCUU | 22 | Noncoding |
| miR1313 | UACCACUGAAAUUAUUGUUCGA | 22 | Noncoding |
| miR1314 | UCGGCCUCGAAUGUUAGGAGAA | 22 | Noncoding |
| miR11529 | UCUUAUCUUUGAAAGUGCUAGC | 22 | Noncoding |
| miR11535 | UGAGCCGCUUUUGGAUGUGACG | 22 | Noncoding |
| miR11550 | UGUGGCGAACAAGUAACUCAUU | 22 | Noncoding |
A very large number of miRNA families, 19 out of the 41 phasiRNA triggers, were found to target NB-LRR genes, including 10 known miRNAs and 9 novel mRNAs (table 1). This set is far larger and more diverse than the triggers of phasiRNAs from NB-LRRs (loci we previously termed phasi-NB-LRRs or pNLs) reported to date, suggesting that the miRNA-pNL network is rather profuse in gymnosperms. Given the remarkable number of NB-LRR triggers and pNLs identified in spruce, our following analyses mainly focused on this expanded pathway.
A Large Set of miRNAs Target NB-LRR Genes and Trigger phasiRNA Production
As observed in several eudicots, NB-LRR genes comprise the largest class of PHAS genes in spruce (Zhai et al. 2011; Xia et al. 2013; Arikit et al. 2014). The network of 19 miRNA triggers and greater than 750 pNLs that we discovered in spruce was the basis for our investigation of the evolutionary significance of this pathway. We examined within NB-LRR genes the target site distribution of 22 miRNA families. As many miRNAs were found to target the encoded TIR domain, we modeled a typical gene encoding a TIR–NB–LRR with its three encoded, conserved domains (TIR, NBS, and LRR), indicating the target-site position for each miRNA (fig. 2A). Among the three domains, the encoded TIR and NBS were heavily targeted, by as many as 19 miRNAs, all of which are 22 nt (fig. 2A), suggesting that these two regions of the transcripts give rise to copious phasiRNAs. Eight miRNAs targeted the region coding for the TIR domain, including miR950, miR951, miR3697, and miR1312. Eleven 22-nt miRNAs targeted the NBS domain region, including the well-known master regulator miR482/miR2118 superfamily. Target sites of only two miRNAs, miR3709 and miR3710, fell into the region coding for the LRR domain, and target sites of a family of 21-nt miRNAs (miR11466) were positioned in the linking region between the encoded NBS and LRR domains (fig. 2A).
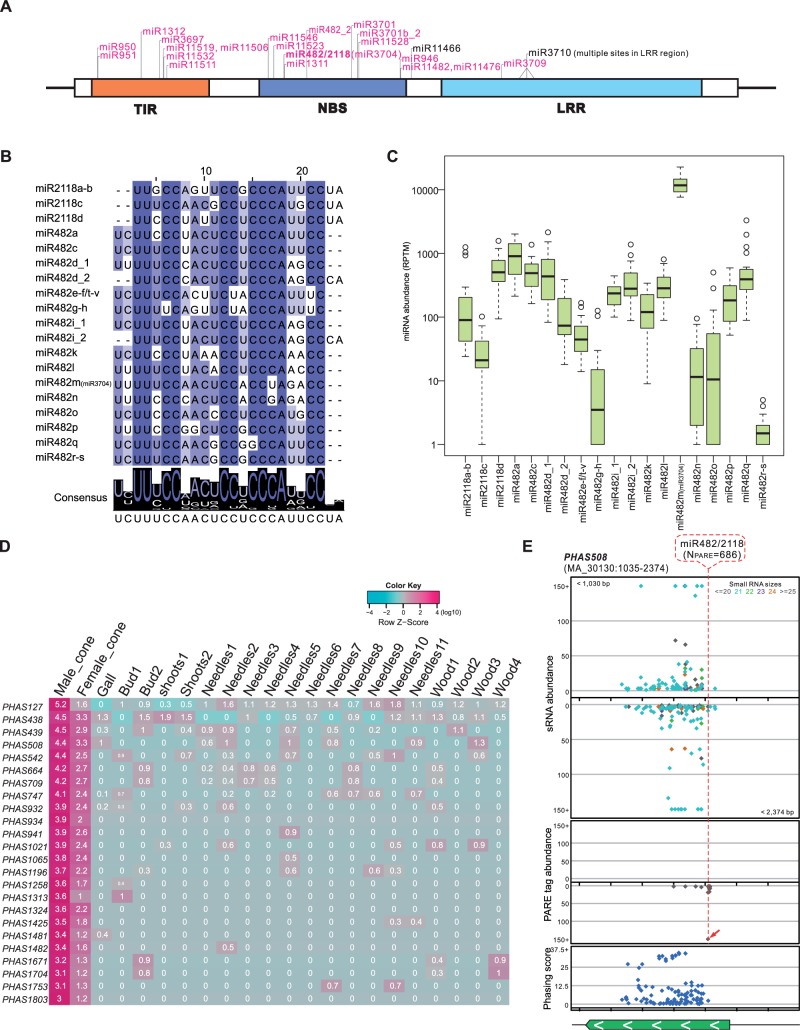
Fig. 2.
An expanded miRNA-NB-LRR-phasiRNA network in Norway spruce. (A) Target site distribution of miRNAs identified in spruce targeting NB-LRRs. A prototypical NB-LRR gene encodes three conserved domains: TIR (Toll/interleukin receptor-like domain), NBS, and an LRR (leucine-rich repeat) region. The 22-nt miRNAs are marked in pink. Multiple target sites for miR3710 (21 nt) are found in the region coding for the LRR domain. (B) Alignment of mature sequences for all members of the miR482/miR2118 superfamily in spruce. The degree of conservation for each nucleotide along the miRNAs is represented by the color, with a dark color denoting a high level of conservation and a light color denoting a low level. The consensus sequence of the alignment is displayed below with sequence logos. (C) Abundance of miR482/miR2118 mature sequences in 22 tissues of spruce. (D) PhasiRNAs of 24 noncoding PHAS loci specifically accumulated in reproductive tissues. Values are displayed in a log10 scale, with the pink color denoting high abundance and green indicating low abundance. Column titles indicate their source tissues, as detailed in supplementary table S1, Supplementary Material online. (E) An example of a noncoding PHAS locus (PHAS508) targeted by miR482/miR2118. Distributions of sRNAs, PARE data, and phasing score of the PHAS508 loci are viewed in different tracks. Dots in different colors in the sRNA track denote sRNAs of different lengths. A transcript annotated in the spruce genome (http://congenie.org/gbrowse) is shown at the bottom with the transcription direction indicated with white arrows. Introns in a gene are indicated by light-orange boxes.
An Expanded miR482/miR2118 Superfamily Triggers phasiRNA Production from Both NB-LRR Genes and Noncoding Transcripts
pNLs are widespread in eudicot species, and a few miRNAs/families have been identified as the triggers of this phasiRNA production (Fei et al. 2013). We observed that miR482/miR2118 is, so far, the only set of pNL triggers conserved in all the eudicot species. The superfamily comprises two types of miRNA sequences: miR482 and miR2118, differing by a 2-nt shift (Shivaprasad et al. 2012). We identified 23 MIR482/MIR2118 genes in spruce by our miRNA annotation pipeline (MIR2118a-b and MIR482a-p/t-x; supplementary table S2, Supplementary Material online). In addition, manual sequence searches and stem-loop structure confirmation identified three more MIR482/MIR2118 genes (MIR482q-s; supplementary table S2, Supplementary Material online); these had an extremely long stem-loop precursor, exceeding the cutoff of precursor length set in our automated annotation pipeline. Twenty-four out of the 26 MIR482/MIR2118 genes encode 19 unique miR482/miR2118 sequences, with MIR482d and i giving rise to both variants (miR482 and miR2118; fig. 2B) and MIR482w and MIR482x generated different mature miRNA sequences, targeting NB-LRR genes as well (supplementary table S2, Supplementary Material online). Among these 24 MIR482/MIR2118 genes, 18 produced only the miR482-type miRNA, and 4 generated only the miR2118-type miRNA (fig. 2B). The 19 miRNA sequences are relatively diverse with only half (12 positions) of the aligned nucleotides highly conserved, but all are 22 nt in length and possess an initial “U” (fig. 2B), canonical features of miRNA triggers of phasiRNAs. Most of these miRNAs showed moderate abundance, in the range of approximately 100 to approximately 1,000 RP20M (Reads per Twenty Million) (fig. 2C and supplementary table S8, Supplementary Material online). miR482m, recorded in miRBase as miR3704, demonstrated a consistently high abundance among all the libraries studied, with an average level exceeding approximately 10,000 RP20M (fig. 2C and supplementary table S8, Supplementary Material online). Copies miR482g/n/o/r/s showed relatively low abundances that varied greatly across libraries (fig. 2C and supplementary table S8, Supplementary Material online).
The miR482/miR2118 superfamily also exists in monocots; in grasses, it targets a large number of noncoding transcripts and triggers subsequent phasiRNA production in reproductive tissues (Johnson et al. 2009; Jeong et al. 2013), clearly distinct from the pNL targets of eudicots (Zhai et al. 2011; Fei et al. 2013). In spruce, the superfamily triggers many pNLs in many tissues (fig. 1D and supplementary table S8, Supplementary Material online). However, we also observed a subset of miR482/miR2118 targeted PHAS loci (24 loci) that displayed tissue-specific enrichment; these phasiRNAs were preferentially produced in reproductive tissues, with abundances in male cones much greater than in female cones (fig. 2D and supplementary table S9, Supplementary Material online). Target cleavage by miR482/miR2118 was confirmed by PARE (supplementary table S9, Supplementary Material online). Of the 24 loci, 21 were apparently noncoding, with no good protein-coding potential as assessed by BLAST (Basic Local Alignment Search Tool), whereas the other three loci potentially encode only very short peptides (<120 amino acids) of low similarity to public proteins, suggesting that they are likely to be noncoding as well. The phasiRNA-producing region is relatively short compared with pNLs, as exemplified in PHAS508 (fig. 2E). We concluded that in spruce, miR482/miR2118 triggers secondary siRNA production from both NB-LRR and noncoding transcripts, with the latter enriched in reproductive tissues, more typical of the grasses.
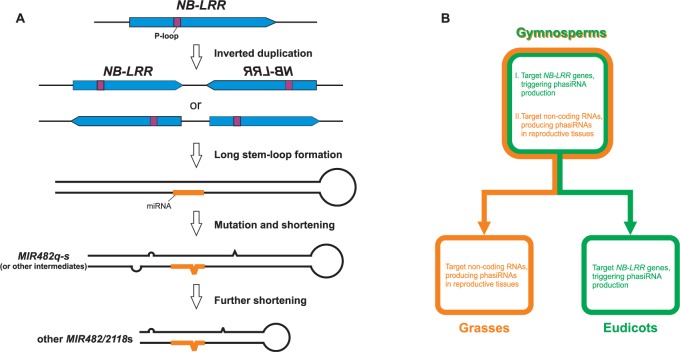
MIR482/MIR2118 Genes Emerged from NB-LRR Genes
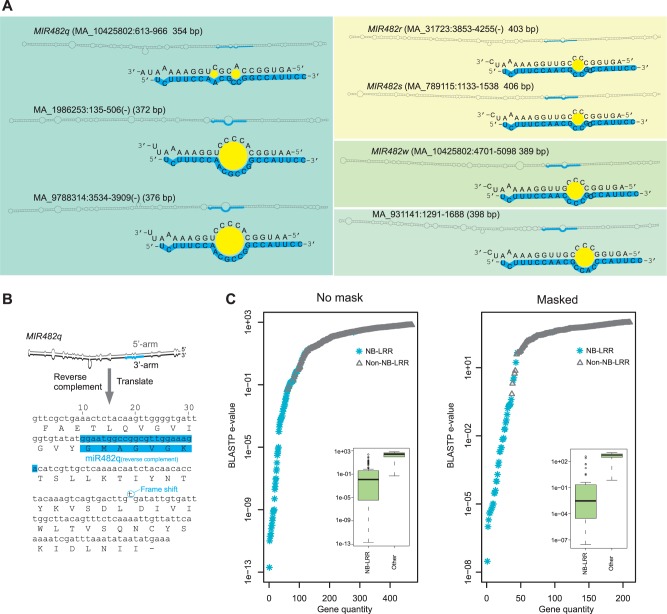
The total of 24 MIR482/MIR2118 genes (not including the two not producing miR482/miR2118) found in the spruce genome is the largest number known so far, in all studied plant species. miR482/miR2118 is only present in seed plants, apparently absent in mosses and ferns (Zhai et al. 2011), suggesting a possible origin in gymnosperms. To assess the evolution of MIR482/MIR2118, we analyzed its precursors. Almost all of these form a stem-loop with a typical less than 200 bp length, excepting three mentioned above (MIR482q–s) which have extremely long stem-loop structures (350 bp; supplementary fig. S3, Supplementary Material online). Several loci in spruce have similarity to these long MIR482 genes, with similar inverted-repeat structures (fig. 3A). Two are like miR482q, but with a diverged miRNA/miRNA* duplex relative to MIR482q (fig. 3A, left). The large loop size, which is rare in canonical plant miRNAs, likely prevents miRNA processing; that is, the mature miRNAs of the miR482/miR2118q were predominantly generated from the locus with a typical duplex structure (MA_10425802:613..966; fig. 3A, left). Two other loci have distinct miR482/miR2118 sequences and moderate middle loops were weakly represented in our data (MIR482w and MA_931141:1291..1688; fig. 3A). Therefore, only the MIR482q locus and the two MIR482r and MIR482s loci have good miRNA precursors (supplementary table S2 and fig. S3, Supplementary Material online), and the other loci might be evolutionary intermediates or devolved loci.
Fig. 3.
MIR482q evolved from a gene encoding a NB-LRR protein. (A) Stem-loop structures for selected copies of MIR482/MIR2118 or putative intermediate MIRNA precursors. miRNA sequences are highlighted by cyan lines, and miRNA/miRNA* duplexes are enlarged with the size of middle loop(s) represented by yellow circles. Stem-loops bearing the same miRNA sequence are indicated with different background colors. (B) The 3′-arm of MIR482q encodes an appreciable peptide. The position of the mature miRNA (its reverse complement) and the translated amino acid sequence are marked with cyan lines. The one nucleotide frame shift allowing for accurate translation is indicated. (C) Distribution of e values from BLASTP (see Materials and Methods) using the peptide encoded by the sequence of the 3′-arm of MIR482q, searching with no mask (left) or masked conditions (right).
We next examined the abnormally long stem-loop sequences of the precursor genes to miR482q and miR482r-s, as prior work has demonstrated the origin of some miRNAs from inverted duplication of coding sequences from their target genes (Allen et al. 2004). We started with miR482q, due to its high abundance compared with miR482r-s (fig. 2C). The reverse complement of the 176-nt long 3′-arm of the MIR482q stem-loop encodes a possible peptide of 57 amino acids, with a single nucleotide frameshift (fig. 3B). A BLASTP search of the peptide sequence against annotated spruce genes found significant similarity to NB-LRR proteins (fig. 3C, left). The e values for the NB-LRR hits were significantly lower than those for non-NB-LRRs (fig. 3C inset). To exclude the possibility that the great similarity to NB-LRRs was mainly obtained from the miRNA/miRNA* region, we masked the miRNA sequence region and repeated the sequence comparison, and the precursors still displayed high sequence similarity to NB-LRRs (fig. 3C, right). Therefore, as exemplified by MIR482q, the data are consistent with an origin for MIR482/MIR2118 genes from NB-LRR genes through inverted duplication and selection for miRNA biogenesis.
Extensive and Dynamic miR390-TAS3 Pathway in Spruce
In plants, the miR390-directed TAS3 pathway is a classical representative of a PHAS network. In previously characterized plant genomes, TAS3 consists of a small gene family (<10 genes), including two types of TAS3 genes: TAS3 genes with two tasiARFs (tasiRNA targeting ARF genes) and TAS3 genes with only one tasiARF (Axtell et al. 2006; Krasnikova et al. 2009; Xia et al. 2012). We characterized 18 TAS3 genes in spruce, an unexpectedly large number (16 from PHAS analysis and 2 from manual sequence comparisons; fig. 4 and supplementary table S10 and file S1, Supplementary Material online); tasiARF-directed cleavage was detected for four ARF genes in spruce (supplementary table S11, Supplementary Material online). Overall, apart from the conserved components that include the miR390 target site and tasiARF motif, the spruce TAS3 gene family showed features indicative of a dynamic history.
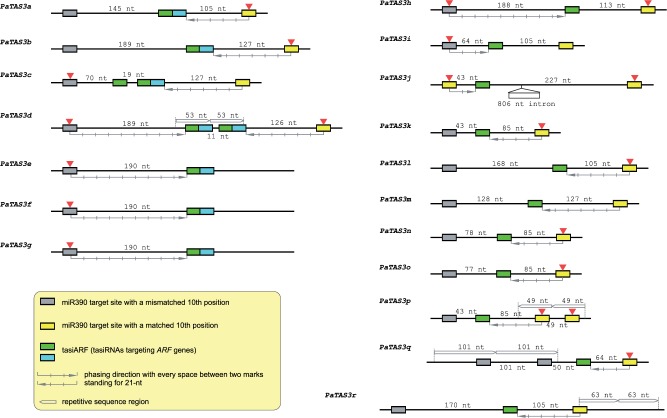
Fig. 4.
TAS3 genes in spruce. TAS3 genes identified in spruce are displayed in diagrams with tasiARFs denoted by green or cyan boxes (indicating different tasiARF sequences) and the miR390 target site in gray or yellow boxes. Target sites cleaved by miR390 are marked with a small red triangle on the top. Internal tandem repeat sequences are shown with long unfilled arrows. The direction of phasing production of tasiARFs is indicated by long gray arrows with hash marks to indicate phasiRNA registers. The distance is indicated between two sequence motifs (miR390 target site or tasiARF), with the miRNA target site measured from the cleavage site (the tenth position of the miRNA). sRNAs within a distance of ±1 nt to a multiple of 21 nt are considered as phased. (For example, the distance between 5′ target site and tasiARF in PaTAS3d is 190 nt, which is 1 nt longer than 9 × 21 nt.).
For the miR390 target site, all the TAS3 genes identified have two miRNA target sites, except PaTAS3q with three miR390 sites (two 5′ sites and one 3′ site) and PaTAS3e/f/g with only a 5′ miR390 target site. The two miR390 sites of most TAS3 genes are arranged in a way resembling the Arabidopsis TAS3 model, that is, the 5′ target site bears a conserved mismatch at the tenth position of miRNA-target pairing and the 3′ site has a tenth position match, critical for miR390-mediated cleavage. The cleavage of 3′ target sites was detected as expected in most cases. However, the 5′ miR390 site containing the tenth mismatch was cleaved in many spruce TAS3 genes (PaTASc-l), and all of them, except PaTAS3c, can serve as a trigger site of tasiARF production. In contrast, the two miR390 target sites of PaTAS3j do not have a tenth position mismatch, and both were cleaved.
The tasiARFs also displayed great diversity. From their precursors, PaTAS3a–g have at least two tasiARFs, with PaTAS3c and PaTAS3d generating three and four tasiARF copies, respectively, whereas PaTAS3h–r generate only one tasiARF. The distance between the miR390 cleavage sites and tasiARFs varied greatly in the spruce TAS3 genes (fig. 4). In addition, tasiARF positions were in phase to either just one miR390 site or both sites, presumably reflecting the production of the tasiARF directed by either the 3′ miR390 site (PaTASa–c, k–r), the 5′ miR390 site (PaTAS3e-j), or by both (PaTAS3d). Interestingly, in PaTAS3d the two sets of tandem tasiARFs are in phase with different miR390 target sites, that is, the 5′ site initiates the production of the 5′ tandem tasiARFs and the 3′ site sets the phasing for the 3′ tandem tasiARFs. For PaTAS3b/i/k/l, although the phasing and cleavage were detected on one side (3′ side for PaTAS3b, k, and l; 3′ side for PaTAS3i), the distance between tasiARF and miR390 cleavage site on the other side is a multiple of 21 nt, implying a possible phasing production of tasiARF from the other direction.
Another feature of note for spruce TAS3 genes is the presence of tandem repeat sequences of varied length within the TAS3 genes. Four spruce TAS3 genes contain tandem repeats within their gene body. A 53-nt repeat in the middle of PaTAS3d generates two sets of tandem tasiARFs. Similarly, a 49-nt repeat in PaTAS3p and a 101-nt repeat in PaTAS3q give rise to duplicated miR390 target sites at the 3′- or 5′-end, respectively. And PaTAS3r has a 63-nt repetitive sequence covering part of the 3′ miR390 target site. Moreover, an 806-nt long intron was found between the 3′ target site and the single tasiARF in PaTAS3j, yet another diverse example in the spruce TAS3 gene family.
In conclusion, spruce TAS3 genes demonstrate divergent features, including a varied number of miR390 target sites and tasiARF sequences, diverse mechanisms of production of the tasiARFs as evidenced in the phasing registers, and the existence of tandemly repetitive sequences or introns in TAS3 genes (fig. 4). These atypical features vary substantially from the canonical “two-hit” TAS3 loci seen in Arabidopsis.
Discussion
miRNAs in Gymnosperms
To date, only 144 MIRNA genes from four gymnosperm species have been deposited in miRBase, a number lagging behind other plant groups. Norway spruce, a conifer that dominates many terrestrial ecosystems in the northern hemisphere, is the first gymnosperm species with full genome sequence (Nystedt et al. 2013). Using stringent criteria, we identified 585 MIRNA genes from the Norway spruce, producing approximately 426 unique miRNA sequences. This is one of the most numerous groups of MIRNAs yet reported from a single plant genome, perhaps consistent with the large genome size of the Norway spruce (∼20 Gb). The large population of miRNAs is attributable to not only the high copy number of conserved miRNAs (e.g., the miR156 family comprises 28 MIR156 genes and the miR397 family contains 21 MIRNA members; supplementary table S2, Supplementary Material online) but also the high number of nonconserved miRNAs (supplementary table S6, Supplementary Material online).
In plants, 22 miRNA families are known to be conserved (Axtell and Bowman 2008; Cuperus et al. 2011), 8 of which are in the common ancestor of all embryophytes (land plants), and the other 14 miRNA families are present in all angiosperm lineages (flowering plants). All 8 embryophyte-conserved miRNAs were found in Norway spruce, and the 14 angiosperm-conserved miRNAs, with the exception of miR827, were also identified in spruce. Thus, we conclude that these miRNAs emerged in the common ancestor of spermatophytes (seed plants, including both gymnosperms and angiosperms). Seventeen of 21 conserved miRNA families maintained conserved target relationships, indicating that the cognate miRNA-target pairs were acquired before the split of gymnosperms and angiosperms and implying the importance of their functions. Several miRNAs of restricted conservation, considered previously as “non- or less-conserved miRNAs,” were also found in spruce, suggesting that early-evolved miRNAs may have been selectively lost in certain lineages during evolution. For instance, miR828 and miR858 and their MYB regulatory functions were thought to be eudicot-specific (Xia et al. 2012; Rock 2013), and miR529 and its SPL targets are present in mosses and monocots but absent in most eudicots (Cuperus et al. 2011; Jeong et al. 2011). For the miR482/miR2118 superfamily, broader analyses show that it is present in almost all the seed plants, with the earliest detectable members in two gymnosperm species, ginkgo and spruce (Zhai et al. 2011; Nystedt et al. 2013). Therefore, we propose that miR482/miR2118 is a conserved miRNA, such as other spermatophyte-conserved miRNAs such as miR172 and miR167.
Apart from the conserved miRNAs, some miRNAs we observed in spruce are also present in other conifer species, such as miR1311–miR1315, miR3709, miR950, and miR951. They may be specific to the Pinaceae or gymnosperms, suggesting potential roles in the Pinaceae or gymnosperm-specific features. We also identified an extraordinarily large number of novel miRNAs in Norway spruce, perhaps more than any single plant genome analyzed to date. Only a small portion of these miRNAs have potential homologs in ginkgo, another gymnosperm, indicating lineage specificity. The extremely large genome size of Norway spruce resulted primarily from accumulation of diverse long-terminal repeat transposable elements, but also the presence of numerous pseudogenes and long noncoding RNAs (Nystedt et al. 2013). Given that 24-nt siRNAs function in transposable element silencing in plants, are expressed at a much lower level in spruce, and are restricted to reproductive tissues, the rapid evolution of new miRNA pathways, possibly in conjunction with downstream PHAS pathways, might compensate to suppress the activity of these sequences in the genome. On the other hand, these genomic elements may provide raw materials to evolve new miRNAs (as discussed below), contributing to the large miRNA population in spruce.
An Expanded miRNA–PHAS–phasiRNA Network in Spruce
PhasiRNAs are a major class of sRNAs in plants. Källman et al. (2013) previously reported that phasiRNAs from NB-LRRs are the main source of sRNAs in Norway spruce, but that study did not assess the overall picture of phasiRNAs in the gymnosperm—that is, how conserved and diverse are PHAS loci in Norway spruce. Here we characterized an impressively high number of PHAS loci (2,061) in spruce, triggered by as many as 41 miRNA families. This is a substantial miRNA–phasiRNA network, greater than others thus far characterized in plants. The three major protein-coding PHAS gene families in eudicots, NB-LRRs, PPRs, and MYBs (Fei et al. 2013), also give rise to profuse phasiRNAs in spruce, indicating that these are ancient features of plant genomes. In fact, these miRNA trigger-PHAS gene relationships are clearly well-established in spruce. For instance, miR482/miR2118 and miR828 are triggers for NB-LRR and MYB phasiRNAs, respectively, as in angiosperms (Zhai et al. 2011; Xia et al. 2012). Three other phasiRNA pathways conserved in plants, miR390-TAS3, miR393-TIR1/AFB, and miR4376-ACA10, were also identified in spruce. On the other hand, some phasiRNA pathways appear to be Pinaceae- or gymnosperm-specific, including 15 miRNAs already in miRBase and 21 newly identified miRNAs (table 1); this indicates that a substantial number of PHAS pathways were likely lost during evolution or gained recently, implying a dynamic feature of PHAS networks in gymnosperm. All these results support that extensive miRNA–phasiRNA networks exist in the gymnosperms, predating the angiosperms.
The NB-LRRs comprise the largest group of PHAS genes in spruce. Around 800 pNL genes were found, producing considerable phasiRNAs. Consistent with such a large number of pNLs, spruce apparently evolved a large MIR482/MIR2118 gene family, consisting of 26 MIRNA genes coding for 19 distinct miR482/miR2118 sequences. It is the largest miR482/miR2118 family yet reported in plants. Another 18 miRNAs in the spruce genome target and trigger phasiRNA production from NB-LRRs. In other plant genomes, miRNAs other than miR482/miR2118 generate pNLs, including miR1507 and miR2109 in Medicago (Zhai et al. 2011), miR1510 in soybean (Arikit et al. 2014), and miR6019 and miR6024–miR6027 in tomato and potato (Li et al. 2012). Yet, none of these genomes has an miRNA–pNL network as extensive as in spruce. The network of NB-LRR genes in spruce is broadly impacted by sRNAs, with the functional importance yet to be determined (as is true for all plants).
Another line of evidence demonstrating the expansive nature of the PHAS network in spruce is its extraordinary number of TAS3 genes, including both known types of TAS3 genes (one or two tasiARFs). In modern plants, the miR390–TAS3–ARF pathway plays a critical role in auxin signaling by producing tasiRNAs that target ARF genes. This pathway is involved in the regulation of leaf morphology, lateral root growth, and developmental timing or patterning (Adenot et al. 2006; Fahlgren et al. 2006; Garcia et al. 2006; Marin et al. 2010; Yifhar et al. 2012; Zhou et al. 2013; Dotto et al. 2014). The TAS3 genes characterized in spruce are of varying size, with varying numbers of miR390 target sites and tasiARFs; tasiARF production is triggered by either miR390 target site (5′ or 3′) or by both, a mechanism inconsistent with the classical “two-hit” model (Axtell et al. 2006).This implies a dynamic evolutionary nature of TAS3 genes. Although the presence of both miR390 and TAS3 traces back to early land plants, such as liverworts and mosses (Axtell et al. 2006; Krasnikova et al. 2013), the function of TAS3 has changed during the course of evolution. In liverworts, TAS3 produces tasiRNAs targeting AP2 genes; in mosses, TAS3 tasiRNAs target both AP2 and ARF genes, whereas in modern plants, TAS3 produces tasiRNA targeting solely ARF genes. How this functional diversification of TAS3 occurred in land plants remains unknown.
Evolution and Function Diversification of miR482/miR2118
The typical evolutionary origins of MIRNAs are partially but perhaps not fully described. Data support several paths for emergence, all involving the divergence and evolutionary tuning of different starting sequences, such as the following: Inverted duplication of target genes (Allen et al. 2004), ancient MIRNA genes (Xia et al. 2013), randomly formed foldback sequences (Felippes et al. 2008), or miniature inverted-repeat transposable elements (Piriyapongsa and Jordan 2008). All except divergence from ancient predecessors require the accumulation of mutations, acquisition of DCL1 as a master processing enzyme, and subsequent production of discrete miRNA species, happening in the context of coevolution with the mRNA targets (Voinnet 2009). In spruce, we discovered sequence similarity of MIR482/MIR2118 precursors and NB-LRR genes, consistent with emergence of miR482/miR2118 through inverted duplications (Allen et al. 2004). In brief, in this model, an NB-LRR gene or fragment is duplicated to form a long, paired, stem-loop structure; this gradually accumulates sequence mutations to become shorter, like MIR482q/r/s or other intermediates, resulting in the recruitment of DCL1 as a miRNA processing enzyme; the shortening of the stem-loop generates a canonical length for an MIRNA precursor (<200 bp, like most MIR482/MIR2118 genes in spruce), ensuring effective processing of the right sRNA, the mature miR482 in this case (fig. 5A). This evolutionary history shows that miRNAs can indeed evolve from target genes, not only giving rise both to completely novel and “young” miRNAs, such as miR162 and miR163 in Arabidopsis (Allen et al. 2004), but also generating added copies of highly conserved miRNAs, such as miR482/miR2118. Conceivably, paralogs of other conserved miRNAs may have originated this way, but the evidence for this, sequence homology between MIRNA gene and target genes, may be obscured by mutation and selection over millions of years.
Fig. 5.
A model of the evolutionary emergence of MIR482/MIR2118 paralogs. (A) A model for the evolutionary emergence of a MIR482/MIR2118 precursor begins with the spontaneous formation of an inverted repeat of an NB-LRR gene or gene fragment through a duplication event that forms a long, well-paired stem-loop structure. This structure, over time, accumulates sequence mutations, which could also involve shortening of the structure. A short and imperfectly-paired stem-loop is a good substrate for DCL1 processing, leading to integration in the miRNA biogenesis pathway, as a canonical MIR482/MIR2118 gene. (B) Functional diversification of MIR482/MIR2118 in seed plants.
The miR482 family targets both NB-LRRs, identified first in Populus trichocarpa (Lu et al. 2005, 2008), and noncoding RNAs, identified first in rice inflorescences (Johnson et al. 2009). Subsequent analyses identified a larger miR482/miR2118 superfamily, and a broader function in dicots in the coordinate regulation of many NB-LRRs by targeting conserved sequences to trigger phasiRNAs (Zhai et al. 2011; Shivaprasad et al. 2012). In grasses, phasiRNA production from noncoding RNAs seems restricted largely to reproductive tissues (Johnson et al. 2009; Song et al. 2010; Vogel et al. 2010). Here we demonstrated that miR482/miR2118 targets both NB-LRRs and noncoding transcripts in Norway spruce, and phasiRNAs from the latter preferentially accumulate in male and female cones. As gymnosperms emerged prior to angiosperms and the monocot/dicot diversification, we infer that miR482/miR2118 performs dual functions, which may have been selectively and differentially retained by dicots and monocots (fig. 5B). Based on phasiRNA presence, miR482/miR2118 in spruce targets hundreds of pNL genes, but only 24 PHAS noncoding RNAs. The pNL count resembles dicots such as tomato, soybean, and peach (i.e., >100 pNLs), whereas the noncoding targets are fewer than in grasses, suggesting a great expansion of the miR482/miR2118-targeted noncoding RNAs occurred later, perhaps during monocot evolution.
In dicots, miR482/miR2118–NB-LRR–phasiRNAs may have a variety of functions, perhaps buffering transcript levels, perhaps acting as a counter–counter–defense system, perhaps limiting fitness costs of overactive resistance, or perhaps other roles (Shivaprasad et al. 2012; Fei et al. 2013; Pumplin and Voinnet 2013). A knockdown of miR472, an miR482/miR2118 family member in Arabidopsis, displayed enhanced disease resistance, whereas plants overexpressing miR472 were more susceptible (Boccara et al. 2014). In grasses, the function of miR482/miR2118-triggered phasiRNAs is unknown. Thus, the dual functions of the miR482/miR2118 in spruce are also largely as-yet unknown, but have clearly been important enough to survive hundreds of millions of years of genome evolution.
Altogether, in this study, we demonstrated the presence of an extensive network of miRNAs and phasiRNAs in the gymnosperm Norway spruce, as evidenced by 1) a multitude of miRNAs which target diverse genes; 2) thousands of PHAS loci, including both coding and noncoding genes, capable of producing phasiRNAs; 3) a large number of miRNAs serving as triggers of phasiRNA production; 4) a remarkable set of miRNAs targeting pNLs and instigating phasiRNA biogenesis; 5) a greatly expanded miR482/miR2118 superfamily, the master regulator of pNLs, and the origins of some paralogs; and 6) the to-date largest and most diverse TAS3 gene family. These results suggested that phasiRNA-based gene regulation is an ancient and broad regulatory strategy adopted in plants, selectively maintained over hundreds of millions of years of plant evolution.
Materials and Methods
sRNA Library Construction and miRNA Annotation
Forty-four sRNA libraries from 22 different tissue samples for spruce were retrieved from the ENA database (http://www.ebi.ac.uk/ena/, last accessed August 8, 2015), project number ERP002476 (Nystedt et al. 2013). miRNA annotation was conducted as in the workflow (supplementary fig. S1, Supplementary Material online). Briefly, the sRNA data were quality-filtered, trimmed of adaptors, and collapsed and counted according to nonredundant sequences; then the distinct sRNA sequences were mapped to the P. abies genome (Pabies 1.0; http://congenie.org/, last accessed August 8, 2015) with no mismatches allowed. Next, the sRNA sequences of ≥10 raw reads, 20–22 nt in length and matching ≤20 genomic loci were retained and subjected to a screen for stem-loop structures, using a modified version of miREAP (http://sourceforge.net/projects/mireap/, last accessed August 8, 2015). The potential miRNAs were classified into known and novel miRNAs by BLAST analysis of sRNA sequences against miRBase version 20, and we applied filters for biases of abundance and strand-matching (as describe above); only miRNAs with miRNA* sequence found in a given library were retained for a final manual check. Stem-loop structures of novel miRNAs are included in supplementary file S2, Supplementary Material online. sRNA data were normalized to RP20M across libraries.
Three tissues of G. biloba “Fastigiata,” leaf, leaf bud, and bark, were collected at the University of Delaware. Total RNA was extracted from the three samples using the Purelink Plant RNA Reagent from Life Technologies (New York) according to the manufacturer’s protocol. sRNA libraries were constructed using the TruSeq Small RNA Sample Preparation Kits (Illumina, Hayward, CA). sRNAs with ≥10 raw reads were retained for identification of putative miRNA homologs in ginkgo versus spruce by sequence comparisons to spruce miRNAs, allowing ≤4 mismatches.
PARE Library Construction and Data Analysis
One-year-old branches were collected from a Norway spruce tree in the University of Delaware Botanic Gardens (Newark, DE). Three tissues, bud, needle, and stem, were separated from the collected branches. Another sample was a mixture pooled with different tissue parts from the branches. Total RNA was extracted from the four samples using the Purelink Plant RNA Reagent from Life Technologies (New York) according to the manufacturer’s protocol. PARE libraries were constructed as previously described (Zhai et al. 2014) and sequenced on the Illumina HiSeq 2500 in the DNA Sequencing & Genotyping Center in the Delaware Biotechnology Institute (Newark, DE).
After adaptor-trimming and genomic mapping, as done for the sRNA data, the CleaveLand pipeline 2.0 (Addo-Quaye et al. 2009) was optimized to analyze the PARE data in collaboration with Targetfinder 1.6 (https://github.com/carringtonlab/TargetFinder?, last accessed August 8, 2015). The alignment score threshold was set to 4.5 for known miRNAs and 5 for novel miRNAs. The model gene file (Pabies1.0_ HC_cds.fna), which contains well-annotated genes of high confidence, and the gene homolog annotation file (pabies_poplar.txt), were obtained from the ConGenIE (http://congenie.org/, last accessed August 8, 2015). PARE data sets were normalized to TP10M (Transcripts per Ten Million).
PHAS Locus Annotation
Annotation of PHAS loci was conducted largely by application of a P-value-based approach, as described in Xia et al. (2013). The P-value cutoff was set to 0.001. Additional filters were applied to retain loci for which 1) sRNA genomic matches (aka “hits”) were ≤10, 2) 21-nt sRNAs accounted for at least 50% of all the reads matched to a given locus, 3) the length of a sequence region producing phasiRNAs was ≥100 bp, and 4) the percentage of in-phase sRNAs was ≥0.3. PHAS annotation was performed separately for each tissue, and results were combined to a nonredundant master list based on the genomic coordinates.
Classification of Coding and Noncoding Loci
The PHAS loci that we identified were classified as “coding” or “noncoding” according to the workflow in supplementary figure S2, Supplementary Material online. Briefly, sequences including the PHAS loci and 50-bp flanking sequences at both ends were collected for each PHAS locus. Sequences failing to encode a single peptide of greater than 100 amino acids were subjected to the calculation of protein-coding potential by CPC (Coding Potential Calculator) (Kong et al. 2007). PHAS loci of coding potential −1 < CP < 0 were then retrieved for comparison by BLAST against the public protein data set UniRef90 (http://www.uniprot.org/help/uniref, last accessed August 8, 2015). Sequences with an e value > 1e-4 were further checked for overlap with annotated genes from spruce (Pabies1.0_HC.gff3, http://congenie.org/). The protein similarity of coding PHAS loci was annotated de novo by BLASTX against UniRef90.
Accession Numbers
All the sRNA data for 44 libraries can be found in the ENA database (http://www.ebi.ac.uk/ena/, last accessed August 8, 2015), project number ERP002476 (Nystedt et al. 2013). Sequence data for the four PARE libraries are deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/, last accessed August 8, 2015) under the accession number of GSE65248.
Supplementary Material
Supplementary files S1 and S2, tables S1–S11, and figures S1–S3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by National Science Fundation Integrative Organismal System award #1257869. The authors are grateful to members of the Meyers lab for helpful discussions and assistance with handling of sequence data.
References
- Addo-Quaye C, Miller W, Axtell MJ. 2009. CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25:130–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouché N, Gasciolli V, Vaucheret H. 2006. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol. 16:927–932. [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. 2005. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121:207–221. [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Sung G-H, Spatafora JW, Carrington JC. 2004. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet. 36:1282–1290. [DOI] [PubMed] [Google Scholar]
- Arikit S, Xia R, Kakrana A, Huang K, Zhai J, Yan Z, Valdés-López O, Prince S, Musket TA, Nguyen HT, et al. 2014. An atlas of soybean small RNAs identifies phased siRNAs from hundreds of coding genes. Plant Cell 26:4584–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Bowman JL. 2008. Evolution of plant microRNAs and their targets. Trends Plant Sci. 13:343–349. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP. 2006. A two-hit trigger for siRNA biogenesis in plants. Cell 127:565–577. [DOI] [PubMed] [Google Scholar]
- Boccara M, Sarazin A, Thiébeauld O, Jay F, Voinnet O, Navarro L, Colot V. 2014. The arabidopsis miR472-RDR6 silencing pathway modulates PAMP- and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog. 10:e1003883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H-M, Chen L-T, Patel K, Li Y-H, Baulcombe DC, Wu S-H. 2010. 22-nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci U S A. 107:15269–15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. 2009. Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol. 25:21–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Carbonell A, Fahlgren N, Garcia-Ruiz H, Burke RT, Takeda A, Sullivan CM, Gilbert SD, Montgomery TA, Carrington JC. 2010. Unique functionality of 22 nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat Struct Mol Biol. 17:997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Fahlgren N, Carrington JC. 2011. Evolution and functional diversification of MIRNA genes. Plant Cell 23:431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG. 2001. Plant pathogens and integrated defence responses to infection. Nature 411:826–833. [DOI] [PubMed] [Google Scholar]
- Dotto MC, Petsch KA, Aukerman MJ, Beatty M, Hammell M, Timmermans MCP. 2014. Genome-wide analysis of leafbladeless1-regulated and phased small RNAs underscores the importance of the TAS3 ta-siRNA pathway to maize development. PLoS Genet. 10:e1004826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC. 2006. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol. 16:939–944. [DOI] [PubMed] [Google Scholar]
- Fei Q, Xia R, Meyers BC. 2013. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25:2400–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felippes FF, Schneeberger K, Dezulian T, Huson DH, Weigel D. 2008. Evolution of Arabidopsis thaliana microRNAs from random sequences. RNA 14:2455–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Collier SA, Byrne ME, Martienssen RA. 2006. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr Biol. 16:933–938. [DOI] [PubMed] [Google Scholar]
- German MA, Pillay M, Jeong D-H, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. 2008. Global identification of microRNA–target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol. 26:941–946. [DOI] [PubMed] [Google Scholar]
- Jeong D-H, Park S, Zhai J, Gurazada SGR, Paoli ED, Meyers BC, Green PJ. 2011. Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 23:4185–4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D-H, Schmidt SA, Rymarquis LA, Park S, Ganssmann M, German MA, Accerbi M, Zhai J, Fahlgren N, Fox SE, et al. 2013. Parallel analysis of RNA ends enhances global investigation of microRNAs and target RNAs of Brachypodium distachyon. Genome Biol. 14:R145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Kasprzewska A, Tennessen K, Fernandes J, Nan G-L, Walbot V, Sundaresan V, Vance V, Bowman LH. 2009. Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 19:1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. 2006. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol. 57:19–53. [DOI] [PubMed] [Google Scholar]
- Källman T, Chen J, Gyllenstrand N, Lagercrantz U. 2013. A significant fraction of 21-nucleotide small RNA originates from phased degradation of resistance genes in several perennial species. Plant Physiol. 162:741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye Z-Q, Liu X-Q, Zhao S-Q, Wei L, Gao G. 2007. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 35:W345–W349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnikova MS, Goryunov DV, Troitsky AV, Solovyev AG, Ozerova LV, Morozov SY. 2013. Peculiar evolutionary history of miR390-guided TAS3-Like genes in land plants. Sci World J. 2013:e924153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnikova MS, Milyutina IA, Bobrova VK, Ozerova LV, Troitsky AV, Solovyev AG, Morozov SY. 2009. Novel miR390-dependent transacting siRNA precursors in plants revealed by a PCR-based experimental approach and database analysis. BioMed Res Int. 2009:e952304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J, Sun H, Kumar P, Baker B. 2012. MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci U S A. 109:1790–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Sun Y-H, Chiang VL. 2008. Stress-responsive microRNAs in Populus. Plant J. 55:131–151. [DOI] [PubMed] [Google Scholar]
- Lu S, Sun Y-H, Shi R, Clark C, Li L, Chiang VL. 2005. Novel and mechanical stress–responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 17:2186–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, Nussaume L, Crespi MD, Maizel A. 2010. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22:1104–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos JL, Bologna NG, Chorostecki U, Palatnik JF. 2010. Identification of microRNA processing determinants by random mutagenesis of Arabidopsis MIR172a precursor. Curr Biol. 20:49–54. [DOI] [PubMed] [Google Scholar]
- Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ, et al. 2008. Criteria for annotation of plant microRNAs. Plant Cell 20:3186–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Kaushik S, Nandety RS. 2005. Evolving disease resistance genes. Curr Opin Plant Biol. 8:129–134. [DOI] [PubMed] [Google Scholar]
- Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin Y-C, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, et al. 2013. The Norway spruce genome sequence and conifer genome evolution. Nature 497:579–584. [DOI] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. 2004. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18:2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piriyapongsa J, Jordan IK. 2008. Dual coding of siRNAs and miRNAs by plant transposable elements. RNA 14:814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumplin N, Voinnet O. 2013. RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat Rev Microbiol. 11:745–760. [DOI] [PubMed] [Google Scholar]
- Rock CD. 2013. Trans-acting small interfering RNA4: key to nutraceuticalsynthesis in grape development? Trends Plant Sci.. 18:601–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad PV, Chen H-M, Patel K, Bond DM, Santos BACM, Baulcombe DC. 2012. A microRNA superfamily regulates nucleotide binding site–leucine-rich repeats and other mRNAs. Plant Cell 24:859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Ammour A, Windels D, Arn-Bouldoires E, Kutter C, Ailhas J, Meins F, Vazquez F. 2011. miR393 and secondary siRNAs regulate expression of the TIR1/AFB2 auxin receptor clade and auxin-related development of Arabidopsis leaves. Plant Physiol. 157:683–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Axtell MJ, Fedoroff NV. 2010. RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr Biol. 20:37–41. [DOI] [PubMed] [Google Scholar]
- Song X, Li P, Zhai J, Zhou M, Ma L, Liu B, Jeong D-H, Nakano M, Cao S, Liu C, et al. 2012. Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J. 69:462–474. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Li Y-F, Jagadeeswaran G. 2012. Functions of microRNAs in plant stress responses. Trends Plant Sci. 17:196–203. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert J-L, Bartel DP, Crété P. 2004. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell. 16:69–79. [DOI] [PubMed] [Google Scholar]
- Vogel JP, Garvin DF, Mockler TC, Schmutz J, Rokhsar D, Bevan MW, Barry K, Lucas S, Harmon-Smith M, Lail K, et al. 2010. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463:763–768. [DOI] [PubMed] [Google Scholar]
- Voinnet O. 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136:669–687. [DOI] [PubMed] [Google Scholar]
- Wang Y, Itaya A, Zhong X, Wu Y, Zhang J, van der Knaap E, Olmstead R, Qi Y, Ding B. 2011. Function and evolution of a microRNA that regulates a Ca2+-ATPase and triggers the formation of phased small interfering RNAs in tomato reproductive growth. Plant Cell 23:3185–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S, Wollmann H, Schneeberger K, Weigel D. 2010. Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr Biol. 20:42–48. [DOI] [PubMed] [Google Scholar]
- Xia R, Meyers BC, Liu Z, Beers EP, Ye S, Liu Z. 2013. MicroRNA superfamilies descended from miR390 and their roles in secondary small interfering RNA biogenesis in eudicots. Plant Cell 25:1555–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R, Zhu H, An Y, Beers EP, Liu Z. 2012. Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol. 13:R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yifhar T, Pekker I, Peled D, Friedlander G, Pistunov A, Sabban M, Wachsman G, Alvarez JP, Amsellem Z, Eshed Y. 2012. Failure of the tomato trans-acting short interfering RNA program to regulate AUXIN RESPONSE FACTOR3 and ARF4 underlies the Wiry leaf syndrome. Plant Cel 24:3575–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS. 2005. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19:2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Arikit S, Simon SA, Kingham BF, Meyers BC. 2014. Rapid construction of parallel analysis of RNA end (PARE) libraries for Illumina sequencing. Methods 67:84–90. [DOI] [PubMed] [Google Scholar]
- Zhai J, Jeong D-H, Paoli ED, Park S, Rosen BD, Li Y, González AJ, Yan Z, Kitto SL, Grusak MA, et al. 2011. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25:2540–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Han L, Fu C, Wen J, Cheng X, Nakashima J, Ma J, Tang Y, Tan Y, Tadege M, et al. 2013. The trans-acting short interfering RNA3 pathway and NO APICAL MERISTEM antagonistically regulate leaf margin development and lateral organ separation, as revealed by analysis of an argonaute7/lobed leaflet1 mutant in Medicago truncatula. Plant Cell 25:4845–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.