Abstract
Coleoid cephalopods show remarkable evolutionary convergence with vertebrates in their neural organization, including (1) eyes and visual system with optic lobes, (2) specialized parts of the brain controlling learning and memory, such as vertical lobes, and (3) unique vasculature supporting such complexity of the central nervous system. We performed deep sequencing of eye transcriptomes of pygmy squids (Idiosepius paradoxus) and chambered nautiluses (Nautilus pompilius) to decipher the molecular basis of convergent evolution in cephalopods. RNA-seq was complemented by in situ hybridization to localize the expression of selected genes. We found three types of genomic innovations in the evolution of complex brains: (1) recruitment of novel genes into morphogenetic pathways, (2) recombination of various coding and regulatory regions of different genes, often called “evolutionary tinkering” or “co-option”, and (3) duplication and divergence of genes. Massive recruitment of novel genes occurred in the evolution of the “camera” eye from nautilus’ “pinhole” eye. We also showed that the type-2 co-option of transcription factors played important roles in the evolution of the lens and visual neurons. In summary, the cephalopod convergent morphological evolution of the camera eyes was driven by a mosaic of all types of gene recruitments. In addition, our analysis revealed unexpected variations of squids’ opsins, retinochromes, and arrestins, providing more detailed information, valuable for further research on intra-ocular and extra-ocular photoreception of the cephalopods.
Introduction
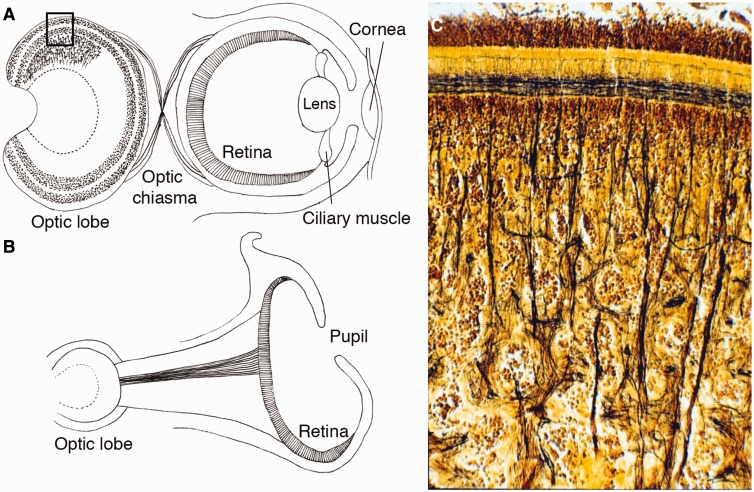
Coleoid cephalopods (squids, cuttlefishes, and octopuses) have been viewed as amazing illustrations of independent origins of complex neural and sensory structures, and possess the most highly centralized brains of any group of invertebrates (Akimushkin 1963; Zullo and Hochner 2011). Three remarkable examples of convergent evolution of neural organization in cephalopods are: (1) eyes and visual systems, including optic lobes (Fig. 1A) (Young 1962); (2) the vertical lobes—a specialized center that controls learning and memory—the analogs of the mammalian hippocampus (Hochner et al. 2006); and (3) the unique vasculature that supports such unprecedented complexity of the invertebrate brain (Abbott and Miyan 1995). Here, by combining comparative transcriptomics and in situ hybridization, we use various molecular markers to trace cephalopods’ molecular innovations in visual systems. The Nautilus genomic data are especially important for understanding molluscan evolution, since this lineage has a simpler cord-like neuronal organization resembling a basic molluscan tetraneury (i.e., the system of four lateral cords and cephalic loops as in chitons) (Moroz 2009), and a simple “pinhole” type of eye (hereafter pinhole eye) (Fig. 1B).
Fig. 1.
Schematic views of camera eyes and pinhole eyes of cephalopods and a photograph of a cephalopod’s optic lobe. (A) Schematic view of the cephalopod camera eye and visual system. The cephalopod retina has a single layer containing rhabdomeric photoreceptors, supporting glia, and small-caliber blood vessels in contrast to the vertebrates’ layered retina with numerous types of neurons. The visual neurons of cephalopods are located outside of the retina. The optic lobes have evolved as layered structures used in processing primary visual information. The optic lobe is the largest lobe of the brain in the all cephalopods and contains approximately 128,940,000 neurons in the brain of Octopus. The organization of the cortical layer superficially resembles that of the deeper layer of the vertebrate retina. (B) The eye of Nautilus is a unique example of a pinhole eye, which lacks lens, cornea, and contractile muscles. Nautilus’ optic lobes are simpler than those of Octopus and lack a granular cell layer. The visual neurons of octopuses and squids have a chiasma between the eye and optic lobe, but a chiasma is absent in Nautilus. (C) A cross-section (Golgi staining) showing a part of the optic lobes of Octopus corresponding to the rectangle region in A. (This figure is available in black and white in print and in color at Integrative and Comparative Biology online.)
The “camera” type of eye (hereafter camera eye) of humans/vertebrates and cephalopods has been described as a classical example of convergent evolution (Carlson 1985), sharing many structural similarities in morphological organization, but independently evolved from a common bilaterian ancestor without camera eyes (Brusca and Brusca 2003). Ogura et al. (2004) used small scale sequencing (Sanger EST) to identify over 1000 non-redundant genes in octopuses eyes. They concluded that 70% of annotated genes are commonly expressed in the eyes of humans and of Octopus, and 96% of these genes date back to the common bilaterian ancestor. What are the roles of these “shared” genes in the independent development of camera eyes?
Gehring and Ikeo (1999) proposed three types of “gene intercalation” leading to a new structure or function: (1) recruitment of novel genes into morphogenetic pathways by the fusion of an enhancer or promoter; (2) the recombination of various coding and regulatory regions of different genes, often called “evolutionary tinkering” or “co-option”; and (3) duplication and divergence of genes. Thus, we applied tissue-specific RNA-seq and comparative genomics to study how the above three categories might contribute to the morphological evolution of cephalopods’ neural and visual systems.
Even more important aspects of convergent morphological evolution are the origins of novel cellular components in complex organs. When considering the transition from a simple prototype of an eye to the camera eye, there is evidence for the concurrent evolution of many novel cellular components such as cornea, lens, muscles, connective tissues, photoreceptors, and primary and secondarily visual neurons, as well as motor and protective circuits (Fig. 1A). All these parts are required to make and support the complex functions of camera eyes. Consequently, we attempted to identify novel or lineage-specific divergence of genes involved in the molecular make up of the cephalopod eyes and visual centers. Our rationale in this article is to categorize cephalopod-specific and eye-specific genes based on comparative genomics. To this end, we present unexpected variation of the opsins and arrestins related to the cephalopod intra-ocular and extra-ocular photoreceptive systems.
Materials and methods
Transcriptome and assemblies
We used the embryonic samples of Idiosepius and Nautilus as well as their adult tissues to capture regulatory genes critical for systemic development of the eye and lens across species. For embryonic eye transcriptomics (RNA-seq) analysis, we utilized assemblies (stage 25 embryos of the pygmy squid, Idiosepius paradoxus and 3-month-old embryos of the chambered nautilus, Nautilus pompilius) obtained by Ogura et al. (2013). For adult Idiosepius and Nautilus, we generated novel sets of RNA-seq data. Tissues of Idiosepius and Nautilus were removed and homogenized in TRIzol reagent (Invitrogen) immediately after the animals were sacrificed. To minimize possible nucleotide polymorphism, we utilized a single individual of Nautilus. However, due to small sizes of Idiosepius, we pooled tissues from several individuals. Total RNAs were isolated according to the manufacture’s protocol, followed by on column DNase treatment using a QIAGEN RNeasy kit. Qualities of the RNAs were tested by Agilent Nanodrop and Agilent 2100 bioanalyzer. The RNA samples were sent to the BGI Inc and short read sequences were obtained by Illumina Hiseq2000 according to the company’s procedures.
FASTQ sequences of Idiosepius or Nautilus were pooled into one dataset and were assembled using the Trinity platform (Grabherr et al. 2011). To obtain normalized intensities of gene expression across tissues (fragments per kilobase per million reads, FPKM), reads from each sample was mapped onto the Trinity assembly with Bowtie (Langmead et al. 2009) and analyzed with RSEM (Li and Dewey 2011) and edgeR (Robinson et al. 2010). In the assembly procedure, variants of putative alternative splicing (sub-components of the Trinity output) were estimated as different contigs, but we merged variants from one sub-component based on the “%comp_fpkm” values of edgeR output. Analytical pipelines on a NIG Cell Innovation program (http://cell-innovation.nig.ac.jp/) were used with the annotation steps to the assembled contigs.
Data from the eyes of Idiosepius were assembled together with data from brain, arm, gonad, and gut. Contigs shorter than 500 bp and FPKM less than 1 were filtered out. Contigs that passed the criteria are used as “the eye genes”. Data from Nautilus eyes were assembled together with data from brain, arm, and siphuncule and processed in the same way. Sequence homology was tested using NCBI BLAST 2.2.30+ (Camacho et al. 2008) after filtering out genes shorter than 500 bp to remove gene fragments having traceability. For comparative analysis, we obtained gene models from two gastropods, the sea hare Aplysia californica (AplCal3.0, GCF_000002075.1, July 2013) and the giant owl limpet Lottia gigantea (Lotgi1, INSDC Assembly GCA_000327385.1, January 2013); the Pacific oyster, Crassostrea gigas (oyster_v9, INSDC Assembly GCA_000297895.1, September 2012); the polychaete annelid, Capitella telata (Capitella teleta v1.0, INSDC Assembly GCA_000328365.1, December 2012); the fly, Drosophila melanogaster (BDGP6, INSDC Assembly GCA_000001215.4) and human (GRCh38, INSDC Assembly GCA_000001405.15, December 2013) from Ensembl. Eye transcriptome data from human fetuses were obtained from an EST analysis by Choy et al. (2006). Choy et al. (2006) listed 4010 human gene models as the fetal eye genes using the previous human genome build. However, 669 genes were missing in the current human genome build (the Ensembl Human Build 38). To compensate, we obtained EST sequences from NCBI (BY794942-BY800475) and used these sequences in the search for homology.
Molecular phylogenetic analysis
The nucleotide sequences obtained in this study are available under the following accession numbers: [DDBJ: LC021432-LC021456] and listed in Supplementary Table S2. For each set of genes (opsins, arrestins, and crystallins), we obtained 97, 16, and 31 sequences from the NCBI and made alignments together with 7, 3, and 14 cephalopod sequences found in this study, respectively. The NCBI accession numbers of the genes are shown in the respective figures.
We used MUSCLE on the EMBL-EBI Web Services to generate a multiple sequence alignment (Edgar 2004; McWilliam et al. 2013). To remove poorly aligned sequences we used TrimAl v1.4.rev15 build[2013-12-17] with -gappyout option (Capella-Gutierrez et al. 2009). Maximum-likelihood inference of phylogenetic trees was inferred using RAxML version 8.0.26 (-f a -No. 1000 -m PROTGAMMAGTR options were applied) (Stamatakis 2014). One thousand bootstrap replicates were performed with the same search options as described above.
In situ hybridization
To generate Idiosepius Tbx20 DIG-labeled RNA targeted probes, we performed RT-PCR using the following primer set (F: ACCAGCCTCGAATTCACATC, R: GGAGGCCCAAATTAGGAAAG). To generate the Idiosepius cDNA, we utilized SMARTer RACE kit (Takara Clontech). The PCR fragments obtained from the RT-PCR were sub-cloned into T-vector (Promega) and used as templates for in vitro transcription using DIG RNA probe synthesis kit (Roche). Whole-mount in situ hybridization was performed using stage 25 embryos of Idiosepius according to the previously published protocol (Yoshida et al. 2010).
Results and discussion
Comparative transcriptomics of eyes and gene recruitment types
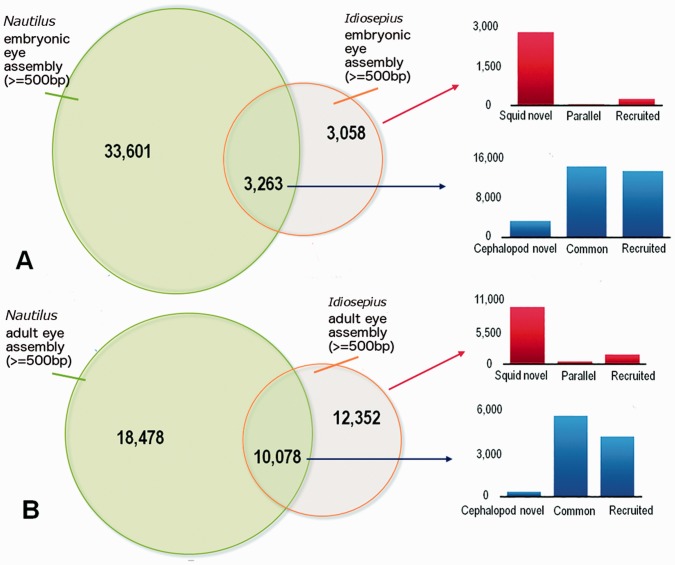
To characterize the scope of genomic differences between eyes of different cephalopods, we compared RNA-seq data between Idiosepius, and those of Nautilus. We analyzed both embryonic (Ogura et al. 2013) and adult eyes (this study). For the embryonic data, we used recently sequenced genomes of two other molluscs, Lottia and Crassostrea (Zhang et al. 2012; Simakov et al. 2013) and the annelid Capitella (Simakov et al. 2013) to test the taxon-specificity of differently expressed genes. Based on TBLASTX and BLASTN homology searches (Fig. 2A; e-value cutoff <1e−5), about 50% of Idiosepius genes (3058 out of 6331) are differentially expressed in squid eyes. Surprisingly, most of these genes (78%; 2385 out of 3058) are novel, apparently squid-specific, and don’t have homologs in any of the animal genomes we analyzed (Aplysia, Lottia, Capitella, Crassostrea, Homo, and Drosophila).
Fig. 2.
Comparsion of gene expressions in the eyes of squids and Nautilus. (A) Venn diagram of eye genes in Idiosepius and in Nautilus embryos (the assembly data obtained from Ogura et al. [2013]). Homology was tested using TBLASTX and BLASTN between Idiosepius and Nautilus and BLASTX against the genome databases of other animals (Aplysia, Lottia, Capitella, Crassostrea, Homo, and Drosophila). Genes differentially expressed between Idiosepius and Nautilus could represent genetic differences between different tissues of the eye, such as lens or cornea. Among the squid genes that have no Nautilus homologs, most of these genes (78%) are labeled as “Squid novel”, are apparently squid-specific, and lack homologs in the genomes of any other animals (no homologs, based on BLASTX search, e-value threshold <1e−5). Genes found commonly in ESTs of human fetal eyes (Choy et al. 2006) are labeled as “parallel”. The rest of the genes are labeled as “recruited” since the genes are commonly found in the genomes of squids and other Bilateria. (B) Venn diagram of eye genes of adult Idiosepius and Nautilus. Homologies were tested in the same manner. As in the genes of embryos, squid-specific genes showed the greatest contribution. (This figure is available in black and white in print and in color at Integrative and Comparative Biology online.)
These novel genes are presumed to be protein coding since 79% (2415 out of 3058) have an open reading frame longer than 100 aa. These data suggest that massive recruitment of novel, protein-coding genes (the type [1] gene recruitment) played important roles in the evolution of the camera eye from the Nautiluses’ pinhole eye.
Only a small portion of the total pool of expressed genes (40 or 1.1%) has homologs in the eye transcriptome of human fetuses (Choy et al. 2006) and might be the subject of parallel evolution between these two lineages. These genes are interesting examples of the type [2] co-option model of the evolution of novelties (Supplementary Table S1).
As shown previously (Ogura et al. 2013), genes commonly expressed in the eyes of Nautilus and Idiosepius such as rhodopsin and eye field transcription factors are thought to be essential for the function and development of photoreceptors. The obtained RNA-seq dataset for the eyes of adult Nautilus and Idiosepius suggests the same trends (Fig. 2B). Nevertheless, more than half of the genes expressed in the eyes of human fetuses do have homologs in the genomes of Nautilus and Idiosepius, but they are not expressed in cephalopod eyes.
Gene recruitments in convergent evolution of lens
In the following two subsections, we will summarize the scope of gene recruitments related to cellular components of the cephalopod camera eyes and brains.
There are remarkable differences between the composition of lens proteins and their regulators in the camera eyes of vertebrates versus cephalopods. The lens proteins of squids are formed by cells surrounding the lens (Tomarev et al. 1997). Our in situ hybridization screen showed that a transcription factor, Six3, is co-localized in the lens-forming cells (Ogura et al. 2013). It is notable that Nautilus doesn’t express Six3 homolog in the equivalent staged embryo.
Pax6 triggers expression of the lens protein in vertebrates (reviewed by Cvekl and Piatigorsky 1996; Cvekl et al. 2004). However, this might not be the case in the cephalopods, since the squid Pax-6 doesn’t co-localize in the lens-forming cells (Tomarev et al. 1997). Instead, regulatory element AP-1/ARE (Tomarev et al. 1994) and miRNAs (miR-124, miR-125b, and let-7) were shown to be involved (Bitel et al. 2012) as the squid-lens’s regulatory mechanisms. Since both AP-1/ARE and the microRNAs are also present in the vertebrates; these are clear examples of type [2] gene recruitments.
Vertebrates’ lenses are composed of proteins with sequence similarity to heat-shock proteins, lactate dehydrogenase, or alcohol dehydrogenases (Piatigorsky 2003). In contrast, crystallins of cephalopods convergently evolved from different classes of proteins (Piatigorsky and Wistow 1989). For example, squids’ lenses are mainly composed of S-crystallins, which are derived from glutathione S-transferase (Tomarev et al. 1995). This is a classic example of type [1] gene recruitment.
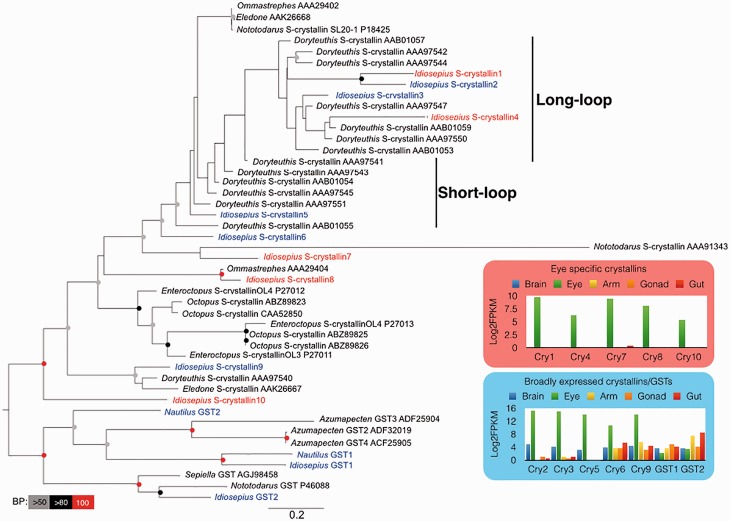
The sequenced Idiosepius transcriptome has mRNAs encoding three full-length and seven partial lens proteins (Fig. 3). The genes show repetitive duplication, and the number of duplications varies across species of cephalopods (Fig. 3). This suggests that the number of crystallin genes differs depending on the squid species, and that phylogenetic clustering does not correlate with expression patterns. For example, Idiosepius crystallin 1 and crystallin 2 showed different expression patterns, even though the genes have a stable sister relationship: crystallin 1 is eye-specific but crystallin 2 is expressed throughout all tissues tested (Fig. 3).
Fig. 3.
Diversity and expressions of squids’ S-crystallins. Maximum-likelihood tree based on amino-acid sequences of S-crystallins and glutathione S-transferases. The best maximum-likelihood tree was inferred using RAxML with the GTR evolutionary model and 1000 bootstrap replicates. The specific crystallins of the eye of Idiosepius are shown in red and the non eye-specific crystallins are shown in blue. Bootstrap support values are shown in the circles at the nodes (Red, 100%; black, 100–80%; gray, 80–50% of 1000 repetitions). Gene-expression patterns of Idiosepius eye-specific and non-specific crystallins are show as the inserts. (This figure is available in black and white in print and in color at Integrative and Comparative Biology online.)
Sweeney et al. (2007) identified long-loop peripherally-located and short-loop centrally-located crystallins. The long-loop crystallins are secondarily derived and produce a high refractive index in the lens (Sweeney et al. 2007). Both crystallin 1 and crystallin 2 make a stable cluster with other long-loop crystallins, which raises the question of why the long-loop crystallins are expressed throughout the body. There are two possible explanations for this. First, the Idiosepius crystallins expressed throughout the body may maintain their original function, protecting cells from oxidative stress. Since some of the lens proteins of squids are known to retain some enzymatic activity (Tomarev et al. 1995), the duplication of genes may be advantageous in protection from UV irradiation or high metabolism, even if the crystallin has low enzymatic activity. Second, evolutionary ratios of the protein region and regulator region might not be equal.
Convergent evolution of vision centers
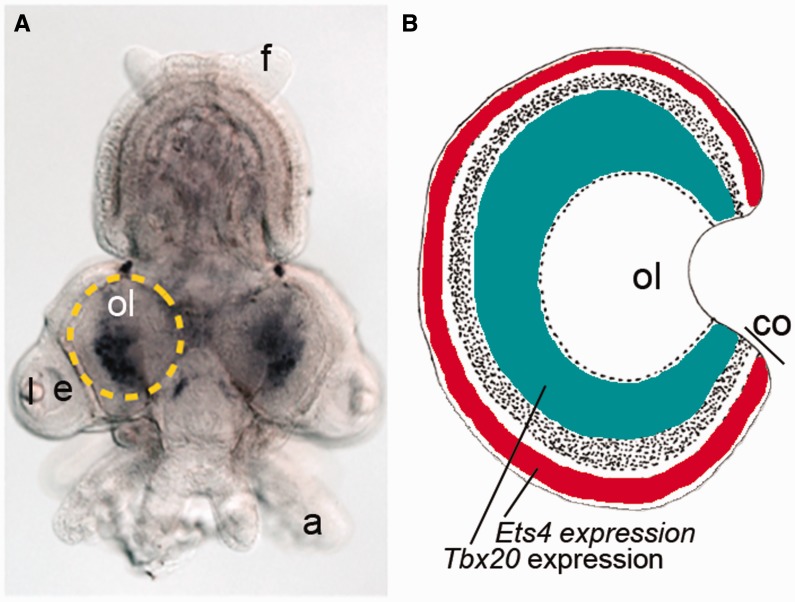
Specialized parts of the brain controlling visual information provide another insight into the evolution of the visual systems of cephalopods. We identified several cephalopod-specific genes differentially expressed in the vision centers. Optic lobes evolved as layered structures used in processing visual information. In fact, the optic lobe is the largest lobe of the brain in all cephalopods and contains approximately 128,940,000 neurons in Octopus (Young 1963). The cortical layer superficially resembles the organization of the deeper layer of the retina in vertebrates (Young 1971). Small amacrine cells and multipolar horizontal cells with spreading dendrites are morphologically similar to their counterparts in vertebrates (Budelmann et al. 1997; Nixon and Young 2003). Nautilus’ optic lobes are simpler than those of Octopus and lack a granular cell layer (Nixon and Young 2003). Our in situ hybridization screen revealed that two transcription factors, Ets4 and Tbx20, were selectively expressed in several different neuronal populations in the optic lobe (Fig. 4) with Ets4 localized in the cortical layer (Yoshida and Ogura 2011). The patterning appears to be a cephalopod novelty since in vertebrates Ets and Tbx genes are used in regulation of mesoderm-derived tissues, such as heart and blood vessels, or those involved in development of the limbs (Maroulakou and Bowe 2000; King et al. 2006; Hoogaars et al. 2007).
Fig. 4.
Gene-expression patterns and unique visual neurons in squids’ optic lobes. (A) Idiosepius Tbx20 expression at stage 25. Dotted circle indicates location of the optic lobe. Posterior side is up. a, arm; e, eye; f, fin; l, lens; ol, optic lobe. (B) Schematic views of squids’ optic lobes. Idiosepius Tbx20 is shown as green whereas Ets4 is labeled as red (see details in Yoshida et al. 2011). co, cortical layers; ol, optic lobe. (This figure is available in black and white in print and in color at Integrative and Comparative Biology online.)
In summary, this is an interesting case of recruitments of homologous genes for completely unrelated systemic functions in cephalopods versus vertebrates. In cephalopods, Ets and Tbx are implemented in the production of neuronal assemblies related to vision, while their function is limited to mesoderm specification in vertebrates. Again, these are examples of type (2) recruitment of genes.
Cephalopod-specific variations of photoreceptors and photoreceptive genes
There are two broad classes of photoreceptors: ciliary and rhabdomeric (Arendt 2003). The deuterostome lineage, including the vertebrates, typically uses ciliary photoreceptors that express c-opsin(s) and have membranous discs modified from a cilium (Arendt 2003; Yau and Hardie 2009). Among protostomes, ciliary photoreceptors were identified in the eyes of larval brachiopods (Passamaneck et al. 2011), adults’ eyes in scallops (Kojima et al. 1997) and in cnidarians (Martin 2002).
Arthropod eyes have classical examples of the rhabdomeric photoreceptors. However, rhabdomeric or r-opsin-expressing retinal ganglion cells were also found in vertebrates (Provencio et al. 2000; Berson et al. 2002) and in spiralians such as the gastropods Aplysia and Lottia (Gastropoda), as well as in polychaete annelids Capitella, and Platynereis (Randel et al. 2013). These data indicate that different classes of photoreceptors are widely distributed across bilaterian lineages. Furthermore, both c-type and r-type opsin classes were identified in the cnidarian, Nematostella (Fig. 5A) (Alvarez 2008), suggesting both deep ancestry and early divergence of two major groups of photoreceptors. What about cephalopods and their complex camera eyes?
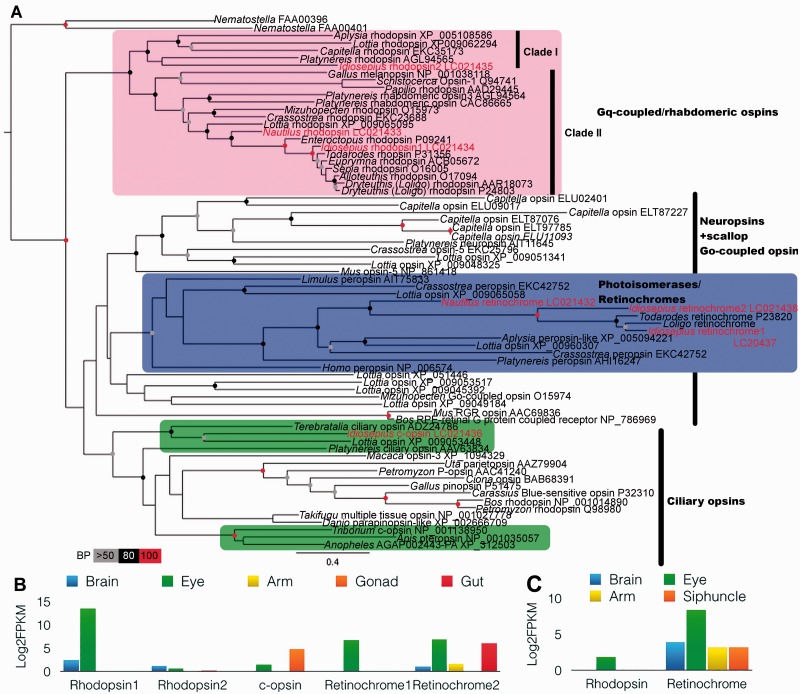
Fig. 5.
Novel opsin isoforms in squid and their expressions across tissues. (A) Maximum-likelihood tree, based on amino-acid sequences of opsins. The best maximum-likelihood tree was inferred using RAxML with the GTR evolutionary model and 1000 bootstrap replicates. Idiosepius and Nautilus opsins are shown in red. The cephalopod opsins are clustered in three groups; rhabdomeric opsins, retinochromes, and invertebrates’ ciliary opsins. Bootstrap support values are shown in the circles at the nodes (Red, 100%; black, 100–80%; gray, 80–50% of 1000 repetitions). (B) Gene-expression patterns of Idiosepius opsins across tissues. (C) Gene-expression patterns of Nautilus opsins across the tissues.
Patterns of distribution of photoreceptors are one of the notable differences between the vertebrates’ and cephalopods’ camera eyes. Cephalopods’ rhabdomeric photoreceptors are characterized by tightly packed microvilli (Budelmann et al. 1997) and are similar to the photoreceptors in the eyes of other protostomes (Arendt 2003; Yau and Hardie 2009). The retina of coleoid cephalopods has a single layer containing rhabdomeric photoreceptors, supporting glia, and small-caliber blood vessels (Budelmann et al. 1997), whereas vertebrates have a layered retina with numerous types of neurons (for a review see Basset and Wallace 2012). In contrast to vertebrates, the primary visual interneurons of cephalopods are located outside of the retina—in the optic lobes (see Convergent evolution of vision centers section).
Diversity of opsins in cephalopods
Our transcriptome-based approach revealed unexpected variations of opsins among all tested species of cephalopods. Specifically, we identified five opsin genes from three types of opsin families (rhabdomeric opsin, ciliary opsin, and retinochrome) in the squids’ eyes (Fig. 5A, B). These findings suggest that there are several photoreceptive types of cells, including two potentially novel subtypes in the camera eyes of squids. As indicated, the co-localization of three receptor types also occurs in vertebrate eyes (ciliary opsin(s) in the photoreceptors, rhabdomeric opsin in intrinsically photosensitive retinal ganglion cells, and retinochrome in retinal pigment epithelia (Lamb 2009)). The finding of c-opsin in the camera eyes of squids is interesting because c-opsin and r-opsin are expressed in different types of cells, and co-localization of both cell types are found in eyes of vertebrates and scallops (Kojima et al. 1997).
Surprisingly, we also found that the ciliary-opsin (c-opsin) of Idiosepius is expressed both in the eyes and gonads. Such expression patterns suggest that c-opsin might have multiple functions in vision, modulation of circadian clock or peripheral photoreception. The genealogical reconstructions suggest that c-opsin of Idiosepius formed a stable cluster with the c-opsin of brachiopod (Terebratalia) (Fig. 5A). The c-opsin of Terebratalia was expressed in the eyes as well as in the brains of larvae and is used in the directional detection of light (Passamaneck et al. 2011).
Diversity of retinochromes
Rhodopsin and retinochrome were commonly found in Nautilus and were typical of rhabdomeric photoreceptors (Fig. 5A, C) (Hara et al. 1995). In addition, we found squid-specific gene-duplications possibly related to properties of the rhabdomeric photoreceptors of the eye. The eyes of Idiosepius express two retinochrome genes, which encode photoisomerase enzymes of 11-cis/all-trans retinal necessary for the formation of functional rhodopsin. Since the molluscan retina lacks structures similar to the pigment epithelium of the vertebrate retina, 11-cis retinal is generated within the photoreceptors by the retinochromes and forms metaretinochrome (Terakita et al. 1989). Idiosepius retinochrome 2 showed broad expression across tissues and is similar to the Nautilus retinochrome (Fig. 5B, C). Idiosepius retinochrome 1 might be the result of gene duplication with eye-specific expression and be responsible for additional visual signaling pathways after the divergence of coleoid cephalopods (i.e., shell-less cephalopods) from nautiloids.
Although the function of three opsin types, and two retinochromes, in squid eyes is unknown, the Idiosepius clade I r-opsin (rhodopsin 2) is also localized in the brain (Fig. 5C). Thus, we do not exclude the possibility of an involvement of opsins and related molecules in other systemic functions. Previous data suggest that most cephalopods are color-blind despite their impressive range of camouflage and body patterns (Brown and Brown 1958; Marshall and Messenger 1996; Mäthger et al. 2006). The camouflage and body patterns of cephalopods are visually controlled, and the expression of rhodopsin in the chromatophore cells suggests an additional mechanism of localized regulation (Mäthger et al. 2010).
Arrestin gene duplications in cephalopod
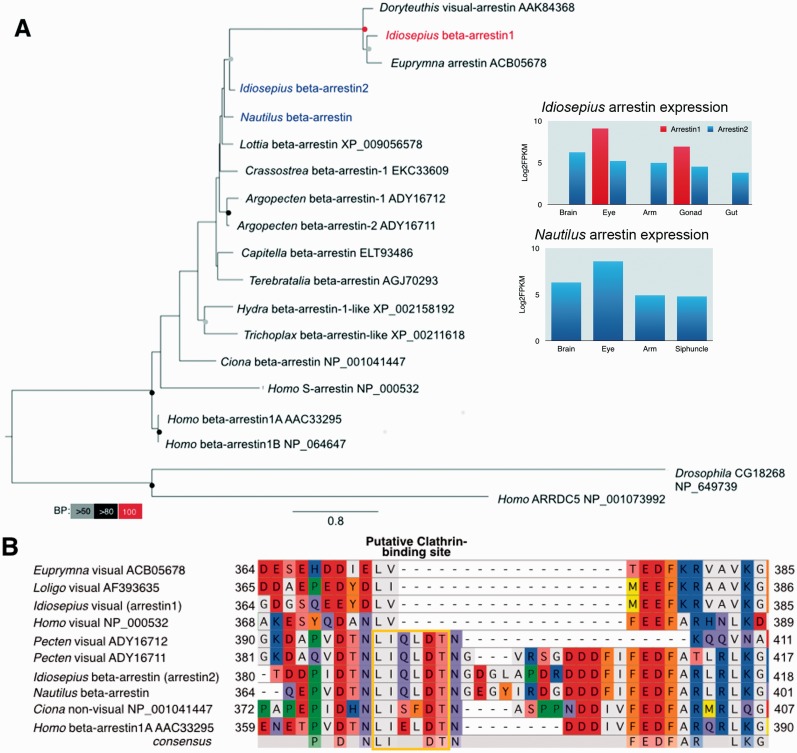
Arrestin is another essential component in visual-signal transduction that prevents further interaction of opsins with G-proteins (reviewed by Luttrell and Lefkowitz 2002). The majority of studied invertebrates have only one arrestin gene (Gurevich and Gurevich 2006). However, we found two arrestin genes from Idiosepius; one is specific to the eye and another is expressed throughout the animal’s tissues as in Nautilus (Fig. 6A). Thus, the expression of arrestin supports specialization of signal transduction in the squids’ visual cells.
Fig. 6.
Novel arrestin isoforms of squid and their expressions across tissues. (A) Maximum-likelihood tree based on amino-acid sequences of arrestins. The best maximum-likelihood tree was inferred using RAxML with the GTR evolutionary model and 1000 bootstrap replicates. A squid’s eye-specific arrestin is shown in red and non-visual arrestins are shown in blue. Bootstrap support values are shown in the circles at the nodes (Red, 100%; black, 100–80%; gray, 80–50% of 1000 repetitions). Gene-expression patterns of Idiosepius and Nautilus arrestins across the tissues are shown in the inserts. (B) Aligned amino-acid sequences of arrestin C-terminal regions. Visual arrestins were independently duplicated in the cephalopods and vertebrates. The squids’ visual arrestins accumulated changes in amino acids after branching from the beta-arrestins, perhaps a reflection of functional specification in the photoreceptor. This is supported by parallel loss of “clathrin-binding motif” in the visual arrestins of the vertebrates and cephalopods. Non-visual arrestins and scallops’ arrestins contain a putative clathrin-biding site, whereas the visual arrestins of squids and humans showed a typical deletion that depleted the clathrin-biding site. (This figure is available in black and white in print and in color at Integrative and Comparative Biology online.)
Similar gene duplications are found in the vertebrates, scallops and fly (Gurevich and Gurevich 2006; Gomez et al. 2011; Shieh et al. 2014). In addition, the visual-arrestins of squids have a shorter C-terminal tail and lack clathrin-binding sites (Fig. 6B). Vertebrates’ and flies’ visual-arrestins also lack a clathrin-binding site which function in mediating internalization, whereas non-visual arrestins maintain the clathrin-binding site (Fig. 6B) (Terakita et al. 2012). This whole process resembles that of vertebrates and suggests that vertebrates and molluscs have duplicated and diverged opsin pathways in parallel.
Molecular parallelism related to convergent morphological evolution
Although many genes are categorized as dominant type [1] recruitment in the camera eyes of cephalopods, we could also identify gene recruitment after duplication or type [3] gene recruitment. There are prominent examples of molecular parallelism that play a major role in the vasculature that support complexity of the brain in cephalopods and vertebrates.
Closed circulatory system
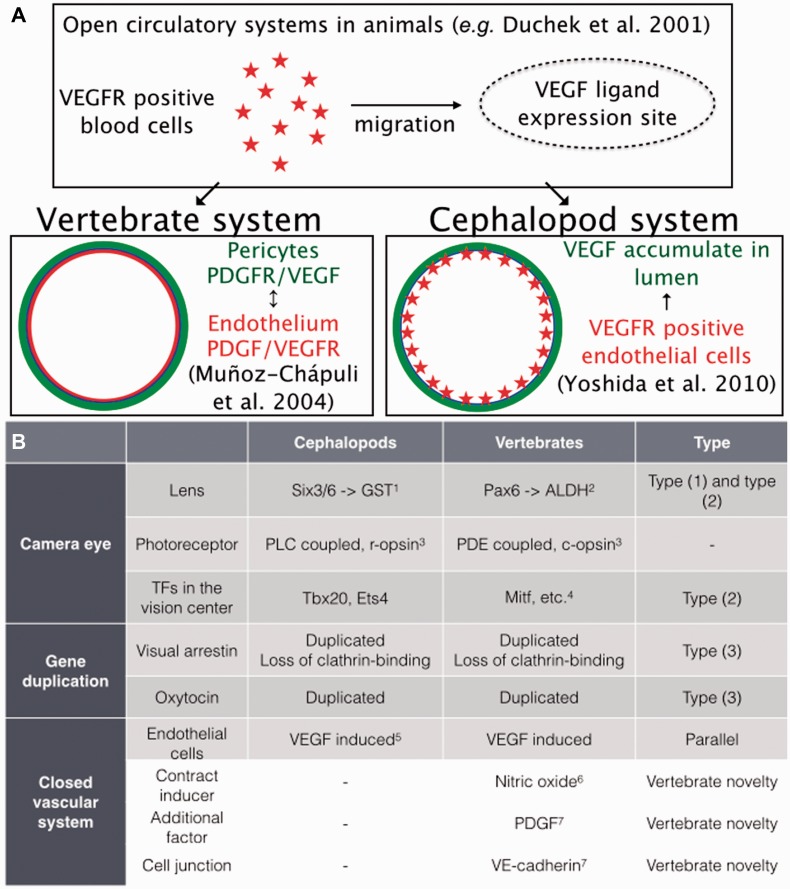
The first example is the evolution of the closed circulatory system. The coleoid cephalopods have distinct blood vessels with capillary-like structures throughout the body (Budelmann et al. 1997). This type of the circulatory system is one of the critical innovations that support a high level of oxygen consumption by the brain. The establishment of closed circulation also relates to the evolution of the endocrine system since a number of neurosecretory cells send a voluminous neuropil inside the vena cava where they form a neurosecretory-type organ (Martin and Voigt 1987; Budelmann et al. 1997). The nerve endings contain immunoreactive substances such as FMRFamide-like, the oxytocin (OT)/vasopressin (VP) family of peptides, proctolin, and α-melanotropin (Martin and Voigt 1987; Reich 1992).
Cephalopods’ blood vessels are composed of three layers, smooth muscle cells, basal membrane, and endothelial cells (ECs) as in vertebrates. On the other hand, the vessels of most invertebrates consist of the basement membranes and myoepithelial cells (reviewed by Muñoz-Chápuli and Pérez-Pomares 2010). The ECs constitute the most important component because the interplay with perivascular cells allows for a finer control of blood flow and blood pressure (Muñoz-Chápuli 2011). The ECs might be evolutionarily related to blood cells because these two types of cells share a molecular marker, vascular endothelial growth factor (VEGF), which is a well-known marker for vertebrates’ ECs (reviewed by Muñoz-Chápuli et al. 2004). In the open circulatory system of insects such as Drosophila, a VEGF receptor ortholog is expressed in the hemocytes that migrate in response to VEGF signals (Duchek et al. 2001). This is analogous to vertebrates, where ECs recruit the VEGF pathway as the main angiogenic signal (Muñoz-Chápuli 2011).
What is observed in the cephalopods? We analyzed VEGFR expression using Idiosepius embryos and found that the squids’ VEGFR was expressed in the developing blood vessels (Yoshida et al. 2010). The expression pattern resembles that of vertebrates and suggests parallel gene-use by the VEGF pathway in the development and maintenance of EC. It is notable that closed vasculature is one of the most obvious examples of convergent morphological features in the cephalopods, and is a remarkable example of molecular parallelism across phyla (Fig. 7A).
Fig. 7.
Summary of genetic recruitment of morphological convergence in cephalopods. (A) Schematic views of convergent recruitment of VEGF pathways to development of vascular endothelial cells. (B) Genes responsible for the morphological convergence and types of genetic recruitment in cephalopods, based on the criteria proposed by Gehring and Ikeo (1999); Type (1) recruitment of novel genes into the morphogenetic pathway by the fusion of an enhancer or promoter; Type (2) the recombination of various coding and regulatory regions of different genes, often called co-option, and (3) duplication and divergence of genes (see the following references for details: 1, Ogura et al. [2013]; 2, Cvekl and Piatigorsky [1996]; 3, Arendt [2003]; 4, Basset and Wallace [2012]; 5, Yoshida et al. [2010]; 6, González-Domenech and Muñoz-Chápuli [2010]; 7, Muñoz-Chápuli et al. [2004]). Nitric oxide in molluscs plays a diversity of roles including control of neural functions with wide-spread expression both in ganglia and at the periphery (e.g. Moroz et al. [2000]; Moroz [2006]; Moroz and Kohn [2011]). However, there is no reported detection of NO production by cells forming blood vessels in mollucs. (This figure is available in black and white in print and in color at Integrative and Comparative Biology online.)
Neuropeptides
The second case is the evolution of the neurohypophysial peptide hormones of the OT/VP superfamily. Vertebrates have two peptide genes called OT and VP. Despite small sequence differences between these two neuropeptides, each has very different physiological functions (reviewed by Gruber 2014). The OT/VP peptides acting in the brain of mammals appear to be critical in neuronal processing for social recognition, maternal behaviors, learning, and memory (reviewed by Gimpl and Fahrenholz 2001). All vertebrate species except cyclostomes contain at least one OT gene and one VP gene (Gwee et al. 2009). Thus, the OT and VP genes have arisen as a result of duplication of a common ancestral gene during the evolution of vertebrates. Surprisingly, the Octopus lineage had a similar peptide-gene duplication event (Kanda et al. 2003; Takuwa-Kuroda et al. 2003). Both Octopus octopressin (OP) and cephalotocin (CT) show different localization in the lobes of the brain (Takuwa-Kuroda et al. 2003). The functional differentiation was supported by duplication of receptors and by the distinct expression patterns of the receptors (Kanda et al. 2005). To date, a comparable co-occurrence of the OT/VP superfamily of peptides has never been demonstrated in other invertebrate groups.
Conclusion
The most influential event in the evolution of cephalopods is considered to be the appearance of vertebrate competitors (Packard 1972). Selective pressures and structural constraints of the cephalopod genomes and body organization may provide a clue to what caused the lineage to undergo substantial evolutionary changes in brain size and in cognition after branching from a chiton-like molluscan ancestor. Molecular components commonly found in camera eyes of vertebrates and cephalopods are the results of massive co-options or of novel genes. However, it appears that the recruitment of genes followed by their duplication, although smaller in scale, e.g., arrestins and OT neuropeptides (Fig. 7B), also played important roles in convergent morphological evolution of cephalopod’s camera-like visual systems.
Supplementary Material
Acknowledgment
The authors thank NIG Cell Innovation program (http://cell-innovation.nig.ac.jp/) for the computational support of assembly and annotation steps.
Funding
This work was supported by Program to Disseminate Tenure Tracking System of the Ministry of Education, Culture, Sports, Science, and Technology; the Japanese Government’s Grant-in-Aid for Young Scientists (B); the National Science Foundation [grant numbers NSF-0744649, NSF CNS-0821622 to L.L.M.]; the National Institute of Health [grant numbers 1R01GM097502, R01MH097062 to L.L.M.]; National Aeronautics and Space Administration (NASA) [grant number NNX13AJ31G to L.L.M.]; McKnight Brain Research, and University of Florida Opportunity Funds. The Society for Integrative and Comparative Biology provided assistance for attending the symposium at which this article was presented.
Supplementary data
Supplementary Data available at ICB online.
References
- Abbott NJ, Miyan JA. Cerebrovascular organization and dynamics in cephalopods. In: Abbott NJ, Williamson R, Maddock L, editors. Cephalopod neurobiology: neuroscience studies in squid, octopus and cuttlefish. London, UK: Oxford University Press; 1995. pp. 459–76. [Google Scholar]
- Akimushkin 1963 Primates of the sea (Primaty Morya, in Russian). Moscow: Gosgeogrizdat. [Google Scholar]
- Alvarez CE. On the origins of arrestin and rhodopsin. BMC Evol Biol. 2008;8:222. doi: 10.1186/1471-2148-8-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt D. Evolution of eyes and photoreceptor cell types. Int J Dev Biol. 2003;47:563–71. [PubMed] [Google Scholar]
- Bassett EA, Wallace VA. Cell fate determination in the vertebrate retina. Trends Neurosci. 2012;35:565–73. doi: 10.1016/j.tins.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Berson DM, Dunn FA, Takao M. Phototransduction by retinal ganglion cells that set the circadian clock. Science. 2002;295:1070–3. doi: 10.1126/science.1067262. [DOI] [PubMed] [Google Scholar]
- Bitel CL, Singh V, Frederikse PH. miR-124, miR-125b, let-7 and vesicle transport proteins in squid lenses in L. pealei. Curr Eye Res. 2012;37:388–94. doi: 10.3109/02713683.2011.635833. [DOI] [PubMed] [Google Scholar]
- Brown PK, Brown PS. Visual pigments of the octopus and cuttlefish. Nature. 1958;182:1288–90. doi: 10.1038/1821288a0. [DOI] [PubMed] [Google Scholar]
- Brusca RC, Brusca GJ. Invertebrates. 2nd ed. Sunderland (MA): Sinauer Associates, Inc; 2003. [Google Scholar]
- Budelmann BU, Schipp R, von Boletzky S. Cephalopoda. In: Harrison FW, Kohn AJ, editors. Microscopic anatomy of invertebrates, Vol. 6A, Mollusca II. New York: Wiley-Liss; 1997. pp. 235–54. [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2008;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–3. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson BA. The first known embryos of the chambered nautilus. Hawaiian Shell News. 1985;33:1. [Google Scholar]
- Choy KW, Wang CC, Ogura A, Lau TK, Rogers MS, Ikeo K, Gojobori T, Lam DS, Pang CP. Genomic annotation of 15,809 ESTs identified from pooled early gestation human eyes. Physiol Genomics. 2006;25:9–15. doi: 10.1152/physiolgenomics.00121.2005. [DOI] [PubMed] [Google Scholar]
- Cvekl A, Piatigorsky J. Lens development and crystallin gene expression: many roles for Pax-6. Bioessays. 1996;18:621–30. doi: 10.1002/bies.950180805. [DOI] [PubMed] [Google Scholar]
- Cvekl A, Yang Y, Chauhan BK, Cveklova K. Regulation of gene expression by Pax6 in ocular cells: a case of tissue-preferred expression of crystallins in lens. Int J Dev Biol. 2004;48:829–44. doi: 10.1387/ijdb.041866ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchek P, Somogyi K, Jékely G, Beccari S, Rørth P. Guidance of cell migration by the Drosophila PDGF/VEGF receptor. Cell. 2001;107:17–26. doi: 10.1016/s0092-8674(01)00502-5. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring WJ, Ikeo K. Pax 6: mastering eye morphogenesis and eye evolution. Trends Genet. 1999;15:371–7. doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
- Gimpl G, Fahrenholz F. The oxytocin receptor system: structure, function, and regulation. Physiol Rev. 2001;81:629–83. doi: 10.1152/physrev.2001.81.2.629. [DOI] [PubMed] [Google Scholar]
- Gomez Mdel P, Espinosa L, Ramirez N, Nasi E. Arrestin in ciliary invertebrate photoreceptors: molecular identification and functional analysis in vivo. J Neurosci. 2011;31:1811–9. doi: 10.1523/JNEUROSCI.3320-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Domenech CM, Muñoz-Chápuli R. Molecular evolution of nitric oxide synthases in metazoans. Comp Biochem Physiol Part D Genomics Proteomics. 2010;5:295–301. doi: 10.1016/j.cbd.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–52. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber CW. Physiology of invertebrate oxytocin and vasopressin neuropeptides. Exp Physiol. 2014;99:55–61. doi: 10.1113/expphysiol.2013.072561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwee PC, Tay BH, Brenner S, Venkatesh B. Characterization of the neurohypophysial hormone gene loci in elephant shark and the Japanese lamprey: origin of the vertebrate neurohypophysial hormone genes. BMC Evol Biol. 2009;9:47. doi: 10.1186/1471-2148-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich EV, Gurevich VV. Arrestins: ubiquitous regulators of cellular signaling pathways. Genome Biol. 2006;7:236. doi: 10.1186/gb-2006-7-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Hara R, Kishigami A, Koshida Y, Horiuchi S, Raj U. Rhodopsin and retinochrome in the retina of a tetrabranchiate cephalopod, Nautilus pompilius. Zool Sci. 1995;12:195–201. [Google Scholar]
- Hochner B, Shomrat T, Fiorito G. The octopus: a model for a comparative analysis of the evolution of learning and memory mechanisms. Biol Bull. 2006;210:308–17. doi: 10.2307/4134567. [DOI] [PubMed] [Google Scholar]
- Hoogaars WM, Barnett P, Moorman AF, Christoffels VM. T-box factors determine cardiac design. Cell Mol Life Sci. 2007;64:646–60. doi: 10.1007/s00018-007-6518-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda A, Satake H, Kawada T, Minakata H. Novel evolutionary lineages of the invertebrate oxytocin/vasopressin superfamily peptides and their receptors in the common octopus (Octopus vulgaris) Biochem J . 2005;387(Pt 1):85–91. doi: 10.1042/BJ20041230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda A, Takuwa-Kuroda K, Iwakoshi-Ukena E, Minakata H. Single exon structures of the oxytocin/vasopressin superfamily peptides of octopus. Biochem Biophys Res Commun. 2003;309:743–8. doi: 10.1016/j.bbrc.2003.08.061. [DOI] [PubMed] [Google Scholar]
- King M, Arnold JS, Shanske A, Morrow BE. T-genes and limb bud development. Am J Med Genet A. 2006;140:1407–13. doi: 10.1002/ajmg.a.31250. [DOI] [PubMed] [Google Scholar]
- Kojima D, Terakita A, Ishikawa T, Tsukahara Y, Maeda A, Shichida Y. A novel Go-mediated phototransduction cascade in scallop visual cells. J Biol Chem. 1997;272:22979–82. doi: 10.1074/jbc.272.37.22979. [DOI] [PubMed] [Google Scholar]
- Lamb TD. Evolution of vertebrate retinal photoreception. Philos Trans R Soc Lond B Biol Sci. 2009;364:2911–24. doi: 10.1098/rstb.2009.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttrell LM, Lefkowitz RJ. The role of beta-arrestins in the termination and transduction of G-protein-coupled receptor signals. J Cell Sci. 2002;115(Pt 3):455–65. doi: 10.1242/jcs.115.3.455. [DOI] [PubMed] [Google Scholar]
- Maroulakou IG, Bowe DB. Expression and function of Ets transcription factors in mammalian development: a regulatory network. Oncogene. 2000;19:6432–42. doi: 10.1038/sj.onc.1204039. [DOI] [PubMed] [Google Scholar]
- Marshall NJ, Messenger JB. Colour-blind camouflage. Nature. 1996;382:408–9. [Google Scholar]
- Martin VJ. Photoreceptors of Cnidarians. Can J Zool. 2002;80:1703–22. [Google Scholar]
- Martin R, Voigt KH. The neurosecretory system of the Octopus vena cava: a neurohemal organ. Experientia. 1987;43:537–43. [Google Scholar]
- Mäthger LM, Barbosa A, Miner S, Hanlon RT. Color blindness and contrast perception in cuttlefish (Sepia officinalis) determined by a visual sensorimotor assay. Vision Res. 2006;46:1746–53. doi: 10.1016/j.visres.2005.09.035. [DOI] [PubMed] [Google Scholar]
- Mäthger LM, Roberts SB, Hanlon RT. Evidence for distributed light sensing in the skin of cuttlefish Sepia officinalis. Biol Lett. 2010;6:600–3. doi: 10.1098/rsbl.2010.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 2013;41(Web Server issue):W597–600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz LL. Localization of putative nitrergic neurons in peripheral chemosensory areas and the central nervoussystem of Aplysia californica. J Comp Neurol. 2006;495:10–20. doi: 10.1002/cne.20842. [DOI] [PubMed] [Google Scholar]
- Moroz LL. On the independent origins of complex brains and neurons. Brain Behav Evol. 2009;74:177–90. doi: 10.1159/000258665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz LL, Kohn AB. Parallel evolution of nitric oxide signaling: diversity of synthesis and memory pathways. Front Biosci (Landmark Ed) 2011;16:2008–51. doi: 10.2741/3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz LL, Norekian TP, Pirtle TJ, Robertson KJ, Satterlie RA. Distribution of NADPH-diaphorase reactivity and effects of nitric oxide on feeding and locomotory circuitry in the pteropod mollusc, Clione limacina. J Comp Neurol. 2000;427:274–84. [PubMed] [Google Scholar]
- Muñoz-Chápuli R. Evolution of angiogenesis. Int J Dev Biol. 2011;55:345–51. doi: 10.1387/ijdb.103212rm. [DOI] [PubMed] [Google Scholar]
- Muñoz-Chápuli R, Pérez-Pomares JM. Origin of the vertebrate endothelial cell lineage: ontogeny and phylogeny. In: Rosenthal N, Harvey RP, editors. Heart development and regeneration. Amsterdam: Elsevier; 2010. pp. 465–86. [Google Scholar]
- Muñoz-Chápuli R, Quesada AR, Medina MA. Angiogenesis and signal transduction in endothelial cells. Cell Mol Life Sci. 2004;61:2224–43. doi: 10.1007/s00018-004-4070-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon M, Young JZ. The brain and lives of cephalopods. Oxford: Oxford University Press; 2003. [Google Scholar]
- Ogura A, Ikeo K, Gojobori T. Comparative analysis of gene expression for convergent evolution of camera eye between octopus and human. Genome Res. 2004;14:1555–61. doi: 10.1101/gr.2268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura A, Yoshida MA, Moritaki T, Okuda Y, Sese J, Shimizu KK, Sousounis K, Tsonis PA. Loss of the six3/6 controlling pathways might have resulted in pinhole-eye evolution in Nautilus. Sci Rep. 2013;3:1432. doi: 10.1038/srep01432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packard A. Cephalopods and fish: the limits of convergence. Biol Rev. 1972;47:241–307. [Google Scholar]
- Passamaneck YJ, Furchheim N, Hejnol A, Martindale MQ, Lüter C. Ciliary photoreceptors in the cerebral eyes of a protostome larva. Evodevo. 2011;2:6. doi: 10.1186/2041-9139-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatigorsky J. Gene sharing, lens crystallins and speculations on an eye/ear evolutionary relationship. Integr Comp Biol. 2003;43:492–9. doi: 10.1093/icb/43.4.492. [DOI] [PubMed] [Google Scholar]
- Piatigorsky J, Wistow GJ. Enzyme/crystallins: gene sharing as an evolutionary strategy. Cell. 1989;57:197–9. doi: 10.1016/0092-8674(89)90956-2. [DOI] [PubMed] [Google Scholar]
- Provencio I, Rodriguez IR, Jiang G, Hayes WP, Moreira EF, Rollag MD. A novel human opsin in the inner retina. J Neurosci. 2000;20:600–5. doi: 10.1523/JNEUROSCI.20-02-00600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randel N, Bezares-Calderón LA, Gühmann M, Shahidi R, Jékely G. Expression dynamics and protein localization of rhabdomeric opsins in Platynereis larvae. Integr Comp Biol. 2013;53:7–16. doi: 10.1093/icb/ict046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich G. A new peptide of the oxytocin/vasopressin family isolated from nerves of the cephalopod Octopus vulgaris. Neurosci Lett. 1992;134:191–4. doi: 10.1016/0304-3940(92)90514-8. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh BH, Kristaponyte I, Hong Y. Distinct roles of arrestin 1 protein in photoreceptors during Drosophila development. J Biol Chem. 2014;289:18526–34. doi: 10.1074/jbc.M114.571224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simakov O, Marletaz F, Cho SJ, Edsinger-Gonzales E, Havlak P, Hellsten U, Kuo DH, Larsson T, Lv J, Arendt D, et al. Insights into bilaterian evolution from three spiralian genomes. Nature. 2013;493:526–31. doi: 10.1038/nature11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney AM, Des Marais DL, Ban YE, Johnsen S. Evolution of graded refractive index in squid lenses. J R Soc Interface. 2007;4:685–98. doi: 10.1098/rsif.2006.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuwa-Kuroda K, Iwakoshi-Ukena E, Kanda A, Minakata H. Octopus, which owns the most advanced brain in invertebrates, has two members of vasopressin/oxytocin superfamily as in vertebrates. Regul Pept. 2003;115:139–49. doi: 10.1016/s0167-0115(03)00151-4. [DOI] [PubMed] [Google Scholar]
- Terakita A, Hara R, Hara T. Retinal-binding protein as a shuttle for retinal in the rhodopsin-retinochrome system of the squid visual cells. Vision Res. 1989;29:639–52. doi: 10.1016/0042-6989(89)90026-6. [DOI] [PubMed] [Google Scholar]
- Terakita A, Kawano-Yamashita E, Koyanagi M. Evolution and diversity of opsins. WIREs Membr Transp Signal. 2012;1:104–11. [Google Scholar]
- Tomarev SI, Chung S, Piatigorsky J. Glutathione S-transferase and S-crystallins of cephalopods: evolution from active enzyme to lens-refractive proteins. J Mol Evol. 1995;41:1048–56. doi: 10.1007/BF00173186. [DOI] [PubMed] [Google Scholar]
- Tomarev SI, Duncan MK, Roth HJ, Cvekl A, Piatigorsky J. Convergent evolution of crystallin gene regulation in squid and chicken: the AP-1/ARE connection. J Mol Evol. 1994;39:134–43. doi: 10.1007/BF00163802. [DOI] [PubMed] [Google Scholar]
- Tomarev SI, Callaerts P, Kos L, Zinovieva R, Halder G, Gehring W, Piatigorsky J. Squid Pax-6 and eye development. Proc Natl Acad Sci USA. 1997;94:2421–6. doi: 10.1073/pnas.94.6.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau KW, Hardie RC. Phototransduction motifs and variations. Cell. 2009;139:246–64. doi: 10.1016/j.cell.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JZ. The optic lobes of Octopus vulgaris. Phil Trans B. 1962;345:10–58. [Google Scholar]
- Young JZ. The number and sizes of nerve cells in octopus. Proc Zool Soc Lond. 1963;140:229–54. [Google Scholar]
- Young JZ. The anatomy of the nervous system of Octopus vulgaris. London: Oxford University Press; 1971. [Google Scholar]
- Yoshida MA, Ogura A. Genetic mechanisms involved in the evolution of the cephalopod camera eye revealed by transcriptomic and developmental studies. BMC Evol Biol. 2011;11:180. doi: 10.1186/1471-2148-11-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida MA, Shigeno S, Tsuneki K, Furuya H. Squid vascular endothelial growth factor receptor: a shared molecular signature in the convergent evolution of closed circulatory systems. Evol Dev. 2010;12:25–33. doi: 10.1111/j.1525-142X.2009.00388.x. [DOI] [PubMed] [Google Scholar]
- Zhang G, Fang X, Guo X, Li L, Luo R, Xu F, Yang P, Zhang L, Wang X, Qi H, et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature. 2012;490:49–54. doi: 10.1038/nature11413. [DOI] [PubMed] [Google Scholar]
- Zullo L, Hochner B. A new perspective on the organization of an invertebrate brain. Commun Integr Biol. 2011;4:26–9. doi: 10.4161/cib.4.1.13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.