ABSTRACT
Due to the spread of resistance, antibiotic exposure receives increasing attention. Ecological consequences for the different niches of individual microbiomes are, however, largely ignored. Here, we report the effects of widely used antibiotics (clindamycin, ciprofloxacin, amoxicillin, and minocycline) with different modes of action on the ecology of both the gut and the oral microbiomes in 66 healthy adults from the United Kingdom and Sweden in a two-center randomized placebo-controlled clinical trial. Feces and saliva were collected at baseline, immediately after exposure, and 1, 2, 4, and 12 months after administration of antibiotics or placebo. Sequences of 16S rRNA gene amplicons from all samples and metagenomic shotgun sequences from selected baseline and post-antibiotic-treatment sample pairs were analyzed. Additionally, metagenomic predictions based on 16S rRNA gene amplicon data were performed using PICRUSt. The salivary microbiome was found to be significantly more robust, whereas the antibiotics negatively affected the fecal microbiome: in particular, health-associated butyrate-producing species became strongly underrepresented. Additionally, exposure to different antibiotics enriched genes associated with antibiotic resistance. In conclusion, healthy individuals, exposed to a single antibiotic treatment, undergo considerable microbial shifts and enrichment in antibiotic resistance in their feces, while their salivary microbiome composition remains unexpectedly stable. The health-related consequences for the gut microbiome should increase the awareness of the individual risks involved with antibiotic use, especially in a (diseased) population with an already dysregulated microbiome. On the other hand, understanding the mechanisms behind the resilience of the oral microbiome toward ecological collapse might prove useful in combating microbial dysbiosis elsewhere in the body.
IMPORTANCE
Many health care professionals use antibiotic prophylaxis strategies to prevent infection after surgery. This practice is under debate since it enhances the spread of antibiotic resistance. Another important reason to avoid nonessential use of antibiotics, the impact on our microbiome, has hardly received attention. In this study, we assessed the impact of antibiotics on the human microbial ecology at two niches. We followed the oral and gut microbiomes in 66 individuals from before, immediately after, and up to 12 months after exposure to different antibiotic classes. The salivary microbiome recovered quickly and was surprisingly robust toward antibiotic-induced disturbance. The fecal microbiome was severely affected by most antibiotics: for months, health-associated butyrate-producing species became strongly underrepresented. Additionally, there was an enrichment of genes associated with antibiotic resistance. Clearly, even a single antibiotic treatment in healthy individuals contributes to the risk of resistance development and leads to long-lasting detrimental shifts in the gut microbiome.
INTRODUCTION
Health care in the 21st century is seriously challenged by the increasing prevalence of bacteria that are resistant to antibiotics. Excessive and incorrect use of antibiotics results in the emergence of both specific-drug-resistant and multidrug-resistant bacterial strains (1), also known as “superbugs.” The occurrence of superbugs is associated with treatment failure, higher morbidity and mortality, and increased health care costs (2). In addition to the emergence of antibiotic-resistant strains, the use of antibiotics is associated with an altered and often less diverse composition of the gut microbiome (3, 4) and with the increased prevalence of ectopic diseases such as asthma, eczema, and inflammatory bowel disease (5–8).
Alarmingly, the use of antibiotics without evidence-based benefit for the patient is widespread. In developing countries, lack of governmental control, low costs, and over-the-counter availability have led to a sharp rise in self-medication with antibiotics (9), where even common cold symptoms such as sore throat and headache are self-treated with antibiotics such as amoxicillin, tetracycline, and ciprofloxacin (10). In developed countries, antibiotics are frequently prescribed before surgery as a prophylactic measure for preventing infection. For instance, in the United Kingdom, one of the highest rates of antibiotic prescriptions in the outpatient population comes from dentists and oral surgeons (11, 12). Systemic antibiotics are commonly prescribed before removal of the third molar (wisdom tooth), periodontal therapy, placement of dental implants, or other surgery in the oral cavity. Although the clinical benefits of these measures are highly debated (13–15), they still form a common practice.
So far, studies that have assessed the ecological impact of antibiotics on the human microbiome are scarce and have focused only on the gut (16–18). Potential negative effects of prophylactic antibiotic use on the oral microbiome have been entirely neglected. Another important aspect that has not been investigated is the impact of systemically administered antibiotics on the microbiome at different niches within an individual. There might be significant collateral damage, for instance, to the gut microbiome due to unnecessary and clinically ineffective preventive antibiotic use before standard oral surgery.
In order to elucidate the impact of systemic antibiotics simultaneously on the oral and the gut microbiome ecology within the same individual, we performed two randomized placebo-controlled clinical trials with four widely used antibiotics. These included four different antibiotic classes with different modes of action, antimicrobial spectra, and pharmacokinetic properties—a lincosamide (clindamycin), a quinolone (ciprofloxacin), a tetracycline (minocycline), and a penicillin (amoxicillin). Lincosamides inhibit bacterial protein synthesis by binding to the 50S ribosomal subunit and hence ultimately inhibit microbial growth (19). The lincosamide clindamycin has a broad spectrum of antimicrobial activity, including Gram-positive aerobes and anaerobes, Gram-negative anaerobes, and selected protozoa and fungi (20). Quinolones are broad-spectrum bactericidal agents active against many Gram-positive and Gram-negative bacteria; they target the bacterial enzymes DNA gyrase and DNA topoisomerase (21), both essential in DNA replication and repair. Tetracyclines bind to the 30S ribosomal subunit and act by inhibiting protein synthesis after uptake into susceptible organisms by active transport and are bacteriostatic rather than bactericidal (22). Finally, the penicillin-type antibiotic amoxicillin is a broad-spectrum beta-lactam antibiotic that interferes with synthesis of the bacterial cell wall peptidoglycan (22).
Our study involved two clinical sites—the Swedish site (Karolinska Institute [KI]) with 30 individuals and the United Kingdom site (Helperby [HP]) with 44 individuals. In either site, subjects were randomized into a group receiving a placebo treatment and into two groups that received orally administered antibiotics. In the KI study, data from which major genera and species richness in the intestinal samples were reported previously (18), the antibiotics used were ciprofloxacin or clindamycin, while in the HP study, subjects received either minocycline or amoxicillin. We analyzed the salivary and fecal microbiomes of healthy adults before and after a standard antibiotic administration at various time points during 1 year. As a reference, we compared the microbiome profiles between the two habitats (feces and saliva) and between the two populations, KI and HP. We then investigated the long-term effects of the various antibiotics on the salivary and fecal microbiomes immediately after the treatment and 1, 2, 4, and 12 months post antibiotic exposure. Metagenome prediction tools were used to identify the differentially affected functional groups in all samples. Furthermore, full metagenome sequencing on selected samples was carried out for the characterization of antibiotic resistance gene (resistome) enrichment.
RESULTS
Of the 74 individuals included, 66 completed the study (Table 1). In the KI study, all volunteers completed the study in the placebo (n = 10) and ciprofloxacin (n = 10) groups, while 9 volunteers remained in the clindamycin group. None of the volunteers experienced any side effects. In the HP study, of the 44 volunteers, 37 completed the study (13 in the placebo group, 14 in the amoxicillin group, and 10 in the minocycline group). All dropouts (n = 1 amoxicillin subject, n = 5 minocycline subjects, and n = 1 placebo subject) were lost to follow-up right after the baseline visit. Among the subjects who completed the HP study, one individual in the amoxicillin group presented with mild diarrhea and one presented with congestion following antibiotic treatment.
TABLE 1 .
Demographic and clinical data of study participants who finished the study, per study center and treatment group
| Center | Group | n | % males | % Caucasian | Avg age, yr (SD) | Avg wt, kg (SD) | Avg ht, cm (SD) |
|---|---|---|---|---|---|---|---|
| KI | Placebo | 10 | 50 | 100 | 26 (4) | 74 (9) | 179 (10) |
| Ciprofloxacin | 10 | 50 | 80 | 26 (3) | 69 (13) | 176 (10) | |
| Clindamycin | 9 | 56 | 100 | 24 (5) | 67 (11) | 175 (9) | |
| HP | Placebo | 13 | 31 | 69 | 30 (5) | 78 (18) | 171 (9) |
| Amoxicillin | 14 | 29 | 79 | 27 (5) | 74 (25) | 175 (8) | |
| Minocycline | 10 | 30 | 90 | 22 (2) | 67 (13) | 171 (8) |
In total, 389 fecal and 391 saliva samples were analyzed by sequencing part of the small subunit of the ribosomal gene (reads available at the SRA of NCBI as SRP057504), yielding 6,825,563 reads after quality filtering with on average 8,751 (standard deviation [SD], 2,601) reads per sample (minimum of 1,736 and maximum of 22,253 reads). The reads were clustered into 1,972 operational taxonomic units (OTUs) at 97% similarity. To allow for between-sample comparisons, the data were randomly subsampled at 1,730 reads/sample.
Microbiome profiles. (i) Comparisons among baseline samples.
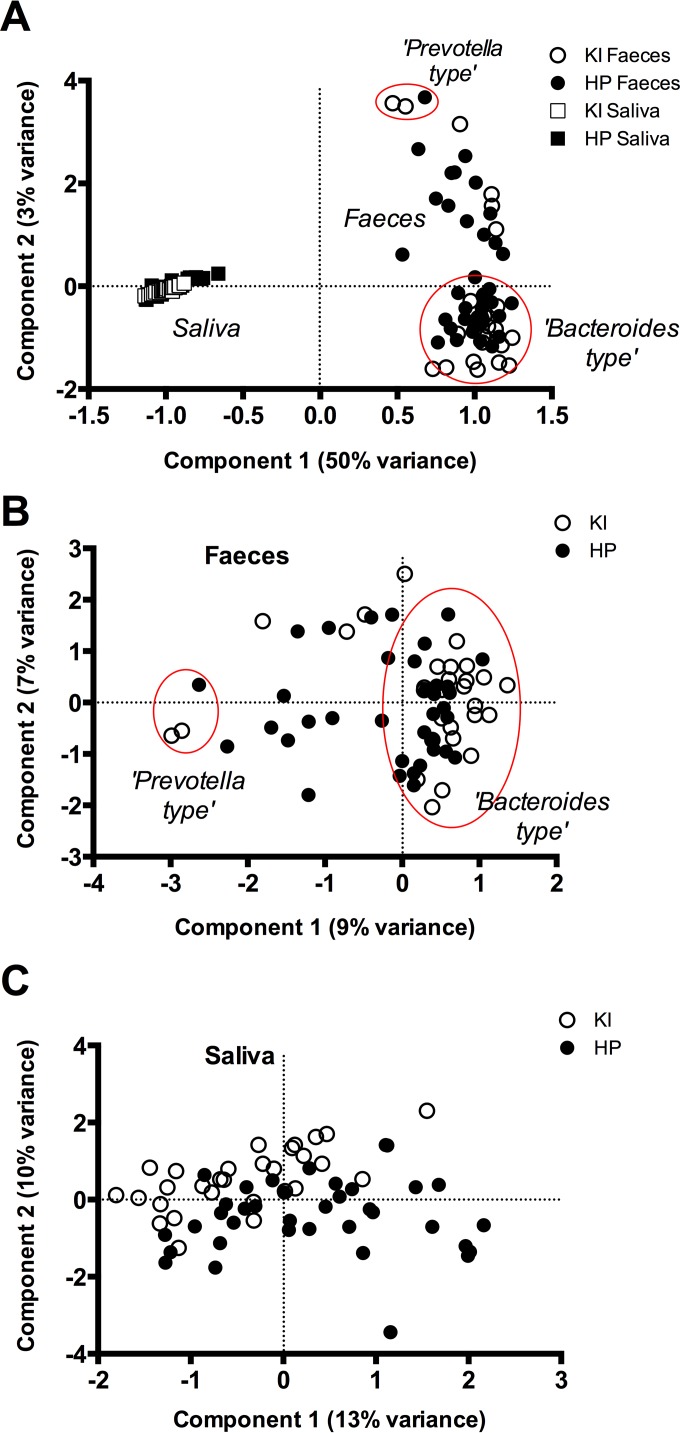
First, the baseline samples from the two habitats—the oral cavity (saliva) and the colon (feces)—were compared. The salivary microbiomes formed distinct clusters away from the fecal microbiomes, and the oral habitats from both KI and HP overlapped in the principal component analysis (PCA) (Fig. 1A). The fecal microbiomes were dichotomized by the second principal component (PC2) into genus Prevotella- and genus Bacteroides-dominated samples (Fig. 1A). Interestingly, the similarity among the salivary microbiomes (average Bray-Curtis similarity, 0.61; SD, 0.07) was significantly higher (P < 0.001, Mann-Whitney test) than that among the fecal samples (average Bray-Curtis similarity, 0.44; SD, 0.08).
FIG 1 .
Comparison of baseline microbiome profiles from both types of samples, saliva and feces (A), and per sample type, feces (B) and saliva (C), by study site, KI (Sweden) and HP (United Kingdom). The PCA plot is based on randomly subsampled and log2-transformed OTU data. The data set included 37 saliva-feces baseline sample pairs from the HP study and 29 from the KI study. The red ellipse highlights the two “types” of fecal samples—Prevotella- and Bacteroides-dominated samples, respectively.
Second, we looked at each of the habitats separately and compared the baseline samples from the two populations—the Swedish (KI) versus the United Kingdom (HP) cohort. The first component of the PCA on the fecal samples (Fig. 1B) separated samples high in genus Prevotella proportion (four OTUs) on the left side of the axis from the samples high in genus Bacteroides (two OTUs) and Alistipes and unclassified Bacteroidales (each a single OTU) at the right side of the axis. The majority of the samples were dominated by the genus Bacteroides. The overall difference between the fecal microbiomes of the two centers was statistically significant (P = 0.0001, F = 2.989, one-way permutational multivariate analysis of variance [PERMANOVA]) and was explained by a higher abundance of OTUs belonging to the unclassified Bacteroidales (1.16% of dissimilarity, similarity percentage [SIMPER]) and Prevotella (1.1% of dissimilarity, SIMPER) in the HP than in the KI data set and a higher abundance of an OTU classified as Bacteroidales Ratan060301c (1% of dissimilarity, SIMPER) in the KI samples.
Although salivary microbiomes showed a higher interindividual similarity than the fecal samples (Fig. 1A), salivary samples clearly differed between the study populations (P = 0.0001, F = 4.925, one-way PERMANOVA), where the second component of the PCA, explaining 10% of the variance, determined this difference (Fig. 1C). The position of the samples (predominantly KI) in the upper part of the plot was determined by six OTUs classified into the genus Prevotella and one OTU classified as Porphyromonas, while OTUs belonging to the genera Rothia and Streptococcus determined the position of the samples (predominantly HP) in the lower part of the plot.
(ii) Effects of antibiotics.
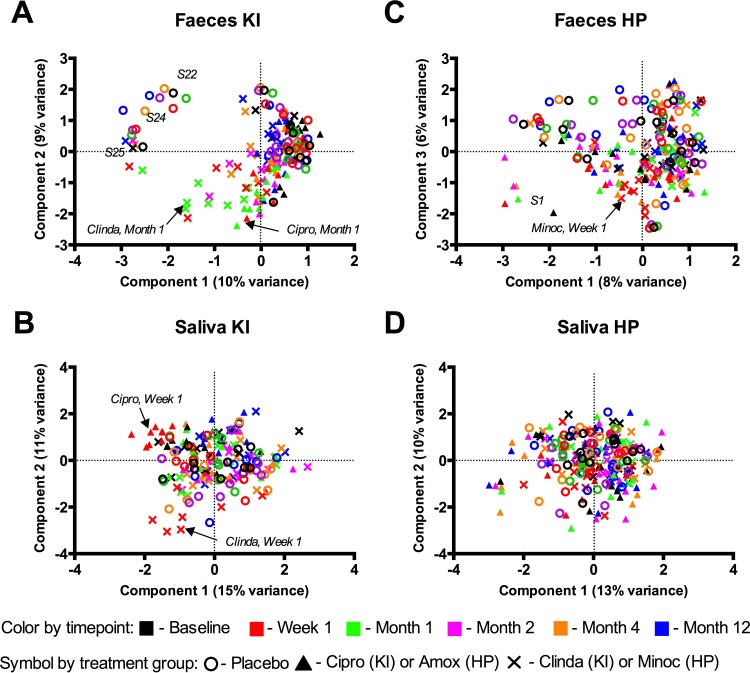
Next, we addressed the effects of each of the four antibiotics on the salivary and fecal microbiomes. A microbial shift was observed in the KI study after both ciprofloxacin and clindamycin exposure at week 1, month 1, and month 2 in feces (Fig. 2A) but at only week 1 in saliva (Fig. 2B). In the HP study, a minor shift in microbiome composition was observed in feces at week 1 in the minocycline group (Fig. 2C), whereas saliva showed no discernible pattern for microbial profile changes (Fig. 2D).
FIG 2 .
Effects of antibiotics on microbiome profiles of feces (A) and saliva (B) from the KI study and feces (C) and saliva (D) from the HP study. The PCA plot is based on log2-transformed OTU data. Different colors indicate different time points; different symbols indicate different treatment groups. Outliers in the KI (A) and HP (C) fecal data sets are highlighted with the respective subject number.
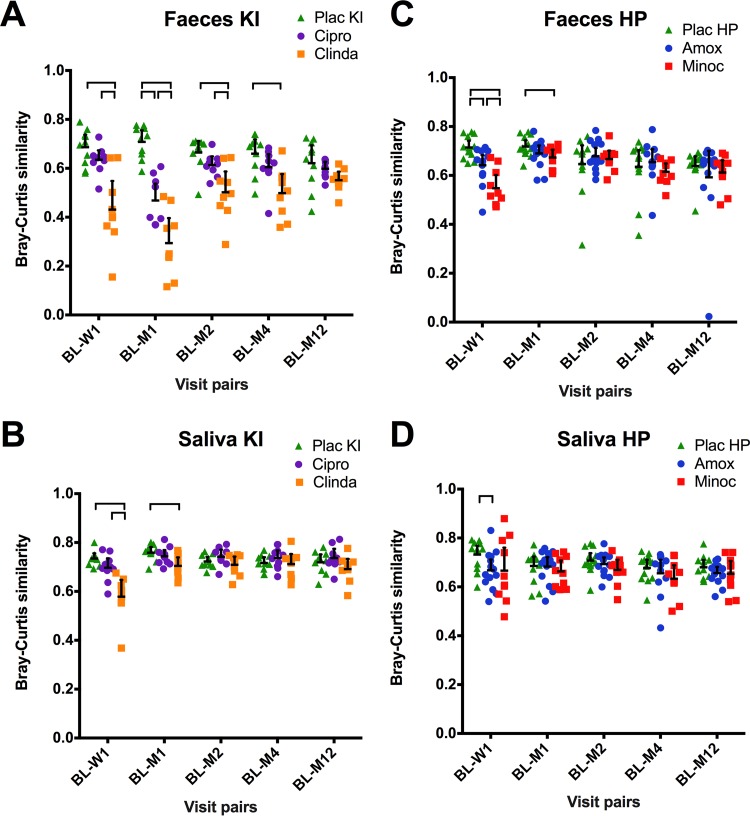
The microbiome shifts were quantified using Bray-Curtis similarity between the baseline sample and each of the other time points within an individual subject. Exposure to clindamycin resulted in the most pronounced and long-lasting change in microbial profiles that remained statistically significant compared to the placebo group for up to 4 months in the fecal samples (Fig. 3A) and up to 1 month in saliva (Fig. 3B). The ciprofloxacin group differed from the placebo group up to 1 month in feces (Fig. 3A). In the HP study, feces from the minocycline group differed from the placebo group up to 1 month (Fig. 3C). Finally, amoxicillin treatment resulted in dissimilarity between baseline and week 1 samples compared to placebo group in both feces (Fig. 3C), and saliva (Fig. 3D).
FIG 3 .
Similarity in microbiome profiles between the baseline (BL) and the other visits (W1, week 1; M1, month 1; M2, month 2; M4, month 4; M12, month 12). The horizontal bar indicates the mean value; the error bar indicates the 95% confidence interval. Bray-Curtis similarities were calculated between the log2-transformed microbiome profiles of the baseline and each of the other time point samples of the respective individual. Brackets connect statistically significantly different groups within each visit pair (P < 0.05; one-way analysis of variance, Games-Howell post hoc test).
The fecal microbial diversity, expressed as a Shannon diversity index, was clearly affected by each of the antibiotics—clindamycin and ciprofloxacin—in the KI study. Compared to the baseline, the fecal microbiome diversity was significantly reduced for up to 4 months in the clindamycin group and even up to 12 months in the ciprofloxacin group (see Fig. S1A in the supplemental material). The salivary microbiome of the same groups showed a short-term reduced diversity only immediately after the exposure to ciprofloxacin (see Fig. S1B). Quite in contrast to the observation in the KI group, in the HP study, the only significant reduction in microbiome diversity in fecal (see Fig. S1C) and saliva (see Fig. S1D) samples was observed directly after the exposure to minocycline. Exposure to amoxicillin had no significant effect on microbiome diversity in either of the two habitats (see Fig. S1C and D).
Next, we explored the effects of exposure to antibiotics on individual microbial taxa. At the OTU level, no OTUs contributed significantly to differences between the samples exposed to antibiotics and their respective placebo (P > 0.05, Welch’s t test, Welch’s inverted confidence interval [CI] method, and Storey false discovery rate [FDR] correction for multiple comparisons). However, lowering the resolution to a higher taxonomic level (genus or higher) allowed identification of taxa that were affected by antibiotics. For fecal samples, exposure to antibiotics resulted in changes in the relative proportions of different genera or higher taxa belonging to nearly all phyla (see Table S1 in the supplemental material). Only the phylum Verrucomicrobia with its sole representative—genus Akkermansia—was not affected by any of the antibiotics. In saliva, the effects were less pronounced, while some phyla, such as Proteobacteria or candidate division TM7, were affected by all or three of the four antibiotics, respectively (see Table S2).
Predictive metagenomics.
A computational approach (PICRUSt) predicts the functional composition of a metagenome using 16S rRNA gene data and a database of reference genomes (23). We applied PICRUSt to our data and validated the outcome with full metagenome data from selected salivary and fecal samples. Predictions based on 16S rRNA gene amplicon data showed a high accuracy compared to the metagenomic shotgun sequencing from both saliva (average Spearman’s rho, 0.92; SD, 0.03) and fecal (average Spearman’s rho, 0.86; SD, 0.03) samples.
(i) Comparison of baseline samples.
Interpopulation observed differences in the microbiome composition at baseline (Fig. 1B and C) were confirmed by differences in the predicted microbiome KEGG orthologous groups (KOs) (see Fig. S2 in the supplemental material), where 1,021 from the predicted 4,737 KOs in feces and 2,119 from the 4,598 predicted KOs in saliva differed significantly in their proportion between the KI and HP populations (P < 0.05, Welch’s t test, Welch’s inverted CI method, and Storey FDR correction for multiple comparisons). When the predicted metagenome data were summarized into functional groups, the fecal baseline samples from KI displayed a significantly higher proportion of bacterial genes encoding motility proteins, ABC transporters, chemotaxis proteins, secretion systems, transcription factors, sporulation proteins, and flagellar assembly proteins than did the samples from HP (see Fig. S2C). The HP fecal samples showed an increased abundance of genes linked to ion channels, biosynthetic activity, and catabolism of different molecules (see Fig. S2C). Among the significantly different KOs in feces, 10 KOs were associated with antibiotic resistance and all but one of these KOs were at a significantly higher proportion in HP samples than in KI samples (see Table S3), suggesting a higher antibiotic resistance gene load in the HP population at the baseline of the study. For saliva, a significantly higher proportion of functional groups related to assimilative processes was found in the KI group, while HP samples had a higher relative abundance of functional groups related to dissimilation (see Fig. S2D). With respect to antibiotic resistance in saliva, 16 of the significantly different KOs between the two centers were associated with antibiotic resistance—with each of the centers showing a higher proportion in half of these KOs (see Table S3).
(ii) Effects of antibiotics.
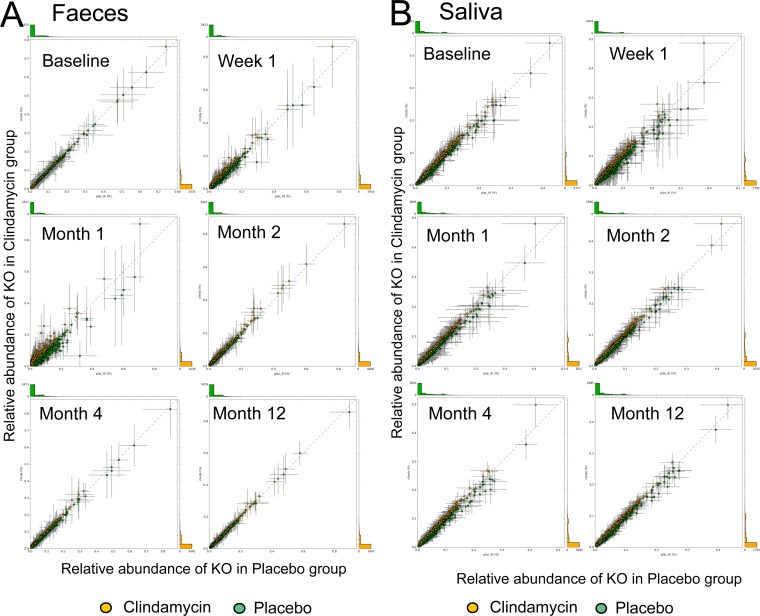
To assess the effects of antibiotics on the predicted metagenomes, we compared KOs from the predicted metagenomes in the antibiotic groups with the predictions from the respective placebo group. At baseline, no differences in relative abundances of the KOs between any of the antibiotic groups and the placebo group were observed. Exposure to antibiotics resulted in significantly different proportions of 4 to 14% of the predicted KOs in fecal samples in all four antibiotic groups, although at different time points. For instance, immediately after clindamycin administration only three of 4,606 KOs significantly differed in their proportion from the placebo group. This number increased to 512 KOs (11% of all KOs) at 1 month after exposure to clindamycin (Fig. 4A). In contrast, in amoxicillin-exposed feces, 706 of 4,968 KOs (14% of all KOs) were significantly different at week 1 compared to baseline, while this significance was lost at month 1. Although exposure to clindamycin had a strong impact on feces, it did not result in significant changes in predicted proportions of KOs in saliva (Fig. 4B). Among all saliva samples, only the predicted metagenomes of the samples collected immediately after exposure to ciprofloxacin showed a significant effect, where 310 of 4,289 KOs (7% of KOs) differed from placebo group at week 1 (see Fig. S3 in the supplemental material).
FIG 4 .
Relative abundance of the predicted KEGG orthologous groups (KOs) in the fecal (A) and salivary (B) samples from the clindamycin group plotted against the samples from the KI placebo group per individual time point. Error lines indicate standard deviations. No significant differences were observed in saliva, while in feces, 3 KOs at 1 week post-antibiotic treatment and 512 of the 4,606 predicted KOs at 1 month post-antibiotic treatment were significantly different in their proportions from the placebo group (P < 0.005, Welch’s t test, Welch’s inverted confidence interval method, and Storey FDR correction for multiple comparisons).
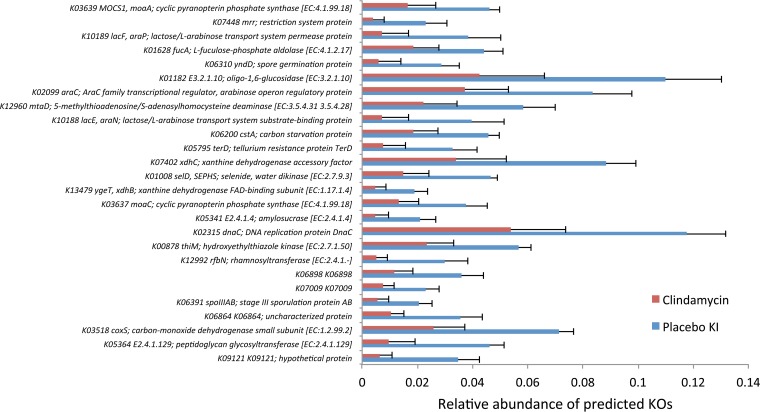
From the analysis of the predicted fecal metagenomes, it could be concluded that 1 month after the exposure to clindamycin, 74 KOs were significantly increased compared to the placebo group, and 438 KOs showed a significant decrease. Reduction in spore formation and lactose-arabinose metabolism were among the most affected functions (Fig. 5). At the same time, a decrease in the fermentative butyrate-producing pathways was observed. Three of the KOs that showed a significant increase in feces after exposure to clindamycin were associated with antibiotic resistance (see Table S4 in the supplemental material).
FIG 5 .
Most significantly affected KOs (26 of 520) in the predicted metagenomes from clindamycin-exposed feces at month 1 compared to the respective placebo group samples. FAD, flavin adenine dinucleotide; SEPHS, selenophosphate synthetase.
One month after exposure to ciprofloxacin, fecal sample metagenome predictions revealed 110 KOs that were significantly increased in their proportion, with 502 KOs being decreased (see Fig. S3 in the supplemental material). Among the most affected KOs were two subunits of the pyruvate ferredoxin oxidoreductase—PorB and PorD—an observation in agreement with a decreased sporeformer community (see Fig. S4). Among the KOs that increased in relative abundance after ciprofloxacin exposure compared to placebo, one KO was associated with antibiotic resistance (see Table S4). Among the KOs that decreased in proportion after ciprofloxacin compared to placebo use were 4 KOs that are associated with antibiotic resistance (see Table S4).
Exposure to minocycline resulted in 102 significantly increased and 81 significantly decreased predicted KO proportions in fecal samples immediately after exposure to antibiotics compared to the placebo group. In general, KOs associated with respiratory processes and amino acid synthesis were reduced and those associated with amino acid degradation and nitrogen metabolism were increased compared to KOs in the placebo group. It is tempting to interpret this observation as being the result of an increase in amino acid abundance of the environment, possibly by lysis of a nonidentified subpopulation. Three of the KOs with increased proportions were associated with antibiotic resistance, while one antibiotic-resistance-associated KO decreased in proportion (see Table S4 in the supplemental material).
Exposure to amoxicillin resulted in 272 significantly increased and 434 significantly decreased predicted KOs in feces immediately after the antibiotic exposure, compared to the placebo group. Nine KOs associated with antibiotic resistance increased after amoxicillin exposure, while four antibiotic-resistance-associated KOs decreased in their proportion (see Table S4 in the supplemental material).
Full metagenome: presence of antibiotic resistance genes.
From each antibiotic group and each sample type, a baseline and a postantibiotic sample were selected that showed the most pronounced change in microbiome profile after exposure to antibiotics. In total, 18 samples were processed for full metagenomic shotgun sequencing. We calculated fold changes in genes associated with antibiotic resistance types present in the Antibiotic Resistance Genes Database (ARDB) (24) between the baseline and post-antibiotic-treatment samples. At baseline, salivary samples from the United Kingdom study (HP) had on average a 1.13-times-higher load of antibiotic resistance genes than those from the Swedish study (KI), while the loads in fecal samples from the two countries were similar (HP/KI ratio = 0.97). After exposure to antibiotics, the antibiotic resistance gene load increased slightly in feces (mean, 1.07; SD, 0.15), while it remained stable in saliva (mean, 0.99; SD, 0.17).
Next, we calculated fold changes in antibiotic resistance genes for individual sample pairs (see Table S5 in the supplemental material). After administration of minocycline (a tetracycline), erythromycin resistance (rRNA adenine N-6-methyltransferase, Erm*) and especially efflux pumps (OprN, Tet*) increased in abundance in both saliva and fecal samples (see Table S5). In saliva, chloramphenicol acetyltransferase (CatB1) and a class A beta-lactamase (BL2f_Sme1) also increased, while in feces resistance to streptomycin (Str), efflux pumps (cml_e7, AdeC, and EmrD), and resistance to aminoglycosides (Aac6Ie, Ant6Ia, and EmrD) were present. The general pattern showed protection against protein synthesis inhibitors (tetracycline, erythromycin, streptomycin, and chloramphenicol).
After treatment with amoxicillin (a penicillin), a general pattern of protection against antibiotics appeared, which included a beta-lactamase (BL2), but also efflux pumps (Mex*, AmrA, and OprN) and protection against erythromycin, aminoglycosides (methyltransferases [Erm*] and aminoglycoside alteration [Aac*]), and trimethoprim (group A drug-insensitive dihydrofolate reductase [Dfr*]). More beta-lactamases (BL*) were present after treatment in feces than in saliva (see Table S5 in the supplemental material).
After exposure to ciprofloxacin (a fluoroquinolone), there was no clearly related signal observed in saliva (see Table S5 in the supplemental material). In feces, the response to ciprofloxacin from two individuals (subjects 6 and 18) differed regarding beta-lactamases and efflux pumps (see Table S5), while it was generally related to the same classes of antibiotics (aminoglycosides, beta-lactams, chloramphenicol, erythromycin, and tetracyclines).
For clindamycin, shifts in the Erm methyltransferases (Erm*) were very prominent in both saliva and feces (see Table S5 in the supplemental material). In saliva, several efflux pumps, including the macrolide-lincosamide-streptogramin B efflux pump (LmrB), appeared. In feces, aminoglycoside modification proteins (Aac* and Aph*) and lincosamide nucleotidyltransferase (LnuA) were more abundant.
In general, the most enhanced resistance genes were related to antibiotics that work intracellularly, such as the Erm genes and the tetracycline efflux and multidrug resistance efflux pumps.
DISCUSSION
Here, we reported that, depending on the type of antibiotics, the effects of a single antibiotic exposure on the human gut microbiome can be very aggravating and prolonged, while ecological consequences of the same exposure on the salivary microbiome are short-lived and superficial. Additionally, based on metagenome predictions and the affected taxa, we can conclude that a single use of antibiotic treatment, especially the lincosamide clindamycin and the quinolone ciprofloxacin, in a healthy population may severely affect short-chain fatty acid (SCFA) production in the gut.
Undesired effects of clindamycin consist of gastrointestinal disturbances, the incidence varying between 2% and 20% (22). Although members of the current study population did not present with any clinically significant side effects, exposure to antibiotics had a significant effect on the gut ecosystem. A month after the exposure to clindamycin, spore-forming, lactose-arabinose metabolism, and fermentative butyrate-producing pathways were among the most significantly reduced functions in the predicted metagenomes. A month after ciprofloxacin use, a significant reduction in pyruvate metabolism was predicted. The SCFA butyrate is synthesized from complex polysaccharides via pyruvate and the acetyl coenzyme A pathways (25). The reduction in the predicted butyrate-producing pathways corresponded with a significant decrease in Faecalibacterium, Subdoligranulum, uncultured Ruminococcaceae, Roseburia, Coprococcus, and uncultured Lachnospiraceae, known as butyrate producers (25). These observations suggest a specific and pronounced negative effect of clindamycin and ciprofloxacin on butyrate production in the gut. Actual measurements of SCFAs in feces are suggested to be unreliable due to their fast uptake and metabolism in the colon by the host and due to different intestinal transit times (26). Production of butyrate has been associated with positive effects on gastrointestinal health by butyrate functioning as an energy source for colonocytes and by inhibiting inflammation, carcinogenesis, and oxidative stress in the gut (26).
The metagenome prediction data indicate that microbiome functions recovered earlier than the microbial community composition following exposure to antibiotics. This supports the importance of functional redundancy within gut microbiota, as suggested previously after a single exposure to ciprofloxacin in three healthy adults (16). However, metagenome predictions also showed that, even without significant effects of antibiotics on microbiome composition, the functions or KEGG orthologous groups might have been significantly affected, as in the case with amoxicillin-exposed fecal samples. The most likely explanation for this phenomenon is the individualized response to this antibiotic at a taxonomic level, meanwhile affecting similar general functionality in the different microbial taxa.
The two study populations that participated in the project differed in their (baseline) microbiome composition. Since the fecal microbiome is greatly shaped by diet (27), it would have been very informative to compare the dietary habits of the two cohorts. Unfortunately, such information was not collected. Even more striking were the differences in the salivary microbiota: the predominant taxa in KI saliva samples were Prevotella spp., as opposed to streptococci in HP samples. The tongue dorsum is shown to be the main habitat for the anaerobic genus Prevotella (28, 29), while streptococcal predominance can be related to either supragingival plaque or nonkeratinized mucosa (28). Since no intraoral clinical examination was performed during this study and the timing of the last personal oral hygiene episode in respect to saliva collection was not recorded, it is not possible to elucidate the exact reasons for the observed differences.
Already prior to exposure to antibiotics in this study, both study populations carried antibiotic resistance genes in their oral and gut microbiomes. This is in line with the report on high levels of antibiotic resistance gene carriage by salivary and fecal commensal bacteria in two healthy individuals without prior exposure to antibiotics for at least a year (30). Both metagenomic predictions for 16S rRNA gene amplicon data and full metagenomic data on selected samples indicated that the United Kingdom (HP) population had a higher antibiotic resistance gene load at the start of the study. These differences may have affected the study outcomes: a relatively weak effect of antibiotics on the gut and oral microbiomes of the HP study population could be influenced by a potentially higher carriage of antibiotic resistance genes at baseline among the study volunteers from the United Kingdom. There has been a significant decline in antibiotic use in Sweden in the last 2 decades, due to efforts put into the Swedish Strategic Programme for the Rational Use of Antimicrobial Agents and Surveillance of Resistance (STRAMA), which started in 1994 (31). Only in 2013, after the end of the current study, was a similar initiative—the English Surveillance Programme for Antimicrobial Utilization and Resistance (ESPAUR) (11)—developed in the United Kingdom. The first ESPAUR report compared the United Kingdom antibiotic consumption rates between 2010 and 2013 and demonstrated an increase in total use of antibiotics by 6%, with the highest increase (32%) of prescriptions issued predominately by dentists (11, 12). In 2012, there were 20.1 defined daily doses (DDD) of antimicrobials for systemic use per 1,000 inhabitants consumed in the United Kingdom outside hospitals (community use) compared to 14.1 DDD in Sweden (12).
Exposure to different antibiotics resulted in an increased abundance of genes associated with antibiotic resistance. Among the antibiotics tested, exposure to amoxicillin resulted in the least discernible effects on the microbiome composition, while these samples had the highest number with antibiotic resistance-associated genes and the most classes that were increased in the predicted metagenomes and in the full metagenomes, respectively, a week after the exposure.
One of the mechanisms of resistance to clindamycin involves acquisition of an erm gene (erythromycin ribosome methylase) usually carried on plasmids or transposons in pathogenic bacteria (19). Comparison of antimicrobial resistance levels in strains isolated 30 years apart (1970s and 2000) showed that Gram-negative Bacteroides species have acquired an ermB gene that originated in Gram-positive bacteria—Clostridium perfringens, Streptococcus pneumoniae, and Enterococcus faecalis (32). Interestingly, our full metagenome samples from an individual exposed to clindamycin (KI subject S15) showed an increase in eight erm genes in feces and two erm genes in saliva. The fecal ermB gene counts of this individual were increased 240-fold, suggesting potential involvement of plasmids or transposons. Whole-genome sequencing of individual strains has revealed that a large proportion of resistance genes in these strains have not evolved within the strain but rather have been acquired by lateral gene transfer, e.g., by plasmids, transposable elements, and integrons (33).
Currently, no other studies have simultaneously assessed effects of different antibiotics on both oral and fecal microbiomes in healthy individuals. In this study, the same exposure to antibiotics resulted in two radically different responses in these two niches of the human body. The salivary microbiome showed unexpected stability, while the microbial ecology of feces was significantly disturbed for up to several months, depending on the type of antibiotics administered. The reasons for these differences are unknown and should be addressed. A potential explanation could be related to the pharmacokinetics of the antibiotics. Another, more intriguing possibility is that the oral microbial ecosystem possesses a higher intrinsic resilience toward stress, including recovery from exposure to antibiotics. This ecosystem has to surmount multiple daily perturbations such as oral hygiene measures, including exposure to topical antimicrobial agents and physical removal by tooth brushing, as well as alterations in temperature and oxygen (34). None of this is applicable to the colon, where more subtle effects, e.g., alterations in dietary habits, would be more common. A recent report did find changes in bacterial communities in both saliva and feces after antibiotic use (35). The study populations and study design, however, were highly different: the individuals assessed by Abeles and coauthors (35) had a medical indication for antibiotic use and received a prolonged (6-week) mixture of broad-spectrum antibiotics, while we focused on healthy individuals and a single antibiotic dose. Unfortunately, the authors did not report the effects of antibiotics after the discontinuation of the antibiotic use, precluding a comparison regarding the long-term effects and potential differences in resilience between the two microbial ecosystems.
In conclusion, we have demonstrated that two European populations differ in their salivary and fecal microbiome compositions and in their antibiotic resistance gene prevalence. The salivary microbiome was far less affected and more resilient toward the exposure to antibiotics than the fecal microbial community in these populations, irrespective of the antibiotic used. Two of the antibiotics—clindamycin and ciprofloxacin—showed a severe and long-term impact on the health-associated butyrate-producing microbial community of the gut.
MATERIALS AND METHODS
Clinical study and 16S rRNA gene amplicon sequencing.
This randomized clinical trial involved two research centers—The Karolinska Institute in Sweden (KI) and Helperby Therapeutics Ltd. in the United Kingdom (HP). Each study was approved by the respective institutional boards and the respective national competent authorities and was registered with the European Union Clinical Trials Register (for details, see Text S1 in the supplemental material). Each center involved healthy volunteers, randomized into two test groups and one control group. At KI, 30 volunteers were randomly assigned to either the ciprofloxacin (Cipro), the clindamycin (Clinda), or the placebo (Plac KI) group. At HP, 44 volunteers were randomly assigned to the amoxicillin (Amox, n = 15), the minocycline (Minoc, n = 15), or the placebo (Plac HP, n = 14) group. Saliva and fecal samples were collected on 6 occasions: immediately before administration of the antibiotic (baseline), immediately after the treatment course was completed (week 1), and 1 month, 2 months, 4 months, and 12 months postdosing. Sample DNA was extracted, 16S rRNA gene amplicons were sequenced, and data were processed as described previously (36–38) (see Text S1).
Metagenomic shotgun sequencing.
Metagenomic libraries were prepared from isolated DNA using the NEB DNA Mastermix Library preparation kit (New England Biolabs), with size selection by eGel (Life Technologies). Libraries were sequenced for 250-bp paired-end reads using the Illumina MiSeq reagent kit V2 on an Illumina MiSeq sequencer. After quality filtering with Trimmomatic (39), the paired and unpaired forward reads were combined and reads with more than 10% ambiguous bases were removed. Next, the reads were screened for human sequences with Best Match Tagger v3.101 (K. Rotmistrovky and R. Agarwala, 2010), after which human reads and duplicate reads were removed.
Metagenome prediction from 16S rRNA gene amplicon data and validation with shotgun data.
Microbial metagenomes were predicted from 16S rRNA gene sequences using PICRUSt (23) according to the pipeline at http://picrust.github.io/picrust/ (see Text S1 in the supplemental material).
Resistome analysis.
Antibiotic resistance genes were downloaded from the Antibiotic Resistance Genes Database (ARDB) (24). The downloaded data set (ardbAnno1.0), containing 7,828 entries, was made nonredundant first (3,002 nonredundant records), and UBLAST from USEARCH v7.0.1090 (40) was used to map the reads to the ARDB proteins (E value threshold of 10, postfiltered to include hits with a maximum E value of 1E−10 inclusive). The results were processed with HUMAnN (41) to assign weights to the proteins. These weights were then normalized by dividing by the total number of filtered (nonhuman) reads and summarized per antibiotic resistance type. Next, fold changes were calculated for all resistance types with a baseline weight larger than 1E−8.
Statistical analyses.
Both alpha and beta diversity statistics on OTU data and predicted metagenome data analyses were performed using PAST software (42) and SPSS version 20.0 and STAMP software (43) (see Text S1 in the supplemental material).
Sequencing data.
Sequences of the small subunit of the ribosomal gene of fecal and saliva samples are available at the SRA of NCBI as SRP057504 .
SUPPLEMENTAL MATERIAL
Supplemental materials and methods. Download
Diversity (Shannon H diversity index) of microbiome profiles per sample type (feces or saliva), study site (KI or HP), and treatment in time (BL, baseline; W1, week 1; M1, month 1; M2, month 2; M4, month 4; M12, month 12). The horizontal mark indicates the mean value; the error bar indicates the 95% CI. Connectors above the data points connect statistically significantly different groups within the respective time point (P < 0.05; one-way analysis of variance; Games-Howell post hoc test). Connectors below the data points connect the samples within one treatment at the time points that differed significantly from the baseline (P < 0.05; general linear model repeated measures test). Download
Relative abundance of the predicted KEGG orthologous groups (KOs) at baseline (error lines indicate standard deviations) in feces (A) and in saliva (B) from the two centers, KI and HP, and the most abundant significantly differently predicted functional groups (L3) in the fecal (C) and salivary (D) baseline samples from the two study sites, HP (n = 37) and KI (n = 29). Download
Relative abundance of the predicted KEGG orthologous groups (KOs) in the fecal (A) and salivary (B) samples from the ciprofloxacin group plotted against the samples from the KI placebo group per individual time point. Error lines indicate standard deviations. In saliva, 310 KOs at 1 week post-antibiotic treatment and, in feces, 612 of the 4,606 predicted KOs at 1 month post-antibiotic treatment were significantly different in their proportions from the placebo group (P < 0.005, Welch’s t test, Welch’s inverted CI method, and Storey FDR correction for multiple comparisons). Download
Most significantly affected KOs (7 of 612) in predicted metagenomes from ciprofloxacin-exposed feces at month 1 compared to the respective placebo group samples. Download
Effects of different antibiotics on taxa in the fecal microbiome. Only genera or higher taxa present in at least 20% of the samples are shown. The colored cells indicate the time points at which a significant difference in the relative abundance of the taxon was observed in the antibiotic group compared to the respective placebo group. Red shows a decrease and green shows an increase compared to the placebo (Mann-Whitney test, P < 0.005). Only taxa in the dark-colored cells remained significant after Bonferroni correction for multiple comparisons. BL, baseline; W1, week 1; M1, month 1; M2, month 2; M4, month 4; M12, month 12. Mean indicates average relative abundance of each taxon per study.
Effects of different antibiotics on taxa in the salivary microbiome. Only genera or higher taxa present in at least 20% of the samples are shown. The colored cells indicate the time points at which a significant difference in the relative abundance of the taxon was observed in the antibiotic group compared to the respective placebo group. Red shows a decrease and green shows an increase compared to the placebo (Mann-Whitney test, P < 0.005). Only taxa in the dark-colored cells remained significant after Bonferroni correction for multiple comparisons. BL, baseline; W1, week 1; M1, month 1; M2, month 2; M4, month 4; M12, month 12. Mean indicates average relative abundance of each taxon per study.
Predicted KOs that are associated with multidrug or antibiotic resistance and that were found in significantly different proportions in both fecal and salivary baseline samples between the two study sites, KI and HP.
Predicted KOs associated with multidrug or antibiotic resistance that were significantly different from the placebo in fecal samples after the use of antibiotics. Proportions of KOs in black significantly increased, while KOs in red decreased after exposure to the respective antibiotic.
Antibiotic classes (Class) in the shotgun sequences of selected fecal and salivary samples that showed at least a 3-fold change (FC) between baseline and postantibiotic samples. Minoc, minocycline; Cipro, ciprofloxacin; Clinda, clindamycin; Amox, amoxicillin. Class abbreviations are as in the ARDB.
ACKNOWLEDGMENTS
This study received financial support from the EU FP7/2007-2013 grant agreement no. HEALTH-F3-2009-241446 and was a part of the project entitled “The effects of antibiotic administration on the emergence and persistence of antibiotic-resistant bacteria in humans and on the composition of the indigenous microbiota at various body sites,” under the acronym ANTIRESDEV (http://www.ucl.ac.uk/antiresdev).
The authors have no competing financial interests.
We are thankful to the clinical teams at Helperby Ltd. and at the Karolinska Institutet for performing the clinical studies. We thank Tony Brooks and Nipurna Jina at UCL for sequencing work and Marloes ter Beek, Hakim Rahaoui, and Jolanda Kool at TNO for sample processing.
A.W., C.E.N., and M.U.R. were responsible for the clinical study at KI; A.R.C., Y.H., and A.S. were responsible for the clinical study at HP; M.W. coordinated the ANTIRESDEV project; D.A.S., W.C., B.J.F.K., M.H., and E.Z. were responsible for the work package on metagenomics; M.J.B. and M.P.M.C. were responsible for sample preparation; M.H. was responsible for the sequencing process; B.W.B. and B.J.F.K. were responsible for resistome data analyses; M.J.T.D.M. was responsible for the predicted metagenome data interpretation; E.Z. performed the overall data analyses; E.Z., B.W.B., and M.J.T.D.M. drafted the manuscript; all authors contributed to the final manuscript.
Footnotes
Citation Zaura E, Brandt BW, Teixeira de Mattos MJ, Buijs MJ, Caspers MPM, Rashid MU, Weintraub A, Nord CE, Savell A, Hu Y, Coates AR, Hubank M, Spratt DA, Wilson M, Keijser BJF, Crielaard W. 2015. Same exposure but two radically different responses to antibiotics: resilience of the salivary microbiome versus long-term microbial shifts in feces. mBio 6(6):e01693-15. doi:10.1128/mBio.01693-15.
REFERENCES
- 1.Cantas L, Shah SQA, Cavaco LM, Manaia CM, Walsh F, Popowska M, Garelick H, Bürgmann H, Sørum H. 2013. A brief multi-disciplinary review on antimicrobial resistance in medicine and its linkage to the global environmental microbiota. Front Microbiol 4:96. doi: 10.3389/fmicb.2013.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alanis AJ. 2005. Resistance to antibiotics: are we in the post-antibiotic era? Arch Med Res 36:697–705. doi: 10.1016/j.arcmed.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Clemente J, Ursell L, Parfrey L, Knight R. 2012. The impact of the gut microbiota on human health: an integrative view. Cell 148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Modi SR, Collins JJ, Relman DA. 2014. Antibiotics and the gut microbiota. J Clin Invest 124:4212–4218. doi: 10.1172/JCI72333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munyaka PM, Khafipour E, Ghia J. 2014. External influence of early childhood establishment of gut microbiota and subsequent health implications. Front Pediatr 2:109. doi: 10.3389/fped.2014.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts SE, Wotton CJ, Williams JG, Griffith M, Goldacre MJ. 2011. Perinatal and early life risk factors for inflammatory bowel disease. World J Gastroenterol 17:743–749. doi: 10.3748/wjg.v17.i6.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira CM, Vieira AT, Vinolo MAR, Oliveira FA, Curi R, Martins FDS. 2014. The central role of the gut microbiota in chronic inflammatory diseases. J Immunol Res 2014:689492. doi: 10.1155/2014/689492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willing BP, Russell SL, Finlay BB. 2011. Shifting the balance: antibiotic effects on host–microbiota mutualism. Nat Rev Microbiol 9:233–243. doi: 10.1038/nrmicro2536. [DOI] [PubMed] [Google Scholar]
- 9.Abdulah R. 2012. Antibiotic abuse in developing countries. Pharm Regul Aff 1:e106. doi: 10.4172/2167-7689.1000e106. [DOI] [Google Scholar]
- 10.Widayati A, Suryawati S, de Crespigny C, Hiller JE. 2011. Self medication with antibiotics in Yogyakarta City Indonesia: a cross sectional population-based survey. BMC Res Notes 4:491. doi: 10.1186/1756-0500-4-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ESPAUR Writing Committee 2014. English surveillance programme for antimicrobial utilization and resistance (ESPAUR). Report 2014 Public Health England, London, United Kingdom: https://www.gov.uk/government/publications/english-surveillance-programme-antimicrobial-utilisation-and-resistance-espaur-report. [Google Scholar]
- 12.European Centre for Disease Prevention and Control. 2014. Surveillance of antimicrobial consumption in Europe 2012. ECDC, European Centre for Disease Prevention and Control, Stockholm, Sweden. [Google Scholar]
- 13.Fernandez y Mostajo M, Zaura E, Crielaard W, Beertsen W. 2011. Does routine analysis of subgingival microbiota in periodontitis contribute to patient benefit? Eur J Oral Sci 119:259–264. doi: 10.1111/j.1600-0722.2011.00828.x. [DOI] [PubMed] [Google Scholar]
- 14.Keenan JR, Veitz-Keenan A. 2015. Antibiotic prophylaxis for dental implant placement? Evid Based Dent 16:52–53. doi: 10.1038/sj.ebd.6401097. [DOI] [PubMed] [Google Scholar]
- 15.Oomens MAE, Forouzanfar T. 2012. Antibiotic prophylaxis in third molar surgery: a review. Oral Surg Oral Med Oral Pathol Oral Radiol 114:e5–e12. doi: 10.1016/j.oooo.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 16.Dethlefsen L, Huse S, Sogin ML, Relman DA. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dethlefsen L, Relman DA. 2011. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A 108:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rashid M-, Zaura E, Buijs MJ, Keijser BJF, Crielaard W, Nord CE, Weintraub A. 2015. Determining the long-term effect of antibiotic administration on the human normal intestinal microbiota using culture and pyrosequencing methods. Clin Infect Dis 60:S77–S84. doi: 10.1093/cid/civ137. [DOI] [PubMed] [Google Scholar]
- 19.Canu A, Leclercq R. 2009. Macrolides and lincosamides, p 211–221. In Mayers DL. (ed), Antimicrobial drug resistance, vol 1 Mechanisms of drug resistance. Humana Press, Springer, New York, NY. [Google Scholar]
- 20.Guay D. 2007. Update on clindamycin in the management of bacterial, fungal and protozoal infections. Expert Opin Pharmacother 8:2401–2444. doi: 10.1517/14656566.8.14.2401. [DOI] [PubMed] [Google Scholar]
- 21.Moudgal VV, Kaatz GW. 2009. Fluoroquinolone resistance in bacteria, p 195–205. In Mayers DL. (ed), Antimicrobial drug resistance, vol 1 Mechanisms of drug resistance. Humana Press, Springer, New York, NY. [Google Scholar]
- 22.Rang HP, Dale MM. 1991. Pharmacology, 2nd ed, p 804–832. Churchill Livingstone, London, United Kingdom. [Google Scholar]
- 23.Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu B, Pop M. 2009. ARDB—antibiotic resistance genes database. Nucleic Acids Res 37:D443–D447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vital M, Howe AC, Tiedje JM. 2014. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio 5:e00889-14. doi: 10.1128/mBio.00889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer R-. 2008. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther 27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 27.Albenberg LG, Wu GD. 2014. Diet and the intestinal microbiome: associations, functions, and implications for health and disease. Gastroenterology 146:1564–1572. doi: 10.1053/j.gastro.2014.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. 2003. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol 30:644–654. doi: 10.1034/j.1600-051X.2003.00376.x. [DOI] [PubMed] [Google Scholar]
- 29.Li K, Bihan M, Methé BA. 2013. Analyses of the stability and core taxonomic memberships of the human microbiome. PLoS One 8:e63139. doi: 10.1371/journal.pone.0063139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sommer MOA, Dantas G, Church GM. 2009. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325:1128–1131. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mölstad S, Erntell M, Hanberger H, Melander E, Norman C, Skoog G, Lundborg CS, Söderström A, Torell E, Cars O. 2008. Sustained reduction of antibiotic use and low bacterial resistance: 10-year follow-up of the Swedish Strama programme. Lancet Infect Dis 8:125–132. doi: 10.1016/S1473-3099(08)70017-3. [DOI] [PubMed] [Google Scholar]
- 32.Shoemaker NB, Vlamakis H, Hayes K, Salyers AA. 2001. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl Environ Microbiol 67:561–568. doi: 10.1128/AEM.67.2.561-568.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 34.Marsh PD, Moter A, Devine DA. 2011. Dental plaque biofilms: communities, conflict and control. Periodontol 2000 55:16–35. doi: 10.1111/j.1600-0757.2009.00339.x. [DOI] [PubMed] [Google Scholar]
- 35.Abeles SR, Ly M, Santiago-Rodriguez TM, Pride DT. 2015. Effects of long term antibiotic therapy on human oral and fecal viromes. PLoS One 10:e0134941. doi: 10.1371/journal.pone.0134941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaura E, Keijser BJ, Huse SM, Crielaard W. 2009. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol 9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kraneveld EA, Buijs MJ, Bonder MJ, Visser M, Keijser BJF, Crielaard W, Zaura E. 2012. The relation between oral Candida load and bacterial microbiome profiles in Dutch older adults. PLoS One 7:e42770. doi: 10.1371/journal.pone.0042770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koopman JE, Röling WFM, Buijs MJ, Sissons CH, ten Cate JM, Keijser BJF, Crielaard W, Zaura E. 2015. Stability and resilience of oral microcosms toward acidification and Candida outgrowth by arginine supplementation. Microb Ecol 69:422–433. doi: 10.1007/s00248-014-0535-x. [DOI] [PubMed] [Google Scholar]
- 39.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 41.Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, Rodriguez-Mueller B, Zucker J, Thiagarajan M, Henrissat B, White O, Kelley ST, Methé B, Schloss PD, Gevers D, Mitreva M, Huttenhower C. 2012. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol 8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hammer O, Harper DAT, Ryan PD. 2001. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron 4:1–9. [Google Scholar]
- 43.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. 2014. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and methods. Download
Diversity (Shannon H diversity index) of microbiome profiles per sample type (feces or saliva), study site (KI or HP), and treatment in time (BL, baseline; W1, week 1; M1, month 1; M2, month 2; M4, month 4; M12, month 12). The horizontal mark indicates the mean value; the error bar indicates the 95% CI. Connectors above the data points connect statistically significantly different groups within the respective time point (P < 0.05; one-way analysis of variance; Games-Howell post hoc test). Connectors below the data points connect the samples within one treatment at the time points that differed significantly from the baseline (P < 0.05; general linear model repeated measures test). Download
Relative abundance of the predicted KEGG orthologous groups (KOs) at baseline (error lines indicate standard deviations) in feces (A) and in saliva (B) from the two centers, KI and HP, and the most abundant significantly differently predicted functional groups (L3) in the fecal (C) and salivary (D) baseline samples from the two study sites, HP (n = 37) and KI (n = 29). Download
Relative abundance of the predicted KEGG orthologous groups (KOs) in the fecal (A) and salivary (B) samples from the ciprofloxacin group plotted against the samples from the KI placebo group per individual time point. Error lines indicate standard deviations. In saliva, 310 KOs at 1 week post-antibiotic treatment and, in feces, 612 of the 4,606 predicted KOs at 1 month post-antibiotic treatment were significantly different in their proportions from the placebo group (P < 0.005, Welch’s t test, Welch’s inverted CI method, and Storey FDR correction for multiple comparisons). Download
Most significantly affected KOs (7 of 612) in predicted metagenomes from ciprofloxacin-exposed feces at month 1 compared to the respective placebo group samples. Download
Effects of different antibiotics on taxa in the fecal microbiome. Only genera or higher taxa present in at least 20% of the samples are shown. The colored cells indicate the time points at which a significant difference in the relative abundance of the taxon was observed in the antibiotic group compared to the respective placebo group. Red shows a decrease and green shows an increase compared to the placebo (Mann-Whitney test, P < 0.005). Only taxa in the dark-colored cells remained significant after Bonferroni correction for multiple comparisons. BL, baseline; W1, week 1; M1, month 1; M2, month 2; M4, month 4; M12, month 12. Mean indicates average relative abundance of each taxon per study.
Effects of different antibiotics on taxa in the salivary microbiome. Only genera or higher taxa present in at least 20% of the samples are shown. The colored cells indicate the time points at which a significant difference in the relative abundance of the taxon was observed in the antibiotic group compared to the respective placebo group. Red shows a decrease and green shows an increase compared to the placebo (Mann-Whitney test, P < 0.005). Only taxa in the dark-colored cells remained significant after Bonferroni correction for multiple comparisons. BL, baseline; W1, week 1; M1, month 1; M2, month 2; M4, month 4; M12, month 12. Mean indicates average relative abundance of each taxon per study.
Predicted KOs that are associated with multidrug or antibiotic resistance and that were found in significantly different proportions in both fecal and salivary baseline samples between the two study sites, KI and HP.
Predicted KOs associated with multidrug or antibiotic resistance that were significantly different from the placebo in fecal samples after the use of antibiotics. Proportions of KOs in black significantly increased, while KOs in red decreased after exposure to the respective antibiotic.
Antibiotic classes (Class) in the shotgun sequences of selected fecal and salivary samples that showed at least a 3-fold change (FC) between baseline and postantibiotic samples. Minoc, minocycline; Cipro, ciprofloxacin; Clinda, clindamycin; Amox, amoxicillin. Class abbreviations are as in the ARDB.