Abstract
DNA damage is a natural hazard of life. The most common DNA lesions are base, sugar, and single-strand break damage resulting from oxidation, alkylation, deamination, and spontaneous hydrolysis. If left unrepaired, such lesions can become fixed in the genome as permanent mutations. Thus, evolution has led to the creation of several highly conserved, partially redundant pathways to repair or mitigate the effects of DNA base damage. The biochemical mechanisms of these pathways have been well characterized and the impact of this work was recently highlighted by the selection of Tomas Lindahl, Aziz Sancar and Paul Modrich as the recipients of the 2015 Nobel Prize in Chemistry for their seminal work in defining DNA repair pathways. However, how these repair pathways are regulated and interconnected is still being elucidated. This review focuses on the classical base excision repair and strand incision pathways in eukaryotes, considering both Saccharomyces cerevisiae and humans, and extends to some important questions and challenges facing the field of DNA base damage repair.
INTRODUCTION
Few systems are as crucial to sustaining life as DNA repair. The genomes in the cells of all organisms are under constant bombardment by genotoxic stresses, both exogenous (e.g. ultraviolet and ionizing radiation, chemical combustion products) and endogenous (e.g. reactive oxygen species, nucleases). These agents can modify the chemical structure of DNA in ways that produce mutations in transcribed RNA and replicated DNA, alter the ability of regulatory elements to be recognized by DNA binding proteins, and lead to cell death by blocking transcription and replication (1,2). Lesions can occur to most parts of the DNA structure, ranging from minor and major chemical modifications, to single-strand breaks and gaps, to full double-strand breaks. Chemical modifications are the most common lesions (3) while double-strand breaks are the most lethal (4). In eukaryotes, these lesions may occur in both nuclear and organellar (mitochondria, chloroplasts) genomes. The constellation of extraordinarily well conserved DNA repair pathways is responsible for removing these lesions and/or mitigating their effects. Properly repairing the common base lesions is important not only to abrogate their immediate impacts, but also to prevent their conversion into more deleterious strand breaks. These repair and tolerance mechanisms have important implications for human health and disease, especially oncogenesis and degenerative disorders associated with aging (5–7). This article reviews the most common and mutagenic classes of lesions and the pathways responsible for their repair, with a focus on the members of the classical base excision repair (BER) pathway first delineated by Tomas Lindahl (8), and recent work that has expanded the impact and roles of this pathway. Finally, the review concludes with a discussion of some current questions and challenges facing the field of DNA base damage repair.
BASE, SUGAR AND SINGLE-STRAND BREAK LESIONS
DNA base lesions, which are chemical modifications to the base of a nucleotide, are the most common type of genomic damage: an estimated 120,000 base lesions occur in the 6.5 Gbp nuclear genome of human liver cells per day (3). Base lesions may be accompanied more rarely by sugar modifications and single-strand breaks. These lesions can have serious consequences for numerous cellular processes, resulting in genomic mutagenesis (i), transcriptional mutagenesis (ii), and disruption of regulatory DNA elements (1). There are four major classes of base lesions: oxidation, deamination, alkylation, and hydrolysis. These lesions are detailed below and are illustrated in Figure 1.
Figure 1.
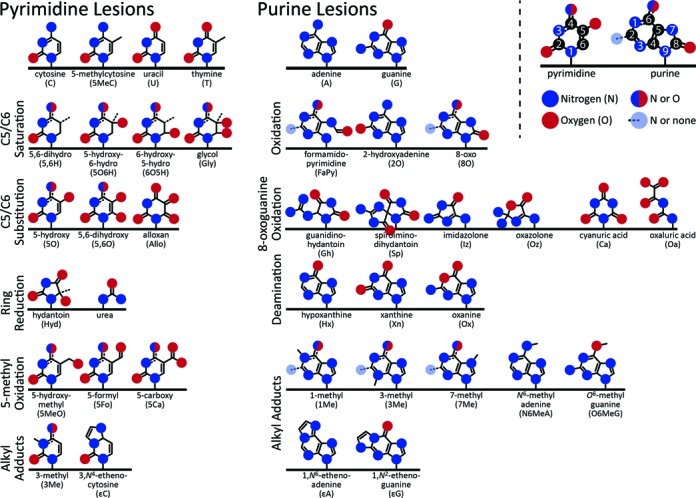
Common base lesions. The basic chemical structures of the common base lesions that occur in DNA grouped by type. The basic nucleotides are shown at the top, and lesions are displayed to indicate the same modification occurring to multiple bases. Hydrogens are omitted. IUPAC numbering for pyrimidines and purines is illustrated at the top right.
Oxidation
Reactive oxygen species (ROS; e.g. hydrogen peroxide, hydroxyl radical, superoxide anion) are critical components of normal signaling pathways (9) yet they are also a significant source of base damage (9). As a result, these chemical species are very carefully regulated, deliberately produced by oxidases and removed by scavengers. ROS can also originate from the environment, both directly and as an aftereffect of the reactions of antioxidants and xenobiotics (10). In addition, hydroxyl radicals can be produced by ultraviolet (UV) radiation in the UV-A band (315–400 nm) (11). Radiolysis of water by ionizing radiation also produces ROS in addition to reactive free protons and electrons which can produce similar sets of base lesions (12). Thus, there are numerous sources of ROS that can lead to formation of oxidative lesions.
Oxidative attack on pyrimidines [cytosine (C), 5-methylcytosine (5MeC), thymine (T)] can result in the saturation of the double bond between pyrimidine carbons 5 and 6 to form hydrate (5-hydroxy-6-hydro, 6-hydroxy-5-hydro) and glycol derivatives. The 5,6-dihydro derivatives are formed exclusively by the free protons and/or electrons generated by ionizing radiation. Carbon 5/6 hydrogens can be substituted to form 5-hydroxy and 5,6-hydroxy derivatives. C and 5MeC saturated lesions are especially prone to deamination, leading to the uracil (U) and T forms of the lesion, respectively. Uracil glycol rapidly decomposes to 5-hydroxyuracil (13–15). An exception to this instability is 6-hydroxy-5-hydrocytosine (13). Further oxidation of C (and U) can also lead to alloxan and then 5-hydroxyhydantoin while further oxidation of T results in 5-hydroxy-5-methylhydantoin. The 5-methyl group of 5MeC or T can be oxidized to 5-hydroxymethyl, 5-formyl, and 5-carboxy C and U derivatives, respectively. The oxidation of the 5-methyl group can be induced enzymatically as part of an active demethylation pathway in eukaryotes (16,17). Finally, any pyrimidine can be further oxidized to urea. Pyrimidine radical reaction mechanisms and products have been reviewed recently (12,18).
Oxidation of purines [adenine (A), guanine (G)] can produce ring-opened formamidopyrimidine derivatives, 8-oxo derivatives, and 2-hydroxyadenine. 8-Oxoguanine can undergo extensive further oxidation to the more mutagenic lesions, guanidinohydantoin, spiroiminohydantoin, imidazolone, oxazolone, cyanuric acid, oxaluric acid and urea. Purine oxidative reaction mechanisms and products are reviewed in (12,18,19).
Deoxyribose in DNA is also vulnerable to oxidation, which usually results in base hydrolysis to form an abasic (apyrimidinic/apurinic, or AP) site and/or a strand break in addition to the modified sugar. Deoxyribose oxidation can also result in a crosslink between the 5′ carbon of the sugar and carbon 8 of an attached purine, forming a cyclic nucleotide (12). There is a report from 1992 of oxidative conversion of deoxyribose to ribose in vivo (20), but no subsequent validation has been reported. An abasic site may also spontaneously form an O-glycosidic bond with an alcohol (21).
Deamination
Deamination is the replacement of a nitrogen atom with an oxygen atom, primarily of exocyclic amines. Both C and 5MeC have an exocyclic amine on carbon 4. Deamination of these bases by a basic molecule produces U or T, respectively (22); deamination of 5MeC to T within the important regulatory CpG repeats likely makes up a large number of the observed mutations in cancer genomes (23). Purine deamination is mediated largely by the signaling radical nitric oxide. Deamination of the exocyclic amine at carbon 6 of A produces hypoxanthine (inosine). Deamination of the exocyclic amine at carbon 2 of G produces xanthine. Uniquely, the internal nitrogen 1 of G can be replaced by oxygen, producing oxanine (24).
Alkylation
Alkylation is one of the less common types of base lesions, but they are often the most mutagenic (25). Methyl groups can be added to any available amine on both pyrimidines and purines as well as to the carbon 6 oxygen or nitrogen of G or A (26). Lipid peroxidation products can also react with an exocyclic amine and an adjacent internal amine in C, A or G to produce exocyclic etheno adducts or other adducts important for colon carcinogenesis including 2-propanoguanine (27,28). Xenobiotic metabolites (resulting from combustion products such as benzo(a)pyrene and other aromatic polyphenols) are highly reactive and thus able to form bulky base adducts (29,30).
Hydrolysis
Hydrolysis of the N-glycosidic bond to generate an abasic site may occur spontaneously, via the action of a DNA N-glycosylase, or as part of a radical reaction mechanism (12). Hydrolysis of the sugar-phosphate backbone can also occur through a radical reaction mechanism, as discussed in Oxidation. The abasic site aldehyde is reactive and may progress to an interstrand crosslink to a purine on the opposing strand (31,32).
Incorporation of damaged nucleotides
While most base damage is introduced into DNA directly, another source of lesions is from the incorporation of damaged bases from the nucleotide pool, particularly U and 8-oxoguanine. Recent studies demonstrated that misincorporation of ribonucleotides in DNA is a relatively common event, at a frequency of ∼4 for every 104 nucleotides inserted in S. cerevisiae (33).
BASE DAMAGE REPAIR PATHWAYS
DNA base lesions pose a challenge for cells on an ongoing basis. The interconnected and overlapping set of pathways collectively known as DNA repair is responsible for either reverting the lesions back to the original form (repair) or otherwise limiting the potential impact of the lesion on cellular function (tolerance). Underscoring their critical role, these pathways are very highly conserved throughout all domains of life, and deleterious mutations in base damage repair genes are associated with neurodegeneration and cancer; for example, inherited mutations in the genes encoding the NTHL1 and MUTYH glycosylases have been linked to colorectal cancer (34–36). Typically, lesions can be repaired through multiple distinct, partially redundant pathways (37,38). The human and budding yeast Saccharomyces cerevisiae pathways are schematized in Figure 2 and reviewed in the following sections.
Figure 2.
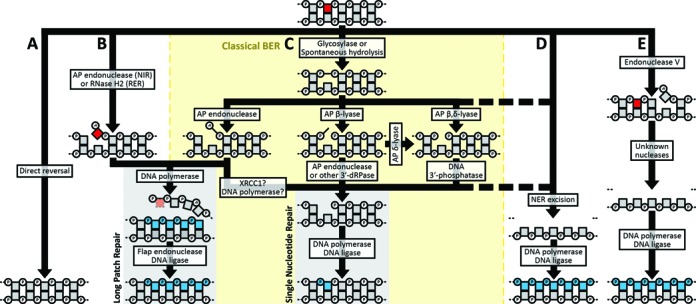
Base, sugar, and single-strand break lesion repair pathways. Base lesions can be processed by: (A) direct reversal; (B) nucleotide incision repair (NIR), ribonucleotide excision repair (RER); (C) classical base excision repair (BER); (D) nucleotide excision repair (NER) or (E) endonuclease V–mediated excision repair. Within the BER pathway (C), multiple semi-redundant pathways are available for processing abasic sites. Pathway choice depends on the lesion, and some lesions can be acted on by multiple pathways. Squares (nucleosides) and connected circles (phosphates) represent a section of a DNA molecule, with the base lesion indicated in red. De novo synthesized bases are shown in blue. The enzyme activity responsible for each step is noted. The dashed segments indicate a fallback pathway. AP = apurinic/apyrimidinic site; dRP = deoxyribose phosphate.
Direct lesion reversal
Certain lesions can be directly removed from a base while leaving the DNA helix intact (Figure 2A). Many alkyl adducts can be removed by the AlkB family of dioxygenases (Human: ALKBH2, ALKBH3; S. cerevisiae: not present). ALKBH2/3 removes alkyl groups (39–41) and exocyclic adducts (42–44) at the nitrogen 1 position of purines and the nitrogen 3 position of pyrimidines. The most highly mutagenic alkylated base, O6-methylguanine, is reversed by O6-methylguanine methyltransferase (human: MGMT; S. cerevisiae: Mgt1), a ‘suicide protein’ which irreversibly transfers the errant methyl group onto itself and is subsequently degraded (45). Photolyases (human: not present; S. cerevisiae: Phr1) are light-dependent enzymes that directly reverse UV-induced pyrimidine dimers (46). Photolyases were lost early in the largely nocturnal mammalian lineage (47). While direct reversal efficiently restores DNA to its original pristine form, the majority of DNA damage is repaired through more general pathways that have the capacity to recognize and repair a spectrum of lesions.
Base excision and strand incision repair
The base excision repair (BER) pathway efficiently corrects most non-bulky DNA base lesions that are not addressable by direct reversal. Thus, BER is responsible for repairing the vast majority of lesions that occur in DNA, and the pathway is active in both nuclei and mitochondria. BER is initiated by the recognition and hydrolysis of the damaged base by a DNA N-glycosylase, leaving an abasic site (48). The glycosylases are reviewed in depth below and their specific substrates are summarized in Figure 3. The BER pathway is illustrated in Figure 2C.
Figure 3.
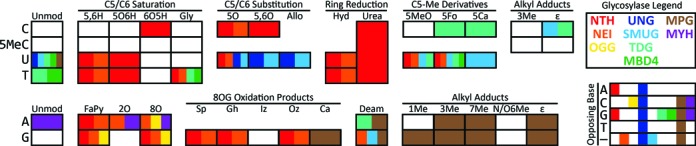
Substrate specificities of base excision repair glycosylases. Each square represents a DNA base lesion, with the row indicating the original base and the column indicating the particular lesion, grouped by shared features. ‘Unmod’ = unmodified. Refer to Figure 1 for lesion abbreviations. Coloration within each box indicates the glycosylase families that recognize the lesion and excise it from DNA, according to the legend at the top-right. The matrix at the bottom-right illustrates the specificity of the glycosylase families with respect to the base opposite from a lesion; — indicates single-stranded DNA. Note that this diagram does not account for differences in enzyme kinetics between or within families. Example: OGG (yellow) excises guanine-derived formamidopyrimidine, 8-oxoadenine, and 8-oxoguanine when these lesions are opposite to cytosine.
Abasic sites can be processed by one of two subpathways. The first subpathway, called single-nucleotide BER (SN-BER; also termed ‘short patch’) is initiated by an AP lyase. The AP lyase cleaves the DNA backbone on the 3′ side of the abasic site by β-elimination (AP β-lyase), which leaves a 3′-deoxyribose phosphate and 5′-phosphate, or by β,δ-elimination (AP β,δ-lyase), which excises the deoxyribose and leaves 3′- and 5′-phosphates (49). Both of these products block DNA polymerase activity. Removal of the 3′-deoxyribose phosphate (dRP) can be catalyzed by as little as a basic tripeptide, but is typically carried out by an AP endonuclease (49). Removal of the 3′-phosphate is catalyzed by the DNA 5′-kinase/3′-phosphatase, polynucleotide kinase/phosphatase (human: PNKP; S. cerevisiae: Tpp1) (50,51). The sequential action of an AP δ-lyase and DNA 3′-phosphatase can also process 3′-dRP as an alternative to AP endonuclease (52,53). The result is a clean 1-nucleotide gap, which is then directly filled by a DNA polymerase and sealed by a DNA ligase (54). The second subpathway, called long patch BER (LP-BER), is initiated by an AP endonuclease (human: APEX1, APEX2; S. cerevisiae: Apn1, Apn2), which cleaves the DNA backbone on the 5′ side of the abasic site (49). This incision leaves a 3′-hydroxyl group that can directly serve as a substrate for DNA polymerase β (human: POLB, S. cerevisiae: POL4). The polymerase fills the removed base and several bases downstream, displacing the strand on the 3′ side of the cut site (55). This displaced strand, which is terminated by the 5′-dRP, is removed by the flap endonuclease (human: FEN1; S. cerevisiae: Rad27) at its base and a DNA ligase seals the nick, leaving a fully repaired segment of DNA (48). An AP endonuclease–cleaved abasic site can also be directed into SN-BER through the 5′-dRPase activity of polymerase β (56). The mechanism controlling the switch between subpathways is unclear though it has been proposed that the human BER scaffold protein XRCC1 plays a role (57,58). Cellular ATP concentration, cell cycle phase, and chemistry of the 5′ terminus may also influence subpathway choice (59,60). The AP lyase activities, which promote SN-BER, are associated with the subset of glycosylases that recognize oxidative lesions (49,61–63). This subpathway may be favored for oxidative lesions which occur in clusters (54). Attempting LP-BER in such a cluster could result in polymerase stalling, mutagenesis, or converting the original simple base lesion into a more serious double-strand break (64,65).
Other processes produce lesions that may enter the BER pathway as intermediates. Bases can spontaneously hydrolyze from the DNA backbone to generate abasic sites with certain base lesions more prone to hydrolysis than other bases. Oxidative attack on deoxyribose can cause base hydrolysis and sugar damage (12), which are effectively processed by the coordinated actions of AP endonuclease, AP lyases and 3′-DNA phosphatases (66,67). Complex single-strand breaks can occur through the action of abortive topoisomerase or DNA ligase activity [via adenosine 5′-monophosphate (AMP)] adjacent to a lesion, which become trapped by covalent linkage to an end-group phosphate. Tyrosyl–DNA phosphodiesterases and aprataxin deadenylase, respectively, are able to reverse these polymerase-blocking groups (68,69). Resolution of these structures allows BER to complete repair. These end-trimming pathways were reviewed in depth recently (70).
Classical BER has been joined by two recently discovered strand incision repair pathways that feed into LP-BER: (i) nucleotide incision repair (NIR) and; (ii) ribonucleotide excision repair (RER) (Figure 2B). NIR is initiated by AP endonuclease, which is able to cleave the DNA backbone 5′ to pyrimidine lesions (71–77). As AP endonucleases are highly abundant, NIR can constitute the major repair activity against a subset of pyrimidine base lesions (74). RER is initiated by the RNase H2 complex (human: RNASEH2A-RNASEH2B-RNASEH2C; S. cerevisiae: Rnh202-Rnh203), which incises the strand 5′ to the misincorporated ribonucleotide (78). RNase H1 (human: RNASEH1; S. cerevisiae: Rnh1) has recently been shown to also incise 3′ to the ribonucleotide, producing a single-nucleotide gap (79), leaving open the possibility of feeding into SN-BER for completion of repair. Advances in RER and other repair pathways for ribonucleotides have been reviewed in depth (80).
The recent broadening of the range of lesions processed through BER, as well as the discovery of strand incision as a major alternate first step in the repair of many classic BER substrates, suggests that the term ‘base excision repair’ is no longer appropriate. Perhaps a more suitable term is ‘base excision and strand incision repair’ (BESIR), which more closely reflects the range of activities of the pathway components. The enzymes most critical for the initiation and processing of lesions in BESIR, the glycosylases, AP endonucleases, end-trimming enzymes, polymerases, and ligases are reviewed in the following sections.
Alkylpurine–DNA glycosylases
The alkylpurine–DNA glycosylases (human: MPG; S. cerevisiae: Mag1) are responsible for excising methylated purines (especially 3-methyladenine) (81–86), purine exocyclic adducts (e.g. 1,N6-ethenoadenine) (87,88), deaminated purines (e.g. hypoxanthine, xanthine, oxanine) (89,90), the 8-oxoguanine oxidation product cyanuric acid (91), uracil (86) and O-glycosidic additions to deoxyribose (21). Many of these substrates can be excised from both double-stranded and single-stranded DNA (86,90). Mag1 can also remove A mispaired with C (87). Intriguingly, MPG is able to catalyze the reverse reaction, forming an N-glycosidic bond with a free base, potentially allowing the correctly-paired base to be directly swapped (92). The biological relevance of this activity would be dependent on the relative concentrations of free bases in the nucleus, and it could be counterproductive if an incorrect base were inserted which was not excisable by MPG.
These glycosylases do not possess lyase activity and their resulting abasic sites can be processed both by SN- and LP-BESIR subpathways (63). Mag1 expression is inducible by the DNA damage checkpoint pathway (93–97) while MPG transcript levels are cell-cycle regulated (98). MPG has been identified both in the nucleus and mitochondria (99) while Mag1 is restricted to nuclei (100). MPG activity is enhanced by XRCC1 and a component of nucleotide excision repair, HR23 (101,102). MPG can bind to PCNA along with APEX1. APEX1 may stimulate MPG turnover by displacing MPG from the abasic site products, though the evidence to support this mechanism is mixed (103–105). MPG can also form a dimer with methylcytosine binding domain protein 1 (MBD1), which sequesters MPG at methylated CpG promoters. Upon alkylative attack on G, MBD1 dissociates from both MPG and the DNA allowing MPG to redistribute throughout the genome (85). This mechanism may provide an alkylation-responsive reservoir for rapid mobilization to repair alkylation damage.
Endonuclease III-like glycosylases
The endonuclease III-like N-glycosylase family (human: NTHL1; S. cerevisiae: Ntg1, Ntg2) is responsible for repairing a wide array of oxidative lesions in double-stranded DNA, primarily oxidized pyrimidines (e.g. 5-hydroxycytosine, cytosine hydrates, thymine glycol) (61,106–116), ring-fragmented purines (108,117–119), and 8-oxoguanine opposite a purine (120,121). There are several differences between these proteins with respect to substrate specificity. For example, Ntg1 and NTHL1 do not process 5-hydroxycytosine as efficiently as Ntg2 processes this lesion (74,106). Ntg1, however, is better at processing cytosine hydrates than is Ntg2. Ntg2 and NTHL1 can excise certain 8-oxoguanine oxidation products while Ntg1 does not (91,119,122). Ntg1 can process dihydrothymine while Ntg2 cannot (114). This family of enzymes possesses AP β-lyase activity which directs lesions into the SN-BESIR subpathway described above through a coordinated reaction mechanism, but this family can also contribute to the processing of abasic sites generated spontaneously or by monofunctional glycosylases (55,62,123,124).
NTHL1 activity on its own is over 100 times slower than is the activity of the bacterial counterpart, endonuclease III (125), and the rate-limiting step is release from the lyase-cleaved abasic site (126). APEX1 enhances release from that product (112). This mechanism may protect the toxic strand-break intermediate in a ‘passing the baton’ or ‘handoff’ substrate channeling mechanism (127). Other binding partners (e.g. XPG, YB-1, XRCC1) enhance the activity of NTHL1 though the precise mechanism behind the enhancement is unknown (102,112,125,128).
Ntg1 and Ntg2 are the result of a genome duplication in the evolutionary history of S. cerevisiae (129,130), and they have segregated certain characteristics that the single original enzyme possessed. NTHL1 and Ntg1 localize to both the nucleus and mitochondria and are involved in repairing both genomes while Ntg2 is strictly nuclear (106,114,131–136). On the other hand, NTHL1 and Ntg2 both contain a conserved iron-sulfur center but this feature is absent from Ntg1 (61,108,137). This iron-sulfur center is redox-active in vivo and has been hypothesized to be involved in DNA damage sensing. In brief, electrons can be transported over long distances through the π-orbitals of the DNA base pair stack between bound redox-active proteins containing iron-sulfur centers, a process called DNA-mediated charge transport. Reduction of the iron-sulfur center allows dissociation of the repair enzyme from the DNA. DNA lesions can disrupt the base stack, retarding charge transport. This condition causes the repair protein to remain bound to the DNA and slide along the helix until it encounters the lesion (138–141). Little is known about the transcriptional regulation of this family of proteins but NTHL1 is upregulated in S phase (118) and the Ntg1 promoter has a conserved (within yeast) promoter element necessary for oxidative stress induction (142).
Endonuclease VIII-like glycosylases
The endonuclease VIII-like N-glycosylase family (Human: NEIL1, NEIL2, NEIL3; S. cerevisiae: not present) is responsible for repairing a wide array of oxidative lesions primarily in single-stranded DNA. Most of the substrates of this enzyme family overlap with those of NTHL1 (74,115,117,119,143–156). NEIL1 and NEIL3 can also process 8-oxoguanine oxidation products in telomere-associated quadruplex DNA (157,158), NEIL3 can process thymine glycol in the same structures and is additionally able to process lesions near strand breaks which are refractory to NTHL1 and the 8-oxoguanine DNA glycosylase, OGG1 (159,160). NEIL1 can excise the internally deaminated G, oxanine (161). NEILs are also involved in repair of some interstrand crosslinks (162).
This family of enzymes possesses AP lyase activity. In contrast to NTHL1, NEIL1 and NEIL2 are AP β,δ-lyases (146,147) while NEIL3 employs the typical β-elimination mechanism (119). The β,δ mechanism directs NEIL1 or NEIL2-excised lesions into the SN subpathway, but there is also evidence that these lesions can undergo LP repair (163). NEIL1 or NEIL2, together with PNK, can provide backup dRPase activity to clean up after other AP β-lyases (53).
NEILs provide specialized repair activities in the cell. NEIL1 participates in pre-replication repair, in which NEIL1 recognizes lesions within the single-stranded region of the replication fork prior to being read by the DNA polymerase. NEIL1 excises the base and creates a strand break, forcing the polymerase to stall and backtrack so that the lesion can be repaired (164,165). NEIL2 associates with RNA polymerase II and CSB to allow repair of lesions encountered as transcription is progressing (143,157,166). There is some evidence from in vitro activity assays, immunoprecipitation, and HEK293 cell culture experiments that NEIL1 and NEIL2 can compensate for one another, but this ability appears to be limited (165,167,168).
NEIL1 is strongly upregulated during S phase (146) as well as under oxidative stress (169). In contrast, expression levels of NEIL2 are constant throughout the cell cycle (147) but are still responsive to oxidative stress (170), perhaps reflecting the distinct roles of these repair factors in replication and transcription. Both NEIL1 and NEIL2 have been found in mitochondria as well as in the nucleus (171,172). NEIL2 has been identified in association with microtubules but the relevance of this interaction is unknown (173). As with NTHL1, NEIL2 activity can be stimulated by the scaffold protein XRCC1 (102). Intriguingly, NEIL1 transcript is subject to RNA editing by the adenosine deaminase ADAR1, which results in a K→R change in the lesion recognition site. The edited form is less efficient at removing thymine glycol, but more efficient at removing 8-oxoguanine oxidation products (174). However, the biological role of this editing is not yet clear.
8-Oxoguanine–DNA glycosylases
The 8-oxoguanine–DNA glycosylase family (human: OGG1; S. cerevisiae: Ogg1) is responsible for excising G oxidation products with intact ring systems including the extremely common 8-oxoguanine and additional modifications, specifically across from C (175–188), and G-derived formamidopyrimidine (176,181,186,188). This family of enzymes possesses an AP β-lyase activity specifically across from a C (63,175). Some weak δ-elimination has also been detected (180) and Ogg1 has a minor dRPase activity, perhaps due to δ-elimination coupled with the Tpp1 3′-phosphatase (52). OGG1 and Ogg1 lyase activity is fairly inefficient compared to their glycosylase activities (182) but OGG1 can be stimulated 5-fold in the presence of APEX1 (183,189,190). OGG1 lyase activity can also be replaced by NEIL1/PNK, which binds abasic sites more strongly than OGG1 (191).
OGG1 is expressed as multiple isoforms resulting from alternative splicing. All isoforms contain a mitochondrial matrix targeting signal (MTS), but only isoform 1a contains a strong nuclear localization signal (NLS) (192). Isoform 1a is primarily localized to nuclei and to the nuclear matrix but has also been detected in small amounts in mitochondria (193,194). All the other isoforms, which differ in the C-terminus, are primarily localized to mitochondria, with isoform 2a forming the majority of the mitochondrial pool, associating with the inner membrane (193,195). As with NEIL2, OGG1 associates with microtubules (196), but the function of this interaction has not been elucidated.
OGG1 expression can be upregulated by treatment with alkylating agents and antioxidants, but not by ROS (190,197,198). OGG1 activity may be stimulated by ribosomal protein S3, which may bring OGG1 and APEX1 to lesions; however, S3 can also bind 8-oxoguanine lesions strongly enough to prevent excision (199,200), which might suggest a role in the nucleolus. OGG1 can also be post-translationally modified. PKC phosphorylates OGG1, though the function of this modification is unknown (194), while ROS-inducible p300-mediated acetylation weakens abasic site binding, thus enhancing APEX1-induced OGG1 activity (201).
Uracil–DNA glycosylases
The uracil–DNA glycosylase superfamily is one of the most highly conserved and diverse families of BESIR enzymes (202). While S. cerevisiae only has one (Ung1), mammals have four (UNG, SMUG1, TDG, MBD4). Mammalian glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and cyclin O (CCNO) have also been reported to have uracil excision activity (203–205). These activities have not been characterized beyond these initial reports, though a 2015 study reported that while GAPDH binds strongly to abasic sites, no uracil excision activity was observed (206). Note that there was some initial confusion in the literature between CCNO and the nuclear UNG isoform UNG2; CCNO was originally named UDG2, which led some early studies of UNG2 to conflate the two proteins.
Uracil–DNA glycosylase (human: UNG; S. cerevisiae: Ung1) is the major enzyme responsible for removing U (resulting from C deamination or misincorporation) (207–215) and oxidized derivatives (e.g. 5-hydroxyuracil, alloxan) (216,217), acting on both double-stranded and single-stranded DNA (207,208). UNG localizes to both the nucleus and mitochondria as separate isoforms from alternate promoters (UNG1: mitochondrial; UNG2: nuclear) (218–222). In contrast, Ung1 is a single isoform that localizes to both compartments (223). UNG1 is the only known human mitochondrial uracil–DNA glycosylase. Both UNG2 and Ung1 are upregulated in S phase (218,224–227). UNG2 is controlled by both cyclin-dependent kinases (228,229) and TP53-dependent phosphatase (230,231) and associates with replication complexes (209,232–234). UNG1 is induced with oxidative stress (235). The expression of both human UNG forms are regulated by several miRNAs (236). With a relatively high KM compared to the other uracil–DNA glycosylases (237), UNG may be specialized for rapidly detecting and excising lesions encountered by the replication fork and for quickly correcting U misincorporated by DNA polymerase.
The other uracil–DNA glycosylase family members have specialized roles. SMUG1 is nuclear and recognizes the same substrates as UNG1 and UNG2 (216,238–241) in addition to U and T oxidation products (e.g. 5-hydroxymethyluracil, 5-formyluracil, alloxan) (216,239–242) with some weak activity for exocyclic adducts of C (e.g. 3,N4-ethenocytosine) (243) and deaminated purines (e.g. oxanine, xanthine) (161,244). SMUG1 overall is less efficient than UNG, but SMUG1 prefers single-stranded DNA 100-fold as compared to double-stranded DNA (238). SMUG1 also has a lower KM than UNG, which gives it an advantage at lower substrate concentrations (237). As a result, SMUG1 is more efficient at repairing rare lesions in nonreplicating chromatin (245). Additionally, SMUG1 has novel activity removing 5-hydroxymethyluracil from rRNA in the nucleolus (246), presumably to perform quality control on rRNAs while they are unfolded and most vulnerable to damage.
TDG is a nuclear mismatch-specific glycosylase which excises U and T (247–249), highly oxidized 5MeC derivatives (e.g. 5-formylcytosine, 5-carboxylcytosine) (250–252), oxidized T derivatives (e.g. 5-hydroxymethyluracil, 5-formyluracil, thymine glycol) (250,253,254), deaminated A (hypoxanthine) (250), and exocyclic C derivatives (255–258). All substrates are recognized primarily when across from G. TDG has a strong preference for a 5′-adjacent G, as is found in CpG islands (259), and has been strongly associated with both DNA methyltransferases (260,261) and transcription activation (262–266). Recently, a set of 5MeC oxidases, the TET dioxygenases, have been discovered which specifically oxidize the 5-methyl group, converting the methylated base into a substrate for TDG (16,17). Thus, TDG has been implicated in active DNA demethylation as well as reducing the mutation rate of these critical genetic control regions (267). TDG is expressed inversely with UNG, degraded when cells enter S phase and upregulated in G2 (268,269), and is translationally regulated by the miRNA miR-29 (270,271). TDG activity is controlled by p300/CBP-SIRT1 acetylation sites which reduce glycosylase activity, adjacent PKCα phosphorylation sites that block acetylation (269,272,273), and transient sumoylation which stimulates abasic site release (274,275).
MBD4 is a recently discovered nuclear mismatch-specific uracil–DNA glycosylase, which is a fusion of a 5MeC binding domain and a uracil–DNA glycosylase domain (276). MBD4 associates with methylated CpG islands (277) and excises U, T and oxidized T derivatives (e.g. 5-formyluracil, thymine glycol) (253,254,278,279) when opposite G. Many other functions have been associated with MBD4 (280). The glycosylase activity of MBD4 has not been fully characterized, but it may be involved in active demethylation similar to TDG (261,281).
MutY family glycosylases
The MutY family is a G mismatch-specific adenine–DNA glycosylase (human: MUTYH; S. cerevisiae: not present). A:G mispairs are the result of a misinsertion of A across from 8-oxoguanine. MUTYH can excise both A and oxidized A (e.g. 2-hydroxyadenine) across from either G or 8-oxoguanine (282–284). MUTYH is present in both nuclei and mitochondria (193,285) and is upregulated in S phase (286). In the nucleus, MUTYH is associated with replication complexes (286,287) so that mispairs can be corrected shortly after replication. There are multiple known isoforms of MUTYH, but their distinct roles are not known (283,285). MUTYH activity is enhanced by the mismatch repair complex MSH2/6 (288) and APEX1 (289), but it also inhibits OGG1 excision across from, and APEX1 excision of, its resulting abasic site (290). As with NTHL1 and Ntg2, MUTYH also contains an iron-sulfur center, and may be regulated by a similar charge transport mechanism (140).
AP endonucleases
AP endonucleases cleave the DNA backbone 5′ to an abasic site or 3′-dRP left behind by AP β-lyase (291–299). AP endonucleases can also recognize oxidized abasic sites (66,67,300). APEX1 and Apn1 have an additional functionality in NIR by directly incising 5′ to oxidized and alkylated pyrimidines (71–77) and they can remove a 3′-terminal lesion with weak exonuclease activity (299,301–304). APEX1 can also cleave at abasic sites within RNA playing a role in RNA quality control (305–307). However, APEX1 has many other known functions due to the redox-sensitive transcription factor domain (reviewed in (308)). Interestingly, while APEX1, APEX2, and Apn2 belong to the exonuclease III family (309), Apn1 belongs to the endonuclease IV family (310).
APEX1 and APEX2 are both upregulated in S phase (311,312) and during DNA damage (313). APEX1 activity is reduced by SIRT1-removed acetylation (314) and CK2-added phosphorylation (315). APEX1 activity is also stimulated by HSP70 binding (316,317), the RAD9-RAD1-HUS1 checkpoint clamp (318), and XRCC1 (319).
APEX1, APEX2, and Apn1 all localize to both the nucleus and mitochondria (294,320–325) while Apn2 is exclusively nuclear (295,296). APEX2 shows the most predominant mitochondrial localization of these enzymes (235). APEX1 and Apn1 make up the majority of AP endonuclease activity in both nuclei and mitochondria with the weaker activities of APEX2 and Apn2 playing minor, backup, or specialized roles (235,294,297,326).
Other end-trimming enzymes
Tyrosyl–DNA phosphodiesterase 1 (human: TDP1; S. cerevisiae: Tdp1) directly catalyzes the release of a cross-linked topoisomerase 1 from the end-group phosphate. PNKP/Tpp1 then removes the 3′-phosphate and adds a 5′-phosphate to the break site. Topoisomerase II lesions are removed by TDP2 (S. cerevisiae: not present), leaving a 5′-phosphate (68). Aprataxin deadenylase (human: APTX; S. cerevisiae: Hnt3) directly removes the 5′-5′ AMP left behind by an aborted ligation (69). TDP1/TDP2/Tdp1 and APTX/Hnt3 have all been found in both the nucleus and mitochondria (327). End-trimming enzymes have been reviewed in depth recently (70).
DNA polymerases
In humans, X-family DNA polymerases are responsible for the replacement of the excised nucleotides in the nucleus, primarily polymerase β (POLB) and secondarily polymerase λ (POLL). These polymerases may be tightly coupled to APEX1 processing of abasic sites by forming a complex with the XRCC1 scaffold protein (328). Both human polymerases β and λ were reviewed in detail recently (329,330). While S. cerevisiae has a POLB homolog, Pol4, it has not been associated with BESIR; Pol4 seems to be involved with gap-filling in double-strand break repair, an activity also associated with polymerase λ (331). Instead, yeast rely on the major replicative polymerase, polymerase δ (Pol3/Pol31/Pol32), to complete repair (332,333). Both human and yeast mitochondrial BESIR appear to utilize the mitochondrial DNA polymerase γ to complete repair of the mitochondrial genome as no X-family polymerases have been identified with mitochondrial localization (334).
DNA ligases
Base excision repair is completed by the action of a DNA ligase. DNA ligase I (human: LIG1, S. cerevisiae: CDC9) is critical for the completion of nuclear BESIR; in yeast this enzyme plays a more general role, localizing to both nuclei and mitochondria (335–337). The activity of human LIG1 is complemented by DNA ligase III (LIG3), which localizes to both nuclei and mitochondria and specializes in SN-BESIR (338). Both LIG1 and LIG3 in humans associate with the XRCC1 scaffold protein (335). The role of ligases in DNA repair has been reviewed in depth recently (339,340).
Endonuclease V-mediated excision repair
Recently, a unique repair pathway initiated by endonuclease V (human: ENDOV; S. cerevisiae: not present) was discovered (341) (Figure 2E). ENDOV recognizes exocyclic-deaminated purines and cuts the DNA backbone 3′ to the nucleotide immediately 3′ to the lesion, preferentially in single-stranded regions. Through an unknown combination of nucleases, a short single-strand gap is created in the region of the nick, which is filled by DNA polymerase and sealed by DNA ligase. The discovery and current knowledge regarding this enzyme family is reviewed in (342).
Nucleotide excision repair
The nucleotide excision repair (NER) pathway corrects bulky chemical base adducts and intrastrand crosslinks such as those produced by UV irradiation which induce helical distortion (Figure 2D). NER can partially compensate for BESIR loss (343), and proteins involved in NER are implicated in the repair of certain BESIR substrates (344–348). NER is active in the nucleus and there is no evidence of mitochondrial NER activity. As there are dozens of copies of the small mitochondrial genome, it may be more efficient to degrade severely damaged mitochondrial DNA molecules and resynthesize new ones than to repair them (reviewed in (349)). NER can be induced generally (global genome repair, GGR) or in response to a stalled RNA polymerase (transcription-coupled repair, TCR). In brief, both pathways involve the dual incision of the strand around the damage, leaving a 24–32 nucleotide gap which is filled by a DNA polymerase and sealed with a ligase (350–353).
OPEN QUESTIONS AND CHALLENGES
The biochemical mechanisms of eukaryotic DNA base damage repair have been elegantly defined by the work of Tomas Lindahl, Aziz Sancar, Paul Modrich and all those who followed. However, much remains to be elucidated regarding the in vivo regulation of the DNA repair pathways, and the interconnections between them and to the core metabolic pathways of the cell. One critical question is how DNA repair pathways could be regulated by partitioning between the nucleus, cytosol, mitochondria, and plastids; recent studies in S. cerevisiae have revealed shifts in localization depending on genotoxic stress conditions termed dynamic compartmentalization (133,134). Another critical issue is to understand how lesions are directed into the multiple, redundant pathways available to them; for example, the choice of incision vs. excision in BESIR, endonuclease or lyase processing of abasic sites, or between single-nucleotide and long-patch BESIR. In addition, novel pathways initiated by endonuclease V and RNase H2 will require focused study to determine how they fit in with the established DNA repair landscape.
A major challenge lies in understanding how cells signal the presence of unrepaired base damage, though some pieces of the puzzle are starting to be identified. Recent work has demonstrated that reactive oxygen species are generated in response to DNA base damage in S. cerevisiae (354), but the biochemical pathway leading to generation of ROS has not yet been defined. Human OGG1 can allosterically recognize free 8-oxoguanine, one of its major reaction products, which counterintuitively leads to activating cellular signaling pathways including those mediated by the Ras, Rac and Rho small GTPases (355–357). Poly(ADP-ribose) is generated at single-strand breaks, including those produced during BESIR, and may protect the break and recruit repair proteins (358). Several recent studies have shown that an NER-generated single-strand gap can be expanded by exonuclease 1, producing an extended gap that activates the ataxia telangiectasia and Rad3-related (ATR) signaling pathway, connecting NER-repairable lesions to the DNA damage cell cycle checkpoints (359–362). The ability of NER-repairable lesions to activate ATR raises the intriguing possibility that other unrepaired base lesions could activate ATR signaling and checkpoint activation either through NER or through an excision intermediate processed by exonuclease 1. Gaining a clear understanding of these putative signaling pathways and their downstream effectors will be an important advance in elucidating the overall regulation of DNA repair.
Along the same lines as base damage signaling pathways, we have an only rudimentary understanding of how base damage repair is regulated at the level of gene expression and/or post-translational modifications. In a number of cases, transcriptional regulation that occurs in response to genomic insult has been documented but whether additional regulatory mechanisms are critical is not known (363). In only a few specific cases has the functional importance of post-translational modifications of DNA repair proteins been defined (364). Research into these questions will be critical to fully understand how cells respond to and deal with potentially deleterious DNA damage.
Another important issue is to determine how DNA repair pathway regulation and deficiencies are related to human health. A few common inherited polymorphisms have been described, including OGG1 S326C (365), which may have subtle impacts on human health. Other rare inherited variants have been reported in NTHL1 and MUTYH with strong associations to colorectal cancer (34,36). Variations in the abundance and localization of BESIR proteins have also been reported in a number of cancers (366–368). The impact of many of these variations on oncogenesis and patient prognosis is not yet clear. Recently, the relationship of metals to inhibition of BESIR enzymes including the NIEL glycosylases has been a topic of investigation, especially as this inhibition may interact with heavy metal exposures and neurological diseases characterized by metal accumulation (369).
A major challenge to studying DNA damage and repair is the lack of precision tools and endpoints. The majority of our current in vivo knowledge of DNA repair pathways relies on genetic manipulations, genotoxic agents, and mutagenic and phenotypic readouts. Genetic manipulations and genotoxic agents shift lesion abundances in broad, nonspecific ways and their effects are scattered randomly throughout the genome. Mutagenic and phenotypic readouts are several levels removed from the lesions, introducing many opportunities for confounding. Ideally, we would want to introduce a defined lesion at specific loci and then be able to observe how those lesions are processed and resolved. Some recently developed tools and methodologies provide steps in this direction. Micro-irradiation is a promising approach, relying on targeted sensitizers and lasers to provide more localized induction of DNA damage (370). One such tool for the induction of targeted oxidative stress is the KillerRed fluorescent protein derivative, which generates singlet oxygen radicals in response to green light (371). KillerRed has been used to target specific genomic regions with oxidative damage allowing the subsequent response to be detected and analyzed (372). Approaches to measure lesions in a more quantitative and specific manner have also been developed. One method releases lesions from genomic samples using glycosylases and then analyzes their frequencies with chromatography/isotope-dilution tandem mass spectrometry (373). Another method called Excision-seq allows the mapping of classes of lesions by releasing damaged bases with a glycosylase from a genomic sample and subjecting the products to massively parallel sequencing (374); other genome-scale techniques have been reviewed in (375). Techniques to analyze the localization of DNA repair proteins have also been developed as localization of DNA repair proteins has emerged as a potentially important level of regulation. One such method is Q-SCAn, which relies on fluorescent marker proteins to quantify the distribution of proteins among subcellular compartments (376). While all of these novel techniques still have their limitations, advances such as these will be important to continue dissecting the details of DNA base damage and repair.
The 2015 Nobel Prize in Chemistry was awarded in recognition of the seminal work that defined the biochemical mechanisms underlying the critical DNA repair pathways. This work spawned a vitally important field of study, which has greatly improved our knowledge of the details and impacts of DNA base damage and repair. However, there are still major challenges in understanding how these pathways are regulated and integrated with one another to ensure genomic integrity.
Footnotes
Present address: Nicholas C. Bauer, Department of Chemistry and Chemical Biology, Harvard University, Cambridge, MA 02138, USA.
FUNDING
National Institutes of Health [P01ES011163 to P.W.D., F31CA168272 to N.C.B.]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Funding for open access charge: National Institutes of Health grants Emory Library resources.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bjelland S., Seeberg E. Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutat. Res. 2003;531:37–80. doi: 10.1016/j.mrfmmm.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Morreall J.F., Petrova L., Doetsch P.W. Transcriptional mutagenesis and its potential roles in the etiology of cancer and bacterial antibiotic resistance. J. Cell. Physiol. 2013;228:2257–2261. doi: 10.1002/jcp.24400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 4.Brown E.J., Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 5.Vermeij W.P., Hoeijmakers J.H.J., Pothof J. Aging: not all DNA damage is equal. Curr. Opin. Genet. Dev. 2014;26:124–130. doi: 10.1016/j.gde.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Madabhushi R., Pan L., Tsai L.-H. DNA damage and its links to neurodegeneration. Neuron. 2014;83:266–282. doi: 10.1016/j.neuron.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malik Q., Herbert K.E. Oxidative and non-oxidative DNA damage and cardiovascular disease. Free Radical Res. 2012;46:554–564. doi: 10.3109/10715762.2012.663913. [DOI] [PubMed] [Google Scholar]
- 8.Lindahl T. My journey to DNA repair. Genomics Proteomics Bioinformatics. 2013;11:2–7. doi: 10.1016/j.gpb.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schieber M., Chandel N.S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014;24:R453-R462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klaunig J.E., Kamendulis L.M., Hocevar B.A. Oxidative stress and oxidative damage in carcinogenesis. Toxicol. Pathol. 2010;38:96–109. doi: 10.1177/0192623309356453. [DOI] [PubMed] [Google Scholar]
- 11.Cadet J., Mouret S., Ravanat J.L., Douki T. Photoinduced damage to cellular DNA: direct and photosensitized reactions. Photochem. Photobiol. 2012;88:1048–1065. doi: 10.1111/j.1751-1097.2012.01200.x. [DOI] [PubMed] [Google Scholar]
- 12.Dizdaroglu M., Jaruga P. Mechanisms of free radical–induced damage to DNA. Free Radical Res. 2012;46:382–419. doi: 10.3109/10715762.2011.653969. [DOI] [PubMed] [Google Scholar]
- 13.Boorstein R.J., Hilbert T.P., Cunningham R.P., Teebor G.W. Formation and stability of repairable pyrimidine photohydrates in DNA. Biochemistry. 1990;29:10455–10460. doi: 10.1021/bi00498a004. [DOI] [PubMed] [Google Scholar]
- 14.Labet V., Morell C., Douki T., Cadet J., Eriksson L.A., Grand A. Hydrolytic deamination of 5,6-dihydrocytosine in a protic medium: a theoretical study. J. Phys. Chem. A. 2010;114:1826–1834. doi: 10.1021/jp9049044. [DOI] [PubMed] [Google Scholar]
- 15.Tremblay S., Wagner J.R. Dehydration, deamination and enzymatic repair of cytosine glycols from oxidized poly(dG-dC) and poly(dI-dC) Nucleic Acids Res. 2008;36:284–293. doi: 10.1093/nar/gkm1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cortellino S., Xu J., Sannai M., Moore R., Caretti E., Cigliano A., Le Coz M., Devarajan K., Wessels A., Soprano D., et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cadet J., Wagner J.R. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jena N.R., Mishra P.C. Formation of ring-opened and rearranged products of guanine: mechanisms and biological significance. Free Radical Biol. Med. 2012;53:81–94. doi: 10.1016/j.freeradbiomed.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 20.Randerath K., Reddy R., Danna T.F., Watson W.P., Crane A.E., Randerath E. Formation of ribonucleotides in DNA modified by oxidative damage in vitro and in vivo. Characterization by 32P-postlabeling. Mutat. Res. 1992;275:355–366. doi: 10.1016/0921-8734(92)90038-q. [DOI] [PubMed] [Google Scholar]
- 21.Admiraal S.J., O'Brien P.J. DNA-N-glycosylases process novel O-glycosidic sites in DNA. Biochemistry. 2013;52:4066–4074. doi: 10.1021/bi400218j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yonekura S., Nakamura N., Yonei S., Zhang-Akiyama Q.M. Generation, biological consequences and repair mechanisms of cytosine deamination in DNA. J. Radiat. Res. 2009;50:19–26. doi: 10.1269/jrr.08080. [DOI] [PubMed] [Google Scholar]
- 23.Temiz N.A., Donohue D.E., Bacolla A., Vasquez K.M., Cooper D.N., Mudunuri U., Ivanic J., Cer R.Z., Yi M., Stephens R.M., et al. The somatic autosomal mutation matrix in cancer genomes. Hum. Genet. 2015;134:851–864. doi: 10.1007/s00439-015-1566-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doi A., Pack S.P., Makino K. Comparison of the molecular influences of NO-induced lesions in DNA strands on the reactivity of polynucleotide kinases, DNA ligases and DNA polymerases. J. Biochemistry. 2010;147:697–703. doi: 10.1093/jb/mvq003. [DOI] [PubMed] [Google Scholar]
- 25.Drablos F., Feyzi E., Aas P.A., Vaagbo C.B., Kavli B., Bratlie M.S., Pena-Diaz J., Otterlei M., Slupphaug G., Krokan H.E. Alkylation damage in DNA and RNA—repair mechanisms and medical significance. DNA Repair. 2004;3:1389–1407. doi: 10.1016/j.dnarep.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Sedgwick B., Bates P.A., Paik J., Jacobs S.C., Lindahl T. Repair of alkylated DNA: recent advances. DNA Repair. 2007;6:429–442. doi: 10.1016/j.dnarep.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Winczura A., Zdzalik D., Tudek B. Damage of DNA and proteins by major lipid peroxidation products in genome stability. Free Radical Res. 2012;46:442–459. doi: 10.3109/10715762.2012.658516. [DOI] [PubMed] [Google Scholar]
- 28.Choudhury S., Dyba M., Pan J., Roy R., Chung F.L. Repair kinetics of acrolein- and (E)-4-hydroxy-2-nonenal-derived DNA adducts in human colon cell extracts. Mutat. Res. 2013;751–752:15–23. doi: 10.1016/j.mrfmmm.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Billet S., Abbas I., Goff J.L., Verdin A., André V., Lafargue P.-E., Hachimi A., Cazier F., Sichel F., Shirali P., et al. Genotoxic potential of polycyclic aromatic hydrocarbons-coated onto airborne particulate matter (PM2.5) in human lung epithelial A549 cells. Cancer Lett. 2008;270:144–155. doi: 10.1016/j.canlet.2008.04.044. [DOI] [PubMed] [Google Scholar]
- 30.Øvrevik J., Arlt V.M., Øya E., Nagy E., Mollerup S., Phillips D.H., Låg M., Holme J.A. Differential effects of nitro-PAHs and amino-PAHs on cytokine and chemokine responses in human bronchial epithelial BEAS-2B cells. Toxicol. Appl. Pharmacol. 2010;242:270–280. doi: 10.1016/j.taap.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 31.Catalano M.J., Liu S., Andersen N., Yang Z., Johnson K.M., Price N.E., Wang Y., Gates K.S. Chemical structure and properties of interstrand cross-links formed by reaction of guanine residues with abasic sites in duplex DNA. J. Am. Chem. Soc. 2015;137:3933–3945. doi: 10.1021/jacs.5b00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price N.E., Johnson K.M., Wang J., Fekry M.I., Wang Y., Gates K.S. Interstrand DNA-DNA cross-link formation between adenine residues and abasic sites in duplex DNA. J. Am. Chem. Soc. 2014;136:3483–3490. doi: 10.1021/ja410969x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nick McElhinny S.A., Watts B.E., Kumar D., Watt D.L., Lundström E.-B., Burgers P.M.J., Johansson E., Chabes A., Kunkel T.A. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. USA. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weren R.D., Ligtenberg M.J., Kets C.M., de Voer R.M., Verwiel E.T., Spruijt L., van Zelst-Stams W.A., Jongmans M.C., Gilissen C., Hehir-Kwa J.Y., et al. A germline homozygous mutation in the base-excision repair gene NTHL1 causes adenomatous polyposis and colorectal cancer. Nat. Genet. 2015;47:668–671. doi: 10.1038/ng.3287. [DOI] [PubMed] [Google Scholar]
- 35.Jeppesen D.K., Bohr V.A., Stevnsner T. DNA repair deficiency in neurodegeneration. Prog. Neurobiol. 2011;94:166–200. doi: 10.1016/j.pneurobio.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brinkmeyer M.K., David S.S. Distinct functional consequences of MUTYH variants associated with colorectal cancer: damaged DNA affinity, glycosylase activity and interaction with PCNA and Hus1. DNA Repair. 2015 doi: 10.1016/j.dnarep.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swanson R.L., Morey N.J., Doetsch P.W., Jinks-Robertson S. Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:2929–2935. doi: 10.1128/mcb.19.4.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doetsch P.W., Morey N.J., Swanson R.L., Jinks-Robertson S. Yeast base excision repair: interconnections and networks. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:29–39. doi: 10.1016/s0079-6603(01)68087-5. [DOI] [PubMed] [Google Scholar]
- 39.Aas P.A., Otterlei M., Falnes P.O., Vagbo C.B., Skorpen F., Akbari M., Sundheim O., Bjoras M., Slupphaug G., Seeberg E., et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 40.Duncan T., Trewick S.C., Koivisto P., Bates P.A., Lindahl T., Sedgwick B. Reversal of DNA alkylation damage by two human dioxygenases. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16660–16665. doi: 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Falnes P.O. Repair of 3-methylthymine and 1-methylguanine lesions by bacterial and human AlkB proteins. Nucleic Acids Res. 2004;32:6260–6267. doi: 10.1093/nar/gkh964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maciejewska A.M., Poznanski J., Kaczmarska Z., Krowisz B., Nieminuszczy J., Polkowska-Nowakowska A., Grzesiuk E., Kusmierek J.T. AlkB dioxygenase preferentially repairs protonated substrates: specificity against exocyclic adducts and molecular mechanism of action. J. Biol. Chem. 2013;288:432–441. doi: 10.1074/jbc.M112.353342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ringvoll J., Moen M.N., Nordstrand L.M., Meira L.B., Pang B., Bekkelund A., Dedon P.C., Bjelland S., Samson L.D., Falnes P.O., et al. AlkB homologue 2-mediated repair of ethenoadenine lesions in mammalian DNA. Cancer Res. 2008;68:4142–4149. doi: 10.1158/0008-5472.CAN-08-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delaney J.C., Smeester L., Wong C., Frick L.E., Taghizadeh K., Wishnok J.S., Drennan C.L., Samson L.D., Essigmann J.M. AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat. Struct. Mol. Biol. 2005;12:855–860. doi: 10.1038/nsmb996. [DOI] [PubMed] [Google Scholar]
- 45.Daniels D.S., Woo T.T., Luu K.X., Noll D.M., Clarke N.D., Pegg A.E., Tainer J.A. DNA binding and nucleotide flipping by the human DNA repair protein AGT. Nat. Struct. Mol. Biol. 2004;11:714–720. doi: 10.1038/nsmb791. [DOI] [PubMed] [Google Scholar]
- 46.Yang W. Surviving the sun: repair and bypass of DNA UV lesions. Protein Sci. 2011;20:1781–1789. doi: 10.1002/pro.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gerkema M.P., Davies W.I., Foster R.G., Menaker M., Hut R.A. The nocturnal bottleneck and the evolution of activity patterns in mammals. Proc. R. Soc. Lond., Ser. B: Biol. Sci. 2013;280:20130508. doi: 10.1098/rspb.2013.0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fortini P., Pascucci B., Parlanti E., D'Errico M., Simonelli V., Dogliotti E. The base excision repair: mechanisms and its relevance for cancer susceptibility. Biochimie. 2003;85:1053–1071. doi: 10.1016/j.biochi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 49.Doetsch P.W., Cunningham R.P. The enzymology of apurinic/apyrimidinic endonucleases. Mutat. Res. 1990;236:173–201. doi: 10.1016/0921-8777(90)90004-o. [DOI] [PubMed] [Google Scholar]
- 50.Wiederhold L., Leppard J.B., Kedar P., Karimi-Busheri F., Rasouli-Nia A., Weinfeld M., Tomkinson A.E., Izumi T., Prasad R., Wilson S.H., et al. AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 51.Deshpande R.A., Wilson T.E. Identification of DNA 3′-phosphatase active site residues and their differential role in DNA binding, Mg2+ coordination, and catalysis. Biochemistry. 2004;43:8579–8589. doi: 10.1021/bi049434n. [DOI] [PubMed] [Google Scholar]
- 52.Sandigursky M., Yacoub A., Kelley M.R., Xu Y., Franklin W.A., Deutsch W.A. The yeast 8-oxoguanine DNA glycosylase (Ogg1) contains a DNA deoxyribophosphodiesterase (dRpase) activity. Nucleic Acids Res. 1997;25:4557–4561. doi: 10.1093/nar/25.22.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grin I.R., Khodyreva S.N., Nevinsky G.A., Zharkov D.O. Deoxyribophosphate lyase activity of mammalian endonuclease VIII-like proteins. FEBS Lett. 2006;580:4916–4922. doi: 10.1016/j.febslet.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 54.Dianov G.L., Souza-Pinto N., Nyaga S.G., Thybo T., Stevnsner T., Bohr V.A. Base excision repair in nuclear and mitochondrial DNA. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:285–297. doi: 10.1016/s0079-6603(01)68107-8. [DOI] [PubMed] [Google Scholar]
- 55.Hanna M., Chow B.L., Morey N.J., Jinks-Robertson S., Doetsch P.W., Xiao W. Involvement of two endonuclease III homologs in the base excision repair pathway for the processing of DNA alkylation damage in Saccharomyces cerevisiae. DNA Repair. 2004;3:51–59. doi: 10.1016/j.dnarep.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 56.Podlutsky A.J., Dianova II, Wilson S.H., Bohr V.A., Dianov G.L. DNA synthesis and dRPase activities of polymerase β are both essential for single-nucleotide patch base excision repair in mammalian cell extracts. Biochemistry. 2001;40:809–813. doi: 10.1021/bi002064s. [DOI] [PubMed] [Google Scholar]
- 57.Hanssen-Bauer A., Solvang-Garten K., Akbari M., Otterlei M. X-ray repair cross complementing protein 1 in base excision repair. Int. J. Mol. Sci. 2012;13:17210–17229. doi: 10.3390/ijms131217210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sokhansanj B.A., Rodrigue G.R., Fitch J.P., Wilson D.M., 3rd A quantitative model of human DNA base excision repair. I. Mechanistic insights. Nucleic Acids Res. 2002;30:1817–1825. doi: 10.1093/nar/30.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petermann E., Keil C., Oei S.L. Roles of DNA ligase III and XRCC1 in regulating the switch between short patch and long patch BER. DNA Repair. 2006;5:544–555. doi: 10.1016/j.dnarep.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 60.Mjelle R., Hegre S.A., Aas P.A., Slupphaug G., Drablos F., Saetrom P., Krokan H.E. Cell cycle regulation of human DNA repair and chromatin remodeling genes. DNA Repair. 2015;30:53–67. doi: 10.1016/j.dnarep.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 61.Sentürker S., Auffret van der Kemp P., You H.J., Doetsch P.W., Dizdaroglu M., Boiteux S. Substrate specificities of the Ntg1 and Ntg2 proteins of Saccharomyces cerevisiae for oxidized DNA bases are not identical. Nucleic Acids Res. 1998;26:5270–5276. doi: 10.1093/nar/26.23.5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meadows K.L., Song B., Doetsch P.W. Characterization of AP lyase activities of Saccharomyces cerevisiae Ntg1p and Ntg2p: implications for biological function. Nucleic Acids Res. 2003;31:5560–5567. doi: 10.1093/nar/gkg749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fortini P., Parlanti E., Sidorkina O.M., Laval J., Dogliotti E. The type of DNA glycosylase determines the base excision repair pathway in mammalian cells. J. Biol. Chem. 1999;274:15230–15236. doi: 10.1074/jbc.274.21.15230. [DOI] [PubMed] [Google Scholar]
- 64.Yang N., Galick H., Wallace S.S. Attempted base excision repair of ionizing radiation damage in human lymphoblastoid cells produces lethal and mutagenic double strand breaks. DNA Repair. 2004;3:1323–1334. doi: 10.1016/j.dnarep.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 65.Yang N., Chaudhry M.A., Wallace S.S. Base excision repair by hNTH1 and hOGG1: a two edged sword in the processing of DNA damage in γ-irradiated human cells. DNA Repair. 2006;5:43–51. doi: 10.1016/j.dnarep.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 66.Xu Y.J., DeMott M.S., Hwang J.T., Greenberg M.M., Demple B. Action of human apurinic endonuclease (Ape1) on C1′-oxidized deoxyribose damage in DNA. DNA Repair. 2003;2:175–185. doi: 10.1016/s1568-7864(02)00194-5. [DOI] [PubMed] [Google Scholar]
- 67.Xu Y.J., Kim E.Y., Demple B. Excision of C-4′-oxidized deoxyribose lesions from double-stranded DNA by human apurinic/apyrimidinic endonuclease (Ape1 protein) and DNA polymerase β. J. Biol. Chem. 1998;273:28837–28844. doi: 10.1074/jbc.273.44.28837. [DOI] [PubMed] [Google Scholar]
- 68.Pommier Y., Huang S.-Y.N., Gao R., Das B.B., Murai J., Marchand C. Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2) DNA Repair. 2014;19:114–129. doi: 10.1016/j.dnarep.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ahel I., Rass U., El-Khamisy S.F., Katyal S., Clements P.M., McKinnon P.J., Caldecott K.W., West S.C. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–716. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- 70.Andres S.N., Schellenberg M.J., Wallace B.D., Tumbale P., Williams R.S. Recognition and repair of chemically heterogeneous structures at DNA ends. Environ. Mol. Mutag. 2015;56:1–21. doi: 10.1002/em.21892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gros L., Ishchenko A.A., Ide H., Elder R.H., Saparbaev M.K. The major human AP endonuclease (Ape1) is involved in the nucleotide incision repair pathway. Nucleic Acids Res. 2004;32:73–81. doi: 10.1093/nar/gkh165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Prorok P., Alili D., Saint-Pierre C., Gasparutto D., Zharkov D.O., Ishchenko A.A., Tudek B., Saparbaev M.K. Uracil in duplex DNA is a substrate for the nucleotide incision repair pathway in human cells. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E3695–3703. doi: 10.1073/pnas.1305624110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Prorok P., Saint-Pierre C., Gasparutto D., Fedorova O.S., Ishchenko A.A., Leh H., Buckle M., Tudek B., Saparbaev M. Highly mutagenic exocyclic DNA adducts are substrates for the human nucleotide incision repair pathway. PLoS One. 2012;7:e51776. doi: 10.1371/journal.pone.0051776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Daviet S., Couve-Privat S., Gros L., Shinozuka K., Ide H., Saparbaev M., Ishchenko A.A. Major oxidative products of cytosine are substrates for the nucleotide incision repair pathway. DNA Repair. 2007;6:8–18. doi: 10.1016/j.dnarep.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 75.Ischenko A.A., Saparbaev M.K. Alternative nucleotide incision repair pathway for oxidative DNA damage. Nature. 2002;415:183–187. doi: 10.1038/415183a. [DOI] [PubMed] [Google Scholar]
- 76.Ishchenko A.A., Ide H., Ramotar D., Nevinsky G., Saparbaev M. α-anomeric deoxynucleotides, anoxic products of ionizing radiation, are substrates for the endonuclease IV-type AP endonucleases. Biochemistry. 2004;43:15210–15216. doi: 10.1021/bi049214+. [DOI] [PubMed] [Google Scholar]
- 77.Ishchenko A.A., Sanz G., Privezentzev C.V., Maksimenko A.V., Saparbaev M. Characterisation of new substrate specificities of Escherichia coli and Saccharomyces cerevisiae AP endonucleases. Nucleic Acids Res. 2003;31:6344–6353. doi: 10.1093/nar/gkg812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sparks Justin L., Chon H., Cerritelli S.M., Kunkel T.A., Johansson E., Crouch R.J., Burgers P.M. RNase H2-initiated ribonucleotide excision repair. Mol. Cell. 2012;47:980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tannous E., Kanaya E., Kanaya S. Role of RNase H1 in DNA repair: removal of single ribonucleotide misincorporated into DNA in collaboration with RNase H2. Scientific Rep. 2015;5:9969. doi: 10.1038/srep09969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vaisman A., Woodgate R. Redundancy in ribonucleotide excision repair: competition, compensation, and cooperation. DNA Repair. 2015;29:74–82. doi: 10.1016/j.dnarep.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chakravarti D., Ibeanu G.C., Tano K., Mitra S. Cloning and expression in Escherichia coli of a human cDNA encoding the DNA repair protein N-methylpurine-DNA glycosylase. J. Biol. Chem. 1991;266:15710–15715. [PubMed] [Google Scholar]
- 82.Engelward B.P., Boosalis M.S., Chen B.J., Deng Z., Siciliano M.J., Samson L.D. Cloning and characterization of a mouse 3-methyladenine/7-methylguanine/3-methylguanine DNA glycosylase cDNA whose gene maps to chromosome 11. Carcinogenesis. 1993;14:175–181. doi: 10.1093/carcin/14.2.175. [DOI] [PubMed] [Google Scholar]
- 83.Roy R., Brooks C., Mitra S. Purification and biochemical characterization of recombinant N-methylpurine-DNA glycosylase of the mouse. Biochemistry. 1994;33:15131–15140. doi: 10.1021/bi00254a024. [DOI] [PubMed] [Google Scholar]
- 84.Xiao W., Penugonde V., Rank G.H. The MAG1* 3-methyladenine DNA glycosylase gene is closely linked to the SPT15 TATA-binding TFIID gene on chromosome V-R in Saccharomyces cerevisiae. Yeast. 1994;10:687–691. doi: 10.1002/yea.320100513. [DOI] [PubMed] [Google Scholar]
- 85.Watanabe S., Ichimura T., Fujita N., Tsuruzoe S., Ohki I., Shirakawa M., Kawasuji M., Nakao M. Methylated DNA-binding domain 1 and methylpurine-DNA glycosylase link transcriptional repression and DNA repair in chromatin. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12859–12864. doi: 10.1073/pnas.2131819100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee C.Y., Delaney J.C., Kartalou M., Lingaraju G.M., Maor-Shoshani A., Essigmann J.M., Samson L.D. Recognition and processing of a new repertoire of DNA substrates by human 3-methyladenine DNA glycosylase (AAG) Biochemistry. 2009;48:1850–1861. doi: 10.1021/bi8018898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Saparbaev M., Kleibl K., Laval J. Escherichia coli, Saccharomyces cerevisiae, rat and human 3-methyladenine DNA glycosylases repair 1,N6-ethenoadenine when present in DNA. Nucleic Acids Res. 1995;23:3750–3755. doi: 10.1093/nar/23.18.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Choudhury S., Adhikari S., Cheema A., Roy R. Evidence of complete cellular repair of 1,N6-ethenoadenine, a mutagenic and potential damage for human cancer, revealed by a novel method. Mol. Cell. Biochem. 2008;313:19–28. doi: 10.1007/s11010-008-9737-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miao F., Bouziane M., O'Connor T.R. Interaction of the recombinant human methylpurine-DNA glycosylase (MPG protein) with oligodeoxyribonucleotides containing either hypoxanthine or abasic sites. Nucleic Acids Res. 1998;26:4034–4041. doi: 10.1093/nar/26.17.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hitchcock T.M., Dong L., Connor E.E., Meira L.B., Samson L.D., Wyatt M.D., Cao W. Oxanine DNA glycosylase activity from mammalian alkyladenine glycosylase. J. Biol. Chem. 2004;279:38177–38183. doi: 10.1074/jbc.M405882200. [DOI] [PubMed] [Google Scholar]
- 91.Dherin C., Gasparutto D., O'Connor T.R., Cadet J., Boiteux S. Excision by the human methylpurine DNA N-glycosylase of cyanuric acid, a stable and mutagenic oxidation product of 8-oxo-7,8-dihydroguanine. Int. J. Radiat. Biol. 2004;80:21–27. doi: 10.1080/09553000310001632976. [DOI] [PubMed] [Google Scholar]
- 92.Admiraal S.J., O'Brien P.J. N-glycosyl bond formation catalyzed by human alkyladenine DNA glycosylase. Biochemistry. 2010;49:9024–9026. doi: 10.1021/bi101380d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu Y., Dai H., Xiao W. UAS(MAG1), a yeast cis-acting element that regulates the expression of MAG1, is located within the protein coding region of DDI1. Mol. Gen. Genet. 1997;255:533–542. doi: 10.1007/s004380050526. [DOI] [PubMed] [Google Scholar]
- 94.Liu Y., Xiao W. Bidirectional regulation of two DNA-damage–inducible genes, MAG1 and DDI1, from Saccharomyces cerevisiae. Mol. Microbiol. 1997;23:777–789. doi: 10.1046/j.1365-2958.1997.2701631.x. [DOI] [PubMed] [Google Scholar]
- 95.Zhu Y., Xiao W. Differential regulation of two closely clustered yeast genes, MAG1 and DDI1, by cell-cycle checkpoints. Nucleic Acids Res. 1998;26:5402–5408. doi: 10.1093/nar/26.23.5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhu Y., Xiao W. Two alternative cell cycle checkpoint pathways differentially control DNA damage–dependent induction of MAG1 and DDI1 expression in yeast. Mol. Genet. Genomics. 2001;266:436–444. doi: 10.1007/s004380100538. [DOI] [PubMed] [Google Scholar]
- 97.Zhu Y., Xiao W. Pdr3 is required for DNA damage induction of MAG1 and DDI1 via a bi-directional promoter element. Nucleic Acids Res. 2004;32:5066–5075. doi: 10.1093/nar/gkh838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bouziane M., Miao F., Bates S.E., Somsouk L., Sang B.C., Denissenko M., O'Connor T.R. Promoter structure and cell cycle dependent expression of the human methylpurine-DNA glycosylase gene. Mutat. Res. 2000;461:15–29. doi: 10.1016/s0921-8777(00)00036-7. [DOI] [PubMed] [Google Scholar]
- 99.van Loon B., Samson L.D. Alkyladenine DNA glycosylase (AAG) localizes to mitochondria and interacts with mitochondrial single-stranded binding protein (mtSSB) DNA Repair. 2013;12:177–187. doi: 10.1016/j.dnarep.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huh W.-K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O'Shea E.K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 101.Miao F., Bouziane M., Dammann R., Masutani C., Hanaoka F., Pfeifer G., O'Connor T.R. 3-Methyladenine-DNA glycosylase (MPG protein) interacts with human RAD23 proteins. J. Biol. Chem. 2000;275:28433–28438. doi: 10.1074/jbc.M001064200. [DOI] [PubMed] [Google Scholar]
- 102.Campalans A., Marsin S., Nakabeppu Y., O'Connor T, Boiteux S., Radicella J.P. XRCC1 interactions with multiple DNA glycosylases: a model for its recruitment to base excision repair. DNA Repair. 2005;4:826–835. doi: 10.1016/j.dnarep.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 103.Xia L., Zheng L., Lee H.W., Bates S.E., Federico L., Shen B., O'Connor T.R. Human 3-methyladenine-DNA glycosylase: effect of sequence context on excision, association with PCNA, and stimulation by AP endonuclease. J. Mol. Biol. 2005;346:1259–1274. doi: 10.1016/j.jmb.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 104.Maher R.L., Vallur A.C., Feller J.A., Bloom L.B. Slow base excision by human alkyladenine DNA glycosylase limits the rate of formation of AP sites and AP endonuclease 1 does not stimulate base excision. DNA Repair. 2007;6:71–81. doi: 10.1016/j.dnarep.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 105.Baldwin M.R., O'Brien P.J. Human AP endonuclease 1 stimulates multiple-turnover base excision by alkyladenine DNA glycosylase. Biochemistry. 2009;48:6022–6033. doi: 10.1021/bi900517y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Alseth I., Eide L., Pirovano M., Rognes T., Seeberg E., Bjoras M. The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Mol. Cell. Biol. 1999;19:3779–3787. doi: 10.1128/mcb.19.5.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dizdaroglu M., Karahalil B., Senturker S., Buckley T.J., Roldan-Arjona T. Excision of products of oxidative DNA base damage by human NTH1 protein. Biochemistry. 1999;38:243–246. doi: 10.1021/bi9819071. [DOI] [PubMed] [Google Scholar]
- 108.Eide L., Bjoras M., Pirovano M., Alseth I., Berdal K.G., Seeberg E. Base excision of oxidative purine and pyrimidine DNA damage in Saccharomyces cerevisiae by a DNA glycosylase with sequence similarity to endonuclease III from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1996;93:10735–10740. doi: 10.1073/pnas.93.20.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Eide L., Luna L., Gustad E.C., Henderson P.T., Essigmann J.M., Demple B., Seeberg E. Human endonuclease III acts preferentially on DNA damage opposite guanine residues in DNA. Biochemistry. 2001;40:6653–6659. doi: 10.1021/bi0028901. [DOI] [PubMed] [Google Scholar]
- 110.Gasparutto D., Ait-Abbas M., Jaquinod M., Boiteux S., Cadet J. Repair and coding properties of 5-hydroxy-5-methylhydantoin nucleosides inserted into DNA oligomers. Chem. Res. Toxicol. 2000;13:575–584. doi: 10.1021/tx000005+. [DOI] [PubMed] [Google Scholar]
- 111.Gasparutto D., Muller E., Boiteux S., Cadet J. Excision of the oxidatively formed 5-hydroxyhydantoin and 5-hydroxy-5-methylhydantoin pyrimidine lesions by Escherichia coli and Saccharomyces cerevisiae DNA N-glycosylases. Biochim. Biophys. Acta. 2009;1790:16–24. doi: 10.1016/j.bbagen.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 112.Marenstein D.R., Chan M.K., Altamirano A., Basu A.K., Boorstein R.J., Cunningham R.P., Teebor G.W. Substrate specificity of human endonuclease III (hNTH1). Effect of human APE1 on hNTH1 activity. J. Biol. Chem. 2003;278:9005–9012. doi: 10.1074/jbc.M212168200. [DOI] [PubMed] [Google Scholar]
- 113.Miyabe I., Zhang Q.M., Kino K., Sugiyama H., Takao M., Yasui A., Yonei S. Identification of 5-formyluracil DNA glycosylase activity of human hNTH1 protein. Nucleic Acids Res. 2002;30:3443–3448. doi: 10.1093/nar/gkf460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.You H.J., Swanson R.L., Harrington C., Corbett A.H., Jinks-Robertson S., Sentürker S., Wallace S.S., Boiteux S., Dizdaroglu M., Doetsch P.W. Saccharomyces cerevisiae Ntg1p and Ntg2p: broad specificity N-glycosylases for the repair of oxidative DNA damage in the nucleus and mitochondria. Biochemistry. 1999;38:11298–11306. doi: 10.1021/bi991121i. [DOI] [PubMed] [Google Scholar]
- 115.Redrejo-Rodriguez M., Saint-Pierre C., Couve S., Mazouzi A., Ishchenko A.A., Gasparutto D., Saparbaev M. New insights in the removal of the hydantoins, oxidation product of pyrimidines, via the base excision and nucleotide incision repair pathways. PLoS One. 2011;6:e21039. doi: 10.1371/journal.pone.0021039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang Q.M., Hashiguchi K., Kino K., Sugiyama H., Yonei S. Ntg1 and Ntg2 proteins as 5-formyluracil-DNA glycosylases/AP lyases in Saccharomyces cerevisiae. Int. J. Radiat. Biol. 2003;79:341–349. doi: 10.1080/0955300032000093119. [DOI] [PubMed] [Google Scholar]
- 117.Kino K., Takao M., Miyazawa H., Hanaoka F. A DNA oligomer containing 2,2,4-triamino-5(2H)-oxazolone is incised by human NEIL1 and NTH1. Mutat. Res. 2012;734:73–77. doi: 10.1016/j.mrfmmm.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 118.Luna L., Bjoras M., Hoff E., Rognes T., Seeberg E. Cell-cycle regulation, intracellular sorting and induced overexpression of the human NTH1 DNA glycosylase involved in removal of formamidopyrimidine residues from DNA. Mutat. Res. 2000;460:95–104. doi: 10.1016/s0921-8777(00)00015-x. [DOI] [PubMed] [Google Scholar]
- 119.Krokeide S.Z., Laerdahl J.K., Salah M., Luna L., Cederkvist F.H., Fleming A.M., Burrows C.J., Dalhus B., Bjoras M. Human NEIL3 is mainly a monofunctional DNA glycosylase removing spiroimindiohydantoin and guanidinohydantoin. DNA Repair. 2013;12:1159–1164. doi: 10.1016/j.dnarep.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bruner S.D., Nash H.M., Lane W.S., Verdine G.L. Repair of oxidatively damaged guanine in Saccharomyces cerevisiae by an alternative pathway. Curr. Biol. 1998;8:393–403. doi: 10.1016/s0960-9822(98)70158-7. [DOI] [PubMed] [Google Scholar]
- 121.Matsumoto Y., Zhang Q.M., Takao M., Yasui A., Yonei S. Escherichia coli Nth and human hNTH1 DNA glycosylases are involved in removal of 8-oxoguanine from 8-oxoguanine/guanine mispairs in DNA. Nucleic Acids Res. 2001;29:1975–1981. doi: 10.1093/nar/29.9.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kim J.E., You H.J., Choi J.Y., Doetsch P.W., Kim J.S., Chung M.H. Ntg2 of Saccharomyces cerevisiae repairs the oxidation products of 8-hydroxyguanine. Biochem. Biophys. Res. Commun. 2001;285:1186–1191. doi: 10.1006/bbrc.2001.5305. [DOI] [PubMed] [Google Scholar]
- 123.Boiteux S., Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair. 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 124.Guillet M., Boiteux S. Origin of endogenous DNA abasic sites in Saccharomyces cerevisiae. Mol. Cell. Biol. 2003;23:8386–8394. doi: 10.1128/MCB.23.22.8386-8394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Klungland A., Hoss M., Gunz D., Constantinou A., Clarkson S.G., Doetsch P.W., Bolton P.H., Wood R.D., Lindahl T. Base excision repair of oxidative DNA damage activated by XPG protein. Mol. Cell. 1999;3:33–42. doi: 10.1016/s1097-2765(00)80172-0. [DOI] [PubMed] [Google Scholar]
- 126.Liu X., Roy R. Truncation of amino-terminal tail stimulates activity of human endonuclease III (hNTH1) J. Mol. Biol. 2002;321:265–276. doi: 10.1016/s0022-2836(02)00623-x. [DOI] [PubMed] [Google Scholar]
- 127.Prasad R., Shock D.D., Beard W.A., Wilson S.H. Substrate channeling in mammalian base excision repair pathways: passing the baton. J. Biol. Chem. 2010;285:40479–40488. doi: 10.1074/jbc.M110.155267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Oyama M., Wakasugi M., Hama T., Hashidume H., Iwakami Y., Imai R., Hoshino S., Morioka H., Ishigaki Y., Nikaido O., et al. Human NTH1 physically interacts with p53 and proliferating cell nuclear antigen. Biochem. Biophys. Res. Commun. 2004;321:183–191. doi: 10.1016/j.bbrc.2004.06.136. [DOI] [PubMed] [Google Scholar]
- 129.Wolfe K.H., Shields D.C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 130.Kellis M., Birren B.W., Lander E.S. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 131.Karahalil B., Bohr V.A., De Souza-Pinto N.C. Base excision repair activities differ in human lung cancer cells and corresponding normal controls. Anticancer Res. 2010;30:4963–4971. [PMC free article] [PubMed] [Google Scholar]
- 132.Doudican N.A., Song B., Shadel G.S., Doetsch P.W. Oxidative DNA damage causes mitochondrial genomic instability in Saccharomyces cerevisiae. Mol. Cell. Biol. 2005;25:5196–5204. doi: 10.1128/MCB.25.12.5196-5204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Griffiths L.M., Swartzlander D., Meadows K.L., Wilkinson K.D., Corbett A.H., Doetsch P.W. Dynamic compartmentalization of base excision repair proteins in response to nuclear and mitochondrial oxidative stress. Mol. Cell. Biol. 2009;29:794–807. doi: 10.1128/MCB.01357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Swartzlander D.B., Griffiths L.M., Lee J., Degtyareva N.P., Doetsch P.W., Corbett A.H. Regulation of base excision repair: Ntg1 nuclear and mitochondrial dynamic localization in response to genotoxic stress. Nucleic Acids Res. 2010;38:3963–3974. doi: 10.1093/nar/gkq108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ikeda S., Kohmoto T., Tabata R., Seki Y. Differential intracellular localization of the human and mouse endonuclease III homologs and analysis of the sorting signals. DNA Repair. 2002;1:847–854. doi: 10.1016/s1568-7864(02)00145-3. [DOI] [PubMed] [Google Scholar]
- 136.O'Rourke T.W., Doudican N.A., Mackereth M.D., Doetsch P.W., Shadel G.S. Mitochondrial dysfunction due to oxidative mitochondrial DNA damage is reduced through cooperative actions of diverse proteins. Mol. Cell. Biol. 2002;22:4086–4093. doi: 10.1128/MCB.22.12.4086-4093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ikeda S., Biswas T., Roy R., Izumi T., Boldogh I., Kurosky A., Sarker A.H., Seki S., Mitra S. Purification and characterization of human NTH1, a homolog of Escherichia coli endonuclease III. Direct identification of Lys-212 as the active nucleophilic residue. J. Biol. Chem. 1998;273:21585–21593. doi: 10.1074/jbc.273.34.21585. [DOI] [PubMed] [Google Scholar]
- 138.Grodick M.A., Segal H.M., Zwang T.J., Barton J.K. DNA-mediated signaling by proteins with 4Fe-4S clusters is necessary for genomic integrity. J. Am. Chem. Soc. 2014;136:6470–6478. doi: 10.1021/ja501973c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Boal A.K., Yavin E., Barton J.K. DNA repair glycosylases with a [4Fe-4S] cluster: a redox cofactor for DNA-mediated charge transport. J. Inorg. Biochem. 2007;101:1913–1921. doi: 10.1016/j.jinorgbio.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lin J.C., Singh R.R., Cox D.L. Theoretical study of DNA damage recognition via electron transfer from the [4Fe-4S] complex of MutY. Biophys. J. 2008;95:3259–3268. doi: 10.1529/biophysj.108.132183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Romano C.A., Sontz P.A., Barton J.K. Mutants of the base excision repair glycosylase, endonuclease III: DNA charge transport as a first step in lesion detection. Biochemistry. 2011;50:6133–6145. doi: 10.1021/bi2003179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yoshitani A., Yoshida M., Ling F. A novel cis-acting element required for DNA damage-inducible expression of yeast DIN7. Biochem. Biophys. Res. Commun. 2008;365:183–188. doi: 10.1016/j.bbrc.2007.10.177. [DOI] [PubMed] [Google Scholar]
- 143.Dou H., Mitra S., Hazra T.K. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J. Biol. Chem. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 144.Grin I.R., Dianov G.L., Zharkov D.O. The role of mammalian NEIL1 protein in the repair of 8-oxo-7,8-dihydroadenine in DNA. FEBS Lett. 2010;584:1553–1557. doi: 10.1016/j.febslet.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hailer M.K., Slade P.G., Martin B.D., Rosenquist T.A., Sugden K.D. Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair. 2005;4:41–50. doi: 10.1016/j.dnarep.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 146.Hazra T.K., Izumi T., Boldogh I., Imhoff B., Kow Y.W., Jaruga P., Dizdaroglu M., Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hazra T.K., Kow Y.W., Hatahet Z., Imhoff B., Boldogh I., Mokkapati S.K., Mitra S., Izumi T. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J. Biol. Chem. 2002;277:30417–30420. doi: 10.1074/jbc.C200355200. [DOI] [PubMed] [Google Scholar]
- 148.Jaruga P., Birincioglu M., Rosenquist T.A., Dizdaroglu M. Mouse NEIL1 protein is specific for excision of 2,6-diamino-4-hydroxy-5-formamidopyrimidine and 4,6-diamino-5-formamidopyrimidine from oxidatively damaged DNA. Biochemistry. 2004;43:15909–15914. doi: 10.1021/bi048162l. [DOI] [PubMed] [Google Scholar]
- 149.Jia L., Shafirovich V., Geacintov N.E., Broyde S. Lesion specificity in the base excision repair enzyme hNeil1: modeling and dynamics studies. Biochemistry. 2007;46:5305–5314. doi: 10.1021/bi062269m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Katafuchi A., Nakano T., Masaoka A., Terato H., Iwai S., Hanaoka F., Ide H. Differential specificity of human and Escherichia coli endonuclease III and VIII homologues for oxidative base lesions. J. Biol. Chem. 2004;279:14464–14471. doi: 10.1074/jbc.M400393200. [DOI] [PubMed] [Google Scholar]
- 151.Krishnamurthy N., Zhao X., Burrows C.J., David S.S. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry. 2008;47:7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.McKibbin P.L., Fleming A.M., Towheed M.A., Van Houten B., Burrows C.J., David S.S. Repair of hydantoin lesions and their amine adducts in DNA by base and nucleotide excision repair. J. Am. Chem. Soc. 2013;135:13851–13861. doi: 10.1021/ja4059469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Takao M., Kanno S., Kobayashi K., Zhang Q.M., Yonei S., van der Horst G.T., Yasui A. A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J. Biol. Chem. 2002;277:42205–42213. doi: 10.1074/jbc.M206884200. [DOI] [PubMed] [Google Scholar]
- 154.Zhang Q.M., Yonekura S., Takao M., Yasui A., Sugiyama H., Yonei S. DNA glycosylase activities for thymine residues oxidized in the methyl group are functions of the hNEIL1 and hNTH1 enzymes in human cells. DNA Repair. 2005;4:71–79. doi: 10.1016/j.dnarep.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 155.Zhao X., Krishnamurthy N., Burrows C.J., David S.S. Mutation versus repair: NEIL1 removal of hydantoin lesions in single-stranded, bulge, bubble, and duplex DNA contexts. Biochemistry. 2010;49:1658–1666. doi: 10.1021/bi901852q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Morland I., Rolseth V., Luna L., Rognes T., Bjoras M., Seeberg E. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002;30:4926–4936. doi: 10.1093/nar/gkf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhou J., Liu M., Fleming A.M., Burrows C.J., Wallace S.S. Neil3 and NEIL1 DNA glycosylases remove oxidative damages from quadruplex DNA and exhibit preferences for lesions in the telomeric sequence context. J. Biol. Chem. 2013;288:27263–27272. doi: 10.1074/jbc.M113.479055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhou J., Fleming A.M., Averill A.M., Burrows C.J., Wallace S.S. The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res. 2015;43:4039–4054. doi: 10.1093/nar/gkv252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dobbs T.A., Palmer P., Maniou Z., Lomax M.E., O'Neill P. Interplay of two major repair pathways in the processing of complex double-strand DNA breaks. DNA Repair. 2008;7:1372–1383. doi: 10.1016/j.dnarep.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 160.Parsons J.L., Kavli B., Slupphaug G., Dianov G.L. NEIL1 is the major DNA glycosylase that processes 5-hydroxyuracil in the proximity of a DNA single-strand break. Biochemistry. 2007;46:4158–4163. doi: 10.1021/bi0622569. [DOI] [PubMed] [Google Scholar]
- 161.Dong L., Meira L.B., Hazra T.K., Samson L.D., Cao W. Oxanine DNA glycosylase activities in mammalian systems. DNA Repair. 2008;7:128–134. doi: 10.1016/j.dnarep.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Couve-Privat S., Mace G., Rosselli F., Saparbaev M.K. Psoralen-induced DNA adducts are substrates for the base excision repair pathway in human cells. Nucleic Acids Res. 2007;35:5672–5682. doi: 10.1093/nar/gkm592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hegde M.L., Theriot C.A., Das A., Hegde P.M., Guo Z., Gary R.K., Hazra T.K., Shen B., Mitra S. Physical and functional interaction between human oxidized base-specific DNA glycosylase NEIL1 and flap endonuclease 1. J. Biol. Chem. 2008;283:27028–27037. doi: 10.1074/jbc.M802712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dou H., Theriot C.A., Das A., Hegde M.L., Matsumoto Y., Boldogh I., Hazra T.K., Bhakat K.K., Mitra S. Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen. The potential for replication-associated repair of oxidized bases in mammalian genomes. J. Biol. Chem. 2008;283:3130–3140. doi: 10.1074/jbc.M709186200. [DOI] [PubMed] [Google Scholar]
- 165.Hegde M.L., Hegde P.M., Bellot L.J., Mandal S.M., Hazra T.K., Li G.M., Boldogh I., Tomkinson A.E., Mitra S. Prereplicative repair of oxidized bases in the human genome is mediated by NEIL1 DNA glycosylase together with replication proteins. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E3090–3099. doi: 10.1073/pnas.1304231110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Banerjee D., Mandal S.M., Das A., Hegde M.L., Das S., Bhakat K.K., Boldogh I., Sarkar P.S., Mitra S., Hazra T.K. Preferential repair of oxidized base damage in the transcribed genes of mammalian cells. J. Biol. Chem. 2011;286:6006–6016. doi: 10.1074/jbc.M110.198796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hegde M.L., Banerjee S., Hegde P.M., Bellot L.J., Hazra T.K., Boldogh I., Mitra S. Enhancement of NEIL1 protein-initiated oxidized DNA base excision repair by heterogeneous nuclear ribonucleoprotein U (hnRNP-U) via direct interaction. J. Biol. Chem. 2012;287:34202–34211. doi: 10.1074/jbc.M112.384032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Muftuoglu M., de Souza-Pinto N.C., Dogan A., Aamann M., Stevnsner T., Rybanska I., Kirkali G., Dizdaroglu M., Bohr V.A. Cockayne syndrome group B protein stimulates repair of formamidopyrimidines by NEIL1 DNA glycosylase. J. Biol. Chem. 2009;284:9270–9279. doi: 10.1074/jbc.M807006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Das A., Hazra T.K., Boldogh I., Mitra S., Bhakat K.K. Induction of the human oxidized base-specific DNA glycosylase NEIL1 by reactive oxygen species. J. Biol. Chem. 2005;280:35272–35280. doi: 10.1074/jbc.M505526200. [DOI] [PubMed] [Google Scholar]
- 170.Kinslow C.J., El-Zein R.A., Rondelli C.M., Hill C.E., Wickliffe J.K., Abdel-Rahman S.Z. Regulatory regions responsive to oxidative stress in the promoter of the human DNA glycosylase gene NEIL2. Mutagenesis. 2010;25:171–177. doi: 10.1093/mutage/gep058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hu J., de Souza-Pinto N.C., Haraguchi K., Hogue B.A., Jaruga P., Greenberg M.M., Dizdaroglu M., Bohr V.A. Repair of formamidopyrimidines in DNA involves different glycosylases: role of the OGG1, NTH1, and NEIL1 enzymes. J. Biol. Chem. 2005;280:40544–40551. doi: 10.1074/jbc.M508772200. [DOI] [PubMed] [Google Scholar]
- 172.Mandal S.M., Hegde M.L., Chatterjee A., Hegde P.M., Szczesny B., Banerjee D., Boldogh I., Gao R., Falkenberg M., Gustafsson C.M., et al. Role of human DNA glycosylase Nei-like 2 (NEIL2) and single strand break repair protein polynucleotide kinase 3′-phosphatase in maintenance of mitochondrial genome. J. Biol. Chem. 2012;287:2819–2829. doi: 10.1074/jbc.M111.272179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Conlon K.A., Miller H., Rosenquist T.A., Zharkov D.O., Berrios M. The murine DNA glycosylase NEIL2 (mNEIL2) and human DNA polymerase β bind microtubules in situ and in vitro. DNA Repair. 2005;4:419–431. doi: 10.1016/j.dnarep.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 174.Yeo J., Goodman R.A., Schirle N.T., David S.S., Beal P.A. RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20715–20719. doi: 10.1073/pnas.1009231107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Aburatani H., Hippo Y., Ishida T., Takashima R., Matsuba C., Kodama T., Takao M., Yasui A., Yamamoto K., Asano M. Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res. 1997;57:2151–2156. [PubMed] [Google Scholar]
- 176.Bjoras M., Luna L., Johnsen B., Hoff E., Haug T., Rognes T., Seeberg E. Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. EMBO J. 1997;16:6314–6322. doi: 10.1093/emboj/16.20.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lu R., Nash H.M., Verdine G.L. A mammalian DNA repair enzyme that excises oxidatively damaged guanines maps to a locus frequently lost in lung cancer. Curr. Biol. 1997;7:397–407. doi: 10.1016/s0960-9822(06)00187-4. [DOI] [PubMed] [Google Scholar]
- 178.Radicella J.P., Dherin C., Desmaze C., Fox M.S., Boiteux S. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Rosenquist T.A., Zharkov D.O., Grollman A.P. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc. Natl. Acad. Sci. U.S.A. 1997;94:7429–7434. doi: 10.1073/pnas.94.14.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shinmura K., Kasai H., Sasaki A., Sugimura H., Yokota J. 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) DNA glycosylase and AP lyase activities of hOGG1 protein and their substrate specificity. Mutat. Res. 1997;385:75–82. doi: 10.1016/s0921-8777(97)00041-4. [DOI] [PubMed] [Google Scholar]
- 181.Dherin C., Radicella J.P., Dizdaroglu M., Boiteux S. Excision of oxidatively damaged DNA bases by the human α-hOgg1 protein and the polymorphic α-hOgg1(Ser326Cys) protein which is frequently found in human populations. Nucleic Acids Res. 1999;27:4001–4007. doi: 10.1093/nar/27.20.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zharkov D.O., Rosenquist T.A., Gerchman S.E., Grollman A.P. Substrate specificity and reaction mechanism of murine 8-oxoguanine-DNA glycosylase. J. Biol. Chem. 2000;275:28607–28617. doi: 10.1074/jbc.M002441200. [DOI] [PubMed] [Google Scholar]
- 183.Sidorenko V.S., Nevinsky G.A., Zharkov D.O. Specificity of stimulation of human 8-oxoguanine-DNA glycosylase by AP endonuclease. Biochem. Biophys. Res. Commun. 2008;368:175–179. doi: 10.1016/j.bbrc.2008.01.076. [DOI] [PubMed] [Google Scholar]
- 184.Boldogh I., Hajas G., Aguilera-Aguirre L., Hegde M.L., Radak Z., Bacsi A., Sur S., Hazra T.K., Mitra S. Activation of ras signaling pathway by 8-oxoguanine DNA glycosylase bound to its excision product, 8-oxoguanine. J. Biol. Chem. 2012;287:20769–20773. doi: 10.1074/jbc.C112.364620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Nash H.M., Bruner S.D., Scharer O.D., Kawate T., Addona T.A., Spooner E., Lane W.S., Verdine G.L. Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr. Biol. 1996;6:968–980. doi: 10.1016/s0960-9822(02)00641-3. [DOI] [PubMed] [Google Scholar]
- 186.van der Kemp P.A., Thomas D., Barbey R., de Oliveira R., Boiteux S. Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which codes for a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine. Proc. Natl. Acad. Sci. U.S.A. 1996;93:5197–5202. doi: 10.1073/pnas.93.11.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Girard P.M., Guibourt N., Boiteux S. The Ogg1 protein of Saccharomyces cerevisiae: a 7,8-dihydro-8-oxoguanine DNA glycosylase/AP lyase whose lysine 241 is a critical residue for catalytic activity. Nucleic Acids Res. 1997;25:3204–3211. doi: 10.1093/nar/25.16.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Karahalil B., Girard P.M., Boiteux S., Dizdaroglu M. Substrate specificity of the Ogg1 protein of Saccharomyces cerevisiae: excision of guanine lesions produced in DNA by ionizing radiation- or hydrogen peroxide/metal ion-generated free radicals. Nucleic Acids Res. 1998;26:1228–1233. doi: 10.1093/nar/26.5.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hill J.W., Hazra T.K., Izumi T., Mitra S. Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res. 2001;29:430–438. doi: 10.1093/nar/29.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Saitoh T., Shinmura K., Yamaguchi S., Tani M., Seki S., Murakami H., Nojima Y., Yokota J. Enhancement of OGG1 protein AP lyase activity by increase of APEX protein. Mutat. Res. 2001;486:31–40. doi: 10.1016/s0921-8777(01)00078-7. [DOI] [PubMed] [Google Scholar]
- 191.Mokkapati S.K., Wiederhold L., Hazra T.K., Mitra S. Stimulation of DNA glycosylase activity of OGG1 by NEIL1: functional collaboration between two human DNA glycosylases. Biochemistry. 2004;43:11596–11604. doi: 10.1021/bi049097i. [DOI] [PubMed] [Google Scholar]
- 192.Monden Y., Arai T., Asano M., Ohtsuka E., Aburatani H., Nishimura S. Human MMH (OGG1) type 1a protein is a major enzyme for repair of 8-hydroxyguanine lesions in human cells. Biochem. Biophys. Res. Commun. 1999;258:605–610. doi: 10.1006/bbrc.1999.0649. [DOI] [PubMed] [Google Scholar]
- 193.Takao M., Aburatani H., Kobayashi K., Yasui A. Mitochondrial targeting of human DNA glycosylases for repair of oxidative DNA damage. Nucleic Acids Res. 1998;26:2917–2922. doi: 10.1093/nar/26.12.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Dantzer F., Luna L., Bjoras M., Seeberg E. Human OGG1 undergoes serine phosphorylation and associates with the nuclear matrix and mitotic chromatin in vivo. Nucleic Acids Res. 2002;30:2349–2357. doi: 10.1093/nar/30.11.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Nishioka K., Ohtsubo T., Oda H., Fujiwara T., Kang D., Sugimachi K., Nakabeppu Y. Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Mol. Biol. Cell. 1999;10:1637–1652. doi: 10.1091/mbc.10.5.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Conlon K.A., Zharkov D.O., Berrios M. Cell cycle regulation of the murine 8-oxoguanine DNA glycosylase (mOGG1): mOGG1 associates with microtubules during interphase and mitosis. DNA Repair. 2004;3:1601–1615. doi: 10.1016/j.dnarep.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 197.Lee M.R., Kim S.H., Cho H.J., Lee K.Y., Moon A.R., Jeong H.G., Lee J.S., Hyun J.W., Chung M.H., You H.J. Transcription factors NF-YA regulate the induction of human OGG1 following DNA-alkylating agent methylmethane sulfonate (MMS) treatment. J. Biol. Chem. 2004;279:9857–9866. doi: 10.1074/jbc.M311132200. [DOI] [PubMed] [Google Scholar]
- 198.Singh B., Chatterjee A., Ronghe A.M., Bhat N.K., Bhat H.K. Antioxidant-mediated up-regulation of OGG1 via NRF2 induction is associated with inhibition of oxidative DNA damage in estrogen-induced breast cancer. BMC Cancer. 2013;13:253. doi: 10.1186/1471-2407-13-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hegde V., Wang M., Deutsch W.A. Human ribosomal protein S3 interacts with DNA base excision repair proteins hAPE/Ref-1 and hOGG1. Biochemistry. 2004;43:14211–14217. doi: 10.1021/bi049234b. [DOI] [PubMed] [Google Scholar]
- 200.Hegde V., Wang M., Deutsch W.A. Characterization of human ribosomal protein S3 binding to 7,8-dihydro-8-oxoguanine and abasic sites by surface plasmon resonance. DNA Repair. 2004;3:121–126. doi: 10.1016/j.dnarep.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 201.Bhakat K.K., Mokkapati S.K., Boldogh I., Hazra T.K., Mitra S. Acetylation of human 8-oxoguanine-DNA glycosylase by p300 and its role in 8-oxoguanine repair in vivo. Mol. Cell. Biol. 2006;26:1654–1665. doi: 10.1128/MCB.26.5.1654-1665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lucas-Lledo J.I., Maddamsetti R., Lynch M. Phylogenomic analysis of the uracil-DNA glycosylase superfamily. Mol. Biol. Evol. 2011;28:1307–1317. doi: 10.1093/molbev/msq318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Meyer-Siegler K., Mauro D.J., Seal G., Wurzer J., deRiel J.K., Sirover M.A. A human nuclear uracil DNA glycosylase is the 37-kDa subunit of glyceraldehyde-3-phosphate dehydrogenase. Proc. Natl. Acad. Sci. USA. 1991;88:8460–8464. doi: 10.1073/pnas.88.19.8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Muller S.J., Caradonna S. Cell cycle regulation of a human cyclin-like gene encoding uracil-DNA glycosylase. J. Biol. Chem. 1993;268:1310–1319. [PubMed] [Google Scholar]
- 205.Muller S.J., Caradonna S. Isolation and characterization of a human cDNA encoding uracil-DNA glycosylase. Biochim. Biophys. Acta. 1991;1088:197–207. doi: 10.1016/0167-4781(91)90055-q. [DOI] [PubMed] [Google Scholar]
- 206.Kosova A.A., Khodyreva S.N., Lavrik O.I. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) interacts with apurinic/apyrimidinic sites in DNA. Mutat. Res.-Fundam. Mol. Mech. Mutag. 2015;779:46–57. doi: 10.1016/j.mrfmmm.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 207.Domena J.D., Mosbaugh D.W. Purification of nuclear and mitochondrial uracil-DNA glycosylase from rat liver. Identification of two distinct subcellular forms. Biochemistry. 1985;24:7320–7328. doi: 10.1021/bi00346a045. [DOI] [PubMed] [Google Scholar]
- 208.Domena J.D., Timmer R.T., Dicharry S.A., Mosbaugh D.W. Purification and properties of mitochondrial uracil-DNA glycosylase from rat liver. Biochemistry. 1988;27:6742–6751. doi: 10.1021/bi00418a015. [DOI] [PubMed] [Google Scholar]
- 209.Krokan H. Preferential association of uracil-DNA glycosylase activity with replicating SV40 minichromosomes. FEBS Lett. 1981;133:89–91. doi: 10.1016/0014-5793(81)80477-2. [DOI] [PubMed] [Google Scholar]
- 210.Otterlei M., Warbrick E., Nagelhus T.A., Haug T., Slupphaug G., Akbari M., Aas P.A., Steinsbekk K., Bakke O., Krokan H.E. Post-replicative base excision repair in replication foci. EMBO J. 1999;18:3834–3844. doi: 10.1093/emboj/18.13.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Slupphaug G., Eftedal I., Kavli B., Bharati S., Helle N.M., Haug T., Levine D.W., Krokan H.E. Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UNG encodes the major uracil-DNA glycosylase. Biochemistry. 1995;34:128–138. doi: 10.1021/bi00001a016. [DOI] [PubMed] [Google Scholar]
- 212.Impellizzeri K.J., Anderson B., Burgers P.M. The spectrum of spontaneous mutations in a Saccharomyces cerevisiae uracil-DNA-glycosylase mutant limits the function of this enzyme to cytosine deamination repair. J. Bacteriol. 1991;173:6807–6810. doi: 10.1128/jb.173.21.6807-6810.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Percival K.J., Klein M.B., Burgers P.M. Molecular cloning and primary structure of the uracil-DNA-glycosylase gene from Saccharomyces cerevisiae. J. Biol. Chem. 1989;264:2593–2598. [PubMed] [Google Scholar]
- 214.Burgers P.M., Klein M.B. Selection by genetic transformation of a Saccharomyces cerevisiae mutant defective for the nuclear uracil-DNA-glycosylase. J. Bacteriol. 1986;166:905–913. doi: 10.1128/jb.166.3.905-913.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Crosby B., Prakash L., Davis H., Hinkle D.C. Purification and characterization of a uracil-DNA glycosylase from the yeast, Saccharomyces cerevisiae. Nucleic Acids Res. 1981;9:5797–5809. doi: 10.1093/nar/9.21.5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.An Q., Robins P., Lindahl T., Barnes D.E. C –> T mutagenesis and γ-radiation sensitivity due to deficiency in the Smug1 and Ung DNA glycosylases. EMBO J. 2005;24:2205–2213. doi: 10.1038/sj.emboj.7600689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Dizdaroglu M., Karakaya A., Jaruga P., Slupphaug G., Krokan H.E. Novel activities of human uracil DNA N-glycosylase for cytosine-derived products of oxidative DNA damage. Nucleic Acids Res. 1996;24:418–422. doi: 10.1093/nar/24.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Haug T., Skorpen F., Aas P.A., Malm V., Skjelbred C., Krokan H.E. Regulation of expression of nuclear and mitochondrial forms of human uracil-DNA glycosylase. Nucleic Acids Res. 1998;26:1449–1457. doi: 10.1093/nar/26.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Nilsen H., Otterlei M., Haug T., Solum K., Nagelhus T.A., Skorpen F., Krokan H.E. Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res. 1997;25:750–755. doi: 10.1093/nar/25.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Otterlei M., Haug T., Nagelhus T.A., Slupphaug G., Lindmo T., Krokan H.E. Nuclear and mitochondrial splice forms of human uracil-DNA glycosylase contain a complex nuclear localisation signal and a strong classical mitochondrial localisation signal, respectively. Nucleic Acids Res. 1998;26:4611–4617. doi: 10.1093/nar/26.20.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Slupphaug G., Markussen F.H., Olsen L.C., Aasland R., Aarsaether N., Bakke O., Krokan H.E., Helland D.E. Nuclear and mitochondrial forms of human uracil-DNA glycosylase are encoded by the same gene. Nucleic Acids Res. 1993;21:2579–2584. doi: 10.1093/nar/21.11.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wittwer C.U., Krokan H. Uracil-DNA glycosylase in HeLa S3 cells: interconvertibility of 50 and 20 kDa forms and similarity of the nuclear and mitochondrial form of the enzyme. Biochim. Biophys. Acta. 1985;832:308–318. doi: 10.1016/0167-4838(85)90264-x. [DOI] [PubMed] [Google Scholar]
- 223.Chatterjee A., Singh K.K. Uracil-DNA glycosylase–deficient yeast exhibit a mitochondrial mutator phenotype. Nucleic Acids Res. 2001;29:4935–4940. doi: 10.1093/nar/29.24.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Gupta P.K., Sirover M.A. Stimulation of the nuclear uracil DNA glycosylase in proliferating human fibroblasts. Cancer Res. 1981;41:3133–3136. [PubMed] [Google Scholar]
- 225.Sirover M.A. Induction of the DNA repair enzyme uracil-DNA glycosylase in stimulated human lymphocytes. Cancer Res. 1979;39:2090–2095. [PubMed] [Google Scholar]
- 226.Slupphaug G., Olsen L.C., Helland D., Aasland R., Krokan H.E. Cell cycle regulation and in vitro hybrid arrest analysis of the major human uracil-DNA glycosylase. Nucleic Acids Res. 1991;19:5131–5137. doi: 10.1093/nar/19.19.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Johnston L.H., Johnson A.L. The DNA repair genes RAD54 and UNG1 are cell cycle regulated in budding yeast but MCB promoter elements have no essential role in the DNA damage response. Nucleic Acids Res. 1995;23:2147–2152. doi: 10.1093/nar/23.12.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Hagen L., Kavli B., Sousa M.M., Torseth K., Liabakk N.B., Sundheim O., Pena-Diaz J., Otterlei M., Horning O., Jensen O.N., et al. Cell cycle-specific UNG2 phosphorylations regulate protein turnover, activity and association with RPA. EMBO J. 2008;27:51–61. doi: 10.1038/sj.emboj.7601958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Muller-Weeks S., Mastran B., Caradonna S. The nuclear isoform of the highly conserved human uracil-DNA glycosylase is an Mr 36,000 phosphoprotein. J. Biol. Chem. 1998;273:21909–21917. doi: 10.1074/jbc.273.34.21909. [DOI] [PubMed] [Google Scholar]
- 230.Lu X., Nguyen T.A., Appella E., Donehower L.A. Homeostatic regulation of base excision repair by a p53-induced phosphatase: linking stress response pathways with DNA repair proteins. Cell Cycle. 2004;3:1363–1366. doi: 10.4161/cc.3.11.1241. [DOI] [PubMed] [Google Scholar]
- 231.Yamaguchi H., Minopoli G., Demidov O.N., Chatterjee D.K., Anderson C.W., Durell S.R., Appella E. Substrate specificity of the human protein phosphatase 2Cδ, Wip1. Biochemistry. 2005;44:5285–5294. doi: 10.1021/bi0476634. [DOI] [PubMed] [Google Scholar]
- 232.Akbari M., Otterlei M., Pena-Diaz J., Aas P.A., Kavli B., Liabakk N.B., Hagen L., Imai K., Durandy A., Slupphaug G., et al. Repair of U/G and U/A in DNA by UNG2-associated repair complexes takes place predominantly by short-patch repair both in proliferating and growth-arrested cells. Nucleic Acids Res. 2004;32:5486–5498. doi: 10.1093/nar/gkh872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ko R., Bennett S.E. Physical and functional interaction of human nuclear uracil-DNA glycosylase with proliferating cell nuclear antigen. DNA Repair. 2005;4:1421–1431. doi: 10.1016/j.dnarep.2005.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Parlanti E., Locatelli G., Maga G., Dogliotti E. Human base excision repair complex is physically associated to DNA replication and cell cycle regulatory proteins. Nucleic Acids Res. 2007;35:1569–1577. doi: 10.1093/nar/gkl1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Akbari M., Otterlei M., Pena-Diaz J., Krokan H.E. Different organization of base excision repair of uracil in DNA in nuclei and mitochondria and selective upregulation of mitochondrial uracil-DNA glycosylase after oxidative stress. Neuroscience. 2007;145:1201–1212. doi: 10.1016/j.neuroscience.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 236.Hegre S.A., Saetrom P., Aas P.A., Pettersen H.S., Otterlei M., Krokan H.E. Multiple microRNAs may regulate the DNA repair enzyme uracil-DNA glycosylase. DNA Repair. 2013;12:80–86. doi: 10.1016/j.dnarep.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 237.Nilsen H., Haushalter K.A., Robins P., Barnes D.E., Verdine G.L., Lindahl T. Excision of deaminated cytosine from the vertebrate genome: role of the SMUG1 uracil-DNA glycosylase. EMBO J. 2001;20:4278–4286. doi: 10.1093/emboj/20.15.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Haushalter K.A., Todd Stukenberg M.W., Kirschner M.W., Verdine G.L. Identification of a new uracil-DNA glycosylase family by expression cloning using synthetic inhibitors. Curr. Biol. 1999;9:174–185. doi: 10.1016/s0960-9822(99)80087-6. [DOI] [PubMed] [Google Scholar]
- 239.Masaoka A., Matsubara M., Hasegawa R., Tanaka T., Kurisu S., Terato H., Ohyama Y., Karino N., Matsuda A., Ide H. Mammalian 5-formyluracil-DNA glycosylase. 2. Role of SMUG1 uracil-DNA glycosylase in repair of 5-formyluracil and other oxidized and deaminated base lesions. Biochemistry. 2003;42:5003–5012. doi: 10.1021/bi0273213. [DOI] [PubMed] [Google Scholar]
- 240.Matsubara M., Masaoka A., Tanaka T., Miyano T., Kato N., Terato H., Ohyama Y., Iwai S., Ide H. Mammalian 5-formyluracil-DNA glycosylase. 1. Identification and characterization of a novel activity that releases 5-formyluracil from DNA. Biochemistry. 2003;42:4993–5002. doi: 10.1021/bi027322v. [DOI] [PubMed] [Google Scholar]
- 241.Matsubara M., Masaoka A., Tanaka T., Terato H., Ohyama Y., Ide H. Identification and characterization of mammalian 5-formyluracil-DNA glycosylase. Nucleic Acids Res. Suppl. 2003:233–234. doi: 10.1093/nass/3.1.233. [DOI] [PubMed] [Google Scholar]
- 242.Boorstein R.J., Cummings A., Jr, Marenstein D.R., Chan M.K., Ma Y., Neubert T.A., Brown S.M., Teebor G.W. Definitive identification of mammalian 5-hydroxymethyluracil DNA N-glycosylase activity as SMUG1. J. Biol. Chem. 2001;276:41991–41997. doi: 10.1074/jbc.M106953200. [DOI] [PubMed] [Google Scholar]
- 243.Krokan H.E., Drablos F., Slupphaug G. Uracil in DNA – occurrence, consequences and repair. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 244.Mi R., Dong L., Kaulgud T., Hackett K.W., Dominy B.N., Cao W. Insights from xanthine and uracil DNA glycosylase activities of bacterial and human SMUG1: switching SMUG1 to UDG. J. Mol. Biol. 2009;385:761–778. doi: 10.1016/j.jmb.2008.09.038. [DOI] [PubMed] [Google Scholar]
- 245.Pettersen H.S., Sundheim O., Gilljam K.M., Slupphaug G., Krokan H.E., Kavli B. Uracil-DNA glycosylases SMUG1 and UNG2 coordinate the initial steps of base excision repair by distinct mechanisms. Nucleic Acids Res. 2007;35:3879–3892. doi: 10.1093/nar/gkm372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Jobert L., Skjeldam H.K., Dalhus B., Galashevskaya A., Vagbo C.B., Bjoras M., Nilsen H. The human base excision repair enzyme SMUG1 directly interacts with DKC1 and contributes to RNA quality control. Mol. Cell. 2013;49:339–345. doi: 10.1016/j.molcel.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 247.Gallinari P., Jiricny J. A new class of uracil-DNA glycosylases related to human thymine-DNA glycosylase. Nature. 1996;383:735–738. doi: 10.1038/383735a0. [DOI] [PubMed] [Google Scholar]
- 248.Neddermann P., Jiricny J. The purification of a mismatch-specific thymine-DNA glycosylase from HeLa cells. J. Biol. Chem. 1993;268:21218–21224. [PubMed] [Google Scholar]
- 249.Wiebauer K., Jiricny J. Mismatch-specific thymine DNA glycosylase and DNA polymerase β mediate the correction of G.T mispairs in nuclear extracts from human cells. Proc. Natl. Acad. Sci. U.S.A. 1990;87:5842–5845. doi: 10.1073/pnas.87.15.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Hardeland U., Bentele M., Jiricny J., Schar P. The versatile thymine DNA-glycosylase: a comparative characterization of the human, Drosophila and fission yeast orthologs. Nucleic Acids Res. 2003;31:2261–2271. doi: 10.1093/nar/gkg344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Maiti A., Drohat A.C. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Zhang L., Lu X., Lu J., Liang H., Dai Q., Xu G.L., Luo C., Jiang H., He C. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat. Chem. Biol. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Liu P., Burdzy A., Sowers L.C. Repair of the mutagenic DNA oxidation product, 5-formyluracil. DNA Repair. 2003;2:199–210. doi: 10.1016/s1568-7864(02)00198-2. [DOI] [PubMed] [Google Scholar]
- 254.Yoon J.H., Iwai S., O'Connor T.R., Pfeifer G.P. Human thymine DNA glycosylase (TDG) and methyl-CpG-binding protein 4 (MBD4) excise thymine glycol (Tg) from a Tg:G mispair. Nucleic Acids Res. 2003;31:5399–5404. doi: 10.1093/nar/gkg730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Borys-Brzywczy E., Arczewska K.D., Saparbaev M., Hardeland U., Schar P., Kusmierek J.T. Mismatch dependent uracil/thymine-DNA glycosylases excise exocyclic hydroxyethano and hydroxypropano cytosine adducts. Acta Biochim. Pol. 2005;52:149–165. [PubMed] [Google Scholar]
- 256.Hang B., Guliaev A.B. Substrate specificity of human thymine-DNA glycosylase on exocyclic cytosine adducts. Chem.-Biol. Interact. 2007;165:230–238. doi: 10.1016/j.cbi.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 257.Jurado J., Maciejewska A., Krwawicz J., Laval J., Saparbaev M.K. Role of mismatch-specific uracil-DNA glycosylase in repair of 3,N4-ethenocytosine in vivo. DNA Repair. 2004;3:1579–1590. doi: 10.1016/j.dnarep.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 258.Sagi J., Perry A., Hang B., Singer B. Differential destabilization of the DNA oligonucleotide double helix by a T.G mismatch, 3,N4-ethenocytosine, 3,N4-ethanocytosine, or an 8-(hydroxymethyl)-3,N4-ethenocytosine adduct incorporated into the same sequence contexts. Chem. Res. Toxicol. 2000;13:839–845. doi: 10.1021/tx000040g. [DOI] [PubMed] [Google Scholar]
- 259.Lari S.U., Al-Khodairy F., Paterson M.C. Substrate specificity and sequence preference of G:T mismatch repair: incision at G:T, O6-methylguanine:T, and G:U mispairs in DNA by human cell extracts. Biochemistry. 2002;41:9248–9255. doi: 10.1021/bi020239n. [DOI] [PubMed] [Google Scholar]
- 260.Li Y.Q., Zhou P.Z., Zheng X.D., Walsh C.P., Xu G.L. Association of Dnmt3a and thymine DNA glycosylase links DNA methylation with base-excision repair. Nucleic Acids Res. 2007;35:390–400. doi: 10.1093/nar/gkl1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Boland M.J., Christman J.K. Characterization of Dnmt3b:thymine-DNA glycosylase interaction and stimulation of thymine glycosylase-mediated repair by DNA methyltransferase(s) and RNA. J. Mol. Biol. 2008;379:492–504. doi: 10.1016/j.jmb.2008.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Chen D., Lucey M.J., Phoenix F., Lopez-Garcia J., Hart S.M., Losson R., Buluwela L., Coombes R.C., Chambon P., Schar P., et al. T:G mismatch-specific thymine-DNA glycosylase potentiates transcription of estrogen-regulated genes through direct interaction with estrogen receptor α. J. Biol. Chem. 2003;278:38586–38592. doi: 10.1074/jbc.M304286200. [DOI] [PubMed] [Google Scholar]
- 263.Chiang S., Burch T., Van Domselaar G., Dick K., Radziwon A., Brusnyk C., Edwards M.R., Piper J., Cutts T., Cao J., et al. The interaction between thymine DNA glycosylase and nuclear receptor coactivator 3 is required for the transcriptional activation of nuclear hormone receptors. Mol. Cell. Biochem. 2010;333:221–232. doi: 10.1007/s11010-009-0223-1. [DOI] [PubMed] [Google Scholar]
- 264.Leger H., Smet-Nocca C., Attmane-Elakeb A., Morley-Fletcher S., Benecke A.G., Eilebrecht S. A TDG/CBP/RARα ternary complex mediates the retinoic acid-dependent expression of DNA methylation-sensitive genes. Genomics Proteomics Bioinformatics. 2014;12:8–18. doi: 10.1016/j.gpb.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Lucey M.J., Chen D., Lopez-Garcia J., Hart S.M., Phoenix F., Al-Jehani R., Alao J.P., White R., Kindle K.B., Losson R., et al. T:G mismatch-specific thymine-DNA glycosylase (TDG) as a coregulator of transcription interacts with SRC1 family members through a novel tyrosine repeat motif. Nucleic Acids Res. 2005;33:6393–6404. doi: 10.1093/nar/gki940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Um S., Harbers M., Benecke A., Pierrat B., Losson R., Chambon P. Retinoic acid receptors interact physically and functionally with the T:G mismatch-specific thymine-DNA glycosylase. J. Biol. Chem. 1998;273:20728–20736. doi: 10.1074/jbc.273.33.20728. [DOI] [PubMed] [Google Scholar]
- 267.Kohli R.M., Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Hardeland U., Kunz C., Focke F., Szadkowski M., Schar P. Cell cycle regulation as a mechanism for functional separation of the apparently redundant uracil DNA glycosylases TDG and UNG2. Nucleic Acids Res. 2007;35:3859–3867. doi: 10.1093/nar/gkm337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Madabushi A., Hwang B.J., Jin J., Lu A.L. Histone deacetylase SIRT1 modulates and deacetylates DNA base excision repair enzyme thymine DNA glycosylase. Biochem. J. 2013;456:89–98. doi: 10.1042/BJ20130670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Morita S., Horii T., Kimura M., Ochiya T., Tajima S., Hatada I. miR-29 represses the activities of DNA methyltransferases and DNA demethylases. Int. J. Mol. Sci. 2013;14:14647–14658. doi: 10.3390/ijms140714647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Zhang P., Huang B., Xu X., Sessa W.C. Ten-eleven translocation (Tet) and thymine DNA glycosylase (TDG), components of the demethylation pathway, are direct targets of miRNA-29a. Biochem. Biophys. Res. Commun. 2013;437:368–373. doi: 10.1016/j.bbrc.2013.06.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Mohan R.D., Litchfield D.W., Torchia J., Tini M. Opposing regulatory roles of phosphorylation and acetylation in DNA mispair processing by thymine DNA glycosylase. Nucleic Acids Res. 2010;38:1135–1148. doi: 10.1093/nar/gkp1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Tini M., Benecke A., Um S.J., Torchia J., Evans R.M., Chambon P. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol. Cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- 274.Hardeland U., Steinacher R., Jiricny J., Schar P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 2002;21:1456–1464. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Smet-Nocca C., Wieruszeski J.M., Leger H., Eilebrecht S., Benecke A. SUMO-1 regulates the conformational dynamics of thymine-DNA glycosylase regulatory domain and competes with its DNA binding activity. BMC Biochem. 2011;12:4. doi: 10.1186/1471-2091-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Owen R.M., Baker R.D., Bader S., Dunlop M.G., Nicholl I.D. The identification of a novel alternatively spliced form of the MBD4 DNA glycosylase. Oncol. Rep. 2007;17:111–116. [PubMed] [Google Scholar]
- 277.Kondo E., Gu Z., Horii A., Fukushige S. The thymine DNA glycosylase MBD4 represses transcription and is associated with methylated p16(INK4a) and hMLH1 genes. Mol. Cell. Biol. 2005;25:4388–4396. doi: 10.1128/MCB.25.11.4388-4396.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Bellacosa A., Cicchillitti L., Schepis F., Riccio A., Yeung A.T., Matsumoto Y., Golemis E.A., Genuardi M., Neri G. MED1, a novel human methyl-CpG-binding endonuclease, interacts with DNA mismatch repair protein MLH1. Proc. Natl. Acad. Sci. U.S.A. 1999;96:3969–3974. doi: 10.1073/pnas.96.7.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Wu P., Qiu C., Sohail A., Zhang X., Bhagwat A.S., Cheng X. Mismatch repair in methylated DNA. Structure and activity of the mismatch-specific thymine glycosylase domain of methyl-CpG-binding protein MBD4. J. Biol. Chem. 2003;278:5285–5291. doi: 10.1074/jbc.M210884200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Sjolund A.B., Senejani A.G., Sweasy J.B. MBD4 and TDG: multifaceted DNA glycosylases with ever expanding biological roles. Mutat. Res. 2013;743–744:12–25. doi: 10.1016/j.mrfmmm.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Laget S., Miotto B., Chin H.G., Esteve P.O., Roberts R.J., Pradhan S., Defossez P.A. MBD4 cooperates with DNMT1 to mediate methyl-DNA repression and protects mammalian cells from oxidative stress. Epigenetics. 2014;9:546–556. doi: 10.4161/epi.27695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.McGoldrick J.P., Yeh Y.C., Solomon M., Essigmann J.M., Lu A.L. Characterization of a mammalian homolog of the Escherichia coli MutY mismatch repair protein. Mol. Cell. Biol. 1995;15:989–996. doi: 10.1128/mcb.15.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Ohtsubo T., Nishioka K., Imaiso Y., Iwai S., Shimokawa H., Oda H., Fujiwara T., Nakabeppu Y. Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic Acids Res. 2000;28:1355–1364. doi: 10.1093/nar/28.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Parker A., Gu Y., Lu A.L. Purification and characterization of a mammalian homolog of Escherichia coli MutY mismatch repair protein from calf liver mitochondria. Nucleic Acids Res. 2000;28:3206–3215. doi: 10.1093/nar/28.17.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Ichinoe A., Behmanesh M., Tominaga Y., Ushijima Y., Hirano S., Sakai Y., Tsuchimoto D., Sakumi K., Wake N., Nakabeppu Y. Identification and characterization of two forms of mouse MUTYH proteins encoded by alternatively spliced transcripts. Nucleic Acids Res. 2004;32:477–487. doi: 10.1093/nar/gkh214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Boldogh I., Milligan D., Lee M.S., Bassett H., Lloyd R.S., McCullough A.K. hMYH cell cycle-dependent expression, subcellular localization and association with replication foci: evidence suggesting replication-coupled repair of adenine:8-oxoguanine mispairs. Nucleic Acids Res. 2001;29:2802–2809. doi: 10.1093/nar/29.13.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Hayashi H., Tominaga Y., Hirano S., McKenna A.E., Nakabeppu Y., Matsumoto Y. Replication-associated repair of adenine:8-oxoguanine mispairs by MYH. Curr. Biol. 2002;12:335–339. doi: 10.1016/s0960-9822(02)00686-3. [DOI] [PubMed] [Google Scholar]
- 288.Gu Y., Parker A., Wilson T.M., Bai H., Chang D.Y., Lu A.L. Human MutY homolog, a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins human MutS homolog 2/human MutS homolog 6. J. Biol. Chem. 2002;277:11135–11142. doi: 10.1074/jbc.M108618200. [DOI] [PubMed] [Google Scholar]
- 289.Yang H., Clendenin W.M., Wong D., Demple B., Slupska M.M., Chiang J.H., Miller J.H. Enhanced activity of adenine-DNA glycosylase (Myh) by apurinic/apyrimidinic endonuclease (Ape1) in mammalian base excision repair of an A/GO mismatch. Nucleic Acids Res. 2001;29:743–752. doi: 10.1093/nar/29.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Tominaga Y., Ushijima Y., Tsuchimoto D., Mishima M., Shirakawa M., Hirano S., Sakumi K., Nakabeppu Y. MUTYH prevents OGG1 or APEX1 from inappropriately processing its substrate or reaction product with its C-terminal domain. Nucleic Acids Res. 2004;32:3198–3211. doi: 10.1093/nar/gkh642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Izumi T., Hazra T.K., Boldogh I., Tomkinson A.E., Park M.S., Ikeda S., Mitra S. Requirement for human AP endonuclease 1 for repair of 3′-blocking damage at DNA single-strand breaks induced by reactive oxygen species. Carcinogenesis. 2000;21:1329–1334. [PubMed] [Google Scholar]
- 292.Seki S., Hatsushika M., Watanabe S., Akiyama K., Nagao K., Tsutsui K. cDNA cloning, sequencing, expression and possible domain structure of human APEX nuclease homologous to Escherichia coli exonuclease III. Biochim. Biophys. Acta. 1992;1131:287–299. doi: 10.1016/0167-4781(92)90027-w. [DOI] [PubMed] [Google Scholar]
- 293.Seki S., Ikeda S., Watanabe S., Hatsushika M., Tsutsui K., Akiyama K., Zhang B. A mouse DNA repair enzyme (APEX nuclease) having exonuclease and apurinic/apyrimidinic endonuclease activities: purification and characterization. Biochim. Biophys. Acta. 1991;1079:57–64. doi: 10.1016/0167-4838(91)90024-t. [DOI] [PubMed] [Google Scholar]
- 294.Hadi M.Z., Wilson D.M., 3rd Second human protein with homology to the Escherichia coli abasic endonuclease exonuclease III. Environ. Mol. Mutag. 2000;36:312–324. [PubMed] [Google Scholar]
- 295.Johnson A.W., Demple B. Yeast DNA diesterase for 3′-fragments of deoxyribose: purification and physical properties of a repair enzyme for oxidative DNA damage. J. Biol. Chem. 1988;263:18009–18016. [PubMed] [Google Scholar]
- 296.Johnson A.W., Demple B. Yeast DNA 3′-repair diesterase is the major cellular apurinic/apyrimidinic endonuclease: substrate specificity and kinetics. J. Biol. Chem. 1988;263:18017–18022. [PubMed] [Google Scholar]
- 297.Bennett R.A. The Saccharomyces cerevisiae ETH1 gene, an inducible homolog of exonuclease III that provides resistance to DNA-damaging agents and limits spontaneous mutagenesis. Mol. Cell. Biol. 1999;19:1800–1809. doi: 10.1128/mcb.19.3.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Unk I., Haracska L., Johnson R.E., Prakash S., Prakash L. Apurinic endonuclease activity of yeast Apn2 protein. J. Biol. Chem. 2000;275:22427–22434. doi: 10.1074/jbc.M002845200. [DOI] [PubMed] [Google Scholar]
- 299.Unk I., Haracska L., Prakash S., Prakash L. 3′-phosphodiesterase and 3′–>5′ exonuclease activities of yeast Apn2 protein and requirement of these activities for repair of oxidative DNA damage. Mol. Cell. Biol. 2001;21:1656–1661. doi: 10.1128/MCB.21.5.1656-1661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Winters T.A., Henner W.D., Russell P.S., McCullough A., Jorgensen T.J. Removal of 3′-phosphoglycolate from DNA strand-break damage in an oligonucleotide substrate by recombinant human apurinic/apyrimidinic endonuclease 1. Nucleic Acids Res. 1994;22:1866–1873. doi: 10.1093/nar/22.10.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Parsons J.L., Dianova I.I., Dianov G.L. APE1-dependent repair of DNA single-strand breaks containing 3′-end 8-oxoguanine. Nucleic Acids Res. 2005;33:2204–2209. doi: 10.1093/nar/gki518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Burkovics P., Szukacsov V., Unk I., Haracska L. Human Ape2 protein has a 3′-5′ exonuclease activity that acts preferentially on mismatched base pairs. Nucleic Acids Res. 2006;34:2508–2515. doi: 10.1093/nar/gkl259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Ishchenko A.A., Yang X., Ramotar D., Saparbaev M. The 3′->5′ exonuclease of Apn1 provides an alternative pathway to repair 7,8-dihydro-8-oxodeoxyguanosine in Saccharomyces cerevisiae. Mol. Cell. Biol. 2005;25:6380–6390. doi: 10.1128/MCB.25.15.6380-6390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Mazouzi A., Vigouroux A., Aikeshev B., Brooks P.J., Saparbaev M.K., Morera S., Ishchenko A.A. Insight into mechanisms of 3′-5′ exonuclease activity and removal of bulky 8,5′-cyclopurine adducts by apurinic/apyrimidinic endonucleases. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E3071–3080. doi: 10.1073/pnas.1305281110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Barnes T., Kim W.C., Mantha A.K., Kim S.E., Izumi T., Mitra S., Lee C.H. Identification of Apurinic/apyrimidinic endonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA. Nucleic Acids Res. 2009;37:3946–3958. doi: 10.1093/nar/gkp275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Kim S.E., Gorrell A., Rader S.D., Lee C.H. Endoribonuclease activity of human apurinic/apyrimidinic endonuclease 1 revealed by a real-time fluorometric assay. Anal. Biochem. 2010;398:69–75. doi: 10.1016/j.ab.2009.11.024. [DOI] [PubMed] [Google Scholar]
- 307.Vascotto C., Fantini D., Romanello M., Cesaratto L., Deganuto M., Leonardi A., Radicella J.P., Kelley M.R., D'Ambrosio C., Scaloni A., et al. APE1/Ref-1 interacts with NPM1 within nucleoli and plays a role in the rRNA quality control process. Mol. Cell. Biol. 2009;29:1834–1854. doi: 10.1128/MCB.01337-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Tell G., Quadrifoglio F., Tiribelli C., Kelley M.R. The many functions of APE1/Ref-1: not only a DNA repair enzyme. Antioxidants Redox Signal. 2009;11:601–620. doi: 10.1089/ars.2008.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Johnson R.E., Torres-Ramos C.A., Izumi T., Mitra S., Prakash S., Prakash L. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 1998;12:3137–3143. doi: 10.1101/gad.12.19.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Popoff S.C., Spira A.I., Johnson A.W., Demple B. Yeast structural gene (APN1) for the major apurinic endonuclease: homology to Escherichia coli endonuclease IV. Proc. Natl. Acad. Sci. U.S.A. 1990;87:4193–4197. doi: 10.1073/pnas.87.11.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Fung H., Bennett R.A., Demple B. Key role of a downstream specificity protein 1 site in cell cycle-regulated transcription of the AP endonuclease gene APE1/APEX in NIH3T3 cells. J. Biol. Chem. 2001;276:42011–42017. doi: 10.1074/jbc.M106423200. [DOI] [PubMed] [Google Scholar]
- 312.Ide Y., Tsuchimoto D., Tominaga Y., Iwamoto Y., Nakabeppu Y. Characterization of the genomic structure and expression of the mouse Apex2 gene. Genomics. 2003;81:47–57. doi: 10.1016/s0888-7543(02)00009-5. [DOI] [PubMed] [Google Scholar]
- 313.Zaky A., Busso C., Izumi T., Chattopadhyay R., Bassiouny A., Mitra S., Bhakat K.K. Regulation of the human AP-endonuclease (APE1/Ref-1) expression by the tumor suppressor p53 in response to DNA damage. Nucleic Acids Res. 2008;36:1555–1566. doi: 10.1093/nar/gkm1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Yamamori T., DeRicco J., Naqvi A., Hoffman T.A., Mattagajasingh I., Kasuno K., Jung S.B., Kim C.S., Irani K. SIRT1 deacetylates APE1 and regulates cellular base excision repair. Nucleic Acids Res. 2010;38:832–845. doi: 10.1093/nar/gkp1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Yacoub A., Kelley M.R., Deutsch W.A. The DNA repair activity of human redox/repair protein APE/Ref-1 is inactivated by phosphorylation. Cancer Res. 1997;57:5457–5459. [PubMed] [Google Scholar]
- 316.Kenny M.K., Mendez F., Sandigursky M., Kureekattil R.P., Goldman J.D., Franklin W.A., Bases R. Heat shock protein 70 binds to human apurinic/apyrimidinic endonuclease and stimulates endonuclease activity at abasic sites. J. Biol. Chem. 2001;276:9532–9536. doi: 10.1074/jbc.M009297200. [DOI] [PubMed] [Google Scholar]
- 317.Mendez F., Sandigursky M., Kureekattil R.P., Kenny M.K., Franklin W.A., Bases R. Specific stimulation of human apurinic/apyrimidinic endonuclease by heat shock protein 70. DNA Repair. 2003;2:259–271. doi: 10.1016/s1568-7864(02)00215-x. [DOI] [PubMed] [Google Scholar]
- 318.Gembka A., Toueille M., Smirnova E., Poltz R., Ferrari E., Villani G., Hubscher U. The checkpoint clamp, Rad9-Rad1-Hus1 complex, preferentially stimulates the activity of apurinic/apyrimidinic endonuclease 1 and DNA polymerase β in long patch base excision repair. Nucleic Acids Res. 2007;35:2596–2608. doi: 10.1093/nar/gkl1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Vidal A.E., Boiteux S., Hickson I.D., Radicella J.P. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. EMBO J. 2001;20:6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Chattopadhyay R., Wiederhold L., Szczesny B., Boldogh I., Hazra T.K., Izumi T., Mitra S. Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells. Nucleic Acids Res. 2006;34:2067–2076. doi: 10.1093/nar/gkl177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Jackson E.B., Theriot C.A., Chattopadhyay R., Mitra S., Izumi T. Analysis of nuclear transport signals in the human apurinic/apyrimidinic endonuclease (APE1/Ref1) Nucleic Acids Res. 2005;33:3303–3312. doi: 10.1093/nar/gki641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Kakolyris S., Kaklamanis L., Giatromanolaki A., Koukourakis M., Hickson I.D., Barzilay G., Turley H., Leek R.D., Kanavaros P., Georgoulias V., et al. Expression and subcellular localization of human AP endonuclease 1 (HAP1/Ref-1) protein: a basis for its role in human disease. Histopathology. 1998;33:561–569. doi: 10.1046/j.1365-2559.1998.00541.x. [DOI] [PubMed] [Google Scholar]
- 323.Tsuchimoto D., Sakai Y., Sakumi K., Nishioka K., Sasaki M., Fujiwara T., Nakabeppu Y. Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucleic Acids Res. 2001;29:2349–2360. doi: 10.1093/nar/29.11.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Ramotar D., Kim C., Lillis R., Demple B. Intracellular localization of the Apn1 DNA repair enzyme of Saccharomyces cerevisiae. Nuclear transport signals and biological role. J. Biol. Chem. 1993;268:20533–20539. [PubMed] [Google Scholar]
- 325.Vongsamphanh R., Fortier P.K., Ramotar D. Pir1p mediates translocation of the yeast Apn1p endonuclease into the mitochondria to maintain genomic stability. Mol. Cell. Biol. 2001;21:1647–1655. doi: 10.1128/MCB.21.5.1647-1655.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Parsons J.L., Dianova I.I., Dianov G.L. APE1 is the major 3′-phosphoglycolate activity in human cell extracts. Nucleic Acids Res. 2004;32:3531–3536. doi: 10.1093/nar/gkh676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Meagher M., Lightowlers R.N. The role of TDP1 and APTX in mitochondrial DNA repair. Biochimie. 2014;100:121–124. doi: 10.1016/j.biochi.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Moor N.A., Vasil'eva I.A., Anarbaev R.O., Antson A.A., Lavrik O.I. Quantitative characterization of protein-protein complexes involved in base excision DNA repair. Nucleic Acids Res. 2015;43:6009–6022. doi: 10.1093/nar/gkv569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Bebenek K., Pedersen L.C., Kunkel T.A. Structure-function studies of DNA polymerase lambda. Biochemistry. 2014;53:2781–2792. doi: 10.1021/bi4017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Beard W.A., Wilson S.H. Structure and mechanism of DNA polymerase beta. Biochemistry. 2014;53:2768–2780. doi: 10.1021/bi500139h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Sterling C.H., Sweasy J.B. DNA polymerase 4 of Saccharomyces cerevisiae is important for accurate repair of methyl-methanesulfonate-induced DNA damage. Genetics. 2006;172:89–98. doi: 10.1534/genetics.105.049254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Blank A., Kim B., Loeb L.A. DNA polymerase delta is required for base excision repair of DNA methylation damage in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 1994;91:9047–9051. doi: 10.1073/pnas.91.19.9047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Prindle M.J., Loeb L.A. DNA polymerase delta in DNA replication and genome maintenance. Environ. Mol. Mutag. 2012;53:666–682. doi: 10.1002/em.21745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Sykora P., Wilson Iii D.M., Bohr V.A. Repair of persistent strand breaks in the mitochondrial genome. Mech. Age. Dev. 2012;133:169–175. doi: 10.1016/j.mad.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Sleeth K.M., Robson R.L., Dianov G.L. Exchangeability of mammalian DNA ligases between base excision repair pathways. Biochemistry. 2004;43:12924–12930. doi: 10.1021/bi0492612. [DOI] [PubMed] [Google Scholar]
- 336.Wu X., Braithwaite E., Wang Z. DNA ligation during excision repair in yeast cell-free extracts is specifically catalyzed by the CDC9 gene product. Biochemistry. 1999;38:2628–2635. doi: 10.1021/bi982592s. [DOI] [PubMed] [Google Scholar]
- 337.Donahue S.L., Corner B.E., Bordone L., Campbell C. Mitochondrial DNA ligase function in Saccharomyces cerevisiae. Nucleic Acids Res. 2001;29:1582–1589. doi: 10.1093/nar/29.7.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Simsek D., Furda A., Gao Y., Artus J., Brunet E., Hadjantonakis A.-K., Van Houten B., Shuman S., McKinnon P.J., Jasin M. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature. 2011;471:245–248. doi: 10.1038/nature09794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Krokan H.E., Bjoras M. Base excision repair. Cold Spring Harb. Perspect. Biol. 2013;5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Ellenberger T., Tomkinson A.E. Eukaryotic DNA ligases: structural and functional insights. Annu. Rev. Biochem. 2008;77:313–338. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Moe A., Ringvoll J., Nordstrand L.M., Eide L., Bjoras M., Seeberg E., Rognes T., Klungland A. Incision at hypoxanthine residues in DNA by a mammalian homologue of the Escherichia coli antimutator enzyme endonuclease V. Nucleic Acids Res. 2003;31:3893–3900. doi: 10.1093/nar/gkg472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Cao W. Endonuclease V: an unusual enzyme for repair of DNA deamination. Cell. Mol. Life Sci. 2013;70:3145–3156. doi: 10.1007/s00018-012-1222-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Evert B.A., Salmon T.B., Song B., Jingjing L., Siede W., Doetsch P.W. Spontaneous DNA damage in Saccharomyces cerevisiae elicits phenotypic properties similar to cancer cells. J. Biol. Chem. 2004;279:22585–22594. doi: 10.1074/jbc.M400468200. [DOI] [PubMed] [Google Scholar]
- 344.D'Errico M., Parlanti E., Teson M., de Jesus B.M., Degan P., Calcagnile A., Jaruga P., Bjoras M., Crescenzi M., Pedrini A.M., et al. New functions of XPC in the protection of human skin cells from oxidative damage. EMBO J. 2006;25:4305–4315. doi: 10.1038/sj.emboj.7601277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Pascucci B., D'Errico M., Parlanti E., Giovannini S., Dogliotti E. Role of nucleotide excision repair proteins in oxidative DNA damage repair: an updating. Biochemistry (Mosc.) 2011;76:4–15. doi: 10.1134/s0006297911010032. [DOI] [PubMed] [Google Scholar]
- 346.Melis J.P., van Steeg H., Luijten M. Oxidative DNA damage and nucleotide excision repair. Antioxid. Redox Signal. 2013;18:2409–2419. doi: 10.1089/ars.2012.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Plosky B., Samson L., Engelward B.P., Gold B., Schlaen B., Millas T., Magnotti M., Schor J., Scicchitano D.A. Base excision repair and nucleotide excision repair contribute to the removal of N-methylpurines from active genes. DNA Repair. 2002;1:683–696. doi: 10.1016/s1568-7864(02)00075-7. [DOI] [PubMed] [Google Scholar]
- 348.Torres-Ramos C.A., Johnson R.E., Prakash L., Prakash S. Evidence for the involvement of nucleotide excision repair in the removal of abasic sites in yeast. Mol. Cell. Biol. 2000;20:3522–3528. doi: 10.1128/mcb.20.10.3522-3528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Alexeyev M., Shokolenko I., Wilson G., LeDoux S. The maintenance of mitochondrial DNA integrity—critical analysis and update. Cold Spring Harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Fousteri M., Mullenders L.H.F. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 2008;18:73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- 351.Evans E., Moggs J.G., Hwang J.R., Egly J.M., Wood R.D. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 1997;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Shivji M.K., Podust V.N., Hubscher U., Wood R.D. Nucleotide excision repair DNA synthesis by DNA polymerase ϵ in the presence of PCNA, RFC, and RPA. Biochemistry. 1995;34:5011–5017. doi: 10.1021/bi00015a012. [DOI] [PubMed] [Google Scholar]
- 353.Moser J., Kool H., Giakzidis I., Caldecott K., Mullenders L.H., Fousteri M.I. Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase IIIα in a cell-cycle-specific manner. Mol. Cell. 2007;27:311–323. doi: 10.1016/j.molcel.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 354.Rowe L.A., Degtyareva N., Doetsch P.W. Yap1: a DNA damage responder in Saccharomyces cerevisiae. Mech. Age. Dev. 2012;133:147–156. doi: 10.1016/j.mad.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.German P., Szaniszlo P., Hajas G., Radak Z., Bacsi A., Hazra T.K., Hegde M.L., Ba X., Boldogh I. Activation of cellular signaling by 8-oxoguanine DNA glycosylase-1–initiated DNA base excision repair. DNA Repair. 2013;12:856–863. doi: 10.1016/j.dnarep.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Hajas G., Bacsi A., Aguilera-Aguirre L., Hegde M.L., Tapas K.H., Sur S., Radak Z., Ba X., Boldogh I. 8-Oxoguanine DNA glycosylase-1 links DNA repair to cellular signaling via the activation of the small GTPase Rac1. Free Radical Biol. Med. 2013;61c:384–394. doi: 10.1016/j.freeradbiomed.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Luo J., Hosoki K., Bacsi A., Radak Z., Hegde M.L., Sur S., Hazra T.K., Brasier A.R., Ba X., Boldogh I. 8-Oxoguanine DNA glycosylase-1-mediated DNA repair is associated with Rho GTPase activation and alpha-smooth muscle actin polymerization. Free Radical Biol. Med. 2014;73:430–438. doi: 10.1016/j.freeradbiomed.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Woodhouse B.C., Dianov G.L. Poly ADP-ribose polymerase-1: an international molecule of mystery. DNA Repair. 2008;7:1077–1086. doi: 10.1016/j.dnarep.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 359.Giannattasio M., Follonier C., Tourrière H., Puddu F., Lazzaro F., Pasero P., Lopes M., Plevani P., Muzi-Falconi M. Exo1 competes with repair synthesis, converts NER intermediates to long ssDNA Gaps, and promotes checkpoint qctivation. Mol. Cell. 2010;40:50–62. doi: 10.1016/j.molcel.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 360.Novarina D., Amara F., Lazzaro F., Plevani P., Muzi-Falconi M. Mind the gap: Keeping UV lesions in check. DNA Repair. 2011;10:751–759. doi: 10.1016/j.dnarep.2011.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Sertic S., Pizzi S., Cloney R., Lehmann A.R., Marini F., Plevani P., Muzi-Falconi M. Human exonuclease 1 connects nucleotide excision repair (NER) processing with checkpoint activation in response to UV irradiation. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13647–13652. doi: 10.1073/pnas.1108547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Lindsey-Boltz L.A., Kemp M.G., Reardon J.T., DeRocco V., Iyer R.R., Modrich P., Sancar A. Coupling of human DNA excision repair and the DNA damage checkpoint in a defined in vitro system. J. Biol. Chem. 2014;289:5074–5082. doi: 10.1074/jbc.M113.542787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Christmann M., Kaina B. Transcriptional regulation of human DNA repair genes following genotoxic stress: trigger mechanisms, inducible responses and genotoxic adaptation. Nucleic Acids Res. 2013;41:8403–8420. doi: 10.1093/nar/gkt635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Almeida K.H., Sobol R.W. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair (Amst.) 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Karahalil B., Bohr V.A., Wilson D.M., 3rd Impact of DNA polymorphisms in key DNA base excision repair proteins on cancer risk. Hum. Exp. Toxicol. 2012;31:981–1005. doi: 10.1177/0960327112444476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Nakabeppu Y. Regulation of intracellular localization of human MTH1, OGG1, and MYH proteins for repair of oxidative DNA damage. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:75–94. doi: 10.1016/s0079-6603(01)68091-7. [DOI] [PubMed] [Google Scholar]
- 367.Koketsu S., Watanabe T., Nagawa H. Expression of DNA repair protein: MYH, NTH1, and MTH1 in colorectal cancer. Hepatogastroenterology. 2004;51:638–642. [PubMed] [Google Scholar]
- 368.Goto M., Shinmura K., Igarashi H., Kobayashi M., Konno H., Yamada H., Iwaizumi M., Kageyama S., Tsuneyoshi T., Tsugane S., et al. Altered expression of the human base excision repair gene NTH1 in gastric cancer. Carcinogenesis. 2009;30:1345–1352. doi: 10.1093/carcin/bgp108. [DOI] [PubMed] [Google Scholar]
- 369.Hegde M.L., Hegde P.M., Holthauzen L.M., Hazra T.K., Rao K.S., Mitra S. Specific Inhibition of NEIL-initiated repair of oxidized base damage in human genome by copper and iron: potential etiological linkage to neurodegenerative diseases. J. Biol. Chem. 2010;285:28812–28825. doi: 10.1074/jbc.M110.126664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Gassman N.R., Wilson S.H. Micro-irradiation tools to visualize base excision repair and single-strand break repair. DNA Repair. 2015;31:52–63. doi: 10.1016/j.dnarep.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Roy A., Carpentier P., Bourgeois D., Field M. Diffusion pathways of oxygen species in the phototoxic fluorescent protein KillerRed. Photochem. Photobiol. Sci. 2010;9:1342–1350. doi: 10.1039/c0pp00141d. [DOI] [PubMed] [Google Scholar]
- 372.Lan L., Nakajima S., Wei L., Sun L., Hsieh C.L., Sobol R.W., Bruchez M., Van Houten B., Yasui A., Levine A.S. Novel method for site-specific induction of oxidative DNA damage reveals differences in recruitment of repair proteins to heterochromatin and euchromatin. Nucleic Acids Res. 2014;42:2330–2345. doi: 10.1093/nar/gkt1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Dizdaroglu M., Coskun E., Jaruga P. Measurement of oxidatively induced DNA damage and its repair, by mass spectrometric techniques. Free Radical Res. 2015;49:525–548. doi: 10.3109/10715762.2015.1014814. [DOI] [PubMed] [Google Scholar]
- 374.Bryan D.S., Ransom M., Adane B., York K., Hesselberth J.R. High resolution mapping of modified DNA nucleobases using excision repair enzymes. Genome Res. 2014;24:1534–1542. doi: 10.1101/gr.174052.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Wyrick J.J., Roberts S.A. Genomic approaches to DNA repair and mutagenesis. DNA Repair. 2015 doi: 10.1016/j.dnarep.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Bauer N.C., Corbett A.H., Doetsch P.W. Automated quantification of the subcellular localization of multicompartment proteins via Q-SCAn. Traffic. 2013;14:1200–1208. doi: 10.1111/tra.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]


