SUMMARY
The RNA-guided DNA endonuclease Cas9 cleaves double-stranded DNA targets with a protospacer adjacent motif (PAM) and complementarity to the guide RNA. Recently, we harnessed Staphylococcus aureus Cas9 (SaCas9), which is significantly smaller than Streptococcus pyogenes Cas9 (SpCas9), to facilitate efficient in vivo genome editing. Here, we report the crystal structures of SaCas9 in complex with a single guide RNA (sgRNA) and its double-stranded DNA targets, containing the 5′-TTGAAT-3′ PAM and the 5′-TTGGGT-3′ PAM, at 2.6 and 2.7 Å resolutions, respectively. The structures revealed the mechanism of the relaxed recognition of the 5′-NNGRRT-3′ PAM by SaCas9. A structural comparison of SaCas9 with SpCas9 highlighted both structural conservation and divergence, explaining their distinct PAM specificities and orthologous sgRNA recognition. Finally, we applied the structural information about this minimal Cas9 to rationally design compact transcriptional activators and inducible nucleases, to further expand the CRISPR-Cas9 genome editing toolbox.
Graphical Abstract
INTRODUCTION
Advances in our understanding of the CRISPR (clustered regularly interspaced short palindromic repeat)-Cas (CRISPR associated) systems (Mojica et al., 2005; Pourcel et al., 2005; Bolotin et al., 2005; Makarova et al., 2006; Barrangou et al., 2007; Brouns et al., 2008; Marraffini and Sontheimer, 2008; Garneau et al., 2010; Deltcheva et al., 2011; Sapranauskas et al., 2011; Jinek et al., 2012; Gasiunas et al., 2012) have propelled the development and applications of Cas9 for genome editing (Cong et al., 2013; Mali et al., 2013; Cho et al., 2013; Hwang et al., 2013; Jiang et al., 2013; Jinek et al., 2013). However, much work remains to understand how Cas9 mediates RNA-guided DNA recognition and cleavage. Previous studies have shown that Cas9 contains two endonuclease domains, HNH and RuvC, which cleave the DNA strands complementary (target DNA strand) and non-complementary (non-target DNA strand) to the guide RNA, respectively (Sapranauskas et al., 2011; Gasiunas et al., 2012; Jinek et al., 2012), and that Cas9-catalyzed DNA cleavage requires the presence of a short sequence, known as a protospacer-adjacent motif (PAM), located immediately downstream of the target DNA sequence (Bolotin et al., 2005; Deveau et al., 2008; Mojica et al., 2009; Garneau et al., 2010; Sapranauskas et al., 2011; Jinek et al., 2012; Gasiunas et al., 2012). The PAM sequences are diverse among the orthologous CRISPR-Cas systems, and the widely used Cas9 from Streptococcus pyogenes (SpCas9) recognizes a 5′-NGG-3′ PAM on the non-target DNA strand.
Structural studies of SpCas9 provided insight into the RNA-guided cleavage mechanism of the Cas9 enzymes. The crystal structures of SpCas9 in its unbound state (Jinek et al., 2014), SpCas9 in complex with a single guide RNA (sgRNA) (Jiang et al., 2015), and SpCas9 in complex with the sgRNA and its DNA target (Nishimasu et al., 2014; Anders et al., 2014) revealed that SpCas9 undergoes significant structural rearrangement upon association with the sgRNA, and subsequently adopts a bilobed architecture, consisting of a recognition (REC) lobe and a nuclease (NUC) lobe, with a central channel enveloping the RNA–DNA heteroduplex (Jinek et al., 2014; Nishimasu et al., 2014). The PAM is recognized by a PAM-interacting (PI) domain, which facilitates the target DNA unwinding and the heteroduplex formation (Nishimasu et al., 2014; Anders et al., 2014). In addition to SpCas9, the crystal structure of Actinomyces naeslundii Cas9 (AnCas9) is also available, but it lacks the sgRNA and the target DNA (Jinek et al., 2014). Thus, additional crystal structures of Cas9 orthologs (Chylinski et al., 2013; Chylinski et al., 2014) bound to nucleic acids are critical to understand the potential mechanistic and structural conservations underlying RNA-guided DNA targeting by Cas9, and to facilitate the development of new Cas9-based genome engineering technologies.
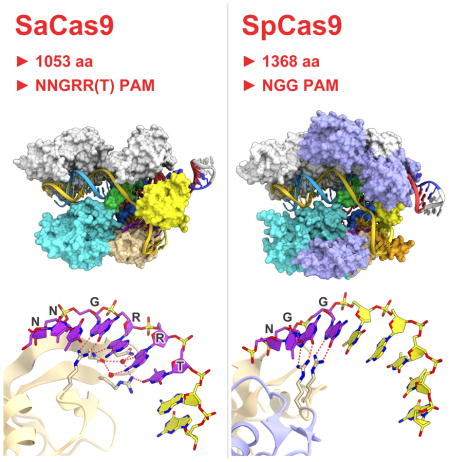
Recently, we harnessed a small Cas9 from Staphylococcus aureus (SaCas9) for eukaryotic genome editing (Ran et al., 2015). Although several Cas9 orthologs can cleave DNA targets in vitro, only SaCas9 and SpCas9 exhibit robust activity when transplanted into mammalian cells. SaCas9 shares only 17% sequence identity with SpCas9, highlighting the structural and functional variations among orthologous CRISPR-Cas9 systems. SaCas9 (1053 amino acid residues) is significantly smaller than SpCas9 (1368 amino acid residues), and thus is easier to deliver to somatic tissues for genome editing (Ran et al., 2015). While our previous attempts to create a smaller version of SpCas9 by rationally removing various domains met with limited success (Nishimasu et al., 2014), the structure of the naturally small SaCas9 may reveal the minimum and essential components of Cas9 enzymes. In addition, SaCas9 recognizes a 5′-NNGRRT-3′ PAM (where R represents a purine (i.e., A or G)), which is distinct from the 5′-NGG-3′ PAM for SpCas9, thereby offering an opportunity to understand the diverse PAM specificities of the Cas9 orthologs.
Here we report the crystal structures of SaCas9 in complex with the sgRNA and its target DNA, containing either the 5′-TTGAAT-3′ PAM (2.6 Å resolution) or the 5′-TTGGGT-3′ PAM (2.7 Å resolution), to facilitate a comparative analysis of the distinct Cas9 orthologs and to provide the structural foundation for the rational design of SaCas9-based genome engineering tools. Our structural and functional data provide insight into the PAM-dependent, RNA-guided DNA cleavage mechanism of SaCas9, as well as the minimal requirements for general Cas9 activity. Furthermore, our data enable the structural comparison between SaCas9 and SpCas9, revealing notable differences in the interactions in their REC lobe–sgRNA scaffolds and PI domain–PAM sequences. These comparative results also highlighted the flexible nature of the RuvC and HNH nuclease domains, and identified a previously uncharacterized and evolutionarily divergent wedge (WED) domain. Given its compact size, SaCas9 holds great potential as a minimum scaffold for genome engineering. The structural and comparative data presented here advance our understanding of the RNA-guided DNA cleavage mechanism, and provide a starting point for the future design of Cas9 variants with expanded target space and improved specificity.
RESULTS
Overall structure of the SaCas9–sgRNA–target DNA complex
We solved the crystal structures of full-length SaCas9 (residues 1–1053; N580A/C946A) in complex with a 73-nucleotide (nt) sgRNA, a 28-nt complementary DNA strand and an 8-nt non-target DNA strand, containing either the 5′-TTGAAT-3′ PAM or 5′-TTGGGT-3′ PAM, at 2.6 and 2.7 Å resolutions, respectively (Figures 1A–1D, Figure S1A and Table S1). Since solvent-exposed cysteine residues may hamper crystallization, we replaced a non-conserved cysteine residue (Cys946) with alanine after confirming that the C946A mutation did not affect the DNA cleavage activity in vivo (Figures S1B–S1D). Based on homology to SpCas9, we mutated one of the putative catalytic residues, Asn580 in the HNH domain, to alanine to prevent the potential cleavage of the target DNA during crystallization. Since the two structures are virtually identical (root-mean-square deviation [rmsd] of 0.2 Å for 1043 equivalent Cα atoms), we will describe the quaternary complex structure containing the 5′-TTGAAT-3′ PAM, unless otherwise stated.
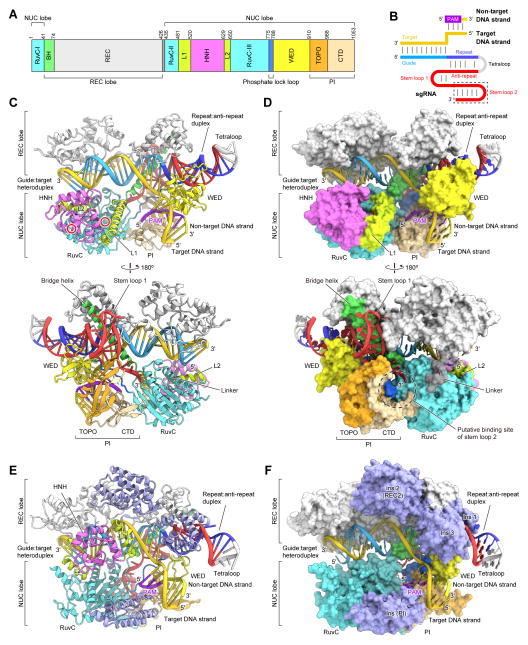
Figure 1. Structure of the SaCas9–sgRNA–target DNA complex.
(A) Domain organization of SaCas9. BH, bridge helix. CTD, C-terminal domain.
(B) Schematic of the sgRNA–target DNA complex. The putative stem loop 2 was truncated for crystallization.
(C and D) Ribbon (C) and surface (D) representations of the SaCas9–sgRNA–target DNA complex. The active sites of the RuvC (Asp10) and HNH (Asn580) domains are indicated by red circles. Molecular graphics images were prepared using CueMol (http://www.cuemol.org).
(E and F) Ribbon (E) and surface (F) representations of the SpCas9–sgRNA–target DNA complex (PDB ID 4UN3). The SpCas9-specific insertions in the REC and PI domains are highlighted in pale blue. In (F), the L1 and L2 linker regions and the HNH domain are omitted for clarity.
See also Figures S1–S3.
SaCas9 adopts a bilobed architecture consisting of a REC lobe (residues 41–425) and a NUC lobe (residues 1–40 and 435–1053) (Figures 1C and 1D). The two lobes are connected by an arginine-rich bridge helix (residues 41–73) and a linker loop (residues 426–434). The NUC lobe consists of the RuvC (residues 1–40, 435–480 and 650–774), HNH (residues 520–628), WED (residues 788–909) and PI (residues 910–1053) domains (Figures 1C and 1D). The PI domain can be divided into a Topoisomerase-homology (TOPO) domain and a C-terminal domain, as in SpCas9 (Jinek et al., 2014). The RuvC domain consists of three separate motifs (RuvC-I–III) and interacts with the HNH and PI domains. The HNH domain is connected to RuvC-II and RuvC-III by the L1 (residues 481–519) and L2 (residues 629–649) linker regions, respectively. The WED and RuvC domains are connected by a ‘phosphate lock’ loop (residues 775–787), as in SpCas9 (Anders et al., 2014). The active site of the HNH domain is distant from the cleavage site in the target DNA strand (the phosphodiester linkage between dC3 and dA4), indicating that the present structure represents the inactive state, as in the cases of the SpCas9–sgRNA–target DNA complex structures (Nishimasu et al., 2014; Anders et al., 2014).
Previous structural studies revealed that SpCas9 undergoes conformational rearrangements upon guide RNA binding, to form the central channel between the REC and NUC lobes (Jinek et al., 2014; Nishimasu et al., 2014; Anders et al., 2014; Jiang et al., 2015). In the absence of the guide RNA, SpCas9 and AnCas9 adopt a closed conformation, where the active site of the HNH domain is covered by the RuvC domain (Figure S2). In contrast, the ternary and quaternary complex structures of SpCas9 adopt an open conformation and have the central channel, which accommodates the guide RNA–target DNA heteroduplex (referred to as the guide:target heteroduplex) (Figures 1E and 1F). The present quaternary complex structure of SaCas9 adopts a similar open conformation to form the central channel, which accommodates the guide:target heteroduplex (Figures 1C and 1D), suggesting that the guide RNA-induced conformational activation is conserved between SaCas9 and SpCas9. A structural comparison between SaCas9 and SpCas9 revealed that, although their overall architectures are similar, there are notable differences in their REC, WED and PI domains, as described in detail below, thereby explaining the significant sequence and size differences of the two Cas9 orthologs (Figures 1C–1F and Figure S3).
Structure of the sgRNA–target DNA complex
The SaCas9 sgRNA consists of the guide region (G1–C20), repeat region (G21–G34), tetraloop (G35–A38), anti-repeat region (C39–C54), stem loop 1 (A56–G68) and single-stranded linker (U69–U73), with A55 connecting the anti-repeat region and stem loop 1 (Figures 2A–2D). U73 at the 3′ end is disordered in the present structure. The guide region (G1–C20) and the target DNA strand (dG1–dC20) form the guide:target heteroduplex, whereas the target DNA strand (dC(−8)–dA(−1)) and the non-target DNA strand (dT1*–dG8*) form a PAM-containing duplex (referred to as the PAM duplex) (Figures 2A and 2B). The repeat (G21–G34) and anti-repeat (C39–C54) regions form a distorted duplex (referred to as the repeat:anti-repeat duplex) via 13 Watson-Crick base pairs (Figures 2A and 2B). The unpaired nucleotides (C30, A43, U44 and C45) form an internal loop (Figure 2C). This distorted structure of the repeat:anti-repeat duplex is precisely recognized by the REC and WED domains, as the insertion of GAU into the repeat region, which eliminated the internal loop, drastically reduced the Cas9-mediated DNA cleavage (Figure 2E).
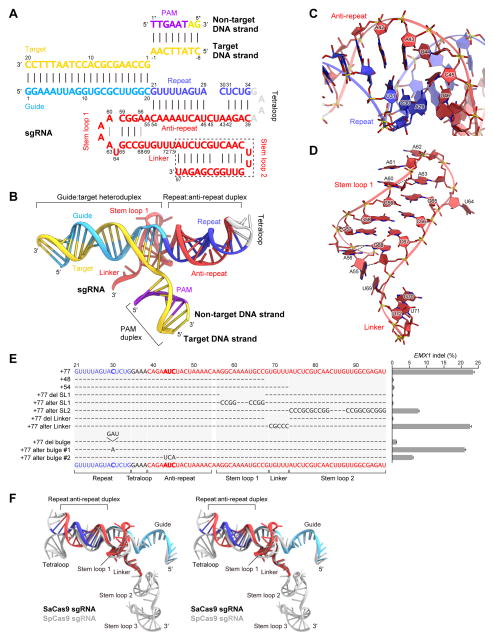
Figure 2. Structure of the sgRNA–target DNA complex.
(A) Nucleotide sequences of the sgRNA and the target DNA. The putative stem loop 2 (dashed box) was truncated for crystallization. U73 is disordered in the structure.
(B) Overview of the sgRNA–target DNA complex.
(C and D) Close-up views of the repeat:anti-repeat duplex (C) and stem loop 1 (D). Key interactions are shown as dashed lines.
(E) Mutational analysis of SaCas9 sgRNA scaffolds. Effects of mutations on the ability to induce indels in the target EMX1 locus were examined. Base changes from the sgRNA (+77) scaffold are shown at the respective positions, with dashes indicating unaltered bases (n = 3, error bars show mean ± S.E.M.).
(F) Superimposition of the sgRNAs of SaCas9 and SpCas9 (PDB ID 4OO8) (stereoview).
See also Figure S4.
Stem loop 1 is formed via three Watson-Crick base pairs (G57:C67–C59:G65) and two non-canonical base pairs (A56:G68 and A60:A63) (Figures 2A and 2D). U64 does not base pair with A60, and is flipped out of the stem loop (Figure 2D). The N1 and N6 of A63 hydrogen bond with the 2′-OH and N3 of A60, respectively. G68 stacks with G57:C67, with the G68 N2 interacting with the backbone phosphate group between A55 and A56. A55 adopts the syn conformation, and its adenine base stacks with U69 (Figure 2D). In addition, the N1 of A55 hydrogen bonds with the 2′-OH of G68, thus stabilizing the basal region of stem loop 1. An adenosine residue immediately after the repeat:anti-repeat duplex is highly conserved among CRISPR-Cas9 systems, and the equivalent adenosine in the SpCas9 sgRNA, A51, also adopts the syn conformation (Nishimasu et al., 2014; Anders et al., 2014) (Figure S4A), suggesting that these adenosine residues play conserved key roles in connecting the repeat:anti-repeat duplex and stem loop 1.
The SpCas9 sgRNA contains three stem loops (stem loops 1–3), which interact with Cas9 and contribute to complex formation (Nishimasu et al., 2014) (Figure 2F and Figure S4A). The sgRNA lacking stem loops 2 and 3 supports SpCas9-catalyzed DNA cleavage in vitro but not in vivo, indicating the importance of stem loops 2 and 3 for the cleavage activity in vivo (Cong et al., 2013; Hsu et al., 2013; Nishimasu et al., 2014; Wright et al., 2015). In contrast, the SaCas9 sgRNA is predicted to contain only two stem loops (stem loops 1 and 2), based on its nucleotide sequence (Figure S4B). As in SpCas9, the sgRNA lacking stem loop 2 supported SaCas9-catalyzed DNA cleavage in vitro but not in vivo (Figure 2E and Figure S4C), suggesting that the secondary structures on the 3′ tracrRNA tail of the sgRNA are critical for successfully harnessing Cas9 for genome editing applications. Although we failed to obtain diffraction-quality crystals of SaCas9 bound to a version of sgRNA containing the full-length tracrRNA, the truncation of stem loop 2 dramatically improved the quality of the crystals. Thus, it remains unknown how stem loop 2 interacts with SaCas9, although it may bind to the positively-charged groove between the RuvC and PI domains, as in SpCas9 (Nishimasu et al., 2014; Anders et al., 2014) (Figure 1D and Figure S4D).
Recognition mechanism of the guide:target heteroduplex
The guide:target heteroduplex is accommodated in the central channel formed between the REC and NUC lobes (Figures 1D and 3). The sugar-phosphate backbone of the PAM-distal region (A3–U6) of the sgRNA interacts with the REC lobe (Thr238, Tyr239, Lys248, Tyr256, Arg314, Asn394 and Gln414) (Figure 3). In SpCas9 and SaCas9, the RNA–DNA base pairing in the 8 bp PAM-proximal ‘seed’ region in the guide:target heteroduplex is critical for Cas9-catalyzed DNA cleavage (Jinek et al., 2012; Hsu et al., 2013; Ran et al., 2015). Consistent with this, the phosphate backbone of the sgRNA seed region (C13–C20) is extensively recognized by the bridge helix (Asn44, Arg48, Arg51, Arg55, Arg59 and Arg60) and the REC lobe (Arg116, Gly117, Arg165, Gly166, Asn169 and Arg209) (Figures 3 and 4A), as in the case of SpCas9 (Nishimasu et al., 2014). In addition, the 2′-OH groups of C15, U16, U17 and G19 interact with the REC lobe (Gly166, Arg208, Arg209 and Tyr211). These structural findings suggest that the sgRNA binds to SaCas9 with its seed region pre-ordered in an A-form conformation for base-pairing with the target DNA strand, as observed in the SpCas9–sgRNA binary complex (Jiang et al., 2015). In addition, the sugar-phosphate backbone of the target DNA strand interacts with the REC lobe (Tyr211, Trp229, Tyr230, Gly235, Arg245, Gly391, Thr392 and Asn419) and the RuvC domain (Leu446, Tyr651 and Arg654) (Figure 3). These structural observations explain the RNA-guided DNA targeting mechanism of SaCas9.
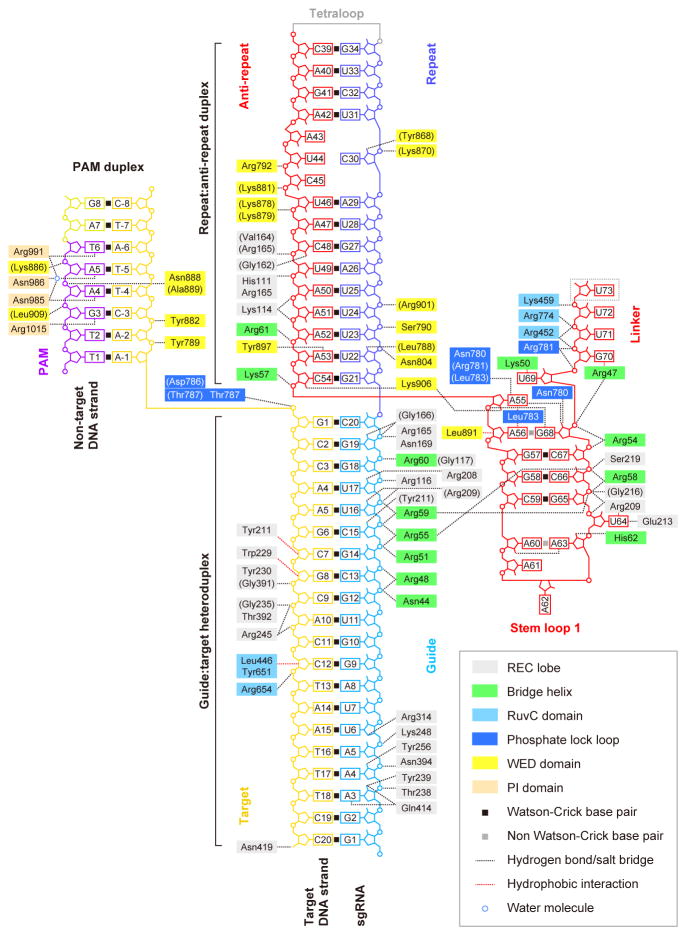
Figure 3. Schematic of nucleic acid recognition by SaCas9.
SaCas9 residues that interact with the sgRNA and the target DNA via their main chain are shown in parentheses. Water-mediated hydrogen-bonding interactions are omitted for clarity.
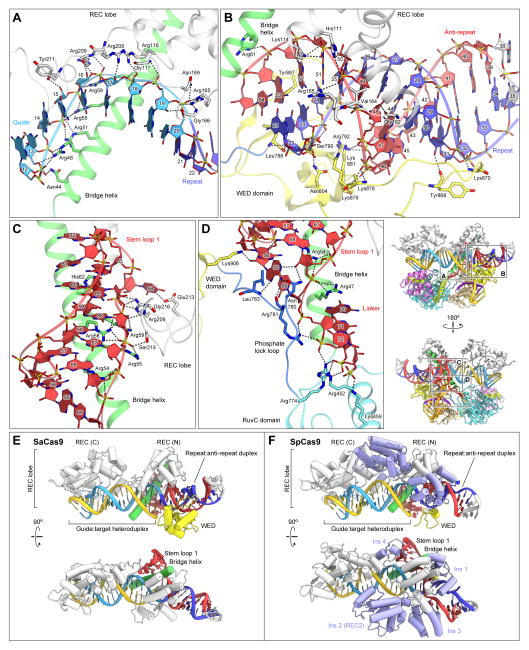
Figure 4. sgRNA recognition mechanism.
(A–D) Recognition of the seed region (A), the repeat:anti-repeat duplex (B), stem loop 1 (C), and the basal region of stem loop 1 (D). Hydrogen bonds and salt bridges are shown as dashed lines. In (A), the target DNA strand is omitted for clarity.
(E and F) Recognition of the sgRNA and target DNA by the REC and WED domains of SaCas9 (E) and SpCas9 (PDB ID 4UN3) (F). The SpCas9-specific insertions are highlighted in pale blue.
See also Figures S5 and S6.
The C-terminal region of the REC lobe interacts with the PAM-distal region of the heteroduplex, whereas the N-terminal region of the REC lobe interacts with the repeat:anti-repeat duplex and the PAM-proximal region of the heteroduplex (Figure 3). Notably, the C-terminal region of the REC lobe of SaCas9 shares structural similarity with those of SpCas9 (PDB ID 4UN3, 26% identity, rmsd of 1.9 Å for 177 equivalent Cα atoms) and AnCas9 (PDB ID 4OGE, 16% identity, rmsd of 3.2 Å for 167 equivalent Cα atoms) (Figure S5). These structural findings suggested that the Cas9 orthologs recognize the PAM-distal region of the guide:target heteroduplex in a similar manner.
Recognition mechanism of the sgRNA scaffold
The repeat:anti-repeat duplex is recognized by the REC and WED domains, primarily through interactions between the protein and the sugar-phosphate backbone (Figures 3 and 4B). Consistent with our data showing that the distorted repeat:anti-repeat duplex is critical for Cas9-catalyzed DNA cleavage (Figure 2E), the internal loop is recognized by the WED domain (Figure 4B). The 2′-OH of C30 hydrogen bonds with Tyr868, and the backbone phosphate groups of U31, C45 and U46 interact with Lys870, Arg792 and Lys881, respectively (Figure 4B). These structural observations explain the structure-dependent recognition of the repeat:anti-repeat duplex by SaCas9.
Stem loop 1 is recognized by the bridge helix and the REC lobe (Figures 3 and 4C). The phosphate backbone of stem loop 1 interacts with the bridge helix (Arg47, Arg54, Arg55, Arg58 and Arg59) and the REC lobe (Arg209, Gly216 and Ser219) (Figure 4C). The 2′-OH of A63 hydrogen bonds with His62. The flipped-out U64 is recognized by Arg209 and Glu213 via stacking and hydrogen-bonding interactions, respectively. A55 is extensively recognized by the phosphate lock loop (Figure 4D). The N6, N7 and 2′-OH of A55 hydrogen bond with Asn780/Arg781, Leu783 and Lys906, respectively. Lys57 interacts with the backbone phosphate group between C54 and A55, and the side chain of Leu783 forms hydrophobic contacts with the nucleobases of A55 and A56. The phosphate backbone of the linker region electrostatically interacts with the RuvC domain (Arg452, Lys459 and Arg774) and the phosphate lock loop (Arg781), and the nucleobase of G70 stacks with the side chain of Arg47 on the bridge helix (Figure 4D).
Structural basis for the orthogonal recognition of sgRNA scaffolds
A comparison of the quaternary complex structures of SaCas9 and SpCas9 revealed that the structurally diverse REC and WED domains recognize distinct structural features of the repeat:anti-repeat duplex, allowing the cognate sgRNAs to be distinguished in a highly specific manner (Figures 4E and 4F and Figure S6). The SaCas9 WED domain has a new fold comprising a twisted five-stranded β-sheet flanked by four α-helices, and is responsible for the recognition of the distorted repeat:anti-repeat duplex, as described above (Figures 4B and 4E and Figures S6A and S6B). In contrast, the SpCas9 WED domain adopts a compact loop conformation, and interacts with the repeat:anti-repeat duplex, which is structurally different from that of the SaCas9 sgRNA (Nishimasu et al., 2014; Anders et al., 2014) (Figure 4F and Figures S6A and S6C). The AnCas9 WED domain has yet another different fold containing three antiparallel β-hairpins (Jinek et al., 2014) (Figure S6A). These structural differences in the WED domains are consistent with the variations in the sgRNA scaffolds among the CRISPR-Cas9 systems (Fonfara et al., 2014; Briner et al., 2014; Ran et al., 2015).
The REC lobes also contribute to the orthogonal recognition of the sgRNA scaffolds. While the REC lobes of SaCas9 and SpCas9 share structural similarity, the SpCas9 REC lobe has four characteristic insertions (Ins 1–4), which are absent in the SaCas9 REC lobe (Figures 4E and 4F and Figures S6B and S6C). Ins 2 (also known as the REC2 domain) does not contact the nucleic acids in the SpCas9 structures and is dispensable for the DNA cleavage activity (Nishimasu et al., 2014), consistent with the absence of Ins 2 in SaCas9 (Figures 4E and 4F). Ins 1 and 3 recognize the SpCas9-specific internal loop in the repeat:anti-repeat duplex (Figure 4F and Figure S6C). In particular, Ins 3 interacts with the flipped-out G43 and U44 in the repeat:anti-repeat duplex in base-specific manners (Figure S6C). In addition, Ins 4 interacts with stem loop 1 of the SpCas9 sgRNA, which is shorter than that of the SaCas9 sgRNA (Figures 4E and 4F and Figures S6B and S6C). Together, these structural observations demonstrate how the Cas9 orthologs recognize their cognate sgRNAs in orthogonal manners, using specific combinations of the structurally diverse REC and WED domains.
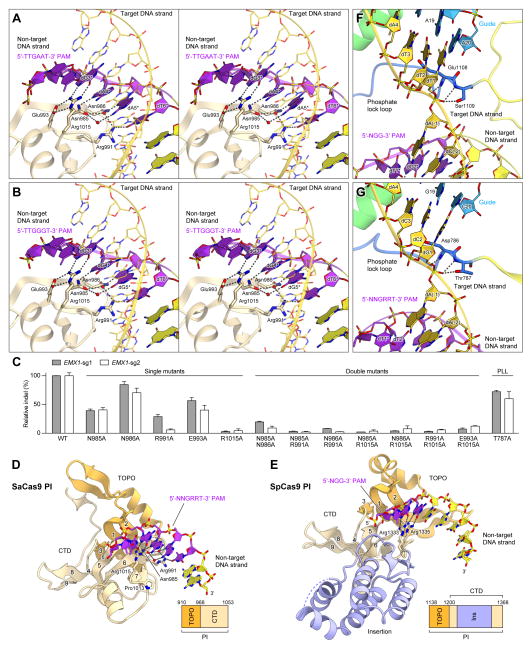
Recognition mechanism of the 5′-NNGRRT-3′ PAM
SaCas9 recognizes the 5′-NNGRRN-3′ PAM, with a preference for a thymine base at the 6th position (Ran et al., 2015), which is distinct from the 5′-NGG-3′ PAM of SpCas9. In the present structures containing either the 5′-TTGAAT-3′ PAM or the 5′-TTGGGT-3′ PAM, the PAM duplex is sandwiched between the WED and PI domains, and the PAM in the non-target DNA strand is read from the major groove side by the PI domain (Figures 5A and 5B). dT1* and dT2* do not directly contact the protein (Figures 5A and 5B). Consistent with the observed requirement for the 3rd G in the 5′-NNGRRT-3′ PAM, the O6 and N7 of dG3* form bidentate hydrogen bonds with the side chain of Arg1015, which is anchored via salt bridges with Glu993 in both complexes (Figures 5A and 5B). In the 5′-TTGAAT-3′ PAM complex, the N7 atoms of dA4* and dA5* form direct and water-mediated hydrogen bonds with Asn985 and Asn985/Asn986/Arg991, respectively (Figure 5A). In addition, the N6 of dA5* forms a water-mediated hydrogen bond with Asn985. Similarly, in the 5′-TTGGGT-3′ PAM complex, the N7 atoms of dG4* and dG5* form direct and water-mediated hydrogen bonds with Asn985 and Asn985/Asn986/Arg991, respectively (Figure 5B). The O6 of dG5* forms a water-mediated hydrogen bond with Asn985. These structural features explain the ability of SaCas9 to recognize the purine nucleotides at positions 4 and 5 in the 5′-NNGRRT-3′ PAM. The O4 of dT6* hydrogen bonds with Arg991 (Figures 5A and 5B), explaining the preference of SaCas9 for the 6th T in the 5′-NNGRRT-3′ PAM. Single alanine mutations of these PAM-interacting residues reduced the cleavage activity in vivo, and double mutations abolished the activity (Figure 5C), confirming the importance of Asn985, Asn986, Arg991, Glu993 and Arg1015 for PAM recognition. In addition, the phosphate backbone of the PAM duplex is recognized from the minor groove side by the WED domain (Tyr789, Tyr882, Lys886, Ans888, Ala889 and Leu909), in a distinct manner from that in SpCas9 (Figure 3). Together, our structural and functional data have revealed the mechanism underlying the relaxed recognition of the 5′-NNGRRT-3′ PAM by SaCas9.
Figure 5. PAM recognition mechanism.
(A and B) Recognition of the 5′-TTGAAT-3′ PAM (A) and the 5′-TTGGGT-3′ PAM (B) (stereoview). The PAM sequences are highlighted in purple. Water molecules are shown as red spheres. Hydrogen bonds are shown as dashed lines.
(C) Mutational analysis of the PAM-interacting residues and the phosphate lock loop (PLL), measured by indel rates at two EMX1 targets (n = 3, error bars show mean ± S.E.M.).
(D and E) PAM recognition by the PI domains of SaCas9 (D) and SpCas9 (PDB ID 4UN3) (E). The PAM sequences are highlighted in purple. Hydrogen bonds are shown as dashed lines. The target DNA strands are omitted for clarity. In (E), the SpCas9-specific insertion is highlighted in pale blue.
(F and G) +1 phosphate recognition by the phosphate lock loop in SpCas9 (PDB ID 4UN3) (F) and SaCas9 (G).
See also Figure S6.
Structural basis for the distinct PAM specificities
A structural comparison of SaCas9, SpCas9 and AnCas9 revealed that, despite the lack of sequence homology, their PI domains share a similar protein fold (Figures 5D and 5E, and Figure S6A). The PI domains consist of the TOPO domain, comprising a three-stranded anti-parallel β-sheet (β1–β3) flanked by several α helices, and the C-terminal domain, comprising a twisted six-stranded anti-parallel β-sheet (β4–β9) (β7 in SpCas9 adopts a loop conformation) (Figures 5D and 5E and Figure S6A). In both SaCas9 and SpCas9, the major groove of the PAM duplex is read by the β5–β7 region in their PI domains (Figures 5D and 5E). The 3rd G in the 5′-NNGRRT PAM-3′ is recognized by Arg1015 in SaCas9 (Figure 5D), whereas the 3rd G in the 5′-NGG-3′ PAM is recognized by Arg1335 in SpCas9 in a similar manner (Figure 5E). However, there are notable differences in the PI domains of SaCas9 and SpCas9, consistent with their distinct PAM specificities. Arg1333 of SpCas9, which recognizes the 2nd G in the 5′-NGG-3′ PAM, is replaced with Pro1013 in SaCas9 (Figures 5D and 5E and Figure S3). In addition, SpCas9 lacks the amino acid residues equivalent to Asn985/Asn986 (β5) and Arg991 (β6) of SaCas9, because the β5–β6 region of SpCas9 is shorter than that of SaCas9 (Figures 5D and 5E and Figure S3). Moreover, Asn985, Asn986, Arg991 and Arg1015 in SaCas9 are replaced with Asp1030, Thr1031, Lys1034 and Lys1061 in AnCas9, respectively (Figure S6A), suggesting that the PAM of AnCas9 is different from those of SaCas9 and SpCas9 (although the sequence remains unknown). Together, these structural findings demonstrate that the distinct PAM specificities of the Cas9 orthologs are primarily defined by the specific differences in the PAM-interacting residues in the PI domains.
Mechanism of target DNA unwinding
In SpCas9, Glu1108 and Ser1109, in the phosphate lock loop, hydrogen bond with the phosphate group between dA(−1) and dT1 in the target DNA strand (referred to as the +1 phosphate), thereby contributing to the target DNA unwinding (Anders et al., 2014) (Figure 5F). The present structure revealed that SaCas9 also has the phosphate lock loop, although it shares limited sequence similarity to that of SpCas9 (Figure 5G and Figure S3). In SaCas9, the +1 phosphate between dA(−1) and dG1, in the target DNA strand, hydrogen bonds with the main-chain amide groups of Asp786 and Thr787 and the side chain of Thr787 in the phosphate lock loop (Figure 5G). These interactions result in the rotation of the +1 phosphate, thereby facilitating base-pairing between dG1 in the target DNA strand and C20 in the sgRNA. Indeed, the SaCas9 T787A mutant showed reduced DNA cleavage activity (Figure 5C), confirming the functional significance of Thr787 in the phosphate lock loop. These observations indicated the conserved molecular mechanism of target DNA unwinding in SaCas9 and SpCas9.
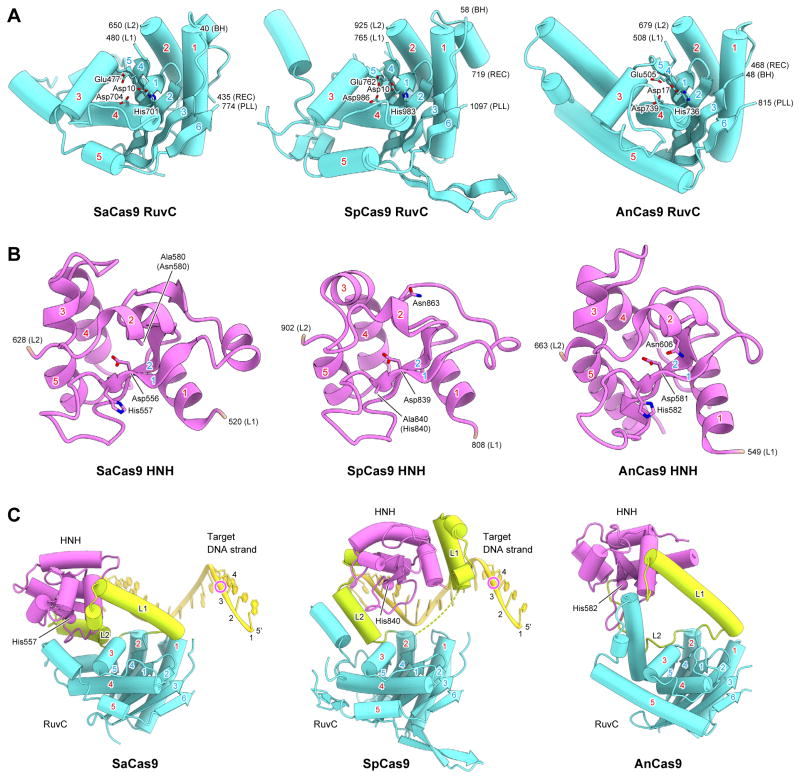
RuvC and HNH nuclease domains
The RuvC domain of SaCas9 has an RNase H fold, and shares structural similarity with those of SpCas9 (PDB ID 4UN3, 26% identity, rmsd of 2.0 Å for 179 equivalent Cα atoms) and AnCas9 (PDB ID 4OGE, 17% identity, rmsd of 3.0 Å for 169 equivalent Cα atoms) (Figure 6A). Asp10, Glu477, His701 and Asp704 of SaCas9 are located at positions similar to those of the catalytic residues of SpCas9 (Asp10, Glu762, His983 and Asp986) and AnCas9 (Asp17, Glu505, His736 and Asp739) (Figure 6A and Figure S3). Indeed, the D10A, E477A, H701A and D704A mutants of SaCas9 exhibited almost no DNA cleavage activity (Figures S7A and S7B), suggesting that the SaCas9 RuvC domain cleaves the non-target DNA strand through a two-metal ion mechanism, as in other RNase H superfamily endonucleases (Gorecka et al., 2013).
Figure 6. RuvC and HNH nuclease domains.
(A and B) Structural comparison of the RuvC (A) and HNH (B) domains of SaCas9, SpCas9 (PDB ID 4UN3) and AnCas9 (PDB ID 4OGE). The conserved α helices and β strands are numbered. The catalytic residues are shown as stick models. BH, bridge helix. PLL, phosphate lock loop.
(C) Comparison of the spatial arrangements between the RuvC and HNH domains in SaCas9, SpCas9 (PDB ID 4UN3) and AnCas9 (PDB ID 4OGE). The Cα atoms of the catalytic histidine residues in the HNH domains are shown as spheres. The cleavage sites of the target DNA strands are indicated by magenta circles. In SpCas9, the disordered region in the L1 linker is indicated by dashed lines.
See also Figure S7.
The HNH domain of SaCas9 has a ββα-metal fold, and shares structural similarity with those of SpCas9 (27% identity, rmsd of 1.8 Å for 93 equivalent Cα atoms) and AnCas9 (18% identity, rmsd of 2.6 Å for 98 equivalent Cα atoms) (Figure 6B). Asp556, His557 and Asn580 of SaCas9 are located at positions similar to those of the catalytic residues of SpCas9 (Asp839, His840 and Asn863) and AnCas9 (Asp581, His582 and Asn606) (Figure 6B and Figure S3). Indeed, the H557A and N580A mutants of SaCas9 almost completely lacked DNA cleavage activity (Figures S7A and S7B), suggesting that the SaCas9 HNH domain cleaves the target DNA strand through a one-metal ion mechanism, as in other ββα-metal endonucleases (Biertumpfel et al., 2007).
A structural comparison of SaCas9 with SpCas9 and AnCas9 revealed that the RuvC and HNH domains are connected by α-helical linkers, L1 and L2, and that notable differences exist in the relative arrangements between the two nuclease domains (Figure 6C). A biochemical study suggested that PAM duplex binding to SpCas9 facilitates the cleavage of the target DNA strand by the HNH domain (Sternberg et al., 2014). However, in the PAM-containing quaternary complex structures of SaCas9 and SpCas9, the HNH domains are distant from the cleavage site of the target DNA strand (Figure 6C). A structural comparison of SaCas9 with Thermus thermophilus RuvC in complex with a Holliday junction substrate (Gorecka et al., 2013) indicated steric clashes between the L1 linker and the modeled non-target DNA strand, bound to the active site of the SaCas9 RuvC domain (Figures S7C and S7D). These observations suggested that the binding of the non-target DNA strand to the RuvC domain may facilitate a conformational change of L1, thereby bringing the HNH domain to the scissile phosphate group in the target DNA strand.
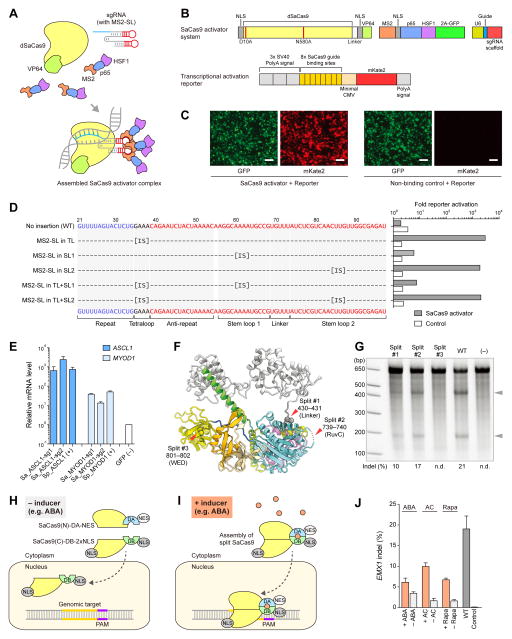
Structure-guided engineering of SaCas9 transcription activators and inducible nucleases
Using the crystal structure of SaCas9, we sought to conduct structure-guided engineering to further expand the CRISPR-Cas9 toolbox, as we have done previously using the SpCas9 crystal structure. Given the similarities in the overall domain organizations of SaCas9 and SpCas9, we initially explored the feasibility of engineering the SaCas9 sgRNA, to develop robust transcription activators. In the SpCas9 structure, the tetraloop and stem loop 2 of the sgRNA are exposed to the solvent (Nishimasu et al., 2014; Anders et al., 2014) (Figure S4D), and permitted the insertion of RNA aptamers into the sgRNA to create robust RNA-guided transcription activators (Konermann et al., 2015). To generate the SaCas9-based activator system, we created a catalytically inactive version of SaCas9 (dSaCas9) by introducing the D10A and N580A mutations to inactivate the RuvC and HNH domains, respectively, and attached VP64 to the C-terminus of dSaCas9 (Figures 7A and 7B). The sgRNA scaffold was modified by the insertion of the MS2 aptamer stem loop (MS2-SL), to allow the recruitment of MS2-p65-HSF1 transcriptional activation modules (Figure 7A). To evaluate the dSaCas9-based activator design, we constructed a transcriptional activation reporter system, consisting of tandem sgRNA target sites upstream of a minimal CMV promoter driving the expression of the fluorescent reporter gene mKate2 (Zhang et al., 2011) (Figure 7B). We included an additional transcriptional termination signal upstream of the reporter cassette, to reduce the background previously observed in a similar reporter (Cong et al., 2012) (Figure 7B). We observed robust activation of mKate2 transcription when we expressed the engineered sgRNA complementary to the target sites, whereas the non-binding sgRNA had no detectable effect (Figure 7C). Based on a screening of different sgRNA designs with this reporter assay, we found that the insertions of MS2-SL into the tetraloop and putative stem loop 2 induced strong activation in our reporter system, whereas the insertion of MS2-SL into stem loop 1 yielded modest activation, consistent with the structural data (Figure 7D). The single insertion of MS2-SL into the tetraloop was the simplest design that yielded strong transcriptional activation. Using this optimal sgRNA design, we further tested the activation of endogenous targets in the human genome. We selected two guides each for the human ASCL1 and MYOD1 genomic loci, and demonstrated that the dSaCas9-based activator system activated both genes to levels comparable to those of the dSpCas9-based activator (Konermann et al., 2013) (Figure 7E). Given that the sgRNAs for SaCas9 and SpCas9 are not interchangeable, the SaCas9-based transcription activator platform complements the SpCas9-based activator systems, by allowing the independent activation of different sets of genes.
Figure 7. Structure-guided engineering of the SaCas9 system and its applications.
(A) Schematic of the three-component SaCas9 activator system.
(B) Design of the dSaCas9-based transcriptional activator and the reporter system. NLS, nuclear localization signal. 2A, 2A self-cleaving peptide. CMV, cytomegalovirus.
(C) Representative fluorescent microscopy images, showing the activation of the reporter gene and the low background level of the newly designed reporter. Scale bar, 100 μm.
(D) Optimization of different fusion scaffolds for the dSaCas9-based activator, using the reporter system. MS2-SL, MS2-binding stem loop. ‘IS’ denotes the insertion site of the MS2-SL.
(E) Transcriptional activation of the endogenous target ASCL1 and MYOD1 genes by dSaCas9-based activators, with previously described dSpCas9-based activators as positive controls.
(F) Structure-guided design of split-SaCas9 systems. Split sites 1 and 2 were designed at flexible linker regions, whereas split site 3 was designed at the β-strand in the WED domain, as a negative control. The sgRNA and target DNA molecules were removed from the original structure for clarity.
(G) Cleavage activity of wild-type (WT) SaCas9 and the three different auto-assembled split-SaCas9 designs.
(H and I) Schematic of the inducible SaCas9 system. NES, nuclear export signal. ABA, abscisic acid. DA, dimerization domain A. DB, dimerization domain B.
(J) Cleavage activity of the wild-type (WT) SaCas9 and three different inducible SaCas9 systems. ‘ABA’ denotes the inducible SaCas9 design based on the abscisic acid sensing system; ‘AC’ denotes the design based on the A/C heterodimerizer; ‘Rapa’ denotes the design based on the FRB/FKBP system (see Experimental Procedures section for additional details).
The SpCas9 structure also facilitated the rational design of split-Cas9s (Zetsche et al., 2015; Wright et al., 2015), which can be further engineered into an inducible system (Zetsche et al., 2015). Our SaCas9 structure revealed several flexible regions in SaCas9 that could likewise serve as potential split sites (Figure 7F). We created three versions of a split-SaCas9, and two of them showed robust cleavage activity at the endogenous EMX1 target locus (Figure 7G). Using the best split design, we then tested inducible schemes based on the abscisic acid (ABA) sensing system (Liang et al., 2011), as well as two versions of the rapamycin-inducible FKBP/FRB system (Banaszynski et al., 2005) (Figures 7H and 7I). All three systems were able to support inducible SaCas9 cleavage activity, demonstrating the possibility of an inducible, split-SaCas9 design; however, further optimization is required to increase its efficiency and reduce its background activity (Figure 7J).
Discussion
The present SaCas9 complex structures allow a detailed structural comparison of Cas9 orthologs bound to nucleic acids, thereby illuminating the conserved structural features. SaCas9 and SpCas9 both adopt a bilobed architecture consisting of the REC and NUC lobes, with the guide:target heteroduplex accommodated between these lobes. In addition, both Cas9 orthologs have a phosphate lock loop, which participates in target DNA unwinding and heteroduplex formation. We also found that the HNH and RuvC domains are connected by the L1 and L2 linkers in both SaCas9 and SpCas9. These flexible linker regions appear to play a role in the inactive-to-active conformational transition of the HNH domain, although further structural and functional studies are required to elucidate the activation mechanism of Cas9 enzymes.
A comparison of the Cas9 orthologs also revealed the structural diversity among the CRISPR-Cas9 systems. In both SaCas9 and SpCas9, the structurally diverse REC and WED domains are responsible for the recognition of sgRNA scaffolds, which have diverse sequences and structures among the CRISPR-Cas9 systems, thereby enabling the orthogonal, species-specific recognition of the sgRNA scaffolds. In addition, the PI domains of SaCas9, SpCas9 and AnCas9 share a similar core fold, but possess different PAM-interacting residues, corresponding to their distinct PAM specificities.
We leveraged our newly obtained structural knowledge to develop SaCas9-based transcriptional activators and inducible SaCas9 systems. This opens the door to new possibilities, including the combinatorial use of SaCas9- and SpCas9-based genome editing and transcriptional regulation systems, to enable simultaneous, inducible editing, activation or repression of multiple endogenous loci. Further applications include the design of a minimal Cas9 enzyme with tailored PAM specificities as well as increased specificity for more versatile genome editing.
EXPERIMENTAL PROCEDURES
Detailed experimental procedures are described in the Supplemental Experimental Procedures.
The full-length S. aureus Cas9 N580A/C946A mutant (residues 1–1053) was expressed in Escherichia coli Rosetta 2 (DE3) (Novagen), and purified by chromatography on Ni-NTA Superflow (QIAGEN), Mono S (GE Healthcare) and HiLoad Superdex 200 16/60 (GE Healthcare) columns. The SeMet-labeled protein was expressed in E. coli B834 (DE3) (Novagen), and purified using a similar protocol as that for the native protein. The sgRNA was transcribed in vitro with T7 RNA polymerase, and purified by 8% denaturing (7 M urea) polyacrylamide gel electrophoresis. The target DNAs were purchased from Sigma-Aldrich. The purified SaCas9 protein was mixed with the sgRNA, target DNA strand and non-target DNA strand (molar ratio, 1:1.5:2.3:3.4), and the SaCas9–sgRNA–target DNA complex was purified by gel filtration chromatography on a Superdex 200 Increase column (GE Healthcare).
The purified SaCas9–sgRNA–target DNA complex (containing either the 5′-TTGAAT-3′ PAM or the 5′-TTGGGT-3′ PAM) was crystallized at 20°C by the hanging-drop vapor diffusion method. Crystals were obtained by mixing 1 μl of the complex solution (A260 nm, 15) and 1 μl of the reservoir solution (10–12% PEG 4,000, 0.75 M NaCl, 0.15 M Na2HPO4 and 0.15 M NaN3). The SeMet-labeled complex (containing the 5′-TTGGGT-3′ PAM) was crystallized under similar conditions. X-ray diffraction data were collected at 100 K on the beamlines BL32XU and BL41XU at SPring-8 (Hyogo, Japan). The structure was determined by the Se-SAD method, using the 3 Å resolution data set from the SeMet-labeled crystal. The final models of the 5′-TTGAAT-3′ PAM complex (2.6 Å resolution) and the 5′-TTGGGT-3′ PAM complex (2.7 ′ resolution) were refined using the native data sets. Molecular graphics images were prepared using CueMol (http://www.cuemol.org).
Around 24 h prior to transfection, human embryonic kidney 293FT cells (Life Technologies) were seeded into 24-well plates (Corning) at a density of 2.5 × 105 cells/well, and transfected at 70–80% confluency using Lipofectamine 2000 (Life Technologies), according to the manufacturer’s recommended protocol. A total of 600 ng DNA was used for each well of the 24-well plate. About 72 h after transfection, the genomic DNA was extracted, and then the genomic modifications were examined using the SURVEYOR assay and targeted deep sequencing, as previously described (Cong et al., 2013).
Supplementary Material
highlights.
Crystal structure of S. aureus Cas9 in complex with sgRNA and target DNA.
Structural basis for the relaxed recognition of the 5′-NNGRRT-3′ PAM.
Conserved and divergent structural features among orthologous CRISPR-Cas9 systems.
Rational design of compact transcriptional activators and inducible nucleases.
Acknowledgments
We thank Tomohiro Nishizawa, Takanori Nakane, Kazuki Kato, Naoki Matsumoto and Rhiannon Macrae for helpful comments on the manuscript. We thank the beam-line staffs at BL32XU and BL41XU of SPring-8, Japan, for assistance with data collection. H.N. is supported by JST, PRESTO, JSPS KAKENHI Grant Numbers 26291010 and 15H01463, and the Platform for Drug Discovery, Informatics, and Structural Life Science from the Ministry of Education, Culture, Sports, Science and Technology. W.X.Y. is supported by T32GM007753 from the National Institute of General Medical Sciences and a Paul and Daisy Soros Fellowship. F.A.R. is a Junior Fellow at the Harvard Society of Fellows. F.Z. is supported by an NIH Director’s Pioneer Award (1DP1-MH100706), the Keck, McKnight, Poitras, Merkin, Vallee, Damon Runyon, Searle Scholars, Klingenstein, and Simons Foundations, Bob Metcalfe, and Jane Pauley. The authors have no conflicting financial interests. O.N. is supported by the Basic Science and Platform Technology Program for Innovative Biological Medicine from the Japan Agency for Medical Research and Development, AMED, and the Council for Science, Technology and Innovation (CSTI), Cross-ministerial Strategic Innovation Promotion Program (SIP), “Technologies for creating next-generation agriculture, forestry and fisheries” (funding agency: Bio-oriented Technology Research Advancement Institution, NARO), and the Platform for Drug Discovery, Informatics, and Structural Life Science from the Ministry of Education, Culture, Sports, Science and Technology. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health. A patent application has been filed related to this work, and the authors plan to make the reagents widely available to the academic community through Addgene and to provide software tools via the Zhang lab Web site (www.genome-engineering.org).
Footnotes
ACCESSION NUMBERS
The atomic coordinates of the SaCas9–sgRNA–target DNA complexes have been deposited in the Protein Data Bank, with the PDB IDs 5CZZ (5′-TTGAAT-3′ PAM) and 5AXW (5′-TTGGGT-3′ PAM).
Supplemental Information includes Supplemental Experimental Procedures, DNA sequences of constructs, sgRNAs and PCR primers used in the study, as well as seven figures and one table, and can be found with this article online.
AUTHOR CONTRIBUTIONS
H.N. solved the crystal structures and performed in vitro cleavage experiments; L.C., W.X.Y., F.A.R., B.Z., and Y.L. performed in vivo functional analyses; A.K. assisted with the plasmid construction; R.I. assisted with the structural analysis; and H.N., L.C., W.X.Y., F.A.R., F.Z., and O.N. wrote the manuscript with help from all authors. H.N., F.Z. and O.N. directed and supervised all of the research.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski LA, Liu CW, Wandless TJ. Characterization of the FKBP•rapamycin•FRB ternary complex. J Am Chem Soc. 2005;127:4715–4721. doi: 10.1021/ja043277y. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Biertumpfel C, Yang W, Suck D. Crystal structure of T4 endonuclease VII resolving a Holliday junction. Nature. 2007;449:616–620. doi: 10.1038/nature06152. [DOI] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- Briner AE, Donohoue PD, Gomaa AA, Selle K, Slorach EM, Nye CH, Haurwitz RE, Beisel CL, May AP, Barrangou R. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol Cell. 2014;56:333–339. doi: 10.1016/j.molcel.2014.09.019. [DOI] [PubMed] [Google Scholar]
- Chylinski K, Le Rhun A, Charpentier E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 2013;10:726–737. doi: 10.4161/rna.24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chylinski K, Makarova KS, Charpentier E, Koonin EV. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:6091–6105. doi: 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Zhou R, Kuo YC, Cunniff M, Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat Commun. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, Le Rhun A, Chylinski K, Makarova KS, Lecrivain AL, Bzdrenga J, Koonin EV, Charpentier E. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:2577–2590. doi: 10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorecka KM, Komorowska W, Nowotny M. Crystal structure of RuvC resolvase in complex with Holliday junction substrate. Nucleic Acids Res. 2013;41:9945–9955. doi: 10.1093/nar/gkt769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. A Cas9–guide RNA complex preorganized for target DNA recognition. Science. 2015;348:1477–1481. doi: 10.1126/science.aab1452. [DOI] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang FS, Ho WQ, Crabtree GR. Engineering the ABA plant stress pathway for regulation of induced proximity. Sci Signal. 2011;4:rs2. doi: 10.1126/scisignal.2001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AV, Sternberg SH, Taylor DW, Staahl BT, Bardales JA, Kornfeld JE, Doudna JA. Rational design of a split-Cas9 enzyme complex. Proc Natl Acad Sci USA. 2015;112:2984–2989. doi: 10.1073/pnas.1501698112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Volz SE, Zhang F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat Biotechnol. 2015;33:139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.