ABSTRACT
Prokaryotes use type IV secretion systems (T4SSs) to translocate substrates (e.g., nucleoprotein, DNA, and protein) and/or elaborate surface structures (i.e., pili or adhesins). Bacterial genomes may encode multiple T4SSs, e.g., there are three functionally divergent T4SSs in some Bartonella species (vir, vbh, and trw). In a unique case, most rickettsial species encode a T4SS (rvh) enriched with gene duplication. Within single genomes, the evolutionary and functional implications of cross-system interchangeability of analogous T4SS protein components remains poorly understood. To lend insight into cross-system interchangeability, we analyzed the VirB8 family of T4SS channel proteins. Crystal structures of three VirB8 and two TrwG Bartonella proteins revealed highly conserved C-terminal periplasmic domain folds and dimerization interfaces, despite tremendous sequence divergence. This implies remarkable structural constraints for VirB8 components in the assembly of a functional T4SS. VirB8/TrwG heterodimers, determined via bacterial two-hybrid assays and molecular modeling, indicate that differential expression of trw and vir systems is the likely barrier to VirB8-TrwG interchangeability. We also determined the crystal structure of Rickettsia typhi RvhB8-II and modeled its coexpressed divergent paralog RvhB8-I. Remarkably, while RvhB8-I dimerizes and is structurally similar to other VirB8 proteins, the RvhB8-II dimer interface deviates substantially from other VirB8 structures, potentially preventing RvhB8-I/RvhB8-II heterodimerization. For the rvh T4SS, the evolution of divergent VirB8 paralogs implies a functional diversification that is unknown in other T4SSs. Collectively, our data identify two different constraints (spatiotemporal for Bartonella trw and vir T4SSs and structural for rvh T4SSs) that mediate the functionality of multiple divergent T4SSs within a single bacterium.
IMPORTANCE
Assembly of multiprotein complexes at the right time and at the right cellular location is a fundamentally important task for any organism. In this respect, bacteria that express multiple analogous type IV secretion systems (T4SSs), each composed of around 12 different components, face an overwhelming complexity. Our work here presents the first structural investigation on factors regulating the maintenance of multiple T4SSs within a single bacterium. The structural data imply that the T4SS-expressing bacteria rely on two strategies to prevent cross-system interchangeability: (i) tight temporal regulation of expression or (ii) rapid diversification of the T4SS components. T4SSs are ideal drug targets provided that no analogous counterparts are known from eukaryotes. Drugs targeting the barriers to cross-system interchangeability (i.e., regulators) could dysregulate the structural and functional independence of discrete systems, potentially creating interference that prevents their efficient coordination throughout bacterial infection.
INTRODUCTION
Occurring in Gram-negative, Gram-positive, and wall-less bacteria, as well as archaea, type IV secretion systems (T4SSs) are primarily utilized for translocating substrates across the cell envelope (1, 2). T4SSs that translocate plasmid (3) and naked DNA (4, 5), as well as genomic islands (6), are major facilitators of bacterial diversification, contributing to the spread of antimicrobial resistance and virulence genes. Other T4SSs translocate nucleoprotein (e.g., oncogenic T-DNA via the vir T4SS of Agrobacterium tumefaciens) or protein substrates directly into eukaryotic cells, wherein these effectors often perturb cell signaling to benefit bacterial survival (7, 8). Recently, it was reported that T4SSs may also be utilized to kill neighboring bacteria via translocation of a proteinaceous effector (9). Typically, T4SSs elaborate surface structures (i.e., pili or specialized adhesins) in a process either coupled to or independent of substrate translocation (10, 11). Thus, T4SSs are extraordinarily diverse in function relative to other bacterial secretion systems (12, 13).
Recently, T4SSs have been classified into eight major groups (14). One well-studied group, P-T4SSs, is typified by the vir T4SS of the pTi plasmid of A. tumefaciens, which encodes 11 scaffold components (VirB1 to VirB11) and a coupling protein (VirD4) that recruits substrates to the secretion channel (15). Tremendous architectural insight has been garnered from recent macromolecular structures generated for other P-T4SSs (16–20), allowing many of the various scaffold components to be arranged into an anatomical model (Fig. 1). Phylogenomics and bioinformatics efforts have shown that the other seven groups of T4SSs contain homologs or analogs to some (or most) of the components of this anatomical model (21), implying that a common T4SS architecture links the prokaryotic cytoplasm to the extracellular milieu. Variations on this theme, particularly regarding the distal region of the translocation channel, are thought to primarily be a consequence of coevolution with cell envelope morphology (22, 23). However, many structural innovations likely correlate with specific functions, e.g., mating pair stabilization during conjugation (24, 25) or pilus interactions with host cells during effector translocation (26).
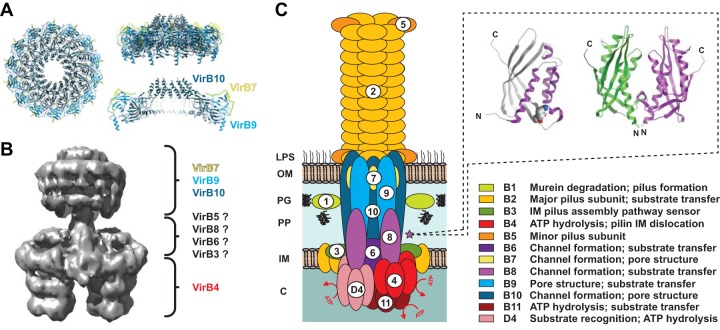
FIG 1 .
Architecture of P-type type IV secretions systems (P-T4SSs). (A) Bird’s-eye (left) and side (right) views of the dodecameric P-T4SS core complex (CC) encoded by plasmid pKM101 of Escherichia coli (PDB ID 3JQO), adapted from the work of Chandran et al. (17). Colors for the CC subunits (VirB7, VirB9, and VirB10) are similar to the model in panel C. (B) Negative-station electron microscopy-generated structure of the P-T4SS encoded by the E. coli R388 conjugative plasmid (EMD-2567) (adapted from the work of Low et al. [19]). Colors for the CC subunits and cytosolic/IM barrels (VirB4) are similar to the model in panel C. The putative positions of other IM channel (IMC) proteins are depicted with questions marks. VirB1, VirB2, VirB11, and VirD4 are not shown, as they were not included in the original structure. (C) General model of the composition of P-T4SSs, with functions for all 12 components (VirB1 to VirB11 and VirD4) listed at bottom right. The purple star depicts the bitopic VirB8 IMC proteins, with a dashed box illustrating the monomeric (left) and dimeric (right) structures for VirB8 C-terminal domains of Agrobacterium tumefaciens (PDB ID 2CC3) (44).
Bacterial genomes may encode multiple divergent T4SSs, e.g., the cag and com P-T4SSs of Helicobacter pylori (27, 28), the vir, vbh, and trw P-T4SSs of certain Bartonella species (29), and the dot/icm (I-T4SS) and lvh (P-T4SS) T4SSs of Legionella species (30, 31). These divergent T4SSs typically have different functions, though cross-system interchangeability is known for L. pneumophila dot/icm and lvh T4SSs. The lvh P-T4SS, which is dispensable for L. pneumophila replication in both amoebae and macrophage hosts (32) but causes virulence phenotypes under conditions that mimic the L. pneumophila aquatic phase (33), was shown to complement certain dot/icm mutants defective in conjugation (32). Furthermore, arrested virulence phenotypes in an L. pneumophila dotA/lvh double mutant were restored via complementation with the lvh coupling protein LvhD4 (34), implying structural and functional resilience in the face of extreme sequence divergence across dot/icm and lvh T4SSs. For other bacteria harboring multiple divergent T4SSs, studies on cross-system interchangeability are lacking, leaving a limited understanding of how distinct T4SSs achieve correct spatiotemporal assembly to execute their specific functions. Such precise regulatory mechanisms must exist, given also that many bacterial genomes are continually bombarded with integrative conjugative elements that often carry T4SS loci (35). If not quickly purged, it is likely that incoming T4SSs with compatibility to the native T4SS(s) either undergo rapid diversifying evolution or acquire mechanisms for differential regulation.
To gain further insight into T4SS cross-system interchangeability, we selected two bacterial genera with multiple divergent analogs of VirB8, a channel protein required for substrate transfer (Fig. 1). First, two or three distinct complete T4SSs are found within the genomes of some Bartonella species. Many of these species encode vir and trw secretion systems, which are highly divergent in sequence and function (36). Some species have an additional vbh T4SS homologous to the vir T4SS (29), wherein the biological function still remains elusive. The Bartonella vir T4SS is involved in protein secretion, with an arsenal of Bartonella effector proteins (Beps) translocated into host cells during infection (8). The trw T4SS is not involved in protein translocation but rather elaborates variable surface pili comprised of different combinations of duplicated VirB2 (TrwL) and VirB5 (TrwJ) homologs (37), with such structures shown to interact with host erythrocytes (38, 39). While these divergent T4SSs are differentially expressed during Bartonella host cell infection (40), the potential for cross-system interchangeability is currently unknown. Second, for species of Rickettsiales, an entirely different strategy has evolved with a single T4SS, the Rickettsiales vir homolog (rvh), which contains duplications of several components, including a VirB8 homolog (RvhB8) (23, 41). The functional significance of these duplicate rvh components is unknown, particularly regarding whether a single rvh T4SS functions during the rickettsial life cycle or if multiple different T4SSs are assembled throughout the complex life cycle (usually involving arthropod and vertebrate hosts) (42).
Herein we report crystal structures for six proteins of the VirB8 family: three VirB8 and two TrwG proteins of several Bartonella species, as well as the first crystal structure of an rvh scaffold component, RvhB8-II of Rickettsia typhi. Comparative structural analysis, in conjunction with biochemical assays, protein modeling, and bioinformatics, leads us to propose two distinct mechanisms (spatiotemporal for Bartonella trw and vir T4SSs and structural for rvh T4SSs) that prevent cross-system interchangeability between multiple divergent T4SSs encoded within single bacterial genomes.
RESULTS
Origin of multiple VirB8 proteins in Bartonella and Rickettsia genomes.
To understand the origin of multiple VirB8-like proteins within Bartonella and Rickettsia, we estimated a phylogeny of selected P-T4SSs. The Bartonella vir and vbh T4SSs share common ancestry, probably arising from whole-system duplication (Fig. 2). Previously, the Bartonella vir and vbh T4SSs were hypothesized to originate via lateral gene transfer (LGT), as they are absent from the ancestral human pathogen Bartonella bacilliformis (29, 36). The Bartonella trw T4SSs are found in a different clade, which includes plasmid-encoded T4SSs of gammaproteobacterial species (Fig. 2). Thus, the Bartonella trw T4SSs were likely acquired via LGT from non-alphaproteobacterial sources, becoming incorporated into the chromosomes, where they evolved to mediate adhesion to host erythrocytes (29, 36).
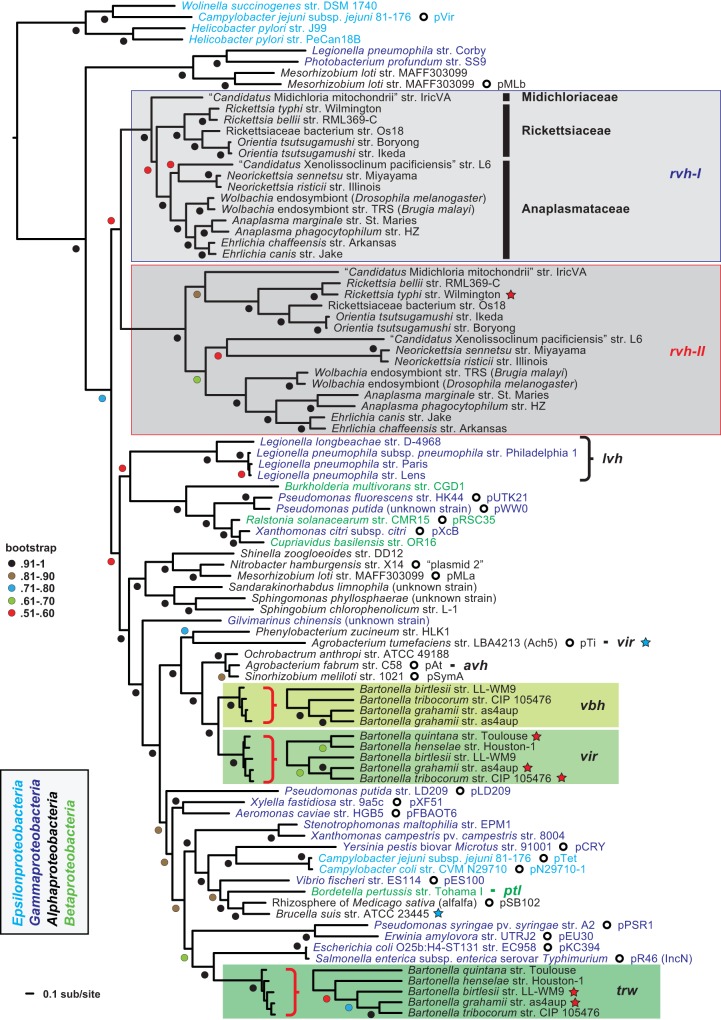
FIG 2 .
Phylogeny estimation of P-type type IV secretion systems (P-T4SSs). Phylogeny was estimated from concatenated alignments of five components (VirB4 and VirB8 to VirB11), except for rvhB-II, which contains homologs to VirB4, VirB8, and VirB9 only (see the text for further details on alignment and data set construction). ML-based phylogeny was estimated with RAxML on the unmasked alignment (LG + gamma + Ι). Branch support was assessed with 1,000 bootstrap pseudoreplications. A tree with specific bootstrap values, as well as additional phylogenies estimated from alternative alignments/models/optimality criteria, is provided in Text S1 in the supplemental material. The tree was rooted with four P-T4SSs from species of Epsilonproteobacteria. rvh-I and rvh-II T4SSs are in light and dark gray, respectively. The Bartonella vir, vbh, and trw T4SSs are colored different shades of green. Red stars depict sequences used to generate VirB8 structures in this study; blue stars depict sequences previously used to determine VirB8 structures. Plasmid-encoded T4SSs are depicted with open circles, with plasmid names provided. NCBI GenBank accession numbers for all proteins are provided in Text S1 in the supplemental material.
The rvh T4SS forms a lineage distinct from other P-T4SSs, though robust support for the relationship of this lineage to other P-T4SSs is lacking (Fig. 2; also see Text S1 in the supplemental material). We considered the paralogous genes encoding RvhB4, RvhB8, and RvhB9 to comprise different structural and functional machines, as previous phylogeny estimation supported these genes as ancient duplications that arose early in Rickettsiales evolution (23, 41). The genes encoding RvhB4-I, RvhB8-I, and RvhB9-I (collectively referred to as rvh-I) are conserved relative to analogs in other P-T4SSs and thus are anticipated to assemble into the rvh T4SS while it functions in protein translocation. Alternatively, the genes encoding RvhB4-II, RvhB8-II, and RvhB9-II (collectively referred to as rvh-II) have evolved atypical features that deviate substantially from counterparts in other P-T4SSs (see Discussion). The structural and functional significance of rvh-II is unknown, though a substantially higher divergence of rvh-II than rvh-I indicates different selective pressures operating on these paralogs (Fig. 2). Remarkably, this odd paralogy within the rvh T4SS is conserved across the three derived families of Rickettsiales (Rickettsiaceae, Anaplasmataceae, and “Candidatus Midichloriaceae”), which comprise a large, diverse assemblage of symbiotic and pathogenic species (43).
Divergent Bartonella VirB8 and TrwG proteins homodimerize via interactions at an NPXG motif highly conserved across T4SSs.
We determined crystal structures of the soluble C-terminal periplasmic domains of three VirB8 and two TrwG proteins of several Bartonella species (see Text S2 in the supplemental material). As expected by the highly conserved sequences of C-terminal periplasmic domains of Bartonella VirB8 (~90%) and TrwG (~90%) proteins (data not shown), the structures within each protein family are highly similar, with 5 α-helices and 4 β-sheets (Fig. 3A and B). Within Bartonella VirB8, the pairwise root mean square deviation (RMSD) values, or measures of the average distance between the atoms of superimposed proteins, range from 0.33 to 0.60 Å for similar Cα atoms, and within Bartonella TrwG, the two structures have a pairwise RMSD value of 0.88 Å. The structures of Bartonella VirB8 and TrwG proteins are also highly similar to each other (Fig. 3C), despite extremely low sequence conservation (~28% identity overall, ~34% identity for C-terminal periplasmic domains) (Fig. 3D; see Text S3 in the supplemental material). The pairwise RMSD values between Bartonella VirB8-TrwG sets range from 1.57 to 1.78 Å. The Bartonella VirB8 and TrwG protein structures are also similar to the Agrobacterium tumefaciens (44) (RMSD ranges of 1.28 to 1.33 Å and 1.52 to 1.63 Å for VirB8 and TrwG, respectively) and Brucella suis (45) (RMSD ranges of 1.73 to 1.78 Å and 0.98 to 1.18 Å for VirB8 and TrwG, respectively) VirB8 protein structures (Fig. 3C). This implies remarkable structural constraints for VirB8 components in the assembly of functional P-T4SSs.
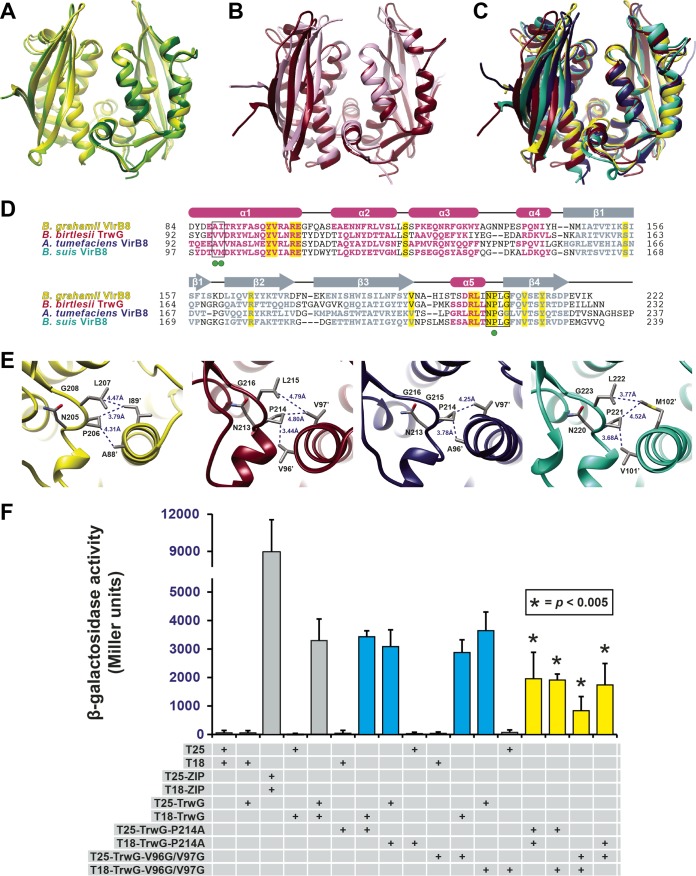
FIG 3 .
Bartonella VirB8 and TrwG proteins form structurally conserved homodimers. (A) Superimposition of the ribbon representations of the VirB8 crystal structures from B. quintana strain Toulouse (dark green) (PDB ID 4LSO), B. grahamii strain as4aup (yellow) (PDB ID 4KZ1) and B. tribocorum strain CIP 105476 (light green) (PDB ID 4MEI). (B) Superimposition of the ribbon representations of the TrwG crystal structures from B. birtlesii strain LL-WM9 (burgundy) (PDB ID 4JF8) and B. grahamii strain as4aup (pink) (PDB ID 4NHF). (C) Superimposition of the ribbon representations for the crystal structures of B. grahamii VirB8, B. birtlesii TrwG, Agrobacterium tumefaciens strain C58 VirB8 (dark blue) (PDB ID 2CC3) (44), and Brucella suis biovar 1 (strain 1330) VirB8 (aquamarine) (PDB ID 2BHM) (45). (D) Sequence alignment of VirB8/TrwG proteins and secondary structure assignment. Sequences were extracted from a larger alignment (see Text S4 in the supplemental material). Sequences are the globular domains depicted in panel C. For each protein, residues involved in the major dimerization site are boxed. Invariant residues are highlighted in yellow. Magenta bars and gray arrows depict the α-helices and β-strands for the VirB8/TrwG structure, with colored residues in the proteins corresponding to these structural features. Green dots mark the residues mutated within the dimerization interface of B. birtlesii TrwG. (E) High-resolution depiction of the major dimerization sites for the proteins illustrated in panels C and D. From left to right: B. grahamii VirB8, B. birtlesii TrwG, A. tumefaciens VirB8, and B. suis VirB8. (F) Analysis of B. birtlesii TrwG-TrwG interactions using the bacterial two-hybrid system. Different plasmid combinations were transformed into adenylate cyclase deficient and cAMP-specific-phosphodiesterase-deficient E. coli strain APE304. After overnight growth in liquid Luria-Bertani medium, β-galactosidase activity was measured and calculated (in Miller units). T25-ZIP and T18-ZIP are positive-interaction control plasmids encoding dimer-forming yeast transcription factor GCN4 (89). Values are from three independent experiments, all analyzed in triplicate. Blue bars depict wild-type-mutant interactions, while yellow bars depict mutant-mutant interactions.
With the exception of monomeric Bartonella quintana VirB8, all of the Bartonella VirB8 and TrwG proteins crystallized as dimers. Similar to the A. tumefaciens and B. suis structures, the dimerization interfaces of Bartonella VirB8 and TrwG structures contain major and minor dimerization sites. The major dimerization site involves helix α1 from one subunit contacting the sharp turn between helix α5 and strand β4 of the other subunit (Fig. 3E). The latter motif, here referred to as the NPXG motif, is the most conserved region of proteins of the VirB8 family and was originally suggested to be critical for VirB8 dimerization (45). Bacterial 2-hybrid (B2H) experiments with selected full-length VirB8 and TrwG proteins (Fig. 3F), as well as glutaraldehyde cross-linking experiments with the corresponding soluble periplasmic domains (see Fig. S1 in the supplemental material), further indicated that a homodimer is the probable functional unit for Bartonella VirB8 and TrwG proteins. To assess the importance of the major dimerization site within the subunit interface, we carried out site-directed mutagenesis of Bartonella birtlesii TrwG to yield mutants in helix α1 (TrwGV96G/V97G) and the NPXG motif (TrwGP214A), which were utilized in a series of B2H assays (Fig. 3F). Both mutations had negligible effects on dimerization when analyzed individually with the wild-type TrwG (TrwGwt) protein (Fig. 3F). However, when both of the fusion constructs contained the mutations, we detected a significant reduction in dimerization (Fig. 3F). Thus, helix α1 and the NPXG motif are central (but not the sole) mediators of B. birtlesii TrwG dimerization and probably function similarly for most other VirB8-like proteins of P-T4SSs, given the high degree of conservation of these structural features.
Bartonella VirB8 and TrwG proteins are capable of forming heterodimers.
The strong structural conservation across Bartonella VirB8 and TrwG proteins suggests that individual VirB8 and TrwG monomers could form a heterodimer. A structural model of the Bartonella grahamii VirB8/TrwG heterodimer illustrates how such a dimer interface might form, with the overall symmetry similar to the VirB8 and TrwG homodimers (Fig. 4A). The formation of this dimerization interface is undoubtedly facilitated by the high degree of sequence conservation of contacting residues within helix α1 and the NPXG motif, four of which are invariant across VirB8 and TrwG sequences (Fig. 4B). Using the B2H assay, we analyzed the potential cross-system interchangeability between B. grahamii VirB8 and TrwG (Fig. 4C). We initially established a reference of interaction strength for B. birtlesii and B. grahamii TrwG homodimers, as well as for the B. grahamii VirB8 homodimer. TrwG interaction signals were much higher than VirB8 homodimerization, possibly reflecting the known biological functions of these divergent systems, i.e., rigid adhesive function of the trw T4SS compared to dynamic effector translocation of the vir T4SS (36). Subsequently, we evaluated the potential for heterodimeric interactions between B. grahamii VirB8 and TrwG proteins. These assays indicate the ability of VirB8 and TrwG to form heterodimers (Fig. 4C), with a strength of interactions not significantly lower than that observed for the B. grahamii VirB8 homodimer. Thus, it appears that in the absence of spatiotemporal barriers (e.g., differential expression), VirB8 and TrwG proteins have the potential to interfere with one another during assembly into their respective T4SSs.
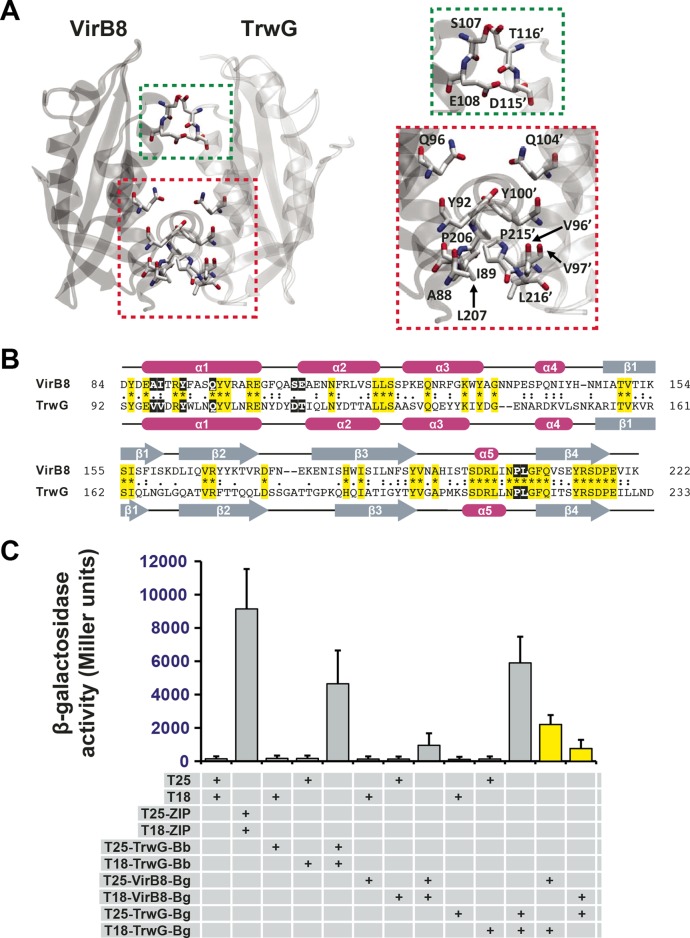
FIG 4 .
In vitro heterodimerization of Bartonella VirB8 and TrwG proteins. (A) Ribbon representation of a modeled heterodimer comprised of the VirB8 (PDB ID 4KZ1) and TrwG (PDB ID 4NHF) crystal structures from B. grahamii strain as4aup. Regions involved in dimerization are boxed and enlarged at right. Green indicates a minor dimerization site involving residues within the loop between α-helices α1 and α2. Red indicates a major dimerization site involving residues of α-helix α1 and the NPXG motif. TrwG residues are denoted with primes. (B) Sequence alignment of B. grahamii VirB8 and TrwG proteins, with secondary structure assignment. Sequences were extracted from a larger alignment (see Text S4 in the supplemental material). Sequences are the globular domains depicted in panel A. For each protein, residues involved in the dimerization interface are within black boxes. Invariant residues are highlighted in yellow. Magenta bars and gray arrows depict the α-helices and β-strands for the VirB8 and TrwG structures. (C) Analysis of B. birtlesii strain LL-WM9 and B. grahamii TrwG-TrwG interactions, the B. grahamii VirB8-VirB8 interaction, and the B. grahamii VirB8-TrwG interaction using the bacterial two-hybrid system. See the legend to Fig. 3F and the text for descriptions of the bacterial two-hybrid assay. Yellow bars depict the interactions tested for the B. grahamii VirB8-TrwG heterodimer.
Duplicate, coexpressed VirB8 proteins encoded within the Rickettsia typhi genome are structurally divergent.
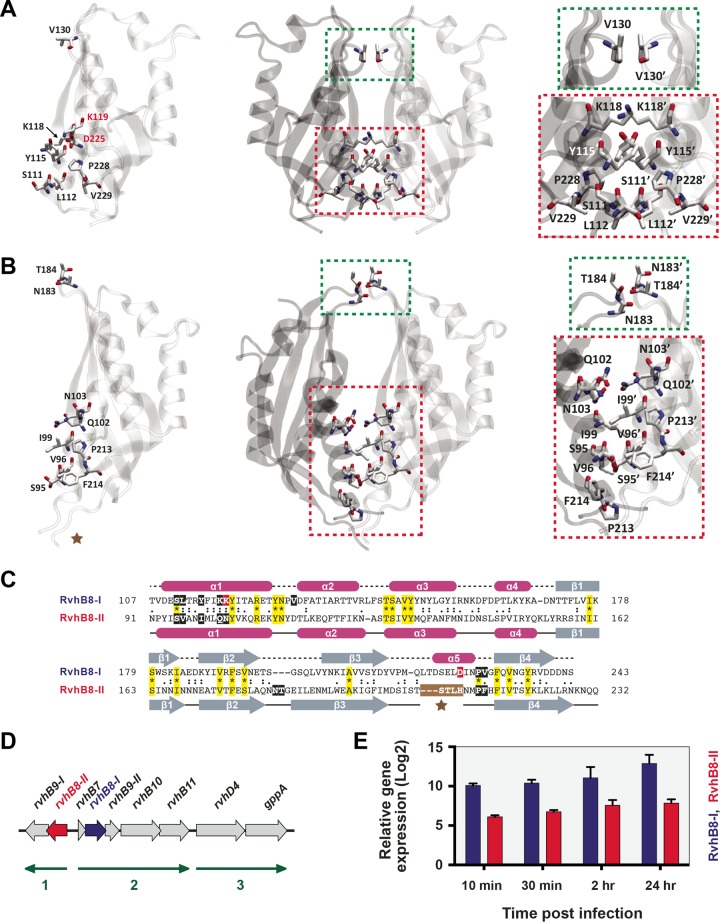
To lend insight into the duplicate VirB8-like proteins encoded within Rickettsia genomes, we set out to determine the crystal structures for these divergent proteins from the murine typhus agent, R. typhi strain Wilmington (RvhB8-I and RvhB8-II, which share only 18% amino acid identity). Attempts to generate a crystal structure for RvhB8-I failed; thus, we modeled both the monomer and dimer of this protein by homology, using a multitemplate approach (Fig. 5A). Because RvhB8-I contains residues conserved in other VirB8 family proteins, such as the NPXG motif (41), its sequence could be reasonably modeled to the Bartonella spp., B. suis, and A. tumefaciens VirB8 structures, as well as the Bartonella TrwG structures. Thus, the RvhB8-I structure is anticipated to fold similarly to these proteins. The protomeric structure of R. typhi RvhB8-I is quite similar to that of Bartonella VirB8 and TrwG proteins, with pairwise RMSD values of 1.46 to 1.64 Å. In contrast, the biological dimeric structure determined for R. typhi RvhB8-II deviates substantially from the RvhB8-I model and all seven VirB8/TrwG dimeric structures, with pairwise RMSD values of 3.20 to 3.79 Å (Fig. 5B). We previously observed that the lack of conservation in the NPXG motif of RvhB8-II, coupled with rapidly evolving sites throughout the protein, indicated a possible divergent structure and function for this protein (41). All other VirB8 and TrwG T4SS proteins with known dimeric structures exhibit the NPXG motif posterior to helix α5 that interacts with helix α1′ of the other protomer. The RvhB8-II sequence differs substantially in this region, and in the structure, this region does not form helix α5. Part of this region is disordered in the crystal structure; one potential explanation could be in situ cleavage of this region during crystallization, although mass spectrometry analysis of the protein stock solution revealed an intact protein (~21 kDa) prior to crystallization (see Text S2 in the supplemental material). The divergent sequence and lack of helix α5 perturbs the major dimerization site to generate an asymmetrical subunit interface, unlike other VirB8 and TrwG structures that had symmetric dimers. To compensate for disruption of the major dimerization site, a minor dimerization site is observed involving residues between β-strands β2 and β3 (Fig. 5C). Glutaraldehyde cross-linking experiments with the soluble periplasmic domains of RvhB8-I and RvhB8-II demonstrated that both proteins form homodimers in solution (see Fig. S1 in the supplemental material). Thus, despite a divergent dimeric structure adopted by RvhB8-II, both VirB8-like proteins of R. typhi appear to form functional dimers.
FIG 5 .
Rickettsia typhi expresses two structurally divergent VirB8-like proteins. (A) Structural model for R. typhi RvhB8-1 (RT0280; YP_067242). (Left) RvhB8-I monomer in ribbon representation with nine residues involved in the dimerization interface shown in stick representation. For clarity, residues colored red are not shown in the dimer. (Center) RvhB8-I dimer in ribbon representation with seven residues involved in the dimerization interface shown in stick representation. Green indicates a minor dimerization site involving residues within the loop between α-helices α1 and α2. Red indicates a major dimerization site involving residues of α-helix α1 and the NPXG motif. (Right) Higher magnification of the RvhB8-I subunit interface. (B) Ribbon representation for the monomer and dimer of the crystal structure (PDB ID 4O3V) of R. typhi RvhB8-II (RT0278; YP_067240). Depiction of dimerization scheme follows the layout shown for RvhB8-I in panel A, except that the minor dimerization site (green) involved residues between β-strands β2 and β3. The brown star denotes the break in the structure. (C) Sequence alignment of RvhB8-I and RvhB8-II proteins and secondary structure assignment. Sequences are those of the globular domains depicted in panels A and B. For each protein, residues involved in the dimerization interface are within black (or red) boxes. Invariant residues are highlighted yellow. Magenta cylinders and gray arrows depict the α-helices and β-strands, respectively, for structures shown in panels A and B. For RvhB8-II, the brown star denotes the break in the structure, with missing residues colored white. (D) rvhB8-I and rvhB8-II are arrayed in tandem operons within the R. typhi genome. The schema shows nucleotide coordinates 351539 to 360917 from the R. typhi strain Wilmington genome (NC_006142) (46). Nine genes are encoded within three predicted operons: 1, rvhB9-I and rvhB8-II (red); 2, rvhB7, rvhB8-I (blue), rvhB9-II, rvhB10, and rvhB11; 3, rvhD4 and gppA (guanosine-5′-triphosphate,3′-diphosphate pyrophosphatase). The direction of transcription for each operon is shown with green arrows. Operons were predicted with fgenesb (90). Other rvh genes are encoded in separate clusters within the R. typhi genome (41). (E) Expression of the R. typhi rvhB8-I and rvhB8-II genes during early host cell infection. RNA was extracted from HeLa cells infected with R. typhi, and gene expression of RT0280 (encoding RvhB8-I) and RT0278 (encoding RvhB8-II) was measured by reverse transcription-quantitative PCR (RT-qPCR). Gene expression was normalized to R. typhi reference genes adr1 and sca5 (2ΔCT). Infections were repeated in triplicate with technical duplicate readings for RT-qPCR. Values are means and standard errors of the means.
The significance of duplicate VirB8-like proteins carried by all species of Rickettsia is unclear. The arrangement of rvhB8-I and rvhB8-II in the R. typhi genome is odd (46), with the genes found in adjacent operons that encode other rvh genes (Fig. 5D). The conservation of these loci across all sequenced Rickettsia genomes implies a conserved function (41). Reverse transcription-quantitative PCR (RT-qPCR) analysis of R. typhi throughout one day of host cell infection indicates that rvhB8-I and rvhB8-II genes are simultaneously expressed (Fig. 5E). Whether RvhB8-I and RvhB8-II both assemble into the same rvh secretion machine remains unknown. A modeled RvhB8-I/RvhB8-II heterodimer suggests that such an interaction would be highly divergent from other VirB8 and TrwG homodimers, as well as the VirB8/TrwG heterodimer (see Text S4 in the supplemental material). Aside from an entirely skewed major dimerization site, there are no residues contacting across the minor dimerization site within the modeled heterodimer. Supporting these observations, B2H assays failed to detect interactions between RvhB8-I and RvhB8-II (data not shown). Furthermore, the N-terminal cytoplasmic and transmembrane-spanning (TMS) regions of RvhB8-I and RvhB8-II are extraordinarily divergent from one another (41). Collectively, if RvhB8-I and RvhB8-II both simultaneously assemble into one rvh T4SS, there is little support for these proteins forming heterodimers.
Divergent RvhB8-II proteins are conserved across species of Rickettsiales but not found in other bacteria.
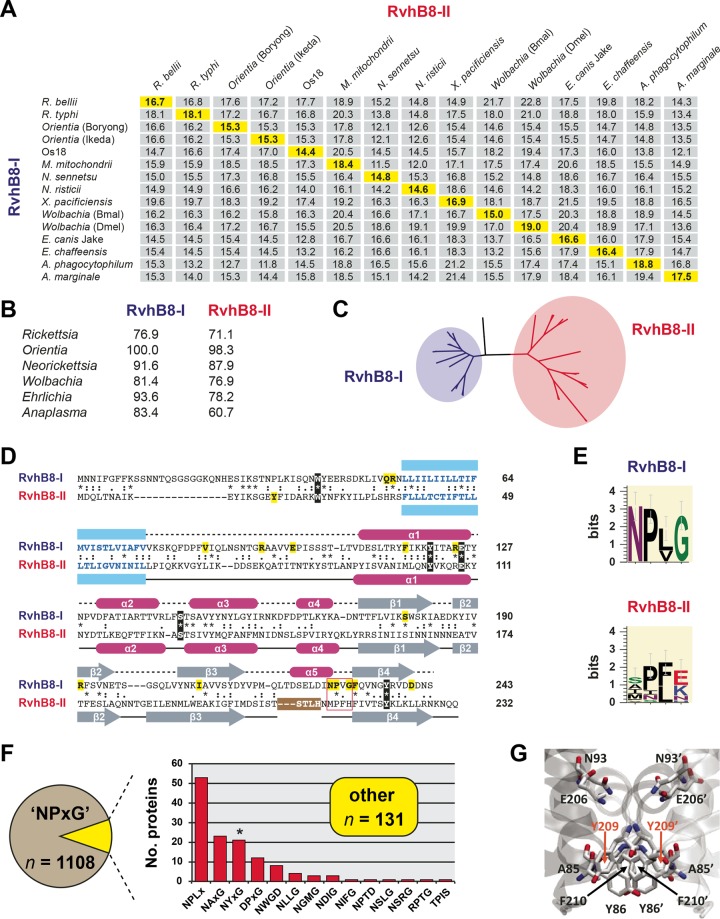
The presence of two VirB8-like genes in all species of Rickettsiales that carry the rvh T4SS prompted us to evaluate if the odd R. typhi RvhB8-II characteristics were present in other RvhB8-II proteins. Analysis of selected rickettsial species revealed that RvhB8-I and RvhB8-II proteins encoded within the same genome never share more than 19% amino acid identity (Fig. 6A, yellow highlighting). Across species within the six major rickettsial genera, RvhB8-I is more conserved relative to RvhB8-II (Fig. 6B). This is further witnessed by phylogeny estimation of RvhB8 proteins, which shows substantially higher divergence of RvhB8-II than RvhB8-I (Fig. 6C). Collectively, these data indicate that an ancient duplication of rvhB8 quickly led to diversification of rvhB8 paralogs under different selective constraints. Considering these results with the similar divergence patterns witnessed for RvhB4 and RvhB9 paralogs (Fig. 2), it is probable that rvh-I and rvh-II contribute differently to the function and regulation of the rvh T4SS.
FIG 6 .
Conserved RvhB8-I and RvhB8-II paralogs are highly divergent from one another. (A) Pairwise divergence between RvhB8-I and RvhB8-II proteins from select rickettsial species. Numbers are amino acid identity (percent) as calculated across a global RvhB8 alignment (see the text for details). Highlighted values on the diagonal depict divergences between paralogs encoded within the same genome. Full species names and NCBI GenBank accession numbers for all proteins are provided in Text S1 in the supplemental material. (B) Across species and strains within the same genus, RvhB8-I is more conserved than RvhB8-II. Numbers are amino acid identity (percent) as described for panel A. Complete percent identity matrices used to estimate protein divergence are provided in Text S5 in the supplemental material. (C) Phylogeny estimation of RvhB8 proteins reveals higher divergence within the RvhB8-II clade than the RvhB8-I clade. ML-based phylogeny was estimated with RAxML on the unmasked global RvhB8 alignment (WAG + gamma + Ι). A complete tree, as well as phylogenies estimated from the masked alignment with other substitution models, is provided in Text S5 supplemental material. (D) Comparison of R. typhi RvhB8-I and RvhB8-II proteins. Sequences were aligned according to structure in SPDBV using “magic fit” followed by “improved fit” algorithms. The alignment shows predicted (top) and solved (bottom) structures for RvhB8-I and RvhB8-II, respectively. Predicted transmembrane-spanning regions (76) are colored blue. Five residues conserved across all RvhB8 proteins are in black, with residues conserved only in RvhB8-I (n = 15) or RvhB8-II (n = 1) highlighted in yellow (see Text S5 in the supplemental material). The NPXG motif is in a red box (see the description of panel E below). For RvhB8-II, the region of proteolysis (STLH) that occurred during crystallization is in a brown box. (E) Composition of the NPXG motif across 15 RvhB8-I (top) and 15 RvhB8-II (bottom) proteins. Sequence logos were generated using WebLogo v.3.3 (77). (F) Analysis of the conservation of the NPXG motif across 1,239 nonredundant proteobacterial VirB8 proteins (excluding Rickettsiales). Proteins lacking the conserved NPXG motif (10.6%) were placed in 14 categories based on their alternative sequences and ranked by their frequency (see Text S4 in the supplemental material for structural modeling of proteins within each category). (G) Example of a canonical interaction across the NPXG motif for Yersinia pestis biovar microtus strain 91001 (NP_995427), which contains the alternative sequence NYFG.
Comparison of full-length RvhB8 paralogs across selected rickettsial species provides further insight into the pattern of rvhB8 diversification (Fig. 6D). Four residues within the periplasmic C-terminal domain are conserved across RvhB8-I and RvhB8-II proteins, as they are in most other VirB8 proteins. One of these residues, a Tyr within β-strand β4, has been implicated in an interaction with VirB4 (47), suggesting that both RvhB8-I and RvhB8-II have the potential to interact with one or both of the RvhB4 paralogs. A Trp residue within the cytoplasmic domain is also conserved across RvhB8-I and RvhB8-II proteins. The functional significance of this Trp residue is unknown, though it demarcates the highly variable N-terminal sequences from a conserved cytoplasmic region proximal to the predicted TMS region (see Text S5 in the supplemental material). RvhB8-I proteins share 15 conserved residues; ten of these are found in the globular C-terminal domain and are also highly conserved across VirB8 proteins from other bacteria. In contrast, RvhB8-II proteins share only one conserved residue, a Tyr residue within the N-terminal cytoplasmic domain. Collectively, RvhB8-I proteins are conserved relative to other VirB8 proteins, while RvhB8-II proteins have retained only a minimal set of the conserved residues that define the VirB8 structural architecture.
Like R. typhi RvhB8-II, the other rickettsial RvhB8-II proteins contain a divergent NPXG motif (Fig. 6E). Specifically, the Asn and Gly residues are replaced with other residues that are not conserved across RvhB8-II homologs, with the Pro residue present only in ~60% of the proteins. This affects the interactions within the major dimerization site, suggesting that all RvhB8-II proteins form atypical dimer interfaces relative to the conserved VirB8 structure. To ascertain how conserved the NPXG motif is and, specifically, whether there is flexibility in this motif in other VirB8 proteins, we evaluated this region in 1,239 nonredundant VirB8 proteins. Only ~10% of these proteins contained divergent NPXG motifs, which were assembled into groups based on the primary sequence of their respective NPXG motifs (Fig. 6F). We then modeled the monomer and dimer structures for at least one representative from each of these groups (see Text S4 in the supplemental material). All of these predicted structures involved conserved contacts within the major dimerization site typical of VirB8 proteins but not RvhB8-II. Even replacement of the Pro residue with bulky aromatic residues does not alter the dimerization interface (Fig. 6G). Altogether, these observations suggest that the structure of RvhB8-II proteins is atypical relative to other VirB8 proteins, with the deviant NPXG motif skewing the dimerization interface, resulting in an asymmetrical dimer.
DISCUSSION
In this study, we set out to identify potential barriers to within-genome T4SS cross-system interchangeability, focusing on the vir and trw T4SSs carried by Bartonella species and the duplication-laden rvh T4SS of Rickettsiales species. Our data imply that these bacteria either rely on robust spatiotemporal regulation or structural constraints to maintain the cooccurrence of these analogous multiprotein complexes.
For Bartonella VirB8 and TrwG proteins, which share overall ~28% identity, the determined structures are highly similar to VirB8 structures previously determined for A. tumefaciens (44) and B. suis (45) (Fig. 3A to D). With the exception of monomeric B. quintana VirB8, all of the Bartonella VirB8 and TrwG proteins crystallized as dimers. Using site-directed mutagenesis of residues within the dimer interface, we demonstrate by B2H assays that contacts between helix α1 of one subunit and the NPXG motif of the other subunit are important for dimerization of B. birtlesii TrwG (Fig. 3F). It is probable that this dimer interface area functions similarly in most other VirB8-like proteins of P-T4SSs, given the high conservation of the NPXG motifs. Strikingly, TrwGwt-TrwGV96G/V97G and TrwGwt-TrwGP214A protein pairs had a strength of interaction that was indistinguishable from those formed by the wild-type proteins. Also, the interaction signal was not completely abolished when mutations were introduced to both of the interacting proteins. These data indicate either that a certain degree of flexibility is allowed across the B. birtlesii TrwG dimer interface or that other areas outside the periplasmic domain of B. birtlesii TrwG, such as the TMS region, are important for the dimerization process. Indeed, an M102R mutation in B. suis VirB8 (Met102 corresponds to the Val97 of B. birtlesii TrwG) (Fig. 3E) abolished the homodimer formation when soluble periplasmic domains were analyzed but had only a partially attenuating effect in an intracellular macrophage survival assay that employed full-length VirB8 (47). The partial M102R mutation-mediated reduction of B. suis VirB8 homodimer formation has also been witnessed in B2H assays with full-length proteins (48). Taken together, our data indicate that the periplasmic domains of VirB8 proteins, specifically via the NPXG motif, are important but not the sole determinants of dimerization.
To further explore the VirB8 dimerization process, we used B2H assays to assess the possibility that the structurally conserved B. grahamii VirB8 and TrwG form heterodimers (Fig. 4C). These assays indicate that, while weaker than TrwG-TrwG interactions, TrwG-VirB8 interactions are not significantly different in strength from VirB8-VirB8 interactions. A generated structural model of the B. grahamii TrwG-VirB8 heterodimer also indicated a similar dimerization interface relative to TrwG and VirB8 homodimers (Fig. 4A). Our data suggest that Bartonella VirB8 and TrwG proteins have the potential to interfere with one another during assembly into their respective T4SSs. It is therefore remarkable that vir and trw T4SSs are under robust spatiotemporal regulation (40). The stringent response factors SpoT and DksA were shown to mediate regulation of the vir T4SS during early infection, with expression of vir and bep loci requiring the dual activation of the alternative sigma factor RpoH1 and the BatR/BatS two-component system (40). Remarkably, SpoT, DksA, and BatR/BatS were shown to negatively regulate the trw T4SS, possibly by moderating the KorA/KorB negative regulator, which is known to control trw expression (37). Taken together, spatiotemporal regulation of vir and trw T4SSs (40) likely prevents cross-system interchangeability, which was shown to be structurally possible at the level of VirB8/TrwG in our current study.
Regarding the rvh T4SS, we observed an entirely different relationship between its two VirB8-like proteins, which share ~16% amino acid identity across selected Rickettsiales species (Fig. 6A). While the sequence of RvhB8-I can be modeled to the structures of VirB8 and TrwG (Fig. 5A), the crystal structure of RvhB8-II revealed a markedly different organization of the dimerization interface. The region equivalent to the NPXG motif is largely disordered and leads to a different arrangement of contacting residues in the major dimerization site (Fig. 5B). Furthermore, relative to other VirB8 and TrwG structures, a unique minor dimerization site is formed across residues between β-strands β2 and β3 in RvhB8-II. This odd structure for RvhB8-II confirms our previous observation that a lack of conservation within the NPXG motif would likely perturb the monomer structure and dimer interface (23, 41). Despite this, the C-terminal domains of both RvhB8-I and RvhB8-II form homodimers in solution (see Fig. S1 in the supplemental material), indicating that each paralog likely forms a functional dimer. This is supported by higher conservation within RvhB8 paralogs (Fig. 6B; see Text S5 in the supplemental material) versus across RvhB8 paralogs (Fig. 6A). A modeled RvhB8-I/RvhB8-II heterodimer (see Text S4 in the supplemental material), combined with a failure to detect interactions between RvhB8-I and RvhB8-II via B2H assays (data not shown) and extraordinary sequence divergence in the N-terminal cytoplasmic/TMS regions across RvhB8 paralogs (see Text S5 in the supplemental material), indicates the unlikely ability of RvhB8-I and RvhB8-II to heterodimerize. Altogether, these data imply that, while highly divergent from one another, RvhB8-I and RvhB8-II proteins are strongly conserved in Rickettsiales genomes, illustrating a bizarre architecture of the rvh T4SS relative to the canonical vir archetypal T4SS.
Unlike the spatiotemporal regulation of Bartonella vir and trw T4SSs, we detected simultaneous expression of rvhB8-I and rvhB8-II genes during R. typhi infection of host cells (Fig. 5E). This suggests that, despite being located in different loci (Fig. 5D), both RvhB8 paralogs are coregulated. While regulators of the Rickettsia rvh T4SS are currently unknown, studies on other rickettsial genera have identified factors that bind the promoters of multiple rvh loci, which are scattered in islets similar to Rickettsia genomes (23). In the case of Ehrlichia chaffeensis, the EcrX regulator coordinately drives expression of all five rvh loci throughout host cell infection, indicating synchronous expression of both RvhB8 paralogs during rickettsial intracellular development (49). For the Wolbachia endosymbiont of Brugia malayi, wBmxR1 and wBmxR2 (homologs of EcrX) were shown to bind upstream of rvhB9-I and rvhB4-II, respectively, with both factors also binding upstream of a larger rvh locus (ribA-rvhB8-I-rvhB9-II-rvhB10-rvhB11-rvhD4-wsp) (50). Within the latter locus, it was determined that rvhB8-I was expressed in an operon with ribA (GTP cyclohydrolase-2), with RvhB8-I detected in B. malayi lysates. A regulator of the rvhB8-II locus was not determined. Additional studies on various rickettsial species have identified gene and protein expression for both RvhB8 paralogs (41), indicating conserved roles for both proteins in rvh T4SS assembly and function.
Despite coexpression, whether RvhB8-I and RvhB8-II both assemble into the same rvh secretion machine remains unknown. As it is probable that the rvh genes in different rickettsial species are uniquely regulated to orchestrate their specific lifestyles and adapt to their specific intracellular niches, it is tempting to speculate that rvh gene duplication underlies stage- and/or host-specific assembly of different rvh components. However, two factors do not favor this hypothesis. First, like RvhB8-II, other Rvh-II proteins (RvhB4-II and RvhB9-II) contain atypical characteristics that should prevent their contribution to functional secretion. RvhB4-II proteins lack conserved ATPase active sites that are known to be critical for VirB4 function (51–53), casting doubt on their ability to provide energy for secretion. The C-terminal domains of RvhB9-II proteins either are absent (Rickettsiaceae) or lack conserved residues relative to VirB9 proteins (Anaplasmataceae and “Candidatus Midichloriaceae”). As the VirB9 C-terminal domain interacts with VirB7 and the C-terminal domain of VirB10 in the dodecameric core complex (16, 17, 54) and also participates in transient interactions with other components (VirB1 and VirB8 to VirB11) (10, 55, 56), it is unlikely that RvhB9-II proteins contribute to core complex formation. Second, the rvh-II loci are not linked in a manner that indicates their efficient and independent regulation relative to the other rvh loci. rvhB8-I and rvhB9-II loci are adjacent within a large rvh islet that includes rvhB10, rvhB11, and rvhD4, while rvhB8-II and rvhB9-I are found in one distinct operon (Rickettsia and Neorickettsia species) or present as separate transcriptional units (all other derived Rickettsiales) (23). Furthermore, the rvhB4-II locus is well separated from all of these loci. Collectively, these characteristics of rvh gene duplication make it difficult to envision a scenario where Rvh-II proteins assemble into a functional T4SS independent of Rvh-I proteins.
With their strict conservation across the derived rickettsial families and strikingly higher rate of evolution than Rvh-I proteins (Fig. 2), it is likely that the functions of Rvh-II proteins are linked. It is also probable that these proteins assemble in some fashion into the rvh T4SS. Despite defective ATPase active sites, RvhB4-II proteins are similar in size to RvhB4-I and other VirB4 proteins. Curiously, the N-terminal domains of RvhB9-II proteins are conserved, indicating that they may make contacts with RvhB4 proteins given that the N-terminal domain of VirB9 interacts with VirB4 in the periplasm near the inner membrane (18). For RvhB8-II proteins, dimers may occupy half of the inner membrane channel (IMC) in conjunction with RvhB8-I dimers, perhaps with an as-yet-unknown interaction across these complexes. In accordance with other P-T4SSs, it can be expected that 12 RvhB8 proteins occupy the IMC, as revealed in the electron-microscopic structure of the R388 P-T4SS VirB3-VirB10 complex (19). However, recent structures for VirB8-like proteins from two conjugative T4SSs of Gram-positive bacterial species, TcpC of Clostridium perfringens (57) and TraM of Enterococcus faecalis (58), revealed trimers as functional units, indicating that the manner by which VirB8 proteins oligomerize in the IMC differs across divergent T4SSs. Furthermore, for I-T4SSs, recent crystal structures of DotI (L. pneumophila) and TraM (IncI plasmid R64) revealed octomers and hexamers, respectively, with DotI also oligomerizing with its truncated paralog DotJ (59). Curiously, like RvhB8-II, the structures of DotI, TraM (R64), TcpC, and TraM (E. faecalis) all lack helix α5, which is highly conserved in RvhB8-I, VirB8, and TrwG structures. Thus, it can be anticipated that RvhB8-II might adopt a different oligomerization scheme relative to RvhB8-I within the rvh IMC. Elucidation of the composition of the IMC within the rvh T4SS will help resolve the structural and functional significance of two divergent RvhB8 proteins.
Conclusion.
Our work here presents the first structural investigation on factors regulating the maintenance of multiple divergent T4SSs (or duplicate T4SS components) within a single bacterium. We identified the ability of VirB8 and TrwG of functionally divergent Bartonella T4SSs (vir and trw) to interact and therefore potentially compete in T4SS assembly. This corroborates previous studies demonstrating tight differential expression of these T4SSs (40). For the Rickettsia rvh T4SS, the simultaneously expressed RvhB8 proteins were shown to be structurally divergent, casting doubt on their ability to form heterodimers. Taken together, our data indicate that two distinct mechanisms (spatiotemporal for Bartonella trw and vir T4SSs, structural for rvh T4SSs) prevent cross-system interchangeability between divergent T4SSs encoded within a single bacterium.
T4SSs are ideal drug targets provided that no analogous counterparts are known from eukaryotes. Compounds inhibiting the transfer of broad-host-range plasmids (60) and the ATPase activity of H. pylori VirB11 (61) prove the efficacy of such therapeutic approaches. As an assembly factor that interacts with half of the T4SS scaffold components, VirB8 has been suggested as an appropriate target for drugs inhibiting its many interactions (62). Indeed, several inhibitors of Brucella abortus VirB8 have been identified, with one compound significantly limiting bacterial growth in vivo (63, 64). Our work in this study expands the modality for drug targeting of T4SSs, applicable to bacterial pathogens that harbor multiple T4SSs. Specifically, drugs targeting the barriers to cross-system interchangeability (i.e., regulators) could dysregulate the structural and functional independence of discrete systems, creating interference that prevents their efficient coordination throughout bacterial infection.
MATERIALS AND METHODS
Phylogeny estimation of P-T4SSs.
For 75 P-T4SSs encoded within diverse proteobacterial genomes, five proteins (VirB4 and VirB8 to VirB11) were selected for analysis. Additionally, second copies of RvhB4, RvhB8, and RvhB9 encoded within Rickettsiales genomes were treated as a minimal T4SS (rvh-II), resulting in 90 P-T4SSs in total (for all NCBI GenBank protein accession numbers, see Text S1 in the supplemental material). Individual proteins were separately aligned with MUSCLE (default parameters) (65) and subsequently concatenated into a single data set (unmasked data set; 3,752 amino acids [aa]). Protein alignments were also trimmed of less-conserved regions using Gblocks (66), with these masked alignments also concatenated into a single data set (masked data set; 818 characters). For both datasets, phylogenies were estimated under maximum likelihood (ML) using RAxML v.7.2.8 (67), implementing a gamma model of rate heterogeneity and estimation of the proportion of invariable sites. Two separate analyses for each data set employed the WAG or LG amino acid substitution model, resulting in four total ML-based phylogeny estimations. For all ML analyses, branch support was assessed with 1,000 bootstrap pseudoreplications.
We also analyzed the masked data set with the CAT substitution model, a nonparametric method for modeling site-specific features of sequence evolution (68, 69). The CAT model, as implemented in PhyloBayes v3.3 (70), accommodates saturation caused by convergences and reversions (71). The strong base compositional biases of Rickettsiaceae genomes (~30% GC) makes the CAT model highly amenable to estimating rickettsial phylogeny (72–74). Two independent Markov chains were run in parallel using PhyloBayes MPI v.1.2e (75) under the CAT-GTR model, with the bipartition frequencies analyzed at various time points using the bpcomp program. For tree building, appropriate burn-in values were determined by plotting the log likelihoods for each chain over sampled generations (time). Analyses were considered complete when the maximum difference in bipartition frequencies between the two chains was less than 0.1. Ultimately, a burn-in value of 1,000, with sampling every 2 trees, was used to build a consensus tree.
Comparative analyses of VirB8 proteins.
For Bartonella and Rickettsiales VirB8 family proteins, predicted TMS regions were determined with TMHMM v.2.0 (76). TMS regions were used to delineate all proteins into NT and CT domains. Bartonella VirB8, VbhB8, and TrwG proteins were aligned with MUSCLE (default parameters), with percent identity matrices used to estimate the divergence across all sequences. The NT and CT domains were also separately aligned to determine their percent divergence relative to full-length proteins.
Rickettsiales RvhB8-I and RvhB8-II sequences were separately aligned, as well as being combined in one global RvhB8 alignment (see Text S5 in the supplemental material), with percent identity matrices used to estimate the divergence across all sequences. Phylogenies were estimated on the global RvhB8 alignment, which included one outgroup VirB8 protein from Yersinia frederiksenii (WP_042562314). Phylogenies of the unmasked (295 aa) and masked (87 aa) alignments were estimated under maximum likelihood (ML) using RAxML, implementing a gamma model of rate heterogeneity and estimation of the proportion of invariable sites. Two separate analyses for each alignment employed the WAG or LG amino acid substitution models, resulting in four total ML-based phylogeny estimations. Branch support was assessed with 1,000 bootstrap pseudoreplications.
A structural alignment of R. typhi RvhB8-I and RvhB8-II was constructed, with conserved residues from the Rickettsiales RvhB8-I and RvhB8-II alignments superimposed to illustrate the divergent selective constraints operating on each paralog. Sequence logos depicting the conservation of the NPXG motif for RvhB8-I and RvhB8-II proteins were generated using WebLogo v.3.3 (77). To gain insight into the conservation of the NPXG motif across a broader set of VirB8 family proteins, BLASTP searches (using A. tumefaciens VirB8 as a query [GenBank no. AHK05288]) were conducted across the NR (all GenBank + RefSeq Nucleotides + EMBL + DDBJ + PDB) database, coupled with a search against the Conserved Domains database (78). Searches were performed with composition-based statistics across four specific databases: (i) Alphaproteobacteria (taxid 28211) excluding Rickettsiales (taxid 766); (ii) Gammaproteobacteria (taxid 1236); (iii) Betaproteobacteria (taxid 28216); and (iv) Deltaproteobacteria (taxid 28221) + Epsilonproteobacteria (taxid 29547). No filter was used. Default matrix parameters (BLOSUM62) and gap costs (existence, 11; extension, 1) were implemented, with an inclusion threshold of 0.005. A maximum of 500 unique VirB8 family proteins per database were retrieved, with all collected sequences aligned using MUSCLE (default parameters) and manually evaluated for composition of the NPXG motif within the VirB8 structure. Sequences lacking the conserved NPXG motif were selected for further analysis (see “Protein modeling” below).
High-throughput protein expression, purification, crystallization, and structure determination.
PCR, cloning, screening, sequencing, expression screening, scale-up, and purification of proteins were performed as described previously (79, 80). DNA templates for PCR amplification were obtained from Donald Bouyer (University of Texas Medical Branch, Galveston, TX, USA) for R. typhi strain Wilmington and from Christoph Dehio (Biozentrum, University of Basel, Basel, Switzerland) for Bartonella tribocorum strain CIP 105476, B. quintana strain Toulouse, B. grahamii strain as4aup, and B. birtlesii strain LL-WM9. Crystal trials, diffraction, and structure solution were performed as described previously (81, 82). Further details and all associated data and summary statistics are provided in a description of the gene-to-structure pipeline (see Text S2 in the supplemental material).
Protein-protein interactions.
For experiments measuring protein-protein interactions, all oligonucleotide primers used for generating constructs, and a description of each plasmid, see Text S6 in the supplemental material.
(i) Cloning of bacterial two-hybrid plasmid constructs. (a) Wild-type genes.
The genes of interest were fused to the 3′ end of the T25 fragment (pKT25) and the 3′ end of the T18 fragment (pUT18c) of the Bordetella pertussis adenylate cyclase. Full-length trwG of B. birtlesii was PCR amplified with oligonucleotides prAPV-36 and prAPV-37 using chromosomal DNA of strain IBS325 as the template. The PCR fragment was digested with BamHI and KpnI and ligated into BamHI- and KpnI-digested pKT25, as well as pUT18c, to acquire prAPV001 and prAPV002, respectively. Full-length trwG of B. grahamii was PCR amplified with oligonucleotides prAPV-52 and prAPV-53 using chromosomal DNA of strain as4aup as the template. The PCR fragment was digested with BamHI and KpnI and ligated into BamHI- and KpnI-digested pKT25, as well as pUT18c, to acquire prAPV003 and prAPV004, respectively. Full-length virB8 of B. grahamii was PCR amplified with oligonucleotides prAPV-54 and prAPV-55 using chromosomal DNA of strain as4aup as the template. The PCR fragment was digested with BamHI and KpnI and ligated into BamHI- and KpnI-digested pKT25, as well as pUT18c, to acquire prAPV005 and prAPV006, respectively.
(b) Mutant genes.
The pAPV001 plasmid encoding T25-TrwG of B. birtlesii was linearized with PCR using mutagenic 5′-phosphorylated oligonucleotide primers prAPV-38 and prAPV-39 with the mutation P214A and prAPV-40 and prAPV-41 with the double mutation V96G/V97G. The PCR products were gel isolated and religated to acquire pAPV-007 and pAPV-008, respectively. After the mutant genes were verified via sequencing, the inserts were shuttle cloned into BamHI- and KpnI-digested pUT18c to acquire pAPV-009 and pAPV-010, respectively.
(ii) Bacterial two-hybrid experiments.
The plasmids were introduced into the adenylate cyclase-deficient and cyclic AMP (cAMP)-specific-phosphodiesterase-deficient Escherichia coli strain APE304 (83) by the polyethylene glycol method. Colonies (5 to 10) from freshly transformed plates were pooled and subcultured overnight in 5 ml of Luria-Bertani medium with 50 µg/ml of kanamycin, 200 µg/ml of ampicillin, and 100 µM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37°C under vigorous shaking (250 rpm). The following day, the strains were diluted either 1:5 or 1:10 in phosphate-buffered saline (PBS) for the measurement of β-galactosidase activities. First, 10 µl of chloroform and 10 µl of 0.1% (wt/vol) sodium dodecyl sulfate (SDS) were added to 1 ml of the diluted bacterial suspensions. Next, 20 µl of the vortexed bacterial suspensions were mixed in triplicate with 180 µl of β-galactosidase substrate solution (1 mg/ml of 2-nitrophenyl β-d-galactopyranoside [catalogue no. N1127; Sigma-Aldrich] in 60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 0.01% [wt/vol] SDS, and 40 mM β-mercaptoethanol). The reaction mixtures were incubated at room temperature for 20 to 40 min. The reactions were stopped by adding 100 µl of 1 M Na2CO3. The end products were measured at 420 nm and 550 nm. Specific β-galactosidase activities (in Miller units) were calculated as follows: 1,000 × [A420 − (1.75 × A550)/t × V × A600], where t is the reaction mixture incubation time, V is the volume of bacterial suspension in the reaction mixture (20 µl), and A600 equals the optical density of the 1:5- or 1:10-diluted bacterial overnight cultures measured at 600 nm.
(iii) Glutaraldehyde cross-linking experiments.
Recombinant N-terminally His-tagged proteins (10 µg in 70 µl of 20 mM HEPES, 300 mM NaCl, 5% glycerol, and 2 mM dithiothreitol [DTT]) were incubated for 20 min at room temperature in the presence of different concentrations of glutaraldehyde (0, 0.005, 0.01, 0.1, 0.5, 1, and 5%). The reactions were quenched by adding 20 µl of 1 M Tris-HCl, pH 7.5. Laemmli loading dye (3×, 45 µl) was added, and the samples were incubated at 95°C for 10 min. Proteins from 5 µl of the samples were separated by SDS-PAGE using 10% SDS gels and transferred onto Protran nitrocellulose transfer membranes (Whatman). The membrane was examined for the His epitope using primary mouse monoclonal anti-His antibody (1:1,000; H1029; Sigma-Aldrich) and secondary HRP-conjugated goat anti-mouse IgG (1:5,000; sc-2005; Santa Cruz Biotechnology) with the enhanced chemiluminescence (ECL) system (SuperSignal West Pico chemiluminescent substrate; Thermo Scientific).
Protein modeling. (i) Building 3D models.
Three-dimensional (3D) structural models were built for R. typhi RvhB8-I and 16 VirB8 family proteins containing divergent NPXG motifs (for complete species names and sequence information, see Text S4 in the supplemental material). 3D models were generated applying the fragment assembly approach of homology modeling (84) using SPDBV/DeepView (85) and the Swiss-Model server (85, 86). Each target sequence was modeled to eight templates, including six VirB8 structures (PDB codes 4MEI from Bartonella tribocorum, 4LSO from Bartonella quintana, 4KZ1 from Bartonella grahamii, 2BHM from Brucella suis [45], 2CC3 from Agrobacterium tumefaciens [44], and 4O3V from Rickettsia typhi) and two TrwG structures (4JF8 from Bartonella birtlesii and chain A of 4NHF from Bartonella grahamii). All structures were downloaded from the Protein DataBank (87). Template structures were structurally aligned using SPDBV/Deep View. The target sequences were computationally aligned to this set of templates and manually curated with the following criteria (position numbers refer to the X-ray structure 4MEI, VirB8 from Bartonella tribocorum; for a full alignment of template structures see Text S4 in the supplemental material). (i) In α-helix 1, there is a semiconserved motif, e.g., EAIT in 4MEI, 4LSO, and 4KZ1. (ii) In position 5 of α-helix 2, an aromatic amino acid can often be found. (iii) In position 8 of α-helix 3, there is often an aromatic residue. In positions 11 and 12 of the same α-helix, there is a tendency to have aromatic residues. (iv) In α-helix 4, there is a tendency to have a proline in position 1. (v) In β-strand 1, a moderately conserved motif exists starting at position 4: hydrophobic-X-hydrophobic-X-X-hydrophobic-polar. In 4MEI, this motif is V151-T152-I153-K154-S155-I156-S157. (vi) In β-strand 2, in many cases there is the triad motif Q-hydrophobic-charged. In 4MEI, it is Q165-V166-R167. (vii) In β-strand 3, alignments were anchored using a moderately conserved motif from positions Ω-4 to Ω-2: aromatic-polar/charged-aromatic. (viii) In the loop between α-helix 5 and β-strand 4, a conserved NPXG motif exists. (ix) Insertions and deletions were moved from inside secondary structure elements to surface loops in order to preserve the fold. This strategy followed the principle that fold is conserved over sequence.
Conserved motifs at the N and C termini were used as main alignment anchors (points 1 and 8 above). Since target sequences share low (or even very low) similarity to the sequences of the templates, only selected criteria for target to template alignment were applied. In some cases (Colwellia psychrerythrea, Hydrogenophaga species, Legionella longbeachae, Legionella pneumophila, Pseudomonas putida, Pseudomonas syringae, and Ralstonia solanacearum), one or several of these alignment “anchors” were missing. This was especially the case for the only moderately conserved motifs in β-strands.
Target to template alignments in SPDBV/DeepView were saved as “projects” and submitted to the Swiss-Model server via the DeepView project mode. Although the overall quality of the models was limited due to low homology, the structures were of sufficient quality for (i) studying structural properties (i.e., neighboring residues, positions of conserved motifs, etc.) and (ii) investigating dimer interfaces.
(ii) Building dimers. (a) Homomeric models.
In SPDBV/DeepView, the dimeric structure 2CC3 from Agrobacterium tumefaciens was used as a template for dimer modeling. Two versions of the same monomeric model were loaded into SPDBV/DeepView as different layers. One of these monomers was fitted to chain A of 2CC3, the other was fitted to chain B. Superposition of the monomers with the respective chains of 2CC3 was optimized using the “Improved Fit” option. The monomers were merged into a new layer to obtain a homodimer.
(b) Heterodimeric models.
For the Rickettsia heterodimeric model, the structure of R. typhi RvhB8-II (4O3V) was used as a template. Chain A corresponded already to RvhB8-II, with chain B replaced by the 3D model generated for RvhB8-I. The TrwG/VirB8 heterodimeric structure for Bartonella grahamii was generated using chains A and B of the X-ray structure 4NHF, which corresponds to the TrwG domain of B. grahamii. Chain B was then replaced by the X-ray structure of B. grahamii VirB8 (4KZ1).
The dimer interface was optimized with SCWRL4 (88). Models were subsequently energy minimized by applying 20 steps of the Steepest Descent algorithm, as implemented in SPDBV/DeepView.
RT-qPCR for RvhB8 proteins.
HeLa cells (~1 × 106 cells) were infected with R. typhi at a multiplicity of infection (MOI) of ~100 and incubated at 34°C with 5% CO2 for 10 min, 30 min, 2 h, and 24 h. At each time point, cells were washed with phosphate-buffered saline (pH 7.4) and RNA was extracted using a Quick-RNA miniprep kit (Zymo Research). IScript reverse transcription supermix for RT-qPCR (Bio-Rad) was used to synthesize cDNA from 500 ng of purified RNA. qPCR was performed with VeriQuest SYBR green qPCR master mix (Affymetrix) in a CFX384 Multicycler (Bio-Rad) (for primer sequences, see Text S6 in the supplemental material). The thermal cycling conditions included 95°C for 3 min followed by 40 cycles of amplification at 95°C for 10 s and 55°C for 60 s. A melting curve analysis was performed to confirm amplification of a single product for each primer pair. A panel of 6 reference genes (see Text S6 in the supplemental material) was tested to determine which genes were stably expressed (geNorm M score ≤ 0.5; Qbase Plus; Biogazelle) with infection. rvhB8-I and rvhB8-II gene expression was normalized to the average cycle threshold (CT) of R. typhi reference genes adr1 and sca5 (2ΔCT).
SUPPLEMENTAL MATERIAL
Associated data pertaining to phylogeny estimations of P-T4SSs. Download
Data and summary statistics for protein expression, purification, crystallization, and structure determination. Download
Relative conservation of Bartonella TrwG, VirB8, and VbhB8 proteins. Download
Structural modeling of VirB8 family proteins. Download
Analyses of RvhB8 proteins across Rickettsiales species. Download
Oligonucleotide primers and plasmids utilized for the Bartonella and Rickettsia experiments. Download
Glutaraldehyde cross-linking experiments. Download
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases (grants R01AI017828, R01AI043006, and R01AI59118 to A.F.A. and contract no. HHSN272200700057C and HHSN272201200025C to P.J.M.) and a grant from the Sigrid Jusélius Foundation (to A.T.P.). S.S.L. and K.E.R-B. were trainees under Institutional Training Grants T32AI007540 and T32AI095190, respectively, from the National Institute of Allergy and Infectious Diseases.
We thank the SSGCID cloning and protein production groups at the Center for Infectious Disease Research and at the University of Washington. For the mass spectrometry analyses we thank members of the University of Washington Medicinal Chemistry Mass Spectrometry Center.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Citation Gillespie JJ, Phan IQH, Scheib H, Subramanian S, Edwards TE, Lehman SS, Piitulainen H, Sayeedur Rahman M, Rennoll-Bankert KE, Staker BL, Taira S, Stacy R, Myler PJ, Azad AF, Pulliainen AT. 2015. Structural insight into how bacteria prevent interference between multiple divergent type IV secretion systems. mBio 6(6):e01867-15. doi:10.1128/mBio.01867-15.
REFERENCES
- 1.Alvarez-Martinez CE, Christie PJ. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev 73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol 59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lawley TD, Klimke WA, Gubbins MJ, Frost LS. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett 224:1–15. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- 4.Hofreuter D, Odenbreit S, Haas R. 2001. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol Microbiol 41:379–391. doi: 10.1046/j.1365-2958.2001.02502.x. [DOI] [PubMed] [Google Scholar]
- 5.Hamilton HL, Domínguez NM, Schwartz KJ, Hackett KT, Dillard JP. 2005. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol Microbiol 55:1704–1721. doi: 10.1111/j.1365-2958.2005.04521.x. [DOI] [PubMed] [Google Scholar]
- 6.Juhas M, Crook DW, Dimopoulou ID, Lunter G, Harding RM, Ferguson DJP, Hood DW. 2007. Novel type IV secretion system involved in propagation of genomic islands. J Bacteriol 189:761–771. doi: 10.1128/JB.01327-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cascales E, Christie PJ. 2003. The versatile bacterial type IV secretion systems. Nat Rev Microbiol 1:137–149. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siamer S, Dehio C. 2015. New insights into the role of Bartonella effector proteins in pathogenesis. Curr Opin Microbiol 23:80–85. doi: 10.1016/j.mib.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Souza DP, Oka GU, Alvarez-Martinez CE, Bisson-Filho AW, Dunger G, Hobeika L, Cavalcante NS, Alegria MC, Barbosa LRS, Salinas RK, Guzzo CR, Farah CS. 2015. Bacterial killing via a type IV secretion system. Nat Commun 6:6453. doi: 10.1038/ncomms7453. [DOI] [PubMed] [Google Scholar]
- 10.Jakubowski SJ, Cascales E, Krishnamoorthy V, Christie PJ. 2005. Agrobacterium tumefaciens VirB9, an outer-membrane-associated component of a type IV secretion system, regulates substrate selection and T-pilus biogenesis. J Bacteriol 187:3486–3495. doi: 10.1128/JB.187.10.3486-3495.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerr JE, Christie PJ. 2010. Evidence for VirB4-mediated dislocation of membrane-integrated VirB2 pilin during biogenesis of the Agrobacterium VirB/VirD4 type IV secretion system. J Bacteriol 192:4923–4934. doi: 10.1128/JB.00557-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voth DE, Broederdorf LJ, Graham JG. 2012. Bacterial type IV secretion systems: versatile virulence machines. Future Microbiol 7:241–257. doi: 10.2217/fmb.11.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallden K, Rivera-Calzada A, Waksman G. 2010. Type IV secretion systems: versatility and diversity in function. Cell Microbiol 12:1203–1212. doi: 10.1111/j.1462-5822.2010.01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guglielmini J, de la Cruz F, Rocha EPC. 2013. Evolution of conjugation and type IV secretion systems. Mol Biol Evol 30:315–331. doi: 10.1093/molbev/mss221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christie PJ. 2004. Type IV secretion: the Agrobacterium VirB/D4 and related conjugation systems. Biochim Biophys Acta 1694:219–234. doi: 10.1016/j.bbamcr.2004.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fronzes R, Schäfer E, Wang L, Saibil HR, Orlova EV, Waksman G. 2009. Structure of a type IV secretion system core complex. Science 323:266–268. doi: 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chandran V, Fronzes R, Duquerroy S, Cronin N, Navaza J, Waksman G. 2009. Structure of the outer membrane complex of a type IV secretion system. Nature 462:1011–1015. doi: 10.1038/nature08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walldén K, Williams R, Yan J, Lian PW, Wang L, Thalassinos K, Orlova EV, Waksman G. 2012. Structure of the VirB4 ATPase, alone and bound to the core complex of a type IV secretion system. Proc Natl Acad Sci U S A 109:11348–11353. doi: 10.1073/pnas.1201428109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Low HH, Gubellini F, Rivera-Calzada A, Braun N, Connery S, Dujeancourt A, Lu F, Redzej A, Fronzes R, Orlova EV, Waksman G. 2014. Structure of a type IV secretion system. Nature 508:550–553. doi: 10.1038/nature13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rivera-Calzada A, Fronzes R, Savva CG, Chandran V, Lian PW, Laeremans T, Pardon E, Steyaert J, Remaut H, Waksman G, Orlova EV. 2013. Structure of a bacterial type IV secretion core complex at subnanometre resolution. EMBO J 32:1195–1204. doi: 10.1038/emboj.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guglielmini J, Neron B, Abby SS, Garcillan-Barcia MP, la Cruz Fd, Rocha EPC. 2014. Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res 42:5715–5727. doi: 10.1093/nar/gku194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhatty M, Laverde Gomez JA, Christie PJ. 2013. The expanding bacterial type IV secretion lexicon. Res Microbiol 164:620–639. doi: 10.1016/j.resmic.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillespie JJ, Brayton KA, Williams KP, Diaz MA, Brown WC, Azad AF, Sobral BW. 2010. Phylogenomics reveals a diverse Rickettsiales type IV secretion system. Infect Immun 78:1809–1823. doi: 10.1128/IAI.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arutyunov D, Arenson B, Manchak J, Frost LS. 2010. F plasmid TraF and TraH are components of an outer membrane complex involved in conjugation. J Bacteriol 192:1730–1734. doi: 10.1128/JB.00726-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silverman PM, Clarke MB. 2010. New insights into F-pilus structure, dynamics, and function. Integr Biol (Camb) 2:25–31. doi: 10.1039/b917761b. [DOI] [PubMed] [Google Scholar]
- 26.Shaffer CL, Gaddy JA, Loh JT, Johnson EM, Hill S, Hennig EE, McClain MS, McDonald WH, Cover TL. 2011. Helicobacter pylori exploits a unique repertoire of type IV secretion system components for pilus assembly at the bacteria-host cell interface. PLoS Pathog 7:e1002237. doi: 10.1371/journal.ppat.1002237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kutter S, Buhrdorf R, Haas J, Schneider-Brachert W, Haas R, Fischer W. 2008. Protein subassemblies of the Helicobacter pylori Cag type IV secretion system revealed by localization and interaction studies. J Bacteriol 190:2161–2171. doi: 10.1128/JB.01341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karnholz A, Hoefler C, Odenbreit S, Fischer W, Hofreuter D, Haas R. 2006. Functional and topological characterization of novel components of the comB DNA transformation competence system in Helicobacter pylori. J Bacteriol 188:882–893. doi: 10.1128/JB.188.3.882-893.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saenz HL, Engel P, Stoeckli MC, Lanz C, Raddatz G, Vayssier-Taussat M, Birtles R, Schuster SC, Dehio C. 2007. Genomic analysis of Bartonella identifies type IV secretion systems as host adaptability factors. Nat Genet 39:1469–1476. doi: 10.1038/ng.2007.38. [DOI] [PubMed] [Google Scholar]
- 30.Vogel JP, Isberg RR. 1999. Cell biology of Legionella pneumophila. Curr Opin Microbiol 2:30–34. doi: 10.1016/S1369-5274(99)80005-8. [DOI] [PubMed] [Google Scholar]
- 31.Segal G, Shuman HA. 1998. How is the intracellular fate of the Legionella pneumophila phagosome determined? Trends Microbiol 6:253–255. doi: 10.1016/S0966-842X(98)01308-0. [DOI] [PubMed] [Google Scholar]
- 32.Segal G, Russo JJ, Shuman HA. 1999. Relationships between a new type IV secretion system and the icm/dot virulence system of Legionella pneumophila. Mol Microbiol 34:799–809. doi: 10.1046/j.1365-2958.1999.01642.x. [DOI] [PubMed] [Google Scholar]
- 33.Bandyopadhyay P, Liu S, Gabbai CB, Venitelli Z, Steinman HM. 2007. Environmental mimics and the Lvh type IVA secretion system contribute to virulence-related phenotypes of Legionella pneumophila. Infect Immun 75:723–735. doi: 10.1128/IAI.00956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bandyopadhyay P, Lang EAS, Rasaputra KS, Steinman HM. 2013. Implication of the VirD4 coupling protein of the Lvh type 4 secretion system in virulence phenotypes of Legionella pneumophila. J Bacteriol 195:3468–3475. doi: 10.1128/JB.00430-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wozniak RAF, Waldor MK. 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol 8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- 36.Dehio C. 2008. Infection-associated type IV secretion systems of Bartonella and their diverse roles in host cell interaction. Cell Microbiol 10:1591–1598. doi: 10.1111/j.1462-5822.2008.01171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seubert A, Hiestand R, de la Cruz F, Dehio C. 2003. A bacterial conjugation machinery recruited for pathogenesis. Mol Microbiol 49:1253–1266. doi: 10.1046/j.1365-2958.2003.03650.x. [DOI] [PubMed] [Google Scholar]
- 38.Vayssier-Taussat M, Le Rhun D, Deng HK, Biville F, Cescau S, Danchin A, Marignac G, Lenaour E, Boulouis HJ, Mavris M, Arnaud L, Yang H, Wang J, Quebatte M, Engel P, Saenz H, Dehio C. 2010. The Trw type IV secretion system of Bartonella mediates host-specific adhesion to erythrocytes. PLoS Pathog 6:e1000946. doi: 10.1371/journal.ppat.1000946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng HK, Le Rhun D, Le Naour E, Bonnet S, Vayssier-Taussat M. 2012. Identification of Bartonella Trw host-specific receptor on erythrocytes. PLoS One 7:e41447. doi: 10.1371/journal.pone.0041447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Québatte M, Dick MS, Kaever V, Schmidt A, Dehio C. 2013. Dual input control: activation of the Bartonella henselae VirB/D4 type IV secretion system by the stringent sigma factor RpoH1 and the BatR/BatS two-component system. Mol Microbiol 90:756–775. doi: 10.1111/mmi.12396. [DOI] [PubMed] [Google Scholar]
- 41.Gillespie JJ, Ammerman NC, Dreher-Lesnick SM, Rahman MS, Worley MJ, Setubal JC, Sobral BS, Azad AF. 2009. An anomalous type IV secretion system in rickettsia is evolutionarily conserved. PLoS One 4:e4833. doi: 10.1371/journal.pone.0004833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gillespie JJ, Kaur SJ, Rahman MS, Rennoll-Bankert K, Sears KT, Beier-Sexton M, Azad AF. 2015. Secretome of obligate intracellular rickettsia. FEMS Microbiol Rev 39:47–80. doi: 10.1111/1574-6976.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gillespie J, Nordberg E, Azad A, Sobral B. 2012. Phylogeny and comparative genomics: the shifting landscape in the genomics era, p 84–141. In Azad AF, Palmer GH (ed), Intracellular pathogens II: Rickettsiales. ASM Press, Washington, DC. doi: 10.1128/9781555817336.ch3. [DOI] [Google Scholar]
- 44.Bailey S, Ward D, Middleton R, Grossmann JG, Zambryski PC. 2006. Agrobacterium tumefaciens VirB8 structure reveals potential protein-protein interaction sites. Proc Natl Acad Sci U S A 103:2582–2587. doi: 10.1073/pnas.0511216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terradot L, Bayliss R, Oomen C, Leonard GA, Baron C, Waksman G. 2005. Structures of two core subunits of the bacterial type IV secretion system, VirB8 from Brucella suis and ComB10 from Helicobacter pylori. Proc Natl Acad Sci U S A 102:4596–4601. doi: 10.1073/pnas.0408927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McLeod MP, Qin X, Karpathy SE, Gioia J, Highlander SK, Fox GE, McNeill TZ, Jiang H, Muzny D, Jacob LS, Hawes AC, Sodergren E, Gill R, Hume J, Morgan M, Fan G, Amin AG, Gibbs RA, Hong C, Yu X-, Walker DH, Weinstock GM. 2004. Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J Bacteriol 186:5842–5855. doi: 10.1128/JB.186.17.5842-5855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paschos A, Patey G, Sivanesan D, Gao C, Bayliss R, Waksman G, O’Callaghan D, Baron C. 2006. Dimerization and interactions of Brucella suis VirB8 with VirB4 and VirB10 are required for its biological activity. Proc Natl Acad Sci U S A 103:7252–7257. doi: 10.1073/pnas.0600862103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sivanesan D, Hancock MA, Villamil Giraldo AM, Baron C. 2010. Quantitative analysis of VirB8-VirB9-VirB10 interactions provides a dynamic model of type IV secretion system core complex assembly. Biochemistry 49:4483–4493. doi: 10.1021/bi902201y. [DOI] [PubMed] [Google Scholar]
- 49.Cheng Z, Wang X, Rikihisa Y. 2008. Regulation of type IV secretion apparatus genes during Ehrlichia chaffeensis intracellular development by a previously unidentified protein. J Bacteriol 190:2096–2105. doi: 10.1128/JB.01813-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Z, Carlow CKS. 2012. Characterization of transcription factors that regulate the type IV secretion system and riboflavin biosynthesis in Wolbachia of Brugia malayi. PLoS One 7:e51597. doi: 10.1371/journal.pone.0051597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berger BR, Christie PJ. 1993. The Agrobacterium tumefaciens virB4 gene product is an essential virulence protein requiring an intact nucleoside triphosphate-binding domain. J Bacteriol 175:1723–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fullner KJ, Stephens KM, Nester EW. 1994. An essential virulence protein of Agrobacterium tumefaciens, VirB4, requires an intact mononucleotide binding domain to function in transfer of T-DNA. Mol Gen Genet 245:704–715. doi: 10.1007/BF00297277. [DOI] [PubMed] [Google Scholar]
- 53.Watarai M, Makino S, Shirahata T. 2002. An essential virulence protein of Brucella abortus, VirB4, requires an intact nucleoside-triphosphate-binding domain. Microbiology 148:1439–1446. doi: 10.1099/00221287-148-5-1439. [DOI] [PubMed] [Google Scholar]
- 54.Bayliss R, Harris R, Coutte L, Monier A, Fronzes R, Christie PJ, Driscoll PC, Waksman G. 2007. NMR structure of a complex between the VirB9/VirB7 interaction domains of the pKM101 type IV secretion system. Proc Natl Acad Sci U S A 104:1673–1678. doi: 10.1073/pnas.0609535104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Höppner C, Carle A, Sivanesan D, Hoeppner S, Baron C. 2005. The putative lytic transglycosylase VirB1 from Brucella suis interacts with the type IV secretion system core components Vir B8, VirB9 and VirB11. Microbiology 151:3469–3482. doi: 10.1099/mic.0.28326-0. [DOI] [PubMed] [Google Scholar]
- 56.Das A, Xie Y-. 2000. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J Bacteriol 182:758–763. doi: 10.1128/JB.182.3.758-763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Porter CJ, Bantwal R, Bannam TL, Rosado CJ, Pearce MC, Adams V, Lyras D, Whisstock JC, Rood JI. 2012. The conjugation protein TcpC from Clostridium perfringens is structurally related to the type IV secretion system protein VirB8 from Gram-negative bacteria. Mol Microbiol 83:275–288. doi: 10.1111/j.1365-2958.2011.07930.x. [DOI] [PubMed] [Google Scholar]
- 58.Goessweiner-Mohr N, Grumet L, Arends K, Pavkov-Keller T, Gruber CC, Gruber K, Birner-Gruenberger R, Kropec-Huebner A, Huebner J, Grohmann E, Keller W. 2013. The 2.5 Å structure of the enterococcus conjugation protein TraM resembles VirB8 type IV secretion proteins. J Biol Chem 288:2018–2028. doi: 10.1074/jbc.M112.428847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuroda T, Kubori T, Thanh Bui X, Hyakutake A, Uchida Y, Imada K, Nagai H. 2015. Molecular and structural analysis of Legionella DotI gives insights into an inner membrane complex essential for type IV secretion. Sci Rep 5:10912. doi: 10.1038/srep10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fernandez-Lopez R, Machón C, Longshaw CM, Martin S, Molin S, Zechner EL, Espinosa M, Lanka E, de la Cruz F. 2005. Unsaturated fatty acids are inhibitors of bacterial conjugation. Microbiology 151:3517–3526. doi: 10.1099/mic.0.28216-0. [DOI] [PubMed] [Google Scholar]
- 61.Hilleringmann M, Pansegrau W, Doyle M, Kaufman S, MacKichan ML, Gianfaldoni C, Ruggiero P, Covacci A. 2006. Inhibitors of Helicobacter pylori ATPase Cagalpha block CagA transport and cag virulence. Microbiology 152:2919–2930. doi: 10.1099/mic.0.28984-0. [DOI] [PubMed] [Google Scholar]
- 62.Baron C. 2006. VirB8: a conserved type IV secretion system assembly factor and drug target. Biochem Cell Biol 84:890–899. doi: 10.1139/o06-148. [DOI] [PubMed] [Google Scholar]
- 63.Paschos A, den Hartigh A, Smith MA, Atluri VL, Sivanesan D, Tsolis RM, Baron C. 2011. An in vivo high-throughput screening approach targeting the type IV secretion system component VirB8 identified inhibitors of Brucella abortus 2308 proliferation. Infect Immun 79:1033–1043. doi: 10.1128/IAI.00993-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith M, Coinçon M, Paschos A, Jolicoeur B, Lavallée P, Sygusch J, Baron C. 2012. Identification of the binding site of Brucella VirB8 interaction inhibitors. Chem Biol 19:1041–1048. doi: 10.1016/j.chembiol.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 65.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 67.Stamatakis A. 2014. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. BioInformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lartillot N. 2004. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol 21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- 69.Lartillot N, Philippe H. 2006. Computing Bayes factors using thermodynamic integration. Syst Biol 55:195–207. doi: 10.1080/10635150500433722. [DOI] [PubMed] [Google Scholar]
- 70.Lartillot N, Lepage T, Blanquart S. 2009. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. BioInformatics 25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- 71.Lartillot N, Brinkmann H, Philippe H. 2007. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol Biol 7(Suppl 1):S4. doi: 10.1186/1471-2148-7-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Driscoll T, Gillespie JJ, Nordberg EK, Azad AF, Sobral BW. 2013. Bacterial DNA sifted from the Trichoplax adhaerens (Animalia: Placozoa) genome project reveals a putative rickettsial endosymbiont. Genome Biol Evol 5:621–645. doi: 10.1093/gbe/evt036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodríguez-Ezpeleta N, Embley TM. 2012. The SAR11 group of alpha-proteobacteria is not related to the origin of mitochondria. PLoS One 7:e30520. doi: 10.1371/journal.pone.0030520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Viklund J, Ettema TJG, Andersson SGE. 2012. Independent genome reduction and phylogenetic reclassification of the oceanic SAR11 clade. Mol Biol Evol 29:599–615. doi: 10.1093/molbev/msr203. [DOI] [PubMed] [Google Scholar]
- 75.Lartillot N, Rodrigue N, Stubbs D, Richer J. 2013. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol 62:611–615. doi: 10.1093/sysbio/syt022. [DOI] [PubMed] [Google Scholar]
- 76.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 77.Crooks GE, Hon G, Chandonia J-M, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res 39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bryan CM, Bhandari J, Napuli AJ, Leibly DJ, Choi R, Kelley A, Van Voorhis WC, Edwards TE, Stewart LJ. 2011. High-throughput protein production and purification at the Seattle Structural Genomics Center for Infectious Disease. Acta Crystallogr Sect F Struct Biol Cryst Commun 67:1010–1014. doi: 10.1107/S1744309111018367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Choi R, Kelley A, Leibly D, Hewitt S, Napuli A, Van Voorhis W. 2011. Immobilized metal-affinity chromatography protein-recovery screening is predictive of crystallographic structure success. Acta Crystallogr F Struct Biol Cryst Commun 67:998–1005. doi: 10.1107/S1744309111017374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Myler P, Stacy R, Stewart L, Staker B, Van Voorhis W, Varani G, Buchko G. 2009. The Seattle Structural Genomics Center for Infectious Disease (SSGCID). Infect Disord Drug Targets 9:493–506. doi: 10.2174/187152609789105687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Begley DW, Hartley RC, Davies DR, Edwards TE, Leonard JT, Abendroth J, Burris CA, Bhandari J, Myler PJ, Staker BL, Stewart LJ. 2011. Leveraging structure determination with fragment screening for infectious disease drug targets: MECP synthase from Burkholderia pseudomallei. J Struct Funct Genomics 12:63–76. doi: 10.1007/s10969-011-9102-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pulliainen AT, Pieles K, Brand CS, Hauert B, Böhm A, Quebatte M, Wepf A, Gstaiger M, Aebersold R, Dessauer CW, Dehio C. 2012. Bacterial effector binds host cell adenylyl cyclase to potentiate Gαs-dependent cAMP production. Proc Natl Acad Sci U S A 109:9581–9586. doi: 10.1073/pnas.1117651109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Greer J. 1981. Comparative model-building of the mammalian serine proteases. J Mol Biol 153:1027–1042. doi: 10.1016/0022-2836(81)90465-4. [DOI] [PubMed] [Google Scholar]
- 85.Guex N, Peitsch MC. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 86.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The Protein Data Bank. Nucleic Acids Res 28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Krivov GG, Shapovalov MV, Dunbrack RL. 2009. Improved prediction of protein side-chain conformations with SCWRL4. Proteins 77:778–795. doi: 10.1002/prot.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Associated data pertaining to phylogeny estimations of P-T4SSs. Download
Data and summary statistics for protein expression, purification, crystallization, and structure determination. Download
Relative conservation of Bartonella TrwG, VirB8, and VbhB8 proteins. Download
Structural modeling of VirB8 family proteins. Download
Analyses of RvhB8 proteins across Rickettsiales species. Download
Oligonucleotide primers and plasmids utilized for the Bartonella and Rickettsia experiments. Download
Glutaraldehyde cross-linking experiments. Download