Significance
Using a rapamycin-based, endoplasmic reticulum (ER) trapping scheme, modified to avoid the problem of an endogenous ER-localized FKBP-binding protein, we demonstrate that Golgi enzymes constitutively recycle back to the ER and that such recycling plays a central role in the maintenance, biogenesis, and inheritance of the Golgi apparatus in mammalian cells. We describe morphological characteristics of the retrograde carriers that ferry Golgi enzymes back to the ER and identify key molecular machinery regulating carrier formation. The study helps resolve the long-standing debate regarding the extent of Golgi enzyme trafficking back to the ER, paving the way for further investigations into the mechanistic details and functional implications of the Golgi's steady-state existence and relationship to the ER.
Keywords: Golgi, recycling, mitosis, rapamycin, ER
Abstract
Whether Golgi enzymes remain localized within the Golgi or constitutively cycle through the endoplasmic reticulum (ER) is unclear, yet is important for understanding Golgi dependence on the ER. Here, we demonstrate that the previously reported inefficient ER trapping of Golgi enzymes in a rapamycin-based assay results from an artifact involving an endogenous ER-localized 13-kD FK506 binding protein (FKBP13) competing with the FKBP12-tagged Golgi enzyme for binding to an FKBP-rapamycin binding domain (FRB)-tagged ER trap. When we express an FKBP12-tagged ER trap and FRB-tagged Golgi enzymes, conditions precluding such competition, the Golgi enzymes completely redistribute to the ER upon rapamycin treatment. A photoactivatable FRB-Golgi enzyme, highlighted only in the Golgi, likewise redistributes to the ER. These data establish Golgi enzymes constitutively cycle through the ER. Using our trapping scheme, we identify roles of rab6a and calcium-independent phospholipase A2 (iPLA2) in Golgi enzyme recycling, and show that retrograde transport of Golgi membrane underlies Golgi dispersal during microtubule depolymerization and mitosis.
The Golgi apparatus is the major processing and sorting station at the crossroads of the secretory pathway (1–4). It receives newly synthesized proteins from the endoplasmic reticulum (ER), processes them using Golgi-specific enzymes, and sorts them to the plasma membrane (PM) and other final destinations. During this process, specialized sorting and transport machinery of the Golgi filter out selected membrane and protein components, returning them back to the ER for continued use. How the Golgi maintains its structure and function amid this ongoing bidirectional membrane trafficking has been a long-standing debate.
A variety of studies have advanced the view that the Golgi apparatus is a dynamic, steady-state system in which resident enzymes continuously recycle back to the ER and return to the Golgi through the same retrograde and anterograde trafficking routes used by other proteins (4–6). This model helps account for the striking dispersal of the Golgi when ER export is blocked (7–9), microtubules are depolymerized (10–12), or cells enter mitosis (13, 14). In each case, Golgi enzymes have been reported to redistribute to the site of transport block in the ER.
An alternative model has been proposed that envisions the Golgi as an autonomous organelle with stable components that provide a template for its growth and division. In this model, Golgi enzymes ordinarily remain localized in the Golgi throughout their lifetime and Golgi dispersal under ER export blockade, microtubule depolymerization, or mitotic entry is due to a reversible breakdown of the Golgi itself into smaller elements or vesicles, without contact with the ER or its associated pathways. One primary support for this model comes from studies examining the relationship of Golgi enzymes and ER proteins using a rapamycin-induced protein heterodimerization assay to trap FKBP12- and FKBP-rapamycin binding domain (FRB)-tagged probes in close proximity (15). Using an FRB-tagged, ER-retained version of invariant chain Iip35 (Ii-FRB) (16, 17) and FKBP12-labeled Golgi enzymes, these studies observed little trapping of Golgi enzymes in the ER in the presence of rapamycin (15), including during Golgi dispersal upon mitotic entry (18).
If representative of normal Golgi enzyme behavior, the ER trapping assay’s results would call into question the wide range of other observations supporting the steady-state Golgi model and its ER dependency. We thus examined the assay in more detail to determine whether it might be underestimating the extent of Golgi enzyme recycling to the ER. Prior work has shown that an endogenous FK506-binding protein, FKBP13, localizes to the lumen of the ER (19, 20), where, in the presence of rapamycin, it forms a ternary complex with FRB-containing proteins (20). We tested the possibility that binding of endogenous FKBP13 to ER-localized Ii-FRB when rapamycin is introduced might explain why no significant trapping of FKBP12-labeled Golgi enzymes in the ER was observed in the prior work. Consistent with this possibility, we found that coexpressing an FKBP12-tagged ER protein and an FRB-tagged Golgi enzyme marker, conditions where endogenous ER-localized FKBP13 would not sequester the ER trap, resulted in complete redistribution of the Golgi marker into the ER within 4 h of rapamycin treatment. In the ER, FRB-Golgi and FKBP12-ER markers underwent FRET, indicating direct binding upon rapamycin-induced redistribution. These data support the Golgi recycling theory by providing evidence of trapping of Golgi enzymes in the ER.
The ability to assess retrograde transport of Golgi enzymes using our modified ER trapping assay enabled us to characterize the pathway while offering additional evidence of its existence. We found that the carriers delivering Golgi enzymes to the ER were tubule-shaped and moved peripherally after extending off from the Golgi. Rab6a and cation-independent phospholipase A2 (iPLA2) were required for their delivery to the ER. Golgi enzyme recycling to the ER was also shown to be involved in Golgi fragmentation during microtubule depolymerization and during mitosis. We anticipate that further use of this modified ER trapping assay will provide more insights into the precise nature of the Golgi’s dependence on the ER.
Results
Ii-FRB Associates with Endogenous, ER-Localized FKBP13 During Rapamycin Treatment.
Rapamycin-based heterodimerization of FKBP12- and FRB-tagged proteins is an elegant way to test whether two proteins come into proximity with each other (21, 22). Pecot and Malhotra (15) used this approach for determining whether Golgi proteins cycle through the ER, attaching FRB to the ER-retained protein Ii (Ii-FRB) and FKBP12 to a Golgi enzyme (Golgi-FKBP12). Because Ii-FRB never leaves the ER, any Golgi-FKBP12 that recycles to the ER should undergo rapamycin-induced heterodimerization and thereafter be “trapped” in the ER. In principle, it should not matter in this assay whether the ER trapping protein (i.e., Ii) is attached to FRB or FKBP12, as long as the Golgi protein is appropriately tagged to create a heterodimer of FRB-FKBP12 under rapamycin treatment. However, prior work has shown that an endogenous FKBP, viz. FKBP13, resides within the ER lumen (19, 23). This finding raises the possibility that the configuration of the ER trapping protein (i.e., FRB-tagged instead of FKBP12-tagged) in the above assay may be problematic, because endogenous FKBP13 might compete with recycling Golgi-FKBP12 for binding to Ii-FRB during rapamycin treatment.
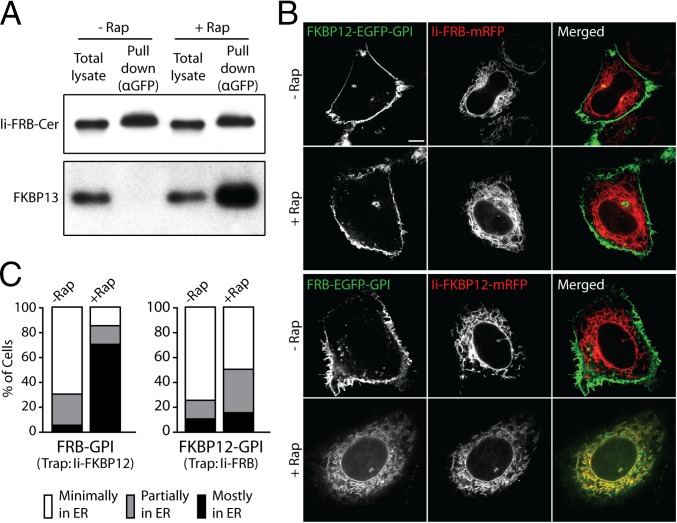
We tested whether such competition occurs in the trapping assay by examining whether endogenous FKBP13 binds to Ii-FRB during rapamycin treatment. Western blot analysis using anti-FKBP13 antibody confirmed that there are significant pools of FKBP13 in the ER of HeLa cells (Fig. 1A). To examine whether endogenous FKBP13 interacts with Ii-FRB, we treated HeLa cells expressing a Cerulean-labeled Ii-FRB construct (Ii-FRB-Cer) with rapamycin for 4 h and then performed a pull-down assay using anti-GFP antibody (which recognizes Cer, a modified GFP). Western blot analysis of the lysate and pull-down fractions revealed that substantial amounts of endogenous FKBP13 were pulled down along with Ii-FRB-Cer by the anti-GFP antibody (Fig. 1A). In cells not treated with rapamycin, the anti-GFP antibody pulled down comparable amounts of Ii-FRB-Cer but failed to pull down detectable amounts of endogenous FKBP13 (Fig. 1A). Thus, endogenous FKBP13 residing in the ER specifically interacts with the FRB domain of Ii-FRB in the presence of rapamycin.
Fig. 1.
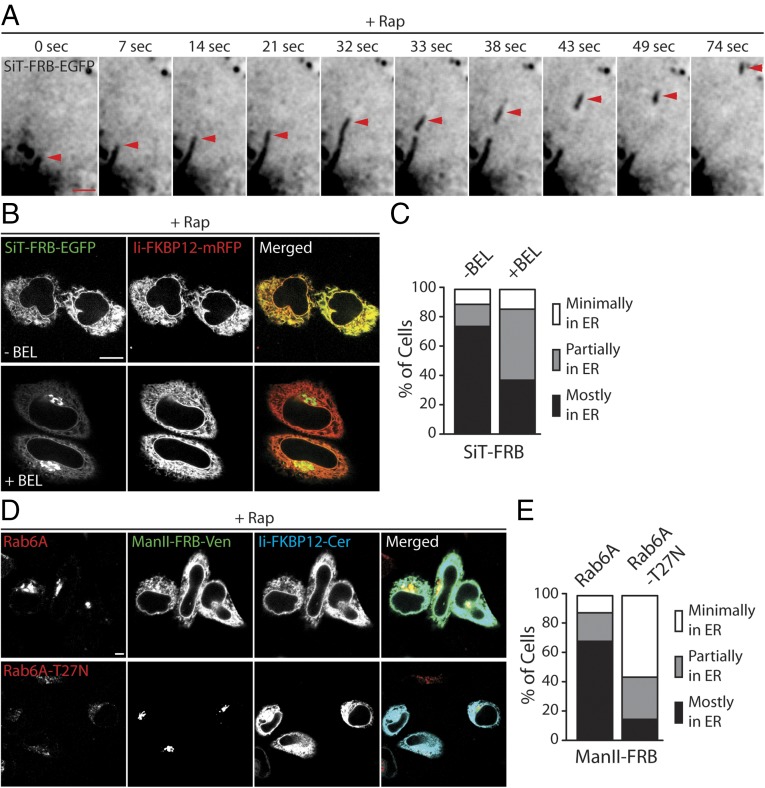
Ii-FRB is not an efficient ER trap for FKBP-tagged proteins due to its interaction with endogenous FKBP13 in the presence of rapamycin (Rap). (A) HeLa cells transiently expressing Ii-FRB-Cer were treated with and without 500 nM rapamycin for 4 h as indicated and subjected to anti-GFP pull-downs. The total lysates and pull-down fractions were analyzed by Western blot for Ii-FRB-Cer and endogenous FKBP13 using anti-GFP and anti-FKBP13 antibodies, respectively. (B) Confocal images of HeLa cells expressing either FKBP12-EGFP-GPI and Ii-FRB-mRFP (rows 1 and 2) or FRB-EGFP-GPI and Ii-FKBP12-mRFP (rows 3 and 4) following incubation with or without rapamycin for 4 h. (Scale bar: 10 μm.) (C) Percentage of cells expressing the indicated ER trap and FRB/FKBP12-tagged GPI-anchored cargo proteins with minimal, partial, and major amounts of EGFP-GPI fluorescence in the ER following 4 h of incubation with rapamycin. At least 50 cells were analyzed for each condition.
Ii-FKBP12, but Not Ii-FRB, Is an Effective ER Trapper.
A consequence of Ii-FRB’s binding to endogenous FKB13 under rapamycin treatment is that Ii-FRB may be an ineffective ER trapper for FKBP12-labeled proteins. We tested this possibility by examining whether Ii-FRB could effectively retain FKBP12 proteins in the ER during rapamycin treatment. As bait for ER retention, we chose the newly synthesized secretory cargo, an EGFP-tagged GPI-anchored protein (EGFP-GPI), given that it originates in the ER before moving to the PM, and therefore should be available for trapping if the assay is effective.
When we coexpressed FKBP12-EGFP-GPI and Ii-FRB-monomeric RFP (mRFP) and examined their distribution in cells treated with rapamycin for 4 h, no significant trapping of FKBP12-EGFP-GPI in the ER was seen (Fig. 1 B and C). Instead, FKBP12-EGFP-GPI trafficked to the PM and accumulated there, whereas Ii-FRB-mRFP labeled the ER, similar to the distributions of these proteins in cells not treated with rapamycin. This result suggested Ii-FRB is an inefficient ER trapper for FKBP12-tagged proteins. We then reversed the configuration of the FRB and FKBP probes on EGFP-GPI and Ii, coexpressing Ii-FKBP12-mRFP and FRB-EGFP-GPI. In cells treated with rapamycin for 4 h, significant ER retention of FRB-EGFP-GPI was now seen, with FRB-EGFP-GPI fluorescence completely overlapping with Ii-FKBP12 fluorescence (Fig. 1 B and C). In the absence of rapamycin, FRB-EGFP-GPI trafficked to the PM, whereas Ii-FKBP12-mRFP remained in the ER. These results suggested that Ii-FKBP12 effectively traps FRB-labeled proteins in the ER under rapamycin treatment. The orientation of FKBP12 and FRB probes thus appears to be critical for successful ER trapping of proteins during rapamycin treatment. The implication of these results is that previous studies using Ii-FRB as a trap for recycling FKBP12-Golgi proteins (15, 18) may have underestimated the extent of Golgi protein cycling through the ER.
Ii-FKBP12 Traps FRB-Tagged Golgi Enzymes in the ER in the Presence of Rapamycin.
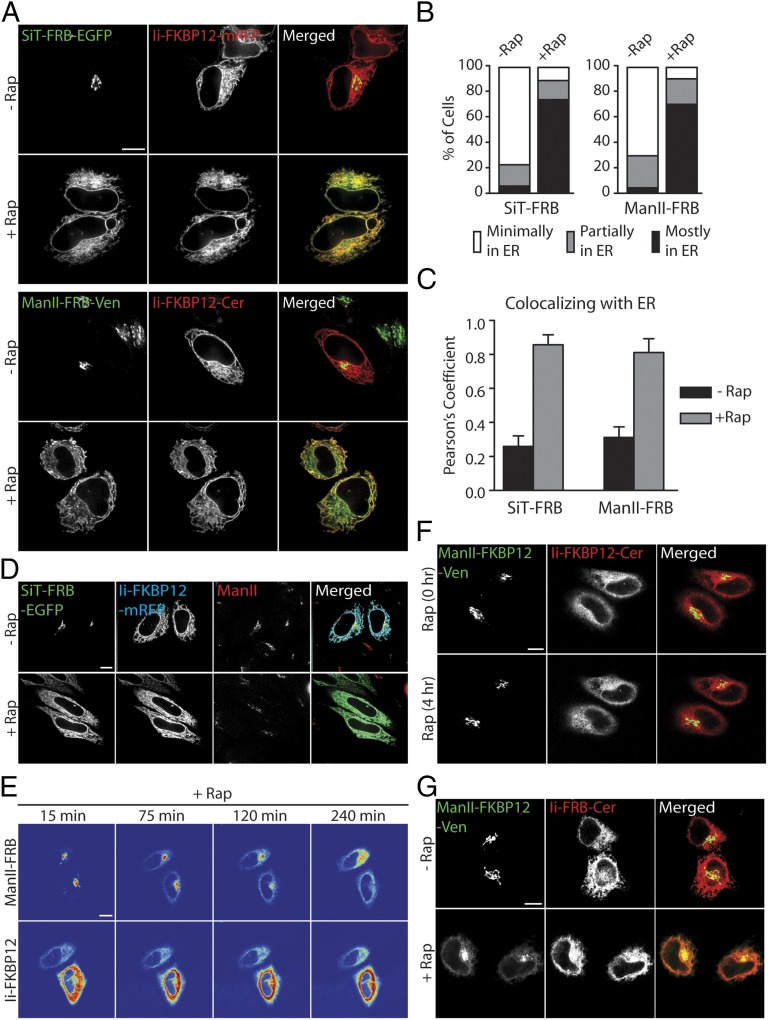
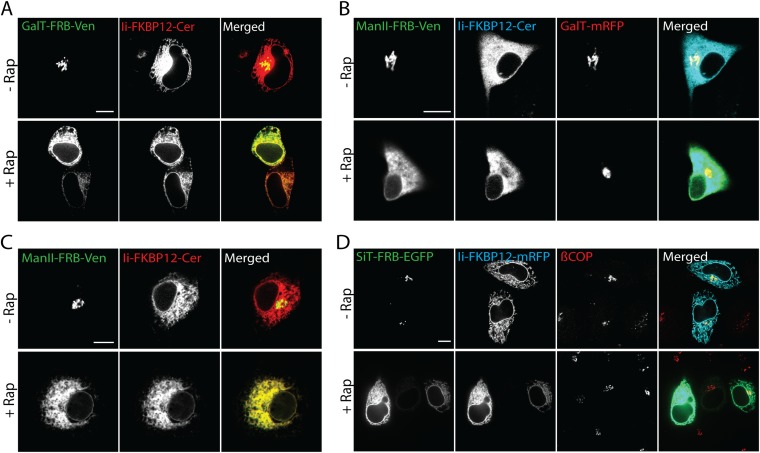
We used the modified ER trapping assay to revisit the question of whether Golgi enzymes cycle through the ER, using Ii-FKBP12 in combination with FRB-tagged Golgi proteins. The Golgi enzymes that we modified with FRB included the Golgi anchoring sequences of mannosidase II (Man II-FRB-Venus) and sialyltransferase (SiT-FRB-EGFP). HeLa cells coexpressing either Ii-FKBP12-Cer and Man II-FRB-Venus or Ii-FKBP12-mRFP and SiT-FRB-EGFP were treated for 4 h with rapamycin (the incubation medium also included cycloheximide to block new protein synthesis). Before rapamycin treatment, the Golgi markers were primarily localized in the Golgi apparatus (appearing as a compact, juxtanuclear structure) and Ii-FKBP12 was dispersed throughout the ER and connecting nuclear envelope (Fig. 2A). Upon addition of rapamycin for 4 h, however, Man II-FRB-Venus and SiT-FRB-EGFP both redistributed into the ER, colocalizing with Ii-FKBP12 (Fig. 2A, merged images). Similar ER trapping was observed in rapamycin-treated cells expressing full-length β-1,4-galactosyl transferase (GalT-FRB-Venus) and Ii-FKBP12-Cer (Fig. S1A). Rapamycin-mediated redistribution of FRB-tagged Golgi enzymes to the ER was also seen in NRK (Fig. S1B) and COS-7 cells (Fig. S1C).
Fig. 2.
Rapamycin-based trapping assay reveals that Golgi enzymes undergo continuous recycling between the Golgi and ER. (A) HeLa cells expressing Ii-FKBP12 tagged with the indicated fluorescent protein and FRB-tagged indicated Golgi enzyme were treated with cycloheximide ± rapamycin for 4 h. (B) Percentage of cells coexpressing the indicated FRB-tagged Golgi enzyme and Ii-FKBP12 with minimal, partial, and major amounts of Golgi enzyme fluorescence in the ER following 4 h of incubation with cycloheximide ± rapamycin. At least 60 cells were analyzed for each condition. (C) Colocalization of the indicated FRB-tagged Golgi enzymes with an ER trap (Ii-FKBP12) following cycloheximide ± rapamycin treatment of cells quantified by Pearson’s colocalization analysis. Data are expressed as mean ± SEM. (D) Distribution of SiT-FRB-EGFP, Ii-FKBP12-mRFP, and endogenous Man II (detected by anti-Man II antibody) in HeLa cells following incubation with cycloheximide ± rapamycin for 4 h. (E) Time-lapse confocal images captured after addition of cycloheximide ± rapamycin to HeLa cells coexpressing Man II-FRB-Venus and Ii-FKBP12-Cer. (F) HeLa cells coexpressing Man II-FKBP12-Venus and Ii-FKBP12-Cer were imaged before and after 4 h of incubation with cycloheximide + rapamycin. (G) Distribution of Man II-FKBP12-Venus and Ii-FRB-Cer in HeLa cells after incubation with cycloheximide ± rapamycin for 4 h. (Scale bars: 10 μm.)
Fig. S1.
Constitutive recycling of Golgi enzymes through the ER revealed by rapamycin (Rap)-based trapping assay. (A) HeLa cells coexpressing GalT-FRB-Venus and Ii-FKBP12-Cer were treated with 100 μg/mL cycloheximide ± 250 nM rapamycin for 4 h. (B) Distribution of Man II-FRB-Venus, Ii-FKBP12-Cer, and GalT-mRFP in NRK cells following incubation with 100 μg/mL cycloheximide ± 250 nM rapamycin for 4 h. (C) COS-7 cells coexpressing Man II-FRB-Venus and Ii-FKBP12-Cer were treated with 100 μg/mL cycloheximide ± 250 nM rapamycin for 4 h. (D) Distribution of SiT-FRB-EGFP, Ii-FKBP12-mRFP, and endogenous βCOP (detected by anti-βCOP antibody) in HeLa cells following incubation with 100 μg/mL cycloheximide ± 250 nM rapamycin for 4 h. (Scale bars: 10 μm.)
When we scored cells based on whether there was minimal, partial, or complete ER redistribution of the Golgi reporter, little or no Golgi reporter fluorescence was left in the juxtanuclear Golgi apparatus after 4 h of the rapamycin treatment (Fig. 2B). Calculation of the Pearson’s colocalization coefficient from deconvoluted images of Ii-FKBP12-Cer and Man II-FRB-Venus or Ii-FKBP12-mRFP and SiT-FRB-EGFP further showed that nearly all 4-h rapamycin-treated cells exhibited a large colocalization coefficient of 0.8 or more, in contrast to cells not treated with rapamycin (Fig. 2C). Therefore, robust trapping of Golgi reporters in the ER upon rapamycin treatment occurred in these cells, supporting the idea that Golgi proteins cycle through the ER constitutively. The ER distribution of the Golgi reporters occurred without affecting the structural integrity of the Golgi apparatus; there was no change in the steady-state distribution of β-COPI, a coat protein regulating intra-Golgi traffic (Fig. S1D), and the distribution of endogenous Golgi enzyme Man II (labeled with antibodies, Fig. 2D) and GalT-mRFP (with no FRB tag) (Fig. S1B) did not change during the experiments.
The time course for FRB-tagged Golgi protein redistribution into the ER upon rapamycin treatment visualized in single cells revealed a slow, continuous loss of Golgi fluorescent signal that was matched by a gain of ER signal over time (Fig. 2E and Movie S1). This redistribution phenotype was consistent with a slow constitutive recycling of Golgi enzymes back to the ER and trapping by Ii-FKBP12-Cer.
When we coexpressed Ii-FKBP12-Cer and Man II-FKBP12-Venus, no trapping of the Golgi constructs in the ER during rapamycin treatment occurred, as expected, because FKBP12 does not dimerize with itself under rapamycin treatment (Fig. 2F and Movie S2). Under these conditions, both Golgi constructs maintained their steady-state distribution as compact, juxtanuclear Golgi-localized proteins during the entire length of image acquisition.
We also looked for rapamycin-induced ER trapping using Ii-FRB as trap for recycling FKBP12-Golgi proteins to reproduce the conditions used by Pecot and Malhotra (15). Under these conditions, Ii-FRB is likely to bind to endogenous FKBP13 in the ER, and therefore be an ineffective trapper (Fig. 1A). Consistent with this possibility, Man II-FKBP12-Venus remained largely localized in the Golgi under these conditions with very little ER trapping (Fig. 2G), similar to the results reported by Pecot and Malhotra (15).
FRB-Golgi Enzymes Trapped in the ER Undergo FRET with Ii-FKBP12.
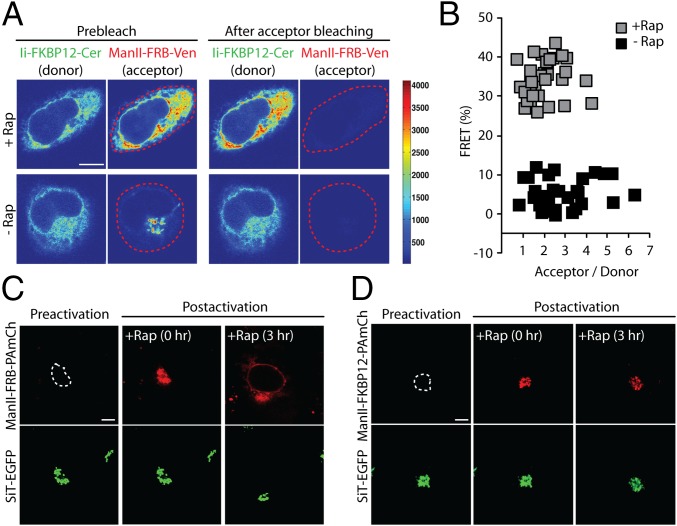
Ii-FKBP12-Cer and Man II-FRB-Venus are topologically optimally oriented for FRET. To verify that the observed ER redistribution of Man II-FRB-Venus under rapamycin treatment was due to trapping by heterodimerization with Ii-FKBP12-Cer, we looked for evidence of FRET between these two markers in rapamycin-treated cells. FRET was quantified from the release of donor (Cer) fluorescence quenching following the irreversible photobleaching of the acceptor (Venus) fluorophore with an intense 514-nm laser pulse. A substantial relief of donor quenching in cells treated with rapamycin was observed, as evidenced by the brighter fluorescence of Ii-FKBP12-Cer after bleaching of Man II-FRB-Venus (Fig. 3A). This fluorescence increase only occurred in rapamycin-treated cells. Quantification of randomly selected cells revealed a mean FRET efficiency of ∼40% after the rapamycin-induced redistribution of Man II-FRB-Venus into the ER, indicating significant interaction between Ii-FKBP12-Cer and Man II-FRB-Venus (Fig. 3B). Untreated cells, by contrast, had a significantly lower average FRET efficiency (<10%) (Fig. 3B). The strong FRET signal confirmed the specificity of the trapping assay, supporting the interpretation that rapamycin-induced redistribution of Man II-FRB into the ER reflects a constitutive recycling pathway followed by Golgi enzymes.
Fig. 3.
FRET and photoactivation-based assays confirm that redistribution of FRB-tagged Golgi enzymes to the ER is due to rapamycin-mediated complexation of recycling enzymes with an FKBP12-tagged ER trap. (A) HeLa cells expressing Ii-FKBP12-Cer and Man II-FRB-Venus were treated with cycloheximide ± rapamycin for 4 h. Confocal images of cells before and after photobleaching of the cellular Man II-FRB-Venus within the region indicated by the dashed line (red). (B) Percentage of FRET between Man II-FRB-Venus and Ii-FKBP12-Cer in HeLa cells imaged according to protocol described in A. (C) Golgi pool of Man II-FRB-PAmCh, transiently coexpressed in HeLa cells with Ii-FKBP12-Cer and SiT-EGFP, was selectively photoactivated within the region indicated by the dashed line. The Golgi region was identified by SiT-EGFP fluorescence. Images of a cell before photoactivation (preactivation), immediately after photoactivation (postactivation, 0 h), and after 3 h of incubation with cycloheximide ± rapamycin (postactivation, 3 h) (D) Golgi pool of Man II-FKBP12-PAmCh in HeLa cells, transiently coexpressed with Ii-FKBP12-Cer and SiT-EGFP, was selectively photoactivated within the region indicated by the dashed line. Images of a cell before photoactivation (preactivation), immediately after photoactivation (postactivation, 0 h), and after 3 h of incubation with cycloheximide ± rapamycin (postactivation, 3 h). (Scale bars: 10 μm.)
ER-Trapped Golgi Enzymes Derive from Preexisting Golgi Structures.
To strengthen this inference further, we used a photoactivatable fluorescent protein, photoactivatable mCherry (PAmCh), attached to Man II-FRB (Man II-FRB-PAmCh) in the ER trapping assay. PAmCh undergoes photoconversion from a dark-to-visible state upon illumination with UV light (24, 25). This photoconversion allows selective highlighting of discrete protein populations within cells. Importantly, molecules that have not been highlighted by selective illumination in the assay remain dark, and are therefore invisible. Thus, proteins that undergo biosynthesis or completion of folding after the photoactivation event will not be visible during the course of an experiment.
We coexpressed Man II-FRB-PAmCh with SiT-EGFP and Ii-FKBP12-Cer. Using SiT-EGFP to identify and demarcate the boundary of the Golgi apparatus, we selectively photoactivated Man II-FRB-PAmCh localized in the Golgi and then treated the cells with rapamycin (Fig. 3C). Man II-FRB-PAmCh signal in the Golgi was slowly depleted with the simultaneous appearance of reticular ER signal that gradually increased in intensity over time. After 3 h, nuclear rim labeling and a fluorescent pattern resembling the ER were clearly detectable, indicating that significant levels of photoactivated Man II-FRB-PAmCh had redistributed to the ER. In contrast to these results, when we photoactivated Man II-FKBP12-PAmCh in cells coexpressing Ii-FKBP12-Cer, conditions where trapping should be ineffective, the Golgi signal remained primarily localized to the Golgi after 3 h of incubation with rapamycin (Fig. 3D).
Because photoactivatable Man II-FRB-PAmCh was used in these experiments, fluorescence signal accumulating in the ER over time during rapamycin treatment could only have originated from the pool of Man II-FRB-PAmCh that was photoactivated in the Golgi (i.e., without photoactivation, Man II-FRB-PAmCh never becomes fluorescent). This result ruled out the possibility that Golgi fluorescence arising in the ER during rapamycin treatment originates from newly synthesized, unfolded Golgi proteins in the ER converting to fluorescent forms. Instead, the data support the view that Golgi enzymes continuously cycle through the ER during their lifetime.
Golgi Enzymes Use Tubule Carriers to Recycle to the ER.
We employed the trapping assay to characterize the transport carriers that mediate the recycling of Golgi enzymes back to the ER. This approach allowed visualization of Golgi enzymes returning to the ER without contamination from ER-derived enzymes moving into the Golgi (i.e., these enzymes would be trapped in the ER). The assay also did not perturb the overall organization and activity of the ER/Golgi system (Fig. 2D and Fig. S1B).
Cells coexpressing SiT-FRB-EGFP and Ii-FKBP12-Cer were treated with rapamycin, and the behavior of SiT-FRB-EGFP in association with the Golgi was then examined using time-lapse confocal imaging (Fig. 4A and Movies S3 and S4). Long tubule elements enriched in SiT-FRB-EGFP were seen extending off the perinuclear Golgi and detaching at variable frequencies. The detached tubules moved outward along straight or curvilinear tracks toward the cell periphery (Movie S4). Often, small, discrete intermediates were seen breaking off from the tips of the extended tubules, but at other times, the tubules themselves broke off from the Golgi. After tracking out to the cell periphery, the tubules disappeared, likely from tubule fusion with the ER, which would quickly disperse SiT-FRB-EGFP signal throughout the large surface area of the ER. The results thus suggested that Golgi enzymes use tubule and globular intermediates to recycle back to the ER.
Fig. 4.
Transport intermediates and regulators of the Golgi-to-ER retrograde pathway mediating cycling of Golgi enzymes. (A) Time-lapse confocal images showing a transport carrier breaking off from a tubule (red arrowhead) originating from the Golgi apparatus in rapamycin-treated cells coexpressing SiT-FRB-EGFP and Ii-FKBP12-mRFP. (Scale bar: 2 μm.) (B) HeLa cells coexpressing SiT-FRB-EGFP and Ii-FKBP12-mRFP were imaged after treatment with cycloheximide + rapamycin ± BEL for 4 h. (Scale bar: 10 μm.) (C) Percentage of cells from B with minimal, partial, and major amounts of SiT fluorescence in the ER following 4 h of incubation with cycloheximide + rapamycin ± BEL. (D) Confocal images of cells coexpressing Man II-FRB-Venus, Ii-FKBP12-Cer, and either myc-tagged WT rab6a (Top) or myc-tagged, dominant-negative, GDP-bound rab6a-T27N (Bottom) following treatment with cycloheximide ± rapamycin for 4 h. (Scale bar: 10 μm.) (E) Percentage of cells coexpressing Man II-FRB-Venus, Ii-FKBP12-Cer, and the indicated rab6a construct, with minimal, partial, and major amounts of Man II fluorescence in the ER following 4 h treatment with cycloheximide + rapamycin. At least 60 cells were analyzed for each condition.
Golgi Enzyme Recycling Requires Cation-iPLA2.
We next investigated what controls the formation of the Golgi enzyme tubule carriers. Previous work has identified iPLA2 as an important regulator of tubules emanating from membrane-bound organelles (26, 27). Preventing iPLA2 activity using a specific inhibitor bromoenol lactone (BEL) both blocks brefeldin A (BFA)-induced Golgi tubulation and prevents tubule-mediated recycling of ERGIC-53 back to the ER (28). We used BEL to test whether the tubule-mediated retrograde transport of Golgi enzymes seen in our ER trapping assay was iPLA2-dependent, and thus inhibitable by BEL treatment.
SiT-FRB-EGFP and Ii-FKBP12-mRFP were coexpressed in cells. After a 15-min pretreatment with 1 μM BEL, the cells were then incubated with rapamycin in the continued presence of BEL for 4 h. As shown in Fig. 4 B and C, a significant fraction of SiT-FRB-EGFP remained localized in the Golgi and did not redistribute into the ER under these conditions. Over the same time period, SiT-FRB-EGFP in rapamycin-treated cells alone had completely relocalized to the ER with little or no fluorescence left in the Golgi region (Fig. 4 B and C). Thus, ER trapping of Golgi enzymes is inhibited by BEL treatment, suggesting Golgi enzyme recycling to the ER is dependent on iPLA2 activity.
Golgi Enzyme Recycling to the ER Is Rab6a-Dependent.
Previous work has implicated rab6a in retrograde transport of Golgi-localized Shiga toxin to the ER (29, 30). We therefore tested whether rab6a activity is required for ER trapping of Golgi enzymes in the rapamycin-induced heterodimerization assay. We transiently transfected myc-tagged versions of WT rab6a (myc-rab6a) and dominant-negative GDP-restricted rab6a (myc-rab6aT27N) and evaluated how their expression affected rapamycin-induced ER trapping of Man II-FRB-Venus by Ii-FKBP12-Cer. The expression of WT and GDP-restricted rab6a was confirmed using a combination of anti-myc polyclonal antibody and Alexa633–anti-rabbit secondary antibody. WT rab6a was mainly localized to the Golgi complex, but the GDP-restricted rab6a appeared diffusely distributed throughout the cell (Fig. 4D, Left).
In cells transfected with WT rab6a, no change in rapamycin-induced Man II-FRB-Venus entrapment in the ER was seen (Fig. 4 D and E), suggesting expression of the protein did not affect Golgi recycling. By contrast, in the presence of rab6a-T27N, Man II-FRB-Venus remained concentrated in a compact, juxtanuclear Golgi-like pattern after 4 h of rapamycin treatment, with no detectable redistribution of the protein to the ER (Fig. 4 D and E). These results suggested that rab6a activity is essential for Golgi enzymes to recycle back to the ER.
Golgi Fragmentation During Microtubule Depolymerization Involves Enzyme Recycling Through the ER.
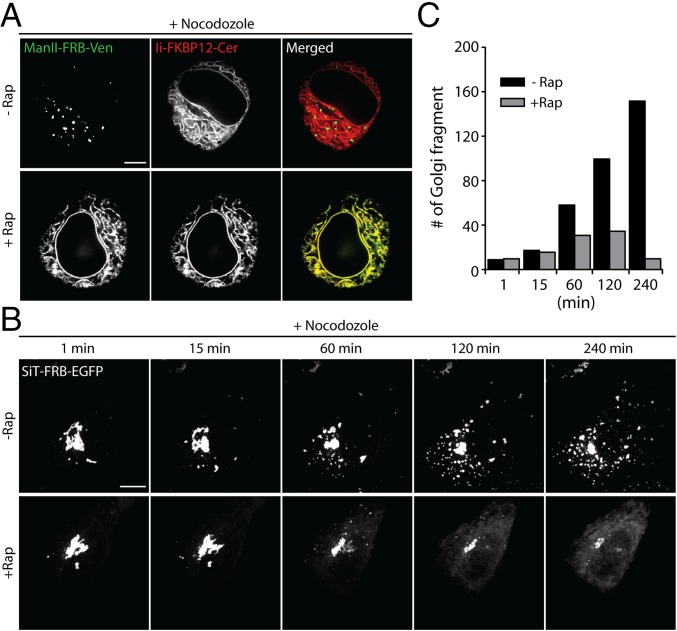
The Golgi is known to fragment into hundreds of smaller elements scattered throughout the cell when microtubules are depolymerized by nocodazole treatment (31, 32). The Golgi fragments are proposed to arise either by de novo generation from recycling Golgi enzymes passing through the ER (11, 12, 33) or by direct Golgi fragmentation without ER involvement (15, 34). To distinguish between these possibilities, we used the ER trapping assay, examining whether or not Golgi fragments arise in nocodazole-treated cells treated with rapamycin. If no fragments appear and Golgi enzymes are trapped in the ER, then the ER-dependent model for nocodazole-induced Golgi fragmentation is supported. On the other hand, if fragments appear and no Golgi enzymes become trapped in the ER, then the ER-independent model for nocodazole-induced Golgi fragmentation is favored.
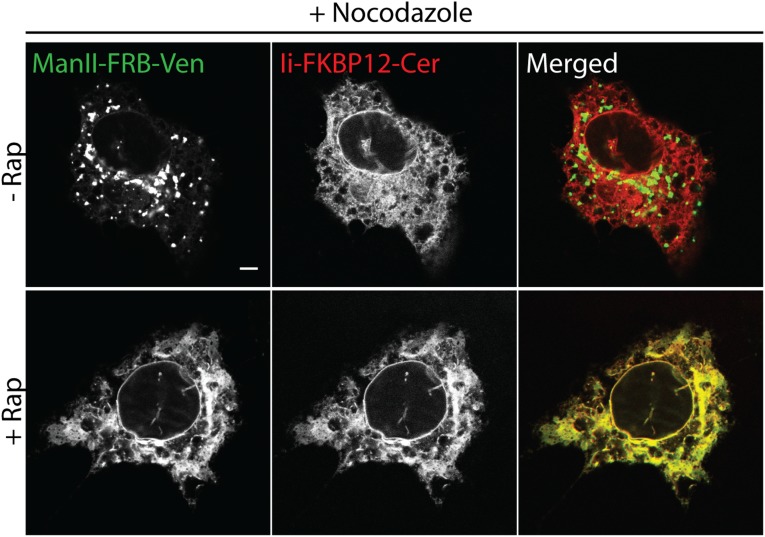
Cells coexpressing either Man II-FRB-Venus and Ii-FKBP12-Cer or SiT-FRB-EGFP and Ii-FKBP12-mRFP were incubated with nocodazole in the presence or absence of rapamycin and imaged after 3 h (Fig. 5 A–C and Fig. S2). In cells treated with nocodazole alone, the labeled Golgi enzymes redistributed from tightly clustered, juxtanuclear structures to scattered small elements distributed throughout the cytoplasm, as reported previously for Golgi enzyme behavior in nocodazole-treated cells (33) (Fig. 5A; no rapamycin). The time-course for fragmentation in cells expressing SiT-FRB-EGFP and Ii-FKBP12-mRFP revealed a slow steady increase in Golgi fragment number with time, reaching over 100 distinct fragments after 4 h of treatment (Fig. 5 B and C; no rapamycin).
Fig. 5.
Golgi fragmentation induced by microtubule depolymerization is mediated by recycling of Golgi enzymes through the ER. (A) Confocal images of HeLa cells coexpressing Man II-FRB-Venus and Ii-FKBP12-Cer following simultaneous treatment with cycloheximide + nocodazole ± rapamycin for 4 h. (B) Time-lapse confocal images of redistribution of Golgi-localized SiT-FRB-EGFP were acquired following treatment of HeLa cells coexpressing SiT-FRB-EGFP and Ii-FKBP12-mRFP with cycloheximide + nocodazole ± rapamycin. Images shown are maximum intensity projections of Z-stacks. (C) Number of discrete SiT-FRB-EGFP containing Golgi fragments in cells shown in B at the indicated time after simultaneous incubation with cycloheximide + nocodazole ± rapamycin. (Scale bars: 10 μm.)
Fig. S2.
Golgi fragmentation in COS-7 cells following depolymerization of microtubules is mediated by recycling of Golgi enzymes through the ER. Images of COS-7 cells coexpressing Man II-FRB-Venus and Ii-FKBP12-Cer following treatment with 100 μg/mL cycloheximide + 5 μg/mL nocodazole ± 250 nM rapamycin for 4 h. (Scale bar: 10 μm.)
When rapamycin was added to the incubation medium, virtually no Golgi fragments appeared during the nocodazole treatment (Fig. 5 A–C; rapamycin). Instead, the Golgi markers shifted from having a juxtanuclear localization to a diffuse ER fluorescence, colocalizing with Ii-FKBP12-mRFP (Fig. 5 A and B; rapamycin). The vast majority of the cells (>80%) showed this phenotype. The slow time course for this redistribution, as shown in cells expressing SiT-FRB-EGFP and Ii-FKBP12-mRFP, matched the rate of ER trapping seen in rapamycin-treated cells alone (Fig. 5 B and C), suggesting nocodazole did not enhance or retard the rate of retrograde transport of the Golgi enzymes. These results support the view that Golgi fragmentation during nocodazole treatment results from recycling of Golgi enzymes back to the ER and reemergence of these enzymes from the ER as distal Golgi elements.
Golgi Fragmentation During Mitosis Involves Enzyme Recycling Through the ER.
The question of how the Golgi apparatus is inherited by daughter cells during mitosis has intrigued researchers for years. Unlike the ER, which is directly segregated as an intact tubular membrane system during mitosis, the Golgi undergoes a choreographed disassembly process (35, 36). Prior studies using conventional imaging approaches have shown Golgi disassembly during mitosis involves redistribution of Golgi enzymes from a perinuclear compact structure into widely dispersed small punctate elements and unresolvable haze. Following completion of mitosis, these scattered elements and the haze disappear and a centrally positioned Golgi composed of stacked cisternae reemerges (14) (13). Two models have been proposed for the composition of this Golgi-derived mitotic material formed during the transition of the cell through mitosis and its role in Golgi reassembly at the end of mitosis. The ER-linked model postulates that the haze and punctae represent either Golgi enzyme resorption into the ER (i.e., haze) or resorption and reemergence at ER exit sites (i.e., puncta). The non–ER-linked model describes the same material as resulting from Golgi disintegration into tiny vesicles and small elements with no communication with the ER (18, 37–40).
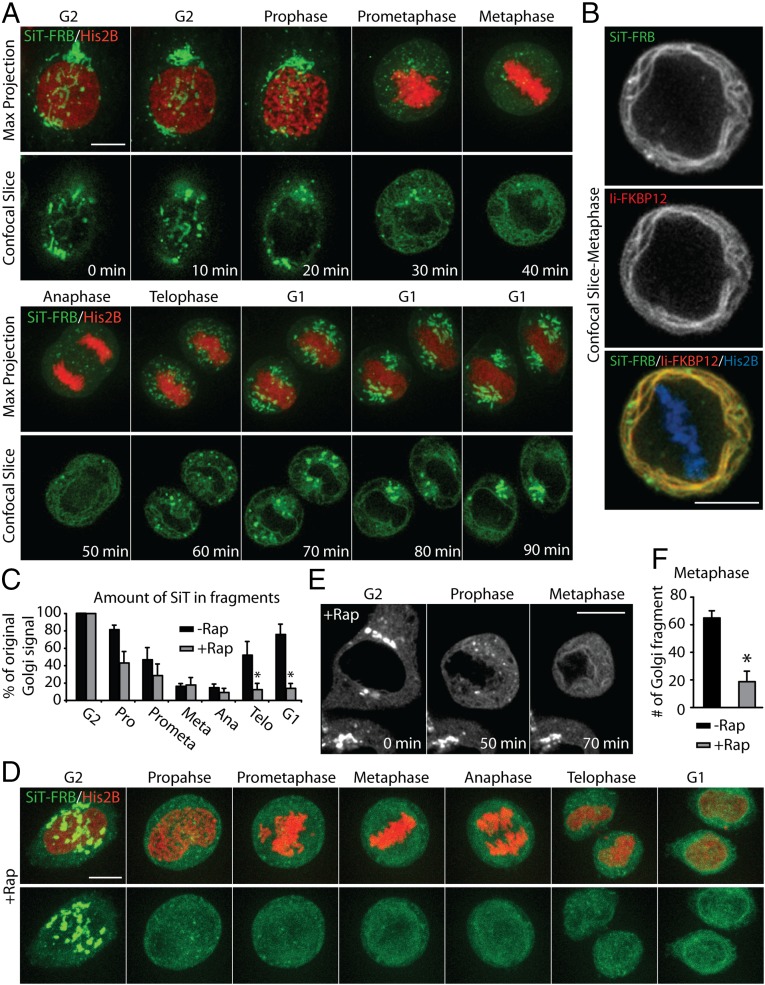
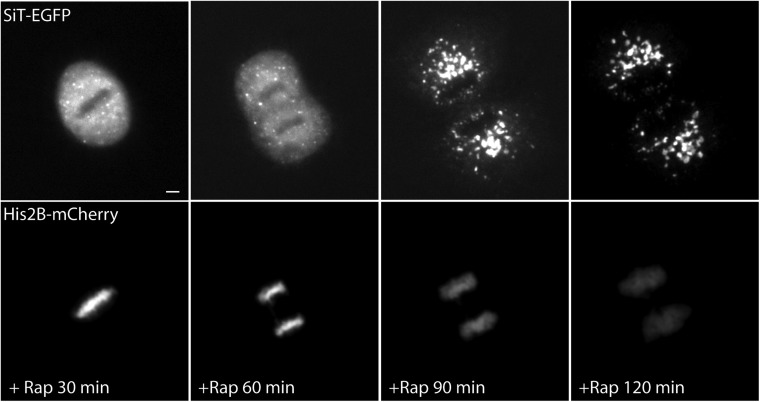
We used the ER trapping assay to distinguish between these two models because mitotic cells treated with rapamycin would have significant ER trapping if the ER resorption model is correct and little ER trapping if the ER-independent model is correct. To visualize the spatial redistribution of Golgi enzymes during mitosis, cells were transfected with SiT-FRB-Venus and Ii-FKBP12-Cer. In addition, we expressed a histone protein tagged with mCherry, H2B-mCh, to highlight chromosomes so that the cell cycle stage could be identified as the cell transitioned through mitosis.
We began by imaging cells undergoing mitosis in the absence of rapamycin (Fig. 6A and Movie S5). As the cell progressed into metaphase, the large centrally located Golgi structure shrank in size and the fluorescence signal of the Golgi enzyme progressively redistributed into structures with distinct morphology: Part of the SiT-FRB-Venus florescence became associated with fine, closely spaced reticular and diffuse structures, whereas the rest of the fluorescence was concentrated into small, discrete elements scattered throughout the cytoplasm. Comparison of the diffuse SiT-FRB-Venus signal with the signal obtained for the ER-resident Ii-FKBP12-Cer revealed a strong colocalization (Fig. 6B), indicating that this nonpunctate SiT-FRB-Venus signal corresponds to the ER. These results support the conclusion that Golgi proteins largely redistribute into the ER in mitotic cells. Most of the small, discrete punctate structures seen in these cells were juxtaposed closely to the ER network, and likely represented ER exit sites (13).
Fig. 6.
Golgi enzymes passage through the ER during mitosis. (A) Time-lapse images of single HeLa cell, transiently coexpressing SiT-FRB-Venus (green), Ii-FKBP12-Cer, and H2B-mCh (red), as it progresses through mitosis. H2B-mCh signal was used to identify the cell cycle stage of the confocal Z-slices maximum projection (Top) and single confocal slices (Bottom) at the indicated time after the start of image acquisition. (B) Distribution of SiT-FRB-Venus, Ii-FKBP12-Cer, and H2B-mCh in a HeLa cell at metaphase. (C) Analysis of SiT-FRB-Venus fluorescence in confocal images of HeLa cells coexpressing Ii-FKBP12-Cer and H2B-mCh and undergoing mitosis in the absence or presence of rapamycin. The percentage of total cellular SiT fluorescence signal in discrete fragments in HeLa cells at indicated stages of mitosis is shown. (D) Time-lapse images of a rapamycin-treated HeLa cell, transiently coexpressing SiT-FRB-Venus (green), Ii-FKBP12-Cer, and H2B-mCh (red), as it undergoes mitosis. (E) Distribution of SiT-FRB-Venus in rapamycin-treated HeLa cells coexpressing Ii-FKBP12-Cer and H2B-mCh at the indicated stages of cell cycle. (F) Number of Golgi fragments in metaphase HeLa cells treated with or without rapamycin. Data are expressed as mean ± SEM. *P < 0.01. (Scale bars: 10 μm.)
We quantified how the fluorescence signal associated with the perinuclear Golgi complex in G2 phase redistributes and partitions between the two mitotic structures during mitotic progression. As the cell transitioned through mitosis, there was a progressive decrease in fluorescence associated with Golgi fragments as increasing amounts of fluorescence became associated with ER-like reticular structures, eventually reaching a minimum in metaphase (Fig. 6C). Eventually, less than 10% of the initial Golgi signal was associated with discrete fragments in metaphase and anaphase. However, the fragment-associated fluorescence started increasing as the cell progressed toward telophase, increasing to ∼50% at telophase and going beyond 80% as the perinuclear Golgi reemerged at G1 (Fig. 6C). This spatiotemporal redistribution of the Golgi enzyme through mitosis strongly suggested that the majority of the Golgi enzymes return to the ER during mitosis.
To test this possibility further, we added rapamycin to cells expressing SiT-FRB-Venus and Ii-FKBP12-Cer as they entered mitosis in late G2. This treatment did not affect the redistribution of SiT-FRB-Venus to ER-associated and disparate Golgi fragments (Fig. 6D and Movie S6). However, there was a significant decrease in the number of punctate structures at different stages of mitosis in the presence of rapamycin (Fig. 6 C–E). The average number of punctae at metaphase decreased from ∼70 to ∼15 when cells underwent mitosis in the presence of rapamycin (Fig. 6F). Concomitantly, the percentage of Golgi fluorescence associated with ER at different stages of mitosis was significantly higher in the presence of rapamycin, with 90% or more of the Golgi signal associated with ER-like structures at anaphase. Finally, SiT-FRB-Venus remained associated with the ER as the cell emerged from mitosis and was not included in the perinuclear Golgi structures that formed at the end of mitosis (Fig. 6D). When cells expressing SiT-EGFP (lacking the FRB domain) were treated with rapamycin as they entered mitosis, SiT-EGFP redistributed into the reassembling Golgi apparatus of daughter cells at the completion of mitosis (Fig. S3), confirming that rapamycin, per se, does not affect the reformation of the Golgi following mitosis. These results support the view that Golgi enzymes redistribute into the ER during mitosis and that they must follow an ER export pathway to reform the Golgi at the end of mitosis.
Fig. S3.
Rapamycin does not affect Golgi reformation following mitosis of HeLa cells. Time-lapse images of a rapamycin (500 nM)-treated HeLa cell, transiently coexpressing SiT-EGFP (Top) and Histone 2B-mCh (Bottom), as it undergoes mitosis. (Scale bar: 10 μm.)
Discussion
For years, the extent to which Golgi enzymes undergo recycling back to the ER has been uncertain despite its significance for understanding the nature of the Golgi’s existence. On the one hand, ER exit site blockers and photobleaching experiments have suggested Golgi constituents continuously cycle through the ER (7, 8). On the other hand, rapamycin-induced ER trapping experiments using Ii-FRB as an ER trap have implied that Golgi enzymes remain localized to the Golgi throughout their lifetime (15). To reconcile these conflicting observations, we used a modified rapamycin trapping assay. Our modified assay used an FKBP12-tagged Ii as an ER trap and FRB-tagged Golgi enzyme marker instead of the reverse configuration used in prior work (i.e., FKBP12-Golgi enzyme and Ii-FRB). This modification was necessary because an endogenous FKBP protein, FKBP13, present in the lumen of the ER, can bind to Ii-FRB in the presence of rapamycin, thereby sequestering the trap and preventing it from interacting with FKBP12-tagged cargo proteins. We confirmed this behavior by showing that endogenous FKBP13 can pull down Ii-FRB in the presence of rapamycin. Furthermore, Ii-FRB does not retain newly synthesized cargo proteins in the ER through rapamycin-induced trapping, whereas the reverse configuration using Ii-FKBP12 does.
Using our modified ER trap assay, we tested whether Ii-FKBP12 could trap FRB-tagged Golgi enzymes in the ER, because this trapping would provide evidence whether or not Golgi enzymes are constitutively retrieved to the ER. Within 4 h of rapamycin treatment, Ii-FKBP12 trapped nearly all Golgi-localized FRB-Golgi enzymes in the ER. Direct redistribution from the Golgi to the ER during rapamycin treatment occurred because selective photoactivation of FRB-tagged, photoactivatable Golgi enzyme in the Golgi resulted in the signal shifting to the ER. In the ER, fluorescent forms of FRB-Golgi enzyme and Ii-FKBP12 underwent FRET, indicating direct binding upon rapamycin-induced redistribution. By contrast, use of Ii-FRB, which we show is an inefficient ER trap, resulted in minimal redistribution of FKBP12-tagged Golgi enzymes during rapamycin treatment. These results are in agreement with prior studies using ER exit site blockers and photobleaching experiments measuring retrograde transport, which suggested that Golgi enzymes undergo slow continuous cycling back to the ER (7, 8). (An additional discussion of Golgi enzyme recycling kinetics is provided in SI Text). Thus, Golgi enzymes continually circulate through the ER, with backward flux of Golgi enzymes back to the ER being a fundamental way the Golgi maintains its existence in the midst of continuous outward flow of secretory cargo to the PM.
The continual revisiting of the ER by Golgi enzymes is likely to play several beneficial roles, including allowing damaged enzymes to refold or undergo degradation through the ER’s quality control system (29, 41, 42). This phenomenon of secondary quality control involving return to the ER from the Golgi could also be important for controlling the positioning and organization of the Golgi itself. Indeed, the time frame of recycling of Golgi enzymes suggests it is a major determinant of the Golgi’s steady-state size and shape. The recycling pathway can also serve as a regulatory circuit to engage cargo molecules and control their distribution at the PM and other post-Golgi organelles. Furthermore, the kinetics of this pathway can be modulated to adjust the steady-state distribution of proteins between the Golgi and ER to maintain homeostasis within the early secretory pathway in the face of changes in the flux of anterograde and retrograde traffic.
One condition in which the Golgi significantly changes its size and shape is during microtubule depolymerization (31). Within a few hours of disrupting microtubules with nocodazole treatment, the Golgi fragments into hundreds of small elements, all closely associated with ER exit sites (33). Using our modified rapamycin trapping assay, we showed that this redistribution requires Golgi recycling through the ER because FRB-Golgi enzymes shifted into the ER rather than relocated to Golgi fragments in rapamycin-treated cells coexpressing Ii-FKBP12. This finding supports previous work that used ER-export blockers and photobleaching experiments to address the mechanism for nocodazole-induced Golgi fragmentation (11, 12, 33), and suggests that microtubules are not necessary for retrograde flux of Golgi enzymes back to the ER. Because microtubules are crucial for delivery of pre-Golgi transport intermediates to the centrosomal region (43), Golgi fragments appearing during nocodazole treatment arise from Golgi enzymes that have returned to the ER, reemerged at ER exit sites, and remodeled into mini-Golgi stacks.
The Golgi also alters its state strikingly during mitosis. Here, the Golgi transitions into fragments and then into a highly dispersed form in metaphase, only to reassemble again into two distinct daughter Golgi systems during cytokinesis (36). Using the modified rapamycin trapping assay, we demonstrated that mitotic Golgi breakdown and reassembly involves the ER as an intermediary at all stages of fragmentation and dispersal. This finding supports prior work using ER exit blockers and photobleaching-based assays that highlighted the role of ER export activity in Golgi reassembly at the end of mitosis (13, 14) and directs attention to mechanisms that alter the rates of anterograde and retrograde pathways between the ER and Golgi in understanding mitotic Golgi disassembly/reassembly pathways. Indeed, the greater speed of rapamycin-induced trapping of Golgi proteins in the ER during mitosis compared with that during interphase (Fig. 5C) suggests retrograde trafficking is greatly accelerated during mitosis.
Given that Golgi enzymes follow a retrograde pathway back to the ER, what mechanism underlies it? Two pathways for Golgi-to-ER transport have been described. In one, KDEL receptor and Rer1p, which contain COPI recognition motifs, function as receptors to bind and retrieve escaped ER resident proteins (44, 45). This pathway is COPI and Arf1 GTPase-dependent (46). A second pathway is Arf1/COPI-independent and relies on Rab6 GTPase, uses tubules rather than vesicles, and carries Shiga and Shiga-like toxins (29, 30, 47, 48). Our results suggest this pathway is used by Golgi enzymes to return to the ER: Not only were tubule intermediates seen carrying Golgi enzymes into the ER during rapamycin-induced ER trapping but no Golgi enzyme trapping occurred in the presence of a dominant-negative rab6a mutant. Our further finding that PLA2 inhibitors retard the retrograde transport of Golgi enzymes is consistent with the tubule character of this pathway and suggests PLA2 could function by introducing lipids like lysophospholipid into the cytoplasmic side of Golgi membranes (49), thereby shifting its planar lipid bilayer into a tubule form.
Recent work by Borgese and coworkers (42) has suggested that positive signals on proteins moving through the Golgi are required for them to escape entering the Rab6-dependent retrograde pathway to the ER. Their data showed that mutation of the di-acidic–based export signal on VSVG led to increased recycling of VSVG back to the ER through a Rab6-dependent retrieval pathway. This result implies that PM-destined cargo lacking explicit export signals may end up spending a significant time cycling between the Golgi and ER before eventually reaching the cell surface. The Rab6 dependence of this retrieval pathway suggests it is the same pathway followed by Golgi enzymes back to the ER. Because Golgi enzymes lack positive export signals, they would continually enter this retrograde pathway.
The rate of entry of cargo into the retrograde pathway is possibly further modulated by interactions with regulators like the coat protein COPI (50). Indeed, recent work by Hsu and coworkers (51) has suggested retrograde transport within the Golgi/ER system is affected by the extent of a cargo protein’s affinity for COPI and the activation state of Cdc42. The idea that COPI–cargo interactions regulate membrane flux within the ER/Golgi system fits well with the explosive retrograde trafficking stimulated by BFA (52). Within seconds of BFA treatment, Arf1 and COPI dissociate from Golgi membranes and massive retrograde trafficking of Golgi enzymes back into the ER occurs via long tubules (52, 53). One possibility is that the retrograde pathway uncovered by our trapping assay represents a regulated version of the rapid retrograde trafficking stimulated by BFA. In this scenario, COPI might play a critical role in suppressing tubule formation and preventing unregulated retrograde trafficking. The interaction of proteins with COPI would restrain their backward flux into the retrograde pathway, whereas release from COPI interactions would lead to their entry into this pathway. Because all membrane proteins reaching the Golgi would be subjected to this system, recycling would be an additional rate-limiting factor for cargo clearance from the Golgi system. Testing these and other ideas regarding the mechanisms and roles of retrograde traffic to the ER is sure to be benefited by the use of the modified ER trapping assay described in this study.
SI Text
Our estimate of Golgi enzyme transport rates back to the ER (t1/2 of ∼2 h) is somewhat slower than the Golgi enzyme transport rates back to the ER reported in prior experiments using ER exit blockers or photobleaching recovery experiments (t1/2 of ∼1 h) (7, 8). This discrepancy could be due to FRB-Golgi enzymes weakly interacting with endogenous FKBP13. Because no ER relocalization of FRB-Golgi enzymes occurred in cells treated with rapamycin without expression of Ii33-FKBP12, any weak binding of endogenous FKBP13 (a luminal protein) to FRB-Golgi enzyme (a membrane protein) likely results in FKBP13 being carried with FRB-Golgi enzyme back to the Golgi, thereby retarding Ii33-FKBP12–mediated trapping of the FRB-Golgi enzymes in the ER. This interference by endogenous FKBP13 would be transient because FKBP12 has a stronger binding affinity for FRB compared with endogenous FKBP13 (23) and we were overexpressing FKBP12-tagged Ii33 relative to FKBP13. The stronger binding affinity of FKBP12 for FRB can also explain the observation that the ER-FRB trap retains FKBP12-Golgi enzyme HeLa cells following BFA treatment and washout (15). BFA causes the entire pool of FKBP12-Golgi enzyme to relocate to the ER. The resulting high concentration of FKBP12-Golgi enzyme in the ER, coupled with the higher binding affinity of FKBP12 for FRB, could result in FKBP12-Gogi enzymes displacing endogenous FKBP13 in the Ii33/FRB/FKBP13 complex. The binding of FKBP12-Golgi enzyme to Ii33-FRB could explain why Golgi enzymes failed to redistribute to the Golgi upon BFA washout in these experiments. However, we found that the effects of BFA in HeLa cells are not completely reversible, which could provide an additional explanation for ER retention of the Golgi enzymes in the previous study.
Experimental Procedures
Detailed methods are provided in SI Experimental Procedures.
Cell Culture and Plasmid Constructs.
Strategies for construction of plasmids are described in SI Experimental Procedures.
All experiments were performed with HeLa cells, unless specified otherwise. Cells were grown in Lab-Tek chambers with no. 1.5 borosilicate cover glasses (Nalge Nunc International) or on no. 1.5 round cover glasses (Warner Instruments) precoated with fibronectin (EMD Millipore). Transfections were performed with FuGENE 6 transfection reagent (Roche). Cells transfected with plasmids were imaged 30–48 h after transient transfection. Unless otherwise indicated, working concentrations of drugs were as follows: 250 nM rapamycin, 100 μg/mL cycloheximide, 1 μM BEL, and 5 μg/mL nocodazole (Sigma). For fixed cell experiments, cells were washed quickly in PBS prewarmed to 37 °C and immediately fixed in 4% (wt/vol) paraformaldehyde and 0.2% (wt/vol) glutaraldehyde (Electron Microscopy Sciences) for 35 min at room temperature in PBS. Fixative was quenched with 50 mM glycine and imaged in PBS. For live cell experiments, cells were imaged either in phenol red-free DMEM (Mediatech, Inc.) within a CO2-controlled stage top incubation chamber or in phenol-red free DMEM supplemented with 25 mM Hepes (pH 7.5).
Biochemistry.
GFP pull-downs were performed on HeLa cells grown to 80% confluency in 10-cm dishes 48 h after transfection with GFP-fusion constructs. The μMACS GFP Isolation Kit (catalog no. 130-091-288; Miltenyi Biotec) was used according to the manufacturer’s recommendations, except that 1% CHAPS, 50 mM Hepes (pH 7.4), and 100 mM NaCl were used for cell lysis and washes to maintain protein interactions.
Fluorescence Microscopy.
Spinning disk confocal images were acquired using either a Marianas spinning disk (Intelligent Imaging Innovations) attached to a Zeiss Observer.Z1 microscope (Carl Zeiss) equipped with a 100× apochromatic, 1.4-N.A. (Carl Zeiss) objective lens or a Nikon Ti-E microscope (Nikon Instruments) equipped with a Yokogawa spinning-disk scan head (no. CSU-X1; Yokogawa) using a 100× apochromatic, 1.49 N.A. oil-immersion objective lens. Confocal images were also collected using Zeiss LSM510 and Zeiss LSM510 META laser-scanning microscopes (Carl Zeiss).
Image Analysis.
Image analysis was performed using custom-written code in MATLAB (MathWorks) and ImageJ (NIH). Details of image analysis strategies are outlined in the SI Experimental Procedures. Deconvolution of spinning disk images was done using Slidebook 5.0 software (Intelligent Imaging Innovations).
SI Experimental Procedures
Cell Culture and Reagents.
HeLa, COS-7, and NRK cells were purchased from the American Type Culture Collection and cultured in phenol red-free DMEM (Corning) supplemented with 10% (vol/vol) FBS and 2 mM glutamine (Invitrogen).
Rabbit antisera for GFP and RFP were made as described (56). Anti-FKBP13 antibody (R&D Systems), anti-Man II antibody (EMD Millipore), anti–β-COP antibody (Abcam), and Alexa Fluor 647 goat anti-rabbit IgG secondary antibody (Life Technologies) were purchased. Rapamycin, cycloheximide, BEL, and nocodazole were purchased from Sigma.
Plasmids.
The cDNAs for FKBP12 and the FRB domain of FRAP1 were synthesized by Genscript and provided in the plasmid pUC57. The FKBP12 cDNA was amplified from the plasmid pUC57-FKBP12 (from Genscript), using the N-terminal primer 5′-GATCGTCGACGATGGGAGTGCAGGTG-3′ containing a SalI site (underlined) and the C-terminal primer 5′-GATCGGATCCCGTTCCAGTTTTAGAAG-3′ containing a BamHI site (underlined). The FRB cDNA was amplified from the plasmid pUC57-FRAP1 (Genscript), using the N-terminal primer 5′-GATCGTCGACGGAGATGTGGCATGAAG-3′ containing a SalI site (underlined) and the C-terminal primer 5′-GATCGGATCCCGCTGCTTTGAGATTCG-3′ containing a BamHI site (underlined). The amplification products were gel-purified, digested with the restriction endonucleases SalI and BamHI, and ligated into a similarly digested pEGFP-N1 containing the A206K monomerizing mutation (5) (Clontech Laboratories, Inc.) to make FKBP-EGFP and FRB-EGFP.
The ER-retained version of invariant chain (Iip35) (16, 17) was amplified from the plasmid, pcDNA-Iip35 (a kind gift from Paul Roche, National Cancer Institute, Bethesda), using the N-terminal primer 5′-ATCGAGCTCGCCACCATGCACAGGAGGAGAAG-3′ containing a SacI site (underlined) and the C-terminal primer 5′-GATCGTCGACATGGGGACTGGGCC-3′ containing a SalI site (underlined). The amplification products were gel-purified, digested with the restriction endonucleases SacI and SalI, and ligated into a similarly digested pVenus-N1 containing the A206K monomerizing mutation (54) (Clontech Laboratories, Inc.) to make Ii-Venus. FKBP-EGFP and FRB-EGFP plasmids were digested with SalI and BamHI to remove the FKBP and FRB cDNAs. These fragments were gel-purified and ligated into similar digested Ii-Venus to produce Ii-FKBP-Venus and Ii-FRB-Venus. Cer versions were made by digesting Ii-FKBP-Venus and Ii-FRB-Venus with XhoI and BamHI. The resulting fragments were gel-purified and ligated into a similarly digested pCer-N1 containing the A206K monomerizing mutation (54) (Clontech Laboratories, Inc.) to produce Ii-FKBP-Cer and Ii-FRB-Cer. Ii-FKBP-mRFP was created by digesting out mRFP from GalT-mRFP with Age1 and Not1 restriction endonucleases and ligating the gel-purified mRFP fragment into similarly digested Ii-FKBP-Cer.
FRB-YFP-GPI and FKBP-YFP-GPI were generated from EGFP-GPI, which is described elsewhere, by polymerase PCR amplifying FRB and FKBP from Ii-FRB-Cer and Ii-FKBP-Cer, respectively; digesting the PCR products with the endonucleases Kpn1 and Age1; and ligating the digested PCR product into similarly digested EGFP-GPI. FRB was amplified using the primers N-terminal primer 5′-GCAGGTACCGGAGATGTGGCATGAAGGCCT-3′ containing a Kpn1 site (underlined) and C-terminal primer 5′-GCTACCGGTAGCTGCTTTGAGATTCGTCGGA-3′ containing an Age1 (underlined) restriction site. FKBP was amplified using the primers N-terminal primer 5′-GCAGGTACCGATGGGAGTGCAGGT GGAAAC-3′ containing a Kpn1 site (underlined) and C-terminal primer 5′-GCTACCGGTAGTTCCAGTTTTAGAAGCTCCA-3′ containing an Age1 restriction site (underlined).
The plasmid construction for the SiT-EGFP, Man II-EGFP, and GalT-EGFP is described elsewhere (55). FKBP-EGFP and FRB-EGFP plasmids were digested with SalI and BamHI to remove the FKBP and FRB cDNAs. These fragments were gel-purified and ligated into similar digested Man II-EGFP, SiT-EGFP, and GalT-EGFP plasmids to produce Man II-FKBP-EGFP, SiT-FKBP-EGFP, GalT-FKBP-EGFP, Man II-FRB-EGFP, SiT-FRB-EGFP, and GalT-FRB-EGFP. The Venus versions of these constructs were made by digesting cDNAs containing the Golgi protein along with the binding domains using NheI and BamHI. The fragments were gel-purified and ligated into similarly digested pVenus-N1 plasmids. Man II-FRB-PAmCh was generated by digesting pN1-PAmCh with Age1 and Bsrg1 and ligating the resulting PAmCh fragment into similarly digested Man II-FRB-Venus. GalT-mRFP was made by digesting pmRFP-N1 with Age1 and Bsrg1 and ligating the resulting mRFP fragment into similarly digested GalT-EGFP.
H2B-mCh was a kind gift from Mike Davidson (Florida State University, Tallahassee, FL), and Myc-rab6a and Myc-rab6a-T27N were generous gifts from Juan Bonifacino (National Institute of Child Health and Human Development, NIH, Bethesda).
Immunofluorescence.
Cells were washed with PBS prewarmed to 37 °C and immediately fixed with 4% (wt/vol) paraformaldehyde in PBS for 20 min at room temperature. Cells were permeabilized with 0.1% Triton X-100 in PBS (10 min), blocked with 10% (vol/vol) bovine serum albumin (Sigma) in PBS (blocking solution) for 30 min, and incubated with primary antibodies in blocking solution for 1 h. Cells were again washed three times with blocking solution before 1 h of incubation with secondary antibodies. Cells were washed three times with PBS and imaged in PBS.
Microscopy.
Selective photoactivation and imaging of the Golgi pool of Man II-FRB-PAmCh was carried out on Zeiss LSM510 laser-scanning microscope with a 63× plan apochromat and 1.4-N.A. objective in multitracking mode. A region of interest (ROI) encompassing the Golgi apparatus, identified by fluorescence of coexpressed SiT-EGFP, was used to photoactivate the Golgi pool of Man II-FRB-PAmCh with a 413-nm laser line, and time-lapse images of the photoactivated protein were taken with a 561-nm laser line.
Three-dimensional (0.5-μm Z-stacks), confocal time-lapse images of nocodazole-treated cells were acquired with a 100× apochromat, 1.49-N.A. oil immersion objective on a Nikon Ti-E microscope (Nikon Instruments) equipped with a Yokogawa spinning-disk scan head (no. CSU-X1; Yokogawa). Time-lapse sequences of images of mitotic cells were captured with a Marianas spinning disk (Intelligent Imaging Innovations) attached to a Zeiss Observer.Z1 microscope (Carl Zeiss) equipped with an apochromat 100×, 1.4-N.A. objective lens (Carl Zeiss).
Image Analysis.
All image analysis was performed using custom-written code in MATLAB (MathWorks) and ImageJ (NIH).
Quantification of ER localization.
The extent of trapping of EGFP-GPI in the ER and the redistribution of Golgi enzymes to the ER were quantified by estimating the fraction of total EGFP-GPI and Golgi enzyme cellular fluorescence, respectively, present in the ER following treatment with the indicated drugs. First, to estimate the total cellular fluorescence (of EGFP-GPI or Golgi enzyme), a ROI representing the cell (cell-ROI) was generated either based on the perimeter of the cell defined by the PM-localized EGFP-GPI or by selecting an area encompassing the entire ER in the ER channel. The mean pixel values of a background region (a region outside the cell) in each channel were subtracted from the mean pixel value of the cell-ROI. The integrated fluorescence intensity of the background-subtracted cell-ROI was used as a measure of the total cellular fluorescence of the EGFP-GPI/Golgi enzyme. Next, the ER-channel image (representing the FKBP12 tagged Ii) was subjected to an image segmentation algorithm to generate a binary mask of the ER. In this ER-mask image, pixels corresponding to the ER have a value of 1, whereas all other pixels have a value of 0. The percentage of total EGFP-GPI or Golgi enzyme fluorescence within the pixels belonging to the ER mask was calculated and used to categorize the retention/redistribution of the proteins in the ER. Cells were classified having minimal, partial, or most of the proteins in the ER as follows:
-
i)
Minimally in ER (less ER signal than PM/Golgi): less than 25% (≤25%) of EGFP-GPI or Golgi enzyme fluorescence in ER pixels
-
ii)
Partially in ER (both ER and PM/Golgi fluorescence): between 25% and 75% (>25% and ≤75%) of EGFP-GPI or Golgi enzyme fluorescence in ER pixels
-
iii)
Mostly in ER (more ER signal than PM/Golgi): greater than 75% (>75%) of EGFP-GPI or Golgi enzyme fluorescence in ER pixels
Pearson’s coefficient.
Pearson’s coefficient between the ER and the Golgi channels was calculated from background-subtracted Z-stack images and for pixels within the ROI representing the cell.
FRET analysis.
Background-subtracted donor images (Ii-FKBP-Cer) before and after photobleaching of the acceptor were used to calculate the FRET efficiency between the Golgi enzymes and the ER trap. A ROI was selected that encompasses the entire ER depicted in the donor channel following photobleaching of the acceptor. The FRET efficiency was calculated for each image from the intensities of the donor before and after photobleaching of the acceptor according to the following equation:
where, and represent the fluorescence intensities of the donor before and after photobleaching of the acceptor, respectively.
Discrete Golgi fragments in cells treated with nocodazole or undergoing mitosis were identified by generating binary masks of the Z-stacks of the Golgi channel images using image segmentation. Using the binary masks, the number of Golgi fragments at different time points was counted and the fraction of original Golgi enzyme fluorescence (at the start of the experiment when the cell is in G2) present in discrete fragments at different stages of mitosis was calculated.
Supplementary Material
Acknowledgments
We thank Juan Bonifacino (US National Institute of Child Health and Development) for sharing the rab6a plasmids and Mike Davidson (Florida State University) for sharing the H2B-mCh plasmid. This work was supported by the Intramural Research Program of the NIH.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1520957112/-/DCSupplemental.
References
- 1.Képès F, Rambourg A, Satiat-Jeunemaître B. Morphodynamics of the secretory pathway. Int Rev Cytol. 2005;242:55–120. doi: 10.1016/S0074-7696(04)42002-6. [DOI] [PubMed] [Google Scholar]
- 2.Nakano A, Luini A. Passage through the Golgi. Curr Opin Cell Biol. 2010;22(4):471–478. doi: 10.1016/j.ceb.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Mellman I, Warren G. The road taken: Past and future foundations of membrane traffic. Cell. 2000;100(1):99–112. doi: 10.1016/s0092-8674(00)81687-6. [DOI] [PubMed] [Google Scholar]
- 4.Altan-Bonnet N, Sougrat R, Lippincott-Schwartz J. Molecular basis for Golgi maintenance and biogenesis. Curr Opin Cell Biol. 2004;16(4):364–372. doi: 10.1016/j.ceb.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 5.Cole NB, Ellenberg J, Song J, DiEuliis D, Lippincott-Schwartz J. Retrograde transport of Golgi-localized proteins to the ER. J Cell Biol. 1998;140(1):1–15. doi: 10.1083/jcb.140.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: Evidence for membrane cycling from Golgi to ER. Cell. 1989;56(5):801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ward TH, Polishchuk RS, Caplan S, Hirschberg K, Lippincott-Schwartz J. Maintenance of Golgi structure and function depends on the integrity of ER export. J Cell Biol. 2001;155(4):557–570. doi: 10.1083/jcb.200107045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miles S, McManus H, Forsten KE, Storrie B. Evidence that the entire Golgi apparatus cycles in interphase HeLa cells: Sensitivity of Golgi matrix proteins to an ER exit block. J Cell Biol. 2001;155(4):543–555. doi: 10.1083/jcb.200103104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puri S, Linstedt AD. Capacity of the golgi apparatus for biogenesis from the endoplasmic reticulum. Mol Biol Cell. 2003;14(12):5011–5018. doi: 10.1091/mbc.E03-06-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole NB, et al. Diffusional mobility of Golgi proteins in membranes of living cells. Science. 1996;273(5276):797–801. doi: 10.1126/science.273.5276.797. [DOI] [PubMed] [Google Scholar]
- 11.Storrie B, et al. Recycling of golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J Cell Biol. 1998;143(6):1505–1521. doi: 10.1083/jcb.143.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drecktrah D, Brown WJ. Phospholipase A(2) antagonists inhibit nocodazole-induced Golgi ministack formation: Evidence of an ER intermediate and constitutive cycling. Mol Biol Cell. 1999;10(12):4021–4032. doi: 10.1091/mbc.10.12.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altan-Bonnet N, et al. Golgi inheritance in mammalian cells is mediated through endoplasmic reticulum export activities. Mol Biol Cell. 2006;17(2):990–1005. doi: 10.1091/mbc.E05-02-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaal KJ, et al. Golgi membranes are absorbed into and reemerge from the ER during mitosis. Cell. 1999;99(6):589–601. doi: 10.1016/s0092-8674(00)81548-2. [DOI] [PubMed] [Google Scholar]
- 15.Pecot MY, Malhotra V. The Golgi apparatus maintains its organization independent of the endoplasmic reticulum. Mol Biol Cell. 2006;17(12):5372–5380. doi: 10.1091/mbc.E06-06-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schutze MP, Peterson PA, Jackson MR. An N-terminal double-arginine motif maintains type II membrane proteins in the endoplasmic reticulum. EMBO J. 1994;13(7):1696–1705. doi: 10.1002/j.1460-2075.1994.tb06434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuwana T, Peterson PA, Karlsson L. Exit of major histocompatibility complex class II-invariant chain p35 complexes from the endoplasmic reticulum is modulated by phosphorylation. Proc Natl Acad Sci USA. 1998;95(3):1056–1061. doi: 10.1073/pnas.95.3.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pecot MY, Malhotra V. Golgi membranes remain segregated from the endoplasmic reticulum during mitosis in mammalian cells. Cell. 2004;116(1):99–107. doi: 10.1016/s0092-8674(03)01068-7. [DOI] [PubMed] [Google Scholar]
- 19.Nigam SK, et al. Localization of the FK506-binding protein, FKBP 13, to the lumen of the endoplasmic reticulum. Biochem J. 1993;294(Pt 2):511–515. doi: 10.1042/bj2940511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin YJ, et al. Molecular cloning of a membrane-associated human FK506- and rapamycin-binding protein, FKBP-13. Proc Natl Acad Sci USA. 1991;88(15):6677–6681. doi: 10.1073/pnas.88.15.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi J, Chen J, Schreiber SL, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273(5272):239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 22.Castellano F, et al. Inducible recruitment of Cdc42 or WASP to a cell-surface receptor triggers actin polymerization and filopodium formation. Curr Biol. 1999;9(7):351–360. doi: 10.1016/s0960-9822(99)80161-4. [DOI] [PubMed] [Google Scholar]
- 23.Walensky LD, et al. The 13-kD FK506 binding protein, FKBP13, interacts with a novel homologue of the erythrocyte membrane cytoskeletal protein 4.1. J Cell Biol. 1998;141(1):143–153. doi: 10.1083/jcb.141.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subach FV, et al. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat Methods. 2009;6(2):153–159. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sengupta P, Lippincott-Schwartz J. Photohighlighting approaches to access membrane dynamics of the Golgi apparatus. Methods Cell Biol. 2013;118:217–234. doi: 10.1016/B978-0-12-417164-0.00013-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Figueiredo P, Drecktrah D, Katzenellenbogen JA, Strang M, Brown WJ. Evidence that phospholipase A2 activity is required for Golgi complex and trans Golgi network membrane tubulation. Proc Natl Acad Sci USA. 1998;95(15):8642–8647. doi: 10.1073/pnas.95.15.8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Figueiredo P, Polizotto RS, Drecktrah D, Brown WJ. Membrane tubule-mediated reassembly and maintenance of the Golgi complex is disrupted by phospholipase A2 antagonists. Mol Biol Cell. 1999;10(6):1763–1782. doi: 10.1091/mbc.10.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Figueiredo P, et al. Phospholipase A2 antagonists inhibit constitutive retrograde membrane traffic to the endoplasmic reticulum. Traffic. 2000;1(6):504–511. doi: 10.1034/j.1600-0854.2000.010608.x. [DOI] [PubMed] [Google Scholar]
- 29.Girod A, et al. Evidence for a COP-I-independent transport route from the Golgi complex to the endoplasmic reticulum. Nat Cell Biol. 1999;1(7):423–430. doi: 10.1038/15658. [DOI] [PubMed] [Google Scholar]
- 30.White J, et al. Rab6 coordinates a novel Golgi to ER retrograde transport pathway in live cells. J Cell Biol. 1999;147(4):743–760. doi: 10.1083/jcb.147.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thyberg J, Moskalewski S. Microtubules and the organization of the Golgi complex. Exp Cell Res. 1985;159(1):1–16. doi: 10.1016/s0014-4827(85)80032-x. [DOI] [PubMed] [Google Scholar]
- 32.Turner JR, Tartakoff AM. The response of the Golgi complex to microtubule alterations: The roles of metabolic energy and membrane traffic in Golgi complex organization. J Cell Biol. 1989;109(5):2081–2088. doi: 10.1083/jcb.109.5.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole NB, Sciaky N, Marotta A, Song J, Lippincott-Schwartz J. Golgi dispersal during microtubule disruption: Regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Mol Biol Cell. 1996;7(4):631–650. doi: 10.1091/mbc.7.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogalski AA, Singer SJ. Associations of elements of the Golgi apparatus with microtubules. J Cell Biol. 1984;99(3):1092–1100. doi: 10.1083/jcb.99.3.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lucocq JM, Warren G. Fragmentation and partitioning of the Golgi apparatus during mitosis in HeLa cells. EMBO J. 1987;6(11):3239–3246. doi: 10.1002/j.1460-2075.1987.tb02641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thyberg J, Moskalewski S. Reorganization of the Golgi complex in association with mitosis: Redistribution of mannosidase II to the endoplasmic reticulum and effects of brefeldin A. J Submicrosc Cytol Pathol. 1992;24(4):495–508. [PubMed] [Google Scholar]
- 37.Axelsson MA, Warren G. Rapid, endoplasmic reticulum-independent diffusion of the mitotic Golgi haze. Mol Biol Cell. 2004;15(4):1843–1852. doi: 10.1091/mbc.E03-07-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jokitalo E, Cabrera-Poch N, Warren G, Shima DT. Golgi clusters and vesicles mediate mitotic inheritance independently of the endoplasmic reticulum. J Cell Biol. 2001;154(2):317–330. doi: 10.1083/jcb.200104073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shima DT, Cabrera-Poch N, Pepperkok R, Warren G. An ordered inheritance strategy for the Golgi apparatus: Visualization of mitotic disassembly reveals a role for the mitotic spindle. J Cell Biol. 1998;141(4):955–966. doi: 10.1083/jcb.141.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warren G. Membrane partitioning during cell division. Annu Rev Biochem. 1993;62:323–348. doi: 10.1146/annurev.bi.62.070193.001543. [DOI] [PubMed] [Google Scholar]
- 41.Storrie B, Pepperkok R, Nilsson T. Breaking the COPI monopoly on Golgi recycling. Trends Cell Biol. 2000;10(9):385–391. doi: 10.1016/s0962-8924(00)01818-3. [DOI] [PubMed] [Google Scholar]
- 42.Fossati M, Colombo SF, Borgese N. A positive signal prevents secretory membrane cargo from recycling between the Golgi and the ER. EMBO J. 2014;33(18):2080–2097. doi: 10.15252/embj.201488367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Presley JF, et al. ER-to-Golgi transport visualized in living cells. Nature. 1997;389(6646):81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- 44.Cosson P, Lefkir Y, Démollière C, Letourneur F. New COP1-binding motifs involved in ER retrieval. EMBO J. 1998;17(23):6863–6870. doi: 10.1093/emboj/17.23.6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cosson P, Letourneur F. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science. 1994;263(5153):1629–1631. doi: 10.1126/science.8128252. [DOI] [PubMed] [Google Scholar]
- 46.Bremser M, et al. Coupling of coat assembly and vesicle budding to packaging of putative cargo receptors. Cell. 1999;96(4):495–506. doi: 10.1016/s0092-8674(00)80654-6. [DOI] [PubMed] [Google Scholar]
- 47.Majeed W, Liu S, Storrie B. Distinct sets of Rab6 effectors contribute to ZW10--and COG-dependent Golgi homeostasis. Traffic. 2014;15(6):630–647. doi: 10.1111/tra.12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith RD, et al. The COG complex, Rab6 and COPI define a novel Golgi retrograde trafficking pathway that is exploited by SubAB toxin. Traffic. 2009;10(10):1502–1517. doi: 10.1111/j.1600-0854.2009.00965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ha KD, Clarke BA, Brown WJ. Regulation of the Golgi complex by phospholipid remodeling enzymes. Biochim Biophys Acta. 2012;1821(8):1078–1088. doi: 10.1016/j.bbalip.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Presley JF, et al. Dissection of COPI and Arf1 dynamics in vivo and role in Golgi membrane transport. Nature. 2002;417(6885):187–193. doi: 10.1038/417187a. [DOI] [PubMed] [Google Scholar]
- 51.Park SY, Yang JS, Schmider AB, Soberman RJ, Hsu VW. Coordinated regulation of bidirectional COPI transport at the Golgi by CDC42. Nature. 2015;521(7553):529–532. doi: 10.1038/nature14457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sciaky N, et al. Golgi tubule traffic and the effects of brefeldin A visualized in living cells. J Cell Biol. 1997;139(5):1137–1155. doi: 10.1083/jcb.139.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klausner RD, Donaldson JG, Lippincott-Schwartz J. Brefeldin A: Insights into the control of membrane traffic and organelle structure. J Cell Biol. 1992;116(5):1071–1080. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296(5569):913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 55.Patterson GH, et al. Transport through the Golgi apparatus by rapid partitioning within a two-phase membrane system. Cell. 2008;133(6):1055–1067. doi: 10.1016/j.cell.2008.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stefanovic S, Hegde RS (2007) Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell 128(6):1147–1159. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.