Abstract
Biofilm disruption and eradication were investigated as a function of nitric oxide- (NO) releasing chitosan oligosaccharide dose with results compared to control (ie non-NO-releasing) chitosan oligosaccharides and tobramycin. Quantification of biofilm expansion/contraction and multiple-particle tracking microrheology were used to assess the structural integrity of the biofilm before and after antibacterial treatment. While tobramycin had no effect on the physical properties of the biofilm, NO-releasing chitosan oligosaccharides exhibited dose-dependent behavior with biofilm degradation. Control chitosan oligosaccharides increased biofilm elasticity, indicating that the scaffold may mitigate the biofilm disrupting power of nitric oxide somewhat. The results from this study indicate that nitric oxide-releasing chitosan oligosaccharides act as dual-action therapeutics capable of eradicating and physically disrupting P. aeruginosa biofilms.
Keywords: nitric oxide, P. aeruginosa, biofilm, rheology
Introduction
Cystic fibrosis (CF) lung disease is caused by defective chloride transport, resulting in thickened, dehydrated mucus with altered biophysical properties such as increased viscoelasticity and osmotic pressure (Button et al. 2012; Matsui et al. 2006). One pathological consequence of the altered CF mucus is the inhibition of mucociliary clearance, ultimately resulting in increased airway inflammation and infection (Cohen & Prince 2012; Gibson et al. 2003). In addition to preventing the removal of pathogens, thickened CF mucus restricts bacterial motility and promotes P. aeruginosa biofilm formation (Matsui et al. 2006). While biofilms are traditionally defined as cooperative communities of bacteria within a protective matrix (Mah & O'Toole 2001), they also constitute viscoelastic materials with well-defined physical and mechanical properties (Lieleg et al. 2011, Zrelli et al. 2013). Strategies for treating P. aeruginosa biofilms and infections in the CF airways to date have focused on reducing bacterial viability through antibiotic treatment, specifically through the use of inhalable tobramycin. Inhaled tobramycin is currently the only antibiotic recommended for both the treatment of initial (Mogayzel et al. 2014) and chronic (Mogayzel et al. 2013) P. aeruginosa infections in patients with CF. While inhaled tobramycin is effective at eradicating bacteria within biofilms, it fails to physically remove the structural remnants of the biofilm from the airways. Any bacteria that survive antibiotic treatment (e.g. persister cells) may initiate biofilm regrowth and the development of antibiotic-resistant infections (Döring et al. 2012, Schultz et al. 2010, Van Acker et al. 2014). As such, degradation of the biofilm and its removal from the airway are essential to preventing recolonization (Jones et al. 2011, Schultz et al. 2010). Physical disruption of the biofilm also increases the anti-biofilm efficacy of co-administered antibiotics, as antibiotic diffusion becomes enhanced in mechanically weakened biofilms (Alipour et al. 2009, Alkawash et al. 2006, Hatch & Schiller 1998). Therefore, an ideal anti-biofilm therapeutic for CF would both eradicate bacteria and physically degrade the biofilm, facilitating clearance from the airway.
In light of the importance of the viscoelastic properties of biofilms, much recent research has focused on quantifying how chemical and antibiotic treatments alter the mechanical properties of biofilms. Lieleg et al. (2011) reported that neither gentamicin, colistin, ofloxacin, ethanol, nor bleach altered the elasticity of P. aeruginosa biofilms when measured via rheometry. In contrast, ciprofloxacin was shown to reduce the elasticity of P. aeruginosa biofilms to that of a viscous fluid (Jones et al. 2011). As each treatment elicits different effects, it is important to probe how antibacterial agents alter biofilm viscoelasticity for the development of any new therapies.
Nitric oxide (NO) is an endogenously produced diatomic free radical with significant antibacterial activity against P. aeruginosa biofilms (Lu et al. 2013, 2014). At sub-bactericidal concentrations, NO has biofilm dispersing properties (Barraud et al. 2006, 2009). The antibacterial efficacy of NO is derived from its ability to exert both nitrosative and oxidative stresses to bacterial membrane components (eg proteins, lipids, DNA) directly or via reactive byproducts including and dinitrogen trioxide and peroxynitrite (Fang 1997; Jones et al. 2010). As the P. aeruginosa biofilm matrix is composed of proteins, extracellular DNA, and polysaccharides, it is likely that NO would alter or disrupt the structural integrity of these biofilms (Flemming & Wingender 2010; Mann & Wozniak 2012). Furthermore, atomic force microscopy has revealed that NO exposure causes structural damage to the membranes of planktonic Gram-negative bacteria, including P. aeruginosa (Deupree & Schoenfisch 2009). To determine the effects of NO on the viscoelastic properties of P. aeruginosa biofilms, macromolecular scaffolds capable of storing and controllably releasing NO were employed to locally deliver NO to bacterial biofilms (Carpenter & Schoenfisch 2012; Riccio & Schoenfisch 2012). Chitosan oligosaccharides represent an attractive scaffold for pulmonary NO delivery due to several attractive properties including biodegradability, tolerability to mammalian cells, and ease of NO donor functionalization (Kean & Thanou 2010, Lu et al. 2014). Herein, the utility of NO-releasing chitosan oligosaccharides to both eradicate and physically alter P. aeruginosa biofilms is evaluated, with comparison to tobramycin.
Materials and methods
Materials
Tobramycin, medium molecular weight chitosan, and 2-methylaziridine were purchased from Sigma Aldrich (St Louis, MO). Sodium methoxide was purchased from Acros Organics (Geel, Belgium). FluoSpheres carboxylate-modified microspheres (1 μm diameter) for use as tracer particles in microrheology experiments were purchased from Molecular Probes (Life Technologies, Carlsbad, CA). Nitric oxide gas was purchased from Praxair (Sanford, NC). Calibration standard NO gas (26.85 ppm, balance N2), nitrogen (N2), and argon were purchased from Airgas National Welders (Durham, NC). P. aeruginosa strain K (PAK) and each of the isogenic mutants (ie ΔflicA, ΔflicApilA, and ΔmucA22) were gifts from Prof. Matthew Wolfgang from the Department of Microbiology and Immunology, University of North Carolina at Chapel Hill. Phosphate buffered saline (PBS) was made with 10 mM sodium phosphate and adjusted to pH 6.5 to more closely resemble the CF airway (Yoon et al. 2006). Luria-Bertani (LB) broth and agar were purchased from Becton, Dickinson & Company (Franklin Lakes, NJ). Biofilm growth medium was prepared by diluting LB Broth 1:4 in water, after which the pH was adjusted to 6.5 using 10 mM sodium phosphate. Milli-Q water with a resistivity of <18.2 mΩ cm and a total organic content of < 6 ppb was prepared by purifying distilled water using a Millipore Milli-Q UV Gradient A-10 system (Bedford, MA). All common laboratory salts, solvents, and reagents were purchased from Fisher Scientific (Pittsburgh, PA). All materials were used without further purification.
Synthesis of water-soluble 2-methylaziridine- modified chitosan oligosaccharides (COS)
Water-soluble chitosan oligosaccharides were synthesized by degrading medium molecular weight chitosan (2.5 g) in 50 ml of hydrogen peroxide (15 wt. %) for 1 h at 85 °C. The resulting oligosaccharides were filtered to remove insoluble oligosaccharides, precipitated with acetone, collected via centrifugation (6,500 × g, 10 min), and dried in vacuo. The viscosity of the chitosan oligosaccharides was determined using a Ubbelohde viscometer in a solution of sodium chloride (0.20 M) and acetic acid (0.10 M) at 25 °C. The classic Mark-Houwink equation (η = 1.81 × 10−3 M0.93) was used to determine the molecular weight (Maghami & Roberts 1988).
2-Methylaziridine was grafted to the chitosan oligosaccharides as previously described (Lu et al. 2014). Briefly, the water-soluble chitosan oligosaccharides (0.5 g) were dissolved in water (10 ml) after which a solution of concentrated hydrochloric acid (27.5 μl), water (250 μl), and 2-methylaziridine (356 μl) was added drop wise at room temperature. The solution was stirred for 5 d at 25 °C followed by 24 h at 70 °C. The 2-methylaziridine-modified chitosan oligosaccharides (COS) were then collected via precipitation in acetone, washed copiously with ethanol, and dried in vacuo at room temperature. 1H NMR data of COS: (400 MHz, D2O, δ): 0.8–1.1 (NH2CH(CH3)CH2NH), 1.9 (C7: CHNHCOCH3), 2.3–2.9 (NH2CH(CH3)CH2NHCH, C2: NH2CH(CH3)CH2NHCH), 3.3–4.0 (C3, C4, C5, C6: OHCH, OCHCH(OH)CH(NH2)CH, OHCH2CH, OHCH2CH), 4.4 (C1: OCH(CHNH2)O).
N-diazeniumdiolate modification of chitosan oligosaccharides (COS-NO)
N-diazeniumdiolate NO donors were formed on the secondary amines of COS via exposure to high pressures of NO gas (Lu et al. 2014). Briefly, 2-methylaziridine-modified chitosan oligosaccharides (45 mg) were dissolved in water (900 μl), methanol (2.10 ml), and sodium methoxide (5.4 M, 75 μl) in 1 dram glass vials. The vials containing the COS solution were placed in a stainless steel reactor. Oxygen was removed from the system by purging with argon via three short purges (15 s, 8 bar) followed by three long purges (10 min, 8 bar). After the final purge, the vessel was filled with NO gas (10 bar) that had been purified in a potassium hydroxide chamber. The solutions were stirred at room temperature for 72 h. Following N-diazeniumdiolate formation, unreacted NO was removed from solution via the same argon purging procedure used to remove oxygen. The resulting solutions of NO-releasing chitosan oligosaccharides (COS-NO) were centrifuged (6,500 × g, 15 min), precipitated with 3 ml of acetone, collected via centrifugation, and dried in vacuo overnight at room temperature. The solid COS-NO was stored in a vacuum-sealed bag at −20°C until use.
Characterization of nitric oxide release
Real-time NO-release kinetics from COS-NO were determined using at a Sievers Nitric Oxide Analyzer (Boulder, CO). Prior to analysis, the instrument was calibrated with air passed through a NO zero filter (0 ppm NO) and 25.87 ppm of NO standard gas (balance N2). Solid COS-NO (1 mg) was added to 30 ml of deoxygenated PBS (37 °C, pH 6.5). Nitric oxide released from COS-NO was carried to the analyzer using nitrogen gas flowing through the solution at rate of 80 ml min−1 Additional nitrogen was supplied to the reaction flask to achieve the required instrument collection rate of 200 ml min−1. Analysis was terminated when NO levels decreased below 10 ppb mg−1 of chitosan oligosaccharide.
Biofilm growth and eradication assays
Frozen cultures of P. aeruginosa were grown overnight in LB broth, diluted 1:100 in 50 ml of fresh LB broth, and grown to mid-log phase (OD600=0.25). Rapidly growing cultures were diluted to 106 CFU ml−1 (1:100 dilution) in biofilm growth medium. Viscous biofilms were grown in 12-well microtiter plates at 37°C for 72 h with gentle shaking (100 rpm). Biofilms appeared as viscous bacterial aggregates (ie microcolonies) floating in the growth medium. These biofilms were mechanically robust (ie they could not be disrupted by vigorous pipetting), indicating the formation of a suitable experimental matrix (Sriramulu et al. 2005). Biofilms were extracted from the growth media via pipetting and then washed by ejection into PBS (pH 6.5). The biofilms were subsequently added to a solution of PBS (pH 6.5) containing COS-NO, COS, or tobramycin and incubated for 18 h at 37 °C with gentle shaking (100 rpm). The biofilms were again washed in PBS (pH 6.5) to remove excess antibacterial agent prior to further analysis.
Following exposure to the test agents and washing, biofilms were plated and enumerated to determine the minimum biofilm eradication concentration (MBEC), defined herein as the minimal concentration of drug required for a 5-log reduction in bacterial viability. Freshly washed biofilms were gently sonicated for 10 min and vortexed to disrupt the matrix. The resulting solutions were serially diluted in PBS, spiral plated on LB Agar, and incubated at 37°C for 24 h. Bacterial colonies were quantified using a Flash & Go colony counter (IUL, Farmingdale, NY). This method has an inherent limit of detection of 2.5×103 CFU ml−1 (Breed & Dotterrer 1916).
Multiple-particle tracking microrheology
Biofilms were grown as described above except with the incorporation of fluorescent tracer particles. Fluorescent tracer particles were diluted 1:1000 from their stock solution (2 wt. %) into the biofilm growth medium prior to the addition of planktonic bacteria. Biofilms were exposed to antibacterial agents (ie COS-NO, COS, or tobramycin) as in the MBEC assays. Following treatment, the biofilms were placed in a transparent sample holder. Specifically, the biofilms were sealed between a glass microscope slide and coverslip with two sheets of parafilm acting as a spacer. The sample holders were sealed on using parafilm to minimize sample evaporation. Tracer particle movement was recorded at 60 frames s−1 for 30 s with a Flea3 grey scale camera (Point Grey, Richmond, Canada) mounted on a Nikon Eclipse TE2000-E inverted microscope at a magnification of 40×. The tracer particle displacement as a function of time was quantified using Video Spot Tracker software (Center for Computer Integrated Systems for Microscopy and Manipulation, University of North Carolina at Chapel Hill).
The mean squared displacement (MSD) of each tracer particle was calculated from the displacement of individual particles as a function of time as previously described (Hill et al. 2014). Briefly, the MSD was determined according to:
| (1) |
where τ represents time lag, ti is the time at the start of the video (0.00 s), and N is the total number of frames in a video (1,800 for all experiments).
Due to biofilm heterogeneity, the MSD of all tracer particles was ensemble averaged to achieve meaningful MSD curves (Hill et al. 2014). For each condition tested, tracer particle displacement was measured in 15 different viewing areas of three separate biofilms, resulting in ensemble averaging of at least 200 particles per treatment. The ensemble averaged MSD was calculated as previously reported (Hill et al. 2014). For clarity, MSD values were analyzed at a lag time (τ) of 0.83 s. At this time point (MSDτ=0.83s), the accuracy of the camera speed (ie small variations in the exact time each frame is collected) does not affect the measurement. Furthermore, this frequency positioned the measurements above the noise floor (1×104 μm2) and minimized the effects of drift. All MSD values are reported as the mean ± standard error of the mean (SEM). Due to the large sampling size, the error is often too small to be visible on the figures presented herein.
Fluorescence microscopy
Following exposure to COS for 18 h (4 mg ml−1), COS-NO (4 mg ml−1), or tobramycin (200 μg ml−1), biofilms were washed and added to 1 ml of PBS (pH 6.5) containing 30 μM PI. The resulting solutions were incubated for 30 min at 37 °C in the dark. Following exposure, 30 μl of each biofilm sample were deposited on a glass slide for wide-field fluorescence imaging. An Olympus iX80 inverted microscope with a CARV light source (Nikon) and Photometrics Quant EM detector was used to image biofilms. Fluorescent PI images were obtained using BP 542 – 582 nm excitation and 604 – 644 nm emission filters. Images were rendered in Image J.
Measurement of distance between nearest neighbor particles
The average distance between nearest neighbor particles was determined to quantify contraction and expansion of the bacterial biofilms (Lu et al. 2006). Distances between nearest neighbor tracer particles were calculated according to the equation:
| (2) |
where R12 represents the distance between beads, x1 and y1 are the location of tracer particle 1, and x2 and y2 are the location of tracer particle 2. The smallest R12 value for each bead was then selected as the distance between nearest neighbor particles.
Results
Water soluble chitosan oligosaccharides were synthesized from chitosan via oxidative degradation (Chang et al. 2001). The molecular weight of the chitosan oligosaccharides was determined to be 4.410 ± 0.037 kDa by the classic Mark-Houwink Equation (Maghami & Roberts 1988). To impart NO-release capabilities, 2-methylaziridine-modified chitosan oligosaccharide scaffolds (COS) were reacted with NO gas at high pressure. The resulting N-diazeniumdiolate-modified chitosan oligosaccharides (COS-NO) released a total of 0.78 ± 0.09 μmol NO mg−1 over a duration of 10.7 ± 1.1 h in PBS (pH 6.5, 37 °C). The half-life of NO release was 0.62 ± 0.08 h (Supplemental material Figure S1, Table S1).
Viscoelastic properties of P. aeruginosa biofilms
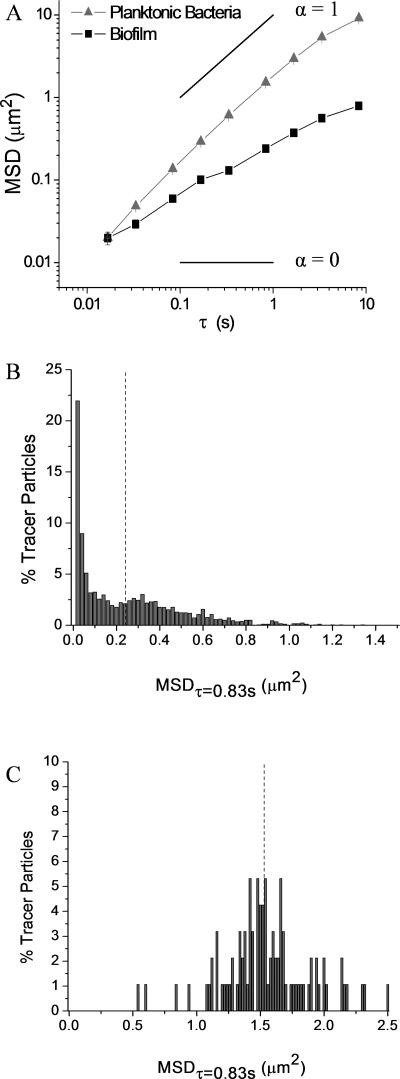
Fluorescent tracer particles were incorporated into P. aeruginosa biofilms during biofilm formation to determine the viscoelastic properties of bacterial biofilms. Tracer particles within the biofilms exhibited decreased diffusion compared to tracer particles in solutions of planktonic bacteria (Figure 1A). The diffusive exponent (α) was derived to approximate viscoelasticity according to MSD ∝ tα. Through this relationship, purely viscous solutions are defined as having diffusive exponents of one, while purely elastic solids exhibit diffusive exponents of zero. Therefore, diffusive exponents of viscoelastic materials range from zero to one (Mason 2000). The displacement of tracer particles in solutions of planktonic bacteria increased linearly with time (α = 1.05 ± 0.13), indicating that they were moving in a purely viscous solution. In contrast, the measured slope for tracer particles within P. aeruginosa biofilms was significantly lower (α = 0.62 ± 0.14), indicating that the medium surrounding the particles was both viscous and elastic. P. aeruginosa biofilms were thus determined to be viscoelastic materials with properties distinct from solutions of planktonic bacteria.
Figure 1.
Microrheology of wildtype P. aeruginosa biofilms. (A) Ensemble average MSD of tracer particles in solutions of planktonic bacteria and biofilms show incorporation of particles into the biofilm. Diffusion coefficients of viscous solutions (α = 1) and elastic solids (α = 0) are superimposed on MSD plots for reference. Distributions of MSDτ=0.83s for tracer particles in (B) biofilms compared to (C) solutions of planktonic bacteria. The mean of each distribution is marked with a vertical dotted line.
Histograms were used to visualize the distribution of the MSDτ=0.83s and to analyze biofilm heterogeneity. The MSDτ=0.83s of tracer particles within P. aeruginosa biofilms was highly heterogeneous, as indicated by the non-Gaussian distribution of MSDτ=0.83s (Figure 1B). The average MSDτ=0.83s was heavily influenced by fast moving particles with the mean (0.24 ± 0.01 μm2) substantially greater than the mode (0.00 – 0.02 μm2). This specific type of heterogeneity indicates that some tracer particle were moving inside large pores or water channels within the biofilms (Melo 2005). Tracer particles in solutions of planktonic bacteria exhibited a Gaussian distribution of MSDτ=0.83s with a mean (1.53 ± 0.04 μm2) and mode (1.40 – 1.68 μm2) approximately equal (Figure 1C). The heterogeneity of the biofilms is further quantified by the non-Gaussian parameter (NGP) (Vorselaars et al. 2007). For mucoid biofilms, the NGP was 1.86 (Figure S2). When treated with increasing concentrations of COS-NO, the NGP decreased and approached zero, indicative of a homogeneous film. In contrast, treatment with COS in the absence of NO resulted in NGP ranging between 10 and 20, consistent with an increasingly heterogeneous biofilm.
Effect of bacterial mobility and EPS on biofilm rheology
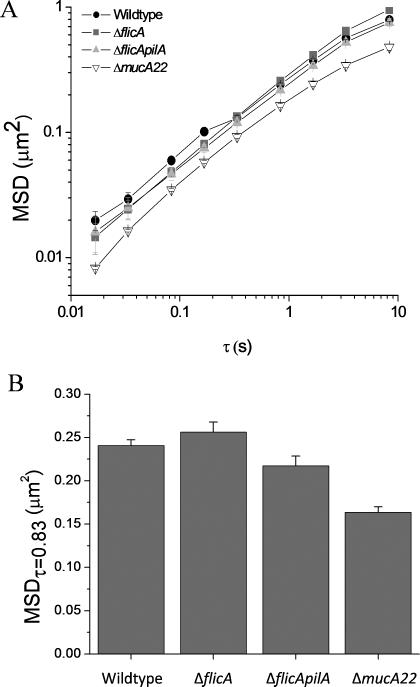
Unlike traditional macro- and microrheology that solely measure the bulk viscoelastic properties of a material, multiple-particle tracking microrheology quantifies the movement of particles within the biofilms. In order to determine whether appendages that impart bacterial motility affected the rheological properties of the biofilms, multiple-particle tracking microrheology was performed using nonmucoid P. aeruginosa strains lacking flagella (ΔflicA) or both flagella and pili (ΔflicApilA). Of note, these strains were isogenic mutants of the wildtype strain used (strain K), and thus should form similar biofilm matrices as the wildtype strain. For the nonmucoid strains evaluated (wildtype, ΔflicA, ΔflicApilA), both the MSD at all time points (Figure 2A) and diffusive exponents were similar (Table 1). Therefore, these appendages neither influence tracer particle movement nor the rheological properties of the biofilm.
Figure 2.
Effects of P. aeruginosa motility and phenotype on biofilm rheology. (A) The MSD of P. aeruginosa biofilms grown with bacterial strains exhibiting different motility appendages and biofilm matrices. (B) MSDτ=0.83s of the same P. aeruginosa biofilms. All MSD were determined by tracking particles in 15 distinct areas of 3 separate biofilms and are plotted as ensemble average MSD ± SEM.
Table 1.
Diffusion exponents (α-values) of P. aeruginosa biofilms with differing phenotypes and motility appendages.
| Strain | Phenotype and characteristics | α |
|---|---|---|
| Wildtype | Nonmucoid | 0.62 ± 0.14 |
| Δ flicA | Nonmucoid, lacking flagella | 0.71 ± 0.04 |
| Δ flicApilA | Nonmucoid, lacking flagella and pili | 0.66 ± 0.02 |
| Mucoid (ΔmucA22) | Mucoid, lacking flagella and pili | 0.58 ± 0.05 |
Mucoid P. aeruginosa biofilms were examined next to confirm that changes in the biofilm matrix altered the biofilm rheological properties. Such bacteria excrete a thicker exopolysaccharide matrix with a greater concentration of alginate than nonmucoid P. aeruginosa (Hentzer et al. 2001, Martin et al. 1993). Compared to the nonmucoid strains, the mucoid P. aeruginosa biofilms exhibited decreased MSD values at all time points (Figure 2A). The MSDτ=0.83s of mucoid biofilms was 0.16 ± 0.01, approximately two-thirds that of nonmucoid biofilms (Figure 2B). However, the diffusive exponents of the mucoid strain were unchanged compared to the nonmucoid wildtype (0.58 ± 0.05 and 0.62 ± 0.14, respectively) (Table 1).
Tobramycin treatment of P. aeruginosa biofilms
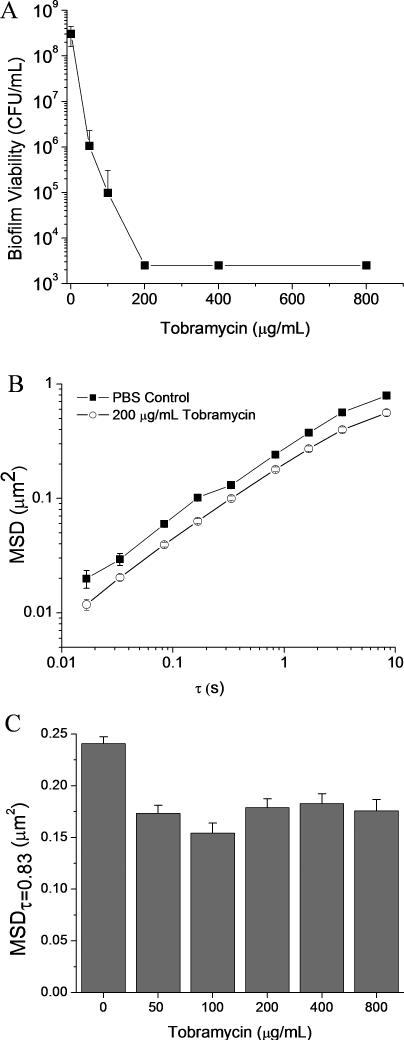
As tobramycin is the current standard for the treatment of P. aeruginosa infections in CF patients (Mogayzel et al. 2013, 2014), the effects of tobramycin on the mechanical properties of P. aeruginosa biofilms were determined as a comparison to NO therapy. Tobramycin was found to eradicate biofilms (ie to reduce bacteria viability by 5 logs over 18 h) at a concentration of 200 μg ml−1 (Figure 3A.).
Figure 3.
Treatment of P. aeruginosa biofilms with tobramycin. (A) Biofilm viability after exposure to tobramycin for 18 h. The MBEC of tobramycin was 200 μg ml−1. (B) MSD values of tracer particles in biofilms grown with wildtype P. aeruginosa following exposure for 18 h to PBS and tobramycin (200 μg ml−1). (C) MSDτ=0.83s of wildtype P. aeruginosa biofilms exposed to tobramycin for 18 h. All MSD were determined by tracking particles in 15 distinct areas of 3 separate biofilms and are plotted as ensemble average MSD ± SEM.
The MSD plots of biofilms treated with tobramycin appeared similar to untreated controls, indicating that tobramycin did not significantly alter the viscoelastic properties of the biofilm (Figure 3B). Over a timescale of 3 decades (0.017 – 3.33 s), no significant differences in the diffusive exponents (α) of untreated and tobramycin-treated biofilms were observed (Table 2). While the temporal dependency of particle diffusion was unchanged by tobramycin treatment, the magnitude of the MSD was reduced at all time points for treated biofilms (Figure 3B). The MSDτ=0.83 ranged from 0.154 ± 0.009 μm2 to 0.183 ± 0.009 μm2, compared to 0.241 ± 0.007 μm2 for untreated biofilms (Figure 3C). This decrease in the MSDτ=0.83 was independent of tobramycin concentration over the range of ¼ – 4x the MBEC (Figure 3C).
Table 2.
Diffusion exponents (α-values) of P. aeruginosa biofilms treated with tobramycin.
| Tobramycin (μg ml−1) | A |
|---|---|
| 0 | 0.62 ± 0.14 |
| 50 | 0.70 ± 0.12 |
| 100 | 0.56 ± 0.02 |
| 200 | 0.66 ± 0.08 |
| 400 | 0.68 ± 0.10 |
| 800 | 0.59 ± 0.06 |
Treatment of P. aeruginosa biofilms with COS and COS-NO
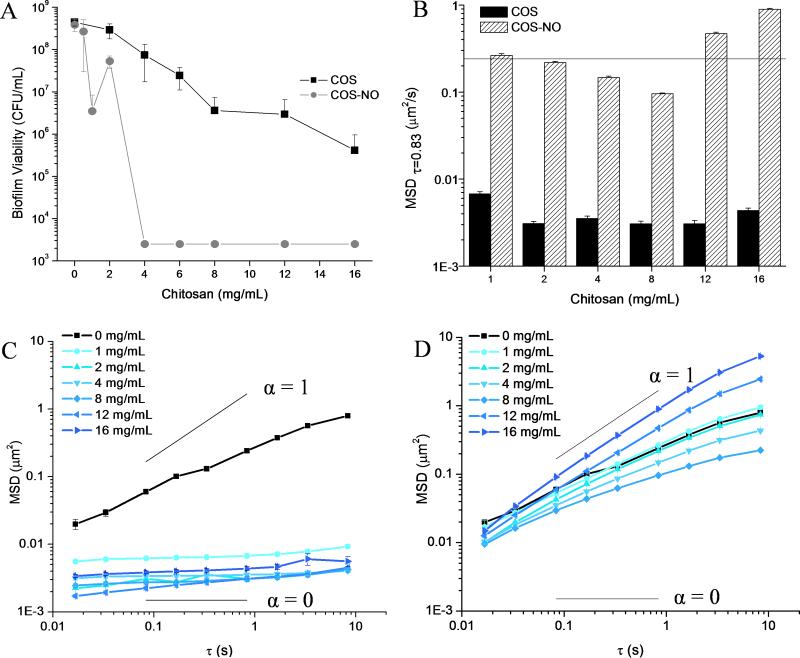
At the largest concentration tested (16 mg ml−1), the COS scaffold decreased bacterial viability by three logs without fully eradicating the P. aeruginosa biofilm (Figure 4A). While chitosan in general is a known antimicrobial (Rabea et al. 2003), the anti-biofilm efficacy of the chitosan oligosaccharides was significantly reduced, as has previously been demonstrated (Jeon et al. 2001). Addition of NO to the COS scaffold improved the anti-biofilm activity further, eradicating biofilms at 4 mg ml−1. This concentration corresponds to a bactericidal NO dose of 3.1 ± 0.4 μmol ml−1 (Figure 4A).
Figure 4.
Treatment of P. aeruginosa biofilms with COS and COS-NO. (A) Biofilm viability following exposure to COS or COS-NO for 18 h. While COS did not fully eradicate biofilms, the MBEC of COS-NO was 4 mg ml−1 (NO dose of 3.1 ± 0.4 μmol). (B) MSDτ=0.83s of biofilms treated with COS and COS-NO. The MSDτ=0.83s of untreated biofilms is indicated as a horizontal line. Note the logarithmic scale of the y-axis. (C) The MSD values of biofilms treated with COS and (D) COS-NO. All MSD were determined by tracking particles in 15 distinct areas of 3 separate biofilms and are plotted as ensemble average MSD ± SEM. Diffusion coefficients of viscous solutions (α = 1) and elastic solids (α = 0) are superimposed on MSD plots for reference.
Despite low biofilm eradication capabilities, the COS scaffold significantly altered the viscoelasticity of P. aeruginosa biofilms. Treatment of biofilms with 2 mg ml−1 of COS decreased the magnitude of the MSDτ=0.83s by two logs (to 0.0031 ± 0.0002 μm2) compared to untreated controls (MSDτ=0.83s = 0.241 ± 0.007 μm2), with no further reduction in the MSDτ=0.83 when the concentration of COS was increased to 16 mg ml−1 (Figure 4B). While the MSD (at all τ) of untreated biofilms exhibited a temporal dependency (revealing partial viscosity), treatment with COS over the concentration range of 1 – 16 mg ml−1 eliminated this time dependence (Figure 4C). The diffusive exponents of COS treated biofilms ranged from 0.03 ± 0.02 to 0.14 ± 0.02, a significant reduction compared to untreated biofilms (0.62 ± 0.14) (Table 3). Treatment with COS clearly transformed P. aeruginosa biofilms into nearly elastic solids with almost no viscous component.
Table 3.
Diffusion exponents (α) of P. aeruginosa biofilms treated with COS and COS-NO.
| Dose (mg ml−1) | COS α | COS-NO α | NO (μmol ml−1) |
|---|---|---|---|
| 0 | 0.62 ± 0.14 | 0.62 ± 0.14 | n/a |
| 1 | 0.07 ± 0.04 | 0.68 ± 0.04 | 0.8 ± 0.1 |
| 2 | 0.10 ± 0.20 | 0.74 ± 0.14 | 1.6 ± 0.2 |
| 4 | 0.03 ± 0.02 | 0.64 ± 0.10 | 3.1 ± 0.4 |
| 8 | 0.07 ± 0.04 | 0.55 ± 0.13 | 6.2 ± 0.7 |
| 12 | 0.14 ± 0.02 | 0.91 ± 0.06 | 9.4 ± 1.1 |
| 16 | 0.11 ± 0.12 | 1.00 ± 0.11 | 12.5 ± 1.4 |
The total NO released from COS-NO at each concentration is also provided.
While the COS scaffold decreased both the MSDτ=0.83s and the slope of the MSD of tracer particles within P. aeruginosa biofilms at all concentrations, NO-releasing COS (COS-NO) induced a dose-dependent response in biofilm viscoelasticity. The MSDτ=0.83s decreased with increasing COS-NO at concentrations ≤ 8 mg ml−1 (Figure 4B, Table 3). Tracer particles in biofilms exposed to COS-NO exhibited a greater MSDτ=0.83s and diffusive exponents than those in biofilms exposed to the same concentrations of the COS scaffold (Figure 4B). At concentrations ≥ 12 mg ml−1 COS-NO, the MSDτ=0.83s was greater than that for the untreated biofilms (Figure 1B Table 3). The increase in MSDτ=0.83s indicates degradation of the viscoelastic properties of the biofilm. As biofilm degradation occurred above the MBEC of COS-NO (4 mg ml−1), destruction of the physical properties of the biofilm required a larger dose of NO (9.4 ± 1.1 μmol ml−1) than biofilm eradication (3.1 ± 0.4 μmol ml−1).
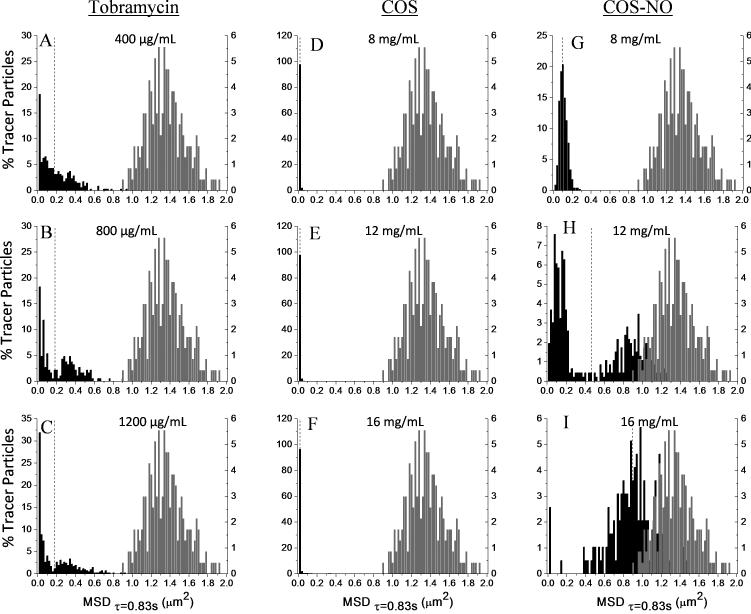
Heterogeneity of treated P. aeruginosa biofilms
Sample heterogeneity has been correlated with increased viscoelasticity for complex biological materials such as sputum (Dawson et al. 2003, Hill et al. 2014). The effect of antibacterial treatment on biofilm heterogeneity was thus evaluated at concentrations in excess of the MBEC. As expected, tobramycin had no observable effect on the heterogeneity of biofilms. While tobramycin treatment decreased the average MSDτ=0.83s, concentrations of tobramycin in excess of the MBEC did not alter the shape of the MSDτ=0.83s distributions (Figure 5A-C) compared to untreated biofilms (Figure 1B); both exhibited MSDτ=0.83s distributions skewed such that the modes were substantially lower than the mean.
Figure 5.
Histograms of biofilm heterogeneity after treatment. The distributions of MSDτ=0.83s of individual tracer particles in (A-C) biofilms treated with tobramycin, (D-F) COS, and (G-I) COS-NO at concentrations above the MBEC values. The mean of each distribution is marked with a vertical dotted line. The distribution of of MSDτ=0.83s of tracer particles in water is denoted in gray on each histogram.
Treatment with COS and COS-NO resulted in opposing effects on the MSDτ=0.83s distributions of P. aeruginosa biofilms. The COS scaffold constrained tracer particle movement, decreasing the width of the MSDτ=0.83s distribution and reducing biofilm heterogeneity (Figure 5D-F). Alternatively, treatment with COS-NO at concentrations above the MBEC resulted in degradation of the biofilm and increased heterogeneity. Treatment with 8 mg ml−1 COS-NO showed little alteration in the MSDτ=0.83s distribution (Figure 5G) compared to the untreated controls (Figure 1B). However, exposure to 12 mg ml−1 COS-NO resulted in a bimodal MSDτ=0.83s distribution (Figure 5H). When treated with 16 mg ml−1 COS-NO, the MSDτ=0.83s distribution was Gaussian and centered around 1 μm2 (Figure 5I).
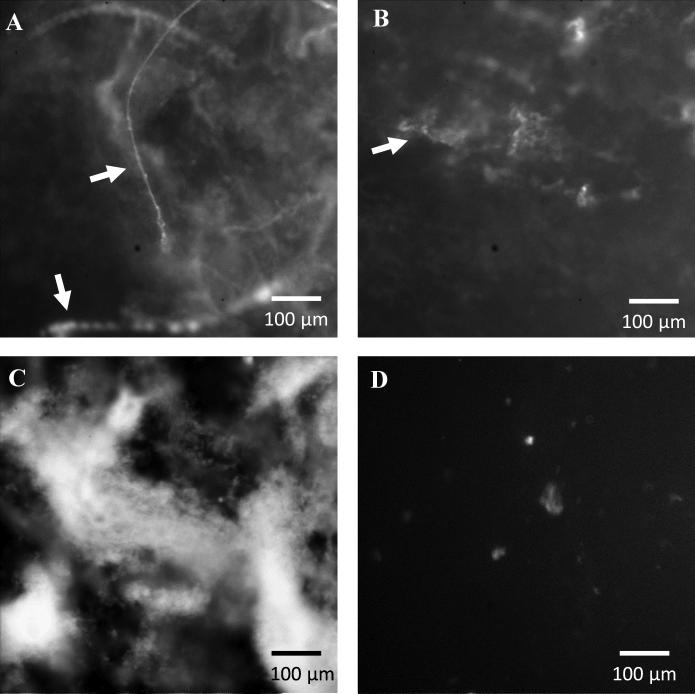
Fluorescence microscopy of treated P. aeruginosa biofilms
P. aeruginosa biofilms were stained with PI following treatment (ie exposure to 200 μg ml−1 tobramycin, 4 mg ml−1 COS, or 4 mg ml−1 COS-NO) to visualize the effects of each on the extracellular DNA (eDNA) matrix. As a membrane impenetrable dye, PI will only stain DNA that has been secreted from the bacteria (ie not intracellular DNA). For the untreated biofilms, eDNA was found in large concentrations throughout the entire biofilm (Figure 6A). As reported previously for mature P. aeruginosa biofilms (Yang et al. 2007), the eDNA was generally localized in large multicellular structures. While tobramycin has been shown to bind to eDNA (Chiang et al. 2013, Mulcahy et al. 2008), this binding did not appear to alter the structure of the eDNA matrix within the biofilm (Figure 6B).
Figure 6.
Fluorescence microscope images of P. aeruginosa biofilms. Biofilms were stained with propidium iodide in order to visualize the intra- and extra-cellular DNA matrix following exposure for 18 h to (A) PBS, (B) tobramycin (200 μg ml−1), (C) COS (4 mg ml−1), and (D) COS-NO (4 mg ml−1). Large extracellular DNA filaments are noted with white arrows.
In contrast to tobramycin, the COS and COS-NO treatments visibly altered the eDNA network of the P. aeruginosa biofilms. Biofilms treated with COS appeared to be highly aggregated. Indeed, highly fluorescent eDNA features dominated the biofilm morphology (Figure 6C). Biofilms treated with COS-NO exhibited almost no fluorescence, indicating that the DNA was damaged to the extent that PI could no longer intercalate into the DNA strands (Figure 6D) (Olive et al. 1994).
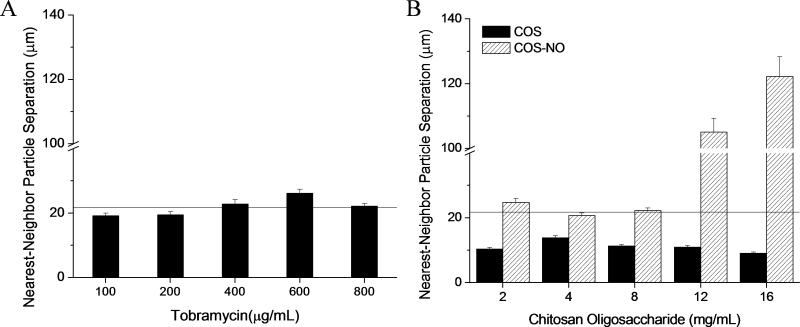
Quantification of biofilm contraction
The average distance between nearest neighbor particles was determined to quantify biofilm expansion and contraction after treatment. For untreated biofilms, the average distance between nearest neighbor particles was 21.7 ± 0.8 μm (indicated by the horizontal line in Figure 6). At concentrations ranging from ½ to 4-times the MBEC, tobramycin exposure did not alter the average distance between nearest neighbor particles compared to the untreated biofilm (Figure 6A), indicating that tobramycin treatment had little effect on the contraction or expansion of the biofilm.
In contrast, COS and COS-NO treatment resulted in significant alterations in the biofilm structure. The COS scaffold reduced the average distance between nearest neighbor tracer particles compared to untreated biofilms with ~50% biofilm contraction that was independent of COS concentration from 1 – 16 mg ml−1 (Figure 6B). Treatment with COS-NO exhibited a dose-dependent response on tracer particle separation (Figure 6B). At concentrations below 8 mg ml−1, treatment with COS-NO had no effect on the distance between nearest neighbor tracer particles. At larger concentrations (ie ≥ 12 mg ml−1), the average particle separation increased significantly. As expected, tracer particle separation increased as the biofilm degraded. The concentration of COS-NO resulting in increased separation of nearest neighbor particles correlated with the concentration required for partial destruction of the viscoelastic properties of the biofilm (12 mg ml−1).
Discussion
An ideal treatment for P. aeruginosa biofilms in CF patients would reduce bacterial viability while physically disrupting the biofilm in order to ease biofilm removal and prevent regrowth. As such, it is important to characterize the effects of antibacterial therapeutics on the viscoelastic properties of P. aeruginosa biofilms. While highly effective at eradicating bacteria, tobramycin does not alter the biophysical properties of P. aeruginosa biofilms. Indeed, the results indicate that treatment with tobramycin neither alters the distance between nearest neighbor tracer particles (Figure 6) nor the diffusive exponents of tracer particles within biofilms (Table 2). Of relevance, a decrease in particle diffusion (ie the MSD at all τ) was observed with tobramycin treatment (Figure 3). While the exact cause is unknown, particle diffusion in biofilms grown with nonmotile (lacking flagella and pili) bacteria did not exhibit such behavior. As such, the loss of bacterial motility upon cell death is not the culprit. As reported previously, the cationic nature of tobramycin results in an association with the negatively charged biofilm matrix (Nichols et al. 1988, Tseng et al. 2013). Jones et al. (2011) noted with macrorheology that the introduction of cations alters the rheological properties of bacteria biofilms. It is thus likely that the small reduction in MSD upon treatment with tobramycin results from an alteration in the biofilm matrix and not decreased bacterial motility.
In comparison to tobramycin, NO released from the chitosan oligosaccharides both eradicated bacteria and degraded the physical properties of the biofilm (Figure 4). At concentrations above the MBEC (ie ≥ 12 mg ml−1), the COS-NO significantly compromised the structural integrity of the biofilms, as evidenced by increases in the MSDτ=0.83s and diffusive exponents of tracer particles within treated biofilms (Figure 4, Table 3). The increased tracer particle movement may be attributed to either an overall loosening of the biofilm or heterogeneous destruction of the biofilm matrix. In the case of uniform biofilm loosening, the diffusion of all particles would increase, shifting the MSDτ=0.83s distributions to larger values without changing the overall shape of the distribution. However, the observed bimodal distribution is consistent with destruction of discrete segments of the biofilm, where some of the tracer particles remained constrained in the biofilm while a second population of tracer particles experienced increased diffusion outside of the biofilm (Figure 5H). Further increasing the concentration of COS-NO resulted a Gaussian MSDτ=0.83s distribution centered around 1 μm2, suggestive of complete biofilm destruction and free diffusion of the tracer particles in PBS (Figure 5I). These results indicate that NO does not cause a gradual loosening of the biofilm but rather destroys segments of the biofilms and is capable of complete biofilm destruction.
While the mechanism of NO-mediated destruction of the biofilm is likely complex and the result of multiple factors, NO has been shown to alter the biological macromolecules that constitute the majority of the biofilm matrix (Carpenter & Schoenfisch 2012, Fang 1997, Flemming & Wingender 2010, Mann & Wozniak 2012). For example, DNA is essential to the formation of P. aeruginosa biofilm (Whitchurch et al. 2002). Indeed, cleavage of DNA by DNase decreases the structural integrity of such biofilms (Tetz et al. 2009). Burney et al. (1999) and Tamir et al. (1996) have reported that exogenous NO both damages and cleaves DNA. It is likely that NO-mediated destruction of DNA would physically degrade P. aeruginosa biofilms. This hypothesis was confirmed using fluorescence microscopy, in which destruction of the eDNA matrix of the P. aeruginosa biofilms was observed at the MBEC (4 mg COS-NO ml−1). However, the viscoelasticity of the biofilm matrix (via multiple-particle tracking microrheology) did not change at this concentration of COS-NO. The discrepancy in these results indicates that other biofilm matrix components (eg exopolysaccharides) may influence the structural integrity of the biofilm matrix more than just eDNA. In fact, NO and its reactive intermediates are known to depolymerize polysaccharides (Duan & Kasper 2011), suggesting NO may reduce biofilm viscoelasticity via damage to alginate, Psl, and Pel structural biofilm polysaccharides (Chew et al. 2014).
While NO represents a promising therapeutic for the physical degradation of P. aeruginosa biofilms, it is currently limited by the effects of the COS scaffold as the chitosan treatment alone results in biofilm contraction regardless of concentration (0.5 – 16 mg ml−1). Such contraction (Figure 6) parallels increased biofilm elasticity and decreased particle diffusion (Table 3, Figure 4), which correlates to physical entanglement of polymers and effective cross-linking (eg covalent or hydrogen bonding, electrostatic interactions) in both ideal polymer and biological systems (Chew et al. 2014, Körstgens et al. 2001, Nielsen 1969). As the P. aeruginosa biofilm matrix is comprised predominantly of anionic macromolecules (Flemming & Wingender 2010), the introduction of cationic chitosan alters the electrostatics of the biofilm and potentially enhances cross-linking. To further improve the efficacy these NO-releasing therapeutics, scaffolds that do not increase the elasticity of bacterial biofilms should be investigated. As the cationic nature of chitosan likely contributes to biofilm cohesion, modifying the chitosan oligosaccharide scaffold with anionic moieties may reduce these effects. Furthermore, the biofilm disrupting capabilities of NO-releasing chitosan oligosaccharides may be improved by adjusting the NO payload. For example, increasing the NO storage should reduce the concentration of COS-NO required for biofilm degradation, thereby minimizing any underlying effects of the scaffold.
Supplementary Material
Figure 7.
Average distance between nearest neighbor particles. Average distances between nearest neighbor particles following treatment with (A) tobramycin and (B) chitosan oligosaccharides. Average separation between tracer particles in untreated biofilms is indicated by a horizontal line. All separation distances were quantified in 15 distinct areas of 3 separate biofilms and are plotted as ensemble average MSD ± SEM.
Acknowledgments
The National Institutes of Health (AI112029) and the National Science Foundation (DMS1100281) supported this work. The authors thank Dr Matthew Wolfgang of the Department of Microbiology and Immunology at the University of North Carolina, Chapel Hill for providing PAK and PAKmucA22 laboratory strains.
Footnotes
The corresponding author declares a competing financial interest. Mark Schoenfisch is co-founder, member of the board of directors, and maintains a financial interest in Novan Therapeutics, Inc., a clinical-stage biotechnology company that is commercializing macromolecular nitric oxide storage and release vehicles for dermatological applications.
References
- Alipour M, Suntres ZE, Omri A. Importance of DNase and alginate lyase for enhancing free and liposome encapsulated aminoglycoside activity against Pseudomonas aeruginosa. J Antimicrob Chemother. 2009;64:317–325. doi: 10.1093/jac/dkp165. [DOI] [PubMed] [Google Scholar]
- Alkawash MA, Soothill JS, Schiller NL. Alginate lyase enhances antibiotic killing of mucoid Pseudomonas aeruginosa in biofilms. APMIS. 2006;114:131–138. doi: 10.1111/j.1600-0463.2006.apm_356.x. [DOI] [PubMed] [Google Scholar]
- Barraud N, Hassett DJ, Hwang S-H, Rice SA, Kjelleberg S, Webb JS. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol. 2006;188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud N, Storey MV, Moore ZP, Webb JS, Rice SA, Kjelleberg S. Nitric oxide-mediated dispersal in single- and multi-species biofilms of clinically and industrially relevant microorganisms. Microb Biotechnol. 2009;2:370–378. doi: 10.1111/j.1751-7915.2009.00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breed RS, Dotterrer W. The number of colonies allowable on satisfactory agar plates. J Bacteriol. 1916;1:321. doi: 10.1128/jb.1.3.321-331.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burney S, Caulfield JL, Niles JC, Wishnok JS, Tannenbaum SR. The chemistry of DNA damage from nitric oxide and peroxynitrite. Mutat Res. 1999;424:37–49. doi: 10.1016/s0027-5107(99)00006-8. [DOI] [PubMed] [Google Scholar]
- Button B, Cai L-H, Ehre C, Kesimer M, Hill DB, Sheehan JK, Boucher RC, Rubinstein M. A periciliary brush promotes the lung health by separating the mucus layer from airway epithelia. Science. 2012;337:937–941. doi: 10.1126/science.1223012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AW, Schoenfisch MH. Nitric oxide release: part II. Therapeutic applications. Chem Soc Rev. 2012;41:3742–3752. doi: 10.1039/c2cs15273h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KLB, Tai M-C, Cheng F-H. Kinetics and products of the degradation of chitosan by hydrogen peroxide. J Agric Food Chem. 2001;49:4845–4851. doi: 10.1021/jf001469g. [DOI] [PubMed] [Google Scholar]
- Chew SC, Kundukad B, Seviour T, Van der Maarel JR, Yang L, Rice SA, Doyle P, Kjelleberg S. Dynamic remodeling of microbial biofilms by functionally distinct exopolysaccharides. mBio. 2014;5:e01536–01514. doi: 10.1128/mBio.01536-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang W-C, Nilsson M, Jensen PØ, Høiby N, Nielsen TE, Givskov M, Tolker-Nielsen T. Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2013;57:2352–2361. doi: 10.1128/AAC.00001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen TS, Prince A. Cystic fibrosis: a mucosal immunodeficiency syndrome. Nat Med. 2012;18:509–519. doi: 10.1038/nm.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson M, Wirtz D, Hanes J. Enhanced viscoelasticity of human cystic fibrotic sputum correlates with increasing microheterogeneity in particle transport. J Biol Chem. 2003;278:50393–50401. doi: 10.1074/jbc.M309026200. [DOI] [PubMed] [Google Scholar]
- Deupree SM, Schoenfisch MH. Morphological analysis of the antimicrobial action of nitric oxide on Gram-negative pathogens using atomic force microscopy. Acta biomater. 2009;5:1405–1415. doi: 10.1016/j.actbio.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring G, Flume P, Heijerman H, Elborn JS, Group CS. Treatment of lung infection in patients with cystic fibrosis: current and future strategies. J Cyst Fibros. 2012;11:461–479. doi: 10.1016/j.jcf.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Duan J, Kasper DL. Oxidative depolymerization of polysaccharides by reactive oxygen/nitrogen species. Glycobiology. 2011;21:401–409. doi: 10.1093/glycob/cwq171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang FC. Perspectives series: host/pathogen interactions. Mechanisms of nitric oxide-related antimicrobial activity. J Clin Invest. 1997;99:2818. doi: 10.1172/JCI119473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H-C, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Gibson RL, Burns JL, Ramsey BW. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am J Respir Crit Care Med. 2003;168:918–951. doi: 10.1164/rccm.200304-505SO. [DOI] [PubMed] [Google Scholar]
- Hatch RA, Schiller NL. Alginate lyase promotes diffusion of aminoglycosides through the extracellular polysaccharide of mucoid Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:974–977. doi: 10.1128/aac.42.4.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentzer M, Teitzel GM, Balzer GJ, Heydorn A, Molin S, Givskov M, Parsek MR. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J Bacteriol. 2001;183:5395–5401. doi: 10.1128/JB.183.18.5395-5401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DB, Vasquez PA, Mellnik J, McKinley SA, Vose A, Mu F, Henderson AG, Donaldson SH, Alexis NE, Boucher RC. A biophysical basis for mucus solids concentration as a candidate biomarker for airways disease. PLOS ONE. 2014;9:e87681. doi: 10.1371/journal.pone.0087681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y-J, Park P-J, Kim S-K. Antimicrobial effect of chitooligosaccharides produced by bioreactor. Carbohydr Polym. 2001;44:71–76. [Google Scholar]
- Jones ML, Ganopolsky JG, Labbé A, Wahl C, Prakash S. Antimicrobial properties of nitric oxide and its application in antimicrobial formulations and medical devices. Appl Microbiol Biotechnol. 2010;88:401–407. doi: 10.1007/s00253-010-2733-x. [DOI] [PubMed] [Google Scholar]
- Jones WL, Sutton MP, McKittrick L, Stewart PS. Chemical and antimicrobial treatments change the viscoelastic properties of bacterial biofilms. Biofouling. 2011;27:207–215. doi: 10.1080/08927014.2011.554977. [DOI] [PubMed] [Google Scholar]
- Kean T, Thanou M. Biodegradation, biodistribution and toxicity of chitosan. Adv Drug Deliv Rev. 2010;62:3–11. doi: 10.1016/j.addr.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Körstgens V, Flemming H, Wingender J, Borchard W. Influence of calcium ions on the mechanical properties of a model biofilm of mucoid Pseudomonas aeruginosa. Water Sci Technol. 2001;43:49–57. [PubMed] [Google Scholar]
- Lieleg O, Caldara M, Baumgärtel R, Ribbeck K. Mechanical robustness of Pseudomonas aeruginosa biofilms. Soft Matter. 2011;7:3307–3314. doi: 10.1039/c0sm01467b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PJ, Conrad JC, Wyss HM, Schofield AB, Weitz DA. Fluids of clusters in attractive colloids. Phys Rev Lett. 2006;96:028306. doi: 10.1103/PhysRevLett.96.028306. [DOI] [PubMed] [Google Scholar]
- Lu Y, Slomberg DL, Schoenfisch MH. Nitric oxide-releasing chitosan oligosaccharides as antibacterial agents. Biomacromolecules. 2014;35:1716–1724. doi: 10.1016/j.biomaterials.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Slomberg DL, Shah A, Schoenfisch MH. Nitric oxide-releasing amphiphilic poly (amidoamine)(PAMAM) dendrimers as antibacterial agents. Biomacromolecules. 2013;14:3589–3598. doi: 10.1021/bm400961r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maghami GG, Roberts GA. Evaluation of the viscometric constants for chitosan. Macromol Chem Phys. 1988;189:195–200. [Google Scholar]
- Mah T-FC, O'Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Micribiol. 2001;9:34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- Mann EE, Wozniak DJ. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol Rev. 2012;36:893–916. doi: 10.1111/j.1574-6976.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D, Schurr M, Mudd M, Govan J, Holloway B, Deretic V. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci USA. 1993;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason TG. Estimating the viscoelastic moduli of complex fluids using the generalized Stokes–Einstein equation. Rheol Acta. 2000;39:371–378. [Google Scholar]
- Matsui H, Wagner VE, Hill DB, Schwab UE, Rogers TD, Button B, Taylor RM, Superfine R, Rubinstein M, Iglewski BH. A physical linkage between cystic fibrosis airway surface dehydration and Pseudomonas aeruginosa biofilms. Proc Natl Acad Sci USA. 2006;103:18131–18136. doi: 10.1073/pnas.0606428103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo L. Biofilm physical structure, internal diffusivity and tortuosity. Water Sci Technol. 2005;52:77–84. [Google Scholar]
- Mogayzel PJ, Naureckas ET, Robinson KA, Brady C, Guill M, Lahiri T, Lubsch L, Matsui J, Oermann CM, Ratjen F. Cystic fibrosis foundation pulmonary guideline. Pharmacologic approaches to prevention and eradication of initial Pseudomonas aeruginosa infection. Ann Am Thorac Soc. 2014;11:1640–1650. doi: 10.1513/AnnalsATS.201404-166OC. [DOI] [PubMed] [Google Scholar]
- Mogayzel PJ, Naureckas ET, Robinson KA, Mueller G, Hadjiliadis D, Hoag JB, Lubsch L, Hazle L, Sabadosa K, Marshall B. Cystic fibrosis pulmonary guidelines: chronic medications for maintenance of lung health. Am J Respir Crit Care Med. 2013;187:680–689. doi: 10.1164/rccm.201207-1160oe. [DOI] [PubMed] [Google Scholar]
- Mulcahy H, Charron-Mazenod L, Lewenza S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLOS Pathogens. 2008;4:e1000213. doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols WW, Dorrington S, Slack M, Walmsley H. Inhibition of tobramycin diffusion by binding to alginate. Antimicrob Agents Chemother. 1988;32:518–523. doi: 10.1128/aac.32.4.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen LE. Cross-linking–effect on physical properties of polymers. J Macromol Sci Polymer Rev. 1969 [Google Scholar]
- Olive P, Banath J, Fjell C. DNA strand breakage and DNA structure influence staining with propidium iodide using the alkaline comet assay. Cytometry. 1994;16:305–312. doi: 10.1002/cyto.990160404. [DOI] [PubMed] [Google Scholar]
- Rabea EI, Badawy ME-T, Stevens CV, Smagghe G, Steurbaut W. Chitosan as antimicrobial agent: applications and mode of action. Biomacromolecules. 2003;4:1457–1465. doi: 10.1021/bm034130m. [DOI] [PubMed] [Google Scholar]
- Riccio DA, Schoenfisch MH. Nitric oxide release: part I. Macromolecular scaffolds. Chem Soc Rev. 2012;41:3731–3741. doi: 10.1039/c2cs15272j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz G, Phillips P, Yang Q, Stewart P. Biofilm maturity studies indicate sharp debridement opens a time-dependent therapeutic window. J Wound Care. 2010;19:320. doi: 10.12968/jowc.2010.19.8.77709. [DOI] [PubMed] [Google Scholar]
- Sriramulu DD, Lünsdorf H, Lam JS, Römling U. Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J Med Microbiol. 2005;54:667–676. doi: 10.1099/jmm.0.45969-0. [DOI] [PubMed] [Google Scholar]
- Tamir S, Burney S, Tannenbaum SR. DNA damage by nitric oxide. Chem Res Toxicol. 1996;9:821–827. doi: 10.1021/tx9600311. [DOI] [PubMed] [Google Scholar]
- Tetz GV, Artemenko NK, Tetz VV. Effect of DNase and antibiotics on biofilm characteristics. Antimicrob Agents Chemother. 2009;53:1204–1209. doi: 10.1128/AAC.00471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng BS, Zhang W, Harrison JJ, Quach TP, Song JL, Penterman J, Singh PK, Chopp DL, Packman AI, Parsek MR. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environmental Microbiol. 2013;15:2865–2878. doi: 10.1111/1462-2920.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Acker H, Van Dijck P, Coenye T. Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol. 2014;22:326–333. doi: 10.1016/j.tim.2014.02.001. [DOI] [PubMed] [Google Scholar]
- Vorselaars B, Lyulin AV, Karatasos K, Michels M. Non-Gaussian nature of glassy dynamics by cage to cage motion. Phys Rev E. 2007;75:011504. doi: 10.1103/PhysRevE.75.011504. [DOI] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487–1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- Yang L, Barken KB, Skindersoe ME, Christensen AB, Givskov M, Tolker-Nielsen T. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiol. 2007;153:1318–1328. doi: 10.1099/mic.0.2006/004911-0. [DOI] [PubMed] [Google Scholar]
- Yoon SS, Coakley R, Lau GW, Lymar SV, Gaston B, Karabulut AC, Hennigan RF, Hwang S-H, Buettner G, Schurr MJ. Anaerobic killing of mucoid Pseudomonas aeruginosa by acidified nitrite derivatives under cystic fibrosis airway conditions. J Clin Invest. 2006;116:436. doi: 10.1172/JCI24684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrelli K, Galy O, Latour-Lambert P, Kirwan L, Ghigo J, Beloin C, Henry N. Bacterial biofilm mechanical properties persist upon antibiotic treatment and survive cell death. New J Phys. 2013;15:125026. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.