Abstract
Purpose
The beta subunit of the rod cyclic nucleotide gated channel B1 (CNGB1) contains a proline/glutamic acid-rich N-terminal domain (GARP), which is also present in rods as a non–membrane-bound protein (GARP1/2). GARP2 and CNGB1 bind to retinal degeneration slow (RDS), which is present in the rims of rod and cone outer segment (OS) layers. Here we focus on the importance of RDS/GARP complexes in OS morphogenesis and stability.
Methods
Retinal structure, function, and biochemistry were assessed in GARP2-Tg transgenic mice crossed onto rds+/+, rds+/−, and rds−/− genetic backgrounds.
Results
GARP2 expression decreased in animals with reduced RDS levels. Overexpression of GARP2 led to abnormalities in disc stacking in GARP2-Tg/rds+/+ and the accumulation of abnormal vesicular structures in GARP2-Tg/rds+/− OS, as well as alterations in RDS-ROM-1 complex formation. These abnormalities were associated with diminished scotopic a- and b-wave amplitudes in GARP2-Tg mice on both the rds+/+ and rds+/− backgrounds. In addition, severe defects in cone function were observed in GARP2-Tg mice on all RDS backgrounds.
Conclusions
Our results indicate that overexpression of GARP2 significantly exacerbates the defects in rod function associated with RDS haploinsufficiency and leads to further abnormalities in OS ultrastructure. These data also suggest that GARP2 expression in cones can be detrimental to cones. RDS/GARP interactions remain under investigation but are critical for both OS structure and function.
Keywords: CNG channel, GARP2, knockout mouse, retinal degeneration slow, transgenic mouse
Visual processing in the vertebrate retina is initiated when light is captured by the outer segment (OS) layers of rod and cone photoreceptors and is converted to an electrical signal. The signal transduction that mediates this process is well characterized, and a key component is the cyclic nucleotide gated (CNG) channel which is present in the OS plasma membrane. The rod CNG channel consists of 3 alpha (α1) subunits (CNGA1) and 1 long beta (β1) subunit (CNGB1),1 whereas the cone CNG channel exhibits a 2A:2B stoichiometry with 2 alpha (α3) CNGA3 and beta (β3) CNGB3 subunits.2 The rod β1subunit (CNGB1) is a product of the Cngb1 gene and contains a unique proline- and glutamic acid-rich region (GARP) in its N terminus.3 Two other soluble proteins, GARP1 and GARP2, are made as splice variants of Cngb14,5 but lack the channel portion and are not membrane bound. GARP2 is ∼75 kDa and is abundant in the rod OS (ROS), while GARP1 is ∼100 kDa3 with much lower abundance in rods. The mean molar ratio of all three Cngb1 protein products is 1:5:26 for GARP1:CNGB1:GARP2, respectively.6 The importance of these proteins for OS function is underscored by animal and human studies. For example, elimination of Cngb1 gene products leads to abnormal trafficking of the CNGA1 channel subunit and rod degeneration as well as structural and functional abnormalities in OSs.3,7 Likewise, a French family with autosomal dominant retinitis pigmentosa has been reported to have a putative pathogenic mutation in a highly conserved glycine in the cGMP binding domain of CNGB1.8
Despite several years of research, the function served by the free GARP proteins is not clear. Studies have suggested that GARP2 can influence phototransduction by binding phosphodiesterase 6 (PDE6) and thus altering cGMP levels.9–11 Additional work using a transgenic mouse line which overexpresses GARP2 demonstrated altered phototransduction gain and slower recovery, suggesting that free GARP could play a role in regulating rod phototransduction.11
Other work has shown that both GARP2 and CNGB1 can bind to the retinal degeneration slow (RDS) protein, also called peripherin-2 (PRPH2).6,12,13 RDS is a tetraspanin protein found in the rim region of rod and cone OS discs and is required for the formation and maintenance of the OS.14,15 Mutations in RDS cause a variety of retinal degenerations including rod-dominant autosomal dominant retinitis pigmentosa (ADRP) and multiple classes of cone-dominant macular dystrophy (MD)16,17 (http://www.retina-international.org/files/sci-news/rdsmut.htm). In addition to interactions with CNGB1/GARP, RDS also interacts with itself and its non-glycosylated homologue, rod outer segment membrane protein-1 (ROM-1), to form a variety of different complexes.18–20 It has been hypothesized that interactions between GARP2/CNGB1 and RDS may contribute to the stability of the OS by linking opposing membranes, helping mediate connections between either the disc rim or the plasma membrane or between adjacent disc rims.12 GARPs are natively unfolded and exist as both monomers and multimeric complexes in a reversible equilibrium.6 Their high degree of intrinsic disorder is consistent with the idea that they may be flexible proteins capable of serving as linkers in the OS. Interestingly, recent data have indicated that GARP2/RDS interactions occur at the base of the OS while CNGB1/RDS and RDS/RDS interactions occur in the inner segment (IS) layer, suggesting that GARP2/RDS interactions may be involved in the structural assembly of the OS.13 In this study our goal was to assess the functional relevance of RDS/GARP interactions in the mammalian retina by evaluating retinal structure and function in the presence of varying ratios of GARP2 and RDS.
Materials and Methods
Ethics Statement and Animal Care and Use
Animal maintenance and experiments were approved by the local Institutional Animal Care and Use Committee (University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA) and conformed to the guidelines on the care and use of animals adopted by the Association for Research in Vision and Ophthalmology (Rockville, MD, USA). GARP2 transgenic mice (referred to here as GARP2-Tg mice) were generated by Dr. Steven Pittler and have been previously described.11 The GARP2 transgene contains the 4.4-kbp mouse opsin promoter, the GARP2 cDNA, and the polyadenylation signal of the mouse protamine gene. A c-Myc epitope was inserted between GARP2 exons 12 and 12a to differentiate the transgenic GARP2 protein from endogenous GARP2.11 GARP2-Tg mice on rds+/− and rds−/− backgrounds were generated by cross-breeding in our facility. Wild-type (referred to as rds+/+), rds+/−, and rds−/− littermates were used as controls. We also used Cngb1−/− mice, provided by Pittler.3 All animals were maintained in a 12-h light, 12-h dark cycle at ∼30 lux).
Antibodies
Various primary antibodies used in Western blotting and in immunofluorescence and are summarized in Supplementary Table S1.
Light and Transmission Electron Microscopy
The methods used for tissue collection, processing, plastic embedding, and transmission electron microscopy (EM) were described previously.21,22 Light microscopy was performed using 0.75-μm plastic embedded sections, and images were captured at ×40 magnification (universal microscope; Zeiss, Wetzlar, Germany). To evaluate outer nuclear layer (ONL) thickness and OS length, images were captured from central superior and inferior regions containing the optic nerve head and at least 3 retinal sections from 3 different eyes/genotype were used. ONL and OS layer thicknesses were measured using Photoshop CS5 software (Adobe, McLean, VA, USA).
Immunofluorescence Labeling
Eyes were harvested, dissected, fixed, and embedded as previously described for paraffin sectioning,23 or cryosectioning.24 Immunostaining was performed as described previously,23,24 using the primary antibodies described in Supplementary Table S1. Appropriate Alexa Fluor-conjugated secondary antibodies (Life Technologies, Grand Island, NY, USA) were used at a dilution of 1:1000 for 1 h at room temperature. Images were captured using a BX-62 microscope (Olympus, Tokyo, Japan) equipped with a spinning disc confocal unit, using a 40× (air, 0.9 numerical aperture [NA]) or a 100× (oil, 1.4 NA) objective. Images were stored and deconvolved (using a nearest-neighbors paradigm) using Slidebook version 4.2.0.3 software (Intelligent Imaging Innovations, Denver, CO, USA). All images shown are single planes.
Electroretinography
Electroretinography (ERG) was performed as previously described.21,22 After mice were dark adapted overnight, they were were anesthetized and eyes were dilated. Electrophysiological function was assessed using the UTAS system (LKC, Gaithersburg, MD, USA). Scotopic ERGs were recorded with a strobe flash stimulus of 157 cd-s/m2 followed by light adaptation for 5 minutes at 29.03 cd/m2 and photopic recording at 157 cd-s/m2 (average 25 flashes). At least five to six mice per genotype were analyzed.
Western Blot Analysis and Velocity Sedimentation
Western blotting (WB) and velocity sedimentation were performed as described previously.23,25 Retinal extracts were prepared using 100 μL of chilled (4°C) solubilization buffer (phosphate-buffered saline [PBS], pH 7.0, containing 1% Triton X-100, 5 mM EDTA, 5 mg/mL N-ethylmaleimide, and a standard protease inhibitor cocktail) per retina. SDS-PAGE and WB were performed using standard protocols under reducing conditions (with dithiothreitol). Non-reducing velocity sedimentation was performed using continuous density gradients of 5% to 20% sucrose and 200 μg of protein per sample. Experiments were repeated 3 to 4 times, and densitometric quantification was done using Image Lab software (Bio-Rad, Temecula, CA, USA).
Statistical Analysis
Graph data are means ± standard errors of the mean (SEM). Differences among genotypes were assessed by 1-way ANOVA with Bonferroni's post hoc pairwise comparison. A P value of <0.05 was considered significant (graph data in Figs. 3, 5, and 6 show significance at *P < 0.05, **P < 0.01, and ***P < 0.001).
Figure 3.
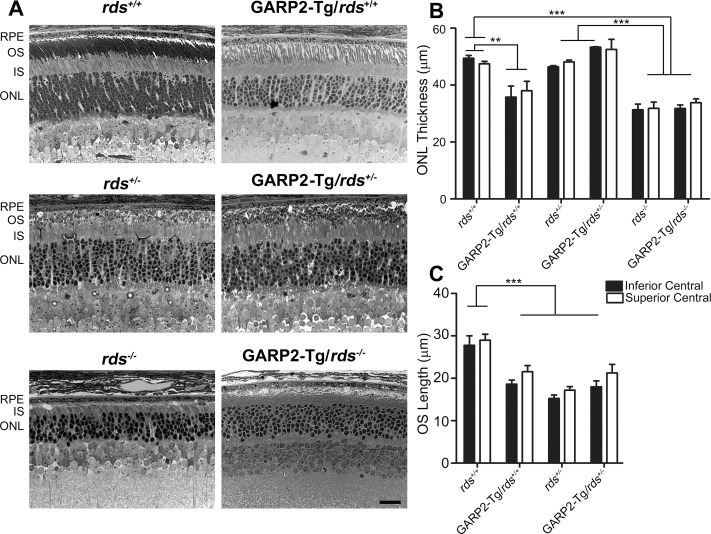
Overexpression of GARP2 protein reduces OS length. Representative brightfield photomicrographs (A) were captured from plastic-embedded retinal sections harvested at P30 from the indicated genotypes. (B) Outer nuclear layer thickness was measured in the central superior and central inferior retina from 3 to 4 eyes per genotype. (C) Outer segment layer thickness was measured in the central superior and central inferior retina from 3 to 4 eyes per genotype. **P < 0.01 and ***P < 0.001 by one-way ANOVA with Bonferroni's post hoc test. Scale bar: 20 μm. RPE, retinal pigment epithelium.
Figure 5.
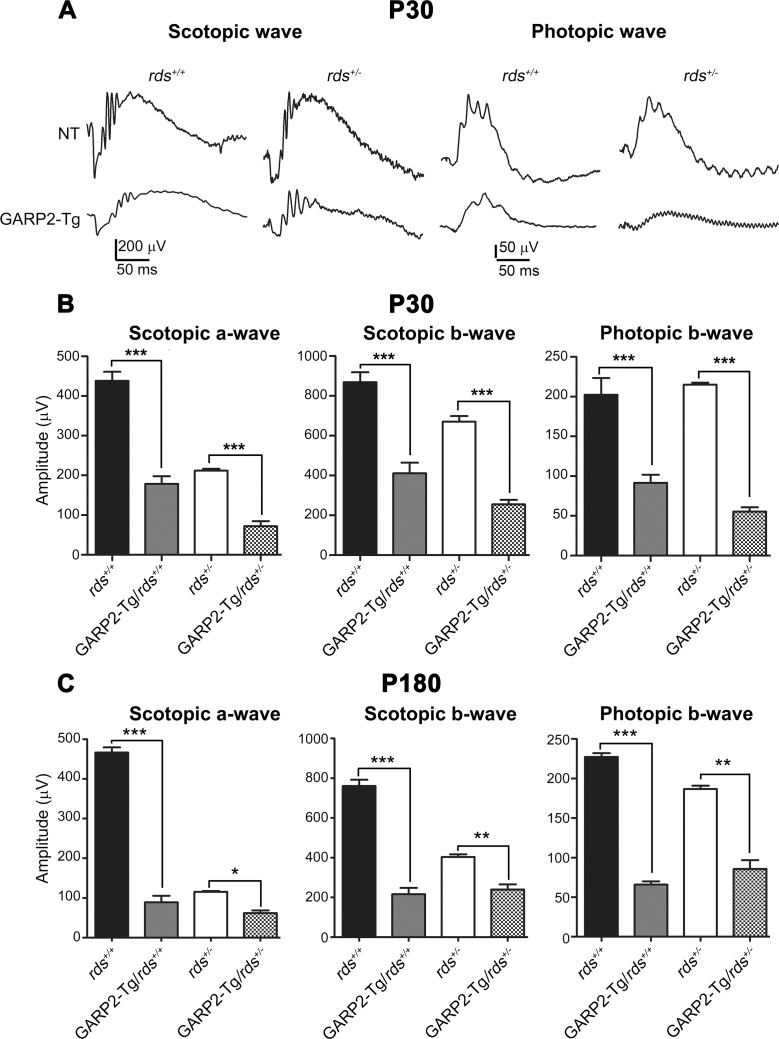
Overexpression of GARP2 causes significant reduction in ERG function. Full-field ERGs were recorded under scotopic (left two panels [A]) or photopic (right two panels [A]) conditions at P30 (A, B) or at P180 (C). Maximum scotopic a- and b-waves and maximum photopic b-waves were plotted for P30 recordings (B) and P180 recordings (C). Data are means ± SEM from 5 to 7 mice per genotype. *P < 0.05, **P < 0.01, and ***P < 0.001 in one-way ANOVA with Bonferroni's post hoc comparison.
Figure 6.
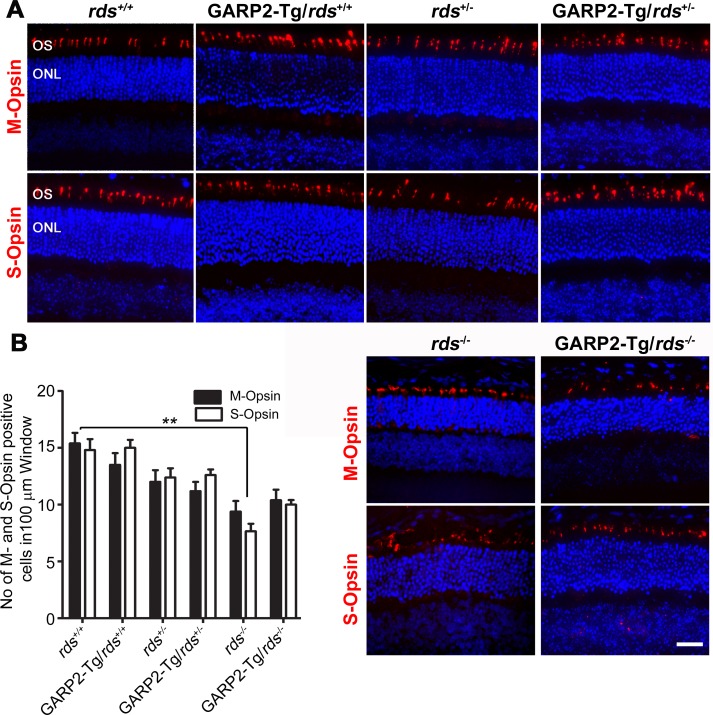
GARP2 expression does not accelerate cone degeneration. (A) Paraffin-embedded retinal sections from indicated genotypes were labeled with anti-M-opsin (top, red) and anti-S-opsin (bottom, red) and were counterstained with DAPI (blue). (B) Numbers of M- and S-opsin cones were counted in a 100-μm window from the central superior and inferior region of the retina, respectively, from P30 transgenic and non-transgenic retinal sections. n = 3 to 6 eyes per genotype. Scale bar: 20 μm.
Results
GARP2 Levels in Different Genetic Backgrounds
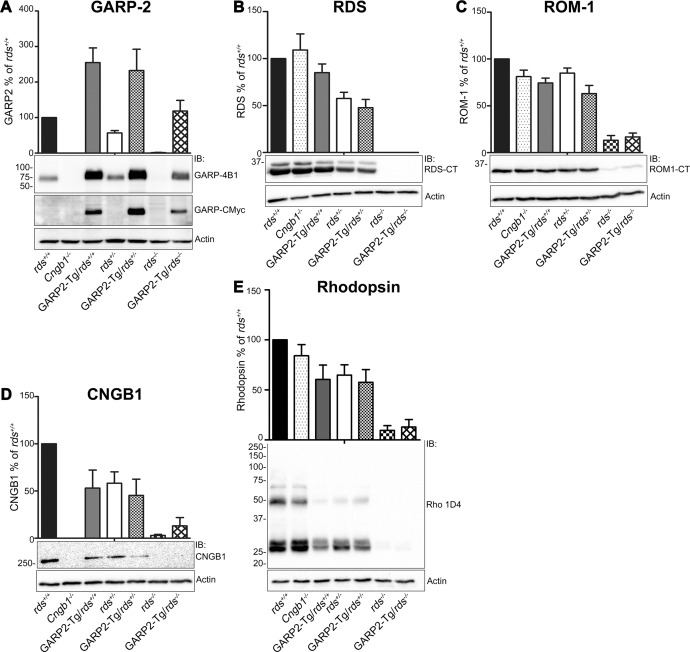
Previously we showed that the RDS-to-rhodopsin ratio in the OS is critical for the formation of proper OS,26 and given the hypothesized role of GARP/RDS complexes in stabilizing OS, we were interested in the joint role of GARP and RDS in forming and maintaining OSs. To begin our assessment of the role of RDS/GARP2 interactions we crossed GARP2-Tg animals onto the rds+/− and rds−/− backgrounds and assessed levels of critical OS proteins by reducing SDS-PAGE/WB at postnatal day 30 (P30). To control for variations in loading, the amount of each protein (i.e., GARP2) in any given genotype was first normalized to that of actin (Fig. 1, bottom panel of each section). To facilitate comparisons across multiple experiments, the amount of protein in each mutant genotype was then expressed as a percentage of the amount of that protein in the wild-type background (rds+/+). Total GARP2 (i.e., endogenous + transgenic) is plotted in Figure 1A (upper panel) with a representative blot below. Transgenic GARP2 protein migrates slightly more slowly than endogenous GARP2 protein due to the c-Myc tag, and the transgenic GARP2 protein alone can be seen on the middle blot of Figure 1A. In non-transgenic animals, GARP2 levels were affected by RDS levels: in rds+/− and rds−/−, GARP2 levels were 57% and 1.5% of rds+/+, respectively. These GARP2 levels virtually mirror RDS levels: RDS levels in the rds+/− were 58% of rds+/+, and RDS was not detected in the rds−/− background (Fig. 1B). This suggests that changing RDS levels changes GARP2 levels, possibly a result of the OS shortening which occurs in the rds+/− background.27 However, the reverse was not the case: GARP2-Tg retinas on the rds+/+ background had 2.5-fold more GARP2 than rds+/+, and CNGB1−/− retinas had no GARP2 (Fig. 1A), yet RDS levels were not significantly different in all three strains (Fig. 1B). Likewise, GARP2-Tg retinas on the rds+/− background had 2.3-fold more GARP2 than rds+/+ and 4-fold more than rds+/−, yet RDS levels were not different in GARP2-Tg/rds+/− compared to rds+/−. ROM-1 levels were similarly unaffected by the presence of excess GARP2 (Fig. 1C).
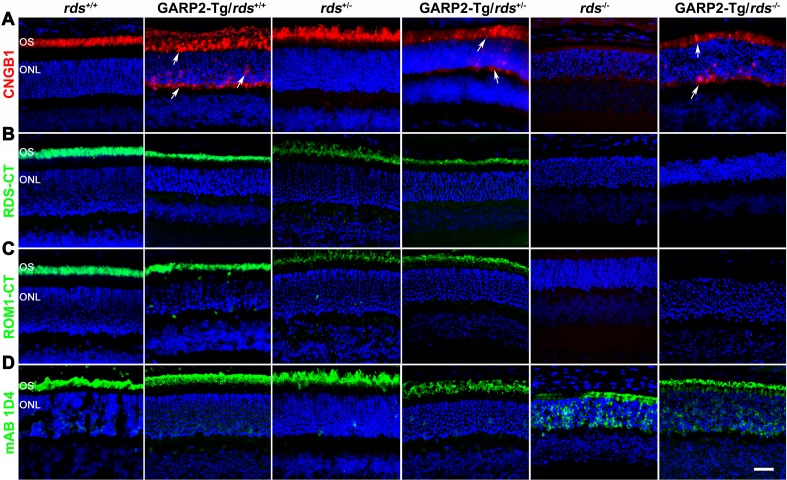
Figure 1.
Levels of OS proteins are altered in the GARP2-Tg retina. Retinal extracts were isolated from P30 rds+/+, GARP2-Tg/rds+/+, rds+/−, GARP2-Tg/rds+/−, rds−/−, and GARP2-Tg/rds−/− and were analyzed by reducing SDS-PAGE and Western blotting. Blots were probed with (A) anti-GARP-4B1 and GARP-CMyc, (B) anti-RDS-CT, (C) anti-ROM1-CT, (D) anti-CNGB1, and (E) monoclonal antibody 1D4 against rhodopsin. Blots were also labeled with actin-HRP as a loading control. Protein was quantified densitometrically and normalized to actin. Levels of OS proteins were measured in 3 to 6 retinas per genotype and are percentages of rds+/+.
We next examined the effect that various levels of GARP2/RDS had on levels of the channel subunit (CNGB1). CNGB1 (Fig. 1D) levels in GARP2-Tg/rds+/+ were reduced to 53% of rds+/+, suggesting that expression of the GARP2 transgene adversely affected expression of the channel. This decrease in CNGB1 in the GARP2-Tg/rds+/+ may be due to the OS shortening previously observed in that genotype,11 although previous work using different CNGB1 antibodies did not find a reduction in CNGB1 levels. Likewise, CNGB1 levels in the rds+/− were 58% of rds+/+, whereas those in the GARP2-Tg/rds+/− were 45% of rds+/+. We also assessed levels of rhodopsin in the presence of various amounts of GARP2 and RDS. Rhodopsin levels were slightly decreased in the GARP2-Tg/rds+/+ versus those in rds+/+, likely again due to OS shortening, but no additional decreases in rhodopsin were seen between the rds+/− and GARP2-Tg/rds+/− (Fig. 1E). All measured proteins were reduced in the rds−/− and the GARP2-Tg/rds−/− (versus rds+/+ controls), likely due to the lack of OS and the ongoing retinal degeneration in the rds−/−.
Mislocalization of GARP2 Does Not Lead to Mislocalization of RDS
We next looked at the effect of altering the GARP2-to-RDS ratio on protein localization. We performed immunofluorescence labeling of OS proteins at P30 (Figs. 2A–2D) in eyes taken from GARP2-Tg and non-transgenic animals on all rds backgrounds. The polyclonal CNGB1 antibody (CNGB1-Ab) recognizes both transgenic GARP2 protein as well as endogenous CNGB1 and GARP1/2, and we observed that in rds+/+ and rds+/−, CNGB1-Ab signal was properly restricted to the OS (Fig. 2A). CNGB1-Ab signal was below the limit of detection in the rds−/−. However, in GARP2-Tg eyes on all three RDS backgrounds, CNGB1-Ab signal accumulated in the IS layer, the ONL, and the outer plexiform layer (OPL), in addition to residing in the OS (Fig. 2A, white arrowhead). We confirmed that the GARP2-Tg protein accumulated throughout the photoreceptor by using a c-Myc antibody specific to the transgene (Supplementary Fig. S1); however because the CNGB1-Ab recognizes all Cngb1 gene products, we could not determine whether the mislocalization seen in sections labeled with CNGB1-Ab arose solely due to GARP2-Tg or whether endogenous CNGB1/GARP1/2 accumulated outside the OS. Given the known interactions between GARP2 and RDS, we next asked whether the IS/ONL/OPL localization of GARP2 protein affected RDS or ROM-1 localization. Immunofluorescence labeling of RDS (Fig. 2B) shows that RDS was found only in the OS layer in both non-transgenic and GARP2-Tg animals, suggesting that RDS localization was not affected by GARP2. ROM-1 localization (Fig. 2C) was similarly unaffected by overexpression of GARP2. We have previously shown that rhodopsin mislocalizes to the ONL in the absence of RDS, likely due to the lack of OS in the rds−/−,25,26 and we found that this rhodopsin distribution was not changed in the presence of excess GARP2 (GARP2-Tg/rds−/−) (Fig. 2D).
Figure 2.
Overexpressed GARP2 accumulates in GARP2-Tg retinas. Paraffin-embedded retinal sections from rds+/+, GARP2-Tg/rds+/+, rds+/−, GARP2-Tg/rds+/−, rds−/−, and GARP2-Tg/rds−/− were labeled with (A) anti-CNGB1 (red), (B) anti RDS-CT (green), (C) anti-ROM1-CT (green), and (D) monoclonal antibody 1D4 against rhodopsin (green) and were all counterstained with DAPI (blue). Arrows indicate mislocalization of CNGB1-Ab immunoreactivity. Scale bar: 20 μm.
Increasing the GARP2-to-RDS Ratio Accelerates Retinal Degeneration
To determine how GARP2 overexpression affected retinal structure and OS organization, we conducted histological analyses at light microscopy and EM levels at P30 in GARP2-Tg and non-transgenic controls on all rds backgrounds. Previously it has been shown that OSs are shorter in GARP2-Tg animals than in the wild-type strain11 and that OSs are likewise shorter and disorganized into abnormal whorls in the rds+/− compared to wild-type.27 Decreasing RDS levels also leads to retinal degeneration.15,27 Representative retinal cross-sections are shown in Figure 3A, with ONL thickness and thickness of the OS layer in the inferior and superior central regions shown in Figures 3B, 3C, respectively. General retinal organization and stratification was not altered by altering GARP2/RDS levels; however ONL thickness was significantly reduced in GARP2-Tg/rds+/+ versus those in rds+/+ (Fig. 3B). Interestingly, ONL thickness was not decreased in the GARP2-Tg/rds+/− compared to either the rds+/− or the rds+/+ strain (Fig. 3B). Likewise, although ONL thickness was decreased as expected in the rds−/− compared to the rds+/+ eyes at P30, there were no differences between GARP2-Tg/rds−/− and rds−/−. These data show that GARP2-Tg accelerates retinal degeneration only in the presence of a full complement of RDS and not when RDS levels are reduced, suggesting that the degenerative effects of excess GARP2 may depend on wild-type levels of RDS.
The ONL thickness results were recapitulated in measurements of OS length (Fig. 3C). As expected based on previous studies,11,27 both the GARP2-Tg/rds+/+ and rds+/− animals had shorter OSs than those of rds+/+ mice (reduced by 19% and 45%, respectively). However, as in our measurements of ONL thickness, there were no differences in OS length between GARP2-Tg/rds+/− and rds+/−. This OS shortening was also visible on low-magnification EM (Fig. 4A).
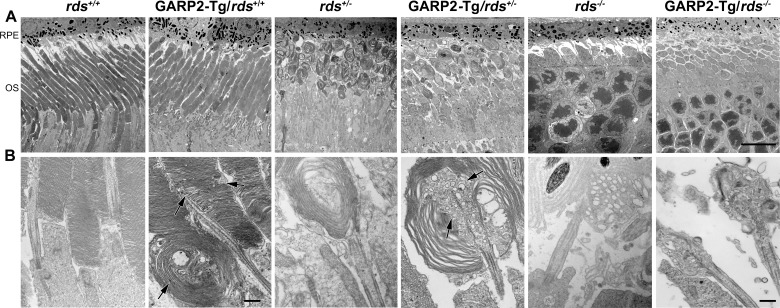
Figure 4.
Overexpression of GARP2 leads to defects in OS ultrastructure. Electron micrographs of the OS-RPE interface was performed using the indicated genotype samples at P30; shown are low-magnification (×3000 [A]) and higher magnification (×30,000 [B]) views. Arrows indicate whorl-shaped OS in GARP2-Tg/rds+/+ and accumulation of vesicular structures in GARP2-Tg/rds+/−, but ultrastructure of GARP2-Tg/rds−/− appeared similar to that of rds−/−. Scale bars: 10 μm (A) and 500 nm (B).
Gross OS ultrastructure (Fig. 4A) was not adversely affected by GARP2 overexpression but was affected by RDS. Both GARP2-Tg/rds+/+ and rds+/+ exhibited nicely stacked discs, whereas GARP2-Tg/rds+/− and rds+/− exhibited OS whorls and GARP2-Tg/rds−/− and rds−/− had no OSs. However, at higher magnification, adverse effects of GARP2 overexpression became apparent (Fig. 4B). OS structures of GARP2-Tg/rds+/+ showed some abnormalities in membrane stacking and, in rare cases, some whorl shapes (Fig. 4B), as well as regions where the disc rims did not elongate all the way to the plasma membrane (Fig. 4B, black arrows, Supplementary Fig. S2A). The regions where discs did not elongate all the way to the plasma membrane were quite frequent, and interspersed throughout the length of the OS (black arrows in Fig. 4B, Supplementary Fig. S2A); however, many OSs exhibited nicely stacked regions of discs (Supplementary Fig. S2, white arrows). Regions of altered disc alignment were also quite common (Supplementary Fig. S2, arrowheads). These occasionally progressed from altered alignment to whorl like structures, but this was quite rare. Most of these features are not quantifiable and cannot be seen on low-magnification EM, but a qualitative idea of the frequency of the whorls can be gleaned by examining low-magnification EM such as Figure 4A (in which no whorl OS are visible in the GARP2-Tg/rds+/+), and Supplementary Figure S2B (whorl OS are indicated with white arrowheads).
In addition, although the OSs of both rds+/− and the GARP2-Tg/rds+/− showed whorl shapes, we also noticed abnormal accumulation of vesicular structures (Fig. 4B, arrows) inside the whorl OS in the GARP2-Tg/rds+/− but not the rds+/−. These data suggest that overexpression of GARP2 leads to subtle ultrastructural abnormalities in rim formation, consistent with the idea that too much GARP2 can interrupt or alter the normal complement of stabilizing RDS-CNGB/GARP2 interactions and thus disrupt rim and rim/plasma membrane alignment.
Overexpression of GARP2 Protein Has Negative Effects on Rod and Cone Function
To evaluate the effects of altering the GARP2-to-RDS protein ratio on retinal function, we conducted full field scotopic (rod) and photopic (cone) ERGs from GARP2-Tg transgenic animals in the rds+/+ and rds+/− backgrounds at P30 and P180. Figure 5A shows representative traces at P30 with quantification in Figures 5B, 5C (**P < 0.01 and ***P < 0.001 in 1-way ANOVA with Bonferroni's post hoc test). A statistically significant 59% reduction in maximum scotopic a-wave was observed in GARP2-Tg/rds+/+ in comparison to rds+/+, and a 66% reduction was seen in GARP2-Tg/rds+/− in comparison to rds+/−. This dramatic GARP2-Tg-associated functional defect was also evident in maximum scotopic b-wave amplitudes (Fig. 5B, middle) and persisted to P180 (Fig. 5C). The reduction in rod function in GARP2-Tg/rds+/− compared with rds+/− is striking as we did not observe accelerated degeneration or OS shortening in a comparison of these two genotypes, and this suggests that the subtle ultrastructural changes we observed in the GARP2-Tg/rds+/− versus rds+/− could have dramatic functional consequences.
We next evaluated photopic ERG responses at P30 and at P180 (Figs. 5A–C, right). The GARP2 transgene is under the control of the mouse rhodopsin promoter, so we did not expect cone function would be affected by overexpression of GARP2. Surprisingly however, we observed significant decreases in maximum photopic b-wave in the presence of the GARP2 transgene on both the rds+/+ and rds+/− backgrounds (compared to non-transgenic counterparts). Although rod-degeneration due to defects in rod-specific proteins can lead to secondary cone degeneration in some cases, the defects in cone function are severe as early as P30, a time point at which only modest rod degeneration (as measured by ONL thickness) was occurring in the GARP2-Tg/rds+/+, and no rod degeneration was found in the GARP2-Tg/rds+/− (Fig. 3B).
Cone Degeneration Is Not Accelerated in GARP2-Tg Animals
The observed deficit in cone function in the GARP2-Tg retina led us to wonder whether there was cone degeneration occurring in GARP2-Tg animals, so we conducted immunofluorescence labeling for S- and M-opsin (Fig. 6A) and counted the number of cone cells in a 100 μm window in the central retina (quantified in Fig. 6B) at P30. We found that though cone numbers were modestly decreased as a result of RDS deficiency (i.e., rds+/+ versus rds−/−) (Fig. 6B), there were no GARP2-associated decreases in cone number in any RDS background (e.g., rds+/+ was not different from GARP2-Tg/rds+/+).
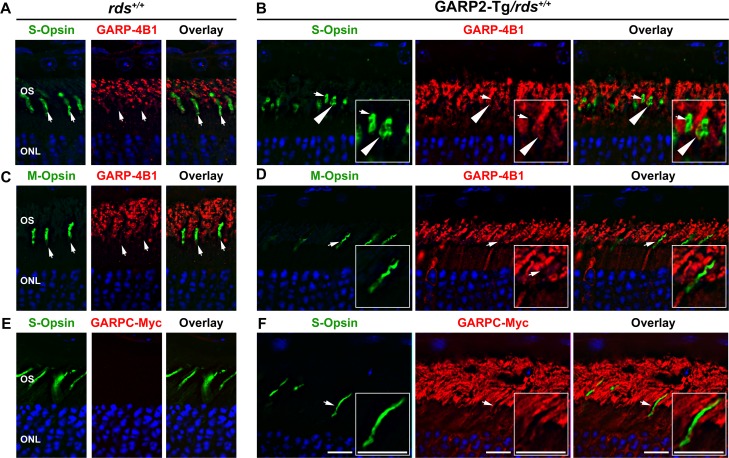
The lack of cone cell loss coupled with functional defects in GARP2-Tg cones led us to hypothesize that overexpression of GARP2 led to a physiological or signal transduction deficit in cones. However such a hypothesis would require expression of the GARP2 transgene in cones. Previous work has shown that a smaller 500-bp fragment of the mouse opsin promoter leads to rod expression and some leaky expression in cones.28,29 Because previous studies did not verify whether the 4.4-kbp mouse opsin promoter used to generate the GARP2-Tg line could drive gene expression in cones,30 we evaluated cone expression of the GARP2 transgene. P30 rds+/+ and GARP2-Tg/rds+/+ retinal sections were colabeled with either S- or M-opsin (Figs. 7A–C, green) and GARP-4B1, which recognizes transgenic GARP2 as well as endogenous CNGB1/GARP2/GARP1 (Figs. 7A, 7B, red) or c-Myc, which recognizes only transgenic GARP2 (Fig. 7C, red). Single planes from a confocal image stack are shown. Although GARP-4B1 recognizes both transgenic and endogenous protein, it can be used to specifically assess transgene expression in cones because endogenous CNGB1/GARP2/GARP1 are not expressed in cones (Figs. 7A, 7C, 7E, arrows and Supplementary Fig. S3A show wild-type cones clearly not labeled with GARP-4B1 or c-Myc). In GARPT2-Tg/rds+/+ retinas, most cone photoreceptors had no detectable GARP2 (Figs. 7B, 7D, 7F, arrows). However, occasionally cone OSs were found in GARP2-Tg/rds+/+ retinas that did label with the GARP-4B1 antibody (Fig. 7A, arrowheads) suggesting there could be a low level of leaky GARP2 expression in cones in transgenic animals. Additional GARP2-Tg/rds+/+ but not rds+/+ cone OSs showing labeling with GARP-4B1 are pictured in Supplementary Figure S3A and S3B (arrowheads). In Supplementary Figures S3A, S3B, GARP-4B1 labeling is shown in red with cone OSs highlighted in green using the cone matrix sheath marker peanut agglutinin-PNA, and in each panel, the bottom row shows magnified insets of the cells highlighted by arrows in the top row. When we repeated this experiment using c-Myc antibody to specifically label transgenic GARP2, similar results were observed: some cone OSs did not express the transgene (Fig. 7F, e.g., arrows) whereas others did (Supplementary Fig. S3C, arrowheads). In many cases, the small size of murine OSs coupled with the high levels of GARP2 expression in adjacent rods made it difficult to determine whether cone cells had any detectable GARP-4B1 immunolabeling. These results suggest that while many cones in GARP2-Tg/rds+/+ do not express any GARP2 transgenic protein; there are cones which exhibit ectopic expression. In addition, co-labeling with cone opsins and c-Myc (to specifically label GARP2-Tg) (Fig. 7C) also confirmed that the GARP2 accumulation we observed in Figure 2 was occurring in rods and not solely due to ectopic expression in cones.
Figure 7.
Most cones in GARP2-Tg retinas do not express the GARP2 protein. Retinal sections from rds+/+ and GARP2-Tg/rds+/+ mice were colabeled with GARP-4B1 (red) and S-opsin (green) (A, B), GARP-4B1 (red), and M-opsin (green) (C, D), and c-Myc (red) and S-opsin (green) in (E, F). Nuclei in all sections were counterstained with DAPI. Arrows indicate cones that do not have detectable GARP-4B1 immunoreactivity. Arrowheads show cones that do display GARP-4B1 immunoreactivity. Areas shown by arrows are expanded in the insets. Scale bar: 10 μM.
RDS:ROM-1 Oligomerization Is Subtly Affected in the GARP2-Tg Retina
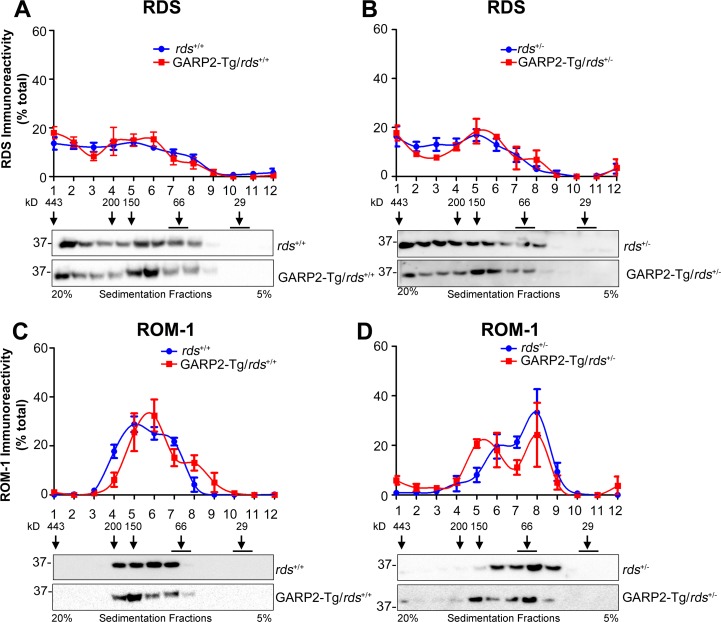
RDS function relies on the formation of a precise complement of homo- and heteromeric (with ROM-1) complexes which can be easily separated by non-reducing sucrose gradient velocity sedimentation.23,25 When RDS complex formation is altered, for example, by mutations in RDS, severe defects in OS structure and function occur.23,31,32 We therefore asked whether overexpression of GARP2, a known RDS binding partner, affected the type of RDS/ROM-1 complexes in the retina. Retinal extracts were fractionated on continuous 5% to 20% non-reducing sucrose gradients and were then separated on reducing SDS-PAGE. Western blots were probed with antibodies for RDS (Figs. 8A, 8B) and ROM-1 (Figs. 8C, 8D). In the wild-type retina, RDS is present as tetramers (fractions 6–8), intermediate oligomers (fractions 4–5), and higher order oligomers (fractions 1–3), whereas ROM-1 was detected only in tetramers and intermediate oligomers. We did not observe any significant alteration in the distribution of RDS complexes in GARP2-Tg retinas on either the rds+/+ or rds+/− backgrounds (Figs. 8A, 8B). However, in the GARP2-Tg/rds+/− retinas, there is a subtle shift in ROM-1 complex formation toward the fractions associated with intermediate sized complexes compared to the non-transgenic rds+/− (Fig. 8D). These data suggest that overexpression of GARP2 could influence the way RDS/ROM-1 heteromers assemble, particularly when RDS levels are decreased (i.e., rds+/−).
Figure 8.
RDS complex formation is not altered in transgenic retinas. Non-reducing sucrose gradient velocity sedimentation was performed in P30 retinal extracts from rds+/+ and GARP2-Tg/rds+/+ mice (A, C) and in rds+/− and GARP2-Tg/rds+/− mice (B, D). Gradient fractions (1–12) were further separated by reducing SDS-PAGE/Western blotting. Resulting blots were probed with anti-RDS-CT (A, B) and anti-ROM1-CT (C, D). The relative percentage of total RDS or ROM-1 in each fraction was assessed densitometrically and plotted as means ± SEM from 3 to 6 independent experiments.
Discussion
RDS and GARP2/CNGB1 have been shown to interact by using multiple approaches and by multiple research groups.6,12,13,33 Although the domains of GARP2/CNGB1 and RDS interaction have not yet been identified, data showing that both CNGB1 and GARP2 can interact with RDS suggest that the GARP region of GARP2/CNGB1 likely mediates the interaction.6 The cytosolic face of RDS consists of a short N terminus and a small cytosolic loop in addition to a lengthy and highly functionalized C terminus characterized by a large degree of intrinsic disorder.34 The C terminus is the site of interaction for some other RDS binding partners, including melanoregulin,35 is involved in RDS OS targeting,36,37 and has the ability to mediate membrane fusion and curvature.38,39 As a result, the C terminus of RDS is the most likely region for GARP2 binding on RDS. Because we observed mislocalization of excess GARP2 throughout the photoreceptor, we wondered whether excessive GARP2 interactions with the RDS C terminus might interfere with RDS OS targeting. However, this was not the case; neither RDS nor ROM-1 showed any signs of mislocalization. This is consistent with recent work suggesting that CNGB1/RDS interactions occur early (i.e., in the inner segment), whereas RDS/GARP2 interactions occur after OS targeting, at or near the base of the OS.13
Despite holding interest for many years, the role of GARP2/RDS interactions remains unknown. It has been hypothesized that GARP2/RDS interactions help to form a connection between the disc membrane and the plasma membrane and contribute to the stabilization of the ROS. The distance between plasma membrane and disc membrane is estimated at 15 to 17 nm by cryo-EM, a technique that preserves structure because no fixatives are used.40,41 Estimates suggest that the maximum length of an intrinsically disordered peptide is 3.8 Å per amino acid, indicating that GARP2 (at 450 amino acids) could reach up to 171 nm in length. This suggests that the CNGB1/RDS complex could reach far enough to span the gap between membranes and to bind directly to RDS. Additionally, knockout of CNGB1 and GARP1/23 severely disrupted plasma/disc membrane interactions, further supporting the hypothesis that CNGB1 and/or GARP1/2 is essential for this interaction. It is also not clear how strong the affinity of RDS is for CNGB1/GARP2. For example, RDS binds ROM-1 with very high affinity, and RDS/ROM-1 complexes are abundant and easily detected in the presence of commonly used detergent (e.g., 1% Triton X-100, CHAPS, and Brij-96). Similarly, GARP2 interacts with PDE6 with high affinity and copurifies with PDE6.10 However, we have observed that RDS/GARP2/CNGB1 interactions are harder to detect by standard immunoprecipitation, and other groups have used more advanced methodologies such as cross-linking12 and bimolecular fluorescence complementation13 to confirm RDS/GARP2 and RDS/CNGB1 interactions. Thus, it is possible that GARP2/RDS interactions are either less abundant or more labile (or both) than other common OS protein/protein interactions.
Additional insight into the function of RDS/GARP2 interactions comes from a recent study by Ritter et al.,13 which reported that interactions between RDS and CNGB1 occur in the IS but RDS and GARP2 interactions occur at the base of the OS at the site of disc morphogenesis. They suggest that GARPS interactions may have some role in disc sizing, and we do observe some abnormally sized and aligned discs in GARP2-Tg overexpressors.13 That RDS is required for disc formation has been known for several decades and that RDS binding partners may regulate disc sizing is likewise accepted. ROM-1 has been hypothesized to play a role in disc sizing,42 and the defect in RDS/ROM-1 complex formation we observe in the presence of excess GARP is a shift in ROM-1 toward larger oligomers. It is thus possible that excess GARP2/RDS interactions alter the type of RDS/ROM-1 complexes that form and thus contribute to structural abnormalities and the minor shifts in ROM-1 complexes we see in sedimentation experiments (Fig. 8).
It is also possible that all of the structural and functional defects we see in animals expressing various GARP2:RDS ratios could be due to independent effects of GARP2 and RDS. RDS haploinsufficiency is known to cause severe structural and functional defects alone,27,43 and recent work has suggested that GARP2 may play a role in regulating rod sensitivity by altering PDE6 activity and cGMP turnover.9–11,44 In addition, our observation that CNGB1 levels were decreased in the presence of excess GARP suggests that CNG channel function or trafficking may be slightly altered in GARP2-Tg animals. Because our CNG antibodies recognize both CNGB1 and GARP, it is not possible to assess CNGB1 alone by immunofluorescence (although the GARP2-TG protein can be visualized by using the c-Myc antibody).
The observation that cone photoreceptor function is reduced in GARP2-Tg animals (on both the rds+/+ and the rds+/−) is one of the most interesting and unexpected outcomes of this study. The 4.4-kbp mouse opsin promoter that was used to make the GARP2-Tg mice has previously shown robust expression in rod photoreceptors, but this early study was unable to conclusively determine whether the promoter drove gene expression in cones as well.30 Given the observation that shorter fragments of the mouse opsin promoter are known to promote expression in both rods and cones and the fact that our results showed a clear functional deficit in GARP2-Tg cones, it was logical to assess GARP2 expression in cones. Our high-magnification immunofluorescence experiment showed that most cone cells had no detectable GARP2, but we did find cone OSs that exhibited some GARP2 immunoreactivity. These data suggest that the 4.4-kbp opsin promoter may have some low level of expression in cones. More importantly, data suggest that a small amount of GARP2 expression in cones could be enough to impair cone function. We did not directly assess cone OS ultrastructure, but we did rarely observe highly abnormal whorl-shaped OSs in the GARP2-Tg/rds+/+, and it is possible that these are cone OSs and appear as either a consequence of low-level ectopic expression in cones or as an indirect consequence of other ongoing changes in retinal morphology. These two possibilities are equally interesting. First, why expression of GARP2 in cones should be so bad for cone function is unclear. The cone CNGB channel subunit (CNGB3) does not have a GARP region, free GARPs are not expressed in wild-type cones, and we previously showed that cone CNGB3 does not interact with RDS.33 If GARP2 in rods is involved in regulating OS structure or maintenance, it is reasonable to imagine that another protein could fill that role in cones and help regulate the open versus closed disc structures present in cones. It is possible that abnormal expression of GARP2 in cones somehow interrupts this interaction. Because GARP2 has also been hypothesized to play an unexplained role in phototransduction,11 it is possible that the adverse effects of GARP2 on cone function are due to interference in the phototransduction process. The second possibility is that defects in cone function in GARP2-Tg animals are secondary abnormalities due to ongoing changes in rods/degeneration. While the very low level of ectopic expression of GARP2-Tg in cones that we observed makes this hypothesis attractive; it is certainly unexpected. While secondary cone degeneration due to loss of rod cells is quite common, it is usually not seen until overall retinal changes are more severe. For example, in the rho−/− retina, rod OS completely fail to form,45 a much worse phenotype than in GARP2-Tg animals, yet cone function remained normal beyond the time points that we observed significant defects in cone function. Certainly further exploration of the effects of GARP2 in or on cones will be an ongoing research goal since it may shed light on differences in the structure and function of the two cell types and the differential role of RDS in them as well as potential interactions between the two cell types during the degenerative process.
In conclusion we showed that increasing GARP2 levels, while decreasing RDS levels, leads to structural and functional defects in photoreceptors. These defects are likely due to a combination of independent and combined factors. For example, GARP2 has a role in regulating phototransduction through effects on PDE, whereas RDS has a role in regulating OS structure. In addition, there are combined effects of the two proteins such as the potential for RDS/GARP2 complexes to be involved in disc sizing and in interactions between the disc and plasma membrane or between adjacent discs. Future work will continue to explore these differences and may elucidate some of the structural and functional differences between rod photoreceptors (which express GARPs) and cone photoreceptors (which do not).
Supplementary Material
Acknowledgments
The authors thank Barb Nagel and Marcellus Banworth for their technical assistance. We also thank Robert Molday, Ph.D., for the provision of reagents as indicated in the text.
Supported by National Institutes of Health Grant R01EY010609 (MIN), by the Oklahoma Center for the Advancement of Science and Technology (SMC), and by NIH Grants R01EY018143-06 and P30EY003039-35 and the Vision Science Research Center (SJP).
Disclosure: D. Chakraborty, None; S.M. Conley, None; M.L. DeRamus, None; S.J. Pittler, None; M.I. Naash, None
References
- 1. Zhong H,, Molday LL,, Molday RS,, Yau KW. The heteromeric cyclic nucleotide-gated channel adopts a 3A:1B stoichiometry. Nature. 2002; 420: 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kaupp UB,, Seifert R. Cyclic nucleotide-gated ion channels. Physiol Rev. 2002; 82: 769–824. [DOI] [PubMed] [Google Scholar]
- 3. Zhang Y,, Molday LL,, Molday RS,, et al. Knockout of GARPs and the beta-subunit of the rod cGMP-gated channel disrupts disk morphogenesis and rod outer segment structural integrity. J Cell Sci. 2009; 122: 1192–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ardell MD,, Bedsole DL,, Schoborg RV,, Pittler SJ. Genomic organization of the human rod photoreceptor cGMP-gated cation channel beta-subunit gene. Gene. 2000; 245: 311–318. [DOI] [PubMed] [Google Scholar]
- 5. Colville CA,, Molday RS. Primary structure and expression of the human beta-subunit and related proteins of the rod photoreceptor cGMP-gated channel. J Biol Chem. 1996; 271: 32968–32974. [DOI] [PubMed] [Google Scholar]
- 6. Batra-Safferling R,, Abarca-Heidemann K,, Korschen HG,, et al. Glutamic acid-rich proteins of rod photoreceptors are natively unfolded. J Biol Chem. 2006; 281: 1449–1460. [DOI] [PubMed] [Google Scholar]
- 7. Huttl S,, Michalakis S,, Seeliger M,, et al. Impaired channel targeting and retinal degeneration in mice lacking the cyclic nucleotide-gated channel subunit CNGB1. J Neurosci. 2005; 25: 130–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bareil C,, Hamel CP,, Delague V,, Arnaud B,, Demaille J,, Claustres M. Segregation of a mutation in CNGB1 encoding the beta-subunit of the rod cGMP-gated channel in a family with autosomal recessive retinitis pigmentosa. Hum Genet. 2001; 108: 328–334. [DOI] [PubMed] [Google Scholar]
- 9. Korschen HG,, Beyermann M,, Muller F,, et al. Interaction of glutamic-acid-rich proteins with the cGMP signalling pathway in rod photoreceptors. Nature. 1999; 400: 761–766. [DOI] [PubMed] [Google Scholar]
- 10. Pentia DC,, Hosier S,, Cote RH. The glutamic acid-rich protein-2 (GARP2) is a high affinity rod photoreceptor phosphodiesterase (PDE6)-binding protein that modulates its catalytic properties. J Biol Chem. 2006; 281: 5500–5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sarfare S,, McKeown AS,, Messinger J,, et al. Overexpression of rod photoreceptor glutamic acid rich protein 2 (GARP2) increases gain and slows recovery in mouse retina. Cell Commun Signal. 2014; 12: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Poetsch A,, Molday LL,, Molday RS. The cGMP-gated channel and related glutamic acid-rich proteins interact with peripherin-2 at the rim region of rod photoreceptor disc membranes. J Biol Chem. 2001; 276: 48009–48016. [DOI] [PubMed] [Google Scholar]
- 13. Ritter LM,, Khattree N,, Tam B,, Moritz OL,, Schmitz F,, Goldberg AF. In situ visualization of protein interactions in sensory neurons: glutamic acid-rich proteins (GARPs) play differential roles for photoreceptor outer segment scaffolding. J Neurosci. 2011; 31: 11231–11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arikawa K,, Molday LL,, Molday RS,, Williams DS. Localization of peripherin/rds in the disk membranes of cone and rod photoreceptors: relationship to disk membrane morphogenesis and retinal degeneration. J Cell Biol. 1992; 116: 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sanyal S,, Zeilmaker GH. Development and degeneration of retina in rds mutant mice: light and electron microscopic observations in experimental chimaeras. Exp Eye Res. 1984; 39: 231–246. [DOI] [PubMed] [Google Scholar]
- 16. Kajiwara K,, Hahn LB,, Mukai S,, Travis GH,, Berson EL,, Dryja TP. Mutations in the human retinal degeneration slow gene in autosomal dominant retinitis pigmentosa. Nature. 1991; 354: 480–483. [DOI] [PubMed] [Google Scholar]
- 17. Wells J,, Wroblewski J,, Keen J,, et al. Mutations in the human retinal degeneration slow (RDS) gene can cause either retinitis pigmentosa or macular dystrophy. Nat Genet. 1993; 3: 213–218. [DOI] [PubMed] [Google Scholar]
- 18. Goldberg AF,, Molday RS. Defective subunit assembly underlies a digenic form of retinitis pigmentosa linked to mutations in peripherin/rds and rom-1. Proc Natl Acad Sci U S A. 1996; 93: 13726–13730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goldberg AF,, Molday RS. Subunit composition of the peripherin/rds-rom-1 disk rim complex from rod photoreceptors: hydrodynamic evidence for a tetrameric quaternary structure. Biochemistry (Mosc). 1996; 35: 6144–6149. [DOI] [PubMed] [Google Scholar]
- 20. Goldberg AF,, Moritz OL,, Molday RS. Heterologous expression of photoreceptor peripherin/rds and Rom-1 in COS-1 cells: assembly interactions, and localization of multisubunit complexes. Biochemistry (Mosc). 1995; 34: 14213–14219. [DOI] [PubMed] [Google Scholar]
- 21. Farjo R,, Skaggs JS,, Nagel BA,, et al. Retention of function without normal disc morphogenesis occurs in cone but not rod photoreceptors. J Cell Biol. 2006; 173: 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stricker HM,, Ding XQ,, Quiambao A,, Fliesler SJ,, Naash MI. The Cys214-->Ser mutation in peripherin/rds causes a loss-of-function phenotype in transgenic mice. Biochem J. 2005; 388: 605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chakraborty D,, Ding XQ,, Conley SM,, Fliesler SJ,, Naash MI. Differential requirements for retinal degeneration slow intermolecular disulfide-linked oligomerization in rods versus cones. Hum Mol Genet. 2009; 18: 797–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chakraborty D,, Conley SM,, Stuck MW,, Naash MI. Differences in RDS trafficking, assembly and function in cones versus rods: insights from studies of C150S-RDS. Hum Mol Genet. 2010; 19: 4799–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chakraborty D,, Ding XQ,, Fliesler SJ,, Naash MI. Outer segment oligomerization of Rds: evidence from mouse models and subcellular fractionation. Biochemistry. 2008; 47: 1144–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chakraborty D,, Conley SM,, Al-Ubaidi MR,, Naash MI. Initiation of rod outer segment disc formation requires RDS. PLoS One. 2014; 9: e98939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cheng T,, Peachey NS,, Li S,, Goto Y,, Cao Y,, Naash MI. The effect of peripherin/rds haploinsufficiency on rod and cone photoreceptors. J Neurosci. 1997; 17: 8118–8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Glushakova LG,, Timmers AM,, Issa TM,, et al. Does recombinant adeno-associated virus-vectored proximal region of mouse rhodopsin promoter support only rod-type specific expression in vivo? Mol Vis. 2006; 12: 298–309. [PubMed] [Google Scholar]
- 29. Cai X,, Conley SM,, Nash Z,, Fliesler SJ,, Cooper MJ,, Naash MI. Gene delivery to mitotic and postmitotic photoreceptors via compacted DNA nanoparticles results in improved phenotype in a mouse model of retinitis pigmentosa. FASEB J. 2010; 24: 1178–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lem J,, Applebury ML,, Falk JD,, Flannery JG,, Simon MI. Tissue-specific and developmental regulation of rod opsin chimeric genes in transgenic mice. Neuron. 1991; 6: 201–210. [DOI] [PubMed] [Google Scholar]
- 31. Conley SM,, Stuck MW,, Burnett JL,, et al. Insights into the mechanisms of macular degeneration associated with the R172W mutation in RDS. Hum Mol Genet. 2014; 23: 3102–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stuck MW,, Conley SM,, Naash MI. The Y141C knockin mutation in RDS leads to complex phenotypes in the mouse. Hum Mol Genet. 2014; 23: 6260–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Conley SM,, Ding XQ,, Naash MI. RDS in cones does not interact with the beta subunit of the cyclic nucleotide gated channel. Adv Exp Med Biol. 2010; 664: 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ritter LM,, Arakawa T,, Goldberg AF. Predicted and measured disorder in peripherin/rds a retinal tetraspanin. Protein Pept Lett. 2005; 12: 677–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boesze-Battaglia K,, Song H,, Sokolov M,, et al. The tetraspanin protein peripherin-2 forms a complex with melanoregulin, a putative membrane fusion regulator. Biochemistry (Mosc). 2007; 46: 1256–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Salinas RY,, Baker SA,, Gospe SM,, III, Arshavsky VYA. Single valine residue plays an essential role in peripherin/rds targeting to photoreceptor outer segments. PLoS One. 2013; 8: e54292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tam BM,, Moritz OL,, Papermaster DS. The C terminus of peripherin/rds participates in rod outer segment targeting and alignment of disk incisures. Mol Biol Cell. 2004; 15: 2027–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boesze-Battaglia K,, Lamba OP,, Napoli AA,, Jr, Sinha S,, Guo Y. Fusion between retinal rod outer segment membranes and model membranes: a role for photoreceptor peripherin/rds. Biochemistry. 1998; 37: 9477–9487. [DOI] [PubMed] [Google Scholar]
- 39. Khattree N,, Ritter LM,, Goldberg AF. Membrane curvature generation by a C-terminal amphipathic helix in peripherin-2/rds a tetraspanin required for photoreceptor sensory cilium morphogenesis. J Cell Sci. 2013; 126: 4659–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nickell S,, Park PS,, Baumeister W,, Palczewski K. Three-dimensional architecture of murine rod outer segments determined by cryoelectron tomography. J Cell Biol. 2007; 177: 917–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gilliam JC,, Chang JT,, Sandoval IM,, et al. Three-dimensional architecture of the rod sensory cilium and its disruption in retinal neurodegeneration. Cell. 2012; 151: 1029–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Clarke G,, Goldberg AF,, Vidgen D,, et al. Rom-1 is required for rod photoreceptor viability and the regulation of disk morphogenesis. Nat Genet. 2000; 25: 67–73. [DOI] [PubMed] [Google Scholar]
- 43. Hawkins RK,, Jansen HG,, Sanyal S. Development and degeneration of retina in rds mutant mice: photoreceptor abnormalities in the heterozygotes. Exp Eye Res. 1985; 41: 701–720. [DOI] [PubMed] [Google Scholar]
- 44. Michalakis S,, Zong X,, Becirovic E,, et al. The glutamic acid-rich protein is a gating inhibitor of cyclic nucleotide-gated channels. J Neurosci. 2011; 31: 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lem J,, Krasnoperova NV,, Calvert PD,, et al. Morphological, physiological, and biochemical changes in rhodopsin knockout mice. Proc Natl Acad Sci U S A. 1999; 96: 736–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.