Abstract
The genus Nairovirus of arthropod-borne bunyaviruses includes the important emerging human pathogen, Crimean–Congo hemorrhagic fever virus (CCHFV), as well as Nairobi sheep disease virus and many other poorly described viruses isolated from mammals, birds, and ticks. Here, we report genome sequence analysis of six nairoviruses: Thiafora virus (TFAV) that was isolated from a shrew in Senegal; Yogue (YOGV), Kasokero (KKOV), and Gossas (GOSV) viruses isolated from bats in Senegal and Uganda; Issyk-Kul virus (IKV) isolated from bats in Kyrgyzstan; and Keterah virus (KTRV) isolated from ticks infesting a bat in Malaysia. The S, M, and L genome segments of each virus were found to encode proteins corresponding to the nucleoprotein, polyglycoprotein, and polymerase protein of CCHFV. However, as observed in Leopards Hill virus (LPHV) and Erve virus (ERVV), polyglycoproteins encoded in the M segment lack sequences encoding the double-membrane-spanning CCHFV NSm protein. Amino acid sequence identities, complement-fixation tests, and phylogenetic analysis indicated that these viruses cluster into three groups comprising KKOV, YOGV, and LPHV from bats of the suborder Yingochiroptera; KTRV, IKV, and GOSV from bats of the suborder Yangochiroptera; and TFAV and ERVV from shrews (Soricomorpha: Soricidae). This reflects clade-specific host and vector associations that extend across the genus.
Introduction
Nairoviruses are arthropod-borne bunyaviruses that are transmitted primarily by ticks and some of which are important pathogens of humans and livestock. Crimean–Congo hemorrhagic fever virus (CCHFV) causes an emerging disease of humans with reported case fatality rates of 3–30%, typically associated with hemorrhage, shock, and multiorgan system failure within 2 weeks of the onset of symptoms.1,2 CCHFV has a distribution that includes parts of Africa, the Middle East, eastern Europe, and Asia, and has been identified as an agent of significant public health concern.2,3 Nairobi sheep disease virus (NSDV) causes hemorrhagic gastroenteritis in sheep and goats with mortality rates of up to 90%.4,5 Together with Ganjam virus, which is considered to be a strain of the same virus, NSDV has a distribution that includes central and east Africa as well as South Asia and China.6,7 At least 50 other nairoviruses have been isolated from ticks or vertebrate hosts and some have been associated with symptoms of disease in humans including fever, headache, and neurological disorders.8 Many of these viruses remain poorly characterized.
The genus Nairovirus (family Bunyaviridae) presently comprises seven species (CCHFV, Dugbe virus, Thiafora virus [TFAV], Qalyub virus, Hughes virus, Sakhalin virus, and Dera Ghazi Khan virus) to which 35 viruses have been assigned on the basis of antigenic cross-reactions.8,9 All nairoviruses within each of the corresponding serogroups are considered to be the same species.10 Like other viruses in the family Bunyaviridae, nairoviruses have enveloped spherical virions and a negative-sense single-stranded RNA genome comprising three segments.10 The L segment encodes a large protein (∼450 kDa) with multiple highly conserved domains associated with replication and transcription, including the RNA-dependent RNA polymerase (RdRp), an ovarian-tumor (OTU)-like cysteine protease domain, and a zinc-finger domain.11–13 The nairovirus M segment encodes a multiple-membrane-spanning polyglycoprotein that is processed by host peptidases to generate the mature envelope glycoproteins (Gn and Gc), a mucin-like protein, and other potential products.14–17 The S segment encodes the viral nucleoprotein (N), subunits of which are organized in a head-to-tail manner to encapsidate the viral genome.18 In CCHFV, the N protein has also been shown to have DNA endonuclease activity19 and a conserved sequence motif that is characteristic of catalytic motif II of N6-adenine-specific DNA methylases.8 The functions of these domains during infection are presently unclear.
In this article, we report the complete genome sequences of six nairoviruses that were isolated from bats and a shrew in Africa, a bat in Central Asia, and ticks feeding on a bat in Southeast Asia. We show that they share a distinctive genome organization that lacks sequences encoding an NSm protein and they cluster phylogenetically according to vertebrate host to form distinct sub-clades within the genus.
Materials and Methods
Description of viruses.
The identification; current taxonomic assignment; date, place, and source of isolation; disease association; and Genbank numbers of the viruses included in this study are shown in Table 1. TFAV from Senegal is serologically related to the previously characterized and sequenced Erve virus (ERVV) from France and each was isolated from a shrew (Crocidura spp.).20–22 TFAV and ERVV were previously assigned to the species Thiafora virus.23 Yogue virus (YOGV) and Kasokero virus (KKOV) are unclassified nairoviruses isolated from Egyptian fruit bats (Rousettus aegyptiacus) in Africa.20,23,24 KKOV was subsequently isolated from sick laboratory workers with an illness characterized by headache, diarrhea, myalgia, and arthralgia.24 Gossas virus (GOSV) was isolated from a free-tailed bat (Tadarida sp.) in Senegal and was originally identified as a rhabdovirus.20,25 However, subsequent sequence analysis has revealed that this was due to laboratory contamination of the sample with Nkolbisson virus (family Rhabdoviridae).26 Keterah virus (KTRV) is an unclassified nairovirus isolated from tick larvae (Argus pusillus) taken from a lesser Asian house bat (Scotophilus temmincki) in Malaysia.27 KTRV is closely related antigenically to Issyk-Kul virus (IKV),20 another unclassified nairovirus first isolated from organs and blood of several bat species (Nyctalus noctula, Myotis blythii, Vespertilio serotinus, and Vespertilio pipistrellus) and bat ticks (Argas vespertilionis) collected in Kyrgyzstan.28 The prototype IKV strain (LEIV-315K) was obtained from pooled organs of a common noctule bat (N. noctula). IKV was subsequently isolated from bats (V. pipistrellus) in Tadzhikistan where it was also associated with an outbreak of febrile illness in humans.29 IKV strain LEZ 86-1784 was obtained from F. Rodhain of Institut Pasteur in Paris. It was isolated from bat ticks (A. vespertilionis) collected near Berlin in Germany in 1986 by H. Sinnecker. Leopards Hill virus (LPHV) was isolated from giant leaf-nosed bats (Hipposideros gigas) in Zambia and shown experimentally to cause hemorrhagic gastroenteritis and severe hepatic disease in mice.30 Genbank accession numbers of partial L gene sequences of other nairoviruses used in phylogenetic analyses are provided in Figure 1 .
Table 1.
Viruses used in this study
| Virus | Strain | Species | Serogroup | Date of isolation | Place of isolation | Species of isolation | Other | Genbank |
|---|---|---|---|---|---|---|---|---|
| Crimean–Congo hemorrhagic fever (CCHFV) | IbAr10200 | Crimean–Congo hemorrhagic fever virus | CCHF | 11.05.1966 | Sokoto, Nigeria | Hyalomma excavatum (ticks) collected from a camel | Severe hemorrhagic fever in humans | NC_005300 |
| NC_005301 | ||||||||
| NC_005303 | ||||||||
| Thiafora (TFAV) | AnD 11411 | Thiafora virus | Thiafora | 17.2.1971 | Bandia, Senegal | Crocidura sp. (shrew) | KR537450 | |
| KR537451 | ||||||||
| KR537452 | ||||||||
| Erve (ERVV) | Brest/An221 | Thiafora virus | Thiafora | 5.5.1982 | Saulges, Mayenne, France | Crocidura russula (white-toed shrew) | Human infections—associated with headache | JF911697 JF911698 |
| JF911699 | ||||||||
| Keterah (KTRV) | P61361 | Not assigned | Keterah | 11.2.1966 | Keterah, Kelantan, Malaysia | Argae pusillus (tick) from Scotophilus kuhlii (temmincki) (lesser Asian yellow house bat) | KR537447 | |
| KR537448 | ||||||||
| KR537449 | ||||||||
| Issyk-Kul (IKV) | LEIV-315K | Not assigned | Keterah | 15.5.1970 | Dzety Oguzsk Region, Kyrgyzstan | Nyctalus noctula (common noctule bat) | Fever in humans | KR537441 |
| KR537442 | ||||||||
| KR537443 | ||||||||
| LEZ 86-787 | 1986 | Germany | Argus vespertlionis (ticks) | |||||
| Gossas (GOSV) | DakAnD 401 | Not assigned | Keterah | 19.11.1964 | Gossas, Senegal | Tadarida sp. (bat) | Formerly classified as a possible rhabdovirus | KR534876 |
| KR534877 | ||||||||
| KR534878 | ||||||||
| Yogue (YOGV) | DakAnD 56 | Not assigned | Yogue | 19.6.1968 | Bandia, Senegal | Rousettus aegyptiacus (Egyptian fruit bat) | KR537453 | |
| KR537454 | ||||||||
| KR537455 | ||||||||
| Kasokero (KKOV) | Z-52963 and Z-52969 | Not assigned | Yogue | XX.8.1977 | Masaka District, Uganda | Rousettus aegyptiacus (Egyptian fruit bat) | Human laboratory cases | KR537444 |
| KR537445 | ||||||||
| KR537446 | ||||||||
| Leopards Hill (LPHV) | 11SB17 | Not assigned | New | 29.11.2011 | Lusaka, Zambia | Hipposideros gigas (leaf-nosed bats) | Hemorrhagic gastroenteritis and severe hepatic disease in mice | NC_025831NC_025832 |
| NC_025833 |
Figure 1.
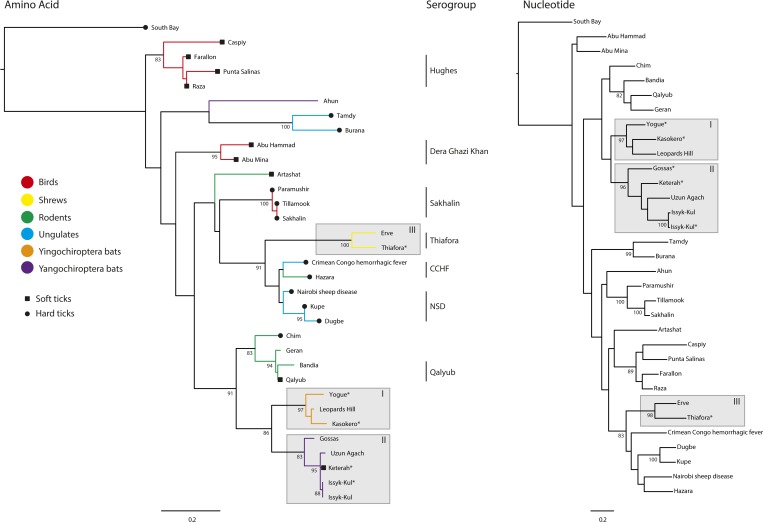
Phylogenetic trees of a partial sequence (466 nucleotides or 154 amino acids) of nairovirus L genes. The tree indicates the sources of isolation of the viruses from vertebrate hosts and hard or soft tick vectors. Current assignments to serogroups (species) are also shown. Genbank accession numbers: South Bay virus (KM048320), Caspiy virus (KF801658), Farallon virus (AY359523), Punte Salinas virus (AY359527), Raza virus (AY359529), Ahun virus (KF170224), Tamdy virus (KF801653), Burana virus (KF801651), Abu Hammad virus (AY357715), Abu Mina virus (AY357716), Artashat virus (KF801650), Paramushir virus (KF801657), Tillamook virus (AY359530), Sakhalin virus (KF801659), Erve virus (AY357719), Thiafora virus (KR537452), Crimean–Congo hemorrhagic fever virus (NC_005301), Hazara virus (AY359524), Nairobi sheep disease virus (KM464726), Kupe virus (EU257628), Dugbe virus (FJ422263), Chim virus (KF801656), Geran virus (KF801649), Bandia virus (AY357717), Qalyub virus (AY359528), Yogue virus (KR537455), Leopards Hill virus (AB842094), Kasokero virus (KR537446), Gossas virus (KR534878), Uzun Agach virus (KJ744032), Keterah virus (KR537449), and Issyk-Kul virus strain LEIV-315K (KF801652 and KR537443).
Growth and passage history.
The six nairoviruses sequenced in this study were obtained from the World Reference Center for Emerging Viruses and Arboviruses at the University of Texas Medical Branch, Galveston, TX and the Institut Pasteur, Paris, France. Each of the viruses was initially isolated by intracranial inoculation of newborn mice and most have been maintained by serial passage in suckling mice (SM).27 Strains and passage histories of viruses sequenced were as follows: GOSV Dak An D401 (SM, BHK-BSR2), IKV strain LEIV-315K (SM7), KKOV strain UgZ52969 (SM8), KTRV strain P6-1361 (SM12), TFAV strain Dak An D11411 (SM6, Vero2), and YOGV strain DakAn D5634 (SM3).
Preparation of samples for sequencing.
For TFAV, fluid medium from a culture of infected Vero cells was used for RNA extraction and sequencing. The source of viral RNA for the other five nairoviruses was filtered homogenates of infected mouse brain that were prepared as 10% suspensions in phosphate-buffered saline (PBS), pH 7.4.
Extraction of viral RNA.
GOSV and TFAV were grown in BHK-BSR and Vero cell monolayers, respectively. Supernatants were harvested and clarified by low-speed centrifugation (2,000 g, 10 minutes at 4°C) once CPE was advanced. For other viruses, RNA was extracted directly from filtered homogenates of infected newborn mouse brains prepared as 10% suspensions in PBS, pH 7.4. To remove remaining cellular debris, all samples (cultured and filtered brain homogenates) were filtered through a 0.45 μM filter (EMD Millipore, Billerica, MA). Clarified supernatants were treated with a cocktail of DNases (14 U Turbo DNase [Ambion, Austin, TX], 20 U Benzonase [EMD Millipore, Billerica, MA], and 20 U RNase One [Promega, Madison, WI]) for 1 hour at 37°C. To reduce volume, 24 mL of supernatant was loaded on top of 8 mL of 30% sucrose (in TEN, pH 7.4) and centrifuged at 150,000 g for 4 hours at 4°C. The pellet was resuspended in 250 μL RNase/DNase and protease-free water (Ambion, Austin, TX). Viral RNA was then extracted using Trizol and resuspended in 50 μL RNase/DNase and protease-free water (Ambion, Austin, TX).
Next-generation sequencing.
Viral RNA (0.1–0.2 μg), quantified by Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific, Pittsburgh, PA), was fragmented by incubation at 94°C for 8 minutes in 19.5 μL of fragmentation buffer (Illumina 15016648). First and second strand synthesis, adapter ligation, and amplification of the library were performed using the TruSeq RNA Sample Preparation kit (Illumina, San Diego, CA) under conditions prescribed by the manufacturer. Reads were assembled using ABySS (Michael Smith Genome Sciences Centre, Vancouver, Canada),31 mapped back to the contigs using bowtie2 (John Hopkins University, Baltimore, MD),32 and visually verified using the Integrated Genomics Viewer (Broad Institute, Boston, MA).33 A total of 1.9, 7.9, 4.1, 6.7, 6.0, and 8.1 million reads were generated for the samples containing GOSV, IKV, KTRV, KKOV, YOGV, and TFAV, respectively. Reads mapping to the virus in each sample comprised ~80,000 (4%), ∼740,000 (9.4%), 213,000 (5.2%), 1,600,000 (24%), 273,000 (4.5%), and 200,000 (2.5%), respectively.
Serological tests.
Antigens used in complement-fixation (CF) tests and for immunizing animals were prepared from infected newborn mouse brains. CF antigens were prepared by the sucrose/acetone extraction method.34 Hyperimmune mouse ascitic fluids were prepared by four intraperitoneal injections, given at weekly intervals, with 10% suspensions of homogenized infected mouse brain in PBS mixed with Freund's complete adjuvant (first injection) or Freund's incomplete adjuvant (subsequent injections). Sarcoma 180 cells were given intraperitoneally 1 day after the final immunization to induce ascites formation. Complement fixation tests were conducted using a microassay described previously35 using two units of guinea pig complement. Titers were recorded as the highest antibody dilutions giving 3+ or 4+ fixation of complement on a scale of 0–4+.
Phylogenetic analysis.
Other than the viruses described earlier, full genome sequences are available for only five of the ∼50 described nairoviruses (CCHFV, Hazara virus, NSDV, Dugbe virus, and Kupe virus). For most of the remaining viruses, only a short sequence fragment (< 450 nt) of a highly conserved region of the L segment is available; therefore, this region was selected for phylogenetic analysis. Amino acid and nucleotide sequence alignments were created from all available sequence data using ClustalW in Geneious v8.1.4 (www.geneious.com36) and refined manually. The resulting sequence alignments included 33 taxa and were 466 nucleotides or 154 amino acids in length. Maximum likelihood phylogenetic trees were constructed using PhyML 3.0,37 employing the HKY85 nucleotide substitution model with invariant sites or the Whelan and Goldman (WAG) model of amino acid substitution, and a combination of nearest neighbor interchange (NNI) and subtree pruning and regrafting branch swapping. The phylogenetic robustness of each node was determined using 1,000 bootstrap replicates and NNI branch swapping.
Results
Genome sequences of TFAV, YOGV, KKOV, GOSV, KTRV, and IKV (resequenced), including the complete coding regions of each of the three segments (L, M, and S), have been deposited in Genbank under accession numbers provided in Table 1. The complete genome sequence was determined for KKOV. For other viruses, only the extreme termini, which are highly conserved and anticomplementary among bunyavirus genome segments, were not determined. The genome sequences of LPHV, ERVV, and CCHFV were obtained from Genbank and used for comparative purposes (Table 1).
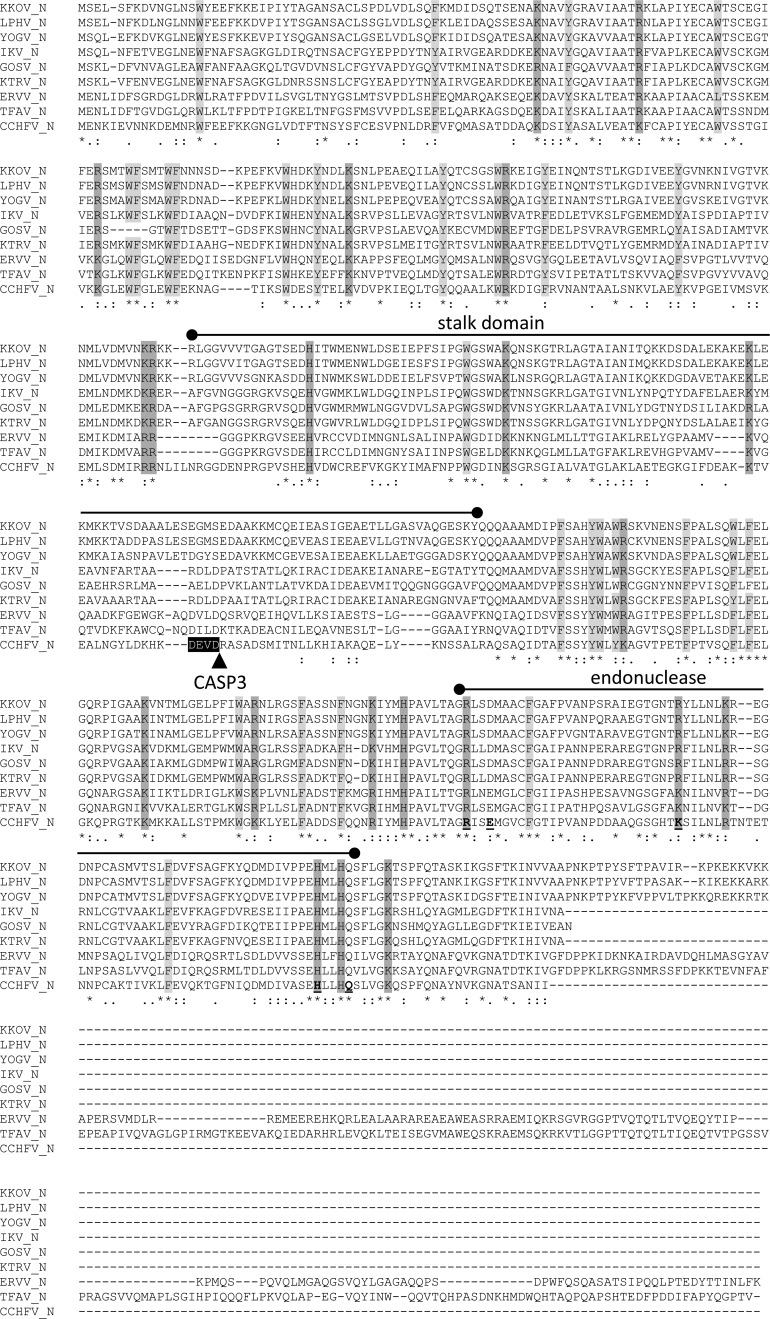
The S RNA segment in each of the viruses contains a single open reading frame encoding a protein with characteristics similar to those of the CCHFV N protein, for which the crystal structure has been resolved.18,19,38,39 The nucleoproteins range in size from 485 to 673 amino acids (estimated 54.6–74.7 kDa). There was good alignment of each protein with the CCHFV N protein across each of the N-terminal and C-terminal components of the head domain and the central stalk domain (Figure 2 ), with amino acid sequence identity ranging from 36.2% to 44.8% (Table 2). The highest amino acid sequence identities across this conserved domain were between KKOV, YOGV, and LPHV (79.3–91.5%), IKV, KTRV, and GOSV (63.3–85.9%), and TFAV and ERVV (70.6%). However, as described previously for ERVV, the TFAV N protein features a long (198 aa) C-terminal extension and, like LPHV, KKOV, and YOGV feature, a short (28 aa) C-terminal extension that is rich in proline and basic residues (Figure 2). The alignment also displayed striking conservation of an unusually high number of large hydrophobic residues (W, Y, F) and of basic residues (K, R, H), many of which have been implicated in RNA binding.19,38 There was also conservation of five of the six residues (R384, E387, K411, H453, and Q457) implicated in DNA binding at the CCHFV endonuclease active site, although conservative substitutions occurred in some of the viruses. Interestingly, a caspase-3 cleavage site identified at the tip of the CCHFV stalk domain18 was not conserved; none of the sequences in this region of the other nairoviruses contain a motif that is known to be recognized by caspase-3.40 In CCHFV, mutations that inhibit cleavage at this site have been shown to enhance viral polymerase activity and it has been proposed that the site is targeted by host cell defences.18,41 Although the site is conserved in all CCHFV strains,41 it is not evident why naturally occurring mutations would not be selected rapidly during infection, allowing the virus to evade the host response.
Figure 2.
ClustalX alignment of the deduced amino acid sequences of the nucleoproteins of nine nairoviruses. Conserved basic (K, R, H) and large hydrophobic (W, Y, F) residues are shaded. The stalk domain and endonuclease domains are marked and the caspase three cleavage site identified only in Crimean-Congo hemorrhagic fever virus (CCHFV) is shown. Identical (*), strongly conserved (:), and weakly conserved (.) residues as assigned in the Gronnet Pam250 matrix are indicated below the alignment.
Table 2.
Amino acid sequence identities (%) of nairovirus N proteins as determined by p-distance estimations in MEGA 6
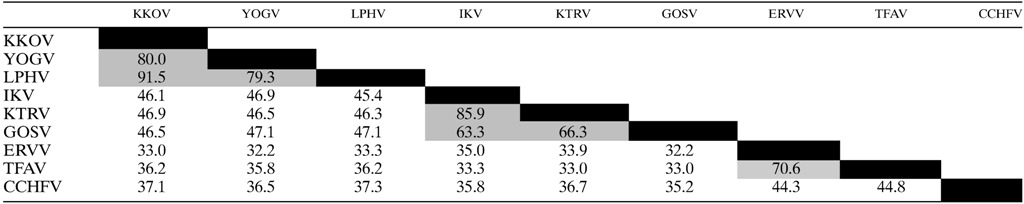
CCHFV = Crimean–Congo hemorrhagic fever virus; ERVV = Erve virus; GOSV = Gossas virus; IKV = Issyk-Kul virus; KKOV = Kasokero virus; KTRV = Keterah virus; LPHV = Leopards Hill virus; TFAV = Thiafora virus; YOGV = Yogue virus.
The aligned regions of each protein up to the end of the CCHFV N protein were compared.
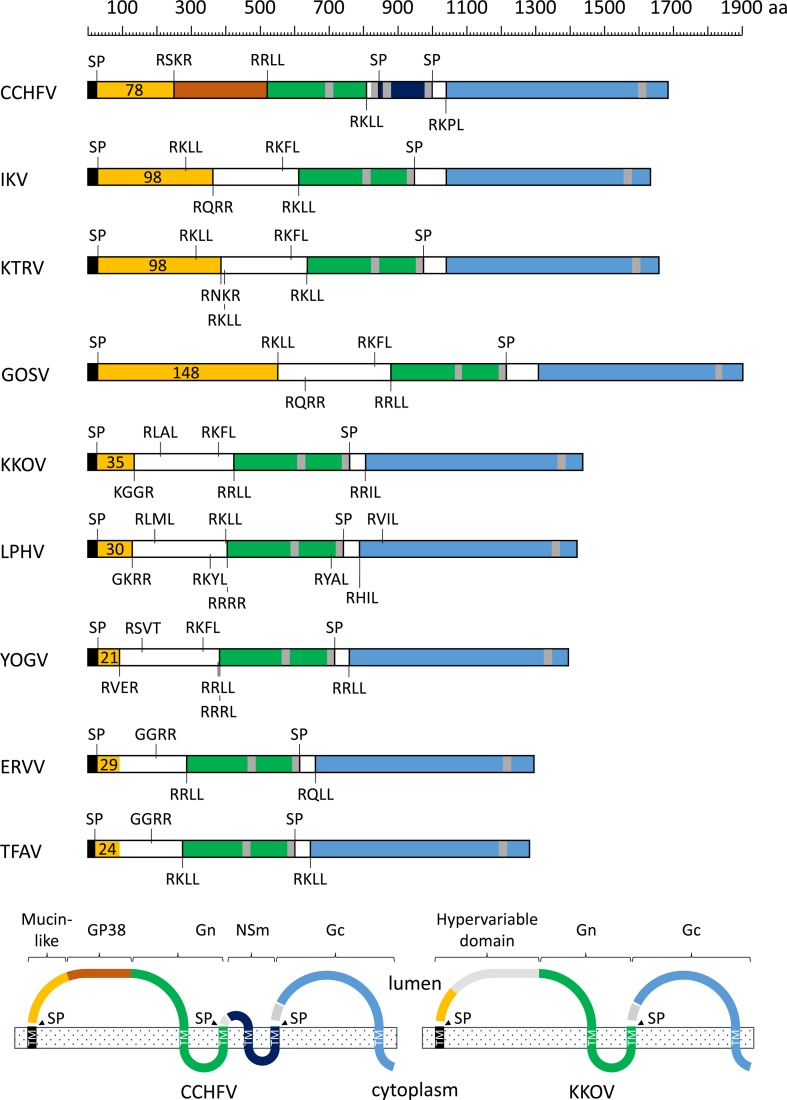
The M RNA segments were each found to encode large proteins with a predicted N-terminal signal peptide and multiple membrane-spanning domains, and displayed extensive but discontinuous homology with the M polyglycoprotein of CCHFV. As in CCHFV, the N-terminal region of each of the nairovirus polyproteins in our data set contains a mucin-like serine/threonine-rich domain with a large number of predicted O-linked and N-linked glycosylation sites that extend up to or beyond predicted (protease) cleavage sites (Figure 3 ). The number of predicted O-glycosylation sites ranged from 21 in YOGV to 148 in GOSV, with large variations in the length of the mucin-like domain contributing primarily to variations in size of the M polyproteins. The mucin-like domains displayed very low amino acid sequence identity, which was detectable only between closely related viruses in the data set. The hypervariable domain of the M polyproteins extends beyond the mucin-like domain into a region that corresponds to the gp38 domain of CCFHV, commencing at one or more furin-like or subtilisin kexin isozyme-1 (SKI-1) protease cleavage sites and terminating at a conserved SKI-1 cleavage site (R[R/K]LL↓) that marks the N-terminus of structural glycoprotein Gn (Figure 3). Although there is little overall sequence identity with CCHFV gp38 across the viruses in the data set, there is recognizable similarity in the C-terminal region of this domain and some conservation of a set of six to eight cysteine residues (Supplemental Figure 1). The Gn glycoproteins extend from the conserved SKI-1 cleavage site to signal peptide cleavage sites that follow the second transmembrane domain in each of the viruses. The Gn glycoproteins share 12 totally conserved cysteine residues and a conserved N-glycosylation site immediately following the second cysteine residue in the predicted ectodomain (Supplemental Figure 2). A second predicted N-glycosylation site immediately following the fifth cysteine residue in the ectodomain does not occur in ERVV, TFAV, or CCHFV. Unlike CCFHV, the Gc glycoproteins of each of the other nairoviruses in the data set commence at the predicted signal peptide cleavage site following the second transmembrane domain and terminate beyond the third transmembrane domain at the C-terminus of the M polyglycoprotein (Figure 3). The predicted Gc ectodomains of these class I transmembrane glycoproteins feature 28 conserved cysteine residues and two cysteine residues that were conserved in all viruses including CCHFV but are not present in ERVV and TFAV. Interestingly, CCFHV, TFAV, and ERVV also have a single (unpaired) cysteine residue that may form an intermolecular disulfide bridge (Supplemental Figure 3). The Gc glycoproteins each have two to three predicted N-glycosylation sites, none of which is totally conserved and multiple short regions (three to five amino acids) of highly conserved sequence. As in CCHFV, predicted subtilisin-like protease cleavage sites in the N-terminal region of each of the viruses in the data set may result in processing of Gc to remove a short polypeptide (45–53 amino acids) containing four conserved cysteine residues. As has been reported previously for LPHV and ERVV,30 the M polyglycoproteins of TFAV, YOGV, KKOV, GOSV, KTRV, and IKV lack the NSm protein of CCHFV due to the absence of an additional double-membrane-spanning region between Gn and Gc.
Figure 3.
Schematic illustration of the structures and predicted membrane topologies of the polyglycoproteins encoded in the M segments of nine nairoviruses. Regions corresponding to the mucin-like domain (orange), gp38 (rust), Gn (green), NSm (dark blue), and Gc (sky blue) are shaded. Predicted signal peptidase cleavage sites (SP) and potential furin-like and SK-1 cleavage sites are shown. The number of predicted O-linked glycosylation sites in the mucin-like domain are also shown.
The L RNA segment of each of the viruses encodes the large multifunctional RNA-dependent RNA polymerase (RdRp) that shares extensive sequence homology with the L proteins of other bunyaviruses. The virus sequences in the data set contained each of the conserved domains (regions 1–4) identified in other bunyaviruses,13 as well as the ovarian tumor (OTU)-like protease domain near the N-terminus (Supplemental Figure 4) that, although detected in various other RNA viruses, is unique to the nairovirus among the Bunyaviridae.11,42 In CCHFV, this OTU-like cysteine protease has been shown to inhibit NF-κB-dependent host innate immune pathways by de-conjugating ubiquitin and the interferon-stimulated gene 15 (ISG15) product from cellular target proteins.43 Like other nairoviruses, the L proteins of the viruses in our data set are significantly larger than those of other bunyaviruses due to an extended N-terminal region containing this domain, an extended C-terminal region and a long extension between conserved regions 2 and 3.8
Phylogenetic analysis of the nucleotide and amino acid sequences of a highly conserved region of the RdRp revealed that the viruses sequenced in this study cluster into three well-supported clades composed of 1) KKOV, YOGV, and LPHV; 2) KTRV, IKV, GOSV, and Uzun Agach virus (UAV); and 3) TFAV and ERVV (Figure 1). Amino acid sequence identities determined from Clustal X alignments of the N, Gn, Gc, and L protein sequences confirmed these relationships with amino acid sequence identities ≥ 59.6% among viruses assigned to each clade (excluding UAV for which only 441 bp of the L gene have been sequenced). In contrast, amino acid sequence identities between viruses representing different clades range from 25.7% to 56.7% (Tables 2–5). Complement-fixation tests conducted using the available antigens and mouse immune ascitic fluids from KKOV, YOGV, KTRV, IKV, and TFAV also supported the assignment of the viruses to these groups (Table 6). As reported previously,24 KKOV is closely related antigenically to YOGV and together these viruses can be considered to form the Kasokero serogroup of which LPHV is also a likely member. Similarly, IKV and KTRV can be considered to form the Issyk-Kul serogroup. The two available strains of IKV (LEIV-315K/Kyrgyzstan/1970 and LEZ 86-1784/Germany/1986) were indistinguishable in the CF test. TFAV was distinct from the other viruses tested here but has previously been shown to be closely related antigenically to ERVV with which it forms the Thiafora serogroup.23
Table 5.
Amino acid sequence identities (%) of nairovirus L proteins as determined by p-distance estimations in MEGA 6
CCHFV = Crimean–Congo hemorrhagic fever virus; ERVV = Erve virus; GOSV = Gossas virus; IKV = Issyk-Kul virus; KKOV = Kasokero virus; KTRV = Keterah virus; LPHV = Leopards Hill virus; TFAV = Thiafora virus; YOGV = Yogue virus.
Table 6.
Complement-fixation tests to detect antigenic relationships between five nairoviruses
IKV = Issyk-Kul virus; KKOV = Kasokero virus; KTRV = Keterah virus; LEIV = IKV strain LEIV; LEZ = IKV strain LEZ; TFAV = Thiafora virus; YOGV = Yogue virus.
Reciprocal of antiserum titer/reciprocal of antigen titer.
Examination of the host associations of each clade revealed some structure by both host and vector across the tree (Figure 1). Clade I viruses were each isolated from bats of the suborder Yingochiroptera (Pteropodidae and Hipposideridae) in Africa; clade II viruses were each isolated from bats of the suborder Yangochiroptera (Vespertilionidae and Molossidae) in Asia and Africa; and clade III viruses were each isolated from shrews (Soricidae) in Africa and Europe.
Discussion
The risks to public health associated with new and emerging viral diseases are well recognized. Most newly emerging viruses are zoonotic and many are transmitted by arthropod vectors.44 The identification and characterization of the myriad of viruses infecting animal reservoirs and vectors will help mitigate disease risks by increasing awareness of potential threats and improve preparedness for future emergence events. New and poorly characterized viruses that may be of particular interest are those that are closely related to and share a similar ecology with known emerging zoonotic pathogens. Here, we report the molecular characterization and phylogenetic assignment of six bunyaviruses (TFAV, YOGV, KKOV, GOSV, KTRV, and IKV) that exist, together with CCHFV, in the genus Nairovirus.
The most striking feature of the genome organization of each of these nairoviruses is the structural arrangement of the M segment-encoded polyglycoprotein. Each contains domains corresponding to the CCHFV mucin-like protein, gp38, Gn, and Gc. However, as reported recently for LPHV and ERVV,30 a double-membrane spanning domain between Gn and Gc, corresponding to the CCHFV NSm protein, was absent from TFAV, YOGV, KKOV, GOSV, KTRV, and IKV. The function of NSm is presently unknown but homologues also occur in the M polyglycoprotein sequences of NSDV, Dugbe virus, Kupe virus, and Hazara virus that cluster phylogenetically with CCHFV. Interestingly, although HAZV is the most closely related virus to CCHFV, both phylogenetically and serologically, it is nonpathogenic in humans and uniquely features a 43 amino acid deletion in the cytoplasmic domain NSm.45 Triple-membrane-spanning NSm proteins are encoded in the M segments of orthobunyaviruses and have been implicated as scaffold proteins involved in virus assembly and morphogenesis.46–48 Although also located between the Gn and Gc glycoproteins, they share no detectable sequence identity with nairovirus double-membrane-spanning NSm proteins.
Amino acid identity comparisons and phylogenetic analyses indicated that the viruses sequenced in the study fell into three groups comprising 1) KKOV, YOGV, and LPHV; 2) KTRV, IKV, and GOSV; and 3) TFAV and ERVV. Phylogenetic analyses also indicated that UAV, which was isolated in 1977 from a lesser mouse-eared bat (M. blythii) in Kazakhstan, falls within the KTRV group. Because of the paucity of biochemical data, nairovirus species are currently assigned according to serogroups.9,10,23 Although serological analysis conducted here was limited by the unavailability of suitable antisera, consideration of cross-reactions in complement-fixation assays and phylogenetic analyses suggests that the groups comprising KKOV, YOGV, and LPHV may be assigned as a single species and that KTRV, IKV, GOSV, and UAV may also constitute a single species. TFAV and ERVV have previously been assigned as a single species based on complement fixation and indirect immunofluorescence data.23 Cross-neutralization tests would be required to confirm these species assignments by current demarcation criteria.
We also observed that there may be a clade-specific association between nairoviruses and specific host taxa. GOSV, IKV, KTRV, and UAV were each isolated from bats of the suborder Yangochiroptera (or from ticks feeding on them), KKOV, YOGV, and LPHV were isolated from bats of the suborder Yingochiroptera,49,50 and TFAV and ERVV were isolated from insectivores (shrews). This correlation extended across the nairovirus phylogeny (Figure 1) with various clades associated with either birds, bats, rodents, insectivores or ungulates. The available data also suggest that specific clades are associated with transmission by either hard ticks (Ixodidae) or soft ticks (Argasidae). For example, the viruses in the large clade comprising the CCHFV, NSDV, and Sakhalin serogroups have been isolated primarily from hard ticks.51–56 Although there are two exceptional reports of the isolation of CCHFV from soft tick species, the circumstances suggest those reports should be viewed with caution52,57 and there is experimental evidence that soft tick species are not competent vectors for CCHFV.58,59 Tamdy virus and closely related Burana virus were also isolated from hard ticks.60 However, with the sole exception of Chim virus,61 viruses in the remaining clades have been isolated only from soft ticks.20,62–66 As observed previously,67 this may indicate a stable ecological association between nairoviruses and their hosts and vectors and suggests that constraints on transmission to vertebrates, including humans, may be limited primarily by vector-feeding preferences and the opportunity for exposure.
Several nairoviruses, including CCHFV, have been implicated in human disease. IKV has caused sporadic cases and epidemics of febrile illness in Central Asia,68 ERVV has been associated with headache and other neurological symptoms in Europe,69 and KKOV has been isolated from laboratory workers who developed a mild-to-severe clinical disease characterized by fever, nausea, myalgia, abdominal pain, and neurological symptoms.24 Further investigations are required to determine the risks to public health from other viruses examined in this study.
Supplementary Material
Table 3.
Amino acid sequence identities (%) of nairovirus Gn proteins as determined by p-distance estimations in MEGA 6
CCHFV = Crimean–Congo hemorrhagic fever virus; ERVV = Erve virus; GOSV = Gossas virus; IKV = Issyk-Kul virus; KKOV = Kasokero virus; KTRV = Keterah virus; LPHV = Leopards Hill virus; TFAV = Thiafora virus; YOGV = Yogue virus.
The sequences were assumed to extend from the conserved protease site (R[R/K]LL) to the signal peptidase cleavage site following the second-last transmembrane domain in the M polyprotein.
Table 4.
Amino acid sequence identities (%) of nairovirus Gc proteins as determined by p-distance estimations in MEGA 6
CCHFV = Crimean–Congo hemorrhagic fever virus; ERVV = Erve virus; GOSV = Gossas virus; IKV = Issyk-Kul virus; KKOV = Kasokero virus; KTRV = Keterah virus; LPHV = Leopards Hill virus; TFAV = Thiafora virus; YOGV = Yogue virus.
The sequences were assumed to extend from the signal peptidase cleavage site following the second-last transmembrane domain to the C terminus of the M polyprotein.
ACKNOWLEDGMENTS
We thank Philippe Despres, Institute Pasteur, Paris, for providing GOSV and Herve Zeller, European Centre for Disease Prevention and Control (ECDC), Stockholm, Sweden, for information on its history.
Footnotes
Financial support: This work was supported in part by NIH contract HHSN272201000040I/HHSN2700004/D4 (Nikos Vasilakis and Robert B. Tesh).
Disclosure: Animal use in this work was done under protocol no. 9505045, approved by the IACUC at the University of Texas Medical Branch.
Authors' addresses: Peter J. Walker, Cadhla Firth, and Kim R. Blasdell, CSIRO Biosecurity, Australian Animal Health Laboratory, Geelong, Victoria, Australia, E-mails: peter.walker@csiro.au, cadhla.firth@csiro.au, and kim.blasdell@csiro.au. Steven G. Widen, Thomas G. Wood, and Robert B. Tesh, Department of Biochemistry and Molecular Biology, University of Texas Medical Branch, Galveston, TX, E-mails: sgwiden@utmb.edu, tgwood@utmb.edu, and rtesh@utmb.edu. Amelia P. A. Travassos da Rosa and Hilda Guzman, Department of Pathology, University of Texas Medical Branch, Galveston, TX, E-mails: aptravas@utmb.edu and hguzman@utmb.edu. Nikos Vasilakis, Department of Pathology and Center for Biodefense and Emerging Infectious Diseases, University of Texas Medical Branch, Galveston, TX, E-mail: nivasila@utmb.edu.
References
- 1.Ergonul O. Crimean-Congo hemorrhagic fever virus: new outbreaks, new discoveries. Curr Opin Virol. 2012;2:215–220. doi: 10.1016/j.coviro.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Ergonul O. Crimean-Congo haemorrhagic fever. Lancet Infect Dis. 2006;6:203–214. doi: 10.1016/S1473-3099(06)70435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bente DA, Forrester NL, Watts DM, McAuley AJ, Whitehouse CA, Bray M. Crimean-Congo hemorrhagic fever: history, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antiviral Res. 2013;100:159–189. doi: 10.1016/j.antiviral.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Davies FG. Nairobi sheep disease. Parassitologia. 1997;39:95–98. [PubMed] [Google Scholar]
- 5.Terpstra C. Nairobi sheep disease. In: Coetzer JAW, Thompson GR, Tustin RC, editors. Infectious Diseases of Livestock with Special Reference to Southern Africa. Cape Town, South Africa: Oxford University Press; 1994. pp. 718–722. [Google Scholar]
- 6.Marczinke BI, Nichol ST. Nairobi sheep disease virus, an important tick-borne pathogen of sheep and goats in Africa, is also present in Asia. Virology. 2002;303:146–151. doi: 10.1006/viro.2002.1514. [DOI] [PubMed] [Google Scholar]
- 7.Gong S, He B, Wang Z, Shang L, Wei F, Liu Q, Tu C. Nairobi sheep disease virus RNA in Ixodid ticks, China, 2013. Emerg Infect Dis. 2015;21:718–720. doi: 10.3201/eid2104.141602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lasecka L, Baron MD. The molecular biology of nairoviruses, an emerging group of tick-borne arboviruses. Arch Virol. 2014;159:1249–1265. doi: 10.1007/s00705-013-1940-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casals J, Tignor GH. The Nairovirus genus: serological relationships. Intervirology. 1980;14:144–147. doi: 10.1159/000149175. [DOI] [PubMed] [Google Scholar]
- 10.Plyusnin A, Beaty BJ, Elliott RM, Goldbach R, Kormelink R, Lundkvist A, Schmaljohn CS, Tesh RB. Family Bunyaviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy. Ninth Report of the International Committee on Taxonomy of Viruses. Amsterdam, The Netherlands: Elsevier Academic Press; 2012. pp. 725–741. [Google Scholar]
- 11.Honig JE, Osborne JC, Nichol ST. Crimean-Congo hemorrhagic fever virus genome L RNA segment and encoded protein. Virology. 2004;321:29–35. doi: 10.1016/j.virol.2003.09.042. [DOI] [PubMed] [Google Scholar]
- 12.Marriott AC, Nuttall PA. Large RNA segment of Dugbe nairovirus encodes the putative RNA polymerase. J Gen Virol. 1996;77:1775–1780. doi: 10.1099/0022-1317-77-8-1775. [DOI] [PubMed] [Google Scholar]
- 13.Kinsella E, Martin SG, Grolla A, Czub M, Feldmann H, Flick R. Sequence determination of the Crimean-Congo hemorrhagic fever virus L segment. Virology. 2004;321:23–28. doi: 10.1016/j.virol.2003.09.046. [DOI] [PubMed] [Google Scholar]
- 14.Vincent MJ, Sanchez AJ, Erickson BR, Basak A, Chretien M, Seidah NG, Nichol ST. Crimean-Congo hemorrhagic fever virus glycoprotein proteolytic processing by subtilase SKI-1. J Virol. 2003;77:8640–8649. doi: 10.1128/JVI.77.16.8640-8649.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez AJ, Vincent MJ, Nichol ST. Characterization of the glycoproteins of Crimean-Congo hemorrhagic fever virus. J Virol. 2002;76:7263–7275. doi: 10.1128/JVI.76.14.7263-7275.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez AJ, Vincent MJ, Erickson BR, Nichol ST. Crimean-Congo hemorrhagic fever virus glycoprotein precursor is cleaved by furin-like and SKI-1 proteases to generate a novel 38-kilodalton glycoprotein. J Virol. 2006;80:514–525. doi: 10.1128/JVI.80.1.514-525.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altamura LA, Bertolotti-Ciarlet A, Teigler J, Paragas J, Schmaljohn CS, Doms RW. Identification of a novel C-terminal cleavage of Crimean-Congo hemorrhagic fever virus preGn that leads to generation of an NSm protein. J Virol. 2007;81:6632–6642. doi: 10.1128/JVI.02730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Dutta S, Karlberg H, Devignot S, Weber F, Hao Q, Tan YJ, Mirazimi A, Kotaka M. Structure of Crimean-Congo hemorrhagic fever virus nucleoprotein: superhelical homo-oligomers and the role of caspase-3 cleavage. J Virol. 2012;86:12294–12303. doi: 10.1128/JVI.01627-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo Y, Wang W, Ji W, Deng M, Sun Y, Zhou H, Yang C, Deng F, Wang H, Hu Z, Lou Z, Rao Z. Crimean-Congo hemorrhagic fever virus nucleoprotein reveals endonuclease activity in Bunyaviruses. Proc Natl Acad Sci USA. 2012;109:5046–5051. doi: 10.1073/pnas.1200808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karabatsos N. International Catalogue of Arboviruses Including Certain other Viruses of Vertebrates. San Antonio, TX: American Society for Tropical Medicine and Hygiene; 1985. [DOI] [PubMed] [Google Scholar]
- 21.Dilcher M, Koch A, Hasib L, Dobler G, Hufert FT, Weidmann M. Genetic characterization of Erve virus, a European nairovirus distantly related to Crimean-Congo hemorrhagic fever virus. Virus Genes. 2012;45:426–432. doi: 10.1007/s11262-012-0796-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chastel C, Main AJ, Richard P, Le Lay G, Legrand-Quillien MC, Beaucournu JC. Erve virus, a probable member of Bunyaviridae family isolated from shrews (Crocidura russula) in France. Acta Virol. 1989;33:270–280. [PubMed] [Google Scholar]
- 23.Zeller HG, Karabatsos N, Calisher CH, Digoutte JP, Cropp CB, Murphy FA, Shope RE. Electron microscopic and antigenic studies of uncharacterized viruses. II. Evidence suggesting the placement of viruses in the family Bunyaviridae. Arch Virol. 1989;108:211–227. doi: 10.1007/BF01310935. [DOI] [PubMed] [Google Scholar]
- 24.Kalunda M, Mukwaya LG, Mukuye A, Lule M, Sekyalo E, Wright J, Casals J. Kasokero virus: a new human pathogen from bats (Rousettus aegyptiacus) in Uganda. Am J Trop Med Hyg. 1986;35:387–392. doi: 10.4269/ajtmh.1986.35.387. [DOI] [PubMed] [Google Scholar]
- 25.Dietzgen RG, Calisher CH, Kurath G, Kuzman IV, Rodriguez LL, Stone DM, Tesh RB, Tordo N, Walker PJ, Wetzel T, Whitfield AE. In: Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses. King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. San Diego, CA: Elsevier; 2012. pp. 654–681. (Rhabdoviridae). [Google Scholar]
- 26.Blasdell KR, Guzman H, Widen SG, Firth C, Wood TG, Holmes EC, Tesh RB, Vasilakis N, Walker PJ. Ledantevirus: a proposed new genus in the Rhabdoviridae has a strong ecological association with bats. Am J Trop Med Hyg. 2015;92:405–410. doi: 10.4269/ajtmh.14-0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varma MG, Converse JD. Keterah virus infections in four species of Argas ticks (Ixodoidea: Argasidae) J Med Entomol. 1976;13:65–70. doi: 10.1093/jmedent/13.1.65. [DOI] [PubMed] [Google Scholar]
- 28.Lvov DK, Karas FR, Timofeev EM, Tsyrkin YM, Vargina SG, Veselovskaya OV, Osipova NZ, Grebenyuk YI, Gromashesvki VL, Steblyanko SN, Fomina KB. “Issyk-Kul” virus, a new arbovirus isolated from bats and Argas (Carios) vespertilionis (Latr., 1802) in the Kirghiz S.S.R. Arch Gesamte Virusforsch. 1973;42:207–209. doi: 10.1007/BF01270841. [DOI] [PubMed] [Google Scholar]
- 29.Lvov DK, Kostiukov MA, Daniiarov OA, Tukhtaev TM, Sherikov BK. Outbreak of arbovirus infection in the Tadzhik SSR due to the Issyk-kul virus (Issyk-kul fever) Vopr Virusol. 1984;29:89–92. [PubMed] [Google Scholar]
- 30.Ishii A, Ueno K, Orba Y, Sasaki M, Moonga L, Hang'ombe BM, Mweene AS, Umemura T, Ito K, Hall WW, Sawa H. A nairovirus isolated from African bats causes haemorrhagic gastroenteritis and severe hepatic disease in mice. Nat Commun. 2014;5:e5651. doi: 10.1038/ncomms6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clarke DH, Casals J. Techniques for hemagglutination and hemagglutination-inhibition with arthropod-borne viruses. Am J Trop Med Hyg. 1958;7:561–573. doi: 10.4269/ajtmh.1958.7.561. [DOI] [PubMed] [Google Scholar]
- 35.Tesh RB, Travassos da Rosa APA, Travassos da Rosa JS. Antigenic relationship among rhabdoviruses infecting terrestrial vertebrates. J Gen Virol. 1983;64:169–176. doi: 10.1099/0022-1317-64-1-169. [DOI] [PubMed] [Google Scholar]
- 36.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 38.Carter SD, Surtees R, Walter CT, Ariza A, Bergeron E, Nichol ST, Hiscox JA, Edwards TA, Barr JN. Structure, function, and evolution of the Crimean-Congo hemorrhagic fever virus nucleocapsid protein. J Virol. 2012;86:10914–10923. doi: 10.1128/JVI.01555-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carter SD, Barr JN, Edwards TA. Expression, purification and crystallization of the Crimean-Congo haemorrhagic fever virus nucleocapsid protein. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012;68:569–573. doi: 10.1107/S1744309112009736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Talanian RV, Quinlan C, Trautz S, Hackett MC, Mankovich JA, Banach D, Ghayur T, Brady KD, Wong WW. Substrate specificities of caspase family proteases. J Biol Chem. 1997;272:9677–9682. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- 41.Karlberg H, Tan YJ, Mirazimi A. Induction of caspase activation and cleavage of the viral nucleocapsid protein in different cell types during Crimean-Congo hemorrhagic fever virus infection. J Biol Chem. 2011;286:3227–3234. doi: 10.1074/jbc.M110.149369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Capodagli GC, McKercher MA, Baker EA, Masters EM, Brunzelle JS, Pegan SD. Structural analysis of a viral ovarian tumor domain protease from the Crimean-Congo hemorrhagic fever virus in complex with covalently bonded ubiquitin. J Virol. 2011;85:3621–3630. doi: 10.1128/JVI.02496-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frias-Staheli N, Giannakopoulos NV, Kikkert M, Taylor SL, Bridgen A, Paragas J, Richt JA, Rowland RR, Schmaljohn CS, Lenschow DJ, Snijder EJ, Garcia-Sastre A, Virgin HW., 4th Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe. 2007;2:404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dowall SD, Findlay-Wilson S, Rayner E, Pearson G, Pickersgill J, Rule A, Merredew N, Smith H, Chamberlain J, Hewson R. Hazara virus infection is lethal for adult type I interferon receptor-knockout mice and may act as a surrogate for infection with the human-pathogenic Crimean-Congo hemorrhagic fever virus. J Gen Virol. 2012;93:560–564. doi: 10.1099/vir.0.038455-0. [DOI] [PubMed] [Google Scholar]
- 46.Shi X, Kohl A, Leonard VH, Li P, McLees A, Elliott RM. Requirement of the N-terminal region of orthobunyavirus nonstructural protein NSm for virus assembly and morphogenesis. J Virol. 2006;80:8089–8099. doi: 10.1128/JVI.00579-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Briese T, Rambaut A, Lipkin WI. Analysis of the medium (M) segment sequence of Guaroa virus and its comparison to other orthobunyaviruses. J Gen Virol. 2004;85:3071–3077. doi: 10.1099/vir.0.80122-0. [DOI] [PubMed] [Google Scholar]
- 48.Lees JF, Pringle CR, Elliott RM. Nucleotide sequence of the Bunyamwera virus M RNA segment: conservation of structural features in the Bunyavirus glycoprotein gene product. Virology. 1986;148:1–14. doi: 10.1016/0042-6822(86)90398-3. [DOI] [PubMed] [Google Scholar]
- 49.Springer MS, Teeling EC, Madsen O, Stanhope MJ, de Jong WW. Integrated fossil and molecular data reconstruct bat echolocation. Proc Natl Acad Sci USA. 2001;98:6241–6246. doi: 10.1073/pnas.111551998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teeling EC, Springer MS, Madsen O, Bates P, O'Brien SJ, Murphy WJ. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- 51.Crabtree MB, Sang R, Miller BR. Kupe virus, a new virus in the family Bunyaviridae, genus Nairovirus, Kenya. Emerg Infect Dis. 2009;15:147–154. doi: 10.3201/eid1502.080851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Horne KM, Vanlandingham DL. Bunyavirus-vector interactions. Viruses. 2014;6:4373–4397. doi: 10.3390/v6114373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Begum F, Wisseman CL, Jr, Casals J. Tick-borne viruses of West Pakistan. II. Hazara virus, a new agent isolated from Ixodes redikorzevi ticks from the Kaghan Valley, W. Pakistan. Am J Epidemiol. 1970;92:192–194. doi: 10.1093/oxfordjournals.aje.a121197. [DOI] [PubMed] [Google Scholar]
- 54.David-West TS. Dugbe virus: a new tick-borne arbovirus from Nigeria. Trans R Soc Trop Med Hyg. 1973;67:438. doi: 10.1016/0035-9203(73)90062-x. [DOI] [PubMed] [Google Scholar]
- 55.Lvov DK, Timofeeva AA, Gromashevski VL, Chervonsky VI, Gromov AI, Tsyrkin YM, Pogrebenko AG, Kostyrko IN. Sakhalin virus—a new arbovirus isolated from Ixodes (Ceratixodes) putus Pick.-Camb. 1878 collected on Tuleniy Island, Sea of Okhotsk. Arch Gesamte Virusforsch. 1972;38:133–138. doi: 10.1007/BF01249662. [DOI] [PubMed] [Google Scholar]
- 56.Lvov DK, Al'khovskii SV, Shchelkanov M, Shchetinin AM, Deriabin PG, Aristova VA, Gitel'man AK, Samokhvalov EI, Botikov AG. Genetic characterization of the Sakhalin virus (SAKV), Paramushir virus (PMRV) (Sakhalin group, Nairovirus, Bunyaviridae), and Rukutama virus (RUKV) (Uukuniemi group, Phlebovirus, Bunyaviridae) isolated from the obligate parasites of the colonial sea-birds ticks Ixodes (Ceratixodes) uriae, White 1852 and I. signatus Birulya, 1895 in the water area of sea of the Okhotsk and Bering sea. Vopr Virusol. 2014;59:11–17. [PubMed] [Google Scholar]
- 57.Manzano-Román R, Díaz-Martín V, de la Fuente J, Pérez-Sánchez R. Soft ticks as pathogen vectors: distribution, surveillance and control. In: Shah MM, editor. Parasitology. Rijeka, Croatia: Intech; 2012. pp. 125–162. [Google Scholar]
- 58.Shepherd AJ, Swanepoel R, Cornel AJ, Mathee O. Experimental studies on the replication and transmission of Crimean-Congo hemorrhagic fever virus in some African tick species. Am J Trop Med Hyg. 1989;40:326–331. doi: 10.4269/ajtmh.1989.40.326. [DOI] [PubMed] [Google Scholar]
- 59.Durden LA, Logan TM, Wilson ML, Linthicum KJ. Experimental vector incompetence of a soft tick, Ornithodoros sonrai (Acari: Argasidae), for Crimean-Congo hemorrhagic fever virus. J Med Entomol. 1993;30:493–496. doi: 10.1093/jmedent/30.2.493. [DOI] [PubMed] [Google Scholar]
- 60.Lvov DK, Sidorova GA, Gromashevsky VL, Kurbanov M, Skvoztsova LM, Gofman YP, Berezina LK, Klimenko SM, Zakharyan VA, Aristova VA, Neronov VM. Virus “Tamdy”—a new arbovirus, isolated in the Uzbee S.S.R. and Turkmen S.S.R. from ticks Hyalomma asiaticum asiaticum Schulee et Schlottke, 1929, and Hyalomma plumbeum plumbeum Panzer, 1796. Arch Virol. 1976;51:15–21. doi: 10.1007/BF01317830. [DOI] [PubMed] [Google Scholar]
- 61.Lvov DK, Sidorova GA, Gromashevskii VL, Skvortsova TM, Berezina LK. Chim virus, a new arbovirus isolated from ixodid and argasid ticks collected in the burrows of great gerbils on the territory of the Uzbek SSR. Vopr Virusol. 1979;24:286–289. [PubMed] [Google Scholar]
- 62.Rodovsky FJ, Stiller D. Descriptive notes on Ornithodoros ticks from gull nests on the Farallon islands and isolation of a variant of Hughes virus. J Parasitol. 1967;53:890–892. [PubMed] [Google Scholar]
- 63.Converse JD, Hoogstraal H, Moussa MI, Evans DE. Soldado virus from Ornithodoros (Alectorobius) maritimus (Ixodoidea: Argasidae) infesting herring gull nests on Puffin Island, northern Wales. Acta Virol. 1976;20:243–246. [PubMed] [Google Scholar]
- 64.Keirans JE, Yunker CE, Clifford CM, Thomas LA, Walton GA, Kelly TC. Isolation of a Soldado-like virus (Hughes group) from Ornithodorus maritimus ticks in Ireland. Experientia. 1976;32:453–454. doi: 10.1007/BF01920791. [DOI] [PubMed] [Google Scholar]
- 65.Converse JD, Moussa MI, Easton ER, Casals J. Punta Salinas virus (Hughes group) from Argas arboreus (Ixodoidea: Argasidae) in Tanzania. Trans R Soc Trop Med Hyg. 1981;75:755–756. doi: 10.1016/0035-9203(81)90175-9. [DOI] [PubMed] [Google Scholar]
- 66.Clerx JP, Bishop DH. Qalyub virus, a member of the newly proposed Nairovirus genus (Bunyavividae) Virology. 1981;108:361–372. doi: 10.1016/0042-6822(81)90444-x. [DOI] [PubMed] [Google Scholar]
- 67.Honig JE, Osborne JC, Nichol ST. The high genetic variation of viruses of the genus Nairovirus reflects the diversity of their predominant tick hosts. Virology. 2004;318:10–16. doi: 10.1016/j.virol.2003.09.021. [DOI] [PubMed] [Google Scholar]
- 68.Alkhovskii SV, Lvov DK, Shchelkanov M, Shchetinin AM, Deriabin PG, Samokhvalov EI, Gitel'man AK, Botikov AG. The taxonomy of the Issyk-Kul virus (ISKV, Bunyaviridae, Nairovirus), the etiologic agent of the Issyk-Kul fever isolated from bats (Vespertilionidae) and ticks Argas (Carios) vespertilionis (Latreille, 1796) Vopr Virusol. 2013;58:11–15. [PubMed] [Google Scholar]
- 69.Treib J, Dobler G, Haass A, von Blohn W, Strittmatter M, Pindur G, Froesner G, Schimrigk K. Thunderclap headache caused by Erve virus? Neurology. 1998;50:509–511. doi: 10.1212/wnl.50.2.509. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.