Abstract
Both yellow and white corn occurs among ancestral open pollinated varieties. More recently, breeders have selected yellow endosperm variants of maize over ancestral white phenotypes for their increased nutritional value resulting from the up-regulation of the Y1 phytoene synthase gene product in endosperm tissue. As a result, diversity within yellow maize lines at the Y1 gene is dramatically decreased as compared to white corn. We analyzed patterns of sequence diversity and linkage disequilibrium in nine low copy regions located at varying distances from the Y1 gene, including a homolog of the barley Mlo gene. Patterns consistent with a selective sweep, such as significant associations of informative single-nucleotide polymorphisms with endosperm color phenotype, linkage disequilibrium, and significantly reduced diversity within the yellow endosperm haplotypes, were observed up to 600 kb downstream of Y1, whereas the upstream region showed a more rapid recovery. The starch branching enzyme 1 (sbe1) gene is the first region downstream of Y1 that does not have a highly conserved haplotype in the yellow endosperm germplasm.
Selective forces acting on allelic variants of genes have a profound effect on local levels of genetic diversity and linkage disequilibria (LD). Positive directional selection leads to reduced variability and increased LD in the respective region (1-6), and the so-called selective sweep regions provide clues to genes that have been subjects of evolutionary forces as well as selection by humans. Recently, Clark et al. (7) characterized a selective sweep in the promoter region of teosinte branched1 (tb1). The sweep extends 60-90 kb upstream of the gene and is indicative of the gene's role in the domestication of maize from teosinte between 6,000 and 10,000 years ago.
The maize Y1 gene on chromosome 6 has undergone recent selection for endosperm color phenotype. A recent study (8) uncovered a dramatic reduction in diversity at this gene for the yellow endosperm maize inbred lines only, over the entire 6-kb gene region. This footprint of selection was characterized by a conserved yellow endosperm haplotype at the 5′ end of the gene with evidence of recombination toward the 3′ end. Strong haplotype conservation at the 5′ end is suggestive of the location of the causal variant associated with the gain-of-function mutation to yellow endosperm and may be indicative of further extension of the selective sweep in the 5′ direction. The presence of recombinants in the 3′ UTR region, however, suggested that the extent of the selective sweep may be limited downstream of the coding region.
The white endosperm lines did not show characteristics of a selective sweep (8), despite the fact that the white endosperm phenotype is also a target of selection due to human taste preference (9). This is because white is the predicted ancestral state of the gene (John Doebley, personal communication), and thus multiple haplotypes are associated with the white endosperm phenotype.
Understanding the boundaries of the selective sweep, defined by the relative levels of diversity, the extent of LD, and the distance at which significant associations with the endosperm phenotype can still be identified, will aid in the understanding of the effects of selection at the molecular and population levels. It will also suggest appropriate approaches for the identification of regions subject to past selection. To this end, we analyzed patterns of diversity at low copy or genic regions in the ≈1.2-megabase (Mb) area surrounding Y1.
Materials and Methods
Plant Material. The germplasm set, consisting of 75 maize inbred lines, was described previously (8) (Table 3, which is published as supporting information on the PNAS web site). Seeds from the intermated B73 × Mo17 mapping population (10) were obtained for genetic mapping purposes. Leaves of 2-week old plants, grown in the greenhouse, were harvested and freeze-dried.
DNA Extractions. Leaf material was either ground in liquid nitrogen by using a mortar and pestle or pulverized through the use of steel balls and a paint shaker. DNA was extracted from the test set by using the DNeasy Maxi-Prep extraction kit (Qiagen, Valencia, CA), following the manufacturer's protocol. For the intermated B73 × Mo17 mapping population, a modified version of the Qiagen protocol (M. Mucha, personal communication) was used; the modifications were intended to make the protocol more rapid and high-throughput. DNA concentrations were determined spectrophotometrically.
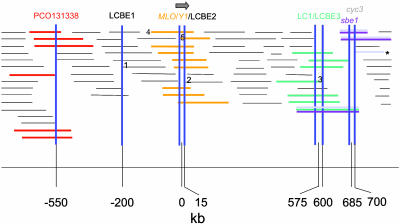
Shotgun Sequencing of Y1-Containing Bacterial Artificial Chromosomes (BAC). The Y1-containing BAC b144b.c06, derived from the maize Mo17 inbred line, was selected for shotgun sequencing (Fig. 1, BAC no. 4). The BAC clones were sheared by nebulization, and purified fragments were cloned into the pBluescript vector and sequenced from each end. The insert size was estimated to be ≈170 kb, and 3,422 sequence reads were obtained, resulting in 10× coverage.
Fig. 1.
The arrangement of the identified low copy regions surrounding the Y1 gene on the BAC physical contig. Horizontal lines indicate individual BAC clones. BACs 1-3 represent the BACs with low copy BAC ends, whereas BAC 4 is the Y1-containing BAC that was shotgun sequenced. BACs 2 and 5 are the BACs whose ends border the Y1 gene. The arrow indicates the orientation of the Y1 gene. BAC clone sizes and physical distances are approximated from the CB map (see Materials and Methods).
Assembly of the Region Surrounding Y1. The sequence reads were assembled by using phred and phrap software (www.phrap.org), and the assemblies were viewed and edited in consed (www.phrap.org). Vector sequences and bacterial contaminants were masked, and pair-mate information was used to make assessments regarding the validity of the assemblies, with the assistance of XGAP (www-gap.dcs.st-and.ac.uk/~gap/Share/xgap.html).
Primers were designed to walk across the sequence gaps by extracting the nonrepetitive ends of the relevant contig sequences and importing them together into the primer 3.0 program [S. Rozen and H. J. Skaletsky (Whitehead Institute for Biomedical Research, Cambridge, MA); code available at www.genome.wi.mit.edu/genome_software/other/primer3.html]. The following conditions were used in the selection of primers: the smallest allowable product size, primer size ≈18 bases, annealing temperature of 55°C, ideal GC of 50%, no more than three consecutive identical nucleotides, and a two-base GC clamp. T3 (5′-AATTAACCCTCACTAAAGGG-3′) and T7 (5′-GTAATACGACTCACTATAGGGC-3′) tags were added to the 5′ ends of the forward and reverse primers, respectively, to facilitate direct sequencing of the PCR products. Table 4, which is published as supporting information on the PNAS web site, lists all primers. PCR was performed by using a Perkin-Elmer 9700 thermocycler under the following conditions: 95°C for 10 min; 10 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min; 35 cycles of 95°C for 30 seconds and 68°C for 1 min; 92°C for 7 min; and then a constant temperature of 4°C. The 25-μl PCR mix consisted of 2 μl of BAC culture diluted 1:1 with 50% glycerol, 10 μM each primer, 5% DMSO, 12.5 μl of Hot Star Taq Master Mix (Qiagen), and sterile water. PCR products (4 μl) were analyzed via agarose gel electrophoresis. PCR products were prepared for sequencing by using exonuclease-I and shrimp alkaline phosphatase (United States Biochemical) and sequenced directly from both the T3 and T7 primers by using an ABI 3700 (PE Applied Biosystems) sequencer and the BigDye Terminators Ver. 3.0 Cycle Sequencing Kit (PE Applied Biosystems).
Assembly Validation. The assembly was verified by comparing electronic digests of the assembled Y1-containing contig sequence with restriction digests of BAC b144b.c6 with BamHI, EcoRI, XhoI, HpaI, and SfiI. Further validation was performed to confirm the orientation of the Y1 gene relative to the BAC contig. Using the overlapping BAC ends as guideposts, particularly the BAC ends from BACs 2 and 5 (Fig. 1) that bordered Y1, primers were designed to perform long-range PCR experiments to bridge the BAC ends to the relevant ends of the Y1 gene. The Expand High-Fidelity system was used with the following reagents: 11.4 μl of water, 2.5 μl of 2.5 mM dNTPs, 0.4 μl of each of the 10 μM primers, 2 μl of BAC DNA (Autogen generated), 2 μl of 10× buffer with 15 mM MgCl2, 1.05 units of the Expand High-Fidelity enzyme combination, and 1 μl of 25 mM MgCl2. The PCR cycling conditions were as follows: 94°C for 2 min; 10 cycles of 94°C for 15 seconds, 55°C for 30 seconds, and 68°C for 8 min; 25 cycles of 94°C for 15 seconds, 55°C for 30 seconds, and 68°C for 8 min with a 5-second increase with each cycle; 72°C for 7 min; and an indefinite hold at 4°C. Bands produced by the long-range PCR were extracted from the gel by using the QiaQuick gel extraction kit and procedure (Qiagen) and then sequenced. Sequencing reads were obtained from the custom primers by using the ABI Prism BigDye Terminators Ver. 3.0 Cycle Sequencing Kit (PE Applied Biosystems) per the manufacturer's protocol.
Identification and PCR of Low Copy Region in the Y1 BAC Contig. The Y1 gene containing BAC contig was identified in the Mo17 physical map (Bailin Li, personal communication) on the basis of overgo probe hybridization data (11). Low copy or genic sequences were identified in the shotgun sequence of BAC 4 and in BAC-end sequences of BACs within the Y1-containing Mo17 contig (Fig. 1). Genic regions were also identified via the placement of ESTs on the BACs through the aforementioned overgo probe-BAC hybridization project. All potential low copy sequences were blasted against sequence databases as well as against a library of repetitive sequences (12). Primers were designed as described above except the product size was selected to be between 400 and 600 bases.
To confirm chromosome 6 location of the amplicons, PCR was first performed on the 10 maize oat addition lines (13) as well as on a positive maize control and a negative oat control by using the conditions described (8) and 20 ng of DNA. If PCR product was present in the negative oat control reaction, then the PCR was reoptimized on Mo17 maize DNA using a gradient thermocycler (Eppendorf). The thermocycler conditions were as above with the exception of the annealing temperature, which ranged from 53°C to 63°C. The annealing temperature resulting in the highest yield of PCR product of expected size, as judged by agarose gel electrophoresis, was then used as the annealing temperature in a subsequent retest of the primers on the maize oat addition lines. Primers were used in subsequent experiments (Table 4) if the oat background was successfully eliminated, and only the chromosome 6 maize oat addition line and positive maize control produced PCR products.
Polymorphism Test and Sequencing. Chromosome 6 specific primers were tested on a subset of maize inbred lines, including B73, Mo17, InbredLo32, PI221788, Ames22443, NC296, PI406108, and PI595531. The conditions for PCR, presequencing cleanup of the products, and sequencing were the same as above, except 50 ng of genomic DNA was used for each reaction. The sequences were aligned and scrutinized for sequence polymorphisms in sequencher (Gene Codes, Ann Arbor, MI). If more than two single-nucleotide polymorphisms (SNPs) were identified on the subset of eight inbred lines, PCR and sequencing were performed on the entire germplasm set. The polymorphisms identified in sequencher were recorded in an excel spreadsheet. Polymorphisms observed in only one of all of the test lines were disregarded unless seen in both forward and reverse sequencing reads. Polymorphisms that could not be typed were designated as N.
Physical Distance Approximation. Distances between Y1 and the surrounding BACs were approximated by using a consensus band (CB) physical map of the Mo17 contig. The CB map uses CB count as the map scale. The distances between genes located on the same contig are estimated by counting the number of CBs between them and multiplying that number by the average band size, which for this map was 1.5 kb (B. Li, personal communication). With the exception of the BAC ends, the positions of which are obvious, the positions of the low copy regions on the CB map were assumed to be the minimum overlap regions of the BACs producing positive overgo probe hybridization signals (see Fig. 1).
DNA Analysis. Association analyses were performed by using Fisher's exact test on informative SNP loci in each respective region. The probability values were corrected by using the Bonferroni method (14) by dividing by the total number of tests performed (n = 133). Descriptive statistics, including π (15), Watterson's estimator of θ (15, 16), and Tajima's D (17), were obtained by using dnasp, Ver. 3 (18). LD between haplotypes was evaluated by defining haplotype states: haplotypes were given a “state” number in order of their frequency of occurrence. For instance, the most common haplotype within a region was given a “1,” the next most common haplotype was a “2,” and so on. The r2 and D′ values were calculated and displayed in tassel (www.maizegenetics.net/bioinformatics/index.htm).
Genetic Mapping. The Y1 gene and the outer low copy regions, PCO131338 and sbe1 were mapped on 279 individuals of the intermated B73 × Mo17 population (10). The B73 inbred line contained a 346-bp Tourist element within the Y1 gene, whereas the Mo17 did not, enabling the indel to be typed using agarose gel electrophoresis. SNPs within sbe1 and PCO131338 were typed by using pyrosequencing (19) and direct DNA sequencing, respectively. Because individual plants were pooled for each recombinant inbred line in the tissue collection stage, heterozygosity of a portion of the recombinant inbred lines in this region was expected. Heterozygous loci occurred in ≈1.7% of the recombinant inbred lines. If individuals were heterozygous at one locus and not at another, then the two loci were assumed to have one-half recombinant occurring between them. Otherwise, individuals having one allele at one locus and the other allele at another locus were designated as having one recombinant between them.
Results
Orienting the Y1 Gene Relative to the Contig. Through phred/phrap assembly, sequencing across gaps, and manual editing in consed, a 95-kb contig containing the Y1 gene was identified (GenBank accession no. AY455286). This contig contained the Y1 gene (nucleotides 39654-45646) and the BAC end sequence of BAC2 (nucleotides 58358-60959), thus orienting the contig with respect to the physical map (Fig. 1). A comparison of the electronic digest of the assembly sequence was consistent with the restriction digests of the BAC using BamHI, EcoRI, XhoI, HpaI, and SfiI. Furthermore, long-range PCR and sequencing of the PCR products confirmed that the Y1 gene was oriented 5′ to 3′ relative to the BAC contig (Fig. 1).
The Gene Content of the Y1 Contig. The 95-kb Y1-containing contig sequence was submitted to Generic Model Organism Database (www.gmod.org) for analysis. The Y1 gene comprises a small uninterrupted gene island consisting of the Y1, an MLO homolog lying immediately downstream of Y1, and an fgenesh-predicted gene with no homology to any known sequence (www.softberry.com, Softberry, Mt. Kisco, NY). A putative RING zinc finger protein, which is homologous (1e-30) to a number of Arabidopsis RING zinc finger proteins, including NP_179337, T51844, AAC68673, AAD32903, AAK44054, and AAL33807, and a maize ornithine carbamoyltransferase homolog with close sequence identity to GenBank no. AF466646 are also located in the immediate downstream region. The remainder of the sequence is largely repetitive. The repetitive sequences include Huck retrotransposons at nucleotides 1-4905, 10845-24204, and 59995-70865, and a Tekay element that precedes the Y1 gene (nucleotides 34848-39708).
Low Copy Regions and Their Estimated Distances from Y1. Low copy regions surrounding Y1 were identified via sequence homology searches of both the shotgun sequences obtained from the Y1-containing BAC4 (Fig. 1) and the BAC end sequences from all of the BACs within the contig. The low copy nature of each was confirmed by chromosome 6-specific PCR amplification from a complete set of maize-oat addition lines (13). Shotgun sequencing of the Y1-containing BAC identified a homolog of the barley MLO gene (20) ≈1 kb from the most common Y1 transcription stop site. This MLO-like gene on chromosome 6 has the highest homology to MLO6 (GenBank no. AY029317), which is present on chromosome 5, with 95% nucleotide sequence identity between the coding regions of the two genes. Primers were designed to amplify the regions corresponding to nucleotides 285-360, 445-681, and 680-737 of the AY029317 MLO6 sequence. Further blast analysis of the shotgun sequencing reads from BAC 4 revealed a 14-3-3 disease resistance homolog (with homology to tomato and Arabidopsis proteins, GenBank nos. X95905 and AF323920, respectively); however, attempts to identify low copy amplicons within this region were unsuccessful.
Three low copy BAC ends were present on the Y1 contig. The first, designated LCBE1 (for low copy BAC end 1), lies ≈200 kb upstream of Y1 and has no significant homologies to anything in the sequence databases. The second low copy BAC end sequence, LCBE2, lies 13 kb downstream of Y1. This sequence has significant homology (e-124) to a putative Arabidopsis ring zinc finger protein (GenBank no. AF078824). The third low copy sequence, designated as LCBE3, is located ≈600 kb downstream of Y1. Although this sequence has homology to a retroelement-related pol gene, suggesting a repetitive nature, we were able to design locus specific PCR primers.
The positioning of EST-derived overgo sequences on the contig (11) facilitated the discovery of additional low copy or genic regions in the area surrounding the Y1 gene. A 40-bp EST probe PCO131338 (from the EST sequence AY107270) with homology to a viral DNA-directed DNA polymerase identified a genic region ≈550 kb upstream of Y1 (Fig. 1). Another low copy region was identified ≈575 kb downstream of Y1 by using an overgo probe designed for a maize cDNA identical to GenBank no. CD0960947. This sequence encodes a hypothetical protein related to a rice genomic sequence (GenBank no. AP002484, nucleotides 60593-60715) and from here on will be referred to as LC1. Two low copy genic regions downstream of Y1 are the cyc3 gene (GenBank no. ZMU10076), which encodes a cell division protein, and sbe1 (GenBank no. AF072724, starch branching enzyme I). Cyc3 and sbe1 are present on the same BACs with the exception of the BAC designated by the asterisk in Fig. 1, which is positive for cyc3 only. Thus, sbe1 is closer to Y1 than cyc3, at an estimated distance of 685 vs. 700 kb between Y1 and cyc3.
Another 40-bp EST probe PCO145715 (from the EST sequence AY106275) identified a genic region ≈1.2 Mb upstream of Y1 (Fig. 1). This EST sequence is similar to an Arabidopsis receptor-like serine/threonine kinase (AC009894) and thus will be denoted as STK. The STK amplicon will be used as a control along with the unlinked PSY2 gene located on chromosome 8 (8). Fig. 1 shows the location of these low copy regions with respect to the BAC contig.
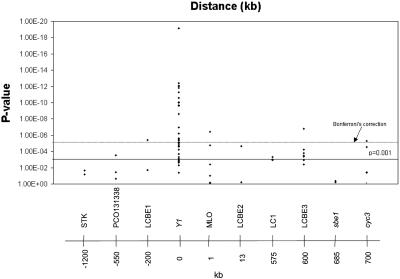
Low Copy Regions: Polymorphism Survey and Association Testing of Informative SNPs. Primers were designed to amplify portions of the identified low copy regions on the entire test set of lines. Polymorphisms were visually identified (Table 5, which is published as supporting information on the PNAS web site), and association tests with endosperm color phenotype were performed for the informative SNPs within each region (Table 6, which is published as supporting information on the PNAS web site). Table 1 lists the types and numbers of polymorphisms for each of the low copy amplicons as well as the P values that resulted from the association tests. Overall, highly significant (P < 0.001) associations with endosperm color phenotype were observed up to 550 kb upstream of Y1 and up to 700 kb downstream. The downstream region also contained a higher proportion of informative SNPs within each tested region that were significantly associated with endosperm color phenotype. For instance, four of four of the informative SNPs in the LC1 region showed highly significant associations, whereas 30 of 34 of the informative SNPs in the highly polymorphic LCBE3 region were significantly associated with the respective phenotype at P < 0.001. After applying a conservative Bonferroni correction for multiple tests, significant associations were found between -200 kb upstream of Y1 and at least 600 kb downstream of Y1 (Fig. 2). Association tests with endosperm color phenotype were also performed for the major haplotypes within each region with very similar results (data not shown).
Table 1. Properties of informative low copy number and genic regions surrounding YI.
| Amplicon | Top BLAST hit | Amplicon size, bp | Distance from Y1 | SNPs | Indels, 1-10 bp | Informative SNPs | P |
|---|---|---|---|---|---|---|---|
| STK | AY 106275; serine/threonine kinase | 705 | − 1.2 Mb | 9 | 1 | 3 | 2NS, 1* |
| PCO131338 | AY 107270; DNA-directed DNA pol | 310 | − 550 kb | 8 | 3 | 3 | 1NS, 1*, 1*** |
| LCBE 1 | No significant hits | 328 | − 200 kb | 5 | 0 | 2 | 1*, 1*** |
| MLO | AY029317; MLO6 homolog | 475 | + 1 kb | 9 | 1 | 8 | 3NS, 1*, 1**, 3*** |
| LCBE2 | AF078824; ring zinc finger protein | 248 | + 13 kb | 9 | 0 | 6 | 1NS, 5*** |
| LC1 | No significant hits | 329 | + 575 kb | 7 | 0 | 4 | 4*** |
| LCBE3 | gag/pol | 421 | + 600 kb | 50 | 3 | 34 | 4**, 30*** |
| sbe 1 | AF072724; starch branching enzyme I | 355 | + 685 kb | 5 | 5 | 2 | 2NS |
| cyc3 | ZMU10076; cyclin III | 535 | + 700 kb | 8 | 6 | 4 | 2*, 2*** |
| PSY2† | Phytoene synthase PSY2 | 1,300 | Chr. 8 | 24 | 15 | 13 | 12NS |
Numbers of SNPs and insertion-deletion polymorphisms as well as results of testing for association with the endoosperm color phenotype are indicated. NS, not significant; asterisks indicate probability (P) values for association with the phenotype ranges: *, 0.10 > P > 0.01; **, 0.01 > P > 0.001; ***, P < 0.001.
Control complicon, location Chi. 8.
Fig. 2.
Significance of associations between informative SNPs within the low copy regions and endosperm color phenotype. P values were calculated by using Fisher's exact test. The points above the solid horizontal line represent SNP association significant at P < 0.001. The dashed line represents P < 0.001 after Bonferroni correction (n = 133). The horizontal axis is not to scale, but distances in kilobases and gene symbols are indicated.
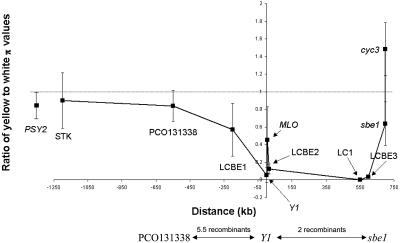
Patterns of Diversity Surrounding Y1. The diversity values obtained for each region are summarized in Table 2. The ratios between the diversity within the yellow endosperm lines vs. the diversity within the whites are not symmetrically distributed with respect to the Y1 gene. In the regions upstream of the gene, the ratio increases to ≈0.6 within 250 kb and reaches values indistinguishable from 1.0 at ≈500 kb (see bars in Fig. 3). Downstream of the gene, the yellow endosperm lines maintain very low diversity (<0.1) up to at least 600 kb, except for an apparent spike of diversity at the Mlo-like gene. Sbe1 and cyc3 are the first regions beyond +600 kb that do not have the characteristic conserved haplotype expected of a selective sweep. The two control regions, STK and PSY2 (8), have yellow/white π diversity ratios of 0.90, which is not significantly different from 1.0, or the value expected for unselected loci.
Table 2. Genetic diversity within the low copy regions surrounding Y1.
| Region | Test set | n | S | π (×10−3) | Θw (×10−3) | D |
|---|---|---|---|---|---|---|
| STK* | All | 73 | 9 | 4.0 (0.45) | 3.2 (1.3) | 0.66 |
| Yellow/orange | 40 | 9 | 3.7 (0.55) | 3.5 (1.5) | 0.17NS | |
| White | 33 | 8 | 4.1 (0.73) | 3.3 (1.5) | 0.74NS | |
| PCO131338 | All | 75 | 8 | 8.0 (0.72) | 6.6 (2.8) | 0.55NS |
| Yellow/orange | 41 | 4 | 6.3 (0.27) | 3.7 (2.1) | 1.7NS | |
| White | 34 | 8 | 7.5 (1.3) | 6.8 (3.0) | 0.31NS | |
| LCBE1† | All | 75 | 4 | 1.4 (0.31) | 3.3 (1.8) | −1.2NS |
| Yellow/orange | 41 | 2 | 0.91 (0.34) | 1.9 (1.4) | −0.96NS | |
| White | 33 | 3 | 1.6 (0.37) | 2.6 (1.6) | −0.85NS | |
| YI* | All | 73 | 89 | 8.4 (0.70) | 6.2 (1.7) | 1.0NS |
| Yellow/orange | 40 | 28 | 0.54 (0.27) | 1.7 (0.58) | −2.4** | |
| White | 33 | 84 | 10.2 (0.56) | 7.0 (2.2) | 1.6NS | |
| MLO‡ | All | 72 | 9 | 7.8 (0.71) | 5.2 (2.1) | 1.3NS |
| Yellow/orange | 40 | 9 | 3.9 (1.1) | 4.7 (2.0) | −0.49NS | |
| White | 32 | 9 | 8.6 (0.72) | 6.3 (2.7) | 1.1NS | |
| LCBE2§ | All | 72 | 9 | 8.7 (1.6) | 7.7 (0.01) | 0.35NS |
| Yellow/orange | 40 | 1 | 1.8 (0.22) | 0.95 (0.0009) | 1.3NS | |
| White | 32 | 9 | 14.6 (0.0032) | 9.3 (0.017) | 1.8NS | |
| LC1 | All | 75 | 7 | 3.4 (0.95) | 5.5 (2.4) | −0.91NS |
| Yellow/orange | 41 | 0 | NA | NA | NA | |
| White | 34 | 7 | 6.5 (1.3) | 6.4 (3.0) | 0.06NS | |
| LCBE3 | All | 75 | 48 | 21.1 (4.8) | 24.9 (7.2) | −0.61NS |
| Yellow/orange | 41 | 1 | 1.3 (0.04) | 0.59 (0.59) | 1.7NS | |
| White | 34 | 47 | 37.4 (5.8) | 29.2 (9.6) | 0.83NS | |
| sbe 1¶ | All | 73 | 5 | 2.9 (0.36) | 3.2 (1.6) | −0.16NS |
| Yellow/orange | 40 | 2 | 2.3 (0.43) | 1.4 (1.1) | 1.1NS | |
| White | 33 | 5 | 3.6 (0.55) | 3.7 (1.9) | 0.051NS | |
| cyc3 | All | 73 | 8 | 4.2 (0.29) | 3.8 (1.6) | 0.29NS |
| Yellow/orange | 40 | 5 | 4.0 (0.22) | 2.6 (1.3) | 1.4NS | |
| White | 33 | 7 | 2.7 (0.49) | 3.7 (1.7) | −0.76NS |
n, number of sequences analyzed; S, number of segregating sites; π, nucleotide diversity per site; Θw, Watterson's estimator; and Tajima's D values are listed for all germplasm analyzed as well as separately for white and yellow germplasm. SD are shown in parentheses. Estimates are based on all nucleotide sites. NA, not applicable. NS, not significant; *, P > 0.10; **, P < 0.001; Θw was calculated by using no recombination assumption (more conservative).
One orange/yellow endosperm line, Ames24575, and one white endosperm line, PI595547, were excluded due to missing data.
One white endosperm line, Ames 22026, was excluded from analysis because it had a 201-bp deletion in the analyzed area.
PI59559, PI595547, and W45 were excluded due to heterozygous polymorphic loci.
Stock6, PI595547, and PI595559 were excluded due to heterozygous polymorphic loci.
PI595547 and PI595559 were excluded due to heterozygous polymorphic loci.
Fig. 3.
Ratios of yellow to white diversity values (πyellow/πwhite) in the surrounding regions of Y1. Standard error bars are plotted. Three of the points on the graph, at positions -550, 0, and 685, were genetically mapped relative to one another, and the number of recombinants between the respective regions is indicated.
Long-Range LD Surrounding Y1. The conserved haplotype within the yellow endosperm lines is indicative of strong LD in the region between Y1 and LCBE3. Because of the nearly complete LD arising from the selective sweep, the entire test set of lines should have an elevated level of LD extending throughout this region; however, previous results within the Y1 gene showed LD between pairs of informative SNPs, as measured by r2, declining to ≈0.1 within 2 kb (8). For the assessment of long-range LD, we chose to evaluate haplotypes. Significant LD was observed from the MLO gene to LCBE3, assuming that the cutoff values for useful levels of LD are 0.1 for r2 and 0.5 for D′ (21) (Fig. 4 a and b, respectively, which is published as supporting information on the PNAS web site).
Genetic Mapping. The Y1 gene and the outer low copy regions, PCO131338 and sbe1, were genetically mapped on 279 individuals of the high-resolution intermated B73 × Mo17 recombinant inbred population (10). The genetic distances in this population are expanded ≈4 fold (10, 22). There were 8.5 recombinants between PCO131338 and Y1, 5 between Y1 and sbe1, and 8.5 between PCO131338 and sbe1, the outer regions of the area in question (for the explanation of scoring method, see Materials and Methods). Because the physical map order is known with high confidence, we were able to identify apparent double recombinants. These are likely due to nonreciprocal events, which are likely between very similar alleles (23). B73 and Mo17 haplotypes are very similar at and around the Y1 gene (8). Elimination of the double recombinants resulted in 5.5 recombinants between PCO131338 and Y1, 2 between Y1 and sbe1, and 7.5 between PCO131338 and sbe1. This genetic map is consistent with the physical map (Fig. 2).
Discussion
The maize ancestor teosinte has white endosperm (John Doebley, personal communication). However, both white and yellow endosperm corn occur among the maize landraces of Mexico. This observation suggests that our ancestors may have identified yellow, carotenoid-containing variants during or after the domestication process. This phenotype most likely resulted from the broadening of phytoene synthase gene expression patterns to include endosperm. In the presence of carotenoid precursor geranylgeranyl diphosphate, this change in gene expression specificity may have been sufficient to produce carotenoids in the endosperm. The yellow endosperm phenotype became popular during the last century, when the nutritional value of carotenoids became known. Human selection resulted in a dramatic reduction in diversity at the Y1 gene, consistent with the introgression of a few independent gain-of-function mutations into the majority of current maize breeding lines (8). Here we asked how far from the gene the effects of selection at Y1 can be detected. To this end, we analyzed genotype-phenotype associations, levels of genetic diversity, and LD at increasing distances from Y1 gene. In addition to the Y1 gene, nine low copy regions, four of which are genic, were included in the analysis. These regions extend ≈700 kb in one direction and 1.2 Mb in the other (Table 1).
Applying a very conservative correction for multiple testing, significant associations are detected at LCBE1, which lies 200 kb upstream of Y1, and up to 700 kb downstream, at cyc3 (Fig. 2). Differences in the yellow/white diversity ratios also indicate that the selective sweep caused by the selection at Y1 shows pronounced asymmetry, extending further downstream of the Y1 gene (Fig. 3). For example, 200 kb upstream of the Y1 gene, yellow/white diversity ratio has increased to 0.8. In contrast, downstream of the Y1 gene, very low diversity ratios (0.1 or less) are exhibited by LCBE2, LC1, and LCBE3, indicating that the selective sweep spans at least 600 kb. The downstream selective sweep ends rather abruptly, with a strong increase in the diversity ratio at sbe1 and especially cyc3. Starch branching enzyme 1 (sbe1) alters starch structure by forming branch points in glucan chains. Reduction of diversity at sbe1 could therefore have an effect on starch structure (24). However, changes in starch structure were not apparent in a knockout mutant of sbe1 (25).
The level and significance of LD between the low copy region haplotypes and Y1 are lower upstream of the Y1 gene (LCBE1, PCO131338, and STK), corroborating the patterns of sequence diversity observed on this side of Y1. The D′ and r2 values for LCBE1/Y1-5′, which are separated by 200 kb, are 0.70 and 0.09, respectively, with highly significant P values, whereas for the PCO131338/Y1-5′ combination, D′ and r2 are 0.51 and 0.05, respectively, with P values of 0.01. Downstream of Y1, the Y1-5′ region shows significant LD with all of the tested regions except sbe1 (Fig. 4).
The asymmetric selective sweep could be attributed to differences in the recombination rates upstream and downstream of Y1. To test this hypothesis, we genetically mapped Y1 and two flanking loci in a biparental mapping population. Recombinants (5.5) were found in the 550-kb interval between PCO131338 and Y1, and two recombinants were identified within the 685 kb between Y1 and sbe1, representing a 2-fold difference that is not significant at the 0.05 level. The genetic to physical distance in the region [0.54 centiMorgan (cM)] is not significantly different from the maize average of 1.2 cM/Mb.
The Y1 gene is bordered on its 3′ end by the MLO, a predicted gene with no known homology to any sequence, the RING zinc finger protein, and an ornithine carbamoyltransferase homolog. This gene island is located on the side of the longer selective sweep. The upstream region of Y1, where the selective sweep drops off within 200 kb, is made up largely of repetitive retrotransposon sequence. Thus, higher apparent gene density downstream does not appear to translate to higher recombination frequency. At the bz1 locus, a gene-rich island on one side of bz1 had a 2-fold greater recombination rate than on the other, where large hypermethylated retrotransposons resided (26).
The MLO gene, located 1 kb downstream from the Y1 termination codon, shows an apparent spike in diversity, with yellow/white diversity ratio close to 0.5, whereas three other regions beyond the MLO show patterns consistent with those observed at Y1, until 685 kb. This diversity ratio at MLO has a high standard error, and the difference may not be significant. A single SNP within the tested MLO region accounts for most of the variation seen in the yellows. This SNP is in LD with a SNP within the Ins2 in the 5′ regulatory region of Y1 (position 1768: ZMU32636), and with the CCA simple sequence repeat within Y1. We proposed that the two variants of Ins2 correspond to two independent introgression of yellow alleles into the modern germplasm (8). The LD between the Ins2 and Mlo polymorphisms indicates that each of the introgressions was linked to a different variant of the MLO gene, thus accounting for the observed increase in diversity within the yellow endosperm lines at this gene.
This study has demonstrated patterns of diversity consistent with the occurrence of a broad selective sweep at the Y1 gene. The selective sweep covering >0.5 Mb is much broader than the domestication-related selective sweep observed at tb1 (7). The extent of LD in the region may be the largest reported to date in maize (27-30). This is presumably the result of the qualitative nature of the trait, the short time since the selection was imposed, and partial genetic isolation of the yellow germplasm after selection (8). We conclude that it is likely that whole-genome scans will be successful in detecting other regions of the maize genome that have been subjected to strong selection by humans. However, the resolution of this method may be limited by the extent of the selective sweep.
Supplementary Material
Acknowledgments
We thank the sequencing and physical map groups at DuPont, especially Phyllis Biddle, Will Krespan, and Karla Butler. We also appreciate helpful technical advice from Ada Ching, Nathan Uhlmann, Thao Dam, Kevin Fengler, and Marianna Faller. K.P. thanks Jim Hawk, Janine Sherrier, and John Doebley for guidance and support. This work was funded by DuPont Crop Genetics.
This report was presented at the international Congress, “In the Wake of the Double Helix: From the Green Revolution to the Gene Revolution,” held May 27-31, 2003, at the University of Bologna, Bologna, Italy. The scientific organizers were Roberto Tuberosa, University of Bologna, Bologna, Italy; Ronald L. Phillips, University of Minnesota, St. Paul, MN; and Mike Gale, John Innes Center, Norwich, United Kingdom. The Congress web site (www.doublehelix.too.it) reports the list of sponsors and the abstracts.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: LD, linkage disequilibrium; SNP, single-nucleotide polymorphism; BAC, bacterial artificial chromosome; CB, consensus band; Mb, megabase.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. for: Y1 contig, AY455286; STK, AY451402-AY451476; PCO131338, AY451477-AY451551; LCBE1, AY451552-AY451626; MLO, AY451627-AY451701; LCBE2, AY451702-AY451776; LC1, AY451777-AY451851; LCBE3, AY459214-AY459288; sbe1, AY453436-AY453510; and cyc3, AY453511-AY453585).
References
- 1.Gilad, Y., Rosenberg, S., Przeworski, M., Lancet, D. & Skorecki, K. (2002) Proc. Natl. Acad. Sci. USA 99, 862-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawabe, A., Yamane, K. & Miyashita, N. T. (2000) Genetics 156, 1339-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Depaulis, F., Brazier, L. & Veuille, M. (1999) Genetics 152, 1017-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabeti, P. C., Reich, D. E., Higgins, J. M., Levine, H. Z., Richter, D. J., Schaffner, S. F., Gabriel, S. B., Platko, J. V., Patterson, N. J., McDonald, G. J., et al. (2002) Nature 419, 832-837. [DOI] [PubMed] [Google Scholar]
- 5.Benassi, V., Depaulis, F., Meghlaoui, G. K. & Veuille, M. (1999) Mol. Biol. Evol. 16, 347-353. [DOI] [PubMed] [Google Scholar]
- 6.Kohn, M. H., Pelz, H. J. & Wayne, R. K. (2000) Proc. Natl. Acad. Sci. USA 97, 7911-7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark, R. M., Linton, E., Messing, J. & Doebley, J. F. (2004) Proc. Natl. Acad. Sci. USA 101, 700-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palaisa, K. A., Morgante, M., Williams, M. & Rafalski, A. (2003) Plant Cell 15, 1795-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poneleit, C. G. (2001) in Specialty Corns, ed. Hallauer, A. R. (CRC, Boca Raton), 2nd Ed.
- 10.Lee, M., Sharopova, N., Beavis, W. D., Grant, D., Katt, M., Blair, D. & Hallauer, A. (2002) Plant Mol. Biol. 48, 453-461. [DOI] [PubMed] [Google Scholar]
- 11.Cone, K. C., McMullen, M. D., Bi, I. V., Davis, G. L., Yim, Y. S., Gardiner, J. M., Polacco, M. L., Sanchez-Villeda, H., Fang, Z., Schroeder, S. G., et al. (2002) Plant Physiol. 130, 1598-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyers, B. C., Tingey, S. V. & Morgante, M. (2001) Genome Res. 11, 1660-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ananiev, E. V., Riera-Lizarazu, O., Rines, H. W. & Phillips, R. L. (1997) Proc. Natl. Acad. Sci. USA 94, 3524-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller, R. G. J. (1991) Simultaneous Statistical Inference (Springer, New York).
- 15.Nei, M. (1987) Molecular Evolutionary Genetics (Columbia Univ. Press, New York).
- 16.Watterson, G. A. (1975) Theor. Popul. Biol. 7, 256-276. [DOI] [PubMed] [Google Scholar]
- 17.Tajima, F. (1989) Genetics 123, 585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rozas, J. & Rozas, R. (1999) Bioinformatics 15, 174-175. [DOI] [PubMed] [Google Scholar]
- 19.Nyren, P., Karamohamed, S. & Ronaghi, M. (1997) Anal. Biochem. 244, 367-373. [DOI] [PubMed] [Google Scholar]
- 20.Buschges, R., Hollricher, K., Panstruga, R., Simons, G., Wolter, M., Frijters, A., van Daelen, R., van der Lee, T., Diergaarde, P., Groenendijk, J., et al. (1997) Cell 88, 695-705. [DOI] [PubMed] [Google Scholar]
- 21.Flint-Garcia, S. A., Thornsberry, J. M. & Buckler, E. S., IV (2003) Annu. Rev. Plant Biol. 54, 357-374. [DOI] [PubMed] [Google Scholar]
- 22.Winkler, C. R., Jensen, N. M., Cooper, M., Podlich, D. W. & Smith, O. S. (2003) Genetics 164, 741-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dooner, H. K. (2002) Plant Cell 14, 1173-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satoh, H., Nishi, A., Yamashita, K., Takemoto, Y., Tanaka, Y., Hosaka, Y., Sakurai, A., Fujita, N. & Nakamura, Y. (2003) Plant Physiol. 133, 1111-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blauth, S. L., Kim, K. N., Klucinec, J., Shannon, J. C., Thompson, D. & Guiltinan, M. (2002) Plant Mol. Biol. 48, 287-297. [DOI] [PubMed] [Google Scholar]
- 26.Yao, H., Zhou, Q., Li, J., Smith, H., Yandeau, M., Nikolau, B. J. & Schnable, P. S. (2002) Proc. Natl. Acad. Sci. USA 99, 6157-6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenaillon, M. I., Sawkins, M. C., Long, A. D., Gaut, R. L., Doebley, J. F. & Gaut, B. S. (2001) Proc. Natl. Acad. Sci. USA 98, 9161-9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Remington, D. L., Thornsberry, J. M., Matsuoka, Y., Wilson, L. M., Whitt, S. R., Doebley, J., Kresovich, S., Goodman, M. M. & Buckler, E. S., IV (2001) Proc. Natl. Acad. Sci. USA 98, 11479-11484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thornsberry, J. M., Goodman, M. M., Doebley, J., Kresovich, S., Nielsen, D. & Buckler, E. S., IV (2001) Nat. Genet. 28, 286-289. [DOI] [PubMed] [Google Scholar]
- 30.Ching, A., Caldwell, K. S., Jung, M., Dolan, M., Smith, O. S., Tingey, S., Morgante, M. & Rafalski, A. J. (2002) BMC Genet. 3, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.