Abstract
Genome-wide association studies (GWAS) have identified several common variants associated with bipolar disorder (BD), but the biological meaning of these findings remains unclear. Integrative genomics—the integration of GWAS signals with gene expression data—may illuminate genes and gene networks that have key roles in the pathogenesis of BD. We applied weighted gene co-expression network analysis (WGCNA), which exploits patterns of co-expression among genes, to brain transcriptome data obtained by sequencing of poly-A RNA derived from postmortem dorsolateral prefrontal cortex from people with BD, along with age- and sex-matched controls. WGCNA identified 33 gene modules. Many of the modules corresponded closely to those previously reported in human cortex. Three modules were associated with BD, enriched for genes differentially expressed in BD, and also enriched for signals in prior GWAS of BD. Functional analysis of genes within these modules revealed significant enrichment of several functionally related sets of genes, especially those involved in the postsynaptic density (PSD). These results provide convergent support for the hypothesis that dysregulation of genes involved in the PSD is a key factor in the pathogenesis of BD. If replicated in larger samples, these findings could point toward new therapeutic targets for BD.
INTRODUCTION
Bipolar disorder (BD) is a debilitating and highly heritable psychiatric disorder whose genetic etiology is largely unknown. Candidate gene, genome-wide association (GWAS), and gene expression studies have all implicated a variety of genes, but a coherent theory of pathogenesis has not yet emerged. Multiple variants in many genes often come together into several gene networks and fewer biological pathways. This points to the powerful strategy, known as ‘integrative genomics' (Schadt, 2006), to address the extreme genetic heterogeneity seen in many common illnesses. This strategy has proven to be of value in interpreting the multigenic signals that have been observed in GWAS, copy number variation, and de novo mutation studies of many common neuropsychiatric diseases (Walsh et al, 2008; Choi et al, 2011; Fromer et al, 2014) but has so far been little studied in BD.
Many of the common genetic variants identified by GWAS lie in the regulatory regions, where they can affect the expression of nearby genes (Maurano et al, 2012). Much of the impact of genetic variation on gene expression is tissue-dependent (Andersson et al, 2014). Non-protein-coding genes that produce regulatory RNAs also appear to have an important role in fine-tuning of gene expression over development and in response to environmental stressors (Barry, 2014). It has also become clear that most genes, especially those expressed in brain, produce a number of distinct messenger RNA molecules, or transcripts, owing to alternative splicing, differential exon usage, and other posttranscriptional modifications (Barry, 2014).
Thus, the integration of GWAS signals with gene expression data requires a sensitive, tissue-specific approach that can assess differences in transcript abundance, noncoding RNAs, and posttranscriptional modification. Next-generation sequencing of RNA, known as RNA sequencing (RNA-seq), addresses many of these needs better than traditional gene expression microarrays, providing a more complete picture of the ‘transcriptome' (McGettigan, 2013).
As a first step toward an integrative genomics strategy in BD, we sequenced RNA derived from postmortem brain obtained from individuals with BD and matched controls. Our initial analysis of these data detected many differentially expressed (DE) genes in BD. These genes have important roles in neuroplasticity, circadian rhythms, and GTPase binding (Akula et al, 2014). When we integrated these data with the results from previous GWAS of BD, we found that DE genes in the GTPase pathway were also enriched for single nucleotide polymorphisms (SNPs) that were associated with BD. This suggested that differential expression of these genes was not just a consequence of BD or its treatment, but also reflected inherited genetic variation associated with disease risk. However, that study was limited by analysis methods that focused on individual genes and transcripts, without regard to their correlated patterns of expression.
Here, we performed a complete re-analysis of the same RNA-seq data using methods that exploit the correlated patterns of expression among groups of genes. We used weighted gene correlation network analysis (WGCNA) (Langfelder and Horvath, 2008), a widely used method that finds modules of highly correlated genes, relates these modules to one another, and tests the influence of sample phenotypes on gene expression correlations. WGCNA has been widely used to identify co-expressed gene networks in various human brain regions (Oldham et al, 2008), animals (Fuller et al, 2007; Langfelder et al, 2012), and in human phenotypes, including schizophrenia (Torkamani et al, 2010), autism (Voineagu et al, 2011), cancer (Clarke et al, 2013), aggressive behavior (Malki et al, 2014), BD (Chen et al, 2013a), and psoriasis (Li et al, 2014). However, aside from one study of a few gene networks (Hong et al, 2013), WGCNA has not yet been applied to the complete brain transcriptome in BD as revealed by RNA-seq.
WGCNA detected a number of robust gene expression modules, several of which were enriched for GWAS signals. Functional analysis of genes within one of these modules revealed significant enrichment of several functionally related sets of genes, especially those involved in the postsynaptic density (PSD). These results provide convergent support for the hypothesis that dysregulation of genes involved in the PSD is a key factor in the pathogenesis of BD. If replicated in larger samples, these findings could point toward new therapeutic targets for BD.
MATERIALS AND METHODS
Samples and RNA-Seq
RNA extracted from the dorsolateral prefrontal cortex of postmortem brains of 11 BD cases and 11 age- and sex-matched controls obtained from the Stanley Medical Research Institute and NIMH Brain Bank was sequenced at the National Institutes of Health Sequencing Center (NISC) using Poly-A selection. Sequencing was performed in two batches: 5 BD cases and 5 controls (NISC1), and 6 cases and 6 controls (NISC2). Owing to technical issues, one BD sample in NISC1 was excluded after initial quality control. Details about sample phenotypes, RNA-seq methods, and extensive quality control procedures are published elsewhere (Akula et al, 2014), and so will only be briefly summarized here. NISC1 and NISC2 cases and controls were randomized within each batch across lanes and sequenced from both ends (paired-end) on Illumina GA-IIx or HiSeq systems, respectively (Illumina Inc, San Diego, CA). Sequencing of cases and controls within each batch was performed in the same run in order to avoid batch-run effects.
TopHat (TopHat version2.0.4; http://tophat.cbcb.umd.edu/) (Trapnell et al, 2009) was used to map the reads to the reference human genome (hg19). A total of 2.3 billion mapped reads from NISC1 and 4.3 billion mapped reads from NISC2 were included in the downstream analyses. HTSeq (http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html) was used to obtain the read counts per gene based on Ensemble gene annotation (http://ftp.ensembl.org/pub/release-67/gtf/homo_sapiens/).
Gene Selection for Co-Expression Analysis
A total of 17 296 genes and 16 919 genes were selected after QC in NISC1 and NISC2, respectively. Of these, 16 571 genes were common to both these datasets and were thus included in the downstream co-expression analysis. Read counts were normalized using DESeq (Anders and Huber, 2010), and the resulting variance-stabilized transformed data were used in the downstream analysis. Weighted gene co-expression network analysis (WGCNA) was used to identify the co-expression modules (sub-networks) in BD (http://labs.genetics.ucla.edu/horvath/CoexpressionNetwork/Rpackages/WGCNA/). The co-expression analysis pipeline is shown is Supplementary Figure S1.
Weighted Gene Co-Expression Network Analysis (WGCNA)
In WGCNA networks, genes are represented as nodes and edges represent the correlation in expression (edge-weight) between gene pairs. The connection strength (adjacency) between two genes is calculated by raising the correlation to a specific power, β, which must be estimated with each dataset. Supplementary Figure S2 shows the relationship between the β and scale free topology fitting index in NISC1 and NISC2. At β=12, networks created by WGNCA showed >75% scale free topology in both datasets, so a value of 12 was used in this study. We used biweight correlation instead of the default Pearson correlation, because it is robust and resistant to outliers (Gaiteri et al, 2014). WGCNA identifies co-expressed genes and puts them into networks or modules. Minimum module size was set to 50 genes per module. Here, we used the terms co-expressed networks and co-expressed modules interchangeably, but we recognize that these terms are sometimes considered distinct (Dong and Horvath, 2007). Genes that did not belong to any module were assigned to a ‘grey' module; this was excluded from further analysis.
First, we identified the modules that were observed in both the NISC1 and NISC2 samples. These are referred to as ‘consensus modules'. Then, we tested whether the connectivity in consensus modules was preserved across both datasets (Langfelder and Horvath, 2007; Langfelder et al, 2011). Default parameters were used when identifying consensus and preserved modules. The R code, along with the data files used in this analysis, can be downloaded from http://intramural.nimh.nih.gov/humangenetics/data.html.
WGCNA calculates a module eigengene value, the first principal component of that module, for every sample. These module eigengene values were tested for correlation (Pearson correlation) with diagnosis of BD; a Student t-test (df=n−2) was used to estimate the statistical significance of the correlation coefficients.
Case and control samples were matched on age and sex. To test for potential impacts of unmatched factors, we used linear regression and ANOVA to test for association between module eigengene values and known biological (smoking and cause of death) and technical (RNA-integrity number and sequencing) variables.
As NISC1 and NISC2 samples were sequenced separately on different platforms, the results were calculated within each sample, and then combined by meta-analysis. Fisher's Chi-square method was used to combine the p-values of preserved modules in NISC1 and NISC2, which generated a meta-p-value under a chi-square distribution with two degrees of freedom. We used the Benjamini false discovery rate for multiple test correction (http://www.sdmproject.com/utilities/?show=FDR). Only preserved modules with Zsummary>10 (Langfelder and Horvath, 2007; Langfelder et al, 2011) whose eigengene was correlated in the same direction with BD in both the NISC1 and NISC2 samples were used in the downstream analysis.
DE Genes in RNA-seq Data
To test which of the modules were significantly enriched with genes that are DE in BD, we compared the genes in each of the modules with the 1225 DE genes reported in our earlier study (gene-level p-value<0.05 in Akula et al, 2014). A hypergeometric p-value was calculated to test the significance of overlap.
GWAS Enrichment Analysis
Postmortem gene expression data alone cannot distinguish between genes whose expression changes are the result of an illness or its treatment and genes that have a role in etiology. As inherited DNA variation is not influenced by illness or treatment, genes that carry inherited variants associated with illness are more likely to lie within causal pathways. Thus, we performed GWAS enrichment analysis in order to help differentiate between ‘causal' and ‘non-causal' gene modules. (Detailed methods can be found in Akula et al, 2014). In short, quasi-independent (r2<0.5) SNPs included in each of two published meta-analysis studies (Psychiatric GWAS Consortium Bipolar Disorder Working Group, 2011; Chen et al, 2013b) were assigned to their closest genes. We calculated the total number of SNPs (N) with p-value<0.05 in all the genes (Noriginal) in a functional category. We then randomly selected N number of SNPs 10 000 times (Nrandom). Lastly, we calculated the number of times Nrandom⩽Noriginal and divided by 10 000 to obtain an empirical GWAS enrichment p-value. The minor allele frequency distributions for the test and random sets were almost identical. This approach accounts for any bias that might be introduced by variable patterns of inter-SNP linkage disequilibrium, minor allele frequency, or gene length. We further validated our GWAS enrichment results by another gene set enrichment analysis program, MAGENTA (nPermutations=10 000) (Segre et al, 2010) which also accounts for gene length bias.
Functional Enrichment Analysis
WGCNA calculates an eigengene-based connectivity (kME or module membership) score for each gene in a module, which is the Pearson correlation between that gene's expression and its corresponding eigengene. Genes with consistent kME values (<0 or >0) in both NISC1 and NISC2 were subjected to functional enrichment analysis by use of the Database for Annotation, Visualization and Integrated Discovery (DAVID) (Huang et al, 2009a, b). The genes represented in the entire transcriptome dataset (n=16 571) were used as background. We used medium to high stringency. Gene ontology (GO) terms with Benjamini q<0.05 were declared significant.
Co-Expression Networks from Microarray Studies
We compared our co-expression modules from RNA-seq with those generated in two previous studies that used microarray data (Oldham et al, 2008; Chen et al, 2013a). We tested the significance of gene overlap between modules by a hypergeometric test, with universe values equal to the number of genes passing QC in both studies. Universe values of 8395 and 13 514 were used for comparisons with the Oldham et al (2008) and Chen et al (2013a) studies, respectively. Comparisons with the Oldham et al (2008) results were limited to modules generated in human cortex. As there is no one-to-one relationship of modules between the microarray and RNA-seq studies, we did not attempt to replicate the findings in Chen et al (2013a), but instead report overlapping modules and their association with BD in both studies.
Cell-Type Enrichment of Co-Expression Networks
Oldham et al (2008) also report gene expression signatures typical of particular cell types in human brain. Their results indicate modules enriched for oligodendrocytes, astrocytes, neurons, and synapses. In order to test whether any of our 21 modules were enriched for gene expression signatures reflecting these cell types, we compared our modules to those from Oldham et al (2008) using a hypergeometric test.
RESULTS
Gene Co-Expression Networks
A total of 33 consensus co-expression modules were detected (Supplementary Figure S3). All 33 modules were highly preserved in both the NISC1 and NISC2 data sets (Zsummary>10; Supplementary Figure S4). The number of genes in each module varied from 74 to 2766, with an average of 446 (Supplementary Table S1). Comparison with previously published data (Oldham et al, 2008) showed good agreement between these modules and those detected in microarray data from human cortex in individuals without psychiatric illness (Supplementary Table S2). Twenty-nine of 33 modules significantly overlapped with modules identified in the Oldham et al (2008) study. This demonstrates that WGCNA can detect robust modules of co-expressed genes across a range of data types and individuals.
Association with BD
Of the 33 preserved modules, 21 were selected for downstream analysis because their eigengene values were correlated with BD in the same direction in both NISC1 and NISC2 (Table 1; Supplementary Table S3 contains the module membership (kME) values for all genes in these 21 modules). The observed association with BD was not explained by differences in age or sex, because samples were matched on these variables. The observed associations were also not explained by differences in known biological (smoking and cause of death) or technical (RNA-integrity number and sequencing depth) covariates (Supplementary Table S4).
Table 1. Twenty-one Gene Co-Expression Modules that Showed Consistent Correlation with Bipolar Disorder (BD).
| Module | BD correlation in NISC1/NISC2 | BD association p-value | BD association FDR | Enrichment of differentially expressed genes | GWAS enrichment |
|---|---|---|---|---|---|
| Yellow | +/+ | 1.94E-03 | 0.017 | 2.20E-16 | ns |
| Light yellow | +/+ | 2.52E-03 | 0.017 | ns | ns |
| Sky blue | +/+ | 2.76E-03 | 0.017 | 1.66E-05 | ns |
| Dark turquoise | −/− | 3.58E-03 | 0.017 | ns | 3.00E-04 |
| Dark grey | +/+ | 3.66E-03 | 0.017 | 3.53E-09 | ns |
| Purple | +/+ | 0.005 | 0.018 | 2.20E-16 | 9.00E-04 |
| Dark orange | −/− | 0.008 | 0.024 | 0.031 | ns |
| Royal blue | +/+ | 0.012 | 0.032 | 0.024 | ns |
| Red | −/− | 0.015 | 0.035 | 2.54E-05 | 8.30E-03 |
| Green | −/− | 0.024 | 0.050 | 2.20E-16 | 5.80E-03 |
| Turquoise | −/− | 0.026 | 0.050 | 1.79E-06 | ns |
| Light cyan | +/+ | 0.036 | ns | ns | 5.30E-03 |
| White | +/+ | 0.049 | ns | ns | ns |
| Saddle brown | +/+ | ns | ns | ns | 9.10E-03 |
| Dark olive green | +/+ | ns | ns | ns | ns |
| Salmon | −/− | ns | ns | ns | 0.034 |
| Pale turquoise | −/− | ns | ns | 0.033 | ns |
| Blue | −/− | ns | ns | ns | ns |
| Dark green | −/− | ns | ns | ns | 0.016 |
| Dark red | +/+ | ns | ns | No overlap | ns |
| Midnight blue | −/− | ns | ns | ns | ns |
Abbreviation: ns, not significant.
‘+' and ‘−' represent increased and decreased gene expression in bipolar disorder, respectively.
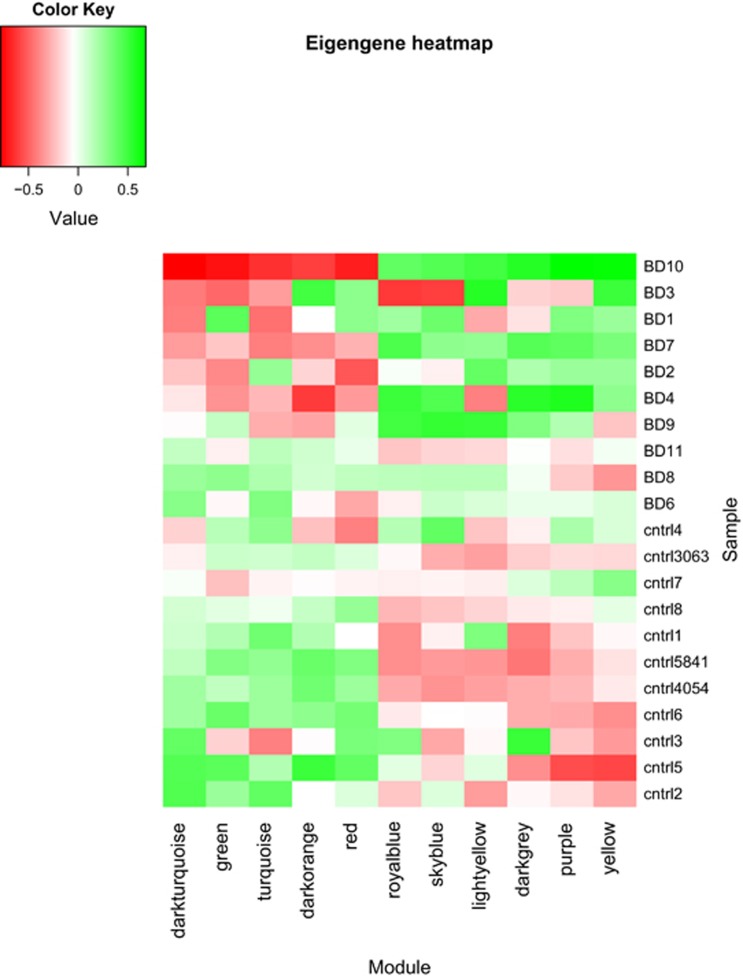
Eleven of the 21 modules were associated with BD at false discovery rate <0.05 (Table 1). The eigengene values for each of these 11 modules are depicted in Figure 1 as a heatmap. This shows that most of the genes within each of five modules (dark turquoise, green, turquoise, dark orange, and red) were downregulated in most of the BD cases we studied, compared with controls. Most of the genes in the remaining six modules (royal blue, sky blue, light yellow, dark grey, purple, and yellow) were upregulated in BD. The module assignments for all genes in the 11 BD-associated modules are depicted in Supplementary Figures S5 and S6.
Figure 1.
Eigengene heatmap. The x-axis shows the modules and the y-axis shows the samples. Red indicates negative, and green indicates positive eigengene values.
Overlap with DE Genes in RNA-seq Data
To assess whether the results of the WGCNA analysis agreed with those of our previous study, we assessed overlap among genes within each of the 21 differentially co-expressed modules identified by WGCNA with genes found to be DE in our previous RNA-seq study (Akula et al, 2014). There was a significant overlap in 10 modules (hypergeometric p-value <0.05), and only 1 module contained no genes previously identified as DE (Table 1). This shows that the WGCNA analysis largely agrees with single gene expression analysis but also identifies additional genes.
Modules Enriched with GWAS Genes (GWAS Enrichment)
In order to distinguish modules containing genes that may have a causal role in BD from those whose differential co-expression may be a consequence of BD or its treatment, we tested genes within each of the 21 modules for evidence of association with BD in previous GWAS. Eight modules were significantly enriched for GWAS-implicated genes (permutation p-value<0.05; Table 1), consistent with a causal role in BD. The remaining modules showed no evidence of GWAS enrichment.
As GWAS enrichment analyses can be biased by gene size, we repeated the analyses with MAGENTA (Segre et al, 2010), which takes gene-length bias into account. Similar results were obtained (Supplementary Table S5). The red and green modules were significantly enriched, and the purple module showed a trend toward enrichment (p<0.1), while the dark turquoise module was not significant in the MAGENTA analysis.
Functional Gene Set Enrichment Analysis
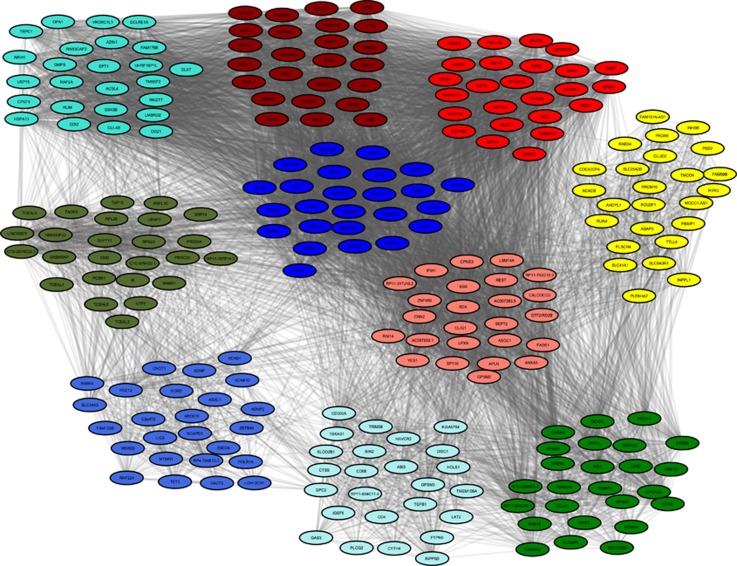
A critical question in this study concerns the potential functional relationships among the implicated genes. In order to explore this question, we performed gene set enrichment analysis in each of the eight gene modules that were consistent with causal involvement in BD. Three of the eight ‘causal' modules (green, red, and salmon) yielded several significantly enriched GO terms, including: cell-cell signaling, PSD, ion transport, synapse, regulation of transcription, and passive transmembrane transporter activity (Table 2). A few specific GO terms, such as PSD, synapse, cation channel activity, and ribosomal subunit were strikingly (>2.5-fold) enriched, whereas most of the other GO terms showed ~twofold enrichment. (Gene significance and module membership values for all the genes in the green module are shown in Supplementary Figures S7 and S8.) Of the remaining 13 modules that were not enriched for GWAS signals, 7 modules (blue, dark olive green, dark red, pale turquoise, royal blue, turquoise, and yellow) yielded significant enrichment for particular GO terms. These included zinc ion binding, defense response, immune system development, response to wounding, proteolysis, and carboxylic acid binding, among others. Most of the immune-related GO terms showed >fivefold enrichment. The enriched GO terms, along with the genes and their respective p-values, are given in Supplementary Table S6. The top 25 genes in each of the modules that yielded significant functional enrichment results are shown in Figure 2.
Table 2. Functional Enrichment of Genes in Co-Expressed Modulesa.
| Category | Module | Term | Fold enrichment | P value | Benjamini Q value |
|---|---|---|---|---|---|
| Causal | Green | GO:0014069~postsynaptic density | 4.6 | 1.09E-04 | 3.30E-03 |
| GO:0045211~postsynaptic membrane | 3.9 | 1.27E-05 | 4.83E-04 | ||
| GO:0045202~synapse | 3.1 | 5.61E-09 | 8.49E-07 | ||
| GO:0030425~dendrite | 2.8 | 8.05E-04 | 2.01E-02 | ||
| GO:0022803~passive transmembrane transporter activity | 2.8 | 1.84E-06 | 4.94E-04 | ||
| GO:0005261~cation channel activity | 2.6 | 3.87E-04 | 1.87E-02 | ||
| Red | GO:0006396~RNA processing | 2.9 | 9.68E-06 | 1.02E-02 | |
| Salmon | GO:0045449~regulation of transcription | 2.3 | 1.31E-05 | 9.09E-03 | |
| Consequential | Blue | GO:0003735~structural constituent of ribosome | 2.8 | 9.97E-06 | 8.14E-03 |
| Dark olive green | GO:0045449~regulation of transcription | 3.6 | 9.45E-04 | 2.18E-02 | |
| GO:0008270~zinc ion binding | 3.2 | 6.38E-03 | 4.99E-02 | ||
| GO:0033279~ribosomal subunit | 16.5 | 1.41E-03 | 4.06E-02 | ||
| Dark red | GO:0044445~cytosolic part | 7.7 | 2.53E-04 | 1.31E-02 | |
| GO:0030529~ribonucleoprotein complex | 3.8 | 2.22E-04 | 1.72E-02 | ||
| Pale turquoise | GO:0002684~positive regulation of immune system process | 17.0 | 5.11E-10 | 1.36E-07 | |
| GO:0045321~leukocyte activation | 16.4 | 9.17E-12 | 4.88E-09 | ||
| GO:0001817~regulation of cytokines | 12.9 | 1.34E-05 | 6.48E-04 | ||
| GO:0002520~immune system development | 10.1 | 4.08E-07 | 3.62E-05 | ||
| GO:0006952~defense response | 8.0 | 1.58E-07 | 1.69E-05 | ||
| GO:0009611~response to wounding | 6.5 | 1.56E-05 | 7.24E-04 | ||
| GO:0009967~positive regulation of signal transduction | 6.4 | 6.65E-04 | 2.12E-02 | ||
| GO:0010033~response to organic substance | 4.0 | 2.81E-04 | 1.03E-02 | ||
| Royal blue | GO:0043167~ion binding | 2.1 | 5.66E-05 | 6.66E-03 | |
| Turquoise | GO:0016879~ligase activity, forming carbon-nitrogen bonds | 2.0 | 6.99E-06 | 3.73E-03 | |
| Yellow | GO:0046395~carboxylic acid catabolic process | 3.7 | 8.19E-05 | 2.66E-02 | |
| GO:0016042~lipid catabolic process | 3.3 | 5.02E-05 | 2.29E-02 | ||
| GO:0031406~carboxylic acid binding | 3.1 | 1.90E-04 | 3.33E-02 | ||
| GO:0006631~fatty acid metabolic process | 3.0 | 3.56E-05 | 4.02E-02 |
For modules with multiple GO terms, only those with fold change >2.5 are presented in this table. See Supplementary Table S6 for full details.
Figure 2.
Gene co-expression networks in bipolar disorder. The top 25 genes in each of the 10 modules correlated with bipolar disorder are illustrated. Thickness of the grey lines are proportional to (absolute) magnitude of the observed gene-gene correlations. Colors correspond to those in Table 2.
RNA-seq Co-Expression Networks Agree with Those Implicated in a Published Microarray-Based WGCNA Analysis
Chen et al (2013a) found 23 co-expression modules that were associated with BD in multiple microarray datasets. We calculated the extent of overlap between these modules and those we found in the RNA-seq data. There was a highly significant overlap between the co-expressed modules in both studies (Supplementary Table S7). Several modules had corrected hypergeometric p-values<0.05, indicating the high reproducibility of the co-expressed gene network structure, even though fewer genes can be detected by microarray.
BD-Associated Co-Expression Networks do not Show Expression Signatures for Specific Cell Types
Comparison with the Oldham et al (2008) data found evidence of significant gene overlap between several modules and genes characteristic of oligodendrocytes, astrocytes, microglia, neurons, glutamatergic neurons, and synaptic proteins (Supplementary Table S2). The strongest gene overlaps (>50%) were observed for modules characteristic of oligodendrocytes.
DISCUSSION
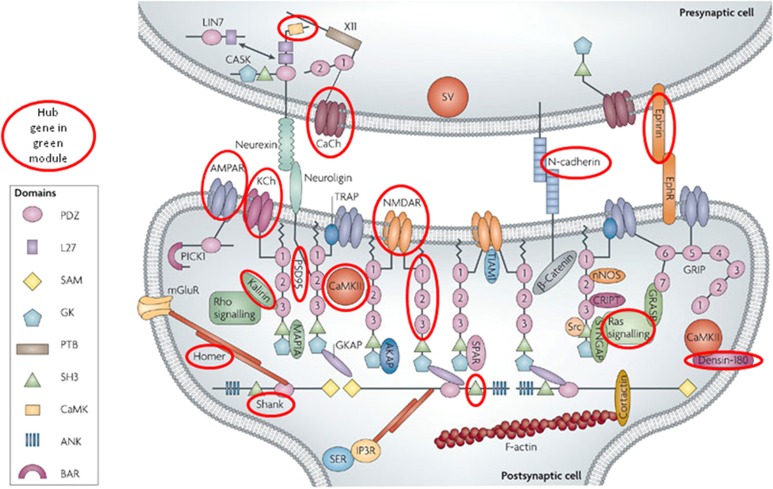
To our knowledge, this is the first study to perform WCGNA analysis on RNA-seq data of the complete brain transcriptome in BD cases. These results provide a high-resolution account of the interacting gene networks in brain that are involved in BD. By incorporating the GWAS signals, we have attempted to distinguish gene modules that have a causal role from those that appear to be a consequence of BD or its treatment. The preserved, differentially co-expressed, GWAS-enriched modules point toward a number of biological pathways as important factors in the pathogenesis of BD. Three modules were associated with BD and enriched for DE genes and BD GWAS signals. Of these, the green module showed a striking (4.6-fold) enrichment for genes involved in the PSD (Figure 3). This finding is noteworthy in light of the several prior studies that have implicated the PSD in BD and other neuropsychiatric disorders (el-Mallakh and Wyatt, 1995; Kristiansen and Meador-Woodruff, 2005; Beneyto and Meador-Woodruff, 2008; Pennington et al, 2008; Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium, 2015). Our results provide independent support for those findings and suggest that genes involved in the PSD are a key factor in the pathogenesis of BD. If replicated in larger samples, our results could point toward new therapeutic targets for BD among the numerous proteins active in the PSD (Feng and Zhang, 2009).
Figure 3.
Post-synaptic and ion channel genes in the green module. The postsynaptic density figure has been adapted from Feng and Zhang (2009). Proteins enclosed in red circles are encoded by one or more genes assigned to the ‘green' module in the present study. CaCh: CACNA1E, CACNA1G, CACNB1, CACNG3; CAMK: CAMK2A, CAMK4; N-cadherin: CDH12, CDH8; PSD95: DLGAP3, DLGAP4; Ephrin: EFNB3, EPHA4, EPHA6; AMPAR: GRIA1, GRIA2, GRIA3; NMDAR: GRIN2B; Homer: HOMER1; Kalirin: KALRN; KCh: KCNA4, KCNB2, KCNC3, KCNG1, KCNH5, KCNIP3, KCNQ2, KCNQ4, KCTD1; Densin-180: LRRC7; PDZ: PDZD2; Ras signaling: RAP2B; SH3: SH3KBP1, SH3PXD2A; Shank: SHANK1, SHANK2.
This study has several limitations. The sample size was relatively small compared with the samples used in previous microarray studies. Nevertheless, we were able to successfully replicate many of the published findings and extend those findings to non-coding genes and previously undiscovered functional gene networks. This reflects the precision and wide dynamic range of high-depth, RNA-seq-based transcriptome data (Iancu et al, 2012, 2014; Zhang et al, 2014). Given the small sample size, subtle biological or technical biases cannot be ruled out. As is inherent in ‘omics' studies where the number of variables far exceeds the number of subjects, the results should be cautiously interpreted until replicated in a larger dataset (Bild et al, 2014). Some of the gene modules that were associated with BD in this study did not reveal recognized functional pathways. This may reflect the limitations inherent in analyses that depend on known relationships between genes in the published literature. This limitation will diminish as more empirical gene–gene relationships are revealed. Other limitations of this study include the focus on only one brain region and lack of cellular resolution. However, a comparison with published cell-specific gene expression signatures (Oldham et al, 2008) suggests that several cell types contribute to these results.
The major strength of this study is the ability to integrate expression data in groups of genes with risk allele data from GWAS. We identified 11 modules that were associated with BD, 4 of which were significantly enriched with GWAS variants. The results for GWAS enrichment were further validated using MAGENTA (Segre et al, 2010), which supported significant GWAS enrichment among genes in the green and red modules. A significant enrichment of miR-137 targets among genes in the red module was observed. A potential causal role for the remaining modules cannot be ruled out, however, because genes in those modules might harbor rare single nucleotide or copy number variants that would not be detectable by GWAS.
As this study relies on the observed co-expression of both protein-coding and non-protein-coding genes in the brain, rather than on genetic relationships apparent in the published literature, it offers a more unbiased account of genetic relationships in the brain. For example, the analysis was able to detect several non-protein-coding genes, such as lncRNAs, that so far are not well understood in the context of brain function but seem to have a key role in tying together otherwise disparate sets of genes involved in BD. Recent research has shown that lncRNAs are highly conserved and have an essential role in synapse formation (Bernard et al, 2010; He et al, 2014) and other key aspects of brain development.
The ‘salmon' module deserves special mention because it contains three replicated GWAS hits for BD: TRANK1, SYNE1, and CACNA1C (Psychiatric GWAS Consortium Bipolar Disorder Working Group, 2011; Chen et al, 2013b; Muhleisen et al, 2014). Contingency table analysis (detailed in Supplementary Table S8) shows that it is highly unlikely that these three genes would fall into the same module by chance (Fisher exact p=0.01). As WGCNA relies on observed co-expression, rather than reports in the literature, it was able to pull together genes like CACNA1C, that are relatively well-studied, with the other genes that are still relatively understudied in the published literature. However, we did not detect differential co-expression of the ‘salmon' module in BD in this sample. This might reflect limited statistical power, the particular anatomical brain region we chose to study, or reliance of the GWAS enrichment strategy on existing studies with limited statistical power.
Several hub genes present in the 11 BD-associated modules overlap with those implicated in other neuropsychiatric disorders. For example, over 100 genes overlap with those within the 108 loci implicated in a recent GWAS of schizophrenia (Supplementary Table S9) (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). Eighteen genes overlap with those implicated in autism (Poultney et al, 2014; Supplementary Table S10). BD, schizophrenia, and autism are all brain disorders, so overlapping findings are perhaps not surprising, but they reaffirm the considerable genetic overlap among these clinically distinct neuropsychiatric disorders.
The strong evidence of immunological enrichment among genes in the modules showing no GWAS enrichment points toward immunological events as a consequence of BD or its treatment. The results suggest that the most genes in the immunologically enriched modules are downregulated in BD, in contrast to the finding of increased expression of immune-related genes reported in an earlier WGCNA analysis in autism (Voineagu et al, 2011). However, the results resonate broadly with a recent report of abnormal activation of peripheral blood monocytes and lymphocytes in BD (Gumieiro et al, 2010).
This study has produced results that are overall consistent with the prior findings, but also implicate novel genes and biological pathways that may contribute to the risk for BD. The integration of RNA-seq-based gene expression data with GWAS data highlights potentially important differences in gene co-expression networks that contain genes harboring risk alleles and those that do not. The current findings may also be valuable for helping to interpret the results of future studies of rare variation in BD. Integration of GWAS with gene co-expression data is a promising approach to better understand the mechanisms of highly genetically heterogeneous neuropsychiatric disorders.
FUNDING AND DISCLOSURE
NA, KHC, and FJM declare no conflict of interest. Over the past three years, JRW has received compensation from F. Hoffmann-La Roche AG, Pfizer Inc., and Nestlé Health Science.
Acknowledgments
We thank Drs Michael B Knable, E Fuller Torrey, Maree J Webster, Serge Weis, and Robert H Yolken at The Stanley Medical Research Institute for sharing brain tissue. We also thank Drs Barbara K Lipska and Joel E Kleinman at NIMH-IRP for providing us RNA from three control brains. Data analysis was performed on the Biowulf high-performance computing platform at the NIH. This study was funded by the Intramural Research Program of the NIMH (ZIAMH002810).
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
Supplementary Material
References
- Akula N, Barb J, Jiang X, Wendland JR, Choi KH, Sen SK et al (2014). RNA-sequencing of the brain transcriptome implicates dysregulation of neuroplasticity, circadian rhythms and GTPase binding in bipolar disorder. Mol Psychiatry 19: 1179–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W (2010). Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M et al (2014). An atlas of active enhancers across human cell types and tissues. Nature 507: 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry G (2014). Integrating the roles of long and small non-coding RNA in brain function and disease. Mol Psychiatry 19: 410–416. [DOI] [PubMed] [Google Scholar]
- Beneyto M, Meador-Woodruff JH (2008). Lamina-specific abnormalities of NMDA receptor-associated postsynaptic protein transcripts in the prefrontal cortex in schizophrenia and bipolar disorder. Neuropsychopharmacology 33: 2175–2186. [DOI] [PubMed] [Google Scholar]
- Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z et al (2010). A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J 29: 3082–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bild AH, Chang JT, Johnson WE, Piccolo SR (2014). A field guide to genomics research. PLoS Biol 12: e1001744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Cheng L, Grennan K, Pibiri F, Zhang C, Badner JA et al (2013. a). Two gene co-expression modules differentiate psychotics and controls. Mol Psychiatry 18: 1308–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DT, Jiang X, Akula N, Shugart YY, Wendland JR, Steele CJ et al (2013. b). Genome-wide association study meta-analysis of European and Asian-ancestry samples identifies three novel loci associated with bipolar disorder. Mol Psychiatry 18: 195–205. [DOI] [PubMed] [Google Scholar]
- Choi KH, Higgs BW, Wendland JR, Song J, McMahon FJ, Webster MJ (2011). Gene expression and genetic variation data implicate PCLO in bipolar disorder. Biol Psychiatry 69: 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke C, Madden SF, Doolan P, Aherne ST, Joyce H, O'Driscoll L et al (2013). Correlating transcriptional networks to breast cancer survival: a large-scale coexpression analysis. Carcinogenesis 34: 2300–2308. [DOI] [PubMed] [Google Scholar]
- Dong J, Horvath S (2007). Understanding network concepts in modules. BMC Syst Biol 1: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Mallakh RS, Wyatt RJ (1995). The Na,K-ATPase hypothesis for bipolar illness. Biol Psychiatry. 37: 235–244. [DOI] [PubMed] [Google Scholar]
- Feng W, Zhang M (2009). Organization and dynamics of PDZ-domain-related supramodules in the postsynaptic density. Nat Rev Neurosci 10: 87–99. [DOI] [PubMed] [Google Scholar]
- Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P et al (2014). De novo mutations in schizophrenia implicate synaptic networks. Nature 506: 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller TF, Ghazalpour A, Aten JE, Drake TA, Lusis AJ, Horvath S (2007). Weighted gene coexpression network analysis strategies applied to mouse weight. Mamm Genome 18: 463–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiteri C, Ding Y, French B, Tseng GC, Sibille E (2014). Beyond modules and hubs: the potential of gene coexpression networks for investigating molecular mechanisms of complex brain disorders. Genes Brain Behav 13: 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumieiro EH, Abrahao M, Jahn RS, Segretto H, Alves MT, Nannmark U et al (2010). Platelet-rich plasma in bone repair of irradiated tibiae of Wistar rats. Acta Cir Bras 25: 257–263. [DOI] [PubMed] [Google Scholar]
- He Z, Bammann H, Han D, Xie G, Khaitovich P (2014). Conserved expression of lincRNA during human and macaque prefrontal cortex development and maturation. RNA 20: 1103–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Chen X, Jin L, Xiong M (2013). Canonical correlation analysis for RNA-seq co-expression networks. Nucleic Acids Res 41: e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, da W, Sherman BT, Lempicki RA (2009. a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, da W, Sherman BT, Lempicki RA (2009. b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Iancu OD, Colville A, Darakjian P, Hitzemann R (2014). Coexpression and cosplicing network approaches for the study of mammalian brain transcriptomes. Int Rev Neurobiol 116: 73–93. [DOI] [PubMed] [Google Scholar]
- Iancu OD, Kawane S, Bottomly D, Searles R, Hitzemann R, McWeeney S (2012). Utilizing RNA-Seq data for de novo coexpression network inference. Bioinformatics 28: 1592–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen LV, Meador-Woodruff JH (2005). Abnormal striatal expression of transcripts encoding NMDA interacting PSD proteins in schizophrenia, bipolar disorder and major depression. Schizophr Res 78: 87–93. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Castellani LW, Zhou Z, Paul E, Davis R, Schadt EE et al (2012). A systems genetic analysis of high density lipoprotein metabolism and network preservation across mouse models. Biochim Biophys Acta 1821: 435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S (2007). Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol 1: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Luo R, Oldham MC, Horvath S (2011). Is my network module preserved and reproducible? PLoS Comput Biol 7: e1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Tsoi LC, Swindell WR, Gudjonsson JE, Tejasvi T, Johnston A et al (2014). Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms. J Invest Dermatol 134: 1828–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malki K, Pain O, Du Rietz E, Tosto MG, Paya-Cano J, Sandnabba KN et al (2014). Genes and gene networks implicated in aggression related behaviour. Neurogenetics 15: 255–266. [DOI] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H et al (2012). Systematic localization of common disease-associated variation in regulatory DNA. Science 337: 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGettigan PA (2013). Transcriptomics in the RNA-seq era. Curr Opin Chem Biol 17: 4–11. [DOI] [PubMed] [Google Scholar]
- Muhleisen TW, Leber M, Schulze TG, Strohmaier J, Degenhardt F, Treutlein J et al (2014). Genome-wide association study reveals two new risk loci for bipolar disorder. Nat Commun 5: 3339. [DOI] [PubMed] [Google Scholar]
- Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium (2015). Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci 18: 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S et al (2008). Functional organization of the transcriptome in human brain. Nat Neurosci 11: 1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington K, Beasley CL, Dicker P, Fagan A, English J, Pariante CM et al (2008). Prominent synaptic and metabolic abnormalities revealed by proteomic analysis of the dorsolateral prefrontal cortex in schizophrenia and bipolar disorder. Mol Psychiatry 13: 1102–1117. [DOI] [PubMed] [Google Scholar]
- Poultney CS, Samocha K, Kou Y, Liu L, Walker S, Singh T et al (2014). Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515: 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Bipolar Disorder Working Group (2011). Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet 43: 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE (2006). Novel integrative genomics strategies to identify genes for complex traits. Anim Genet 37(Suppl 1): 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511: 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre AV, Groop L, Mootha VK, Daly MJ, Altshuler D (2010). Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet 6: pii: e1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkamani A, Dean B, Schork NJ, Thomas EA (2010). Coexpression network analysis of neural tissue reveals perturbations in developmental processes in schizophrenia. Genome Res 20: 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S et al (2011). Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474: 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CA, Morrow EM, Rubenstein JL (2008). Autism and brain development. Cell 135: 396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S et al (2014). An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34: 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.