Abstract
Three-dimensional genome structure plays an important role in gene regulation. Globally, chromosomes are organized into active and inactive compartments while, at the gene level, looping interactions connect promoters to regulatory elements. Topologically associating domains (TADs), typically several hundred kilobases in size, form an intermediate level of organization. Major questions include how TADs are formed and how they are related to looping interactions between genes and regulatory elements. Here we performed a focused 5C analysis of a 2.8 Mb chromosome 7 region surrounding CFTR in a panel of cell types. We find that the same TAD boundaries are present in all cell types, indicating that TADs represent a universal chromosome architecture. Furthermore, we find that these TAD boundaries are present irrespective of the expression and looping of genes located between them. In contrast, looping interactions between promoters and regulatory elements are cell-type specific and occur mostly within TADs. This is exemplified by the CFTR promoter that in different cell types interacts with distinct sets of distal cell-type-specific regulatory elements that are all located within the same TAD. Finally, we find that long-range associations between loci located in different TADs are also detected, but these display much lower interaction frequencies than looping interactions within TADs. Interestingly, interactions between TADs are also highly cell-type-specific and often involve loci clustered around TAD boundaries. These data point to key roles of invariant TAD boundaries in constraining as well as mediating cell-type-specific long-range interactions and gene regulation.
Keywords: CFTR, gene regulation, chromatin looping, topologically associating domain
Introduction
The three-dimensional (3D) structure of chromosomes is thought to play a critical role in gene regulation.1, 2 At the nuclear level, individual chromosomes occupy their own territories,3, 4 although some intermingle where they touch.5 Larger chromosomes tend to be positioned more peripherally, whereas smaller, gene-dense chromosomes are preferentially located near other small chromosomes in the center of the nucleus.6, 7 Chromosomes themselves are compartmentalized so that active (open) and inactive (closed) chromatin domains are spatially separated. In Hi-C data this is apparent through the detection of several A-type and B-type compartments:8, 9, 10, 11 large chromatin domains (up to several megabases) that alternate along the length of the chromosomes. A-type compartments represent active regions of chromosomes as assessed by gene expression and the presence of chromatin features such as DNaseI sensitivity and the presence of active histone modifications (H3K4Me3, H3K27Ac). B-type compartments typically display little or no transcription and are composed of closed chromatin. A-type compartments might represent transcription factories where active genes cluster together, whereas inactive chromatin is partitioned to repressed nuclear sites, such as the nuclear periphery.12, 13, 14, 15
At a considerably smaller scale, chromatin organization plays a direct role in the regulation of gene expression through looping interactions between gene promoters and their distal regulatory elements, including enhancers and CTCF-bound insulator-like elements. Such locus-specific looping interactions mostly occur on a scale of a few kilobases up to 1 Mb.11, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 This is consistent with genetic analyses and functional studies that show that most regulatory elements act on a length scale of several hundred kilobases.29, 30
Recently, an additional feature of chromatin organization was described at intermediate length scales: topologically associating domains (TADs).31, 32, 33 TADs are defined as contiguous chromatin domains that display relatively high levels of self-association, separated by boundaries. Loci located in adjacent TADs interact much less frequently than those within the same TAD, suggesting that TAD boundaries act as physical insulators. TADs range in size from several hundred kilobases to a few megabases and are found across cell types and across species.31, 32 TAD boundaries often correlate with CTCF sites, and a subset of TAD-like domains have been shown to form loops where the two domain boundaries both associate with CTCF and interact with each other.11 Similarly, in C. elegans, TAD boundaries on the X chromosome directly interact with each other, and these interactions depend on the dosage-compensation complex.34 CTCF has been proposed to act as an insulator, preventing communication between elements located on either side of its binding site. Consistent with an insulation role, TAD boundaries reduce physical contacts between loci located in adjacent TADs. Supporting the idea of TAD boundaries as insulators, deletion of a TAD boundary region causes the two neighboring TADs to partially intermingle.32 TADs seem to be invariant between the small set of cell types studied to date,31, 32, 35 although differences in the internal organization of TADs have been observed in different cell lines.24, 32, 35 TADs are also structurally modulated during the cell cycle; they disappear in mitosis and reform in early G1.36, 37
Several lines of evidence indicate that TADs are important for appropriate regulation of gene expression. First, genes located within a TAD can show more correlation in their expression during cell differentiation than do genes located in different TADs.32 Second, domains of histone modifications and Lamin association, both features related to the expression status of genes, correlate with a subset of TADs.32, 38 Third, an enhancer sensor approach allowed the identification of functional domains that represent target regions of enhancers. These domains correlate well with TADs, suggesting that regulatory elements can act on entire TADs as a structural unit.30
A major question is whether TADs are defined by their boundaries only or whether their formation is determined or facilitated by looping interactions between loci, e.g., between promoters and enhancers, located inside TADs.39 Thus, a key question is whether TADs act upstream of chromatin looping or whether TADs are, at least in part, driven by looping interactions within them. Another question is whether and how TADs interact with each other, e.g., to form larger compartments,40 and whether specific elements are involved. In order to examine the relationship between TAD structure and promoter-enhancer looping interactions in more depth, we analyzed a 2.8 Mb region containing the cystic fibrosis transmembrane conductance regulator gene (CFTR [MIM: 602421]) on chromosome 7. We and others have previously used 3C to identify several regulatory elements that are located up to 200 kb from the CFTR promoter and that directly loop and interact with the promoter and with each other.18, 21 Here we analyzed the chromosome conformation of a 2.8 Mb region around CFTR by using 5C41 to identify TADs and the presence of specific looping interactions between genes and distal regulatory elements. The 5C data reveals six TAD boundaries that are located at the same positions in all cell types, indicating that they are conserved not only in different tissues but also between cancer cell lines. In contrast, looping interactions between gene promoters and distal elements are highly cell-type specific and occur most frequently within invariant TADs. Interestingly, interactions between TADs are much less frequent but are also highly cell-type specific and involve loci clustered near TAD boundaries. Our data support a model where TAD boundaries play critical roles in controlling long-range chromatin interactions both within and between TADs.
Material and Methods
Cell Culture
All cell lines were grown with antibiotic (1% penicillin-streptomycin). GM12878 lymphoblastoid cells (Coriell Cell Repositories) were grown in RPMI 1640 medium supplemented with 2 mM L-glutamine and 15% fetal bovine serum (FBS). HepG2 hepatocellular carcinoma cells (from the American Type Culture Collection [ATCC]) were grown in MEMα with 10% FBS. Caco2 colorectal adenocarcinoma cells (ATCC) were grown in MEMα with 20% FBS. Calu3 lung adenocarcinoma cells (ATCC) were grown in ATCC-formulated E-MEM with 10% FBS. Capan1 pancreas adenocarcinoma cells (ATCC) were grown in IMDM with 20% FBS. Cell densities were maintained as recommended, and Accutase (Life Technologies) was used for detaching adherent cells from plates.
Chromosome Conformation Capture Carbon Copy
5C Experiments
Chromosome conformation capture carbon copy (5C) was carried out as previously described.25, 41 We investigated a 2.8 Mb region on chromosome 7 (hg18 chr7: 115597757–118405450) containing the ENCODE region ENm001.42 The 5C experiment was designed to interrogate looping interactions between HindIII fragments containing transcription start sites (TSSs) and any other HindIII restriction fragment (distal fragments) in the target region. Libraries were generated for five cell lines: Caco2, Calu3, Capan1, GM12878, and HepG2; there were two biological replicates for each line.
5C Probe Design
5C probes were designed at HindIII restriction sites (AAGCTT) with previously developed 5C primer-design tools and made publicly available online at our My5C website.43 Probes were designed on the basis of the ENCODE manual region 1 (ENM001) design,25 and additional probes were placed throughout the region when appropriate. We also added probes to extend the analysis to include a 700 kb gene desert region located directly adjacent to ENM001. All probe locations can be found in Table S1, available with this article online. Probe settings were as follows: U-BLAST, 3; S-BLAST, 100; 15-MER, 3,000; MIN_FSIZE, 250; MAX_FSIZE, 20,000; OPT_TM, 65; and OPT_PSIZE, 40. We designed 74 reverse 5C probes and 605 forward 5C probes.
Generation of 5C Libraries
Chromosome conformation capture (3C) was performed with HindIII restriction enzyme as previously described44, 45 for Caco2, Calu3, Capan1, GM12878, and HepG2 cells with two biological replicates for each cell line. The 3C libraries were then interrogated by 5C.41, 46 We analyzed the region by pooling all probes for a final concentration of 0.5 fmol/μl. In total, 75 reverse probes and 605 forward probes were pooled for a possible 44,770 interactions. We included control probes in other regions as follows: Enm002 (Chr5), 45 reverse probes; Enm004 (Chr22), 46 reverse probes; Enm008 (Chr16), 28 probes; Enr311 (Chr14), 67 reverse probes; Enr112 (Chr13), 53 reverse probes; Enr113 (Chr4), 53 forward probes; Enr131 (Chr2), 65 forward probes; Enr232 (Chr9), 50 forward probes; Enr233 (Chr15), 52 forward probes; and Enr332 (Chr11), 42 forward probes. All probes in these ENCODE regions are the same as those previously published.25
5C was performed as described25 with the following changes: ten ligation reactions were performed for each 5C library, each containing an amount of 3C template that represents 400,000 genome equivalents and 2 fmol of each primer.
5C Read Mapping
Sequencing data were obtained from an Illumina GAIIx machine and processed by a custom pipeline for mapping and assembly of 5C interactions, as previously described.25, 43 We used an updated version of the Novoalign mapping algorithm (V2.07.11).
Measures regarding the 5C library quality, mapping efficiency, and other mapping statistics are available in Table S2. Table S3 summarizes the read depth of each 5C library. Pearson correlation coefficients between the biological replicates are available in Table S4.
5C Bias Correction
5C experiments involve a number of steps that can differ in efficiency, thereby introducing biases in efficiency of detection of interactions. These biases could be due to differences in the efficiency of crosslinking, the efficiency of restriction digestion (related to crosslinking efficiency), the efficiency of ligation (related to fragment size), the efficiency of 5C probes (related to annealing and PCR amplification), or the efficiency of DNA sequencing (related to base composition). All of these potential biases—several of which (for example, crosslinking efficiency, PCR amplification, base-composition-dependent sequencing efficiency) are common to other approaches, such as chromatin immunoprecipitation—will have an impact on the overall efficiency with which long-range interactions for a given locus (restriction fragment) can be detected. We implemented the following steps to estimate and correct for such technical biases.
Probe Filtering—Cis-Purge
Not all probes are represented equally in our 5C dataset as a result of over- and under-performance in the assay. As the first step in our data correction pipeline, we remove probes that perform significantly differently than the overall set. The relative performance of each probe is determined as follows. First, a global average relationship between interaction frequency and genomic distance is calculated via Loess smoothing for each dataset. Interaction profiles anchored on each probe across the 2.8 Mb region are then compared to this global average. If the individual Loess is more or less than 0.85 of the scaled Z score distance (a measurement of the number of standard deviations a data point is from the mean) from the average global Loess, the probe is flagged as problematic. If a probe is flagged as problematic in more than 40% of the datasets, it is removed from downstream analysis from all datasets. Using this threshold, we removed 34 probes from downstream analysis (Table S5).
Singleton Removal
As we examined the 5C datasets, we noticed several instances when the interaction between two probes was much higher than neighboring interactions by an order or magnitude or more. Although there were not many of these “blowouts,” we removed them from the dataset to avoid problems downstream during peak calling. Thus, we removed any interaction that had a Z score of 12 or more, resulting in the removal of 44 individual interactions (out of 44,770 interrogated interactions) from downstream analysis (Table S6). To calculate the Z score of a given data point, we used the following equation:
where the Z score (Z) is the read count (x) minus the population mean read count (μ) divided by the standard deviation (σ). This measurement was calculated for the given genomic distance between the corresponding loci, meaning the average and standard deviation of the read count were calculated only for points separated by the same genomic distance as the data point (x).
Coverage Correction
Once the outlier probes and blowout interactions were removed from the 5C dataset, the profiles of each probe were normalized so that they could be quantitatively compared to each other. Here, we slightly modified a previous method25 such that we used only local (cis) chromatin interaction data (within the 2.8 Mb CFTR region). First, all 5C datasets were read-normalized (each interaction value was divided by the number of reads obtained for that dataset). Second, to determine how probes might perform differently as a result of non-biological technical biases (see above), we combined all ten 5C datasets. Next, a global average relationship between interaction frequency and genomic distance was calculated with Loess smoothing, and the interaction profile detected with each probe was compared to this average. In the absence of any technical detection bias, we assumed that the overall profile of each probe was similar to the dataset-wide average profile. For each probe we then calculated a correction factor by which the profile of that probe should be lifted or lowered in order to match the average Loess profile from the entire combined dataset. We then calculated this correction factor as follows: we first converted interactions into Z scores by using the Loess values for the corresponding genomic distance as average and the standard deviation around that average at the corresponding genomic distance (as described above and in Sanyal et al.25). Zeros were excluded. We then calculated the average Z score for each row and column of the interaction map (corresponding to all interactions detected with individual forward and reverse 5C probes) but left out the top and bottom 5% of values. This calculation yielded average Z score values for each probe, and we used these as correction factors. To correct individual interactions, detected by pairs of probes, we combined correction factors as follows. The corresponding averaged Z score values of the two probes were summed. The corrected interaction frequency was then calculated as
The Loess value and standard deviation were calculated for each genomic distance as described above and in Sanyal et al.25
We used this bias-correction approach to correct each of the ten individual read-normalized datasets to produce the final bias-corrected datasets. By combining the datasets before correction to calculate correction factors, we reduced the risk of overcorrecting certain probes that were truly giving high biological signals in one cell line. When the datasets were combined, this biologically high signal was averaged with the other lower signals, giving that interaction a less stringent correction. If correction factors were calculated for each dataset separately, the interaction profile would be penalized for giving such a high interaction and would be corrected too harshly.
Data corrections did not change the overall structure of the data. In fact, analysis of specific probes from raw and corrected data show few differences in their profiles, indicating that probes display only minor differences in detection efficiency. Furthermore, domains detected with the corrected 5C interaction maps are very similar to those obtained with a completely independently obtained high-resolution Hi-C interaction map.11
Insulation Index
To define regions of our dataset that contain TAD boundaries, we calculated an insulation score along the locus.34, 47 This method is based on the concept that TAD boundaries act as physical insulators that prevent or inhibit interaction across them. First, 5C data were binned at 100 kb with a 10× (10 kb) step size. Next, we calculated for each bin the combined number of observed interactions across it by summing all interactions between loci located up to 250 kb upstream and loci located up to 250 kb downstream of the bin. This sum was then calculated for each bin along the 2.8 Mb locus, then divided by the average sum of all bins to give insulation scores. We plotted insulation scores along the locus to obtain an insulation profile. (Figure S11). Local minima in this profile represent bins that display the largest insulation and thus indicate the positions of TAD boundaries. We detected local minima in the insulation profiles by identifying the bins with the lowest insulation scores in a local 490 kb window. We then set the midpoint of this low-value bin as the boundary.
Peak Calling
To detect statistically significant looping interactions at the restriction-fragment level, we applied a “5C peak calling” algorithm as described before25 with the following modifications. We called peaks on three different subsets of the data—all the data, intra-TAD data, and inter-TAD data (Figures S14–S16 and Table S7). Peaks called on intra-TAD and inter-TAD data were called after the TAD boundaries were defined as described above. Peak calling for intra-TAD and inter-TAD interactions was done separately because the background signal is overall higher within TADs than between TADs. Thus, by performing peak calling separately for intra-TAD and inter-TAD signals (and by using different background estimations for the two sets of interactions), we avoid calling many false positives for intra-TAD signals (that tend to be higher) and suffering false negatives for inter-TAD signals (that tend to be of lower frequency). Peaks were defined as signals that are significantly higher than expected. Expected values were calculated as follows: for peak calling for the complete dataset, we calculated the average interaction frequency for each genomic distance by using Loess smoothing (alpha value 0.01). This provides a weighted average and a weighted standard deviation at each genomic distance. For peak calling within or between TADs separately, we calculated the average interaction frequency for each genomic distance by using Loess smoothing with only intra-TAD or inter-TAD data, respectively (alpha value 0.01). We assume the large majority of interactions were not significant looping interactions, and therefore we interpret this weighted average as the expected 5C signal for a given genomic distance. We then transformed observed 5C signals into a Z score by calculating the (observed value − expected value)/standard deviation as described earlier, where the observed value is the detected 5C signal for a specific interaction, the expected value is the calculated weighted average of 5C signals for a specific genomic distance, and the standard deviation is the calculated weighted standard deviation of 5C signals for the corresponding genomic distance. Once the Z scores were calculated, their distribution was fit to a Weibull distribution. p values were calculated for each Z score and transformed into q values for false discovery rate (FDR) analysis. We used the “qvalue” package from R (qvalue.cal [siggenes]) to compute the q values for the given set of p values determined from the fit to the Weibull distribution. We used a stringent FDR threshold of 0.001%. We chose this threshold because it was the most stringent FDR at which all known “gold standard” looping interactions in the CFTR locus were detected in the appropriate cell lines by previous 3C studies, and these interactions were not deemed significant in cell lines that do not express the genes and were shown by 3C to not display these long-range interactions.18, 21 We called peaks in each 5C biological replicate separately and then took only the peaks that intersect across replicates as our final list of significant looping interactions. Using this cutoff, the fraction of peaks that we observed in both replicates was comparable to that in our previous 5C studies.25 We note that interactions that were statistically significant in only one replicate were still significantly more frequent in the second replicate than were interactions that were not significant in either replicate (Figure S13), similar to results from our previously published 5C studies.25 This suggests that these interactions may in fact be bona fide looping interactions but that the typically low signal-to-noise ratio in these experiments prevented their reproducible detection. By limiting our analysis to long-range interactions that were statistically significant at a very stringent FDR in both replicates we restricted our analysis to the strongest signals, but might have introduced false negatives.
RT-PCR
Gene expression levels were determined with qRT-PCR. Three technical replicates and three biological replicates were performed for each cell line. Gene expression levels were analyzed with a StepOnePlus instrument (Applied Biosystems) with the Power SYBR Green RNA-to-Ct 1-step kit (Life Technologies). Results were normalized to HPRT as an internal control. Any results with a Ct value higher than 34 were considered “not expressed.” RT-PCR primers were designed in neighboring exons with the Primer3 tool. We assayed primers for effectiveness by checking their titration ability and whether they gave a single melt curve. Primers used for this experiment can be found in Table S8.
Results
Generation of 5C Chromatin Interaction Maps in Five Cell Lines
To determine the relationship between the location of TADs and the presence of chromatin looping interactions between genes and regulatory elements, we applied 5C,41 the first method that combines 3C with a variant of hybrid capture. 5C is particularly well suited for such analysis as it allows cost-effective simultaneous high-resolution (single restriction fragment) detection of looping interactions25 and analysis of large chromosomal domains to identify TADs.32 We applied 5C to analyze the conformation of a 2.8 Mb domain on human chromosome 7 (Figure 1A). We chose this region because it is centered on the cystic fibrosis transmembrane conductance regulator (CFTR) gene, where we and others previously identified several cell-type-specific looping interactions between the CFTR promoter and distal enhancers and CTCF-bound elements.18, 21 Additionally, the region is sufficiently large to cover several TADs.31
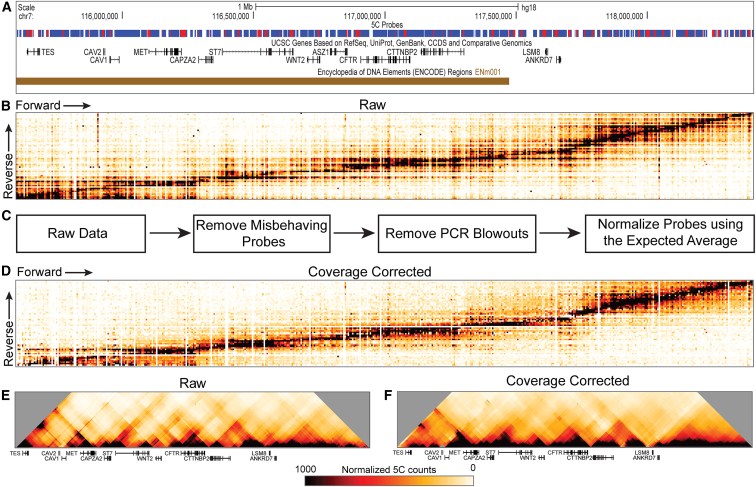
Figure 1.
Generation of 5C Chromatin Interaction Maps
(A) 5C region showing probe design. Red fragments represent reverse probes. Blue fragments represent forward probes.
(B) Raw 5C data plotted as all reverse primers versus all forward primers. Color scale: white to orange to red to black, where white represents no detected interaction frequency and black represents the highest interaction frequency.
(C) Steps in our data-correction pipeline. Misbehaving probes are removed first, then PCR blowouts are removed. The final step normalizes all probes to each other (coverage correction). For full details, see Material and Methods.
(D) 5C data after coverage correction. White stripes running through the heatmap indicate misbehaving primers that were removed in the correction steps.
(E) Binned heatmap of raw data, before they were run through the correction pipeline.
(F) Binned final 5C data after data correction. Data are binned in 100 kb windows with a 10x step. All heatmaps are plotted on the same scale throughout the paper, unless indicated.
We used a 5C capture probe set (Figure 1A) that places reverse probes on gene promoters and forward probes on the remaining restriction fragments in the region. This design allows capturing of long-range interactions between gene promoters and surrounding chromatin, e.g., distal enhancers. This design contains 74 reverse probes and 605 forward probes for a possible 44,770 interrogated interactions.
We selected a panel of cell types, including three cell lines (Caco2, Calu3, and Capan1) known to express CFTR, to study. These cell lines are derived from different locations in the body: Caco2 cells are colon-derived colorectal adenocarcinoma cells; Calu3 cells are lung-derived adenocarcinoma cells; and Capan1 cells are pancreas-derived adenocarcinoma cells. It is likely that distinct cell-type-specific enhancers are involved in the expression of CFTR in these diverse tissues. We also included two cell lines that do not express this gene: the lymphoblastoid cell line GM12878 and the liver-derived hepatocellular carcinoma cell line HepG2.
Figure 1B shows raw 5C data obtained from GM12878 cells. Reverse probes are plotted as rows and forward probes as columns in interaction heatmaps. Each intersection between a reverse and forward probe represents a measured interaction frequency between two genomic loci (restriction fragments). As expected, neighboring genomic regions interact with each other frequently, creating a black “diagonal” through the middle of the heatmap. As the genomic distance between fragments increases, the interaction frequency predictably decreases.44, 48
5C data were corrected for detection biases as we did previously,25 with some modifications (see Material and Methods). First, we removed data obtained with probes that performed aberrantly (in that they reported interaction frequencies that were either too high or too low; see Figure 1C; Material and Methods). Second, individual interactions were removed when deemed to be outliers (Figure 1C and Material and Methods); and third, data were corrected for any remaining minor variations in probe efficiency. The corrected data are displayed in Figure 1D. Figures 1E and 1F show the same data as Figures 1B and 1D, but here the chromatin interaction maps are binned and display the 5C region versus itself (all heatmaps in this paper are binned in 100 kb bins with a 10 kb step size). Raw and corrected data for all cell lines and replicates can be found in Figures S1–S10. Pearson correlation analysis showed that replicates of the same cell line are highly correlated and also tend to be more correlated with each other than with replicates from different cell lines, as expected (Table S4).
Identification of TADs
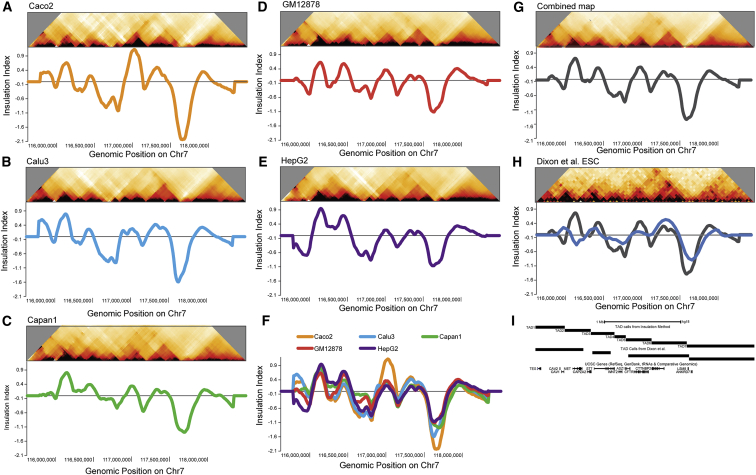
Hundreds of kilobases in size, TADs are consecutive regions wherein loci associate and mix more frequently with each other than with loci located in adjacent TADs.31, 32 Visual inspection of the binned 5C interaction maps indicates that TAD structures are readily detected as triangles of strong self-association along the region (Figures 1E and 1F and Figure 2). We employed a straightforward, previously published approach to quantify the pattern of TAD signals along the locus.34, 47 The approach is based on the observation that TAD boundaries represent loci across which few long-range chromatin associations, i.e., associations between loci located upstream and downstream of the boundary, occur.31 We quantified the relative frequency of interactions occurring across each bin throughout the 2.8 Mb region.34 We refer to this number as the insulation score of a genomic location. We then plotted these scores along the region to obtain an “insulation profile” for each cell line (Figures 2A–2E; Figure S11; see Material and Methods). Minima in the insulation profile represent TAD boundaries, across which interactions occur at a low frequency. Insulation profiles and the locations of TAD boundaries were not dependent on the size of the window used for calculation of the insulation score (Figure S11). Because we used binned data with a step size of 10 kb, the resolution of minima detection is around 10 kb+/− 10 kb (as shown in Crane et al.34). Figures 2A–2E show binned 5C interaction maps for all cell lines and their insulation profiles plotted below. Strikingly, although the amplitude of the signal varies, the insulation profiles of all five cell lines are overall very similar in that TAD boundaries are present at the same locations (Figure 2F and Figure S11). Furthermore, we compared our 5C data to previously published Hi-C data in human embryonic stem cell lines (ESCs)31 (Figure 2H). The overall TAD organization in ESCs as detected by Hi-C and quantified by our insulation score approach is again very similar to the organization we detected by 5C (Figure 2H). We note that there are some differences (e.g., the region around position 116,250,000 bp). These could represent real differences in TAD boundary positions between ESCs and differentiated cells, or they could reflect experiment-dependent variations in interaction patterns, e.g., as a result of the lower resolution of the Hi-C data.31
Figure 2.
Detection of TADs via Insulation Profiles
(A–E) Heatmaps and corresponding insulation profiles for each cell line: Caco2, Calu3, Capan1, GM12878, and HepG2. Minima in the profiles represent boundaries between two TADs.
(F) The insulation profiles for all five cell lines are plotted in one graph.
(G) The heatmap is a combined map of all our data. Below is the average insulation profile of all the data.
(H) Our insulation index method run on human embryonic stem cell Hi-C data from Dixon et al., 2012.31 This heatmap is binned as in the Dixon et al. paper with 40 kb bins. Its color scale is from 0 to 35.
(I) The 5C region TAD calls based on our insulation profile are compared with the TAD calls made with the method in Dixon et al.
These results confirm and extend earlier observations that TADs are similar, but not always identical, in different cell types.31, 32 Because the insulation profiles are extremely similar between the five differentiated cell lines studied here (Figure 2F and Figure S11), we created a consensus insulation profile by calculating the average profile across all five differentiated cell lines to use for downstream analysis (Figure 2G). Six TAD boundaries define seven TADs in the regions (Figure 2I).
Previous studies have shown that, in general, TAD boundaries are enriched in gene promoters and CTCF sites, as well as in several other features.11, 31, 49, 50 In order to determine whether the TAD boundaries we identify here also contain promoters and CTCF sites, we examined the TAD boundary regions to determine what features were present. We noticed that the TAD boundaries in this region were not very close to gene promoters, but rather were near the 3′ ends of genes. The midpoint of the boundary between TADs 1 and 2 is around 10 kb from the 3′ end of the Caveolin 1 gene (CAV1 [MIM: 601047]). The closest promoter is 45 kb away. Additionally, the boundary between TADs 2 and 3 occurs 9 kb from the 3′ end of the Capping Protein Alpha 2 gene (CAPZA2 [MIM: 601571]), and the boundary between TADs 3 and 4 occurs 3.5 kb from the 3′ end of the Suppressor of Tumorigenicity 7 gene (ST7 [MIM: 600833]). Also, the boundary between TADs 4 and 5 occurs within the Ankyrin Repeat, SAM, and Basic Leucine Zipper Domain-containing 1 gene (ASZ1 [MIM: 605797]), 7 kb from its termination site but 57 kb from its promoter. The boundary between TADs 5 and 6 contains the 3′ end of the Coractin-binding Protein 2 gene (CTTNBP2 [MIM: 609772]). Located 13.5 kb from the LSM8 Protein gene promoter (LSM8 [MIM: 607288]), the boundary between TADs 6 and 7 is the only one that is closer to a gene promoter than to a gene end. Thus, although there is genome-wide enrichment of TAD boundaries near gene promoters, we conclude that this is not the case for every TAD boundary and suggest that gene promoters are not essential for TAD boundary formation. We do find that all six boundaries are located very close to CTCF sites (< 10–20 kb, which is within the resolution of the binned 5C data used for boundary calling), as reported before,31 and that many of these sites are at or near the 3′ end of genes (Figure S11). Furthermore, we observed that the TADs we identify here closely align with a set of CTCF-CTCF loops detected by high-resolution Hi-C followed by extremely deep sequencing.11 Thus, our data suggest that CTCF binding, and not promoter sequences, contributes to TAD boundary formation. However, we also note that many CTCF sites are found within TADs as well, indicating that CTCF is not sufficient for boundary formation, consistent with previous studies.32, 50, 51, 52
TAD Positions Are Not Affected by Cell Type-Specific Gene Expression
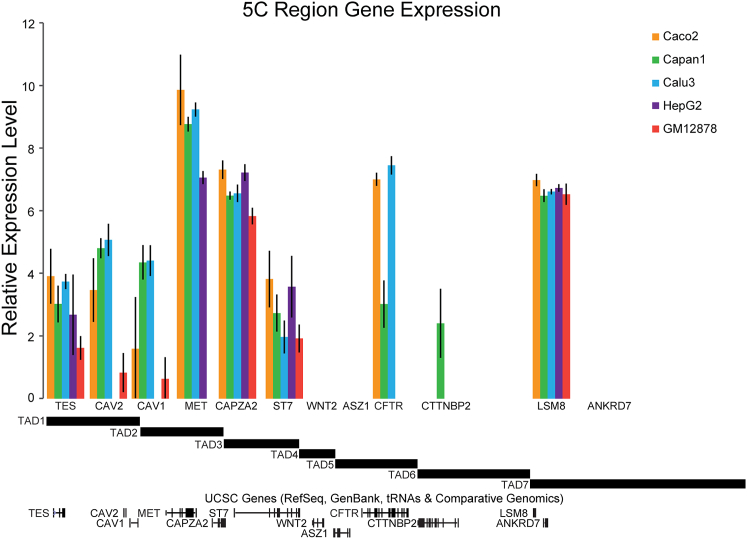
To investigate the relationship between TADs and gene expression, we measured the expression level of all genes in the 2.8 Mb region in the five cell lines studied (Figure 3). Interestingly, we found that several TADs (TADs 4–6) were transcriptionally silent in some cell lines but displayed transcription of at least one gene in other cell lines. Yet TAD boundaries are the same whether or not the TAD contains an expressed gene. This is particularly well illustrated in GM12878 and HepG2 cells where TADs 4, 5, and 6 are transcriptionally inactive, but the same set of TAD boundaries that separate them is present in cells that do express genes located in these TADs. On the other hand, TADs 1 and 2 have at least one gene active in each cell line, but the precise set of genes that is active differs between different cell lines, and TAD boundaries are invariant as well. Together, these observations indicate that TAD boundaries occur irrespective of gene transcription and are not determined by the expression status of genes located within TADs.
Figure 3.
Gene Expression within the 5C Region Is Not Related to TAD Structure
Relative gene expression is plotted for the five cell types studied. The TAD structure of the locus is shown below the graph for reference.
Identification of Long-Range Looping Interactions
Next, we set out to identify specific and statistically significant long-range interactions such as promoter-enhancer contacts throughout the 2.8 Mb domain. Previously, we developed and applied a statistical methodology to identify pair-wise interactions that occur between individual restriction fragments in a 5C dataset significantly more frequently than expected.25 The approach first uses the complete dataset to calculate the baseline contact frequency of pairs of loci (restriction fragments) as expected on the basis of their genomic site separation. Then, individual interactions between pairs of loci are identified that are significantly above this baseline. Importantly, and as we have stated before,25 not all statistically significant interactions identified in this manner represent specific point-to-point looping contacts: a pair of loci can also interact more frequently than expected when they are brought into relatively close proximity as a result of looping between nearby sites, e.g., when they are located within a loop formed by a different pair of loci11, 25, 53 (see below for discussion).
One limitation of previous analyses of long-range looping interactions25, 54 is that they did not take the presence of TADs into account when calculating the expected baseline interaction frequency, which biases detection of intra-TAD looping because of the overall increased interaction frequency within these domains. Here we further refined our long-range detection approach by taking into consideration the presence of TADs. Specifically, we calculated the expected baseline interactions separately for interactions occurring within and between TADs. For comparison, we also performed our peak-calling approach on the entire dataset while ignoring TADs, as we did previously (Material and Methods and Sanyal et al.25). When an FDR of 0.001% is used, the two different approaches give similar but not identical sets of statistically significant looping interactions, indicating that the presence of TADs has some impact on our ability to detect statistically significant signals (Figure S17). For our analyses, we used the set of significant interactions that were detected in two independent biological replicates when we explicitly took TADs into consideration (Figures S15 and S16).
Overall, we found that reproducibility of statistically significant interactions is very comparable to our previous 5C studies.24, 25 Interestingly, intra-TAD looping interactions are more reproducible that inter-TAD interactions, possibly because these tend to display much higher interaction frequencies (see below; Figures S15 and S16).
Loci located within a TAD generally interact more frequently than pairs of loci separated by the same genomic distance but located in different TADs.11, 31, 32, 49, 54 We can visualize this for our 5C dataset by plotting all intra-TAD and inter-TAD interactions as a function of their genomic site separation (Figure 4A and Figure S12). This plot shows that at a given genomic distance, any intra-TAD interactions occur more frequently than inter-TAD interactions. We also note that, up to several hundred kilobases, the interaction frequency decays with genomic distance as a power law with a slope of around −0.5. As we have shown before,36 this slope is consistent with the formation of arrays of consecutive, non-overlapping chromatin loops, suggesting similar consecutive loop formation at that length scale within TADs (see below).
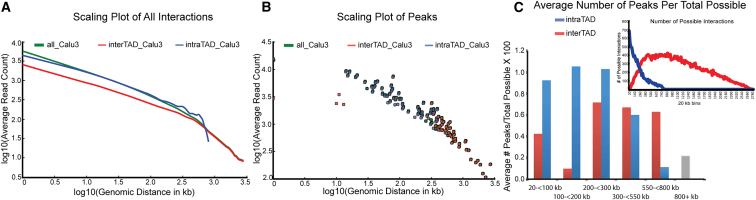
Figure 4.
Significant and Strong Long-Range Interactions Occur Mostly within TADs
(A) Scaling plot showing the average read count versus genomic distance in Calu3 cells. Interactions from the whole dataset are green, interactions from the intra-TAD space are blue, and interactions from the inter-TAD space are red.
(B) Scaling plot showing only significant long-range interactions in the three different spaces in Calu3 cells. Coloring is the same as in (C). Scaling plots for all cell lines are shown in Figure S12.
(C) The number of significant interactions (across all cell types) is plotted for different genomic distances; only the intra-TAD and inter-TAD data are used. Significant long-range inter-TAD interactions between loci separated by less than 20 kb are not included because these represent regions around TAD boundaries that are particularly frequently involved in interactions (see Figure 6F). Inset: The total number of interrogated interactions for intra-TAD (blue) and inter-TAD (red) datasets is plotted versus genomic distance.
As a result, the interaction frequency of statistically significant looping interactions within TADs is also higher than the interaction frequency of statistically significant inter-TAD looping interactions, even when they involve loci separated by the same genome distance (Figure 4B and Figure S12).
We see that the majority of significant inter-TAD interactions occur over distances of 200 kb and greater, whereas intra-TAD interactions are mostly evenly spread between distances of 20–300 kb (Figure 4C). To explore this further, we corrected for differences in numbers of interrogated inter-TAD and intra-TAD interactions at different genomic distances (Figure 4C, inset) by calculating the percentage of interrogated inter- or intra-TAD interactions that were statistically significant as a function of genomic distance between the loci. Note that our peak calling approach corrects for differences in the baseline frequency of interaction within and between TADs. Strikingly, we found that, up to approximately 200 kb, significant long-range interactions occur almost exclusively within TADs. In absolute numbers, for loci separated by 20 kb up to 200 kb, we found that 184 out of 192 significant interactions occurred between pairs of loci located within the same TAD. Furthermore, these intra-TAD interactions were relatively strong. Inter-TAD interactions were observed mostly for loci separated by more than 200 kb. These interactions were much less strong than intra-TAD interactions (Figure 4). Combined, these results show that frequent looping interactions between promoters and distal loci occur mostly within TADs and over several hundred kilobases, consistent with the enhancer-promoter connectivity predicted by independent computational methods,55 whereas much less frequent, but statistically significant, interactions occur between loci located in different TADs, and those involve loci separated by much larger (>200 kb) distances.
When we compared looping interactions among the five cell lines, we found that the majority of interactions were cell-type specific, a minority of interactions were observed in two or more cell lines, and only a handful of interactions were observed in all five cell lines. This holds for interactions within TADs and for interactions that occur between TADs (see below). This is consistent with other 5C analyses.24, 25 Thus, looping interactions within and between TADs are highly tissue specific, whereas TAD boundaries are largely cell-type invariant.
CFTR Promoter-Enhancer Loops Occur within a Single TAD
We next focused our analysis on long-range interactions of the CFTR gene. Previous studies have identified a number of putative cell-type-specific regulatory elements within and flanking CFTR (see reviews56, 57). These were identified as DNase I hypersensitive sites (DHSs), often representing nucleosome-free DNA sequences that can interact with transcription factors. These elements are spread out over several hundred kilobases, making it difficult to predict what gene(s) they regulate. Interestingly, we found that all these previously identified elements were located within a single TAD. Some of these elements, located 44 kb and 35 kb upstream of the promoter, in introns 1 and 11, and +202 kb downstream from the TSS (the +15.6 kb DHS in58), have been shown to act as enhancer elements when tested in luciferase assays and contain chromatin marks typically associated with enhancer activity in CFTR-expressing cells.18, 21, 59, 60 3C studies revealed that several of these enhancer elements (in introns 1 and 11 and +202 kb downstream from the TSS) directly loop to the CFTR promoter, and to each other specifically in cells that express CFTR, but not (or much less frequently) in cells that do not express CFTR.18, 21, 59, 60 These studies strongly suggest that these elements regulate CFTR. The CFTR promoter was also found to interact with several CTCF-bound sites located ∼21 and ∼80 kb upstream of the gene in Caco2 cells, but not in non-expressing cells even though CTCF is bound to the same elements in those cells.18
The 5C analysis presented here reproduced most of the previously identified looping interactions between the CFTR promoter and distal regulatory elements, validating the approach. First, in all three CFTR-expressing cells (Caco2, Capan1, and Calu3), we detected significant interaction frequencies between the CFTR promoter and the known CFTR enhancer located within intron 11 (108 kb downstream of the TSS) (Figures 5M–5O). In cells that do not express CFTR, this interaction is either strongly reduced (GM12878) (Figure 5P) or not significant (HepG2) (Figure 5Q).
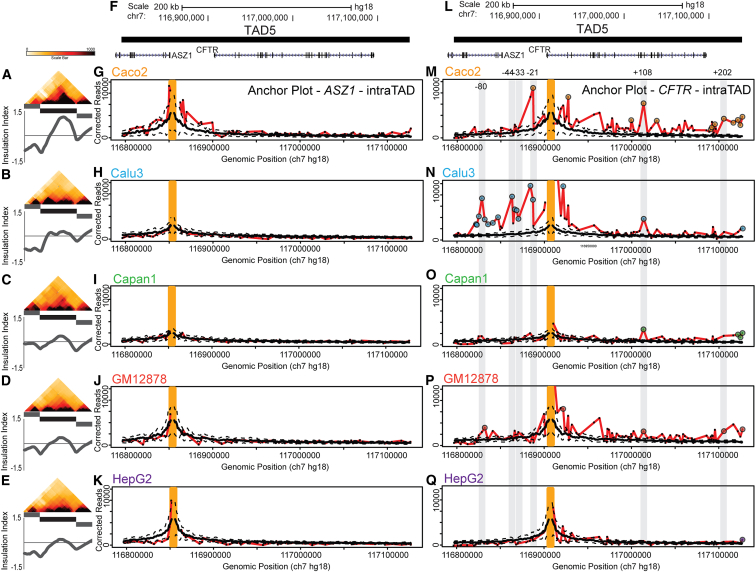
Figure 5.
CFTR Regulatory Elements Are Contained within One TAD
(A–E) Zoomed-in view of the 5C chromatin interaction map. TAD5 is highlighted by the black bar. Insulation profiles for each heatmap section are displayed below each heatmap.
(F and L) Genome browser snapshot of TAD5, showing the two genes ASZ1 and CFTR located in the TAD.
(G–K) 3C-style chromatin interaction profile anchored on the ASZ1 promoter (probe REV_404). This gene is not expressed in any of the five cell lines.
(M–Q) 3C-style chromatin-interaction profile anchored on the CFTR promoter (REV_421). This gene is expressed in Caco2, Calu3, and Capan1 cell lines. For (G–K) and (M–Q), the orange bar represents the location of the anchoring interaction. The dotted black lines represent the Loess standard deviation, and the thick black line represents the Loess average. The red line represents the interactions of the genomic fragment indicated by the orange line with all other interrogated fragments in the region. Significant interactions are indicated by large circles. Grey lines and genomic coordinates indicate locations (kb from the promoter) of known CFTR enhancer elements.
Second, in Caco2 cells the CFTR promoter interacts with a region that falls just downstream of the gene (202 kb downstream of the promoter) and contains a known enhancer, consistent with previous 3C studies performed with this cell line18, 21 (Figure 5M). This interaction is not observed in CFTR-expressing Calu3 and Capan1 cells or in the non-expressing GM12878 and HepG2 cells.
Third, in Calu3 cells the promoter loops to several additional sites that are located upstream of the promoter and that had not been shown to engage in looping interactions before. We note that in Calu3 cells the entire region up to around 100 kb upstream is interacting frequently, and statistically significantly, with the CFTR promoter. Several obvious peaks in the interaction profile stand out. The other weaker, but statistically significant, interactions probably represent indirect interactions that are brought into relatively close proximity with the CFTR promoter as a result of the other prominent looping interactions present in the region. Interestingly, one prominent peak in the Calu3 interaction profile involves sites located ∼35–44 kb upstream of the promoter. These sites contain a pair of previously identified lung-specific CFTR enhancers.59, 60 These elements are active in lung-derived Calu3 cells, as indicated by the presence of DNaseI-hypersensitive sites.61
Finally, in Calu3 and Caco2 cells, but not in Capan1 cells, the active CFTR promoter engages in looping interactions with CTCF sites upstream of the promoter (Figures 5M and 5N). In both cell lines the promoter loops to a site that is located ∼21 kb upstream and binds the CTCF protein, consistent with 3C experiments.18, 21 Another significant interaction occurs at and around a site that is located ∼80 kb upstream and that also corresponds to a CTCF-bound element. We previously used 3C to show that this site weakly interacts with the expressed CFTR promoter in some cell lines.18
From these analyses we conclude that the CFTR promoter engages in several cell-type-specific long-range looping interactions with cell-type-specific distal enhancers that are active in the corresponding cell line and that are all located within the CFTR TAD. The promoter also interacts with several distal CTCF-bound elements, and it seems these elements, located within the TAD, appear not to prevent looping of the promoter with enhancer elements located more distally. The role of these cell-type-specific interactions with otherwise tissue-invariant CTCF sites is currently not known (see Discussion). Interestingly, we note that in all cell lines studied, the CFTR promoter is interacting with the right boundary of its TAD. As described below, TAD boundaries often engage in long-range interactions, including with elements located in other TADs. The relevance of these interactions is currently not known.
We note that ASZ1, located in the same TAD as CFTR, is inactive in all cell lines and does not engage in any significant long-range looping interactions (Figures 5G–5K). Thus, distal elements that interact with the CFTR promoter do not interact with other (inactive) promoters in the domain.
Previous studies have shown that the internal organization of TADs depends on expression status.32, 38 Therefore, we were interested to determine the impact of the diverse intra-TAD looping interactions in the five cell lines on the appearance of the overall binned chromatin interaction maps of the TAD. Figures 5A–5E show zoomed-in views of TAD5, with the insulation score of this zoomed-in region plotted below. TAD5 is indicated as the black box, and the neighboring TADs 4 and 6 are indicated as gray boxes. The numerous significant looping interactions between the CFTR promoter and elements located downstream of the gene promoter in Caco2 cells (Figure 5M) lead to the presence of a dark triangle located at the right within TAD5 (Figure 5A). The numerous significant interactions present between the CFTR promoter and the upstream region in Calu3 cells lead to the black triangle located toward the left part of TAD5 (Figure 5B). The relatively flat 3C profiles and limited looping in Capan1, GM12878, and HepG2 cells are represented by lack of a clear intra-TAD structure in their corresponding heatmaps (Figures 5C–5E). Although the looping and interaction patterns within TAD5 differ between cell lines, the boundary regions of this TAD are clearly defined in all cell lines. Thus, TAD boundaries are invariant, and the internal organization of TADs is dependent on the pattern of cell-type-specific looping interactions between promoters and regulatory elements within the domain.
Long-Range Looping Interactions within Other TADs in the Region
We detected statistically significant long-range looping interactions within the other TADs. Interaction profiles for all genes are shown in Figures S18–S24. We found that active gene promoters were more likely to be involved in long-range interactions than promoters that were not expressed (Figures 6G and 6H), consistent with our earlier findings.25 For instance, CTTNBP2 is only expressed in Capan1 cells, and it is engaged in long-range looping interactions only in that cell type (Figure S23). However, the correlation between expression and looping is not always absolute. For example, CAV2 interacts with several elements throughout the TAD in Calu3 and Capan1 cells that both express the gene (Figure S18). Yet no looping interactions were detected in Caco2 and GM12878 cells even though the gene is expressed in those cell types (Figure S18). There could be technical reasons for this (e.g., no 5C probe was included for a restriction fragment that contains Caco2 or GM12878-specific elements or the interaction is simply missed due to false-negatives). Alternatively, in some cell types it might be that no distal looping interactions are required for gene activation.
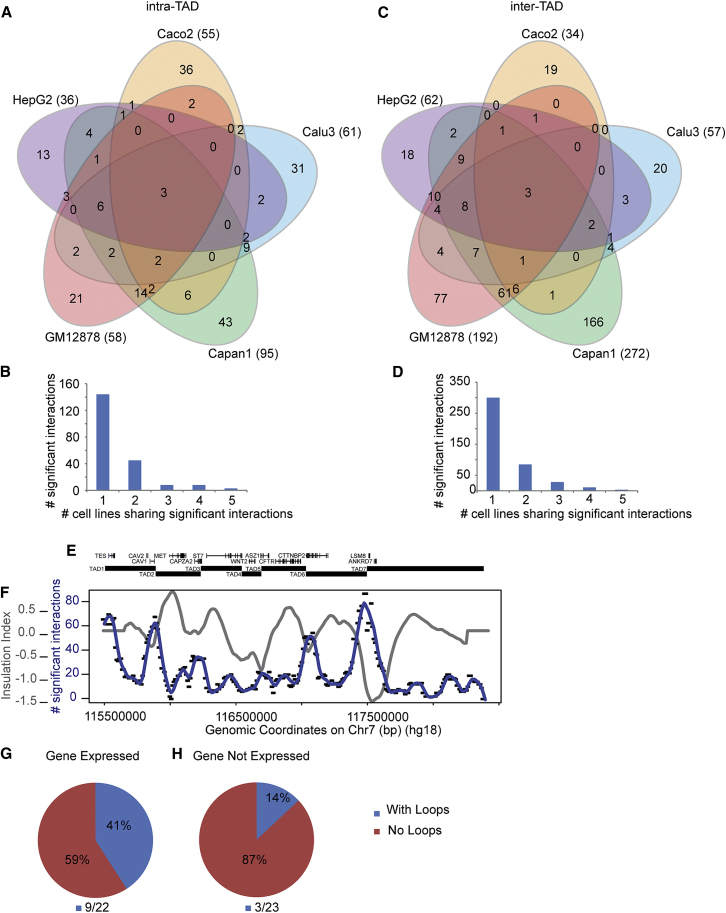
Figure 6.
Most Interactions Are Cell-Type Specific
(A) Venn diagram showing all significant interactions detected in the five cell types in the intra-TAD space.
(B) Bar graph displaying how many significant intra-TAD interactions are shared between cell types.
(C) Venn diagram showing all significant interactions detected in the five cell types in inter-TAD space.
(D) Bar graph displaying how many significant inter-TAD interactions are shared between cell types.
(E) Diagram of the region under study; genes and TADs are indicated, and the diagram is aligned with the plot in (F).
(F) Plot displaying the average insulation profile (gray) and the average number of significant inter-TAD interactions for loci along the region (blue line, 100 kb window, step size 10 kb). TAD boundaries (minima in insulation profile) display high levels of inter-TAD interactions.
(G) Pie chart showing the number of expressed genes that show significant looping interactions in our datasets.
(H) Pie chart showing the number of non-expressed genes that show significant looping in our datasets.
On the basis of previous analyses from our lab and those of others,11, 24, 25, 54 and the detailed analysis of the CFTR locus described above, it is likely that the looping interactions in these TADs also involve putative gene regulatory elements, e.g., enhancers, and architectural elements such as CTCF-bound elements. Indeed, many of the looping interactions overlap sites that are bound by CTCF or that contain chromatin features that indicate the presence of regulatory elements. In GM12878 cells, for which there is information on the locations of predicted regulatory elements generated by the ENCODE consortium,62, 63 we find that 99 HindIII fragments that were interrogated in our 5C analysis overlap predicted enhancers. Out of this set, our 5C study identified 26 that are engaged in statistically significant interactions with promoters. In this cell type the fraction of statistically significant looping elements that involve distal enhancers is 20% (26 out of 129 fragments that display significant interactions), which is comparable to our earlier larger-scale analysis of long-range chromatin interactions.25 Whether any of these elements are functional enhancers is currently not known. Furthermore, we find that nine out of 25 interrogated CTCF-bound insulator elements show statistically significant long-range interactions in GM12878 cells, again comparable to results from our earlier studies.
Long-Range Interactions between TADs
Our analysis also identified significant long-range looping interactions between loci located in different TADs. As mentioned above, these interactions tend to be much longer range (between loci separated by more than several hundred kb), to display much lower contact frequency, and to be less reproducible (Figure 4C and Figures S15 and S16). Interestingly, these interactions were again highly cell-type specific, as were the interactions within TADs (Figures 6A–6D). In addition, we found that many of the inter-TAD interactions occur in zones. For instance, the CFTR promoter interacts with an entire region located at the boundary between TADs 5 and 6 and with a region near the boundary between TADs 6 and 7 (Figure S22), and these regions of elevated interactions are readily seen in the chromatin interaction map (Figures S1 and S2). Thus, inter-TAD interactions can appear as interaction zones, rather than point-to-point looping interactions. Strikingly, when we examined which loci engage in such inter-TAD interactions, we found a strong correlation with TAD boundaries (Figures 6E and 6F). Thus, while the positions of TAD boundaries are invariant, loci located near them engage in highly cell-type-specific, but rather weak, long-range associations with loci located in other TADs.
Discussion
TADs Do Not Depend on Gene Expression or Intra-TAD Looping between Promoters and Distal Elements
An obvious feature of our data is the set of clearly defined TAD boundaries present in all cell lines we studied. We identified six boundaries defining seven TADs in our region of study. We noticed that a majority (5/6) of these TAD boundaries were located very close to the 3′ end of genes, in contrast to earlier findings that TAD boundaries are marked by gene promoters.31, 38, 49 Thus, although boundaries are enriched for promoters genome-wide, they do not define them. We did find that all TAD boundaries were very close to CTCF-bound sites, confirming the critical role of this protein in boundary formation.
TADs were present in all cell lines we examined, regardless of gene expression status or the presence of significant long-range looping interactions (Figure 3). For example, in HepG2 and GM12878 cells a set of four adjacent genes (Wingless-type MMTV Integration Site Family, Member 2 [WNT2 (MIM: 147870)], ASZ1, CFTR and CTTNBP2) spanning three TADs were not expressed and do not engage in looping within the TADs. Yet, the boundaries between TADs 4, 5, and 6 were still clearly demarcated in both these cell lines. Thus, TADs are not maintained or determined by gene expression within them or by looping interactions between promoters and distal elements. A particularly clear example of this phenomenon is provided by the CFTR-containing TAD5 (Figure 5), which shows that although looping interactions inside that TAD differ between the five cell lines, the TAD boundaries remain constant. We note that a recent study showed that at least a subset of TADs represent looped structures that result from interactions between their boundaries, and these mostly involve CTCF sites.11 Given that many CTCF sites are bound across cell types, these interactions might explain the cell-type-invariant nature of some TADs.
Chromatin Looping Is Correlated with Expression, but not all Actively Transcribed Genes Have Looping Interactions
We have shown that expressed genes have significantly more looping interactions than non-expressed genes (Figures 6G and 6H), consistent with earlier findings.25 However, it is clearly not the case that all expressed genes have looping interactions. Interestingly, some expressed genes have looping interactions in certain cell lines but not in others. For example, the CAV2 promoter is engaged in looping interactions in Calu3 and Capan1 lines, but not in Caco2 or GM12878 lines, even though it is expressed in all four lines. Similarly, MET Protooncogene (MET [MIM: 164860]) is expressed in all cell lines studied except GM12878 and shows looping interactions in all except Caco2 cells.
Looping between Promoters and Distal Elements Occurs Mostly within TADs
We found that looping interactions between loci separated by up to several hundred kilobases occurred mostly within TADs and only very rarely between TADs. It is important to emphasize that our peak-calling approach explicitly took the presence and location of TADs into account by using different expected background interaction frequencies within and between TADs. Thus, the finding that significant looping interactions mostly occur within TADs is not simply due to the fact that interaction frequencies are generally higher within TADs. Statistically significant looping interactions between restriction fragments located in different TADs were mostly observed for loci separated by larger genomic distances (>300–500 kb), i.e., at distances that are larger than the average TAD, even though many inter-TAD interactions at smaller genomic distances were interrogated. These significant inter-TAD interactions are much lower in frequency, indicating they occur in fewer cells in the population. Our results confirm and extend previous work that had indicated that TADs correspond to regulatory domains responsive to specific enhancers.30, 40
Cell-Type-Specific Looping between the Promoter and Distal Elements in the CFTR Locus
We focused our analysis on CFTR. We examined three cell lines (Caco2, Calu3, and Capan1) that express the gene and two cell lines (GM12878 and HepG2) that do not express the gene (Figure 3). Of the lines that do not express CFTR, HepG2 displays no intra-TAD looping between the CFTR promoter and any element except for one located just at the TAD boundary, and this long-range interaction is present in all five cell lines examined (Figure 5). This region of the CFTR locus has been shown previously to be an area of elevated interaction frequency in several cell lines,18 and is located very close to the TAD boundary. Interestingly, in GM12878 cells the CFTR promoter is inactive but does engage in some looping interactions, e.g., with the known CFTR enhancers in intron 11 (+108 kb) and just downstream of the gene at +202 kb18, 21 (Figure 5P). It also interacts with an upstream element known to bind CTCF. However, the frequency of these interactions is much lower than in cells that do express CFTR. These three interactions with potential regulatory elements might indicate a poised conformation of CFTR in GM12878 cells.64 Indeed, in lymphoblastoid cells a DNaseI-hypersensitive site is present at the enhancer in intron 11 (ENCODE data42).
Focusing on the three cell lines (Caco2, Calu3, and Capan1) that express CFTR in our study, we note that three of the four looping interactions we and others previously detected in Caco2 cells by using conventional 3C are also present in our 5C data (the −21 kb site, intron 11 (+108 kb), and the +202 kb site)18, 21 (Figures 5M–5O). These interactions occur with relatively high frequency, both in comparison to interactions with directly adjacent chromatin and in comparison to interactions in non-CFTR-expressing cells. Two of those elements, intron 11 and the +202 kb site, are known CFTR intestinal enhancers.18, 21 The −21 kb site is a CTCF-binding element and has been proposed to play a structural role in the 3D conformation of the locus.65, 66 We also observe a significant looping interaction between the CFTR promoter and the DHS in intron 11 in Calu3 cells, although the interaction is weaker than in Caco2 cells. This is interesting because the intron 11 enhancer was shown to be inactive in 16HBE14o- airway epithelial cells (but active in colon cells) in reporter assays.59 Importantly, in lung-derived Calu3 cells the enhancer in intron 11 does contain a DHS, suggesting that in these cells the element is active.61
Calu3 cells display a CFTR intra-TAD looping pattern that is strikingly different from that of Caco2 and Capan1. Significant interactions occur upstream of the CFTR promoter. The entire region is lifted above background, and three clear peaks are present. Two of these peaks are known CTCF binding sites (−21 kb and −80 kb).42, 65, 66 The middle peak is situated at a pair of known lung-specific CFTR enhancers, located 35–44 kb upstream of the promoter.60 We hypothesize that the entire upstream region displays statistically significant elevated interaction frequencies in Calu3 cells as a result of the three strong looping interactions with the CFTR promoter. Looping between the CFTR promoter and these distal elements would also bring their neighboring fragments into close proximity with the promoter, as predicted from polymer rings.67 Alternately, there could be more unknown lung-specific enhancers located in this upstream region.
Interactions between TADs
We also detected statistically significant interactions between loci located in different TADs. Interestingly, inter-TAD interactions are as cell-type specific as the looping interactions within TADs. However, these interactions also differ in at least two ways from looping interactions within TADs. First, looping interactions between TADs are much weaker (fewer reads) than looping interactions within TADs (Figures 4C and 4D), in part because they occur over larger genomic distances. They are also less reproducible between replicates (Figure 4; Figures S15 and S16). This can mean that they occur in fewer cells in the population.40 One interpretation is that intra-TAD looping interactions occur in more cells in the population and thus are stronger and more likely to be biologically relevant, e.g., to be involved in regulating gene expression, than inter-TAD looping interactions. Second, inter-TAD looping interactions often involve loci located at or near TAD boundaries, whereas looping interactions within TADs can occur throughout these domains. The roles, if any, of these inter-TAD interactions in gene regulation are currently not known.
TAD Boundaries Have Multiple Roles in Organizing Long-Range Looping Interactions
The results presented here reveal important roles for TAD boundaries in organizing long-range looping interactions within and between TADs. First, TAD boundaries act as physical insulators that prevent frequent interaction across them. Interactions between loci located within a TAD are therefore more frequent than interactions between loci located in different TADs. In addition, significant looping interactions between genes and regulatory elements occur mostly and with higher interaction frequencies between loci located within the same TAD. In the case of CFTR, we found that all its known regulatory elements that the CFTR promoter loops with were contained within the same TAD. We propose that TADs represent domains of gene regulation that are at least partially physically insulated from regulatory input from other regions outside the TAD. This is consistent with other studies that predicted promoter-enhancer pairings via independent approaches and found that they occur mostly within TADs.54, 55 Also, functional assays have shown that TADs correspond to regulatory domains controlled by sets of enhancers to regulate genes located within them.30
Here we uncover another role for TAD boundaries in modulating long-range interactions. Loci located near TAD boundaries often engage in significant interactions with loci located in different TADs. These interactions tend to be much longer-range than interactions within TADs, and consequently they are at least an order of magnitude weaker and probably more stochastic in the cell population (Figure 4D). Possibly these interactions are not specific chromatin loops between defined elements but play an architectural role. We previously proposed that cell-type-invariant TADs assemble into higher-order structures such as A- and B-type compartments that are cell-type specific and related to chromatin status.40 Possibly such higher-order assemblies are driven by cell-type-specific interactions between loci at or near TAD boundaries. Roles for boundaries in organizing higher-order chromosomal compartments in the nucleus was also proposed on the basis of Hi-C studies in Drosophila.49
Interestingly, we note that condensin-dependent long-range interactions between TAD boundaries were recently also found along the X chromosome in C. elegans hermaphrodites,34 and more generally that such interactions were found between CTCF-bound sites at domain boundaries.11 Studies in Drosophila also found that boundaries of chromatin domains are often involved in long-range interactions,49 suggesting this may be a general phenomenon. Looping between boundaries can contribute to physical insulation.53
In summary, we propose that TAD boundaries play two important roles: first, they constrain frequent regulatory looping interactions between gene promoters and gene regulatory elements within TADs. Second, they are involved in weaker, longer-range interactions with other TADs, possibly leading to formation of higher-order chromatin architectures such as A-type and B-type compartments.
Acknowledgments
Work in the Dekker lab is supported by the National Human Genome Research Institute (grants HG003143, HG007010) and the Human Frontier Science Project Organization. J.D. is an investigator of the Howard Hughes Medical Institute.
Published: January 7, 2016
Footnotes
Supplemental Data include 24 figures and eight tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.12.002.
Accession Numbers
All 5C probes, raw 5C interaction data, and processed 5C data (corrected 5C data and peak called data) are publicly available at GEO (GEO: GSE75634).
Web Resources
The URLs for data presented herein are as follows:
Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo/
My5C Online Tool, http://my5C.umassmed.edu
Novocraft Technologies, http://novocraft.com
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
Supplemental Data
References
- 1.Bickmore W.A., van Steensel B. Genome architecture: Domain organization of interphase chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Fraser P., Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 3.Bolzer A., Kreth G., Solovei I., Koehler D., Saracoglu K., Fauth C., Müller S., Eils R., Cremer C., Speicher M.R., Cremer T. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cremer T., Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 5.Branco M.R., Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Croft J.A., Bridger J.M., Boyle S., Perry P., Teague P., Bickmore W.A. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanabe H., Müller S., Neusser M., von Hase J., Calcagno E., Cremer M., Solovei I., Cremer C., Cremer T. Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc. Natl. Acad. Sci. USA. 2002;99:4424–4429. doi: 10.1073/pnas.072618599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y., McCord R.P., Ho Y.J., Lajoie B.R., Hildebrand D.G., Simon A.C., Becker M.S., Alt F.W., Dekker J. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imakaev M., Fudenberg G., McCord R.P., Naumova N., Goloborodko A., Lajoie B.R., Dekker J., Mirny L.A. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat. Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., Aiden E.L. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sexton T., Schober H., Fraser P., Gasser S.M. Gene regulation through nuclear organization. Nat. Struct. Mol. Biol. 2007;14:1049–1055. doi: 10.1038/nsmb1324. [DOI] [PubMed] [Google Scholar]
- 13.Iborra F.J., Pombo A., Jackson D.A., Cook P.R. Active RNA polymerases are localized within discrete transcription “factories” in human nuclei. J. Cell Sci. 1996;109:1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- 14.Schoenfelder S., Sexton T., Chakalova L., Cope N.F., Horton A., Andrews S., Kurukuti S., Mitchell J.A., Umlauf D., Dimitrova D.S. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat. Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guelen L., Pagie L., Brasset E., Meuleman W., Faza M.B., Talhout W., Eussen B.H., de Klein A., Wessels L., de Laat W., van Steensel B. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 16.Amano T., Sagai T., Tanabe H., Mizushina Y., Nakazawa H., Shiroishi T. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev. Cell. 2009;16:47–57. doi: 10.1016/j.devcel.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 17.Baù D., Sanyal A., Lajoie B.R., Capriotti E., Byron M., Lawrence J.B., Dekker J., Marti-Renom M.A. The three-dimensional folding of the α-globin gene domain reveals formation of chromatin globules. Nat. Struct. Mol. Biol. 2011;18:107–114. doi: 10.1038/nsmb.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gheldof N., Smith E.M., Tabuchi T.M., Koch C.M., Dunham I., Stamatoyannopoulos J.A., Dekker J. Cell-type-specific long-range looping interactions identify distant regulatory elements of the CFTR gene. Nucleic Acids Res. 2010;38:4325–4336. doi: 10.1093/nar/gkq175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lettice L.A., Heaney S.J., Purdie L.A., Li L., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 20.Murrell A., Heeson S., Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 21.Ott C.J., Blackledge N.P., Kerschner J.L., Leir S.H., Crawford G.E., Cotton C.U., Harris A. Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc. Natl. Acad. Sci. USA. 2009;106:19934–19939. doi: 10.1073/pnas.0900946106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miele A., Bystricky K., Dekker J. Yeast silent mating type loci form heterochromatic clusters through silencer protein-dependent long-range interactions. PLoS Genet. 2009;5:e1000478. doi: 10.1371/journal.pgen.1000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palstra R.J., Tolhuis B., Splinter E., Nijmeijer R., Grosveld F., de Laat W. The beta-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- 24.Phillips-Cremins J.E., Sauria M.E., Sanyal A., Gerasimova T.I., Lajoie B.R., Bell J.S., Ong C.T., Hookway T.A., Guo C., Sun Y. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanyal A., Lajoie B.R., Jain G., Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tolhuis B., Palstra R.J., Splinter E., Grosveld F., de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 27.Vernimmen D., De Gobbi M., Sloane-Stanley J.A., Wood W.G., Higgs D.R. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J. 2007;26:2041–2051. doi: 10.1038/sj.emboj.7601654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright J.B., Brown S.J., Cole M.D. Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Mol. Cell. Biol. 2010;30:1411–1420. doi: 10.1128/MCB.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleinjan D.A., van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Symmons O., Uslu V.V., Tsujimura T., Ruf S., Nassari S., Schwarzer W., Ettwiller L., Spitz F. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 2014;24:390–400. doi: 10.1101/gr.163519.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N.L., Meisig J., Sedat J. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., Hoichman M., Parrinello H., Tanay A., Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 34.Crane E., Bian Q., McCord R.P., Lajoie B.R., Wheeler B.S., Ralston E.J., Uzawa S., Dekker J., Meyer B.J. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015;523:240–244. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dixon J.R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J.E., Lee A.Y., Ye Z., Kim A., Rajagopal N., Xie W. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naumova N., Imakaev M., Fudenberg G., Zhan Y., Lajoie B.R., Mirny L.A., Dekker J. Organization of the mitotic chromosome. Science. 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dekker J. Two ways to fold the genome during the cell cycle: insights obtained with chromosome conformation capture. Epigenetics Chromatin. 2014;7:25. doi: 10.1186/1756-8935-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Dily F., Baù D., Pohl A., Vicent G.P., Serra F., Soronellas D., Castellano G., Wright R.H., Ballare C., Filion G. Distinct structural transitions of chromatin topological domains correlate with coordinated hormone-induced gene regulation. Genes Dev. 2014;28:2151–2162. doi: 10.1101/gad.241422.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giorgetti L., Galupa R., Nora E.P., Piolot T., Lam F., Dekker J., Tiana G., Heard E. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell. 2014;157:950–963. doi: 10.1016/j.cell.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gibcus J.H., Dekker J. The hierarchy of the 3D genome. Mol. Cell. 2013;49:773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dostie J., Richmond T.A., Arnaout R.A., Selzer R.R., Lee W.L., Honan T.A., Rubio E.D., Krumm A., Lamb J., Nusbaum C. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Birney E., Stamatoyannopoulos J.A., Dutta A., Guigó R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., ENCODE Project Consortium. NISC Comparative Sequencing Program. Baylor College of Medicine Human Genome Sequencing Center. Washington University Genome Sequencing Center. Broad Institute. Children’s Hospital Oakland Research Institute Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lajoie B.R., van Berkum N.L., Sanyal A., Dekker J. My5C: web tools for chromosome conformation capture studies. Nat. Methods. 2009;6:690–691. doi: 10.1038/nmeth1009-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 45.Naumova N., Smith E.M., Zhan Y., Dekker J. Analysis of long-range chromatin interactions using Chromosome Conformation Capture. Methods. 2012;58:192–203. doi: 10.1016/j.ymeth.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferraiuolo M.A., Sanyal A., Naumova N., Dekker J., Dostie J. From cells to chromatin: capturing snapshots of genome organization with 5C technology. Methods. 2012;58:255–267. doi: 10.1016/j.ymeth.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lajoie B.R., Dekker J., Kaplan N. The Hitchhiker’s guide to Hi-C analysis: practical guidelines. Methods. 2015;72:65–75. doi: 10.1016/j.ymeth.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dekker J., Marti-Renom M.A., Mirny L.A. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hou C., Li L., Qin Z.S., Corces V.G. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol. Cell. 2012;48:471–484. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Bortle K., Nichols M.H., Li L., Ong C.T., Takenaka N., Qin Z.S., Corces V.G. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 2014;15:R82. doi: 10.1186/gb-2014-15-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zuin J., Dixon J.R., van der Reijden M.I., Ye Z., Kolovos P., Brouwer R.W., van de Corput M.P., van de Werken H.J., Knoch T.A., van IJcken W.F. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci. USA. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sofueva S., Yaffe E., Chan W.C., Georgopoulou D., Vietri Rudan M., Mira-Bontenbal H., Pollard S.M., Schroth G.P., Tanay A., Hadjur S. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 2013;32:3119–3129. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doyle B., Fudenberg G., Imakaev M., Mirny L.A. Chromatin loops as allosteric modulators of enhancer-promoter interactions. PLoS Comput. Biol. 2014;10:e1003867. doi: 10.1371/journal.pcbi.1003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin F., Li Y., Dixon J.R., Selvaraj S., Ye Z., Lee A.Y., Yen C.A., Schmitt A.D., Espinoza C.A., Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen Y., Yue F., McCleary D.F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V.V., Ren B. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCarthy V.A., Harris A. The CFTR gene and regulation of its expression. Pediatr. Pulmonol. 2005;40:1–8. doi: 10.1002/ppul.20199. [DOI] [PubMed] [Google Scholar]
- 57.Ott C.J., Blackledge N.P., Leir S.H., Harris A. Novel regulatory mechanisms for the CFTR gene. Biochem. Soc. Trans. 2009;37:843–848. doi: 10.1042/BST0370843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nuthall H.N., Moulin D.S., Huxley C., Harris A. Analysis of DNase-I-hypersensitive sites at the 3′ end of the cystic fibrosis transmembrane conductance regulator gene (CFTR) Biochem. J. 1999;341:601–611. [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Z., Leir S.H., Harris A. Immune mediators regulate CFTR expression through a bifunctional airway-selective enhancer. Mol. Cell. Biol. 2013;33:2843–2853. doi: 10.1128/MCB.00003-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Z., Leir S.H., Harris A. Oxidative stress regulates CFTR gene expression in human airway epithelial cells through a distal antioxidant response element. Am. J. Respir. Cell Mol. Biol. 2015;52:387–396. doi: 10.1165/rcmb.2014-0263OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Z., Ott C.J., Lewandowska M.A., Leir S.H., Harris A. Molecular mechanisms controlling CFTR gene expression in the airway. J. Cell. Mol. Med. 2012;16:1321–1330. doi: 10.1111/j.1582-4934.2011.01439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ernst J., Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ghavi-Helm Y., Klein F.A., Pakozdi T., Ciglar L., Noordermeer D., Huber W., Furlong E.E. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014;512:96–100. doi: 10.1038/nature13417. [DOI] [PubMed] [Google Scholar]
- 65.Blackledge N.P., Carter E.J., Evans J.R., Lawson V., Rowntree R.K., Harris A. CTCF mediates insulator function at the CFTR locus. Biochem. J. 2007;408:267–275. doi: 10.1042/BJ20070429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gosalia N., Neems D., Kerschner J.L., Kosak S.T., Harris A. Architectural proteins CTCF and cohesin have distinct roles in modulating the higher order structure and expression of the CFTR locus. Nucleic Acids Res. 2014;42:9612–9622. doi: 10.1093/nar/gku648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rippe K. Making contacts on a nucleic acid polymer. Trends Biochem. Sci. 2001;26:733–740. doi: 10.1016/s0968-0004(01)01978-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.