ABSTRACT
Endogenous retroviruses (ERVs) are remnants of ancestral retroviral infections of germ cells. Retroviral endogenization is an adaptation process for the host genome, and ERVs are gradually attenuated or inactivated by mutation. However, some ERVs that have been “domesticated” by their hosts eventually gain physiological functions, such as placentation or viral resistance. We previously reported the discovery of Refrex-1, a soluble antiretroviral factor in domestic cats that specifically inhibits infection by feline leukemia virus subgroup D (FeLV-D), a chimeric virus of FeLV, and a feline ERV, ERV-DC. Refrex-1 is a truncated envelope protein (Env) encoded by both ERV-DC7 and ERV-DC16 proviral loci. Here, we reconstituted ancestral and functional Env from ERV-DC7 and ERV-DC16 envelope genes (env) by inducing reverse mutations. Unexpectedly, ERV-DC7 and ERV-DC16 full-length Env (ERV-DC7 fl and ERV-DC16 fl), reconstructed by removing stop codons, did not produce infectious viral particles. ERV-DC7 fl and ERV-DC16 fl were highly expressed in cells but were not cleaved into surface subunits (SU) and transmembrane subunits, nor were they incorporated into virions. G407R/N427I-A429T and Y431D substitutions within the SU C-terminal domain of ERV-DC7 fl and ERV-DC16 fl, respectively, caused these dysfunctions. The residues glycine 407 and tyrosine 431 are relatively conserved among infectious gammaretroviruses, and their substitution causes the same dysfunctions as the tested retroviruses. Our results reveal that specific mutations within the SU C-terminal domain suppressed Env cleavage and incorporation into virions and indicate that these mutations contributed to the domestication of Refrex-1 through multistep events that occurred in the postintegration period.
IMPORTANCE Domestic cats are colonized with various exogenous retroviruses (exRVs), such as feline leukemia virus (FeLV), and their genomes contain numerous ERVs, some of which are replication-competent proviruses. The feline hosts, exRVs, and ERVs have complicated genetic interactions and provide an interesting field model for triangular relationships: recombination between FeLV and ERV-DC, which is a feline ERV, generated FeLV-D, a chimeric virus, and FeLV-D is restricted by Refrex-1, an antiretroviral factor corresponding to truncated Env of ERV-DC7 and ERV-DC16. Here, we reconstructed ancestral, functional Env from ERV-DC7 and ERV-DC16 env by inducing reverse mutations to elucidate how Refrex-1 was generated from its ancestor. Our results reveal that they were repeatedly inactivated by mutations preventing Env maturation. Our results provide insights into how ERVs were “domesticated” by their hosts and identify the mutations that mediated these evolutions. Notably, experiments that restore inactivated ERVs might uncover previously unrecognized features or properties of retroviruses.
INTRODUCTION
Endogenous retroviruses (ERVs) are present in all vertebrate genomes and are thought to be the remnants of ancestral germ line infections by exogenous retroviruses (exRVs) (1). ERVs make up a significant fraction of the mammalian genome (for example, 8 to 10% of the human or mouse genomes) and are transmitted in a Mendelian fashion (2, 3). Once retroviruses invade a host genome, they increase their copy numbers in the host genome by repeated reinfection or retrotransposition in germ line cells (4–6). During the process of endogenization, retroviruses become “ERV-like” through virus-host coevolution. They are gradually attenuated or inactivated by preinsertional mutations that occur during viral replications and by postinsertional mutations that occur during host genome replication. Nucleotide changes in the long terminal repeats (LTR) diminish the transcription activity of the virus, and amino acid changes in the viral genes disrupt or modify the functions of their coding proteins (7–12).
Retroviral envelope proteins (Env) are composed of a trimer of heterodimers formed between the surface subunit (SU) and the transmembrane subunit (TM). The SUs of gammaretroviruses are composed of two globular domains, the N-terminal and C-terminal domains, and they mediate viral attachment to target cells through viral receptor recognition and binding (13, 14). The TM tethers Env on membranes, and its fusion peptide mediates viral entry through fusion between the viral envelope and the cell membrane. Env is first synthesized as a SU-TM precursor polypeptide in the rough endoplasmic reticulum and then transported into the trans-Golgi network (TGN), where it is cleaved into a SU and a TM at the cleavage motif (R-X-K/R-R↓Y) by cellular proteases (15–20). Env cleavage may be an essential process for Env maturation because it enables the TM fusion peptide to change the conformational position it acquired during membrane fusion.
Some ERVs are domesticated by their hosts and eventually gain physiological functions, such as placentation or viral resistance, in exchange for the loss of their ancestral, viral properties (21–29). For example, the Friend virus susceptibility4 (Fv4) gene, which is an endogenous defective provirus that resembles the ecotropic murine leukemia virus (MLV), encodes a restriction factor against ecotropic MLVs (30–32). The Env of Fv4 binds to viral receptors and protects the cells expressing it from infection by ecotropic MLVs. However, the Env of Fv4 lacks fusogenicity owing to a substitution in its TM fusion peptide, and it has lost the capacity to produce infectious virions (33, 34). Thus, domesticated ERVs seem to be strictly regulated by their hosts, and dysregulation of them can result in negative effects for the hosts (35–37).
Domestic cats (Felis silvestris catus) are colonized with various exRVs, such as feline leukemia virus (FeLV), feline immunodeficiency virus (FIV), and feline foamy virus (FFV), and their genomes also contain various ERVs, such as endogenous FeLV (enFeLV), ERV-DCs, and RD-114. These exRVs and ERVs genetically interact with each other. FeLV is a highly pathogenic gammaretrovirus that widely circulates among domestic cats through grooming or bites (38, 39). ERV-DCs are endogenous gammaretroviruses of domestic cats that are closely related to MLV and FeLV (40).
Previously, we molecularly cloned 13 proviral loci of ERV-DCs and characterized them (40): ERV-DCs are classified into genotypes I, II, and III, and most are insertionally polymorphic loci. Those results indicated that ERV-DCs invaded feline genomes several times within the past few million years. Some ERV-DCs have intact open reading frames (ORFs) for gag, pol, and the envelope gene (env). Notably, ERV-DC10 (the q12–q21 region on chromosome C1) and ERV-DC18 (q14 on chromosome D4), both of which belong to genotype III, are replication competent in cultured cells and can infect a broad range of cells, including human cells. Furthermore, we identified FeLV-D, a novel subgroup of FeLV that has an infection tropism different from that of other FeLVs, and reported that this is a recombinant virus between FeLV and ERV-DC genotype I in the env region (40, 41). FeLV can be categorized into several FeLV subgroups based on their viral receptor usage and interference capacities. For example, FeLV-A typically uses THTR-1 (42), FeLV-B typically uses Pit-1 (43) and Pit-2 (44), and FeLV-C typically uses FLVCR-1 (45, 46). Receptors for FeLV-D or for ERV-DCs have not yet been identified. Importantly, ERV-DCs can still impact the lives of their hosts, both through their potential viral activity and through their contribution to the emergence of recombinant viruses.
We recently reported the discovery of Refrex-1, which is a feline soluble restriction factor against ERV-DCs and FeLV-D (41). All of the ERV-DCs and FeLV-D fall into two receptor interference groups, and Refrex-1 specifically inhibits the group that includes ERV-DC genotype I and FeLV-D. Refrex-1 is a truncated ERV-DC Env, and it includes a signal peptide and a SU N-terminal domain, which is also called a receptor binding domain, but it lacks a TM because of a premature stop codon present in the middle of the env ORF. As indicated by its structure, Refrex-1 is efficiently secreted from cells and blocks viral infection, probably by receptor interference. There are two forms of Refrex-1, encoded by ERV-DC7 and ERV-DC16, and both belong to ERV-DC genotype II. ERV-DC7 and ERV-DC16 are fixed in the feline genome, unlike the other ERV-DCs, which are insertionally polymorphic (40). Thus, Refrex-1 seems to be a restriction factor that has been acquired by cats through ERV domestication during the evolutional arms race between hosts and ERV-DCs.
Refrex-1 was generated from the ancestral Env of ERV-DC7 and ERV-DC16 in exchange for the loss of their ancestral viral properties. During this process, the mutations that alter protein function, such as premature codons, may have contributed to this loss. Here, we reconstituted functional, ancestral ERV-DC7 and ERV-DC16 Env by inducing reverse mutations into their env genes in order to analyze their original properties and speculate about the process of their domestication. We removed their premature and latent stop codons and constructed full-length Env, termed ERV-DC7 full-length (fl) and ERV-DC16 fl. However, we found that they could not produce infectious viral particles. This study discovered that some mutations in the SU C-terminal domain prevent Env cleavage and incorporation into virions, as is common among gammaretroviruses, and our results indicate that these mutations contributed to the process of ERV inactivation and domestication.
MATERIALS AND METHODS
Cell cultures.
HEK293T (47, 48), NIH 3T3 (49), and AH927 (50) cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS). HEK293T cells infected with FeLV-A/Glasgow-1 (51), FeLV-B/Gardner-Arnstein (52), FeLV-D/c33 (41), or ERV-DC10 (40) were also cultured in DMEM with 10% FCS, as were GPLac cells (40), which are an env-negative packaging cell line containing a LacZ-coding retroviral vector.
In this study, cells stably expressing Env were established through dilution cloning (53). Briefly, HEK293T cells were transfected with pFUΔss ERV-DC14, pFUΔss ERV-DC7 fl, or pFUΔss ERV-DC16 fl and selected in medium containing 200 μg/ml of zeocin (InvivoGen, San Diego, CA) for 1 week, and then single cell clones were obtained via the limiting dilution method. After screening by immunoblotting, HEK293T cells stably expressing ERV-DC14 Env, ERV-DC7 fl, or ERV-DC16 fl were established. These cells were also cultured in DMEM with 10% FCS.
Construction of expression plasmids.
In this study, the vector pFUΔss (40) was used for the construction of expression plasmids. All PCRs were conducted with KOD FX Neo DNA polymerase (Toyobo, Osaka, Japan).
We constructed pFUΔss Fr-MLV Env and pFUΔss KoRV-A_1 Env, which are env expression plasmids of Friend murine leukemia virus (Fr-MLV) and koala retrovirus A (KoRV), respectively. The env fragments of Fr-MLV and KoRV-A were PCR amplified from the Fr-MLV clone 57 plasmid (54) and from koala bone marrow genomic DNA, respectively, by using the primers FrMLV env 1S and FrMLV env 1R and the primers KoRV env eco 1S and KoRV env eco 1R, respectively. These env fragments were inserted into a pFUΔss vector through EcoRI restriction sites. Accession numbers for the env nucleotide sequences of KoRV-A_1 are presented below.
pFUΔss ERV-DC7 fl and pFUΔss ERV-DC16 fl, which encode the full-length Env proteins of ERV-DC7 and ERV-DC16, respectively, were constructed as follows. We first used PCR to amplify the env regions corresponding to the full-length ORF from templates of PCR-Blunt ERV-DC7 (Kyoto-SPF1) (41) and ERV-DC16 (Kyoto-SPF1) (41) with primer pairs Fe-207S–Fe-222R and Fe-209S–Fe-222R, respectively. These PCR products were inserted into pFUΔss vectors through BamHI restriction sites and named pFUΔss ERV-DC7 env and pFUΔss ERV-DC16 env. We next removed premature or latent stop codons in ERV-DC7 and ERV-DC16 by site-directed mutagenesis following the manufacturer's protocol and using a QuikChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). The primers used for site-directed mutagenesis are summarized in Table 1.
TABLE 1.
Expression plasmids constructed by site-directed mutagenesis in this study
| Expression plasmid | Template plasmid | Forward primer | Reverse primer |
|---|---|---|---|
| pFUΔss ERV-DC7 fl | pFUΔss ERV-DC7 env | Fe-246S | Fe-218R |
| Fe-247S | Fe-219R | ||
| pFUΔss ERV-DC16 fl | pFUΔss ERV-DC16 env | Fe-248S | Fe-220R |
| Fe-249S | Fe-221R | ||
| pFUΔss DC14/G407R | pFUΔss DC14 | Fe-344S | Fe-281R |
| pFUΔss DC14/N427I | pFUΔss DC14 | Fe-345S | Fe-282R |
| pFUΔss DC14/N427T | pFUΔss DC14 | Fe-378S | Fe-411R |
| pFUΔss DC14/A429T | pFUΔss DC14 | Fe-346S | Fe-283R |
| pFUΔss DC14/Y431D | pFUΔss DC14 | Fe-358S | Fe-395R |
| pFUΔss DC14/G457S | pFUΔss DC14 | Fe-347S | Fe-284R |
| pFUΔss DC14/N427I-A429T | pFUΔss DC14 | Fe-382S | Fe-413R |
| pFUΔss DC7 fl/R407G | pFUΔss ERV-DC7 fl | Fe-362S | Fe-401R |
| pFUΔss DC7 fl/ | |||
| R407G-I427N-T429A | pFUΔss DC7/r401-472 | Fe-376S | Fe-409R |
| pFUΔss DC16 fl/D431Y | pFUΔss ERV-DC16 fl | Fe-363S | Fe-402R |
| pFUΔss Fr-MLV/407R | pFUΔss Fr-MLV Env | Fr-MLV mu1 S | Fr-MLV mu1 R |
| pFUΔss Fr-MLV/431D | pFUΔss Fr-MLV Env | Fr-MLV mu5 S | Fr-MLV mu5 R |
| pFUΔss FeLV-A/407R | pFUΔss A5 | Fe-379S | Fe-412R |
| pFUΔss FeLV-A/431D | pFUΔss A5 | Fe-380S | Fe-413R |
| pFUΔss ERV-DC10/407R | pFUΔss DC10 | Fe-344S | Fe-281R |
| pFUΔss ERV-DC10/431D | pFUΔss DC10 | Fe-377S | Fe-410R |
| pFUΔss KoRV-A/407R | pFUΔss KoRV-A_1 Env | KoRV mu1 S | KoRV mu1 R |
| pFUΔss KoRV-A/431D | pFUΔss KoRV-A_1 Env | KoRV mu5 S | KoRV mu5 R |
| pFUΔss KoRV-A-Myc/407R | pFUΔss KoRV-A-Myc | KoRV mu1 S | KoRV mu1 R |
| pFUΔss KoRV-A-Myc/431D | pFUΔss KoRV-A-Myc | KoRV mu5 S | KoRV mu5 R |
Primer sequences are shown in Table S1 in the supplemental material.
pFUΔss DC14-Myc, pFUΔss DC7 fl-Myc, pFUΔss DC16 fl-Myc, and pFUΔss KoRV-A-Myc encoding Env fused to a Myc tag in the C terminus of their TM were constructed. These were amplified by PCR from the templates pFUΔss DC14 (41), pFUΔss ERV-DC7 fl, pFUΔss ERV-DC16 fl, and pFUΔss KoRV-A_1 Env with the primer pairs Fe-207S–ERV-DC GI,II Env Myc R, Fe-207S–ERV-DC GI,II Env Myc R, Fe-209S–ERV-DC GI,II Env Myc R, and KoRV 3S–KoRV 4R, respectively, and inserted into pFUΔss vectors through EcoRI or BamHI restriction sites.
Expression plasmids encoding Env chimera proteins were constructed using ERV-DC7 fl and ERV-DC14 Env. pFUΔss DC7/-262 was constructed by replacing the 3′ NheI-BamHI env fragment of pFUΔss ERV-DC7 fl with that of pFUΔss DC14. pFUΔss DC7/-300, pFUΔss DC7/-400, pFUΔss DC7/-472, pFUΔss DC7/401-472, and pFUΔss DC7/r401-472 were constructed by using the GeneArt seamless cloning and assembly enzyme mix (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The templates and primers used for the construction of these chimeras are summarized Table 2. The expression plasmids encoding Env chimeras were also constructed using ERV-DC16 fl and ERV-DC14 Env. pFUΔss DC16/-262 was constructed by replacing the 3′ NheI-BamHI env fragment of pFUΔss ERV-DC16 fl with that of pFUΔss DC14. pFUΔss DC16/-300, pFUΔss DC16/-400, pFUΔss DC16/-472, and pFUΔss DC16/401-472 were constructed with the same kit and method used for pFUΔss DC7/-300, pFUΔss DC7/-400, pFUΔss DC7/-472, pFUΔss DC7/401-472, and pFUΔss DC7/r401-472. The templates and primers used for the construction of these chimeras are also summarized in Table 2.
TABLE 2.
Expression plasmids constructed by the seamless cloning method in this study
| Expression plasmid | Template plasmid | Forward primer | Reverse primer |
|---|---|---|---|
| pFUΔss DC7/-300 | pFUΔss ERV-DC7 fl | Fe-327S | Fe-267R |
| pFUΔss DC14 | Fe-328S | Fe-266R | |
| pFUΔss DC7/-400 | pFUΔss ERV-DC7 fl | Fe-327S | Fe-268R |
| pFUΔss DC14 | Fe-329S | Fe-266R | |
| pFUΔss DC7/-472 | pFUΔss ERV-DC7 fl | Fe-327S | Fe-269R |
| pFUΔss DC14 | Fe-330S | Fe-266R | |
| pFUΔss DC7/401-472 | pFUΔss DC14 | Fe-327S | Fe-268R |
| pFUΔss DC7/-472 | Fe-329S | Fe-266R | |
| pFUΔss DC7/r401-472 | pFUΔss DC7/-400 | Fe-327S | Fe-269R |
| pFUΔss ERV-DC7 fl | Fe-330S | Fe-266R | |
| pFUΔss DC16/-300 | pFUΔss ERV-DC16 fl | Fe-327S | Fe-267R |
| pFUΔss DC14 | Fe-328S | Fe-266R | |
| pFUΔss DC16/-400 | pFUΔss ERV-DC16 fl | Fe-327S | Fe-268R |
| pFUΔss DC14 | Fe-329S | Fe-266R | |
| pFUΔss DC16/-472 | pFUΔss ERV-DC16 fl | Fe-327S | Fe-269R |
| pFUΔss DC14 | Fe-330S | Fe-266R | |
| pFUΔss DC16/401-472 | pFUΔss DC14 | Fe-327S | Fe-268R |
| pFUΔss DC16/-472 | Fe-329S | Fe-266R |
Primer sequences are shown in Table S1 in the supplemental material.
The site-directed mutagenesis method described above was also used to construct expression plasmids of Env point mutants: pFUΔss DC14/G407R, pFUΔss DC14/N427I, pFUΔss DC14/N427T, pFUΔss DC14/A429T, pFUΔss DC14/Y431D, pFUΔss DC14/G457S, and pFUΔss DC14/N427I-A429T; pFUΔss DC7 fl/R407G and pFUΔss DC7 fl/R407G-I427N-T429A; pFUΔss DC16 fl/D431Y; pFUΔss Fr-MLV/407R and pFUΔss Fr-MLV/431D; FeLV-A/407R and FeLV-A/431D; ERV-DC10/407R and ERV-DC10/431D; KoRV-A/407R and KoRV-A/431D; and KoRV-A-Myc/407R and KoRV-A-Myc/431D. The primers and templates used for site-directed mutagenesis are summarized in Table 1, and the sequences of the PCR primers used in this study are summarized in Table S1 in the supplemental material.
Transfection.
HEK 293T and GP Lac cells were transfected with plasmids using ScreenFect A (Wako, Osaka, Japan) or polyethylenimine Max (Polysciences Inc., Warrington, PA) according to each manufacturer's instructions.
Preparation of LacZ-carrying Env pseudotyped viruses.
GP Lac cells were transfected with env expression plasmids to produce LacZ-carrying Env-pseudotyped viruses. After 48 h, cell culture supernatants were collected, filtered through a 0.45-μm-pore-size filter, and stored at −80°C.
Viral infection and titration.
Target cells (HEK293T, NIH 3T3, or AH927 cells in 24-well plates) were inoculated with 250 μl of Env pseudotyped viruses and cultured in medium containing 8 μg/ml of Polybrene. After 48 h, cells were stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), and single-cycle infectivity was titrated by counting blue-stained nuclei under the microscope.
Immunoblotting.
Viral particles were pelleted from 5 ml of viral supernatants in a Beckman MLS-50 rotor (29,000 × g, 90 min, 4°C). Viral pellets were resuspended in 30 μl of phosphate-buffered saline (PBS) and analyzed. Immunoblotting was performed as previously described (41). Primary antibodies were goat anti-FeLV SU (gp70) (National Cancer Institute [NCI], Frederick, MD), goat anti-Rauscher MLV SU antibody (NCI), goat anti-Rauscher MLV CA (p30) antibody (NCI), mouse anti-β-actin (Santa Cruz Biotechnology, Santa Cruz, CA), and horseradish peroxidase (HRP)-conjugated anti-c-Myc monoclonal antibody (Wako, Osaka, Japan). Secondary antibodies were HRP-conjugated anti-goat IgG antibody (Santa Cruz Biotechnology) and HRP-conjugated anti-mouse IgG antibody (Cell Signaling Technology).
Fluorescence-activated cell sorting (FACS) analysis.
HEK 293T cells were seeded in 6-well plates and transfected with env expression plasmids. After 48 h, cells were harvested with PBS containing 0.2% EDTA and washed with PBS containing 0.1% bovine serum albumin (BSA). Some of the cells were permeabilized with 90% methanol at 4°C for 90 min. Goat anti-FeLV SU (NCI) or mouse anti-β-actin (Santa Cruz Biotechnology) was used as the primary antibody, and HRP-conjugated anti-goat IgG antibody (Santa Cruz Biotechnology) or HRP-conjugated anti-mouse IgG antibody (GE Healthcare Japan, Tokyo, Japan) was used as the secondary antibody. Cells were treated with each antibody for 30 min at 4°C. Finally, cells were fixed with 4% formaldehyde and analyzed with a BD Accuri C6 flow cytometer (Becton, Dickinson and Company, Franklin Lakes, NJ).
Interference assay.
Interference assays using preinfected cells were performed as previously described (40). Target cells (HEK 293T cells persistently infected with each virus in 24-well plates) were inoculated with 250 μl of Env pseudotyped viruses and cultured in medium containing 8 μg/ml Polybrene. Additionally, HEK293T cells stably expressing Env were inoculated with Env-pseudotyped viruses of FeLV-B/Gardner-Arnstein, FeLV-D/Ty26, or ERV-DC10 and cultured in the same medium. After 48 h, cells were stained with X-Gal, and single-cycle infectivity was titrated by counting blue-stained nuclei under the microscope.
Phylogenetic and genetic analysis.
The following amino acid sequences were used to construct a phylogenetic tree for Env: Friend MLV (accession number CAA26561.1), Moloney MLV (AAB59943.1), xenotropic murine leukemia virus-related virus (XMRV; ADU55752.1), FeLV subgroup A/61E (FeLV-A/61E; AAA93093.1), endogenous FeLV AGTT locus (enFeLV/AGTT; AY364318.1), ERV-DC8, -DC10, -DC14 (AB674443.1, AB674444.1, and AB674445.1, respectively), porcine endogenous retrovirus subgroup A (PERV-A; EU789636.1), gibbon ape leukemia virus (GALV; AAA46811.1), KoRV-A_1, ERV-DC7 fl, and ERV-DC16 fl. Multiple alignments of the above amino acid sequences were generated by using MUSCLE (55). The maximum-likelihood method was used to construct a phylogenetic tree. As an amino acid substitution model, a JTT matrix-based model (56) with discrete (five categories) gamma-distributed rate variation and inferred proportion of invariable sites (+G, +I; α = 1.5861, invar = 0.191146) was used, which was selected on the basis of its Bayesian information criterion (BIC) score. All positions containing gaps were eliminated from the analysis. The tree robustness was evaluated by the bootstrap method (1,000 times).
We also constructed a phylogenetic tree for ERV-DC7 env obtained from 22 domestic cats (39) by using the maximum-likelihood method. We first used PCR to amplify ERV-DC7 env from the chromosomal DNA of domestic cats with the primers Fe-186S and Fe-53R, designed for the pol region and 3′ flanking region, respectively, and directly determined their sequences. We used DNA samples from male cats because the ERV-DC7 locus is present on the X chromosome, and therefore males have only a single allele. The accession numbers for the nucleotide sequences of ERV-DC7 env are included in the following section. As a nucleotide substitution model, a Kimura 2-parameter model (57) with discrete gamma-distributed rate variation (+G, α = 0.0500) was used, which was selected on the basis of its BIC score. All positions containing gaps were eliminated from the analysis. The tree robustness was evaluated by the bootstrap method (1,000 times). All programs used for phylogenetic analyses were packaged in MEGA6 (58). A bootscanning analysis was conducted by using Simplot to detect possible recombination events (59). To estimate an ancestral natural selection on the noncoding region of ERV-DC7 env, corresponding to the region between the premature stop codon of Refrex-1 and the original env stop codon, approximate nonsynonymous/synonymous substitution rate (dN/dS) ratios on the noncoding region of the ERV-DC7 env were calculated by the Nei-Gojobori method (60) with Fisher's exact test, and this calculation was conducted by using KaKs_calculator (61).
Statistical tests.
The results of infection assays and dN/dS ratio calculations were considered statistically significant if their P values were <0.05 by Student's t test and Fisher's exact test, respectively. The difference in the number of polymorphic sites between the Refrex-1 coding region and the noncoding region of ERV-DC7 env was considered statistically significant with P values of <0.05 by a one-tailed Fisher's exact test.
Nucleotide sequence accession numbers.
The sequences reported here have been deposited in DDBJ/EMBL/GenBank under accession numbers LC033970 and LC011888 to LC011907.
RESULTS
Reconstruction of full-length Env from Refrex-1.
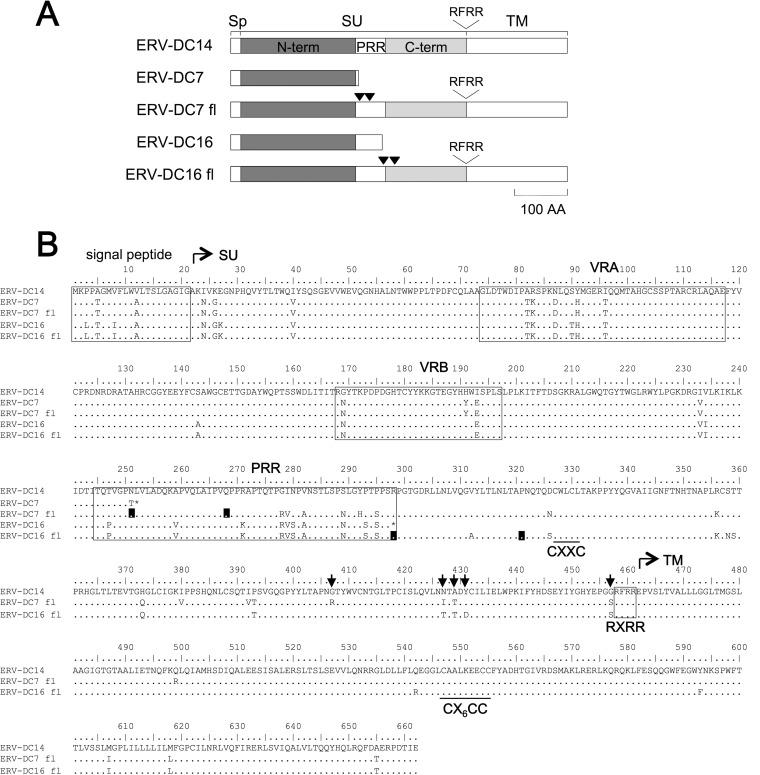
ERV-DC7 and ERV-DC16 env both encode Refrex-1, a truncated Env that has a length of 251 or 297 amino acids, respectively (Fig. 1), and consequently, these Env proteins do not retain the ability to form viral particles. In this study, we attempted to reconstitute functional, ancestral Env from the env of ERV-DC7 and ERV-DC16, both of which were cloned from Kyoto-SPF1 (41), by inducing reverse mutations. First, we removed premature and latent stop codons from the ERV-DC7 and ERV-DC16 env by site-directed mutagenesis and reconstructed full-length Env based on the sequence of ERV-DC14, which has a functional Env and also belongs to genotype I. We used a genotype I sequence for the reconstruction of ERV-DC7 and ERV-DC16 Env, even though they are both genotype II, because no functional full-length genotype II Env proteins have been identified yet. In ERV-DC7 env, codons 249 and 268 were altered from GG_ to GGT and from TAG to CAG, respectively, and in ERV-DC16 env, codons 298 and 321 were altered from TGA to CGA and from CC_ to CCA, respectively. Each codon position is counted from the first ATG of the signal peptide. These full-length Env proteins were named ERV-DC7 full-length (fl) and ERV-DC16 fl, and Fig. 1B shows their amino acid sequences. The sequences of ERV-DC7 fl and ERV-DC16 fl are 94.9% and 93.2% identical, respectively, to ERV-DC14 Env.
FIG 1.
Reconstruction of full-length Env from Refrex-1. ERV-DC7 and ERV-DC16 env encode truncated Env because of the premature stop codons present in the middle of their env ORFs. Their full-length Env (ERV-DC7 fl and ERV-DC16 fl) were constructed by removing the premature and latent stop codons. (A) Schematic representation of ERV-DC7 fl and ERV-DC16 fl. Sp, signal peptide; SU, surface subunit; TM, transmembrane subunit; N-term, SU N-terminal domain; PRR, proline-rich region; C-term, SU C-terminal domain. RFRR is the cleavage motif. Inverted triangles indicate the positions of stop codons. (B) Amino acid sequences of ERV-DC7 fl and ERV-DC16 fl. VRA, variable region A; VRB, variable region B. CXXC and CX6CC are sites of covalent interaction. Black boxes indicate positions where reverse mutations were induced for the removal of stop codons. Arrows indicate the positions of residues 407, 427, 429, 431, and 457.
Lack of infectious viral particle production by ERV-DC7 fl and ERV-DC16 fl.
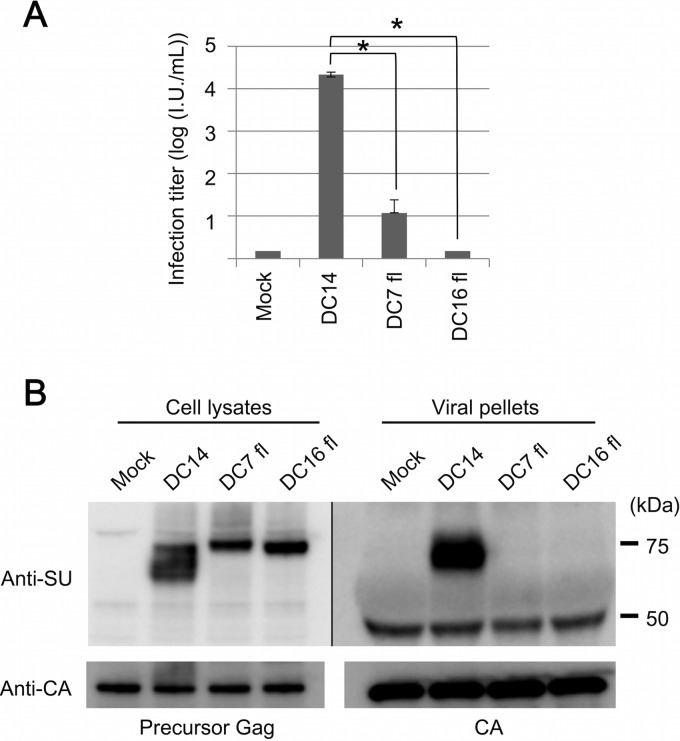
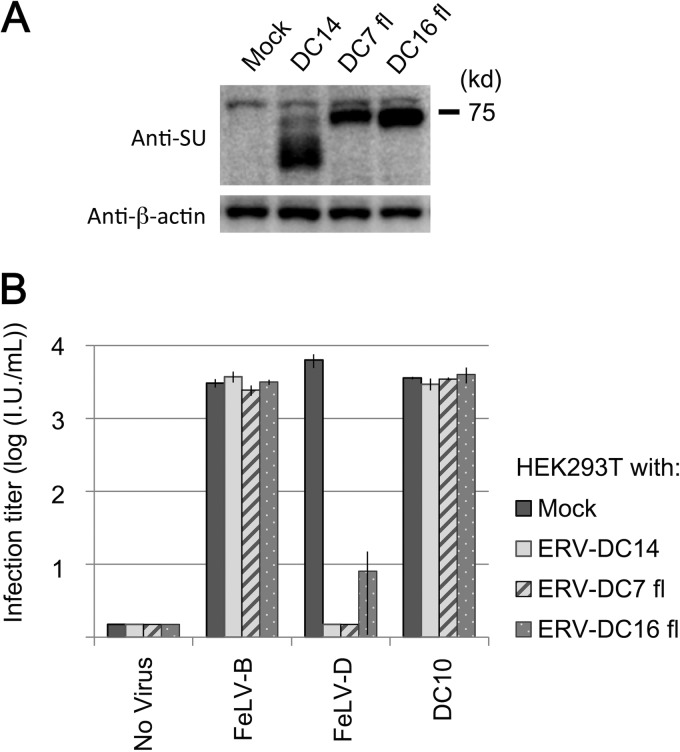
Next, we attempted to produce and analyze Env-pseudotyped viruses of ERV-DC7 fl and ERV-DC16 fl. GP Lac cells, which are env-negative packaging cells, were transfected with empty plasmids or expression plasmids of ERV-DC14 Env, ERV-DC7 fl, or ERV-DC16 fl. Cell lysates were prepared from the transfectants 48 h posttransfection, and the culture supernatants were collected, filtered, and stored at −80°C as viral supernatants. First, we conducted infection assays using the above-described Env-pseudotyped viruses. Fresh HEK293T cells were inoculated with the viruses, and after 48 h, they were stained with X-Gal and their infectivities were titrated. As shown in Fig. 2A, ERV-DC14 Env pseudotyped virus could efficiently infect HEK293T cells, and its viral titer was >104, whereas ERV-DC7 fl and ERV-DC16 fl Env pseudotyped viruses could barely infect HEK293T cells, and their viral titers were 101 and <1, respectively, which are both approximately 1,000-fold less than the viral titer of ERV-DC14 Env (P < 0.001).
FIG 2.
Evaluation of the ability of ERV-DC7 fl and ERV-DC16 fl to produce infectious virions. (A) Infection assay with LacZ-carrying Env pseudotyped viruses. GP Lac cells were transfected with empty or expression plasmids of ERV-DC14 Env, ERV-DC7 fl, or ERV-DC16 fl and induced to produce their pseudotyped viruses. Fresh HEK293T cells were inoculated with these viruses, and after 48 h, X-Gal-positive cells were counted as infectious units (I.U.). *, P < 0.001 (Student's t test). (B) Immunoblotting analysis of GP Lac cells expressing ERV-DC14 Env, ERV-DC7 fl, or ERV-DC16 fl. Viral pellets were prepared by centrifugation (29,000 × g, 90 min, 4°C) from cell culture supernatants. Env and Gag CA or Pr65 Gag precursor proteins were detected with anti-FeLV SU (gp70) and anti-MLV CA antibody, respectively. The filter exposure time was different between the left and right images.
We next conducted immunoblotting analyses using lysates from Env-expressing cells. ERV-DC Env were detected with a goat anti-FeLV SU (gp70) antibody, which cross-reacts with ERV-DC Env SU, as previously described (40). As shown in Fig. 2B (left), Env species of approximately 75 kDa and smaller were detected in the lysate of ERV-DC14 Env-expressing cells, whereas only one Env species, of approximately 75 kDa, was detected in cells expressing ERV-DC7 fl or ERV-DC16 fl Env. Pr65 Gag precursor protein was detected in all samples with a goat anti-Rauscher MLV CA antibody.
Lastly, we used an anti-FeLV SU antibody to attempt to detect Env in viral pellets prepared from the supernatants (Fig. 2B, right). Env, approximately 70 kDa, was detected in viral pellets of ERV-DC14 Env, whereas Env was not detected in the ERV-DC7 fl or ERV-DC16 fl viral pellets. Gag CA protein was detected in all samples with an anti-MLV CA antibody. Thus, we unexpectedly uncovered a dysfunction in ERV-DC7 fl and ERV-DC16 fl. These Env proteins did not participate in the production of infectious viral particles and were not incorporated into virions, even though they were both highly expressed in cells.
ERV-DC7 fl and ERV-DC16 fl Env are cleavage-defective.
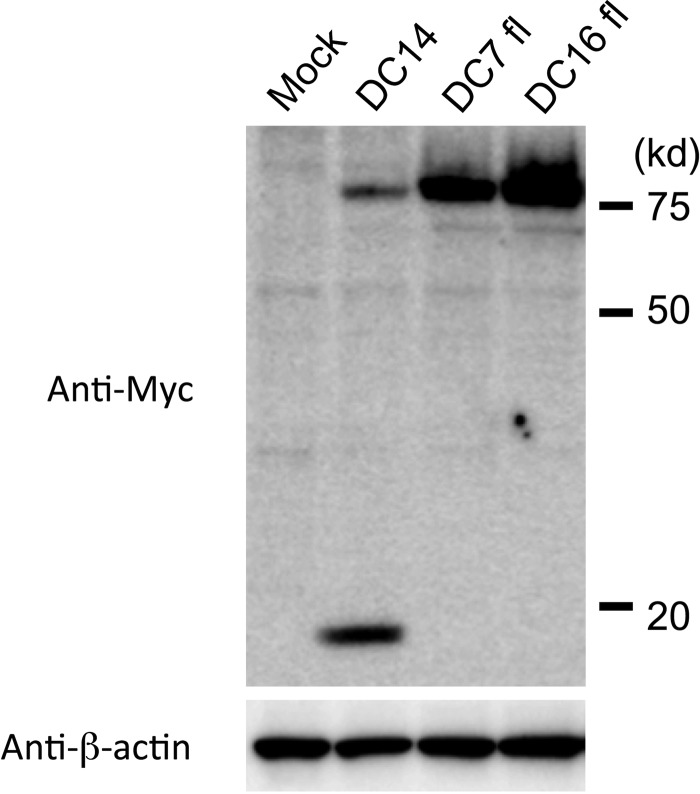
Retroviral Env proteins mature through the cleavage of SU-TM precursor proteins by cellular proteases, such as furin or closely related enzymes (15–20). We regarded the 75-kDa Env species as corresponding to Env precursors and the smaller Env species as corresponding to a cleaved SU subunit. We suspected that the dysfunctions of ERV-DC7 fl and ERV-DC16 fl were caused by defects in their cleavage (Fig. 2B, left). Therefore, we constructed expression plasmids of Env fused with a Myc epitope sequence in the C terminus of the TM (DC14-Myc, DC7 fl-Myc, and DC16 fl-Myc) to determine whether these Env are successfully cleaved. HEK293T cells were transfected with these plasmids, and after 48 h, fusion proteins were detected with an anti-c-Myc antibody (Fig. 3). In cells expressing DC14-Myc, two bands were detected, 80 kDa and 20 kDa, corresponding to the Env precursor-Myc and TM-Myc fusion proteins, respectively, whereas only Env precursor-Myc was detected in cells expressing DC7 fl-Myc or DC16 fl-Myc. Thus, it was confirmed that ERV-DC7 fl and ERV-DC16 fl are not cleaved into SU and TM.
FIG 3.
Evaluation of the ability of ERV-DC7 fl and ERV-DC16 fl Env to undergo cleavage. HEK293T cells were transfected with empty or expression plasmids of DC14-Myc, DC7 fl-Myc, or DC16 fl-Myc, which were Env fused to a Myc tag in the C terminus of the TM. The Myc fusion proteins, both precursor Myc (80 kDa) and TM-Myc (20 kDa), and β-actin were detected in cell lysates with anti-c-Myc and anti-β-actin antibodies, respectively.
ERV-DC7 fl and ERV-DC16 fl cell surface expression.
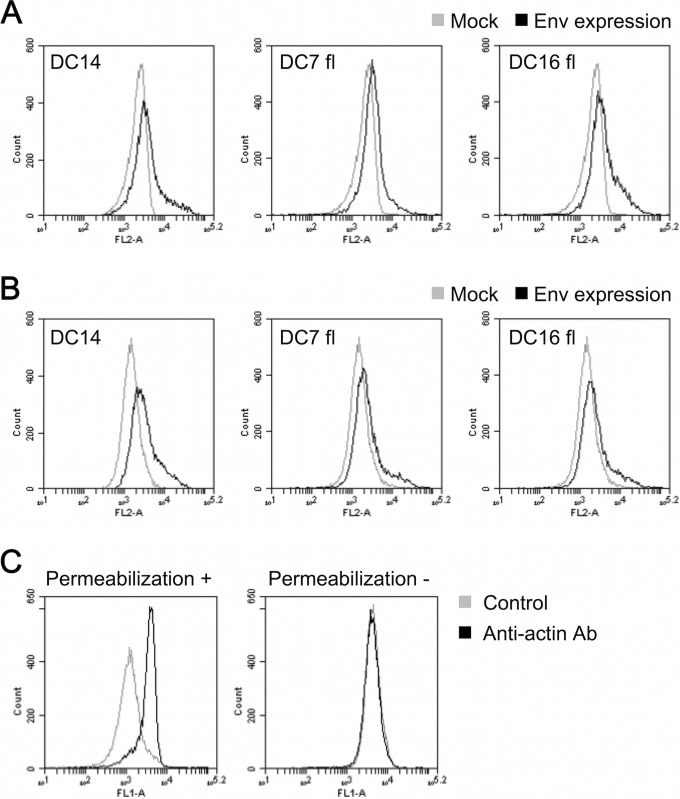
Several uncleaved Env mutants have been previously reported, and some of these were defective in membrane trafficking toward the cell surface (11, 62, 63). To determine if ERV-DC7 fl and ERV-DC16 fl shared this defect, we used FACS analysis to test if ERV-DC7 fl and ERV-DC16 fl were transported normally to the cell surface. HEK293T cells were transfected with empty or expression plasmids containing ERV-DC14 Env, ERV-DC7 fl, or ERV-DC16 fl. After 48 h, the cells were harvested and divided into two samples, one of which was permeabilized with 90% methanol (Fig. 4A) and the other of which was left unpermeabilized (Fig. 4B). The samples were stained with anti-FeLV SU antibody and phycoerythrin (PE)-conjugated anti-goat IgG antibody as the primary and secondary antibodies, respectively, and then the fluorescence signals were detected by a flow cytometer (Fig. 4A and B). All Env-expressing cells increased their signals compared with the mock-transfected cells in both permeabilized and nonpermeabilized samples. These results suggest that ERV-DC7 fl and ERV-DC16 fl are transported to and expressed on the cell surface. To confirm the validity of this assay, we attempted to detect the β-actin present in the cytoplasm in both permeabilized and nonpermeabilized samples of mock-transfected cells. The permeabilized samples increased their fluorescent signals compared with control (without primary antibody) samples, whereas nonpermeabilized samples had fluorescent signals similar to those of the control samples (Fig. 4C).
FIG 4.
FACS analysis detection of ERV-DC7 fl and ERV-DC16 fl cell surface expression. (A and B) Detection of Env on the inside and surfaces of cells. HEK293T cells expressing ERV-DC14 Env, ERV-DC7 fl, or ERV-DC16 fl were permeabilized with 90% methanol (A) or left unpermeabilized (B), and the intracellular (A) and cell surface (A and B) Env proteins were stained with goat anti-FeLV SU and PE-conjugated anti-goat IgG antibody. Fluorescent signals were detected by the FL-2 channel of a flow cytometer. Histograms of Env-expressing cells (black) and mock-transfected cells (gray) are overlaid in each graph. The x axis shows signal intensity in FL-2; the y axis shows cell counts. (C) Staining of β-actin in permeabilized (left) or nonpermeabilized (right) samples of mock-transfected cells with mouse anti-β-actin and fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG antibodies. Histograms of the cells treated with anti-β-actin antibody (Ab) and without antibody (control) are overlaid in each graph. The x axis shows signal intensity in FL-1; the y axis shows cell counts.
Receptor interference caused by ERV-DC7 fl and ERV-DC16 fl.
Receptor interference is a retroviral phenomenon in which, through Env-receptor interaction, the Env expressed in cells prevent superinfection of viruses using the same receptor (28). In this study, we examined whether ERV-DC7 fl and ERV-DC16 fl could interact with their receptor by evaluating their receptor interference capacities. For this purpose, HEK293T cells stably expressing ERV-DC14 Env, ERV-DC7 fl, or ERV-DC16 fl were established through single-cell cloning. Expression of Env was confirmed by immunoblotting with an anti-FeLV SU antibody (Fig. 5A). Fresh HEK293T cells and cells stably expressing Env were inoculated with pseudotyped viruses of FeLV-B/Gardner-Arnstein, FeLV-D/Ty26, or ERV-DC10, and after 48 h, their infectivities were titrated. As shown in Fig. 5B, cells stably expressing ERV-DC14 Env, ERV-DC7 fl, or ERV-DC16 fl were specifically resistant to FeLV-D infection. These results suggest that ERV-DC7 fl and ERV-DC16 fl interact with their viral receptor.
FIG 5.
Evaluation of ERV-DC7 fl and ERV-DC16 fl receptor interference. Cells stably expressing ERV-DC14 Env, ERV-DC7 fl, or ERV-DC16 fl were established through single-cell cloning. (A) Immunoblotting analysis of these cells. Env and β-actin were detected with anti-FeLV SU and anti-β-actin antibodies, respectively. (B) Interference assay in which cells stably expressing Env were inoculated with Env pseudotyped viruses of mock (empty vector), FeLV-B/Gardner-Arnstein, FeLV-D/Ty26, or ERV-DC10. After 48 h, X-Gal-positive cells were counted as infectious units (I.U.).
Determination of the mutations responsible for the dysfunctions of ERV-DC7 fl and ERV-DC16 fl.
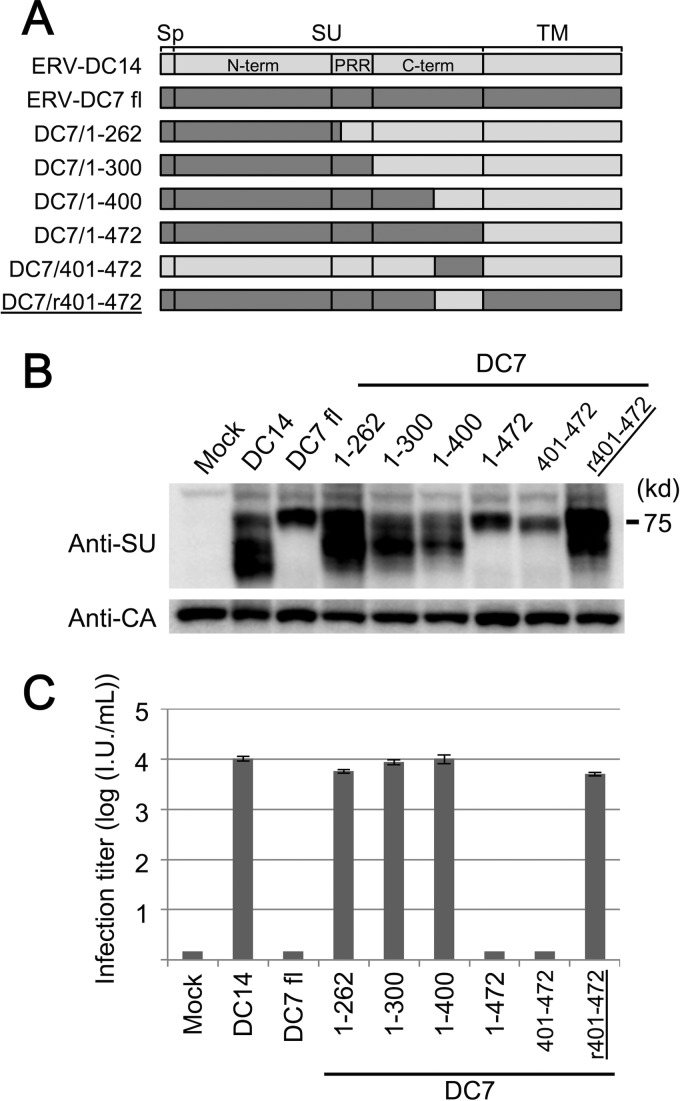
The above results revealed ERV-DC7 fl and ERV-DC16 fl dysfunctions in Env cleavage and incorporation into virions, so we next attempted to determine the region responsible for these dysfunctions. We first constructed a series of expression plasmids with Env chimeras made of ERV-DC7 fl or ERV-DC14 Env, as shown in Fig. 6A. These Env chimeras were expressed in GP Lac cells, and after 48 h, cell lysates and culture supernatants were harvested. Immunoblotting of the cell lysates showed that Env chimeras of DC7/1-262, DC7/1-300, and DC7/1-400 were cleavage competent, whereas Env chimeras of DC7/1-472 and DC7/401-472 were defective (Fig. 6B). Similarly, the results of infection assays showed that Env pseudotyped viruses of DC7/1-262, DC7/1-300, and DC7/1-400 were infectious, whereas those of DC7/1-472 and DC7/401-472 were not infectious (Fig. 6C). Furthermore, the chimeric Env of DC7/r401-472 was cleavage competent, and its pseudotyped virus was infectious (Fig. 6B and C). Immunoblotting analyses and infection assays using chimeras of ERV-DC16 fl and ERV-DC14 Env produced the same results (data not shown). Our findings indicate that the mutations responsible for the observed ERV-DC7 fl and ERV-DC16 fl dysfunctions are commonly present in the region within residues 401 to 472, which corresponds to the C terminus of the SU C-terminal domain.
FIG 6.
Determination of the region responsible for ERV-DC7 fl dysfunction. (A) Schematic representation of expression constructs coding recombinant Env composed of ERV-DC7 fl and ERV-DC14 Env. The names of constructs indicate the range of amino acid sequences from ERV-DC7 fl (the only exception is DC7/r401-472, whose name indicates the range from ERV-DC14 Env). Sp, signal peptide; SU, surface subunit; TM, transmembrane subunit; N-term, SU N-terminal domain; PRR, proline-rich region; C-term, SU C-terminal domain. (B) Immunoblotting analysis, in which Env proteins were expressed in GP Lac cells and detected in the cell lysates with anti-FeLV SU antibody. (C) Infection assay using Env-pseudotyped viruses. Fresh HEK293T cells were inoculated with pseudotyped viruses of chimeric Env, and after 48 h, X-Gal-positive cells were counted as infectious units (I.U.).
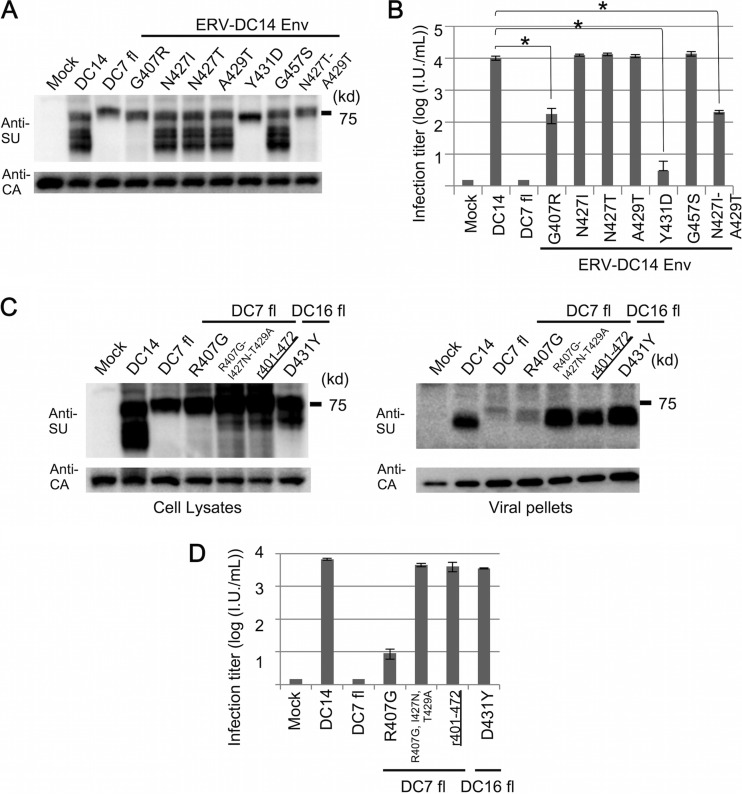
Within the region of residues 401 to 472, ERV-DC7 fl and ERV-DC16 fl each possess four amino acid substitutions compared with ERV-DC14 Env. The substitutions A429T and G457S occurred in both ERV-DC7 fl and ERV-DC16 fl, whereas G407R and N427I or N427T and Y431D independently occurred in ERV-DC7 fl or ERV-DC16 fl, respectively (Fig. 1A). We constructed six mutants of ERV-DC14 Env, each bearing a single mutation (DC14/G407R, DC14/N427I, DC14/N427T, DC14/A429T, DC14/Y431D, and DC14/G457S), and attempted to determine which mutations were responsible for the dysfunctions observed in ERV-DC7 fl and ERV-DC16 fl. These mutants were expressed in GP Lac cells, and after 48 h, cell lysates and culture supernatants were harvested and analyzed.
Immunoblotting of cell lysates showed that Env mutants of DC14/N427I, DC14/N427T, DC14/A429T, and DC14/G457S were all cleavage competent, whereas those of DC14/G407R and DC14/Y431D were cleavage defective (Fig. 7A). Similarly, infection assay results showed that Env pseudotyped viruses of DC14/N427I, DC14/N427T, DC14/A429T, and DC14/G457S were highly infectious, whereas those of DC14/G407R and DC14/Y431D had a lower viral titer than wild-type ERV-DC14 Env (Fig. 7B). Next, we constructed and analyzed ERV-DC7 fl and ERV-DC16 fl Env mutants bearing R407G and D431Y, respectively. As shown Fig. 7C and D, the mutant of ERV-DC16 fl bearing D431Y recovered its cleavage capacity and infectivity, but DC7 fl/R407G did not.
FIG 7.
Determination of mutations responsible for ERV-DC7 fl and ERV-DC16 fl dysfunctions. (A) Immunoblotting analyses of ERV-DC14 Env mutants. We constructed ERV-DC14 Env mutants, each bearing a single mutation (DC14/G407R, DC14/N427I, DC14/N427T, DC14/A429T, DC14/Y431D, and DC14/G457S) or double mutations (DC14/N427I-A429T). These mutants were expressed in GP Lac cells, and an anti-FeLV SU antibody was used for detection of Env in the cell lysates. (B) Infection assay using pseudotyped viruses of ERV-DC14 Env mutants. Fresh HEK293T cells were inoculated with these pseudotyped viruses, and after 48 h, X-Gal-positive cells were counted as infectious units (I.U.). *, P < 0.001 (Student's t test). (C) Immunoblotting analysis of ERV-DC7 fl and ERV-DC16 fl Env mutants. We constructed ERV-DC7 fl Env mutants bearing either a single mutation (DC7 fl/R407G) or triple mutations (DC7 fl/R407G-I427N-T429A) and ERV-DC16 fl Env mutants bearing a single mutation (DC16 fl/D431Y). These mutants or a chimera of DC7/r401-472 was expressed in GP Lac cells, and an anti-FeLV SU antibody was used for detection of Env in the cell lysates and viral pellets. (D) Infection assay using pseudotyped viruses of ERV-DC7 fl and ERV-DC16 fl Env mutants. Fresh HEK293T cells were inoculated with these pseudotyped viruses, and after 48 h, X-Gal-positive cells were counted as infectious units (I.U.).
This result suggests that there are additional mutations responsible for the observed dysfunctions of ERV-DC7 fl, so we constructed and analyzed ERV-DC14 Env mutants bearing double mutations. Eventually, we discovered that inducing the double mutations of N427I and A429T also prevented cleavage of ERV-DC14 Env (Fig. 7A and B). Finally, we constructed triple mutants of ERV-DC7 fl bearing the mutations R407G, I427N, and T429A and analyzed them. Immunoblotting and infection assays showed that its function was recovered to nearly the level of the DC7/r401-472 chimera, which bears the mutations R407G, I427N, T429A, and S457G (Fig. 7C and D). In summary, we identified the mutations responsible for the observed dysfunctions of ERV-DC7 fl and ERV-DC16 fl and then succeeded in reconstructing functional, ancestral Env from ERV-DC7 env and from ERV-DC16 env.
Receptor usage of the ancestor of ERV-DC7 and ERV-DC16.
As shown above, we successfully reconstituted functional, ancestral Env of ERV-DC7 and ERV-DC16 (here renamed ERV-DC7 rec and ERV-DC16 rec, respectively), which have the minimal reverse mutations R407G-I427N-T429A and D431Y, respectively. While we do not know whether these functional constructs are truly ancestral, they specifically lack the mutations that prevent Env from having its original functions. We next performed an interference assay to determine the receptor usage of ERV-DC7 rec and ERV-DC16 rec. HEK293T cells, uninfected or preinfected with FeLV-A/Glasgow-1, FeLV-B/Gardner-Arnstein, FeLV-D/Ty26, or ERV-DC10, were inoculated with the following Env pseudotyped viruses: mock (empty plasmids), ERV-DC14 Env, ERV-DC7 rec, ERV-DC16 rec, FeLV-A/Glasgow-1, FeLV-B/Gardner-Arnstein, FeLV-D/ON-T, ERV-DC10, or amphotropic MLV/4070A. After 48 h of infection, the cells were stained with X-Gal, and their infectivities were titrated. As shown Table 3, preinfection with FeLV-D specifically interfered with infection by the pseudotyped viruses of ERV-DC7 rec and ERV-DC16 rec as well as with that of ERV-DC14 and FeLV-D. This interference allowed us to identify the receptor interference group of the ancestor of ERV-DC7 and ERV-DC16; namely, they belong to the group that includes ERV-DC genotype I and FeLV-D.
TABLE 3.
Viral interference assay using LacZ-carrying Env pseudotyped viruses
| Env pseudotype | Titer (IU/ml) in HEK293T cells preinfected with: |
||||
|---|---|---|---|---|---|
| No virus | FeLV-A | FeLV-B | FeLV-D | ERV-DC10 | |
| Mock | <1 | <1 | <1 | <1 | <1 |
| ERV-DC14 Env | 7.3 × 103 | 5.8 × 103 | 8.1 × 103 | <1 | 7.2 × 103 |
| ERV-DC7 rec | 3.1 × 103 | 2.7 × 103 | 3.0 × 103 | <1 | 3.1 × 103 |
| ERV-DC16 rec | 2.3 × 103 | 3.5 × 103 | 3.9 × 103 | <1 | 2.7 × 103 |
| FeLV-A/Glasgow-1 | 4.1 × 103 | <1 | 3.8 × 103 | 3.2 × 103 | 2.6 × 103 |
| FeLV-B/Gardner-Arnstein | 1.2 × 104 | 1.2 × 104 | <1 | 1.7 × 104 | 1.5 × 104 |
| FeLV-D/ON-T | 7.8 × 103 | 6.1 × 103 | 9.4 × 103 | <1 | 7.4 × 103 |
| ERV-DC10 | 3.7 × 103 | 2.6 × 103 | 2.1 × 103 | 2.8 × 103 | <1 |
| Amphotropic MLV/4070A | 2.1 × 104 | 1.8 × 104 | 2.2 × 104 | 1.9 × 104 | 1.8 × 104 |
HEK293T cells were inoculated with Env-pseudotyped viruses. Mock, empty plasmid. X-Gal-positive cells were counted as infectious units (IU) at 48 h postinfection.
Dysfunction caused by mutations within the SU C-terminal domain among gammaretroviruses.
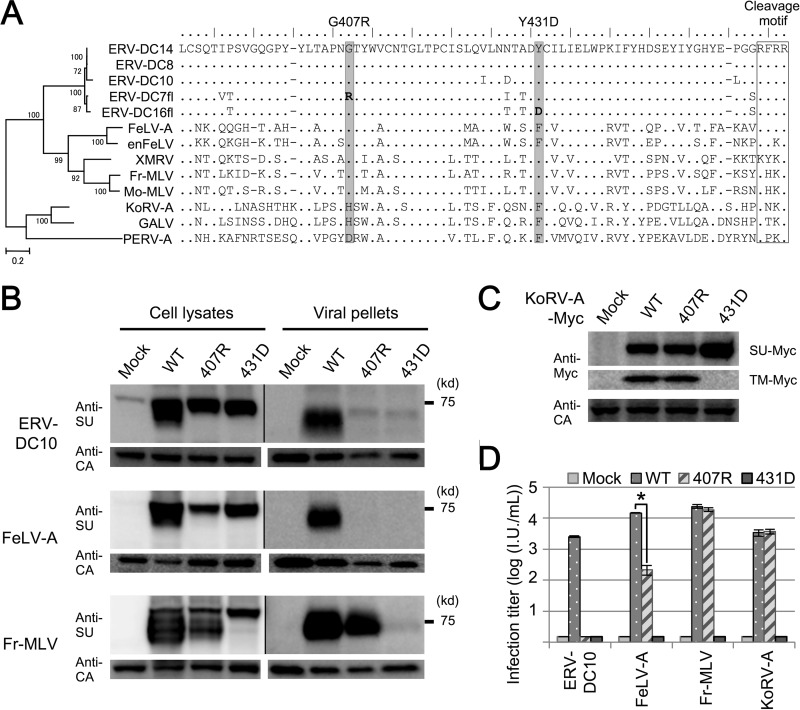
Analyses using chimeras and mutants of Env revealed that single substitutions of G407R and Y431D in the SU C-terminal domain caused Env dysfunctions in ERV-DC14. Next, we investigated if these mutations can cause the same dysfunctions in other gammaretroviruses. Figure 8A shows a phylogenetic tree and the SU C terminus amino acid sequences of several gammaretroviruses. Glycine 407, a mutation which is responsible for the observed dysfunctions of ERV-DC7 fl, is conserved within ERV-DCs, FeLV, XMRV, and MLV. Tyrosine 431, a mutation in which is responsible for the observed dysfunctions of ERV-DC16 fl, is conserved within ERV-DCs, XMRV, and MLV, and it commonly changes to phenylalanine in FeLV, PERV-A, GALV, and KoRV. We next constructed Env expression plasmids of ERV-DC10, FeLV, Fr-MLV, and KoRV-A bearing 407R or 431D and, further, constructed expression plasmids for Myc-tagged wild-type Env of KoRV-A or its mutants. Immunoblotting analyses with anti-FeLV SU antibody and anti-MLV SU antibody revealed that, in ERV-DC10 and FeLV-A, Env cleavages and incorporations were prevented by both 407R and 431D, whereas in Fr-MLV, they were prevented only by 431D (Fig. 9B). Moreover, immunoblotting analyses with anti-Myc antibody showed that the cleavage of Myc-tagged KoRV Env was prevented only by 431D (Fig. 9C). Similarly, the results of infection assays using these pseudotyped viruses revealed that the transduction activities of ERV-DC10 and FeLV-A Env were suppressed by both 407R and 431D, whereas those of Fr-MLV and KoRV-A were suppressed only by 431D (Fig. 9D). Furthermore, FACS analyses showed that, as with ERV-DC7 fl and ERV-D16 fl, 407R and 431D did not affect the membrane trafficking toward the cell surface in Fr-MLV Env (data not shown). Thus, these substitutions within the SU C-terminal domain caused the same dysfunctions in other gammaretroviruses as those observed in ERV-DC7 fl and ERV-DC16 fl.
FIG 8.
Dysfunction caused by mutations within the SU C-terminal domain in gammaretroviruses. (A) A phylogenetic tree of Env in gammaretroviruses constructed by the maximum-likelihood method is shown on the left. The percentages of branch junctions indicate their bootstrap values (1,000 times). Amino acid sequences of the gammaretrovirus SU C terminus regions are also presented. The positions of G407R and Y431D are shaded. (B) Immunoblotting analysis of GP Lac cells expressing wild-type (WT) Env or the Env mutants described for panel A. We constructed expression plasmids for Env mutants of ERV-DC10, FeLV-A/Glasgow-1, and Fr-MLV/clone 57 and expressed them in GP Lac cells. After 48 h, we harvested the cell lysates and viral pellets from the culture supernatants and analyzed them. ERV-DC10 and FeLV-A Env were detected with an anti-FeLV SU antibody, and Fr-MLV Env proteins were detected with an anti-MLV SU antibody. Pr65 Gag precursor and Gag CA were detected with an anti-MLV CA antibody. The exposure time of the filter was different for the cell lysates and viral pellets. (C) Immunoblotting analysis of GP Lac cells expressing Myc-fused WT or mutant (407R or 431D) KoRV-A_1 Env. Lysates of these cells were harvested, and Env precursor-Myc and TM-Myc were detected with an anti-Myc antibody. Gag CA was detected with an anti-MLV CA antibody. (D) Infection assay using pseudotyped viruses of WT or mutant Env. Fresh HEK293T cells were inoculated with these viral supernatants, and after 48 h, X-Gal-positive cells were counted as infectious units (I.U.). *, P < 0.001 (Student's t test).
FIG 9.
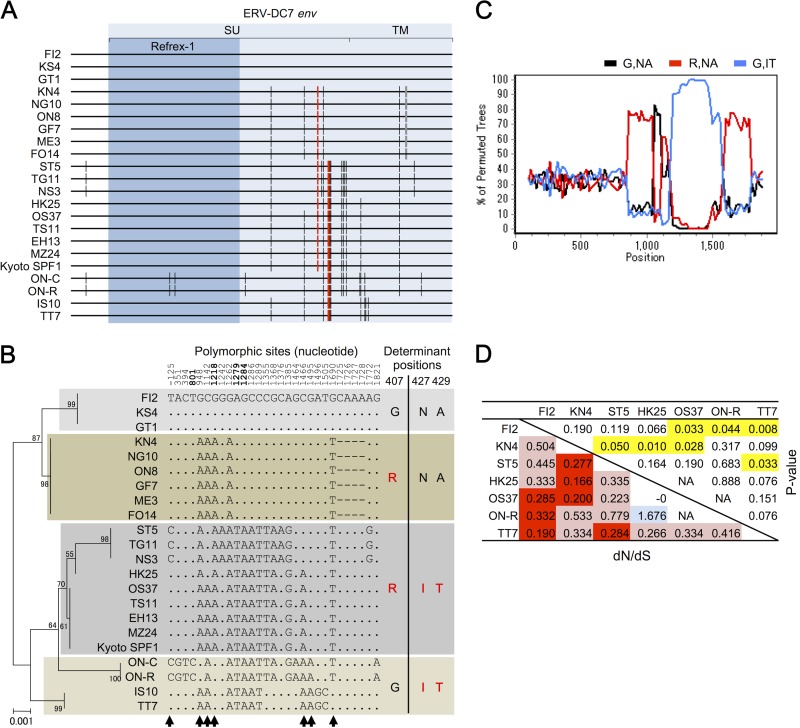
Sequence diversity of ERV-DC7 env within domestic cats. Sequences of ERV-DC7 env, other than Kyoto SPF1 and IS10, were determined from 20 domestic cats in Japan. (A) Schematic representation of ERV-DC7 env sequences from 22 individuals. The env and Refrex-1 coding regions are in light blue and blue, respectively. The black vertical bars indicate nucleotide substitution compared with the FI2 sequence. The red vertical bars indicate nucleotide substitutions at 1218, 1279, and 1284, which are responsible for the amino acid substitutions G407R, N427I, and A429T, respectively. The gray bars indicate a deletion within the region from position 1725 to 1728. (B) A phylogenetic tree of ERV-DC7 env sequences constructed by the maximum-likelihood method is shown on the left. The percentages of branch junctions indicate their bootstrap values (1,000 times). In the middle are excerptions of polymorphic sites from the ERV-DC7 env sequence and its upstream region. The numbers indicate nucleotide positions (negative values indicate the region upstream of env), and bold numbers depict the nucleotide positions generating a latent stop codon and causing amino acid substitutions at positions 407, 427, and 429. Arrows indicate the sites that conflict with the consensus tree (left). The original alignment of ERV-DC7 is shown in Fig. S1 in the supplemental material. On the right are variations of the amino acids at positions 407 and 427 to 429. Amino acids suppressing Env cleavage are in red. (C) Possible recombination events were detected by bootscanning analysis. The sequences of the R,IT group were set as the query, and those of the R,NA and G,IT groups were set as the parental pairs. (D) Estimation of the ancestral natural selection on the noncoding region of ERV-DC7 env, corresponding to the region between the premature stop codon of Refrex-1 and the original env stop codon. Pairwise approximate dN/dS ratios in the noncoding regions between alleles were calculated by the Nei-Gojobori method followed by a Fisher exact test. The lower left portion of the matrix presents the dN/dS ratios between alleles. Red indicates statistically significant dN/dS ratios of <1, and light red indicates dN/dS ratios of <1 that did not reach statistical significance. Light blue indicates a dN/dS ratios of >1 that did not reach statistical significance. The upper right portion of the matrix presents the P values calculated by a Fisher exact test. Yellow cells indicate P values of <0.05.
Sequence diversity of ERV-DC7 env within domestic cats.
We previously identified two molecular clones of ERV-DC7, IS10 (40) and Kyoto SPF1 (41), and in this study, we used Kyoto SPF1 for the reconstitution of its ancestral Env. However, we noticed that, surprisingly, ERV-DC7 IS10 did not possess a G407R substitution, which is one of the determinants of ERV-DC7 fl dysfunction. We then ascertained the sequences of ERV-DC7 env, other than Kyoto SPF1 and IS10, from 20 domestic cats in Japan and observed their sequence diversity (π = 0.004). We identified seven alleles of ERV-DC7 env from a total of 22 individuals (Fig. 8A and B, left).
All of the alleles had the same premature stop codon in the middle of env and encoded a truncated Env (Refrex-1), but a total of 24 single nucleotide polymorphisms (SNPs) and four deletion sites were present within the whole env sequence. The SNPs included 11 synonymous substitutions, 12 nonsynonymous substitutions, and 1 latent nonsense mutation (Fig. 8B, middle). The Refrex-1-coding region of ERV-DC7 env had 1 synonymous and 1 nonsynonymous substitution. Interestingly, the percentages of the polymorphic sites in this region were significantly lower than those in the noncoding region of ERV-DC7 original env, both for the total substitutions and for the synonymous substitutions, as determined by using a one-tailed Fisher's exact test (P < 0.01 and P < 0.05, respectively). Further, G407R and N427I-A429T, the determinants of ERV-DC7 fl dysfunction, were not fixed in the population of domestic cats, and four combination variants were identified: 407G and 427N-429A (G,NA), 407R and 427N-429A (R,NA), 407G and 427I-429T (G,IT), and 407R and 427I-429T (R,IT) (Fig. 8B, right). Moreover, in addition to the nucleotides described above, columns of nucleotides at −125, 948, 1142, 1466, 1495, and 1690 support tree topologies that are different from the consensus one. These topological conflicts suggest that recombination (allelic gene conversion) or parallel substitution events have occurred since the integration of ERV-DC7, and the results of a bootscanning analysis also indicate that several recombination events have occurred between these alleles (Fig. 8C).
We calculated the nonsynonymous/synonymous substitution rate (dN/dS) ratios between ERV-DC7 env allele pairs to gain insight into whether their Refrex-1 noncoding regions had undergone natural selection during the period between the integration of ERV-DC7 and its env truncation (Fig. 8D). For these calculations, we used the sequences of a ERV-DC7 env noncoding region, which corresponds to the region between the premature stop codon of Refrex-1 and the original env stop codon. The dN/dS ratios were significantly lower than 1, based on a Fisher exact test, in some allele pairs: FI2-Kyoto SPF1, FI2-ON-R, FI2-TT7, KN4-ST5, KN4-HK25, KN4-Kyoto SPF1, and ST5-TT7. This result indicates that the sequences of the ERV-DC7 env noncoding region had undergone a purifying selection between its integration and its truncation.
DISCUSSION
We previously discovered Refrex-1, a soluble restriction factor for ERV-DC genotype I and FeLV-D (41), and found that Refrex-1 is a truncated Env encoded by both ERV-DC7 and ERV-DC16, members of ERV-DC genotype II. Refrex-1 appears to be a domesticated ERV, like mouse Fv1 and Fv4 (23, 28, 29, 31). In this study, we induced several reverse mutations into ERV-DC7 and ERV-DC16 env and reconstituted full-length functional, ancestral Env (named ERV-DC7 rec and ERV-DC16 rec). As mentioned earlier, we are unable to prove that these functional constructs are truly ancestral, but, importantly, they do not have the mutations that prevent the original properties of Env.
According to our phylogenetic analyses, genotype II split off before the divergence between genotype I and genotype III (Fig. 9A). This group contains ERV-DC7 locus, which, judging from a genetic analysis of LTRs, are the oldest ERV-DC locus (40). These results indicate that genotype II is the most ancient ERV-DC group; therefore, the reconstruction of ERV-DC7 and ERV-DC16 Env enabled us to investigate the ancestral properties of ERV-DC Env.
Receptor interference assays showed that preinfection with FeLV-D, which is classified in the same interference group as ERV-DC14, specifically interfered with infections by Env pseudotyped viruses of ERV-DC7 rec and ERV-DC16 rec (Table 3). This work, together with another recent study (41), revealed that ERV-DCs have two receptor interference groups; one includes genotypes I and II, and the other includes genotype III. These results were expected given that Refrex-1 inhibits ERV-DC genotype I and FeLV-D but not genotype III (41). Genotype II is the most ancient ERV-DC group, and therefore, genotype III may have been generated from genotype II by switching its receptor usage from its ancestral one. Based on their tolerance to Refrex-1, this receptor switch might have been caused by selection pressure from this truncated Env.
In the process of reconstituting the ancestral Env, we unexpectedly discovered that full-length ERV-DC7 and ERV-DC16 Env (named ERV-DC7 fl and ERV-DC16 fl), made by the removal of premature or latent stop codons, did not produce infectious virions when expressed in packaging cells that also expressed Moloney murine leukemia virus (Mo-MLV) gag and pol (Fig. 2A). This lack of infectivity appeared to be the result of dysfunctions in Env cleavage and incorporation into virions (Fig. 3 and 2B, respectively). Analysis of Env chimeras and mutants revealed that the mutations G407R plus N427I-A429T and the mutation Y431D present within the C terminus of the SU C-terminal domain were the mutations responsible for ERV-DC7 fl and ERV-DC16 fl dysfunctions, respectively. These substitutions are present in positions distant from the cleavage motif in the amino acid sequences, so they may prevent Env cleavage by changing the three-dimensional (3D) structure of Env and decreasing the accessibility of host proteases to the cleavage site. Substitutions from a hydrophobic amino acid to a charged one, such as G407R and Y431D, typically appear to have a strong impact on protein folding. These conformational changes do not affect Env trafficking to the cell surface or receptor binding, based on the results from our FACS analysis and the interference capacities of these mutants (Fig. 4 and 5, respectively).
Glycine 407 and tyrosine or phenylalanine 431 were relatively conserved among gammaretroviruses; the former was conserved in MLV, FeLV, and ERV-DCs, and the latter was conserved in MLV, FeLV, ERV-DCs, PERV-A, GaLV, and KoRV. Substitutions of these amino acids caused the same dysfunctions in Fr-MLV, FeLV-A, ERV-DC10, and KoRV-A as those observed in ERV-DC7 fl and ERV-DC16 fl. This suggests that the 3D structure and/or folding pathway of the SU C-terminal domains is conserved in gammaretroviruses.
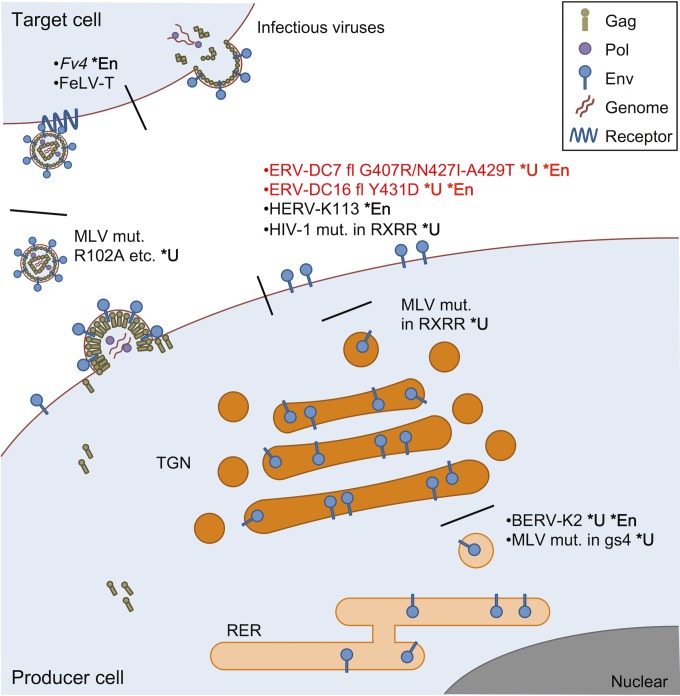
In previous studies, several uncleaved Env mutants have been reported in various retroviruses (16–19, 62–64). The properties of uncleaved Env are different from each other, and they appear to be determined by the positions of the mutations within Env (Fig. 10). For example, uncleaved Env mutants of Mo-MLV bearing mutations within their SU N-terminal domains, such as R102D or R124E, were transported normally to the cell surface and incorporated into virions, but they lacked receptor binding abilities (64). Thus, Env cleavage does not appear to be essential for Env incorporation, at least in Mo-MLV. Our study showed that uncleaved Env bearing substitutions within the C terminus of the SU C-terminal domain were transported normally to the cell surface but were not incorporated into virions. This indicates that the SU C-terminal domain is related to Env incorporation after the stage of transport to the cell surface. Env incorporation is likely carried out by interactions between the Env cytoplasmic tail (CT) and the gag matrix (MA) segment, so a more detailed investigation is still needed (15, 65, 66).
FIG 10.
Various Env dysfunctions caused by mutations within its SU. BERV-K2 Env and Fr-MLV Env mutants with mutations within glycosylation site 4 (gs4) are not cleaved because their mutation prevents intracellular trafficking from the rough endoplasmic reticulum (RER) to the trans-Golgi network (TGN), where Env cleavage occurs (11, 63). Uncleaved Mo-MLV Env mutants with mutations within the cleavage motif (RXRR) are trapped in the TGN without transport to the cell surface (62). ERV-DC7 fl and ERV-DC16 fl, both of which are full length but harbor substitutions of G407R/N427I-A549T and Y431D, respectively, and Env mutants of HIV-1 with mutations within the cleavage motif are cleavage defective, and they can be transported to the cell surface but are not incorporated into virions (17). HERV-K113 Env can also reach the cell surface, but it cannot be incorporated into virions (68). Uncleaved Mo-MLV Env mutants with mutations within the N-terminal domain, such as R102A, are incorporated into virions, but they do not bind to viral receptors (64). Fv4 Env can be incorporated into virions and bind to viral receptors, but it does not undergo membrane fusion owing to a substitution in its TM fusion peptide (34, 69). FeLV-T Env is incorporated into virions and successfully binds its receptor, but it is unable to undergo fusion because of mutations within its PHQ motif, which mediates receptor binding and membrane fusion (70, 71). The defective fusion of FeLV-T Env is recovered by the presence of FeLIX (70). *U, uncleaved Env; *En, ERV Env.
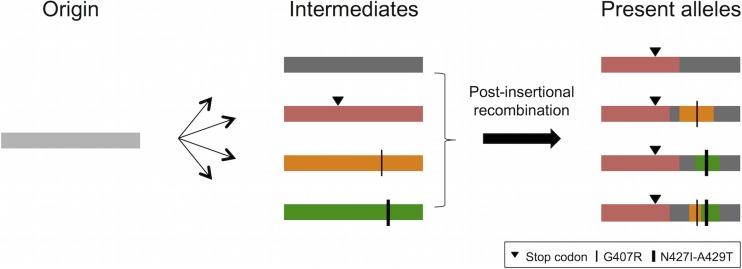
The substitutions G407R, N427I-A429T, and Y431D are responsible for some Env dysfunctions, and they likely contributed to the past inactivation of ERV-DC7 and ERV-DC16 loci, even though upstream premature stop codons present in the modern truncated Env currently prevent them from exerting their effects. The fact that the mutations that caused the same dysfunctions independently and repeatedly occurred in ERV-DC7 env and ERV-DC16 env supports this hypothesis. ERV-DC7 fl and ERV-DC16 fl possess receptor interference capacities and were able to protect the cells from infection with FeLV-D (Fig. 5B). Furthermore, the dN/dS ratios for the sequences of the ERV-DC7 env noncoding region, corresponding to the region between the premature stop codon of Refrex-1 and the original env stop codon, are significantly lower than 1 in some allele pairs (Fig. 8D). This result indicates that these sequences of the noncoding region had undergone purifying selection during the period between ancestral integration and env truncations. In the process of their domestication, full-length but dysfunctional Env proteins, such as ERV-DC7 fl and ERV-DC16 fl, may have existed as intermediates, and, like Refrex-1, they might have also contributed to viral resistance (Fig. 11).
FIG 11.
Schematic representation of the proposed model for the evolution of ERV-DC7 env (Refrex-1). The origin of ERV-DC7 was generated by the integration of an ancestral retrovirus. Postinsertional mutations were accumulated in this origin, and several alleles arose from it as intermediates. Some were inactivated by the mutations that suppressed Env functions, such as premature or latent stop codons, G407R, and N427I-A429T. Present alleles were generated by multiple recombination events among these intermediate alleles. The premature stop codon may have been distributed and fixed within the population of domestic cats through these processes.
A sequence analysis of ERV-DC7 env from 22 cats in Japan revealed that at least seven alleles of ERV-DC7 env are present in the population of domestic cats. This diversity among alleles was generated by postintegrated substitutions. ERV-DC7 is one of the oldest ERV-DC loci, which is probably the reason that it has such diversity (40). Surprisingly, G407R and N427I-A429T, the determinants of ERV-DC7 fl dysfunction, were not fixed in the cat population, and there were four combination variants (G,NA, R,NA, G,IT, and R,IT).
In addition to those described above, some SNPs that were observed within several alleles support tree topologies that are different from both the consensus tree topology and the tree topologies supported by one another. These topological conflicts are explained by the presence of recombination (allelic gene conversion) or parallel substitution events that occurred since its integration. Some of them may have been generated by recombination among alleles, as indicated by the results of a bootscanning analysis (Fig. 8C). Furthermore, the Refrex-1 coding region of ERV-DC7 env has a significantly lower percentage of polymorphic sites than the noncoding region, not only for the total substitutions but also for the synonymous substitutions, based on a one-tailed Fisher's exact test (P < 0.01 and P < 0.05, respectively).
One possible hypothesis is that the all of the present ERV-DC7 env alleles were generated through replacement of the Refrex-1 coding region; intermediate alleles arose from the original ERV-DC7 insertion via an accumulation of mutations, and then their Refrex-1 regions were replaced by the one that had acquired the Refrex-1 stop codon by gene conversion (Fig. 11). This replacement of the Refrex-1 region may have swept up mutations that had originally accumulated in this region of intermediate alleles. Determining the ERV-DC7 and ERV-DC16 sequences from wild cats that belong to the genus Felis will lead to further elucidation of the evolution of Refrex-1.
This study revealed that ERV-DC7 and ERV-DC16 env were repeatedly inactivated by several mutations. ERV-DC7 env has a premature and a latent stop codon and the substitutions G407R and N427I-A429T. Similarly, ERV-DC16 env also has a premature and a latent stop codon and the substitution Y431D. It is interesting that two similar molecules of Refrex-1 are present and that they occurred in parallel through the acquisition of similar mutations during their domestications. Furthermore, this study indicates that the ERV-DC7 env region has undergone several multistep events, such as gene mutations like parallel substitutions and recombination among alleles, since its integration. We hypothesize that ERV genes gradually evolved into host genes and that several intermediate alleles have contributed to this process through the independent acquisitions of mutations and recombination events, such as host gene evolutions (67) (Fig. 11).
Finally, the strategy of reconstituting inactivated ERVs and determining the mutations responsible for their dysfunctions can help to discover previously unrecognized features or properties of retroviruses. In our case, the results of this study uncovered the vulnerability of the SU C-terminal domain in gammaretroviruses. Adding to this knowledge may lead to new strategies for establishing an effective retrovirus remedy or for developing efficient retroviral vectors.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the Saitama Children's Zoo (Saitama, Japan) for providing the koala tissue.
This study was partly supported by JSPS KAKENHI grant number 15H04602.
Funding Statement
This study was partly supported by JSPS KAKENHI grant number 15H04602.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01904-15.
REFERENCES
- 1.Herniou E, Martin J, Miller K, Cook J, Wilkinson M, Tristem M. 1998. Retroviral diversity and distribution in vertebrates. J Virol 72:5955–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 4.Belshaw R, Katzourakis A, Paces J, Burt A, Tristem M. 2005. High copy number in human endogenous retrovirus families is associated with copying mechanisms in addition to reinfection. Mol Biol Evol 22:814–817. doi: 10.1093/molbev/msi088. [DOI] [PubMed] [Google Scholar]
- 5.Belshaw R, Pereira V, Katzourakis A, Talbot G, Paces J, Burt A, Tristem M. 2004. Long-term reinfection of the human genome by endogenous retroviruses. Proc Natl Acad Sci U S A 101:4894–4899. doi: 10.1073/pnas.0307800101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costas J. 2002. Characterization of the intragenomic spread of the human endogenous retrovirus family HERV-W. Mol Biol Evol 19:526–533. doi: 10.1093/oxfordjournals.molbev.a004108. [DOI] [PubMed] [Google Scholar]
- 7.Spodick DA, Ghosh AK, Parimoo S, Roy-Burman P. 1988. The long terminal repeat of feline endogenous RD-114 retroviral DNAs: analysis of transcription regulatory activity and nucleotide sequence. Virus Res 9:263–283. doi: 10.1016/0168-1702(88)90035-4. [DOI] [PubMed] [Google Scholar]
- 8.Ghosh AK, Roy-Burman P. 1989. Characterization of enhancer elements and their mutations in the long terminal repeat of feline endogenous RD-114 proviruses. J Virol 63:4234–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliveira NM, Satija H, Kouwenhoven IA, Eiden MV. 2007. Changes in viral protein function that accompany retroviral endogenization. Proc Natl Acad Sci U S A 104:17506–17511. doi: 10.1073/pnas.0704313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee YN, Bieniasz PD. 2007. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog 3:e10. doi: 10.1371/journal.ppat.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakaya Y, Miyazawa T. 2014. Dysfunction of bovine endogenous retrovirus K2 envelope glycoprotein is related to unsuccessful intracellular trafficking. J Virol 88:6896–6905. doi: 10.1128/JVI.00288-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozak CA. 2015. Origins of the endogenous and infectious laboratory mouse gammaretroviruses. Viruses 7:1–26. doi: 10.3390/v7010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fass D, Davey RA, Hamson CA, Kim PS, Cunningham JM, Berger JM. 1997. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science 277:1662–1666. doi: 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- 14.Barnett AL, Davey RA, Cunningham JM. 2001. Modular organization of the Friend murine leukemia virus envelope protein underlies the mechanism of infection. Proc Natl Acad Sci U S A 98:4113–4118. doi: 10.1073/pnas.071432398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murakami T. 2012. Retroviral env glycoprotein trafficking and incorporation into virions. Mol Biol Int 2012:682850. doi: 10.1155/2012/682850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freed EO, Risser R. 1987. The role of envelope glycoprotein processing in murine leukemia virus infection. J Virol 61:2852–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubay JW, Dubay SR, Shin HJ, Hunter E. 1995. Analysis of the cleavage site of the human immunodeficiency virus type 1 glycoprotein: requirement of precursor cleavage for glycoprotein incorporation. J Virol 69:4675–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong JY, Dubay JW, Perez LG, Hunter E. 1992. Mutations within the proteolytic cleavage site of the Rous sarcoma virus glycoprotein define a requirement for dibasic residues for intracellular cleavage. J Virol 66:865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodman LJ, Kain SR, Firestone GL. 1993. Trafficking of wild-type and an endoproteolytic-site mutant of the mouse mammary tumor virus glycoprotein. J Biol Chem 268:2329–2336. [PubMed] [Google Scholar]
- 20.Hallenberger S, Bosch V, Angliker H, Shaw E, Klenk HD, Garten W. 1992. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature 360:358–361. doi: 10.1038/360358a0. [DOI] [PubMed] [Google Scholar]
- 21.Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, Keith JC Jr, McCoy JM. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 22.Lilly F. 1967. Susceptibility to two strains of Friend leukemia virus in mice. Science 155:461–462. doi: 10.1126/science.155.3761.461. [DOI] [PubMed] [Google Scholar]
- 23.Best S, Le Tissier P, Towers G, Stoye JP. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 24.Benit L, De Parseval N, Casella JF, Callebaut I, Cordonnier A, Heidmann T. 1997. Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J Virol 71:5652–5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jung YT, Lyu MS, Buckler-White A, Kozak CA. 2002. Characterization of a polytropic murine leukemia virus proviral sequence associated with the virus resistance gene Rmcf of DBA/2 mice. J Virol 76:8218–8224. doi: 10.1128/JVI.76.16.8218-8224.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu T, Yan Y, Kozak CA. 2005. Rmcf2, a xenotropic provirus in the Asian mouse species Mus castaneus, blocks infection by polytropic mouse gammaretroviruses. J Virol 79:9677–9684. doi: 10.1128/JVI.79.15.9677-9684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dupressoir A, Lavialle C, Heidmann T. 2012. From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta 33:663–671. doi: 10.1016/j.placenta.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 28.Nethe M, Berkhout B, van der Kuyl AC. 2005. Retroviral superinfection resistance. Retrovirology 2:52. doi: 10.1186/1742-4690-2-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malfavon-Borja R, Feschotte C. 2015. Fighting fire with fire: endogenous retrovirus envelopes as restriction factors. J Virol 89:4047–4050. doi: 10.1128/JVI.03653-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikeda H, Odaka T. 1983. Cellular expression of murine leukemia virus gp70-related antigen on thymocytes of uninfected mice correlates with Fv-4 gene-controlled resistance to Friend leukemia virus infection. Virology 128:127–139. doi: 10.1016/0042-6822(83)90324-0. [DOI] [PubMed] [Google Scholar]
- 31.Ikeda H, Laigret F, Martin MA, Repaske R. 1985. Characterization of a molecularly cloned retroviral sequence associated with Fv-4 resistance. J Virol 55:768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikeda H, Sugimura H. 1989. Fv-4 resistance gene: a truncated endogenous murine leukemia virus with ecotropic interference properties. J Virol 63:5405–5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masuda M, Yoshikura H. 1990. Construction and characterization of the recombinant Moloney murine leukemia viruses bearing the mouse Fv-4 env gene. J Virol 64:1033–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor GM, Gao Y, Sanders DA. 2001. Fv-4: identification of the defect in Env and the mechanism of resistance to ecotropic murine leukemia virus. J Virol 75:11244–11248. doi: 10.1128/JVI.75.22.11244-11248.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trejbalova K, Blazkova J, Matouskova M, Kucerova D, Pecnova L, Vernerova Z, Heracek J, Hirsch I, Hejnar J. 2011. Epigenetic regulation of transcription and splicing of syncytins, fusogenic glycoproteins of retroviral origin. Nucleic Acids Res 39:8728–8739. doi: 10.1093/nar/gkr562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bjerregaard B, Holck S, Christensen IJ, Larsson LI. 2006. Syncytin is involved in breast cancer-endothelial cell fusions. Cell Mol Life Sci 63:1906–1911. doi: 10.1007/s00018-006-6201-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu H, Liu T, Zhao Z, Chen Y, Zeng J, Liu S, Zhu F. 2014. Mutations in 3′-long terminal repeat of HERV-W family in chromosome 7 upregulate syncytin-1 expression in urothelial cell carcinoma of the bladder through interacting with c-Myb. Oncogene 33:3947–3958. doi: 10.1038/onc.2013.366. [DOI] [PubMed] [Google Scholar]
- 38.Hartmann K. 2011. Clinical aspects of feline immunodeficiency and feline leukemia virus infection. Vet Immunol Immunopathol 143:190–201. doi: 10.1016/j.vetimm.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe S, Kawamura M, Odahara Y, Anai Y, Ochi H, Nakagawa S, Endo Y, Tsujimoto H, Nishigaki K. 2013. Phylogenetic and structural diversity in the feline leukemia virus env gene. PLoS One 8:e61009. doi: 10.1371/journal.pone.0061009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anai Y, Ochi H, Watanabe S, Nakagawa S, Kawamura M, Gojobori T, Nishigaki K. 2012. Infectious endogenous retroviruses in cats and emergence of recombinant viruses. J Virol 86:8634–8644. doi: 10.1128/JVI.00280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ito J, Watanabe S, Hiratsuka T, Kuse K, Odahara Y, Ochi H, Kawamura M, Nishigaki K. 2013. Refrex-1, a soluble restriction factor against feline endogenous and exogenous retroviruses. J Virol 87:12029–12040. doi: 10.1128/JVI.01267-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mendoza R, Anderson MM, Overbaugh J. 2006. A putative thiamine transport protein is a receptor for feline leukemia virus subgroup A. J Virol 80:3378–3385. doi: 10.1128/JVI.80.7.3378-3385.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeuchi Y, Vile RG, Simpson G, O'Hara B, Collins MK, Weiss RA. 1992. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J Virol 66:1219–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson MM, Lauring AS, Robertson S, Dirks C, Overbaugh J. 2001. Feline Pit2 functions as a receptor for subgroup B feline leukemia viruses. J Virol 75:10563–10572. doi: 10.1128/JVI.75.22.10563-10572.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tailor CS, Willett BJ, Kabat D. 1999. A putative cell surface receptor for anemia-inducing feline leukemia virus subgroup C is a member of a transporter superfamily. J Virol 73:6500–6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quigley JG, Burns CC, Anderson MM, Lynch ED, Sabo KM, Overbaugh J, Abkowitz JL. 2000. Cloning of the cellular receptor for feline leukemia virus subgroup C (FeLV-C), a retrovirus that induces red cell aplasia. Blood 95:1093–1099. [PubMed] [Google Scholar]
- 47.Graham FL, Smiley J, Russell WC, Nairn R. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol 36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 48.Kim CH, Oh Y, Lee TH. 1997. Codon optimization for high-level expression of human erythropoietin (EPO) in mammalian cells. Gene 199:293–301. doi: 10.1016/S0378-1119(97)00384-3. [DOI] [PubMed] [Google Scholar]
- 49.Jainchill JL, Aaronson SA, Todaro GJ. 1969. Murine sarcoma and leukemia viruses: assay using clonal lines of contact-inhibited mouse cells. J Virol 4:549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rasheed S, Gardner MB. 1980. Characterization of cat cell cultures for expression of retrovirus, FOCMA and endogenous sarc genes, p 393–400. In Hardy WD Jr, Essex M, McClelland AJ (ed), Proceedings of the Third International Feline Leukemia Virus Meeting. Elsevier/North-Holland Publishing Co., New York, NY. [Google Scholar]
- 51.Stewart MA, Warnock M, Wheeler A, Wilkie N, Mullins JI, Onions DE, Neil JC. 1986. Nucleotide sequences of a feline leukemia virus subgroup A envelope gene and long terminal repeat and evidence for the recombinational origin of subgroup B viruses. J Virol 58:825–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nunberg JH, Williams ME, Innis MA. 1984. Nucleotide sequences of the envelope genes of two isolates of feline leukemia virus subgroup B. J Virol 49:629–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Freshney RI. 2010. Culture of animal cells: a manual of basic technique and specialized applications, 6th ed, p 208–211 Wiley-Blackwell, Hoboken, NJ. [Google Scholar]
- 54.Koch W, Hunsmann G, Friedrich R. 1983. Nucleotide sequence of the envelope gene of Friend murine leukemia virus. J Virol 45:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. [DOI] [PubMed] [Google Scholar]
- 57.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 58.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nei M, Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3:418–426. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Z, Li J, Zhao XQ, Wang J, Wong GK, Yu J. 2006. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4:259–263. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Apte S, Sanders DA. 2010. Effects of retroviral envelope-protein cleavage upon trafficking, incorporation, and membrane fusion. Virology 405:214–224. doi: 10.1016/j.virol.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 63.Li Z, Pinter A, Kayman SC. 1997. The critical N-linked glycan of murine leukemia virus envelope protein promotes both folding of the C-terminal domains of the precursor polyprotein and stability of the postcleavage envelope complex. J Virol 71:7012–7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zavorotinskaya T, Albritton LM. 1999. Failure to cleave murine leukemia virus envelope protein does not preclude its incorporation in virions and productive virus-receptor interaction. J Virol 73:5621–5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu X, Yuan X, Matsuda Z, Lee TH, Essex M. 1992. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J Virol 66:4966–4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lucas TM, Lyddon TD, Grosse SA, Johnson MC. 2010. Two distinct mechanisms regulate recruitment of murine leukemia virus envelope protein to retroviral assembly sites. Virology 405:548–555. doi: 10.1016/j.virol.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kitano T, Noda R, Takenaka O, Saitou N. 2009. Relic of ancient recombinations in gibbon ABO blood group genes deciphered through phylogenetic network analysis. Mol Phylogenet Evol 51:465–471. doi: 10.1016/j.ympev.2009.02.023. [DOI] [PubMed] [Google Scholar]
- 68.Dewannieux M, Blaise S, Heidmann T. 2005. Identification of a functional envelope protein from the HERV-K family of human endogenous retroviruses. J Virol 79:15573–15577. doi: 10.1128/JVI.79.24.15573-15577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takeda A, Matano T. 2007. Inhibition of infectious murine leukemia virus production by Fv-4 env gene products exerting dominant negative effect on viral envelope glycoprotein. Microbes Infect 9:1590–1596. doi: 10.1016/j.micinf.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 70.Anderson MM, Lauring AS, Burns CC, Overbaugh J. 2000. Identification of a cellular cofactor required for infection by feline leukemia virus. Science 287:1828–1830. doi: 10.1126/science.287.5459.1828. [DOI] [PubMed] [Google Scholar]
- 71.Zavorotinskaya T, Albritton LM. 1999. Suppression of a fusion defect by second site mutations in the ecotropic murine leukemia virus surface protein. J Virol 73:5034–5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.