ABSTRACT
Bacterial interspecies interactions play clinically important roles in shaping microbial community composition. We observed that Corynebacterium spp. are overrepresented in children free of Streptococcus pneumoniae (pneumococcus), a common pediatric nasal colonizer and an important infectious agent. Corynebacterium accolens, a benign lipid-requiring species, inhibits pneumococcal growth during in vitro cocultivation on medium supplemented with human skin surface triacylglycerols (TAGs) that are likely present in the nostrils. This inhibition depends on LipS1, a TAG lipase necessary for C. accolens growth on TAGs such as triolein. We determined that C. accolens hydrolysis of triolein releases oleic acid, which inhibits pneumococcus, as do other free fatty acids (FFAs) that might be released by LipS1 from human skin surface TAGs. Our results support a model in which C. accolens hydrolyzes skin surface TAGS in vivo releasing antipneumococcal FFAs. These data indicate that C. accolens may play a beneficial role in sculpting the human microbiome.
IMPORTANCE
Little is known about how harmless Corynebacterium species that colonize the human nose and skin might impact pathogen colonization and proliferation at these sites. We show that Corynebacterium accolens, a common benign nasal bacterium, modifies its local habitat in vitro as it inhibits growth of Streptococcus pneumoniae by releasing antibacterial free fatty acids from host skin surface triacylglycerols. We further identify the primary C. accolens lipase required for this activity. We postulate a model in which higher numbers of C. accolens cells deter/limit S. pneumoniae nostril colonization, which might partly explain why children without S. pneumoniae colonization have higher levels of nasal Corynebacterium. This work narrows the gap between descriptive studies and the needed in-depth understanding of the molecular mechanisms of microbe-microbe interactions that help shape the human microbiome. It also lays the foundation for future in vivo studies to determine whether habitat modification by C. accolens could be promoted to control pathogen colonization.
INTRODUCTION
Streptococcus pneumoniae (pneumococcus) is a major cause of pneumonia, septicemia, meningitis, otitis media, and sinusitis in children and adults worldwide. The World Health Organization estimates that each year S. pneumoniae leads to more than 1 million deaths, the majority of which occur in developing countries and are of children under the age of 5 years (1, 2). More than 20,000 deaths annually have been estimated in the United States (3–5). Although most individuals colonized with S. pneumoniae do not develop pneumococcal infection, colonization greatly increases the risk of, and is a prerequisite for, both infection and transmission. Children are the primary reservoir, with those between 6 months and 7 years of age having the highest rate of nasal colonization (40 to 60% on average in developed countries), and this rate decreases in adults. Colonization rates show extensive variation depending on multiple factors, such as geographical strain epidemiology, health status, socioeconomic environment, day care use, etc. (1, 6). Because S. pneumoniae can be part of the healthy microbiota without causing disease, it is considered a pathobiont—a commensal bacterium that is also a pathogen (7).
A challenge to controlling pneumococcus in humans is that there are over 92 known capsular serotypes of S. pneumoniae and these display different antigenic properties and biological behaviors (8–12). Introduction of conjugated vaccines, which are effective in infants, against the most commonly invasive serotypes has led to a significant reduction of severe infections, i.e., invasive pneumococcal disease (13, 14). In contrast, conjugate vaccines have been less effective in controlling milder, noninvasive pneumococcal disease, such as middle ear infections (otitis media). In addition, while vaccinated individuals are uncommonly colonized by vaccine serotypes, the ability of nonvaccine pneumococcal serotypes to occupy the niche vacated by vaccine serotypes (serotype replacement) means that overall pneumococcal colonization rates remain unchanged (15–17). Thus, an increased understanding of S. pneumoniae colonization dynamics, especially in children, is important for developing new approaches to prevent pneumococcal colonization.
The pediatric nasal passages are the primary reservoir for S. pneumoniae. Pneumococcus is present from where the nostrils (anterior nares) open onto the skin of the nasal vestibules (18, 19), a habitat complete with sweat and sebaceous glands, through the mucosal surfaces of more posterior sections of the nasal cavity to the nasopharynx (see Fig. S1 in the supplemental material). (Henceforth, we refer to the nostrils and nasal vestibules simply as the nostrils.) In prepubertal children, many other bacteria cocolonize these nasal surfaces with pneumococcus, including other members of the phylum Firmicutes, along with members of the phyla Proteobacteria, Bacteroidetes, and Actinobacteria (18, 20, 21). We and others (18, 19) have hypothesized that pneumococcal interactions with commensal bacteria impact the ability of pneumococcus to colonize, proliferate, and persist. If so, then defining pediatric nasal microbiota in the presence and absence of pneumococcal colonization might reveal potential clinically relevant interactions that affect pneumococcal colonization dynamics. For example, Pettigrew and colleagues observed that children free of nostril pneumococcal colonization had a greater relative abundance of Corynebacterium spp. than did children with pneumococcal colonization (18). In a separate cohort, Pettigrew et al. found that children harboring low levels of Streptococcus spp. had high levels of Corynebacterium spp. (19). These observations led the authors to speculate that commensal Corynebacterium spp. may play a protective role against pneumococcus. Determining the mechanism(s) that underlies such antagonism is of particular interest. Because there is no established animal model for commensal Corynebacterium spp. colonization, we chose to approach identification of a potential mechanism(s) by using in vitro cocultivation.
Although non-diphtheriae Corynebacterium spp., along with pneumococcus, commonly colonize the skin and nasal passages (see Fig. S1 in the supplemental material), their function(s) in these complex microbial communities is poorly understood (21–31). Among the Corynebacterium spp., a subset require an exogenous source of free fatty acids (FFAs) for growth because they lack a fatty acid synthase (32). Culture media for these fatty acid synthase-deficient Corynebacterium spp. commonly incorporate Tween 80 (Tw80; polyoxyethylenesorbitan mono-oleate), a synthetic lipid that contains an ester of oleic acid, a monounsaturated 18-carbon fatty acid (32). Many bacterial lipases and esterases can hydrolyze the ester bond in Tw80, releasing free oleic acid (33). Growth of C. accolens on Tw80 suggests that lipids containing oleic acid might serve as a source of required FFAs in vivo. However, to our knowledge, the host lipids from which fatty acid-requiring Corynebacterium spp. harvest FFAs in vivo have not been thoroughly investigated, nor have the genes required for such harvest.
In humans, unlike other animals tested to date, e.g., mice and rats (34, 35), triacylglycerols (TAGs) constitute a large percentage (40 to 60%) of the total lipid content of sebum and a slightly lower percentage of skin surface lipids (SSLs). Thus, TAGs are a major source of FFAs on skin (36, 37). TAGs consist of three fatty acid moieties, each linked via an ester bond to a glycerol backbone. Recent analyses using high-resolution chromatography coupled to mass spectrometry reveal an amazing diversity of TAGs on human skin (37). This diversity arises from variations in both the combination and the composition of the fatty acid moieties, which on human skin reportedly range in length from 12 to 18 carbon atoms with differing degrees of saturation (36, 37). Lipases act primarily on esters of water-insoluble fatty acids with chain lengths greater than nine carbon atoms. Microbiota- and host-produced lipases are expected to hydrolyze sebum and skin surface TAGs, including those coating the human nostrils, releasing FFAs. These FFAs are likely used by lipid-requiring Corynebacterium spp. such as C. accolens that colonize human nasal and skin surfaces.
In this study, we present data for pediatric nasopharyngeal bacterial microbiota that corroborate the finding that Corynebacterium spp. are overrepresented in children free of pneumococcal nasal colonization (18). Based on these data, we hypothesized that Corynebacterium spp. might directly antagonize pneumococcus during in vitro cocultivation. We found that, in vitro, C. accolens, a lipid-requiring species, uses representative human skin surface TAGs to satisfy this nutritional requirement and, in doing so, releases FFAs that inhibit pneumococcal growth. Our genetic analyses indicate that hydrolysis of a representative skin surface TAG depends on the C. accolens lipase LipS1. In addition, we have shown that many of the FFAs that would likely be released by C. accolens LipS1 from human skin and nostril surface TAGs inhibit pneumococcus in vitro. Thus, based on our data, we hypothesize that this release of antipneumococcal FFAs from human TAGs represents a mechanism by which C. accolens antagonizes pneumococcus in vivo and thus shapes human nasal microbiota in a manner that contributes to pneumococcal colonization resistance.
RESULTS
Corynebacterium species are overrepresented in nasopharyngeal samples from children who are free of pneumococcal colonization compared to samples from those with pneumococcal colonization.
To test the hypothesis that nasopharyngeal microbiota differ in children with and without pneumococcal nasopharyngeal colonization, we performed 16S rRNA gene sequencing on a small subset of samples collected as part of a large epidemiological study of pneumococcal colonization in eastern Massachusetts (16, 17, 38, 39). Data were analyzed using linear discriminant analysis effect size (LEfSe) (40) to compare the microbiota composition of children positive and negative for culture-proven S. pneumoniae nasopharyngeal colonization. This revealed that the genera Corynebacterium and Dolosigranulum were overrepresented in children negative for pneumococcal colonization by cultivation (Fig. 1, dark gray bars; see also Fig. S2 in the supplemental material), whereas Streptococcus was overrepresented in children positive for pneumococcal colonization (Fig. 1, light gray bar; see also Fig. S2). Our results from nasopharyngeal swabs complement published data from 16S rRNA gene studies of bacterial microbiota of nostril swabs from a different cohort of children that were analyzed using a different technique (18, 19), suggesting these data are robust. One potential explanation for these data is that there is direct antagonism between Corynebacterium spp. and S. pneumoniae in the human nose. However, these data do not indicate a mechanism for this hypothetical microbiological antagonism.
FIG 1 .
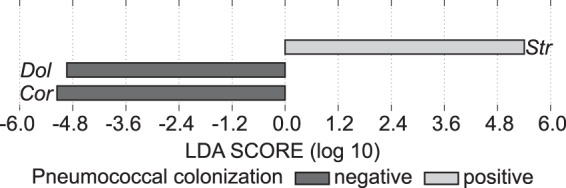
Nasopharyngeal Corynebacterium spp. are overrepresented in children without S. pneumoniae nasal colonization. Shown is a linear discriminant analysis (LDA) plot of the nasopharyngeal bacterial genera overrepresented when S. pneumoniae was absent (dark gray) or present (light gray) by cultivation. Abbreviations indicate the following genera: Str, Streptococcus; Dol, Dolosigranulum; Cor, Corynebacterium. Data were obtained from 27 nasopharyngeal swabs collected from healthy children aged 6 months to 7 years. Swabs used to inoculate medium for S. pneumoniae cultivation were frozen and later used to harvest nucleic acids, which were analyzed by 16S rRNA V1-3 tag pyrosequencing. Sequences were analyzed as described in Materials and Methods.
Interestingly, at 6 weeks of age, Corynebacterium spp. and Dolosigranulum spp. are also overrepresented in the nasopharyngeal microbiota of breastfed infants compared to formula-fed infants (41). In these breast-fed infants, the Corynebacterium spp. operational taxonomic units (OTUs) (identified by sequencing the V5-V7 region of the 16S rRNA gene) are reportedly most homologous with C. pseudodiphtheriticum and C. propinquum or with C. accolens. Metagenomic or cultivation-dependent studies are needed to conclusively determine which species of Corynebacterium are most common in the nasal microbiota of children younger than 7 years. However, given the available data, we focused our analyses on C. accolens, since this species is commonly isolated from/detected in adult noses (nostrils, nasal mucosa, and nasopharynx) (28–30), is present in the nasopharynx of young infants (41), and is rarely reported in association with infection (e.g., a PubMed search for “accolens AND infection” yielded four reports clearly implicating C. accolens in human infection, which is fewer than for C. pseudodiphtheriticum or C. propinquum). All of these are desirable characteristics for any potential nasal probiotic (42).
Representative human skin surface TAGs support the growth of C. accolens, a fatty acid-requiring species.
To test the hypothesis that there is direct antagonism between C. accolens and pneumococcus, we first established conditions under which both species will grow. To satisfy the fatty acid requirement of C. accolens, we opted to identify physiologically relevant lipids, i.e., SSLs that support cultivation of C. accolens. Oleic and linoleic acids are detected in nasal fluid (43) as well as on human skin (36, 37, 44). Triolein (Fig. 2A) and trilinolein (Fig. 2B) contain esters of oleic acid (18:1 cis-9) and linoleic acid (18:2 cis-9,12), respectively, and serve as models for human sebum and skin surface TAGs (34, 45–47). Therefore, we tested these two commercially available, representative human skin surface TAGs for the ability to support the growth of Corynebacterium sp. KPL1818, a primary nostril isolate from a healthy adult that we identified as C. accolens (see Fig. S3 in the supplemental material). (Henceforth, Corynebacterium sp. KPL1818 is referred to as C. accolens.) C. accolens grew robustly in brain heart infusion (BHI) broth supplemented with 100 µg/ml of either triolein or trilinolein but poorly, if at all, in BHI broth supplemented with solvent alone (Fig. 2C). Having identified these TAGs as physiologically relevant sources of fatty acids for C. accolens, we used one as a medium supplement for all subsequent experiments (in place of Tw80).
FIG 2 .
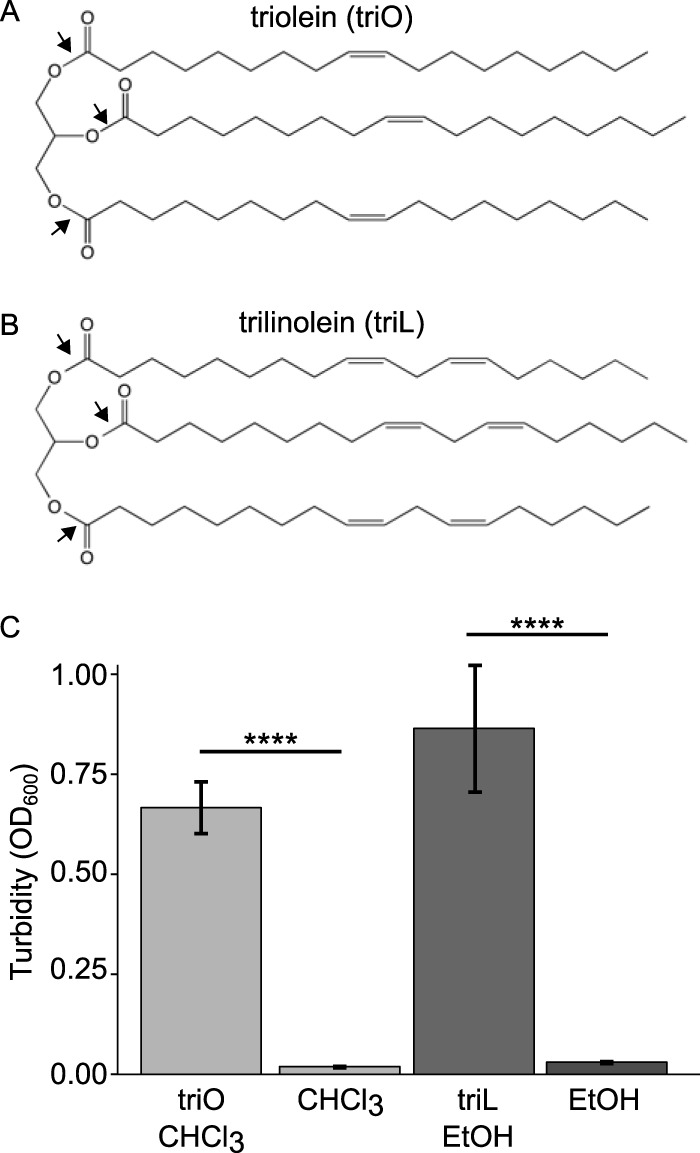
Model human skin surface TAGs triolein (triO) and trilinolein (triL) support the growth of the fatty acid-requiring species C. accolens. Black arrows indicate the oleic acid esters in (A) triolein and the linoleic esters in (B) trilinolein. (C) The mean turbidity (OD600) after 48 h of shaken aerobic growth at 37°C demonstrated increased C. accolens growth in BHI broth supplemented with one of the TAGs compared to solvent-only controls (n = 9). Data were analyzed using an ANOVA and Tukey’s multiple-comparisons test. Error bars represent standard errors of the means. ****, P ≤ 0.0001.
C. accolens inhibits pneumococcal growth during cocultivation.
To test the hypothesis that C. accolens produces a diffusible activity that antagonizes S. pneumoniae, we developed a solid agar medium-based assay that permits side-by-side cocultivation of C. accolens and S. pneumoniae, where growth antagonism is detected as zones of inhibition (ZOI). For this assay, 5 mg of either triolein or trilinolein was thinly spread on the surface of 10 ml of BHI agar, since neither one is readily soluble in aqueous-based agar medium, and served as the source of FFA for C. accolens (oleic or linoleic acid, respectively). In addition, the medium was supplemented with catalase to mitigate potential S. pneumoniae-produced hydrogen peroxide (H2O2) inhibition of Corynebacterium for two reasons. First, in vivo, other human and bacterial catalases present on the surface of the nasal passages likely contribute to detoxification of pneumococcus-produced H2O2, as occurs in saliva (48–50). Second, data indicate that H2O2-mediated killing in vitro does not necessarily reflect interactions in vivo. For example, although S. pneumoniae-produced H2O2 kills Staphylococcus aureus in vitro, data from animal models and pediatric upper respiratory tract epidemiological studies that show a negative association between S. pneumoniae and S. aureus cannot be explained by H2O2-mediated killing of cooccurring bacteria (51–54). Similarly, although pneumococcus-produced H2O2 kills Haemophilus influenzae in vitro, this effect does not explain in vivo animal model observations, which show H. influenzae-induced clearance of S. pneumoniae in a nasopharyngeal colonization mouse model (55, 56). Finally, to compensate for the differential growth rate of the two species, C. accolens was grown for 42 h prior to adjacent inoculation of S. pneumoniae 603.
On agar medium with either triolein or trilinolein as the FFA source, C. accolens inhibited S. pneumoniae (Fig. 3A and C). In contrast, in the absence of exogenous TAG (solvent alone), C. accolens grew poorly and did not inhibit S. pneumoniae (Fig. 3B and D). These results indicate that C. accolens produces an extracellular, diffusible activity that inhibits S. pneumoniae growth. C. accolens strains DSM 44278T and DSM 44279 also inhibited pneumococcal growth in either this assay or a simpler membrane-based assay (see Table S1 in the supplemental material). Having established that this inhibitory phenotype is not strain specific, we used C. accolens KPL1818 for all further experiments, as it was isolated directly from a human nostril.
FIG 3 .
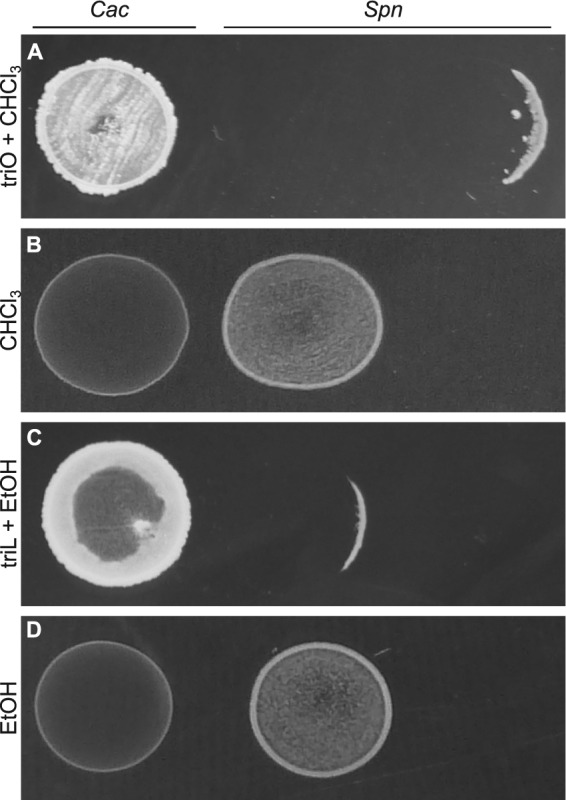
C. accolens inhibits S. pneumoniae. C. accolens KPL1818 (Cac) inhibited S. pneumoniae 603 (Spn) growth on BHIC agar spread with 50 µl of a 100-mg/ml solution of (A) triolein solubilized in chloroform (triO + CHCl3) or (C) trilinolein emulsified in ethanol (triL + EtOH), but not on medium spread with 50 µl of solvent alone: (B) chloroform (CHCl3) or (D) EtOH. Representative images are shown (n = 3). To permit visualization of partial inhibition of the pneumococcal spot, S. pneumoniae was inoculated at various distances from C. accolens, as shown by the varied sizes of ZOIs, which are likely due to differences in diffusion of the antipneumococcal molecule(s).
Methanol extracts of C. accolens cell-free conditioned medium exhibit antipneumococcal activity.
Because many antibacterial molecules are soluble in methanol, we hypothesized that the diffusible antipneumococcal activity produced by C. accolens (Fig. 3A and C; see also Table S1 in the supplemental material) might be extractable with methanol from medium conditioned by growth of C. accolens in the absence of pneumococcus. C. accolens produced antipneumococcal activity with both triolein and trilinolein supplementation (Fig. 3A and C), and we selected triolein as the medium supplement for further experimentation. We prepared crude methanol extracts of cell-free conditioned LB agarose medium spread with triolein (CFCM). For ease of harvesting CFCM, C. accolens was grown atop 0.2-μm polycarbonate membranes that were removed prior to extraction. Concentrated aliquots of crude CFCM methanol extracts killed pneumococcus, whereas equivalent amounts of control extracts of unconditioned sterile medium did not (Table 1). These data indicate that C. accolens produces an antipneumococcal activity that is extractable in methanol and does so in the absence of pneumococcus under the growth conditions of this assay.
TABLE 1 .
Methanol extracts of C. accolens KPL1818 CFCM inhibit S. pneumoniae 603 and contain free oleic acid
| Methanol extract source (growth medium) |
ZOI (mm)a | Free oleic acid concn (µM)b |
|---|---|---|
|
C. accolens CFCM (LB agarose triolein) |
11.7 ± 3.8 | 1,099.40 ± 422.98 |
| Sterile medium (LB agarose triolein) |
0.00 ± 0.00 | 6.07 ± 12.15 |
The mean ZOI ± SD produced in a disk-diffusion assay versus S. pneumoniae 603. The ZOI includes the disk diameter (6 mm). Extracts from each biological replicate (n = 4) were assayed in three technical replicates and results were then averaged.
The mean ± SD concentration of oleic acid found in methanol extracts. For samples in which no free oleic acid was detected, a value of 0 was used for calculating the mean and SD and for statistical analysis.
Extracts of C. accolens CFCM are enriched in free oleic acid.
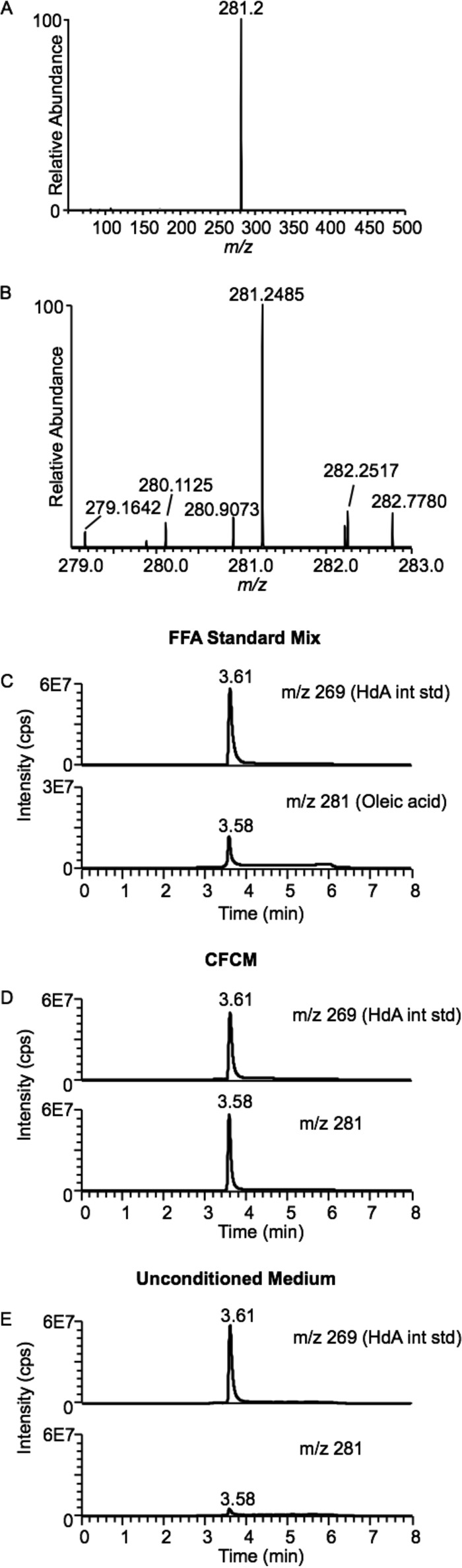
To identify the compound(s) with antipneumococcal activity, we compared qualitative mass spectrometry (MS) analysis results for methanol extracts from CFCM to those for unconditioned medium (LB agarose triolein) in search of compounds uniquely present or enriched in C. accolens CFCM. Mass spectrometric analysis of methanol extracts of CFCM revealed that oleic acid was the most abundant compound uniquely enriched in the CFCM extracts (Fig. 4). Compounds enriched in CFCM versus unconditioned medium were identified by performing limited mass scanning in negative ion mode and subtracting the spectrum for unconditioned medium from that for CFCM. One large novel peak with m/z 281 was identified in CFCM (Fig. 4A). Using high-resolution mass spectrometric analysis, the measured mass-to-charge ratio of this compound was 281.2485 (Fig. 4B), differing from the calculated mass of the [M-H]− ion of oleic acid of m/z 281.2486 by less than 5 ppm, consistent with the compound being oleic acid. In gradient reversed-phase ultraperformance liquid chromatography, the CFCM m/z 281 compound perfectly coeluted with authentic free oleic acid (Fig. 4C and D), further confirming that the compound present in CFCM is oleic acid. The concentrations of free oleic acid in CFCM extract were consistently greater than those observed in extracts of unconditioned medium (CFCM versus LB agarose triolein, two-tailed paired t test, P < 0.05) (Table 1; Fig. 4D versus E). These data clearly indicate that C. accolens releases free oleic acid from triolein, likely via lipase activity, and raise the possibility that free oleic acid might be responsible for the antipneumococcal activity produced by C. accolens.
FIG 4 .
Oleic acid is enriched in methanolic extracts of C. accolens CFCM. We used mass spectrometric analysis to identify compounds uniquely enriched in C. accolens CFCM. Methanolic extracts were analyzed by direct line infusion into a triple-quadrupole mass spectrometer operating in negative-ion scanning mode from m/z 50 to m/z 500. (A) To identify novel compounds present in the CFCM, the limited mass spectrum of the methanolic extract from unconditioned medium was subtracted from the spectrum for C. accolens CFCM. (B) Limited mass scanning of the novel m/z 281 peak present in CFCM, determined via high-resolution mass spectrometry, which gave a measured mass of m/z 281.2485, consistent with the calculated mass of oleic acid (m/z 281.2486). (C) An FFA standard mixture containing authentic oleic acid and heptadecanoic acid (HdA), as an internal standard, was analyzed by reversed-phase ultraperformance liquid chromatography coupled to tandem mass spectrometry, operating in multiple reaction mode monitoring transition of m/z 281 to m/z 281 at 10 eV for HdA and m/z 269 to m/z 269 at 10 eV for oleic acid, respectively. (D) CFCM with HdA added as an internal standard, analyzed under the same chromatographic conditions. (E) Unconditioned medium with HdA added as an internal standard, analyzed under the same chromatographic conditions. Representative spectra (A and B) or chromatographs (C, D, and E) are shown.
Authentic FFAs that can be released from human skin/nasal TAGs inhibit pneumococcal growth.
Based on the combination of our observation that C. accolens released free oleic acid from triolein (Fig. 4; Table 1) and previous reports that some FFAs inhibit bacterial growth (57–60), we hypothesized that C. accolens-liberated FFAs inhibit pneumococcal growth during both cocultivation (Fig. 3A and C) and exposure to CFCM extracts (Table 1). We tested this in a disk diffusion assay and observed that both authentic oleic and linoleic acids, which would be liberated from triolein and trilinolein, respectively, inhibited the growth of S. pneumoniae 603 (serotype 6B) and five other tested S. pneumoniae strains (TIGR4, DBL5, WU2, DSM 24048, and DSM 11868, which represent capsular serotypes 4, 5, 3, 19F, and 23F, respectively) (Table 2). These strains were selected, in part, because they represent a range of encapsulation capacities, from low encapsulation to heavy encapsulation. Next, we expanded the panel of FFAs tested to include other commercially available FFAs that are detected on human skin or in human nasal secretions (36, 37, 43, 44, 61). This included sapienic acid, which is the most abundantly detected FFA in sebum and SSLs (36, 37). We based these experiments on the hypothesis that C. accolens also liberates these other FFAs from the diversity of human skin surface TAGs (36, 37) via a lipase(s). Sapienic, palmitioleic, lauric, and myristic acids also inhibited pneumococcus, whereas stearic and palmitic acids did not inhibit pneumococcus under the assay conditions used (Table 2). The only difference between stearic and oleic acids is that stearic acid is a fully saturated 18-carbon FFA and thus is more difficult to emulsify in solvent at room temperature. Palmitic acid is similarly a saturated 16-carbon FFA. It is possible these two saturated 16- and 18-carbon FFAs do not diffuse from the filter disk and/or that they do not kill encapsulated pneumococcus at the quantities tested. (Both are reported to kill the unencapsulated mutant strain R6 [59].) Arsic and colleagues report that palmitic and stearic acids do not inhibit S. aureus, which leads us to speculate that these might not inhibit encapsulated pneumococcus (62). Several of the observed antipneumococcal FFAs (Table 2) have been reported to inhibit other microbes (58, 61), including S. pneumoniae (57, 59, 63). Here, we demonstrated that six serotypes are inhibited by a range of FFAs which can be released from human skin/nasal TAGs, and we predict that these FFAs inhibit additional pneumococcal serotypes.
TABLE 2 .
Antipneumococcal activity of FFAs reported on human skin or in human nasal fluid
| Test FFAa | FFA description | ZOIb (mm) for S. pneumoniae strain (serotype) |
|||||
|---|---|---|---|---|---|---|---|
| 603 (6B) | TIGR4 (4) | DBL5 (5) | WU2 (3) | DSM 24048 (19F) | DSM 11868 (23F) | ||
| Oleic | 18:1 cis-9 | 33.0 ± 3.0 | 24.0 ± 3.6 | 24.3 ± 4.0 | 35.7 ± 3.8 | 20.3 ± 4.0 | 30.0 ± 4.4 |
| Linoleic | 18:2 cis-9,12 | 30.3 ± 1.5 | 28.7 ± 3.5 | 25.0 ± 3.0 | 30.0 ± 6.6 | 20.3 ± 3.2 | 26.0 ± 6.1 |
| Stearic | 18:0 | ND | ND | ND | ND | ND | ND |
| Sapienic | 16:1 cis-6 | 29.3 ± 5.8 | 28.7 ± 4.2 | 24.7 ± 3.5 | 33.7 ± 5.7 | 25.3 ± 6.7 | 28.7 ± 5.7 |
| Palmitoleic | 16:1 cis-9 | 19.0 ± 1.7 | 20.3 ± 2.9 | 19.0 ± 3.0 | 24.3 ± 0.6 | 17.7 ± 0.6 | 18.0 ± 3.6 |
| Palmitic | 16:0 | ND | ND | ND | ND | ND | ND |
| Myristic | 14:0 | 11.3 ± 0.6 | 10.0 ± 0.0 | 10.0 ± 0.0 | 9.7 ± 0.6 | 10.0 ± 0.0 | 10.3 ± 0.6 |
| Lauric | 12:0 | 20.7 ± 0.6 | 14.3 ± 6.7 | 8.3 ± 0.6 | 15.3 ± 1.2 | 11.3 ± 1.5 | 16.0 ± 2.6 |
| Ethanol | Negative control | ND | ND | ND | ND | ND | ND |
One hundred micrograms of each FFA in 10 µl of 200-proof ethanol was pipetted onto a 6-mm filter disk that was placed on a lawn of S. pneumoniae.
The diameter of the ZOI (mean ± standard deviation; n = 3) was measured after overnight incubation at 37°C with 5% CO2. ND, ZOI not detected.
Identification of a putative C. accolens TAG lipase.
TAG lipases preferentially hydrolyze ester bonds to release fatty acids with chain lengths longer than nine carbon atoms from TAGs (64). Based on this, we hypothesized that the enzyme required for C. accolens to hydrolyze triolein and trilinolein, releasing free oleic (C18:1) and linoleic (C18:2) acids, respectively, might be an extracellular TAG lipase. To identify candidate TAG lipases, we queried the draft genome of C. accolens KPL1818 (RefSeq accession NZ_AXMA00000000) against the Conserved Domain Database (CDD) of NCBI (65). We searched for C. accolens proteins that aligned to the LIP family (pfam03583), the Lipase_2 family (pfam01674), or the LipA/EstA cluster of orthologous groups (COG1075). We focused on these groups because characterized TAG lipases from other common human skin- and nostril-associated bacteria, i.e., S. aureus, S. epidermidis, and Proprionibacterium acnes (45, 66–68) and those putatively identified in other Corynebacterium spp. (69, 70) align to at least one of these groups. This search revealed two candidate TAG lipases in C. accolens KPL1818: WP_034657805.1 and WP_023029886.1, here abbreviated WP_03 and WP_02.
The CDD analysis predicted that WP_03 belongs to the LIP protein family (E value, 3.39e−36), whereas WP_02 aligned to both the LipA/EstA COG (E value, 4.88e−40) and the Lipase_2 family (E value, 1.08e−08). Both proteins contain the canonical lipase motif G-X-S-X-G, complete with the predicted active site serine residue (64). SignalP analysis (version 4.1) (71) of the two protein sequences predicted that only WP_03 is secreted via the general secretion pathway, similar to other skin-associated bacterial TAG lipases (66–68). It is possible that WP_02 is secreted by mechanisms that are not dependent on the general secretion pathway. However, based on the CDD and SignalP analyses, we hypothesized that WP_03 is the primary extracellular C. accolens TAG lipase, and we proceeded to test this by using a genetic approach.
C. accolens lipS1 (WP_034657805.1) is necessary for growth when triolein is the only source of fatty acids.
A C. accolens KPL1818 mutant strain with an in-frame deletion of WP_03 did not grow in BHI supplemented with triolein, whereas the wild type did (Fig. 5A). These results indicated that WP_03 is required for hydrolysis of the TAG triolein; therefore, we named this gene lipS1, in adherence with existing nomenclature for Corynebacterium TAG lipases predicted to be members of the LIP family (69, 70). In contrast to its inability to grow in medium supplemented with triolein, the lipS1-deficient mutant grew comparably to the wild type in medium supplemented with Tw80 (see Fig. S4A in the supplemental material). This growth on Tw80 is likely due to esterase activity, since some esterases can hydrolyze the ester of oleic acid (C18:1) in Tw80 but not in a TAG (33); in TAGs, esterases preferentially hydrolyze ester bonds linking water-soluble fatty acids of chain lengths less than 10 carbon atoms (<C10) to the glycerol backbone (64).
FIG 5 .
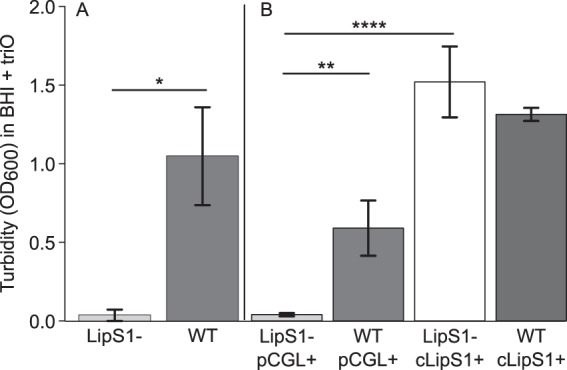
C. accolens LipS1 is necessary for growth on triolein (triO) as the sole source of fatty acids. (A) A C. accolens mutant strain (LipS1−) harboring an in-frame deletion in lipS1 did not grow in BHI supplemented with triolein (0.1 mg/ml), whereas the wild-type (WT) did. Growth was measured as the mean OD600 following 48 h of aerobic growth with shaking at 37°C. Data were analyzed using a Welch two-sample t test. n = 3. Error bars represent standard deviations. *, P ≤ 0.05. (B) The lipase-deficient mutant carrying the empty vector (LipS1− pCGL+) did not grow in BHI supplemented with triolein; however, the complemented mutant (LipS1− cLipS1+) did. For comparison, the empty vector and complemented vector controls for the parental strain (WT pCGL+ and WT cLipS1+, respectively) are included. Data (n = 3) were analyzed using an ANOVA and Tukey’s multiple-comparisons test. Error bars represent standard deviations. **, P ≤ 0.01; ****, P ≤ 0.0001.
Complementation of the lipS1 deletion with lipS1 (with a His6 tag) expressed from its native promoter on a multicopy plasmid (see Fig. S5 in the supplemental material) restored the ability of C. accolens to grow to wild-type levels with triolein as the sole source of fatty acids (Fig. 5B). In contrast, there were no growth differences among these strains in medium supplemented with Tw80 (see Fig. S4B in the supplemental material). Again, this is likely due to esterase activity encoded by another gene(s). In addition to identifying the primary C. accolens lipase, this work demonstrates that C. accolens is genetically tractable.
Among Corynebacterium strains with an assigned species, LipS1 homologs are detected only in C. accolens.
To determine whether homologs of LipS1 are present in other Corynebacterium spp., we queried the nonredundant database at NCBI with the predicted 464-amino-acid sequence of LipS1 via BLASTP (72). We restricted our search to the Corynebacterium genus (taxid 1716) using a ≥96% protein identity to LipS1 as the cutoff for homology. LipS1 was detected in the C. accolens type strain (98% identity) which inhibited pneumococcal growth (see Table S1 in the supplemental material). LipS1 was also detected in the C. accolens primary nostril isolate KPL1855 (73) and in ATCC 49726, suggesting that these strains likely possess the ability to hydrolyze TAGs to release antipneumococcal FFAs. Based on this bioinformatic analysis and the genetic data described above, we conclude that LipS1 is a primary TAG lipase that C. accolens uses to harvest FFAs from host skin surface TAGs.
DISCUSSION
To our knowledge, this work provides the first evidence that C. accolens, a commensal Corynebacterium species that colonizes the nose, can inhibit S. pneumoniae. The goal of identifying antagonistic interactions between commensal nasal bacteria and S. pneumoniae is to lay the foundation for future translational research aimed at preventing pneumococcal colonization and subsequent infection. As illustrated here, we approached this by starting with data on the community composition of the pediatric nasal microbiota (Fig. 1) (18, 19). We then used a reductionist approach to test our hypothesis that growth of the medically important pathobiont S. pneumoniae is antagonized by C. accolens, a commensal bacterium that is rarely reported in conjunction with infection and, thus, might have potential as a probiotic. Liquid chromatography-mass spectroscopy (LC/MS) of C. accolens CFCM and disk-diffusion assays with both CFCM extracts and authentic FFAs demonstrated that FFAs released from skin surface TAGs by C. accolens inhibit pneumococcal growth. We also showed that FFAs present on skin and in nasal secretions (36, 37, 43, 44, 61) inhibit pneumococcus (Table 2). The antibacterial activity of FFAs is well documented, and oleic acid inhibits pneumococcal growth through membrane depolarization and cell rupture (57). Overall, our results lay the foundation for future testing of C. accolens as a potential skin and nasal probiotic. There is already a precedent for testing Corynebacterium spp. as probiotics for the eradication of nasal colonization by the pathobiont S. aureus (31, 74). Our results also pave the way for future testing of representative human skin surface TAGs as potential prebiotics, the addition of which is predicted to result in decreased levels of FFA-susceptible pathogenic streptococcal species, such as S. pneumoniae and Streptococcus pyogenes (60). Since colonization is a prerequisite for infection with these pathobionts, eradication or a decrease in the level of colonization could prevent infection.
The discovery that C. accolens releases antibacterial FFAs from human TAGs provides an impetus for further investigations. For example, because C. accolens grows poorly in the absence of an exogenous FFA source (Fig. 3), we cannot exclude the possibility that in the presence of a fatty acid source, C. accolens produces another compound(s) that also contributes to pneumococcal growth inhibition. It remains unknown whether other Corynebacterium spp. inhibit pneumococcal growth via the release of antibacterial FFAs and/or via an as-yet-undiscovered alternative mechanism. Further, future investigations will determine what species of Corynebacterium are most commonly found in the nasal microbiota of children 6 months to 7 years of age, as it is not yet known how common natural C. accolens nasal colonization is in this age group. Finally, the absence, or low levels, of skin surface TAGs in other mammals tested to date (34, 35), including mice and rats, limits the testing of our model (Fig. 6) in animals. In addition, although animal models exist for S. pneumoniae nasal colonization, there is not yet an established model for nasal colonization by commensal Corynebacterium spp. However, the use of some TAGs, including both trilinolein and triolein (i.e., triglycerides) in some cosmetics (75) opens up the possibility for future testing directly on human skin.
FIG 6 .
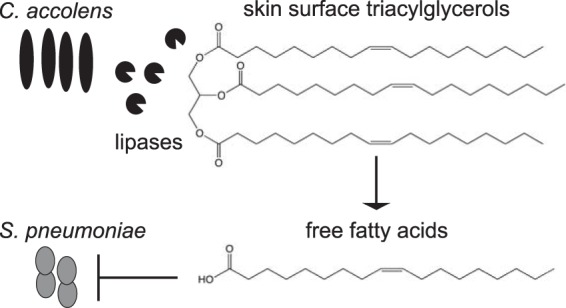
One model is that C. accolens modifies the nasal and/or skin habitat, making it inhospitable for S. pneumoniae. C. accolens produces a lipase, likely extracellular, that releases FFAs from host TAGs. Some of these FFAs have antibacterial activity and might impede the colonization and proliferation of S. pneumoniae and possibly other FFA-susceptible colonizers.
Based on the in vitro mechanistic data presented, we developed a model that addresses the potential relevance of our findings to the in vivo ecology of C. accolens and S. pneumoniae and generates testable hypotheses for future research. In this model (Fig. 6), the C. accolens TAG lipase LipS1, which is likely to be extracellular, hydrolyzes skin surface TAGs, releasing antibacterial FFAs, many of which inhibit pneumococcus. This current model might partly explain the greater relative abundance of nasal Corynebacterium spp. in children who lack pneumococcal nasal colonization (Fig. 1) (18). The recent identification of an OTU associated with 6-week-old breast-fed infants that resembles C. accolens raises the possibility that C. accolens might be present in many young infants (41). Collectively, Corynebacterium spp. are widely distributed across various skin sites, as well as on mucosal surfaces of the upper respiratory tract (18, 21–31). In addition, other pathobionts are also susceptible to FFAs, e.g., S. aureus (45, 59, 76) and S. pyogenes (60). Thus, our current model in which Corynebacterium spp. contribute to modifying the local habitat via release of antibacterial FFAs might be generalizable to other sites where Corynebacterium spp. are in higher relative abundance, such that the local concentration of FFAs reaches a level that can shape bacterial community structure.
By demonstrating that C. accolens liberates FFAs from human skin surface TAGs and identifying LipS1 as a primary C. accolens TAG lipase, this work expands the repertoire of bacterial contributors to FFAs on human skin. Historically, FFA release from skin surface TAGs is attributed to both human-produced and bacteria-produced lipases (77). In support of a role for host lipases, a recent study revealed that in vitro human sebocytes produce FFAs in the absence of bacteria (78). The role of P. acnes-produced lipases was initially based on a 1971 paper in which Marples and colleagues showed that a 99% reduction in the P. acnes population on adult forehead skin corresponded to a 50% decrease in FFA content, thus establishing that P. acnes (formerly known as Corynebacterium acnes) plays a significant role in regulating FFA content on human skin (79). Follow-up studies have identified the P. acnes TAG lipases (68, 80, 81). However, the findings of our study and others are consistent with lipases from C. accolens and other skin microbes also playing an important role in liberating FFAs from SSLs, e.g., Staphylococcus epidermidis and S. aureus both produce TAG lipases (45, 66, 67). Interestingly, some strains of P. acnes (82) and S. aureus (45) are also susceptible to the toxic effects of some FFAs released from human sebum and skin surface TAGs, which might limit the role of these bacteria in FFA release.
We speculate that the ability of Corynebacterium spp. to release FFAs from host surface TAGs is especially important in populations and at sites where Propionibacterium spp. are less abundant than Corynebacterium spp., such as skin surfaces of prepubertal children (18, 21). For example, the genus Corynebacterium is the most abundant member of the Actinobacteria in the microbiota of the pediatric nostril (18, 21) and of the pediatric nasopharynx, a mucosal surface (20, 41); although communities at both sites are dominated by the phyla Firmicutes, Proteobacteria, and Bacteroidetes. In these settings, Corynebacterium spp. might play an important role, along with host lipases, in regulating epithelial surface FFA content.
In contrast to prepubertal children, the nostril microbiota of adults under 65 years of age does not commonly contain pneumococcus and is often dominated by the phyla Actinobacteria, in particular Corynebacterium and Propionibacterium spp., and Firmicutes, in particular Staphylococcus spp. (21–31). This composition appears to carry over into the mucosal surfaces of the nasal cavity and nasopharynx (29, 83, 84), although the mucosal surface of the nasal cavity seems to host a greater diversity of bacterial genera than the nostrils (29). These observations raise the possibility that increased sebum production beginning with puberty (85) gives an advantage to microbes in the nostrils, and on other oily or moist skin sites, that both secrete TAG lipases and are relatively resistant to FFAs, which would fit with bacterial community composition at these sites in adults (21). It also raises the possibility that topical application of TAGs in circumstances where TAG production is naturally lower might sculpt a microbial community dominated by Corynebacterium and/or Propionibacterium spp.
In addition to impacting S. pneumoniae directly, the C. accolens release of FFAs might impact the host immune system, leading to indirect effects on the composition of the bacterial microbiota. For example, using a human sebocyte cell culture line, Gallo and colleagues show that lauric, palmitic, and oleic acids increased expression of the human beta-defensin-2 gene (hBD-2). The hBD-2 antimicrobial peptide (AMP) is found on skin (86) and is antibacterial toward pneumococcus (87), suggesting an additional indirect mechanism by which release of FFAs might contribute to protection against S. pneumoniae colonization.
Treating the surfaces of human body sites as individual ecosystems implies the existence of a complex network of both microbe-microbe and microbe-host interactions (42). Based on our findings, we predict that on human surfaces C. accolens functions as a mutualist, and not merely as a commensal, by altering the local host surface environment in a manner that can partially shape the composition of nasal and skin microbiota and by helping to protect against colonization by pathobionts, such as S. pneumoniae. This interaction is part of a growing list of examples of skin/nasal commensals impacting pathobiont growth/behavior (73, 88–90), and the outcome of this interaction could have important implications for the human host. We predict that a greater understanding of the molecular mechanisms underlying commensal-pathobiont interactions, in particular those that occur between Corynebacterium spp. and pathobionts belonging to the phylum Firmicutes, will facilitate the development of alternative approaches to interfere with pathobiont colonization and/or infection.
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial strains and plasmids are listed in Table S2 in the supplemental material, and additional growth information is in Text S1 (a further description of our study’s materials and methods) in the supplemental material. As the base medium for all coculture assays, 10-ml aliquots of BHI agar containing 200 U/ml of bovine liver catalase (BHIC; EMD Millipore, Billerica, MA) was added to 100-mm by 15-mm petri dishes. Calculation for the addition of catalase was based on the following spec sheet information: activity of ≥5,000 U per mg dry weight. On the day of use, one of the following was spread onto the agar medium: 50 µl of 100-mg/ml glyceryl trioleate (triolein; catalog number T7140; Sigma-Aldrich) solubilized in chloroform; 50 µl of 100-mg/ml glyceryl trilinoleate (trilinolein; catalog number T9517; Sigma-Aldrich) emulsified in ethanol; 50 µl of chloroform or 50 µl of ethanol. (Sapienic acid is abundant on human skin [61], but we were unable to locate a commercially available TAG containing sapienic acid.) For consistent results, we observed that it is best to use agar medium that has been prepared within 1 to 2 days of use.
Ethics statement.
Under a protocol amendment approved by the Harvard Pilgrim Health Care Human Studies Committee, and with approval from the Forsyth Institute Institutional Review Board, nasopharyngeal swabs were analyzed from children >6 months and <7 years of age. These samples were collected as part of an ongoing study of S. pneumoniae colonization, as described in references 16, 17, 38, and 39.
Identification of pediatric nasopharyngeal bacterial microbiota.
We received a randomly selected subset of ~200 of the ~1,000 nasopharyngeal swabs (calcium alginate) collected from October 2008 to April 2009 as part of a study of pneumococcal colonization in Massachusetts (16, 17, 38). For each swab, we received information on the age group (<2 years or 2 to ≤7 years) of the participant and whether the swab was positive for S. pneumoniae by cultivation. Swabs had been originally shipped overnight and inoculated onto sterile agar medium and then stored in STGG (skim-milk, tryptone, glucose, and glycerol) with 10% glycerol at −70°C. In 2010, to harvest DNA, swabs were thawed, removed from the storage medium, and aseptically cut from the remaining wire applicator into a PowerSoil tube. To maximize recovery of bacterial cells, the storage medium was then centrifuged at 17,000 × g for 10 to 20 min, after which most supernatant was discarded, leaving behind 50 µl. The remaining 50 µl was vortexed to resuspend any bacterial cells before adding 45 µl to the PowerSoil tube containing the swab. DNA was harvested with the MoBio PowerSoil DNA isolation kit per the Human Microbiome Project protocol (section 7.8.2) (28) with minor modifications, including bead beating in a FastPrep24 for 30 s at 5 m/s for more rapid cell lysis.
From the ~200 DNA samples, we selected a representative subset of 45 that had been amplified by PCR with ARISA primers 1406F-FAM and 125R (91). We submitted these, along with the negative control of a processed sterile swab, for 16S rRNA gene V1-V3 titanium pyrosequencing at the Broad Institute (Cambridge, MA). Sequencing and processing from raw sequences to an OTU table was done as described in reference 92. Twenty-seven samples, of which nine were culture-positive for pneumococcus, had >1,000 high-quality sequences each (minimal number of sequence/sample, 1,503). After filtering in QIIME (93) (using the MacQIIME 1.4 package), first to remove samples with <1,000 sequences and second, to remove OTU sequences that were not present more than once in one sample, sequences were subsampled to the minimum of 1,503 reads/sample. Data from these 27 samples were analyzed using linear discriminant analysis effect size (LEfSe) (40) with pneumococcal colonization by cultivation as class and age group (<2 years versus 2 to <7 years) as the subclass. Sequences (pre-chimera check) are accessible in the short read archive (SRA) under biosample accessions SAMN04323356 through SAMN04323382.
Chemical structures.
Chemical structures were produced using ChemDraw Professional version 15 (PerkinElmer).
Growth assays.
C. accolens KPL1818 cells were transferred from BHI Tw80 agar medium into sterile 1× phosphate-buffered saline (PBS) and vortexed for 2 min. Cells were diluted to an optical density at 600 nm (OD600) of approximately 0.3 to 0.4, 25 µl of which was added to 25 ml BHI broth and vortexed. Aliquots of 5 ml were then dispensed into 15-ml Falcon tubes (Corning Life Sciences) to ensure an equal inoculum for each biological replicate. We then added 5 µl of one of the following to each tube: 100 mg/ml of triolein in chloroform, 100 mg/ml of trilinolein in ethanol, chloroform, or ethanol, resulting in a final concentrations of 112.9395 µM and 113.7165 µM for triolein and trilinolein, respectively. The tubes were incubated aerobically at 37°C with shaking at 200 rpm. Sterile medium controls for all four types of final media were included. Following 48 h of incubation, the OD600 was recorded using sterile BHI without additives as a reference medium. After confirming that the sterile medium controls were not turbid, measurements of BHI containing TAG were used to correct the OD600 values for the corresponding growth tube, since the TAGs have slight absorbtion at OD600. Data represent nine biological replicates carried out on four separate days. Analysis of variance (ANOVA) was performed, and P values were adjusted for multiple testing by using Tukey’s test.
Side-by-side cocultivation assay.
C. accolens cells were resuspended in 1× PBS to an OD600 of 0.3 to 0.4, after which a 5-µl aliquot was inoculated onto the appropriate solid medium (see above). C. accolens was then grown for ~42 h at 37°C, after which a 5-µl suspension of S. pneumoniae in 1× PBS (OD600 of 0.3 to 0.4) was inoculated adjacent to C. accolens. We also developed the alternative membrane-based assay for C. accolens excretion of antipneumococcal compounds (described in the materials and methods of Text S1 in the supplemental material). For both assays, to show that in the absence of C. accolens BHI supplemented with triolein, trilinolein, chloroform, or ethanol supported pneumococcal growth, S. pneumoniae was inoculated onto each medium alone that had been prepped in exactly the same manner. Plates were held at room temperature for ~4 to 6 h after pneumococcal inoculation and then incubated overnight at 37°C with 5% CO2; this minimized pneumococcal autolysis prior to observation and photographic documentation of results.
Methanol extracts of C. accolens CFCM and sterile medium controls.
We used LB SeaKem LE agarose (Lonza) spread with triolein for methanol extraction experiments. Triolein is not soluble in aqueous solution, so 50 µl of triolein (100 mg/ml in chloroform) was added to each plate and spread with a sterile spreader to generate as homogenous a layer as possible. Plates were dried for ≥30 min before use to ensure evaporation of residual solvent. After drying, a sterile, 0.2 μm, 47-mm-diameter polycarbonate membrane (EMD Millipore) was placed atop each agarose plate using sterile tweezers. Using a sterile swab, freshly grown C. accolens KPL1818 cells were transferred from BHI Tw80 agar to the membrane and a lawn was spread. The membrane permits passage between cells and medium of nutrients, waste products, and secreted compounds/enzymes. Agarose plates were incubated for 72 h at 37°C, after which the membranes (and cells) were removed and discarded. After incubation with or without cells, agarose medium was moved to a sterile glass bottle to which 75 ml of high-performance liquid chromatography/Optima-grade methanol (Fisher Scientific) was added. The bottles were gently agitated and placed at 4°C overnight (~17 h). Following extraction at 4°C, the methanol was decanted into 50-ml Falcon tubes and spun at 5,000 × g for 10 min to pellet any residual agarose or debris. A single, 1-ml aliquot of each clarified methanol extract was reserved for LC/MS analysis. The remainder of each methanol extract was concentrated using a Büchi Rotavapor R-215 to a volume of ≤0.5 ml, adjusting the vacuum pressure as needed to maintain a relatively constant evaporation rate. Sterile medium controls were incubated alongside the experimental plates and processed the same to demonstrate that FFAs were not abundant in unconditioned medium. The approximate “fold concentrate” of each extract was calculated by dividing the starting volume of agar (25 ml) by the remaining volume of methanol extract (e.g., 25 ml agarose/0.5 ml methanol extract = 50× concentrated).
Disk-diffusion assays with methanol extracts.
Aliquots of 20 µl of an ~50×-concentrated extract were assayed for antipneumococcal activity. Methanol was added to bring the final volume of each aliquot to 20 µl. Each extract was then placed on a 6-mm sterile filter disk, dried for ≥20 min, and then placed on a freshly plated lawn of S. pneumoniae 603 on BHIC agar medium. Lawns were prepared by transferring pneumococcal cells with a sterile swab from CNA agar onto BHIC agar. For consistency, the BHIC medium was prepared 2 days prior to use by dispensing 10-ml aliquots of medium into 100-mm by 15-mm petri dishes. Plates were incubated overnight at 37°C in a 5% CO2 incubator overnight. The ZOI was measured as the smallest diameter of the area of no growth. If a ZOI was not uniform, the shortest diameter of no growth was reported.
LC/MS analysis of methanol extracts.
Methanolic extracts were initially analyzed using a ThermoFinnigan Quantum electrospray ionization triple-quadrupole mass spectrometer operating in negative-ion mode. Data were analyzed by using Xcaliber 2.0.7 software (Thermo Scientific). Extracts from unconditioned medium, and cell-free conditioned media (CFCM) were introduced by direct line infusion for several minutes for each and the limited mass spectrum from m/z 50 to 500 recorded continuously. The spectrum of unconditioned medium was subtracted from the spectrum for CFCM using the Xcaliber software to identify peaks in the spectrum unique to CFCM. CFCM and unconditioned medium from four different preparations were analyzed by this method, and the resulting subtracted spectra were highly similar. High-resolution mass analysis of CFCM was performed on a ThermoScientific Orbitrap XL mass spectrometer operating in negative-ion mode and equipped with an IonMax source housing, a standard electrospray ionization probe, and a 50-µm-inner diameter high-flow metal needle kit. The Orbitrap was calibrated in negative mode over a mass range of m/z 265 to 1,780 by using a mixture of SDS, taurocholic acid, and Ultramark-1621 (Thermo-Pierce). Resolution at m/z 500 was 30,000 and the maximum injection time was 100 ms. For determination of the concentration of oleic acid in methanolic extracts, 3.7 nmol of heptadecanoic acid (HdA; a C17:0 fatty acid) was added to 60 µl of extracts or authentic synthetic oleic acid and then analyzed via LC/MS. Ultraperformance liquid chromatography was performed with an Acquity UPLC apparatus (Waters) coupled to a Quantum mass spectrometer operating in multiple reaction mode monitoring transition of m/z 281 to m/z 281 at 10 eV for HdA and m/z 269 to m/z 269 at 10 eV for oleic acid, respectively. The volume of injection was 10 µl. Mobile phase A was 1 mM triethylammonium acetate in water, and mobile phase B was 0.1% acetic acid in methanol. Extracts were chromatographed on a Kinetex C8 column (50 by 2.1 mm; 2.6 µm; Phenomenex) with a constant flow of 250 µl/min. After a 0.5-min hold at 1% mobile phase B, the solvent gradient was ramped to 99% B over 2 min, then held at 99% B for another 2.5 min, and returned to 1% B over 1 min and held at 1% B for 2 min prior to starting the next injection. The concentration of oleic acid was calculated from the ratio of the peak area for the m/z 281 peak at 3.58 min to the peak area for the m/z 269 peak at 3.60 min, multiplied by 3.7 nmol (amount of internal standard added), and divided by 60 µl (volume of extract used). In comparing the concentrations of oleic acid detected in CFCM versus sterile medium controls, we considered the control and the experimental sample as paired samples, to account for any possible batch effects of media that were prepared on separate days. We analyzed these data with a two-tailed, paired t test (at a P level of <0.05).
Fatty acid susceptibility.
We purchased FFAs from Sigma-Aldrich, except for sapienic acid, which was purchased from Matreya (Pleasant Gap, PA). For each FFA, 10 µl of a stock solution of 10 mg/ml in 200-proof ethanol (100 µg total) was pipetted onto a sterile, 6-mm filter disk (Whatman). The control in each experiment was a disk spotted with 10 µl of 200-proof ethanol. Disks were dried for ≥40 min prior to placement on a freshly plated lawn of S. pneumoniae prepared and incubated as described above. The ZOI was measured and documented as described above. For consistency, 25-ml aliquots of the BHIC medium were dispensed into 100-mm by 15-mm petri dishes.
Construction of an in-frame deletion of lipS1.
Regions extending approximately 1 kb adjacent to the 5′ and 3′ ends of the putative TAG lipase gene, from nucleotides 281230 to 282573, encoded in the KPL1818 genome (nucleotide GenBank accession AXMA01000001.1), were PCR amplified from C. accolens KPL1818 genomic DNA with primers designed such that the 5′ fragment ended with a 21-bp random scar sequence and a GC content of 54% and the 3′ fragment started with the same 21-bp scar sequence, followed by the stop codon (oKL516-519 [see Table S3 in the supplemental material]). After digestion with PstI followed by treatment with calf intestinal phosphatase for 1 h each at 37°C (both NEB), pJSC232 (94), which replicates in Escherichia coli and not in Corynebacterium spp., was cleaned up by using the QIAquick PCR purification kit. The two PCR fragments were purified, assembled with PstI-opened pJSC232 by Gibson assembly (95), and then transformed into NEB 5-alpha competent E. coli cells according to the manufacturer’s instructions. Clones on LB-kanamycin plates were screened using primers flanking the insertion site (oKL520-521 and also oKL516-519 [Table S3]). The resulting pJSC232-derived deletion plasmid (pSB121) was purified and verified by PCR and restriction enzyme digestion. Competent cells were prepared, and pSB121 was introduced into C. accolens KPL1818 by electroporation as previously described (96), except that the growth temperature was 37°C and all media contained 1% Tw80 (see Text S1 in the supplemental material for more details regarding our materials and methods). Transformants that had integrated the nonreplicating plasmid in the genome by a single homologous recombination event were selected using the kanamycin resistance marker on the vector backbone. Kan-resistant transformants were then grown nonselectively in BHI-sorbitol and subsequently plated on BHI agar–1% Tw80 with 5% sucrose, which should permit growth of only clones that have lost the sacB-containing vector backbone (i.e., a negative selection marker for sucrose sensitivity) by a second homologous recombination event. Successful deletions were identified by PCR with primers oKL522 and oKL523 (see Table S3) that anneal outside the regions of homology used in the deletion construct, and correct deletion was verified by sequencing (Macrogen USA).
Construction of the lipS1 complementation vector pLB502.
The complementation vector pLB502 was constructed using the Gibson assembly method and the Corynebacterium spp.-E. coli shuttle vector pCGL0243 (97) as detailed in Text S1 in the supplemental material. To ensure that the promoter region was included, we cloned 949 bp upstream of the predicted start codon listed with NCBI, along with the predicted ORF for lipS1. In addition to the ORF, a His6 tag fused to the C terminus was introduced in-frame immediately downstream of the ORF, followed by the stop codon (see Fig. S5 in the supplemental material). Gibson reactions were transformed into E. coli NEB 5-alpha cells, transformants were selected on LB agar containing 40 µg/ml of kanamycin, and Kan-resistant transformants were screened by PCR with primers oKL381 and oKL382 (see Table S3 in the supplemental material), which amplify the multiple-cloning site in pCGL0243, to detect candidates harboring pLB502. A single PCR-positive clone was inoculated into LB broth with kanamycin and grown at 37°C overnight with shaking at 200 rpm. Plasmid (pLB502) purified from this culture (or cultures from the frozen stocks) was sequenced to verify the correct insert (see Table S3 for primers).
Complementation of the lipS1-deficient mutant strain.
Plasmids pCGL0243 and pLB502 were transformed separately into both wild-type C. accolens KPL1818 and an isogenic lipS1-deficient mutant (KPL2503), generating the following strains: KPL2489, KPL2478, KPL2504, and KPL2505 (see Table S2 in the supplemental material). Electroporations were performed using one of two methods: (i) as described above with the exception that transformants were selected for on BHI Tw80 sorbitol plates with 20 µg/ml kanamycin, or (ii) as described above with the exceptions that heat shock was not performed, the outgrowth medium did not contain sorbitol, and transformants were selected on BHI Tw80 with 20 µg/ml kanamycin. With both electroporation methods, Tw80 was used to support the growth of lipid-requiring C. accolens, because FFA can be liberated from Tw80 by an esterase. Successful transformation of C. accolens with each plasmid was verified by PCR amplification of the plasmid’s multiple-cloning site (oKL562 and oKL563 [see Table S3 in the supplemental material]).
Growth of C. accolens KPL1818 mutants with triolein or Tw80 as the sole source of fatty acid.
Colonies of each strain were resuspended separately in sterile 1× PBS to an OD600 of 0.3 prior to inoculating 5 µl of the suspension into 5 ml of BHI broth supplemented with 5 µl of a 100-mg/ml triolein solution or 1% Tw80; medium for strains with pCGL0243 or pLB502 included 20 µg/ml kanamycin. Cultures were grown at 37°C with shaking (200 rpm), and the OD600 was measured after 48 h. Each independent growth experiment (n = 3) included a sterile medium control. Data comparing growth of the LipS1− mutant and the wild-type strain were analyzed using a Welch two-sample t test. Data comparing growth of strains carrying pCGL0243 or pLB502 were analyzed via an ANOVA and Tukey’s test when applicable.
Statistical analyses.
As described above, ANOVA and t tests, with corrections for multiple comparisons if applicable (Tukey test), were performed using R version 3.2.0 (RStudio version 0.98.1103) and GraphPad Prism version 6.01 (La Jolla, CA). Figures were plotted in R, and indicators of significance (asterisks and bars) were added using Inkscape version 0.91.
SUPPLEMENTAL MATERIAL
Supplemental Materials and Methods. Download
Basic anatomy of the human nasal passages. Pneumococcus colonizes the nose from the nostrils to the nasopharynx. The nostrils (indicated by the arrow), also known as the anterior nares, are the entrance to the nasal passages and open onto the skin-covered surface of the nasal vestibules. The skin lining the nasal vestibules includes sweat and sebaceous glands. Posterior to the nasal vestibules, the epithelial surface of the nasal cavity transitions to a mucosal surface. Respiratory epithelium, including cilia, lines the posterior segment of the nasal cavity and the nasopharynx, at the top of the back of the throat. Download
Relative abundance plots for each of the three bacterial genera that are overrepresented when S. pneumoniae is absent (class 0) or present (class 1) via cultivation. Subclasses are shown as follows: A, in red, <2 years old; B, in green, 2 to <7 years. (A) Streptococcus; (B) Dolosigranulum; (C) Corynebacterium. Plots were generated using LEfSE from the same data used to generate Fig. 1. The solid and dashed lines indicate the mean and the median relative abundance levels, respectively. Each bar represents a nasopharyngeal (NP) sample from one volunteer. Download
Dendrogram representing phylogenetic relationship of KPL1818 with C. accolens and other Corynebacterium spp. The dendrogram was constructed using the maximum-likelihood method with 1,000 bootstrap replications of ClustalW-aligned partial rpoB sequences (446-bp fragment between primers C2700F and C3130R) (Khamis A, Raoult D, La Scola B, J Clin Microbiol 42:3925–3931, doi:10.1128/JCM.42.9.3925-3931.2004). Branch support is indicated by the value at each node. D. cinnamea was used as an outgroup. Download
C. accolens lipS1 is not necessary for growth when Tween 80 (Tw80) is the sole source of fatty acids. (A) A C. accolens mutant (LipS1-) harboring an in-frame deletion in lipS1 grew comparably to wild type (WT) in BHI supplemented with 1% Tw80. Growth in A and B was measured as the mean OD600 following 48 h of aerobic growth shaking at 37°C. Data were analyzed using a Welch two-sample t test, n = 3, and error bars represent SD. (B) The lipase-deficient mutant carrying the empty vector (LipS1- pCGL+) also grew comparably to wild type (WT) in BHI supplemented with 1% Tw80 as did the complemented mutant (LipS1- cLipS1+). For comparison, the empty vector and complemented vector controls for the parental strain (WT pCGL+ and WT cLipS1+, respectively) are included. Data (n = 3) were analyzed using ANOVA. Error bars represent SD. Download
LipS1 from C. accolens KPL1818. Shown at the top are the predicted open reading frame and corresponding translation for the His6-tagged LipS1 (WP_034657805.1), as annotated in NCBI. The predicted signal sequence (orange bar), canonical G-X-S-X-G motif (red bar), His6 tag (gray bar), and stop codon (blue bar) are highlighted. The sequence cloned into pLB502 contains an additional 949 bp 5′ of the predicted start codon shown. Download
Additional C. accolens strains tested for inhibition of S. pneumoniae 603.
Bacterial strains and plasmids used in this study
Primers designed for this study
ACKNOWLEDGMENTS
We are deeply indebted to Grace M. Lee, Jonathan A. Finkelstein, their colleagues (especially, Maya Dutta-Linn), and the participants in their long-term study of pneumococcal colonization for supplying specimens used in this study (NIAID R01 AI066304). Pyrosequencing was done at the Broad Institute, and we thank Dirk Gevers for processing sequences from raw reads to a QIIME OTU table and Ashlee M. Earl for invaluable advice and support on sample submission. We thank Richard Malley for S. pneumoniae strains, Hung Ton-that for pCGL0243, Jeffery S. Cox for pJSC232, and Jan Claesen for advice on gene deletion in C. accolens. Brett J. Pellock, Andrew J. Collins, Adam C. Silver, Isabel F. Escapa, and Erin A. Gontang provided constructive critiques on the manuscript, to its benefit. Thanks to members of the Lemon Lab and to Mary Ellen Davey, Jorge Frias-Lopez, and their labs for input.
Conception and design for this study were by K.P.L., L.B., S.D.B., and S.S.D. Acquisition, analysis, and interpretation of data were performed by L.B., S.D.B., B.H.Y., S.S.D., and K.P.L. Drafting or revising the article was performed by L.B., S.D.B., S.S.D., and K.P.L. All authors read and approved the final manuscript.
Funding Statement
HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID) provided funding to collect pediatric nasopharyngeal swabs under grant number AI066304 to Jonathan A. Finkelstein. These funders did not have any role in study design, data collection, data analysis, or the decision to submit the work for publication. Harvard Catalyst Pilot Grant provided funding to Katherine P. Lemon under grant number NIH grant 1 UL1 RR 025758-02 and financial contributions from participating institutions.
Footnotes
Citation Bomar L, Brugger SD, Yost BH, Davies SS, Lemon KP. 2016. Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. mBio 7(1):e01725-15. doi:10.1128/mBio.01725-15.
REFERENCES
- 1.Bogaert D, de Groot R, Hermans P. 2004. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis 4:144–154. doi: 10.1016/S1473-3099(04)00938-7. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization 2012. Pneumococcal vaccines. WHO position paper, 2012 recommendations. Vaccine 30:4717–4718. doi: 10.1016/j.vaccine.2012.04.093. [DOI] [PubMed] [Google Scholar]
- 3.Drijkoningen JJC, Rohde GGU. 2014. Pneumococcal infection in adults: burden of disease. Clin Microbiol Infect 20(Suppl 5):45–51. doi: 10.1111/1469-0691.12461. [DOI] [PubMed] [Google Scholar]
- 4.Huang SS, Johnson KM, Ray GT, Wroe P, Lieu TA, Moore MR, Zell ER, Linder JA, Grijalva CG, Metlay JP, Finkelstein JA. 2011. Healthcare utilization and cost of pneumococcal disease in the United States. Vaccine 29:3398–3412. doi: 10.1016/j.vaccine.2011.02.088. [DOI] [PubMed] [Google Scholar]
- 5.Weycker D, Strutton D, Edelsberg J, Sato R, Jackson LA. 2010. Clinical and economic burden of pneumococcal disease in older US adults. Vaccine 28:4955–4960. doi: 10.1016/j.vaccine.2010.05.030. [DOI] [PubMed] [Google Scholar]
- 6.García-Rodríquez JA, Fresnadillo Martínez MJ. 2002. Dynamics of nasopharyngeal colonization by potential respiratory pathogens. J Antimicrob Chemother 50:59–73. doi: 10.1093/jac/dkf506. [DOI] [PubMed] [Google Scholar]
- 7.Mazmanian SK, Round JL, Kasper DL. 2008. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 8.Hausdorff WP, Bryant J, Paradiso PR, Siber GR. 2000. Which pneumococcal serogroups cause the most invasive disease: implications for conjugate vaccine formulation and use, part I. Clin Infect Dis 30:100–121. doi: 10.1086/313608. [DOI] [PubMed] [Google Scholar]
- 9.Babl FE, Pelton SI, Theodore S, Klein JO. 2001. Constancy of distribution of serogroups of invasive pneumococcal isolates among children: experience during 4 decades. Clin Infect Dis 32:1155–1161. doi: 10.1086/319750. [DOI] [PubMed] [Google Scholar]
- 10.Hausdorff W, Feikin D, Klugman K. 2005. Epidemiological differences among pneumococcal serotypes. Lancet Infect Dis 5:83–93. http://www.thelancet.com/journals/laninf/article/PIIS1473-3099(05)01280-6/abstract. [DOI] [PubMed] [Google Scholar]
- 11.Weinberger DM, Trzciński K, Lu Y, Bogaert D, Brandes A, Galagan J, Anderson PW, Malley R, Lipsitch M. 2009. Pneumococcal capsular polysaccharide structure predicts serotype prevalence. PLoS Pathog 5:e1000476. doi: 10.1371/journal.ppat.1000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hathaway LJ, Brugger SD, Morand B, Bangert M, Rotzetter JU, Hauser C, Graber WA, Gore S, Kadioglu A, Mühlemann K. 2012. Capsule type of Streptococcus pneumoniae determines growth phenotype. PLoS Pathog 8:e1002574. doi: 10.1371/journal.ppat.1002574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention 2008. Invasive pneumococcal disease in children 5 years after conjugate vaccine introduction—eight states, 1998–2005. MMWR Morb Mortal Wkly Rep 57:144–148. [PubMed] [Google Scholar]
- 14.Whitney CG, Farley MM, Hadler J, Harrison LH, Bennett NM, Lynfield R, Reingold A, Cieslak PR, Pilishvili T, Jackson D, Facklam RR, Jorgensen JH, Schuchat A. 2003. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N Engl J Med 348:1737–1746. doi: 10.1056/NEJMoa022823. [DOI] [PubMed] [Google Scholar]
- 15.Jackson LA, Neuzil KM, Yu O, Benson P, Barlow WE, Adams AL, Hanson CA, Mahoney LD, Shay DK, Thompson WW. 2003. Effectiveness of pneumococcal polysaccharide vaccine in older adults. N Engl J Med 348:1747–1755. doi: 10.1056/NEJMoa022678. [DOI] [PubMed] [Google Scholar]
- 16.Lee GM, Kleinman K, Pelton SI, Hanage W, Huang SS, Lakoma M, Dutta-Linn M, Croucher NJ, Stevenson A, Finkelstein JA. 2014. Impact of 13-valent pneumococcal conjugate vaccination on Streptococcus pneumoniae carriage in young children in Massachusetts. J Pediatr Infect Dis Soc 3:23–32. doi: 10.1093/jpids/pit057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wroe PC, Lee GM, Finkelstein JA, Pelton SI, Hanage WP, Lipsitch M, Stevenson AE, Rifas-Shiman SL, Kleinman K, Dutta-Linn MM, Hinrichsen VL, Lakoma M, Huang SS. 2012. Pneumococcal carriage and antibiotic resistance in young children before 13-valent conjugate vaccine. Pediatr Infect Dis J 31:249–254. doi: 10.1097/INF.0b013e31824214ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laufer AS, Metlay JP, Gent JF, Fennie KP, Kong Y, Pettigrew MM. 2011. Microbial communities of the upper respiratory tract and otitis media in children. mBio 2:e00245-10. doi: 10.1128/mBio.00245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pettigrew MM, Laufer AS, Gent JF, Kong Y, Fennie KP, Metlay JP. 2012. Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl Environ Microbiol 78:6262–6270. doi: 10.1128/AEM.01051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bogaert D, Keijser B, Huse S, Rossen J, Veenhoven R, van Gils E, Bruin J, Montijn R, Bonten M, Sanders E. 2011. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS One 6:e17035. doi: 10.1371/journal.pone.0017035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh J, Conlan S, Polley EC, Segre JA, Kong HH. 2012. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med 4:77. doi: 10.1186/gm378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao Z, Tseng C-H, Pei Z, Blaser MJ. 2007. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci U S A 104:2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. 2009. Bacterial community variation in human body habitats across space and time. Science 326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, NISC Comparative Sequencing Program, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frank DN, Feazel LM, Bessesen MT, Price CS, Janoff EN, Pace NR. 2010. The human nasal microbiota and Staphylococcus aureus carriage. PLoS One 5:e10598. doi: 10.1371/journal.pone.0010598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemon K, Klepac-Ceraj V, Schiffer HK, Brodie EL, Lynch SV, Kolter R. 2010. Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. mBio 1(3):e00129-10. doi: 10.1128/mBio.00129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wos-Oxley ML, Plumeier I, von Eiff C, Taudien S, Platzer M, Vilchez-Vargas R, Becker K, Pieper DH. 2010. A poke into the diversity and associations within human anterior nare microbial communities. ISME J 4:839–851. doi: 10.1038/ismej.2010.15. [DOI] [PubMed] [Google Scholar]
- 28.Human Microbiome Project Consortium 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan M, Pamp S, Fukuyama J, Hwang P, Cho D, Holmes S, Relman D. 2013. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe 14:631–640. doi: 10.1016/j.chom.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasmussen TT, Kirkeby LP, Poulsen K, Reinholdt J, Kilian M. 2000. Resident aerobic microbiota of the adult human nasal cavity. APMIS 108:663–675. doi: 10.1034/j.1600-0463.2000.d01-13.x. [DOI] [PubMed] [Google Scholar]
- 31.Uehara Y, Nakama H, Agematsu K, Uchida M, Kawakami Y, Abdul Fattah ASM, Maruchi N. 2000. Bacterial interference among nasal inhabitants: eradication of Staphylococcus aureus from nasal cavities by artificial implantation of Corynebacterium sp. J Hosp Infect 44:127–133. doi: 10.1053/jhin.1999.0680. [DOI] [PubMed] [Google Scholar]
- 32.Bernard K. 2012. The genus Corynebacterium and other medically relevant coryneform-like bacteria. J Clin Microbiol 50:3152–3158. doi: 10.1128/JCM.00796-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akao T, Kusaka T, Kobashi K. 1981. Two esterases released from Mycobacterium smegmatis for the hydrolysis of long chain acyl-CoAs and Tween. J Biochem 90:1661–1669. [DOI] [PubMed] [Google Scholar]
- 34.Nicolaides N, Fu HC, Rice GR. 1968. The skin surface lipids of man compared with those of eighteen species of animals. J Invest Dermatol 51:83–89. [DOI] [PubMed] [Google Scholar]
- 35.Webster GF, Ruggieri MR, McGinley KJ. 1981. Correlation of Propionibacterium acnes populations with the presence of triglycerides on nonhuman skin. Appl Environ Microbiol 41:1269–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Camera E, Ludovici M, Galante M, Sinagra J-, Picardo M. 2010. Comprehensive analysis of the major lipid classes in sebum by rapid resolution high-performance liquid chromatography and electrospray mass spectrometry. J Lipid Res 51:3377–3388. doi: 10.1194/jlr.D008391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michael-Jubeli R, Bleton J, Baillet-Guffroy A. 2011. High-temperature gas chromatography-mass spectrometry for skin surface lipids profiling. J Lipid Res 52:143–151. doi: 10.1194/jlr.D008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yildirim I, Hanage WP, Lipsitch M, Shea KM, Stevenson A, Finkelstein J, Huang SS, Lee GM, Kleinman K, Pelton SI. 2010. Serotype specific invasive capacity and persistent reduction in invasive pneumococcal disease. Vaccine 29:283–288. doi: 10.1016/j.vaccine.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanage WP, Bishop CJ, Huang SS, Stevenson AE, Pelton SI, Lipsitch M, Finkelstein JA. 2011. Carried pneumococci in Massachusetts children: the contribution of clonal expansion and serotype switching. Pediatr Infect Dis J 30:302–308. doi: 10.1097/INF.0b013e318201a154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60 http://www.genomebiology.com/2011/12/6/R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biesbroek G, Bosch AATM, Wang X, Keijser BJF, Veenhoven RH, Sanders EAM, Bogaert D. 2014. The impact of breastfeeding on nasopharyngeal microbial communities in infants. Am J Respir Crit Care Med 190:298–308. doi: 10.1164/rccm.201401-0073OC. [DOI] [PubMed] [Google Scholar]
- 42.Lemon KP, Armitage GC, Relman DA, Fischbach MA. 2012. Microbiota-targeted therapies: an ecological perspective. Sci Transl Med 4:137rv5. doi: 10.1126/scitranslmed.3004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Do TQ, Moshkani S, Castillo P, Anunta S, Pogosyan A, Cheung A, Marbois B, Faull KF, Ernst W, Chiang SM, Fujii G, Clarke CF, Foster K, Porter E. 2008. Lipids including cholesteryl linoleate and cholesteryl arachidonate contribute to the inherent antibacterial activity of human nasal fluid. J Immunol 181:4177–4187. doi: 10.4049/jimmunol.181.6.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicolaides N. 1974. Skin lipids: their biochemical uniqueness. Science 186:19–26. doi: 10.1126/science.186.4158.19. [DOI] [PubMed] [Google Scholar]
- 45.Cadieux B, Vijayakumaran V, Bernards MA, McGavin MJ, Heinrichs DE. 2014. Role of lipase from community-associated methicillin-resistant Staphylococcus aureus strain USA300 in hydrolyzing triglycerides into growth-inhibitory free fatty acids. J Bacteriol 196:4044–4056. doi: 10.1128/JB.02044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gribbon EM, Cunliffe WJ, Holland KT. 1993. Interaction of Propionibacterium acnes with skin lipids in vitro. J Gen Microbiol 139:1745–1751. doi: 10.1099/00221287-139-8-1745. [DOI] [PubMed] [Google Scholar]
- 47.Stefaniak AB, Harvey CJ, Wertz PW. 2010. Formulation and stability of a novel artificial sebum under conditions of storage and use. Int J Cosmet Sci 32:347–355. doi: 10.1111/j.1468-2494.2010.00561.x. [DOI] [PubMed] [Google Scholar]
- 48.Nagler RM, Klein I, Zarzhevsky N, Drigues N, Reznick AZ. 2002. Characterization of the differentiated antioxidant profile of human saliva. Free Radic Biol Med 32:268–277. doi: 10.1016/S0891-5849(01)00806-1. [DOI] [PubMed] [Google Scholar]
- 49.Pruitt KM, Kamau DN, Miller K, Månsson-Rahemtulla B, Rahemtulla F. 1990. Quantitative, standardized assays for determining the concentrations of bovine lactoperoxidase, human salivary peroxidase, and human myeloperoxidase. Anal Biochem 191:278–286. doi: 10.1016/0003-2697(90)90220-4. [DOI] [PubMed] [Google Scholar]
- 50.Uehara Y, Agematsu K, Kikuchi K, Matsuzaki S, Imai S, Takamoto M, Sugane K, Sugiura T, Konishi Y, Yoshino N, Takeuchi S, Seo H, Kuramoto S, Sugai M. 2006. Secretory IgA, salivary peroxidase, and catalase-mediated microbicidal activity during hydrogen peroxide catabolism in viridans streptococci: pathogen coaggregation. J Infect Dis 194:98–107. doi: 10.1086/504439. [DOI] [PubMed] [Google Scholar]
- 51.Lijek RS, Weiser JN. 2012. Co-infection subverts mucosal immunity in the upper respiratory tract. Curr Opin Immunol 24:417–423. doi: 10.1016/j.coi.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Margolis E. 2009. Hydrogen peroxide-mediated interference competition by Streptococcus pneumoniae has no significant effect on Staphylococcus aureus nasal colonization of neonatal rats. J Bacteriol 191:571–575. doi: 10.1128/JB.00950-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Melles DC, Bogaert D, Gorkink RFJ, Peeters JK, Moorhouse MJ, Ott A, van Leeuwen WB, Simons G, Verbrugh HA, Hermans PWM, van Belkum A. 2007. Nasopharyngeal co-colonization with Staphylococcus aureus and Streptococcus pneumoniae in children is bacterial genotype independent. Microbiology 153:686–692. doi: 10.1099/mic.0.2006/002279-0. [DOI] [PubMed] [Google Scholar]
- 54.Regev-Yochay G, Lipsitch M, Basset A, Rubinstein E, Dagan R, Raz M, Malley R. 2009. The pneumococcal pilus predicts the absence of Staphylococcus aureus co-colonization in pneumococcal carriers. Clin Infect Dis 48:760–763. doi: 10.1086/597040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pericone CD, Overweg K, Hermans PWM, Weiser JN. 2000. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect Immun 68:3990–3997. doi: 10.1128/IAI.68.7.3990-3997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lysenko ES, Ratner AJ, Nelson AL, Weiser JN. 2005. The role of innate immune responses in the outcome of interspecies competition for colonization of mucosal surfaces. PLoS Pathog 1:e1. doi: 10.1371/journal.ppat.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clementi EA, Wilhelm KR, Schleucher J, Morozova-Roche LA, Hakansson AP. 2013. A complex of equine lysozyme and oleic acid with bactericidal activity against Streptococcus pneumoniae. PLoS One 8:e80649. doi: 10.1371/journal.pone.0080649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fischer CL, Drake DR, Dawson DV, Blanchette DR, Brogden KA, Wertz PW. 2012. Antibacterial activity of sphingoid bases and fatty acids against Gram-positive and Gram-negative bacteria. Antimicrob Agents Chemother 56:1157–1161. doi: 10.1128/AAC.05151-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parsons JB, Yao J, Frank MW, Jackson P, Rock CO. 2012. Membrane disruption by antimicrobial fatty acids releases low-molecular-weight proteins from Staphylococcus aureus. J Bacteriol 194:5294–5304. doi: 10.1128/JB.00743-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Speert DP, Wannamaker LW, Gray ED, Clawson CC. 1979. Bactericidal effect of oleic acid on group A streptococci: mechanism of action. Infect Immun 26:1202–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Drake DR, Brogden KA, Dawson DV, Wertz PW. 2008. Thematic review series: skin lipids. Antimicrobial lipids at the skin surface. J Lipid Res 49:4–11. doi: 10.1194/jlr.R700016-JLR200. [DOI] [PubMed] [Google Scholar]
- 62.Arsic B, Zhu Y, Heinrichs DE, McGavin MJ. 2012. Induction of the staphylococcal proteolytic cascade by antimicrobial fatty acids in community acquired methicillin resistant Staphylococcus aureus. PLoS One 7:e45952. doi: 10.1371/journal.pone.0045952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kabara JJ, Swieczkowski DM, Conley AJ, Truant JP. 1972. Fatty acids and derivatives as antimicrobial agents. Antimicrob Agents Chemother 2:23–28. doi: 10.1128/AAC.2.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jaeger K, Ransac S, Dijkstra BW, Colson C, van Heuvel M, Misset O. 1994. Bacterial lipases. FEMS Microbiol Rev 15:29–63. doi: 10.1111/j.1574-6976.1994.tb00121.x. [DOI] [PubMed] [Google Scholar]
- 65.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res 39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Farrell AM, Foster TJ, Holland KT. 1993. Molecular analysis and expression of the lipase of Staphylococcus epidermidis. J Gen Microbiol 139:267–277. doi: 10.1099/00221287-139-2-267. [DOI] [PubMed] [Google Scholar]
- 67.Longshaw CM, Farrell AM, Wright JD, Holland KT. 2000. Identification of a second lipase gene, gehD, in Staphylococcus epidermidis: comparison of sequence with those of other staphylococcal lipases. Microbiology 146:1419–1427. doi: 10.1099/00221287-146-6-1419. [DOI] [PubMed] [Google Scholar]
- 68.Miskin JE, Farrell AM, Cunliffe WJ, Holland KT. 1997. Propionibacterium acnes, a resident of lipid-rich human skin, produces a 33 kDa extracellular lipase encoded by gehA. Microbiology 143:1745–1755. doi: 10.1099/00221287-143-5-1745. [DOI] [PubMed] [Google Scholar]
- 69.Schröder J, Maus I, Meyer K, Wördemann S, Blom J, Jaenicke S, Schneider J, Trost E, Tauch A. 2012. Complete genome sequence, lifestyle, and multi-drug resistance of the human pathogen Corynebacterium resistens DSM 45100 isolated from blood samples of a leukemia patient. BMC Genomics 13:141. doi: 10.1186/1471-2164-13-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schröder J, Maus I, Trost E, Tauch A. 2011. Complete genome sequence of Corynebacterium variabile DSM 44702 isolated from the surface of smear-ripened cheeses and insights into cheese ripening and flavor generation. BMC Genomics 12:545. doi: 10.1186/1471-2164-12-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 72.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic Local Alignment Search Tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 73.Wollenberg MS, Claesen J, Escapa IF, Aldridge KL, Fischbach MA, Lemon KP. 2014. Propionibacterium-produced coproporphyrin III induces Staphylococcus aureus aggregation and biofilm formation. mBio 5:e01286-14. doi: 10.1128/mBio.01286-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kiryukhina NV, Melnikov VG, Suvorov AV, Morozova YA, Ilyin VK. 2013. Use of Corynebacterium pseudodiphtheriticum for elimination of Staphylococcus aureus from the nasal cavity in volunteers exposed to abnormal microclimate and altered gaseous environment. Probiotics Antimicrob Prot 5:233–238. doi: 10.1007/s12602-013-9147-x. [DOI] [PubMed] [Google Scholar]
- 75.Rabasco Alvarez AM, González Rodríguez ML. 2000. Lipids in pharmaceutical and cosmetic preparations. Grasas Aceites 51:74–96. doi: 10.3989/gya.2000.v51.i1-2.409. [DOI] [Google Scholar]
- 76.Chen CH, Wang Y, Nakatsuji T, Liu YT, Zouboulis CC, Gallo RL, Zhang L, Hsieh MF, Huang CM. 2011. An innate bactericidal oleic acid effective against skin infection of methicillin-resistant Staphylococcus aureus: a therapy concordant with evolutionary medicine. J Microbiol Biotechnol 21:391–399. http://www.jmb.or.kr/journal/viewJournal.html?year=2011&vol=21&num=4&page=391. [PubMed] [Google Scholar]
- 77.Nicolaides N, Wells GC. 1957. On the biogenesis of the free fatty acids in human skin surface fat. J Invest Dermatol 29:423–433. [DOI] [PubMed] [Google Scholar]
- 78.Zouboulis CC, Seltmann H, Neitzel H, Orfanos CE. 1999. Establishment and characterization of an immortalized human sebaceous gland cell line (SZ95). J Invest Dermatol 113:1011–1020. doi: 10.1046/j.1523-1747.1999.00771.x. [DOI] [PubMed] [Google Scholar]
- 79.Marples RR, Downing DT, Kligman AM. 1971. Control of free fatty acids in human surface lipids by Corynebacterium acnes. J Invest Dermatol 56:127–131. doi: 10.1111/1523-1747.ep12260695. [DOI] [PubMed] [Google Scholar]
- 80.Bek-Thomsen M, Lomholt HB, Scavenius C, Enghild JJ, Brüggemann H. 2014. Proteome analysis of human sebaceous follicle infundibula extracted from healthy and acne-affected skin. PLoS One 9:e107908. doi: 10.1371/journal.pone.0107908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ingham E, Holland KT, Gowland G, Cunliffe WJ. 1981. Partial purification and characterization of lipase (EC 3.1.1.3) from Propionibacterium acnes. J Gen Microbiol 124:393–401. doi: 10.1099/00221287-124-2-393. [DOI] [PubMed] [Google Scholar]
- 82.Nakatsuji T, Kao MC, Fang J, Zouboulis CC, Zhang L, Gallo RL, Huang C. 2009. Antimicrobial property of lauric acid against Propionibacterium acnes: its therapeutic potential for inflammatory acne vulgaris. J Invest Dermatol 129:2480–2488. doi: 10.1038/jid.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, Bushman FD, Collman RG. 2011. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med 184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Charlson ES, Chen J, Custers-Allen R, Bittinger K, Li H, Sinha R, Hwang J, Bushman FD, Collman RG. 2010. Disordered microbial communities in the upper respiratory tract of cigarette smokers. PLoS One 5:e15216. doi: 10.1371/journal.pone.0015216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cotterill JA, Cunliffe WJ, Williamson B, Bulusu L. 1972. Age and sex variation in skin surface lipid composition and sebum excretion rate. Br J Dermatol 87:333–341. doi: 10.1111/j.1365-2133.1972.tb07419.x. [DOI] [PubMed] [Google Scholar]
- 86.Nakatsuji T, Kao MC, Zhang L, Zouboulis CC, Gallo RL, Huang C. 2010. Sebum free fatty acids enhance the innate immune defense of human sebocytes by upregulating beta-defensin-2 expression. J Invest Dermatol 130:985–994. doi: 10.1038/jid.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee H, Andalibi A, Webster P, Moon S, Teufert K, Kang S, Li J, Nagura M, Ganz T, Lim DJ. 2004. Antimicrobial activity of innate immune molecules against Streptococcus pneumoniae, Moraxella catarrhalis and nontypeable Haemophilus influenzae. BMC Infect Dis 4:12. doi: 10.1186/1471-2334-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, Agata T, Mizunoe Y. 2010. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465:346–349. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 89.Lo C, Lai Y, Liu Y, Gallo RL, Huang C. 2011. Staphylococcus aureus hijacks a skin commensal to intensify its virulence: immunization targeting beta-hemolysin and CAMP factor. J Invest Dermatol 131:401–409. doi: 10.1038/jid.2010.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shu M, Wang Y, Yu J, Kuo S, Coda A, Jiang Y, Gallo RL, Huang C. 2013. Fermentation of Propionibacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus. PLoS One 8:e55380. doi: 10.1371/journal.pone.0055380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fisher MM, Triplett EW. 1999. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl Environ Microbiol 65:4630–4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Morrow AL, Lagomarcino AJ, Schibler KR, Taft DH, Yu Z, Wang B, Altaye M, Wagner M, Gevers D, Ward DV, Kennedy MA, Huttenhower C, Newburg DS. 2013. Early microbial and metabolomic signatures predict later onset of necrotizing enterocolitis in preterm infants. Microbiome 1:13. doi: 10.1186/2049-2618-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Converse SE, Cox JS. 2005. A protein secretion pathway critical for Mycobacterium tuberculosis virulence is conserved and functional in Mycobacterium smegmatis. J Bacteriol 187:1238–1245. doi: 10.1128/JB.187.4.1238-1245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gibson DG, Young L, Chuang R, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 96.Eggeling L, Bott M. 2005. Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, FL. [Google Scholar]
- 97.Reyes O, Guyonvarch A, Bonamy C, Salti V, David F, Leblon G. 1991. “Integron”-bearing vectors: a method suitable for stable chromosomal integration in highly restrictive corynebacteria. Gene 107:61–68. doi: 10.1016/0378-1119(91)90297-O. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Materials and Methods. Download
Basic anatomy of the human nasal passages. Pneumococcus colonizes the nose from the nostrils to the nasopharynx. The nostrils (indicated by the arrow), also known as the anterior nares, are the entrance to the nasal passages and open onto the skin-covered surface of the nasal vestibules. The skin lining the nasal vestibules includes sweat and sebaceous glands. Posterior to the nasal vestibules, the epithelial surface of the nasal cavity transitions to a mucosal surface. Respiratory epithelium, including cilia, lines the posterior segment of the nasal cavity and the nasopharynx, at the top of the back of the throat. Download
Relative abundance plots for each of the three bacterial genera that are overrepresented when S. pneumoniae is absent (class 0) or present (class 1) via cultivation. Subclasses are shown as follows: A, in red, <2 years old; B, in green, 2 to <7 years. (A) Streptococcus; (B) Dolosigranulum; (C) Corynebacterium. Plots were generated using LEfSE from the same data used to generate Fig. 1. The solid and dashed lines indicate the mean and the median relative abundance levels, respectively. Each bar represents a nasopharyngeal (NP) sample from one volunteer. Download
Dendrogram representing phylogenetic relationship of KPL1818 with C. accolens and other Corynebacterium spp. The dendrogram was constructed using the maximum-likelihood method with 1,000 bootstrap replications of ClustalW-aligned partial rpoB sequences (446-bp fragment between primers C2700F and C3130R) (Khamis A, Raoult D, La Scola B, J Clin Microbiol 42:3925–3931, doi:10.1128/JCM.42.9.3925-3931.2004). Branch support is indicated by the value at each node. D. cinnamea was used as an outgroup. Download
C. accolens lipS1 is not necessary for growth when Tween 80 (Tw80) is the sole source of fatty acids. (A) A C. accolens mutant (LipS1-) harboring an in-frame deletion in lipS1 grew comparably to wild type (WT) in BHI supplemented with 1% Tw80. Growth in A and B was measured as the mean OD600 following 48 h of aerobic growth shaking at 37°C. Data were analyzed using a Welch two-sample t test, n = 3, and error bars represent SD. (B) The lipase-deficient mutant carrying the empty vector (LipS1- pCGL+) also grew comparably to wild type (WT) in BHI supplemented with 1% Tw80 as did the complemented mutant (LipS1- cLipS1+). For comparison, the empty vector and complemented vector controls for the parental strain (WT pCGL+ and WT cLipS1+, respectively) are included. Data (n = 3) were analyzed using ANOVA. Error bars represent SD. Download
LipS1 from C. accolens KPL1818. Shown at the top are the predicted open reading frame and corresponding translation for the His6-tagged LipS1 (WP_034657805.1), as annotated in NCBI. The predicted signal sequence (orange bar), canonical G-X-S-X-G motif (red bar), His6 tag (gray bar), and stop codon (blue bar) are highlighted. The sequence cloned into pLB502 contains an additional 949 bp 5′ of the predicted start codon shown. Download
Additional C. accolens strains tested for inhibition of S. pneumoniae 603.
Bacterial strains and plasmids used in this study
Primers designed for this study