Abstract
The lifespan and activity of proteins depend on protein quality control systems formed by chaperones and proteases that ensure correct protein folding and prevent the formation of toxic aggregates. We previously found that the Arabidopsis thaliana J-protein J20 delivers inactive (misfolded) forms of the plastidial enzyme deoxyxylulose 5-phosphate synthase (DXS) to the Hsp70 chaperone for either proper folding or degradation. Here we show that the fate of Hsp70-bound DXS depends on pathways involving specific Hsp100 chaperones. Analysis of individual mutants for the four Hsp100 chaperones present in Arabidopsis chloroplasts showed increased levels of DXS proteins (but not transcripts) only in those defective in ClpC1 or ClpB3. However, the accumulated enzyme was active in the clpc1 mutant but inactive in clpb3 plants. Genetic evidence indicated that ClpC chaperones might be required for the unfolding of J20-delivered DXS protein coupled to degradation by the Clp protease. By contrast, biochemical and genetic approaches confirmed that Hsp70 and ClpB3 chaperones interact to collaborate in the refolding and activation of DXS. We conclude that specific J-proteins and Hsp100 chaperones act together with Hsp70 to recognize and deliver DXS to either reactivation (via ClpB3) or removal (via ClpC1) depending on the physiological status of the plastid.
Author Summary
In this paper we report a relatively simple mechanism by which plant chloroplasts deal with inactive forms of DXS, the main rate-determining enzyme for the production of plastidial isoprenoids relevant for photosynthesis and development. We provide evidence supporting that particular members of the Hsp100 chaperone family contribute to either refold or degrade inactive DXS proteins specifically recognized by the J-protein adaptor J20 and delivered to Hsp70 chaperones. Our results also unveil a J-protein-based mechanism for substrate delivery to the Clp complex, the main protease in the chloroplast stroma. Together, this work allows a better understanding of how chloroplasts get rid of damaged DXS (and potentially other proteins), which should contribute to take more informed decisions in future approaches aimed to manipulate the levels of plastidial metabolites of interest (including vitamins, biofuels, or drugs against cancer and malaria) in crop plants.
Introduction
Organelles like mitochondria and plastids play fundamental roles in all eukaryotic organisms. In particular, plastids were acquired by a symbiosis between photosynthetic cyanobacteria and eukaryotic cells. Today, plastids (like mitochondria) are intimately integrated into the metabolism of plant cells but they still remain as separate functional entities that regulate their own biochemistry by relatively independent mechanisms. An important part of this regulation relies on the effective control of plastidial enzyme activities. Most of the enzymes required for plastidial metabolism are encoded by nuclear genes, synthesized in precursor form in the cytosol, and transported into plastids using energy-dependent import machineries [1]. Following import, specific proteases cleave the transit peptides and complex networks of plastidial chaperones ensure proper folding, assembly, or suborganellar targeting of the mature proteins. Chaperones and proteases are also essential components of the protein quality control (PQC) system that promotes the stabilization, refolding, or degradation of mature proteins that lose their native conformation and activity after metabolic perturbations or environmental challenges such as excess light, temperature peaks, oxidative stress or nutrient starvation [2,3]. While plant plastids contain many groups of prokaryotic-like chaperones (such as Hsp70 and Hsp100) and proteases (including Clp, Lon, Deg, and FstH), their specific targets and PQC-related roles remain little studied [1–4].
Due to the presence of plastids, plants have biochemical pathways that are not found in other eukaryotic kingdoms. For example, isoprenoid precursors are produced by the methylerythritol 4-phosphate (MEP) pathway in bacteria and plant plastids, whereas animals and fungi synthesize these essential metabolites using a completely unrelated pathway which is also used by plants to produce cytosolic and mitochondrial isoprenoids [5,6]. MEP-derived isoprenoids include compounds essential for photosynthesis (such as carotenoids and the side chain of chlorophylls, tocopherols, plastoquinone and phylloquinones) and growth regulation (including the hormones gibberellins, cytokinins, strigolactones and abscisic acid). Many plastidial isoprenoids also have nutritional and economic relevance [6]. All MEP pathway enzymes are located in the plastid stroma [5,7]. While transcriptional regulation of genes encoding biosynthetic enzymes is known to exert a coarse control of the MEP pathway, fine-tuning of metabolic flux appears to rely on post-transcriptional or/and post-translational regulation of enzyme levels and activity [8–12]. This is most evident for deoxyxylulose 5-phosphate synthase (DXS), the homodimeric enzyme that catalyzes the first step of the pathway. Metabolic control analysis calculations confirmed that DXS is the enzyme with the highest flux control coefficient (i.e. the main rate-determining step) of the MEP pathway [13]. Consistent with this prime regulatory role, DXS activity is tightly regulated by several post-translational mechanisms [10–12]. In particular, DXS enzymatic activity is allosterically inhibited by MEP pathway products [14,15], which also repress DXS protein accumulation [8,14,16–18]. Mathematical modeling recently showed that the post-translational control of DXS protein abundance and enzyme activity is crucial for the adjustment of the MEP pathway flux to persistent changes in environmental conditions, such as substrate supply or product demand [18]. Despite the central relevance of this type of regulation, little is known about the molecular mechanisms behind it.
We previously showed that the Arabidopsis thaliana J-protein J20 interacts with inactive forms of DXS to deliver them to the Hsp70 chaperone for eventual activation (which involves folding or refolding) or degradation (which involves unfolding) [19]. However, the particular protease involved and the specific components of the two J20-dependent antagonistic pathways remained unknown. Here we show that DXS is primarily degraded by the Clp protease complex through a pathway involving J20 and Hsp100 chaperones of the ClpC type. We also demonstrate that Hsp70 can physically interact with ClpB3, another plastidial Hsp100 chaperone, to promote the activation of non-functional DXS enzymes.
Results and Discussion
DXS appears to be primarily degraded by the stromal Clp protease
The main protease families involved in the degradation of terminally damaged or surplus proteins in plastids are Clp, Lon, Deg, and FtsH, all of them of prokaryotic origin [3,4]. We and others have previously shown that Arabidopsis mutants with a decreased activity of the stromal Clp protease complex display an accumulation of several MEP pathway enzymes, including DXS [20–24]. However, whether other plastidial proteases involved in PQC networks could also contribute to DXS degradation in the stroma remains unexplored. Several functional Lon homologues are found in Arabidopsis, but only Lon1 [25] and Lon4 [26] have been localized to chloroplasts, where they are attached to the stromal side of thylakoids. The Deg gene family in Arabidopsis contains 16 members, with 5 of them experimentally confirmed to be localized in chloroplasts [27]. From these, the isoforms Deg1, Deg5 and Deg8 were found in the thylakoid lumen, whereas Deg2 and Deg7 were detected in the stromal side [28,29]. FtsH proteases are encoded by 12 genes in Arabidopsis, and 9 of them can be found in chloroplasts [30]. The four major chloroplast isomers (FtsH2, FtsH5, FtsH8 and FtsH1, in order of abundance) have been shown to reside in the thylakoid membrane with their catalytic domain facing the stromal side [31–33].
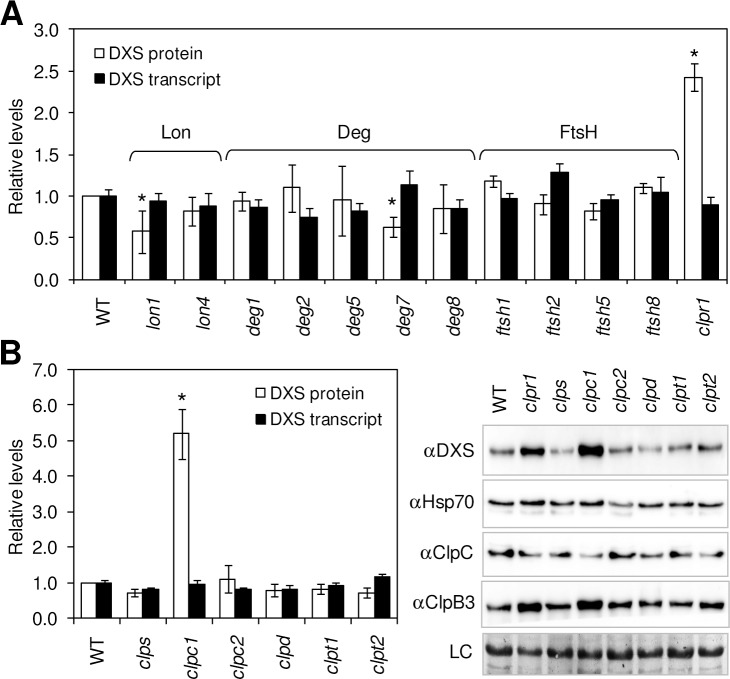
DXS protein levels were examined by immunoblot analysis in Arabidopsis wild-type (WT) plants and single mutants defective in plastidial Lon, Deg, or FtsH isomers (Fig 1 and S1 Table). As a control, we included the Clp protease mutant clpr1, which displays a reduction of other subunits of the Clp proteolytic core [34] but increased DXS protein levels [20]. As shown in Fig 1A, DXS protein levels in the analyzed mutants were similar to those in WT plants with only three exceptions. Lines defective in Lon1 and Deg7 showed a decreased accumulation of the protein compared to the WT, whereas DXS levels were only increased in the clpr1 mutant (Fig 1A). No changes in DXS transcript levels were detected in any of the mutant lines (Fig 1A). Since enzyme levels would be expected to be post-translationally increased (but not decreased) in the mutants impaired in DXS-degrading proteases, we conclude that the Clp complex is likely the primary protease for DXS removal. The contribution of Lon, Deg, or FtsH proteases, however, cannot be fully discarded as we only tested mutants for individual isoforms of those other proteases and it is possible that different isoforms may have redundant functions.
Fig 1. DXS is primarily degraded by the Clp protease.
Columns represent DXS protein and transcript levels in 10-day-old WT plants and single mutants defective in the indicated subunits of plastidial proteases (A) or regulatory components of the Clp protease complex (B). Data correspond to the mean and SEM values of n≥3 independent experiments and are represented relative to the levels in WT plants. Asterisks mark statistically significant differences (t test: p<0.05) relative to WT samples. Representative images of immunoblot analyses with the indicated antibodies and a Coomassie blue staining of the blots (loading control, LC) are also shown in (B).
ClpC chaperones are required for the degradation of DXS by the Clp protease
Clp proteases are found in almost all bacteria and endosymbiotic organelles (mitochondria and plastids). In bacteria (S1 Fig), they are formed by a barrel-like catalytic core of two heptameric rings of proteolytic subunits (ClpP) and a dynamically interacting hexameric ring of Hsp100 chaperones (ClpA and ClpX in Escherichia coli; ClpC, ClpX, and ClpE in Bacillus subtilis) that unfold substrates for translocation into the proteolytic chamber [35]. Additionally, interaction of Hsp100 members with adaptor proteins (such as ClpS and SspB in E. coli and ClpS, MecA, McsB, and others in B. subtilis) enhance or expand substrate specificity [35–37]. In plant plastids, the Clp protease is more complex [38,39] but the basic components are conserved (S1 Fig). It presents a protease core (formed by two heptameric rings of plastome encoded ClpP1 and nuclear-encoded ClpP3-P6 and ClpR1-R4 subunits) stabilized by two plant-specific subunits (ClpT1-T2). The Arabidopsis homologues of the bacterial ClpA and ClpC unfolding chaperones are ClpC1, ClpC2, and ClpD. A ClpS adaptor is also found in chloroplasts [40], where it might form a plant-specific binary adaptor complex with the ClpF protein [41]. The possibility of other pathways delivering proteins to the Clp protease, however, remains open.
As shown above (Fig 1A), DXS levels increase in mutants defective in Clp protease activity such as clpr1 [20]. If DXS is targeted to the Clp protease for degradation, we would also expect a post-translational upregulation of DXS enzyme levels in mutants impaired in the adaptors and chaperones that deliver the protein to the Clp catalytic core. A systematic analysis of such mutants (clps, clpc1, clpc2, clpd, clpt1, and clpt2) showed that only those defective in ClpC1 accumulated higher levels of DXS protein than WT plants (Fig 1B and S2 Fig). Quantification of DXS-encoding transcripts in the same mutant lines showed WT levels in all cases (Fig 1B), confirming that the observed accumulation of DXS polypeptides in ClpC1-defective lines was not a consequence of increased gene expression.
It has been proposed that the two Arabidopsis ClpC paralogs ClpC1 and ClpC2 perform similar if not identical functions in the chloroplast [42]. However, proteolytic assays with known Clp protease substrates only showed a greatly reduced degradation rate in clpc1 plants [42], which showed the strongest reduction in total ClpC content (Fig 1B and S2 Fig). Estimation of DXS degradation rates upon treating WT and mutant plants with the protein synthesis inhibitor cycloheximide also showed a slower proteolytic removal of DXS polypeptides in clpc1 mutants (Fig 2A). As expected, a defective Clp catalytic core in the clpr1 mutant led to similarly reduced DXS degradation rates (Fig 2A), again supporting our conclusion that DXS is a target for this proteolytic complex. To confirm whether DXS might be a ClpC1 substrate, tagged versions of the Arabidopsis proteins (DXS-GFP and ClpC1-MYC) were overproduced in Nicotiana benthamiana leaves by agroinfiltration and co-immunoprecipitation assays were next performed. As shown in Fig 2B, these assays confirmed that DXS and ClpC1 can indeed interact. Together, we conclude that DXS might be mainly unfolded by ClpC1 for degradation by the Clp proteolytic core.
Fig 2. J20 and ClpC1 are required for normal DXS degradation.
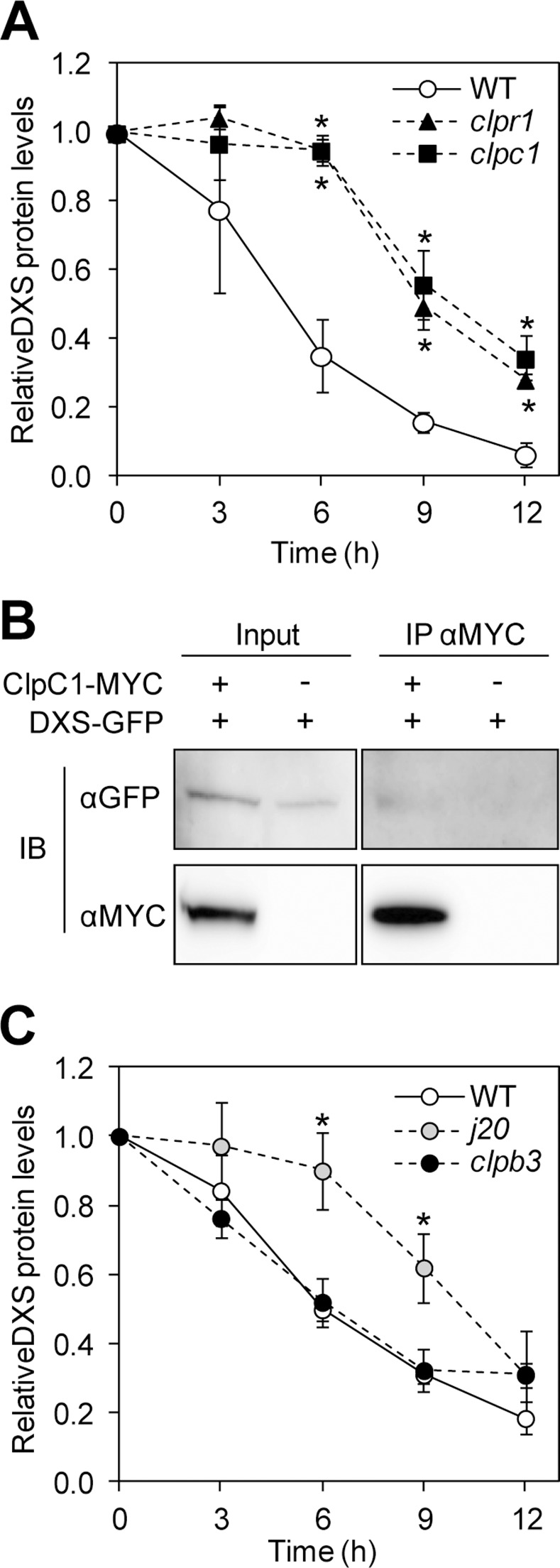
(A) WT plants and mutants defective in ClpR1 or ClpC1 were grown for one week on medium lacking the protein synthesis inhibitor cycloheximide and then treated with the inhibitor for the indicated times. DXS protein levels detected by immunoblot analysis are represented relative to those before treatment. (B) Protein extracts from Nicotiana benthamiana plants transiently producing DXS-GFP alone or together with a MYC-tagged ClpC1 protein were used for immunoprecipitation (IP) with anti-MYC antibodies (αMYC) and further immunoblot (IB) analysis with anti-GFP or anti-MYC sera. Immunoblot analyses of the extracts before immunoprecipitation (Input samples) are also shown. (C) Protein degradation rates of DXS in WT plants and mutants defective in J20 or ClpB3. The experiment was performed as described in (A). Mean and SEM of n≥3 experiments are shown in (A) and (C). Asterisks mark statistically significant differences (t test: p<0.05) relative to WT samples.
J20 together with Hsp70 might deliver DXS to ClpC chaperones for eventual degradation
Recent results have shown that client proteins of the stromal Clp protease are recognized and delivered to ClpC chaperones by ClpS and ClpF adaptors [40,41]. While DXS might actually be a target of ClpS in bacteria [43], a wild-type phenotype in terms of DXS protein levels was observed in Arabidopsis plants defective in the proposed chloroplast adaptors (Fig 1B) [40,41]. Although ClpC could possibly directly deliver client proteins to the Clp protease without the need of an adaptor, we reasoned that further substrate specificity should be achieved using an alternative ClpS/ClpF-independent adaptor system.
Our previous work showed that inactive forms of DXS are recognized by J20, a J-protein adaptor that delivers them to the Hsp70 chaperone [19]. Computational analysis of the Arabidopsis DXS monomer with the Aggrescan3D algorithm revealed the presence of several aggregation-prone clusters (S3 Fig). Consistent with the conclusion that DXS tends to aggregate and that J20 prevents its aggregation, GFP-tagged DXS proteins accumulate in plastidial speckles that are larger in j20 plants (S4 Fig) [19]. In addition, the endogenous DXS enzymes are less accessible to proteinase K cleavage in the j20 mutant (S4 Fig), again suggesting that DXS aggregation is increased in the absence of J20, likely because the delivery of aggregated (and hence inactive) DXS proteins to the Hsp70 chaperone is impaired. The main role of Hsp70 is actually to prevent the formation of toxic aggregates of damaged proteins and, together with Hsp100 chaperones, promote their solubilization [44–50]. However, Hsp70 chaperones also facilitate the transfer of irreparably damaged client proteins to proteolytic systems [49,51–53]. For example, cytosolic Hsp70 is involved in the degradation of Arabidopsis protein clients by the eukaryotic 26S proteasome [51].
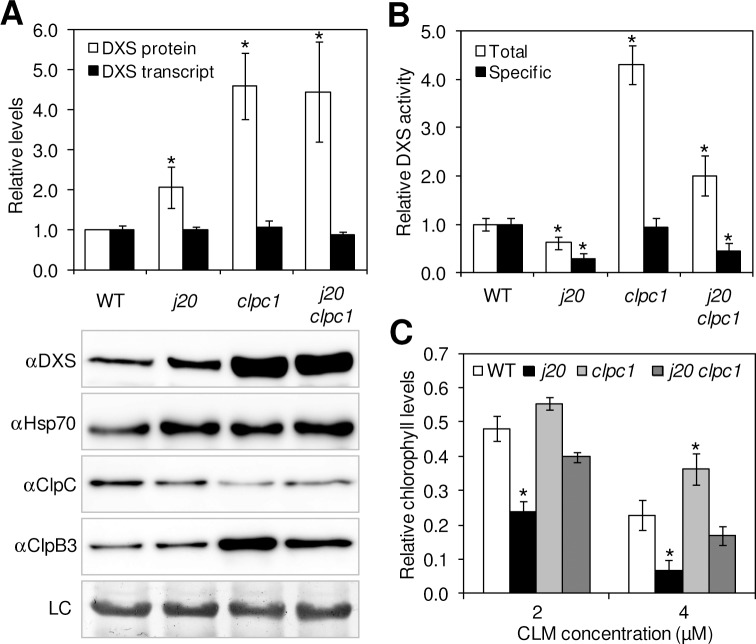
Despite the absence of conserved domains for direct interactions between Hsp70 and ClpC-type Hsp100 proteins (S5 Fig) [36,45,46], co-immunoprecipitation experiments showed that both chaperones can be found together in the chloroplast envelope [54,55]. It is therefore possible that Hsp70 and ClpC might interact either directly (using unidentified chaperone binding motifs) or indirectly (via third partners) to participate in PQC events at the stromal side of the inner envelope membrane [1,42,56,57]. Because in Arabidopsis the two plastidial isoforms of Hsp70 (Hsp70.1 and Hsp70.2) and ClpC (ClpC1 and ClpC2) are also found in the stroma [42,58], we reasoned that Hsp70 and ClpC proteins might collaborate to deliver DXS to the Clp protease using J20 as an adaptor. Interestingly, overexpression of J20 in transgenic Arabidopsis plants leads to decreased DXS protein levels, whereas loss of J20 function causes a reduced degradation rate of the enzyme (Fig 2C) [19]. Since both the J20 adaptor and ClpC chaperones are involved in the control of DXS degradation, we next tested whether they might function in the same pathway. We followed a genetic strategy based on comparing the DXS accumulation phenotype of single mutants defective in either J20 or ClpC1 with that of double j20 clpc1 mutants (Fig 3). All three mutants accumulated higher levels of DXS proteins (but not transcripts) compared to WT plants. In particular, DXS levels increased ca. 2-fold in j20 plants and 4-fold in single clpc1 and double j20 clpc1 mutants (Fig 3A). The absence of an additive or synergistic phenotype in the double mutant supports the conclusion that J20 and ClpC1 actually function in the same pathway delivering DXS to degradation in Arabidopsis plastids. Such a ClpS/ClpF-independent pathway could potentially be functioning for other plastidial clients of J-proteins. However, the lack of bona-fide substrates for other plastidial J-proteins prevents to experimentally testing this possibility at the moment.
Fig 3. J20 and ClpC1 participate in the same pathway for DXS degradation.
(A) Quantification of DXS protein and transcript levels in 10-day-old WT and mutant plants defective in J20, ClpC1, or both. Representative images of immunoblot analyses with the indicated antibodies and a loading control are also shown. (B) DXS activity levels in the indicated lines represented as total or specific (i.e. relative to the amount of DXS protein) values. (C) CLM resistance of the lines shown represented as the amount of chlorophyll remaining in plants grown at the indicated concentrations of the inhibitor. Levels are represented relative to those in WT plants in (A) and (B) or to untreated samples in (C). Data correspond to the mean and SEM values of n≥3 independent experiments and asterisks mark statistically significant differences (t test: p<0.05) relative to WT samples.
ClpB3 contributes to activation of J20-delivered DXS proteins by interaction with plastidial Hsp70 chaperones
The results described above suggest that damaged DXS polypeptides might be recognized by J20 and then delivered to Hsp70 and ClpC chaperones for unfolding and degradation by the Clp proteolytic core. But unlike that observed for J20-defective mutants [19], DXS accumulates in a mostly active form in clpc1 mutants (Fig 3B). Thus, measurement of DXS activity in plant extracts showed increased total activity but unchanged specific activity (i.e. relative to protein levels) in clpc1 compared to WT controls (Fig 3B). Estimation of DXS activity in planta by quantification of the resistance to clomazone (CLM), a specific DXS inhibitor [19,59,60] further supported the presence of higher DXS activity levels (i.e. increased resistance to the inhibitor) in the clpc1 mutant, opposite to the increased sensitivity detected in the case of j20 plants (Fig 3C and S6 Fig). To reconcile these results, we propose that loss of J20 causes an accumulation of aggregated (i.e. enzymatically inactive) DXS because the protein cannot be normally reactivated (refolded) or discarded (degraded). By contrast, accumulation of active DXS enzyme when ClpC activity decreases might be due to the existence of a functional pathway to disaggregate and refold the excess protein that cannot be degraded in the clpc1 mutant. The participation of J20 in such a putative reactivation pathway is supported by the observation that loss of J20 function in degradation-impaired (ClpC1-defective) j20 clpc1 plants results in a higher proportion of inactive DXS enzyme (Fig 3B) and hence a reduced resistance to CLM (Fig 3C and S6 Fig) compared to the single clpc1 mutant.
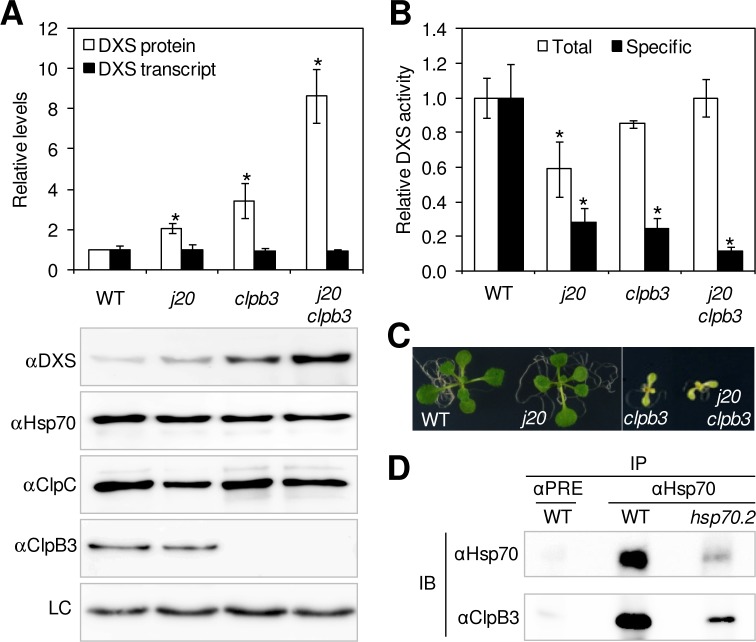
Work in different systems has shown that Hsp70 can be assisted by Hsp100 chaperones of the ClpB type in the solubilization of toxic aggregates of damaged proteins [45–50,61–64]. ClpB3 is the only ClpB-type chaperone found in Arabidopsis plastids [44]. Unlike the rest of plastidial Hsp100 chaperones present in this plant (ClpC1, ClpC2, and ClpD), ClpB3 lacks the IGF motif (or ClpP-loop) required for interaction with proteolytic subunits of the Clp core but it harbors a domain responsible for the interaction with Hsp70 chaperones (S5 Fig) [36,45,46]. Interestingly, the levels of ClpB3 were increased in mutants defective in Clp protease subunits, including ClpC1 [21,22,24,40,65] (Figs 1B and 3A and S2 Fig), suggesting that ClpB3 might contribute to mitigate protein folding stress caused by a defective Clp protease activity. In agreement, impairment of both ClpB3 and Clp protease activity results in a seedling lethal phenotype [22]. Based on these data, we speculated that ClpB3 might also participate in the DXS reactivation pathway mediated by J20 and Hsp70 chaperones. To evaluate this possibility, we first analyzed DXS protein levels and activity in ClpB3-defective Arabidopsis plants (Fig 4). If ClpB3 promotes DXS protein disaggregation (and hence activation), it was expected that clpb3 mutants would show a transcription-independent accumulation of inactive forms of DXS, assuming that the degradation rate of J20-delivered proteins would remain constant. Indeed, clpb3 plants showed a WT rate of DXS degradation (Fig 2C) but an enhanced accumulation of DXS enzyme without changes in transcript levels (Fig 4A). Also as predicted by our model, the specific activity of the DXS protein found in the ClpB3-defective mutant was much lower than that measured in WT plants (Fig 4B). Loss of both ClpB3 and J20 activities in the double j20 clpb3 mutant resulted in an even higher accumulation (Fig 4A) of mostly inactive DXS protein (Fig 4B), presumably because the absence of J20 prevents the targeting of non-functional enzymes to ClpC for eventual degradation by the Clp protease. The dramatic phenotype displayed by single clpb3 and double j20 clpb3 mutant plants (Fig 4C) [66] prevented the reliable quantification of their CLM resistance. In any case, the available data suggests that when the proteolytic degradation of inactive (e.g. aggregated) forms of DXS delivered to the Clp protease by J20 via ClpC is impaired (e.g. in clpr1 and clpc1 mutants), an increase in ClpB3 levels promotes the disaggregation and activation of the enzyme, eventually resulting in higher levels of enzymatically active DXS. When J20 activity is missing, however, inactive DXS forms cannot be properly reactivated via ClpB3 (as deduced from the similar levels of DXS protein but lower proportion of active enzyme found in the double j20 clpc1 mutant compared to the single clpc1 line; Fig 3) or degraded via ClpC (as deduced from the increased levels of inactive DXS protein present in double j20 clpb3 plants compared to the single clpb3 mutant; Fig 4).
Fig 4. J20, Hsp70 and ClpB3 participate in the same pathway for DXS reactivation.
(A) Quantification of DXS protein and transcript levels in 10-day-old WT and mutant plants defective in J20, ClpB3, or both. Representative images of immunoblot analyses with the indicated antibodies and a loading control are also shown. (B) DXS activity levels in the indicated lines represented as total or specific (i.e. relative to the amount of protein) values. Levels in (A) and (B) are represented relative to those in WT plants and correspond to the mean and SEM values of n≥3 independent experiments. Asterisks mark statistically significant differences (t test: p<0.05) relative to WT samples. (C) Representative picture of individual 10-day-old plants of the indicated lines. (D) Immunoprecipitation of ClpB3 with anti-Hsp70 antibodies. Protein extracts from WT or Hsp70.2-defective plants were used for immunoprecipitation (IP) with preimmune serum (αPRE) or an anti-Hsp70 antibody (αHsp70) and further immunoblot (IB) analysis with anti-Hsp70 (as a control) or anti-ClpB3 sera.
As described above, the mechanistic basis for the collaboration between J20, Hsp70, and ClpC chaperones is currently unknown. However, the presence of a Hsp70-binding motif in the amino acid sequence of ClpB3 (S5 Fig) suggests that plastidial Hsp70 isoforms might be able to directly interact with ClpB3 to synergistically activate damaged DXS proteins recognized by the J20 adaptor. In agreement with this possibility, the ClpB3 protein was efficiently immunoprecipitated from WT extracts using an anti-Hsp70 serum (Fig 4D). When a similar experiment was performed with the Arabidopsis hsp70.2 mutant, previously shown to contain lower amounts of plastidial Hsp70 proteins than the WT [58], the level of immunoprecipitated ClpB3 protein was concomitantly decreased (Fig 4D). These results confirm that plastidial Hsp70 isoforms can be found together with ClpB3 in Arabidopsis chloroplasts, providing a mechanistic frame for the observed collaboration between these two families of chaperones in the J20-mediated activation of DXS.
The fate of aggregated DXS proteins recognized by J20 and delivered to Hsp70 likely depends on the relative abundance of particular Hsp100 chaperones
The results described above are consistent with a model involving the participation of ClpB3 and ClpC1 on opposite pathways resulting in either reactivation or degradation, respectively, of inactive DXS proteins recognized by the Hsp70 adaptor J20. Under normal growth conditions, the levels of ClpB3 transcripts and protein are lower than those of ClpC1 (S7 Fig) [42,67]. However, ClpB3 transcript levels have been shown to strongly increase upon exposure to high temperatures [66,68,69] whereas virtually no changes in RNA or protein levels have been detected for ClpC1 or ClpC2 in response to heat or other types of stress, including cold, drought, salt, and oxidative stress [66,70]. The ratio between plastidial ClpB3 and ClpC1 chaperones (and hence the potential capacity to reactivate damaged or/and aggregated DXS polypeptides) could therefore increase when plants are challenged with at least some types of stress (S7 Fig).
DXS-derived isoprenoids such as carotenoids and tocopherols protect plants against oxidative stress, whereas others (including chlorophylls and prenylated quinones) are essential for photosynthesis. Therefore, a decreased production of these isoprenoids (e.g. upon down-regulating DXS activity) is expected to trigger a stress response. We observed that a specific reduction in DXS activity in Arabidopsis WT plants germinated and grown in the presence of CLM caused an increased accumulation of ClpB3 but not ClpC chaperones compared to controls grown in the absence of inhibitor (Fig 5). A similar ClpB3 protein accumulation response was also observed in mutants with a defective MEP pathway (Fig 5). As previously observed [8,14,16–18], the pharmacological or genetic blockage of the pathway also resulted in increased accumulation of DXS protein. Most interestingly, the DXS and ClpB3 accumulation response was detected as soon as 5 hours after reducing the MEP pathway flux by treatment with specific inhibitors (Fig 5). We therefore conclude that stress situations (including those causing a decreased DXS activity and/or MEP pathway flux) could rapidly trigger an increased accumulation of ClpB3, but not ClpC chaperones, likely aimed to promote the reactivation pathway that would keep DXS enzymes in an enzymatically active condition. Furthermore, our data show that ClpB3 levels are more prone to change compared to those of ClpC proteins, suggesting that ClpB3 concentration might be a major factor regulating the fate of inactive DXS polypeptides recognized by J20 and delivered to Hsp70.
Fig 5. Accumulation of ClpB3 but not ClpC chaperones increases when DXS activity or MEP pathway flux decrease.
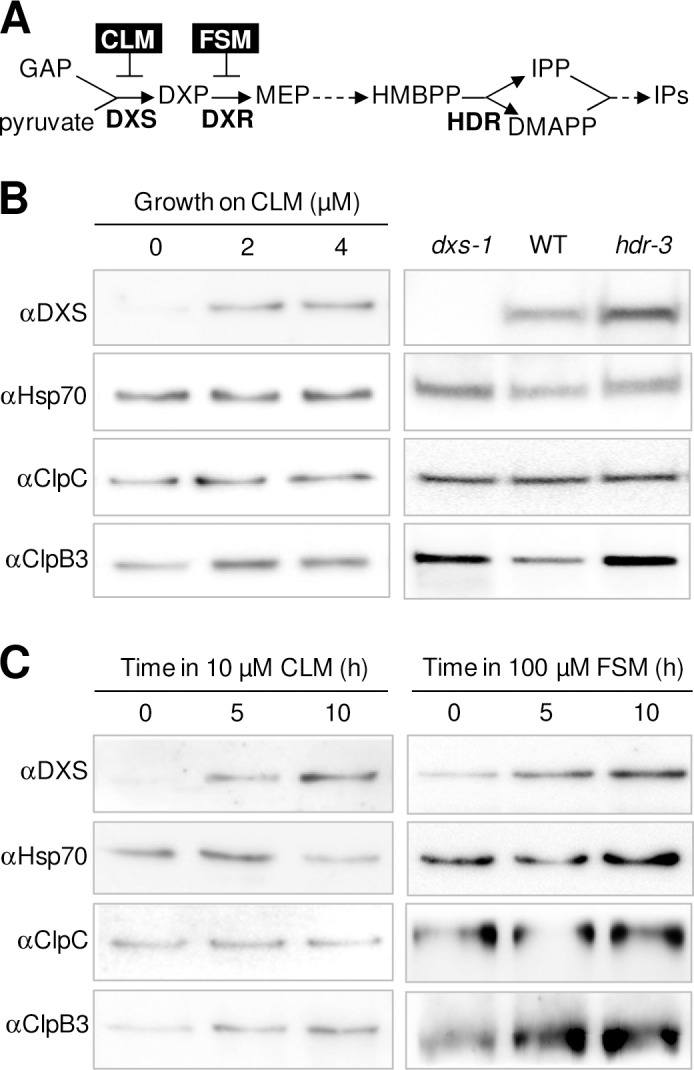
(A) Schematic representation of the MEP pathway. GAP, glyceraldehyde 3-phosphate; DXP, deoxyxylulose 5-phosphate; MEP, methylerythritol 4-phosphate; HMBPP, hydroxymethylbutenyl diphosphate; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; IPs, isoprenoids. Enzymes are represented in bold: DXS, DXP synthase; DXR, DXP reductoisomerase; HDR, HMBPP reductase. Inhibitors are boxed: CLM, clomazone; FSM, fosmidomycin. (B) Immunoblot analyses with the indicated antibodies of WT plants germinated and grown for 18 days on media supplemented with the indicated concentrations of CLM (left panels) or mutants defective in DXS (dxs-1) or HDR (hdr-3) grown for 10 days on media with no inhibitors (right panels). (C) Immunoblot analyses with the indicated antibodies of WT plants germinated and grown for 7 days on media with no inhibitors and then transferred for the indicated times to media supplemented with CLM (left panels) or FSM (right panels).
A model for the role of J20, Hsp70, and Hsp100 chaperones in the regulation of DXS enzyme levels and activity in plastids
Based on the presented data, we propose a model for the regulation of DXS enzyme levels and activity by different types of plastidial chaperones (Fig 6). According to this model, J20 (a plastidial member of the J-domain protein family, also known as J-proteins or Hsp40 co-chaperones) acts as an adaptor providing substrate specificity [19]. In particular, J20 delivers inactive DXS proteins to Hsp70 chaperones that would next act together with particular Hsp100 proteins to either degrade (ClpC1) or reactivate (ClpB3) the enzyme (Fig 6A). J20 might recognize DXS polypeptides that remain unfolded after plastid import or become misfolded by ordinary perturbations and eventually aggregate (S3 and S4 Figs), a process that would render the protein more insoluble and enzymatically inactive. Under normal growth conditions, most DXS proteins remain soluble but some are indeed found associated to the insoluble fraction (Fig 6B). This might be due to the relative low levels of ClpB3 relative to ClpC1 (S7 Fig) [42,67]. In agreement, a further reduction in ClpB3 levels (e.g. in the clpb3 mutant) results in a higher proportion of DXS protein associated to the insoluble fraction (Fig 6B) and hence inactive (Fig 4). By contrast, an enhanced accumulation of ClpB3 takes place in stress situations (Fig 5 and S7 Fig) or when Clp protease function is impaired (Figs 1B and 3A and S2 Fig) [21,22,24,40,65], likely aimed to mitigate general protein folding stress. In the case of DXS, a reduced degradation rate in the clpc1 mutant (Fig 2A) results in increased levels of active (soluble) enzyme (Figs 3 and 6B) likely because a higher accumulation of ClpB3 prevents DXS aggregation. Similar to that proposed in other systems [47–50,61–64], ClpB3 directly interacts with Hsp70 to synergistically perform this role (Fig 4). The observed changes in DXS protein levels and solubility appear to be highly specific, as the next enzyme of the MEP pathway (Fig 5A), deoxyxylulose 5-phosphate reductoisomerase (DXR), was found to be essentially soluble in WT and Hsp100-defective mutants (Fig 6B) and to remain unchanged in J20-defective plants [19].
Fig 6. Model of the molecular pathways determining the fate of DXS in plastids.
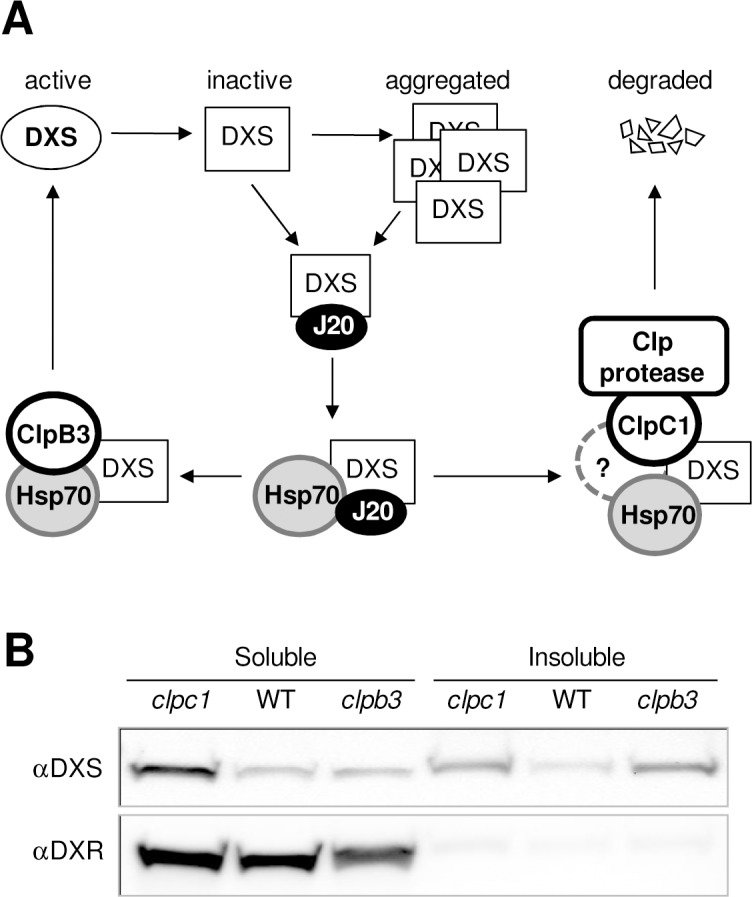
(A) The J-protein J20 facilitates the recognition of damaged (misfolded) or aggregated DXS proteins and their delivery to the Hsp70 chaperone. Then, interaction with the Hsp100 chaperone ClpB3 can synergistically contribute to refold the enzyme back to its active form. Alternatively, Hsp70 can deliver the inactive DXS protein to the Clp protease via ClpC1 for unfolding and degradation. (B) Distribution of DXS and DXR proteins in soluble and insoluble fractions in WT plants and mutants defective in any of the two Hsp100 chaperones that can determine the fate of J20-targeted DXS protein. Representative immunoblots are shown. Ratios of soluble vs. insoluble DXS protein levels (mean±SEM of at least 3 independent experiments) are as follows: WT, 1.84±0.05; clpc1, 1.99±0.26; clpb3, 0.36±0.08.
Interaction with CHIP, a co-chaperone that functions as an E3 ubiquitin ligase, converts Hsp70 from a protein-folding machine into a degradation factor that targets unfolded substrates for degradation by the eukaryotic 26S proteosome [51,71]. Based on genetic evidence (Fig 3) and published results that ClpC and Hsp70 chaperones can be found together in plastid complexes [54,55], we propose that Hsp70 and ClpC chaperones could somehow interact (either directly or by means of unidentified partners) to deliver client proteins like DXS to the Clp catalytic complex. In summary, our model (Fig 6A) proposes that collaboration of Hsp70 with Hsp100 chaperones might deliver inactive (misfolded or/and aggregated) forms of DXS (and potentially many other plastidial proteins recognized by specific J-proteins, the substrate adaptors for Hsp70) to either refolding (via ClpB3) or degradation (via ClpC chaperones). The seedling lethal phenotype of double mutants with no ClpB3 and Clp protease activity [22] illustrates the key relevance of these two seemingly antagonistic pathways for plant life. We speculate that taking a specific pathway (i.e. deciding whether to repair or degrade the protein) might depend on the relative abundance of these Hsp100 partners, particularly as a consequence of changes in ClpB3 levels. The main reason behind the existence of such sophisticated and expensive pathways for the regulation of DXS levels and activity is likely to be the major role demonstrated for this enzyme in the control of the MEP pathway flux [10,11,13]. Future work should next determine how the collaboration of different sets of plastidial chaperone types, and hence the fate of the client protein, is specifically regulated.
Materials and Methods
Plant material, treatments and constructs
Arabidopsis thaliana mutant lines used here are indicated in S1 Table (all in the Columbia background). Sibling lines expressing 35S:DXS-GFP in WT and j20 backgrounds were previously generated [19]. Seeds were surface-sterilized and germinated on solid Murashige and Skoog (MS) medium supplemented with 1% sucrose. Plants were grown under long day conditions as described [19]. For cycloheximide experiments, seeds were germinated on top of a sterile disc of synthetic fabric (SefarNitex 03-100/44). At day 7, the disc with the seedlings was transferred to fresh medium supplemented with 100 μM cycloheximide and samples were collected at different times afterwards (up to 12h) for immunoblot analysis. Inhibition of protein synthesis with cycloheximide had no visual effect on treated seedlings at the times used for the experiment (S8 Fig). Treatments with MEP pathway inhibitors were performed by transferring discs with 7-day-old seedlings to fresh medium supplemented with 10 μM clomazone (CLM) or 100 μM fosmidomycin (FSM). CLM resistance was estimated by quantifying chlorophyll levels in the presence of increasing concentrations of the inhibitor as described [60].
For transient expression and co-immunoprecipitation assays, an Arabidopsis full-length cDNA encoding ClpC1 without the stop codon was PCR-amplified, cloned into the pDONOR207 vector (Invitrogen), and subcloned into the Gateway vector pGWB417 to be expressed under the 35S promoter with a C-terminal MYC epitope (35S:ClpC1-MYC construct). A 35S:DXS-GFP construct was available in the lab [19]. Transient expression of these constructs was carried out by agroinfiltration of Nicotiana benthamiana leaves using the Agrobacterium GV3101 strain. Samples for immunoprecipitation were collected after 3 days.
Protein extraction, immunoprecipitation and detection
Protein extracts were obtained from whole plants and used for immunoprecipitation assays or/and immunoblot analysis as described [19]. For the separation of soluble and insoluble (with protein aggregates) fractions, native protein extracts were obtained in a buffer containing 100 mM Tris-HCl pH7.9, 10 mM MgCl2, 1% (v/v) glycerol, and 20 μl/ml protease inhibitor cocktail (Sigma). After centrifugation for 10 min at 10.000 xg, the supernatant was collected as the soluble fraction. The pellet was washed with fresh buffer and centrifuged again. The obtained pellet fraction was then resuspended in denaturing TKMES buffer [19] and centrifuged again to collect the supernatant as the insoluble fraction. In all cases, protein concentration was determined using the Bio-Rad protein assay. For immunoblot assays, antibodies raised against DXS and DXR [19], GFP (Life Technologies), MYC (Millipore), and chloroplast Hsp70, ClpC, and ClpB proteins (Agrisera) were diluted 1:500 for DXS, 1:7,000 for DXR, 1:1,000 for GFP and MYC, 1:6,000 for Hsp70, 1:2,000 for ClpC, and 1:3,000 for ClpB. The total amount of protein loaded per lane was calculated for each particular antibody to remain in the linear range (S9 Fig). Chemiluminescent signals were visualized using a LAS-4000 (Fujifilm) image analyzer and quantified with Quantity One (Bio-Rad). Student´s t test was used to assess statistical significance of quantified differences.
Protease accessibility assays
For protease accessibility assays, protein extracts from 10-day-old WT and j20 seedlings containing 30 μg of total protein were incubated for 5 min at 37°C with increasing concentrations of Proteinase K (Invitrogen). After stopping the reaction with SDS-PAGE loading buffer, extracts were used for immunoblot analysis.
DXS activity assays
DXS enzyme activity measurements were carried out as described [19,72]. Specific activity was calculated by dividing the total activity measured in extracts with the amount of DXS protein found in the corresponding sample.
RNA isolation and qPCR analysis
RNA isolation, cDNA synthesis, and qPCR experiments were performed as described [19] using the APT1 (At1g27450) gene for normalization.
Prediction of aggregation propensity
The Aggrescan3D algorithm [73] was used to analyze protein aggregation propensity. Predictions were performed in static mode using a distance of aggregation analysis of 10 Å. The Arabidopsis DXS structure was modelled using Swiss-Model [74] on top of the 2.40 Å resolution E. coli DXS structure with PDB code 2O1S. Residues 72 to 707 of the Arabidopsis DXS monomer, sharing a sequence identity of 41.08% with the E. coli protein, were structurally aligned and modelled. The interface of the generated homodimer was evaluated with PDBePISA (http://www.ebi.ac.uk/pdbe/pisa/) rendering an area of 8892 Å and a predicted dissociation ΔG for the dimer of 51.2 kcal/mol (close to those of the template E. coli crystal structure, which exhibits an interface of 7970 Å and a dissociation ΔG of 59.1 kcal/mol).
Supporting Information
(PDF)
LC, loading control.
(PDF)
The protein surface is colored according to Aggrescan3D score in gradient from red (high predicted aggregation propensity) to white (negligible impact on protein aggregation) to blue (high predicted solubility). The figure was generated with PyMOL using the aggregation propensities encoded in the temperature factor column of the A3D.pdb output.
(PDF)
(A) Confocal microscopy analysis of GFP (green) and chlorophyll (red) fluorescence distribution within the chloroplast. Representative images obtained with the same confocal parameters from siblings harboring the same T-DNA insertion with the 35S:DXS-GFP construct in a wild-type (WT) or J20-defective (j20) background are shown. (B) Analysis of DXS protein abundance by immunoblot analysis of protein extracts from WT and j20 plants incubated with the indicated concentrations (μg/ml) of proteinase K. Representative blots and quantitative data corresponding to the mean and SE values of n = 4 independent experiments are shown. Asterisks mark statistically significant differences (t test: p<0.05) relative to WT samples.
(PDF)
Alignment was performed using Clustal Omega (www.ebi.ac.uk/Tools/msa/clustalo) with protein sequences of Arabidopsis thaliana ClpD (At5g51070), ClpC1 (At5g50920), ClpC2 (At3g48870), and ClpB3 (At5g15450), Bacillus subtilis ClpC (AAA19233), and Escherichia coli ClpB (EDV64786). The generated alignment was then edited with Bioedit (http://www.mbio.ncsu.edu/bioedit/page2.html). The domain responsible for the interaction of E. coli ClpB with Hsp70 is boxed in blue. The tripeptide loop shown to be required for interaction with ClpP subunits of the Clp protease complex is boxed in red.
(PDF)
Representative pictures of plants of WT, single mutant, and double mutant lines germinated and grown for 10 days in the presence of the indicated concentrations of CLM are shown.
(PDF)
Data were obtained from the Arabidopsis eFP browser at www.bar.utoront.ca and correspond to the gene expression map of Arabidopsis development (A) and abiotic stress treatments (B).
(PDF)
WT plants were grown on top of a sterile disc of synthetic fabric for 7 days. After transferring the disc with the seedlings to fresh medium supplemented with 100 μM cycloheximide, pictures of the same individual were taken at the indicated times.
(PDF)
(PDF)
(PDF)
Acknowledgments
We are grateful to H.M. Li, W. Sakamoto, L. Zhang, and P. Hatzopoulos for providing Arabidopsis lines and T. Nakagawa for the pGWB417 vector. Technical support from M. Rosa Rodríguez-Goberna and members of the CRAG services is greatly appreciated.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by grants from the European Commission FP7 (TiMet and CarotenActors), Ministerio de Economia y Competitividad (BIO2011-23680 and BIO2014-59092-P), Programa iberoamericano de ciencia y tecnología para el desarrollo. (Ibercarot), and Generalitat de Catalunya (2014SGR-1434 and XRB) to MRC, and PhD fellowships from the Mexican Consejo Nacional de Ciencia y Tecnologia (421688 and beca-complemento) to EL. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Flores-Perez U, Jarvis P (2013) Molecular chaperone involvement in chloroplast protein import. Biochim Biophys Acta 1833: 332–340. 10.1016/j.bbamcr.2012.03.019 [DOI] [PubMed] [Google Scholar]
- 2.Trosch R, Muhlhaus T, Schroda M, Willmund F (2015) ATP-dependent molecular chaperones in plastids—More complex than expected. Biochim Biophys Acta 10.1016/j.bbabio.2015.01.002 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 3.van Wijk KJ (2015) Protein maturation and proteolysis in plant plastids, mitochondria, and peroxisomes. Annu Rev Plant Biol 66: 75–111. 10.1146/annurev-arplant-043014-115547 [DOI] [PubMed] [Google Scholar]
- 4.Kato Y, Sakamoto W (2010) New insights into the types and function of proteases in plastids. Int Rev Cell Mol Biol 280: 185–218. 10.1016/S1937-6448(10)80004-8 [DOI] [PubMed] [Google Scholar]
- 5.Rodríguez-Concepción M, Boronat A (2002) Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol 130: 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouvier F, Rahier A, Camara B (2005) Biogenesis, molecular regulation and function of plant isoprenoids. Prog Lipid Res 44: 357–429. [DOI] [PubMed] [Google Scholar]
- 7.Joyard J, Ferro M, Masselon C, Seigneurin-Berny D, Salvi D, et al. (2009) Chloroplast proteomics and the compartmentation of plastidial isoprenoid biosynthetic pathways. Mol Plant 2: 1154–1180. 10.1093/mp/ssp088 [DOI] [PubMed] [Google Scholar]
- 8.Guevara-Garcia A, San Roman C, Arroyo A, Cortes ME, Gutierrez-Nava ML, et al. (2005) Characterization of the Arabidopsis clb6 Mutant Illustrates the Importance of Posttranscriptional Regulation of the Methyl-D-Erythritol 4- Phosphate Pathway. Plant Cell 17: 628–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laule O, Furholz A, Chang HS, Zhu T, Wang X, et al. (2003) Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci U S A 100: 6866–6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodríguez-Concepción M, Boronat A (2015) Breaking new ground in the regulation of the early steps of plant isoprenoid biosynthesis. Curr Opin Plant Biol 25: 17–22. 10.1016/j.pbi.2015.04.001 [DOI] [PubMed] [Google Scholar]
- 11.Hemmerlin A (2013) Post-translational events and modifications regulating plant enzymes involved in isoprenoid precursor biosynthesis. Plant Sci 203–204: 41–54. 10.1016/j.plantsci.2012.12.008 [DOI] [PubMed] [Google Scholar]
- 12.Banerjee A, Sharkey TD (2014) Methylerythritol 4-phosphate (MEP) pathway metabolic regulation. Nat Prod Rep 31: 1043–1055. 10.1039/c3np70124g [DOI] [PubMed] [Google Scholar]
- 13.Wright LP, Rohwer JM, Ghirardo A, Hammerbacher A, Ortiz-Alcaide M, et al. (2014) Deoxyxylulose 5-Phosphate Synthase Controls Flux through the Methylerythritol 4-Phosphate Pathway in Arabidopsis. Plant Physiol 165: 1488–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghirardo A, Wright LP, Bi Z, Rosenkranz M, Pulido P, et al. (2014) Metabolic flux analysis of plastidic isoprenoid biosynthesis in poplar leaves emitting and nonemitting isoprene. Plant Physiol 165: 37–51. 10.1104/pp.114.236018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banerjee A, Wu Y, Banerjee R, Li Y, Yan H, et al. (2013) Feedback inhibition of deoxy-D-xylulose-5-phosphate synthase regulates the methylerythritol 4-phosphate pathway. J Biol Chem 288: 16926–16936. 10.1074/jbc.M113.464636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez-Villalon A, Gas E, Rodriguez-Concepcion M (2009) Phytoene synthase activity controls the biosynthesis of carotenoids and the supply of their metabolic precursors in dark-grown Arabidopsis seedlings. Plant J 60: 424–435. 10.1111/j.1365-313X.2009.03966.x [DOI] [PubMed] [Google Scholar]
- 17.Han M, Heppel SC, Su T, Bogs J, Zu Y, et al. (2013) Enzyme inhibitor studies reveal complex control of methyl-D-erythritol 4-phosphate (MEP) pathway enzyme expression in Catharanthus roseus. PLoS One 8: e62467 10.1371/journal.pone.0062467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pokhilko A, Bou-Torrent J, Pulido P, Rodriguez-Concepcion M, Ebenhoh O (2015) Mathematical modelling of the diurnal regulation of the MEP pathway in Arabidopsis. New Phytol 206: 1075–1085. 10.1111/nph.13258 [DOI] [PubMed] [Google Scholar]
- 19.Pulido P, Toledo-Ortiz G, Phillips MA, Wright LP, Rodriguez-Concepcion M (2013) Arabidopsis J-Protein J20 Delivers the First Enzyme of the Plastidial Isoprenoid Pathway to Protein Quality Control. Plant Cell 25: 4183–4194. 10.1105/tpc.113.113001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flores-Pérez U, Sauret-Güeto S, Gas E, Jarvis P, Rodríguez-Concepción M (2008) A Mutant Impaired in the Production of Plastome-Encoded Proteins Uncovers a Mechanism for the Homeostasis of Isoprenoid Biosynthetic Enzymes in Arabidopsis Plastids. Plant Cell 20: 1303–1315. 10.1105/tpc.108.058768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J, Rudella A, Ramirez Rodriguez V, Zybailov B, Olinares PD, et al. (2009) Subunits of the plastid ClpPR protease complex have differential contributions to embryogenesis, plastid biogenesis, and plant development in Arabidopsis. Plant Cell 21: 1669–1692. 10.1105/tpc.108.063784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zybailov B, Friso G, Kim J, Rudella A, Rodriguez VR, et al. (2009) Large scale comparative proteomics of a chloroplast Clp protease mutant reveals folding stress, altered protein homeostasis, and feedback regulation of metabolism. Mol Cell Proteomics 8: 1789–1810. 10.1074/mcp.M900104-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudella A, Friso G, Alonso JM, Ecker JR, van Wijk KJ (2006) Downregulation of ClpR2 leads to reduced accumulation of the ClpPRS protease complex and defects in chloroplast biogenesis in Arabidopsis. Plant Cell 18: 1704–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J, Olinares PD, Oh SH, Ghisaura S, Poliakov A, et al. (2013) Modified Clp protease complex in the ClpP3 null mutant and consequences for chloroplast development and function in Arabidopsis. Plant Physiol 162: 157–179. 10.1104/pp.113.215699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rigas S, Daras G, Tsitsekian D, Hatzopoulos P (2012) The multifaceted role of Lon proteolysis in seedling establishment and maintenance of plant organelle function: living from protein destruction. Physiol Plant 145: 215–223. 10.1111/j.1399-3054.2011.01537.x [DOI] [PubMed] [Google Scholar]
- 26.Ostersetzer O, Kato Y, Adam Z, Sakamoto W (2007) Multiple intracellular locations of Lon protease in Arabidopsis: evidence for the localization of AtLon4 to chloroplasts. Plant Cell Physiol 48: 881–885. [DOI] [PubMed] [Google Scholar]
- 27.Schuhmann H, Adamska I (2012) Deg proteases and their role in protein quality control and processing in different subcellular compartments of the plant cell. Physiol Plant 145: 224–234. 10.1111/j.1399-3054.2011.01533.x [DOI] [PubMed] [Google Scholar]
- 28.Haussuhl K, Andersson B, Adamska I (2001) A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. Embo J 20: 713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun X, Peng L, Guo J, Chi W, Ma J, et al. (2007) Formation of DEG5 and DEG8 complexes and their involvement in the degradation of photodamaged photosystem II reaction center D1 protein in Arabidopsis. Plant Cell 19: 1347–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner R, Aigner H, Funk C (2012) FtsH proteases located in the plant chloroplast. Physiol Plant 145: 203–214. 10.1111/j.1399-3054.2011.01548.x [DOI] [PubMed] [Google Scholar]
- 31.Sakamoto W, Zaltsman A, Adam Z, Takahashi Y (2003) Coordinated regulation and complex formation of yellow variegated1 and yellow variegated2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15: 2843–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu F, Park S, Rodermel SR (2004) The Arabidopsis FtsH metalloprotease gene family: interchangeability of subunits in chloroplast oligomeric complexes. Plant J 37: 864–876. [DOI] [PubMed] [Google Scholar]
- 33.Rodrigues RA, Silva-Filho MC, Cline K (2011) FtsH2 and FtsH5: two homologous subunits use different integration mechanisms leading to the same thylakoid multimeric complex. Plant J 65: 600–609. 10.1111/j.1365-313X.2010.04448.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koussevitzky S, Stanne TM, Peto CA, Giap T, Sjogren LL, et al. (2007) An Arabidopsis thaliana virescent mutant reveals a role for ClpR1 in plastid development. Plant Mol Biol 63: 85–96. [DOI] [PubMed] [Google Scholar]
- 35.Kirstein J, Moliere N, Dougan DA, Turgay K (2009) Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat Rev Microbiol 7: 589–599. 10.1038/nrmicro2185 [DOI] [PubMed] [Google Scholar]
- 36.Levchenko I, Seidel M, Sauer RT, Baker TA (2000) A specificity-enhancing factor for the ClpXP degradation machine. Science 289: 2354–2356. [DOI] [PubMed] [Google Scholar]
- 37.Dougan DA, Reid BG, Horwich AL, Bukau B (2002) ClpS, a substrate modulator of the ClpAP machine. Mol Cell 9: 673–683. [DOI] [PubMed] [Google Scholar]
- 38.Nishimura K, van Wijk KJ (2014) Organization, function and substrates of the essential Clp protease system in plastids. Biochim Biophys Acta 10.1016/j.bbabio.2014.11.012 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 39.Clarke AK (2012) The chloroplast ATP-dependent Clp protease in vascular plants—new dimensions and future challenges. Physiol Plant 145: 235–244. 10.1111/j.1399-3054.2011.01541.x [DOI] [PubMed] [Google Scholar]
- 40.Nishimura K, Asakura Y, Friso G, Kim J, Oh SH, et al. (2013) ClpS1 is a conserved substrate selector for the chloroplast Clp protease system in Arabidopsis. Plant Cell 25: 2276–2301. 10.1105/tpc.113.112557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishimura K, Apitz J, Friso G, Kim J, Ponnala L, et al. (2015) Discovery of a Unique Clp Component, ClpF, in Chloroplasts: A Proposed Binary ClpF-ClpS1 Adaptor Complex Functions in Substrate Recognition and Delivery. Plant Cell 27:2677–2691. 10.1105/tpc.15.00574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sjogren LL, Tanabe N, Lymperopoulos P, Khan NZ, Rodermel SR, et al. (2014) Quantitative analysis of the chloroplast molecular chaperone ClpC/Hsp93 in Arabidopsis reveals new insights into its localization, interaction with the Clp proteolytic core, and functional importance. J Biol Chem 289: 11318–11330. 10.1074/jbc.M113.534552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ninnis RL, Spall SK, Talbo GH, Truscott KN, Dougan DA (2009) Modification of PATase by L/F-transferase generates a ClpS-dependent N-end rule substrate in Escherichia coli. Embo J 28: 1732–1744. 10.1038/emboj.2009.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh A, Grover A (2010) Plant Hsp100/ClpB-like proteins: poorly-analyzed cousins of yeast ClpB machine. Plant Mol Biol 74: 395–404. 10.1007/s11103-010-9682-8 [DOI] [PubMed] [Google Scholar]
- 45.Kim YI, Levchenko I, Fraczkowska K, Woodruff RV, Sauer RT, et al. (2001) Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nat Struct Biol 8: 230–233. [DOI] [PubMed] [Google Scholar]
- 46.Doyle SM, Hoskins JR, Wickner S (2007) Collaboration between the ClpB AAA+ remodeling protein and the DnaK chaperone system. Proc Natl Acad Sci U S A 104: 11138–11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zolkiewski M (1999) ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. A novel multi-chaperone system from Escherichia coli. J Biol Chem 274: 28083–28086. [DOI] [PubMed] [Google Scholar]
- 48.Haslberger T, Weibezahn J, Zahn R, Lee S, Tsai FT, et al. (2007) M domains couple the ClpB threading motor with the DnaK chaperone activity. Mol Cell 25: 247–260. [DOI] [PubMed] [Google Scholar]
- 49.Kampinga HH, Craig EA (2010) The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 11: 579–592. 10.1038/nrm2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miot M, Reidy M, Doyle SM, Hoskins JR, Johnston DM, et al. (2011) Species-specific collaboration of heat shock proteins (Hsp) 70 and 100 in thermotolerance and protein disaggregation. Proc Natl Acad Sci U S A 108: 6915–6920. 10.1073/pnas.1102828108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee S, Lee DW, Lee Y, Mayer U, Stierhof YD, et al. (2009) Heat shock protein cognate 70–4 and an E3 ubiquitin ligase, CHIP, mediate plastid-destined precursor degradation through the ubiquitin-26S proteasome system in Arabidopsis. Plant Cell 21: 3984–4001. 10.1105/tpc.109.071548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hayes SA, Dice JF (1996) Roles of molecular chaperones in protein degradation. J Cell Biol 132: 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nordhues A, Miller SM, Muhlhaus T, Schroda M (2010) New insights into the roles of molecular chaperones in Chlamydomonas and Volvox. Int Rev Cell Mol Biol 285: 75–113. 10.1016/B978-0-12-381047-2.00002-5 [DOI] [PubMed] [Google Scholar]
- 54.Su PH, Li HM (2010) Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22: 1516–1531. 10.1105/tpc.109.071415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi LX, Theg SM (2010) A stromal heat shock protein 70 system functions in protein import into chloroplasts in the moss Physcomitrella patens. Plant Cell 22: 205–220. 10.1105/tpc.109.071464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flores-Pérez Ú, Bédard J, Tanabe N, Lymperopoulos P, Clarke AK, Jarvis P (2015) Functional analysis of the Hsp93/ClpC chaperone at the chloroplast envelope. Plant Physiol pii: pp.01538.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Inoue H, Li M, Schnell DJ (2013) An essential role for chloroplast heat shock protein 90 (Hsp90C) in protein import into chloroplasts. Proc Natl Acad Sci U S A 110: 3173–3178. 10.1073/pnas.1219229110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Su PH, Li HM (2008) Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermotolerance of germinating seeds. Plant Physiol 146: 1231–1241. 10.1104/pp.107.114496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matsue Y, Mizuno H, Tomita T, Asami T, Nishiyama M, et al. (2010) The herbicide ketoclomazone inhibits 1-deoxy-D-xylulose 5-phosphate synthase in the 2-C-methyl-D-erythritol 4-phosphate pathway and shows antibacterial activity against Haemophilus influenzae. J Antibiot (Tokyo) 63: 583–588. [DOI] [PubMed] [Google Scholar]
- 60.Perello C, Rodriguez-Concepcion M, Pulido P (2014) Quantification of plant resistance to isoprenoid biosynthesis inhibitors. Methods Mol Biol 1153: 273–283. 10.1007/978-1-4939-0606-2_20 [DOI] [PubMed] [Google Scholar]
- 61.Glover JR, Lindquist S (1998) Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94: 73–82. [DOI] [PubMed] [Google Scholar]
- 62.Goloubinoff P, Mogk A, Zvi AP, Tomoyasu T, Bukau B (1999) Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc Natl Acad Sci U S A 96: 13732–13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seyffer F, Kummer E, Oguchi Y, Winkler J, Kumar M, et al. (2012) Hsp70 proteins bind Hsp100 regulatory M domains to activate AAA+ disaggregase at aggregate surfaces. Nat Struct Mol Biol 19: 1347–1355. 10.1038/nsmb.2442 [DOI] [PubMed] [Google Scholar]
- 64.Rosenzweig R, Moradi S, Zarrine-Afsar A, Glover JR, Kay LE (2013) Unraveling the mechanism of protein disaggregation through a ClpB-DnaK interaction. Science 339: 1080–1083. 10.1126/science.1233066 [DOI] [PubMed] [Google Scholar]
- 65.Stanne TM, Sjogren LL, Koussevitzky S, Clarke AK (2009) Identification of new protein substrates for the chloroplast ATP-dependent Clp protease supports its constitutive role in Arabidopsis. Biochem J 417: 257–268. 10.1042/BJ20081146 [DOI] [PubMed] [Google Scholar]
- 66.Lee U, Rioflorido I, Hong SW, Larkindale J, Waters ER, et al. (2007) The Arabidopsis ClpB/Hsp100 family of proteins: chaperones for stress and chloroplast development. Plant J 49: 115–127. [DOI] [PubMed] [Google Scholar]
- 67.Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, et al. (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3: e1994 10.1371/journal.pone.0001994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Myouga F, Motohashi R, Kuromori T, Nagata N, Shinozaki K (2006) An Arabidopsis chloroplast-targeted Hsp101 homologue, APG6, has an essential role in chloroplast development as well as heat-stress response. Plant J 48: 249–260. [DOI] [PubMed] [Google Scholar]
- 69.Rasmussen S, Barah P, Suarez-Rodriguez MC, Bressendorff S, Friis P, et al. (2013) Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiol 161: 1783–1794. 10.1104/pp.112.210773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng B, Halperin T, Hruskova-Heidingsfeldova O, Adam Z, Clarke AK (2002) Characterization of Chloroplast Clp proteins in Arabidopsis: Localization, tissue specificity and stress responses. Physiol Plant 114: 92–101. [DOI] [PubMed] [Google Scholar]
- 71.Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM (2001) The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol 3: 100–105. [DOI] [PubMed] [Google Scholar]
- 72.Wright LP, Phillips MA (2014) Measuring the activity of 1-deoxy-D-xylulose 5-phosphate synthase, the first enzyme in the MEP pathway, in plant extracts. Methods Mol Biol 1153: 9–20. 10.1007/978-1-4939-0606-2_2 [DOI] [PubMed] [Google Scholar]
- 73.Zambrano R, Jamroz M, Szczasiuk A, Pujols J, Kmiecik S, Ventura S (2015) AGGRESCAN3D (A3D): server for prediction of aggregation properties of protein structures. Nucleic Acids Res 43: W306–313. 10.1093/nar/gkv359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bordoli L, Schwede T (2012) Automated protein structure modeling with SWISS-MODEL Workspace and the Protein Model Portal. Methods Mol Biol 857: 107–136. 10.1007/978-1-61779-588-6_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
LC, loading control.
(PDF)
The protein surface is colored according to Aggrescan3D score in gradient from red (high predicted aggregation propensity) to white (negligible impact on protein aggregation) to blue (high predicted solubility). The figure was generated with PyMOL using the aggregation propensities encoded in the temperature factor column of the A3D.pdb output.
(PDF)
(A) Confocal microscopy analysis of GFP (green) and chlorophyll (red) fluorescence distribution within the chloroplast. Representative images obtained with the same confocal parameters from siblings harboring the same T-DNA insertion with the 35S:DXS-GFP construct in a wild-type (WT) or J20-defective (j20) background are shown. (B) Analysis of DXS protein abundance by immunoblot analysis of protein extracts from WT and j20 plants incubated with the indicated concentrations (μg/ml) of proteinase K. Representative blots and quantitative data corresponding to the mean and SE values of n = 4 independent experiments are shown. Asterisks mark statistically significant differences (t test: p<0.05) relative to WT samples.
(PDF)
Alignment was performed using Clustal Omega (www.ebi.ac.uk/Tools/msa/clustalo) with protein sequences of Arabidopsis thaliana ClpD (At5g51070), ClpC1 (At5g50920), ClpC2 (At3g48870), and ClpB3 (At5g15450), Bacillus subtilis ClpC (AAA19233), and Escherichia coli ClpB (EDV64786). The generated alignment was then edited with Bioedit (http://www.mbio.ncsu.edu/bioedit/page2.html). The domain responsible for the interaction of E. coli ClpB with Hsp70 is boxed in blue. The tripeptide loop shown to be required for interaction with ClpP subunits of the Clp protease complex is boxed in red.
(PDF)
Representative pictures of plants of WT, single mutant, and double mutant lines germinated and grown for 10 days in the presence of the indicated concentrations of CLM are shown.
(PDF)
Data were obtained from the Arabidopsis eFP browser at www.bar.utoront.ca and correspond to the gene expression map of Arabidopsis development (A) and abiotic stress treatments (B).
(PDF)
WT plants were grown on top of a sterile disc of synthetic fabric for 7 days. After transferring the disc with the seedlings to fresh medium supplemented with 100 μM cycloheximide, pictures of the same individual were taken at the indicated times.
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.