Light interacts via photosynthesis with the carbon dioxide signaling and controls gene expression crucial to oceanic production in a marine diatom.
Abstract
Our previous study showed that three CO2/cAMP-responsive elements (CCRE) CCRE1, CCRE2, and CCRE3 in the promoter of the chloroplastic β-carbonic anhydrase 1 gene in the marine diatom Phaeodactylum tricornutum (Pptca1) were critical for the cAMP-mediated transcriptional response to ambient CO2 concentration. Pptca1 was activated under CO2 limitation, but the absence of light partially disabled this low-CO2-triggered transcriptional activation. This suppression effect disappeared when CCRE2 or two of three CCREs were replaced with a NotI restriction site, strongly suggesting that light signal cross-talks with CO2 on the cAMP-signal transduction pathway that targets CCREs. The paralogous chloroplastic carbonic anhydrase gene, ptca2 was also CO2/cAMP-responsive. The upstream truncation assay of the ptca2 promoter (Pptca2) revealed a short sequence of −367 to −333 relative to the transcription-start site to be a critical regulatory region for the CO2 and light responses. This core-regulatory region comprises one CCRE1 and two CCRE2 sequences. Further detailed analysis of Pptca2 clearly indicates that two CCRE2s are the cis-element governing the CO2/light response of Pptca2. The transcriptional activation of two Pptcas in CO2 limitation was evident under illumination with a photosynthetically active light wavelength, and an artificial electron acceptor from the reduction side of PSI efficiently inhibited Pptcas activation, while neither inhibition of the linear electron transport from PSII to PSI nor inhibition of ATP synthesis showed an effect on the promoter activity, strongly suggesting a specific involvement of the redox level of the stromal side of the PSI in the CO2/light cross talk.
Marine diatoms are one of the most successful groups of photoautotrophic organisms in the ocean and are responsible for up to one-fifth of annual global carbon fixation, playing also an important role in global elemental cycles (Tréguer et al., 1995; Falkowski et al., 2000). The acquisition of CO2 in diatoms is sustained by a CO2-concentrating mechanism (CCM), which works to supply high concentration of CO2 to the carbon-fixation enzyme, Rubisco in diatoms. The CCM helps diatoms to overcome the problems of low CO2 concentration in seawater and a relatively high Km[CO2] value of diatom Rubisco (Badger et al., 1998). The CCM in diatoms is composed of an active uptake system of both CO2 and HCO3− from the surrounding seawater (Burkhardt et al., 2001; Rost et al., 2003; Trimborn et al., 2008; Hopkinson et al., 2011; Matsuda et al., 2011), and of the system to mobilize the accumulated dissolved-inorganic carbon (DIC) to Rubisco (Matsuda et al., 2011; Hopkinson et al., 2013). The latter process is facilitated by an enzyme, carbonic anhydrase (CA; Hopkinson et al., 2011; Matsuda et al., 2011; Samukawa et al., 2014) and may also involve, in some species of marine diatoms, a C4-like biochemical conversion of DIC into organic acids (Reinfelder, 2011). Only a few molecular data are so far available for the diatom CCM. A mammalian-type solute-carrier protein family 4 (SLC4) functions as a Na+-dependent plasma membrane type HCO3− transporter in the raphid pennate diatom, Phaeodactylum tricornutum (Nakajima et al., 2013). Many other putative transporter genes, which apparently belong to subclasses of SLC4 and SLC26, were also found in the genomes of P. tricornutum and the multipolar centric diatom, Thalassiosira pseudonana, suggesting that these genes share the common origin with mammals and heterokonts (Nakajima et al., 2013).
Numerous CAs, a critical CCM component, were also found in the genomes of P. tricornutum and T. pseudonana and most of them have been localized (Harada et al., 2005; Tanaka et al., 2005; Tachibana et al., 2011; Samukawa et al., 2014). Interestingly, there are significant differences in the location of CAs between these two species. That is, there are pyrenoidal CAs but no cytosolic nor external CA in P. tricornutum (Tachibana et al., 2011), while, in contrast, there are cytosolic and external CAs but no pyrenoidal CA in T. pseudonana (Samukawa et al., 2014). These data suggest an occurrence of a significant diversity in the mechanism of the diatom CCMs (Samukawa et al., 2014).
Many components of the CCM are suppressed under elevated CO2 conditions but induced (or derepressed) at the transcriptional level when cells of diatoms, cyanobacteria, and green algae are exposed to a limited CO2 around or below the atmospheric level within a few hours (Kaplan et al., 1980; Miller et al., 1990; Colman and Rotatore, 1995; Matsuda and Colman, 1995; Johnston and Raven, 1996; Matsuda et al., 2001; Kucho et al., 2003; Vance and Spalding, 2005; Ma et al., 2011). This indicates that algal cells can perceive CO2 concentration as a signal and quickly acclimate to changing CO2 concentrations. In the cyanobacterium Synechocystis sp. PCC6803, the cytoplasmic-membrane protein (cmp) ABCD operon, which encodes a Bicarbonate Transporter 1, is induced upon interaction of Cytoplasmic-membrane protein Regulator (CmpR) to the promoter under low CO2 conditions (Omata et al., 2001; Nishimura et al., 2008) and this interaction on the promoter is apparently stimulated by 2-phosphoglycolate (2-PG), an initial photorespiratory metabolite (Nishimura et al., 2008). In contrast, the other CO2-responsive operons (e.g. sodium bicarbonate transporter (sbt)A/sbtB, NAD(P)H dehydrogenase (ndh)F3/ndhD3/CO2 hydration protein Y/open reading frame 133, and modified NDH-1 (mnh)D1/mnhD2) are repressed by CO2-concentrating mechanism Regulator/NAD(P)H dehydrogenase Regulator under high CO2 conditions (Wang et al., 2004; Price et al., 2008). On the other hand, no common mechanism of CO2 response is found in eukaryotic algae. The CO2-response mechanism has been well studied in the green alga, Chlamydomonas reinhardtii using the transcriptional regulation system of a periplasmic CA, Cah1 and the mitochondrial CA (mtCA) genes, both of which are highly responsive to the ambient CO2 concentration (Fujiwara et al., 1990; Eriksson et al., 1998). The Cah1 promoter is regulated positively and negatively by enhancer and silencer regions. The enhancer region is controlled by two critical elements denoted enhancer-element consensus (Kucho et al., 2003), to which a class of basic-ZIP transcription factors, enhancer-element-consensus binding proteins, interact (Yoshioka et al., 2004). A zinc finger protein, CCM1/Cia5, has been found to be involved in the up-regulation of most low-CO2-inducible genes, strongly suggesting that CCM1/Cia5 is a master regulator for the transcriptional response of the Chlamydomonas CCM, but the mechanism of the initial CO2 signal perception is still unknown (Fukuzawa et al., 2001; Xiang et al., 2001).
The high photosynthetic affinity for DIC was induced concomitantly with the capacity to take up DIC upon transferring diatom cells from high CO2 to atmospheric CO2 (Johnston and Raven, 1996; Burkhardt et al., 2001; Matsuda et al., 2001), strongly suggesting that DIC uptake and internal CO2 flow toward Rubisco are strengthened under CO2 limitation in diatoms. It is also pointed out that acclimation to the extreme CO2 limitation (below 2 mL m−3) induces a distinct high-affinity state of the CCM in P. tricornutum (Matsuda et al., 2001). Similarly, in the green alga, C. reinhardtii, cells appear to take different strategies for CO2 acquisition in high CO2, atmospheric level CO2, and subatmospheric level CO2 (Spalding et al., 2002; Vance and Spalding, 2005; Yamano et al., 2010). However, the mechanism to respond differentially to a spectrum of CO2 condition is still unclear in any algal cells. The pyrenoidal β-CAs (PtCA1 and PtCA2) and plasma membrane type HCO3− transporter (PtSLC4-2) in P. tricornutum were induced under atmospheric CO2 at the transcriptional level (Harada and Matsuda, 2005; Nakajima et al., 2013). Three paralogous SLC4 type genes, ptSLC4-1, ptSLC4-2, and ptSLC4-4 were also induced under atmospheric CO2 (Nakajima et al., 2013), suggesting that these CCM factors are under the control of a CO2-signaling mechanism.
The function of the CO2-responsive ptca1 promoter (Pptca1) has been studied in detail (Harada et al., 2006; Ohno et al., 2012). Three cis-elements denoted CO2/cAMP-responsive elements (CCRE1, CCRE2, and CCRE3), which are tandemly aligned between −86 and −42 relative to the transcription-start site, are critical motifs for Pptca1 to respond to changes in CO2 concentration via the second messenger, cAMP (Ohno et al., 2012). CCRE1, CCRE2, and CCRE3 are an analogous sequence (TGACGT/C) and located in an inverted direction to each other with 12 to 15 bps of spacer sequences (Ohno et al., 2012). CCRE2 is the core element, which works with either CCRE1 or CCRE3, suggesting that two elements possessing the complementary sequence may cooperate to recruit specific transcription factors (Ohno et al., 2012). Indeed, a basic zipper (bZIP) protein PtbZIP11, which resembles the mammalian cAMP-responsive element-binding protein/activation transcription factor, is detected as a Trans-acting factor, which specifically binds to CCREs (Ohno et al., 2012). However, the upstream signal transduction mechanism of the diatom CO2-response is still unclear, and factors controlling the Pptca1 promoter activity await further investigations.
In photosynthetic organisms, CO2 signals often cross talk with light signals that may regulate multiple stages of gene expression. It has been reported that CCMs are regulated by light and CO2 with a variety of sensitivity depending on organisms. In cyanobacteria, light is an absolute requisite for the induction of the CCM under CO2 limitation (Omata et al., 2001; Nishimura et al., 2008). As described above, 2-PG seems to be a cofactor for the inducer, CmpR, strongly suggesting the participation of the photorespiratory metabolism for the transcriptional regulation of the CCM factors (Nishimura et al., 2008). Indeed, transcriptions of cyanobacterial CCM factors cmpA, ndhF3, and sbtA were found to be enhanced by strong light (Hihara et al., 2001; McGinn et al., 2003, 2004; Woodger et al., 2003). In contrast, regulation of CCMs in green algae does not necessarily depend on light. In the green alga, Chlorella ellipsoidea, the CCM was fully derepressed in low CO2 in the dark (Matsuda and Colman, 1995), while in C. reinhardtii, CCM induction was strongly light-dependent, although a trace amount of transcript of CCM factors such as that of Cah1 was detected in low CO2 in the dark (Rawat and Moroney, 1995). The photorespiratory pathway appeared to be a minor factor in the CCM regulation of C. reinhardtii (Suzuki et al., 1990; Vance and Spalding, 2005), but the light signal is more likely to cross talk with the CO2 signal independently of the balance between the C2 and C3 carbon metabolisms. Similarly to C. reinhardtii, the CCM regulation in diatoms is highly dependent of light as well as CO2; that is, the CO2-responsive regulation of Pptcas activities in P. tricornutum requires light (Harada et al., 2006). However, the mechanisms linking light signaling to the CO2 signaling pathway are yet to be elucidated.
In this study, the relationship of light to the transcriptional regulation of two pyrenoidal β-type CA genes, ptca1 and ptca2, was studied in P. tricornutum. The critical cis-elements identified to respond to light in promoters of these CA genes were identical to those required for CO2 response. The involvements of the light wavelength, photosystems (PSs) and ATP synthesis in their transcriptional regulation, were also examined and the results strongly suggest that the function of Pptcas is deeply related to photosynthetic light and to the function of the acceptor side of the PSI.
RESULTS
Requirement of Light for CO2-Responsive Regulation of the Activities of ptca1 and ptca2 Promoters
Our previous studies have demonstrated that the endogenous ptca1 and ptca2 showed, respectively, about 25 and 3.5 times increase in their transcript levels by acclimating from 5% CO2 to air (Harada and Matsuda, 2005; Harada et al., 2006) and the reporter construct of Pptca1 fused with the β-glucuronidase (GUS) gene, uidA, was a quantitative indicator for the CO2 response of Pptca1 in transformant cells expressing this fusion reporter construct (Harada et al., 2005, 2006; Sakaue et al., 2008, Ohno et al., 2012). We thus used this GUS reporter system to identify the critical element required for switching Pptcas activity in response to light/CO2 by means of sequence manipulations of heterologously introduced promoter constructs.
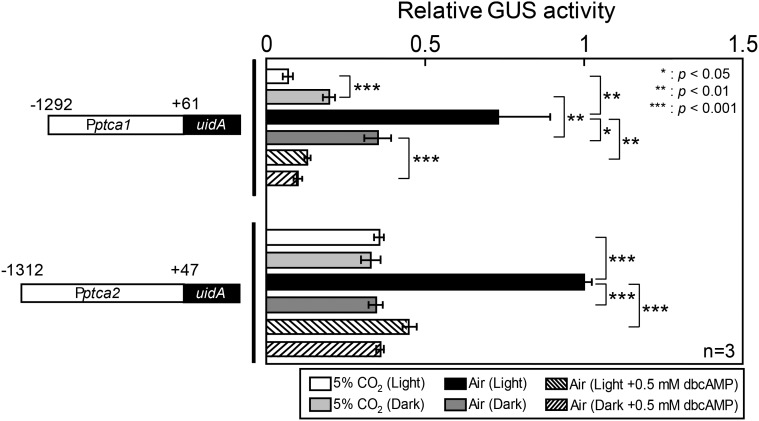
Both Pptca1(−1292−+61)::uidA and Pptca2(−1312−+47)::uidA constructs expressed in P. tricornutum cells showed a clear CO2 response in the light; i.e. GUS levels were about 8 and 2.5 times in air, respectively, in transformants carrying Pptca1(−1292−+61)::uidA and Pptca2(−1312−+47)::uidA compared to those of 5% CO2 (Fig. 1, black and white bars). The GUS expression in air in the light was suppressed in the presence of a permeable cAMP analog, 0.5 mm dibutyryl cAMP (dbcAMP; Fig. 1). In the absence of light, the Pptca1-driven expression of GUS in 5% CO2 was about 2.5 times that of 5% CO2 in the light (Fig. 1). The derepression of Pptca1 in air was also greatly diminished to about 50% in the dark compared to that in the light (Fig. 1). These data suggest that light is required for Pptca1 to achieve a full dynamic range of CO2 response (i.e. to be fully repressed and derepressed). In contrast, the level of expression of Pptca2-driven uidA in the dark was maintained stably at the basal 5% CO2 level both under 5% CO2 and air conditions (Fig. 1), suggesting that derepression of Pptca2 is strongly dependent on both limitation of CO2 and light, and either high CO2 or the absence of light can fully repress this promoter. This is in sharp contrast to the case of Pptca1 in which CO2 and light exhibited an additive effect on the promoter regulation. The dbcAMP treatment did not show notable difference under changing light conditions and worked repressively to both Pptca1 and Pptca2 (Fig. 1), indicating that the increase in the intracellular cAMP level constitutes the basal repression signal, and light would participate in this signaling pathway. Also these data suggest that two paralogous pyrenoidal β-CAs are under slightly different expression controls.
Figure 1.
Promoter activity of ptcas reported by GUS in cells grown in changing CO2 and light conditions in the absence or presence of dbcAMP. Isolated upstream genomic DNA sequence of ptca1 (−1292 to +61: Pptca1) and ptca2 (−1312 to +47: Pptca2) were fused with the GUS reporter gene, uidA (left half); GUS activity relative to the value in 2 d air-acclimated cells in the light (black bar in Pptca2) were shown for 5% CO2 cells or 2 d air-acclimated cells (right half). Value is mean ± sd of three replicates. Statistical significance was determined by t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
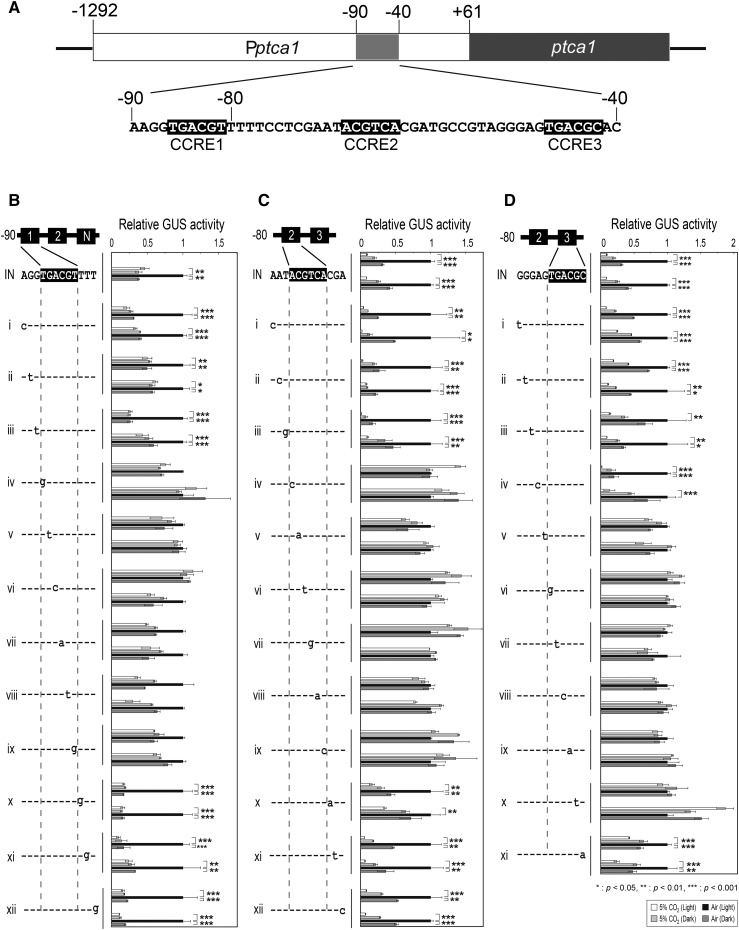
Determination of Critical CO2/Light-Responsive Elements in the Core-Regulatory Region of Pptca1
We tested light response of the Pptca1 core-regulatory region by a series of single nucleotide replacements of three CCREs on an assumption that the critical light responsive element was associated with the core-CO2-regulatory region and deeply related to the primary control under the CO2/cAMP signal (Fig. 2). Each point-mutation construct, which was ligated with uidA at +61 bps relative to the transcription-start site, was transformed into P. tricornutum cells by microprojectile bombardment. Obtained transformants revealed a variety of specific GUS activity levels per total protein (data not shown), presumably due to the copy number and/or positioning effects of genes integrated to the genome. However, CO2/light-response of GUS expression was stably observed in each transformant and thus switching of the Pptca1 activity in response to CO2/light was quantified with some accuracy. The results in Figure 2, B to D, showed the apparent loss of the repression of the manipulated promoters in the dark (Fig. 2B, iv–ix; Fig. 2C, iv–ix; Fig. 2D, v–x), while other reporter constructs showed a clear repression under the dark and air conditions (Fig. 2, B–D). The identified light-responsive element of Pptca1 was identical to the CO2/cAMP-responsive elements, CCRE1, CCRE2, and CCRE3. Interestingly, one-base replacements adjacent only to the CCRE1 sequence gave an equivalent repression of promoter in the absence of light to that of 5% CO2 in the light, apparently dissipating the additive effect of CO2 and light, and thus regulation of Pptca1 resembled that of Pptca2 (Fig. 2B, i, iii, and x–xii). As the promoter constructs in Fig. 2B also lack in the CCRE3 sequence, these data suggest that adjacent sequences of CCRE1 or the lack of CCRE3 may alter the dependency of Pptca1 to CO2 and light.
Figure 2.
CO2 and light responses of one-base-replaced promoter. A, A schematic drawing of core-regulatory region of Pptca1. B, One base replacement assay for the CCRE1 sequence; CCRE3 sequence was replaced by NotI site to avoid CCRE3 substituting for the function of CCRE1. C, D, One base replacement assay, respectively, for CCRE2 and CCRE3; CCRE1 sequence was removed for the same reason as described in (B). Cells were grown in 5% CO2 or air in the absence or presence of light. Vertical bar indicates the dataset for each construct. Two independent clones of transformants carrying each manipulated promoter-reporter construct were selected for a set of reporter assays. Value is mean ± sd of three replicates. Statistical significance was determined by t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
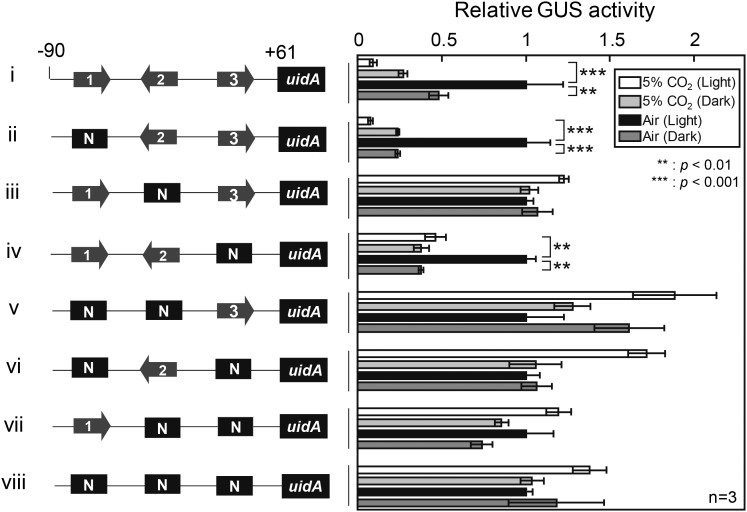
Three CCREs in Pptca1(−90 − +61) were manipulated in a series of combinations, and light response in P. tricornutum cells was evaluated under 5% CO2 and air conditions (Fig. 3). The replacement of CCRE2 with the NotI restriction site completely abolished the light response of Pptca1 as well as CO2 response (Fig. 3, iii), while the single NotI replacement of either CCRE1 or CCRE3 did not affect CO2 or light response (Fig. 3, ii and iv). However, the NotI replacement of more than two CCREs abolished both CO2 and light responses (Fig. 3, v–viii), in agreement with our previous CO2-response model of the CCRE1, CCRE2, and CCRE3 that is, CCRE2 provides a core platform for the Trans-acting factor, PtbZIP11 cooperating with either CCRE1 or CCRE3 (Ohno et al., 2012). These data indicate that light response elements completely overlap with CCRE1, CCRE2, and CCRE3, strongly suggesting the occurrence of the light and CO2 cross talk in the cAMP-signal transduction pathway at the upstream processes toward CCREs. The regulation of Pptca1 containing CCRE1 and CCRE2 with NotI replacement of CCRE3 resembled that of Pptca2 (Fig. 1, iv and Fig. 3, iv). Together with the result in Fig. 2B, the promoter regulation without CCRE3 may confer the Pptca2 type sensitivity for CO2 and light.
Figure 3.
Determination of critical cis-element combination for light response. Pptca1 core-regulatory region (−90 to +61) was fused with uidA and CCREs were replaced by NotI site with a set of different combinations and a GUS assay was performed. Cells were grown in 5% CO2 or air in the absence or presence of light. Value is mean ± sd of three replicates. Statistical significance was determined by t test (**, P < 0.01; ***, P < 0.001).
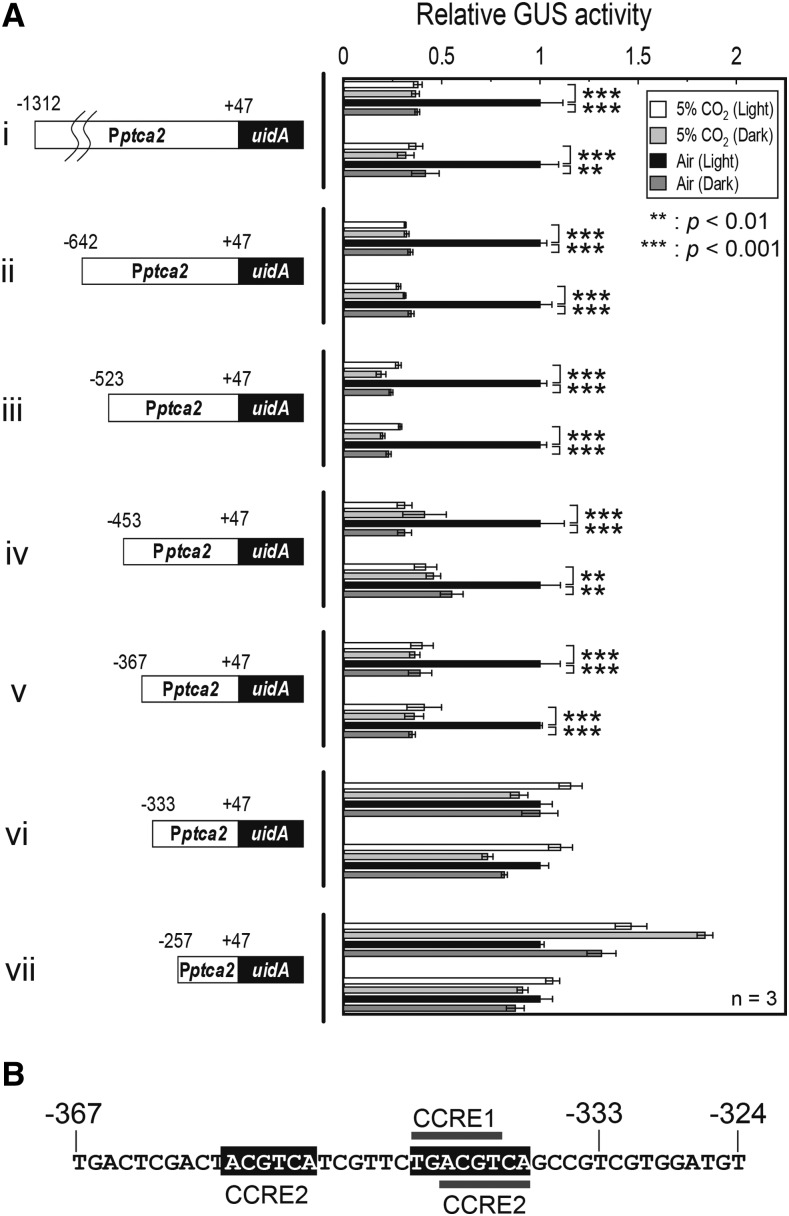
Truncation Assay of Pptca2
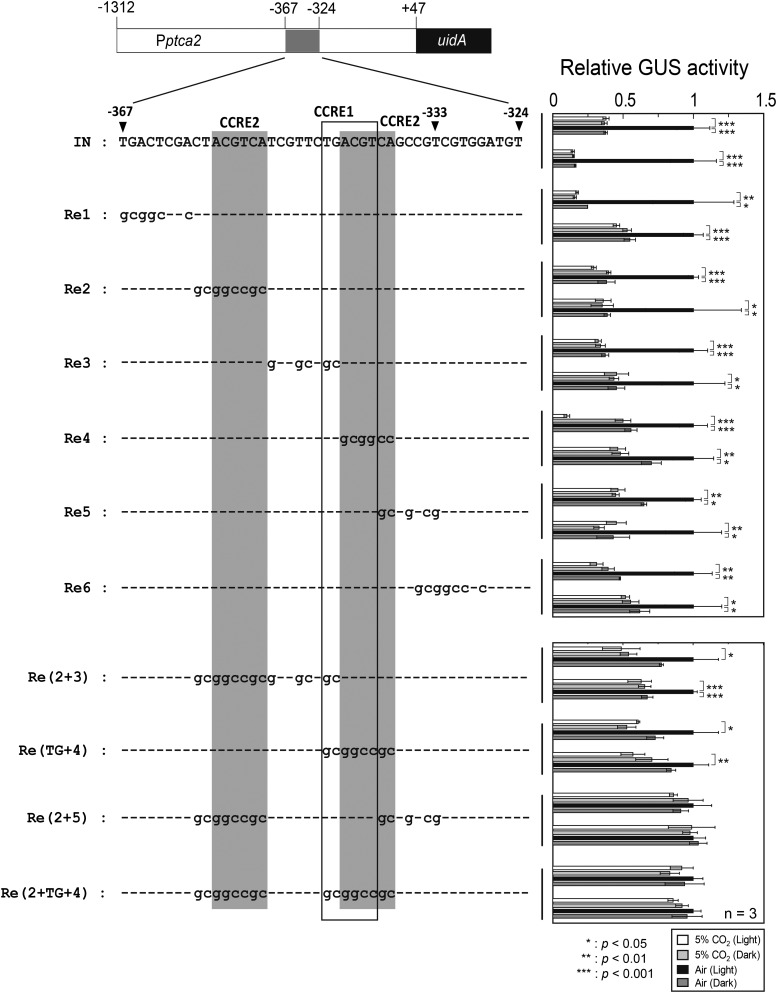
PtCA2 is a paralogous CA to PtCA1, with an approximately 80% amino acid sequence identity whose transcription is also CO2-responsive, and they colocalize at the pyrenoid of P. tricornutum (Harada and Matsuda, 2005; Tanaka et al., 2005; Tachibana et al., 2011). Both CAs are under redox regulation by the function of diatom plastidic thioredoxins (Kikutani et al., 2012) and the function of the promoters of both CA genes are responsive to CO2 via the function of cAMP (Fig. 1; Harada et al., 2006; Ohno et al., 2012). These striking similarities raised a question why PtCA1 and PtCA2 locate together and how these enzymes differ in function. This consideration prompted us to investigate the difference in transcriptional control of the ptca2 gene from that of the ptca1 gene. The isolated promoter region of ptca2 was from +47 to −1312 bps relative to the transcription-start site. This sequence was ligated to the uidA reporter gene (Fig. 4, i) and a series of upstream truncations were engineered to this reporter construct at −642, −523, −453, −367, −333, and −257 relative to the transcription-start site (Fig. 4A, ii–vii). The reporter assay was carried out using these truncated promoter-reporter constructs after transformation into P. tricornutum cells. As a result, the promoters possessing the upstream sequence beyond −367 bps relative to the transcriptional-start site clearly maintained the responses to both CO2 and light (Fig. 4A, i–v), while the promoters truncated at −333 completely lost the response to either CO2 or light (Fig. 4A, vi and vii), indicating that the critical CO2/light-responsive region of Pptca2 resides between −367 and −333. Indeed, the sequence of this region comprised a couple of CCRE2 sequences, ACGTCA (at −355 to −350, and −343 to −338), and a CCRE1 sequence, TGACGT (at −345 to −340; Fig. 4B), strongly indicative that this region is under the control of the cAMP-signal transduction pathway.
Figure 4.
Truncation assay of Pptca2 to screen the critical regulatory region for CO2 and light responses. A, A series of upstream truncation was carried out on Pptca2 (−1312 to +47) and each truncated sequence was fused with uidA (left half) and the GUS reporter assay was done with each transformant (right half). Cells were grown in 5% CO2 or air in the absence or presence of light. Vertical bar indicates the dataset for each construct. Two independent clones of transformants carrying each manipulated promoter-reporter construct were selected for a set of reporter assays. Value is mean ± sd of three replicates. Statistical significance was determined by t test (**, P < 0.01; ***, P < 0.001). B, The primary structure of the newly found core-regulatory region of Pptca2, which contains one CCRE1 and two CCRE2 sequences.
Sequence-Replacement Assay to Determine the cis-Elements for CO2/Light Response in the Core-Regulatory Region of Pptca2
The CCRE1 and CCRE2 sequences in the short region of about 30 bps of Pptca2 were singly or concurrently replaced by the NotI restriction sequence and the CO2/light response of manipulated Pptca2 was evaluated by the GUS reporter assay (Fig. 5). Introduction of the NotI restriction site to the region adjacent to the CCREs cluster gave no effect on the CO2/light response of Pptca2 (Fig. 5, Re1 and Re6). A single insertion of the NotI restriction site to either CCRE1 or CCRE2 also showed little effect on the Pptca2 function (Fig. 5, Re2, Re3, and Re5). In the construct Re4, the overlapping part of CCRE1 and the second CCRE2 were replaced by the NotI sequence but CO2 response was retained in this manipulation (Fig. 5, Re4). Complete replacement of both CCRE1 and the second CCRE2 largely dissipated the CO2/light response; it is thus suggested that a small part of CCRE1 (the first TG sequence) may have some function [Fig. 5, Re(TG+4)]. In contrast, concurrent NotI insertions into CCREs showed a significant dissipation effect on the CO2/light responses of Pptca2. That is, a disruption of one of two CCRE2s with the CCRE1 disruption greatly diminished the CO2/light response of Pptca2 (Fig. 5, Re(2+3) and Re(TG+4)). A concurrent NotI insertion to two CCRE2s completely abolished the CO2/light response (Fig. 5, Re(2+5)). Similarly, disruption of all CCREs completely abolished the CO2/light response of Pptca2 (Fig. 5, Re(2+TG+4)).
Figure 5.
Sequence-replacement assay to determine the cis-elements for CO2/light response in the core-regulatory region of Pptca2. CCRE1 and CCRE2 sequences and adjacent sequences within the core-regulatory region of Pptca2 were replaced by NotI restriction site (left half), and the GUS reporter assay was done with each transformant (right half). Cells were grown in 5% CO2 or air in the absence or presence of light. Vertical bar indicates the dataset for each construct. Two independent clones of transformants carrying each manipulated promoter-reporter construct were selected for a set of reporter assays. Value is mean ± sd of three replicates. Statistical significance was determined by t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Light Wavelength Required to Maintain the Activities of Pptca1 and Pptca2 in Air
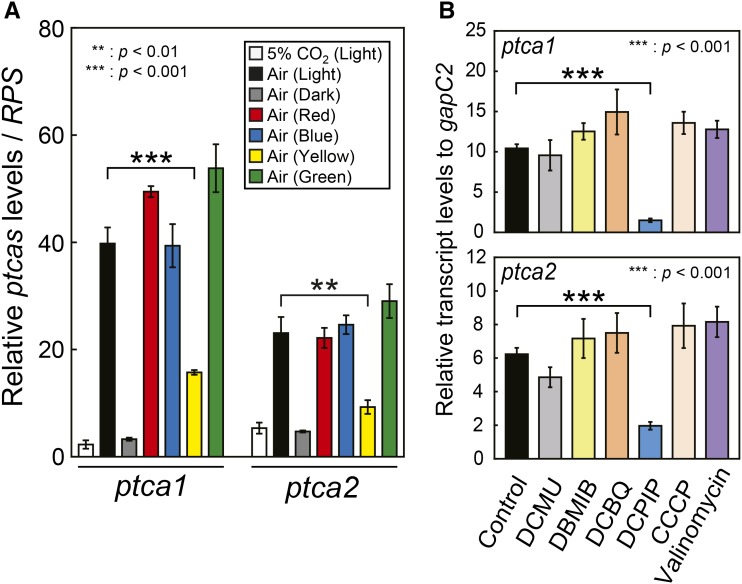
Illumination at the photosynthetic photon flux density (PPFD) of 80 μmol m−2 s−1 of three types of monochromatic light, blue (455 nm), green (520 nm), and red (635 nm; Supplemental Fig. S1A), gave an equivalent photosynthetic O2 evolution rate to that under the white fluorescent light of the same PPFD at a saturated DIC condition (5 mM) using a cell culture of OD730 at 0.2, indicating that blue and green light are both photosynthetically active as well as red light. On the other hand, yellow light (595 nm) showed little activation on photosynthesis (Supplemental Fig. S1, B and C). The capacities of these four monochromatic light to maintain the CO2-responsive transcriptional regulation of the endogenous Pptca1 and Pptca2 were tested by quantitative RT-PCR using ribosomal protein S1 (RPS) for a reference gene (Fig. 6A). The glyceraldehyde-3-phosphate dehydrogenase C2 gene (gapC2) was not used in this experiment, as changes in light wavelength gave a significant effect on the transcript level of gapC2. Blue, green, and red light revealed an equivalent stimulation on both Pptca1 and Pptca2 activities in air to that of white light, while yellow light showed a significantly small stimulatory effect (30–40% compared to other light) on these two promoter activities (Fig. 6A). The responses of two ptcas to different light wavelengths strongly suggest that the main switching function of these CO2-responsive promoters in response to CO2 and light is deeply related to photosynthetic light.
Figure 6.
Changes in transcript levels of endogenous ptca1 and ptca2 under different light wavelength and inhibitor treatments for the PS and ATP synthesis. A, 5% CO2-grown cells were acclimated to air under illumination with different wavelengths of light for 2 d and quantitative RT-PCR was carried out by RPS as an internal control. Value is mean ± sd of three replicates. Statistical significance was determined by t test (**, P < 0.01; ***, P < 0.001). B, Transcript levels of endogenous ptca1 and ptca2 in air-grown cells in the light were compared among cells treated with PS inhibitors (DCMU, DBMIB, DCBQ, and DCPIP) and ATP synthesis inhibitors (CCCP and Valinomycin). The levels of transcript were determined by quantitative RT-PCR as a relative amount to the gapC2 transcript. Value is mean ± sd of three replicates. Statistical significance was determined by t test (***, P < 0.001).
Changes in Transcript Levels of Endogenous ptca1 and ptca2 under Inhibitor Treatments against the PSs and ATP Synthesis
PtCA1 and PtCA2 are posttranslationally regulated by redox levels via thioredoxins in response to the energy state of the PSI and PSII (Kikutani et al., 2012), which probably functions as a part of the fine regulation systems of CO2 supply to Rubisco at the pyrenoid under CO2-limited condition where the expressions of ptcas are derepressed (Matsuda and Kroth, 2014). This prompted us to investigate the existence of a regulation system of the derepressed level of the ptcas transcripts by physiological state relating to redox conditions in the chloroplast. For this purpose, we employed a quantitative RT-PCR assay to detect differences in the derepressed levels of the endogenous ptcas transcripts.
P. tricornutum cells were treated with inhibitors for the photosystems or ATP synthesis. Photosystem inhibitors used were 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), a specific blocker of electron flow to plastoquinone (PQ) in PSII (Izawa, 1980); 2,5-dibromo-6-isopropyl-3-methyl-1,4-benzoquinone (DBMIB), a blocker of the Qo site of the cytochrome b6f complex (Farineau et al., 1984; Trebst, 2007); 2,5-dichloro-1,4-benzoquinone (DCBQ), an artificial electron acceptor from QB in PSII (Graan and Ort, 1986); and 2,6-dichlorophenolindophenol (DCPIP), a strong oxidizing reagent at the acceptor side of PSI (Izawa, 1980; Supplemental Fig. S2A). The ATP synthesis inhibitor, carbonyl cyanide m-chlorophenylhydrazone (CCCP), is a universal uncoupler of proton-motive force and ATP synthase (Heytler and Prichard, 1962; Terada, 1981; Supplemental Fig. S2B), and Valinomycin is a specific potassium ionophore that dissipates membrane potential, Δψ but not ΔpH (Harold, 1969; Gómez-Puyou and Gómez, 1977; Yildiz et al., 1994; Supplemental Fig. S2C). The growth rate of cells slowed down in 2 d treatment with 0.1 μm DCMU, 3 μm DBMIB, or 2 μm DCBQ, while DCPIP up to 50 μm did not reveal any growth inhibition over 4 d of culture (Supplemental Fig. S3A). Photosynthetic O2 evolution under short time exposure to inhibitors showed significantly different response from that of growth response (Supplemental Fig. S3B; Supplemental Table S1). The quantities 0.1 μm and 1 μm of DCMU and DBMIB respectively were highly toxic to PSII and no O2 evolution was observed (Supplemental Fig. S3B; Supplemental Table S1). On the other hand, DCBQ showed a clear inhibitory effect on O2 evolution at 10 μm but below this concentration, there was no significant inhibition (Supplemental Fig. S3B; Supplemental Table S1). DCPIP showed a linear concentration dependency of inhibition to O2 evolution from 5 μm over 100 μm (Supplemental Fig. S3B; Supplemental Table S1). In the presence of 2.5 μm CCCP, there was little O2 evolution, while 2 μm Valinomycin inhibited the maximum photosynthesis to about 60% of the nontreated cells (data not shown). From these preliminary experiments, we set the dose of each inhibitor at the level below the acute toxic level (0.025 μM DCMU, 1 μM DBMIB, 2 μM DCBQ, 10 μM DCPIP, 0.5 μM CCCP, and 2 μM Valinomycin) and after 2 d of treatment, total RNAs were extracted from the inhibitor-treated cells and transcript levels of endogenous ptca1 and ptca2 were analyzed with quantitative RT-PCR.
The transcript levels of both ptca1 and ptca2 in cells treated with DCMU, DBMIB, DCBQ, CCCP or Valinomycin exhibited marginal differences compared to those of nontreated cells (Fig. 6B), suggesting the absence of influence on the transcriptional control mechanism in Pptca1 and Pptca2 from either processes between the PSII and PQ pool or photophosphorylation. In sharp contrast to the above drug treatments, cells treated with DCPIP showed drastic decreases in transcript levels of ptca1 and ptca2, respectively, to about 10% and 28% that of the control cells (Fig. 6B). Considering the low toxicity on growth of the used concentration of DCPIP, which did not show significant inhibition on growth even at 50 μM (Supplemental Fig. S3A), this repressive effect of DCPIP on accumulations of the ptca1 and ptca2 transcripts looked extremely efficient with little metabolic damage, strongly suggesting the critical requirement of a high reduced level of the PSI acceptor side for a full derepression of transcription of both Pptca1 and Pptca2 in air. Photosynthetic affinity for DIC was slightly inhibited in cells treated with 10 μM DCPIP (Supplemental Fig. S3B), which is presumably a result of repression of both ptcas by DCPIP.
DISCUSSION
In this study, the light-responsive cis-elements in Pptca1 were newly determined. Also, the structure and the function of Pptca2 were determined and compared in detail with those of Pptca1. We found that Pptca1 and Pptca2 were essentially controlled by a similar switching mechanism of transcription, which was mediated by a second messenger cAMP, in response to CO2 and light. But in contrast, it was also strongly suggested that there were some fine-tuning systems at the transcriptional level that might differentiate the function of these two paralogous pyrenoidal CAs, PtCA1 and PtCA2, when they were derepressed under air and illuminated conditions.
The CCRE2 sequences identified in the core-regulatory regions of both Pptca1 and Pptca2 were shown to be essential to repress the promoter activity in response to CO2, and light was required for proper switching function. As indicated previously, CCRE2 was the core cis-element that worked either with CCRE1 or CCRE3 in Pptca1 (Ohno et al., 2012) and these functional combinations of cis-elements were also responsible for the light response of this promoter. Light response of Pptca2 also revealed a tight relation to CO2 response and the disruption of CCREs directly caused a disappearance of light response (Figs. 4 and 5). These results strongly suggest the existence of the CO2 and light cross talk at upstream signal transduction pathways toward CCREs both in Pptca1 and Pptca2. However, there were some differences in the alignment of CCREs between Pptca1 and Pptca2 (Figs. 2A and 4B). Our previous study demonstrated by in vitro gel shift assay that all CCREs were the specific target of a diatom basic ZIP transcription factor, PtbZIP11 (Ohno et al., 2012), strongly suggesting that CCREs in the core-regulatory region of Pptca2 are also targeted by PtbZIP11. But the difference in the alignment of CCREs probably conferred some differences in the transcriptional regulations in response to CO2 and light; that is, transcriptional activation of Pptca2 in response to the decrease in CO2 was strongly light-dependent and either CO2 or the absence of light can maximally down-regulate the promoter, while CO2 and light have additive effect on the regulation of Pptca1 (Fig. 1). One of the interesting structural features clarified in this study is that the manipulated Pptca1 only with CCRE1 and CCRE2 altered its CO2/light response to that similar to Pptca2 (Figs. 2 and 3). Interestingly, the structure of Pptca2 does not have CCRE3 sequence either (Figs. 4B and 5), although the alignment of CCREs between two promoters is not similar and the detail is yet to be made clear.
A requirement of light for the activity of CO2-responsive promoters is commonly observed in the regulation of algal CCM factors but the molecular mechanism of the CO2/light cross talk in eukaryotic algae is yet to be elucidated. In bacterial systems, in contrast, the mechanism of CO2/light cross talk is relatively clear. Physiological studies have demonstrated that cyanobacterial CCM is regulated in response to total DIC in the medium presumably corresponding to the availability of substrate inorganic carbon for photosynthesis (Mayo et al., 1986). In fact, a recent molecular study has revealed that 2-PG directly stimulated the binding of CmpR to the CO2-responsive promoter of the HCO3− transporter gene operon, cmpABCD in the cyanobacterium, Synechocystis sp. PCC 6803, strongly suggesting that CO2/light cross talk occurs at the C2/C3 photosynthetic carbon flux mediated by the accumulation of the initial photorespiratory metabolite (Nishimura et al., 2008). In contrast, regulation of CCM in eukaryotic algae is relatively complicated; that is, cells respond specifically to CO2 concentration to control CCM levels even though they possess high HCO3− uptake capacity. In the green algae, C. reinhardtii and C. ellipsoidea, a physiological determinant of the CCM level was CO2 in the bulk media but not HCO3− nor total DIC (Matsuda and Colman, 1995; Bozzo and Colman, 2000; Vance and Spalding, 2005). In C. ellipsoidea, even light was not required for a regulation of the CCM expression (Matsuda and Colman, 1995) but in contrast, the CCM expression in C. reinhardtii was strongly blue light-dependent (Dionisio-Sese et al., 1990). In the marine diatom, P. tricornutum, also the similar signaling role of CO2 was reported in the regulation of the CCM expression (Matsuda et al., 2001). This evidence strongly suggests the occurrence of some direct signal reception mechanisms for CO2 and light in eukaryotic algae, which may cross talk independently of photosynthesis. The role of the cAMP analog in CO2/light signals to control Pptcas in P. tricornutum strongly suggests the involvement of cAMP in this cross talk (Fig. 1).
Furthermore, all photosynthetically active-monochromic-light wavelength (red, blue, and green) conferred a derepression on both Pptcas with equivalent efficiency to the normal light, while weakly photosynthetically active-light wavelength (yellow) showed a weak derepression activity to Pptcas (Fig. 6A; Supplemental Fig. S1). This apparent synchronization of the promoter activity with the photosynthesis strongly suggests an involvement of photosynthetic process to the CO2/light signaling, although an involvement of a direct sensing of the light wavelength by specific photoreceptors is not ruled out.
The light-dependent chloroplastic metabolism and the redox state seemed to work to control the transcript levels of ptca1 and ptca2 in a mode similar to each other. Interestingly, accumulation levels of both ptcas transcripts showed a drastic decrease by the treatment with low dose of DCPIP, which is a specific oxidizing reagent for the acceptor side of the PSI and would enhance the proton gradient across the thylakoid membrane by accepting electron from the PSI with high affinity or by releasing proton at the thylakoid lumen (Mühlbauer and Eichacker, 1998). However, the absence of a notable effect of electron transport inhibitors from the PSII to PSI and of the ΔpH inhibitor on the ptcas transcription strongly suggests that the PQ pool level or proton gradient is not involved in mediating light signal, but the redox state of the PSI acceptor side would be critical in this process. The PSI acceptor side is extremely multifunctional in sorting electron flow into a number of different metabolic pathways and the CO2/cAMP signaling pathway might be one of the targets of this sorting system, integrating the light signal into the promoter function based upon CCREs and PtbZIP11. Although the mechanism of this process awaits further investigation, the results of this study may provide an initial clue to clarify it.
MATERIALS AND METHODS
Cells and Culture Conditions
The marine diatom Phaeodactylum tricornutum Bohlin (UTEX 642) was obtained from the University of Texas Culture Collection and was grown in artificial seawater, which was supplemented with half-strength Guillard’s “f” solution (F/2ASW; Guillard and Ryther, 1962; Harrison et al., 1980) under continuous illumination of photosynthetic photon flux density (PPFD) of 50 μmol m−2 s−1 and constant aeration with 5% CO2 or ambient air (0.039% CO2) at 20°C. In acclimation experiments, 5% CO2-grown cells were harvested by centrifugation at 3,500g for 5 min at room temperature, washed twice with CO2-free F/2ASW, and allowed to acclimate to air-aerated F/2ASW for 1–2 d. In some experiments, 0.5 mm dibutyryl cAMP (dbcAMP; Sigma-Aldrich, St. Louis, MO), 0.025 μM 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), 1 μM 2,5-dibromo-3-metyl-6-isopropyl-pbenzoquinone (DBMIB), 2 μM 2,6-dichlorobenzoquinone (DCBQ), 10 μM 2,6-dichlorophenolindophenol (DCPIP), 0.5 μM carbonylcyanide m-chlorophenyl-hydrazone (CCCP), and 2 μM Valinomycin were added during the acclimation to air.
Preparation of Transformation Constructs
The genomic DNA was extracted from air-grown P. tricornutum cells with a modified method as described by Sambrook and Russell (2001). The promoter region of ptca2 (−1312 to +47 relative to the transcription-start site) was amplified on the template genomic DNA with high-fidelity PrimeSTAR HS DNA polymerase (TaKaRa Bio, Kusatsu, Shiga, Japan) and a primer pair: Fw: 5′-CGCAATGAGCATCTCGACGGCACCAATCGCT-3′; Rv: 5′-GGTAGAACAGTAAACTAGGTTGGTGTACCACA-3′ and was phosphorylated. To construct the plasmid fused with uidA gene, a vector pFcpApGUS (Harada et al., 2005) was double-digested with EcoRI and NdeI to remove the promoter region of fucoxanthin chlorophyll (Chl) a/c binding protein gene (fcpA) and blunt-ended with T4 DNA polymerase. A phosphorylated PCR product was inserted into blunt-ended pFcpApGUS and sequenced (pPtCA2pGUS).
A series of upstream truncated constructs of Pptca1 (−1292 to +61 and −90 to +61 relative to the transcription-start site) were used as described by Ohno et al. (2012), while Pptca2 (−642 to +47, −523 to +47, −453 to +47, −367 to +47, −333 to +47, and −257 to +47 relative to the transcription-start site) were amplified by the inverse-PCR method using pPtCA2pGUS as a template with a set of primers shown in Supplemental Table S2. These constructs were phosphorylated, self-ligated, and sequenced.
To generate one-base replacement constructs of Pptca1, PCR reactions were carried out targeting the sequence −71 to −60 and −52 to −42 using the vector pFcpApGUS containing Pptca1 (−90 to +61), and targeting the sequence −86 to −81 using the vector pFcpApGUS containing Pptca1 (−90 to +61) replacing the sequence −52 to −42 with NotI restriction site. A set of linker scan constructs with a NotI restriction linker site was synthesized by a modified inverse-PCR method using the vector plasmid pPtCA2pGUS as a template. Primer sets were designed to generate a NotI restriction site (GCGGCCGC) at desired locations by self-ligation (Supplemental Table S3). In some experiments, NotI substitutions were introduced into −358 to −351 and −350 to −343 or −344 to −337 or −338 to −331 relative to the transcription-start site using the aforementioned vector plasmids pFcpApGUS containing upstream truncated Pptca2 (−1312 to +47) as templates.
Transformation of P. tricornutum
Air-grown cells of P. tricornutum were harvested at the midlogarithmic growth phase (optical density at 730 nm of 0.2–0.4). Approximately 5 × 107 cells were spotted as a plaque of 2.5 cm diameter on the surface of the F/2ASW agar plate. A 500 μg tungsten microcarrier (Japan New Metals, Toyonaka, Osaka, Japan; particle size: 0.79 μm) was coated with 1 μg of plasmid DNA containing 1 m CaCl2 and 16 mm spermidine. The PDS-1000/He Biolistic Particle Delivery System (Bio-Rad Laboratories, Hercules, CA) was used for microprojectile bombardment of the microcarrier. The bombardment was done at 10.7 MPa to the cells in the chamber under a negative pressure of 91.4 kPa with a target distance of 6 cm. Bombarded cells were maintained on the plate for 1 d in the dark and were then suspended in 300 μL of F/2ASW. This cell suspension was plated on F/2ASW agar plates containing 100 μg mL−1 Zeocin (Invitrogen, Carlsbad, CA) and allowed to form colonies for 3–4 weeks under continuous illumination.
Screening of GUS-Expressing Transformants
Zeocin-resistant clones were suspended in 60 μL of GUS extraction buffer (50 mm sodium phosphate, pH 7.0; 10 mm β-mercaptoethanol, 0.1% w/v sodium lauroyl sarcosine; and 0.1% v/v Triton X-100) containing 1 mm 5-bromo-4-chloro-3-indolyl-β-d-GlcA cyclohexyl ammonium salt (X-GlucA; Sigma-Aldrich) on 96-well plates and were incubated overnight at 37°C. Transformants expressing GUS, which displayed the dark blue-colored chromogenic reaction product of X-GlucA, were inoculated and used for further experiments.
Quantification of GUS Activity
Cells at different acclimation stages were harvested by centrifugation at 3,000g at 20°C, resuspended in 0.3–1.0 mL of GUS extraction buffer, and disrupted by sonicator (Ultrasonic disruptor model no. UD-201; TOMY Digital Biology, Tokyo, Japan), followed by five cycles of disruption for 10 s, a pause for 30 s in an ice bath, and then centrifugation at 15,400g for 5 min at 4°C. Supernatant (lysate, 18 μL) was added to 882 μL of GUS reaction buffer [10 mm p-nitrophenyl β-d-glucuronide (PNPG) in 50 mm sodium phosphate buffer, pH 7.0] and incubated at 37°C. The reaction was terminated by the addition of 80 μL 0.5 m Na2CO3 to 200 μL reaction solutions at every 10 min after starting the reaction, and the absorbance of p-nitrophenol (PNP; Wako Chemicals USA, Richmond, VA) released from the substrate was measured at 405 nm. Protein concentrations in lysate were measured by the Bradford method (Bio-Rad Laboratories) according to the manufacturer’s protocol. The standard curve of PNP was drawn with 0.2–8.0 nmol PNP, which gave a range of A405 from 0.011 to 0.454. Concentrations of lysate protein in the reaction mixture were adjusted to produce PNP within the range of the above standard curve in the reaction for 4 min. GUS activity was calculated as the rate of PNP production per min per mg lysate protein.
Determination of Photosynthetic Parameters under Illumination of Monochromatic Light
Air-grown cells at the midlogarithmic growth phase were harvested by centrifugation at 1,400g at room temperature and washed three times with CO2-free F/2ASW. Cells were then suspended in the same F/2ASW at a final Chla concentration of 10 μg mL−1. The rate of photosynthetic O2 evolution was measured with a Clark-type oxygen electrode (Hansatech Instruments, Pentney, King’s Lynn, Norfolk, UK; and Qubit Systems, Kingston, Ontario, Canada). A quantity of 1.5 mL of cell suspension was placed in an O2-electrode chamber and illuminated with a fiber illuminator at PPFD of 40 μmol m−2 s−1 under aeration with N2 for 10 min until cells reached to the CO2-compensation point, which was determined by the gas chromatography method. For some inhibitor experiments, DCMU, DBMIB, DCBQ, or DCPIP were added to a chamber for 10 min before the measurement. PPFDs of illumination or LED bulbs were then increased to 250 μmol m−2 s−1, and a known amount of DIC was added as a form of NaHCO3 to the desired final concentrations to cell culture. The rate of O2 evolution at each DIC concentration was plotted against DIC concentration. Chla concentration was calculated with spectrophotometer according to Jeffrey and Humphrey (1975). Based on the plot, photosynthetic parameters were determined by the least-squares method.
Quantifications of the Endogenous ptca1 and ptca2 Transcripts
Total RNAs were extracted using NucleoSpin RNA (Macherey-Nagel, Bethlehem, PA) from P. tricornutum cells grown under illumination of monochromatic light or treated with inhibitors to PS or ATP synthesis for 2 d under an air condition according to the manufacture’s protocol. Each single-strand cDNA was synthesized from 1 μg total RNA using an Oligo (dT)20 (Toyobo, Osaka, Japan) and reverse transcriptase, ReverTra Ace (Toyobo). Quantitative RT-PCR was carried out with Thermal Cycler Dice Real Time System II (TaKaRa) and GeneAce SYBR qPCR Mixα No ROX (Nippon Gene, Tokyo, Japan) under PCR conditions as follows: heating at 95°C for 10 min followed by 45 cycles of denaturing at 95°C for 30 s, annealing, and elongation at 60°C for 1 min. To standardize the transcript level, ribosomal protein S1 (RPS) or endogenous glyceraldehyde-3-phosphate dehydrogenase C2 (gapC2) genes were used as the constitutive maker genes. Quantitative RT-PCR was carried out by a set of primers shown in Supplemental Table S4. The standard curves of ptca1, ptca2, RPS, and gapC2 for Quantitative RT-PCR were drawn with a known amount of template using plasmids containing the ptca1, ptca2, RPS, and gapC2 cDNAs. Each transcript level was calculated using each standard curve from the values of the threshold cycle (Ct) that were the intersection point of the threshold and the amplification curve. The levels of the ptca1 and ptca2 transcripts were normalized by the level of the RPS or gapC2 transcripts.
Sequence data from this article can be found in the JGI Genome Portal or the GenBank/EMBL data libraries under accession numbers: RPS (protein ID: 10847), gapC2 (GenBank accession no. AF63805), ptca1 (GenBank accession no. AAL07493), Ptca2 (GenBank accession no. BAD67442).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Diatom photosynthesis under monochromatic light.
Supplemental Figure S2. Diagrams of functional mechanisms of inhibitors for the PSs and ATP synthesis.
Supplemental Figure S3. Growth and photosynthetic profiles under different doses of inhibitors for the PS.
Supplemental Table S1. Effects of photosynthesis inhibitors and ATP synthesis inhibitors on photosynthesis parameters.
Supplemental Table S2. Primer sequences used to make Pptca2 truncated constructs.
Supplemental Table S3. Primer sequences used to make Pptca2 linker-scan constructs.
Supplemental Table S4. Primer sequences for quantitative RT-PCR.
Supplementary Material
Acknowledgments
We thank Ms. Nobuko Higashiuchi and Keiji Goto for their technical assistance, and Ms. Miyabi Inoue for her skillful secretarial aid.
Glossary
- bZIP
basic zipper protein
- CCM
CO2-concentrating mechanism
- CCCP
carbonyl cyanide m-chlorophenylhydrazone
- CCRE
CO2/cAMP-responsive elements
- Chl
chlorophyll
- cmp
cytoplasmic-membrane protein
- CmpR
Cytoplasmic-membrane protein Regulator
- dbcAMP
dibutyryl cAMP
- DBMIB
2,5-dibromo-6-isopropyl-3-methyl-1,4-benzoquinone
- DCBQ
2,5-dichloro-1,4-benzoquinone
- DCPIP
2,6-dichlorophenolindophenol
- DIC
dissolved-inorganic carbon
- gapC2
glyceraldehyde-3-phosphate dehydrogenase C2 gene
- mnh
modified NDH-1
- ndh
NAD(P)H dehydrogenase
- 2-PG
2-phosphoglycolate
- PQ
plastoquinone
- PS
photosystem
- RPS
ribosomal protein S1
- sbt
sodium bicarbonate transporter
References
- Badger MR, Andrews TJ, Whitney SM, Ludwig M, Yellowlees DC, Leggat W, Price GD (1998) The diversity and co-evolution of Rubisco, plastids, pyrenoids and chloroplast-based CO2-concentrating mechanisms in the algae. Can J Bot 76: 1052–1071 [Google Scholar]
- Bozzo GG, Colman B (2000) The induction of inorganic carbon transport and external carbonic anhydrase in Chlamydomonas reinhardtii is regulated by external CO2 concentration. Plant Cell Environ 23: 1137–1144 [Google Scholar]
- Burkhardt S, Amoroso G, Riebesell U, Sültemeyer D (2001) CO2 and HCO3− uptake in marine diatoms acclimated to different CO2 concentrations. Limnol Oceanogr 46: 1378–1391 [Google Scholar]
- Colman B, Rotatore C (1995) Photosynthetic inorganic carbon uptake and accumulation in two marine diatoms. Plant Cell Environ 18: 919–924 [Google Scholar]
- Dionisio-Sese ML, Fukuzawa H, Miyachi S (1990) Light-induced carbonic anhydrase expression in Chlamydomonas reinhardtii. Plant Physiol 94: 1103–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M, Villand P, Gardeström P, Samuelsson G (1998) Induction and regulation of expression of a low-CO2-induced mitochondrial carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol 116: 637–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowski P, Scholes RJ, Boyle E, Canadell J, Canfield D, Elser J, Gruber N, Hibbard K, Högberg P, Linder S, Mackenzie FT, Moore B III, et al. (2000) The global carbon cycle: a test of our knowledge of earth as a system. Science 290: 291–296 [DOI] [PubMed] [Google Scholar]
- Farineau J, Bottin H, Garab G (1984) Effect of dibromothymoquinone (DBMIB) on reduction rates of Photosystem I donors in intact chloroplasts. Biochem Biophys Res Commun 120: 721–725 [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Fukuzawa H, Tachiki A, Miyachi S (1990) Structure and differential expression of two genes encoding carbonic anhydrase in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 87: 9779–9783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuzawa H, Miura K, Ishizaki K, Kucho KI, Saito T, Kohinata T, Ohyama K (2001) Ccm1, a regulatory gene controlling the induction of a carbon-concentrating mechanism in Chlamydomonas reinhardtii by sensing CO2 availability. Proc Natl Acad Sci USA 98: 5347–5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Puyou A, Gómez L (1977) The use of ionophores and channel formers in the study of the function of biological membranes. Curr Top Bioenerg 6: 221–257 [Google Scholar]
- Graan T, Ort DR (1986) Detection of oxygen-evolving photosystem II centers inactive in plastoquinone reduction. Biochem Biophys 852: 320–330 [Google Scholar]
- Guillard RRL, Ryther JH (1962) Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Can J Microbiol 8: 229–239 [DOI] [PubMed] [Google Scholar]
- Harada H, Nakajima K, Sakaue K, Matsuda Y (2006) CO2 sensing at ocean surface mediated by cAMP in a marine diatom. Plant Physiol 142: 1318–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Nakatsuma D, Ishida M, Matsuda Y (2005) Regulation of the expression of intracellular β-carbonic anhydrase in response to CO2 and light in the marine diatom Phaeodactylum tricornutum. Plant Physiol 139: 1041–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Matsuda Y (2005) Identification and characterization of a new carbonic anhydrase in the marine diatom Phaeodactylum tricornutum. Can J Bot 83: 909–916 [Google Scholar]
- Harold FM. (1969) Antimicrobial agents and membrane function. Adv Microb Physiol 4: 45–104 [Google Scholar]
- Harrison PJ, Waters RE, Taylor FJR (1980) A broad spectrum artificial seawater medium for coastal and open ocean phytoplankton. J Phycol 16: 28–35 [Google Scholar]
- Heytler PG, Prichard WW (1962) A new class of uncoupling agents—carbonyl cyanide phenylhydrazones. Biochem Biophys Res Commun 7: 272–275 [DOI] [PubMed] [Google Scholar]
- Hihara Y, Kamei A, Kanehisa M, Kaplan A, Ikeuchi M (2001) DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13: 793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkinson BM, Dupont CL, Allen AE, Morel FMM (2011) Efficiency of the CO2-concentrating mechanism of diatoms. Proc Natl Acad Sci USA 108: 3830–3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkinson BM, Meile C, Shen C (2013) Quantification of extracellular carbonic anhydrase activity in two marine diatoms and investigation of its role. Plant Physiol 162: 1142–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa S. (1980) Acceptors and donors for chloroplast electron transport. Methods Enzymol 69: 413–434 [Google Scholar]
- Jeffrey SW, Humphrey GF (1975) New spectrometric equations for determining chlorophylls a, b, c1, and c2 in higher plants, algae, and natural phytoplankton. Biochem Biophys Pflanz 167: 191–194 [Google Scholar]
- Johnston AM, Raven JA (1996) Inorganic carbon accumulation by the marine diatom Phaeodactylum tricornutum. Eur J Phycol 31: 285–290 [Google Scholar]
- Kaplan A, Badger MR, Berry JA (1980) Photosynthesis and the intracellular inorganic carbon pool in the blue-green alga Anabaena variabilis: response to external CO2 concentration. Planta 149: 219–226 [DOI] [PubMed] [Google Scholar]
- Kikutani S, Tanaka R, Yamazaki Y, Hara S, Hisabori T, Kroth PG, Matsuda Y (2012) Redox regulation of carbonic anhydrases via thioredoxin in chloroplast of the marine diatom Phaeodactylum tricornutum. J Biol Chem 287: 20689–20700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucho K, Yoshioka S, Taniguchi F, Ohyama K, Fukuzawa H (2003) Cis-acting elements and DNA-binding proteins involved in CO2-responsive transcriptional activation of Cah1 encoding a periplasmic carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol 133: 783–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Pollock SV, Xiao Y, Cunnusamy K, Moroney JV (2011) Identification of a novel gene, CIA6, required for normal pyrenoid formation in Chlamydomonas reinhardtii. Plant Physiol 156: 884–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Colman B (1995) Induction of CO2 and bicarbonate transport in the green alga Chlorella ellipsoidea: I. Time course of induction of the two systems. Plant Physiol 108: 247–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Hara T, Colman B (2001) Regulation of the induction of bicarbonate uptake by dissolved CO2 in the marine diatom, Phaeodactylum tricornutum. Plant Cell Environ 24: 611–620 [Google Scholar]
- Matsuda Y, Kroth PG (2014) Carbon fixation in diatoms. In Hohmann-Marriott MF, ed, The structural basis of biological energy generation, Vol. 39 Springer Netherlands, Dordrecht, pp 335–362 [Google Scholar]
- Matsuda Y, Nakajima K, Tachibana M (2011) Recent progresses on the genetic basis of the regulation of CO2 acquisition systems in response to CO2 concentration. Photosynth Res 109: 191–203 [DOI] [PubMed] [Google Scholar]
- Mayo WP, Williams TG, Birch DG, Turpin DH (1986) Photosynthetic adaptation by Synechococcus leopoliensis in response to exogenous dissolved inorganic carbon. Plant Physiol 80: 1038–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinn PJ, Price GD, Badger MR (2004) High light enhances the expression of low-CO2-inducible transcripts involved in the CO2-concentrating mechanism in Synechocystis sp. PCC6803. Plant Cell Environ 27: 615–626 [Google Scholar]
- McGinn PJ, Price GD, Maleszka R, Badger MR (2003) Inorganic carbon limitation and light control the expression of transcripts related to the CO2-concentrating mechanism in the cyanobacterium Synechocystis sp. strain PCC6803. Plant Physiol 132: 218–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AG, Espie GS, Canvin DT (1990) Physiological aspects of CO2 and HCO3− transport by cyanobacteria: a review. Can J Bot 68: 1291–1302 [Google Scholar]
- Mühlbauer SK, Eichacker LA (1998) Light-dependent formation of the photosynthetic proton gradient regulates translation elongation in chloroplasts. J Biol Chem 273: 20935–20940 [DOI] [PubMed] [Google Scholar]
- Nakajima K, Tanaka A, Matsuda Y (2013) SLC4 family transporters in a marine diatom directly pump bicarbonate from seawater. Proc Natl Acad Sci USA 110: 1767–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Takahashi Y, Yamaguchi O, Suzuki H, Maeda S, Omata T (2008) Mechanism of low CO2-induced activation of the cmp bicarbonate transporter operon by a LysR family protein in the cyanobacterium Synechococcus elongatus strain PCC 7942. Mol Microbiol 68: 98–109 [DOI] [PubMed] [Google Scholar]
- Ohno N, Inoue T, Yamashiki R, Nakajima K, Kitahara Y, Ishibashi M, Matsuda Y (2012) CO2-cAMP-responsive cis-elements targeted by a transcription factor with CREB/ATF-like basic zipper domain in the marine diatom Phaeodactylum tricornutum. Plant Physiol 158: 499–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omata T, Gohta S, Takahashi Y, Harano Y, Maeda S (2001) Involvement of a CbbR homolog in low CO2-induced activation of the bicarbonate transporter operon in cyanobacteria. J Bacteriol 183: 1891–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Badger MR, Woodger FJ, Long BM (2008) Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J Exp Bot 59: 1441–1461 [DOI] [PubMed] [Google Scholar]
- Rawat M, Moroney JV (1995) The regulation of carbonic anhydrase and ribulose-1, 5-bisphosphate carboxylase/oxygenase activase by light and CO2 in Chlamydomonas reinhardtii. Plant Physiol 109: 937–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinfelder JR. (2011) Carbon concentrating mechanisms in eukaryotic marine phytoplankton. Annu Rev Mar Sci 3: 291–315 [DOI] [PubMed] [Google Scholar]
- Rost B, Riebesell U, Burkhardt S, Sültemeyer D (2003) Carbon acquisition of bloom-forming marine phytoplankton. Limnol Oceanogr 48: 55–67 [Google Scholar]
- Sakaue K, Harada H, Matsuda Y (2008) Development of gene expression system in a marine diatom using viral promoters of a wide variety of origin. Physiol Plant 133: 59–67 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 6.4–6.11 [Google Scholar]
- Samukawa M, Shen C, Hopkinson BM, Matsuda Y (2014) Localization of putative carbonic anhydrases in the marine diatom, Thalassiosira pseudonana. Photosynth Res 121: 235–249 [DOI] [PubMed] [Google Scholar]
- Spalding MH, Van K, Wang Y, Nakamura Y (2002) Acclimation of Chlamydomonas to changing carbon availability. Funct Plant Biol 29: 221–230 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Marek LF, Spalding MH (1990) A photorespiratory mutant of Chlamydomonas reinhardtii. Plant Physiol 93: 231–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Allen AE, Kikutani S, Endo Y, Bowler C, Matsuda Y (2011) Localization of putative carbonic anhydrases in two marine diatoms, Phaeodactylum tricornutum and Thalassiosira pseudonana. Photosynth Res 109: 205–221 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Nakatsuma D, Harada H, Ishida M, Matsuda Y (2005) Localization of soluble β-carbonic anhydrase in the marine diatom Phaeodactylum tricornutum. Sorting to the chloroplast and cluster formation on the girdle lamellae. Plant Physiol 138: 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada H. (1981) The interaction of highly active uncouplers with mitochondria. Biochim Biophys Acta 639: 225–242 [DOI] [PubMed] [Google Scholar]
- Trebst A. (2007) Inhibitors in the functional dissection of the photosynthetic electron transport system. Photosynth Res 92: 217–224 [DOI] [PubMed] [Google Scholar]
- Tréguer P, Nelson DM, Van Bennekom AJ, Demaster DJ, Leynaert A, Quéguiner B (1995) The silica balance in the world ocean: a reestimate. Science 268: 375–379 [DOI] [PubMed] [Google Scholar]
- Trimborn S, Lundholm N, Thoms S, Richter KU, Krock B, Hansen PJ, Rost B (2008) Inorganic carbon acquisition in potentially toxic and non-toxic diatoms: the effect of pH-induced changes in seawater carbonate chemistry. Physiol Plant 133: 92–105 [DOI] [PubMed] [Google Scholar]
- Vance P, Spalding MH (2005) Growth, photosynthesis and gene expression in Chlamydomonas over a range of CO2 concentrations and CO2/O2 ratios: CO2 regulates multiple acclimation states. Can J Bot 83: 796–809 [Google Scholar]
- Wang HL, Postier BL, Burnap RL (2004) Alterations in global patterns of gene expression in Synechocystis sp. PCC 6803 in response to inorganic carbon limitation and the inactivation of ndhR, a LysR family regulator. J Biol Chem 279: 5739–5751 [DOI] [PubMed] [Google Scholar]
- Woodger FJ, Badger MR, Price GD (2003) Inorganic carbon limitation induces transcripts encoding components of the CO2-concentrating mechanism in Synechococcus sp. PCC7942 through a redox-independent pathway. Plant Physiol 133: 2069–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Zhang J, Weeks DP (2001) The Cia5 gene controls formation of the carbon concentrating mechanism in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 98: 5341–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T, Tsujikawa T, Hatano K, Ozawa S, Takahashi Y, Fukuzawa H (2010) Light and low-CO2-dependent LCIB-LCIC complex localization in the chloroplast supports the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Cell Physiol 51: 1453–1468 [DOI] [PubMed] [Google Scholar]
- Yildiz FH, Davies JP, Grossman AR (1994) Characterization of sulfate transport in Chlamydomonas reinhardtii during sulfur-limited and sulfur-sufficient growth. Plant Physiol 104: 981–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka S, Taniguchi F, Miura K, Inoue T, Yamano T, Fukuzawa H (2004) The novel Myb transcription factor LCR1 regulates the CO2-responsive gene Cah1, encoding a periplasmic carbonic anhydrase in Chlamydomonas reinhardtii. Plant Cell 16: 1466–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.