Abstract
Various studies in the past have revealed that molluscs can produce a wide range of rather complex N-glycan structures, which vary from those occurring in other invertebrate animals; particularly methylated glycans have been found in gastropods, and there are some reports of anionic glycans in bivalves. Due to the high variability in terms of previously described structures and methodologies, it is a major challenge to establish glycomic workflows that yield the maximum amount of detailed structural information from relatively low quantities of sample. In this study, we apply differential release with peptide:N-glycosidases F and A followed by solid-phase extraction on graphitized carbon and reversed-phase materials to examine the glycome of Volvarina rubella (C. B. Adams, 1845), a margin snail of the clade Neogastropoda. The resulting four pools of N-glycans were fractionated on a fused core RP-HPLC column and subject to MALDI-TOF MS and MS/MS in conjunction with chemical and enzymatic treatments. In addition, selected N-glycan fractions, as well as O-glycans released by β-elimination, were analyzed by porous graphitized carbon-LC-MS and MSn. This comprehensive approach enabled us to determine a number of novel modifications of protein-linked glycans, including N-methyl-2-aminoethylphosphonate on mannose and N-acetylhexosamine residues, core β1,3-linked mannose, zwitterionic moieties on core Galβ1,4Fuc motifs, additional mannose residues on oligomannosidic glycans, and bisubstituted antennal fucose; furthermore, typical invertebrate N-glycans with sulfate and core fucose residues are present in this gastropod.
Molluscs represent one of the largest groups of animals on the planet; there is an estimated 200,000 species, which vary in morphology from gastropods (snails) through to cephalopods (octopus) and live in a range of marine, aquatic, and terrestrial environments (1). Many molluscs are familiar due to their shells or being seafood. Less appreciated is perhaps their ecological role as filter feeders or scavengers and their being an indicator for water quality (2–4); also, some molluscs are intermediate hosts for pathogens such as viruses or schistosomes (5, 6).
In glycobiological terms, the most studies on molluscs have been structural characterizations of the N-glycans on hemocyanins of a range of gastropods, such as from keyhole limpet (Megathura crenulata; KLH is an often-used carrier protein for immunization), Lymnaea stagnalis, Helix pomatia, and Rapana venosa (7–10). Furthermore, glycans from cephalopod rhodopsins, proteins of bivalves involved in biomineralization, or whole snail viscera have also been analyzed. Including our recent study on the hemocytes and plasma of the eastern oyster (Crassostrea virginica), the variety of modifications of N-glycans in these organisms is immense and includes branched fucose residues, glucuronylation, sulfation, methylation, core xylose, and galactosylation of core fucose as well as LacdiNAc and blood-group-like motifs (11–15). On the other hand, there is only scattered information regarding the biosynthesis of mollusc N-glycan epitopes, based on assay of some fucosyl-, xylosyl-, N-acetylglucosaminyltransferases, and N-acetylgalactosaminyltransferases (16–18); also, probably only two mollusc glycosyltransferases have ever been characterized in recombinant form (19, 20).
The high variability and lack of predictability of mollusc glycomes mean that a suitable glycomic workflow has to be employed that takes account of the maxim “expect the unexpected.” Thereby, in comparison to mammalian glycomes with known major components, the analyses of those of lower eukaryotes can present major challenges. In the past, mollusc glycans from either a single glycoprotein or from tissue were very often analyzed in any single study by one or two methods (e.g. GC-MS and NMR or MALDI-TOF MS/MS of HPLC-fractionated N-glycans, LC-MS/MS of glycopeptides, or GC-MS and MSn of permethylated N-glycans; see references above). In some cases, chemical and enzymatic treatments were employed. Here, we have sought to maximize the potential of off-line MALDI-TOF MS and MS/MS by prefractionating N-glycans first on the basis of whether they can be released by peptide:N-glycosidase A or F (the former being able to remove glycans containing core α1,3-fucose (21)) and then using solid-phase extraction on nonporous graphitized carbon (for an initial separation of anionic from neutral glycans (22)) and on a reversed-phase resin (which aids enrichment of glycans with substitutions of core α1,6-fucose). Subsequent use of a fused core reversed-phase (RP)-HPLC column (23) resulted in high-resolution separation into fractions containing either a single or very few glycan species that facilitated further MS-based analyses; as this RP column offers isomeric/isobaric separation, HPLC fractionation was a prerequisite for the definition of the individual N-glycan structures. Furthermore, the residual glycopeptides (posttreatment with peptide:N-glycosidases) were subject to β-elimination to release the O-glycans followed by LC-MS.
On the basis of these considerations, we have examined the N- and O-glycomes of a margin snail (Volvarina rubella), a species of carnivorous and scavenging marine gastropod first described as Marginella rubella in 1845 (24). Using off-line LC-MALDI-TOF MS and on-line LC-ESI-MS, we reveal a particularly complex N-glycome encompassing a range of oligomannosidic, paucimannosidic, core-modified, and complex (up to triantennary) N-linked oligosaccharides with also a number of anionic and zwitterionic modifications, which are also present on O-glycans. Although some of these features are also found on N-glycans or lipid-linked glycans of other species, the majority of the ∼100 structures are described here for the first time.
EXPERIMENTAL PROCEDURES
Biological Material and Glycan Preparation
Adult margin snails, cultivated on a shrimp-based diet, were obtained from the “Haus des Meeres—Aqua Terra Zoo,” a public aquarium in Vienna, Austria. The species was determined as being V. rubella (C. B. Adams, 1845 Mollusca: Gastropoda: Muricoidea: Marginellidae) (25, 26). The snails (3.4 g wet weight; 1.2 g after lyophilization) were washed once with deionized water and stored at −80 °C. After thawing, the material was heat inactivated for 10 min in boiling water and lyophilized prior to grinding in liquid nitrogen. The powder was suspended in deionized water and the pH adjusted with ammonium carbonate buffer to 8.2. CaCl2 was added to a final concentration of 0.5 mm before addition of 2 mg Thermolysin (Promega, Madison, WI). Proteolysis was allowed to proceed for 2 h at 70 °C.
The glycopeptides were purified using standard laboratory protocols (27) prior to release with PNGase F (Roche) overnight at 37 °C. After cation exchange chromatography on Dowex, the unbound material was subject to solid-phase extraction on nonporous graphitized carbon (NPGC; ENVIcarb, Supelco, Bellefonte, PA) and eluted with 40% acetonitrile1 or 40% acetonitrile with 0.1% TFA. A further solid-phase extraction step was performed on a C18 reversed-phase resin (LiChroprep; Merck, Darmstadt, Germany), and the glycans were eluted with water and with stepwise increases in the methanol concentration (15%, 40%, 100% (v/v)). Glycan-containing fractions, as judged by MALDI-TOF MS, were fluorescently labeled with 2-aminopyridine. The remaining glycopeptides were gel filtrated (Sephadex G25) prior to incubation with PNGase A (recombinant form prepared in house) overnight at 37 °C. The glycans released with this enzyme underwent the same purification and fluorescent-labeling steps as for the PNGase F released ones; however, no anionic glycans were detected in this pool. The residual glycopeptides after PNGase A digestion were subjected to reductive β-elimination with 0.5 m NaBH4 and 50 mm NaOH at 50 °C overnight (see Fig. 1A for workflow summary). Released O-glycans were cleaned up as previously described prior to LC-MS/MS analysis (28).
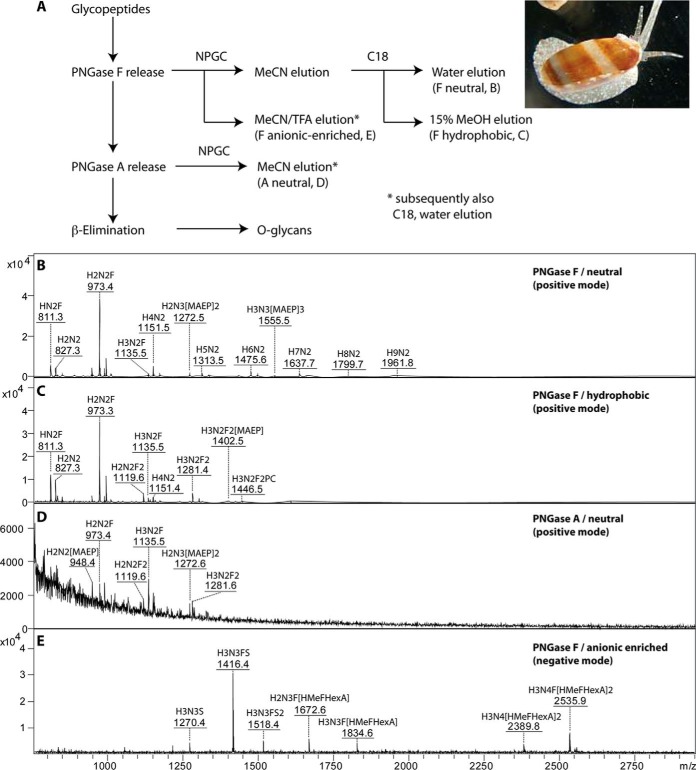
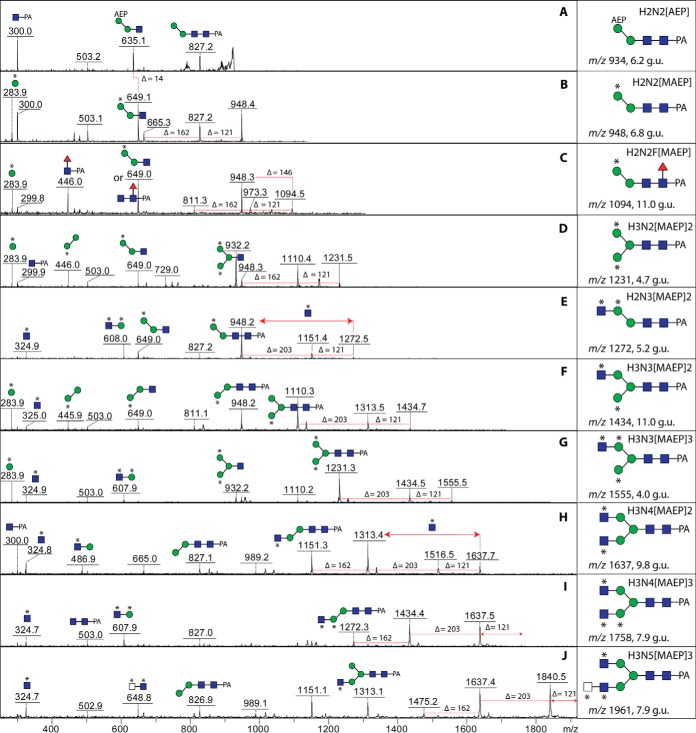
Fig. 1.
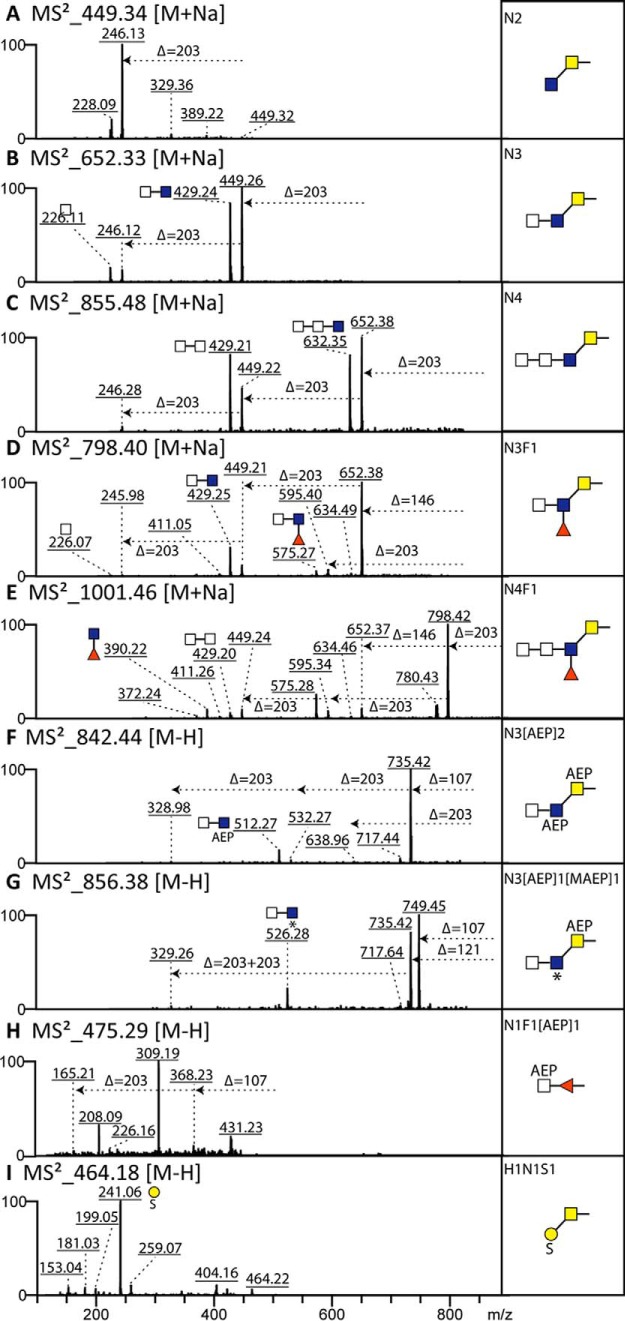
Mass spectrometry of V. rubella N-glycan pools. (A) Glycomic workflow for the analysis of V. rubella N- and O-glycans. (B-E) The four N-glycan pools derived from two-step solid-phase extraction (nonporous graphitized carbon and reversed phase) were pyridylaminated prior to analysis by MALDI-TOF MS in either positive or negative modes. Glycans as [M+H]+ or [M-H]− (in the case of m/z 1518 as [M-2H+Na]−) are annotated with abbreviated compositions and intensities are in arbitrary units: F, fucose; H, hexose; N, N-acetylhexosamine; PC, phosphorylcholine; MAEP, N-methyl-2-aminoethylphosphonate; HMe, methylhexose; HexA, hexuronic acid; S, sulfate. (B) PNGase F neutral, nonporous graphitized carbon (NPGC) acetonitrile (MeCN) elution followed by C18 water elution; (C) PNGase F hydrophobic, NPGC acetonitrile elution followed by C18 15% methanol elution; (D) PNGase A, NPGC acetonitrile elution followed by C18 water elution; (E) PNGase F anionic-enriched, nonporous graphitized carbon (NPGC) acetonitrile/TFA elution followed by C18 water elution.
N-glycan Fractionation
The pyridylaminated N-glycans were fractionated by HPLC using a Kinetex 5 μm RP-column (XB-C18 100A, 250 × 4.6 mm; Phenomenex®, Torrance, CA), and a gradient of methanol in 0.1 m ammonium acetate, pH 4, up to 16.5% over 44 min was applied at a flow rate of 0.8 ml/min as follows: 0–30 min, 0–9% methanol; 30–35 min, 9–12% methanol; 35–40 min, 12–16.5% methanol; 40–44 min, 16.5% methanol; and 44–50 min, return to 0% methanol. Lyophilized HPLC fractions were dissolved in water and subject to MALDI-TOF MS. A partial dextran hydrolysate (Sigma-Aldrich, St. Louis, MO; subsequently pyridylaminated) was used to calibrate the RP-HPLC column in terms of glucose units (g.u.) as previously described (29); the masses of selected peaks of this linear glucose polymer were verified by MALDI-TOF MS after RP-HPLC (e.g. 6 and 10 g.u. eluting at 20 and 26 mins; m/z 1069.6 and 1717.8 as [M+H]+).
MALDI-TOF Mass Spectrometry
Free glycans and pyridylaminated glycans were analyzed in positive and negative-ion modes using either Bruker Autoflex Speed or UltrafleXtreme instruments (both with 1000 Hz Smartbeam™-II lasers) with 6-aza-2-thiothymine as matrix; calibration was performed using a Bruker peptide standard. MS/MS was performed by laser-induced dissociation (precursor ion selector was generally ±0.6%). The detector voltage was normally set at 1977 V for MS and 2133 V for MS/MS; 1000–2000 shots from different regions of the sample spots were summed. Spectra were processed with the manufacturer's software (Bruker Flexanalysis 3.3.80) using the SNAP algorithm with a signal/noise threshold of 6 for MS (unsmoothed) and 3 for MS/MS (four-times smoothed). Glycan MS and MS/MS spectra (∼2700 in total) were manually interpreted on the basis of the masses of the predicted component monosaccharides, the differences of mass in glycan series, fragmentation patterns, and results of enzymatic and chemical treatments. For the ∼100 structures (see the Supplemental Table 1 for a summary of evidence for each glycan), the minimum criterion for inclusion in the Supplemental Table 1 was an interpretable MALDI-TOF MS/MS spectrum. Furthermore, examples for each core and antennal motif were verified by digestion data; comparison was also made to elution, in terms of glucose units, with previous data on glycans of other species. About 20 representative structures are also supported by LC-MSn data. Calculated theoretical masses were verified using GlycoWorkbench 2.0. The deviation between calculated and observed m/z values was typically 0.1–0.2 Da.
LC-ESI Mass Spectrometry
Selected RP-HPLC fractions of pyridylaminated N-glycans were also analyzed by online LC-MS/MS using a 10 cm × 150 μm inner diameter column, prepared in-house, containing 5 μm porous graphitized carbon (PGC) particles coupled to an LTQ ion trap mass spectrometer (Thermo Scientific, Waltham, MA). Glycans were eluted using a linear gradient from 0–40% acetonitrile in 10 mm ammonium bicarbonate over 40 min at a flow rate of 10 μl/min. The eluted N-glycans were detected in negative-ion mode with an electrospray voltage of 3.5 kV, capillary voltage of -33.0 V, and capillary temperature of 300 °C. Specified ions were isolated for MSn fragmentation by collision-induced dissociation with the collision energy set to 30%. Air was used as a sheath gas, and mass ranges were defined dependent on the specific structure to be analyzed. The data were processed using the Xcalibur software (version 2.0.7, Thermo Scientific). Glycans were identified from their MS/MS spectra by manual annotation according to the nomenclature of Domon and Costello (30).
Exoglycosidase and Hydrofluoric Acid Treatment
Aliquots of the isolated HPLC fractions were, based on results of HPLC elution and MALDI-TOF MS and MS/MS data, subject to targeted exoglycosidase digestion and chemical treatment. Either α-mannosidases (jack bean from Sigma, Aspergillus α1,2-specific from Prozyme (Hayward, CA), Xanthomonas α1,2/3-specific from NEB or Xanthomonas α1,6-specific from NEB (Ipswich, MA)), β-mannosidase (H. pomatia from Sigma), α-fucosidases (bovine kidney α1,6-specific from Sigma, Corynebacterium α1,2-specific from Takara (Shiga, Japan), or microbial α1,2-specific from Megazyme (Bray, Ireland)), β-galactosidases (recombinant Aspergillus niger or oryzae; prepared in-house (31), Xanthomonas β1,3-specific from NEB or Bacillus fragilis β1,4-specific from NEB), α-galactosidase (green coffee bean from Sigma), or β-hexosaminidases (recombinant Apis mellifera FDL1, prepared in-house (32), or jack bean from Sigma) were used for further treatment of the sample in 25 mm ammonium acetate, pH 5 (pH 8 for α1,2-fucosidase from Takara), at 37 °C for 24 or 48 h (or 2 h for FDL). The FDL (fused lobes) hexosaminidase is under these conditions specific for the β1,2-GlcNAc linked to the α1,3-mannose residue of the trimannosyl N-glycan core.
For removal of methylaminoethylphosphonate, phosphorylcholine, or branched fucose residues, selected fractions were dried and incubated for 48 h at 0 °C with 3 μl 48% (v/v) hydrofluoric acid prior to evaporation in a centrifugal concentrator (33). The samples were diluted in water and re-evaporated, before redissolving once again. The chemically or enzymatically treated fractions were subject to MALDI-TOF MS and MS/MS (as above) without further purification.
RESULTS
Overall N-glycome of a Marine Margin Snail
The analytical strategy for V. rubella was based on selective release and fractionation of different classes of N-glycans from thermolysin-generated glycopeptides. We, thereby, modified a procedure we previously used for Dictyostelium N-glycans (22). Using two different peptide:N-glycosidases (PNGase F then PNGase A; the latter able to remove glycans with core α1,3-fucose) and by solid-phase extraction on nonporous graphitized carbon and reversed-phase resin, we obtained a total of four different pools of N-glycans (PNGase A-released and “neutral,” “hydrophobic,” and “anionic-enriched” PNGase F-released fractions; see also Fig. 1A). The free glycan pools were separately fluorescently labeled with 2-aminopyridine prior to MALDI-TOF MS and HPLC.
The first impression of the PNGase A and neutral-enriched PNGase F digests (both eluted with acetonitrile from the graphitized carbon and with water from the C18 column) is of a set of paucimannosidic and oligomannosidic N-glycans typical for invertebrates (Hex2–9HexNAc2 and Hex1–3HexNAc2Fuc1–2; Figs. 1B and 1D). The PNGase A pool contains relatively low amounts of difucosylated species (m/z 1119 and 1281 as [M+H]+); preliminary MS/MS of glycans in the entire pool, confirmed later on individual fractions, showed the occurrence of m/z 592 Y1-fragments suggestive for disubstitution of the core GlcNAc by both α1,3 and α1,6-fucose. Additionally, the overall mass spectra of both of these pools indicate the presence of glycans whose masses (m/z 948, 1272, and 1555 as [M+H]+) could not be accounted for by typical monosaccharide compositions. Only in later experiments did it become clear that these are carrying the zwitterionic N-methyl-2-aminoethylphosphonate moiety (Δm/z 121).
The hydrophobic neutral PNGase F-released glycans (eluted with 15% methanol from the C18 solid-phase material) were primarily fucosylated (Fig. 1C). Unexpected also, difucosylated glycans were present in this pool (also of m/z 1119 and 1281 as [M+H]+); preliminary MS/MS, however, showed the presence of m/z 754 Y1-fragments indicative of a structural difference as compared with the isomeric glycans in the PNGase A pool. Two unusual difucosylated glycans with zwitterionic modifications (as later determined; m/z 1402 and 1446 as [M+H]+) were also obvious in this pool.
Finally, the anionic-enriched PNGase F-released pool that was eluted with acetonitrile supplemented with 0.1% trifluoroacetic acid from graphitized carbon and then with water from the C18 column contained two basic types of N-glycans (Fig. 1E): sulfated species (m/z 1270, 1416, and 1518 as [M-H]− or [M-2H+Na]−) and glycans of m/z 1672, 1834, 2389, and 2536 estimated to contain a modification of 498 Da, which, as these are anionic species easily detected in the negative mode, could correspond to a composition of Hex1Me1Fuc1HexA1. In order to reveal more about the structures of the glycans in this and the other pools, they were applied to a fused core reversed-phase HPLC column (Kinetex XB-C18), which was chosen due to its increased resolution as compared with other RP-HPLC columns in a recent study (33). All fractions were then subject to MALDI-TOF MS and MS/MS analyses in conjunction with chemical and enzymatic treatments; example structures were also analyzed by LC-MSn. Some of the observed structures can, in terms of elution time (glucose units, g.u.), fragmentation patterns, and digestion data (see also Supplemental Table), be directly compared with those in our recent study on N-glycans of the nematode Pristionchus pacificus (33). Thereby, the ability of RP columns to distinguish isomers of oligomannosidic or core fucosylated glycans or to separate glycans that only differ in terms of which antenna is elongated is in accordance to previously published data (22, 29, 34, 35). In the case of acidic glycans, the decrease in RP-HPLC retention time correlates with the number of anionic moieties (36, 37).
Smaller Mono- and Difucosylated N-glycans
As mentioned above, a number of lower molecular mass glycans carrying one or two fucose residues were observed in the overall MALDI-TOF MS spectra of the PNGase A (neutral) and PNGase F (hydrophobic) pools. RP-HPLC of the PNGase A pool resulted in separation of glycans across a range of elution times (Fig. 2A); the early eluting monofucosylated species (5.2–5.5 g.u.; Hex2–3HexNAc2Fuc1; m/z 973 and 1135 as [M+H]+) were concluded to be core α1,3-fucosylated due to the comparison to Pristionchus N-glycans (33) and the general trend for this modification to result in lower retention on RP-HPLC columns (35). The most dominant peak on the RP-HPLC of this pool (8.4 g.u.) contained difucosylated glycans (Hex2–3HexNAc2Fuc2; m/z 1119 and 1281), which yielded MS/MS spectra showing the fragment of m/z 592, Fuc2GlcNAc1-PA, being indicative of difucosylation of the core GlcNAc-PA (Fig. 3A). Furthermore, the elution time was compatible to previous data for such glycans on the same column (33) with a retention time between those of glycans with solely core α1,3- or α1,6-fucose (38); the linkage of the mannose residues was verified using specific α1,2/3- or α1,6-mannosidases (see Supplemental Table).
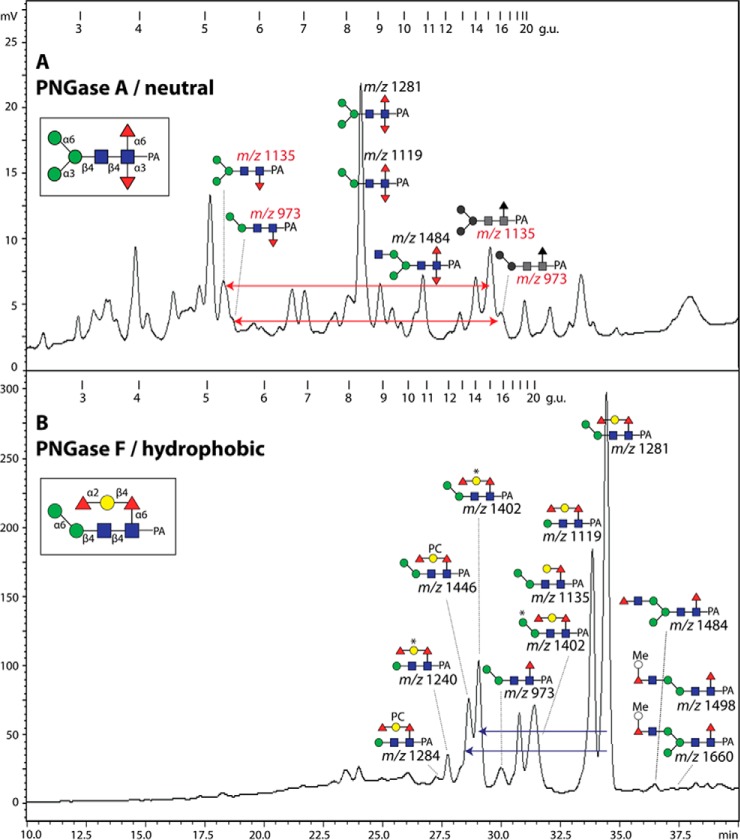
Fig. 2.
RP-HPLC of PNGase A neutral and PNGase F hydrophobic N-glycan pools from V. rubella. (A) PNGase A released glycan pool fractionated on the Kinetex XB-C18 RP-HPLC column; only those fucosylated glycans specific to this pool are annotated according to the nomenclature of the Consortium for Functional Glycomics. The remaining peaks in this chromatogram are due to remnants of standard oligomannosidic and core α1,6-fucosylated glycans which are otherwise dominating the PNGase F neutral pool. (B) PNGase F released hydrophobic glycan pool fractionated on the Kinetex XB-C18 RP-HPLC column with annotations, whereby an asterisk (*) indicates the presence of methylaminoethylphosphonate, PC phosphorylcholine and Me a methyl group. The glycans were all in their pyridylaminated (PA) forms and example structures with all linkages are shown in insets. The external calibration with glucose units (3–20 g.u.) is indicated and the fluorescence intensities (320/400 nm) are shown in terms of mV. The red arrows in panel A indicate the shift in retention of the core α1,3- as opposed to the core α1,6-fucosylated isomeric structures; the blue arrows in panel B show the effect on elution of the zwitterionic modifications. The two structures in gray (16 g.u.) indicate the elution positions of the core α1,6-fucosylated isomers in the PNGase F digest.
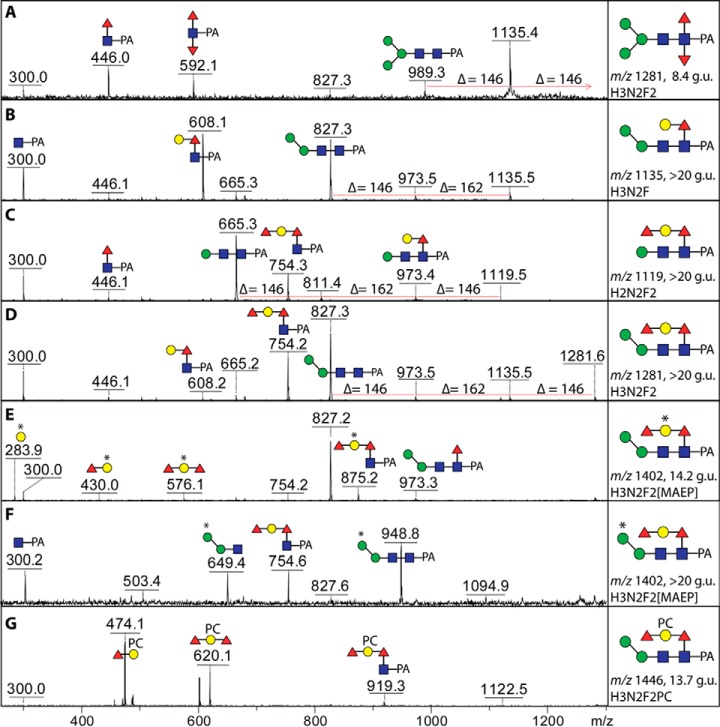
Fig. 3.
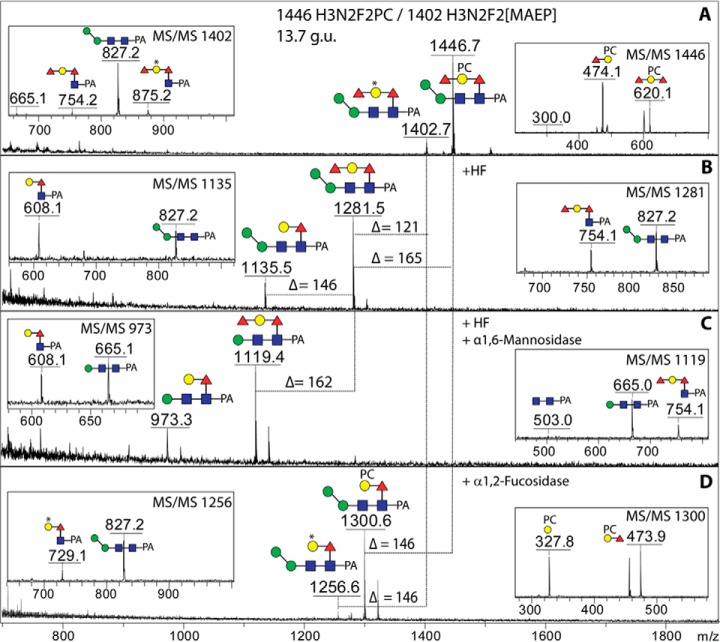
MS/MS of N-glycans with various modifications of core fucose. Positive-ion mode MALDI-TOF MS/MS spectra of the protonated pyridylaminated N-glycans from either the PNGase A (A) or PNGase F hydrophobic (B-G) pools are annotated with the predicted structures of the selected fragments. The major fragments are either due to loss of the fucose modification on the reducing-terminal GlcNAc (e.g. m/z 665 in C and 827 in B, D, and E) or are Y1-ions with the entire modification on the GlcNAc-PA (m/z 592, 608 and 754 in A, B, C, D, and F); otherwise, the core fragment lacking the GlcNAc-PA or a B-fragment is only visible if a zwitterionic modification (methylaminoethylphosphonate, * or MAEP, or phosphorylcholine, PC) is present (e.g. m/z 576, 649, and 620 in E, F, and G). Note there are two isomeric glycans of m/z 1281 and m/z 1402 of different retention time showing alternative positions of either the second fucose and a hexose (A and D) or of the methylaminoethylphosphonate modification (E and F).
As expected from the solid-phase C18 elution properties, the hydrophobic PNGase F-released pool contained rather late-eluting glycans (9 g.u. and above; Fig. 2B). A minor amount of monofucosylated glycans carrying core α1,6-fucose was present; among these, fragmentation resulted not only in observation of m/z 446 (Fuc1GlcNAc1-PA) but also of m/z 608 fragment ions (Hex1Fuc1GlcNAc1-PA; Fig. 3B) of a type observed with nematode N-glycans previously shown to have β1,4-galactosylated core α1,6-fucose (33, 39) and verified by glycosidase digestions (see Supplemental Table). The tendency of core α1,6-fucose to result in late elution on RP-HPLC first noted by Tomiya et al. (29) and also observed for in vitro products of the relevant core fucosyltransferase (40) is even more pronounced for glycans with galactose substitutions of the fucose residue.
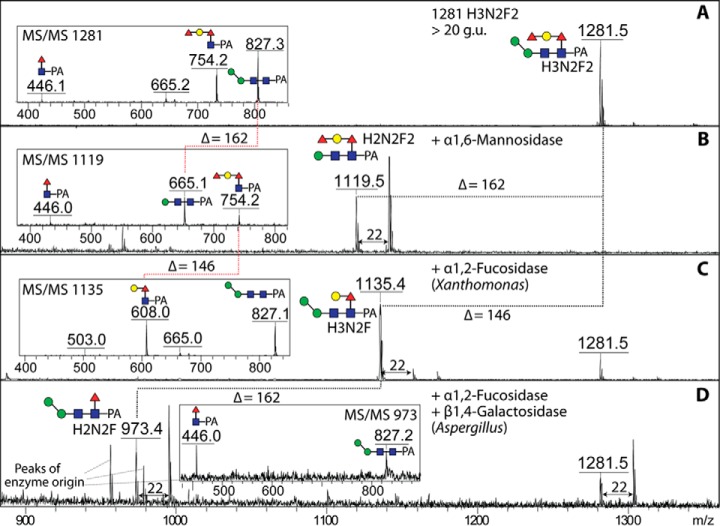
The majority of glycans, though, in the hydrophobic pool were difucosylated. However, as this is a PNGase F-released pool, the typical core difucosylation was not expected. Indeed, MS/MS of these glycans did not reveal the presence of any m/z 592 fragments (Fuc2GlcNAc1-PA; see above): on the other hand, fragments of m/z 754 (Figs. 3C, 3D, and 3F), reminiscent of ones previously found in Caenorhabditis elegans (39), suggested that core α1,6-fucose was capped with a hexose and a fucose. In the case of Hex3HexNAc2Fuc2-PA (>20 g.u.; m/z 1281), use of a specific mannosidase proved that the single α-mannose is 1,6-linked and led to loss of the m/z 827 MS/MS fragment (Fig. 4B), whereas elucidation of the extended core modification was based on other glycosidase digests: α1,2-fucosidase removed one residue and resulted in loss of the m/z 754 fragment and the gain of one at m/z 608 (Fig. 4C). Subsequent digestion with a recombinant fungal β1,4-preferring galactosidase (31) resulted in loss of a further residue and another shift in the MS/MS pattern yielding a fragment of m/z 446 (Fig. 4D). Thus, this form of Hex3HexNAc2Fuc2 is concluded to carry a Fucα1,2Galβ1,4Fucα1,6-modification on the core proximal GlcNAc residue. Some glycans in the anionic pool with antennal modifications also possessed these extended core fucose modifications (see below).
Fig. 4.
Exoglycosidase digestion of a core modified N-glycan. The pyridylaminated glycan of m/z 1281 from the hydrophobic PNGase F-released pool (A; see also Fig. 3D for full MS/MS) was treated with either α1,6-mannosidase (B), α1,2-fucosidase (C) or a combination of α1,2-fucosidase and β1,4-galactosidase (D) prior to positive-ion mode MALDI-TOF MS. Insets show the changes in fragmentation patterns of the core region upon digestion. Although the sodiated ions were dominant in two digests (shown by Δ = 22), the protonated forms were those subject to MS/MS.
Five N-glycans in the hydrophobic pool had compositions that suggested the presence of a nonsugar modification. The mass difference of 165 Da between the glycans of m/z 1284 and 1446 as compared with those of m/z 1119 and 1281 was suggestive for the presence of phosphorylcholine, which is also a known modification of nematode N-glycans (33). Our standard procedure to analyze glycans modified with a phosphodiester is to treat them with hydrofluoric acid (22); in the case of the m/z 1446 glycan (Hex3HexNAc2Fuc2PC1-PA; 13.7 g.u.), there was indeed a loss of 165 Da, and the unusual fragments of m/z 474 and 620 (interpreted as being Fuc1–2Hex1PC1; Fig. 3G) were no longer observed (compare MS/MS of m/z 1446 and m/z 1281 in Figs. 5A and 5B). Some loss of a fucose residue also occurred, which appeared to be due to concomitant removal of the terminal core α1,2-fucose from the galactosylated core α1,6-fucose. Specific mannosidase and fucosidase digests aided the definition of the structure (Figs. 5C and 5D); based on the m/z 328 fragment (Hex1PC1; MS/MS of m/z 1300) after fucosidase treatment, we conclude that phosphorylcholine is linked to the galactose residue of the core modification. Interestingly, the glycan of overlapping elution (m/z 1402, mainly eluting in the next fraction of 14.2 g.u.; Fig. 2B) was also sensitive to the same treatments and the loss of 121 Da after hydrofluoric acid treatment was an indication for the presence of a phosphoester; further examples of glycans, including two isomers of m/z 1402 (Figs. 3E and 3F), with this 121 Da modification are discussed below.
Fig. 5.
Chemical and enzymatic digestion of core modified zwitterionic N-glycans. The pyridylaminated glycans of m/z 1402 and 1446 from the hydrophobic PNGase F-released pool (A; see Fig. 3E and 3G for the full MS/MS) were subject to hydrofluoric acid treatment (B) followed by α1,6-mannosidase treatment (C) or to microbial α1,2-fucosidase treatment alone (D) prior to positive-ion mode MALDI-TOF MS. Hydrofluoric acid (HF) removes the zwitterionic modifications (phosphorylcholine or methylaminoethylphosphonate) fully but only partially removes the α1,2-fucose, which on the other hand is released to a higher degree with the bovine fucosidase (not shown) and completely by the α1,2-fucosidase. Shifts in m/z as well as segments of the MS/MS spectra are shown.
Standard Paucimannosidic Glycans
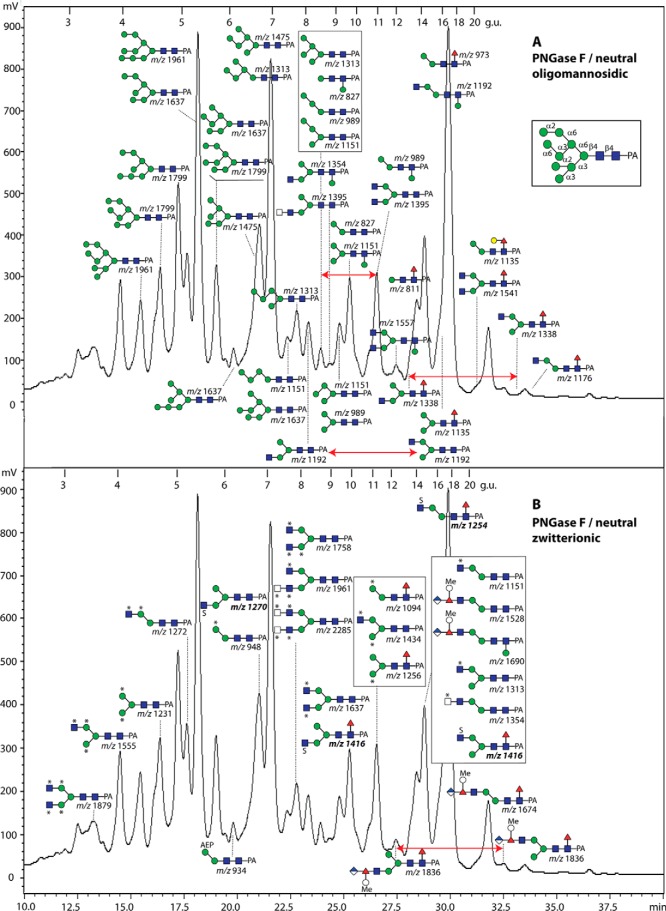
The largest pool of glycans in terms of total fluorescence was of the neutral-enriched PNGase F digest. RP-HPLC of this pool resulted in some 40 glycan-containing fractions (Fig. 6). The later-eluting RP-HPLC fractions (>11 g.u.) contain some typical monofucosylated species (Hex1–3HexNAc2–4Fuc1-PA), which display major MS/MS protonated fragments at m/z 446 (GlcNAc1Fuc1-PA). The elution times are indicative of α1,6-fucosylation as compared with previous studies using the same column and gradient (33) and literature values for such glycans (29). Based on its retention time, the major m/z 973 glycan (16 g.u.) was assumed to be Manα1,6Manβ1,4GlcNAcβ1,4(Fucα1,6)GlcNAc-PA. Supporting this assignment, α1,6-mannosidase and bovine α-fucosidase removed one residue each to result in products of m/z 811 and 827, respectively (see Supplemental Table). Furthermore, there are minor amounts of Hex3HexNAc3Fuc0–1-PA glycans (m/z 1192 and 1338) with the third HexNAc residue on either the α1,3- or α1,6-mannose residue as judged by the retention times in comparison to previous studies (33) as well as the α1,2/3-mannosidase and FDL hexosaminidase sensitivities (see Supplemental Table). The contrasting retention times resulting from extension of the different antennae are exemplified (red arrows, Fig. 6A) for the glycans of m/z 1192, 1338, and 1395, with elongation of the α1,3-arm causing earlier elution as compared with the effect of substitution of the α1,6-arm (41).
Fig. 6.
RP-HPLC of the PNGase F neutral-enriched pool of N-glycans from V. rubella. The glycans in the respective acetonitrile and water elutions from the serial NPGC and C18 solid-phase extraction steps (see Fig. 1) were applied to the Kinetex XB-C18 column; both chromatograms are of the same run but for simplicity are annotated respectively with the (A) oligo- and paucimannosidic or (B) zwitterionic and monoacidic pyridylaminated glycan structures. Annotated glycans in the same fraction are in their order of abundance (uppermost being more abundant as judged by MALDI-TOF MS of the individual fraction). An example anomalous oligomannosidic structure annotated with the linkages is shown in the inset; m/z values in bold are structures solely observed in negative-ion mode; *, methylaminoethylphosphonate; Me, methyl; S, sulfate. The external calibration with glucose units (3–20 g.u.) is indicated and the fluorescence intensities (320/400 nm) are shown in terms of mV. The red arrows indicate the contrasting retention times of m/z 1192, 1338, 1395, and 1836 resulting from extension of the different antennae, with elongation of the α1,3-arm causing earlier elution as compared with the effect of substitution of the α1,6-arm.
Typical also of invertebrates are nonfucosylated paucimannosidic glycans such as Man2–5GlcNAc2; other than m/z 827 glycan (Man2GlcNAc2-PA; at 9.8 g.u.), being also sensitive to α1,6-mannosidase and so lacking the core α1,3-mannose, additional glycans of this class were of rather low amounts as compared with insects and nematodes. On the other hand, one of the three major peaks contained a glycan with m/z 1313 (Hex5HexNAc2-PA) at an unusual elution position (7.2 rather than 8.5 g.u. for the standard isomer) and was proven by LC-MSn to also have no lower arm α1,3-mannose (see below). The tendency for glycans of this gastropod to lack this residue is also shared with monoantennary glycans with “complex” antennae (e.g. m/z 1528 and 1674 in the anionic pool; see below).
Unusual Isomers of Oligomannosidic Glycans
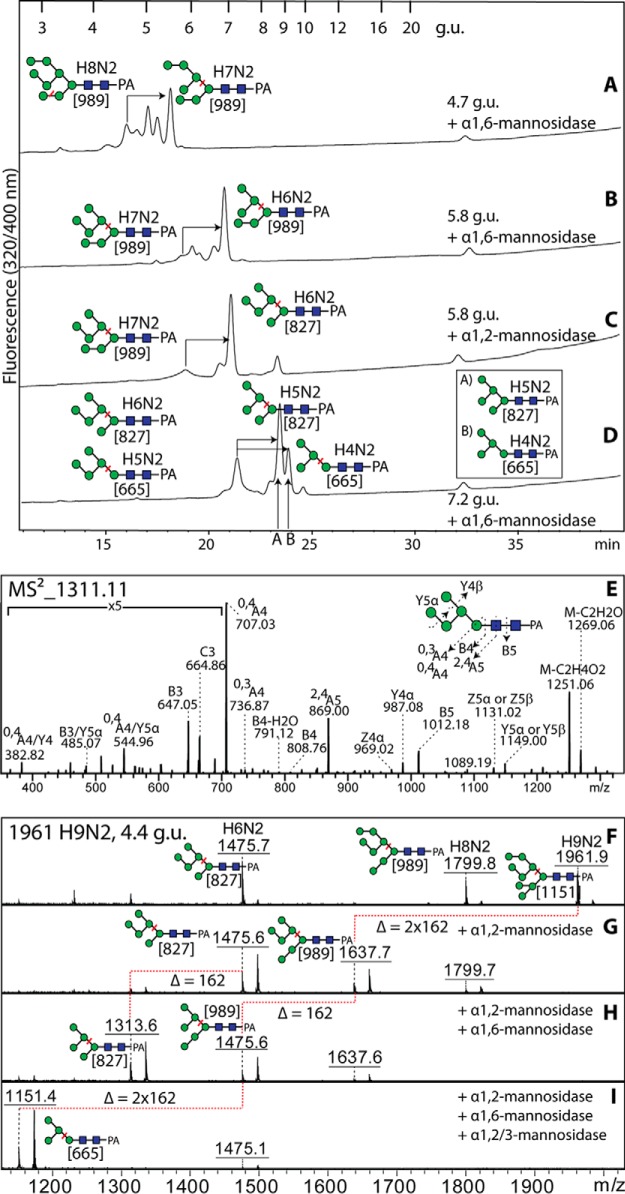
In addition to fractions in the neutral-enriched PNGase F pool (4–9 g.u.; Fig. 6A) containing the normal oligo- and paucimannosidic glycans (Hex3–9HexNAc2-PA; m/z 989–1961, [M+H]+), as based on their coelution with known oligomannosidic glycans and similar fragmentation (see Supplemental Table), the multiplicity of further elution positions for these masses raised suspicions as to the exact compositions and structures of many of these glycans. For instance, there were three elution positions for glycans with m/z 1799 (Hex8HexNAc2-PA; 4.7, 5.0, and 5.9 g.u.). The 5.0 and 5.9 g.u. elution positions would correspond to the standard Man8B and Man8A isomers (respectively, with the “middle” or “lower” mannoses of Man9 having been removed by endoplasmic reticulum mannosidase or by a Golgi mannosidase I) as found in Dictyostelium or Pristionchus (22, 33). The earlier eluting form of Hex8HexNAc2-PA (4.7 g.u.), however, does not correspond to any standard isomer. After proving loss of seven residues with jack bean α-mannosidase, this glycan was digested with linkage-specific mannosidases. Surprisingly, the Xanthomonas α1,6-mannosidase removed one mannose, resulting in a major product of m/z 1637 coeluting with a standard form of Man7GlcNAc2-PA (5.4 g.u.; still with m/z 989 as the major MS/MS Y3β fragment that correlates, in our hands, with the length of the lower arm; Fig. 7A). Based on the specificity of this enzyme (42) and our experience that it otherwise does not digest any standard oligomannosidic glycan without pretreatment with other specific mannosidases (43), it is concluded that the 4.7 g.u. glycan has a terminal α1,6-mannose linked to an unsubstituted mannose.
Fig. 7.
Analysis of unusual oligomannosidic N-glycans by HPLC and LC-MS/MS. (A-D) Aliquots of the 4.7, 5.8, and 7.2 g.u. fractions (see Fig. 6A) were digested with α1,2- or α1,6-specific mannosidases prior to being rechromatographed by RP-HPLC; the shifts in elution time with respect to the original nondigested structures are indicated by arrows and the structures of the original and digested structures are shown, together with the m/z of the key Y3 positive-ion mode MALDI-TOF MS/MS fragment (665, 827, or 989 in square brackets), respectively, on the left- and right-hand side of the product HPLC peak; coelution with standard Man5GlcNAc2 or a Man4GlcNAc2 (prepared by an α1,2/3-mannosidase digest of standard Man5GlcNAc2; see structures in inset) is indicated with the letters A and B. (E) LC-MS2 in negative-ion mode of an unusual Man5GlcNAc2 isomer (7.2 g.u.); MS3 verified the position of the α1,6-mannose (see text). (F-I) MALDI-TOF MS analysis of the 4.4 g.u. fraction containing unusual Man8,9GlcNAc2 isomers before and after digestion with specific mannosidases; the coeluting Man6GlcNAc2 glycan is an earlier-eluting epimer (core ManNAc-PA as reduction artifact) of the structure in the major 7.2 g.u. fraction.
For an aberrantly eluting form of Hex7HexNAc2-PA (m/z 1637; 5.8 g.u.), treatment with both α1,6- or α1,2-specific mannosidases was performed. While both mannosidases removed one mannose from the glycan (Figs. 7B and 7C), the shift in the major MS/MS Y3β-fragment from m/z 989 to m/z 827 was only observed with the α1,2-specific mannosidase. These data were considered to indicate that the accessible α1,6-mannose linked to an unsubstituted mannose was on the upper or middle arms, whereas the α1,2-mannose is on the lower arm; this would rule out that the α1,6-mannose is a lower arm “outer chain” modification as in fungi (43). Final examples of the unusual oligomannosidic glycans are forms of Hex5–6HexNAc2 (7.2 g.u.), which were also susceptible to α1,6-mannosidase digestion and shifted to coeluting with known Man4–5GlcNAc2-PA (two peaks of around 9 g.u. with no alteration in the major MS/MS fragment; Fig. 7D). The Hex5–6HexNAc2 glycans were, on the other hand, resistant to α1,2-mannosidase but lost in total four or five residues upon jack bean α-mannosidase digestion; however, only the Hex6HexNAc2 was sensitive to α1,2/3-mannosidase, compatible with its fragmentation showing the presence of a lower arm (see Supplemental Table).
In order to locate the extra α1,6-mannose residue more exactly, LC-ESI-MS/MS in the negative-ion mode was performed on one of these glycans (Man5GlcNAc2-PA; [M-H]− ions at m/z 1311; Fig. 7E), which is proposed to have an α1,6-mannose linked to the middle or upper arm and be devoid of the lower arm (α1,3-mannose). Fragmentation ions at m/z 869 and 1012 were assigned, respectively, as products of the 2,4A cross-ring cleavage of the penultimate GlcNAc (2,4A5) and glycosidic cleavage within the chitobiose (B5). Cross-ring cleavages of β-mannose (0,3A4 ions at m/z 737 and 0,4A4 ions at m/z 707) together with “D ions” at m/z 809/791 (mass of 6-antenna plus β-Man (44) indicate that this N-glycan is devoid of a lower arm but has four mannoses on the upper arm. The presence of B3/C3 ions at m/z 665/647 but the lack of ions at m/z 503 (mass of three mannoses plus H2O) suggests that the four mannoses are branched rather than linear. The presence of fragment ions at m/z 485 but none at m/z 467 suggests that two mannoses are linked to the C3 of the α1,6-mannose (also confirmed by MS3, data not shown) while only one mannose is linked to the C6 of the α1,6-mannose. Thus, this verifies that the free α1,6-mannose is on the middle (B) arm. Analogously, a form of Man4GlcNAc2 as well as another Man5GlcNAc2 isomer also possessed this “middle” α1,6-mannose as judged by α1,6-mannosidase digests (see Supplemental Table).
A further unusual oligomannosidic modification is the presence of an additional α1,3-mannose on the lower arm of two early-eluting Man8–9GlcNAc2 isomers. Specific mannosidases were employed to serially digest these structures. First Aspergillus α1,2-mannosidase removed one or two residues with a shift in the fragmentation pattern and α1,6-mannosidase a further residue (Figs. 7F-7H). Finally, α1,2/3-mannosidase treatment resulted in a product of m/z 1151 (Fig. 7I) displaying a major Y3β-fragment ion of m/z 665, suggesting that loss of the lower arm only occurred when a mannosidase capable of hydrolyzing α1,3-linkages was employed.
Mannosylation of the Reducing Terminus
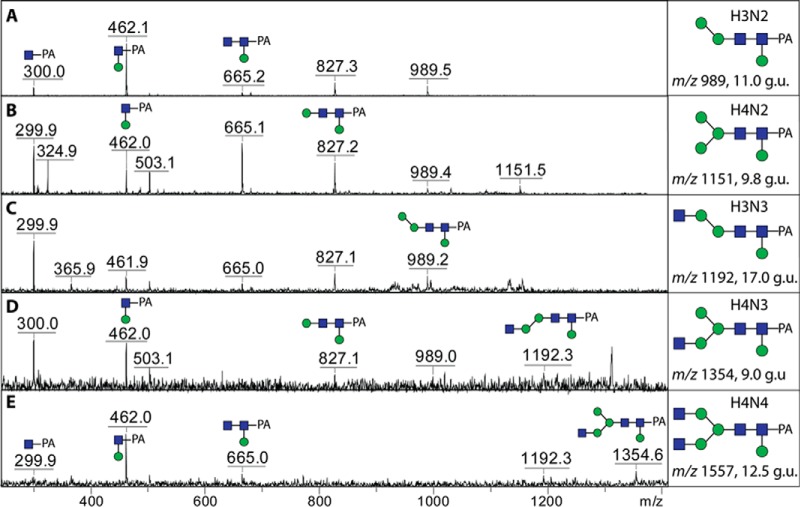
Another unexpected feature of the neutral-enriched PNGase F pool were glycans with major MS/MS Y1-fragment ions at m/z 462 (Hex1GlcNAc1-PA, Fig. 8); as these glycans contain two core GlcNAc residues, it can be ruled out that this fragment results from an endoglycosidase product. We have previously observed a similar feature for glycans from the eastern oyster and assumed this may be indicative of a hexose attached to the reducing terminal GlcNAc residue, but the low amounts precluded a fuller analysis (12); some of these oyster glycans were also core α1,6-fucosylated, so the assumption was that the “extra” core hexose was 1,3-linked. However, due to the larger amounts of such core hexosylated glycans in the margin snail, we were able to test their sensitivity to a number of hexosidases (α- and β-galactosidases and α- and β-mannosidases); incubations of an Hex3HexNAc2-PA (m/z 989 glycan; 11 g.u.) with Xanthomonas α1,6-mannosidase and H. pomatia β-mannosidase both removed one hexose residue, but only the latter treatment resulted in a loss of the m/z 462 fragment (Figs. 9 A-9C). Thus, we assumed (as the 2 and 4 positions of the proximal core GlcNAc are substituted with an N-acetyl moiety and the distal GlcNAc) that, in analogy to the oyster glycans, this β-mannose is 3-linked. In contrast to the oyster, though, the core hexose residue in the snail is present on some hybrid and biantennary glycans lacking core fucose. Interestingly, the effect of core β1,3-mannosylation on retention time is relatively minor (a shift of ca. 1 g.u. later than the “parent” structure) as compared with core α1,3-fucosylation (ca. 4 g.u. earlier). Furthermore, the presence of the core β1,3-mannosylated glycans in the PNGase F pool, as opposed to the absence of core α1,3-fucosylated ones, means that the action of PNGase F is not inhibited per se by a 3-substitution of the reducing terminal GlcNAc.
Fig. 8.
MALDI-TOF MS/MS of core mannosylated N-glycans. The positive-ion mode MALDI-TOF MS/MS spectra of selected glycans displaying an m/z 462 fragment (Hex1HexNAc1-PA) are shown along with the putative structures; their elution positions are given in glucose units (see also Fig. 6A). The digestion of the Hex3HexNAc2-PA glycan (m/z 989; A) with mannosidases is shown in Fig. 9 along with LC-MSn spectra.
Fig. 9.
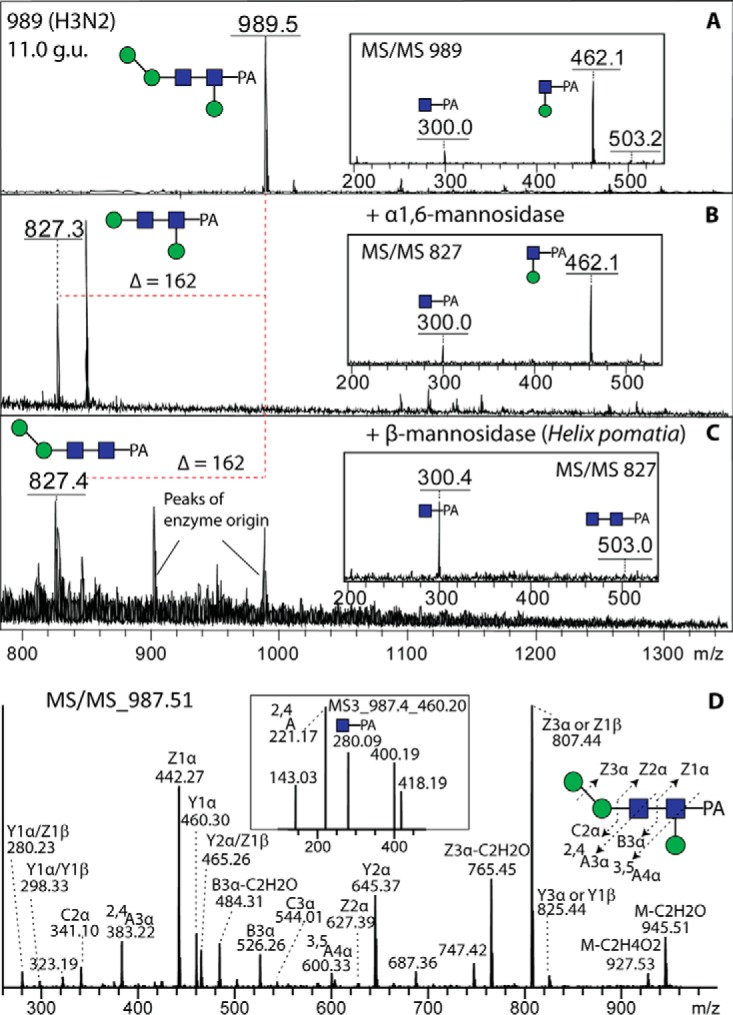
Determination of the β1,3-mannose linkage by MALDI-TOF MS and LC-MS. An anomalous Hex3HexNAc2-PA glycan (11.0 g.u., see Figs. 6A and 8A) was digested with specific α1,6- and β-mannosidases (B and C) prior to positive-ion mode MALDI-TOF MS. Alterations in the core fragments (i.e. the effect on the m/z 462 fragment) are shown in the insets. The same glycan was also subject to LC-MSn in negative-ion mode (D); MS3 of the m/z 460 fragment ion (inset) reveals a 2,4A cross-ring fragment supportive of the β1,3-mannose modification of the reducing terminal GlcNAc-PA.
A specific proof for β1,3-mannosylation in V. rubella was delivered by LC-ESI-MS/MS data of Hex3HexNAc2-PA (Fig. 9D). In negative-ion mode, the MS/MS spectra were dominated with Y and Z ions. The presence of Y1α/Z1α ions at m/z 460 and 442, together with B3α ions at m/z 526, indicates that one hexose is linked to the proximal GlcNAc. Fragmentation ions at m/z 600 were interpreted as being derived from 3,5A cleavage of the proximal GlcNAc, indicating that the hexose is linked to the C3 position of this residue, whereas those at m/z 221 in MS3 of Y1α are compatible with 2,4A cleavage of the proximal GlcNAc, which further confirms the C3 substitution of this residue. Taken together, these data indicate that this N-glycan from V. rubella is substituted with β1,3-mannose on the proximal core GlcNAc. This modification is also present on some anionic glycans with an antennal branched fucose residue (e.g. m/z 1690 and 1852; see below).
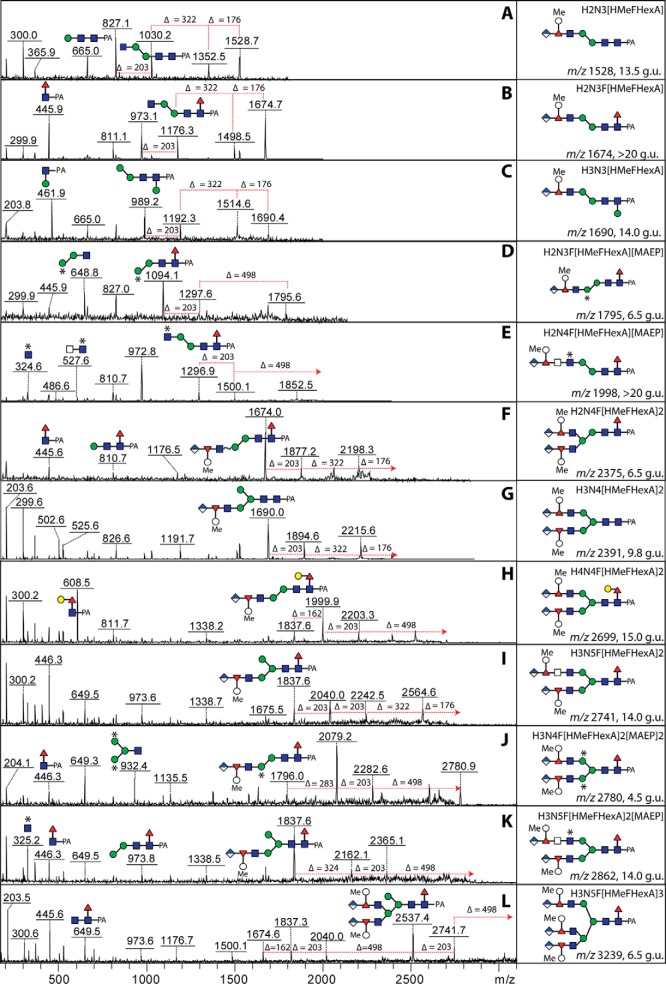
Zwitterionic N-glycans
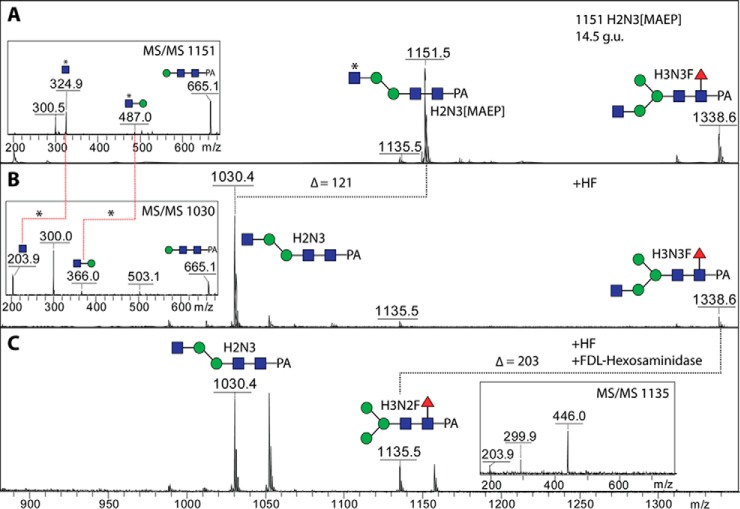
The neutral-enriched PNGase F-released pool also contained a number of glycans (see Fig. 6B for the annotated chromatogram) whose compositions and fragmentation patterns did not match known glycans, despite even sharing the same mass with oligomannosidic glycans (e.g. m/z 1151 and 1313 at 14.5 g.u., 1637 at 9.8 g.u., and 1961 at 7.9 g.u.). Even though some individual fragment ions were identical, relatively strong m/z 325 fragments, losses of 121 Da and strong negative-ion mode signals were observed indicative of an unusual modification (Fig. 10). As a modification of 121 Da is unlikely to be a monosaccharide, we considered the possibility that this may be a zwitterionic modification other than phosphoethanolamine (Δm/z 123) as found in Trichomonas N-glycans (27) or phosphorylcholine (Δm/z 165) as found in some core-modified glycans described above (see Fig. 5). Based on the literature on invertebrate N-glycans and glycolipids, we considered that this modification is N-methyl-2-aminoethylphosphonate (Δm/z 121), which has been previously found on glycolipids of one mollusc (45) as well as in unmethylated form (i.e. 2-aminoethylphosphonate, Δm/z 107) on locust N-glycans (46). As with standard phosphodiesters, hydrofluoric acid also removes this phosphonate from glycans, resulting in loss of 121 Da and of the m/z 284 and/or 325 fragments (Hex[MAEP] or HexNAc[MAEP]; see example in Fig. 11B). The treatment appears to leave behind putative monoantennary, pseudohybrid, and biantennary structures. In the case of an m/z 1151 glycan (Hex2HexNAc3[MAEP]-PA; 14.5 g.u.), incubation in series with hydrofluoric acid and a recombinant branch-specific β-N-acetylhexosaminidase did not result in removal of the nonreducing terminal GlcNAc despite hydrolysis of the phosphonate. Due to previous data on the specificity of the insect FDL hexosaminidase (32), this is indicative that the glycan carries the phosphonate-modified GlcNAc on the α1,6-arm (Fig. 11C). For the pseudohybrid glycans Hex3HexNAc3[MAEP]1–3-PA, the upper arm modification with GlcNAc was indicated by the ability to remove one mannose from the lower arm with the α1,2/3-mannosidase after hydrofluoric acid treatment (see Supplemental Table).
Fig. 10.
MALDI-TOF MS/MS of zwitterionic N-glycans. Ten example positive-ion mode MALDI-TOF MS/MS of N-glycans modified with (A) one aminoethylphosphonate (AEP) and one (B and C), two (D, E, F, and H) or three (G, I, and J) methylaminoethylphosphonate (MAEP) residues present in different RP-HPLC fractions (see Fig. 6B) are shown annotated with proposed structures for the individual fragments. Note that the glycans of m/z 1637 and 1961 are isobaric with standard oligomannosidic glycans (Man7,9GlcNAc2-PA). Digestion of a further methylaminoethylphosphonate-modified N-glycan (m/z 1151, isobaric with Man4GlcNAc2-PA) is shown in Fig. 11. An asterisk indicates modification of a Man or GlcNAc by methylaminoethylphosphonate.
Fig. 11.
Chemical treatment of a methylaminoethylphosphonate-modified N-glycan. The RP-HPLC glycan fraction of 14.5 g.u. (see Fig. 6B) was treated with hydrofluoric acid (B) and then further incubated with the branch-specific honeybee FDL β-hexosaminidase (C) prior to positive-ion mode MALDI-TOF MS. While the m/z 1151 glycan was sensitive to hydrofluoric acid, subsequent incubation with FDL only removed a GlcNAc from the coeluting m/z 1338 structure, indicating that the nonreducing terminal GlcNAc residues of the two structures are on different arms. An asterisk indicates the modification by methylaminoethylphosphonate (MAEP).
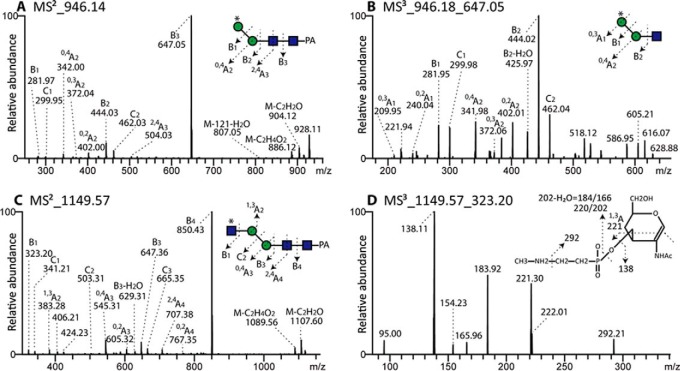
In one case of these zwitterionic glycans (m/z 934), we had a modification with aminoethylphosphonate (Δm/z 107; see Fig. 10A); otherwise, up to four methylaminoethylphosphonate modifications per glycan (on different monosaccharide residues) were detected. Thereby, it is noteworthy that a number of methylaminoethylphosphonate-containing fragments (m/z 325, 446, 608, 649, and 1151) are isobaric with more familiar fragments (Hex2, GlcNAc1Fuc1-PA, GlcNAc1Fuc1Hex1-PA, GlcNAc2Fuc1-PA, and Hex4HexNAc2-PA), which is an indication of the care required in interpreting solely on the basis of MS/MS without orthogonal evidence from digests. The MS/MS spectra, however, show protonated fragments resulting from modifications of hexose at m/z 284 and 446 (Hex1–2[MAEP]1; Figs. 10 B-10D and 10F) and modifications of N-acetylglucosamine at m/z 325 and 486 (Hex0–1HexNAc1[MAEP]1; Figs. 10 E-10J). Bisubstitution of HexHexNAc and HexNAc2 motifs result, respectively, in fragments at m/z 608 (Hex1HexNAc1[MAEP]2; Figs. 10 E, 10G, and 10I) and m/z 649 (HexNAc2[MAEP]2; Fig. 10J). In general, the modification with methylaminoethylphosphonate resulted in an earlier retention time than the corresponding neutral “backbone”: e.g. compare the biantennary m/z 1395 glycan eluting at 11 g.u. with the forms with two, three, and four methylaminoethylphosphonate residues (m/z 1637, 1758, and 1879) at 9.8, 7.9, and 3.4 g.u. respectively (Fig. 6B and Supplemental Table); the trend to earlier elution on the Kinetex column with an increasing number of zwitterionic residues is shared with phosphorylcholine-modified nematode glycans (33). Further evidence for the modification with methylaminoethylphosphonate and its exact location on mannose and N-acetylglucosamine residues came from LC-ESI-MS/MS data.
In negative-ion mode LC-MS of Hex2HexNAc2[MAEP]1 (Fig. 12A), fragment ions at m/z 282 (B1) and 300 (C1) suggest either a C4- or a C6-substitution of the terminal mannose with methylaminoethylphosphonate. Cross-ring cleavage of β-mannose (0,2A2 ions at m/z 402, 0,3A2 ions at m/z 372, and 0,4A2 ions at m/z 342) indicates that the terminal mannose is on the upper arm, in keeping with the α1,6-mannosidase sensitivity after hydrofluoric acid treatment. The MS3 spectrum of the dominant B3 ion (m/z 647, Fig. 12B) shows two cross-ring cleavages of substituted mannose (0,2A1 ions at m/z 240, 0,2A1-H2O ions at m/z 222 and 0,3A1 ions at m/z 210), which meant that the exact position of the methylaminoethylphosphonate (4 or 6 substitution) could not be defined.
Fig. 12.
LC-MSn of methylaminoethylphosphonate-modifed N-glycans. Two selected zwitterionic N-glycans (6.8 and 14.5 g.u.; compare with MALDI-TOF MS data in Figs. 10B and 11A) were analyzed by negative-ion mode MS2 and MS3. MS3 of the m/z 946 structure (0,3A ion at 210; B) suggests either a 4- or 6-linkage of the zwitterionic modification to mannose, whereas MS3 (1,3A ion at 221; D) of the m/z 1149 glycan suggests modification of the C3 of the nonreducing terminal GlcNAc. An asterisk indicates modification of a Man or GlcNAc by methylaminoethylphosphonate (MAEP); the presence of this moiety on the upper arm is supported by specific glycosidase treatments after its removal by hydrofluoric acid (see Supplemental Table).
Regarding the GlcNAc substituted with methylaminoethylphosphonate, MS/MS spectrum of a glycan with the composition of Hex2HexNAc3[MAEP]1-PA ([M-H]− ions at m/z 1149, Fig. 12C) can be easily misinterpreted as the isobaric Hex4HexNAc2-PA because the B1 and C1 ions (m/z 323 and 341) can be interpreted as either HexNAc1[MAEP]1 or Hex2. Cross-ring cleavage of β-mannose (0,2A3 ions at m/z 605, 0,4A3 ions at m/z 545) suggests that this N-glycan lacks the lower arm, which correlates with the FDL-hexosaminidase insensitivity after hydrofluoric acid treatment. However, MS3 of B1 ions (Fig. 12D) supports the presence of substituted GlcNAc rather than substituted mannose and the fragment ions at m/z 221, assigned as 1,3A cleavage of GlcNAc, indicates that the MAEP is linked to the C3 of GlcNAc. As mentioned above, the 121 Da modification (concluded to be methylaminoethylphosphonate) was also found on galactose of the Fucα1,2Galβ1,4Fucα1,6-modification of the core GlcNAc residues (Figs. 3E and 5) while a second isomer of m/z 1402 was modified on the mannose as deduced from the MS/MS (Fig. 3F).
Sulfated N-glycans
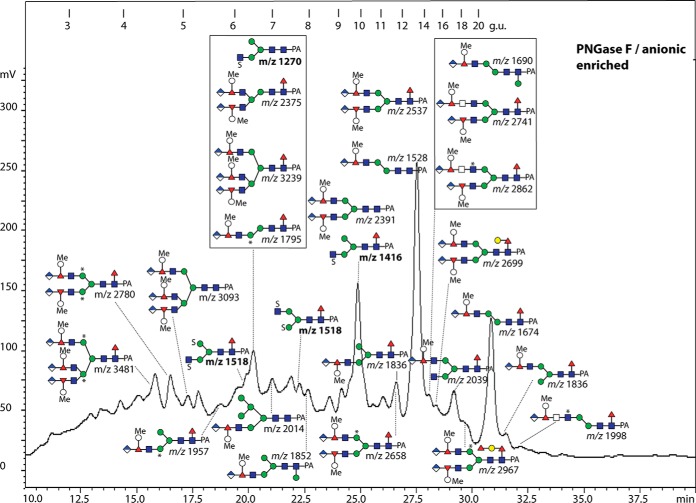
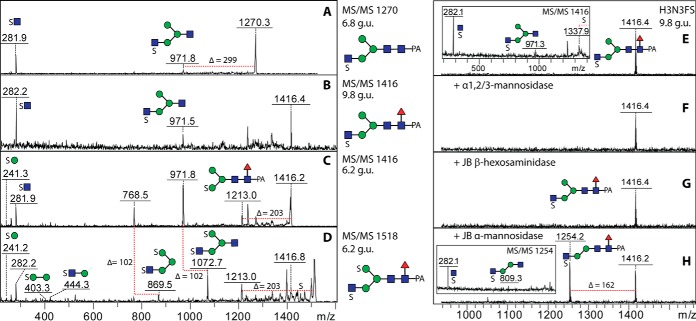
The anionic-enriched PNGase F-released glycan pool, accounting for some 15% of the total, contained a range of glycans detected in negative-ion mode, which eluted between 4 and >20 g.u. (Fig. 13; see also Fig. 1E). MS/MS of some of these yielded fragments at m/z 241 or 282 (Figs. 14A-14D). This was suggestive for the presence of sulfate on hexose and N-acetylhexosamine; based on previous experience with Dictyostelium glycans (22), phosphorylation can be ruled out, as hydrofluoric acid treatment did not result in loss of 80 Da (data not shown). In order to locate the sulfate moiety more exactly, one Hex3HexNAc3Fuc1S1 glycan (m/z 1416; 9.8 g.u.) was incubated with nonspecific and 1,2/3-specific α-mannosidases and with jack bean hexosaminidase; as only digestion with jack bean mannosidase occurred (Figs. 14E-14H), a modification with sulfated GlcNAc is assumed for the α1,3-arm (see also Supplemental Table). Another isomer of Hex3HexNAc3Fuc1S1 eluted later (15 g.u.) and so is concluded to have the “opposite” configuration (i.e. GlcNAc on the α1,6-arm), which would be in keeping with the long-established elution effects of isomeric glycans on RP-HPLC (29). Nonfucosylated and double-sulfated variants were also detected that eluted earlier than the fucosylated mono-sulfated form (e.g. 6.8 and 6.2 g.u.; m/z 1270 as [M-H]− and 1518 as [M-2H+Na]−), whereby sulfation of one mannose and one N-acetylglucosamine is concluded in the latter case (fragments of 241, 282, 403, and 444; Figs. 14C and 14D). While sulfation of N-acetylglucosamine is known in mammals (47), sulfation of mannose is known for a lobster N-glycan (48).
Fig. 13.
RP-HPLC of the PNGase F anionic-enriched pool of N-glycans from V. rubella. The glycans in the acetonitrile/trifluoroacetic acid elution from the NPGC solid-phase extraction step (followed by a second purification on C18; see Fig. 1) were applied to the Kinetex XB-C18 column. The external calibration with glucose units (3–20 g.u.) is indicated, the fluorescence intensity (320/400 nm) is shown in mV and the m/z values in bold are structures detected solely in negative-ion mode; *, methylaminoethylphosphonate; Me, methyl; S, sulfate.
Fig. 14.
Analysis of sulfated N-glycans. (A-D) Negative-ion mode MALDI-TOF MS/MS of sulfated N-glycans present in three different RP-HPLC fractions of the anionic-enriched pool (see Fig. 13) are shown with interpretation of the fragmentation patterns; the disulphated glycan at 6.2 g.u. was detected also in the [M-H-SO3]− and [M-2H+Na]− forms (C and D), whereby the former is due to random in source loss of one sulfate from either a hexose or N-acetylhexosamine residue. (E-H) Analysis by negative-ion mode MALDI-TOF MS of exoglycosidase digestions of the m/z 1416 glycan (9.8 g.u.) showing resistance to α1,2/3-mannosidase and jack bean hexosaminidase but sensitivity to nonspecific jack bean α-mannosidase; insets in panels E and H show alterations in the occurrence of key fragments.
Branched Fucose Modifications
In addition to the sulfated glycans, the anionic-enriched pool was rather dominated by glycans with unusual compositions (see discussion of Fig. 1E above), which eluted in a broad range from the RP-HPLC column (Fig. 13). Upon fragmentation in the positive-ion mode, serial loss of 176 and 322 (i.e. 146 + 176) or a loss of 498 (i.e. 322 + 176) en bloc was observed (Fig. 15). In the lower mass range, fragments indicative of the various other aforementioned modifications (core fucose, core mannose, galactosylated core fucose, and methylaminoethylphosphonate; e.g. at m/z 446, 462, 608, and 325, respectively) were present, which suggested that the 498 Da modification (hypothesized to be Hex1Me1Fuc1HexA1) was present on a wide range of glycan types in the margin snail. The efficient ionization in negative mode was considered to be compatible with the presence of a hexuronic acid (176 Da). Interestingly, in the hydrophobic pool, some very late-eluting potential biosynthetic intermediates of these glycans were observed (Fig. 2B; m/z 1484, 1498, and 1660). Due to no ionization in the negative mode, the high retention on reversed-phase and analyses after hydrofluoric acid (loss of 146 or 322) or bovine α-fucosidase (loss of one or two fucose residues; see Supplemental Table), these glycans are assumed to lack the glucuronic acid moiety but have either terminal α-fucose or methylhexose-substituted α-fucose. Therefore, it is also concluded that the second 176 Da unit of the acidic 498 Da modification is a methylated hexose.
Fig. 15.
MALDI-TOF MS/MS of N-glycans with anionic substitution of antennal fucose. Twelve example positive-ion mode MALDI-TOF MS/MS of N-glycans in various RP-HPLC fractions of the anionic-enriched pool (see Fig. 13) are shown together with annotations indicating the presence of one (A-E), two (F-K), or three (L) antennal bisubstituted fucose motifs; four of the glycans are also modified with methylaminoethylphosphonate (MAEP, position of modification indicated by an asterisk). Example MS/MS, digests and LC-MSn spectra of related glycans are shown in Figs. 16 and 17.
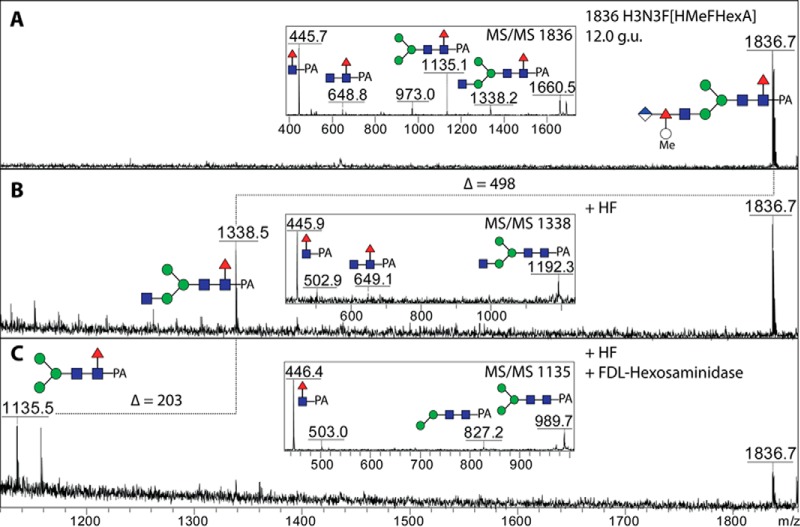
As fucose was apparently a component of this modification, we tested the effect of hydrofluoric acid, and indeed, a loss of 498 Da en bloc could be observed (see example treatment of the 12 g.u. fraction; Fig. 16B). The degree of removal (∼40%) would be compatible with an α1,4-linkage of fucose to the underlying GlcNAc residue; the C2 is occupied by the N-acetyl function, while previous experience with treatment of nematode glycans (49) with hydrofluoric acid indicates that a C3-linked fucose would probably be completely removed and C6-linked fucose not at all. The chemically treated product was then susceptible to the action of the FDL hexosaminidase (Fig. 16C), which is specific, under the conditions used, for removal of GlcNAc from the α1,3-arm (32).
Fig. 16.
Chemical and enzymatic treatment of an N-glycan modified with a branched fucose residue. The RP-HPLC glycan fraction of 12 g.u. (see Fig. 13) was treated with hydrofluoric acid (B) and then further incubated with the branch-specific honeybee FDL β-hexosaminidase (C) prior to positive-ion mode MALDI-TOF MS. Alterations in the structure were also monitored by MS/MS (see insets for the nondigested and digested forms). About 40% of the antennal modification was removed by hydrofluoric acid (a percentage consistent with α1,4-fucosylation) to yield a nonreducing terminal GlcNAc, which is concluded to be on the α1,3-arm due to its sensitivity toward the FDL β-hexosaminidase.
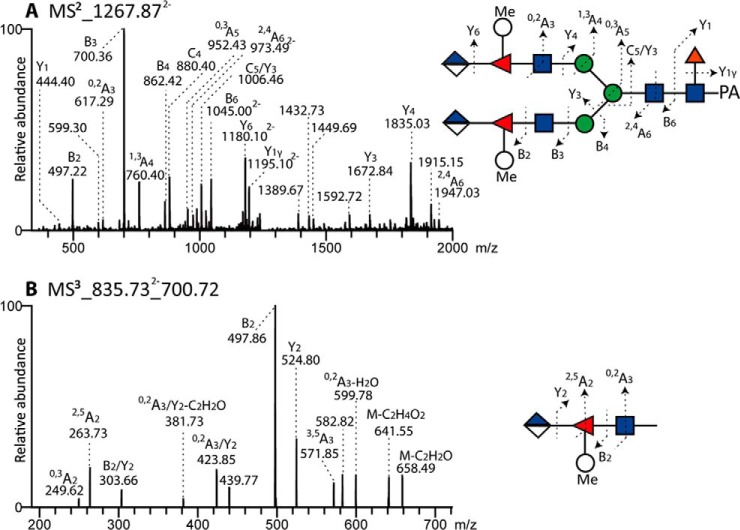
Further proof for the proposed position of the branched fucose came from LC-ESI-MS/MS data on two other glycans; the cross-ring cleavage of GlcNAc (0,2A3-H2O, and 0,2A3 ions at m/z 599/617) is compatible with either a 4- or 6-substituted GlcNAc (Fig. 17A). As the bond between Fuc and GlcNAc was sensitive to hydrofluoric acid, we rather conclude that the Fuc residue is linked to C4 of GlcNAc. MS3 of B3 ions at m/z 700 yielded ions at m/z 249 (0,3A of Fuc) and 263 (2,5A of Fuc) compatible with one 176 Da unit being linked to C4 of Fuc and another to C2 of Fuc (Fig. 17B). Thus, considering all the evidence, as well as comparisons to the literature on branched fucose in other molluscs (15), a possible model is that the 498 Da modification consists of an α1,4-linked fucose modified in turn by a 2-linked methylhexose and a 4-linked glucuronic acid. We further conclude that up to three such units can be present on triantennary glycans (e.g. m/z 3239; Fig. 15L). The definition of the triantennary backbone is based on analogy to the multiantennary glycans with branched fucose found in the blue mussel (15). Some monosubstituted forms were present in the standard “neutral” pool
Fig. 17.
LC-MSn analysis of an N-glycan modified with branched fucose. (A) The RP-HPLC fraction of the anionic-enriched pool (13.5 g.u; see Fig. 13, m/z 2537 as [M+H]+) containing an N-glycan with the potential composition of Hex3HexNAc4Fuc1[MeHexFucHexA]2 was analyzed by negative-ion mode LC-MS2 of the [M-2H]2- ion. (B) The anionic N-glycan with the composition Hex2HexNAc3Fuc1[MeHexFucHexA] (> 20 g.u.; see Fig. 13, m/z 1674 as [M+H]+) was analyzed by LC-MSn; the MS3 of the fragment ion at m/z 700 revealed cross-ring cleavages indicative of the presence of a 2,4-disubstituted fucose 4- or 6-linked to the antennal GlcNAc residue.
Mucin and O-fucose-Type O-glycans
LC-ESI-MS analysis of the β-eliminated O-glycans indicated the presence of a variety of fucose-, sulfate- and (methyl)aminoethylphosphonate-modified di- and trisaccharides (Fig. 18). The majority of the detected structures are, as judged by retention time and fragmentation, based on the mucin core 1 (Galβ1,3GalNAc) and core 3 (GlcNAcβ1,3GalNAc) structures (50); however, also two O-fucose glycans, of the type familiar from Notch and blood clotting factors (51), were also found. The basic form of core 1 is the most common O-glycan detected in V. rubella; it was also observed in sulfated form (Fig. 18I). Less common were the core 3 structures, but these encompassed fucosylated (HexNAc2–4Fuc1), extended (HexNAc2–3), and zwitterionic (HexNAc3Me0–1AEP1–2) forms (Figs. 18B-18G). One of the O-fucose glycans was also modified with a zwitterion (AEP-HexNAc-Fuc), which is probably based on GlcNAcβ1,3Fuc (Fig. 18H).
Fig. 18.
Analysis of V. rubella O-glycans by LC-MS/MS. O-glycans released by reductive β-elimination were analyzed in both positive-ion (A-E) and negative-ion mode (F-I). HexNAc-rich (up to 4) O-glycans were detected in the margin snail (A, B, and C). Fucosylated HexNAc-rich O-glycans were also found (D and E). As in the case of some N-glycans, O-glycans from the margin snail were also modified with aminoethylphosphonate (AEP) and methylaminoethylphosphonate (MAEP; F and G). In addition, the AEP-HexNAc motif was also found on O-Fuc (panel H). Cross-ring cleavage of HexNAc (0,2AHexNAc/0,2AHexNAc-H2O, m/z 226/208) suggested that AEP is linked to either C4 or C6 of HexNAc (H). However, core 1 (not shown) and sulfated core 1 (panel I) were the two most dominant O-glycans isolated from the margin snail.
DISCUSSION
Neutral N-glycans of V. rubella
The data presented here on the margin snail, V. rubella (C. B. Adams, 1845), indicate an unprecedented complexity and variety in an invertebrate N-glycome comprising some 100 structures (see Supplemental Table). Thereby, some “typical” invertebrate features, such as nonfucosylated paucimannosidic forms, core α1,3-fucosylation and core α1,3/6-difucosylation are of relatively low abundance and xylosylation or methylation of oligomannosidic glycans, found in terrestial and freshwater gastropods (13), are not at all present. Particularly unusual among the neutral N-glycan modifications of V. rubella is the β1,3-mannosylation of the core GlcNAc and the peripheral α1,6-mannosylation: the former may be the same as the previously described core hexosylation of oyster hemocyte N-glycans (12), whereas the latter is more akin to fungal “outer chains,” but the position of the α1,6-mannose is unusual in that it “replaces” the middle “B” α1,2-mannose that is removed first by the so-called ER mannosidase I. The most dominant N-glycan found in our analysis of the V. rubella N-glycome has a “humble” Man2GlcNAc2Fuc1 structure, and, like a number of the smaller glycans in the margin snail, it lacks the “lower” arm α1,3-mannose, in keeping with a greater degree of processing by mannosidases in “lower” eukaryotes than in vertebrates; the loss of the α1,3-mannose as observed here is known from insects and plants (38, 52), while the bias in nematodes is toward removal of the α1,6-mannose (33).
Other than the standard core α1,3- and α1,6-fucosylation, consistent with its status as an invertebrate and aforementioned core β1,3-mannosylation, complex extensions of the core α1,6-fucose are also observed in V. rubella: Not only was galactose linked to the fucose, but the galactose was often, in turn, substituted with α1,2-fucose and a zwitterion (see below). Such core modifications are not without precedent: Galactosylation of core fucose (“GalFuc”) is known from cephalopod rhodopsins and nematodes (11, 33), whereas Fucα1,2Galβ1,4Fuc occurs in mutant Caenorhabditis strains (39, 53). Other substitutions of the GalFuc motif include another galactose or a methylhexose as on keyhole limpet hemocyanin or in nematodes and Planaria (39, 54–56); however, a zwitterionic modification of core GalFuc has never, to our knowledge, been previously described.
Anionic and Zwitterionic N-glycans of V. rubella
The anionic N-glycans can be divided into two groups: those modified with sulfate or with a trisaccharide motif (HexA[MeHex]Fuc). Even though sulfate has been previously found on N-glycans, the linkage here is to mannose, as in Dictyostelium (22), or N-acetylglucosamine and not to galactose, as in the oyster (12). On the other hand, disubstituted fucose has been previously found in two other marine molluscs. In the shell-forming fluid of the blue mussel, a so-called valence epitope with the structure 4-O-methyl-GlcA1,4(GlcNAc1,3)Fuc1,4GlcNAc was described (15), while for Rapana venosa (thomasiana) hemocyanins MeHex(HexNAc)Fuc1,4GlcNAc or HexA(HexNAc)Fuc1,4GlcNAc motifs were proposed (57–59). In our case, the anionic HexA(MeHex)Fuc1,4GlcNAc modification with 2,4-disubstition of the α1,4-linked fucose is very similar, and based on analogy to the mussel glycan, it may actually be GlcA1,4(MeHex1,2)Fuc1,4GlcNAc.
The zwitterionic glycans of the margin snail are generally modified with methylaminoethylphosphonate, but some examples of phosphorylcholine substitution were also found. Whereas both methylaminoethylphosphonate and aminoethylphosphonate are known as a modification of mollusc glycolipids (45, 60) and aminoethylphosphonate is a component of jellyfish O-glycans (61) and protozoan glycolipid anchors (62), phosphorylcholine is a component of protein- and lipid-linked glycans from a range of species including nematodes, cestodes, annelids, and bacteria (63–66). However, in V. rubella, the phosphorylcholine is linked to the galactose residue of the core GlcNAc modification and not to antennal GlcNAc as on the N-glycans of nematodes or cestodes. On the other hand, the closest “homologue” to the modification of N-glycans with methylaminoethylphosphonate is the presence of the nonmethylated form (aminoethylphosphonate) on both peripheral mannose and GlcNAc residues of the locust apolipophorin III (46). Thereby, the described linkages (both C6) on the locust glycans contrast to the probable mixture of substitutions to mannose, galactose, GlcNAc, and potentially GalNAc on both neutral and anionic N-glycan “backbones” of the margin snail.
Not only the range of modifications but the size of the N-glycans of V. rubella is noteworthy: We can propose structures with up to 20 residues (with masses of 3500 Da). This is probably not the upper limit (due to the limits of confident detection) but is of a range not typically associated with invertebrate glycans, albeit N-glycans of similar or larger masses are present in oysters (12), insects (37), and trematodes (67). This is due to the presence of both complex modifications and multiple branches, the latter probably being products of GlcNAc-TIV or V homologues catalyzing the formation of the triantennary structures found in these invertebrates.
O-glycans of V. rubella
The O-glycans detected by LC-ESI-MS in this study are, on the other hand, not so complex: We have detected either the O-GalNAc or O-Fuc types. The latter is represented by variants of GlcNAcβ1,3Fuc, which is the product of the fringe β1,3-N-acetylglucosaminyltransferase (68), but in contrast to insects (69), the “ionic” modification of O-fucose found in the margin snail is (methyl)aminoethylphosphonate rather than glucuronic acid. For the core 1 and core 3 type mucin-type O-glycans, fucose, sulfate, and (methyl)aminoethylphosphonate were among the modifications, the latter modification being akin to the linkage of aminoethylphosphonate directly to the reducing-terminal GalNAc found in jellyfish (61). There are few studies regarding the O-glycans of gastropods: Two examples, however, showed the presence of a bisubstituted core GalNAc on either a cone snail conopeptide toxin (70) or in whole snail tissues (71). This contrasts with only monosubstituted GalNAc being observed in the present study on V. rubella.
Analytical Considerations for Mollusc Glycomics
Key to all these analyses, especially of the N-glycans, was the differential release and solid-phase extraction yielding four pools prior to use of fused-core RP-HPLC. This reduced the number of glycans per fraction and so simplified the interpretation of fragmentation and digestion patterns as well as overcame suppression effects which may occur in complex mixtures of glycans of different types and amounts. As discussed in the introduction, many different procedures have been used to analyze mollusc glycomes in the past; here, we have brought together various methods used in different previously published and ongoing studies.
Porous and nonporous graphitized carbon was first used with mammalian glycans to desalt and separate neutral and sialylated glycans by elution with acetonitrile followed by acetonitrile supplemented with trifluoroacetic acid (72, 73). C18-based solid-phase extraction is usually used as a “clean-up” step, but not to separate types of glycans. Only recently was the ability of C18 to separate oligomannosidic from fucosylated N-glycans described, albeit under different conditions (74); independently, based on the concept that 10–15% (v/v) methanol is required to elute N-glycans with non- and monosubstituted core α1,6-fucose from typical C18 RP-HPLC columns, we tested use of a step-gradient during solid-phase extraction. As we successfully used graphitized carbon to enrich sulfated and methylphosphorylated N-glycans from Dictyostelium away from the neutral structures (22), we adopted the approach of using NPGC and C18 in series prior to RP-HPLC. In the case of V. rubella, the glycans carrying sulfate or glucuronylated fucose were primarily in the “anionic” pool, whereas the “hydrophobic” pool is very much biased toward structures with core Fucα1,2Galβ1,4Fucα1,6 motifs. The subsequent use of RP-HPLC facilitates the separation of isomeric and isobaric structures, a strategy established for some 30 years to resolve complex, paucimannosidic, and oligomannosidic glycans from mammalian and other sources (29).
Another aspect, which we consider important, is the optimized mix of methods on individual RP-HPLC fractions. The ability to “deconstruct” glycans using hydrofluoric acid and exoglycosidases, in addition to MSn, enables us to make further conclusions as to the composition and linkages, especially when dealing with unknown structures and lacking defined standards. Hydrofluoric acid has proven useful in the determination of linkages with phosphodiesters, phosphonates, galactofuranose, and fucose in previous studies from our own and other laboratories (22, 27, 45, 75–77), whereas linkage-specific glycosidases (such as the α1,6-mannosidase, α1,2-fucosidase, β1,4-galactosidase, and FDL β-hexosaminidase used here) are another tool for determining the nature of the isomeric status of the oligosaccharide structures. Glycosidases and hydrofluoric acid indeed were key to determining the structure of, e.g. the methylaminoethylphosphonate-substituted N-glycans in this study as, based on MS spectra alone, these structures could have been easily misinterpreted as pauci- or oligomannosidic N-glycans. As one can expect that an organism should be able, as part of glycoprotein turnover in the lysosomes, to degrade its own glycans, it will be of interest to find further glycan-degrading enzymes capable of removing, e.g. glucuronic acid and methylhexose residues.
CONCLUSION
The present study exemplifies that there is, in comparison to insects or plants, no mollusc-specific glycomic characteristic signature. On the contrary, the majority of the N- and O-glycans described has for V. rubella have not been described for any other organism, although aspects of their structures (branched fucose, zwitterionic modifications, and elongated core modifications) have been found in other organisms (e.g. also in nematodes) or on other types of glycoconjugate (e.g. on glycolipids). This mix of highly modified glycans must have a biological function, which is difficult to elucidate considering the lack of genetic and microarray tools for gastropods in general. However, it is known that molluscs interact with other species in a glycan-dependent manner, e.g. Biomphalaria and Schistosoma or Crassostrea and Perkinsus (78, 79), in a way that can involve a degree of glycomimickry. Considering that parasites often hijack systems of innate immunity, it may be that the unusual glycans of molluscs may have initially evolved as a defense mechanism in the absence of an adaptive immune response (which is present in vertebrates) or as a means for interactions with symbionts. The wide variability of mollusc glycomes may then reflect subtle differences in genetic and epigenetic systems enabling the evolution and adaptation of the “sugar” coating of their cells: As a specific glycan may be less essential than a specific protein, alteration of glycan modifications is a potential “red queen” strategy (“running to stay in the same place”) for a group of species to survive (80). However, support for such a conclusion will need a more systematic set of analyses on related and less related mollusc species than has been performed to date.
Supplementary Material
Acknowledgments
We thank Dr. Niclas Karlsson for access to the LTQ mass spectrometer, which was obtained by a grant from the Swedish Research Council (342–2004-4434), Megazyme for the kind gift of the microbial α1,2-specific fucosidase (E-FUCM), Dr. Jorick Vanbeselaere and Dr. Shi Yan for performing some analyses, Dr. Ján Mucha for access to the UltrafleXtreme MALDI-TOF MS at the Slovak Academy of Sciences, and Dr. Iain Wilson for support and discussions.
Footnotes
Author contributions: K.P. designed the research; B.E. and C.J. performed the research; D.A. contributed new reagents or analytic tools; B.E., C.J., and K.P. analyzed data; and K.P. wrote the paper.
* This work was funded by the Austrian Fonds zur Förderung der wissenschaftlichen Forschung (FWF; grant P25058) to K.P. C.J. was supported by the Knut and Alice Wallenberg Foundation.
 This article contains supplemental material Supplemental Table.
This article contains supplemental material Supplemental Table.
1 The abbreviations used are:
- FDL
- fused lobes
- ESI
- electrospray ioniziation
- HF
- hydrofluoric acid
- MAEP
- methylaminoethylphosphonate
- MeOH
- methanol
- MeCN
- acetonitrile
- NPGC
- non-porous graphitized carbon
- PA
- pyridylamino
- PC
- phosphorylcholine
- PNGase
- peptide:N-glycosidase
- RP
- reversed-phase
- TFA
- trifluoroacetic acid.
REFERENCES
- 1.Ponder W. F., and Lindberg D. R. (2008) Phylogeny and evolution of the Mollusca, University of California Press, Berkeley [Google Scholar]
- 2.Volety A. K., Haynes L., Goodman P., and Gorman P. (2014) Ecological condition and value of oyster reefs of the Southwest Florida shelf ecosystem. Ecological Indicators 44, 108–119 [Google Scholar]
- 3.Salanki J., Farkas A., Kamardina T., and Rozsa K. S. (2003) Molluscs in biological monitoring of water quality. Toxicol. Lett. 140–141, 403–410 [DOI] [PubMed] [Google Scholar]
- 4.Zuykov M., Pelletier E., and Harper D. A. (2013) Bivalve mollusks in metal pollution studies: From bioaccumulation to biomonitoring. Chemosphere 93, 201–208 [DOI] [PubMed] [Google Scholar]
- 5.Tian P., Engelbrektson A. L., and Mandrell R. E. (2008) Seasonal tracking of histo-blood group antigen expression and norovirus binding in oyster gastrointestinal cells. J. Food Prot. 71, 1696–1700 [DOI] [PubMed] [Google Scholar]
- 6.Bayne C. J. (2009) Successful parasitism of vector snail Biomphalaria glabrata by the human blood fluke (trematode) Schistosoma mansoni: A 2009 assessment. Mol. Biochem. Parasitol. 165, 8–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geyer H., Wuhrer M., Resemann A., and Geyer R. (2005) Identification and characterization of keyhole limpet hemocyanin N-glycans mediating cross-reactivity with Schistosoma mansoni. J. Biol. Chem. 280, 40731–40748 [DOI] [PubMed] [Google Scholar]
- 8.Van Kuik J. A., Sijbesma R. P., Kamerling J. P., Vliegenthart J. F., and Wood E. J. (1987) Primary structure determination of seven novel N-linked carbohydrate chains derived from hemocyanin of Lymnaea stagnalis. 3- O-methyl-D-galactose and N-acetyl-D-galactosamine as constituents of xylose-containing N-linked oligosaccharides in an animal glycoprotein. Eur. J. Biochem. 169, 399–411 [DOI] [PubMed] [Google Scholar]
- 9.Lommerse J. P., Thomas-Oates J. E., Gielens C., Préaux G., Kamerling J. P., and Vliegenthart J. F. G. (1997) Primary structure of 21 novel monoantennary and diantennary N- linked carbohydrate chains from aD-hemocyanin of Helix pomatia. Eur. J. Biochem. 249, 195–222 [DOI] [PubMed] [Google Scholar]
- 10.Siddiqui N. I., Idakieva K., Demarsin B., Doumanova L., Compernolle F., and Gielens C. (2007) Involvement of glycan chains in the antigenicity of Rapana thomasiana hemocyanin. Biochem. Biophys. Res. Commun. 361, 705–711 [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y., Iwasa T., Tsuda M., Kobata A., and Takasaki S. (1997) A novel monoantennary complex-type sugar chain found in octopus rhodopsin: Occurrence of the Galβ1→4Fuc group linked to the proximal N-acetylglucosamine residue of the trimannosyl core. Glycobiology 7, 1153–1158 [DOI] [PubMed] [Google Scholar]
- 12.Kurz S., Jin C., Hykollari A., Gregorich D., Giomarelli B., Vasta G. R., Wilson I. B., and Paschinger K. (2013) Haemocytes and plasma of the eastern oyster (Crassostrea virginica) display a diverse repertoire of sulphated and blood group A-modified N-glycans. J. Biol. Chem. 288, 24410–24428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutternigg M., Bürgmayr S., Pöltl G., Rudolf J., and Staudacher E. (2007) Neutral N-glycan patterns of the gastropods Limax maximus, Cepaea hortensis, Planorbarius corneus, Arianta arbustorum and Achatina fulica. Glycoconj J. 24, 475–489 [DOI] [PubMed] [Google Scholar]
- 14.Lehr T., Geyer H., Maass K., Doenhoff M. J., and Geyer R. (2007) Structural characterization of N-glycans from the freshwater snail Biomphalaria glabrata cross-reacting with Schistosoma mansoni glycoconjugates. Glycobiology 17, 82–103 [DOI] [PubMed] [Google Scholar]
- 15.Zhou H., Hanneman A. J., Chasteen N. D., and Reinhold V. N. (2013) Anomalous N-glycan structures with an internal fucose branched to GlcA and GlcN residues isolated from a mollusk shell-forming fluid. J. Proteome. Res. 12, 4547–4555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Tetering A., Schiphorst W. E., van den Eijnden D. H., and van Die I. (1999) Characterization of a core α1→3-fucosyltransferase from the snail Lymnaea stagnalis that is involved in the synthesis of complex-type N-glycans. FEBS Lett. 461, 311–314 [DOI] [PubMed] [Google Scholar]
- 17.Mulder H., Dideberg F., Schachter H., Spronk B. A., De Jong-Brink M., Kamerling J. P., and Vliegenthart J. F. G. (1995) In the biosynthesis of N-glycans in connective tissue of the snail Lymnaea stagnalis of incorporation GlcNAc by β2GlcNAc-transferase I is an essential prerequisite for the action of β2GlcNAc-transferase II and β2Xyl-transferase. Eur. J. Biochem. 232, 272–283 [DOI] [PubMed] [Google Scholar]
- 18.Neeleman A. P., and van de Eijnden D. H. (1996) α-Lactalbumin affects the acceptor specificity of Lymnaea stagnalis albumen gland UDP-GalNAc:GlcNAc β-R β1→4-N-acetylgalactosaminyltransferase: Synthesis of GalNAc β1→4Glc. Proc. Natl. Acad. Sci. U.S.A. 93, 10111–10116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bakker H., Agterberg M., van Tetering A., Koeleman C. A., van den Eijnden D. H., and van Die I. (1994) A Lymnaea stagnalis gene, with sequence similarity to that of mammalian β1→4-galactosyltransferases, encodes a novel UDP-GlcNAc:GlcNAc β-R β1→4-N-acetylglucosaminyltransferase. J. Biol. Chem. 269, 30326–30333 [PubMed] [Google Scholar]
- 20.Taus C., Lucini C., Sato T., Furukawa K., Grabherr R., and Staudacher E. (2013) Expression and characterization of the first snail-derived UDP-N-acetyl-α-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase. Glycoconj J. 30, 825–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tretter V., Altmann F., and März L. (1991) Peptide-N4-(N-acetyl-β-glucosaminyl)asparagine amidase F cannot release glycans with fucose attached α1→3 to the asparagine-linked N-acetylglucosamine residue. Eur. J. Biochem. 199, 647–652 [DOI] [PubMed] [Google Scholar]
- 22.Hykollari A., Balog C. I., Rendić D., Braulke T., Wilson I. B., and Paschinger K. (2013) Mass spectrometric analysis of neutral and anionic N-glycans from a Dictyostelium discoideum model for human congenital disorder of glycosylation CDG IL. J. Proteome. Res. 12, 1173–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ali I., Al-Othman Z. A., Nagae N., Gaitonde V. D., and Dutta K. K. (2012) Recent trends in ultra-fast HPLC: new generation superficially porous silica columns. J. Sep. Sci. 35, 3235–3249 [DOI] [PubMed] [Google Scholar]
- 24.Adams C. B. (1845) Specierum novarum conchyliorum, in Jamaica repertorum, synopsis Proc. Boston Soc. Natural History 2, 1–17 [Google Scholar]
- 25.Cossignani T. (2006) Marginellidae and Cystiscidae of the World L'Informatore Piceno, Ancona. Online publication [Google Scholar]
- 26.Rosenberg G. (2015) Volvarina rubella (Adams C. B., 1845). http://www.marinespecies.org/aphia.php?p=taxdetails&id=474072
- 27.Paschinger K., Hykollari A., Razzazi-Fazeli E., Greenwell P., Leitsch D., Walochnik J., and Wilson I. B. H. (2012) The N-glycans of Trichomonas vaginalis contain variable core and antennal modifications. Glycobiology 22, 300–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulz B. L., Packer N. H., and Karlsson N. G. (2002) Small-scale analysis of O-linked oligosaccharides from glycoproteins and mucins separated by gel electrophoresis. Anal. Chem. 74, 6088–6097 [DOI] [PubMed] [Google Scholar]
- 29.Tomiya N., Kurono M., Ishihara H., Tejima S., Endo S., Arata Y., and Takahashi N. (1987) Structural analysis of N-linked oligosaccharides by a combination of glycopeptidase, exoglycosidases, and high-performance liquid chromatography. Anal. Biochem. 163, 489–499 [DOI] [PubMed] [Google Scholar]
- 30.Domon B., and Costello C. E. (1988) A systematic nomenclature for carbohydrate fragmentations in Fab-MS MS spectra of glycoconjugates. Glycoconjugate J. 5, 397–409 [Google Scholar]
- 31.Dragosits M., Pflugl S., Kurz S., Razzazi-Fazeli E., Wilson I. B., and Rendić D. (2014) Recombinant Aspergillus β-galactosidases as a robust glycomic and biotechnological tool. Appl. Microbiol. Biotechnol. 98, 3553–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dragosits M., Yan S., Razzazi-Fazeli E., Wilson I. B., and Rendić D. (2015) Enzymatic properties and subtle differences in the substrate specificity of phylogenetically distinct invertebrate N-glycan processing hexosaminidases. Glycobiology 25, 448–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan S., Wilson I. B., and Paschinger K. (2015) Comparison of RP-HPLC modes to analyse the N-glycome of the free-living nematode Pristionchus pacificus. Electrophoresis 36, 1314–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomiya N., Lee Y. C., Yoshida T., Wada Y., Awaya J., Kurono M., and Takahashi N. (1991) Calculated two-dimensional sugar map of pyridylaminated oligosaccharides: Elucidation of the jack bean α-mannosidase digestion pathway of Man9GlcNAc2. Anal. Biochem. 193, 90–100 [DOI] [PubMed] [Google Scholar]
- 35.Tomiya N., and Takahashi N. (1998) Contribution of component monosaccharides to the coordinates of neutral and sialyl pyridylaminated N-glycans on a two- dimensional sugar map. Anal. Biochem. 264, 204–210 [DOI] [PubMed] [Google Scholar]
- 36.Hykollari A., Dragosits M., Rendić D., Wilson I. B., and Paschinger K. (2014) N-glycomic profiling of a glucosidase II mutant of Dictyostelium discoideum by '‘off-line’' liquid chromatography and mass spectrometry. Electrophoresis 35, 2116–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurz S., Aoki K., Jin C., Karlsson N. G., Tiemeyer M., Wilson I. B., and Paschinger K. (2015) Targetted release and fractionation reveal glucuronylated and sulphated N- and O-glycans in larvae of dipteran insects. J. Proteomics 126, 172–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kubelka V., Altmann F., Staudacher E., Tretter V., März L., Hård K., Kamerling J. P., and Vliegenthart J. F. G. (1993) Primary structures of the N-linked carbohydrate chains from honeybee venom phospholipase A2. Eur. J. Biochem. 213, 1193–1204 [DOI] [PubMed] [Google Scholar]
- 39.Yan S., Bleuler-Martinez S., Plaza Gutierrez D. F., Künzler M., Aebi M., Joachim A., Razzazi-Fazeli E., Jantsch V., Geyer R., Wilson I. B., and Paschinger K. (2012) Galactosylated fucose epitopes in nematodes: increased expression in a Caenorhabditis mutant associated with altered lectin sensitivity and occurrence in parasitic species. J. Biol. Chem. 287, 28276–28290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paschinger K., Staudacher E., Stemmer U., Fabini G., and Wilson I. B. H. (2005) Fucosyltransferase substrate specificity and the order of fucosylation in invertebrates. Glycobiology 15, 463–474 [DOI] [PubMed] [Google Scholar]
- 41.Tomiya N., Awaya J., Kurono M., Endo S., Arata Y., and Takahashi N. (1988) Analyses of N-linked oligosaccharides using a two-dimensional mapping technique. Anal. Biochem. 171, 73–90 [DOI] [PubMed] [Google Scholar]
- 42.Wong-Madden S. T., and Landry D. (1995) Purification and characterization of novel glycosidases from the bacterial genus Xanthomonas. Glycobiology 5, 19–28 [DOI] [PubMed] [Google Scholar]
- 43.Hykollari A., Eckmair B., Voglmeir J., Jin C., Yan S., Vanbeselaere J., Razzazi-Fazeli E., Wilson I. B. H., and Paschinger K. (2015) More than just oligomannose: An N-glycomic comparison of Penicillium species. Mol. Cell Proteomics 1, 73–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harvey D. J. (2005) Fragmentation of negative ions from carbohydrates: Part 3. Fragmentation of hybrid and complex N-linked glycans. J. Am. Soc. Mass Spectrom. 16, 647–659 [DOI] [PubMed] [Google Scholar]
- 45.Hayashi A., and Matsubara T. (1989) A new homolog of phosphonoglycosphingolipid, N-methylaminoethylphosphonyltrigalactosylceramide. Biochim. Biophys. Acta 1006, 89–96 [Google Scholar]
- 46.Hård K., Van Doorn J. M., Thomas-Oates J. E., Kamerling J. P., and Van der Horst D. J. (1993) Structure of the Asn-linked oligosaccharides of apolipophorin III from the insect Locusta migratoria. Carbohydrate- linked 2-aminoethylphosphonate as a constituent of a glycoprotein. Biochemistry 32, 766–775 [DOI] [PubMed] [Google Scholar]
- 47.Gallego R. G., Blanco J. L., Thijssen-van Zuylen C. W., Gotfredsen C. H., Voshol H., Duus J. Ø., Schachner M., and Vliegenthart J. F. (2001) Epitope diversity of N-glycans from bovine peripheral myelin glycoprotein P0 revealed by mass spectrometry and nano probe magic angle spinning 1H NMR spectroscopy. J. Biol. Chem. 276, 30834–30844 [DOI] [PubMed] [Google Scholar]
- 48.van Kuik J. A., Breg J., Kolsteeg C. E. M., Kamerling J. P., and Vliegenthart J. F. G. (1987) Primary structure of the acidic carbohydrate chain of hemocyanin from Panulirus interruptus. FEBS Lett. 221, 150–154 [Google Scholar]
- 49.Paschinger K., and Wilson I. B. H. (2015) Two types of galactosylated fucose motifs are present on N-glycans of Haemonchus contortus. Glycobiology 25, 585–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schachter H., and Williams D. (1982) Biosynthesis of mucus glycoproteins. Adv. Exp. Med. Biol. 144, 3–28 [DOI] [PubMed] [Google Scholar]
- 51.Shao L., and Haltiwanger R. S. (2003) O-fucose modifications of epidermal growth factor-like repeats and thrombospondin type 1 repeats: Unusual modifications in unusual places. Cell Mol. Life Sci. 60, 241–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Kuik J. A., Hoffmann R. A., Mutsaers J. H. G. M., Van Halbeek H., Kamerling J. P., and Vliegenthart J. F. G. (1986) A 500-MHz 1H-NMR study on the N-linked carbohydrate chain of bromelain. Glycoconjugate J. 3, 27–34 [Google Scholar]
- 53.Yan S., Jin C., Wilson I. B. H., and Paschinger K. (2015) Comparisons of Caenorhabditis fucosyltransferase mutants reveal a multiplicity of isomeric N-glycan structures. J. Proteome Res. in press, 10.1021/acs.jproteome.1025b00746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanneman A. J., Rosa J. C., Ashline D., and Reinhold V. N. (2006) Isomer and glycomer complexities of core GlcNAcs in Caenorhabditis elegans. Glycobiology 16, 874–890 [DOI] [PubMed] [Google Scholar]
- 55.Wuhrer M., Robijn M. L., Koeleman C. A., Balog C. I., Geyer R., Deelder A. M., and Hokke C. H. (2004) A novel Gal(β1–4)Gal(β1–4)Fuc(α1–6)-core modification attached to the proximal N-acetylglucosamine of keyhole limpet haemocyanin (KLH) N-glycans. Biochem. J. 378, 625–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paschinger K., Razzazi-Fazeli E., Furukawa K., and Wilson I. B. H. (2011) Presence of galactosylated core fucose on N-glycans in the planaria Dugesia japonica. J. Mass Spectrom 46, 561–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gielens C., Idakieva K., Van den Bergh V., Siddiqui N. I., Parvanova K., and Compernolle F. (2005) Mass spectral evidence for N-glycans with branching on fucose in a molluscan hemocyanin. Biochem. Biophys. Res. Commun. 331, 562–570 [DOI] [PubMed] [Google Scholar]
- 58.Sandra K., Dolashka-Angelova P., Devreese B., and Van Beeumen J. (2007) New insights in Rapana venosa hemocyanin N-glycosylation resulting from on-line mass spectrometric analyses. Glycobiology 17, 141–156 [DOI] [PubMed] [Google Scholar]
- 59.Dolashka P., Velkova L., Shishkov S., Kostova K., Dolashki A., Dimitrov I., Atanasov B., Devreese B., Voelter W., and Van Beeumen J. (2010) Glycan structures and antiviral effect of the structural subunit RvH2 of Rapana hemocyanin. Carbohydr. Res. 345, 2361–2367 [DOI] [PubMed] [Google Scholar]
- 60.Satake M., and Miyamoto E. (2012) A group of glycosphingolipids found in an invertebrate: Their structures and biological significance. Proc. Japan Acad. Ser. B. Phys. Biolog. Sci. 88, 509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Urai M., Nakamura T., Uzawa J., Baba T., Taniguchi K., Seki H., and Ushida K. (2009) Structural analysis of O-glycans of mucin from jellyfish (Aurelia aurita) containing 2-aminoethylphosphonate. Carbohydr Res. 344, 2182–2187 [DOI] [PubMed] [Google Scholar]
- 62.Serrano A. A., Schenkman S., Yoshida N., Mehlert A., Richardson J. M., and Ferguson M. A. J. (1995) The lipid structure of the glycosylphosphatidylinositol-anchored mucin-like sialic acid acceptors of Trypanosoma cruzi changes during parasite differentiation from epimastigotes to infective metacyclic trypomastigote forms. J. Biol. Chem. 270, 27244–27253 [DOI] [PubMed] [Google Scholar]
- 63.Paschinger K., Gutternigg M., Rendić D., and Wilson I. B. (2008) The N-glycosylation pattern of Caenorhabditis elegans. Carbohydr Res 343, 2041–2049 [DOI] [PubMed] [Google Scholar]
- 64.Paschinger K., Gonzalez-Sapienza G. G., and Wilson I. B. H. (2012) Mass spectrometric analysis of the immunodominant glycan epitope of Echinococcus granulosus antigen Ag5. Int. J. Parasitol. 42, 279–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sugita M., Fujii H., Dulaney J. T., Inagaki F., Suzuki M., Suzuki A., and Ohta S. (1995) Structural elucidation of two novel amphoteric glycosphingolipids from the earthworm, Pheretima hilgendorfi. Biochim. Biophys. Acta 1259, 220–226 [DOI] [PubMed] [Google Scholar]
- 66.Lysenko E., Richards J. C., Cox A. D., Stewart A., Martin A., Kapoor M., and Weiser J. N. (2000) The position of phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae affects binding and sensitivity to C-reactive protein-mediated killing. Mol. Microbiol. 35, 234–245 [DOI] [PubMed] [Google Scholar]
- 67.Wuhrer M., Koeleman C. A., Fitzpatrick J. M., Hoffmann K. F., Deelder A. M., and Hokke C. H. (2006) Gender-specific expression of complex-type N-glycans in schistosomes. Glycobiology 16, 991–1006 [DOI] [PubMed] [Google Scholar]
- 68.Moloney D. J., Panin V. M., Johnston S. H., Chen J., Shao L., Wilson R., Wang Y., Stanley P., Irvine K. D., Haltiwanger R. S., and Vogt T. F. (2000) Fringe is a glycosyltransferase that modifies Notch. Nature 406, 369–375 [DOI] [PubMed] [Google Scholar]
- 69.Aoki K., Porterfield M., Lee S. S., Dong B., Nguyen K., McGlamry K. H., and Tiemeyer M. (2008) The diversity of O-linked glycans expressed during Drosophila melanogaster development reflects stage- and tissue-specific requirements for cell signaling. J. Biol. Chem. 283, 30385–30400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hocking H. G., Gerwig G. J., Dutertre S., Violette A., Favreau P., Stöcklin R., Kamerling J. P., and Boelens R. (2013) Structure of the O-glycosylated conopeptide CcTx from Conus consors venom. Chemistry 19, 870–879 [DOI] [PubMed] [Google Scholar]
- 71.Stepan H., Pabst M., Altmann F., Geyer H., Geyer R., and Staudacher E. (2012) O-Glycosylation of snails. Glycoconjugate J. 29, 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Packer N. H., Lawson M. A., Jardine D. R., and Redmond J. W. (1998) A general approach to desalting oligosaccharides released from glycoproteins. Glycoconjugate Journal 15, 737–747 [DOI] [PubMed] [Google Scholar]
- 73.Chu C. S., Niñonuevo M. R., Clowers B. H., Perkins P. D., An H. J., Yin H., Killeen K., Miyamoto S., Grimm R., and Lebrilla C. B. (2009) Profile of native N-linked glycan structures from human serum using high performance liquid chromatography on a microfluidic chip and time-of-flight mass spectrometry. Proteomics 9, 1939–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin C. H., Kuo C. W., Jarvis D. L., and Khoo K. H. (2014) Facile removal of high mannose structures prior to extracting complex type N-glycans from de-N-glycosylated peptides retained by C18 solid phase to allow more efficient glycomic mapping. Proteomics 14, 87–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morelle W., Bernard M., Debeaupuis J. P., Buitrago M., Tabouret M., and Latgé J. P. (2005) Galactomannoproteins of Aspergillus fumigatus. Eukaryot. Cell 4, 1308–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schneider P., and Ferguson M. A. J. (1995) Microscale analysis of glycosylphosphatidylinositol structures. Methods Enzymol. 250, 614–630 [DOI] [PubMed] [Google Scholar]
- 77.Haslam S. M., Coles G. C., Morris H. R., and Dell A. (2000) Structural characterisation of the N-glycans of Dictyocaulus viviparus: discovery of the Lewisx structure in a nematode. Glycobiology 10, 223–229 [DOI] [PubMed] [Google Scholar]
- 78.Adema C. M., and Loker E. S. (2015) Digenean-gastropod host associations inform on aspects of specific immunity in snails. Dev. Comp. Immunol. 48, 275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tasumi S., and Vasta G. R. (2007) A galectin of unique domain organization from hemocytes of the Eastern oyster (Crassostrea virginica) is a receptor for the protistan parasite Perkinsus marinus. J. Immunol. 179, 3086–3098 [DOI] [PubMed] [Google Scholar]
- 80.Varki A. (2006) Nothing in glycobiology makes sense, except in the light of evolution. Cell 126, 841–845 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.