A loss-of-function mutation in a key enzyme of GA biosynthesis underlies alpine dwarfism.
Abstract
Alpine dwarfism is widely observed in alpine plant populations and often considered a high-altitude adaptation, yet its molecular basis and ecological relevance remain unclear. In this study, we used map-based cloning and field transplant experiments to investigate dwarfism in natural Arabidopsis (Arabidopsis thaliana) accessions collected from the Swiss Alps. A loss-of-function mutation due to a single nucleotide deletion in gibberellin20-oxidase1 (GA5) was identified as the cause of dwarfism in an alpine accession. The mutated allele, ga5-184, was found in two natural Arabidopsis populations collected from one geographic region at high altitude, but was different from all other reported ga5 null alleles, suggesting that this allele has evolved locally. In field transplant experiments, the dwarf accession with ga5-184 exhibited a fitness pattern consistent with adaptation to high altitude. Across a wider array of accessions from the Swiss Alps, plant height decreased with altitude of origin, but fitness patterns in the transplant experiments were variable and general altitudinal adaptation was not evident. In general, our study provides new insights into molecular basis and possible ecological roles of alpine dwarfism, and demonstrates the importance of the GA-signaling pathway for the generation of ecologically relevant variation in higher plants.
Adaptation to environmental conditions is one of the evolutionary processes that can lead to lineage divergence and the generation of biodiversity (Savolainen et al., 2013). Local adaptation within species may evolve in spatially heterogeneous environments if the strength of divergent selection among populations overrides other evolutionary forces (e.g. genetic drift, gene flow), and is manifested in an increased fitness of local genotypes in the local habitat when compared with foreign genotypes in the local habitat and when compared with fitness of the local genotypes in foreign habitats (Kawecki and Ebert, 2004; Savolainen et al., 2013). It is often a challenge for classic studies of local adaptation to identify traits mediating local adaptation and the underlying genetic architectures. To date, our knowledge on the molecular basis of local adaptation remains remarkably limited, even though this topic has been extensively studied in recent decades (Bergelson and Roux, 2010; Barrett and Hoekstra, 2011). A particular difficulty in studying the molecular basis of local adaptation comes from the need to combine meaningful field experiments together with relevant molecular studies to provide solid evidence on the adaptive value of phenotypes and associated genotypes. Therefore, a complete study of molecular mechanisms of adaptation ideally requires (1) identifying candidate traits under natural selection or relevant for adaptation, (2) isolating genes/loci that influence candidate traits, and (3) quantifying the fitness (adaptive value) conferred by different alleles under natural conditions (Bergelson and Roux, 2010). In the model plant Arabidopsis (Arabidopsis thaliana) and its relatives, as well as in several animal systems, great advances in detecting the molecular basis of adaptation have been made (e.g. Linnen et al., 2009; Fournier-Level et al., 2011; Hancock et al., 2011). Nevertheless, discussions on adaptionist storytelling or of producing molecular spandrels have been recurrent (for review, see Barrett and Hoekstra, 2011).
Alpine habitats are characterized by a fine mosaic of heterogeneous and often extreme environmental conditions (Körner, 2003; Byars et al., 2007; Pico, 2012). Along altitudinal gradients, changes in environmental conditions are often steep, and plants growing in alpine areas have developed a variety of morphological and physiological adaptations that allow them to cope with extreme conditions. Alpine plant dwarfism, that is, reduced plant stature with increasing altitude, is one of the most common characteristics observed in plant populations originating from high altitudes (Clausen et al., 1948; Körner, 2003). Alpine dwarfism is thought to help alpine plants take advantage of the higher ambient temperature close to the soil surface, allocate more resources to reproduction, decrease damage from strong wind, and reduce evaporation (Turesson, 1922; Körner, 2003). Therefore, alpine dwarfism is widely considered as adaptive in plants (Turesson, 1922; Clausen et al., 1948; Körner, 2003; Byars et al., 2007; Gonzalo-Turpin and Hazard, 2009). However, whether alpine dwarfism is indeed adaptive and how it is genetically controlled remain unknown in many species. This may be partially attributable to the limited genomic resources that are available for alpine plant species, which are usually nonmodel organisms. Hence, studying the molecular basis of this ecologically important trait in model organisms such as Arabidopsis can give much-needed insights (Bergelson and Roux, 2010).
Dwarfism and semidwarfism are important and well-studied traits in agriculture because they help overcome lodging and thus substantially contributed to the green revolution in the last century, the unprecedented increase in crop yields due to the adoption of genetically improved crop varieties (Peng et al., 1999; Khush, 2001). It has repeatedly been found in a diversity of plants that dwarfism and semidwarfism were due to deficiencies in either signaling or biosynthesis of GA (e.g. Spray et al., 1996; Peng et al., 1999; Monna et al., 2002). Dwarfism and dwarfing alleles have also been reported in natural Arabidopsis accessions (Barboza et al., 2013), and other recent studies have shown evidence of population differentiation and climatic adaptation along altitudinal gradients in this model plant (Montesinos et al., 2009; Méndez-Vigo et al., 2011, 2013; Montesinos-Navarro et al., 2011; Pico, 2012; Suter et al., 2014; Luo et al., 2015). In this study, we have investigated plant dwarfism in natural Arabidopsis accessions collected in the Swiss Alps (Supplemental Table S1) and identified a loss-of-function mutation in gibberellin20-oxidase1 (GA5, also called GA20ox1) as the cause of dwarfism in an Arabidopsis accession collected from high altitude (2,012 m above sea level [a.s.l.]). In field transplant experiments, this accession displayed fitness patterns consistent with altitudinal adaptation. Across a larger set of regional accessions, including 10 further accessions without the dwarfing mutation, plant height decreased with altitude of origin; however, this pattern could not be tied to adaptive differentiation along altitude.
RESULTS
Map-Based Cloning of an Alpine Dwarfing Gene
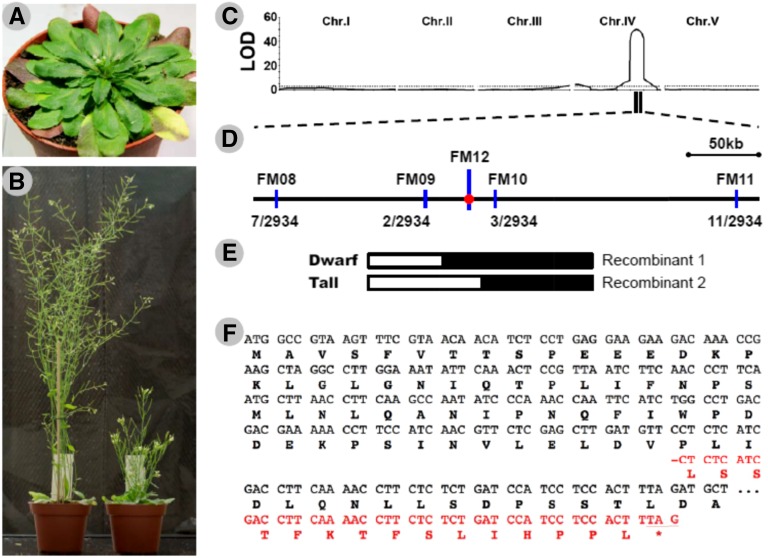
To study the molecular basis underlying alpine dwarfism, we surveyed the trait in accessions propagated from natural Arabidopsis populations originating from 800 to 2,700 m a.s.l. in the Swiss Alps (Supplemental Fig. S1; Supplemental Table S1). We identified accessions in an alpine population (named SAO) collected from an altitude of 2,012 m, showing a stable dwarf phenotype in our greenhouse experiments (Figs. 1, A and B, and 2, C and D). To investigate the genetic basis underlying dwarfism in SAO, QTL mapping was performed using an F2 population derived from Col-0 (mother, a reference line without dwarfism) and SAO (father, with dwarfism; Fig. 2D). A major QTL locus on chromosome 4 that explained a large proportion (62%) of the phenotypic variation of plant height was identified (Fig. 1C). The analysis was followed by fine mapping, and the major QTL was located within a 54-kb genomic region (Fig. 1, D and E) that contains a well-known dwarfing gene (GA5, locus tag At4g25420). The functional copy of GA5 encodes 377 amino acids and controls a key step of GA biosynthesis in vivo, whereas a loss-of-function mutant in GA5 exhibited plant dwarfism (Koornneef and van der Veen, 1980; Xu et al., 1995). The GA5 locus in SAO was sequenced, and a 1-bp deletion at position 184 of the open reading frame (ga5-184) was identified (Supplemental Fig. S2). This deletion induced a frameshift and consequently a premature stop codon that resulted in a truncated protein (Fig. 1F). Given that this truncated protein has only 76 amino acids and has lost most functional motifs of GA5 (Xu et al., 1995), ga5-184 is likely to be a loss-of-function mutation.
Figure 1.
Map-based cloning of the dwarfing gene. A, An SAO plant showing the morphology when the first flower opens in the greenhouse. B, Adult plants of Columbia (Col-0; left) and SAO (right) in the greenhouse. C, Logarithm of odds (LOD) maps of quantitative trait locus (QTL) analysis for total plant height in an F2 population derived from a cross between Col-0 (mother) and SAO (father). The dashed line indicates the position of the genome-wide significance threshold (LOD = 3.0). Chr., Chromosome. D, High-resolution linkage map of the GA5 region generated with 1,467 F2 plants of three near-isogenic lines. The number of detected recombinants between each indicated marker (blue lines) and FM12 (a molecular marker located in the GA5 gene shown by the red point) and the total number of examined chromosomes are shown under the map. E, Recombinants identified from the near-isogenic line populations delimit the causal locus to the 54-kb region between markers FM09 and FM10. White and black boxes indicate homozygous chromosomal segments from Col-0 and SAO, respectively. F, The beginning portion of the coding region and deduced amino acid sequences of the Arabidopsis GA5 gene. The sequence from Col-0 is shown in black, and the SAO frameshift sequence between a 1-bp deletion and the resulting stop codon is shown in red. −, The 1-bp deletion at position 184 of the SAO ga5 gene (ga5-184); *, the premature stop codon caused by the deletion.
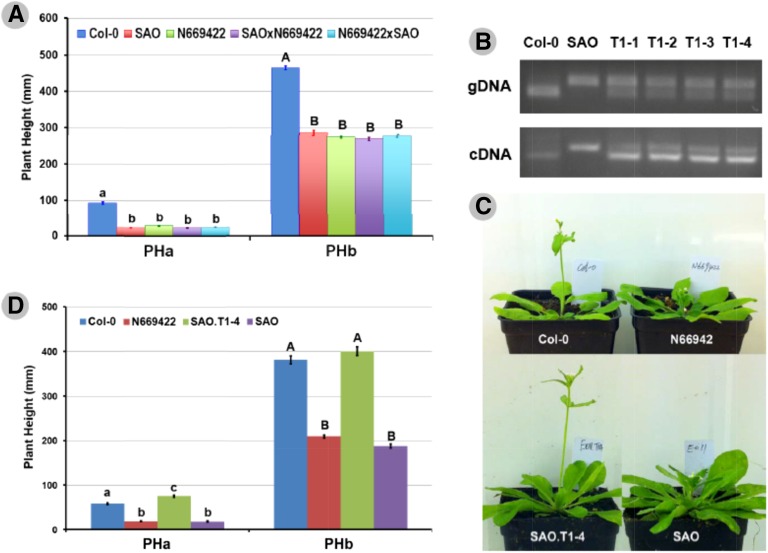
Figure 2.
Validation of the isolated dwarfing gene. A, Allelism tests between SAO and N669422 (a transferred DNA insertional mutant of GA5). Means ± se of plant height when the first flower opens (PHa) and plant height toward the end of growth (PHb) of indicated genotypes are shown. Letters above bars indicate statistical significance of pairwise differences within each group, as examined by Tukey’s post hoc tests. B, Validation of transformation of the functional GA5 copy from Col-0 into ga5-184 background using a cleaved-amplified polymorphic sequence (CAPS) marker in both genomic and transcriptional levels (for more details, see “Materials and Methods”). Templates for PCR are indicated on the left, whereas genotypes are shown above. gDNA, Genomic DNA; cDNA, complementary DNA. C, Morphology of plants when the first flower opens in different genetic backgrounds as indicated. Plants were germinated and grown under the same conditions, and photos were taken at different dates due to different flowering times. D, Measurements of PHa and PHb of indicated genetic backgrounds. Letters above bars indicate statistical significance of pairwise differences within each group, as examined by Tukey’s post hoc tests.
Distribution of the Dwarfing Allele
We directly sequenced the region containing the mutation described earlier in 16 to 26 accessions from each of 15 Swiss populations (Supplemental Table S2) and found that (1) in the SAO population, 15 out of 17 accessions carried ga5-184; (2) in another population (named SF3) from 1,792 m (a.s.l.) in the same area, 16 out of 26 accessions carried this mutation; (3) the accessions carrying the ga5-184 allele exhibit dwarf phenotypes under greenhouse conditions (Supplemental Table S2); and (4) the ga5-184 allele was not present in other investigated populations. As a second step, we examined the ga5-184 mutation in 58 worldwide natural accessions (Supplemental Table S3; accessions with altitudinal information were chosen) and did not find the ga5-184 mutation. Third, we determined that the ga5-184 allele is different from previously reported haplotypes of GA5 representing multiple independent loss-of-function mutations (Barboza et al., 2013; Supplemental Fig. S3). We also failed to detect the ga5-184 mutation in 855 genomes (all available genomes on November 12, 2013) in the Arabidopsis 1001 Genomes database. Together, this suggests that the ga5-184 allele likely evolved locally in the Saas Fee region of the Swiss Alps where SAO and SF3 were collected.
Validation of the Isolated Dwarfing Gene
Allelism tests were performed on 93 F1 plants originating from reciprocal crosses between SAO (individual with dwarfism, same as below) and N669422 (a homozygous transferred DNA insertional line targeting the GA5 gene). The F1 plants were grown together with parental accessions including Col-0, SAO, and N669422 in the greenhouse, and validated by visual observation on leaf morphology and flowering time compared with their parental plants. When compared with Col-0 (the background line), N669422 exhibited an obvious dwarf phenotype (Fig. 2A), suggesting the effectiveness of GA5 gene silencing induced by the T-DNA insertion. SAO and N669422 had highly similar morphology in various developmental stages, and failed to complement each other in F1 plants (Fig. 2A).
In addition, a complementation test was performed by creating transgenic plants. The functional GA5 copy from Col-0 was cloned and then transferred into the ga5-184 background. The derived transgenic plants carried both the ga5-184 and wild-type (Col-0) alleles at the genomic and transcriptional levels (Fig. 2B), indicating success of our transformation experiment. These transgenic plants showed a similar morphology to Col-0 but no dwarfism as SAO or N669422 plants (Fig. 2, C and D). These results indicate that the functional GA5 allele from Col-0 rescues the dwarf phenotype induced by ga5-184. Thus, we conclude that the loss-of-function mutation of ga5-184 causes dwarfism in our alpine accessions SAO and SF3.
Transplant Experiments
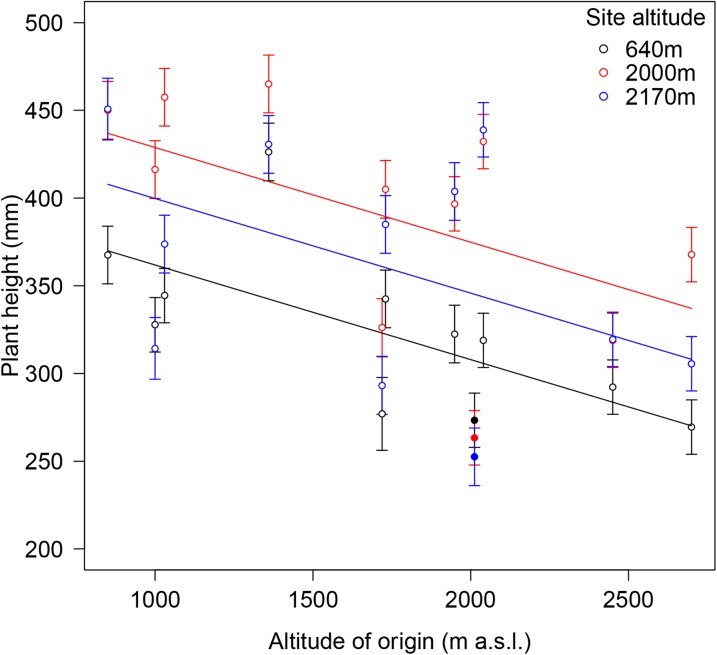
We investigated trait expression and performance in 11 Swiss accessions of Arabidopsis, including an accession from SAO with the dwarfing allele described earlier. The second accession with this allele, SF3, was not included in this experiment as the allele was detected only afterward. Eight to nine individuals of each accession were transplanted into each of three field sites, one at low altitude (640 m a.s.l.) and two at high altitude (2,000 and 2,170 m a.s.l.; for more details, see “Materials and Methods”). An altitudinal cline analysis of plant height using a mixed model with accession as a random effect showed that plant height decreased with altitude of origin, regardless of the transplant sites (the interaction between site and altitude of origin was not significant at the significance level of α = 0.05; Fig. 3; Table I). This suggests that a decrease in plant height with altitude of origin is mostly genetically, and not environmentally, determined. Plant height nonetheless exhibited significant overall differences between transplant sites: plants were smallest at the low site at 640 m, intermediate at the highest at 2,170 m, and tallest at 2, 000 m (Fig. 3; Supplemental Table S4). Interestingly, this constitutes a counter-gradient pattern where genotypic effects oppose environmental effects (e.g. Gonzalo-Turpin and Hazard, 2009). The SAO accession carrying the dwarfing ga5-184 allele, however, did not show this pattern; only in this accession, plant height did not vary considerably with transplant sites, suggesting that plant height is less plastic in this accession than in the other accessions.
Figure 3.
Altitudinal clines in plant height in 11 Arabidopsis accessions transplanted into field sites at 640, 2,000, and 2,170 m a.s.l. Means ± 1 se of plant height together with significant regression lines are shown. The SAO accession with a ga5-184 dwarfing allele is represented as black, red, and blue circles.
Table I. Mixed model analyses of altitudinal clines in plant height and local adaptation using total seed number as a fitness proxy in 11 Arabidopsis accessions collected from altitudes from 820 to 2,700 m in the Swiss Alps.
Accessions were transplanted into experimental sites at 640, 2,000, and 2,170 m a.s.l. Effects were tested using likelihood ratio tests between nested models with the fixed effects site, altitude of origin (alt.), and the interaction of these factors.
| Model (Fixed Effects)a | Degrees of Freedom | Model Comparison | Plant Height |
Total Seed No.b |
||
|---|---|---|---|---|---|---|
| Deviance | P | Deviance | P | |||
| 1: alt. + site + alt. * site | 8 | 2,943 | 533.4 | |||
| 2: alt. + site | 6 | 2 vs. 1 | 2,946 | 0.2330 | 546.0 | 0.0018 |
| 3: Site | 5 | 3 vs. 2 | 2,951 | 0.0301 | −c | |
| 4: Altitude | 4 | 4 vs. 2 | 3,015 | <9.4*10−16 | −c | −c |
All models included accession as a random effect.
Log transformed.
No further model reduction because of significant interaction term.
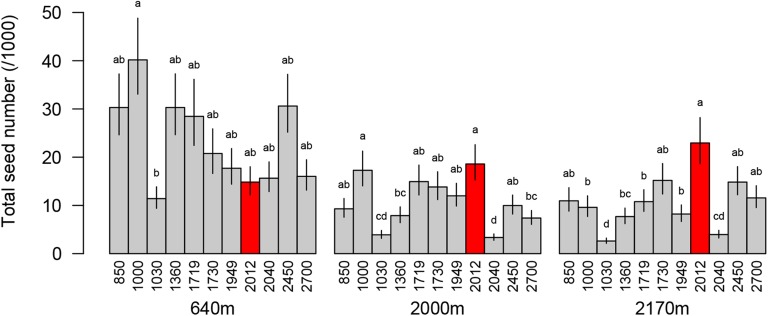
We tested for altitudinal adaptation by relating a fitness proxy, total seed number, to altitude, site, and the altitude-by-site interaction using mixed models with accession as a random effect. Although the interaction was statistically significant, the altitude of origin did not have a significant effect on total seed number at any of the sites (Supplemental Table S5; Table I), suggesting that there is no general pattern of altitudinal adaptation in the accessions studied here. We further explored patterns in total seed number at the three sites using ANOVA. Here, differences in total seed number between genotypes also depended on the site (significant interaction term;Fig. 4; Table II). Tukey tests within sites at an overall significance level of α = 0.05 showed that the SAO accession grouped with the lowest performing accessions at the low-altitude site but was among the best performing accessions at the higher sites (Fig. 4). Indeed, this dwarf accession appeared to increase total seed number with increasing altitude, unlike all other accessions (Fig. 4). These patterns are consistent with adaptation to high altitude of the SAO population.
Figure 4.
Total seed number (means ± 1 se) as a fitness proxy in 11 Arabidopsis accessions collected from different altitudes and transplanted into field sites at 640, 2,000, and 2,170 m a.s.l. Bars with different letters are significantly different (Tukey’s test), and accessions are ordered by altitude of origin. The SAO accession with a ga5-184 dwarfing allele is represented as red bars.
Table II. ANOVA of a fitness proxy, total seed number, in 11 Arabidopsis accessions collected from altitudes from 820 to 2,700 m a.s.l. in the Swiss Alps and transplanted into experimental sites at 640, 2,000, and 2,170 m a.s.l.
| Degrees of Freedom | Mean Square | F Value | P (>F) | |
|---|---|---|---|---|
| Accession | 10 | 5.042 | 14.75 | <2.2*10−16 |
| Site | 2 | 21.18 | 61.96 | <2.2*10−16 |
| Accession * site | 20 | 1.033 | 3.023 | 3.0 *10−5 |
| Residuals | 240 | 0.3418 |
Selection differentials for plant height were significant and positive for the low-altitude site and one of the higher altitude sites (β = 0.34 [95% bootstrap confidence interval: 0.27, 0.42] and β = 0.21 [0.06, 0.39] for the 640- and 2,000-m site, respectively; Supplemental Table S6), whereas the selection differential was not significantly different from zero at the highest site at 2,170 m as its confidence interval included zero (β = 0.03122 [−0.1452, 0.2025]; Supplemental Table S6).
DISCUSSION
Although alpine dwarfism is commonly observed across diverse alpine plant species, its molecular basis remains poorly investigated. In this study, we report that alpine dwarfism in natural Arabidopsis accessions is caused by a loss-of-function mutation in GA5 resulting from a single nucleotide deletion. Plant dwarfism is due to deficiency in either signaling or biosynthesis of GA across diverse plant species (e.g. Spray et al., 1996; Peng et al., 1999; Monna et al., 2002). GA promotes plant growth through cell expansion and proliferation by interacting with the GA-Gibberellin Insensitive Dwarf1-DELLA module in vivo (Sun, 2011). A reduction of GA levels and signaling, therefore, constrains plant growth (Achard et al., 2006; Colebrook et al., 2014). Mutations in GA5 were previously identified as a cause of small stature in worldwide accessions of Arabidopsis (Barboza et al., 2013), and a regulation of dwarfism by GAs has been proposed for an arctic plant species, Stellaria longipes (Emery et al., 2001; Kurepin et al., 2006). The new ga5-184 allele reported here was found in two high-altitude populations from the same region, SF3 (1,792 m a.s.l.) and SAO (2,012 m a.s.l.), suggesting that the evolution of this dwarfing allele is unlikely an isolated and transient event. In contrast to other regional accessions, SAO exhibited a signature of altitudinal adaptation in field transplant experiments (SF3 was not included in field experiments): SAO individuals were among the best performing accessions at the high-altitude transplant sites (local versus foreign criterion; Kawecki and Ebert, 2004), and SAO had higher seed output at the high-altitude transplant sites compared with the low-altitude site (home versus away criterion; Kawecki and Ebert, 2004). However, from our data, we cannot make inferences on whether altitudinal adaptation in SAO was caused by the dwarf phenotype. This would require comparing individuals with and without the dwarfing allele in the same genetic background under contrasting altitudinal conditions. Nonetheless, our study provides direct molecular evidence for the molecular causes of alpine dwarfism and further supports the involvement of GA-signaling genes in the evolution of alpine dwarfism in higher plants.
Our transplant experiments further included a wide range of Swiss accessions collected at altitudes between 850 and 2,700 m a.s.l. Plant height declined with altitude of origin, irrespective of the transplant sites. In the array of accessions used here, only one accession carried the dwarfing allele of ga5-184, suggesting that the variation in plant height across all accessions is complicated by other factors. A decrease in plant height with altitude is expected if altitudinal selection led to trait divergence; however, it is also consistent with a role of genetic drift (Leinonen et al., 2013; Luo et al., 2015). Indeed, analyses of a fitness proxy, total seed number, revealed that accessions did not generally perform better at an altitude close to their altitude of origin. This implies that either genetic drift or selective agents unrelated to altitude may have contributed to performance differences. Nonetheless, the plant height decrease with altitude of origin is consistent with patterns of phenotypic selection identified in our transplant experiments: at low altitude, there was strong selection against small-stature phenotypes (positive selection differential for plant height), whereas at high altitude, we found either weaker selection against small-stature phenotypes or a nonsignificant selection differential. However, for these analyses, we only considered fitness effects arising from the juvenile life history stage onward and cannot exclude differences between accessions at earlier life history stages, such as germination and early establishment (Donohue et al., 2010).
One of the major challenges in studies of local adaptation is to conduct joint analyses combining meaningful ecological and solid molecular experiments. Here, we used recently collected regional accessions for forward-genetic analyses and for field transplant experiments, and uncovered a balanced, albeit complicated, picture of the generation of alpine dwarfism. A locally evolved dwarfing allele, ga5-184, was found in an alpine accession exhibiting signs of altitudinal adaptation. A decrease of plant height with altitude of origin across a larger set of accessions, however, could not be attributed to altitudinal adaptation. Overall, our study provides new insights into the molecular basis of alpine dwarfism and shows that modifications of the GA-signaling pathway are important for the generation of ecologically relevant variation.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Information on Arabidopsis (Arabidopsis thaliana) accessions used in this study is presented in Supplemental Table S1 and Supplemental Figure S1. Seeds were sown and kept at 4°C for 4 to 5 d to break seed dormancy and were germinated and grown at 22°C/20°C (day/night) under long-day light conditions (16 h of light/8 h of dark) for 7 d. The plants were then vernalized at 4°C under short-day light conditions (8 h of light/16 h of dark) for 6 weeks and were subsequently transplanted into individual pots or field sites as described below.
QTL Mapping, Map-Based Cloning, and Sequencing
The 373 F2 individuals derived from a cross between Col-0 (mother) and SAO (father) were grown and used for phenotypic measurement of plant height in the greenhouse. These plants were genotyped using 31 fluorescent-labeled microsatellite markers (markers were selected from the following Web site according to their genomic positions: http://www.inra.fr/internet/Produits/vast/msat.php) across the Arabidopsis genome. Linkage maps were constructed using JoinMap version 4 (Van Ooijen and Voorrips, 2001), and QTL analysis was performed with MapQTL version 5 (Van Ooijen, 2004). To improve the normality of phenotypic data, plant height was box-cox transformed. QTL mapping was initially performed on transformed data with interval mapping followed by composite interval mapping, referred to as multiple QTL mapping in MapQTL version 5, where markers outside the test interval in the genome were used as cofactors to increase the power and precision of QTL identification. The significant cofactors for each multiple QTL mapping model were determined through an iterative automatic cofactor selection. The genome-wide LOD significance threshold was obtained from permutation tests with 1,000 replicates as implemented in MapQTL version 5. The percentage of variance explained for each QTL was estimated at the LOD peak using MapQTL version 5.
For fine mapping, we backcrossed three F2 individuals that exhibited the dwarf phenotype (SAO-like) to Col-0 and obtained BC1F1 lines. With marker-assisted analyses, the BC1F1 lines were further backcrossed to Col-0 to generate BC2F1 plants; subsequently, BC2F2 populations were obtained. In total, 1,467 BC2F2 individuals were used for the fine-mapping analyses with five PCR-based genetic markers (primers in Supplemental Table S7).
To detect possible mutations in the GA5 gene in SAO and other populations, one to three overlapping genomic fragments were amplified from genomic DNA (primers in Supplemental Table S7), sequenced using a 3730xl DNA Analyzer (Applied Biosystems, Inc.) and analyzed using Sequencer version 4.0 (Gene Codes Corp.). In addition, the GA5 coding sequences from 855 genomes were obtained on November 12, 2013, from the Arabidopsis 1001 Genomes project (http://signal.salk.edu/atg1001/3.0/gebrowser.php) and analyzed.
Allelism Tests and Transgenic Experiments
To perform allelism tests, 47 and 46 F1 plants generated by reciprocal genetic crosses between SAO and N669422 were grown together with eight individuals of each of the involved parents (i.e. Col-0, SAO, and N669422). Plant height at two developmental stages (i.e. the beginning and the end of flowering) was measured and analyzed.
To conduct the transgenic complementation test, a functional GA5 copy was amplified by PCR from complementary DNA derived from Col-0 (GA5_Col-0; primers in Supplemental Table S7), cloned into the binary vector of pCAMBIA1300, confirmed by direct sequencing, and transferred to Agrobacterium tumefaciens. Subsequently, transformation of Arabidopsis into the ga5-184 background was performed according to the floral dip method (Clough and Bent, 1998). Given that the mutation of ga5-184 eliminated a restriction enzyme site (i.e. MnlI), we designed a CAPS marker (primers in Supplemental Table S7) to distinguish the GA5_Col-0 haplotype from ga5-184. Genomic DNA and total RNA were isolated from various individuals, including transgenic plants, and used for the validation by the CAPS maker. In addition, phenotypic investigation of plant heights with various genetic backgrounds was conducted to validate the functional complementation.
Field Experiments and Trait Measurements
Transplant experiments were performed in the Swiss Alps (in the area of Flims, Switzerland) at one low-altitude site (named S1:640; altitude, 640 m a.s.l.; coordinates, 46° 52′ 23.45′′ N/9° 31′ 07.01′′ E) and two high-altitude sites (S2:2000: altitude, 2,000 m; coordinates, 46° 53′ 16.04′′ N/9° 29′ 22.11′′ E; and S3:2170: altitude, 2,170 m; coordinates, 46° 51′ 52.11′′ N/9° 14′ 11.83′′ E).
To reduce the logistic pressure of performing field experiments in high mountains, we had to control the size of our experimental populations. Therefore, single-seed descents (accessions generated from seeds of a single plant from each population) were transplanted into the field sites. In brief, eight or nine seedlings of each population/accession were randomly transplanted into a rectangular block in each site and with a distance of 10 cm between direct neighbors 7 d after germination, as described previously (Luo and Widmer, 2013). We used the same artificial soil for the experiments (i.e. the same soil was filled in blocks and pots) and performed the transplantation so that it coincided with the growth season of local plant populations as possible. The dates for transplantation were April 21, 2011; May 30, 2011; and June 16, 2011 for S1:640, S2:2000, and S3:2170, respectively.
Toward the end of plant growth, the numbers of nondispersed and dispersed siliques were counted and used to estimate the harvesting index (i.e. HI, the percentage of harvested seeds in total seed no.; HI = nondispersed/total silique no.) in each individual. Afterward, aboveground material of each plant was harvested, stored in an envelope, and dried in a climate chamber at 25°C for 1 week. Plant height was measured, and subsequently, all available seeds of each plant were harvested and cleaned. For each plant, (1) a small proportion of seeds (usually 15–25 mg) were weighed (W1) using an analytical balance and counted (S1) using a seed-counting machine, and (2) total seed weight (TSW) was obtained by weighing all the seeds with the analytical balance, and then total seed number (TSN) was calculated as TSN = (S1*TSW/W1)/HI. Overall, mortality in the field transplant experiments was low (for the measurement of plant height, three out of 279 individuals were not available, whereas for total seed number, four out of 279 individuals were not available). Therefore, we used the total seed number of surviving individuals as a fitness proxy for the statistical analysis (below).
Statistical Analyses of Phenotypic Data
We used mixed models with accession as a random effect to investigate (1) altitudinal clines in plant height, (2) altitudinal adaptation using total seed number as a fitness proxy, and (3) selection on plant height. Analyses were carried out with the lmer command in the package lme4 in R version 3.0.2 (Bates et al., 2013; R Core Team, 2013). The altitudinal cline model for plant height and the local adaptation model had altitude, site, and the altitude-by-site interaction as fixed effects. Selection analysis models on relative seed number (within sites) had standardized plant height (within sites), site, and the plant height-by-site interaction as fixed effects. Confidence intervals of the selection differentials were calculated using bootstrapping with the boot package for R (Canty and Ripley, 2014). In each of these analyses, we determined the significance of fixed effects by a series of likelihood ratio tests between nested models. Post hoc tests were conducted in the package multcomp using false discovery rate control for the overall significance level of α = 0.05 to correct for multiple comparisons (Hothorn et al., 2008). Adaptation to altitude was further investigated with ANOVA on total seed number with accession, site, and the site-by-accession interaction as explanatory factors, followed by post hoc comparisons of accessions within sites with Tukey’s tests at an overall significance level of α = 0.05 using standard analyses in R. For analyses of altitudinal adaptation, total seed number was log transformed to improve the model fit and the distribution of residuals.
Sequence data of the ga5-184 allele reported in this article can be found in the GenBank/EMBL data libraries under accession number KP123605.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The geographical locations in a Swiss map showing the collection sites of 11 Swiss Arabidopsis accessions and three field experimental sites in this study.
Supplemental Figure S2. Sequence alignment between SAO and Col-0.
Supplemental Figure S3. Schematic comparison between the ga5-184 haplotype in SAO/SF3 and other loss-of-function haplotypes reported in Barboza et al. (2013).
Supplemental Table S1. Information of natural Arabidopsis accessions from the Swiss Alps used in this study.
Supplemental Table S2. Plant dwarfism observed in the greenhouse is associated with the presence of ga5-184.
Supplemental Table S3. Information for 58 worldwide Arabidopsis accessions used in this study.
Supplemental Table S4. Multiple comparisons of plant height in Arabidopsis (11 accessions) at three transplant sites, S1 at 640, S2 at 2,000, and S3 at 2,170 m a.s.l., in a mixed model analysis including accession as a random factor and altitude of origin and site as fixed factors.
Supplemental Table S5. Mixed model analysis of altitude of origin effects on total seed number in 11 accessions of Arabidopsis at three transplant sites.
Supplemental Table S6. Selection analysis for plant height in 11 Arabidopsis accessions at each of three transplant sites at different altitudes.
Supplemental Table S7. Primers used for fine mapping, direct sequencing, and transgene in this study.
Supplementary Material
Acknowledgments
We thank F. Berger, B. Blattmann, M. Frei, and the Genetic Diversity Center at ETH Zurich for technical support; Drs. Q. Long, T. Tsuchimatsu, and J. Levine for a valuable discussion on the earlier versions of the article; and the editor, Dr. Jeff J. Doyle, and two anonymous reviewers who provided critical and helpful comments that improved the quality of the article.
Glossary
- a.s.l.
above sea level
- LOD
logarithm of odds
- QTL
quantitative trait locus
- CAPS
cleaved-amplified polymorphic sequence
- HI
harvesting index
Footnotes
This work was supported by Jiangsu Normal University (grant no. 13XLR031 for doctoral teachers), the Natural Science Foundation of Jiangsu Province (grant no. BK20141146), the Priority Academic Program Development of Jiangsu Higher Education Institutions, and the Competence Center Environment and Sustainability of the ETH Domain.
Articles can be viewed without a subscription.
References
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Barboza L, Effgen S, Alonso-Blanco C, Kooke R, Keurentjes JJ, Koornneef M, Alcázar R (2013) Arabidopsis semidwarfs evolved from independent mutations in GA20ox1, ortholog to green revolution dwarf alleles in rice and barley. Proc Natl Acad Sci USA 110: 15818–15823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RD, Hoekstra HE (2011) Molecular spandrels: tests of adaptation at the genetic level. Nat Rev Genet 12: 767–780 [DOI] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B, Walker S (2013) lme4: linear mixed-effects models using Eigen and S4. R package version 1.0-5. http://CRAN.R-project.org/package=lme4
- Bergelson J, Roux F (2010) Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. Nat Rev Genet 11: 867–879 [DOI] [PubMed] [Google Scholar]
- Byars SG, Papst W, Hoffmann AA (2007) Local adaptation and cogradient selection in the alpine plant, Poa hiemata, along a narrow altitudinal gradient. Evolution 61: 2925–2941 [DOI] [PubMed] [Google Scholar]
- Canty A, Ripley B (2014) boot: Bootstrap R (S-Plus) Functions. R package version 1.3-13
- Clausen J, Keck DD, Heisey WM (1948) Experimental studies on the nature of species III: Environmental responses of climatic races of Achillea. Carnegie Institute of Washington, D.C., USA [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colebrook EH, Thomas SG, Phillips AL, Hedden P (2014) The role of gibberellin signalling in plant responses to abiotic stress. J Exp Biol 217: 67–75 [DOI] [PubMed] [Google Scholar]
- Donohue K, de Casas RR, Burghardt L, Kovach K, Willis CG (2010) Germination, post-germination adaptation, and species ecological ranges. Annu Rev Ecol Evol Syst 41: 293–319 [Google Scholar]
- Emery RJN, Pearce DW, Pharis RP, Reid DM, Chinnappa CC (2001) Stem elongation and gibberellins in alpine and prairie ecotypes of Stellaria longipes. Plant Growth Regul 35: 17–29 [Google Scholar]
- Fournier-Level A, Korte A, Cooper MD, Nordborg M, Schmitt J, Wilczek AM (2011) A map of local adaptation in Arabidopsis thaliana. Science 334: 86–89 [DOI] [PubMed] [Google Scholar]
- Gonzalo-Turpin H, Hazard L (2009) Local adaptation occurs along altitudinal gradient despite the existence of gene flow in the alpine plant species Festuca eskia. J Ecol 97: 742–751 [Google Scholar]
- Hancock AM, Brachi B, Faure N, Horton MW, Jarymowycz LB, Sperone FG, Toomajian C, Roux F, Bergelson J (2011) Adaptation to climate across the Arabidopsis thaliana genome. Science 334: 83–86 [DOI] [PubMed] [Google Scholar]
- Hothorn T, Bretz F, Westfall P (2008) Simultaneous inference in general parametric models. Biom J 50: 346–363 [DOI] [PubMed] [Google Scholar]
- Kawecki TJ, Ebert D (2004) Conceptual issues in local adaptation. Ecol Lett 7: 1225–1241 [Google Scholar]
- Khush GS. (2001) Green revolution: the way forward. Nat Rev Genet 2: 815–822 [DOI] [PubMed] [Google Scholar]
- Koornneef M, van der Veen JH (1980) Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) heynh. Theor Appl Genet 58: 257–263 [DOI] [PubMed] [Google Scholar]
- Körner C. (2003) Alpine plant life: functional plant ecology of high mountain ecosystems. Second edition Springer, Berlin, Germany [Google Scholar]
- Kurepin LV, Mancell L, Reid DM, Pharis RP, Chinnappa CC (2006) Possible roles for ethylene and gibberellin in the phenotypic plasticity of an alpine population of Stellaria longipes. Can J Bot 84: 1101–1109 [Google Scholar]
- Leinonen PH, Remington DL, Leppälä J, Savolainen O (2013) Genetic basis of local adaptation and flowering time variation in Arabidopsis lyrata. Mol Ecol 22: 709–723 [DOI] [PubMed] [Google Scholar]
- Linnen CR, Kingsley EP, Jensen JD, Hoekstra HE (2009) On the origin and spread of an adaptive allele in deer mice. Science 325: 1095–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Widmer A (2013) Herkogamy and its effects on mating patterns in Arabidopsis thaliana. PLoS ONE 8: e57902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Widmer A, Karrenberg S (2015) The roles of genetic drift and natural selection in quantitative trait divergence along an altitudinal gradient in Arabidopsis thaliana. Heredity (Edinb) 114: 220–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez-Vigo B, Gomaa NH, Alonso-Blanco C, Picó FX (2013) Among- and within-population variation in flowering time of Iberian Arabidopsis thaliana estimated in field and glasshouse conditions. New Phytol 197: 1332–1343 [DOI] [PubMed] [Google Scholar]
- Méndez-Vigo B, Picó FX, Ramiro M, Martínez-Zapater JM, Alonso-Blanco C (2011) Altitudinal and climatic adaptation is mediated by flowering traits and FRI, FLC, and PHYC genes in Arabidopsis. Plant Physiol 157: 1942–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monna L, Kitazawa N, Yoshino R, Suzuki J, Masuda H, Maehara Y, Tanji M, Sato M, Nasu S, Minobe Y (2002) Positional cloning of rice semidwarfing gene, sd-1: rice “green revolution gene” encodes a mutant enzyme involved in gibberellin synthesis. DNA Res 9: 11–17 [DOI] [PubMed] [Google Scholar]
- Montesinos A, Tonsor SJ, Alonso-Blanco C, Picó FX (2009) Demographic and genetic patterns of variation among populations of Arabidopsis thaliana from contrasting native environments. PLoS ONE 4: e7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos-Navarro A, Wig J, Pico FX, Tonsor SJ (2011) Arabidopsis thaliana populations show clinal variation in a climatic gradient associated with altitude. New Phytol 189: 282–294 [DOI] [PubMed] [Google Scholar]
- Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, et al. (1999) ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400: 256–261 [DOI] [PubMed] [Google Scholar]
- Picó FX. (2012) Demographic fate of Arabidopsis thaliana cohorts of autumn- and spring-germinated plants along an altitudinal gradient. J Ecol 100: 1009–1018 [Google Scholar]
- R Core Team (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria. http://www.R-project.org/
- Savolainen O, Lascoux M, Merilä J (2013) Ecological genomics of local adaptation. Nat Rev Genet 14: 807–820 [DOI] [PubMed] [Google Scholar]
- Spray CR, Kobayashi M, Suzuki Y, Phinney BO, Gaskin P, MacMillan J (1996) The dwarf-1 (dt) Mutant of Zea mays blocks three steps in the gibberellin-biosynthetic pathway. Proc Natl Acad Sci USA 93: 10515–10518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP. (2011) The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol 21: R338–R345 [DOI] [PubMed] [Google Scholar]
- Suter L, Rüegg M, Zemp N, Hennig L, Widmer A (2014) Gene regulatory variation mediates flowering responses to vernalization along an altitudinal gradient in Arabidopsis. Plant Physiol 166: 1928–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turesson G. (1922) The genotypical response of the plant species to the habitat. Hereditas 3: 211–350 [Google Scholar]
- Van Ooijen JW. (2004) MapQTL 5, software for the mapping of quantitative trait loci in experimental populations. Wageningen, Netherlands: Kyazma BV [Google Scholar]
- Van Ooijen JW, Voorrips RE (2001) Joinmap(R) version 3.0: Software for the calculation of genetic linkage maps. Plant Research International, Wageningen, The Netherlands [Google Scholar]
- Xu YL, Li L, Wu K, Peeters AJ, Gage DA, Zeevaart JA (1995) The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proc Natl Acad Sci USA 92: 6640–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.