Abstract
Leaf rust is an important disease, threatening wheat production annually. Identification of resistance genes or QTLs for effective field resistance could greatly enhance our ability to breed durably resistant varieties. We applied a genome wide association study (GWAS) approach to identify resistance genes or QTLs in 338 spring wheat breeding lines from public and private sectors that were predominately developed in the Americas. A total of 46 QTLs were identified for field and seedling traits and approximately 20–30 confer field resistance in varying degrees. The 10 QTLs accounting for the most variation in field resistance explained 26–30% of the total variation (depending on traits: percent severity, coefficient of infection or response type). Similarly, the 10 QTLs accounting for most of the variation in seedling resistance to different races explained 24–34% of the variation, after correcting for population structure. Two potentially novel QTLs (QLr.umn-1AL, QLr.umn-4AS) were identified. Identification of novel genes or QTLs and validation of previously identified genes or QTLs for seedling and especially adult plant resistance will enhance understanding of leaf rust resistance and assist breeding for resistant wheat varieties. We also developed computer programs to automate field and seedling rust phenotype data conversions. This is the first GWAS study of leaf rust resistance in elite wheat breeding lines genotyped with high density 90K SNP arrays.
Introduction
Wheat leaf rust, caused by the fungus Puccinia triticina Eriks, is a threat to world wheat production. Identification of resistance genes using molecular markers is an important step toward marker assisted selection and resistance breeding. To date, there have been more than 70 leaf rust resistance genes identified, the majority of which confer leaf rust resistance in the seedling stage and are race-specific [1]. Identification of resistance loci or major QTLs that confer adult plant or field resistance against leaf rust will enhance our ability to develop leaf rust resistant wheat varieties.
One way to identify leaf rust resistance QTLs, is through association mapping (AM). Association mapping has the potential of accommodating wide collections of germplasm and due to historic recombination, AM studies generally have better mapping resolution compared to bi-parental mapping [2]. With decreasing sequencing costs and the advent of high throughput genotyping assays [3–5], association mapping using high density genome wide markers, also known as genome wide association study (GWAS), has become more widely available to plant researchers.
Association mapping for various traits in a number of crop species, including rice, corn, soybean, wheat, barley, tomato and potato have been conducted [2, 6]. Association mapping studies were also conducted for various rust disease traits in wheat [7–13]. Maccaferri et al [7] reported association mapping of leaf rust resistance using mostly SSR markers and a collection of durum wheat (Triticum turgidum L. var. durum, tetraploid AABB), but not common wheat (allohexaploid AABBDD). Kertho et al [13] explored resistance QTLs in a collection of winter wheat landraces, but the study focused on seedling resistance, with no data on field resistance. Few GWAS studies were conducted for mapping leaf rust resistance loci in both seedling and adult plant stages, and in elite bread wheat cultivars or breeding lines. Furthermore, to our knowledge, no study has utilized the recently developed high density iSelect 90K SNP array [5] for GWAS analysis of leaf rust resistance in bread wheat.
The main feature of this mapping panel is that a large amount of the germplasm possesses resistance to leaf rust and in some cases specific genes providing the resistance are known. It is possible that such a panel will allow us to identify resistance alleles that are normally not detected due to low allele frequency. This study aims to validate known genomic loci effective to leaf rust resistance and to identify novel genes or QTLs that are effective against the leaf rust pathogen in the seedling and (or) adult plant stages. Meanwhile, this study also explores the genetic architecture and phenotypic correlations for seedling and adult plant resistance and discusses ways to implement our research results in plant breeding and genetics efforts.
Materials and Methods
Plant Materials
A total of 381 spring wheat breeding lines derived from commercial and public breeding sectors were selected to form a leaf rust association mapping (AM) panel. These lines were from different countries of North, Central and South America, as well as some parts of Africa, Europe and Asia. Public sector contributors included the University of Minnesota (MN), North Dakota State University (NDSU), South Dakota State University (SDSU), the United States Department of Agriculture, Agricultural Research Service (USDA-ARS), International Center for Maize and Wheat Improvement (CIMMYT), Uruguay, Argentina, Brazil, and Chile as well as some European and Asian countries. Private sector contributors included Syngenta (Basel, Switzerland), Limagrain (Puy-de-Dome, France) and Westbred (Monsanto Co, St. Louis, MO). The AM panel consists of wheat lines with leaf rust resistance to one or multiple races. Over 70 Thatcher near isogenic lines (NILs) [14] with one or more known leaf rust resistance genes were also included to assist with gene postulations.
Leaf Rust Disease Phenotyping
The 381 plants were grown under greenhouse conditions and inoculated at the first leaf seedling stage, 7d after planting, in three independent experiments, each using a different race or combination of races. Races of P. triticina included race 1 BBBDB (Long and Kolmer 1989) which is widely avirulent to many Lr genes in wheat; race CA1.2 (BBBQD), a race that is virulent to most durum wheat cultivars and avirulent to most bread wheat cultivars; and a mixture of six races of MHDSB, MFPSB, MLDSB, TBBGJ, TFGJQ and TFBGQ common in North America. The six race mixture was also used to inoculate plants in the field. Disease phenotypes were scored 10–12 d after inoculation, according to the 0 to 4 infection type (IT) scale developed by Stakman et al. [15]. The phenotype data was converted to a linearized 0 to 9 scales using a custom Perl script and analyzed using R [16] to derive summary statistics. The program for seedling rust score conversion adopted the scales developed by Zhang et al [10] with slight modifications. Specifically, if it is a simple reading such as “1+” or “2-”, the original 0–9 scale proposed by Zhang et al was used; if it is a complex reading such as “;13+”, the readings were first split into simple readings such as “;” “1” “3+”, then the first reading was weighted double, all readings were converted to 0–9 scales, and arithmetic means were calculated. The program is fully automated and can take data tables with practically unlimited number of columns or trait values. With slight modifications, this program can be used to convert other types of text based categorical data into numeric scales (https://github.com/umngao/rust_scores_conversion).
The 381 lines (with four check cultivars Thatcher, Tom, Verde and Knudson) were planted in single rows in the field and inoculated with the race mixture (see above), approximately one month after planting when the entries were starting to tiller. A mixture of the susceptible cultivars Max, Little Club, Thatcher and Morocco were planted perpendicular to the entries and were inoculated with the race mixture to uniformly infect the entries. Phenotyping (rust scoring) was done approximately one month post inoculation after anthesis on flag leaves of adult plants. Phenotypes were collected in four years and two locations (2012 in Crookston, MN and 2012–2015 in St. Paul, MN). Leaf rust data was scored using the percentage of diseased leaf area (“Field.SEV” or severity) using a modified Cobb scale [17] and response (“Field.IT” or infection type) based on pustule sizes of uredinia spores and amount of necrosis and chlorosis, on a scale of resistant to susceptible response that was converted to a numeric 0 to 1 scale [18]. The phenotypic data were automatically converted to three measures (severity, response and coefficient of infection) using another custom Perl script for field rust score conversion (https://github.com/umngao/rust_scores_conversion), whereas “severity” (Field.SEV) = percentage of diseased leaf area; “response” (Field.IT) = a numeric scale of 0 to 1. It is worth noting that field. IT was converted to a 0–1 linearized scale, while seedling infection types against race1, race.CA1.2 or race.Mix were converted to a 0–9 linearized scales [18]. Coefficient of infection (Field.COI) equals to the product of “severity” and “response”. All of these measures were used in association mapping, with COI values being the primary trait for selection of most significant QTLs, as it combines the information from both severity and response or IT for field resistance.
Phenotypic values were further adjusted based on a mixed linear model, with environments (each year and location combination was counted as one environment for a total of five environments: CrK12, StP12, StP13, StP14, StP15) as random effects, and genotypes as having fixed effects. Best linear unbiased estimates (BLUE) combined the trait values across years. Heading and flowering dates were also collected and initially included into the mixed model analysis. Because significant effects of heading or flowering on overall leaf rust disease severity for this particular dataset were not detected, they were later dropped from the model. Mixed model analysis was performed using the “lme4” package of the open source statistical language or software environment R [16].
DNA Extraction and Genotyping
DNA was collected from seedling plants using a Qiagen (Venlo, Limberg, Netherlands) Biosprint method, and genotyped with the Illumina’s iSelect 90K SNP array at the USDA-ARS Biosciences Research lab in Fargo, North Dakota, USA. Forty-two of the 381 lines were phenotyped, but not genotyped, thus not included into GWAS analysis. The data generated were called and curated using Illumina GenomeStudio software, and uploaded to the T3 database (http://triticeaetoolbox.org/wheat/). SNPs were filtered and converted to Hapmap format using a custom Perl script incorporating the criteria for: minor allele frequency (MAF) greater than 0.05; missing data less than 20% (only two lines had missing data greater than 15%) and chromosome positions previously mapped in the wheat 90K consensus map [5]. A molecular marker (csLV34) [19] tightly associated to Lr34, was used to genotype this panel, and the genotyped results were included in GWAS analysis.
Population Structure (Q), Kinship (K) and Linkage Disequilibrium (LD) Analysis
Population structure (Q) was analyzed using a model based clustering method (admixture models with correlated allele frequencies) in STRUCTURE [20] v2.3.4, and principal component analysis (PCA) using the statistical software R. An LD-based pruning method implemented in the PLINK software [21] v1.09 was used to prune the total filtered marker set (18,924) using the command line option of “—indep-pairwise 100 5 0.2” under Linux environment. The pruned (or filtered) set of SNP markers (1309) were used for structure analysis. The reason for using pruned markers rather than the whole set of markers is that STRUCTURE assumes loci are at linkage equilibrium within sub-populations.
Ten independent STRUCTURE runs were conducted for each specified K (number of subpopulations, from 2 to 10), with 20,000 burn-in length and 40,000 Markov chain Monte Carlo (MCMC) iterations under Linux environment. The most likely number of clusters (K) was chosen based on the ΔK method [22], implemented in a web-based informatics tool “Structure Harvester” [23]. The method estimates ΔK based on the rate of change in the log probability between successive K values. Clumpp [24] software v1.1 was used to consolidate STRUCTURE runs and derive the Q matrix used in AM with mixed linear models (MLM). DISTRUCT v1.1 [25] was used to plot the population Q matrix bar graph. The fixation index (Fst) of subpopulations was obtained through STRUCTURE run outputs.
TASSEL [26] v4.3.13 was used to derive a population Kinship matrix based on the scaled IBS (identity by state) method using the complete set of markers that passed quality filtering. Previous research has shown that using the complete set of markers is useful to control genome-wide error rate and performs much better than pedigree based methods for kinship estimates [27]. Linkage disequilibrium (LD) was calculated using TASSEL. LD decay curves for each sub-genome (A, B, D) were fitted using a non-linear model described by Marroni et al. [28]. The LD values (R2) were also used to assist QTL block determination (see next section for details). Some LD blocks or adjacent regions were selected using PLINK software tool set [21] and visualized using Haploview software [29].
Genome Wide Association Study (GWAS) Using MLM (QK), QGLM and G-Model Analysis
Association mapping based on mixed linear model (MLM) and generalized linear model with population structure as a covariate (QGLM) analysis was conducted primarily using TASSEL [26], a JAVA based open source software for linkage and association analysis. MLM model results were further validated using the Genome Association and Prediction Integrated Tool (GAPIT) [30] under open source R environment. The GAPIT tool does not report QGLM values. We also explored the use of a Fortran software or program developed by Dr. Rex Bernardo (University of Minnesota) for GWAS analysis using genome wide markers to control for background variations in a multiple regression setting (known as G-model) (http://bernardo-group.org/books-and-software/) [31]. For this study, we focused primarily on MLM results from TASSEL outputs, but the results from GAPIT and G-models were used in a supporting manner.
The Mixed linear model (MLM) approach [32] was used in GWAS analysis of leaf rust resistance. The mixed linear model for GWAS can be specified as follows: y = Xβ + Qv + Zu +e, where y is the phenotype values (either BLUE adjusted or environment specific values), β and v are fixed effects due to marker and population structures respectively, u is a vector of random effects due to the portion of breeding values not accounted for by the markers. X, Q and Z are incidence matrices that related y to β, v and u. The covariance matrix of breeding value “u” is the product of kinship matrix (commonly designated as A or K) and Vg, the portion of additive variance that is not accounted for by the marker under test [33]. Considering the size of the population in this study, we did not use the “compressed MLM” ability of GAPIT or Tassel which groups individuals based on phenotypes [34].
P-values, R2 and marker effects were extracted from GWAS results. The false discovery rate (FDR) adjusted p-values used in GAPIT were found to be highly stringent. Researchers have debated that correcting for marker effects based on both population structure Q and kinship K (which in turn were often calculated based on marker data as well) could be over-correcting and might result in a need for relaxed p-value levels such as 0.001 [11, 35] or alternative ways to correct for background variations [31]. Some researchers have developed or adopted alternative multiple test correction methods such as “simpleM”, a variant of the Bonferroni correction [36, 37]. The SimpleM [36] approach applies a Bonferroni correction to the effective number of independent tests (Meff). In this study, marker–wise significance was based on three criteria (p<0.05, p<0.001, and SimpleM) with increasing stringency. For QTL nomination, a p-value of less than 0.001 (detected in at least one environment) was required. For count-based analysis of previously designated QTLs using vennCounts and vennDiagram functions from LIMMA (linear model for microarrays, a Bioconductor package of R), a p-value cutoff of 0.05 was used to capture the global similarity or dissimilarity between traits or environments. An effective number of tests (Meff) were calculated for each chromosome individually and the SimpleM level was determined as p-value = 7.72E-5, derived from 0.1 divided by the total number of Meff for all chromosomes (n = 1295).
R2 values from LD analysis, in conjunction with genetic distances were used to assign co-segregating or adjacent significant markers into a unique QTL block based on the criteria of R2 value greater than 0.2 and genetic distance less than 10 cM, with a few exceptions: QTLs 1B_1 and 4A_3 both had markers that are more than 10 cM apart, yet were almost completely linked (LD R2≈1).
Most Significant Markers, Stepwise Regression, and Postulation of Gene Names
For each LD block or QTL as defined above, markers with the most significant p-values or largest R2 were extracted as representative markers for each marker-trait association. The most significant 10 QTLs for each trait (field disease resistance, seedling resistance against Race 1, CA1.2, and the race mixture) were summarized in more detail and the cumulative effects of significant markers were assessed using a multiple regression approach (considering additive effects only) with population structure as a covariate.
Stepwise regressions were performed on significant markers to identify the loci that were independently associated with disease resistance for each trait, respectively, with population structure Q as covariate. For field BLUE trait values, a p-value cutoff of 0.005 was required for inclusion in the stepwise regression (the selected markers also passed the 0.001 threshold for at least one of the tested environments). For seedling traits, if less than 10 loci passed the 0.001 threshold (e.g. QTLs resistant to race CA1.2), the most significant 10 loci were used, roughly equivalent to a p-value cutoff of 0.1 percentile. For both forward and backward selection (in stepwise regression of the multiple regression models), a p-value of 0.05 determined whether including or dropping a given QTL from the model was appropriate.
The most significant QTLs were postulated based on position, LD, infection type and pedigree or donor line information. All sequences associated with 90K SNPs were blasted against the wheat genome survey sequences v2 (http://www.wheatgenome.org/) using command line version of BLAST+ v2.2.8 under Linux environment. A few target SNP sequences were also used to BLAST against a newer version of wheat genome build [38] and reference Barley genome sequences [39] for candidate gene annotation. The criterion was percent identity greater than 95% and only the best hit was taken if there were multiple BLAST hits. The BLAST hit information was used to explore candidate genes for certain QTLs and to assist optimal chromosome arm assignments for certain SNPs. For this study, candidate genes or QTLs were postulated or designated for those loci whose p-values (derived from either MLM or QGLM analysis) surpassed the simpleM Bonferroni correction threshold. The candidate QTLs identified through G-model generally have higher significance levels (strongly exceeding the simpleM threshold).
Results
Phenotype Data and Results
For seedling plants, more than 50% of the tested lines were completely resistant (immune, with IT = 0) against leaf rust race 1 or race CA1.2 (Fig 1). This suggests the presence of resistance genes in many of the tested lines. Approximately 70% of the highly resistant lines are resistant to both race 1 and race CA1.2, likely suggesting that these lines possess major genes that are effective against both races. The median seedling leaf rust disease score was 0 (the completely immune type) for resistance to race 1 and CA1.2 and 6 (equivalent to IT of 2+) for resistance to the field race mixture. The mean disease severity of the race mixture was higher than the two single races by 2.8 and 3.4 respectively, on a 0–9 disease scale. The ANOVA analysis for treatment effects (or inoculation method) p-value was less than 2.2E-16. The higher disease severity for race mixtures was expected, as the field race mixtures contain races that are more virulent than race 1 and race CA1.2. Nonetheless, approximately 40% of the tested lines were resistant (with IT of 2+ or less) to the field race mixtures at the seedling stage, suggesting that these lines are broadly resistant to common North American races and also may be resistant in field trials.
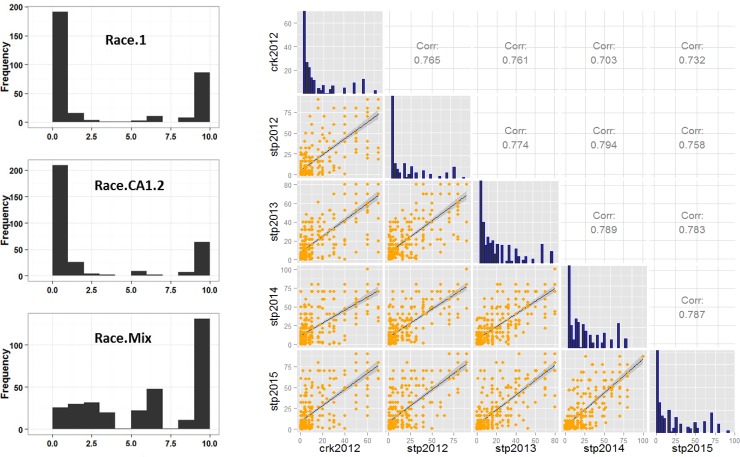
Fig 1. Seedling disease levels and field disease (coefficient of infection, COI) correlations.
(a) Left (black and white) panel showing seedling disease distributions; X-axis represents linearized 0–9 scales; (b) Right (orange and blue) panel showing field disease correlations. Diagonals are histogram for each environment (Crookston CRK12,Saint Paul StP12-15).
Disease severity varied between years of field plot tests, with the year 2012 displaying the lowest leaf rust severity. Nonetheless, the year to year correlations are still high, with an average Pearson correlation coefficient of R = 0.77 (Fig 1), similar to those observed for a stripe rust study [40]. After a mixed model adjustment, we estimated the heritability (h2) of leaf rust disease trait (for coefficient of infection, COI) to be 0.92. The overall disease distribution among the 381 lines was highly skewed toward resistance types (Fig 1B, diagonal histograms). Coefficient of infections (COI) for 69% of the lines was less than 20 (on a 0–100 scale). These results suggest that most of the breeding lines used in this AM study contain effective resistance genes or QTLs against multiple leaf rust races under field environments.
The overall Pearson’s correlations among measured traits are listed in Table 1. Seedling plant infection types to single races (race 1 and race CA1.2) were moderately correlated with each other (0.63). We also observed a moderate correlation between seedling and field response (IT) to race mixtures (R = 0.57). Our results showed a high correlation (R> = 0.88) among field trait values calculated using different methods (COI, severity, and response or infection type IT). The field response and response to single races are less well correlated (R<0.29), which was expected as race 1 and CA1.2 are highly avirulent, while the mixture of race used for the seedling and field plot tests is virulent to a number of Lr genes.
Table 1. Phenotype correlations among field and seedling traits (Race.1, Race.CA1.2, Race.Mix, Field.COI, FIELD.SEV, FIELD.IT).
| Race.1 | Race.CA1.2 | Race.Mix | Field.COI | Field.SEV | Field.IT | ||
|---|---|---|---|---|---|---|---|
| Race.1 | - | ||||||
| Race.CA1.2 | 0.63 | - | |||||
| Race.Mix | 0.41 | 0.31 | - | ||||
| Field.COI | 0.20 | 0.29 | 0.50 | - | |||
| Field.SEV | 0.18 | 0.25 | 0.51 | 0.98 | - | ||
| Field.IT | 0.17 | 0.22 | 0.57 | 0.90 | 0.88 | - | |
Genome Wide SNP Coverage and Linkage Disequilibrium (LD) Based on 90K SNP Arrays
A total of 18,925 markers passed the quality filters (18,924 of them are SNP array based markers). On average, there are 1.4 markers per centimorgan (cM) (S1 Fig). Marker density on D genome chromosomes was much lower compared to A and B genomes: one marker every 2.6 cM compared to one marker every 0.74–0.97 cM. This is roughly proportional to the total mapped markers for each genome [5].
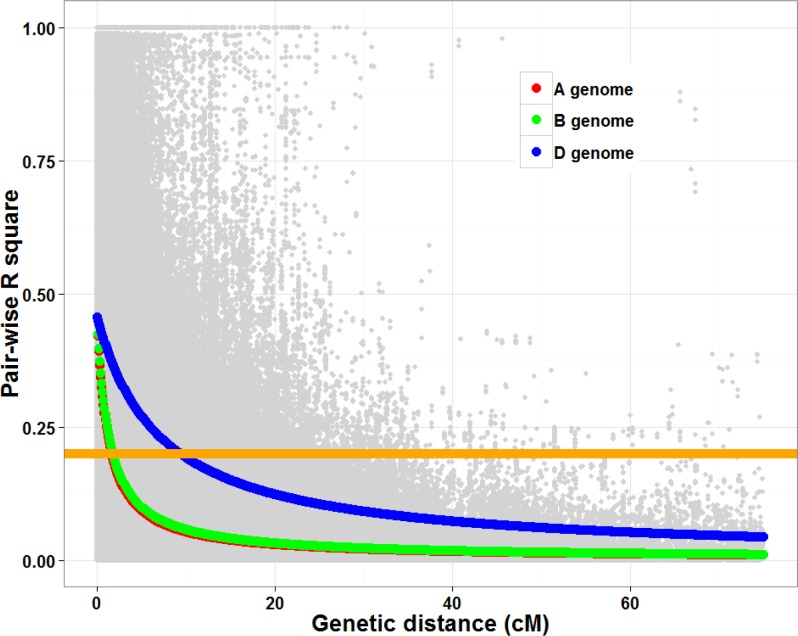
We found that the LD decays (as defined by R2 declining to below 0.2) at around 1.5–1.7 cM for A and B genomes, but extends to more than 8 cM in D genome chromosomes (Fig 2), consistent with previous findings [5]. The high LD in D genome chromosomes has the practical effect of reducing the number of markers required to detect significant marker traits associations.
Fig 2. Linkage disequilibrium based on 18924 SNP markers.
For color fitted LD decay lines: red represents A genome; green represents B genome; blue represents D genome. Orange bar indicates LD decay level (R2 = 0.2).
Population Structures of the Leaf Rust Association Mapping Panel and Correlation with Leaf Rust Phenotype in the Field
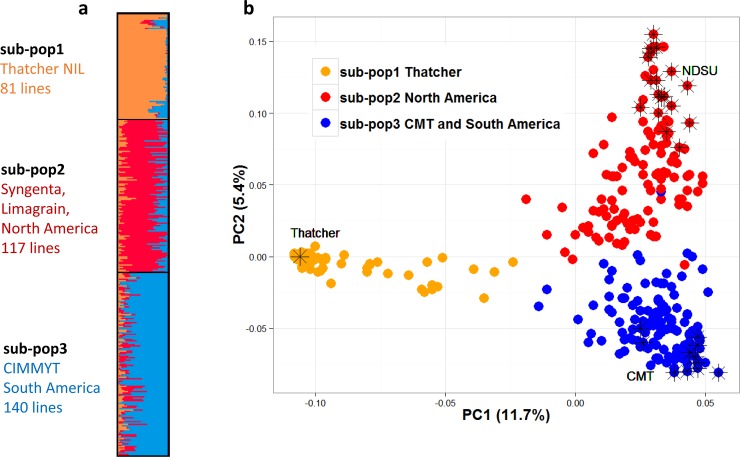
For model based analysis using STRUCTURE [20], the optimal number of K was determined to be 3 based on the ΔK method [22]. In this study, ΔK value equals 380 for K = 3 and the remaining tested K’s all have a much smaller ΔK value (mean = 2, max = 5). Similarly, for PCA based analysis, we found that the total variance explained by each principal component (PC) drops sharply for the first three PCs (from 11.7 to 5.4 to 3.3), and levels after PC3. The first three PCs capture over 20% of the total variance. Given the results from both model based and PCA analysis, we used K = 3 for constructing the Q matrix in the mixed model marker trait association analysis in both TASSEL and GAPIT. The population structure derived from the model–based and PCA–based approaches were very similar (Fig 3). GWAS results (identified QTLs and p-values for markers) using different version of Q3 matrices (either model based or PCA based) agreed with each other: The correlation among sets of p-values and marker effects were both over 0.9 for the field BLUE trait, and the correlation among set of R2 values was over 0.96 for the same trait (Field.COI).
Fig 3. Population structure: model based approach and PCA approach.
(a) model based approach for population structure analysis. (b) PCA approach for population structure analysis. Orange: sub-population 1; Red: sub-population 2; Blue: sub-population 3. Asterisks (*) on PCA plot indicate accessions that are typical of each sub-population. Sub-population 3 also has lines from North America. Both sub-pop2 and sub-pop 3 might include lines from Asia, Europe etc.
STRUCTURE analysis revealed three sub-populations: i) Sub-population 1 (81 lines) consisted of almost entirely Thatcher near isogenic lines (NILs) with a few exceptions; ii) Sub-population 2 (117 lines) consisted of mostly (~70%) North American lines including genotypes from South Dakota State University, University of Minnesota, North Dakota State University, Syngenta and Limagrain; iii) The majority (with only one exception) of CIMMYT and South American lines were in sub-population 3 (140 lines). However, sub-population 3 also contained some North American lines. A few European, African and Asian lines were also present in Sub-populations 2 and 3. The fixation index (Fst) values for the three sub-populations are 0.80, 0.43 and 0.12 respectively, suggesting that sub-population 3 has a much higher divergence within itself. Different sub-populations are associated with different resistance levels and the differences are highly significant (p < 1.8E-5). Sub-population 1 (Thatcher near isogenic lines) has the highest disease (mean 33.4), followed by sub-population 3 (South America, mean 18.0) and sub-population 2 (North America, mean 9.9) (Fig 4).
Fig 4. Different sub populations of this AM panel are associated with different levels of leaf rust disease severity (measured using coefficient of infection Field.COI).
Marker Trait Associations by MLM (QK), QGLM and G-Model Analysis
After trimming the genotype and phenotype datasets, 18,925 markers and 338 lines were included in the GWAS analysis. As an exploratory analysis, GWAS (MLM) for plant flowering date phenotype analysis revealed a single genomic locus at 90cM (long arm) of chromosome 5A using GAPIT with default settings (S2 Fig). The marker trait association p-value was highly significant (p < 1E-10). We hypothesize that this locus is Vrn-A1, consistent with previous mapping results [4]. The successful detection of Vrn-A1 suggests that the MLM (with population structure Q and kinship K) model fits the data. We further utilized quantile-quantile (QQ) plots to examine the MLM model fitness for leaf rust traits. The expected–log10P value and the observed–log10P value distributions follow the X = Y diagonal line until it curves at the end (S3 Fig). These results further suggest that our MLM models fit the data.
After fitting the MLM using TASSEL [26], a total of 333 markers were identified to be significantly (p < 0.001) associated with leaf rust resistance for at least one of the field or seedling traits (S1 Table) We combined adjacent markers based on LD and genetic distance information, and were able to obtain 46 unique QTLs (Table 2).
Table 2. A total of 46 QTLs were identified that were significantly associated with field or seedling leaf rust disease resistant (p.MLM < 0.001).
| QTL (gene)a | Chr | Position (90K) | Num SNPs | Crk 2012 | StP 2012 | StP 2013 | StP 2014 | StP 2015 | Lr.COI | Lr.SEV | Lr.IT | Race.1 | Race CA1.2 | Race Mix |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1A_3 | 1A | 148.99–151.22 | 6 | 4.43E-03 | 6.89E-05 | 2.01E-03 | 8.55E-05 | 6.34E-04 | 1.11E-04 | 8.63E-04 | 2.35E-03 | - | - | 3.55E-03 |
| 1B_1 | 1B | 43.66–64.89 | 88 | 1.17E-02 | 3.39E-04 | 2.09E-03 | 6.23E-04 | 2.56E-03 | 1.07E-03 | 2.01E-03 | 4.78E-05 | - | 9.21E-03 | 3.77E-04 |
| 1B_t1 | 1B | 81.95–82.86 | 2 | 1.96E-02 | 7.36E-04 | 5.27E-03 | 8.63E-04 | 8.03E-03 | 1.62E-03 | 1.17E-03 | 1.49E-02 | 9.03E-03 | - | - |
| 1B_2 | 1B | 84.43–85.57 | 11 | - | - | - | - | - | - | - | - | 7.84E-05 | 1.96E-02 | - |
| 1D_1 | 1D | 3.5–8.71 | 4 | 1.46E-02 | 7.02E-03 | 4.90E-03 | 5.87E-03 | 7.72E-03 | 1.80E-03 | 3.55E-03 | 1.31E-04 | - | 1.05E-02 | 1.46E-03 |
| 1D_2 | 1D | 44.69–44.69 | 2 | - | - | 9.64E-03 | - | 1.48E-02 | 1.76E-02 | 2.67E-02 | 4.48E-04 | - | - | - |
| 1D_t1 | 1D | 45.44–45.44 | 1 | - | - | - | - | - | - | - | - | 3.85E-04 | - | - |
| 1D_3 | 1D | 88.85–89.58 | 5 | 2.45E-02 | - | 5.10E-03 | 1.49E-02 | - | 1.44E-02 | 2.81E-02 | - | 2.97E-02 | 3.96E-04 | - |
| 2A_1 | 2A | 20.14–20.14 | 1 | - | - | - | - | - | - | - | - | 3.64E-02 | - | 2.58E-04 |
| 2A_2 | 2A | 101.97–101.97 | 1 | - | - | - | - | 3.28E-04 | 3.66E-02 | - | 1.61E-02 | - | 2.21E-02 | - |
| 2A_3 | 2A | 108.46–108.46 | 1 | 1.69E-02 | 4.64E-04 | 3.69E-03 | 1.78E-03 | 1.19E-02 | 4.38E-04 | 1.54E-03 | 2.60E-03 | 3.94E-02 | 5.61E-03 | - |
| 2B_2 | 2B | 88.44–97.26 | 24 | 3.45E-06 | 1.95E-02 | 2.88E-03 | 7.75E-03 | 1.20E-04 | 4.52E-04 | 5.97E-04 | 7.80E-04 | - | 9.08E-05 | 4.47E-02 |
| 2B_3 | 2B | 102.28–108.35 | 15 | 1.57E-03 | 4.29E-04 | 4.61E-03 | 3.45E-04 | 1.75E-03 | 7.55E-04 | 1.38E-03 | 2.13E-03 | - | - | - |
| 2D_1 | 2D | 18.22–18.22 | 1 | - | - | 6.82E-03 | 1.12E-02 | 9.26E-03 | 4.84E-03 | 3.48E-04 | 5.73E-04 | - | - | - |
| 3A_1 | 3A | 86.16–87.78 | 3 | - | 8.17E-04 | 9.32E-04 | 5.56E-05 | 9.54E-03 | 1.50E-03 | 3.72E-04 | 4.15E-02 | - | - | 3.11E-02 |
| 3A_t2 | 3A | 169.89–169.89 | 1 | 1.25E-03 | 1.50E-03 | 4.96E-03 | 2.96E-03 | 1.73E-03 | 4.31E-04 | 7.47E-04 | 1.19E-03 | - | - | 1.16E-03 |
| 3B_t2 | 3B | 51.07–51.08 | 2 | 3.09E-02 | - | 5.14E-03 | 3.50E-02 | 1.32E-02 | 1.82E-02 | - | 1.34E-03 | - | - | 6.69E-04 |
| 3B_1 | 3B | 139.62–139.62 | 3 | - | 1.29E-02 | 4.62E-05 | 4.14E-02 | - | 8.23E-03 | 3.06E-02 | 3.53E-02 | - | - | - |
| 4A_1 (umn-4AS) | 4A | 37.05–37.05 | 2 | 7.72E-03 | 9.45E-04 | 8.60E-05 | 3.50E-04 | 2.20E-03 | 4.17E-05 | 4.74E-05 | 9.64E-05 | 3.24E-02 | - | - |
| 4A_2 | 4A | 48.52–48.84 | 60 | 1.34E-02 | - | 7.26E-05 | - | 2.34E-02 | 1.69E-02 | 4.00E-02 | 3.22E-02 | - | 6.04E-03 | 2.89E-02 |
| 4A_3 | 4A | 51.7–53.13 | 7 | 1.39E-03 | 1.24E-03 | 8.79E-03 | 1.18E-04 | 3.01E-02 | 6.67E-04 | 1.40E-03 | 3.69E-03 | - | - | - |
| 4A_t1 | 4A | 73–73 | 1 | 2.57E-02 | 6.18E-03 | 2.89E-02 | 5.72E-04 | 1.31E-02 | 1.43E-03 | 9.18E-03 | 2.34E-03 | - | - | - |
| 4A_t2 | 4A | 139.97–144.38 | 4 | - | 3.09E-02 | 4.26E-02 | 7.16E-03 | 1.53E-02 | 1.41E-02 | 4.17E-03 | 3.24E-04 | 5.59E-04 | 7.94E-03 | - |
| 4B_1 | 4B | 15.91–15.91 | 1 | - | - | 1.91E-02 | - | 6.01E-03 | 7.47E-03 | 9.71E-03 | 7.71E-04 | - | - | 4.02E-03 |
| 4B_3 | 4B | 74.62–90.07 | 10 | 5.64E-05 | 2.77E-03 | 1.73E-03 | 1.99E-04 | 1.07E-02 | 1.56E-04 | 1.02E-04 | 1.35E-03 | - | 2.44E-02 | 2.93E-03 |
| 4D_1 | 4D | 70.59–70.59 | 1 | 7.58E-03 | 6.42E-03 | 5.06E-03 | 6.95E-03 | 1.35E-02 | 1.82E-03 | 2.78E-03 | 2.26E-04 | 2.34E-02 | 1.90E-02 | 2.31E-04 |
| 5B_1 | 5B | 39.4–39.4 | 8 | - | 1.67E-03 | 5.73E-03 | 2.41E-03 | 4.05E-04 | 9.90E-04 | 1.51E-03 | 4.03E-02 | - | - | - |
| 5B_2 | 5B | 49.01–49.65 | 2 | 6.71E-04 | 8.99E-04 | 2.34E-02 | 5.95E-06 | 2.07E-03 | 1.69E-04 | 6.78E-04 | 1.14E-03 | 5.08E-05 | - | - |
| 5B_t1 | 5B | 119.54–119.54 | 1 | 9.68E-04 | 3.26E-02 | 2.05E-02 | - | - | 3.58E-02 | 3.46E-02 | - | - | - | - |
| 5D_1 (Lr1) | 5D | 203.88–204.58 | 6 | - | - | - | - | - | - | - | - | 1.17E-04 | 3.74E-02 | - |
| 6A_1 | 6A | 25.86–27.15 | 2 | 4.60E-04 | - | 2.22E-02 | 8.55E-03 | 7.57E-03 | 1.32E-03 | 5.59E-03 | 3.16E-03 | - | - | - |
| 6A_t1 | 6A | 48.09–48.09 | 1 | 4.38E-03 | 4.46E-03 | 1.70E-02 | 9.81E-03 | 8.01E-04 | 4.97E-04 | 5.57E-04 | 3.28E-03 | - | - | - |
| 6A_2 | 6A | 100.62–100.62 | 1 | - | - | - | - | - | - | - | - | 2.23E-04 | 1.24E-02 | - |
| 6A_3 | 6A | 119.64–119.64 | 1 | 1.56E-02 | - | - | - | - | - | - | - | - | - | 2.28E-04 |
| 6B_1 | 6B | 16.76–16.76 | 1 | - | - | - | - | - | - | - | - | 9.43E-04 | - | - |
| 6B_3 | 6B | 66.36–66.36 | 2 | - | 8.10E-03 | - | 3.04E-03 | 2.84E-02 | 1.36E-02 | 1.24E-02 | 1.14E-02 | 2.33E-03 | 5.05E-04 | 7.02E-04 |
| 6B_4 (Lr3) | 6B | 118.99–122.92 | 31 | - | - | - | - | - | - | - | - | 6.02E-07 | 8.78E-05 | 8.81E-03 |
| 7A_3 | 7A | 125.47–125.47 | 1 | 9.59E-04 | 2.90E-02 | - | - | 3.24E-02 | 1.25E-02 | 2.01E-02 | - | - | - | - |
| 7B_1 | 7B | 58.17–58.63 | 3 | - | 3.60E-04 | 6.11E-04 | 7.56E-03 | 2.32E-02 | 1.57E-02 | 2.54E-02 | 2.51E-03 | - | 4.03E-03 | - |
| 7B_2 | 7B | 66.62–71.33 | 8 | - | - | - | - | - | - | - | - | - | 1.22E-04 | - |
| 7B_t1 | 7B | 73.79–73.79 | 1 | 4.73E-02 | 1.47E-02 | 1.59E-02 | 2.77E-02 | 9.50E-03 | 3.88E-03 | 2.53E-03 | 5.60E-04 | - | - | - |
| 7B_3 | 7B | 76.31–76.31 | 2 | - | - | - | - | - | - | - | - | 2.09E-04 | - | - |
| 7B_t2 | 7B | 89.13–89.13 | 2 | 1.90E-02 | 9.54E-03 | 3.54E-03 | - | 6.00E-04 | 2.60E-03 | 4.48E-03 | 3.74E-03 | - | - | - |
| 7B_4 | 7B | 166.99–166.99 | 1 | 1.89E-04 | 2.58E-04 | 7.93E-04 | 1.42E-03 | 2.35E-03 | 5.03E-04 | 1.66E-03 | 6.36E-03 | - | - | - |
| 7D_1 (Lr34) | 7D | 35.00–35.00b | 1 | 8.16E-05 | 7.55E-03 | 2.15E-02 | - | 3.15E-02 | 3.51E-03 | 2.03E-03 | - | - | 3.22E-02 | - |
| 7D_2 | 7D | 169.51–169.51 | 1 | 2.86E-03 | 1.15E-02 | - | - | 6.53E-04 | 2.91E-03 | 3.91E-03 | 2.32E-02 | - | - | - |
aPostulated loci names are given (within parentheses) if strong evidences to support such postulations were obtained.
bThe approximate Lr34 position on the 90K map is based on blast hit information on a recent chromosome 7D scaffold build published by Chapman et al. [38].
Columns “CrK2012”, “StP2012”- “StP2015” indicate GWAS p-values for individual field environments in Crookston and St Paul, MN.
Columns “Lr.COI”, “Lr.SEV”, “Lr.IT” indicate GWAS p-values for BLUE estimated field traits (coefficient of infection, severity and infection type).
Underlined p-values are those that surpass the simpleM Bonferroni correction threshold.
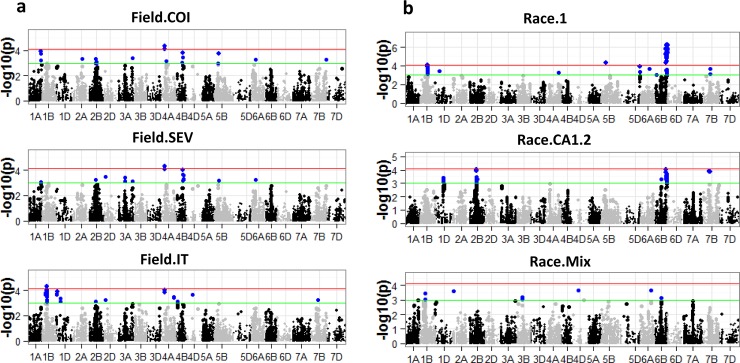
Manhattan plots for the BLUE estimated field traits across different field environments (2012–2015) are shown in Fig 5. A majority of the field QTLs were detected under multiple environments (Table 2 and Fig 6). Similar to MLM analysis, exploratory analysis using QGLM model (population structure + generalized linear model) also revealed the known flowering locus at 5AL (data not shown). The QGLM p-values for leaf rust traits are generally more significant compared to MLM analysis (Table 3).
Fig 5.
Manhattan plots for field (a) and seedling (b) traits. Red line indicates SimpleM bonferroni corrected p-value threshold for significance (7.7x10-5); Green line indicates p-value of 1x10-3.
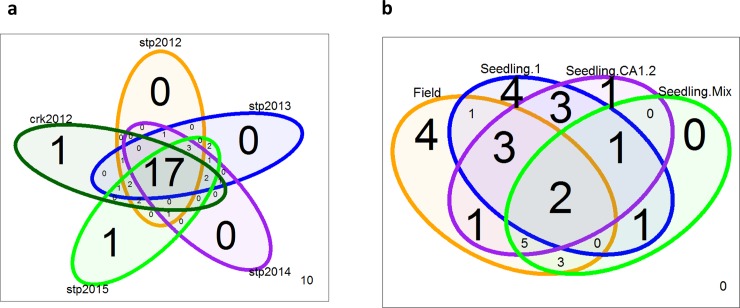
Fig 6.
Venn diagrams (a) Overlap between QTLs under different field environments. (b) Overlap between field and seedling QTLs.
Table 3. Most significant (top 10) QTLs and representative SNPs for field and seedling traits.
| Trait | QTL | Marker | blast | Chr | Position | p.MLM | p.QGLM | RSQ | eff | R | Sub pop1 | Sub pop2 | Sub pop3 | postulation | reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Race.1 | 1B_2 | IWB13336 | NA | 1B | 85.57 | 7.84E-05 | 1.39E-06 | 0.05 | 2.76 | C | 78 | 99 | 108 | Lr26? | Kolmer 2003 |
| Race.1 | 1D_t1 | IWB14612 | 1DS_1885467 | 1D | 45.44 | 3.85E-04 | 2.77E-05 | 0.04 | 2.48 | A | 77 | 107 | 101 | Lr42? | Liu et al 2013 |
| Race.1 | 4A_t2 | IWB3569 | 4AL_v2_7176180 | 4A | 144.38 | 5.59E-04 | 1.08E-05 | 0.04 | 2.32 | A | 10 | 107 | 83 | Lr28? | Bipinraj et al 2011 |
| Race.1 | 5B_2 | IWB9055 | NA | 5B | 49.65 | 5.08E-05 | 1.44E-06 | 0.06 | 2.35 | G | 76 | 101 | 78 | QLr. cdl-5BL? | Kolmer 2015 |
| Race.1 | 5D_1 | IWB53861 | NA | 5D | 203.88 | 1.17E-04 | 2.32E-05 | 0.05 | -2.58 | A | 3 | 16 | 30 | Lr1 | Kolmer 2003 |
| Race.1 | 6A_2 | IWB625.2 | NA | 6A | 100.62 | 2.23E-04 | 5.76E-06 | 0.05 | 1.97 | G | 75 | 57 | 87 | Lr64? | - |
| Race.1 | 6B_1 | IWB65148 | NA | 6B | 16.76 | 9.43E-04 | 4.82E-06 | 0.04 | 2.51 | G | 9 | 114 | 102 | - | - |
| Race.1 | 6B_3 | IWB65914 | 6BL_4378239 | 6B | 66.36 | 2.33E-03 | 3.06E-04 | 0.03 | 1.83 | G | 43 | 110 | 112 | Lr9? | Kolmer 2003 |
| Race.1 | 6B_4 | IWB3292 | 6BL_4278271 | 6B | 122.92 | 6.02E-07 | 1.18E-07 | 0.09 | -3.43 | C | 4 | 14 | 23 | Lr3 | Kolmer 2003 |
| Race.1 | 7B_3 | IWA306 | 7BL_6747122 | 7B | 76.31 | 2.09E-04 | 9.56E-07 | 0.05 | 3.16 | G | 75 | 115 | 115 | Lr14? | Kolmer 2003 |
| Race.CA1.2 | 1B_1 | IWB19584 | NA | 1B | 63.91 | 9.21E-03 | 3.24E-04 | 0.02 | -2.46 | G | 4 | 3 | 20 | QLr.cimmyt-1BS | Rosewarne 2012 |
| Race.CA1.2 | 1D_3 | IWB35520 | 1DL_2290849 | 1D | 89.58 | 3.96E-04 | 5.16E-08 | 0.04 | 2.42 | A | 79 | 113 | 94 | Lr21? | |
| Race.CA1.2 | 2A_3 | IWA3151 | 2AL_6426630 | 2A | 108.46 | 5.61E-03 | 2.97E-05 | 0.03 | -1.40 | T | 4 | 49 | 104 | Lr11.Lr38? | Darino 2015 |
| Race.CA1.2 | 2B_2 | IWB37811 | 2BS_5186722 | 2B | 93.47 | 9.08E-05 | 8.75E-08 | 0.05 | 1.91 | G | 73 | 47 | 82 | Lr13.Lr23.Lr16? | Oelke & Kolmer 2005 |
| Race.CA1.2 | 4A_2 | IWB40915 | 4AS_v2_6008166 | 4A | 48.52 | 6.04E-03 | 1.13E-04 | 0.03 | 1.95 | T | 79 | 116 | 95 | - | - |
| Race.CA1.2 | 4A_t2 | IWB3569 | 4AL_V2_7176180 | 4A | 144.38 | 7.94E-03 | 1.86E-04 | 0.02 | 1.57 | A | 10 | 107 | 83 | Lr28? | Bipinraj et al 2011 |
| Race.CA1.2 | 6B_3 | IWB65914 | 6BL_4378239 | 6B | 66.36 | 5.05E-04 | 1.16E-06 | 0.04 | 1.89 | G | 43 | 110 | 112 | Lr9? | Kolmer 2003 |
| Race.CA1.2 | 6B_4 | IWB6474 | NA | 6B | 119.73 | 8.78E-05 | 1.53E-05 | 0.06 | 2.27 | G | 9 | 110 | 95 | Lr3 | Kolmer 2003 |
| Race.CA1.2 | 7B_1 | IWB39492 | 7BS_3168118 | 7B | 58.17 | 4.03E-03 | 1.60E-02 | 0.03 | 1.76 | A | 72 | 111 | 113 | Lr72? | Herrera-Foessel et al. 2014 |
| Race.CA1.2 | 7B_2 | IWB68484 | 7BS_3079273 | 7B | 66.62 | 1.22E-04 | 3.19E-07 | 0.05 | 4.06 | T | 80 | 111 | 126 | Lr14? | Singh et al. 2009 |
| Race.Mix | 1A_3 | IWB48030 | 1AL_3976804 | 1A | 149.82 | 3.55E-03 | 5.99E-04 | 0.03 | 1.72 | G | 10 | 113 | 103 | QLr.umn-1AL | * |
| Race.Mix | 1B_1 | IWB19584 | NA | 1B | 63.91 | 3.77E-04 | 2.67E-07 | 0.05 | -3.44 | G | 4 | 3 | 20 | QLr.cimmyt-1BS | Rosewarne et al 2012 |
| Race.Mix | 1D_1 | IWB44021 | 1DS_1912623 | 1D | 8.71 | 1.46E-03 | 9.51E-07 | 0.04 | -3.22 | T | 4 | 3 | 17 | Lr42? | Liu et al 2013 |
| Race.Mix | 2A_1 | IWB74529 | NA | 2A | 20.14 | 2.58E-04 | 4.56E-06 | 0.05 | 2.10 | C | 79 | 36 | 121 | Lr17? | Kolmer 2003 |
| Race.Mix | 3A_t2 | IWB34789 | 3AL_4449581 | 3A | 169.89 | 1.16E-03 | 3.57E-05 | 0.04 | 1.57 | G | 8 | 61 | 105 | QLr.fcu-3AL? | Chu et al. 2009 |
| Race.Mix | 3B_t2 | IWB74350 | 3B_10762316 | 3B | 51.07 | 6.69E-04 | 8.56E-05 | 0.04 | 1.74 | A | 73 | 83 | 85 | - | - |
| Race.Mix | 4B_3 | IWB72129 | 4BL_7035179 | 4B | 86.55 | 2.93E-03 | 4.75E-06 | 0.03 | 1.55 | G | 76 | 80 | 103 | Lr30? | Draz et al 2015 |
| Race.Mix | 4D_1 | IWB17540 | 4DS_2288313 | 4D | 70.59 | 2.31E-04 | 1.78E-07 | 0.05 | -3.35 | T | 6 | 3 | 19 | ? | |
| Race.Mix | 6A_3 | IWA7764 | 6AL_5772638 | 6A | 119.64 | 2.28E-04 | 1.13E-06 | 0.06 | 2.12 | C | 74 | 60 | 83 | - | - |
| Race.Mix | 6B_3 | IWB11702 | 6BL_4398818 | 6B | 66.36 | 7.02E-04 | 5.60E-05 | 0.04 | 1.86 | T | 43 | 111 | 125 | Lr9? | Kolmer 2003 |
| Field.COI | 1A_3 | IWB48030 | 1AL_3976804 | 1A | 149.82 | 1.11E-04 | 7.23E-06 | 0.05 | 12.98 | G | 10 | 113 | 103 | QLr.umn-1AL | * |
| Field.COI | 2A_3 | IWA3151 | 2AL_6426630 | 2A | 108.46 | 4.38E-04 | 1.17E-06 | 0.04 | -10.14 | T | 4 | 49 | 104 | Lr11? | Darino et al 2015 |
| Field.COI | 2B_2 | IWB22236 | 2BS_5202128 | 2B | 88.44 | 4.52E-04 | 6.65E-06 | 0.04 | 10.02 | T | 13 | 79 | 104 | Lr13.Lr23.Lr16? | Oelke & Kolmer 2005 |
| Field.COI | 3A_t2 | IWB34789 | 3AL_4449581 | 3A | 169.89 | 4.31E-04 | 8.19E-06 | 0.04 | 9.48 | G | 8 | 61 | 105 | QLr.fcu-3AL? | Chu et al. 2009 |
| Field.COI | 4A_1 | IWB59410 | 4AS_v2_5925149 | 4A | 37.05 | 4.17E-05 | 7.73E-06 | 0.05 | 11.23 | T | 7 | 74 | 99 | QLr.umn-4AS | * |
| Field.COI | 4A_3 | IWB7998 | NA | 4A | 51.7 | 6.67E-04 | 1.11E-04 | 0.04 | -9.63 | T | 38 | 38 | 16 | - | - |
| Field.COI | 4B_3 | IWB7278 | 4BL_6967384 | 4B | 78.96 | 1.56E-04 | 6.47E-06 | 0.04 | 13.46 | T | 81 | 108 | 103 | QLr.cimmyt-4BL? | William et al 2006 |
| Field.COI | 5B_2 | IWB39735 | 5BL_10794137 | 5B | 49.01 | 1.69E-04 | 6.69E-06 | 0.04 | 20.17 | C | 80 | 115 | 124 | QLr. cdl-5BL? | Kolmer 2015 |
| Field.COI | 6A_t1 | IWB40242 | NA | 6A | 48.09 | 4.97E-04 | 3.26E-04 | 0.04 | -11.69 | T | 4 | 10 | 61 | - | - |
| Field.COI | 7B_4 | IWB64015 | 7BL_6699942 | 7B | 166.99 | 5.03E-04 | 9.98E-05 | 0.04 | 10.87 | T | 75 | 107 | 81 | Lr68 | Herrera-Foessel et al 2012 |
Column "Marker", underlining indicates SNPs detected by or significant in stepwise regression models.
Columns "p.MLM" and "p.QGLM" reflect p-values from the MLM and QGLM methods, underlining indicates p-values passing the simpleM threshold.
Column R indicates resistance (or favorable) allele.
Columns “RSQ” and “eff” indicate marker R2 and effects based on MLM models.
Columns “Sub pop1”, “Sub pop2” and “Sub pop3” indicate number of favorable allele within each sub-populations. Column "postulation" are postulated genes or QTLs based on position, infection type, and donor parents or pedigree information. Some of the loci names were adopted from Li et al [41]. The postulated gene or QTLs followed by questions marks "?" are primarily based on position and infection type (pedigree or donor line information was unobtainable).
Column "reference" shows literature reports that support our loci postulation. Dash "-" signs indicate that the corresponding p-values are below the simpleM threshold and loci identity were not postulated. Asterisks "*" indicate potentially novel loci identified through this study.
We used the GAPIT tool to validate QTLs detected by TASSEL. A total of 44 out of 46 QTLs detected by TASSEL (96%) were also detected using GAPIT (S2 Table). By relaxing the p-value threshold slightly from 0.0010 to 0.0013, all of the 46 QTLs (100%) were significantly detected by GAPIT (S2 Table). We also explored the use of G-model [31] to do GWAS analysis, the p-value used in multiple regression fitting is 0.000001 (highly significant and far surpassing the simpleM threshold), and a total of 108 markers were identified for the Field.COI trait. Many of these markers share similar positions with those identified through MLM and QGLM analysis (S3 Table). Overall, our GWAS results showed high agreement across multiple environments and between various methods of QTL detection.
Gene or QTL Postulations for the Most Significant (Top 10) QTLs for Seedling and Field Resistance
Combining the most significant 10 QTLs (based on MLM p-values, abbreviated as p.MLM) for each trait (Race1, Race.CA1.2, Race.Mix, Field.COI) resulted in a total of 29 unique QTLs. Some of the loci were detected for multiple traits, thus the unique number of QTLs for the four traits is 29 instead of 40 (Table 3). We found that a number of loci were likely known or previously identified loci that are involved in rust resistance. For example, QTL 5D_1 (203.88 cM) located on long arm of chromosome 5D (Table 3), approximately the same location as Lr1. This locus was detected only in the seedling stage, which is consistent with Lr1 being not effective to races present in the field (environments in which the locus was not detected were represented with a dash “-” in Table 2). Thatcher near isogenic line RL6003 (known to possess Lr1) has the favorable allele. This evidence suggests that QTL 5D_1 is Lr1. One of the most significant loci for seedling resistance against race 1 and CA1.2 was QTL 6B_4, which was located on long arm of chromosome 6B (122.92cM, Table 3, Fig 5). Thatcher near isogenic lines RL6002 and RL6042 were known to possess Lr3 [42], and both of these lines possess the favorable SNP allele (C). This evidence suggests that QTL 6B_4 is Lr3. Our data show that the Lr68 representing contig 7BL_6748067 [43] was positioned at 171 cM based on BLAST hit information, roughly at the same location of QTL 7B_4 (166.99 cM). QTL 7B_4 is effective at the adult plant stage only (Table 2). All these results suggest that QTL 7B_4 (SNP IWB64015) represents Lr68.
Besides Lr1, Lr3, Lr68, a number of quantitative trait loci (Table 2) were mapped to approximately the locations of loci Lr26 (QTL 1B_2) [29], Lr42 (1D_t1) [44], Lr21 (1D_3), Lr17(2A_1), [42], Lr11 (2A_3) [45], Lr28 (4A_t2) [46], Lr14 (7B_3) [47], QLr.cimmyt-1BS (1B_1) [48], QLr.cdl-5BL (5B_2) [49], and possibly Lr30 and others [50, 51]. However, due to the lack of pedigree and donor line genotype information (within this panel), and also the near fixation of some alleles within sub-population 1 (i.e., majority of Thatcher NILs have the same allele), more genetic experiments are needed to further confirm these postulations, especially for those that are postulated based primarily on position.
We also designated two potentially novel loci (Table 3) that are associated with different seedling and field rust traits with p-values surpassing the simpleM threshold (7.72E-5). QTL 1A_3 (QLr.umn-1AL) was mapped to position 149.8 cM on chromosome 1AL for both field BLUE and seedling race mixture traits (Table 3). Chromosome 1AL is not associated with adult leaf rust resistance [41]. It is also not likely to be Lr59 [52] because Lr59 is located on an alien introgression that is unlikely to be present in this AM panel. QTL 4A_1 (QLr.umn-4AS), highly associated with field resistance (both p.MLM and p.QGLM surpassing the simpleM threshold), was represented by two SNP markers (IWB13323 and IWB59410), located at 37 cM of 4AS (Fig 5 and Table 2). No known QTLs or genes are present on chromosome 4AS that confer resistance to leaf rust.
Trait Genetic Architecture Revealed by Most Significant QTLs and by Loci Selected Using Stepwise Regression Models
The top 10 QTLs for field resistance were all detected in at least three field environments (p.MLM < 0.001), or all of the five environments (CrK12, StP12, StP13, StP14, StP15) when using less stringent p.MLM cutoffs of 0.02 (S1 Table and Table 2). Our results also show that QTLs associated with the seedling race mixture trait had a higher overlap with QTLs detected in the field (Fig 6, Table 2), consistent with the finding that the two traits are moderately correlated (R > 0.5, Table 1). QTL 1B_1 (likely QLr.cimmyt-1BS, Table 3) is highly effective (p.MLM = 3.77E-4, p.QGLM = 2.67E-7) against seedling race mixtures, but is not among the most significant ones for BLUE estimated coefficient of infection (COI) trait (Field.COI), despite a moderately significant p-value (p.MLM = 1.07E-3) (Table 2). Interestingly, it is the most significant QTL for field response type (Field.IT) trait (p.MLM = 4.78E-5, Table 2). This observation corresponds with the field response type values (Field.IT) having the highest correlation with seedling IT values against race mixtures (Table 1), likely reflecting the biology that both field response type (Field.IT) and seedling infection type are partially measured by pustule sizes of leaf rust fungus uredinia spores. These results suggest that although field response type (Field.IT) and field severity (Field.SEV) or COI (Field.COI) have high correlations between each other (r > = 0.88), the exact ranks and significance levels of effective loci may differ. Overall, the high number of common loci shared between seedling and field traits suggest that race mixtures can be used to screen for field resistance against the same race mixtures at the seedling stage.
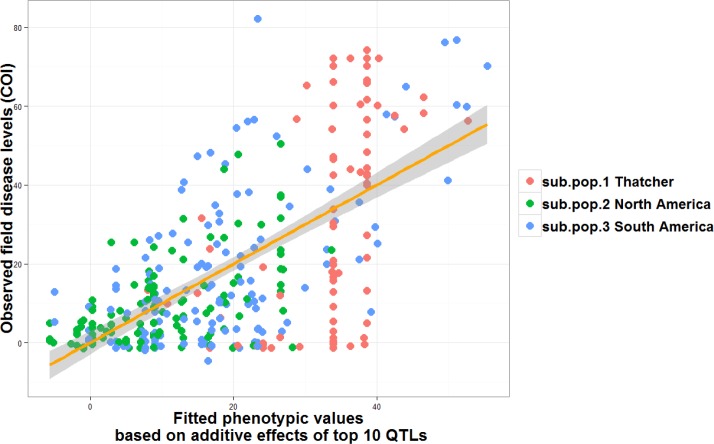
The main contributor genes or loci for seedling resistance included QTLs 6B_3 (likely Lr9), 6B_4 (Lr3), 5D_1 (Lr1), and 2B_2 (likely one of Lr13, Lr16 or Lr23) (Table 3). The 10 most significant seedling QTLs (Table 3) explained a total of 29.4%, 24.3% and 34.3% phenotypic variations for seedling race 1, seedling race CA1.2, and seedling race mixtures, respectively, after subtracting variance due to population structure. The main contributor loci or genes for field resistance include 4A_1 (QLr.umn-4AS), 1A_3 (QLr.umn-1AL), 2B_2 (likely one of Lr13, Lr16 or Lr23) and 5B_2 (likely QLr.cdl-5BL) [49] and 7B_4 (likely Lr68). We plotted the cumulative additive effects of the 10 most significant QTLs (Table 3) in each line of the TCAP leaf rust AM population with their observed phenotype in the field (Fig 7). The selected 10 QTLs for field resistance (measured as COI) (Table 3) explained over 26% of the total phenotypic variation (excluding the amount of contribution derived from population structure). The 10 most significant for field traits (severity and response type) explained 26% and 30% of the phenotypic variations (S4 Fig). Three of the 10 field QTLs (QLr.umn-1AL, QLr.umn-4AS and Q.Lr.cimmyt-4BL) together explained 12% of phenotypic variation (after removing population structure effects). QLr.umn-4AS is the only QTL whose p.MLM and p.QGLM values both pass the simpleM threshold for field resistance. A Haploview [29] of this region is provided (S5 Fig). BLAST search using flanking loci sequence information revealed a possible candidate gene, belonging to the family of ras proteins. One family member of this family was previously identified to be involved in resistance against wheat stripe rust pathogen [53].
Fig 7. Scatterplot of phenotypic values versus genotypic fitted values using markers representing top ten QTLs for field resistance.
We utilized stepwise regression [13, 54] to identify the minimal number of markers that are independently associated with leaf rust resistance for each trait. Our results indicate that, overall, the most significant markers are often included by stepwise regression to account for phenotypic variation (Table 3). However, there are cases where markers with less significance levels were included. For example, the Lr34 marker (csLv34, Table 2) was associated with the leaf rust severity trait (p = 2.03E-3), but its rank of significance is only at 15th (among all 46 loci). However, it was included as one of the seven markers (IWB19584.1B_1, IWA3151.2A_3, IWA6877.3A_1, IWB59410.4A_1, IWB39735.5B_2, IWB40242.6A_t1, cslv34.7D_1) to account for a significant portion of variation (1.5% out of a total of 25%) in the multiple regression model for leaf severity (Field.SEV), but not for leaf response type (Field.IT). These results suggest that leaf rust severity and response types are probably controlled by different genetic loci, and is consistent with the role of Lr34 as a gene mostly effective for APR (as response type likely share more common loci with seedling resistance).
Overall, the leaf rust phenotype for field and seedling plants both suggest the presence of resistance alleles in a large percentage of lines within this AM population (Fig 1). The number of favorable alleles and disease resistance are significantly correlated (p<0.05). Our GWAS results confirmed the presence of multiple favorable alleles in a high percentage of individuals within the AM population. We identified a subset of cultivars or lines that possess favorable alleles for 6 of the 7 QTLs obtained through stepwise regression (see above paragraph for regression model components) for field resistance measured by coefficient of infection (COI). As expected, their field evaluation often showed “trace” resistance (Field.Sev < 5, S4 Table). Adult plant leaf rust resistance breeding could potentially be rapidly improved by selecting lines with complimentary resistance genes or QTLs as parents.
Discussion
Population Structure and Its Relationship with Leaf Rust Disease Levels and GWAS Results
For field data, the observation that sub-population 1 (Thatcher NILs) was associated with higher disease (Fig 4) was not unexpected. Thatcher is susceptible to leaf rust and the Thatcher near isogenic lines carry single resistance genes that are mostly ineffective against multiple race mixtures as tested in the field. Sub-pop 2 (mostly North American lines) were more resistant than the other two sub-populations. It remains to be explored whether this is partially due to the better adaptation of sub-pop 2 in Northern environments (such as day length), or more likely, because the lines have been selected for leaf rust resistance with the races that are common in North America [55]. The South American lines were selected for inclusion in the panel based on their resistance to predominant races in South America. Sub-population 3 (mostly CIMMYT and South American lines, but some North American lines as well, including lines from MN, North Star Genetics, and Limagrain) has the largest range of phenotypic variation, reflecting its more diverse geographical origins and genetic compositions (lower Fst values compared to the other two sub-populations).
Collectively, both origin of the lines and structure analysis based on molecular markers suggest that the true population structures were well captured in our GWAS analysis. It is worth noting that some of the alleles were (nearly) fixed in sub-population 1, as this population consists of almost entirely Thatcher near isogenic lines, which means that they share a significant portion of exactly the same genome. Thus, the presence of favorable marker alleles (particularly in this sub-population) might not indicate the presence of resistant genes. Nonetheless, QTL 5D_1 (Lr1) and QTL 6B_4 (Lr3) alleles were detected only in a few Thatcher NILs, and our evidence suggests that these are SNPs that are in high LD (or may even be diagnostic) with the gene. We also explored the effects of removing sub-population 1 on GWAS analysis results, and found that the most significant loci were mostly not affected. For example, QTL 5D_1 (Lr1) and 6B_4 (Lr3), 4A_1 (QLr.umn-4AS), 1A_3 (QLr.umn-1AL) were again detected (results not shown), adding further support that these are true loci conferring resistance to the leaf rust disease.
Comparisons between Models
The difference between the two models (MLM QK and QGLM) used in this study lies in whether Kinship was included into the model analysis. As expected, the p-values derived from QGLM model are generally more significant compared to MLM (QK) model (Table 3), as no kinship relatedness was factored into the model analysis. We further noticed that the p-value differences between MLM (QK) and QGLM models are more pronounced for seedling traits than for field traits (Table 3). It was known that for this panel, many related cultivars or lines possess certain seedling leaf rust resistance genes. For example, the NDSU cultivars Faller and Glenn and the MN cultivar RB07 all possess Lr21 [56]. Thus, kinship itself could be correlated with the presence or absence of certain seedling leaf rust genes. Correcting for kinship within each sub-population could result in lower statistical power to detect those true QTLs or genes under these circumstances. The QQ plot (S3 Fig) shows that for some traits such as the seedling disease levels against race mixtures, the upper corner dots (higher significance level p-values) curved downwards instead of upwards, possibly indicating that there might be over-correction in the MLM (QK) model analysis. It might make more sense to use p-values derived from QGLM analysis to assist QTL or gene detection under these circumstances. Overall, the QGLM models and MLM (QK) models reveal a similar set of markers. As an exploratory analysis, the G-model revealed QTLs that are often at the same positions of the MLM or QGLM models. The two putatively novel loci (QLr.umn-1AL, QLr.umn-4AS) were repeatedly detected using either one of the models or the G-model, suggesting the robustness of these associations and likelihood that these associations represent relevant Lr resistance loci.
Comparison of Seedling and Field Resistance Genes or QTLs
Although seedling resistance QTLs or genes against race 1 are largely non-overlapping with field resistance loci (Fig 6), we found a high percentage of common loci for field and seedling resistant against race CA1.2 and race mixtures (68 and 80 percent for race CA1.2 and race mixtures respectively) (Fig 6) with consistent marker effects. These results suggest that the seedling resistance genes we detected also contribute to field resistance.
This study also detected some resistance effects in the same regions as the known race non-specific resistance genes (Lr34, and possibly Lr68) that are effective in the adult plant stage. Gene Lr34 encodes an ABC transporter and is effective against multiple pathogen species including leaf rust (Puccinia triticina), stripe rust (P. striiformis) and powdery mildew (Blumeria graminis) [57]. Our results are consistent with the previous discovery that Lr34 is more effective in the adult plant stage rather than the seedling stage.
One of the major QTLs for field resistance (QLr.umn-4AS) was detected in every field environment but not at the seedling stage. Similarly, a few other loci (such as 7B_4, 4B_3 and 5B_2, possibly representing Lr68, QLr.cimmyt-4BL, QLr.cdl-5BL) were also detected for APR but not for seedling resistance. The presence of multiple adult plant APR only loci suggest that field and seedling resistance differ considerably, despite a moderate correlation and the presence of multiple common loci (Table 1 and Fig 5B) between the field and seedling resistance.
Potential Applications in Future Research
The wheat 90K SNP array is among the highest density genotyping platforms available for wheat researchers [5] and has a much improved coverage of the genome than the 9K SNP array [4]. Various research projects have been published using this genotyping platform [58–61]. The availability of high density consensus maps [5] coupled with the rapid progress in genome-wide sequencing efforts [38, 62] will greatly enhance our ability to dissect important agronomic traits such as leaf rust resistance.
The QTLs or genes identified or validated in this study were associated with sequence based markers which could be more efficiently anchored to reference genomes [62] than traditional markers such as simple sequence repeats (SSRs). Assays such as KASP [63, 64] can be developed based on closely linked SNP markers (such as QLr.umn-4AS, 5D_1.Lr1, 7B_4.Lr3 loci) to provide more high-throughput genotyping and marker assisted breeding. Contextual genome sequences around target SNPs might provide direct insights into the genetic composition of trait of interest. Accessions with a high percentage of leaf rust resistance alleles could serve as parental breeding lines to enable more efficient breeding, especially for adult plant resistance.
Conclusions
We conducted a genome-wide association study on a population consisting of mostly breeding lines with known seedling or adult plant resistance. This study is among the first GWAS studies that utilizes the wheat iSelect 90K SNP array to explore leaf rust resistance QTLs. A large percentage of lines were associated with multiple resistance alleles or QTLs for leaf rust resistance. The 10 most significant QTLs accounted for 24–34% of phenotypic variation for each trait analyzed. Compared to single races, leaf rust reaction to race mixtures in the seedling test best resembled field resistance, suggesting that field resistance can be partially screened in the seedling stage using race mixtures. Identification of novel QTLs (such as QLr.umn-1AL, QLr.umn-4AS) with field resistance against leaf rust could enhance our understanding of leaf rust resistance and provide new resources of leaf rust resistance. Identification of a subset of lines with a high percentage of favorable alleles (based on SNP marker information) may serve as valuable parental materials for further resistance breeding.
Supporting Information
Number of unique markers is defined as the number of markers that are at different positions of the consensus map (Wang et al 2014).
(PPTX)
(PPTX)
(PPTX)
(PPTX)
Picture showing relative position and degree of LD in the region.
(PPTX)
(CSV)
(CSV)
(XLSX)
(XLSX)
Acknowledgments
We thank Amy Fox (USDA-ARS, Cereal Disease Laboratory) for inoculation preparation for both field and seedling plants. We thank Kun Xiao (USDA-ARS, St. Paul, MN), Paul Mihalyov (Washington State University, Pullman, WA), Dr. Matthew N. Rouse (USDA-ARS, St. Paul) for testing the Perl scripts for rust score conversions. We thank Drs. Katherine Frels and Xiaofei Zhang (UMN) for reviewing an earlier version of this manuscript and Dr. Rex Bernardo (UMN) for thoughtful discussions on the data analysis and sharing of the G-model software for our exploratory GWAS analysis. Computing resources from the Minnesota Supercomputing Institute at the University of Minnesota are greatly appreciated.
Data Availability
T3 database (http://triticeaetoolbox.org/wheat/).
Funding Statement
This study is part of the Triticeae Coordinated Agriculture Project (TCAP, www.triticeaecap.org), funded by the United States Department of Agriculture National Institute of Food and Agriculture (USDA-NIFA) grant no. 2011-68002-30029.
References
- 1.McIntosh RA, Dubcovsky J, Rogers WJ, Morris C, Appels R, Xia XC. Catalogue of gene symbols for wheat 2013–2014 supplement 2013. Available: http://www.shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2013.pdf.
- 2.Zhu C, Gore M, Buckler ES, Yu J. Status and Prospects of Association Mapping in Plants. The Plant Genome. 2008;1(1):5–20. 10.3835/plantgenome2008.02.0089 [DOI] [Google Scholar]
- 3.Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, et al. A Robust, Simple Genotyping-by-Sequencing (GBS) Approach for High Diversity Species. PLoS One. 2011;6(5). e19379 10.1371/journal.pone.0019379 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavanagh CR, Chao SM, Wang SC, Huang BE, Stephen S, Kiani S, et al. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(20):8057–62. 10.1073/pnas.1217133110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, et al. Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnology Journal. 2014;12:787–96. 10.1111/pbi.12183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rafalski JA. Association genetics in crop improvement. Current Opinion in Plant Biology. 2010;13(2):174–80. 10.1016/j.pbi.2009.12.004 . [DOI] [PubMed] [Google Scholar]
- 7.Maccaferri M, Sanguineti MC, Mantovani P, Demontis A, Massi A, Ammar K, et al. Association mapping of leaf rust response in durum wheat. Mol Breed. 2010;26(2):189–228. 10.1007/s11032-009-9353-0 . [DOI] [Google Scholar]
- 8.Yu L-X, Lorenz A, Rutkoski J, Singh RP, Bhavani S, Huerta-Espino J, et al. Association mapping and gene-gene interaction for stem rust resistance in CIMMYT spring wheat germplasm. Theor Appl Genet. 2011;123(8):1257–68. 10.1007/s00122-011-1664-y . [DOI] [PubMed] [Google Scholar]
- 9.Letta T, Olivera P, Maccaferri M, Jin Y, Ammar K, Badebo A, et al. Association Mapping Reveals Novel Stem Rust Resistance Loci in Durum Wheat at the Seedling Stage. Plant Genome. 2014;7(1). 10.3835/plantgenome2013.08.0026 . [DOI] [Google Scholar]
- 10.Zhang DD, Bowden RL, Yu JM, Carver BF, Bai GH. Association Analysis of Stem Rust Resistance in US Winter Wheat. PLoS One. 2014;9(7). e103747 10.1371/journal.pone.0103747 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zegeye H, Rasheed A, Makdis F, Badebo A, Ogbonnaya FC. Genome-Wide Association Mapping for Seedling and Adult Plant Resistance to Stripe Rust in Synthetic Hexaploid Wheat. PLoS One. 2014;9(8). e105593 10.1371/journal.pone.0105593 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maccaferri M, Zhang J, Bulli P, Abate Z, Chao S, Cantu D, et al. A Genome-Wide Association Study of Resistance to Stripe Rust (Puccinia striiformis f. sp. tritici) in a Worldwide Collection of Hexaploid Spring Wheat (Triticum aestivum L.). G3: Genes|Genomes|Genetics. 2015. 10.1534/g3.114.014563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kertho A, Mamidi S, Bonman JM, McClean PE, Acevedo M. Genome-Wide Association Mapping for Resistance to Leaf and Stripe Rust in Winter-Habit Hexaploid Wheat Landraces. PLoS One. 2015;10(6). ARTN e0129580 10.1371/journal.pone.0129580 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McIntosh RA, Wellings CR, Park RF. Wheat Rusts: An Atlas of Resistance Genes. Dordrecht, the Netherlands: Kluwer Academic Publishers; 1995. [Google Scholar]
- 15.Stakman EC, Stewart DM, Loegering WQ. Identification of physiologic races of Puccinia graminis var. tricici: United States Department of Agricultura., Agricultural Research Service; 1962. [Google Scholar]
- 16.R Core Team. R: A language and environment for statistical computing Vienna, Australia: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 17.Peterson RR, Campbell AB, Hannah AE. A diagrammatic scale for estimating rust intensity on leaves and stems of cereals. Canadian Journal of Research. 1948;26c(5):496–500. [Google Scholar]
- 18.Stubbs RW, Prescott JM, Saari EE, Dubin HJ. Cereal disease methodology mannual. Mexico, DF: CIMMYT; 1986. 46 p. [Google Scholar]
- 19.Lagudah ES, McFadden H, Singh RP, Huerta-Espino J, Bariana HS, Spielmeyer W. Molecular genetic characterization of the Lr34/Yr18 slow rusting resistance gene region in wheat. Theor Appl Genet. 2006;114(1):21–30. 10.1007/s00122-006-0406-z . [DOI] [PubMed] [Google Scholar]
- 20.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–59. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. 10.1086/519795 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology. 2005;14(8):2611–20. 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- 23.Earl D, vonHoldt B. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genet Resour. 2012;4(2):359–61. 10.1007/s12686-011-9548-7 [DOI] [Google Scholar]
- 24.Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23(14):1801–6. Epub 2007/05/09. 10.1093/bioinformatics/btm233 . [DOI] [PubMed] [Google Scholar]
- 25.Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Notes. 2004;4(1):137–8. 10.1046/j.1471-8286.2003.00566.x . [DOI] [Google Scholar]
- 26.Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23(19):2633–5. 10.1093/bioinformatics/btm308 [DOI] [PubMed] [Google Scholar]
- 27.Eu-ahsunthornwattana J, Miller EN, Fakiola M, Jeronimo SMB, Blackwell JM, Cordell HJ, et al. Comparison of Methods to Account for Relatedness in Genome-Wide Association Studies with Family-Based Data. PLoS Genet. 2014;10(7):e1004445 10.1371/journal.pgen.1004445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marroni F, Pinosio S, Zaina G, Fogolari F, Felice N, Cattonaro F, et al. Nucleotide diversity and linkage disequilibrium in Populus nigra cinnamyl alcohol dehydrogenase (CAD4) gene. Tree Genetics & Genomes. 2011;7(5):1011–23. 10.1007/s11295-011-0391-5 [DOI] [Google Scholar]
- 29.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. 10.1093/bioinformatics/bth457 . [DOI] [PubMed] [Google Scholar]
- 30.Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, et al. GAPIT: genome association and prediction integrated tool. Bioinformatics. 2012;28(18):2397–9. 10.1093/bioinformatics/bts444 [DOI] [PubMed] [Google Scholar]
- 31.Bernardo R. Genomewide Markers for Controlling Background Variation in Association Mapping. Plant Genome. 2013;6(1):-. 10.3835/plantgenome2012.11.0028 [DOI] [Google Scholar]
- 32.Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006;38(2):203–8. [DOI] [PubMed] [Google Scholar]
- 33.Bernardo R. Breeding for quantitative traits in plants. 2nd ed: Stemma Press; 2010. 390 p. [Google Scholar]
- 34.Zhang Z, Ersoz E, Lai C-Q, Todhunter RJ, Tiwari HK, Gore MA, et al. Mixed linear model approach adapted for genome-wide association studies. Nat Genet. 2010;42(4):355–60. 10.1038/ng.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pasam R, Sharma R, Malosetti M, van Eeuwijk F, Haseneyer G, Kilian B, et al. Genome-wide association studies for agronomical traits in a world wide spring barley collection. BMC Plant Biology. 2012;12(1):16 10.1186/1471-2229-12-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao XY, Stamier J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008;32(4):361–9. 10.1002/gepi.20310 . [DOI] [PubMed] [Google Scholar]
- 37.Hirsch CN, Foerster JM, Johnson JM, Sekhon RS, Muttoni G, Vaillancourt B, et al. Insights into the Maize Pan-Genome and Pan-Transcriptome. Plant Cell. 2014;26(1):121–35. 10.1105/tpc.113.119982 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chapman JA, Mascher M, Buluc A, Barry K, Georganas E, Session A, et al. A whole-genome shotgun approach for assembling and anchoring the hexaploid bread wheat genome. Genome Biology. 2015;16 UNSP 26 10.1186/s13059-015-0582-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayer KFX, Waugh R, Langridge P, Close TJ, Wise RP, Graner A, et al. A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012;491(7426):711–+. 10.1038/Nature11543 . [DOI] [PubMed] [Google Scholar]
- 40.Maccaferri M, Zhang J, Bulli P, Abate Z, Chao S, Cantu D, et al. A genome-wide association study of resistance to stripe rust (Puccinia striiformis f. sp. tritici) in a worldwide collection of hexaploid spring wheat (Triticum aestivum L.). G3: Genes|Genomes|Genetics. 2015;5(3):449–65. 10.1534/g3.114.014563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Z, Lan C, He Z, Singh RP, Rosewarne GM, Chen X, et al. Overview and Application of QTL for Adult Plant Resistance to Leaf Rust and Powdery Mildew in Wheat. Crop Science. 2014;54(5):1907–25. 10.2135/cropsci2014.02.0162 . [DOI] [Google Scholar]
- 42.Kolmer JA. Postulation of leaf rust resistance genes in selected soft red winter wheats. Crop Science. 2003;43(4):1266–74. . [Google Scholar]
- 43.Jordan K, Wang S, Lun Y, Gardiner L-J, MacLachlan R, Hucl P, et al. A haplotype map of allohexaploid wheat reveals distinct patterns of selection on homoeologous genomes. Genome Biology. 2015;16(1):48 10.1186/s13059-015-0606-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu ZL, Bowden RL, Bai GH. Molecular Markers for Leaf Rust Resistance Gene Lr42 in Wheat. Crop Science. 2013;53(4):1566–70. 10.2135/cropsci2012.09.0532 . [DOI] [Google Scholar]
- 45.Darino MA, Dieguez MJ, Singh D, Ingala LR, Pergolesi MF, Park RF, et al. Detection and location of Lr11 and other leaf rust resistance genes in the durably resistant wheat cultivar Buck Poncho. Euphytica. 2015:1–13. 10.1007/s10681-015-1486-0 [DOI] [Google Scholar]
- 46.Bipinraj A, Honrao B, Prashar M, Bhardwaj S, Rao S, Tamhankar S. Validation and identification of molecular markers linked to the leaf rust resistance gene Lr28 in wheat. J Appl Genet. 2011;52(2):171–5. 10.1007/s13353-010-0026-9 . [DOI] [PubMed] [Google Scholar]
- 47.Singh D, Simmonds J, Park RF, Bariana HS, Snape JW. Inheritance and QTL mapping of leaf rust resistance in the European winter wheat cultivar 'Beaver'. Euphytica. 2009;169(2):253–61. 10.1007/s10681-009-9959-7 . [DOI] [Google Scholar]
- 48.Rosewarne GM, Singh RP, Huerta-Espino J, Herrera-Foessel SA, Forrest KL, Hayden MJ, et al. Analysis of leaf and stripe rust severities reveals pathotype changes and multiple minor QTLs associated with resistance in an Avocet x Pastor wheat population. Theor Appl Genet. 2012;124(7):1283–94. 10.1007/s00122-012-1786-x . [DOI] [PubMed] [Google Scholar]
- 49.Kolmer JA. A QTL on chromosome 5BL in wheat enhances leaf rust resistance of Lr46. Mol Breed. 2015;35(2). 74 10.1007/s11032-015-0274-9 . [DOI] [Google Scholar]
- 50.Draz IS, Abou-Elseoud MS, Kamara A-EM, Alaa-Eldein OA-E, El-Bebany AF. Screening of wheat genotypes for leaf rust resistance along with grain yield. Annals of Agricultural Sciences. 2015;60(1):29–39. 10.1016/j.aoas.2015.01.001. [DOI] [Google Scholar]
- 51.Chu CG, Friesen TL, Xu SS, Faris JD, Kolmer JA. Identification of novel QTLs for seedling and adult plant leaf rust resistance in a wheat doubled haploid population. Theor Appl Genet. 2009;119(2):263–9. 10.1007/s00122-009-1035-0 . [DOI] [PubMed] [Google Scholar]
- 52.Marais GF, McCallum B, Marais AS. Wheat leaf rust resistance gene Lr59 derived from Aegilops peregrina. Plant Breeding. 2008;127(4):340–5. 10.1111/j.1439-0523.2008.01513.x [DOI] [Google Scholar]
- 53.Liu F, Guo J, Bai P, Duan Y, Wang X, Cheng Y, et al. Wheat TaRab7 GTPase Is Part of the Signaling Pathway in Responses to Stripe Rust and Abiotic Stimuli. PLoS One. 2012;7(5):e37146 10.1371/journal.pone.0037146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS Results: A Review of Statistical Methods and Recommendations for Their Application. Am J Hum Genet. 2010;86(1):6–22. 10.1016/j.ajhg.2009.11.017 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huerta-Espino J, Singh RP, Germán S, McCallum BD, Park RF, Chen WQ, et al. Global status of wheat leaf rust caused by Puccinia triticina. Euphytica. 2011;179(1):143–60. 10.1007/s10681-011-0361-x [DOI] [Google Scholar]
- 56.Kolmer JA, Anderson JA. First Detection in North America of Virulence in Wheat Leaf Rust (Puccinia triticina) to Seedling Plants of Wheat with Lr21. Plant Dis. 2011;95(8):1032–. 10.1094/pdis-04-11-0275 [DOI] [PubMed] [Google Scholar]
- 57.Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino J, McFadden H, et al. A Putative ABC Transporter Confers Durable Resistance to Multiple Fungal Pathogens in Wheat. Science. 2009;323(5919):1360–3. 10.1126/science.1166453 [DOI] [PubMed] [Google Scholar]
- 58.Thavamanikumar S, Dolferus R, Thumma BR. Comparison of Genomic Selection Models to Predict Flowering Time and Spike Grain Number in Two Hexaploid Wheat Doubled Haploid Populations. G3 (Bethesda). 2015;5(10):1991–8. Epub 2015/07/25. 10.1534/g3.115.019745 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Babiker EM, Gordon TC, Chao S, Newcomb M, Rouse MN, Jin Y, et al. Mapping resistance to the Ug99 race group of the stem rust pathogen in a spring wheat landrace. Theor Appl Genet. 2015;128(4):605–12. 10.1007/s00122-015-2456-6 [DOI] [PubMed] [Google Scholar]
- 60.Guttieri MJ, Baenziger PS, Frels K, Carver B, Arnall B, Wang S, et al. Prospects for Selecting Wheat with Increased Zinc and Decreased Cadmium Concentration in Grain. Crop Science. 2015. [Google Scholar]
- 61.Bajgain P, Rouse M, Bulli P, Bhavani S, Gordon T, Wanyera R, et al. Association mapping of North American spring wheat breeding germplasm reveals loci conferring resistance to Ug99 and other African stem rust races. BMC Plant Biology. 2015;15(1):249 10.1186/s12870-015-0628-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Consortium TIWGS. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. 2014;345(6194). 10.1126/science.1251788 [DOI] [PubMed] [Google Scholar]
- 63.Semagn K, Babu R, Hearne S, Olsen M. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): overview of the technology and its application in crop improvement. Mol Breed. 2014;33(1):1–14. 10.1007/s11032-013-9917-x . [DOI] [Google Scholar]
- 64.Gao L, Kielsmeier-Cook J, Bajgain P, Zhang X, Chao S, Rouse M, et al. Development of genotyping by sequencing (GBS)- and array-derived SNP markers for stem rust resistance gene Sr42. Mol Breed. 2015;35(11):1–13. 10.1007/s11032-015-0404-4 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Number of unique markers is defined as the number of markers that are at different positions of the consensus map (Wang et al 2014).
(PPTX)
(PPTX)
(PPTX)
(PPTX)
Picture showing relative position and degree of LD in the region.
(PPTX)
(CSV)
(CSV)
(XLSX)
(XLSX)
Data Availability Statement
T3 database (http://triticeaetoolbox.org/wheat/).