Abstract
Aims: The ferric uptake regulator (Fur) is the main transcriptional regulator of genes involved in iron homeostasis in most prokaryotes. FurA from Anabaena sp. PCC 7120 contains five cysteine residues, four of them arranged in two redox-active CXXC motifs. The protein needs not only metal but also reducing conditions to remain fully active in vitro. Through a mutational study of the cysteine residues present in FurA, we have investigated their involvement in metal and DNA binding. Results: Residue C101 that belongs to a conserved CXXC motif plays an essential role in both metal and DNA binding activities in vitro. Substitution of C101 by serine impairs DNA and metal binding abilities of FurA. Isothermal titration calorimetry measurements show that the redox state of C101 is responsible for the protein ability to coordinate the metal corepressor. Moreover, the redox state of C101 varies with the presence or absence of C104 or C133, suggesting that the environments of these cysteines are mutually interdependent. Innovation: We propose that C101 is part of a thiol/disulfide redox switch that determines FurA ability to bind the metal corepressor. Conclusion: This mechanism supports a novel feature of a Fur protein that emerges as a regulator, which connects the response to changes in the intracellular redox state and iron management in cyanobacteria. Antioxid. Redox Signal. 24, 173–185.
Introduction
Acquiring and maintaining adequate intracellular iron levels are major challenges that face most organisms, since insufficient iron impairs the correct function of essential iron proteins, whereas excess free iron can be deleterious to biomolecules due to its high reactivity, producing reactive-free radicals. The control of intracellular iron concentration in many bacteria is mainly operated by the transcriptional regulator Fur protein (11). The current model of action for this protein establishes that, under iron rich conditions, a dimer of Fur binds to specific DNA sequences (“iron boxes”) located in the promoters of iron responsive genes using Fe2+ as corepressor and prevents their transcription (3). However, this model does not take into account certain aspects of Fur-mediated regulation revealed by recent studies. In this sense, Fur participates in gene activation and iron-independent regulation of a variety of genes involved in distinct functions (6, 36).
Innovation.
Ferric uptake regulator (Fur) proteins control intracellular iron concentration in most bacteria. The results of this study show for the first time for a Fur homologue that thiol/disulfide interconversion controls the activity of cyanobacterial FurA. The reduced FurA can bind its metal corepressor and hence DNA conversely. When FurA is oxidized it loses the metal and dissociates from DNA. The FurA thiol/disulfide exchange responds to alteration in the cellular redox potential. Thus, the binding of FurA to the metal corepressor mediated by its redox state may provide the basis of a concerted mechanism of iron homeostasis in response to the redox state of the cytosol in Anabaena.
The crystal structures of diverse Fur family members confirm the basic folding firstly described for the Pseudomonas aeruginosa Fur homologue, even though each Fur subclass responds to different signals (42). It consists of two well-defined domains: an N-terminal DNA binding domain and a C-terminal dimerization region. The amino acid sequences of many Fur homologues include two potential metal binding motifs, a conserved HHXHXXCXXC signature and another less conserved C-terminal CXXC motif. In fact, two or three metal atoms whose coordination involves amino acids of these motifs are observed in the crystal structures of several Fur homologues (1, 6, 9, 34, 39, 47, 48). Usually, one of the metal binding sites accommodates the regulatory metal atom, while another site has a structural character. Regarding the third site, it seems to play a role in stabilizing the dimeric form of the regulator (9).
The regulatory metal binding site appears to be conserved in all Fur and Fur-like proteins, although it shows some variability in the coordination depending on the Fur homologue and the metal. The structural metal binding site, in some Fur homologues, involves regular tetrahedral coordination of a zinc atom by four cysteine residues belonging to the two CXXC sequences present in the previously mentioned metal binding motifs (9, 28, 34). In fact, zinc coordination by the four cysteines of both CXXC motifs is suggested to be critical to stabilize the dimeric structure in Bacillus subtilis PerR (48). However, the existence of these CXXC sequences does not ensure the binding of structural metal. In this sense, the crystal structure of Nur, obtained under reducing conditions, indicates that both cysteines of the two CXXC motifs do not coordinate zinc, although this regulator maintains its DNA binding activity (1). In addition, Vibrio cholerae Fur structure does not reveal any metal binding site involving equivalent cysteines present in its primary sequence (47).
In the filamentous nitrogen-fixing cyanobacterium Anabaena sp. PCC 7120, three different Fur proteins have been identified: FurA (all1691), FurB (all2473), and FurC (alr0957) (21). FurA is a multifaceted protein that controls iron homeostasis and modulates an extensive regulon involved in a variety of cell functions (13, 14, 15). It also fulfills the features of a heme sensor protein whose interaction with this cofactor negatively affects its DNA binding ability in vitro (23, 41).
The FurA primary sequence contains five cysteines located at the C-terminal domain (C101, C104, C133, C141, and C144), four of them arranged in two CXXC motifs (C101VKC104 and C141PKC144) (4). However, these CXXC motifs do not appear to coordinate a structural metal atom in FurA since metal analysis and electrospray ionization mass spectrometry revealed that neither zinc nor other metals are present in the purified protein (20). Interestingly, recent data have demonstrated that FurA exhibits disulfide reductase activity in vitro based on these CXXC motifs (5).
Previous results from our laboratory indicated that recombinant FurA exists in solution in several discrete disulfide-linked oligomeric species, being monomers and dimers the major species (20), and that four out of five cysteines are not reactive. In fact, determination of free thiols indicated that only one cysteine was accessible to solvent (20). Different data suggest the relevance of cysteines and their redox status in recombinant Anabaena FurA function. On one hand, thiol reducing agents enhance the affinity of FurA for its target DNA sequences in vitro (22). On the other hand, FurA reductase activity occurs depending on the redox state of cysteines present in its CXXC motifs (5). Finally, the FurA–heme interaction that involves residue C141 from one of the CXXC motifs undergoes redox-mediated ligand switching (41). Therefore, considering the apparent dependence of FurA activity on its oxidation state and the lack of a structural metal involving its two CXXC motifs, it can be envisaged that at least one redox-active cysteine would be required for the function of this protein. We therefore sought to identify the redox-active cysteine(s) in FurA to deepen its/their involvement in the operation mechanism of this regulator. For this purpose, mutations in the cysteines present in FurA have been performed to analyze their implication in metal and DNA interactions.
Our results evidence that a single cysteine belonging to one of the CXXC motifs of FurA is essential for both metal and DNA binding in vitro. This reactive cysteine, identified as C101, is responsible for the requirement of reducing agents by recombinant FurA to maintain optimal DNA binding activity. In addition, we have demonstrated that the redox state (thiol or disulfide) of C101 is modulated by C104, its partner in the CXXC motif, compromising the ability of FurA to bind the metal corepressor necessary for DNA binding.
Results
Substitution of C101 by serine impairs the ability of Anabaena FurA to bind DNA
To determine whether single or multiple cysteines are important for the activity of Anabaena FurA in vitro, each of the five cysteines (C101, C104, C133, C141, and C144) was mutated to serine. The mutations were confirmed by DNA sequencing, and after expression in Escherichia coli, FurA variants were purified with a yield comparable to that obtained for the wild-type protein (40).
Mutant proteins were assayed for DNA binding activity in a standard gel mobility shift assay (EMSA). Experiments were performed in the presence and absence of dithiothreitol (DTT) and/or metal, using recombinant wild-type FurA as a control. Mn2+ was used as a surrogate for Fe2+ for the in vitro analyses of FurA because ferrous iron readily oxidizes to Fe3+ in air.
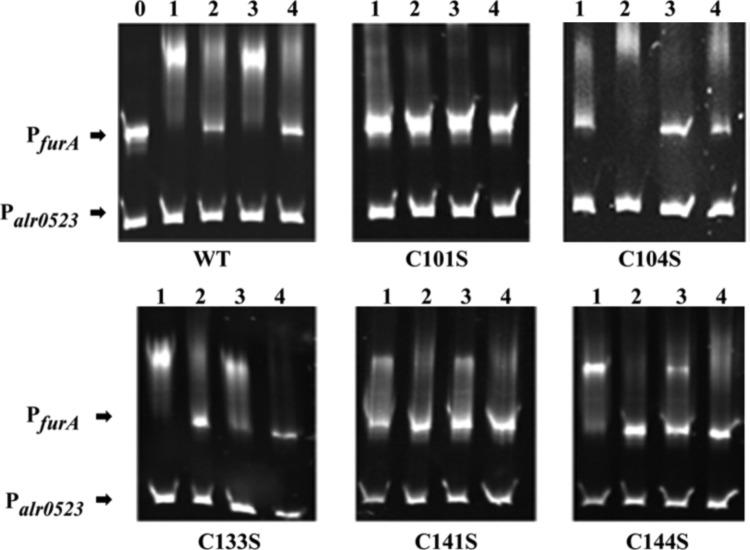
Analysis of single cysteine mutants showed that C133S exhibited lower activity than wild-type FurA in the presence of Mn2+ or DTT (Fig. 1). Besides, C133S-DNA binding ability was negligible in the absence of both effectors (Fig. 1). Activity of C141S was also lower compared with FurA in the four conditions tested, although its dependence on metal and reducing conditions resembled to that of wild-type FurA (Fig. 1). Moreover, the highest activity of C144S was observed in the presence of DTT.
FIG. 1.
DNA binding activity of FurA (ferric uptake regulator) and its single cysteine mutants. Electrophoretic mobility shift assays show the ability of FurA wild-type or cysteine mutant proteins to bind its own promoter (PfurA) in the presence of a nonspecific DNA control (Palr0523) in vitro. One hundred nanograms of DNA fragments free (lane 0) or mixed with 300 nM FurA proteins were separated on 6% native PAGE. Lane 1: binding assay in the presence of 1 mM DTT and 100 μM Mn2+; lane 2: only in the presence of 100 μM Mn2+; lane 3: only in the presence of 1 mM DTT; lane 4: in the absence of both DTT and Mn2+. DTT, dithiothreitol.
However, the most outstanding differences with respect to wild-type FurA were found in EMSA assays with mutants C101S and C104S. As shown in Figure 1, the absence of the C104 residue produced a protein that only needed metal to specifically bind DNA. However, as observed for the C101S mutant, the absence of the C101 residue prevented the binding of the protein to the consensus sequence in any assayed conditions (Fig. 1). According to these results, C101 seems to be essential for the sequence-specific FurA-DNA binding in vitro.
The loss of DNA binding ability of the C101S mutant is not due to a structural change
To know whether the loss of DNA binding activity observed for the C101S variant was associated with an important change in secondary or tertiary structures of FurA after mutation, circular dichroism (CD), Fourier transform infrared spectroscopy (FT-IR), and nuclear magnetic resonance spectroscopy (1H 1D-NMR) experiments of wild-type FurA and C101S variant were performed.
Based on CD analysis, the substitution of C101 by serine caused small changes in the shape of the spectra at 222 nm, which could be due to the absorbance of cysteine residues, occurring in this region (31, 50) (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/ars). The results obtained by FT-IR indicated that the content in α-helix and ß-sheet was similar in the wild-type and C101S proteins (Supplementary Table 1). Finally, the 1H 1D-NMR spectra of both species were identical (Supplementary Fig. S2), indicating the absence of major changes in the secondary and tertiary structures of the wild-type protein and its mutants. All these results strongly suggest that the inability of C101S to bind DNA is not due to a change in the protein secondary or tertiary structures after mutation of C101.
The reduced state of the C101 is essential for Mn2+ coordination by wild-type FurA
Structural and site-directed mutagenesis data reported in the literature indicated that cysteines are involved in metal binding sites in different Fur and Fur-like homologues (1, 6, 9, 30, 34, 39). After ruling out a structural change as the cause of the loss of activity of the C101S mutant, the ability of FurA cysteine mutants to bind the metal corepressor was assessed by isothermal titration calorimetry (ITC). Taking into account the importance of redox conditions and the presence of metal corepressor to allow purified FurA-DNA interaction (22), the binding of Mn2+ to wild-type FurA was studied by ITC, either in the presence or absence of DTT.
At acidic pH (pH 4), FurA did not interact with Mn2+, even in the presence of 1 mM DTT (not shown). At basic pH, FurA is an aggregation-prone species and this tendency rises with increasing protein concentrations (20). Therefore, the measurements were performed at basic pH in the presence of 50 mM arginine, whose use has been described as an assistant for correct folding of previously denatured proteins (37, 46) and it seems to minimize intermolecular noncovalent interactions (2).
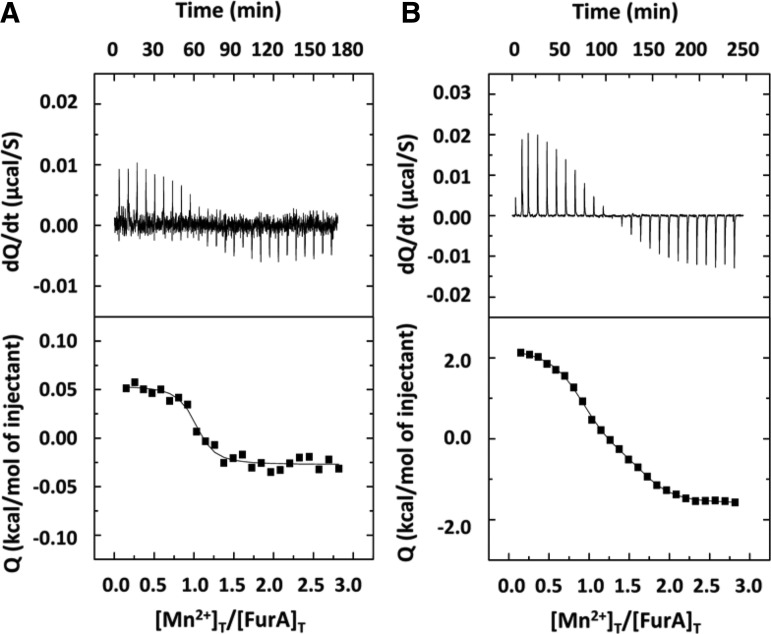
Figure 2 shows the results of titrations of reduced and oxidized wild-type FurA with MnCl2 in the presence of arginine. In the curve for oxidized wild-type FurA, the interaction is characterized by a small enthalpic contribution (Fig. 2A). Fitting the data to a model with a single binding site led to a dissociation constant (Kd) value of 0.28 μM for the interaction of Mn2+ and wild-type FurA. However, addition of DTT increased about 20 times the heat effect (Fig. 2B) and the analysis required fitting the data to a model with two independent binding sites, providing Kd values of 50 nM and 0.84 μM, respectively. These data indicated that the higher affinity metal binding site disappears when cysteines are in the disulfide state, suggesting that cysteines are involved in metal binding in FurA.
FIG. 2.
ITC experiment following Mn2+ binding to oxidized or reduced FurA. Top sections show raw data obtained on a MicroCal VP-ITC calorimeter at 25°C. Aliquots of 300 μM MnCl2 were added over 20 μM FurA in 50 mM Tris/HCl, 50 mM arginine, and pH 9 in the absence (A) or presence (B) of 1 mM DTT. Lower sections show the ITC isotherms of the data integrated and fitted to a single-site model (A) or two independent-site models (B). ITC, isothermal titration calorimetry.
To investigate the location of the two Mn2+ sites in FurA, metal binding affinities were determined for all single cysteine FurA mutants under reducing conditions (Supplementary Fig. S3). Four out of five mutants (C104S, C133S, C141S, and C144S) showed Mn2+ binding affinities in the same order of magnitude as that of the wild-type protein (Table 1), in contrast to C101S variant where the mutation of C101 led to lose the Mn2+ higher affinity site. The lack of the higher affinity site was also observed in a double mutant C101/133S (not shown). However, major changes were not observed regarding the second Mn2+ binding site. Thus, C101 residue is clearly important for FurA to coordinate Mn2+ in vitro. Moreover, since there are no significant changes in the secondary or tertiary structures of C101S, we suggest that the loss of the higher affinity metal binding site is largely responsible for the inability of C101S to bind DNA.
Table 1.
Dissociation Constants (Kd) Estimated by ITC Analyses for the Interaction of FurA Proteins with Mn2+, Fitting the Experimental Data to the Theoretical Equation Based on One (Kd1) or Two (Kd2) Binding Site Models
| Protein | Site 1 (nM) | Site 2 (μM) |
|---|---|---|
| FurA | Kd1 = 50 | Kd2 = 0.8 |
| C101S | — | Kd2 = 0.5 |
| C104S | Kd1 = 14 | Kd2 = 3.4 |
| C133S | Kd1 = 47 | Kd2 = 3.0 |
| C141S | Kd1 = 32 | Kd2 = 10.2 |
| C144S | Kd1 = 71 | Kd2 = 1.8 |
Relative errors: 10–15% for Kd, 5% for ΔH and–TΔS, and 2% for ΔG.
Fur, ferric uptake regulator; ITC, isothermal titration calorimetry.
According to the results shown so far, we can conclude that the higher affinity metal binding site is lost in oxidized FurA and the C101 residue is essential for the binding of the metal atom. Therefore, it appears that, in purified FurA, C101 is part of a disulfide bridge and its redox state is responsible for the protein to coordinate the metal corepressor, since C101S mutant cannot bind DNA in the presence of metal under reducing conditions (Fig. 1).
Recombinant FurA contains two intramolecular disulfide bridges and one free cysteine
Since sulfenic acids can serve as intermediates between thiols and disulfides, we tested the possibility that FurA cysteines could be oxidized to sulfenic acids using 7-chloro-4-nitrobenz-2-oxa-1,3-diazole (NBD-Cl). This electrophilic compound reacts with thiols and sulfenic acids, but the conjugation products have different absorption wavelengths. Upon reaction of NBD-Cl with thiol groups, an absorption peak at about 420 nm can be registered, whereas the peak shifts to 347 nm when the compound reacts with sulfenic acids (38). This approach showed that no sulfenylation occurs in prereduced FurA untreated or treated with H2O2 upon exposure to NBD-Cl (Supplementary Fig. S4).
Trapping experiments of free thiol groups were performed to determine the presence of disulfide bonds in the recombinant wild-type FurA. The thiol modifying agent 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS) reacts with free cysteines in the protein increasing its molecular weight in 540 Da per AMS molecule added (8).
As observed in Figure 3, reduced wild-type FurA displayed five free cysteines with a size shift of 2700 Da. Two bands were observed in the oxidized state, one corresponding to FurA dimers with an intermolecular disulfide bridge (no AMS molecules) and another band corresponding to monomers showing a weight consistent with the uptake of one AMS molecule. A control of recombinant FurA with no DTT subjected to AMS alkylation showed an identical band pattern to that of oxidized FurA (not shown), confirming that the purified protein is obtained in the oxidized state. According to these results, recombinant wild-type FurA monomers contain two intramolecular disulfide bridges and a single free cysteine that is prone to forming an intermolecular disulfide bond with the free cysteine of another monomer. This is in good agreement with previous data obtained by analyzing the free thiol content in FurA with the Ellman's reagent (20).
FIG. 3.
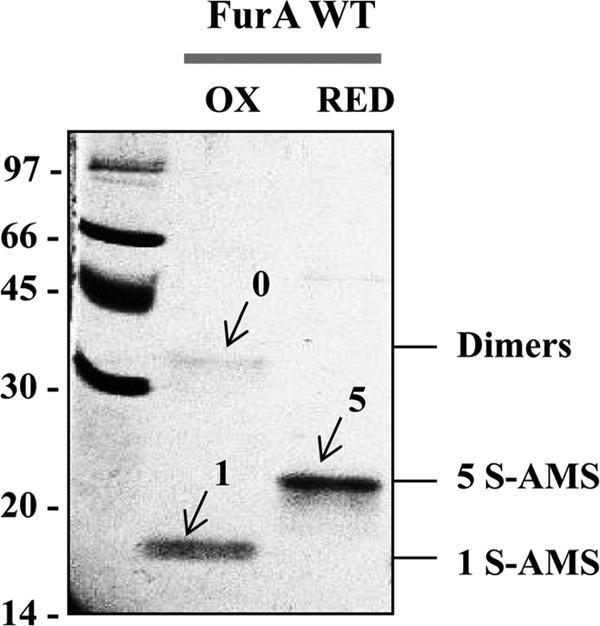
In vitro redox state of FurA. DTT treatment, free thiol alkylation with AMS, and electrophoretic analysis of FurA were performed as indicated in the Materials and Methods section. The resulting proteins resolved on 12.5% nonreducing SDS-PAGE gels were stained with Coomassie blue. The presence of dimers and monomers with 1 (1 S-AMS) or 5 (5 S-AMS) free thiols alkylated with AMS is indicated. OX, oxidized protein; RED, reduced protein; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
C101 seems to be responsible for dimerization of recombinant wild-type FurA
Sensitivity of recombinant FurA and its cysteine mutants to reducing conditions were characterized using nonreducing sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). FurA mutant proteins preincubated with 10 mM DTT were predominantly found as monomers, although a weak band appeared at the position expected for dimers, likely resulting of hydrophobic interactions (Fig. 4). Thus, reduced proteins contained a very little amount of intermolecularly cross-linked dimeric species, confirming that dimerization of wild-type FurA and cysteine mutant species was mainly due to intermolecular disulfide bridges. The differences in the hydrophobicity of the resulting proteins after mutation of cysteines, which are supposed to be in the surface of the protein, could account for the trimer bands observed after reduction with DTT (33).
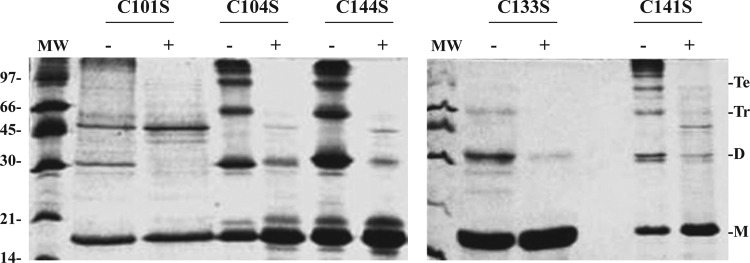
FIG. 4.
Nonreducing electrophoretic analyses of the oligomeric state of FurA and its single cysteine mutants. The 17% nonreducing SDS-PAGE gels stained with Coomassie blue show the oligomerization via disulfide bonds of different FurA variants preincubated with 10 mM DTT as reducing agent (+) or without any treatment (−). MW, molecular weight markers (97, 66, 45, 30, 20, and 14 kDa). The presence of monomers (M), dimers (D), trimers (Tr), and tetramers (Te) is indicated.
In contrast, in the absence of DTT, the C101S variant migrates mainly as a monomeric species and no tetramer band is observed, unlike the rest of mutants (Fig. 4), suggesting that the formation of tetramers in purified wild-type FurA is dependent on the prior existence of dimers in vitro. From this finding, it follows that in C101S variant, intermolecular disulfide bond formation is disfavored with respect to the other cysteine mutants and it is reasonable to assume that the four cysteines in this variant (C104, C133, C141, and C144) form intramolecular disulfide bridges.
Considering the oxidation state of the cysteines in FurA and the inability of the C101S mutant to bind DNA in any conditions tested (Fig. 1), we suggest that the low DNA binding activity of FurA with metal but not DTT could be attributed to its inability to bind the metal corepressor, as a consequence of the formation of a disulfide bridge between C101 from different monomers, preventing metal binding.
Characterization of intramolecular disulfide bridges in wild-type FurA monomers
The behavior shown by FurA and its C101S mutant led us to consider that residues C104, C133, C141, and C144 in recombinant wild-type FurA are involved in intramolecular disulfide bonds.
Analyzing the position of these residues in the primary sequence of FurA contributed to shed some light on the possible match. Taking into account that the formation of a disulfide bond between the cysteines of a CXXC motif is strongly favored when this sequence is within an N-terminal helical position (26), we used the PSIPRED secondary structure prediction server to assess whether the CXXC sequences in FurA were helical (25, 29). According to this method, C141 and C144 are part of a CXXC motif (C141PKC144) predicted to be helical, and therefore, both residues are likely to form a disulfide bridge (Supplementary Fig. S5).
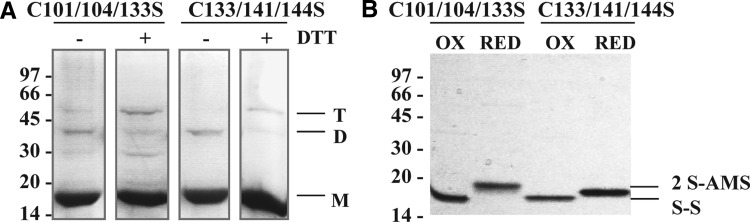
Several evidences contributed to support the existence of an intramolecular disulfide bond between C141 and C144. First of all, electrophoretic analyses in the presence or absence of DTT were carried out to evaluate the contribution of disulfide bridges to the oligomerization of the triple C101/104/133S mutant. Results shown in Figure 5A indicated that, in the absence of DTT, monomers were the predominant form in this mutant. Moreover, AMS trapping experiments shown in Figure 5B indicated that the remaining cysteines (C141 and C144) were disulfide linked when this mutant was oxidized. Furthermore, the motifs C142KEC145 in Helicobacter pylori Fur or C105KNC108 in the Campylobacter jejuni homologue, which are equivalent to Anabaena FurA C141PKC144 motif, are also localized in a α-helix according to the crystal structures of both regulators (6, 9). However, in H. pylori or C. jejuni Fur, these cysteines are reduced since they coordinate the structural metal. Finally, Anabaena FurA C141PKC144 sequence is similar to the CXXC motif of E. coli DsbA (C30PHC33), where these cysteines remain naturally oxidized forming a disulfide bridge (44, 52).
FIG. 5.
Disulfide bonds in FurA triple cysteine mutants. (A) Nonreducing SDS-PAGE and Coomassie blue staining of C101/104/133S and C133/141/144S FurA variants with (+) or without (−) 10 mM DTT treatment. The presence of monomers (M), dimers (D), and trimers (T) is indicated. (B) Free thiol alkylation experiments with AMS of oxidized (OX) or reduced (RED) FurA triple cysteine mutants resolved on a nonreducing 12.5% SDS-PAGE gel and stained with Coomassie. The bands named S-S correspond to the presence of an intramolecular disulfide bond in both oxidized proteins, whereas the bands named 2 S-AMS indicate the presence of two reduced thiols alkylated with two AMS molecules (shift size 1080 Da).
Accordingly, if C141 and C144 are intramolecularly disulfide bonded and C101 is mainly involved in the dimerization of FurA, C104 and C133 would be responsible for the second intramolecular disulfide bridge observed in the AMS trapping experiments with FurA. To gather experimental evidence on the existence of this intramolecular disulfide bond, thiol trapping experiments were performed with a triple C101/141/144S mutant containing C104 and C133.
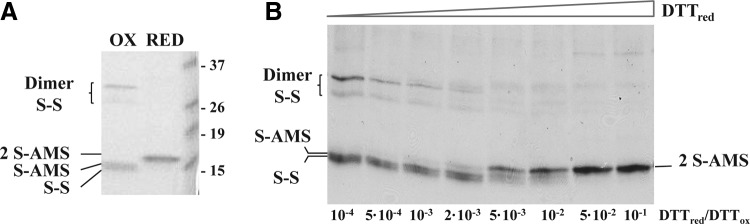
The thiol content in this mutant was monitored with AMS using different reduced DTT/oxidized DTT, trans-4,5-Dihydroxy-1,2-dithiane (DTTred/DTTox) ratios. As observed in Figure 6, only one band was observed under the most reducing conditions (2S-AMS) that corresponded to the addition of two AMS molecules. This result indicated that the protein was completely reduced. However, several bands were observed as the DTTred/DTTox ratio diminished. These bands corresponded to different species namely oxidized protein (S-S), protein with one AMS molecule (S-AMS), and different intermolecularly disulfide-bonded dimer combinations (Dimers S-S) that disappeared along the titration. This experiment clearly demonstrated that, in the triple C101/C141/C144S mutant, C104 and C133 can form an intramolecular disulfide bond in vitro and that intramolecularly oxidized cysteines coexist with reduced and intermolecularly oxidized cysteines.
FIG. 6.
C104 and C133 intramolecular disulfide bond. (A) Fully oxidized (OX) or reduced (RED) FurA triple mutant C101/141/144S was obtained as indicated in the Materials and Methods section and subjected to AMS treatment to be used as controls. (B). Oxidized and reduced fractions of the FurA triple mutant C101/141/144S were obtained by incubation with variable DTTred/DTTox ratios and subjected to thiol trapping treatment with AMS. Proteins were resolved by nonreducing SDS-PAGE and Coomassie stained. The oxidized protein contained an intramolecular bond (band S-S), although some molecules exhibit a free thiol that reacted with one AMS (band S-AMS) or was intermolecularly disulfide bridged in dimers (band Dimer S-S). The reduced protein reacted with 2 AMS molecules (band 2 S-AMS).
The mutational study presented here suggests that recombinant dimeric FurA contains an intermolecular disulfide bridge C101-C101 and two intramolecular bonds C104-C133 and C141-C144 in vitro. Interestingly, previous data obtained in our laboratory indicated the occurrence of a redox-active disulfide bond between C101 and C104 (5). Moreover, nonreducing SDS-PAGE and AMS trapping experiments using the triple C133/141/144S mutant retaining these cysteines confirmed the presence of an intramolecular bridge C101-C104 in the protein (Fig. 5A, B). All together, we conclude that cysteines C101, C104, and C133 of FurA can form disulfide bonds in different combinations, namely C101-C104 or C104-C133.
The redox status of C101 is modulated by C104
Mutations of any of these cysteines probably intramolecularly bridged in wild-type FurA (C104-C133 or C101-C104) would release its partner to form a different disulfide bond, either intra- or intermolecularly.
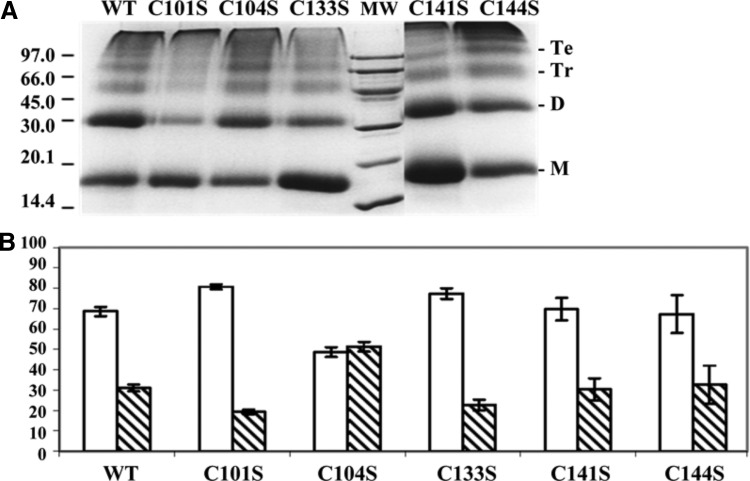
This possibility prompted us to analyze the behavior of FurA and its single cysteine mutants by cross-linking experiments. Results shown in Figure 7 indicate that mutant C104S has a high tendency to dimerize, suggesting that residues C133 and C101 are not prone to forming an intramolecular disulfide bridge. Proof of this is that the C104S variant requires only metal to reach maximal DNA binding activity (Fig. 1), and therefore, C101 is reduced in this mutant. These results suggest that dimers observed in the C104S mutant, unlike in wild-type FurA, would involve residues C133 from two monomers and C101 would be free to interact with metal and hence with DNA. On the contrary, mutant C133S shows a low tendency to dimerize (Fig. 7), suggesting that C101 can be disulfide bridged to C104 in this protein. As expected, C133S needed both metal and reducing conditions to display maximal DNA binding activity (Fig. 1), suggesting that C101 was actually intramolecularly disulfide bonded in this mutant and not available to directly coordinate the metal corepressor.
FIG. 7.
Oligomerization of FurA and its single cysteine mutants. (A) Proteins were dark incubated with 0.5% glutaraldehyde, resolved on a 17% reducing SDS-PAGE gel, and stained with Coomassie blue. Monomers (M), dimers (D), trimers (Tr), and tetramers (Te), as well as the MW, are indicated. (B) The ratios of monomers (white bars) and dimers (striped bars) exhibited by wild-type FurA and the cysteine mutants are represented. Band intensities from the Coomassie-stained gels were quantified by measuring the density of the bands on Gel Doc 2000 (Bio-Rad) and analyzed by Multi-Analyst software (Bio-Rad). Values in bar graphs represent means ± standard deviations for three separate experiments performed.
Previous results presented along with this article have shown that mutant C101S contained a lower amount of dimeric species than the rest of single cysteine mutants (Fig. 4 and 7), suggesting that C104 and C133 tend to form an intramolecular bond in vitro. This mutant is unable to bind DNA, since it lacks C101 to coordinate the metal corepressor (Fig. 1).
In summary, the above results indicate that the redox state of C101 is affected by the presence or absence of C104. When C104 is present in a mutant lacking C133 (C133S), C101 is oxidized, but C101 is reduced in a mutant lacking C104 (C104S), although C133 is present. Taking into account that C104 can form intramolecular disulfide bonds with either C101 or C133, a mechanism of Anabaena FurA activation–deactivation based on an intramolecular disulfide interchange can be envisaged. When C101 is bound to C104, FurA is inactive because it is unable to bind the metal corepressor and hence DNA. If C133 forms a disulfide bridge with C104, then C101 is released from the C101-C104 disulfide bond, and consequently, FurA is activated.
FurA thiol/disulfide redox state responds to cellular redox alterations
To determine whether thiol/disulfide couples in FurA respond to changes in intracellular redox conditions, AMS trapping experiments were used to analyze the redox states of FurA cysteines in Anabaena PCC 7120 cultures at different growth phases or subjected to redox stress.
Under early exponential growth conditions, FurA was mainly reduced. Stationary growth conditions, which typically involve bacterial protein oxidation as a consequence of nutritional or oxidative stresses (10), entailed the conversion of most of the cysteines to the disulfide state (Supplementary Fig. S6).
When cells in early exponential phase were subjected to oxidative stress generated by the addition of 0.5 mM H2O2, the proportion of reduced cysteines decreased. Estimates by densitometry indicated that around 60% of the fully reduced protein converted to forms bearing three or only one reduced cysteines (Supplementary Fig. S6). Thus, FurA thiol/disulfide state responds to changes in Anabaena sp. PCC 7120 intracellular redox conditions.
Discussion
The lack of structural Zn2+ in Anabaena FurA provides its cysteines with a great reactivity, not detected in other family members (5). Cysteines in FurA play a key role in the different functions, including the interactions with its metal corepressor and DNA. In this study, the status of FurA cysteines, as well as their contribution to the activity of this global regulator, has been investigated.
Our data suggest that four out of five cysteines in recombinant FurA are engaged in the formation of two intramolecular disulfide bridges, C104-C133 and C141-C144. The remainder cysteine (C101) easily oxidizes in the air, leading to the formation of an intermolecular disulfide bond (C101-C101) that appears to be responsible for most dimers observed in recombinant FurA. This behavior is comparable to that shown by P. aeruginosa Fur, where C92, the only cysteine in the protein, forms intermolecular disulfide bonds on air oxidation (32). Interestingly, this residue aligns with C101 in Anabaena FurA (Fig. 8). However, the formation of (C101-C101) dimers in Anabaena FurA leads to the loss of DNA binding ability in vitro, unlike Pseudomonas Fur where the presence of dimers is batch dependent and does not affect DNA binding (32).
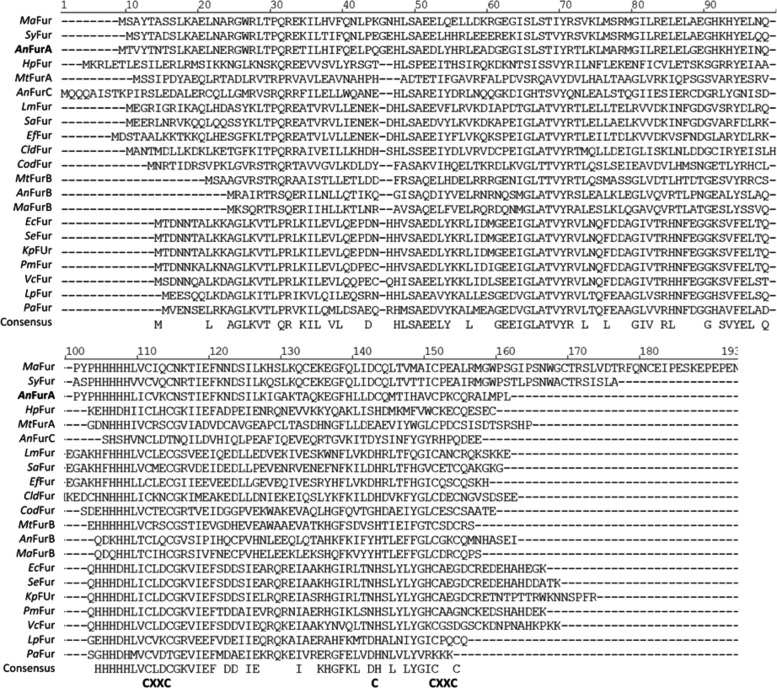
FIG. 8.
Multiple sequence alignment of Fur proteins present in different bacteria. The abbreviations correspond to the following microorganisms: Ma: Microcystis aeruginosa PCC 7806, Sy: Synechocystis PCC 6803, An: Anabaena PCC 7120, Hp: Helicobacter pylori 26695, Mt: Mycobacterium tuberculosis H37Rv, Lm: Listeria monocytogenes EGDe, Sa: Staphylococcus aureus ATCC BAA-39, Ef: Enterococcus faecalis ATCC 29212, Cld: Clostridium difficile ATCC 43255, Cod: Corynebacterium diphtheriae ATCC 13812, Ec: Escherichia coli BL21, Se: Salmonella enterica Typhimurium LT2, Kp: Klebsiella pneumoniae ATCC 43816, Pm: Proteus mirabilis HI 4320, Vc: Vibrio cholerae CECT512, Lp: Legionella pneumophila Philadelphia-1, Pa: Pseudomonas aeruginosa ATCC 27853.
The behavior of recombinant wild-type FurA in vitro is consistent with our in vivo results showing that this protein is mainly a monomer with a single free cysteine in the cytoplasm of Anabaena sp. PCC 7120 cultures at the stationary phase, suggesting that two intramolecular disulfide bridges are also formed in vivo (5).
The FurA C101 residue is highly conserved in Fur homologues and it belongs to a CXXC (C101VKC104) motif present in a large number of Fur proteins as well (Fig. 8). However, the importance of this cysteine varies among different homologues. For instance, it is essential for the functionality of E. coli and H. pylori Fur proteins (9, 27, 39), while P. putida Fur lacks cysteines (32). Our results indicate that C101 and its particular redox state play an essential role in the coordination of the metal corepressor, which ultimately controls FurA-DNA binding activity in vitro. In fact, ITC experiments show that reduced recombinant FurA contains two metal binding sites of different affinities. The high affinity site (Kd 50 nM) disappears when recombinant wild-type FurA is oxidized. The C101S variant also loses the high affinity metal binding site under reducing conditions, suggesting an essential role for C101 in metal coordination, conditional on its redox state. Parallel EMSA experiments show that, when C101 is missing, FurA does not interact with DNA even in the presence of metal under reducing conditions, thus connecting FurA-DNA binding and its ability to interact with metal through C101.
According to our results, oxidized Anabaena FurA has a unique Mn2+ binding site with a Kd value of 0.28 μM, assumed to correspond to the low affinity metal binding site in reduced FurA (0.84 μM). This second metal binding site could be equivalent to the third metal binding site that, aside from the regulatory and structural ones, has been detected in the crystal structures of H. pylori Fur or B. subtilis Zur, where it is proposed to stabilize protein dimers (9, 35).
Many Fur proteins also contain a second pair of cysteines close to the C-terminus of its primary sequence, in some cases as part of a second CXXC motif. The existence of two CXXC motifs in the primary sequence of Fur has been related to the existence of zinc (6, 9, 12, 28, 48). In Anabaena FurA, as occurs in Streptomyces coelicolor Nur, both CXXC motifs are present but they do not coordinate a structural metal (20). Regarding FurA, as previously mentioned, in vivo results show that two intramolecular disulfide bridges are formed (5). It indicates that the presence of intramolecular disulfide bridges is important for the protein activity in vivo.
According to our mutational study in vitro, different intramolecular disulfide bonds can be found in Anabaena FurA, involving cysteines C101, C104, and C133. On one hand, residues C101 and C104 belonging to the C101VKC104 motif can be disulfide bridged in a mutant lacking C133 residue. On the other hand, C133 seems to be disulfide bonded to C104 in C101S, while C101 appears not to be disulfide bridged to residue C133 in C104S.
Taking into account a recent definition of redox switch (45), it is not unreasonable to speculate about a mechanism of FurA functioning based on a thiol–disulfide redox switch, involving these cysteines and controlling the redox state of C101. In this way, the oxidation of the C101VKC104 motif would result in the loss of FurA metal binding ability, causing a reduction of DNA binding affinity with consequent derepression of regulated genes. It does not exclude the existence of a second thiol–disulfide switch functioning either independently or in coordination with the previous one, affecting the cysteines of the second C141PKC144 motif. These cysteines form a redox-sensitive disulfide bridge in a triple C101/104/133S mutant containing C141 and C144 (5). C141 is also involved in Anabaena FurA–heme coordination (41).
In this regard, a recent study has described a thiol–disulfide redox switch that controls the interaction between heme and the ligand binding domain of Rev-erbb (18). Moreover, an NMR study of different single and double mutants of the heme oxygenase HO-2 involved in signaling and regulatory processes demonstrates that, analogous to our observations, the redox state of a given cysteine out of the three cysteines in HO-2 is sensitive to the absence of one or both cysteines (49).
The finding that different disulfide-bonded forms of FurA exist in vivo, generally with two and sometimes one or even no intramolecular disulfide bridges, whose ratio depends on the redox state of the cytoplasm of the cyanobacterium (Supplementary Fig. S6) (5), supports the idea that disulfide shuffling in FurA is important to control its function. In this sense, according to our results, an early exponential phase Anabaena culture contains FurA mainly in the reduced state. Exposure of this culture to oxidative stress causes the oxidation of FurA cysteines in vivo and presumably its inactivation. This is confirmed by different studies that show that transcription of FurA-repressed target genes, namely isiA and the flavodoxin-encoding gene isiB is induced under oxidative stress conditions in different cyanobacterial strains (19, 51). In particular, Yousef et al. show that transcription of major iron-regulated genes, such as isiA, isiB, and irpA among others, is induced by oxidative stress within a few minutes by treatment of cells with hydrogen peroxide or methyl viologen (51). Thus, it appears that the cysteines of FurA form a physiological relevant redox rheostat that responds to the intracellular redox potential.
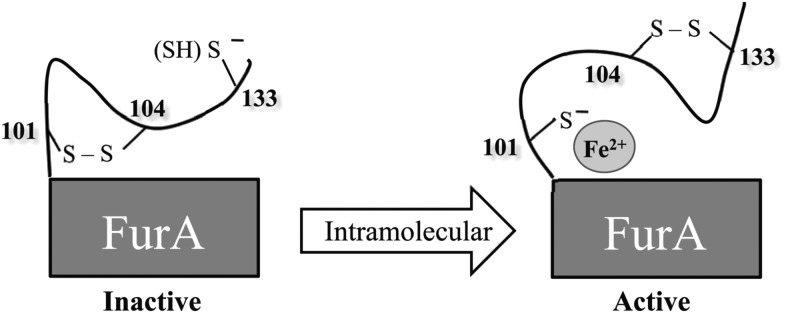
The FurA thiol-based switch mechanism that we propose is new for this kind of metalloregulators and likely specific for cyanobacterial Fur, since it relies on the presence of C133, only conserved in Fur homologues from cyanobacteria (Fig. 8). C133 would be responsible for maintaining C104 in the oxidized state, avoiding the formation of a C104-C101 disulfide bond that would lead to the inactivation of FurA. The reverse process would be necessary to activate FurA again (Fig. 9). The mechanism also explains the lack of structural zinc in this regulator, since C101 and C104 are part of the redox switch and cannot be found simultaneously reduced to coordinate zinc.
FIG. 9.
Proposed mechanism of the redox-mediated FurA activity. When the protein is transcriptionally active, C133 and C104 form an intramolecular disulfide bond and C101 can coordinate the metal corepressor. Upon an intracellular redox status, a thiol–disulfide interchange would generate a new intramolecular disulfide bond between C101 and C104. This oxidation event would simultaneously occur with metal release and FurA inactivation. The reverse process would be necessary to activate FurA again.
This behavior described for FurA is comparable to that shown by transglutaminase 2 (TG2), a protein tightly regulated through three identified allosteric modifiers: Ca2+ ions, guanine nucleotides, and an allosteric disulfide bond whose thiol/disulfide exchange is proposed to control the conversion of the inactive TG2 to the active form (7).
It would be expected that a redox stimulus would account for a disulfide bond exchange in Anabaena FurA that would trigger some sort of change, allowing the protein to bind or release the metal corepressor and in turn DNA. Cleavage and formation of the corresponding disulfide bridges should be reversible so that the perturbation of the balance between reduced and disulfide-bonded forms could be the key of FurA regulation. The way by which the expected disulfide bridge between C104 and C133 would reform to activate FurA is an important issue. The presence of CXXC motifs in different redox states in FurA suggests that this regulator could be a redox partner of thioredoxin. However, we did not retrieve any interaction between FurA and thioredoxin or another partner entailing thiol–disulfide exchange using pull-down experiments complemented with bacterial two hybrid assays (5). Instead, we detected some interacting partners of FurA, namely the hypothetical protein All1140 that contains an Ami_3 domain related to N-acetylmuramoyl-L-alanine amidase activity, the histone-like DNA binding protein HU, and the phosphoribulokinase enzyme. Up to now, no redox partners of FurA have been identified, but work is currently underway to determine the implication of different photosynthetic electron carriers in the disulfide bond exchange in this cyanobacterial regulator.
Materials and Methods
Protein preparation
The furA wild-type gene (all1691) from Anabaena sp. PCC 7120 was cloned into pET28a (Novagen: Merck, Darmstadt, Germany), as previously described (4). Several pET28a-furA variants containing cysteine-to-serine substitutions were generated by site-directed mutagenesis or purchased from Mutagenex, Inc. (Piscataway, NJ). DNA sequences were confirmed by DNA sequencing. All the proteins were overexpressed and purified using the high-recovery one-step purification method previously described (40). Proteins were spectrophotometrically quantified using the experimental molar extinction coefficient at 276 nm 13,760 M−1 cm−1 (20).
Spectroscopic analyses
Ultraviolet–visible (UV-vis) absorption spectra were recorded on a Cary 100 UV-Vis double beam spectrophotometer (Agilent Technologies, Santa Clara, CA).
CD spectra were collected on a Jasco J815 spectropolarimeter (Jasco International Co. LTD., Tokyo, Japan) fitted with a thermostated cell holder and interfaced with a Peltier unit. Spectra were acquired at a scan speed of 50 nm/min with a response time of 2 s and averaged over four scans at 25°C. Far-UV measurements were performed using 10 μM of FurA proteins in 10 mM sodium acetate buffer pH 4 in 0.1 cm pathlength quartz cells (Hellma, Forest Hills, NY). All spectra were corrected by subtracting the proper baseline. The molar ellipticity ([θ]) was expressed in deg·cm2·dmol−1.
For FT-IR studies, the proteins (250 μg) were lyophilized and dissolved in 30 μL of deuterated water. No pH corrections were applied for isotope effects. Protein samples were placed between a pair of CaF2 windows separated by a 50-mm-thick spacer, in a Harrick demountable cell at 25°C. Spectra were acquired on a Bruker FT-IR spectrophotometer (Brucker Optics, Inc., Billerica, MA), equipped with a DTGS detector, and thermostated with a Braun water bath. The cell container was filled with dry air. Five hundred scans per sample were taken, averaged, apodized with a Happ–Genzel function, and Fourier transformed to give a final resolution of 2 cm−1. The signal-to-noise ratio of the spectra was better than 10.000:1. Baselines were subtracted, and contributions of side chains removed as described (16, 17). Only in those cases where there were no distortions in the spectra, the resulting difference spectra were used for analysis. To determine the amount of secondary structure components, the Amide I band in D2O was decomposed into its constituents by curve fitting, using a line shape that was a combination of Gaussian and Lorentzian functions (43).
The NMR experiments were acquired on a Bruker Avance DRX-500 spectrometer. Samples of FurA proteins (0.3–1.4 mM) were prepared in a 10 mM deuterated sodium acetate buffer pH 4 in 5-mm NMR tubes at 25°C. Spectra were acquired using 16 K data points, averaged over 512 scans and with a spectral width of 7801.69 Hz (13 ppm).
Isothermal titration calorimetry
Metal binding to FurA proteins was assayed in a high-sensitivity isothermal titration VP-ITC microcalorimeter (MicroCal, Northampton, MA). Protein samples and reference solutions were properly degassed and carefully loaded into the cells to avoid bubble formation during stirring. Experiments were performed with 20 μM freshly prepared FurA protein solutions in 50 mM Tris buffer and 50 mM arginine pH 9 at 25°C. Samples were titrated with 300 μM MnCl2 in the calorimetric cell. Control experiments were performed under the same experimental conditions. The heat due to the binding reaction was obtained as the difference between the reaction heat and the corresponding heat of dilution, the latter estimated as an adjustable parameter in the analysis. The association constant and the enthalpy change were obtained through nonlinear regression of experimental data to a model for either a single ligand binding site or two independent ligand binding sites.
Electrophoretic mobility shift assays
Gel retardation assays were performed as previously described (22) using the promoter region of furA (PfurA) as target DNA and the promoter of alr0523 (Palr0523) as nonspecific DNA. Both promoters were obtained by PCR amplification using oligonucleotides 5′-CTCGCCTAGCAATTTAACAAC-3′ and 5′-GCCTTGAGCGAAGTATTTGTG-3′ for PfurA and 5′-GTCTGTATGGATTAACACTATC-3′ and 5′-CTGCCTGTTCAACTACTTTGG-3′ for Palr0523 and then purified by the GFX PCR DNA and Gel Band Purification Kit (GE Healthcare, Buckinghamshire, United Kingdom). A 300 nM protein solution was incubated with 100 ng for each target and nonspecific DNA regions in the reaction buffer containing 10 mM Bis-Tris pH 7.5, 40 mM KCl, 5% glycerol, and 0.05 mg/mL bovine serum albumin for 20 min before being loaded on native 6% PAGE gels. Depending on the tested conditions, 1 mM DTT and/or 100 μM MnCl2 were added to the reaction mixtures.
AMS trapping of reduced thiols
Proteins (25 μM) were incubated for 2 h at 30°C in a redox buffer containing 100 mM trans-4,5-dihydroxy-1,2,dithane (oxidized DTT, DTTox; Sigma-Aldrich, St. Louis, MO) and 1 μM or 20 mM (DTTred; Sigma-Aldrich) to obtain oxidized or reduced FurA proteins, respectively. Reactions were stopped by adding 5% (v/v) trichloroacetic acid (TCA; Sigma-Aldrich), and proteins were precipitated by centrifugation at 13,300 g for 10 min at 4°C. After washing with cold acetone, air-dried protein pellets were resuspended in 10 μL of thiol modification buffer containing 50 mM Tris/HCl pH 7.5, 1% (w/v) SDS (Sigma-Aldrich), and 20 mM AMS (Molecular Probes; Life Technologies Ltd., Paisley, United Kingdom). Proteins in nonreducing Laemmli buffer were resolved on 12.5% (w/v) polyacrylamide [30:0.8 (w/w) acrylamide/bisacrylamide] SDS-PAGE gels and stained with Coomassie Blue R-250 (Sigma-Aldrich) (24).
For in vivo trapping experiments, 10 mL of exponentially growing Anabaena cultures was treated with 0.5 mM hydrogen peroxide for 30 min. A culture without treatment was used as control. Both cultures were then incubated with TCA to a final concentration of 1% (v/v) for 30 min. Proteins were precipitated by centrifugation, washed with cold acetone, and resuspended in 25 μL of thiol modification buffer. After 1 h of treatment, samples were subjected to 12.5% nonreducing SDS-PAGE and revealed by Western blotting with rabbit polyclonal antibodies raised against FurA.
Analysis of the oligomeric state of FurA proteins in vitro
Oligomeric species were analyzed using glutaraldehyde as a chemical crosslinker. Proteins (40 μM) were incubated in the dark with 0.5% (v/v) glutaraldehyde (Sigma-Aldrich) for 30 min, and reactions stopped by adding Laemmli buffer and boiling. Samples were finally subjected to 17% SDS-PAGE and stained with Coomassie Blue R-250. The effect of the reducing agent was assessed by incubating 40 μM protein solutions with 10 mM DTT for 30 min. Nonreducing Laemmli buffer was added to the samples before boiling. Proteins were separated by 17% SDS-PAGE followed by Coomassie Blue R-250 staining.
Supplementary Material
Abbreviations Used
- [θ]
molar ellipticity
- AMS
4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid
- CD
circular dichroism
- D
dimers
- DTT
dithiothreitol
- DTTox
oxidized DTT, trans-4,5-Dihydroxy-1,2-dithiane
- DTTred
reduced DTT
- EMSA
electrophoretic mobility shift assay
- FT-IR
Fourier transform infrared spectroscopy
- Fur
ferric uptake regulator
- ITC
isothermal titration calorimetry
- Kd
dissociation constant
- M
monomers
- MW
molecular weight markers
- NBD-Cl
7-chloro-4-nitrobenz-2-oxa-1,3-diazole
- NMR
nuclear magnetic resonance
- OX
oxidized protein(s)
- PAGE
polyacrylamide gel electrophoresis
- Palr0523
promoter region of the alr0523 gene
- PfurA
promoter region of the furA gene
- RED
reduced protein(s)
- SDS
sodium dodecyl sulfate
- TCA
trichloroacetic acid
- Te
tetramers
- TG2
transglutaminase 2
- Tr
trimers
- UV-vis
ultraviolet–visible region
Acknowledgments
This work was supported by grants CTQ2011-24393 and CSD2008-0005 from the Spanish Ministry of Science and Innovation and Prometeo 2013/18 from Generalitat Valenciana to J.L.N., Spanish Ministry of Economy and Competitiveness BFU2012-31458 to M.F.F., Spanish Ministry of Science and Innovation BFU2010-19451 to A.V-C., Miguel Servet Program from Instituto de Salud Carlos III (CP07/00289 to O.A.), Fondo de Investigaciones Sanitarias (PI10/00186 to O.A.), Diputación General de Aragón—Spain (PI044/09 and Protein Targets Group B89 to A.V-C., Digestive Pathology Group B01 to O.A., and Structural Biology Group B18 to M.L.P.). L.B-M. was recipient of a FPU predoctoral fellowship from the Spanish Ministry of Education. V. C. S-E. was recipient of a predoctoral fellowship from the Government of Aragon.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.An YJ, Ahn BE, Han AR, Kim HM, Chung KM, Shin JH, Cho YB, Roe JH, and Cha SS. Structural basis for the specialization of Nur, a nickel-specific Fur homolog, in metal sensing and DNA recognition. Nucleic Acids Res 37: 3442–3451, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arakawa T, Ejima D, Tsumoto K, Obeyama N, Tanaka Y, Kita Y, and Timasheff SN. Suppression of protein interactions by arginine: a proposed mechanism of the arginine effects. Biophys Chem 127: 1–8, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Bagg A. and Neilands JB. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26: 5471–5477, 1987 [DOI] [PubMed] [Google Scholar]
- 4.Bes MT, Hernandez JA, Peleato ML, and Fillat MF. Cloning, overexpression and interaction of recombinant Fur from the cyanobacterium Anabaena PCC 7119 with isiB and its own promoter. FEMS Microbiol Lett 194: 187–192, 2001 [DOI] [PubMed] [Google Scholar]
- 5.Botello-Morte L, Bes MT, Heras B, Fernandez-Otal A, Peleato ML, and Fillat MF. Unraveling the redox properties of the global regulator FurA from Anabaena sp. PCC 7120: disulfide reductase activity based on its CXXC motifs. Antioxid Redox Signal 20: 1396–1406, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butcher J, Sarvan S, Brunzelle JS, Couture JF, and Stintzi A. Structure and regulon of Campylobacter jejuni ferric uptake regulator Fur define apo-Fur regulation. Proc Natl Acad Sci U S A 109: 10047–10052, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook KM. and Hogg PJ. Post-translational control of protein function by disulfide bond cleavage. Antioxid Redox Signal 18: 1987–2015, 2013 [DOI] [PubMed] [Google Scholar]
- 8.Denoncin K, Nicolaes V, Cho SH, Leverrier P, and Collet JF. Protein disulfide bond formation in the periplasm: determination of the in vivo redox state of cysteine residues. Methods Mol Biol 966: 325–336, 2013 [DOI] [PubMed] [Google Scholar]
- 9.Dian C, Vitale S, Leonard GA, Bahlawane C, Fauquant C, Leduc D, Muller C, de Reuse H, Michaud-Soret I, and Terradot L. The structure of the Helicobacter pylori ferric uptake regulator Fur reveals three functional metal binding sites. Mol Microbiol 79: 1260–1275, 2011 [DOI] [PubMed] [Google Scholar]
- 10.Dukan S. and Nystrom T. Bacterial senescence: stasis results in increased and differential oxidation of cytoplasmic proteins leading to developmental induction of the heat shock regulon. Genes Dev 12: 3431–3441, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Escolar L, Perez-Martin J, and de Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol 181: 6223–6229, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiorini F, Stefanini S, Valenti P, Chiancone E, and De Biase D. Transcription of the Listeria monocytogenes fri gene is growth-phase dependent and is repressed directly by Fur, the ferric uptake regulator. Gene 410: 113–121, 2008 [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez A, Angarica VE, Sancho J, and Fillat MF. The FurA regulon in Anabaena sp. PCC 7120: in silico prediction and experimental validation of novel target genes. Nucleic Acids Res 42: 4833–4846, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez A, Bes MT, Barja F, Peleato ML, and Fillat MF. Overexpression of FurA in Anabaena sp. PCC 7120 reveals new targets for this regulator involved in photosynthesis, iron uptake and cellular morphology. Plant Cell Physiol 51: 1900–1914, 2010 [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez A, Bes MT, Valladares A, Peleato ML, and Fillat MF. FurA is the master regulator of iron homeostasis and modulates the expression of tetrapyrrole biosynthesis genes in Anabaena sp. PCC 7120. Environ Microbiol 14: 3175–3187, 2012 [DOI] [PubMed] [Google Scholar]
- 16.Goormaghtigh E, Raussens V, and Ruysschaert JM. Attenuated total reflection infrared spectroscopy of proteins and lipids in biological membranes. Biochim Biophys Acta 1422: 105–185, 1999 [DOI] [PubMed] [Google Scholar]
- 17.Guldenhaupt J, Adiguzel Y, Kuhlmann J, Waldmann H, Kotting C, and Gerwert K. Secondary structure of lipidated Ras bound to a lipid bilayer. FEBS J 275: 5910–5918, 2008 [DOI] [PubMed] [Google Scholar]
- 18.Gupta N. and Ragsdale SW. Thiol-disulfide redox dependence of heme binding and heme ligand switching in nuclear hormone receptor rev-erb{beta}. J Biol Chem 286: 4392–4403, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Havaux M, Guedeney G, Hagemann M, Yeremenko N, Matthijs HC, and Jeanjean R. The chlorophyll-binding protein IsiA is inducible by high light and protects the cyanobacterium Synechocystis PCC6803 from photooxidative stress. FEBS Lett 579: 2289–2293, 2005 [DOI] [PubMed] [Google Scholar]
- 20.Hernandez JA, Bes MT, Fillat MF, Neira JL, and Peleato ML. Biochemical analysis of the recombinant Fur (ferric uptake regulator) protein from Anabaena PCC 7119: factors affecting its oligomerization state. Biochem J 366: 315–322, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernandez JA, Lopez-Gomollon S, Bes MT, Fillat MF, and Peleato ML. Three fur homologues from Anabaena sp. PCC7120: exploring reciprocal protein-promoter recognition. FEMS Microbiol Lett 236: 275–282, 2004 [DOI] [PubMed] [Google Scholar]
- 22.Hernandez JA, Lopez-Gomollon S, Muro-Pastor A, Valladares A, Bes MT, Peleato ML, and Fillat MF. Interaction of FurA from Anabaena sp. PCC 7120 with DNA: a reducing environment and the presence of Mn(2+) are positive effectors in the binding to isiB and furA promoters. Biometals 19: 259–268, 2006 [DOI] [PubMed] [Google Scholar]
- 23.Hernandez JA, Peleato ML, Fillat MF, and Bes MT. Heme binds to and inhibits the DNA-binding activity of the global regulator FurA from Anabaena sp. PCC 7120. FEBS Lett 577: 35–41, 2004 [DOI] [PubMed] [Google Scholar]
- 24.Inaba K, Takahashi YH, and Ito K. Reactivities of quinone-free DsbB from Escherichia coli. J Biol Chem 280: 33035–33044, 2005 [DOI] [PubMed] [Google Scholar]
- 25.Indu S, Kumar ST, Thakurela S, Gupta M, Bhaskara RM, Ramakrishnan C, and Varadarajan R. Disulfide conformation and design at helix N-termini. Proteins 78: 1228–1242, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iqbalsyah TM, Moutevelis E, Warwicker J, Errington N, and Doig AJ. The CXXC motif at the N terminus of an alpha-helical peptide. Protein Sci 15: 1945–1950, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacquamet L, Aberdam D, Adrait A, Hazemann JL, Latour JM, and Michaud-Soret I. X-ray absorption spectroscopy of a new zinc site in the fur protein from Escherichia coli. Biochemistry 37: 2564–2571, 1998 [DOI] [PubMed] [Google Scholar]
- 28.Jacquamet L, Traore DA, Ferrer JL, Proux O, Testemale D, Hazemann JL, Nazarenko E, El Ghazouani A, Caux-Thang C, Duarte V, and Latour JM. Structural characterization of the active form of PerR: insights into the metal-induced activation of PerR and Fur proteins for DNA binding. Mol Microbiol 73: 20–31, 2009 [DOI] [PubMed] [Google Scholar]
- 29.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292: 195–202, 1999 [DOI] [PubMed] [Google Scholar]
- 30.Katigbak J. and Zhang Y. Iron binding site in a global regulator in bacteria—ferric uptake regulator (Fur) protein: structure, mossbauer properties, and functional implication. J Phys Chem Lett 2012: 3503–3508, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelly SM. and Price NC. The use of circular dichroism in the investigation of protein structure and function. Curr Protein Pept Sci 1: 349–384, 2000 [DOI] [PubMed] [Google Scholar]
- 32.Lewin AC, Doughty PA, Flegg L, Moore GR, and Spiro S. The ferric uptake regulator of Pseudomonas aeruginosa has no essential cysteine residues and does not contain a structural zinc ion. Microbiology 148: 2449–2456, 2002 [DOI] [PubMed] [Google Scholar]
- 33.Lostao A, Peleato ML, Gomez-Moreno C, and Fillat MF. Oligomerization properties of FurA from the cyanobacterium Anabaena sp. PCC 7120: direct visualization by in situ atomic force microscopy under different redox conditions. Biochim Biophys Acta 1804: 1723–1729, 2010 [DOI] [PubMed] [Google Scholar]
- 34.Lucarelli D, Russo S, Garman E, Milano A, Meyer-Klaucke W, and Pohl E. Crystal structure and function of the zinc uptake regulator FurB from Mycobacterium tuberculosis. J Biol Chem 282: 9914–9922, 2007 [DOI] [PubMed] [Google Scholar]
- 35.Ma Z, Gabriel SE, and Helmann JD. Sequential binding and sensing of Zn(II) by Bacillus subtilis Zur. Nucleic Acids Res 39: 9130–9138, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miles S, Carpenter BM, Gancz H, and Merrell DS. Helicobacter pylori apo-Fur regulation appears unconserved across species. J Microbiol 48: 378–386, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orsini G. and Goldberg ME. The renaturation of reduced chymotrypsinogen A in guanidine HCl. Refolding versus aggregation. J Biol Chem 253: 3453–3458, 1978 [PubMed] [Google Scholar]
- 38.Pan J. and Carroll KS. Chemical biology approaches to study protein cysteine sulfenylation. Biopolymers 101: 165–172, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pecqueur L, D'Autreaux B, Dupuy J, Nicolet Y, Jacquamet L, Brutscher B, Michaud-Soret I, and Bersch B. Structural changes of Escherichia coli ferric uptake regulator during metal-dependent dimerization and activation explored by NMR and X-ray crystallography. J Biol Chem 281: 21286–21295, 2006 [DOI] [PubMed] [Google Scholar]
- 40.Pellicer S, Bes MT, Gonzalez A, Neira JL, Peleato ML, and Fillat MF. High-recovery one-step purification of the DNA-binding protein Fur by mild guanidinium chloride treatment. Process Biochem 45: 292–296, 2010 [Google Scholar]
- 41.Pellicer S, Gonzalez A, Peleato ML, Martinez JI, Fillat MF, and Bes MT. Site-directed mutagenesis and spectral studies suggest a putative role of FurA from Anabaena sp. PCC 7120 as a heme sensor protein. FEBS J 279: 2231–2246, 2012 [DOI] [PubMed] [Google Scholar]
- 42.Pohl E, Haller JC, Mijovilovich A, Meyer-Klaucke W, Garman E, and Vasil ML. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol Microbiol 47: 903–915, 2003 [DOI] [PubMed] [Google Scholar]
- 43.Pozo-Dengra J, Martinez-Rodriguez S, Contreras LM, Prieto J, Andujar-Sanchez M, Clemente-Jimenez JM, Las Heras-Vazquez FJ, Rodriguez-Vico F, and Neira JL. Structure and conformational stability of a tetrameric thermostable N-succinylamino acid racemase. Biopolymers 91: 757–772, 2009 [DOI] [PubMed] [Google Scholar]
- 44.Quan S, Schneider I, Pan J, Von Hacht A, and Bardwell JC. The CXXC motif is more than a redox rheostat. J Biol Chem 282: 28823–28833, 2007 [DOI] [PubMed] [Google Scholar]
- 45.Ragsdale SW. and Yi L. Thiol/Disulfide redox switches in the regulation of heme binding to proteins. Antioxid Redox Signal 14: 1039–1047, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rudolph R. and Lilie H. In vitro folding of inclusion body proteins. FASEB J 10: 49–56, 1996 [PubMed] [Google Scholar]
- 47.Sheikh MA. and Taylor GL. Crystal structure of the Vibrio cholerae ferric uptake regulator (Fur) reveals insights into metal co-ordination. Mol Microbiol 72: 1208–1220, 2009 [DOI] [PubMed] [Google Scholar]
- 48.Traore DA, El Ghazouani A, Ilango S, Dupuy J, Jacquamet L, Ferrer JL, Caux-Thang C, Duarte V, and Latour JM. Crystal structure of the apo-PerR-Zn protein from Bacillus subtilis. Mol Microbiol 61: 1211–1219, 2006 [DOI] [PubMed] [Google Scholar]
- 49.Varfaj F, Lampe JN, and Ortiz de Montellano PR. Role of cysteine residues in heme binding to human heme oxygenase-2 elucidated by two-dimensional NMR spectroscopy. J Biol Chem 287: 35181–35191, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woody RW. Circular dichroism. Methods Enzymol 246: 34–71, 1995 [DOI] [PubMed] [Google Scholar]
- 51.Yousef N, Pistorius EK, and Michel KP. Comparative analysis of idiA and isiA transcription under iron starvation and oxidative stress in Synechococcus elongatus PCC 7942 wild-type and selected mutants. Arch Microbiol 180: 471–483, 2003 [DOI] [PubMed] [Google Scholar]
- 52.Zapun A, Bardwell JC, and Creighton TE. The reactive and destabilizing disulfide bond of DsbA, a protein required for protein disulfide bond formation in vivo. Biochemistry 32: 5083–5092, 1993 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.