The short inflorescence stem phenotype of a chloroplast lipid mutant is caused by jasmonic acid overproduction, demonstrating a direct link between the biosynthesis of this lipid and jasmonic acid.
Abstract
DIGALACTOSYLDIACYLGLYCEROL SYNTHASE1 (DGD1) is a chloroplast outer membrane protein responsible for the biosynthesis of the lipid digalactosyldiacylglycerol (DGDG) from monogalactosyldiacylglycerol (MGDG). The Arabidopsis thaliana dgd1 mutants have a greater than 90% reduction in DGDG content, reduced photosynthesis, and altered chloroplast morphology. However, the most pronounced visible phenotype is the extremely short inflorescence stem, but how deficient DGDG biosynthesis causes this phenotype is unclear. We found that, in dgd1 mutants, phloem cap cells were lignified and jasmonic acid (JA)-responsive genes were highly upregulated under normal growth conditions. The coronative insensitive1 dgd1 and allene oxide synthase dgd1 double mutants no longer exhibited the short inflorescence stem and lignification phenotypes but still had the same lipid profile and reduced photosynthesis as dgd1 single mutants. Hormone and lipidomics analyses showed higher levels of JA, JA-isoleucine, 12-oxo-phytodienoic acid, and arabidopsides in dgd1 mutants. Transcript and protein level analyses further suggest that JA biosynthesis in dgd1 is initially activated through the increased expression of genes encoding 13-lipoxygenases (LOXs) and phospholipase A-Iγ3 (At1g51440), a plastid lipase with a high substrate preference for MGDG, and is sustained by further increases in LOX and allene oxide cyclase mRNA and protein levels. Our results demonstrate a link between the biosynthesis of DGDG and JA.
INTRODUCTION
The galactolipids monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG), the major lipids of chloroplast membranes, are important for photosynthesis and are completely absent in most nonphotosynthetic organisms (Härtel et al., 1997; Dörmann and Benning, 2002; Steffen et al., 2005; Joyard et al., 2010; Boudière et al., 2014; Fujii et al., 2014). MGDG is synthesized by the enzyme MGD synthase, which catalyzes the transfer of a galactose from UDP-galactose onto the diacylglycerol (DAG) backbone. The enzyme DGD synthase then transfers a second galactose from UDP-galactose onto MGDG to form DGDG. Although Arabidopsis thaliana is a 16:3 plant and about half of its MGDG molecules have C16 fatty acids at the sn-2 position, the predominant fraction of DGDG is derived from the eukaryotic pathway and has C18:3 fatty acids at both the sn-1 and sn-2 positions (Joyard et al., 2010). In Arabidopsis, DGD synthase is encoded by two genes, DGD1 and DGD2. A mutant with a null mutation in the DGD1 gene, dgd1, has been isolated, and its DGDG content was found to be more than 90% lower than in the wild type (Dörmann et al., 1995), indicating that DGD1 is the major functional isoform. Studies on DGD2 have shown that it is mainly involved in DGDG biosynthesis under phosphate-limiting conditions and is also responsible for synthesizing the residual DGDG in the dgd1 mutant (Kelly and Dörmann, 2002; Kelly et al., 2003).
The dgd1 mutant has several additional phenotypes. It shows a small reduction in total chlorophyll content and a corresponding reduction in photosynthetic quantum yield. It also has an altered chloroplast morphology, with a rounded envelope enclosing bent thylakoid membranes and large thylakoid-free stromal areas; interestingly, the most pronounced phenotype is stunted growth, in particular, extremely short inflorescence stems, short petioles, and ruffled leaves (Dörmann et al., 1995). Complementation of the dgd1 single mutant or the dgd1 dgd2 double mutant with a bacterial glucosyl transferase, thereby replacing DGDG with glucosyl-galactosyl diacylglycerol (GGDG), does not fully rescue the photosynthesis defect, showing that the galactose moiety in DGDG has specific functions in photosynthesis and cannot be replaced by glucose, but the bacterial glucosyl transferase does restore growth and chloroplast morphology (Hölzl et al., 2006, 2009). Therefore, it was suggested that the altered chloroplast morphology is the primary cause of growth retardation. However, how an altered chloroplast morphology could result in short inflorescence stems and ruffled leaves is not clear.
Jasmonic acid (JA) is an important hormone for both regular developmental processes, such as pollen maturation, and wound- and pathogen-induced defense responses (Wasternack and Hause, 2013). JA and its precursors, 12-oxo-phytodienoic acid (OPDA) and dinor-12-oxo-phytodienoic acid (dn-OPDA), belong to the lipid class of oxylipins. JA is synthesized by the allene oxide synthase (AOS) branch of the oxylipin pathway from 18:3 and 16:3 fatty acids released from plastid lipids by lipases (Feussner and Wasternack, 2002), although the identities of these lipases are still a matter of debate (Ellinger et al., 2010; Wasternack and Hause, 2013). The 18:3 and 16:3 fatty acids are sequentially metabolized by lipoxygenase (LOX), AOS, and allene oxide cyclase (AOC) to OPDA and dn-OPDA, respectively, which are exported from the plastids into the peroxisome for the remaining steps of JA biosynthesis. JA is then transported back into the cytosol and is conjugated to the amino acid isoleucine to form the active hormone JA-Ile. The complex of JA-Ile with its receptor CORONATIVE INSENSITIVE1 (COI1) targets JAZMONATE ZIM DOMAIN transcription repressors for proteasome degradation and induces the expression of genes required for defense signaling and also for a positive feedback response, leading to more JA production (Acosta and Farmer, 2010). In Arabidopsis, an additional pathway may exist for the biosynthesis of OPDA and dn-OPDA, in which the 18:3 and 16:3 acyl chains on galactolipids are directly converted into OPDA and dn-OPDA, producing arabidopsides (galactolipids containing esterified OPDA and dn-OPDA), which are proposed to be storage compounds that allow the rapid release of OPDA and dn-OPDA upon wounding (Mosblech et al., 2009; Acosta and Farmer, 2010; Wasternack and Hause, 2013).
We set out to identify the cause of the reduced inflorescence stem elongation in the dgd1 mutant and found that the visible phenotypes of dgd1 were mostly caused by JA overproduction and could be uncoupled from the altered chloroplast morphology. Elevated levels of JA and JA-Ile accumulated in both the dgd1 single mutants and the coi1-30 dgd1-1 double mutant lacking the COI1-dependent positive feedback loop, whereas the accumulation of OPDA and arabidopsides was COI1-dependent. Transcript abundance analyses suggested that the initial JA biosynthesis is activated through increased levels of mRNAs coding for LOX and a specific lipase that prefers MGDG as a substrate. High levels of other oxylipins in the dgd1 single mutants are maintained by further increases in LOX and AOC mRNA and protein levels.
RESULTS
dgd1 Mutants Have Lignified Phloem Cap Cells
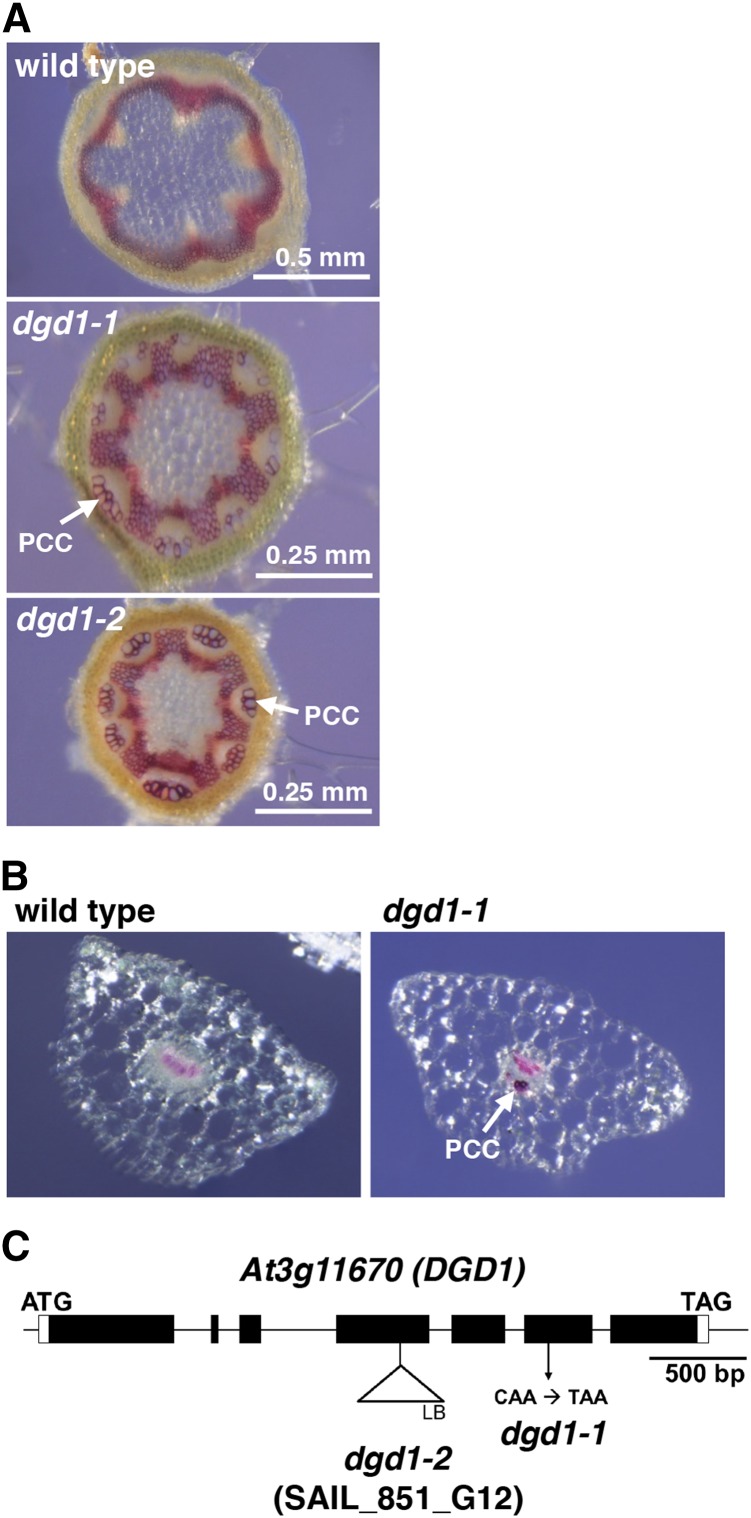
To investigate the cause of the short inflorescence stem phenotype (Supplemental Figure 1), we stained stem cross sections of the dgd1 mutant for lignin with phloroglucinol to allow easy observation of the positions of the xylem and fibers to check for structural abnormalities. We used the previously described dgd1 mutant allele (Dörmann et al., 1995). We also obtained a new dgd1 allele (see below); the original allele and the new allele are hereafter referred to as dgd1-1 and dgd1-2, respectively. As shown in Figure 1A, in the wild-type stem, red lignin staining was only detected in the xylem and interfascicular fibers, whereas in the dgd1-1 mutant, interestingly, an extra group of cells was also lignified. These cells were located at the outermost region of the phloem, immediately beneath the cortex, and have been referred to as phloem fibers or phloem cap cells (Zhong et al., 2000; Altamura et al., 2001). We followed the latter nomenclature since, in Arabidopsis, these cells are usually large and thin-walled. Because the dgd1-1 plants also had extremely short petioles compared with the wild type (Supplemental Figure 1), we also stained dgd1-1 petiole cross sections for lignin and observed lignified phloem cap cells (Figure 1B). To confirm that the lignification phenotype was not restricted to this particular allele, we examined the dgd1-2 mutant produced by T-DNA insertion (Figure 1C) and found that it had the same short inflorescence stem, short petiole, and ruffled leaf phenotypes (Supplemental Figure 1) and reduced DGDG content (Table 1) as the original mutant. Stem cross sections showed that its phloem cap cells were also lignified (Figure 1A).
Figure 1.
Both of the dgd1 Mutant Alleles Have Lignified Phloem Cap Cells.
(A) Inflorescence stem cross sections stained with phloroglucinol. Plants were grown for 10 d on MS plates and then transferred to soil for another 25 d. The dgd1 mutant sections are shown at a 2-fold higher magnification than the wild type. PCC, phloem cap cell.
(B) Petiole cross sections from 40-d-old plants stained with phloroglucinol.
(C) Schematic representation of the mutation positions of the dgd1-1 and dgd1-2 alleles. The black boxes, connecting lines between boxes, and white boxes represent, respectively, exons, introns, and the 5′ and 3′ untranslated regions, respectively.
Table 1. Galactolipid Composition of the Wild Type and Various Mutants.
| Plant | MGDG | DGDG |
|---|---|---|
| Wild type | 33.5 ± 8.3 | 6.2 ± 1.4 |
| aos | 30.5 ± 8.9 | 5.8 ± 1.2 |
| coi1-30 | 30.6 ± 5.6 | 6.1 ± 1.0 |
| dgd1-1 | 28.4 ± 3.8 | 0.5 ± 0.2 |
| dgd1-2 | 24.0 ± 3.4 | 0.4 ± 0.04 |
| aos dgd1-1 | 29.1 ± 3.5 | 0.5 ± 0.1 |
| aos dgd1-2 | 31.9 ± 4.3 | 0.5 ± 0.2 |
| coi1-30 dgd1-1 | 31.0 ± 5.4 | 0.6 ± 0.1 |
Plants were grown on 16-h-light/8-h-dark cycles for 10 d on MS plates and then transferred to soil for another 10 d. Means ± se of at least three independent plant batches are shown. Values are relative peak areas (%).
Since ectopic lignification in phloem cap cells has been observed in other mutants, including deetiolated3 (det3, encoding V-ATPase subunit C; Newman et al., 2004), ectopic lignification1 (eli1, encoding cellulose synthase CESA3; Caño-Delgado et al., 2003), broomhead (encoding eukaryotic release factor eRF1; Petsch et al., 2005), cpk28 (encoding a calcium-dependent protein kinase; Matschi et al., 2013), ectopic deposition of lignin in pith1 (elp1, encoding a chitinase; Zhong et al., 2000), and wrky12 (encoding a WRKY transcription factor; Wang et al., 2010), we analyzed the expression of these genes in the dgd1-1 mutant and found no reduction in expression of any of these genes compared with the wild type (Supplemental Figure 2).
Auxins and Ethylene Are Not the Cause of the dgd1 Visible Phenotypes
At least three hormones, auxins, ethylene, and JA, have been shown to stimulate secondary cell wall growth or ectopic lignification. Auxins induce xylem differentiation (Fukuda, 2004; Schuetz et al., 2013), and the amount of indole-3-acetonitrile, the precursor of the auxin indole-3-acetic acid, is highly increased in dgd1-1 (Fiehn et al., 2000). Therefore, we examined whether auxin levels in the phloem cap cell region were higher than in the wild type by crossing into the dgd1-1 mutant the DR5-GUS construct, a reporter gene encoding GUS driven by the synthetic auxin-responsive promoter DR5 (Ulmasov et al., 1995). In seedling aerial tissues of both wild-type and dgd1-1 plants (Supplemental Figure 3A, top panels), GUS staining was mainly detected in the periphery of the leaf blades, and no staining was detected in the petioles. In inflorescence stem cross sections of older plants (Supplemental Figure 3A, bottom panels), GUS staining in wild-type plants was mainly detected in the parenchyma cells adjacent to the xylem, while in dgd1-1 plants, only very faint GUS staining was detected in the same region, and no staining was detected in the phloem cap cell region. This result suggests that auxin levels are not increased in the phloem cap cell region of dgd1.
In the det3 and eli1 mutants, which show lignification of the phloem cap cells, the ethylene and JA signaling pathways are activated (Caño-Delgado et al., 2003; Brüx et al., 2008). Therefore, we first blocked the ethylene signaling pathway by crossing the dgd1 mutant with the ethylene insensitive2 (ein2) mutant and found that the double mutant, dgd1-1 ein2, was indistinguishable in visible appearance from the dgd1-1 mutant (Supplemental Figure 3B). Expression of the ethylene-responsive genes EBP and ETR2 was also not induced in the dgd1-1 mutant (Supplemental Figure 3C). These results show that activation of the ethylene signaling pathway is not the cause of the dgd1 phenotypes. We then examined the effect of JA on the mutants.
JA Biosynthesis and JA-Responsive Genes Are Highly Upregulated, and High Levels of Oxylipins Are Seen in dgd1 under Normal Growth Conditions
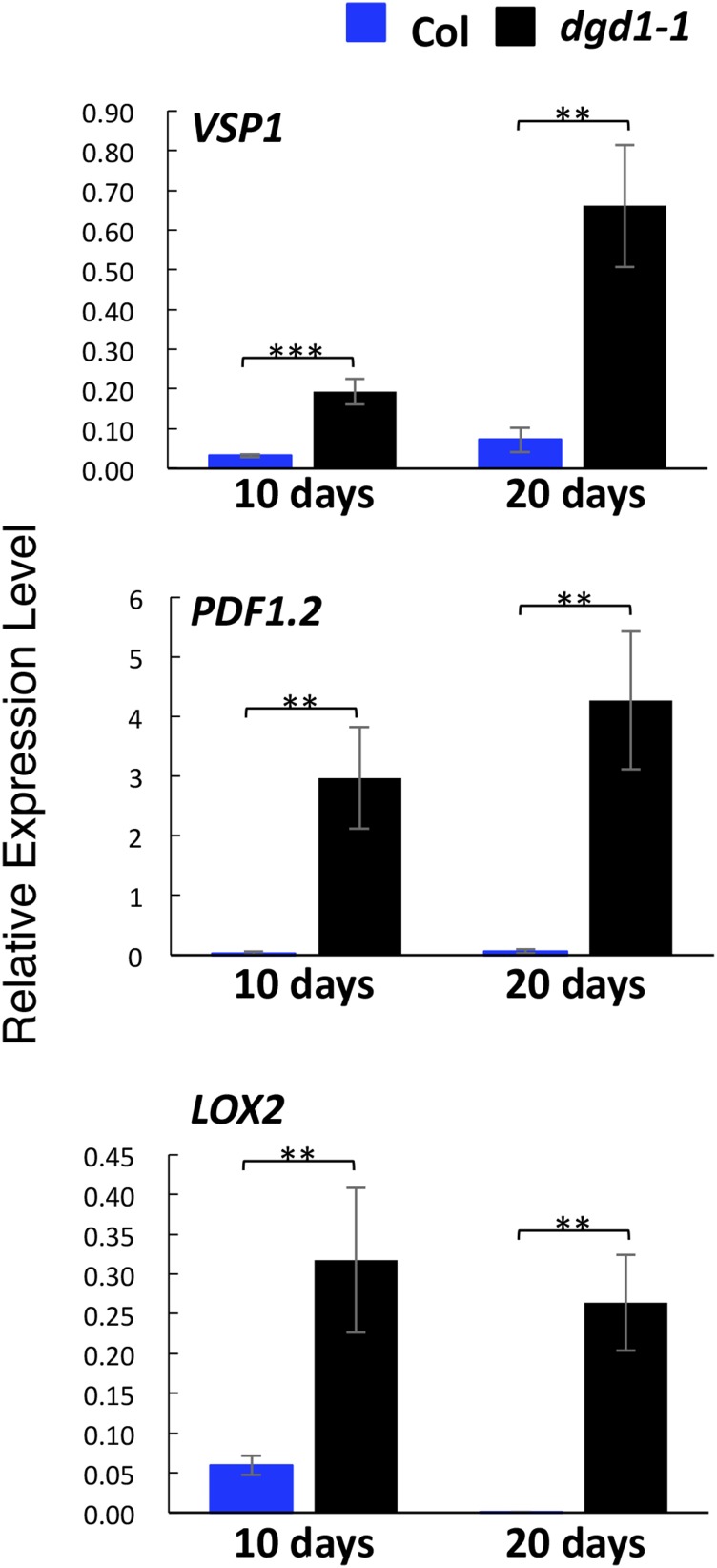
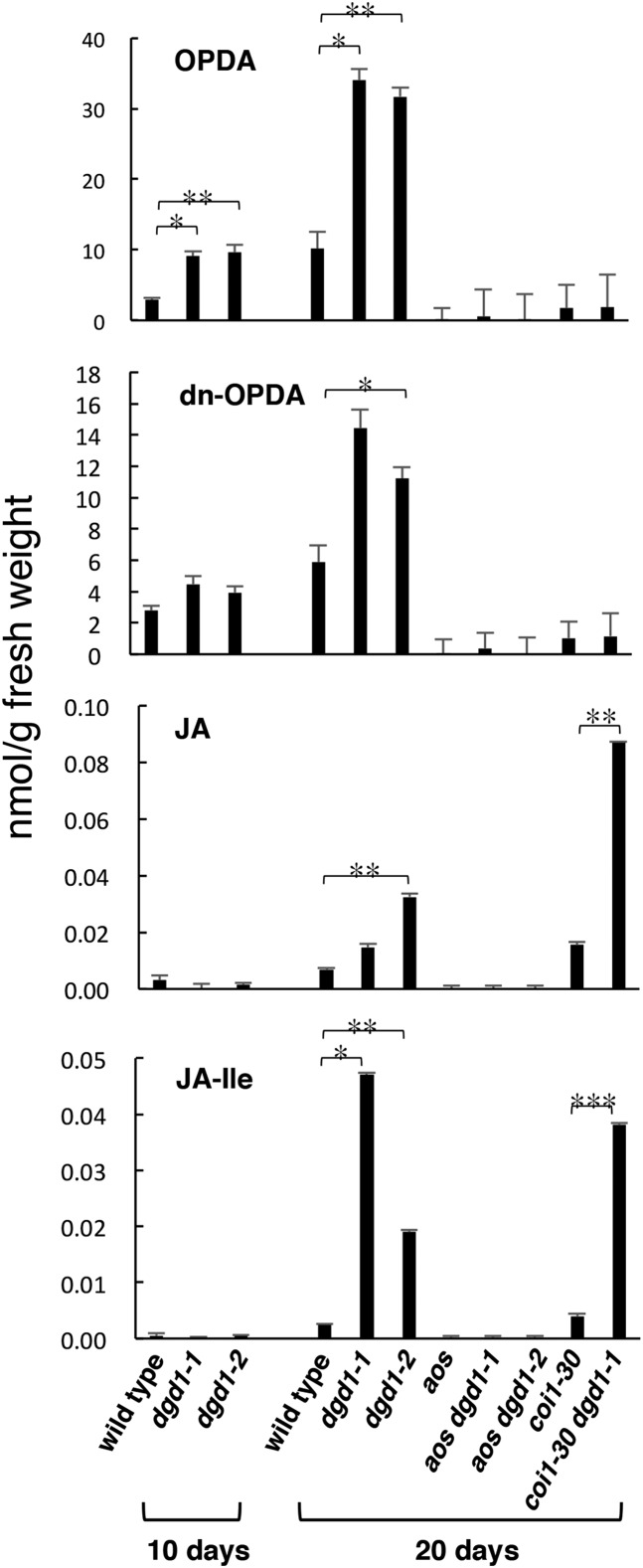
JA has been shown to stimulate secondary growth, and exogenous application of JA causes phloem fiber formation (Sehr et al., 2010). Methyl jasmonate treatment of Arabidopsis cells results in the increased expression of genes involved in monolignol biosynthesis and in increased monolignol and oligolignol production (Pauwels et al., 2008). JA biosynthesis has also been shown to be necessary for hypocotyl growth inhibition in the det3 and cpk28 mutants (Brüx et al., 2008; Matschi et al., 2015). Furthermore, overexpression of the lipase DONGLE (DGL) results in JA overproduction and generates visible phenotypes of short inflorescence stems and ruffled leaves, very similar to those in dgd1 (Hyun et al., 2008). Therefore, we examined whether JA signaling was activated in dgd1. Quantitative RT-PCR analyses showed that levels of mRNAs coding for two JA-responsive genes, VSP1 and PDF1.2, and for a JA biosynthesis gene, LOX2, were all highly upregulated in both 10- and 20-d-old dgd1-1 mutant seedlings (Figure 2). We then measured the content of various major oxylipins in these plants. As shown in Figure 3, after 10 d of growth, both dgd1 mutant alleles already had high OPDA levels compared with the wild type, and at day 20, the difference increased markedly and high levels of dn-OPDA, JA, and JA-Ile were also seen in the mutants.
Figure 2.
JA Biosynthesis and Signaling Genes Are Highly Activated in the dgd1-1 Mutant.
Plants were grown on MS plates for 10 d, then one set was harvested and the other was moved to soil and grown for another 10 d. Total RNA was then isolated, and the levels of expression of the three indicated genes were analyzed by quantitative RT-PCR and expressed relative to UBQ10 gene expression. Means ± se for three independent plant batches are shown. Significance levels are as follows: **P < 0.01 and ***P < 0.001 (Student’s t test). Col, wild-type ecotype Columbia.
Figure 3.
The Two dgd1 Single Mutants and the coi1-30 dgd1-1 Double Mutant Have High Oxylipin Levels under Normal Conditions.
Plants were grown on MS plates for 10 d, then one set was harvested and the other was moved to soil and grown for another 10 d, after which the levels of the indicated oxylipins were measured. Means ± se for at least three independent plant batches are shown. Significance levels are as follows: *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t test).
Mutations in Genes Involved in JA Biosynthesis or Signaling Can Rescue the Short Inflorescence Stem and Phloem Cap Lignification Phenotypes of dgd1
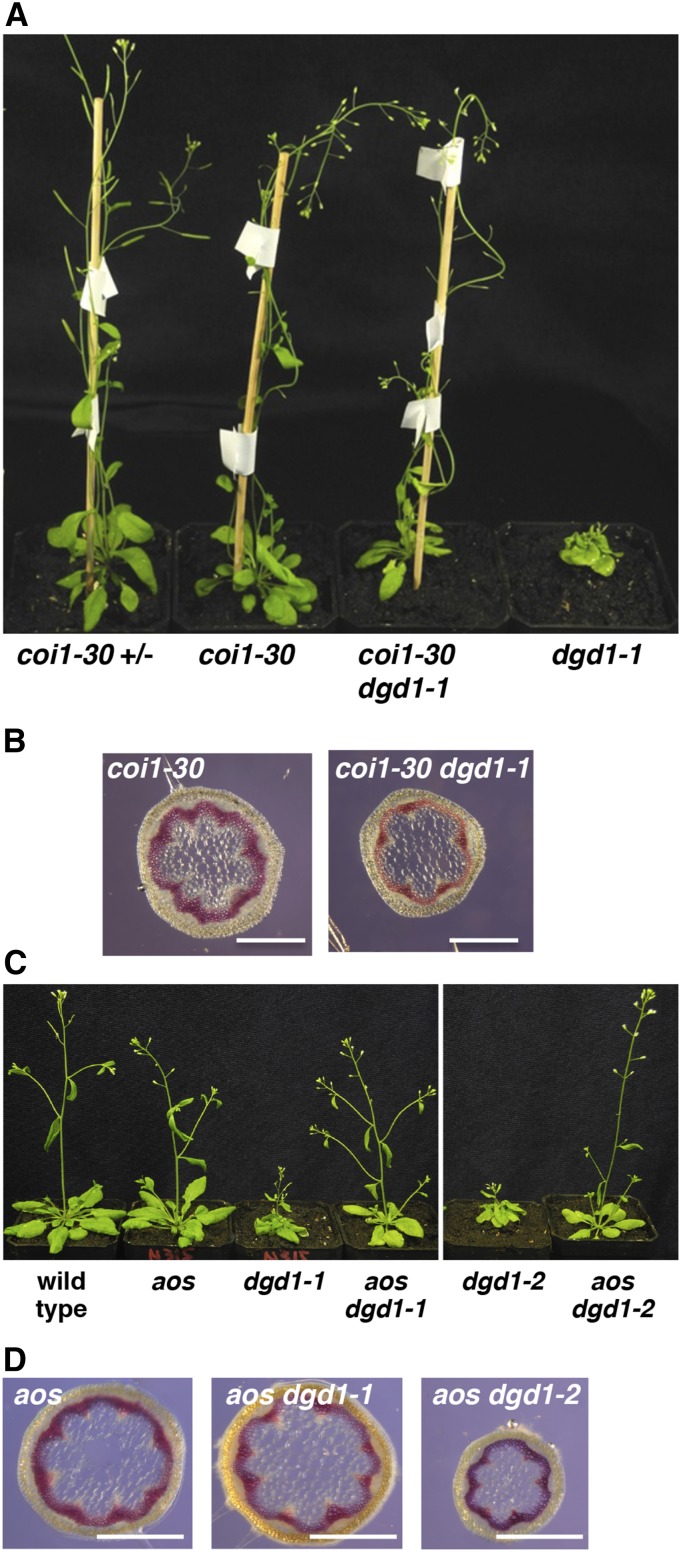
To examine whether the dgd1 visible phenotypes and phloem cap cell lignification were caused by activation of the JA signaling pathway, we crossed the dgd1-1 mutant with a coi1 mutant (the coi1-30 allele produced by T-DNA insertion [SALK_035548]; Mosblech et al., 2011; Yang et al., 2012) and found that the coi1-30 dgd1-1 double mutant had the same general appearance as the coi1-30 single mutant (Figure 4A). Furthermore, inflorescence stem cross sections showed that phloem cap cells in the double mutant were also not lignified (Figure 4B). To exclude the possibility that the rescue by the coi1 mutation was due to inactivation of some JA-independent COI1 activity, we also crossed the dgd1-1 and dgd1-2 mutants with an AOS knockout mutant (von Malek et al., 2002) and found that the short inflorescence stem, ruffled leaf (Figure 4C), and lignified phloem cap cell (Figure 4D) phenotypes were absent in the aos dgd1-1 and aos dgd1-2 double mutants, showing that blocking JA biosynthesis resulted in a general alleviation of most dgd1 visible phenotypes. However, further quantifications are required to assess if all growth defects are fully rescued.
Figure 4.
The coi1 dgd1 and aos dgd1 Double Mutants No Longer Exhibit the Short Inflorescence Stem and Lignification Phenotypes.
(A) Plants of the indicated genotypes after 40 d of growth; the coi1-30 heterozygous plant (coi1-30 +/−) was used as a wild-type control.
(B) Plants of the indicated genotypes were grown for 40 d, then inflorescence stem cross sections were stained with phloroglucinol. Bars = 0.5 mm.
(C) Plants of the indicated genotypes after 31 d of growth.
(D) Plants of the indicated genotypes were grown for 35 d, then inflorescence stem cross sections were stained with phloroglucinol. Bars = 0.5 mm.
We then measured oxylipin levels in the double mutants. As shown in Figure 3, the 20-d-old aos dgd1 double mutants showed no OPDA- or dn-OPDA-related oxylipin production, but, interestingly, in the 20-d-old coi1-30 dgd1-1 double mutant, high levels of JA and JA-Ile accumulated, showing that JA was overproduced even in the absence of COI1 for positive feedback induction of JA biosynthesis. However, no accumulation of OPDA or dn-OPDA occurred in the coi1-30 dgd1-1 double mutant, showing that the high-level accumulation of OPDA and dn-OPDA observed in the dgd1 single mutants was COI1-dependent.
Lipidomics Analyses of the Mutants
We next performed detailed lipidomics analyses to verify that rescue of the dgd1 visible phenotypes was not due to the coi1 and aos mutations somehow increasing the DGDG content in the double mutants. As shown in Table 1, the aos and coi1-30 single mutants had the same MDGD and DGDG contents as the wild type (for all lipids analyzed, see Supplemental Table 1), whereas all three double mutants, aos dgd1-1, aos dgd1-2, and coi1-30 dgd1-1, which did not show the dgd1 visible phenotypes, had less than 10% of wild-type DGDG levels. In addition, when we analyzed the fatty acid compositions of the lipids (Table 2; Supplemental Data Set 1), the three double mutants and the two dgd1 single mutants were found to have decreased MGDG (16:3/18:3) levels and increased MGDG (18:3/18:3) levels (Table 2), similar to previously reported findings for dgd1-1 (Xu et al., 2008).
Table 2. Fatty Acid Compositions of the Major Forms of MGDG and DGDG.
| Plant | MGDG (16:3/18:3) | MGDG (18:3/18:3) | DGDG (16:3/18:3) | DGDG (18:3/18:3) |
|---|---|---|---|---|
| Wild type | 13.1 ± 3.1 | 12.5 ± 3.6 | 0.50 ± 0.06 | 4.1 ± 1.1 |
| aos | 13.1 ± 4.1 | 10.1 ± 3.1 | 0.64 ± 0.11 | 3.8 ± 0.8 |
| coi1-30 | 13.3 ± 2.4 | 10.7 ± 1.6 | 0.66 ± 0.05 | 4.1 ± 0.7 |
| dgd1-1 | 6.3 ± 1.6 | 17.2 ± 1.8 | 0.05 ± 0.03 | 0.3 ± 0.2 |
| dgd1-2 | 5.6 ± 1.4 | 13.7 ± 1.9 | 0.05 ± 0.01 | 0.3 ± 0.03 |
| aos dgd1-1 | 8.3 ± 0.7 | 16.5 ± 2.2 | 0.08 ± 0.005 | 0.4 ± 0.1 |
| aos dgd1-2 | 7.8 ± 0.1 | 18.4 ± 2.7 | 0.06 ± 0.03 | 0.4 ± 0.2 |
| coi1-30 dgd1-1 | 8.4 ± 0.5 | 17.9 ± 4.2 | 0.08 ± 0.02 | 0.4 ± 0.05 |
Plants were grown on 16-h-light/8-h-dark cycles for 10 d on MS plates and then transferred to soil for another 10 d. Means ± se of at least three independent plant batches are shown. Values are relative peak areas (%).
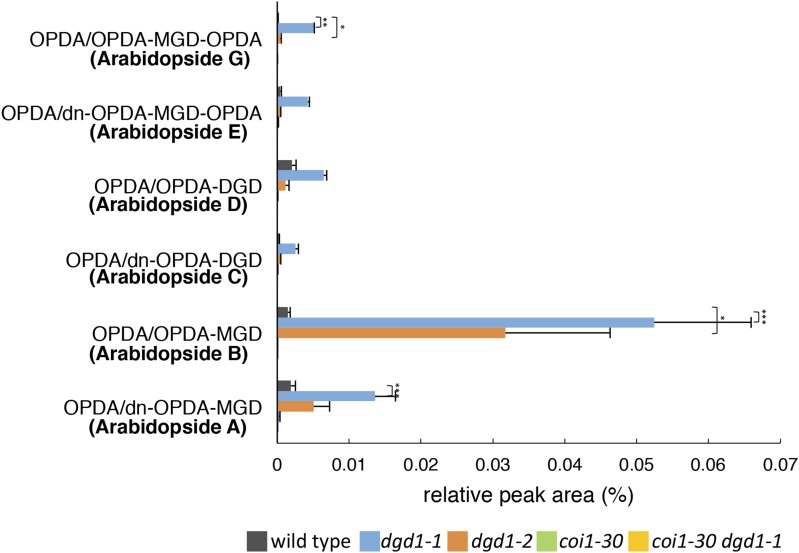
We also measured the profiles of arabidopsides, which are galactolipids containing up to three OPDA and/or dn-OPDA molecules. Arabidopsides are present at very low levels under normal growth conditions, and it has been reported that, upon wounding, the most abundant arabidopside is arabidopside A (MGDG esterified with one OPDA moiety and one dn-OPDA moiety) (Kourtchenko et al., 2007; Ibrahim et al., 2011; Vu et al., 2012). As shown in Figure 5, in the absence of wounding and under normal growth conditions, all arabidopside forms were present at very low levels in the wild type, whereas in the dgd1 single mutants, high levels of both arabidopside B (MGDG esterified with two OPDA) and arabidopside A were seen, with arabidopside B levels being higher than arabidopside A levels. Even levels of arabidopside G (MGDG esterified with three OPDA), which are normally extremely low, were increased significantly in the dgd1 mutants. This result is in agreement with the dgd1 mutants having higher levels of MGDG (18:3/18:3) than MGDG (16:3/18:3) (Table 2) and suggests that some excess MDGD may be converted into the corresponding arabidopsides in the dgd1 mutants. In addition, arabidopside levels were not increased in the coi1-30 dgd1-1 double mutant, showing that, like the high-level accumulation of OPDA and dn-OPDA, the much higher arabidopside levels in the dgd1 single mutants are COI1-dependent.
Figure 5.
The Two dgd1 Single Mutants, but Not the coi1-30 dgd1-1 Double Mutant, Have Increased Levels of Arabidopsides.
Plants were grown on MS plates for 10 d and then moved to soil for another 10 d, after which the levels of the five major arabidopsides were measured. Means ± se for at least three independent plant batches are shown. Results for plants with the aos mutation were close to the detection limits and are not shown. Significance levels are as follows: *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t test).
Reduced Photosynthesis and Altered Chloroplast Morphology Are Not Caused by Increased JA Levels
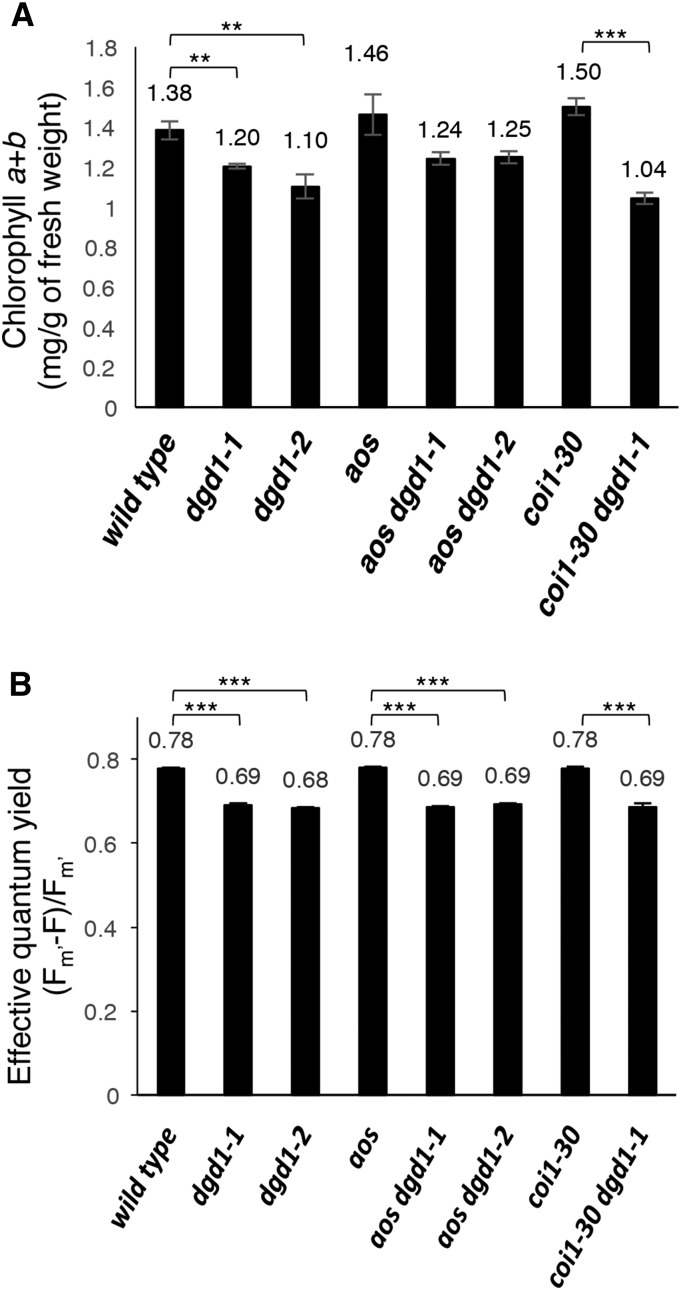
Although the coi1-30 dgd1-1, aos dgd1-1, and aos dgd1-2 double mutants no longer exhibited short inflorescence stems and ruffled leaves, their leaves still appeared pale green, like the dgd1 single mutants. Therefore, we measured the chlorophyll content (Figure 6A) and PSII quantum yield (Figure 6B) and found that both were reduced in the single and double mutants compared with the wild type.
Figure 6.
The coi1-30 dgd1-1 and aos dgd1 Double Mutants Still Have Reduced Chlorophyll Levels and Photosynthetic Capacity.
(A) Plants were grown on MS plates for 20 d, then leaves were harvested for chlorophyll determination.
(B) Plants were grown on MS plates for 10 d, then the PSII quantum yield was measured.
Means ± se for at least three independent plant batches are shown. Significance levels are as follows: **P < 0.01 and ***P < 0.001 (Student’s t test).
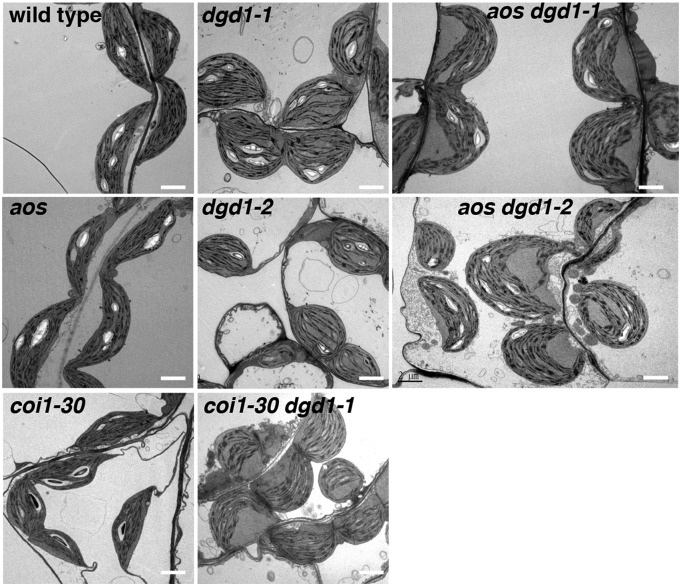
Another major phenotype reported for the dgd1-1 mutant is altered chloroplast morphology. When observed by electron microscopy, instead of being spindle-shaped, like wild-type chloroplasts, dgd1-1 chloroplast envelopes are usually more rounded, but the thylakoids remain elongated, resulting in bent thylakoids and large thylakoid-free stromal areas (Dörmann et al., 1995; Hölzl et al., 2009). As shown in Figure 7, the dgd1-2 allele showed the same phenotype as the dgd1-1 allele, whereas chloroplasts in the aos and coi1-30 mutants had the same morphology as the wild-type chloroplasts, and all double mutants exhibited rounded chloroplasts with bent thylakoids, as in the dgd1 mutants. This result shows that the altered chloroplast morphology in dgd1 is not rescued by eliminating JA signaling or production.
Figure 7.
Leaf Chloroplast Morphology of the Wild Type and Various Mutants Shown by Electron Microscopy.
Plants were grown on MS plates for 10 d and then moved to soil for another 16 d. Bars = 2 µm.
Altered Chloroplast Morphology in the vipp1 Mutant Does Not Elicit a JA Response
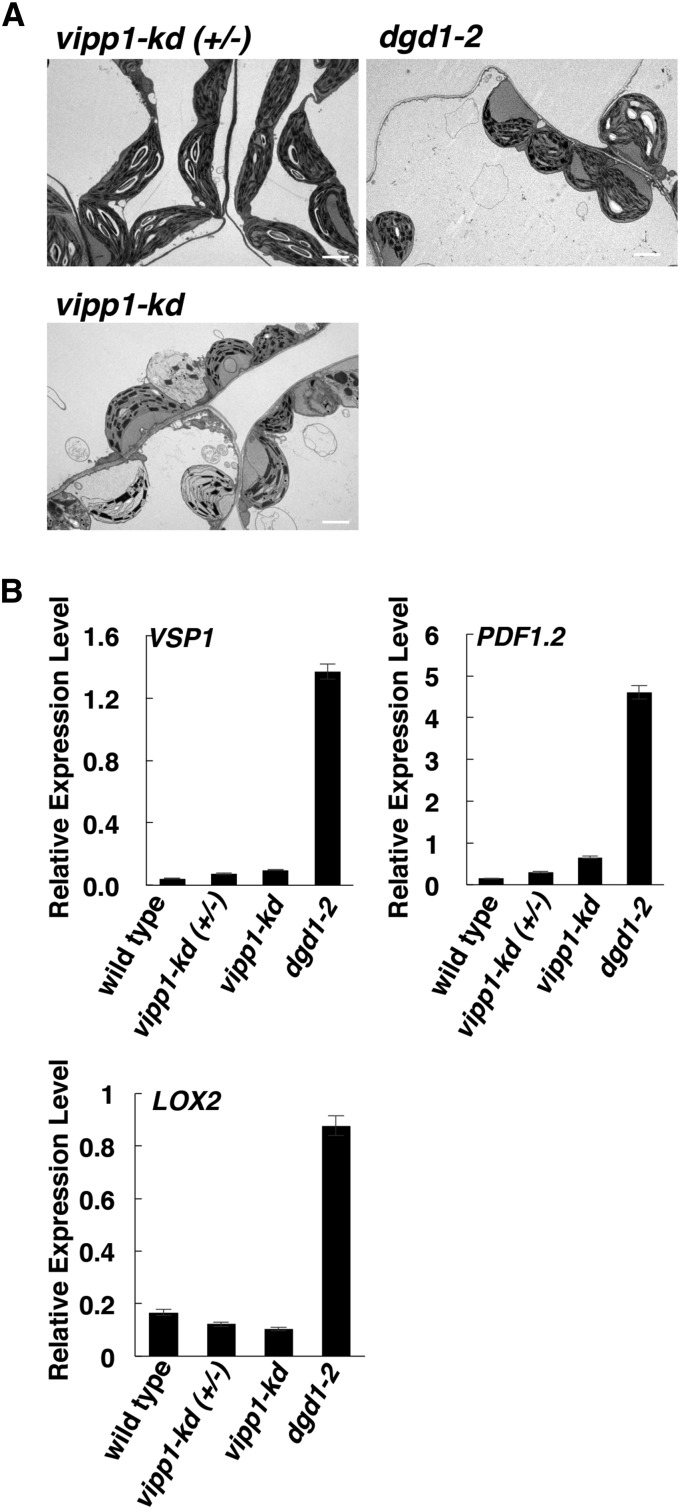
Our results showed that, in dgd1 mutants, activation of JA signaling caused the visible phenotypes but not the altered chloroplast morphology. However, it was still possible that the altered chloroplast morphology resulted in a stress condition that induced JA production in the dgd1 mutants. Therefore, we examined this possibility by analyzing another mutant with a similar altered chloroplast morphology. The vipp1 (vesicle-inducing protein in plastids1) knockdown mutant has rounded chloroplasts with bent thylakoid membranes and large thylakoid-free stromal areas (Zhang et al., 2012), features very similar to those in the dgd1 mutants. For our analyses of the dgd1 mutants shown in Figure 7, plants were grown on Murashige and Skoog (MS) plates for 10 d and then moved to soil for another 16 d. For analyses of the vipp1 mutants, vipp1 and dgd1-2 plants were also grown for 26 d but on MS plates throughout, as the vipp1 knockdown mutant cannot survive on soil (Zhang et al., 2012). Under these conditions, the dgd1-2 mutant still showed altered chloroplast morphology (Figure 8A) and activation of the JA-responsive genes VSP1, PDF1.2, and LOX2 (Figure 8B), whereas the vipp1 knockdown mutant plants showed a similarly altered chloroplast morphology but no induction of expression of the JA-responsive genes. These data suggest that OPDA and JA overproduction in the dgd1 mutants is not caused by altered chloroplast morphology.
Figure 8.
The vipp1 Knockdown Mutant Shows Altered Chloroplast Morphology but No Activation of JA Signaling.
Plants were grown for 26 d on MS plates, then morphology and JA-responsive gene transcript levels were examined.
(A) Morphology of chloroplasts from vipp1 knockdown mutant (vipp1-kd) plants and heterozygous control [vipp1-kd (+/−)] plants. Chloroplasts of dgd1-2 plants grown under the same conditions are shown for comparison.
(B) Levels of expression of the JA-responsive genes analyzed by quantitative RT-PCR and normalized to UBQ10 gene expression. Values are means ± sd of three technical replicates.
JA Biosynthesis Might Be Initially Activated through Increased Levels of Transcripts of PLA-Iγ3 and LOXs
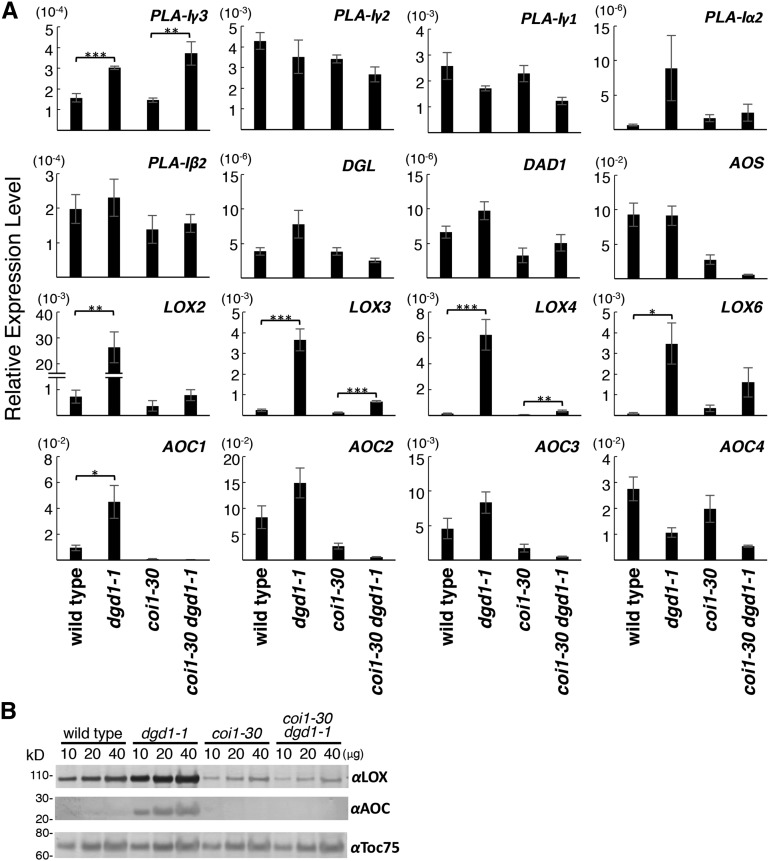
To further understand how JA biosynthesis is activated in the dgd1 mutants, we analyzed the levels of mRNAs for genes encoding enzymes involved in the first three chloroplast-localized steps of JA biosynthesis, namely linoleate 13-LOX (LOX2, LOX3, LOX4, and LOX6), AOS, and AOC (AOC1 to AOC4). We also analyzed the levels of mRNAs coding for the seven class I phospholipase A1 (PLA1) members, including DEFECTIVE IN ANTHER DEHISCENCE1 (DAD1; Ishiguro et al., 2001) and DGL (Hyun et al., 2008). Class I PLA1 members have a predicted chloroplast-targeting transit peptide and are implicated in generating the 18:3 fatty acid serving as the substrate for 13-LOX (Ishiguro et al., 2001; Seo et al., 2009; Ellinger et al., 2010). We measured transcript levels in the wild type, the dgd1-1 and coi1-30 single mutants, and the coi1-30 dgd1-1 double mutant, as comparison of the gene expression profile of the coi1-30 dgd1-1 double mutant with that of the dgd1-1 single mutant would allow us to distinguish the immediate responses caused by the dgd1 mutation from the subsequent responses induced by the COI1-mediated positive feedback loop. As shown in Figure 9A, transcript levels for the four LOX genes were all slightly higher in the coi1-30 dgd1-1 double mutant than in the coi1-30 single mutant, the increases in LOX3 and LOX4 transcript levels being significant. Transcript levels for all four genes were markedly higher in the dgd1-1 single mutant than in the wild type or the coi1-30 dgd1-1 double mutant, suggesting that the dgd1 mutation initially resulted in a slight induction and that the COI1-mediated positive feedback loop resulted in a further induction of LOX gene expression. AOC1 transcript levels in the dgd1-1 single mutant were increased in a COI1-dependent manner, but no significant induction of expression of the other AOCs or the AOS gene was seen. Interestingly, of the transcript levels for the seven class I PLAs, only those for PLA-Iγ3 were increased significantly in the dgd1-1 and coi1-30 dgd1-1 mutants. The increase was actually higher in the coi1-30 dgd1-1 double mutant, suggesting that PLA-Iγ3 activation was due to the dgd1 mutation and not related to COI1. When the activity of the seven class I PLAs was analyzed previously using MGDG, DGDG, phosphatidylcholine (PC), and triacylglycerol (TAG) as substrates, PLA-Iγ3 was found to have one of the highest activities and to be the only PLA with a high substrate preference for MGDG (Seo et al., 2009). A mutation in this gene reduces basal JA and OPDA levels but had no effect on JA biosynthesis in wounded leaves (Ellinger et al., 2010). These data suggest that, under nonwounded conditions, some MGDG molecules are cleaved by PLA-Iγ3 to release fatty acids that lead to basal JA production.
Figure 9.
Levels of Expression of Genes Encoding Enzymes for the Initial Steps of JA Biosynthesis.
Plants of the indicated genotypes were grown on MS plates for 10 d, then moved to soil and grown for another 10 d before being harvested for total RNA isolation and protein extraction.
(A) Levels of expression of the indicated genes analyzed by quantitative RT-PCR and expressed relative to UBQ10 gene expression. Means ± se for at least three independent plant batches are shown. Significantly higher expression in dgd1-1 or coi1-30 dgd1-1 compared with the wild type or coi1-30, respectively, is indicated as follows: *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t test).
(B) Total proteins (10, 20, or 40 μg, indicated above the lanes) were analyzed by SDS-PAGE, followed by immunoblotting with antiserum against cucumber lipid body LOX, Arabidopsis AOC, or pea Toc75.
We then analyzed LOX and AOC protein levels by immunoblotting using rabbit antisera raised against LOX from cucumber (Cucumis sativus) lipid bodies or Arabidopsis AOC. Both antisera recognize multiple LOX and AOC forms in Arabidopsis leaves (Hause et al., 2000; Berger et al., 2001; Stenzel et al., 2003). The chloroplast outer membrane protein-import channel Toc75 (Schnell et al., 1994) was also analyzed as a loading control. As shown in Figure 9B, the coi1-30 mutation resulted in reduced levels of LOX, as shown previously (Benedetti et al., 1995), and LOX and AOC levels were highly increased in the dgd1-1 mutant but not in the coi1-30 dgd1-1 double mutant. These results suggest that, through the COI1-mediated positive feedback loop, the dgd1 mutation has induced a high-level stable accumulation of LOX and AOC proteins, which maintained high-level oxylipin production in the mutant plants.
DISCUSSION
Our data show that the dgd1 mutation leads to JA overproduction, which results in short inflorescence stems and lignification of phloem cap cells. The aos dgd1 and coi1-30 dgd1-1 double mutants appeared almost wild type, with only a small reduction in chlorophyll content and photosynthesis, suggesting that, without JA-COI1-mediated growth inhibition, a 90% reduction in DGDG content only has a small effect on plant growth. It is well known that JA inhibits cell cycle progression (Swiatek et al., 2002, 2004; Pauwels et al., 2008; Noir et al., 2013), and growth inhibition is also achieved through crosstalk of JA with other phytohormones, such as auxins, gibberellins, and brassinosteroids (Ren et al., 2009; Kazan and Manners, 2012; Yang et al., 2012). JA also inhibits cell expansion, for example, in petals (Brioudes et al., 2009). In the dgd1 mutants, JA-COI1-mediated growth inhibition seemed most severe in tissues with vascular bundles; the inflorescence stems were extremely short, while in the leaf, the petioles were very short and the major veins did not elongate sufficiently, but the leaf blade still increased in size, resulting in the ruffled appearance. It is possible that, in addition to suppressed cell division, lignification of the phloem cap cells further restricts the expansion of vascular bundles and pith, resulting in severe inhibition of the elongation of inflorescence stems and major veins. It is not known why oxylipin overproduction causes lignification only in phloem cap cells. Perhaps this group of cells can develop into phloem fibers even in wild-type plants and, therefore, secondary cell wall biosynthesis can be induced more easily in these cells.
MDGD, not DGDG, is suggested to be the primary substrate for arabidopside production because, upon mechanical or freeze-thaw wounding, high levels of arabidopsides are seen, in particular those synthesized from MDGD (Ibrahim et al., 2011; Nilsson et al., 2012; Vu et al., 2012). Our data here suggest that, when the major route for the conversion of MDGD to DGDG is blocked by the dgd1 mutation, some resulting excess MDGD is converted to JA, supporting the idea that, in addition to being a substrate for arabidopside production, MDGD could also be the primary substrate for JA production. In agreement with this suggestion, expression of the gene MGD1, coding for the major MGDG synthase, is upregulated by wounding and methyl jasmonate (Kobayashi et al., 2009). However, when rice (Oryza sativa) MGDG synthase (OsMGD) was overexpressed in tobacco (Nicotiana tabacum), no JA overproduction was observed, but, unlike in the dgd1 mutants, which have a greatly increased MGDG:DGDG ratio, levels of both MGDG and DGDG were increased in the OsMGD-overexpressing plants, leading to a reduced MGDG:DGDG ratio (Wang et al., 2014). It is likely that, in these OsMGD-overexpressing plants, DGDG biosynthesis was adjusted to maintain the MGDG:DGDG ratio. Interestingly, DGD1 is localized at the outer membrane of plastids, while MGD1 is localized at the inner membrane, and most of the enzymes for OPDA biosynthesis may also be attached to the stromal side of the inner envelope membrane (Froehlich et al., 2001; Joyard et al., 2010). Conversion of MGDG to DGDG by DGD1, therefore, might rapidly channel some of the MGDG to the outer membrane. This may help in maintaining a correct MGDG:DGDG ratio at the inner membrane and in diverting some MGDG away from the OPDA-synthesizing enzymes and, therefore, may help prevent JA overproduction.
To further understand how JA biosynthesis is activated in the dgd1 mutants, we measured the transcript levels of genes responsible for the initial steps of JA biosynthesis. Our results showed that PLA-Iγ3 and LOX transcript levels were increased in the coi1-30 dgd1-1 double mutant. Since LOX catalyzes the first step of oxylipin formation leading to JA biosynthesis, it is reasonable that LOX is the first enzyme to show increased expression when levels of its substrate are increased, as may be the case in the dgd1 mutant. While all four LOX genes contribute to wound-induced JA formation (Caldelari et al., 2011; Chauvin et al., 2013), they also have different functions. For example, LOX2 is required to generate the high levels of JA seen proximal to a wound (Schommer et al., 2008; Glauser et al., 2009), and LOX3 and LOX4 are required for male fertility (Caldelari et al., 2011). Nonetheless, their responses to the dgd1 mutation were very similar, all showing a weak induction of expression in the coi1-30 dgd1-1 double mutant but a very high level of induction in the dgd1-1 single mutant, although only LOX3 and LOX4 transcript levels in the coi1-30 dgd1-1 double mutant were significantly different from those in the coi1-30 single mutant. It is possible that the promoters of the LOX3 and LOX4 genes may be more responsive to the dgd1 mutation.
There seem to be stimuli- and pathway-specific lipases to generate fatty acid substrates for JA production, but these have not been clearly identified. DAD1 is required for male fertility but displays a 4-fold higher substrate preference for PC over MDGD (Ishiguro et al., 2001). Overexpression of DGL results in JA overproduction in leaves (Hyun et al., 2008), but DGL shows much higher activity toward DGDG than MGDG (Hyun et al., 2008) and is localized in the cytosol (Ellinger et al., 2010). A knockout mutant of PLA-Iγ1 reduces initial wound-induced JA formation, but JA levels reach nearly wild-type levels at 60 min after wounding (Ellinger et al., 2010). Our transcript abundance analyses showed that PLA-Iγ3 was the only PLA gene showing upregulated expression in both the dgd1-1 and coi1-30 dgd1-1 mutants. A PLA-Iγ3 knockout mutant was shown previously to have normal levels of wound-induced JA production but a greater than 50% reduction in basal levels of JA, OPDA, and dn-OPDA (Ellinger et al., 2010), suggesting that lipid cleavage by PLA-Iγ3 can indeed lead to JA production under normal conditions. Furthermore, PLA-Iγ3 is the only lipase shown to have a specific substrate preference for MDGD (Seo et al., 2009).
The exact mechanism for the activation of JA biosynthesis in the dgd1 mutant remains to be investigated. We hypothesize that the increased MGDG:DGDG ratio in dgd1 chloroplasts may induce an increased level of PLA-Iγ3, the lipase with a substrate preference for MGDG. This would result in an initial increase in JA and JA-Ile production, as observed in the coi1-30 dgd1-1 mutant. However, we have only observed increased PLA-Iγ3 transcript levels. Whether the protein level and activity of PLA-Iγ3 are increased in the dgd1 mutants remain to be investigated. The causal relationship between increased MGDG:DGDG ratio and JA production also needs to be experimentally tested by directly manipulating the MGDG:DGDG ratio and then studying its effect on JA production. Other induction mechanisms for JA biosynthesis also need to be considered. For example, a reduction in DGDG content itself may alter the biophysical properties of the membranes, or the altered chloroplast morphology may trigger stress signals different from that trigged by vipp1, and activate the OPDA biosynthesis enzymes.
In the dgd1 mutant with an intact COI1 positive feedback loop, in addition to the increased levels of PLA-Iγ3 and LOX transcripts seen in the coi1-30 dgd1-1 mutant, further induction of the expression of LOX genes and LOX proteins, increased expression of AOC1 and AOC proteins, and accumulation of OPDA, dn-OPDA, and arabidopsides A and B were seen. These increases are most likely the result of positive feedback induction after the COI1-JA-Ile interaction. This may also explain how a small increase in MGDG (18:3/18:3) levels can result in a large increase in OPDA and dn-OPDA production in the dgd1 mutants, as the small excess of MGDG (18:3/18:3) may only result in the initial increase in JA and JA-Ile levels, as in the coi1-30 dgd1-1 mutant, but subsequent activation of the COI1 positive feedback loop would result in the further activation of other enzymes in the oxylipin biosynthesis pathway.
It has been shown that replacing DGDG with GGDG in the dgd1 mutant can restore plant growth and chloroplast shape but not photosynthesis (Hölzl et al., 2006, 2009). Together with our data, this shows that the dgd1 phenotypes are caused by three factors. First, the reduced photosynthesis and chlorophylls are caused directly by reduced DGDG levels (Hölzl et al., 2006, 2009). Indeed, DGDG molecules have been found in the crystal structure of cyanobacterial PSII (Guskov et al., 2009). Second, the visible phenotypes of dgd1 are caused by increased JA levels, as shown in this study. The rescue of visible phenotypes in GGDG-complemented dgd1 plants (Hölzl et al., 2006, 2009) is most likely due to conversion of the excess MGDG into GGDG, thereby preventing its conversion to JA. Third, the altered chloroplast morphology can be rescued by GGDG but not by blocking JA signaling. MGDG is wedge-shaped and thus has non-bilayer-forming characteristics, whereas DGDG is a bilayer-forming lipid and the MGDG:DGDG ratio may be critical for the shape of chloroplast membranes. It is possible that GGDG can replace DGDG in maintaining chloroplast shape due to its similar bilayer-forming property. Together, these data highlight the many functions of chloroplast membrane lipids and call attention to the need for caution when analyzing phenotypes of chloroplast lipid mutants. GGDG-complemented dgd1 plants provide an important tool for studying the exact function of the galactose moiety in DGDG. Similarly, the aos dgd1 double mutants established here could provide a tool for studying the direct effect of reduced DGDG levels on plant growth without activation of the JA signaling pathway.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana dgd1-2 mutant allele (SAIL_851_G12) was obtained from the ABRC (http://abrc.osu.edu/). The coi1-30 (SALK_035548; Mosblech et al., 2011; Yang et al., 2012) and ein2 (SAIL_265_D03) mutants were gifts of Hsu-Liang Hsieh and Long-Chi Wang, respectively. Seeds of Arabidopsis were sterilized and plated on MS agar medium containing 2% sucrose and grown in growth chambers with a light intensity of 71 μmol m−2 s−1 (cool-white fluorescent light bulbs) and 16-h-light/8-h-dark conditions at 22°C. Unless stated otherwise, in experiments involving adult plants, 10-d-old seedlings were transferred from MS agar plates to soil for further growth. Seeds were obtained from homozygous aos mutant plants by spraying the flowers with methyl jasmonate. Seeds from coi1-30 heterozygous plants and coi1-30 heterozygous dgd1-1 homozygous plants were plated, and a small piece of leaf tissue was then cut from each plant for genotyping to identify coi1-30 homozygous plants and coi1-30 dgd1-1 double mutant plants. Genotypes of all mutant lines were confirmed by DNA sequencing or genomic PCR using a Phire Plant Direct PCR Kit (Thermo Scientific); the primers used are listed in Supplemental Table 2.
Quantitative RT-PCR and Immunoblot Analyses
Total plant RNA was extracted using TriPure Isolation Reagent (Roche Diagnostics), and genomic DNA contamination was removed with DNase I (Thermo Scientific). First-strand cDNAs were synthesized using Maxima H Minus Reverse Transcriptase (Thermo Scientific) and RNA isolated from seedlings of the indicated age. Quantitative RT-PCR was performed using a LightCycler system (Roche Diagnostics) and a LightCycler-FastStart DNA Master SYBR Green I Kit (Roche Diagnostics). Each PCR mixture contained 50 ng of cDNA and 0.5 mM of each primer pair. The initial denaturing step of 10 min was followed by 30 to 50 PCR cycles of 95°C for 10 s, 60°C for 5 s, and 72°C for 1 s per 25 bp of the expected product. After the PCR, the melting temperature was tested. Quantification was performed using LightCycler 480 software version 1.5.1.62. Normalization was performed using UBQ10 transcript levels. The primers used are listed in Supplemental Table 2.
For immunoblot studies, the aboveground tissues of 20-d-old seedlings were harvested and snap-frozen in liquid nitrogen and then ground to powder in liquid nitrogen. Total proteins were extracted and analyzed by SDS-PAGE (NuPAGE 4-12% gradient gel system; Invitrogen) followed by immunoblotting with rabbit antiserum against cucumber (Cucumis sativus) lipid body LOX (1:1000 dilution; Hause et al., 2000; Berger et al., 2001), Arabidopsis AOC (1:5000 dilution; Stenzel et al., 2003), or pea (Pisum sativum) Toc75 (1:2000 dilution; Tu et al., 2004) and alkaline phosphatase-coupled goat anti-rabbit IgG antibodies (1:5000 dilution; Jackson ImmunoResearch). Bound antibodies were detected using the nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate colorimetric system.
Lignin and GUS Staining
Petioles, or inflorescence stems immediately above the rosette leaves, from plants of the indicated age were cut and embedded in 7% agarose. Then, 100-µm-thick sections were prepared using a vibratome, stained with 5% phloroglucinol-HCl solution (freshly prepared by mixing equal volumes of 10% phloroglucinol in 95% ethanol and 37% HCl), and observed with a dissection microscope with dark-field illumination. For GUS staining of seedlings shown in Supplemental Figure 3, tissues were fixed in 0.3% formaldehyde in 50 mM phosphate buffer, stained with 1 mM X-Gluc in phosphate buffer at 37°C overnight, and cleared in 95% ethanol. Stems from stained seedlings were then cut, embedded, and sectioned as described above.
Hormone and Lipid Analyses
For phytohormone analyses, 100 mg of snap-frozen sample was extracted in a two-phase partitioning system using a mixture of tert-butyl methyl ether:methanol:water (4.2:1.25:1, v/v/v) and analyzed on an HPLC/nanoelectrospray ionization-tandem mass spectrometry system (Iven et al., 2012). Internal standards were used for quantifications. For lipid analyses, 200 mg of snap-frozen sample was extracted in a mixture of 2-propanol, hexane, and water. Analysis of glycerolipids and nonpolar lipids was performed on an ultra-HPLC-nanoelectrospray ionization-tandem mass spectrometry system (Tarazona et al., 2015), as described below.
Lipid species were separated using the Acquity UPLC system (Waters) equipped with an Acquity UPLC HSS T3 column (100 mm × 1 mm, 1 µm; Waters); aliquots (2 μL) were injected in partial loop with needle overfill mode at a flow rate of 0.1 mL/min and a separation temperature of 35°C. For chromatography, the following solvent mixtures used: methanol:20 mM ammonium acetate (3:7, v/v) containing 0.1% (v/v) acetic acid (A) and tetrahydrofurane:methanol:20 mM ammonium acetate (6:3:1, v/v/v) containing 0.1% (v/v) acetic acid (B). Different linear binary ultra-HPLC elution gradients were used for the different lipid classes. For all lipids except TAG, elution was performed using a start condition of 65% B for glycerolipid analysis, 80% B for lysolipid analysis, and 50% B for DAG analysis maintained for 2 min, followed by a linear increase to 100% B over 8 min, then 100% B for 2 min, followed by reequilibration to start conditions over 4 min. For TAG, 100% B was used and the chromatographic run was for 8 min.
Chip-based nanoelectrospray ionization was achieved using a TriVersa Nanomate (Advion) with 5-µm i.d. nozzles at a flow rate of 218 nL/min and a voltage of 1.3 kV. The electrospray current was set to 70 nA, and the ions were infused into a 4000 QTRAP tandem mass spectrometer (AB Sciex). Targeted molecular species analysis was performed in multiple reaction monitoring mode. Target precursor ions were [M+NH4]+ for DAG, TAG, and arabidopsides; [M−H]− for phosphatidylethanolamine, phosphatidylglycerol, phosphatidylinositol, phosphatidylserine, and sulphoquinovosyldiacylglycerol, including their lyso species; and [M−H+CH3CO2H]− for PC, MGDG, and DGDG, including their lyso species. For all lipid species except arabidopsides, the target single reaction monitoring (SRM) transitions were diagnostic for the molecular species acyl chain composition, either by a fatty acid-associated neutral loss in positive ion mode (nonpolar lipids) or by the formation of fatty acyl-related fragments in negative ion mode (glycerolipids). For arabidopsides, target SRM transitions were diagnostic for the molecular species head groups by the formation of head group-related fragments in positive ion mode (Ibrahim et al., 2011). The dwell time was 20 ms for all SRM transitions. Ion focusing and collision energy were optimized to maximize detector response.
Chlorophyll and Photosynthesis Measurements
Chlorophyll content was measured as described previously (Lichtenthaler, 1987). All measurements were performed in triplicate using three independent batches of 20-d-old plant samples. Chlorophyll fluorescence was determined using the IMAGING-PAM MAXI version chlorophyll fluorometer (Walz). To determine the effective PSII quantum yield, fluorescence emissions Fm′ and F of 10-d-old light-adapted (71 μmol m−2 s−1) seedlings on MS agar medium were measured, and the quantum yield was calculated as (Fm′ − F)/Fm′.
Transmission Electron Microscopy
Leaves from 26-d-old plants were cut into 1- × 1-mm pieces that were then fixed in a solution of 2.5% glutaraldehyde and 4% paraformaldehyde for 4 h, washed with 0.1 M sodium cacodylate, and fixed with 1% OsO4 for another 1 h. The samples were dehydrated in graded concentrations of ethanol (30, 50, 70, 85, 95, and 100%), transferred to 1,2-propylene oxide, and infiltrated with a series of EPON 812 (EMS) solutions (25, 50, 75, and 100% in 1,2-propylene oxide). The resin was polymerized at 70°C for 16 h and sectioned. Images of the chloroplast structure were observed and captured using a Tecnai G2 Spirit TWIN electron microscope (FEI) and DigitalMicrograph acquisition software (Gatan).
Accession Numbers
Sequence data for this article can be found in the GenBank/EMBL data libraries under the following accession numbers: DGD1 (At3g11670), LOX2 (At3g45140), PDF1.2 (At5g44420), COI1 (At2g39940), AOS (At5g42650), CESA3 (At5g05170), eRF1 (At5g47880), ELP1 (At1g05850), WRKY12 (At2g44745), DET3 (At1g12840), CPK28 (At5g66210), EBP (At3g16770), ETR2 (At3g23150), DAD1 (At2g44810), DGL (At1g05800), PLA-Iγ3 (At1g51440), PLA-Iγ2 (At2g30550), PLA-Iγ1 (At1g06800), PLA-Iβ2 (At4g16820), and PLA-Iα2 (At2g31690).
Supplemental Data
Supplemental Figure 1. Visible Phenotypes of the Wild Type and the Two dgd1 Mutants.
Supplemental Figure 2. Expression of Genes, Mutation of Which Results in Ectopic Lignification.
Supplemental Figure 3. The dgd1 Phenotypes Are Not Caused by Activation of Auxin or Ethylene Signaling.
Supplemental Table 1. Lipid Composition of the Wild Type and Various Mutant Plants.
Supplemental Table 2. Primers Used for Quantitative RT-PCR and Genotyping.
Supplemental Data Set 1. Fatty Acid Composition of All Lipids Analyzed.
Supplementary Material
Acknowledgments
We thank Su-Ping Tsay for assistance with electron microscopy, Wataru Sakamoto for providing vipp1 knockdown mutant seeds, Hsu-Liang Hsieh for providing coi1-30 mutant seeds, Long-Chi Wang for providing ein2 mutant (SAIL_265_D03) seeds, Bettina Hause for providing the antiserum against Arabidopsis AOC and the IMB English Editing Core, and Tom Barkas for English editing. This work was supported by the Ministry of Science and Technology, Taiwan (Grants NSC100-2321-B-001-011, NSC101-2918-I-001-012, and MOST104-2321-B-001-021 to H.-m.L.), and Academia Sinica of Taiwan (to H.-m.L.).
AUTHOR CONTRIBUTIONS
H.-m.L. and I.F. designed the research. H.-m.L., Y.-T.L., L.-J.C., and C.H. performed the experiments. H.-m.L., Y.-T.L., L.-J.C., C.H., and I.F. analyzed the data. H.-m.L. wrote the article.
Glossary
- MGDG
monogalactosyldiacylglycerol
- DGDG
digalactosyldiacylglycerol
- GGDG
glucosyl-galactosyl diacylglycerol
- JA
jasmonic acid
- OPDA
12-oxo-phytodienoic acid
- dn-OPDA
dinor-12-oxo-phytodienoic acid
- JA-Ile
jasmonic acid-isoleucine
- MS
Murashige and Skoog
- PC
phosphatidylcholine
- TAG
triacylglycerol
- DAG
diacylglycerol
- SRM
single reaction monitoring
Footnotes
Articles can be viewed without a subscription.
References
- Acosta I.F., Farmer E.E. (2010). Jasmonates. The Arabidopsis Book 8: e0129, doi/10.1199/tab.0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altamura M., Marco Possenti M., Antonella Matteucci A., Simona Baima S., Ida Ruberti I., Morelli G. (2001). Development of the vascular system in the inflorescence stem of Arabidopsis. New Phytol. 151: 381–389. [Google Scholar]
- Benedetti C.E., Xie D., Turner J.G. (1995). Coi1-dependent expression of an Arabidopsis vegetative storage protein in flowers and siliques and in response to coronatine or methyl jasmonate. Plant Physiol. 109: 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S., Weichert H., Porzel A., Wasternack C., Kühn H., Feussner I. (2001). Enzymatic and non-enzymatic lipid peroxidation in leaf development. Biochim. Biophys. Acta 1533: 266–276. [DOI] [PubMed] [Google Scholar]
- Boudière L., Michaud M., Petroutsos D., Rébeillé F., Falconet D., Bastien O., Roy S., Finazzi G., Rolland N., Jouhet J., Block M.A., Maréchal E. (2014). Glycerolipids in photosynthesis: composition, synthesis and trafficking. Biochim. Biophys. Acta 1837: 470–480. [DOI] [PubMed] [Google Scholar]
- Brioudes F., Joly C., Szécsi J., Varaud E., Leroux J., Bellvert F., Bertrand C., Bendahmane M. (2009). Jasmonate controls late development stages of petal growth in Arabidopsis thaliana. Plant J. 60: 1070–1080. [DOI] [PubMed] [Google Scholar]
- Brüx A., Liu T.Y., Krebs M., Stierhof Y.D., Lohmann J.U., Miersch O., Wasternack C., Schumacher K. (2008). Reduced V-ATPase activity in the trans-Golgi network causes oxylipin-dependent hypocotyl growth inhibition in Arabidopsis. Plant Cell 20: 1088–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldelari D., Wang G., Farmer E.E., Dong X. (2011). Arabidopsis lox3 lox4 double mutants are male sterile and defective in global proliferative arrest. Plant Mol. Biol. 75: 25–33. [DOI] [PubMed] [Google Scholar]
- Caño-Delgado A., Penfield S., Smith C., Catley M., Bevan M. (2003). Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J. 34: 351–362. [DOI] [PubMed] [Google Scholar]
- Chauvin A., Caldelari D., Wolfender J.L., Farmer E.E. (2013). Four 13-lipoxygenases contribute to rapid jasmonate synthesis in wounded Arabidopsis thaliana leaves: a role for lipoxygenase 6 in responses to long-distance wound signals. New Phytol. 197: 566–575. [DOI] [PubMed] [Google Scholar]
- Dörmann P., Benning C. (2002). Galactolipids rule in seed plants. Trends Plant Sci. 7: 112–118. [DOI] [PubMed] [Google Scholar]
- Dörmann P., Hoffmann-Benning S., Balbo I., Benning C. (1995). Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell 7: 1801–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger D., Stingl N., Kubigsteltig I.I., Bals T., Juenger M., Pollmann S., Berger S., Schuenemann D., Mueller M.J. (2010). DONGLE and DEFECTIVE IN ANTHER DEHISCENCE1 lipases are not essential for wound- and pathogen-induced jasmonate biosynthesis: Redundant lipases contribute to jasmonate formation. Plant Physiol. 153: 114–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feussner I., Wasternack C. (2002). The lipoxygenase pathway. Annu. Rev. Plant Biol. 53: 275–297. [DOI] [PubMed] [Google Scholar]
- Fiehn O., Kopka J., Dörmann P., Altmann T., Trethewey R.N., Willmitzer L. (2000). Metabolite profiling for plant functional genomics. Nat. Biotechnol. 18: 1157–1161. [DOI] [PubMed] [Google Scholar]
- Froehlich J.E., Itoh A., Howe G.A. (2001). Tomato allene oxide synthase and fatty acid hydroperoxide lyase, two cytochrome P450s involved in oxylipin metabolism, are targeted to different membranes of chloroplast envelope. Plant Physiol. 125: 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S., Kobayashi K., Nakamura Y., Wada H. (2014). Inducible knockdown of MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE1 reveals roles of galactolipids in organelle differentiation in Arabidopsis cotyledons. Plant Physiol. 166: 1436–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda H. (2004). Signals that control plant vascular cell differentiation. Nat. Rev. Mol. Cell Biol. 5: 379–391. [DOI] [PubMed] [Google Scholar]
- Glauser G., Dubugnon L., Mousavi S.A., Rudaz S., Wolfender J.L., Farmer E.E. (2009). Velocity estimates for signal propagation leading to systemic jasmonic acid accumulation in wounded Arabidopsis. J. Biol. Chem. 284: 34506–34513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guskov A., Kern J., Gabdulkhakov A., Broser M., Zouni A., Saenger W. (2009). Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride. Nat. Struct. Mol. Biol. 16: 334–342. [DOI] [PubMed] [Google Scholar]
- Härtel H., Lokstein H., Dörmann P., Grimm B., Benning C. (1997). Changes in the composition of the photosynthetic apparatus in the galactolipid-deficient dgd1 mutant of Arabidopsis thaliana. Plant Physiol. 115: 1175–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause B., Weichert H., Höhne M., Kindl H., Feussner I. (2000). Expression of cucumber lipid-body lipoxygenase in transgenic tobacco: Lipid-body lipoxygenase is correctly targeted to seed lipid bodies. Planta 210: 708–714. [DOI] [PubMed] [Google Scholar]
- Hölzl G., Witt S., Gaude N., Melzer M., Schöttler M.A., Dörmann P. (2009). The role of diglycosyl lipids in photosynthesis and membrane lipid homeostasis in Arabidopsis. Plant Physiol. 150: 1147–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölzl G., Witt S., Kelly A.A., Zähringer U., Warnecke D., Dörmann P., Heinz E. (2006). Functional differences between galactolipids and glucolipids revealed in photosynthesis of higher plants. Proc. Natl. Acad. Sci. USA 103: 7512–7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y., et al. (2008). Cooperation and functional diversification of two closely related galactolipase genes for jasmonate biosynthesis. Dev. Cell 14: 183–192. [DOI] [PubMed] [Google Scholar]
- Ibrahim A., Schütz A.L., Galano J.M., Herrfurth C., Feussner K., Durand T., Brodhun F., Feussner I. (2011). The alphabet of galactolipids in Arabidopsis thaliana. Front. Plant Sci. 2: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro S., Kawai-Oda A., Ueda J., Nishida I., Okada K. (2001). The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13: 2191–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iven T., König S., Singh S., Braus-Stromeyer S.A., Bischoff M., Tietze L.F., Braus G.H., Lipka V., Feussner I., Dröge-Laser W. (2012). Transcriptional activation and production of tryptophan-derived secondary metabolites in Arabidopsis roots contributes to the defense against the fungal vascular pathogen Verticillium longisporum. Mol. Plant 5: 1389–1402. [DOI] [PubMed] [Google Scholar]
- Joyard J., Ferro M., Masselon C., Seigneurin-Berny D., Salvi D., Garin J., Rolland N. (2010). Chloroplast proteomics highlights the subcellular compartmentation of lipid metabolism. Prog. Lipid Res. 49: 128–158. [DOI] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2012). JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 17: 22–31. [DOI] [PubMed] [Google Scholar]
- Kelly A.A., Dörmann P. (2002). DGD2, an Arabidopsis gene encoding a UDP-galactose-dependent digalactosyldiacylglycerol synthase is expressed during growth under phosphate-limiting conditions. J. Biol. Chem. 277: 1166–1173. [DOI] [PubMed] [Google Scholar]
- Kelly A.A., Froehlich J.E., Dörmann P. (2003). Disruption of the two digalactosyldiacylglycerol synthase genes DGD1 and DGD2 in Arabidopsis reveals the existence of an additional enzyme of galactolipid synthesis. Plant Cell 15: 2694–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Nakamura Y., Ohta H. (2009). Type A and type B monogalactosyldiacylglycerol synthases are spatially and functionally separated in the plastids of higher plants. Plant Physiol. Biochem. 47: 518–525. [DOI] [PubMed] [Google Scholar]
- Kourtchenko O., Andersson M.X., Hamberg M., Brunnström A., Göbel C., McPhail K.L., Gerwick W.H., Feussner I., Ellerström M. (2007). Oxo-phytodienoic acid-containing galactolipids in Arabidopsis: jasmonate signaling dependence. Plant Physiol. 145: 1658–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler H. (1987). Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 148: 350–382. [Google Scholar]
- Matschi S., Hake K., Herde M., Hause B., Romeis T. (2015). The calcium-dependent protein kinase CPK28 regulates development by inducing growth phase-specific, spatially restricted alterations in jasmonic acid levels independent of defense responses in Arabidopsis. Plant Cell 27: 591–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matschi S., Werner S., Schulze W.X., Legen J., Hilger H.H., Romeis T. (2013). Function of calcium-dependent protein kinase CPK28 of Arabidopsis thaliana in plant stem elongation and vascular development. Plant J. 73: 883–896. [DOI] [PubMed] [Google Scholar]
- Mosblech A., Feussner I., Heilmann I. (2009). Oxylipins: structurally diverse metabolites from fatty acid oxidation. Plant Physiol. Biochem. 47: 511–517. [DOI] [PubMed] [Google Scholar]
- Mosblech A., Thurow C., Gatz C., Feussner I., Heilmann I. (2011). Jasmonic acid perception by COI1 involves inositol polyphosphates in Arabidopsis thaliana. Plant J. 65: 949–957. [DOI] [PubMed] [Google Scholar]
- Newman L.J., Perazza D.E., Juda L., Campbell M.M. (2004). Involvement of the R2R3-MYB, AtMYB61, in the ectopic lignification and dark-photomorphogenic components of the det3 mutant phenotype. Plant J. 37: 239–250. [DOI] [PubMed] [Google Scholar]
- Nilsson A.K., Fahlberg P., Ellerström M., Andersson M.X. (2012). Oxo-phytodienoic acid (OPDA) is formed on fatty acids esterified to galactolipids after tissue disruption in Arabidopsis thaliana. FEBS Lett. 586: 2483–2487. [DOI] [PubMed] [Google Scholar]
- Noir S., Bömer M., Takahashi N., Ishida T., Tsui T.L., Balbi V., Shanahan H., Sugimoto K., Devoto A. (2013). Jasmonate controls leaf growth by repressing cell proliferation and the onset of endoreduplication while maintaining a potential stand-by mode. Plant Physiol. 161: 1930–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., Morreel K., De Witte E., Lammertyn F., Van Montagu M., Boerjan W., Inzé D., Goossens A. (2008). Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. USA 105: 1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsch K.A., Mylne J., Botella J.R. (2005). Cosuppression of eukaryotic release factor 1-1 in Arabidopsis affects cell elongation and radial cell division. Plant Physiol. 139: 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C., Han C., Peng W., Huang Y., Peng Z., Xiong X., Zhu Q., Gao B., Xie D. (2009). A leaky mutation in DWARF4 reveals an antagonistic role of brassinosteroid in the inhibition of root growth by jasmonate in Arabidopsis. Plant Physiol. 151: 1412–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell D.J., Kessler F., Blobel G. (1994). Isolation of components of the chloroplast protein import machinery. Science 266: 1007–1012. [DOI] [PubMed] [Google Scholar]
- Schommer C., Palatnik J.F., Aggarwal P., Chételat A., Cubas P., Farmer E.E., Nath U., Weigel D. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz M., Smith R., Ellis B. (2013). Xylem tissue specification, patterning, and differentiation mechanisms. J. Exp. Bot. 64: 11–31. [DOI] [PubMed] [Google Scholar]
- Sehr E.M., Agusti J., Lehner R., Farmer E.E., Schwarz M., Greb T. (2010). Analysis of secondary growth in the Arabidopsis shoot reveals a positive role of jasmonate signalling in cambium formation. Plant J. 63: 811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo Y.S., Kim E.Y., Kim J.H., Kim W.T. (2009). Enzymatic characterization of class I DAD1-like acylhydrolase members targeted to chloroplast in Arabidopsis. FEBS Lett. 583: 2301–2307. [DOI] [PubMed] [Google Scholar]
- Steffen R., Kelly A.A., Huyer J., Dörmann P., Renger G. (2005). Investigations on the reaction pattern of photosystem II in leaves from Arabidopsis thaliana wild type plants and mutants with genetically modified lipid content. Biochemistry 44: 3134–3142. [DOI] [PubMed] [Google Scholar]
- Stenzel I., Hause B., Miersch O., Kurz T., Maucher H., Weichert H., Ziegler J., Feussner I., Wasternack C. (2003). Jasmonate biosynthesis and the allene oxide cyclase family of Arabidopsis thaliana. Plant Mol. Biol. 51: 895–911. [DOI] [PubMed] [Google Scholar]
- Swiatek A., Azmi A., Stals H., Inzé D., Van Onckelen H. (2004). Jasmonic acid prevents the accumulation of cyclin B1;1 and CDK-B in synchronized tobacco BY-2 cells. FEBS Lett. 572: 118–122. [DOI] [PubMed] [Google Scholar]
- Swiatek A., Lenjou M., Van Bockstaele D., Inzé D., Van Onckelen H. (2002). Differential effect of jasmonic acid and abscisic acid on cell cycle progression in tobacco BY-2 cells. Plant Physiol. 128: 201–211. [PMC free article] [PubMed] [Google Scholar]
- Tarazona, P., Feussner, K., and Feussner, I. (2015). Enhanced plant lipidomics method based on multiplexed LC-MS reveals additional insights into cold and drought-induced membrane remodeling. Plant J. 84: 621–633. [DOI] [PubMed]
- Tu S.L., Chen L.J., Smith M.D., Su Y.S., Schnell D.J., Li H.M. (2004). Import pathways of chloroplast interior proteins and the outer-membrane protein OEP14 converge at Toc75. Plant Cell 16: 2078–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Liu Z.B., Hagen G., Guilfoyle T.J. (1995). Composite structure of auxin response elements. Plant Cell 7: 1611–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Malek B., van der Graaff E., Schneitz K., Keller B. (2002). The Arabidopsis male-sterile mutant dde2-2 is defective in the ALLENE OXIDE SYNTHASE gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta 216: 187–192. [DOI] [PubMed] [Google Scholar]
- Vu H.S., Tamura P., Galeva N.A., Chaturvedi R., Roth M.R., Williams T.D., Wang X., Shah J., Welti R. (2012). Direct infusion mass spectrometry of oxylipin-containing Arabidopsis membrane lipids reveals varied patterns in different stress responses. Plant Physiol. 158: 324–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Avci U., Nakashima J., Hahn M.G., Chen F., Dixon R.A. (2010). Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc. Natl. Acad. Sci. USA 107: 22338–22343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Uddin M.I., Tanaka K., Yin L., Shi Z., Qi Y., Mano J., Matsui K., Shimomura N., Sakaki T., Deng X., Zhang S. (2014). Maintenance of chloroplast structure and function by overexpression of the rice MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE gene leads to enhanced salt tolerance in tobacco. Plant Physiol. 165: 1144–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C., Hause B. (2013). Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. (Lond.) 111: 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Moellering E.R., Fan J., Benning C. (2008). Mutation of a mitochondrial outer membrane protein affects chloroplast lipid biosynthesis. Plant J. 54: 163–175. [DOI] [PubMed] [Google Scholar]
- Yang D.L., et al. (2012). Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 109: E1192–E1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Kato Y., Otters S., Vothknecht U.C., Sakamoto W. (2012). Essential role of VIPP1 in chloroplast envelope maintenance in Arabidopsis. Plant Cell 24: 3695–3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Ripperger A., Ye Z.H. (2000). Ectopic deposition of lignin in the pith of stems of two Arabidopsis mutants. Plant Physiol. 123: 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.