Abstract
In this overview, we provide an update on recent progress made in understanding the mechanisms of action, physiological functions, and roles in disease of retinoic acid related orphan receptors (RORs). We are particularly focusing on their roles in the regulation of adaptive and innate immunity, brain function, retinal development, cancer, glucose and lipid metabolism, circadian rhythm, metabolic and inflammatory diseases and neuropsychiatric disorders. We also summarize the current status of ROR agonists and inverse agonists, including their regulation of ROR activity and their therapeutic potential for management of various diseases in which RORs have been implicated.
Keywords: retinoic acid-related orphan receptor, RORγ, RORα, RORβ, immunity, Th17 cells, innate lymphoid cells, autoimmune disease, retina, brain, cancer, metabolism, glucose homeostasis, lipid metabolism, insulin sensitivity, diabetes, circadian clock, agonists, cholesterol biosynthesis, autism, agonists, antagonist, transcription
1. Introduction
The retinoic acid-related orphan receptors alpha, beta, and gamma (RORα-γ, RORA-C or NR1F1–3) constitute a subfamily of nuclear receptors that function as ligand-dependent transcription factors [1–3]. By using different promoters and/or alternative splicing, each ROR gene produces several isoforms that vary only at their N-terminus. The RORa gene generates four isoforms, RORα1–4, while RORb and RORc each generate two isoforms [4–10]. Most isoforms exhibit a distinct tissue-specific pattern of expression and regulate different biological processes and target genes. For example, the expression of RORγ2, commonly referred to as RORγt, is restricted to several immune cell types, while RORγ1 is only expressed in various peripheral tissues, including liver, adipose tissue, skeletal muscle, and kidney [1, 7, 8, 11–14]. RORs are critical in the regulation of many physiological processes, including immunity, circadian rhythm, embryonic development, and several metabolic pathways, and have been implicated in several pathologies associated with those processes.
RORs exhibit a domain structure that is typical of nuclear receptors and contain an N-terminal domain, a highly conserved DNA-binding domain (DBD) consisting of two C2-C2 zinc finger motifs, a ligand-binding domain (LBD), and a hinge domain spacing the DBD and LBD [1]. The RORs regulate gene transcription by binding as monomers to ROR response elements (ROREs) consisting of the RGGTCA consensus preceded by an A/T-rich sequence in the regulatory regions of target genes [6, 15]. The ability to bind ROREs is shared with several other nuclear receptors, including the transcriptional repressors Rev-Erbα and Rev-Erbβ (NR1D1–2) [16]. By competing for RORE binding, these receptors can antagonize each other’s effects on transcription. For example, crosstalk between RORs and Rev-Erbs plays a role in the transcriptional regulation of a number of metabolic and clock genes [9, 16– 25].
Relatively little is known about posttranslational modifications and upstream signaling pathways that modulate ROR transcription activity. Protein kinase A (PKA) has been reported to activate RORα4, and although PKA phosphorylates RORα4 at Ser99, mutation of this site had little influence on RORα4 transcriptional activity [26], while phosphorylation of RORα4 at Thr128 by ERK2 enhances its transcriptional activity [27]. PGE2/PKCα-dependent phosphorylation of RORα has been reported to attenuate Wnt target gene expression in colon cancer cells [28], while sumoylation of RORα enhanced its transcriptional activity [29]. A recent study demonstrated that the deubiquitinase, DUB, interacts with and stabilizes the ubiquitin ligase UBR5 in response to TGF-β signaling [30]. This results in an increase in RORγt ubiquitination that leads to reduced RORγt stability and diminished transactivation of RORγt target genes in T-helper type 17 (Th17) cells. Another study reported that the protein deacetylase, Sirtuin 1 (SIRT1), deacetylates RORγt and increases its transcriptional activity, thereby enhancing Th17 generation [31].
Reports showing that cholesterol and cholesterol sulfate, as well as a series of other small molecules, were able to bind the LBD of RORs and modulate its transcriptional activity indicated that RORs function as ligand-dependent transcription factors [2, 25, 32, 33]. Recently, several intermediates of the cholesterol biosynthetic pathway were reported to act as endogenous agonists of RORγ [34, 35]. These studies revealed that RORγ transcriptional activity and the physiological processes it regulates, can be controlled by changes in the intracellular pool of these sterol intermediates. In addition, these discoveries raised the possibility that ROR ligands might be valuable in the development of new therapeutic strategies for diseases in which RORs are implicated, including various inflammatory and metabolic diseases and neuropsychiatric disorders. In this review, we summarize several areas of ROR research in which recently significant progress has been made.
2. RORs in Adaptive Immunity
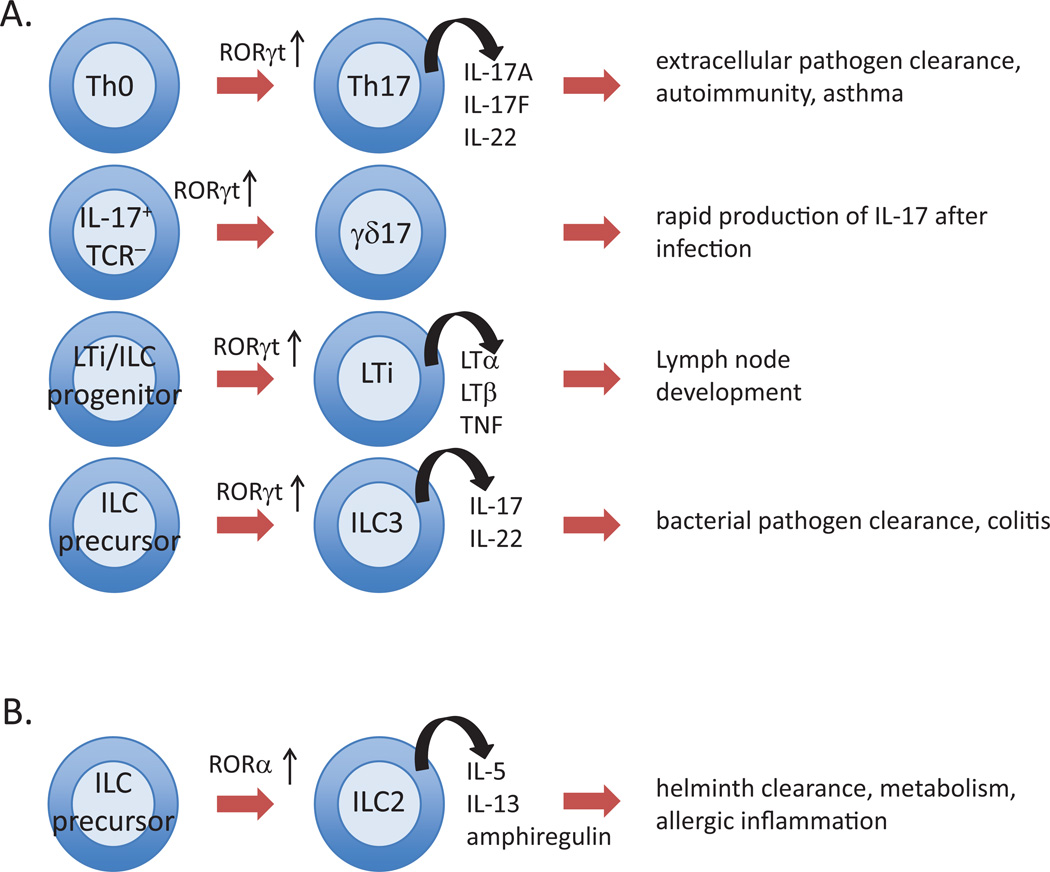
The innate and adaptive immune systems are highly integrated and serve to protect the host from being overwhelmed by pathogen invasion. Innate immune responses are immediate and utilize germline-encoded receptors to recognize and respond to pathogens, whereas adaptive immunity is a delayed response that requires expansion of a small number of cells bearing antigen-specific receptors on the surface of lymphocytes. Genetically modified mice lacking RORα or RORγt have revealed that each receptor plays a key role in the development of several immune cells and are critical for some immune responses (Figure 1).
Figure 1. Multiple functions of RORs in lymphocyte development.
A. Roles of RORγt in the development of Th17 cells, γδ-17 T cells, lymphoid tissue inducer (LTi) cells and innate lymphoid cells 3 (ILC3) cells. (B) Role of RORα in the development of ILC2 cells. RORα has also a role in the regulation of Th17 cells.
Lymphoid progenitor cells undergo several stages of differentiation in the thymus prior to becoming mature T cells. These stages can be identified in part by display of CD4 and CD8 on the cell surface. RORγt is selectively expressed in T cells that display both CD4 and CD8, typically called double positive (DP) cells. RORγt is required in these cells for expression of the anti-apoptotic gene Bcl-XL [36–40]. Bcl-XL expression is repressed in DP thymocytes of RORγ null mice, resulting in accelerated apoptosis in vivo and in vitro. Consequently, thymi of RORγ null mice have reduced numbers of DP cells and their descendants, including single positive (SP) mature CD4+ T helper cells (Th) and CD8+ cytotoxic cells.
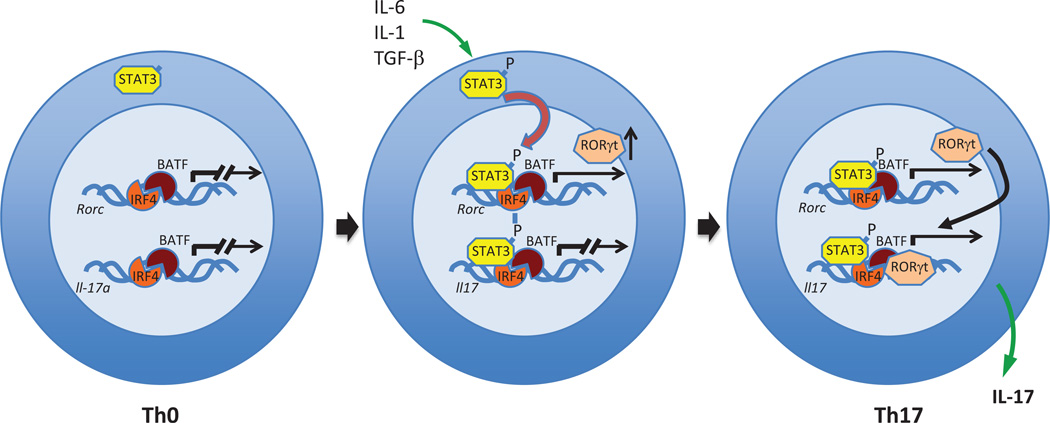
Mature, but naïve CD4+T (Th0) cells can be differentially polarized to produce the cytokines characteristic of Th1, Th2 and Th17 cells [1, 41]. RORγt is required for the development of Th17 cells [12, 13, 42–45], whose name reflects their ability to produce the cytokines IL-17A and IL-17F, as well as IL-21 and IL-22. Like RORγt, RORα can also contribute to Th17 development and acts synergistically with RORγt in this regard [13, 44]. Th17 cells protect against extracellular pathogens, but are also associated with various diseases, such as rheumatoid arthritis, inflammatory bowel disease, multiple sclerosis and asthma [46, 47]. Forced expression of RORγt is sufficient for the induction of several Th17-associated genes, including Il17, Il22, Ccr6, and the IL-23 receptor (Il23r) [12, 48, 49]. Combinations of promoter and chromatin immunoprecipitation (ChIP) analysis, and cistrome mapping showed that RORγt is recruited to ROREs in several Th17 marker genes, including Il17a, Il17f, Irf4 and Il23r, and directly regulate their transcription [43, 44, 50]. However, in addition to RORγt, several other transcriptional factors are necessary to induce the full Th17 differentiation program, including BATF [51], IRF4 [52], and STAT3 [53]. Recent advances in ChIP and RNA-seq technologies have shed light on the hierarchy and order of transcription factor occupancy during Th17 differentiation [54, 55]. In Th0 cells, the transcription factors, IRF4 and BATF, are cooperatively bound to overlapping sites in chromatin near several genes, including Rorc and Il17, thereby increasing chromatin accessibility to other transcription factors (Figure 2). In response to TGFβ and IL-6, STAT3 becomes phosphorylated (pSTAT3) and moves to the nucleus, where it binds to chromatin and induces expression of Rorc. RORγt and pSTAT3 then cooperate with IRF4 or BATF and other factors to increase expression of Th17-associated genes, including Il17a, Il17f, Il23r, Ccl20 and Il1r1 [55]. Thus, IRF4 and BATF have broad and self-reinforcing effects on chromatin remodeling, whereas RORγt specifically regulates a relatively small number of key Th17-associated genes in a manner potentially responsive to environmental cues. In addition to transcriptional control by IRF4/BATF/STAT4, the PI3K-Akt-mTORC1-S6K1/1 cell signaling axis has also been linked to the control of Th17 differentiation by RORγt [56]. Activation of PI3K-Akt-mTORC1 induces ribosomal protein S6 kinase β2 (RPS6KB2) expression that subsequently promotes the nuclear localization of RORγt and RORγt-mediated Th17 differentiation.
Figure 2. RORγt-dependent induction of Th17 differentiation and Th17-associated genes.
In Th0 cells, IRF4 and BATF are bound to chromatin near Th17-associated genes, but the loci are transcriptionally silent. Upon exposure to cytokines, such as IL-6, STAT-3 becomes phosphorylated and transfers to the nucleus, where it binds DNA near IRF4 and BATF and induces Rorc transcription. RORγt can then join the IRF4/BATF/STAT3 transcription factor complex and induce expression of Th17-associated genes, such as Il17 and Il23r.
The differentiation of Th0 cells into Treg and Th17 cells is dependent on the balance between the level of expression of Foxp3 and RORγt, respectively. Through its interaction with RORγt, Foxp3 inhibits RORγt function and promotes Treg differentiation [1, 57, 58]. This balance is controlled by the concentration of specific cytokines in the environmental milieu. Foxp3 expression and consequent Treg development is favored in cultures containing high levels of TGF-β, IL-2 and retinoic acid, whereas Th17 development is promoted by low amounts of TGF-β in combination with the proinflammatory cytokines, IL-6 and IL-1 [58– 60]. IL-1 can repress the suppressor of cytokine signaling 3 (SOCS3), an inhibitor of STAT3 phosphorylation [61], thereby increasing Rorc expression. Th17 cells share an overlapping developmental program with that of inducible regulatory T cells (iTregs) [62]. In the small intestine, a number of RORγt+Foxp3+ T cells do not produce IL-17, but instead express IL-10. These RORγt+ Tregs develop outside the thymus and require gut microbiota for their development. In addition, dietary vitamin A favors the generation of these RORγt+ Tregs over that of Th17 cells [62]. RORγt+ Treg cells regulate Th2 cells - but not Th1 or Th17 cells -through a CTLA4-dependent regulation of CD80 and CD86 on dendritic cells.
Th17 and IL17 have been implicated in several inflammatory and autoimmune diseases. Mice lacking RORγ are partially protected against the development of diseases, including autoimmune diseases such as experimental autoimmune encephalomyelitis (EAE) and type II collagen-induced arthritis, as well as allergen-induced lung inflammation [12, 44, 58, 63]. Mice lacking both RORα and RORγ are greatly protected from EAE [44]. Although IL-17A, IL-17F and IL-22 are the signature cytokines of Th17 cells, they appear not to be sufficient for pathogenicity in EAE [64, 65]. In this model, RORγt-dependent production of granulocyte macrophage colony stimulating factor (GM-CSF) is reported to drive the effector phase of neuroinflammation [66, 67]. However, the molecular requirements of pathogenicity might depend on the disease model because either IL-17A or IL-17F is required for Th17 cell-mediated intestinal inflammation [68]. Together, these studies raise the possibility that RORγ antagonists might be useful in the management of autoimmune disease.
3. RORs in Innate Immunity
Like conventional αβ T cell receptor (TCR)+ cells, T cells expressing the γ and δ TCR chains (γδ T cells) develop in the thymus, but they have a more limited repertoire than αβ TCR+ cells and lack major histocompatibility complex (MHC) restriction [69]. Many γδ T cells express IL-17 and are thus termed γδ-17 T cells [70, 71]. Unlike αβ TCR Th17 cells, which acquire effector functions only after encountering their cognate antigens in peripheral tissue, many γδ-17 cells express IL-17 very early during their development in the thymus, even prior to TCR rearrangement [70, 71]. Thus, γδ-17 cells have the potential to be a major cell source of IL-17, especially during the early phases of disease, prior to the development of antigen-specific Th17 cells [14, 72]. There are some commonalities, but also differences in the molecular pathways leading to γδ-17 and Th17 cells. For both cell types, RORγt is critically important. Thus, mice deficient for RORγt lack γδ-17 cells in peripheral organs and lymphoid tissues [12]. However, the induction of RORγt in Th17 cells requires the canonical c-Rel-dependent NF-kB pathway, whereas γδ-17 cells require RelB and the noncanonical NF-kB pathway [73]. In addition, IRF4 is required for the induction of Th17 cells, but this transcription factor is dispensable for the development of γδ-17 T cells.
RORα and RORγ also play a critical role in the generation of innate lymphoid cells (ILCs). ILCs are a heterogeneous population of cells that possess the typical lymphoid cell morphology, but lack some cell surface molecules typically seen on lymphocytes [74]. In particular, ILCs lack TCRs and the associated CD3 complex found on conventional T cells. Consequently, ILCs cannot recognize specific antigens, and instead respond to cytokines produced during innate immune responses. ILCs have been classified into three groups, based on their cytokine production profiles and the transcription factors that regulate their development [75]. The cytokines produced by each of these groups mirrors those produced by specific T helper (Th) cell types: Group 1 ILCs and Th1 cells produce IFN-γ, Group 2 ILCs and Th2 cells produce IL-5 and IL-13, and Group 3 ILCs and Th17 cells produce IL-17A, IL17F and IL-22.
RORγt is required for the development of all ILC3s, a heterogeneous population of cells that also depends on IL-7 for their development. The first discovered member of the Group 3 ILCs is the lymphoid tissue inducer (LTi) cell, a type of CD4+CD3− cell that displays transmembrane lymphotoxin α1β2 [76, 77]. These cells are required for the development of secondary lymphoid organs, Peyer’s patches, and intestinal lymphoid follicles [78–81]. RORγt-deficient mice lack these cells and therefore do not develop secondary lymphoid organs [1, 36, 37, 82]. Recently, retinoic acid (RA) was found to control LTi cell maturation upstream of RORγt by positively regulating RORγt expression directly through the recruitment of RA receptors (RARs) to the promoter region of RORγt [83]. Impairment of LTi maturation in cells defective in RA signaling can be rescued by the exogenous expression of RORγt. More recently, other ILC3 subpopulations have been identified that, like Th17 cells, are abundant in the gut and can produce IL-17A, IL-17F, IL-22, GM-CSF, and TNF, suggesting their importance for clearing extra-cellular pathogens. Gata-3 is critical for the development of gut RORγt+ ILC3s subsets [84]. At least some ILC3s can transition to ILC1 cells [85], reminiscent of Th17 cell conversion to a Th1-like phenotype. This transition, which is accompanied by elevated levels of TBX21, is driven by cytokine signals in the cellular milieu.
Group 2 ILCs are the most homogeneous group of the ILCs and are dependent on RORα, but not RORγt, for their development [86, 87]. They display the cell surface markers IL-7Ra (CD127), IL-2RA (CD25), Sca-1, KLRG1 and the IL-33 receptor, ST2. ILC2s are defined based on their ability to produce an array of type 2 cytokines, including IL-4, IL-5, IL-9, and IL-13, as well the cytokine, amphiregulin, and are implicated in helminth clearance and allergic inflammation [88]. Several single nucleotide polymorphisms (SNPs) within the RORA gene are associated with increased susceptibility to asthma [89–91], and RORα null mice and ILC2-deficient mice generated by RORα-deficient bone marrow transplants have reduced type 2 cytokine production and partial protection from airway hyper-reactivity [87, 92]. RORα expression was significantly upregulated in patients with therapy-resistant asthma [93]. RORα expression was also found to be significantly elevated in skin from patients with atopic dermatitis (AD), while in an experimental model of AD-like inflammation RORα-deficient mice exhibit a profound deficit in ILC2 cells and significantly reduced allergic skin inflammation [88]. Together, these observations indicate that RORα plays a critical role in ILC2 cell lineage determination and control of allergy-induced inflammation in multiple tissues.
RORα also contributes to immune function in the intestinal epithelium by controlling the diurnal regulation of several pathogen recognition receptors, including Nod2 and various Toll-like receptors [94]. The expression of these genes in the intestine is reduced in RORα-deficient mice, particularly when RORα expression is at its highest and bound to promoter regions of these genes. Other genes whose diurnal expression is directly controlled by RORα include interleukin-1 receptor-associated kinase 1 (IRAK1), Toll-interleukin 1 receptor domain containing adaptor protein (TIRAP), and the clock genes, Bmal1 and nuclear factor, interleukin 3 regulated (NFIL3 or E4BP4). The decreased expression of RORα in the intestine of microbiota-depleted mice further provides additional evidence of a relationship between RORα, commensal bacteria and diurnal regulation of immune-related genes in the gut [94].
4. ROR Functions in Brain and Retina
RORα is highly expressed and developmentally regulated in several regions of the brain, including cerebellar Purkinje cells and the thalamus [1, 95–97]. Genetically modified mice, in which RORα is disrupted, and RORα-deficient staggerer (RORαsg/sg) mice display severe cerebellar ataxia due to cerebellar neurodegeneration [98–100]. Further characterization of these mice revealed that RORα plays a critical role in the regulation of the survival and differentiation of Purkinje cells from embryonic development throughout adulthood [98, 100–103]. Deletion of RORα in Purkinje cells between postnatal days 10–21 revealed that continued expression of RORα is necessary after neuronal maturation to maintain mature morphological and innervation characteristics in the adult Purkinje cells [103]. Loss of Purkinje cells was also observed in >6 months-old male RORα+/sg mice and to a much smaller degree in RORα+/sg females. The marked, age-related Purkinje cell death in RORα+/sg male mice has been linked to a premature decrease in circulating sex steroids, which have been shown to be neuroprotective, and appeared not to be due to changes in cerebellar neurosteroids [104]. Genome-wide gene expression studies showed that RORα regulates the expression of a number of genes linked to Purkinje cell maturation, particularly dendritic differentiation, and the glutamatergic pathway [98]. ChIP analysis demonstrated that RORα directly controls the transcription of several of these genes, including the glutamate transporter Slc1a6, the calmodulin inhibitor Pcp4, and the IP3 receptor (Itpr1). Several of the RORα target genes were found to be also down-regulated in Ski family transcriptional corepressor 2 (Skor2)-deficient mice; however, RORα expression was not altered in these mice [105]. It was proposed that RORα and Skor2 cooperate in regulating Purkinje cell differentiation and gene expression. In Purkinje cells, RORα also directly regulates the expression of sonic hedgehog (Shh)[98], which is required for the proliferation and survival of cerebellar granule precursor cells through its activation of Gli transcription factors. The degeneration of cerebellar granule cells observed in RORα-deficient mice can at least in part be attributed to this loss in Shh production.
Accumulating evidence indicates a role for RORα in several neuropsychiatric disorders, including autism spectrum and bipolar disorder (ASD), schizophrenia, depression, and posttraumatic stress syndrome [106–117]. Studies demonstrating that the expression of RORA was reduced in sections of cerebellum and cortex of autistic subjects and observations showing differential methylation of the RORA gene in lymphoblastoid cells from autistic and nonautistic siblings supported a role of RORα in the development of ASD [107]. This, together with reports showing reduced number and size of Purkinje cells (PC) in the majority of cerebellar specimens from ASD patients, as was observed in RORα-deficient mice (Chugani 2014), is consistent with a link between RORα, its regulation of Purkinje cell survival and differentiation, and ASD. A connection between RORα and ASD is further supported by genome-wide ChIP-Seq analysis showing that in human neuronal cells, RORα was associated with the promoter region of a number of ASD-associated genes, including ataxin binding protein (A2BP1), neuroligin 1 (NLGN1), and aromatase (CYP19A1) [118, 119]. Several of the genes down regulated in RORα-deficient neuronal cells, including aromatase, were also found to be repressed in the frontal brain of individuals with ASD. The positive regulation by RORα of aromatase, which converts testosterone to estrogen, is intriguing because estrogen has been reported to enhance RORα expression and to exhibit a neuroprotective effect, as reported for RORα. Thus, RORα, aromatase, and estrogen might be part of a positive regulatory pathway. Therefore, reduced expression of aromatase and estrogen production in RORα-deficiency might excerbate Purkinje cell death and enhance the risk for ASD. Loss of Purkinje cells was also observed in >6 months-old male RORα+/sg mice and to a much lesser degree in RORα+/sg females. The marked, age-related Purkinje cell death in RORα+/sg male mice was linked to a premature decrease in circulating sex steroids, which have been shown to be neuroprotective, and not due to changes in cerebellar neurosteroids [104]. Since both RORα and estrogen exhibit neuroprotective effects, this pathway might help to explain why RORα+/sg female mice are less susceptible to Purkinje cell loss during aging. Further indications for a link between RORα and ASD came from a recent study demonstrating that the microRNA MIR137, which has been implicated in ASD and schizophrenia, targets the 5’-UTR of RORα [108]. RORα has also been reported to have neuroprotective functions in neurons and astrocytes during hypoxia [120, 121]. Protecting brain cells from the damaging effects of injury and stress might be an important function of RORα and be relevant to several brain disorders.
RORβ is highly expressed in the suprachiasmatic nucleus, the retina and pineal gland, and has been implicated in the regulation of circadian, motor, and visual functions [3, 122, 123]. RORβ deficient mice displayed motor (“duck gait,” hind paw clasping reflex), olfactory deficits, reduced anxiety and learned helplessness-related behaviors, and alterations in circadian behavior [124]. Examination of RORβ expression during embryonic and postnatal development of the mouse neocortex showed that after E16.5 RORβ transcripts increasingly localized to the primary sensory areas and reached peak expression at P10 with strongest expression in the primary somatosensory, auditory, and visual areas [125, 126]. This developmental pattern of expression was similar to that reported for rat neocortex [127]. A possible connection was found between RORβ expression levels and the control of cytoarchitectural patterning of neocortical neurons during mouse development [128]. GWAS studies revealed an association between a series of RORB genetic variants with schizophrenia, and bipolar I disorder [113, 116, 129, 130] and between RORβ expression in the temporal cortex and verbal intelligence [131]. Similarly, a syndrome characterized by moderate facial dysmorphy, mental retardation, epilepsy, speech delay, and autistic behaviour in patients with a 9q21 deletion at the RORB locus identified RORβ as a strong candidate for this neurological disorder [132–134]. Prenatal ethanol exposure has been reported to lead to neurobiological damage in early development. Newborns prenatally exposed to alcohol show neuroanatomical defects in the neocortex and an abnormal neocortical expression pattern of RORβ that is associated with mental and intellectual dysfunction, and behavioral and motor deficits [135].
Recent studies showed that the RORβ1 and RORβ2 isoforms exhibit distinct patterns of expression during retinal development [136, 137]. RORβ was shown to play a critical role in regulating retinal progenitor proliferation and differentiation [123, 137–139]. RORβ1-deficient mice lack amacrine and horizontal interneurons, cells important for the integration of visual information, suggesting that RORβ1 is critical for the differentiation of retinal progenitors into these interneurons [136]. This involves direct transcriptional regulation of Ptf1a, a key factor required for the generation of amacrine and horizontal cells, by RORβ1. Re-expression of RORβ1 was able to rescue amacrine differentiation in RORβ null mice. Retinal progenitors can also differentiate into two morphologically, developmentally, and functionally distinct photoreceptors, rods and cones. Rods function in dim light, while cones mediate daylight, and in most mammals, color vision. Mice lacking RORβ lose rods, but overproduce primitive S cones that lack outer segments [139]. RORβ1 and RORβ2 control rod cell differentiation through its transcriptional regulation of neural retina leucine zipper factor (NRL), a transcription factor that promotes the differentiation of retinal progenitors into the rod cell lineage, while suppressing the cone cell lineage [137, 139, 140]. The lack of rod photoreceptors in RORβ null mice is in part due to the loss of NRL expression. This can be reversed by re-expression of NRL in these mice [139]. RORβ2 is expressed in rod photoreceptors and its transcription was shown to be directly regulated by NRL. Thus, RORβ2 and NRL form two positive feedback loops that synergistically promote the commitment to a rod cell lineage [137]. In addition to these roles, RORβ-deficient mice fail to induce S opsin appropriately during postnatal cone development, suggesting a function for RORβ in morphological maturation of cone photoreceptors [123]. RORβ was found to activate the S opsin gene (Opn1sw) expression through binding sites in the upstream promoter region.
5. Role of RORs in Cancer
Very little is known about the role of RORβ and RORγ in cancer. Initial analysis of RORγ knockout mice revealed that these mice develop T-cell lymphomas within the first months after birth that rapidly metastasize to liver and spleen [141]. The mechanism underlying the development of thymic lymphomas in RORγ null mice has yet to be elucidated. A recent study reported that low levels of RORγ mRNA expression in somatotroph adenomas was associated with reduced E-cadherin expression and increased epithelial mesenchymal transition (EMT), and increased tumor size and invasiveness [142]. Similarly, higher expression of RORγ correlated with longer metastasis-free survival in breast cancer [143]. With respect to RORβ and cancer, a recent study reported that RORB is overexpressed in primary leiomyosarcomas, the most common type of uterine sarcoma [144]. In contrast, RORB expression was found to be highly down-regulated in both serous and endometrioid types of endometrial cancer [145].
A number of studies reported that RORα expression is significantly down-regulated during tumor development and progression, while expression of exogenous RORα inhibited cell proliferation and tumor growth [146–151]. Reduced RORα expression has been observed in colorectal and mammary carcinomas and found to be associated with poorer prognosis in hepatocellular and breast carcinoma patients [146, 147, 150, 152, 153]. In addition, silencing RORα in mammary epithelial cells significantly enhanced cell proliferation in ductal epithelial cells and promoted side branching of mammary ducts, suggesting that RORα has an important role in mammary gland branching morphogenesis [150]. Conversely, restoring RORα expression in cultured breast cancer cells was shown to inhibit cell migration and suppress tumor growth and metastasis in nude mice. This was accompanied by enhanced expression of semaphorin 3F (SEMA3F), a tumor suppressor that inhibits tumor growth and invasiveness. RORα was shown to regulate SEMA3F transcription directly through ROREs in its promoter region [150]. A different study indicated a role for SPARC, which is critical in the regulation of cell growth and adhesion, in the anti-tumor and anti-proliferation effects of RORα in human hepatoma cells [154]. SPARC was found to be a direct target gene of RORα. Treatment of colon carcinoma HCT116 cells with DNA-damage agents led to a p53-dependent increase in RORα expression that is directly mediated through functional p53 response elements in the RORa promoter [155]. RORα itself stabilized p53 by inhibiting its ubiquitination and enhanced p53 transcription in a HAUSP/Usp7-dependent manner leading to increased apoptosis. The connection between RORα and p53 was supported by a report demonstrating that treatment of hepatocellular carcinoma cells with an RORα agonist enhanced p53 stability [156]. This increase was shown to involve elevated SOX4 transcription, a gene critically involved in MDM2-dependent regulation of p53 stability. Another role for RORα in cancer was revealed by a recent study demonstrating that RORα expression was decreased in tumor tissues compared to adjacent normal tissues in human hepatocellular carcinoma patients and that this was associated with a change in glucose metabolism [157]. RORα was shown to inhibit pyruvate dehydrogenase kinase 2 (PDK2) expression and phosphorylation, thereby promoting aerobic glycolysis rather than oxidative phosphorylation, whereas reduced RORα expression in tumor cells promotes oxidative phosphorylation and tumor cell growth.
Several studies revealed a role for a noncanonical RORα pathway in cancer that does not involve RORE binding, but in which RORα functions as a transcriptional cofactor. In colon carcinoma cells, RORα was shown to bind β-catenin directly and inhibit β-catenin-mediated transcriptional activation of the target genes, cyclin D1 (CCND1) and c-myc (MYC), resulting in repression of cell proliferation and migration [158]. RORα was found to be recruited to the lymphoid enhancer-binding factor 1 (LEF1)-binding sites of the LEF1-target gene, CCND1, together with LEF1 and β-catenin. RORα interacted with residues within the armadillo repeat domains of β-catenin, which function as binding sites for a subset of coactivators. The interaction of RORα with β-catenin required the N-terminus of RORα and was dependent on the phosphorylation of Ser35 by protein kinase Cα (PKCα) activated by the noncanonical Wnt pathway. Interestingly, in many colorectal carcinomas phosphorylation of RORα was reduced. In another noncanonical pathway, RORα interacts with the heptad repeat and marked box region of the transcription factor E2F1 and suppressed E2F1-regulated transcription and cell cycle progression in epithelial cells [159]. In mammary ducts, RORα levels inversely correlated with the expression of E2F1 target genes and cell proliferation. Binding of RORα was shown to inhibit E2F1 acetylation and its DNA-binding activity by enhancing its interaction with histone deacetylase 1 (HDAC1). Knockdown of HDAC1 or inhibition of HDAC activity partially reversed the repression of E2F1 activity by RORα. In contrast to its growth inhibitory effects, RORα was shown to enhance the proliferation in mammary carcinoma MCF7 cells and significantly induced the expression of aromatase mRNA by binding an RORE in the aromatase promoter region [160]. It was proposed that the increase in aromatase expression by RORα accelerates the local production of estrogen, which then enhances the proliferation of breast cancer cells.
6. RORs in the Regulation of Metabolism
Both RORα and RORγ have been implicated in the control of energy homeostasis and the regulation of several lipid and glucose metabolic genes [3, 22, 24, 161–169]. Regulation of energy homeostasis is a complex process that involves multiple interrelated glucose and lipid metabolic pathways in many organs and is controlled by the circadian clock, gut microbiota, and by the endocrine, immune and nervous systems [170–173]. This has made it difficult to determine whether the metabolic changes observed in ROR-deficiency are cause or effect. RORα-deficient (staggerer) mice were shown to be protected against high fat diet (HFD)-induced metabolic syndrome as indicated by reduced weight gain, adiposity and hepatic steatosis, and improved insulin sensitivity [161, 174, 175]. Adipocytes in RORα-deficient mice fed a high fat diet accumulated considerably less lipid and the infiltration of inflammatory macrophages and expression of several inflammatory genes, including interleukin 6 (Il6), Toll-like receptor 8 (Trl8), and chemokine (C-C motif) ligand 8 (Ccl8), were greatly diminished [174]. Interleukin-1 receptor antagonist (Il1rn) was among the genes most dramatically repressed in white adipose tissue (WAT) of RORα-deficient mice. This gene has been implicated in the regulation of obesity and insulin resistance, suggesting that the reduced susceptibility to metabolic syndrome in RORα-deficient mice might at least in part be attributed to Il1rn repression [174, 176]. WAT-associated inflammation plays a critical role in the development of metabolic syndrome [172, 177]. The reduced inflammation observed in RORα-deficient mice might be in part responsible for the improved insulin sensitivity in these mice. A role for RORα in the regulation of insulin sensitivity is supported by a study showing an association between a single nucleotide polymorphism in RORα (rs7164773) and an increased risk for type 2 diabetes in the Mexico Mestizo population [178]. A recent study showed that the expression of several thermogenic genes, such as uncoupling protein 1 (Ucp1) and deiodinase 2 (Dio2), markers of brown adipose tissue (BAT), was enhanced in adipose tissue from RORα-deficient mice. This was associated with increased expression of the histone-lysine N-methyltransferase 1 (Ehmt1), a gene that controls BAT specification and maintenance [175, 179]. The greater cold-tolerance of RORα-deficient mice appears to be related to the increased expression of these genes, leading to increased oxygen consumption and heat generation from lipid oxidation that likely contributes to the improved energy homeostasis and insulin-sensitivity observed in these mice. Both RORα and RORγ have been shown to be induced during adipocyte differentiation in 3T3-L1 cells [180]; however, exogenous expression of RORα inhibits adipocyte differentiation in 3T3-L1 cells, as indicated by the reduced induction of fatty acid binding protein 4 (Fabp4), perilipin 1 (Plin1) and fatty acid synthase (Fasn) [181].
In addition to WAT, loss of RORα induces changes in gene expression in macrophages and liver. Disruption of RORα in macrophages leads to diminished expression of cholesterol 25-hydroxylase (Ch25h), which converts cholesterol to 25-hydroxycholesterol, and reduced phagocytosis [182, 183]. Interestingly, addition of 25-hydroxycholesterol was able to reverse the inhibition of phagocytosis in RORα-deficient macrophages suggesting a link between oxysterol metabolism and the regulation of phagocytosis. In the liver, the expression of a large number of genes related to lipid and glucose metabolism were found to be down-regulated in RORα-deficient mice fed a HFD [174]. These included phosphoenolpyruvate carboxykinase (Pepck) and glucose-6 phosphatase (G6pc), which play a role in gluconeogenesis, fibroblast growth factor 21 (Fgf21), which is an important regulator of glucose and lipid homeostasis, and genes involved in triglyceride synthesis and storage, such as glycerol-3-phosphate acyltransferase (Gpam), perilipin 2 (Plin2), monoacylglycerol O-acyltransferase 1 (Mogat1), and cell death-inducing DFFA-like effector a (Cidea) [154, 166, 174, 184]. In addition, the hepatic expression of several genes involved in sterol and bile acid metabolism, including cytochrome P450 8b1 (Cyp8b1), Cyp7b1, and sulfotransferase Sul1b1 were significantly diminished in RORα-deficient mice [153, 167, 174, 185]. However, the hepatic expression of sulfotransferase Sult1e1 was found to be dramatically induced in both male and female RORα-, but not in RORγ-deficient mice, whereas Sult2a1, known to sulfonate bile acids, hydroxysteroid dehydroepiandrosterone, and related androgens, was increased in both RORα- and RORγ-deficient mice, but only in female mice [167]. In contrast, in cultured human hepatocytes and hepatoma HepG2 cells, exogenous expression of RORα induced SULT2A1, while RORα knockdown with siRNAs decreased its expression [153]. Moreover, overexpression of RORα inhibited LXR and SREBP expression as well as lipid accumulation in these cells [186]. Adenovirus-mediated overexpression of RORα in liver also reduced triglyceride levels in mice fed a high fat diet. The cause of the discrepancy between the observations in RORα-deficient mice and those in HepG2 and liver overexpressing RORα has yet to be understood. ChIP and promoter analysis indicated that many metabolic genes, including G6pc, Adfp, Cyp7b1, citrate synthase (Cs), Cyp2c8, Fgf21, secreted protein, acidic, cysteine-rich (Sparc), Sult1b1, and Sult2a1, were directly regulated by RORα in HepG2 cells [153, 154, 166, 174, 185, 187, 188]. RORα cistrome data [165] revealed that in liver, RORα was recruited to ROREs in several genes important in glucose homeostasis and lipid metabolism, including G6pc, Fasn, Pepck1, Apoa1, and Elovl3, indicating that RORα positively regulates the transcription of these metabolic genes by binding ROREs in their regulatory region.
RORα-deficient mice also display metabolic changes in skeletal muscle that are accompanied by alterations in the expression of several genes [169]. Glucose uptake in skeletal muscle of RORα-deficient mice was enhanced and found to be associated with increased phosphatidylinositol 3-kinase signaling and Glut4 expression [161, 169]. Expression of a dominant-negative RORα in skeletal muscle C2C12 cells and in skeletal muscle in mice was reported to down-regulate the expression of carnitine palmitoyltransferase-1 (Cpt1), caveolin 3 (Cav3), and Abca1 encoding proteins involved in β-oxidation and cholesterol homeostasis, and of Srebp1c and its downstream targets, Fas and Scd1/2l, which are involved in lipogenesis [163, 189]. Promoter analysis indicated that Cav3 and Cpt1 were directly regulated by RORα. Expression of a dominant-negative RORα in skeletal muscle induced mild hyperglycemia and glucose intolerance and attenuated insulin-mediated phosphorylation of Akt2. The latter contrasts with the increase in Akt2 expression and phosphorylation observed in RORα-deficient sg/sg mice.
RORγ also plays a role in the regulation of glucose metabolism and insulin sensitivity [164, 165, 168, 190, 191]. RORγ-deficient mice were significantly more insulin sensitive and glucose tolerant than WT mice. The euglycemic clamp test revealed that hepatic glucose production was considerably reduced in RORγ-deficient mice, whereas ectopic expression of RORγ in RORγ-deficient liver tissue or primary hepatocytes increased glucose production [165]. Moreover, the conversion of exogenously administered pyruvate to glucose was significantly lower in RORγ−/−. The reduced hepatic gluconeogenesis in RORγ-deficient mice may be at least partly responsible for the improved insulin sensitivity and glucose tolerance observed in these mice [165, 190]. Loss of RORγ significantly decreased peak expression of several glucose (e.g., G6pase, Pklr, Glut2, PPARδ) and lipid (e.g., Insig2a, Elovl3, Cyp8b1, Sult1e1) metabolic genes [165, 167, 168, 192]. Conversely, exogenous expression of RORγ in RORγ−/− liver tissue by adenovirus significantly increased the expression of G6pase, Pepck, Gck, Gckr, Pparδ, Pcx, and Klf15. [165]. Together, these observations indicated that RORγ is an important modulator of hepatic gluconeogenesis and glycolysis. ChIP-Seq analysis not only uncovered the consensus sequence of the in vivo RORE, but also revealed that RORγ is recruited to the regulatory region of a number of metabolic genes involved in glycolysis and gluconeogenesis, including G6pase, Pepck, Pklr, Pparδ, Gck, Gckr, Glut2, Gys2, Dlat, Pcx, and Klf15 [165]. These data indicated that RORγ positively regulates the transcription of these metabolic genes by binding ROREs in their regulatory region. Promoter analysis further supported that the expression of several of these genes was directly regulated by RORγ. The observations further suggested that the decreased expression of these genes is at least in part responsible for the reduced gluconeogenesis and lower glycogen accumulation and consequently for the improved insulin sensitivity and glucose tolerance observed in RORγ null mice. A role for RORγ in the regulation of insulin resistance is supported by studies showing that the level of RORγ expression positively correlates with adiposity and insulin resistance in human obese patients [190, 191]. These observations suggest that RORγ antagonists might be beneficial in controlling glucose homeostasis and in the management of metabolic diseases.
In addition to gluconeogenesis, RORγ regulates hepatic lipid metabolism. Loss of RORγ reduced the expression of a number of lipid metabolic genes, including the insulin-induced gene 2a (Insig2a), elongation of very long chain fatty acids-like (Elovl3), Sult2a1, Cyp7b1, and Cyp8b1 [153, 167, 168, 185]. ChIP and promoter analysis showed that several of these genes are directly regulated by RORγ. The changes in the expression of these genes were associated reduced levels of triglycerides, cholesterol, and bile acids in liver and blood in RORγ-deficient mice fed a HFD. Lipid and glucose metabolic genes are under a complex control and involve regulation by other transcription factors, including several nuclear receptors, such as Rev-Erb, PPAR, LXR, and CAR. Since some of these receptors interact with similar binding sites, the transcriptional control of several of lipid and glucose metabolic genes likely involves interplay between different nuclear receptor signaling pathways. The best known example of this is the competition of Rev-Erbs with RORs for the same binding sites. Comparison of the RORα and RORγ cistromes from liver indicated that although many genes were selectively regulated by either RORα or RORγ, several genes, including G6pc, Apoa2, Elovl5, and Cry1, were regulated by both RORα and RORγ, indicating some redundancy between the two RORs in regulating these genes [165].
7. RORs and Circadian Rhythm
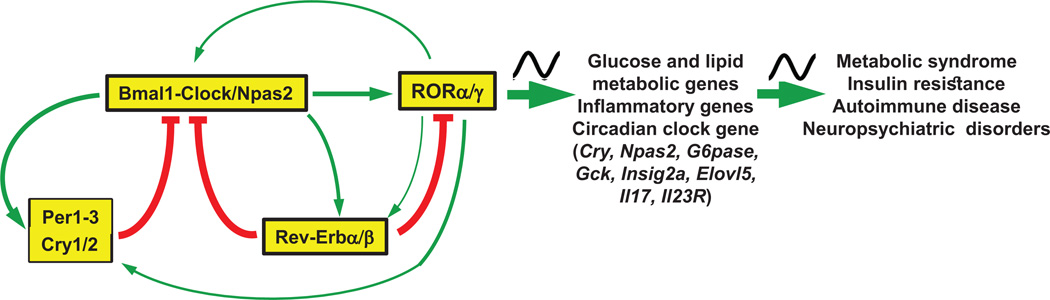
It has been well established that the regulation of the circadian rhythm is interconnected with the diurnal control of behavior, metabolic activities, immune responses, and many other physiological functions. For example, the circadian clock has been shown to regulate the diurnal expression of many lipid and glucose metabolic genes as well as immune response genes [170, 193–195]. It therefore not surprising that disruption of the circadian rhythm has been linked to increased risk for metabolic diseases, including obesity, diabetes, and liver steatosis, as well as several inflammatory and neuropsychiatric disorders [170, 171, 196–201]. In mammals, the suprachiasmatic nucleus (SCN) serves as the central circadian pacemaker that integrates light-dark cycle input and synchronizes the autonomous oscillators in peripheral tissues [170, 171, 196, 197]. The molecular clock machinery consists of several transcription/translation feedback loops in which the heterodimeric complex consisting of brain and muscle ARNT-like (Bmal1) and circadian locomotor output cycles kaput (Clock) or its paralog neuronal PAS domain protein 2 (Npas2) form the positive regulatory loop of the oscillator, whereas two cryptochrome (Cry) and three period proteins (Per) are part of the negative control mechanism. The nuclear receptors Rev-Erbα and β (NR1D1/2) further regulate the core loop by repressing the transcription of several clock genes, including Bmal1, Clock and Npas2 (Figure 3).
Figure 3. RORα and RORγ function as intermediaries between the circadian clock and its regulation of glucose/lipid metabolic and inflammatory gene expression.
RORs are linked to the circadian clock at different levels: a) ROR expression is regulated by the circadian clock machinery, including Bmal1, Clock, Rev-Erbs and Cry1; b) RORs are involved in the modulation of clock gene expression, including Npas2, Clock and Rev-Erb, and participate in the regulation of the rhythmic expression of glucose and lipid metabolic genes as well as inflammatory genes; c) Deficiency in RORα or RORβ causes changes in the circadian behavior, which might be linked to neuropsychiatric disorders, while deficiency in RORγ leads to increased insulin sensitivity and glucose tolerance and a lower risk of developing diabetes.
RORs are associated with the circadian clock at several different levels (Figure 3). First, RORs exhibit a rhythmic pattern of expression in several tissues. In particular, RORγ exhibits a robust oscillatory pattern of expression in liver, brown adipose tissue (BAT), pancreatic β cells, kidney, and small intestines (jejunum), with peak expression around Zeitgeber Time (ZT) 16–18, whereas RORα exhibits no to moderate oscillation in the SCN and several other tissues [94, 192, 202–205]. RORβ2 displays a rhythmic expression pattern in mouse SCN, pineal gland and retina, with a maximum at ZT18 [122, 203, 206, 207]. Several studies showed that RORa and RORc are regulated by Bmal1/Clock and RevErb. This is supported by data indicating that Bmal1, Clock and Rev-Erbα/β were recruited to the E-box and RORE, respectively, in the proximal RORc promoter in mouse liver [192, 208, 209]. Moreover, Bmal1 and Clock were able to induce activation of the RORc promoter in reporter assays [192, 210]. The RORc gene contains two E-box binding sites for Bmal/Clock [192, 204, 208, 211]. Mutation of either E1 or E2 significantly reduced the activation, while the double mutation totally abolished this induction by Clock/Bmal1. The activation of the RORc promoter by Clock/Bmal1 was repressed by Cry1 and correlated with changes in chromatin accessibility at the RORc promoter. Rev-Erbs, rather than Bmal1, regulate the rhythmic expression of RORc [210]. This is supported by data showing that in Bmal1 KO mice, the hepatic expression of RORγ is greatly enhanced particularly at ZT4–8, thereby largely abolishing the robust rhythmic expression pattern of RORγ [210]. The increase in RORγ mRNA expression appeared largely due to the loss of RevErb expression in Bmal1 KO liver, which subsequently abolished the repression of RORc by Rev-Erb at ZT4–8.
A second association between RORs and the circadian clock is their participation in the diurnal regulation of a number of clock genes, including Bmal1, Npas2, Clock, Rev-Erbα, and Cry1 [1, 3, 24, 171, 192, 202, 211, 221]. Exogenous expression of RORγ, as well as of RORα, in Hepa1–6 cells enhanced the endogenous expression of Cry1, Bmal1, E4bp4, Clock, Npas2, and Rev-Erbα, whereas treatment with an RORγ antagonist inhibited their induction [24, 192, 214]. ROREs have been identified in these clock genes [24, 192, 211, 214, 218]. Reporter gene and mutation analysis indicated that RORs are involved in the transcriptional regulation of these genes [24, 192, 214–217]. Rev-Erbs, which can compete with RORs for RORE binding, inhibited this activation. ChIP-Seq and ChIP-QPCR analyses further supported the association of RORs with these ROREs in vivo, consistent with the conclusion that these clock genes are directly regulated by RORs. The transcriptional mediator, RIP140, has been shown to be recruited by RORα to the Bmal1 promoter, suggesting that it is involved in mediating the transactivation of Bmal1 by RORα [222].
One might predict that the rhythmic expression of RORs leads to a rhythmic expression of ROR target genes. Indeed, several studies demonstrated that, in addition to clock genes, RORγ also participates in the diurnal regulation of several metabolic genes. Loss of RORγ significantly decreased peak expression of several glucose (e.g., G6pase, Pklr, Glut2, PPARδ) and lipid (e.g., Insig2a, Elovl3, Cyp8b1, Sult1e1) metabolic genes [165, 167, 168, 192]. ChIP analysis showed a ZT-dependent association of RORγ with ROREs in several of these genes. The transcriptional mediator, Prospero-related homeobox 1 (Prox1), which functions as a co-repressor of RORs as well as several other nuclear receptors, was shown to participate in the diurnal regulation of hepatic lipid/glucose metabolism by RORs [223, 224]. RORγ-deficient mice exhibited a significantly greater insulin sensitivity and glucose tolerance than WT mice particularly at ZT4–6. Moreover, the conversion of exogenously administered pyruvate to glucose was significantly lower in RORc−/− mice particularly at ZT4–6. Together these findings suggested that RORγ participates in the diurnal regulation of hepatic lipid metabolism, gluconeogenesis and insulin sensitivity. These studies further suggest that RORγ functions as an intermediary between the circadian clock machinery and its regulation of glucose and lipid metabolism.
Recent observations uncovered a connection between RORs and the circadian control of immune functions. RORγt was found to play a role in the diurnal regulation of Th17 differentiation by the circadian clock [225]. In Th17 cells, RORγt is expressed at significantly higher levels at daytime than at nighttime. This diurnal pattern of expression was found to be related to an increase in the daytime expression of Rev-Erb by Bmal1/Clock, which results in repression of NFIL3 transcription. Since NFIL3 functions as a repressor of RORγt transcription, its repression during daytime alleviates its inhibition of RORγt transcription leading to enhanced RORγt expression. Another study demonstrated that in the ileum, RORα regulates the diurnal expression of several genes associated with TLR signaling [94]. Analysis of gene expression profiles of mucosal biopsies from healthy individuals and patients with inflammatory bowel diseases (IBD) showed that the expression of several circadian genes, including ARNTL2. NPAS2, PER1, and RORA, was upregulated in IBD patients, consistent with a role for these proteins in this pathophysiology [226]. Together, these studies indicate that RORα and RORγ function as a link between the circadian clock and its regulation of various inflammatory pathways and provide a possible mechanism by which disruption of the circadian rhythm is associated with an increased risk of inflammatory diseases.
Clinical studies have indicated an important association between abnormalities in circadian rhythms and patients with mood and neuropsychiatric disorders. Alterations in circadian behavior observed in mice deficient in either RORα or RORβ receptor [122, 124, 216] and associations between SNPs in RORA and RORB with an increased risk for several neuropsychiatric disorders, including autism spectrum (ASD) and bipolar disorder, schizophrenia, depression, and posttraumatic stress syndrome [106–117, 129, 130], would be consistent with a link between disturbance in the circadian rhythm and these pathologies.
8. ROR (ant)agonists
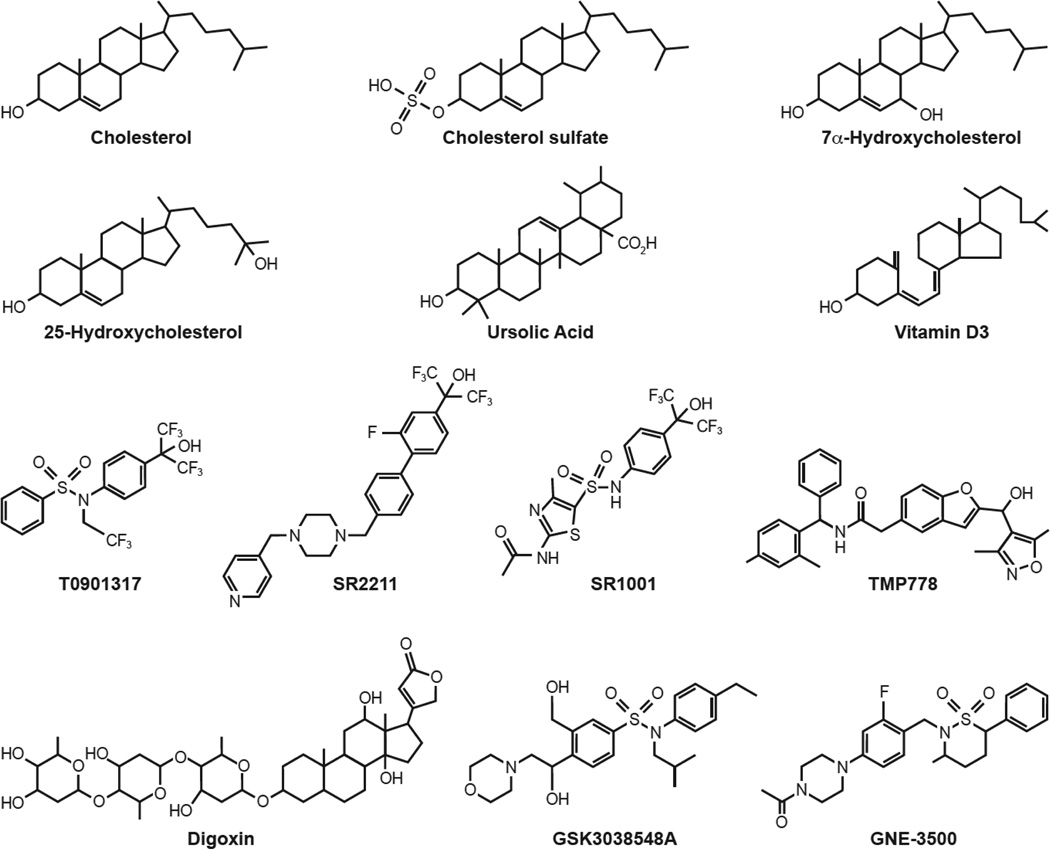
There has long been debate about whether RORs function as constitutively active receptors or whether their activity is regulated by (endogenous) ligands that function as an agonist or active antagonist (referred to as inverse agonist) or neutral antagonist [227]. Kallen, Stehlin-Gaon, and co-workers provided the first evidence for the hypothesis that RORs function as ligand-dependent transcription factors [33, 228, 229]. Crystal structure analysis revealed that cholesterol and cholesterol sulfate (Figure 4) bind the ligand-binding pocket of RORα and act as RORα agonists [33, 228]. Similarly, several retinoids were found to interact with the ligand-binding pocket of RORβ and to function as inverse agonists of RORβ as well as RORγ [229]. Subsequent studies identified a series of oxysterols as ligands for RORα and RORγ [25, 32, 230–233]. For example, 7α-hydroxycholesterol and 24(R)-hydroxycholesterol (Figure 4) were shown to function as inverse agonists, while 25-hydroxycholesterol, 20(α)-hydroxycholesterol, 22(R)-hydroxycholesterol, and 7α and 7β 27-hydroxycholesterol act as agonists in mammalian cells. A search for additional ROR ligands led to the discovery of a number of other small molecule modulators of RORγ [25, 32, 234–242]. The synthetic LXR agonist T0901317 was found to interact with both RORα and RORγ and to act as an inverse agonist [234]. Through chemical modification of T0901317, Burris and Griffin and coworkers identified a series of related ROR agonists and inverse agonists, such as SR2211 and SR1001 (Figure 4), which do not bind LXR [25, 237, 243]. Some of these compounds interacted with both RORα and RORγ, while others were RORα- or RORγ-selective [25, 234, 237]. Ursolic acid, a pentacyclic triterpene acid found in many plants, and several vitamin D metabolites, including 20-hydroxyvitamin D, were shown to exhibit RORγ antagonist activity [35, 244, 245]. Evidence was provided suggesting that these vitamin D metabolites were able to bind the RORγ LBD. A high throughput screen for RORγ ligands led to the identification of the cardiac glycoside, digoxin (Figure 4), and several of its analogs as RORγ antagonists [246]. Subsequently, other investigators set out to discover additional RORγ ligands [32]. This led to the identification of several series of high affinity RORγ inverse agonists, including various sulfonamides, such as GSK3038548A and GNE-3500 (Figure 4)[43, 240–242, 247, 248]. For a comprehensive review of small molecule ligands that interact with and modulate ROR receptors, we refer to several recent reviews [2, 25, 32, 249].
Figure 4. Chemical structure of several RORα/γ inverse agonists and (ant)agonists.
T0901317, SR1001, and 7α-hydroxycholesterol function as inverse agonists of both RORα and RORγ; cholesterol, cholesterol sulfate, and 25-hydroxycholesterol act as RORα and/or RORγ agonists; all other compounds have been reported to function as an inverse agonist or antagonist of RORγ.
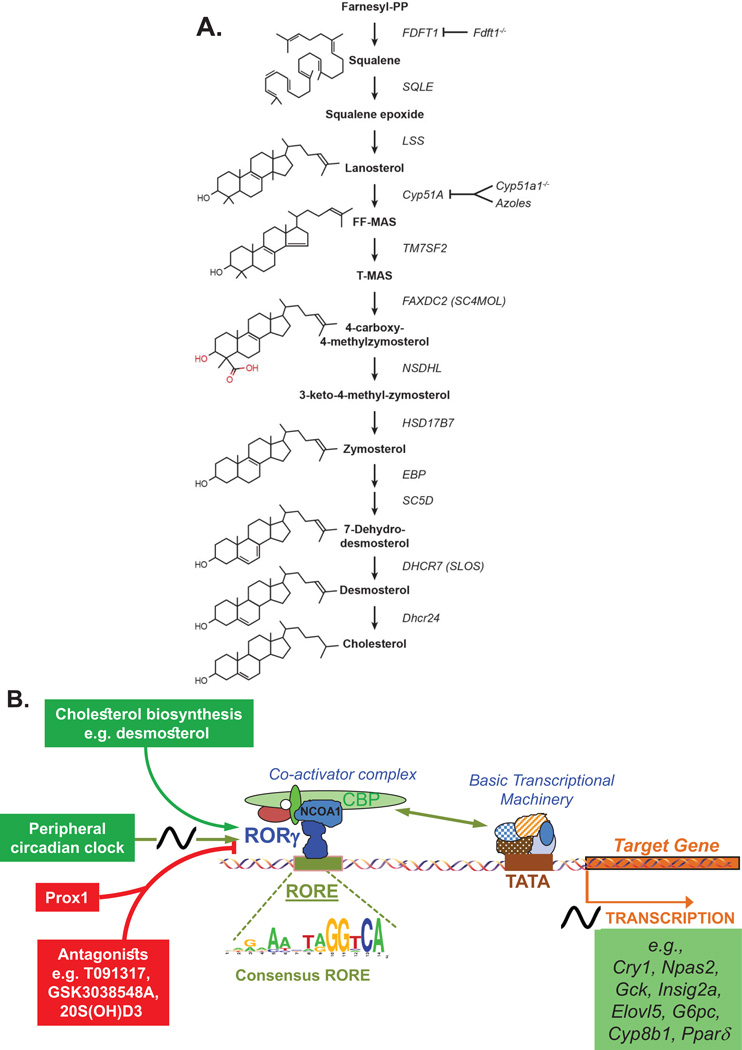
Recently, the connection between sterols and their modulation of ROR activity was further strengthened by studies showing a link between the cholesterol biosynthetic pathway (Figure 5A) and the regulation of RORγ activity [34, 35]. These studies demonstrated that several intermediates of the cholesterol biosynthetic pathway were able to function as endogenous agonists of RORγt. Zymosterol and desmosterol were among the most effective sterols activating RORγ, exhibiting EC50s of 0.11 and 0.08 µM, respectively, while cholesterol exhibited a much lower affinity for RORγ. These sterols enhanced RORγ transcriptional activity as well as the recruitment of coactivators. In addition, these sterols were able to enhance Th17 differentiation and increase the expression of IL-17A [34, 35]. Characterization of lipid-bound RORγ complexes immunoprecipitated from mammalian cells supported the concept that cholesterol biosynthetic intermediates function as endogenous RORγ ligands [35]. The connection between RORγ and sterol metabolism was further supported by studies showing that changes in the expression of enzymes involved in the cholesterol biosynthetic pathway were able to modulate RORγ activity. For example, RORγ transcriptional activity was lost in Fdft1-deficient cells lacking squalene synthase, an enzyme acting upstream in the cholesterol biosynthetic pathway [35]. Treatment with azole-type fungicides, such as ketoconazole and clotrimazole, which inhibit the sterol 14α-demethylase cytochrome P450, Cyp51a1, an enzyme upstream in the cholesterol biosynthetic pathway (Figure 5A), caused a dramatic reduction in zymosterol and desmosterol levels and a decrease in RORγ-mediated transactivation, Th17 differentiation, and IL-17 expression [34, 250]. Moreover, RORγ-mediated transactivation is greatly diminished in mammalian cells made deficient in Cyp51a1 by shRNA knockdown or germline deletion. Interestingly, several physiological processes that were impaired in RORγ-deficient mice were also affected in Cyp51a1−/− mice [35]; branchial lymph node anlagen were absent in 75% of Cyp51a1−/− mice and the number of IL17RA+ and CD4+ Lti cells was reduced. In a separate study, mice deficient in the mitochondrial sterol 27-hydroxylase (Cyp27A1), a key enzyme in bile acid synthesis and the production of 27-hydroxy cholesterol, exhibit a reduction in CD4+ and γδ+ T cells and a reduced capacity for Th17 differentiation [233]. These similarities in phenotypic changes are consistent with a link between the cholesterol biosynthetic pathway and RORγ activation. The role of cholesterol synthesis and RORγ activity in Th17 cells was further supported by observations showing that Th17 differentiation is associated with increased cholesterol uptake and biosynthesis and an accumulation of desmosterol that subsequently enhances RORγt activation and Th17 differentiation. In addition, activation of the TCR pathway, which results in activation of SREBP in favor of sterol-sulfate and cholesterol synthesis, might synergize with RORγ in promoting Th17 differentiation and IL-17 synthesis [34, 251]. Together, these studies suggest that changes in the cholesterol biosynthetic pathway and the level of cholesterol intermediates by diet or cholesterol-lowering drugs might control RORγ activation and as a consequence influence physiological processes regulated by RORγ, including Th17 differentiation. For example, an increase in Th17 cells and IL-17A under hypercholesterolemic conditions might at least in part be due to an increase in endogenous sterol levels and their subsequent activation of RORγt, while low cholesterol diet might do the inverse. This hypothesis is supported by a study reporting that patients with chronic hepatitis C, which is associated with increased levels of Th17 cells, when placed on a normocaloric, low cholesterol diet showed a significant reduction in Th17 cells and IL-17 levels [252]. Furthermore, treatment with statins, inhibitors of cholesterol synthesis, lead to a reduction in Th17 differentiation and IL-17 production [253].
Figure 5. Metabolites of the cholesterol biosynthetic pathway function as endogenous RORγ agonists.
A. Shown is a schematic view of the cholesterol synthetic pathway. Zymosterol and desmosterol are among the RORγ agonists with the highest affinity. Deficiency in Fdft1 or Cyp51A1, enzymes acting upstream in the cholesterol biosynthetic pathway, inhibit the synthesis of downstream RORγ agonists subsequently leading to reduced RORγt activation and Th17 differentiation. FDT1, Farnesyl-Diphosphate Farnesyltransferase 1; SQLE, Squalene Epoxidase; LSS, Lanosterol Synthase; TM7SF2, Transmembrane 7 Superfamily Member 2 (C-14 Sterol Reductase); FAXDC2/SC4MOL, Fatty Acid Hydroxylase Domain Containing 2/Methylsterol Monooxygenase 1; NSDHL, NAD(P) Dependent Steroid Dehydrogenase-Like; HSD17B7, Hydroxysteroid (17-Beta) Dehydrogenase 7; EBP, Emopamil Binding Protein (Sterol Isomerase); SC5D, Sterol-C5-Desaturase; DHCR7, 7-Dehydrocholesterol Reductase; DHCR24, 24-Dehydrocholesterol Reductase. B. Schematic view of RORγ-mediated transcriptional activation of target genes by endogenous sterol agonists and its inhibition by antagonists. The circadian clock regulates RORγ expression and as a consequence the expression of RORγ target genes. Prox1 modulates RORγ transcriptional activity. The in vivo consensus RORE derived from ChIP-Seq analysis using liver tissue and an anti-RORγ antibody, is shown.
Several of the sulfated conjugates, such as desmosterol sulfate, have also been shown to activate RORγ at levels twofold higher than the unsulfated sterols. In this context, it is interesting to note that Th17 cell differentiation is accompanied with an increase in the expression of the sulfotransferase, Sult2B1, and reduced expression of the sulfotransferase, STS [34]. This would be consistent with increased synthesis of desmosterol sulfate and RORγ activation and stimulation of Th17 differentiation. Oxysterols exhibit a much lower affinity for RORγ and appear to play a lesser role in modulating RORγ activity in Th17 cells; however, this may depend on the cell type and the type of oxysterol. Interestingly, Cyp7b1 and 3β-hydroxysteroid dehydrogenases (3βHSDs), which are involved in the hydroxylation or dehydrogenation of sterols, have been reported to be regulated by RORα and RORγ [167, 185] and therefore might affect the formation of certain (oxy)sterols and as a consequence the activation of RORγ [233].
Many of the ROR (ant)agonists have been shown to bind the ligand-binding pocket within the LBD of RORs [2, 32, 33, 228–232]. As has been reported for other nuclear receptors, agonist binding induces a conformational change in the ROR LBD and realignment of helix 12 that allows release of co-repressor complexes and promotes recruitment of co-activator complexes, which then mediate the transcriptional activation by RORs [1]. Particularly, the PLYKELF sequence within the C-terminal activation function (AF) plays a critical role in ROR transactivation activity and mutations in or deletion of this motif result in a dominant-negative ROR [254, 255]. Conversely, binding of ROR antagonists and inverse agonists, such as 25-hydroxycholesterol, inhibits the interaction with co-activators and promote interaction with co-repressors (Figure 5B). A number of co-repressors and coactivators have been identified that mediate ROR-dependent transcriptional activation, including NCOR, SRC1/2, and RIP140 [36, 222, 256].
ROR inverse agonists can inhibit ROR-induced transcriptional activation through different mechanisms. Certain inverse agonists, such as TMP920, have been reported to inhibit RORγ binding to ROREs, whereas the ability of RORγ to bind DNA target sites was mostly preserved with other inverse agonists, such as TMP778 and GSK805 [43]. Interestingly, RORγ cistrome analysis revealed that the latter compounds stabilized RORγt binding to a number of new genomic sites [43]. The distinct effects by various ligands are likely related to the induction of different conformational changes in RORγ that influence its affinity for different ROREs as well as its interaction with other transcriptional mediators. In addition to Th17 related genes, such as Il17 [35, 43, 244, 246], RORγ inverse agonists have been reported to inhibit the expression of a number of RORγ target genes, including the clock genes, Bmal1, Cry1, and Npas2, and several glucose and lipid metabolic genes, such as G6pase, Insig2a, Elovl3, Gck, and PPARδ [24, 165, 168, 192]. RORα inverse agonists and agonists were shown to, respectively, suppress or induce the expression of the RORα target genes, G6pase, Fgf21, CS, and Npas2 [166, 187, 231].
9. Summary
The clear evidence that RORγ activity is regulated by endogenous ligands suggests that this is likely the case also for RORα and RORβ. The regulation of RORγ activity by intermediates of the cholesterol biosynthetic pathway suggests that RORγ and the physiological processes controlled by RORγ can be influenced by environmental factors that affect this pathway, including cholesterol-rich or -low diets, environmental agents, such as the azole-type fungicides, and drugs that control cholesterol levels, such as lovastatin. Most importantly, by inhibiting RORγ transcriptional activity and thereby reducing Th17 generation and IL-17A/F production, RORγ inverse agonists may provide a novel strategy in the treatment of various pathologies in which RORγ is implicated, including inflammatory, metabolic, endocrine, and autoimmune diseases [1, 2, 13, 25, 257, 258]. Similarly, RORα antagonists might affect pathologies by inhibiting the generation of ILC2 cells and other physiological functions and be useful in the management of inflammatory, metabolic, and neuropsychiatric disorders [1, 2, 13, 25, 108, 113– 117, 174]. This concept is supported by reports showing that by inhibiting Th17 differentiation and IL-17 production, RORγ inverse agonists suppress Th17 responses in mice and ameliorate the development of experimental autoimmune encephalomyelitis and imiquimod-induced cutaneous inflammation [43, 244, 246, 259]. The beneficial effects of RORγ antagonists may not only be mediated through the inhibition of IL-17A and IL-17F synthesis in Th17 cells, but also by repressing the synthesis of these and other cytokines in RORγt+ innate lymphoid cells (ILC3), and RORγt+ γδ T cells, which also play a critical role in several autoimmune and inflammatory diseases [34, 77, 260, 261]. Attenuating RORα/γ activity by antagonist treatment might also be beneficial for the management of metabolic diseases, including metabolic syndrome and insulin resistance [161, 165, 174, 178, 190, 191]. Recently, the RORα/γ inverse agonist SR1001 was shown to suppress insulitis and prevent hyperglycemia in a mouse model of type 1 diabetes [262]. Together, these studies reinforce the potential of ROR antagonists in the management of autoimmune disease, neuropsychiatric and metabolic disorders, and other pathologies.
Acknowledgments
This research was supported by the Intramural Research Program of the NIEHS, NIH (Z01-ES-100486).
References
- 1.Jetten AM. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nuclear Receptor Signaling. 2009;7:e003. doi: 10.1621/nrs.07003. Article ID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solt LA, Burris TP. Action of RORs and their ligands in (patho)physiology. Trends Endocrinol Metab. 2012;23:619–627. doi: 10.1016/j.tem.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jetten AM, Kang HS, Takeda Y. Retinoic acid-related orphan receptors α and γ: key regulators of lipid/glucose metabolism, inflammation, and insulin sensitivity. Front Endocrinol (Lausanne) 2013;4:1–8. doi: 10.3389/fendo.2013.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giguère V, Tini M, Flock G, Ong E, Evans RM, Otulakowski G. Isoform-specific amino-terminal domains dictate DNA-binding properties of ROR α, a novel family of orphan hormone nuclear receptors. Genes Dev. 1994;8:538–553. doi: 10.1101/gad.8.5.538. [DOI] [PubMed] [Google Scholar]
- 5.André E, Gawlas K, Becker-André M. A novel isoform of the orphan nuclear receptor RORβ is specifically expressed in pineal gland and retina. Gene. 1998;216:277–283. doi: 10.1016/s0378-1119(98)00348-5. [DOI] [PubMed] [Google Scholar]
- 6.Giguère V, McBroom LD, Flock G. Determinants of target gene specificity for ROR α 1: monomeric DNA binding by an orphan nuclear receptor. Mol Cell Biol. 1995;15:2517–2526. doi: 10.1128/mcb.15.5.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He YW, Deftos ML, Ojala EW, Bevan MJ. RORγ t, a novel isoform of an orphan receptor, negatively regulates Fas ligand expression and IL-2 production in T cells. Immunity. 1998;9:797–806. doi: 10.1016/s1074-7613(00)80645-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirose T, Smith RJ, Jetten AM. ROR γ: the third member of ROR/RZR orphan receptor subfamily that is highly expressed in skeletal muscle. Biochem Biophys Res Commun. 1994;205:1976–1983. doi: 10.1006/bbrc.1994.2902. [DOI] [PubMed] [Google Scholar]
- 9.Medvedev A, Chistokhina A, Hirose T, Jetten AM. Genomic structure and chromosomal mapping of the nuclear orphan receptor ROR γ (RORC) gene. Genomics. 1997;46:93–102. doi: 10.1006/geno.1997.4980. [DOI] [PubMed] [Google Scholar]
- 10.Villey I, de Chasseval R, de Villartay JP. RORγT, a thymus-specific isoform of the orphan nuclear receptor RORγ / TOR, is up-regulated by signaling through the pre-T cell receptor and binds to the TEA promoter. Eur J Immunol. 1999;29:4072–4080. doi: 10.1002/(SICI)1521-4141(199912)29:12<4072::AID-IMMU4072>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 11.Eberl G, Littman DR. The role of the nuclear hormone receptor RORγt in the development of lymph nodes and Peyer’s patches. Immunol Rev. 2003;195:81–90. doi: 10.1034/j.1600-065x.2003.00074.x. [DOI] [PubMed] [Google Scholar]
- 12.Ivanov II BS, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 13.Jetten AM. Immunology: A helping hand against autoimmunity. Nature. 2011;472:421–422. doi: 10.1038/472421a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lockhart E, Green AM, Flynn JL. IL-17 production is dominated by γδ T cells rather than CD4 T cells during Mycobacterium tuberculosis infection. J Immunol. 2006;177:4662–4669. doi: 10.4049/jimmunol.177.7.4662. [DOI] [PubMed] [Google Scholar]
- 15.Medvedev A, Yan ZH, Hirose T, Giguère V, Jetten AM. Cloning of a cDNA encoding the murine orphan receptor RZR/ROR γ and characterization of its response element. Gene. 1996;181:199–206. doi: 10.1016/s0378-1119(96)00504-5. [DOI] [PubMed] [Google Scholar]
- 16.Downes M, Burke LJ, Muscat GE. Transcriptional repression by Rev-erbA α is dependent on the signature motif and helix 5 in the ligand binding domain: silencing does not involve an interaction with N-CoR. Nucleic Acids Res. 1996;24:3490–3498. doi: 10.1093/nar/24.18.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Retnakaran R, Flock G, Giguère V. Identification of RVR, a novel orphan nuclear receptor that acts as a negative transcriptional regulator. Mol Endocrinol. 1994;8:1234–1244. doi: 10.1210/mend.8.9.7838156. [DOI] [PubMed] [Google Scholar]
- 18.Forman BM, Chen J, Blumberg B, Kliewer SA, Henshaw R, Ong ES, Evans RM. Cross-talk among ROR α 1 and the Rev-erb family of orphan nuclear receptors. Mol Endocrinol. 1994;8:1253–1261. doi: 10.1210/mend.8.9.7838158. [DOI] [PubMed] [Google Scholar]
- 19.Austin S, Medvedev A, Yan ZH, Adachi H, Hirose T, Jetten AM. Induction of the nuclear orphan receptor RORγ during adipocyte differentiation of D1 and 3T3-L1 cells. Cell Growth Differ. 1998;9:267–276. [PubMed] [Google Scholar]
- 20.Bois-Joyeux B, Chauvet C, Nacer-Chérif H, Bergeret W, Mazure N, Giguère V, Laudet V, Danan JL. Modulation of the far-upstream enhancer of the rat α-fetoprotein gene by members of the ROR α, Rev-erb α and Rev-erb β groups of monomeric orphan nuclear receptors. DNA Cell Biol. 2000;19:589–599. doi: 10.1089/104454900750019344. [DOI] [PubMed] [Google Scholar]
- 21.Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1α integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 22.Duez H, Staels B. The nuclear receptors Rev-erbs and RORs integrate circadian rhythms and metabolism. Diab Vasc Dis Res. 2008;5:82–88. doi: 10.3132/dvdr.2008.0014. [DOI] [PubMed] [Google Scholar]
- 23.Dussault I, Giguère V. Differential regulation of the N-myc proto-oncogene by ROR α and RVR, two orphan members of the superfamily of nuclear hormone receptors. Mol Cell Biol. 1997;17:1860–1867. doi: 10.1128/mcb.17.4.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeda Y, Kang HS, Angers M, Jetten AM. Retinoic acid-related orphan receptor γ directly regulates neuronal PAS domain protein 2 transcription in vivo. Nucleic Acids Res. 2011;39:4769–4782. doi: 10.1093/nar/gkq1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kojetin DJ, Burris TP. REV-ERB and ROR nuclear receptors as drug targets. Nat Rev Drug Discov. 2014;13:197–216. doi: 10.1038/nrd4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ermisch M, Firla B, Steinhilber D. Protein kinase A activates and phosphorylates RORα4 in vitro and takes part in RORα activation by CaMK-IV. Biochem Biophys Res Commun. 2011;408:442–446. doi: 10.1016/j.bbrc.2011.04.046. [DOI] [PubMed] [Google Scholar]
- 27.Lechtken A, Hörnig M, Werz O, Corvey N, Zündorf I, Dingermann T, Brandes R, Steinhilber D. Extracellular signal-regulated kinase-2 phosphorylates RORα4 in vitro. Biochem Biophys Res Commun. 2007;358:890–896. doi: 10.1016/j.bbrc.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 28.Shin D, Kim IS, Lee JM, Shin SY, Lee JH, Baek SH, Cho KH. The hidden switches underlying RORα-mediated circuits that critically regulate uncontrolled cell proliferation. J Mol Cell Biol. 2014;6:338–348. doi: 10.1093/jmcb/mju023. [DOI] [PubMed] [Google Scholar]
- 29.Hwang EJ, Lee JM, Jeong J, Park JH, Yang Y, Lim JS, Kim JH, Baek SH, Kim KI. SUMOylation of RORα potentiates transcriptional activation function. Biochem Biophys Res Commun. 2009;378:513–517. doi: 10.1016/j.bbrc.2008.11.072. [DOI] [PubMed] [Google Scholar]
- 30.Rutz S, Kayagaki N, Phung QT, Eidenschenk C, Noubade R, Wang X, Lesch J, Lu R, Newton K, Huang OW, Cochran AG, Vasser M, Fauber BP, DeVoss J, Webster J, Diehl L, Modrusan Z, Kirkpatrick DS, Lill JR, Ouyang W, Dixit VM. Deubiquitinase DUBA is a post-translational brake on interleukin-17 production in T cells. Nature. 2015;518:417–421. doi: 10.1038/nature13979. [DOI] [PubMed] [Google Scholar]
- 31.Lim HW, Kang SG, Ryu JK, Schilling B, Fei M, Lee IS, Kehasse A, Shirakawa K, Yokoyama M, Schnölzer M, Kasler HG, Kwon HS, Gibson BW, Sato H, Akassoglou K, Xiao C, Littman DR, Ott M, Verdin E. SIRT1 deacetylates RORγt and enhances Th17 cell generation. J Exp Med. 2015;212:607–617. doi: 10.1084/jem.20132378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fauber BP, Magnuson S. Modulators of the nuclear receptor retinoic acid receptor-related orphan receptor-γ (RORγ or RORc) J Med Chem. 2014;57:5871–5892. doi: 10.1021/jm401901d. [DOI] [PubMed] [Google Scholar]
- 33.Kallen JA, Schlaeppi JM, Bitsch F, Geisse S, Geiser M, Delhon I, Fournier B. X-ray structure of the hRORα LBD at 1.63 A: structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORα. Structure. 2002;10:1697–1707. doi: 10.1016/s0969-2126(02)00912-7. [DOI] [PubMed] [Google Scholar]
- 34.Hu X, Wang Y, Hao L-Y, Liu X, Lesch CA, Sanchez BM, Wendling JM, Morgan RW, Aicher TD, Carter LL, Toogood PL, Glick GD. Sterol metabolism controls T(H)17 differentiation by generating endogenous RORγ agonists. Nat Chem Biol. 2015;11:141–147. doi: 10.1038/nchembio.1714. [DOI] [PubMed] [Google Scholar]
- 35.Santori FR, Huang P, van de Pavert SA, Douglass EF, Jr, Leaver DJ, Haubrich BA, Keber R, Lorbek G, Konijn T, Rosales BN, Rozman D, Horvat S, Rahier A, Mebius RE, Rastinejad F, Nes WD, Littman DR. Identification of natural RORγ ligands that regulate the development of lymphoid cells. Cell Metab. 2015;21:286–297. doi: 10.1016/j.cmet.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurebayashi S, Ueda E, Sakaue M, Patel DD, Medvedev A, Zhang F, Jetten AM. Retinoid-related orphan receptor γ (RORγ) is essential for lymphoid organogenesis and controls apoptosis during thymopoiesis. Proc Natl Acad Sci USA. 2000;97:10132–10137. doi: 10.1073/pnas.97.18.10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun Z, Unutmaz D, Zou YR, Sunshine MJ, Pierani A, Brenner-Morton S, Mebius RE, Littman DR. Requirement for RORγ in thymocyte survival and lymphoid organ development. Science. 2000;288:2369–2373. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- 38.Eberl G, Littman DR. Thymic origin of intestinal αβ T cells revealed by fate mapping of RORγt+ cells. Science. 2004;305:248–251. doi: 10.1126/science.1096472. [DOI] [PubMed] [Google Scholar]
- 39.He YW, Beers C, Deftos ML, Ojala EW, Forbush KA, Bevan MJ. Down-regulation of the orphan nuclear receptor ROR γ t is essential for T lymphocyte maturation. J Immunol. 2000;164:5668–5674. doi: 10.4049/jimmunol.164.11.5668. [DOI] [PubMed] [Google Scholar]
- 40.Jetten AM, Joo JH. Retinoid-related Orphan Receptors (RORs): Roles in Cellular Differentiation and Development. Adv Dev Biol. 2006;16:313–355. doi: 10.1016/S1574-3349(06)16010-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrington LE, Mangan PR, Weaver CT. Expanding the effector CD4 T-cell repertoire: the Th17 lineage. Curr Opin Immunol. 2006;18:349–356. doi: 10.1016/j.coi.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 42.Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, Ma L, Schluns K, Tian Q, Watowich SS, Jetten AM, Dong C. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448:480–483. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- 43.Xiao S, Yosef N, Yang J, Wang Y, Zhou L, Zhu C, Wu C, Baloglu E, Schmidt D, Ramesh R, Lobera M, Sundrud MS, Tsai PY, Xiang Z, Wang J, Xu Y, Lin X, Kretschmer K, Rahl PB, Young RA, Zhong Z, Hafler DA, Regev A, Ghosh S, Marson A, Kuchroo VK, OR R. Small-molecule, Small-molecule RORγt antagonists inhibit T helper 17 cell transcriptional network by divergent mechanisms. Immunity. 2014;40:477–489. doi: 10.1016/j.immuni.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, Watowich SS, Tian Q, Jetten AM, Dong C. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR α and ROR γ. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong C. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat Rev Immunol. 2008;8:337–348. doi: 10.1038/nri2295. [DOI] [PubMed] [Google Scholar]
- 46.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 47.Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, Dong C. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ano S, Morishima Y, Ishii Y, Yoh K, Yageta Y, Ohtsuka S, Matsuyama M, Kawaguchi M, Takahashi S, Hizawa N. Transcription factors GATA-3 and RORγt are important for determining the phenotype of allergic airway inflammation in a murine model of asthma. J Immunol. 2013;190:1056–1065. doi: 10.4049/jimmunol.1202386. [DOI] [PubMed] [Google Scholar]
- 49.Manel N, Unutmaz D, Littman DR. The differentiation of human T(H)-17 cells requires transforming growth factor-β and induction of the nuclear receptor RORγt. Nat Immunol. 2008;9:641–649. doi: 10.1038/ni.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang F, Meng G, Strober W. Interactions among the transcription factors Runx1, RORγt and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat Immunol. 2008;9:1297–1306. doi: 10.1038/ni.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schraml BU, Hildner K, Ise W, Lee WL, Smith WA, Solomon B, Sahota G, Sim J, Mukasa R, Cemerski S, Hatton RD, Stormo GD, Weaver CT, Russell JH, Murphy TL, Murphy KM. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 2009;460:405–409. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brüstle A, Heink S, Huber M, Rosenplänter C, Stadel-mann C, Yu P, Arpaia E, Mak TW, Kamradt T, Lohoff M. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol. 2007;8:958–966. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- 53.Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, Dong C. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem. 2007;282:9358–9363. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- 54.Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, Newberry KM, Meadows S, Greenfield A, Yang Y, Jain P, Kirigin FK, Birchmeier C, Wagner EF, Murphy KM, Myers RM, Bonneau R, Littman DR. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sundrud MS, Trivigno C. Identity crisis of Th17 cells: many forms, many functions, many questions. Semin Immunol. 2013;25:263–272. doi: 10.1016/j.smim.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 56.Kurebayashi Y, Nagai S, Ikejiri A, Ohtani M, Ichiyama K, Baba Y, Yamada T, Egami S, Hoshii T, Hirao A, Matsuda S, Koyasu S. PI3K-Akt-mTORC1-S6K1/2 axis controls Th17 differentiation by regulating Gfi1 expression and nuclear translocation of RORγ. Cell Reports. 2012;1:360–373. doi: 10.1016/j.celrep.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 57.Zhou L, Lopes JE, Chong MM, Ivanov II R. TGF-β-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORγt function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 59.Mangan PR, Harrington LE, O’Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, Weaver CT. Transforming growth factor-β induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 60.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFβ in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 61.Basu R, Whitley SK, Bhaumik S, Zindl CL, Schoeb TR, Benveniste EN, Pear WS, Hatton RD, Weaver CT. IL-1 signaling modulates activation of STAT transcription factors to antagonize retinoic acid signaling and control the TH17 cell-iTreg cell balance. Nat Immunol. 2015;16:286–295. doi: 10.1038/ni.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohnmacht C, Park JH, Cording S, Wing JB, Atarashi K, Obata Y, Gaboriau-Routhiau V, Marques R, Dulauroy S, Fedoseeva M, Busslinger M, Cerf-Bensussan N, Boneca IG, Voehringer D, Hase K, Honda K, Sakaguchi S, Eberl G. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORγt+ T cells. Science. 2015;349:989–993. doi: 10.1126/science.aac4263. [DOI] [PubMed] [Google Scholar]
- 63.Tilley SL, Jaradat M, Stapleton C, Dixon D, Hua X, Erikson CJ, McCaskill JG, Chason KD, Liao G, Jania L, Koller BH, Jetten AM. Retinoid-related orphan receptor γ controls immunoglobulin production and Th1/Th2 cytokine balance in the adaptive immune response to allergen. J Immunol. 2007;178:3208–3218. doi: 10.4049/jimmunol.178.5.3208. [DOI] [PubMed] [Google Scholar]
- 64.Haak S, Croxford AL, Kreymborg K, Heppner FL, Pouly S, Becher B, Waisman A. IL-17A and IL-17F do not contribute vitally to autoimmune neuroinflammation in mice. J Clin Invest. 2009;119:61–69. doi: 10.1172/JCI35997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kreymborg K, Etzensperger R, Dumoutier L, Haak S, Rebollo A, Buch T, Heppner FL, Renauld JC, Becher B. IL-22 is expressed by Th17 cells in an IL-23-dependent fashion, but not required for the development of autoimmune encephalomyelitis. J Immunol. 2007;179:8098–8104. doi: 10.4049/jimmunol.179.12.8098. [DOI] [PubMed] [Google Scholar]
- 66.Codarri L, Gyülvészi G, Tosevski V, Hesske L, Fontana A, Magnenat L, Suter T, Becher B. RORγt drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat Immunol. 2011;12:560–567. doi: 10.1038/ni.2027. [DOI] [PubMed] [Google Scholar]
- 67.El-Behi M, Ciric B, Dai H, Yan Y, Cullimore M, Safavi F, Zhang GX, Dittel BN, Rostami A. The encephalitogenicity of T(H)17 cells is dependent on IL-1-and IL-23-induced production of the cytokine GM-CSF. Nat Immunol. 2011;12:568–575. doi: 10.1038/ni.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leppkes M, Becker C, Ivanov S, II, Hirth S, Wirtz S, Neufert C, Pouly S, Murphy AJ, Valenzuela DM, Yancopoulos GD, Becher B, Littman DR, Neurath MF. RORγ-expressing Th17 cells induce murine chronic intestinal inflammation via redundant effects of IL-17A and IL-17F. Gastroenterology. 2009;136:257–267. doi: 10.1053/j.gastro.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 69.Kabelitz D, Peters C, Wesch D, Oberg HH. Regulatory functions of γδ T cells. Int Immunopharmacol. 2013;16:382–387. doi: 10.1016/j.intimp.2013.01.022. [DOI] [PubMed] [Google Scholar]
- 70.Haas JD, Ravens S, Düber S, Sandrock I, Oberdörfer L, Kashani E, Chennupati V, Föhse L, Naumann R, Weiss S, Krueger A, Förster R, Prinz I. Development of interleukin-17-producing γδ T cells is restricted to a functional embryonic wave. Immunity. 2012;37:48–59. doi: 10.1016/j.immuni.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 71.Hayes SM, Laird RM. Genetic requirements for the development and differentiation of interleukin-17-producing γδ T cells. Crit Rev Immunol. 2012;32:81–95. doi: 10.1615/critrevimmunol.v32.i1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sutton CE, Lalor SJ, Sweeney CM, Brereton CF, Lavelle EC, Mills KH. Interleukin-1 and IL-23 induce innate IL-17 production from γδ T cells, amplifying Th17 responses and autoimmunity. Immunity. 2009;31:331–341. doi: 10.1016/j.immuni.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 73.Powolny-Budnicka I, Riemann M, Tänzer S, Schmid RM, Hehlgans T, Weih F, Rel A. RelA and RelB transcription factors in distinct thymocyte populations control lymphotoxin-dependent interleukin-17 production in γδ T cells. Immunity. 2011;34:364–374. doi: 10.1016/j.immuni.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 74.Walker JA, Barlow JL, McKenzie AN. Innate lymphoid cells-how did we miss them? Nat Rev Immunol. 2013;13:75–87. doi: 10.1038/nri3349. [DOI] [PubMed] [Google Scholar]
- 75.Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, Powrie F, Vivier E. Innate lymphoid cells-a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 76.Mebius RE. Organogenesis of lymphoid tissues. Nat Rev Immunol. 2003;3:292–303. doi: 10.1038/nri1054. [DOI] [PubMed] [Google Scholar]
- 77.Cording S, Medvedovic J, Cherrier M, Eberl G. Development and regulation of RORγt(+) innate lymphoid cells. FEBS Lett. 2014;588:4176–4181. doi: 10.1016/j.febslet.2014.03.034. [DOI] [PubMed] [Google Scholar]
- 78.Mebius RE, Rennert P, Weissman IL. Developing lymph nodes collect CD4+CD3− LTβ+ cells that can differentiate to APC, NK cells, and follicular cells but not T or B cells. Immunity. 1997;7:493–504. doi: 10.1016/s1074-7613(00)80371-4. [DOI] [PubMed] [Google Scholar]
- 79.McKenzie AN, Spits H, Eberl G. Innate lymphoid cells in inflammation and immunity. Immunity. 2014;41:366–374. doi: 10.1016/j.immuni.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 80.Diefenbach A, Colonna M, Koyasu S. Development, differentiation, and diversity of innate lymphoid cells. Immunity. 2014;41:354–365. doi: 10.1016/j.immuni.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Eberl G, Marmon S, Sunshine MJ, Rennert PD, Choi Y, Littman DR. An essential function for the nuclear receptor RORγ(t) in the generation of fetal lymphoid tissue inducer cells. Nat Immunol. 2004;5:64–73. doi: 10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- 82.Kiss EA, Diefenbach A. Role of the Aryl Hydrocarbon Receptor in Controlling Maintenance and Functional Programs of RORγt(+) Innate Lymphoid Cells and Intraepithelial Lymphocytes. Front Immunol. 2012;3:124. doi: 10.3389/fimmu.2012.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van de Pavert SA, Ferreira M, Domingues RG, Ribeiro H, Molenaar R, Moreira-Santos L, Almeida FF, Ibiza S, Barbosa I, Goverse G, Labão-Almeida C, Godinho-Silva C, Konijn T, Schooneman D, O’Toole T, Mizee MR, Habani Y, Haak E, Santori FR, Littman DR, Schulte-Merker S, Dzierzak E, Simas JP, Mebius RE, Veiga-Fernandes H. Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature. 2014;508:123–127. doi: 10.1038/nature13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Serafini N, Klein Wolterink RG, Satoh-Takayama N, Xu W, Vosshenrich CA, Hendriks RW, Di Santo JP. Gata3 drives development of RORγt+ group 3 innate lymphoid cells. J Exp Med. 2014;211:199–208. doi: 10.1084/jem.20131038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bernink JH, Peters CP, Munneke M, te Velde AA, Meijer SL, Weijer K, Hreggvidsdottir HS, Heinsbroek SE, Legrand N, Buskens CJ, Bemelman WA, Mjösberg JM, Spits H. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol. 2013;14:221–229. doi: 10.1038/ni.2534. [DOI] [PubMed] [Google Scholar]
- 86.Wong SH, Walker JA, Jolin HE, Drynan LF, Hams E, Camelo A, Barlow JL, Neill DR, Panova V, Koch U, Radtke F, Hardman CS, Hwang YY, Fallon PG, McKenzie AN. Transcription factor RORα is critical for nuocyte development. Nat Immunol. 2012;13:229–236. doi: 10.1038/ni.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Halim TY, MacLaren A, Romanish MT, Gold MJ, McNagny KM, Takei F. Retinoic-acid-receptor-related orphan nuclear receptor α is required for natural helper cell development and allergic inflammation. Immunity. 2012;37:463–474. doi: 10.1016/j.immuni.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 88.Salimi M, Barlow JL, Saunders SP, Xue L, Gutowska-Owsiak D, Wang X, Huang LC, Johnson D, Scanlon ST, McKenzie AN, Fallon PG, Ogg GS. A role for IL-25 and IL-33-driven type-2 innate lymphoid cells in atopic dermatitis. J Exp Med. 2013;210:2939–2950. doi: 10.1084/jem.20130351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ramasamy A, Kuokkanen M, Vedantam S, Gajdos ZK, Couto Alves A, Lyon HN, Ferreira MA, Strachan DP, Zhao JH, Abramson MJ, Brown MA, Coin L, Dharmage SC, Duffy DL, Haahtela T, Heath AC, Janson C, Kähönen M, Khaw KT, Laitinen J, Le Souef P, Lehtimäki T, Madden PA, Marks GB, Martin NG, Matheson MC, Palmer CD, Palotie A, Pouta A, Robertson CF, Viikari J, Widen E, Wjst M, Jarvis DL, Montgomery GW, Thompson PJ, Wareham N, Eriksson J, Jousilahti P, Laitinen T, Pekkanen J, Raitakari OT, O’Connor GT, Salomaa V, Jarvelin MR, Hirschhorn JN. Genome-wide association studies of asthma in population-based cohorts confirm known and suggested loci and identify an additional association near HLA. PLoS One. 2012;7:e44008. doi: 10.1371/journal.pone.0044008. Article ID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, von Mutius E, Farrall M, Lathrop M, Cookson WO. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Melén E, Kho AT, Sharma S, Gaedigk R, Leeder JS, Mariani TJ, Carey VJ, Weiss ST, Tantisira KG. Expression analysis of asthma candidate genes during human and murine lung development. Respir Res. 2011;12:86–95. doi: 10.1186/1465-9921-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jaradat M, Stapleton C, Tilley SL, Dixon D, Erikson CJ, McCaskill JG, Kang HS, Angers M, Liao G, Collins J, Grissom S, Jetten AM. Modulatory role for retinoid-related orphan receptor α in allergen-induced lung inflammation. Am J Respir Crit Care Med. 2006;174:1299–1309. doi: 10.1164/rccm.200510-1672OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Persson H, Kwon AT, Ramilowski JA, Silberberg G, Söderhäll C, Orsmark-Pietras C, Nordlund B, Konradsen JR, de Hoon MJ, Melén E, Hayashizaki Y, Hedlin G, Kere J, Daub CO. Transcriptome analysis of controlled and therapy-resistant childhood asthma reveals distinct gene expression profiles. J Allergy Clin Immunol. 2015;136:638–648. doi: 10.1016/j.jaci.2015.02.026. [DOI] [PubMed] [Google Scholar]
- 94.Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153:812–827. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 95.Nakagawa Y, O’Leary DD. Dynamic patterned expression of orphan nuclear receptor genes RORα and RORβ in developing mouse forebrain. Dev Neurosci. 2003;25:234–244. doi: 10.1159/000072271. [DOI] [PubMed] [Google Scholar]
- 96.Sashihara S, Felts PA, Waxman SG, Matsui T. Orphan nuclear receptor ROR α gene: isoform-specific spatiotemporal expression during postnatal development of brain. Brain Res Mol Brain Res. 1996;42:109–117. doi: 10.1016/s0169-328x(96)00118-0. [DOI] [PubMed] [Google Scholar]
- 97.Matsui T, Sashihara S, Oh Y, Waxman SG. An orphan nuclear receptor, mROR α, and its spatial expression in adult mouse brain. Brain Res Mol Brain Res. 1995;33:217–226. doi: 10.1016/0169-328x(95)00126-d. [DOI] [PubMed] [Google Scholar]
- 98.Hamilton BA, Frankel WN, Kerrebrock AW, Hawkins TL, FitzHugh W, Kusumi K, Russell LB, Mueller KL, van Berkel V, Birren BW, Kruglyak L, Lander ES. Disruption of the nuclear hormone receptor RORα in staggerer mice. Nature. 1996;379:736–739. doi: 10.1038/379736a0. [DOI] [PubMed] [Google Scholar]
- 99.Steinmayr M, André E, Conquet F, Rondi-Reig L, Delhaye-Bouchaud N, Auclair N, Daniel H, Crépel F, Mariani J, Sotelo C, Becker-André M. staggerer phenotype in retinoid-related orphan receptor α-deficient mice. Proc Natl Acad Sci USA. 1998;95:3960–3965. doi: 10.1073/pnas.95.7.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dussault I, Fawcett D, Matthyssen A, Bader JA, Giguère V. Orphan nuclear receptor ROR α-deficient mice display the cerebellar defects of staggerer. Mech Dev. 1998;70:147–153. doi: 10.1016/s0925-4773(97)00187-1. [DOI] [PubMed] [Google Scholar]
- 101.Boukhtouche F, Doulazmi M, Frederic F, Dusart I, Brugg B, Mariani J. RORα, a pivotal nuclear receptor for Purkinje neuron survival and differentiation: from development to ageing. Cerebellum. 2006;5:97–104. doi: 10.1080/14734220600750184. [DOI] [PubMed] [Google Scholar]
- 102.Doulazmi M, Frédéric F, Capone F, Becker-André M, Delhaye-Bouchaud N, Mariani J. A comparative study of Purkinje cells in two RORα gene mutant mice: staggerer and RORα(−/−) Brain Res Dev Brain Res. 2001;127:165–174. doi: 10.1016/s0165-3806(01)00131-6. [DOI] [PubMed] [Google Scholar]
- 103.Chen XR, Heck N, Lohof AM, Rochefort C, Morel MP, Wehrlé R, Doulazmi M, Marty S, Cannaya V, Avci HX, Mariani J, Rondi-Reig L, Vodjdani G, Sherrard RM, Sotelo C, Dusart I. Mature Purkinje cells require the retinoic acid-related orphan receptor-α (RORα) to maintain climbing fiber mono-innervation and other adult characteristics. J Neurosci. 2013;33:9546–9562. doi: 10.1523/JNEUROSCI.2977-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Janmaat S, Akwa Y, Doulazmi M, Bakouche J, Gautheron V, Liere P, Eychenne B, Pianos A, Luiten P, Groothuis T, Baulieu EE, Mariani J, Sherrard RM, Frédéric F. Age-related Purkinje cell death is steroid dependent: RORα haplo-insufficiency impairs plasma and cerebellar steroids and Purkinje cell survival. Age (Dordr) 2011;33:565–578. doi: 10.1007/s11357-010-9203-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nakatani T, Minaki Y, Kumai M, Nitta C, Ono Y. The c-Ski family member and transcriptional regulator Corl2/Skor2 promotes early differentiation of cerebellar Purkinje cells. Dev Biol. 2014;388:68–80. doi: 10.1016/j.ydbio.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 106.Etain B, Jamain S, Milhiet V, Lajnef M, Boudebesse C, Dumaine A, Mathieu F, Gombert A, Ledudal K, Gard S, Kahn JP, Henry C, Boland A, Zelenika D, Lechner D, Lathrop M, Leboyer M, Bellivier F. Association between circadian genes, bipolar disorders and chronotypes. Chronobiol Int. 2014;31:807–814. doi: 10.3109/07420528.2014.906445. [DOI] [PubMed] [Google Scholar]
- 107.Nguyen A, Rauch TA, Pfeifer GP, Hu VW. Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain. FASEB J. 2010;24:3036–3051. doi: 10.1096/fj.10-154484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Devanna P, Vernes SC. A direct molecular link between the autism candidate gene RORa and the schizophrenia candidate MIR137. Sci Rep. 2014;4:3994. doi: 10.1038/srep03994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Miller MW, Wolf EJ, Logue MW, Baldwin CT. The retinoid-related orphan receptor α (RORA) gene and fear-related psychopathology. J Affect Disord. 2013;151:702–708. doi: 10.1016/j.jad.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Amstadter AB, Sumner JA, Acierno R, Ruggiero KJ, Koenen KC, Kilpatrick DG, Galea S, Gelernter J. Support for association of RORA variant and post traumatic stress symptoms in a population-based study of hurricane exposed adults. Mol Psychiatry. 2013;18:1148–1149. doi: 10.1038/mp.2012.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Logue MW, Baldwin C, Guffanti G, Melista E, Wolf EJ, Reardon AF, Uddin M, Wildman D, Galea S, Koenen KC, Miller MW. A genome-wide association study of post-traumatic stress disorder identifies the retinoid-related orphan receptor α (RORA) gene as a significant risk locus. Mol Psychiatry. 2013;18:937–942. doi: 10.1038/mp.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Partonen T. Clock gene variants in mood and anxiety disorders. J Neural Transm Vienna. 2012;119:1133–1145. doi: 10.1007/s00702-012-0810-2. [DOI] [PubMed] [Google Scholar]
- 113.Lai YC, Kao CF, Lu ML, Chen HC, Chen PY, Chen CH, Shen WW, Wu JY, Lu RB, Kuo PH. Investigation of associations between NR1D1, RORA and RORB genes and bipolar disorder. PLoS One. 2015;10:e0121245. doi: 10.1371/journal.pone.0121245. Article ID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Haerian BS, Sha’ari HM, Tan HJ, Fong CY, Wong SW, Ong LC, Raymond AA, Tan CT, Mohamed Z. RORA gene rs12912233 and rs880626 polymorphisms and their interaction with SCN1A rs3812718 in the risk of epilepsy: a case-control study in Malaysia. Genomics. 2015;105:229–236. doi: 10.1016/j.ygeno.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 115.Soria V, Martinez-Amoros E, Escaramis G, et al. Differential association of circadian genes with mood disorders: CRY1 and NPAS2 are associated with unipolar major depression and CLOCK and VIP with bipolar disorder. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2010;35:1279–1289. doi: 10.1038/npp.2009.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Geoffroy PA, Lajnef M, Bellivier F, Jamain S, Gard S, Kahn JP, Henry C, Leboyer M, Etain B. Genetic association study of circadian genes with seasonal pattern in bipolar disorders. Sci Rep. 2015;5:10232. doi: 10.1038/srep10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Maglione JE, Nievergelt CM, Parimi N, Evans DS, Ancoli-Israel S, Stone KL, Yaffe K, Redline S, Tranah GJ. Associations of PER3 and RORA Circadian Gene Polymorphisms and Depressive Symptoms in Older Adults. Am J Geriatr Psychiatry. 2015;23:1075–1087. doi: 10.1016/j.jagp.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sarachana T, Xu M, Wu RC, Hu VW. Sex hormones in autism: androgens and estrogens differentially and reciprocally regulate RORA, a novel candidate gene for autism. PLoS One. 2011;6:e17116. doi: 10.1371/journal.pone.0017116. Article ID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sarachana T, Hu VW. Differential recruitment of coregulators to the RORA promoter adds another layer of complexity to gene (dys) regulation by sex hormones in autism. Mol Autism. 2013;4:39. doi: 10.1186/2040-2392-4-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jolly S, Journiac N, Naudet F, Gautheron V, Mariani J, Vernet-der Garabedian B. Cell-autonomous and non-cell-autonomous neuroprotective functions of RORα in neurons and astrocytes during hypoxia. J Neurosci. 2011;31:14314–14323. doi: 10.1523/JNEUROSCI.1443-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Boukhtouche F, Vodjdani G, Jarvis CI, Bakouche J, Staels B, Mallet J, Mariani J, Lemaigre-Dubreuil Y, Brugg B. Human retinoic acid receptor-related orphan receptor α1 overexpression protects neurones against oxidative stress-induced apoptosis. J Neurochem. 2006;96:1778–1789. doi: 10.1111/j.1471-4159.2006.03708.x. [DOI] [PubMed] [Google Scholar]
- 122.Schaeren-Wiemers N, André E, Kapfhammer JP, Becker-André M. The expression pattern of the orphan nuclear receptor RORβ in the developing and adult rat nervous system suggests a role in the processing of sensory information and in circadian rhythm. Eur J Neurosci. 1997;9:2687–2701. doi: 10.1111/j.1460-9568.1997.tb01698.x. [DOI] [PubMed] [Google Scholar]
- 123.Srinivas M, Ng L, Liu H, Jia L, Forrest D. Activation of the blue opsin gene in cone photoreceptor development by retinoid-related orphan receptor β. Mol Endocrinol. 2006;20:1728–1741. doi: 10.1210/me.2005-0505. [DOI] [PubMed] [Google Scholar]
- 124.Masana MI, Sumaya IC, Becker-Andre M, Dubocovich ML. Behavioral characterization and modulation of circadian rhythms by light and melatonin in C3H/HeN mice homozygous for the RORβ knockout. AmJ Physiol Regul Integr Comp Physiol. 2007;292:R2357–R2367. doi: 10.1152/ajpregu.00687.2006. [DOI] [PubMed] [Google Scholar]
- 125.Dye CA, El Shawa H, Huffman KJ. A lifespan analysis of intraneocortical connections and gene expression in the mouse I. Cereb Cortex. 2011;21:1311–1330. doi: 10.1093/cercor/bhq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dye CA, El Shawa H, Huffman KJ. A lifespan analysis of intraneocortical connections and gene expression in the mouse II. Cereb Cortex. 2011;21:1331–1350. doi: 10.1093/cercor/bhq213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hirokawa J, Watakabe A, Ohsawa S, Yamamori T. Analysis of area-specific expression patterns of RORβ, ER81 and Nurr1 mRNAs in rat neocortex by double in situ hybridization and cortical box method. PLoS One. 2008;3:e3266. doi: 10.1371/journal.pone.0003266. Article ID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jabaudon D, Shnider SJ, Tischfield DJ, Galazo MJ, Macklis JD. RORβ induces barrel-like neuronal clusters in the developing neocortex. Cereb Cortex. 2012;22:996–1006. doi: 10.1093/cercor/bhr182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McGrath CL, Glatt SJ, Sklar P, Le-Niculescu H, Kuczenski R, Doyle AE, Biederman J, Mick E, Faraone SV, Niculescu AB, Tsuang MT. Evidence for genetic association of RORB with bipolar disorder. BMC Psychiatry. 2009;9:70. doi: 10.1186/1471-244X-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mansour HA, Talkowski ME, Wood J, Chowdari KV, McClain L, Prasad K, Montrose D, Fagiolini A, Friedman ES, Allen MH, Bowden CL, Calabrese J, El-Mallakh RS, Escamilla M, Faraone SV, Fossey MD, Gyulai L, Loftis JM, Hauser P, Ketter TA, Marangell LB, Miklowitz DJ, Nierenberg AA, Patel J, Sachs GS, Sklar P, Smoller JW, Laird N, Keshavan M, Thase ME, Axelson D, Birmaher B, Lewis D, Monk T, Frank E, Kupfer DJ, Devlin B, Nimgaonkar VL. Association study of 21 circadian genes with bipolar I disorder, schizoaffective disorder, and schizophrenia. Bipolar Disord. 2009;11:701–710. doi: 10.1111/j.1399-5618.2009.00756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ersland KM, Christoforou A, Stansberg C, Espeseth T, Mattheisen M, Mattingsdal M, Hardarson GA, Hansen T, Fernandes CP, Giddaluru S, Breuer R, Strohmaier J, Djurovic S, Nöthen MM, Rietschel M, Lundervold AJ, Werge T, Cichon S, Andreassen OA, Reinvang I, Steen VM, Le Hellard S. Gene-based analysis of regionally enriched cortical genes in GWAS data sets of cognitive traits and psychiatric disorders. PLoS One. 2012;7:e31687. doi: 10.1371/journal.pone.0031687. Article ID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Baglietto MG, Caridi G, Gimelli G, Mancardi M, Prato G, Ronchetto P, Cuoco C, Tassano E. RORB gene and 9q21.13 microdeletion: report on a patient with epilepsy and mild intellectual disability. Eur J Med Genet. 2014;57:44–46. doi: 10.1016/j.ejmg.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 133.Boudry-Labis E, Demeer B, Le Caignec C, Isidor B, Mathieu-Dramard M, Plessis G, George AM, Taylor J, Aftimos S, Wiemer-Kruel A, Kohlhase J, Annerén G, Firth H, Simonic I, Vermeesch J, Thuresson AC, Copin H, Love DR, Andrieux J. A novel microdeletion syndrome at 9q21.13 characterised by mental retardation, speech delay, epilepsy and characteristic facial features. Eur J Med Genet. 2013;56:163–170. doi: 10.1016/j.ejmg.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 134.Lal D, Ruppert AK, Trucks H, Schulz H, de Kovel CG, Kasteleijn-Nolst Trenité D, Sonsma AC, Koeleman BP, Lindhout D, Weber YG, Lerche H, Kapser C, Schankin CJ, Kunz WS, Surges R, Elger CE, Gaus V, Schmitz B, Helbig I, Muhle H, Stephani U, Klein KM, Rosenow F, Neubauer BA, Reinthaler EM, Zimprich F, Feucht M, Møller RS, Hjalgrim H, De Jonghe P, Suls A, Lieb W, Franke A, Strauch K, Gieger C, Schurmann C, Schminke U, Nürnberg P, Sander T. Burden analysis of rare microdeletions suggests a strong impact of neurodevelopmental genes in genetic generalised epilepsies. PLoS Genet. 2015;11:e1005226. doi: 10.1371/journal.pgen.1005226. Article ID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.El Shawa H, Abbott CW, III, Huffman KJ. Prenatal ethanol exposure disrupts intraneocortical circuitry, cortical gene expression, and behavior in a mouse model of FASD. J Neurosci. 2013;33:18893–18905. doi: 10.1523/JNEUROSCI.3721-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liu H, Kim SY, Fu Y, Wu X, Ng L, Swaroop A, Forrest D. An isoform of retinoid-related orphan receptor β directs differentiation of retinal amacrine and horizontal interneurons. Nat Commun. 2013;4:1813. doi: 10.1038/ncomms2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fu Y, Liu H, Ng L, Kim JW, Hao H, Swaroop A, Forrest D. Feedback induction of a photoreceptor-specific isoform of retinoid-related orphan nuclear receptor β by the rod transcription factor NRL. J Biol Chem. 2014;289:32469–32480. doi: 10.1074/jbc.M114.605774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chow L, Levine EM, Reh TA. The nuclear receptor transcription factor, retinoid-related orphan receptor β, regulates retinal progenitor proliferation. Mech Dev. 1998;77:149–164. doi: 10.1016/s0925-4773(98)00135-x. [DOI] [PubMed] [Google Scholar]
- 139.Jia L, Oh EC, Ng L, Srinivas M, Brooks M, Swaroop A, Forrest D. Retinoid-related orphan nuclear receptor RORβ is an early-acting factor in rod photoreceptor development. Proc Natl Acad Sci USA. 2009;106:17534–17539. doi: 10.1073/pnas.0902425106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kautzmann MA, Kim DS, Felder-Schmittbuhl MP, Swaroop A. Combinatorial regulation of photoreceptor differentiation factor, neural retina leucine zipper gene NRL, revealed by in vivo promoter analysis. J Biol Chem. 2011;286:28247–28255. doi: 10.1074/jbc.M111.257246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ueda E, Kurebayashi S, Sakaue M, Backlund M, Koller B, Jetten AM. High incidence of T-cell lymphomas in mice deficient in the retinoid-related orphan receptor RORγ. Cancer Res. 2002;62:901–909. [PubMed] [Google Scholar]
- 142.Lekva T, Berg JP, Heck A, Lyngvi Fougner S, Olstad OK, Ringstad G, Bollerslev J, Ueland T. Attenuated RORC expression in the presence of EMT progression in somatotroph adenomas following treatment with somatostatin analogs is associated with poor clinical recovery. PLoS One. 2013;8:e66927. doi: 10.1371/journal.pone.0066927. Article ID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cadenas C, van de Sandt L, Edlund K, Lohr M, Hellwig B, Marchan R, Schmidt M, Rahnenführer J, Oster H, Hengstler JG. Loss of circadian clock gene expression is associated with tumor progression in breast cancer. Cell Cycle. 2014;13:3282–3291. doi: 10.4161/15384101.2014.954454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Davidson B, Abeler VM, Førsund M, Holth A, Yang Y, Kobayashi Y, Chen L, Kristensen GB, Shih IM, Wang TL. Gene expression signatures of primary and metastatic uterine leiomyosarcoma. Hum Pathol. 2014;45:691–700. doi: 10.1016/j.humpath.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Risinger JI, Allard J, Chandran U, Day R, Chandramouli GV, Miller C, Zahn C, Oliver J, Litzi T, Marcus C, Dubil E, Byrd K, Cassablanca Y, Becich M, Berchuck A, Darcy KM, Hamilton CA, Conrads TP, Maxwell GL. Gene expression analysis of early stage endometrial cancers reveals unique transcripts associated with grade and histology but not depth of invasion. Front Oncol. 2013;3:139. doi: 10.3389/fonc.2013.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhu Y, McAvoy S, Kuhn R, Smith DI. RORA, a large common fragile site gene, is involved in cellular stress response. Oncogene. 2006;25:2901–2908. doi: 10.1038/sj.onc.1209314. [DOI] [PubMed] [Google Scholar]
- 147.Kottorou AE, Antonacopoulou AG, Dimitrakopoulos FI, Tsamandas AC, Scopa CD, Petsas T, Kalofonos HP. Altered expression of NFY-C and RORA in colorectal adenocarcinomas. Acta Histochem. 2012;114:553–561. doi: 10.1016/j.acthis.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 148.Moretti RM, Montagnani Marelli M, Sala A, Motta M, Limonta P. Activation of the orphan nuclear receptor RORα counteracts the proliferative effect of fatty acids on prostate cancer cells: crucial role of 5-lipoxygenase. Int J Cancer. 2004;112:87–93. doi: 10.1002/ijc.20387. [DOI] [PubMed] [Google Scholar]
- 149.Moretti RM, Marelli MM, Motta M, Polizzi D, Monestiroli S, Pratesi G, Limonta P. Activation of the orphan nuclear receptor RORα induces growth arrest in androgen-independent DU 145 prostate cancer cells. Prostate. 2001;46:327–335. doi: 10.1002/1097-0045(20010301)46:4<327::aid-pros1040>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 150.Xiong G, Wang C, Evers BM, Zhou BP, Xu R. RORα suppresses breast tumor invasion by inducing SEMA3F expression. Cancer Res. 2012;72:1728–1739. doi: 10.1158/0008-5472.CAN-11-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Du J, Xu R. RORα, a potential tumor suppressor and therapeutic target of breast cancer. Int J Mol Sci. 2012;13:15755–15766. doi: 10.3390/ijms131215755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Fu RD, Qiu CH, Chen HA, Zhang ZG, Lu MQ. Retinoic acid receptor-related receptor α (RORα) is a prognostic marker for hepatocellular carcinoma. Tumour Biol. 2014;35:7603–7610. doi: 10.1007/s13277-014-2007-9. [DOI] [PubMed] [Google Scholar]
- 153.Ou Z, Shi X, Gilroy RK, Kirisci L, Romkes M, Lynch C, Wang H, Xu M, Jiang M, Ren S, Gramignoli R, Strom SC, Huang M, Xie W. Regulation of the human hydroxysteroid sulfotransferase (SULT2A1) by RORα and RORγ and its potential relevance to human liver diseases. Mol Endocrinol. 2013;27:106–115. doi: 10.1210/me.2012-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chauvet C, Vanhoutteghem A, Duhem C, Saint-Auret G, Bois-Joyeux B, Djian P, Staels B, Danan JL. Control of gene expression by the retinoic acid-related orphan receptor α in HepG2 human hepatoma cells. PLoS One. 2011;6:e22545. doi: 10.1371/journal.pone.0022545. Article ID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kim H, Lee JM, Lee G, Bhin J, Oh SK, Kim K, Pyo KE, Lee JS, Yim HY, Kim KI, Hwang D, Chung J, Baek SH. DNA damage-induced RORα is crucial for p53 stabilization and increased apoptosis. Mol Cell. 2011;44:797–810. doi: 10.1016/j.molcel.2011.09.023. [DOI] [PubMed] [Google Scholar]
- 156.Wang Y, Solt LA, Kojetin DJ, Burris TP. Regulation of p53 stability and apoptosis by a ROR agonist. PLoS One. 2012;7:e34921. doi: 10.1371/journal.pone.0034921. Article ID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Byun JK, Choi YK, Kang YN, Jang BK, Kang KJ, Jeon YH, Lee HW, Jeon JH, Koo SH, Jeong WI, Harris RA, Lee IK, Park KG. Retinoic acid-related orphan receptor α reprograms glucose metabolism in glutamine-deficient hepatoma cells. Hepatology. 2015;61:953–964. doi: 10.1002/hep.27577. [DOI] [PubMed] [Google Scholar]
- 158.Lee JM, Kim IS, Kim H, Lee JS, Kim K, Yim HY, Jeong J, Kim JH, Kim JY, Lee H, Seo SB, Kim H, Rosenfeld MG, Kim KI, Baek SH. RORα attenuates Wnt/β-catenin signaling by PKCα-dependent phosphorylation in colon cancer. Mol Cell. 2010;37:183–195. doi: 10.1016/j.molcel.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 159.Xiong G, Xu R. RORα binds to E2F1 to inhibit cell proliferation and regulate mammary gland branching morphogenesis. Mol Cell Biol. 2014;34:3066–3075. doi: 10.1128/MCB.00279-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Odawara H, Iwasaki T, Horiguchi J, Rokutanda N, Hirooka K, Miyazaki W, Koibuchi Y, Shimokawa N, Iino Y, Takeyoshi I, Koibuchi N. Activation of aromatase expression by retinoic acid receptor-related orphan receptor (ROR) α in breast cancer cells: identification of a novel ROR response element. J Biol Chem. 2009;284:17711–17719. doi: 10.1074/jbc.M109.009241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lau P, Fitzsimmons RL, Pearen MA, Watt MJ, Muscat GE. Homozygous staggerer (sg/sg) mice display improved insulin sensitivity and enhanced glucose uptake in skeletal muscle. Diabetologia. 2011;54:1169–1180. doi: 10.1007/s00125-011-2046-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lau P, Fitzsimmons RL, Raichur S, Wang SC, Lechtken A, Muscat GE. The orphan nuclear receptor, RORα, regulates gene expression that controls lipid metabolism: staggerer (SG/SG) mice are resistant to diet-induced obesity. J Biol Chem. 2008;283:18411–18421. doi: 10.1074/jbc.M710526200. [DOI] [PubMed] [Google Scholar]
- 163.Raichur S, Fitzsimmons RL, Myers SA, Pearen MA, Lau P, Eriksson N, Wang SM, Muscat GE. Identification and validation of the pathways and functions regulated by the orphan nuclear receptor, ROR α1, in skeletal muscle. Nucleic Acids Res. 2010;38:4296–4312. doi: 10.1093/nar/gkq180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Raichur S, Lau P, Staels B, Muscat GE. Retinoid-related orphan receptor γ regulates several genes that control metabolism in skeletal muscle cells: links to modulation of reactive oxygen species production. J Mol Endocrinol. 2007;39:29–44. doi: 10.1677/jme.1.00010. [DOI] [PubMed] [Google Scholar]
- 165.Takeda Y, Kang HS, Freudenberg J, DeGraff LM, Jothi R, Jetten AM. Retinoic acid-related orphan receptor γ (RORγ): a novel participant in the diurnal regulation of hepatic gluconeogenesis and insulin sensitivity. PLoS Genet. 2014;10:e1004331. doi: 10.1371/journal.pgen.1004331. Article ID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang Y, Solt LA, Burris TP. Regulation of FGF21 expression and secretion by retinoic acid receptor-related orphan receptor α. J Biol Chem. 2010;285:15668–15673. doi: 10.1074/jbc.M110.102160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kang HS, Angers M, Beak JY, Wu X, Gimble JM, Wada T, Xie W, Collins JB, Grissom SF, Jetten AM. Gene expression profiling reveals a regulatory role for ROR α and ROR γ in phase I and phase II metabolism. Physiol Genomics. 2007;31:281–294. doi: 10.1152/physiolgenomics.00098.2007. [DOI] [PubMed] [Google Scholar]
- 168.Takeda Y, Kang HS, Lih FB, Jiang H, Blaner WS, Jetten AM. Retinoid acid-related orphan receptor γ, RORγ, participates in diurnal transcriptional regulation of lipid metabolic genes. Nucleic Acids Res. 2014;42:10448–10459. doi: 10.1093/nar/gku766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Fitzsimmons RL, Lau P, Muscat GE. Retinoid-related orphan receptor α and the regulation of lipid homeostasis. J Steroid Biochem Mol Biol. 2012;130:159–168. doi: 10.1016/j.jsbmb.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 170.Bass J. Circadian topology of metabolism. Nature. 2012;491:348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 171.Duez H, Staels B. Nuclear receptors linking circadian rhythms and cardiometabolic control. Arterioscler Thromb Vasc Biol. 2010;30:1529–1534. doi: 10.1161/ATVBAHA.110.209098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 173.Moullé VS, Picard A, Le Foll C, Levin BE, Magnan C. Lipid sensing in the brain and regulation of energy balance. Diabetes Metab. 2014;40:29–33. doi: 10.1016/j.diabet.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 174.Kang HS, Okamoto K, Takeda Y, Beak JY, Gerrish K, Bortner CD, DeGraff LM, Wada T, Xie W, Jetten AM. Transcriptional profiling reveals a role for RORα in regulating gene expression in obesity-associated inflammation and hepatic steatosis. Physiol Genomics. 2011;43:818–828. doi: 10.1152/physiolgenomics.00206.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lau P, Tuong ZK, Wang SC, Fitzsimmons RL, Goode JM, Thomas GP, Cowin GJ, Pearen MA, Mardon K, Stow JL, Muscat GE. Rorα deficiency and decreased adiposity are associated with induction of thermogenic gene expression in subcutaneous white adipose and brown adipose tissue. Am J Physiol Endocrinol Metab. 2015;308:E159–E171. doi: 10.1152/ajpendo.00056.2014. [DOI] [PubMed] [Google Scholar]
- 176.Somm E, Henrichot E, Pernin A, Juge-Aubry CE, Muzzin P, Dayer JM, Nicklin MJ, Meier CA. Decreased fat mass in interleukin-1 receptor antagonist-deficient mice: impact on adipogenesis, food intake, and energy expenditure. Diabetes. 2005;54:3503–3509. doi: 10.2337/diabetes.54.12.3503. [DOI] [PubMed] [Google Scholar]
- 177.Nishimura S, Manabe I, Nagasaki M, Eto K, Yamashita H, Ohsugi M, Otsu M, Hara K, Ueki K, Sugiura S, Yoshimura K, Kadowaki T, Nagai R. CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat Med. 2009;15:914–920. doi: 10.1038/nm.1964. [DOI] [PubMed] [Google Scholar]
- 178.Gamboa-Meléndez MA, Huerta-Chagoya A, Moreno-Macías H, Vázquez-Cárdenas P, Ordóñez-Sánchez ML, Rodríguez-Guillén R, Riba L, Rodríguez-Torres M, Guerra-García MT, Guillén-Pineda LE, Choudhry S, Del Bosque-Plata L, Canizales-Quinteros S, Pérez-Ortiz G, Escobedo-Aguirre F, Parra A, Lerman-Garber I, Aguilar-Salinas CA, Tusié-Luna MT. Contribution of common genetic variation to the risk of type 2 diabetes in the Mexican Mestizo population. Diabetes. 2012;61:3314–3321. doi: 10.2337/db11-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nagano G, Ohno H, Oki K, Kobuke K, Shiwa T, Yoneda M, Kohno N. Activation of classical brown adipocytes in the adult human perirenal depot is highly correlated with PRDM16-EHMT1 complex expression. PLoS One. 2015;10:e0122584. doi: 10.1371/journal.pone.0122584. Article ID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Austin S, Medvedev A, Yan ZH, Adachi H, Hirose T, Jetten AM. Induction of the nuclear orphan receptor RORγ during adipocyte differentiation of D1 and 3T3-L1 cells. Cell Growth Differ. 1998;9:267–276. [PubMed] [Google Scholar]
- 181.Ohoka N, Kato S, Takahashi Y, Hayashi H, Sato R. The orphan nuclear receptor RORα restrains adipocyte differentiation through a reduction of C/EBPβ activity and perilipin gene expression. Mol Endocrinol. 2009;23:759–771. doi: 10.1210/me.2008-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tuong ZK, Lau P, Yeo JC, Pearen MA, Wall AA, Stanley AC, Stow JL, Muscat GE. Disruption of Rorα1 and cholesterol 25-hydroxylase expression attenuates phagocytosis in male Rorαsg/sg mice. Endocrinology. 2013;154:140–149. doi: 10.1210/en.2012-1889. [DOI] [PubMed] [Google Scholar]
- 183.Pathak P, Li T, Chiang JY. Retinoic acid-related orphan receptor α regulates diurnal rhythm and fasting induction of sterol 12α-hydroxylase in bile acid synthesis. J Biol Chem. 2013;288:37154–37165. doi: 10.1074/jbc.M113.485987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kumar N, Kojetin DJ, Solt LA, Kumar KG, Nuhant P, Duckett DR, Cameron MD, Butler AA, Roush WR, Griffin PR, Burris TP. Identification of SR3335 (ML-176): a synthetic RORα selective inverse agonist. ACS Chem Biol. 2011;6:218–222. doi: 10.1021/cb1002762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wada T, Kang HS, Angers M, Gong H, Bhatia S, Khadem S, Ren S, Ellis E, Strom SC, Jetten AM, Xie W. Identification of oxysterol 7α-hydroxylase (Cyp7b1) as a novel retinoid-related orphan receptor α (RORα) (NR1F1) target gene and a functional cross-talk between RORα and liver X receptor (NR1H3) Mol Pharmacol. 2008;73:891–899. doi: 10.1124/mol.107.040741. [DOI] [PubMed] [Google Scholar]
- 186.Kim EJ, Yoon YS, Hong S, Son HY, Na TY, Lee MH, Kang HJ, Park J, Cho WJ, Kim SG, Koo SH, Park HG, Lee MO. Retinoic acid receptor-related orphan receptor α-induced activation of adenosine monophosphate-activated protein kinase results in attenuation of hepatic steatosis. Hepatology. 2012;55:1379–1388. doi: 10.1002/hep.25529. [DOI] [PubMed] [Google Scholar]
- 187.Crumbley C, Wang Y, Banerjee S, Burris TP. Regulation of expression of citrate synthase by the retinoic acid receptor-related orphan receptor α (RORα) PLoS One. 2012;7:e33804. doi: 10.1371/journal.pone.0033804. Article ID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Chen Y, Coulter S, Jetten AM, Goldstein JA. Identification of human CYP2C8 as a retinoid-related orphan nuclear receptor target gene. J Pharmacol Exp Ther. 2009;329:192–201. doi: 10.1124/jpet.108.148916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lau P, Nixon SJ, Parton RG, Muscat GE. RORα regulates the expression of genes involved in lipid homeostasis in skeletal muscle cells: caveolin-3 and CPT-1 are direct targets of ROR. J Biol Chem. 2004;279:36828–36840. doi: 10.1074/jbc.M404927200. [DOI] [PubMed] [Google Scholar]
- 190.Meissburger B, Ukropec J, Roeder E, Beaton N, Geiger M, Teupser D, Civan B, Langhans W, Nawroth PP, Gasperikova D, Rudofsky G, Wolfrum C. Adipogenesis and insulin sensitivity in obesity are regulated by retinoid-related orphan receptor γ. EMBO Mol Med. 2011;3:637–651. doi: 10.1002/emmm.201100172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Tinahones FJ, Moreno-Santos I, Vendrell J, Chacon MR, Garrido-Sanchez L, García-Fuentes E, Macias-González M. The retinoic acid receptor-related orphan nuclear receptor γ1 (RORγ1): a novel player determinant of insulin sensitivity in morbid obesity. Obesity (Silver Spring) 2012;20:488–497. doi: 10.1038/oby.2011.267. [DOI] [PubMed] [Google Scholar]
- 192.Takeda Y, Jothi R, Birault V, Jetten AM. RORγ directly regulates the circadian expression of clock genes and downstream targets in vivo. Nucleic Acids Res. 2012;40:8519–8535. doi: 10.1093/nar/gks630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Narasimamurthy R, Hatori M, Nayak SK, Liu F, Panda S, Verma IM. Circadian clock protein cryptochrome regulates the expression of proinflammatory cytokines. Proc Natl Acad Sci USA. 2012;109:12662–12667. doi: 10.1073/pnas.1209965109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. Article ID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, Hirota T, Nusinow DA, Sun X, Landais S, Kodama Y, Brenner DA, Montminy M, Kay SA. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 197.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15:R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 198.Maury E, Ramsey KM, Bass J. Circadian rhythms and metabolic syndrome: from experimental genetics to human disease. Circ Res. 2010;106:447–462. doi: 10.1161/CIRCRESAHA.109.208355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, Ellisman MH, Panda S. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15:848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Dibner C, Schibler U. Circadian timing of metabolism in animal models and humans. J Intern Med. 2015;277:513–527. doi: 10.1111/joim.12347. [DOI] [PubMed] [Google Scholar]
- 201.Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, Lopez JP, Philipson LH, Bradfield CA, Crosby SD, JeBailey L, Wang X, Takahashi JS, Bass J. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Akashi M, Takumi T. The orphan nuclear receptor RORα regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol. 2005;12:441–448. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- 203.Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, Iino M, Shigeyoshi Y, Hashimoto S. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 204.Mongrain V, Ruan X, Dardente H, Fortier EE, Cermakian N. Clock-dependent and independent transcriptional control of the two isoforms from the mouse Rorγ gene. Genes Cells. 2008;13:1197–1210. doi: 10.1111/j.1365-2443.2008.01237.x. [DOI] [PubMed] [Google Scholar]
- 205.Mühlbauer E, Bazwinsky-Wutschke I, Wolgast S, Labucay K, Peschke E. Differential and day-time dependent expression of nuclear receptors RORα, RORβ, RORγ and RXRα in the rodent pancreas and islet. Mol Cell Endocrinol. 2013;365:129–138. doi: 10.1016/j.mce.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 206.André E, Conquet F, Steinmayr M, Stratton SC, Porciatti V, Becker-André M, Disruption of retinoid-related orphan receptor β changes circadian behavior. causes retinal degeneration and leads to vacillans phenotype in mice. EMBO J. 1998;17:3867–3877. doi: 10.1093/emboj/17.14.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Sumi Y, Yagita K, Yamaguchi S, Ishida Y, Kuroda Y, Okamura H. Rhythmic expression of ROR β mRNA in the mice suprachiasmatic nucleus. Neurosci Lett. 2002;320:13–16. doi: 10.1016/s0304-3940(02)00011-3. [DOI] [PubMed] [Google Scholar]
- 208.Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. Article ID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, Liddle C, Auwerx J, Downes M, Panda S, Evans RM. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. Redundant function of REV-ERBα and β and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. Article ID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 212.Kumaki Y, Ukai-Tadenuma M, Uno KD, Nishio J, Masumoto KH, Nagano M, Komori T, Shigeyoshi Y, Hogenesch JB, Ueda HR. Analysis and synthesis of high-amplitude Cis-elements in the mammalian circadian clock. Proc Natl Acad Sci USA. 2008;105:14946–14951. doi: 10.1073/pnas.0802636105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Nakajima Y, Ikeda M, Kimura T, Honma S, Ohmiya Y, Honma K. Bidirectional role of orphan nuclear receptor RORα in clock gene transcriptions demonstrated by a novel reporter assay system. FEBS Lett. 2004;565:122–126. doi: 10.1016/j.febslet.2004.03.083. [DOI] [PubMed] [Google Scholar]
- 214.Crumbley C, Wang Y, Kojetin DJ, Burris TP. Characterization of the core mammalian clock component, NPAS2, as a REV-ERBα/RORα target gene. J Biol Chem. 2010;285:35386–35392. doi: 10.1074/jbc.M110.129288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Matsumura R, Matsubara C, Node K, Takumi T, Akashi M. Nuclear receptor-mediated cell-autonomous oscillatory expression of the circadian transcription factor, neuronal PAS domain protein 2 (NPAS2) J Biol Chem. 2013;288:36548–36553. doi: 10.1074/jbc.M113.517235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 217.Guillaumond F, Dardente H, Giguère V, Cermakian N. Differential control of Bmal1 circadian transcription by REVERB and ROR nuclear receptors. J Biol Rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 218.Delerive P, Chin WW, Suen CS. Identification of Reverb(α) as a novel ROR(α) target gene. J Biol Chem. 2002;277:35013–35018. doi: 10.1074/jbc.M202979200. [DOI] [PubMed] [Google Scholar]
- 219.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 220.Raspé E, Duez H, Mansén A, Fontaine C, Fiévet C, Fruchart JC, Vennström B, Staels B. Identification of Rev-erbα as a physiological repressor of apoC-III gene transcription. J Lipid Res. 2002;43:2172–2179. doi: 10.1194/jlr.m200386-jlr200. [DOI] [PubMed] [Google Scholar]
- 221.Solt LA, Kojetin DJ, Burris TP. The REV-ERBs and RORs: molecular links between circadian rhythms and lipid homeostasis. Future Med Chem. 2011;3:623–638. doi: 10.4155/fmc.11.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Poliandri AH, Gamsby JJ, Christian M, Spinella MJ, Loros JJ, Dunlap JC, Parker MG. Modulation of clock gene expression by the transcriptional coregulator receptor interacting protein 140 (RIP140) J Biol Rhythms. 2011;26:187–199. doi: 10.1177/0748730411401579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Takeda Y, Jetten AM. Prospero-related homeobox 1 (Prox1) functions as a novel modulator of retinoic acid-related orphan receptors α- and γ-mediated transactivation. Nucleic Acids Res. 2013;41:6992–7008. doi: 10.1093/nar/gkt447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Dufour CR, Levasseur MP, Pham NH, Eichner LJ, Wilson BJ, Charest-Marcotte A, Duguay D, Poirier-Héon JF, Cermakian N, Giguère V. Genomic convergence among ERRα, PROX1, and BMAL1 in the control of metabolic clock outputs. PLoS Genet. 2011;7:e1002143. doi: 10.1371/journal.pgen.1002143. Article ID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Yu X, Rollins D, Ruhn KA, Stubblefield JJ, Green CB, Kashiwada M, Rothman PB, Takahashi JS, Hooper LV. TH17 cell differentiation is regulated by the circadian clock. Science. 2013;342:727–730. doi: 10.1126/science.1243884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Palmieri O, Mazzoccoli G, Bossa F, Maglietta R, Palumbo O, Ancona N, Corritore G, Latiano T, Martino G, Rubino R, Biscaglia G, Scimeca D, Carella M, Annese V, Andriulli A, Latiano A. Systematic analysis of circadian genes using genome-wide cDNA microarrays in the inflammatory bowel disease transcriptome. Chronobiol Int. 2015;32:903–916. doi: 10.3109/07420528.2015.1050726. [DOI] [PubMed] [Google Scholar]
- 227.Klein ES, Pino ME, Johnson AT, Davies PJ, Nagpal S, Thacher SM, Krasinski G, Chandraratna RA. Identification and functional separation of retinoic acid receptor neutral antagonists and inverse agonists. J Biol Chem. 1996;271:22692–22696. doi: 10.1074/jbc.271.37.22692. [DOI] [PubMed] [Google Scholar]
- 228.Kallen J, Schlaeppi JM, Bitsch F, Delhon I, Fournier B. Crystal structure of the human RORα Ligand binding domain in complex with cholesterol sulfate at 2.2 A. J Biol Chem. 2004;279:14033–14038. doi: 10.1074/jbc.M400302200. [DOI] [PubMed] [Google Scholar]
- 229.Stehlin-Gaon C, Willmann D, Zeyer D, Sanglier S, Van Dorsselaer A, Renaud JP, Moras D, Schüle R. All-trans retinoic acid is a ligand for the orphan nuclear receptor ROR β. Nat Struct Biol. 2003;10:820–825. doi: 10.1038/nsb979. [DOI] [PubMed] [Google Scholar]
- 230.Jin L, Martynowski D, Zheng S, Wada T, Xie W, Li Y. Structural basis for hydroxycholesterols as natural ligands of orphan nuclear receptor RORγ. Mol Endocrinol. 2010;24:923–929. doi: 10.1210/me.2009-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Wang Y, Kumar N, Crumbley C, Griffin PR, Burris TP. A second class of nuclear receptors for oxysterols: Regulation of RORα and RORγ activity by 24S-hydroxycholesterol (cerebrosterol) Biochim Biophys Acta. 2010;1801:917–923. doi: 10.1016/j.bbalip.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Wang Y, Kumar N, Solt LA, Richardson TI, Helvering LM, Crumbley C, Garcia-Ordonez RD, Stayrook KR, Zhang X, Novick S, Chalmers MJ, Griffin PR, Burris TP. Modulation of retinoic acid receptor-related orphan receptor α and γ activity by 7-oxygenated sterol ligands. J Biol Chem. 2010;285:5013–5025. doi: 10.1074/jbc.M109.080614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Soroosh P, Wu J, Xue X, Song J, Sutton SW, Sablad M, Yu J, Nelen MI, Liu X, Castro G, Luna R, Crawford S, Banie H, Dandridge RA, Deng X, Bittner A, Kuei C, Tootoonchi M, Rozenkrants N, Herman K, Gao J, Yang XV, Sachen K, Ngo K, Fung-Leung WP, Nguyen S, de Leon-Tabaldo A, Blevitt J, Zhang Y, Cummings MD, Rao T, Mani NS, Liu C, McKinnon M, Milla ME, Fourie AM, Sun S. Oxysterols are agonist ligands of RORγt and drive Th17 cell differentiation. Proc Natl Acad Sci USA. 2014;111:12163–12168. doi: 10.1073/pnas.1322807111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Kumar N, Solt LA, Conkright JJ, Wang Y, Istrate MA, Busby SA, Garcia-Ordonez RD, Burris TP, Griffin PR. The benzenesulfoamide T0901317 [N-(2,2,2-trifluoroethyl)-N-[4-[2,2,2-trifluoro-1-hydroxy-1-(trifluoromethyl)ethyl]phenyl]-benzenesulfonamide] is a novel retinoic acid receptor-related orphan receptor-α/γ inverse agonist. Mol Pharmacol. 2010;77:228–236. doi: 10.1124/mol.109.060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Kinzel O, Gege C, Steeneck C, Kleymann G, Hoffmann T. Seven-membered sulfonamides as modulators of RAR-related orphan receptor-γ (RORγ, NR1F3) 2013 WO 2013/064231. [Google Scholar]
- 236.Steeneck C, Kinzel O, Gege C, Kleymann G, Hoffmann T. Pyrrolo carboxamides as modulators of orphan nuclear receptor RAR-related orphan receptor-γ (RORγ, NR1F3) activity and for the treatment of chronic inflammatory and autoimmune diseases. 2013 WO 2013/079223. [Google Scholar]
- 237.Solt LA, Kumar N, Nuhant P, Wang Y, Lauer JL, Liu J, Istrate MA, Kamenecka TM, Roush WR, Vidović D, Schürer SC, Xu J, Wagoner G, Drew PD, Griffin PR, Burris TP. Suppression of TH17 differentiation and autoimmunity by a synthetic ROR ligand. Nature. 2011;472:491–494. doi: 10.1038/nature10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Glick G, Toogood P, Romero A, Vanhuis C, Aicher T, Kaub C, Mattson M, Thomas W, Stein K, Krigh-Jespersen E, Wang Z. Tetrahydroquinoline and related bicyclic compounds for inhibition of RORγ activity and treatment of disease. 2012 WO 2012/064744. [Google Scholar]
- 239.Birault V, Campbell A, Harrison S, Le J. Sulfonamide compounds and their use in the modulation of retinoid-related orphan receptor. 2013 WO 2013/045431. [Google Scholar]
- 240.Wang Y, Cai W, Zhang G, Yang T, Liu Q, Cheng Y, Zhou L, Ma Y, Cheng Z, Lu S, Zhao YG, Zhang W, Xiang Z, Wang S, Yang L, Wu Q, Orband-Miller LA, Xu Y, Zhang J, Gao R, Huxdorf M, Xiang JN, Zhong Z, Elliott JD, Leung S, Lin X. Discovery of novel N-(5-(arylcarbonyl)thiazol-2-yl)amides and N-(5-(arylcarbonyl)thiophen-2-yl)amides as potent RORγt inhibitors. Bioorg Med Chem. 2014;22:692–702. doi: 10.1016/j.bmc.2013.12.021. [DOI] [PubMed] [Google Scholar]
- 241.Fauber BP, René O, Burton B, Everett C, Gobbi A, Hawkins J, Johnson AR, Liimatta M, Lockey P, Norman M, Wong H. Identification of tertiary sulfonamides as RORc inverse agonists. Bioorg Med Chem Lett. 2014;24:2182–2187. doi: 10.1016/j.bmcl.2014.03.038. [DOI] [PubMed] [Google Scholar]
- 242.van Niel MB, Fauber BP, Cartwright M, Gaines S, Killen JC, René O, Ward SI, de Leon Boenig G, Deng Y, Eidenschenk C, Everett C, Gancia E, Ganguli A, Gobbi A, Hawkins J, Johnson AR, Kiefer JR, La H, Lockey P, Norman M, Ouyang W, Qin A, Wakes N, Waszkowycz B, Wong H. A reversed sulfonamide series of selective RORc inverse agonists. Bioorg Med Chem Lett. 2014;24:5769–5776. doi: 10.1016/j.bmcl.2014.10.037. [DOI] [PubMed] [Google Scholar]
- 243.Kumar N, Lyda B, Chang MR, Lauer JL, Solt LA, Burris TP, Kamenecka TM, Griffin PR. Identification of SR2211: a potent synthetic RORγ-selective modulator. ACS Chem Biol. 2012;7:672–677. doi: 10.1021/cb200496y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Xu T, Wang X, Zhong B, Nurieva RI, Ding S, Dong C. Ursolic acid suppresses interleukin-17 (IL-17) production by selectively antagonizing the function of RORγ t protein. J Biol Chem. 2011;286:22707–22710. doi: 10.1074/jbc.C111.250407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Slominski AT, Kim TK, Takeda Y, Janjetovic Z, Brozyna AA, Skobowiat C, Wang J, Postlethwaite A, Li W, Tuckey RC, Jetten AM. RORα and ROR γ are expressed in human skin and serve as receptors for endogenously produced noncalcemic 20-hydroxy- and 20,23-dihydroxyvitamin D. FASEB J. 2014;28:2775–2789. doi: 10.1096/fj.13-242040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Huh JR, Leung MW, Huang P, Ryan DA, Krout MR, Malapaka RR, Chow J, Manel N, Ciofani M, Kim SV, Cuesta A, Santori FR, Lafaille JJ, Xu HE, Gin DY, Rastinejad F, Littman DR. Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORγt activity. Nature. 2011;472:486–490. doi: 10.1038/nature09978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Yang T, Liu Q, Cheng Y, Cai W, Ma Y, Yang L, Wu Q, Orband-Miller LA, Zhou L, Xiang Z, Huxdorf M, Zhang W, Zhang J, Xiang JN, Leung S, Qiu Y, Zhong Z, Elliott JD, Lin X, Wang Y. Discovery of tertiary amine and indole derivatives as potent RORγt inverse agonists. ACS Med Chem Lett. 2014;5:65–68. doi: 10.1021/ml4003875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Fauber BP, René O, Deng Y, DeVoss J, Eidenschenk C, Everett C, Ganguli A, Gobbi A, Hawkins J, Johnson AR, La H, Lesch J, Lockey P, Norman M, Ouyang W, Summerhill S, Wong H. Discovery of 1-4-[3-fluoro-4-((3s,6r)-3-methyl-1,1-dioxo-6-phenyl-[1,2]thiazinan-2-ylmethyl)-phenyl]-piperazin-1-yl-ethanone (GNE-3500): a potent, selective, and orally bioavailable retinoic acid receptor-related orphan receptor C (RORc or RORγ) inverse agonist. J Med Chem. 2015;58:5308–5322. doi: 10.1021/acs.jmedchem.5b00597. [DOI] [PubMed] [Google Scholar]
- 249.Marciano DP, Chang MR, Corzo CA, Goswami D, Lam VQ, Pascal BD, Griffin PR. The therapeutic potential of nuclear receptor modulators for treatment of metabolic disorders: PPARγ, RORs, and Rev-erbs. Cell Metab. 2014;19:193–208. doi: 10.1016/j.cmet.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 250.Kojima H, Muromoto R, Takahashi M, Takeuchi S, Takeda Y, Jetten AM, Matsuda T. Inhibitory effects of azole-type fungicides on interleukin-17 gene expression via retinoic acid receptor-related orphan receptors α and γ. Toxicol Appl Pharmacol. 2012;259:338–345. doi: 10.1016/j.taap.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Kidani Y, Elsaesser H, Hock MB, Vergnes L, Williams KJ, Argus JP, Marbois BN, Komisopoulou E, Wilson EB, Osborne TF, Graeber TG, Reue K, Brooks DG, Bensinger SJ. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat Immunol. 2013;14:489–499. doi: 10.1038/ni.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Maggio R, Viscomi C, Andreozzi P, D’Ettorre G, Viscogliosi G, Barbaro B, Gori M, Vullo V, Balsano C. Normocaloric low cholesterol diet modulates Th17/Treg balance in patients with chronic hepatitis C virus infection. PLoS One. 2014;9:e112346. doi: 10.1371/journal.pone.0112346. Article ID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Ulivieri C, Baldari CT. Statins: from cholesterol-lowering drugs to novel immunomodulators for the treatment of Th17-mediated autoimmune diseases. Pharmacol Res. 2014;88:41–52. doi: 10.1016/j.phrs.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 254.Kurebayashi S, Nakajima T, Kim SC, Chang CY, McDonnell DP, Renaud JP, Jetten AM. Selective LXXLL peptides antagonize transcriptional activation by the retinoid-related orphan receptor RORγ. Biochem Biophys Res Commun. 2004;315:919–927. doi: 10.1016/j.bbrc.2004.01.131. [DOI] [PubMed] [Google Scholar]
- 255.Lau P, Bailey P, Dowhan DH, Muscat GE. Exogenous expression of a dominant negative RORα1 vector in muscle cells impairs differentiation: RORα1 directly interacts with p300 and myoD. Nucleic Acids Res. 1999;27:411–420. doi: 10.1093/nar/27.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Moraitis AN, Giguère V, Thompson CC. Novel mechanism of nuclear receptor corepressor interaction dictated by activation function 2 helix determinants. Mol Cell Biol. 2002;22:6831–6841. doi: 10.1128/MCB.22.19.6831-6841.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Huh JR, Littman DR. Small molecule inhibitors of RORγt: targeting Th17 cells and other applications. Eur J Immunol. 2012;42:2232–2237. doi: 10.1002/eji.201242740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Elloso MM, Gomez-Angelats M, Fourie AM. Targeting the Th17 pathway in psoriasis. J Leukoc Biol. 2012;92:1187–1197. doi: 10.1189/jlb.0212101. [DOI] [PubMed] [Google Scholar]
- 259.Skepner J, Ramesh R, Trocha M, Schmidt D, Baloglu E, Lobera M, Carlson T, Hill J, Orband-Miller LA, Barnes A, Boudjelal M, Sundrud M, Ghosh S, Yang J. Pharmacologic inhibition of RORγt regulates Th17 signature gene expression and suppresses cutaneous inflammation in vivo. J Immunol. 2014;192:2564–2575. doi: 10.4049/jimmunol.1302190. [DOI] [PubMed] [Google Scholar]
- 260.Hwang YY, McKenzie AN. Innate lymphoid cells in immunity and disease. Adv Exp Med Biol. 2013;785:9–26. doi: 10.1007/978-1-4614-6217-0_2. [DOI] [PubMed] [Google Scholar]
- 261.Pantelyushin S, Haak S, Ingold B, Kulig P, Heppner FL, Navarini AA, Becher B. Rorγt+ innate lymphocytes and γδ T cells initiate psoriasiform plaque formation in mice. Journal of Clinical Investigation. 2012;122:2252–2256. doi: 10.1172/JCI61862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Solt LA, Banerjee S, Campbell S, Kamenecka TM, Burris TP. ROR inverse agonist suppresses insulitis and prevents hyperglycemia in a mouse model of type 1 diabetes. Endocrinology. 2015;156:869–881. doi: 10.1210/en.2014-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]