Abstract
Recent advances in laboratory sciences hold a promise for a ‘leap forward’ in understanding the aetiology of complex human diseases, notably cancer, potentially providing an evidence base for prevention. For example, remarkable advances in epigenomics have an important impact on our understanding of biological phenomena and importance of environmental stressors in complex diseases. Environmental and lifestyle factors are thought to be implicated in the development of a wide range of human cancers by eliciting changes in the epigenome. These changes, thus, represent attractive targets for biomarker discovery intended for the improvement of exposure and risk assessment, diagnosis and prognosis and provision of short-term outcomes in intervention studies. The epigenome can be viewed as an interface between the genome and the environment; therefore, aberrant epigenetic events associated with environmental exposures are likely to play an important role in the onset and progression of different human diseases. The advent of powerful technologies for analysing epigenetic patterns in both cancer tissues and normal cells holds promise that the next few years will be fundamental for the identification of critical cancer- and exposure-associated epigenetic changes and for their evaluation as new generation of biomarkers. Here, we discuss new opportunities in the current age of ‘omics’ technologies for studies with prospective design and associated biospecimens that represent exciting potential for characterising the epigenome as a key component of the fetal exposome and for understanding causal pathways and robust predictors of cancer risk and associated environmental determinants during in utero life. Such studies should improve our knowledge concerning the aetiology of childhood cancer and identify both novel biomarkers and clues to causation, thus, providing an evidence base for cancer prevention.
Introduction
With an annual incidence rate ~150 per million children in developed countries and supposedly lower rates in developing countries, childhood cancer (CC) is relatively rare. In addition, for some common cancer types, such as acute lymphoblastic leukaemia, survival rates have dramatically improved in developed countries. However, overall incidence of cancer in children and adolescents has been steadily increasing in most countries, and the burden of disease in many countries is substantial (1), particularly that it is still the leading cause of disease-related death among children and adolescents (ages 1–19 years) in many countries. In addition, among cancer survivors, several disease-related late effects have been described, including an increased risk of secondary malignancy and social disadvantages. Therefore, despite the successes in treatment, identification of preventable risk factors and high risk groups as well as understanding the natural history of CC remain the preferred options for successful prevention.
About 5% of all CCs are caused by an inherited mutation and even in subtypes like retinoblastoma (a cancer of the eye that develops mainly in children) or acute lymphoblastic leukaemia, in which genetic factors attribute to higher risks, only 25–40% of cases would exhibit genetic alterations (NCI, USA). For example, retinoblastoma is associated with an inherited mutation in the RB1 tumour suppressor gene (2) in 25–30% of cases, and retinoblastoma accounts for only ~3% of all cancers in children. Genetic mutations that cause cancer can arise during the development of a foetus in the womb. For example, one in every 100 children is born with a genetic abnormality that increases risk for leukaemia, although only one child in 8000 with that abnormality actually develops leukaemia (3,4).
A number of modifiable risk factors have been linked with CC. It is likely that the effects of these risk factors are mediated through gene regulatory pathways including epigenetic mechanisms. Adaptive responses during in utero life may also include epigenetic changes (including DNA methylation) in different developmental pathways (such as production and expansion of somatic stem/progenitor cells, metabolic changes and production of and sensitivity to hormones), a combination of which may alter normal development of tissues and organs. These epigenetic changes and other related molecular alterations may be evident at birth and thus could constitute powerful mechanism-based biomarkers that could be exploited in epigenetic-based interventions (5). For example, the epigenetic mechanisms could provide attractive targets for prenatal modulation of different processes related to disease risk in childhood and adulthood (6). Furthermore, with the development of epigenomic high-throughput arrays and deep sequencing-based profiling, it is now possible to perform comprehensive analysis of the epigenome and other ‘omes’ of cord blood samples collected prospectively at birth and analysed later in life after cancers developed and explore potential links between ‘omics’ measures, exposures and CC. These technologies together with robust protocols to analyse the blood epigenome offer new avenues to conduct epigenome-wide association studies (EWAS), either alone or as a part of the ‘exposome’ characterisation (7), in a similar way to genome-wide studies. Association studies in humans between early-life environmental exposures and epigenome-wide changes in DNA methylation are summarised in Table 1.
Table 1.
Association studies in humans between early-life environmental exposures and epigenome-wide changes in DNA methylation
| Reference | (8) | (9) | (10) | (11) | (12) | (13) | (14) | (15) | Hernandez-Vargas in preparation |
|---|---|---|---|---|---|---|---|---|---|
| Exposurea | Smoking | Smoking | Arsenic | Cadmium | Smoking | Smoking | Smoking | Smoking | Aflatoxin B1 |
| Locationb | USA | Norway + USA | USA | Bangladesh | USA | Norway | UK | USA + Norway | Gambia |
| Tissuec | Placenta | Cord blood | Cord blood | Cord blood + 4.5 years | Fetal lung + placenta | Heel blood at birth | Cord blood + 7 years + 17 years | Blood 5–7 years | Infant’s blood (3–6 m) |
| Sample size | 36 | 1062 | 134 | 127 | 85/85 | 889 | 790 | 527 | 115 |
| Array Platformd | HM27 | HM450 | HM450 | HM450 | HM450 | HM450 | HM450 | HM27 | HM450 |
| White blood celle | – | Cell fractionation (n = 21) | Houseman | – | – | Houseman | Houseman | Houseman | Houseman |
| Covariatesf | Gender, birth weight, gestational age | Gender | Gender | Gender, birth weight | Age | Gender, birth weight, gestational age | Gender | Gender | Gender |
| Diff Methg | 38 (10% delta) | 26 | 0 | 0 | Lung = 0; Placenta = 2 | 110 | 28 | 26 | 71 |
| Validation | Bisulfite seq | – | Pyroseq | – | Pyroseq | – | – | Pyroseq | |
| PubMed Identifier (PMID) | 21937876 | 22851337 | 23757598 | 23644563 | 25482056 | 24906187 | 25552657 | 24964093 | – |
aCotinin was used as a surrogate in most studies of association with smoking.
bMain country of origin of the samples used for profiling.
cSome studies included samples several months or years after birth in the form of peripheral blood.
dIllumina Infinium 27K (HM27) or 450K (HM450).
eStrategy used to account for blood cell subpopulations. Houseman’s algorithm is originally published in PMID: 22568884.
fNewborn covariates considered in the analyses.
gNumber of differentially methylated (Diff Meth) sites associated with exposure and with a P value < 0.05 after correction for multiple comparisons.
In addition, there is accumulating evidence that fetal life and early childhood might have an important effect on health in adulthood, too. Early exposure to poor diet, lack of physical activity, tobacco smoke and other environmental exposures can alter child’s growth pattern and may result in altered metabolism, obesity and risk of chronic disease in adulthood. Epigenetic changes in the regulation of genes have been evoked as important mechanisms and could condition rapid growth and childhood obesity through premature changes in hormonal profiles and early maturation. However, the role of specific nutrients and environmental exposure during fetal life and early childhood on epigenetic changes remain unclear. New developments in epigenomics and exposomics can be applied in well-characterised ongoing birth cohorts to evaluate the impact of early exposure on intermediate markers of cancer.
Evidence for prenatal origin of CC
Despite the fact that overall incidence of cancer in children and adolescents has been steadily increasing worldwide (1,16), there have been limited advances in our understanding of the causes and molecular mechanism underlying these malignancies. Evidence from epidemiological studies suggests that environmental ‘exposures’ experienced in utero may influence the risk of developing diseases in childhood (17). Various studies have linked, though with insufficient evidence, exposure to infectious agents, parental smoking, pesticides and maternal folic acid intake to CC aetiology (3,4). Compelling evidence also suggests a link between high birth weight and early-life neoplasia. Most of this evidence relates to childhood leukaemia, the largest subgroup of CC (3,4). That the relevant timing for the effect of these exposures on CC includes fetal life is supported by data from neonatal blood spots, which show that the initiating events for leukaemia occur during fetal development (18,19). However, despite the potential importance of ascertaining whether these exposures might truly be preventable causes of CC, the evidence base remains fairly weak. Apart from the reproducible association of high birth weight with leukaemia in children, for which prospective evidence exists, the majority of associations in observational studies have relied on retrospective evidence, often linked to recall bias, and attributing risks to rare genetic events in only few CC subtypes. Furthermore, the mechanisms by which such exposures might predispose to CC are not well understood. As for prospectively designed cohorts, particularly those involving follow-up on environmental data, they have often been restricted to statistically underpowered sample sizes. Similarly, for disease aetiology, evidence linking environmental factors to CC is lacking or conflicting, apart from the effect of ionising radiation that was based on incidental findings from World War II atomic bombs and accidents at nuclear power plants. Therefore, studying CC in multiple cohorts worldwide is crucial to reach sufficient sample sizes and data on environmental causes. If coupled to prospective designs with available biospecimens and questionnaire data and other ‘physical’ metrics, such studies afford new opportunities for taking a ‘leap forward’ in understanding causal pathways in the current age of ‘omics’ technologies to identify robust predictors of cancer risk.
Changes in the epigenome during in utero life and predisposition to CC
Epigenetic mechanisms play the key role in the establishment and stable propagation of gene activity states over cell generations. Epigenetic mechanisms are believed to play a critical role in modulating the gene expression programme in response to endogenous cues and environmental exposures (20,21). In contrast to the genome, the epigenome is dynamic owing to its plasticity and altered epigenetic states may represent stable fingerprints of environmental exposure. Experimental and epidemiological studies provided the evidence of the effects of environmental exposures on the epigenome (20,21). DNA methylation, histone modifications and non-coding RNAs are the main epigenetic mechanisms that control the gene expression programme during development and cell differentiation. DNA methylation is the most extensively studied epigenetic mark in both normal and cancer cells and a profound deregulation of the methylome is a common event in human malignancies (22).
Previous studies suggested that a ‘window of vulnerability’ exists during in utero development, within which maternal factors and exposures may alter the fetal epigenome, increasing susceptibility to childhood diseases. Interestingly, the frequent chromosomal rearrangements and their fusion proteins in childhood leukaemia usually target or recruit epigenetic modifiers, such as histone acetyl transferases, histone deacetylases and DNA methyltransferases (23). Additionally, chromosomal translocations, the most consistent abnormality in acute lymphoblastic leukaemia (24), may themselves be induced by epigenetic changes, particularly alterations in methylation of CpG-rich areas (hot spots), which increase susceptibility to DNA breaks (25). In this sense, methylome deregulation may act as an early step in leukaemogenesis and may cause persistent chromatin alterations that provoke long-lasting effects (26). Furthermore, if alteration in the methylome occurs in stem/progenitor cells, the epigenetic deregulation would be predicted to be carried across cell generations with the potential to be manifested later as disease in a manner consistent with the epigenetic progenitor model of cancer progression (27).
Aberrant DNA methylation and cancer
Aberrant DNA methylation is observed in virtually all types of cancer and involved in various steps of tumour development (22); therefore, a precise map and eventual understanding of the methylome changes associated with cancer onset and progression are fundamental to improving our abilities to successfully diagnose, treat and prevent CC. The molecular mechanisms involving DNA methylation may modulate the gene expression programme in response to environmental exposures, and it has been suggested that the epigenome functions as an interface between the genome and the environment (21,28,29). The effects of environmental agents on the methylome have been either demonstrated experimentally using different animal and cellular models or inferred from epidemiological studies. Although the epigenome is dynamic owing to the reversible and plastic nature of epigenetic states (30–32), an altered methylome may represent a stable signature of environmental exposure (21,33). In addition, recent studies indicate that blood-based DNA methylation testing may provide a potentially useful biomarker for cancer diagnostics. This can be explained by two alternative rationales, which are not mutually exclusive: (i) DNA methylation alterations can be induced during in utero development (by environmental factors), and that these changes may be propagated as constitutional epimutations in all tissues, although they may constitute the basis for increased cancer risk later in life only in certain tissues and (ii) DNA methylation patterns in blood cells (as a surrogate tissue) may reflect epigenome alterations induced by environmental stimuli that constitute cancer risk factors in other tissues.
Analysing the methylome of cord blood cells using prospective birth cohorts
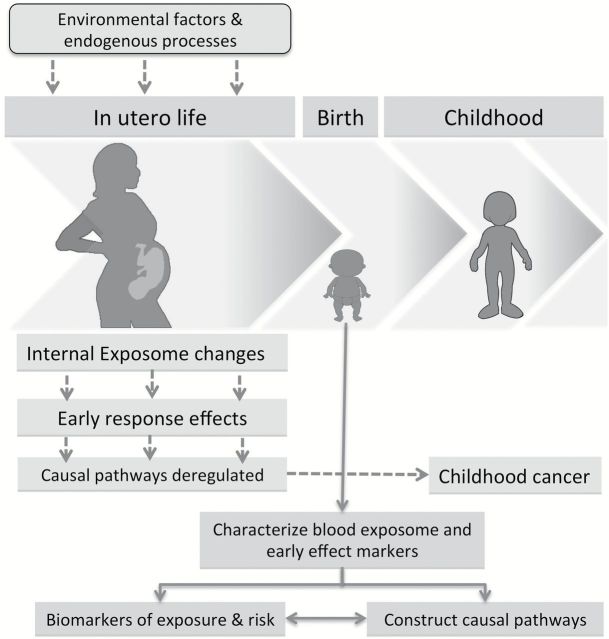
Changes in the epigenome during in utero life (either as a result of maternal exposure or stochastic events) may be at the heart of developmental programming of CC (Figure 1). Accepting the limitations afforded by the incidence of CC, this can best be tested by investigating the epigenetic profile of an infant’s cord blood in existing studies with (i) clinically defined incidence of CC and (ii) data on maternal environmental exposures in utero (Figure 1). Such associations have not been studied previously because both biospecimens collected at birth and follow-up for CC among those sampled are necessary. This material can only be provided by prospective cohort studies of sufficient size that have collected biospecimens on a large fraction of subjects at birth. Such studies are extremely rare and in isolation are generally underpowered to examine associations of this nature.
Figure 1.
In utero exposures and the concept of a developmental origin of CC. This concept suggests that susceptibility to childhood and adult diseases is strongly influenced through adaptive responses, including the deregulation of the epigenome, to in utero conditions. These responses may include changes in different developmental pathways (such as production and expansion of somatic stem/progenitor cells, metabolic changes and production of and sensitivity to hormones), a combination of which may alter normal development of tissues and organs. These changes could persist throughout postnatal life and constitute the bases for differential susceptibility to disease.
While there has been research on the association of maternal exposures with epigenetic signatures (9) (Table I), no study has extended this to obtain evidence on CC occurrence in the infants of these mothers. This is because to date there has been no prospective cohort large enough to provide a meaningful number of incident CCs that could be included in such an examination. Only with the recent establishment of the International Childhood Cancer Cohort Consortium (I4C), which combines the efforts of a growing number of mother/child cohorts internationally (34), has such a possibility existed. Currently, there are ~500000 births in the I4C database for which data have been collected on maternal exposures during pregnancy, biospecimens collected at birth and incident CC ascertained during early life (Table 2) (www.mcri.edu.au/research/research-projects/i4c/). With ~800 CC cases (including ~250 leukaemias), I4C is uniquely positioned to investigate the link between early-life exposures, neonatal epigenetic profile and development of CC.
Table 2.
Maternal factors and risk exposures during pregnancy for six I4C cohorts that are currently participating in data sharing and pooling
| Variable | ALSPAC | CPP | DNBC | JPS | MoBa | TIHS |
|---|---|---|---|---|---|---|
| Age | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Maternal age | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Paternal age, years | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Education | ||||||
| Maternal education, years | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Paternal education, years | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Marital Status | ||||||
| Single, married, divorced, living together… | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Occupation | ||||||
| Maternal occupation | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Paternal occupation | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Smoking | ||||||
| Maternal, prenatal smoking | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Passive smoking, prenatal | ✓ | ✖ | ✓ | ✓ | ✓ | ✓ |
| Alcohol | ||||||
| Maternal, prenatal alcohol consumption | ✓ | ✖ | ✓ | ✖ | ✓ | ✓ |
| Maternal Adiposity | ||||||
| Maternal prepregnancy body mass index, kg/m2 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Maternal pregnancy weight gain, kg | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Diabetes | ||||||
| Maternal diabetes mellitus (DM) | ✓ | ✓ | ✓ | ✓ | ✓ | ✖ |
| Paternal DM | ✓ | ✓ | ✓ | ✓ | ✓ | ✖ |
| Reproductive History | ||||||
| Parity (number of prior live births) | ✓ | ✓ | ✓ | ✓ | ✓ | ✖ |
| Prior miscarriages | ✓ | ✓ | ✓ | ? | ✓ | ✖ |
| Radiation Exposure | ||||||
| X-ray exposure, prenatal | ✓ | ✓ | ✖ | ✓ | ✓ | ✖ |
ALSPAC, The Avon Longitudinal Study of Parents and Children, UK; CPP, The Collaborative Perinatal Project cohort, USA; DNBC, The Danish National Birth Cohort, Denmark; JPS, The Jerusalem Perinatal Study, Israel; MoBa, The Norwegian Mother & Child Cohort Study, Norway; TIHS, The Tasmanian Infant Health Survey, Australia.
CC heterogeneity is an important consideration in these studies and may weaken some correlations between epigenetic signatures and cancer predisposition but may itself be an important criterion allowing the identification (if existing) of epigenetic signatures common across some CC subtypes but not others. For example, such signatures may be associated with birth weight pathways, particularly considering that increased birth weight associates with increased risk for many CC types (our unpublished data). Large-scale prospective studies offer the opportunity of investigating such possibilities, especially that causal evidence for CC is minimal. Importantly, the heterogeneous CC design allows some cancer types to serve as positive controls for some signatures and negative controls for others, relative to other tumour categories, as we have recently observed (our unpublished data). Hence, it is equally important to know, e.g. whether some biomarkers specific for childhood leukaemia would also be associated or not with central nervous system tumours, and a heterogenous CC design of samples (altogether homogeneously analysed for methylation biomarkers) would serve as a good strategy to address such questions. A recent study suggested that common mutations could be observed among childhood diseases of intrinsically different origins, such as fibrodysplasia ossificans, which turns a child’s muscle into bone, and diffuse intrinsic pontine glioma, an incurable paediatric brainstem tumour, which exhibits an epigenetic mutation in histone H3 in 90% of patients (35). The early onset of CC in life allows much less time for accumulating mutations than is the case of adult tumours and, hence, childhood tumours and diseases may be less divergent mechanistically than their adult equivalents. Moreover, different CC subtypes may cluster together not necessarily because of common epigenetic mechanisms but due to common environmental causes. It is well established that some exposure factors, such as radiation or smoking, associate with several distinct tumour types even in adulthood, and thus, a large-scale heterogeneous CC approach offers an opportunity to test whether similar scenarios could exist across different tumour types in children. Therefore, considering CC heterogeneity as a limitation is arguable in such studies, and this heterogeneity can instead be considered as an advantage, specifically if the study is well designed and sufficiently powered.
Power calculation and statistical analysis in prospective epigenomic studies of CC
The recent availability of high-throughput epigenomic analyses offers new possibilities to address previously untested hypothesis on the link between DNA methylation variation and disease susceptibility. Specifically, bead array epigenomic analyses of cord blood samples and neonatal blood spots have shown that DNA methylation variation can be reliably detected at birth (9,36). Such studies allow the evaluation of associations between levels of methylation in a very large number of loci and the risk of CC and/or exposures. However, the large number of tested loci (p) in methylome-wide technologies and the constrained number of prospective samples available for analysis (n) oblige the use of less direct approaches than the simple site-by-site methods used in GWAS. Strategic choices are necessary to extract value from the available samples.
Methods of dealing with the n < p situation in omics have been surveyed (37). They note in particular the highly correlated nature of this data, which makes the use of variable selection techniques less suitable. It should be noted that this is not purely the result of common biological pathways or functions but is also a result of finite sample size: it is not possible to have more than n uncorrelated variables in a sample of size n. Dimensional reduction techniques are well adapted to extracting simpler structure from such data, but one is forced to choose between unsupervised (principal component analysis [PCA] related) and supervised (partial least squares [PLS] related) approaches. Using an unsupervised method will extract the most important sources of variation, but these may not be related to disease status. Supervised methods can detect which components of variation are associated with case status but introduce a difficult multiple comparison problem (not discussed in ref. 37), which needs to be corrected for by Bonferroni or false discovery rate (FDR) methods, for example. Evaluating significance levels using PLS-type methods can only be performed by computationally intensive permutation or bootstrapping methods, which must be repeated for all outcomes tested. In contrast, PCA-type reductions are blind to case status and, therefore, do not result in any increased multiple testing burden beyond that associated with the number of retained components. A further advantage of unsupervised methods is that the definition of principal components can be based on control subjects alone: controls are far more plentiful in a CC cohort and their biological samples can be more readily used for exploratory analyses. Furthermore, many of the control-based methylome clusters may often be embedded in cases, with only their magnitudes of variation being different in the diseased tissues. This is particularly prominent in prospectively designed studies, in which cases, similar to controls, also represent normal tissues.
As an example of power calculations, we consider the association of CC with the ~485000 methylation sites analysed by HM450K bead array (Illumina), comparing 250 prospective cases to 500 controls. Using continuously valued predictors, the power is strongly affected by the variability across controls. Using pilot data from I4C cord blood samples assayed via the Illumina bead array technology, we found that the beta values (log-odds of methylation) had standard deviations (SDs) approximately independent of means. The 75th, 90th, 95th and 99th centiles of SD across the sites were estimated at 0.355, 0.443, 0.530 and 0.971, respectively (our unpublished data), lower than that found in healthy adult samples. With an FDR of 5%, testing each site for association, effect sizes (difference in beta) needed for minimum 80% power to find a single associated site would be, respectively, 0.17, 0.21, 0.25 and 0.46 depending on the level of noise at the site.
In contrast, given n samples, there are at most n independent linear combinations of methylation sites, and these explain 100% of the variance in the samples. So if we restrict to 250 components in the above comparison, the multiple comparison penalty is far smaller and, hence, the power is considerably enhanced: the same expected differences in beta would give > 99% power, or 80% power would be obtained with differences of 0.125, 0.156, 0.187 and 0.342. In particular, one avoids redundant tests of correlated variables. Similar arguments can be made for diet or other questionnaire-based exposures.
Methylation marks can also be biologically clustered, e.g. by promoter region and by gene. This has the advantage of being more easily interpreted than a principal component, but the disadvantage is that the number of genes is larger and there will be correlation between gene-average methylation values. Proper statistical analysis, covering early phases of study design to more downstream stages after data acquisition, is crucial, particularly in prospectively designed studies in which small-to-moderate effect sizes are expected and/or in which cancer heterogeneity is an important factor.
DNA methylome profiling suitable for large-scale studies
To characterise the methylome in neonatal blood samples, methylome profiling in blood samples of CC cases and controls using a large sample of subjects from prospective cohorts needs to be performed. A wide range of approaches is now available for assessing patterns of DNA methylation in normal and cancer cells. With the advent of high-throughput and genome-wide profiling technologies, it is now possible to study methylation profiles at both genome-wide scale and high resolution. All methods for DNA methylation analysis are based on one of three techniques: bisulfite conversion, affinity enrichment of methylated DNA and digestion with methylation-sensitive restriction enzymes (38–43). Combining these techniques with DNA microarrays and high-throughput sequencing has made the mapping of DNA methylation feasible on a genome-wide scale (41,42).
The large number of CpG sites to be studied in the large sample set in this study means that a fast, parallel, relatively inexpensive technology for DNA methylation analysis with high degree of automation is necessary. The most high-resolution and scalable protocols use bisulfite conversion of unmethylated DNA in combination with high-throughput sequencing or microarray analysis as the read-out (44). Epigenomic approaches involving microarrays fulfil these conditions. Among the major microarray-based methods is BeadArray/Infinium chemistry (Illumina) that allows for genome-wide analysis of methylation at single-base resolution (45,46). This versatile approach can be applied to a modest number of loci (multiples of 96) up to a whole-genome array with the 450K Infinium Methylation BeadChip. A strength of the technique is that it provides quantitative evaluation of specific cytosines and can process many samples in parallel (45,47). The 450K Methylation BeadChip allows the simultaneous interrogation of >450000 CpG sites, spanning all RefSeq genes (including micro RNA genes). Accurately identifying aberrant methylation using the Infinium 450K technology requires the establishment of a reliable preprocessing procedure to lower measurement errors and the development of advanced computational and statistical methods that can cope with the large number of measurements. Despite the availability of a large set of processing methods, accurate processing of Infinium 450K data remains difficult due to the lack of a reference method and because of several underestimated confounding parameters such as cross-reactive probes, probes containing common single nucleotide polymorphism or differences between the Infinium I and Infinium II probe types. To eliminate measurement variations due to different types of external parameters, including ‘batch effects’, various normalisation methods have been developed and compared (48).
In addition to the microarray-based assays, whole-genome bisulfite sequencing (WGBS) and reduced representation bisulfite sequencing (RRBS) provide comparable accuracy but differ substantially in terms of their genome-wide coverage, robustness towards quality samples, susceptibility to batch effects and cost (45,49). WGBS covers the vast majority of CpGs in the human genome, making it the technology of choice for large-scale reference epigenome projects such as BLUEPRINT (50) and IHEC (51). However, costs per sample makes WGBS currently unfeasible for large cohort studies (and will for the foreseeable future remain limited to relatively small sample numbers). In contrast, the Illumina Infinium 450K microarray is relatively affordable and widely used for measuring DNA methylation in large sample cohorts. However, because of the hybridisation step, the assay is much more susceptible to batch effects than ‘digital’ sequencing-based methods such as WGBS and RRBS.
RRBS provides a promising alternative to both WGBS and the Infinium assay, as it combines the robustness of bisulfite sequencing with a selection step that restricts the analysis on some of the most ‘informative’ CpGs in the genome. RRBS covers 5–10% of all CpGs by sequencing only a defined 1% of the human genome. Thus, the statistical power for detecting small differences in DNA methylation is substantially improved compared with both WGBS and the Infinium assay because the deeper coverage per CpG directly translates into higher quantitative accuracy of the measurement. Despite the enrichment for CpG-rich regions, the relatively short recognition motif CCGG results in significant coverage of genomic regions that are not particularly CpG-rich, including introns and gene deserts. Finally, RRBS also provides extensive information on non-CpG methylation (52), and it can be combined with additional preprocessing steps in order to measure 5-hydroxymethylation alongside the more classical DNA methylation patterns (53). Therefore, RRBS appears to be the suitable assay for studying DNA methylation in a large cohort of samples, some of which are available only as formalin-fixed paraffin-embedded tissue blocks.
Challenges associated with epigenomic analysis of neonatal blood samples
Blood samples obtained at birth (cord blood or neonatal blood spots) represent an attractive material for characterising the fetal exposome; however, the reliable profiling of DNA methylome in these samples (particularly blood spots) has proven to be technically challenging (54). In particular, the amount and quality of DNA in archived blood spots is limited and, therefore, requires considerable efforts to develop a reliable and robust methodology that would allow sensitive detection of genome-wide DNA methylome changes in blood samples collected at birth. I4C allows analysis of methylome using DNA obtained from cord blood samples and blood spots collected within I4C cohorts. As DNA from blood spots is available in limited quantities, it is important to optimise DNA extraction methods from these samples. We have recently tested several DNA extraction protocols and optimised the methods that yield optimal quantity and quality of DNA, particularly suited for methylome-wide studies. The optimised strategies were successfully tested on blood spots from several cohorts with satisfactory results (55); therefore, our protocols may prove highly useful in DNA methylome analyses.
Integrative biostatistics and identification of biomarker candidates of CC and associated in utero factors
Using the DNA methylation data generated in the methylome-wide screens, it is possible to identify methylation markers significantly and consistently associated with CC. Furthermore, statistical analyses need to be refined to explore possible biological links of these markers to known cancer risk factors and to explore the use of these markers for the purpose of CC risk prediction. A variety of statistical approaches, including multiple regression models, can be applied to interrogate risk factor variables and their role in determining methylation status. In all analyses, careful control of type I error and FDR need to be implemented. These analyses should (i) identify methylation markers that can be measured in cord blood and blood spot DNA and are significantly associated with CC risk, overall or by CC subtype; (ii) identify plausible mechanisms underlying disease development, by inferences about specific gene loci involved, and through analyses of methylation markers in blood samples taken at birth as potential intermediates between CC risk factors (including parental smoking, infections during first trimester of pregnancy, pesticides, herbicides, alcohol, diet, demographic data, data on the pregnancy and data on the baby) and CC as an outcome, overall and by cancer subtype and (iii) examine the use of blood-based methylation markers for improvement of CC risk prediction models (by CC subtype) based on standard epidemiologic risk factor information.
Using bioinformatics and high-throughput procedures to analyse the methylome data of DNA from cord blood samples, a selection can be made of candidate methylation markers that are measurable in blood samples and have the highest likelihood of being related to CC. Conditional logistic regression models may be used, testing for the association of CC risk to each of the markers individually and accounting for the case–control matching by cohort centre. Further models can incorporate additional CC risk factors and/or their interaction terms with methylation markers to explore possible confounding or effect modification. I4C provides the following data that may be quantified in the form of total environmental load score in utero to enable quantitative correlations (Table 2): (i) Environmental exposure: parental smoking, infections during first trimester of pregnancy, pesticides, herbicides, alcohol, diet (folic acid, vitamins, bioflavonoids, phytoestrogens) and ionising radiation; (ii) Demographic data: socioeconomic status, parental age and parental occupation; (iii) Data on the pregnancy: gestational age, mother’s parity, placenta weight, obstetric variables (gestational diabetes, preeclampsia, abruptio placentae) and (iv) Data on the baby: gender, birth weight and birth length. For children who developed acute lymphoblastic leukaemia, data on immune-phenotyping and karyotyping may be retrieved.
Comprehensive statistical models could be also used to examine the overall risk prediction potential by the methylation markers combined, using selected statistical procedures for inclusion of only those markers that contribute significantly to risk prediction. Overall model performance for CC risk prediction could be expressed in terms of increments in the C-statistic (area under the receiver operating characteristic curve) and integrated discrimination improvement statistics. Statistical overfitting may be adjusted for by N-fold cross-validation. Finally, the incremental risk discrimination by methylation markers can be examined in a statistical prediction model that includes classical epidemiological risk factor information (as in the well-known model by Gail (56), plus a predefined score based on genetic polymorphisms) (57).
Cross-omics approaches to characterise the fetal exposome
The exposome refers to the totality of exposures to which an individual is subjected, from conception onwards (7,58,59). By performing the simultaneous and comprehensive search of large numbers of potential targets without prior hypotheses, omics technologies provide opportunities for the discovery of a new generation of biomarkers of exposure and disease risk, potentially linked to mechanistic pathways (7,60). Therefore, methylome analysis of neonatal blood samples of large mother:child cohorts may be combined with other ‘omics’ analyses in the context of multidisciplinary initiative aiming to characterise the fetal exposome. Other omics may include those devised to analyse small molecules, reactive electrophiles, and large molecules derived from both exogenous and endogenous exposures. These methods have not yet been applied to characterise the fetal exposome, although cord blood specimens should provide a suitable vehicle for doing so and also for interrogating other components of the exposome (transcriptomic, epigenomics and proteomic changes) in the same samples.
The efforts aimed at characterising the fetal exposome could be based on prospectively collected cord blood samples (including DNA, RNA and plasma/serum) of CC cases and controls from the I4C cohorts for which high-quality epidemiological and clinical data are available. In addition to the methylome profiling based on DNA, the fetal exposome characterisation may take advantage of other new powerful technologies for analysis in an integrated manner of the transcriptome (based on RNA), metabolome and adductome (using plasma or serum, whichever is available) and infectome (relying on DNA to check for the ‘presence’ of viral genes and RNA to assess whether identified viruses are transcriptionally ‘active’).
The fetal exposome data inevitably consist of very high-dimensional intercorrelated explanatory variables. A priori knowledge of interesting groups of markers (like gene sets, biochemical pathways) is sparse, so data reduction must be data-driven, and can be done by using a cross-validation-based methodology (61). The exposome concept is relatively new, and deciphering its molecular components may very likely be feasible with the current age of omics but remains an interesting challenge requiring the close collaboration among epidemiologists, biostatisticians/bioinformaticians and laboratory scientists.
Conclusions and perspectives
Building on materials obtained from a consortium of the largest birth cohorts globally, it is now possible to test the association between prospectively collected exposure data and exposome measures obtained from biospecimens collected at birth and subsequent incident CC. Using both the exposure and exposome measures, the associations for those risk factors for which some evidence has already emerged from previous case–control and biological studies can now be explored in detail, but also new risk factors may be identified. The identification of an exposome features at birth associated with CC risk and their in utero determinants may change our paradigm of tumourigenesis and increase our knowledge about the underlying mechanisms and potential developmental origins of CC. This may also allow us to evaluate identified novel biomarkers in new longitudinal mother:child cohorts that facilitate studying molecular changes and cancer risk throughout the life course from birth up to adulthood (7,14,62 –64). This should also provide opportunities for additional mechanistic studies that may provide insights into the development of malignancies and other diseases later in life. A high-resolution characterisation of the fetal methylome and the early-life exposome should improve our knowledge concerning the aetiology of CC and identify both novel biomarkers and clues to causation, thus, providing an evidence base for cancer prevention.
Funding
The work in the Epigenetics Group at the International Agency for Research on Cancer (Lyon, France) is supported by grants from EU FP7 (TRANS201301184), National Institutes of Health/National Cancer Institute (1R01CA172460-01A1), Institut National du Cancer (INCA 2014-154), France; the Bill and Melinda Gates Foundation (OPP1066947); l’Agence Nationale de Recherche Contre le Sida et Hépatites Virales (ANRS 12328, France); l’Association pour la Recherche sur le Cancer (ARC SFI12101201777), France and la Ligue Nationale (Française) Contre le Cancer (EGE/43/27), France (to Z.H.) and by the IARC Postdoctoral Fellowship and Marie Curie Actions-People-COFUND (to A.G.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Acknowledgements
We thank the International Childhood Cancer Cohort Consortium for contributing to the coordination between participating cohorts. This manuscript is dedicated to the participating children and their families.
Conflict of interest statement: None declared.
References
- 1. Steliarova-Foucher E., Stiller C., Kaatsch P., Berrino F., Coebergh J. W., Lacour B., Parkin M. (2004) Geographical patterns and time trends of cancer incidence and survival among children and adolescents in Europe since the 1970s (the ACCISproject): an epidemiological study. Lancet, 364, 2097–2105. [DOI] [PubMed] [Google Scholar]
- 2.Dommering, C.J., Mol, B.M., Moll, A.C., Burton, M., Cloos, J., Dorsman, J.C., Meijers-Heijboer, H., van der Hout, A.H. (2014). RB1 mutation spectrum in a comprehensive nationwide cohort of retinoblastoma patients. J Med Genet., 51, 366–374. 10.1136/jmedgenet-2014-102264. [DOI] [PubMed]
- 3. Belson M., Kingsley B., Holmes A. (2007) Risk factors for acute leukemia in children: a review. Environ. Health Perspect., 115, 138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gluckman P. D., Hanson M. A., Cooper C., Thornburg K. L. (2008) Effect of in utero and early-life conditions on adult health and disease. N. Engl. J. Med., 359, 61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee H. S., Herceg Z. (2014) The epigenome and cancer prevention: a complex story of dietary supplementation. Cancer Lett., 342, 275–284. [DOI] [PubMed] [Google Scholar]
- 6. Lee H. S., Barraza-Villarreal A., Hernandez-Vargas H., Sly P. D., Biessy C., Ramakrishnan U., Romieu I., Herceg Z. (2013) Modulation of DNA methylation states and infant immune system by dietary supplementation with ω-3 PUFA during pregnancy in an intervention study. Am. J. Clin. Nutr., 98, 480–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wild C. P., Scalbert A., Herceg Z. (2013) Measuring the exposome: a powerful basis for evaluating environmental exposures and cancer risk. Environ. Mol. Mutagen., 54, 480–499. [DOI] [PubMed] [Google Scholar]
- 8. Suter M., Ma J., Harris A., Patterson L., Brown K. A., Shope C., Showalter L., Abramovici A., Aagaard-Tillery K. M. (2011) Maternal tobacco use modestly alters correlated epigenome-wide placental DNA methylation and gene expression. Epigenetics, 6, 1284–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Joubert B. R., Håberg S. E., Nilsen R. M., et al. (2012) 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ. Health Perspect., 120, 1425–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Koestler D. C., Avissar-Whiting M., Houseman E. A., Karagas M. R., Marsit C. J. (2013) Differential DNA methylation in umbilical cord blood of infants exposed to low levels of arsenic in utero. Environ. Health Perspect., 121, 971–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kippler M., Engström K., Mlakar S. J., Bottai M., Ahmed S., Hossain M. B., Raqib R., Vahter M., Broberg K. (2013) Sex-specific effects of early life cadmium exposure on DNA methylation and implications for birth weight. Epigenetics, 8, 494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chhabra D., Sharma S., Kho A. T., et al. (2014) Fetal lung and placental methylation is associated with in utero nicotine exposure. Epigenetics, 9, 1473–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Markunas C. A., Xu Z., Harlid S., Wade P. A., Lie R. T., Taylor J. A., Wilcox A. J. (2014) Identification of DNA methylation changes in newborns related to maternal smoking during pregnancy. Environ. Health Perspect., 122, 1147–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Richmond R.C., Simpkin A.J., Woodward G., et al. (2014) Prenatal exposure to maternal smoking and offspring DNA methylation across the lifecourse: findings from the Avon Longitudinal Study of Parents and Children (ALSPAC). Hum Mol Genet. 2014 December 30. pii: ddu739. [Epub ahead of print], PubMed PMID: 25552657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Breton C. V., Siegmund K. D., Joubert B. R., et al. (2014) Prenatal tobacco smoke exposure is associated with childhood DNA CpG methylation. PLoS One, 9, e99716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Magrath I., Steliarova-Foucher E., Epelman S., Ribeiro R. C., Harif M., Li C. K., Kebudi R., Macfarlane S. D., Howard S. C. (2013) Paediatric cancer in low-income and middle-income countries. Lancet. Oncol., 14, e104–e116. [DOI] [PubMed] [Google Scholar]
- 17. Dolinoy D. C., Huang D., Jirtle R. L. (2007) Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc. Natl. Acad. Sci. U. S. A., 104, 13056–13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Smith M. T., McHale C. M., Wiemels J. L., et al. (2005) Molecular biomarkers for the study of childhood leukemia. Toxicol. Appl. Pharmacol., 206, 237–245. [DOI] [PubMed] [Google Scholar]
- 19. Greaves M. (2005) In utero origins of childhood leukaemia. Early Hum. Dev., 81, 123–129. [DOI] [PubMed] [Google Scholar]
- 20. Herceg Z., Vaissière T. (2011) Epigenetic mechanisms and cancer: an interface between the environment and the genome. Epigenetics, 6, 804–819. [DOI] [PubMed] [Google Scholar]
- 21. Feil R., Fraga M. F. (2011) Epigenetics and the environment: emerging patterns and implications. Nat. Rev. Genet., 13, 97–109. [DOI] [PubMed] [Google Scholar]
- 22. Shen H., Laird P. W. (2013) Interplay between the cancer genome and epigenome. Cell, 153, 38–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Uribesalgo I., Di Croce L. (2011) Dynamics of epigenetic modifications in leukemia. Brief. Funct. Genomics, 10, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Teitell M. A., Pandolfi P. P. (2009) Molecular genetics of acute lymphoblastic leukemia. Annu. Rev. Pathol., 4, 175–198. [DOI] [PubMed] [Google Scholar]
- 25. Tsai A. G., Lu H., Raghavan S. C., Muschen M., Hsieh C. L., Lieber M. R. (2008) Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell, 135, 1130–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Natoli G. (2010) Maintaining cell identity through global control of genomic organization. Immunity, 33, 12–24. [DOI] [PubMed] [Google Scholar]
- 27. Feinberg A. P., Ohlsson R., Henikoff S. (2006) The epigenetic progenitor origin of human cancer. Nat. Rev. Genet., 7, 21–33. [DOI] [PubMed] [Google Scholar]
- 28. Herceg Z. (2011) Epigenetic changes induced by environment and diet in cancer. In Nriagu J. O. (ed.), Encyclopedia of Environmental Health, Vol. 12. Elsevier, Burlington, pp. 582–589. [Google Scholar]
- 29. Herceg Z. (2007) Epigenetics and cancer: towards an evaluation of the impact of environmental and dietary factors. Mutagenesis, 22, 91–103. [DOI] [PubMed] [Google Scholar]
- 30. Milosavljevic A. (2011) Emerging patterns of epigenomic variation. Trends Genet., 27, 242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Biron V. L., McManus K. J., Hu N., Hendzel M. J., Underhill D. A. (2004) Distinct dynamics and distribution of histone methyl-lysine derivatives in mouse development. Dev. Biol., 276, 337–351. [DOI] [PubMed] [Google Scholar]
- 32. Barouki R., Gluckman P. D., Grandjean P., Hanson M., Heindel J. J. (2012) Developmental origins of non-communicable disease: implications for research and public health. Environ. Health, 11, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Herceg Z., Lambert M. P., van Veldhoven K., Demetriou C., Vineis P., Smith M. T., Straif K., Wild C. P. (2013) Towards incorporating epigenetic mechanisms into carcinogen identification and evaluation. Carcinogenesis, 34, 1955–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brown R. C., Dwyer T., Kasten C., Krotoski D., Li Z., Linet M. S., Olsen J., Scheidt P., Winn D. M. (2007) Cohort profile: the International Childhood Cancer Cohort Consortium (I4C). Int. J. Epidemiol., 36, 724–730. [DOI] [PubMed] [Google Scholar]
- 35. Delude, C. M. (2014) Shared mutation for two childhood diseases. J. Natl. Cancer Inst., 106(7). pii dju222. 10.1093/jnci/dju222. [DOI] [PubMed] [Google Scholar]
- 36. Beyan H., Down T. A., Ramagopalan S. V., et al. (2012) Guthrie card methylomics identifies temporally stable epialleles that are present at birth in humans. Genome Res., 22, 2138–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chadeau-Hyam M., Campanella G., Jombart T., Bottolo L., Portengen L., Vineis P., Liquet B., Vermeulen R. C. (2013) Deciphering the complex: methodological overview of statistical models to derive OMICS-based biomarkers. Environ. Mol. Mutagen., 54, 542–557. [DOI] [PubMed] [Google Scholar]
- 38. Brena R. M., Auer H., Kornacker K., Plass C. (2006) Quantification of DNA methylation in electrofluidics chips (Bio-COBRA). Nat. Protoc., 1, 52–58. [DOI] [PubMed] [Google Scholar]
- 39. Callinan P. A., Feinberg A. P. (2006) The emerging science of epigenomics. Hum. Mol. Genet., 15, R95–R101. [DOI] [PubMed] [Google Scholar]
- 40. Ushijima T. (2005) Detection and interpretation of altered methylation patterns in cancer cells. Nat. Rev. Cancer, 5, 223–231. [DOI] [PubMed] [Google Scholar]
- 41. Esteller M. (2007) Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet., 8, 286–298. [DOI] [PubMed] [Google Scholar]
- 42. Zilberman D. (2007) The human promoter methylome. Nat. Genet., 39, 442–443. [DOI] [PubMed] [Google Scholar]
- 43. Ammerpohl O., Martín-Subero J. I., Richter J., Vater I., Siebert R. (2009) Hunting for the 5th base: techniques for analyzing DNA methylation. Biochim. Biophys. Acta, 1790, 847–862. [DOI] [PubMed] [Google Scholar]
- 44. Laird P. W. (2010) Principles and challenges of genomewide DNA methylation analysis. Nat. Rev. Genet., 11, 191–203. [DOI] [PubMed] [Google Scholar]
- 45. Bock C., Tomazou E. M., Brinkman A. B., et al. (2010) Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat. Biotechnol., 28, 1106–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Beck S. (2010) Taking the measure of the methylome. Nat. Biotechnol., 28, 1026–1028. [DOI] [PubMed] [Google Scholar]
- 47. Ladd-Acosta C., Pevsner J., Sabunciyan S., et al. (2007) DNA methylation signatures within the human brain. Am. J. Hum. Genet., 81, 1304–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu M. C., Joubert B. R., Kuan P. F., Håberg S. E., Nystad W., Peddada S. D., London S. J. (2014) A systematic assessment of normalization approaches for the Infinium 450K methylation platform. Epigenetics, 9, 318–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Harris R. A., Wang T., Coarfa C., et al. (2010) Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat. Biotechnol., 28, 1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Adams D., Altucci L., Antonarakis S. E., et al. (2012) BLUEPRINT to decode the epigenetic signature written in blood. Nat. Biotechnol., 30, 224–226. [DOI] [PubMed] [Google Scholar]
- 51. Satterlee J. S., Schübeler D., Ng H. H. (2010) Tackling the epigenome: challenges and opportunities for collaboration. Nat. Biotechnol., 28, 1039–1044. [DOI] [PubMed] [Google Scholar]
- 52. Ziller M. J., Müller F., Liao J., et al. (2011) Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet., 7, e1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Booth M. J., Branco M. R., Ficz G., Oxley D., Krueger F., Reik W., Balasubramanian S. (2012) Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science, 336, 934–937. [DOI] [PubMed] [Google Scholar]
- 54. Wong N., Morley R., Saffery R., Craig J. (2008) Archived Guthrie blood spots as a novel source for quantitative DNA methylation analysis. Biotechniques, 45, 423–424, 426, 428 passim. [DOI] [PubMed] [Google Scholar]
- 55. Ghantous A., Saffery R., Cros M. P., Ponsonby A. L., Hirschfeld S., Kasten C., Dwyer T., Herceg Z., Hernandez-Vargas H. (2014) Optimized DNA extraction from neonatal dried blood spots: application in methylome profiling. BMC Biotechnol., 14, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gail M. H. (2011) Personalized estimates of breast cancer risk in clinical practice and public health. Stat. Med., 30, 1090–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hüsing A., Canzian F., Beckmann L., et al. (2012) Prediction of breast cancer risk by genetic risk factors, overall and by hormone receptor status. J. Med. Genet., 49, 601–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wild C. P. (2005) Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol. Biomarkers Prev., 14, 1847–1850. [DOI] [PubMed] [Google Scholar]
- 59. Rappaport S. M., Smith M. T. (2010) Epidemiology. Environment and disease risks. Science, 330, 460–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rappaport S. M. (2012) Biomarkers intersect with the exposome. Biomarkers, 17, 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. van der Laan M.J., Hubbard A.E., Pajouh S.K. (2013) Statistical Inference for Data Adaptive Target Parameters. U.C. Berkeley Division of Biostatistics Working Paper Series. The Berkeley Electronic Press. [Google Scholar]
- 62. Vrijheid M., Slama R., Robinson O., et al. (2014) The human early-life exposome (HELIX): project rationale and design. Environ. Health Perspect., 122, 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Khoury M. J., Lam T. K., Ioannidis J. P., et al. (2013) Transforming epidemiology for 21st century medicine and public health. Cancer Epidemiol. Biomarkers Prev., 22, 508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee, K.W., Richmond, R., Hu, P., et al. (2015). Prenatal exposure to maternal cigarette smoking and DNA methylation epigenome-wide association in a discovery sample of adolescents and replication in an independent cohort at birth through 17 years of age. Environ Health Perspect., 123, 193–199. 10.1289/ehp.1408614. [DOI] [PMC free article] [PubMed]