Abstract
To incorporate ontological concepts in natural language processing (NLP) it is often necessary to combine simple concepts into complex concepts (post-coordination). This is especially true in consumer language, where a more limited vocabulary forces consumers to utilize highly productive language that is almost impossible to pre-coordinate in an ontology. Our work focuses on recognizing an important case for post-coordination in natural language: spatial relations between disorders and anatomical structures. Consumers typically utilize such spatial relations when describing symptoms. We describe an annotated corpus of 2,000 sentences with 1,300 spatial relations, and a second corpus of 500 of these relations manually normalized to UMLS concepts. We use machine learning techniques to recognize these relations, obtaining good performance. Further, we experiment with methods to normalize the relations to an existing ontology. This two-step process is analogous to the combination of concept recognition and normalization, and achieves comparable results.
Introduction
Medical ontologies are fundamentally limited in the number of concepts and terms they contain. The range of potential problems, treatments, tests, biological processes, and other concepts encountered in medicine is far too vast to encode directly as concepts in an ontology. Instead, ontologies generally rely on attributes and relations to handle on-the-fly concept creation. Further, for a given concept, it is often impossible to enumerate all the textual strings that can be used to refer to that concept in natural language. The ontological solution for this problem is known as post-coordination, where simple concepts are composed (or coordinated) into a more complex concept. Making full use of an ontology when interpreting unstructured text therefore necessitates natural language processing (NLP) techniques.
The need to perform post-coordination is particularly strong in consumer language. Non-experts lack the breadth of vocabulary that experts often use to describe medical concepts. Instead, consumers often rely on lengthy descriptions with a reduced vocabulary when describing symptoms or conditions. This results in an increased need for post-coordination. Consider the following three examples of consumer language:
I am experiencing pain in my left leg.
They measured his blood pressure and found it to be above acceptable levels.
During the accident I sustained an injury to the back of my head and neck.
The first example corresponds to the UMLS concept Pain in left leg (C0564822 in UMLS 2014AB), yet the presence of the word “my” in the middle of the phrase prevents the concept from being recognized by a simple lexicon lookup. Instead, the concept Pain (C0030193) and Left leg (C0230443) need to be composed to find the pre-coordinated concept in UMLS. In the second example, the patient has Hypertension (C0020538), but this has to be inferred from the result of the high measurement. This is an example of where consumer language would likely differ from that of medical professionals, who might simply state, “He has hypertension.” In the third example, no concept in UMLS corresponds to the problem “injury to the back of the head and neck”. Instead, the problem concept Injury (C3263722), the direction Back (C0205095), and anatomical locations Head (C0018670) and Neck (C0027530) must be post-coordinated into an on-the-fly concept.
Unfortunately, short of complete natural language understanding, there is no general-purpose NLP technique for performing post-coordination. Rather, methods must be used that target specific domains or linguistic phenomena. In this paper, we limit our scope to a vital phenomena for understanding consumer-described symptoms and conditions: the spatial relationships between disorders and anatomical locations. These are generally expressed in one of two ways: (i) a noun compound (e.g., arm pain), or (ii) a grammatical relation between an indicator term such as a spatial preposition (e.g., in, on, at) and two or more concepts. While noun compounds describing spatial relationships are quite common, they are often pre-coordinated in an ontology and are easier to automatically recognize due to their contiguous nature. The grammatical relations describing spatial relationships are more likely to require special handling and are more difficult to automatically recognize, and hence are the focus of this work. Examples (1) and (3) above demonstrate the second type of spatial relation. While Example (2) contains a spatial relation (above), it does not describe the relationship between the concepts of interest here and is therefore outside the scope of this paper.
In this work, we describe both manually annotated datasets and automatic NLP methods to extract disorder-anatomy relations and normalize them to their appropriate UMLS CUIs. Specifically, the contributions of this paper are:
A manually annotated dataset based on the Spatial Role Labeling (SpRL) schema[1] to extract spatial relations between disorders and anatomical locations.
An automatic SpRL method based on supervised machine learning.
A manually annotated dataset normalizing the extracted spatial relations to UMLS CUIs.
An evaluation of several existing automatic methods for normalizing text to CUIs, customized to handle the extracted spatial relations.
A discussion of the fundamental limitations to this task based on error analysis of the above automatic methods.
Further, both annotated datasets are being made publicly available via the National Library of Medicine website.
Background
While not as well studied as temporal language in medical text, spatial language–especially spatial relations–has nonetheless received considerable attention. Rindflesch et al.[2] describes how syntax relationships are crucial to the semantic interpretation of anatomical relationships in cardiac catheterization reports. In particular, they focus on handcrafted rules for recognizing arterial branching relations as well as the locations of stenosis. Their method relies heavily on the structured data source University of Washington Digital Anatomist[3] (UWDA), part of the Foundational Model of Anatomy[4] (FMA). Closer to our approach are methods that utilize machine learning (ML) to relate disorders with their anatomical locations. Roberts et al.[5] find relations between an inflammation term and its anatomical location within radiology reports. Their method is able to recognize a single layer of relation nesting–that is, when two spatial relations combine into a single conceptual relation (e.g., “inflammation on the wall of the gallbladder”). Dligach et al.[6] similarly recognize anatomical locations for disorders. Their method is designed to operate on any type of disorder, but they do not recognize nested relations. So in the phrase “skin tumor removed from behind his left ear”, the anatomical location for the tumor would be the entire phrase “behind his left ear”. The lack of nesting can present several difficulties, however. From a practical perspective, longer phrases are more difficult to recognize automatically. From a linguistic perspective, the phrase still requires further semantic interpretation. In contrast to these two ML methods, our approach can operate on any disorder term and places no restriction on the depth of nesting, and then goes a step further by providing a semantic interpretation. Thus the method could understand a complex spatial phrase such as “skin tumor on the side of the elbow of his left arm”.
Also, unlike any of the previous approaches, our focus is on consumer language instead of clinical language. Consumer language has been explored in the past as a means of supporting health information seeking[7,8,9] and making medical terms more comprehensible[10] for consumers. This work fits more in the former category, where a semantic understanding of consumer language enables systems that connect consumers with health information resources. In previous work with consumer language, we have focused on co-reference[11] and question classification[12,13].
Outside of medicine, spatial relations have received more attention. Several schemas have been proposed for natural language, including SpatialML[14], SpRL[1], and ISO-Space[15]. Of these, ISO-Space is the most recent and likely the best developed, especially for representing highly-specified geographic descriptions. For under-specified relations, however, ISO-Space and SpRL are largely interchangeable. For our purposes here, therefore, we use SpRL due to its relative simplicity in the knowledge that our annotations could easily be transformed to ISO-Space in the future should the need arise. SpRL was utilized in two SemEval tasks, in 2012[16] and 2013[17], while a hybrid SpRL/ISO-Space representation was used in the SemEval 2014 SpaceEval task[18]. Successful approaches to these tasks have combined supervised ML, syntactic parsing, and integration of real-world knowledge[19].
Post-coordination is generally studied in the context of ontologies and medical coding. For instance, Oniki et al.[20] discuss best practices for pre- and post-coordinating concepts when creating a structured knowledge source based on Clinical Element Models (CEMs). Conversely, Dhombres et al.[21] employ post-coordination when utilizing a structured knowledge source, SNOMED CT, to extend its phenotype coverage. The best known NLP system to perform post-coordination is SemRep[22], which utilizes MetaMap’s[23] option to identify and normalize short phrases. Sem-Rep then identifies relations (e.g., treatment for, location of) that in many cases can be thought of as post-coordinations. However, SemRep does not, nor does any NLP application we are aware of, attempt to normalize these relations back into an ontology to determine if there is an equivalent pre-coordinated concept as described in this paper.
Instead, the most similar NLP task to the one presented in this paper is the combination of concept recognition and concept normalization.[24,25] In this comparison, concept recognition would be analogous to SpRL relation extraction, while concept normalization would be analogous to relation normalization. In theory, an approach similar to concept recognition/normalization could be used here, but would likely perform poorly for two reasons: (a) concept recognizers typically use sequential classifiers, which perform poorly with long concept spans such as those in Examples (1)–(3) above, and (b) concept normalizers attempt to use all the words in the concept, but the relation structure recognizes that some of the words aren’t relevant (e.g., “my” in Example (1)). In the Discussion, we provide some insights as to how comparable our results are to the state-of-the-art in concept recognition/normalization.
Methods
We begin by describing the two sets of annotated data. Next, we describe the process of automatically extracting spatial relations from text. Finally, we describe how these relations are automatically normalized to UMLS concepts.
A. Data
In this section, we describe two different manually annotated data sets: (1) a set of spatial relations in natural language text, and (2) a set of normalizations from the extracted spatial relations to UMLS concepts. Both datasets are publicly available from the U.S. National Library of Medicine (NLM) website.1
Spatial Relations
To gather a set of consumer-written texts likely to contain spatial relations, we started with a large set of emails and online form requests sent to the NLM customer service team. Every year, NLM receives over 40,000 such requests, several thousand of which are manually classified by NLM staff as consumer health questions pertaining to diseases, conditions, and therapies. From these, we extracted 1,976 sentences containing (1) at least one term from UMLS in the Disorder semantic group,[26] (2) at least one term from UMLS in the Anatomy semantic group, and (3) a preposition between one UMLS term of each type. Next, two medical experts–an MD/PhD (LR) and medical librarian (SS)–double-annotated spatial relations using a simplified Spatial Role Labeling (SpRL) schema.[1] SpRL defines a spatial relation between three main elements:
SpatialIndicator: The word or phrase (typically a preposition) that acts as a trigger for the spatial relation.
Trajector: The object whose spatial position is being described.
Landmark: The location of the Trajector.
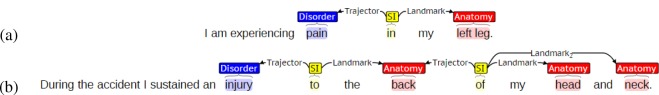
In our case, the Landmark is almost always an anatomical location from UMLS (hereafter, an Anatomy annotation), while the Trajector is usually a Disorder but can be an Anatomy as well. Figure 1 shows how two of the example phrases from the Introduction would be annotated in SpRL using Brat[27]. In the simple example in Figure 1(a), the Spatialindicator “in” connects the Trajector “pain” with the Landmark “left leg”. In the more complex example in Figure 1(b), there are two connected spatial relations: (i) a Disorder-Anatomy relation connects the Trajector “injury” to its Landmark “back”, then (ii) an Anatomy-Anatomy relation connects “back”, this time a Trajector, to two Landmarks, “head” and “neck”. The annotators were allowed to manually add or edit Disorder or Anatomy terms. Approximately 7% of Disorders were manually created, while around 8% of Anatomys were manually created. Since the focus of this paper is relation extraction and normalization, we do not address automatic concept extraction. The UMLS and manual terms are distinguished in the publicly available data, though no distinction is made between them in the experiments below.
Figure 1:
Spatial Role Labeling for Examples (1) and (3).
From the 1,976 sentences, a total of 1,291 SpRL relations were annotated. To assess agreement, we consider one annotator’s labels as the “gold” set and measure the other annotator’s labels against this with F1-measure (if the annotators were switched, F1 would be the same, while precision and recall would be reversed). Using this method, the inter-annotator agreement has an F1 of 88.9 for Spatialindicators, 81.5 for Trajectors, 86.2 for Landmarks, and 72.67 for the complete relation (i.e., exact match for every argument). The annotators reported their agreement improving over time, where many of the disagreements were the result of ungrammatical and medically incorrect consumer language.
Concept Normalization
Instead of uninterrupted text spans, here we consider relations to refer to concepts. Any text not part of a Spatialindicator, Trajector, or Landmark–such as the word “my” in Example (1)–is not considered part of the concept. Further, we consider the fully-nested relation of all Disorders and Anatomys connected through some SpRL relation–such as merging the Disorder-Anatomy and Anatomy-Anatomy relations in Example (3) into one nested spatial relation. The nested structure of these two examples would be:
| (1) pain | (3) injury |
| in: left leg | to: back of: head of: neck |
This nested structure is then used to normalize relations to an ontology. We used 514 such nested relations from the SpRL relations above and manually normalized them to UMLS concepts as follows. The same two annotators as above double-annotated both the nested relations as well as parts of the relations. Normalizing parts of a relation to UMLS enables two useful applications: (1) post-coordination of concepts when the full relation has no UMLS concept, and (2) assignment of codes to parts of a larger concept to specify its deep structure. For the two examples above, the annotated items would be:
| (1) phrase: pain in left leg | (3) phrase: injury to back of head and neck | term: back |
| term: pain | phrase: back of head and neck | term: head |
| term: left leg | term: injury | term: neck |
Here, a “phrase” is the result of a relation, while “term” is an individual Disorder or Anatomy. We only distinguish for the purpose of evaluation. For each phrase/term, the UMLS Terminology Services (UTS) API2 is queried for potential concepts belonging to either the Disorder or Anatomy semantic groups. Both the words and approximate matching functions were used, since in our exploratory experiments both these had better recall than the other matching functions. The top 10 results per matching function were retrieved, de-duplicated, and sorted by CUI instead of relevance rank to reduce biasing the annotators toward choosing the top result. The annotators then chose the correct normalization or indicated that none of the concepts were proper normalizations. Table 1 shows an example of how one phrase and one term were annotated.
Table 1:
Examples of the candidate concepts provided to the annotators for normalization (2nd column), as well as the proper normalization selected by the annotators (3rd column).
| Phrase or Term | Candidate Concepts | Gold Concept | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phrase: tumors in her brain | C0006118 [Neoplastic Process]: Brain Neoplasms | ⇐ | |||||||||||
| C0007798 [Neoplastic Process]: Cerebral Ventricle Neoplasms | |||||||||||||
| C0025149 [Neoplastic Process]: Medulloblastoma | |||||||||||||
| C0025286 [Neoplastic Process]: Meningioma | |||||||||||||
| C0153633 [Neoplastic Process]: Malignant neoplasm of brain | |||||||||||||
| C0220603 [Neoplastic Process]: childhood brain tumor | |||||||||||||
| C0220624 [Neoplastic Process]: adult brain tumor | |||||||||||||
| C0220650 [Neoplastic Process]: Metastatic malignant neoplasm to brain | |||||||||||||
| C0496899 [Neoplastic Process]: Benign neoplasm of brain, unspecified | |||||||||||||
| C0677866 [Neoplastic Process]: Brain Stem Neoplasms | |||||||||||||
|
| |||||||||||||
| Term: tumors | C0000735 [Neoplastic Process]: Abdominal Neoplasms | ⇐ | |||||||||||
| C0001429 [Neoplastic Process]: Adenolymphoma | |||||||||||||
| C0001624 [Neoplastic Process]: Adrenal Gland Neoplasms | |||||||||||||
| C0003463 [Neoplastic Process]: Anus Neoplasms | |||||||||||||
| C0003614 [Neoplastic Process]: Appendiceal Neoplasms | |||||||||||||
| C0005695 [Neoplastic Process]: Bladder Neoplasm | |||||||||||||
| C0005967 [Neoplastic Process]: Bone neoplasms | |||||||||||||
| C0006118 [Neoplastic Process]: Brain Neoplasms | |||||||||||||
| C0006160 [Neoplastic Process]: Brenner Tumor | |||||||||||||
| C0027651 [Neoplastic Process]: Neoplasms | |||||||||||||
| C0877578 [Neoplastic Process]: Treatment related secondary malignancy | |||||||||||||
| C3844254 [Finding]: No proper value is applicable in this context… | |||||||||||||
| C3844255 [Finding]: A valid date value is provided in item Date… | |||||||||||||
| C3844256 [Finding]: A proper value is applicable but not known… | |||||||||||||
There were a total of 1,747 annotated phrases and terms. Around 66% of the annotations had a concept normalization (i.e., a pre-coordinated concept in UMLS). Phrases (relations) were far more likely to not have a valid normalization. 74% of the phrases had no normalization, and thus require post-coordination of their component terms to be properly interpreted. By comparison, only 15% of the individual Disorder and Anatomy terms have no normalization. About half of the terms without a normalization are the result of manually creating Disorder and Anatomy terms that were not in UMLS. The rest are the result of none of the candidate normalizations being judged valid by the annotators. Inter-annotator agreement was fairly good at 84.8%.
B. Spatial Role Extraction
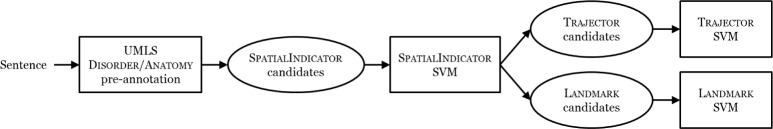
Figure 2 presents the architecture of our ML-based SpRL relation extractor. We utilize a pipeline method where Disorders and Anatomys (from their respective UMLS semantic groups) are pre-annotated. Next, Spatialindi-Cators are extracted. Then, for each SpatialIndicator, Trajectors and Landmarks are identified. These steps are described in detail below except for the pre-annotation, which is described in the Data section.
Figure 2:
Automatic Spatial Role Labeling Architecture
For candidate SpatialIndicators, we consider all prepositions, adverbs, and participles. Some light verbs (such as have and get) can be indicators, but not sufficiently often to merit their inclusion here. We use Stanford CoreNLP[28] to perform part-of-speech tagging. The primary goal in candidate selection is recall: incorrect indicators can still be filtered out by the classifier, but missing indicators cannot be recovered. This recall-based technique achieves 98% recall with a precision of 14%. This precision is sufficiently high to ensure the training data is not overly imbalanced, thus giving the machine learning classifier below the opportunity to filter out most of the incorrect candidates. A binary support vector machine[29] (SVM) then classifies candidates as positive or negative. The primary consideration in deciding whether a word is actually a SpatialIndicator is its context, especially its relationship with nearby Disorder and Anatomy terms. With that in mind, the features used by this SVM are shown in Table 2(a).
Table 2:
Features used by the (a) SpatialIndicator, (b) Trajector, and (c) Landmark methods.
| (a) SpatialIndicator features | (b) Trajector features |
|---|---|
|
fI1 The SpatialIndicator candidate’s words. fI2 The part-of-speech of the candidate. fI3 If the next token is also a SpatialIndicator candidate. fI4 The word distance to the nearest Anatomy. fI5 The word distance to the nearest Disorder. fI6 The dependency distance to the nearest Anatomy. fI7 The dependency distance to the nearest Disorder. fI8 The words between the candidate and the nearest Anatomy. fI9 The words between the candidate and the nearest Disorder. fI10 The dependency path between the candidate and the nearest Anatomy. fI11 The dependency path between the candidate and the nearest Disorder. fI12 The types of the UMLS terms on the left and right of the candidate, along with the candidate itself. For example, Example (1) would be Disorder-in-Anatomy. Example (3) contains both Disorder-to-Anatomy and Anatomy-of-Anatomy. |
fT1 The Trajector candidate’s words. fT2 The word distance between the candidate and its SpatialIndicator. fT3 The dependency distance between the candidate and its SpatialIndicator. fT4 The words between the candidate and its SpatialIndicator. fT5 The dependency path between the candidate and its SpatialIndicator. |
| (c) Landmark features | |
|
fL1 The Landmark candidate’s parts-of-speech. fL2 The equivalent of fT2. fL3 The equivalent of fT3. fL4 The equivalent of fT4. fL5 The equivalent of fT5. |
Since Spatialindicators are the central element of an SpRL relation, candidate Trajectors and Landmarks are defined in terms of the Disorder/Anatomy concept and the Spatialindicator (e.g., in Example (3), “head” is a valid Landmark for “of” but not for “to”). For a given SpatialIndicator, all Disorder and Anatomy terms within its sentence are considered as candidate Trajectors and Landmarks (a Disorder is rarely a Landmark, but this is possible). Then, two binary SVMs (one Trajector, one Landmark) classify candidates as positive or negative. The primary consideration for a positive Trajector/Landmark is its relationship with its SpatialIndicator, especially its syntactic relationship (using syntactic dependencies obtained from CoreNLP). With this in mind, the Trajector features are shown in Table 2(b), while the Landmark features are shown in Table 2(c).
Post-processing
We then consider two post-processing modules, one for SpatialIndicators and one for both Trajectors and Landmarks. The NoArgFilter prunes any SpatialIndicators that do not have either a Trajector or Landmark. Since most SpRL relations have all three elements, this filter is essentially a voting strategy where, if the two argument classifiers agree an indicator has no argument, then it is likely that the indicator classifier erred. Then, the AddCoords heuristic adds a new Trajector or Landmark if an existing Trajector or Landmark is separated by a concept of the same type (Disorder/Anatomy) by an and or an or. This helps identify errors where only one item in a coordination (e.g., “head and neck”) is marked as an argument.
C. Spatial Relation Normalization
Given a sentence containing at least one SpRL relation, the same process as described in the Data section is used to obtain the fully-nested relations as well as the sub-parts of the nested relation. For the purpose of this paper, we are not proposing any new methods for normalizing concepts. Instead, we evaluate baseline and existing methods for concept normalization, and customize these methods for the spatial relation structure. Since existing concept normalization methods expect a word sequence instead of a relation, we use all the words in any element of the relation just as for generating concept candidates from UTS. We also filter out any concepts that do not belong to the Disorder or Anatomy semantic groups.
We evaluated the following methods:3
tfidf_pref: (Baseline) Ranks the candidates (returned by the UTS method described in the Data section) using TF-IDF scored cosine similarity between the relation and the concept’s preferred name. Document frequencies are derived from a recent version of PubMed Central (PMC).
tfidf_all: (Baseline) The same as tfidf_pref, but uses every concept name instead of just the preferred name.
UTS[exact]: The top result for the UTS exact matching function.
UTS[approximate]: Same except for the UTS approximate function.
UTS[left trunc]: Same except for the UTS leftTruncation function.
UTS[right_trunc]: Same except for the UTS rightTruncation function.
UTS[words]: Same except for the UTS words function.
UTS[norm_words]: Same except for the UTS normalizedWords function.
UTS[norm_str]: Same except for the UTS normalizedString function.
UTS[vote]: A combination of the above seven UTS methods using a simple voting strategy. Ties are broken by choosing the concept with the lower CUI number.
Here, the TF-IDF based methods provide simple baselines based on a standard similarity metric. They should work well when relations have a high word overlap with the valid concept, but they do not utilize UMLS in any way. Instead, the UTS API utilizes the structure of UMLS to return candidate normalizations, performing its own internal ranking (though we are not aware of any way in which the internal scores are exposed through the API). In contrast to the systems evaluated in the ShARe/CLEF 2013 task[24], UTS is not limited simply to Disorder concepts, and is thus more appropriate for our evaluation, which includes many Anatomy concepts.
Results
The SpRL method is evaluated using 10-fold cross validation. The results are shown in Table 3. The best SpatialIndicator performance (including the NoArgFilter) has an F1 of 91.0, while the best Trajector classifier (without the AddCoords heuristic) has an F1 of 84.9, and the best Landmark classifier (with the AddCoords heuristic) has an F1 of 86.0. The post-processing NoArgFilter produced a significant gain in precision, from 87.3 to 96.8, for almost no cost to recall. The AddCoords heuristic, however, had only a small effect. In many cases, the classifiers were able to find coordinated Trajectors and Landmarks without the AddCoords heuristic, which makes sense as the syntactic dependency features should be identical for both sides of the coordination. Overall, these results are quite good, even a bit high compared to similar relation extraction tasks.
Table 3:
SpRL Results
| P | R | F1 | |
|---|---|---|---|
| SpatialIndicator + NoArgFilter | 87.3 96.8 |
86.0 85.8 |
86.7 91.0 |
| Trajector + AddCoords | 88.9 87.6 |
81.3 82.0 |
84.9 84.7 |
| Landmark + AddCoords | 89.3 88.7 |
82.6 83.5 |
85.8 86.0 |
The results of the concept normalization methods are shown in Table 4. The baseline methods (tfidf_pref and tfidf_all) performed poorly compared to the best UTS methods. Given that the UTS methods have a better understanding of the UMLS structure and can utilize it for retrieval (e.g., understanding synonymy), they perform quite a bit better and should form the baseline for any future work. However, the individual UTS methods varied significantly, both by method and by the type of concept they were normalizing. UTS[exact] had the best F1 on individual terms (79.0), but performed quite poorly with the relation phrases (35.3). UTS[approximate] performed decently, but well below the best methods for each concept type. This confirmed our pre-existing observations when experimenting with the methods, as the UTS[approximate] method does a good job of retrieving the correct candidate in the top 10, but a poor job of ranking the correct candidate first. It is therefore good for annotation, but not ideal as a run-time system. The other method used in annotation, UTS[words], performed better, but was still outperformed by other methods. UTS[left_trunc] and UTS[right_trunc] where quite poor in general, followed by UTS[norm_words]. UTS[norm_str] was the most consistent method, and also the best performing method for phrases (73.8). As might be expected when forming an ensemble method from highly noisy constituent methods, UTS[vote] was unable to outperform the best individual methods. For future work, methods should be compared either to UTS[norm_str], or to the use of UTS[exact] for individual terms and UTS[norm_str] for phrases.
Table 4:
UMLS Normalization Results
| Method | Overall | Phrases | Terms | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acc. | P | R | F1 | Acc. | P | R | F1 | Acc. | P | R | F1 | |
| tfidf_pref | 61.8 | 49.8 | 57.7 | 53.5 | 77.0 | 41.0 | 59.9 | 48.7 | 54.4 | 51.6 | 57.4 | 54.3 |
| tfidf_all | 40.6 | 21.9 | 25.3 | 23.5 | 72.6 | 29.7 | 43.4 | 35.3 | 24.9 | 20.3 | 22.6 | 21.4 |
|
| ||||||||||||
| UTS[exact] | 79.4 | 79.2 | 71.6 | 75.2 | 79.3 | 94.3 | 21.7 | 35.3 | 79.5 | 78.7 | 79.2 | 79.0 |
| UTS[approximate] | 69.3 | 63.1 | 66.0 | 64.5 | 80.8 | 57.6 | 64.5 | 60.9 | 63.6 | 64.0 | 66.2 | 65.1 |
| UTS[left_trunc] | 44.0 | 23.1 | 22.8 | 23.0 | 75.6 | 40.9 | 17.8 | 24.8 | 28.5 | 22.0 | 23.6 | 22.8 |
| UTS[right_trunc] | 44.8 | 26.5 | 26.1 | 26.3 | 78.9 | 71.7 | 21.7 | 33.3 | 28.1 | 24.6 | 26.8 | 25.7 |
| UTS[words] | 74.9 | 67.0 | 76.7 | 71.6 | 84.7 | 60.7 | 84.2 | 70.5 | 70.2 | 68.2 | 75.6 | 71.7 |
| UTS[norm_words] | 57.1 | 62.3 | 42.1 | 50.3 | 73.2 | 25.0 | 0.7 | 1.3 | 49.1 | 62.5 | 48.5 | 54.6 |
| UTS[norm_str] | 78.6 | 72.4 | 76.1 | 74.2 | 89.4 | 75.3 | 72.4 | 73.8 | 73.2 | 72.0 | 76.7 | 74.3 |
| UTS[vote] | 76.4 | 73.3 | 71.6 | 72.4 | 81.9 | 92.5 | 32.4 | 47.8 | 73.8 | 72.4 | 77.6 | 74.9 |
Discussion
Using the system described above, many of the detailed concepts found within consumer text can be mapped into an ontology containing either pre-coordinated terms or that allows for spatial post-coordination. Given the informal language often utilized in consumer text, such a method is often necessary to gain a more complete semantic understanding. We now discuss the general types of errors made by the system, how some of these errors illustrate limitations to our approach, and how they motivate future work on this task.
As is typical with consumer-authored text, many of the system’s errors were the result of ungrammatical and error-prone language. This includes misspellings (e.g., “ifection” instead of infection, “boh legs” instead of both legs), missing punctuation (e.g., “pain cramping, redness and swelling”), and incorrect tense (e.g., “fingers are stick together by her skin”). These errors are compounded by the fact that many of the consumer information requests were submitted by non-native English speakers, resulting in text that can be difficult to understand (SpatialIndicators in bold):
(4) i had got sunburn effect before 3 year on my both hands.
(5) i have pain on ma left side under the ribis n difficult on urinating seeming as if threz something blocking is it connected to the kidneys
Other errors were the result of using UMLS directly as a lexicon. While this removes the need to run a noisy concept recognition system, and while the UMLS terms should be far easier to normalize, using UMLS directly introduces noise as well. As stated above, between 7–8% of the Disorder and Anatomy terms needed to be manually created (thus indicating recall issues), but precision issues are an effect as well. For instance, mrs (in the phrase “mrs. p gupta”) and mm (in the phrase “1–3 mm”) are both marked as Disorders, and could easily result in mistaken Trajectors. In many cases, the resulting errors were relatively innocuous, as in the following:
(6) …she developed water behind her eye balls…
Here, UMLS does not contain the full phrase “eye balls” (though it does contain “eye ball”, but we performed no stemming for relation extraction). Instead, “eye” is selected as the Landmark. Also see Example (8) below.
As with many relation extraction approaches, long distance relations often prove problematic. Consider the example:
(7) The cancer cells are in the fluid, around the lung, and in the trachea lymph nodes…
Here, “cancer cells” is the Trajector for all three SpatialIndicators, but the classifier fails to identify it as the Trajector for the second and third indicators. The dependency parser should attach all three prepositions to the verb “are”, but instead it attaches “around” and the second “in” to the noun “fluid”. Thus, the dependency paths are atypical for Trajectors, and the classifier misses this argument. Prepositional phrase attachment is a very common source of syntax parsing errors, and the choice of attaching the preposition to the nearer word is typical.
Another common problem is Spatialindicators missing either a Trajector or a Landmark. For instance:
(8) Is matastatic non small cell lung cancer able to spread to the spine and brain in as little as 1 month?
Here, the SpatialIndicator “to” should have a Trajector (“non small cell lung cancer”) and two Landmarks (“spine” and “brain”). While the classifier found both Landmarks, it missed the Trajector. With very few exceptions, all Spatialindicators should have a Trajector. But doing this through a post-processing heuristic by simply forcing the nearest concept to be the Trajector hurts performance. In this case, for example, the nearest concept is actually “able”, which is a Finding in UMLS. Instead, in future work, we plan to integrate more intelligent post-processing to enforce constraints such as “Every SpatialIndicator should have at least 1 Trajector” directly on top of the output of each classifier.
Since the normalization methods evaluated here are either baseline (TF-IDF) or existing (UTS) methods, we omit a detailed error discussion and instead provide some insights on what may enable future methods to improve on normalization performance. The fundamental issue with the evaluated methods is that they consider the concept as a phrase, not as a relation, and thus miss out on potentially useful information. When normalizing a relation, a very similar process to that of post-coordination can be used to identify potentially pre-coordinated concepts. That is, identify the individual argument terms (which almost all of the UTS methods do with higher performance than the full phrases), then identify valid pre-coordinations. For instance, the phrase “fracture on his thigh bone” results in the Trajector fracture with Landmark thigh bone. The UTS[words] method identified the concept C0840234 (Fracture of bone in neoplastic disease; pelvic region and thigh), which is overly specific because it indicates the cause of the fracture (a bone tumor). Through relations in UMLS, it can be determined that C0840234 is a pathological fracture and thus too specific, avoiding this incorrect normalization. This type of method would clearly require a significant understanding of the UMLS relation structure and is beyond the scope of this work, but this work does provide the required NLP methods to enable such ontological methods by extracting and structuring spatially related concepts from text. A second, but also fundamental limitation of the normalization approaches used here is the lack of context employed. The context surrounding a relation likely provides valuable clues as to its proper interpretation.
As addressed in the Background section, this normalization process is largely analogous to the types of concept normalization performed in the CLEF task[24], though that task is limited to Disorders. While evaluating an automatic system in that task (e.g., DNorm[25]) would be an unfair comparison based on the type of data, some discussion on the results of the CLEF task and that of this work might provide some useful insights. The CLEF task evaluated systems in two different ways. First, an end-to-end evaluation (“strict”) that required systems to correctly recognize disorder concepts and then normalize the disorders to their UMLS CUIs. Since errors could occur at either the recognition or normalization stage, the results of this evaluation essentially form a lower bound to normalization performance. A second evaluation (“relaxed”) then measured performance of normalization on only those disorders that were correctly recognized. Since the correctly recognized disorders were likely to be shorter terms resembling those in the training data, the relaxed evaluation essentially forms an upper bound to normalization performance. The best system on the strict measure, DNorm, achieved a strict accuracy of 58.9% and a relaxed accuracy of 89.5%. By comparison, in our data the UTS[exact] method has an accuracy of 79.4%. This suggests that the UTS method, though far simpler than DNorm, is not likely to be substantially worse, but would likely nonetheless be outperformed by DNorm if that system were to be tailored to our spatial relations. Due to the complexity of such a project, we leave this idea to future work.
Conclusion
This paper described a method for extracting and normalizing spatial relations between disorders and anatomical structures. While the method might still generalize to physician and other clinical notes, our focus was on consumer language. We have described two new annotated corpora for these tasks: (1) a corpus of spatial relations from consumer health requests, and (2) a set of annotated concept normalizations for many of the relations from the first corpus. A machine learning-based method is used to automatically perform spatial relation extraction on the first corpus. Then, a set of baseline and existing methods are used to automatically perform concept normalization on the second corpus. Both achieve good performance, though we have identified several areas of future work to improve upon these results.
Acknowledgments
This work was supported by the intramural research program at the U.S. National Library of Medicine, National Institutes of Health. The authors would also like to thank Parisa Kordjamshidi, Olivier Bodenreider, and Halil Kilicoglu for their valuable input.
Footnotes
More details on the UTS methods can be found on the project website: https://uts.nlm.nih.gov/doc/devGuide/webservices/metaops.html
References
- 1.Kordjamshidi P, Otterlo M van, Moens M-F. LREC. 2010. Spatial Role Labeling: Task Definition and Annotation Scheme; pp. 413–420. [Google Scholar]
- 2.Rindflesch TC, Bean CA, Sneiderman CA. AMIA Annu Symp Proc. 2000. Argument Identification for Arterial Branching Predications Asserted in Cardiac Catheterization Reports; pp. 704–708. [PMC free article] [PubMed] [Google Scholar]
- 3.Rosse C, Mejino JL, Modayur BR, Jakobovits R, Hinshaw KP, Brinkley JF. Motivation and organizational principles for anatomical knowledge representation: The Digital Anatomist Symbolic Knowledge Base. J Am Med Inform Assoc. 1998;5(1):17–40. doi: 10.1136/jamia.1998.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosse C, Mejino JL. A reference ontology for biomedical informatics: the Foundational Model of Anatomy. J Biomed Inform. 2003;36(6):489–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Roberts K, Rink B, Harabagiu SM, Scheuermann RH, Toomay S, Browning T, Bosler T, Peshock R. AMIA Annu Symp Proc. 2012. Learning Approach for Identifying Anatomical Locations of Actionable Findings in Radiology Reports. [PMC free article] [PubMed] [Google Scholar]
- 6.Dligach D, Bethard S, Becker L, Miller T, Savova GK. Discovering body site and severity modifiers in clinical texts. J Am Med Inform Assoc. 2013;21(3):448–454. doi: 10.1136/amiajnl-2013-001766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCray AT, Ide NC, Loane RR, Tse T. MEDINFO. 2004. Strategies for Supporting Consumer Health Information Seeking; pp. 1152–1156. [PubMed] [Google Scholar]
- 8.Jadhav A, Andrews D, Fiksdal A, Kumbamu A, McCormick JB, Mistano A, Nelsen L, Ryu E, Sheth A, Wu S, Pathak J. Comparative Analysis of Online Health Queries Originating from Personal Computers and Smart Devices on a Consumer Health Information Portal. J Med Internet Res. 16(7):2014. doi: 10.2196/jmir.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui L, Tao S, Zhang G-Q. AMIA Annu Symp Proc. 2014. A Semantic-based Approach for Exploring Consumer Health Questions Using UMLS; pp. 432–441. [PMC free article] [PubMed] [Google Scholar]
- 10.Goryachev S, Zeng-Treitler Q, Smith CA, Browne AC, Divita G. AMIA Annu Symp Proc. 2008. Making Primarily Professional Terms More Comprehensible to the Lay Audience. [PubMed] [Google Scholar]
- 11.Kilicoglu H, Fiszman M, Demner-Fushman D. Interpreting Consumer Health Questions: The Role of Anaphora and Ellipsis. BioNLP [Google Scholar]
- 12.Roberts K, Masterton K, Fiszman M, Kilicoglu H, Demner-Fushman D. LREC. 2014. Annotating Question Decomposition on Complex Medical Questions; pp. 2598–2602. [Google Scholar]
- 13.Roberts K, Kilicoglu H, Fiszman M, Demner-Fushman D. AMIA Annu Symp Proc. 2014. Automatically Classifying Question Types for Consumer Health Questions. [PMC free article] [PubMed] [Google Scholar]
- 14.Mani I, Hitzeman J, Richer J, Harris D, Quimby R, Wellner B. SpatialML: Annotation Scheme, Corpora, and Tools. LREC. 2008 [Google Scholar]
- 15.Pustejovsky J, Moszkowicz JL, Verhagen M. ISO-ACL SIGSEM Workshop on Interoperable Semantic Annotation. 2011. ISO-Space: The Annotation of Spatial Information in Language; pp. 1–9. [Google Scholar]
- 16.Kordjamshidi P, Bethard S, Moens M-F. SemEval-2012 Task 3: Spatial Role Labeling. SemEval. 2012 [Google Scholar]
- 17.Kolomiyets O, Kordjamshidi P, Moens M-F, Bethard S. SemEval. 2013. SemEval-2013 Task 3: Spatial Role Labeling; pp. 255–262. [Google Scholar]
- 18.Pustejovsky J, Kordjamshidi P, Moens M-F, Yocum Z. SemEval-2015 Task 8: SpaceEval. 2015. http://alt.qcri.org/semeval2015/task8/
- 19.Roberts K, Harabagiu SM. UTD-SpRL: A Joint Approach to Spatial Role Labeling. SemEval. 2012 [Google Scholar]
- 20.Oniki TA, Coyle JF, Parker CG, Huff SM. Lessons learned in detailed clinical modeling at Intermountain Healthcare. J Am Med Inform Assoc. 2014;21(6):1076–1081. doi: 10.1136/amiajnl-2014-002875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dhombres F, Winnenburg R, Case JT, Bodenreider O. MEDINFO. Extending the coverage of phenotypes in SNOMED CT through post-coordination. page In Submission. [PMC free article] [PubMed] [Google Scholar]
- 22.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J Biomed Inform. 2003;36(6):462–477. doi: 10.1016/j.jbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 23.Aronson AR, Lang F-M. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010;17:229–236. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pradhan S, Elhadad N, South BR, Martinez D, Christensen L, Vogel A, Suominen H, Chapman WW, Savova G. Evaluating the state of the art in disorder recognition and normalization of the clinical narrative. J Am Med Inform Assoc. 2015;22(1):143–154. doi: 10.1136/amiajnl-2013-002544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leaman R, Khare R, Lu Z. CLEF 2013 Working Notes. 2013. NCBI at 2013 ShARe/CLEF eHealth Shared Task: Disorder Normalization in Clinical Notes with DNorm. [Google Scholar]
- 26.McCray AT, Burgun A, Bodenreider O. Aggregating UMLS Semantic Types for Reducing Conceptual Complexity. MEDINFO. 2001;84(1):216–220. [PMC free article] [PubMed] [Google Scholar]
- 27.Stenetorp P, Pyysalo S, Topić G, Ohta T, Ananiadou S, Tsujii J. BRAT: a web-based tool for NLP-assisted text annotation. EACL. 2012:102–107. [Google Scholar]
- 28.Manning CD, Surdeanu M, Bauer J, Finkel J, Bethard SJ, McClosky D. The Stanford CoreNLP Natural Language Processing Toolkit. ACL: System Demonstrations. 2014:55–60. [Google Scholar]
- 29.Fan R-E, Chang K-W, Hsieh C-J, Wang X-R, Lin C-J. LIBLINEAR: A Library for Large Linear Classification. Journal of Machine Learning Research. 2008;9:1871–1874. [Google Scholar]