Abstract
Currently, the lack of a universal and specific marker of clonality hampers the diagnosis and classification of chronic expansions of natural killer (NK) cells. Here we investigated the utility of flow cytometric detection of aberrant/altered NK-cell phenotypes as a surrogate marker for clonality, in the diagnostic work-up of chronic lymphoproliferative disorders of NK cells (CLPD-NK). For this purpose, a large panel of markers was evaluated by multiparametric flow cytometry on peripheral blood (PB) CD56low NK cells from 60 patients, including 23 subjects with predefined clonal (n = 9) and polyclonal (n = 14) CD56low NK-cell expansions, and 37 with CLPD-NK of undetermined clonality; also, PB samples from 10 healthy adults were included. Clonality was established using the human androgen receptor (HUMARA) assay. Clonal NK cells were found to show decreased expression of CD7, CD11b and CD38, and higher CD2, CD94 and HLADR levels vs. normal NK cells, together with a restricted repertoire of expression of the CD158a, CD158b and CD161 killer-associated receptors. In turn, NK cells from both clonal and polyclonal CLPD-NK showed similar/overlapping phenotypic profiles, except for high and more homogeneous expression of CD94 and HLADR, which was restricted to clonal CLPD-NK. We conclude that the CD94hi/HLADR+ phenotypic profile proved to be a useful surrogate marker for NK-cell clonality.
Keywords: natural killer cells, NK cells, immunophenotype, clonality, CLPD-NK
INTRODUCTION
Chronic lymphoproliferative disorders (CLPD) of natural killer (NK) cells (CLPD-NK) are a relatively rare and heterogeneous group of diseases characterized by a persistent (> 6 months) increase of mature-appearing NK cells (> 2 × 109/L) in peripheral blood (PB), in the absence of a clearly identifiable cause [1, 2, 3]. Expanded NK cells usually display a large granular lymphocyte (LGL) morphology and a typical surface-membrane CD3−, CD2+, CD16+, CD56low and CD57−/+ immunophenotype [4, 5]. From the clinical point of view, most patients present with an indolent course in the absence of any symptoms [1, 6, 7, 8]; without treatment, the number of circulating PB NK cells usually remains stable for long periods of time, and some cases have even been reported to show spontaneous regression [9, 10]. Actually, CLPD-NK have recently emerged as a new (provisional) entity in the WHO2008 (World Health Organization 2008) classification of lymphoid malignancies [11], mostly because of the need for distinguishing such CLPD-NK cases from more aggressive NK-cell leukemias/lymphomas [12].
The WHO2008 category of CLPD-NK comprises a broad spectrum of NK-cell expansions, ranging from reactive to monoclonal/neoplastic disorders, which have been previously denominated as chronic NK-cell lymphocytosis, chronic NK-large granular leukemia or NK-cell LGL lymphocytosis, among other terms [7, 13, 14]. Hence, a major challenge for the diagnosis of CLPD-NK remains the ability to assess the reactive (polyclonal/oligoclonal) vs. (mono)clonal nature of the expanded NK cells, due to the lack of an universal and specific marker for NK-cell clonality. Therefore, because clonal NK cells from CLPD-NK typically do not show immunoglobulin or T-cell receptor gene rearrangements and their karyotype is normal in most cases [10, 13, 15], alternative methods have been used as surrogate markers for NK-cell clonality –including detection of EBV+ NK cells by Southern blotting [16, 17, 18, 19, 20], analysis of restriction fragment length polymorphisms (RLFP) and restricted (or aberrant) expression of a single isoform of KIR receptors [21, 22, 23, 24], the assessment of the pattern of inactivation of the X-chromosome (e.g. the human androgen receptor assay –HUMARA-) [25, 26] and more recently, the presence of STAT3 mutations [27]; however, these approaches only proved to work in a fraction of patients with chronic NK-cell expansions [18, 19, 20, 23, 24], or they showed inconclusive results [22, 27, 28, 29]. Therefore, since a universal and specific marker for NK-cell clonality is still lacking, precise definition of those immunophenotypic profiles specifically associated with NK-cell clonality, could contribute to the distinction between reactive and clonal CLPD-NK.
In the present study we investigated the immunophenotypic profile of expanded NK cells from 23 females (selected from a total of 60 patients) with predefined monoclonal and polyclonal CLPD of CD56low NK cells vs. that of normal PB NK cells, using a large panel of 26 markers analyzed by multiparameter flow cytometry; ultimately, we aimed at identifying aberrant immunophenotypes that could be used as surrogate markers for NK cell clonality. Overall, our results showed that despite multiple immunophenotypic differences existed between normal and expanded CD56low NK cells, the distinction between monoclonal and polyclonal NK cells mostly relied on a strong and more homogeneous pattern of expression of HLADR and CD94, typically restricted to clonal CLPD-NK.
RESULTS
Clinical and laboratory characteristics of patients with monoclonal vs. polyclonal expansions of PB CD56low NK-cells
From the 23 female patients who were heterozygous for the HUMARA assay, 9 were classified as having monoclonal and 14 as polyclonal NK-cell expansions. Analysis of somatic mutations of the signal transducer and activator of transcription 3 and 5b genes (STAT3 and STAT5b) were consistent with the HUMARA assay, as the hotspot D661Y (c.1981G > T) mutation at the SH2 (exon 21) domain of STAT3 was detected in 1/4 cases carrying clonal NK cells, while no mutations were found in any of the polyclonal cases screened for STAT3 and STAT5b genes (n = 5).
From the clinical point of view, no statistically significant differences were found between the polyclonal and monoclonal cases. Briefly, both groups showed indolent disease, in the absence of organomegalies, recurrent infections, associated neoplasias and/or autoimmune disorders, except for two monoclonal CD56low NK-cell cases, who presented skin lesions, associated in one of them with immune thrombocytopenia (Table 1).
Table 1. Clinical and laboratory characteristics of subjects with monoclonal vs. polyclonal expansions of PB CD56low NK cells vs. normal CD56low NK cells.
| Healthy adults (n = 10) | Cases with expanded/aberrant CD56low NK cells | P-value | ||
|---|---|---|---|---|
| Polyclonal cases (n = 14) | Monoclonal cases (n = 9) | |||
| Age (years)* | 47 ± 24 (27–87) | 55 ± 16 (42–83) | 54 ± 26 (25–81) | NS |
| Reason for consulting: | ||||
| Routine blood analysis | NA | 83% | 80% | NS |
| General symptoms | NA | 17% | 20% | NS |
| Physical examination: | ||||
| Skin lesions | 0% | 0% | 25% | NS |
| Associated autoimmune diseases | 0% | 0% | 13%**§ | NS |
| Laboratory parameters: | ||||
| Leukocytosis (>10 × 109/L) | 0% | 17% | 25% | NS |
| Lymphocytosis (>5 × 109/L) | 0% | 33% | 38% | 0.04a |
| Lymphocytosis (>3 × 109/L) | 0% | 58% | 100% | ≤ 0.05a,b,c,d |
| Neutropenia (<1.5 × 109/L) | 0% | 8% | 13% | NS |
| Anemia (<100g/L) | 0% | 17% | 0% | NS |
| Thrombocytopenia (<100×109/L) | 0% | 25% | 13%§ | NS |
| PB cell counts* | ||||
| Hemoglobin (g/L) | 135 ± 17 (160–97) | 124 ± 26 (88–186) | 139 ± 13 (114–159) | NS |
| N. of platelets (× 109/L) | 233 ± 61 (159–327) | 240 ± 120 (79–476) | 218 ± 86 (83–346) | NS |
| WBC count (× 109/L) | 6.1 ± 3.1 (4.5–8.2) | 7.1 ± 3.4 (3.9–15.5) | 8.7 ± 3.6 (4.2–15.4) | NS |
| N. of neutrophils (× 109/L) | 3.6 ± 0.8 (2.4–4.9) | 2.6 ± 1.6 (0.9–7.4) | 3.2 ± 2.4 (0.3–8.3) | 0.02a |
| N. of lymphocytes (× 109/L) | 1.9 ± 0.7 (0.7–2.8) | 3.8 ± 1.9 (1.6–7) | 4.7 ± 1.3 (3.1–6.6) | ≤ 0.01a,b,c |
| N. of CD56lowNK cells (× 109/L) | 0.25 ± 0.1 (0.1–0.4) | 1.2 ± 0.6 (0.6–2.2) | 2.2 ± 1.2 (0.6–4) | ≤ 0.001a,b,c |
| % of CD56low NK cells* | ||||
| From all PB leucocytes | 4.2 ± 1.3 (2–6) | 20 ± 7 (7–33) | 28 ± 12 (14–43) | ≤ 0.001a,b,c |
| From PB lymphocytes | 13 ± 13 (8–19) | 38 ± 10 (20–50) | 55 ± 11 (38–67) | ≤ 0.003a,b,c,d |
Results expressed as percentage of cases or (*) as mean ± one standard deviation (range).
P-values correspond to the following group comparisons:
healthy adults vs. both polyclonal plus monoclonal CD56low NK-cell expansions;
healthy adults vs. polyclonal CD56low NK-cell expansions;
healthy adults vs. monoclonal CD56low NK-cell expansions;
polyclonal vs. monoclonal CD56low NK-cell expansions.
PB: peripheral blood. NA: not applicable. NS: no statistically significant differences found (p > 0.05).PB: peripheral blood. WBC: white blood cells.
None of the cases studied showed lymphadenopathies, hepatosplenomegaly and/or associated neoplasias.
One case diagnosed with immune thrombocytopenia.
Despite the above referred similarities, all cases with monoclonal CD56low NK cells had lymphocytosis > 3 × 109/L at presentation, while this occurred in only 58% of cases with polyclonal CD56low NK cells (p = 0.05). As expected, increased lymphocyte counts in both groups were at the expense of an increased number of PB CD56low NK cells (p ≤ 0.001 vs. healthy donors), in both absolute and relative numbers (Table 1); of note, the increase in the percentage of NK cells from all PB lymphocytes, was significantly higher among those patients in whom clonality was confirmed vs. polyclonal cases (Table 1).
Immunophenotypic features of monoclonal vs. normal/reactive polyclonal PB CD56low NK cells
Overall, expanded CD56low NK cells from both monoclonal (n = 9) and polyclonal (n = 14) cases showed a similar immunophenotypic profile, which was clearly different from that of normal PB CD56low NK cells from healthy controls: increased levels of CD2, CD57, CD94, HLADR, granzyme B and perforin, and lower expression of CD7, CD11b, CD38 and the CD161 and CD158b killer receptors (p < 0.05) (Figure 1 and Supplementary Figure S1). Despite this, expanded CD56low NK cells from monoclonal cases showed significantly greater levels of both CD94 and HLADR, with also increased percentages of both CD94+ and HLADR+ NK cells, versus polyclonally expanded CD56low NK cells (p < 0.04) (Figure 1 and Table 2). In addition, clonal CD56low NK cells showed a pattern of expression of KIR molecules different from that of polyclonal CD56low NK cells; accordingly, significantly lower percentages of CD158a+ and CD158b+cells, together with a higher intensity of expression of CD158e were found in the former group (Figure 1). In more detail, CD56low NK cells from most monoclonal cases did not express any of the KIR molecules investigated (≤7% CD56low NK cells were found to be positive for CD158a/b/e in 6/9 monoclonal vs. 0/12 polyclonal cases; p < 0.05), while CD161 expression was usually restricted either to virtually all or ≤ 5% clonal cells (Table 2). In contrast, patients with a polyclonal expansion of CD56low NK cells usually showed predominant expression of one KIR (>50% of CD56low NK cells) molecule in 6/11 polyclonal cases vs. 1/9 monoclonal cases (Table 2 and Supplementary Figure S1). Clonal CD56low NK cells also showed significantly lower levels of granzyme B (mean fluorescence intensity -MFI- of granzyme B of 160 vs. 98 for cases with expanded polyclonal vs. clonal NK cells; p = 0.03). In turn, whereas two cases having clonal CD56low NK cells were CD2−, clonal cases showed overall significantly higher levels of CD2 and lower amounts of CD38/cell vs. both normal and expanded polyclonal NK cells (p ≤ 0.002); finally, CD11c levels were greater in clonal vs. polyclonally expanded CD56low NK cells (p = 0.01) (Figure 1).
Figure 1. Immunophenotypic characteristics of expanded monoclonal versus expanded polyclonal and normal adult peripheral blood CD56low NK cells.
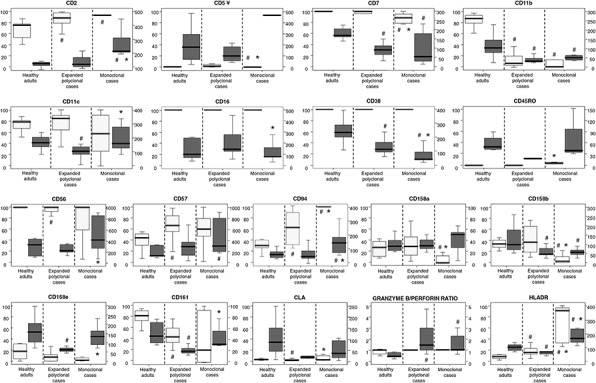
Clonality was assessed in all patients included in the analyses displayed (n = 23) by the HUMARA assay. CD56low NK cells from the three groups of subjects showed a similar positive reactivity for CD8, CD45RA and CD45, while the expression of CD3, CD25, CD27, CD28 and CD197 (CCR7) was systematically negative. Results are expressed for each marker as percentage of positive cells (light gray boxes, referred to the scales at the left of the plots) and mean fluorescence intensity - MFI- of positive cells - arbitrary relative linear units scaled from 0 to 104 cells (dark gray boxes, referred to the scales at the right of the plots). Notches boxes represent 25th and 75th percentile values; the line in the middle and vertical lines correspond to the median value and the minimum-maximum values (without the extreme values and outliers), respectively. ¥ Please note that the high expression levels of CD5 for clonal cases corresponds only to one case in which 1% of CD56low NK cells were CD5+ with a MFI of 482. # P-value < 0.05 vs. CD56low NK cells from the control group (healthy adults).*P-value < 0.05 vs. expanded polyclonal CD56low NK cells.
Table 2. Expression of the CD158a/b/e killer-immunoglobulin-like receptors (KIR) and the CD94 and CD161 C-type lectin-like receptors on peripheral blood CD56low NK cells from patients with clonal (n = 9) and polyclonal (n = 12*) expansions of CD56low NK cells.
| Percentage of CD56low NK cells expressing each marker | |||||
|---|---|---|---|---|---|
| CD158a | CD158b | CD158e | CD94 | CD161 | |
| Clonal#1 | 20 | 58 | 6 | 100 | 40 |
| Clonal#2 | 8 | 10 | 8 | 100 | 100 |
| Clonal#3 | 39 | 20 | 46 | 100 | 87 |
| Clonal#4 | 0 | 1 | 1 | 100 | 0 |
| Clonal#5 | 0 | 1 | 1 | 100 | 5 |
| Clonal#6 | 0 | 3 | 2 | 100 | 0 |
| Clonal#7 | 1 | 2 | 2 | 100 | 2 |
| Clonal#8 | 0 | 0 | 0 | 100 | 0 |
| Clonal#9 | 0 | 7 | 0 | 100 | 100 |
| Median | 0 | 3 | 2 | 100 | 5 |
| Interquartile range | 0–14 | 1–15 | 0.5–7 | 100–100 | 0–93 |
| Minimum-maximum values | 0–39 | 0–58 | 0–46 | 100–100 | 0–100 |
| Polyclonal#1 | 73 | 23 | 9 | 100 | 57 |
| Polyclonal#2 | 10 | 10 | 0 | 81 | 35 |
| Polyclonal#5 | 14 | n.a. | 9 | 47 | 32 |
| Polyclonal#6 | 27 | 28 | 20 | 42 | 25 |
| Polyclonal#7 | 28 | 15 | 11 | 40 | 78 |
| Polyclonal#8 | 46 | 37 | 26 | 34 | 75 |
| Polyclonal#9 | 3 | 43 | 0 | 56 | n.a. |
| Polyclonal#10 | 75 | 15 | 6 | 65 | 61 |
| Polyclonal#11 | 58 | 73 | 0 | 75 | 62 |
| Polyclonal#12 | 6 | 76 | 0 | 80 | 20 |
| Polyclonal#13 | 3 | 60 | 18 | 76 | 77 |
| Polyclonal#14 | 29 | 70 | 0 | 72 | 37 |
| Median | 27.5 | 37 | 7.5 | 68.5 | 57 |
| Interquartile range | 7–55 | 15–70 | 0–16.3 | 43.3–79 | 32–75 |
| Minimum-maximum values | 3–75 | 23–76 | 0–26 | 34–100 | 25–78 |
| p-values | 0.007 | 0.003 | n.s. | <0.0001 | n.s. |
n.a.: not available; n.s.: Not statistically significant differences (p > 0.05) between clonal and polyclonal cases.
In two out of the 14 cases with expanded polyclonal CD56low NK cells, data about more than one killer receptor was not available, and therefore, these two cases were excluded from Table 2.
In contrast, CD56low NK cells from all three groups of subjects displayed stable and similar reactivity for CD8, CD45RA and CD45 (Figure 1), in the absence of CD3, CD25, CD27, CD28 and CD197 (CCR7).
Identification of the most discriminating immunophenotypic markers for the classification of CD56low NK cells
Unsupervised hierarchical clustering analysis based on 30 immunophenotypic parameters confirmed the existence of three different, but partially overlapping groups of CD56low NK cells, such unique phenotypic profiles being associated with expanded clonal and polyclonal NK cells, plus normal PB NK cells, respectively (Figure 2). The most relevant parameters for the discrimination between monoclonal NK cells from both control (normal) and expanded polyclonal NK cells included three overexpressed markers (HLADR, CD94 and CD45RO) and four down-regulated protein-associated variables (% CD158a+, % CD161+, % CD11b+ and CD38 MFI) (Figure 2) among the former group.
Figure 2. Unsupervised hierarchical clustering analysis of the immunophenotypic pattern of expanded monoclonal, expanded polyclonal and normal adult peripheral blood (PB) CD56low NK cells.
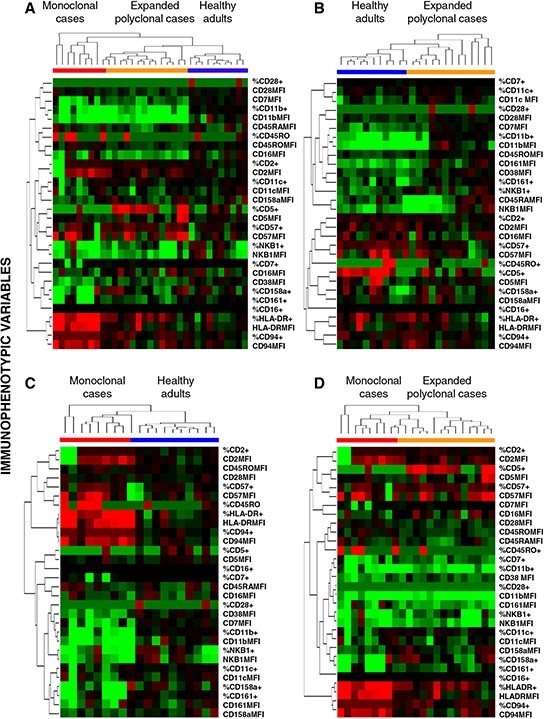
Trees were generated based on the expression patterns observed on PB CD56low NK cells for the 30 immunophenotypic variables included in the study, generated using the Cluster and TreeView software programs. Panel A. shows the clustering trees of the three groups included in the study, while Panels B, C. and D. show the trees generated for the following analyses: healthy adults vs. expanded polyclonal NK-cell cases (B); expanded monoclonal NK-cell cases vs. healthy adults (C) and; expanded monoclonal vs. expanded polyclonal patient groups (D), respectively. For each given marker, immunophenotypic data is expressed both as percentage of positive cells and as mean fluorescence intensity of positive cells –arbitrary relative linear units scaled from 0 to 104cells–, except for CD38 and CD45RA, which are expressed only as MFI values (for a total of 30 immunophenotypic variables). Columns show the expression levels for a single sample; rows represent the relative levels of expression for a single immunophenotypic parameter, centered at the geometric mean of the expression levels observed for all samples. Red and green colors indicate values higher or lower than the mean, respectively; color intensity represents the magnitude of the deviation from the mean; black color represents no significant change in the expression levels for that sample vs. the mean.
A predictive model (PM) based on the above referred discriminating parameters was then built and evaluated by applying it to the analysis of the 33 samples of the training set (10 healthy adult donors plus 9 monoclonal female cases plus 14 polyclonal female cases; Figure 3, Panel A). Based on this model, all but two samples (31/33; 94%), corresponding to CD94low/HLADR−/low clonal CD56low NK cells, were correctly classified, with a probability of correct classification of 81% for clonal cases and 98% for polyclonal/normal CD56low NK-cell samples. Subsequently, the same PM was used to classify the remaining 37 patient samples with expansions of CD56low NK cells, in which the HUMARA assay could not been performed and/or was not informative for the assessment of clonality. From these 37 cases, 11 were classified as being clonal and 26 as polyclonal/normal, with a probability of being correctly classified of 90% and 91%, respectively.
Figure 3. Identification of the most relevant phenotypic markers contributing to the discrimination between normal/polyclonal and monoclonal peripheral blood CD56low NK cells.
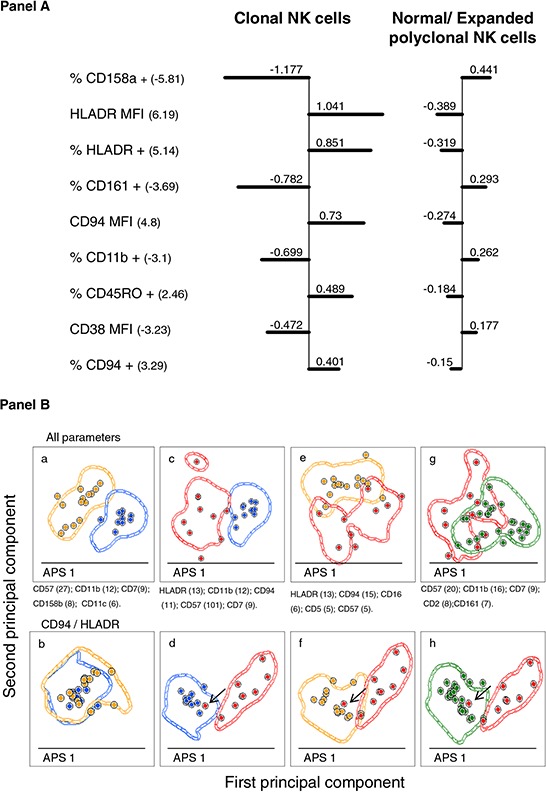
Panel A. illustrates the “Nearest-shrunken centroid” analysis, showing the shrunken differences for the 7 immunophenotypic markers having at least 1 non-zero difference, selected as significant by SAM analysis. Left-sided and right-sided bars indicate lower and higher expression in each group with respect to the overall centroid, respectively. Markers are ordered from the greatest to the smallest difference. Numbers in brackets indicate d-Values: positive d-Values denote over-expression on clonal CD56low NK cells, while negative d-Values indicate under-expression on clonal CD56low NK cells. Panel B. shows pairwise PCA-based comparisons of the phenotypes of expanded monoclonal, expanded polyclonal and normal adult peripheral blood CD56low NK cells. APS -Automated Population Separator function included in the INFINICYT software, corresponding to the principal components 1 (X axis) and 2 (y axis)- views showing the discrimination between CD56low NK cells from: healthy adults (blue circles) vs. reactive polyclonal (orange circles) cases (a and b); healthy adults (blue circles) vs. clonal (red circles) cases (c and d); clonal (red circles) vs. polyclonal (orange circles) (e and f); and clonal (red circles) vs. both healthy adults and reactive (green circles) cases (g and h). Each circle represents one single case (median expression observed for all the phenotypic parameters evaluated). Contour lines in these plots represent one standard deviation curves. Panels a, c, e and g show mean principal component 1 vs. principal component 2 values for each case, considering all phenotypic parameters analyzed; the first four parameters contributing to the best discrimination between the two groups (ordered according to their percentage of contribution) are displayed at the bottom of each plot. Panels b, d, f and h show mean principal component 1 vs. principal component 2 values for each case, but considering only the CD94 and HLADR parameters. The arrow denotes the only misclassified case.
To further confirm the value of the above described phenotypic markers for the discrimination between monoclonal vs. polyclonal CD56low NK cells, the PCA (principal component analysis) function of the INFINICYT software, was used. Despite clonal cases could not be clustered in a single group by PCA, they clearly separated from both normal and expanded polyclonal cases when phenotypic parameters were considered in the analysis (Figure 3, Panel Bg); such discrimination was even more clear on PCA when only the two most informative markers (HLADR and CD94) were used (Figure 3, Panel Bh). Actually, only one clonal case did not cluster separately from the group with expanded polyclonal CD56low NK cells, due to the fact that NK cells from this discordant case had low CD94 expression levels, in the absence of HLADR. The INFINICYT reference data set generated upon excluding this case, was then used to classify the remaining 37 patient samples containing CD56low NK cells of undetermined/unknown clonality (Supplementary Figure S2); then, each tested case was evaluated by the “fixed” PCA function and assigned to its nearest reference entity in the principal component (PC)1 vs. PC2 (APS plot in the INFINICYT software) space. Based on this approach, 11 cases were classified as monoclonal and 26 as normal/polyclonal cases, with a 100% concordance between the PCA-based and the PM-based classifications.
Follow-up of CLPD-NK cases predicted to be clonal vs. polyclonal on immunophenotypic grounds
Clinical and hematological data were collected after long-term follow-up (median of 34 months, ranging from 3 to 156 months) in a subset of 21/37 CLPD-NK cases in which both the HUMARA assay and the analysis of somatic mutations of STAT3/STAT5b were not performed and/or were not informative (9 classified as clonal and 12 as polyclonal by the predictive models). Interestingly, in all 9 cases with undetermined clonality who were predicted to have clonal NK-cells, the absolute lymphocyte count remained over the normal range after a median follow-up of 29 months; even more, in 2/9 cases the lymphocyte count increased (from 4.3 to 10.2 × 109/l and from 4.8 to 8.4 × 109/l). Of note, one of these later patients died 4 years after first diagnosis, although the actual cause of death was unknown. In contrast, in 58% (7/12) of cases predicted to be polyclonal by the statistical predictive models described below, the lymphocyte count decreased and returned to normal values after a median follow-up of 31 months, while it remained stable and high in the other 5 cases. Most of these "polyclonal" CLPD-NK patients (n = 10/12) had no clinical symptoms at follow-up; the two symptomatic cases corresponded to two patients diagnosed with cancer who died due to their tumors, after NK-cell counts had decreased toward normal values.
DISCUSSION
CLPD-NK has been recognized as a new provisional diagnostic entity in the WHO2008 classification of hematopoietic tumors, these cases being clearly different from aggressive mature NK-cell neoplasms such as extranodal NK/T cell lymphoma, nasal type and aggressive NK cell leukemia (ANKL) [11, 30, 31]. CLPD-NK usually shows an indolent clinical behavior; however, far from being a homogeneous group of diseases, CLPD-NK encompasses truly leukemic cases, some of which even show transformation to ANKL [32, 33], and reactive conditions, which might likely represent the extreme pole of an immunologic response to unknown stimuli [10]. In contrast to chronic expansions of clonal vs. reactive B and T cells, which can be more easily distinguished based on the analysis of the immunoglobulin/T-cell receptor gene rearrangements, CLPD of clonal vs. polyclonal NK cells are rather difficult to diagnose, among other reasons, because of the lack of a universal and specific marker for NK- cell clonality. This also concerns detection of STAT3 somatic mutations which have recently been identified as a recurrent genetic abnormality in patients with CLPD-NK, since they are only present in between 18% and 30% of cases by conventional sequencing techniques [27, 34], in line with our findings. The challenge that assessment of clonality represents in CLPD-NK, together with the rarity of NK-cell tumors, undoubtedly have hampered our ability to standardize the diagnostic procedures for this entity, and hence, the clinical management of CLPD-NK patients.
At present, it is well-established that in most hematological malignancies (e.g. acute leukemias, B- and T-cell CLPD and plasma cell disorders) neoplastic cells from virtually every patient display aberrant phenotypes, that can be used for the distinction of such tumor cells from their normal counterparts [35, 36,]. Of note, such aberrant phenotypes are not exclusive of malignant leukemias and lymphomas, as they can be also found at similar frequencies in benign conditions, e.g. monoclonal B-cell lymphocytosis (MBL) [37] and monoclonal gammapathy of undetermined significance (MGUS) [38]. Consequently, the "leukemia-associated phenotypes" can also be considered to represent markers for clonality. Based on this hypothesis, here we investigated for the first time the utility of a broad panel of immunophenotypic markers assessed by flow cytometry, as surrogate markers for the classification of CLPD of CD56low NK cells into clonal vs. polyclonal cases. For this purpose, we comparatively analyzed the immunophenotypic profile of PB CD56low NK cells from female patients in whom the clonal vs. polyclonal nature of the expanded cells was previously known (e.g. by the HUMARA X-chromosome inactivation assay performed on highly purified cells) to that of normal adult PB CD56low NK cells. Phenotypic analyses included a large panel of NK-cell-associated markers, consisting of signaling (e.g. CD2, CD5, CD7, CD8) and adhesion molecules (e.g CD11b and CD11c), activation-related markers (e.g. CD38, CD45RO and HLADR), IL2 receptors (e.g. CD25 and CD122), maturation-associated proteins (e.g. CD57), cytotoxic enzymes (e.g. granzyme B and perforin) and receptors involved in the recognition of target cells (e.g. CD16, the CD158a, CD158b and CD158e natural-killer immunoglobulin-like receptors –KIR- and the CD94 and CD161 C-type lectin-like receptors), among other molecules. Based on the above referred markers, expanded CD56low NK cells from both monoclonal and polyclonal patients showed an overlapping immunophenotypic profile consistent with that of late-stage activated NK cells [5], which was clearly different from the one displayed by normal CD56low NK cells. These findings support the hypothesis that both clonal and polyclonally expanded NK cells could emerge as a consequence of chronic antigen stimuli. In this regard, CD56low NK cells from CLPD patients showed higher expression of CD57, granzyme B and perforin than normal control cells, together with a CD2high/CD7low phenotypic pattern, upregulation of CD94 and HLADR and lower expression of CD11b, CD38 and CD161; while the former marker profile is typical of late-maturation stage cells, the later is usually featured on activated NK cells [5].
Recent studies have evaluated the expression profile of natural cytotoxicity receptors [24] and different killer cell immunoglobulin-like receptors [21, 31], showing a skewed repertoire of KIR receptors on the expanded cells from CLPD-NK patients [24, 29]; in turn, others have reported bright CD94 expression on pathological cells from CLPD-NK patients [24, 29]. However, despite the biological findings and the interest of these reports, in none of them the monoclonal vs. polyclonal nature of the expanded NK cells was investigated, in contrast to what was achieved here. Thus, the potential contribution of immunophenotyping to the differential diagnosis between expanded clonal and polyclonal CD56low NK cells in CLPD-NK, would be of utmost clinical relevance, specially if we take into account that there were no significant clinical differences between the two patient groups, in line with previous observations [7, 8, 40]. In this regard, and despite the similarities between the expanded clonal and polyclonal NK cells, detailed analysis of the pattern of expression of individual markers already highlighted significant differences between the two patient groups, including a greater expression of CD94 and HLADR (both in terms of percentage of cells and expression levels/cell of both markers), as well as CD2, together with lower positivity for the CD158a/b/e KIR molecules, CD38 and granzyme B in expanded clonal vs. polyclonal CD56low NK cells. Of note, expanded CD56low NK cells from most monoclonal cases lacked on expression of all KIR investigated, in contrast to normal/polyclonal NK cells from both reactive cases and healthy controls; these findings support and extend on previous studies that reported a skewed KIR repertoire for LGL-NK cell leukemias vs. normal NK cells [24]; in addition, here we show for the first time that most expanded CD56low NK cells from individual reactive/polyclonal cases usually expressed one single (exceptionally two) KIR molecules, a feature that others have considered as a surrogate marker for clonality [24]. Further studies including a higher number of KIR and other killer receptor molecules analyzed on larger series of patients with CD56low NK-cell expansions vs. healthy donors would be required, to definitively clarify this apparent discrepancy.
In order to identify a more robust NK-cell phenotypic profile that could be prospectively applied to the diagnosis of clonality in CLPD-NK patients, multivariate analyses were performed. As a result, unsupervised hierarchical clustering analysis showed that the most relevant parameters contributing to the distinction between monoclonal vs. both expanded polyclonal and normal CD56low NK cells included overexpression of HLADR and CD94, together with CD45RO, and downregulation of CD158a, CD38, CD11b and CD161. Based on these markers, cases were segregated into three groups, polyclonal/reactive CLPD-NK cases being located between the normal and the monoclonal NK-cell groups. Based on these results, a predictive model was constructed using the above markers and the training set of cases; such model provided to properly classify all but two clonal CLPD-NK cases, whose CD56low NK cells did not express high levels of HLADR and CD94. These observations also indicated that the phenotypic profile that would mostly contribute to the diagnosis of clonality of CLPD-NK strongly relies on the existence of a strong and more homogeneous expression of both HLADR and CD94, in such a way that when clonal NK cells show a different HLADR/CD94 profile, a high overlap with the phenotype of polyclonal (particularly reactive) NK cells might exist. This was further confirmed by PCA, which showed that the two most useful phenotypic parameters for the identification of clonal vs. normal/polyclonal CD56low NK cells were HLADR and CD94. Actually, only one clonal case did not cluster separately from the expanded polyclonal CD56low NK-cell group in the PCA, such a case showing a CD94low/HLADRlow immunophenotypic profile.
More interestingly, when the two predictive models were applied for the prospective classification of the 37 CLPD-NK cases in which the HUMARA assay could not be performed and/or it was not informative, they showed a 100% concordance for the classification of this group of cases into the monoclonal (n = 11) and the polyclonal (n = 26) NK-cell categories. Follow-up of most of these patients was highly concordant with the predicted monoclonal vs. polyclonal nature of the expanded PB CD56low NK cells at diagnosis. Thus, in two patients predicted to have clonal NK cells the absolute number of lymphocytes was increased with time, while in all other cases from this group kept stable over the normal range; in contrast, the lymphocyte count returned to normal values in near two thirds of cases predicted to be polyclonal and remained stable in the other third.
Altogether, these results provide the basis for prospective usage of altered NK-cell immunophenotypes, in the absence of other universal markers for NK-cell clonality, as a surrogate marker for the distinction between clonal and reactive NK-cell expansions in the diagnostic work-up of CLPD-NK. The most discriminating phenotypic profile that contributed to such distinction consist of a strong and more homogeneous expression of CD94 and HLADR and, to a lesser extent also, downregulation of CD158a, CD38, CD11b and CD161.
MATERIALs AND METHODS
Patients and samples
A total of 60 patients who showed increased numbers of circulating PB CD56low NK cells, referred to the Cytometry Service of the University of Salamanca (NUCLEUS) during a 12-year period of time, were studied. Median age of the patients −29 males and 31 females- was of 65 years (range: 25 to 86 years). Inclusion criteria were: presence of increased numbers of PB CD56low NK cells (>2 × 109/L and/or > 20% NK cells from all circulating lymphocytes), for more than 6 months, or identification of phenotypically aberrant CD56low NK cells, regardless of their actual numbers. An additional group of 10 adult healthy volunteers (3 males and 7 females; median age of 47 years, ranging from 27 to 83 years) with normal blood cell counts, in the absence of any apparent or known disease was also included in the study.
PB samples from both the patients and the controls were collected into tubes containing K3-EDTA, after written informed consent was given by each subject, according to the Declaration of Helsinki. The study was approved by the Ethics Committee of the University Hospital of Salamanca (Salamanca, Spain).
Flow cytometry immunophenotypic studies
Enumeration of PB lymphoid cell subsets, including CD56low NK cells, was performed using the LymphoclonalTM reagent (Cytognos SL, Salamanca, Spain), which contains a mixture of seven antibodies (Ab) [fluorescein isothiocyanate (FITC)/phycoerythrin (PE)/peridin chlorophyll protein-cyanin 5.5 (PerCP-Cy5.5)/allophycocyanin (APC)]: anti-human immunoglobulin light chain λ (Igλ) + CD8/anti-human Igκ + CD56/CD19 + CD4/CD3. Further immunophenotypic characterization of PB CD56low NK cells was performed using the following 4-color combinations of fluorochrome-conjugated -FITC/PE/PE-cyanin 5 (PE-Cy5) or PerCP-Cy5.5/APC- monoclonal Ab (MAb): CD7/CD5/CD56/CD3; CD2/CD28/CD56/CD3; CD57/CD11c/CD56/CD3; CD38/CD11b/CD56/CD3; CD94/HLA-DR/CD56/CD3; CD45RA/CD197/CD56/CD3; CD27/CD45RO/CD56/CD3; CD16/CD158e (NKB1)/CD56/CD3; CLA/CD158b (NKAT2)/CD56/CD3; CD158a (NKAT1)/CD161/CD56/CD3; CD122/CD25/CD56/CD3 and cytoplasmic (Cy) perforin/Cy granzyme B/CD56/CD3. The sources and specificities of the MAb reagents used have been previously described in detail [26, 28]. Cell staining was performed on whole blood samples, using a well-established stain-and-then-lyse method, as previously reported [26, 28]. Combined staining for surface antigens and intracellular molecules was performed using the Fix & PermTM reagent kit (Invitrogen, Carlsbad, CA, USA), according to the recommendations of the manufacturer. Data acquisition was performed immediately after completion of sample preparation, in a FACSCalibur flow cytometer -Becton Dickinson Biosciences (BD), San Jose, CA, USA- using the CellQUESTTM software program (BD). For data analysis, the INFINICYTTM software program (Cytognos), was used. In each case, CD56low NK cells were defined as those CD56low/CD3− events included in a broad forward (FSC) vs. sideward light scatter (SSC) lymphocyte region. For each antigen analyzed, both the percentage of positive CD56low NK cells and its intensity of expression -reflected by the MFI expressed in arbitrary relative linear units scaled from 0 to 104-, were evaluated.
HUMARA assay
In order to confirm the monoclonal vs. polyclonal nature of the expanded/aberrant CD56low NK cells, the pattern of inactivation of the human androgen receptor coded in chromosome X was studied in 25/31 female patients. The methylation status of the human androgen receptor gene was evaluated with a HUMARA polymerase chain reaction (PCR)-based assay [25, 41, 42, 43] for paired fractions of purified CD56low NK cells (mean purity of 98% ± 1.2%; range: 94% to 99%) and the corresponding NK-cell depleted leukocyte fraction from the same subject (n = 25). NK cells were purified from mononuclear PB cells (obtained after Ficoll-PaqueTM density gradient centrifugation) by a single immunomagnetic depletion step using the Human NK-Cell Enrichment Kit (StemStepTM, StemCell Technologies Inc, Vancouver, BC, Canada) and an AutoMACS magnetic cell separator (Miltenyi Biotec, Bergisch Gladbach, Germany), strictly following the recommendations of the manufacturer. Purified populations were used to extract genomic DNA (gDNA) using the QIAamp mini- or micro-DNA extraction Kits (QIAGEN, Valencia, CA). Digestion of genomic DNA was performed with the HapII methylation-sensitive restriction endonuclease and subsequent PCR amplification of the methylated (inactivated) alleles, as previously described [25, 41, 42]. According to previous reports, a case was considered to be monoclonal when the corrected allele ratio was of ≤0.33 or ≥3, indicating that one of the parental alleles was represented at an excess of ≥50% vs. the other allele [43].
Analysis of STAT3 and STAT5b somatic mutations
STAT3 and STAT5b gene mutations were analyzed on gDNA extracted from highly purified CD56low NK cells -according to the techniques described above- from a total of 9 cases which had also been evaluated by the HUMARA assay, from which 4 corresponded to monoclonal and 5 to polyclonal cases. Previously reported PCR primers were used to amplify known somatic mutational hotspot regions in the STAT3 and STAT5b genes, located at STAT3 SH2 exons 19–20 and 21, as well as at STAT5b SH2 exon 16 [27, 34, 44]. The amplified products were sequenced by conventional techniques at the Genomic Unit of the Cancer Research Center (IBMCC, USAL-CSIC, Salamanca, Spain); all mutations were screened by bidirectional sequencing and scored as pathogenic mutations on the basis of the observation that they were not detected in paired cell subsets other than NK cells. The sequences were analyzed using the Chromas Lite Sequencing Software (South Brisbane, Australia).
Statistical methods
Conventional statistics –nonparametric Kruskal-Wallis and Mann-Whitney U tests for continuous variables, or the Pearson's χ2 and Fisher exact tests for categorical variables– were performed using the SPSS software program (IBM SPSS Statistics, IBM, Armonk, NY, USA). P values ≤ 0.05 were considered to be associated with statistical significance.
Multivariate analysis of markers differentially expressed in normal and reactive vs. clonal CD56low NK cells was performed in four consecutive steps. Only healthy subjects (n = 10) and those female patients found to be heterozygous for the HUMARA assay (n = 23 female cases tested; 9 monoclonal and 14 polyclonal) were included in the analysis. Prior to the analysis, data about the immunophenotype of CD56low NK cells – percentage of cells positive for each individual marker, as well as the MFI values of CD56low NK cells positive for each marker (n = 30 variables)– was normalized using the mean value obtained for each individual variable in normal PB NK cells and logarithmically (base 2) transformed. Afterward (step 1), both unsupervised and supervised hierarchical clustering analyses based on the average-linkage method and an Euclidean metric [45] were applied (Cluster 3.0 and Tree View software, Stanford University, Stanford, CA, USA) [46]. In a second step, a Significant Analysis of Microarrays (SAM) algorithm was built -with the same 30 variables used in step 1- in order to identify those markers showing a statistically significantly different expression in the clonal (n = 9) vs. polyclonal classes (n = 14 cases plus 10 healthy controls). Data was permutated over 100 cycles by using the two-class (unpaired) format and the statistically relevant variables (T-test) were selected, based on the lowest false discovery ratio (FDR < 2.2) and the q-value (0) [47] obtained. In the following step (third step), those variables identified to be significantly associated with different classes were used to build a class predictor (weighted voting algorithm with a leave-one-out cross validation), that included the smallest combination of variables for correct classification of CD56low NK-cell expansions as monoclonal vs. polyclonal. A training set containing all above referred 33 cases (10 healthy adult donors plus 9 monoclonal female cases plus 14 polyclonal female cases) was used to train the classifier, and the accuracy of the predictor was subsequently tested in a set including all the 60 patients studied and who presented with abnormal NK-cell numbers and/or phenotypes, using the nearest shrunken centroids method (PAM software; University of Stanford, Stanford, CA, USA) [48]. In the last (fourth) step, principal component analysis (PCA) was applied to flow cytometric data from individual cells, in order to confirm the value of the individual variables selected for correct classification of monoclonal vs. polyclonal nature of a given expansion of CD56low NK cells. For this purpose, a reference data set was built with data from healthy controls (n = 10) and those patients proved to have polyclonal (n = 14) and monoclonal (n = 9) NK-cell expansions by the HUMARA assay, using the merge and calculation functions of the INFINICYTTM software, as previously described [49]. Unsupervised PCA was then performed for the reference database and data was then visualized in a 2-dimensional plot defined by the first (PC1) vs. second (PC2) principal component (APS plot in the INFINICYT software) [50, 51] for the distinction among different clusters of NK cells.
SUPPLEMENTARY FIGURES
ACKNOWLEDGMENTS AND FUNDING
The authors thank the following medical doctors for their assistance in collecting samples and clinical data from patients: I. Alberca, M. Alcocer, J.M. Alonso, I. Alvarez-Twose, M. Barbon, M.C. Berges, R. Butron, M. Cordero, J. Galende, A. Garcia de Coca, J.R. Garcia-Talavera P. Giraldo, M. Gonzalez, L. Guerras, I. Marchante, G. Martin, O. Martinez, N. Mendez, M. Mejido, D. Monego, F. Ortega, J.M. Perdiguer, F. Puente, J.A. Queizan, M. Romero, A. Rubio, J. Sabas, F. Salas, L.A. Silvestre, A. Smucler.
This work has been partially supported by the following grants: FIS 02/1244-FEDER, DTS 15/00119-FEDER, RTICC RD06/0020/0035-FEDER and RTICC RD12/0036/0048-FEDER from the Fondo de Investigación Sanitaria, Instituto de Salud Carlos III, Ministerio de Economía y Competitividad, Madrid, Spain; SA103/03 and SA079U14 from the Consejería de Educación, Junta de Castilla y León, Valladolid, Spain. The research activities of the EuroFlow Consortium were supported by the European Commission (grant STREP EU-FP6, LSHB-CT-2006–018708, entitled ‘Flow cytometry for fast and sensitive diagnosis and follow-up of hematological malignancies’).
Footnotes
CONFLICTS OF INTEREST
The authors declare that they have no competing interests; therefore, the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author's contributions
PB performed research, data analysis and wrote the manuscript; MJ-A, MLS and ACGM contributed with expert technical assistance in the experiments; MDT, AL performed data analysis; MBV and AP provided patient samples and clinical data; QL contributed to design the InfinicytTM software and performed data analysis; ML, AWL, SB and JvD contributed to the design of the InfinicytTM software and to test its utility for the analysis of NK cells; AO and JA designed research, analyzed data and wrote the manuscript. All authors have read and approved the manuscript.
REFERENCES
- 1.Loughran TP., Jr Clonal diseases of large granular lymphocytes. Blood. 1993;82:1–14. [PubMed] [Google Scholar]
- 2.Lamy T, Loughran TP., Jr Current concepts: large granular lymphocyte leukemia. Blood Reviews. 1999;13:230–240. doi: 10.1054/blre.1999.0118. [DOI] [PubMed] [Google Scholar]
- 3.Semenzato G, Marino F, Zambello R. State of the art in natural killer cell malignancies. International Journal of Laboratory Hematology. 2012;34:117–128. doi: 10.1111/j.1751-553X.2011.01374.x. [DOI] [PubMed] [Google Scholar]
- 4.Mori KL, Egashira M, Oshimi K. Differentiation stage of natural killer cell-lineage lymphoproliferative disorders based on phenotypic analysis. British Journal of Haematology. 2001;115:225–228. doi: 10.1046/j.1365-2141.2001.03038.x. [DOI] [PubMed] [Google Scholar]
- 5.Lima M, Almeida J, Teixeira MA, Santos AH, Queirós ML, Fonseca S, Moura J, Gonçalves M, Orfao A, Pinto Ribeiro AC. Reactive phenotypes after acute and chronic NK-cell activation. Journal of Biological Regulators and Homeostatic Agent. 2004;18:331–334. [PubMed] [Google Scholar]
- 6.Oshimi K, Yamada O, Kaneko T, Nishinarita S, Lizuka Y, Urabe A, Inamori T, Asano S, Takahashi S, Hattori M. Laboratory findings and clinical courses of 33 patients with granular lymphocyte-proliferative disorders. Leukemia. 1993;7:782–788. [PubMed] [Google Scholar]
- 7.Tefferi A, Li CY, Witzig TE, Dhodapkar MV, Okuno SH, Phyliky RL. Chronic natural killer cell lymphocytosis: a descriptive clinical study. Blood. 1994;84:2721–2725. [PubMed] [Google Scholar]
- 8.Semenzato G, Zambello R, Starkebaum G, Oshimi K, Loughran TP., Jr The lymphoproliferative disease of granular lymphocytes: updated criteria for diagnosis. Blood. 1997;89:256–260. [PubMed] [Google Scholar]
- 9.Oshimi K. Progress in understanding and managing natural killer-cell malignancies. Brithish Journal of Haematology. 2007;139:532–544. doi: 10.1111/j.1365-2141.2007.06835.x. [DOI] [PubMed] [Google Scholar]
- 10.Zambello R, Semenzato G. Large granular lymphocyte disorders: new etiopathogenetic clues as a rationale for innovative therapeutic approaches. Haematologica. 2009;94:1341–1345. doi: 10.3324/haematol.2009.012161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villamor N, Morice WG, Chan WC, Fourcar K. Chronic lymphoproliferative disorders of NK cells. In: Swerdlow SH, Harris NL, Jaffe EJ, Pirelli SA, Stein H, Thiele J, Vardiman J, editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4. International Agency for Research of Cancer (IARC); Lyon, France: 2008. pp. 274–275. [Google Scholar]
- 12.Lim MS, de Leval L, Quintanilla-Martinez L. Commentary on the 2008 WHO classification of mature T- and NK-cell neoplasms. Journal of Hematopathology. 2009;2:65–73. doi: 10.1007/s12308-009-0034-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tien HF, Su IJ, Tang JL, Liu MC, Lee FY, Chen YC, Chuang SM. Clonal chromosomal abnormalities as direct evidence for clonality in nasal T/natural killer cell lymphomas. British Journal of Haematology. 1997;97:621–625. doi: 10.1046/j.1365-2141.1997.752711.x. [DOI] [PubMed] [Google Scholar]
- 14.Wong KF, Chan JK, Kwong YL. Identification of del(6)(q21q25) as a recurring chromosomal abnormality in putative NK cell lymphoma/leukaemia. British Journal of Haematology. 1997;98:922–926. doi: 10.1046/j.1365-2141.1997.3223139.x. [DOI] [PubMed] [Google Scholar]
- 15.Oshimi K. Leukemia and lymphoma of natural killer lineage cells. International Journal of Hematology. 2003;78:18–23. doi: 10.1007/BF02983235. [DOI] [PubMed] [Google Scholar]
- 16.Kawa-Ha K, Ishihara S, Ninomiya T, Yumura-Yagi K, Hara J, Murayama F, Tawa A, Hirai K. CD3-negative lymphoproliferative disease of granular lymphocytes containing Epstein-Barr viral DNA. Journal of Clinical Investigation. 1989;84:51–55. doi: 10.1172/JCI114168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satoh M, Oyama N, Akiba H, Ohtsuka M, Iwatsuki K, Kaneko F. Hypersensitivity to mosquito bites with natural-killer cell lymphocytosis: the possible implication of Epstein-Barr virus reactivation. European Journal of Dermatology. 2002;12:381–384. [PubMed] [Google Scholar]
- 18.Siu LL, Chan JK, Kwong YL. Natural killer cell malignancies: clinicopathologic and molecular features. Histology and Histopathology. 2002;17:539–554. doi: 10.14670/HH-17.539. [DOI] [PubMed] [Google Scholar]
- 19.Choi YL, Moriuchi R, Osawa M, Iwama A, Makishima H, Wada T, Kisanuki H, Kaneda R, Ota J, Koinuma K, Ishikawa M, Takada S, Yamashita Y, Oshimi K, Mano H. Retroviral expression screening of oncogenes in natural killer cell leukemia. Leukemia Research. 2005;29:943–949. doi: 10.1016/j.leukres.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Oshimi K, Kawa K, Nakamura S, Suzuki R, Suzumiya J, Yamaguchi M, Kameoka J, Tagawa S, Imamura N, Ohshima K, Kojya S, Iwatsuki K, Tokura Y, Sato E, Sugimori H. NK-cell neoplasms in Japan. Hematology. 2005;10:237–245. doi: 10.1080/10245330400026162. [DOI] [PubMed] [Google Scholar]
- 21.Zambello R, Falco M, Della Chiesa M, Trentin L, Carollo D, Castriconi R, Cannas G, Carlomagno S, Cabrelle A, Lamy T, Agostini C, Moretta A, Semenzato G, Vitale M. Expression and function of KIR and natural cytotoxicity receptors in NK-type lymphoproliferative diseases of granular lymphocytes. Blood. 2003;102:1797–1805. doi: 10.1182/blood-2002-12-3898. [DOI] [PubMed] [Google Scholar]
- 22.Morice WG, Kurtin PJ, Leibson PJ, Tefferi A, Hanson CA. Demonstration of aberrant T-cell and natural killer-cell antigen expression in all cases of granular lymphocytic leukaemia. British Journal of Haematology. 2003;120:1026–1036. doi: 10.1046/j.1365-2141.2003.04201.x. [DOI] [PubMed] [Google Scholar]
- 23.Epling-Burnette PK, Painter JS, Chaurasia P, Bai F, Wei S, Djeu JY, Loughran TP., Jr Dysregulated NK receptor expression in patients with lymphoproliferative disease of granular lymphocytes. Blood. 2004;103:3431–3439. doi: 10.1182/blood-2003-02-0400. [DOI] [PubMed] [Google Scholar]
- 24.Fischer L, Hummel M, Burmeister T, Schwartz S, Thiel E. Skewed expression of natural-killer (NK)-associated antigens on lymphoproliferations of large granular lymphocytes (LGL) Hematological Oncology. 2006;24:78–85. doi: 10.1002/hon.777. [DOI] [PubMed] [Google Scholar]
- 25.Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. The American Journal of Human Genetics. 1992;51:1229–1239. [PMC free article] [PubMed] [Google Scholar]
- 26.Lima M, Almeida J, Montero AG, Teixeira MA, Queirós ML, Santos AH, Balanzategui A, Estevinho A, Algueró MC, Barcena P, Fonseca S, Amorim ML, Cabeda JM, Pinho L, Gonzalez M, San Miguel J, Justiça B, Orfao A. Clinicobiological, immunophenotypic, and molecular characteristics of monoclonal CD56-/+dim chronic natural killer cell large granular lymphocytosis. American Journal of Pathology. 2004;165:1117–1127. doi: 10.1016/s0002-9440(10)63373-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jerez A, Clemente MJ, Makishima H, Koskela H, Leblanc F, Peng Ng K, Olson T, Przychodzen B, Afable M, Gomez-Segui I, Guinta K, Durkin L, Hsi ED, McGraw K, Zhang D, Wlodarski MW, Porkka K, Sekeres MA, List A, Mustjoki S, Loughran TP, Maciejewski JP. STAT3 mutations unify the pathogenesis of chronic lymphoproliferative disorders of NK cells and T-cell large granular lymphocyte leukemia. Blood. 2012;120:3048–3057. doi: 10.1182/blood-2012-06-435297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lima M, Teixeira MA, Queiros ML, Leite M, Santos AH, Justiça B, Orfao A. Immunophenotypic characterization of normal blood CD56+lo versus CD56+hi NK-cell subsets and its impact on the understanding of their tissue distribution and functional properties. Blood Cells Molecules and Diseases. 2001;27:731–743. doi: 10.1006/bcmd.2001.0443. [DOI] [PubMed] [Google Scholar]
- 29.Zambello R, Teramo A, Barilà G, Gattazzo C, Semenzato G. Activating KIRs in Chronic Lymphoproliferative Disorder of NK Cells: Protection from Viruses and Disease Induction? Frontiers in Immunology. 2014;72:1–5. doi: 10.3389/fimmu.2014.00072. 5 Art. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan JK, Quintanilla-Martinez L, Ferry JA, Peh SC. Extranodal NK/T cell-lymphoma, nasal type. In: Swerdlow SH, Harris NL, Jaffe EJ, Pirelli SA, Stein H, Thiele J, Vardiman J, editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4. International Agency for Research of Cancer (IARC); Lyon, France: [Google Scholar]
- 31.Li C, Tian Y, Wang J, Zhu L, Huang L, Wang N, Xu D, Cao Y, Li J, Zhou J. Abnormal immunophenotype provides a key diagnostic marker: a report of 29 cases of de novo aggressive natural killer cell leukemia. Translational Research. 2014;63:565–577. doi: 10.1016/j.trsl.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 32.Ohno Y, Amakawa R, Fukuhara S, Huang CR, Kamesaki H, Amano H, Imanaka T, Takahashi Y, Arita Y, Uchiyama T. Acute transformation of chronic large granular lymphocyte leukemia associated with additional chromosome abnormality. Cancer. 1989;64:63–67. doi: 10.1002/1097-0142(19890701)64:1<63::aid-cncr2820640111>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 33.Matsubara A, Matsumoto M, Takada K, Hato T, Hasegawa H, Tamai T, Yasukawa M, Fujita S. Acute transformation of chronic large granular lymphocyte leukemia into an aggressive form associated with preferential organ involvement. Acta Haematologica. 1994;91:206–210. doi: 10.1159/000204336. [DOI] [PubMed] [Google Scholar]
- 34.Rajala HL, Olson T, Clemente MJ, Lagström S, Ellonen P, Lundan T, Hamm DE, Zaman SA, Lopez Marti JM, Andersson EI, Jerez A, Porkka K, Maciejewski JP, Loughran TP, Mustjoki S. The analysis of clonal diversity and therapy responses using STAT3 mutations as a molecular marker in large granular lymphocytic leukemia. Haematologica. 2015;100:91–99. doi: 10.3324/haematol.2014.113142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macedo A, Orfao A, Vidriales MB, López-Berges MC, Valverde B, González M, Caballero MD, Ramos F, Martínez M, Fernández-Calvo J, San Miguel JF. Characterization of aberrant phenotypes in acute myeloblastic leukemia. Annals of Hematology. 1995;70:89–194. doi: 10.1007/BF01700374. [DOI] [PubMed] [Google Scholar]
- 36.Bahia DM, Yamamoto M, Chauffaille M de L, Kimura EY, Bordin JO, Filgueiras MA, Kerbauy J. Aberrant phenotypes in acute myeloid leukemia: a high frequency and its clinical significance. Haematologica. 2001;86:801–806. [PubMed] [Google Scholar]
- 37.Sanchez ML, Almeida J, Vidriales B, López-Berges MC, García-Marcos MA, Moro MJ, Corrales A, Calmuntia MJ, San Miguel JF, Orfao A. Incidence of phenotypic aberrations in a series of 467 patients with B chronic lymphoproliferative disorders: basis for the design of specific four-color stainings to be used for minimal residual disease investigation. Leukemia. 2002;16:1460–1469. doi: 10.1038/sj.leu.2402584. [DOI] [PubMed] [Google Scholar]
- 38.Rawstron AC, Bennett FL, O'Connor SJ, Kwok M, Fenton JA, Plummer M, de Tute R, Owen RG, Richards SJ, Jack AS, Hillmen P. Monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. New England Journal of Medicine. 2008;359:575–583. doi: 10.1056/NEJMoa075290. [DOI] [PubMed] [Google Scholar]
- 39.Ocqueteau M, Orfao A, Almeida J, Bladé J, González M, García-Sanz R, López-Berges C, Moro MJ, Hernández J, Escribano L, Caballero D, Rozman M, San Miguel JF. Immunophenotypic characterization of plasma cells from monoclonal gammopathy of undetermined significance patients. Implications for the differential diagnosis between MGUS and multiple myeloma. American of Journal Pathology. 1998;152:1655–1665. [PMC free article] [PubMed] [Google Scholar]
- 40.Rabbani GR, Phyliky RL, Tefferi A. A long-term study of patients with chronic natural killer cell lymphocytosis. British Journal of Haematology. 1999;106:960–966. doi: 10.1046/j.1365-2141.1999.01624.x. [DOI] [PubMed] [Google Scholar]
- 41.Paradis V, Laurent A, Flejou JF, Vidaud M, Bedossa P. Evidence for the polyclonal nature of focal nodular hyperplasia of the liver by the study of X-chromosome inactivation. Hepatology. 1997;26:891–895. doi: 10.1002/hep.510260414. [DOI] [PubMed] [Google Scholar]
- 42.Wu Y, Basir Z, Kajdacsy-Balla A, Strawn E, Macias V, Montgomery K, Guo SW. Resolution of clonal origins for endometriotic lesions using laser capture microdissection and the human androgen receptor (HUMARA) assay. Fertility and Sterility. 2003;79(Suppl 1):710–717. doi: 10.1016/s0015-0282(02)04821-5. [DOI] [PubMed] [Google Scholar]
- 43.Kopp P, Jaggi R, Tobler A, Borisch B, Oestreicher M, Sabacan L, Jameson JL, Fey MF. Clonal X-inactivation analysis of human tumours using the human androgen receptor gene (HUMARA) polymorphism. a non-radioactive and semiquantitative strategy applicable to fresh and archival tissue. Molecular and Cellular Probes. 1997;11:217–228. doi: 10.1006/mcpr.1997.0099. [DOI] [PubMed] [Google Scholar]
- 44.Rajala HL, Eldfors S, Kuusanmäki H, van Adrichem AJ, Olson T, Lagström S, Andersson EI, Jerez A, Clemente MJ, Yan Y, Zhang D, Awwad A, Ellonen P, Kallioniemi O, Wennerberg K, Porkka K, Maciejewski JP, Loughran TP, Jr, Heckman C, Mustjoki S. Discovery of somatic STAT5b mutations in large granular lymphocytic leukemia. Blood. 2013;121:4541–4550. doi: 10.1182/blood-2012-12-474577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michaels GS, Carr DB, Askenazi M, Fuhrman S, Wen X, Somogyi R. Cluster analysis and data visualization of large-scale gene expression data. Pacific Symposium on Biocomputing. 1998:42–53. [PubMed] [Google Scholar]
- 46.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pedreira CE, Costa ES, Almeida J, Fernandez C, Quijano S, Flores J, Barrena S, Lecrevisse Q, Van Dongen JJ, Orfao A. A probabilistic approach for the evaluation of minimal residual disease by multiparameter flow cytometry in leukemic B-cell chronic lymphoproliferative disorders. Cytometry A. 2008;73:1141–1150. doi: 10.1002/cyto.a.20638. [DOI] [PubMed] [Google Scholar]
- 50.Pedreira CE, Costa ES, Barrena S, Lecrevisse Q, Almeida J, van Dongen JJ, Orfao A. Generation of flow cytometry data files with a potentially infinite number of dimensions. Cytometry A. 2008;73:834–846. doi: 10.1002/cyto.a.20608. [DOI] [PubMed] [Google Scholar]
- 51.Costa ES, Pedreira CE, Barrena S, Lecrevisse Q, Flores J, Quijano S, Almeida J, Garcia-Macias MC, Bottcher S, Van Dongen JJ, Orfao A. Automated pattern-guided principal component analysis vs expert-based immunophenotypic classification of B-cell chronic lymphoproliferative disorders: a step forward in the standardization of clinical immunophenotyping. Leukemia. 2010;24:1927–1933. doi: 10.1038/leu.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


