Abstract
We present SEEK (http://seek.princeton.edu), a query-based search engine across very large transcriptomic data collections, including thousands of human data sets from almost 50 microarray and next-generation sequencing platforms. SEEK uses a novel query-level cross-validation-based algorithm to automatically prioritize data sets relevant to the query and a robust search approach to identify query-coregulated genes, pathways, and processes. SEEK provides cross-platform handling, multi-gene query search, iterative metadata-based search refinement, and extensive visualization-based analysis options.
The accumulation of human gene expression data in public repositories, such as The Cancer Genome Atlas1 and Gene Expression Omnibus2, offers unprecedented opportunities for data-driven characterization of biological pathways that underlie human diseases. Unsupervised, exploratory approaches are particularly suitable for data-driven discovery and in settings with insufficient or biased training data. However, traditional unsupervised methods, such as clustering and bi-clustering3,4, do not readily extend to compendia containing thousands of data sets from different expression technologies and platforms. Query-based search can enable biomedical researchers to effectively explore and analyze the large collection of expression data sets to identify co-expressed genes in order to explore functional relationships, and make inferences about pathway function with regard to query genes of interest. However, existing search approaches are limited to smaller compendia in model organisms5,6 or, in human, to identifying similar arrays7 or performing gene-level search on a single microarray platform8.
We present SEEK (Search-based Exploration of Expression Kompendia), a robust, cross-platform search system capable of handling very large compendia of human expression data across multiple expression platforms, including microarray and next-generation sequencing (NGS) technologies, and automatically prioritizing data sets relevant to the user’s single or multi-gene query to identify genes co-regulated with the query in informative data sets. SEEK provides biomedical researchers with a systems-level, unbiased exploration of diverse human pathways, tissues, and diseases represented in the entire heterogeneous human compendium. The system integrates thousands of data sets on-the-fly using a novel cross-validation-based data set-weighting algorithm, which robustly identifies relevant data sets and leverages them to retrieve genes co-regulated with the query. It supports sophisticated biological search contexts defined by multi-gene queries and enables cross-platform analysis, with the current compendium including 155,025 experiments spanning 5,210 data sets from 41 different microarray and RNASeq platforms (Fig. 1a and Supplementary Data 1). It has been implemented in a user-friendly, interactive web-interface (http://seek.princeton.edu), which includes expression visualization and interpretation modules (Fig. 1a). This interface facilitates hypothesis generation by providing 1) intuitive expression visualizations of the retrieved co-expressed genes, 2) explorations of individual data sets to establish associations between co-expressed genes and biological variables, and 3) further refinement of the search results such as limiting data sets to a specific tissue (e.g. brain or kidney) or disease (e.g. primary tumor or non-cancerous disease).
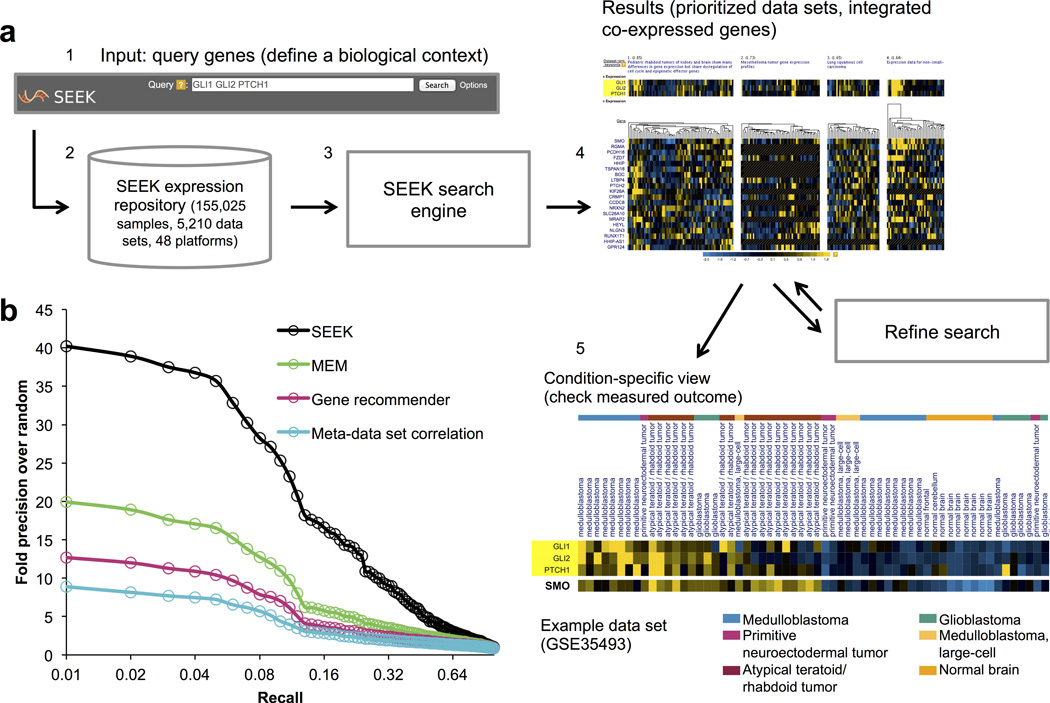
Figure 1. The SEEK system overview and systematic functional evaluation.
(a) The system overview. Users begin by defining a query gene set of interest. SEEK can easily accommodate gene sets as small as 1–2 genes and as large as 100 genes (step 1). The SEEK search engine searches the entire compendium, and returns genes that are co-expressed with the query and the top relevant data sets (steps 2, 3). The web user-interface provides visualizations of gene co-expressions across data sets (step 4), and enables users to iteratively refine their search (Fig. 2) and further analyze the results through condition-specific view (step 5). The latter allows users to check possible associations with the measured outcomes in order to interpret the co-expressed genes (Supplementary Note 3). (b) Gene retrieval evaluations across 995 diverse GO biological process terms, for each of SEEK, MEM, Gene recommender, and meta-data set correlation algorithms (Supplementary Note 1). Queries of diverse sizes (2–20 genes) were selected randomly among each term’s genes to evaluate the precision of retrieving the remaining genes in each term. Individual term performances (Supplementary Data 2) and additional detailed comparative evaluations (Supplementary Figs. 1, 2) are provided.
The search algorithm (Methods) allows multi-gene queries and includes a gene hubbiness9,10 correction procedure, a novel cross-validation data set weighting method, and finally a summarization procedure to calculate the final score for each gene. Prior to applying the search algorithm, the data compendium is pre-processed to make correlation distributions comparable across data sets, and then a hubbiness-correction procedure is applied to remove biases caused by generically well-coexpressed genes not specific to the user’s area of interest that is defined by the query. The data set weighting algorithm then prioritizes relevant data sets based on the query. The intuition of this approach is to up-weight data sets where a subset of the query genes can retrieve the remaining query genes well based on normalized, hubbiness-corrected co-expression in that data set (cross-validation-based weighting). This approach is effective even when not all query genes are co-expressed. Finally, the integrated gene scores are calculated based on the data set weights and genes’ co-expression patterns in each data set to provide a final gene ranking.
SEEK is accurate and robust in a large-scale gene-retrieval assessment across a diverse array of biological contexts. Specifically, we constructed over 129,000 queries spanning 995 human GO biological process gene-sets (by choosing subsets of genes from each process) and evaluated the ability of the algorithm to retrieve the remaining genes in the process (Methods). This set-up was designed to simulate realistic situations where the query genes are biologically coherent, but are not necessarily well co-expressed, and users are interested in identifying genes functionally related to the query (in this case, members of the same biological process). SEEK’s performance is robust across a wide range of pathways (Supplementary Data 2), and it consistently outperforms previous search approaches, including the only query-based human search system MEM8, Gene recommender6 (not available for human as a resource), and meta-data set correlations (Fig. 1b and Supplementary Note 1). Furthermore, our evaluation demonstrates that SEEK’s support for multi-gene queries enhances the algorithm’s ability to effectively weight relevant data sets from the compendium (Supplementary Fig. 1a), and that the algorithm is robust to query noise (Supplementary Fig. 2).
Importantly, our evaluation demonstrates the benefits of robust search of a compendium with thousands of expression data sets, as SEEK’s performance improves with the inclusion of more microarray and RNASeq data sets in the compendium, assessed by sub-sampling our large compendium to create smaller subsets (Supplementary Fig. 3 and Supplementary Data 3). Furthermore, being able to integrate the full scale of the existing human gene expression data allows the approach to support focused queries covering diverse areas of biology (Supplementary Fig. 4), providing strong performance across diverse processes including erythrocyte differentiation (44-fold increase of precision over random (FIOR) at 10% recall) and glutamate signaling (104-fold) (Supplementary Fig. 4). In contrast, using the most relevant single data set for the same query yields weak performance of just 3 and 6 FIOR for the two processes respectively, demonstrating the value of using the entire compendium.
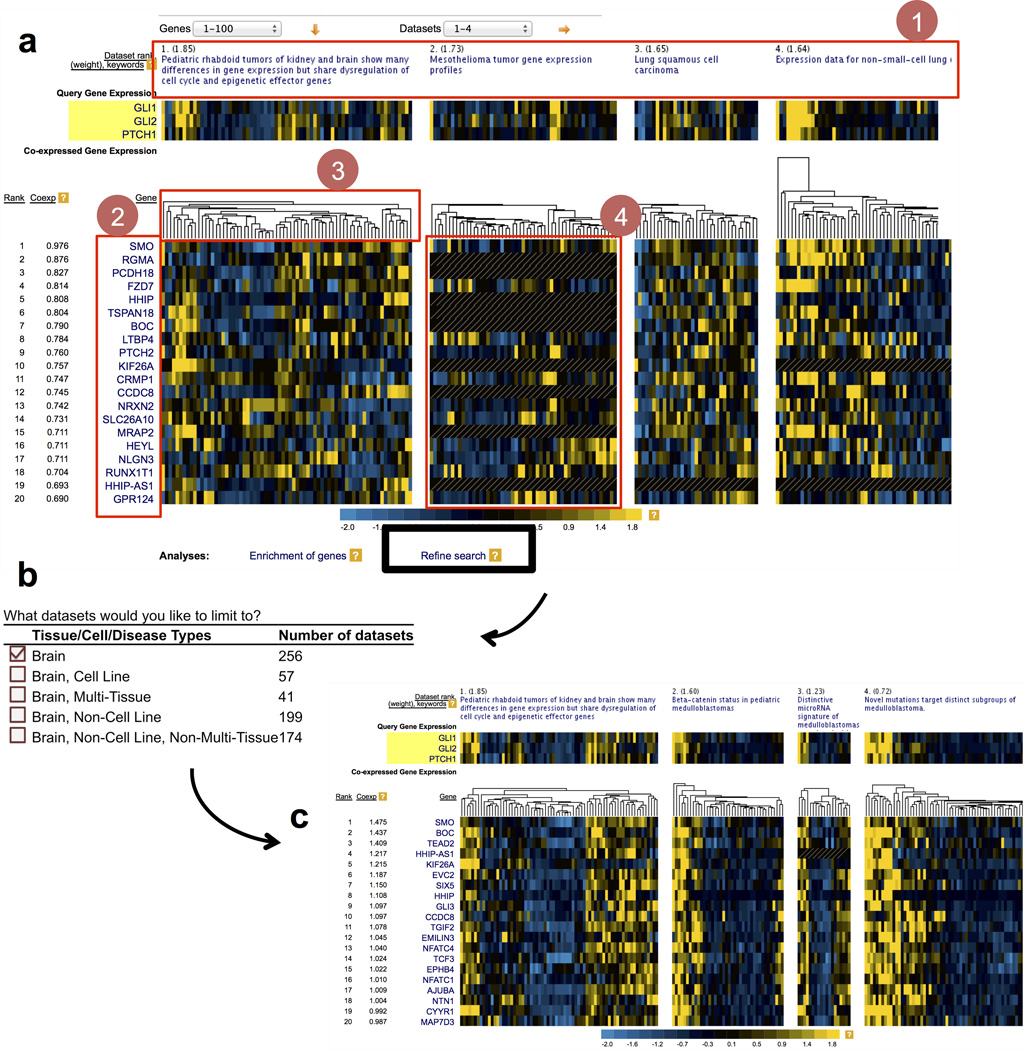
We illustrate the power of SEEK and multi-gene queries by using SEEK to identify genes dysregulated in the Hedgehog (Hh) pathway and the corresponding tissues and disease states where the Hh pathway is hyper-activated. We use Hh genes GLI1, GLI2, and PTCH1 as the query, where transcription factors GLI1 and GLI2 have been suggested as pathway markers of Hh signaling11. By examining this query in the context 5 of a large-compendium of expression data sets (Fig. 2a and Supplementary Fig. 5), we observe a wide prevalence of aberrant Hh-signaling across many diseased tissues (Supplementary Fig. 5). The top ranked data sets have substantially higher weights, indicating the presence of a strong query-related signal in these data (Supplementary Fig. 5) and appear to be more specific to the Hh query than to random queries (Supplementary Fig. 6a). These highly weighted data sets include studies of tumors with previously documented connections to aberrant Hh signaling, such as medulloblastoma, where over-activation of Hh has been documented12,13, human germ cell tumors, where Hh pathway mutations have been linked to aberrant Hh activation in human germ cells14, and malignant rhabdoid tumors15,16, where mutations have been found to lead to Hh signaling activation16. Thus, SEEK correctly identifies data sets relevant to the Hh signaling and helps explore the important role of the Hh pathway in a wide array of cancer types. The data set weighting leads to accurate retrieval of other genes in the Hh pathway, including Hh pathway signaling receptors and their associated genes SMO, PTCH2, HHIP, BOC17, and the Cos2 homolog KIF718 (Fig. 2a and Supplementary Fig. 6b) as well as additional genes associated with Hh dysregulations in cancer (Supplementary Note 2).
Figure 2. Search results for the Hedgehog (Hh) query (GLI1, GLI2, PTCH1) and search refinement.
(a) Data sets prioritized and genes retrieved for the query in the main result page, expression view. The result is retrieved from the Hh query after a global compendium search. The top ranked data sets (1) and the co-expressed gene list (2) are indicated. Conditions in each data set are hierarchically clustered in real-time according to the expression values of the top genes retrieved from the search (3). The expression heat-map of the genes in one of the data sets is shown in (4). (b) Illustration of the search refinement function. Refine Search enables users to narrow the scope of their search based on a powerful and broad set of selection criteria including tissue, cell-type, or disease categories, platforms, or rank of data sets from initial search (Supplementary Note 3). (c) The final results after limiting the search scope to brain data sets. Brain-specific co-expressions are noted in this case with higher co-expression scores to the query and better groupings of conditions than the initial search. SEEK also has alternative view modes such as co-expression view, and condition-specific view (Supplementary Note 3).
The SEEK interface can visualize the abovementioned results including the top-ranked data sets, genes, and their co-expression profiles, using flexible and interactive visualizations (an example of the Hh query is provided in Fig. 2a). The main search result page provides users with the ability to perform extensive follow-up analyses, including functional analysis of results with a co-expression view that summarizes the query and retrieved genes’ co-expression across 50 data sets at a time (Supplementary Note 3). The users can also examine the behavior of any gene in a given data set in detail through a condition-specific view (Fig. 1a), where they can examine associations between co-expressed genes and treatments or outcomes based on data set metadata. An additional post-search analysis, the search refinement function, allows users to iteratively refine their search by limiting the scope of the query search to data sets of a specific disease or tissue of interest, e.g. brain or non-cancer data, if the user wishes to further reveal disease-state or tissue-specific co-expression patterns (Fig. 2b, c). Refine Search currently provides customized search over not only the 2,685 cancer data sets of various tissue origins, but also almost 2,000 non-cancer data sets, including nearly 280 stem cell, over 100 neurodegenerative disease, and 1,400 various immune and other cell type related data sets.
Thus, the proposed search approach is effective in enabling robust and accurate search over very large and diverse human expression compendia by defining specific biological questions based on multi-gene queries. Such compendium-wide search is powerful as it identifies and combines relevant information across many data sets, each representing a mixture of signals from diverse pathways affected by disease, environmental factors, and clinical or experimental treatments. SEEK is based on measuring co-expressions, which minimizes biases toward prior knowledge, and accurately extracts functional information without need to explicitly model outcome variables such as treatment and control experiments (Fig. 1b and prior works5,6,8,19). The use of co-expression thus enables the robust integration of a large number of data sets from diverse tissues, cell lines, and disease origins, generated from diverse platforms, and it can be extended to make functional comparisons across organisms. A key challenge here is that the search results can be polluted by batch effects20, poor quality data sets, or even good quality data sets irrelevant to the user’s query context. Yet the detailed, targeted correction of these issues in each data set or modeling of each outcome variable is impossible in the context of a very large, multi-platform compendium. SEEK’s data set weighting algorithm addresses this challenge, by enabling multi-gene query support for constructing expressive search contexts, and using a discriminative algorithm for identifying which data sets are relevant and accurate in representing query-related biological processes. This algorithm thus automatically down-weights low-quality data sets (e.g. with severe batch effects) (Supplementary Fig. 7 and Supplementary Note 4) and provides accurate retrieval of functionally related genes and data sets (Fig. 1b and Supplementary Figs. 1, 2).
In summary, SEEK enables biomedical researchers to tap the enormous potential of existing expression data in a flexible, integrative, and interactive way. SEEK allows users to prioritize existing data sets. To our knowledge, SEEK is the first search system that addresses the challenges of cross-platform and cross-data set integrated search in human, integrating the large diversity of microarray and RNASeq platforms, thus fully utilizing the expression compendium, and is the largest scale integration of human transcriptomic data to date. We demonstrated that the ability to effectively search such a large and diverse compendium is important, and that search performance benefits from leveraging more diverse data. As such, we plan to regularly update SEEK’s compendium as new microarray and RNASeq data sets become publicly available.
Online methods
Data preparation and correlation normalization
SEEK assembles its human gene expression compendium by obtaining data sets from NCBI’s Gene Expression Omnibus (GEO) database2 and the Cancer Genome Atlas (TCGA)1. The compendium consists of data sets from 41 platforms including 32 platforms from Affymetrix, Agilent, and Illumina, and 9 RNA sequencing platforms (Supplementary Data 1). These platforms were chosen based on the number of available data sets and the availability of raw data to perform consistent processing for each platform. The data sets were processed consistently by applying platform-specific procedures on their raw measurements (Supplementary Note 5 and Supplementary Data 4) to remove the systematic differences among data sets21. The normalized data sets containing gene-level expression values can be accessed through the SEEK website.
The next step of data processing is calculating the Pearson correlations rd(x, y) between all pairs of genes x and y in each data set d. As correlation values arising from different genome-wide distributions are not directly comparable across data sets, a Fisher transform procedure22 is applied to convert the distribution of correlations to a normal-like distribution:
where fd(x, y) is the Fisher-transformed score. Then, the data are translated to z-scores for standardization:
where avg(fd) is the average of fd for all (x, y) pairs, and std(fd) is the standard deviation of fd.
The normalization procedure has been used in previous studies5,23, and has been found successful in transforming most correlation distributions that are generated from different platforms and technologies into a comparable normal distribution with mean 0 and variance 1 (Supplementary Fig. 8). Note that SEEK also works well with other correlation measures, such as Spearman and bicor24 (Supplementary Fig. 9). We found that the normalized Pearson correlation provides performance better or comparable to that of Spearman and bicor in the search setting, likely because the normalization procedure and the SEEK algorithm itself reduce the effects of outliers in search performance (Supplementary Fig. 9).
Search algorithm
The search algorithm takes two inputs 1) a set of query genes Q = {q1, …, qx}, and 2) the set of correlation z-scores containing the query: zd(g, q), for each data set d in the data compendium D, for all genes q in Q and for all genes g in the genome G. The outputs of the algorithm are a prioritized list of data sets and co-expressed genes relevant to Q.
The search algorithm consists of four steps. The first step is to load pre-computed z-scores of Pearson correlations (in the normalization step above) containing the query across D.
The second step is to perform hubbiness correction on each data set d. The correction procedure is motivated by the observation that hubby9,10 or well-connected genes in the co-expression network represent global, well-co-expressed processes25, and can contaminate the search results regardless of query composition due to the effect of unbalanced gene connectivity in a scale-free co-expression network9,10,26–28, and can lead to non-specific results in search or clustering approaches. To avoid the bias created by hubby genes that are not related to the user’s query or pathway of interest, our method corrects each gene g’s correlation to q in each data set d:
| (Eq. 1) |
where z̃ is the hubbiness-corrected z-score. By subtracting g’s average correlation from the correlation of (g, q), we expect the resulting score to emphasize g’s co-expression specifically with the query rather than its general connectivity. The control of co-expression hubbiness enables the detection of specific biological signals in the data that would otherwise be swamped by broad co-expression patterns of the most well-connected genes.
The third step performs cross-validation-based data set weighting. The goal is to rank data sets based on each data set’s relevance to the query5. The result will be the first output of the search system and will also be used to compute the final gene-score vector for the last step. The main idea is to upweight data sets where a subset of the query genes can retrieve the remaining query genes well based on normalized, hubbiness-corrected co-expression in that data set. Thus, it is analogous in spirit to the cross-validation procedures commonly used in machine learning, where a subset of the standard (in this case query) “hides” from the system to assess how well the method can predict these hidden genes.
To describe the weighting method, we first introduce some notations. The data set d is implicit in each formula below and omitted for brevity, thus z̃(g, q) is the corrected z-score for g to a query gene q in Q in data set d. Let Rq = (g(1), g(2), g(3), …, g(r)) be the sequence of genes at rank 1, 2, 3, …, r obtained from ordering genes by decreasing z̃(g, q). That is, Rq satisfies: z̃(g(1), q) ≥ z̃(g(2), q) ≥ z̃(g(3), q)… Let r(t, Rq) be the rank of gene t in the ranking Rq minus 1 (for example, r(g(1), Rq) = 0), and let p < 1be a rate parameter, which we set at 0.99 based on empirical analysis (Supplementary Fig. 10). Then the weight of the data set is
| (Eq. 2) |
The weighting formula performs cross-validations on q in the set Q. The goal is to detect which query genes q can best retrieve the remainder query Q – q; such q’s have a high contribution to w. We shorten r(t, Rq) in Eq. 2 as r(t). The exact form of this expression for weight (i.e. sum of pr(t)) is inspired by rank-biased precision29, and is adapted to our setting to robustly measure the effectiveness of the data set in retrieving Q – q. Here, p < 1 is the rate parameter in rank-biased precision, and is the parameter of geometric distribution, since r(t) assumes discrete values. When it is employed, pr(t) upweights ranks for genes t in the set Q – q that are high in the rank list (i.e., r(t) is small), which intuitively emphasizes those genes in the query that are highly co-expressed to each other. The measure has the desired property of upweighting pairs of query genes that are well correlated while not allowing the correlations between the uninformative, non-coherent part of the query to affect the weight of the data set because the query genes may only be partially co-expressed in a given data set. Compared to previous methods5, our method gains robustness to heterogeneous query signals, because the reward on the highly coherent query genes far outweighs the damaging effect of a few non-coherent query genes, which are poorly ranked to other query genes, have high r(t), and have scores pr(t) tending to zero.
The last step of the algorithm calculates the final integrated gene scores to generate a master ranking of co-expressed genes that is the second output of the system (in addition to data set relevance weighting). We obtain the gene-to-query score matrix MG,D, where the entry Mg,d is the average corrected z-score of gene g to the query in data set d:
With the data set weight vector from the previous step w = [w1, w2, …], a simple formulation of the final gene-score vector F is given by:
Although previous research had some success with this formulation5, our findings show that it works well only in the presence of complete gene information with no missing genes in MG,D. When there are heterogeneous sources of data in the compendium (e.g. different microarray and RNAseq platforms), the confounding factor of missing genes and partial gene rankings must be accounted for. Our approach is to modify the procedure above by employing threshold parameters to exclude a data set from weighting if it does not contain enough query genes, and exclude a gene from the final ranking if it is not assayed by a sufficient number of data sets in the compendium (Supplementary Note 6).
The pseudocode for the entire SEEK search algorithm can be found in Supplementary Note 6. The algorithm is robust to query composition (Supplementary Figs. 1, 2) and data set quality, including automatically down weighting data sets with substantial batch effects (Supplementary Note 4 and Supplementary Fig. 7).
For single-gene queries, the search algorithm performs the same steps above except that in the data set weighting step the algorithm assigns equal weight to all data sets. Thus, for single-gene queries, the search system will treat each data set equally and retrieve genes that are generally correlated with the query in the hubbiness-corrected space. If users wish to perform their single-gene searches in a tissue-specific or disease-specific manner, they can manually define a category of data sets using the extensive “Refine Search” interface on the SEEK website, which will restrict D in the search system input.
Estimating the significance of gene scores
We estimate a P value for each retrieved gene by comparing the integrated score of each gene with scores from a pool of 10,000 randomly generated queries with diverse query sizes varying from 1 to 100 genes. The random pool allows SEEK to estimate the significance of gene-score as well as evaluate the specificity of that gene to the query genes (as opposed to random queries). For a given gene g and its final co-expression score SQ(g) generated from the user’s query Q, the P value of g is estimated as the number of random queries R in which SR(g) > SQ(g) divided by the random pool size.
Algorithm and interface implementations
The SEEK algorithm is implemented in C++ and has been integrated into the open-source C++ Sleipnir library, enabling other computational users to use and expand SEEK without website tie-in30. The backend employs the efficient data structures from the Sleipnir library to facilitate the process of handling large query sets of over 100 genes without memory overflow. SEEK’s jobs are parallelized to make full use of the multiprocessor resources and their processing power. The SEEK web server is constructed with some of the latest web technologies including JQuery and Qtip2 libraries. Dynamic pages are generated with Java servlets running behind the Apache Tomcat server on a Red Hat CentOS Linux operating system. In addition, Ajax technology is deployed to send and retrieve data from the server asynchronously such that users can receive instant feedback on their gene enrichment analysis, expression zoom-in function, and data set selection module without having to leave or refresh the page.
Metadata processing
SEEK categorizes data sets into tissue and disease groups by mining the description, title, and sample level characteristic fields in data sets’ metadata. The text mining procedure utilizes the UMLS MetaThesaurus31 and BRENDA32 controlled vocabularies to extract predefined concept names that are present in the individual fields. To ensure that tissue groups are accurate, we manually reviewed annotations to the frequently appearing terms generated by text mining. Similarly, we formed additional ‘meta’ data set groups, such as cancer and non-cancer groups and the multi-tissue profiling group (Supplementary Data 5), to provide users with the ability to limit their search to such groups under the “Refine Search” feature of the website.
Large-scale functional evaluation setup
We conducted a comprehensive evaluation of SEEK in comparison with existing algorithms Gene recommender, MEM, and meta-data set correlation search (Supplementary Note 1). We tested each system’s ability to retrieve genes from the same biological process given some chosen genes from the process as queries. For the evaluation, we partitioned the genes in each of the 995 GO biological process terms (Supplementary Data 2) into a query building set and a testing set. The query building set consists of a random sample of 25 genes from each term if the term has more than 40 genes, or else it is made of half of the number of genes in the term. Queries were formed by repeatedly sampling genes from the set, so that each query size has 10 different queries of that size represented, and we iteratively generated queries for sizes 2, 3, 4, … up to Q genes, where Q=0.8|query building set|. The testing set consists of the remaining genes in the term (after subtracting the query building set), and is used for evaluating the queries’ retrieval results. A precision recall (PR) curve is computed on a per-query basis, averaged over all queries of a term, and finally averaged over all evaluated terms to derive an overall system performance plot for each method. Fold precision over random is calculated at an indicated recall (10%), and uses a random ranking of genes where genes’ rank positions are shuffled. By selecting genes randomly from each process in building the queries, we mimic the situation in which the query genes are functionally related but not well co-expressed. By keeping the two sets (query building and testing) separate in the evaluation, we can reduce the performance variation between the queries of the same size within a process.
For building gold standard GO gene-set for evaluation, we used gene annotations with experimental evidence codes (IMP, IGI, IPI, IDA, IEP, EXP) as well as TAS (traceable author statements) and NAS (non-traceable author statement). To select the GO slim set (Supplementary Data 3) used for studying the effect of compendium size, we carefully examined the title and description of the GO terms in the context of the GO hierarchy and arrived at a non-redundant subset of GO terms that are both specific enough to be informative, and diverse enough to represent the hierarchy, similar to our approach in33.
To evaluate SEEK’s performance as a function of the query size, we pooled together previously built biological process queries from 995 processes, then binned them by query size (2–20 genes). We examined 3 categories of biological processes based on process size: 20–40 genes, 40–100 genes, and 100–300 genes. Performance refers to the fold precision over random at 10% recall (fold PR10%) in using each query to retrieve remaining genes from its corresponding process.
To evaluate the search system’s robustness to noisy query genes, we selected over 1,800 5-gene and 10-gene queries from 90 KEGG pathways with 50–100 genes per pathway. Each pathway had 10 queries selected of each query size. We established a “no noise” case, where each query was purely made of genes belonging to the same KEGG pathway, and a noisy case, where 1-, 2-, 4- random genes were added to each query. The fraction (fold PR@10% of each noisy query) / (fold PR@10% of the corresponding “no noise” query) was calculated, where fold PR@10% refers to the performance of retrieving KEGG pathway genes using the queries.
Supplementary Material
Acknowledgments
The work was supported in part by the NIH award R01 HG005998 and partially supported by the NSF CAREER award (DBI-0546275) and NIH award R01 GM071966 to O.G.T. The project was also partially supported in part by the NIH awards T32 HG003284 and P50 GM071508. M.C. was supported by NSF awards CCF 1218687 and CCF 1302518. O.G.T. is a Senior Fellow of the Canadian Institute For Advanced Research in the Genetic Networks group.
We thank members of the Troyanskaya lab for comments and inputs about SEEK in the regular lab meetings, and Qibo Zhu for critically reading the manuscript. We thank the volunteers from Princeton and other universities, including the Canadian Institute for Advanced Research Genetic Networks meetings attendees for testing the SEEK web interface and providing valuable feedback.
Footnotes
Authors’ contributions
Q.Z. and O.G.T. wrote the manuscript. Q.Z., O.G.T., K.L., M.C. designed the algorithm. Q.Z. implemented the search backend and frontend, and performed evaluations. A.K.W., D.C.C., A.K., R.Z., M.R.A., C.S.G. contributed ideas, performed analyses, and edited the manuscript. A.T., L.A.B., Q.Z. performed data and metadata processing. V.N.K., O.G.T. contributed ideas in the biological study. O.G.T., K.L., M.C. conceived the study and gave guidance.
References
- 1.Cancer T, Atlas G. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanay A, Sharan R, Kupiec M, Shamir R. Revealing modularity and organization in the yeast molecular network by integrated analysis of highly heterogeneous genomewide data. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2981–2986. doi: 10.1073/pnas.0308661100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hibbs Ma, et al. Exploring the functional landscape of gene expression: directed search of large microarray compendia. Bioinformatics. 2007;23:2692–2699. doi: 10.1093/bioinformatics/btm403. [DOI] [PubMed] [Google Scholar]
- 6.Owen AB, Stuart J, Mach K, Villeneuve AM, Kim S. A Gene Recommender Algorithm to Identify Coexpressed Genes in C. elegans. Genome Res. 2003;13:1828–1837. doi: 10.1101/gr.1125403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zinman GE, Naiman S, Kanfi Y, Cohen H, Bar-Joseph Z. ExpressionBlast: mining large, unstructured expression databases. Nat. Methods. 2013;10:925–926. doi: 10.1038/nmeth.2630. [DOI] [PubMed] [Google Scholar]
- 8.Adler P, et al. Mining for coexpression across hundreds of datasets using novel rank aggregation and visualization methods. Genome Biol. 2009;10:R139. doi: 10.1186/gb-2009-10-12-r139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barabási A-L, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 10.Han J-DJ, et al. Evidence for dynamically organized modularity in the yeast protein–protein interaction network. Nature. 2004;430 doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 11.Kimura H, Stephen D, Joyner A, Curran T. Gli1 is important for medulloblastoma formation in Ptc1+/− mice. Oncogene. 2005;24:4026–4036. doi: 10.1038/sj.onc.1208567. [DOI] [PubMed] [Google Scholar]
- 12.Oliver TG, et al. Transcriptional profiling of the Sonic hedgehog response: a critical role for N-myc in proliferation of neuronal precursors. Proc. Natl. Acad. Sci. U. S. A. 2003;100:7331–7336. doi: 10.1073/pnas.0832317100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berman D, Karhadkar S, Hallahan A. Medulloblastoma growth inhibition by hedgehog pathway blockade. Science. 2002;297:1559–1561. doi: 10.1126/science.1073733. [DOI] [PubMed] [Google Scholar]
- 14.Carpenter D, et al. Characterization of two patched receptors for the vertebrate. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13630–13634. doi: 10.1073/pnas.95.23.13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oue T, Yoneda A, Uehara S. Increased expression of the hedgehog signaling pathway in pediatric solid malignancies. J. Pediatr. Surg. 2010;45:387–392. doi: 10.1016/j.jpedsurg.2009.10.081. [DOI] [PubMed] [Google Scholar]
- 16.Jagani Z, Mora-Blanco E, Sansam C. Loss of the tumor suppressor Snf5 leads to aberrant activation of the Hedgehog-Gli pathway. Nat. Med. 2010;16:1429–1433. doi: 10.1038/nm.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen M. The hedgehog signaling network. Am. J. Med. Genet. Part A. 2003;123A:5–28. doi: 10.1002/ajmg.a.20495. [DOI] [PubMed] [Google Scholar]
- 18.Cheung HO-L, et al. The kinesin protein Kif7 is a critical regulator of Gli transcription factors in mammalian hedgehog signaling. Sci. Signal. 2009;2:ra29. doi: 10.1126/scisignal.2000405. [DOI] [PubMed] [Google Scholar]
- 19.Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P. Coexpression analysis of human genes across many microarray data sets. Genome Res. 2004;14:1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leek J, Scharpf R, Bravo H. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 21.Ramasamy A, Mondry A, Holmes CC, Altman DG. Key issues in conducting a meta-analysis of gene expression microarray datasets. PLoS Med. 2008;5:e184. doi: 10.1371/journal.pmed.0050184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fisher R. Frequency distribution of the values of the correlation coefficient in samples from an indefinitely large population. Biometrika. 1915;10:507–521. [Google Scholar]
- 23.Huttenhower C, et al. Exploring the human genome with functional maps. Genome Res. 2009;19:1093–1106. doi: 10.1101/gr.082214.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song L, Langfelder P, Horvath S. Comparison of co-expression measures: mutual information, correlation, and model based indices. BMC Bioinformatics. 2012;13:328. doi: 10.1186/1471-2105-13-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 26.Horvath S, Dong J. Geometric interpretation of gene coexpression network analysis. PLoS Comput. Biol. 2008;4:e1000117. doi: 10.1371/journal.pcbi.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruan J, Dean AK, Zhang W. A general co-expression network-based approach to gene expression analysis: comparison and applications. BMC Syst. Biol. 2010;4:8. doi: 10.1186/1752-0509-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xulvi-Brunet R, Li H. Co-expression networks: graph properties and topological comparisons. Bioinformatics. 2010;26:205–214. doi: 10.1093/bioinformatics/btp632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moffat A, Zobel J. Rank-biased precision for measurement of retrieval effectiveness. ACM Trans. Inf. Syst. 2008;27:1–27. [Google Scholar]
- 30.Huttenhower C, Schroeder M, Chikina MD, Troyanskaya OG. The Sleipnir library for computational functional genomics. Bioinformatics. 2008;24:1559–1561. doi: 10.1093/bioinformatics/btn237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gremse M, Chang A, Schomburg I. The BRENDA Tissue Ontology (BTO): the first all-integrating ontology of all organisms for enzyme sources. Nucleic Acids Res. 2011;39:D507–D513. doi: 10.1093/nar/gkq968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myers C, Barrett D, Hibbs M. Finding function: evaluation methods for functional genomic data. BMC Genomics. 2006;7:187. doi: 10.1186/1471-2164-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.