Abstract
Staphylococcus aureus is known as a frequent colonizer of the skin and mucosa. Among bacterial factors involved in colonization are adhesins such as the microbial surface components recognizing adhesive matrix molecules (MSCRAMMs). Serine aspartate repeat containing protein D (SdrD) is involved in adhesion to human squamous cells isolated from the nose. Here, we identify Desmoglein 1 (Dsg1) as a novel interaction partner for SdrD. Genetic deletion of sdrD in S. aureus NCTC8325-4 through allelic replacement resulted in decreased bacterial adherence to Dsg1- expressing HaCaT cells in vitro. Complementary gain-of-function was demonstrated by heterologous expression of SdrD in Lactococcus lactis, which increased adherence to HaCaT cells. Also ectopic expression of Dsg1 in HEK293 cells resulted in increased adherence of S. aureus NCTC8325-4 in vitro. Increased adherence of NCTC8325-4, compared to NCTC8325-4ΔsdrD, to the recombinant immobilized Dsg1 demonstrated direct interaction between SdrD and Dsg1. Specificity of SdrD interaction with Dsg1 was further verified using flow cytometry and confirmed binding of recombinant SdrD to HaCaT cells expressing Dsg1 on their surface. These data demonstrate that Dsg1 is a host ligand for SdrD.
Staphylococcus aureus is a human commensal that frequently colonizes the human skin and mucosa, either for long or short periods throughout life1,2. It is also an important cause of several life-threatening infections. The ability of S. aureus to efficiently colonize the host epithelium, invade tissues and survive within host cells is regulated through numerous adhesive and invasive factors3,4.
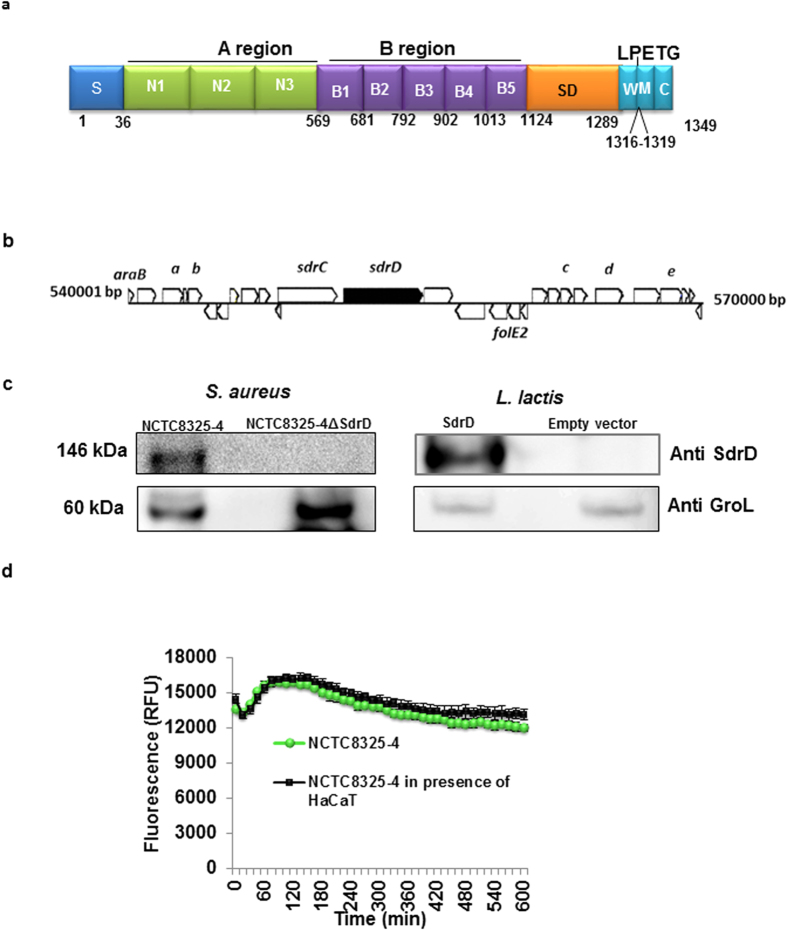
S. aureus expresses a panel of cell-wall anchored adhesins including the microbial surface components recognizing adhesive matrix molecule (MSCRAMM) families, which target extracellular matrix proteins and other molecules on host cells5,6,7,8. The Clumping factor (Clf) and Serine aspartate repeat containing protein (Sdr) families of MSCRAMMs share structural features. They contain N-terminal signal peptide followed by an A region (divided into distinct sub-domains called N1, N2 and N3), two to five B repeats, an R domain (Ser-Asp repeats), a LPXTG cell wall-anchoring motif, a hydrophobic membrane spanning region, and a cytoplasmic C-terminal end8,9 (Fig. 1a).
Figure 1. sdrD gene localization and expression in S. aureus NCTC8325-4.
(a) Schematic representation of SdrD domain structure in S. aureus NCTC8325-4 based on UniProtKB. S, signal sequence; A region composed of N1, N2 and N3; B repeats composed of B1 to B5; SD, serine-aspartate acid repeat region; W, wall-spanning fragment; LPETG, cell wall anchoring motif; M, transmembrane domain; C, cytoplasmic domain. (b) sdrC and sdrD is located between ORFs encoding hypothetical proteins according to annotation is from KEGG Genome map. Gene and protein name based on UniProtKB: araB, ribonuclokinase; a: SAOUHSC_00536, Branched-chain-amino-acid aminotransferase; b: SAOUHSC_00538, Haloacid dehalogenase-like hydrolase; sdrC, serine-aspartate repeat-containing protein C; sdrD, serine-aspartate repeat-containing protein D; folE2, GTP cyclohydrolase FolE2; c: SAOUHSC_00554, SIS domain protein; d: SAOUHSC_00556, Proline/betaine transporter; e: SAOUHSC_00558, Acetyl-CoA acetyltransferase (c) Immunoblot using SdrD A-region and GroL antibodies on cell lysate S. aureus NCTC8325-4 or its isogenic mutant NCTC8325-4ΔsdrD and L.lactis with pMG36e-SdrD (SdrD) or pMG36e (empty vector). (d) sdrD promoter activity in DMEM supplemented with FBS without agitation in the absence ( ) or presence (■) of HaCaT cells using S. aureus NCTC8325-4 harbouring sdrD-GFP reporter construct. Data expressed as mean ± standard deviation (SD) of an individual experiment.
) or presence (■) of HaCaT cells using S. aureus NCTC8325-4 harbouring sdrD-GFP reporter construct. Data expressed as mean ± standard deviation (SD) of an individual experiment.
Bacteria can target intercellular junctions on host cells to promote adhesion and/or internalization during colonization and infection10,11,12. Desmosomes are the main type of adhesive disk shaped intercellular junctions, abundant in tissues exposed to mechanical stress, such as skin and heart13. The desmosomal cadherins, desmogleins (Dsgs) consist of extracellular, transmembrane and cytoplasmic domains14. The extracellular domain is calcium-dependent and composed of 4 cadherin repeats (EC 1–4), and the extracellular anchor domain (EA), while the intracellular tail binds to adaptors connected to the intermediate filaments of host cytoskeleton15. The expression pattern of desmoglein isoforms 1–4 in human epidermis is highly variable16,17,18. These expression patterns impact on the barrier function19 and also the differentiation of the epithelial tissues20.
Dsg1 is expressed throughout all the nucleated cell layers of human epidermis18 and recent evidence suggests that either Dsg1 or Dsg3 can compensate for the adhesive functions of each other in different epithelia tissues21,22. Dsg1 has been shown to mediate keratinocyte differentiation in the epidermis23. In addition, Dsg1 plays a key role in the pathogenesis of three different dermatological conditions including; pemphigus foliaceus, staphylococcal scalded skin syndrome and striate palmoplantar keratoderma (reviewed in24). It has also been demonstrated that mutations in the dsg1 gene results in severe dermatitis, multiple allergies and metabolic wasting25.
The role of MSCRAMMs in S. aureus colonization and infection has been demonstrated previously (reviewed in8). Several of the MSCRAMMs are involved in attachment of S. aureus to squamous epithelial cells26,27,28 and keratinocytes29 in vitro as well as promoting nasal colonization in mice27,30 and humans31. SdrD specifically promotes adherence of bacteria to desquamated nasal epithelial cells, harvested from human donors26. Thus, we hypothesize that SdrD may promote colonization through interaction with particular host molecules. The aim of this study was to identify a host ligand for S. aureus SdrD and to investigate the potential effect of this interaction on S. aureus colonization of host cells.
Results
sdrD gene localization and expression in S. aureus NCTC8325-4
The sdrD gene encodes LPXTG-anchored protein of 1349 amino acids that is composed of an anterior A region (residues 36–568), a medial B region (residues 569–1123) and a posterior SD repeat R region (residues 1124–1289) (Fig. 1a). The sdr locus of S. aureus consists of sdrC, sdrD and/or sdrE, but may not all be present in the same strain6,7. From the annotation, the sdr locus of NCTC8325-4 contains sdrC and sdrD (Fig. 1b).
By allelic replacement, a NCTC8325-4ΔsdrD mutant was created. Bacterial growth was not significantly affected by deletion of sdrD (P < 0.05). Furthermore, both strains were haemolytic as visible on the blood agar plates (see supplementary Fig. S1). A heterologous expression system was constructed by expressing SdrD in the non-invasive bacterium L.lactis. Immunoblot analysis showed that the expression of SdrD was detected when sdrD gene was present in S. aureus and L.lactis (Fig. 1c, upper lane). The presence of bacterial lysate was proven by immunoblot of GroL (Fig. 1c, lower lane).
The expression pattern of sdrD in NCTC8325-4 was assayed in eukaryotic cell culture medium (DMEM supplemented with FBS) in the absence or presence of HaCaT cells by use of a sdrD-specific GFP reporter construct. Immediately after inoculation, the expression of GFP showed a slight decrease, followed by an increase (Fig. 1d). The expression of the reporter was similar in the presence or absence HaCaT cells. Expression of SdrD in presence of HaCaT was detected in NCTC8325-4, but not in its isogenic mutant as confirmed by immunoblot (data not shown).
The presence of SdrD promotes adhesion of S. aureus in HaCaT cells
To investigate the contribution of SdrD in adherence to human keratinocytes, S. aureus NCTC8325-4 and the isogenic mutant NCTC8325-4ΔsdrD were incubated with confluent layers of HaCaT cells. Unbound bacteria were removed by washing and adherent bacteria were quantified by plating serial dilutions. The presence of SdrD promoted better adherence of NCTC8325-4 to HaCaT cells, as the isogenic mutant showed two-fold reduction in adherence (P < 0.05) (Fig. 2a). S. aureus internalization into HaCaT cells has been demonstrated previously32,33. However, the internalized bacteria did not exceed 0.8% of the adhered bacteria (results not shown).
Figure 2. SdrD mediates adherence of S. aureus and L. lactis to HaCaT cells.
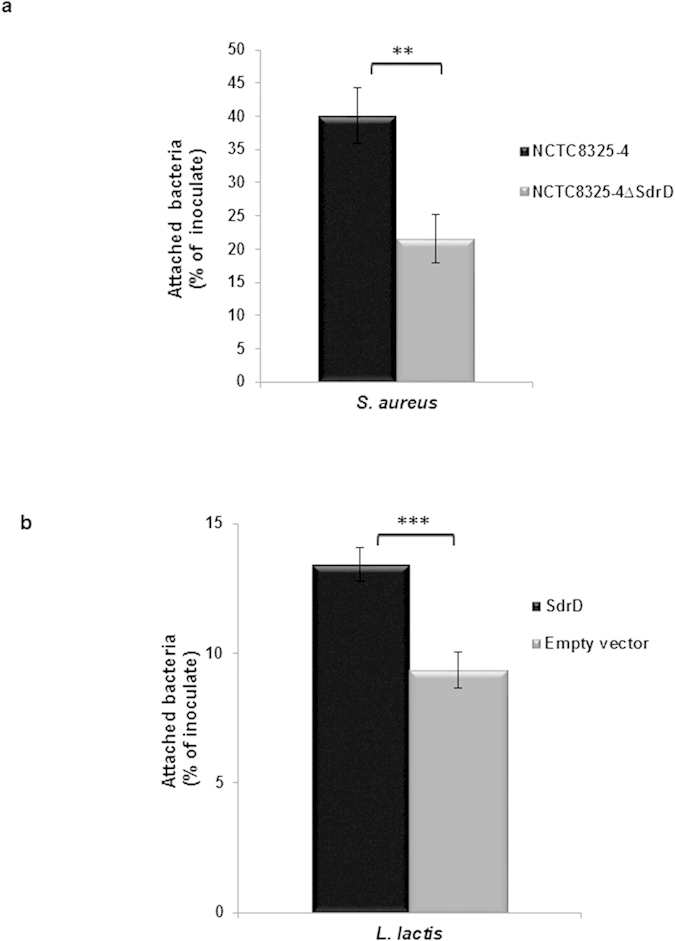
Adherence of (a) S. aureus NCTC8325-4 and its isogenic mutant NCTC8325-4ΔsdrD and (b) L. lactis with pMG36e-sdrD (SdrD) or pMG36e (empty vector) to HaCaT cells. The number of inoculated bacteria was arbitrarily set as 100% and the number of attached bacteria represented as the mean percentage of inoculate. Data represent means ± SEM of 4 independent experiments. Statistical analysis was performed by Student’s t-test. Significant differences are indicated by two (P < 0.01), or three (P < 0.001) asterisks (*).
S. aureus expresses MSCRAMMs which might cause some functional redundancy in adherence assays8. Therefore, sdrD was cloned into a lactococcal vector (pMG36e) allowing heterologous expression in L. lactis. In adhesion assay performed using L. lactis, a significant increase was observed when L. lactis expressed SdrD compared to L. lactis carrying only empty vector (Fig. 2b). Through this approach, the redundancy challenges associated with S. aureus adhesins is avoided, and the effect of SdrD on bacterial adhesion could be observed. These results confirm that SdrD contributes to the S. aureus adhesion to HaCaT cells.
The presence of SdrD promotes S. aureus attachment to human desmoglein 1
To identify the host interaction partner within human skin, a yeast-two hybrid screen was performed using GAL4-SdrD A-region as bait in human reconstituted skin library. By this approach, desmoglein 1 (Dsg1) was identified as a putative interaction partner.
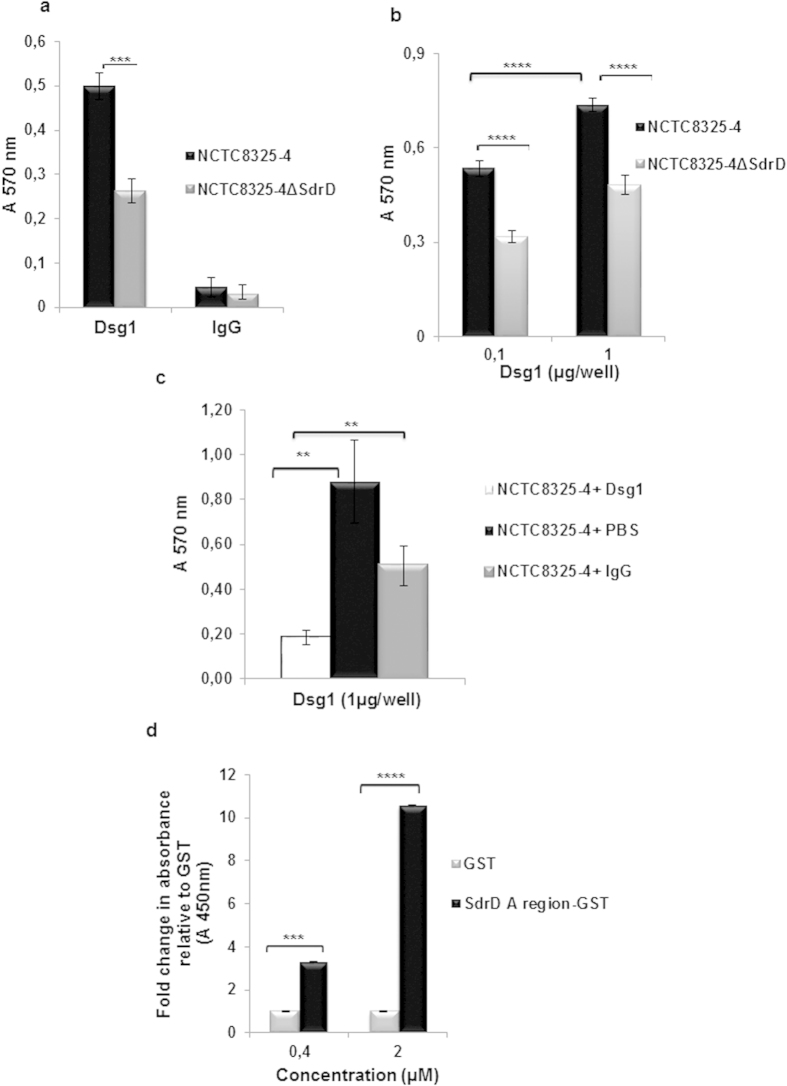
To assess Dsg1 as binding partner, we first studied the ability of SdrD to facilitate the adherence of S. aureus to Dsg1-coated microtiter plates. Wells were coated with Dsg1 Fc-chimera, IgG1 Fc or buffer only and the adherence of S. aureus NCTC8325-4 and its isogenic SdrD mutant were compared. S. aureus NCTC8325-4 adhered significantly better to immobilized Dsg1 than NCTC8325-4ΔsdrD (p < 0.05) (Fig. 3a). Similarly, when the amount of Dsg1 was raised from 0.1 to 1 μg/well, the bacterial adhesion was increased (Fig. 3b).
Figure 3. Dsg1 is a host ligand for SdrD.
(a) S. aureus NCTC8325-4 and the isogenic mutant NCTC8325-4ΔsdrD were added to wells coated with Dsg1-Fc chimera or control IgG1 Fc. Bacterial adherence was measured by staining with crystal violet and measurement of the absorbance at 570 nm. (b) Binding of S. aureus to Dsg1 is concentration dependent. Different concentrations of Dsg1 were coated on ELISA plates and bacterial adherence was evaluated as described in a. (c) S. aureus NCTC8325-4 was pre-incubated with IgG1 Fc or Dsg1-Fc chimera and added to Dsg1-coated ELISA wells (1 μg/well). Bacterial adherence was evaluated as described in legend a. (d) Wells were coated with GST-SdrD-A region and GST before adding of Dsg1. Adherence of Dsg1 was determined using Dsg1-specific antibodies, followed by HRP-conjugated secondary antibody. The measurement of absorbance at 450 nm was carried out using an ELISA reader (VERSAmax, USA). The absorbance value for GST was arbitrarily 1 and the absorbance in presence of SdrD-A region is represented as fold change. Data represent means ± SEM of at least 3 independent experiments. Statistical analysis was performed by Student’s t-test. Significant differences are indicated by ns (no statistical significance), two (P < 0.01), three (P < 0.001) or four (P < 0.0001) asterisks (*).
If bacteria expressing SdrD bind Dsg1 in solution, one would expect that the bacterium is coated with the protein and bind less efficiently to the immobilized Dsg1. Competition-based studies were performed where S. aureus NCTC8325-4 was pre-incubated with Dsg1, PBS or IgG1 Fc (the two latter as negative controls) before being allowed to adhere to immobilized Dsg1. As shown in Fig. 3c, pre-incubation of S. aureus NCTC8325-4 with Dsg1 considerably reduced its adhesion ability to immobilized Dsg1. This reduction in adherence was not seen or was much less when cells were pre-incubated with PBS or IgG1 Fc, respectively, thus showing that the binding between SdrD and Dsg1 is specific.
Finally, the interaction was evaluated using purified proteins. Thus, the microtiter plate was coated with two concentrations of immobilized GST-SdrD (A-region), prior to addition of recombinant Dsg1-protein. The binding of Dsg1 was thereafter evaluated using Dsg1-specific antibodies. As seen in Fig. 3d, recombinant Dsg-1 bound in a dose-dependent manner to immobilized SdrD (A region) protein. There was no significant binding between GST and Dsg1. All these results suggest a direct interaction between SdrD and Dsg1 in vitro.
Recombinant SdrD binds to HaCaT cells expressing Dsg1 and ectopic Dsg1 expression promotes SdrD-mediated S. aureus adhesion to HEK293 cells
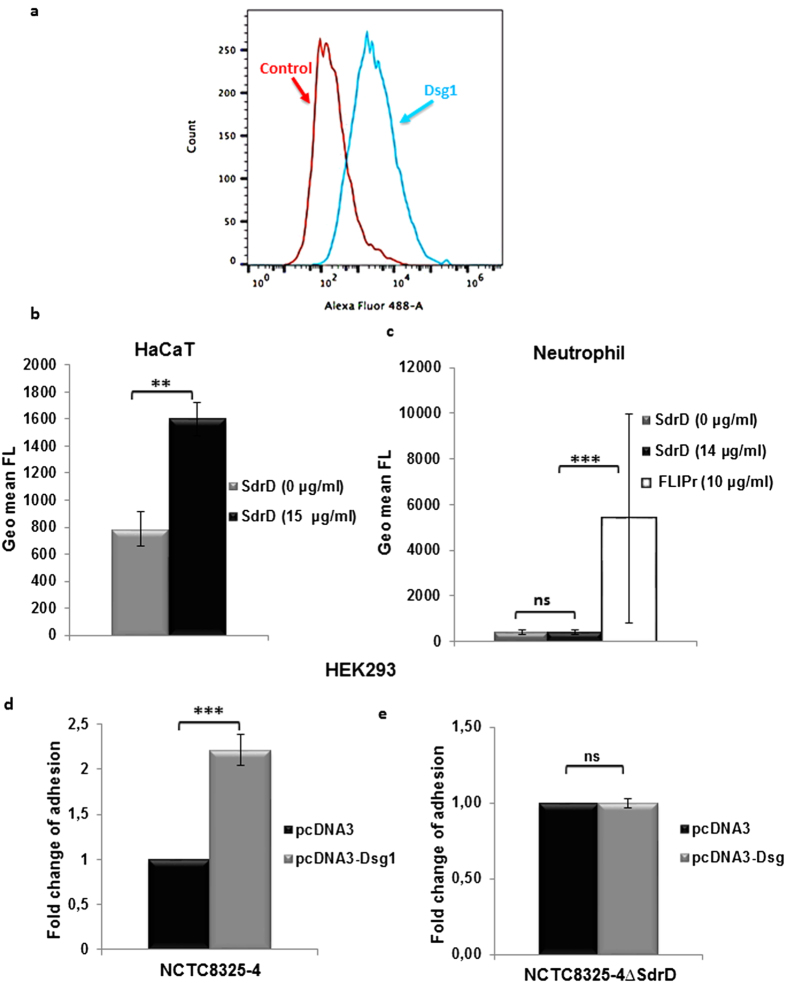
HaCaT cells express Dsg1, which is predominantly located in the upper layer of epidermis34. Expression of Dsg1 on the surface was confirmed by flow cytometry using Dsg1-specific antibody followed by detection with an Alexa-488 conjugated secondary antibody (Fig. 4a). To verify specificity of SdrD for Dsg1, recombinant full-length SdrD (see supplementary Fig. S2) were added to HaCaT cells and neutrophils. As shown in Fig. 4b, full length SdrD significantly bound to HaCaT cells expressing Dsg1 on their surface but not to neutrophils (Fig. 4c), which do not express Dsg123,35. On the other hand, S. aureus formyl peptide receptor inhibitor (FLIPr) bound neutrophils as expected36. Thus, SdrD specifically targets host cells expressing Dsg1.
Figure 4. SdrD specifically binds and promotes S. aureus adherence to the host cells expressing Dsg1.
(a) Dsg1 is expressed surface of HaCaT cells as measured by flow cytometry (blue histogram). Control antibody binding to cells in red histogram. (b) Binding of his-tagged SdrD (full length) to HaCaT cells as detected with anti-his-FITC by flow cytometry. The results are presented as geometric mean of the fluorescence intensity (FL). (c) Binding of his-tagged SdrD (full length) or FLIPr (positive control) to neutrophils measured as indicated in the figure legend b. (d) Ectopic expression of Dsg1 in HEK293 promotes adherence of NCTC8325-4 but (e) not NCTC8325-4ΔsdrD. The cells transfected with empty vector were arbitrarily set as 1, and the fold change of adherence in pcDNA3-Dsg1 transfected is represented as fold change. Data represent means ± SEM of at least 3 independent experiments. Statistical analysis was performed by Student’s t-test. Significant differences are indicated by ns (no statistical significance), two (P < 0.01), or three (P < 0.001) asterisks (*).
Next, we assessed whether ectopic Dsg1 expression can promote S. aureus NCTC8325-4 adhesion to host cells. HEK293 cells were transfected with pcDNA3-Dsg1 (encoding Dsg1) or the control plasmid pcDNA3 and their binding to NCTC8325-4 or the isogenic NCTC8325-4ΔsdrD (negative control) was assessed. The number of adhered S. aureus NCTC8325-4 was significantly increased in HEK293 cells transfected with pcDNA3-Dsg1 compared to HEK293 cells with the control plasmid pcDNA3 (Fig. 4d). Such an increase was not seen when assayed with the isogenic mutant (Fig. 4e). A direct comparison, using a five-fold increased MOI, confirmed that Dsg1 contributes in SdrD-mediated S. aureus attachment to the host cells (Supplementary Fig. S3).
Taken together, these results confirm that SdrD contributes significantly to adherence of S. aureus to host cells using Dsg1 as its interaction partner.
Discussion
Host adhesion molecules such as cadherins, integrins or immunoglobulin-related cell adhesion molecules (ICAMs) are connected to the host cytoskeleton, and are often used as targets for bacterial adhesion as well as internalization12. S. aureus binds specifically integrins through a fibronectin-bridge, while Haemophilus influenzae is an example of a bacterium that binds ICAMs. The EC1 domain of E-cadherin is a ligand for Listeria InlA12, while desmosomal cadherin Dsg2 is the receptor for Adenovirus37. In this study, we demonstrate for the first time that S. aureus SdrD interacts directly with Dsg1. The interaction was confirmed using several approaches including yeast two hybrid (Hybrigenics) and ELISA based assays using purified recombinant SdrD and Dsg1 proteins. Additionally, disruption of sdrD in S. aureus NCTC8325-4 attenuated adhesion to immobilized Dsg1 and to human keratinocytes. However, there is a high functional redundancy among MSCRAMMs8. Since interaction with keratinocytes is not completely lost by deletion of SdrD, our data suggest that Dsg1 may also interact with other S. aureus adhesion molecules (Fig. 3a,b). Complementary, ectopic surface expression of Dsg1 in HEK293 cells significantly increased adhesion of SdrD-expressing S. aureus and purified full-length SdrD protein binds to HaCaT cells that expressed Dsg1. All these results provide evidence for a direct interaction between SdrD and Dsg1, which improves bacterial adherence to keratinocytes.
Colonization is a well-known risk factor for development of staphylococcal infections, and adherence of S. aureus to the host cells promotes colonization38. The molecular aspects of colonization include interaction between bacterial adhesins and host molecules. Clumping factor B has recently been found to support colonization through its binding to loricrin39. Loricrin is highly expressed in stratum corneum40, and the interaction between loricrin and ClfB may be especially important in the initial phase of colonization. ClfB has previously also been found to interact with keratin 8 and 10 (reviewed in8), and cytokeratin 10 is expressed in the suprabasal layer of epidermis41. Surprisingly, bacteria are present in all layers of human epidermis and even sub-epidermal compartment of human skin42, Here we have established that the SdrD–Dsg1 interaction promotes adhesion to keratinocytes. The desmosomal structure of which Dsg1 is part of, is connected to the keratins18, and it is tempting to speculate that both SdrD and ClfB may be of importance later during the colonization throught their interaction with Dsg1 and cytokeratin 10, respectively. However, although SdrD is shown to contribute to bacterial adhesion to human squamous cells isolated from the nose26, and its expression increases during nasal colonization43, it is not known whether SdrD-Dsg1 interaction promotes nasal colonization.
Keratinocytes constitute a large part of the epidermis44, and Dsg1 contributes to maintaining the structural epidermal integrity in stratified epidermal cells45. Dsg1 is targeted by S. aureus exfoliative toxin, resulting in loss of cell-cell adhesion in the epidermis46. Moreover, in the immune disease pemphigus, the tissue integrity is disrupted by autoantibodies against Dsg1/347. One hypothesis is that SdrD-Dsg1 interaction may also affect the structural integrity of the epidermis. However, subsequent molecular mechanism of this interaction remains to be investigated.
In addition, Dsg1 influences keratinocyte differentiation via its influence on the Ras intracellular signalling25. Thus, another hypothesis is that SdrD-Dsg1 interaction influences the ability of keratinocytes to effectively differentiate into respective cellular structures within the epidermal-stratified structure. Dsg1 also influences the Rho signalling pathway48, an essential pathway affecting cytoskeleton and overall migration of cells49. This scenario could possibly influence the ability of S. aureus to invade deep tissues and subsequently enter into the bloodstream. SdrD expression is regulated in the presence of whole human blood50. Thus this interaction might be essential for the invasion and subsequent migration of the bacterium from the site of colonization.
In conclusion, we have identified Dsg1 as a novel host ligand for S. aureus SdrD and demonstrated the importance of this interaction in the promotion of S.aureus adhesion to host cells. This interaction may initiate further biological processes, which could influence the pathogenesis and progression of S. aureus infections. A deeper understanding of the molecular mechanism of this interaction may aid in the development of anti-virulence strategies/therapeutics to combat S. aureus colonization of the epidermis.
Materials and Methods
Bacterial strains, mammalian cell lines and growth conditions
Staphylococcus aureus subsp. aureus NCTC8325-4 was used for allelic replacement and cloning of srdD for expression constructs. Lactococcus lactis MG1363 was used for heterologous expression assays51. S. aureus was cultured in tryptic soy broth (TSB) at 37 °C with shaking at 220 rpm. L. lactis MG1363 was cultured in SMG17 (M17 broth supplemented with 0.5M sucrose (Sigma Aldrich, Germany) and 0.5% glucose (Sigma Aldrich, Germany) at 30 °C, without shaking. For adherence assays, overnight bacterial cultures were diluted 1:100 in appropriate culture medium and grown to OD600nm = 0.7–0.8 for S. aureus, and to OD600nm = 0.5–0.6 for L. lactis, before the bacteria were pelleted, washed twice in 1 × PBS (37 °C) and in DMEM media supplemented with 10% FBS.
HaCaT cells, a human keratinocyte cell line52 and HEK293 cells, a human embryonic kidney cell line, were purchased from PromoCell (Germany) and European collection of Cell Cultures (Porton Down, UK), respectively. The cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma Aldrich, Germany), supplemented with 10% (v/v) fetal bovine serum (FBS) (Invitrogen Life Technologies, USA), penicillin (100 units/ml), and streptomycin 100 μg/ml (Sigma Aldrich, Germany) in a CO2 incubator (5% CO2) at 37 °C.
Neutrophils were freshly isolated from heparinized venous blood of healthy volunteers (see section for ethical approval) using Histopaque (Sigma Aldrich, Germany)-Ficoll-paque (GE Healthcare, Sweden) gradient centrifugation. Neutrophils were kept in RPMI 1640 (Gibco, Life Technologies, UK), supplemented with 0.05% human serum albumin (HSA) (Sanquin, Amsterdam, The Netherlands).
Genetic manipulation of S. aureus
Markerless precise allelic replacement of sdrD was performed in S. aureus NCTC8325-4 using previously described methods53 with minor modifications54. Briefly, DNA fragments 1050 bp upstream and 1025 bp downstream of sdrD were amplified using primers Up For sdrD + attB1, Up Rev sdrD, Down For sdrD and Down Rev sdrD + attB2 (Table 1). The upstream and downstream PCR products were fused by PCR using Up For sdrD + attB1 and Down Rev sdrD + attB2 primers. The fusion construct was subcloned into a temperature sensitive plasmid, pKOR155, using Gateway BP clonase II enzyme mix (Invitrogen, USA) and transformed into S. aureus NCTC8325-4 through electroporation. A temperature shifting and antisense counter selection resulted in precise allelic replacement of sdrD. Deletion of sdrD was confirmed by PCR using primers sdrD KO confirm For + sdrD KO confirm Rev and sdrD Int For + sdrD Int Rev (Table 1), and by Western blot.
Table 1. Primers used in this study.
| Primers | Sequence (5′- ′3) → | Use | Origin |
|---|---|---|---|
| Up For sdrD + attB1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTGACTCGGATAGCGACTCAGAC | Generating fusion construct | This work |
| Up Rev sdrD | CAAGGACCTGGGTCATATTGTATAGATTACTCCTAAT TCATC | Generating and sequencing fusion construct | This work |
| Down For sdrD | GATGAATTAGGAGTAATCTATACAATATG ACCCAGGTCCTT G | Generating and sequencing fusion construct | This work |
| Down Rev sdrD + attB2 | GGGGACCACTTTGTACAAGAAAGCTGGGTCGTAGC CAACCGGAATATTG | Generating fusion construct | This work |
| pKOR1 For | AGCTCCAGATCCATATCCTTC | Sequencing fusion construct | This work |
| pKOR1 Rev | CACACAGGAAACAGCTATGAC | Sequencing fusion construct | This work |
| sdrD KO confirm For | CGGTGGATTATTCGCGGC | To confirm isogenic mutant | This work |
| sdrD KO confirm Rev | CACATTTTGAAGATATGCCGTGTTG | To confirm isogenic mutant | This work |
| sdrD Int For | CGAGTGATAAAGTTGATATGCAGC | To confirm isogenic mutant | This work |
| sdrD Int Rev | AGCCTCTGTTGATGATGGCTGTAC | To confirm isogenic mutant | This work |
| sdrD- prom For | CGCCTGCAGCCAGGTCCATGTGGCCTGGTT | Generating reporter construct | This work |
| sdrD- prom Rev | CGCGGTACCCAA ATT TTTAAATAATACAAT TGTTTTAAATACAAAAAT | Generating reporter construct | This work |
| sdrD- pMG36e For | AAAATCTAGATGAATTAGGAGTAATCTAATGCT | Generating heterologous construct | This work |
| sdrD- pMG36e Rev | TTCACTCGAGCGCCTCATATAAGTTTTATTCCGT | Generating heterologous construct | This work |
| Full Dsg1-F | TAGTCCAAGCTTATGGACTGGAGTTTCTTC | Generating eukaryotic expression construct of full length Dsg1 | This work |
| Full Dsg1-R | TTGTATGGATCCCTACTTGCTATATTGCAC | Generating eukaryotic expression construct of full length Dsg1 | This work |
| SdrD-A- F | AACTGTCAAGGATTCTTAGTAGGTACAAC | Amplification of SdrD-A region | This work |
| SdrD-A-R | ACGTAACGTAAGATCTTTAGGTTTGTAAATACC | Amplification of SdrD-A region | This work |
| SdrD- Full length-_F | ATAGCGGCCGCTGTTTCTGGTAATGCTTTTGCTTTTGCTTTATTGTGATGG | Amplification of full length SdrD | This work |
| SdrD- Full length- R | CGCGGATCCGCAGAAAGTACTAATAAAGAATTGAACGAA | Amplification of full length SdrD | This work |
Heterologous expression of SdrD in L. lactis
For heterologous expression of SdrD in L. lactis MG1363, sdrD open reading frame from NCTC8325-4 genomic DNA was amplified using primers sdrD- pMG36e For and sdrD- pMG36e Rev (Table 1). The PCR product was digested using XbaI and XhoI, and ligated to the corresponding sites of pMG36e56, yielding pMG36e-SdrD. pMG36e control or pMG36e-SdrD constructs were transformed into L. lactis M1363 through electroporation (100-Ω resistance, 25 μF capacitance and 2.5 kV voltage). Transformants were selected and maintained in M17 medium (Oxoid, UK) containing 0.5% glucose and 10 μg/ml erythromycin. The expression of SdrD in the transformed L.lactis M1363 was evaluated by Western blot using antibodies against the SdrD-A region (kind gift from Dr. Elisabet Josefsson).
Expression and purification of recombinant SdrD
Prokaryotic expression constructs for recombinant SdrD were created as follows: The sdrD gene of S.aureus NCTC 8325-4 was amplified by PCR using either SdrD- Full length-F and SdrD- Full length-R or SdrD-A-F and SdrD-A-R (Table 1) primers. The PCR products with full-length or the A-region of SdrD were ligated to the corresponding sites of pRSETB vector (Invitrogen) or pGEX4T-1 (GE healthcare), respectively. Both plasmids were expressed in E. coli BL21 (DE3) and BL21, respectively. Briefly, recombinant full-length SdrD was expressed upon overnight incubation at 20 °C using Luria broth (LB) media supplemented with ampicillin and glucose. IPTG was added to a final concentration of 1mM, and the induction occurred overnight. Recombinant full-length SdrD was purified from pRSETB-SdrD by Norwegian Structural Biology Center (http://norstruct.uit.no) using His Trap column. According to the company, the purity of the full-length His-tagged SdrD protein is 85%. A selected fraction (supplementary Fig. S2, marked with red) was sent for mass spectrometry analysis and SdrD (gi 44685714) was detected as one of the protein hits (http://www.matrixscience.com/cgi/master_results.pl?file=..%2Fdata%2F20141209%2FFTgmOeamL.dat). GST-SdrD (A-region) was purified from pGEX4T-1-SdrD-A according to the manufacturer’s instructions.
Immunoblot
Immunoblot analysis was carried out on bacterial lysates. Cells of S. aureus and L. lactis were pelleted, dissolved in B-PER (Thermo Scientific, UK) and treated with 20 μg/ml lysostaphin (Sigma Aldrich, Germany) for 30 min at room temperature and 10 mg/ml lysozyme (Sigma Aldrich, Germany) for 1 hour at 37 °C, respectively. Bacterial lysates were freeze-thawed three times, followed by sonication. Aliquots of the bacterial lysates were used for western blot. The expression of SdrD was evaluated by immunoblot using antibodies against the SdrD-A region (kind gift from Dr. Elisabet Josefsson) as primary antibody and polyclonal Swine Anti Rabbit immunoglobulin (DAKO, Denmark) as secondary antibody. The GroL antibody (kind gift from Nikola Zlatkov Kolev) was used as control.
sdrD expression in the absence and presence of eukaryotic cells
An sdrD-GFP -reporter construct was generated by amplifying the sdrD promoter region by PCR from genomic DNA of S. aureus NCTC8325-4 using primers sdrD prom For and sdrD prom Rev (Table 1). The PCR product was ligated into the corresponding sites of pCM2957 upstream of GFP and transformed into S. aureus NCTC8325-4 by electroporation as previously described54. Expression of sdrD-GFP in S. aureus NCTC8325-4 was assessed in the absence or presence of HaCaT cells in DMEM supplemented with 10% FBS (without agitation). HaCaT cells were seeded at a density of 2 × 104 cells/well into 96 wells microtiter plate (Corning, USA) in DMEM 10% FBS and infected with S. aureus sdrD-GFP reporter strain. For this purpose, an overnight culture of S. aureus NCTC8325-4 harbouring the sdrD-reporter construct was diluted 1:100 into pre-warmed tryptic soy broth (TSB, Sigma Aldrich, Germany), incubated at 37 °C with agitation, and harvested at OD600nm = 0.7. The bacterial cells were pelleted, washed in phosphate saline buffer (PBS, pH 7.5), and diluted in DMEM 10% FBS. Fluorescence was measured using Synergy H1 Hybrid Reader (BioTek, USA) with excitation/emission of 488/520 nm. A control sample of non-transformed S. aureus NCTC8325-4 was included for background correction.
Yeast two-hybrid screen
A yeast-two hybrid service was ordered from Hybrigenics (http://www.hybrigenics-services.com/). Briefly, GAL4-SdrD-A-region was used as bait in a human reconstituted skin library.
S. aureus NCTC8325-4 and NCTC8325-4ΔsdrD adherence and inhibition of binding to immobilized recombinant Dsg1
Adherence of S. aureus NCTC8325-4 and S. aureus NCTC8325-4ΔsdrD to immobilized recombinant Dsg1 was principally evaluated as described previously39. Briefly, microtiter plates (Nunc, Denmark) were coated with coupling buffer (100 mM sodium carbonate (Sigma Aldrich, Germany), pH 9.6) containing 1 μg/well recombinant human Dsg1-Fc chimera (R&D systems, USA) or Recombinant human IgG1 Fc (R&D systems, USA) and incubated overnight at 4 °C. Wells were blocked with 0.05% (w/v) bovine serum albumin (BSA) (Sigma Aldrich, Germany) for 2 h at 37 °C and washed with PBS supplemented with 0.05% (v/v) Tween (PBST). S. aureus NCTC8325-4 and NCTC8325-4ΔsdrD were grown overnight in TSB medium. The next day, bacteria were diluted and grown to an OD600nm = 0.7–0.8 in TSB, washed in PBS, and suspended in PBS at OD600nm = 0.8–1. Thereafter, 100 μl of bacterial suspension was added to the Dsg1-coated or IgG1 Fc coated wells. The plates were incubated for 2 h at 37 °C, washed with PBST, and bound cells were fixed with paraformaldehyde (4% w/v) (Sigma Aldrich, Germany) for 20–30 min and stained with crystal violet (0.5% v/v, 100 μl per well) for 1 min followed by PBS and acetic acid (5% v/v) washes. The absorbance was measured at 570 nm in an ELISA plate reader (VERSAmax, USA). Inhibition of S. aureus binding to immobilized recombinant Dsg1 was evaluated as described previously39 by preincubating NCTC8325-4 and NCTC8325-4ΔsdrD with recombinant protein Dsg1-Fc (10 μg/ml), IgG1 Fc (10 μg/ml) or PBS for 30 min at room temperature.
Ligand binding assay
ELISA was used to analyse the ability of SdrD-A region to bind Dsg1 as described previously58 with minor modifications. Briefly, different concentrations of purified SdrD-A region were added to wells of a 96-well microtiter plate (Nunc) and the plate was incubated overnight at 4 °C. Wells were washed three times with PBS containing 0.05% Tween-20 (Sigma Aldrich, Germany), blocked with 0.05% or 1% (wt/vol) filter sterilized BSA in PBS for 2h at 37 °C. Wells were re-washed and Dsg1-Fc chimera (10 μg/ml) (R&D systems, USA) were added, and GST was used as a control. The adhered Dsg1 was determined using Dsg1471–499 antibodies (Abgent, USA), followed by HRP-conjugated polyclonal Swine Anti Rabbit immunoglobulin (DAKO, Denmark). The measurement of the absorbance at 450 nm was carried out in an ELISA reader (VERSAmax, USA).
Expression of Dsg1 on the surface of HaCaT cells
To detect the expression of Dsg1 on the surface of HaCaT cells, HaCaT cells were incubated for 40 min at 4 °C with anti-Dsg1 antibodies (R&D systems, Minneapolis, MN), which is directed against the Dsg1 extracellular domain. After washing, cells were incubated for 30 min at 4 °C with the secondary antibody (Alexa 488® Donkey anti goat, Thermo scientific, UK). Flow cytometry analyses were performed using FACS Fortessa (BD Bioscience).
Binding of recombinant SdrD protein to eukaryotic cells
HaCaT cells were detached by treating with 1 × Citric Saline solution, thereafter fixed with 4% paraformaldehyde for 15 min. After washing, cells were blocked with 10% fetal bovine serum (FBS) in PBS for 15 min at room temperature. Cells were incubated for 1 hour at 4 °C with 15 μg/ml SdrD in PBS supplemented with 5% FBS (Gibco, Life Technologies, UK). Human neutrophils were incubated on ice for 30 min at 4 °C with 14 μg/ml with full-length SdrD in RPMI/HSA or 10 ug/mg FLIPr (S. aureus formyl peptide receptor inhibitor, a kind gift from Prof. Jos van Strijp). Bound his-tagged SdrD and FLIPr were detected using anti-his-FITC monoclonal antibody (LS Bio Sciences, USA/Abcam, UK). Flow cytometry was performed using BD Biosciences FACS Fortessa (HaCaT) and BD Bioscience FACS Calibur (Neutrophils). The fluorescence intensity (FL) of 10,000-gated cells was measured for each sample and geometric mean FL was calculated using FlowJo software (Flowjo, Ashland, OR).
Transfection
Human full-length Dsg1 in pLKpac was a kind gift from Dr Wahl59. The full-length dsg1 was amplified by PCR using primers Full Dsg1-F and Full Dsg1-R (Table 1), digested and sub-cloned into corresponding BamHI/HindIII site of pcDNA3 (Invitrogen). The presence of dsg1 gene in the vector was confirmed by sequencing. HEK293 cells were seeded in 24-well plates at the concentration of 8.0–10.0 × 104 cells/well. After 24 hours, transfection with pcDNA3-Dsg1 or control plasmid pcDNA3 was carried out using METAFECTENE® PRO (Biontex, USA) according to the manufacturer’s instructions. The total amount of DNA used in transfection was kept constant by adding CT-DNA (Calf thymus DNA, Invitrogen, USA). A GFP reporter plasmid (pEGFP-C2) was used for monitoring transfection efficiency in each experiment. Transfection efficiency was routinely >80% for HEK293 cells.
Growth curves of S. aureus NCTC8325-4 and NCTC8325-4ΔsdrD
S. aureus NCTC8325-4 and NCTC8325-4ΔsdrD bacteria were grown overnight in Tryptic Soy Broth (TSB). The next day, bacteria were re-grown to OD600nm = 0.6 in TSB, washed, and resuspended in TSB or DMEM supplemented with 10% FCS (DMEM-FCS) at 1 × 105 CFU/ml. Bacteria dilutions were added to 100-well Bioscreen Honeycomb plates and growth was monitored by OD600nm measurements every 30 min under shaking conditions using Bioscreen C MBR machine at 37 °C.
Haemolytic activity of S. aureus NCTC8325-4 and NCTC8325-4ΔsdrD
S. aureus NCTC8325-4 and NCTC8325-4ΔsdrD were streaked on the TSA plate containing 5% sheep blood and incubated overnight at 37 °C to assess their haemolytic capacity after 18 hours. The plate was tranferred to 4 °C, and evaluated again after 48 hours.
Adhesion assay
The adhesion assay was carried out in triplicate as previously described32. Briefly, HaCaT cells or HEK293 cells were seeded into 24-well plates at confluent concentration of approximately 1.3–1.5 × 105 cells per well in DMEM 10% FBS. HEK293 cells were transfected with indicated plasmids. The day after, the prepared bacterial strains were added to HaCaT cells and HEK293 cells at multiplicity of infection (MOI) 250 and 10 or 50, respectively. The plates were incubated for 90 min at 37 °C in a 5% CO2–95% air atmosphere. Non-adhered bacterial cells were washed off, and the adhered bacteria were quantified by serial dilution plating after trypsinizing (trypsin-EDTA, Sigma Aldrich, Germany) and lysing of HaCaT cells using PBS containing 0.1% Triton X-100 (Sigma Aldrich, Germany).
Ethical Approval
Human neutrophils were isolated from freshly drawn human blood in accordance with ethical principles of the Helsinki Declaration. The blood donors provided written informed consent, and the medical ethics committee of the University Medical Center Utrecht (Utrecht, The Netherlands) approved the used protocol.
Statistical analysis
At least three biological replicates were done for the adhesion assays and ELISA-based experiments. The specific growth rate (k = ((log10 N − log10 N0) 2.303)/(t − t0)) of S. aureus NCTC8325-4 and NCTC8325-4ΔsdrD exponential phase were used to compare the growth of wild type versus isogenic mutant. Statistical analysis was carried out on the data of pooled experiments. Student’s t-test in Excel was used for determination of statistically significant differences between groups (P <0.05). Excel was used for generating of graphs.
Additional Information
How to cite this article: Askarian, F. et al. The interaction between Staphylococcus aureus SdrD and desmoglein 1 is important for adhesion to host cells. Sci. Rep. 6, 22134; doi: 10.1038/srep22134 (2016).
Supplementary Material
Acknowledgments
This project was funded by The Northern Norway Regional Health Authority/UiT-The Artic University of Norway “miljøstøtte” to JS and NWO-VIDI grant (91713303) to NvS. We thank Dr. Wahl, Dr. Elisabet Josefsson, Dr. Reindert Nijland, Nikola Zlatkov Kolev and Prof. Jos van Strijp for providing pLKpac-Dsg1 construct, SdrD A-region antibody, pCM29 construct, GroL antibody and recombinant FliPr, respectively.
Footnotes
Author Contributions F.A., C.A. and M.J. designed the experiments and prepared the manuscript. F.A. and C.A. performed the experiments. F.A. generated constructs and expressed full-length SdrD. N.v.S. contributed to the preparation of the isogenic mutant and reporter construct in S. aureus. D.B.D. generated the heterologous expression construct in L. lactis, while I.P. performed adhesion assays using the heterologous expression construct. A.M.H. and J.S. contributed to experimental design and intellectual input. All authors reviewed and approved the manuscript.
References
- Cole A. M. et al. Determinants of Staphylococcus aureus nasal carriage. Clin Diagn Lab Immunol 8, 1064–1069 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim H. F. L. et al. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 364, 703–705 (2004). [DOI] [PubMed] [Google Scholar]
- Lowy F. D. Staphylococcus aureus Infections. New Engl J Med 339, 520–532 (1998). [DOI] [PubMed] [Google Scholar]
- Ricklin D. et al. A molecular insight into complement evasion by the staphylococcal complement inhibitor protein family. J Immunology 183, 2565–2574 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti J. M. & Hook M. Microbial adhesins recognizing extracellular matrix macromolecules. Curr Opin Cell Biol 6, 752–758 (1994). [DOI] [PubMed] [Google Scholar]
- Josefsson E. et al. Three new members of the serine-aspartate repeat protein multigene family of Staphylococcus aureus. Microbiology-Uk 144, 3387–3395 (1998). [DOI] [PubMed] [Google Scholar]
- Sabat A. et al. Distribution of the serine-aspartate repeat protein-encoding sdr genes among nasal-carriage and invasive Staphylococcus aureus strains. J Clin Microbiol 44, 1135–1138 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J., Geoghegan J. A., Ganesh V. K. & Hook M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol 12, 49–62 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Ge J., Liu B., Hu Y. & Yang M. Structures of SdrD from Staphylococcus aureus reveal the molecular mechanism of how the cell surface receptors recognize their ligands. Protein & Cell 4, 277–285 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrijan L. & Lipozencic J. Adhesion molecules in keratinocytes. Clin Dermatol 29, 427–431 (2011). [DOI] [PubMed] [Google Scholar]
- Bonazzi M. & Cossart P. Impenetrable barriers or entry portals? The role of cell-cell adhesion during infection. J Cell Biol 195, 348–357 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck C. R., Agerer F., Muenzner P. & Schmitter T. Cellular adhesion molecules as targets for bacterial infection. Eur J Cell Biol 85, 235–242 (2006). [DOI] [PubMed] [Google Scholar]
- Berika M. & Garrod D. Desmosomal adhesion in vivo. Cell Commun Adhes 21, 65–75 (2014). [DOI] [PubMed] [Google Scholar]
- Schwarz M. A., Owaribe K., Kartenbeck J. & Franke W. W. Desmosomes and hemidesmosomes: constitutive molecular components. Ann Rev Cell Biol 6, 461–491 (1990). [DOI] [PubMed] [Google Scholar]
- Nekrasova O. & Green K. J. Desmosome assembly and dynamics. Trends Cell Biol 23, 537–546 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amagai M., Koch P. J., Nishikawa T. & Stanley J. R. Pemphigus vulgaris antigen (Desmoglein 3) is localized in the lower epidermis, the site of blister formation in patients. J Invest Dermatol 106, 351–355 (1996). [DOI] [PubMed] [Google Scholar]
- Bazzi H. et al. Desmoglein 4 is expressed in highly differentiated keratinocytes and trichocytes in human epidermis and hair follicle. Differentiation 74, 129–140 (2006). [DOI] [PubMed] [Google Scholar]
- Shirakata Y., Amagai M., Hanakawa Y., Nishikawa T. & Hashimoto K. Lack of mucosal involvement in pemphigus foliaceus may be due to low expression of desmoglein 1. J Invest Dermatol 110, 76–78 (1998). [DOI] [PubMed] [Google Scholar]
- Elias P. M. et al. Desmoglein isoform distribution affects stratum corneum structure and function. J Cell Biol 153, 243–249 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Stanley J. R. & Cotsarelis G. Desmoglein isotype expression in the hair follicle and its cysts correlates with type of keratinization and degree of differentiation. J Invest Dermatol 120, 1052–1057 (2003). [DOI] [PubMed] [Google Scholar]
- Wu H. et al. Protection against pemphigus foliaceus by desmoglein 3 in neonates. New Engl J Med 343, 31–35 (2000). [DOI] [PubMed] [Google Scholar]
- Hanakawa Y., Matsuyoshi N. & Stanley J. R. Expression of desmoglein 1 compensates for genetic loss of desmoglein 3 in keratinocyte adhesion. J Invest Dermatol 119, 27–31 (2002). [DOI] [PubMed] [Google Scholar]
- Hammers C. M. & Stanley J. R. Desmoglein-1, differentiation, and disease. J Clin Invest 123, 1419–1422 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelov L. et al. Desmoglein 1 deficiency results in severe dermatitis, multiple allergies and metabolic wasting. Nat Genet 45, 1244–U1201, doi: 10.1038/Ng.2739 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amagai M. & Stanley J. R. Desmoglein as a target in skin disease and beyond. J Invest Dermatol 132, 776–784 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan R. M., Miajlovic H. & Foster T. J. Surface proteins that promote adherence of Staphylococcus aureus to human desquamated nasal epithelial cells. BMC Microbiol 9, 22, doi: 10.1186/1471-2180-9-22 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S. R. et al. Identification of in vivo-expressed antigens of Staphylococcus aureus and their use in vaccinations for protection against nasal carriage. J Infect Dis 193, 1098–1108 (2006). [DOI] [PubMed] [Google Scholar]
- O’Brien L. M., Walsh E. J., Massey R. C., Peacock S. J. & Foster T. J. Staphylococcus aureus clumping factor B (ClfB) promotes adherence to human type I cytokeratin 10: implications for nasal colonization. Cell Microbiol 4, 759–770 (2002). [DOI] [PubMed] [Google Scholar]
- Mempel M. et al. Role of Staphylococcus aureus surface-associated proteins in the attachment to cultured HaCaT keratinocytes in a new adhesion assay. J Invest Dermatol 111, 452–456 (1998). [DOI] [PubMed] [Google Scholar]
- Schaffer A. C. et al. Immunization with Staphylococcus aureus clumping factor B, a major determinant in nasal carriage, reduces nasal colonization in a murine model. Infect Immun 74, 2145–2153 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim H. F. et al. Key role for clumping factor B in Staphylococcus aureus nasal colonization of humans. PLoS Med 5, e17 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintarak S., Whawell S. A., Speight P. M., Packer S. & Nair S. P. Internalization of Staphylococcus aureus by human keratinocytes. Infect Immun 72, 5668–5675 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A. M., Potter U., Meenan N. A., Potts J. R. & Massey R. C. Staphylococcus aureus keratinocyte invasion is dependent upon multiple high-affinity fibronectin-binding repeats within FnBPA. PLoS One 6, e18899 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning M. F. et al. The expression of desmoglein isoforms in cultured human keratinocytes is regulated by calcium, serum, and protein kinase C. Exp Cell Res 239, 50–59 (1998). [DOI] [PubMed] [Google Scholar]
- Schafer S., Koch P. J. & Franke W. W. Identification of the ubiquitous human desmoglein, Dsg2, and the expression catalogue of the desmoglein subfamily of desmosomal cadherins. Exp Cell Res 211, 391–399 (1994). [DOI] [PubMed] [Google Scholar]
- Prat C., Bestebroer J., de Haas C. J., van Strijp J. A. & van Kessel K. P. A new staphylococcal anti-inflammatory protein that antagonizes the formyl peptide receptor-like 1. J Immunol 177, 8017–8026 (2006). [DOI] [PubMed] [Google Scholar]
- Wang H. J. et al. Desmoglein 2 is a receptor for adenovirus serotypes 3, 7, 11 and 14. Nat Med 17, 96–U273, doi: 10.1038/nm.2270 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidenmaier C., Goerke C. & Wolz C. Staphylococcus aureus determinants for nasal colonization. Trends Microbiol 20, 243–250 (2012). [DOI] [PubMed] [Google Scholar]
- Mulcahy M. E. et al. Nasal colonisation by Staphylococcus aureus depends upon clumping factor B binding to the squamous epithelial cell envelope protein loricrin. PLoS Pathog 8, e1003092 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nithya S., Radhika T. & Jeddy N. Loricrin - an overview. J Oral Maxillofac Pathol 19, 64–68 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe P. A. & Lee C. H. Defining keratin protein function in skin epithelia: epidermolysis bullosa simplex and its aftermath. J Invest Dermatol 132, 763–775 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T. et al. The microbiome extends to subepidermal compartments of normal skin. Nat Commun 4, 1431 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins A. et al. Differential Expression and Roles of Staphylococcus aureus Virulence Determinants during Colonization and Disease. Mbio 6, e02272–14, doi: 10.1128/mBio.02272-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestle F. O., Di Meglio P., Qin J. Z. & Nickoloff B. J. Skin immune sentinels in health and disease. Nat Rev Immunol 9, 679–691 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley J. R. & Amagai M. Pemphigus, bullous impetigo, and the staphylococcal scalded-skin syndrome. N Engl J Med 355, 1800–1810 (2006). [DOI] [PubMed] [Google Scholar]
- Nishifuji K., Sugai M. & Amagai M. Staphylococcal exfoliative toxins: “Molecular scissors” of bacteria that attack the cutaneous defense barrier in mammals. J Dermatol Sci 49, 21–31 (2008). [DOI] [PubMed] [Google Scholar]
- Kitajima Y. 150(th) anniversary series: Desmosomes and autoimmune disease, perspective of dynamic desmosome remodeling and its impairments in pemphigus. Cell Commun Adhes 21, 269–280 (2014). [DOI] [PubMed] [Google Scholar]
- Dubash A. D. et al. The GEF Bcr activates RhoA/MAL signaling to promote keratinocyte differentiation via desmoglein-1. J Cell Biol 202, 653–666 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami T. et al. Inhibition of Rho-associated kinases disturbs the collective cell migration of stratified TE-10 cells. Biol Res 48, 48 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitkiewicz I., Babiak I. & Hryniewicz W. Characterization of transcription within sdr region of Staphylococcus aureus. Antonie Van Leeuwenhoek 99, 409–416 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasson M. J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol 154, 1–9 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukamp P. et al. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol 106, 761–771 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae T. & Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55, 58–63 (2006). [DOI] [PubMed] [Google Scholar]
- Askarian F. et al. A Staphylococcus aureus TIR domain protein virulence factor blocks TLR2-mediated NF-kappaB signaling. J Innate Immun 6, 485–498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Sorge N. M. et al. Methicillin-resistant Staphylococcus aureus bacterial nitric-oxide synthase affects antibiotic sensitivity and skin abscess development. J Biol Chem 288, 6417–6426 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Guchte M., van der Vossen J. M., Kok J. & Venema G. Construction of a lactococcal expression vector: expression of hen egg white lysozyme in Lactococcus lactis subsp. lactis. Appl Environ Microbiol 55, 224–228 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surewaard B. G. et al. Staphylococcal alpha-phenol soluble modulins contribute to neutrophil lysis after phagocytosis. Cell Microbiol 15, 1427–1437 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S. R. et al. Iron-regulated surface determinant protein A mediates adhesion of Staphylococcus aureus to human corneocyte envelope proteins. Infect Immun 77, 2408–2416 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl J. K. 3rd. Generation of monoclonal antibodies specific for desmoglein family members. Hybrid Hybridomics 21, 37–44 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.