Abstract
The African trypanosome, Trypanosoma brucei, is a unicellular parasite causing African Trypanosomiasis (sleeping sickness in humans and nagana in animals). Due to some of its unique properties, it has emerged as a popular model organism in systems biology. A predictive quantitative model of glycolysis in the bloodstream form of the parasite has been constructed and updated several times. The Silicon Trypanosome (SilicoTryp) is a project that brings together modellers and experimentalists to improve and extend this core model with new pathways and additional levels of regulation. These new extensions and analyses use computational methods that explicitly take different levels of uncertainty into account. During this project, numerous tools and techniques have been developed for this purpose, which can now be used for a wide range of different studies in systems biology.
1 Introduction
Trypanosoma brucei, a unicellular bloodstream parasite transmitted by tsetse flies, is the causative agent of African Trypanosomiasis (sleeping sickness in humans and nagana in animals; [1]). In recent years, it has emerged as a popular model organism in systems biology [2], due to the wealth of information obtained in biomedical studies and the many advantages it offers: reproducible cultivation methods exist and quantitative analysis is well-established. A quantitative mathematical model of central energy metabolism in the bloodstream form of the parasite was one of the first predictive kinetic models of a complex biological system [3], and this model has been iteratively updated after experimental testing and validation [4–8], yielding one of the most extensively curated kinetic metabolic models currently available.
The Silicon Trypanosome (SilicoTryp) is a project that brings together computational systems biologists and trypanosome experts to address multi-level regulation of trypanosome physiology on a new scale. Using a Bayesian approach that explicitly takes multiple levels of uncertainty into account, the project aims at re-analysing the core model of glycolysis and extends it in two directions: metabolic extensions towards new pathways (“horizontal” extensions) and inclusion of transcription and translation (“vertical” extensions). Explicitly including our uncertainty, both in the parameter values and the topologies of the different extensions, allows us not only to quantify our level of confidence in the results, but also to determine, in an unbiased manner, pitfalls and inconsistencies that might have gone unnoticed if the modelling was done without uncertainty awareness.
The horizontal extensions initially focus on the pathways controlling redox balance in trypanosomes: the trypanothione pathway, which is critical for parasite survival in the host, and the pentose phosphate pathway that links the trypanothione pathway to glycolysis by providing the necessary NADPH. Adding these two pathways requires knowledge of the kinetic parameters of all enzymes involved. In order to obtain the most accurate parameter values for these additional enzymes, a new buffer was developed to mimic as closely as possible the conditions in the cytosol of the parasite, as has previously been done for yeast [9]. The first enzyme characterised in this buffer was trypanothione synthetase, the final enzyme of the trypanothione biosynthesis pathway, using a method that integrates laboratory experimentation and modelling to explore alternative model topologies [10].
In addition to these metabolic extensions, the SilicoTryp project aims at a vertical extension of the model by including additional levels of control: transcription and translation. In trypanosomes, transcription rates (with very few exceptions) play little role in the regulation of mRNA concentrations since transcription by RNA polymerase II is not regulated for individual genes [11]. Instead, regulation is achieved at the levels of pre-mRNA and mRNA processing and degradation. These levels of control are modelled based on the previously published ordinary differential equations (ODE) model of phosphoglycerate kinase (PGK) transcription and translation [12]. The necessary parameters are acquired using deep sequencing (RNAseq). These estimates will be particularly uncertain, compared to metabolic enzyme parameters, and this provides an additional motivation for using a fully uncertainty-aware modelling strategy.
The uncertainty-aware modelling requires a rigorous method for updating our initial (prior) beliefs on the parameter values of the models, when new data becomes available. For this purpose, it was necessary to develop a computationally efficient Monte Carlo variance reduction method (Papamarkou et al. in press). Two types of large-scale experimental data are generated within the SilicoTryp project to drive the iterative updating process: steady-state concentration of metabolites and metabolic fluxes were measured using liquid-chromatography mass spectrometry (LC-MS) in standard conditions, and a collection of RNAi mutants were constructed, to examine the behaviour of the system under a variety of perturbations. The mutants in this collection will be characterised at the metabolome, transcriptome and proteome level, providing new data to further refine the extended model and reduce our uncertainty on model topology and parameter values.
2 Uncertainty in the glycolysis model
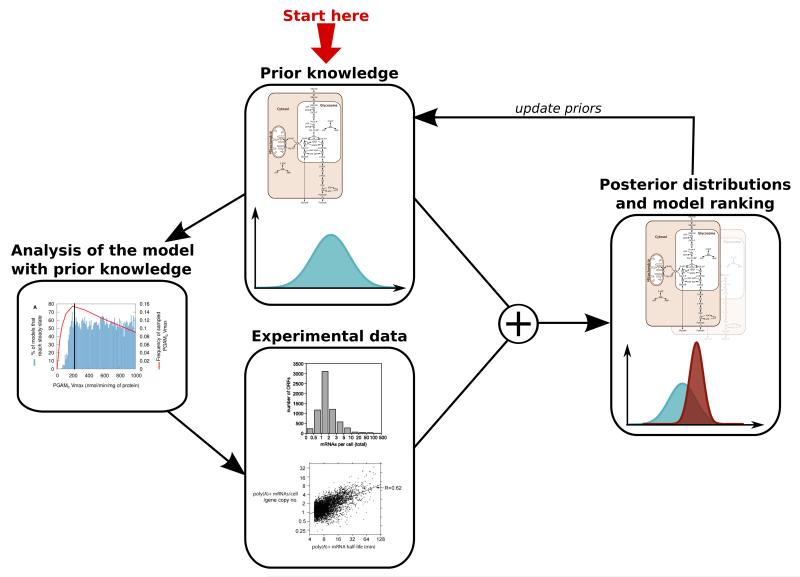
SilicoTryp explicitly deals with uncertainty at multiple levels (experimental data, kinetic parameter values, model topology) in every aspect of the project. It aims at applying the general framework of Figure 1 [13,14].
Figure 1. General Bayesian framework for explicitly handling uncertainty in the SilicoTryp project.
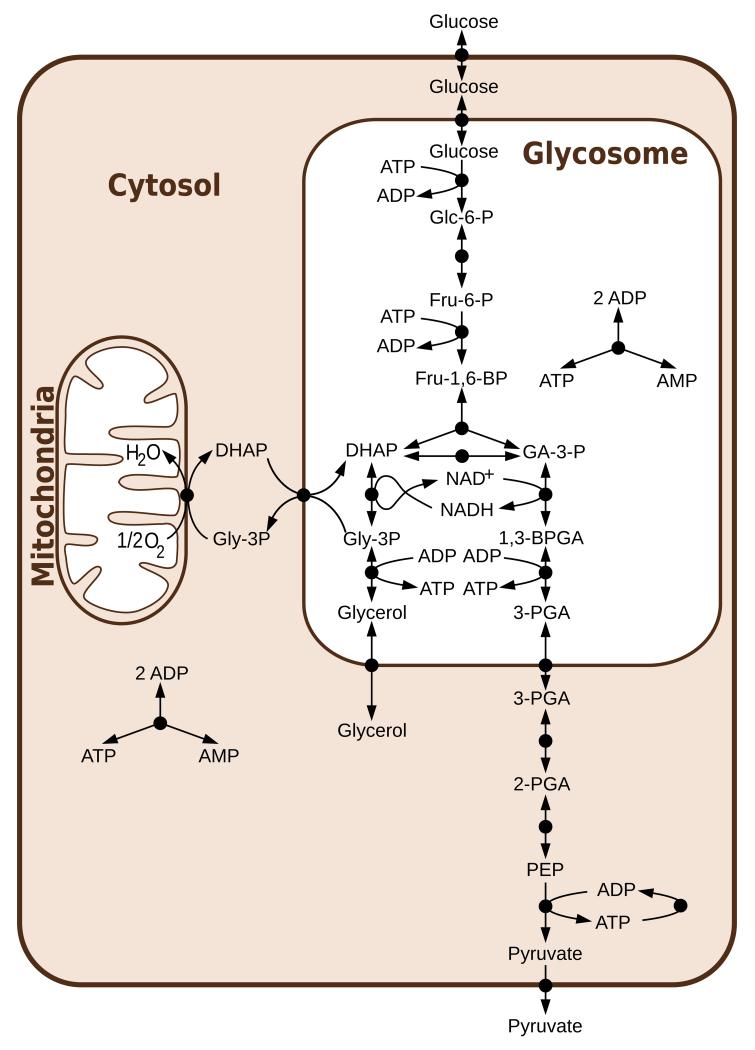
The T. brucei glycolysis model, as previously published [3–8](Figure 2), used a single set of parameters values. Some basic uncertainty analysis was included by scanning a range of values for selected parameters individually [3] or comparing two alternative topologies [4], but the combined effect of uncertainty in all parameters was not evaluated systematically, nor were the plausible distributions of parameter values explicitly defined. Therefore, an important step in the SilicoTryp project was to collect information on the (experimental and biological) uncertainty of all parameter values in the previously published model, but also on the uncertainty of the topology of the metabolic network.
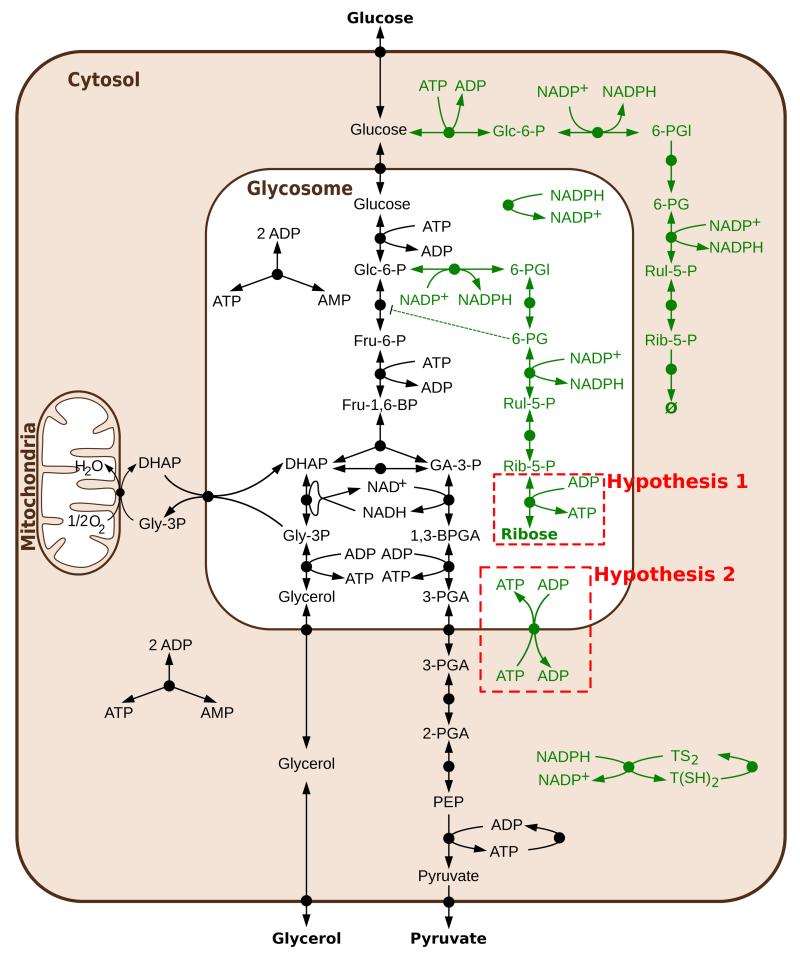
Figure 2. Glycolysis in T. brucei as previously published [7].
Abbreviations: Glc-6-P = glucose 6-phosphate, Fru-6-P = fructose 6-phosphate, Fru-1,6-BP = fructose 1,6-bisphosphate, DHAP = dihydroxyacetone phosphate, GA-3-P = glyceraldehyde 3-phosphate, Gly-3-P = glycerol 3-phosphate, 1,3-BPGA = 1,3-bisphosphoglycerate, 3-PGA = 3-phosphoglycerate, 2-PGA = 2-phosphoglycerate, PEP = phosphoenolpyruvate. Reprinted from [15], under the Creative Commons Attribution License.
2.1 Parameter uncertainty in the glycolysis model
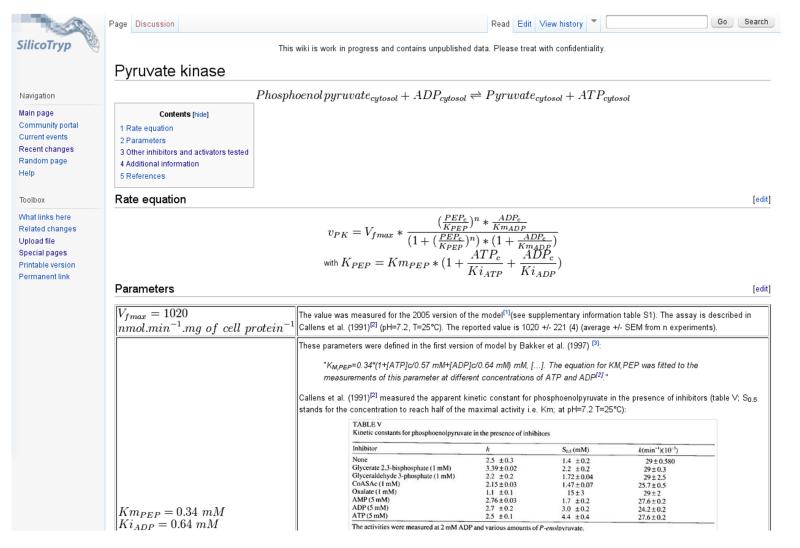
In order to evaluate uncertainty on the parameter values of the glycolysis model, a Mediawiki-based website was created, the SilicoTryp wiki (http://silicotryp.ibls.gla.ac.uk/wiki). In this wiki, the sources and calculations underlying the values for every parameter of the glycolysis model were documented (see example in Figure 3). This detailed source of documentation allowed us to define probability distributions for each parameter, hence representing our prior beliefs on what the true parameter values are [15].
Figure 3. Example page of the SilicoTryp wiki.
Chemical and rate equations are described, and sources for each parameter are documented in detail.
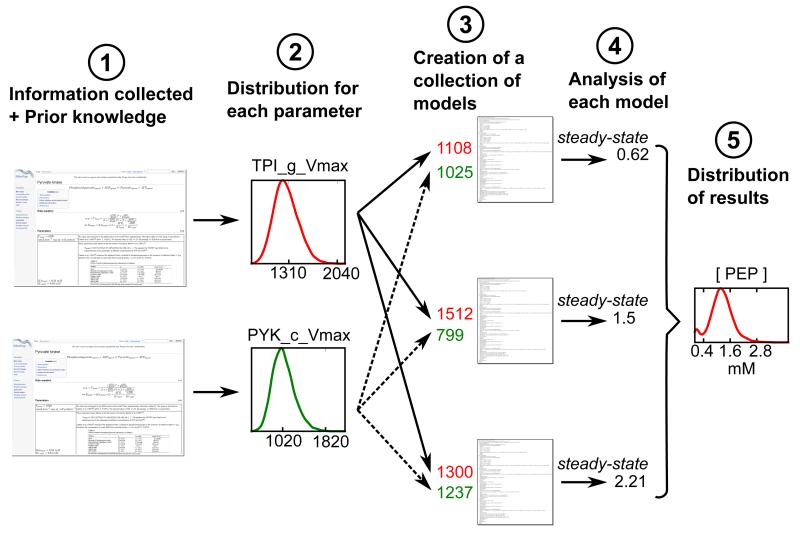
Using these distributions, sets of plausible parameters were sampled. A large collection of variant models, each with a different set of sampled parameters, was then simulated, allowing us to quantify confidence intervals for different model properties, such as steady-state concentrations and fluxes or control coefficients resulting from the parameter uncertainty (see Figure 4). This strategy allowed us not only to gain knowledge on how confident we are about the model predictions, but also to detect fragilities of the model in an unbiased way, highlighting areas of the model behaviour that are in contradiction with experimental data [15]. We showed, for example, that only in 40% of our sampled models is the control over the glucose consumption flux mostly held by the glucose transporter, as described before with the fixed parameter model [4,16]. In the equally plausible scenarios represented by the remaining 60% of the models, this control is shared between several other glycolytic enzymes, and which enzymes share control over the glucose consumption flux depends on the parameter sets considered [15].
Figure 4. Evaluation of parameter uncertainties and their effect on the model results.
. Reprinted with permission from [17]. Copyright 2013 American Chemical Society.
2.2 Topological uncertainty in the glycolysis model
In addition to evaluating the effect of parameter value uncertainty, we explored the effect of topological uncertainty in the glycolysis model [14]. Although T. brucei central energy metabolism has been exceptionally well studied for a long time, some areas of its network connectivity remain controversial. Indeed, the previously published model considered the glycosomes, organelles where most of the glycolytic enzymes are localised (see Figure 2), as impermeable compartments into which all required metabolites need to be transported via specific transporters. This hypothesis was reinforced by both theoretical [5] and experimental [8] findings that mislocalising the glycolytic enzymes to the cytosol causes a lethal accumulation of sugar phosphate. This confirmation of theoretical predictions by experimental results has been considered one of the major successes of the T. brucei glycolysis model.
However, recently, size-specific pores have been discovered in the glycosomal membrane [18]. These pores are essentially chemically non-selective holes and their presence implies that small metabolites can diffuse freely across the membrane, while only the largest molecules would be retained inside the glycosomes. This creates additional potential links in the metabolic network, between cytosol and glycosome, which are not yet fully understood, but need to be considered in the metabolic modelling.
Another aspect of topological uncertainty is due to the fact that we know that enzymes are imported into the glycosome fully folded [19,20]; of course these might be fully sequestered in transit by the protein import machinery, but there is also a concrete possibility that there could be some residual activity of glycosomal glycolytic enzymes in the cytosol. Cytosolic activity of the relevant enzymes has so far been poorly characterised and is generally neglected in modelling.
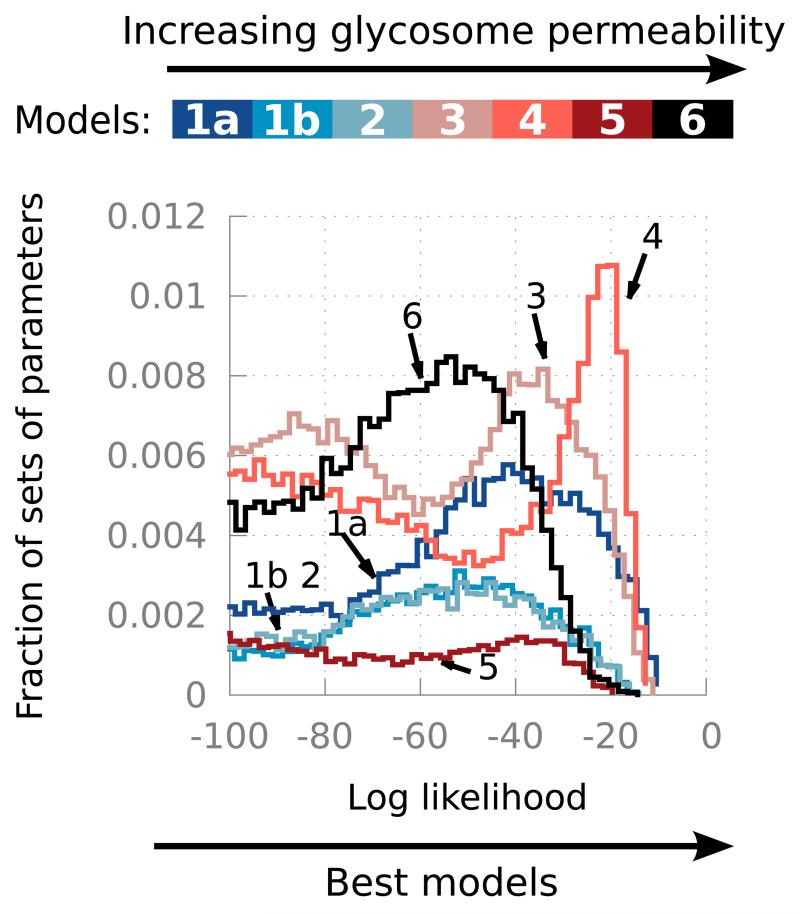
To test the effect of increasing permeability of the glycosome, a series of models was constructed [21]. These models have increasingly permeable glycosomes, ranging from the impermeable glycosomes of the previous models (model 1a has a similar topology as the models published since 1999 [4], model 1b has a similar topology as the model of 1997 [3]) to glycosomes permeable to all metabolites up to the size of ATP (model 6). For each of these models, the uncertainty on the parameter values was taken into account as previously [15]. This was then extended by also including the uncertainty on the percentage of activity of the glycosomal enzymes in the cytosol. The resulting collection of models was simulated, and results were compared to experimental data (metabolite concentrations and fluxes) by computing log-likelihoods: the higher the log-likelihoods the better the match between simulations and experiments (see Figure 5). Our results show that, quite surprisingly, the semi-permeable models that let metabolites smaller than fructose 6-phosphate (model 3) or smaller than fructose 1,6-bisphosphate (model 4) freely diffuse, agree with the experimental data at least as well as the impermeable model (model 1a). This is consistent with the idea that only compartmentation of ATP is required. Contrary to the common intuition that organelles like the glycosome function as tighly sealed compartments, at least for glycosomal glycolysis a certain degree of leakiness for smaller metabolites appears to be fully compatible with its function.
Figure 5. Log-likelihood (goodness of fit to experimental data) of models with increasing glycosome permeability and cytosolic activities of the glycosomal enzymes.
The higher the log-likelihood, the better the match between simulations and experimental results. Each model is simulated with a range of parameter sets that describes our uncertainty about the parameter values. See [21] for details. Modified with permission from [21]. Copyright 2013 John Wiley and Sons.
In addition, the analysis allowed us to show that, for all topologies with a semi-permeable glycosome tested, the fraction of residual cytosolic activity of some glycolytic enzymes probably plays a crucial role in explaining parasite physiology.
3 Metabolic extensions of the model
With the uncertainty-aware model of central carbon metabolism established, the SilicoTryp project could aim at including the trypanothione and pentose phosphate pathways and the associated extra levels of uncertainty into the model.
3.1 The pentose phosphate pathway
In T. brucei, the pentose phosphate pathway is localised in both the cytosol and the glycosome where its presence breaks the bound-phosphate balance that was thought to be essential [5, 8]. This phosphate “leak” needs to be solved either by finding a viable model that does not need bound-phosphate to be balanced in the glycosome, or by adding reactions to restore this balance. One solution within each of these two categories of solutions was tested (Kerkhoven, Achcar et al., under review).
The first solution tested maintained the bound-phosphate balance by adding ribokinase to the glycosome (hypothesis 1 of Figure 6). In the context of impermeable glycosomes, ribokinase would work in the direction of producing ATP and ribose. To be able to model this solution, the enzyme was characterised biochemically (Kerkhoven, Achcar et al., under review). However, despite being sufficient theoretically, the results showed that this enzyme alone cannot maintain the bound-phosphate balance. The second solution modelled involves breaking the bound-phosphate balance by introducing an ATP/ADP translocator. However, modelling shows that this translocator would need to be very tightly regulated for the parasite to be viable. This makes it unlikely that such an antiporter alone could represent a solution and more reactions are probably involved in the regulation of the bound-phosphate balance.
Figure 6. Two hypotheses tested to solve the topology of the pentose phosphate pathway.
In black, the reactions of the glycolysis model, in green the reactions introduced to model the pentose phosphate pathway. Abbreviations not present in Figure 2: 6-PGl = 6-phosphogluconolactone, 6-PG = 6-phosphogluconate, Rul-5-P = ribulose 5-phosphate, Rib-5-P = ribose 5-phosphate, TS2 = trypanothione disulfide, T(SH)2 = trypanothione.
Despite the topological uncertainty that remains, both of these model versions can already be used to some extent. Both of them handle oxidative stress in the same way, and the models have been used to test possible mechanisms to explain why the third enzyme of the PPP, 6-phosphogluconate dehydrogenase (from 6-PG to Rul-5-P in Figure 6), is essential for parasite survival. The resulting predictions were then tested experimentally and it was shown that a positive feedback loop related to 6-PG inhibition of phosphoglucose isomerase was not responsible for the 6-phosphogluconate dehydrogenase essentiality in T. brucei (Kerkhoven, Achcar et al., under review), in contrast to the situation in many other organisms.
3.2 The trypanothione pathway
3.2.1 The pathway topology
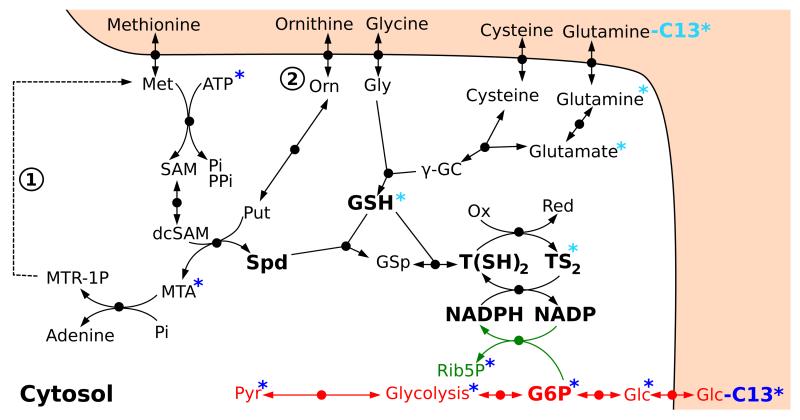
In order to extend the model with the trypanothione pathway, a main contributor to redox balance in T. brucei, we first had to determine the stochiometric map of this pathway. We constructed a stochiometric map of the trypanothione pathway based on the literature and LC-MS data (see Figure 7). Cells were grown in the presence of 50% 13C-labelled glucose over 48 hours, after which their metabolites were extracted and analysed using LC-MS (Kim, Achcar et al., in preparation). The partially labelled metabolome allowed us to rule out the presence of the methionine salvage pathway (marked 1 in Figure 7) in T. brucei. Indeed, if this pathway – recycling methylthioadenosine (MTA) into methionine – was occuring, we should detect some four carbon labelled methionine. However, methionine is clearly detected as fully unlabelled, indicating that the pathway is not complete (Kim, Achcar et al., in preparation).
Figure 7. Stochiometric map of the trypanothione pathway.
The blue asterisks indicate which metabolites are found C13-labelled when all glucose carbons are 13C-labelled. The turquoise asterisks indicate which metabolites are labelled when glutamine is labelled. The methionine salvage pathway, marked as 1, does not seem to be active in T. brucei (see text). The origin of ornithine, marked as 2, remains uncertain. Abbreviations: Orn = ornithine, Gly = glycine, Met = Methionine, γ-GC = γ-glutamylcysteine, SAM = S-adenosylmethionine, dcSAM = decarboxylated S-adenosylmethonine, Spd = spermidine, GSp = glutathionylspermidine, MTA = methylthioadenosine, MTR-1P = methylthioribose 1-phosphate, Glc = glucose, G6P = glucose 6-phosphate, Pyr = pyruvate, TS2 = trypanothione disulfide, T(SH)2 = trypanothione.
One question that remains open is the origin of ornithine in vivo. Previous data indicated that trypanosomes lack arginase [22], the enzyme that produces ornithine from arginine, which is the common source of ornithine in eukaryotic cells including other parasites [23]. It was also shown that in the presence of ornithine, T. brucei is capable of using it directly, and therefore it was hypothesised that the cells use ornithine directly from the medium. However, measurements of the metabolite composition in the growth medium over time (Kim, Achcar et al., in preparation; see below) showed that the ornithine concentration is quite low in the growth medium ( [24], see below section “Perturbations: RNAi cell lines”) and does not change over time (staying between 15 and 20 μM in the medium; it was measured as 54.4 ± 16.1 μM by Martens-Lobenhoffer et al. in blood [25], i.e. the same order of magnitude). This suggests that there probably is another route to the production of ornithine in T. brucei, and its origin is currently being examined.
3.2.2 Uniform assay conditions
Usually, enzyme activities are measured under non-physiological optimum conditions. For a reliable in silico model and to reduce our uncertainty about the parameter values, the data should preferably be obtained under conditions that mimic the milieu in which the pathway is active in the cell [9]. The importance of physiological conditions to measure metabolic functions was recently demonstrated: the use of an “in vivo-like” assay medium yielded enzyme kinetic parameters that substantially improved a computational model of yeast glycolysis [26]. Such a buffer had not been used for T. brucei before. Based on the available literature, we therefore developed a new phosphate buffer system at pH 7.0 in which measurements are done at 37 ° C (Table 1), mimicking the physiological environment of the enzymes in the cytosol of bloodstream-form parasites [10].
Table 1. Characteristics of the in vivo-like buffer system for the cytosol of bloodstream T. brucei.
3.2.3 A detailed focus on one enzyme: Trypanothione synthetase
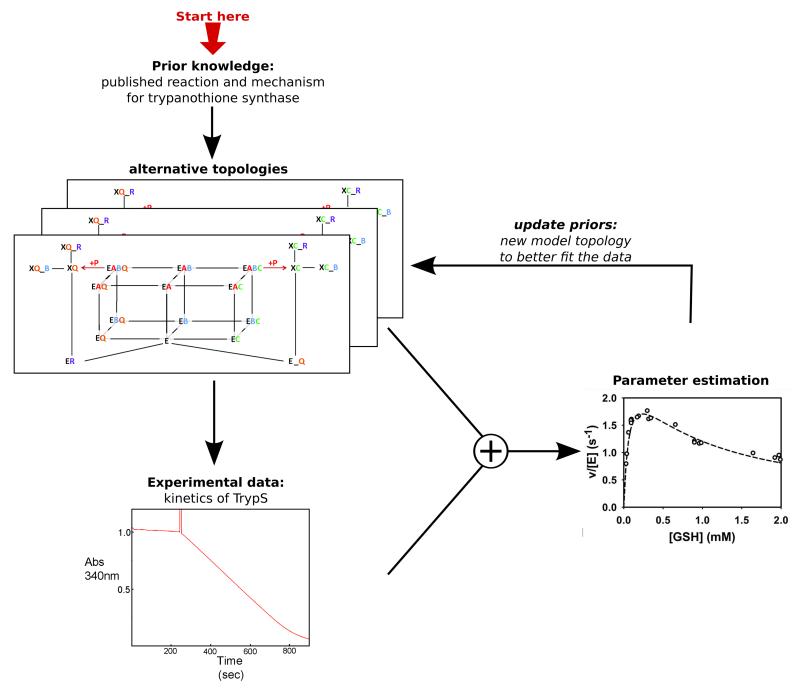
We first employed our in vivo-like assay to characterise kinetically the enzyme of the final step in trypanothione (T(SH)2) synthesis. T. brucei trypanothione synthetase (TryS) generates T(SH)2 from glutathione (GSH) and spermidine (Spd) in two consecutive ATP-dependent reactions [32, 33]. In the first step, glutathionylspermidine (Gsp) is formed, which is then combined with a second GSH to yield T(SH)2. In vitro, TryS also displays amidase activity and can, in total, catalyze five different reactions: ATP hydrolysis, formation of Gsp and T(SH)2 as well as hydrolysis of the conjugates to regenerate GSH and Spd. As the mechanism of TryS is rather complex, a rate equation could not be easily formulated. To obtain a deeper insight in the exact molecular mechanism of the synthetase reactions of TryS, we formulated alternative kinetic models with different topologies of the catalytic cycle. Each elementary reaction step was modelled by a mass-action kinetic equation. The model parameters were then fitted by evolutionary programming to the extensive matrix of steady-state data obtained for different substrate/product combinations using our in vivo-like buffer. In the process of building the model, a strong interaction between experimental and modelling e orts proved to be essential. New enzyme kinetic experiments were designed to provide data to help discriminate between different models. Moreover, when the fit to the data was suboptimal model topology was adjusted in an iterative cycle (see Figure 8).
Figure 8. Workflow applied for dissecting the kinetics mechanism of trypanothione synthetase.
Modified with permission from [10]. Copyright 2013 American Society for Biochemistry and Molecular Biology.
The best model describes the full kinetic profile of TryS and is also able to predict time profiles of (intermediate) product formation that were not used in the fitting of the parameters, and thus serve as validation data [10]. It also provides a mechanism for inhibition by the substrate glutathione and the product trypanothione. In our in vivo-like assay, some of the kinetic constants proved markedly different from earlier measurements in other buffer systems [33, 34]. Major alterations were a three-fold higher Ki value for GSH, a rise of the Km value for Spd from 139 μM to 687 μM and a reduction of the kcat from 5.2 to 2.8 s−1. These observations are further demonstration of the importance of mimicking the physiological environment.
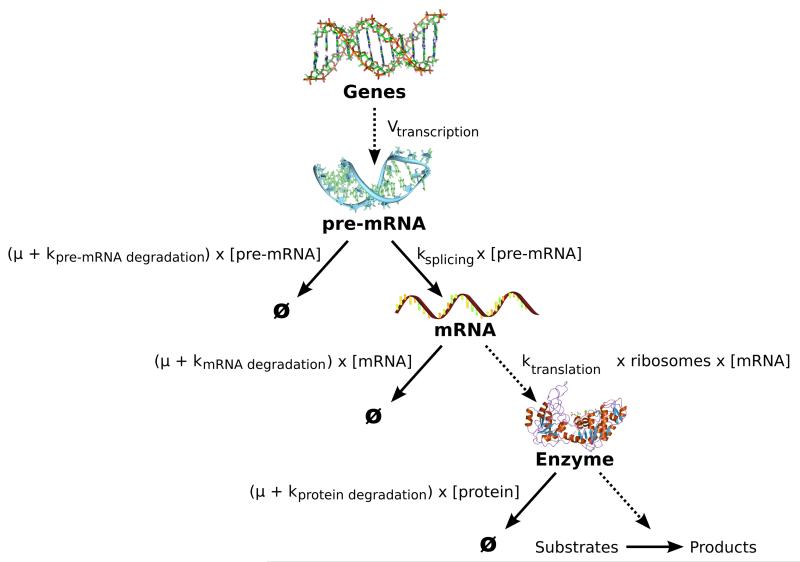
4 Transcription/Translation extensions of the model
One of the aims of the SilicoTryp project is to include transcription and translation in the model, resulting in dynamic enzyme levels, instead of using fixed Vmax values for all enzymes. This will be done using a module of four differential equations for each enzyme (see Figure 9), based on the previously published model of phosphoglycerol kinase transcription and translation [12]. Regulation of enzyme concentrations in T. brucei is typically achieved by controlling the pre-mRNA processing rate (ksplicing in Figure 9), the translation rate (ktranslation) and/or the degradation rates. To measure the pre-mRNA and mRNA degradation and processing rates, RNAseq experiments were done, and are currently being analysed.
Figure 9. Transcription/Translation module that will be added for every enzyme of the model.
The module is based on the model published by Haanstra et al. [12]. The rate constants of the various processes involved are indicated by k, μ is the specific growth rate of the trypanosomes, and ribosomes represents the number of ribosomes per molecule of mRNA.
4.1 The pre-mRNA processing rates
Genes in trypanosomes are transcribed as polycistronic precursor messages (pre-mRNA) that are destined for either trans-splicing into mature mRNA or degradation. In order to measure the rate of pre-mRNA processing for each gene, cells were treated with Actinomycin D for 5 minutes to inhibit mRNA transcription. RNA was then extracted and, after being depleted from rRNA, 2 biological replicates and 2 controls were subjected to transcriptome-wide deep sequencing (RNAseq, an average of 50 million reads of length 50 mer, using Illumina HiSeq 2000). A decrease in pre-mRNA read counts in treated cells will be due to the splicing of the pre-mRNA in the absence of transcription. The ratio of pre-mRNA read counts (treated cells over untreated) was then used to estimate the rate of mRNA processing. A difficulty in differentiating between reads from pre-mRNA sequences and those due to spurious transcription events in the same region, led us to limit our analysis to the 20 nucleotides immediately upstream of known splice acceptor sites. Although this had the drawback of limiting the number of examined regions, this filter increases our confidence about the origin of the reads.
Preliminary results indicate that 50% of the transcripts have a half-life of less than 2 min, but some showed much longer apparent half-lives. We speculate that this is due to persistence of bi-cistronic RNAs, perhaps in the cytoplasm (Fadda et al. in prep.).
4.2 mRNA decay rates
In order to measure mRNA degradation rates, cells were treated with both Actinomycin D and Sinefungin to inhibit both transcription and splicing. RNA was extracted at several time points after transcription inhibition, depleted from rRNA, and subjected to deep sequencing (an average of 25 million 75mer reads, or 200 million 50mer reads, using Illumina GAIIX and HiSeq 2000 respectively). Since all trypanosome mRNAs carry the same 39-nt spliced leader (SL) at the 5′-end, the abundance of the SL can serve as a proxy for the abundance of total mRNA. This allowed us to normalise the read counts using the total mRNA decay pattern across the same time points. Then, amounts of total mRNA were measured relative to the steady-state [35], and mRNA half-lives were first estimated using an exponential model.
It has been shown, however, that in a transcription inhibition experiment, while the majority of transcripts decay exponentially, others exhibit a slow-to-fast or fast-to-slow decay pattern. In our work we see the same pattern. Therefore, we used a Markov chain model developed earlier by Deneke et al. [36] to analyse the data for transcripts that do not show exponential kinetics. Transcripts exhibiting non-exponential decay are presumed to be degraded in a multi-step reaction, in which more than one rate-limiting step exists, and where the age of the transcript becomes relevant to its rate of degradation. This is now further studied with a particular focus on the mRNAs encoding enzymes of the glycolytic and redox pathways, many of which show non-exponential decay.
The mRNA kinetic data are furthermore being complemented with information on the translation rates for each transcript, using a ribosome profiling [37] strategy. The number of ribosomes on each mRNA is determined by treating cell lysates with RNase to digest the mRNA except those stretches protected by ribosomes. The protected RNA fragments are then purified and sequenced, and the rate of translation estimated by assuming a linear relationship between the ribosome number on a transcript and the rate of translation.
5 Experimental data to reduce model uncertainty and refine predictions
In order to reduce our uncertainty about both model topology and parameter values, further experimental data is needed, in addition to the transcription dynamics work described above. Data collection within SilicoTryp focused initially on steady-state metabolite concentrations and fluxes, which are directly informative for model refinement.
In addition, a collection of RNAi mutants was constructed with the aim of providing additional information on the system’s behaviour upon perturbation.
5.1 Measurements of concentrations and fluxes
Intracellular and extracellular concentrations and fluxes were measured using LC-MS. However, mass spectrometry intensities are not always linearly proportional to the concentrations due to ion suppression effects [38]. In addition, variations in instrumental response and degradation of metabolites of interest during sample preparation can also result in a biased quantitative result [39]. To overcome this problem, we need to spike a fixed amount of the 13C-labelled metabolite into each calibration sample [40]. Being chemically identical but having a different mass, this compound will be similarly ion-suppressed within the matrix entering the mass spectrometer, but distinguished from unlabelled compound since its mass is slightly different. Therefore, the ratios of intensities of the 12C-metabolite over the 13C-metabolite are directly proportional to the concentrations of 12C-metabolite. Using a calibration curve, the concentration of the metabolite can be determined in any sample if the same amount of 13C-metabolite that has been used for the calibration curve is added to the sample. However, 13C-labelled compounds are expensive and di cult to obtain. Therefore, uniformly (U)-13C-labelled E. coli metabolite extracts, obtained by growing E. coli in M9 minimal medium with (U)-13C-labelled glucose as the sole carbon source [41], were used as standards in the SilicoTryp project. This approach also offers the advantage that it potentially allows simultaneous quantification of all metabolites that can be detected in both E. coli and T. brucei.
Using this method, we quantified 44 intracellular metabolites and 32 metabolites in the growth medium over a 57-hour time course (starting from the low cell density of 3800 cells/ml to maintain the cells in exponential growth throughout the time course). Based on the 32 medium metabolite dynamics, we could quantify the fluxes of 6 metabolites that are constantly produced or consumed by the cells using the absolute concentrations and the cell densities at the same time points. Seven metabolites did not show any significant changes in concentration over time; for example, ornithine levels did not seem to change in the spent medium, which was unexpected since previous work had demonstrated that ornithine is taken up by the cells. 19 metabolites, all amino acids or nucleotides, showed changes of concentrations that are not compatible with constant production or consumption by the cells, but are compatible with the presence of peptidases and nucleosidases which degrade proteins and nucleic acids to their corresponding monomers. Indeed, it is known that T. brucei excretes peptidases and nucleosidases into the medium [42–44].
From these results, and the 13C-labelling experiments (see above), we could, for example, determine that about 2–3% of the pyruvate produced by cells grown in the simplified medium ( [24], see below section “Perturbations: RNAi cell lines”) is converted to alanine before being excreted. These results can now be used to extend the model and reduce our uncertainty about topology and parameter values.
5.2 Perturbations: RNAi cell lines
In order to provide information on the effect of reduced level of selected enzymes on the levels of mRNA, protein and the metabolome, we generated 28 stable RNA interference (RNAi) cell lines: 8 of them targetting genes of the polyamine pathway (the part of the trypanothione pathway that leads to the production of spermidine in Figure 7), 12 of the redox metabolism and 8 of the pentose phosphate pathway (see Table 2). The RNAi cell lines were generated in monomorphic 2T1 Trypanosoma brucei brucei, selected among other available cell lines as it provides a unique target sequence at a single ribosomal RNA locus, and has been validated for robust inducible expression and has a higher RNAi efficiency [45]. To confirm the specific down regulation of these enzymes at the protein level, RNAi cell lines where antibodies are available were selected, induced and tested by Western blot. So far, all RNAi lines tested by Western blot show down regulation of the protein of interest. The effects of ablating some of these enzymes have already been published, but most remain to be fully characterised (see Table 2). From the pentose phosphate pathway, only the glucose 6-phosphate 1-dehydrogenase [46] and the 6-phosphogluconate dehydrogenase [47] have been characterised previously.
Table 2.
List of RNA-i mutants constructed and the previously published phenotypes.
| Gene ID | Protein | Growth reduction/Phenotype | Ref. |
|---|---|---|---|
| Polyamine pathway | |||
|
| |||
| Tb09.v1.0380 | Spermidine synthase | Spermidine auxotrophy, depletion of T(SH)2 and cell death | [48] |
| Tb11.01.5300 | Ornithine decarboxylase | Depletion of reduced thiols and cell growth arrest | [49] |
| Tb927.6.4410 | S-adenosylmethionine decarboxylase | 60-90% | [50] |
| Tb927.6.4470 | Prozyme, activator of SAMDC | 100% | [50] |
| Tb927.6.4840 | Methionine adenosyltransferase | No effect | [50] |
| Tb927.6.4890 | S-adenosylmethionine synthase | Unknown | [50] |
| Tb927.8.1910 | Acetylornithine deacetylase | No effect | [50] |
|
| |||
| Pentose Phosphate Pathway and related enzymes | |||
|
| |||
| Tb927.10.2490 | Glucose-6-phosphate 1-dehydrogenase | Growth arrest, cell death | [46] |
| Tb11.02.4200 | 6-phosphogluconolactonase | None | [50] |
| Tb09.211.3180 | 6-phosphogluconate dehydrogenase | 70-100% | [50], * |
| Tb927.8.6170 | Transketolase | None | [50] |
| Tb927.10.12210 | Ribulose-5-phosphate 3-epimerase | None | [50] |
| Tb11.03.0090 | Ribokinase | 70-80% | [50] |
| Tb927.4.1350 | Glyoxalase II | 20-80% | [50, 51] |
| Tb11.01.0700 | Ribose 5-phosphate isomerase | >90% | [50] |
|
| |||
| Redox metabolism | |||
|
| |||
| Tb927.3.3760 | Tryparedoxin | Growth arrest and enhanced sensitivity against hydrogen peroxide | [50, 52] |
| Tb09.160.2020 | Thioredoxin | None | [50, 53] |
| Tb927.2.4370 | Trypanothione synthetase | Depletion of Gsp and T(SH)2, growth arrest, loss of viability, enhanced sensitivity against hydroperoxides and drugs | [32, 54] |
| Tb927.7.4000 | Glutathione synthetase | 10-80% | [50] |
| Tb927.10.12370 | Gamma-glutamylcysteine synthetase | Depletion of GSH and T(SH)2, cell death and loss of virulence | [55] |
| Tb927.10.10390 | Trypanothione reductase | Growth arrest, loss of viability and virulence | [56] |
| Tb11.01.7560 | Glutathione peroxidase, putative | >90% | [50] |
| Tb11.12.0016 | Glutathionylspermidine synthetase | No effect | [50] |
| Tb09.160.4250 | Peroxiredoxin-type tryparedoxin peroxidase (cytosolic) | Loss of viability and enhanced sensitivity against hydrogen peroxide | [50, 57] |
| Tb927.4.2450 | Protein disulfide isomerase I | 15-60% | [50] |
| Tb927.3.4240 | Thioredoxin, putative | 40-80% | [50] |
| Tb927.5.950 | Monothiol glutaredoxin 3 | 50-100% | [50] |
| Tb927.8.1990 | Peroxiredoxin-type tryparedoxin peroxidase (mitochondrial) | None | [50, 57] |
= Kerkhoven, Achcar et al., under review
Most of the evalution of phenotypes in previous work has been carried out in a medium containing a vast excess of many metabolites. Comparing fresh and spent medium metabolomes revealed that most metabolites, however, are not consumed by trypanosomes. We therefore created a new medium from which those metabolites not consumed by trypanosomes were removed [24]. Parasites grew equally well in this medium, which better reflects serum concentrations of many metabolites. Remarkably, experiments in the simplified medium revealed phenotypic effects that were hidden in the classical medium. For example, T. brucei bloodstream forms became over 100-fold more sensitive to antifolate drugs in the absence of the high concentrations of folate that are present in the classical medium [24].
The individual stable RNAi cell lines available now for each of our main genes of interest will allow us to confirm these results and also measure the changes in metabolites upon depletion of any particular enzyme as well as the effects, if any, on mRNA or protein levels of other enzymes. These perturbation responses will then inform the model debugging and refinement process.
6 Conclusion
The Silicon Trypanosome project aimed at extending the core glycosolysis model of the unicellular parasite Trypanosoma brucei with the trypanothione pathway, and the translation and transcription of the enzymes involved. All of this work required newly developed modelling methods that explicitly take the multiple levels of biological and uncertainty into account. Hence, we first documented in detail the sources of the parameters of the existing core glycolysis model. This detailed information allowed us to re-evaluate this model taking our uncertainty about the parameter values into account [15]. In addition, we also questioned the topology of this model by analysing and comparing alternative possibilities [21]. These analyses formalised what our current levels of uncertainty are regarding the core glycosolysis model. In the future, we aim at reducing these uncertainties using Bayesian inference methods, which use new experimental data to update our belief on parameter values and model topology (see Figure 1). Because of the complexity of the models we are dealing with, we developed a new state-of-the-art variance reduction method specifically for this purpose (Papamarkou et al., in press).
Although modelling attempts have been made for part of this pathway both in T. brucei [58] and T. cruzi [59], working towards the introduction of the trypanothione pathway introduced a new set of challenges. First, introducing the pentose phosphate pathway, the link between glycolysis and the trypanothione pathway, revealed new uncertainties about the topology of this pathway in T. brucei. We were nevertheless able to predict relevant behaviours of the system based on the alternative versions of the model (Kerkhoven, Achcar et al. under review). Then, to be able to introduce the trypanothione pathway per se, we again had to clarify some topological questions. We were able to do so using 13C-labelled metabolites and LC-MS. For example, we showed that methylthioadenosine is not recycled to methionine in T. brucei. However, deciphering the origin of ornithine in vivo will require further investigation. In addition to investigating the topology of this pathway, new kinetic parameters of the enzymes involved have to be measured. To reduce our uncertainty about these new kinetics, we designed a new in vivo-like buffer. Using this new assay system and a method based on a strong interaction between modellers and experimentalists, we deciphered in detail the complex mechanism of the trypanothione synthetase, the last enzyme of the trypanothione biosynthesis pathway [10].
Introducing the possibility of dynamic regulation to these pathways at the transcriptional and translational levels requires new sets of parameters that have to be measured using specific methodologies. These regulatory steps will be introduced to the models using a module of 4 differential equations based on the previously published model of PGK transcription and translation [12]. We designed appropriate methodology to measure the parameters needed based on deep sequencing (RNAseq).
All of these new additions introduced many new parameters and additional topological uncertainties. To enable reduction of these uncertainties using a Bayesian methodology, we collected additional data about metabolite concentrations and fluxes, using a new method based on LC-MS data. Intracellular and extracellular concentrations and fluxes were collected in the wild type cell line. In the future, we also plan to collect metabolite concentrations in mutant cell lines to produce new information about the system under perturbation. Therefore, 28 RNAi cell lines targetting critical enzymes in our system were constructed and verified. We will now be able to grow these cell lines in uniform controlled conditions to characterise the metabolomic and transcriptomic response to these perturbations.
This work opens many novel perspectives: our main aim now is to apply our newly developed Bayesian inference methods to our models and data, starting with small models and gradually increasing model size. While the first small models will be analysed, new enzymes kinetics will be acquired, but also the deep sequencing data will be analysed and the RNAi mutants studied. Each additional data-set acquired will allow us to reduce our uncertainty on some part of the model. We argue that the conceptual framework we are developing here to study the metabolism of T. brucei, including the explicit quantitative consideration of topological and parameter uncertainty, can be applied to study any biological system and offers powerful new approaches to enable the robust and predictive simulation of cellular function.
Acknowledgements
This work was supported by the Scottish Universities Life Sciences Alliance (SULSA), the Federal Ministry of Education and Research (Germany), the Netherlands Organisation for Scientific Research (NWO, The Netherlands), and the Biotechnology and Biological Sciences Research Council (BBSRC, United Kingdom), within the framework of the SysMO2-funded SilicoTryp project. BMB received a Rosalind Franklin Fellowship from the University of Groningen.
References
- [1].Barrett MP, Burchmore RJS, Stich A, Lazzari JO, Frasch AC, Cazzulo JJ, Krishna S. The trypanosomiases. Lancet. 2003 Nov;362(9394):1469–1480. doi: 10.1016/S0140-6736(03)14694-6. [DOI] [PubMed] [Google Scholar]
- [2].Barrett MP, Bakker BM, Breitling R. Metabolomic systems biology of trypanosomes. Parasitology. 2010 Aug;137(9):1285–1290. doi: 10.1017/S003118201000017X. [DOI] [PubMed] [Google Scholar]
- [3].Bakker BM, Michels PAM, Opperdoes FR, Westerhoff HV. Glycolysis in bloodstream form Trypanosoma brucei can be understood in terms of the kinetics of the glycolytic enzymes. J. Biol. Chem. 1997 Feb;272(6):3207–3215. doi: 10.1074/jbc.272.6.3207. [DOI] [PubMed] [Google Scholar]
- [4].Bakker BM, Michels PAM, Opperdoes FR, Westerhoff HV. What controls glycolysis in bloodstream form Trypanosoma brucei? J Biol Chem. 1999 May;274(21):14551–14559. doi: 10.1074/jbc.274.21.14551. [DOI] [PubMed] [Google Scholar]
- [5].Bakker BM, Mensonides FI, Teusink B, van Hoek P, Michels PAM, Westerhoff HV. Compartmentation protects trypanosomes from the dangerous design of glycolysis. Proc. Natl. Acad. Sci. U.S.A. 2000 Feb;97(5):2087–2092. doi: 10.1073/pnas.030539197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Helfert S, Estévez AM, Bakker B, Michels P, Clayton C. Roles of triosephosphate isomerase and aerobic metabolism in Trypanosoma brucei. Biochem J. 2001 Jul;357(Pt 1):117–125. doi: 10.1042/0264-6021:3570117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Albert MA, Haanstra JR, Hannaert V, Van Roy J, Opperdoes FR, Bakker BM, Michels PAM. Experimental and in silico analyses of glycolytic flux control in bloodstream form Trypanosoma brucei. J. Biol. Chem. 2005 Aug;280(31):28306–28315. doi: 10.1074/jbc.M502403200. [DOI] [PubMed] [Google Scholar]
- [8].Haanstra JR, van Tuijl A, Kessler P, Reijnders W, Michels PAM, Westerhoff HV, Parsons M, Bakker BM. Compartmentation prevents a lethal turbo-explosion of glycolysis in trypanosomes. Proc. Natl. Acad. Sci. U.S.A. 2008 Nov;105(46):17718–17723. doi: 10.1073/pnas.0806664105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].van Eunen K, Bouwman J, Daran-Lapujade P, Postmus J, Canelas AB, Mensonides FIC, Orij R, Tuzun I, van den Brink J, Smits GJ, van Gulik WM, Brul S, Heijnen JJ, de Winde JH, de Mattos MJT, Kettner C, Nielsen J, Westerhoff HV, Bakker BM. Measuring enzyme activities under standardized in vivo-like conditions for systems biology. FEBS J. 2010 Feb;277(3):749–760. doi: 10.1111/j.1742-4658.2009.07524.x. [DOI] [PubMed] [Google Scholar]
- [10].Leroux AE, Haanstra JR, Bakker BM, Krauth-Siegel RL. Dissecting the catalytic mechanism of Trypanosoma brucei trypanothione synthetase by kinetic analysis and computational modelling. J. Biol. Chem. 2013 Jun; doi: 10.1074/jbc.M113.483289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Siegel TN, Gunasekera K, Cross GAM, Ochsenreiter T. Gene expression in Trypanosoma brucei: lessons from high-throughput RNA sequencing. Trends Parasitol. 2011 Oct;27(10):434–441. doi: 10.1016/j.pt.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Haanstra JR, Stewart M, Luu VD, van Tuijl A, Westerhoff HV, Clayton C, Bakker BM. Control and regulation of gene expression: quantitative analysis of the expression of phosphoglycerate kinase in bloodstream form Trypanosoma brucei. J. Biol. Chem. 2008 Feb;283(5):2495–2507. doi: 10.1074/jbc.M705782200. [DOI] [PubMed] [Google Scholar]
- [13].Vyshemirsky V, Girolami MA. Bayesian ranking of biochemical system models. Bioinformatics. 2008 Mar;24(6):833–839. doi: 10.1093/bioinformatics/btm607. [DOI] [PubMed] [Google Scholar]
- [14].Xu TR, Vyshemirsky V, Gormand A, von Kriegsheim A, Girolami M, Baillie GS, Ketley D, Dunlop AJ, Milligan G, Houslay MD, Kolch W. Inferring signaling pathway topologies from multiple perturbation measurements of specific biochemical species. Sci. Signal. 2010 Mar;3(113):ra20. doi: 10.1126/scisignal.2000517. [DOI] [PubMed] [Google Scholar]
- [15].Achcar F, Kerkhoven EJ, Bakker BM, Barrett MP, Breitling R, The SilicoTryp Consortium Dynamic modelling under uncertainty: The case of Trypanosoma brucei energy metabolism. PLoS Comput Biol. 2012 Jan;8(1):e1002352. doi: 10.1371/journal.pcbi.1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bakker BM, Walsh MC, ter Kuile BH, Mensonides FIC, Michels PAM, Opperdoes FR, Westerhoff HV. Contribution of glucose transport to the control of the glycolytic flux in Trypanosoma brucei. Proc Natl Acad Sci U S A. 1999 Aug;96(18):10098–10103. doi: 10.1073/pnas.96.18.10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Breitling R, Achcar F, Takano E. Modeling challenges in the synthetic biology of secondary metabolism. ACS Synth Biol. 2013 Jul;2(7):373–378. doi: 10.1021/sb4000228. [DOI] [PubMed] [Google Scholar]
- [18].Gualdron-López M, Vapola MH, Miinalainen IJ, Hiltunen JK, Michels PAM, Antonenkov VD. Channel-forming activities in the glycosomal fraction from the bloodstream form of Trypanosoma brucei. PLoS ONE. 2012 Apr;7(4):e34530. doi: 10.1371/journal.pone.0034530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Häusler T, Stierhof YD, Wirtz E, Clayton C. Import of a DHFR hybrid protein into glycosomes in vivo is not inhibited by the folate-analogue aminopterin. J Cell Biol. 1996 Jan;132(3):311–324. doi: 10.1083/jcb.132.3.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Michels RAM, Moyersoen J, Krazy H, Galland N, Herman M, Hannaert V. Peroxisomes, glyoxysomes and glycosomes (review) Mol Membr Biol. 2005 Jan;22(1-2):133–145. doi: 10.1080/09687860400024186. [DOI] [PubMed] [Google Scholar]
- [21].Achcar F, Barrett MP, Breitling R. Explicit consideration of topological and parameter uncertainty gives new insights into a well-established model of glycolysis. 2013 Feb; doi: 10.1111/febs.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Vincent IM, Creek DJ, Burgess K, Woods DJ, Burchmore RJS, Barrett MP. Untargeted metabolomics reveals a lack of synergy between nifurtimox and eflornithine against Trypanosoma brucei. PLoS Negl Trop Dis. 2012;6(5):e1618. doi: 10.1371/journal.pntd.0001618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Vincendeau P, Gobert AP, Daulouède S, Moynet D, Djavad Mossalayi M. Arginases in parasitic diseases. Trends in Parasitology. 2003 Jan;19(1):9–12. doi: 10.1016/s1471-4922(02)00010-7. [DOI] [PubMed] [Google Scholar]
- [24].Creek DJ, Nijagal B, Kim DH, Rojas F, Matthews KR, Barrett MP. Metabolomics guides rational development of a simplified cell culture medium for drug screening against Trypanosoma brucei. Antimicrob. Agents Chemother. 2013 Jun;57(6):2768–2779. doi: 10.1128/AAC.00044-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Martens-Lobenhoffer J, Postel S, Tröger U, Bode-Böger SM. Determination of ornithine in human plasma by hydrophilic interaction chromatography–tandem mass spectrometry. 2007 Aug;855(2):271–275. doi: 10.1016/j.jchromb.2007.04.045. [DOI] [PubMed] [Google Scholar]
- [26].van Eunen K, Kiewiet JAL, Westerhoff HV, Bakker BM. Testing biochemistry revisited: how in vivo metabolism can be understood from in vitro enzyme kinetics. PLoS Comput. Biol. 2012;8(4):e1002483. doi: 10.1371/journal.pcbi.1002483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nolan DP, Voorheis HP. Hydrogen ion gradients across the mitochondrial, endosomal and plasma membranes in bloodstream forms of Trypanosoma brucei solving the three-compartment problem. Eur. J. Biochem. 2000 Aug;267(15):4601–4614. doi: 10.1046/j.1432-1327.2000.01476.x. [DOI] [PubMed] [Google Scholar]
- [28].Thissen JA, Wang CC. Maintenance of internal pH and an electrochemical gradient in Trypanosoma brucei. Exp. Parasitol. 1991 Apr;72(3):243–251. doi: 10.1016/0014-4894(91)90143-k. [DOI] [PubMed] [Google Scholar]
- [29].Moreno B, Urbina JA, Oldfield E, Bailey BN, Rodrigues CO, Docampo R. 31P NMR spectroscopy of Trypanosoma brucei, Trypanosoma cruzi, and Leishmania major. evidence for high levels of condensed inorganic phosphates. J. Biol. Chem. 2000 Sep;275(37):28356–28362. doi: 10.1074/jbc.M003893200. [DOI] [PubMed] [Google Scholar]
- [30].Nolan DP, Voorheis HP. Factors that determine the plasma-membrane potential in bloodstream forms of Trypanosoma brucei. Eur. J. Biochem. 2000 Aug;267(15):4615–4623. doi: 10.1046/j.1432-1327.2000.01477.x. [DOI] [PubMed] [Google Scholar]
- [31].Fuad FAA, Fothergill-Gilmore LA, Nowicki MW, Eades LJ, Morgan HP, McNae IW, Michels PAM, Walkinshaw MD. Phosphoglycerate mutase from Trypanosoma brucei is hyperactivated by cobalt in vitro, but not in vivo. Metallomics. 2011 Dec;3(12):1310–1317. doi: 10.1039/c1mt00119a. [DOI] [PubMed] [Google Scholar]
- [32].Comini MA, Guerrero SA, Haile S, Menge U, Lünsdorf H, Flohé L. Validation of Trypanosoma brucei trypanothione synthetase as drug target. Free Radic. Biol. Med. 2004 May;36(10):1289–1302. doi: 10.1016/j.freeradbiomed.2004.02.008. [DOI] [PubMed] [Google Scholar]
- [33].Oza SL, Ariyanayagam MR, Aitcheson N, Fairlamb AH. Properties of trypanothione synthetase from Trypanosoma brucei. Mol. Biochem. Parasitol. 2003 Sep;131(1):25–33. doi: 10.1016/s0166-6851(03)00176-2. [DOI] [PubMed] [Google Scholar]
- [34].Torrie LS, Wyllie S, Spinks D, Oza SL, Thompson S, Harrison JR, Gilbert IH, Wyatt PG, Fairlamb AH, Frearson JA. Chemical validation of trypanothione synthetase: a potential drug target for human trypanosomiasis. J. Biol. Chem. 2009 Dec;284(52):36137–36145. doi: 10.1074/jbc.M109.045336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fadda A, Färber V, Droll D, Clayton C. The roles of 3′-exoribonucleases and the exosome in trypanosome mRNA degradation. RNA. 2013 Jul;19(7):937–947. doi: 10.1261/rna.038430.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Deneke C, Lipowsky R, Valleriani A. Complex degradation processes lead to non-exponential decay patterns and age-dependent decay rates of messenger RNA. PLoS ONE. 2013;8(2):e55442. doi: 10.1371/journal.pone.0055442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009 Apr;324(5924):218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Annesley TM. Ion suppression in mass spectrometry. Clin. Chem. 2003 Jul;49(7):1041–1044. doi: 10.1373/49.7.1041. [DOI] [PubMed] [Google Scholar]
- [39].Vuckovic D. Current trends and challenges in sample preparation for global metabolomics using liquid chromatography-mass spectrometry. Anal Bioanal Chem. 2012 Jun;403(6):1523–1548. doi: 10.1007/s00216-012-6039-y. [DOI] [PubMed] [Google Scholar]
- [40].Mashego MR, Wu L, Van Dam JC, Ras C, Vinke JL, Van Winden WA, Van Gulik WM, Heijnen JJ. MIRACLE: mass isotopomer ratio analysis of u-13C-labeled extracts. a new method for accurate quantification of changes in concentrations of intracellular metabolites. Biotechnol. Bioeng. 2004 Mar;85(6):620–628. doi: 10.1002/bit.10907. [DOI] [PubMed] [Google Scholar]
- [41].Kiefer P, Portais JC, Vorholt JA. Quantitative metabolome analysis using liquid chromatography-high-resolution mass spectrometry. Anal. Biochem. 2008 Nov;382(2):94–100. doi: 10.1016/j.ab.2008.07.010. [DOI] [PubMed] [Google Scholar]
- [42].Knowles G, Black SJ, Whitelaw DD. Peptidase in the plasma of mice infected with Trypanosoma brucei brucei. Parasitology. 1987 Oct;95(Pt 2):291–300. doi: 10.1017/s0031182000057747. [DOI] [PubMed] [Google Scholar]
- [43].Bossard G, Cuny G, Geiger A. Secreted proteases of trypanosoma brucei gambiense: Possible targets for sleeping sickness control? Biofactors. 2013 Mar; doi: 10.1002/biof.1100. [DOI] [PubMed] [Google Scholar]
- [44].Geiger A, Hirtz C, Bécue T, Bellard E, Centeno D, Gargani D, Rossignol M, Cuny G, Peltier JB. Exocytosis and protein secretion in trypanosoma. BMC Microbiol. 2010;10:20. doi: 10.1186/1471-2180-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Alsford S, Kawahara T, Glover L, Horn D. Tagging a T. brucei RRNA locus improves stable transfection efficiency and circumvents inducible expression position effects. Mol. Biochem. Parasitol. 2005 Dec;144(2):142–148. doi: 10.1016/j.molbiopara.2005.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Cordeiro AT, Thiemann OH, Michels PAM. Inhibition of Trypanosoma brucei glucose-6-phosphate dehydrogenase by human steroids and their effects on the viability of cultured parasites. Bioorg. Med. Chem. 2009 Mar;17(6):2483–2489. doi: 10.1016/j.bmc.2009.01.068. [DOI] [PubMed] [Google Scholar]
- [47].Hanau S, Rippa M, Bertelli M, Dallocchio F, Barrett MP. 6-phosphogluconate dehydrogenase from Trypanosoma brucei. kinetic analysis and inhibition by try-panocidal drugs. Eur. J. Biochem. 1996 Sep;240(3):592–599. doi: 10.1111/j.1432-1033.1996.0592h.x. [DOI] [PubMed] [Google Scholar]
- [48].Xiao Y, McCloskey DE, Phillips MA. RNA interference-mediated silencing of ornithine decarboxylase and spermidine synthase genes in Trypanosoma brucei provides insight into regulation of polyamine biosynthesis. Eukaryotic Cell. 2009 May;8(5):747–755. doi: 10.1128/EC.00047-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Fairlamb AH, Henderson GB, Bacchi CJ, Cerami A. In vivo effects of difluoromethylornithine on trypanothione and polyamine levels in bloodstream forms of Trypanosoma brucei. Mol. Biochem. Parasitol. 1987 Jun;24(2):185–191. doi: 10.1016/0166-6851(87)90105-8. [DOI] [PubMed] [Google Scholar]
- [50].Alsford S, Turner DJ, Obado SO, Sanchez-Flores A, Glover L, Berriman M, Hertz-Fowler C, Horn D. High-throughput phenotyping using parallel sequencing of RNA interference targets in the african trypanosome. Genome Res. 2011 Jun;21(6):915–924. doi: 10.1101/gr.115089.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wendler A, Irsch T, Rabbani N, Thornalley PJ, Krauth-Siegel RL. Glyoxalase II does not support methylglyoxal detoxification but serves as a general trypanothione thioesterase in african trypanosomes. Mol. Biochem. Parasitol. 2009 Jan;163(1):19–27. doi: 10.1016/j.molbiopara.2008.09.005. [DOI] [PubMed] [Google Scholar]
- [52].Comini MA, Krauth-Siegel RL, Flohé L. Depletion of the thioredoxin homologue tryparedoxin impairs antioxidative defence in african trypanosomes. Biochem. J. 2007 Feb;402(1):43–49. doi: 10.1042/BJ20061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Schmidt A, Clayton CE, Krauth-Siegel RL. Silencing of the thioredoxin gene in trypanosoma brucei brucei. Mol. Biochem. Parasitol. 2002 Dec;125(1-2):207–210. doi: 10.1016/s0166-6851(02)00215-3. [DOI] [PubMed] [Google Scholar]
- [54].Ariyanayagam MR, Oza SL, Guther MLS, Fairlamb AH. Phenotypic analysis of trypanothione synthetase knockdown in the african trypanosome. Biochem. J. 2005 Oct;391(Pt 2):425–432. doi: 10.1042/BJ20050911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Huynh TT, Huynh VT, Harmon MA, Phillips MA. Gene knockdown of gamma-glutamylcysteine synthetase by RNAi in the parasitic protozoa Trypanosoma brucei demonstrates that it is an essential enzyme. J. Biol. Chem. 2003 Oct;278(41):39794–39800. doi: 10.1074/jbc.M306306200. [DOI] [PubMed] [Google Scholar]
- [56].Krieger S, Schwarz W, Ariyanayagam MR, Fairlamb AH, Krauth-Siegel RL, Clayton C. Trypanosomes lacking trypanothione reductase are avirulent and show increased sensitivity to oxidative stress. Mol. Microbiol. 2000 Feb;35(3):542–552. doi: 10.1046/j.1365-2958.2000.01721.x. [DOI] [PubMed] [Google Scholar]
- [57].Wilkinson SR, Horn D, Prathalingam SR, Kelly JM. RNA interference identifies two hydroperoxide metabolizing enzymes that are essential to the bloodstream form of the african trypanosome. J. Biol. Chem. 2003 Aug;278(34):31640–31646. doi: 10.1074/jbc.M303035200. [DOI] [PubMed] [Google Scholar]
- [58].Gu X, Reid D, Higham DJ, Gilbert D. Mathematical modelling of polyamine metabolism in bloodstream-form Trypanosoma brucei: an application to drug target identification. PLoS ONE. 2013;8(1):e53734. doi: 10.1371/journal.pone.0053734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Olin-Sandoval V, González-Chávez Z, Berzunza-Cruz M, Martínez I, Jasso-Chávez R, Becker I, Espinoza B, Moreno-Sánchez R, Saavedra E. Drug target validation of the trypanothione pathway enzymes through metabolic modelling. 2012;279(10):1811–1833. doi: 10.1111/j.1742-4658.2012.08557.x. [DOI] [PubMed] [Google Scholar]