Abstract
Background
HNA-3a alloantibodies can cause severe transfusion-related acute lung injury (TRALI). The frequency of the single nucleotide polymorphisms (SNPs) indicative of the two clinically relevant HNA-3a/b antigens are known in many populations. In the present study, we determined the full length nucleotide sequence of common SLC44A2 alleles encoding the choline transporter-like protein-2 (CTL2) that harbors HNA-3a/b antigens.
Study design and methods
A method was devised to determine the full length coding sequence and adjacent intron sequences from genomic DNA by 8 polymerase chain reaction (PCR) amplifications covering all 22 SLC44A2 exons. Samples from 200 African American, 96 Caucasian, 2 Hispanic and 4 Asian blood donors were analyzed. We developed a decision tree to determine alleles (confirmed haplotypes) from the genotype data.
Results
A total of 10 SNPs were detected in the SLC44A2 coding sequence. The non-coding sequences harbored an additional 28 SNPs (1 in the 5’-untranslated region (UTR); 23 in the introns; and 4 in the 3’-UTR). No SNP indicative of a non-functional allele was detected. The nucleotide sequences for 30 SLC44A2 alleles (haplotypes) were confirmed. There may be 66 haplotypes among the 604 chromosomes screened.
Conclusions
We found 38 SNPs, including 1 novel SNP, in 8192 nucleotides covering the coding sequence of the SLC44A2 gene among 302 blood donors. Population frequencies of these SNPs were established for African Americans and Caucasians. Because alleles encoding HNA-3b are more common than non-functional SLC44A2 alleles, we confirmed our previous postulate that African American donors are less likely to form HNA-3a antibodies compared to Caucasians.
Introduction
Transfusion-related acute lung injury (TRALI) is a life-threatening complication1-3 and reportedly the leading cause of transfusion-related mortality. Antibodies against Human Leukocyte Antigens (HLA) class I and class II or Human Neutrophil Antigens (HNAs) are frequently implicated in the pathogenesis of TRALI.4-6 Anti-HNA-3a is more often associated with severe and fatal TRALI reactions than other HNA or HLA antibodies.5,7-9
HNA-3a and HNA-3b antigens are carried on the 68–72 kDa choline transporter-like protein-2 (CTL2).10 CTL2 is a multi-transmembraneous glycoprotein expressed on neutrophils, lymphocytes, platelets, kidney, spleen, placental cells and inner ear cells.7 The SLC44A2 (Solute Carrier Protein 44A2) gene encoding the CTL2 protein is located on chromosome 19p13.2 and consists of 22 exons. There are 2 isoforms of the full length cDNA, which differ only by the amino acids encoded by their respective first exon.11-14 The HNA-3a/b antigens are expressed on the first extracellular loop of CTL2 protein and are caused by the non-synonymous single nucleotide polymorphism (SNP) G>A in exon 7 at position 461 (rs2288904) in the mRNA isoform 1 (NM_020428.3) leading to an amino acid substitution of Arg154 (HNA-3a, SLC44A2*1) to Gln154 (HNA-3b, SLC44A2*2) in protein isoform 1 (NP_065161.3).10,15,16 The Leu153Phe (457C>T; rs147820753) has been reported to yield false negative genotyping results for HNA-3a if the specific primer encompasses this variation.17
The frequency of the SNP indicative of the SLC44A2*1 and SLC44A2*2 alleles is known for Caucasian18,19, African American18, Han Chinese20, Japanese21, Tunisian22, Brazilian23, Thai24,25 and other populations.26-30 Chu et al.31 applied an algorithmic approach to predict HNA antigens from whole genome sequencing (WGS) data. It was possible to impute frequency statistics of known variants unambiguously and explore unknown variants in WGS data.
Using a decision tree, we defined SLC44A2 alleles that were termed “confirmed haplotypes”, if sufficient experimental evidence was established. Most of the remaining putative haplotypes may represent actual alleles, but required further confirmation. In our previous study we postulated that African Americans may be less likely to form an anti-HNA-3a;18 to prove this postulate, we aimed to confirm the lack of any prevalent non-functional SLC44A2 allele (“HNA-3 null”) in the population. We determined the nucleotide sequence of all 22 exons and large adjacent intronic segments and established haplotypes in a cohort of African American and Caucasian blood donors. Our approach allowed detecting non-functional SLC44A2 alleles and proved our previous postulate18 to be correct.
Materials and Methods
Blood samples
Ethylenediaminetetraacetic acid (EDTA) blood samples from 200 African American, 96 Caucasian, 2 Hispanic and 4 Asian blood donors were collected with written informed consent at the NIH Blood Bank and DNA extracted (EZ1 DNA blood kit on the BioRobot EZ1 Workstation; Qiagen, Valencia, CA).
Primers and SLC44A2 gene amplification
The primers (Eurofins MWG Operon; Huntsville, AL) for amplification (Table S1) and sequencing (Table S2) were designed using the Primer3 web resource.32 All 22 exons and their adjacent intronic sequences of the SLC44A2 gene were amplified (DNA Engine Tetrad 2 Peltier Thermal Cycler; Bio-Rad, Hercules, CA) using only 8 reactions (Table S1).
Nucleotide sequencing
The PCR products were purified and sequenced as previously described.18 Nucleotide sequences were aligned (CodonCode Aligner; CodonCode, Dedham, MA) to NCBI RefSeq NC_000019.9 (range 10,713,121..10,755,235) and nucleotide positions defined using the first nucleotide of the coding sequence (CDS) of NM_020428.3 or NP_065161.3.
Determination of alleles
The SLC44A2 alleles (haplotypes) were determined using the Markov Chain based haplotyper MaCH 1.0.33 Briefly, the software starts by randomly generating a pair of haplotypes, compatible with observed genotypes, for each sampled individual. These initial haplotypes are then refined using Hidden Markov Model (HMM)-based iterations that describe the haplotype pair as an imperfect mosaic of the other haplotypes. After a number of iterations, typically 20 to 100, the consensus haplotypes are constructed by merging the haplotypes sampled in each round.
Computational modeling of amino acid substitutions
Polymorphism Phenotyping algorithm (PolyPhen-2),34 Protein Variation Effect Analyzer (PROVEAN)35 and Sorting Tolerant From Intolerant (SIFT)36 were used to predict the functional impact of non-synonymous amino acid substitutions.
Statistical description
95% confidence intervals (CI) for allele frequencies were calculated using the Poisson distribution.37 The Fisher’s exact test was performed to compare the allele (haplotype) frequency distributions between African Americans and Caucasians; because of the multiple testing, we applied the Bonferroni and Bonferroni-Holm38 multiple comparison corrections.
Results
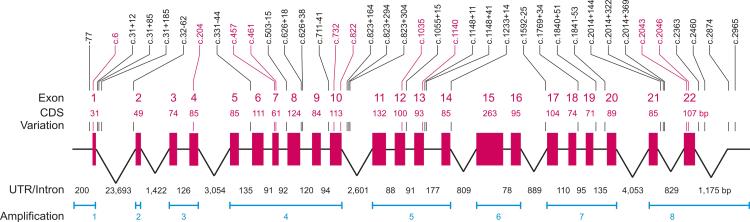
We designed a sequencing strategy and determined the sequence of 2115 nucleotides of the SLC44A2 coding sequence (CDS) in the 22 exons, 200 bp of the 5’-UTR, 1175 bp of the 3’-UTR and 4702 bp of the 21 introns adjacent to the exons (Fig. 1). More than 2 million nucleotides were sequenced in a random survey using 302 blood donor samples to describe the genetic variability of the SLC44A2 gene and define a large set of its prevalent alleles.
Figure 1. Schematic representation of the SLC44A2 gene.
The SLC44A2 gene is located at chromosome 19p13.1 and consists of 22 exons encoding a protein of 706 amino acids. The exons (red boxes) are shown schematically along with their 5’-UTR, 3’-UTR and 21 introns (black lines). The locations of 8 amplification reactions covering at least the complete SLC44A2 coding sequence (CDS) are indicated (blue). All exons and introns smaller than 200 base pair (bp) are at the same scale. Introns longer than 200 bp are scaled logarithmically. The positions of the 38 variations (SNPs) are indicated (│) in the exons (red) and introns (black).
Variations in SLC44A2 gene
In the 8192 nucleotides of the SLC44A2 gene, which were sequenced, we observed a total of 38 SNPs (Fig. 1). Among these variations, 10 were in the CDS, 1 in 5’-UTR, 4 in 3’-UTR and 23 in the 21 introns (Table S3 and Table S4). Two of the variations in CDS were non-synonymous while 8 were synonymous changes. Only 1 variation located in the 5’-UTR was novel and not present in the dbSNP or NHLBI Exome Sequencing Project (ESP) databases. No non-sense or splice site mutation was detected in the analyzed samples.
Genotype patterns
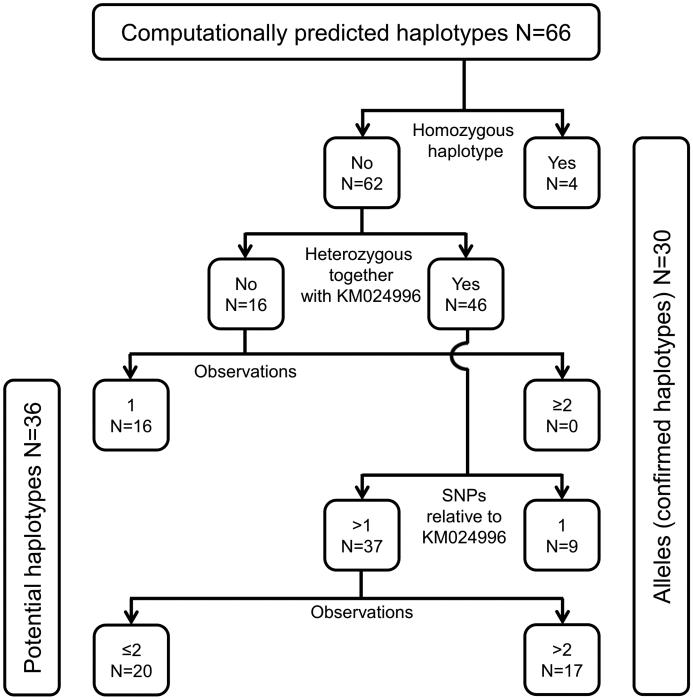
We tabulated the genotype, which comprises the combination of two haplotypes, for all 302 donors (Table S6). Overall, 107 distinct genotype patterns were found. Based on this genotype information, the predicted haplotypes were computed by a software program (MaCH 1.0). We used a decision tree to determine confirmed haplotypes (SLC44A2 alleles) and potential haplotypes (Fig. 2).
Figure 2. Decision tree to determine SLC44A2 alleles.
The computationally predicted haplotypes (alleles) were sorted according to 4 criteria. SLC44A2 alleles (confirmed haplotypes) need to fulfill at least one of the following criteria: 1) homozygous observation, 2) observation in at least 2 samples, 3) observed at least once heterozygously together with KM024996 and harboring only 1 SNP relative to KM024996 or 4) observed at least twice heterozygously together with KM024996. All other haplotypes were considered potential haplotypes requiring additional confirmatory testing. KM024996 is the prevalent SLC44A2 allele (wild type, see Table 1). Data is shown for all haplotypes computed for the 302 donors.
SLC44A2 alleles
Applying the decision tree rules (Fig. 2), 30 SLC44A2 alleles were confirmed which allowed to calculate the allele frequencies in the populations (Table 1).37 Beside the prevalent SLC44A2*1 allele KM024996, the alleles KM024997, KM024998 and KM025025 were observed in homozygous form providing direct sequence evidence. One SNP each identified 9 alleles that occured heterozygously together with KM024996. Other evidence allowed to confirm the remaining 17 alleles for a total of 30 alleles (Fig. 2). An additional 36 haplotypes were computed which, however, would require more observations or nucleotide sequence analysis for confirmation (Table S5).
Table 1.
SLC44A2 alleles (confirmed haplotypes) and their frequencies in African Americans and Caucasians
| Allele number | Allele (haplotype)* | GenBank No. | Allele frequency | |||||
|---|---|---|---|---|---|---|---|---|
| African American | Caucasian | |||||||
| Observed (n) | Mean† | 95% CI‡ | Observed (n) | Mean† | 95% CI‡ | |||
| 1 | TGCGCCTGCGTTTCGCTTTACCCCGCCCCCTGGCTGGT | KM024996 | 219¶ | 54.8% | 47.4-62.3 | 76¶ | 39.6% | 30.6-49.1 |
| 2 | ------CA-ACCG---CC--T------T-------A-G | KM024997 | 10¶ | 2.5% | 1.3-4.4 | 25¶ | 13% | 8.7-18.8 |
| 3 | ---A--CA-ACCG---CC--T------ T-------A-G | KM024998 | 3¶ | 0.8% | 0.2-2.0 | 11¶ | 5.7% | 2.8-9.9 |
| 4 | ------CA--CCG---CC-----AA--T-------A-G | KM024999 | 16 | 4% | 2.4-6.3 | 1 | 0.5% | 0.03-2.8 |
| 5 | ------CA--CCG----C-----AA--T-------A-G | KM025000 | 4 | 1% | 0.3-2.4 | 0 | 0% | 0-1.7 |
| 6 | -A----C---CCG----C---------T-T-----A-- | KM025001 | 4 | 1% | 0.3-2.4 | 0 | 0% | 0-1.7 |
| 7 | ----T---------------T------T-------A-G | KM025002 | 6¶ | 1.5% | 0.6-3.2 | 22¶ | 11.5% | 7.2-16.8 |
| 8 | --------------------T------T-------A-G | KM025003 | 2 | 0.5% | 0.09-1.7 | 2 | 1% | 0.18-2.9 |
| 9 | --------------------T-------A-G | KM025004 | 6 | 1.5% | 0.6-3.2 | 0 | 0% | 0-1.7 |
| 10 | ------------------CG-TA---------A--A-- | KM025005 | 16 | 4% | 2.4-6.3 | 0 | 0% | 0-1.7 |
| 11 | ------------------CG-TA---------A-CA-- | KM025006 | 4 | 1% | 0.3-2.4 | 0 | 0% | 0-1.7 |
| 12 | -------------------G-TA---------A--A-- | KM025007 | 5 | 1.2% | 0.5-2.8 | 0 | 0% | 0-1.7 |
| 13 | ---------------------------T--C-A--A-- | KM025008 | 6 | 1.5% | 0.6-3.2 | 0 | 0% | 0-1.7 |
| 14 | ----T--------------------T-T-------A-- | KM025009 | 0¶ | 0% | 0-0.8 | 7¶ | 3.6% | 1.7-7.2 |
| 15 | -------------------------T-T-------A-- | KM025010 | 7 | 1.8% | 0.8-3.4 | 2 | 1% | 0.18-2.9 |
| 16 | ---------------------------T-------A-- | KM025011 | 6 | 1.5% | 0.6-3.2 | 1 | 0.5% | 0.03-2.8 |
| 17 | --T-T--------------------------------- | KM025012 | 1 | 0.2% | 0.01-1.3 | 3 | 1.6% | 0.4-4.2 |
| 18 | ----T-----------------------G--------- | KM025013 | 4 | 1% | 0.3-2.4 | 7 | 3.6% | 1.7-7.2 |
| 19 | -----T-------------------------A------ | KM025014 | 7 | 1.8% | 0.8-3.4 | 1 | 0.5% | 0.03-2.8 |
| 20 | --------------------------T---------A- | KM025015 | 5 | 1.2% | 0.5-2.8 | 0 | 0% | 0-1.7 |
| 21 | A------------------------------------- | KM025016 | 3 | 0.8% | 0.2-2.0 | 0 | 0% | 0-1.7 |
| 22 | -A------------------------------------ | KM025017 | 9 | 2.2% | 1.1-4.2 | 0 | 0% | 0-1.7 |
| 23 | ---A---------------------------------- | KM025018 | 4 | 1% | 0.3-2.4 | 1 | 0.5% | 0.03-2.8 |
| 24 | ----T----- ---------------------------- | KM025019 | 3¶ | 0.8% | 0.2-2.0 | 15¶ | 7.8% | 4.2-12.4 |
| 25 | -----T-------------------------------- | KM025020 | 1 | 0.2% | 0.01-1.3 | 0 | 0% | 0-1.7 |
| 26 | --------T---------------------------- | KM025021 | 1 | 0.2% | 0.01-1.3 | 2 | 1% | 0.18-2.9 |
| 27 | ------------------C------------------ | KM025022 | 3 | 0.8% | 0.2-2.0 | 0 | 0% | 0-1.7 |
| 28 | ----------------------------G--------- | KM025023 | 2 | 0.5% | 0.09-1.7 | 3 | 1.6% | 0.4-4.2 |
| 29 | -----------------------------------A-- | KM025024 | 2 | 0.5% | 0.09-1.7 | 0 | 0% | 0-1.7 |
| 30 | ------------------------------------A- | KM025025 | 5 | 1.2% | 0.5-2.8 | 7 | 3.6% | 1.7-7.2 |
| Potential haplotypes | See Table S5 | 36 | 6 | |||||
| Total | 400 | 192 | ||||||
The nucleotides at the 38 SNP positions with variations are shown in 5‘ to 3‘ orientation (see Figure 1 and Table S6). The remaining 8131 nucleotide positions that we determined had no variation relative to the reference sequence KM024996. The SNP G/A (bold) determines the HNA-3a/b antigen polymorphism.
Number of observed alleles (n)/Total number of alleles (African American=400 and Caucasian=192).
95% confidence interval (CI), Poisson distribution, two sided.37
Statistically significant difference by the Fisher’s exact test, two sided (p<0.0017); Bonferroni multiple comparison correction, n=30, 0.05/30=0.0017. The same alleles show statistically significant differences when the Bonferroni-Holm correction is applied.
The 2 confirmed alleles KM024997 and KM024998 were encoding HNA-3b (Table 1) while 2 of the 36 potential haplotypes were encoding HNA-3b (Table S5). The HNA-3b variant was found to occur only with a signature haplotype “C-A-C-C-G” consisting of c.204C, c.331-44A, c.503-15C, c.626+18C and c.626+38G variants distributed over 3825 nucleotides.
Population frequencies
We calculated the allele frequencies (Table 1) and the SNP frequencies (Table S3 and S4) in both of our cohorts. The allele frequency of 6 alelles were statistically significant between African American and Caucasians (Table 1). With the exception of 1 SNP (rs74795234) in Caucasians, the remaining 37 SNPs were in accordance with the Hardy–Weinberg equilibrium (HWE). We compared our SNP frequencies with the data from the NHLBI Exome Sequencing Project (ESP) (Table 2).39
Table 2.
Exonic SNPs identified in the current study and the Exome Sequencing Project (ESP)*
| SNPs detected |
Nucleotide read length (base pairs) |
Total number of nucleotides sequenced (base pairs) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Exons |
Individuals tested |
||||||||||
| Study | Non- synonymous |
Synonymous | All | Intron | Total | Exons | Introns | Exons | Introns | Total | |
| Actual number | |||||||||||
| Current study | 2 | 8 | 10 | 23 | 33 | 302 | 2115 | 4635 | 638730 | 1399770 | 2028500 |
| ESP | 51 | 35 | 86 | 85 | 171 | 6503 | 2115 | 2100 | 13753845 | 13656300 | 27410145 |
| Normalized SNP detection rate† | |||||||||||
| Current study | 1.00 | 4.00 | 5.00 | 5.25 | 5.19 | ||||||
| ESP | 1.18 | 0.81 | 1.99 | 1.99 | 1.99 | ||||||
NHLBI Exome Sequencing Project39
Normalized SNP detection rate: Number of SNPs detected × (319365 nucleotides/number of nucleotides sequenced)
Non-functional alleles
No SNP encoding a non-sense mutation and no deletion or insertion encoding a frame-shift mutation was observed. Non-functional alleles occured with less than 1 in 122 chromosomes (<0.82%) in African Americans (<1 in 58 chromosomes and <1.71% in Caucasians; upper limit of 95% confidence interval, Poisson distribution).37
Effect on protein structure
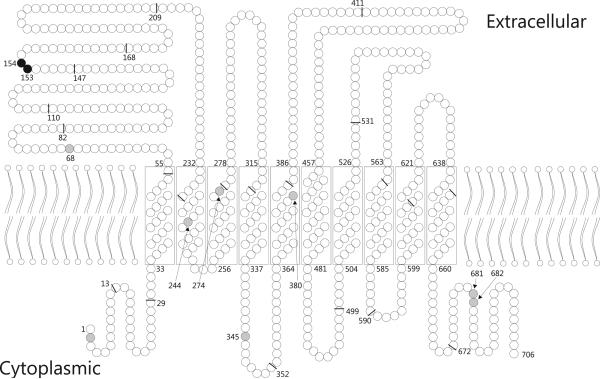
The Arg154Gln (rs2288904) amino acid substitution responsible for the HNA-3a/b polymorphism is located in the first extracellular loop of the mature CTL2 protein (Fig. 3).15 Computational modeling predicted structural changes induced by the Arg154Gln substitution to be neutral (Table 3). However, Leu153Phe (rs147820753) showed inconsistant predictions with the 3 modeling tools.
Figure 3. Schematic model of the CTL2 protein.
There are 2 non-synonymous variant (black circles) and 8 synonymous variant positions (grey circles). The exon boundaries in the cDNA, as reflected in the amino acid sequence, are indicated (black bars).
Table 3.
Amino acid substitution and predicted effect on protein structure
| Bioinformatics program and computational analysis results |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Variant |
Protein |
PolyPhen2 † |
PROVEAN ‡ |
SIFT ¶ |
|||||
| dbSNP reference no |
Nucleotide change | Amino acid substitution * |
Classifi- cation |
Score | Classifi- cation |
Score | Classifi- cation |
Score | MIC |
| rs147820753 | c.457C>T | Leu153Phe | benign | 0.092 | neutral | −2.45 | damaging | 0.02 | 2.62 |
| rs2288904 | c.461G>A | Arg154Gln | benign | 0.0 | neutral | 0.51 | tolerated | 1.00 | 2.62 |
relative to NCBI Reference Sequence NP_065161.3
score 0.00–0.452 = benign, 0.453–0.956 = possibly damaging, 0.957–1.00 = probably damaging
score >−2.5 = neutral, score ≤−2.5 = deleterious
score ≤0.05 = damaging, >0.05 = tolerated; MIC = median sequence information (range 0 to 4.32)
Asian and Hispanic
The 2 Hispanic samples were both heterozygous for the alleles KM024996 + KM024497 and KM024996 + KM025018. Two out of 4 Asian samples were homozygous for the prevalent KM024996 allele, while the other 2 samples were heterozygous for the alleles KM024996 + KM024497.
Discussion
The aim of this study was to determine common alleles of the SLC44A2 gene in African American and Caucasian populations. We sequenced 8192 nucleotides of the SLC44A2 gene and identified a number of recurrent alleles (haplotypes) in both populations. This is the first study to systematically categorize the various SNPs known to occur in the SLC44A2 gene into a large set of prevalent alleles. The frequencies of some SLC44A2 alleles differed significantly between the 2 populations. We excluded the possibility that non-functional SLC44A2 alleles occur frequently in African Americans; their combined frequency is less than 1 in 122 chromosomes (upper limit of the 95% confidence interval according to the Poisson distribution).
We developed a sensitive and specific screening assay to genotype the SLC44A2 gene in a high-throughput setting. Our decision tree can be applied to any population.30 A computational approach to predict HNA3 antigens from next generation sequencing or WGS data31 will require a method for detecting carriers of low prevalence alleles. This can be best achieved if detailed haplotype information, particularly for confirmed alleles, is known. Our present results will aid in such studies.
Based on the 38 analyzed SNPs, the algorithm of the MaCH program computed a total of 60 haplotypes in African Americans and 22 haplotypes in Caucasians. Applying a decision tree (Fig. 2), we confirmed 30 SLC44A2 alleles (Table 1). Most of the remaining 36 potential SLC44A2 haplotypes (Table S5) are probably also extant; due to the intrinsic limitations of the haplotype computation, we would recommend confirmatory testing before entering the latter 36 haplotypes in definite allele databases. The common wild type KM024996 allele accounted for approximately half of the chromosomes (Table 1). A large number of low frequency alleles were due to one or two rare SNPs in the KM024996 allele (Table 1, alleles no. 17 to 30).
The total number of haplotypes computationally observed in African Americans was greater than in Caucasians. Alleles shared in both populations (16/66=24%), considered to be the oldest haplotypes, account for 93% of those occuring in Caucasians but only for 74% in African Americans. These observations were compatible with the out-of-Africa bottleneck hypothesis (serial founder effects) and proposed admixture between Neanderthals and modern humans in Eurasia.40,41
The dbSNP database42 lists more than 1000 variations for the SLC44A2 gene while NHLBI Exome Sequencing Project (ESP)39 identifies 108 variations in Caucasians and 98 variations in African Americans. In the present study, we observed 38 variations in the African American cohort and 23 variations in the Caucasian cohort. The variations described in dbSNP or NHLBI ESP databases, not observed in our study, may be rare variants that our screening panel did not have adequate power to detect, or were not polymorphic in the populations we studied. We also did not detect any variant associated with a non-functional SLC44A2 allele in our study, which is consistent with the fact that no such allele has been reported in the literature or dbSNP database. However, one non-sense mutation c.1885C>T (p.Gln629Ter) has been reported in a renal cell carcinoma (sample name TCGA-A3-3365-01; Catalogue of somatic mutations in cancer (COSMIC) database).43
Our observed SLC44A2 variant frequencies (Table S3 and S4) were comparable to the NHLBI ESP project data,39 but the normalized SNP detection rate for synonymous variants was not (Table 2): our rate for synonymous substitutions was higher than for non-synonymous substitutions, a discrepancy not observed in the NHLBI ESP study. However, the difference in the overall SNP detection rate (5.19 in this study vs. 1.99 in NHLBI ESP) was explained by the fact that increasing the sample size as well as decreasing the length of the analyzed nucleotide sequence are expected to yield diminishing returns of additional SNPs.
The common CTL2 protein structure has been predicted by computer simulation.12 Using a similar approach, we applied 3 common bioinformatic tools to predict the effect of amino acid substitutions on the protein structure. The 3 tools gave a consistent prediction for the Arg154Gln variant but an inconsistent one for the Leu153Phe variant (Table 3). This exemplified that bioinformatic pathogenicity predictions for non-synonymous variations are not always reliable and should be interpreted with caution especially when the actual 3-dimensional protein structure is unknown, as it is in the case for the CTL2 protein.
The 2 alleles KM024997 and KM024998, encoding HNA-3b and known to be associated with TRALI, were more frequently observed in Caucasians than African Americans. We did not detect any non-functional SLC44A2 alleles in either population. Hence, we proved that African American donors are less likely than Caucasian donors to induce TRALI in transfusion recipients due to donor-related HNA-3a antibodies, as had been postulated before.18
Supplementary Material
Acknowledgments
QC was supported by a grant “Jiangsu Health International Exchange Program”, by grant no. RC2011088 from the Jiangsu Province Medical Elite Program, and by grant no. 81101734 from the National Natural Science Foundation of China. Part of the study was done at NIH in 2012 by QC as Visiting Senior Research Scientist, by SA in partial fulfillment of her study of molecular medicine (University of Ulm), and by KL as part of his Summer Internship program. We thank Franz F. Wagner for support with the statistics; Harvey G. Klein for critical review; Sherry L. Sheldon and Sharon D. Adams for sample coordination; and the staff of the HLA laboratory in the Laboratory Services Section for DNA extraction.
This research was supported by the Intramural Research Program of the NIH Clinical Center.
Footnotes
Conflict of interest disclosure: The authors have no competing interests relevant to this article.
Statement of Disclaimer: The views expressed do not necessarily represent the view of the National Institutes of Health, the Department of Health and Human Services, or the U.S. Federal Government.
Authorship contribution: PS and WAF conceived the study. Initial experiments were designed and performed by MJH and PS. The experimental part of this study was completed by QC, KS, SA and KL. Data were analyzed by QC, KS and WAF, who also wrote the manuscript.
Web resources
COSMIC database (http://cancer.sanger.ac.uk/cosmic)
dbSNP database, Build ID: 138 Phase I (http://www.ncbi.nlm.nih.gov/SNP/)
Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP; 09, 2013) (http://evs.gs.washington.edu/EVS/)
International Society Blood Transfusion (ISBT) (http://www.isbtweb.org/)
MaCH (http://csg.sph.umich.edu/abecasis/MACH/)
PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/)
Primer3 software, version 4.0.0 (http://bioinfo.ut.ee/primer3-0.4.0/)
PROVEAN (http://provean.jcvi.org/seq_submit.php)
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1. SLC44A2 amplification primers and PCR conditions
Table S2. SLC44A2 sequencing primers
Table S3. Genetic variations identified in the SLC44A2 exons
Table S4. Genetic variations detected in the SLC44A2 introns
Table S5. SLC44A2 potential haplotypes and their frequencies in African Americans and Caucasians
Table S6. SLC44A2 genotypes observed in African American, Caucasian, Hispanic and Asian samples
References
- 1.Barnard RD. Indiscriminate transfusion: a critique of case reports illustrating hypersensitivity reactions. N Y State J Med. 1951;51:2399–402. [PubMed] [Google Scholar]
- 2.Popovsky MA, Moore SB. Diagnostic and pathogenetic considerations in transfusion-related acute lung injury. Transfusion. 1985;25:573–7. doi: 10.1046/j.1537-2995.1985.25686071434.x. [DOI] [PubMed] [Google Scholar]
- 3.Webert KE. Splitting versus lumping: reconsidering the definition of transfusion-related acute lung injury. Transfusion. 2015;55:927–9. doi: 10.1111/trf.13067. [DOI] [PubMed] [Google Scholar]
- 4.Middelburg RA, Van Stein D, Briët E, et al. The role of donor antibodies in the pathogenesis of transfusion-related acute lung injury: a systematic review. Transfusion. 2008;48:2167–76. doi: 10.1111/j.1537-2995.2008.01810.x. [DOI] [PubMed] [Google Scholar]
- 5.Reil A, Keller-Stanislawski B, Gunay S, et al. Specificities of leucocyte alloantibodies in transfusion-related acute lung injury and results of leucocyte antibody screening of blood donors. Vox Sang. 2008;95:313–7. doi: 10.1111/j.1423-0410.2008.01092.x. [DOI] [PubMed] [Google Scholar]
- 6.Peters AL, Van Stein D, Vlaar AP. Antibody-mediated transfusion-related acute lung injury; from discovery to prevention. Br J Haematol. 2015 doi: 10.1111/bjh.13459. doi: 10.1111/bjh.13459. [DOI] [PubMed] [Google Scholar]
- 7.Davoren A, Curtis BR, Shulman IA, et al. TRALI due to granulocyte-agglutinating human neutrophil antigen-3a (5b) alloantibodies in donor plasma: a report of 2 fatalities. Transfusion. 2003;43:641–5. doi: 10.1046/j.1537-2995.2003.00374.x. [DOI] [PubMed] [Google Scholar]
- 8.Kopko PM, Marshall CS, MacKenzie MR, et al. Transfusion-related acute lung injury: Report of a clinical look-back investigation. JAMA. 2002;287:1968–71. doi: 10.1001/jama.287.15.1968. [DOI] [PubMed] [Google Scholar]
- 9.Silliman CC, Curtis BR, Kopko PM, et al. Donor antibodies to HNA-3a implicated in TRALI reactions prime neutrophils and cause PMN-mediated damage to human pulmonary microvascular endothelial cells in a two-event in vitro model. Blood. 2007;109:1752–5. doi: 10.1182/blood-2006-05-025106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greinacher A, Wesche J, Hammer E, et al. Characterization of the human neutrophil alloantigen-3a. Nat Med. 2010;16:45–8. doi: 10.1038/nm.2070. [DOI] [PubMed] [Google Scholar]
- 11.O'Regan S, Traiffort E, Ruat M, et al. An electric lobe suppressor for a yeast choline transport mutation belongs to a new family of transporter-like proteins. Proc Natl Acad Sci U S A. 2000;97:1835–40. doi: 10.1073/pnas.030339697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nair TS, Kozma KE, Hoefling NL, et al. Identification and characterization of choline transporter-like protein 2, an inner ear glycoprotein of 68 and 72 kDa that is the target of antibody-induced hearing loss. J Neurosci. 2004;24:1772–9. doi: 10.1523/JNEUROSCI.5063-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kommareddi PK, Nair TS, Thang LV, et al. Isoforms, expression, glycosylation, and tissue distribution of CTL2/SLC44A2. Protein J. 2010;29:417–26. doi: 10.1007/s10930-010-9268-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beyer LA, Galano MM, Nair TS, et al. Age-related changes in expression of CTL2/SLC44A2 and its isoforms in the mouse inner ear. Hear Res. 2011;282:63–8. doi: 10.1016/j.heares.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curtis BR, Cox NJ, Sullivan MJ, et al. The neutrophil alloantigen HNA-3a (5b) is located on choline transporter-like protein 2 and appears to be encoded by an R>Q154 amino acid substitution. Blood. 2010;115:2073–6. doi: 10.1182/blood-2009-11-248336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bougie DW, Peterson JA, Kanack AJ, et al. Transfusion-related acute lung injury-associated HNA-3a antibodies recognize complex determinants on choline transporter-like protein 2. Transfusion. 2014;54:3208–15. doi: 10.1111/trf.12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flesch BK, Reil A, Bux J. Genetic variation of the HNA-3a encoding gene. Transfusion. 2011;51:2391–7. doi: 10.1111/j.1537-2995.2011.03155.x. [DOI] [PubMed] [Google Scholar]
- 18.Huvard MJ, Schmid P, Stroncek DF, et al. Frequencies of SLC44A2 alleles encoding human neutrophil antigen-3 variants in the African American population. Transfusion. 2012;52:1106–11. doi: 10.1111/j.1537-2995.2011.03396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cardoso SP, Chong W, Lucas G, et al. Determination of human neutrophil antigen-1, -3, -4 and -5 allele frequencies in English Caucasoid blood donors using a multiplex fluorescent DNA-based assay. Vox Sang. 2013;105:65–72. doi: 10.1111/vox.12016. [DOI] [PubMed] [Google Scholar]
- 20.He J, Zhang W, Wang W, et al. Genotyping of human neutrophil antigens by polymerase chain reaction sequence-based typing. Blood Transfus. 2014;12(Suppl 1):s292–8. doi: 10.2450/2013.0308-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuhashi M, Tsuno NH, Kawabata M, et al. The frequencies of human neutrophil alloantigens among the Japanese population. Tissue Antigens. 2012;80:336–40. doi: 10.1111/j.1399-0039.2012.01930.x. [DOI] [PubMed] [Google Scholar]
- 22.Abid S, Zili M, Bouzid L, et al. Gene frequencies of human neutrophil antigens in the Tunisian blood donors and Berbers. Tissue Antigens. 2001;58:90–2. doi: 10.1034/j.1399-0039.2001.580204.x. [DOI] [PubMed] [Google Scholar]
- 23.Norcia AM, Sugano EY, Chiba AK, et al. Human neutrophil alloantigen-1a, -1b, -2, -3a and -4a frequencies in Brazilians. Tissue Antigens. 2009;74:404–7. doi: 10.1111/j.1399-0039.2009.01357.x. [DOI] [PubMed] [Google Scholar]
- 24.Changsri K, Tobunluepop P, Songthammawat D, et al. Human neutrophil alloantigen genotype frequencies in Thai blood donors. Blood Transfus. 2014;12(Suppl 1):s286–91. doi: 10.2450/2013.0161-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nathalang O, Intharanut K, Siriphanthong K, et al. Risk estimation of HNA-3 incompatibility and alloimmunization in Thai populations. PLoS ONE. 2015;10:e0116905. doi: 10.1371/journal.pone.0116905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hauck B, Philipp A, Eckstein R, et al. Human neutrophil alloantigen genotype frequencies among blood donors with Turkish and German descent. Tissue Antigens. 2011;78:416–20. doi: 10.1111/j.1399-0039.2011.01779.x. [DOI] [PubMed] [Google Scholar]
- 27.Bowens KL, Sullivan MJ, Curtis BR. Determination of neutrophil antigen HNA-3a and HNA-3b genotype frequencies in six racial groups by high-throughput 5' exonuclease assay. Transfusion. 2012;52:2368–74. doi: 10.1111/j.1537-2995.2012.03600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen KR, Koelbaek MD, Varming K, et al. Frequencies of HNA-1, HNA-3, HNA-4, and HNA-5 in the Danish and Zambian populations determined using a novel TaqMan real time polymerase chain reaction method. Tissue Antigens. 2012;80:249–53. doi: 10.1111/j.1399-0039.2012.01912.x. [DOI] [PubMed] [Google Scholar]
- 29.Lopes LB, Baleotti W, Jr., Suzuki RB, et al. HNA-3 gene frequencies in Brazilians and a new polymerase chain reaction-restriction fragment length polymorphism method for HNA-3a/3b genotyping. Transfusion. 2014;54:1619–21. doi: 10.1111/trf.12493. [DOI] [PubMed] [Google Scholar]
- 30.Chen Q, Liu Z, Xiao J, et al. Genotype frequencies of Human Neutrophil Antigen-3 (HNA-3) polymorphisms in the Han, Tibetan, and Yi populations of China. Transfusion. 2014;54(Suppl. 2):217A–8A. doi: 10.1111/trf.13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chu HT, Lin H, Tsao TT, et al. Genotyping of human neutrophil antigens (HNA) from whole genome sequencing data. BMC Med Genomics. 2013;6:31. doi: 10.1186/1755-8794-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Untergasser A, Cutcutache I, Koressaar T, et al. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Willer CJ, Ding J, et al. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Meth. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi Y, Sims GE, Murphy S, et al. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–74. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sachs L. Angewandte Statistik - Anwendung statistischer Methoden. 7th Springer-Verlag; Berlin: 1992. pp. 446–7. [Google Scholar]
- 38.Holm S. A simple sequentially rejective multiple test procedure. Scand J Statistics. 1979;6:65–70. [Google Scholar]
- 39.Tennessen JA, Bigham AW, O’Connor TD, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–9. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Labuda D, Yotova V, Lefebvre J-F, et al. X-linked MTMR8 diversity and evolutionary history of Sub-Saharan populations. PLoS ONE. 2013;8:e80710. doi: 10.1371/journal.pone.0080710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gomez F, Hirbo J, Tishkoff SA. Genetic variation and adaptation in Africa: Implications for human evolution and disease. Cold Spring Harbor Perspectives in Biology. 2014;6 doi: 10.1101/cshperspect.a008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sherry ST, Ward M-H, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forbes SA, Beare D, Gunasekaran P, et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–11. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.