Abstract
The current era of targeted treatment has accelerated the interest in studying gene-treatment, gene-gene and gene-environment interactions using statistical models in the health sciences. Interactions are incorporated into models as product terms of risk factors. The statistical significance of interactions is traditionally examined using a likelihood ratio test (LRT). Epidemiological and clinical studies also evaluate interactions in order to understand the prognostic and predictive values of genetic factors. However, it is not clear how different types and magnitudes of interaction effects are related to prognostic and predictive values. The contribution of interaction to prognostic values can be examined via improvements in the area under the receiver operating characteristic curve due to the inclusion of interaction terms in the model (ΔAUC). We develop a resampling based approach to test the significance of this improvement and show that it is equivalent to LRT. Predictive values provide insights into whether carriers of genetic factors benefit from specific treatment or preventive interventions relative to non-carriers, under some definition of treatment benefit. However, there is no unique definition of the term treatment benefit. We show that DeltaAUC and relative excess risk due to interaction (RERI) measure predictive values under two specific definitions of treatment benefit. We investigate the properties of LRT, ΔAUC and RERI using simulations and illustrate these approaches using published data on MC1R and sun exposure in melanoma.
Keywords: area under the receiver operating characterisitc curve, likelihood ratio test, relative excess risk due to interaction (RERI), resampling, treatment benefit
Introduction
During the past decade, the field of oncology has made considerable advances in the discovery of cancer-related genes [Sawyers 2008]. These successes have led to an explosion of research on preventive interventions and anticancer therapies focused on specific patient populations, and have led to the hypothesis that carriers of certain genetic variants are more likely to benefit from specialized treatments or interventions than non-carriers [Lerman et al. 2007; Amado et al. 2008; Karapetis et al. 2008]. This hypothesis has propelled investigations of targeted treatments through studies of gene-treatment, gene-environment, and gene-gene interactions using statistical models. One of the key objectives of these studies is to understand the prognostic and predictive values of a genetic factor such as a germline genetic variant or a somatic mutation [Sawyers 2008]. The objective of this paper is to examine the relationship between interaction terms in statistical models (henceforth referred to as statistical interactions or, simply, interactions) and prognostic and predictive values.
The term prognostic value refers to a genetic factor’s ability to project the natural history of disease in relation to another factor (such as treatment or environmental exposure or another genetic factor; henceforth referred to as treatment) by discriminating between good versus bad prognosis, thereby providing insights into whom to treat with novel modalities [Sawyers 2008; Italiano 2011]. The prognostic value is measured as the area under the receiver operating characteristic curve (AUC; [Pepe et al. 2013]) of a statistical model that includes the genetic factor and treatment as explanatory variables. An important issue is whether the effect of treatment on the outcome changes according to the level of the genetic factor of interest, and how this change impacts the prognostic value. Changes in treatment effects according to a genetic factor are incorporated into statistical models as gene-treatment interaction terms. The issue then is to determine whether incorporating gene-treatment interactions into a statistical model improves the prognostic value.
Often we use the likelihood ratio test (LRT) to examine the significance of interaction terms in statistical models [Agresti 2002]. When there are no interactions, we say that the model is additive. The presence of interactions depends upon the scale on which disease risk is measured [Satagopan and Elston 2013]. If interactions exist when modeling data under a clinically meaningful scale of risk, then including them in the model is anticipated to improve the prognostic value of a genetic factor. This is equivalent to the model’s ability to explain the variation in disease risk and, thus, discriminate between low-risk and high-risk individuals [for example, Khoury et al. 2004; Moore and Williams 2009; Sun et al. 2014]. This improvement is measured as the difference between the AUCs of two models: an additive model and a model including interaction terms. In this paper, we denote this difference as ΔAUC. Some studies noted that the inclusion of interactions did not lead to a considerable increase in the magnitude of ΔAUC [Aschard et al. 2012]. This raises several issues: (i) When an interaction is statistically significant, should its inclusion in a model be expected to improve ΔAUC by a considerable magnitude? (ii) Would a small ΔAUC be statistically significant when there is a statistically significant interaction? and (iii) What is the role of interaction on prognostic values? In this paper, we examine these questions by comparing the operating characteristics of LRT with those of a test for H0 : ΔAUC = 0 against HA : ΔAUC ≠ 0. Since an additive model is nested within a model that includes interactions, the DeLong test for H0 [DeLong et al. 1988] is not valid [Vickers et al. 2011]. Other recent studies have proposed valid tests for ΔAUC when evaluating the incremental increase in AUC due to the inclusion of a new biomarker, without focusing on interactions in statistical models [Pepe et al. 2013; Seshan et al. 2013]. These tests are not applicable to our setting where the focus is on interactions. Therefore, we develop a novel resampling procedure for testing H0 : ΔAUC = 0 in the interaction setting.
The term predictive value refers to a genetic factor’s ability to project the benefit of treatment under a suitable definition of the term benefit [Sawyers 2008; Italiano 2011]. When the magnitude of treatment benefit changes with the level of the genetic factor, a gene-treatment interaction can be included in a relevant statistical model to represent this change. Unlike prognostic values that are typically measured using AUC, there is no standardized way to measure treatment benefit and, hence, predictive values in the health sciences. Epidemiologists examine public health benefits of treatment using difference in the outcome (such as disease risk or response rate) between treated and untreated individuals. This difference can be calculated separately according to the levels of a genetic factor. The treatment benefit in a level of the genetic factor compared with the baseline level is referred to as the relative excess risk due to interaction (RERI; [Rothman et al. 2008]), and can be taken as a measure of predictive value. In contrast, recent clinical studies have examined predictive values of genetic factors in a two-step process: first, the statistical significance of the interaction effect in a model was examined using the LRT; next, the odds ratios for treatment were examined at each level of the genetic factor to identify the level(s) showing considerable risk reduction associated with treatment [Lerman et al. 2007; Amado et al. 2008; Karapetis et al. 2008]. Here we examine the role of interactions on the properties of these two methods for measuring predictive values.
Our paper is organized as follows. In Section 2, we first establish the notations, define interaction effect, and describe how this definition relates to AUC, ΔAUC and RERI. We describe the concepts in the main text and provide details of the hypothesis test for ΔAUC in the context of interactions in the Supplementary Material. In Section 3, we conduct simulations under a variety of parametric configurations to investigate the role of interactions on prognostic and predictive values. We illustrate these methods using published data from a study of melanoma in Section 4, and conclude the paper with a discussion in Section 5.
Methods
We consider the setting of N independent individuals having a binary disease outcome (for example, affected and unaffected) and two categorical risk factors (which we shall refer to as genetic factor and treatment). The disease outcome of a person having the j-th level of genetic factor and k-th level of treatment has a Bernoulli distribution with probability πjk, which is modeled via logistic regression as:
| (1) |
where μ is the baseline risk, βj and δk are referred to as the main effects of the j-th level of the genetic factor and k-th level of treatment, and γjk are referred to as the interaction effects. Denoting 0 as the baseline level, we set β0 = 0 = δ0 and γ0k = 0 = γj0 for all j and k. This model can be expanded easily to accommodate covariates, additional risk factors, and higher order interaction terms. When γjk = 0 in Equation (1), we refer to the resulting model as an additive model. The probability πjk is also referred to as disease risk corresponding to the j-th level of the genetic factor and k-th level of treatment.
Interaction effect
Before proceeding further, it will be useful to explain interactions in relation to the scale of the outcome. Denote ηjk as the outcome summary for the j-th level of the genetic factor and the k-th level of treatment. The quantity ηjk − ηj0 measures the difference in the outcome summaries between the k-th level of treatment and its baseline level when the genetic factor is fixed at the j-th level. For each j = 0, ···, L1 − 1 and k = 0, ···, L2 − 1, we define the interaction effect, denoted ωjk, as the difference in the outcome summary between the k-th and the baseline levels of treatment when the genetic factor is at the j-th level, compared to when the genetic factor is at its baseline level [Scheffe 1999]:
| (2) |
The null hypothesis of no interaction is H0 : ωjk = 0 for all j = 1, ···, L1 − 1 and k = 1, ···, L2 − 1. The choice of a summary measure for ηjk leads to different frameworks for evaluating interactions [Wang et al. 2010].
Suppose we define ηjk as the logarithm of disease odds i.e., . It follows from Equation (1) that ωjk = γjk, which is also referred to as multiplicative interaction. Testing the null hypothesis of no interaction is equivalent to testing H0 : γjk = 0 for all j and k in Equation (1), which can be done using the LRT. Under H0, the LRT has an asymptotic central chi squared distribution with (L1 − 1) × (L2 − 1) degrees of freedom.
Suppose we define ηjk as the disease odds i.e., . [Note that, when the disease is rare, ηjk ≈ πjk, which is the disease risk.] Using Equations (1) and (2), the interaction effect can be written as:
| (3) |
We shall write RERIjk = exp{βj + δk + γjk} − exp{βj} − exp{δk} + 1, which is referred to as the relative excess risk due to interaction corresponding to the j-th and k-th levels of the two factors of interest [Rothman et al. 2008]. When η00 ≠ 0, testing the null hypothesis of no interaction in this setting is equivalent to testing RERIjk = 0, which is also referred to as a test for additive interaction. When γjk = 0, we have RERIjk = exp{βj + δk} − exp{βj} − exp{δk} + 1, and RERIjk → 0 when βj → 0 or δj → 0 Thus, under an additive logistic regression model, we will have RERIjk ≠ 0 when the magnitudes of the effects of the genetic factor and treatment are not negligible. We can test H0 : RERIjk = 0 against HA : RERIjk ≠ 0 using a confidence interval method [Zou 2008] or a likelihood ratio test [Han et al. 2012]. Here we use the confidence interval method.
Interactions and prognostic value
Denote AUC1 as the AUC of the model given by Equation (1). Further, denote AUC0 as the AUC of the additive model (i.e., Equation 1 with γjk = 0). The contribution of interactions to the prognostic value can be measured as ΔAUC = AUC1 − AUC0. The theoretical value of AUC can be written by postulating a normal distribution for the logarithm of disease risk in the general population (i.e., log-normal distribution for disease risk) with some mean and variance σ2. Under this approach, the theoretical value of AUC is , where Φ(.) is the cumulative probability of a standard normal distribution. When the risk factors are independent and are centered and scaled to have mean 0 and variance 1, using Equation (1) we obtain and , where and (see Supplementary Material A). Therefore, we have:
| (4) |
From Equation (4), ΔAUC = 0 if and only if γjk = 0 for all j and k. Since Φ(.) is a strictly monotonic function, this observation suggests that testing H0 : ΔAUC = 0 is equivalent to testing H0 : γjk = 0 for all j and k.
These observations suggest that, the properties of LRT for testing the null hypothesis of no multiplicative interaction are likely to be equivalent to those of a test for H0 : ΔAUC = 0. Our observations also align with those of (Pepe et al.) [2013] who demonstrated the equivalence of several null hypotheses when testing the significance of the incremental improvement in AUC due to the inclusion of a new biomarker in a statistical model. Here we have shown the equivalence between two hypothesis tests in the context of interactions. The LRT for testing H0 : γjk = 0 for all j and k is readily available in most statistical software packages. However, a statistic for testing H0 : ΔAUC = 0 in the context of interactions is needed.
We develop a resampling-based approach for this test. Briefly, this is a parametric bootstrap approach and proceeds as follows. For binary outcomes, interaction measures the association between gene and treatment in the affected individuals relative to the unaffected individuals. Our approach is to first estimate this association in the observed data set by fitting a model for the genetic factors given the disease outcome and treatment. This model will provide estimates of the magnitudes of association between gene and treatment in the affected and unaffected individuals. Next, we generate data sets under the null hypothesis of no interaction via a resampling procedure using these estimated effects. Retaining the disease outcome and treatment assignment as observed, we randomly sample genetic factors for each individual such that the association between gene and treatment among affected and unaffected individuals is the same (i.e., the interaction effect is 0). We estimate ΔAUC for this data set. This is one realization from the null distribution of ΔAUC. We repeat this sampling procedure multiple times and estimate the p-value as the proportion of these ΔAUCs that are larger than that for the observed data. Detailed steps for this resampling procedure are provided in Supplementary Material B.
Interactions and predictive value
The predictive value depends upon the definition of treatment benefit. For the j-th level of genetic factor, denote Bjk as the benefit of the k-th level of treatment compared to the baseline treatment level. We consider two definitions of Bjk.
First, for the j-th level of the genetic factor, we define Bjk in terms of odds ratio: Bjk = {πjk/(1 − πjk)} / {πj0/(1 − πj0)}. From Equation (1), we have Bjk = exp {δk + γjk}. We define the predictive value of the j-th level of the genetic factor for the k-th level of treatment, denoted Pjk, as treatment benefit for the j-th level of the genetic factor relative to its baseline level: Pjk = Bjk/B0k. It then follows from Equation (1) that Pjk = exp {γjk} i.e., the multiplicative interaction is a measure of predictive value in this setting. When Pjk = 1, it means that the the benefits of the k-th level of treatment is the same for the j-th and baseline levels of the genetic factor. The null hypothesis that the j-th level of the genetic factor does not predict treatment benefit is given by H0 : Pjk = 1. The null hypothesis that no level of the genetic factor predicts treatment benefit is H0 : Pjk = 1 for all j and k. Under the above definition of treatment benefit, this is equivalent to the null hypothesis of no multiplicative interaction: H0 : γjk = 0 for all j and k i.e., H0ΔAUC = 0.
Next, for the j-th level of the genetic factor, suppose we define Bjk as the excess disease odds for the k-th level compared to the baseline level of treatment i.e., Bjk = {πjk/(1 − πjk)} − {πj0/(1 − πj0)}. We define Pjk as the excess treatment benefit for the j-th level compared to the baseline level of the genetic factor i.e., Pjk = Bjk − B0k. Under this definition of treatment benefit, it follows from Equation (1) that Pjk ∝ RERIjk. The null hypothesis that the j-th level of the genetic factor does not predict treatment benefit is: H0 : Pjk = 0 i.e., H0 : RERIjk = 0.
Taken together, these observations suggest the following results: (i) H0 : γjk = 0 for all j and k and H0 : ΔAUC = 0 are equivalent; (ii) this is also equivalent to the null hypothesis of no predictive value of a (genetic) risk factor when treatment benefit is defined based on odds ratios; (iii) H0 : RERIjk = 0 is equivalent to the null hypothesis that the j-th level of the genetic risk factor does not predict the benefits of the k-th level of treatment, when treatment benefit is defined based on differences between disease odds; and (iv) when γjk = 0 for some j and k in Equation (1), RERIjk ≠ 0 only when both βj and δk are not small. We performed simulation studies to assess the operating characteristics of the above tests. Table 1 lists the hypotheses being tested.
Table 1.
Hypotheses being tested
| Objective | Statistic | H0 | H1 | ||
|---|---|---|---|---|---|
| Interaction Effect | Likelihood Ratio | γjk = 0a | γjk= 0b | ||
| Prognostic / Predictive | ΔAUC | AUC1− AUC0 = 0 | AUC1− AUC0 ≠ 0 | ||
| Predictive | RERIjk |
|
|
for all j and k in Equation (1)
for at least one j and k in Equation (1)
Simulation study
We simulated a genetic factor X with L1 levels and a treatment Z with L2 levels for N independent individuals, and generated their disease risk using the following model:
| (5) |
where μ is the baseline risk; I(.) is an indicator variable taking value 1 when the condition in the parentheses is true and taking value 0 otherwise; ; β and δ are the main effects of the genetic factor and treatment, respectively; γ is the interaction effect; px = P(X ≥ C1); and pz = P(Z ≥ C2). The gene and treatment contribute to disease risk only when their levels are at least C1 and C2, respectively. Further, G and T denote the standardized gene and treatment variables, respectively. In our simulations we also generated data under different magnitudes of correlations (denoted ρ) between X and Z, and under two types of interactions - quantitative and qualitative (Supplementary Figure S1). When the interaction is quantitative, the magnitude of the treatment effect, but not its direction, differs according to the level of the genetic factor. In the presence of a qualitative interaction, both the magnitude and direction of the treatment effect differ according to the level of the genetic factor. In such cases where the direction of treatment effect changes, we expect that omitting the interaction term from the model may result in incorrect prognostic and predictive values, potentially leading to inappropriate decisions about treatment benefits. We chose the parameters of Equation (5) such that the theoretical AUCs were set at some desired values and ΔAUC ranged between 0.05 and 0.15. Further details of the simulation setup, model, and parametric configurations are given in the Supplementary Material C and Supplementary Table S1.
We analyzed the simulated data sets using Equation (1) and using an additive model. While the true risk model was based on Equation (5), we analyzed the data without assuming knowledge of the true model generating the disease. We estimated AUC0, AUC1, and ΔAUC, and tested the significance of ΔAUC using our proposed resampling procedure. We also tested the significance of interactions using a LRT. We also estimated RERI for various levels of the genetic risk factor. Under each parametric configuration, we calculated type I error and power of the LRT, and for the ΔAUC and RERI tests.
A common aim of clinical and epidemiological studies is to increase the prognostic value of models by including novel genetic factors and/or interactions of the genetic factors with other risk factors. However, few studies have reported an increase in AUC of considerable magnitude. In order to obtain insights into the properties of the estimated AUCs, we also fitted the true risk model (Equation 5) to our simulated data and calculated the AUCs, which we refer to as the attainable AUCs. Note that the estimated AUCs are obtained via non-parametric ranking procedure. The attainable AUCs are also obtained in a similar manner and, thus, serve as a benchmark in our comparison of the estimated ΔAUC against its theoretical value.
Results
Type I error
The type I errors of LRT and the test for ΔAUC were similar under all parametric configurations considered (Table 2). In general, the type I error of the test of ΔAUC was controlled around the nominal value of 0.05 for all values of N and ρ. There was a slight departure from the nominal rate of 0.05 for LRT (and ΔAUC) in some scenarios, which has also been previously reported in other simulation studies [Allison et al. 1999]. The Type I error for RERI was maintained at 0.05 when the effect of at least one of the risk factors was small (for example when δ = 0.1). The probability presented in Table 2 is the probability of rejecting the null that RERI = 0 under the assumption that γjk = 0. However, when both the risk factors have non-negligible main effects (e.g. β = 0.75, δ = 0.6) we are in a setting where RERI ≠ 0. As expected in this specific scenario we did not reject H0 : RERI = 0, and hence the type I error was considerable larger than the nominal value of 0.05 (type I error of 0.70 when ρ = 0, β = 0.75, δ = 0.60, and 500 affected and 500 unaffected individuals).
Table 2.
Type I errors of ΔAUC test, LRT, and RERI test. The sample sizes are 500 and 200 affected individuals and an equal number of unaffected individuals. The correlations between the genetic factor and treatment are ρ = 0 and ρ = 0.50. The estimated type I errors are shown under various true values of the main effects β and δ (note: γ = 0). In our simulations, the genetic factor had 3 levels and treatment had 2 levels. Hence, we report two RERIs: RERI11 and RERI21.
| Number affected | β | δ | ρ = 0 | ρ = 0.50 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| ΔAUC | LRT | RERI11 | RERI21 | ΔAUC | LRT | RERI11 | RERI21 | |||
| 500 | 0.15 | 0.1 | 0.03 | 0.05 | 0.07 | 0.06 | 0.07 | 0.07 | 0.04 | 0.03 |
| 0.35 | 0.1 | 0.07 | 0.07 | 0.06 | 0.07 | 0.04 | 0.04 | 0.06 | 0.06 | |
| 0.55 | 0.1 | 0.08 | 0.06 | 0.03 | 0.03 | 0.07 | 0.06 | 0.05 | 0.07 | |
| 0.75 | 0.1 | 0.02 | 0.03 | 0.05 | 0.03 | 0.04 | 0.09 | 0.06 | 0.08 | |
| 0.95 | 0.1 | 0.02 | 0.05 | 0.03 | 0.03 | 0.02 | 0.03 | 0.04 | 0.03 | |
| 0.75 | 0.6 | 0.01 | 0.04 | 0.01 | 0.7 | 0.02 | 0.09 | 0.03 | 0.35 | |
|
| ||||||||||
| 200 | 0.15 | 0.1 | 0.03 | 0.04 | 0.04 | 0.03 | 0.06 | 0.08 | 0.01 | 0.02 |
| 0.35 | 0.1 | 0.03 | 0.02 | 0.05 | 0.03 | 0.05 | 0.10 | 0.03 | 0.03 | |
| 0.55 | 0.1 | 0.04 | 0.08 | 0.02 | 0.04 | 0.05 | 0.08 | 0.04 | 0.04 | |
| 0.75 | 0.1 | 0.03 | 0.02 | 0.04 | 0.02 | 0.03 | 0.02 | 0.00 | 0.01 | |
| 0.95 | 0.1 | 0.05 | 0.07 | 0.01 | 0.01 | 0.05 | 0.05 | 0.01 | 0.02 | |
| 0.75 | 0.6 | 0.01 | 0.05 | 0.01 | 0.13 | 0.02 | 0.02 | 0.00 | 0.04 | |
Power
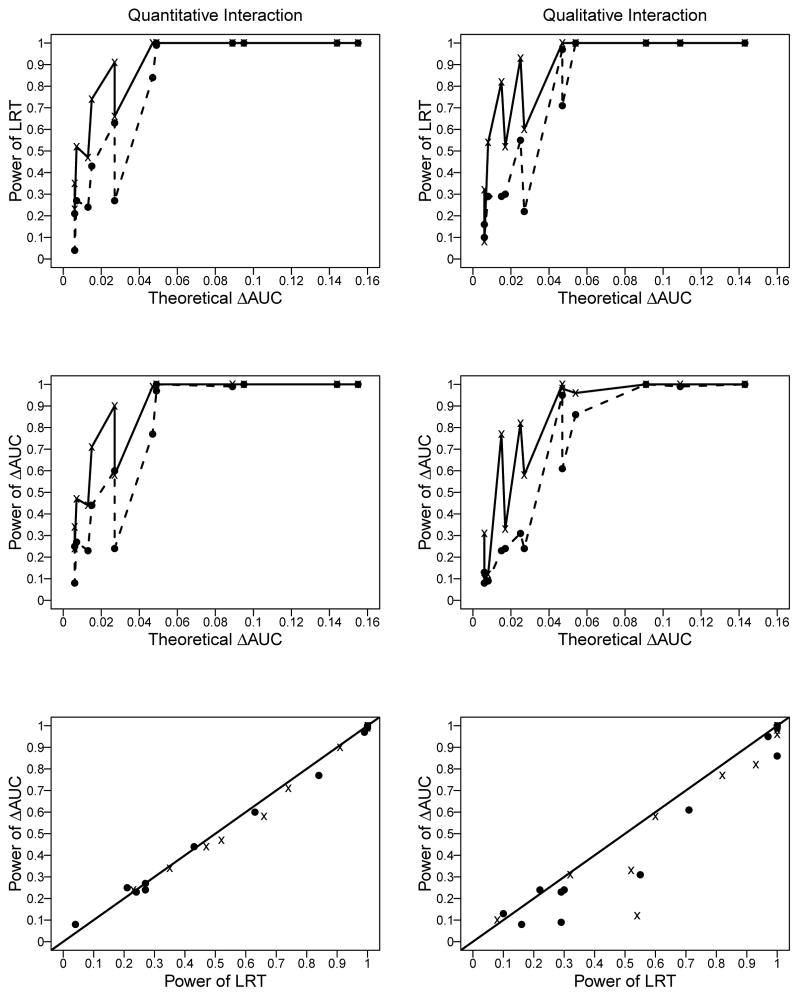
Figure 1 shows the power of LRT and the ΔAUC test for data simulated with ρ = 0. As expected, power increased with sample size. Further, power increased as the theoretical value of ΔAUC increased. The plots indicate that even with a sample size of 200, we can attain power of 1 to detect a ΔAUC of 0.05. However, whether a ΔAUC = 0.05 increase is clinically meaningful is a topic of debate (see Discussion). Note that ΔAUC = AUC1 − AUC0, where AUC0 is the AUC under the null hypothesis that interactions do not contribute to the prognostic value. For a given value of ΔAUC, the power also depended upon the magnitude of AUC0. For example, detecting ΔAUC = 0.05 when AUC0 = 0.55 had power of 0.77 and 0.61 under quantitative and qualitative interactions respectively; whereas, the power increased to 0.97 and 0.95 under these two types of interactions when AUC0 = 0.65. The last row of Figure 1 suggests that, in general, LRT and ΔAUC tests had similar power, although ΔAUC test had slightly lower power than LRT under a few parametric configurations. Power for simulations with ρ = 0.5 showed similar patterns (detailed results not shown).
Figure 1.
Power of the likelihood ratio test (LRT; top row) and the resampling-based ΔAUC test (middle row), and a comparison of the powers of these two tests with the 45-degree line shown as a benchmark (bottom row). The left and right columns correspond to results for data simulated based on quantitative and qualitative interactions, respectively. These results are shown for sample sizes of 500 cases and 500 controls (bold lines in the top and middle rows with points in all the panels shown as “x”) and 200 cases and 200 controls (dashed lines in the top and middle rows with points in all the panels shown as a closed circle)
Estimated and attained AUCs
The estimated and attained AUCs and ΔAUCs are shown in Table 3 for sample size of 1000 individuals (500 affected). In general, the estimated ΔAUC values were smaller than the theoretical ΔAUC values. This was particularly the case for large value of theoretical AUC0 (for example, AUC0 = 0.7). The attainable ΔAUCs were closer to the estimated ΔAUC than to the theoretical value. These observations suggest that the inclusion of interactions in a model may not increase ΔAUC by a considerable magnitude even when the interaction effects (and, hence, the prognostic and predictive value) are significantly different from 0. This can happen particularly when AUC of the additive model is already large (for example, theoretical AUC0 ≈ 0.70 and ΔAUC ≈ 0.10 in our simulations). Similar results were obtained for sample size of 400 individuals (200 affected; Supplementary Table S2).
Table 3.
Theoretical, estimated and attained AUCs and ΔAUC shown for simulations with 500 affected and 500 unaffected individuals. The first column shows the true values of β, δ, and γ used for generating disease risk from Equation (5).
| ρ = 0 | ρ = 0.5 | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| (β, δ, γ) | AUC | AUC0 | AUC1 | ΔAUC | AUC0 | AUC1 | ΔAUC |
| Quantitative Interactions | |||||||
| (0.15, 0.10, 0.30) | theoretical | 0.551 | 0.598 | 0.05 | 0.561 | 0.583 | 0.02 |
| attainable | 0.559 (0.017) | 0.596 (0.016) | 0.04 | 0.555 (0.018) | 0.572 (0.017) | 0.02 | |
| estimated | 0.561 (0.018) | 0.596 (0.017) | 0.03 | 0.561 (0.014) | 0.579 (0.015) | 0.02 | |
| (0.15, 0.10, 0.70) | theoretical | 0.551 | 0.695 | 0.14 | 0.561 | 0.663 | 0.10 |
| attainable | 0.551 (0.016) | 0.666 (0.013) | 0.12 | 0.578 (0.017) | 0.608 (0.015) | 0.03 | |
| estimated | 0.559 (0.016) | 0.669 (0.013) | 0.11 | 0.578 (0.014) | 0.607 (0.013) | 0.03 | |
| (0.55, 0.10, 0.50) | theoretical | 0.654 | 0.703 | 0.05 | 0.666 | 0.693 | 0.03 |
| attainable | 0.657 (0.016) | 0.691 (0.014) | 0.03 | 0.638 (0.015) | 0.659 (0.015) | 0.02 | |
| estimated | 0.656 (0.018) | 0.69 (0.017) | 0.03 | 0.638 (0.014) | 0.658 (0.013) | 0.02 | |
| (0.75, 0.10, 0.90) | theoretical | 0.703 | 0.798 | 0.10 | 0.715 | 0.778 | 0.06 |
| attainable | 0.69 (0.02) | 0.753 (0.014) | 0.06 | 0.663 (0.015) | 0.694 (0.014) | 0.03 | |
| estimated | 0.689 (0.018) | 0.753 (0.013) | 0.06 | 0.663 (0.017) | 0.696 (0.015) | 0.03 | |
|
| |||||||
| Qualitative Interactions | |||||||
| (0.15, 0.10, −0.30) | theoretical | 0.551 | 0.598 | 0.05 | 0.561 | 0.605 | 0.04 |
| attainable | 0.537 (0.017) | 0.576 (0.015) | 0.04 | 0.559 (0.017) | 0.582 (0.014) | 0.02 | |
| estimated | 0.54 (0.018) | 0.581 (0.016) | 0.04 | 0.559 (0.016) | 0.583 (0.016) | 0.02 | |
| (0.15, 0.10, −0.70) | theoretical | 0.551 | 0.694 | 0.14 | 0.561 | 0.687 | 0.13 |
| attainable | 0.536 (0.022) | 0.667 (0.014) | 0.13 | 0.562 (0.031) | 0.66 (0.015) | 0.10 | |
| estimated | 0.54 (0.022) | 0.67 (0.016) | 0.13 | 0.562 (0.037) | 0.665 (0.014) | 0.10 | |
| (0.55, 0.10, −0.50) | theoretical | 0.654 | 0.701 | 0.05 | 0.666 | 0.708 | 0.04 |
| attainable | 0.64 (0.017) | 0.668 (0.016) | 0.03 | 0.624 (0.015) | 0.652 (0.014) | 0.03 | |
| estimated | 0.642 (0.019) | 0.672 (0.018) | 0.03 | 0.625 (0.015) | 0.652 (0.015) | 0.03 | |
| (0.75, 0.10, −0.90) | theoretical | 0.703 | 0.794 | 0.09 | 0.715 | 0.793 | 0.08 |
| attainable | 0.72 (0.017) | 0.749 (0.015) | 0.03 | 0.634 (0.02) | 0.698 (0.015) | 0.06 | |
| estimated | 0.718 (0.016) | 0.748 (0.015) | 0.03 | 0.636 (0.016) | 0.698 (0.014) | 0.06 | |
Data Example
We illustrate the proposed concepts and methods using published data from an epidemiology study of melanoma [Kricker et al. 2010]. In this study, the outcome is binary, denoting the presence/absence of a second primary melanoma. Sun exposure and variants in the pigmentation gene MC1R are among the important known risk factors for melanoma. Table 4 shows the melanoma data with MC1R as a binary variable denoting the presence/absence of the R allele (i.e., red hair color variant) and sun exposure measured as a binary variable in terms of: (i) beach and water activities from age 15; (ii) average annual lifetime ambient ultraviolet (UV) exposure; and (iii) early life ambient UV exposure.
Table 4.
Application in melanoma (Kricker et al). MC1R and each of the three types of sun exposures have L1 = L2 = 2 levels. For the levels of MC1R, “No R” denotes that there is no variant predisposing the individuals to red hair color, and “Any R” denotes the presence of at least one such variant. The columns SPM and MPM denote the total number of individuals with single and multiple primary melanomas, respectively. The two columns on the right denote the probability of predictive value calculated via the bootstrap procedure. The notation PRERI denote max {P (RERI > 0), P (RERI < 0)}.
| MC1R | Sun Exposure | SPM | MPM | LRT (p-value) | ΔAUC (p-value) | RERI (95% CI) | Bootstrap predictive values
|
|
|---|---|---|---|---|---|---|---|---|
| P(ΔAUC > 0) | PRERI | |||||||
| Beach and water activities from age 15 | ||||||||
| No R | None | 248 | 68 | 4.60 | 0.003a | −0.4038 | 0.29 | 0.89 |
| Any | 733 | 380 | (0.03) | (0.02) | (−1.314, 0.151) | |||
| Any R | None | 236 | 129 | |||||
| Any | 644 | 438 | ||||||
|
| ||||||||
| Average annual lifetime ambient UV | ||||||||
| No R | 369–848 KJ/m2 | 573 | 179 | 0.003 | 0b | 0.4818 | 0.05 | 0.93 |
| 849–1500 KJ/m2 | 359 | 249 | (0.95) | (0.94) | (−0.183, 1.121) | |||
| Any R | 369–848 KJ/m2 | 495 | 212 | |||||
| 849–1500 KJ/m2 | 350 | 336 | ||||||
|
| ||||||||
| Early life ambient UV | ||||||||
| No R | 371–853 KJ/m2 | 563 | 193 | 0.052 | 0c | 0.2539 | 0.04 | 0.82 |
| 854–1520 KJ/m2 | 387 | 241 | (0.82) | (0.90) | (−0.324, 0.785) | |||
| Any R | 371–853 KJ/m2 | 483 | 236 | |||||
| 854–1520 KJ/m2 | 374 | 320 | ||||||
AUC0 = 0.557, AUC1 = 0.560
AUC0 ≈ AUC1 ≈ 0.619
AUC0 ≈ AUC1 ≈ 0.593
For each data set, we fitted logistic regression models to the outcome in relation to MC1R and sun exposure and tested the null hypothesis of no interaction between gene and sun exposure using a LRT, and tested H0 : ΔAUC = 0. We also calculated RERI and tested H0 : RERI = 0. In clinical studies, it is meaningful to refer to preditive values in terms of probability that a genetic factor projects treatment benefit [Sawyers 2008]. In our setting, this is equivalent to the probability that ΔAUC and RERI exceed certain threshold. To obtain insights into this, we calculated P(ΔAUC > 0) and max {P(RERI > 0), P(RERI < 0)} (since RERI can be positive or negative). We generated 10,000 bootstrap samples of the data, estimated ΔAUC and RERI for each sampled data set and calculated P(ΔAUC > 0) as the fraction of data sets for which the estimated ΔAUC was positive, and estimated max {P(RERI > 0), P(RERI < 0)} as the maximum of the fraction of data sets having negative and positive estimates of RERI (see Table 4 and Supplementary Figure S2).
There was a significant interaction between MC1R and sun exposure from beach and water activity (LRT = 4.60, d.f = 1, p-value = 0.03). The magnitude of the estimated ΔAUC was small (AUC1 = 0.557, AUC0 = 0.560, ΔAUC = 0.003). However, as expected, it was significantly different from 0 (p-value = 0.021). This suggests that interaction contributed significantly to the prognostic value. Further, when treatment benefit was defined in terms of odds ratios, this result suggests that MC1R significantly predicts the benefits of reducing sun exposure from beach and water activity. In particular, the benefit of reducing sun exposure was 1.244 (= 438 × 236/(644 × 129)) for carriers of an R variant and 1.891 (= 380 × 248/(733 × 68)) among non-carriers. These results suggest that non-carriers had significantly higher benefit in terms of reduced risk of second primary melanoma associated with reducing sun exposure than carriers (estimated predictive value is P11 = 1.52 = 1.891/1.244). However deciding whether this benefit is clinically actionable will require more rigorous investigations based on further studies (see Discussion). The estimated value of RERI was −0.4038 (95% confidence interval: −1.314, 0.151), which was not significantly different from 0. This suggests that, when treatment benefit was defined in terms of excess disease odds (or excess disease risk), reduction in melanoma risk due to reducing sun exposure from beach and water activity was higher for non-carriers than carriers of an R variant, although this was not statistically significant. The bootstrap estimates of predictive values were P(ΔAUC > 0) = 0.29 and max {P(RERI > 0), P(RERI < 0)} = 0.89. Since LRT and the test for ΔAUC were significant, we would, in principle, expect the estimated P(ΔAUC > 0) to be large. The seemingly small estimated probability of 0.29 may be due to the small number of non-carrier cases without sun exposure from beach and water activities (68 multiple primary melanomas). Even though RERI was not significantly different from 0, our bootstrap approach showed that the predictive value of MC1R was approximately 89% when treatment benefit was defined in terms of differences between disease odds.
There was no significant interaction between MC1R and annual average lifetime UV (LRT = 0.0034, d.f = 1, p-value = 0.95). As expected interaction did not contribute to the prognostic value (ΔAUC ≈ 0, p-value = 0.94). When treatment benefit was defined in terms of odds ratios, the benefits of reducing annual average lifetime UV were 2.24 and 2.22 among carriers and non-carriers, respectively, of an R variant. Hence, the predictive value was P11 = 1.01 = 2.24/2.22. The estimated value of RERI was 0.4818 (95% CI: −0.183, 1.121), suggesting that MC1R does not significantly predict the benefits of reducing annual average lifetime UV when benefit is defined in terms of differences between disease odds. Based on the bootstrap approach, P(ΔAUC > 0) = 0.05 and max {P(RERI > 0), P(RERI < 0)} = 0.93. Thus, even though RERI was not significantly different from 0, the bootstrap estimate of predictive probability based on RERI was greater than 90%. Similar results were obtained for the interaction between MC1R and early life ambient UV (Table 4).
Discussion
In studies of targeted treatment/intervention, often the goal is to understand how treatment benefits vary according to genetic predisposition [Lerman et al. 2007; Keedy et al. 2011]. This has accelerated the investigations of gene-treatment, gene-environment, and gene-gene interactions using statistical models in the health sciences. Interactions depend upon the scale on which the outcome is measured [Wang et al. 2010; Satagopan and Elston 2013]. In this paper we have examined the relationship between interactions (measured on two different scales) and prognostic and predictive values of (genetic) risk factors.
Specifically, we examined whether including interaction terms in the model improves the prognostic value i.e., increases ΔAUC by a significant magnitude. We showed that testing the null hypothesis that ΔAUC = 0 is equivalent to testing the null hypothesis of no multiplicative interaction, and developed a resampling approach to test the statistical significance of ΔAUC in relation to interactions. Although previous work has demonstrated the equivalence of several null hypotheses when evaluating whether a new biomarker has significant prognostic value [Pepe et al. 2013], we have shown the equivalence of two null hypotheses in the specific context of interactions. Kerr and Pepe [2011] studied the properties of receiver operating characteristic (ROC) curves in the context of interactions by evaluating ROC curves separately among carriers and non-carriers of the genetic factor of interest by fitting an additive logistic regression model. Our work is distinct from this in that it focuses specifically on how interaction terms in logistic regression models relate to ΔAUC.
It has been noted that including interactions in a model does not increase the AUC by a considerable magnitude [Aschard et al. 2012]. To obtain insights into this, we used simulation studies, which showed that, even when the true disease risk model is known, the attainable ΔAUCs are considerably smaller than the theoretical ΔAUCs. Our simulations also show that the power to reject H0 : ΔAUC ≠ 0 of a certain magnitude in the context of interactions also depends upon the magnitude of AUC0, which also aligns with recent work that examined model performance in the context of evaluating a new biomarker [Kerr et al. 2012]. To obtain insights into this, consider, for example, the case where AUC0 = 0.70. The theoretical value of σ0 is . Then, a 10% increase in σ0 will result in a 2% increase in AUC since . We will need 55% and 100% increases in σ0 to attain ≈ 10% and 15% increases in AUC when AUC0 = 0.70. Whether such increases in the magnitude of σ0 can be achieved through the inclusion of interactions in a model is an important question, but one that will require investigations in a broad range of real data sets.
While ΔAUC is a commonly used measure for evaluating prognostic values, there is no consensus on a measure of predictive values. The RERI is an important statistic in epidemiology for evaluating additive interactions, and is a measure of predictive value when treatment benefit is defined in terms of difference between disease odds or disease risk [Rothman et al. 2008]. However, recent epidemiology studies have reported predictive values of genetic factors based on the statistical significance of interactions without specifically reporting a measure of treatment benefit. For example, (Lerman et al.) [2007] used the statistical significance of a multiplicative interaction between bupropion treatment and the CYP2B6 gene in a logistic regression model to show that carriers of a variant allele may be more vulnerable to abstinance symptoms and smoking relapse. Here we have shown that multiplicative interaction (equivalently, ΔAUC) is a measure of predictive value when treatment benefit is defined based on odds ratios.
An important practical question for future study is: what values of ΔAUC and RERI would be clinically meaningful? Addressing this question is outside the scope of our work. Further investigations based on rigorous study designs with validation data sets are needed to evaluate clinically actionable magnitudes of prognostic and predictive values and compare their properties with other emerging measures in the setting of interactions in statistical models.
Supplementary Material
Acknowledgments
This work was supported by research grants R01CA137420 and P30CA008748 from the National Cancer Institute, USA, and grant UL1RR024996 from the Clinical and Translational Science Center at Weill Cornell Medical College, New York, USA.
Footnotes
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Agresti A. Categorical data analysis. New York: Wiley; 2002. [Google Scholar]
- Allison DB, Neale MC, Zannolli R, Schord NJ, Amos CI, Blangero J. Testing the robustness of the likelihood-ratio test in a variance-component quantitative-trait locimapping procedure. Am J Hum Genet. 1999;65:531–544. doi: 10.1086/302487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- Aschard H, Chen J, Cornelis MC, Chibnik LB, Karlson EW, Kraft P. Inclusion of gene-gene and gene-environment interactions unlikely to dramatically improve risk prediction for complex diseases. Am J Hum Genet. 2012;90:962–972. doi: 10.1016/j.ajhg.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- Han SS, Rosenberg PS, Garcia-Closas M, Figueroa JD, Silverman D, Chanock SJ, Rothman N, Chatterjee N. Likelihood ratio test for detecting gene (g)-environment (e) interactions under an additive risk model exploiting ge independence for case-control data. Am J Epid. 2012;176:1060–1067. doi: 10.1093/aje/kws166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiano A. Prognostic or predictive? it’s time to get back to definitions! Journal of Clinical Oncology. 2011;29:4718. doi: 10.1200/JCO.2011.38.3729. [DOI] [PubMed] [Google Scholar]
- Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- Keedy VL, Temin S, Somerfield MR, Beasley MB, McShane LM, DHJ, Milton DT, Strawn JR, Wakelee HA, Giaccone G. American society of clinical oncology provisional clinical opinion: epidermal growth factor receptor (EGFR) mutation testing for patients with advanced non-small-cell lung cancer considering first-line egfr tyrosine kinase inhibitor therapy. J Clin Oncol. 2011;29:2121–2127. doi: 10.1200/JCO.2010.31.8923. [DOI] [PubMed] [Google Scholar]
- Kerr KF, Bansal A, Pepe MS. Further insight into the incremental value of new markers: the interpretation of performance measures and the importance of clinical context. Am J Epid. 2012;176:482–487. doi: 10.1093/aje/kws210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr KF, Pepe MS. Joint modeling, covariate adjustment, and interaction: contrasting notions in risk prediction models and risk prediction performance. Epidemiology. 2011;22:805–812. doi: 10.1097/EDE.0b013e31823035fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury MJ, Yang Q, Gwinn M, Little J, Flanders WD. An epidemiologic assessment of genomic profiling for measuring susceptibility to common diseases and targeting interventions. Genet Med. 2004;6:38–47. doi: 10.1097/01.gim.0000105751.71430.79. [DOI] [PubMed] [Google Scholar]
- Kricker A, Armstrong BK, Goumas C, Kanetsky P, Gallagher RP, Begg CB, Millikan RC, Dwyer T, Rosso S, Marrett LD, Thomas NE, Berwick M GEM Study Group. Mc1r genotype may modify the effect of sun exposure on melanoma risk in the gem study. Cancer Causes Control. 2010;21:2137–2147. doi: 10.1007/s10552-010-9633-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman CE, Schnoll RA, Munafo MR. Genetics and smoking cessation. Am J Prev Med. 2007;33:S398–S405. doi: 10.1016/j.amepre.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JH, Williams SM. Epistasis and its implications for personal genetics. Am J Hum Genet. 2009;85:309–320. doi: 10.1016/j.ajhg.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepe MS, Kerr KF, Longton G, Wang Z. Testing for improvement in prediction model performance. Stat Med. 2013;32:1467–1482. doi: 10.1002/sim.5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman KJ, Greenland S, Lash TL. Modern epidemiology. New York: Lippincott WIlliams & Wilkins; 2008. [Google Scholar]
- Satagopan JM, Elston RC. Evaluation of removable statistical interaction for binary traits. Stat Med. 2013;32:1164–1190. doi: 10.1002/sim.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- Scheffe H. The Analysis of Variance. John Wiley & Sons; 1999. [Google Scholar]
- Seshan VE, Gonen M, Begg CB. Comparing roc curves derived from regression models. Stat Med. 2013;32:1483–1493. doi: 10.1002/sim.5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Lu Q, Mukherjee S, Crane PK, Elston R, Ritchie MD. Analysis pipeline for the epistasis search - statistical versus biological filtering. Front Genet. 2014;5:106. doi: 10.3389/fgene.2014.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers AJ, Cronin AM, Begg CB. One statistical test is sufficient for assessing new predictive markers. BMC Med Res Methodol. 2011;11:13. doi: 10.1186/1471-2288-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Elston RC, Zhu X. The meaning of interaction. Hum Hered. 2010;70:269–277. doi: 10.1159/000321967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou GY. On the estimation of additive interaction by use of the four-by-two table and beyond. Am J Epid. 2008;168:212–224. doi: 10.1093/aje/kwn104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.