Abstract
The PHB-domain protein podocin maintains the renal filtration barrier and its mutation is an important cause of hereditary nephrotic syndrome. Podocin and its Caenorhabditis elegans orthologue MEC-2 have emerged as key components of mechanosensitive membrane protein signalling complexes. Whereas podocin resides at a specialized cell junction at the podocyte slit diaphragm, MEC-2 is found in neurons required for touch sensitivity. Here, we show that the ubiquitin ligase Ubr4 is a key component of the podocin interactome purified both from cultured podocytes and native glomeruli. It colocalizes with podocin and regulates its stability. In C. elegans, this process is conserved. Here, Ubr4 is responsible for the degradation of mislocalized MEC-2 multimers. Ubiquitylomic analysis of mouse glomeruli revealed that podocin is ubiquitylated at two lysine residues. These sites were Ubr4-dependent and were conserved across species. Molecular dynamics simulations revealed that ubiquitylation of one site, K301, do not only target podocin/MEC-2 for proteasomal degradation, but may also affect stability and disassembly of the multimeric complex. We suggest that Ubr4 is a key regulator of podocyte foot process proteostasis.
Introduction
Mutations in the kidney-expressed PHB-domain protein podocin are a major cause of renal failure in childhood (1,2). The membrane protein podocin is specifically expressed in podocytes, the visceral epithelial cells of the kidney. Podocytes are post-mitotic, highly arborized cells that maintain the kidney filtration barrier (3). Recent studies have demonstrated that podocin and its orthologue MEC-2 in Caenorhabditis elegans are part of high-molecular-weight multi-protein complexes at the plasma membrane that contain ion channels, transmembrane signalling proteins, lipid-modifying proteins as well as signalling adapters (4). These multi-protein complexes are mechanosensitive, mechanical strain is transmitted into channel opening of associated ion channels (5–7). To serve as mechanoreceptors, these multi-protein complexes must be expressed at very specific locations, the slit diaphragm of podocytes in the case of podocin and touch channel punctae in mechanosensitive neurons in the case of MEC-2. Podocin/MEC-2 multimerize and bind cholesterol of the plasma membrane to form megadalton protein–lipid supercomplexes required as platforms for the assembly of the signalling complexes (5).
Mutation of genes often leads to misfolded proteins and affects stability. The ubiquitin–proteasome pathway is one of the major pathways of protein degradation. This system relies on recruitment of one of the >500 E3 ubiquitin ligases to their target proteins and often occurs in a tightly controlled manner at specific subcellular localizations (8,9). An alternative degradation pathway, in particular important for membrane proteins, is the lysosomal-dependent pathways. For only a few proteins mutated in human disease, the molecular control of degradation is elucidated. The aim of this study was to unravel the mechanism of podocin degradation. To this end, we identified the N-recognin Ubr4 as podocin interacting protein. We show that Ubr4 ubiquitylates podocin/MEC-2 to counteract expression of mistargeted protein multimers. Surprisingly, degradation is determined by ubiquitylation of two specific lysine residues in the PHB domain of the proteins by influencing protein folding and complex stability.
Results
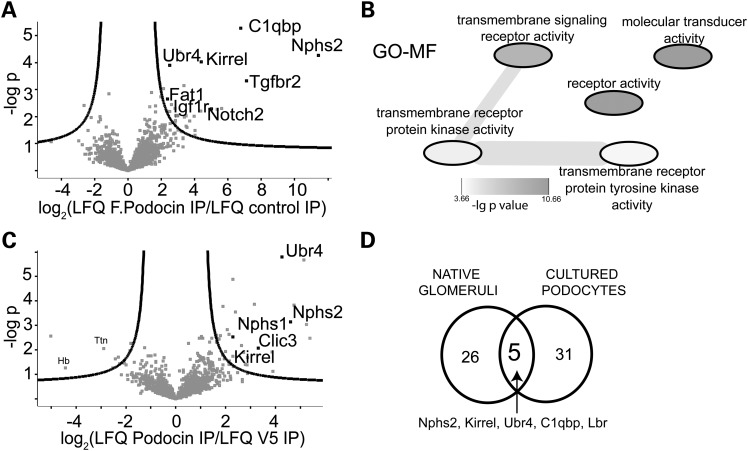
To identify the ubiquitin ligase complex responsible for controlling podocin stability, FLAG-tagged podocin was stably expressed in podocyte cell lines and precipitated to analyse the podocin interactome by a label-free liquid chromatography–mass spectrometry (LC-MS/MS) approach (10,11). Among >2000 identified and 1651 quantified proteins, 36 proteins passed stringent criterions for being significant interactors, among these podocin and the known podocin interactors Neph1 and Fat1 (Fig. 1A). The interactome also contained several signalling receptors, among these podocin and Notch2, TGF-β receptor 2 and Igf1-receptor. All interactors are listed in Table 1, they showed an enrichment of GO-terms associated with signalling and receptor activity (Fig. 1B). The quantitative MS approach revealed Ubr4 as the only annotated E3-ubiquitin ligase connected to the complex. Stoichiometry analysis (12) showed that Ubr4 is a low-abundant binding partner (Supplementary Material, Fig. S1). To further validate the interactions endogenous podocin was precipitated from native glomerular protein lysates using a podocin C-terminus-directed antibody (Fig. 1C). Similarly, the complex again contained the known podocin interactors Neph1 (Gene name: Kirrel) and Nephrin (Nphs1), and also the ubiquitin ligase Ubr4 (Fig 1C, Table 2). A comparison of the interactomes confirmed that the known interactor Neph1 and Ubr4 are core components of the podocin multi-protein complex (Fig. 1D). Interacting proteins in both systems were mainly membrane proteins and were functionally related to vesicle trafficking.
Figure 1.
Discovery of the podocin interactome and Ubr4 as a key interactor of podocin. (A) Analysis of the interactome of podocin expressed in podocytes. Podocytes were lysed, and FLAG-tagged podocin was pulled down using anti-FLAG antibody. The pull-down was analysed by MS/MS, and compared with a control pull-down. The negative decadic logarithm of the P-value (−logP) of the LFQ intensities is plotted against the log2-fold change between podocin pull-down and control pull-down. The proteins beyond the line are significant interactors (FDR = 0.2, s0 = 1). Ubr4 is a significant interactor. (B) Overrepresented GO-MF terms in the interacting proteins when compared with non-interacting proteins. Overrepresentation was analysed using a Fisher exact test (FDR < 0.05). GO-MF analysis of the podocin interactome revealed a strong association with signalling molecules. (C) Analysis of the podocin interactome purified from native glomeruli. Glomeruli from one mouse were lysed, and podocin was purified using an anti-podocin antibody. The pull-down was analysed by MS/MS and compared with a control (anti-V5) pull-down. The negative decadic logarithm of the P-value (−log P) of the LFQ intensities is plotted against the log2-fold change between podocin pull-down and control pull-down. Ubr4 was identified as a significant interactor. (D) Venn diagram comparing of significant interactors from glomeruli and cultured podocytes. Ubr4 and the known interactor Neph1 (Kirrel) are core components of the podocin interactome.
Table 1.
The table indicates proteins determined to be interactors of FLAG-tagged podocin expressed in podocytes
| Gene names | Uniprot | Protein names | t-test difference [log2 (LFQ FLAG podocin IP/LFQ control IP)] | −Log P-values |
|---|---|---|---|---|
| Nphs2 | Q91X05 | Podocin | 11.42 | 4.27 |
| Tgfbr2 | Q62312 | TGF-β receptor type-2 | 7.11 | 3.31 |
| C1qbp | Q8R5L1 | Complement component 1 Q subcomponent-binding protein, mitochondrial | 6.78 | 5.27 |
| Ifitm3 | Q9CQW9 | Interferon-induced transmembrane protein 3 | 5.62 | 2.30 |
| Epha2 | Q03145 | Ephrin type-A receptor 2 | 5.34 | 2.26 |
| Notch2 | G5E8J0 | Neurogenic locus notch homolog protein 2 | 5.01 | 2.29 |
| Ptprf | A2A8L5 | Receptor-type tyrosine-protein phosphatase F | 4.41 | 4.03 |
| Axl | Q6PE80 | Tyrosine-protein kinase receptor UFO | 4.38 | 2.19 |
| Irgm1 | Q60766 | Immunity-related GTPase family M protein 1 | 4.03 | 1.30 |
| H2-K1 | P01901 | H-2 class I histocompatibility antigen, K-B-α chain | 4.01 | 1.48 |
| Kirrel | Q80W68 | Kin of IRRE-like protein 1 | 3.97 | 2.01 |
| Steap2 | Q8BWB6 | Metalloreductase STEAP2 | 3.71 | 1.84 |
| Plxnb2 | B2RXS4 | Plexin-B2 | 3.56 | 1.59 |
| Myof | Q69ZN7 | Myoferlin | 3.49 | 1.64 |
| Ptprk | P35822 | Receptor-type tyrosine-protein phosphatase kappa | 3.48 | 2.03 |
| Tfrc | Q62351 | Transferrin receptor protein 1 | 3.39 | 1.98 |
| Atp2a2 | O55143 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | 3.37 | 3.14 |
| H2-Q8 | P14430 | H-2 class I histocompatibility antigen, Q8-α chain | 3.26 | 1.43 |
| Unc5b | Q8K1S3 | Netrin receptor UNC5B | 3.19 | 1.45 |
| Lbr | Q3U9G9 | Lamin-B receptor | 3.13 | 4.39 |
| Dnajb6 | O54946 | DnaJ homolog subfamily B member 6 | 3.00 | 1.50 |
| Slc25a4 | P48962 | ADP/ATP translocase 1 | 2.99 | 2.03 |
| Igf1r | E9QNX9 | Tyrosine-protein kinase receptor | 2.93 | 2.04 |
| Tmem176b | Q9R1Q6 | Transmembrane protein 176B | 2.90 | 2.02 |
| Arf4 | P61750 | ADP-ribosylation factor 4 | 2.88 | 2.04 |
| Erbb2 | P70424 | Receptor tyrosine-protein kinase erbB-2 | 2.86 | 1.85 |
| Slc20a1 | Q61609 | Sodium-dependent phosphate transporter 1 | 2.83 | 1.79 |
| Bag2 | Q91YN9 | BAG family molecular chaperone regulator 2 | 2.82 | 2.30 |
| Fat1 | F2Z4A3 | 2.62 | 2.45 | |
| Atp5c1 | Q8C2Q8 | ATP synthase subunit-γ | 2.54 | 1.73 |
| Ubr4 | A2AN08 | E3 ubiquitin-protein ligase UBR4 | 2.52 | 3.89 |
| Slc25a5 | P51881 | ADP/ATP translocase 2 | 2.50 | 2.19 |
| Slc25a24 | Q8BMD8 | Calcium-binding mitochondrial carrier protein SCaMC-1 | 2.38 | 2.65 |
| Rps18 | P62270 | 40S ribosomal protein S18 | 2.33 | 2.21 |
| Erlin2 | Q8BFZ9 | Erlin-2 | 2.13 | 3.03 |
| Rps3 | P62908 | 40S ribosomal protein S3 | 1.97 | 2.73 |
Podocin was purified using antibodies recognizing the anti-FLAG epitope. Protein LFQ in podocin pull-downs was compared with a control pull-down. N = 4 (control IP) or 5 (FLAG IP) independent biological and technical replicates.
Table 2.
The table indicates proteins determined to be interactors of podocin expressed native glomeruli
| Gene names | Uniprot | Protein names | t-test difference [log2 (LFQ podocin IP/LFQ control IP)] | −Log P-values |
|---|---|---|---|---|
| Vapa | Q9WV55 | Vesicle-associated membrane protein-associated protein A | 5.39 | 2.47 |
| Ppa2 | D3Z636 | Inorganic pyrophosphatase 2, mitochondrial | 5.25 | 3.03 |
| Sept9 | A2A6U3 | Septin-9 | 5.14 | 5.68 |
| Dctn2 | Q99KJ8 | Dynactin subunit 2 | 4.74 | 3.82 |
| Nphs2 | Q91X05 | Podocin | 4.61 | 3.14 |
| Ubr4 | A2AN08 | E3 ubiquitin-protein ligase UBR4 | 4.26 | 5.79 |
| Cbx3 | Q9DCC5 | Chromobox protein homolog 3 | 4.03 | 3.14 |
| C1qbp | Q8R5L1 | Complement component 1 Q subcomponent-binding protein, mitochondrial | 3.90 | 1.12 |
| Bicd2 | Q921C5 | Protein bicaudal D homolog 2 | 3.87 | 1.90 |
| Dctn3 | Q9Z0Y1 | Dynactin subunit 3 | 3.32 | 2.06 |
| Lbr | Q3U9G9 | Lamin-B receptor | 3.17 | 3.87 |
| Nek9 | Q8K1R7 | Serine/threonine-protein kinase Nek9 | 3.13 | 3.16 |
| Vac14 | Q80WQ2 | Protein VAC14 homolog | 2.88 | 1.69 |
| Pkd2 | O35245 | Polycystin-2 | 2.86 | 1.39 |
| Gstz1 | Q9WVL0 | Maleylacetoacetate isomerase | 2.79 | 1.39 |
| Ppa1 | Q9D819 | Inorganic pyrophosphatase | 2.73 | 1.90 |
| Itm2b | O89051 | Integral membrane protein 2B | 2.69 | 3.17 |
| Akap1 | O08715 | A-kinase anchor protein 1, mitochondrial | 2.64 | 2.17 |
| Nphs1 | Q9QZS7 | Nephrin | 2.40 | 3.55 |
| Ndufa4 | Q62425 | NADH dehydrogenase [ubiquinone] 1-α subcomplex subunit 4 | 2.30 | 2.53 |
| Tubb2b | Q9CWF2 | Tubulin β-2B chain | 2.30 | 1.74 |
| Slc25a12 | Q8BH59 | Calcium-binding mitochondrial carrier protein Aralar1 | 2.29 | 4.89 |
| Kirrel | Q80W68 | Kin of IRRE-like protein 1 | 2.15 | 1.55 |
| Tufm | Q8BFR5 | Elongation factor Tu, mitochondrial | 2.14 | 2.62 |
| Slc25a10 | Q9QZD8 | Mitochondrial dicarboxylate carrier | 2.07 | 2.82 |
| Rhpn1 | E9Q7Q7 | Rhophilin-1 | 1.99 | 1.93 |
| Sept11 | Q8C1B7 | Septin-11 | 1.89 | 3.77 |
| Dld | O08749 | Dihydrolipoyl dehydrogenase, mitochondrial | 1.80 | 2.58 |
| Atp5o | Q9DB20 | ATP synthase subunit O, mitochondrial | 1.74 | 2.43 |
| Kank2 | Q8BX02 | KN motif and ankyrin repeat domain-containing protein 2 | 1.69 | 2.45 |
| Slc25a13 | Q9QXX4 | Calcium-binding mitochondrial carrier protein Aralar2 | 1.63 | 2.59 |
Podocin was purified using antibodies recognizing the podocin C-terminus. Protein LFQ in podocin pull-downs was compared with a control IP; n = 4 independent biological and technical replicates.
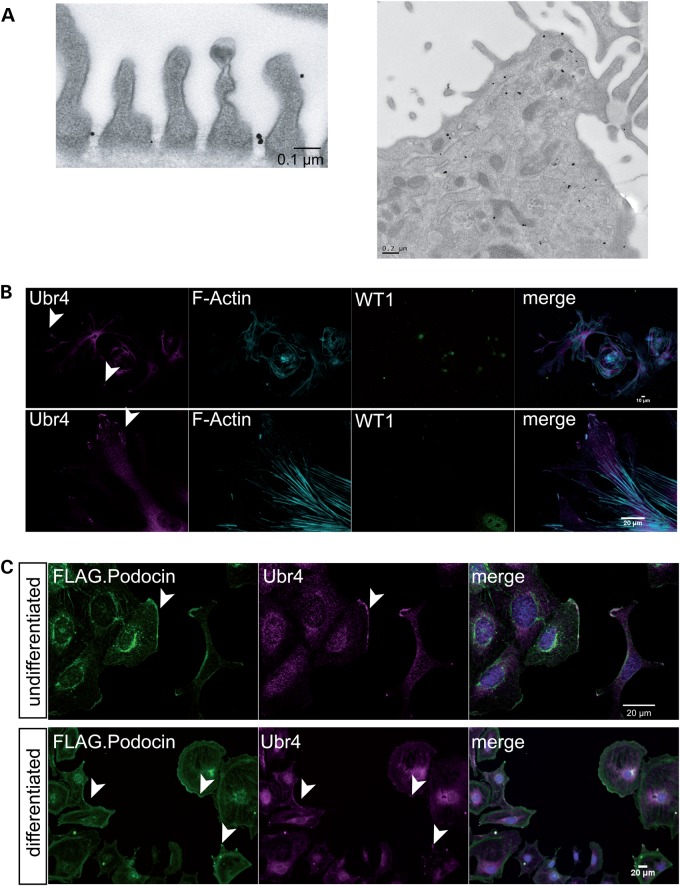
We next assessed the distribution of Ubr4 in podocytes. Interestingly, Ubr4 was localized at the slit diaphragm as demonstrated by immunogold localization in native podocytes in murine kidneys suggesting a role directly at the sites of the podocin protein complexes (Fig. 2A, left panel). It was also found in vesicular structures and the cytoplasm (Fig. 2A, right panel). In glomerular outgrowth cultures containing primary podocytes [positive for the podocyte-specific nuclear protein WT-1], we found that Ubr4 not only localized to the cell body, but also to the leading edge of migrating podocytes (Fig 2B). These results could be confirmed in established cell culture systems. In podocyte cell lines transduced with FLAG-podocin, the identical cell line used for initial discovery of Ubr4, we also found a colocalization of Ubr4 and podocin (Fig. 2C).
Figure 2.
Ubr4 localizes to the slit diaphragm and leading edges of podocytes. (A) Immunogold labelling of Ubr4 in native mouse glomeruli. There was positive immunogold signal for Ubr4 at the slit diaphragm spanning between two cell processes. The right picture demonstrates a podocyte cell body with positive immunogold labelling in cytoplasma vesicles, multi-vesicular bodies and ER. (B) Immunostaining of Ubr4 in a glomerular outgrowth culture. The Ubr4 signal was detected at leading edges of podocytes. (C) Immunostaining of Ubr4 and F.Podocin in cultured mouse podocytes revealed an overlap in both undifferentiated (33°C) and differentiated (37°C) podocytes. Nuclei were counterstained with DAPI. An overlap is marked by white staining in the merged channel. Scale bars indicate 20 µm.
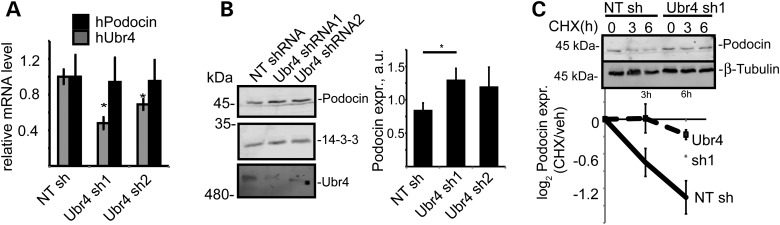
To investigate whether the E3 ubiquitin ligase Ubr4 may regulate podocin protein levels, we generated stable UBR4 knock-down human podocyte cell lines by expression of shRNAs. These cells showed a significant down-regulation of UBR4 protein as well as mRNA (Fig. 3A and B). Analysis of the endogenous protein levels revealed that endogenous podocin protein levels were increased in the cell lines depleted of UBR4 while the level of podocin mRNA was not affected by the UBR4 shRNAs (Fig. 3A and B). Consistent with a role of UBR4 in regulating podocin abundance, knock-down of UBR4 also enhanced podocin protein stability when protein synthesis was blocked using cycloheximide (CHX) (Fig. 3C). Immunolocalization of endogenously expressed podocin confirmed association with membrane ruffle-like structures and leading edges in wild-type (WT) podocytes (Supplementary Material, Fig. S2). Podocin was not found in these structures in the Ubr4 knock-down cell lines [membrane ruffling could not be observed in Ubr4 knock-down cell lines as previously described (13)]. We, thus, hypothesized that Ubr4 may also counteract expression of mistargeted protein multimers.
Figure 3.
The effect of Ubr4 knock-down on cultured human podocytes expressing podocin. (A) mRNA expression of Ubr4 and podocin was measured using quantitative RT-PCR. A non-targeting (NT) shRNA served as control. n = 5 replicates, P < 0.05. (B) Protein expression of podocin was increased in Ubr4 knock-down cells. Whole-cell lysates were analysed using immunoblotting for podocin. N = 5 replicates, P < 0.01 in a two-tailed t-test. (C) Pulse chase assay with CHX incubation. Podocytes were incubated using CHX (c = 100 ng/ml) for the indicated time periods. Whole-cell protein lysates were analysed using immunoblotting and membranes were probed with an anti-podocin antibody. Stability of podocin was increased in Ubr4 knock-down cells. n = 5 replicates. Asterisk indicates significance (P < 0.05) in a two-tailed t-test. All error bars in this figure represent SEM.
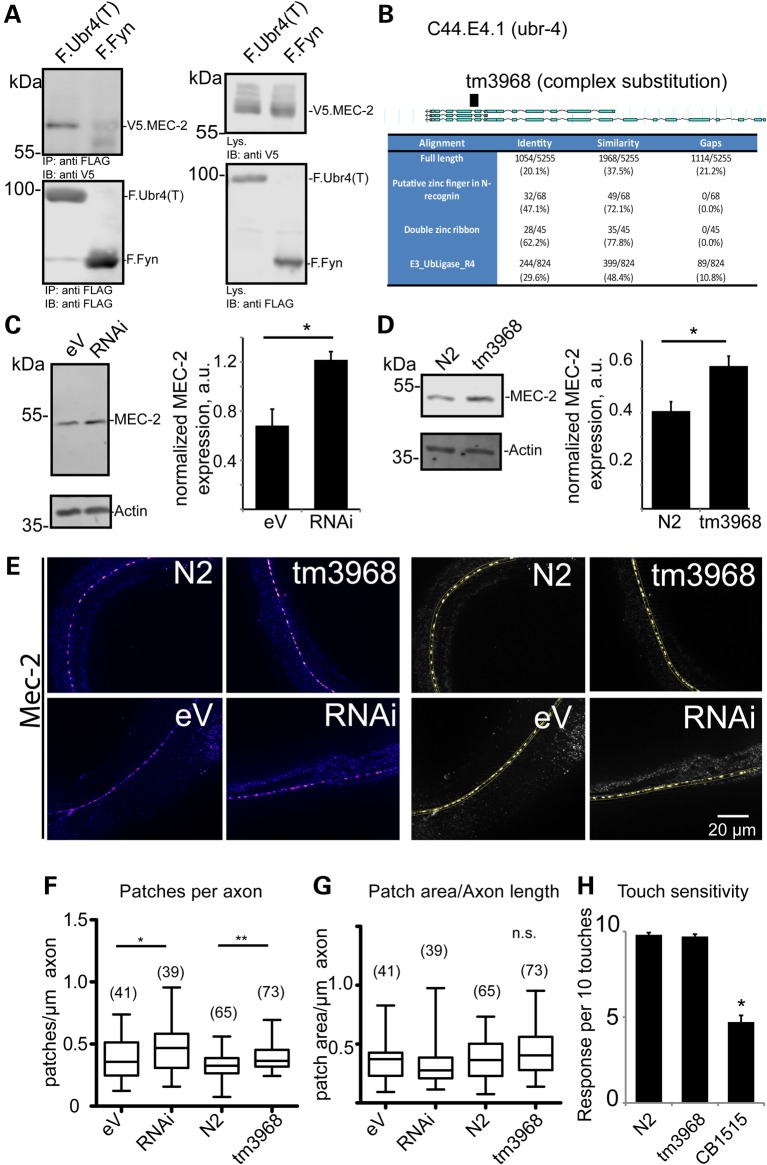
To assess the role of Ubr4 in targeting podocin multi-protein complexes in vivo, we used the C. elegans system as a model. The podocin-associated slit-diaphragm multi-protein complex is conserved in evolution and required for touch sensitivity in C. elegans (5). The podocin orthologue in C. elegans is MEC-2 that, similarly to podocin, orchestrates a mechanosensitive multi-protein complex, in this case required for touch activation of touch-sensitive neurons. As shown for podocin, MEC-2 also interacted with Ubr4 (Fig. 4A, Supplementary Material, Fig. S3). C44E4.1 is the putative nematode orthologue of Ubr4 (Fig. 4B). Knock-down of C44E4.1 (ubr-4) using RNAi in a worm system that allows efficient knock-down of neuronal proteins in vivo resulted in a significant increase of MEC-2 abundance (Fig. 4C). Next, an ubr-4 knock-out worm (tm3968) containing a deletion and frameshift in the fourth intron of C44E4.1 was analysed. Again, MEC-2 abundance was increased (Fig. 4D). MEC-2 mRNA was not affected in both systems (Supplementary Material, Fig. S4), suggesting a posttranslational mechanism. MEC-2 multi-protein supercomplexes can be visualized as touch channel punctae using immunostaining. We performed immunofluorescence analyses using MEC-2 antibodies in WT and ubr-4 knock-down and mutant worms and quantified the number of MEC-2-positive punctae along a large number of axons (Fig. 4E). Interestingly, these analyses revealed that the number of MEC-2 punctae was significantly increased in both ubr-4 mutant and knock-down worms without affecting the punctae size (Fig. 4F and G) whereas touch sensitivity was not affected (Fig. 4H). These data indicated that ubr-4 regulates protein expression and degradation of redundant MEC-2 multimers.
Figure 4.
The effect of mutation or knock-down of the Ubr4 C. elegans orthologue, C44E4.1 (ubr-4), on the C. elegans podocin orthologue, MEC-2. (A) An C-terminal truncation of Ubr4 interacts with MEC-2. FLAG-tagged Ubr4 truncation (containing the last 800 amino acids of human Ubr4), and V5-tagged MEC-2 were transfected in HEK293T cells. FLAG-tagged Fyn served as a control protein. Proteins were immunoprecipitated using an anti-FLAG pull-down lysates were separated using SDS–PAGE. Blots were probed with V5 and FLAG antibody. (B) Identification of the Ubr4 orthologue in C. elegans as C44E4.1, now termed ubr-4. A strain carrying a mutated allele (tm3968) was utilized for further studies. (C) The effect of knock-down of ubr-4 (RNAi) on MEC-2 abundance in nematode when compared with control RNAi [empty vector (eV)]. MEC-2 abundance was measured using immunoblotting and densitometry (n = 5 and P < 0.01 in a two-tailed t-test). (D) The effect of ubr-4 mutation (tm3968) on MEC-2 abundance when compared with wild-type (N2) worms. MEC-2 abundance was measured using immunoblotting (n = 5 and P < 0.01 in a two-tailed t-test). (E) Immunostaining of MEC-2 in ubr-4 mutant and knock-down worms. The left panel shows MEC-2-positive punctae in C. elegans heads. The right panel demonstrates thresholding analysis to quantify MEC-2-positive punctae. (F, G) Quantification of patch number and patch size. The number of quantified worm images (three independent grownups) is given in brackets. (H) Touch response of worms lacking ubr-4. Reaction of worms in response to 10 gentle touches was measured. No effect on touch sensitivity was found. The mutant CB1515 (Mec-10) served as a touch-hyposensitive control (14).
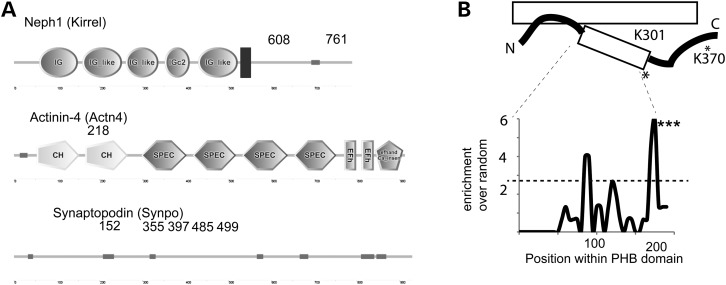
Next, we aimed for the identification of Ubr4-dependent ubiquitylation sites in vivo. Here, we used an anti-K-ε-Gly-Gly remnant antibody to enrich for ubiquitylated peptides after tryptic digestions (15–17). We freshly isolated glomeruli from mice and utilized this technique to identify endogenously ubiquitylated sites (15). We discovered 714 endogenous ubiquitylation sites within the glomerular sample, among these many important glomerular proteins such as Neph1 (Kirrel), actinin-4 (Actn4) and synaptopodin (Synpo) (Fig. 5A). Results of these analyses are deposited in a kidney systems biology repository (https://helixweb.nih.gov/ESBL/Database/GloUR/) and some features of the data set are highlighted in Supplementary Material, Figure S5. Consistent with a previous deep-mapping ubiquitylomic study (17), we found endogenous podocin ubiquitylation at the lysines K301 and K370 (Fig. 5B). No other podocin ubiquitylation sites have been described in large-scale studies at this point. Bioinformatic strategies have recently been developed to pinpoint specific areas within protein domains to prioritize the importance of post-translational modifications (18). The method calculates enrichment over random of collected MS/MS evidence of an ubiquitylation site within a certain domain across numerous species (18). We performed such an enrichment analysis for the PHB domain (spanning from residues 125–301, according to the PFAM database). We found that ubiquitylation evidence clustered significantly at the C-terminal part of the PHB domain in close vicinity to the discovered ubiquitylation site K301 (Fig. 5B).
Figure 5.
Ubiquitylomic analysis of native glomeruli. (A) Cartoon of Neph1 (Kirrel), Actn4 and Synpo proteins. Among > 600 ubiquitylation sites, ubiquitylation sites on Neph1, Actn4 and Synpo were discovered. The numbers indicate the lysine residues. (B) Proteomic discovery of the podocin ubiquitylation sites K301 and K370 (asterisk) in native glomeruli. The cartoon depicts membrane model of podocin. The podocin ubiquitylation site K301 localizes at the distal part of the PHB domain (spanning residues 125–301 according to PFAM). The distal part of the PHB domain is significantly enriched for ubiquitylation sites based on mass spectrometric evidence (***P = 0.0006).
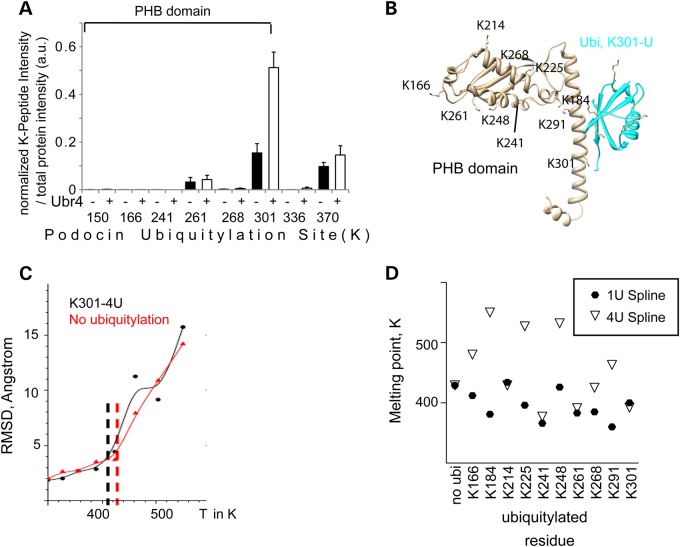
We assumed that Ubr4 might be involved in ubiquitylating podocin at the residues K301 and/or K370. To test this hypothesis, we used an independent approach expressing podocin and Ubr4 in HEK293T cells followed by protein denaturation, podocin immunoprecipitation, tryptic digestion and LC-MS/MS to detect ubiquitylation sites of podocin (Fig. 6A). The intensity (MS1) of the peptide containing the K-ε-Gly-Gly-site was normalized by the total intensity of all peptides belonging to the protein. At baseline conditions, K301 and K370 were already the most predominant ubiquitin remnants discovered on podocin confirming the in vivo findings (Fig. 6A). Moreover, most of the ubiquitylation sites localized to the PHB domain. Ubr4 coexpression specifically increased the ubiquitylation at the podocin K301 site, and this increase was in part mediated by increased K48 and K63 ubiquitylation (Supplementary Material, Fig. S6A). Immunoblot analysis confirmed the increase, since K48-dependent ubiquitylation was increased in the presence of Ubr4. This was again partially dependent on the K301 and K370 sites (Supplementary Material, Fig. S6B).
Figure 6.
Ubr4 ubiquitylates podocin at K301 to unfold the protein. (A) Analysis of the ubiquitylation of expressed podocin in the presence (black columns) and absence (white columns) of overexpressed Ubr4 by proteomics (n = 5 experiments). Cells were incubated with MG132 (10 µm, 1 h). Podocin was purified using an anti-FLAG antibody, and immunoprecipitates were analysed using LC-MS/MS. Ubiquitylation sites of podocin were identified as K-ε-Gly-Gly ubiquitin remnant peptides. MS1 intensities of site containing ubiquitylation sites were normalized against total protein intensity (based on quantification of 47 unique peptides). The majority of all ubiquitylation sites reside within the PHB domain. Error bars indicate SEM. (B) The model of the podocin PHB domain with an attached monoubiquitylation chain at the residue K301. K301 corresponds to K142. The PHB domain (grey) was modelled using the crystal structure template PDB 3bk6, the ubiquitin residue (cyan) was based on PDB 1ubq. (C) Melting curve of podocin ubiquitylation in the presence or absence of a K48-linked ubiquitin chain (4U) as determined by replica exchange molecular dynamics simulation. (D) Melting point determination of mono- and polyubiquitylated podocin by second derivative measurements. The melting points of each ubiquitylated proteoform are presented for the monoubiquitylated (1U spline, closed circles) and the polyubiquitylated (4U spline, open triangles) form of each proteoform. The residues K241, K261 and K301 (when attached to a mono- or polyubiquitylated residue) unfold the protein more easily when compared with ubiquitylation at other sites. See Supplementary Material, Figs. S8–S12 for further details of molecular dynamics simulation.
Given the known promiscuity of ubiquitylation processes, we asked whether there was a biophysical explanation for this strongly site-specific ubiquitylation. In fact, it has been postulated that attachment of ubiquitin chains may cause local defolding of the protein around the ubiquitylation site, and a strong defolding is favourable for the degradation of a protein (19–21). We postulated that ubiquitylation would be particularly important for such a compact structural domain as the PHB domain. This effect is dependent on the localization of the ubiquitylation site and can be predicted using an in silico molecular dynamics technology capable of predicting folding effects using both quantum mechanics and classical physics (19). This technology allows to investigate alterations in podocin conformation (2,22). To this end, we performed replica exchange molecular dynamics simulations with ubiquitylation at every potential ubiquitylation site (Supplementary Material, Figs S7 and S8). The root-mean-square deviation (RMSD) of atomic positions was measured at the core of the PHB domain in response to attaching a monoubiquitin (1u) or a K48-linked polyubiquitylation chain (4u) at each available lysine residue (Fig. 6B and C, Supplementary Material, Figs. S7–S12). These analyses revealed that the K301–4u (K48 linked) podocin protein was among the most easily to unfold (together with polyubiquitylated K241, K261), suggesting that K301 ubiquitylation may not only cause degradation, but specifically affect unfolding of the podocin monomeric proteins. Thus, evolutionary analyses, proteomics and in silico experiments predict a high importance of the site K301 when compared with the other sites within the PHB domain (Fig. 6D).
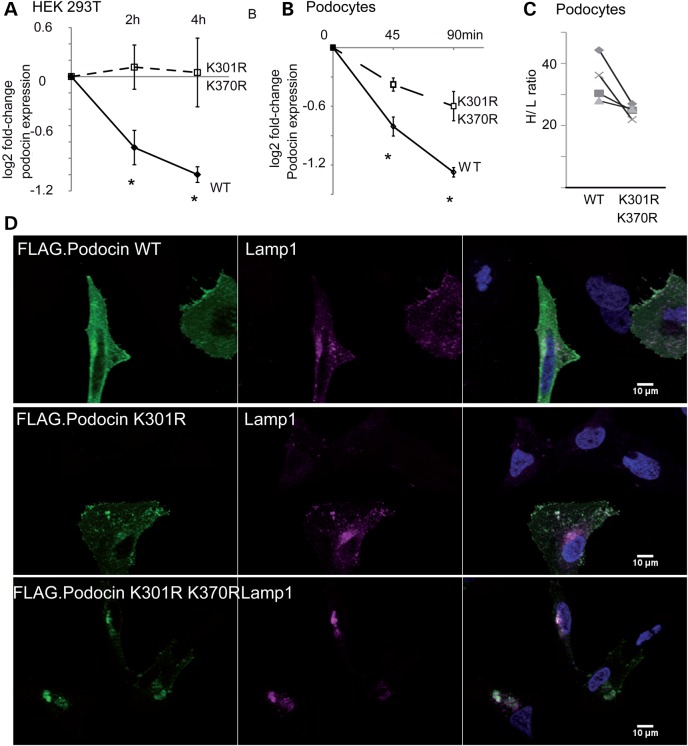
If this hypothesis was true, mutation of the specific lysine residues should affect protein stability. Therefore, we next investigated whether abolishing these sites was in fact sufficient to induce stability. Assays using CHX to block protein translation revealed that protein stability of the K301R or K301R/K370R mutant was significantly increased when compared with the WT podocin protein (Fig. 7A). We also generated stable podocyte cell lines expressing FLAG-tagged WT or K301R–K370R form of the protein. We found that in this system, both proteasomal and lysosomal inhibition significantly decreased podocin WT degradation (Supplementary Material, Fig. S13). When analysing the mutated protein, we found that the WT protein was less stable (Fig. 7B), and we confirmed this finding with a pulsed-SILAC approach. We subjected both differentiated podocyte cell cultures to medium-containing isotopic labelled arginine (13C615N4; Arg-10) and lysine (13C615N2; Lys-8) (Fig. 7C). Consistently, we observed less incorporation of heavy isotopes when both lysines were mutated to arginine. We also analysed whether the mutated podocin would localize differently in transiently transfected HeLa cells. A strong preference of the overexpressed mutated protein to form large, LAMP1-positive aggregates were observed (Fig. 7D). Moreover, a colocalization was already observed with a K301R mutation (Fig. 7D). Taken together, these data suggest that ubiquitylation of specific lysine residues may represent an important principle in proteostasis of podocin/MEC-2 protein supercomplexes.
Figure 7.
The Ubr4-dependent residues regulate podocin stability. (A) Measurement of podocin protein stability when expressed in HEK293T cells. Cells were incubated with CHX (c = 100 ng/ml) for the indicated periods. Densitometry quantification of WT and mutant (K301R and K370R) podocin are depicted as logarithmic ratios (n = 5; P < 0.01 in a two-tailed t-test). (B) Measurement of podocin protein stably expressed in podocyte cell lines. Podocytes were incubated with CHX for the indicated periods of time ratios (n = 5; P < 0.01 in a two-tailed t-test). Podocin expression was analysed using immunoblotting for FLAG. (C) Pulse labelling of podocytes with medium containing heavy isotope labelled amino acids. Podocin WT and mutant (K301R/K370R) was purified using an anti-FLAG antibody. Incorporation of heavy isotope labelled amino acids into podocin WT and mutant was measured. Each symbol represents a separate replicate of SILAC ratio quantification per protein. P = 0.05 in a two-tailed t-test. The following 10 peptides were analysed across the experiment: AASESLR, APAATATVVDVDEVR, ARPDAGAER, DMFIMEIDAVCYYR, LGHLLPGR, LPAGLQHSLAVEAEAQR, MAAEILSGTPAAVQLR, SLTEILLER, VALDAVTCIWGIK and VVQEYER. (D) Localization of FLAG-tagged podocin and podocin mutant forms in HeLa cells. Podocin and podocin mutants were transfected in HeLa cells. Podocin was stained using an anti-FLAG antibody. Nuclei were counterstained with DAPI. Podocin with K301R and K301R–K370R substitution colocalized with cotransfected Lamp1, a lysosome marker. Altered podocin localization was independent of co-transfection of Lamp1 (data not shown).
Discussion
The unique morphology of the podocyte slit diaphragm requires a tightly regulated local protein homeostasis. Ubiquitylation plays a major role in turnover of slit-diaphragm proteins, including podocin (23–25). In this manuscript, we discover and functionally characterize the molecular correlates of the podocin protein degradation mechanism. Podocin stability was largely dependent on Ubr4 (Figs 1, 3, 4 and 6). Ubr4 is an E3 ubiquitin ligase that was initially discovered as an N-recognin (13,26). It might also function as an adaptor for other E3 ubiquitin ligases (27). Very recent studies revealed that Ubr4 is essential for proteasomal, autophagosomal and lysosomal degradation of several cytoplasmatic and membrane proteins (27–30). It is also required for membrane ruffling and actin cytoskeleton organization (13). Ubr4 is recruited to tyrosine phosphorylated EGF receptor (31). Receptor tyrosine kinase environments are largely similar to the slit-diaphragm protein complex, which is orchestrated by tyrosine phosphorylation of Fn3 domain-containing proteins (32,33). In addition, Ubr4 has been implicated in various functions including localization and turnover of membrane channels in neurons or axon guidance (27,34). Knock-out of this protein in mice causes an embryonically lethal phenotype, mainly via disruption of vascular development and yolk sack defects (29). Interestingly, when reducing or ablating this multi-functional protein in the model organism C. elegans, we found a viable and normally developing worm with increased MEC-2 abundance (without affecting RNA levels). In both worm strains, the number of neuronal, MEC-2-positive touch punctae was increased. If the Ubr4 orthologue was only involved in the mere protein degradation, we would have expected more intensely stained or maybe larger punctae. However, the in vivo data in the model organism C. elegans suggested that Ubr4 is responsible for the turnover of excessive or mistargeted membrane proteins and that this process is closely linked to subcellular localization.
Podocin was found to be ubiquitylated on the conserved residues K301 and K370 in this and another large-scale ubiquitylomic study (17). These residues are also the predominant ubiquitylation sites when this protein is expressed in cell culture systems (Fig. 6A), and Ubr4 even increases its ubiquitylation further. When we induced point mutations in these residues, we saw an effect on subcellular localization and protein degradation, indicating that these specific residues are of importance for the stability and potentially localization of the protein. Computational biophysical analysis using replica exchange molecular dynamics simulations for the PHB domain with ubiquitylation at every potential ubiquitylation site revealed that K301 mono- and polyubiquitylation specifically affects podocin stability by allowing unfolding of the domain (Fig. 6B–D). These data clearly indicated that K301 ubiquitylation may not only cause degradation, but specifically affects unfolding of the podocin monomeric proteins. Podocin and all PHB domain proteins form large megadalton complexes mediated by the PHB domain (5,22). Therefore, it is important to realize that only some parts of the PHB domain may be not accessible to ubiquitylation [e.g. K268 is a predicted part of the PHB–PHB interaction surface (35)]. These data support the concept that ubiquitylation has site-dependent effects. Taken together, this study identified important components and biophysical principles of the podocin/MEC-2 protein degradation machinery, revealing tight control both on a subcellular and on a molecular level. Now, future studies should elucidate if the podocin protein degradation mechanism pointed out here is altered in pathophysiological conditions. To this end, knock-in mouse models of podocin mutant forms, and inducible, podocyte-specific knock-out models of Ubr4 could be analysed.
This study has two ancillary findings. First, this study provides an initial draft of the podocin interactome—both native and in cell culture (Fig. 1). It is strongly dominated by membrane proteins, and interacts with signalling receptors such as Notch, Tgfbr2 and Igf-receptor, all of which are of extraordinary importance for podocyte function and survival (36–38). It will be interesting to test whether podocin provides an optimal signalling environment for these proteins and if its mutation alters the signal transduction pathways of these three receptors. In native glomeruli, podocin also interacts with its known slit-diaphragm partners, such as Nephrin and Neph1. Intriguingly, Kank2, a gene mutated in focal segemental glomerular sclerosis (FSGS) in humans (39), had high affinity to podocin in this analysis. Secondly, this study provides an initial analysis of native ubiquitylation sites on glomerular proteins (Fig. 4). These identifications include one ubiquitylation site on the slit-diaphragm protein Neph1 (Kirrel) and one on the ACTN4 protein, a protein mutated and degraded in forms of hereditary FSGS (40,41). It will be important to elucidate the functional consequences of these and other recently discovered post-translational modifications in vivo (42), and to dissect their interplay. These data may also help to elucidate specific protein degradation pathways of proteins which may be first molecular targets for rescuing decay of proteins in renal genetic disease.
Concise Methods
Cell culture
HEK293T cells were maintained at 37° in DMEM and 10% fetal bovine serum (Sigma). HeLa cells were maintained at 37° in DMEM and 10% serum. Murine podocytes (line obtained from Dr Stuart Shankland, Seattle) were cultivated on Collagen-I-coated dishes at 33° as previously described (43,44). Human podocytes (45) (line obtained from Dr Moin Salem, Bristol) were cultivated in the presence of 10% serum and 1 × insulin–transferrin–selenium supplement (Sigma). To allow for differentiation, both podocyte cell lines were cultivated at 37° (10 days). The cell lines are characterized in the respective references and are standard podocyte cell culture models. All of the cell lines were tested mycoplasma negative during this study. No cell-line cross-contamination was reported in the respective databases. Glomerular outgrowth cultures containing native podocytes were generated by plating decapsulated glomeruli on Collagen-I-coated dishes and growing for 7–10 days until the appearance of star-shaped cells with extensions as described previously (46).
Isolation of murine glomeruli
All animal studies were approved by the responsible local and federal committees according to the German Tierschutzgesetz (LANUV, NRW). Murine glomeruli were isolated using the magnetic bead perfusion technique (47). The kidneys of 8-week male Bl6N mice were perfused with 1 ml of Hanks buffered saline solution (HBSS) containing 200 µl of tosyl-activated magnetic beads (Invitrogen). Then, glomeruli were digested as previously described (48) and subjected to sieving (100 µm mesh size). Glomeruli were extracted using a magnet, washed with ice-cold HBSS buffer and purity was verified microscopically (>97% glomeruli). Then, the pellet was suspended either in modified RIPA buffer for IP analysis or in 8 m urea buffer for analysis of ubiquitin remnants. After homogenization using a glass homogenizer, the sample was centrifuged at 16 000g for 20 min and the supernatant was subjected to tryptic digestion or immunoprecipitation.
Interactome analysis
For interactome analysis, we used a protocol previously developed for endogenous immunoprecipitations (10,11). Briefly, cells or tissue lysates were homogenized in modified RIPA buffer (1% IGPal, 150 mm NaCl, 0.25% sodium deoxycholate and 50 mm Tris) supplemented with 1 × protease inhibitor mix with EDTA (Roche). Next, lysates were centrifuged for 10 min at 16 000g. 800–1000 µl of glomerular lysates was incubated with primary antibody (anti-Podocin, Sigma, 1:200) for 30 min and subsequently with 100 µl micro-Protein G beads (Miltenyi). For control IPs, the same amounts of lysates were incubated in parallel with an anti-V5 polyclonal rabbit antibody (Sigma). For purifying of stably expressed FLAG epitopes, lysates were directly incubated with anti-DYKDDDDK micro-beads (Miltenyi). A cell line stably expressing GFP served as control for the same incubation with anti-DYKDDDDK micro-beads. Lysates incubated with micro-beads (and antibodies) were then subjected to magnetic column purification. Three washes were performed using wash buffer 1 (containing 50 mm Tris, 150 mm NaCl, 5% glycerol and 1% IgPal). Next, columns were washed five times with wash buffer 2 (containing 50 mm Tris, 150 mm NaCl). Then, columns were subjected to in-column tryptic digestion containing 7.5 mm ammonium bicarbonate, 2 m urea, 1 mm DTT and 5 ng/ml Trypsin. Then, digests were eluted using two times 50 µl of elution buffer 1 containing 2 m urea, 7.5 mm Ambic, and 4 mm IAA. Digests were incubated over night at room temperature with mild shaking in the dark. Samples were stage-tipped the next day as previously described (49).
Electron microscopy
Mice were deeply anesthetized with sodium pentobarbital and transcardially perfused using 4% paraformaldehyde and 0.05% glutaraldehyde in 0.1 m PB. The kidneys were removed and post-fixed in the same fixative (overnight at 4°C). After post-fixation, tissue blocks were washed in 0.1 m PB, afterwards horizontal vibratome sections (50 µm) of the kidney were cut. For pre-embedding immunogold labelling vibratome sections were first incubated with anti-ubr4 antibody (1:50 or 1:100, overnight. at 4°C) followed by nanogold-coupled secondary antibody (1:100, overnight. at 4°C, Nanoprobes, USA). Finally, silver enhancement was performed for 8 min on ice (HQsilver, Nanoprobes, USA). After treatment with OsO4 (0.5% for 30 min), the sections were stained with uranyl acetate (1 in 70% ethanol for 30 min), dehydrated and flat-embedded in epoxy resin (Durcupan ACM, Fluka, Sigma-Aldrich, Gillingham, UK). 40 nm ultrathin sections were cut and analysed using an 80 kV Zeiss Leo transmission electron microscope.
Immunoprecipitation
Immunoprecipitation was performed as previously described (11). Briefly, HEK293T cells were harvested using IP buffer [1% Triton X-100, 20 mm Tris–HCl (pH 7.5), 50 mm NaCl, 50 mm NaF, 15 mm Na4P2O7, 2 mM Na3VO4 and protease inhibitors]. After 15 min of incubation on ice, lysates were centrifuged (20 000×g, 15 min, 4°C). The supernatant was used for further procedures. Fifty microlitres of the supernatant were kept as the ‘lysate’ and boiled with 1 × Laemmli buffer, and the remaining supernatant was incubated at 4°C with anti-FLAG-agarose beads (M2, Sigma) for 1 h. The beads were washed five times extensively with 1 ml of IP buffer and proteins were eluted by boiling in 2 × Laemmli buffer. Lysates and immunoprecipitated samples were subjected to immunoblot.
Plasmids, transfection and transduction
pRK5-HA-Ubiquitin-K48 and WT was a gift from Ted Dawson [Addgene plasmid #17605, Plasmid #17608)] (50). The full-length Ubr4 expression construct was kindly provided by Dr Yong-Tae Kwon (Pittsburgh), and the Ubr4(D) construct, containing an C-terminal truncation of the Ubr4 (pRK7) was obtained from Dr Zhang (Fudan University, Shanghai). Lamp1-mCherry fusion protein was obtained from Astrid Schauss (University Cologne). Constructs cloned into a pcDNA 6 Vector (5 µg, F.Podocin) or into a pcDNA 3 Vector (10 µg, full-length Ubr4.V5, HA.Ubiquitin) were transfected using the Calcium phosphate method. GFP served as a transfection control in a separate dish.
For generation of stable podocyte cell lines overexpressing podocin, lentiviral gene transfer was utilized. The vector utilized for this was F.Podocin cloned into pLenti6.3. For knock-down studies, two commercial high-titre virus stocks of lentivirus expressing Ubr4 shRNAs (Sigma) were used. After incubation with virus for 48 h, the cells were incubated in virus-free medium for 24 h and then selected using puromycin or blasticidin according to the resistance casettte of the transduced vector for 48–96 h or all control cells died.
Quickchange mutagenesis was performed to introduce indicated point mutations into podocin. The following primer sequences were used for mutagenesis:
- K301R
- FP: GCTGCAGAAGGGGAAAGGGCTGCTTCAGAGTCCC
- RP: GGGACTCTGAAGCAGCCCTTTCCCCTTCTGCAGC
- K370R
- FP: CCCAAGTTCCTCCAGACCAGTTGAACC
- RP: GGTTCAACTGGTCTGGAGGAACTTGGG
Primary antibodies
| Target | Type | Species | Company | Cat. No. | Application | Dilution |
|---|---|---|---|---|---|---|
| Ubr4 | Polyclonal | Rabbit | Abcam | ab86738 | Electron microscopy | 1:50, 1:100 |
| Immunofluorescence | 1:400 | |||||
| Immunoblot | 1:500 | |||||
| Ubr4 | Polyclonal | Rabbit | Abcam | ab114109 | Immunofluorescence | 1:400 |
| Podocin | Polyclonal | Rabbit | Sigma | P0372 | Immunoprecipitation | 1:200 |
| Immunoblot | 1:2000 | |||||
| Immunofluorescence | 1:400 | |||||
| WT1 | Monoclonal | Mouse | Santa Cruz | sc-7385 | Immunofluorescence | 1:400 |
| Polyclonal | Goat | Santa Cruz | sc-15421 | Immunofluorescence | 1:400 | |
| Flag | Polyclonal | Rabbit | Sigma | F7425 | Immunoblot | 1:2000 |
| Monoclonal | Mouse | Sigma | F3165 | Immunofluorescence | 1:400 | |
| Immunoblot | 1:2000 | |||||
| V5 | Monoclonal | Mouse | Serotec | mca1360 | Immunoblot | 1:10000 |
| Mec-2 | Polyclonal | Rabbit | gift from Martin Chalfie | Immunofluorescence | 1:200 | |
| Immunoblot | 1:200 | |||||
| HA-tag | Polyclonal | Rabbit | Covance | PRB-101P | Immunoblot | 1:500 |
| Tubulin | Monoclonal | Mouse | Sigma | T0198 | Immunoblot | 1:1000 |
| Actin | Monoclonal | Mouse | Millipore | MAB1501R | Immunoblot | 1:1000 |
| 14-3-3 | Polyclonal | Rabbit | Santa Cruz | sc-629 | Immunoblot | 1:1000 |
Secondary antibodies and other dyes
| Antibody | Company | Cat. No. | Application | Dilution |
|---|---|---|---|---|
| Goat anti-rabbit, HRP coupled | Cayman | 10004301 | Immunoblot, ECL | 1:25 000 |
| Goat anti-mouse, HRP coupled | Cayman | 10004302 | Immunoblot, ECL | 1:25 000 |
| Donkey anti-rabbit, IRDye® 680RD | Licor | 926-68073 | Immunoblot, Licor | 1:25 000 |
| Donkey anti-mouse 800 IRDye® 800 RD | Licor | 926-32280 | Immunoblot, Licor | 1:25 000 |
| Donkey anti-rabbit, Cy3 | Jackson ImmunoResearch | 711-165-152 | Immunofluorescence | 1:400 |
| Donkey anti-rabbit, A488 | Jackson ImmunoResearch | 711-546-152 | Immunofluorescence | 1:400 |
| Donkey anti-mouse Cy 3 | Jackson ImmunoResearch | 715-165-150 | Immunofluorescence | 1:400 |
| Donkey anti-mouse A488 | Jackson ImmunoResearch | 715-545-150 | Immunofluorescence | 1:400 |
| Donkey anti-goat A488 | Jackson ImmunoResearch | 705-545-003 | Immunofluorescence | 1:400 |
| Alexa flour 568 phalloidin | Invitrogen | A12380 | Immunofluorescence | 1:400 |
| Alexa flour 488 phalloidin | Invitrogen | A12379 | Immunofluorescence | 1:400 |
| Phalloidin, 647 | Dyomics | 647P1-33 | Immunofluorescence | 1:400 |
| DAPI | Invitrogen | D1306 | Immunofluorescence | 1:10 000 |
| Nanogold goat anti-rabbit | Nanoprobes | #2004 | Electron microscopy | 1:100 |
Immunoblot
Lysates were run on a 6–10% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) and blotted onto a PVDF membrane. Blots were either developed using fluorescence-based detection of proteins (Licor Odyssey developer) or using enhanced chemoluminiscence (Fusion developer). For fluorescence-based detection, samples were blotted ‘semi-dry’ on a PVDF low-fluorescence membrane, blocked with one-time Rothiblock (Roth) and incubated in antibody diluted in PBST. Secondary antibodies (Licor) were directed anti-rabbit (wavelength, 680 nm) or mouse (wavelength, 800 nm). For chemoluminiscence-based detection, membranes were blocked using 5% bovine serum albumin and incubated with antibody in TBST. For detecting full-length Ubr4, a 6% SDS–PAGE was used and the transfer was performed overnight in a tank blot at 4°. Western blots were densitometrically quantified using the image studio lite version 4.0 (Licor) for fluorescence-based protein detection, and imageJ/FiJi for chemoluminiscence-based detection as previously described (22).
For every experiment and biological replicates, the same western blot method/development method/quantification method was used.
Detection of podocin ubiquitylation in cell culture by MS and western blot
Transfected HEK293T cells were lysed in 2% SDS, 50 mm Tris, pH = 8 and subjected to boiling for 10 min. Next, lysates were diluted using a dilution buffer (10 mm Tris–HCl, pH 8.0, 150 mm NaCl, 2 mm EDTA, 1% Triton) to a final concentration of <0.1% SDS. Then, lysates were subjected to centrifugation (16 000g, 10 min), and the supernatant was subjected to immunoprecipitation using M2-FLAG beads (Sigma). After 1.5 h, beads were washed three times with 1 ml of ubiquitylation wash buffer (10 mm Tris–HCl, pH 8.0, 1 M NaCl, 1 mm EDTA, 1% NP-40) and were then eluted using two times Laemmli buffer.
For the detection of ubiquitylation residues by MS, lysates were incubated with 50 µl of Miltenyi µbeads followed by magnetic µMacs column purification (Miltenyi). Then, columns were washed one time with dilution buffer, two times with ubiquitylation wash buffer and five times with 150 mm NaCl, 50 mm Tris. Then, bound proteins were subjected to tryptic digestion by in column digestion as described in the ‘Interactome analysis’ section. Then peptides were stage-tipped and subjected to LC-MS/MS analysis.
Immunofluorescence
Immunofluorescence analysis of PFA-fixed cells was performed as previously described (51). MEC-2 staining on Day 1 adult hermaphrodites was performed using a modified Finney–Ruvkun protocol (52). Worms were prepared for antibody staining as described previously (53). Strains were then incubated with a rabbit antibody detecting the N-terminus of MEC-2 (kind gift of Professor Martin Chalfie; dilution 1:200 in antibody buffer). Several washing steps were followed by incubation with a Cy3-labelled secondary antibody (Jackson Immunoresearch, West Grove, PA, USA) (1:400 dilution in antibody buffer). After extensive washing the worms were subsequently counterstained with DAPI (Invitrogen) and mounted on slides with Prolong Gold Antifade Reagent (Invitrogen). Images were captured with an Axiovision microscope with apotome function enabled using a water immersion objective. The Axiovision software was used to record (all from Carl Zeiss MicroImaging GmbH). zvi images were further processed using the ImageJ/Fiji software version 1.46 (NIH) (54,55) and the ‘magic montage’ macro. Appropriate negative controls were performed.
Worm strains
All strains were grown and maintained according to standard methods on OP50-containing NGM agar plates 17. Following worm strains were used: WT N2 and CB1515 received from CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440) and the strain containing the tm3968 allele received from Mitani lab, Japan.
Caenorhabditis elegans RNAi feeding experiment
RNAi experiments were performed as described by Fire et al. (56,57). RNAi clones for C44E4.1 or empty vector were used as contained in the library established by Julie Ahringer (58). All clones used were confirmed by sequencing. Twenty WT N2 L4 hermaphrodites were placed on 6 cm NGM plates with ampicillin and IPTG spotted with HT115 bacteria expressing RNAi clone of interest. Day 1 adult F1 progeny of these hermaphrodites were examined. Each n represents a biologically independent, separated group.
Caenorhabditis elegans touch assay
Day 1 adult hermaphrodites were tested for gentle-touch response as described by Chalfie and Sulston (59). Each animal was touched 10 times by a thin hair at the anterior part of the body. Stopping or backward movements were scored and shown as touch response out of 10 touches. The data were generated in three independent experiments using ≥10 worms each time for each data point. Statistical evaluation was done using a one-way analysis of variance (ANOVA), and a threshold P < 0.01 was considered significant.
Worm lysates for immunoblot analysis
Of note, 100 Day 1 adult hermaphrodites were picked in 50 ul of M9 buffer followed by sonication for three times for 10 s on ice with 10 s breaks using a Branson digital signifier (Danbury, CT, USA) at 10% amplitude. Two times Laemmli buffer was added to the worm lysate and samples were heated at 95°C for 10 min. Lysates were analysed by immunoblot.
Quantification of Mec-2 signal
Fiji/ImageJ (54,55) was used to count Mec-2-positive punctae along axons. The pictures were analysed in their native, zvi format without adjustment of contrast. First, suitable axon regions were marked manually. This linear region was extended to a width of 1.9 µm (20 pixel) and after background subtraction (ImageJ rolling ball/sliding paraboloid algorithm) and thresholding (ImageJ MaxEntropy algorithm) Mec-2-positive spots within this region were counted. This analysis was performed in a blind fashion.
qPCR of mRNA expression in cell culture and C. elegans
RNA from Day 1 adult hermaphrodites (C. elegans) was prepared by phenol extraction using Qiagen RNeasy kit. RNA was reverse transcribed using the SuperScript III First-Strand Synthesis System (Life Technologies, Carlsbad, CA, USA). Ten nanograms of cDNA were used per PCR reaction. Real-time PCR was performed using the Taqman Universal Master Mix (Life Technologies). The Data Assist software package version 3.01 (Life Technologies) was used for data analysis. P-values were calculated using a two-tailed Student's t-test. The following primer pairs and probes were used for qPCR analysis:
- tba-1:
- Forward primer: GAT CTC TGC TGA CAA GGC TTA C
- Reverse primer: TGG ATC GCA CTT CAC CAT TT
- Probe: /56-FAM/ATC ACC AAC /ZEN/AGT TGC TTC GAG CCA /3IABkFQ/
- rpl-32:
- Forward primer: CGT CCT TCC AAA CGG ATA CA
- Reverse primer: AGA CTC CAT GTC CGA TCT CT
- Probe: /56-FAM/CCA AAA TGT /ZEN/TAA GGA TTT GGA CAT GCT CC/3IABkFQ/
- mec-2:
- Forward primer: GGT AGT GCA AGA GTA CGA AAG AG
- Reverse primer: GAC AAT GAA GAA TAT ACC TGG TCC T
- Probe: /56-FAM/ATC TTC CGA /ZEN/CTT GGT CGC CTG ATG /3IABkFQ/
For cell culture studies, RNA was prepared by harvesting cells in Qiazol, (Qiagen), and RNA was subsequently using phenol-chloroform preparation. One millilitre trizol was mixed with 200 µl chloroform followed by 15 min centrifugation at 4000 rpm. The aqueous phase was removed. 1.5 × volume isopropanol was added, and the mixed sample was incubated at RT for 10 min. After additional centrifugation step, the RNA pellet was briefly dried on air and was dissolved with water. Quality was checked by measuring absorption in a nanodrop (Thermo Scientific) and reverse transcription was performed using the superscript III kit as described above. qPCR was performed using SYBR Green mastermixes, 50 ng of cDNA and published and validated primers for podocin, actin and UBR4.
The primer sequences were:
- UBR4:
- Forward primer: TGGACAAGCAATCAGCAGTTCT
- Reverse primer: GCACCCAGCAATTGGAAATT
- NPHS2 (Podocin):
- Forward primer: gct gcc gtt cag ctt cga
- Reverse primer: taa aac cac agt gga agg ctt ct
- ActB:
- Forward primer: ggacttcgagcaagagatgg
- Reverse primer: agcactgtgttggcgtacag
Enrichment for peptides with ubiquitin remnants
Ubiquitin remnant peptides were isolated as previously described with some modifications (15). After tryptic digestion, 3–4 mg of peptide samples were desalted using SepPak columns and were fractionated using a basic reverse phase column (XBridge BEH300 C18 3.5 μm 4.6 × 250 Column, Waters). Then, samples were pooled non-contiguously according to a previously described scheme (15). Ubiquitylated peptides were pulled down using the PTM-scan ubiquitin remnant antibody (10 µl slurry solution/fraction, Cell Signalling) exactly as previously described (15). In alternative experiments, 2 mg of peptides were directly subjected to immunoprecipitation using the PTM-scan ubiquitin remnant antibody (10 µl slurry solution). After acidic elution of the peptides, stage-tip (49) clean-up was performed.
Pulse labelling of differentiated podocytes
Podocytes expressing podocin WT, podocin K301R–K370R were seeded at the same density and were differentiated at 37° for 10 days and serum starved for 24 h. The medium was washed of and the cells were switched to a medium containing exclusively heavy isotope-labelled arginine (13C615N4; Arg-10) and lysine (13C6; Lys-6) (Thermo) for 24 h. Proteins were purified using a FLAG magnetic column and subjected to in column-digestion and LC-MS/MS analysis.
Liquid chromatography–mass spectrometry/mass spectrometry
Peptide separation was carried out using nLC coupled to a Q Exactive Plus tandem mass spectrometer (Thermo Scientific). An in-house packed 50 cm column with 1.7 μm C18 beads (Dr Maisch GmbH) was utilized within a column oven. A binary buffer system consisting of solution A: 0.1% formic acid and B: 80% acetonitrile, 0.1% formic acid was used. Linear gradients from 7 to 38% B in 60–240 min were applied with a following increase to 80% B for 5 min and a re-equilibration to 5% B. Peptides were sprayed into a quadrupole–orbitrap-based mass spectrometer, the Q Exactive plus (60). MS1 spectra were acquired using 1E6 as an AGC target. The maximal injection time was 20 ms and resolution was 70 000 resolution (mass range, 200–1200 mz−1). MS/MS spectra of the top 10 most intense peaks were obtained by higher-energy collisional dissociation fragmentation. Resolution for MS/MS spectra was set to 35 000 at 200 mz−1, AGC target to 5E5, max injection time to 120 ms and the isolation window to 1.3 Th.
Bioinformatic analysis
Samples were processed using MaxQuant V 1.5.0.1 (61) using default settings with experiment-specific modifications. For analysis of lysine ubiquitylation, K-ε-Gly-Gly was put as a variable modification for samples enriched for peptides with ubiquitin remnants. For analysis of the SILAC-pulsed podocytes, arginine (+10) and lysine (+6) were added as second multiplicity label. Match between run was enabled. iBAQ was calculated for all IP samples. The MaxQuant label-free quantification (LFQ) was enabled for all samples (62). Perseus (v. 1.4.0.1 and v. 1.5.2.4) (63) was used for further analysis. To correct for multiple testing, an approach similar to SAM was utilized (64). Enriched GO-terms within the interactors where determined using a Fisher exact test (FDR = 0.05). The GO terms were cleared for redundant annotations using the ReViGo software with default options (65). Cytoscape was utilized for network visualization (66). The IP data sets were analysed as previously described (11). Complex stoichiometry was calculated based on iBAQ parameters as previously described (11,12). Enrichment for PTMs within the PHB domain was performed using the PTMfunc database as previously described (18,22).
Visualization, modelling and molecular dynamics simulations
The podocin (Uniprot Q91X05) with a sequence:
MDSRARSSSREAHGRSSRSSSRDDKKAKAGRGSRGRARPDAGAERQSTGRTATRGEPRAPAATATVVDVDEVRGPGEEGTEVVALLESERPEEGIKPSGLGACEWLLVLASLIFIIMTFPFSIWFCIKVVQEYERVIIFRLGHLLPGRAKGPGLFFFLPCLDTYHKVDLRLQTLEIPFHEVVTKDMFIMEIDAVCYYRMENASLLLSSLAHVSKAIQFLVQTTMKRLLAHRSLTEILLERKSIAQDVKVALDAVTCIWGIKVERTEIKDVRLPAGLQHSLAVEAEAQRQAKVRVIAAEGEKAASESLRMAAEILSGTPAAVQLRYLHTLQSLSTEKPATVVLPLPFDMLSLLSSPGNRAQGSINYPSSSKPVEPLNPKKKDSPML was homology modelled with SWISS-MODEL (67–69). Residues 160–328 structure was modelled with the template PDB 3bk6 chain A. There are 10 lysine residues in this segment. By numbering starting from residue 160 as 1, these 10 lysines are: 7, 25, 55, 66, 82, 89, 102, 109, 132 and 142, which correspond to residues 166, 184, 214, 225, 241, 248, 261, 268, 291 and 301 in mouse podocin. The ubiquitin structure was based on PDB 1ubq. The k48-pUb, ubiquitin-chain tetramer, the structure was based on PDB 2o6v. The monomer ubiquitin or the tetramer ubiquitin was linked to a specific lysine side chain through the C-terminal glycine, as seen in Supplementary Material, Figures S7 and S8. To examine the stability of podocin under various types of ubiquitylation, we performed replica-exchange simulations (70) at eight temperatures, 300 K, 327 K, 356 K, 389 K, 424 K, 462 K, 504 K and 550 K. Replica-exchange simulations maintain distribution balance between different temperatures and can produce temperature-dependent conformational change more reliably. The CHARMM force field (71) is used to calculate interaction energies. The SCPISM solvation model (72) is used to describe solvent effect. CHARMM (73) is used for the replica-exchange simulations. We use the RMSD of the core part of podocin, residues 10–110, as related to the initial structure to measure the conformational change during the replica-exchange simulations. The initial structures are shown in Supplementary Material, Figures S7 and S8, respectively. The results are shown in Supplementary Material, Figures S9 and S10 for mono (U)—and poly-(4U) ubiquitylations. To objectively describe the conformational changing behaviour, we define a melting point as the temperature where the second derivative of the RMSD curve has the first maximum. Supplementary Material, Figures S11 and S12 show the second derivatives of the curves shown in Supplementary Material, Figures S9 and S10, respectively. As can be seen, K301U has lower or same melting point than most other mono-ubiquitylation species, and K301U4 has lower melting point than most of other K48-poly-ubiquitylation species.
Statistics
Statistical analysis was performed using one-way ANOVA, Wilcoxon ranked test, and two-tailed paired or unpaired t-test where appropriate and as indicated in the figure legends. Each n is an independent, biological and technical replicate (performed on another day). The only exception was the quantitative analysis of the immunofluorescence signal in C. elegans, where worms (n as indicated) from three different grownups were quantified in a blind manner. Statistical analysis was performed using the Perseus software, Graph pad (Prism) suite or Microsoft Excel. A P-value of <0.05 was considered significant unless indicated. Correction for multiple testing was performed as indicated in the ‘bioinformatics’ section. No means to predetermine sample size were utilized. The number of replicates, however, followed the common practice and replicates in the field (4–5 replicates for label-free proteomic analyses, 5–6 biological replicates for quantitative measurements of RNA and protein expression). Variance was estimated to be similar between the experimental group. No quantitative data were excluded, unless the following criteria were met: IF data—positive staining in the negative control. Western blotdata—no specific staining or high-background signal. MS data—insufficient ESI spray conditions or dropping MS performance.
Supplementary Material
Funding
T.B. was supported by the DFG (BE2212 and SFB 635). M.M.R. was supported by the DFG (UoC Postdoc Grant, Exzellenzinitiative) and by intramural grants (Stiftungsgelder). X.W. and B.R.B. were supported by the Intramural Research Programs of National Heart, Lung, and Blood Institute (Z01 HL001027-30). I.S. was supported by the Alexander von Humboldt Foundation. Following worm strains were used: Wild-type N2 and CB1515 received from CGC which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440) and strain TM3968 received from the NBRP and the Mitani Lab, Japan.
Supplementary Material
Acknowledgements
The authors thank our colleagues Yong-Tae Kwon (University of Pittsburgh), Qunying Lei (Fudan University, Shanghai), Martin Chalfie (Columbia University, New York) and Astrid Schauss (Cologne) for providing reagents and helpful discussions. The authors thank Pedro Beltrao (EBI, Cambridge) for help with the PTM-enrichment analysis. The authors thank Ruth Herzog, Astrid Wilbrand-Hennes, Rene Grandjean, S. Nestel and B. Joch for technical help. The authors thank Stuart Shankland (Seattle) and Moin Saleem (Bristol) for providing podocyte cell lines and the Nephrolab Cologne for helpful contributions and discussions.
Conflict of Interest statement. None declared.
References
- 1.Boute N., Gribouval O., Roselli S., Benessy F., Lee H., Fuchshuber A., Dahan K., Gubler M.C., Niaudet P., Antignac C. (2000) NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat. Genet., 24, 349–354. [DOI] [PubMed] [Google Scholar]
- 2.Tory K., Menyhárd D.K., Woerner S., Nevo F., Gribouval O., Kerti A., Stráner P., Arrondel C., Huynh Cong E., Tulassay T. et al. (2014) Mutation-dependent recessive inheritance of NPHS2-associated steroid-resistant nephrotic syndrome. Nat. Genet., 46, 299–304. [DOI] [PubMed] [Google Scholar]
- 3.Pavenstädt H., Kriz W., Kretzler M. (2003) Cell biology of the glomerular podocyte. Physiol. Rev., 83, 253–307. [DOI] [PubMed] [Google Scholar]
- 4.Huber T.B., Schermer B., Benzing T. (2007) Podocin organizes ion channel-lipid supercomplexes: implications for mechanosensation at the slit diaphragm. Nephron Exp. Nephrol., 106, e27–e31. [DOI] [PubMed] [Google Scholar]
- 5.Huber T.B., Schermer B., Müller R.U., Höhne M., Bartram M., Calixto A., Hagmann H., Reinhardt C., Koos F., Kunzelmann K. et al. (2006) Podocin and MEC-2 bind cholesterol to regulate the activity of associated ion channels. Proc. Natl Acad. Sci. USA, 103, 17079–17086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodman M.B., Ernstrom G.G., Chelur D.S., O'Hagan R., Yao C.A., Chalfie M. (2002) MEC-2 regulates C. elegans DEG/ENaC channels needed for mechanosensation. Nature, 415, 1039–1042. [DOI] [PubMed] [Google Scholar]
- 7.Zhang S., Arnadottir J., Keller C., Caldwell G.A., Yao C.A., Chalfie M. (2004) MEC-2 is recruited to the putative mechanosensory complex in C. elegans touch receptor neurons through its stomatin-like domain. Curr. Biol., 14, 1888–1896. [DOI] [PubMed] [Google Scholar]
- 8.Saez I., Vilchez D. (2014) The mechanistic links between proteasome activity, aging and age-related diseases. Curr. Genomics, 15, 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagy V., Dikic I. (2010) Ubiquitin ligase complexes: from substrate selectivity to conjugational specificity. Biol. Chem., 391, 163–169. [DOI] [PubMed] [Google Scholar]
- 10.Hubner N.C., Mann M. (2011) Extracting gene function from protein-protein interactions using Quantitative BAC InteraCtomics (QUBIC). Methods San Diego Calif, 53, 453–459. [DOI] [PubMed] [Google Scholar]
- 11.Kohli P., Bartram M.P., Habbig S., Pahmeyer C., Lamkemeyer T., Benzing T., Schermer B., Rinschen M.M. (2014) Label-free quantitative proteomic analysis of the YAP/TAZ interactome. Am. J. Physiol. Cell Physiol., 306, C805–C818. [DOI] [PubMed] [Google Scholar]
- 12.Smits A.H., Jansen P.W.T.C., Poser I., Hyman A.A., Vermeulen M. (2013) Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res., 41, e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakatani Y., Konishi H., Vassilev A., Kurooka H., Ishiguro K., Sawada J., Ikura T., Korsmeyer S.J., Qin J., Herlitz A.M. (2005) p600, a unique protein required for membrane morphogenesis and cell survival. Proc. Natl Acad. Sci. USA, 102, 15093–15098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang M., Chalfie M. (1994) Gene interactions affecting mechanosensory transduction in Caenorhabditis elegans. Nature, 367, 467–470. [DOI] [PubMed] [Google Scholar]
- 15.Udeshi N.D., Mertins P., Svinkina T., Carr S.A. (2013) Large-scale identification of ubiquitination sites by mass spectrometry. Nat. Protoc., 8, 1950–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Povlsen L.K., Beli P., Wagner S.A., Poulsen S.L., Sylvestersen K.B., Poulsen J.W., Nielsen M.L., Bekker-Jensen S., Mailand N., Choudhary C. (2012) Systems-wide analysis of ubiquitylation dynamics reveals a key role for PAF15 ubiquitylation in DNA-damage bypass. Nat. Cell Biol., 14, 1089–1098. [DOI] [PubMed] [Google Scholar]
- 17.Wagner S.A., Beli P., Weinert B.T., Schölz C., Kelstrup C.D., Young C., Nielsen M.L., Olsen J.V., Brakebusch C., Choudhary C. (2012) Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol. Cell. Proteomics, 11, 1578–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beltrao P., Albanèse V., Kenner L.R., Swaney D.L., Burlingame A., Villén J., Lim W.A., Fraser J.S., Frydman J., Krogan N.J. (2012) Systematic functional prioritization of protein posttranslational modifications. Cell, 150, 413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagai T., Levy Y. (2010) Ubiquitin not only serves as a tag but also assists degradation by inducing protein unfolding. Proc. Natl Acad. Sci. USA, 107, 2001–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braun B.C., Glickman M., Kraft R., Dahlmann B., Kloetzel P.M., Finley D., Schmidt M. (1999) The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat. Cell Biol., 1, 221–226. [DOI] [PubMed] [Google Scholar]
- 21.Martin A., Baker T.A., Sauer R.T. (2008) Protein unfolding by a AAA+ protease is dependent on ATP-hydrolysis rates and substrate energy landscapes. Nat. Struct. Mol. Biol., 15, 139–145. [DOI] [PubMed] [Google Scholar]
- 22.Rinschen M.M., Wu X., König T., Pisitkun T., Hagmann H., Pahmeyer C., Lamkemeyer T., Kohli P., Schnell N., Schermer B. et al. (2014) Phosphoproteomic analysis reveals regulatory mechanisms at the kidney filtration barrier. J. Am. Soc. Nephrol., 10.1681/ASN.2013070760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gödel M., Ostendorf B.N., Baumer J., Weber K., Huber T.B. (2013) A novel domain regulating degradation of the glomerular slit diaphragm protein podocin in cell culture systems. PLoS One, 8, e57078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tossidou I., Teng B., Drobot L., Meyer-Schwesinger C., Worthmann K., Haller H., Schiffer M. (2010) CIN85/RukL is a novel binding partner of nephrin and podocin and mediates slit diaphragm turnover in podocytes. J. Biol. Chem., 285, 25285–25295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin C.-L., Lee P.-H., Hsu Y.-C., Lei C.-C., Ko J.-Y., Chuang P.-C., Huang Y.-T., Wang S.-Y., Wu S.-L., Chen Y.-S. et al. (2014) MicroRNA-29a promotion of nephrin acetylation ameliorates hyperglycemia-induced podocyte dysfunction. J. Am. Soc. Nephrol., 25, 1698–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tasaki T., Mulder L.C.F., Iwamatsu A., Lee M.J., Davydov I.V., Varshavsky A., Muesing M., Kwon Y.T. (2005) A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol. Cell. Biol., 25, 7120–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parsons K., Nakatani Y., Nguyen M.D. (2015) p600/UBR4 in the central nervous system. Cell. Mol. Life Sci., 72, 1149–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong J.H., Kaustov L., Coyaud E., Srikumar T., Wan J., Arrowsmith C., Raught B. (2015) KCMF1 (potassium channel modulatory factor 1) Links RAD6 to UBR4 (ubiquitin N-recognin domain-containing E3 ligase 4) and lysosome-mediated degradation. Mol. Cell. Proteomics, 14, 674–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tasaki T., Kim S.T., Zakrzewska A., Lee B.E., Kang M.J., Yoo Y.D., Cha-Molstad H.J., Hwang J., Soung N.K., Sung K.S. et al. (2013) UBR box N-recognin-4 (UBR4), an N-recognin of the N-end rule pathway, and its role in yolk sac vascular development and autophagy. Proc. Natl Acad. Sci. USA, 110, 3800–3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin R., Tao R., Gao X., Li T., Zhou X., Guan K.-L., Xiong Y., Lei Q.-Y. (2013) Acetylation stabilizes ATP-citrate lyase to promote lipid biosynthesis and tumor growth. Mol. Cell, 51, 506–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong J., Taylor P., Moran M.F. (2014) Proteomic analysis of the epidermal growth factor receptor (EGFR) interactome and post-translational modifications associated with receptor endocytosis in response to EGF and stress. Mol. Cell. Proteomics, 13, 1644–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benzing T. (2004) Signaling at the slit diaphragm. J. Am. Soc. Nephrol., 15, 1382–1391. [DOI] [PubMed] [Google Scholar]
- 33.Jin J., Sison K., Li C., Tian R., Wnuk M., Sung H.-K., Jeansson M., Zhang C., Tucholska M., Jones N. et al. (2012) Soluble FLT1 binds lipid microdomains in podocytes to control cell morphology and glomerular barrier function. Cell, 151, 384–399. [DOI] [PubMed] [Google Scholar]
- 34.Belzil C., Ramos T., Sanada K., Colicos M.A., Nguyen M.D. (2014) p600 stabilizes microtubules to prevent the aggregation of CaMKIIα during photoconductive stimulation. Cell. Mol. Biol. Lett., 19, 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brand J., Smith E.S.J., Schwefel D., Lapatsina L., Poole K., Omerbašić D., Kozlenkov A., Behlke J., Lewin G.R., Daumke O. (2012) A stomatin dimer modulates the activity of acid-sensing ion channels. EMBO J., 31, 3635–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Welsh G.I., Hale L.J., Eremina V., Jeansson M., Maezawa Y., Lennon R., Pons D.A., Owen R.J., Satchell S.C., Miles M.J. et al. (2010) Insulin signaling to the glomerular podocyte is critical for normal kidney function. Cell Metab.,12, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niranjan T., Bielesz B., Gruenwald A., Ponda M.P., Kopp J.B., Thomas D.B., Susztak K. (2008) The Notch pathway in podocytes plays a role in the development of glomerular disease. Nat. Med., 14, 290–298. [DOI] [PubMed] [Google Scholar]
- 38.Schiffer M., Bitzer M., Roberts I.S., Kopp J.B., ten Dijke P., Mundel P., Böttinger E.P. (2001) Apoptosis in podocytes induced by TGF-beta and Smad7. J. Clin. Invest.,108, 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gee H.Y., Zhang F., Ashraf S., Kohl S., Sadowski C.E., Vega-Warner V., Zhou W., Lovric S., Fang H., Nettleton M. et al. (2015) KANK deficiency leads to podocyte dysfunction and nephrotic syndrome. J. Clin. Invest.,125, 2375–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaplan J.M., Kim S.H., North K.N., Rennke H., Correia L.A., Tong H.Q., Mathis B.J., Rodríguez-Pérez J.C., Allen P.G., Beggs A.H. et al. (2000) Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat. Genet., 24, 251–256. [DOI] [PubMed] [Google Scholar]
- 41.Bartram M.P., Habbig S., Pahmeyer C., Höhne M., Weber L.T., Thiele H., Altmüller J., Kottoor N., Wenzel A., Krueger M. et al. (2016) Three-layered proteomic characterization of a novel ACTN4 mutation unravels its pathogenic potential in FSGS. Hum. Mol. Genet., 25, 1152–1164. [DOI] [PubMed] [Google Scholar]
- 42.Rinschen M.M., Benzing T., Limbutara K., Pisitkun T. (2015) Proteomic analysis of the kidney filtration barrier—problems and perspectives. Proteomics Clin. Appl., 10.1002/prca.201400201. [DOI] [PubMed] [Google Scholar]
- 43.Griffin S.V., Hiromura K., Pippin J., Petermann A.T., Blonski M.J., Krofft R., Takahashi S., Kulkarni A.B., Shankland S.J. (2004) Cyclin-dependent kinase 5 is a regulator of podocyte differentiation, proliferation, and morphology. Am. J. Pathol., 165, 1175–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brinkkoetter P.T., Olivier P., Wu J.S., Henderson S., Krofft R.D., Pippin J.W., Hockenbery D., Roberts J.M., Shankland S.J. (2009) Cyclin I activates Cdk5 and regulates expression of Bcl-2 and Bcl-XL in postmitotic mouse cells. J. Clin. Invest.,119, 3089–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saleem M.A., O'Hare M.J., Reiser J., Coward R.J., Inward C.D., Farren T., Xing C.Y., Ni L., Mathieson P.W., Mundel P. (2002) A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J. Am. Soc. Nephrol., 13, 630–638. [DOI] [PubMed] [Google Scholar]
- 46.Ohse T., Pippin J.W., Vaughan M.R., Brinkkoetter P.T., Krofft R.D., Shankland S.J. (2008) Establishment of conditionally immortalized mouse glomerular parietal epithelial cells in culture. J. Am. Soc. Nephrol., 19, 1879–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takemoto M., Asker N., Gerhardt H., Lundkvist A., Johansson B.R., Saito Y., Betsholtz C. (2002) A new method for large scale isolation of kidney glomeruli from mice. Am. J. Pathol., 161, 799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boerries M., Grahammer F., Eiselein S., Buck M., Meyer C., Goedel M., Bechtel W., Zschiedrich S., Pfeifer D., Laloë D. et al. (2013) Molecular fingerprinting of the podocyte reveals novel gene and protein regulatory networks. Kidney Int., 83, 1052–1064. [DOI] [PubMed] [Google Scholar]
- 49.Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem., 75, 663–670. [DOI] [PubMed] [Google Scholar]
- 50.Lim K.L., Chew K.C.M., Tan J.M.M., Wang C., Chung K.K.K., Zhang Y., Tanaka Y., Smith W., Engelender S., Ross C.A. et al. (2005) Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J. Neurosci., 25, 2002–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rinschen M.M., Pahmeyer C., Pisitkun T., Schnell N., Wu X., Maaß M., Bartram M.P., Lamkemeyer T., Schermer B., Benzing T. et al. (2014) Comparative phosphoproteomic analysis of mammalian glomeruli reveals conserved podocin C-terminal phosphorylation as a determinant of slit diaphragm complex architecture. Proteomics, 10.1002/pmic.201400235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Finney M., Ruvkun G. (1990) The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell, 63, 895–905. [DOI] [PubMed] [Google Scholar]
- 53.Bharill P., Ayyadevara S., Alla R., Shmookler Reis R.J. (2013) extreme depletion of pip3 accompanies the increased life span and stress tolerance of PI3K-null C. elegans mutants. Front. Genet.,4, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schneider C.A., Rasband W.S., Eliceiri K.W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Timmons L., Court D.L., Fire A. (2001) Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene, 263, 103–112. [DOI] [PubMed] [Google Scholar]
- 57.Fire A., Albertson D., Harrison S.W., Moerman D.G. (1991) Production of antisense RNA leads to effective and specific inhibition of gene expression in C. elegans muscle. Dev. Camb. Engl.,113, 503–514. [DOI] [PubMed] [Google Scholar]
- 58.Kamath R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M. et al. (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature, 421, 231–237. [DOI] [PubMed] [Google Scholar]
- 59.Chalfie M., Sulston J. (1981) Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev. Biol., 82, 358–370. [DOI] [PubMed] [Google Scholar]
- 60.Michalski A., Damoc E., Hauschild J.-P., Lange O., Wieghaus A., Makarov A., Nagaraj N., Cox J., Mann M., Horning S. (2011) Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol. Cell. Proteomics, 10, M111.011015, doi: 10.1074/mcp.M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol., 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- 62.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics, 13, 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cox J., Mann M. (2012) 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics, 13(Suppl. 16), S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tusher V.G., Tibshirani R., Chu G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA, 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Supek F., Bošnjak M., Škunca N., Šmuc T. (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One, 6, e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res., 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Cassarino T.G., Bertoni M., Bordoli L. et al. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res., 42, W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bordoli L., Kiefer F., Arnold K., Benkert P., Battey J., Schwede T. (2009) Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc., 4, 1–13. [DOI] [PubMed] [Google Scholar]
- 69.Guex N., Peitsch M.C., Schwede T. (2009) Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis, 30(Suppl. 1), S162–S173. [DOI] [PubMed] [Google Scholar]
- 70.Wu X., Hodoscek M., Brooks B.R. (2012) Replica exchanging self-guided Langevin dynamics for efficient and accurate conformational sampling. J. Chem. Phys., 137, 044106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.MacKerell A.D., Bashford D., Bellott M., Dunbrack R.L., Evanseck J.D., Field M.J., Fischer S., Gao J., Guo H., Ha S. et al. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B, 102, 3586–3616. [DOI] [PubMed] [Google Scholar]
- 72.Hassan S.A., Mehler E.L. (2002) A critical analysis of continuum electrostatics: the screened Coulomb potential—implicit solvent model and the study of the alanine dipeptide and discrimination of misfolded structures of proteins. Proteins, 47, 45–61. [DOI] [PubMed] [Google Scholar]
- 73.Brooks B.R., Brooks C.L. III, Mackerell A.D. Jr, Nilsson L., Petrella R.J., Roux B., Won Y., Archontis G., Bartels C., Boresch S. et al. (2009) CHARMM: the biomolecular simulation program. J. Comput. Chem., 30, 1545–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.