Abstract
The replication of Sindbis virus (SIN) profoundly affects the metabolism of infected vertebrate cells. One of the main events during SIN infection is the strong inhibition of translation of cellular mRNAs. In this study, we used a combination of approaches, including the study of SIN replication in PKR−/− mouse embryo fibroblasts or in the presence of an excess of catalytically inactive PKR. We show that the PKR-dependent inhibition of translation is not the only and most likely not the major pathway mediating translational shutoff during SIN infection. The PKR-independent mechanism strongly affects the translation of cellular templates, whereas translation of SIN subgenomic RNA is resistant to inhibition, and this leads to a benefit for viral replication. Our findings suggest that both PKR-dependent and non-PKR-dependent mechanisms of SIN-induced translational shutoff can be manipulated by using SIN replicons expressing mutated SIN nsP2 or kinase-defective PKR. Specifically, we show that expression of heterologous genes from SIN-based and most likely other alphavirus-based replicons can be increased by downregulating both the PKR-dependent and PKR-independent translational shutoffs.
Alphaviruses are a widely distributed group of significant human and animal pathogens. Some of them, including Venezuelan, eastern, and western equine encephalitis viruses, cause serious febrile illness and encephalitis (19). Recent epidemics of Venezuelan equine encephalitis and O'nyong-nyong viruses indicate that alphaviruses are an important public health threat (22, 33, 45).
Alphaviruses circulate in nature by continuous transmission between mosquitoes and susceptible vertebrate hosts (35). In insect vectors, they cause lifelong chronic infection characterized by the presence of virus in high titers in salivary glands that does not appear to strongly affect mosquito viability. Consistent with this finding, many alphaviruses develop a moderate cytopathic effect (CPE) in cultured mosquito cells in the early stages of infection and then establish persistent or chronic infection (20). In contrast, vertebrate hosts show acute disease characterized by a high viremia required for virus transmission to mosquitoes before clearance by the immune system (17, 18). In cultured vertebrate cells, alphaviruses rapidly replicate to high titers and develop complete CPE within 24 to 48 h postinfection. In spite of a distinct heterogeneity in the sequences of both structural and nonstructural proteins, all of the known members of the genus are believed to have similar structures of the viral particles and similar replication strategies. Sindbis virus (SIN) is one of the least pathogenic alphaviruses, but its study has revealed highly valuable information about the mechanism of RNA replication and virus interaction with host cells that is generally applicable to other alphaviruses (41).
SIN has a single-stranded RNA genome that is ∼11.5 kb in length and has positive polarity (40). The 5′ two-thirds of the genome encodes four nonstructural proteins (nsP1 to nsP4) forming, together with cellular factors, a replicative enzyme complex (RdRp). This complex sequentially changes its composition at different stages of virus replication, producing the full-length minus-strand copy of the genome that functions as a template for the synthesis of new viral genomes and the subgenomic 26S RNA. The RNA (ca. 4 kb) is identical to the 3′ one-third of the genome and is translated into structural proteins forming infectious viral particles.
SIN replication strongly affects the metabolism of infected vertebrate cells. The major virus-induced changes include inhibition of both transcription and translation of cellular mRNAs (11, 14). Between 4 and 8 h postinfection, the synthesis of host cell proteins becomes 5- to 10-fold less efficient, and a few hours later, SIN-infected cells lose their integrity and die. For many cell types, SIN-induced CPE is accompanied by apoptotic changes (25).
Virus-mediated translational shutoff is an event that has been described for many infections (21, 27, 28, 31), but most likely the phenomenon is achieved by a variety of mechanisms. In spite of great progress during the last few years in understanding alphavirus replication, one of the critical questions about alphavirus-host cell interactions, the mechanism of translational shutoff, remains obscure. It was previously demonstrated that the synthesis of SIN structural proteins is dispensable for triggering the inhibition of translation (11, 13). Upon delivery into the cells, SIN replicons (the self-replicating RNAs encoding only the RdRp-forming nonstructural proteins) downregulate the translation of cellular RNA templates as efficiently as replicating virus. Based on a widely accepted hypothesis, the translational shutoff can be explained by activation of the double-stranded RNA (dsRNA)-activated protein kinase (PKR) (16) by newly synthesized SIN dsRNA that is likely present in the replicative RNA intermediates. These dsRNAs bind to PKR and lead to a conformational change(s) in the protein, inducing its kinase activity, which mediates autophosphorylation and phosphorylation of the translation initiation factor eIF2α, resulting in inefficient initiation of translation (15, 30, 46). In addition to its effect on translation, the activation of PKR was also postulated to induce signaling cascades, leading to development of apoptosis (5, 6, 43, 44).
To elucidate PKR functions during SIN infection, we examined changes in cellular translation and translational machinery proceeding in different cells infected with a variety of recombinant SINs and SIN-based replicons. We analyzed the effects of overexpression of the wild-type (wtPKR) and dominant-negative mutant (mutPKR) forms of PKR on the inhibition of translation of cellular and virus-specific RNAs. Our data indicate that SIN-specific translational shutoff is determined by at least two mechanisms, one of which is independent of PKR. This inhibition of translation strongly affects the translation of cellular mRNAs, but translation of SIN-specific RNAs remains very efficient.
MATERIALS AND METHODS
Cell cultures.
BHK-21 cells were kindly provided by Paul Olivo (Washington University, St. Louis, Mo.). NIH 3T3 cells were obtained from the American Type Culture Collection (Manassas, Va.). These cell lines were maintained at 37°C in alpha minimum essential medium (alpha MEM) supplemented with 10% fetal bovine serum (FBS) and vitamins. Wild-type (wt) mouse embryo fibroblasts (MEFs), derived from C57BL/6 mice, and fibroblasts lacking the PKR gene (PKR−/− MEFs) (47), derived from mice with the PKR gene deleted, were propagated in Dulbecco's MEM supplemented with 10% FBS.
Plasmid constructs.
Standard recombinant-DNA techniques were used for all plasmid constructions. Maps and sequences are available from the authors upon request. Plasmids encoding the wtSIN and SIN/G viral genomes and having an additional subgenomic promoter (SP) driving the expression of a codon-optimized green fluorescent protein (GFP) have been described elsewhere (14). They differed only in the nsP2 coding sequence, in that Pro726 (CCA) in pwtSIN was replaced by Gly (GGA) in pSIN/G. A plasmid encoding the SIN replicon with GFP, pSINrep/GFP, was kindly provided by Nicolas Ruggli (N. Ruggli and C. M. Rice, unpublished data). The pSINrep/HuPKR and pSINrep/mutHuPKR plasmids encoding SIN replicons with the genes for wt human PKR and human PKR with a single substitution (K296 → R) (1) cloned under the control of the SP were kindly provided by Eugene Agapov (E. Agapov and C. M. Rice, unpublished data).
The pSINrep/wtPKR/GFP and pSINrep/mutPKR/GFP plasmids were designed by cloning the BamHI-XhoI fragment of pSINrep/GFP into MluI-XhoI-digested pSINrep/HuPKR and pSINrep/mutHuPKR. (Both the BamHI and MluI sites were filled in using T4 DNA polymerase.) The resulting pSINrep/wtPKR/GFP and pSINrep/mutPKR/GFP plasmids contained the SP6 promoter, followed by nucleotides (nt) 1 to 7646 of the SIN genome, TCTAGT, a 1,813-nt-long sequence encoding the entire PKR (starting from the initiating ATG), the CCCGGGGGATCCTAGACGCG sequence, nt 7335 to 7646 of the SIN genome (encoding the second SP), a TCTAGAGCTTGCCGCCACC sequence, a 720-nt-long fragment encoding GFP, AGCGGCCC, and a 352-nt sequence containing the SIN 3′ untranslated region (UTR), poly(A), followed by an XhoI restriction site. The pSINrep/G/wtPKR/GFP and pSINrep/G/mutPKR/GFP plasmids had essentially the same sequence, but Pro726-encoding CCA in the nsP2 gene was replaced by GGA, encoding Gly. pSIN/Toto, including the entire genome of SIN Toto1101, has been described elsewhere (32). The pSIN/wtPKR and pSIN/mutPKR plasmids, including viral genomes with an additional SP driving the expression of wt human PKR and its mutated variant with a K296 → R substitution, were designed by cloning the BamHI-XhoI fragment (the BamHI site was filled in using T4 DNA polymerase) of pSIN/Toto into MluI-XhoI-digested pSINrep/HuPKR and pSINrep/mutHuPKR (the MluI site was filled in using T4 DNA polymerase). The resulting pSIN/wtPKR and pSIN/mutPKR plasmids contained the SP6 promoter followed by nt 1 to 7646 of the SIN genome, TCTAGT, a 1,813-nt-long sequence encoding the entire PKR (starting from the initiating ATG), the CCCGGGGGATCCTAGACGCG sequence, and nt 7335 to 11740 of the SIN Toto1101 genome, including the SP, and the entire viral subgenomic RNA, including the 3′ UTR and poly(A), followed by an XhoI restriction site. Schematic representations of all of the constructs are shown (see Fig. 1 and 4 to 7). pSINrep/C/GFP contained the SP6 promoter, followed by nt 1 to 8447 of the SIN genome, which included not only the sequence encoding the nsPs and the SP, but the entire capsid coding sequence and the first nine nucleotides of E3 as well, followed by one extra codon (GTC), the GFP-encoding sequence (fused in frame with the capsid gene), and the SIN 3′ UTR derived from pSINrep/GFP. The pSINrep/wtPKR/C/GFP plasmid, encoding replicon with two SPs driving the expression of wt human PKR and the SIN capsid fused in frame with GFP, was designed by cloning the BamHI-XhoI fragment (the BamHI site was filled in using T4 DNA polymerase) of pSINrep/C/GFP into MluI-XhoI-digested pSINrep/HuPKR (the MluI site was filled in using T4 DNA polymerase). pDH-BB(delSL2)-Csin and pDH-BB(delSL2)-(CΔ3rrv) included the genomes of helper RNAs that had essentially the same structure as the previously described DH-BB-Csin and DH-BB (CΔ3rrv) helpers (9) expressing the SIN capsid and SIN glycoproteins, respectively. However, their 5′ termini did not contain tRNAAsp and started from 425 nt of the SIN genome with nt 47 to 152 deleted (10). This deletion increased the levels of their replication and packaging activities, particularly when the helpers were used for packaging of replicons with mutated nsP2.
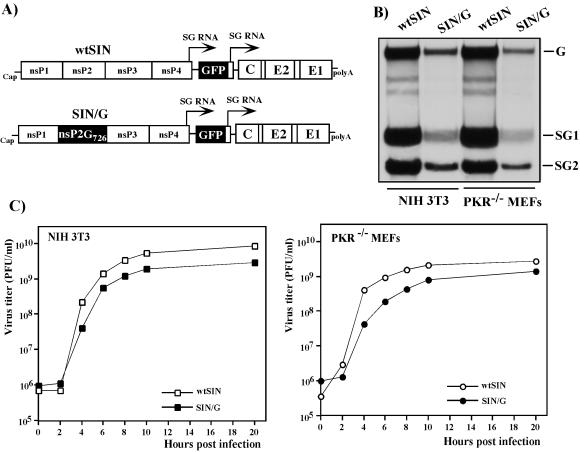
FIG. 1.
Schematic representation of double subgenomic viral genomes and analysis of viral RNA replication and virus growth in NIH 3T3 cells and PKR−/− MEFs. (A) In both genomes, the first SP was driving transcription of GFP-encoding RNA and the second was driving the expression of natural 26S subgenomic RNA encoding SIN structural genes derived from the SIN TE12 strain. The SIN/G variant differed from wtSIN by one amino acid in nsP2, P726 → G. (B) NIH 3T3 cells and PKR−/− MEFs in six-well Costar plates were infected with wtSIN and SIN/G variants at an MOI of 20 PFU/cell. At 5 h postinfection, the media were replaced by 0.8 ml of alpha MEM supplemented with 10% FBS, dactinomycin (1 μg/ml), and [3H]uridine (20 μCi/ml). After 3 h of incubation at 37°C, RNAs were isolated and analyzed by agarose gel electrophoresis as described in Materials and Methods. G, SG1, and SG2 indicate the positions of genomic RNA and subgenomic RNAs 1 and 2, respectively. (C) NIH 3T3 cells and PKR−/− MEFs in six-well Costar plates were infected with wtSIN and SIN/G variants at an MOI of 20 PFU/cell. At the indicated times, the media were replaced and virus titers were determined as described in Materials and Methods.
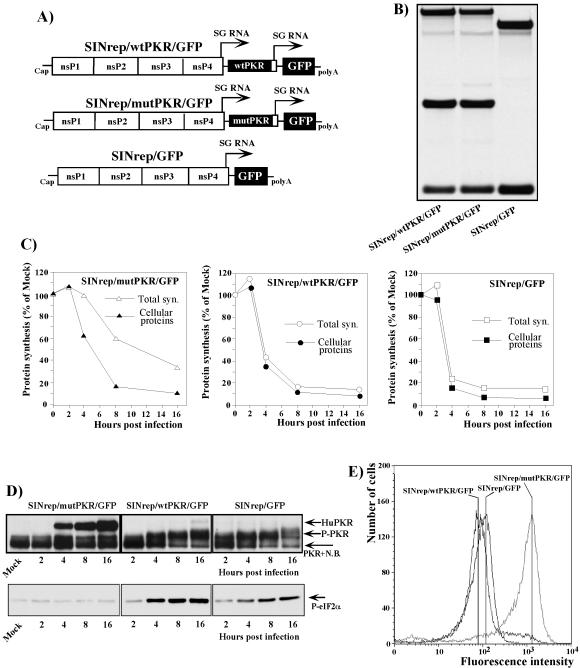
FIG. 4.
Schematic representation of wtSIN-based replicons and analysis of their RNA replication and transcription, protein synthesis, and phosphorylation of PKR and eIF2α in the infected cells. (A) All three replicons contained an SP driving the expression of GFP. The SINrep/wtPKR/GFP and SINrep/mutPKR/GFP constructs also had a second SP driving the expression of wt human PKR and human PKR with a single substitution (K296 → R), respectively. (B) NIH 3T3 cells (5 × 105) were infected with packaged replicons at an MOI of 20 infectious units/cell. At 2 h postinfection, the media were replaced by the same media supplemented with dactinomycin (1 μg/ml) and [3H]uridine (20 μCi/ml). After 4 h of incubation at 37°C, RNAs were isolated and analyzed as described in Materials and Methods. (C) NIH 3T3 cells (5 × 105 cells/well) in six-well Costar plates) were infected with packaged replicons at an MOI of 20 infectious units/cell. Proteins were pulse-labeled with [35S]methionine at the indicated times and analyzed on sodium dodecyl sulfate-10% polyacrylamide gels. The gels were dried and analyzed on a Storm 860 PhosphorImager as described in Materials and Methods. The levels of synthesis of cellular proteins and total protein synthesis (Total syn.) in the infected cells were evaluated as described in the legend to Fig. 3. One of the reproducible experiments is shown. Mock indicates mock-infected cells. (D) Phosphorylation of PKR (P-PKR) and eIF2α (P-eIF2α) in replicon-infected cells was analyzed as described in the legend to Fig. 3. N.B. indicates the position of a minor protein band that nonspecifically binds anti-PKR antibodies in all tested mouse cells. HuPKR indicates the position of human PKR. (E) Analysis of GFP expression in cells infected with wtSIN-based replicons. NIH 3T3 cells (1.5 × 106) were infected with packaged replicons at an MOI of 20 infectious units/cell. At 12 h postinfection, the cells were fixed with 1% formaldehyde in PBS for 10 min and analyzed by flow cytometry on a FACS Vantage (Becton Dickinson).
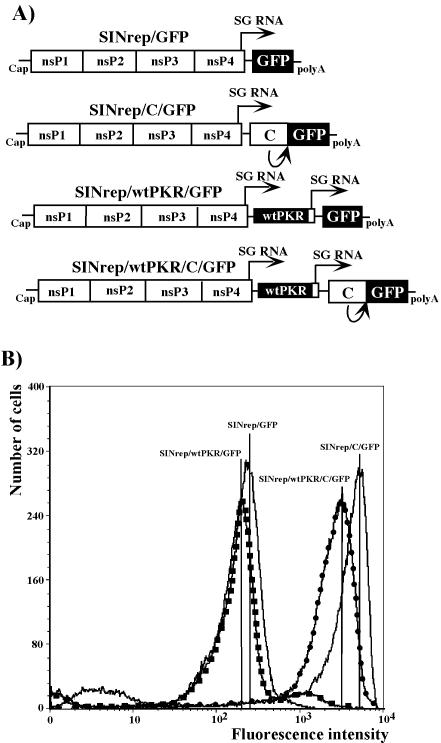
FIG. 7.
Schematic representation of SIN-based replicons and analysis of GFP expression in infected cells. (A) All replicons contained SP driving the expression of GFP. Subgenomic RNAs of SINrep/C/GFP and SINrep/wtPKR/C/GFP encoded fusion proteins containing the sequence of the entire capsid protein followed by four extra amino acids and GFP. SINrep/wtPKR/C/GFP and SINrep/wtPKR/GFP expressed wt human PKR from the second SP. (B) NIH 3T3 cells (1.5 × 106) were infected with packaged replicons at an MOI of 20 infectious units/cell. At 12 h postinfection, the cells were fixed in 1% formaldehyde in PBS for 10 min and analyzed by flow cytometry on a FACS Vantage (Becton Dickinson).
RNA transcriptions.
Plasmids were purified by centrifugation in CsCl gradients. Before the transcription reaction, the viral genomes, replicon, or helper genomes coding plasmids were linearized by XhoI digestion. RNAs were synthesized by SP6 RNA polymerase in the presence of cap analog. The yields and integrity of transcripts were analyzed by gel electrophoresis under nondenaturing conditions. Aliquots of transcription reaction mixtures were used for electroporation without additional purification.
RNA transfections.
Electroporation of BHK-21 cells was performed under previously described conditions (26). For rescuing wtSIN, SIN/G, SIN/Toto, SIN/wtPKR, and SIN/mutPKR viruses, usually 1 μg of in vitro-synthesized viral genome RNA was electroporated into the cells (26), and then they were seeded into 100-mm-diameter dishes and incubated until the CPE was observed. These virus stocks were used for all of the experiments without additional passaging. Replicons with the wt sequence of the nonstructural proteins (SINrep/GFP, SINrep/wtPKR/GFP, and SINrep/mutPKR/GFP) were packaged by coelectroporation of 4 to 5 μg of in vitro-synthesized replicon RNA and 6 μg of the helpers DH-BB(delSL2)-Csin and DH-BB(delSL2)-(CΔ3rrv) into BHK-21 cells. The packaged replicons were harvested after CPE development (usually 24 h posttransfection). Replicons with a P726 → G mutation in nsP2 (SINrep/G/GFP, SINrep/G/wtPKR/GFP, and SINrep/G/mutPKR/GFP) were packaged by coelectroporation of 4 μg of their in vitro-synthesized RNAs and 6 μg of SIN capsid- and SIN glycoprotein-encoding helper RNAs. The packaged replicons were harvested 30 h postelectroporation.
Virus titers were determined using a standard plaque assay on BHK-21 cells (24). For measuring the titers of packaged replicons, BHK-21 cells were seeded into six-well Costar plates at a concentration of 5 × 105 cells/well. Four hours later, the cells were infected with different dilutions of the packaged replicons, and after 1 h of incubation at 37°C in a 5% CO2 incubator, they were overlaid with 2 ml of 0.5% Ultra-Pure agarose (Invitrogen)-containing MEM supplemented with 3% FBS. The numbers of infected cells were estimated by counting the GFP-positive cells under an inverted UV microscope and were used to calculate the titers.
Infectious-center assay.
In standard experiments, 1 μg of in vitro-synthesized, full-length RNA transcripts of SIN viral genomes was used per electroporation (26). Tenfold dilutions of electroporated BHK-21 cells were seeded in six-well Costar plates containing subconfluent naïve cells. After 1 h of incubation at 37°C in a 5% CO2 incubator, the cells were overlaid with 2 ml of MEM containing 0.5% Ultra-Pure agarose supplemented with 3% FBS. The plaques were stained with crystal violet after 2 days of incubation at 37°C.
Viral replication analysis.
BHK-21 cells or PKR−/− MEFs were seeded at a concentration of 5 × 105 cells/35-mm-diameter dish. After 4 h of incubation at 37°C, the monolayers were infected at various multiplicities of infection (MOI), as indicated in the figure legends, for 1 h, washed three times with phosphate-buffered saline (PBS), and overlaid with 1 ml of complete medium. At appropriate times postinfection, the media were replaced by fresh media, and the virus titers in the harvested samples were determined by plaque assay of BHK-21 cells (24).
Analysis of protein synthesis.
NIH 3T3 cells or MEFs were seeded into six-well Costar plates at a concentration of 5 × 105 cells/well. After 4 h of incubation at 37°C in 5% CO2, they were infected at an MOI of 20 PFU of virus/cell or 20 infectious units of packaged replicons/cell in 500 μl of alpha MEM supplemented with 1% FBS at room temperature for 1 h with continuous shaking. The medium was then replaced by corresponding complete medium, and incubation continued at 37°C. At 2, 4, 8, and 16 h postinfection, the cells were washed three times with PBS and then incubated for 30 min at 37°C in 0.8 ml of Dulbecco's MEM lacking methionine and supplemented with 0.1% FBS and 20 μCi of [35S]methionine/ml. After incubation, the cells were scraped into the medium, pelleted by centrifugation, and dissolved in 100 μl of standard protein loading buffer. The protein concentrations in the samples were measured using SYPRO Ruby protein dye (Molecular Dynamics) according to the manufacturer's instructions, and equal amounts of proteins were loaded onto sodium dodecyl sulfate-10% polyacrylamide gels. After electrophoresis, the gels were dried, autoradiographed, and analyzed on a Storm 860 PhosphorImager (Molecular Dynamics).
The amount of radioactivity detected in the protein band corresponding to actin or the radioactivity in the entire fragment of the lane between the SIN-specific glycoprotein bands and the SIN capsid was used to evaluate the residual host cell protein synthesis. The results were normalized to the amounts of radioactivity detected in the same fragments of the lane representing uninfected cells. The two ways of testing host cell protein synthesis generated very similar results.
Total protein synthesis in the infected cells was evaluated by measuring the radioactivity in the entire lane on the gel. The results were normalized to the radioactivity in uninfected cells and represented the residual synthesis of host cell proteins and all the proteins encoded by viral or replicon genomes.
Analysis of PKR and eIF2α phosphorylation.
PKR and eIF2α phosphorylation were always tested in parallel with analysis of protein synthesis in the infected cells. NIH 3T3 cells or PKR−/− MEFs were seeded into 60-mm-diameter dishes at a concentration of 1.5 × 106 cells/dish and infected for 4 h with viruses or packaged replicons at an MOI of 20 PFU/cell or 20 infectious units/cell in 500 μl of alpha MEM supplemented with 1% FBS at room temperature for 1 h with continuous shaking. Then, the medium was replaced by corresponding complete medium, and incubation continued at 37°C. At 2, 4, 8, and 16 h postinfection, the cells were washed with PBS, scraped into PBS, pelleted by centrifugation, dissolved in 1% sodium dodecyl sulfate, and heated for 5 min at 95°C in the presence of phosphatase inhibitor cocktails 1 and 2 (Sigma, St. Louis, Mo.) used at concentrations recommended by the manufacturer. Protein concentrations were measured with a BCA Protein Assay kit (Pierce), and 10 μg of protein from each sample, including the uninfected cells, was loaded onto sodium dodecyl sulfate-10% polyacrylamide gels in a standard loading buffer. After transfer, the nitrocellulose was always stained with 0.5% Ponceau S (Fisher) in 1% acetic acid to control the quality of the transfer and additionally to confirm that all of the samples contained equal amounts of proteins. To probe PKR and phosphorylated eIF2α, anti-PKR (Santa Cruz Biotechnology) and anti-phospho-eIF2α (Ser51) (Cell Signaling Technology) antibodies, diluted 1:1,000, were used. Horseradish peroxidase-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology and diluted 1:5,000. The ECL Western Blotting Detection Reagent was purchased from Amersham Pharmacia Biotech and used according to the manufacturer's recommendations.
RNA analysis.
Cells were infected at the MOIs indicated in the figure legends with viruses or packaged replicons, and SIN-specific RNAs were labeled with [3H]uridine as described in the legend to Fig. 1. RNAs were isolated from the cells by TRIzol reagent (Gibco-BRL, Bethesda, Md.), as recommended by the manufacturer, denatured with glyoxal in dimethyl sulfoxide, and analyzed by agarose gel electrophoresis under previously described conditions (4).
RESULTS
SIN replicates equally efficiently in PKR-expressing and PKR-negative cells.
To elucidate the role of PKR during alphavirus infection, we used two modified versions of SIN, wtSIN and SIN/G, described in a previous study (14) (Fig. 1A). Both wtSIN and SIN/G had nonstructural genes and cis-acting RNA elements derived from SIN Toto1101 (32); the structural genes were derived from a mouse-adapted SIN TE12 variant (29). The only difference between the two viruses was a single point mutation. Proline in the 726 position of wtSIN nsP2 was replaced with glycine in SIN/G. This mutation reduced SIN/G RNA replication and its ability to cause translational shutoff in BHK-21 and NIH 3T3 cells (8, 14). In addition to the SP that drives the transcription of the subgenomic 26S RNA encoding the structural proteins, the genomes of these viruses contained a second SP that controls the expression of a GFP-encoding subgenomic mRNA (Fig. 1B). The expression of GFP was used to confirm the infection of all the cells in the monolayers. Both wtSIN and SIN/G stocks were generated by transfection of the in vitro-synthesized RNAs into BHK-21 cells and had similar titers exceeding 109 PFU/ml.
In initial experiments to compare levels of virus growth, RNA replication levels, and inhibition of translation of cellular mRNAs, we used the immortalized wt MEFs and PKR−/− MEFs (47). However, the wt MEFs were found to be less susceptible to SIN infection and required a 5- to 10-fold higher MOI for infection of the cells than PKR−/− MEFs and NIH 3T3 cells. It was particularly difficult to initiate productive replication of the SIN/G mutant in wt MEFs. The GFP-negative cells were still present even after infection with SIN/G at an MOI higher than 100 PFU/cell, and this factor made it very difficult to interpret the data in the experiments that included quantitative analysis of translational shutoff in the infected cells, the event that we were mainly interested in. We did not detect marked differences in cell reactions (microarray data not shown) or rates of virus replication in wt MEFs compared to the NIH 3T3 cells (data not shown). Thus, in order to use the same MOI in all of the experiments and to link the results to those of previous studies, NIH 3T3 cells were used as the PKR+/+ counterpart of PKR−/− MEFs.
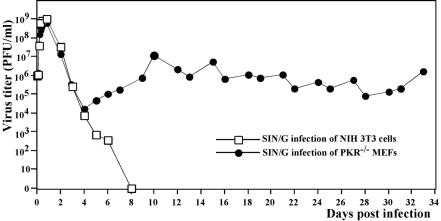
Both wtSIN and SIN/G demonstrated efficient growth in NIH 3T3 cells and PKR−/− MEFs (Fig. 1C), and the RNA of each virus was synthesized equally efficiently in both cell types. However, SIN/G genome replication and transcription of the subgenomic RNAs were 5- to 10-fold less efficient than for wtSIN (Fig. 1B). The abilities of SIN/G and wtSIN to cause CPE strongly differed. Infection of the NIH 3T3 cells with wt SIN led to complete CPE within 24 h postinfection, and PKR−/− MEFs infected with the virus demonstrated profound morphological changes within 8 h postinfection, followed by complete destruction of cells after the next 8 h of incubation. This accelerated CPE observed in the absence of PKR was not associated with any significant change in virus production (Fig. 1C), although an increase in virus release was detected during the first 3 to 4 h. SIN/G, in contrast, did not cause CPE in either cell line, in spite of infection of all of the cells (shown by GFP expression) at 4 to 8 h postinfection and release of high titers of virus (Fig. 1C). The NIH 3T3 cells inhibited replication of this mutant and eventually cleared the infection (Fig. 2). In contrast, the PKR−/− MEFs were incapable of stopping SIN/G replication and continued to produce virus for >30 days (Fig. 2). In a few days, the virus lost the ability to express GFP (a common event during passaging of double subgenomic SIN), but staining with SIN E2-specific antibodies easily detected the presence of SIN glycoproteins in >90% of the cells (data not shown). Detailed investigations of the mechanisms of cell death and virus persistence were beyond the goal of this study, and the subjects were not further explored.
FIG. 2.
Growth of SIN/G in NIH 3T3 cells and PKR−/− MEFs. Subconfluent cells in six-well Costar plates (5 × 105 cells/well) were infected with SIN/G at an MOI of 20 PFU/cell. At the indicated times, the media were replaced and virus titers were determined as described in Materials and Methods. For PKR−/− MEFs, after 7 days of incubation, virus titers were determined at the indicated times, when the cells were split at a dilution of 1:3.
The data indicated that SIN variants could replicate with similar efficiencies in both PKR-positive and PKR-negative cells. PKR had some regulatory functions in SIN infection, and its antiviral effect was characterized by slower CPE progression in NIH 3T3 cells infected with wtSIN than in PKR−/− MEFs infected with the same virus. PKR-positive cells, but not PKR−/− MEFs, could also abrogate replication of SIN/G.
wtSIN, but not an nsP2 mutant, causes translational shutoff in both PKR+/+ and PKR−/− cells.
One of PKR's functions is phosphorylation of eIF2α in response to replication of different viruses, particularly RNA viruses synthesizing dsRNA genomes or replicative intermediates (16). To test changes in cellular translation, we infected the NIH 3T3 cells and PKR−/− MEFs with wtSIN and SIN/G and examined the levels of PKR and eIF2α phosphorylation. We also analyzed the synthesis of cellular proteins and total protein synthesis (which includes translation of both SIN structural and nonstructural proteins) at different times postinfection.
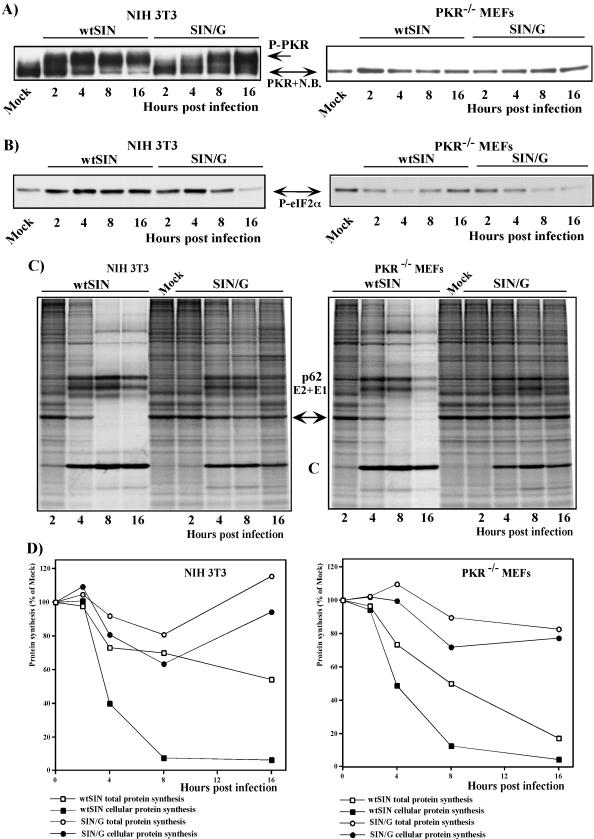
Within 4 to 8 h postinfection, wtSIN induced the phosphorylation of PKR in the NIH 3T3 cells (Fig. 3A), leading to a detectable increase in phosphorylation of eIF2α (Fig. 3B). The latter could explain the downregulation of translation of cellular mRNAs (Fig. 3C and D), but the efficient translational shutoff caused by wtSIN in PKR−/− MEFs could not be explained the same way, because of the absence of PKR in this cell line and an undetectable increase in eIF2α phosphorylation (Fig. 3A and B) during wtSIN infection.
FIG.3.
Protein synthesis and phosphorylation of PKR and eIF2α in virus-infected cells. (A and B) NIH 3T3 cells or PKR−/− MEFs (1.5 × 106 cells in 60-mm-diameter dishes) were infected with wtSIN or SIN/G at an MOI of 20 PFU/cell. At the indicated times, cells were harvested and 10 μg of protein from each sample was separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis. After transfer, the nitrocellulose membranes were stained with Ponceau S as described in Materials and Methods to control the quality of transfer and processed by anti-PKR (A) or anti-phospho-eIF2α (Ser51) (B) antibody. The results represent one of three reproducible experiments. N.B. indicates a minor protein band that nonspecifically binds anti-PKR antibodies in all tested mouse cells. Mock indicates mock-infected cells. (C and D) NIH 3T3 cells and PKR−/− MEFs (5 × 105 cells/well) in six-well Costar plates) were infected with wtSIN and SIN/G variants at an MOI of 20. Proteins were pulse-labeled with [35S]methionine at the indicated times and analyzed on sodium dodecyl sulfate-10% polyacrylamide gels. The gels were dried and autoradiographed (C) or analyzed on a Storm 860 PhosphorImager (D) as described in Materials and Methods. The levels of synthesis of cellular proteins were determined by measuring radioactivity in the protein band corresponding to actin (indicated by the arrow) and were normalized to the radioactivity in the actin band in uninfected cells. Total protein synthesis in the infected cells was evaluated by measuring the radioactivity in the entire lane on the gel, and the results were normalized to the radioactivity in uninfected cells. One of the reproducible experiments is shown.
The SIN/G mutant did not cause profound translational shutoff in NIH 3T3 cells (Fig. 3C and D), normally seen with wt SIN. In spite of lower, but still very efficient, SIN/G RNA replication (Fig. 1B and C), these PKR-positive cells efficiently translated cellular mRNAs. In NIH 3T3 cells infected with mutant virus, PKR phosphorylation proceeded, reaching a level similar to that observed during wtSIN infection (Fig. 3A). However, despite the presence of phosphorylated PKR in SIN/G-infected NIH 3T3 cells, the phosphorylation of eIF2α initially increased to the level detected in wt SIN-infected cells (Fig. 3B) at early times (2 to 4 h) postinfection but returned to the level of uninfected cells by 16 h postinfection (confirmed in multiple experiments) (Fig. 3B). These kinetics of eIF2α phosphorylation correlated with very limited, but detectable, translational shutoff in SIN/G-infected NIH 3T3 cells (Fig. 3C and D).
Similar to NIH 3T3 cells, the PKR−/− MEFs infected with the SIN/G mutant exhibited inefficient inhibition of cellular translation, and total translation also remained almost unchanged (Fig. 3C and D). In the case of PKR−/− MEFs, but not of PKR-positive cell lines, the deficit of translational shutoff could be explained by a lack of eIF2α phosphorylation (Fig. 3B). At 8 to 16 h postinfection with SIN/G, the PKR−/− MEFs reproducibly contained even less phosphorylated eIF2α than did uninfected cells.
Taken together, the data indicated that there was no exact correlation between eIF2α phosphorylation and inhibition of translation in SIN-infected cells, and the inhibition of translation in SIN-infected cells could not be explained by activation of a PKR-dependent mechanism only.
Inhibition of PKR phosphorylation does not arrest translational shutoff in SIN-infected cells.
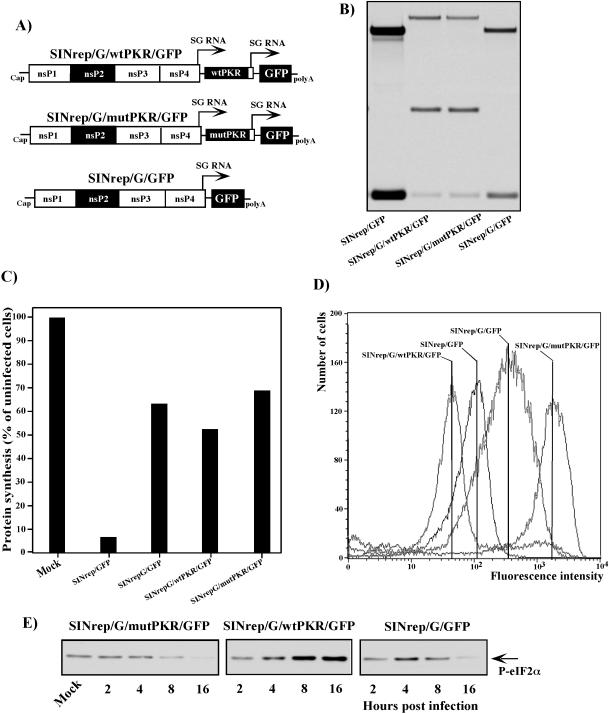
To distinguish between the possibilities that the translational shutoff in SIN-infected cells is completely independent of PKR phosphorylation or that PKR activation has an effect on downregulation of translation and works synergistically with another mechanism(s), we tested the effects of overexpression of wtPKR or its kinase-defective form (which acts as a dsRNA-binding decoy) on translational shutoff during SIN infection. To achieve this, we designed a set of SIN replicons (Fig. 4A). All of them contained the SP driving the transcription of the subgenomic RNA encoding GFP, and in addition, two of the replicons, SINrep/wtPKR/GFP and SINrep/mutPKR/GFP, encoded the wt form of human PKR or a dominant-negative mutant of the enzyme that is capable of binding to dsRNA but lacks the kinase activity because of the single point mutation K296 → R (1). Heterologous human PKR was intentionally used for the experiments described below, because it had an electrophoretic mobility different from that of murine PKR. However, the human form of PKR was previously shown to function efficiently in murine cells (1, 2). Both PKR genes were positioned under the control of a second SP in these replicons (Fig. 4A).
When the replicons and the helper RNAs were cotransfected into BHK-21 cells, they produced similar titers of replicon-containing viral particles, approaching 2 × 109 infectious units/ml. These stocks were used to infect NIH 3T3 cells at an MOI of 20 infectious units/cell, and the inhibition of translation was analyzed together with phosphorylation of PKR and eIF2α. All three replicons had the same level of replication and transcription of the subgenomic RNAs (Fig. 4B). They also caused similar levels of translational shutoff that developed at comparable rates (Fig. 4C). Replication of SINrep/GFP caused phosphorylation of both PKR and eIF2α (Fig. 4D) to levels that were even higher than in wtSIN-infected cells (Fig. 3A and B). The additional expression of wtPKR by SINrep/wtPKR/GFP did not change the phosphorylation of endogenous murine PKR, which was already high in SINrep/GFP replicon-infected cells, but it did increase the phosphorylation of eIF2α (Fig. 4D). In contrast, expression of mutant PKR by SINrep/mutPKR/GFP strongly affected the phosphorylation of murine PKR. The level of its phosphorylation was indistinguishable from that observed in uninfected cells. Consequently, no change in the phosphorylation of eIF2α was detected (Fig. 4D). Nevertheless, SINrep/mutPKR/GFP replication caused translational shutoff very efficiently (Fig. 4C), supporting the idea that another mechanism besides PKR and eIF2α phosphorylation is involved in the inhibition of translation in the cells containing replicating SIN-specific RNAs encoding wt nsPs.
These replicons demonstrated obvious differences in the ability to express GFP, indicating that PKR phosphorylation played a significant role in inhibition of translation. SINrep/mutPKR/GFP expressed 20-fold more GFP than did SINrep/GFP, which reproducibly expressed between two- and fourfold more of this protein than did SINrep/wtPKR/GFP (Fig. 4E). In SINrep/mutPKR/GFP replicon-infected cells, not only GFP-encoding subgenomic RNA, but also viral genome RNA and PKR-encoding subgenomic RNAs, were translated better (data not shown; see also the difference between total and cellular protein synthesis in Fig. 4C) than their counterparts in SINrep/GFP- and SINrep/wtPKR/GFP-infected cells. In the latter case, the expression of wt human PKR was barely detectable (Fig. 4D). To rule out the possibility of a cloning error, the pSINrep/wtPKR/GFP plasmid was also sequenced and found to have a 5′ UTR sequence in the PKR-encoding subgenomic RNA and a PKR sequence identical to those in the SINrep/mutPKR/GFP replicon (except for the presence of Lys in the 296 position). We believe that overexpressed wt human PKR functioned efficiently, not only in the inhibition of translation of GFP-encoding RNA, but also in downregulating its own translation from SIN-encoded subgenomic RNA. Despite being present in smaller amounts than mutant PKR in SINrep/mutPKR/GFP replicon-infected cells, wtPKR was catalytically active and had a stronger effect on translation than its catalytically inactive form, which works by stoichiometric binding to dsRNA.
These data clearly show that replicon-encoded mutant PKR was capable of functioning as a decoy to downregulate phosphorylation of endogenous murine PKR. Its expression strongly upregulated translation of SIN replicon-encoded RNAs, but this mutant PKR was not able to completely prevent the inhibition of cellular RNA translation in SIN replicon-infected cells. As a result, total protein synthesis in SINrep/mutPKR/GFP-infected cells was more than twofold lower than before infection (Fig. 4C).
PKR-dependent translational shutoff proceeds in cells infected with SIN replicons encoding the mutated nsP2.
In the next experiments, we determined if PKR-dependent inhibition of translation is a universal reaction and if it played a significant role in regulating translation in cells infected with SIN replicons expressing a mutant form of nsP2 (Fig. 5A). Similar to SIN/G, this mutation makes replicons incapable of inducing profound translational shutoff. The constructs had the same design as the replicons shown in Fig. 4A but had a point mutation, P726 → G, in the nsP2, as in the SIN/G viral genome. All of these replicons replicated 5- to 10-fold less efficiently than wt SINrep/GFP (Fig. 5B) and did not cause strong inhibition of host cell protein synthesis (Fig. 5C) to the level observed in the wt replicon SINrep/GFP-infected cells. Nevertheless, SINrep/G/GFP caused detectable eIF2α phosphorylation at an early time postinfection (Fig. 5E). Then, in a manner similar to that of SIN/G (Fig. 3B), the amount of phosphorylated eIF2α dropped by 16 h postinfection to a level that was lower than that seen in uninfected cells (Fig. 5E). Overexpression of wtPKR caused a marked increase in eIF2α phosphorylation, whereas expression of mutant PKR abolished phosphorylation of this translation initiation factor. The expression of GFP correlated with eIF2α phosphorylation. Despite a lower level of replication, SINrep/G/GFP expressed fourfold more GFP than did the wt replicon, SINrep/GFP, most likely because of the absence of general translational shutoff due to the nsP2 mutation (Fig. 5D), and SINrep/G/mutPKR/GFP produced fivefold more GFP than did SINrep/G/GFP. (The latter finding was likely the result of inhibition of eIF2α phosphorylation early after infection.) The expression of wtPKR by the SINrep/G/wtPKR/GFP replicon inhibited GFP production almost fivefold compared to SINrep/G/GFP replicon-infected cells (Fig. 5D). This result correlated with the increase in eIF2α phosphorylation (Fig. 5E).
FIG. 5.
Schematic representation of SIN/G-based replicons and analysis of their RNA replication and transcription, protein synthesis, and phosphorylation of eIF2α in the infected cells. (A) The replicons had essentially the same design as the constructs described in the legend to Fig. 4 but differed from the wtSIN-based constructs by one amino acid in nsP2, P726 → G. (B) NIH 3T3 cells (5 × 105) were infected with packaged replicons at an MOI of 20 infectious units/cell. At 2 h postinfection, the media were replaced by the same media supplemented with dactinomycin (1 μg/ml) and [3H]uridine (20 μCi/ml). After 4 h of incubation at 37°C, RNAs were isolated and analyzed as described in Materials and Methods. (C) NIH 3T3 cells (5 × 105 cells/well) in six-well Costar plates) were infected with packaged replicons at an MOI of 20 infectious units/cell. Proteins were pulse-labeled with [35S]methionine 16 h postinfection and analyzed on sodium dodecyl sulfate-10% polyacrylamide gels. The gels were dried and analyzed on a Storm 860 PhosphorImager as described in Materials and Methods. The levels of synthesis of cellular proteins in the infected cells were evaluated as described in the legend to Fig. 3. Mock indicates mock-infected cells. (D) NIH 3T3 cells (1.5 × 106) were infected with packaged replicons at an MOI of 20 infectious units/cell. At 12 h postinfection, the cells were fixed by 1% formaldehyde in PBS for 10 min and analyzed by flow cytometry on a FACS Vantage (Becton Dickinson). (E) Phosphorylation of eIF2α (P-eIF2α) in replicon-infected cells was analyzed as described in the legend to Fig. 3.
These data indicated that a lack of profound translational shutoff in the cells infected with SIN replicons or SIN expressing a mutated form of nsP2 is most probably the result of the inability of these SIN-specific RNAs to induce PKR-independent inhibition of translation. Their replication did cause detectable phosphorylation of eIF2α at early times postinfection, which could be inhibited by expression of mutant PKR, leading in turn to higher expression of the replicon-encoded heterologous gene. On the other hand, these data showed that overexpression of the wtPKR strongly increased eIF2α phosphorylation and decreased the level of heterologous protein expression, but this phosphorylated eIF2α did not downregulate host cell protein synthesis (Fig. 5C) to the level found in the wt virus- (Fig. 3D) or wtSIN replicon-infected cells (Fig. 4C). This fact suggests that in SIN-infected cells, PKR-dependent translational shutoff does universally develop but most likely does not cause strong inhibition of translation in the entire cell.
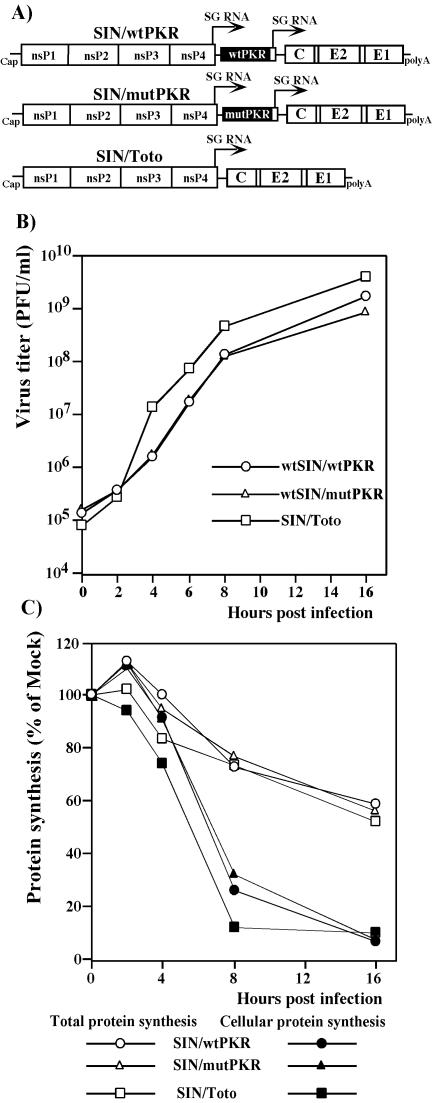
Overexpression of wt or mutated PKR does not affect replication of SIN.
Numerous viruses have developed mechanisms to inhibit PKR activation. However, SIN replication in NIH 3T3 cells induced PKR phosphorylation, but the virus was able to propagate to high titers (Fig. 1C). To further examine the effects of PKR phosphorylation on SIN replication, we designed double subgenomic viral genomes (Fig. 6A) expressing wtPKR or the kinase-defective dominant-negative mutant form of PKR from a second subgenomic RNA. In an infectious-center assay, all three in vitro-synthesized RNAs had similar specific infectivities (∼106 PFU/μg), indicating that the recombinant viruses were viable and unimpaired in the ability to initiate replication. Viral stocks generated by RNA electroporation of BHK-21 cells were used to compare general biological characteristics of SIN/Toto, SIN/mutPKR, and SIN/wtPKR viruses in vitro. After infection of NIH 3T3 cells, the viruses exhibited similar levels of genome RNA replication and subgenomic mRNA transcription (data not shown), growth rates (Fig. 6B), and kinetics of inhibition of host cell protein synthesis (Fig. 6C). Remarkably, translations of viral structural proteins from the subgenomic RNAs were essentially the same for all constructs (data not shown; see also total protein synthesis, shown in Fig. 6C) regardless of the overexpression of the wt or mutant form of PKR. The high level of translation of the viral subgenomic RNA encoding the structural proteins, in spite of both PKR-dependent and PKR-independent translational shutoff in SIN-infected cells, was likely due to the unique RNA element, translational enhancer, present in the capsid coding sequence in SIN subgenomic RNA (13). Even at late times postinfection, almost 60% of the capacity of the translational apparatus was still utilized for the translation of the SIN subgenomic RNA (see the total protein synthesis shown in Fig. 6C).
FIG. 6.
Schematic representation of double subgenomic viral genomes and analysis of virus growth and protein synthesis in infected NIH 3T3 cells. (A) All three viral genomes expressed the same structural proteins derived from SIN Toto1101. The SIN/wtPKR and SIN/mutPKR constructs contained a second SP driving the expression of wt human PKR and human PKR with a single substitution (K296 → R),respectively. (B) NIH 3T3 cells in six-well Costar plates were infected with SIN at an MOI of 5 PFU/cell. At the indicated times, the media were replaced and virus titers were determined as described in Materials and Methods. (C) NIH 3T3 cells (5 × 105 cells/well) in six-well Costar plates) were infected with viruses at an MOI of 5 PFU/cell. Proteins were pulse-labeled with [35S]methionine at the indicated times and analyzed on sodium dodecyl sulfate-10% polyacrylamide gels. The gels were dried and analyzed on a Storm 860 PhosphorImager as described in Materials and Methods. The levels of synthesis of cellular proteins and total protein synthesis in the infected cells were evaluated as described in the legend to Fig. 3.
To confirm the critical role of the translational enhancer, we designed two more SIN replicons: SINrep/C/GFP, with a wt sequence of the nonstructural proteins and expressing GFP from the subgenomic RNA, and SINrep/wtPKR/C/GFP, also expressing the wt human PKR from the second SP (Fig. 7A). The GFP-encoding gene was fused in frame with the capsid- coding sequence, which contains the translation enhancer element. Because of its autoprotease activity, the capsid cleaved itself co- and posttranslationally, generating GFP with only four extra amino acids upstream of its natural initiating methionine. The NIH 3T3 cells infected with SINrep/C/GFP produced almost 20-fold more GFP then did cells infected with SINrep/GFP at the same MOI (Fig. 7B). The additional expression of wtPKR in cis by SINrep/wtPKR/C/GFP did not have a deleterious effect on GFP expression. We always detect some decrease in protein production when two subgenomic RNAs, instead of one, are transcribed from SIN replicons. However, the possibility of a small effect of expressed wtPKR cannot be ruled out.
These results demonstrated that by using the translation enhancer element in the amino-terminal part of the capsid-encoding gene, SIN has developed a very efficient means of replicating under conditions of both PKR-dependent (host cell-induced) and PKR-independent (virus- or host cell-induced) translational shutoff that takes place in infected vertebrate cells.
DISCUSSION
The alpha/beta interferon (IFN-α/β)-dependent innate immune response is one of the most important mechanisms that control virus replication on both the cellular and the organism levels. One of the best-characterized components of the cellular response is PKR activation, which is induced by dsRNA synthesized in the cells during virus replication. After binding to dsRNA, PKR changes its conformation, undergoes autophosphorylation and dimerization, and phosphorylates eIF2α (30, 38, 46). This, in turn, leads to a decrease in translation initiation. However, in the present study, we demonstrated that the PKR−/− MEFs, in which PKR is not present and eIF2α is not phosphorylated, exhibit the same downregulation of translation as PKR-positive cells after infection with SIN. Moreover, the NIH 3T3 cells infected with SIN overexpressing a mutated form of nsP2, the SIN/G mutant, exhibit phosphorylation of PKR to the level observed upon infection with wtSIN, but eIF2α phosphorylation and translational shutoff are inefficient. By 16 h postinfection, protein synthesis in SIN/G-infected cells is at the same level as in uninfected cells, and phosphorylation of eIF2α is no longer detectable (Fig. 3). However, even at this late time point, PKR remains highly phosphorylated, apparently due to RNA replication, but does not cause inhibition of cellular translation by an eIF2α-mediated mechanism to the level observed in wtSIN-infected NIH 3T3 cells (Fig. 3).
This finding strongly suggests the existence of another, PKR-independent, mechanism that plays a dominant role in SIN-specific inhibition of host mRNA translation. This mechanism is not based on competition between viral subgenomic RNAs present at high concentrations and cellular mRNAs, because as was previously found (11-13) and as confirmed in the present work, the replicon subgenomic RNAs encoding heterologous genes recruit cellular translational machinery inefficiently (Fig. 4, 5 and 7) and are not translated at the same rate as SIN 26S RNA, despite having the same 5′ and 3′ UTRs and being present in very high concentrations.
Nevertheless, the role of PKR in SIN-specific inhibition of translation cannot be ignored, and PKR activation is likely a universal event during SIN infection. Both SIN replicons expressing a kinase-defective mutant form of human PKR and containing either the wt sequence of the nonstructural proteins or nsP2 with a P726 → G mutation demonstrated undetectable phosphorylation of the authentic murine PKR and eIF2α, and this in turn strongly increased the translation of the GFP-encoding RNA, transcribed from the replicon's second SP (Fig. 4 and 5).
The results of our study open the possibility of experimental manipulation with both mechanisms of SIN-induced translation inhibition to achieve more efficient expression of heterologous proteins from SIN replicons. Indeed, (i) the PKR-dependent component of translational shutoff could be inhibited by the excessive expression of a dominant-negative mutant form of PKR, functioning as a decoy that binds to dsRNA but is incapable of eIF2α phosphorylation (Fig. 4); (ii) activation of the PKR-independent mechanism could be avoided by using SIN replicons expressing a mutated form of nsP2 (these replicons produce almost 4-fold more heterologous protein than did the wtSIN-based replicons, despite a 5- to 10-fold-lower level of RNA replication and transcription of the subgenomic RNA [Fig. 5]); and finally, (iii) both PKR-dependent and PKR-independent components of translation inhibition could be eluded by using replicons with the mutation in nsP2 and expressing a mutant form of human PKR. The latter construct (which also replicated less efficiently than the wt replicons) produced 20-fold more GFP than the wtSIN genome-based replicons and 4-fold more than its counterpart with the nsP2 mutation but not expressing mutPKR (Fig. 5).
PKR-mediated pathways undoubtedly represent one of the important components of the cellular response to virus replication (3, 38). It was previously demonstrated that PKR−/− mice are more susceptible to vesicular stomatitis virus (39), Bunyamwera virus (42), encephalomyocarditis virus (48), and herpes simplex virus type 1 (23). It was also suggested that an early anti-SIN response develops more efficiently in PKR-positive cells (34). In agreement with these data, PKR−/− MEFs infected with wtSIN demonstrated earlier virus release and faster CPE than the NIH 3T3 cells and wt MEFs. These cells are also incapable of clearing the poorly cytopathic SIN nsP2 mutant, SIN/G (Fig. 2). Upon infection with this virus, PKR−/− MEFs released IFN-α/β to almost the same concentration as NIH 3T3 cells (data not shown) and activated the same set of genes in response to virus replication in microarray-based gene expression profiling experiments (data not shown). They were also capable of efficiently developing an antiviral state in response to IFN-α/β pretreatment (data not shown). However, in contrast to their PKR-positive counterparts, the PKR−/− cells remained persistently infected, like some other cell types with defects in IFN-α/β signaling or infected in the presence of anti-IFN antibodies (14).
Based on our data, we hypothesize that the inhibition of translation observed in SIN-infected cells develops by two mechanisms, of which the PKR-independent pathway determines translational shutoff to a greater extent. However, we are only beginning to understand its nature. This mechanism is likely not unique to alphaviruses. For example, after IFN treatment, translation of dengue virus RNA is inhibited through a PKR-independent mechanism as well (7).
Both PKR-dependent and PKR-independent mechanisms of translational shutoff have minimal effects on the translation of natural SIN subgenomic RNA. This RNA contains a translational enhancer, represented by a GC-rich stem-loop structure, in the capsid coding sequence (12) and remains efficiently translated by modified cellular translational machinery (Fig. 6 and 7). By developing this structural element in the capsid coding sequence, SIN, like some other alphaviruses, successfully overcomes the negative effect of the inhibition of translation (12, 36, 37), and viral structural proteins are synthesized very efficiently, even in the late stages of infection. Even overexpression of wtPKR in cis does not affect SIN replication (Fig. 6) or expression of the heterologous gene, if it is fused with the capsid coding sequence (Fig. 7).
In addition to enhancing our understanding of alphavirus-host cell interactions, our results have practical applications. By using the approaches uncovered in this study, it might be possible to increase the expression of the heterologous genes from the SIN-based replicons and, most likely, from other alphavirus-based replicons to augment their ability to produce heterologous proteins.
Acknowledgments
We thank Peter Mason and Scott Weaver for critical reading of the manuscript.
This work was supported by Public Health Service grants AI053135 (R.G. and I.F.) and AI24134 (C.M.R.).
REFERENCES
- 1.Barber, G. N., R. Jagus, E. F. Meurs, A. G. Hovanessian, and M. G. Katze. 1995. Molecular mechanisms responsible for malignant transformation by regulatory and catalytic domain variants of the interferon-induced enzyme RNA-dependent protein kinase. J. Biol. Chem. 270:17423-17428. [DOI] [PubMed] [Google Scholar]
- 2.Barber, G. N., M. Wambach, S. Thompson, R. Jagus, and M. G. Katze. 1995. Mutants of the RNA-dependent protein kinase (PKR) lacking double-stranded RNA binding domain I can act as transdominant inhibitors and induce malignant transformation. Mol. Cell. Biol. 15:3138-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogdan, C. 2000. The function of type I interferons in antimicrobial immunity. Curr. Opin. Immunol. 12:419-424. [DOI] [PubMed] [Google Scholar]
- 4.Bredenbeek, P. J., I. Frolov, C. M. Rice, and S. Schlesinger. 1993. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J. Virol. 67:6439-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chawla-Sarkar, M., D. J. Lindner, Y. F. Liu, B. R. Williams, G. C. Sen, R. H. Silverman, and E. C. Borden. 2003. Apoptosis and interferons: role of interferon-stimulated genes as mediators of apoptosis. Apoptosis 8:237-249. [DOI] [PubMed] [Google Scholar]
- 6.Der, S. D., Y. L. Yang, C. Weissmann, and B. R. Williams. 1997. A double-stranded RNA-activated protein kinase-dependent pathway mediating stress-induced apoptosis. Proc. Natl. Acad. Sci. USA 94:3279-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diamond, M. S., and E. Harris. 2001. Interferon inhibits dengue virus infection by preventing translation of viral RNA through a PKR-independent mechanism. Virology 289:297-311. [DOI] [PubMed] [Google Scholar]
- 8.Frolov, I., E. Agapov, T. A. Hoffman, Jr., B. M. Prágai, M. Lippa, S. Schlesinger, and C. M. Rice. 1999. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J. Virol. 73:3854-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frolov, I., E. Frolova, and S. Schlesinger. 1997. Sindbis virus replicons and Sindbis virus: assembly of chimeras and of particles deficient in virus RNA. J. Virol. 71:2819-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frolov, I., R. Hardy, and C. M. Rice. 2001. cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA 7:1638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frolov, I., and S. Schlesinger. 1994. Comparison of the effects of Sindbis virus and Sindbis virus replicons on host cell protein synthesis and cytopathogenicity in BHK cells. J. Virol. 68:1721-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frolov, I., and S. Schlesinger. 1996. Translation of Sindbis virus mRNA: analysis of sequences downstream of the initiating AUG codon that enhance translation. J. Virol. 70:1182-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frolov, I., and S. Schlesinger. 1994. Translation of Sindbis virus mRNA: effects of sequences downstream of the initiating codon. J. Virol. 68:8111-8117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frolova, E. I., R. Z. Fayzulin, S. H. Cook, D. E. Griffin, C. M. Rice, and I. Frolov. 2002. Roles of nonstructural protein nsP2 and alpha/beta interferons in determining the outcome of Sindbis virus infection. J. Virol. 76:11254-11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galabru, J., and A. Hovanessian. 1987. Autophosphorylation of the protein kinase dependent on double-stranded RNA. J. Biol. Chem. 262:15538-15544. [PubMed] [Google Scholar]
- 16.Gale, M., Jr., and M. G. Katze. 1998. Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol. Ther. 78:29-46. [DOI] [PubMed] [Google Scholar]
- 17.Griffin, D. E. 2001. Alphaviruses, p. 917-962. In P. M. Howley (ed.), Fields virology, 4th ed. Lippincott, Williams and Wilkins, New York, N.Y.
- 18.Griffin, D. E., and R. T. Johnson. 1977. Role of the immune response in recovery from Sindbis virus encephalitis in mice. J. Immunol. 118:1070-1075. [PubMed] [Google Scholar]
- 19.Johnston, R. E., and C. J. Peters. 1996. Alphaviruses, p. 843-898. In P. M. Howley (ed.), Fields virology, 3rd ed. Raven Press, New York, N.Y.
- 20.Karpf, A. R., and D. T. Brown. 1998. Comparison of Sindbis virus-induced pathology in mosquito and vertebrate cell cultures. Virology 240:193-201. [DOI] [PubMed] [Google Scholar]
- 21.Katze, M. G., and R. M. Krug. 1990. Translational control in influenza virus-infected cells. Enzyme 44:265-277. [DOI] [PubMed] [Google Scholar]
- 22.Kiwanuka, N., E. J. Sanders, E. B. Rwaguma, J. Kawamata, F. P. Ssengooba, R. Najjemba, W. A. Were, M. Lamunu, G. Bagambisa, T. R. Burkot, L. Dunster, J. J. Lutwama, D. A. Martin, C. B. Cropp, N. Karabatsos, R. S. Lanciotti, T. F. Tsai, and G. L. Campbell. 1999. O'nyong-nyong fever in south-central Uganda, 1996-1997: clinical features and validation of a clinical case definition for surveillance purposes. Clin. Infect. Dis. 29:1243-1250. [DOI] [PubMed] [Google Scholar]
- 23.Leib, D. A., M. A. Machalek, B. R. Williams, R. H. Silverman, and H. W. Virgin. 2000. Specific phenotypic restoration of an attenuated virus by knockout of a host resistance gene. Proc. Natl. Acad. Sci. USA 97:6097-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemm, J. A., R. K. Durbin, V. Stollar, and C. M. Rice. 1990. Mutations which alter the level or structure of nsP4 can affect the efficiency of Sindbis virus replication in a host-dependent manner. J. Virol. 64:3001-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levine, B., Q. Huang, J. T. Isaacs, J. C. Reed, D. E. Griffin, and J. M. Hardwick. 1993. Conversion of lytic to persistent alphavirus infection by the bcl-2 cellular oncogene. Nature 361:739-742. [DOI] [PubMed] [Google Scholar]
- 26.Liljeström, P., S. Lusa, D. Huylebroeck, and H. Garoff. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 65:4107-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lodish, H. F., and M. Porter. 1980. Translational control of protein synthesis after infection by vesicular stomatitis virus. J. Virol. 36:719-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lodish, H. F., and M. Porter. 1981. Vesicular stomatitis virus mRNA and inhibition of translation of cellular mRNA—is there a P function in vesicular stomatitis virus? J. Virol. 38:504-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lustig, S., A. Jackson, C. S. Hahn, D. E. Griffin, E. G. Strauss, and J. H. Strauss. 1988. Molecular basis of Sindbis virus neurovirulence in mice. J. Virol. 62:2329-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nanduri, S., F. Rahman, B. R. Williams, and J. Qin. 2000. A dynamically tuned double-stranded RNA binding mechanism for the activation of antiviral kinase PKR. EMBO J. 19:5567-5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porter, A. G. 1993. Picornavirus nonstructural proteins: Emerging roles in virus replication and inhibition of host cell functions. J. Virol. 67:6917-6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice, C. M., R. Levis, J. H. Strauss, and H. V. Huang. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: Mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J. Virol. 61:3809-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rivas, F., L. A. Diaz, V. M. Cardenas, E. Daza, L. Bruzon, A. Alcala, O. De la Hoz, F. M. Caceres, G. Aristizabal, J. W. Martinez, D. Revelo, F. De la Hoz, J. Boshell, T. Camacho, L. Calderon, V. A. Olano, L. I. Villarreal, D. Roselli, G. Alvarez, G. Ludwig, and T. Tsai. 1997. Epidemic Venezuelan equine encephalitis in La Guajira, Colombia, 1995. J. Infect. Dis. 175:828-832. [DOI] [PubMed] [Google Scholar]
- 34.Ryman, K. D., L. J. White, R. E. Johnston, and W. B. Klimstra. 2002. Effects of PKR/RNase L-dependent and alternative antiviral pathways on alphavirus replication and pathogenesis. Viral Immunol. 15:53-76. [DOI] [PubMed] [Google Scholar]
- 35.Scott, T. W., and S. C. Weaver. 1989. Eastern equine encephalomyelitis virus: epidemiology and evolution of mosquito transmission. Adv. Virus Res. 37:277-328. [DOI] [PubMed] [Google Scholar]
- 36.Sjöberg, E. M., and H. Garoff. 1996. The translation-enhancing region of the Semliki Forest virus subgenome is only functional in the virus-infected cell. J. Gen. Virol. 77:1323-1327. [DOI] [PubMed] [Google Scholar]
- 37.Sjöberg, E. M., M. Suomalainen, and H. Garoff. 1994. A significantly improved Semliki Forest virus expression system based on translation enhancer segments from the viral capsid gene. BioTechnology 12:1127-1131. [DOI] [PubMed] [Google Scholar]
- 38.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 39.Stojdl, D. F., N. Abraham, S. Knowles, R. Marius, A. Brasey, B. D. Lichty, E. G. Brown, N. Sonenberg, and J. C. Bell. 2000. The murine double-stranded RNA-dependent protein kinase PKR is required for resistance to vesicular stomatitis virus. J. Virol. 74:9580-9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strauss, E. G., C. M. Rice, and J. H. Strauss. 1984. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology 133:92-110. [DOI] [PubMed] [Google Scholar]
- 41.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Streitenfeld, H., A. Boyd, J. K. Fazakerley, A. Bridgen, R. M. Elliott, and F. Weber. 2003. Activation of PKR by Bunyamwera virus is independent of the viral interferon antagonist NSs. J. Virol. 77:5507-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takizawa, T., C. Tatematsu, and Y. Nakanishi. 2002. Double-stranded RNA-activated protein kinase interacts with apoptosis signal-regulating kinase 1. Implications for apoptosis signaling pathways. Eur. J. Biochem. 269:6126-6132. [DOI] [PubMed] [Google Scholar]
- 44.Tan, S. L., and M. G. Katze. 1999. The emerging role of the interferon-induced PKR protein kinase as an apoptotic effector: a new face of death? J. Interferon Cytokine Res. 19:543-554. [DOI] [PubMed] [Google Scholar]
- 45.Weaver, S. C., R. Salas, R. Rico-Hesse, G. V. Ludwig, M. S. Oberste, J. Boshell, R. B. Tesh, et al. 1996. Re-emergence of epidemic Venezuelan equine encephalomyelitis in South America. Lancet 348:436-440. [DOI] [PubMed] [Google Scholar]
- 46.Wu, S., and R. J. Kaufman. 1997. A model for the double-stranded RNA (dsRNA)-dependent dimerization and activation of the dsRNA-activated protein kinase PKR. J. Biol. Chem. 272:1291-1296. [DOI] [PubMed] [Google Scholar]
- 47.Yang, Y. L., L. F. Reis, J. Pavlovic, A. Aguzzi, R. Schafer, A. Kumar, B. R. Williams, M. Aguet, and C. Weissmann. 1995. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 14:6095-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeung, M. C., D. L. Chang, R. E. Camantigue, and A. S. Lau. 1999. Inhibitory role of the host apoptogenic gene PKR in the establishment of persistent infection by encephalomyocarditis virus in U937 cells. Proc. Natl. Acad. Sci. USA 96:11860-11865. [DOI] [PMC free article] [PubMed] [Google Scholar]