A short sequence motif that regulates mesophyll-specific accumulation of multiple enzymes is used in C4 photosynthesis.
Abstract
C4 photosynthesis is a complex phenotype that allows more efficient carbon capture than the ancestral C3 pathway. In leaves of C4 species, hundreds of transcripts increase in abundance compared with C3 relatives and become restricted to mesophyll (M) or bundle sheath (BS) cells. However, no mechanism has been reported that regulates the compartmentation of multiple enzymes in M or BS cells. We examined mechanisms regulating CARBONIC ANHYDRASE4 (CA4) in C4 Gynandropsis gynandra. Increased abundance is directed by both the promoter region and introns of the G. gynandra gene. A nine-nucleotide motif located in the 5′ untranslated region (UTR) is required for preferential accumulation of GUS in M cells. This element is present and functional in three additional 5′ UTRs and six 3′ UTRs where it determines accumulation of two isoforms of CA and pyruvate,orthophosphate dikinase in M cells. Although the GgCA4 5′ UTR is sufficient to direct GUS accumulation in M cells, transcripts encoding GUS are abundant in both M and BS. Mutating the GgCA4 5′ UTR abolishes enrichment of protein in M cells without affecting transcript abundance. The work identifies a mechanism that directs cell-preferential accumulation of multiple enzymes required for C4 photosynthesis.
INTRODUCTION
The ability to restrict synthesis of specific proteins to individual cell types is essential to the development of multicellular organisms. Understanding the molecular mechanisms generating and maintaining this cell specificity in gene expression has therefore attracted wide interest (Brady et al., 2007; Heintzman et al., 2009). In plants, leaves differentiate into a number of cell types that together allow efficient photosynthesis, and this differentiation is particularly apparent in plants that use the C4 assimilatory pathway. In contrast to the majority of angiosperms in which CO2 is fixed directly into three-carbon compounds by the enzyme Rubisco in many cell types (a process termed C3 photosynthesis), C4 species have acquired a carbon concentrating mechanism that almost always relies on compartmentation of gene expression between mesophyll (M) and bundle sheath (BS) cells. By repositioning the photosynthetic process between these cells, C4 species generate a metabolic pathway that concentrates CO2 around Rubisco and therefore increases the efficiency of photosynthesis (Hatch and Slack, 1970).
In C4 leaves, CO2 is initially converted to bicarbonate by carbonic anhydrases (CAs) and assimilated to form four-carbon acids by an alternative carboxylase, phosphoenolpyruvate carboxylase. This fixation by phosphoenolpyruvate carboxylase occurs specifically in M cells and produces high concentrations of four-carbon acids. These acids then diffuse into BS cells where they are decarboxylated and refixed by Rubisco. All of the enzymes operating in C4 metabolism are already present within C3 leaves, but they are present within both M and BS cells and in most cases are much less abundant than in leaves of C4 species (Aubry et al., 2011). The evolution of a two-celled C4 pathway therefore requires the recruitment of mechanisms restricting C4 photosynthesis enzymes to M or BS cells, as well as mechanisms that increase the abundance of these enzymes in C4 compared with C3 leaves (Hibberd and Covshoff, 2010). As C4 photosynthesis is present within at least 60 independent lineages of angiosperm (Sage et al., 2011), the repeated evolution of these gene regulatory networks represents one of the most striking examples of convergence within biology.
A number of mechanisms responsible for the patterns of gene expression in C4 leaves have been characterized. For example, the increased abundance of C4 enzymes can be conferred by elements within the promoter (Matsuoka et al., 1994; Gowik et al., 2004; Kajala et al., 2012; Wiludda et al., 2012), intron (Nomura et al., 2005b), or untranslated regions (UTRs) (Ali and Taylor, 2001; Wiludda et al., 2012). Similarly, cell specificity can be generated by the promoter (Matsuoka et al., 1994; Gowik et al., 2004), exon (Brown et al., 2011), or UTRs (Patel et al., 2004; Kajala et al., 2012). As these studies focused on a number of genes isolated from phylogenetically distinct C4 species, they suggest that the mechanisms regulating genes of the C4 pathway vary both within and between independent C4 lineages. In the majority of cases, different sequences generate increased abundance and cell specificity for the same gene (Marshall et al., 1997; Akyildiz et al., 2007; Kajala et al., 2012; Wiludda et al., 2012), suggesting that the evolution of these two traits is not coordinated. Most mechanisms identified to date are also unique to each C4 lineage studied, although a homologous mechanism has been co-opted to generate BS specificity for multiple genes encoding malic enzyme within maize (Zea mays) and Gynandropsis gynandra (formerly designated as Cleome gynandra; Brown et al., 2011). However, other C4 genes appear to be regulated by nonhomologous mechanisms (Williams et al., 2012). This seems surprising, as comparative transcriptomics of congeneric C3 and C4 species suggest that increased or decreased expression may have evolved for thousands of genes (Bräutigam et al., 2011; Gowik et al., 2011). Furthermore, transcriptomics of M and BS cells from C4 species maize and Setaria viridis suggests that over 5000 transcripts are differentially abundant between the two cell types (Li et al., 2010a; Chang et al., 2012; John et al., 2014). High-throughput experimental studies have therefore highlighted a large gap in current knowledge about how two-celled C4 photosynthesis is both established and maintained. Increased abundance and cell specificity appears to have evolved for thousands of transcripts within C4 leaves, yet mechanisms coordinating the regulation of multiple transcripts mostly remain uncharacterized. Identifying these mechanisms is important for understanding how a system as complex as the C4 leaf could evolve.
CA is encoded by a multigene family, and previous studies have observed very low CA activity in BS cells of multiple C4 species from independent lineages (Burnell and Hatch, 1988). For low levels of CA to be maintained, multiple CA isoforms must be preferentially restricted to M cells. Despite this, only individual CA genes have been studied to date. For example, in Flaveria, loss of a transit peptide repositions a highly abundant chloroplastic CA into the cytosol (Tanz et al., 2009), while in G. gynandra, M specificity of CA4 is mediated by the UTRs (Kajala et al., 2012). To better understand the precise alterations required to recruit CA genes into C4 photosynthesis, we examined GgCA4, which is preferentially expressed in M cells of the C4 model species G. gynandra (Kajala et al., 2012). We report two molecular events associated with the evolution of increased expression conferred by GgCA4, as well as a posttranscriptional mechanism conferred by a short RNA motif that generates preferential accumulation in M cells compared with BS cells. We further establish that relatively limited expression in BS cells of a carbonic anhydrase enzyme, CA2, as well as a third C4 enzyme, PPDK, is mediated by the same motif. This is the first characterized mechanism coordinating the accumulation of multiple C4 pathway enzymes in M cells.
RESULTS
Multiple Mechanisms Underlie the Evolution of Increased CA4 Activity in C4 Leaves
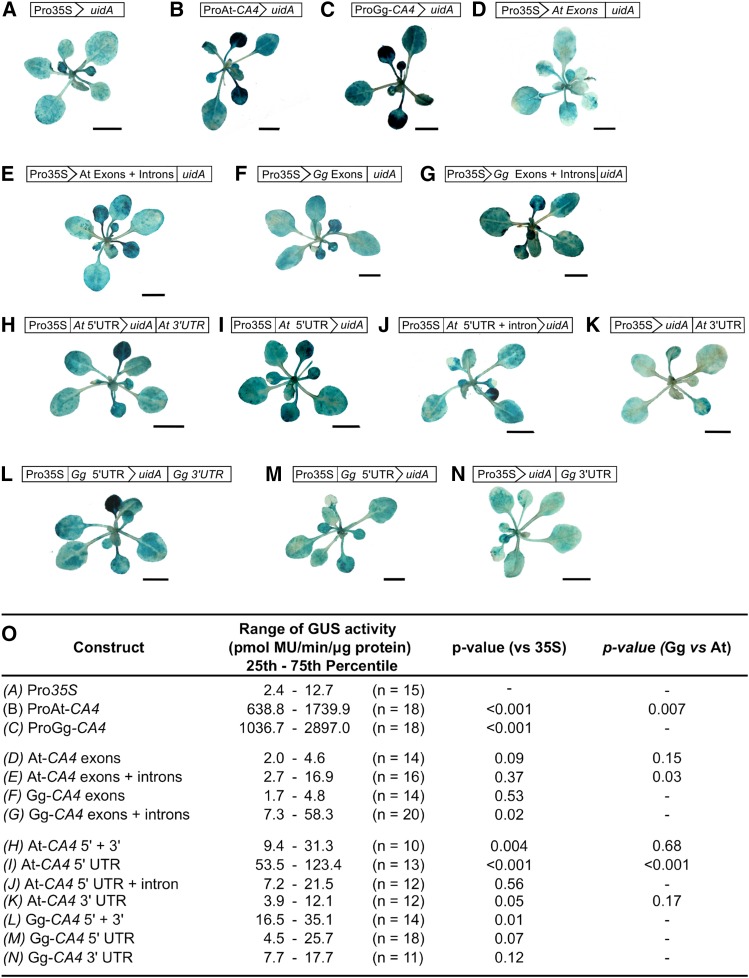
The promoters of many C4 genes confer strong, light-activated expression in leaves of C4 monocotyledons and dicotyledons (Sheen, 1999). In maize and Flaveria, this recruitment to photosynthetic metabolism evolved via the acquisition of cis-elements that are recognized by preexisting trans-acting factors in C3 leaves (Ku et al., 1999; Nomura et al., 2000; Akyildiz et al., 2007; Engelmann et al., 2008). Despite this, it is unclear whether these increases in expression compared with orthologous C3 genes are mediated by one or multiple evolutionary events. We therefore investigated whether the G. gynandra CA4 gene evolved cis-elements that are recognized by preexisting trans-factors in the C3 relative, and if this was the case, how many evolutionary events likely underlie this increased expression. To test this, promoter, exon, intron, and UTR sequences from the CA4 gene were isolated from both the C3 model species Arabidopsis thaliana and its closest C4 relative G. gynandra, fused to the uidA gene encoding the GUS reporter (Figure 1) and used to transform Arabidopsis. The localization and activity of GUS were then determined to characterize mechanisms that have evolved in cis to increase CA4 abundance in C4 leaves. Promoter regions were defined as the entire intergenic region between the transcriptional start site of CA4 and the upstream locus (AT1G70420 in Arabidopsis). In G. gynandra, genome walking isolated a 695-bp open reading frame with 74% identity to AT1G70420 that is present 1023 bp upstream of the translation start site of Gg-CA4. This implies that this region is syntenic between Arabidopsis and G. gynandra. Histological staining for GUS activity revealed that the promoter regions of At-CA4 and Gg-CA4 are both sufficient to generate increased activity compared with the control CaMV35S promoter (Figures 1A to 1C). This increase in activity was most evident in young leaves. Quantitative fluorometric assays (Jefferson et al., 1987) established that GUS activities directed by the promoters of At-CA4 and Gg-CA4 were 200- and 380-fold higher than the CaMV35S control, respectively (Figure 1O). The high-level expression of CA4 conferred by the promoter from Arabidopsis indicates that the ancestral C3 state is for strong expression. However, the promoter from G. gynandra has evolved to increase CA4 expression almost 2-fold further (P = 0.007). The intergenic regions upstream of At-CA4 and Gg-CA4 are 2019 and 1023 bp, respectively, so it is possible that cis-elements reducing expression in the ancestral state were lost in the evolution of C4 photosynthesis.
Figure 1.
Promoter and Intron Sequences Generate Increased Abundance of Gg-CA4.
(A) to (N) Promoter, coding region, and UTR sequences of At-CA4 and Gg-CA4 were fused to the uidA reporter encoding GUS. Exon and intron sequences were tested by fusing the genomic coding region to uidA. Exon sequences alone were tested by fusing the spliced open reading frame from cDNA to uidA. Multiple independent T1 lines (represented by n) were assayed for each construct. Representative images were selected from a minimum of six independent T1 lines. All seedlings were stained for 24 h, except (B) and (C), which were imaged after 6 h of staining. Bars = 5 mm.
(O) GUS activity was quantified by measuring the rate of 4-methylumbeliferone (MU) synthesis in at least 11 independent lines for each construct. Both the promoter and introns of Gg-CA4 directed increased GUS activity compared with either the CaMV35S control or homologous sequences from At-CA4. P values are derived from two-tailed Student's t tests.
Neither the exons nor introns of At-CA4 had a statistically significant impact on GUS accumulation in Arabidopsis compared with the CaMV35S control (Figures 1D, 1E, and 1O). However, the expression conferred by exons and introns combined compared with exons alone was marginally higher (P = 0.06, Student's two-tailed t test). Alternatively, introns of GgCA4 led to increased activity of the reporter compared with the CaMV35S control (P = 0.02, Student's two-tailed t test) (Figures 1F, 1G, and 1O). The combined intron length of Gg-CA4 is much shorter than At-CA4 (Kajala et al., 2012), so the enhanced expression conferred by Gg-CA4 introns may have evolved via the loss of cis-elements that act to reduce expression in the ancestral C3 state. Quantification of GUS activity directed by At-CA4 UTRs also identified two additional sequences reducing expression. First, GUS activity directed by the At-CA4 5′ UTR alone was higher than when both UTRs were present (P = 0.003, Student's two-tailed t test) (Figures 1H, 1I, and 1O), suggesting that the 3′ UTR of At-CA4 negatively regulates expression. Second, the 5′ UTR of At-CA4 contains a 625-bp intron that also acts to repress expression compared with the spliced 5′ UTR (P = 0.0002, Student's two-tailed t test) (Figures 1I, 1J, and 1O). Interestingly, this intron is absent from the 5′ UTR of Gg-CA4. However, as both UTRs of Gg-CA4 and At-CA4 direct similar levels of GUS activity (P = 0.68), loss of these elements within UTRs appears not to have led directly to the overall increased expression of CA4 in the C4 lineage. We therefore conclude that increased abundance of CA4 in leaves of G. gynandra likely evolved via two events: loss of cis-elements within the promoter as well as loss of elements within introns that act to reduce CA4 expression in the ancestral C3 leaf.
CA4 UTRs Contain a cis-Element That Also Regulates Other C4 Transcripts
We previously demonstrated that either the 5′ or 3′ UTR from CA4 from both G. gynandra and Arabidopsis is sufficient to generate M specificity within leaves of G. gynandra (Kajala et al., 2012). To test whether this is also the case for the other highly expressed CA gene in leaves of G. gynandra (Bräutigam et al., 2011), we isolated UTRs from Gg-CA2, as well as the homologous UTRs from Arabidopsis. We fused these UTRs to the uidA reporter, with the CaMV35S promoter driving expression of the fusion, and used microprojectile bombardment (Supplemental Figure 1) to determine if the UTRs were sufficient to generate M specificity. In our microprojectile bombardment assay, discrete foci of GUS activity represent independent transformation events. For the BS, GUS staining was always restricted to individual cells, allowing us to count GUS-positive BS cells independently. For the M, GUS staining sometimes spread from the highly expressing transformed cell to adjacent M cells (Supplemental Figure 1B). In these instances, only one M cell was counted, to eliminate false positives from spreading of the GUS stain. The CaMV35S promoter alone directed GUS accumulation in equal numbers of M and BS cells (595 and 613 cells, respectively, totalled across all replicates; Supplemental Table 1). This high efficiency and transformation rate effectively control against variation in transgene expression caused by the insertion location, as each individual transformation event has a very low statistical effect. Fusion of the 5′ UTRs of Gg-CA2 and At-CA2 did not affect the ratio of M and BS cells accumulating GUS (Supplemental Figure 2), but the 3′ UTRs of both homologs were sufficient to generate strong preferential accumulation in M cells (Supplemental Figure 2). This result supported the hypothesis that sequences within CA UTRs coordinate M specificity for multiple functionally related transcripts. These data combined with previous work (Kajala et al., 2012) indicated that four 5′ UTRs and six 3′ UTRs from CA2, CA4, and PPDK derived from both C4 G. gynandra and C3 Arabidopsis are capable of preferentially directing accumulation in M cells.
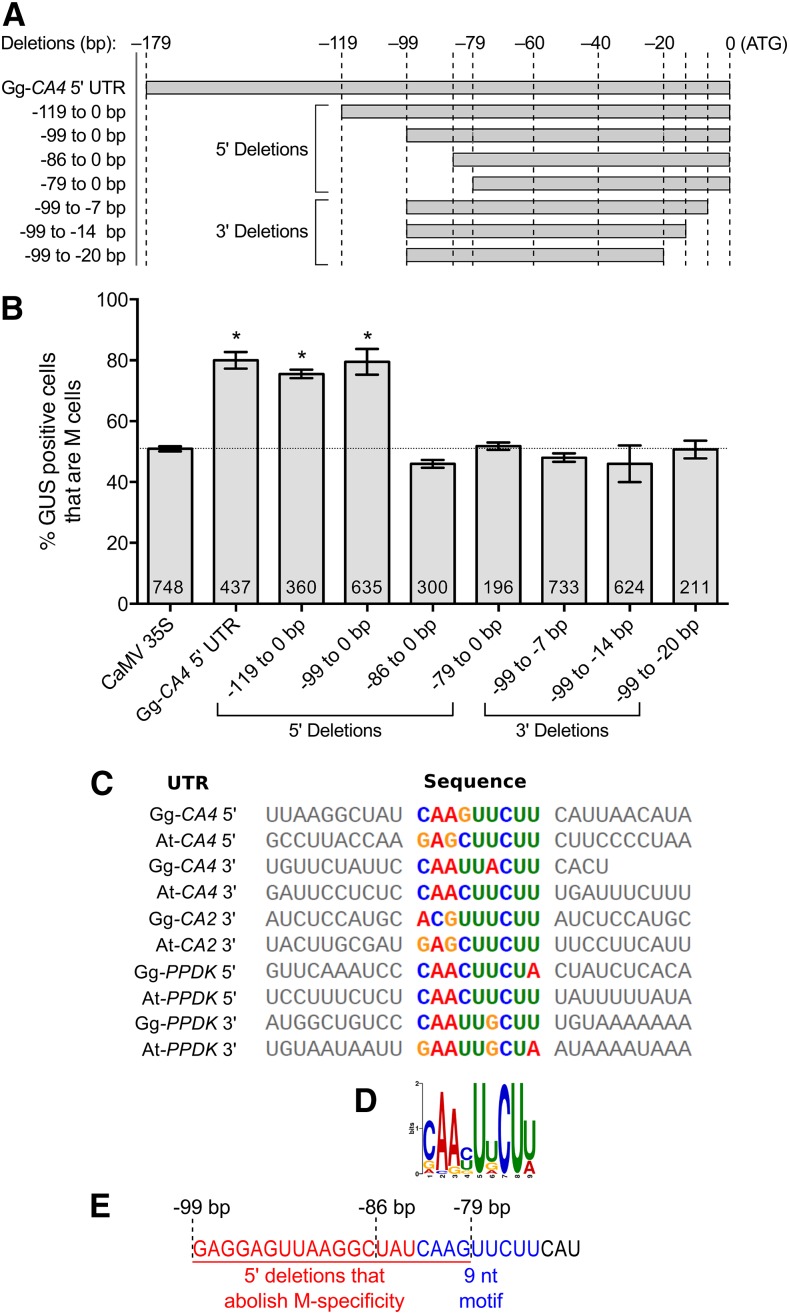
To gain insight into the mechanisms responsible for generating this M specificity, two independent approaches were taken. First, a deletion analysis on the 5′ UTR of Gg-CA4 was used to identify sequences necessary for M specificity (Figure 2A). Second, computational motif detection was undertaken to identify cis-elements that may be present in all of the Arabidopsis and G. gynandra UTRs demonstrated to generate M specificity. Deletion analysis was performed by generating 7- to 20-bp incremental deletions from either the 5′ or 3′ end of the UTR and testing the effect of each deletion on M specificity using microprojectile bombardment. We identified a 13-nucleotide region from −99 to −86 bp, as well as a seven-nucleotide sequence from the 3′ end, that were individually necessary for M specificity (Figure 2B).
Figure 2.
A cis-Element for M-Preferential Expression Identified by Two Independent Approaches.
(A) Deletion analysis was performed on the 5′ UTR of Gg-CA4. Truncated UTRs missing sequence from the 5′ or 3′ ends were fused to the uidA reporter.
(B) The number of M cells expressing GUS after microprojectile bombardment of each deletion construct is expressed as a percentage of all GUS positive cells observed. Numbers within histogram bars represent the number of independently transformed cells for each construct. The 5′ and 3′ deletions identified 13 and 7 bp necessary for M specificity, respectively.
(C) to (E) Computational prediction identified a nine-nucleotide motif present in all 10 UTRs (C) with three completely conserved nucleotides (D). This motif (blue font) is in close proximity to the 13-bp region identified as necessary by deletion analysis (red font) (E). Asterisks denote statistical significance compared with the control (P < 0.005, two-tailed Student's t test), and error bars denote one se.
Computational motif prediction using 10 UTR sequences from both Arabidopsis and G. gynandra identified a nine-nucleotide motif present in all 10 UTRs (Figure 2C). Within this motif, three nucleotides were conserved in all ten UTRs, while a further two were conserved between at least seven sequences (Figures 2C and 2D). This nine-nucleotide motif overlaps with the 5′ deletions that abolished the M-preferential expression of GUS, suggesting that this motif and nucleotides directly upstream may be critical for directing accumulation of Gg-CA4 in M cells (Figure 2E).
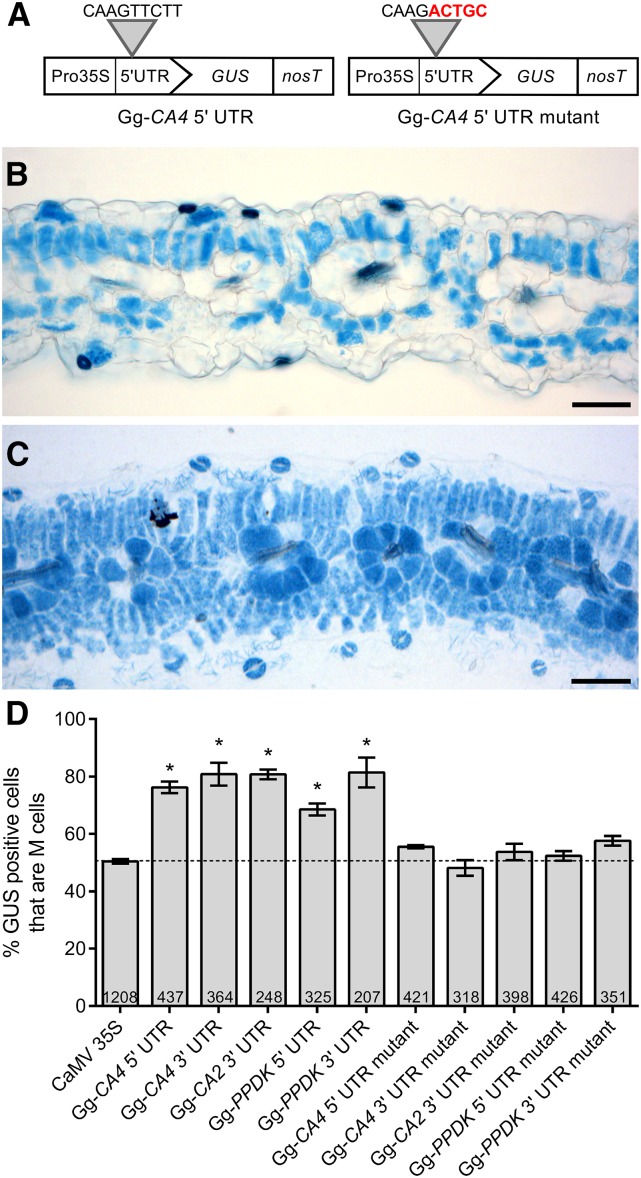
To verify that this nine-nucleotide cis-element is required for preferential GUS accumulation in M cells, site-directed mutagenesis was used to alter the last five nucleotides of the motif, which contained the three completely conserved nucleotides (Figure 3A). Stable transformants of G. gynandra confirmed that whereas a 99-nucleotide truncation of Gg-CA4 5′ UTR was sufficient to generate M specificity (Figure 3B), the mutated UTR sequence directed GUS expression in both M and BS cells (Figure 3C). This was observed across three independent transgenic lines for each construct (Supplemental Figure 3). Substitution of the same five nucleotides within 5′ and 3′ UTRs of other transcripts was also sufficient to abolish M specificity in all cases (Figure 3D). This is the second cis-element directing M specificity in C4 leaves to be identified after MEM1 (Mesophyll Enhancing Module 1), which is present in the ppcA promoter of C4 Flaveria species (Gowik et al., 2004; Akyildiz et al., 2007). We therefore named this nine-nucleotide motif present in Gg-CA4 as MEM2.
Figure 3.
A Five-Nucleotide Sequence Is Necessary for M-Specific Accumulation of Multiple C4 Transcripts.
Site-directed mutagenesis was used to alter five nucleotides (red font) within the Gg-CA4 5′ UTR (A). Gray arrows denote the position of the mutated nucleotides. Histological GUS staining of transverse leaf sections from stable transgenic G. gynandra lines showed that the 99-bp Gg-CA4 5′ UTR generated M-specific accumulation of GUS (B). When the five nucleotides predicted to be important were mutated, GUS was present in M and BS cells (C). Transverse sections in (B) and (C) are representative images from three independent transgenic lines each. The same five-nucleotide motif was mutated in additional 5′ and 3′ UTRs. The number of M cells expressing GUS after microprojectile bombardment of each mutant construct is expressed as a percentage of all GUS-positive cells observed (D). Numbers within histogram bars represent the number of independently transformed cells for each construct. Asterisks denote statistical significance compared with the control (P < 0.005, two-tailed Student's t test), and error bars denote one se. Bars = 100 µm.
MEM2 Does Not Impact Abundance of Transcripts in M or BS Cells
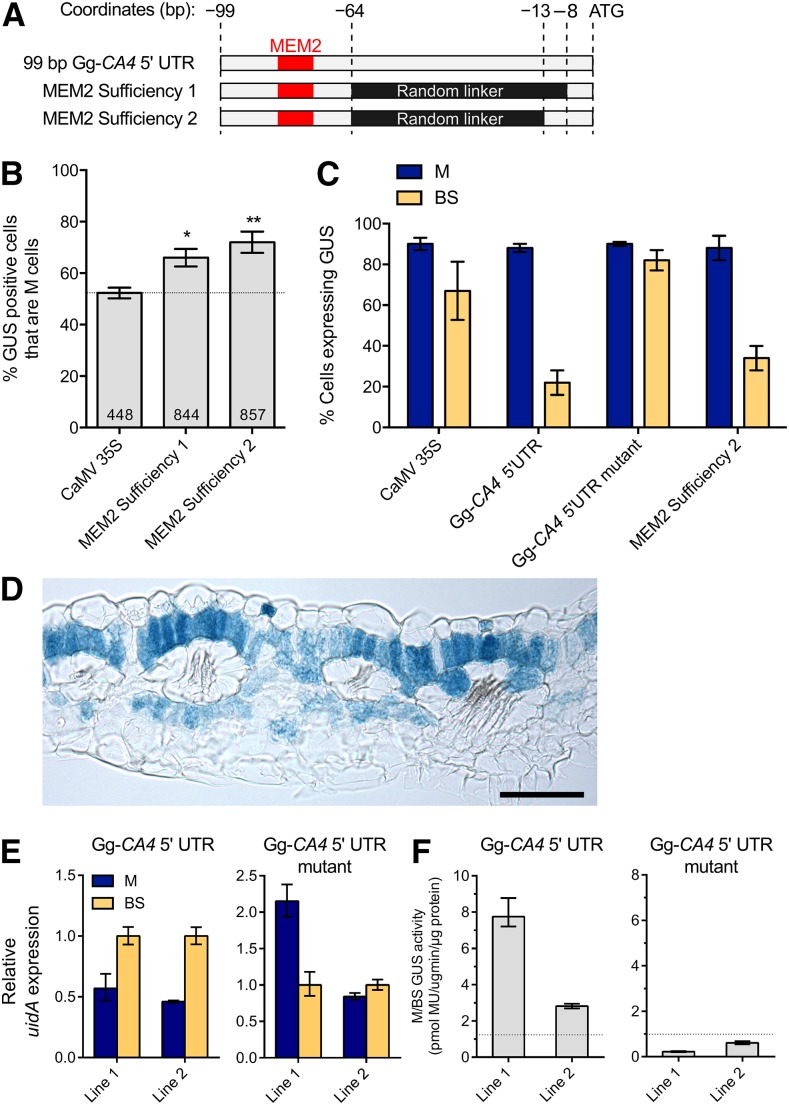
As MEM2 is a common element in multiple UTRs that direct M-preferential expression, we sought to define the minimal sequence sufficient for preferential accumulation of GUS in M cells. We observed that deletions from the 5′ or the 3′ ends of the minimal Gg-CA4 5′ UTR both disrupted M specificity. We therefore hypothesized that the 5′ UTR region directly upstream of the start codon is also important for MEM2 function. In eukaryotic genes, this region is important for recognition of the translation start codon AUG by the ribosome and translation initiation complex (Jackson et al., 2010). To test the importance of this sequence in the generation of M specificity by the Gg-CA4 5′ UTR, we generated two chimeric constructs, fusing a short 35-bp sequence around MEM2 to 8- or 13-bp sequences from immediately upstream of the translational start codon. To reflect the distance between these sequences in the endogenous UTR, randomly generated linker sequences of either 57 or 52 bp in length were inserted between the two (Figure 4A). Microprojectile bombardment suggested that both constructs were sufficient to direct M-preferential expression, but the effect was stronger when the larger (13 bp) sequence upstream of the translational start site was included (Figure 4B). Further evidence supporting these results was obtained by generating two independent G. gynandra stable lines expressing this chimeric construct. To assess the extent to which this construct generated M specificity, the percentage of M and BS cells expressing GUS was calculated by examining 22 transverse sections of GUS-stained leaves from these stable lines. Although there was some variability between sections, on average MEM2 combined with 13 nucleotides upstream of the start codon led to a reduction in GUS accumulation in BS cells, similar to the unmodified Gg-CA4 5′ UTR alone (Figure 4C). A representative example of a transverse section is shown in Figure 4D, and further examples of both transgenic lines are shown in Supplemental Figure 3. We therefore conclude that MEM2 is sufficient to direct strong preferential expression in M cells when combined with a second element, the sequence immediately 5′ of the start codon from the same UTR.
Figure 4.
MEM2 Confers Equal Transcript Abundance in M and BS Cells.
(A) and (B) Two chimeric constructs were synthesized by placing the nucleotides preceding the start codon of Gg-CA4 downstream of MEM2 (red; [A]) and tested using microprojectile bombardment (B). Numbers within histogram bars represent the number of independently transformed cells for each construct. Results from microprojectile bombardment were validated by generating three stable transgenic lines. Error bars represent one se. *P < 0.01, **P < 0.001, two-tailed Student's t test.
(C) The percentage of GUS positive M and BS cells was quantified in stable lines expressing the Gg-CA4 5′ UTR (three lines), Gg-CA4 5′ UTR mutant (three lines), MEM2 Sufficiency 2 (two lines), and the CaMV35S promoter (four lines) fused to uidA. Error bars represent one se.
(D) A representative transverse section expressing MEM2 Sufficiency 2. Bar = 100 µm.
(E) RT-qPCR quantification of uidA transcripts from M and BS cells expressing uidA under the control of the endogenous and mutated Gg-CA4 5′ UTR. Error bars represent one se. Three technical replicates were performed for each line and cell type.
(F) The activity of GUS measured by the rate of MU synthesis in the same cell fractions, expressed as fold enrichment in M cells versus BS cells. Error bars represent one se. Three technical replicates were performed.
Given the importance of nucleotides 5′ of the start codon in the initiation of translation (Jackson et al., 2010), we sought to test the hypothesis that MEM2 acts posttranscriptionally. To investigate this, we examined the abundance of Gg-CA4 transcripts in M and BS cells from cell-specific transcriptome data sets (Aubry et al., 2014) (Supplemental Table 2). Gg-CA4 transcripts were highly and similarly abundant in both M and BS cells (log2 fold change M:BS = −0.05, P value = 0.73, Fisher's exact test). We next used the stable transformants containing uidA fused to the endogenous and mutated versions of the Gg-CA4 5′ UTR to investigate whether MEM2 affected the abundance of uidA transcripts and GUS protein in each cell type. M and BS preparations were generated from these lines and both RNA and protein isolated. RT-qPCR on two transcripts known to be enriched in either cell type (PHOSPHOENOLPYRUVATE CARBOXYLASE [PPC], enriched in M cells, and NAD-DEPENDENT MALIC ENZYME2 [NADME2], enriched in BS cells) indicated that there was clear enrichment of the desired cell type in both M and BS fractions from leaves expressing the Gg-CA4 5′ UTR fused to GUS (Supplemental Figure 4). In our BS preparations from leaves expressing the mutated Gg-CA4 5′ UTR, the PPC marker for M cells was clearly depleted, suggesting that the BS preparation had minimal contamination from M cells. However, we did detect the BS marker NADME in the M preparations from these leaves, suggesting that enrichment of M cells was less successful in these lines (Supplemental Figure 4). We note that this does not affect the conclusions drawn about transcript or protein abundance in M or BS cells from lines expressing the unmutated Gg-CA4 5′ UTR.
We found that uidA transcripts were not enriched in the M preparation and tended in fact to be more abundant the BS when driven by the Gg-CA4 5′ UTR (Figure 4E). This implies that the Gg-CA4 5′ UTR does not generate M specificity by reducing the abundance of transcripts in BS cells. In lines expressing the MEM2-mutated UTR, uidA expression in M preparations was more variable between lines, but transcripts were still abundant in both M and BS preparations. This variability may be due to differential integrity of RNA between samples. Together, these data suggest that MEM2 does not generate M-preferential expression by altering the abundance of transcripts in a cell-type-specific manner. As GUS transcripts were not enriched in our isolated M preparation, it is likely that MEM2 functions posttranscriptionally to enrich the quantity of GUS protein translated within M cells compared with BS cells. To confirm this, the activity of GUS in the same cell-type fractions used for RT-qPCR was determined using the quantitative 4-methylumbulliferone-glucuronide (MUG) assay. GUS activity was highly enriched in M preparations compared with BS preparations when driven by the endogenous Gg-CA4 5′ UTR (Figure 4F). Conversely, when driven by the mutated UTR sequence, the activity of GUS was much reduced in M cells and marginally higher in BS preparations. These quantitative assays are consistent with our qualitative observation of GUS accumulation in transverse sections (Figures 3B and 3C) and suggest that when MEM2 is mutated, transcripts in both M and BS cells are translated to a similar extent, so that GUS is present in both cell types. However, when the MEM2 sequence is intact, the abundant transcripts in BS cells are translated to a much lesser extent than in M cells, leading to M-specific accumulation of GUS. We observed that mutation of MEM2 does not affect the rate of translation in vitro (Supplemental Figure 5) or the overall activity of GUS in Arabidopsis leaves (Supplemental Table 3). We therefore propose that regulating translation is not a general function of the MEM2 motif in all cells, but specific to its role in generating M specificity in the C4 G. gynandra leaf.
DISCUSSION
Evolutionary Events Underlying the Recruitment of CA4 into C4 Photosynthesis
The evolution of C4 photosynthesis from the C3 ancestral state requires mechanisms generating increased abundance as well as cell specificity of a large number of enzymes (Marshall et al., 1997; Patel et al., 2004; Gowik et al., 2004; Wiludda et al., 2012). A recent study has suggested that the evolution of increased abundance can be complex, with multiple sequences within promoters and 5′ UTRs contributing to the expression of C4 genes (Wiludda et al., 2012). We examined mechanisms underlying the evolution of CA4, an enzyme catalyzing the first step of the C4 pathway. Our data support the finding that the evolution of increased expression and cell specificity of C4 genes is complex, and in the case of CA4 sequences within promoters, 5′ and 3′ UTRs and introns all contribute to the evolution of increased expression of Gg-CA4 relative to At-CA4. Similar to the PHOSPHOENOLPYRUVATE CARBOXYLASE A and GLYCINE DECARBOXYLASE P-SUBUNIT A (GLDPA) genes of Flaveria bidentis (Gowik et al., 2004; Wiludda et al., 2012), we demonstrate that increased expression of Gg-CA4 evolved via changes to cis-elements upstream of the transcriptional start site. However, we extend this analysis to show that sequences within introns and interactions between 5′ and 3′ UTRs can also generate increased CA4 abundance. In maize, representing an independent C4 lineage, introns direct increased expression of genes encoding NADP-dependent malic enzyme and aspartate aminotransferase (Nomura et al., 2005a, 2005b). Together, our data and these previous studies suggest that multiple alterations in cis to noncoding regions are a primary route through which the increased abundance of C4 pathway enzymes evolved in multiple C4 lineages. We also found that the UTRs of Gg-CA4 confer an increase in translational efficiency in vitro, as well as an increase in GUS accumulation in vivo, but that these effects are independent of the MEM2 element that restricts expression to M cells. UTRs of NADP-ME1 from F. bidentis (Ali and Taylor, 2001) and RbcS from Amaranthus hypochondriacus (Patel et al., 2004) are also sufficient to generate increased translational efficiency and so these combined data indicate that this appears to be an important mechanism that has been recruited repeatedly to regulate expression of C4 genes. As we found that UTRs of the C3 homolog At-CA4 were sufficient to increase accumulation of GUS, this likely represents an ancestral character present in C3 plants.
Together, these findings allow us to hypothesize evolutionary events underlying the recruitment of CA4 to C4 photosynthesis (Supplemental Figure 6). In Arabidopsis, which is assumed to represent the ancestral C3 state, the promoter of CA4 directs strong expression; however, in C4 G. gynandra, the promoter and introns increase CA4 expression further (Supplemental Figure 6). As both the promoter and combined introns of Gg-CA4 are half as long as the homologous sequences from At-CA4, we suggest that the loss of cis-elements that reduce expression is a likely evolutionary scenario for this mechanism. Our data suggest that in leaves of C4, G. gynandra M-preferential accumulation of CA4 is generated by posttranscriptional regulation mediated by the both the 5′ and 3′ UTR of CA4 transcripts. This is consistent with the fact that CA4 transcripts are equally abundant between M and BS cells of G. gynandra leaves (Supplemental Table 2) and that the Gg-CA4 5′ UTR directs equal transcript abundance in M and BS cells (Figure 4E). Combining MEM2 with the nucleotides preceding the start codon from Gg-CA4 is sufficient to confer accumulation of GUS in M cells. In eukaryotes, these nucleotides play an important role in recognizing the proper translation start site by the translation initiation complex (Jackson et al., 2010). Interestingly, both 5′ and 3′ UTRs are bound in this complex at the initiation of translation (Tarun and Sachs, 1995; Wilkie et al., 2003; Jackson et al., 2010). One possibility is that M specificity evolved via the recruitment of trans-acting factors recognizing MEM2 within 5′ and 3′ UTRs at this complex within M or BS cells (Supplemental Figure 6). However, although we have defined the 5′ UTR sequences sufficient to direct M-preferential accumulation (Figures 4A to 4D), we have not yet defined if adding the MEM2 element to a heterologous 3′ UTR is sufficient to confer M-specific translation in the same way. This future experiment will be an important step in validating the roles and interactions between 5′ and 3′ UTRs containing MEM2 in generating M specificity. In addition, it is also currently unclear whether MEM2 functions to promote translation in M cells or repress translation in BS cells. Understanding this will be key in providing insight into the trans-acting factors that recognize MEM2.
Our data suggest that the evolution of increased abundance and M-preferential expression of Gg-CA4 evolved independently of each other and via distinct molecular mechanisms (Supplemental Figure 6). Mutation of MEM2 is sufficient to abolish M-preferential expression but has no effect on the levels of GUS activity conferred in cis in a C3 background (Supplemental Table 3). Conversely, increased abundance is conferred in cis by the Gg-CA4 promoter region (Figure 1C). This promoter sequence is unlikely to be important in generating M specificity, as Gg-CA4 transcripts are equally abundant between M and BS cells (Supplemental Table 2). Computational modeling of the convergent evolutionary events generating C4 photosynthesis in a wide variety of lineages predicts that increased abundance and cell specificity also evolved independently for at least five additional C4 pathway enzymes (Williams et al., 2013). It therefore appears that the decoupling of changes to enzyme abundance and cell type localization may be a general trend in C4 evolution, occurring in multiple lineages and in the evolution of both enzymes localized to either BS or M cells.
MEM2: A Motif Coordinating Cell-Type-Specific Accumulation of Multiple Enzymes
Deletion analysis and computational detection identified MEM2, a nine-nucleotide element present within the 5′ and 3′ UTRs of multiple Arabidopsis and G. gynandra transcripts. Several studies from yeast, animals, and plants have identified functionally related groups of mRNAs whose regulation is coordinated by posttranscriptional processes (Keene, 2007; Zhu et al., 2007; Staiger and Köster, 2011). Furthermore, some regulons have also been shown to share small sequence-specific or structural elements that confer similar rates of processing or decay (Chan et al., 2005; Goodarzi et al., 2012). As hundreds of transcripts can be coordinated by small elements within transcripts (Goodarzi et al., 2012), CA2, CA4, and PPDK could belong to a much larger suite of transcripts whose regulation is coordinated by MEM2 in C4 leaves.
MEM2 also represents a rare example of an element that performs the same function in either the 5′ or the 3′ UTR of transcripts. Other elements present in either 5′ or 3′ UTR are known, such as the iron-responsive elements present in animals and single-celled eukaryotes (Leipuviene and Theil, 2007). In contrast to MEM2, however, iron-responsive elements perform different functions when present in the 5′ or 3′ UTRs (Muckenthaler et al., 2008). The adaptive significance of possessing multiple copies of MEM2 within transcripts is not yet clear, but both UTRs of PPDK and CA4 are known to confer stronger M-preferential expression when combined compared with the 5′ or 3′ alone (Kajala et al., 2012). It is therefore possible that UTRs containing MEM2 function additively.
Distinct C4 Pathway Enzymes Share a Common Regulator
Since the discovery of the C4 pathway (Hatch and Slack, 1966), only one mechanism that regulates the expression of multiple genes in specific cells has been reported (Brown et al., 2011), and in that case, the specific cis-element was not identified. In addition, although Brown et al. (2011) reported a shared mechanism regulating the expression of multiple genes, they all encoded subunits of malic enzymes. MEM2 represents a cis-element that coordinates the regulation of transcripts encoding distinct C4 pathway enzymes. Interestingly, three M-specific and two BS-specific genes in three C4 monocot species may also share similar cell-type-enriched histone modifications (Heimann et al., 2013). These modifications include trimethylation of H3K4 residues as well as histone acetylation, both of which correlate with active transcription. This suggests transcriptional control at least in part contributes to the cell-type-specific accumulation of multiple enzymes in monocotyledons.
The complexity of C4 metabolism currently represents a considerable challenge for attempts to engineer C4 photosynthesis into C3 crop species (Hibberd et al., 2008). The discovery of MEM2 is therefore a key finding as it identifies a mechanism that coordinates the cell-specific synthesis of multiple C4 enzymes. Defining the exact method by which MEM2 is recognized in C4 leaves is an important next step. It is possible that a microRNA (miRNA) is the trans-acting factor that recognizes MEM2, as many miRNAs act at the ribosome to repress translation (Li et al., 2013). However, we were unable to find any known miRNAs that matched MEM2 or surrounding sequences within UTRs of CA2, CA4, or PPDK in Arabidopsis or G. gynandra. It is also possible that an RNA binding protein may recognize MEM2 in G. gynandra bundle sheath cells. PUF proteins recognize cis-elements present in mRNA that are typically within 3′ UTRs, of a similar size to MEM2, and are dependent on secondary structure (Tadauchi et al., 2001; Francischini and Quaggio, 2009; Li et al., 2010b; Filipovska et al., 2011). However, PUF proteins typically recognize a sequence beginning with 5′ UGUR (whereby R represents A or G) (Lu et al., 2009), which is not part of the conserved MEM2 sequence or directly upstream in any of the MEM2-containing UTRs. A PUF recognition domain that does not require a target sequence containing UGUR at the 5′ end has been engineered (Campbell et al., 2014), suggesting that the evolution of alternative recognition sites is at least possible, if unlikely.
Another large family of RNA binding proteins in Arabidopsis is the PENTATRICOPEPTIDE REPEAT (PPR) proteins, which appear not to be restricted to any consensus in target sequence, but are mostly active in chloroplasts and mitochondria (Barkan and Small, 2014). The MEM2 consensus is similar to that bound by RNA editing PPR proteins ORGANELLE TRANSCRIPT PROCESSING84 (Hammani et al., 2009) and MITOCHONDRIAL EDITING FACTOR14 (Verbitskiy et al., 2011; Barkan et al., 2012), and so it is possible that MEM2 is recognized by a related protein, which lost organellar targeting. However, as fewer than 1% of Arabidopsis PPR proteins are known to localize outside of the nucleus (Colcombet et al., 2013), this seems unlikely. The MEM2 sequence itself therefore provides few clues into the likely trans-acting factors that recognize MEM2 in BS cells. Identifying the trans-acting factor recognizing MEM2 is therefore an important aim for future research. MEM2 is the first cis-element identified that is necessary to direct cell-type-specific accumulation of multiple C4 enzymes. Our study suggests that recruitment of this preexisting cis-element parsimoniously explains how multiple C4 enzymes evolved the same localization. Its discovery therefore represents an important breakthrough toward understanding how a complex phenotype such as C4 photosynthesis can evolve over relatively short evolutionary timescales.
METHODS
Plant Growth
Sterilized Arabidopsis thaliana ecotype Col-0 seeds were spread on plates containing 0.5× Murashige and Skoog (MS) basal salts and 1% (w/v) agar, pH 5.8, then stratified for 48 h at 4°C in the absence of light. Following stratification, seeds were transferred to a long-day growth room at 22°C, relative humidity of 65%, ambient CO2 concentration, and a light intensity of 200 μmol m−2 s−1 PFD for 7 d. Seedlings were then transferred to a 1:1 mixture of Levington’s M3 potting compost:fine vermiculite. Gynandropsis gynandra seeds were germinated in the dark at 30°C for 30 h on a bed of wet filter papers. Seeds were then transferred to a medium containing 1× MS, 1% (w/v) sucrose, and 1% (w/v) agar. Seedlings were then grown in 16 h light/8 h dark, at 20°C, ambient CO2 concentration, and a light intensity of 200 μmol m−2 s−1 PFD.
Vector Construction and Stable Plant Transformation
cDNA sequences were obtained using 5′ and 3′ RACE, coding regions were amplified from both cDNA and genomic DNA, and promoter regions were isolated by genome walking as described previously (Kajala et al., 2012). UTR reporter constructs containing the uidA gene (encoding GUS) were generated by ligation of UTRs into a modified vector containing a gfp:uidA:nosT cassette (Brown et al., 2011). The 3′ UTRs were inserted between uidA and nosT. The 5′ UTRs were fused to the CaMV35S promoter by PCR and inserted in front of the cassette. Vectors containing promoter and coding regions were assembled into the same cassette using Gibson assembly (Gibson et al., 2009). Two constructs were synthesized using the coding regions of Gg-CA4 and At-CA4. First, the genomic coding sequence containing all exons and introns intact were fused to the open reading frame of uidA, to test regulation conferred by exons and introns combined. Second, the spliced open reading frame of Gg-CA4 and At-CA4 was amplified from cDNA and fused to uidA to test regulation conferred in the absence of introns.
Assembled constructs were used for microprojectile bombardment and also placed in binary vectors to generate stable Arabidopsis or G. gynandra transformants. Site-directed mutagenesis was performed using the Quickchange method. Chimeric constructs were generated by site-directed insertion or fusion of sequences by overlapping PCR. UTRs and mutated UTRs fused to uidA were ligated into the pTNT vector (Promega) for in vitro transcription. Transgenic Arabidopsis Col-0 plants were generated by floral dipping (Clough and Bent, 1998) using Agrobacterium tumefaciens strain GV3101. Primary transformants were identified by selection on 50 μg mL−1 kanamycin for 7 d. Stable transformation of G. gynandra was performed as previously described (Newell et al., 2010), with modifications to the cocultivation medium (MS, 3% [w/v] sucrose, 1 mg/L thidiazuron, 0.2 mg/L indole-3-acetic acid, and 100 μM acetosyringone), regeneration medium (MS, 3% [w/v] sucrose, 1 mg/L thidiazuron, and 0.2 mg/L indole-3-acetic acid), and shoot regeneration medium (MS, 3% [w/v] sucrose, 1 mg/L BAP, and 0.2 mg/L indole-3-acetic acid).
Microprojectile Bombardment of G. gynandra
Transient expression of the constructs in G. gynandra was achieved by microprojectile bombardment using a Bio-Rad PDS-1000/He particle delivery system as described by Brown et al. (2011). G. gynandra seeds were incubated at 30°C for 30 h on moistened filter paper to achieve uniform germination. G. gynandra seeds were transferred to 0.5× MS medium supplemented with 1% (w/v) sucrose and 1% (w/v) agar in sterile conditions. Seedlings were then grown in a growth room with 16-h days, at 22°C, relative humidity of 65%, ambient CO2 concentration, and a light intensity of 200 μmol m−2 s−1 PFD. After 14 d of growth, seedlings were prepared for microprojectile bombardment by removing all root tissue below the medium surface, leaving only the stems, cotyledons, and two-expanding leaves, with the primary leaflet measuring ∼110 mm from base to tip and 75 to 80 mm across at the widest point (Supplemental Figure 1A). Leaf size was found to be an important factor determining the ratio of mesophyll and bundle transformed using the ubiquitously expressed CaMV35S control.
To transform individual cells within G. gynandra leaves, 350 ng M-17 tungsten particles (1.1-μm diameter; Bio-Rad) were washed with 100% ethanol and resuspended in ultrapure water. Then, 1.5 μg of plasmid DNA was adhered to the tungsten particles as described by Patel et al. (2006), adding DNA to the tungsten particles while vortexing at a slow speed by a Vortex-Genie 2 (Scientific Industries). After addition of the DNA, 50 μL 2.5 M calcium chloride (Fisher Scientific) and 10 μL 100 mM spermidine (Sigma-Aldrich) were added to the particle suspension to facilitate binding of the DNA to the tungsten particles. The tungsten-DNA suspension was incubated for 10 min on ice, with frequent agitation to prevent pelleting of the tungsten particles. The particles were then pelleted by centrifugation at 6000 rpm for 2 s and washed once and resuspended in 100 μL 100% ethanol. Immediately prior to the bombardment of G. gynandra, 10 μL aliquots of tungsten/DNA were transferred to plastic macrocarriers (Bio-Rad) and the ethanol allowed to evaporate. Three macrocarriers were used for each transformation. After microprojectile bombardment (Bio-Rad PDS-1000/He particle delivery system), seedlings were placed upright in a sealed Petri dish, with the base of their stems immersed in 0.5× MS medium. The abaxial leaf surface was placed on Whatman grade 1 filter paper moistened with 0.5× MS to prevent leaves from drying. Seedlings were left for 40 h after bombardment prior to staining for GUS activity.
GUS Assays and in Vitro Translation Assay
Staining for GUS activity was performed on Arabidopsis or G. gynandra plants as described (Jefferson et al., 1987). Tissue was fixed in 3:1 ethanol/acetic acid for 30 min at room temperature, and chlorophyll was cleared using 70% (v/v) ethanol at 37°C for 24 h and then 5% (w/v) NaOH at 37°C for 2 h. G. gynandra seedlings subjected to microprojectile bombardment were incubated in X-GlcA solution for 24 h. Arabidopsis plants transformed with constructs containing the CaMV35S promoter were incubated in X-GlcA solution for 18 h. At least six independent T1 stable transgenic lines were stained for each construct. Plants derived from independent T1 lines transformed with constructs containing the At-CA4 or Gg-CA4 promoters were incubated for 6 h at 37°C. Tissue was then fixed in 3:1 ethanol/acetic acid for 30 min at room temperature, and chlorophyll was cleared using 70% (v/v) ethanol at 37°C for 24 h and then 5% (w/v) NaOH at 37°C for 2 h. Transverse leaf sections were obtained from at least three independent T1 transgenic G. gynandra lines by embedding fresh leaf tissue in 5% (w/v) agarose and isolating 60-µm sections using a vibratome. Sections were stained for GUS activity for 1 and 2 h.
The activity of GUS was also quantified by measuring the rate of MUG conversion to 4-methylumbulliferone (MU) as described (Jefferson et al., 1987). Soluble protein was extracted from transgenic Arabidopsis plants by freezing in liquid nitrogen and maceration, followed by addition of an extraction buffer (50 mM NaH2PO4, 0.007% [v/v] β-mercaptoethanol, and 0.01% [v/v] Triton X-100). Extracts underwent centrifugation for 5 min at 4°C. Diluted protein extracts were incubated with 1 mM MUG at 37°C for 5, 10, 20, and 30 min in a 96-well plate. GUS activity was terminated at the end of each time point by the addition of 200 mM Na2CO3, and MU fluorescence was measured by exciting at 365 nm and measuring emission at 455 nm, averaging five light pulses for each well. The concentration of MU/unit fluorescence in each sample was interpolated using a concentration gradient of MU from 1.5 to 800 μM MU. MU/unit fluorescence was found to be linear over this range. Pilot experiments were performed with two to three independent T1 transgenic lines expressing each transgenic construct to determine the optimal protein dilution required to retain linearity. Subsequent assays were performed using between 10 and 20 independent T1 lines.
In vitro transcription was initiated from the T7 promoter using a MEGAShortScript T7 Kit (Ambion), with the addition of 1 unit of RNase OUT (Invitrogen). Reactions were incubated for 2 h at 37°C and transcripts were purified using MEGAClear, and the integrity of transcribed products was verified using a Bioanalyzer 2100 (Agilent Technologies). Quantitative in vitro translation was performed using a Fluorotect GreenLys in vitro translation labeling system (Promega) in conjunction with a wheat germ extract (Promega). Time-course and concentration gradient experiments were performed to ensure fluorescent protein production was linear and unsaturated relative to incubation time and RNA concentration, respectively. After these optimization steps, reactions were incubated for 45 min at 25°C, with 200 nM RNA templates, using equimolar concentrations of each transcript.
RNA-Seq Analysis and qPCR Analysis of GUS Accumulation in G. gynandra Transgenic Lines
RNA-seq data from G. gynandra bundle sheath and mesophyll fractions extracted by laser capture microdissection (SRA066236; Aubry et al., 2014) were quality trimmed and the adapters removed using Trimmomatic v0.30 (Bolger et al., 2014). Reads shorter than 60 bp after trimming were discarded. The remaining reads were aligned against the CA4 gene using TopHat v2 (Kim et al., 2013). Gene and transcript expression was quantified using RSEM v1.2.7 (Li and Dewey, 2011).
Isolation of M extracts from G. gynandra was performed as described previously (Covshoff et al., 2013) and BS cells according to Markelz et al. (2003). For each replicate, between six and eight leaves representing 350 to 400 mg of tissue were initially rolled to extract M sap and then blended to separate BS strands. To quantify GUS activity, sap containing M cells was transferred directly into extraction buffer consisting of 100 mM NaH2PO4 (Sigma-Aldrich), 0.007% (v/v) β-mercaptoethanol (Fisher Scientific), and 0.01% (v/v) Triton X-100 (Sigma-Aldrich). Ground bundle sheath powder was resuspended in extraction buffer and samples were spun down at 13,000g for 5 min and the pellet discarded. Protein concentration was determined using the Qubit Protein Assay Kit (ThermoFisher Scientific), and the MUG assay was performed immediately as described in the previous section. To quantify transcript abundance in M and BS preparations, samples were extracted using the mirVana miRNA isolation kit (Ambion), stopping the protocol after the extraction of total RNA (prior to the optional purification of small RNAs). Total RNA was eluted in RNase-free water and samples treated with DNase-I (Promega). Although leaf rolling gives RNA of good quality (Covshoff et al., 2013; John et al., 2014), due to the small quantities of RNA obtained, we note that variability in the integrity of some transcripts may exist between samples. qPCR was performed as described by Aubry et al. (2014) with the exception that data are presented as an average of four replicates. Primer sequences were as follows: GUS (forward, 5′-ACCTCGCATTACCCTTACGCTGAA-3′, reverse, 5′-GCCGACAGCAGCAGTTTCATCAAT-3′), Actin7 (forward, 5′-TCCGACCCGATGTGATGTTATGGT-3′, reverse: 5′-CAATCACTTTCCGGCTGCAACCAA-3′).
cis-Element Prediction
cis-element detection was performed using the Multiple Em for Motif Elucidation (MEME) suite v.4.8.1. (Bailey et al., 2009). Unaligned transcribed UTR sequences were used as the input with the following conditions imposed: (1) no limit to the minimum or maximum motif length, (2) a minimum of one motif present in each UTR in the data set, and (3) motif detection was not applied to the complementary strand. The extent of conservation of nucleotides within detected motifs was plotted using the MEME suite.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Gg-CA4, NCBI GenBank Accession KU517022; Gg-CA2, NCBI GenBank Accession KU517023; Gg-PPDK, NCBI GenBank Accession KU517024; At-CA4, AT1G70410; At-CA2, AT5G14740; and At-PPDK, AT4G15530.
Supplemental Data
Supplemental Figure 1. Transformation of G. gynandra M and BS cells.
Supplemental Figure 2. 3′ UTRs of Gg-CA2 and At-CA2 generate M specificity.
Supplemental Figure 3. Representative transverse sections from stable transgenic lines.
Supplemental Figure 4. Quantification of marker transcripts in isolated M and BS cells.
Supplemental Figure 5. Mutation of MEM2 does not affect translation in vitro.
Supplemental Figure 6. Hypothesis for mechanisms regulating the abundance and cell specificity of CA4.
Supplemental Table 1. Total number of transformed M and BS cells analyzed for each construct.
Supplemental Table 2. Gg-CA4 transcript abundance measured by transcriptome sequencing of M and BS cells.
Supplemental Table 3. GUS activity conferred by mutated Gg-CA4 UTRs Arabidopsis seedlings.
Supplementary Material
Acknowledgments
We thank Steve Kelly (Plant Sciences, Oxford) for advice. We also thank the BBSRC for funding.
AUTHOR CONTRIBUTIONS
B.P.W., S.J.B., and J.M.H. conceived the study and designed experiments. B.P.W., S.J.B., I.R.-L., J.K., S.A., and S.S. performed the experiments. B.P.W. and S.J.B. analyzed the data and produced figures. J.M.H. supervised the whole study and wrote the article together with B.P.W. and S.J.B.
Glossary
- M
mesophyll
- BS
bundle sheath
- CA
carbonic anhydrase
- UTR
untranslated region
- MUG
4-methylumbulliferone-glucuronide
- miRNA
microRNA
- MS
Murashige and Skoog
- MU
4-methylumbulliferone
Footnotes
Articles can be viewed online without a subscription.
References
- Akyildiz M., Gowik U., Engelmann S., Koczor M., Streubel M., Westhoff P. (2007). Evolution and function of a cis-regulatory module for mesophyll-specific gene expression in the C4 dicot Flaveria trinervia. Plant Cell 19: 3391–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S., Taylor W.C. (2001). Quantitative regulation of the Flaveria Me1 gene is controlled by the 3′-untranslated region and sequences near the amino terminus. Plant Mol. Biol. 46: 251–261. [DOI] [PubMed] [Google Scholar]
- Aubry S., Brown N.J., Hibberd J.M. (2011). The role of proteins in C3 plants prior to their recruitment into the C4 pathway. J. Exp. Bot. 62: 3049–3059. [DOI] [PubMed] [Google Scholar]
- Aubry S., Kelly S., Kümpers B.M.C., Smith-Unna R.D., Hibberd J.M. (2014). Deep evolutionary comparison of gene expression identifies parallel recruitment of trans-factors in two independent origins of C4 photosynthesis. PLoS Genet. 10: e1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37: W202– W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Rojas M., Fujii S., Yap A., Chong Y.S., Bond C.S., Small I. (2012). A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 8: e1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Small I. (2014). Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65: 415–442. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady S.M., Orlando D.A., Lee J.-Y., Wang J.Y., Koch J., Dinneny J.R., Mace D., Ohler U., Benfey P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806. [DOI] [PubMed] [Google Scholar]
- Bräutigam A., et al. (2011). An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiol. 155: 142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N.J., Newell C.A., Stanley S., Chen J.E., Perrin A.J., Kajala K., Hibberd J.M. (2011). Independent and parallel recruitment of preexisting mechanisms underlying C4 photosynthesis. Science 331: 1436–1439. [DOI] [PubMed] [Google Scholar]
- Burnell J.N., Hatch M.D. (1988). Low bundle sheath carbonic anhydrase is apparently essential for effective c(4) pathway operation. Plant Physiol. 86: 1252–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell Z.T., Valley C.T., Wickens M. (2014). A protein-RNA specificity code enables targeted activation of an endogenous human transcript. Nat. Struct. Mol. Biol. 21: 732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.S., Elemento O., Tavazoie S. (2005). Revealing posttranscriptional regulatory elements through network-level conservation. PLOS Comput. Biol. 1: e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.M., Liu W.Y., Shih A.C.C., Shen M.N., Lu C.H., Lu M.Y.J., Yang H.W., Wang T.Y., Chen S.C.C., Chen S.M., Li W.H., Ku M.S. (2012). Characterizing regulatory and functional differentiation between maize mesophyll and bundle sheath cells by transcriptomic analysis. Plant Physiol. 160: 165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Colcombet J., Lopez-Obando M., Heurtevin L., Bernard C., Martin K., Berthomé R., Lurin C. (2013). Systematic study of subcellular localization of Arabidopsis PPR proteins confirms a massive targeting to organelles. RNA Biol. 10: 1557–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covshoff S., Furbank R.T., Leegood R.C., Hibberd J.M. (2013). Leaf rolling allows quantification of mRNA abundance in mesophyll cells of sorghum. J. Exp. Bot. 64: 807–813. [DOI] [PubMed] [Google Scholar]
- Engelmann S., Wiludda C., Burscheidt J., Gowik U., Schlue U., Koczor M., Streubel M., Cossu R., Bauwe H., Westhoff P. (2008). The gene for the P-subunit of glycine decarboxylase from the C4 species Flaveria trinervia: analysis of transcriptional control in transgenic Flaveria bidentis (C4) and Arabidopsis (C3). Plant Physiol. 146: 1773–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipovska A., Razif M.F.M., Nygård K.K.A., Rackham O. (2011). A universal code for RNA recognition by PUF proteins. Nat. Chem. Biol. 7: 425–427. [DOI] [PubMed] [Google Scholar]
- Francischini C.W., Quaggio R.B. (2009). Molecular characterization of Arabidopsis thaliana PUF proteins--binding specificity and target candidates. FEBS J. 276: 5456–5470. [DOI] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A. III, Smith H.O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Goodarzi H., Najafabadi H.S., Oikonomou P., Greco T.M., Fish L., Salavati R., Cristea I.M., Tavazoie S. (2012). Systematic discovery of structural elements governing stability of mammalian messenger RNAs. Nature 485: 264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowik U., Bräutigam A., Weber K.L., Weber A.P.M., Westhoff P. (2011). Evolution of C4 photosynthesis in the genus Flaveria: how many and which genes does it take to make C4? Plant Cell 23: 2087–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowik U., Burscheidt J., Akyildiz M., Schlue U., Koczor M., Streubel M., Westhoff P. (2004). cis-Regulatory elements for mesophyll-specific gene expression in the C4 plant Flaveria trinervia, the promoter of the C4 phosphoenolpyruvate carboxylase gene. Plant Cell 16: 1077–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K., Okuda K., Tanz S.K., Chateigner-Boutin A.-L., Shikanai T., Small I. (2009). A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. Plant Cell 21: 3686–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch M.D., Slack C.R. (1966). Photosynthesis by sugar-cane leaves. A new carboxylation reaction and the pathway of sugar formation. Biochem. J. 101: 103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch M.D., Slack C.R. (1970). Photosynthetic CO2-fixation pathways. Annu. Rev. Plant Physiol. 21: 141–162. [Google Scholar]
- Heimann L., Horst I., Perduns R., Dreesen B., Offermann S., Peterhänsel C. (2013). A Common histone modification code on C4 genes in maize and its conservation in Sorghum and Setaria italica. Plant Physiol. 162: 456–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N.D., et al. (2009). Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd J.M., Covshoff S. (2010). The regulation of gene expression required for C4 photosynthesis. Annu. Rev. Plant Biol. 61: 181–207. [DOI] [PubMed] [Google Scholar]
- Hibberd J.M., Sheehy J.E., Langdale J.A. (2008). Using C4 photosynthesis to increase the yield of rice - rationale and feasibility. Curr. Opin. Plant Biol. 11: 228–231. [DOI] [PubMed] [Google Scholar]
- Jackson R.J., Hellen C.U., Pestova T.V. (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11: 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John C.R., Smith-Unna R.D., Woodfield H., Covshoff S., Hibberd J.M. (2014). Evolutionary convergence of cell-specific gene expression in independent lineages of C4 grasses. Plant Physiol. 165: 62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajala K., Brown N.J., Williams B.P., Borrill P., Taylor L.E., Hibberd J.M. (2012). Multiple Arabidopsis genes primed for recruitment into C4 photosynthesis. Plant J. 69: 47–56. [DOI] [PubMed] [Google Scholar]
- Keene J.D. (2007). RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 8: 533–543. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzburg S.L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M.S.B., Agarie S., Nomura M., Fukayama H., Tsuchida H., Ono K., Hirose S., Toki S., Miyao M., Matsuoka M. (1999). High-level expression of maize phosphoenolpyruvate carboxylase in transgenic rice plants. Nat. Biotechnol. 17: 76–80. [DOI] [PubMed] [Google Scholar]
- Leipuviene R., Theil E.C. (2007). The family of iron responsive RNA structures regulated by changes in cellular iron and oxygen. Cell. Mol. Life Sci. 64: 2945–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., et al. (2010a). The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 42: 1060–1067. [DOI] [PubMed] [Google Scholar]
- Li S., et al. (2013). MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153: 562–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Quon G., Lipshitz H.D., Morris Q. (2010b). Predicting in vivo binding sites of RNA-binding proteins using mRNA secondary structure. RNA 16: 1096–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Dolgner S.J., Hall T.M. (2009). Understanding and engineering RNA sequence specificity of PUF proteins. Curr. Opin. Struct. Biol. 19: 110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markelz N.H., Costich D.E., Brutnell T.P. (2003). Photomorphogenic responses in maize seedling development. Plant Physiol. 133: 1578–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall J.S., Stubbs J.D., Chitty J.A., Surin B., Taylor W.C. (1997). Expression of the C4 Me1 gene from Flaveria bidentis requires an interaction between 5′ and 3′ sequences. Plant Cell 9: 1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka M., Kyozuka J., Shimamoto K., Kano-Murakami Y. (1994). The promoters of two carboxylases in a C4 plant (maize) direct cell-specific, light-regulated expression in a C3 plant (rice). Plant J. 6: 311–319. [DOI] [PubMed] [Google Scholar]
- Muckenthaler M.U., Galy B., Hentze M.W. (2008). Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu. Rev. Nutr. 28: 197–213. [DOI] [PubMed] [Google Scholar]
- Newell C.A., Brown N.J., Liu Z., Pflug A., Gowik U., Westhoff P., Hibberd J.M. (2010). Agrobacterium tumefaciens-mediated transformation of Cleome gynandra L., a C(4) dicotyledon that is closely related to Arabidopsis thaliana. J. Exp. Bot. 61: 1311–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M., Higuchi T., Ishida Y., Ohta S., Komari T., Imaizumi N., Miyao-Tokutomi M., Matsuoka M., Tajima S. (2005a). Differential expression pattern of C4 bundle sheath expression genes in rice, a C3 plant. Plant Cell Physiol. 46: 754–761. [DOI] [PubMed] [Google Scholar]
- Nomura M., Higuchi T., Katayama K., Taniguchi M., Miyao-Tokutomi M., Matsuoka M., Tajima S. (2005b). The promoter for C4-type mitochondrial aspartate aminotransferase does not direct bundle sheath-specific expression in transgenic rice plants. Plant Cell Physiol. 46: 743–753. [DOI] [PubMed] [Google Scholar]
- Nomura M., et al. (2000). The evolution of C4 plants: acquisition of cis-regulatory sequences in the promoter of C4-type pyruvate, orthophosphate dikinase gene. Plant J. 22: 211–221. [DOI] [PubMed] [Google Scholar]
- Patel M., Corey A.C., Yin L.-P., Ali S., Taylor W.C., Berry J.O. (2004). Untranslated regions from C4 amaranth AhRbcS1 mRNAs confer translational enhancement and preferential bundle sheath cell expression in transgenic C4 Flaveria bidentis. Plant Physiol. 136: 3550–3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M., Siegel A.J., Berry J.O. (2006). Untranslated regions of FbRbcS1 mRNA mediate bundle sheath cell-specific gene expression in leaves of a C4 plant. J. Biol. Chem. 281: 25485–25491. [DOI] [PubMed] [Google Scholar]
- Sage R.F., Christin P.-A., Edwards E.J. (2011). The C(4) plant lineages of planet Earth. J. Exp. Bot. 62: 3155–3169. [DOI] [PubMed] [Google Scholar]
- Sheen J. (1999). C4 gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 187–217. [DOI] [PubMed] [Google Scholar]
- Staiger D., Köster T. (2011). Spotlight on post-transcriptional control in the circadian system. Cell. Mol. Life Sci. 68: 71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadauchi T., Matsumoto K., Herskowitz I., Irie K. (2001). Post-transcriptional regulation through the HO 3′-UTR by Mpt5, a yeast homolog of Pumilio and FBF. EMBO J. 20: 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanz S.K., Tetu S.G., Vella N.G.F., Ludwig M. (2009). Loss of the transit peptide and an increase in gene expression of an ancestral chloroplastic carbonic anhydrase were instrumental in the evolution of the cytosolic C4 carbonic anhydrase in Flaveria. Plant Physiol. 150: 1515–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun S.Z. Jr., Sachs A.B. (1995). A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 9: 2997–3007. [DOI] [PubMed] [Google Scholar]
- Verbitskiy D., Härtel B., Zehrmann A., Brennicke A., Takenaka M. (2011). The DYW-E-PPR protein MEF14 is required for RNA editing at site matR-1895 in mitochondria of Arabidopsis thaliana. FEBS Lett. 585: 700–704. [DOI] [PubMed] [Google Scholar]
- Wilkie G.S., Dickson K.S., Gray N.K. (2003). Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci. 28: 182–188. [DOI] [PubMed] [Google Scholar]
- Williams B.P., Aubry S., Hibberd J.M. (2012). Molecular evolution of genes recruited into C4 photosynthesis. Trends Plant Sci. 17: 213–220. [DOI] [PubMed] [Google Scholar]
- Williams B.P., Johnston I.G., Covshoff S., Hibberd J.M. (2013). Phenotypic landscape inference reveals multiple evolutionary paths to C4 photosynthesis. eLife 2: e00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiludda C., Schulze S., Gowik U., Engelmann S., Koczor M., Streubel M., Bauwe H., Westhoff P. (2012). Regulation of the photorespiratory GLDPA gene in C(4) flaveria: an intricate interplay of transcriptional and posttranscriptional processes. Plant Cell 24: 137–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Dong C.-H., Zhu J.-K. (2007). Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr. Opin. Plant Biol. 10: 290–295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.