Abstract
The platinum drugs, cisplatin, carboplatin, and oxaliplatin, prevail in the treatment of cancer,, but new platinum agents have been very slow to enter the clinic. Recently, however, there has been a surge of activity, based on a great deal of mechanistic information, aimed at developing non-classical platinum complexes that operate via mechanisms of action distinct from those of the approved drugs. The use of nanodelivery devices has also grown and many different strategies have been explored to incorporate platinum warheads into nanomedicine constructs. In this review, we discuss these efforts to create the next generation of platinum anticancer drugs. The introduction provides the reader with a brief overview of the use, development, and mechanism of action of the approved platinum drugs to provide the context in which more recent research has flourished. We then describe approaches that explore non-classical platinum(II) complexes with trans geometry and with a monofunctional coordination mode, polynuclear platinum(II) compounds, platinum(IV) prodrugs, dual-treat agents, and photoactivatable platinum(IV) complexes. Nanodelivery particles designed to deliver platinum(IV) complexes will also be discussed, including carbon nanotubes, carbon nanoparticles, gold nanoparticles, quantum dots, upconversion nanoparticles, and polymeric micelles. Additional nanoformulations including supramolecular self-assembled structures, proteins, peptides, metal-organic frameworks, and coordination polymers will then be described. Finally, the significant clinical progress made by nanoparticle formulations of platinum(II) agents will be reviewed. We anticipate that such a synthesis of disparate research efforts will not only help to generate new drug development ideas and strategies, but also reflect our optimism that the next generation of platinum cancer drugs is about to arrive.
1. Introduction
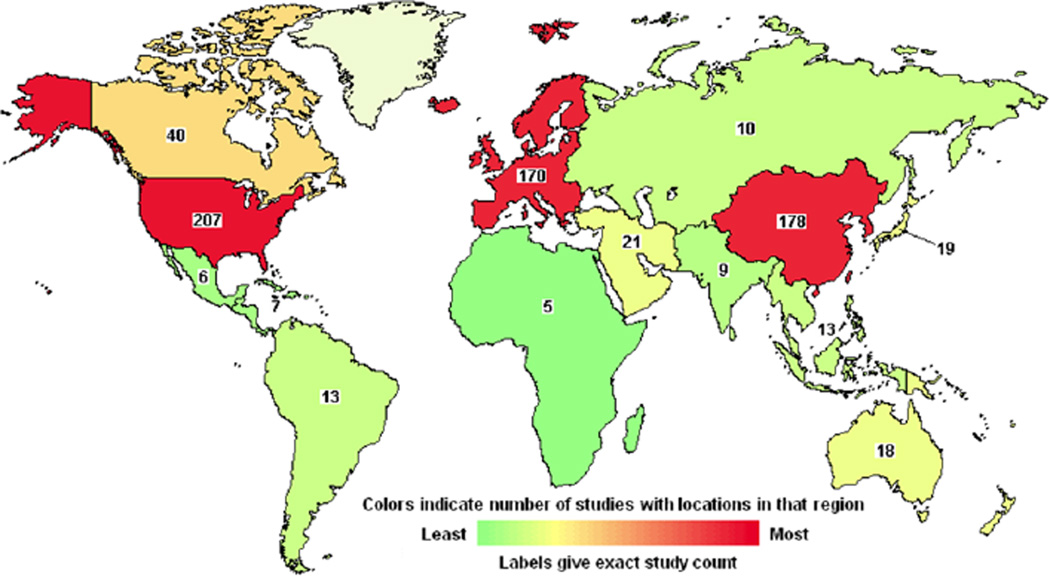
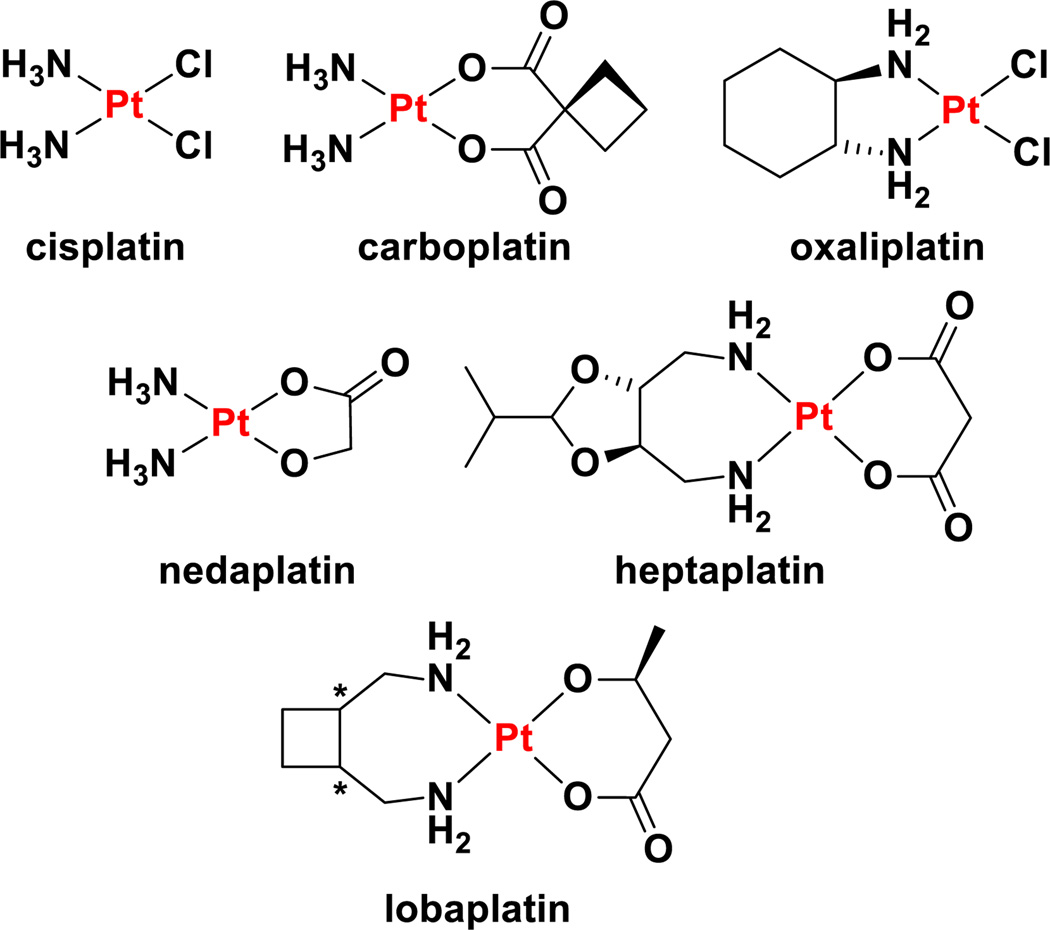
Platinum anticancer agents represent one of the great success stories in the field of medicinal inorganic chemistry. They highlight the confluence of serendipity and rational design in drug development. Three platinum-containing drugs are approved worldwide for treating cancer in humans, namely, cisplatin, carboplatin, and oxaliplatin (section 3.1). An additional three are approved for use in specific countries and they are nedaplatin, lobaplatin, and heptaplatin (section 3.1). Despite having been introduced to the market almost 40 years ago, platinum complexes remain among the most widely used anticancer chemotherapeutics. One important mark of the success of the platinum drugs is the fact that, since the introduction of cisplatin into the treatment regimen of testicular cancer patients, cure rates for this disease have exceeded 95%.1 The clinical relevance of these drugs is further underscored by the fact that carboplatin is listed as a complementary item on the World Health Organization’s Model List of Essential Medicines.2 Moreover, in the 2009 Ambulatory Care Drug Database maintained by the U.S. Center for Disease Control and Prevention, platinum complexes as a class were listed in the medical charts of American patients with a frequency surpassed only by five other anticancer drugs (methotrexate, raloxifene, medroxyprogesterone, tamoxifen, and leuprolide).3 The clinical trials database maintained by the U.S. National Institutes of Health, which lists over 186000 clinical trials in over 180 countries, cites cisplatin as a component in more active clinical trials than any other anticancer agent (Figure 1).4 Similar trends hold for the European Union Clinical Trial Register, which is maintained by the EMA and lists over 25000 trials with a European clinical trials database (EudraCT) protocol,5 as well as the International Clinical Trials Registry Platform of the WHO.6 Despite the widespread use of these drugs, a new platinum agent has not received worldwide approval in over a decade. Research activity into new platinum anticancer agents has remained intense, however,7 as this review will demonstrate.
Figure 1.
NIH-registered clinical trials involving cisplatin in various parts of the world as of 2015. The numbers reflect only those trials that are open and whose activity has been verified by the NIH within the past two years. Graphic generated using search tools from www.clinicaltrials.gov.
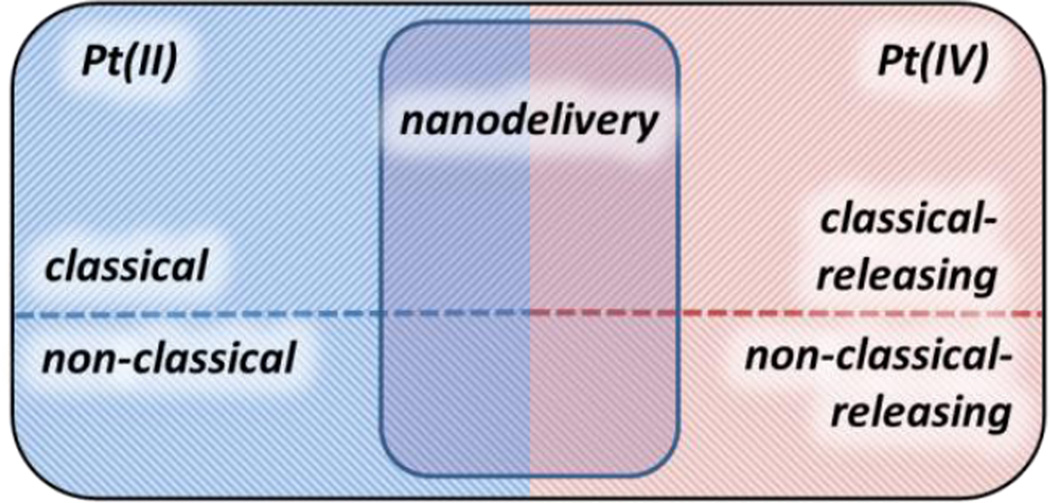
Our discussion begins with a brief description of the mechanism of action of the classical platinum drugs. For more comprehensive treatments, the reader is referred to several excellent reviews and monographs.7–12 Here we provide sufficient mechanistic background information for appreciating the discussions that follow. We then discuss platinum(II) complexes that lead to cancer cell death by the same mechanism as the three classical platinum drugs but which enjoy enhanced activity owing to molecular targeting. This section is followed by coverage of platinum(II) complexes that operate by different mechanisms. Because a very large number of complexes have been prepared that fall in these two categories, emphasis is given to those having validated targeting properties and/or well established mechanism. Coverage of platinum(IV) prodrugs that release cisplatin, carboplatin, oxaliplatin, or a close analogue upon reduction in the cell follows next. A subset of these complexes not only release an active platinum molecule, but also may also provide an additional bioactive substance that may function in a manner orthogonal to that of the platinum(II) agent, serving as “dual-threat” drug candidates. A small number of platinum(IV) complexes appear to act by mechanisms distinct from that of the prodrug family and they are covered next. An extensive treatment of the nanodelivery of platinum complexes is then provided, with a focus on two nanoparticulate formulations that have shown the greatest progress in clinical trials.13 The organization of this review thus reflects the structures and mechanisms of the compounds (Figure 2).
Figure 2.
Schematic summary of the topics discussed in this review.
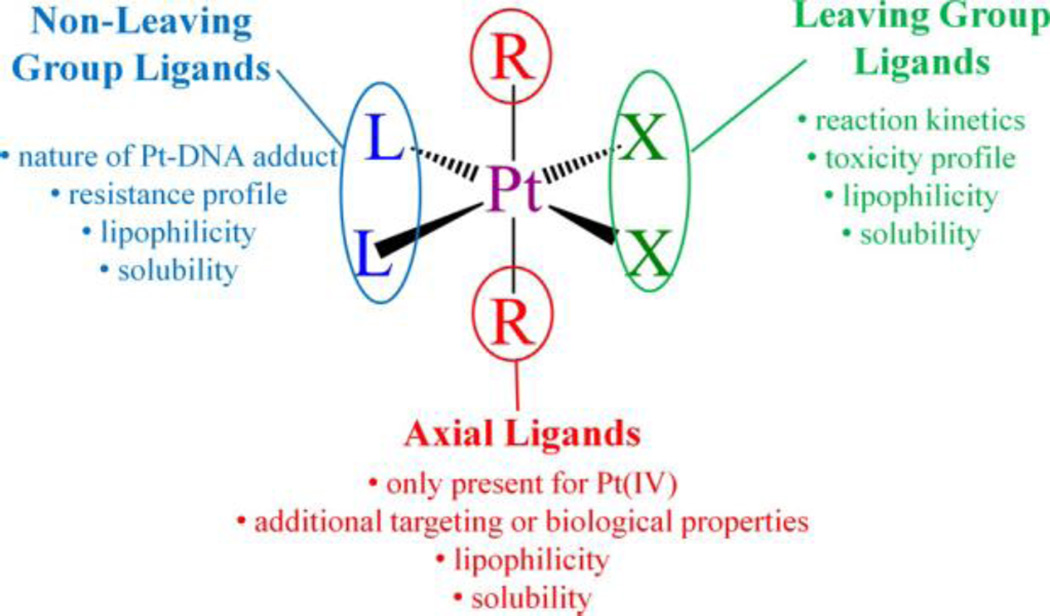
2. Mechanism of Action
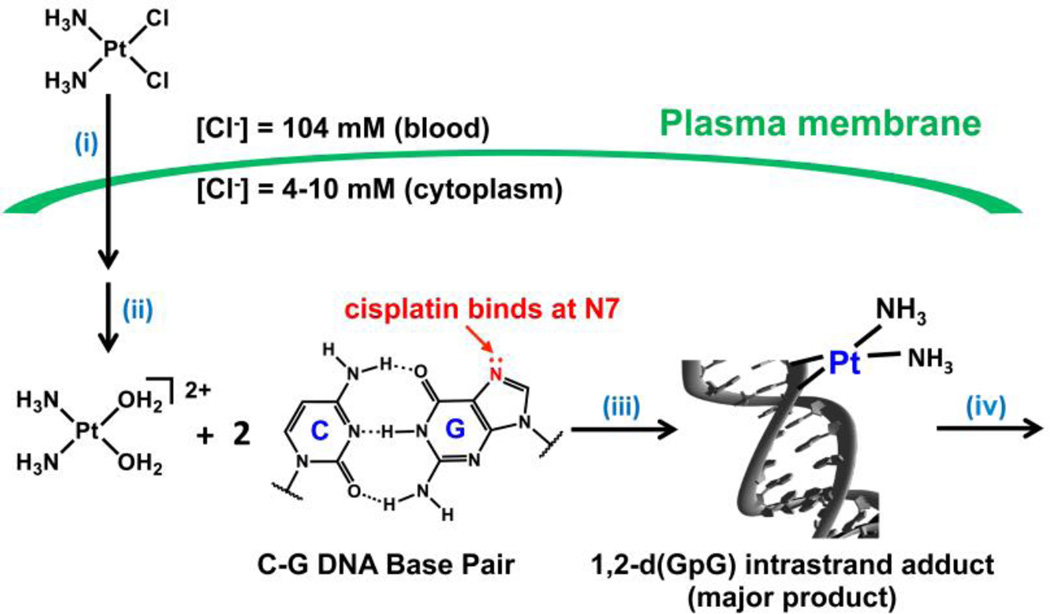
The mechanism by which the classical platinum drugs elicit an anticancer effect has been the subject of decades of investigation. The synthesis of the multitudes of experiments and trials conducted by chemists, biologists, and physicians has produced a consistent framework under which we can explain the data that have been obtained from compounds analogous to cisplatin.14,15 The analogy extends to include those platinum complexes that are neutral and square-planar with cis am(m)ine ligands and cis anionic ligands. The am(m)ine ligands can be chelating or non-chelating and are referred to as the “nonleaving group ligands” because, as described below, they remain bound to the metal center throughout the course of the mechanism. In contrast, the anionic “leaving group” ligand(s), which can be monodentate anionic or chelating dianionic fragments, are so called because they leave the platinum(II) coordination sphere. We include the caveat that, like any mechanism, the one presented below cannot be proved but rather has so far stood the test of time. The generalized mechanism of action involves four key steps (Figure 3): (i) cellular uptake, (ii) aquation/activation, (iii) DNA binding, and (iv) cellular processing of DNA lesions leading to apoptosis.15
Figure 3.
The four steps of the mechanism of cisplatin and, by extension, related platinum anticancer drugs. (i) Cellular uptake, (ii) aquation/activation, (iii) DNA binding, and (iv) cellular processing of DNA lesions leading to apoptosis. Reproduced from reference 15. Copyright © 2015, The Royal Society.
The cellular uptake of cisplatin has long been investigated with oftentimes seemingly contradictory results.16 The two pathways by which this molecule is most likely to be taken up are passive diffusion through the plasma membrane and active transport mediated by membrane proteins. The small size of cisplatin, along with it planar shape, have long been cited as support for a passive diffusion mechanism and indeed other properties of its cellular accumulation are consistent with uptake via this pathway. For instance, the uptake does not saturate with increasing concentration and is proportional to the administered concentration.17–19 Moreover, structural analogues of cisplatin do not inhibit uptake of the drug.20 Conversely, data have also been obtained that support an active transport mechanism. For instance, cisplatin uptake can be specifically stimulated and has been linked to expression levels of copper transporters.16,21,22 In a similar manner, oxaliplatin efficacy has been linked to expression of organic cation transporters (OCTs).23 Also, reactive aldehydes can inhibit cisplatin uptake, presumably by interacting with membrane proteins.24 The current model posits a combination of both passive and active transport, but the relative importance of these pathways and the extent to which they influence each other remains to be determined.
The square-planar geometry of cisplatin facilitates associative ligand substitution and, as will be discussed in the next section, such substitution is necessary for it to form the DNA lesions that characterize its activity. Cisplatin can undergo a ligand substitution event prior to DNA binding in which a chloride ligand is replaced with a water molecule. Such aquation is suppressed in the bloodstream, where the chloride ion concentration is high (≈100 mM) but occurs more readily in the cytoplasm, where the chloride ion concentration drops lower than 20 mM.25 In the presence of these lower salt concentrations, the half-life of the aquation reaction producing cis-[Pt(NH3)2Cl(H2O)]+ is approximately two hours. The positive charge on the platinum complex may help attract it to the negatively charged DNA molecule in the nucleus. Carboplatin and oxaliplatin feature chelating ligands opposite the firmly bound am(m)ine groups. These chelating ligands are substituted by water much more slowly and solutions of these two drugs are stable to aquation over a period of weeks to months.26–30
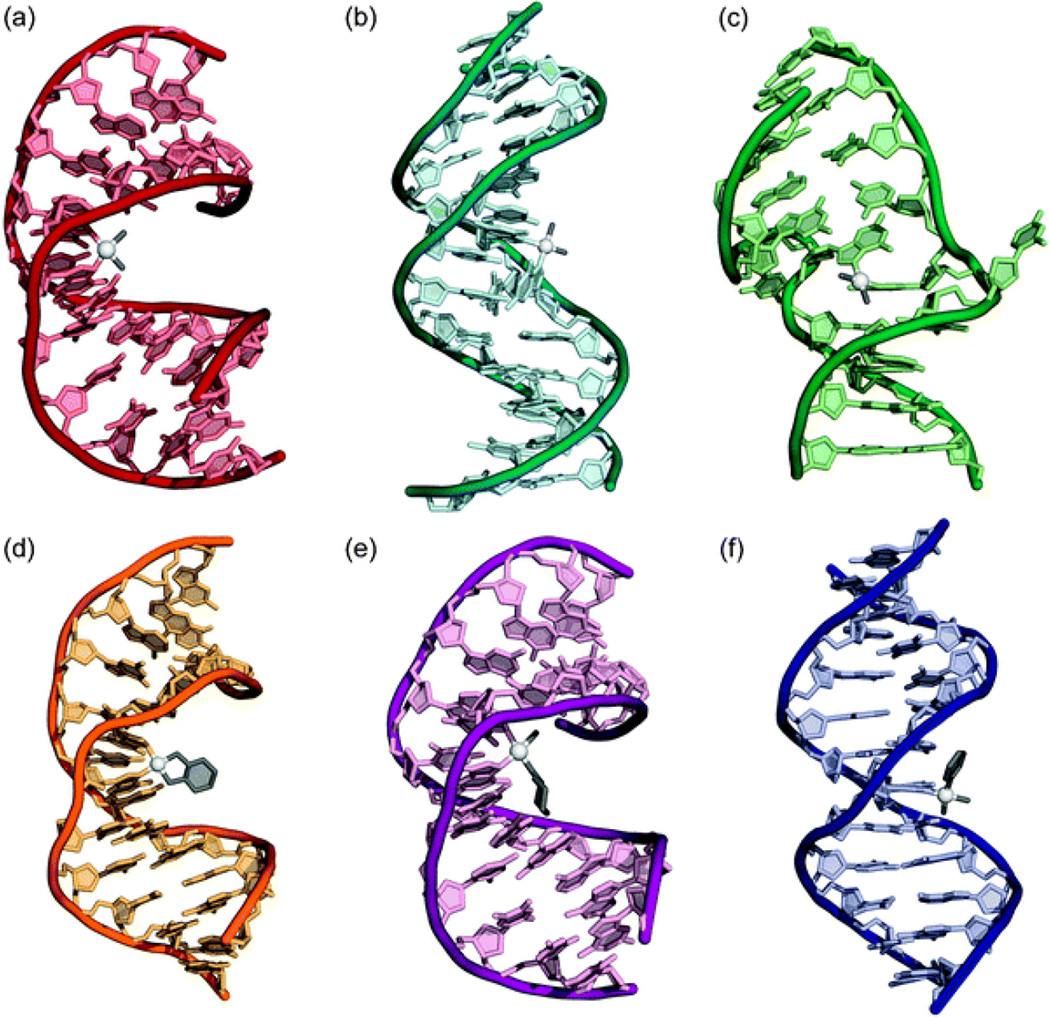
Aquated cisplatin can enter the nucleus and undergo substitution of the water ligand for a heterocylic DNA base. The strongest early evidence that confirmed DNA as the primary target of platinum drugs was the sensitivity of cells deficient in DNA repair to treatment with these compounds.31 Although decades of research have supported the hypothesis that nuclear DNA is the functional target of platinum drugs, other interactions, notably those with proteins and RNA, have been proposed to play a role as well.32–34 The most nucleophilic positions on DNA are the N7 sites of deoxyguanosine residues, and these are the residues that are preferentially platinated. 195Pt NMR spectroscopic monitoring experiments revealed that cisplatin first forms monofunctional adducts on DNA, that is, it forms only one covalent bond to the genomic polymer.35 In a distinct second reaction, the remaining chloride ligand is substituted for a second guanine base, forming a cross-link on the DNA. Such cross-links can occur between deoxyguanosines on the same strand or on different strands, giving rise to intrastrand and interstrand DNA cross-links, respectively. The 1,2-d(GpG) intrastrand cross-link is the most prevalent lesion (65%), but 1,2-(ApG) (25%) and 1,3-d(GpTpG) (10%) intrastrand cross-links also form along with small amounts of GG interstrand cross-links.36,37 Similar cross-links are formed by carboplatin and oxaliplatin, although the relative proportions vary.38,39 These DNA adducts distort the structure of DNA in a drastic and characteristic manner. Early studies provided evidence of bending and unwinding of the double helix upon platination.40 Atomic level details of the structures of many of the adducts formed by different platinum anticancer are now known (Figure 3).
Figure 3.
The structures of double-stranded DNA adducts of different platinum anticancer agents as determined by X-ray crystallography or NMR spectroscopy. (a) Cisplatin 1,2-d(GpG) intrastrand cross-link (PDB 1AIO). (b) Cisplatin 1,3-d(GpTpG) intrastrand cross-link (PDB 1DA4). (c) Cisplatin interstrand cross-link (PDB 1A2E). (d) Oxaliplatin 1,2-d(GpG) intrastrand cross-link (PDB 1PG9). (e) Satraplatin 1,2-d(GpG) intrastrand cross-link (PDB 1LU5). (f) cDPCP monofunctional adduct (PDB 3CO3). Reproduced from reference 41. Copyright © 2009, The Royal Society of Chemistry.
Cells whose DNA has been damaged in this way arrest at the G2/M transition of the cell cycle and attempt to repair this damage.10 Cisplatin lesions are most effectively removed by the nucleotide excision repair machinery and enhanced repair of this sort can lead cells to resist platinum treatment.42,43 The repair machinery must, however, be able to access the damage and binding of proteins to the lesion can shield it from repair. Curiously, the distortion that a cisplatin lesion induces in the DNA double helix fortuitously resembles that which is recognized by the high-mobility group (HMG) box proteins.44 The HMGB proteins, one of the most abundant proteins in the nucleus,45 display a particularly great affinity for the 1,2-d(GpG) intrastrand cross-link.46 The ability of these proteins to shield platinum adducts from repair may contribute to the sensitivity of certain cancer cells to cisplatin, but the previously ignored redox state dependence of the platinated DNA-protein interaction confounds an interpretation of the results present in the literature.47 Interestingly, the ability of cisplatin to cure testicular cancer may be related to the fact that testes cells express the HMGB4 isoform of this protein,48 and experiments along this line of investigation are ongoing. If the cell is unable to repair platinum-DNA damage, the expression of proapoptotic proteins increases, prompting the release of cytochrome c and the activation of intracellular caspases.10 These proteases effectively degrade the cell in a process of programmed cell death known as apoptosis. One of the main mechanisms by which the cell is signaled to trigger apoptosis in response to platinum treatment is the inhibition of transcription past platinum lesions.41
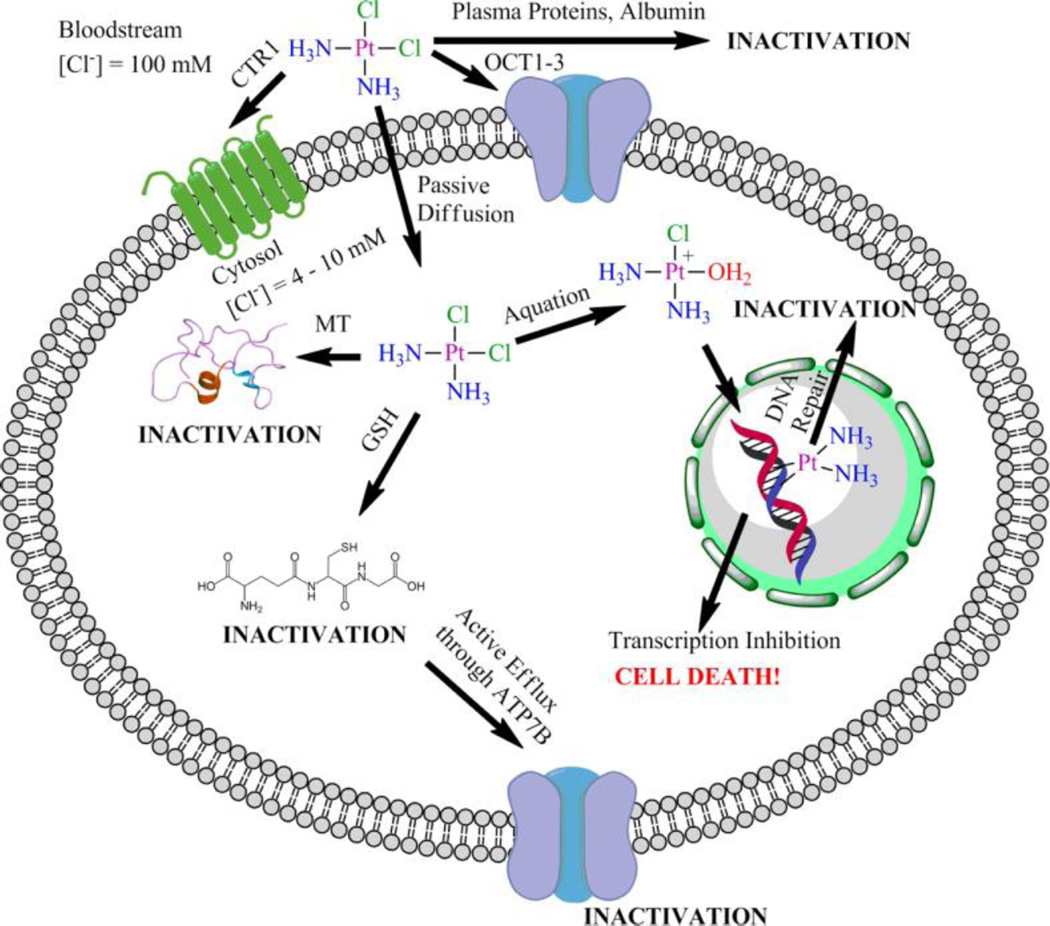
The ability of a platinum drug to elicit this ultimate cell-killing response relies on its ability to proceed through these mechanistic steps unhindered. In reality, a number of deactivation pathways exist that can sequester platinum complexes or otherwise prevent them from causing apoptosis (Figure 4).49 Because the current platinum drugs are all administered intravenously, blood components can interact with the metal centers. Notably, human serum albumin (HSA, the most abundant protein in the human bloodstream, contains a cysteine residue that can interact with systemically administered metal complexes.50 In accordance with hard-soft acid-base theory, the soft platinum(II) metal center will form stable complexes with ligands presenting soft donor atoms, such as sulfur. The main interaction of cisplatin with HSA, however, appears to involve sulfur-donors other than cysteine thiols, namely the thioether side chains of methionine residues.51
Figure 4.
The paths travelled by cisplatin before and after entering the cell. Attention is drawn to instances where deactivation/sequestration can occur. Reproduced from 49. Copyright © 2013, The American Chemical Society.
Once inside the cell, sulfur-rich metallothioneins can sequester platinum complexes as can glutathione.42 As part of the cellular detoxification program, dedicated export pumps removed glutathione adducts from the cytoplasm. Overexpression of these pumps, such as ATP7B, has been implicated in cisplatin-resistance.
3. Platinum(II) compounds with a mechanism of action similar to that of cisplatin
3.1. Approved platinum drugs
The first experiments showing that platinum complexes could have anticancer activity are rooted in the serendipitous observations made by Rosenberg and coworkers when studying the influence of electric fields on bacterial cell division.52–55 We refer the interested reader to a detailed and highly readable account that Rosenberg compiled of the experiments and circumstances that led to the 20th century clinical use of cisplatin (Chart 1),56 a compound whose synthesis had been reported over a century earlier.57 These works led to the first human patient being treated with cisplatin in 1971 and approval for marketing in 1978, first in Canada and soon after in the United States and then elsewhere across the world.58 It is currently used primarily to treat testicular, ovarian, and bladder cancers, but has also been used in the treatment of head and neck cancers, lung cancer, malignant pleural mesothelioma, neuroblastoma, tumors of the brain, and esophageal and cervical cancers.59 The subsequent discovery of newer platinum complexes that would come to be approved for clinical use relied less on serendipity and more on systematic, targeted investigations. It is interesting to note that the need for large amounts of precious metal starting materials, a situation atypical in traditional medicinal chemistry, led to the involvement of precious metal refining companies in the drug discovery process. For instance, the initial clinical development of cisplatin was fostered by a collaboration between the National Cancer Institute (NCI), Johnson Matthey, and Engelhard Industries, the latter two being precious metal companies based in the United Kingdom and Unites States, respectively.56 Johnson Matthey continued to pursue a research program into platinum anticancer agents and, in collaboration with Bristol-Myers, the Institute for Cancer Research, and the Royal Marsden Hospital, developed carboplatin (Chart 1).60 The success of carboplatin, originally known as JM8, has largely been driven by its favorable toxicity profile.61 This feature derives directly from alteration in reactivity at the metal center because of the chelating nature of the leaving group ligand and, potentially, the conformation that this ligand assumes.12 Carboplatin is used primarily to treat ovarian cancer but has also found use in treating retinoblastomas, neuroblastomas, nephroblastomas, and brain tumors, as well as cancers of the head and neck, endometrium, cervix, testes, breast, lung, and bladder.62
Chart 1.
Chemical structures of clinically-approved and marketed platinum anticancer drugs.
The discovery and development of nedaplatin (Chart 1) by Shionogi Pharmaceutical Company has been carried out entirely in Japan, and this is the only country in which it has regulatory approval, granted in 1995.58,63 This drug, initially referred to as 254-S, features cis ammine nonleaving group ligands as in the case of both cisplatin and carboplatin. The chelating leaving group ligand is glycolate, which confers greater water solubility (10 mg mL−1) than the two chloride ligands of cisplatin (2.5 mg mL−1). Nedaplatin is primarily used to treat cancers of the head and neck and esophagus as well as small cell lung cancer and non-small cell lung cancer.13,61 A number of clinical trials exploring the expanded use of nedaplatin are ongoing.13,63
Heptaplatin was developed by Sunkyong Industry Research Center in Korea under the name SKI 2053R. It was entered into clinical trials in the 1990s and received approval from the Korean Food and Drug Administration in 1999. It is marketed under the name SunPla for the treatment of gastric cancer and was the first new drug to be developed in Korea. The compound features malonate as a chelating leaving group ligand, reminiscent of carboplatin, as well as a chelating 2-(1-methylethyl)-1,3-dioxolane-4,5-dimethanamine. The nonleaving group ligand forms a seven-membered chelate ring, giving the drug its generic name. The two stereocenters in the ligand have R stereochemistry, and although we have not been able to find any publications that describe the reason as to why this stereoisomer was chosen, by analogy to oxaliplatin (vide infra) the opposite enantiomer and meso compound likely have lower activity.
Lobaplatin can be viewed as a derivative of heptaplatin in which a cyclobutane ring is fused to the seven-membered chelate ring as opposed to a functionalized dioxolane. Lobaplatin, however, is formulated as a racemic mixture of the R, R and S, S enantiomers of the nonleaving group ligand. Because only S-lactate is used as the leaving group ligand, diastereomers are formed.64 The compound was initially developed by ASTA Medica in Germany under the name D-19466, but was later acquired by the German company Zentaris AG, a subsidiary of the Canadian biopharmaceutical company Æterna Zentaris. Zentaris eventually sold Hainan Tianwang (Chang’an) International Pharmaceutical the rights to manufacture and market the drug in China.65 Although clinical trials were initially carried out in Europe, the United States, Australia, Brazil, and South Africa examining patients with a range of different cancers, regulatory approval was only obtained in China. Lobaplatin is approved primarily for the treatment of chronic myelogenous leukaemia but is also used in patients suffering from small cell lung cancer and metastatic breast cancer.65 Although literature sources and press releases describing the sale of the rights to lobaplatin in 2003 indicated that, at that time (2003), lobaplatin had already received regulatory approval,13,65,66 the Chinese FDA State Food and Drug Administration Database lists the approval year as 2010.67
Oxaliplatin is the most recent platinum anticancer drug to have gained international approval for marketing.68 This drug, occasionally referred to as l-OHP (note that the “l” refers to the use of the levorotatory chiral ligand in the preparation of the drug and is not an “L” indicating absolute stereochemistry) was first synthesized in Japan, but was subsequently developed in France. First approved in and subsequently in the United States, oxaliplatin is a component of the front-line combination chemotherapy treatment for colon cancer.7 Oxaliplatin features a chelating oxalate leaving group ligand and a chelating R, R-diaminocyclohexane (DACH) nonleaving group ligand.69 DACH ligands have long been investigated as components in platinum anticancer agents.60 In the case of oxaliplatin, empirical evidence revealed that the R, R stereoisomer was more effective than the enantiomeric S, S isomer or the related meso compound with cis amine groups.70 The origin of the greater activity of the R, R isomer came to light in later crystallographic studies that revealed this isomer preferentially forms a hydrogen bond between a pseudoequatorial NH hydrogen atom of the R, R-DACH ligand and the O6 atom of the 3′-dG of the platinated d(GpG) lesion.71 The exceptional activity of oxaliplatin in colon cancer has been linked to the ability of this drug to act as a substrate for the OCTs and the overexpression of these membrane proteins is observed in a large proportion of colon cancer patients.23
It can be appreciated that there are many commonalities that exist between the approved platinum drugs. Accordingly, the mechanisms by which these complexes induce cancer cell death have broad parallels to the general mechanism outlined above.10,11,14 The differences in the molecular structures of these drugs induce slight modulations in the mechanism but the general path appears to be similar. For example, carboplatin aquates at a rate different from that of cisplatin, limiting off target toxicity, and activation by carbonate has been implicated in its activity, but {Pt(NH3)2}2+ adducts analogous to those formed by cisplatin ultimately lead to transcription inhibition and apoptosis. Oxaliplatin may exploit an alternative uptake pathway, viz. active transport by OCTs, but again forms DNA cross-links, inhibiting nucleic acid polymerases and initiating apoptosis. The difference in the nonleaving group ligand results in a structurally distinct class of DNA adducts that are repaired and recognized at different rates, contributing to a distinct spectrum of action, but the DNA lesions ultimately trigger the same cell killing pathways.
3.2. The next generation of cisplatin-like platinum(II) complexes
In designing the next generation of platinum anticancer agents, many researchers are seeking to make increasingly drastic perturbations to the general molecular framework shared by these drugs in the hopes of uncovering novel mechanisms of cell killing, altering the spectrum of activity and rendering new cancers susceptible to platinum therapy. Such endeavors will be described in subsequent sections of this review. In this section, we will describe efforts to create novel platinum(II) complexes that are structurally similar to the approved drugs and are expected to operate via a comparable mechanism of action. As described above, early medicinal chemistry efforts produced many compounds of the form cis-PtA2X2 where the A group is ammine or a substituted ammine and X is an anionic ligand or X2 is a chelating dianionic ligand. A comprehensive review of all of these compounds is beyond the scope of this review, for indeed the SciFinder search tool maintained by the Chemical Abstracts Service lists over 4700 distinct compounds with this general formulation that are classified as anti-tumor agents. We suspect that this number is most likely a significant underestimate of the true extent of development that has occurred across both academia and industry. In one single report, for instance, the products of over 3600 reactions that prepared square-planar diam(m)neplatinum(II) complexes were screened for transcription inhibition activity using high-throughput methods.72
As described above, the sheer number of cisplatin derivatives precludes a detailed and comprehensive discussion of all the strategies that have been explored. We have chosen instead to focus in depth on the inclusion of targeting units into a platinum(II) agent of known anticancer activity. Such efforts seek to finally realize the magische Kugel that Ehrlich sought over 100 years ago.73 This conception of a drug as a magic bullet that seeks out its target of its own accord is well-matched with constructs bearing targeting units that direct platinum warheads to cancer cells by interacting with receptors that are overexpressed on the surfaces of these cells.74 The concept can be extended to encompass targeting of the tumor as a whole instead of cancer cells themselves by seeking proteins expressed on angiogenic blood vessels or allowing selective activation within the acidic or hypoxic tumor microenvironment.75 Finally, targeting can also take place at the subcellular level, whereby platinum can be directed to specific organelles to elicit distinct biological effects. Targeting of all of these sorts can also be applied to platinum(IV) complexes and nanoparticle delivery devices, as well, which will be discussed in subsequent sections.
3.2.1. Sugar targeting
Carbohydrates can engage in an intricate array of hydrogen bonding interactions, a feature of these molecules that is exploited in biological systems to achieve high fidelity recognition.76 This recognition has also been proposed as a paradigm for drug targeting.77 Another facet of sugar biology can be exploited for drug targeting, namely the enhanced uptake of glucose by cancer cells.78 In order to sustain the uncontrolled cell division that is characteristic of cancer, malignant cells require much greater levels of nutrients, in particular glucose.79 The need for glucose is further compounded by the altered metabolic state in which many cancer cells exist, a manifestation of the Warburg effect, more details of which are provided in Section 6.80 This enhanced uptake of glucose relies on the overexpression of glucose membrane transporters, such as GLUT1–4, and has been widely exploited in the use of 18F 2-fluoro-2-deoxy-D-glucose as an agent for positron emission tomography imaging of tumors.81,82 Although many example of platinum complexes bound to a variety of sugars are known, as will be described below, little evidence has been accumulated to suggest that these carbohydrate motifs have played a role in enhancing the activity of the anticancer agent by interacting with a specific receptor.
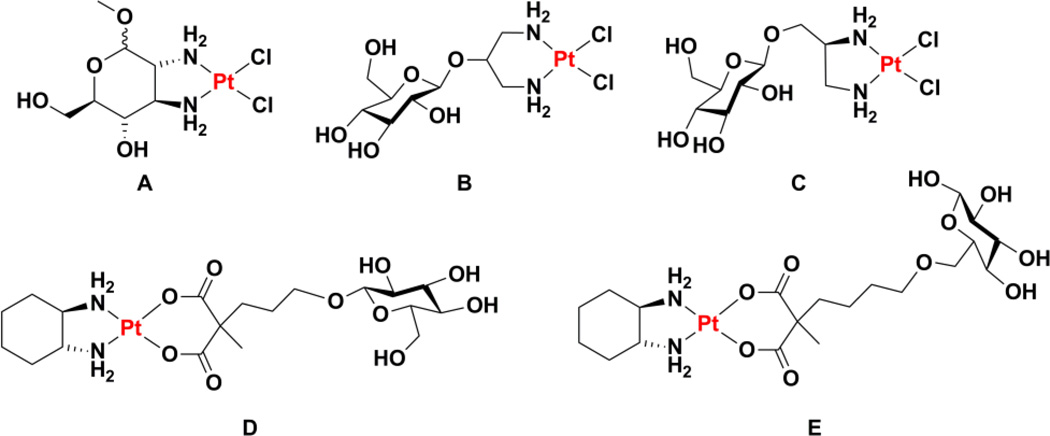
Using aminosugars, simple analogues of cisplatin were prepared in which the ammine ligands are replaced. Early studies demonstrated that complexes featuring coordination of 2-amino-2-deoxy-D-glucopyranose to platinum could be synthesized, but demonstrated no significant anticancer activity.83–87 Using 2,3-diaminosugars, complexes analogous to oxaliplatin were formed and found to have promising activity in vitro and in animal models. For instance, PtCl2(2,3-diamino-2,3-dideoxy-D-glucose) (Chart 2A) was able to more than triple the survival time of mice bearing sarcoma 180 when given as a 50 mg kg−1 i.p. injection.88 Although cisplatin can have a similar effect at a much lower dose (8 mg kg−1) this latter value approached the MTD (13 mg kg−1). Substitution of the halide leaving group ligands in the diaminodideoxyglucose platinum(II) complexes for oxalate or malonate produced less active species and studies investigating the reactivity of these compounds with dGMP are consistent with the slower rate of reaction expected from a chelating leaving group ligand.89 A similar reduction in activity was observed by incorporating the CBDCA ligand of carboplatin into the platinum(II) complex of methyl 2,3-diamino-2,3-dideoxy-L-xylopyranoside.90 Although one of the justifications for pursuing 2,3-diaminoglucose complexes is the similarity between the 2,3-diaminoglucose and the 1,2-diaminocyclohexane of oxaliplatin, it is important to note that the conformation of the D-glucopyranose ring results in the diamine chelate ring adopting a λ conformation. The stereochemistry of the R, R-DACH in oxaliplatin results in a δ conformation of this chelate ring.69 This feature of oxaliplatin has been suggested to be the origin of the greater activity of complexes of R, R-DACH as opposed to S, S-DACH.70,71,91 Although this discrepancy is noted in passing in one publication,92 its effects are clearly seen, but not discussed, in later work.89 We anticipate that the use of L-glucose in the preparation of the diaminoglucose would result in a more active platinum complex as a result of its ability to more closely mimic oxaliplatin. We note that the use of L-glucose may appear to run contrary to the motivation of using a sugar that can be recognized by the cellular uptake machinery, but it can be inferred from the results of the experiments presented above and those that follow that modifications to the structure of the glucose as drastic as substitution of alcohols for amines and their chelation of a metal will most likely inhibit any specific recognition and transport to an equivalent or greater extent than the use of the mirror image of the natural glucose enantiomer.
Chart 2.
Chemical structures of cugar-conjugated platinum(II) complexes.
We also note briefly that an interesting pair of enantiomeric platinum complexes bearing chiral 2,3-diaminocamphore ligands also investigated and one enantiomer was similarly found to be significantly more cytotoxic than the other in in vitro assays. The amino substituents were, however, arranged cis to one another (R, S; chirality at the camphor 1-position prevents the cis-diamino compound from being a meso compound) precluding an analysis of whether the trend of greater activity for R, R persists in this system as well.93
A linker can also be inserted between the sugar unit and the platinum-binding amine, as in the case of cis-dichloro[(2-β-D-glucopyranosidyl)propane-1,3-diamine]platinum(II) (Chart 2B).94 The glycosylation was found to increase water solubility without compromising anticancer activity. In an analogous system, a (2S)-2,3-diamino-1-propanol linker was attached to D-glucose (Chart 2C), L-glucose, D-galactose, D-xylose, or D-mannose.95 The authors found a distinct difference between the activities of the D and L glucose conjugates and suggest that interaction with a specific receptor may play a role in the greater activity of the D-glucose conjugate. Despite platinum-sugar conjugates having been explored for almost two decades by the time this report95 was published, it appears to be one of the earliest instances in which interaction of the sugar moiety with a specific receptor is proposed to enhance activity. Linkage through an ethylenediamine was also carried out with D-galactose and D-ribose, but the activity of the complexes was not investigated.96
Glucose can alternatively be incorporated into a leaving group ligand such as malonate, although in one early study no increase in activity was observed over the analogous carboplatin.97,98 A larger set of complexes with a ranges of different sugars similarly incorporated into a malonate leaving group ligand were prepared, but the results of the biological assays with these complexes have yet to be released.99 For the sake of brevity, the remainder of the discussion of sugar conjugates will focus on those complexes for which experiments have been done to characterize the mechanism of uptake. The reader interested in other platinum-glucose conjugates is referred to a recent excellent review.100
The first experimental evidence that inclusion of a glucose unit actually exploits the glucose receptor to enhance cellular uptake was presented for a platinum(II) complex bearing a DACH nonleaving group ligand and a glucose-functionalized malonate leaving group ligand (Chart 2D).101 In vitro cytotoxicity assays in the presence of phlorizin, an inhibitor of the glucose transporter GLUT1, indicated that the inhibitor decreased the efficacy of the platinum-glucose conjugate, consistent with a model in which GLUT1 mediates uptake of the complex.101 In the studies listed above, glucose was never attached to the platinum center through the 6 position, perhaps because of the synthetic difficulty of carrying out this modification. Analysis of the crystal structure of a bacterial homologue of GLUT1 bound to D-glucose revealed, however, that the hydroxyl group at this position is the only one that does not have hydrogen bonding interactions with protein side chains.102 Platinum(II) complexes with a DACH nonleaving group ligand and a malonate leaving group ligand attached to glucose at the 6 position via a linker of variable length were prepared and shown to be taken up selectively by GLUT1 (Chart 2E).103 Studies with different GLUT1 inhibitors confirmed that cellular uptake was dependent on glucosylation and directly impacted cell-killing efficacy. An interesting effect of chain length on uptake via GLUT1 was observed and modelling studies indicate that an overly long linker between the glucose and the platinum inhibits the ability of the protein to undergo the conformational change required to transport the construct across the cell membrane. The organic cation transporters were also found to play a role in the uptake and efficacy of the most potent of the glucoconjugates prepared.103
3.2.2. Steroid targeting: estrogen and testosterone
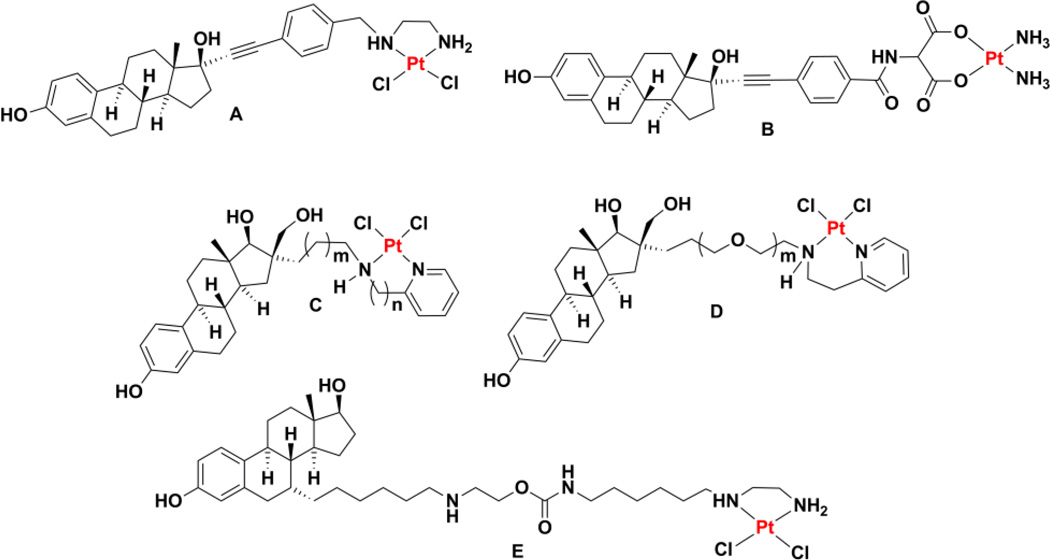
Another class of targeted platinum(II) complexes comprises those in which a steroid unit has been incorporated into the nonleaving group ligand. These steroids can act as targeting units that direct the platinum agent to tissues expressing the cognate steroid receptor. The estrogen receptor (ER), for instance, is an established oncology target because this protein is overexpressed on the surfaces of some cancers, particularly breast cancer, where it stimulates cell proliferation.104–107 The monoclonal antibody trastuzumab, also known as Herceptin, binds to and interferes with the signaling of this receptor.108 It is important to note that in addition to the classical estrogen receptor, now designated ERα, a novel estrogen receptor, ERβ, has been more recently discovered.109–111 This ERβ receptor may even play an important role in cancer progression by exhibiting antiangiogenic and antiproliferative properties.112 Linkage of a steroid unit capable of interacting with the ER to a platinum center can influence the anticancer activity of the metal complex by either interfering with the biological function of the receptor or by permitting enhanced uptake of the platinum complex, leading to an increase in DNA platination and a greater chance of apoptotic cell death. Several recent reviews have comprehensively summarized the early developments in this area.74,113,114 One of the earliest studies that investigated platinum complexes conjugated to steroids involved platinum complexes with phosphine nonleaving group ligands and a leaving group catecholate ligand functionalized with steroids for use in metalloimmunoassays. Although tested in vitro for cell killing activity, they did not fare better than cisplatin.115–117 Although dozens of platinum(II) complexes bearing estrogen derivatives at either the leaving group or nonleaving group ligands have been reported,113 most have not been tested to ensure that an interaction with the ER is operative and leads to enhanced activity via one of the two mechanisms described above. In one report that does take such measures, two compounds cis-dichloro[N-(4-(17-ethynylestradiolyl)-benzyl)-ethylenediamine]platinum(II) (Chart 3A) and cis-diamino[2-(4-(17-ethynylestradiolyl)-benzoylamino)-malonato]platinum(II) (Chart 3B), were found to agonize the ER at low concentrations, leading to enhanced proliferation, but exhibited cytotoxicity at higher concentrations.118 Such a bimodal effect renders these particular compounds unsuitable for further investigation as cytotoxic anticancer agents.
Chart 3.
Chemical structures of estrogen receptor ligands tethered to platinum(II) complexes.
A series of estradiol conjugates were prepared that were linked to the platinum center through a spacer attached to an N-functionalized 2-aminoalkylpyridyl chelate (Chart 3C). Although they did not show any apparent enhanced toxicity in ER+ cells as compared to those that are ER−, most compounds in the series bound to ERα with very high affinity.119–121 Some members of the series also demonstrated high affinity for ERβ.122 One difficulty with this series of complexes is that, as the length of the aliphatic chain that links the estradiol and the platinum complex is increased, the solubility of the complexes drastically decreases. Use of a poly(ethylene glycol) (PEG) chain (Chart 3D), however, allows the length to be varied without compromising solubility.123 Enhanced potency in ER+ cell lines was observed for certain PEG chain lengths and these results were rationalized using molecular modeling methods. This steroid-targeting strategy was also applied to derivatives of carboplatin and oxaliplatin.124 Recently, this strategy was extended with the design and synthesis of a 17β-acetyl-testosterone conjugate linked to the platinum center through the 7α position.125 In the same way that estrogen units can target cancer cells expressing the ER, testosterone can target platinum to cancer cells expressing the androgen receptor (AR). Activity was observed in both androgen receptor AR+ and AR– prostate cancer cell lines.125 Mechanistic studies established that the compounds induce S-phase arrest and double-stranded DNA breaks. Antitumor studies using a chick chorioallantoic membrane xenograft assay confirmed the ability of these compounds to inhibit tumor growth.
Before leaving the discussion of estrogen- and testosterone-targeted platinum(II) agents, we highlight a study showing that a platinum(II) complex with an ethylenediamine nonleaving group ligand functionalized with a ligand for the ER could maintain its ability to interact with the estrogen receptor even after binding to a 16-mer DNA duplex (Chart 3E).126 Although the steroid conjugate binds DNA with lesser facility than an analogue lacking an ER ligand, the former is more toxic to cells. Enhanced toxicity was observed in cell lines deficient in DNA repair, strongly suggesting that DNA damage is the means by which cell death is induced. The authors propose that DNA repair shielding or steroid receptor hijacking may be operative.
Non-steroidal estrogen mimics have also been linked to platinum compounds to elicit the same effect. The first compounds of this sort to be prepared, a series of seteroisomers of dichloro[1,2-bis(4-hydroxyphenyl)ethylenediamine]platinum(II), competed with estrogen for interaction with the ER, but were toxic to ER+ and ER− cells alike.127 Subsequent substitution at the 2 and 6 positions of the two phenyl rings with chlorine atoms, a substitution that had been shown to increase the affinity of the ligand for the ER,128 produced a set of complexes that not only interacted with the ER but also selectively killed ER+ mammary carcinoma cells.129 Variations on the substitutions of the nitrogen atoms and ring carbon atoms can influence estrogenicity and cytotoxicity, but often in a mutually exclusive manner.130
3.2.3. Steroid targeting: bile acids
Platinum(II) complexes have also been conjugated to members of the steroid acids known as bile acids in an effort to target compounds to the liver because hepatic epithelial cells express a number of transport proteins that take up bile salts from the bloodstream.131 The first work in this area appears to be described in a set of papers describing the preparation of a series of platinum(II) complexes bound by a DACH ligand and two bile acids (e.g. hyodeoxycholate, Chart 4A).132,133 The lability of monodentate carboxylates bound to platinum(II) complexes almost certainly assures that, upon dissolution, the complex will very rapidly form a distribution of aquated species in dynamic equilibrium. Although activity was observed in these studies, the research does not appear to have been pursued further by these authors.
Chart 4.
Chemical structures of bile-acid tethered platinum(II) agents.
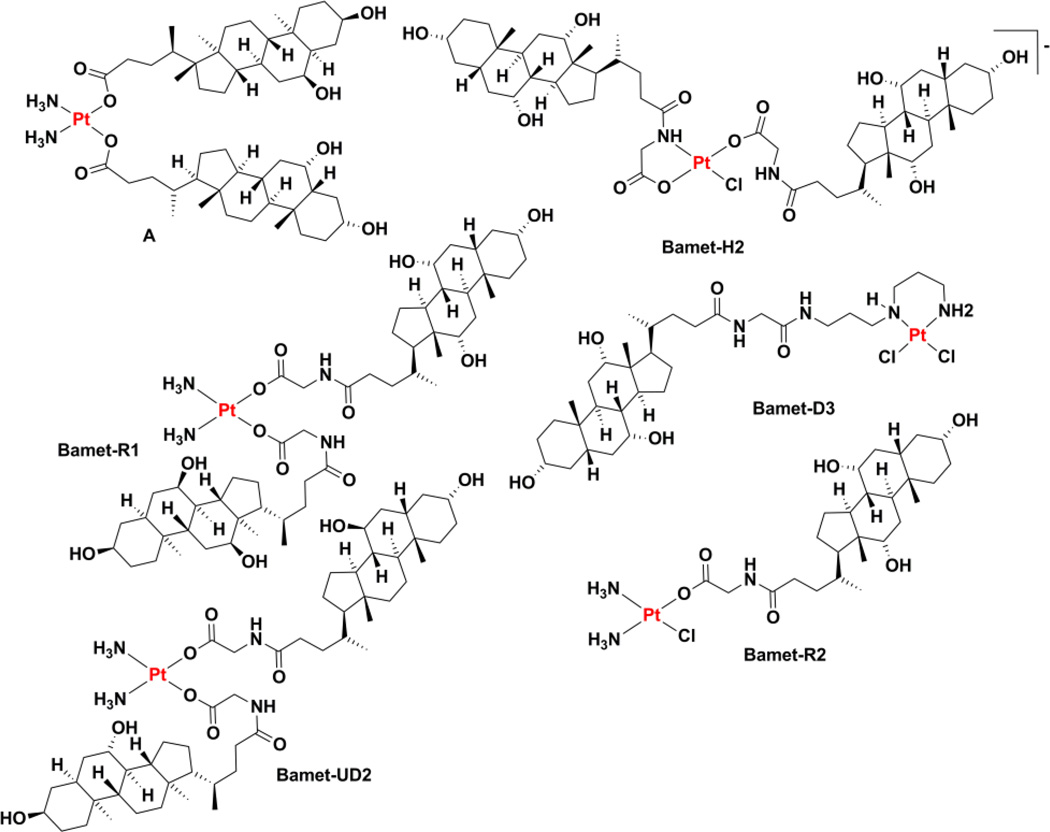
A related series of compounds named Bamet, a portmanteau of bile acid and metal, was prepared that also features bile acids attached to a platinum warhead. The first generation complex, Bamet-H2 (Chart 4), was simply prepared by allowing sodium cholylglycinate to react with tetrachloroplatinate(II). The product, formulated as a sodium salt on the basis of conductivity data, was cytostatic against L1210 murine leukemia and demonstrated enhanced uptake by the liver as compared to cisplatin.134 The compounds known Bamet-R1 and Bamet-R2 (Chart 4) were prepared by treating cisplatin with sodium cholylglycinate.135–137 The resulting complexes are presumably able to then form cisplatin cross-links following cellular uptake and shedding of the bile acid via aquation.138 Although active, the potencies of these compounds, and those of the related bis(ursodeoxycholate) complex Bamet-UD2 (Chart 4) and [(cis-dichloro(3,3-amminepropylammine)propyl)glycocholamideplatinum(II)] (Bamet-D3, Chart 4), were less than that of cisplatin in cultured cell lines tested in vitro.139,140 Related complexes with trans geometry were even less active.141 Given the propensity of bile acids to form liposomes, it is not surprising that a liposomal formulation of Bamet-R2 was readily prepared.136 In a parallel line of investigation, the ethylenediamine analogues of these complexes were studied because they are fluorescent and have increased emission upon binding to DNA and release of the leaving group ligands.142–145 The mechanism of action and many of the in vivo properties of these compounds, notably those related to their special affinity for liver tissue (hepatic organotropism), have been reviewed.146 Early studies with rat hepatocytes and isolated rat livers confirmed that Bamet-R2 is taken up by the pathway naturally used for bile acid uptake and is secreted into the bile.147 These cholephilic characteristics were also observed in experiments with live rats.148 Close to a decade of subsequent preclinical experiments have produced a set of results suggesting that these compounds may merit clinical investigation for the treatment of hepatic malignancy.146
A bile acid with a chelating dicarboxylate motif bound to a cis-diammineplatinum(II) fragment was explored as an orally administered anticancer agent.149 Preliminary in vitro assays revealed activity in cultured murine hepatoma cells. A syngeneic orthotopic rat model of hepatocellular carcinoma confirmed that the complex had antitumor activity.149 An alternative means of linking a bile acid to a platinum(II) center involves conjugation to the nonleaving group ligands, a strategy that has afforded complexes demonstrating activity in cultured cells via a mechanism of action similar to that of cisplatin.150,151 Other terpenoids, the class of molecules to which steroids and bile acids belong, have also been conjugated to platinum(II) complexes in an effort to direct the cytotoxic agent to cancer cells,152–155 although in some instances it remains to be determined whether the enhanced cellular uptake observed for these complexes arises from specific interactions with membrane receptors or if their inherent lipophilicity simply enhances passive membrane diffusion.
3.2.4. Steroid-related targeting
The peripheral benzodiazepine receptor, also known as TSPO,156 is thought to be involved in regulating the transport of cholesterol and the synthesis of steroids, although recent evidence has called this latter role into question.157 The protein has been suggested as a viable target for directing cancer therapeutics158 and it is overexpressed in a number of tumor tissues.159–161 Platinum(II) complexes chelated by a functionalized bidentate thiazolylimidazopyridine were found to interact strongly with this receptor.162 Although these complexes can be taken up by cultured cancer cells that express TSPO, they exhibit weak anticancer activity. Alteration of the thiazolyl ring to generate a monodentate ligand and addition of NH3 to the vacated coordination site, produced complexes with enhanced potency.163 Radioligand binding assays confirmed the ability of the complex to interact with, and presumably be taken up by, TSPO and microscopic studies confirmed that treatment with the platinum complex induced apoptosis.
3.2.5. Folate targeting
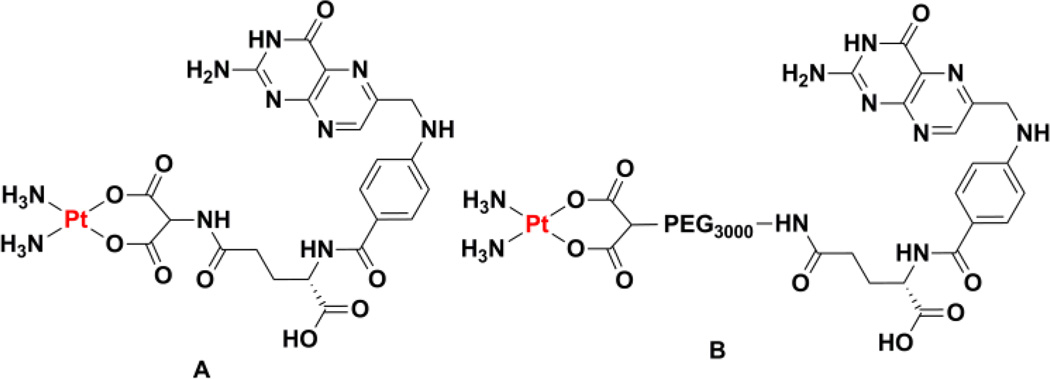
A number of different cancer cell lines and cells derived from the tumors of patients display an overexpression of a glycoprotein that acts as a folate receptor (FR)164. Folic acid contains the pteroic acid unit and is vital to a number of central biochemical pathways, including those related to DNA synthesis. In order to satisfy rapid cell growth, folate uptake is enhanced in cancer cells and the folate motif could conceivably be used to target a platinum complex to them.165 In comparison to the use of sugars and steroids, however, folates have seen significantly less use as targeting agents of platinum complexes. An early study of the interaction of cisplatin with cellular folates involved the isolation of the complex that results from substitution of the chloride ligands with tetrahydrofolate.166 Although this complex was found to be an inhibitor of dihydrofolate reductase and the folate transport system, the lack of labile coordination sites suggests that it will not be able to operate as a cytotoxic agent in a manner analogous to that of cisplatin.166 This proposal is supported by that fact that L1210 cells were treated with concentrations of the complex in excess of 200 µM to observed inhibition of folate transport, but no significant cell killing was reported. A systematic preparation of a variety of cisplatin and carboplatin derivatives bearing a folate unit conjugated to either the nonleaving group ligands or the leaving group ligands (Chart 5A) established much of the chemistry required to prepared complexes with the capacity to target the FR.167 Unfortunately, the low water solubility of these molecules prohibited their use in biological experiments. The use of a PEG spacer between a dicarboxylate chelator and a folic acid unit (Chart 5B) affords enhanced water solubility and mechanistic studies showed that the conjugate is taken up by folate receptor-mediated endocytosis.168 The conjugate was, however, less potent than carboplatin, a feature that appears to stem directly from the formation of fewer platinum-DNA adducts. As will be described in Section 8, folate targeting has been successfully used to direct platinum-loaded nanoparticle drug delivery vehicles to FR-expressing cancer cells.
Chart 5.
Chemical structures of folate-targeted platinum(II) complexes.
3.2.6. Peptide targeting
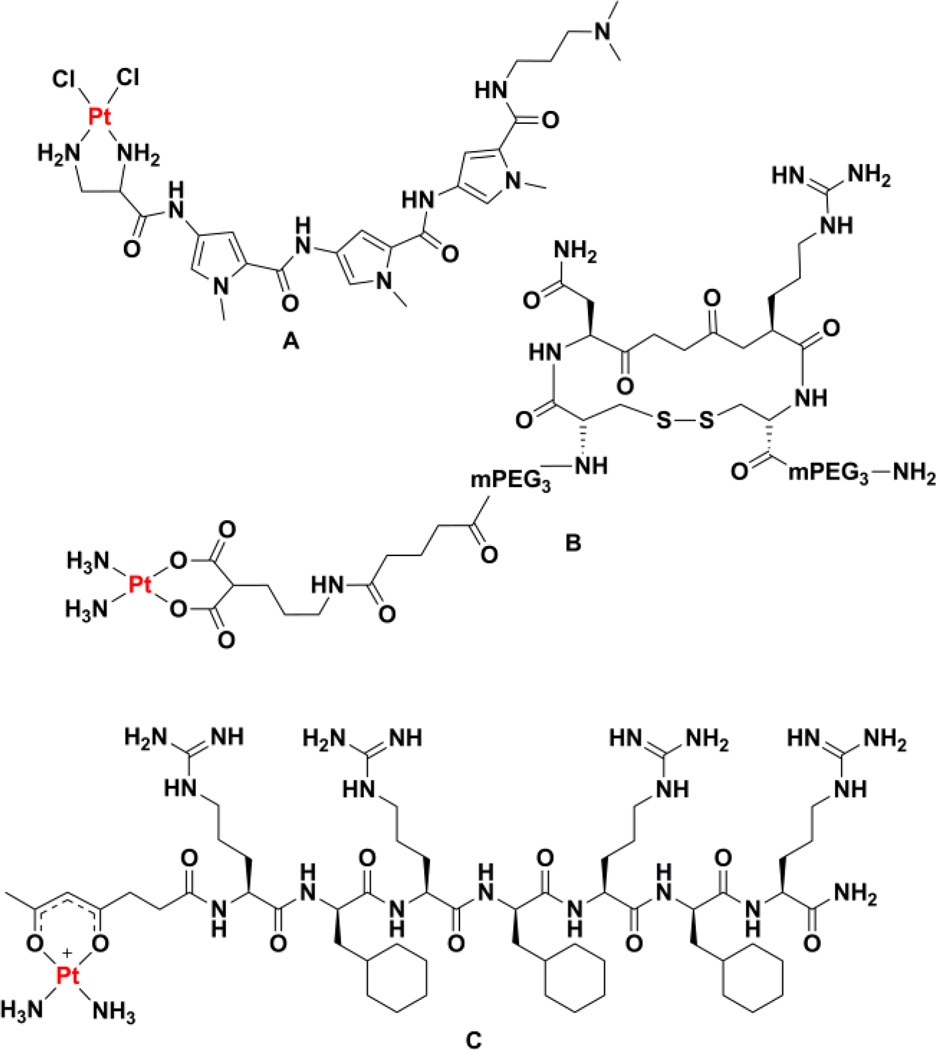
The earliest report of a platinum(II)-peptide conjugate involved attaching a platinum warhead to derivatives of the minor groove binding agents netropsin and distamycin (Chart 6A).169 The peptide was able to enhance platination of poly(dA) tracts over poly(dG) tracts, the latter of which are preferentially platinated by cisplatin. The ability to explore a much wider range of platinum(II)-peptide conjugates arose when synthetic strategies compatible with solid-phase peptide systhesis were developed.170–173 This chemistry involved linking the platinum complex to the peptide via a functionalized ethylenediamine nonleaving group ligand. The platinum chemistry could be performed on the solid support and the complex survived the deprotection and cleavage reactions. This technology was used to screen dozens of platinum(II)-peptide conjugates in vitro for anticancer activity, although no significant hits were obtained in this screen.174 Although these peptide conjugates exhibit a reduced capacity to platinate DNA, and consequently reduced potency, the DNA sequence specificity of platination is generally unaltered with different peptide sequences.175 We note briefly, however, that use of individual charged amino acids, such as ornithine, lysine, or arginine, in place of the nonleaving group ligands can alter this specificity.176 Highly complex peptide architectures can be conjugated to platinum(II) using these methods, including conjugates octreotide analogues.177
Chart 6.
Chemical structures of platinum(II) complexes tethered to peptides.
Although the opportunities offered by peptide-based targeting are great, instances in which targeting has been successfully achieved with platinum(II) conjugates are few. More examples exist with platinum(IV) constructs and nanodelivery vehicles as described below. One well characterized example involves the use of a malonate nonleaving group ligand attached to a PEGylated cyclic peptide via a linker (Chart 6B).178 The cyclic peptide, c(CNRGC), presents the Asn-Gly-Arg sequence that targets the CD13 receptor overexpressed on the surface of certain cancer cells.179 The targeted complex was more toxic to prostate cancer cells expressing CD13 than non-targeted carboplatin and competition assays confirmed that the complex is taken up via interaction with CD13. Fluorescence microscopy studies and DNA fragmentation assays are consistent with an apoptotic mechanism of action.
Another example involves the subcellular targeting of active platinum(II) units to the mitochondria. Following work on the anticancer activity of cationic platinum(II) complexes with ammine nonleaving group ligands and β-diketonate leaving group ligands,180 an analogous complex with a succinylacetonate ligand was prepared. The pedant carboxylate of this complex was used to form an amide bond to the N-terminus of a mitochondrial penetrating peptide (Chart 6C).181 The decapeptide, r(Fxr)3 where r is D-arginine and Fx is L-cyclohexylalanine, is non-toxic, protease resistant, and should localize to mitochondria because of its lipophilic and cationic nature.182,183 The conjugation of the platinum complex to the peptide was carried out on the solid support and survived trifluoroacetic acid-mediated cleavage. Fluorescence microscopy confirmed localization of the conjugate to the mitochondria of cultured ovarian cancer cells and PCR amplification studies indicate that, in contrast to treatment with cisplatin, mitochondrial DNA is platinated while nuclear DNA is not. Even though the location of platination differs, the platinum-peptide conjugate was still able to induce apoptosis. Mouse embryonic fibroblasts that are deficient in their proofreading of mitochondrial DNA were more sensitive to the treatment, an observation consistent with a shift in the target of the platinum complex from nuclear DNA to mitochondrial DNA.
Although nucleic acids are not typically used to prepare targeted platinum(II) complexes because of the inherent reactivity of the platinum center with the nitrogenous DNA bases, a peptide nucleic acid (PNA)-plaintum(II) conjugate has been reported.184 The nucleic acid sequence of the PNA conjugate, which lacks d(GpG) units, can target a complementary mRNA sequence in such a way as to present the pendant platinum(II) center to a GpG unit on the target mRNA sequence. Evidence supporting cross-linking of the PNA and the target mRNA were obtained suggesting that this strategy could be used to enhance antisense therapy.
4. Platinum(II) compounds with a mechanism of action different from that of cisplatin
Early structure-activity relationship (SAR) studies found that subtle modifications to cisplatin geometry led to drastic changes in chemotherapeutic activity. Whereas cisplatin displays excellent activity against some cancer types, trans-diamminedichloroplatinum(II), the trans isomer of cisplatin, lacks activity.60,185 Two major factors are thought to contribute to the contrasting activities of the cis and trans stereoisomers. The trans disposition of the two chloride ligands in trans-diamminedichloroplatinum(II) renders them kinetically labile in comparison to those of the cis isomer, and thus susceptible to undesirable side reactions with extra- and intra-cellular biomolecules.186–189 Additionally, the major cytotoxic DNA lesion formed by cisplatin, the 1,2-intrastrand cross-link between adjacent purine bases, is stereochemically inaccessible to trans-diamminedichloroplatinum(II). Instead, trans-diamminedichloroplatinum(II) forms 1,3-intrastand cross-links (ca. 28%) and monofunctional adducts (ca. 60%), which undergo conversion to interstrand cross-links (ca. 12%), preferentially between guanine and a complementary cytosine.190 In cells, however, very few interstrand cross-links are formed because of the slow transformation of monofunctional adducts and 1,3-intrastand cross-links to interstrand cross-links.191 DNA adducts formed by trans-diamminedichloroplatinum(II) do not halt DNA replication as efficiently as those formed by cisplatin and are prone to effective DNA repair.192
4.1. Trans complexes
The discrepancy in cisplatin and trans-diamminedichloroplatinum(II) activity led to the early belief that only platinum complexes with cis leaving groups were endowed with antitumor activity.60 The development of biologically active trans-diamminedichloroplatinum(II) analogues, however, has dispelled this notion.186–189 There are now several examples in the literature of active trans-platinum complexes. These complexes can be divided into the following sub-types; (i) trans-platinum(II) complexes with heteroaromatic ligands, (ii) trans-platinum(II) complexes with iminoether ligands, and (iii) trans-platinum(II) complexes with asymmetric aliphatic amine ligands.
4.1.1. trans-Platinum(II) complexes with heteroaromatic ligands
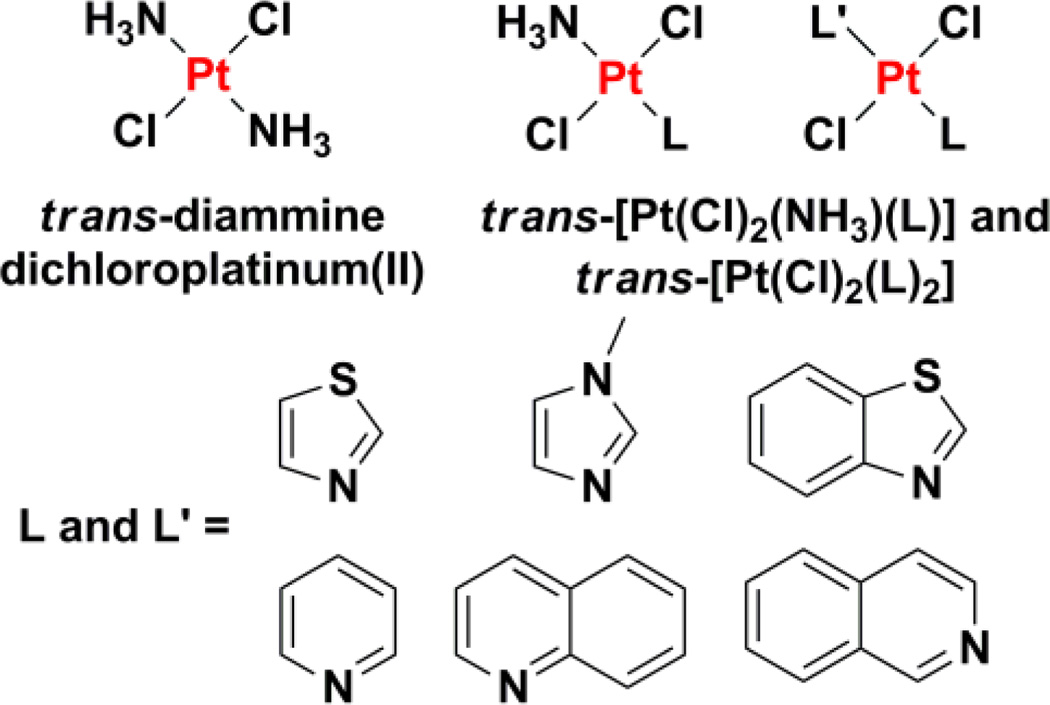
Substitution of the ammine ligand(s) in trans-diamminedichloroplatinum(II) with bulky, planar N-donor ligands affords trans-platinum(II) complexes with high in vitro cytotoxicity, equivalent to their corresponding cis-isomers and cisplatin.193–195 Some analogues, such as trans-[PtCl2(NH3)L] and trans-[PtCl2L2] where L = pyridine, quinolone, isoquinoline, thiazole, or benzothiazole (Chart 7), display therapeutically significant activities in cisplatin- and oxaliplatin-resistant cell lines.195,196 According to NCI human tumor panel screening studies and COMPARE algorithm197 analyses, trans-platinum(II) complexes of this type exhibit a spectrum of activity that differs significantly from that of any other anticancer agent in the NCI database.196 Their unique cytotoxicity profiles are attributed to their structural and DNA-binding properties. For instance, the weakly trans-directing aromatic heterocyclic ligands reduce the kinetic liability of trans chloride groups and thus prevent deactivation by sulfur-rich biomolecules, a common detoxification pathway for cisplatin.186 Additionally, the type and distribution of DNA lesions induced by such trans-platinum(II) complexes is distinctly different from those of cisplatin, trans-diamminedichloroplatinum(II), and other cis-platinum(II) agents.198,199 The presence of bulky planar ligands increases the propensity for monofunctional adduct formation and subsequent interstrand cross-linking. Monofunctional adducts formed by members of the trans-[PtCl2(NH3)L] series depicted in Chart 7 on short duplex DNA induce conformational changes similar to those produced by cisplatin.200 In vitro studies in cultured breast cancer cells showed that the complexes formed DNA-topoisomerase I cross-links capable of triggering DNA strand breaks and apoptosis.186,201 Such ternary DNA-protein cross-links are not observed for cisplatin201 and therefore could explain, in part, the distinctive cellular response evoked by trans-platinum(II) complexes with bulky planar ligands.
Chart 7.
Chemical structures of biologically inactive and active trans-platinum(II) agents.
4.1.2. trans-Platinum(II) complexes with iminoether ligands
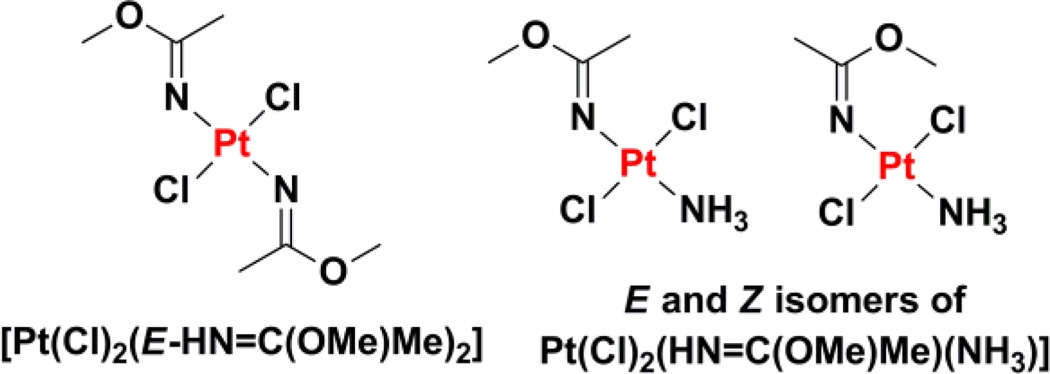
trans-Diamminedichloroplatinum(II) analogues with one or two iminoether ligands exhibit promising biological activity.202–204 Iminoether ligands exist as isomers because of different possible E and Z configurations about the C=N bond. As a result, iminoether complexes of platinum(II) produce not only cis and trans isomers, but also exhibit additional isomersism at the coordinated ligand.205,206 The trans-EE-[PtCl2(HN=C(OMe)Me)2] complex was the first member of the series to be studied in detail (Chart 8). This complex displays inhibitory effects against a panel of cancer cell lines comparable to those of cisplatin.206 Furthermore, this compound exhibits no cross-resistance with cisplatin in ovarian cancer cells and substantial in vivo activity in P388 leukemia-bearing mice.207,208 The cellular uptake and degree of DNA platination was significantly higher for iminoether bearing trans-platinum(II) complexes compared to cisplatin, and suggested that DNA was the main intracellular target.208 The complexes form stable monofunctional adducts with duplex DNA,209,210 which bend the DNA backbone axis toward the minor groove.211 As a result of this form of conformational distortion, the monofunctional adducts were not recognized by HMGB proteins, and thus were readily removed by NER.211 Conversion of the monofunctional adducts into DNA-protein cross-links, however, produced lesions that were able to bypass NER, inhibit DNA polymerases, and lead to cell death. SAR studies established that trans-platinum(II) analogues with one iminoether ligand were less toxic than those with two such ligands.204 Within the trans-[PtCl2(HN=C(OMe)Me)(NH3)] series (Chart 8), the E configuration exhibited greater inhibitory potency against cancer cells than the Z configuration, indicating that iminoether ligand configuration is a major determinant of activity. To systematically study the effect of iminoether ligand conformation on trans-platinum(II) activity, trans-platinum(II) complexes bearing cyclic iminoether ligands mimicking the E and Z configuration were prepared. Cyclic ligands avoid complications that arise from the isomerization between E and Z configurations encountered in acyclic iminoether compounds.212 Mechanistic studies found that, like trans-diamminedichloroplatinum(II), the trans-[PtCl2(HN=C(OMe)Me)(NH3)] series formed monofunctional adducts that developed into interstrand cross-links between adjacent guanine and cytosine bases.204,213 The trans-[PtCl2(HN=C(OMe)Me)(NH3)] lesions have been likened to a flexible hinge, inducing different structural effects on DNA than the more rigid trans-diamminedichloroplatinum(II) lesion.204 More recently another generation of platinum complexes mimicking iminoether derivatives were investigated. These trans-platinum(II) complexes bearing one or two ketamine ligands (acetonimine) exhibited micromolar toxicity against cancer cells and circumvented cisplatin resistance in ovarian cancer cell lines (A2780cisR and 41McisR).214
Chart 8.
Chemical structures of trans-platinum(II) agents with one or two iminoether ligands.
4.1.3. trans-Platinum(II) complexes with asymmetric aliphatic amine ligands
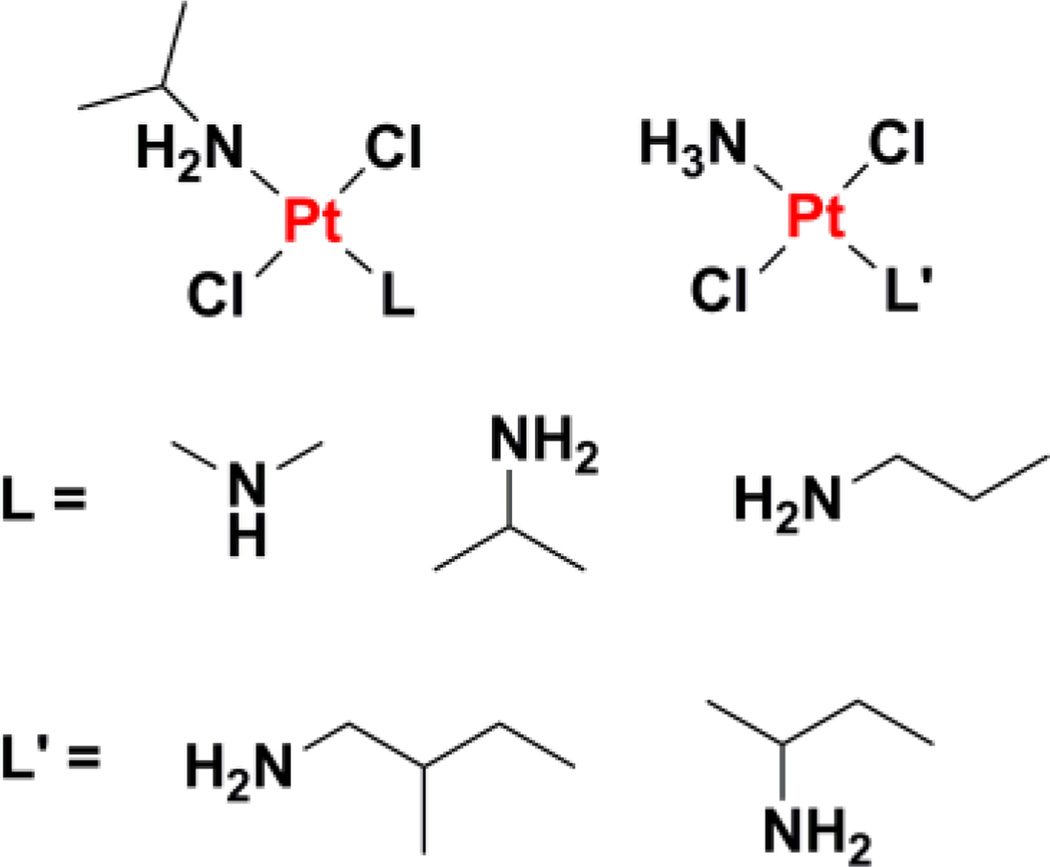
Aliphatic amine ligands have been employed to generate asymmetric platinum(II) agents such as trans-[PtCl2(isopropylamine)L] where L = dimethylamine, isopropylamine, or propylamine (Chart 9), that display potency against cancer cells with multifactorial cisplatin resistance and ras oncogene overexpression.215,216 In a similar fashion to trans-diamminedichloroplatinum(II), trans-[PtCl2(isopropylamine)(dimethylamine)] forms interstrand cross-links between guanine and a complementary cytosine but the quantity of lesions formed is 3-fold higher for the former.217 The ability of this complex to form interstrand cross-links over a relatively short period of time is claimed to be the major contribution to overcoming cisplatin and ras-related resistance. trans-Platinum(II) complexes with one aliphatic amine ligand, such as trans-[PtCl2(NH3)L] where L = 2-methyl-butylamine or sec-butylamine, have been prepared with the aim of improving the water solubility of the parent di-aliphatic amine complexes.218 The second generation complexes retained the cytotoxicity profile of the original series, including the ability to form efficient interstand cross-links and bypass cisplatin resistance.
Chart 9.
Chemical structures of trans-platinum(II) agents with one or two aliphatic amine ligands.
4.2. Polynuclear compounds
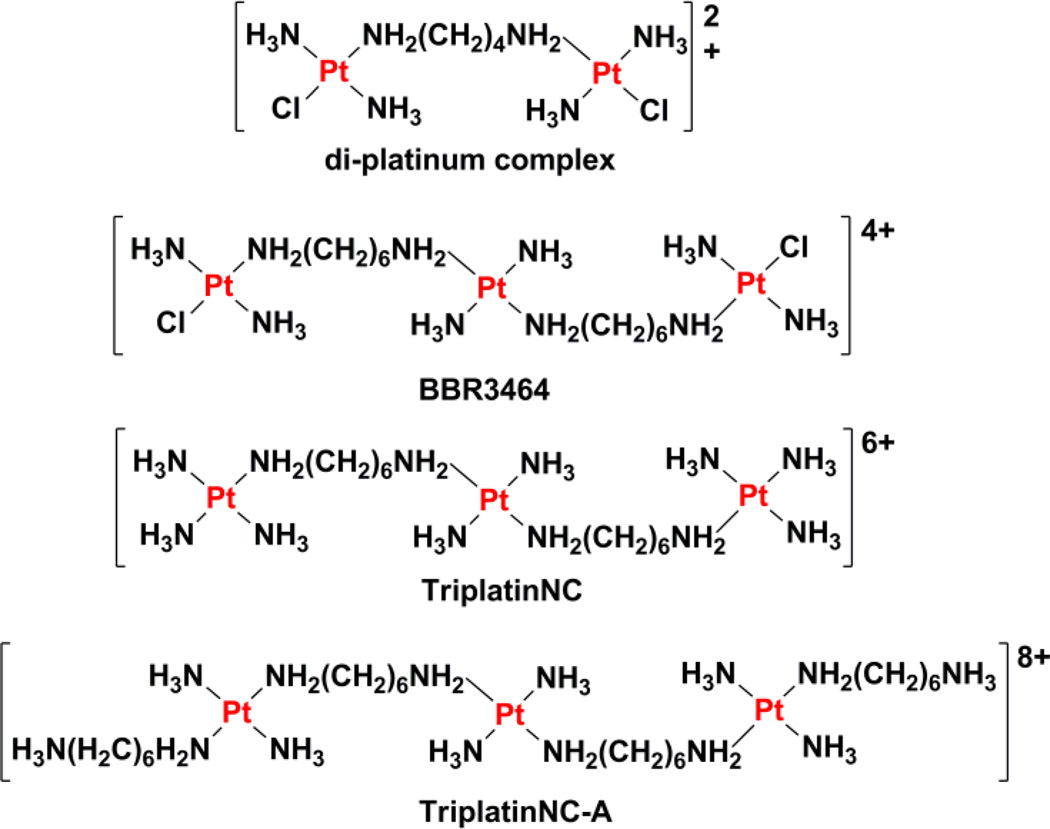
Polynuclear platinum agents that share similarities with trans-platinum(II) complexes account for another class of pharmacologically active platinum-based anticancer agents.219–222 Such compounds contain trans-[Pt(NH3)2Cl] units with bridging alkanediamine linkers of various lengths, designed to facilitate long-distance, flexible intrastrand and interstrand cross-links, which are unattainable by traditional mononuclear platinum(II) agents like cisplatin and trans-diamminedichloroplatinum(II) (Chart 10). The di-platinum complex trans-[(PtCl(NH3)2)2µ-(H2N(CH2)4NH2)]Cl2 forms 1,2-, 1,3-, and 1,4-interstand cross-links between guanines on opposite strands.223,224 In 1,3- and 1,4-cross-links, the guanines are separated by one and two base pairs, respectively, whereas the 1,2-cross-link is formed between guanines on neighboring base pairs.223 These unconventional DNA adducts enable the di-platinum complex to overcome cisplatin resistance in ovarian cancer cells.221 In order to improve the DNA binding ability of the di-platinum(II) complex, tri-nuclear platinum(II) complexes were prepared by incorporating a third platinum center within the alkanediamine linker.225 After systematic SAR studies, [trans-diamminechloroplatinum(II)][µ-trans-diamminedihexanediamineplatinum(II)] nitrate (BBR3464) was selected for preclinical development. BBR3464 is taken up in large amounts by cancer cells, and forms characteristic DNA cross-links, which mediate its cytotoxic effect.226,227 The major DNA adduct formed is the 1,4-interstand cross-link. This lesion induces directional bending of the DNA helical axis and local unwinding of the helix. Moreover, this lesion evades removal by NER. The DNA lesions formed by BBR3464 were not detected by antibodies raised against cisplatin-modified DNA but were recognized by antibodies raised against trans-diamminedichloroplatinum(II)-modified DNA, indicating that BBR3464 may exhibit greater mechanistic similarities with trans-platinum(II) complexes.225 Other studies claim that BBR3464 induces DNA damage by triggering B-to-Z and B-to-A transitions in DNA conformation.228,229 In vitro and in vivo toxicity studies showed that BBR3464 is able to kill cancer cells up to three orders of magnitude better than cisplatin, and to overcome cisplatin resistance in several types of cancer.219,226, 230–232 The fact that BBR3464 acts independently of p53, a tumor suppressor protein that is mutated, defective, or inactivated in several cancers, suggested that it holds great therapeutic potential. The major drawback of BBR3464 is its inherent systemic toxicity. Phase I trials in humans revealed that the MTD is as little as 0.12 mg m−2 day−1 on a daily, five-dose schedule.233 Upon escalating the dose to 0.17 mg m−2 day−1, severe myelosuppression and gastro-intestinal toxicity were observed. On the other hand, low urinary excretion and low nephrotoxicity were reported. Phase II trials in patients with ovarian cancer, small cell lung cancer, non-small cell lung cancer, gastric, and gastro-esophageal adenocarcinoma, produced mixed results.234–237 Although BBR3464 displayed a distinct lack of activity in gastric and small cell lung cancers, it showed better activity in non-small cell lung cancer patients and advanced ovarian cancer patients failing platinum-taxane regimens. In the latter case, BBR3464 displayed hints of activity deserving of further evaluation (16 partial responses out of 79 patients). The results of two Phase II trials launched over 10 years ago by Theradex, for the treatment of patients with locally advanced or metastatic pancreatic cancer and small cell lung cancer have yet to be realeased. We would also like to highlight that BBR3464 was the most recent novel platinum complex to have begun clinical trials. No new small molecule platinum agents have been introduced into clinical trials since 1999.
Chart 10.
Chemical structures of di- and tri-nuclear platinum agents. The pendent aliphatic groups of TriplatinNC-A are shown in the protonated state, raising the overall charge of the complex to 8+.
Non-coordinating multinuclear platinum(II) agents, based on BBR3464, have been reported to exhibit strong DNA binding affinity and anticancer activity. These agents were generated by replacing the labile chloride ligands on BBR3464 with ammine groups (TriplatinNC) or extended amine groups (TriplatinNC-A) (Chart 10).238 X-ray crystallographic studies revealed that TriplatinNC and TriplatinNC-A bind non-covalently (via electrostatic and hydrogen-bonding interactions) to DNA. The terminal platinum units form discrete amine-phosphate-ammine binding motifs called “phosphate clamps” within the minor groove, which induce B-to-A and B-to-Z conformational changes in canonical DNA sequences.238–240 The conformational change cooperatively enhances the interaction of minor-groove binders like Hoechst 33258, and remains unperturbed in the presence of intercalators, such as ethidium bromide.241,242 Recent work has shown that TriplatinNC and TriplatinNC-A can also condense DNA and induce aggregation of small transfer RNA molecules, owing to the highly cationic nature of the platinum complexes.243,244 Furthermore, these agents inhibit topoisomerase-I-mediated relaxation of supercoiled DNA. In light of these findings, the biological mechanism of action of TriplatinNC and TriplatinNC-A is thought to involve nucleic acid condensation or aggregation, with inhibitory effects on topoisomerase-I enzymatic activity.243 Cellular studies showed that TriplatinNC and TriplatinNC-A display micromolar toxicity against cisplatin-sensitive and cisplatin-resistant ovarian cancer cells.245 The ability of the agents to overcome cisplatin-resistance was accredited to their high cellular accumulation, presumably because of their cationic nature, and their unique mode of binding to DNA. Molecular biology assays showed that the downstream cellular responses evoked by TriplatinNC and TriplatinNC-A in mastocytoma cells were markedly different.246 While TriplatinNC induces caspase-mediated apoptosis reliant on p53 and BAX (a pro-apoptotic protein) function, in a similar manner to cisplatin and BBR3464, TriplatinNC-A induces cell death in a manner that is independent of p53- or BAX-status. Given the role of p53 and BAX in cisplatin-mediated cell death, TriplatinNC-A could hold significant clinical value if the results can be recapitulated with in vivo models.
4.3. Non-covalent binding
Mononuclear platinum(II) compounds that bind to DNA non-covalently have also shown promising antineoplastic properties and are gaining increasing support as potential alternatives to conventional platinum drugs.247–252 Metallointercalators with π-conjugated heterocyclic ligands, such as bipyridine, terpyridine, and phenanthroline, utilise π-π stacking and dipole-dipole interactions to intercalate between base pairs in double-stranded DNA.253–255 Metallointercalators can unwind, bend, and distort DNA topology, and it is their structural effect on DNA that is thought to mediate their antiproliferative properties. Systematic studies on charged platinum(II) complexes of general formula [Pt(IL)(AL)]2+, where IL is an intercalating ligand and AL is an ancillary ligand, have yielded some of the most promising results (Chart 11).251 Combinations of phenanthroline-based ligands (IL), and 1,2-diaminocycloalkane ligands (AL) gave impressive cytotoxicities against L1210 murine leukaemia cells. Detailed SAR studies using different R, R- and S, S-ancillary ligands revealed that chirality was a major determinant of toxicity.252 In terms of metallointercalators with diaminocyclopentane (DACP) ligands, R, R enantiomers are more potent than S, S enantiomers whereas the reverse enantiomeric specificity was observed for metallointercalators with DACH ligands.248,252 The latter observation is in stark contrast to the activity of clinically administered oxaliplatin, which contains a R, R-DACH ligand.70 Within the PHENSS/RR series (made up of 1,10-phenanthroline and DACH ligands), the S, S enantiomer (PHENSS) exhibited one order of magnitude greater toxicity against leukaemia cells than the corresponding R, R enantiomer and cisplatin. Given the encouraging biological activity of PHENSS, the compound was evaluated in PC3 xenograft mouse models. In vivo studies found that PHENSS was relatively non-toxic, and somewhat effective at reducing tumor growth over a period of 20 days as compared to saline controls.250 Unfortunately, statically significant results were not obtained because the studies were carried out with a small number of mice. More detailed studies are needed to determine the complete in vivo potential of PHENSS. Recently, the 56MESS/RR series, made up of 5,6-dimethyl-1,10-phenanthroline and DACH ligands, has emerged as a highly promising anticancer candidate. Strikingly, the S, S enantiomer (56MESS) displays nano-molar toxicity toward leukaemia cells. Although this compound interacts with DNA, the significance of DNA as an important cellular target has been questioned.256,257 Comprehensive mechanistic analysis of 56MESS in Madin Darby Canine Kidney (MDCK) cells revealed an increase expression of the mitochondria-associated protein labeled by MTC02, cell cycle arrest in synchronised and non-synchronised cells, and caspase-independent cell death. Collectively these observations suggest that the mechanism of cytotoxic action involves mitochondrial and cell cycle proteins rather than DNA.257
Chart 11.
Chemical structure of platinum(II) complexes that bind to DNA through non-covalent interactions.
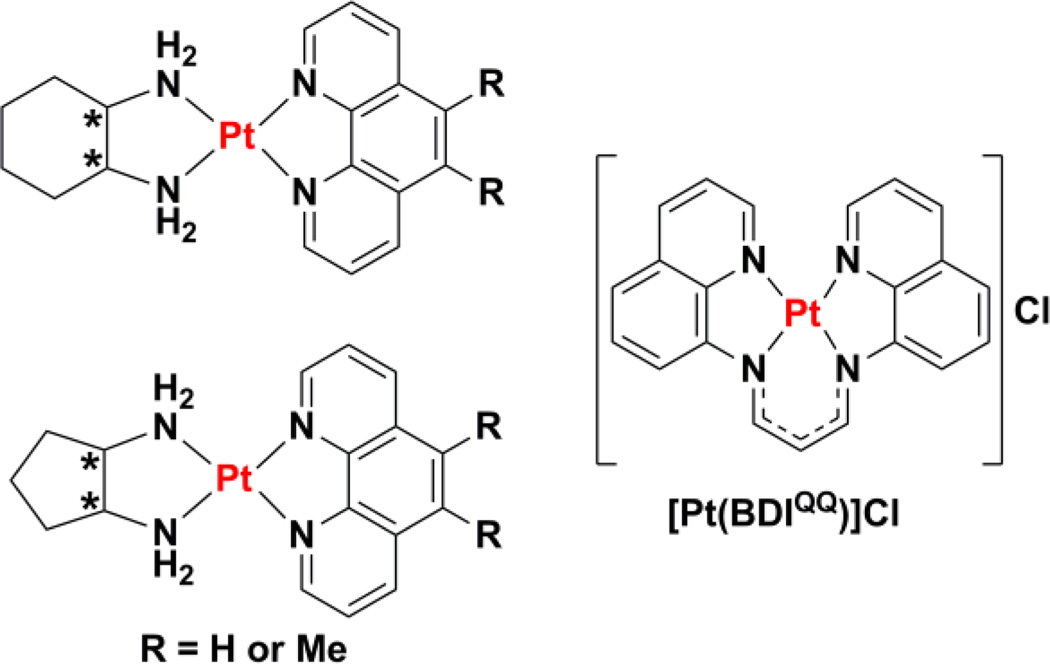
The cytotoxic potential of a planar platinum(II) complex bearing a tetradentate ligand, BDIQQH, was reported recently.258 In aqueous buffer, the platinum(II) complex [Pt(BDIQQ)]Cl (Chart 11), forms aggregates, but in the presence of DNA, the aggregates disperse yielding single molecules capable of intercalating between base pairs and unwinding DNA. [Pt(BDIQQ)]Cl exhibits selective toxicity for cancer cells over normal fibroblast cells, and no cross-resistance with cisplatin in ovarian cancer cells. In a manner uncharacteristic of platinum(II) complexes, [Pt(BDIQQ)]Cl acts in a dual-threat manner.259 As well as attacking DNA in cells, which enriches p53 and BAX levels and subsequently induces mitochondria-mediated apoptosis, Pt(BDIQQ)]Cl also accumulates in mitochondria because of its lipophilic and cationic properties and causes direct mitochondrial damage. The latter mechanism proceeds independently of p53 and therefore allows [Pt(BDIQQ)]Cl to exhibit equal toxicity in p53-negative and p53-postive cells. Because p53 activity is abrogated in many cancers,260 the p53-independence of [Pt(BDIQQ)]Cl is thought to be highly desirable in terms of preclinical development.
4.4. Monofunctional complexes
Mononuclear monofunctional platinum(II) complexes represent another class of very propitious anticancer agents. These complexes contain only one labile ligand and are expect to form only one covalent bond to DNA. Unlike the complexes described above, which can form monofunctional adducts as well as bifunctional adducts, the class of compounds described here is designed to form at most one bond to DNA. Early studies found monofunctional platinum(II) complexes such as [Pt(NH3)3Cl]+ and [Pt(dien)Cl]+ to be inactive both in vitro and in vivo.60,261 These results were in agreement with the prevailing viewpoint at the time, which stated that only neutral, square-planar platinum(II) complexes with a pair of inert ligands in a cis arrangement could have anticancer activity. This belief was overturned, in part, by work conducted by Engelhard Industries, which showed that monofunctional platinum(II) complexes of the form cis-[Pt(NH3)2(Am)Cl]+, where Am is an aromatic N-heterocyclic amine, inhibited tumor cell growth in vitro and in L1210 and P388 mouse leukemia models.262 Two platinum(II) complexes, cis-[Pt(NH3)2(9-aminoacridine)Cl]+ and cis-[Pt(NH3)2(chloroquine)Cl]+, capable of binding to DNA both in a monofunctional covalent manner and via intercalation were prepared following these studies,263 but unfortunately murine sarcoma 180 ascites (S180a) screens found both complexes to have high dose-limiting toxicity (5 mg kg−1), and any useful antineoplastic activity was masked by this systemic toxicity. The in vivo properties of these complexes were not investigated further.
Further studies with cis-[Pt(NH3)2(Am)Cl]+ complexes investigated the possibility that an ammine or Am group could be lost upon DNA binding, allowing bifunctional coordination.264 Careful analysis of NMR spectra and enzymatic digestion data on the products of the reaction of these complexes with d(GpG) and dG revealed the formation of monofunctional adducts only with no evidence for the release of the ammine or Am groups.264 This result was supported by the fact that monoclonal antibodies capable of detecting bifunctional DNA lesions did not recognize cis-[Pt(NH3)2(Am)Cl]+ induced DNA adducts. Electrophoretic mobility shift assays showed that the adducts induced by cis-[Pt(NH3)2(N3-cytosine)Cl]+ had minimal structural effects, in fact, the DNA helix remained rod-like after treatment.265 Subsequent studies on the interaction of a structurally similar complex, cis-[Pt(NH3)2(4-bromopyridine)Cl]+ with supercoiled DNA revealed that monofunctional adducts not only bent DNA less but also unwound DNA less than traditional bifunctional complexes.266 A further vindication of the differing structural effects of monofunctional and bifunctional adducts was shown by the ability of HMGB proteins to recognize cisplatin modified DNA but not cis-[Pt(NH3)2(N3-cytosine)Cl]+ platinated DNA.267
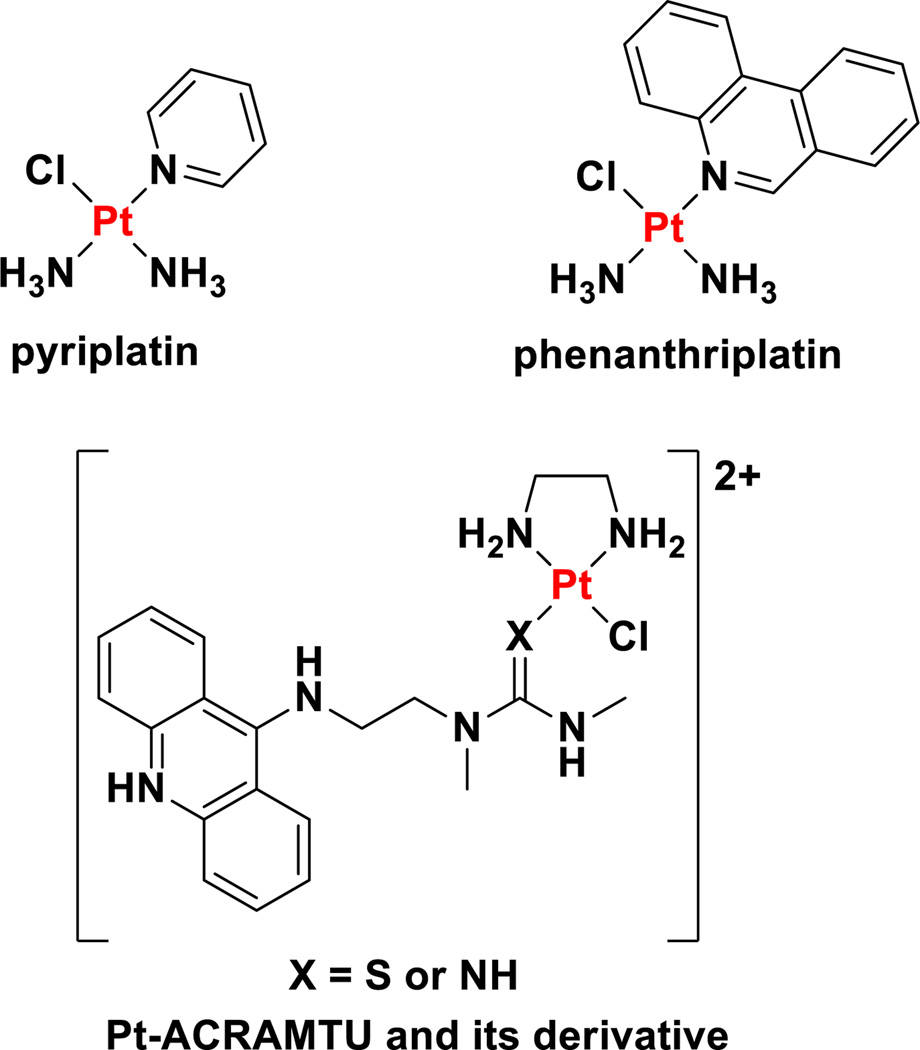
In spite of the aforementioned work on monofunctional platinum(II) complexes, no notable results of experiments with these compounds were reported for almost two decades, until a re-examination of these cationic monofunctional platinum(II) complexes arose unexpectedly from studies on the role of OCTs in the cellular uptake and activity of oxaliplatin.23,268 These studies uncovered the fact that cis-[Pt(NH3)2(pyridine)Cl]+ (pyriplatin, Chart 12), a monofunctional, cationic platinum(II) compound, displayed excellent substrate compatibility with organic cationic transporters 1 and 2.269 Cells with high OCT expression were more sensitive to pyriplatin treatment than those with low OCT expression. Moreover, the differential toxicity in pairs of cell lines with high and low OCT expression was up to 137-fold for pyriplatin as compared to toxicity enhancements of up to 53-fold for oxaliplatin.269 Electrophoretic mobility shift assays showed that monofunctional DNA adducts formed by pyriplatin did not significantly unwind duplex DNA. Structural determination of a DNA dodecamer duplex site-specifically platinated with pyriplatin at the N7 site of a deoxyguanosine residue also revealed minimal perturbations to the structure of the DNA double helix. In fact, hydrogen bonding between the platinated guanine base and the complementary cytosine base was completely intact.269 In vitro studies in HOP-62 lung adenocarcinoma cells revealed that pyriplatin damages DNA, blocks cell cycle progression at the G2/M phase, and prompts apoptotic cell death.270 Monofunctional lesions formed by pyriplatin are repaired by NER, however, not with the same fidelity as bifunctional lesions.271 NCI cytotoxicity screening studies revealed that pyriplatin exhibits a novel spectrum of activity compared to other platinum agents in the NCI database. The poor potency of this compound relative to conventional platinum-based drugs, however, motivated a search for more active analogues. Elucidation of the X-ray crystal structure of RNA polymerase II stalled at a monofunctional pyriplatin-DNA adduct directed the strategy for making improved analogues.272 This crystal structure indicated that larger N-heterocyclic ligands could more effectively block the progression of RNA polymerase II, leading to improved transcription inhibition and cytotoxicity. The pyridine in pyriplatin was therefore rationally substituted for more expansive N-heterocycles. This process eventually led to the development and discovery of cis-[Pt(NH3)2(phenanthridine)Cl]+ (phenanthriplatin, Chart 12).273
Chart 12.
Chemical structures of monofunctional platinum(II) complexes.
According to the NCI cytotoxicity screening assay, phenanthriplatin exhibited a unique cancer cell-killing profile compared to all other platinum agents held in the NCI archives.273 Unlike pyriplatin, the potency of phenanthriplatin toward cultured cancer cells is therapeutically relevant and significantly higher than that of cisplatin. Upon loss of the chloride ligand, phenanthriplatin forms monofunctional adducts with simple guanine bases as well as duplex DNA. Phenanthriplatin-DNA adducts introduce steric hindrance within the major groove and thus stall RNA polymerase II. Kinetic studies on site-specifically platinated DNA showed that the insertion of CTP opposite the platinated guanine by RNA polymerase II occurs in an error-free manner, but further mRNA synthesis along the template DNA strand is halted.274 Phenanthriplatin-DNA adducts also inhibit DNA polymerases. DNA polmerase η, a translesion synthase capable of bypassing 1,2-intrastand cross-links formed by cisplatin, is able to insert the correct nucleotide opposite the phenanthriplatin-bound guanine, but is unable to proceed any further.275 The detailed mechanism of DNA polmerase η inhibition was inferred from X-ray crystallographic data, which suggested that the diastereoselectivity imposed on the adduct by the phenanthridine ring may play a significant role in blocking polymerase progression.275,276 Studies with Escherichia coli resembling those conducted by Rosenberg, showed that akin to cisplatin treatment, phenanthriplatin induced filamentous cell growth.277 Monofunctional platinum(II) complexes with little biological activity in cultured cancer cells were not able to replicate this result. Phenanthriplatin-mediated filamentous E. coli growth resulted from the bacterial SOS response, indicative of DNA damage. So far, data acquired for phenanthriplatin in cultured systems suggest that its anticancer activity is exerted through interaction with DNA.
Platinum(II) complexes with tethered acridine units represent another important class of DNA-targeting anticancer agents. Such complexes contain a platinum moiety capable of forming monofunctional-DNA adducts and a planar acridine motif capable of intercalating between base pairs (Chart 12).278–280 A semi-rigid linker is usually employed to promote platination of DNA bases directly adjacent to the intercalation site. An early example of this series, Pt-ACRAMTU, [PtCl(ethane-1,2-diamine)(ACRAMTU)] where ACRAMTU=1-[2-(acridin-9-ylamino)ethyl]-1,3-dimethylthiourea, contains a chloride leaving group and an ACRAMTU group coordinated via a Pt-S bond, disposed cis to each other.279 This arrangement was hypothesised to enable rapid DNA adduct formation without the need for rate-limiting aquation. The NMR solution structure of a site-specifically modified octamer containing a Pt-ACRAMTU adduct, revealed that platination of a guanine within the major groove did not result in large structural changes. Intercalation of the appended acridine unit, however, did lengthen (6.62 Å) and unwind (twist, 15.4°) DNA.281 The authors propose that rapid intercalation precedes platination, and that this mechanism is responsible for moving platinum away from DNA sites targeted by conventional platinum(II) agents. Clonogenic growth and cell proliferation studies showed that Pt-ACRAMTU and its derivatives were very active and display sub-micromolar IC50 values against several cancer cell lines.282–287 Polymerase stop assays and flow cytometric assays showed that the hybrid adducts inhibited RNA polymerase II and DNA synthesis.285,286,288 Inhibition of DNA synthesis led to S phase cell cycle arrest as opposed to G2/M phase cell cycle arrest, as is customary following cisplatin treatment.288 Although Pt-ACRAMTU maintained sub-micromolar activity in cell lines with aberrant p53 and k-ras expression, because of their inability to stop tumor growth in corresponding tumor mouse models, preclinical development was halted and other Pt-ACRAMTU analogues have been subsequently investigated.289 The most promising second generation derivative was developed by replacing the thiourea donor on Pt-ACRAMTU with an amidine nitrogen.290,291 This modification accelerated DNA binding, increased cancer cell toxicity by two orders of magnitude (nanomolar IC50 values), and inhibited tumor growth in vivo.290,291 Studies in non-small cell lung cancer cells (NCI-H460) suggest that their impressive cytotoxicities can be attributed to rapid intracellular accumulation, DNA adduct formation, and less efficient removal of the DNA adducts. Unfortunately the amidine-based complexes exhibit extremely high systemic toxicity in vivo; the maximum tolerated dose determined for one of the most potent complexes being 0.5 mg kg−1. It is possible that the development of delivery agents could allow for safe delivery while maintaining potency.
4.5. Other examples
Bone and other calcified tissues show a great propensity to take up bisphosphonates, a property that has been exploited in the design of a number of drugs that target bone diseases.292–294 The ability of the bisphosphonate unit to chelate calcium confers this targeting ability. In an effort to develop cisplatin analogues that selectively target bone tissue, a series of platinum(II) complexes were prepared with the chelating 2-amino(m)ethylpyridine ligand functionalized at the amine with a bisphosphonate unit.295 In vitro cytotoxicity assays reveal that these complexes are less active than cisplatin. Flow cytometric studies suggest that cisplatin and the bisphosphonate complexes differentially influence the membrane permeability that is assayed to probe apoptosis. In conjunction with a lack of observed DNA binding, the authors interpret these results to mean that an alternative cell killing mechanism may be operative. It is also possible to load platinum complexes with bisphosphonate leaving group ligands on a solid matrix for implantation and release at the site of disease. Many of the developments in this area have recently been reviewed.296
A series of platinum complexes with pyrophosphate ligands in place of the traditional nonleaving group ligands afforded complexes that did not bind to DNA but in some cases were more active that the parent drug from which they were derived.297 The authors accumulate evidence highlighting the distinctions between the biological effects of the classical platinum drugs and the pyrophosphate complexes, but no mechanistic model has yet to be proposed.298
Platinum(II) complexes with diethyl[(methylsulfinyl)methyl]phosphonate as the non-leaving group ligands demonstrate low potency killing but in addition to DNA binding, as revealed by replication mapping experiments, these complexes were also found to be potent inhibitors of matrix metalloproteinases.299 Similarly, inhibition of matrix metalloproteinase 3 (MMP-3) was observed with other platinum complexes in which three labile ligands are present in the platinum coordination sphere. The authors present evidence indicating that the platinum binds to a key histidine residue of the enzyme.300 A crystal structure of matrix MMP-3, also known as stromelysin-1, confirmed binding to this histidine.301
5. Platinum(IV) prodrugs that release classical platinum(II) anticancer agents
The anticancer potential of platinum(IV) agents has been recognized since the discovery of the medicinal potential of cisplatin,55 but their clinical significance has only been realized more recently. The physicochemical properties of platinum(IV) agents differ greatly from their platinum(II) counterparts. Unlike square-planar platinum(II) complexes, platinum(IV) complexes generally adopt octahedral geometries (Figure 5). The saturated coordination sphere of platinum(IV) is more resistant to ligand substitution than four-coordinate platinum(II) centers, and thus limits unwanted side reactions with biomolecules prior to DNA binding. The two extra ligands afforded by the low-spin d6 platinum(IV) center provide a means to impart and fine-tune desirable biological properties such as lipophilicity, redox stability, specific targeting, orthogonal bioactivity, and improved cellular uptake. The additional ligands also facilitate attachment to nanoparticles and other forms of carriers systems, a detailed discussion of which will be provided later.
Figure 5.
The composition of platinum(IV) prodrugs. Adapted from reference302. Copyright © 2014, The American Chemical Society.
Interpretation of the reactivity of platinum(IV) complexes with DNA and other biomolecules requires consideration of their rates of substitution and reduction. Although platinum(IV) complexes can platinate DNA in their oxidized form, the formation of cytotoxic lesions via ligand substitution requires weeks.303 Given that platinum agents are typically cleared from the body in a matter of hours, direct platinum(IV)-biomolecule coordination is unlike to be of clinical importance.304 Reduction of the platinum(IV) center to platinum(II), in concert with the loss of two ligands, is thought to be essential for the anticancer activity of these agents. Specifically, the canonical mechanism of reduction involves the loss of the two axial ligands (Figure 5). The resulting divalent form, usually cisplatin or a related derivative, binds to DNA, inhibits transcription and replication, and induces apoptosis. The reduction process is dependent on the composition of the platinum(IV) agents as well as the biological reducing agent involved. A convenient measure of reduction can be obtained from voltammetric experiments, but because the platinum(IV)-to-platinum(II) reduction is irreversible, a standard redox potential cannot be obtained as the mid-point potential from a typical cyclic voltammogram. Instead, the peak potential, Ep, for the cathodic wave obtained in a cyclic voltammetric measurement is typically quoted. Care must be taken in interpreting these values, however, because the cathodic peak potential of an irreversible process is not dictated solely by the thermodynamics of reduction, but also by the heterogeneous charge transfer rate constant at the electrode surface and is influenced by the scan rate.305 The relative cathodic peak potentials of structurally related complexes are, however, typically correlated to the relative facility with which the metal center is reduced and have been correlated with reduction rates in solution.306 Early quantitative SAR studies showed that the nature of the axial ligands has a stronger influence on reduction rates than the equatorial ligands.307 Within the cis-[Pt(en)Cl2X2] series where X is an anionic axial ligand, the chloride species (Ep = −4 mV) is much more susceptible to reduction than the carboxylate (Ep = −250–350 mV) or hydroxide (Ep = −664 mV) derivatives.308,309 On the other hand, systematic variation of equatorial amine ligands in complexes of general formula cis-[PtCl4(NHR2)2] where R = alkyl and aromatic hydrocarbons, did not have a considerable effect on the reduction rates.310 Platinum(IV) reduction by small biomolecules such as ascorbate and glutathione can occur by inner- or outer-sphere electron-transfer mechanisms.311–314 The reduction path taken is highly dependent on the nature of the ligands coordinated to the metal. Because inner-sphere electron-transfer requires the formation of a chemical bridge between the participating species, platinum(IV) agents with halide or hydroxide ligands trans to a good leaving group are particularly suited to undergo reduction by this mechanism.313 Differences in ability to form such bridges can even override trends in reduction rate expected on the basis of differences in ligand-to-metal electron donating ability.315 Bimolecular outer-sphere electron-transfer processes are generally slower than inner-sphere electron-transfers but can provide a viable reduction mechanism for platinum(IV) agents in which ligands that are capable of forming a bridge to an electron transfer agent are trans to firmly bound ligands.
Ormaplatin (also known as tetraplatin), tetrachloro(trans-1,2-diaminocyclohexane)platinum(IV) (Chart 13) was one of the first platinum(IV) agents to undergo clinical trials. Ormaplatin is rapidly reduced to the corresponding dichloro(trans-1,2-diaminocyclohexane)platinum(II) form in tissue culture medium (t1/2 = 5–15 min) and undiluted rat plasma (t1/2 = 3 s).316 The active platinum(II) species is similar to oxaliplatin, however, it contains both R, R and S, S isomers. Ormaplatin displayed in vitro and in vivo activity against some cisplatin-resistant cancers and was taken forward to clinical trials commissioned by NCI and UpJohn.317–321 Various doses, dose patterns, and modes of administration (intravenous and intraperitoneal) were investigated in six Phase I clinical trials, however, no Phase II clinical trials have been planned.321–323 Ormaplatin was found to induce severe neurotoxicity at the MTD, and in some cases a safe MTD could not be determined. Toxicity is thought to arise from fast reduction to the active platinum(II) form as a consequence of the axial chloride ligands.
Chart 13.
Chemical structures of platinum(IV) agents that have undergone clinical trials.
Another notable platinum(IV) complex to have undergone clinical trials is iproplatin (also known as JM9 and CHIP), cis, trans, cis-dichlororidodihydroxidobis(isopropylamine)platinum(IV) (Chart 13).324 Iproplatin is structurally similar to ormaplatin in the sense that it contains two equatorial chloride groups which are cis to each other. Carbon-14 labelling studies showed that the mechanism of action of iproplatin involves the reduction of the platinum(IV) center to platinum(II) followed by covalent bond formation with DNA.325 Iproplatin is less prone to reduction and deactivation by biological reducing agents than ormaplatin, presumably because of the presence of hydroxide axial ligands, allowing less hindered distribution throughout the body. Another advantage of iproplatin is its very high water solubility (44.1 mM), which allows simpler formulation and administration. Iproplatin is one of the most clinically studied platinum agents to have not been approved for marketing, with 38 clinical trials ranging from Phase I to III having been concluded, many of which were commissioned by Johnson Matthey and Bristol Myers. Phase I studies revealed that the dose-limiting toxic effect was myelosuppression, which, in one study involving children, was partly correlated with the amount of prior therapy chemo- and radiotherapy received.326 The same study recommended intravenous doses of 324 mg m−2 over 2 h every 3–4 weeks for Phase II trials in children. Other studies proposed doses of 45–65 mg m−2 and 95 mg m−2 for patients treated on a five-times daily schedule every three weeks and a four-times weekly schedule with two-week break periods, respectively.327 Phase II trials were carried out in patients with a variety of different cancer types328–332 and Phase III trials were conducted in ovarian cancer patients and those with metastatic epidermoid carcinoma of the head and neck.333,334 The ultimate conclusion of these studies was that iproplatin did not exhibit overall effectiveness that surpassed that of cisplatin or carboplatin and no further trials were undertaken.
Satraplatin, trans, cis,cis-bis(acetato)amminecyclohexylaminedichloroplatinum(IV) (Chart 13) was the first orally active platinum agent to be reported.335,336 Satraplatin was rationally designed such that the lipophilicity and stability were suitable for oral administration. The half-life of reduction of satraplatin by 5 mM ascorbate is 50 min, which is an adequate time for absorption by the gastrointestinal mucosa in the platinum(IV) form once ingested.337 Upon entry into the bloodstream, satraplatin undergoes reduction to give six distinct platinum(II) species. Ammine(cyclohexylamine)dichloroplatinum(II), derived from the loss of two acetate ligands, is the major metabolite and also exhibits the most potent anticancer activity.338 In preclinical studies, satraplatin exhibited a better toxicity profile than cisplatin, and showed activity in cisplatin-resistant human tumor cell lines.335 Similarly to cisplatin, satraplatin acts through the formation DNA cross-links, DNA distortion, and subsequent inhibition of DNA transcription and replication. The ability of satraplatin to overcome cisplatin resistance is thought to arise from the asymmetric nature of the DNA lesions, which unlike cisplatin adducts, can evade recognition by DNA repair proteins.339–341 In vivo studies in mice bearing murine ADJ/PC6 plasmacytoma, which we note was the same model used to identify carboplatin as a viable alternative to cisplatin,342 showed satraplatin to exhibit markedly superior antitumor efficacy relative to cisplatin, carboplatin, and ormaplatin.335 Furthermore, in four ovarian carcinoma xenograft models of varying cisplatin and carboplatin resistance, satraplatin displayed activity similar to that of cisplatin and carboplatin, which were administered intravenously, and far superior to intraperitoneal administration ormaplatin. In rodents, the dose-limiting toxicity of satraplatin was myelosuppression. Encouragingly, less hepatotoxicity and fewer gastrointestinal effects were observed as compared to treatment with cisplatin or carboplatin. The favorable toxicity profile and preclinical antitumor activity of satraplatin prompted several development companies to begin Phase I clinical trials. In the first Phase I study, satraplatin was administered at doses ranging from 60–170 mg m−2 as a single oral dose.343,344 The pharmacokinetics data suggested that gastrointestinal absorption was being saturated, preventing the MTD from being reached. To improve absorption into the bloodstream, patients were treated on a five-times daily schedule with lower doses (30–140 mg m−2).345 The dose-limiting toxicities were thrombocytopenia and neutropenia and in about 10% of the patients treated, nausea, vomiting, and diarrhea were also observed. Based on the Phase I studies, doses of 100–120 and 45–50 mg m−2 were recommended for repeated daily dosing for 5 and 14 days, respectively, in Phase II/III trials.346–348 Several Phase II/III trials have been carried out to determine the efficacy of satraplatin alone and in combination other active agents. A Phase II study on metastatic NSCLC patients, in which satraplatin was administered as single daily 120 mg m−2 doses for 5 days on 3 week cycles failed to provide any objective responses.349 Nevertheless 46% of the patients were noted to express some palliation. A more advanced Phase II study on patients with small-cell lung cancer and squamous cell head and neck cancer, with escalated doses of satraplatin produced a response rate of 38%, similar to that observed with cisplatin.343 Encouragingly this study found no signs of severe neurotoxicity or nephrotoxicity. Other Phase II studies in patients with relapsed ovarian cancer and advanced/recurrent squamous cancer of the cervix, produced clinically beneficial or partial rates of response in several patients.350,351 The former study noted that the most common form of toxicity was neutropenia and thrombocytopenia. Satraplatin has also been heavily studied as a potential second-line chemotherapeutic for patients with metastatic castration-resistant prostate cancer (CRPC).352,353 Treatment with 120 mg m−2 satraplatin daily for 5 days, used in patients with CRPC that had undergone front-line hormone therapy, resulted in 62% of patients expressing stable disease or partial response. Follow-up Phase III trials commissioned by Bristol-Myers Squibb, assessed the capability of satraplatin as a front-line chemotherapeutic in combination with prednisone.354 This study was terminated after only 50 of the intended cohort of 380 patients were treated because satraplatin in combination with prednisone was found to be less efficacious than prednisone alone. Following this setback, satraplatin was acquired by GPC Biotech, who set up a multicenter, multinational, double-blind, placebo-controlled Phase III trial called Satraplatin and Prednisone Against Refractory Cancer (SPARC) with 950 patients.355,356 The aim of the trial was to compare treatments of satraplatin and prednisone against placebo and prednisone, as second-line therapy for patients who had received a cytotoxic agent, which in some cases was docetaxel. The study found that the satraplatin/prednisone combinations led to a 36% decrease in pain progression and an improvement in progression free survival rates. The positive outcome of the SPARC trial prompted GPC Biotech to file for FDA approval, however, this claim was rejected on the grounds that overall survive was not significantly improved and that more than half the patients in the study had received prior docetaxel treatment.357,358
6. Dual-threat platinum(IV) prodrugs that release classical platinum(II) anticancer agents
In all of the examples discussed above, the platinum(IV) agent undergoes intracellular reduction to produce an active platinum(II) species and two biologically innocent groups. The inclusion of one or two biologically active ligands within the platinum(IV) scaffold can produce dual-threat platinum(IV) agents. The biologically active ligands are typically chosen to have non-DNA targets to limit cross-resistance with the DNA-targeting platinum(II) species released.
Ethacraplatin comprises a cisplatin equatorial core axially coordinated to two ethacrynic acid ligands through their carboxylic acid groups (Chart 14).359,360 Ethacrynic acid and its glutathione adduct are a potent inhibitors of glutathione-S-transferase (GST), an enzyme which aids the detoxification of platinum agents by catalyzing their conjugation to glutathione.361 Upon intracellular reduction, ethacraplatin releases cisplatin and two equivalents of ethacrynic acid. Ethacrynic acid inhibits GST and reverses platinum drug resistance, enabling ethacraplatin to inhibit the growth of cisplatin-resistant breast, lung, and colon cancer cells more effectively than cisplatin alone. The platinum(IV) divalproate complex (VAAP) is another example of a dual-threat agent (Chart 14).362 Upon reduction, VAAP generates cisplatin and two equivalents of valproic acid, a potent histone deacetylase (HDAC) inhibitor which stimulates differentiation and apoptosis in cancer cells. VAAP displays strong synergistic cytotoxicity, up to two orders of magnitude higher than cis, cis, trans-diaminedichlorodihydroxoplatinum(IV) alone or in combination with valproic acid. Furthermore, VAAP-loaded poly(ethylene glycol)-block-poly(caprolactone) nanoparticles (see Section 8) display pharmacologically relevant blood circulation times, with high tumor accumulation and significant inhibitory effects in lung adenocarcinoma xenograft mouse models. More recently, platinum(IV) derivatives of oxaliplatin with one or two valproate axial ligands have been prepared with the aim of developing VAAP derivatives with more favorable toxicity and tolerability profiles.363
Chart 14.
Chemical structure of dual-treat platinum(IV) agents.
Normal cells generate energy within the mitochondria, in the form of ATP, via the citric acid cycle and oxidative phosphorylation. Under the hypoxic conditions present in many tumor tissues, however, cancer cells obtain a larger proportion of their energy by aerobic glycolysis. This phenomenon is known as the Warburg effect.80 The difference in metabolic pathways operational in normal and cancer cells has been exploited by mitaplatin, a platinum(IV) complex designed to selectively kill cancer cells over non-malignant cells (Chart 14).364 Mitaplatin consists of two dichloroacetate (DCA) ligands appended to a cisplatin core. Upon reduction in cancer cells, DCA inhibits pyruvate dehydrogenase kinase (PDK), which in turn, reduces the flux of metabolites through aerobic glycolysis and restores normal mitochondrial function. This process promotes apoptosis by releasing cytochrome c from mitochondria and translocating apoptosis-inducing factor (AIF) to the nucleus. At the same time, free cisplatin induces DNA damage in the usual manner and prompts apoptotic cell death. The concerted action of cisplatin and DCA allows mitaplatin to kill lung carcinoma cells (A549) more readily than normal lung fibroblasts (MRC5) in co-cultured in vitro systems. Subsequent work showed that this mechanism of action was able to overcome cisplatin-resistance in human epidermoid adenocarcinoma and hepatoma cancer cells.365 Detailed biophysical studies investigating the aqueous chemistry of mitaplatin and related platinum(IV) complexes with axial haloacetate ligands, found that, contrary to the typical dogma that platinum(IV) prodrugs are inert to ligand substitution, these the axial ligands of these complexes can be substituted for hydroxide under biologically relevant conditions.366 Isotopic labelling studies revealed that the hydrolysis proceeds via the attack of a hydroxide ion on the platinum(IV) center, and not at the carbonyl of the haloacetate. Importantly, at physiological pH, however, the half-life of hydrolysis is much longer than the rate at which mitaplatin is cleared from the blood of rodents367 and so this process likely has little influence on the in vivo effects of this compound.
One of the major disadvantages of mitaplatin is the widely differing effective doses of the reduced products, DCA and cisplatin, which alone exhibit their characteristic activities at millimolar and micromolar concentrations, respectively. In order to achieve a better match in activity between the intracellular reduction products, DCA was replaced with a vitamin E analog, α-tocopherol succinate (α-TOS).368 α-TOS inhibits the anti-apoptotic proteins Bcl-2 and Bcl-xL in the micromolar range, thereby inducing mitochondria-mediated apoptotic cell death at comparable concentrations to cisplatin. Platinum(IV) complexes comprising cisplatin attached to one or two α-TOS ligands were prepared. The di-substituted derivative was non-toxic, presumably because of its high lipophilicity and susceptibility to entrapment inside the cell membrane. Contrastingly, the mono-substituted derivative (Chart 15A) exhibited potency 7–25 times greater than that of cisplatin across several tumor cell lines. Mechanistic studies revealed that this complex induces nuclear DNA damage and simultaneous mitochondrial membrane depolarization because of inhibition of Bcl-xL-Bax protein-protein interactions.
Chart 15.
Chemical structure of dual-treat platinum(IV) agents bearing vitamin E (A) or estrogen (B) derivatives.
In ER+ breast cancer cells, addition of estrogen increases HMGB1 expression. HMGB1 inhibits repair of cisplatin-induced DNA lesions by preventing DNA repair proteins from accessing the site of damage.369 Thus, co-treatment of unconjugated estrogen and cisplatin enhances the activity of cisplatin.370 Incorporation of two axially coordinated estrogen moieties into cisplatin enabled the preparation of cisplatin-estrogen conjugates capable of concurrently releasing both groups (Chart 15B).371 Given that the estrogen units were modified with ester groups, hydrolysis to generate free estradiol is a prerequisite for activity. ER+ MCF-7 cells treated with the platinum(IV)-estrogen complex displayed an increase in HMGB1 expression to a degree similar to that observed following treatment with estradiol. The activity of the estradiol potentiates the activity of cisplatin. As predicted based on the construct design, HMGB1 levels in ER− HCC-1937 cells were less affected and the IC50 values were comparatively higher (up to 1.8-fold).
Platinum(IV) agents with axially coordinated bioactive peptides have been designed and prepared with the aim of targeted drug delivery to tumors. Several mono- and di-functionalized platinum(IV) complexes with tri- and pentapetides capable of binding to αvβ3 and αvβ5 integrins and aminopeptidase N (APN) on the surface of cancer cells have been prepared.372 Integrins and APN are highly expressed in tumor-induced angiogenesis, and thus the platinum(IV) conjugates are able to selectively target angiogenic tumor cells over primary proliferating endothelial cells. The anti-proliferative effect of the platinum(IV)-peptide conjugates decreased upon co-incubation with αvβ3- and αvβ5-specific peptides and transfection with β3 integrin siRNA, confirming that their activity was mediated by the recognition of specific integrins on the cancer cell membrane surface. A very recent report describes the enhancement that can be obtained by using an axial ligand that displays multiple units of the cyclic c(RGDfK) integrin-targeting peptide.373 In this work, a picoplatin prodrug displaying a tertrameric c(RDGfK) motif was able to accumulate in cells overexpressing αVβ3 and αVβ5 integrins resulting in a 10-fold enhancement in cytotoxicity over cells that do not express these membrane proteins. A cyclic RGD motif was also used in the construction of a theranostic platinum(IV) complex capable of targeting αVβ3 integrins, releasing cisplatin upon reduction, and reporting on the activation of apoptosis using an aggregation-induced emission fluorophore conjugated to a caspase-3 sensitive Asp-Glu-Val-Asp (DEVD) peptide.374 The two different peptides were attached to the two axial positions of a cisplatin prodrug.
Platinum(IV) complexes with analogues of neurotensin and somatostatin have also been reported with the aim of targeting cancer cell lines expressing the corresponding receptors.375 Cytotoxicity studies revealed that, although potency was improved compared to the non-targeting platinum(IV) precursor, cellular uptake was non-specific, presumably because the receptors were unable to recognize the peptides once tethered to platinum(IV). Fusion of cell penetrating peptides (CPPs) such as a TAT (Trans-acting Activator of Transcription) protein fragment with platinum(IV) centers has yielded very active conjugates. The oxaliplatin-TAT monoconjugate displays 39-fold higher potency against cultured cancer cells than the corresponding platinum(IV) analogue with no targeting peptide(s).376 Large peptides like chlorotoxin (CTX) have also been attached to platinum(IV) in a 1:1 ratio for targeting purposes.377 CTX, a 36-amino-acid peptide found in the venom of the deathstalker scorpion,378 binds to functional proteins like matrix metalloproteinase 2 (MMP2), annexin A2, and chloride ion channels, which are overexpressed on certain cancer cell surfaces. The platinum(IV)-chlorotoxin conjugate exhibits higher in vitro toxicity against human cervical HeLa cells than the non-targeted platinum(IV) building block. The difference in potency is attributed to the targeting of annexin A2 and chloride ion channels present on the cell surface of HeLa cells.
7. Platinum(IV) complexes with non-cisplatin-like mechanisms of action
Before it became established that most anticancer-active platinum(IV) complexes function as prodrugs that undergo reduction to release active platinum(II) agents, alternative mechanisms of action for platinum(IV) were envisioned. Because of the steric restraints imposed by the DNA double helix, it seemed unlikely that DNA cross-linking of the cisplatin type was operative,15 although modelling studies suggest that such adducts may be able to form.379 Monofunctional adducts could also conceivably form,380,381 but the generally slow kinetics of ligand substitution at platinum(IV) argue against this process being of great biological relevance. A series of studies have, however, appeared describing the platinum(IV)-mediated oxidation of guanine to 8-oxo-guanine. The first report of this reactivity came from the observation that incubation of Pt(DACH)Cl4 with GMP or dGMP led to the formation of Pt(DACH)Cl2.382 The oxidation of guanine was confirmed as was the ability of the oxidation to occur with guanine bases in double-stranded oligonucleotides. The reactions typically occurred on the order of days and theoretical calculations suggest that the reaction proceeds via a cyclic 5′,8-phosphodiester intermediate.383,384 Analysis of kinetic data revealed that platinum(II) centers can catalyze the reaction, which is initiated by intermolecular nucleophilic attack, e.g. by phosphate.385,386 The platinum(IV) complex cis,cis,trans-[Pt(NH3)2Cl2(OH)2] was reported to cleave double-stranded DNA but this reactivity was subsequently shown to arise from molecules of H2O2 that co-crystallized with the platinum(IV) complex.387,388
Photoactivated chemotherapy offers temporal and spatial control over drug activation and has shown potential for the treatment of several cancers including those of the skin, lung, brain, and esophagus. The activated toxic species is produced by irradiation only where it is required, allowing tumors to be targeted specifically. This approach is advantageous over other therapies such as surgery, radiotherapy, and conventional chemotherapy because, ideally, normal tissue is not affected and the treatment can be repeated as often as required. In an attempt to increase selectivity and lower systemic toxicity of platinum agents, photoactivatable platinum(IV) prodrugs have been developed.389,390 Two main classes of photoactivatable platinum(IV) have been reported in the literature thus far, diiodo- and diazido-platinum(IV) complexes.
The first generation of photoactivatable platinum(IV) complexes were based on iodide as the reducing ligand and ethylenediamine as the nonleaving group (Chart 16).391,392 A bidentate ligand was chosen to prevent photo-induced isomerisation, which could lead to the formation of thermodynamically stable but potentially inactive trans congeners. cis-Diiodoplatinum(IV) complexes exhibit dissociative LMCT/d-d excited states that can be populated by excitation from visible light. It was postulated that population of these states could trigger photoreduction and photosubstitution at the platinum(IV) center. The resulting platinum(II) species were then expected to coordinate to DNA bases and induce apoptosis. The first diiodo-platinum(IV) complex reported was prepared with axial chloride ligands.391 Although this complex was able to irreversibly platinate DNA upon irradiation at 375 nm, a similar effect was also observed in the dark, probably because of facile reduction owing to the poor ability of chloride ligands to stabilize the 4+ oxidation state of the metal. In order to prevent chemical reduction in the dark, hydroxide-, acetate- and methysulfonate-based ligands were introduced at the axial position.392 As anticipated, the modified platinum(IV) complexes had better dark-stability, the methylsulfonate complex being the most stable with a half-life of 72h. Upon irradiation at 375 nm, the complexes underwent photoreduction, giving platinum species with differing DNA platination propensities. Photolysis of the platinum(IV)-acetate complex generated the highest number of platinum-DNA adducts. 1D- and 2D-NMR experiments indicated that photoreduction to the corresponding platinum(II) species was necessary for DNA binding.393 In vitro studies with TCCSUP bladder cancer cells with and without 1.5 h of irradiation showed that the photolysis products were more cytotoxic that the parent platinum(IV) complexes, however, the toxicity differential was not as high as expected.391,392 A statistically significant difference between dark and light IC50 values was only observed for the platinum(IV)-acetate complex. To understand the underlying reasons for the high toxicity of the diiodo-platinum(IV) complexes in the dark, biophysical studies were conducted with sulfur-rich biomolecules.394 NMR analysis showed that glutathione and N-acetylcysteine rapidly reduced the complexes to the reactive platinum(II) form via an inner-sphere mechanism similar to ormaplatin reduction. Given their fast reduction rates in the presence of biologically relevant thiols, diiodo-platinum(IV) complexes were deemed unsuitable for development as photoactivable drugs.
Chart 16.
Chemical structures of photoactivable platinum(II)-diiodo complexes.
Because several transition metal azide complexes are known to be light-sensitive and undergo photosubstitution and photoreduction reactions, the iodide ligands in the aforementioned complexes were substituted for azide ligands to prepare a second generation of photoactivatable platinum(IV) prodrugs. The earliest example was trans-[Pt(N3)2(CN)4]2−, which upon irradiation into the 302 nm LMCT band led to trans elimination of the azide ligands via azidyl radical formation and reduction to platinum(II).395 The two radicals can rapidly decompose in aqueous solution to produce molecular nitrogen. This process prevents re-oxidation of the platinum center, unlike halogen-based radicals that do not decompose in water and instead interact with the metal center to regenerate the starting material. Early biologically active platinum(IV)-diazide complexes were prepared with the aizde ligands disposed cis to one another, trans to ammine or ethylenediamine non-leaving groups, and cis to axially-coordinated hydroxide ligands (Chart 17).396 Unlike the diiodo-platinum(IV) complexes, the azide-bearing complexes were stable toward hydrolysis for up to 90 days and did not react readily with glutathione, in the dark, over the course of several weeks. Sophisticated NMR and biophysical experiments showed that photoreduction was dependent on the non-leaving groups present and the wavelength of light used for irradiation.397,398 Irradiation with blue and UV light triggered the formation of many platinum(II) and platinum(IV) species, indicating that photoreduction was not the sole photochemical process taking place. Indeed, in this system, irradiation is thought to promote photosubstitution, photoisomerisation, and photoaquation. Cytotoxicity studies with 5637 human bladder cancer cells showed that irradiation significantly enhanced the potency of the complexes, from > 300 µM to ca. 50 µM.397 Experiments with cisplatin-resistant 5637 cells indicated that the diazido-platinum(IV) complexes displayed no cross-resistance with cisplatin. Although in vitro assays indicate that the diazido-platinum(IV) complexes can bind DNA upon irradiation and inhibit RNA synthesis,399 fluorescence microscopy studies showed none of the typical signs of apoptosis. This finding implies that the irradiated diazido-platinum(IV) complexes may induce cell death in a manner different from cisplatin.
Chart 17.
Chemical structures of photoactivable cis- and trans-platinum(II)-diazido complexes.
Diazido-platinum(IV) complexes containing azide ligands in a trans arrangement displayed more favorable electronic properties than their cis congeners (Chart 17).398,400 For instance, within the [Pt(N3)2(OH)2(NH3)2] series, the LMCT band was shifted toward the visible region for the trans isomer, allowing activation with tissue-penetrating light. Also, trans,trans,trans-[Pt(N3)2(OH)2(NH3)2] is stable under biological conditions and upon irradiation with red light binds readily to DNA bases and induces toxicity in human HaCaT keratinocytes to a degree similar to cisplatin. Impressive phototoxicity was also observed against cisplatin-resistant cell lines. More recently, diazido-platinum(IV) complexes with higher photocytotoxicity have been prepared by replacing one or two NH3 ligands with pyridine, methylamine, or thiazole (Chart 17).401–403 These complexes are resistant to hydrolysis and reduction in the dark and only become active upon irradiation with UVA or blue light. The photolysis products are highly toxic toward cancer cells and display no cross-resistance with cisplatin in ovarian carcinoma cells. Toxicity is attributed to the formation of a novel combination of mono- and bi-functional DNA adducts, primarily with guanine and cytosine, that unwind DNA. In depth biophysical studies showed that the trans,trans,trans-[Pt(N3)2(OH)2(methylamine)(pyridine)] complex induced oxidation of guanine upon irradiation.404 This unexpected result is thought to arise from the reaction of singlet oxygen and platinum-nitrene intermediates. Extensive fluorescence experiments ruled out singlet oxygen generation from dissolve dioxygen or water. The most plausible source of singlet oxygen was the axially coordinated hydroxide groups. Guanine oxidation is a form of mutagenic DNA damage and so this process could be a contributing factor in the mechanism of action of trans-diazido-platinum(IV) complexes. As singlet oxygen generation and subsequent guanine oxidation do not require any exogenous source of oxygen in this system, it could be applied to target cancer cells that reside in hypoxic niches.
8. Nanodelivery of platinum(IV) complexes
As described above, one prominent paradigm in the design of platinum(IV) anticancer agents is that of a prodrug bearing equatorial ligands identical to those of a platinum(II) complex with established anticancer activity and axial ligands chosen to either modulate the physicochemical properties of the compound or confer additional biological activity. The main impetus for altering the physicochemical properties of the complex is the attendant change that occurs in pharmacological activity. An alternative motive that guides the development of some platinum(IV) prodrugs is the desire to incorporate the complex into a drug delivery device, particularly those with nanoscale dimensions.
Nanodelivery of biologically active agents is a blossoming field at the intersection of materials science, engineering, medicine, and chemistry. The advantages to be gained from any drug delivery system, macroscopic or nanoscale, include the ability to reduce the systemic dose but increase the amount of active agent that reaches the target site. A needle used to inject a drug intratumorally is an example of a macroscopic drug delivery system. Broadly defined, nanoscale drug delivery is the use of any object with dimensions in the nanometer regime to transport pharmaceutically active agents. Nanoparticles can often be engineered to have properties such as sustained circulation, affording any cargo that they transport an enhanced retention in the bloodstream.405 In the nanodelivery of anticancer agents, the main advantages of using nanoparticles relates to their ability to target tumor tissue in either an active or passive manner.406,407 Active targeting can be realized in a manner similar to that described above for small molecules. If the surface of the nanoparticle is decorated with a ligand for a receptor expressed selectively on the surface of cancer cells, then the particle is more likely to be taken up by those cells via receptor-mediated endocytosis.408 Multivalent effects that arise from the presence of multiple targeting unit on the surface of a nanoparticle can also enhance this mode of uptake.409 Passive targeting arises directly from the ability of nanoscale object in sustained circulation to accumulate in tumor tissue over time. This phenomenon, known as the enhanced permeation and retention (EPR) effect, occurs because the tumor vasculature is inherently leaky and the tumor tissue is poorly irrigated by the lymphatic system (Figure 6).410 As a result, nanoparticles with dimensions in the 50–200 nm range can readily extravasate into the tumor interstitial space (permeation) and remain there (retention) releasing their contents into the extracellular space of the tumor microenvironment or being taken up by cancer cells.
Figure 6.
Schematic representation of the accumulation of nanoparticles in tumor tissues as a result of the enhanced permeation and retention effect. Reproduced from reference411. Copyright © 2014, A. M. Jhaveri and V. P. Torchilin (Creative Commons Attribution License).
One broad strategy in the nanodelivery of platinum anticancer agents involves the use of platinum(IV) synthons similar to those used to conjugate platinum(IV) centers to peptides or bioactive small molecules, as described above. In particular, cisplatin prodrugs with axial succinate ligands have enabled the functionalization of a variety of nanoscale objects using simple ester- and amide-bond forming reactions. A range of different platinum(IV) complexes and nanomaterials have been used for this purpose.405,409,412 The following is a review of the sytems that have appeared in the peer-review literature grouped according to the nanomaterial that is used as the delivery vector.
8.1. Carbon-based materials
Carbon nanotubes have been extensively investigated as drug delivery vehicles and a number of platinum(IV) prodrug-containing constructs have been prepared.413 One early example of such platinum(IV) prodrug delivery was the use of single walled carbon nanotubes (SWCNTs) to ferry a cytotoxic platinum payload into cancer cells.414 The SWCNTs were rendered biocompatible and water dispersible via the non-covalent binding of phospholipid-PEG-amine groups. The phospholipid interacted with the nanotube surface, the PEG chain acted as a spacer, and the pendent amine functional group provided a reactive handle through which to couple the pendent carboxylic acid of cis, cis, trans-[Pt(NH3)2Cl2(OEt)(O2CCH2CH2CO2H)] via an amide-bond forming reaction (Chart 18). An average of 65 platinum centers could be conjugated to each SWCNT. Fluorescence microscopy was used to confirm that cultured testicular cancer cells take up the SWCNTs conjugated to both the platinum(IV) prodrug and a fluorescent reporter molecule, and trap the nanotubes within endosomes. The platinum is then released into the rest of the cell. Subsequent studies with this nanotube system used the disuccinate complex, cis, cis, trans-[Pt(NH3)2Cl2(O2CCH2CH2CO2H)2] to allow conjugation to not only the functionalized carbon nanotube, but also a targeting unit, folic acid (Chart 18).415 A number of human cancer cells, including those forming tumors in patients with ovarian, breast, lung, kidney, and colon cancer, overexpress the folate receptor. Indeed, immunohistochemical methods were used to establish that < 90% of ovarian cancers overexpress the folate receptor;416 a result which was more recently confirmed using a quantitative radioligand binding assay.417 Inclusion of the targeting unit was able to selectively direct the platinum-bearing SWCNT longboats to FR+ human choriocarcinoma (JAR) and nasopharyngeal carcinoma (KB) cells as opposed to FR− human testicular carcinoma cells, which typically display marked sensitivity to cisplatin.415
Chart 18.
Carbon-based delivery systems for platinum(IV) prodrugs including single-walled carbon nanotubes (SWCNTs), multi-walled carbon nanotubes (MWCNTs), and spherical carbon nanoparticles.
In addition to conjugation to the nanotube surface, the internal cavities of these structures provide attractive opportunities for drug delivery. Multi-walled carbon nanotubes (MWCNTs), which typically have larger internal diameters than SWCNTs, were loaded with the hydrophobic cisplatin prodrug cis,cis,trans-[Pt(NH3)2Cl2(O2CC6H5)2] via nano-extraction over a period of multiple days (Chart 18).418,419 After extensive washing, the surface of the nanotubes bore no platinum, as confirmed by energy dispersive X-ray spectroscopy, but the tubes had been loaded with the platinum complex to a degree quantified by atomic emission spectroscopic measurements of incinerated samples. The construct did not release any platinum unless a reducing agent was present. Ascorbic acid, for instance, is capable of reducing the prodrug, reversing its hydrophobicity and allowing release of cisplatin. The activity of this construct was further enhanced by functionalizing the surface of the nanoparticle with a fluorescent rhodamine dye prior to loading with the prodrug.420 This dye served as a targeting agent, directing the nanotubes to mitochondria. Although unloaded rhodamine-functionalized multi-walled nanotubes had little effect on the viability of cultured cells and did not appear to disrupt mitochondrial function, coencapsulation of the cisplatin prodrug with axial benzoate ligands and 3-bromopyruvate, a compound used to perturb the altered metabolism of cancer cells,421 afforded enhanced anticancer activity in vitro. A decrease in the mitochondrial membrane potential was observed. In vivo studies of the biodistribution of the platinum-loaded multi-walled nanotubes without any surface modification revealed that, in mice, the platinum(IV)-loaded construct decreased levels of platinum in the liver and kidney as compared to treatment with cisplatin.422 Accumulation in the lungs, however, was increased. Analysis of histological slices and cytokine levels indicate that no inflammation or abnormal immune response occurred.
As mentioned above, multi-walled nanotubes were chosen for these encapsulation-based constructs because they typically have larger internal cavities than SWCNT. The explicit dependence of platinum(IV) prodrug release on the diameter of the MWCNTs was recently probed.423 Smaller MWCNTs, once loaded with the prodrug, release platinum more slowly, as expected given the smaller size of the opening through which it must diffuse in order to escape. In addition to delivering a prodrug that only releases cisplatin, dual-threat prodrugs can also be loaded into MWCNTs. A prodrug was designed that is capable of releasing one equivalent of doxorubicin for every equivalent of cisplatin released (Chart 18).424 This feat was achieved by forming an amide bond between the amine group of doxorubicin and the succinate of cis,cis,trans-[Pt(NH3)2Cl2(O2CC6H5)(O2CCH2CH2CO2H)]. The final complex is sufficiently hydrophobic to be encapsulated within the nanotubes and importantly releases the two chemotherapeutic agents at relative concentrations close to those administered during combination chemotherapy. The integrin-targeting peptide c(RGDfK) was used to functionalize the nanotubes to provide an active targeting mechanism. One potential complication observed by the authors is that reduction of the platinum(IV) center does not release doxorubicin, but rather the succinyl amide derivative of the drug, which alters it subcellular distribution.
Carbon nanoparticles are a recent addition to repertoire of carbon-based nanoscale objects. This fluorescent material was first isolated as a side product in the arc-discharge synthesis of SWCNTs,425 but subsequent studies have led to their production by a variety of methods including the hydrothermal treatment of orange juice426 and nitric acid digestion of candle soot.427 Upon surface passivation, these materials exhibit striking photophysical properties428 and they have been exploited for a variety of biological applications.429 Very recently, the photoactivatable platinum(IV) azide complex cis, trans, cis-[Pt(N3)2(OH)2(NH3)(3-NH2py)] was conjugated to a carboxylate-functionalized carbon nanoparticle (Chart 18).430 Folic acid was also conjugated to the particle via a diaminoethane linker. These surface modifications did not alter the structure of the nanoparticles as revealed by transmission electron microscopy and photoelectron spectroscopy confirmed the elemental composition of the construct. UV irradiation leads to photoreduction of the platinum(IV) species, which was proposed to arise not just from direct population of the excited state of the platinum complex, but also via photoinduced electron transfer from the carbon nanoparticles. In vitro studies exploited the inherent luminescence of these nanoparticles to monitor preferential cellular uptake by FR+ cells. Cytotoxicity assays confirmed the capacity of this construct to kill cultured cancer cells upon irradiation with UV light.
A related carbon-based nanoparticle delivery system comprised PEGylated nanosized graphene oxide conjugated to cis, cis, trans-[Pt(NH3)2Cl2(OH)(O2CCH2CH2CO2H)] and an apoptosis sensing peptide.431 One motivation for using nanosized graphene oxide as the delivery platform was the established ability of this material to absorb near-IR light and release the energy as heat, a property that has been explored for photothermal therapy applications.432,433 In vivo studies involving a murine breast cancer xenograft model confirmed that near-IR irradiation of tumors following intravenous administration of the construct completely inhibited tumor growth.
8.2. Gold nanoparticles
Gold nanoparticles provide another nanodelivery platform to which platinum(IV) prodrugs can be covalently conjugated. The pendent carboxylate of a platinum(IV)-succinate complex similar to those described above can be conjugated via amide-bond-forming reactions to gold nanoparticles that are functionalized with thiolated, dodecylamine-terminated 28-mer oligonucleotides.434 The specific platinum complex delivered was cis,cis,trans-[Pt(NH3)2Cl2(OH)(O2CCH2CH2CO2H)] (Chart 19A) and the characteristic surface plasmon visible absorption band of the nanoparticles was used to confirm that they remained dispersed and did not aggregate. Fluorescence microscopy was used in conjunction with fluorescently labelled nanoparticles to monitor the progression of the constructs from vesicles to the cytosol. The R-C18 antibody, which was raised against the 1,2-d(GpG) intrastrand DNA cross-link,435 was then used to detect the formation of this cytotoxic adduct, confirming that the platinum released from the nanoparticle construct is able carry out the steps of the cell killing mechanism of cisplatin.
Chart 19.
Chemical structures of platinum(IV) prodrugs used in the preparation of gold nanoparticle delivery constructs.
Another system employing gold nanoparticles involved the use of a cisplatin prodrug functionalize with an axial ligand bearing a terminal adamantly unit capable of interacting with β-cyclodextrin. Using this characteristic non-covalent interaction, cis, cis, trans-[Pt(NH3)2Cl2(OH)(O2CCH2CH2C(O)NHCH2(C10H14))] (Chart 19C) was loaded onto gold nanoparticles that had been surface-functionalized with thiolated β-cyclodextrin.436 The host-guest interaction was studied in solution using NMR spectroscopy but in vitro cytotoxicity studies revealed that the nanoconstruct was less active than cisplatin itself, perhaps reflecting an inhibition of the platinum(IV) reduction event that is required for cyctoxicity.
Glutathione-stabilized gold nanoparticles were used to prepare a platinum-bearing, targeted drug delivery system. The use of glutathione as the surface passivating agent of the nanoparticles allow for conjugation to the pendent reactive groups of this tri-peptide. The cisplatin prodrug cis, cis, trans-[Pt(NH3)2Cl2(O2CCH2CH2CO2H)2] (Chart 19B) was conjugated to the surface, as was the neuropilin-1 receptor-targeting peptide, CRGDK. In vitro studies confirmed that delivery was enhanced in cells that express high levels of the neuropilin-1 receptor as compared to those that express low levels of it.
Gold nanorods, particles with one dimension significantly longer than those of gold nanoparticles, have also been investigated for their drug delivery capabilities.437 The cisplatin prodrug cis, cis, trans-[Pt(NH3)2Cl2(OH)(O2CCH2CH2CO2H)] (Chart 19A) was conjugated to the surface of PEGylated gold nanorods, whose PEG chains were terminally modified with amine groups.438 The conjugation afforded a stable construct, as determined by probing the surface plasmon electronic absorption, and provided enhanced cellular uptake and cytotoxicity in cultured cancer cells. Subsequent studies showed that this nanoparticle delivery strategy circumvents resistance that arises from lowered expression levels of the copper transporter CTR1 and decreases the interaction of the platinum complexes with biological deactivation agents, such as metallothionein and glutathione.439
8.3. Inorganic nanoparticles
Conjugation of a platinum(IV) prodrug to a nanoparticle can provide benefits that extend beyond those simply related to delivery. The photoactivatable trans platinum(IV) complexes described above are often limited by the need to absorb high energy light in order to be activated, although as described earlier, advancements have been made in this regard through judicious ligand choice. In an alternative strategy, trans, trans, trans-[Pt(N3)2(NH3)(py)(O2CCH2CH2CO2H)2] (Chart 20A) was conjugated to a core-shell upconversion nanoparticle.440 These nanoparticles, composed of a core of NaYF4 doped with ytterbium(III) and thulium(III) surrounded by a shell of NaGdF4 doped with ytterbium(III), are capable of absorbing 980 nm laser light and emitting radiation with wavelengths of 291, 346, and 363 nm. Irradiation of buffered suspensions of the platinum-bearing, PEGylated nanoparticle construct for only 30 min was able to release approximately half of the conjugated platinum. In vitro cyctotoxicity studies confirmed that the platinum released by 980 nm irradiation is competent to kill cancer cells. In a mouse xenograft model of murine hepatocarcinoma, tumor-bearing mice received an intratumoral injection of the nanoparticle construct. The tumor site was then irradiated with no light, UV-light, or 980 nm light. The UV light is capable of activating the platinum complex but has weaker tissue penetration than the near-IR light, which is able to activate the platinum(IV) complex via upconversion luminescence. Consequently, the tumors in the mice irradiated with UV light grew to a greater extent than those irradiated with 980 nm light. In fact, the average tumor size in the latter group did not increase, even over the course of two weeks. In a related system, silica-coated upconversion nanoparticles of NaYF4 doped with ytterbium(III) and thulium(III) were conjugated to trans, trans, trans-[Pt(N3)2(OH)(O2CCH2CH2CO2H)(py)2] (Chart 20B), through a bridging peptide, as well as a fluorescent apoptosis-sensing peptide.441 Near-IR irradiation of this construct was able to activate the platinum(IV) prodrug and induce apoptosis in cultured cancer cells that were both cisplatin-sensitive and cisplatin-resistant.
Chart 20.
Chemical structures of platinum(IV) complexes conjugated to inorganic nanoparticles including lanthanide-based upconversion nanoparticles, quantum dots, iron oxide nanoparticles, and layered double hydroxides.
Platinum(IV) prodrugs with cis or trans azide ligands are designed to exhibit photoreactivity, which has been exploited in the constructs described above. It is widely known, however, that many platinum(IV) complexes without azide or iodide ligands are sensitive to photodecomposition. In order to exploit this reactivity as a route toward photoactivation of general platinum(IV) prodrugs, quantum dots were investigated as photosensitizers. Quantum dots, semi-conductor nanoparticles, have exceptional electronic properties that vary with the size of the nanostructure and have been extensively explored as photosensitizers for photodynamic therapy.442 In a proof-of-concept study, PtCl4(bpy), where byp is 2,2′-bipyridine, was suspended in organic solvent with CdSe/ZnS core-shell quantum dots and irradiated with 530 nm light.443 PtCl2(bpy) was released as monitored by electronic absorption spectroscopy. 1H NMR spectroscopic measurements indicate that the hydrophobic platinum complex interacts with the hydrophobic surface of the quatum dots and the authors suggest that this interaction facilitates a photoinduced electron transfer from the dot to the platinum complex.
To render the system more biologically relevant, micelles packed with the CdSe/ZnS quantum dots were prepared by addition of phospholipids and PEG2000.444 The photosensitized reduction of a more relevant prodrug, namely cis, cis, trans-[Pt(NH3)2Cl2(O2CCH2CH2CO2H)2] (Chart 20C), was investigated. As in the earlier study, NMR spectroscopic measurements revealed an interaction between the prodrug and the quantum dots. When a colloidal suspension of the self-assembled micellar structures in an aqueous solution of the prodrug was irradiated with either 480 or 630 nm light, the platinum(IV) complex was reduced with loss of the two axial ligands. Control studies confirmed that the presence of the quantum dots was required for efficient photoreduction and X-ray photoelectron spectroscopic studies of the platinum 4f5/2 and 4f7/2 peaks confirmed that after irradiation the platinum was pesent in the 2+ oxidation state. In vitro cytotoxicity assays showed that neither the prodrug (IC50 ≈ 500 µM) nor the quantum dot-filled micelles alone displayed significant toxicity but that nanomolar concentrations of the dots combined with irradiation led to an IC50 of 25 µM. Extensive theoretical calculations were carried out to probe the mechanism photoactivation of cisplatin prodrugs by quantum dots. The results were consistent with a model in which the platinum(IV) complex interacts with the nanoparticle surface and computation of the electronic coupling between the donor and acceptor indicates that the electrons injected into the platinum(IV) complex produce an excited state that leads specifically to dissociation of the succinate ligands.445
Although we have not found any reports of platinum(IV) prodrugs conjugated to quantum dot drug-delivery vehicles, we anticipate that such a development is forthcoming. The authors of these quantum dot photoactivation studies, have however, prepared a lanthanide upconversion nanoparticle construct in which cis, cis, trans-[Pt(NH3)2Cl2(O2CCH2CH2CO2H)2] (Chart 20C) is conjugated to a phospholipid-functionalized PEG chain. This platinated polymer was used to prepare stable aqueous suspensions of thulium(III)-doped NaYF4:Yb(III) upconversion nanoparticles that are functionalized with the platinum(IV) complex.446 Irradiation of this construct with 980 nm light not only led to release of succinate, as expected because of the photolabilization of the axial ligands, but also reduced all of the platinum centers to the platinum(II) oxidation state as determined by XPS.
Upconversion nanoparticles have also been used as a nanodelivery vehicle in which the lanthanide-based luminescence is not implicated in the release or activity of the platinum agent. A cisplatin-releasing platinum(IV) prodrug was conjugated to the surface of a poly(ethyleneimine)-coated NaYF4 nanoparticle doped with ytterbium(III) and erbium(III) via a succinate axial ligand.447 The nanoparticle was further functionalized with a folic acid targeting group. Cellular uptake of the nanoparticles was monitored using the inherent luminenscent properties of the nanoparticles and cisplatin was released upon intracellular reduction.
Hydrophobic iron oxide nanoparticles can be encapsulated in gelatin to enhance their water solubility. The amine functionalities on gelatin allowed the platinum (IV) prodrug cis, cis, trans-[Pt(NH3)2Cl2(O2CCH2CH2CO2H)2] (Chart 20C) to be conjugated to the surface along with a fluorescent marker, fluorescein isothiocyanate.448 Release of platinum was not explicitly measured but rather was inferred from the photometrically quantitated release of the fluorophore. The release could be enhanced by the presence of an undefined pancreatic enzyme, which we suppose to be trypsin. The superparamagnetic properties of the nanoparticles could be used to generate T2-weighted magnetic resonance images showing contrast in the tumor region following intratumoral injection.
Layered double hydroxides are inorganic materials in which positively charged layers are interspersed with loosely associated charge-balancing anions.449 These anions in the interlayer space can often be readily exchanged. Indeed, cis, cis, trans-[Pt(NH3)2Cl2(O2CCH2CH2CO2H)2] (Chart 20C) was loaded into layered double hydroxide nanoparticles of the formula [Mg2+0.66Al3+0.34(OH)2][Cl− 0.34]·2.7H2O simply by incubating the material in an pH 8 aqueous solution of the prodrug for one day. Platinum loading did not change the morphology of the particles, although it did cause an approximate doubling in the average diameter of the particles as determined by dynamic light scattering. Platinum incorporation was measured using atomic absorption spectroscopy and a slight decrease in the zeta potential of the material was taken as corroborating evidence of the inclusion of the negatively-charged succinate-bearing prodrug into the material. The nanoparticle construct was more effective at killing cancer cells than cisplatin alone and the former demonstrated reduced toxicity in non-cancerous immortalized cell lines. Mechanistic studies confirmed that the construct acted via a mechanism analogous to that of cisplatin.
8.4. Coordination Polymers
In an alternative strategy, the disuccinate complex described above in the preparation of the folate-targeted SWCNT was used to create coordination polymers that precipitated from solution as nanoparticles.450 The unit cross-linking the carboxylate functional groups of different platinum complexes was the Tb3+ ion (Chart 21A). ICP-MS and thermal gravimetric analysis measurements confirmed that the empirical formula of the coordination polymer was Tb2(PtIV)3(H2O)12 where PtIV represents cis, cis, trans-[Pt(NH3)2Cl2(O2CCH2CH2CO2H)2]. The stability of the nanoparticles in suspension could be significantly enhanced with a silica shell coating. Moreover, a silyl-derived c(RGDfK) could be grafted to the silica surface of the coated nanoparticles, which targeted the construct to cells that express the αVβ3 integrin, such as HT-29, preferentially over those that do not, such as MCF-7.
Chart 21.
Depiction of the formation of coordination polymers using metal units to link platinum(IV) prodrugs bearing axial ligands with coordinating motifs.
A variation on this theme appeared in the report of nanoscale coordination polymers formed from platinum(IV) prodrugs bearing pendent phophonates and Zn2+ ions.451 The platinum(IV) prodrugs featured either a cisplatin or oxaliplatin equatorial core and axial phosphonylcarbamate ligands. The phosphonate moiety permits self-assembly with Zn2+ to form the extended coordination polymer network which precipitates from solution in nanoparticulate form (Chart 21B). The particles were stabilized and rendered biocompatible by PEGylation using a phospholipid, cholesterol, and a PEGylated phospholipid. Fluorescently labelled analogues of these particles were observed to enter into the cell by fluorescence microscopy and in vitro experiments with cultured cancer cells confirmed the ability of the construct to induce the DNA damage characteristic of the parent platinum drugs and trigger apoptosis. Further in vitro studies with inhibitors of endocytosis and fluorescent dyes that selectively localize to endosomal and lysosomal compartments confirmed that uptake occurred through energy-dependent endocytotic processes. Pharmacokinetics studies in mice revealed that the nanoparticle formulation provided blood circulation times that were more than 40-fold greater than those of the parent drugs. In mouse xenograft models of non-small cell lung cancer and pancreatic cancer, these constructs were able to inhibit tumor growth significantly more than the parent platinum(II) drugs. In a very recent development in this delivery platform, the cisplatin-delivering nanoconstruct was prepared using an alternative lipid to afford nanoparticles with an overall positive charge concentrated near the surface of the particle but below the surface of the outer PEG layer.452 Negatively charged siRNAs were then loaded into the particle via electrostatic interactions. Three distinct genes were targeted by the siRNAs for silencing: survivin, bcl-2, and p-gp. The construct demonstrated the ability to release active platinum agents and the siRNAs in a controlled fashion, all of which were able to carry out their intended biological functions in vitro. In a mouse xenograft model of ovarian cancer, the co-delivery enhanced the anticancer activity of the platinum agent as evidence by inhibition of tumor growth, reduced expression of the silenced proteins in tumor tissue, and increased evidence of apoptosis in tumor cells.
8.5. Metal-organic frameworks
As an extension of the Pt-Tb coordination polymer work that was described above, platinum conjugates of nanosized metal-organic frameworks (MOFs) were prepared.453 In these instances, the platinum complex does not act as a structural component of the coordination polymer, which is instead formed from a first-row transition metal and an amino-functionalized terephthalate. The non-platinum metal center combines with the aromatic dicarboxylate to form the extended 3-dimensional MOF structure. Iron was used to form a nanoparticulate MOF and the same platinum-(IV) prodrug used to for the initial SWCNT conjugates, cis, cis, trans-[Pt(NH3)2Cl2(OEt)(O2CCH2CH2CO2H)], was attached to the pendent amine following activation with 1,1-carbonyldiimidazole.454 As with the Pt-Tb coordination polymer, aqueous stability of the nanoparticles was enhanced with a coating of amorphous silica, although an alternative chemistry using Na2SiO3 as the silica source needed to be employed to avoid decomposition of the particles. The silica shell also provided a more controlled release of platinum from the particle. No significant enhancement in activity was obtained, however, in in vitro cytotoxicity tests. In a subsequent iteration of this drug-delivery strategy, a zirconium(IV) containing MOF from the UiO series was prepared using aminotriphenyldicarboxylate.455 The similarity of the organic bridging ligands to the amino-functionalized terephthalate in the system above, would suggest that the nanoscale MOF could be post-synthetically modified with cis, cis, trans-[Pt(NH3)2Cl2(OEt)(O2CCH2CH2CO2H)] using amide bond forming reactions, but the authors instead simply incubate the platinum complex with the MOF to load the nanostructure through non-covalent interactions. This mode of encapsulation was confirmed using 1H NMR spectroscopy. The survivin/Bcl-2/P-gp siRNA cocktail described above was also loaded into the nanoscale MOF and the encapsulation was proposed to proceed through coordination of the sugar-phosphate backbone to the zirconium centers. Protection of the encapsulated siRNAs from degradation by nucleases was observed and the combined delivery of the platinum agent and the siRNAs provided a chemotherapeutic enhancement of over 10-fold in in vitro cytotoxicity assays.
8.6. Polysiloxane
One research effort that seeks to maintain the philosophy of using the platinum(IV) prodrug as an integral constituent of the polymer that forms the nanoparticle, while improving upon the stability of the systems that feature the nanoscale coordination polymers, involves the preparation of polysilsesquioxane nanoparticles.456 These nanoparticles are formed from the anionic reverse microemulsion base-catalyzed sol-gel polymerization of platinum(IV) prodrugs bearing axial ligands with pendent trialkoxysilanes (Chart 22). The nanoparticles functioned as effective controlled-release agents of oxaliplatin, releasing 80% of the encapsulated payload over the course of two days upon incubation with cysteine. PEGylation was used to enhance biocompatibility of the nanoparticles, which were further functionalized with anisamid to target the sigma receptor. This opioid receptor is overexpressed on the surfaces of many types of cancers cells.457 An integrin-targeting RGD unit could also be attached to the nanoparticle surface. These targeting agents were successful in enhancing efficacy both in vitro and in vivo. A cisplatin-delivering analogue of this construct was also developed and tested for its ability to improve upon cisplatin-mediated tumor growth inhibition when administered in conjunction with radiation therapy.458 In vitro and in vivo studies, the latter in a xenograft model of non-small cell lung cancer, suggest that this cisplatin-delivering polysilsesquioxane nanoparticle can offer significant improvements over conventional chemoradiation therapy using cisplatin.
Chart 22.
Depiction of the polymerization of platinum(IV) prodrugs bearing axial ligands with pendent trialkoxysilanes to form platinum-containing polysilsesquioxane nanoparticles.
8.7. Polymeric micelles
A highly successful platform that has been exploited in the nanodelivery of platinum(IV) prodrugs comprises polymeric micellar nanoparticles that are formed from the self-assembly of amphiphilic block copolymers. When an organic solution of such polymers, which contain a hydrophobic block and a hydrophilic block, is added to water, the hydrophobic portions of the chains cluster to form a hydrophobic core, which is surrounded by a shell formed from the hydrophilic portions of the copolymer chains. If this self-assembly process occurs in the presence of a hydrophobic drug-like molecule, it can be encapsulated within the core of the nanoparticle, which then serves as a controlled-release drug delivery device. As opposed to the constructs described above, in which the axial ligands of the platinum(IV) prodrug were chosen so as to permit covalent conjugation, the axial ligands of the prodrug can be used to tune hydrophobicity, a key parameter in nanoencapsulation. An alternative strategy, more akin to that used in the delivery devices described in previous subsections, involves covalent conjugation of the platinum(IV) complex to the polymer backbone and subsequent nanoparticle formation. Examples of these two strategies will now be treated sequentially.
8.7.1. Polymer micelles: Non-covalent encapsulation
One copolymer system that has been extensively investigated is poly(lactic-co-glycolic acid)-block-poly(ethylene glycol) or PLGA-PEG (Chart 23), in which the statistical copolymer PLGA serves as the hydrophobic block and PEG as the hydrophilic block. PLGA is a biocompatible, biodegradable polymer that is approved for use by the FDA for a variety of biomedical devices.459 The safety of PEG has long been investigated and the FDA has declare that it is generally recognized as safe.460,461 PLGA nanoparticles have been widely explored for drug delivery applications459 and the current popularity of the PLGA-PEG block copolymer has led a number of commercial vendors to offer PLGA-PEG with a variety of block sizes and variations in the relative ratio of lactic acid to glycolic acid in the PLGA block. These parameters influence the properties of the nanoparticles formed from the polymer and recent work has even investigated the influence that the ordering of the lactic acid and glycolic acid units within the PLGA block.462,463
Chart 23.
Platinum complexes encapsulated within polymeric micelles using non-covalent interactions. The complexes are shown next to the polymer (blue) that was used to make the nanoparticle. In the case of the PLGA nanoparticle, PEGylated lipids were used to stabilize the particles formed from the non-amphiphilic polymer.
The platinum(IV) prodrug cis,cis,trans-[Pt(NH3)2Cl2(O2CCH2CH2CH2 CH2CH3)2] (Chart 23A), which bears an equatorial cisplatin core and axial hexanoate ligands, was successfully encapsulated within the hydrophobic core of a nanoparticle formed from PLGA-PEG-COOH. This polymer is a derivative of PLGA-PEG in which the exposed end of the PEG chain is functionalized with a carboxylic acid. This pendent carboxylic acid was then used to conjugate a targeting unit to the surface of the platinum-loaded nanoparticle. An RNA aptamer that could recognize the prostate-specific membrane antigen (PSMA) was used and provided selective targeting of the construct to LNCaP prostate cancer cells that abundantly express this membrane protein. PSMA is highly expressed in many prostate tumors, particularly in the metastatic and hormone-refractory forms.464 Fluorescence microscopy confirmed that the construct was taken up by endocytosis and immunofluorescence imaging using the R-C18 antibody confirmed formation of 1,2-d(GpG) intrastrand DNA cross-links. The construct exhibited sub-micromolar IC50 values in cultured human prostate cancer cells. Subsequent studies with Swiss albino mice and Sprague Dawley rats demonstrated that this prodrug-loaded nanoparticle construct has enhanced pharmacokinetics, biodistribution, and tolerability as compared to cisplatin.465 Using a murine model of prostate cancer in which BALB/c nude mice were injected with cultured LNCaP cells to form a subcutaneous xenograft, the nanoparticle construct was able to provide an equivalent degree of reduction in tumor size as a three-fold higher molar dose of cisplatin. The enhanced activity was attributed to a combination of passive targeting of tumor tissue via the EPR effect, active targeting of the PMSA expressing LNCaP cells by the aptamer conjugated to the surface, and prolonged residence of the platinum species in the blood. This nanoparticle platform was also used to prepare platinum-loaded constructs that were functionalized with the cyclic pentapeptide c(RGDfK).466 This unit allowed the nanoparticles to target angiogenic blood vessels in an orthotopic breast cancer xenograft model. In this model, the nanoparticle was more efficacious and better tolerated than cisplatin.
A series of complexes of the form cis,cis,trans-[Pt(NH3)2Cl2(O2C(CH2)nCH3)2] (Chart 23A), of which the hexanoate complex described above is a member, was prepared to systematically investigate the effect of the length of the methylene chain of the alkyl carboxylate axial ligands on nanoencapsulation.467 This study revealed that increasing the length of the chain increases platinum loading into the nanoparticle but also increases the propensity for aggregation and macroscopic precipitation. An optimal balance was struck using a 4:6 w/w mixture of the PLGA-PEG polymer and the decanoate complex cis,cis,trans-[Pt(NH3)2Cl2(O2C(CH2)8CH3)2]. This complex was then used as the prodrug component of a nanoparticle platform designed to deliver cisplatin and siRNAs capable of suppressing the function of REV1 and REV3L, which are involved in the process of error-prone translesion DNA synthesis.468 Such translesion synthesis can contribute to cisplatin resistance in tumors.469 The nanoparticle construct was formed from an interaction of PLGA-PEG, a cationic lipid, the platinum prodrug, and the siRNA. The PLGA block interacted with the cationic lipid to form a polymer matrix in which the prodrug was suspended and the PEG block provided an outer shell. The nanoparticles were formulated using a double emulsion strategy (vide infra) that allowed the PLGA-lipid matrix to surround an aqueous core in which the siRNA molecules were dissolved. Sustained release of the platinum and RNA was achieved and the released siRNAs were able to decrease expression of their target genes both in vitro and in vivo. In a LNCaP xenograft model, inclusion of the siRNA was able to successfully render the tumors more susceptible to platinum-based therapy.
A platform for preparing platinum(IV) prodrugs that can be readily conjugated to another chemical moiety using strain-promoted azide-alkyne cycloaddition, one of the so-called “copper-free click chemistries,” was recently developed.470 As a proof of principle, a platinum(IV) prodrug with axial ligands displaying pendent azide units was coupled to a functionalized azadibenzocyclooctyne (ADIBO) (Chart 23D). The significant increase in lipophilicity upon reaction with the strained cyclooctyne prompted the authors to investigate the nanoencapsulation of this complex in PLGA-PEG-based nanoparticles. The ADIBO-functionalized cisplatin prodrug encapsulated far better than the unmodified, azide-terminated platinum complex. This platform also holds significant promise with regards to conjugation of platinum prodrugs to a variety of other nanodelivery devices, targeting units, bioactive molecules, and reported beacons. Indeed, coupling of the platinum(IV) azide-bearing complex to a cyclooctyne-modified triphenylphosphonium salt afforded a platinum(IV) prodrug that targets the mitochondria.471 This complex was then encapsulated with a PLGA-PEG nanoparticle which was itself functionalized with a triphenylphosphonium salt. In vitro studies confirmed the ability of this construct to accumulate in the mitochondria of cultured cancer cells, disrupt their altered mitochondrial metabolism, and induce cell death. The ability of the triphenylphosphonium-derivatized nanoparticles to penetrate the blood-brain barrier led the authors to investigate the potency of this construct in neuroblastoma cells and they found that it was approximately 17-fold more active than cisplatin.
Instead of using synthetic high polymers, nanoparticles formed from polymers of natural origin have also been explored for the delivery of platinum(IV) prodrugs. The association of platinum(IV) complexes with discrete folded proteins will be dealt with below. Silk fibroin (Chart 23) is the fibrous protein component of the silk made by spiders, silkworks, and other insects. This biocompatible, biodegradable material has been successfully employed in a range of biomedical applications from sutures to three-dimensional tissue scaffolding,472,473 and silk fibroin can be formed into nanoparticles for drug delivery.474 In order to improve upon an initial nanoparticle design in which cisplatin was loaded into silk fibroin nanoparticles via coordination of the platinum(II) complex to the polymer,475 a new construct was very recently reported in which the hydrophobic prodrug cis,cis,trans-[Pt(NH3)2Cl2(O2CC6H5)2] (Chart 23G) was encapsulated within such nanoparticles.476 Unlike many of the other polymer-based delivery systems described here, the platinum loading is not accomplished simultaneously with nanoparticle formation, but rather a dried sample of preformed nanoparticles are suspended in DMSO solution of the platinum complex to load the prodrug into the nanoparticles. TEM images support the internalization of the nanoparticle constructs and in vitro assay indicate that the construct is effective at killing cultured cancer cells. Flow cytometric methods were used to carry out cell cycle analyses and propidium iodide straining assays that confirm a mechanism of action similar to that of cisplatin is operative.
Although increasing the lipophilicity of a platinum(IV) prodrug by increasing the hydrophobic character of the axial ligands is an effective way to influence nanoencapsulation, this strategy may not be applicable to dual-threat complexes in which the axial ligands, selected to elicit a particular biological response, render the complex hydrophilic. For instance, mitaplatin (Chart 23A) is relatively water soluble and does not readily encapsulate within PLGA-PEG using conventional nanoprecipitation techniques. Moreover, changing any of the ligands to increase lipophilicity could compromise the activity of the platinum(II) or DCA species released. Instead, an alternative encapsulation strategy, which had previously been employed to encapsulate hydrophilic species like proteins, was investigated.477 Using a water-in-oil-in-water double-emulsion solvent evaporation strategy, mitaplatin could be encapsulated within nanoparticles formed from PLGA-PEG.367 This nanoencapsulation formulation afforded mitaplatin an increased residence time in the bloodstream and decreased accumulation in the kidneys without negatively impacting anticancer activity in a mouse xenograft model of triple-negative breast cancer.
It is possible that a dual-threat complex may fortuitously have properties such that the axial ligands permit facile incorporation within the hydrophobic core of a polymeric micelle. Such is the case for canthaplatin (Chart 23B),478 a cisplatin prodrug in which the axial ligands are derivatives of the protein phosphatase 2A inhibitor demethylcantharidin.479 The Boc-protected pipirazinyl groups on the axial canthaplatin-derived ligands allow the prodrug to be readily encapsulated in PLGA-PEG nanoparticles affording a construct that is taken up via endocytosis, decreases the efficiency of DNA repair by inhibiting protein phosphatase 2A, and releases cisplatin. The enhancement of the efficacy of the cisplatin as a result of the inhibited DNA repair was confirmed in vitro and in vivo with a mouse xenograft model of lung cancer.
In another instance, a cisplatin prodrug in which a paclitaxel derivative was installed at one axial position through a platinum-coordinated glutaric acid (Chart 23F), but no significant enhancement over co-treatment with the un-encapsulated species was observed.480 The platinum(IV) prodrug VAAP (see Section 8) was encapsulated within the a poly(caprolactone)-PEG polymeric nanoparticle (Chart 23E).362 The hydrophobic valproate axial ligands not only permit encapsulation, but upon reductive release are capable of acting as histone deacetylase inhibitors. This enzyme inhibitory activity, which can potentiate the activity of cisplatin,481 was observed in in vitro experiments using VAAP, as described above. In mice, the nanoparticle formulation significantly enhanced retention of platinum in the bloodstream. Murine xenograft studies with a human lung cancer model, did not reveal any significant enhancement in tumor reduction in the nanoparticle-treated branch as compared to tumor-bearing animals treated with un-encapsulated VAAP, but encapsulation did result in enhanced platinum accumulation in the tumor and reduced kidney toxicity, as assessed by histological analysis of renal tissue.
8.7.2. Polymer micelles: Covalent conjugation
Instead of relying on non-covalent interactions to associate a platinum complex with a polymer nanoparticle, the axial ligands of a platinum(IV) prodrug can be covalently linked to a polymer chain using well-established coupling chemistry. The first example of covalent conjugation of a platinum(IV) complex to a polymer chain for subsequent formation of a polymeric micelle exploited the ability of a cisplatin prodrug with axial levulinate ligands to react with the end group of a hydrazine-terminated poly(ethylene glycol)-block-poly(L-lactic acid) (PLA-PEG).482 The hydrazine group caps the hydrophobic block and in the resulting platinated polymer the complex is linked to the macromolecule through a hydrazone. The polymer chains can self-assemble into micelles with the platinum buried in the hydrophobic core. The rate of platinum release from the nanoparticle construct varied with pH because of the acid-lability of the hydrazone linkage and efficacy was demonstrated in vitro using cultured cancer cells.
In a very recent variant on this theme, the termini of the hydrophobic blocks of a PLA-PEG copolymer were cross-linked via esterification with the succinate ligands of cis, cis, trans [Pt(NH3)2Cl2(O2CCH2CH2CO2H)2].483 These conjugates formed micelles in aqueous solution that were observed to undergo a thermoreversible sol-gel transition, forming hydrogels at 37 °C. These semi-solids were capable of releasing platinum in a controlled fashion, without an initial burst, over the course of two months. The platinum is mainly released in a micellar form, which was shown to be taken up by endocytosis and demonstrates enhanced toxicity in cultured cancer cells as compared to cisplatin.
The use of platinum(IV) complexes with succinate axial ligands to conjugate prodrugs to the polymer backbone via ester- or amide-forming reactions is a recurring theme within this class of constructs. In the first report of this strategy, cis,cis,trans-[Pt(NH3)2Cl2(OH)(O2CCH2CH2CO2H)] was conjugated to a PLA derivative bearing pendent hydroxyl functional groups.484 Using hydrodynamic flow focusing,485 nanoparticles comprising a blend of this platinated polymer and PLGA-PEG-COOH were formed. If the nanoparticle formation was carried out in the presence of docetaxel, then this molecule was encapsulated within the hydrophobic core of the nanoparticles, affording a construct that can deliver two chemotherapeutics via orthogonal release mechanisms. The docetaxel is released by passive diffusion from the nanoparticle, whereas the stable covalent bond of the platinum complex to the polymer chain permits release of the platinum only upon reduction of the metal center. Surface modification of the assembled nanoparticle with a PSMA-targeting aptamer allowed for enhanced uptake by cultured prostate cancer cells. A similar construct was developed in which the drug delivered along with the platinum was irinotecan.486 A particularly low polydispersity among the nanoparticles was again achieved by carrying out the nanoprecipitation using microfluidic devices. Targeting with the PSMA aptamer, again allowed for enhanced cellular uptake in cells expressing this antigen. Inclusion of irinotecan in the particles provided an enhancement over nanoparticles containing only the cisplatin prodrug.
A similar covalent conjugation strategy was used to prepare a nanoparticle construct that delivers the monofunctional complex phenanthriplatin.487 A platinum(IV) derivative of phenanthriplatin bearing a single pendent carboxylate was prepared and conjugated to the hydroxyl-modified PLA. Standard nanoprecipitation techniques were used to prepare nanoparticles of this platinated polymer blended with PLGA-PEG. Using in vitro assays, this nanoparticle construct was shown to protect the platinum species from deactivation by biological nucleophiles. In a mouse xenograft model of prostate cancer, the construct outperformed both treatment with an equimolar dose of a double-emulsion nanoparticle formulation of the parent platinum(II) complex, i.e. unmodified phenanthriplatin, and treatment with a 10-fold higher molar dose of cisplatin.
In the study described above in which PLGA-PEG particles were designed to release both cisplatin and docetaxel, the cisplatin prodrug was covalently conjugated to the polymer backbone and the docetaxel was non-covalently encapsulated within the nanoparticle core. In an alternative strategy, two separate polymer chains were covalently modified with different anticancer agents, an oxaliplatin prodrug bearing a pendent carboxylate and daunomycin.488 The former can be released by reduction and the latter by hydrolysis. The novel amphiphilic block copolymer used for this study was poly(lactide-co-2-methyl-2-carboxyl-propylene carbonate)-block-poly(ethylene glycol). Composite nanoparticles were formed from combinations of the two polymer chains, with the ratio of the two anticancer agents tuned by changing the relative amounts of the two modified chains during nanoparticle formation. Facile variation of this ratio allowed the authors to readily interrogate synergistic effects in vitro and in vivo. A similar construct was investigated in which the platinum agent was a cisplatin prodrug and the second anticancer agent was a paclitaxel conjugate.489 This polymer platform was also be used to prepare a conjugate with cis,cis,trans-[Pt(NH3)2Cl2(OH)(O2CCHCl2)], a platinum(IV) prodrug designed to release cisplatin and DCA, much like mitaplatin. This strategy provides an alternative to the double emulsion strategy described above to permit nanodelivery of the hydrophilic complex.490
Using the same poly(lactide-co-2-methyl-2-carboxyl-propylene carbonate)-block-poly(ethylene glycol), a nanoparticle construct was prepared in which the platinum(IV) prodrug conjugated to the polymer backbone displays the photoactivatable cis-diazide motif.491 Fluorescence microscopy studies were used to confirm that the nanoparticles were taken up via endocytosis and the authors emphasize that this route of cellular uptake provides a means of circumventing resistance related to expression levels of copper transporters, proteins whose role in the activity of and resistance to cisplatin has been extensively studied.21 The particles, which are stable in the dark, exhibit fast release of platinum upon irradiation with UV light. A variety of pharmacokinetic parameters were evaluated in Chinese KM mice, which indicated that treatment with the nanoparticle construct followed by UV irradiation resulted in much lower systemic toxicity that treatment with cisplatin.
In addition to the diblock copolymers described above, a triblock amphiphilic copolymer was also developed to deliver platinum(IV) prodrugs as micellar nanoparticles. This polymer is non-toxic and biodegradable, comprising a methoxy-terminated poly(ethylene glycol) block, a poly(ε-caprolactone) block, and a poly(L-lysine) block.492,493 This polymer can be platinated with cis,cis,trans-[Pt(NH3)2Cl2(OH)(O2CCH2CH2CO2H)] through amide bond forming reactions with the lysine side chains and the platinated polymer self-assembles into micellar nanoparticles. These particles can release platinum(IV) complexes upon exposure to acidic conditions or platinum(II) complexes upon reduction. In vitro assays confirmed that the nanoparticles are taken up via endocytosis affording enhanced intracellular platinum accumulation and cytotoxicity. A series photoactivatable platinum(IV) complex with cis azide ligands trans to either cis amines or a chelating R, R-diaminocyclohexane (DACH) were also conjugated to the polymer.494 These constructs were stable in the dark, but upon irradiation with UV light they released cytotoxic platinum(II) species. Following studies with cultured cancer cells that verified the light-induced cytotoxic activity of the construct, the nanoparticle functionalized with cis,trans-[Pt(DACH)(N3)2(OH)(O2CCH2CH2CO2H)], the most active compound from the in vitro studies, was carried forward for testing in a xenograft model of murine hepatocarcinoma. Enhanced tumor growth inhibition was observed when mice were injected intratumorally with the construct and the tumor was irradiated with UV light for 1 h. An additional hour of UV irradiation was carried out 5 day post-injection. The combination of nanoparticle treatment and UV irradiation was found to cause less of an effect on body weight than treatment with an equivalent dose of oxaliplatin.
Using this same polymer system, a nanoparticle construct was developed that is capable of releasing carboplatin upon reduction of the platinum(IV) center.495 The carboplatin prodrug was attached to the polymer backbone through an axial succinate ligand. In the same report, an alternative method of nanoparticle-mediated co-delivery of cisplatin and DCA was described. As opposed to mitaplatin, which bears axial DCA ligands, a platinum(IV) complex was prepared by hydrogen peroxide oxidation of cis-[Pt(NH3)2(DCA)2]. Subsequent derivatization afforded an axial succinate, through which the complex could be tethered to the polymer backbone using amide bond forming reactions. An enhancement in the activity is reported as compared to the carboplatin delivering polymer or co-treatment with unencapsulated carboplatin and DCA, but this result is unsurprising given the great kinetic inertness of the chelating cyclobutanedicarboxylate ligand of carboplatin as compared to the monodentate carboxylate DCA. Dinulcear platinum complexes, analogous to those described in Section 4,496 were also delivered via covalent conjugation of the corresponding platinum(IV) prodrugs to the polymer backbone.497 Compounds with both ammine and R, R-DACH non-leaving group ligands were prepared bearing axial carboxylates. Coupling to the polymer was achieved though amide bond forming reactions with the lysine amines.
Very recently, another construct based on this polymer was reported.498 Camplatin, a platinum(IV) prodrug derived from oxidized cisplatin and camphoric anhydride, was conjugated to the pendent amine groups of the poly(L-lysine) block. Quantification of the mRNA levels of Bcl-2 and Bax, revealed that treatment with the nanoparticle encapsulated camplatin decreased production of these proteins in culture ovarian cancer cells.
Although micellar nanoparticles formed from amphiphilic block copolymers of the types described above appear to be stable enough to maintain their structural integrity and elicit characteristic biological responses, particularly in vivo, the classical theory of micelles states that in solution an equilibrium is present between the self-assembled structure and de-assembled unimers. Chemical cross-linking of the polymer chain within the core or the shell can be carried out to improve micellar stability.499 Overly stable micelles, however, can prevent efficient release of encapsulated active agents, and so stimuli-responsive cross-linked micelles have been developed that are able to release a cytotoxic payload upon, for instance, entry into the acidic microenvironment of the tumor.500 A platinum-delivering polymeric micelle based on a poly[(2-(2-methoxyethoxy)ethyl methacrylate)-co-(N-methacryloxy-3-azidopropylamide)]-block-poly(N-(2-hydroxypropyl)methacrylamide) block copolymer was prepared.501 The pendent azide groups were functionalized with both a platinum(IV) prodrug, cis, cis, trans-[Pt(NH3)2Cl2(OH)(O2CCH2CH2CONHCH2CCH)], and a near-IR dye, cypate, for combination platinum/photothermal therapy. The attachment of the warheads to the polymer backbone was accomplished using Cu-catalyzed alkyne-azide cycloaddition. The authors of the study described above in which strain-promoted alkyne-azide cycloaddition was used to functionalize platinum(IV) complexes noted that their use of the strain-promotion strategy was because of the propensity for Cu-catalyzed click reactions to reduce the platinum(IV) centers. In this instance, the successful use of the latter coupling strategy may stem from the use of pentamethyldiethylenetriamine as a Cu-chelating agent. Chemical cross-linking of the cores of the micelles formed from these functionalized polymers was carried out using a cystamine cross-linker, producing a construct that is sensitive to reducing environments, such as the cytoplasm of cancer cells. In vitro studies confirmed the ability of the construct to release its cargo in a controlled fashion and elicit chemotherapy/photothermal therapy synergy.
The platinum complex itself can act as the chemical cross-linking agent. Following functionalization of the axial succinate ligands of cis, cis, trans-[Pt(NH3)2Cl2(O2CCH2CH2CONHCH2CCH)2] with ethylenediamine, the platinum complex was used to cross-link the cores of polymeric micelles formed from poly(oligo(ethylene glycol)methyl ether methacrylate)-block-poly(styrene-co-3-isopropenyl-α,α-dimethylbenzyl isocyanate).502 The facile reaction between the isocyante groups on the polymer and the pendent amine groups of the platinum complex afforded controlled-release nanoparticles. The same complex was used without ethylenediamine functionalization to cross-link the core of the biodegradable polymer monomethoxyl poly(ethylene glycol)-block-poly(L-lysine) via reaction of the lysine amine groups with the carboxylic acids of the prodrug.503 Analogously, the core-cross-linked micelles formed from the triblock copolymer monomethoxyl poly(ethylene glycol)-block-poly(ε-caprolactone)-block-poly(L-lysine) were also prepared.504
As a variation on tethering a platinum(IV) complex to a polymer backbone, the metal complex can itself act as a monomer for polymerization. In one instance, cis, cis, trans-[Pt(NH3)2Cl2(O2CCH2CH2CONHCH2CCH)2] was used as a monomer for condensation polymerization with ethylenediamine or piperazine.505 The corresponding condensation with diols proved unfruitful, but reaction of cis, cis, trans-[Pt(NH3)2Cl2(OH)2] with a bifunctional anhydride, such as cyclobutane tetraacetic anhydride, was able to give polyesters that could be furthers PEGylated to greatly enhance water solubility. The lack of blocky character in these polymers precluded their self-assembly into micellar nanoparticle structures. Another example of this strategy was the use of a cisplatin prodrug with pendent norbornenyl units at the axial positions that could serve to cross-link norbornene-terminated PEG chains, some of which were functionalized with either camptothecin or doxorubicin.506 The linkages to the three drug-derivatives were all chosen so as to release their payload under different circumstance, viz. reduction, hydrolysis, or UV-irradiation. The platform design also readily allows for variation in the relative amounts of the three different chemotherapeutics.
A less common motif for platinum(IV) delivery using polymer nanoparticles is that in which the platinum complexes are not buried within the particle, but rather are displayed on the surface in a manner somewhat analogous to that exhibited by the non-polymeric constructs described in this section (e.g. SWCNTs, gold nanoparticles, etc.). Conjugation of cis, cis, trans-[Pt(NH3)2Cl2(OH)2] to the succinate terminus of the amphiphilic molecule α-tocopheryl-PEG1000-succinate, which is conceptually identical to conjugation of cis, cis, trans-[Pt(NH3)2Cl2(OH)(O2CCH2CH2CO2H)] to the hydrophilic portion of α-tocopheryl-PEG1000, produces a polymer that can self-assemble into micelles that will present the platinum(IV) complex on its surface. As observed in many other systems, even though the platinum center is putatively attached to the nanostructure through a stable covalent linkage, a substantial burst release is observed followed by a longer period of more sustained controlled release. In a subsequent development, this nanoparticle platform was stabilized by addition of α-tocopheryl-PEG1000-succinylpoly(lactic acid). Inclusion of α-tocopheryl-PEG1000-succinate in the formulation provides surface-exposed carboxylic acids to which the monoclonal antibody trastuzumab (Herceptin) could be conjugated.507 This targeted nanoparticle, perhaps because of the difference in nanoparticle formation, shows a much more controlled release profile than its predecessor. The nanoparticle could also be prepared with docetaxel encapsulated. In in vitro assays the combination-delivering nanoparticle was more efficacious than either of the drugs in isolation.
8.8. Other supramolecular systems
The cisplatin-releasing platinum(IV) prodrug cis, cis, trans-[Pt(NH3)2Cl2(OH)(O2CCH2CH2CO2H)] was conjugated to a phosphorylated oligopeptide that can act as a substrate for phosphatases.508 The molecule is designed to remain monomeric in circulation but upon entering the tumor, whose cells overexpress various phosphatases, enzyme-catalyzed dephosphorylation will trigger self-assembly into supramolecular nanofibers. This self-assembly process is proposed to enhance retention of the prodrug at the site of disease. In vitro studies of the construct confirmed its ability to self-assemble into β-sheet-like structures following action of alkaline phosphatase. The hydrogel formed by these nanofibers provided controlled release of active platinum(II) species upon incubation with chemical reductants. In vitro cytotoxicity studies confirmed that the compound can kill cultured murine and human cancer cells via induction of apoptosis. Mouse xenograft studies with subcutaneously grown murine breast cancer tumors demonstrated that the construct exhibited enhanced tumor accumulation, comparable tumor growth inhibition, and lower systemic toxicity as compared to treatment with cisplatin.
Supramolecular cages formed from metal center vertices and rigid organic linkers have long been investigated as for their interesting chemical properties.509 These constructs are related to the MOFs described above, but form discrete structures as opposed to extended networks. An octahedral hexanuclear cage was formed from six platinum(II) ethylenediammine units and four 2,4,6-tris(2-pyridyl)-s-triazine molecules (Figure 7). The metal centers form the vertices of octahedron and the triazine units cover four faces of the polyhedron. The remaining four faces are open and provide access to the interior of the structure. The platinum(IV) prodrug cis, cis, trans-[PtCl2(NH3)2(OC(O)NHC10H15)(O2CCH2CH2CO2H)] (Figure 7) was designed to have one axial adamantyl unit and one trans succinate in order to act as a guest for the cage because four adamantyl groups can be loaded into the cage through the four open faces.510 The succinate ligand extends to the solvent and may provide enhanced water solubility and a reactive handle for further functionalization, although such functionalization was not explored in this proof-of-principle study. The host-guest complex exhibited significantly enhanced intracellular accumulation and DNA platination as compared to the un-encapsulated prodrug.
Figure 7.
Formation of a supramolecular drug delivery device driven by host-guest interactions between a platinum(IV) prodrug and a platinum(II) cage. Reproduced from reference510. Copyright © 2015, The Royal Society of Chemistry.
8.9. Proteins
In addition to the peptide targeting systems described in Section 6, entire proteins have been used as drug delivery devices. The α-helical right handed coiled coil (RHCC) is a 20 kDa portion of the tetrabrachion surface complex of Staphylothermus marinus.511 This extremophile colonizes exceedingly harsh environments, and the RHCC is correspondingly able to withstand extreme pH, boiling temperatures, high pressures, and high salt concentrations.512 The hydrophobic pockets of this tetramer were found to bind cisplatin and this platinum-loaded sell-assembled polypeptide structure was investigated as a drug-delivery vehicle.513 The protein-based construct did not elicit a significant immune response in mice, but the short half-life of the construct posed a significant challenge to further development. The authors subsequently investigated the ability of platinum(IV) species to be delivered by this tetramer.514 The chemical identity of the platinum species used is not indicated in the paper but it is referred to as “PtCl4” and we surmise that it is cis-[Pt(NH3)2Cl4] as an extension of the earlier cisplatin work. The construct is taken up by clathrin-mediated endocytosis and shown to be effective in propagated cell lines and in primary glioblastoma cells obtained from adult glioblastoma patients. Immunoblotting analyses indicated that apoptotic pathways were triggered by the construct. As compared to treatment with the un-encapsulated platinum(IV) complex, intratumoral injection of the construct was better able to inhibit the growth of subcutaneous xenograft and intracerebral orthotopic tumors in mice.
Serum albumin is the most abundant protein in human blood and any intravenously administered drug will inevitably encounter it. In many cases this interaction may serve to sequester and deactivate the compound. In an effort to capitalize on the ability of human serum albumin (HSA) to act as a drug delivery vehicle, a platinum(IV) prodrug was designed to mimic the form of the fatty acids that this protein is known to bind. The complex cis, cis, trans-[Pt(NH3)2Cl2(O2CCH2CH2CO2H)(OC(O)NH(CH2)15)CH3] interacts with HSA in a non-covalent, well-defined manner (Figure 8).515 A 1:1 complex of the platinum(IV) prodrug and HSA forms spontaneously on mixing and is sufficiently robust that is can be purified by fast protein liquid chromatography. Fluorescence quenching and modelling studies suggest that the complex is buried beneath the surface of the protein and this encapsulation inhibits reduction by ascorbic acid. Significant enhancement in blood stability as compared to cisplatin or satraplatin was consequently realized.
Figure 8.
A) A platinum(IV) prodrug designed to mimic a fatty acid. B) The modelled complex of the platinum(IV) prodrug in human serum albumin. Adapted from reference515. Copyright © 2014, The American Chemical Society.
9. Nanodelivery of platinum(II)
A great number of nanoconstructs designed to deliver platinum(II) complexes have also been described. These have recently been reviewed extensively by a number of different authors74,114,516–522 and so we will not give an exhaustive account of such constructs here. We would be remiss, however, if in a review of the next generation of platinum anticancer drugs we did not provide an overview of the significant clinical progress that has been made with certain macromolecular or self-assembled construct that directly incorporate platinum(II) species.
9.1. ProLindac
A number of platinated polymers have been prepared in which the leaving group ligands of either cisplatin/carboplatin or oxaliplatin have been replace by a chelating motif attached to the polymer backbone.520 In one such construct, the chloride ligands of cisplatin were replaced by an O-N chelate from the pendent tetrapeptide arms of a functionalized poly(N-(2-hydroxypropyl)methacrylamide) (HPMA), a hydrophilic, non-toxic, non-immunogenic polymer that persists for extended periods of time in circulation.523 The resulting platinum-conjugate, AP5280, was enrolled in a Phase I clinical trial by Access Pharmaceuticals, but the lack of significant response led the company to discontinue its development.524 AP5346, also known as ProLindac, is the oxaliplatin analogue of this construct in which the platinum is chelated by an amidomalonate attached to the HPMA via a triglycine spacer.525 Following promising preclinical studies,526 ProLindac was used in a Phase I clinical trial in which treatment was tolerated well and patients experienced no significant impact on blood cell counts.527 A subsequent Phase I/II trial in patients with advanced ovarian cancer showed that ProLindac treatment was again tolerated well and able to elicit an effect similar to that of oxaliplatin alone.526 In the ProLindac treated patients, no signs of acute neurotoxicity were observed, a significant finding given that this toxicity is dose-limiting in most oxaliplatin regimens. A Phase II trial in which ProLindac and pactitaxel are together used the second-line treatment of pre-treated advanced ovarian cancer began in 2010.526 The complete results of this study have not yet been released.
9.2. Lipoplatin
Liposomes are self-assembled vesicular structures composed of a lipid bilayer. They are attractive vehicles for drug delivery because they can encapsulate hydrophilic compounds in their aqueous lumen or hydrophobic compounds within the bilayer itself.528 Surface functionalization with PEG can produce so-called Stealth® liposomes,529 which demonstrate enhanced circulation by avoiding clearance from the bloodstream. The typical size of liposomes, approximately 100 nm diameters, suggests that these objects can accumulate in tumor tissue via the EPR effect.530 Clinical validation of the liposomal drug delivery strategy was realized with the approval of a liposomal formulation of doxorubicin. More recently, a liposomal formulation of vincristine also received approval for use in the United States. The first liposomal formulation of cisplatin to be tested in clinical trials was initially developed by SEQUUS Pharmaceuticals. The earliest preclinical data for the construct, referred to either as SPI-77 or SPI-077, appear in a press release from 1996 and the first published clinical data appeared in 1998.531–533 The data acquired in Phase I and Phase II clinical trials, concluding in 2001, showed a lack of improved efficacy as compared to treatments using standard cisplatin.521 In that same year, however, another liposomal cisplatin preparation with a different formulation began clinical trials. Known as lipoplatin, this 110 nm diameter nanoparticle has an aqueous core loaded with cisplatin that is contained by a liposomal vesicle comprising soy phosphatidyl choline, cholesterol, dipalmitoyl phosphatidyl glycerol, and methoxy-poly(ethylene glycol)-distearoyl phosphatidylethanolamine (Figure 9).534
Figure 9.
Artistic rendition of lipoplatin. The cisplatin core is shown as a blue, roughly spherical ball surrounded by a vesicular lipid bilayer. PEG chains protrude from the surface of the liposome. Adapted from reference534. Copyright © 2012, G. P. Stathopoulos and T. Boulikas (Creative Commons Attribution License).
Lipoplatin was first validated in preclinical models and subsequently in a range of clinical studies including three Phase III trials. Lipoplatin has been used in a clinical setting on non-small cell lung cancer but has also been investigated in cancers of the breast, pancreas, and head and neck. A recent review by the developers of lipoplatin provides a comprehensive overview of the clinical progress that this investigational drug has made.534 Most recently, the results of a Phase III clinical trial with 202 patients were analysed and the authors concluded that lipoplatin in combination with paclitaxel produces a response rate in non-squamous NSCLC patients that statistically greater than treatment with cisplatin and paclitaxel. Moreover, nephrotoxicity, the dose-limiting toxicity of cisplatin, was greatly reduced. Regulon, the company developing lipoplatin has announced that the EMA has granted it approval to launch a Phase III clinical trial with 884 patients testing the efficacy of lipoplatin and pemetrexed versus cisplatin and pemetrexed as a first-line therapy against non-squamous non-small cell lung cancer. Two other Phase III trials are underway in Europe. One, started in 2006, is comparing lipoplatin and paclitaxel versus cisplatin and paclitaxel as front line treatment of advanced epithelial ovarian cancer. The other, launched in 2012, is investigating lipoplatin and gemcitabine versus gemcitabine as a first-line treatment in inoperable, locally advanced or metastatic pancreatic cancer. With the progress that lipoplatin has already made, it seems poised on the brink of becoming the next platinum drug and could serve as an excellent validation and motivation for those researchers and companies seeking to develop nanodelivery devices to enhance platinum-based anticancer therapy. The trajectory of satraplatin, however, provides a cautionary tale highlighting that even encouraging progress made during clinical trials does not guarantee regulatory approval.
10. Summary and outlook
In this review we have highlighted the work done to generate the next generation of platinum drugs. Although the scientific literature presents evidence that a significant effort continues in the area of preparing cisplatin derivatives that are expected to function via a similar mechanism of action, the research efforts of the community have been turning steadily towards the development of molecules that deviate increasingly in structure and mechanism. The efforts we have highlighted here include the incorporation of targeting agents into the molecular scaffolds of classical platinum(II) complexes, non-classical platinum(II) scaffolds that elicit biological effects distinct from those of the approved platinum drugs, and oxidation of active the platinum(II) complexes to platinum(IV) prodrugs that can be reductively released. The axial ligands of the platinum(IV) prodrug can be chosen so as to tune physico-chemical properties, unleash an orthogonal biological response, or facilitate incorporation into a drug delivery device. The use of nanoscale drug delivery devices is a particularly explosive area of research. Liposomes were one of the earliest nanoscale platforms to be developed for drug delivery, and the clinical approval of doxorubicin and vincristine liposomal formulations validates these research efforts. Lipoplatin, a liposomal formulation of cisplatin, has also progressed well in clinical trials and may indeed become the next platinum-based drug. We anticipate that the renewed interest in developing platinum agents, particularly in nanoparticle formulations, by researchers around the world will generate an increased flow of these platinum drug candidates into the development pipeline, ushering in the next generation of platinum drugs.
Table 1.
Clinically approved platinum anticancer agents.
| Generic Name |
Research Name |
Trade Name |
Approval Granted |
Tenure of approval |
|---|---|---|---|---|
| Cispatin | CDDP | Platinol | 1978 | Global |
| Carboplatin | JM8 | Paraplatin | 1989 | Global |
| Oxaliplatin | l-OHP | Eloxatin | 2002 | Global |
| Nedaplatin | 254-S | Aqupla アクプラ |
1995 | Japan |
| Heptaplatin | SKI 2053R | Sunpla 선플라 |
1999 | Korea |
| Lobaplatin | D-19466 | 洛鉑 | 2010† | China |
See main test for discussion
Acknowledgments
This work was supported by NCI grant XX. Y.-R. Zheng is thanked for his assistance is navigating the Chinese FDA Database.
References
- 1.Howlader N, Noone AM, Krapcho M, Neyman N, Aminou R, Waldron W, Altekruse SF, Kosary CL, Ruhl J, Tatalovich Z, Cho H, Mariotto A, Eisner MP, Lewis DR, Chen HS, Feuer EJ, Cronin KA. Bethesda, MD: National Cancer Institute; 2012. [Google Scholar]
- 2.World Health Organization. WHO Model List of Essential Medicines. 2013 [Google Scholar]
- 3.Centers for Disease Control and Prevention. [Accessed: 2015];Ambulatory Care Drug Database System. http://www.cdc.gov/nchs/ahcd/ahcd_database.htm.
- 4.National Institutes of Health. [Accessed: 2015]; www.clinicaltrials.gov.
- 5.Haug C, Gøtzsche PC, Schroeder TV. N Engl. J. Med. 2005;353:2811–2812. doi: 10.1056/NEJMe058280. [DOI] [PubMed] [Google Scholar]
- 6.W. H. Organization. [Accessed: 2015];International Clinical Trials Registry Platform (ICTRP) < http://www.who.int/ictrp/en/>.
- 7.Kelland L. Nat. Rev. Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 8.Jamieson ER, Lippard SJ. Chem. Rev. 1999;99:2467–2498. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]
- 9.Kelland LR, Farrell N. Platinum-Based Drugs in Cancer Therapy. Totowa, N.J.: Humana Press; 2000. [Google Scholar]
- 10.Wang D, Lippard SJ. Nat. Rev. Drug Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 11.Jung Y, Lippard SJ. Chem. Rev. 2007;107:1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 12.Dabrowiak JC. Metals in Medicine. Hoboken: Wiley; 2009. pp. 109–147. [Google Scholar]
- 13.Wheate NJ, Walker S, Craig GE, Oun R. Dalton Trans. 2010;39:8113–8127. doi: 10.1039/c0dt00292e. [DOI] [PubMed] [Google Scholar]
- 14.Dhar S, Lippard SJ. Platinum and Other Heavy Metal Compounds in Cancer Chemotherapy. 2009:135–147. [Google Scholar]
- 15.Johnstone TC, Suntharalingam K, Lippard SJ. Philos. Trans. R. Soc., A. 2015;373:20140185. doi: 10.1098/rsta.2014.0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gately DP, Howell SB. Br. J. Cancer. 1993;67:1171–1176. doi: 10.1038/bjc.1993.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gale GR, Morris CR, Atkins LM, Smith AB. Cancer Res. 1973;33:813–818. [PubMed] [Google Scholar]
- 18.Ogawa M, Gale GR, Keirn SS. Cancer Res. 1975;35:1398–1401. [PubMed] [Google Scholar]
- 19.Binks SP, Dobrota M. Biochem. Pharmacol. 1990;40:1329–1336. doi: 10.1016/0006-2952(90)90400-f. [DOI] [PubMed] [Google Scholar]
- 20.Andrews PA, Mann SC, Velury S, Howell SB. In: Platinum and Other Metal Coordination Compounds in Cancer Chemotherapy. Nicolini M, editor. Vol. 54. Springer US: 1988. pp. 248–254. [Google Scholar]
- 21.Howell SB, Safaei R, Larson CA, Sailor MJ. Mol. Pharmacol. 2010;77:887–894. doi: 10.1124/mol.109.063172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abada P, Howell SB. Met.-Based Drugs. 2010;2010:317581. doi: 10.1155/2010/317581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang S, Lovejoy KS, Shima JE, Lagpacan LL, Shu Y, Lapuk A, Chen Y, Komori T, Gray JW, Chen X, Lippard SJ, Giacomini KM. Cancer Res. 2006;66:8847–8857. doi: 10.1158/0008-5472.CAN-06-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dornish JM, Pettersen EO. Biochem. Pharmacol. 1990;39:309–318. doi: 10.1016/0006-2952(90)90030-o. [DOI] [PubMed] [Google Scholar]
- 25.Reishus JW, Martin DS., Jr J Am. Chem. Soc. 1961;83:2457–2462. [Google Scholar]
- 26.Frey U, Ranford JD, Sadler PJ. Inorg. Chem. 1993;32:1333–1340. [Google Scholar]
- 27.Jerremalm E, Videhult P, Alvelius G, Griffiths WJ, Bergman T, Eksborg S, Ehrsson H. J Pharm. Sci. 2002;91:2116–2121. doi: 10.1002/jps.10201. [DOI] [PubMed] [Google Scholar]
- 28.Di Pasqua AJ, Goodisman J, Kerwood DJ, Toms BB, Dubowy RL, Dabrowiak JC. Chem. Res. Toxicol. 2006;19:139–149. doi: 10.1021/tx050261s. [DOI] [PubMed] [Google Scholar]
- 29.Junker A, Farinotti R. Ann. Pharmacother. 2009;43:390–391. doi: 10.1345/aph.1L122. [DOI] [PubMed] [Google Scholar]
- 30.Di Pasqua AJ, Kerwood DJ, Shi Y, Goodisman J, Dabrowiak JC. Dalton Trans. 2011;40:4821–4825. doi: 10.1039/c0dt01758b. [DOI] [PubMed] [Google Scholar]
- 31.Brouwer J, van de Putte P, Fichtinger-Schepman AMJ, Reedijk J. Proc. Natl. Acad. Sci. U.S.A. 1981;78:7010–7014. doi: 10.1073/pnas.78.11.7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Casini A, Reedijk J. Chem. Sci. 2012;3:3135–3144. [Google Scholar]
- 33.Mezencev R. Curr. Cancer Drug Targets. 2014;14:794–816. doi: 10.2174/1568009614666141128105146. [DOI] [PubMed] [Google Scholar]
- 34.Osborn MF, White JD, Haley MM, DeRose VJ. ACS Chem. Biol. 2014;9:2404–2411. doi: 10.1021/cb500395z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bancroft DP, Lepre CA, Lippard SJ. J Am. Chem. Soc. 1990;112:6860–6871. [Google Scholar]
- 36.Fichtinger-Schepman AMJ, van der Veer JL, Den Hartog JHJ, Lohman PHM, Reedijk J. Biochemistry. 1985;24:707–713. doi: 10.1021/bi00324a025. [DOI] [PubMed] [Google Scholar]
- 37.Kartalou M, Essigmann JM. Mutat. Res. 2001;478:1–21. doi: 10.1016/s0027-5107(01)00142-7. [DOI] [PubMed] [Google Scholar]
- 38.Blommaert FA, van Dijk-Knijnenburg HCM, Dijt FJ, den Engelse L, Baan RA, Berends F, Fichtinger-Schepman AMJ. Biochemistry. 1995;34:8474–8480. doi: 10.1021/bi00026a031. [DOI] [PubMed] [Google Scholar]
- 39.Woynarowski JM, Chapman WG, Napier C, Herzig MCS, Juniewicz P. Mol. Pharmacol. 1998;54:770–777. doi: 10.1124/mol.54.5.770. [DOI] [PubMed] [Google Scholar]
- 40.Cohen GL, Bauer WR, Barton JK, Lippard SJ. Science. 1979;203:1014–1016. doi: 10.1126/science.370979. [DOI] [PubMed] [Google Scholar]
- 41.Todd RC, Lippard SJ. Metallomics. 2009;1:280–291. doi: 10.1039/b907567d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rabik CA, Dolan ME. Cancer Treat. Rev. 2007;33:9–23. doi: 10.1016/j.ctrv.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graf N, Ang WH, Zhu G, Myint M, Lippard SJ. Chem Bio Chem. 2011;12:1115–1123. doi: 10.1002/cbic.201000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bianchi ME, Beltrame M, Paonessa G. Science. 1989;243:1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- 45.Thomas JO, Travers AA. Trends Biochem. Sci. 2001;26:167–174. doi: 10.1016/s0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- 46.Pil PM, Lippard SJ. Science. 1992;256:234–237. doi: 10.1126/science.1566071. [DOI] [PubMed] [Google Scholar]
- 47.Park S, Lippard SJ. Biochemistry. 2011;50:2567–2574. doi: 10.1021/bi2000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park S, Lippard SJ. Biochemistry. 2012;51:6728–6737. doi: 10.1021/bi300649v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnstone TC, Wilson JJ, Lippard SJ. Inorg. Chem. 2013;52:12234–12249. doi: 10.1021/ic400538c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Esposito BP, Najjar R. Coord. Chem. Rev. 2002;232:137–149. [Google Scholar]
- 51.Ivanov AI, Christodoulou J, Parkinson JA, Barnham KJ, Tucker A, Woodrow J, Sadler PJ. J Biol. Chem. 1998;273:14721–14730. doi: 10.1074/jbc.273.24.14721. [DOI] [PubMed] [Google Scholar]
- 52.Rosenberg B, Vancamp L, Krigas T. Nature. 1965;205:698–699. doi: 10.1038/205698a0. [DOI] [PubMed] [Google Scholar]
- 53.Rosenberg B, Van Camp L, Grimley EB, Thomson AJ. J Biol. Chem. 1967;242:1347–1352. [PubMed] [Google Scholar]
- 54.Rosenberg B, Renshaw E, Vancamp L, Hartwick J, Drobnik J. J Bacteriol. 1967;93:716–721. doi: 10.1128/jb.93.2.716-721.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosenberg B, VanCamp L, Trosko JE, Mansour VH. Nature. 1969;222:385–386. doi: 10.1038/222385a0. [DOI] [PubMed] [Google Scholar]
- 56.Rosenberg B. In: Cisplatin: Chemistry and Biochemistry of a Leading Anticancer Drug. Lippert B, editor. Zürich: Verlag Helvetica Chimica Acta; 1999. pp. 1–27. [Google Scholar]
- 57.Peyrone M. Justus Liebigs Ann. Chem. 1844;51:1–29. [Google Scholar]
- 58.Lebwohl D, Canetta R. Eur. J. Cancer. 1998;34:1522–1534. doi: 10.1016/s0959-8049(98)00224-x. [DOI] [PubMed] [Google Scholar]
- 59.US National Library of Medicine. [Accessed: 2015];Cisplatin Injection. < http://www.nlm.nih.gov/medlineplus/druginfo/meds/a684036.html>.
- 60.Cleare MJ, Hoeschele JD. Bioinorg. Chem. 1973;2:187–210. [Google Scholar]
- 61.Alberts DS, Dorr RT. Oncologist. 1998;3:15–34. [PubMed] [Google Scholar]
- 62.US National Library of Medicine. [Accessed: 2015];Carboplatin Injection. < http://www.nlm.nih.gov/medlineplus/druginfo/meds/a684036.html>.
- 63.Shimada M, Itamochi H, Kigawa J. Cancer Manage. Res. 2013;5:67–76. doi: 10.2147/CMAR.S35785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Welink J, Boven E, Vermorken JB, Gall HE, van der Vijgh WJF. Clin. Cancer Res. 1999;5:2349–2358. [PubMed] [Google Scholar]
- 65.Drugs in R & D. 2003;4:369–372. doi: 10.2165/00126839-200304060-00008. [DOI] [PubMed] [Google Scholar]
- 66. , 2003. [Google Scholar]
- 67.Chinese State Food and Drug Administration. [Accessed: 2015];State Food and Drug Administration Database: Lobaplatin. < sfda.gov.cn>.
- 68.Graham J, Muhsin M, Kirkpatrick P. Nat. Rev. Drug Discov. 2004;3:11–12. doi: 10.1038/nrd1601. [DOI] [PubMed] [Google Scholar]
- 69.Johnstone TC. Polyhedron. 2014;67:429–435. doi: 10.1016/j.poly.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kidani Y, Inagaki K, Iigo M, Hoshi A, Kuretani K. J Med. Chem. 1978;21:1315–1318. doi: 10.1021/jm00210a029. [DOI] [PubMed] [Google Scholar]
- 71.Spingler B, Whittington DA, Lippard SJ. Inorg. Chem. 2001;40:5596–5602. doi: 10.1021/ic010790t. [DOI] [PubMed] [Google Scholar]
- 72.Ziegler CJ, Silverman AP, Lippard SJ. J Biol. Inorg. Chem. 2000;5:774–783. doi: 10.1007/s007750000170. [DOI] [PubMed] [Google Scholar]
- 73.Strebhardt K, Ullrich A. Nat. Rev. Cancer. 2008;8:473–480. doi: 10.1038/nrc2394. [DOI] [PubMed] [Google Scholar]
- 74.Wang X, Guo Z. Chem. Soc. Rev. 2013;42:202–224. doi: 10.1039/c2cs35259a. [DOI] [PubMed] [Google Scholar]
- 75.Tannock IF, Rotin D. Cancer Res. 1989;49:4373–4384. [PubMed] [Google Scholar]
- 76.Dam TK, Brewer CF. Glycobiology. 2009;20:270–279. doi: 10.1093/glycob/cwp186. [DOI] [PubMed] [Google Scholar]
- 77.Vyas SP, Singh A, Sihorkar V. Crit. Rev. Ther. Drug Carrier Syst. 2001;18:76. [PubMed] [Google Scholar]
- 78.Storr T, Thompson KH, Orvig C. Chem. Soc. Rev. 2006;35:534–544. doi: 10.1039/b514859f. [DOI] [PubMed] [Google Scholar]
- 79.Shaw RJ. Curr. Opin. Cell Biol. 2006;18:598–608. doi: 10.1016/j.ceb.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 80.Heiden MGV, Cantley LC, Thompson CB. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lindholm P, Minn H, Leskinenkallio S, Bergman J, Ruotsalainen U, Joensuu H. J Nucl. Med. 1993;34:1–6. [PubMed] [Google Scholar]
- 82.Delbeke D. J Nucl. Med. 1999;40:591–603. [PubMed] [Google Scholar]
- 83.Thiel G, Beck W. Z Naturforsch. 1983;38B:1081–1093. [Google Scholar]
- 84.Nagel Y, Beck W. Z Naturforsch. 1985;40B:1181–1187. [Google Scholar]
- 85.Kuduk-Jaworska J, Jeżowska-Trzebiatowska B. Inorg. Chim. Acta. 1986;123:209–212. [Google Scholar]
- 86.Haroutounian SA, Georgiadis MP, Bailar JC. Inorg. Chim. Acta. 1986;124:137–139. [Google Scholar]
- 87.Pill T, Polborn K, Beck W. Chemische Berichte. 1990;123:11–17. [Google Scholar]
- 88.Tsubomura T, Yano S, Kobayashi K, Sakurai T, Yoshikawa S. J Chem. Soc., Chem. Commun. 1986;459 [Google Scholar]
- 89.Berger I, Nazarov AA, Hartinger CG, Groessl M, Valiahdi S-M, Jakupec MA, Keppler BK. Chem Med Chem. 2007;2:505–514. doi: 10.1002/cmdc.200600279. [DOI] [PubMed] [Google Scholar]
- 90.Hanessian S, Wang J. Can. J. Chem. 1993;71:886–895. [Google Scholar]
- 91.Kidani Y, Inagaki K, Tsukagoshi S. Gann. 1976;67:921–922. [PubMed] [Google Scholar]
- 92.Tsubomura T, Ogawa M, Yano S, Kobayashi K, Sakurai T, Yoshikawa S. Inorg. Chem. 1990;29:2622–2626. [Google Scholar]
- 93.Montaña ÁM, Bernal FJ, Lorenzo J, Farnós C, Batalla C, Prieto MJ, Moreno V, Avilés FX, Mesas JM, Alegre M-T. Bioorg. Med. Chem. 2008;16:1721–1737. doi: 10.1016/j.bmc.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 94.Chen Y, Heeg MJ, Braunschweiger PG, Xie W, Wang PG. Angew. Chem., Int. Ed. 1999;38:1768–1769. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1768::AID-ANIE1768>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 95.Mikata Y, Shinohara Y, Yoneda K, Nakamura Y, Brudziñska I, Tanase T, Kitayama T, Takagi R, Okamoto T, Kinoshita I, Doe M, Orvig C, Yano S. Bioorg. Med. Chem. Lett. 2001;11:3045–3047. doi: 10.1016/s0960-894x(01)00623-0. [DOI] [PubMed] [Google Scholar]
- 96.De Almeida MV, Cesar ET, Fontes APS, Felício EdCA. J Carbohydr. Chem. 2000;19:323–329. [Google Scholar]
- 97.Möker J, Thiem J. Eur. J. Org. Chem. 2009;2009:4842–4847. [Google Scholar]
- 98.Möker J, Salge-Bartels U, Thiem J. J Carbohydr. Chem. 2012;31:702–710. [Google Scholar]
- 99.Möker J, Thiem J. Carbohydr. Res. 2012;348:14–26. doi: 10.1016/j.carres.2011.08.024. [DOI] [PubMed] [Google Scholar]
- 100.Hartinger CG, Nazarov AA, Ashraf SM, Dyson PJ, Keppler BK. Curr. Med. Chem. 2008;15:2574–2591. doi: 10.2174/092986708785908978. [DOI] [PubMed] [Google Scholar]
- 101.Liu P, Lu Y, Gao X, Liu R, Zhang-Negrerie D, Shi Y, Wang Y, Wang S, Gao Q. Chem. Commun. 2013;49:2421–2423. doi: 10.1039/c3cc38589b. [DOI] [PubMed] [Google Scholar]
- 102.Sun L, Zeng X, Yan C, Sun X, Gong X, Rao Y, Yan N. Nature. 2012;490:361–366. doi: 10.1038/nature11524. [DOI] [PubMed] [Google Scholar]
- 103.Patra M, Johnstone TC, Suntharalingam K, Lippard SJ. Manuscript in preparation. 2015 [Google Scholar]
- 104.Knight WA, Livingston RB, Gregory EJ, McGuire WL. Cancer Res. 1977;37:4669–4671. [PubMed] [Google Scholar]
- 105.Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, Press MF. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 106.Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 107.Carlsson J, Nordgren H, Sjostrom J, Wester K, Villman K, Bengtsson NO, Ostenstad B, Lundqvist H, Blomqvist C. Br. J. Cancer. 2004;90:2344–2348. doi: 10.1038/sj.bjc.6601881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Scott AM, Wolchok JD, Old LJ. Nat. Rev. Cancer. 2012;12:278–287. doi: 10.1038/nrc3236. [DOI] [PubMed] [Google Scholar]
- 109.Gustaftson J-Å. Trends Pharmacol. Sci. 2003;24:479–485. doi: 10.1016/S0165-6147(03)00229-3. [DOI] [PubMed] [Google Scholar]
- 110.Koehler KF, Helguero LA, Haldosén L-A, Warner M, Gustafsson J-Å. Endocr. Rev. 2005;26:465–478. doi: 10.1210/er.2004-0027. [DOI] [PubMed] [Google Scholar]
- 111.Dahlman-Wright K, Cavailles V, Fuqua SA, Jordan VC, Katzenellenbogen JA, Korach KS, Maggi A, Muramatsu M, Parker MG, Gustafsson J-Å. Pharmacol. Rev. 2006;58:773–781. doi: 10.1124/pr.58.4.8. [DOI] [PubMed] [Google Scholar]
- 112.Hartman J, Lindberg K, Morani A, Inzunza J, Strom A, Gustafsson JA. Cancer Res. 2006;66:11207–11213. doi: 10.1158/0008-5472.CAN-06-0017. [DOI] [PubMed] [Google Scholar]
- 113.Gust R, Beck W, Jaouen G, Schönenberger H. Coord. Chem. Rev. 2009;253:2742–2759. [Google Scholar]
- 114.Gabano E, Ravera M, Osella D. Curr. Med. Chem. 2009;16:4544–4580. doi: 10.2174/092986709789760661. [DOI] [PubMed] [Google Scholar]
- 115.Gandolfi O, Cais M, Dolcetti G, Ghedini M, Modiano A. Inorg. Chim. Acta. 1981;56:127–133. [Google Scholar]
- 116.Gandolfi O, Blum J. Inorg. Chim. Acta. 1983;80:103–106. [Google Scholar]
- 117.Gandolfi O, Blum J, Mandelbaum-Shavit F. Inorg. Chim. Acta. 1984;91:257–261. [Google Scholar]
- 118.Gabano E, Cassino C, Bonetti S, Prandi C, Colangelo D, Ghiglia A, Osella D. Org. Biomol. Chem. 2005;3:3531–3539. doi: 10.1039/b507716h. [DOI] [PubMed] [Google Scholar]
- 119.Descôteaux C, Provencher-Mandeville J, Mathieu I, Perron V, Mandal SK, Asselin É, Bérubé G. Bioorg. Med. Chem. Lett. 2003;13:3927–3931. doi: 10.1016/j.bmcl.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 120.Gagnon V, St-Germain M-È, Descôteaux C, Provencher-Mandeville J, Parent S, Mandal SK, Asselin E, Bérubé G. Bioorg. Med. Chem. Lett. 2004;14:5919–5924. doi: 10.1016/j.bmcl.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 121.Perron V, Rabouin D, Asselin É, Parent S, Gaudreault RC, Bérubé G. Bioorganic Chemistry. 2005;33:1–15. doi: 10.1016/j.bioorg.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 122.Descôteaux C, Leblanc V, Belanger G, Parent S, Asselin E, Bérubé G. Steroids. 2008;73:1077–1089. doi: 10.1016/j.steroids.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 123.Provencher-Mandeville J, Descôteaux C, Mandal SK, Leblanc V, Asselin É, Bérubé G. Bioorg. Med. Chem. Lett. 2008;18:2282–2287. doi: 10.1016/j.bmcl.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 124.Saha P, Descôteaux C, Brasseur K, Fortin S, Leblanc V, Parent S, Asselin É, Bérubé G. Eur. J. Med. Chem. 2012;48:385–390. doi: 10.1016/j.ejmech.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 125.Fortin S, Brasseur K, Morin N, Asselin É, Bérubé G. Eur. J. Med. Chem. 2013;68:433–443. doi: 10.1016/j.ejmech.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 126.Kim E, Rye PT, Essigmann JM, Croy RG. J Inorg. Biochem. 2009;103:256–261. doi: 10.1016/j.jinorgbio.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wappes B, Jennerwein M, Von Angerer E, Schoenenberger H, Engel J, Berger M, Wrobel KH. J Med. Chem. 1984;27:1280–1286. doi: 10.1021/jm00376a009. [DOI] [PubMed] [Google Scholar]
- 128.Hartmann RW, Heindl A, Schwarz W, Schoenenberger H. J Med. Chem. 1984;27:819–824. doi: 10.1021/jm00373a001. [DOI] [PubMed] [Google Scholar]
- 129.Karl J, Gust R, Spruss T, Schneider MR, Schoenenberger H, Engel J, Wrobel KH, Lux F, Haeberlin ST. J Med. Chem. 1988;31:72–83. doi: 10.1021/jm00396a012. [DOI] [PubMed] [Google Scholar]
- 130.Gust R, Niebler K, Schönenberger H. J Med. Chem. 2005;48:7132–7144. doi: 10.1021/jm050186i. [DOI] [PubMed] [Google Scholar]
- 131.Trauner M, Boyer JL. Physiol. Rev. 2003;83:633–671. doi: 10.1152/physrev.00027.2002. [DOI] [PubMed] [Google Scholar]
- 132.Maeda M, Suga T, Takasuka N, Hoshi A, Sasaki T. Cancer Lett. 1990;55:143–147. doi: 10.1016/0304-3835(90)90024-r. [DOI] [PubMed] [Google Scholar]
- 133.Maeda M, Tanaka S, Ida H, Takasuka N, Uehera N, Hoshi A. Cancer Lett. 1993;70:57–64. doi: 10.1016/0304-3835(93)90075-k. [DOI] [PubMed] [Google Scholar]
- 134.Criado JJ, Herrera MC, Palomero MF, Medarde M, Rodriguez E, Marin JJG. J Lipid Res. 1997;38:1022–1032. [PubMed] [Google Scholar]
- 135.Criado JJ, Domínguez MF, Medarde M, Fernández ER, Macías RIR, Marín JJG. Bioconjugate Chem. 2000;11:167–174. doi: 10.1021/bc9901088. [DOI] [PubMed] [Google Scholar]
- 136.Briz O, Serrano MA, Macias RIR, Marin JJG. Int. J. Cancer. 2000;88:287–292. [PubMed] [Google Scholar]
- 137.Criado JJ, Macias RIR, Medarde M, Monte MJ, Serrano MA, Marin JJG. Bioconjugate Chem. 1997;8:453–458. doi: 10.1021/bc970061v. [DOI] [PubMed] [Google Scholar]
- 138.Marin JJG, Macias RIR, Criado JJ, Bueno A, Monte MJ, Serrano MA. Int. J. Cancer. 1998;78:346–352. doi: 10.1002/(SICI)1097-0215(19981029)78:3<346::AID-IJC15>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 139.Martinez-Diez MC, Larena MG, Serrano MA, Macias RIR, Izco-Basurko I, Marin JJG. Anticancer Res. 2000;20:3315–3321. [PubMed] [Google Scholar]
- 140.Larena MG, Martinez-Diez MC, Macias RIR, Dominguez MF, Serrano MA, Marin JJG. J Drug Targeting. 2002;10:397–404. doi: 10.1080/1061186021000001841. [DOI] [PubMed] [Google Scholar]
- 141.Almendral-Parra MJ, Alonso A, Criado JJ, Manzano JL, Rodríguez-Fernández E. Inorg. Chim. Acta. 2011;376:651–654. [Google Scholar]
- 142.Criado JJ, Fernández ER, Manzano JL, Alonso A, Barrena S, Medarde M, Pelaez R, Tabernero MD, Orfao A. Bioconjugate Chem. 2005;16:275–282. doi: 10.1021/bc049788r. [DOI] [PubMed] [Google Scholar]
- 143.Alonso A, Almendral MJ, Curto Y, Criado JJ, Rodríguez E, Manzano JL. J Fluoresc. 2007;17:390–400. doi: 10.1007/s10895-007-0211-3. [DOI] [PubMed] [Google Scholar]
- 144.Pérez-Andrés M, Benito JJ, Rodríguez-Fernández E, Corradetti B, Primo D, Manzano JL, Orfao A, Criado JJ. Dalton Trans. 2008;6159 doi: 10.1039/b807965j. [DOI] [PubMed] [Google Scholar]
- 145.González M, Bartolomé R, Matarraz S, Rodríguez-Fernández E, Manzano JL, Pérez-Andrés M, Orfao A, Fuentes M, Criado JJ. J Inorg. Biochem. 2012;106:43–45. doi: 10.1016/j.jinorgbio.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 146.Marin JJG, Romero MR, Vallejo M, Monte MJ. Cancer Ther. 2005;3:57–64. [Google Scholar]
- 147.Macias RIR, Monte MJ, El-Mir MY, Villanueva GR, Marin JJG. J Lipid Res. 1998;39:1792–1798. [PubMed] [Google Scholar]
- 148.Macias R. J Controlled Release. 1999;57:161–169. doi: 10.1016/s0168-3659(98)00114-x. [DOI] [PubMed] [Google Scholar]
- 149.Barbara C, Orlandi P, Bocci G, Fioravanti A, Di Paolo A, Natale G, Del Tacca M, Danesi R. Eur. J. Pharmacol. 2006;549:27–34. doi: 10.1016/j.ejphar.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 150.Paschke R, Kalbitz J, Paetz C. Inorg. Chim. Acta. 2000;304:241–249. [Google Scholar]
- 151.Paschke R, Kalbitz J, Paetz C, Luckner M, Mueller T, Schmoll H-J, Mueller H, Sorkau E, Sinn E. J Inorg. Biochem. 2003;94:335–342. doi: 10.1016/s0162-0134(03)00024-2. [DOI] [PubMed] [Google Scholar]
- 152.Schobert R, Biersack B, Dietrich A, Grotemeier A, Müller T, Kalinowski B, Knauer S, Voigt W, Paschke R. J Med. Chem. 2007;50:1288–1293. doi: 10.1021/jm061379o. [DOI] [PubMed] [Google Scholar]
- 153.Bernhardt G, Biersack B, Bollwein S, Schobert R, Zoldakova M. Chem. Biodivers. 2008;5:1645–1659. doi: 10.1002/cbdv.200890152. [DOI] [PubMed] [Google Scholar]
- 154.Biersack B, Dietrich A, Zoldakova M, Kalinowski B, Paschke R, Schobert R, Mueller T. J Inorg. Biochem. 2011;105:1630–1637. doi: 10.1016/j.jinorgbio.2011.08.028. [DOI] [PubMed] [Google Scholar]
- 155.Emmerich D, Vanchanagiri K, Baratto LC, Schmidt H, Paschke R. Eur. J. Med. Chem. 2014;75:460–466. doi: 10.1016/j.ejmech.2014.01.031. [DOI] [PubMed] [Google Scholar]
- 156.Papadopoulos V, Baraldi M, Guilarte TR, Knudsen TB, Lacapere J-J, Lindemann P, Norenberg MD, Nutt D, Weizman A, Zhang M-R, Gavish M. Trends Pharmacol. Sci. 2006;27:402–409. doi: 10.1016/j.tips.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 157.Morohaku K, Pelton SH, Daugherty DJ, Butler WR, Deng W, Selvaraj V. Endocrinology. 2014;155:89–97. doi: 10.1210/en.2013-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Galiegue S, Tinel N, Casellas P. Curr. Med. Chem. 2003;10:1563–1572. doi: 10.2174/0929867033457223. [DOI] [PubMed] [Google Scholar]
- 159.Miettinen H, Kononen J, Haapasalo H, Helen P, Sallinen P, Harjuntausta T, Helin H, Alho H. Cancer Res. 1995;55:2691–2695. [PubMed] [Google Scholar]
- 160.Maaser K, Grabowski P, Sutter AP, Hopfner M, Foss HD, Stein H, Berger G, Gavish M, Zeitz M, Scherubl H. Clin. Cancer Res. 2002;8:3205–3209. [PubMed] [Google Scholar]
- 161.Veenman L, Levin E, Weisinger G, Leschiner S, Spanier I, Snyder SH, Weizman A, Gavish M. Biochem. Pharmacol. 2004;68:689–698. doi: 10.1016/j.bcp.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 162.Margiotta N, Ostuni R, Ranaldo R, Denora N, Laquintana V, Trapani G, Liso G, Natile G. J Med. Chem. 2007;50:1019–1027. doi: 10.1021/jm0612160. [DOI] [PubMed] [Google Scholar]
- 163.Margiotta N, Denora N, Ostuni R, Laquintana V, Anderson A, Johnson SW, Trapani G, Natile G. J Med. Chem. 2010;53:5144–5154. doi: 10.1021/jm100429r. [DOI] [PubMed] [Google Scholar]
- 164.Weitman SD, Lark RH, Coney LR, Fort DW, Frasca V, Zurawski VR, Kamen BA. Cancer Res. 1992;52:3396–3401. [PubMed] [Google Scholar]
- 165.Sudimack J, Lee RJ. Adv. Drug Delivery Rev. 2000;41:147–162. doi: 10.1016/s0169-409x(99)00062-9. [DOI] [PubMed] [Google Scholar]
- 166.Vitols KS, Montejano Y, Duffy T, Pope L, Grundler G, Huennekens FM. Adv. Enzyme Regul. 1987;26:17–27. doi: 10.1016/0065-2571(87)90004-5. [DOI] [PubMed] [Google Scholar]
- 167.Gabano E, Ravera M, Cassino C, Bonetti S, Palmisano G, Osella D. Inorg. Chim. Acta. 2008;361:1447–1455. [Google Scholar]
- 168.Aronov O, Horowitz AT, Gabizon A, Gibson D. Bioconjugate Chem. 2003;14:563–574. doi: 10.1021/bc025642l. [DOI] [PubMed] [Google Scholar]
- 169.Lee M, Simpson JE, Burns AJ, Kupchinsky S, Brooks N, Hartley JA, Kelland LR. Medicinal Chemistry Research. 1996;6:365–371. [Google Scholar]
- 170.Robillard MS, Valentijn ARPM, Meeuwenoord NJ, van der Marel GA, van Boom JH, Reedijk J. Angewandte Chemie. 2000;39:3096–3099. doi: 10.1002/1521-3773(20000901)39:17<3096::aid-anie3096>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 171.Robillard MS, van Alphen S, Meeuwenoord NJ, Jansen BAJ, van der Marel GA, van Boom JH, Reedijk J. New J. Chem. 2005;29:220. [Google Scholar]
- 172.van Zutphen S, Stone E, van Rijt S, Robillard M, van der Marel G, Overkleeft H, den Dulk H, Brouwer J, Reedijk J. J Inorg. Biochem. 2005;99:2032–2038. doi: 10.1016/j.jinorgbio.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 173.Damian MS, Hedman HK, Elmroth SKC, Diederichsen U. Eur. J. Org. Chem. 2010;2010:6161–6170. [Google Scholar]
- 174.Robillard MS, Bacac M, van den Elst H, Flamigni A, van der Marel GA, van Boom JH, Reedijk J. J Comb. Chem. 2003;5:821–825. doi: 10.1021/cc030011z. [DOI] [PubMed] [Google Scholar]
- 175.Robillard MS, Davies NP, van der Marel GA, van Boom JH, Reedijk J, Murray V. J Inorg. Biochem. 2003;96:331–338. doi: 10.1016/s0162-0134(03)00180-6. [DOI] [PubMed] [Google Scholar]
- 176.Rijal K, Bao X, Chow CS. Chem. Commun. 2014;50:3918. doi: 10.1039/c3cc49035a. [DOI] [PubMed] [Google Scholar]
- 177.Barragán F, Moreno V, Marchán V. Chem. Commun. 2009:4705–4707. doi: 10.1039/b909698a. [DOI] [PubMed] [Google Scholar]
- 178.Ndinguri MW, Solipuram R, Gambrell RP, Aggarwal S, Hammer RP. Bioconjugate Chem. 2009;20:1869–1878. doi: 10.1021/bc900065r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Colombo G, Curnis F, De Mori GMS, Gasparri A, Longoni C, Sacchi A, Longhi R, Corti A. J Biol. Chem. 2002;277:47891–47897. doi: 10.1074/jbc.M207500200. [DOI] [PubMed] [Google Scholar]
- 180.Wilson JJ, Lippard SJ. J Med. Chem. 2012;55:5326–5336. doi: 10.1021/jm3002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wisnovsky SP, Wilson JJ, Radford RJ, Pereira MP, Chan MR, Laposa RR, Lippard SJ, Kelley SO. Chem. Biol. 2013;20:1323–1328. doi: 10.1016/j.chembiol.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Fonseca SB, Pereira MP, Mourtada R, Gronda M, Horton KL, Hurren R, Minden MD, Schimmer AD, Kelley SO. Chem. Biol. 2011;18:445–453. doi: 10.1016/j.chembiol.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 183.Horton KL, Pereira MP, Stewart KM, Fonseca SB, Kelley SO. ChemBioChem. 2012;13:476–485. doi: 10.1002/cbic.201100415. [DOI] [PubMed] [Google Scholar]
- 184.Dodd DW, Damjanovski S, Hudson RHE. Nucleosides, Nucleotides Nucleic Acids. 2011;30:257–263. doi: 10.1080/15257770.2011.580290. [DOI] [PubMed] [Google Scholar]
- 185.Cleare MJ, Hoeschele JD. Platinum Metals Rev. 1973;17:2–13. [Google Scholar]
- 186.Farrell N. Met. Ions Biol. Syst. 1996;32:603–639. [PubMed] [Google Scholar]
- 187.Pérez JM, Fuertes MA, Alonso C, Navarro-Ranninger C. Crit. Rev. Oncol. Hemat. 2000;35:109–120. doi: 10.1016/s1040-8428(00)00053-6. [DOI] [PubMed] [Google Scholar]
- 188.Coluccia M, Natile G. Anti-Cancer Agents Med. Chem. 2007;7:111–123. doi: 10.2174/187152007779314080. [DOI] [PubMed] [Google Scholar]
- 189.Aris SM, Farrell NP. Eur. J. Inorg. Chem. 2009;2009:1293–1302. doi: 10.1002/ejic.200801118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Brabec V, Leng M. Proc. Natl. Acad. Sci. U.S.A. 1993;90:5345–5349. doi: 10.1073/pnas.90.11.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bernal-Mendez E, Boudvillain M, Gonzalez-Vilchez F, Leng M. Biochemistry. 1997;36:7281–7287. doi: 10.1021/bi9703148. [DOI] [PubMed] [Google Scholar]
- 192.Leng M, Brabec V. IARC Sci. Publ. 1994:339–348. [PubMed] [Google Scholar]
- 193.Farrell N, Ha TTB, Souchard JP, Wimmer FL, Cros S, Johnson NP. J Med. Chem. 1989;32:2240–2241. doi: 10.1021/jm00130a002. [DOI] [PubMed] [Google Scholar]
- 194.Farrell N, Kelland LR, Roberts JD, Van Beusichem M. Cancer Res. 1992;52:5065–5072. [PubMed] [Google Scholar]
- 195.Kelland LR, Barnard CF, Mellish KJ, Jones M, Goddard PM, Valenti M, Bryant A, Murrer BA, Harrap KR. Cancer Res. 1994;54:5618–5622. [PubMed] [Google Scholar]
- 196.Murphy N RFF, Aguila A, Okada M, Balis FM, Fojo T. Proc. Am. Assoc. Cancer Res. 2005;46:4109. [Google Scholar]
- 197.Paull KD, Shoemaker RH, Hodes L, Monks A, Scudiero DA, Rubinstein L, Plowman J, Boyd MR. J Natl. Cancer Inst. 1989;81:1088–1092. doi: 10.1093/jnci/81.14.1088. [DOI] [PubMed] [Google Scholar]
- 198.Zakovska A, Novakova O, Balcarova Z, Bierbach U, Farrell N, Brabec V. Eur. J. Biochem. 1998;254:547–557. doi: 10.1046/j.1432-1327.1998.2540547.x. [DOI] [PubMed] [Google Scholar]
- 199.Brabec V, Neplechova K, Kasparkova J, Farrell N. J Biol. Inorg. Chem. 2000;5:364–368. doi: 10.1007/pl00010665. [DOI] [PubMed] [Google Scholar]
- 200.Brabec V. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:1–68. doi: 10.1016/s0079-6603(02)71040-4. [DOI] [PubMed] [Google Scholar]
- 201.Farrell N, Povirk LF, Dange Y, DeMasters G, Gupta MS, Kohlhagen G, Khan QA, Pommier Y, Gewirtz DA. Biochem. Pharmacol. 2004;68:857–866. doi: 10.1016/j.bcp.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 202.Coluccia M, Nassi A, Loseto F, Boccarelli A, Mariggio MA, Giordano D, Intini FP, Caputo P, Natile G. J Med. Chem. 1993;36:510–512. doi: 10.1021/jm00056a012. [DOI] [PubMed] [Google Scholar]
- 203.Coluccia M, Boccarelli A, Mariggio MA, Cardellicchio N, Caputo P, Intini FP, Natile G. Chem. Biol. Interact. 1995;98:251–266. doi: 10.1016/0009-2797(95)03650-4. [DOI] [PubMed] [Google Scholar]
- 204.Intini FP, Boccarelli A, Francia VC, Pacifico C, Sivo MF, Natile G, Giordano D, De Rinaldis P, Coluccia M. J Biol. Inorg. Chem. 2004;9:768–780. doi: 10.1007/s00775-004-0572-x. [DOI] [PubMed] [Google Scholar]
- 205.Cini R, Caputo PA, Intini FP, Natile G. Inorg. Chem. 1995;34:1130–1137. [Google Scholar]
- 206.Boccarelli A, Coluccia M, Intini FP, Natile G, Locker D, Leng M. Anticancer Drug Des. 1999;14:253–264. [PubMed] [Google Scholar]
- 207.Coluccia M, Nassi A, Boccarelli A, Giordano D, Cardellicchio N, Intini FP, Natile G, Barletta A, Paradiso A. Int. J. Oncol. 1999;15:1039–1044. doi: 10.3892/ijo.15.5.1039. [DOI] [PubMed] [Google Scholar]
- 208.Coluccia M, Nassi A, Boccarelli A, Giordano D, Cardellicchio N, Locker D, Leng M, Sivo M, Intini FP, Natile G. J Inorg. Biochem. 1999;77:31–35. doi: 10.1016/s0162-0134(99)00139-7. [DOI] [PubMed] [Google Scholar]
- 209.Brabec V, Vrana O, Novakova O, Kleinwachter V, Intini FP, Coluccia M, Natile G. Nucleic Acids Res. 1996;24:336–341. doi: 10.1093/nar/24.2.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Zaludova R, Zakovska A, Kasparkova J, Balcarova Z, Vrana O, Coluccia M, Natile G, Brabec V. Mol. Pharmacol. 1997;52:354–361. doi: 10.1124/mol.52.3.354. [DOI] [PubMed] [Google Scholar]
- 211.Novakova O, Kasparkova J, Malina J, Natile G, Brabec V. Nucleic Acids Res. 2003;31:6450–6460. doi: 10.1093/nar/gkg863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Liu Y, Sivo MF, Natile G, Sletten E. Met. Based Drugs. 2000;7:169–176. doi: 10.1155/MBD.2000.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Leng M, Locker D, Giraud-Panis M-J, Schwartz A, Intini FP, Natile G, Pisano C, Boccarelli A, Giordano D, Coluccia M. Mol. Pharmacol. 2000;58:1525–1535. doi: 10.1124/mol.58.6.1525. [DOI] [PubMed] [Google Scholar]
- 214.Boccarelli A, Intini FP, Sasanelli R, Sivo MF, Coluccia M, Natile G. J Med. Chem. 2006;49:829–837. doi: 10.1021/jm050986t. [DOI] [PubMed] [Google Scholar]
- 215.Montero EI, Diaz S, Gonzalez-Vadillo AM, Perez JM, Alonso C, Navarro-Ranninger C. J Med. Chem. 1999;42:4264–4268. doi: 10.1021/jm991015e. [DOI] [PubMed] [Google Scholar]
- 216.Pérez JM, Montero EI, González AM, Alvarez-Valdés A, Alonso C, Navarro-Ranninger C. J Inorg. Biochem. 1999;77:37–42. doi: 10.1016/s0162-0134(99)00143-9. [DOI] [PubMed] [Google Scholar]
- 217.Montero EI, Perez JM, Schwartz A, Fuertes MA, Malinge JM, Alonso C, Leng M, Navarro-Ranninger C. Chem Bio Chem. 2002;3:61–67. doi: 10.1002/1439-7633(20020104)3:1<61::AID-CBIC61>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 218.Prokop R, Kasparkova J, Novakova O, Marini V, Pizarro AM, Navarro-Ranninger C, Brabec V. Biochem. Pharmacol. 2004;67:1097–1109. doi: 10.1016/j.bcp.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 219.Billecke C, Finniss S, Tahash L, Miller C, Mikkelsen T, Farrell NP, Bogler O. Neuro. Oncol. 2006;8:215–226. doi: 10.1215/15228517-2006-004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Farrell N, Qu Y, Bierbach U, Valsecchi M, Mentab E. Cisplatin. Verlag Helvetica Chimica Acta: 2006. pp. 477–496. [Google Scholar]
- 221.Farrell N, Qu Y, Hacker MP. J Med. Chem. 1990;33:2179–2184. doi: 10.1021/jm00170a021. [DOI] [PubMed] [Google Scholar]
- 222.Farrell N. Cancer Invest. 1993;11:578–589. doi: 10.3109/07357909309011676. [DOI] [PubMed] [Google Scholar]
- 223.Zou Y, Van Houten B, Farrell N. Biochemistry. 1994;33:5404–5410. doi: 10.1021/bi00184a007. [DOI] [PubMed] [Google Scholar]
- 224.Yang D, van Boom SS, Reedijk J, van Boom JH, Farrell N, Wang AH. Nat. Struct. Biol. 1995;2:577–578. doi: 10.1038/nsb0795-577. [DOI] [PubMed] [Google Scholar]
- 225.Brabec V, Kasparkova J, Vrana O, Novakova O, Cox JW, Qu Y, Farrell N. Biochemistry. 1999;38:6781–6790. doi: 10.1021/bi990124s. [DOI] [PubMed] [Google Scholar]
- 226.Perego P, Caserini C, Gatti L, Carenini N, Romanelli S, Supino R, Colangelo D, Viano I, Leone R, Spinelli S, Pezzoni G, Manzotti C, Farrell N, Zunino F. Mol. Pharmacol. 1999;55:528–534. [PubMed] [Google Scholar]
- 227.Roberts JD, Peroutka J, Beggiolin G, Manzotti C, Piazzoni L, Farrell N. J Inorg. Biochem. 1999;77:47–50. doi: 10.1016/s0162-0134(99)00137-3. [DOI] [PubMed] [Google Scholar]
- 228.McGregor TD, Balcarova Z, Qu Y, Tran MC, Zaludova R, Brabec V, Farrell N. J Inorg. Biochem. 1999;77:43–46. doi: 10.1016/s0162-0134(99)00136-1. [DOI] [PubMed] [Google Scholar]
- 229.McGregor TD, Bousfield W, Qu Y, Farrell N. J Inorg. Biochem. 2002;91:212–219. doi: 10.1016/s0162-0134(02)00398-7. [DOI] [PubMed] [Google Scholar]
- 230.Pratesi G, Perego P, Polizzi D, Righetti SC, Supino R, Caserini C, Manzotti C, Giuliani FC, Pezzoni G, Tognella S, Spinelli S, Farrell N, Zunino F. Br. J. Cancer. 1999;80:1912–1919. doi: 10.1038/sj.bjc.6690620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Orlandi L, Colella G, Bearzatto A, Abolafio G, Manzotti C, Daidone MG, Zaffaroni N. Eur. J. Cancer. 2001;37:649–659. doi: 10.1016/s0959-8049(00)00445-7. [DOI] [PubMed] [Google Scholar]
- 232.Colella G, Pennati M, Bearzatto A, Leone R, Colangelo D, Manzotti C, Daidone MG, Zaffaroni N. Br. J. Cancer. 2001;84:1387–1390. doi: 10.1054/bjoc.2001.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sessa C, Capri G, Gianni L, Peccatori F, Grasselli G, Bauer J, Zucchetti M, Viganò L, Gatti A, Minoia C, Liati P, Van den Bosch S, Bernareggi A, Camboni G, Marsoni S. Ann. Oncol. 2000;11:977–983. doi: 10.1023/a:1008302309734. [DOI] [PubMed] [Google Scholar]
- 234.Calvert AH, Thomas H, Colombo N, Gore M, Earl H, Sena L, Camboni G, Liati P, Sessa C. Eur. J. Cancer. 2001;37(Supplement 6):S260. [Google Scholar]
- 235.Scagliotti G, Novello S, Crinò L, De Marinis F, Tonato M, Noberasco C, Selvaggi G, Massoni F, Gatti B, Camboni G. Lung Cancer. 2003;41(Supplement 2):S223. [Google Scholar]
- 236.Jodrell DI, Evans TR, Steward W, Cameron D, Prendiville J, Aschele C, Noberasco C, Lind M, Carmichael J, Dobbs N, Camboni G, Gatti B, De Braud F. Eur. J. Cancer. 2004;40:1872–1877. doi: 10.1016/j.ejca.2004.04.032. [DOI] [PubMed] [Google Scholar]
- 237.Hensing TA, Hanna NH, Gillenwater HH, Gabriella Camboni M, Allievi C, Socinski MA. Anticancer Drugs. 2006;17:697–704. doi: 10.1097/01.cad.0000215054.62942.7f. [DOI] [PubMed] [Google Scholar]
- 238.Qu Y, Harris A, Hegmans A, Petz A, Kabolizadeh P, Penazova H, Farrell N. J Inorg. Biochem. 2004;98:1591–1598. doi: 10.1016/j.jinorgbio.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 239.Komeda S, Moulaei T, Woods KK, Chikuma M, Farrell NP, Williams LD. J Am. Chem. Soc. 2006;128:16092–16103. doi: 10.1021/ja062851y. [DOI] [PubMed] [Google Scholar]
- 240.Mangrum JB, Farrell NP. Chem. Commun. 2010;46:6640–6650. doi: 10.1039/c0cc01254h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Harris A, Qu Y, Farrell N. Inorg. Chem. 2005;44:1196–1198. doi: 10.1021/ic048356p. [DOI] [PubMed] [Google Scholar]
- 242.Prisecaru A, Molphy Z, Kipping RG, Peterson EJ, Qu Y, Kellett A, Farrell NP. Nucleic Acids Res. 2014;42:13474–13487. doi: 10.1093/nar/gku1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Malina J, Farrell NP, Brabec V. Angew. Chem., Int. Ed. 2014;53:12812–12816. doi: 10.1002/anie.201408012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Malina J, Farrell NP, Brabec V. Inorg. Chem. 2014;53:1662–1671. doi: 10.1021/ic402796k. [DOI] [PubMed] [Google Scholar]
- 245.Harris AL, Yang X, Hegmans A, Povirk L, Ryan JJ, Kelland L, Farrell NP. Inorg. Chem. 2005;44:9598–9600. doi: 10.1021/ic051390z. [DOI] [PubMed] [Google Scholar]
- 246.Harris AL, Ryan JJ, Farrell N. Mol. Pharmacol. 2006;69:666–672. doi: 10.1124/mol.105.018762. [DOI] [PubMed] [Google Scholar]
- 247.Brodie CR, Collins JG, Aldrich-Wright JR. Dalton Trans. 2004:1145–1152. doi: 10.1039/b316511f. [DOI] [PubMed] [Google Scholar]
- 248.Kemp S, Wheate NJ, Buck DP, Nikac M, Collins JG, Aldrich-Wright JR. J Inorg. Biochem. 2007;101:1049–1058. doi: 10.1016/j.jinorgbio.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 249.Fisher DM, Bednarski PJ, Grunert R, Turner P, Fenton RR, Aldrich-Wright JR. ChemMedChem. 2007;2:488–495. doi: 10.1002/cmdc.200600211. [DOI] [PubMed] [Google Scholar]
- 250.Fisher DM, Fenton RR, Aldrich-Wright JR. Chem. Commun. 2008:5613–5615. doi: 10.1039/b811723c. [DOI] [PubMed] [Google Scholar]
- 251.Krause-Heuer AM, Grunert R, Kuhne S, Buczkowska M, Wheate NJ, Le Pevelen DD, Boag LR, Fisher DM, Kasparkova J, Malina J, Bednarski PJ, Brabec V, Aldrich-Wright JR. J Med. Chem. 2009;52:5474–5484. doi: 10.1021/jm9007104. [DOI] [PubMed] [Google Scholar]
- 252.Garbutcheon-Singh KB, Leverett P, Myers S, Aldrich-Wright JR. Dalton Trans. 2013;42:918–926. doi: 10.1039/c2dt31323e. [DOI] [PubMed] [Google Scholar]
- 253.Jennette KW, Lippard SJ, Vassiliades GA, Bauer WR. Proc. Natl. Acad. Sci. U.S.A. 1974;71:3839–3843. doi: 10.1073/pnas.71.10.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Long EC, Barton JK. Accounts of Chemical Reseach. 1990;23:271–273. [Google Scholar]
- 255.Wu YS, Koch KR, Abratt VR, Klump HH. Arch. Biochem. Biophys. 2005;440:28–37. doi: 10.1016/j.abb.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 256.Davis KJ, Carrall JA, Lai B, Aldrich-Wright JR, Ralph SF, Dillon CT. Dalton Trans. 2012;41:9417–9426. doi: 10.1039/c2dt30217a. [DOI] [PubMed] [Google Scholar]
- 257.Garbutcheon-Singh KB, Myers S, Harper BW, Ng NS, Dong Q, Xie C, Aldrich-Wright JR. Metallomics. 2013;5:1061–1067. doi: 10.1039/c3mt00023k. [DOI] [PubMed] [Google Scholar]
- 258.Hope JM, Wilson JJ, Lippard SJ. Dalton Trans. 2013;42:3176–3180. doi: 10.1039/c2dt32462h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Suntharalingam K, Wilson JJ, Lin W, Lippard SJ. Metallomics. 2014;6:437–443. doi: 10.1039/c3mt00364g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Muller PAJ, Vousden KH. Nature Cell Biology. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 261.Macquet J-P, Butour J-L. J Natl. Cancer Inst. 1983;70:899–905. [PubMed] [Google Scholar]
- 262.Hollis LS, Miller AV, Amundsen AR, Stern EW, Sundquist WI, Toni J, Burstyn JN, Heiger W, Lippard SJ. J Inorg. Biochem. 1989;36:153. [Google Scholar]
- 263.Sundquist WI, Bancroft DP, Lippard SJ. J Am. Chem. Soc. 1990;112:1590–1596. [Google Scholar]
- 264.Hollis LS, Sundquist WI, Burstyn JN, Heigerbernays WJ, Bellon SF, Ahmed KJ, Amundsen AR, Stern EW, Lippard SJ. Cancer Res. 1991;51:1866–1875. [PubMed] [Google Scholar]
- 265.Bellon SF, Lippard SJ. Biophys. Chem. 1990;35:179–188. doi: 10.1016/0301-4622(90)80007-t. [DOI] [PubMed] [Google Scholar]
- 266.Keck MV, Lippard SJ. J Am. Chem. Soc. 1992;114:3386–3390. [Google Scholar]
- 267.Donahue BA, Augot M, Bellon SF, Treiber DK, Toney JH, Lippard SJ, Essigmann JM. Biochemistry. 1990;29:5872–5880. doi: 10.1021/bi00476a032. [DOI] [PubMed] [Google Scholar]
- 268.Johnstone TC, Park GY, Lippard SJ. Anticancer Res. 2014;34:471–476. [PMC free article] [PubMed] [Google Scholar]
- 269.Lovejoy KS, Todd RC, Zhang S, McCormick MS, D'Aquino JA, Reardon JT, Sancar A, Giacomini KM, Lippard SJ. Proc. Natl. Acad. Sci. U.S.A. 2008;105:8902–8907. doi: 10.1073/pnas.0803441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Lovejoy KS, Serova M, Bieche I, Emami S, D'Incalci M, Broggini M, Erba E, Gespach C, Cvitkovic E, Faivre S, Raymond E, Lippard SJ. Mol. Cancer Ther. 2011;10:1709–1719. doi: 10.1158/1535-7163.MCT-11-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Zhu G, Myint M, Ang WH, Song L, Lippard SJ. Cancer Res. 2012;72:790–800. doi: 10.1158/0008-5472.CAN-11-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Wang D, Zhu G, Huang X, Lippard SJ. Proc. Natl. Acad. Sci. U.S.A. 2010;107:9584–9589. doi: 10.1073/pnas.1002565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Park GY, Wilson JJ, Song Y, Lippard SJ. Proc. Natl. Acad. Sci. U.S.A. 2012;109:11987–11992. doi: 10.1073/pnas.1207670109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Kellinger MW, Park GY, Chong J, Lippard SJ, Wang D. J Am. Chem. Soc. 2013;135:13054–13061. doi: 10.1021/ja405475y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Gregory MT, Park GY, Johnstone TC, Lee Y-S, Yang W, Lippard SJ. Proc. Natl. Acad. Sci. U.S.A. 2014;111:9133–9138. doi: 10.1073/pnas.1405739111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Johnstone TC, Lippard SJ. J Am. Chem. Soc. 2014;136:2126–2134. doi: 10.1021/ja4125115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Johnstone TC, Alexander SM, Lin W, Lippard SJ. J Am. Chem. Soc. 2014;136:116–118. doi: 10.1021/ja411742c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Martins ET, Baruah H, Kramarczyk J, Saluta G, Day CS, Kucera GL, Bierbach U. J Med. Chem. 2001;44:4492–4496. doi: 10.1021/jm010293m. [DOI] [PubMed] [Google Scholar]
- 279.Baruah H, Rector CL, Monnier SM, Bierbach U. Biochem. Pharmacol. 2002;64:191–200. doi: 10.1016/s0006-2952(02)01107-3. [DOI] [PubMed] [Google Scholar]
- 280.Baruah H, Barry CG, Bierbach U. Curr. Top. Med. Chem. 2004;4:1537–1549. doi: 10.2174/1568026043387313. [DOI] [PubMed] [Google Scholar]
- 281.Baruah H, Wright MW, Bierbach U. Biochemistry. 2005;44:6059–6070. doi: 10.1021/bi050021b. [DOI] [PubMed] [Google Scholar]
- 282.Ackley MC, Barry CG, Mounce AM, Farmer MC, Springer BE, Day CS, Wright MW, Berners-Price SJ, Hess SM, Bierbach U. J Biol. Inorg. Chem. 2004;9:453–461. doi: 10.1007/s00775-004-0541-4. [DOI] [PubMed] [Google Scholar]
- 283.Hess SM, Mounce AM, Sequeira RC, Augustus TM, Ackley MC, Bierbach U. Cancer Chemother. Pharmacol. 2005;56:337–343. doi: 10.1007/s00280-004-0987-7. [DOI] [PubMed] [Google Scholar]
- 284.Hess SM, Anderson JG, Bierbach U. Bioorg. Med. Chem. Lett. 2005;15:443–446. doi: 10.1016/j.bmcl.2004.10.049. [DOI] [PubMed] [Google Scholar]
- 285.Guddneppanavar R, Saluta G, Kucera GL, Bierbach U. J Med. Chem. 2006;49:3204–3214. doi: 10.1021/jm060035v. [DOI] [PubMed] [Google Scholar]
- 286.Guddneppanavar R, Choudhury JR, Kheradi AR, Steen BD, Saluta G, Kucera GL, Day CS, Bierbach U. J Med. Chem. 2007;50:2259–2263. doi: 10.1021/jm0614376. [DOI] [PubMed] [Google Scholar]
- 287.Ma Z, Saluta G, Kucera GL, Bierbach U. Bioorg. Med. Chem. Lett. 2008;18:3799–3801. doi: 10.1016/j.bmcl.2008.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Smyre CL, Saluta G, Kute TE, Kucera GL, Bierbach U. ACS Med. Chem. Lett. 2011;2:870–874. doi: 10.1021/ml2001888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Guddneppanavar R, Bierbach U. Anticancer Agents Med. Chem. 2007;7:125–138. doi: 10.2174/187152007779313991. [DOI] [PubMed] [Google Scholar]
- 290.Choudhury J, Rao L, Bierbach U. J Biol. Inorg. Chem. 2011;16:373–380. doi: 10.1007/s00775-010-0733-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Ma Z, Choudhury JR, Wright MW, Day CS, Saluta G, Kucera GL, Bierbach U. J Med. Chem. 2008;51:7574–7580. doi: 10.1021/jm800900g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Lin JH. Bone. 1996;18:75–85. doi: 10.1016/8756-3282(95)00445-9. [DOI] [PubMed] [Google Scholar]
- 293.Stresing V, Daubiné F, Benzaid I, Mönkkönen H, Clézardin P. Cancer Lett. 2007;257:16–35. doi: 10.1016/j.canlet.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 294.Cremers S, Papapoulos S. Bone. 2011;49:42–49. doi: 10.1016/j.bone.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 295.Xue Z, Lin M, Zhu J, Zhang J, Li Y, Guo Z. Chem. Commun. 2010;46:1212–1214. doi: 10.1039/b922222g. [DOI] [PubMed] [Google Scholar]
- 296.Iafisco M, Margiotta N. J Inorg. Biochem. 2012;117:237–247. doi: 10.1016/j.jinorgbio.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 297.Bose RN, Maurmann L, Mishur RJ, Yasui L, Gupta S, Grayburn WS, Hofstetter H, Salley T. Proc. Natl. Acad. Sci. U.S.A. 2008;105:18314–18319. doi: 10.1073/pnas.0803094105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Moghaddas S, Majmudar P, Marin R, Dezvareh H, Qi C, Soans E, Bose RN. Inorg. Chim. Acta. 2012;393:173–181. [Google Scholar]
- 299.Sasanelli R, Boccarelli A, Giordano D, Laforgia M, Arnesano F, Natile G, Cardellicchio C, Capozzi MAM, Coluccia M. J Med. Chem. 2007;50:3434–3441. doi: 10.1021/jm061435l. [DOI] [PubMed] [Google Scholar]
- 300.Arnesano F, Boccarelli A, Cornacchia D, Nushi F, Sasanelli R, Coluccia M, Natile G. J Med. Chem. 2009;52:7847–7855. doi: 10.1021/jm900845t. [DOI] [PubMed] [Google Scholar]
- 301.Belviso BD, Caliandro R, Siliqi D, Calderone V, Arnesano F, Natile G. Chem. Commun. 2013;49:5492. doi: 10.1039/c3cc41278d. [DOI] [PubMed] [Google Scholar]
- 302.Wilson JJ, Lippard SJ. Chem. Rev. 2014;114:4470–4495. doi: 10.1021/cr4004314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Dolman RC, Deacon GB, Hambley TW. J Inorg. Biochem. 2002;88:260–267. doi: 10.1016/s0162-0134(01)00360-9. [DOI] [PubMed] [Google Scholar]
- 304.Creaven PJ, Madajewicz S, Pendyala L, Mittelman A, Pontes E, Spaulding M, Arbuck S, Solomon J. Cancer Treat Rep. 1983;67:795–800. [PubMed] [Google Scholar]
- 305.McCormick MC, Keijzer K, Polavarapu A, Schultz FA, Baik M-H. J Am. Chem. Soc. 2014;136:8992–9000. doi: 10.1021/ja5029765. [DOI] [PubMed] [Google Scholar]
- 306.Wilson JJ, Lippard SJ. Inorg. Chem. 2011;50:3103–3115. doi: 10.1021/ic2000816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Hambley TW, Battle AR, Deacon GB, Lawrenz ET, Fallon GD, Gatehouse BM, Webster LK, Rainone S. J Inorg. Biochem. 1999;77:3–12. doi: 10.1016/s0162-0134(99)00133-6. [DOI] [PubMed] [Google Scholar]
- 308.Ellis LT, Er HM, Hambley TW. Aust. J. Chem. 1995;48:793–806. [Google Scholar]
- 309.Choi S, Filotto C, Bisanzo M, Delaney S, Lagasee D, Whitworth JL, Jusko A, Li CR, Wood NA, Willingham J, Schwenker A, Spaulding K. Inorg. Chem. 1998;37:2500–2504. [Google Scholar]
- 310.Peloso A. J Chem. Soc., Dalton Trans. 1984:249–254. [Google Scholar]
- 311.Lemma K, Berglund J, Farrell N, Elding LI. J Biol. Inorg. Chem. 2000;5:300–306. doi: 10.1007/pl00010658. [DOI] [PubMed] [Google Scholar]
- 312.Lemma K, Shi T, Elding LI. Inorg. Chem. 2000;39:1728–1734. doi: 10.1021/ic991351l. [DOI] [PubMed] [Google Scholar]
- 313.Lemma K, Sargeson AM, Elding LI. J Chem. Soc., Dalton Trans. 2000:1167–1172. [Google Scholar]
- 314.Lemma K, House DA, Retta N, Elding LI. Inorg. Chim. Acta. 2002;331:98–108. [Google Scholar]
- 315.Zhang JZ, Wexselblatt E, Hambley TW, Gibson D. Chem. Commun. 2012;48:847–849. doi: 10.1039/c1cc16647f. [DOI] [PubMed] [Google Scholar]
- 316.Gibbons GR, Wyrick S, Chaney SG. Cancer Res. 1989;49:1402–1407. [PubMed] [Google Scholar]
- 317.Rose WC, Schurig JE, Huftalen JB, Bradner WT. Cancer Treat Rep. 1982;66:135–146. [PubMed] [Google Scholar]
- 318.Anderson WK, Quagliato DA, Haugwitz RD, Narayanan VL, Wolpert-DeFilippes MK. Cancer Treat. Rep. 1986;70:997–1002. [PubMed] [Google Scholar]
- 319.Rahman A, Roh JK, Wolpert-DeFilippes MK, Goldin A, Venditti JM, Woolley PV. Cancer Res. 1988;48:1745–1752. [PubMed] [Google Scholar]
- 320.Parker RJ, Vionnet JA, Bostick-Bruton F, Reed E. Cancer Res. 1993;53:242–247. [PubMed] [Google Scholar]
- 321.Schilder RJ, LaCreta FP, Perez RP, Johnson SW, Brennan JM, Rogatko A, Nash S, McAleer C, Hamilton TC, Roby D, et al. Cancer Res. 1994;54:709–717. [PubMed] [Google Scholar]
- 322.O'Rourke TJ, Weiss GR, New P, Burris HA, 3rd, Rodriguez G, Eckhardt J, Hardy J, Kuhn JG, Fields S, Clark GM, et al. Anticancer Drugs. 1994;5:520–526. doi: 10.1097/00001813-199410000-00002. [DOI] [PubMed] [Google Scholar]
- 323.Tutsch KD, Arzoomanian RZ, Alberti D, Tombes MB, Feierabend C, Robins HI, Spriggs DR, Wilding G. Invest. New Drugs. 1999;17:63–72. doi: 10.1023/a:1006223100561. [DOI] [PubMed] [Google Scholar]
- 324.Pendyala L, Cowens JW, Chheda GB, Dutta SP, Creaven PJ. Cancer Res. 1988;48:3533–3536. [PubMed] [Google Scholar]
- 325.Pendyala L, Arakali AV, Sansone P, Cowens JW, Creaven PJ. Cancer Chemoth. Pharm. 1990;27:248–250. doi: 10.1007/BF00685722. [DOI] [PubMed] [Google Scholar]
- 326.Bramwell VH, Crowther D, O'Malley S, Swindell R, Johnson R, Cooper EH, Thatcher N, Howell A. Cancer Treat Rep. 1985;69:409–416. [PubMed] [Google Scholar]
- 327.Paolozzi FP, Gaver R, Poiesz BJ, Louie A, DiFino S, Comis RL, Newman N, Ginsberg S. Invest. New Drugs. 1988;6:199–206. doi: 10.1007/BF00175398. [DOI] [PubMed] [Google Scholar]
- 328.Sessa C, Vermorken J, Renard J, Kaye S, Smith D, ten Bokkel Huinink W, Cavalli F, Pinedo H. J Clin. Oncol. 1988;6:98–105. doi: 10.1200/JCO.1988.6.1.98. [DOI] [PubMed] [Google Scholar]
- 329.Clavel M, Monfardini S, Gundersen S, Kaye S, Siegenthaler P, Renard J, van Glabbeke M, Pinedo HM. Eur. J. Cancer Clin. Oncol. 1988;24:1345–1348. doi: 10.1016/0277-5379(88)90226-x. [DOI] [PubMed] [Google Scholar]
- 330.Petrelli NJ, Creaven PJ, Herrera L, Mittelman A. Cancer Chemother. Pharmacol. 1989;23:61–62. doi: 10.1007/BF00258461. [DOI] [PubMed] [Google Scholar]
- 331.Meisner DJ, Ginsberg S, Ditch A, Louie A, Newman N, Comis R, Poiesz B. American Journal of Clinical Oncology. 1989;12:129–131. doi: 10.1097/00000421-198904000-00008. [DOI] [PubMed] [Google Scholar]
- 332.de Wit R, Tesselaar M, Kok TC, Seynaeve C, Rodenburg CJ, Verweij J, Helle PA, Stoter G. Eur. J. Cancer. 1991;27:1383–1385. doi: 10.1016/0277-5379(91)90015-6. [DOI] [PubMed] [Google Scholar]
- 333.Anderson H, Wagstaff J, Crowther D, Swindell R, Lind MJ, McGregor J, Timms MS, Brown D, Palmer P. Eur. J. Cancer Clin. Oncol. 1988;24:1471–1479. doi: 10.1016/0277-5379(88)90338-0. [DOI] [PubMed] [Google Scholar]
- 334.Echo DV. Phase II/III Comparison of the Platinum Compounds CBDCA vs CHIP vs CACP for Recurrent or Metastatic Epidermoid Carcinoma of the Head and Neck. Accessed. [Google Scholar]
- 335.Kelland LR, Abel G, McKeage MJ, Jones M, Goddard PM, Valenti M, Murrer BA, Harrap KR. Cancer Res. 1993;53:2581–2586. [PubMed] [Google Scholar]
- 336.Bhargava A, Vaishampayan UN. Expert Opin Investig Drugs. 2009;18:1787–1797. doi: 10.1517/13543780903362437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Kelland LR. Expert Opin. Investig. Drugs. 2000;9:1373–1382. doi: 10.1517/13543784.9.6.1373. [DOI] [PubMed] [Google Scholar]
- 338.Raynaud FI, Mistry P, Donaghue A, Poon GK, Kelland LR, Barnard CF, Murrer BA, Harrap KR. Cancer Chemother. Pharmacol. 1996;38:155–162. doi: 10.1007/s002800050464. [DOI] [PubMed] [Google Scholar]
- 339.Fink D, Nebel S, Aebi S, Zheng H, Cenni B, Nehme A, Christen RD, Howell SB. Cancer Res. 1996;56:4881–4886. [PubMed] [Google Scholar]
- 340.Vaisman A, Lim SE, Patrick SM, Copeland WC, Hinkle DC, Turchi JJ, Chaney SG. Biochemistry. 1999;38:11026–11039. doi: 10.1021/bi9909187. [DOI] [PubMed] [Google Scholar]
- 341.Wei M, Cohen SM, Silverman AP, Lippard SJ. J Biol. Chem. 2001;276:38774–38780. doi: 10.1074/jbc.M106374200. [DOI] [PubMed] [Google Scholar]
- 342.Harrap KR. Cancer Treat. Rev. 1985;12:21–33. doi: 10.1016/0305-7372(85)90015-5. [DOI] [PubMed] [Google Scholar]
- 343.Choy H, Park C, Yao M. Clin. Cancer Res. 2008;14:1633–1638. doi: 10.1158/1078-0432.CCR-07-2176. [DOI] [PubMed] [Google Scholar]
- 344.McKeage MJ, Raynaud F, Ward J, Berry C, O'Dell D, Kelland LR, Murrer B, Santabarabara P, Harrap KR, Judson IR. J Clin. Oncol. 1997;15:2691–2700. doi: 10.1200/JCO.1997.15.7.2691. [DOI] [PubMed] [Google Scholar]
- 345.Kurata T, Tamura T, Sasaki Y, Fujii H, Negoro S, Fukuoka M, Saijo N. Jpn. J. Clin. Oncol. 2000;30:377–384. doi: 10.1093/jjco/hyd102. [DOI] [PubMed] [Google Scholar]
- 346.Beale P, Raynaud F, Hanwell J, Berry C, Moore S, Odell D, Judson I. Cancer Chemother. Pharmacol. 1998;42:142–148. doi: 10.1007/s002800050797. [DOI] [PubMed] [Google Scholar]
- 347.Sessa C, Minoia C, Ronchi A, Zucchetti M, Bauer J, Borner M, de Jong J, Pagani O, Renard J, Weil C, D'Incalci M. Ann. Oncol. 1998;9:1315–1322. doi: 10.1023/a:1008441416790. [DOI] [PubMed] [Google Scholar]
- 348.Vouillamoz-Lorenz S, Buclin T, Lejeune F, Bauer J, Leyvraz S, Decosterd LA. Anticancer Res. 2003;23:2757–2765. [PubMed] [Google Scholar]
- 349.Judson I, Cerny T, Epelbaum R, Dunlop D, Smyth J, Schaefer B, Roelvink M, Kaplan S, Hanauske A. Ann. Oncol. 1997;8:604–606. doi: 10.1023/a:1008245709924. [DOI] [PubMed] [Google Scholar]
- 350.Squibb Bristol-Myers. In: Squibb B-M, editor. 1998. vol. Accession No. 910068667. [Google Scholar]
- 351.Trudeau M, Stuart G, Hirte H, Drouin P, Plante M, Bessette P, Dulude H, Lebwohl D, Fisher B, Seymour L. Gynecol. Oncol. 2002;84:327–331. doi: 10.1006/gyno.2001.6409. [DOI] [PubMed] [Google Scholar]
- 352.Figg WD, Chau CH, Madan RA, Gulley JL, Gao R, Sissung TM, Spencer S, Beatson M, Aragon-Ching J, Steinberg SM, Dahut WL. Clin. Genitourin. Cancer. 2013;11:229–237. doi: 10.1016/j.clgc.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Vaishampayan UN, Fontana J, Heilbrun LK, Smith D, Heath E, Dickow B, Figg WD. Urol. Oncol. 2014;32:31 e25–33 e25. doi: 10.1016/j.urolonc.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Sternberg CN, Whelan P, Hetherington J, Paluchowska B, Slee PH, Vekemans K, Van Erps P, Theodore C, Koriakine O, Oliver T, Lebwohl D, Debois M, Zurlo A, Collette L. Oncology. 2005;68:2–9. doi: 10.1159/000084201. [DOI] [PubMed] [Google Scholar]
- 355.Petrylak D, Sartor O, Witje F. Proc. Am. Soc. Clin. Oncol. 2007 [Google Scholar]
- 356.Sternberg CN, Petrylak D, Witjes F. Proc. Am. Soc. Clin. Oncol. 2007;25:5019. [Google Scholar]
- 357.Inc GB. In: Advisory Committee Briefing Document. U. F. a. D. Administration, editor. 2007. [Google Scholar]
- 358.E. M. Agency, editor. 2008. pp. 1–37. [Google Scholar]
- 359.Ang WH, Khalaila I, Allardyce CS, Juillerat-Jeanneret L, Dyson PJ. J Am. Chem. Soc. 2005;127:1382–1383. doi: 10.1021/ja0432618. [DOI] [PubMed] [Google Scholar]
- 360.Parker LJ, Italiano LC, Morton CJ, Hancock NC, Ascher DB, Aitken JB, Harris HH, Campomanes P, Rothlisberger U, De Luca A, Lo Bello M, Ang WH, Dyson PJ, Parker MW. Chemistry. 2011;17:7806–7816. doi: 10.1002/chem.201100586. [DOI] [PubMed] [Google Scholar]
- 361.Townsend DM, Tew KD. Oncogene. 2003;22:7369–7375. doi: 10.1038/sj.onc.1206940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Yang J, Sun X, Mao W, Sui M, Tang J, Shen Y. Mol. Pharmaceutics. 2012;9:2793–2800. doi: 10.1021/mp200597r. [DOI] [PubMed] [Google Scholar]
- 363.Novohradsky V, Zerzankova L, Stepankova J, Vrana O, Raveendran R, Gibson D, Kasparkova J, Brabec V. J Inorg. Biochem. 2014;140:72–79. doi: 10.1016/j.jinorgbio.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 364.Dhar S, Lippard SJ. Proc. Natl. Acad. Sci. U.S.A. 2009;106:22199–22204. doi: 10.1073/pnas.0912276106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Xue X, You S, Zhang Q, Wu Y, Zou GZ, Wang PC, Zhao YL, Xu Y, Jia L, Zhang X, Liang XJ. Mol. Pharm. 2012;9:634–644. doi: 10.1021/mp200571k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Wexselblatt E, Yavin E, Gibson D. Angew. Chem., Int. Ed. 2013;52:6059–6062. doi: 10.1002/anie.201300640. [DOI] [PubMed] [Google Scholar]
- 367.Johnstone TC, Kulak N, Pridgen EM, Farokhzad OC, Langer R, Lippard SJ. ACS Nano. 2013;7:5675–5683. doi: 10.1021/nn401905g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Suntharalingam K, Song Y, Lippard SJ. Chem. Commun. 2014;50:2465–2468. doi: 10.1039/c3cc48740g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Huang JC, Zamble DB, Reardon JT, Lippard SJ, Sancar A. Proc. Natl. Acad. Sci. U.S.A. 1994;91:10394–10398. doi: 10.1073/pnas.91.22.10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.He Q, Liang CH, Lippard SJ. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5768–5772. doi: 10.1073/pnas.100108697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Barnes KR, Kutikov A, Lippard SJ. Chem. Biol. 2004;11:557–564. doi: 10.1016/j.chembiol.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 372.Mukhopadhyay S, Barnés CM, Haskel A, Short SM, Barnes KR, Lippard SJ. Bioconjugate Chem. 2008;19:39–49. doi: 10.1021/bc070031k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Massaguer A, González-Cantó A, Escribano E, Barrabés S, Artigas G, Moreno V, Marchán V. Dalton Trans. 2015;44:202–212. doi: 10.1039/c4dt02710h. [DOI] [PubMed] [Google Scholar]
- 374.Yuan Y, Chen Y, Tang BZ, Liu B. Chem. Commun. 2014;50:3868–3870. doi: 10.1039/c3cc49516g. [DOI] [PubMed] [Google Scholar]
- 375.Gaviglio L, Gross A, Metzler-Nolte N, Ravera M. Metallomics. 2012;4:260–266. doi: 10.1039/c2mt00171c. [DOI] [PubMed] [Google Scholar]
- 376.Abramkin S, Valiahdi SM, Jakupec MA, Galanski M, Metzler-Nolte N, Keppler BK. Dalton. Trans. 2012;41:3001–3005. doi: 10.1039/c2dt12024k. [DOI] [PubMed] [Google Scholar]
- 377.Graf N, Mokhtari TE, Papayannopoulos IA, Lippard SJ. J Inorg. Biochem. 2012;110:58–63. doi: 10.1016/j.jinorgbio.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.DeBin JA, Strichartz GR. Toxicon. 1991;29:1403–1408. doi: 10.1016/0041-0101(91)90128-e. [DOI] [PubMed] [Google Scholar]
- 379.Hall MD, Hambley TW. Coord. Chem. Rev. 2002;232:49–67. [Google Scholar]
- 380.Vrana O, Brabec V, Kleinwachter V. Anti-Cancer Drug Des. 1986;1:95–109. [PubMed] [Google Scholar]
- 381.Brabec V, Vrana O, Kleinwachter V. Studia Biophysica. 1986;114:199–207. [Google Scholar]
- 382.Choi S, Delaney S, Orbai L, Padgett EJ, Hakemian AS. Inorg. Chem. 2001;40:5481–5482. doi: 10.1021/ic015549t. [DOI] [PubMed] [Google Scholar]
- 383.Choi S, Cooley RB, Hakemian AS, Larrabee YC, Bunt RC, Maupas SD, Muller JG, Burrows CJ. J Am. Chem. Soc. 2004;126:591–598. doi: 10.1021/ja038334m. [DOI] [PubMed] [Google Scholar]
- 384.Choi S, Cooley RB, Voutchkova A, Leung CH, Vastag L, Knowles DE. J Am. Chem. Soc. 2005;127:1773–1781. doi: 10.1021/ja045194n. [DOI] [PubMed] [Google Scholar]
- 385.Choi S, Vastag L, Leung C-H, Beard AM, Knowles DE, Larrabee JA. Inorg. Chem. 2006;45:10108–10114. doi: 10.1021/ic061243g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Choi S, Personick ML, Bogart JA, Ryu D, Redman RM, Laryea-Walker E. Dalton Trans. 2011;40:2888. doi: 10.1039/c0dt00822b. [DOI] [PubMed] [Google Scholar]
- 387.Vollano JF, Blatter EE, Dabrowiak JC. J Am. Chem. Soc. 1984;106:2732–2733. [Google Scholar]
- 388.Johnstone TC, Lippard SJ. J Biol. Inorg. Chem. 2014;19:667–674. doi: 10.1007/s00775-014-1109-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Bednarski PJ, Mackay FS, Sadler PJ. Anti-Cancer Agents Med. Chem. 2007;7:75–93. doi: 10.2174/187152007779314053. [DOI] [PubMed] [Google Scholar]
- 390.Smith NA, Sadler PJ. Photoactivatable metal complexes: from theory to applications in biotechnology and medicine. 2013 doi: 10.1098/rsta.2012.0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Kratochwil NA, Bednarski PJ, Mrozek H, Vogler A, Nagle JK. Anti-Cancer Drug Des. 1996;11:155–171. [PubMed] [Google Scholar]
- 392.Kratochwil NA, Zabel M, Range K-J, Bednarski PJ. J Med. Chem. 1996;39:2499–2507. doi: 10.1021/jm9509105. [DOI] [PubMed] [Google Scholar]
- 393.Kratochwil NA, Parkinson JA, Bednarski PJ, Sadler PJ. Angew. Chem., Int. Ed. 1999;38:1460–1463. doi: 10.1002/(SICI)1521-3773(19990517)38:10<1460::AID-ANIE1460>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 394.Kratochwil NA, Guo Z, del Socorro Murdoch P, Parkinson JA, Bednarski PJ, Sadler PJ. J Am. Chem. Soc. 1998;120:8253–8254. [Google Scholar]
- 395.Vogler A, Kern A, Hüttermann J. Angew. Chem., Int. Ed. 1978;17:524–525. [Google Scholar]
- 396.Müller P, Schröder B, Parkinson JA, Kratochwil NA, Coxall RA, Parkin A, Parsons S, Sadler PJ. Angew. Chem., Int. Ed. 2003;115:349–353. doi: 10.1002/anie.200390110. [DOI] [PubMed] [Google Scholar]
- 397.Bednarski PJ, Grunert R, Zielzki M, Wellner A, Mackay FS, Sadler PJ. Chem. Biol. 2006;13:61–67. doi: 10.1016/j.chembiol.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 398.Mackay FS, Woods JA, Moseley H, Ferguson J, Dawson A, Parsons S, Sadler PJ. Chemistry. 2006;12:3155–3161. doi: 10.1002/chem.200501601. [DOI] [PubMed] [Google Scholar]
- 399.Kasparkova J, Mackay FS, Brabec V, Sadler PJ. J Biol. Inorg. Chem. 2003;8:741–745. doi: 10.1007/s00775-003-0474-3. [DOI] [PubMed] [Google Scholar]
- 400.Ronconi L, Sadler PJ. Dalton. Trans. 2011;40:262–268. doi: 10.1039/c0dt00546k. [DOI] [PubMed] [Google Scholar]
- 401.Pracharova J, Zerzankova L, Stepankova J, Novakova O, Farrer NJ, Sadler PJ, Brabec V, Kasparkova J. Chem Res Toxicol. 2012;25:1099–1111. doi: 10.1021/tx300057y. [DOI] [PubMed] [Google Scholar]
- 402.Westendorf AF, Woods JA, Korpis K, Farrer NJ, Salassa L, Robinson K, Appleyard V, Murray K, Grunert R, Thompson AM, Sadler PJ, Bednarski PJ. Mol. Cancer Ther. 2012;11:1894–1904. doi: 10.1158/1535-7163.MCT-11-0959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Zhao Y, Woods JA, Farrer NJ, Robinson KS, Pracharova J, Kasparkova J, Novakova O, Li H, Salassa L, Pizarro AM, Clarkson GJ, Song L, Brabec V, Sadler PJ. Chemistry. 2013;19:9578–9591. doi: 10.1002/chem.201300374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Zhao Y, Farrer NJ, Li H, Butler JS, McQuitty RJ, Habtemariam A, Wang F, Sadler PJ. Angew. Chem., Int. Ed. 2013;52:13633–13637. doi: 10.1002/anie.201307505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Farokhzad OC, Langer R. ACS Nano. 2009;3:16–20. doi: 10.1021/nn900002m. [DOI] [PubMed] [Google Scholar]
- 406.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nat. Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 407.Ruoslahti E, Bhatia SN, Sailor MJ. The Journal of Cell Biology. 2010;188:759–768. doi: 10.1083/jcb.200910104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Iversen T-G, Skotland T, Sandvig K. Nano Today. 2011;6:176–185. [Google Scholar]
- 409.Davis ME, Chen Z, Shin DM. Nat. Rev. Drug Discov. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 410.Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. J Controlled Release. 2000;65:271–284. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- 411.Jhaveri AM, Torchilin VP. Front. Pharmacol. 2014;5 doi: 10.3389/fphar.2014.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Barry NPE, Sadler PJ. ACS Nano. 2013;7:5654–5659. doi: 10.1021/nn403220e. [DOI] [PubMed] [Google Scholar]
- 413.Wong BS, Yoong SL, Jagusiak A, Panczyk T, Ho HK, Ang WH, Pastorin G. Adv. Drug Delivery Rev. 2013;65:1964–2015. doi: 10.1016/j.addr.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 414.Feazell RP, Nakayama-Ratchford N, Dai H, Lippard SJ. J Am. Chem. Soc. 2007;129:8438–8439. doi: 10.1021/ja073231f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Dhar S, Liu Z, Thomale J, Dai HJ, Lippard SJ. J Am. Chem. Soc. 2008;130:11467–11476. doi: 10.1021/ja803036e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Garin-Chesa P, Campbell I, Saigo PE, Lewis JL, Jr, Old LJ, Rettig WJ. Am. J. Pathol. 1993;142:557–567. [PMC free article] [PubMed] [Google Scholar]
- 417.Parker N, Turk MJ, Westrick E, Lewis JD, Low PS, Leamon CP. Anal. Biochem. 2005;338:284–293. doi: 10.1016/j.ab.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 418.Li J, Yap SQ, Chin CF, Tian Q, Yoong SL, Pastorin G, Ang WH. Chem. Sci. 2012;3:2083–2087. [Google Scholar]
- 419.Yudasaka M, Ajima K, Suenaga K, Ichihashi T, Hashimoto A, Iijima S. Chem. Phys. Lett. 2003;380:42–46. [Google Scholar]
- 420.Yoong SL, Wong BS, Zhou QL, Chin CF, Li J, Venkatesan T, Ho HK, Yu V, Ang WH, Pastorin G. Biomaterials. 2014;35:748–759. doi: 10.1016/j.biomaterials.2013.09.036. [DOI] [PubMed] [Google Scholar]
- 421.Ko YH, Smith BL, Wang Y, Pomper MG, Rini DA, Torbenson MS, Hullihen J, Pedersen PL. Biochem. Biophys. Res. Commun. 2004;324:269–275. doi: 10.1016/j.bbrc.2004.09.047. [DOI] [PubMed] [Google Scholar]
- 422.Li J, Pant A, Chin CF, Ang WH, Ménard-Moyon C, Nayak TR, Gibson D, Ramaprabhu S, Panczyk T, Bianco A, Pastorin G. Nanomedicine: Nanotechnology, Biology and Medicine. 2014;10:1465–1475. doi: 10.1016/j.nano.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 423.Muzi L, Ménard-Moyon C, Russier J, Li J, Chin CF, Ang WH, Pastorin G, Risuleo G, Bianco A. Nanoscale. 2015;7:5383–5394. doi: 10.1039/c5nr00220f. [DOI] [PubMed] [Google Scholar]
- 424.Chin CF, Yap SQ, Li J, Pastorin G, Ang WH. Chem. Sci. 2014;5:2265–2270. [Google Scholar]
- 425.Xu X, Ray R, Gu Y, Ploehn HJ, Gearheart L, Raker K, Scrivens WA. J Am. Chem. Soc. 2004;126:12736–12737. doi: 10.1021/ja040082h. [DOI] [PubMed] [Google Scholar]
- 426.Sahu S, Behera B, Maiti TK, Mohapatra S. Chem. Commun. 2012;48:8835–8837. doi: 10.1039/c2cc33796g. [DOI] [PubMed] [Google Scholar]
- 427.Liu H, Ye T, Mao C. Angew. Chem., Int. Ed. 2007;46:6473–6475. doi: 10.1002/anie.200701271. [DOI] [PubMed] [Google Scholar]
- 428.Sun Y-P, Zhou B, Lin Y, Wang W, Shiral Fernando KA, Pathak P, Meziani MJ, Harruff BA, Wang X, Wang H, Luo PG, Yang H, Kose ME, Chen B, Veca LM, Xie S-Y. J Am. Chem. Soc. 2006;128:7756–7757. doi: 10.1021/ja062677d. [DOI] [PubMed] [Google Scholar]
- 429.Zhu S, Meng Q, Wang L, Zhang J, Song Y, Jin H, Zhang K, Sun H, Wang H, Yang B. Angew. Chem., Int. Ed. 2013;52:3953–3957. doi: 10.1002/anie.201300519. [DOI] [PubMed] [Google Scholar]
- 430.Yang X-D, Xiang H-J, An L, Yang S-P, Liu J-G. New J. Chem. 2015;39:800–804. [Google Scholar]
- 431.Li J, Lyv Z, Li Y, Liu H, Wang J, Zhan W, Chen H, Chen H, Li X. Biomaterials. 2015;51:12–21. doi: 10.1016/j.biomaterials.2015.01.074. [DOI] [PubMed] [Google Scholar]
- 432.Robinson JT, Tabakman SM, Liang Y, Wang H, Sanchez Casalongue H, Vinh D, Dai H. J Am. Chem. Soc. 2011;133:6825–6831. doi: 10.1021/ja2010175. [DOI] [PubMed] [Google Scholar]
- 433.Zhang W, Guo Z, Huang D, Liu Z, Guo X, Zhong H. Biomaterials. 2011;32:8555–8561. doi: 10.1016/j.biomaterials.2011.07.071. [DOI] [PubMed] [Google Scholar]
- 434.Dhar S, Daniel WL, Giljohann DA, Mirkin CA, Lippard SJ. J Am. Chem. Soc. 2009;131:14652–14653. doi: 10.1021/ja9071282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Liedert B, Pluim D, Schellens J, Thomale J. Nucleic Acids Res. 2006;34:e47. doi: 10.1093/nar/gkl051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Shi Y, Goodisman J, Dabrowiak JC. Inorg. Chem. 2013;52:9418–9426. doi: 10.1021/ic400989v. [DOI] [PubMed] [Google Scholar]
- 437.Alkilany AM, Thompson LB, Boulos SP, Sisco PN, Murphy CJ. Adv. Drug Delivery Rev. 2012;64:190–199. doi: 10.1016/j.addr.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 438.Min YZ, Mao CQ, Xu DC, Wang J, Liu YZ. Chem. Commun. 2010;46:8424–8426. doi: 10.1039/c0cc03108a. [DOI] [PubMed] [Google Scholar]
- 439.Min Y, Mao C-Q, Chen S, Ma G, Wang J, Liu Y. Angew. Chem., Int. Ed. 2012;51:6742–6747. doi: 10.1002/anie.201201562. [DOI] [PubMed] [Google Scholar]
- 440.Dai Y, Xiao H, Liu J, Yuan Q, Ma Pa, Yang D, Li C, Cheng Z, Hou Z, Yang P, Lin J. J Am. Chem. Soc. 2013;135:18920–18929. doi: 10.1021/ja410028q. [DOI] [PubMed] [Google Scholar]
- 441.Min Y, Li J, Liu F, Yeow EKL, Xing B. Angew. Chem., Int. Ed. 2014;53:1012–1016. doi: 10.1002/anie.201308834. [DOI] [PubMed] [Google Scholar]
- 442.Bakalova R, Ohba H, Zhelev Z, Nagase T, Jose R, Ishikawa M, Baba Y. Nano Lett. 2004;4:1567–1573. [Google Scholar]
- 443.Gomez Blanco N, Maldonado CR, Mareque-Rivas JC. Chem. Commun. 2009:5257–5259. doi: 10.1039/b910000h. [DOI] [PubMed] [Google Scholar]
- 444.Maldonado CR, Gómez-Blanco N, Jauregui-Osoro M, Brunton VG, Yate L, Mareque-Rivas JC. Chem. Commun. 2013;49:3985–3987. doi: 10.1039/c3cc39104c. [DOI] [PubMed] [Google Scholar]
- 445.Infante I, Azpiroz JM, Gomez Blanco N, Ruggiero E, Ugalde JM, Mareque-Rivas JC, Salassa L. The Journal of Physical Chemistry C. 2014;118:8712–8721. [Google Scholar]
- 446.Ruggiero E, Hernández-Gil J, Mareque-Rivas JC, Salassa L. Chem. Commun. 2015;51:2091–2094. doi: 10.1039/c4cc07960d. [DOI] [PubMed] [Google Scholar]
- 447.Dai Y, Kang X, Yang D, Li X, Zhang X, Li C, Hou Z, Cheng Z, Ma Pa, Lin J. Adv. Healthcare Mater. 2013;2:562–567. doi: 10.1002/adhm.201200234. [DOI] [PubMed] [Google Scholar]
- 448.Cheng Z, Dai Y, Kang X, Li C, Huang S, Lian H, Hou Z, Ma P, Lin J. Biomaterials. 2014;35:6359–6368. doi: 10.1016/j.biomaterials.2014.04.029. [DOI] [PubMed] [Google Scholar]
- 449.Duan X, Evans DG, editors. Layered double hydroxides. Berlin ; New York: Springer; 2005. [Google Scholar]
- 450.Rieter WJ, Pott KM, Taylor KML, Lin W. J Am. Chem. Soc. 2008;130:11584–11585. doi: 10.1021/ja803383k. [DOI] [PubMed] [Google Scholar]
- 451.Liu D, Poon C, Lu K, He C, Lin W. Nat. Commun. 2014;5:4182. doi: 10.1038/ncomms5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.He C, Liu D, Lin W. Biomaterials. 2015;36:124–133. doi: 10.1016/j.biomaterials.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Huxford RC, Della Rocca J, Lin W. Curr. Opin. Chem. Biol. 2010;14:262–268. doi: 10.1016/j.cbpa.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Taylor-Pashow KML, Della Rocca J, Xie Z, Tran S, Lin W. J Am. Chem. Soc. 2009;131:14261–14263. doi: 10.1021/ja906198y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.He C, Lu K, Liu D, Lin W. J Am. Chem. Soc. 2014;136:5181–5184. doi: 10.1021/ja4098862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Della Rocca J, Huxford RC, Comstock-Duggan E, Lin W. Angew. Chem., Int. Ed. 2011;50:10330–10334. doi: 10.1002/anie.201104510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Aydar E. Cancer Res. 2004;64:5029–5035. doi: 10.1158/0008-5472.CAN-03-2329. [DOI] [PubMed] [Google Scholar]
- 458.Della Rocca J, Werner ME, Kramer SA, Huxford-Phillips RC, Sukumar R, Cummings ND, Vivero-Escoto JL, Wang AZ, Lin W. Nanomedicine: Nanotechnology, Biology, and Medicine. 2015;11:31–38. doi: 10.1016/j.nano.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Jain RA. Biomaterials. 2000;21:2475–2490. doi: 10.1016/s0142-9612(00)00115-0. [DOI] [PubMed] [Google Scholar]
- 460.Working PK, Newman MS, Johnson J, Cornacoff JB. In: Poly(ethylene glycol): Chemistry and Biological Applications. Harris JM, Zalipsky S, editors. Vol. 680. American Chemical Society; 1997. pp. 45–57. [Google Scholar]
- 461.. [Google Scholar]
- 462.Stayshich RM, Meyer TY. J Am. Chem. Soc. 2010;132:10920–10934. doi: 10.1021/ja102670n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Li J, Stayshich RM, Meyer TY. J Am. Chem. Soc. 2011;133:6910–6913. doi: 10.1021/ja200895s. [DOI] [PubMed] [Google Scholar]
- 464.Reiter RE, Gu ZN, Watabe T, Thomas G, Szigeti K, Davis E, Wahl M, Nisitani S, Yamashiro J, Le Beau MM, Loda M, Witte ON. Proc. Natl. Acad. Sci. U.S.A. 1998;95:1735–1740. doi: 10.1073/pnas.95.4.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Dhar S, Kolishetti N, Lippard SJ, Farokhzad OC. Proc. Natl. Acad. Sci. U.S.A. 2011;108:1850–1855. doi: 10.1073/pnas.1011379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Graf N, Bielenberg DR, Kolishetti N, Muus C, Banyard J, Farokhzad OC, Lippard SJ. ACS Nano. 2012;6:4530–4539. doi: 10.1021/nn301148e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Johnstone TC, Lippard SJ. Inorg. Chem. 2013;52:9915–9920. doi: 10.1021/ic4010642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Gan GN, Wittschieben JP, Wittschieben BØ, Wood RD. Cell Research. 2008;18:174–183. doi: 10.1038/cr.2007.117. [DOI] [PubMed] [Google Scholar]
- 469.Xie K, Doles J, Hemann MT, Walker GC. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20792–20797. doi: 10.1073/pnas.1011412107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Pathak RK, McNitt CD, Popik VV, Dhar S. Chem.–Eur. J. 2014;20:6861–6865. doi: 10.1002/chem.201402573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Marrache S, Pathak RK, Dhar S. Proc. Natl. Acad. Sci. U.S.A. 2014;111:10444–10449. doi: 10.1073/pnas.1405244111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Murphy AR, Kaplan DL. J Mater. Chem. 2009;19:6443–6450. doi: 10.1039/b905802h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Kim U-J, Park J, Kim HJ, Wada M, Kaplan DL. Biomaterials. 2005;26:2775–2785. doi: 10.1016/j.biomaterials.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 474.Mathur AB, Gupta V. Nanomedicine. 2010;5:807–820. doi: 10.2217/nnm.10.51. [DOI] [PubMed] [Google Scholar]
- 475.Qu J, Liu Y, Yu Y, Li J, Luo J, Li M. Mater. Sci. Eng., C. 2014;44:166–174. doi: 10.1016/j.msec.2014.08.034. [DOI] [PubMed] [Google Scholar]
- 476.Lozano-Pérez AA, Gil AL, Pérez S, Cutillas N, Meyer H, Pedreño MI, Aznar-Cervantes SD, Janiak C, Cenis JL, Ruiz J. Dalton Trans. 2015 doi: 10.1039/c5dt00378d. In press. [DOI] [PubMed] [Google Scholar]
- 477.Cohen S, Yoshioka T, Lucarelli M, Hwang LH, Langer R. Pharm. Res. 1991;8:713–720. doi: 10.1023/a:1015841715384. [DOI] [PubMed] [Google Scholar]
- 478.Zhou D, Cong Y, Qi Y, He S, Xiong H, Wu Y, Xie Z, Chen X, Jing X, Huang Y. Biomater. Sci. 2015;3:182–191. doi: 10.1039/c4bm00305e. [DOI] [PubMed] [Google Scholar]
- 479.Reithofer MR, Valiahdi SM, Galanski M, Jakupec MA, Arion VB, Keppler BK. Chem. Biodivers. 2008;5:2160–2170. doi: 10.1002/cbdv.200890197. [DOI] [PubMed] [Google Scholar]
- 480.Aryal S, Jack Hu C-M, Fu V, Zhang L. J Mater. Chem. 2012;22:994–999. [Google Scholar]
- 481.Lin C-T, Lai H-C, Lee H-Y, Lin W-H, Chang C-C, Chu T-Y, Lin Y-W, Lee K-D, Yu M-H. Cancer Sci. 2008;99:1218–1226. doi: 10.1111/j.1349-7006.2008.00793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Aryal S, Hu C-MJ, Zhang L. ACS Nano. 2010;4:251–258. doi: 10.1021/nn9014032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Shen W, Luan J, Cao L, Sun J, Yu L, Ding J. Biomacromolecules. 2015;16:105–115. doi: 10.1021/bm501220a. [DOI] [PubMed] [Google Scholar]
- 484.Kolishetti N, Dhar S, Valencia PM, Lin LQ, Karnik R, Lippard SJ, Langer R, Farokhzad OC. Proc. Natl. Acad. Sci. U.S.A. 2010;107:17939–17944. doi: 10.1073/pnas.1011368107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Karnik R, Gu F, Basto P, Cannizzaro C, Dean L, Kyei-Manu W, Langer R, Farokhzad OC. Nano Lett. 2008;8:2906–2912. doi: 10.1021/nl801736q. [DOI] [PubMed] [Google Scholar]
- 486.Valencia PM, Pridgen EM, Perea B, Gadde S, Sweeney C, Kantoff PW, Bander NH, Lippard SJ, Langer R, Karnik R, Farokhzad OC. Nanomedicine. 2013;8:687–698. doi: 10.2217/nnm.12.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Park GY, Gadde S, Johnstone TC, Gagnon PG, Hasjia AK, Farokhzad OC, Lippard SJ. Manuscript in preparation [Google Scholar]
- 488.Xiao H, Li W, Qi R, Yan L, Wang R, Liu S, Zheng Y, Xie Z, Huang Y, Jing X. J Controlled Release. 2012;163:304–314. doi: 10.1016/j.jconrel.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 489.Xiao H, Song H, Yang Q, Cai H, Qi R, Yan L, Liu S, Zheng Y, Huang Y, Liu T, Jing X. Biomaterials. 2012;33:6507–6519. doi: 10.1016/j.biomaterials.2012.05.049. [DOI] [PubMed] [Google Scholar]
- 490.Xiao H, Yan L, Zhang Y, Qi R, Li W, Wang R, Liu S, Huang Y, Li Y, Jing X. Chem. Commun. 2012;48:10730–10732. doi: 10.1039/c2cc34297a. [DOI] [PubMed] [Google Scholar]
- 491.Du R, Xiao H, Guo G, Jiang B, Yan X, Li W, Yang X, Zhang Y, Li Y, Jing X. Colloids Surf., B. 2014;123:734–741. doi: 10.1016/j.colsurfb.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 492.Xiao H, Qi R, Liu S, Hu X, Duan T, Zheng Y, Huang Y, Jing X. Biomaterials. 2011;32:7732–7739. doi: 10.1016/j.biomaterials.2011.06.072. [DOI] [PubMed] [Google Scholar]
- 493.Qi R, Liu S, Chen J, Xiao H, Yan L, Huang Y, Jing X. J Controlled Release. 2012;159:251–260. doi: 10.1016/j.jconrel.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 494.Xiao H, Noble GT, Stefanick JF, Qi R, Kiziltepe T, Jing X, Bilgicer B. J Controlled Release. 2014;173:11–17. [PubMed] [Google Scholar]
- 495.Song H, Xiao H, Zhang Y, Cai H, Wang R, Zheng Y, Huang Y, Li Y, Xie Z, Liu T, Jing X. J Mater. Chem. B. 2013;1:762–772. doi: 10.1039/c2tb00206j. [DOI] [PubMed] [Google Scholar]
- 496.Farrell N. In: Platinum-Based Drugs in Cancer Therapy. Kelland LR, Farrell NP, editors. Humana Press; 2000. pp. 321–338. [Google Scholar]
- 497.Xiao H, Song H, Zhang Y, Qi R, Wang R, Xie Z, Huang Y, Li Y, Wu Y, Jing X. Biomaterials. 2012;33:8657–8669. doi: 10.1016/j.biomaterials.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 498.Qi R, Xiao H, Wu S, Li Y, Zhang Y, Jing X. J Mater. Chem. B. 2015;3:176–179. doi: 10.1039/c4tb01146e. [DOI] [PubMed] [Google Scholar]
- 499.O'Reilly RK, Hawker CJ, Wooley KL. Chem. Soc. Rev. 2006;35:1068–1083. doi: 10.1039/b514858h. [DOI] [PubMed] [Google Scholar]
- 500.Fleige E, Quadir MA, Haag R. Adv. Drug Delivery Rev. 2012;64:866–884. doi: 10.1016/j.addr.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 501.Han Y, Li J, Zan M, Luo S, Ge Z, Liu S. Polymer Chemistry. 2014;5:3707–3718. [Google Scholar]
- 502.Duong HTT, Huynh VT, de Souza P, Stenzel MH. Biomacromolecules. 2010;11:2290–2299. doi: 10.1021/bm100396s. [DOI] [PubMed] [Google Scholar]
- 503.Hou J, Shang J, Jiao C, Jiang P, Xiao H, Luo L, Liu T. Macromol. Biosci. 2013;13:954–965. doi: 10.1002/mabi.201300057. [DOI] [PubMed] [Google Scholar]
- 504.Song H, Wang R, Xiao H, Cai H, Zhang W, Xie Z, Huang Y, Jing X, Liu T. Eur. J. Pharm. Biopharm. 2013;83:63–75. doi: 10.1016/j.ejpb.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 505.Yang J, Liu W, Sui M, Tang J, Shen Y. Biomaterials. 2011;32:9136–9143. doi: 10.1016/j.biomaterials.2011.08.022. [DOI] [PubMed] [Google Scholar]
- 506.Liao L, Liu J, Dreaden EC, Morton SW, Shopsowitz KE, Hammond PT, Johnson JA. J Am. Chem. Soc. 2014;136:5896–5899. doi: 10.1021/ja502011g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Mi Y, Zhao J, Feng S-S. J Controlled Release. 2013;169:185–192. doi: 10.1016/j.jconrel.2013.01.035. [DOI] [PubMed] [Google Scholar]
- 508.Liu H, Li Y, Lyu Z, Wan Y, Li X, Chen H, Chen H, Li X. J Mater. Chem. B. 2014;2:8303–8309. doi: 10.1039/c4tb01563k. [DOI] [PubMed] [Google Scholar]
- 509.Cook TR, Zheng Y-R, Stang PJ. Chem. Rev. 2013;113:734–777. doi: 10.1021/cr3002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Zheng Y-R, Suntharalingam K, Johnstone TC, Lippard SJ. Chem. Sci. 2015;6:1189–1193. doi: 10.1039/c4sc01892c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Stetefeld J, Jenny M, Schulthess T, Landwehr R, Engel J, Kammerer RA. Nat. Struct. Biol. 2000;7:772–776. doi: 10.1038/79006. [DOI] [PubMed] [Google Scholar]
- 512.Peters J, Baumeister W, Lupas A. J Mol. Biol. 1996;257:1031–1041. doi: 10.1006/jmbi.1996.0221. [DOI] [PubMed] [Google Scholar]
- 513.Eriksson M, Hassan S, Larsson R, Linder S, Ramqvist T, Lövborg H, Vikinge T, Figgemeier E, Müller J, Stetefeld J, Dalianis T, Özbek S. Anticancer Res. 2009;29:11–18. [PubMed] [Google Scholar]
- 514.Thanasupawat T, Bergen H, Hombach-Klonisch S, Krcek J, Ghavami S, Del Bigio MR, Krawitz S, Stelmack G, Halayko A, McDougall M, Meier M, Stetefeld J, Klonisch T. Nanomedicine: Nanotechnology, Biology and Medicine. 2015 doi: 10.1016/j.nano.2015.01.014. In press. [DOI] [PubMed] [Google Scholar]
- 515.Zheng Y-R, Suntharalingam K, Johnstone TC, Yoo H, Lin W, Brooks JG, Lippard SJ. J Am. Chem. Soc. 2014;136:8790–8798. doi: 10.1021/ja5038269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Haxton KJ, Burt HM. J Pharm. Sci. 2009;98:2299–2316. doi: 10.1002/jps.21611. [DOI] [PubMed] [Google Scholar]
- 517.Bryde S, M. de Kroon AIP. Future Med. Chem. 2009;1:1467–1480. doi: 10.4155/fmc.09.112. [DOI] [PubMed] [Google Scholar]
- 518.Liu D, He C, Wang A, Lin W. Int. J. Nanomed. 2013;3309 doi: 10.2147/IJN.S38354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Butler JS, Sadler PJ. Curr. Opin. Chem. Biol. 2013;17:175–188. doi: 10.1016/j.cbpa.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 520.Oberoi HS, Nukolova NV, Kabanov AV, Bronich TK. Adv. Drug Delivery Rev. 2013;65:1667–1685. doi: 10.1016/j.addr.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Zalba S, Garrido MJ. Expert Opin. Drug Delivery. 2013;10:829–844. doi: 10.1517/17425247.2013.778240. [DOI] [PubMed] [Google Scholar]
- 522.Kieler-Ferguson HM, Fréchet JMJ, Szoka FC. Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol. 2013;5:130–138. doi: 10.1002/wnan.1209. [DOI] [PubMed] [Google Scholar]
- 523.Ulbrich K, Šubr V. Adv. Drug Delivery Rev. 2010;62:150–166. doi: 10.1016/j.addr.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 524.Rademaker-Lakhai JM, Terret C, Howell SB, Baud CM, de Boer RF, Pluim D, Beijnen JH, Schellens JHM, Droz J-P. Clin. Cancer Res. 2004;10:3386–3395. doi: 10.1158/1078-0432.CCR-03-0315. [DOI] [PubMed] [Google Scholar]
- 525.Sood P, Thurmond KB, II, Jacob JE, Waller LK, Silva GO, Stewart DR, Nowotnik DP. Bioconjugate Chem. 2006;17:1270–1279. doi: 10.1021/bc0600517. [DOI] [PubMed] [Google Scholar]
- 526.Nowotnik DP, Cvitkovic E. Adv. Drug Delivery Rev. 2009;61:1214–1219. doi: 10.1016/j.addr.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 527.Campone M, Rademaker-Lakhai JM, Bennouna J, Howell SB, Nowotnik DP, Beijnen JH, Schellens JHM. Cancer Chemother. Pharmacol. 2007;60:523–533. doi: 10.1007/s00280-006-0397-0. [DOI] [PubMed] [Google Scholar]
- 528.Ait-Oudhia S, Mager D, Straubinger R. Pharmaceutics. 2014;6:137–174. doi: 10.3390/pharmaceutics6010137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Immordino ML, Dosio F, Cattel L. Int. J. Nanomed. 2006;1:297–315. [PMC free article] [PubMed] [Google Scholar]
- 530.Petschauer JS, Madden AJ, Kirschbrown WP, Song G, Zamboni WC. Nanomedicine. 2015;10:447–463. doi: 10.2217/nnm.14.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Business Wire. New York: 1996. [Google Scholar]
- 532.Amantea MA, DeMario MD, Schwartz G, Vogelzang NJ, Tonda M, Pendyala L, Ratain MJ. Ann. Oncol. 1998;9(suppl2) 121 #465. [Google Scholar]
- 533.Working PK, Newman MS, Amantea MA. Ann. Oncol. 1998;9(suppl 2) 53 #201. [Google Scholar]
- 534.Stathopoulos GP, Boulikas T. J Drug Delivery. 2012;2012:581363. doi: 10.1155/2012/581363. [DOI] [PMC free article] [PubMed] [Google Scholar]