SUMMARY
Antimicrobial peptides (AMPs), also known as host defense peptides, are small naturally occurring microbicidal molecules produced by the host innate immune response that function as a first line of defense to kill pathogenic microorganisms by inducing deleterious cell membrane damage. AMPs also possess signaling and chemoattractant activities and can modulate the innate immune response to enhance protective immunity or suppress inflammation. Human pathogens have evolved defense molecules and strategies to counter and survive the AMPs released by host immune cells such as neutrophils and macrophages. Here, we review the various mechanisms used by human bacterial pathogens to resist AMP-mediated killing, including surface charge modification, active efflux, alteration of membrane fluidity, inactivation by proteolytic digestion, and entrapment by surface proteins and polysaccharides. Enhanced understanding of AMP resistance at the molecular level may offer insight into the mechanisms of bacterial pathogenesis and augment the discovery of novel therapeutic targets and drug design for the treatment of recalcitrant multidrug-resistant bacterial infections.
Keywords: antimicrobial peptide, antibiotic resistance, bacterial pathogenesis, bacterial resistance, cathelicidin, defensin, host defense peptide, infectious disease, innate immunity, invasive disease, mechanism of action, mechanism of resistance, membrane modification, membrane permeability
INTRODUCTION
Antimicrobial peptides (AMPs) are small (<10 kDa) soluble host defense peptides that play an important role in the mammalian innate immune response, helping to prevent infection by inhibiting pathogen growth on skin and mucosal surfaces and subsequent dissemination to normally sterile sites. These natural antibiotics are produced by many cell types including epithelial cells, leukocytes (neutrophils, macrophages, dendritic cells and mast cells), platelets, endothelial cells and adipocytes in response to tissue damage or infectious stimuli, and are found in body fluids and secretions including saliva, urine, sweat, and breast milk. To date, more than 2,000 AMPs have been identified from a wide variety of organisms including bacteria, insects, plants, amphibians, birds, reptiles and mammals including humans (1, 2). Whereas prokaryotic AMPs are produced as a competitive strategy to facilitate the acquisition of nutrients and promote niche colonization (3), AMPs produced by higher organisms are generally conceived to carry out immune defense functions. In humans, the principal AMPs are hydrophobic molecules composed of ~10–50 amino acid residues with a net positive charge, which exhibit varying degrees of broad-spectrum bioactivity against Gram-positive and Gram-negative bacteria, fungi, protozoan parasites, and certain enveloped viruses (4, 5). AMPs may be expressed constitutively or induced in response to infection (e.g. pro-inflammatory cytokines, toll-like receptor (TLR) signaling) (6), and are commonly produced as pro-peptides that undergo subsequent proteolytic processing to the mature bioactive peptide (7). AMPs with central roles in host defense are active at micromolar to nanomolar concentrations and facilitate microbial killing through perturbation of the cytoplasmic membrane (8). Several important human pathogens display significant resistance to AMPs, which appears to play a key role in their potential to produce serious invasive infections.
AMPs can be classified into four main groups according to their secondary structure: 1) α-helical peptides, 2) β-sheet peptides, 3) loop peptides, and 4) extended peptides (1, 9). The two major AMP families in mammals are the cathelicidins and the defensins (Table 1). In their mature form, cathelicidins are often α-helical cationic AMPs that do not contain cysteine residues. LL-37 is the sole human cathelicidin (10). Defensins are β-sheet-stabilized peptides classified as either α- or β-defensins according to the pattern formed by three disulphide bridges. α-defensins are primarily produced by neutrophils and intestinal Paneth cells, while β-defensins are expressed by epithelial tissues in the respiratory, gastrointestinal and urinary tracts (11, 12). Mammalian defensins produced by human epithelial and immune cells are cysteine-rich peptide ~30–40 amino acid residues in length (13). Humans produce six α-defensins: HNP 1-4 are found in the azurophilic granules of neutrophil granulocytes (14), while human α-defensins HD-5 and HD-6 are expressed in Paneth cells located in the small intestine (15) and female urogenital tract (16) (Table 1). Six human β-defensins, HBD-1 through HBD-6, have been identified and are expressed by epithelial cells, monocytes, macrophages and dendritic cells (11, 17). Cathelicidins are found in skin cells, gastrointestinal cells, neutrophils and myeloid bone marrow cells (18) (Table 1). Activated platelets produce additional groups of cationic chemokine-related AMPs called thrombocidins and kinocidins (19-21).
Table 1.
Human antimicrobial peptides and murine cathelicidin mCRAMP.
| Class | Peptide | Gene | Species | Producing Cells | Reference(s) | Amino Acid Sequence* |
|---|---|---|---|---|---|---|
| α-defensins | HNP-1 | DEFA1 | Human | Azurophilic granules of neutrophil granulocytes | (14) | AC1YC2RIPAC3IAGERRYGTC2IYQGRLWAFC3C1 |
| HNP-2 | DEFA1 | Human | C1YC2RIPAC3IAGERRYGTC2IYQGRLWAFC3C1 | |||
| HNP-3 | DEFA3 | Human | DC1YC2RIPAC3IAGERRYGTC2IYQGRLWAFC3C1 | |||
| HNP-4 | DEFA4 | Human | VC1SC2RLVFC3RRTELRVGNC2LIGGVSFTYC3C1TRV | |||
| HD-5 | DEFA5 | Human | Paneth cells in small intestine and female urogenital tract | (15, 16) | ATC1YC2RTGRC3ATRESLSGVC2EISGRLYRLC3C1R | |
| HD-6 | DEFA6 | Human | AFTC1HC2RRSC3YSTEYSYGTC2TVMGINHRFC3C1L | |||
| β-defensins | HBD-1 | DEFB1 | Human | Epithelial cells, monocytes, macrophages and dendritic cells | (11, 17) | DHYNC1VSSGGQC2LYSAC3PIFTKIQGTC2YRGKAKC1C3K |
| HBD-2 | DEFB4 | Human | GIGDPVTC1LKSGAIC2HPVFC3PRRYKQIGTC2GLPGTKC1C3KKP | |||
| HBD-3 | DEFB103 | Human | GIINTLQKYYC1RVRGGRC2AVLSC3LPKEEQIGKC2STRGRKC1C3RRKK | |||
| HBD-4 | DEFB104 | Human | ELDRIC1GYGTARC2RKKC3RSQEYRIGRC2PNTYAC1C3LRK | |||
| Cathelicidins | LL-37 | CAMP | Human | Skin cells, gastrointestinal cells, neutrophils and myeloid bone marrow cells | (18) | LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES |
| mCRAMP | Cnlp | Murine | GLLRKGGEKIGEKLKKIGQKIKNFFQKLVPQPEQ | |||
| Others | C18G | n/a | Synthetic | n/a | (253) | ALYKKLLKKLLKSAKKLG |
Numbers denote cysteine residues involved in disulfide bonds.
Abbreviations: HNP-1, human neutrophil defensin 1; HNP-2, human neutrophil defensin 2; HNP-3, human neutrophil defensin-3; HNP-4, human neutrophil defensin-4; HD-5, human α-defensin-5; HD-6, human α-defensin-6; HBD-1, human β-defensin 1, HBD-2, human β-defensin 2; HBD-3, human β-defensin-3; HBD-4, human β-defensin-4; LL-37, human cathelicidin; mCRAMP, murine cathelicidin-related peptides; C18G, α-helical peptide derived from the carboxy terminus of platelet factor IV. Modified from Gruenheid and Moual 2012 (229).
These prototypical AMPs have a net positive charge to facilitate interaction with the net negative charge of bacterial surfaces (22). While cationic peptides comprise the largest class of AMPs, certain anionic peptides such as dermcidin produced by eccrine sweat glands, also contribute to host epithelial defense (23). In addition to charge, other factors influencing AMP spectrum and mechanism of action include size, amino acid composition, structural conformation, amphipathicity, and hydrophobicity (24). A primary mechanism of AMP action is through electrostatic interaction with the anionic phospholipid headgroups in the outer bacterial cytoplasmic membrane or cell wall components (22, 25). Upon penetration of the outer membrane or cell wall, AMP insertion into the cytoplasmic membrane causes membrane rupture and cell death (11).
Three general modes of AMP action have been proposed to explain the membrane disruption: 1) the “barrel-stave” mechanism where AMPs directly integrate into the target membrane forming membrane-spanning pores (26); 2) the toroidal-pore mechanism where AMPs form membrane-spanning pores with intercalated lipids inducing a curvature in the membrane (27); and 3) the “carpet” mechanism where AMPs at high concentration accumulate on the cell surface and dissolve the cell membrane in a detergent-like manner without forming membrane-spanning pores (28). In addition to cell membrane perturbation, some AMPs may exert downstream antimicrobial effects by inhibiting the bacterial DNA, RNA, or protein synthesis machinery or biosynthesis of cell wall components (29, 30). Nisin, an AMP commonly used in the food industry as a preservative, is a member of the bacteriocin or lantibiotic family of AMPs that inhibits the biosynthesis of teichoic acid (TA) and lipoteichoic acid (LTA) in Gram-positive bacteria (31). Another bacteriocin, mersacidin, inhibits cell wall peptidoglycan biosynthesis and is active against methicillin-resistant Staphylococcus aureus (32). Some eukaryotic defensins target the lipid II biosynthesis pathway, an essential component of peptidoglycan, to inhibit cell wall biosynthesis. Several AMPs inhibit nucleic acid biosynthesis including buforin II (33), indolicidin (34), and puroindoline (35). Human neutrophil peptide 1 (HNP-1), also known as human α-defensin 1, inhibits cell wall, DNA and protein synthesis (36).
Genetic animal models have established an essential role for AMPs in the innate immune system. For example, mice deficient in the murine cathelicidin (mCRAMP) suffer more severe necrotic skin lesions than wild-type (WT) littermates following subcutaneous infection with Streptococcus pyogenes (group A Streptococcus; GAS), a Gram-positive human pathogen (37, 38). GAS are killed less efficiently by whole blood and mast cells isolated from mCRAMP knockout mice (37, 38), and Salmonella enterica serovar Typhimurium (S. Typhimurium) proliferate better within macrophages of mCRAMP knockout mice (39). Cathelicidin-deficient mice are likewise more susceptible to Escherichia coli urinary tract infection (40), meningococcal septicemia (41), Pseudomonas aeruginosa keratitis (42), Klebsiella pneumoniae lung infection (43) and Helicobacter pylori gastritis (23254369), while mice deficient in β-defensin production show impaired defense against P. aeruginosa (44) or Fusarium solani keratitis (45). In gain-of-function analyses, transgenic mice overexpressing porcine cathelicidin were more resistant to bacterial skin infection (46), while transgenic expression of the human defensin-5 in mouse Paneth cells provided enhanced defense against S. Typhimurium enteritis (47).
Beyond their direct antimicrobial activities, AMPs including cathelicidins have also been reported to modulate cytokine production, apoptosis, functional angiogenesis, or wound repair by stimulating keratinocyte migration and proliferation (48-50). Serving as an important link between the innate and adaptive immune system, AMPs may induce the expression of cytokines and chemokines (51, 52), exert direct chemotactic action on neutrophils, macrophages, immature dendritic cells, mast cells, monocytes, and T lymphocytes (53-55), and stimulate histamine release from mast cell to promote neutrophil migration to the site of infection (56).
The resistance mechanisms employed by commensals or microbial pathogens to combat AMPs have been intensively studied over the past two decades. This chapter highlights current information on the direct and indirect mechanisms of action used by human pathogenic bacteria to counteract AMPs, including surface charge alteration, external sequestration by secreted or surface-associated molecules, energy-dependent membrane efflux pumps, peptidase degradation, and the downregulation of AMP expression by host cells. Perturbation of these AMP resistance mechanisms may impair bacterial colonization capacity and reduce virulence in animal infection models. Understanding the molecular mechanisms of AMP resistance may identify novel targets for intervention in difficult to treat bacterial infections.
BACTERIAL AMP RESISTANCE MECHANISMS
Bacterial Surface Charge Modification Increases AMP Resistance
The cationic nature of human AMPs such as defensins and cathelicidin LL-37 provide an electrostatic affinity for bacterial cell surfaces, which are composed of negatively charged hydroxylated phospholipids including phosphatidylglycerol (PG), cardiolipin (also known as diphosphatidylglycerol), and phosphatidylserine (57). In contrast, mammalian and eukaryotic cell membranes contain neutral lipids (phosphatidylcholine, phosphatidylethanolamine, sphingomyelin) and sterols (cholesterol, ergosterol), and carry a net neutral charge that allows selectivity of AMPs for the mostly anionic bacterial cell membranes (3, 57). The amphipathic structure of AMPs resulting from the separation of charged or polar and hydrophobic moieties within the molecule enables their integration into the lipid bilayer of Gram-positive and Gram-negative bacteria, fungi, or viruses, and the formation of destabilizing transmembrane pores that induce cell rupture and death (58, 59).
While Gram-positive bacteria lack an outer membrane, AMP access to the cytoplasmic membrane is inhibited by a thick peptidoglycan-containing cell wall cross-linked with polymers of TA or LTA. In Gram-negative bacteria, AMPs must traverse the outer membrane envelope composed of negatively charged lipopolysaccharide (LPS; up to 70% of the outer membrane) (60), and the periplasmic space beneath the outer membrane, which contains a thin peptidoglycan matrix. Surface-associated proteins and large capsular polysaccharides also hinder AMP access to the cytoplasmic membrane.
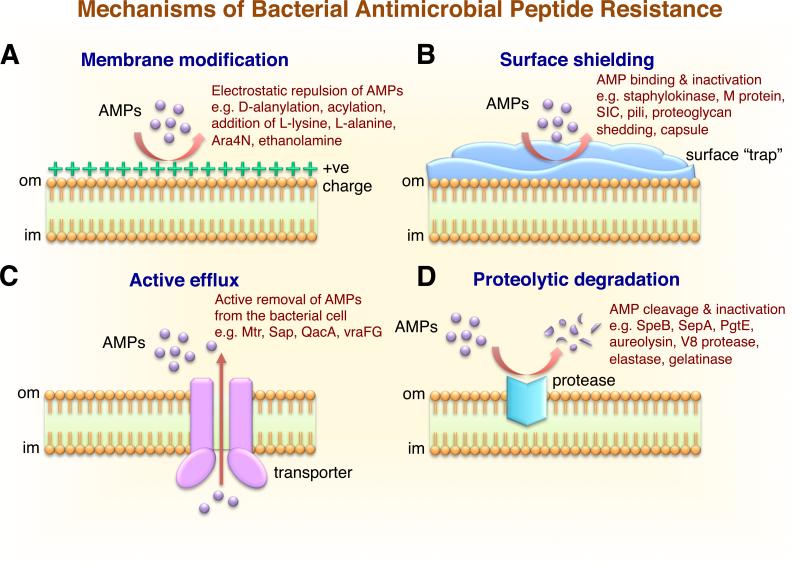
One common AMP-resistance strategy used by Gram-positive and Gram-negative bacteria is to increase their net positive surface charge through modification with cationic molecules, resulting in the electrostatic repulsion of cationic AMPs, thus preventing access to and disruption of the cytoplasmic membrane (Table 2). Several Gram-negative bacteria reduce the net negative charge of LPS lipid A through the addition of 4-amino-4-deoxy-L-arabinose (L-Ara4N), phosphoethanolamine (pEtN), or palmitoyl groups (61). Lipid A is negatively charged and consists of two glucosamine units with free phosphate groups linked to four or more acyl chains (62). Lipid A acylation coordinated by the PhoPQ regulatory system masks the negative surface charge in Gram-negative human pathogens such as E. coli, Salmonella spp., Yersinia enterocolitica, Haemophilus influenzae, K. pneumoniae, and Legionella pneumophila (3, 63). Some Gram-positive pathogens alter their surface charge through the modification of TAs composed of linear anionic glycopolymers of polyglycerol phosphate and polyribitol phosphate linked by phosphodiester bonds. LTAs are non-covalently inserted into the cell membrane with a glycolipid anchor, while wall teichoic acids (WTAs) are covalently attached to the peptidoglycan cell wall by a glycosidic bridge (64, 65). TAs play important roles in bacterial virulence, the adherence and invasion of host cells, biofilm formation (65), antimicrobial resistance (66-68), and activation of the immune response (69, 70). The D-alanylation of teichoic acids by the dlt operon and integration of L-lysine into PG by membrane protein multipeptide resistance factor (MprF) are common strategies employed by Gram-positive bacteria to reduce the negative surface charge and enhance AMP resistance (65, 71-73). D-alanylation of TAs is only known to occur in the bacterial Firmicutes phylum (65).
Table 2.
Bacterial antimicrobial peptide resistance mechanisms.
| AMP Resistance Mechanism | AMP Resistance Phenotype | Gene(s) | Target AMPs | Bacteria* | Reference(s) |
|---|---|---|---|---|---|
| Cell surface alterations | D-alanylation of lipoteichoic acid and teichoic acid in bacterial cell wall |
dlt operon dltA |
Cecropin B, colistin, gallidermin, HNP1-3, indolicidin, mCRAMP, magainin II, nisin, polymyxin B, protegrin 1, 3 and 5, tachyplesin 1 and 3, daptomycin, vancomycin |
Staphylococcus aureus Listeria monocytogenes Group B Streptococcus Group A Streptococcus Streptococcus pneumoniae Streptococcus suis Enterococcus faecalis Bacillus anthracis Bacillus cereus Clostridium difficile |
(66-68, 71, 72, 83, 254-259) |
| Addition of L-lysine or L-alanine to phosphatidylglycerol in cell membrane |
mprF
lysC lysX PA0920 |
Arenicin-1, CAP18, gallidermin, HBD-3, HNP1-3, LL-37, lugworm beta-sheet peptide, lysozyme, magainin II, melittin, nisin, NK-2, polymyxin B, protamine, protegrin 3 and 5, tachyplesin 1, vancomycin |
Staphylococcus aureus
Bacillus anthracis Listeria monocytogenes Mycobacterium tuberculosis Pseudomonas aeruginosa |
(73, 81, 82, 85, 87, 91, 93-95) | |
| Synthesis and extension of lipooligosaccharide |
lpxA
lgtF galT cstII |
Crp4, Fowl-1, HD-5, LL-37, polymyxin B |
Neisseria meningitidis
Campylobacter jejuni |
(113-115) | |
| Addition of ethanolamine (pEtN) to lipid A |
waaF
lpxEHP cj0256 pmrC lptA |
LL-37, protegrin 1, polymyxin B |
Helicobacter pylori
Campylobacter jejuni S. Typhimurium Neisseria gonorrhoeae Neisseria meningitidis |
(101, 108, 110, 111, 260) | |
| Addition of aminoarabinose to lipid A in LPS | pmr genes | C18G, HBD-2, polymyxin B, protegrin 1, synthetic protegrin analogs |
S. Typhimurium
Proteus mirabilis Pseudomonas aeruginosa Klebsiella pneumoniae |
(100, 103, 105, 106) | |
| Acylation of lipid A in LPS |
pagP
rcp htrB msbB lpxM |
C18G, colistin, CP28, HBD-2, LL-37, magainin II, mCRAMP, protegrin 1, PGLa, polymyxin B and E |
Salmonella spp.
Legionella pneumophila Haemophilus influenzae Vibrio cholerae Klebsiella pneumoniae |
(63, 117, 118, 120, 121) | |
| Phosphorylcholine in LPS | licD | LL-37 | Haemophilus influenzae | (119) | |
| Synthesis of polysaccharide capsule |
cps
siaD sia operon ica genes cap hasABC |
HBD-1 and 3, HNP-1 and 2, lactoferrin, polymyxin B, protamine, mCRAMP, CRAMP-18, HNP-1 and 2, LL-37, protegrin 1, polymyxin B, β-defensin-1, 2 and 3 |
Klebsiella pneumoniae
Neisseria meningitidis Staphylococcus epidermidis Streptococcus pneumoniae Streptococcus pyogenes |
(113, 139-143, 146) | |
| PCN-binding protein PBP1a | ponA | HNP-1, LL-37, mCRAMP | Group B Streptococcus | (96) | |
| Mycolic acid synthesis | kasB | HNP-1, protamine, lysozyme | Mycobacterium marinum | (99) | |
| Production of carotenoids | crtOPQMN | HNP-1, thrombin-induced platelet microbicidal proteins (tPMPs), polymyxin B | Staphylococcus aureus | (261-263) | |
| Binding and inactivation | Staphylokinase | sak | HNP-1 and 2 | Staphylococcus aureus | (122, 123) |
| M1 surface protein | emm1 | LL-37 | Group A Streptococcus | (130) | |
| SIC protein | sic | LL-37, α-defensins, lysozyme | Group A Streptococcus | (124, 125) | |
| Shedding of host proteoglycans | lasA | LL-37, HNP-1 |
Pseudomonas aeruginosa Enterococcus faecalis Group A Streptococcus |
(136, 264) | |
| PilB | pilB | LL-37, mCRAMP, polymyxin B | Group B Streptococcus | (132) | |
| LciA | lciA | Lactococcin A | Lactococcus lactis | (265, 266) | |
| LanI lipoproteins | lanI | Lantibiotics |
Lactococcus lactis
Bacillus subtilis |
(267-269) | |
| Active efflux | ATP-dependent efflux system | mtr genes | LL-37, mCRAMP, PC-8, TP-1, protegrin-1 (PG1) |
Neisseria gonorrhoeae
Neisseria meningitidis |
(111, 168, 170, 270) |
| K+-linked efflux pump | sap | Protamine | S. Typhimurium | (167) | |
| Plasmid-encoded efflux pump | qacA | Rabbit thrombin-induced platelet microbicidal protein (tPMP) | Staphylococcus aureus | (177) | |
| VraFG ABC transporter | vraFG | nisin, colistin, bacitracin, vancomycin, indolicidin, LL-37, hBD3 |
Staphylococcus aureus
Staphylococcus epidermidis |
(155, 271-274) | |
| Proteolytic degradation | Elastase | lasB | LL-37 | Pseudomonas aeruginosa | (181) |
| Gelatinase | gelE | LL-37 | Enterococcus faecalis | (181, 275) | |
| Metalloproteinase |
zapA
aur degP |
LL-37, lactoferricin |
Proteus mirabilis
Staphylococcus aureus Escherichia coli |
(181, 183, 184, 192) | |
| Cysteine protease |
speB
ideS |
LL-37 | Group A Streptococcus | (181, 182) | |
| Surface protease | pgtE | C18G | S. Typhimurium | (188) | |
| Gingipains (serine proteases) | rgpA/B | Cecropin B | Porphyromonas gingivalis | (193) | |
| Aureolysin | aur | LL-37 | Staphylococcus aureus | (183, 276) | |
| V8 protease | sspA | LL-37 | Staphylococcus aureus | (183) | |
| SepA protease | sepA | Dermcidin | Staphylococcus epidermidis | (277, 278) | |
| Alteration of Host Processes | Downregulate AMP transcription | mxiE | LL-37, human beta-defensin-1, human beta-defensin HBD-3 |
Shigella dysenteriae
Shigella flexneri S. Typhimurium Neisseria gonorrhoeae |
(225, 226, 279, 280) |
| Stimulation of host cysteine proteases cathepsins | Unknown | HBD-2, HBD-3 | Pseudomonas aeruginosa | (227, 281) | |
| Regulatory Networks | Two-component regulator | phoP/phoQ | Defensins, protamine |
S. Typhimurium
Pseudomonas aeruginosa |
(200, 205) |
| Two-component regulator | pmrA/pmrB | Defensins, polymyxin B |
S. Typhimurium
Pseudomonas aeruginosa |
(100, 206) | |
| Thermoregulated transcription factor | prfA | Defensins | Listeria monocytogenes | (224) |
Not all bacteria are resistant to the CAMP indicated; please see reference for specific resistance profile.
Abbreviations: C18G, α-helical peptide derived from the carboxy terminus of platelet factor IV; CAP18, cationic LPS-binding protein 18 from rabbit; CP28, α-helical synthetic cationic peptide based on the cecropin-mellitin hybrid peptide CEME; mCRAMP and CRAMP-18, murine cathelicidin-related peptides; Crp4, murine homologous to human α-defensin-5; Fowl-1, heterophil-derived cathelicidin homolog fowlicidin-1; HBD-1, human β-defensin 1, HBD-2, human β-defensin 2; HBD-3, human β-defensin-3; HD-5, human α-defensin-5; HNP-1, human neutrophil defensin 1; HNP-2, human neutrophil defensin 2; HNP-3, human neutrophil defensin-3; LL-37, human cathelicidin, C-terminal part of the human cationic antimicrobial protein (hCAP-18); NK-2, α-helical fragment of mammalian NK-lysin; PGLa, peptide starting with a glycine and ending with a leucine amide from magainin peptide family. Modified from Anaya-Lopez et al. (3).
D-alanylation of Cell Wall Teichoic Acids
S. aureus, a major Gram-positive human pathogen, is the etiologic agent of abscesses, cellulitis, osteomyelitis, septic arthritis, septicemia and endocarditis. S. aureus resists killing by human AMPs through the D-alanylation of cell wall TA. Incorporation of D-alanyl esters into the cell wall by the action of four proteins encoded by the dltABCD operon exposes a positively charged amino group, reducing the net negative charge of TAs, and diminishing the electrostatic attraction between cationic AMPs and the bacterial cell envelope (66-68, 72, 74, 75) (Fig. 1A). D-alanine is activated by D-alanyl carrier protein ligase (Dcl; encoded by dltA) and delivered to D-alanine carrier protein (Dcp; encoded by dltC) with assistance from chaperone protein DltD (encoded by dltD). The putative transmembrane protein DltB (encoded by dltB) is thought to facilitate transfer of the D-alanyl-Dcp complex across the cytoplasmic membrane (65). The D-alanylation of TA is also dependent upon environmental factors such as temperature, pH and salt (e.g. NaCl) concentration (76, 77). Transcriptional regulators of TA D-alanylation have been identified for several species, including Bacillus subtilis (global transcriptional regulators AbrB and Spo0A) (78), group B Streptococcus (GBS; two-component system DltRS) (79), and S. aureus (global regulators Agr and Rot, two-component system ArlRS) (20). In a recently proposed model, the increased density of the peptidoglycan sacculus resulting from cell wall D-alanylation may also sterically hinder AMP access to the cell membrane and contribute to AMP resistance (80). As a consequence, the cell wall of a GBS mutant lacking dltA was less compact and more permeable to AMPs than the WT parent strain (80). However, additional research is required to ascertain whether or not this mechanism applies to other Gram-positive species.
Figure 1.
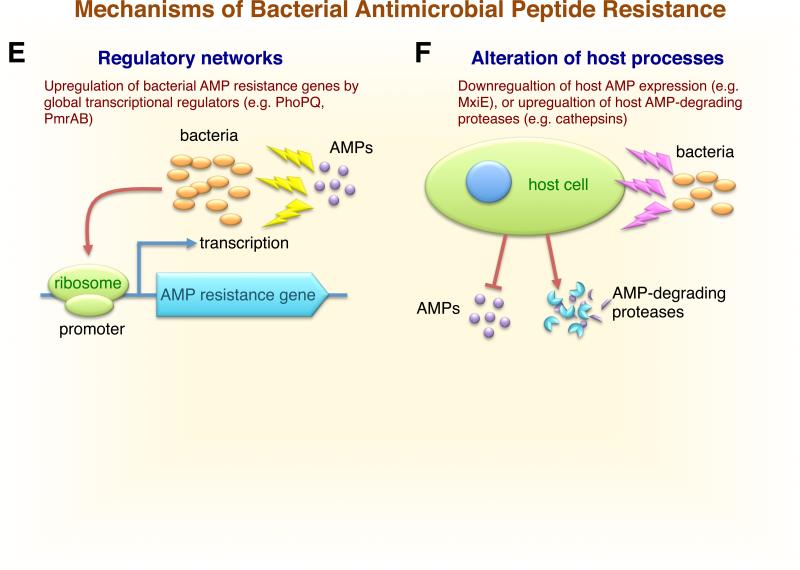
Schematic representation of the multiple resistance mechanisms developed by bacteria to overcome host antimicrobial peptides. A) Modification of the bacterial outer membrane. Bacterial resistance to cationic antimicrobial peptides is mediated by alterations in surface charge. Gram-positive bacteria: D-alanine modification of cell wall techoic acid (dlt), L-lysine (mprf), or L-alanine modification of phosphatidylglycerol (mprf). Gram-negative bacteria: aminoarabinose or acylation modifications of lipid A in LPS (pmr, pagP), or addition of ethanolamine to lipid A (pmrC, lptA). The increased positive charge on bacterial surface repels cationic AMPs. B) Shielding of the bacterial surface through the trapping and inactivation of AMPs in the extracellular milieu enhances resistance and pathogenicity. Surface-associated capsule traps AMP (e.g. K. pneumoniae cps operon), surface protein binds AMP (e.g. GAS M1 protein, GBS PilB pilus protein), secreted protein binds AMP (e.g. GAS SIC protein or S. aureus staphylokinase), or bacterial proteases release host proteoglycans to block AMP (e.g. P. aeruginosa LasA). C) Membrane efflux pumps function by translocating the AMP out of cell (e.g. Neisseria spp. Mtr, S. Typhimurium Sap, S. aureus QacA, and Staphylococcus spp. VraFG). D) Degradation and inactivation of AMPs by bacterial proteases (e.g. GAS SpeB protease, S. epidermidis SepA, S. Typhimurium PgtE, S. aureus aureolysin and V8 protease, P. aeruginosa elastase, and E. faecalis gelatinase). E) Bacterial exposure to AMPs upregulates the expression of AMP-resistance genes through global gene regulatory networks (e.g. S. Typhimurium and P. aeruginosa PhoPQ and PmrAB). F) Alteration of host processes by bacteria, including the downregulation of host AMP production (e.g. Shigella spp. transcriptional factor MxiE), or the upregulation and activation of host AMP-degrading proteases (e.g. P. aeruginosa). Abbreviations: om, bacterial outer membrane; im, bacterial inner membrane.
Compared to WT strains, S. aureus dltA null mutants and dltA, dltB, and dltD mutants of Staphylococcus xylosusare deficient in D-alanine esters of LTAs are hypersensitive to human α-defensins and cathelicidin due to an increase in negative surface charge and enhanced AMP binding (66, 72, 75). Furthermore, the overexpression of dlt in WT S. aureus enhances AMP resistance by increasing the cell surface positive charge (72). An S. aureus dltA mutant has reduced adherence to artificial surfaces, diminished biofilm formation, and reduced virulence in murine infection models (75, 81, 82). Several Gram-positive human pathogens have dlt operons, including GAS (71), GBS (66), Streptococcus pneumoniae (68), Enterococcus faecalis (67), Listeria monocytogenes (83), and B. subtilis. Inactivation of dltA in these and other Gram-positive species enhances sensitivity to human α-defensins and cathelicidin LL-37 (66, 71, 72, 83). Correspondingly, LTA D-alanylation is required for full virulence in mouse models of GAS (71), L. monocytogenes (83), and GBS infection (66). D-alanylation promotes GAS neutrophil intracellular survival as well as L. monocytogenes in vivo whole blood survival and in vitro adherence to macrophages, hepatocytes and epithelial cells (83). In Lactobacillus, TA D-alanylation plays an important role in establishing gastrointestinal tract colonization (84).
Aminoacylation with L-Lysine or L-Alanine
The multiple peptide resistance factor MprF (also known as LysS), encoded by the mprF gene, is highly conserved ~97 kDa integral membrane protein found in both Gram-positive and Gram-negative bacteria. MprF possesses a conserved C- terminal hydrophilic cytoplasmic domain and a large N-terminal flippase domain (85) that reduces the net negative surface charge of Gram-positive bacteria by incorporating L-lysine or L-alanine into cell wall PG (82, 85, 86). MprF is a lysine-substituted phophatidylglcerol (L-PG) synthase that alters surface charge through the formation of a positively charged membrane phospholipid (82, 87) (Fig. 1A). Both the N- and C-terminal domains of MprF are necessary for AMP resistance in S. aureus (85) and mprF gene expression is controlled by the ApsRSX regulator (88) A S. aureus mutant strain lacking mprF has an increase in negative surface charge compared to WT, is more sensitive to killing by a broad range of bacterial and mammalian AMPs, including neutrophil defensins (82), and is less virulent in mouse infection models (75, 81, 82).
Aminoacylation of PG by MprF homologues increases AMP resistance in multiple bacterial species, including Clostridium perfringens, E. faecalis, P. aeruginosa, Mycobacterium tuberculosis, Bacillus anthracis, B. subtilis, Enterococcus faecium, and L. monocytogenes (89). C. perfringens, a Gram-positive spore-forming bacterium and common cause of foodborne illness, expresses two mprF genes designated mprF1 and mprF2, which encode for alanyl phosphatidylglycerol synthase (A-PGS) and lysylphosphatidylglycerol synthase (L-PGS), respectively (86, 90). In M. tuberculosis, the addition of positively charged amino acid L-lysine to PG is encoded by the lysX gene encoding for MprF homolog LysX, and is essential for resistance to cationic antibiotics and AMPs (91, 92). Lysinylation of PG has also been described for L. monocytogenes (93), P. aeruginosa (94), and B. anthracis (95).
Additional cell wall modifications in Gram-positive bacteria have also been reported to influence AMP resistance. In GBS, the ponA gene encodes for penicillin-binding protein 1a (PBP1a) and promotes resistance to human cathelicidin and defensins (96). The pgm gene, encoding a phosphoglucomutase, contributes to AMP resistance in porcine pathogen Bordetella bronchiseptica and the fish pathogen Streptococcus iniae (97, 98). In Mycobacterium marinum, mutation of the kasB gene, encoding beta-ketoacyl-acyl carrier protein synthase B (KasB) reduces growth in human macrophages and bacterial survival in the presence of human defensins (99).
Modification of LPS with L-Ara4N or pEtN
The outer membrane of Gram-negative bacteria is composed of lipid A, an anionic dimer of glucosamine linked to fatty acid chains and flanked by polar phosphate groups synthesized on the cytoplasmic surface of the inner membrane by highly conserved enzymes. The lipid A moiety has an attached core polysaccharide and species specific side-chain “O” polysaccharides (62). Modification of this complex, known as LPS, with amine substituents L-Ara4N or pEtN reduces the net negative surface charge and AMP affinity, thereby promoting AMP resistance in Gram-negative bacteria such as Salmonella spp., important human pathogens and the causative agents of enteric/typhoid fever (Fig. 1A). In S. Typhimurium, two-component regulatory system PmrAB plays an important role in sensing extracellular cationic AMPs in vivo, and coordinates the expression of pmrC to decorate lipid A with ethanolamine, and pmrEHFIJKLM for the attachment of positively charged of L-Ara4N to the 4’-phosphate group of the lipid A backbone, which together reduce the net negative charge of lipid A and enhance resistance to cationic AMPs (100, 101). All genes except for pmrM are required for the addition of L-Ara4N and increased resistance to cationic AMPs in S. Typhimurium (100). S. Typhimurium lacking the LPS modifying enzyme PmrA are more sensitive to AMPs and have reduced virulence in a murine model of enteric infection (100, 102). L-Ara4N modification of LPS enhances AMP resistance of several Gram-negative species including Proteus mirabilis, responsible for urinary tract infections (103); Yersinia pseudotubercolosis, a causative agent of enterocolitis (104); K. pneumoniae, a human lung pathogen (105); and P. aeruginosa (106), associated with chronic airway infections in cystic fibrosis patients (107).
In the Gram-negative pathogen H. pylori, an etiologic agent of peptic ulcers and increased gastrointestinal cancer risk, the addition of pEtN to dephosphorylated lipid A of LPS increases AMP resistance and reduces TLR4–mediated activation of the innate immune system (108, 109). Mutation of the H. pylori lpxEHP genes disrupted direct attachment of pEtN to the disaccharide backbone of lipid A, increased net negative charge of LPS and concomitantly reduced the minimum inhibitory concentration (MIC) of polymyxin B, a bacterial-derived AMP, by 25-fold compared to WT (108). In Neisseria gonorrhoeae, the lptA gene catalyzes addition of pEtN to lipid A and is necessary for polymyxin B resistance and survival in humans (110). Similarly, mutagenesis of lptA in Neisseria meningitidis decreased resistance to polymyxin B, protegrin-1 (PG1) and LL-37 (111). Deletion of the lpxA gene encoding an enzyme in the lipid A biosynthesis pathway of N. meningitidis, abolishes lipooligosaccharide (LOS) production and increases sensitivity to cationic AMPs (112, 113). Similarly, mutation of waaF, cstII, galT or lgtF genes in Campylobacter jejuni results in LOS truncation and hypersensitivity to AMPs including polymyxin B, human α-defensin-5 (HD-5), and the murine HD-5 homologue Crp4 (114, 115).
Acylation and Phosphorylcholination of LPS
The pagP gene encoding acetyltransferase PagP in the outer membrane of S. Typhimurium acylates lipid A and increases AMP (C18G, pGLa, and PG1) resistance by reducing outer membrane permeability (63, 116) (Fig. 1A). Inactivation of the pagP homologue rcp in respiratory tract pathogen L. pneumophila reduces growth rate, AMP resistance, intracellular survival, and mouse lung colonization (117). LOS acylation by the H. influenzae htrB gene product is required for resistance to human AMP β-defensin 2 (HBD-2) (118). Addition of phosphorylcholine (ChoP) to the oligosaccharide portion of LPS promotes H. influenzae resistance to human cathelicidin LL-37 (119), conceivably through the cell surface exposure of the positively charged quaternary amine on choline to promote electrostatic repulsion (Fig. 1A). Inactivation of the lpxM gene in K. pneumoniae, which encodes an enzyme necessary for secondary acylation of immature lipid A, increases sensitivity to α-helical cationic AMPs through enhanced outer membrane permeability (120). In pathogenic Vibrio cholerae strain El Tor, the msbB gene is required for full acylation of the lipid A moiety and resistance to cationic AMPs (121).
Trapping of AMPs by Surface Molecules
Proteins and polysaccharides associated with the bacterial surface or secreted into the extracellular milieu may directly bind AMPs (Fig. 1B), thereby blocking access to the cytoplasmic membrane target of action and the formation of lytic pores. Another indirect AMP neutralization strategy employed by bacterial pathogens involves the release of the bound AMP from the bacterial surface (Table 2).
Surface-associated Proteins, Secreted Proteins and Polysaccharides
Plasminogen is the inactive form of plasmin, a host serine protease involved in the degradation of blood clots and tissue remodeling. S. aureus secretes a plasminogen activating protein known as staphylokinase (SK). The accumulation of active plasmin activity on the S. aureus cell surface promotes host tissue invasion and dissemination to normally sterile sties (122). SK binds and inactivates mCRAMP and α-defensins released from human neutrophils including HNP 1-3 (122, 123) (Fig. 1B), reducing AMP activity against S. aureus by more than 80%. Further, S. aureus strains expressing SK are more resistant to killing by α-defensins in a mouse model of arthritis, and the addition of purified SK to SK-deficient strains enhanced survival in the presence of α-defensin in vitro (123). The secreted hydrophilic GAS protein streptococcal inhibitor of complement (SIC) binds and inactivates human LL-37, α-defensin and lysozyme to promote bacterial survival (Fig. 1B) (124-126). A sic knockout mutant in the highly invasive M1T1 GAS genetic background was more sensitive to killing by AMPs, and shows diminished virulence in animal infection models (124, 125).
The M protein of GAS, encoded by the emm gene, is a major cell wall-anchored coiled-coil protein required for resistance to opsonophagocytosis, adherence to host cells, and full virulence in animal models of GAS infection (127). The C-terminal region of M protein is highly conserved and contains the canonical LPXTG well wall anchor motif. GAS is classified into emm types according to the nucleotide sequence of the hypervariable N-terminal region. Currently, there are more than 200 known GAS serotypes and the M1 GAS serotype is the most frequently isolated serotype from invasive GAS infections worldwide (128, 129). Mutation of the emm1 gene, encoding M1 protein, significantly increased the sensitivity to LL-37 or mCRAMP compared to WT (130), while the heterologous expression of M1 protein in serotype M49 GAS or Lactococcus lactis enhanced LL-37 resistance. The trapping of LL-37 through the hypervariable extracellular N-terminal domain of M protein impedes LL-37 access to the cell membrane and promotes bacterial survival in LL-37-containing neutrophil extracellular traps (NETs) (Fig. 1B) (130). In GBS, surface-associated penicillin-binding protein-1a and the PilB surface pilus protein promotes adherence to host cells and resistance to cathelicidin AMPs through surface sequestration of LL-37 and mCRAMP in vitro (131, 132). Inactivation of pilB in GBS also reduces virulence in a mouse infection model (132).
Serological classification of streptococci in groups is based upon expression of unique carbohydrate antigens in the bacterial cell wall (133) known to play a structural role in cell wall biogenesis (134). Approximately 50% of the GAS cell wall by weight is made up of a single polysaccharide molecule termed the group A carbohydrate (GAC) antigen. All strains of GAS express GAC, composed of a polyrhamnose core with an immunodominant N-acetylglucosamine (GlcNAc) side chain (134). Inactivation of the gacI gene, encoding for a glycosyltransferase, abolished expression of the GlcNAc side chain in serotype M1 GAS. The gacI mutant was more susceptible to killing within NETs and to human cathelicidin LL-37, a component of neutrophil specific granules important for intracellular killing and deployed within NETs (135). Similarly, the gacI mutant had reduced growth in human serum and was hypersensitive to killing by the antimicrobial releasate from thrombin-activated human platelets. Loss of the GlcNAc epitope on GAC attenuated GAS virulence in a rabbit model of pulmonary infection, and a mouse model of systemic infection (135). In studies with purified WT and mutant GAC, the GlcNAc side chain was shown to impede LL-37 interaction with the underlying polyrhamnose core (135).
The active shedding of negatively charged surface exposed proteoglycans on host epithelial cells by proteases from bacterial pathogens is another resistance mechanism to trap and inactivate AMPs in tissues (Fig. 1B). Proteases secreted by GAS, E. faecalis and P. aeruginosa degrade decorin and release dermatan sulfate, which can bind and inactivate human α defensin HNP-1 (136). Syndecan-1, a proteoglycan derived from the degradation of heparan sulfate, is released from the host cell surface by P. aeruginosa virulence factor LasA to bind and impede AMP function (137). S. epidermidis synthesizes polysaccharide intercellular adhesin (PIA), a positively charged extracellular matrix polymer encoded by the ica gene locus (icaADBC) and the cap gene (138), to enhance electrostatic repulsion and resistance to cationic AMPs LL-37 and human β-defensin-3 (HBD-3) (139-141).
Capsular Polysaccharides
Several bacterial pathogens express surface capsules composed of high molecular mass polysaccharides that promote in vivo survival and trap cationic AMPs to impede interactions with the microbial cell surface (Fig. 1B). The hyaluronan capsule of GAS promotes survival in NETs through enhanced resistance to LL-37 (142). In K. pneumoniae, the cps capsule biosynthesis operon is transcriptionally upregulated in the presence of AMPs to enhance resistance to polymyxin B, protamine sulfate, defensin-1, β-defensin-1, and lactoferrin (143). The capsule of K. pneumoniae prevents engagement of TLR 2 and 4 and subsequent activation of the nuclear factor-κB (NF-κb) and mitogen-activated protein kinases (MAPK) pathways to inhibit the expression of human β-defensins (144). Administration of capsular polysaccharide extracts from S. pneumoniae serotype 3 and P. aeruginosa enhanced the resistance of non-encapsulated K. pneumoniae to α-defensin HNP-1 and polymyxin B, suggesting that the release of capsule from the bacterial surface promotes the trapping of AMPs to prevent access to the site of action (145). Further, polymyxin B and HNP-1 also stimulate the release of capsule from the S. pneumoniae cell surface to sequester AMPs and increase AMP resistance (145). Studies with encapsulated WT and nonencapsulated serotype B mutant N. meningitidis demonstrate that capsule promotes resistance to protegrins, α- and β-defensins, polymyxin B, and cathelicidins LL-37 and mCRAMP (146). Moreover, the release of capsule from the surface of N. meningitidis is reported to promote resistance to LL-37 (113), and sub-lethal concentrations of AMP induce capsule biosynthesis (113, 146). Other bacterial species shield AMP targets with surface polymers. For example, LOS expression in C. jejuni increases LL-37, α-defensins and polymyxin B resistance (114). P. aeruginosa biofilms produce alginate polysaccharide, a polymer of β-D-manuronate and α-L-guluronate, to sequester and induce AMP conformational changes and peptide aggregation to prevent AMP access to the cell membrane (147).
Efflux Systems for AMP Resistance
Well-studied for their prominent role in resistance to pharmaceutical antibiotics, certain adenosine triphosphate (ATP)-binding cassette (ABC) driven efflux pumps are used by human bacterial pathogens to resist AMPs through the extrusion of AMPs from the cell membrane site of action to the extracellular environment (148) (Fig. 1C). Three major classes of ABC transporter systems play a role in AMP resistance, including 1) three-component ABC-transporters, 2) two-component ABC-transporters, and 3) single protein multidrug-resistance transporters (149). Several three-component ABC transporters implicated in AMP resistance have been described in Gram-positive species, including NisFEG (L. lactis) (150), SpaFEG (B. subtilis) (151), and CprABC (Clostridium difficile) (152, 153) (Table 2). Common two-component systems involved in AMP resistance include the BceAB transporter system identified in B. subtilis (154), S. aureus (155), L. lactis (156), S. pneumoniae (157) and L. monocytogenes (158), and the BcrAB(C) transporter identified in some species of Bacillus (159), Enterococcus (160), Clostridium (161) and Streptococcus (162). The energy-driven efflux pumps RosA/RosB and AcrAB are required for polymyxin B resistance in Y. enterocolitica and K. pneumoniae, respectively (163, 164). In Y. enterocolitica, RosA and RosB upregulate the ros locus and are necessary and sufficient for resistance to cationic AMPs. In K. pneumoniae, AacrAB also enhances resistance to α- and β-defensins (164). The MefE/Mel efflux pump contributes to LL-37 resistance in S. pneumoniae (165), while the TrkA and SapG potassium transport proteins in Vibrio vulnificus and S. Typhimurium, respectively, are essential for cationic AMP resistance (166, 167).
The energy-dependent MtrCDE efflux pump is a member of the resistance-nodulation-division (RND) efflux family. In the pathogens N. gonorrhoeae and N. meningitidis, MtrCDE is involved in actively transporting AMPs out of the bacterial cytoplasm and periplasmic space to promote resistance to LL-37, mCRAMP, PC-8, TP-1 and PG1 (61, 111, 168). In addition, the Mtr efflux pump increases resistance to β-lactam and macrolide antibiotics and in vivo resistance to innate immune clearance (61, 168, 169). MtrCDE is necessary for N. gonorrhoeae colonization in a mouse model of genital tract infection (170), and inactivation of mtrC in Haemophilus ducreyi induces hypersensitivity to β-defensins and human LL-37 (171). The sapABCDF operon encoding ABC importer Sap (‘sensitive to antimicrobial peptides’) in S. Typhimurium enhances resistance to protamine, bee-derived AMP melittin, and crude extracts from human neutrophil granule extracts (167, 172, 173). The Sap transporter also contributes to AMP resistance in other Gram-negative species, including H. influenzae (172) and H. ducreyi (174). Deletion of the S. Typhimurium yejF gene from the yejABEF operon encoding an ABC-type peptide import system, reduced resistance to polymyxin B, melittin, protamine, and human β-defensins 1 and 2 (175). In S. aureus, single protein efflux pump QacA, encoded on naturally occurring plasmid pSK1, belongs to the major facilitator superfamily of transport proteins and uses proton motive force to extrude substrates (176). QacA promotes resistance to rabbit platelet AMP and host-derived thrombin-induced platelet microbicidal protein (tPMP-1) (177), and may also induce secondary changes in membrane fluidity to promote AMP resistance (178). Increased resistance to tPMP-1 in S. aureus is correlated with in vivo survival in animal infection models, and endocarditis in humans (177, 179).
Inactivation of AMPs by Proteolytic Degradation
AMPs are relatively resistant to proteolytic degradation by surface-associated or secreted proteases produced by bacterial pathogens (180). However, some bacterial proteases with broad substrate specificity promote disease pathogenesis by efficiently cleaving and inactivating AMPs (Fig. 1D). The human AMP LL-37 is cleaved into nonfunctional breakdown products by proteases expressed by several human pathogens including E. faecalis (metallopeptidase gelatinase) (181), GAS (broad-spectrum cysteine protease SpeB) (182), S. aureus (aureolysin) (183), and P. mirabilis (50 kDa metalloprotease) (184). Aureolysin inactivates LL-37 by cleaving the C-terminal peptide bonds between the Arg19-Ile20, Arg23-Ile24 and Leu31-Val32 (183), and promotes survival within the LL-37 rich environment of macrophage phagolysosomes (185). The GAS protease inhibitor α2-macroglobulin (α2M) binds broad-spectrum cysteine protease SpeB to the cell surface with the help of surface-associated G-related α2M-binding (GRAB) protein to facilitate LL-37 cleavage and bacterial survival (186, 187). The metalloprotease ZapA, a major virulence factor of P. mirabilis that degrades antibodies, extracellular matrix molecules, and complement components C1q and C3, also contributes to AMP resistance by cleaving human β-defensin 1, LL-37 and PG1 (184). The elastase of P. aeruginosa completely degrades and inactivates LL-37, promoting survival in an ex vivo wound fluid model (181). The S. Typhimurium pgtE gene that encodes for outer membrane protease PgtE, enhances resistance to LL-37 and C18G, an α-helical cationic AMP (188). Plasminogen-activating streptokinase secreted by GAS results in the accumulation of cell surface plasmin activity capable of degrading LL-37 (189). In Burkholderia cenocepacia, ZmpA and ZmpB zinc-dependent metalloproteases cleave and inactivate AMPs LL-37 and β-defensin 1, respectively (190). High-level expression of outer membrane protease OmpT of enterohemorrhagic E. coli (EHEC) promotes AMP resistance through the efficient degradation of LL-37 at dibasic sites (191). Proteases secreted by other pathogens also efficiently cleave and inactivate AMPs, including B. anthracis (LL-37), Porphyromonas gingivalis (α- and β-defensins, cecropin B), and Prevotella spp. (brevinin) (192-196) (Table 2).
Regulatory Networks and AMP Resistance
Bacterial pathogens use two-component regulatory systems to modulate gene expression in response to extracellular metal ion concentrations, metabolic requirements, growth phase, or to subvert the host innate immune response mounted by neutrophils or macrophages within host tissue, resulting in the up- or down-regulation of genes necessary for survival and disease progression. Several pathogens achieve maximal resistance to AMPs through the coordinated transcriptional up-regulation of AMP resistance factors (Fig. 1E). PhoPQ is a well-studied two-component system in S. Typhimurium that responds to changes in magnesium ion (Mg2+) concentration, pH and the presence of cationic AMPs (20, 197) (Table 2). Sensor kinase PhoP directly or indirectly coordinates the expression of >100 genes in S. Typhimurium encoding for proteins involved in Mg2+ transport (MgtA and MgtCB), transcriptional regulators important for intracellular macrophage survival (SlyA), oxidative stress resistance (RpoS), LPS modification by amino arabinose (PmrAB), and lipid A acylation (PagP) to reduce the fluidity and permeability of the bacterial membrane and enhance AMP resistance (20, 188, 198). Consequently, PhoPQ plays a role in modifying LPS surface charge (63), in enhancing macrophage resistance through the up-regulation of the AMP-degrading outer membrane protease PgtE (188, 199-201), and is required for full virulence in a mouse model of gastrointestinal infection (167). Additional S. Typhimurium transcriptional factors associated with resistance to bacterially derived AMP polymyxin B include virK, somA, and rcsC (202). PhoPQ homologs have been identified in other Gram-negative pathogens, including Yersinia pestis, Shigella flexneri, and P. aeruginosa (203). Mutant strains of Y. pestis deficient in PhoPQ are more sensitive to AMPs and neutrophil intracellular killing (204). In P. aeruginosa, the presence of AMPs or divalent cations activates the PhoPQ and PmrAB systems to enhance resistance to cationic AMPs such as LL-37 and polymyxin B (205-207). The two-component system PmrAB in P. aeruginosa co-ordinates the incorporation of positively charged L-Ara4N subunits into LPS and promotes AMP resistance through electrostatic repulsion (106, 206).
Upon encountering bacteria at the site of infection, NETs are released to help trap and kill the bacteria. NETs are a composed of DNA backbone and antimicrobial effectors such as histones, granule proteases and AMPs (in particular cathelicidin) that promote microbe killing (208, 209). Degradation of the DNA scaffold by secreted bacterial DNAses promotes NET escape and survival for several bacterial pathogens including GAS (210-212), S. pneumoniae (213), GBS (214) and S. aureus (215). Subinhibitory concentrations of exogenous DNA promote P. aeruginosa AMP resistance through the chelation of divalent cations and the resultant upregulation of AMP resistance genes (216). In S. Typhimurium, extracellular DNA also induces pmr expression and AMP resistance (217).
The D-alanylation of techoic acid by the dlt operon is regulated by the agr locus in S. aureus and promotes AMP resistance (Dunman et al., 2001). Exposure of S. aureus to AMPs activates the VraSR and VraDE operons involved in resistance to AMPs and cell wall-targeting antibiotics such as bacitracin (28). Human β-defensin (HBD-3) triggers the upregulation of the cell wall stress response pathway in S. aureus to counteract HBD-3-induced perturbation of peptidoglycan synthesis (13). Exposure of S. aureus to sub-lethal concentrations of magainin 2 and gramicidin D promotes to resistance to these AMPs through the enhancement of membrane rigidity (218). Changes in membrane fluidity induced by incorporation of longer chain unsaturated fatty acids into the lipid bilayer (resulting in increased membrane fluidity), or carotenoid staphyloxanthin pigment (resulting in increased membrane rigidity), promotes S. aureus resistance to platelet-derived AMPs (tPMPs), or polymyxin B and human neutrophil defensin 1, respectively (219, 220). While the precise resistance mechanism has yet to be determined, a significant increase or reduction in membrane fluidity may hinder AMP insertion into the cellular membrane (89, 221). In L. monocytogenes, an increase in the concentration of membrane saturated fatty acids and phophatidylethanolamine, and a decrease in phophatidylglycerol concentration, reduces the fluidity of the cell membrane to promote nisin resistance (222, 223). PrfA, a temperature-regulated transcription factor in L. monocytogenes, contributes to defensin resistance (224).
Modulation of Host AMP Production by Bacterial Pathogens
While low levels of AMPs are produced by epithelial and host immune cells at baseline, AMP expression is typically dramatically upregulated in response to bacterial infection. Some bacterial pathogens resist AMP-mediated innate immune clearance by interfering with, or suppressing, host AMP expression levels (Fig. 1F). Shigella spp. are Gram-negative rods capable of causing life-threatening invasive human infections such as bacillary dysentery. Shigella dysenteriae and S. flexneri downregulate the expression of LL-37 and β-defensin-1 in intestinal epithelial cells during early infection through a mechanism dependent on transcriptional factor MxiE and the type III secretion system to promote bacterial survival, colonization and invasion of the gastrointestinal tract (225, 226) (Table 2). P. aeruginosa, a human pathogen commonly isolated from the lungs of cystic fibrosis patients, induces the expression of host cysteine proteases cathepsins B, L and S to cleave and inactivate β-defensins 2 and 3 and thwart AMP-mediated clearance of the bacteria in airway fluid (227). Enterotoxigenic E. coli (ETEC) and V. cholerae exotoxins reportedly repress the expression of host cell HBD-1 and LL-37 (228), while N. gonorrhoeae downregulates the expression of AMP genes (229). Burkholderia spp. are human pathogens associated with opportunistic infections in cystic fibrosis patients and chronic granulomatous disease (230). The high level AMP resistance exhibited by this Genus has been attributed to the constitutive incorporation of L-Ara4N into the LPS molecule (230, 231). Alternative sigma factor RpoE coordinates Burkholderia gene expression under stress conditions and contributes to AMP resistance in a temperature-dependent manner (230, 232).
Concluding Remarks and Future Directions
AMPs are present in most organisms and are an ancient and diverse group of naturally occurring anti-infective molecules that play an integral part in the host innate immune defense against bacterial infection. Bacterial AMP resistance mechanisms have evolved as a result of selection pressures from direct competition among species (bacteriocins) and during host-pathogen interactions (innate defense AMPs). Human bacterial pathogens have evolved a broad diversity of intrinsic or inducible AMP-defense mechanisms to promote survival, colonization, and subsequent dissemination to normally sterile sites within the body to cause life-threatening invasive syndromes. Bacterial pathogens with intrinsic high-level resistance to AMPs, such as S. aureus and Salmonella spp. can bypass normally effective mucosal defenses and are consequently among the leading causes of causes of deep tissue and systemic infections. AMP resistance is mediated by a variety of different molecular mechanisms including net cell surface charge alteration, efflux, restricting AMP access to their targets, and proteolytic cleavage of AMPs. Bacterial mutants sensitive to AMPs in in vitro assays are attenuated for virulence in systemic animal infection models. An improved comprehension of AMP modes of action, resistance mechanisms and host pathogen interactions may inspire the development of alternative antibacterial therapeutics that target the cell wall, efflux pumps, or AMP-inactivating proteases, ultimately enhancing bacterial sensitivity to the AMPs of the host innate immune system. Understanding the interaction between conventional antibiotics and endogenous AMPs can also lead to improved therapeutic strategies for drug-resistant pathogens. An action of beta-lactam antibiotics to sensitize methicillin-resistant S. aureus (MRSA) and vancomycin-resistant Enterococcus spp. (VRE) to killing by human cathelicidin LL-37 and cationic peptide antibiotic daptomycin has shown promise in synergy studies and small clinical series in patients with previously recalcitrant infections (233, 234).
The emergence of antibiotic resistant microbes through the excessive and inappropriate use of conventional antibiotics is a critical public health threat responsible for high morbidity and rates and significant socioeconomic costs worldwide. Moreover, the antibiotic development pipelines of the major pharmaceutical companies have steadily declined over the past 20 years. Consequently, there is considerable interest in alternative therapeutic approaches to facilitate the fight against multidrug-resistant pathogens, including the development of novel broad-spectrum AMPs against bacteria, fungi, protozoa and enveloped viruses (30, 235). Importantly, the AMP mechanism of action is very rapid at concentrations close to the MIC, in comparison to conventional antibiotics (236). In recent years, intensive research and has led to the establishment of several bioinformatics tools and databases (e.g. APD2, CAMP, iAMP-2L) to identify and isolate new AMP classes and to elucidate their structure, function and biological activity (237). However, prolonged in vitro exposure of bacteria to sub-lethal AMP concentrations (238), and pre-clinical trials with naturally occurring cationic AMPs have detected resistant strains, indicating that optimization of AMP composition and structures are required to enhance stability and efficacy (237). Cross-resistance to AMPs with disparate modes of action has also been reported. For example, S. aureus is resistant to pexiganan and cross-resistant to HNP-1 (239). S. aureus isolates resistant to daptomycin, a cyclic lipopeptide antibiotic that associates with Ca2+ to form a cationic complex (240), are also more resistant host defense AMPs with diverse mechanisms of action, including HNP-1, polymyxin B, and tPMPs (241). Human pathogens resistant to nisin, an AMP used as a food preservative (L. monocytogenes, Streptococcus bovis) (242, 243), and colistin, also known as polymyxin E (Acinetobacter baumannii, P. aeruginosa, Brevundimonas diminuta, Ochrobactrum anthropic, K. pneumoniae) (244, 245) have recently been reported.
The transfer of broad-spectrum resistance mechanisms between bacteria and the development of resistance against our own host defense peptides remain valid concerns moving forward with the development of AMPs for clinical use (246, 247). Systemic toxicity and decreased blood and/or serum activity of natural peptides have significantly hampered clinical AMP development and provided the impetus for de novo designed peptide sequences (1). To this end, multiple new classes of AMPs have been reported (e.g. mimetic peptides, hybrid peptides, peptide congeners, stabilized AMPs, peptide conjugates, immobilized peptides) with potential application in medicine, veterinary medicine, and agriculture (248). Rationally designed synthetic AMPs have recently been demonstrated to be active against antibiotic-resistant A. baumannii and K. pneumoniae (249). Synthetic peptides could also be designed to resist bacterial and host proteases through the incorporation of D-amino acids (229). While pathogenic bacteria have successfully evolved AMP-resistance mechanisms, resistance to a broad range of AMPs has not yet occurred. Enhanced microbicidal activity of phagocytic cells and enhanced resistance to bacterial infection in vivo has been achieved by genetic or pharmacological augmentation of transcriptional regulator hypoxia-inducible factor (HIF) (250, 251), which regulates the expression of human and murine cathelicidin at the transcriptional level (250, 252). Combination therapy with AMPs and classical antibiotics that target more than one site of action, such as the inhibition of cell wall synthesis coupled with cell membrane disruption, may help to combat the increasing emergence of multidrug-resistant microbes associated with challenging and deadly microbial infections.
ACKNOWLEDGEMENT
The authors thank Anna Henningham, University of California San Diego School of Medicine, for the critical reading of this manuscript and many helpful suggestions.
FUNDING
This work was supported by the National Health and Medical Research Council of Australia (APP1033258 to J.N.C.), and the National Institutes of Health (AI093451, AR052728, AI077780, AI052453, and HD071600 to V.N.).
Abbreviations
- α2M
α2-macroglobulin
- ABC
adenosine triphosphate-binding cassette
- AMPs
antimicrobial peptides
- A-PGS
alanyl phosphatidylglycerol synthase
- L-Ara4N
4-amino-4-deoxy-L-arabinose
- ATP
adenosine triphosphate
- CAMP
cathelicidin antimicrobial peptide
- ChoP
phosphorylcholine
- Dcl
D-alanyl carrier protein ligase
- Dcp
D-alanine carrier protein
- EHEC
enterohemorrhagic Escherichia coli
- ETEC
enterotoxigenic Escherichia coli
- GAC
group A carbohydrate
- GAS
group A Streptococcus
- GBS
group B Streptococcus
- GlcNAc
N-acetylglucosamine
- GRAB
protein G-related α2M-binding protein
- HBD 1-6
human β-defensin 1-6
- HD 5-6
human α-defensin 5-6
- HIF
hypoxia-inducible factor
- HNP 1-4
human neutrophil peptide 1-4
- KasB
beta-ketoacyl-acyl carrier protein synthase B
- LL-37
human cathelicidin
- LOS
lipooligosaccharide
- L-PG
lysine-substituted phophatidylglcerol
- LPGS
lysylphosphatidylglycerol synthase
- LPS
lipopolysaccharide
- LTA
lipoteichoic acid
- MAPK
mitogen-activated protein kinases
- mCRAMP
murine cathelicidin-related antimicrobial peptide
- MIC
minimum inhibitory concentration
- MprF
membrane protein multipeptide resistance factor
- MRSA
methicillin-resistant S. aureus
- NETs
neutrophil extracellular traps
- NF-κb
nuclear factor-κB
- PBP1a
penicillin-binding protein 1a
- pEtN
phosphoethanolamine
- PG
phosphatidylglycerol
- PG1
protegrin-1
- PIA
polysaccharide intercellular adhesion
- RND
resistance-nodulation-division efflux family
- Sap
sensitive to antimicrobial peptides ABC importer
- SIC
streptococcal inhibitor of complement
- SK
staphylokinase
- SpeB
streptococcal pyrogenic exotoxin B
- TA
teichoic acid
- TLR
toll-like receptor
- tPMP-1
thrombin-induced platelet microbicidal protein
- TP-1
tachyplesin-1
- VRE
vancomycin-resistant Enterococcus
- WT
wild-type
- WTAs
wall teichoic acids
REFERENCES
- 1.Steckbeck JD, Deslouches B, Montelaro RC. Antimicrobial peptides: new drugs for bad bugs? Expert Opin Biol Ther. 2014;14:11–14. doi: 10.1517/14712598.2013.844227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Francesco A, Favaroni A, Donati M. Host defense peptides: general overview and an update on their activity against Chlamydia spp. Expert Rev Anti Infect Ther. 2013;11:1215–1224. doi: 10.1586/14787210.2013.841450. [DOI] [PubMed] [Google Scholar]
- 3.Anaya-Lopez JL, Lopez-Meza JE, Ochoa-Zarzosa A. Bacterial resistance to cationic antimicrobial peptides. Crit Rev Microbiol. 2013;39:180–195. doi: 10.3109/1040841X.2012.699025. [DOI] [PubMed] [Google Scholar]
- 4.Jenssen H, Hamill P, Hancock RE. Peptide antimicrobial agents. Clin Microbiol Rev. 2006;19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakatsuji T, Gallo RL. Antimicrobial peptides: old molecules with new ideas. J Invest Dermatol. 2012;132:887–895. doi: 10.1038/jid.2011.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinheiro da Silva F, Machado MC. Antimicrobial peptides: clinical relevance and therapeutic implications. Peptides. 2012;36:308–314. doi: 10.1016/j.peptides.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 7.Morrison G, Kilanowski F, Davidson D, Dorin J. Characterization of the mouse beta defensin 1, Defb1, mutant mouse model. Infect Immun. 2002;70:3053–3060. doi: 10.1128/IAI.70.6.3053-3060.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guralp SA, Murgha YE, Rouillard JM, Gulari E. From design to screening: a new antimicrobial peptide discovery pipeline. PLoS One. 2013;8:e59305. doi: 10.1371/journal.pone.0059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen LT, Haney EF, Vogel HJ. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011;29:464–472. doi: 10.1016/j.tibtech.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Lehrer RI, Ganz T. Cathelicidins: a family of endogenous antimicrobial peptides. Curr Opin Hematol. 2002;9:18–22. doi: 10.1097/00062752-200201000-00004. [DOI] [PubMed] [Google Scholar]
- 11.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 12.Ayabe T, Satchell DP, Wilson CL, Parks WC, Selsted ME, Ouellette AJ. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat Immunol. 2000;1:113–118. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- 13.Yount NY, Yeaman MR. Peptide antimicrobials: cell wall as a bacterial target. Ann N Y Acad Sci. 2013;1277:127–138. doi: 10.1111/nyas.12005. [DOI] [PubMed] [Google Scholar]
- 14.Ganz T, Lehrer RI. Antimicrobial peptides of leukocytes. Curr Opin Hematol. 1997;4:53–58. doi: 10.1097/00062752-199704010-00009. [DOI] [PubMed] [Google Scholar]
- 15.Jones DE, Bevins CL. Paneth cells of the human small intestine express an antimicrobial peptide gene. J Biol Chem. 1992;267:23216–23225. [PubMed] [Google Scholar]
- 16.Quayle AJ, Porter EM, Nussbaum AA, Wang YM, Brabec C, Yip KP, Mok SC. Gene expression, immunolocalization, and secretion of human defensin-5 in human female reproductive tract. Am J Pathol. 1998;152:1247–1258. [PMC free article] [PubMed] [Google Scholar]
- 17.Duits LA, Ravensbergen B, Rademaker M, Hiemstra PS, Nibbering PH. Expression of beta-defensin 1 and 2 mRNA by human monocytes, macrophages and dendritic cells. Immunology. 2002;106:517–525. doi: 10.1046/j.1365-2567.2002.01430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosciuczuk EM, Lisowski P, Jarczak J, Strzalkowska N, Jozwik A, Horbanczuk J, Krzyzewski J, Zwierzchowski L, Bagnicka E. Cathelicidins: family of antimicrobial peptides. A review. Mol Biol Rep. 2012;39:10957–10970. doi: 10.1007/s11033-012-1997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeaman MR. Platelets in defense against bacterial pathogens. Cell Mol Life Sci. 2010;67:525–544. doi: 10.1007/s00018-009-0210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koprivnjak T, Peschel A. Bacterial resistance mechanisms against host defense peptides. Cell Mol Life Sci. 2011;68:2243–2254. doi: 10.1007/s00018-011-0716-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwakman PH, Krijgsveld J, de Boer L, Nguyen LT, Boszhard L, Vreede J, Dekker HL, Speijer D, Drijfhout JW, te Velde AA, Crielaard W, Vogel HJ, Vandenbroucke-Grauls CM, Zaat SA. Native thrombocidin-1 and unfolded thrombocidin-1 exert antimicrobial activity via distinct structural elements. J Biol Chem. 2011;286:43506–43514. doi: 10.1074/jbc.M111.248641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 23.Senyurek I, Paulmann M, Sinnberg T, Kalbacher H, Deeg M, Gutsmann T, Hermes M, Kohler T, Gotz F, Wolz C, Peschel A, Schittek B. Dermcidin-derived peptides show a different mode of action than the cathelicidin LL-37 against Staphylococcus aureus. Antimicrob Agents Chemother. 2009;53:2499–2509. doi: 10.1128/AAC.01679-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gennaro R, Zanetti M. Structural features and biological activities of the cathelicidin-derived antimicrobial peptides. Biopolymers. 2000;55:31–49. doi: 10.1002/1097-0282(2000)55:1<31::AID-BIP40>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 25.Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 26.Ehrenstein G, Lecar H. Electrically gated ionic channels in lipid bilayers. Q Rev Biophys. 1977;10:1–34. doi: 10.1017/s0033583500000123. [DOI] [PubMed] [Google Scholar]
- 27.Matsuzaki K, Murase O, Fujii N, Miyajima K. An antimicrobial peptide, magainin 2, induced rapid flip-flop of phospholipids coupled with pore formation and peptide translocation. Biochemistry. 1996;35:11361–11368. doi: 10.1021/bi960016v. [DOI] [PubMed] [Google Scholar]
- 28.Pietiainen M, Francois P, Hyyrylainen HL, Tangomo M, Sass V, Sahl HG, Schrenzel J, Kontinen VP. Transcriptome analysis of the responses of Staphylococcus aureus to antimicrobial peptides and characterization of the roles of vraDE and vraSR in antimicrobial resistance. BMC Genomics. 2009;10:429. doi: 10.1186/1471-2164-10-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Straus SK, Hancock RE. Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: comparison with cationic antimicrobial peptides and lipopeptides. Biochim Biophys Acta. 2006;1758:1215–1223. doi: 10.1016/j.bbamem.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 31.Muller A, Ulm H, Reder-Christ K, Sahl HG, Schneider T. Interaction of type A lantibiotics with undecaprenol-bound cell envelope precursors. Microb Drug Resist. 2012;18:261–270. doi: 10.1089/mdr.2011.0242. [DOI] [PubMed] [Google Scholar]
- 32.Islam MR, Nagao J, Zendo T, Sonomoto K. Antimicrobial mechanism of lantibiotics. Biochem Soc Trans. 2012;40:1528–1533. doi: 10.1042/BST20120190. [DOI] [PubMed] [Google Scholar]
- 33.Cho JH, Sung BH, Kim SC. Buforins: histone H2A-derived antimicrobial peptides from toad stomach. Biochim Biophys Acta. 2009;1788:1564–1569. doi: 10.1016/j.bbamem.2008.10.025. [DOI] [PubMed] [Google Scholar]
- 34.Subbalakshmi C, Sitaram N. Mechanism of antimicrobial action of indolicidin. FEMS Microbiol Lett. 1998;160:91–96. doi: 10.1111/j.1574-6968.1998.tb12896.x. [DOI] [PubMed] [Google Scholar]
- 35.Haney EF, Petersen AP, Lau CK, Jing W, Storey DG, Vogel HJ. Mechanism of action of puroindoline derived tryptophan-rich antimicrobial peptides. Biochim Biophys Acta. 2013;1828:1802–1813. doi: 10.1016/j.bbamem.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 36.Lehrer RI, Barton A, Daher KA, Harwig SS, Ganz T, Selsted ME. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J Clin Invest. 1989;84:553–561. doi: 10.1172/JCI114198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Nardo A, Vitiello A, Gallo RL. Cutting edge: mast cell antimicrobial activity is mediated by expression of cathelicidin antimicrobial peptide. J Immunol. 2003;170:2274–2278. doi: 10.4049/jimmunol.170.5.2274. [DOI] [PubMed] [Google Scholar]
- 38.Nizet V, Ohtake T, Lauth X, Trowbridge J, Rudisill J, Dorschner RA, Pestonjamasp V, Piraino J, Huttner K, Gallo RL. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414:454–457. doi: 10.1038/35106587. [DOI] [PubMed] [Google Scholar]
- 39.Rosenberger CM, Gallo RL, Finlay BB. Interplay between antibacterial effectors: A macrophage antimicrobial peptide impairs intracellular Salmonella replication. Proc Natl Acad Sci USA. 2004;101:2422–2427. doi: 10.1073/pnas.0304455101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chromek M, Slamova Z, Bergman P, Kovacs L, Podracka L, Ehren I, Hokfelt T, Gudmundsson GH, Gallo RL, Agerberth B, Brauner A. The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection. Nat Med. 2006;12:636–641. doi: 10.1038/nm1407. [DOI] [PubMed] [Google Scholar]
- 41.Bergman P, Johansson L, Wan H, Jones A, Gallo RL, Gudmundsson GH, Hokfelt T, Jonsson AB, Agerberth B. Induction of the antimicrobial peptide CRAMP in the blood-brain barrier and meninges after meningococcal infection. Infect Immun. 2006;74:6982–6991. doi: 10.1128/IAI.01043-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar A, Gao N, Standiford TJ, Gallo RL, Yu FS. Topical flagellin protects the injured corneas from Pseudomonas aeruginosa infection. Microbes Infect. 2010;12:978–989. doi: 10.1016/j.micinf.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kovach MA, Ballinger MN, Newstead MW, Zeng X, Bhan U, Yu FS, Moore BB, Gallo RL, Standiford TJ. Cathelicidin-related antimicrobial peptide is required for effective lung mucosal immunity in Gram-negative bacterial pneumonia. J Immunol. 2012;189:304–311. doi: 10.4049/jimmunol.1103196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Augustin DK, Heimer SR, Tam C, Li WY, Le Due JM, Evans DJ, Fleiszig SM. Role of defensins in corneal epithelial barrier function against Pseudomonas aeruginosa traversal. Infect Immun. 2011;79:595–605. doi: 10.1128/IAI.00854-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kolar SS, Baidouri H, Hanlon S, McDermott AM. Protective role of murine beta-defensins 3 and 4 and cathelin-related antimicrobial peptide in Fusarium solani keratitis. Infect Immun. 2013;81:2669–2677. doi: 10.1128/IAI.00179-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee PH, Ohtake T, Zaiou M, Murakami M, Rudisill JA, Lin KH, Gallo RL. Expression of an additional cathelicidin antimicrobial peptide protects against bacterial skin infection. Proc Natl Acad Sci USA. 2005;102:3750–3755. doi: 10.1073/pnas.0500268102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522–526. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- 48.Niyonsaba F, Ushio H, Nakano N, Ng W, Sayama K, Hashimoto K, Nagaoka I, Okumura K, Ogawa H. Antimicrobial peptides human beta-defensins stimulate epidermal keratinocyte migration, proliferation and production of proinflammatory cytokines and chemokines. J Invest Dermatol. 2007;127:594–604. doi: 10.1038/sj.jid.5700599. [DOI] [PubMed] [Google Scholar]
- 49.Zanetti M. Cathelicidins, multifunctional peptides of the innate immunity. J Leukoc Biol. 2004;75:39–48. doi: 10.1189/jlb.0403147. [DOI] [PubMed] [Google Scholar]
- 50.Koczulla R, von Degenfeld G, Kupatt C, Krotz F, Zahler S, Gloe T, Issbrucker K, Unterberger P, Zaiou M, Lebherz C, Karl A, Raake P, Pfosser A, Boekstegers P, Welsch U, Hiemstra PS, Vogelmeier C, Gallo RL, Clauss M, Bals R. An angiogenic role for the human peptide antibiotic LL-37/hCAP-18. J Clin Invest. 2003;111:1665–1672. doi: 10.1172/JCI17545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elssner A, Duncan M, Gavrilin M, Wewers MD. A novel P2X7 receptor activator, the human cathelicidin-derived peptide LL37, induces IL-1 beta processing and release. J Immunol. 2004;172:4987–4994. doi: 10.4049/jimmunol.172.8.4987. [DOI] [PubMed] [Google Scholar]
- 52.Davidson DJ, Currie AJ, Reid GS, Bowdish DM, MacDonald KL, Ma RC, Hancock RE, Speert DP. The cationic antimicrobial peptide LL-37 modulates dendritic cell differentiation and dendritic cell-induced T cell polarization. J Immunol. 2004;172:1146–1156. doi: 10.4049/jimmunol.172.2.1146. [DOI] [PubMed] [Google Scholar]
- 53.Territo MC, Ganz T, Selsted ME, Lehrer R. Monocyte-chemotactic activity of defensins from human neutrophils. J Clin Invest. 1989;84:2017–2020. doi: 10.1172/JCI114394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kurosaka K, Chen Q, Yarovinsky F, Oppenheim JJ, Yang D. Mouse cathelin-related antimicrobial peptide chemoattracts leukocytes using formyl peptide receptor-like 1/mouse formyl peptide receptor-like 2 as the receptor and acts as an immune adjuvant. J Immunol. 2005;174:6257–6265. doi: 10.4049/jimmunol.174.10.6257. [DOI] [PubMed] [Google Scholar]
- 55.Niyonsaba F, Iwabuchi K, Someya A, Hirata M, Matsuda H, Ogawa H, Nagaoka I. A cathelicidin family of human antibacterial peptide LL-37 induces mast cell chemotaxis. Immunology. 2002;106:20–26. doi: 10.1046/j.1365-2567.2002.01398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niyonsaba F, Someya A, Hirata M, Ogawa H, Nagaoka I. Evaluation of the effects of peptide antibiotics human beta-defensins-1/-2 and LL-37 on histamine release and prostaglandin D(2) production from mast cells. Eur J Immunol. 2001;31:1066–1075. doi: 10.1002/1521-4141(200104)31:4<1066::aid-immu1066>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 57.Lohner K. New strategies for novel antibiotics: peptides targeting bacterial cell membranes. Gen Physiol Biophys. 2009;28:105–116. doi: 10.4149/gpb_2009_02_105. [DOI] [PubMed] [Google Scholar]
- 58.Gutsmann T, Hagge SO, Larrick JW, Seydel U, Wiese A. Interaction of CAP18-derived peptides with membranes made from endotoxins or phospholipids. Biophys J. 2001;80:2935–2945. doi: 10.1016/S0006-3495(01)76259-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oren Z, Lerman JC, Gudmundsson GH, Agerberth B, Shai Y. Structure and organization of the human antimicrobial peptide LL-37 in phospholipid membranes: relevance to the molecular basis for its non-cell-selective activity. Biochem J. 1999;341:501–513. [PMC free article] [PubMed] [Google Scholar]
- 60.Schmidtchen A, Pasupuleti M, Malmsten M. Effect of hydrophobic modifications in antimicrobial peptides. Adv Colloid Interface Sci. 2014;205:265–274. doi: 10.1016/j.cis.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 61.Guilhelmelli F, Vilela N, Albuquerque P, Derengowski Lda S, Silva-Pereira I, Kyaw CM. Antibiotic development challenges: the various mechanisms of action of antimicrobial peptides and of bacterial resistance. Front Microbiol. 2013;4:353. doi: 10.3389/fmicb.2013.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raetz CR, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in Gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo L, Lim KB, Poduje CM, Daniel M, Gunn JS, Hackett M, Miller SI. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- 64.Brown S, Santa Maria JP, Jr., Walker S. Wall teichoic acids of Gram-positive bacteria. Annu Rev Microbiol. 2013;67:313–336. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neuhaus FC, Baddiley J. A continuum of anionic charge: structures and functions of D-alanyl-teichoic acids in Gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67:686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Poyart C, Pellegrini E, Marceau M, Baptista M, Jaubert F, Lamy MC, Trieu-Cuot P. Attenuated virulence of Streptococcus agalactiae deficient in D-alanyl-lipoteichoic acid is due to an increased susceptibility to defensins and phagocytic cells. Mol Microbiol. 2003;49:1615–1625. doi: 10.1046/j.1365-2958.2003.03655.x. [DOI] [PubMed] [Google Scholar]
- 67.Fabretti F, Theilacker C, Baldassarri L, Kaczynski Z, Kropec A, Holst O, Huebner J. Alanine esters of enterococcal lipoteichoic acid play a role in biofilm formation and resistance to antimicrobial peptides. Infect Immun. 2006;74:4164–4171. doi: 10.1128/IAI.00111-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kovacs M, Halfmann A, Fedtke I, Heintz M, Peschel A, Vollmer W, Hakenbeck R, Bruckner R. A functional dlt operon, encoding proteins required for incorporation of D-alanine in teichoic acids in gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J Bacteriol. 2006;188:5797–5805. doi: 10.1128/JB.00336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morath S, Geyer A, Hartung T. Structure-function relationship of cytokine induction by lipoteichoic acid from Staphylococcus aureus. J Exp Med. 2001;193:393–397. doi: 10.1084/jem.193.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grangette C, Nutten S, Palumbo E, Morath S, Hermann C, Dewulf J, Pot B, Hartung T, Hols P, Mercenier A. Enhanced antiinflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc Natl Acad Sci USA. 2005;102:10321–10326. doi: 10.1073/pnas.0504084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kristian SA, Datta V, Weidenmaier C, Kansal R, Fedtke I, Peschel A, Gallo RL, Nizet V. D-alanylation of teichoic acids promotes group A Streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J Bacteriol. 2005;187:6719–6725. doi: 10.1128/JB.187.19.6719-6725.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Gotz F. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem. 1999;274:8405–8410. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 73.Andra J, Goldmann T, Ernst CM, Peschel A, Gutsmann T. Multiple peptide resistance factor (MprF)-mediated Resistance of Staphylococcus aureus against antimicrobial peptides coincides with a modulated peptide interaction with artificial membranes comprising lysyl-phosphatidylglycerol. J Biol Chem. 2011;286:18692–18700. doi: 10.1074/jbc.M111.226886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peschel A. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 2002;10:179–186. doi: 10.1016/s0966-842x(02)02333-8. [DOI] [PubMed] [Google Scholar]
- 75.Kristian SA, Lauth X, Nizet V, Goetz F, Neumeister B, Peschel A, Landmann R. Alanylation of teichoic acids protects Staphylococcus aureus against Toll-like receptor 2-dependent host defense in a mouse tissue cage infection model. J Infect Dis. 2003;188:414–423. doi: 10.1086/376533. [DOI] [PubMed] [Google Scholar]
- 76.Heptinstall S, Archibald AR, Baddiley J. Teichoic acids and membrane function in bacteria. Nature. 1970;225:519–521. doi: 10.1038/225519a0. [DOI] [PubMed] [Google Scholar]
- 77.MacArthur AE, Archibald AR. Effect of culture pH on the pere-alanine ester content of lipoteichoic acid in Staphylococcus aureus. J Bacteriol. 1984;160:792–793. doi: 10.1128/jb.160.2.792-793.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Perego M, Glaser P, Minutello A, Strauch MA, Leopold K, Fischer W. Incorporation of D-alanine into lipoteichoic acid and wall teichoic acid in Bacillus subtilis. Identification of genes and regulation. J Biol Chem. 1995;270:15598–15606. doi: 10.1074/jbc.270.26.15598. [DOI] [PubMed] [Google Scholar]
- 79.Poyart C, Lamy MC, Boumaila C, Fiedler F, Trieu-Cuot P. Regulation of D-alanyl-lipoteichoic acid biosynthesis in Streptococcus agalactiae involves a novel two-component regulatory system. J Bacteriol. 2001;183:6324–6334. doi: 10.1128/JB.183.21.6324-6334.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saar-Dover R, Bitler A, Nezer R, Shmuel-Galia L, Firon A, Shimoni E, Trieu-Cuot P, Shai Y. D-alanylation of lipoteichoic acids confers resistance to cationic peptides in group B Streptococcus by increasing the cell wall density. PLoS Pathog. 2012;8:e1002891. doi: 10.1371/journal.ppat.1002891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kristian SA, Durr M, Van Strijp JA, Neumeister B, Peschel A. MprF-mediated lysinylation of phospholipids in Staphylococcus aureus leads to protection against oxygen-independent neutrophil killing. Infect Immun. 2003;71:546–549. doi: 10.1128/IAI.71.1.546-549.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peschel A, Jack RW, Otto M, Collins LV, Staubitz P, Nicholson G, Kalbacher H, Nieuwenhuizen WF, Jung G, Tarkowski A, van Kessel KP, van Strijp JA. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with L-lysine. J Exp Med. 2001;193:1067–1076. doi: 10.1084/jem.193.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Abachin E, Poyart C, Pellegrini E, Milohanic E, Fiedler F, Berche P, Trieu-Cuot P. Formation of D-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol Microbiol. 2002;43:1–14. doi: 10.1046/j.1365-2958.2002.02723.x. [DOI] [PubMed] [Google Scholar]
- 84.Walter J, Loach DM, Alqumber M, Rockel C, Hermann C, Pfitzenmaier M, Tannock GW. D-alanyl ester depletion of teichoic acids in Lactobacillus reuteri 100-23 results in impaired colonization of the mouse gastrointestinal tract. Environ Microbiol. 2007;9:1750–1760. doi: 10.1111/j.1462-2920.2007.01292.x. [DOI] [PubMed] [Google Scholar]
- 85.Ernst CM, Staubitz P, Mishra NN, Yang SJ, Hornig G, Kalbacher H, Bayer AS, Kraus D, Peschel A. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 2009;5:e1000660. doi: 10.1371/journal.ppat.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Staubitz P, Neumann H, Schneider T, Wiedemann I, Peschel A. MprF-mediated biosynthesis of lysylphosphatidylglycerol, an important determinant in staphylococcal defensin resistance. FEMS Microbiol Lett. 2004;231:67–71. doi: 10.1016/S0378-1097(03)00921-2. [DOI] [PubMed] [Google Scholar]
- 87.Nishi H, Komatsuzawa H, Fujiwara T, McCallum N, Sugai M. Reduced content of lysyl-phosphatidylglycerol in the cytoplasmic membrane affects susceptibility to moenomycin, as well as vancomycin, gentamicin, and antimicrobial peptides, in Staphylococcus aureus. Antimicrob Agents Chemother. 2004;48:4800–4807. doi: 10.1128/AAC.48.12.4800-4807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Izadpanah A, Gallo RL. Antimicrobial peptides. J Am Acad Dermatol. 2005;52:381–390. doi: 10.1016/j.jaad.2004.08.026. quiz 391-382. [DOI] [PubMed] [Google Scholar]
- 89.Nawrocki KL, Crispell EK, McBride SM. Antimicrobial peptide resistance mechanisms of Gram-positive bacteria. Antibiotics. 2014;3:461–492. doi: 10.3390/antibiotics3040461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Roy H, Ibba M. RNA-dependent lipid remodeling by bacterial multiple peptide resistance factors. Proc Natl Acad Sci USA. 2008;105:4667–4672. doi: 10.1073/pnas.0800006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maloney E, Stankowska D, Zhang J, Fol M, Cheng QJ, Lun S, Bishai WR, Rajagopalan M, Chatterjee D, Madiraju MV. The two-domain LysX protein of Mycobacterium tuberculosis is required for production of lysinylated phosphatidylglycerol and resistance to cationic antimicrobial peptides. PLoS Pathog. 2009;5:e1000534. doi: 10.1371/journal.ppat.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maloney E, Lun S, Stankowska D, Guo H, Rajagoapalan M, Bishai WR, Madiraju MV. Alterations in phospholipid catabolism in Mycobacterium tuberculosis lysX mutant. Front Microbiol. 2011;2:19. doi: 10.3389/fmicb.2011.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thedieck K, Hain T, Mohamed W, Tindall BJ, Nimtz M, Chakraborty T, Wehland J, Jansch L. The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol Microbiol. 2006;62:1325–1339. doi: 10.1111/j.1365-2958.2006.05452.x. [DOI] [PubMed] [Google Scholar]
- 94.Klein S, Lorenzo C, Hoffmann S, Walther JM, Storbeck S, Piekarski T, Tindall BJ, Wray V, Nimtz M, Moser J. Adaptation of Pseudomonas aeruginosa to various conditions includes tRNA-dependent formation of alanyl-phosphatidylglycerol. Mol Microbiol. 2009;71:551–565. doi: 10.1111/j.1365-2958.2008.06562.x. [DOI] [PubMed] [Google Scholar]
- 95.Samant S, Hsu FF, Neyfakh AA, Lee H. The Bacillus anthracis protein MprF is required for synthesis of lysylphosphatidylglycerols and for resistance to cationic antimicrobial peptides. J Bacteriol. 2009;191:1311–1319. doi: 10.1128/JB.01345-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hamilton A, Popham DL, Carl DJ, Lauth X, Nizet V, Jones AL. Penicillin-binding protein 1a promotes resistance of group B Streptococcus to antimicrobial peptides. Infect Immun. 2006;74:6179–6187. doi: 10.1128/IAI.00895-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.West NP, Jungnitz H, Fitter JT, McArthur JD, Guzman CA, Walker MJ. Role of phosphoglucomutase of Bordetella bronchiseptica in lipopolysaccharide biosynthesis and virulence. Infect Immun. 2000;68:4673–4680. doi: 10.1128/iai.68.8.4673-4680.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Buchanan JT, Stannard JA, Lauth X, Ostland VE, Powell HC, Westerman ME, Nizet V. Streptococcus iniae phosphoglucomutase is a virulence factor and a target for vaccine development. Infect Immun. 2005;73:6935–6944. doi: 10.1128/IAI.73.10.6935-6944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gao LY, Laval F, Lawson EH, Groger RK, Woodruff A, Morisaki JH, Cox JS, Daffe M, Brown EJ. Requirement for kasB in Mycobacterium mycolic acid biosynthesis, cell wall impermeability and intracellular survival: implications for therapy. Mol Microbiol. 2003;49:1547–1563. doi: 10.1046/j.1365-2958.2003.03667.x. [DOI] [PubMed] [Google Scholar]
- 100.Gunn JS, Ryan SS, Van Velkinburgh JC, Ernst RK, Miller SI. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar Typhimurium. Infect Immun. 2000;68:6139–6146. doi: 10.1128/iai.68.11.6139-6146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tamayo R, Choudhury B, Septer A, Merighi M, Carlson R, Gunn JS. Identification of cptA, a PmrA-regulated locus required for phosphoethanolamine modification of the Salmonella enterica serovar Typhimurium lipopolysaccharide core. J Bacteriol. 2005;187:3391–3399. doi: 10.1128/JB.187.10.3391-3399.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gunn JS. Bacterial modification of LPS and resistance to antimicrobial peptides. J Endotoxin Res. 2001;7:57–62. [PubMed] [Google Scholar]
- 103.McCoy AJ, Liu H, Falla TJ, Gunn JS. Identification of Proteus mirabilis mutants with increased sensitivity to antimicrobial peptides. Antimicrob Agents Chemother. 2001;45:2030–2037. doi: 10.1128/AAC.45.7.2030-2037.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Marceau M, Sebbane F, Collyn F, Simonet M. Function and regulation of the Salmonella-like pmrF antimicrobial peptide resistance operon in Yersinia pseudotuberculosis. Adv Exp Med Biol. 2003;529:253–256. doi: 10.1007/0-306-48416-1_49. [DOI] [PubMed] [Google Scholar]
- 105.Cheng HY, Chen YF, Peng HL. Molecular characterization of the PhoPQ-PmrD-PmrAB mediated pathway regulating polymyxin B resistance in Klebsiella pneumoniae CG43. J Biomed Sci. 2010;17:60. doi: 10.1186/1423-0127-17-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Moskowitz SM, Ernst RK, Miller SI. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J Bacteriol. 2004;186:575–579. doi: 10.1128/JB.186.2.575-579.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ernst RK, Yi EC, Guo L, Lim KB, Burns JL, Hackett M, Miller SI. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science. 1999;286:1561–1565. doi: 10.1126/science.286.5444.1561. [DOI] [PubMed] [Google Scholar]
- 108.Tran AX, Whittimore JD, Wyrick PB, McGrath SC, Cotter RJ, Trent MS. The lipid A 1-phosphatase of Helicobacter pylori is required for resistance to the antimicrobial peptide polymyxin. J Bacteriol. 2006;188:4531–4541. doi: 10.1128/JB.00146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cullen TW, Giles DK, Wolf LN, Ecobichon C, Boneca IG, Trent MS. Helicobacter pylori versus the host: remodeling of the bacterial outer membrane is required for survival in the gastric mucosa. PLoS Pathog. 2011;7:e1002454. doi: 10.1371/journal.ppat.1002454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lewis LA, Choudhury B, Balthazar JT, Martin LE, Ram S, Rice PA, Stephens DS, Carlson R, Shafer WM. Phosphoethanolamine substitution of lipid A and resistance of Neisseria gonorrhoeae to cationic antimicrobial peptides and complement-mediated killing by normal human serum. Infect Immun. 2009;77:1112–1120. doi: 10.1128/IAI.01280-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tzeng YL, Ambrose KD, Zughaier S, Zhou X, Miller YK, Shafer WM, Stephens DS. Cationic antimicrobial peptide resistance in Neisseria meningitidis. J Bacteriol. 2005;187:5387–5396. doi: 10.1128/JB.187.15.5387-5396.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Albiger B, Johansson L, Jonsson AB. Lipooligosaccharide-deficient Neisseria meningitidis shows altered pilus-associated characteristics. Infect Immun. 2003;71:155–162. doi: 10.1128/IAI.71.1.155-162.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jones A, Georg M, Maudsdotter L, Jonsson AB. Endotoxin, capsule, and bacterial attachment contribute to Neisseria meningitidis resistance to the human antimicrobial peptide LL-37. J Bacteriol. 2009;191:3861–3868. doi: 10.1128/JB.01313-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Keo T, Collins J, Kunwar P, Blaser MJ, Iovine NM. Campylobacter capsule and lipooligosaccharide confer resistance to serum and cationic antimicrobials. Virulence. 2011;2:30–40. doi: 10.4161/viru.2.1.14752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Naito M, Frirdich E, Fields JA, Pryjma M, Li J, Cameron A, Gilbert M, Thompson SA, Gaynor EC. Effects of sequential Campylobacter jejuni 81-176 lipooligosaccharide core truncations on biofilm formation, stress survival, and pathogenesis. J Bacteriol. 2010;192:2182–2192. doi: 10.1128/JB.01222-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bishop RE, Gibbons HS, Guina T, Trent MS, Miller SI, Raetz CR. Transfer of palmitate from phospholipids to lipid A in outer membranes of Gram-negative bacteria. EMBO J. 2000;19:5071–5080. doi: 10.1093/emboj/cdd507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Robey M, O'Connell W, Cianciotto NP. Identification of Legionella pneumophila rcp, a pagP-like gene that confers resistance to cationic antimicrobial peptides and promotes intracellular infection. Infect Immun. 2001;69:4276–4286. doi: 10.1128/IAI.69.7.4276-4286.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Starner TD, Swords WE, Apicella MA, McCray PB., Jr. Susceptibility of nontypeable Haemophilus influenzae to human beta-defensins is influenced by lipooligosaccharide acylation. Infect Immun. 2002;70:5287–5289. doi: 10.1128/IAI.70.9.5287-5289.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lysenko ES, Gould J, Bals R, Wilson JM, Weiser JN. Bacterial phosphorylcholine decreases susceptibility to the antimicrobial peptide LL-37/hCAP18 expressed in the upper respiratory tract. Infect Immun. 2000;68:1664–1671. doi: 10.1128/iai.68.3.1664-1671.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Clements A, Tull D, Jenney AW, Farn JL, Kim SH, Bishop RE, McPhee JB, Hancock RE, Hartland EL, Pearse MJ, Wijburg OL, Jackson DC, McConville MJ, Strugnell RA. Secondary acylation of Klebsiella pneumoniae lipopolysaccharide contributes to sensitivity to antibacterial peptides. J Biol Chem. 2007;282:15569–15577. doi: 10.1074/jbc.M701454200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Matson JS, Yoo HJ, Hakansson K, Dirita VJ. Polymyxin B resistance in El Tor Vibrio cholerae requires lipid acylation catalyzed by MsbB. J Bacteriol. 2010;192:2044–2052. doi: 10.1128/JB.00023-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Braff MH, Jones AL, Skerrett SJ, Rubens CE. Staphylococcus aureus exploits cathelicidin antimicrobial peptides produced during early pneumonia to promote staphylokinase-dependent fibrinolysis. J Infect Dis. 2007;195:1365–1372. doi: 10.1086/513277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jin T, Bokarewa M, Foster T, Mitchell J, Higgins J, Tarkowski A. Staphylococcus aureus resists human defensins by production of staphylokinase, a novel bacterial evasion mechanism. J Immunol. 2004;172:1169–1176. doi: 10.4049/jimmunol.172.2.1169. [DOI] [PubMed] [Google Scholar]
- 124.Frick IM, Akesson P, Rasmussen M, Schmidtchen A, Bjorck L. SIC, a secreted protein of Streptococcus pyogenes that inactivates antibacterial peptides. J Biol Chem. 2003;278:16561–16566. doi: 10.1074/jbc.M301995200. [DOI] [PubMed] [Google Scholar]
- 125.Pence MA, Rooijakkers SH, Cogen AL, Cole JN, Hollands A, Gallo RL, Nizet V. Streptococcal inhibitor of complement promotes innate immune resistance phenotypes of invasive M1T1 group A Streptococcus. J Innate Immun. 2010;2:587–595. doi: 10.1159/000317672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fernie-King BA, Seilly DJ, Davies A, Lachmann PJ. Streptococcal inhibitor of complement inhibits two additional components of the mucosal innate immune system: secretory leukocyte proteinase inhibitor and lysozyme. Infect Immun. 2002;70:4908–4916. doi: 10.1128/IAI.70.9.4908-4916.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Walker MJ, Barnett TC, McArthur JD, Cole JN, Gillen CM, Henningham A, Sriprakash KS, Sanderson-Smith ML, Nizet V. Disease manifestations and pathogenic mechanisms of group A Streptococcus. Clin Microbiol Rev. 2014;27:264–301. doi: 10.1128/CMR.00101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cole JN, Barnett TC, Nizet V, Walker MJ. Molecular insight into invasive group A streptococcal disease. Nat Rev Microbiol. 2011;9:724–736. doi: 10.1038/nrmicro2648. [DOI] [PubMed] [Google Scholar]
- 129.Steer AC, Law I, Matatolu L, Beall BW, Carapetis JR. Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect Dis. 2009;9:611–616. doi: 10.1016/S1473-3099(09)70178-1. [DOI] [PubMed] [Google Scholar]
- 130.Lauth X, von Kockritz-Blickwede M, McNamara CW, Myskowski S, Zinkernagel AS, Beall B, Ghosh P, Gallo RL, Nizet V. M1 protein allows group A streptococcal survival in phagocyte extracellular traps through cathelicidin inhibition. J Innate Immun. 2009;1:202–214. doi: 10.1159/000203645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jones AL, Mertz RH, Carl DJ, Rubens CE. A streptococcal penicillin-binding protein is critical for resisting innate airway defenses in the neonatal lung. J Immunol. 2007;179:3196–3202. doi: 10.4049/jimmunol.179.5.3196. [DOI] [PubMed] [Google Scholar]
- 132.Maisey HC, Quach D, Hensler ME, Liu GY, Gallo RL, Nizet V, Doran KS. A group B streptococcal pilus protein promotes phagocyte resistance and systemic virulence. FASEB J. 2008;22:1715–1724. doi: 10.1096/fj.07-093963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lancefield RC. The antigenic complex of Streptococcus haemolyticus: I. Demonstration of a type-specific substance in extracts of Streptococcus haemolyticus. J Exp Med. 1928;47:91–103. doi: 10.1084/jem.47.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.McCarty M. The lysis of group A hemolytic streptococci by extracellular enzymes of Streptomyces albus. II. Nature of the cellular substrate attacked by the lytic enzymes. J Exp Med. 1952;96:569–580. doi: 10.1084/jem.96.6.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.van Sorge NM, Cole JN, Kuipers K, Henningham A, Aziz RK, Kasirer-Friede A, Lin L, Berends ET, Davies MR, Dougan G, Zhang F, Dahesh S, Shaw L, Gin J, Cunningham M, Merriman JA, Hutter J, Lepenies B, Rooijakkers SH, Malley R, Walker MJ, Shattil SJ, Schlievert PM, Choudhury B, Nizet V. The classical lancefield antigen of group A Streptococcus is a virulence determinant with implications for vaccine design. Cell Host Microbe. 2014;15:729–740. doi: 10.1016/j.chom.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schmidtchen A, Frick IM, Bjorck L. Dermatan sulphate is released by proteinases of common pathogenic bacteria and inactivates antibacterial alpha-defensin. Mol Microbiol. 2001;39:708–713. doi: 10.1046/j.1365-2958.2001.02251.x. [DOI] [PubMed] [Google Scholar]
- 137.Park PW, Pier GB, Preston MJ, Goldberger O, Fitzgerald ML, Bernfield M. Syndecan-1 shedding is enhanced by LasA, a secreted virulence factor of Pseudomonas aeruginosa. J Biol Chem. 2000;275:3057–3064. doi: 10.1074/jbc.275.5.3057. [DOI] [PubMed] [Google Scholar]
- 138.Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Gotz F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20:1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 139.Vuong C, Kocianova S, Voyich JM, Yao Y, Fischer ER, DeLeo FR, Otto M. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem. 2004;279:54881–54886. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- 140.Vuong C, Voyich JM, Fischer ER, Braughton KR, Whitney AR, DeLeo FR, Otto M. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol. 2004;6:269–275. doi: 10.1046/j.1462-5822.2004.00367.x. [DOI] [PubMed] [Google Scholar]
- 141.Kocianova S, Vuong C, Yao Y, Voyich JM, Fischer ER, DeLeo FR, Otto M. Key role of poly-gamma-DL-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis. J Clin Invest. 2005;115:688–694. doi: 10.1172/JCI23523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cole JN, Pence MA, von Kockritz-Blickwede M, Hollands A, Gallo RL, Walker MJ, Nizet V. M protein and hyaluronic acid capsule are essential for in vivo selection of covRS mutations characteristic of invasive serotype M1T1 group A Streptococcus. mBio. 2010;1 doi: 10.1128/mBio.00191-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Campos MA, Vargas MA, Regueiro V, Llompart CM, Alberti S, Bengoechea JA. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect Immun. 2004;72:7107–7114. doi: 10.1128/IAI.72.12.7107-7114.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Moranta D, Regueiro V, March C, Llobet E, Margareto J, Larrarte E, Garmendia J, Bengoechea JA. Klebsiella pneumoniae capsule polysaccharide impedes the expression of beta-defensins by airway epithelial cells. Infect Immun. 2010;78:1135–1146. doi: 10.1128/IAI.00940-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Llobet E, Tomas JM, Bengoechea JA. Capsule polysaccharide is a bacterial decoy for antimicrobial peptides. Microbiology. 2008;154:3877–3886. doi: 10.1099/mic.0.2008/022301-0. [DOI] [PubMed] [Google Scholar]
- 146.Spinosa MR, Progida C, Tala A, Cogli L, Alifano P, Bucci C. The Neisseria meningitidis capsule is important for intracellular survival in human cells. Infect Immun. 2007;75:3594–3603. doi: 10.1128/IAI.01945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chan C, Burrows LL, Deber CM. Helix induction in antimicrobial peptides by alginate in biofilms. J Biol Chem. 2004;279:38749–38754. doi: 10.1074/jbc.M406044200. [DOI] [PubMed] [Google Scholar]
- 148.Piddock LJ. Multidrug-resistance efflux pumps - not just for resistance. Nat Rev Microbiol. 2006;4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 149.Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev. 2008;72:317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Stein T, Heinzmann S, Solovieva I, Entian KD. Function of Lactococcus lactis nisin immunity genes nisI and nisFEG after coordinated expression in the surrogate host Bacillus subtilis. J Biol Chem. 2003;278:89–94. doi: 10.1074/jbc.M207237200. [DOI] [PubMed] [Google Scholar]
- 151.Stein T, Heinzmann S, Dusterhus S, Borchert S, Entian KD. Expression and functional analysis of the subtilin immunity genes spaIFEG in the subtilin-sensitive host Bacillus subtilis MO1099. J Bacteriol. 2005;187:822–828. doi: 10.1128/JB.187.3.822-828.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Suarez JM, Edwards AN, McBride SM. The Clostridium difficile cpr locus is regulated by a noncontiguous two-component system in response to type A and B lantibiotics. J Bacteriol. 2013;195:2621–2631. doi: 10.1128/JB.00166-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.McBride SM, Sonenshein AL. Identification of a genetic locus responsible for antimicrobial peptide resistance in Clostridium difficile. Infect Immun. 2011;79:167–176. doi: 10.1128/IAI.00731-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mascher T, Margulis NG, Wang T, Ye RW, Helmann JD. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol Microbiol. 2003;50:1591–1604. doi: 10.1046/j.1365-2958.2003.03786.x. [DOI] [PubMed] [Google Scholar]
- 155.Meehl M, Herbert S, Gotz F, Cheung A. Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2007;51:2679–2689. doi: 10.1128/AAC.00209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kramer NE, van Hijum SA, Knol J, Kok J, Kuipers OP. Transcriptome analysis reveals mechanisms by which Lactococcus lactis acquires nisin resistance. Antimicrob Agents Chemother. 2006;50:1753–1761. doi: 10.1128/AAC.50.5.1753-1761.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Majchrzykiewicz JA, Kuipers OP, Bijlsma JJ. Generic and specific adaptive responses of Streptococcus pneumoniae to challenge with three distinct antimicrobial peptides, bacitracin, LL-37, and nisin. Antimicrob Agents Chemother. 2010;54:440–451. doi: 10.1128/AAC.00769-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mandin P, Fsihi H, Dussurget O, Vergassola M, Milohanic E, Toledo-Arana A, Lasa I, Johansson J, Cossart P. VirR, a response regulator critical for Listeria monocytogenes virulence. Mol Microbiol. 2005;57:1367–1380. doi: 10.1111/j.1365-2958.2005.04776.x. [DOI] [PubMed] [Google Scholar]
- 159.Podlesek Z, Comino A, Herzog-Velikonja B, Zgur-Bertok D, Komel R, Grabnar M. Bacillus licheniformis bacitracin-resistance ABC transporter: relationship to mammalian multidrug resistance. Mol Microbiol. 1995;16:969–976. doi: 10.1111/j.1365-2958.1995.tb02322.x. [DOI] [PubMed] [Google Scholar]
- 160.Manson JM, Keis S, Smith JM, Cook GM. Acquired bacitracin resistance in Enterococcus faecalis is mediated by an ABC transporter and a novel regulatory protein, BcrR. Antimicrob Agents Chemother. 2004;48:3743–3748. doi: 10.1128/AAC.48.10.3743-3748.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Charlebois A, Jalbert LA, Harel J, Masson L, Archambault M. Characterization of genes encoding for acquired bacitracin resistance in Clostridium perfringens. PLoS One. 2012;7:e44449. doi: 10.1371/journal.pone.0044449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tsuda H, Yamashita Y, Shibata Y, Nakano Y, Koga T. Genes involved in bacitracin resistance in Streptococcus mutans. Antimicrob Agents Chemother. 2002;46:3756–3764. doi: 10.1128/AAC.46.12.3756-3764.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bengoechea JA, Skurnik M. Temperature-regulated efflux pump/potassium antiporter system mediates resistance to cationic antimicrobial peptides in Yersinia. Mol Microbiol. 2000;37:67–80. doi: 10.1046/j.1365-2958.2000.01956.x. [DOI] [PubMed] [Google Scholar]
- 164.Padilla E, Llobet E, Domenech-Sanchez A, Martinez-Martinez L, Bengoechea JA, Alberti S. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob Agents Chemother. 2010;54:177–183. doi: 10.1128/AAC.00715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zahner D, Zhou X, Chancey ST, Pohl J, Shafer WM, Stephens DS. Human antimicrobial peptide LL-37 induces MefE/Mel-mediated macrolide resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 2010;54:3516–3519. doi: 10.1128/AAC.01756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chen YC, Chuang YC, Chang CC, Jeang CL, Chang MC. A K+ uptake protein, TrkA, is required for serum, protamine, and polymyxin B resistance in Vibrio vulnificus. Infect Immun. 2004;72:629–636. doi: 10.1128/IAI.72.2.629-636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Parra-Lopez C, Lin R, Aspedon A, Groisman EA. A Salmonella protein that is required for resistance to antimicrobial peptides and transport of potassium. EMBO J. 1994;13:3964–3972. doi: 10.1002/j.1460-2075.1994.tb06712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shafer WM, Qu X, Waring AJ, Lehrer RI. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc Natl Acad Sci USA. 1998;95:1829–1833. doi: 10.1073/pnas.95.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Veal WL, Nicholas RA, Shafer WM. Overexpression of the MtrC-MtrD-MtrE efflux pump due to an mtrR mutation is required for chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. J Bacteriol. 2002;184:5619–5624. doi: 10.1128/JB.184.20.5619-5624.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Jerse AE, Sharma ND, Simms AN, Crow ET, Snyder LA, Shafer WM. A gonococcal efflux pump system enhances bacterial survival in a female mouse model of genital tract infection. Infect Immun. 2003;71:5576–5582. doi: 10.1128/IAI.71.10.5576-5582.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rinker SD, Trombley MP, Gu X, Fortney KR, Bauer ME. Deletion of mtrC in Haemophilus ducreyi increases sensitivity to human antimicrobial peptides and activates the CpxRA regulon. Infect Immun. 2011;79:2324–2334. doi: 10.1128/IAI.01316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Mason KM, Munson RS, Jr., Bakaletz LO. A mutation in the sap operon attenuates survival of nontypeable Haemophilus influenzae in a chinchilla model of otitis media. Infect Immun. 2005;73:599–608. doi: 10.1128/IAI.73.1.599-608.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Parra-Lopez C, Baer MT, Groisman EA. Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. EMBO J. 1993;12:4053–4062. doi: 10.1002/j.1460-2075.1993.tb06089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mount KL, Townsend CA, Rinker SD, Gu X, Fortney KR, Zwickl BW, Janowicz DM, Spinola SM, Katz BP, Bauer ME. Haemophilus ducreyi SapA contributes to cathelicidin resistance and virulence in humans. Infect Immun. 2010;78:1176–1184. doi: 10.1128/IAI.01014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Eswarappa SM, Panguluri KK, Hensel M, Chakravortty D. The yejABEF operon of Salmonella confers resistance to antimicrobial peptides and contributes to its virulence. Microbiology. 2008;154:666–678. doi: 10.1099/mic.0.2007/011114-0. [DOI] [PubMed] [Google Scholar]
- 176.Saidijam M, Benedetti G, Ren Q, Xu Z, Hoyle CJ, Palmer SL, Ward A, Bettaney KE, Szakonyi G, Meuller J, Morrison S, Pos MK, Butaye P, Walravens K, Langton K, Herbert RB, Skurray RA, Paulsen IT, O'Reilly J, Rutherford NG, Brown MH, Bill RM, Henderson PJ. Microbial drug efflux proteins of the major facilitator superfamily. Curr Drug Targets. 2006;7:793–811. doi: 10.2174/138945006777709575. [DOI] [PubMed] [Google Scholar]
- 177.Kupferwasser LI, Skurray RA, Brown MH, Firth N, Yeaman MR, Bayer AS. Plasmid-mediated resistance to thrombin-induced platelet microbicidal protein in staphylococci: role of the qacA locus. Antimicrob Agents Chemother. 1999;43:2395–2399. doi: 10.1128/aac.43.10.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bayer AS, Kupferwasser LI, Brown MH, Skurray RA, Grkovic S, Jones T, Mukhopadhay K, Yeaman MR. Low-level resistance of Staphylococcus aureus to thrombin-induced platelet microbicidal protein 1 in vitro associated with qacA gene carriage is independent of multidrug efflux pump activity. Antimicrob Agents Chemother. 2006;50:2448–2454. doi: 10.1128/AAC.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bayer AS, Cheng D, Yeaman MR, Corey GR, McClelland RS, Harrel LJ, Fowler VG., Jr. In vitro resistance to thrombin-induced platelet microbicidal protein among clinical bacteremic isolates of Staphylococcus aureus correlates with an endovascular infectious source. Antimicrob Agents Chemother. 1998;42:3169–3172. doi: 10.1128/aac.42.12.3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shinnar AE, Butler KL, Park HJ. Cathelicidin family of antimicrobial peptides: proteolytic processing and protease resistance. Bioorg Chem. 2003;31:425–436. doi: 10.1016/s0045-2068(03)00080-4. [DOI] [PubMed] [Google Scholar]
- 181.Schmidtchen A, Frick IM, Andersson E, Tapper H, Bjorck L. Proteinases of common pathogenic bacteria degrade and inactivate the antibacterial peptide LL-37. Mol Microbiol. 2002;46:157–168. doi: 10.1046/j.1365-2958.2002.03146.x. [DOI] [PubMed] [Google Scholar]
- 182.Johansson L, Thulin P, Sendi P, Hertzen E, Linder A, Akesson P, Low DE, Agerberth B, Norrby-Teglund A. Cathelicidin LL-37 in severe Streptococcus pyogenes soft tissue infections in humans. Infect Immun. 2008;76:3399–3404. doi: 10.1128/IAI.01392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sieprawska-Lupa M, Mydel P, Krawczyk K, Wojcik K, Puklo M, Lupa B, Suder P, Silberring J, Reed M, Pohl J, Shafer W, McAleese F, Foster T, Travis J, Potempa J. Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob Agents Chemother. 2004;48:4673–4679. doi: 10.1128/AAC.48.12.4673-4679.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Belas R, Manos J, Suvanasuthi R. Proteus mirabilis ZapA metalloprotease degrades a broad spectrum of substrates, including antimicrobial peptides. Infect Immun. 2004;72:5159–5167. doi: 10.1128/IAI.72.9.5159-5167.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kubica M, Guzik K, Koziel J, Zarebski M, Richter W, Gajkowska B, Golda A, Maciag-Gudowska A, Brix K, Shaw L, Foster T, Potempa J. A potential new pathway for Staphylococcus aureus dissemination: the silent survival of S. aureus phagocytosed by human monocyte-derived macrophages. PLoS One. 2008;3:e1409. doi: 10.1371/journal.pone.0001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Nyberg P, Rasmussen M, Bjorck L. alpha2-Macroglobulin-proteinase complexes protect Streptococcus pyogenes from killing by the antimicrobial peptide LL-37. J Biol Chem. 2004;279:52820–52823. doi: 10.1074/jbc.C400485200. [DOI] [PubMed] [Google Scholar]
- 187.Rasmussen M, Muller HP, Bjorck L. Protein GRAB of Streptococcus pyogenes regulates proteolysis at the bacterial surface by binding alpha2-macroglobulin. J Biol Chem. 1999;274:15336–15344. doi: 10.1074/jbc.274.22.15336. [DOI] [PubMed] [Google Scholar]
- 188.Guina T, Yi EC, Wang H, Hackett M, Miller SI. A PhoP-regulated outer membrane protease of Salmonella enterica serovar Typhimurium promotes resistance to alpha-helical antimicrobial peptides. J Bacteriol. 2000;182:4077–4086. doi: 10.1128/jb.182.14.4077-4086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ly D, Taylor JM, Tsatsaronis JA, Monteleone MM, Skora AS, Donald CA, Maddocks T, Nizet V, West NP, Ranson M, Walker MJ, McArthur JD, Sanderson-Smith ML. Plasmin(ogen) acquisition by group A Streptococcus protects against C3b-mediated neutrophil killing. J Innate Immun. 2014;6:240–250. doi: 10.1159/000353754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kooi C, Sokol PA. Burkholderia cenocepacia zinc metalloproteases influence resistance to antimicrobial peptides. Microbiology. 2009;155:2818–2825. doi: 10.1099/mic.0.028969-0. [DOI] [PubMed] [Google Scholar]
- 191.Thomassin JL, Brannon JR, Gibbs BF, Gruenheid S, Le Moual H. OmpT outer membrane proteases of enterohemorrhagic and enteropathogenic Escherichia coli contribute differently to the degradation of human LL-37. Infect Immun. 2012;80:483–492. doi: 10.1128/IAI.05674-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ulvatne H, Haukland HH, Samuelsen O, Kramer M, Vorland LH. Proteases in Escherichia coli and Staphylococcus aureus confer reduced susceptibility to lactoferricin B. J Antimicrob Chemother. 2002;50:461–467. doi: 10.1093/jac/dkf156. [DOI] [PubMed] [Google Scholar]
- 193.Devine DA, Marsh PD, Percival RS, Rangarajan M, Curtis MA. Modulation of antibacterial peptide activity by products of Porphyromonas gingivalis and Prevotella spp. Microbiology. 1999;145:965–971. doi: 10.1099/13500872-145-4-965. [DOI] [PubMed] [Google Scholar]
- 194.Carlisle MD, Srikantha RN, Brogden KA. Degradation of human alpha- and beta-defensins by culture supernatants of Porphyromonas gingivalis strain 381. J Innate Immun. 2009;1:118–122. doi: 10.1159/000181015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Bachrach G, Altman H, Kolenbrander PE, Chalmers NI, Gabai-Gutner M, Mor A, Friedman M, Steinberg D. Resistance of Porphyromonas gingivalis ATCC 33277 to direct killing by antimicrobial peptides is protease independent. Antimicrob Agents Chemother. 2008;52:638–642. doi: 10.1128/AAC.01271-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Thwaite JE, Hibbs S, Titball RW, Atkins TP. Proteolytic degradation of human antimicrobial peptide LL-37 by Bacillus anthracis may contribute to virulence. Antimicrob Agents Chemother. 2006;50:2316–2322. doi: 10.1128/AAC.01488-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Chamnongpol S, Cromie M, Groisman EA. Mg2+ sensing by the Mg2+ sensor PhoQ of Salmonella enterica. J Mol Biol. 2003;325:795–807. doi: 10.1016/s0022-2836(02)01268-8. [DOI] [PubMed] [Google Scholar]
- 198.Guo L, Lim KB, Gunn JS, Bainbridge B, Darveau RP, Hackett M, Miller SI. Regulation of lipid A modifications by Salmonella Typhimurium virulence genes phoP-phoQ. Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 199.Ernst RK, Guina T, Miller SI. How intracellular bacteria survive: surface modifications that promote resistance to host innate immune responses. J Infect Dis. 1999;179(Suppl 2):S326–330. doi: 10.1086/513850. [DOI] [PubMed] [Google Scholar]
- 200.Ernst RK, Guina T, Miller SI. Salmonella Typhimurium outer membrane remodeling: role in resistance to host innate immunity. Microbes Infect. 2001;3:1327–1334. doi: 10.1016/s1286-4579(01)01494-0. [DOI] [PubMed] [Google Scholar]
- 201.Garcia Vescovi E, Soncini FC, Groisman EA. The role of the PhoP/PhoQ regulon in Salmonella virulence. Res Microbiol. 1994;145:473–480. doi: 10.1016/0923-2508(94)90096-5. [DOI] [PubMed] [Google Scholar]
- 202.Detweiler CS, Monack DM, Brodsky IE, Mathew H, Falkow S. virK, somA and rcsC are important for systemic Salmonella enterica serovar Typhimurium infection and cationic peptide resistance. Mol Microbiol. 2003;48:385–400. doi: 10.1046/j.1365-2958.2003.03455.x. [DOI] [PubMed] [Google Scholar]
- 203.Prost LR, Daley ME, Bader MW, Klevit RE, Miller SI. The PhoQ histidine kinases of Salmonella and Pseudomonas spp. are structurally and functionally different: evidence that pH and antimicrobial peptide sensing contribute to mammalian pathogenesis. Mol Microbiol. 2008;69:503–519. doi: 10.1111/j.1365-2958.2008.06303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.O'Loughlin JL, Spinner JL, Minnich SA, Kobayashi SD. Yersinia pestis two-component gene regulatory systems promote survival in human neutrophils. Infect Immun. 2010;78:773–782. doi: 10.1128/IAI.00718-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Macfarlane EL, Kwasnicka A, Hancock RE. Role of Pseudomonas aeruginosa PhoP-phoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology. 2000;146:2543–2554. doi: 10.1099/00221287-146-10-2543. [DOI] [PubMed] [Google Scholar]
- 206.McPhee JB, Lewenza S, Hancock RE. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol. 2003;50:205–217. doi: 10.1046/j.1365-2958.2003.03673.x. [DOI] [PubMed] [Google Scholar]
- 207.McPhee JB, Bains M, Winsor G, Lewenza S, Kwasnicka A, Brazas MD, Brinkman FS, Hancock RE. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J Bacteriol. 2006;188:3995–4006. doi: 10.1128/JB.00053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Amulic B, Cazalet C, Hayes GL, Metzler KD, Zychlinsky A. Neutrophil function: from mechanisms to disease. Annu Rev Immunol. 2012;30:459–489. doi: 10.1146/annurev-immunol-020711-074942. [DOI] [PubMed] [Google Scholar]
- 209.von Kockritz-Blickwede M, Nizet V. Innate immunity turned inside-out: antimicrobial defense by phagocyte extracellular traps. J Mol Med. 2009;87:775–783. doi: 10.1007/s00109-009-0481-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Buchanan JT, Simpson AJ, Aziz RK, Liu GY, Kristian SA, Kotb M, Feramisco J, Nizet V. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr Biol. 2006;16:396–400. doi: 10.1016/j.cub.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 211.Sumby P, Barbian KD, Gardner DJ, Whitney AR, Welty DM, Long RD, Bailey JR, Parnell MJ, Hoe NP, Adams GG, Deleo FR, Musser JM. Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proc Natl Acad Sci USA. 2005;102:1679–1684. doi: 10.1073/pnas.0406641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Walker MJ, Hollands A, Sanderson-Smith ML, Cole JN, Kirk JK, Henningham A, McArthur JD, Dinkla K, Aziz RK, Kansal RG, Simpson AJ, Buchanan JT, Chhatwal GS, Kotb M, Nizet V. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat Med. 2007;13:981–985. doi: 10.1038/nm1612. [DOI] [PubMed] [Google Scholar]
- 213.Beiter K, Wartha F, Albiger B, Normark S, Zychlinsky A, Henriques-Normark B. An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr Biol. 2006;16:401–407. doi: 10.1016/j.cub.2006.01.056. [DOI] [PubMed] [Google Scholar]
- 214.Derre-Bobillot A, Cortes-Perez NG, Yamamoto Y, Kharrat P, Couve E, Da Cunha V, Decker P, Boissier MC, Escartin F, Cesselin B, Langella P, Bermudez-Humaran LG, Gaudu P. Nuclease A (Gbs0661), an extracellular nuclease of Streptococcus agalactiae, attacks the neutrophil extracellular traps and is needed for full virulence. Mol Microbiol. 2013;89:518–531. doi: 10.1111/mmi.12295. [DOI] [PubMed] [Google Scholar]
- 215.Berends ET, Horswill AR, Haste NM, Monestier M, Nizet V, von Kockritz-Blickwede M. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J Innate Immun. 2010;2:576–586. doi: 10.1159/000319909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Mulcahy H, Charron-Mazenod L, Lewenza S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008;4:e1000213. doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Johnson L, Horsman SR, Charron-Mazenod L, Turnbull AL, Mulcahy H, Surette MG, Lewenza S. Extracellular DNA-induced antimicrobial peptide resistance in Salmonella enterica serovar Typhimurium. BMC Microbiol. 2013;13:115. doi: 10.1186/1471-2180-13-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Shireen T, Singh M, Das T, Mukhopadhyay K. Differential adaptive responses of Staphylococcus aureus to in vitro selection with different antimicrobial peptides. Antimicrob Agents Chemother. 2013;57:5134–5137. doi: 10.1128/AAC.00780-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Bayer AS, Prasad R, Chandra J, Koul A, Smriti M, Varma A, Skurray RA, Firth N, Brown MH, Koo SP, Yeaman MR. In vitro resistance of Staphylococcus aureus to thrombin-induced platelet microbicidal protein is associated with alterations in cytoplasmic membrane fluidity. Infect Immun. 2000;68:3548–3553. doi: 10.1128/iai.68.6.3548-3553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Mishra NN, Liu GY, Yeaman MR, Nast CC, Proctor RA, McKinnell J, Bayer AS. Carotenoid-related alteration of cell membrane fluidity impacts Staphylococcus aureus susceptibility to host defense peptides. Antimicrob Agents Chemother. 2011;55:526–531. doi: 10.1128/AAC.00680-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Subczynski WK, Wisniewska A. Physical properties of lipid bilayer membranes: relevance to membrane biological functions. Acta Biochim Pol. 2000;47:613–625. [PubMed] [Google Scholar]
- 222.Verheul A, Russell NJ, Van THR, Rombouts FM, Abee T. Modifications of membrane phospholipid composition in nisin-resistant Listeria monocytogenes Scott A. Appl Environ Microbiol. 1997;63:3451–3457. doi: 10.1128/aem.63.9.3451-3457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Crandall AD, Montville TJ. Nisin resistance in Listeria monocytogenes ATCC 700302 is a complex phenotype. Appl Environ Microbiol. 1998;64:231–237. doi: 10.1128/aem.64.1.231-237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Lopez-Solanilla E, Gonzalez-Zorn B, Novella S, Vazquez-Boland JA, Rodriguez-Palenzuela P. Susceptibility of Listeria monocytogenes to antimicrobial peptides. FEMS Microbiol Lett. 2003;226:101–105. doi: 10.1016/S0378-1097(03)00579-2. [DOI] [PubMed] [Google Scholar]
- 225.Islam D, Bandholtz L, Nilsson J, Wigzell H, Christensson B, Agerberth B, Gudmundsson G. Downregulation of bactericidal peptides in enteric infections: a novel immune escape mechanism with bacterial DNA as a potential regulator. Nat Med. 2001;7:180–185. doi: 10.1038/84627. [DOI] [PubMed] [Google Scholar]
- 226.Sperandio B, Regnault B, Guo J, Zhang Z, Stanley SL, Jr., Sansonetti PJ, Pedron T. Virulent Shigella flexneri subverts the host innate immune response through manipulation of antimicrobial peptide gene expression. J Exp Med. 2008;205:1121–1132. doi: 10.1084/jem.20071698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Taggart CC, Greene CM, Smith SG, Levine RL, McCray PB, Jr., O'Neill S, McElvaney NG. Inactivation of human beta-defensins 2 and 3 by elastolytic cathepsins. J Immunol. 2003;171:931–937. doi: 10.4049/jimmunol.171.2.931. [DOI] [PubMed] [Google Scholar]
- 228.Chakraborty K, Ghosh S, Koley H, Mukhopadhyay AK, Ramamurthy T, Saha DR, Mukhopadhyay D, Roychowdhury S, Hamabata T, Takeda Y, Das S. Bacterial exotoxins downregulate cathelicidin (hCAP-18/LL-37) and human beta-defensin 1 (HBD-1) expression in the intestinal epithelial cells. Cell Microbiol. 2008;10:2520–2537. doi: 10.1111/j.1462-5822.2008.01227.x. [DOI] [PubMed] [Google Scholar]
- 229.Gruenheid S, Le Moual H. Resistance to antimicrobial peptides in Gram-negative bacteria. FEMS Microbiol Lett. 2012;330:81–89. doi: 10.1111/j.1574-6968.2012.02528.x. [DOI] [PubMed] [Google Scholar]
- 230.Loutet SA, Valvano MA. Extreme antimicrobial Peptide and polymyxin B resistance in the genus Burkholderia. Front Microbiol. 2011;2:159. doi: 10.3389/fmicb.2011.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Cox AD, Wilkinson SG. Ionizing groups in lipopolysaccharides of Pseudomonas cepacia in relation to antibiotic resistance. Mol Microbiol. 1991;5:641–646. doi: 10.1111/j.1365-2958.1991.tb00735.x. [DOI] [PubMed] [Google Scholar]
- 232.Loutet SA, Mussen LE, Flannagan RS, Valvano MA. A two-tier model of polymyxin B resistance in Burkholderia cenocepacia. Environ Microbiol Rep. 2011;3:278–285. doi: 10.1111/j.1758-2229.2010.00222.x. [DOI] [PubMed] [Google Scholar]
- 233.Dhand A, Bayer AS, Pogliano J, Yang SJ, Bolaris M, Nizet V, Wang G, Sakoulas G. Use of antistaphylococcal beta-lactams to increase daptomycin activity in eradicating persistent bacteremia due to methicillin-resistant Staphylococcus aureus: role of enhanced daptomycin binding. Clin Infect Dis. 2011;53:158–163. doi: 10.1093/cid/cir340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Sakoulas G, Bayer AS, Pogliano J, Tsuji BT, Yang SJ, Mishra NN, Nizet V, Yeaman MR, Moise PA. Ampicillin enhances daptomycin- and cationic host defense peptide-mediated killing of ampicillin- and vancomycin-resistant Enterococcus faecium. Antimicrob Agents Chemother. 2012;56:838–844. doi: 10.1128/AAC.05551-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 236.Wiesner J, Vilcinskas A. Antimicrobial peptides: the ancient arm of the human immune system. Virulence. 2010;1:440–464. doi: 10.4161/viru.1.5.12983. [DOI] [PubMed] [Google Scholar]
- 237.Tavares LS, Silva CS, de Souza VC, da Silva VL, Diniz CG, Santos MO. Strategies and molecular tools to fight antimicrobial resistance: resistome, transcriptome, and antimicrobial peptides. Front Microbiol. 2013;4:412. doi: 10.3389/fmicb.2013.00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Peschel A, Sahl HG. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat Rev Microbiol. 2006;4:529–536. doi: 10.1038/nrmicro1441. [DOI] [PubMed] [Google Scholar]
- 239.Habets MG, Brockhurst MA. Therapeutic antimicrobial peptides may compromise natural immunity. Biol Lett. 2012;8:416–418. doi: 10.1098/rsbl.2011.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Jung D, Rozek A, Okon M, Hancock RE. Structural transitions as determinants of the action of the calcium-dependent antibiotic daptomycin. Chem Biol. 2004;11:949–957. doi: 10.1016/j.chembiol.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 241.Mishra NN, McKinnell J, Yeaman MR, Rubio A, Nast CC, Chen L, Kreiswirth BN, Bayer AS. In vitro cross-resistance to daptomycin and host defense cationic antimicrobial peptides in clinical methicillin-resistant Staphylococcus aureus isolates. Antimicrob Agents Chemother. 2011;55:4012–4018. doi: 10.1128/AAC.00223-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Gravesen A, Jydegaard Axelsen AM, Mendes da Silva J, Hansen TB, Knochel S. Frequency of bacteriocin resistance development and associated fitness costs in Listeria monocytogenes. Appl Environ Microbiol. 2002;68:756–764. doi: 10.1128/AEM.68.2.756-764.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Gravesen A, Ramnath M, Rechinger KB, Andersen N, Jansch L, Hechard Y, Hastings JW, Knochel S. High-level resistance to class IIa bacteriocins is associated with one general mechanism in Listeria monocytogenes. Microbiology. 2002;148:2361–2369. doi: 10.1099/00221287-148-8-2361. [DOI] [PubMed] [Google Scholar]
- 244.Menuet M, Bittar F, Stremler N, Dubus JC, Sarles J, Raoult D, Rolain JM. First isolation of two colistin-resistant emerging pathogens, Brevundimonas diminuta and Ochrobactrum anthropi, in a woman with cystic fibrosis: a case report. J Med Case Rep. 2008;2:373. doi: 10.1186/1752-1947-2-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Antoniadou A, Kontopidou F, Poulakou G, Koratzanis E, Galani I, Papadomichelakis E, Kopterides P, Souli M, Armaganidis A, Giamarellou H. Colistin-resistant isolates of Klebsiella pneumoniae emerging in intensive care unit patients: first report of a multiclonal cluster. J Antimicrob Chemother. 2007;59:786–790. doi: 10.1093/jac/dkl562. [DOI] [PubMed] [Google Scholar]
- 246.Huddleston JR. Horizontal gene transfer in the human gastrointestinal tract: potential spread of antibiotic resistance genes. Infect Drug Resist. 2014;7:167–176. doi: 10.2147/IDR.S48820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Napier BA, Band V, Burd EM, Weiss DS. Colistin heteroresistance in Enterobacter cloacae is associated with cross-resistance to the host antimicrobial lysozyme. Antimicrob Agents Chemother. 2014;58:5594–5597. doi: 10.1128/AAC.02432-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Costa F, Carvalho IF, Montelaro RC, Gomes P, Martins MC. Covalent immobilization of antimicrobial peptides (AMPs) onto biomaterial surfaces. Acta Biomater. 2011;7:1431–1440. doi: 10.1016/j.actbio.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 249.Deslouches B, Steckbeck JD, Craigo JK, Doi Y, Mietzner TA, Montelaro RC. Rational design of engineered cationic antimicrobial peptides consisting exclusively of arginine and tryptophan, and their activity against multidrug-resistant pathogens. Antimicrob Agents Chemother. 2013;57:2511–2521. doi: 10.1128/AAC.02218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Peyssonnaux C, Datta V, Cramer T, Doedens A, Theodorakis EA, Gallo RL, Hurtado-Ziola N, Nizet V, Johnson RS. HIF-1alpha expression regulates the bactericidal capacity of phagocytes. J Clin Invest. 2005;115:1806–1815. doi: 10.1172/JCI23865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Okumura CY, Hollands A, Tran DN, Olson J, Dahesh S, von Kockritz-Blickwede M, Thienphrapa W, Corle C, Jeung SN, Kotsakis A, Shalwitz RA, Johnson RS, Nizet V. A new pharmacological agent (AKB-4924) stabilizes hypoxia inducible factor-1 (HIF-1) and increases skin innate defenses against bacterial infection. J Mol Med. 2012;90:1079–1089. doi: 10.1007/s00109-012-0882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Nizet V, Johnson RS. Interdependence of hypoxic and innate immune responses. Nat Rev Immunol. 2009;9:609–617. doi: 10.1038/nri2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Darveau RP, Blake J, Seachord CL, Cosand WL, Cunningham MD, Cassiano-Clough L, Maloney G. Peptides related to the carboxyl terminus of human platelet factor IV with antibacterial activity. J Clin Invest. 1992;90:447–455. doi: 10.1172/JCI115880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Peschel A, Vuong C, Otto M, Gotz F. The D-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrob Agents Chemother. 2000;44:2845–2847. doi: 10.1128/aac.44.10.2845-2847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Abi Khattar Z, Rejasse A, Destoumieux-Garzon D, Escoubas JM, Sanchis V, Lereclus D, Givaudan A, Kallassy M, Nielsen-Leroux C, Gaudriault S. The dlt operon of Bacillus cereus is required for resistance to cationic antimicrobial peptides and for virulence in insects. J Bacteriol. 2009;191:7063–7073. doi: 10.1128/JB.00892-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Cox KH, Ruiz-Bustos E, Courtney HS, Dale JB, Pence MA, Nizet V, Aziz RK, Gerling I, Price SM, Hasty DL. Inactivation of DltA modulates virulence factor expression in Streptococcus pyogenes. PLoS One. 2009;4:e5366. doi: 10.1371/journal.pone.0005366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Fisher N, Shetron-Rama L, Herring-Palmer A, Heffernan B, Bergman N, Hanna P. The dltABCD operon of Bacillus anthracis sterne is required for virulence and resistance to peptide, enzymatic, and cellular mediators of innate immunity. J Bacteriol. 2006;188:1301–1309. doi: 10.1128/JB.188.4.1301-1309.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Fittipaldi N, Sekizaki T, Takamatsu D, Harel J, Dominguez-Punaro Mde L, Von Aulock S, Draing C, Marois C, Kobisch M, Gottschalk M. D-alanylation of lipoteichoic acid contributes to the virulence of Streptococcus suis. Infect Immun. 2008;76:3587–3594. doi: 10.1128/IAI.01568-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Collins LV, Kristian SA, Weidenmaier C, Faigle M, Van Kessel KP, Van Strijp JA, Gotz F, Neumeister B, Peschel A. Staphylococcus aureus strains lacking D-alanine modifications of teichoic acids are highly susceptible to human neutrophil killing and are virulence attenuated in mice. J Infect Dis. 2002;186:214–219. doi: 10.1086/341454. [DOI] [PubMed] [Google Scholar]
- 260.Cullen TW, Trent MS. A link between the assembly of flagella and lipooligosaccharide of the Gram-negative bacterium Campylobacter jejuni. Proc Natl Acad Sci USA. 2010;107:5160–5165. doi: 10.1073/pnas.0913451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Pelz A, Wieland KP, Putzbach K, Hentschel P, Albert K, Gotz F. Structure and biosynthesis of staphyloxanthin from Staphylococcus aureus. J Biol Chem. 2005;280:32493–32498. doi: 10.1074/jbc.M505070200. [DOI] [PubMed] [Google Scholar]
- 262.Clauditz A, Resch A, Wieland KP, Peschel A, Gotz F. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect Immun. 2006;74:4950–4953. doi: 10.1128/IAI.00204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Liu GY, Essex A, Buchanan JT, Datta V, Hoffman HM, Bastian JF, Fierer J, Nizet V. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J Exp Med. 2005;202:209–215. doi: 10.1084/jem.20050846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Park PW, Pier GB, Hinkes MT, Bernfield M. Exploitation of syndecan-1 shedding by Pseudomonas aeruginosa enhances virulence. Nature. 2001;411:98–102. doi: 10.1038/35075100. [DOI] [PubMed] [Google Scholar]
- 265.Diep DB, Havarstein LS, Nes IF. Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J Bacteriol. 1996;178:4472–4483. doi: 10.1128/jb.178.15.4472-4483.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Diep DB, Skaugen M, Salehian Z, Holo H, Nes IF. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc Natl Acad Sci USA. 2007;104:2384–2389. doi: 10.1073/pnas.0608775104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Klein C, Entian KD. Genes involved in self-protection against the lantibiotic subtilin produced by Bacillus subtilis ATCC 6633. Appl Environ Microbiol. 1994;60:2793–2801. doi: 10.1128/aem.60.8.2793-2801.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Kuipers OP, Beerthuyzen MM, Siezen RJ, De Vos WM. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur J Biochem. 1993;216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 269.Saris PE, Immonen T, Reis M, Sahl HG. Immunity to lantibiotics. Antonie Van Leeuwenhoek. 1996;69:151–159. doi: 10.1007/BF00399420. [DOI] [PubMed] [Google Scholar]
- 270.Warner DM, Shafer WM, Jerse AE. Clinically relevant mutations that cause derepression of the Neisseria gonorrhoeae MtrC-MtrD-MtrE Efflux pump system confer different levels of antimicrobial resistance and in vivo fitness. Mol Microbiol. 2008;70:462–478. doi: 10.1111/j.1365-2958.2008.06424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Kawada-Matsuo M, Yoshida Y, Zendo T, Nagao J, Oogai Y, Nakamura Y, Sonomoto K, Nakamura N, Komatsuzawa H. Three distinct two-component systems are involved in resistance to the class I bacteriocins, Nukacin ISK-1 and nisin A, in Staphylococcus aureus. PLoS One. 2013;8:e69455. doi: 10.1371/journal.pone.0069455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Li M, Cha DJ, Lai Y, Villaruz AE, Sturdevant DE, Otto M. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol Microbiol. 2007;66:1136–1147. doi: 10.1111/j.1365-2958.2007.05986.x. [DOI] [PubMed] [Google Scholar]
- 273.Hiron A, Falord M, Valle J, Debarbouille M, Msadek T. Bacitracin and nisin resistance in Staphylococcus aureus: a novel pathway involving the BraS/BraR two-component system (SA2417/SA2418) and both the BraD/BraE and VraD/VraE ABC transporters. Mol Microbiol. 2011;81:602–622. doi: 10.1111/j.1365-2958.2011.07735.x. [DOI] [PubMed] [Google Scholar]
- 274.Falord M, Karimova G, Hiron A, Msadek T. GraXSR proteins interact with the VraFG ABC transporter to form a five-component system required for cationic antimicrobial peptide sensing and resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2012;56:1047–1058. doi: 10.1128/AAC.05054-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Thurlow LR, Thomas VC, Narayanan S, Olson S, Fleming SD, Hancock LE. Gelatinase contributes to the pathogenesis of endocarditis caused by Enterococcus faecalis. Infect Immun. 2010;78:4936–4943. doi: 10.1128/IAI.01118-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Sabat A, Kosowska K, Poulsen K, Kasprowicz A, Sekowska A, van Den Burg B, Travis J, Potempa J. Two allelic forms of the aureolysin gene (aur) within Staphylococcus aureus. Infect Immun. 2000;68:973–976. doi: 10.1128/iai.68.2.973-976.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Lai Y, Villaruz AE, Li M, Cha DJ, Sturdevant DE, Otto M. The human anionic antimicrobial peptide dermcidin induces proteolytic defence mechanisms in staphylococci. Mol Microbiol. 2007;63:497–506. doi: 10.1111/j.1365-2958.2006.05540.x. [DOI] [PubMed] [Google Scholar]
- 278.Cheung GY, Rigby K, Wang R, Queck SY, Braughton KR, Whitney AR, Teintze M, DeLeo FR, Otto M. Staphylococcus epidermidis strategies to avoid killing by human neutrophils. PLoS Pathog. 2010;6:e1001133. doi: 10.1371/journal.ppat.1001133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Bergman P, Johansson L, Asp V, Plant L, Gudmundsson GH, Jonsson AB, Agerberth B. Neisseria gonorrhoeae downregulates expression of the human antimicrobial peptide LL-37. Cell Microbiol. 2005;7:1009–1017. doi: 10.1111/j.1462-5822.2005.00530.x. [DOI] [PubMed] [Google Scholar]
- 280.Salzman NH, Chou MM, de Jong H, Liu L, Porter EM, Paterson Y. Enteric Salmonella infection inhibits Paneth cell antimicrobial peptide expression. Infect Immun. 2003;71:1109–1115. doi: 10.1128/IAI.71.3.1109-1115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Yanagi S, Ashitani J, Imai K, Kyoraku Y, Sano A, Matsumoto N, Nakazato M. Significance of human beta-defensins in the epithelial lining fluid of patients with chronic lower respiratory tract infections. Clin Microbiol Infect. 2007;13:63–69. doi: 10.1111/j.1469-0691.2006.01574.x. [DOI] [PubMed] [Google Scholar]