Abstract
Adhesion between biological surfaces, which is typically the result of molecular binding between receptors on one surface and ligands on another, plays a fundamental role in biology and is key to the infection mechanisms of certain viruses, including influenza. The physiological outcome of adhesion depends on both the number of bound cells (or viruses, or other biological particles) and the properties of the adhesion interface that is formed, including the equilibrium number of receptor-ligand connections. Here, we introduce a quantitative model for biological adhesion by adapting thermodynamic models developed for the related problem of multivalent molecular binding. In our model, adhesion affinity is approximated by a simple, analytical expression involving the numbers of ligands and receptors at the interface. Our model contains only two fitting parameters and is simple to interpret. When applied to the adhesion between the hemagglutinin ligands on influenza viruses and the sialic acid receptors on biosensors or on host cells, our model generates adhesion affinities consistent with experimental measurements performed over a range of numbers of receptors, and provides a semiquantitative estimate of the affinity range of the hemagglutinin-sialic acid interaction necessary for the influenza virus to successfully infect host cells. The model also provides a quantitative explanation for the experimental finding that a mutant avian virus gained transmissibility in mammals despite the mutations conferring only a less than twofold increase in the affinity of its hemagglutinin for mammalian receptors: the model predicts an order-of-magnitude improvement in adhesion to mammalian cells. We also extend our model to describe the competitive inhibition of adhesion: the model predicts that hemagglutinin inhibitors of relatively modest affinity can dramatically reduce influenza virus adhesion to host cells, suggesting that such inhibitors, if discovered, may be viable therapeutic agents against influenza.
Introduction
Adhesion between two cells, or between a virus and a cell, mediates diverse biological phenomena including pathogen recognition of host cells (1, 2, 3), cell trafficking (4, 5, 6, 7), and cell signaling (8, 9). Such adhesion typically involves the simultaneous binding of multiple copies of ligands on one of the contact surfaces with multiple copies of receptors on the other (10, 11), and at biological interfaces there are often hundreds to thousands of such ligand-receptor pairs. Influenza virus, for example, in its first step of infection, attaches to vertebrate host cells through the binding between multiple hemagglutinin (HA) trimers on the viral surface and multiple copies of the N-acetylneuraminic acid (also known as sialic acid (SA)) moiety on the host-cell surface (12). Although the binding between a single pair of HA and SA molecules is weak (equilibrium dissociation constant KD ∼ 1 mM) (13), the large number of HA and SA molecules present at the contact surface collectively ensures that the virus adheres to the host cell with high affinity. This strong adhesion is thus a consequence of multivalent (also referred to as polyvalent) interactions, where valency refers to the number of simultaneous connections between one kind of particle and another.
The physiological outcome of adhesion depends on both the number of bound cells (or viruses, or other biological particles) and the properties of the adhesion interface that is formed, such as the contact surface area, the distance of cell-cell separation, and the equilibrium number of receptor-ligand connections. Thermodynamic models have been developed to study the properties of the individual adhesion interface (14, 15, 16, 17, 18). Such models, however, do not calculate the fraction of bound cells in a given condition (15). Full characterization of adhesion requires models that can estimate the affinity of adhesion and thus the population of bound cells, which can now be measured experimentally. In this study we develop such a model.
Many biological processes involve modulating the strength of multivalent adhesion, either by changing the number of available receptors or ligands at the interface or by changing the binding affinity of each receptor-ligand pair. Quantitative characterization of such modulation requires a model that accurately estimates the adhesion affinity from the parameters of the underlying receptor-ligand binding.
Several empirical models of multivalent adhesion have been reported in the literature (11). One widely used approach approximates the dissociation constant of adhesion, KD,ad, as (KD)m, where m is referred to as the multiplicity, the value of which depends on the number of adhesive receptor and ligand molecules at the contact surface (19). To our knowledge, however, no theoretical model has been proposed to estimate m from the number of receptors, NR, the number of ligands, NL, and KD: It can only be measured empirically at discrete, static numbers of receptors and/or ligands. As suggested above, it is often necessary to consider adhesion for changing numbers of receptors and ligands and for a range of binding affinities. The empirical approach is typically difficult to apply in these problems.
Thermodynamic models have been developed for the related problem of binding between molecules with multiple binding sites, such as in multivalent host-guest binding (20, 21, 22, 23, 24, 25). These models relate the avidity of multivalent binding to the numbers of available binding sites on the multivalent molecules (receptors and ligands or host and guest molecules) and to the KD of individual pairs of binding sites. These models have enjoyed great success treating molecular systems in which there are <∼10 binding site pairs, but they have not been applied to adhesion between larger biological particles. Adhesion between larger particles (e.g., between a virus and a host cell) often involves a much larger contact interface, which incurs nonspecific interactions, such as electrostatic repulsion between membranes, that can significantly affect the adhesion affinity.
In this work, we extend previous thermodynamic models of multivalent molecular binding to propose a simple model of the phenomenon of multivalent adhesion, in part by introducing a fitting parameter to account for the contribution of nonspecific interactions. We also introduce approximations appropriate for the large number of adhesive connections, resulting in a simple equation that estimates KD,ad from KD, NR, and NL. Our model establishes a clear relationship between KD,ad and the equilibrium number of receptor-ligand connections, the latter being a key quantity in the groundbreaking model developed by Bell and co-workers (14, 15, 16, 17, 18).
To demonstrate the utility of this approach, we use our model to study adhesion of influenza virus to SA-conjugated biosensors and to host cells. The influenza virus exploits two mechanisms to modulate its adhesion to host cells: 1) changing the number of receptors to spread from cell to cell (26, 27), and 2) changing the affinity of receptor-ligand binding to adapt from avian to mammalian hosts (28, 29, 30). We apply our model to the analysis of both mechanisms of modulation. Our model accurately reproduces the experimentally measured relationship between adhesion and the number of available SA molecules conjugated to biosensors, and quantitatively explains the observed sensitivity of adhesion affinity to small changes in the HA-SA binding affinity. Our model also provides a quantitative technique for analyzing the functional interplay between HA-SA binding affinity and the catalytic efficiency of neuraminidase, and yields a reasonable estimate for the range of affinity of HA-SA binding that leads to a viable virus.
We also extend our model to the study of competitive inhibition of multivalent adhesion. A competitive inhibitor of the adhesive molecules—against either the receptor or the ligand—reduces the number of available connection sites, thus weakening or abolishing adhesion (31). The efficacy of such an inhibitor depends not only on its affinity for its molecular target, but also on how the adhesion affinity changes with respect to the numbers of available receptors and ligands. Our model permits a quantitative analysis of the competitive inhibition of multivalent adhesion. When applied to influenza, our model indicates that it may be possible to effectively inhibit the adhesion of influenza virus to host cells using a molecule with only modest affinity for HA. It might thus be feasible to take existing modest inhibitors of HA, or discover new ones, and develop them as therapeutic agents against influenza infection.
Materials and Methods
Please see the Supporting Material.
Results and Discussion
Theoretical model
Our model estimates KD,ad of adhesion from the KD of individual receptor-ligand binding and the numbers of receptors, NR, and ligands, NL, at the adhesion interface. The result, which we derive later in this section, is
| (1) |
where is the equilibrium number of connections formed between receptors and ligands, and is obtained from the equilibrium equation
| (2) |
Here, K0 is the 0-valency dissociation constant between the two surfaces when no connections are formed, and Veff is the effective molar volume in which receptor-ligand binding occurs. K0 and Veff are difficult to measure directly, but can be estimated by fitting to adhesion measurements at varying values of NR and/or NL. Typically, KD can be measured by standard binding assays or calorimetry, and NR and NL can be measured by electron microscopy or fluorescence. In the specific case of HA-SA-mediated adhesion by the influenza virus, the HA-SA binding affinity has been measured by nuclear magnetic resonance (13) and microscale thermophoresis (MST) (19), and the number of sialic acids on the cell surface has been measured by cleavage experiments (32).
We first derive Eq. 1 by the method of statistical mechanics, and then give a simple interpretation of the result at the end. At the adhesion interface, individual connections between receptors and ligands form and break in a thermodynamic equilibrium, and the number of connections at a given instant, n, fluctuates between 0 and min (NR, NL). We assume that the connections form and break independently and that the Gibbs free energy change of forming a connection between a given ligand and receptor is ΔG = RTln(KDVeff), with R being the gas constant and T the temperature. The molar volume Veff accounts for the adjustment of the reaction condition, because the binding between the receptors and the ligands is restrained in the narrow confine of the adhesive interface, and both the receptors and the ligands are tethered to their respective surfaces. Our model in this form ignores potential cooperativity in receptor-ligand binding (25), in which the binding free energy of a given receptor-ligand pair depends on the number of existing connections. Such cooperativity, however, can be included in our model at the cost of extra parameters, as discussed in a subsequent section.
At thermal equilibrium, the probability that n connections are formed is given by
| (3) |
where
| (4) |
is the grand canonical partition function. In the first line of Eq. 3, the binomials
count the numbers of ways to choose n receptors out of a total NR and n ligands out of a total NL to form n connections. The value n! stands for the number of ways to pair the n chosen receptors and ligands. The factor 1/(NR!NL!) takes into account that the receptors are indistinguishable from one another and so are the ligands. In situations where the receptors are attached to fixed locations on a surface (such as experiments in which the receptors are tethered to a surface through streptavidin-biotin linkers), thus becoming distinguishable, NR! should be removed from the denominator. The value pn, however, remains unchanged. Equation 3 is an extension of the Klotz equation, first developed for the binding of ligands to receptors with multiple binding sites (33). The exact number of receptors and ligands at the contact surface varies among individual biological particles; in our model we take NR to be the average number of receptors on receptor-bearing particles, and NL to be the average number of ligands on ligand-bearing particles.
The above receptor-ligand binding equilibrium is part of the overall equilibrium of adhesion between two biological surfaces C and V,
| (5) |
where CVn stands for the microstate of the adhesion surface where n connections are formed between the receptors and the ligands. The 0-valency dissociation constant, K0, accounts for the contribution of nonspecific interactions to the adhesion affinity.
The dissociation constant of adhesion is thus given by
| (6) |
Given KD, NR, NL, K0, and Veff, one can calculate KD,ad from Eqs. 3, 4, and 6 using a computer program. When NR and NL are large, however, we can introduce approximations that greatly simplify the expression of KD,ad and remove the factorials in the equation.
Under conditions where reasonably large numbers of connections form, the grand canonical partition function Z can be well approximated by the maximum term in the summand,
| (7) |
where is the number at which pn is at its maximum. The value thus satisfies , so
| (8) |
which implies that is the equilibrium number of connections formed between the receptors and the ligands confined within the adhesion interface. Applying the Stirling approximation ln x! = xlnx – x to lnp0 = −ln(ZNR!NL!), we have
| (9) |
where the last line is due to Eq. 8. Substituting p0 above in Eq. 6, we arrive at our expression for KD,ad in Eq. 1.
Equations 1 and 2 can also be derived by considering the following imaginary thermodynamic process (because the free energy change of adhesion is the difference between two state functions and is independent of the connecting thermodynamic path). The reaction of adhesion can be approximately decomposed into two steps: 1) the attachment of the two surfaces without the formation of any receptor-ligand connections, and 2) the establishment of the equilibrium of receptor-ligand binding at the interface. The total free energy of adhesion is thus
| (10) |
where is the free energy of forming receptor-ligand connections, and is given by
| (11) |
where the second term on the right-hand side is the entropic contribution due to the number of ways to form connections out of NR receptors and NL ligands. Equations 10 and 11, with the Stirling approximation and the same algebra as in Eq. 9, lead to Eq. 1.
Finally, we derive an approximate expression for the so-called 0-valency dissociation constant K0, which is valid when the contribution of nonspecific interactions to this quantity can be neglected. Suppose that the total numbers of receptors and ligands exposed on the respective biological particles are MR and ML, out of which the NR receptors and the NL ligands that form the connections are the subsets that happen to lie within the interface. The value K0 can be related to the fractions NR/MR and NL/ML, and the effective molar volume Veff, as follows.
The reaction to form one connection between the two biological particles is
| (12) |
If we ignore the nonspecific interactions between the two biological particles, however, this monovalent reaction is equivalent to the reaction of individual receptor-ligand binding, where the receptor and ligand concentrations are MR and ML times the concentrations of the respective biological particles. As a result, we have
| (13) |
and
| (14) |
If the fractions NR/MR and NL/ML are known, Veff becomes the only free parameter in our model. In this work, we treat K0 as a free fitting parameter, and we check its estimated value against sensible guesses of the fractions.
We have so far developed the model assuming that one particle of V adheres to one particle of C. Different types of biological particles, however, often have disparate sizes. If C is much larger than V, many particles of V can adhere to one particle of C, each occupying a nonoverlapping site of adhesion and making a number of adhesive molecules on C, which we can call M∗, unavailable to other particles of V. If the total number of adhesive molecules on one particle of C is M, the number of particles of V adhering to one particle of C at adhesion equilibrium can be shown to be
| (15) |
where S = M/M∗ is the maximum number of particles of V that can be accommodated on the surface of one particle of C. Equation 15 is equivalent to the Langmuir equation, S being the number of sites of adhesion, and K′D,ad ≡ SKD,ad being the per-site dissociation constant of adhesion. Correspondingly, the per-site 0-valency dissociation constant is K′0 = SK0. Clearly, K′D,ad/K′0 = KD,ad/K0, and within our model this ratio still follows from Eq. 1. The value K0 follows from Eq. 14; note that K′0 is also given by Eq. 14 if we reinterpret MR (or ML, if the ligands rather than receptors lie on C) as the number of adhesive molecules per site of adhesion (i.e., the number of adhesive molecules on C made unavailable to other particles of V by adhesion of one particle of V). Below, we use the more convenient per-site dissociation constants K′0 and K′D,ad, but we switch notation by dropping the prime symbols.
Our model in its simple form (Eqs. 1 and 2) assumes that only one type of receptor and one type of ligand are present at the interface, and that the binding affinity between any receptor-ligand pair is the same. Real cells, however, may present diverse sets of different receptor types, each with a different binding affinity for the ligand. A vertebrate cell, for example, may present a range of different glycans that terminate with the SA moiety, and each type of glycan may have a different binding affinity for the HA molecule (34, 35, 36). It is straightforward to extend our model to account for such heterogeneity. If the number of receptors with binding affinity between K and K + dK is N(K)dK, the dissociation constant of adhesion in our model is given by
| (16) |
where (K)dK, satisfying the equilibrium equation
| (17) |
is the equilibrium number of receptor-ligand connections with binding affinity between K and K + dK, and = ∫(K)dK is the total equilibrium number of connections formed between all receptors and the ligand molecules. The value can be determined from the following equation:
| (18) |
Equations 16–18 (derived in the Supporting Material) describe a cell’s properties of adhesion once the distribution of binding affinity N(K) is known for the receptors on the cell. Equations 1 and 2 are a special case of Eqs. 16–18, with N(K) = δ(K − KD). Our model, thus generalized, can be used to analyze the variability in adhesion affinity in a population of cells due to the fluctuations in the expression levels of different receptors.
Properties of the model
The expression of pn and the partition function Z (Eqs. 3 and 4, without the approximations introduced in our model) can also be derived by equations of mass action, as has been done in the development of previous models of multivalent molecular binding (23), if the combinatorial factors of pairing any receptor molecule with any ligand molecule are properly accounted for. The exact expression of Z (Eq. 4) can be used as is, but our approximation leads to a simple analytical expression of the adhesion affinity (Eq. 1), making apparent its relationship to the other variables. The expression in Eq. 1 makes it easy to include NR and NL, which are discrete variables in the exact solution, as continuous parameters in fitting to experimental adhesion data, when their values are unknown. It also relates the adhesion affinity to the equilibrium number of receptor-ligand connections, , in a simple formula.
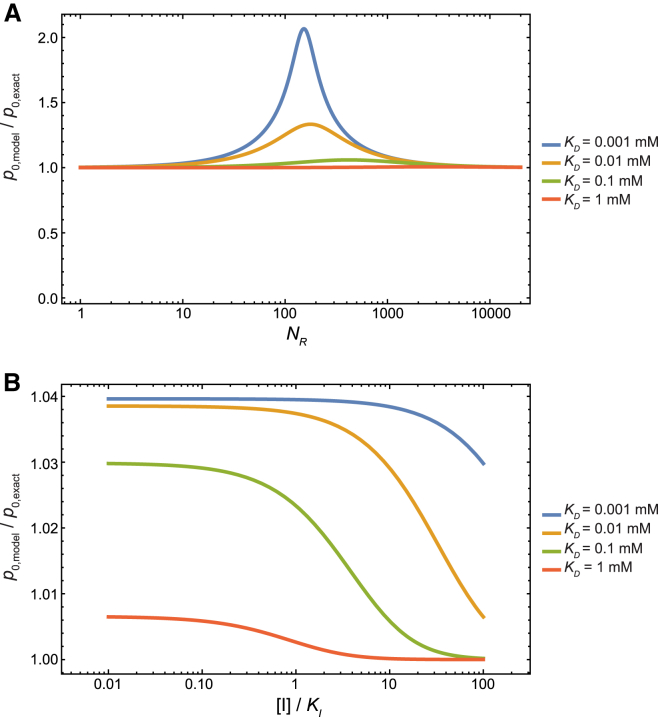
Although Eq. 1 is derived by applying the Stirling approximation at large values of NR, NL, and , Eq. 1 remains a good approximation even at small values of these quantities. Fig. 1 A plots the ratio p0/p0,exact as NR is varied between 1 and 20,000, where p0,exact is given by Eq. 3. In the case of weak receptor-ligand affinities (KD ∼ 1 mM), as in the case of cell-influenza adhesion, the ratio remains very close to 1 throughout this range of NR. At stronger affinity, KD = 0.001 mM, our model can overestimate the adhesion affinity by a factor of up to ∼2, which may still be acceptable for most applications.
Figure 1.
The accuracy of our model. (A) The ratio p0/p0,exact as NR is varied between 1 and 20,000, for different receptor-ligand affinities. The other model parameters are NL = 150, Veff = 2.7 × 103 mM−1. (B) The ratio p0/p0,exact as the concentration of an inhibitor of the ligand is varied. The parameters NL and Veff have the same values as in (A), and NR = 2000.
The strength of adhesion can be modulated by changing the available number of receptors (or ligands) at the adhesion interface. Consider the adhesion between viruses and cells: the fraction of bound virus particles is
| (19) |
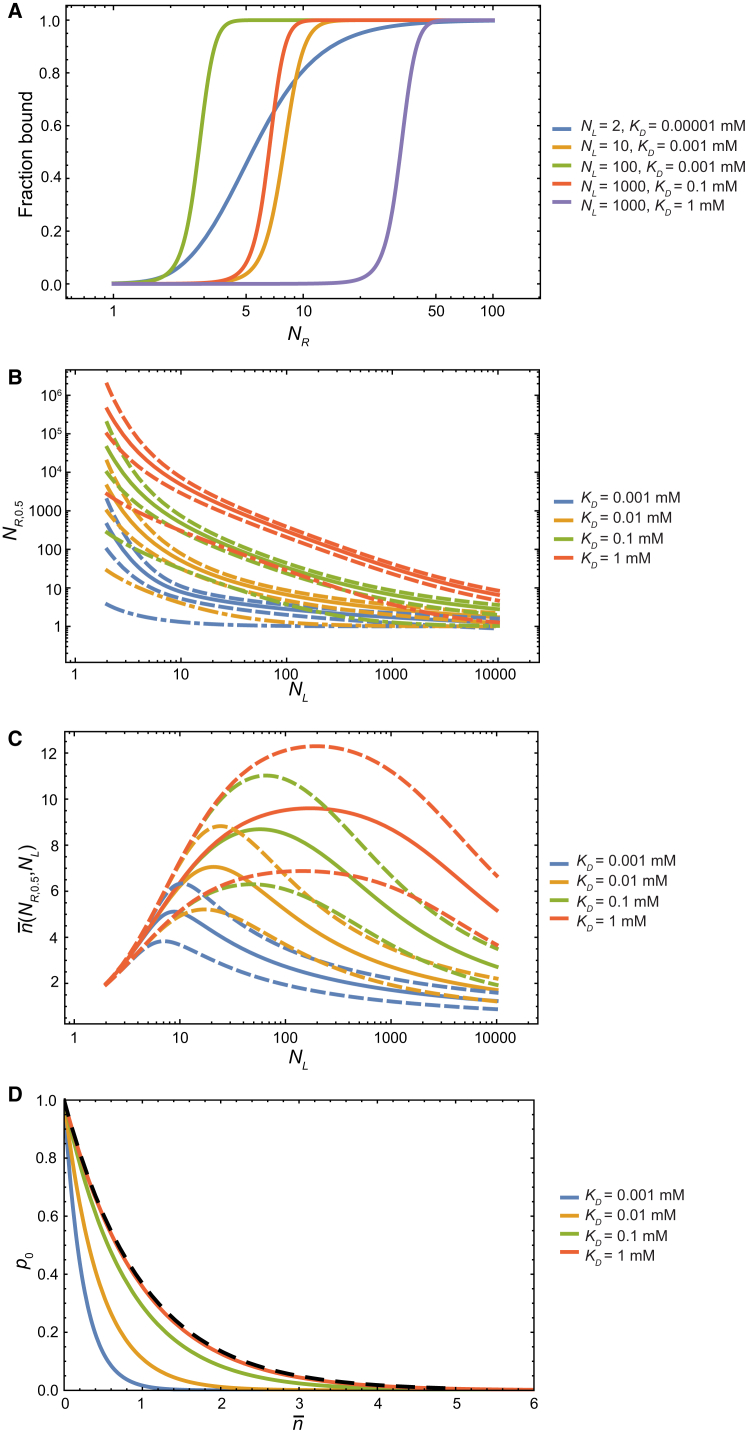
where [cell] is the concentration of the cells. The value f changes with the adhesion affinity, and thus with the number of receptors NR. Fig. 2 A plots f against NR, holding the number of ligands, NL, at various fixed values. These adhesion isotherms are steep at large values of NL. By solving Eq. 19 for NR for any given f, our model can be used to estimate NR,f, the number of receptors at which that fraction of the virus particles is bound (Fig. 2 B). At high values of NL, the bound fractions increase quickly from 5 to 95%, with little change in the number of receptors.
Figure 2.
Modulation of multivalent adhesion by changing the number of receptors at the adhesion interface. (A) The fraction of bound virus particles as a function of the number of receptors at the adhesion interface. (B) NR,0.5 (solid lines), NR,0.95 (dashed lines above), and NR,0.05 (dashed lines below) values (see text) as a function of the number of ligands at the adhesion surface. The gap between the two dashed lines narrows as NL increases, indicating that at higher valencies it takes a smaller increase in the number of receptors to produce the same increase in the adhesion affinity. The number of receptors below which the equilibrium number of connections becomes no greater than 1 ( ≤ 1) is also shown (dash-and-dot lines). (C) The equilibrium number of connections, , when the fraction of bound virus particles is 0.5 (solid lines), 0.95 (dashed lines), and 0.05 (dashed lines below). (D) The probability, p0, of spontaneously breaking all receptor-ligand connections plotted against the equilibrium number of connections, , at varying numbers of receptors, NR, while the number of ligands is held constant, at NL = 150. The black dashed curve corresponds to the limit p0 = exp(−). Each color of lines (B–D) corresponds to a different receptor-ligand binding affinity KD. The parameters used here are [cell] = 4.0 × 10−10 mM, Veff = 2.7 × 103 mM−1, and K0 = 1.0 × 10−5 mM; the derivation of these parameter values is explained in the discussion of cell-influenza adhesion.
To break apart an adhesion interface, it is not necessary to reduce the equilibrium number of receptor-ligand connections to = 0; when becomes small enough, thermal fluctuations can spontaneously break all connections—so that the instantaneous number of connections is 0—which abolishes the adhesion. Without specific receptor-ligand connections, cells (and viruses) tend to repel each other, due to similar charges on their surfaces (15), such that K0/[cell] >> 1. Consequently, adhesion can be disrupted even when p0 is still very small, and a few receptor-ligand connections persist in binding equilibrium at the interface. At different values of KD and NL, can take different values when half of the virus particles are bound (Fig. 2 C). Conversely, the number of receptors at which most of the virus particles are unbound (f = 0.05) differs from the value below which ≤ 1 (Fig. 2 B). Our model will thus predict different and likely more accurate conditions under which adhesion is abolished than previous models, which consider that condition to occur when = 0 (14).
If the affinity between the receptor and the ligand is sufficiently weak, the equilibrium number of connections is small (i.e., << min(NR, NL)), and Eq. 9 can be well approximated by p0 ≈ exp(−). Under such conditions, the adhesion affinity is (KD,ad)−1 ∝ exp() ≈ 1 + ≈ , thus approximately agreeing with a previous suggestion that took adhesion affinity to be proportional to (16). At moderate receptor-ligand affinities, however, p0, and thus the adhesion affinity, cannot be estimated from alone (Fig. 2 D).
Adhesion of influenza virus
We now apply our model to the analysis of the binding of influenza virus. The influenza virus uses its surface glycoprotein HA to bind to terminal SA moieties of carbohydrate chains on the host-cell surface. SA is frequently referred to as the receptor, and HA as the ligand. HA of avian influenza (bird flu) strains preferentially binds to SA that is in α2,3 linkage to its neighboring sugar (galactose) in carbohydrate chains (characteristic of cells in the avian enteric tracts), whereas HA of influenza strains transmissible in mammals, including humans, needs to bind to SA in α2,6 linkage to galactose (characteristic of human trachea airway epithelia) (28, 29). Mutations in the HA sequence can cause a change in its binding preference from α2,3 linkage to α2,6 linkage, enabling transmissibility of the virus from birds to mammals (30), as was demonstrated in highly pathogenic H5N1 influenza strains (37, 38).
Adhesion to SA-conjugated biosensors
We will first use our model to analyze the adhesion of influenza virus to SA-conjugated biosensors. This simplified experimental system, which measures the binding of free virus to immobilized SA, has been used to give insights relevant to the attachment of influenza virus to vertebrate cells and, in particular, to help address the question of the species specificity of different viral strains.
Recently, Xiong et al. (19) measured the binding affinities between HA in various influenza strains and SA with α2,3 and α2,6 linkages. The authors also measured the adhesion affinities of the whole viruses of these strains (except for a ferret-transmissible mutant of an H5N1 virus) to SA attached to biosensors, and they demonstrated that a modest change in the receptor-ligand binding affinity is magnified to a much larger change in the viral adhesion to the cell surface. Using the empirical model KD,ad = (KD)m, where the value of m depends on the SA concentration, and where its value at each SA concentration is estimated from ln(KD,ad)/ln(KD) using data at multiple HA concentrations, the authors predicted the binding isotherms of the ferret transmissible mutant virus and suggested that the small changes in the HA-SA binding affinities were sufficient to switch the binding preference of the virus from avian receptors to human receptors.
We analyzed the adhesion measurements by Xiong et al. (19) using Eq. 1. To validate our model, we fit the two parameters, K0 and Veff, to three of the measured adhesion isotherms and used the parameters to predict the remaining three measured isotherms. These isotherms relate the amount of attached viral particles and the relative sugar loading (i.e., the amount of SA as a fraction of the amount at maximum loading) on the surface. The fractional saturation of the biosensor by the virus is given by
| (20) |
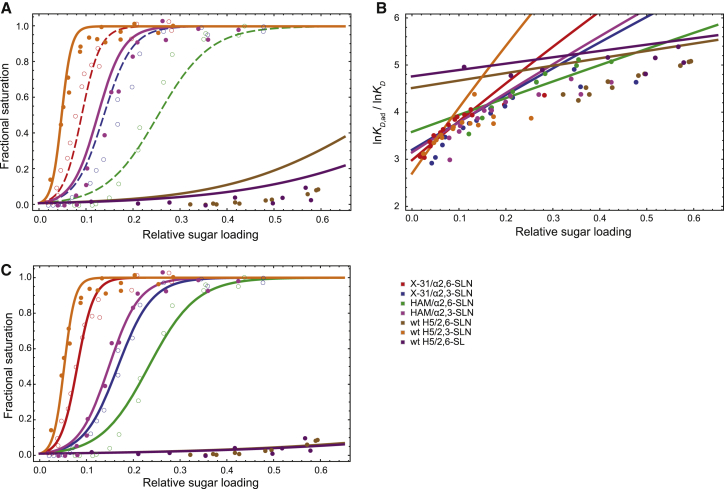
where [virus] = 10−7 mM is the solution concentration of the virus used in the experiment, and KD,ad varies according to Eq. 1 with respect to the relative sugar loading. The values of the fitted parameters are summarized in Table 1, and the resulting adhesion isotherms are shown in Fig. 3 A. To compare our model to the empirical model KD,ad = (KD)m, which is used by Xiong et al. (19) in their analysis of the data, we computed the multiplicity m = lnKD,ad/lnKD at different relative sugar loading values for each HA-SA pair, using the KD,ad value estimated from the experiment (by Eq. 19) and from our model (by Eq. 1). The results are shown in Fig. 3 B. As expected, the multiplicity varies with both the relative sugar loading and the individual KD; this variation is reasonably well captured by our model.
Table 1.
Values of the model parameters for influenza virus-host cell adhesion
| NHA | NSA | K0 (mM) | Veff (mM−1) |
|---|---|---|---|
| 3 × 50 | 2000 | 1.0 ± 0.5 × 10−5 | 2.7 ± 0.3 × 103 |
The parameters K0 and Veff are fitted to three experimentally measured isotherms. NHA is the number of HA monomers, and NSA is the number of SA receptors at maximum sugar loading. The number of ligands in our model is then NL = NHA, and the number of receptors NR = NSA × relative sugar loading. In principle, NHA and NSA can be treated as fitting parameters as well. The fitting problem then becomes underrestrained, however, given the small amount of isotherm data, and there are large statistical uncertainties in the fitted NHA, NSA, and K0 parameters. Instead, we used rough estimates of NHA and NSA from previous publications. It has been estimated (19) that 50 HA trimers per virus participate in forming adhesive connections, and thus NHA ≈ 3 × 50. The estimate for NSA ranges from 600 to 4500 (19, 32, 39). We used an intermediate value. Different values of NSA used in fitting result in compensatory values of Veff, but the consequent isotherms are little changed. More accurate measurements of NHA and NSA can help determine more reliably the parameters K0 and Veff. We have also fit the parameters K0 and Veff using the exact solution (Eqs. 3 and 4), obtaining the same values.
Figure 3.
The agreement between our model and experimental measurements of adhesion. (A) Isotherms of adhesion between influenza virus and biosensors loaded with SA. (Lines) Isotherms predicted by our model; (circles) experimental results. Different colors correspond to different combinations of viral strains and α2,3-SA or α2,6-SA. (Dashed lines and open circles) The three isotherms used for fitting the model parameters; (solid lines) predictions of isotherms for which experimental measurements are available. (B) Multiplicity of adhesion, m = lnKD,ad/lnKD, as a function of the relative sugar loading for different HA-SA pairs. The variation of m across different relative sugar-loading values is reasonably well captured by our model. (C) Computed and measured isotherms of adhesion, with KD values refitted to each individual isotherm, using the parameter values of K0 and Veff in Table 1. The fractional saturation predicted by our model is in good agreement with the experimental measurements, with fitted KD values close to those measured by MST. X-31, HAM, and wild-type (wt) H5 are different influenza strains, each with a different HA sequence and thus different binding affinities for sialic acids (19) (Table 2). α2,3-SLN (α2,3-linked sialyl lactosamine), α2,6-SLN (α2,6-linked sialyl lactosamine), and α2,6-SL (α2,6-linked sialyl lactose) are different terminal sialic acid moieties.
The KD values of HA-SA binding, used in the fitting and prediction, are taken from the MST measurements by Xiong et al. (19) and are listed in Table 2. To further assess the quality of our model, we took the K0 and Veff values from the fitting and refit the dissociation constant, KD, of each individual HA-SA binding to the corresponding adhesion isotherm (Fig. 3 C). The KD values obtained from the adhesion isotherms are in good agreement with the values determined from MST (Table 2). This suggests that our model may be used to estimate the binding affinities from adhesion experiments. Such an approach may be advantageous when the adhesion affinity is easier to measure than the receptor-ligand binding affinity (e.g., when the receptor-ligand binding is too weak to be detected by the binding assays).
Table 2.
The dissociation constants, KD, of HA-SA binding for various viral strains and SA
| Viral Strain | SA | KD (mM) (MST) | KD (mM) (Refitted) |
|---|---|---|---|
| X-31 | α2,6-SLN | 2.1 ± 0.3 | 1.85 ± 0.04 |
| X-31 | α2,3-SLN | 3.2 ± 0.6 | 3.88 ± 0.07 |
| HAM | α2,6-SLN | 5.9 ± 0.7 | 5.4 ± 0.2 |
| HAM | α2,3-SLN | 2.9 ± 0.3 | 3.44 ± 0.08 |
| wt H5 | α2,6-SLN | 17 ± 3 | 34 ± 4 |
| wt H5 | α2,3-SLN | 1.1 ± 0.2 | 1.20 ± 0.03 |
| wt H5 | α2,6-SL | 21 ± 6 | 37 ± 6 |
The last column lists the KD values estimated by refitting them to the virus adhesion isotherms, using the K0 and Veff parameters shown in Table 1.
We can check that the fitted value of K0 is reasonable by using Eq. 14. Assuming that 10% of the virus surface is in contact with the cell surface (19) (i.e., NHA/MHA ≈ 0.1), we can estimate the fraction of the SA molecules per adhesion site that lie within contact interface to be NSA/MSA = K0 Veff MHA/NHA ≈ 0.3, a reasonable estimate for the SA-loaded surfaces considered in this work, which are relatively flat compared to the virus—more of the available SA molecules per site of adhesion on the cell thus lie within the interface.
Adhesion to cells
Our model can be used to analyze the implications of multivalent adhesion in the biological function of influenza virus and the competitive inhibition of influenza adhesion needed to effectively reduce influenza infection. Unlike biosensors, which are conjugated with a pure form of SA-terminated oligosaccharide, a real vertebrate cell presents a diverse set of SA-terminated glycans with different affinities for HA molecules. To characterize the adhesion of influenza virus to real cells, we ideally need the distribution of binding affinities, N(K), of the surface glycans to the HA molecules as input for the model. We also ideally need to fit the values of K0 and Veff to a series of measured adhesion affinities between the cells and the influenza virus. The necessary experimental data are not available, however, and so we make some reasonable assumptions and approximations to enable us to apply our model to cell-influenza adhesion. Although some quantitative predictions of our model will improve as new experimental data yield more realistic parameters, some of the predictions will remain essentially unchanged with respect to changes in experimental inputs.
In the following examples, our model is applied to determine the condition at which adhesion is dramatically altered (e.g., the value of KD above which the adhesion is effectively abolished). Here, we discuss why such a condition is insensitive to the value of K0, which may be different between adhesion to cells and adhesion to biosensors because of the difference in the nonspecific interactions of the different adhesion surfaces. According to Eq. 19, the population of bound virus changes rapidly around KD,ad ≈ [cell]. The predicted parameter θ—which may be KD, NR, NL, or the concentration of hemagglutinin inhibitor (see below)—corresponding to this condition thus changes with K0 as dθ/dlnK0 = −(dlnKD,ad/dθ)−1. In our model, because dlnKD,ad/dθ is large (e.g., a small change in KD can lead to orders-of-magnitude change in KD,ad, as discussed below), dθ/dlnK0 is small. This insensitivity to the value of K0 will be demonstrated in the later discussion on the viable range of HA-SA binding. We will thus continue to use its value determined above from the biosensor experiments.
The parameter Veff appears as a scaling factor of KD, adjusting the affinity for the confined geometry in which receptor binding takes place. As such, the predictions of our model would remain valid, and the adhesion curves would look identical, for a different value of V′eff, if we simply replace the binding affinity KD with K′D = KDVeff/V′eff. The value of Veff in Table 1 will be used in the subsequent discussions, but it will be straightforward to update the conclusions should new experimental measurements on cell-to-influenza virus adhesion permit the determination of a more accurate value of Veff for cells.
Experimental data for the distribution of binding affinity N(K) is not yet available. We will first apply our model using the assumption that all SA molecules on a cell bind to the HA molecule with the same binding affinity; later, we examine the sensitivity of the results to changes in the distribution of binding affinity.
Multivalency amplifies the weak binding between HA and SA to achieve strong affinity between the influenza virus and the host cell. Assuming that the host cells are closely packed against one another, with an individual diameter of 20 μm, the cell density is then approximately [cell] = 4 × 10−10 mM. We can estimate the fraction of virus particles that are bound to the host cells using Eq. 19. The red curve in Fig. 4 shows how the fraction of virus particles bound to host cells changes with HA-SA binding affinity. Even when the affinity is as weak as KD = KD,max ≈ 10 mM (KD,max is determined by solving for KD in Eq. 19 with f = 0.5), the majority of virus particles are still bound to host cells. The fraction of bound virus particles increases steeply (by >100-fold) as KD decreases from 20 to 5 mM, allowing the virus to gain affinity to and thus to infect new host species by small changes in its HA sequence.
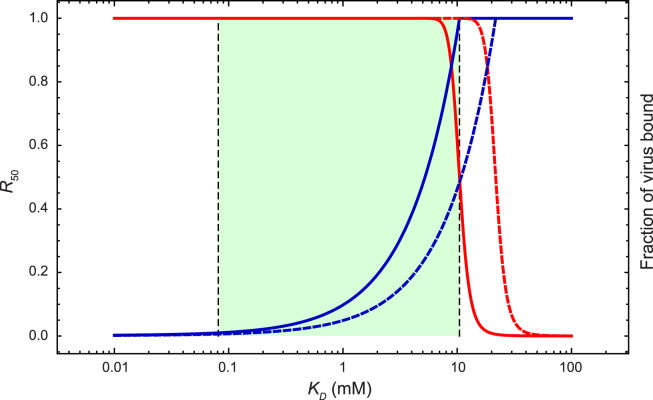
Figure 4.
Dependence of virus adhesion on HA-SA binding affinity. (Red curve) Fraction of virus particles bound to host cells versus KD of HA-SA binding. (Dark-blue curve) R50 (see text) versus KD. (Light-green region) Interval of viable KD values such that the virus can 1) attach sufficiently to and infect cells and 2) release itself, with the help of neuraminidase, from the cells to spread the infection. The upper bound of the region corresponds to the KD value at which half of the viruses attach to cells; the lower bound corresponds to the KD value at which 99% of all SA moieties must be removed in order for half of the viruses to detach from the cells. (Solid curves) Results from Eqs. 1 and 2, which assume that all SA receptors on the cell bind to the HA molecule with the same affinity KD; (dashed curves) results from Eqs. 16–18, which assume that the binding free energy is uniformly distributed between RTln(KD/10) and RTln(KD10).
A nascent influenza virus emerging from a host cell must detach itself from that cell to spread the infection to other cells. To overcome the strong multivalent adhesion, the virus uses neuraminidase to cleave the SA moieties at the adhesion interface (26, 27), reducing the number of SA molecules available and thus the affinity of adhesion. The stronger the binding between HA and SA, the more SA neuraminidase needs to cleave for effective release of the virus. We define a quantity R50, which is the factor by which the population of SA molecules on the surface needs to be reduced for half of the virus particles to be unbound. The blue curve in Fig. 4 plots R50 against KD. At KD = KD,min ≈ 0.08 mM, R50 = 0.01 (KD,min is determined by solving for KD in Eq. 19 with f = 0.5 and NR = R50 NSA = 0.01 NSA), which implies that neuraminidase will need to remove 99% of all SA moieties to release half of the virus particles from the host cell. The enzymatic efficiency of neuraminidase thus imposes a lower limit of viable KD in HA-SA binding. On the other hand, the virus can compensate for inefficient neuraminidase, and develop resistance to neuraminidase inhibitors (40), with mutations in HA that decrease HA-SA affinity (41, 42).
Our model suggests that KD falls within a range that satisfies the dual requirements that the virus be able to attach to host cells and, with the aid of neuraminidase, to detach from them. Values of the parameters—namely the percentage of SA moieties neuraminidase can remove before the destruction of the immobilized virus on the cell surface and the fraction of nascent viral particles that need to detach for successful establishment of influenza within a host—are unfortunately not known, but using the speculative values discussed above yields a permissible range of KD from our model of KD,min = 0.08 to KD,max = 10 mM. This estimated range is in line with what has been measured experimentally (12, 19). The specific range estimate can vary depending on the assumed values of the parameters in our model. This dependence on the parameters—including the discrete NR and NL—is easy to analyze: differentiating Eq. 19 using the chain rule, then substituting the numerical values of the current parameters, we obtain
| (21) |
and
| (22) |
which suggest that our estimated range of viable KD has relative errors comparable to those in the estimates of NHA and NSA, but is insensitive to the parameters K0 and [cell]. Increasing K0 by a factor of 1000, for example, changes the estimated viable range by only a factor of ∼2, to KD,min = 0.04 mM and KD,max = 6 mM. Although the predicted values of KD,min and KD,max depend on Veff, the width of the viable range (i.e., KD,max/KD,min) does not.
Different cells will have different SA densities on their surfaces, and thus different numbers of receptors at the interface. The fraction of bound virus particles, however, changes steeply over a narrow range of NR, and remains essentially constant at ∼0 for NR below this range and at ∼1 for NR above this range (Fig. 2 A). This allows us to divide the cells into two populations, one with an NR above the midpoint of the transition range and the other with an NR below it, and to consider only the adhesion of virus to the first population. The adhesion can thus be analyzed using a cell concentration adjusted by the fraction of the first population. As shown above in Eqs. 21 and 22, the predictions of our model are insensitive to the value of [cell], suggesting that the predictions of our model will remain mostly valid despite the variability in SA densities among different cells. We present in the Supporting Material a detailed analysis of viral adhesion to a population of cells with varying numbers of interface receptors; KD,max obtained from this analysis is essentially the same as that calculated for adhesion to cells with a uniform number of interface receptors.
The above analysis assumes that all SA molecules on the cell surface bind to the HA molecule with the same binding affinity KD. On the surface of a real cell, SA moieties in different glycans may bind to HA with different affinities KD. We now discuss the effect of such heterogeneity.
We examine a simple, concrete example, in which the HA-SA binding affinity spans two orders of magnitude from Kmin to Kmax = 100 Kmin, the binding free energy is uniformly distributed from RTlnKmin to RTlnKmax, and the total number of SA molecules is NSA. The distribution is then N(K) = NSA(ln(Kmax/Kmin))−1 K−1. Defining the mean affinity K′D ≡ , the value of K′D then determines the distribution N(K) (e.g., K′D = 1 mM signifies that the binding affinity ranges from Kmin = 0.1 mM to Kmax = 10 mM, with the binding free energy uniformly distributed in the corresponding range). We can compute the fraction of bound viral particles and the R50 parameter at different values of K′D (the dashed blue and red curves, respectively, in Fig. 4). Compared to the case of homogeneous receptors, the viable range of the mean affinity is shifted by a factor of ∼2, but the width of the range is similar (K′D,max/K′D,min = 137 versus KD,max/KD,min = 129). It will be straightforward to incorporate more realistic estimates of N(K) in our model as innovative experimental techniques, such as glycan microarrays (43), more comprehensively characterize the HA-binding properties of diverse glycans on real cells.
HA of avian influenza preferentially binds SA of α2,3 linkage, whereas HA of human influenza preferentially binds SA of α2,6 linkage. Due to multivalency in the adhesion, a small difference in the binding affinity can be amplified to a big difference in biological adhesion. The X-31 HA (an extensively studied HA construct), for instance, binds α2,6 SA with KD = 2.1 ± 0.3 mM, and it binds α2,3 SA with KD = 3.2 ± 0.6 mM (13). The ability of the whole virus to adhere to cells, in contrast, can differ drastically between cells with surface SA of α2,3 linkage and those of α2,6 linkage: Assuming the parameters in Table 1, the ratio of the adhesion constants is = 106 ± 3. As another example, the wild-type HA of the avian strain A/Vietnam/1194/2004 binds α2,6 SA with KD = 17 ± 3 mM (19). A mutant HA was identified in a strain derived from A/Vietnam/1194/2004 that is transmissible in ferrets. This mutant HA, which contains a handful of point mutations, binds α2,6 SA with KD = 12 ± 2.5 mM (19). According to our model, this 1.4-fold increase in the binding affinity will lead to a much larger increase in the adhesion affinity, as = 101.1 ± 0.9, and thus potentially a much higher fraction of virus particles bound to the mammalian cells. Our model thus quantitatively predicts that small changes in the HA-SA binding affinities might be sufficient to switch the binding preference of the virus from avian cells to human cells, and that a small gain in the hemagglutinin’s affinity for human receptors may sufficiently increase the virus’s adhesion to human cells to enable human infection.
There has been a longstanding interest in developing HA inhibitors as potential therapeutic agents against influenza (31, 44, 45). A competitive inhibitor I of HA forms complexes HAI in the reaction
| (23) |
where KI = [HA][I]/[HAI] is the equilibrium dissociation constant, and reduces the number of available HA by the number of complexes formed, NHAI. The presence of the inhibitor can be included in our model through the following set of simultaneous equations for NHAI and :
| (24) |
and
| (25) |
Solving for NHAI in terms of NHA − and KI /[I] from the second line of Eq. 24, and substituting it into Eq. 25, we can show that KD,ad has the same expression as in Eq. 1, but with the value of determined by Eq. 24.
The above equations can be derived from the partition function of the system
| (26) |
where is the excess chemical potential of the HA-inhibitor complex. In the above, we assume that the inhibitor is in excess such that the free inhibitor concentration [I] does not depend on the number of HA-inhibitor complexes formed. The probability of spontaneously breaking all HA-SA connections (i.e., n = 0) is then
| (27) |
By replacing Z with the largest term in the summand of Eq. 26, which corresponds to m = NHAI and n = in Eq. 24, and applying the Stirling approximation, we arrive at Eq. 25. The approximation is quite accurate: the ratio of the approximate value and the exact value of p0 stays close to 1 across a large range of inhibitor concentrations (Fig. 1 B).
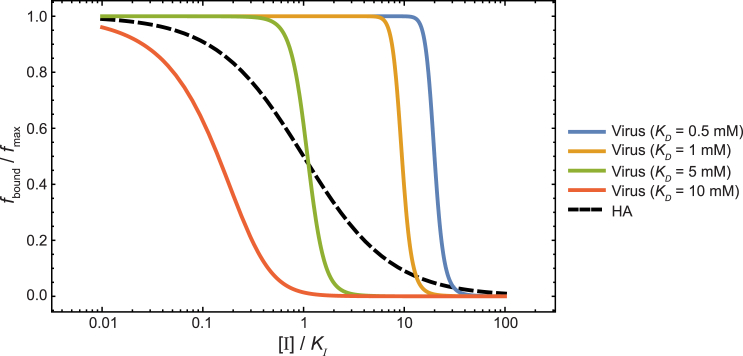
Inhibition of the multivalent adhesion depends on inhibitor concentration in a very different manner than inhibition of monovalent binding, as shown in Fig. 5, in which the bound fraction of virus particles, normalized by the maximum bound fraction (at [I] = 0), is plotted against the inhibitor concentration [I]/KI. The normalized fraction of HA bound to SA at different inhibitor concentrations, which is given by fbound/fmax = 1/(1 + [I]/KI), if the inhibitor binds to HA with much higher affinity than SA (i.e., KI << KD), is also shown to contrast the difference between inhibition of monovalent binding and of multivalent adhesion. Whereas the normalized fraction of HA bound with SA decreases gradually with increasing [I]/KI, reaching half at [I] = KI, the normalized fraction of bound virus drops precipitously at different [I]/KI values, depending on the affinity between HA and SA. In viral strains with high HA-SA affinity KD = 0.5 mM, adhesion is half-inhibited only when [I]/KI = 20. On the other hand, once [I]/KI exceeds that value, the normalized fraction steeply decreases, reaching fbound/fmax = 0.001 at [I]/KI = 66. The inhibitor concentrations [I]/KI required to reduce the virus adhesion to given fbound/fmax values are computed for different HA-SA affinities and shown in Fig. 6.
Figure 5.
Inhibition of multivalent adhesion and of monovalent binding. (Solid lines) The normalized fractions of bound viruses at different inhibitor concentrations for viruses of different HA-SA binding affinities. (Dashed line) Fraction of HA bound to SA at different inhibitor concentrations.
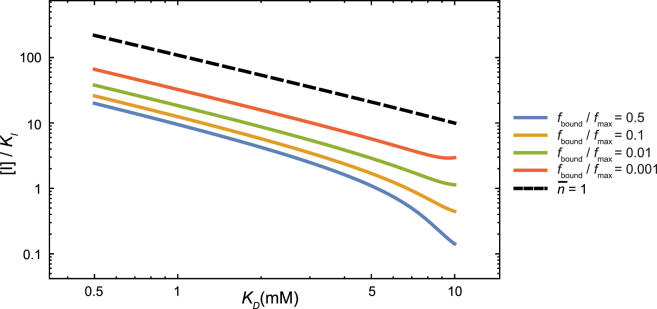
Figure 6.
Inhibitor concentrations required to reduce the virus adhesion to given normalized fractions for different HA-SA affinities. Even when the affinity of receptor-ligand binding is high, a moderate inhibitor concentration relative to the inhibition constant is sufficient to drastically reduce viral adhesion. (Dashed line) Inhibitor concentration required to reduce the equilibrium number of connections, , to <1. The inhibitor concentration required for ≤ 1 is greater than that required to reduce the fraction of bound virus by 1000-fold, which suggests that adhesion can be effectively inhibited before ≈ 0.
Our analysis above suggests that inhibitors of multivalent adhesion should be assessed differently than inhibitors of monovalent binding. Consider, for example, the challenges facing the search for effective HA inhibitors: the receptor-binding pocket of HA is very shallow, making it difficult for any small molecule to achieve high-affinity (i.e., nanomolar) binding. It has been hypothesized that a strong HA inhibitor is required to counter the strong adhesion due to multivalency, yet to our knowledge no quantitative estimate has been made as to the inhibitor potency required for effective inhibition of adhesion. According to our model, a relatively weak HA inhibitor with good bioavailability can effectively inhibit virus attachment. At a typical plasma concentration of [I] = 10 μM, for example, an inhibitor of submicromolar affinity (KI = 0.1 μM) is predicted to reduce the virus binding by ∼3000-fold, even for viral strains with HA of high SA-binding affinities (KD ∼ 0.5 mM).
The role of binding cooperativity in multivalent adhesion
Equation 1 is predicated on the constancy of the affinity of individual receptor-ligand binding throughout the adhesion interface. The binding affinity of a receptor-ligand pair, however, may vary with the position of the receptor-ligand pair within the interface, and it may also be affected by existing receptor-ligand connections. The latter dependence is often referred to as cooperativity. Here we discuss the implications of cooperativity and position-dependent affinity in multivalent adhesion.
In general, the free energy change of forming n labeled and distinguishable receptor-ligand connections can be written as
| (28) |
where ri is the position of the ith receptor-ligand pair, and g(r1,r2,···,rn) is the free energy change of forming n connections in the corresponding positions. (We note that because Eq. 28 refers to distinguishable receptor-ligand connections, there is no factor of 1/n! on the right-hand side.) Without better experimental and theoretical characterization of the microscopic details of the adhesion interface, the function G(n) is not known for realistic biological adhesion interfaces, and it is necessary to make some approximations in our analysis.
The study of adhesion is most often concerned with the conditions under which adhesion is established or abolished, corresponding to conditions where the bound fraction is f ≈ 0.5. We will show that the effect of cooperativity on such conditions diminishes as the numbers of receptors and ligands at the adhesion interface become large. Under such conditions, the probability, p0, of spontaneously breaking all receptor-ligand connections cannot be vanishingly small, and, according to the above analysis, the equilibrium number of connections must be small (Fig. 2). If NR and NL are large, << min(NR, NL), the small number of formed receptor-ligand pairs will almost never be close to each other, and thus the binding affinity of one pair will not be affected by other pairs. In such cases, we can ignore the multibody terms in g, and approximate it as
| (29) |
and
| (30) |
where ΔG = −RTln(∫drexp(−g1(r)/RT)) is a constant. The details of g1(r) determine Veff, the value of which is obtained by parameter fitting in our model. Equation 1, and thus all the quantitative results of our model, remain unchanged when the receptor-ligand affinity varies with the position in the adhesion interface.
To quantify the effect of cooperativity, we consider a special and commonplace case, which is pertinent to the study of influenza-cell adhesion: cooperativity within oligomeric receptors or ligands. The effect of such cooperativity on the equilibrium number of receptor-ligand connections at the cell-cell adhesion interface has been previously investigated (18); here we study its effect on adhesion affinity. HA is a homotrimer. Each monomer can individually bind an SA molecule, and its affinity may depend on the number of SA molecules already bound to the same HA trimer. If the binding of k = 1,2,3 SA molecules to the same HA trimer corresponds to the free energy change of ΔGk, the probability of breaking all connections is given by
| (31) |
where is the equilibrium number of HA trimers bound to k = 1,2,3 SA molecules, is the equilibrium number of SA molecules bound to HA, and = NHA/3 is the total number of HA trimers. The values of are given by the following equilibrium condition:
| (32) |
Equations 31 and 32, derived in the Supporting Material, account for the cooperativity in the binding of SA molecules to different monomers within a HA trimer. In the absence of cooperativity, ΔG3/3 = ΔG2/2 = ΔG1 = ΔG, and Eqs. 31 and 32 can be shown to reduce to Eq. 1.
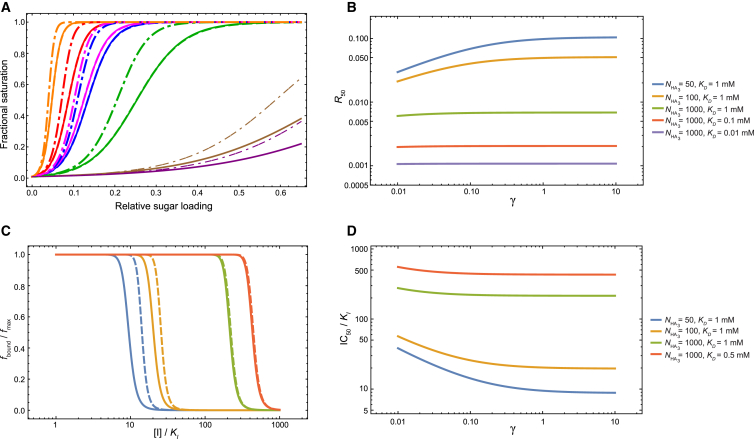
We introduced into our model the cooperativity between SA molecules binding to the same HA trimer by the coefficient γ, such that the binding affinity of SA to the first HA monomer is KD Veff, but the subsequent binding affinities to the remaining two HA monomers in the same trimer are KD Veff γ (i.e., ). Attempting to derive the parameter γ by fitting the model to the experimental isotherms yields large statistical uncertainty on its value (the standard error for ln γ is 600), suggesting that these experimental isotherms do not provide sufficient information to infer the presence or the extent of the cooperativity embodied by our model. The fitted model (parameters given in the caption of Fig. 7 A) exhibits almost identical isotherms, as does the model without cooperativity (Fig. 7 A).
Figure 7.
The effect of binding cooperativity within the same HA trimer on the isotherms and the inhibition of adhesion. (A) Adhesion isotherms for the viral strains considered in this work. (Solid lines) Model without cooperativity (i.e., γ = 1), with the fit parameters in Table 1. (Dashed lines) Model refit with cooperativity, which yielded a value of γ with large statistical uncertainty. The parameters from the refitting are Veff = 2.6 × 103 mM−1, K0 = 1.2 × 10−5, and γ = 72. (Dashed lines are barely visible because they are almost on top of the solid lines, which indicates that the variations of the model that do and do not consider binding cooperativity produce very similar fits to the experiment.) (Dash-and-dot lines) Isotherms generated with the parameters in Table 1, but with γ = 0.1. (B) The dependence of R50 on the cooperativity parameter γ. (C) The effect of cooperativity on the inhibition of adhesion. (Solid lines) Inhibition of adhesion if there is no cooperativity (i.e., γ = 1); (dashed lines) inhibition of adhesion if the binding affinities of the second and third SA molecules are 10-fold stronger than that of the first SA molecule to the same HA trimer (γ = 0.1). (D) The dependence of IC50 values ([I]/KI at which the fraction of bound cells is half that in absence of the inhibitor) on cooperativity. The values of NSA, Veff, and K0 in Table 1 are used for (B)–(D). In (C), the cell concentration is as estimated in the text ([cell] = 4 × 10−10 mM). (B–D) Effect of cooperativity diminishes with the number of HA trimers.
To explore the extent to which cooperativity can affect the adhesion isotherms, we computed R50 as a function of γ, for various and KD values, holding the other parameters in our model to the corresponding values in Table 1 (Fig. 7 B). As expected, the effect of cooperativity diminishes as the number of HA trimers increases; when = 1000, the effect of cooperativity is negligible. At the assumed value = 50 for the influenza virus, cooperativity has a noticeable effect: R50 = 0.030 for γ = 0.01, R50 = 0.069 for γ = 0.1, and R50 = 0.098 for γ = 1 (KD = 1 mM). The adhesion isotherms of the various viral strains are plotted for γ = 0.1 as dash-and-dot lines in Fig. 7 A.
Cooperativity within the trimer can be similarly considered in the inhibition of adhesion. Denoting as the equilibrium number of HA trimers that have i = 0,1,2,3 inhibitors bound, and as the equilibrium number of HA trimers that have i inhibitors and j SA molecules bound, the probability of breaking all HA-SA connections is
| (33) |
with the following equilibrium condition:
| (34) |
The derivation of Eqs. 33 and 34 is outlined in the Supporting Material.
The binding cooperativity shifts the inhibition curve to higher inhibitor concentrations: given the parameters in Table 1, with = NHA/3 = 50, a typical HA-SA binding affinity of KD = 1 mM, and a cooperativity coefficient γ = 0.1, the inhibitor concentration necessary to reduce the adhesion by half is 1.5 times the concentration necessary if there is no cooperativity (Fig. 7 C). This shift, however, all but vanishes at large numbers of trimers (Fig. 7, C and D).
In all the discussions above, we have assumed that the HA inhibitor has the same inhibition constant against HA anywhere in the adhesion interface that it does against HA in the solution, which is likely a valid assumption for small-molecule HA inhibitors. If the inhibitor binding to HA is diminished or enhanced inside the adhesion interface, our model would underestimate or overestimate, respectively, the inhibitor concentration required to inhibit virus-cell adhesion.
Conclusions
Multivalent adhesion at biological surfaces often involves a very large number of simultaneous receptor-ligand connections, commonly on the order of hundreds to thousands. The adhesion affinity is the result of both receptor-ligand binding and nonspecific interactions. We have proposed a model for the adhesion affinity that takes into account both contributions. Our model gives a simple expression for the adhesion affinity by taking advantage of approximations appropriate for large numbers of receptors and ligands. We have shown that these approximations are accurate for a wide range of parameters common in biological adhesion.
We have applied our model to the analysis of the adhesion between influenza virus and SA-conjugated biosensors, and between influenza virus and vertebrate cells. Compared to experimental measurements, our model has reasonable quantitative accuracy using just two fitting parameters. Our model quantifies the high amplification that results from the high valency characteristic of many examples of biological adhesion: the high valency not only amplifies the weak receptor-ligand binding to produce high-affinity adhesion but also turns gradual changes in the conditions (e.g., receptor-ligand affinity, the number of receptors or ligands, and the concentration of competitive inhibitors) into abrupt transitions in adhesion. Analysis of the relationship between adhesion and receptor-ligand affinity in our model provides quantitative support for the notion that the range of observed HA-SA affinities is a natural consequence of the functional requirements for the influenza virus. When used to study the inhibition of adhesion by competitive inhibitors of HA, our model suggests that it might be feasible to inhibit adhesion with a relatively weak (0.1 μM in dissociation constant) HA inhibitor. Our model may aid the analysis of experiments and the design of therapeutic strategies involving cell-cell and cell-virus adhesion in general.
Author Contributions
H.X. and D.E.S. designed research; H.X. performed research and analyzed results; and H.X. and D.E.S. wrote the article.
Acknowledgments
We thank David Borhani, Robert Dirks, and Michael Eastwood for helpful discussions and critical readings of the article, and Rebecca Bish-Cornelissen and Berkman Frank for editorial assistance.
Editor: David Piston.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supporting Material is available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)01122-4.
Contributor Information
Huafeng Xu, Email: huafeng.xu@deshawresearch.com.
David E. Shaw, Email: david.shaw@deshawresearch.com.
Supporting Material
References
- 1.Chan D.C., Kim P.S. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 2.Stehle T., Harrison S.C. Crystal structures of murine polyomavirus in complex with straight-chain and branched-chain sialyloligosaccharide receptor fragments. Structure. 1996;4:183–194. doi: 10.1016/s0969-2126(96)00021-4. [DOI] [PubMed] [Google Scholar]
- 3.Wiley D.C., Skehel J.J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- 4.Harris T.J., Tepass U. Adherens junctions: from molecules to morphogenesis. Nat. Rev. Mol. Cell Biol. 2010;11:502–514. doi: 10.1038/nrm2927. [DOI] [PubMed] [Google Scholar]
- 5.Harrison O.J., Jin X., Honig B. The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure. 2011;19:244–256. doi: 10.1016/j.str.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ley K. The role of selectins in inflammation and disease. Trends Mol. Med. 2003;9:263–268. doi: 10.1016/s1471-4914(03)00071-6. [DOI] [PubMed] [Google Scholar]
- 7.Rosen S.D., Bertozzi C.R. The selectins and their ligands. Curr. Opin. Cell Biol. 1994;6:663–673. doi: 10.1016/0955-0674(94)90092-2. [DOI] [PubMed] [Google Scholar]
- 8.Grakoui A., Bromley S.K., Dustin M.L. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285:221–227. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- 9.Hynes R.O. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 10.Collins B.E., Paulson J.C. Cell surface biology mediated by low affinity multivalent protein-glycan interactions. Curr. Opin. Chem. Biol. 2004;8:617–625. doi: 10.1016/j.cbpa.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Mammen M., Choi S.-K., Whitesides G.M. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 12.Skehel J.J., Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 13.Sauter N.K., Bednarski M.D., Wiley D.C. Hemagglutinins from two influenza virus variants bind to sialic acid derivatives with millimolar dissociation constants: a 500-MHz proton nuclear magnetic resonance study. Biochemistry. 1989;28:8388–8396. doi: 10.1021/bi00447a018. [DOI] [PubMed] [Google Scholar]
- 14.Bell G.I. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 15.Bell G.I., Dembo M., Bongrand P. Cell adhesion. Competition between nonspecific repulsion and specific bonding. Biophys. J. 1984;45:1051–1064. doi: 10.1016/S0006-3495(84)84252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell G.I., Torney D.C. On the adhesion of vesicles by cell adhesion molecules. Biophys. J. 1985;48:939–947. doi: 10.1016/S0006-3495(85)83857-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dembo M., Bell G.I. The thermodynamics of cell adhesion. Curr. Top. Membr. Trans. 1987;29:71–89. [Google Scholar]
- 18.Dustin M.L., Starr T., Whitty A. Quantification and modeling of tripartite CD2-, CD58FC chimera (alefacept)-, and CD16-mediated cell adhesion. J. Biol. Chem. 2007;282:34748–34757. doi: 10.1074/jbc.M705616200. [DOI] [PubMed] [Google Scholar]
- 19.Xiong X., Coombs P.J., Gamblin S.J. Receptor binding by a ferret-transmissible H5 avian influenza virus. Nature. 2013;497:392–396. doi: 10.1038/nature12144. [DOI] [PubMed] [Google Scholar]
- 20.Hammes G.G., Wu C.-W. Kinetics of allosteric enzymes. Annu. Rev. Biophys. Bioeng. 1974;3:1–33. doi: 10.1146/annurev.bb.03.060174.000245. [DOI] [PubMed] [Google Scholar]
- 21.Huskens J., Mulder A., Reinhoudt D.N. A model for describing the thermodynamics of multivalent host-guest interactions at interfaces. J. Am. Chem. Soc. 2004;126:6784–6797. doi: 10.1021/ja049085k. [DOI] [PubMed] [Google Scholar]
- 22.Huskens J., van Bekkum H., Peters J.A. A convenient spreadsheet approach to the calculation of stability constants and the simulation of kinetics. Comput. Chem. 1995;19:409–416. [Google Scholar]
- 23.Kitov P.I., Bundle D.R. On the nature of the multivalency effect: a thermodynamic model. J. Am. Chem. Soc. 2003;125:16271–16284. doi: 10.1021/ja038223n. [DOI] [PubMed] [Google Scholar]
- 24.Klotz I.M. Ligand-receptor complexes: origin and development of the concept. J. Biol. Chem. 2004;279:1–12. doi: 10.1074/jbc.X300006200. [DOI] [PubMed] [Google Scholar]
- 25.Perlmutter-Hayman B. Cooperative binding to macromolecules. A formal approach. Acc. Chem. Res. 1986;19:90–96. [Google Scholar]
- 26.Palese P., Tobita K., Compans R.W. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology. 1974;61:397–410. doi: 10.1016/0042-6822(74)90276-1. [DOI] [PubMed] [Google Scholar]
- 27.Wagner R., Matrosovich M., Klenk H.-D. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev. Med. Virol. 2002;12:159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 28.Couceiro J.N., Paulson J.C., Baum L.G. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium; the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 1993;29:155–165. doi: 10.1016/0168-1702(93)90056-s. [DOI] [PubMed] [Google Scholar]
- 29.Kuiken T., Holmes E.C., Grenfell B.T. Host species barriers to influenza virus infections. Science. 2006;312:394–397. doi: 10.1126/science.1122818. [DOI] [PubMed] [Google Scholar]
- 30.Stevens J., Blixt O., Wilson I.A. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 31.Matrosovich M., Klenk H.-D. Natural and synthetic sialic acid-containing inhibitors of influenza virus receptor binding. Rev. Med. Virol. 2003;13:85–97. doi: 10.1002/rmv.372. [DOI] [PubMed] [Google Scholar]
- 32.Rosenberg S.A., Einstein A.B., Jr. Sialic acids on the plasma membrane of cultured human lymphoid cells. Chemical aspects and biosynthesis. J. Cell Biol. 1972;53:466–473. doi: 10.1083/jcb.53.2.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klotz I.M., Walker F.M., Pivan R.B. The binding of organic ions by proteins. J. Am. Chem. Soc. 1946;68:1486–1490. doi: 10.1021/ja01212a030. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe T., Kiso M., Kawaoka Y. Characterization of H7N9 influenza A viruses isolated from humans. Nature. 2013;501:551–555. doi: 10.1038/nature12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chandrasekaran A., Srinivasan A., Sasisekharan R. Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nat. Biotechnol. 2008;26:107–113. doi: 10.1038/nbt1375. [DOI] [PubMed] [Google Scholar]
- 36.de Vries R.P., Zhu X., Paulson J.C. Hemagglutinin receptor specificity and structural analyses of respiratory droplet-transmissible H5N1 viruses. J. Virol. 2014;88:768–773. doi: 10.1128/JVI.02690-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herfst S., Schrauwen E.J., Fouchier R.A. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imai M., Watanabe T., Kawaoka Y. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stray S.J., Cummings R.D., Air G.M. Influenza virus infection of desialylated cells. Glycobiology. 2000;10:649–658. doi: 10.1093/glycob/10.7.649. [DOI] [PubMed] [Google Scholar]
- 40.von Itzstein M. The war against influenza: discovery and development of sialidase inhibitors. Nat. Rev. Drug Discov. 2007;6:967–974. doi: 10.1038/nrd2400. [DOI] [PubMed] [Google Scholar]
- 41.Blick T.J., Sahasrabudhe A., McKimm-Breschkin J.L. The interaction of neuraminidase and hemagglutinin mutations in influenza virus in resistance to 4-guanidino-Neu5Ac2en. Virology. 1998;246:95–103. doi: 10.1006/viro.1998.9194. [DOI] [PubMed] [Google Scholar]
- 42.Kaverin N.V., Gambaryan A.S., Kropotkina E.A. Postreassortment changes in influenza A virus hemagglutinin restoring HA-NA functional match. Virology. 1998;244:315–321. doi: 10.1006/viro.1998.9119. [DOI] [PubMed] [Google Scholar]
- 43.Rillahan C.D., Paulson J.C. Glycan microarrays for decoding the glycome. Annu. Rev. Biochem. 2011;80:797–823. doi: 10.1146/annurev-biochem-061809-152236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Machytka D., Kharitonenkov I., Egge H. Methyl α-glycoside of n-thioacetyl-d-neuraminic acid: a potential inhibitor of influenza A virus. A 1H NMR study. FEBS Lett. 1993;334:117–120. doi: 10.1016/0014-5793(93)81694-u. [DOI] [PubMed] [Google Scholar]
- 45.Toogood P.L., Galliker P.K., Knowles J.R. Monovalent sialosides that bind tightly to influenza A virus. J. Med. Chem. 1991;34:3138–3140. doi: 10.1021/jm00114a025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.