OVERVIEW
Ross and Thompson’s 1910 report of periodic spikes of Trypanosoma gambiense parasitemia (1), made possible by the relatively large size of the parasite which allowed quantitation using light microscopy, was seminal in understanding how pathogens persist and led to later studies defining how trypanosomes and numerous other pathogens use antigenic variation to evade host immunity and clearance. Antigenic variation is a strategy used by a broad diversity of microbial pathogens, from small RNA viruses, notably the Human Immunodeficiency Virus (HIV), to large eukaryotic parasites with multiple chromosomes, illustrated by trypanosomal and malarial parasites, to persist within mammalian hosts (2,3). Using a variety of genetic mechanisms to generate antigenic variants, immune evasion results in the infected host serving as a microbial reservoir for subsequent transmission. Unlike respiratory and gastrointestinal pathogens that have essentially continual opportunities for transmission, arthropod vector-borne and sexually transmitted pathogens have episodic transmission opportunities. Correspondingly, both vector-borne and sexually transmitted agents are overrepresented among antigenically variant pathogens (3,4).
Pathogens utilize a breadth of genetic mechanisms to generate antigenic variants (3,4). The large progeny size and minimal proofreading capacity allow viruses to utilize random mutation to generate variants. In contrast, antigenically variant bacteria have evolved mechanisms utilizing a stable genome that safeguards progeny fitness. In this chapter, we focus on three well-characterized, highly antigenically variant bacterial pathogens: Anaplasma, Borrelia, and Neisseria. Collectively, these pathogens represent a diversity of bacterial genetic mechanisms utilized to create variation, structural and antigenic changes required for in host persistence, and escape phenotypes. While the focus is on intrahost antigenic variation, as opposed to evolution of new strain variants at the population level, the potential for the same mechanisms used to allow escape from immunity within the individual to allow escape at the population level is also presented.
ANAPLASMA
Introduction
In the same year, 1910, that Ross reported the aforementioned observation of recurrent, episodic spikes in trypanosome parasitemia, Arnold Theiler (later Sir Theiler) published a monograph in which he described the microscopically detectable appearance, disappearance, and then re-appearance of organisms “resembling to a certain extent, the bacteria” in the blood of infected cattle (5). Although 75 years would pass before molecular techniques would define the cyclic waves of bacteremia, Theiler clearly understood that the pathogen, which he termed Anaplasma for its lack of eosinophilic cytoplasm as observed in other tick-borne agents (which at that time were predominantly protozoa), persisted in the host and served as a reservoir for onward transmission (6). In the past decade the availability of complete genome sequences has driven discovery of the genetic mechanisms used to generate Anaplasma marginale, and later A. ovis and A. phagocytophilum, antigenic variants that escape immune detection and allow persistence. Although the occurrence of outer membrane structural variation resulting in immune evasion is common to numerous bacterial pathogens, study of Anaplasma marginale has uncovered two broadly applicable findings that will be the focus here. The first is the mechanism of segmental gene conversion to exponentially expand the capacity of a small, stable bacterial genome to generate thousands of outer membrane protein (OMP) variants. The second is the relevance of gene conversion and its permissiveness for allelic duplication and generation of novel OMP-encoding alleles that permits strain superinfection—essentially immune evasion at the level of the host population rather that within an individual. Strikingly, both these findings also apply broadly to African trypanosomes, highlighting the principle of convergent evolution and unifying the original observations made by Ross and Theiler 75 years ago.
The genus Anaplasma includes A. marginale, the type species, A. bovis, A. ovis, A. phagocytophilum, and A. platys (7). A. marginale and A. phagocytophilum are well-described tick-borne pathogens while the other three recognized species remain less studied and characterized, although there is evidence all members of the genus persist in their respective reservoir hosts and are tick-transmitted. A. marginale infects both wild and domestic ruminants while A. phagocytophilum infects multiple mammalian species (7). The severity of disease varies by host species and ranges from inapparent infection to severe acute febrile syndromes that progress to severe morbidity and mortality. A. marginale infections in most wild ruminants and Bos indicus breeds are asymptomatic or cause only mild disease. Acute infection of Bos taurus breeds commonly results in severe morbidity and can progress to fatal disease. A. phagocytophilum is also asymptomatic in many reservoir species but is responsible for Human Granulocytic Anaplasmosis, Canine and Equine Granulocytic Anaplasmosis, and Tick-borne Fever in ruminants (7). Antigenic variation, immune evasion, persistence, and transmission have been most completely studied in A. marginale and is the focus here. However, detailed studies, including complete genome sequencing, of A. phagocytophilum have supported common genetic mechanisms underlying antigenic variation (8–10) as have more limited studies for A. ovis and A. platys (11–12).
A. marginale persistence and transmission
Following initial tick-borne transmission into an immunologically naïve host, A. marginale replicates to >108 organisms per ml of blood (reviewed in 13). In surviving animals, the immune response controls this acute phase bacteremia but does not result in clearance but rather persistence (Fig. 1). Bacteremia levels decrease concomitant with the detection of IgG antibodies directed against A. marginale OMPs and resolve into a cyclical pattern composed of bacteremic waves ranging between 102–107 organisms/ml (14). Epidemiologically, persistence provides the reservoir for subsequent tick-borne transmission. As tick populations vary in presence, abundance, and activity seasonally and by macro- and micro-climatic variables, transmission opportunities are episodic rather than continuous. Consequently, persistence in the animal host is required for long-term survival of A. marginale and achieving persistence is a primary evolutionary force shaping the organism’s genome. Importantly, ticks that acquisition feed on infected hosts during the lower bacteremia levels associated with persistence still successfully acquire A. marginale (15–17). Subsequent replication within the tick salivary gland at the time of transmission feeding to a new host makes up for the initially lower number of organisms ingested and efficient onward transmission is achieved (17).
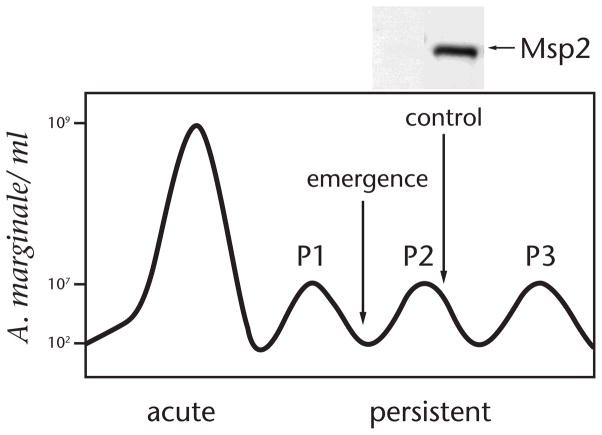
Figure 1. Emergence of Msp2 antigenic variants during cyclic A. marginale bacteremia.
A. marginale replicates to >108 organisms per ml during acute infection; the immune response does not completely clear the infection which persists in a series of sequential bacteremia peaks. Organisms in each peak express an antigenically variant immunodominant surface protein, Msp2, which is not recognized by existing antibody at the time of emergence (as illustrated by the IgG immunoblot for the peak 2 variant). Immune recognition of the variant results in clearance followed by emergent of novel variants in peak 3. Original data from ref. 14, 27.
Cyclic bacteremia and Omp variation
A combination of bioinformatics, immunologic, and proteomic approaches has identified ~60 Omps in A. marginale (18–22). Of these, two closely related proteins, Major Surface Protein (Msp)-2 and Msp3, are both highly abundant and highly immunodominant (12,22,23). The two proteins are similar in structure with a central surface exposed region flanked by hydrophobic membrane domains (24,25). Msp2 and Msp3 share a common C-terminus and likely were derived from a single ancestral gene. This is supported by the presence of only Msp2 in the closely related A. phagocytophilum but with an allelic repertoire that incorporates the complexity of the A. marginale Msp2/Msp3. In contrast to the other identified Omps in A. marginale, which are encoded by single copy genes and are invariant within a strain and during infection of the host, the surface exposed domains of Msp2 and Msp3 are encoded by multiple chromosomal alleles and recombination during infection generates structural and antigenic variants (18,26). Studies by French et al., established that the variants expressed in sequential cyclic bacteremic peaks were both structurally and antigenically unique; variants were unrecognized by existing antibody at the time of their emergence and replicated to form a new bacteremic peak (Fig. 1) (27). Using quantitative analysis, Brayton et al., confirmed that both Msp2 and Msp3 specific variants emerged in sequential peaks with dramatic loss of the previously predominant variant (28).
Antibody responses are preferentially directed against the central surface exposed domains, representing the structurally and antigenically unique regions of Msp2/3 and designated as the hypervariable region (HVR) (29,30). Correspondingly, there is minimal antibody recognition of the conserved membrane domains (29,30). The variant-specific IgG antibody responses are highly dynamic with antibody against the HVR being remarkably short-lived as compared to the responses against other conserved A. marginale Omps (30). This short-lived response was unexpected given the richness of CD4+ T cell epitopes in both variant-specific HVRs and the flanking conserved domains (29,31), the latter would be predicted to provide abundant T cell help for HVR-directed B cell responses due to the principle of linked recognition. However a series of studies conducted by Wendy Brown and colleagues demonstrated that Msp2-specific CD4+ T cell responses, which can be strongly induced by immunization, were suppressed during A. marginale bacteremia and only restored following antibiotic-mediated clearance of the organism (32–34). Although the mechanism of suppression remains incompletely defined, these findings suggest that the ability of Msp2 to evade immune recognition and clearance involves both the structural variation and immunologic modulation.
Genomic structure and mechanisms underlying A. marginale antigenic variation
Although Msp2/3 antigenic variation had been identified prior to A. marginale genome sequencing, the completion of the first strain sequenced, the St. Maries strain, fully identified the allelic repertoire responsible for variation and allowed dissection of the mechanisms for long-term persistence (18). While only a single full-length Msp2/3 is expressed at one time, using a single operon-linked expression site, multiple alleles encoding unique HVRs are maintained within widely dispersed chromosomal loci (Fig. 2A). The non-expression site alleles are each composed of a variable sequence encoding a unique HVR flanked by highly conserved 5′ and 3′ sequences that are identical to those of the single expression site copy (26). These alleles lack elements for transcription and translation and are silent in their loci: recombination into the expression site, mediated by the conserved flanking sequences, is required for expression of the unique HVR. Recombination, which occurs by gene conversion and retains the allelic donor unchanged, results in the expression site encoding a structurally and antigenically unique HVR, replacing the pre-existing Msp2/3 and allowing evasion of immune clearance and replication to form a new bacteremic peak. Detailed tracking of variant progression over time revealed that there is no set order in use of the potential donor alleles, each is capable of being recombined to the expression site and generating a replication competent A. marginale variant (35). The position of the locus itself relative to the expression site or origin of replication has no directive effect on variant progression, rather the encoded HVR itself is deterministic, consistent with the importance of immune escape (36).
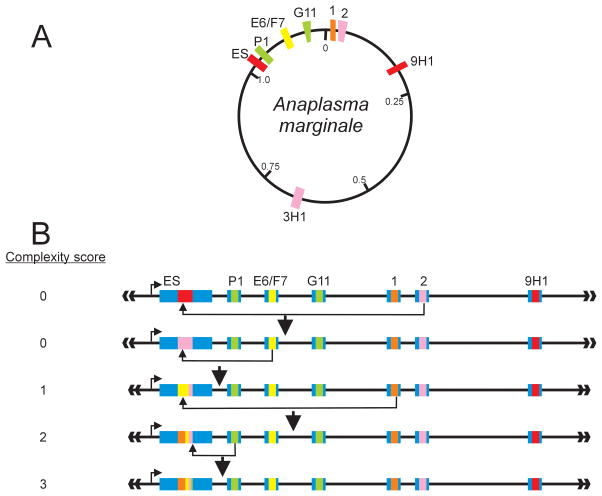
Figure 2. Generation of A. marginale Msp2 variants through gene conversion.
A) Genomic structure of the A. marginale msp2 donor and expression site loci. The 1.2-Mb genome is a single circular chromosome and encodes a single Msp2 expression site (ES). Multiple msp2 donor alleles, silent in their chromosomal loci, encode unique hypervariable regions (HVR), which are expressed only when recombined into the expression site. The allelic repertoire shown is for the St. Maries strain; alleles encoding a unique HVR are shown in different colors; duplicated alleles (2, 3H1; 1, 9H1) are shown in the same color. The expression site msp2 variant, in this example, corresponds to that encoded by the full 9H1 (or 1) allele. B) Gene conversion generates unique msp2 expression site variants. Sequential rounds of recombination result in replacement of the existing expression site copy by either a whole allelic donor sequence (as shown in the first gene conversion event in which allele 2 is the donor) or progressive modification of the existing expression site copy by segmental gene conversion. Over time, this process of segmental gene conversion generates complex expression site mosaics derived from multiple allelic donors and is reflected in a complexity score. The light blue regions flanking the expression site represent the conserved 5′ and 3′ domains; identical, truncated domains flank each donor allele and direct recombination. Figure and legend is from ref. 41 with permission.
Notably the A. marginale genome contains approximately 10–12 total unique msp2 and msp3 donor alleles (18,37), too few to account for the number of bacteremic peaks observed and required for years of persistence. This paradox was resolved by a series of detailed studies that identified segmental gene conversion as a combinatorial mechanism to exponentially expand the potential repertoire (Fig. 2B) (35,38,39). Recombination of an oligonucleotide segment derived from the donor allele hvr into the existing expression site hvr generates a novel, mosaic expression site msp2 not previously represented anywhere in the genome. Two further keys to understanding this mechanism were definition of how recombination was guided and how a mosaic could maintain minimal Msp2 structure. Futse et al., demonstrated that an anchoring mechanism in which only either the 5′ or the 3′ conserved flanking sequence was required for recombination of a segment (35). The lack of a requirement for internal sequence identity allows tremendous expansion in the number of possible variants—assuming these would maintain a protein structure required for at least minimal fitness. Examination of several hundred mosaic HVRs identified stretches of 2–10 highly conserved amino acids, including conserved cysteines, which serve as structural “tethers” to maintain overall Msp2 structure (40). Thus the HVR is itself composed of hypervariable microdomains that provide the diversity underlying antigenic variation with overall Msp2 structure maintained by the conserved tether elements. The resulting combinatorial diversity in the variant repertoire is estimated at 45 variants (four micro-domains generated by segmental gene conversion utilizing a minimum of 5 unique alleles per genome), consistent with continual antigenic variation.
Competing selective pressures for immune escape and variant fitness
Unlike the Msp2 variants generated from recombination of the intact hvr encoded by a donor allele, mosaic variants generated by segmental gene conversion exist only in the expression site and only transiently—due to the unidirectional recombination which results in the loss of the expression site copy. As a consequence, mosaic variants are not under long-term selection for maximal structural fitness but only for short-term ability to evade immune recognition and clearance. In the absence of an adaptive immune response in the host, variants generated by recombination of the intact hvr from the chromosomal alleles (designated as “simple variants”) have a competitive advantage over mosaics generated by segmental gene conversion (“complex variants”). This is supported by two lines of evidence—both of which have relevance for pathogenesis and epidemiology. The first is that acute bacteremia in a newly infected host is composed of simple variants; over time as the repertoire of simple variants is expressed and sequentially cleared, mosaics generated by segmental gene conversion appear and predominate (35). As the infection continues to persist, variants become increasingly complex with mosaics assembled with oligonucleotide segments derived from multiple alleles (Fig. 3A). This selection for complex variants occurs only under selective pressure of the adaptive immune response—direct inoculation into an immunologically naïve host results in acute bacteremia, again composed of simple variants (Fig. 3B) (41). The high level bacteremia in acute infection reflects, at least in part, the core fitness of simple variants with a 102–103 reduction in fitness as complex variants emerge. The second supporting observation is that upon natural acquisition feeding by ixodid ticks on a persistently infected host, ingested complex variants are rapidly lost from the A. marginale and simple variants predominate in the tick midgut and, importantly, in the salivary gland upon onward transmission feeding (Fig. 3C, D) (41–43). These tfindings underlie the paradox whereby a persistently infected host cannot clear its existing infection—due to sequential assembly of new mosaic variants by segmental gene conversion—but is immune to a new infection with the same strain, which expresses simple variants previously encountered. Importantly this provides the framework for understanding antigenic variation at the level of the host population and how population immunity drives divergence in strain structure.
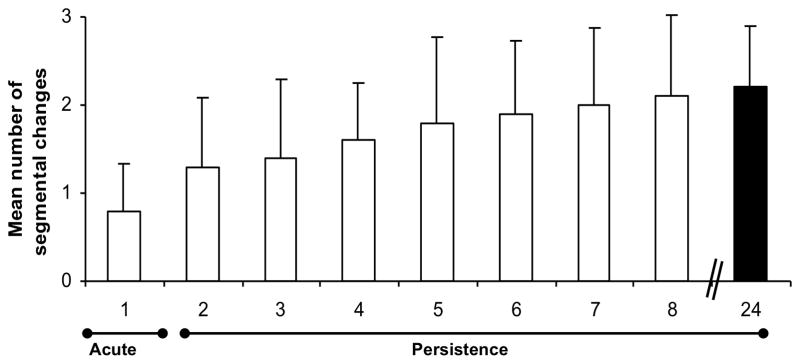
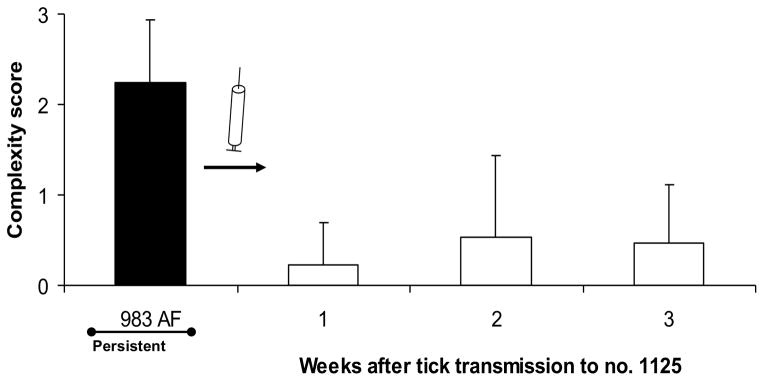
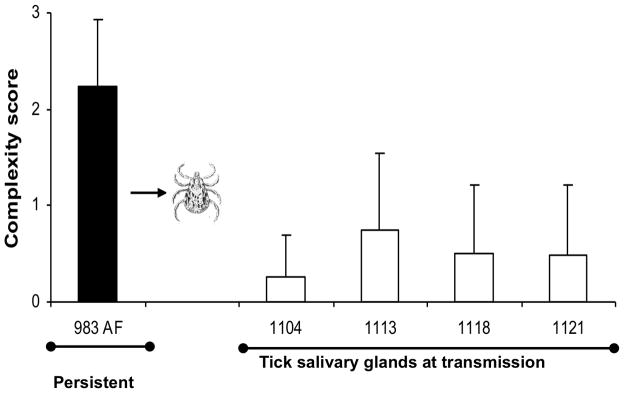
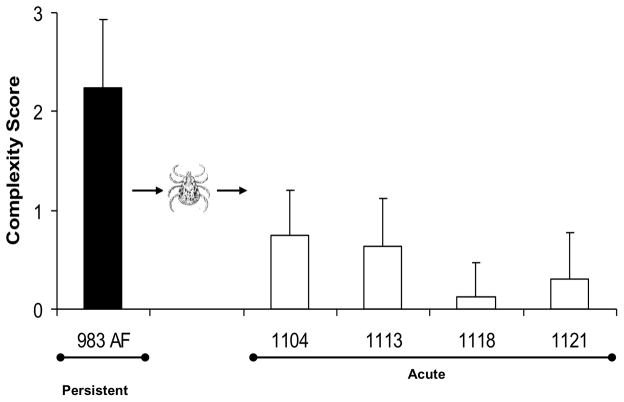
Figure 3. Complex variants are favored only under selection pressure of the adaptive immune response.
A) Development of complex Msp2 variants during persistent infection; complexity, measured by the number of expression site segments derived from different donor alleles (see Fig. 2B) is plotted on the y axis and duration of infection (in months) is shown on the x axis. At 24 months of infection (solid bar), this animal was used both as the source for direct transmission by intravenous inoculation of A. marginale into immunogically naïve animals (Fig. 3B) or for acquisition feeding of Dermacentor andersoni for tick transmission (Figs. 3C and 3D). B) Expression of simple Msp2 variants following direct inoculation of complex variants into an immunologically naïve animal. Solid bar, complexity score of A. marginale variants in persistently infected calf 983. These were inoculated intravenously into immunologically naïve calf 1125. Open bars, complexity of the Msp2 variants emergent during the 3 weeks of acute bacteremia. C] Expression of simple Msp2 variants in the tick salivary gland. Solid bar, complexity score of A. marginale Msp2 variants in persistently infected calf 983 during the acquisition feeding of ticks. Open bars, complexity of the variants in the salivary glands of ticks subsequently transmission fed on each of four calves (calves 1104, 1113, 1118, and 1121). D] Expression of simple Msp2 variants following tick transmission to immunologically naïve animals. Solid bar, complexity score of A. marginale msp2 variants in persistently infected calf 983 during the acquisition feeding of ticks. Open bars, complexity of the variants arising during acute infection following tick transmission to each of four immunologically naïve calves (1104, 1113, 1118, and 1121). Data, figure, and legend from ref. 41 with permission.
Population immunity as a driver of strain superinfection
Strain superinfection occurs when a second, genetically distinct pathogen strain infects a host that has already been infected with and mounted an immune response to a primary strain. Perhaps best understood in viruses, as illuminated by studies with HIV and Hepatitis C Virus (44,45), the principles can be more broadly applied to more complex pathogens, including bacteria and protozoa. Following initial identification of A. marginale strain superinfection under conditions of natural transmission (46), Rodriguez et al. identified that superinfecting strains differed in their msp2 allelic repertoire (47) and Futse et al., demonstrated that even a single unique allele was sufficient to allow a second strain to superinfect and that strain superinfection required sole expression of the unique allele as an antigenically distinct Msp2 (48). This was in marked contrast to non-superinfecting strains that shared only identical alleles.
The complete msp2 allelic repertoire was determined for multiple strains using a combination of targeted and whole genome sequencing (48). Unlike the overall A. marginale genome, which is considered to be “closed core” with a high level of conservation in gene content and within individual genes (47), alleles encoding Msp2/3 differed as much between superinfecting strain pairs as they did within a strain (48)--the latter being the basis for continual antigenic variation within an individual infected host. This led to the hypothesis that immunity at the population level was selecting for strain divergence, specifically in the variant-encoding alleles, with the capacity to evade pre-existing immunity raised by a primary, existing strain. Three lines of evidence have supported this hypothesis. The first is that strain-specific variation in primary structure of encoded simple variants is reflected in the lack of immune recognition—allowing evasion of immunity raised by the primary strain. Successful transmission of the secondary strain, which expresses simple variants in the tick vector, requires that one of the simple variants be sufficiently different to allow initial replication without immune recognition (48). Second, once established, superinfecting strains continue to generate, via segmental gene conversion, complex variants that reflect the greater level of overall antigenic diversity as compared to single strain primary infection (40). Third, the incidence of superinfection increases with the overall prevalence of A. marginale infection (40,49). At low levels of infection prevalence and consequent population immunity, there is a large proportion of immunologically naïve hosts and therefore minimal selective pressure for divergence. However, as infection prevalence rises—especially towards saturation as occurs in the tropics—the selective pressure markedly increases (40,49). A “newly emergent” strain, defined as having at least one unique allele, whether derived de novo or exogenously introduced, has a clear selective advantage as the host population is immunologically naïve to the new strain (Fig. 4).
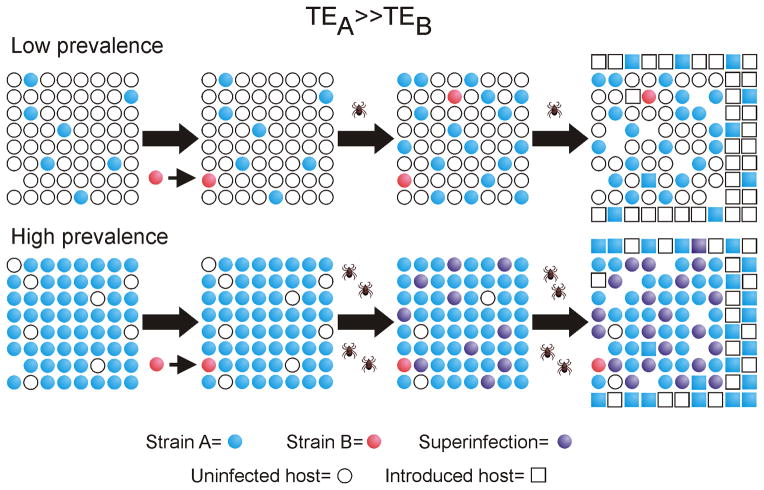
Figure 4. A. marginale strain superinfection is favored under the selective pressure of high population immunity.
Circles indicate the existing animal population at To: white represent uninfected and immunologically naïve hosts; blue represents hosts carrying strain A; orange, strain B; and purple represents hosts superinfected with strains A and B. Squares represent individual hosts introduced to the population by birth or immigration. The intrinsic transmission fitness is greater for strain A than strain B (TEA ≫ TEB). Under conditions of low prevalence of infection (and hence low population immunity), strain A predominates. Following introduction of strain B, its transmission is at a strong disadvantage and there is minimal selective pressure for strain B superinfection. Consequently, strain A predominance is maintained over time. Under conditions of high prevalence of infection (and high population immunity), strain A is predominant but there is strong selective pressure for strain B superinfection. Strain A transmission is favored for newly introduced naïve hosts and thus remains predominant but accompanied by prevalent superinfection. Figure and legend from ref. 54 with permission.
This duality of A. marginale antigenic variation for persistence both within the individual host and at the level of the host population role of interaction is consistent with propagation of self. Not surprisingly, this duality has also been recently observed in other highly antigenically variable bacterial pathogens, including but not limited to Borrelia burgdorferi (50), which is discussed in a subsequent section of this chapter.
Knowledge Gaps
Several significant knowledge gaps remain regarding mechanisms and selection determinants of A. marginale strain structure. How a new strain evolves from an existing one is not understood; both the rate of allelic change and mechanism(s) of change are unknown. One clue comes from the presence of exactly duplicated alleles in different loci within a strain (18,37). This suggests that the duplicate allele could serve as a template for change, essentially experimentation to result in a sufficiently diverse allele to allow superinfection, without jeopardizing the existing complement of alleles that allow antigenic variation within the individual host. This is supported by identification of two unique alleles in a second strain that are in the identical loci occupied by duplicated alleles in the first strain (37). The mechanism by which new sequence is introduced is also currently uncertain. Although 99% of variants map completely to one or more donor alleles, novel expression site variants characterized by mutation, insertions, and deletions at sites of segmental recombination have been identified (35). Gene conversion in reverse, resulting in an expression site sequence replacing a pre-existing allelic hvr, could generate de novo allelic change with “successful” alleles being retained in the genome. Finally, the trade-off between variant diversity that allows strain superinfection and maintaining minimal structure for bacterial fitness represents a clear knowledge gap with major epidemiological implications. Unchecked pressure only for allelic divergence would be expected to result in strain “chaos”—with dozens or more unique strains within a defined host population. However study of naturally occurring infections identifies a much more limited strain structure, often defined by a predominant strain (40,46,51–53). The current hypothesis is that there is a dynamic balance between allelic divergence to allow superinfection and retention of minimal protein structure for bacterial fitness (54). Only under conditions of high population immunity do the lower fitness strains compete successfully and only within the structural limits of maintaining minimal function (Fig. 4). Addition of new hosts, by birth or exogenous introduction, favors the most fit strain, maintaining its predominance. Testing of this hypothesis under conditions of natural transmission is proposed as a needed and potentially illuminating set of experiments with broad relevance.
BORRELIA
Introduction
The genus Borrelia represents a genetically unique and highly separated lineage within the Spirochaetes branch of the bacterial kingdom (55,56). These pathogenic spirochetes are obligated to a life cycle that requires transmission by hematophagous arthropods and long-term maintenance within a vertebrate host. All members of the Borrelia genus are defined by an unusual genome that consists of a linear chromosome and multiple linear plasmids containing covalently-closed hairpin ends. Borrelia hermsii and Borrelia burgdorferi are two well-studied members that exemplify relapsing fever and Lyme disease agents, respectively (55,57,58). A number of Borrelia species have been implicated in causing either the epidemic or endemic form of relapsing fever (56). Borrelia recurrentis is the only representative species responsible for causing epidemic relapsing fever, which is transmitted by the human body louse, Pediculus humanus. The remaining species are agents of the endemic form of relapsing fever, and are primarily transmitted by soft-bodied ticks of the genus Ornithodoros. Both forms of the disease are characterized by recurrent spikes of high fever that correlate with periods of bacteremia that can reach levels as high as 108 spirochetes per milliliter of blood (59). Each febrile episode corresponds to spirochetes that produce a different form of the Vlp or Vsp lipoprotein through an antigenic variation system that is crucial to the success of relapsing fever borreliae as pathogens (60,61). An effective host humoral response against Vlp/Vsp reduces the bacterial load in the blood, which brings about the resolution of fever in the host (62,63). New vlp/vsp variants arise in the host at a frequency of 10−3 to 10−4 per cell per generation (59), which in turn proliferate and seed the next wave of spirochetemia.
The multisystem disease known as Lyme disease (or Lyme borreliosis) is the most prevalent vector-borne infection affecting humans in both North America and Europe (64,65). Borrelia burgdorferi, Borrelia garinii and Borrelia afzelii are the causative bacterial agents of the disease, and are transmitted by hard-bodied ticks of the genus Ixodes. Infected ticks transmit Lyme disease borreliae to humans during feeding, which can result in a localized infection (erythema migrans) at the site of the tick bite. Disseminated and chronic stages of infection follow; these are characterized by neurological, cardiological, and arthritic manifestations of disease. Infection with Lyme disease borreliae can last from months to years due to avoidance of the host immune response, and key to its successful evasion tactics is recombination at the vls locus located at the telomeric end of a 28-kilobase linear plasmid (4,66,67). Gene conversion events within the vls locus result in sequence variations of the VlsE surface lipoprotein that in turn alter its antigenic properties and provide the spirochete with the ability to evade the host’s antibody-mediated response (67–69). Although similarities and differences exist between the antigenic variation systems of relapsing fever and Lyme disease borreliae, this chapter will focus on the more well-studied vls system of the Lyme disease spirochete.
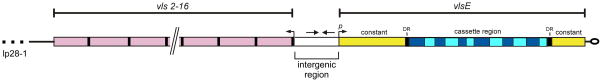
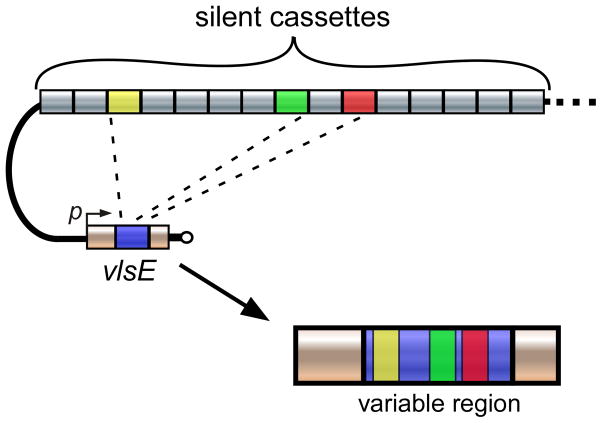
A schematic of the vls locus from the B. burgdorferi B31 strain is shown in Figure 5. The locus consists of the vlsE expression site that encodes the 35 kDa lipoprotein, which is located 82 basepairs (bp) from the right telomere end of the linear plasmid, lp28-1 (66–68). The locus also includes a tandem array of 15 silent cassettes (vls2-16; each approximately 500bp in length) that are oriented in the opposite direction of the vlsE gene. A short intergenic region (~160 bp) separates the vlsE locus and the silent cassettes, and this intergenic space contains a near-perfect inverted repeat of a 51 bp sequence capable of forming a highly stable DNA stem loop (70). In addition, a portion of the promoter sequence required for vlsE expression is located within this inverted repeat (70). The vlsE expression region is comprised of a central variable cassette (Fig. 5, blue region) that is flanked by constant regions (yellow). At the junction of the variable and constant regions are 17 bp direct repeats. With the exception of the final cassette, 17 bp direct repeat sequences are also found at either end of the silent cassettes. The vlsE cassette region exhibits roughly 90% sequence identity with each of the silent cassettes (67), and most of the sequence differences reside in six variable regions (Fig. 5, light blue) that are flanked by six highly conserved, or invariant, sequences (dark blue).
Figure 5. The vls locus of B. burgdorferi B31.
Illustration showing the arrangement of the vls expression site, vlsE, and the contiguous array of 15 silent cassettes comprising the vls locus on the right telomeric end of lp28-1. The six variable regions of the central vlsE cassette are colored light blue, while the six invariant regions are colored dark blue. The black bars flanking the vlsE cassette region and silent cassettes represent the 17 bp direct repeats. The silent cassettes (vls2-16) are not drawn to scale. Arrows positioned at the beginning of vlsE and silent cassettes indicate the respective orientations. The arrows located within the intergenic region denote the inverted DNA repeat. DR, 17 bp direct repeat; p, vlsE promoter. Figure adapted from ref. 191 with permission.
The vls locus in immune evasion and persistence
A number of experimental findings over the years have provided strong evidence implicating the importance of VlsE antigenic variation for B. burgdorferi persistence. The first studies involved determining the kinetics of gene conversion, and recombination events were observed as early as four days after infection of mice and continued to occur throughout infection (69). However, assessing the exact rate of variation has proven difficult; this is because vlsE antigenic switching only occurs during mammalian infections (71), and B. burgdorferi is not readily sampled from the blood. VlsE variation occurs to such a degree that each clone examined from a single mouse skin biopsy can have a different vlsE sequence after only 28 days post infection, and the sequence changes primarily occur within the six variable regions. Early work also demonstrated that antibodies specific for the variable regions of VlsE were produced during experimental infection of mice (72).
More evidence for the role of the vls system in immune evasion came from studies involving the vls-resident plasmid, lp28-1 (73,74). Clones lacking lp28-1 were shown to exhibit an infectivity phenotype whereby these spirochetes were able to disseminate to tissue sites but were unable to persist in the murine host. Notably, these same clones are capable of long-term survival in severe-combined immunodeficient (SCID) mice that lack an effective antibody response (75,76). Lp28-1-deficient isolates also grow normally in a dialysis membrane chamber implanted in the peritoneal cavity of rats, where exposure to either antibodies or immune cells is restricted (76). Finally, immunocompetent mice infected with an lp28-1 minus strain complemented with only the vlsE gene (sans the vls silent cassettes) are able to clear infection, demonstrating that it is not the mere presence of VlsE that provides the capacity for persistent infection, but rather the ability to undergo vls antigenic variation to produce VlsE variants (77). Although these studies provided a strong indication that lp28-1, and presumably the vls locus, are required for persistence only in the presence of an effective humoral immune response, definitive evidence for the role of VlsE antigenic variation in immune evasion remained elusive until the generation of a genetic deletion of the vls locus.
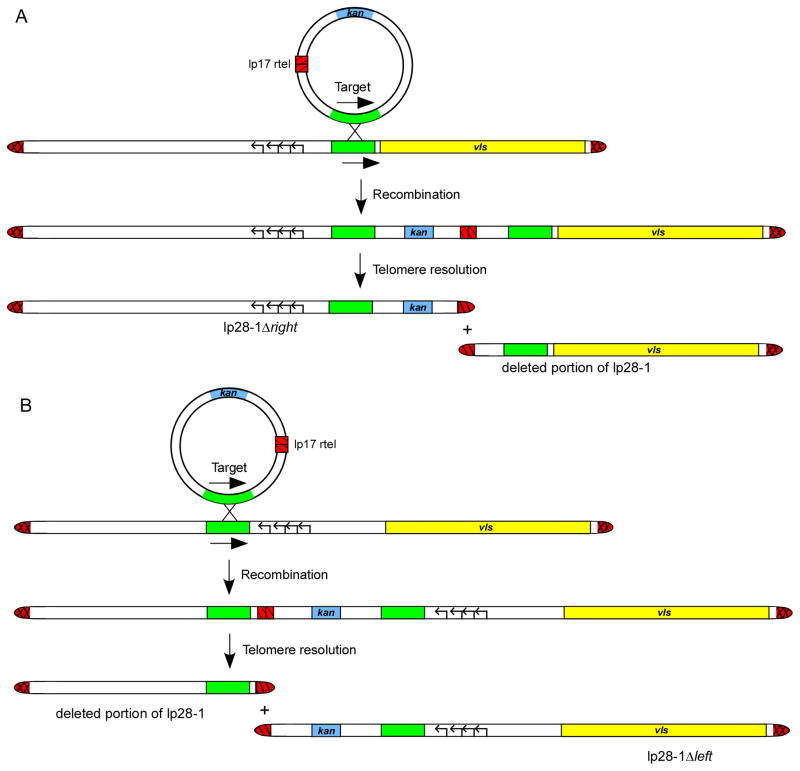
The construction of a vls knockout mutant in B. burgdorferi was made possible by taking advantage of the linear nature of lp28-1 (Fig. 6A). The method used involves integration of a plasmid containing a replicated telomere within a chosen linear plasmid of B. burgdorferi (78–80). The end result of the internal placement of the replicated telomere is deletion of DNA through the action of the Borrelia telomere resolvase, ResT (80,81). Spirochetes that lost the vls locus due to telomere-mediated removal were completely cleared from immunocompetent C3H mice by day 21 post infection (78), matching the phenotype observed with clones that lacked the lp28-1 plasmid. Consistent with the findings that lp28-1 is not required for persistent infection in the absence of an adaptive immune response (75–77), the vls deletion mutant exhibited long-term survival in SCID mice, thereby confirming the hypothesis that vls recombination functions to evade the humoral immune response in the mouse host (66,67).
Figure 6. Telomere-mediated deletion of the vls locus in B. burgdorferi.
A) Construction strategy for the generation of the vls deletion mutant clone of B. burgdorferi B31. The target sequence for insertion (green) is chosen so that only the vls locus would be deleted from lp28-1 plasmid. The genes encoding plasmid maintenance proteins that have been previously shown to allow autonomous replication are shown as black-lined arrows. B) Schematic of the construction strategy for the lp28-1Δleft plasmid. The target sequence for insertion (green) was chosen so that only the genes encoding proteins that allow autonomous replication of lp28-1 (shown as arrows arranged from left to right), and the right side of lp28-1 would remain. Hairpin telomeres are shown as red hatched regions. Figure adapted from ref. 78 with permission.
Targeted deletion was also utilized to obtain lp28-1 mutants containing mainly the vls locus and the necessary genes for autonomous replication of the plasmid (Fig. 6B). The only other potential genes retained on the lp28-1 mutant plasmid were eight very small open reading frames (bbf19-22 and bbf27-30) predicted to potentially encode for proteins consisting of only 82 amino acids or less. Infectivity experiments showed that these B. burgdorferi clones are fully infectious and persistent in immunocompetent mice, providing further evidence that the silent vls cassettes and vlsE are involved in spirochete persistence (78). Moreover, these mutant clones carried out vlsE recombination in immunocompetent C3H mice, indicating that protein factors required for antigenic switching are likely not carried on lp28-1, but encoded elsewhere.
The remaining presence of the small ORF regions bbf19-bbf22 and bbf27- bbf30 left open the possibility that the loss of genes within these loci could have some role in the intermediate infectivity phenotype exhibited by the lp28-1-deficient clones, and not due solely to an absence of the vls locus. A study published by Embers et al. reported that expression of the immunodominant outer surface protein C (OspC) by an lp28-1-deficient B. burgdorferi clone was abnormally high in vivo, suggesting that down-regulation of this protein is impaired (82). Previous studies have shown that OspC is downregulated in B. burgdorferi shortly after establishing infection in the animal host, and it has been suggested that this provides the spirochete with a mechanism to avoid clearance mediated by anti-OspC antibodies (83–85). The overall conclusion from the Embers et al. study was that failure of OspC repression by lp28-1-deficient spirochetes renders them susceptible to immune-mediated clearance, which could potentially be responsible in part for the intermediate infectivity phenotype associated with these B. burgdorferi clones. Thus, the possibility was raised that one or more genes involved in ospC repression may be present on the lp28-1 plasmid. However, recently generated mutants lacking either region bbf19-22 or bbf27-30 were found to be capable of persistent infection of immunocompetent C3H mice for up to 91 days (Hove and Bankhead, unpublished data). Together with previously published results involving additional lp28-1 deletion mutants, this raises the likelihood that the vls locus is likely the only lp28-1-resident genetic system responsible for persistence during infection of the mammalian host.
Mechanism of recombination
Although gene conversion has been implicated in vlsE antigenic variation, little is known about the exact mechanism of recombination and the proteins involved in this process. Since the discovery of the vls system almost two decades ago, progress towards elucidating the mechanistic details of recombinational switching has been impeded by two factors. First, switching does not occur in either culture or the tick vector, but instead requires passage through a mammalian host (66), and second, the vls locus is genetically unstable when cloned in E. coli (67,86).
The proposed model for vlsE antigenic switching involves a non-reciprocal gene conversion mechanism whereby segments within the vlsE central cassette region are replaced by sections of varied length and location from the silent cassettes (Fig. 7). Only the cassette region of vlsE displays sequence variation resulting from gene conversion; the sequence and organization of the silent cassettes remain unaltered (69). An elaborate study by Norris and colleagues examined 1,399 clones isolated from various tissue sites of mice infected from 4 to 365 days (87). Detailed analysis of the vlsE sequence changes found that the vls antigenic variation system promotes both short and long recombination events within each cassette region. Along with the gene conversion events, template-independent changes also occur that lead to additional sequence variation of vlsE. The end result of these accumulated changes is a new vlsE sequence with a mosaic structure, and the total combined potential for sequence variation makes the vlsE switching process one of the most dynamic antigenic variation systems known.
Figure 7. Overview of vlsE antigenic switching in B. burgdorferi.
Variant-specific segments act as a source of DNA for nonreciprocal recombination events with the vlsE expression locus. Through this process, segments of the variable region (blue) are replaced by sections of varied length and location from the donor sequences. In the example shown, three sequential gene conversion events (represented by dashed lines) occur within each expression site through recombination with the colored donor sections (yellow, green or red) to generate a new expression site sequence with a mosaic structure. Figure adapted from ref. 4 with permission.
Surprisingly, the RecA protein does not seem to be required for the generation of VlsE antigenic variants, despite being involved in DNA repair and homologous recombination in the spirochete (88). This is in sharp contrast to other well-studied antigenic variation systems that are RecA-dependent, such as the pilE locus of Neisseria gonorrhoeae (89). Similarly, disruption of a large number of genes involved in DNA recombination, repair, and replication ruled out the involvement of their respective encoded proteins in vlsE switching (90,91). However, these same studies did find that the RuvAB complex of B. burgdorferi is required for vls antigenic variation, presumably by promoting branch migration of Holliday junctions during vlsE recombination (90,91). Interestingly, while B. burgdorferi contains genes that encode the branch-migration complex RuvAB, these spirochetes are noticeably lacking the gene for the Holliday junction resolvase, RuvC. In fact, no recognizable ortholog exists in the B. burgdorferi genomic sequence, which suggests that a specialized Holliday junction resolvase may be expressed in the spirochete. This raises the question as to how the Lyme disease pathogen promotes recombination at the vls locus with such a limited involvement of the usual collection of DNA recombination proteins.
One possible answer is that the vlsE antigenic switching process may involve the formation of G-quadruplex DNA structures. A study by Wahlia and Chaconas found that G4 DNA could be formed by G-runs located in the 17bp direct repeats of the vlsE gene (92). A G-quadruplex DNA structure has previously been shown to have a role in pilin antigenic variation in N. gonorrhoeae by acting as a signal for DNA strand nicking (93,94). However, this process involves recombination proteins that are either known to not exist in B. burgdorferi, or have been shown to not have a role in antigenic switching. Moreover, the G-runs found in the vls locus are not separated by short stretches of DNA similar to that typically found with G4 DNA. Thus, it remains unclear whether G-quadruplex formation has an actual role in vlsE recombination, or if inter-molecular interactions between the G-runs of the direct repeats promote gene conversion through an entirely different mechanism.
The VlsE lipoprotein
The crystal structure of the recombinant-variant VlsE outer surface lipoprotein presents a three-dimensional protein fold that is substantially different from most other solved protein structures (95). Despite this unique configuration, analysis of the VlsE protein structure did reveal a resemblance to the variable surface protein of the relapsing fever agent, Borrelia hermsii. The VlsE monomer is also quite similar to the overall protein fold of the OspC dimer structure, with both presenting membrane distal alpha-helical bundles to the host environment (95,96). The overall primary structure of VlsE consists of N-terminal and C-terminal constant domains that flank the central cassette region. The loops containing the variable regions are localized at the distal surface of the protein, while the C terminus is positioned close to the N terminus that is attached to the bacterial lipid outer membrane. Gel filtration chromatography of VlsE showed that that the protein is primarily monomeric in solution, although the observed interface in the crystal structure raises the possibility that VlsE could exist as a dimer on the bacterial cell surface.
The observed position of the variable regions at the distal surface of VlsE confirms their accessibility to antibodies. In contrast to the variant regions, the invariant regions of VlsE have very limited surface exposure. Despite the apparent inaccessibility to antibodies, a strong humoral response is mounted against the conserved regions of VlsE (most notably invariant region 6). In fact, VlsE-based recombinant proteins or synthetic peptides are used for the immunodiagnosis of Lyme disease (97–101). The function of VlsE independent of its antigenic variation properties is not currently known. There have been previous indications that VlsE may be involved in tissue tropism, including reports that VlsE production is elevated during mammalian infection (102) with higher levels observed in spirochetes recovered from joint and skin tissues than from heart tissue (103). However, a recent study found no obvious differences in the amino acid sequence of VlsE variants recovered from different tissue sites, suggesting the absence of any VlsE role in tissue tropism (87).
Knowledge Gaps
Past research involving B. burgdorferi antigenic variation has focused on either analyzing vlsE sequence changes, the dynamics of those changes, or the role of vlsE switching in B. burgdorferi pathogenesis. However, studies that address the fundamental question of how antigenic variation occurs in B. burgdorferi have been seriously lacking. In addition, the use of molecular approaches to study vls recombination has been limited. In fact, despite the widespread nature of this effective pathogenic strategy, few molecular details of the recombinational switching process have been reported for any organism.
A number of aspects regarding the molecular details of VlsE antigenic variation remain unknown, and will likely be the focus of future experimental studies. For example, the subtelomeric positioning of the vls locus is a common feature associated with many antigenic variation systems found in both eubacterial and protozoal pathogens (104,105). The reasons behind this phenomenon are still unclear and could potentially be of great significance to the overall mechanism of antigenic variation. The importance of the 17 bp direct repeats that demarcate the vlsE variable region and flank the silent cassettes is also currently unknown. Although it was originally thought that these sequences might play a role in recombinational switching (67), sequence analysis showed that these repeats are not well conserved across Borrelia species and even within some species, casting doubt on a proposed role in recombination (106). However, the recent finding that G4 DNA can be formed at the guanine-rich 17bp direct repeats may warrant further investigation into the potential mechanistic importance of these sequences for vls gene conversion. Nevertheless, any potential roles of these direct repeat elements for vlsE recombination during host infection has not been directly investigated. Also remaining to be determined is whether the vlsE-silent cassette intergenic region is required for vls antigenic variation. One intriguing idea is that the stem loop-forming potential of the intergenic region between vlsE and the silent cassette array functions to deliver the cassette copies to vlsE during the gene conversion process. Finally, a study demonstrated that antigenic switching does not occur with a trans copy of vlsE during infection, suggesting a possible cis requirement for the vls recombination mechanism (77). However, the trans copy of vlsE was carried on a circular shuttle vector, as opposed to the linear lp28-1 plasmid in which vlsE normally resides, and whether this may have inhibited recombination between the shuttle plasmid and lp28-1 will require additional studies.
A role for the VlsE protein other than providing an antigenic disguise is not currently known, but it has been proposed that the protein might function in other forms of immune evasion (107,108). An elegant study by Tilly, Bestor and Rosa demonstrated that VlsE and OspC may share similar but distinct roles during host infection (109). Unfortunately, the exact function(s) of the OspC protein also remains undefined, and thus exactly what common role these two lipoproteins may share is still a mystery. Based on previous studies, it has been speculated that they may serve to protect the pathogen from host defenses. Alternatively, they may aid in stabilizing the bacterial structure during host infection similar to that recently demonstrated for OspA and OspB in lipid rafts on the spirochete membrane (110,111). Though OspC was not found to be associated with these lipid rafts (111), high levels of either OspC or VlsE may still function in some fashion as an essential component for overall stability of the bacterial outer membrane.
Although a number of other surface proteins exist that are immunogenic, VlsE is the only known B. burgdorferi antigen that exhibits variation of its surface epitope. A daunting question regarding B. burgdorferi immune escape has been how such a feat is accomplished through sequence variation of this single lipoprotein, despite the presence of a substantial number of additional antigens residing on the bacterial surface. Many pathogens utilize their antigenically variable proteins in a number of ways as an evasion strategy, and several models have been suggested for how VlsE might become the primary target for the host immune response. In other pathogen systems, such as that found in Trypanosoma brucei, the variable surface protein hides other antigens by essentially coating the surface of the organism (112,113).
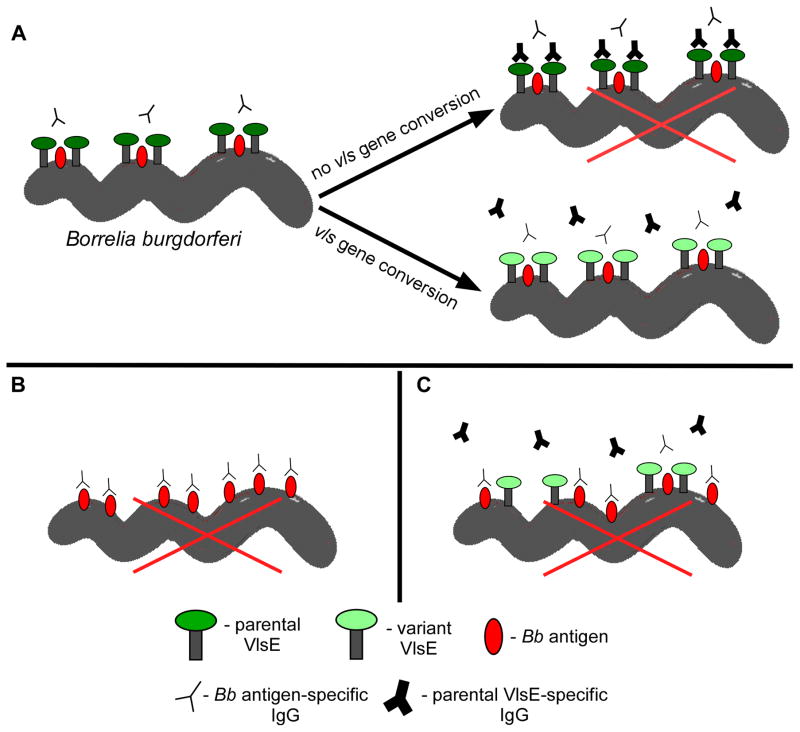
It is possible that VlsE may act as a shield to obscure the epitopes of other surface antigens. In fact, crystallography data suggests that the binding of VlsE to other proteins on the membrane surface may block antibody binding to the lateral surface of VlsE (95). This in turn may protect other surface antigens that are tightly juxtaposed to VlsE from antibody recognition (Fig. 8A). A precedent for this type of interaction has been demonstrated in studies with the Borrelia protein P66, in which the protein is protected from proteolytic cleavage in Lyme Borrelia expressing high levels of the outer surface protein OspA (114).
Figure 8. Model for VlsE-mediated protection of B. burgdorferi surface antigens.
A) Shortly after host infection, upregulation of vlsE expression leads to surface localization of the encoded lipoprotein. Interaction of VlsE with other proteins results in a complex that functions to shield epitopes of these surface antigens. Continued vls gene conversion leading to production of VlsE variants is necessary to avoid killing by antibodies raised against the parental and subsequent VlsE variants, allowing for sustained epitope masking. Absence (panel B) or low expression (panel C) of VlsE allows binding of neutralizing antibodies to B. burgdorferi surface antigens that ultimately leads to spirochete death (denoted by a large red X). A legend indicating the identity of the various molecular cartoon depictions is provided at the bottom of the figure.
With respect to VlsE-mediated shielding, experiments utilizing either in vitro-grown or host-adapted wild-type B. burgdorferi were conducted to determine whether VlsE expression could provide Lyme disease spirochetes with the capacity to reinfect mice (115). The levels of VlsE expression have been shown to be upregulated 32-fold during host infection relative to those measured in vitro (102). Unlike the highly susceptible in vitro-grown spirochetes, B. burgdorferi that have adapted within the animal host have been demonstrated to be relatively invulnerable to the protective effects of immune sera (116,117). The study showed that cultured (low VlsE-expressing) wild-type B. burgdorferi are unable to reinfect mice, while host-adapted (high VlsE-expressing) wild-type spirochetes are fully competent for host reinfection (115). Additional experiments involving wild-type and VlsE mutant clones found that only the wild type could reinfect mice initially infected and cleared of spirochetes devoid of VlsE. In other words, the immune response of these mice was sufficient to prevent reinfection by a VlsE-deficient clone, but could not block reinfection by spirochetes capable of expressing variable VlsE (see Fig. 8B). It was also shown that SCID mice treated with immune sera generated to a VlsE-deficient mutant were resistant to infection by this same clone, but could be successfully challenged by host adapted wild-type B31 spirochetes expressing VlsE (115). The finding that passively-transferred antibodies developed to non-VlsE surface antigens can provide immunity against the VlsE-deficient clone, but were not borreliacidal to wild-type spirochetes, may hint at a possible VlsE-mediated shielding mechanism. Finally, VlsE expression is known to be highly reduced while in the tick environment, and in agreement with findings described above, mice fed with wild type-infected ticks are immune to reinfection (Rogovskyy and Bankhead, unpublished report). Together, these data indicate that the adaptation state of infecting spirochetes, likely due to its respective effects on VlsE expression, can greatly influence B. burgdorferi evasion from the host antibody-mediated response (see Fig. 8C). Whether this VlsE-mediated immune avoidance system involves epitope shielding will require further investigation.
It has also been proposed that VlsE might be a T-cell-independent antigen that could directly stimulate certain B cell subsets (78,108). The resulting humoral response generated by VlsE may serve to override antibody production against other potential surface antigens. It has been observed that vls mutant B. burgdorferi clones complemented with a non-switchable form of vlsE are cleared faster in immunocompetent mice relative to non VlsE-complemented mutants (78). It is conceivable that this outcome could be the result of direct stimulation of B cells by VlsE, and this modulating ability could result in more effective clearance of these spirochetes due to the absence of VlsE antigenic variation in those clones. In support of this, a recent study found that sera derived from nude mice infected with wild-type B. burgdorferi contained anti-VlsE T-cell independent antibodies at sufficient levels to prevent infection by in vitro-grown wild-type B. burgdorferi, but were at inadequate quantities to prevent infection by the VlsE-deficient mutant clone. This finding is suggestive that the T-cell independent antibody response is directed primarily to VlsE, with subdominant titers of these antibodies against non-VlsE surface antigens as demonstrated by their inability to prevent infection by the VlsE-deficient mutant clone.
Finally, because survival of the Lyme pathogen in nature is completely dependent on its enzootic life cycle, it would be interesting to assess whether the variant-generating capacity of the vls system is a must for the Lyme pathogen to be efficiently and successfully perpetuated throughout the its life cycle. Although never tested, the expected outcome would be that antigenic variation of VlsE is required for persistence in the natural reservoir host. Also unknown, and potentially more interesting, are the effects of vls mutation on the ability of B. burgdorferi to be acquired, persist, and transmitted to naïve mice by Ixodes ticks. In all, continued pursuit towards identifying the mechanism responsible for B. burgdorferi immune escape has important implications in the development of vaccines against the pathogen and other Borrelia species. If the protective effects of VlsE can be minimized using therapeutics, then an effective vaccine can be developed and used in conjunction to prevent infection or reinfection by the Lyme disease spirochete. Additionally, such knowledge could lead to the development of novel strategies for targeting Lyme disease Borrelia in the tick vector and/or reservoir host. Overall, such future studies will significantly advance our knowledge of immune evasion by B. burgdorferi, and could in turn have broad implications for other animal and human pathogens.
NEISSERIA
Introduction
The Genus Neisseria contains group of Gram-negative, proteobacterial species, most of which colonize mucosal surfaces of mammals, but are commensal organisms that do not normally elicit pathology or cause disease (118). Whether they have a mutualistic relationship with their hosts has not been established, but considering their long co-evolution with their host it is likely. There are 11 species that colonize humans and these species appear to be host-restricted since they are not known to inhabit any other natural ecological niche. It is unknown whether a progenitor of the human–restricted Neisseria colonized a human and that the 11 different species evolved from this one progenitor or whether a progenitor colonized one or more different hosts and then transferred and adapted to life exclusively within humans. While all of these organisms have the ability to colonize mucosal surfaces and grow, only two members of the genus are considered to be frank pathogens with serious sequelae of morbidity and mortality (118). The pathogenic Neisseria are Neisseria gonorrhoeae (the gonococcus or Gc), the sole causative agent of gonorrhea and Neisseria meningitidis (the meningococcus or Mc), the major cause of bacterial meningitis in teenagers and young adults. While these organisms are true pathogens that have the ability to cause disease in otherwise healthy people, they represent pathogens that have evolved from and share many of the colonization determinants with the commensal Neisseria and can behave either as nonpathogenic (commensal or asymptomatic) or pathogenic organisms when colonizing different individuals.
N. gonorrhoeae was the first member of the genus described and is a unique member of the genus since it mainly colonizes the human genital tract, and not the nasopharynx or oral cavity. A disease similar to gonorrhea has been described throughout human history (119). Gonorrhea is still prevalent worldwide today and the rise in antibiotic resistance makes gonorrhea a major public health concern (120). Gonococcal infection usually results in symptomatic disease in men, with an obvious purulent exudate and painful urination; but it is more linked to asymptomatic infections in women, with a less obvious exudate and little or no pain during uncomplicated infection (121,122). However, there are no general population studies that measure the frequency of asymptomatic infection in either gender. In the developed world, a significant portion of infections are found within populations of men who have sex with men (123). Of the human-adapted Neisseria, Gc more often elicits inflammation than the other species. While the reasons for this are unknown, it is a reasonable hypothesis that inflammation increases sexual transmission.
N. meningitidis, like the commensal Neisseria, colonizes the nasopharynx and usually behaves similarly to the commensal Neisseria (124). However, in a small number of colonized people (<1%) the meningococcus can leave the nasopharynx and go systemic to cause meningococcemia or bacterial meningitis (125). Localized outbreaks of meningococcal disease occur when young adults from different geographical regions come in close contact with one another (e.g., dormitories or military barracks) or during the dry season in sub-Saharan Africa (126). These epidemiological correlations suggest that there are both immunological and environmental factors that can contribute to meningococcal disease. However, since many outbreaks are clonal in nature, it is postulated that there are hyper-invasive Mc clones that are more able to go systemic and/or cross the blood brain barrier (124). While many genes or gene products have been correlated with hyper-invasive lineages, there is no single virulence factor that correlates with systemic disease (127–130). While the frequency of invasive disease is low relative to commensal carriage, the high level of mortality associated with Mc meningitis makes this the most dangerous member of the genus.
Gc and Mc appear to have evolved from a common progenitor since they show greater than 90% identity in their core genomic sequences and are closely related (131). Not surprisingly, due to their different sites of colonization, transmission and pathogenesis, each species has unique defining genetic determinants (e.g., the capsule of Mc), but the genetic similarities between these species are more impressive than their differences (132). One genetic characteristic that differentiates the pathogenic from the commensal Neisseria is the greater amount of phenotypic variation that is found in the pathogenic species. For reasons that are not well understood, the commensal Neisseria have low potential for variation, Mc has extensive variation, but does not have the vast capacity that Gc encodes. One plausible hypothesis is that both Gc and Mc use variation for functional reasons not needed by the commensal Neisseria, while as an STI, Gc requires a greater amount variation for immune evasion.
Phase and Antigenic Variation in the Neisseria
The pathogenic Neisseria are notable in the number of genes that can undergo stochastic variation of gene expression. The types of stochastic variation that have been described in the pathogenic Neisseria include: ON/OFF phase variation, the reversible expression or nonexpression of gene products; and antigenic variation, the expression of multiple antigenic forms of a gene product or structure. There are three major surface structures that all can undergo both phase and antigenic variation; the lipooligosaccharide (LOS) glycan structure that constitutes the outer layer of the outer membrane (133), the family of outer membrane adhesins called opacity proteins (134), and the Type IV pilus (135). The properties and mechanism of variation of each of these important antigenically-variable surface structures will be discussed in detail below.
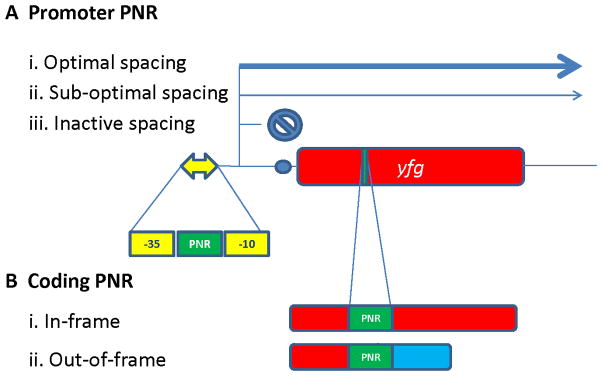
There are also 50–100 genes in each pathogenic Neisseria that undergo simple ON/OFF phase variation (136). Many of the phase variable genes are involved in pathogenesis, but there are other functions controlled by phase variation and not all of the gene products have a known function. The phase variable genes mostly alter expression through changes in polynucleotide repeats (PNR) located either in the promoter of the gene or within the coding sequence (Fig. 9). The number of nucleotides within the PNR can change +/−1 during DNA replication through a process called slipped-strand mispairing (137). If the PNR is located within the promoter region of a gene, the size of the PNR therefore sets whether the −10 and −35 elements of the promoter are within optimal distance of one another (high expression), sub-optimal distance (moderate expression), or non-functional distance (no expression) (Fig. 9A). If the PNR is within a coding region, the change in repeat number can alter the translational reading frame and either allows expression of the full length gene product, or with a frame-shift that alters reading frame this usually results in a premature stop codon (Fig. 9B). This large number of phase variable genes with the pathogenic Neisseria creates a situation where every population is a heterogeneous mix of phenotypic variants. This is one reason why genetic complementation is so critical in laboratory studies using mutants.
Figure 9. Polynucleotide repeat (PNR)-based phase variation in Neisseria.
A) PNR within a promoter of your favorite gene (yfg). The yellow double headed arrow represents the promoter with a −35 and −10 sequence and a PNR in between these promoter elements. The blue circle shows a ribosome binding site to initiate translation. (i) When the PNR repeat number is in the optimal spacing, the promoter initiates maximal transcription as long as other regulatory signals do not prevent expression. (ii) When the PNR changes +/−1 nucleotide, the spacing between the −35 and −10 elements is not optimal but there is still transcriptional initiation to produce a low level of gene expression. (iii) When the PNR number increases or decreases due to a spacing that does not allow RNA polymerase to effectively interact with the promoter, there is essentially no transcription. B) PNR within a coding sequence. (i) The PNR length maintains the reading frame to translate the entire gene product. (ii) Change in the PNR length by +/− 1 or 2 changes the reading frame downstream of the PNR and usually terminates translation prematurely. It is possible that this change in reading frame could result in a different protein product.
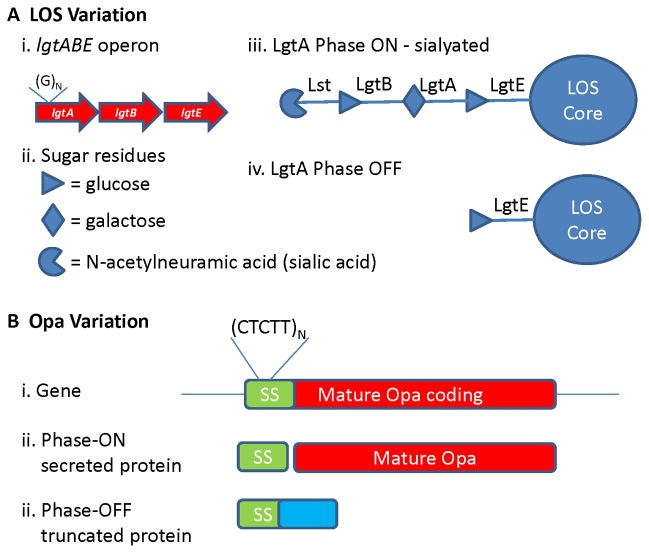
LOS Variation
As Gram negative bacteria, members of the Neisseria have two phospholipid bilayer membranes. The outer membrane is an asymmetric bilayer with the outer leaflet being comprised of LOS molecules. LOS is similar to the lipopolysaccharide (LPS) expressed by many Gram negative bacteria but does not have the long extended O-antigen polymeric sugars characteristic of LPS (133). The LOS has many functions for Neisseria including the barrier function of the outer membrane, immuno-stimulatory endotoxin activity, and providing complex glycosylation substrates that are recognized as being similar to host glycans and binding host lectins (138). LOS is a major component of the outer membrane vesicles that are the basis of experimental vaccines (139), and an important immune-stimulatory component of the new Mc group B vaccine Bexsero (140).
In the pathogenic Neisseria, there are several invariant glycosyltransferases that help build the LOS glycan structure and a few phase variable glycosyltransferases that each can be ON or OFF in different cells (141–143). The glycan structure of the LOS is defined by the combination of the phase variable glycosyltransferases that are producing each enzyme. Phase variation of the LOS biosynthetic glycosyltransferase is mediated by changes in PNRs (polyG) found within the coding region of each variable gene allowing for a simple ON/OFF alteration in expression of the protein. The effect of these variable expression genes on the bacteria surface is amplified by the further modification of some but not all glycan structures by sialylation (141,142,144,145) (Figure 10A). LOS variation constitutes a powerful way to use replication error to create multiple sugar structures to vary the bacterial surface. The reasons why Mc retains LOS variation despite having a distinct polysaccharide capsule covering the outer surface is not well-established, but the existence of acapsular strains and the ability of outer membrane proteins to engage host cell receptors suggests that the capsule is not a complete barrier to the bacterial surface (146).
Figure 10. LOS and Opa phase variation.
A) LOS variation. Shown is one of the variable LOS biosynthetic gene clusters. (i) The ltgA glycosyltransferase gene has a guanine PNR repeat within the coding sequence that can change repeat number +/−1. (ii) Sugars residues added by each transglycosylase. (iii) When the ltgA PNR is in the correct reading frame, the glycosyltransferase is produced to add a galactose residue to the acceptor glucose. This galactose is then a substrate for the LgtB glycosyltransferase to add a glucose, which is then a substrate for the sialyltransferase (Lst). (iv) When the lgtA PNR changes number and causes a frame shift, this form of the LOS cannot be modified or sialylated. B) Opa variation. (i) A representative opa gene showing the pentamer repeat within the signal sequence (SS). (ii) Phase-ON when the pentamer repeat number encodes an in-frame protein, the full protein is translated and the signal sequence is cleaved during secretion. (iii) Phase-OFF when the pentamer repeat changes number to be out-of-frame and an altered protein coding sequence is produces (blue), leading to a premature stop codon.
Opa Variation
The pathogenic Neisseria can express a family of outer membrane proteins called opacity (Opa) proteins because some of these proteins cause change in colony opacity when cells are grown on agar plates (134,147). Mc encodes 4–5 variable opa loci, N. Gc encodes 11–13 opa loci, while commensal Neisseria encode one to two opa loci. Opa proteins are beta-barrel outer membrane proteins that mainly act as adhesins. Different Opa variants bind to different member of the human CEACAM receptor family or to heparin sulphate (148–150). It is possible that Opa protein variation is driven more by receptor specificity rather than by immune selection but this has never been directly tested.
Each opa locus is independently transcribed and each locus is under independent phase variation control. There is a pentamer nucleotide repeat (CTCTT) contained within the DNA segment that encodes the protein secretion signal sequence (134,151). During DNA replication, this pentamer repeat will add or remove one repeat and put the mature Opa protein coding sequence in or out of the reading frame of the initiating AUG (Fig. 10B). Therefore, each Neisseria cell can have any combination of Opa proteins expressed in a stochastic manner. The frequency of Opa phase variation is influenced by promoter strength and the rate is larger when the repeat length is longer (152,153).
Pilin Variation
Both the pathogenic and commensal Neisseria express a Type IV pilus. The type IV pilus is an important colonization factor that also plays many roles in the pathogenesis of disease. Pili help mediate adherence to host cells and tissues (154), twitching motility (155), resistance to PMN killing mechanisms (156), and genetic transfer (157). Only the pathogenic Neisseria have the ability to antigenically vary the pilus, with all Gc strains having the ability and most Mc strains, however, a small subset of Mc strains expressing a nonvariable pilus that is more similar to the commensal type IV pilus but still can cause invasive disease (158).
In the pathogenic Neisseria, pilus expression and antigenicity is modulated by many different mechanisms. There is transcriptional regulation of pilus expression in Mc (159), and if there is transcriptional regulation of pilus expression in Gc, it is by different unknown mechanisms. There is stochastic phase variation of pilus expression that can result in three major levels of expression. Most clinical isolates of Gc are piliated expressing many pili per cell (160,161). Since Mc do not express the pilus-dependent colony morphology characteristic of Gc, it is less certain whether Mc piliation is also strongly selected for in vivo. It is likely that loss of full piliation occurs in both Gc and Mc infections but this has not been extensively studied. Loss of piliation would allow individual bacterial cells to escape from bacterial communities (either microcolonies or biofilms) to move to different anatomical sites or to different hosts.
Both Gc and Mc can lose a majority of pilus expression through phase variation of the pilus-associated pilC genes (162). Both species have two independent pilC loci that each have a PNR in their coding sequences that allow then to vary ON and OFF. Expression of either the pilC1 or pilC2 locus allows for full piliation (162), although in Mc there have been different reported different phenotypes for the PilC1 and PilC2 pili, but this has been disputed (163). However, even when both pilC genes are phased OFF, there remains a low level of pilus expression and pilus expression can be restored by inactivating the PilT traffic ATPase (164,165). Low piliation variants can also result from pilin antigenic variation reactions that result in a full length pilin protein but one that is either inefficiently assembled into pili or cannot maintain an extended pilus conformation (166,167). True OFF phase variation of pilus expression is only achieved when Gc incorporates a silent copy sequence encoding a nonsense codon (89,166,168), but not all Gc isolates have silent copies with stop codons and silent copy-encoded stop codons have not been reported for Mc. Therefore, all of Mc pilus phase variation and a majority of Gc phase variation occurs between full piliation and poor piliation and not between ON and OFF.
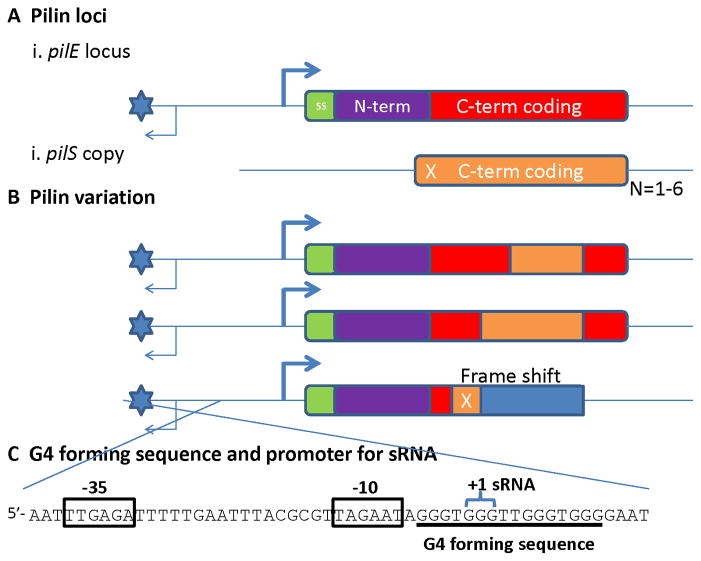
Most of the work on the mechanisms of pilin antigenic variation has been done using Gc, and while the central mechanisms are likely to be shared between Gc and Mc, this section will exclusively feature results from Gc and the relationship to Mc is assumed but has not always been tested. Both Gc strains MS11 and FA1090 have been used to study pilin antigenic variation and have 15 and 19 silent copies, respectively, arranged in different loci both upstream and downstream of the expression locus (169,170). The silent copies differ from the expression locus in that the silent copies are missing the conserved N-terminal coding sequences, the ribosome binding site and the transcriptional promoter (Fig. 11A). Thus, although the silent copies do not produce a functional protein, they function to supply variant sequences to pilE. Transcription of the pilE gene is not required for pilin antigenic variation, disproving the hypothesis that transcription could differentiate the donor silent copies from the recipient pilE locus (171).
Figure 11. Neisserial pilin antigenic variation.
A) The pilin loci. (i) The pilin expression locus (pilE) has a promoter sequence to initiate transcription, a 7 amino acid signal sequence (SS – green); an absolutely-conserved, N-terminal constant coding sequence (N-term, purple) and a variable carboxy-terminal coding sequence (C-term, red). The G4 forming sequence is represented by the blue star and the sRNA transcript by the thin arrow. (ii) Shown is a variant pilS gene copy that has variant pilin C-terminal coding sequences (C-term, orange), but cannot express a protein product. This pilS copy has a frame shift in the 5′-coding sequences (X). Each pilS locus can have one to six repeated pilS copies (N=1–6). B) Pilin variation. Nonreciprocal transfer of variant pilS copy sequences into the pilE locus produces pilin variants. The pilS sequences that are transferred can be from any part of the pilS copy that overlaps with the pilE sequence as long as there are regions of microhomology at the ends of the transferred sequence (not shown). Some (but not all) pilS copies encode a frame shift in their 5′ potential coding sequences that can result in an altered reading frame (blue), a premature stop codon and a nonpiliated variant. C) The G4 forming sequence and promoter for the cis-acting sRNA. The sequence shown is on the bottom strand of the cartoons in parts A and B. The sRNA promoter is indicated by the boxed −10 and −35 sequences. The sRNA required for pilin antigenic variation initiates (+1 sRNA) within the second set of Gs in the G4 forming sequence.
Pilin antigenic variation results from the nonreciprocal transfer of variant pilin coding sequences from one of the silent copies to the pilE locus (Fig. 11B); thus the silent copies do not normally change during pilin antigenic variation. However, this may not be true for the silent copies that are located immediately upstream of the pilE gene (unpublished observations). The segment of DNA that is transferred can be any part of the silent copy that aligns with the pilE sequence (168,172), and the changes can be as small as one nucleotide or the replacement of the entire sequence by the donor pilS copy sequence (168,172). Many pilE variants have sequence information from two silent donors and some have three or more. The transferred sequence is always bordered by regions of micro homology that can be as small as 3 bp (168,172). The frequency of variants can be from 3–15% depending on the species and strain, and not all silent copies recombine at the same frequency (168,173,174). However, the rules that govern the frequency of each donor silent copy used have not been determined. The multiple donor silent copies located at positions on either side of the recipient pilE gene creates an issue for modeling how gene conversion can occur within a bacterial chromosome. This led to the hypothesis that recombination might occur between sister chromosomes and it was found that both Gc and Mc have two chromosomal copies, while the most closely related commensal that does not undergo pilin antigenic variation (Neisseria lactamica) has one chromosomal copy (175,176). These pathogenic Neisseria are still genetically haploid since Gc cannot maintain different alleles at the same locus (176).
Pilin antigenic variation relies on the general recombination and repair systems common to most bacterial species. The process is absolutely dependent on having functional RecA, RecO, and RecR recombinases, and shows reduced frequencies when RecQ, RecJ, Rep, RecX, RdgC, RuvABC or RecG are inactivated by mutation (89,177–182). The partial phenotypes of some of these factors may be due to redundancy (e.g., the helicases RecQ and Rep), an enhancing role on another factor (e.g., RecX on RecA), or the processing of different substrates that can lead to antigenic variation (not known). In addition, the Holliday Junction processing helicases RuvAB and RecG are both individually required for pilin antigenic variation, but a double mutant with recG and any one of the ruvABC genes produces synthetic lethality only when pilin antigenic variation can occur (183). Thus, cells that do not initiate pilin antigenic variation can survive, as do cells that have deleted the pilE locus (183). Inactivation of many of the recombinases required for pilin antigenic variation can also prevent the synthetic lethality ((183) and unpublished data). Inactivation of mismatch correction increases the frequency of pilin antigenic variation (and also PilC phase variation) (184) and loss of mismatch correction also increases the level of synthetic lethality (Rotman, E. and Seifert H.S., in preparation).
There are several sequences required for pilin antigenic variation. The first identified was called the Sma/Cla repeat since it often has sites for the Sma I and Cla I restriction endonucleases (185). This repeat is present at the 3′ end of all pilin loci, both pilE and each pilS locus, regardless of the number of pilS copies in that locus. It has been suggested that the Sma/Cla repeat binds an unknown recombinase (186), but it is likely that the Sma/Cla sequence provides an extended region of 3′-homology for recombination during pilin antigenic variation. The second site that has been suggested to be important for pilin antigenic variation is the cys2 sequence within the pilE and pilS copies. This sequence encodes an invariant 10 amino acid segment of the pilin protein that contains one of the two conserved cysteines. If the ~30 bp conserved cys2 region is interrupted with an antibiotic resistance cassette or a 10 bp linker, normal pilin antigenic variation is disrupted, even though the same 10 bp linker inserted into the neighboring hypervariable loop sequences has no effect on the reactions (187). It has not been determined whether the cys2 sequence is important to provide a stretch of homology during recombination or plays a more active role in the process.
The most important cis-acting sequence that has been described is the pilE-associated guanine quartet (G4) forming sequence (93). The existence of this element was suggested by transposon insertions that totally inactivated pilin antigenic variation (171,182). These transposons were located upstream of the pilE gene, were not within an open reading frame, and did not alter pilin expression (171,182). Saturation mutagenesis of the region containing the transposon insertions showed that single point mutations in any one of 12 G-C bps disrupt pilin antigenic variation (93) (Fig. 11). Mutation of the four T-A bp within the region of G-C bp or sequences immediately adjacent to this GC-rich segment had no effect on pilin antigenic variation (93). The G-rich strand was predicted to form a G4 structure and this prediction was confirmed by a series of in vitro assays (93). The structure of this G4 determined by Nuclear Magnetic Resonance Spectroscopy predicted that the G4 structure could bind to RecA and this was confirmed experimentally (94). Interestingly, other G4 forming sequences precisely replacing the pilE G4 sequence could not mediate pilin antigenic variation and did not bind RecA (93,94). Whether RecA binding to the pilE G4 structure is important for pilin antigenic variation has not been determined. Activity of the G4 forming sequence was also found to depend on a promoter element that was located between the G4 forming sequence and the pilE that transcribes the G-rich strand the opposite direction from the pilE gene (188). The transcript produced initiates within the second set of Gs within the G4 forming sequence but this transcript is of low abundance and does not appear to have a defined 3′-end (188). It is likely that this transcript allows the G4 structure to form by melting the DNA duplex and occluding the C-rich strand through the formation of an R-loop. What molecular steps follow these reactions are not known. Expression of the G4 structure and transcript at an ectopic locus does not rescue a promoter mutation showing this element must be present in-cis (188). Moreover, the G4 forming sequence and associated transcript only work in the normal orientation at the normal location in the chromosome. If the direction is reversed or G-rich and C-rich strand are exchanged, antigenic variation cannot proceed (188). One hypothesis that might explain the orientation is that the G4 structure is on the lagging strand while the putative R-loop is on the leading strand and that these elements acts by specifically interacting with the replication apparatus. A replication interrupting structure may be a common mechanism of initiating recombination-based diversity generation since other G-rich sequences have been subsequently proposed to have roles in Borellia (92) and Treponema pallidum antigenic variation (189). When replication stalls at the G-rich site, a programed recombination/repair process occurs to initiate each phenotypic variation process.
Knowledge Gaps
The three major variation systems expressed by the pathogenic Neisseria species strongly suggests that they are under strong selection for stochastically produced subpopulations. While we assume that this selection is immune-based, it is almost certain that the initial evolution of these diversity generating systems was based on functional alterations (e.g., different Opa receptors) and not immune selection. As many issues in biology, these diversity generation systems are used for more than one function. Regardless of whether it is functional, immune selection or both, the existence and conservation of these systems also suggest that the pathogenic Neisseria function as populations to allow subpopulations to be selected for rather than regulation that effects the entire population.
What is the most notable about Neisseria antigenic variation is that these are mostly mucosal pathogens, while many of the other antigenically variable organisms are blood-born and exposed to antibody and phagocyte surveillance regularly. It is possible that the ability to continually infect the high-risk, core group of the sexually active population is the driving selection behind Gc variation. However, there are reports that Gc infection is immune-suppressive (190), which begs the question of why such a large amount of antigenic variation is necessary. The case for immune selection is less obvious for Mc with its largely commensal lifestyle and smaller capacity for variation. With Mc it may be functional selection that remains the driving force for phenotypic variation and whether phenotypic variation contributes to invasive disease remains unanswered.
While there have been many advances into understanding the mechanisms behind the gene conversion reactions that underlie pilin antigenic variation, there are many open questions remaining. Are the mechanisms used by Mc identical to those used by Gc? Does recombination occur between diploid chromosomes? Does the G4 sRNA play a direct role in the process? Is there a factor that facilitates G4 structure formation? Does G4 structure formation promote recombination through inhibiting replication, binding a recombinase, or both? What other sites are required to complete this high-frequency, gene conversion process? Is this a homologous recombination process using heteroduplex, an annealing process, or both? Answering these questions will increase our understanding of the possible mechanisms allow all of the phenotypic variation in prokaryotes and eukaryotes that rely on recombination.
References
- 1.Ross T, Thomson D. A case of sleeping sickness studied by precise enumerative methods: regular periodic increase of the parasites disclosed. Ann Trop Med Parasitol. 1910;4:261–264. [Google Scholar]
- 2.Holland J, Spindler K, Horodyski F, Grabau E, Nichol S, VandePol S. Rapid evolution of RNA genomes. Science. 1982;215:1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- 3.Deitsch KW, Lukehart SA, Stringer JR. Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nature Rev Microbiol. 2009;7:493–503. doi: 10.1038/nrmicro2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palmer GH, Bankhead T, Lukehart SA. Nothing is permanent but change: Antigenic variation in persistent bacterial pathogens. Cell Microbiol. 2009;11:1697–1705. doi: 10.1111/j.1462-5822.2009.01366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Theiler A. Report of the Government Veterinary Bacteriologist, 1908–1909. Transvaal, South Africa: 1910. Anaplasma marginale (gen. spec. nov.). The marginal points in the blood of cattle suffering from a specific disease. [Google Scholar]
- 6.Palmer GH. Sir Arnold Theiler and the discovery of anaplasmosis: A centennial perspective. Onderstepoort J Vet Res. 2009;76:75–79. doi: 10.4102/ojvr.v76i1.68. [DOI] [PubMed] [Google Scholar]
- 7.Dumler JS, Barbet AF, Bekker CPJ, Dasch GA, Palmer GH, Ray SC, Rikihisa Y, Rurangirwa FR. Reorganization of genera in the Families Rickettsiaceae and Anaplasmataceae in the Order Rickettsiales; unification of some Species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia, and Ehrlichia with Neorickettsia; descriptions of six new species combinations; and designation of Ehrlichia equi and “HGE agent” as subjective synonyms of Ehrlichia phagocytophila. Int J Sys Evol Microbiol. 2001;51:2145–2165. doi: 10.1099/00207713-51-6-2145. [DOI] [PubMed] [Google Scholar]
- 8.Dunning Hotopp JC, Lin M, Madupu R, Crabtree J, Angiuoli SV, Eisen JA, Seshadri R, Ren Q, Wu M, Utterback TR, Smith S, Lewis M, Khouri H, Zhang C, Niu H, Lin Q, Ohashi N, Zhi N, Nelson W, Brinkac LM, Dodson RJ, Rosovitz MJ, Sundaram J, Daugherty SC, Davidsen T, Durkin AS, Gwinn M, Haft DH, Selengut JD, Sullivan SA, Zafar N, Zhou L, Benahmed F, Forberger H, Halpin R, Mulligan S, Robinson J, White O, Rikihisa Y, Tettelin H. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2006;2:e21. doi: 10.1371/journal.pgen.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Granquist EG, Stuen S, Crosby L, Lundgren AM, Alleman AR, Barbet AF. Variant-specific and diminishing immune responses towards the highly variable MSP2(P44) outer membrane protein of Anaplasma phagocytophilum during persistent infection in lambs. Vet Immunol Immunopathol. 2010;133:117–124. doi: 10.1016/j.vetimm.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rejmanek D, Foley P, Barbet A, Foley J. Antigenic variability in Anaplasma phagocytophilum during chronic infection of a reservoir host. Microbiol. 2012;158:2632–2641. doi: 10.1099/mic.0.059808-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai TH, Orellana NG, Yuasa Y, Rikihisa Y. Cloning of the major outer membrane protein expression locus in Anaplasma platys and seroreactivity of a species-specific antigen. J Bacteriol. 2011;193:2924–2930. doi: 10.1128/JB.00082-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palmer GH, Abbott JR, French DM, McElwain TF. Persistence of Anaplasma ovis infection and conservation of the msp2 and msp3 multigene families within the genus Anaplasma. Infect Immun. 1998;66:6035–6039. doi: 10.1128/iai.66.12.6035-6039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmer GH, Brown WC, Rurangirwa FR. Antigenic variation in the persistence and transmission of the ehrlichia Anaplasma marginale. Microb Infect. 2000;2:167–176. doi: 10.1016/s1286-4579(00)00271-9. [DOI] [PubMed] [Google Scholar]
- 14.French DF, McElwain TF, McGuire TC, Palmer GH. Expression of Anaplasma marginale Major Surface Protein 2 variants during persistent cyclic rickettsemia. Infect Immun. 1998;66:1200–1207. doi: 10.1128/iai.66.3.1200-1207.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eriks IS, Stiller D, Palmer GH. Impact of persistent Anaplasma marginale rickettsemia on tick infection and transmission. J Clin Microbiol. 1993;31:2091–2096. doi: 10.1128/jcm.31.8.2091-2096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scoles GA, Broce AB, Lysyk TJ, Palmer GH. Relative efficiency of biological transmission of Anaplasma marginale (Rickettsiales: Anaplasmataceae) by Dermacentor andersoni Stiles (Acari: Ixodidae) compared to mechanical transmission by the stable fly, Stomoxys calcitrans (L.) (Diptera: Muscidae) J Med Entomol. 2005;42:668–675. doi: 10.1603/0022-2585(2005)042[0668:REOBTO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 17.Ueti MW, Knowles DP, Davitt CM, Scoles GA, Baszler TV, Palmer GH. Quantitative differences in salivary pathogen load during tick transmission underlie strain-specific variation in transmission efficiency of Anaplasma marginale. Infect Immun. 2009;77:70–75. doi: 10.1128/IAI.01164-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brayton KA, Kappmeyer LS, Herndon DR, Dark MJ, Tibbals DL, Palmer GH, McGuire TC, Knowles DP. Complete genome sequencing of Anaplasma marginale reveals that the surface is skewed to two superfamilies of outer membrane proteins. Proc Natl Acad Sci USA. 2005;102:844–849. doi: 10.1073/pnas.0406656102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noh SM, Brayton KA, Knowles DP, Agnes JT, Dark MJ, Brown WC, Baszler TV, Palmer GH. Differential expression and sequence conservation of the Anaplasma marginale msp2 gene superfamily outer membrane proteins. Infect Immun. 2006;74:3471–3479. doi: 10.1128/IAI.01843-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noh SM, Brayton KA, Brown WC, Norimine J, Munske GR, Davitt CM, Palmer GH. Composition of the surface proteome of Anaplasma marginale and its role in protective immunity induced by outer membrane immunization. Infect Immun. 2008;76:2219–2226. doi: 10.1128/IAI.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palmer GH, Brown WC, Noh SM, Brayton KA. Genome-wide screening and identification of antigens for rickettsial vaccine development. FEMS Immunolo Med Microbiol. 2012;64:115–119. doi: 10.1111/j.1574-695X.2011.00878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmer GH, Barbet AF, Kuttler KL, McGuire TC. Detection of an Anaplasma marginale common surface protein present in all stages of infection. J Cin MIcro. 1986;23:1078–1083. doi: 10.1128/jcm.23.6.1078-1083.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGuire TC, Davis WC, Brassfield AL, McElwain TF, Palmer GH. Identification of Anaplasma marginale long-term carrier cattle by detection of serum antibody to isolated MSP-3. J Clin Microbiol. 1991;29:788–793. doi: 10.1128/jcm.29.4.788-793.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmer GH, Eid G, Barbet AF, McGuire TC, McElwain TF. The immunoprotective Anaplasma marginale major surface protein-2 (MSP-2) is encoded by a polymorphic multigene family. Infect Immun. 1994;62:3808–3816. doi: 10.1128/iai.62.9.3808-3816.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meeus PFM, Brayton KA, Palmer GH, Barbet AF. Conservation of a gene conversion mechanism in two distantly related paralogues of Anaplasma marginale. Mol Microbiol. 2003;47:633–643. doi: 10.1046/j.1365-2958.2003.03331.x. [DOI] [PubMed] [Google Scholar]
- 26.Brayton KA, Knowles DP, McGuire TC, Palmer GH. Efficient use of small genome to generate antigenic diversity in tick-borne ehrlichial pathogens. Proc Natl Acad Sci USA. 2001;98:4130–4135. doi: 10.1073/pnas.071056298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.French DM, Brown WC, Palmer GH. Emergence of Anaplasma marginale antigenic variants during persistent rickettsemia. Infect Immun. 1999;67:5834–5840. doi: 10.1128/iai.67.11.5834-5840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brayton KA, Meeus PFM, Barbet AF, Palmer GH. Simultaneous variation of the immunodominant outer membrane proteins MSP2 and MSP3 during Anaplasma marginale persistence in vivo. Infect Immun. 2003;71:6627–6632. doi: 10.1128/IAI.71.11.6627-6632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abbott JR, Palmer GH, Howard CJ, Hope JC, Brown WC. Anaplasma marginale major surface protein 2 CD4+ T-cell epitopes are evenly distributed in conserved and hypervariable regions (HVR), whereas linear B-cell epitopes are predominantly located in the HVR. Infect Immun. 2004;72:7360–7366. doi: 10.1128/IAI.72.12.7360-7366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhuang Y, Futse JE, Brown WC, Brayton KA, Palmer GH. Maintenance of antibody to pathogen epitopes generated by segmental gene conversion is highly dynamic during long-term persistent infection. Infect Immun. 2007;75:5185–5190. doi: 10.1128/IAI.00913-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown WC, Brayton KA, Styer CM, Palmer GH. The hypervariable region of Anaplasma marginale major surface protein 2 (MSP2) contains multiple immunodominant CD4+ T lymphocyte epitopes that elicit variant-specific proliferative and IFN-γ responses in MSP2 vaccinates. J Immunol. 2003;170:3790–3798. doi: 10.4049/jimmunol.170.7.3790. [DOI] [PubMed] [Google Scholar]
- 32.Han S, Norimine J, Palmer GH, Mwangi W, Lahmers KK, Brown WC. Rapid deletion of antigen-specific CD4+ T 1 cells following infection represents a strategy of immune evasion and persistence for Anaplasma marginale. J Immunol. 2008;181:7759–7769. doi: 10.4049/jimmunol.181.11.7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han S, Norimine J, Brayton KA, Palmer GH, Scoles GA, Brown WC. Anaplasma marginale infection with persistent high load bacteremia induces a dysfunctional memory CD4+ T lymphocyte response but sustained high IgG titers. Clin Vacc Immunol. 2010;17:1881–1890. doi: 10.1128/CVI.00257-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turse JE, Scoles GA, Deringer JR, Fry LM, Brown WC. Immunization-induced Anaplasma marginale-specific T-lymphocyte responses impaired by A. marginale infection are restored after eliminating infection with tetracycline. Clin Vacc Immunol. 2014;21:1369–1375. doi: 10.1128/CVI.00246-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Futse JE, Brayton KA, Knowles DP, Palmer GH. Structural basis for segmental gene conversion in generation of Anaplasma marginale outer membrane protein variants. Mol Microbiol. 2005;57:212–221. doi: 10.1111/j.1365-2958.2005.04670.x. [DOI] [PubMed] [Google Scholar]
- 36.Futse JE, Brayton KA, Nydam SD, Palmer GH. Generation of antigenic variants by gene conversion: Evidence for recombination fitness selection at the locus level in Anaplasma marginale. Infect Immun. 2009;71:6627–6632. doi: 10.1128/IAI.00348-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dark MJ, Herndon D, Kappmeyer LS, Gonzales MP, Nordeen E, Palmer GH, Knowles DP, Brayton KA. Conservation in the face of diversity: multistrain analysis of an intracellular bacterium. BMC Genomics. 2009;10:16–28. doi: 10.1186/1471-2164-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barbet AF, Lundgren A, Yi J, Rurangirwa FR, Palmer GH. Antigenic variation of Anaplasma marginale by expression of MSP2 sequence mosaics. Infect Immun. 2000;68:6133–6138. doi: 10.1128/iai.68.11.6133-6138.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brayton KA, Palmer GH, Lundgren A, Yi J, Barbet AF. Antigenic variation of Anaplasma marginale msp2 occurs by combinatorial gene conversion. Mol Microbiol. 2002;43:1151–1159. doi: 10.1046/j.1365-2958.2002.02792.x. [DOI] [PubMed] [Google Scholar]
- 40.Ueti MW, Tan Y, Broschat SL, Castañeda Ortiz EJ, Camacho-Nuez M, Mosqueda JJ, Scoles GA, Grimes M, Brayton KA, Palmer GH. Expansion of variant diversity associated with high prevalence of pathogen strain superinfection under conditions of natural transmission. Infect Immun. 2012;80:2354–2360. doi: 10.1128/IAI.00341-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmer GH, Futse JE, Leverich CK, Knowles DP, Rurangirwa FR, Brayton KA. Selection for simple Msp2 variants during Anaplasma marginale transmission to immunologically naïve animals. Infect Immun. 2007;75:1502–1506. doi: 10.1128/IAI.01801-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Löhr CV, Rurangirwa FR, McElwain TF, Stiller D, Palmer GH. Specific expression of Anaplasma marginale major surface protein 2 salivary gland variants occurs in the midgut and is an early event during tick transmission. Infect Immun. 2002;70:114–120. doi: 10.1128/IAI.70.1.114-120.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rurangirwa FR, Stiller DS, French DM, Palmer GH. Restriction of Major Surface Protein 2 (MSP2) variants during tick transmission of the ehrlichiae Anaplasma marginale. Proc Natl Acad Sci USA. 1999;96:3171–3176. doi: 10.1073/pnas.96.6.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith DM, Richman DD, Little SJ. HIV superinfection. J Infect Dis. 2005;192:438–444. doi: 10.1086/431682. [DOI] [PubMed] [Google Scholar]
- 45.Blackard JT. HCV superinfection and reinfection. Antiv Ther. 2012;17:1443–1448. doi: 10.3851/IMP2460. [DOI] [PubMed] [Google Scholar]
- 46.Palmer GH, Knowles DP, Rodriguez JL, Gnad DP, Hollis LC, Marston T, Brayton KA. Stochastic transmission of multiple genotypically distinct Anaplasma marginale strains within an endemic herd. J Clin Microbiol. 2004;42:5381–5384. doi: 10.1128/JCM.42.11.5381-5384.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez JL, Palmer GH, Knowles DP, Brayton KA. Distinctly different msp2 pseudogene repertoires in Anaplasma marginale strains that are capable of superinfection. Gene. 2005;361:127–132. doi: 10.1016/j.gene.2005.06.038. [DOI] [PubMed] [Google Scholar]
- 48.Futse JE, Brayton KA, Dark MJ, Knowles DP, Palmer GH. Superinfection as a driver of genomic diversification in antigenically variant pathogens. Proc Natl Acad Sci USA. 2008;105:2123–2127. doi: 10.1073/pnas.0710333105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vallejo Esquerra E, Herndon DR, Alpirez Mendoza F, Mosqueda J, Palmer GH. Anaplasma marginale superinfection attributable to pathogen strains with distinct genomic backgrounds. Infect Immun. 2014;82:5286–5292. doi: 10.1128/IAI.02537-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rogovsky AS, Bankhead T. Bacterial heterogeneity is a requirement for host superinfection by the Lyme disease spirochete. Infect Immun. 2014;82:4542–4552. doi: 10.1128/IAI.01817-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palmer GH, Rurangirwa FR, McElwain TF. Strain composition of the ehrlichia Anaplasma marginale within persistently infected cattle, a mammalian reservoir for tick-transmission. J Clin Microbiol. 2001;39:631–635. doi: 10.1128/JCM.39.2.631-635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De la Fuente J, Passos LM, Van Den Bussche RA, Ribeiro MF, Facury-Filho EJ, Kocan KM. Genetic diversity and molecular phylogeny of Anaplasma marginale isolates from Minas Gerais, Brazil. Vet Parasitol. 2004;121:307–316. doi: 10.1016/j.vetpar.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 53.Ruybal P, Moretta R, Perez A, Petrigh R, Zimmer P, Alcarez E, Echaide I, Torioni de Echaide S, Kocan KM, de la Fuente J, Farber M. Genetic diversity of Anaplasma marginale in Argentina. Vet Parasitol. 2009;162:176–180. doi: 10.1016/j.vetpar.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 54.Palmer GH, Brayton KA. Antigenic variation and transmission fitness as drivers of bacterial strain structure. Cell Microbiol. 2013;15:1969–1975. doi: 10.1111/cmi.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paster BJ, Dewhirst FE, Weisburg WG, Tordoff LA, Fraser GJ, Hespell RB, Stanton TB, Zablen L, Mandelco L, Woese CR. Phylogenetic analysis of the spirochetes. J Bacteriol. 1991;173:6101–6109. doi: 10.1128/jb.173.19.6101-6109.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Radolf JD, Samuels DS. Borrelia: molecular biology, host interaction, and pathogenesis. Caister Academic Press; Norfolk, UK: 2010. [Google Scholar]
- 57.Ras NM, Lascola B, Postic D, Cutler SJ, Rodhain F, Baranton G, Raoult D. Phylogenesis of relapsing fever Borrelia spp. Int J Sys Bacteriol. 1996;46:859–865. doi: 10.1099/00207713-46-4-859. [DOI] [PubMed] [Google Scholar]
- 58.Schwan TG, Raffel SJ, Schrumpf ME, Porcella SF. Diversity and distribution of Borrelia hermsii. Emerg Inf Dis. 2007;13:436–442. doi: 10.3201/eid1303.060958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stoenner HG, Dodd T, Larsen C. Antigenic variation of Borrelia hermsii. J Exp Med. 1982;156:1297–1311. doi: 10.1084/jem.156.5.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barbour AG, Restrepo BI. Antigenic variation in vector-borne pathogens. Emerg Inf Dis. 2000;6:449–457. doi: 10.3201/eid0605.000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Plasterk RH, Simon MI, Barbour AG. Transposition of structural genes to an expression sequence on a linear plasmid causes antigenic variation in the bacterium Borrelia hermsii. Nature. 1985;318:257–263. doi: 10.1038/318257a0. [DOI] [PubMed] [Google Scholar]
- 62.Barbour AG, Bundoc V. In vitro and in vivo neutralization of the relapsing fever agent Borrelia hermsii with serotype-specific immunoglobulin M antibodies. Infect Immun. 2001;69:1009–1015. doi: 10.1128/IAI.69.2.1009-1015.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Connolly SE, Benach JL. Cutting edge: the spirochetemia of murine relapsing fever is cleared by complement-independent bactericidal antibodies. J Immunol. 2001;167:3029–3032. doi: 10.4049/jimmunol.167.6.3029. [DOI] [PubMed] [Google Scholar]
- 64.Barbour AG. Borrelia: a diverse and ubiquitous genus of tick-borne pathogens. In: Scheld MW, Craig WA, Hughes JM, editors. Emerging Infections 5. American Society for Microbiology; Washington, D.C: 2001. pp. 153–173. [Google Scholar]
- 65.Steere AC, Coburn J, Glickstein L. The emergence of Lyme disease. J Clin Invest. 2004;113:1093–1101. doi: 10.1172/JCI21681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Norris SJ. Antigenic variation with a twist - the Borrelia story. Mol Microbiol. 2006;60:1319–1322. doi: 10.1111/j.1365-2958.2006.05204.x. [DOI] [PubMed] [Google Scholar]
- 67.Zhang JR, Hardham JM, Barbour AG, Norris SJ. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell. 1997;89:275–285. doi: 10.1016/s0092-8674(00)80206-8. [DOI] [PubMed] [Google Scholar]
- 68.Zhang JR, Norris SJ. Genetic variation of the Borrelia burgdorferi gene vlsE involves cassette-specific, segmental gene conversion. Infect Immun. 1998;66:3698–3704. doi: 10.1128/iai.66.8.3698-3704.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang JR, Norris SJ. Kinetics and in vivo induction of genetic variation of vlsE in Borrelia burgdorferi. Infect Immun. 1998;66:3689–3697. doi: 10.1128/iai.66.8.3689-3697.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hudson CR, Frye JG, Quinn FD, Gherardini FC. Increased expression of Borrelia burgdorferi vlsE in response to human endothelial cell membranes. Mol Microbiol. 2001;41:229–239. doi: 10.1046/j.1365-2958.2001.02511.x. [DOI] [PubMed] [Google Scholar]
- 71.Indest KJ, Howell JK, Jacobs MB, Scholl-Meeker D, Norris SJ, Philipp MT. Analysis of Borrelia burgdorferi vlsE gene expression and recombination in the tick vector. Infect Immun. 2001;69:7083–7090. doi: 10.1128/IAI.69.11.7083-7090.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McDowell JV, Sung SY, Hu LT, Marconi RT. Evidence that the variable regions of the central domain of VlsE are antigenic during infection with Lyme disease spirochetes. Infect Immun. 2002;70:4196–4203. doi: 10.1128/IAI.70.8.4196-4203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Labandeira-Rey M, Skare JT. Decreased infectivity in Borrelia burgdorferi strain B31 is associated with loss of linear plasmid 25 or 28-1. Infect Immun. 2001;69:446–455. doi: 10.1128/IAI.69.1.446-455.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Purser JE, Norris SJ. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc Natl Acad Sci U S A. 2000;97:13865–13870. doi: 10.1073/pnas.97.25.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Labandeira-Rey M, Seshu J, Skare JT. The absence of linear plasmid 25 or 28-1 of Borrelia burgdorferi dramatically alters the kinetics of experimental infection via distinct mechanisms. Infect Immun. 2003;71:4608–4613. doi: 10.1128/IAI.71.8.4608-4613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Purser JE, Lawrenz MB, Caimano MJ, Howell JK, Radolf JD, Norris SJ. A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol Microbiol. 2003;48:753–764. doi: 10.1046/j.1365-2958.2003.03452.x. [DOI] [PubMed] [Google Scholar]
- 77.Lawrenz MB, Wooten RM, Norris SJ. Effects of vlsE complementation on the infectivity of Borrelia burgdorferi lacking the linear plasmid lp28-1. Infect Immun. 2004;72:6577–6585. doi: 10.1128/IAI.72.11.6577-6585.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bankhead T, Chaconas G. The role of VlsE antigenic variation in the Lyme disease spirochete: persistence through a mechanism that differs from other pathogens. Mol Microbiol. 2007;65:1547–1558. doi: 10.1111/j.1365-2958.2007.05895.x. [DOI] [PubMed] [Google Scholar]
- 79.Beaurepaire C, Chaconas G. Mapping of essential replication functions of the linear plasmid lp17 of B. burgdorferi by targeted deletion walking. Mol Microbiol. 2005;57:132–142. doi: 10.1111/j.1365-2958.2005.04688.x. [DOI] [PubMed] [Google Scholar]
- 80.Chaconas Hairpin telomere and genome plasticity in Borrelia: all mixed up in the end. Mol Microbiol. 2005;58:625–635. doi: 10.1111/j.1365-2958.2005.04872.x. [DOI] [PubMed] [Google Scholar]
- 81.Kobryn K, Chaconas G. ResT, a telomere resolvase encoded by the Lyme disease spirochete. Mol Cell. 2002;9:195–201. doi: 10.1016/s1097-2765(01)00433-6. [DOI] [PubMed] [Google Scholar]
- 82.Embers ME, Alvarez X, Ooms T, Philipp MT. The failure of immune response evasion by linear plasmid 28-1-deficient Borrelia burgdorferi is attributable to persistent expression of an outer surface protein. Infect Immun. 2008;76:3984–3991. doi: 10.1128/IAI.00387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fung BP, McHugh GL, Leong JM, Steere AC. Humoral immune response to outer surface protein C of Borrelia burgdorferi in Lyme disease: role of the immunoglobulin M response in the serodiagnosis of early infection. Infect Immun. 1994;62:3213–3221. doi: 10.1128/iai.62.8.3213-3221.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liang FT, Brown EL, Wang T, Iozzo RV, Fikrig E. Protective niche for Borrelia burgdorferi to evade humoral immunity. Am J Pathol. 2004;165:977–985. doi: 10.1016/S0002-9440(10)63359-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu Q, Seemanapalli SV, McShan K, Liang FT. Constitutive expression of outer surface protein C diminishes the ability of Borrelia burgdorferi to evade specific humoral immunity. Infect Immun. 2006;74:5177–5184. doi: 10.1128/IAI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Casjens S, Palmer N, Van Vugt R, Huang WH, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, Haft D, Hickey E, Gwinn M, White O, Fraser CM. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 87.Coutte L, Botkin DJ, Gao L, Norris SJ. Detailed analysis of sequence changes occurring during vlsE antigenic variation in the mouse model of Borrelia burgdorferi infection. PLoS Pathog. 2009;5:e1000293. doi: 10.1371/journal.ppat.1000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liveris D, Mulay V, Sandigursky S, Schwartz I. Borrelia burgdorferi vlsE antigenic variation is not mediated by RecA. Infect Immun. 2008;76:4009–4018. doi: 10.1128/IAI.00027-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koomey M, Gotschlich EC, Robbins K, Bergstrom S, Swanson J. Effects of recA mutations on pilus antigenic variation and phase transitions in Neisseria gonorrhoeae. Genetics. 1987;117:391–398. doi: 10.1093/genetics/117.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dresser AR, Hardy PO, Chaconas G. Investigation of the genes involved in antigenic switching at the vlsE locus in Borrelia burgdorferi: an essential role for the RuvAB branch migrase. PLoS Pathog. 2009;5:e1000680. doi: 10.1371/journal.ppat.1000680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lin T, Gao L, Edmondson DG, Jacobs MB, Philipp MT, Norris SJ. Central role of the Holliday junction helicase RuvAB in vlsE recombination and infectivity of Borrelia burgdorferi. PLoS Pathog. 2009;5:e1000679. doi: 10.1371/journal.ppat.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Walia R, Chaconas G. Suggested role for G4 DNA in recombinational switching at the antigenic variation locus of the Lyme disease spirochete. PLoS One. 2013;8:e57792. doi: 10.1371/journal.pone.0057792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cahoon LA, Seifert HS. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science. 2009;325:764–767. doi: 10.1126/science.1175653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kuryavyi V, Cahoon LA, Seifert HS, Patel DJ. RecA-binding pilE G4 sequence essential for pilin antigenic variation forms monomeric and 5′ end-stacked dimeric parallel G-quadruplexes. Structure. 2012;20:2090–2102. doi: 10.1016/j.str.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Eicken C, Sharma V, Klabunde T, Lawrenz MB, Hardham JM, Norris SJ, Sacchettini JC. Crystal structure of Lyme disease variable surface antigen VlsE of Borrelia burgdorferi. J Biol Chem. 2002;277:21691–21696. doi: 10.1074/jbc.M201547200. [DOI] [PubMed] [Google Scholar]
- 96.Zuckert WR. A call to order at the spirochaetal host-pathogen interface. Mol Microbiol. 2013;89:207–211. doi: 10.1111/mmi.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bacon RM, Biggerstaff BJ, Schriefer ME, Gilmore RD, Jr, Philipp MT, Steere AC, Wormser GP, Marques AR, Johnson BJ. Serodiagnosis of Lyme disease by kinetic enzyme-linked immunosorbent assay using recombinant VlsE1 or peptide antigens of Borrelia burgdorferi compared with 2-tiered testing using whole-cell lysates. J Infect Dis. 2003;187:1187–1199. doi: 10.1086/374395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Embers ME, Jacobs MB, Johnson BJ, Philipp MT. Dominant epitopes of the C6 diagnostic peptide of Borrelia burgdorferi are largely inaccessible to antibody on the parent VlsE molecule. Clin Vaccine Immunol. 2007;14:931–936. doi: 10.1128/CVI.00075-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Embers ME, Wormser GP, Schwartz I, Martin DS, Philipp MT. Borrelia burgdorferi spirochetes that harbor only a portion of the lp28-1 plasmid elicit antibody responses detectable with the C6 test for Lyme disease. Clin Vaccine Immunol. 2007;14:90–93. doi: 10.1128/CVI.00266-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liang FT, Steere AC, Marques AR, Johnson BJ, Miller JN, Philipp MT. Sensitive and specific serodiagnosis of Lyme disease by enzyme-linked immunosorbent assay with a peptide based on an immunodominant conserved region of Borrelia burgdorferi vlsE. J Clin Microbiol. 1999;37:3990–3996. doi: 10.1128/jcm.37.12.3990-3996.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schulte-Spechtel U, Lehnert G, Liegl G, Fingerle V, Heimerl C, Johnson BJ, Wilske B. Significant improvement of the recombinant Borrelia-specific immunoglobulin G immunoblot test by addition of VlsE and a DbpA homologue derived from Borrelia garinii for diagnosis of early neuroborreliosis. J Clin Microbiol. 2003;41:1299–1303. doi: 10.1128/JCM.41.3.1299-1303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liang FT, Yan J, Mbow ML, Sviat SL, Gilmore RD, Mamula M, Fikrig E. Borrelia burgdorferi changes its surface antigenic expression in response to host immune responses. Infect Immun. 2004;72:5759–5767. doi: 10.1128/IAI.72.10.5759-5767.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Crother TR, Champion CI, Wu XY, Blanco DR, Miller JN, Lovett MA. Antigenic composition of Borrelia burgdorferi during infection of SCID mice. Infect Immun. 2003;71:3419–3428. doi: 10.1128/IAI.71.6.3419-3428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Barry JD, Ginger ML, Burton P, McCulloch R. Why are parasite contingency genes often associated with telomeres? Int J Parasitol. 2003;33:29–45. doi: 10.1016/s0020-7519(02)00247-3. [DOI] [PubMed] [Google Scholar]
- 105.Kraemer SM, Smith JD. A family affair: var genes, PfEMP1 binding, and malaria disease. Curr Opin Microbiol. 2006;9:374–380. doi: 10.1016/j.mib.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 106.Wang D, Botkin DJ, Norris SJ. Characterization of the vls antigenic variation loci of the Lyme disease spirochaetes Borrelia garinii Ip90 and Borrelia afzelii ACAI. Mol Microbiol. 2003;47:1407–1417. doi: 10.1046/j.1365-2958.2003.03386.x. [DOI] [PubMed] [Google Scholar]
- 107.Liang FT, Jacobs MB, Bowers LC, Philipp MT. An immune evasion mechanism for spirochetal persistence in Lyme borreliosis. J Exp Med. 2002;195:415–422. doi: 10.1084/jem.20011870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Philipp MT, Bowers LC, Fawcett PT, Jacobs MB, Liang FT, Marques AR, Mitchell PD, Purcell JE, Ratterree MS, Straubinger RK. Antibody response to IR6, a conserved immunodominant region of the VlsE lipoprotein, wanes rapidly after antibiotic treatment of Borrelia burgdorferi infection in experimental animals and in humans. J Infect Dis. 2001;184:870–878. doi: 10.1086/323392. [DOI] [PubMed] [Google Scholar]
- 109.Tilly K, Bestor A, Rosa PA. Lipoprotein succession in Borrelia burgdorferi: similar but distinct roles for OspC and VlsE at different stages of mammalian infection. Mol Microbiol. 2013;89:216–227. doi: 10.1111/mmi.12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.LaRocca TJ, Crowley JT, Cusack BJ, Pathak P, Benach J, London E, Garcia-Monco JC, Benach JL. Cholesterol lipids of Borrelia burgdorferi form lipid rafts and are required for the bactericidal activity of a complement-independent antibody. Cell Host Microbe. 2010;8:331–342. doi: 10.1016/j.chom.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Toledo A, Crowley JT, Coleman JL, LaRocca TJ, Chiantia S, London E, Benach JL. Selective association of outer surface lipoproteins with the lipid rafts of Borrelia burgdorferi. mBio. 2014;5:e00899–00814. doi: 10.1128/mBio.00899-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dzikowski R, Templeton TJ, Deitsch K. Variant antigen gene expression in malaria. Cell Microbiol. 2006;8:1371–1381. doi: 10.1111/j.1462-5822.2006.00760.x. [DOI] [PubMed] [Google Scholar]
- 113.Taylor JE, Rudenko G. Switching trypanosome coats: what’s in the wardrobe? Trends Genet. 2006;22:614–620. doi: 10.1016/j.tig.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 114.Bunikis J, Barbour AG. Access of antibody or trypsin to an integral outer membrane protein (P66) of Borrelia burgdorferi is hindered by Osp lipoproteins. Infect Immun. 1999;67:2874–2883. doi: 10.1128/iai.67.6.2874-2883.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rogovskyy AS, Bankhead T. Variable VlsE is critical for host reinfection by the Lyme disease spirochete. PLoS One. 2013;8:e61226. doi: 10.1371/journal.pone.0061226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Barthold SW. Specificity of infection-induced immunity among Borrelia burgdorferi sensu lato species. Infect Immun. 1999;67:36–42. doi: 10.1128/iai.67.1.36-42.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.de Silva AM, Fikrig E, Hodzic E, Kantor FS, Telford SR, 3rd, Barthold SW. Immune evasion by tickborne and host-adapted Borrelia burgdorferi. J Infect Dis. 1998;177:395–400. doi: 10.1086/514200. [DOI] [PubMed] [Google Scholar]
- 118.Virji M. Pathogenic neisseriae: surface modulation, pathogenesis and infection control. Nat Rev Microbiol. 2009;7:274–286. doi: 10.1038/nrmicro2097. [DOI] [PubMed] [Google Scholar]
- 119.Morton RS. Gonorrhoea in Earlier Times. In: Morton RS, editor. Gonorrhoea. Vol. 9. W.B. Saunders; London: 1977. pp. 1–24. [Google Scholar]
- 120.Ndowa FJ, Ison CA, Cole MJ, Lusti-Narasimhan M. Gonococcal antimicrobial resistance: challenges for public health control. Sex Transm Infect. 2013;89(Suppl 4):iv3–4. doi: 10.1136/sextrans-2013-051395. [DOI] [PubMed] [Google Scholar]
- 121.Kellogg DS, Jr, Thayer JD. Virulence of gonococci. Ann Rev Med. 1969;20:323–328. doi: 10.1146/annurev.me.20.020169.001543. [DOI] [PubMed] [Google Scholar]
- 122.Morse SA. The biology of the gonococcus. CRC Crit Rev Microbiol. 1978;7:93–189. doi: 10.3109/10408417909083071. [DOI] [PubMed] [Google Scholar]
- 123.Lewis DA. The role of core groups in the emergence and dissemination of antimicrobial-resistant N gonorrhoeae. Sex Transm Infect. 2013;89(Suppl 4):iv47–51. doi: 10.1136/sextrans-2013-051020. [DOI] [PubMed] [Google Scholar]
- 124.Caugant DA, Maiden MC. Meningococcal carriage and disease--population biology and evolution. Vaccine. 2009;27(Suppl 2):B64–70. doi: 10.1016/j.vaccine.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pizza M, Rappuoli R. Neisseria meningitidis: pathogenesis and immunity. Curr Opin Microbiol. 2014;23c:68–72. doi: 10.1016/j.mib.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 126.Rouphael NG, Stephens DS. Neisseria meningitidis: biology, microbiology, and epidemiology. Methods Mol Biol. 2012;799:1–20. doi: 10.1007/978-1-61779-346-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Callaghan MJ, Jolley KA, Maiden MC. Opacity-associated adhesin repertoire in hyperinvasive Neisseria meningitidis. Infect Immun. 2006;74:5085–5094. doi: 10.1128/IAI.00293-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Urwin R, Russell JE, Thompson EA, Holmes EC, Feavers IM, Maiden MC. Distribution of surface protein variants among hyperinvasive meningococci: implications for vaccine design. Infect Immun. 2004;72:5955–5962. doi: 10.1128/IAI.72.10.5955-5962.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bille E, Zahar JR, Perrin A, Morelle S, Kriz P, Jolley KA, Maiden MC, Dervin C, Nassif X, Tinsley CR. A chromosomally integrated bacteriophage in invasive meningococci. J Exp Med. 2005;201:1905–1913. doi: 10.1084/jem.20050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Harrison OB, Maiden MC, Rokbi B. Distribution of transferrin binding protein B gene (tbpB) variants among Neisseria species. BMC Microbiol. 2008;8:66. doi: 10.1186/1471-2180-8-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bennett JS, Jolley KA, Earle SG, Corton C, Bentley SD, Parkhill J, Maiden MC. A genomic approach to bacterial taxonomy: an examination and proposed reclassification of species within the genus Neisseria. Microbiol. 2012;158:1570–1580. doi: 10.1099/mic.0.056077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Snyder LA, Saunders NJ. The majority of genes in the pathogenic Neisseria species are present in non-pathogenic Neisseria lactamica, including those designated as ‘virulence genes’. BMC Genomics. 2006;7:128. doi: 10.1186/1471-2164-7-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang G, Meredith TC, Kahne D. On the essentiality of lipopolysaccharide to Gram-negative bacteria. Curr Opin Microbiol. 2013;16:779–785. doi: 10.1016/j.mib.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stern A, Brown M, Nickel P, Meyer TF. Opacity genes in Neisseria gonorrhoeae: control of phase and antigenic variation. Cell. 1986;47:61–71. doi: 10.1016/0092-8674(86)90366-1. [DOI] [PubMed] [Google Scholar]
- 135.Swanson J, Kraus SJ, Gotschlich EC. Studies on gonococcus infection. I. Pili and zones of adhesion: their relation to gonococcal growth patterns. J Exp Med. 1971;134:886–906. doi: 10.1084/jem.134.4.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Snyder LA, Butcher SA, Saunders NJ. Comparative whole-genome analyses reveal over 100 putative phase-variable genes in the pathogenic Neisseria spp. Microbiol. 2001;147:2321–2332. doi: 10.1099/00221287-147-8-2321. [DOI] [PubMed] [Google Scholar]
- 137.Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 138.Lo H, Tang CM, Exley RM. Mechanisms of avoidance of host immunity by Neisseria meningitidis and its effect on vaccine development. Lancet Infect Dis. 2009;9:418–427. doi: 10.1016/S1473-3099(09)70132-X. [DOI] [PubMed] [Google Scholar]
- 139.Moran EE, Burden R, Labrie JE, 3rd, Wen Z, Wang XM, Zollinger WD, Zhang L, Pinto VB. Analysis of the bactericidal response to an experimental Neisseria meningitidis vesicle vaccine. Clin Vacc Immunol. 2012;19:659–665. doi: 10.1128/CVI.00070-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Granoff DM. Review of meningococcal group B vaccines. Clin Infect Dis. 2010;50:S54–S65. doi: 10.1086/648966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gotschlich EC. Genetic locus for the biosynthesis of the variable portion of Neisseria gonorrhoeae lipooligosaccharide. J Exp Med. 1994;180:2181–2190. doi: 10.1084/jem.180.6.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Danaher RJ, Levin JC, Arking D, Burch CL, Sandlin R, Stein DC. Genetic basis of Neisseria gonorrhoeae lipooligosaccharide antigenic variation. J Bacteriol. 1995;177:7275–7279. doi: 10.1128/jb.177.24.7275-7279.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yang QL, Gotschlich EC. Variation of gonococcal lipooligosaccharide structure is due to alterations in poly-G tracts in lgt genes encoding glycosyl transferases. J Exp Med. 1996;183:323–327. doi: 10.1084/jem.183.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kahler CM, Stephens DS. Genetic basis for biosynthesis, structure, and function of meningococcal lipooligosaccharide (endotoxin) Crit Rev Microbiol. 1998;24:281–334. doi: 10.1080/10408419891294216. [DOI] [PubMed] [Google Scholar]
- 145.Burch CL, Danaher RJ, Stein DC. Antigenic variation in Neisseria gonorrhoeae: production of multiple lipooligosaccharides. J Bacteriol. 1997;179:982–986. doi: 10.1128/jb.179.3.982-986.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Johswich KO, Zhou J, Law DK, St Michael F, McCaw SE, Jamieson FB, Cox AD, Tsang RS, Gray-Owen SD. Invasive potential of nonencapsulated disease isolates of Neisseria meningitidis. Infect Immun. 2012;80:2346–2353. doi: 10.1128/IAI.00293-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bhat KS, Gibbs CP, Barrera O, Morrison SG, Jahnig F, Stern A, Kupsch EM, Meyer TF, Swanson J. The opacity proteins of Neisseria gonorrhoeae strain MS11 are encoded by a family of 11 complete genes. Mol Microbiol. 1991;5:1889–1901. doi: 10.1111/j.1365-2958.1991.tb00813.x. [DOI] [PubMed] [Google Scholar]
- 148.Gray-Owen SD, Lorenzen DR, Haude A, Meyer TF, Dehio C. Differential Opa specificities for CD66 receptors influence tissue interactions and cellular response to Neisseria gonorrhoeae. Mol Microbiol. 1997;26:971–980. doi: 10.1046/j.1365-2958.1997.6342006.x. [DOI] [PubMed] [Google Scholar]
- 149.Dehio C, Gray-Owen SD, Meyer TF. The role of neisserial Opa proteins in interactions with host cells. Trends Microbiol. 1998;6:489–495. doi: 10.1016/s0966-842x(98)01365-1. [DOI] [PubMed] [Google Scholar]
- 150.Sadarangani M, Pollard AJ, Gray-Owen SD. Opa proteins and CEACAMs: pathways of immune engagement for pathogenic Neisseria. FEMS Microbiol Rev. 2011;35:498–514. doi: 10.1111/j.1574-6976.2010.00260.x. [DOI] [PubMed] [Google Scholar]
- 151.Murphy GL, Connell TD, Barritt DS, Koomey M, Cannon JG. Phase variation of gonococcal protein II: regulation of gene expression by slipped-strand mispairing of a repetitive DNA sequence. Cell. 1989;56:539–547. doi: 10.1016/0092-8674(89)90577-1. [DOI] [PubMed] [Google Scholar]
- 152.Belland RJ, Morrison SG, van der Ley P, Swanson J. Expression and phase variation of gonococcal P.II genes in Escherichia coli involves ribosomal frameshifting and slipped- strand mispairing. Mol Microbiol. 1989;3:777–786. doi: 10.1111/j.1365-2958.1989.tb00226.x. [DOI] [PubMed] [Google Scholar]
- 153.Belland RJ, Morrison SG, Carlson JH, Hogan DM. Promoter strength influences phase variation of neisserial opa genes. Mol Microbiol. 1997;23:123–135. doi: 10.1046/j.1365-2958.1997.1971556.x. [DOI] [PubMed] [Google Scholar]
- 154.Virji M, Heckels JE. The role of common and type-specific pilus antigenic domains in adhesion and virulence of gonococci for human epithelial cells. J Gen Micro. 1984;130:1089–1095. doi: 10.1099/00221287-130-5-1089. [DOI] [PubMed] [Google Scholar]
- 155.Koomey M, Fox R, Brossay L, Hebe’rt J. The gonococcal PilT protein plays an essential role in pilus-associated phenotypes of twitching motility and natural competence for transformation. Proceedings of the 9th Pathogenic Neiseria Conference. 1996:64–65. [Google Scholar]
- 156.Stohl EA, Dale EM, Criss AK, Seifert HS. Neisseria gonorrhoeae metalloprotease NGO1686 is required for full piliation, and piliation is required for resistance to H2O2- and neutrophil-mediated killing. mBio. 2013;4:e00399–13. doi: 10.1128/mBio.00399-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Biswas GD, Sox T, Blackman E, Sparling PF. Factors affecting genetic transformation of Neisseria gonorrhoeae. J Bacteriol. 1977;129:983–992. doi: 10.1128/jb.129.2.983-992.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wormann ME, Horien CL, Bennett JS, Jolley KA, Maiden MC, Tang CM, Aho EL, Exley RM. Sequence, distribution and chromosomal context of class I and class II pilin genes of Neisseria meningitidis identified in whole genome sequences. BMC Genomics. 2014;15:253. doi: 10.1186/1471-2164-15-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.De Reuse H, Taha MK. RegF, an SspA homologue, regulates the expression of the Neisseria gonorrhoeae pilE gene. Res Microbiol. 1997;148:289–303. doi: 10.1016/S0923-2508(97)81585-9. [DOI] [PubMed] [Google Scholar]
- 160.Kellogg DS, Jr, Cohen IR, Norins LC, Schroeter AL, Reising G. Neisseria gonorrhoeae. II. Colonial variation and pathogenicity during 35 months in vitro. J Bacteriol. 1968;96:596–605. doi: 10.1128/jb.96.3.596-605.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kellogg DS, Jr, Peacock WL, Deacon WE, Brown L, Pirkle CI. Neisseria gonorrhoeae. I. Virulence genetically linked to clonial variation. J Bacteriol. 1963;85:1274–1279. doi: 10.1128/jb.85.6.1274-1279.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jonsson AB, Nyberg G, Normark S. Phase variation of gonococcal pili by frameshift mutation in pilC, a novel gene for pilus assembly. EMBO J. 1991;10:477–488. doi: 10.1002/j.1460-2075.1991.tb07970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Morand PC, Drab M, Rajalingam K, Nassif X, Meyer TF. Neisseria meningitidis differentially controls host cell motility through PilC1 and PilC2 components of type IV pili. PLoS One. 2009;4:e6834. doi: 10.1371/journal.pone.0006834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rudel T, Boxberger HJ, Meyer TF. Pilus biogenesis and epithelial cell adherence of Neisseria gonorrhoeae pilC double knock-out mutants. Mol Microbiol. 1995;17:1057–1071. doi: 10.1111/j.1365-2958.1995.mmi_17061057.x. [DOI] [PubMed] [Google Scholar]
- 165.Wolfgang M, Park HS, Hayes SF, van Putten JP, Koomey M. Suppression of an absolute defect in type IV pilus biogenesis by loss- of-function mutations in pilT, a twitching motility gene in Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1998;95:14973–14978. doi: 10.1073/pnas.95.25.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Swanson J, Bergstrom S, Robbins K, Barrera O, Corwin D, Koomey JM. Gene conversion involving the pilin structural gene correlates with pilus+ in equilibrium with pilus- changes in Neisseria gonorrhoeae. Cell. 1986;47:267–276. doi: 10.1016/0092-8674(86)90449-6. [DOI] [PubMed] [Google Scholar]
- 167.Long CD, Madraswala RN, Seifert HS. Comparisons between colony phase variation of Neisseria gonorrhoeae FA1090 and pilus, pilin, and S-pilin expression. Infect Immun. 1998;66:1918–1927. doi: 10.1128/iai.66.5.1918-1927.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Criss AK, Kline KA, Seifert HS. The frequency and rate of pilin antigenic variation in Neisseria gonorrhoeae. Mol Microbiol. 2005;58:510–519. doi: 10.1111/j.1365-2958.2005.04838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Meyer TF, Billyard E, Haas R, Storzbach S, So M. Pilus genes of Neisseria gonorrheae: chromosomal organization and DNA sequence. Proc Natl Acad Sci USA. 1984;81:6110–6114. doi: 10.1073/pnas.81.19.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Snodgrass TL, Dempsey JAF, Cannon JG. The repertoire of silent pilin gene copies and their chromosomal location is similar but not identical in gonococcal strains FA1090 and MS11. Neisseria 94--Proceedings from the Ninth International Pathogenic Neisseria Conference; Winchester, England. 1994. pp. 113–115. [Google Scholar]
- 171.Kline KA, Criss AK, Wallace A, Seifert HS. Transposon mutagenesis identifies sites upstream of the Neisseria gonorrhoeae pilE gene that modulate pilin antigenic variation. J Bacteriol. 2007;189:3462–3470. doi: 10.1128/JB.01911-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Howell-Adams B, Seifert HS. Molecular models accounting for the gene conversion reactions mediating gonococcal pilin antigenic variation. Mol Microbiol. 2000;37:1146–1158. doi: 10.1046/j.1365-2958.2000.02067.x. [DOI] [PubMed] [Google Scholar]
- 173.Rohrer MS, Lazio MP, Seifert HS. A real-time semi-quantitative RT-PCR assay demonstrates that the pilE sequence dictates the frequency and characteristics of pilin antigenic variation in Neisseria gonorrhoeae. Nucleic Acids Res. 2005;33:3363–3371. doi: 10.1093/nar/gki650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Helm RA, Seifert HS. Frequency and rate of pilin antigenic variation of Neisseria meningitidis. J Bacteriol. 2010;192:3822–3823. doi: 10.1128/JB.00280-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tobiason DM, Seifert HS. The obligate human pathogen, Neisseria gonorrhoeae, is polyploid. PLoS Biol. 2006;4:1069–1078. doi: 10.1371/journal.pbio.0040185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tobiason DM, Seifert HS. Genomic content of Neisseria species. J Bacteriol. 2010;192:2160–2168. doi: 10.1128/JB.01593-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mehr IJ, Seifert HS. Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation, and DNA repair. Mol Microbiol. 1998;30:697–710. doi: 10.1046/j.1365-2958.1998.01089.x. [DOI] [PubMed] [Google Scholar]
- 178.Mehr IJ, Long CD, Serkin CD, Seifert HS. A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics. 2000;154:523–532. doi: 10.1093/genetics/154.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Stohl EA, Seifert HS. The recX gene potentiates homologous recombination in Neisseria gonorrhoeae. Mol Microbiol. 2001;40:1301–1310. doi: 10.1046/j.1365-2958.2001.02463.x. [DOI] [PubMed] [Google Scholar]
- 180.Skaar EP, Lazio MP, Seifert HS. Roles of the recJ and recN genes in homologous recombination and DNA repair pathways of Neisseria gonorrhoeae. J Bacteriol. 2002;184:919–927. doi: 10.1128/jb.184.4.919-927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kline KA, Seifert HS. Role of the Rep helicase gene in homologous recombination in Neisseria gonorrhoeae. J Bacteriol. 2005;187:2903–2907. doi: 10.1128/JB.187.8.2903-2907.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sechman EV, Rohrer MS, Seifert HS. A genetic screen identifies genes and sites involved in pilin antigenic variation in Neisseria gonorrhoeae. Mol Microbiol. 2005;57:468–483. doi: 10.1111/j.1365-2958.2005.04657.x. [DOI] [PubMed] [Google Scholar]
- 183.Sechman EV, Kline KA, Seifert HS. Loss of both Holliday junction processing pathways is synthetically lethal in the presence of gonococcal pilin antigenic variation. Mol Microbiol. 2006;61:185–193. doi: 10.1111/j.1365-2958.2006.05213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Criss AK, Bonney KM, Chang RA, Duffin PM, LeCuyer BE, Seifert HS. Mismatch correction modulates mutation frequency and pilus phase and antigenic variation in Neisseria gonorrhoeae. J Bacteriol. 2010;192:316–325. doi: 10.1128/JB.01228-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Segal E, Hagblom P, Seifert HS, So M. Antigenic variation of gonococcal pilus involves assembly of separated silent gene segments. Proc Natl Acad Sci USA. 1986;83:2177–2181. doi: 10.1073/pnas.83.7.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wainwright LA, Frangipane JV, Seifert HS. Analysis of protein binding to the Sma/Cla DNA repeat in pathogenic Neisseriae. Nucl Acids Res. 1997;25:1362–1368. doi: 10.1093/nar/25.7.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Howell-Adams B, Wainwright LA, Seifert HS. The size and position of heterologous insertions in a silent locus differentially affect pilin recombination in Neisseria gonorrhoeae. Mol Microbiol. 1996;22:509–522. doi: 10.1046/j.1365-2958.1996.00128.x. [DOI] [PubMed] [Google Scholar]
- 188.Cahoon LA, Seifert HS. Transcription of a cis-acting, noncoding, small RNA is required for pilin antigenic variation in Neisseria gonorrhoeae. PLoS Pathog. 2013;9:e1003074. doi: 10.1371/journal.ppat.1003074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Giacani L, Brandt SL, Giacani L, Brandt SL, Puray-Chavez M, Reid TB, Godornes C, Molini BJ, Benzler M, Hartig JS, Lukehart SA, Centurion-Lara A. Comparative investigation of the genomic regions involved in antigenic variation of the TprK antigen among treponemal species, subspecies, and strains. J Bacteriol. 2012;194:4208–4225. doi: 10.1128/JB.00863-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Liu Y, Liu W, Russell MW. Suppression of host adaptive immune responses by Neisseria gonorrhoeae: role of interleukin 10 and type 1 regulatory T cells. Mucosal Immunol. 2014;7:165–176. doi: 10.1038/mi.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bankhead T. Antigenic Variation of VlsE in Borrelia burgdorferi. In: Embers ME, editor. The Pathogenic Spirochetes. Springer; New York: 2012. pp. 113–124. [Google Scholar]