Abstract
In recent years, the etiology of human disease has greatly improved with the inclusion of epigenetic mechanisms, in particular as a common link between environment and disease. However, for most diseases we lack a detailed interpretation of the epigenetic regulatory pathways perturbed by environment and causal mechanisms. Here, we focus on recent findings elucidating nutrient-related epigenetic changes linked to obesity. We highlight studies demonstrating that obesity is a complex disease linked to disruption of epigenetically regulated metabolic pathways in the brain, adipose tissue and liver. These pathways regulate (1) homeostatic and hedonic eating behaviors (2) adipocyte differentiation and fat accumulation, and (3) energy expenditure. By compiling these data we illustrate that obesity-related phenotypes are repeatedly linked to disruption of critical epigenetic mechanisms that regulate of key metabolic genes. These data are supported by genetic mutation of key epigenetic regulators and many of the diet induced epigenetic mechanisms of obesity are also perturbed by exposure to environmental toxicants. Identifying similarly perturbed epigenetic mechanisms in multiple experimental models of obesity strengthens the translational applications of these findings. We also discuss many of the ongoing challenges to understanding the role of environmentally-induced epigenetic pathways in obesity and suggest future studies to elucidate these roles. This assessment illustrates our current understanding of molecular pathways of obesity that are susceptible to environmental perturbation via epigenetic mechanisms. Thus, it lays the groundwork for dissecting the complex interactions between diet, genes, and toxicants that contribute to obesity and obesity-related phenotypes.
Keywords: Epigenetics, nutrition, toxicant, obesity, metabolism, diet
1. Introduction
Environmental exposure contributes significantly to disease risk [1]. Accumulating evidence from dietary and genetic models of nutrient perturbation, links the disruption of critical metabolic pathways to development of disease. Obesity was recently classified as a disease with strong links to environmental exposures. According to the World Health Organization (WHO), worldwide over 600 million adults and 42 million children are obese, figures which have more than doubled since 1980. While obesity is most often associated with overconsumption of calories (overnutrition) combined with lowered energy expenditure, recent studies show that obesity is a highly complex disease with both causal and contributory links to metabolic dysfunction. Development of obesity is influenced by multiple pathways, including (1) hedonic and homeostatic eating behaviors that are directly regulated by the brain, (2) energy expenditure in tissues such as liver, fat and muscle (3) and changes in adiposity mediated by adipocyte differentiation and lipid accumulation in adipose tissues. Homeostatic eating is need-based and replenishes energy storage, while hedonic eating is reward-driven and motivated by desire for palatable food [2]. As a causal factor, obesity is linked to increased risk of diseases such as diabetes [3, 4], cardiovascular disease (CVD) [5–7], cancer [8–10], arthritis [11, 12], infertility [13, 14], and mental health disorders [15, 16]. Importantly, new studies show that many of these pathways are regulated by epigenetic mechanisms that are disrupted by environmental factors. Here, we compile these findings in order to highlight common mechanisms of epigenetic perturbation linked to obesity as potential targets for prevention, diagnoses or treatment.
Epigenetic machinery regulates gene expression by orchestrating the levels and interactive activity of highly conserved genomic processing including DNA methylation, post-translational histone modification, and non-coding RNA activity [17]. In mammals, methylation at CpG dinucleotides to generate 5-methylcytosine (5mC) is catalyzed by DNA methyltransferases (DNMTs) including DNMT1, DNMT3A and DNMT3B. DNMT3A and DNMT3B enzymes mediate de novo DNA methylation primarily during embryonic development [18] while DNMT1 maintains DNA methylation states in all mitotically dividing cells [19]. Methylated DNA is often considered a primary epigenetic mark that can then be recognized and bound by methyl binding proteins. These include methyl CpG binding protein 2 (MECP2) to trigger subsequent epigenetic changes by histone modulators [20, 21]. The lysine residues of histone N-terminal tails within DNA bound nucleosomes are modified by methylation, acetylation, phosphorylation, ubiquitination, sumoylation and biotinylation [17, 22–24]. On the other hand, non-coding RNA include functionally distinct classes of untranslated RNA including micro RNA (miRNA), long noncoding RNA (IncRNA), piwi-interacting RNA (piRNA), and small nucleolar RNA (snoRNA) [25]. Of equal importance to the factors required to establish and maintain DNA and histone modifications and non-coding RNA levels and functions, are the factors required to erase or deplete these epigenetic states. This is required for the resetting or alteration of epigenetic programs necessary for reprogramming cell fate towards germline or somatic lineages and for cellular differentiation and response to environmental stimuli. These epigenetic erasers include histone demethylases and deacetylases and putative DNA demethylases. For example, the ten-eleven translocation (TET) proteins convert 5mC into 5-hydroxymethylcytosine (5hmC) but remain under investigation as to whether this is a functional modification or merely a step towards demethylation [26].
The role of epigenetic mechanisms in gene expression is defined by many factors including cellular or genomic location of the acting mechanism, the developmental timing of establishment/maintenance of the mechanism and co-regulation by multiple co-acting epigenetic mechanisms. Multiple mechanisms co-regulate different loci at different stages of development to establish, maintain or change the transcriptional outcome of the cell. Despite ongoing research to fully elucidate these concurrent mechanisms, some general functions have been assigned to commonly occurring epigenetic states. DNA hypermethylation at promoter regions is most often correlated with transcriptional repression, however hypermethylation at intra- or intergenic regions may reflect either an active or repressed state. Likewise, the transcriptional regulation of modified histones depends on the type of modification and the location of the lysine modified. For example, acetylated histones primarily mark actively transcribed regions while the function of methylated histones depends on the location of lysine residue, e.g. trimethylation of histone H3 lysine 9 (H3K9me3) marks transcriptional repression and H3K4me3 are enriched around transcription start site of active genes [27]. On the other hand, non-coding RNA function is primarily determined by the sequence encoded, post-transcriptional processing and secondary structure of the RNA, cellular transport and target (RNA, DNA, or protein) of the non-coding RNA and can result in either upregulation or downregulation of the target at the transcriptional or posttranscriptional level [25].
Epigenetic regulatory mechanisms are reported to play a role in all of the obesity related pathways mentioned above, including food intake, energy expenditure and adiposity. Diet has been shown to play an important, albeit complex, role in determining these epigenetic states. Some dietary compounds act directly on epigenetic machinery while other nutrients act indirectly. For example, retinoid X receptors (RXRs), activated by vitamin A metabolite retinoid acid, regulate transcription in adipocytes by directly binding to promoters of histone modifier genes such as SET domain bifurcated 1 (Setdb1) and SET domain containing protein 8 (Setd8), both lysine methyltransferases [28–30]. On the other hand, intake of niacin and methyl donor nutrients such as folate and choline determine cellular availability of methyl groups and thus are proposed to act by limiting DNA and histone methylation reactions [31, 32].
There is a significant and growing body of work highlighting the role of epigenetic mechanisms in obesity. Here we focus on a few specific studies and well characterized pathways of obesity to elucidate the role of altered nutrient availability in causing obesity via epigenetic mechanisms. We also discuss a potential overlapping or aggregate role for nutrition in susceptibility to obesity linked to environmental pollutants since it is well known that diet can indeed alter the effect of toxic compounds. One example is the role of dietary calcium, vitamin D, iron, fat and phosphorus in inhibiting gastrointestinal lead absorption in the gastrointestinal tract (as reviewed in [33]). Another example is the role of diet-induced adiposity in influencing the availability of circulating toxicants via toxicant sequestration in adipose tissue. Studies show that body fat composition is inversely correlated with circulating plasma levels of lipophilic persistent organic pollutants (POPs) and rapid weight loss is correlated with increased plasma levels of POPs [34–36]. Furthermore, nutrient availability can interfere with the levels of key enzymes required for toxicant metabolism. Such interactions influence metabolism required for toxicant inactivation and excretion [37–41] and metabolism required for toxicant activation, thus altering toxicity [42–44]. For example, Cytochrome p450 (Cyp450) enzymes have been shown to play critical roles in metabolizing POPs [35, 45]. Cyp450 enzyme levels are altered by many nutrients including dietary protein [46, 47], fat [48], sugar [49], vitamin C [50, 51], vitamin E [39], folate[40], betaine [52], iodine and selenium [53]. These findings highlight the potential for aggregate effects of diet-toxicant interactions and offer the promise of dietary intervention of diseases related to toxicant exposure. In order to bring these research findings to practice and maximize public health benefits we must first understand more about the mechanisms responsible. Here, we focus on obesity because of the abundance of research available, but we hold the expectation that this paradigm likely applies to other environmentally based diseases.
2.1 Molecular pathways of obesity regulated by nutrition
2.1.1 Proopiomelanocortin methylation and epi-regulation of proopiomelanocortin neuron
Proopiomelanocortin is a precursor to several hormones related to appetite regulation, energy homeostasis, sexual behavior and various reward pathways. While it is expressed at high levels in the pituitary, it is also found in other cells/tissues primarily in other parts of the brain such as the hypothalamus, skin cells and reproductive systems, and relatively accurate levels can be measured in peripheral blood [54–56]. Proopiomelanocortin neurons are nerve cells in the arcuate nucleus of hypothalamus that express proopiomelanocortin protein. Hypothalamic proopiomelanocortin neurons inhibit satiety, thus altered proopiomelanocortin gene expression is most commonly associated with either obesity or underweight phenotypes although studies suggest there may be inverse relationships depending on the cell/tissue type where it is measured [56]. Generally, decreased hypothalamus expression of proopiomelanocortin or its downstream products is highly correlated with weight gain/obesity while increased expression is associated with weight loss [57–59].
Association studies have identified several SNPs in or near the proopiomelanocortin locus associated with weight related phenotypes [60–62]. There is also strong evidence of additional epigenetic regulation of both proopiomelanocortin expression via DNA methylation of two CpG islands within the gene promoter and body [56, 63]. Recent studies propose DNA methylation levels at the proopiomelanocortin promoter in cord blood serves as an important early predictive marker of metabolic syndrome later in life [64]. Despite the strong correlation between DNA methylation at proopiomelanocortin, gene expression and phenotypic variation, mechanisms of epigenetic regulation of proopiomelanocortin expression remain unclear. Most recently a study by Zhang and colleagues [65] use cultured cells to show that hypermethylation at the proopiomelanocortin promoter and within the binding site [66] for proximal specificity protein 1 (SP1), blocked formation of the SP1-promoter complex required for activation of proopiomelanocortin. Interestingly, they also showed using a lactation-staged mouse model this aberrant methylation at the proopiomelanocortin promoter is induced by maternal dietary supplementation of conjugated linoleic acids (CLAs). Similarly, the hypothalamic proopiomelanocortin SP1 cis-element was hypermethylated in a neonatal overfeeding model induced by rearing rats in small litters. This caused a blunted response of proopiomelanocortin neurons to leptin signaling [67]. In contrast, both gestational and post-weaning high folate diet in a rat model resulted in hypomethylation of the proopiomelanocortin promoter in the hypothalamus of offspring, decreased food intake and lower weight [68]. Two separate studies examined the effects of a maternal high fat diet in a rat model and showed consistent results of hypermethylation at the hypothalamic proopiomelanocortin promoter associated with disrupted energy homeostasis leading to increased food intake and subsequent weight gain of exposed pups [69, 70]. Although limited in mechanistic potential, human studies seemingly mirror these effects and a recent study showed that caloric restriction induced weight gain or weight loss is associated with changes in proopiomelanocortin gene body methylation in leukocytes [71]. To further validate the importance of DNA methylation at the proopiomelanocortin locus in fat accumulation, a study by Wang and colleagues [72] cleverly uses a genetic mouse model carrying a proopiomelanocortin-neuron-specific knockout of Mecp2, a major epigenetic regulator that acts by binding directly to methylated DNA. MECP2 is a known antagonist of SP1-activated transcription [73], therefore as expected, the Mecp2−/− mouse model exhibited increased food intake and subsequent increased fat mass, which was linked to increased DNA methylation at the proopiomelanocortin promoter and decreased proopiomelanocortin expression [72]. This finding highlights the importance of methylation status at the SP1 binding site in regulating proopiomelanocortin expression required for downstream phenotypic effects.
The hypothalamus utilizes a self-regulating mechanism of energy homeostasis, which relies on plasticity of the adaptive neuron in the feeding circuitry and requires rapid cell-to-cell interactions [74]. Cell-surface protein polysialylation is critical for modulating synaptic adaptation for neural cells [75]. High fat diet in the mouse induces recruitment of histone lysine acetyltransferase 8 to activate transcription of polysialylation gene ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 (St8sia4) in hypothalamus. This signal promotes the plasticity of proopiomelanocortin neuron in acutely responding to high fat diet in mouse [76, 77]. Upregulation of polyasialytion through histone acetylation partially offsets the effect of proopiomelanocortin hypermethylation caused by high fat diet. Histone lysine acetyltransferase 8 silencing in adult mice leads to accelerated weight gain in response to a high fat diet [76]. The diet induced antagonistic changes between proopiomelanocortin neuron plasticity and proopiomelanocortin expression demonstrates a self-regulatory mechanism for weight control achieved though distinct epigenetic machineries.
The role of miRNA in neuronal control of energy homeostasis is still poorly understood. However in a recent study, a conditional deletion of Dicer1 (encodes the endoribonuclease required for generating mature miRNA) specifically in mouse forebrain neurons results in severe hyperphagic obesity, and decreased proopiomelanocortin mRNA in the hypothalamus [78]. Further investigation shows that this phenotype occurs likely via upregulation of the PI3K–Akt-mTOR pathway. Downregulation of this pathway attenuates adiposity in the Dicer1 knockout mice. The authors used the miTALOS tool to identify miRNA that were most likely to play a role in the observed phenotype based on high expression in the hypothalamus and predicted targets within the PI3K–Akt-mTOR pathway[79]. Depletion of miR-103 was shown to upregulate the pathway while injection of excess miR-103 mimic into the arcuate nucleus was shown to reduce body weight changes, food intake and expression levels of the gene encoding for catalytic gamma polypeptide of phosphoinositide-3 kinase (Pik3cg) within the PI3K–Akt-mTOR pathway [78]. Whether there are direct links specifically between miR-103 and proopiomelanocortin remains unclear.
2.1.2 Adipose leptin signaling in homeostasis circuitry
Leptin serves as a direct signal from white adipose tissue to the neural network, allowing the brain to sense stored nutrients and thus regulate energy homeostasis [80]. Surprisingly, elevated serum leptin is usually positively correlated with obesity despite leptin being an appetite suppressant and thus anti-obesity hormone [81]. Diet-induced obesity models demonstrate that the elevated leptin signals in obesity states is actually due to leptin resistance in obese subjects and that the pathway is only upregulated as a means of compensating for this resistant phenotype [82]. Both appetite-promoting (orexigenic) peptides (i.e. neuropeptide Y (NPY)) and appetite-inhibitory (anorexigenic) peptides (i.e. proopiomelanocortin peptides) are regulated by leptin in order to prevent obesity [83]. For example, subcutaneous injections of leptin in humans [84]and mice [85] with genetic leptin deficiency leads to reduced food intake, fat mass and body weight. This is partly because leptin administration activates proopiomelanocortin neurons in the hypothalamus via leptin receptor and consequently reduces body weight by inhibiting food intake [81]. Leptin signaling in the hypothalamus proopiomelanocortin neurons depends on the nerve growth factor Nur77, which subsequently activates signal transducer and activator of transcription 3 (Stat3) [86]. The downstream STAT3-activated nescient helix-loop-helix 2 in leptin signaling pathway is required for transcriptional activation of proopiomelanocortin processing enzyme proprotein convertase 1 [87]. Most importantly, STAT3 binds to the cis-regulatory elements on proopiomelanocortin promoter together with SP1 to activate its transcription and diet-induced leptin resistance was usually associated with impaired STAT3 signaling cascade in proopiomelanocortin neurons [66, 88, 89].
Evidence suggests that leptin signaling pathway from adipose tissue is regulated by epigenetic mechanisms. Rhett syndrome patients that carry loss of function mutation in Mecp2 exhibit increased plasma leptin concentration [90]. Furthermore, knockout of Mecp2 in hypothalamic Sim1-expressing neurons elicits hyperphagic and obese phenotypes in mice, with increased serum leptin concentration [91]. Meanwhile, knockout of the de novo DNA methyltransferase 3 (Dnmt3a) in the mouse hypothalamus caused obesity, increased food intake and higher plasma leptin levels [92]. Diet-induced obesity models demonstrate the role of epigenetic modulators in controlling leptin function in adipose tissue. DNA methylation at the leptin promoter regulates transcription in a complex way. Maternal high fat diet is associated with leptin promoter hypomethylation [93]. Interestingly, this effect is similar to that of maternal undernourishment, which causes 11 CpGs in the leptin promoter to become hypomethylated [94]. However, serum leptin level was oppositely regulated by maternal high fat diet and undernourishment, with the prior model having increased serum leptin [93, 94]. The seemingly contradictory observation is consistent with another study from Shen and colleagues [95]. Using a diet-induced-obesity model, they show that obesity induces adipocyte leptin mRNA levels despite seemingly enriched repressive epigenetic states at the leptin promoter, including enriched binding of DNMT1 and methyl-CpG binding domain protein 2 (MBD2) and reduced enrichment of the active acetylated histone marks and RNA polymerase II at the promoter. In the same study, DNA methylation at the leptin promoter was positively correlated with leptin mRNA level in adipose tissue of diet-induced obese mice, whereas the correlation remained negative in the control [95]. Most likely, leptin mRNA quantity is not only determined by rate of transcription, but also, in this case, substantially affected by mRNA turnover influenced by dietary compounds such as protein and sugar [96]. High fat and sugar diet in rat models induced hypo- and hypermethylation of several CpGs in adipocyte leptin promoter. These changes were reversible at some of the CpGs by switching to regular diet [97]. Overall, these findings demonstrate that when animals are fed an obesogenic diet, adipocyte leptin transcription is inhibited primarily through leptin promoter hypermethylation. However, an added effect of reduced leptin mRNA turnover may actually cause accumulation of cellular leptin transcripts. On the other hand, hypermethylation of hypothalamus proopiomelanocortin promoter blocks transcriptional activation by trans-acting factors STAT3 and SP1, thus elevating circulating leptin levels. In this manner, diet-induced epigenetic changes can lead to increased caloric intake despite high leptin levels.
MiRNA play a significant role in leptin pathways. Hyperphagic obesity caused by deletion of Dicer1 (discussed above) is correlated with increased plasma leptin levels [78]. In a separate study, Viesti and colleagues [100] investigated the role of miRNAs in increased leptin in subcutaneous fat of obese human participants compared to non-obese fat. Although there was not a significant correlation between leptin levels and any of the miRNAs measured in this study, they reported that higher levels of leptin receptor in obese individuals was correlated with lower levels of MiR-145. Interestingly, a separate study using mouse models showed increased levels of both MiR-145 and genetically linked and co-expressed miR-143 in the liver in response to obesity induced by either high fat or genetic mutation (loss-of-function mutation in leptin receptor, db/db) [101]. In the leptin receptor mutant samples, MiR-145 was increased compared to controls in liver, skeletal muscle and pancreas while miR-143 was increased in heart and pancreas but decreased in white adipose tissue. MiR-145 levels measured in white and brown adipose tissue of the leptin receptor mutant were not significantly different. Conditional overexpression of miR-143 in the liver did not affect body weight or plasma leptin levels but did result in impaired glucose tolerance and insulin sensitivity. Similarly overexpressed miR-145 did not affect body weight, plasma leptin levels, glucose or insulin pathways [101]. These findings demonstrate that up or downregulation of miRNAs found to be associated with obesity and obesity-related molecular pathways may not be sufficient to cause obesity or may require tissue- or temporal-specificity to exert effect on adiposity.
Several rodent models demonstrate that MiR-200a is also very important for proper leptin signaling. Benoit and colleagues reported that compared to untreated controls, rats treated with a leptin inhibitor during postnatal development exhibited higher weight in addition to leptin and insulin resistance in the hypothalamus during adulthood [102]. MiR-200a was among 34 miRNAs upregulated in the leptin inhibited hypothalamus. Using a leptin deficient genetic mouse model of obesity, Crepin and colleagues demonstrated that reduced leptin levels in the hypothalamus of deficient animals resulted in up-regulation of miR-200a, miR-200b and miR-429 [103]. Leptin treatment in these animals resulted in downregulation of these miRNAs, upregulation of the leptin receptor, reduction in body weight and restoration of insulin sensitivity. Overexpression of the miR-200a precursor in a neuroblastoma cell line impaired insulin and leptin signaling suggesting that changes in this miRNA level are sufficient to induce obesity-related metabolic pathways. A separate study showed that the correlation between obesity, miR-200a, and leptin is timing dependent [104]. When compared to control fed rats, rats fed a high fat diet (45% fat) post weaning showed reduced levels of hypothalamic miR-200a after two weeks on the diet and also after 1 month but this difference was alleviated by 3 months on the diet. Opposite but similar time dependent changes were observed with other miRNAs assayed. For example, miR-132 and MiR-145 exhibited minimal differences between high fat and control diet at 2 weeks and 1 month but significant upregulation was observed at 3 months. The authors used the ingenuity pathway analysis software to demonstrate that several of the miRNA affected were interrelated with each other and genes related to insulin, leptin and adiponectin signaling. Of importance, treatment with leptin by IP injection over a 48hr period resulted in a significant decrease of MiR-145 and miR-132 compared to untreated animals and the effect was exacerbated by fasting. MiR-200a was also seemingly downregulated in leptin treated animals but results were not significantly different. MiR-200a is implicated in several types of cancer, however, its direct role in leptin signaling remains under investigation. MiR-132 was previously implicated in glucose homeostasis in isolated rodent and human islets[105] and has been shown to be differentially expressed in both visceral fat and whole blood between obese and non-obese human participants [106].
There are very few human studies investigating epigenetic mechanisms regulating the leptin pathway. One study of importance shows that in comparing human subjects in weight management programs, weight loss response was correlated with adipose leptin mRNA level and DNA methylation at non-leptin loci [98]. Leptin was not reported among these epigenetically perturbed loci, however, they did include imprinted genes and genes associated with body weight and insulin secretion. Changes at these genes were sufficient to distinguish low and high responders among overweight and obese human subjects. In another study, female subjects with baseline hypomethylation at adipocyte leptin promoter, showed better weight loss response to low-calorie diet, though leptin mRNA levels did not differ [99]. We can use what has been characterized in the animal models to further characterize these responses in humans.
2.1.3 DNA methylation in dopamine neurons in regulating reward circuitry
Food intake is primarily controlled by two subdivisions of the brain: hypothalamus regulates homeostatic needs for food, and central reward circuitry (projection from ventral tegmental area and substantia nigra to striatum, nucleus accumbens and prefrontal cortex) regulates hedonic eating behaviors [83, 107, 108]. While leptin signaling influences proopiomelanocortin and NPY neurons in the hypothalamus for maintaining energy homeostasis, dopamine neurons within the mesocorticolimbic system are major regulators of the reward pathway [109]. To be noted, hypothalamus dopamine signaling is a regulator of energy homeostasis [110]. Obesity is linked to compulsive eating and defective dopamine system [111, 112]. Tyrosine hydroxylase is the rate-limiting enzyme in synthesizing dopamine [113], and dopamine transporter solute carrier family 6 (SLC6A3) is essential for re-absorbing dopamine from synaptic cleft. Two families of dopamine receptors (D1 like and D2 like) differ with each other in function and brain distribution [114].
Mice on prolonged high fat diet showed differential regulation of tyrosine hydroxylase and dopamine transporter in hypothalamus and ventral tegmental area: transcription of both were down-regulated in ventral tegmental area with increased level of promoter DNA methylation, while opposite changes of mRNA levels and DNA methylation were observed in hypothalamus [107]. Lack of DNMT3A in mouse hypothalamic neuron cells showed similar epigenetic alteration with high fat diet on tyrosine hydroxylase promoter methylation, suggesting a role of DNMT3A in maintaining dopamine function in hypothalamus [92]. The findings indicate concomitant obesogenic epigenetic changes by high fat diet in one of the two pathways controlling food intake, which promotes homeostatic eating despite lowered desire for palatability. A maternal high fat diet mouse model [115] showed more severe obesogenic changes in brain, where both homeostatic and hedonic eating are increased with hypomethylation in dopamine transporter but not tyrosine hydroxylase promoter. In contrast, a maternal protein restriction model revealed a slightly different epigenetic and reward-response change in brain. Tyrosine hydroxylase and dopamine transporter were overexpressed in all brain regions including ventral tegmental area and hypothalamus, and the dopaminergic neuron developmental regulator cyclin-dependent kinase inhibitor 1C underwent ~50% decrease in DNA methylation in its promoter region with consistent increase in expression throughout the brain [116]. The differential regulation of dopamine-associated genes indicates that epigenetic changes depend on timing of dietary perturbation and the function of the targeted pathway.
Overall, in the reward circuitry, tyrosine hydroxylase functions to generate dopamine and dopamine transporter functions to remove dopamine from synapse to quench the signal. In this way, tyrosine hydroxylase and dopamine transporter regulate hedonic eating behaviors. To further understand the differences in diet-induced epigenetic changes in tyrosine hydroxylase and dopamine transporter promoters, the role of dopamine receptors should be investigated. The distinct distribution of dopamine receptors in the brain partially determines the role of dopamine in each regions of the brain, as D1 like (D1 and D5) and D2 like (D2, D3 and D4) receptors induce opposite intracellular signal transduction cascades [114]. D1 [117], D2 and D4 [118] receptors are differentially regulated by diets high in fat and/or sugar. Interestingly, the effects differ between obesity-prone and -resistant animals. D2 receptor in reward circuitry was down regulated in obese rat. Also, D2 knockdown exacerbated diet-induced deficits in rewards system and increased animals’ desire for palatable food [111]. Since dopamine receptors are essential for transmitting dopamine signals to the receiving cells, the dopamine -induced intracellular cascade determines the outcome of dopamine signaling. However, mechanisms that directly regulate dopamine receptors have not been reported.
2.1.4 Adipose tissue differentiation and fat accumulation
Adipogenesis in adipose tissue is controlled by a variety of transcription regulators, primarily the CCAAT enhancer-binding protein (C/EBP) family and the nuclear receptor peroxisome proliferator-activated receptor gamma (PPARG) [119]. The extent of fat accumulation in white adipose tissue is positively correlated with the adipose expression of fatty acid synthase [120], the promoter of which is collectively bound and regulated by transcriptional regulators, sterol regulatory element binding transcription factor 1 (SREBF1) and other factors [121]. Adipocyte fatty acid synthase also contributes to development of diet-induced obesity through PPARG-mediated adipogenesis [122]. Adiposity regulated by fatty acid synthase as well as abovementioned transcription factors is influenced by dietary compounds including folate [32], Vitamin D [123, 124], Vitamin A [30] and obesogenic diets [125].
Histone methylation exerts a critical role for preadipocyte differentiation through marking PPARG promoter state as reviewed by Okamura et. al [29]. Epigenetic regulation of PPARG and CCAAT/enhancer binding protein (C/EBP) alpha (CEBPA) involves many different histone modifiers during different stages of adipogenesis. Pparg promoter is subject to bivalent histone modification with concurrent localization of H3K4me3 and H3K27me3 in embryonic fibroblasts [126]. Upon adipocyte differentiation, fundamental changes in chromatin state occur at the Pparg promoter that are determined by H3K4 methyltransferases MLL3 and MLL4 [127], PAX interacting (with transcription-activation domain) protein 1 (PAXIP1) [128] and the H3K27 demethylase KDM6A [129]. Furthermore, PPARG mediates adiposity by regulating the H4K20 mono-methyltransferase Setd8 [28] and possibly in a feedback loop mechanism [29]. Genome-wide analysis reveals that the majority of PPARG targets during adipogesis are also bound by CEBPA in mouse preadipocyte 3T3-L1 [130]. It is noteworthy that PPARG and CEBPA are mutually stimulating [29] and both are repressed by the H3K9 tri-methyltransferase the suppressor of variegation 3–9 homolog 1 (SUV39H1) in early stages of adipogenesis [131]. C/EBP families including CEBPA are required for inducing expression of SREBF1 in maturing adipocytes [132] and linking PPARG/C/EBP mediated adiposity to fatty acid synthase dependent fat accumulation. Human adipose tissues of obese subjects consistently have higher fatty acid synthase mRNA and protein levels in visceral and subcutaneous fat tissue [120].
PPARG determines adipogenesis by regulating two histone methyltransferases Setdbl and Setd8, which generate repressive histone marks H3K9me3 and active H3K20 methylation marks, respectively. Both diet-induced obese and genetically predisposed obese mouse models exhibit decreased Setdbl and increased Setd8 mRNA in visceral fat [28]. To be noted, PPARG itself and CEBPA are targeted by SETD8 for H4K20me1 -marked activation, indicating a potential positive feedback loop for PPARG and its downstream adipogenic targets, including its brown adipose tissue isoform [28]. PPARG forms a heterodimer with RXRs to bind to promoters and activate transcription. Retinoic acid, an active vitamin A derivative, triggers cellular signal cascade via RXRs. Mice on a vitamin A deficient diet exhibit elevated adiposity and increased adipose Pparg, Srebfl and Cebpa mRNA levels [30]. Last but not least, in a chicken preadipocyte model, interactions between PPARG, CEBPA and fatty acid synthase are epigenetically influenced by folate such that Cebpa promoter undergoes hypomethylation upon folate depletion [32].
MiR-27a and miR-27b have been specifically implicated in adipogenesis. In a comparison of obese and non-obese participants, miR27a was shown to be significantly upregulated in the visceral fat of obese individuals [100]. Kang et al used adipose-derived stem cells to demonstrate that adipocyte differentiation triggers the down regulation of prohibitin, miR-27a and miR– 27b [137]. In agreement with previous reports [138], the authors showed that overexpression of miR-27a and miR-27b inhibits adipocyte differentiation. In addition, by using a luciferase reporter assay they demonstrated that the function of miR-27a and miR-27b in adipocyte differentiation is partly mediated by a direct interaction with prohibitin and subsequent impairment of mitochondrial function. The same research group previously demonstrated that prohibitin silencing induces downregulation of mouse Pparg [139]. However, a separate study used a luciferase reporter assay in HeLa cells to show that miR-27a targets human PPARG 3’UTR to negatively regulate its transcription and that overexpression of miR-27a in mouse preadipocytes can inhibit adipocyte differentiation by repressing Pparg expression[140]. In addition, miR-27a levels were lower in mature adipocytes of high fat diet induced obese mice compared to lean mice.
2.1.5 NNMT drains methyl donor pool and regulates energy expenditure
Nicotinamide N-methyltransferase (NNMT) catalyzes N-methylation of nicotinamide using S-adenosyl-L-methionine (SAM) and yields 1-methylnicotinamide and S-adenosyl-L-homocysteine (SAH). Extensive studies link NNMT function to Parkinson’s disease [141], hyperhomocysteinemia [142], cancer [143, 144], insulin resistance [145, 146] and obesity [147]. NNMT activity in white adipose tissue is upregulated by high fat diet while not affected in liver, despite that NNMT was primarily expressed in liver [142]. This indicates a role of adipose NNMT in diet-induced obesity. Emerging evidence suggests that NNMT contributes to increased risk for insulin resistance and type 2 diabetes associated with visceral adiposity [146].
Elevated NNMT activity leads to depletion of SAM, the donor of methyl groups for DNA and histone methylation. Decreased availability of methyl group can potentially directly affect epigenetic pathways. Indeed, maternal supplementation of the NNMT substrate nicotinomide induced global hypomethylation in both placenta and fetal liver, which was prevented by co-supplementation of the methyl donor nutrient betaine [31]. Induction of NNMT in cancer cells results in global H3K9 and H3K27 hypomethylation. These defective histone marks were restored upon Nnmt knockdown [143].
Kraus and colleagues demonstrated that Nnmt knockdown in white adipose tissue and liver protects against diet-induced obesity and insulin resistance in mice. The leanness was caused by elevated energy expenditure rather than changes in food intake [147]. Polyamine flux contributes to energy expenditure and SAM provides the starting materials for this pathway after removal of methyl groups. Ornithine decarboxylase and spermidine-spermine N-acetyltransferase are rate-limiting enzymes in polyamine metabolism [148]. In the diet-induced obesity model, Nnmt knockdown activated expression of ornithine decarboxylase and spermidine-spermine N-acetyltransferase as well as polyamine flux in white adipose tissue through increasing global H3K4me1/2/3 levels. In adipocyte cell culture, enrichment of methylated H3K4 was increased at ornithine decarboxylase and spermidine-spermine N-acetyltransferase genes upon NNMT inhibition [147]. NNMT is also involved in development of insulin resistance and type 2 diabetes as demonstrated in glucose transporter type 4 (Glut4) knock-out and overexpressed, and genetically predisposed diabetic (db/db) mouse models, where white adipocyte Nnmt was upregulated in diabetic conditions [147]. Treatments containing niacin were reported to trigger insulin resistance in patients with hyperlipidemia [149, 150], however the mechanisms are still unknown. The widespread effects of NNMT on epigenetic pathways may serve as a tentative explanation for insulin resistance caused by niacin (and its derivatives nicotinamide and nicotinic acid) usage, although further investigation is required.
2.2. Role of environmental toxicants in nutrient-regulated obesity pathways
2.2.1 BisphenolA (BPA)
BPA is an endocrine disrupting compound (EDC) found in many commonly used products and thus a majority of the world’s population is exposed to varying levels [151, 152]. A critical breakthrough in understanding nutrient-toxicant interactions was established through a study published by Dolinoy and colleagues in 2007 [153] showing that perturbation of mouse coat color induced by Bisphenol A (BPA) exposure is rescued by methyl donor nutrient supplementation (folic acid, choline, betaine and vitamin B12). This was not the first demonstration of dietary intervention as a means of alleviating toxicant effects. However, this was the first demonstration of the role of epigenetic state (DNA methylation) at the agouti viable yellow (Avy) locus as the intermediate link between toxicant exposure, diet and phenotypic outcome. The mechanism of epigenetic rescue by methyl donor nutrients remains unclear but has been shown to occur in several other studies [154–157]. The commonly accepted paradigm is that methyl donor nutrient intake determines the availability of subsequent methyl groups generated through the one carbon metabolism pathway.
Recent studies link BPA to obesity through several different mechanisms [158, 159]. Cell culture studies show that BPA at concentrations >10uM increases murine adipocyte differentiation and decreases global DNA methylation compared to controls [160]. Mackay and colleagues [161] recently used a CD-1 mouse model to show that maternal consumption of a maximum of 7.2 µg/kg/day on average of BPA (well below the EPA reference dose of 50µg/kg/dy) resulted in higher weight of both male and female offspring. Significantly higher caloric intake was observed in female offspring only while the males exhibited greater energy expenditure. Although BPA treated female offspring had increased plasma leptin levels, males were unaffected. Also, in BPA-exposed female offspring there was no increased proopiomelanocortin expression in the arcuate nucleus, which normally would have signalled excess energy, satiety and subsequent reduced food intake. Interestingly, most of these effects of BPA were only observed when mice were exposed in conjunction with a high fat diet, demonstrating the aggregate effect of diet. The sexually dimorphic nature of these effects is likely due to the estrogenic properties of BPA. Indeed, females but not males, exposed to BPA exhibited increased estrogen receptor alpha (ERα) mRNA expression in the hypothalamus compared to controls. As a potential mechanism, a separate study shows that in utero BPA exposure alters methylation status at ERα and ERβ[162].
BPA exposure has also been linked to epigenetic perturbation at imprinted loci in the mouse [163]. Of particular interest, Susiarjo and colleagues reported that maternal BPA exposure during pregestation, gestation and lactation at a dose of 10mg/kg/day results in a gain of methylation at the DMR1 of the Igf2 gene. This was correlated with loss of imprinted monoallelic expression of Igf2 and Igf2 overexpression. A follow up study showed that these epigenetic changes coincide with increased body fat, insulin resistance and glucose intolerance only in adult male offspring [164]. In contrast to the study by Mackay and colleagues described above [161], females were seemingly unaffected. Interestingly, F2 male offspring inherited the increase in Igf2 expression and Igf2 DMR1 methylation changes as well as the metabolic phenotypes from their sires. Use of a targeted genetic mutant mouse model, H193.8/+ mice [165], showed that of loss of Igf2 imprinting and overexpression is sufficient to result in glucose intolerance phenotype in the male but not female mice. These gender specific phenotypes may also be linked to the estrogenic properties of BPA as speculated above. The explanation for the opposite nature of the gender bias in the two studies remains unclear although it is well known that BPA does not always have a linear dose response curve and the difference may be explained by amount and timing of dosage.
2.2.2 Alcohol
Fetal alcohol exposure is linked to increased feeding behaviors and obesity in childhood [166, 167]. As discussed above, proopiomelanocortin promoter methylation regulates proopiomelanocortin expression and increased proopiomelanocortin promoter methylation is linked to obesity. Fetal alcohol exposure in a rat model results in proopiomelanocortin promoter hypermethylation and downregulated proopiomelanocortin expression in the hypothalamus persisting until the F3 generation [168]. Surprisingly, these changes were not correlated with weight differences as has been shown in dietary models of perturbed proopiomelanocortin [169]. Further assessment by the same research group revealed that alcohol-induced proopiomelanocortin repression is alleviated by dietary choline supplementation during gestation [157]. Based on this finding, the authors propose that alcohol-induced methylation changes at proopiomelanocortin likely involve disruption of the one carbon metabolism pathway and subsequent limitation of methyl groups. In this way, choline may rescue by increasing methyl group availability thus compensating for alcohol-induced disruption of folate-derived methyl groups [157]. However, this “limited methyl group” hypothesis is contradicted by the apparent gain of methylation induced by alcohol at the proopiomelanocortin promoter and choline-induced hypomethylation required for restoration of gene expression. Therefore, the mechanism of alcohol-induced proopiomelanocortin promoter hypermethylation and the choline dependent rescue is likely via a separate or indirect pathway.
More evidence of an indirect role comes in a recent study by the same group showing that fetal alcohol exposure in rats increases MECP2 levels and MECP2 binding to the proopiomelanocortin locus in the hypothalamus [170]. Mecp2 knockdown via targeted shRNA to the brain alleviated the effects of fetal alcohol exposure on proopiomelanocortin expression in the hypothalamus. We can speculate that the increased MECP2 binding at the proopiomelanocortin promoter is the result of a combination of hypermethylation at the proopiomelanocortin promoter and increased MECP2 protein availability leading to increased recruitment of MECP2 to the locus and increased proopiomelanocortin repression. The mechanistic link between choline supplementation and the effects of MECP2 remains unclear, however, several studies show that choline supplementation also effectively rescues some of the phenotypic and molecular effects associated with Mecp2 depletion in Rhett syndrome models [171–173]. All together these finding suggest a complex diametric role for choline in MECP2 mediated epigenetic mechanisms such that choline supplementation appears to rescue the effects of both Mecp2 depletion and overexpression.
2.2.3 Persistent organic pollutants
Persistent organic pollutants (POPs) are classified on the basis of having slow degradation process such that they persist in a toxic form in the environment or body in a way that may lead to accumulating and/or chronic exposure. These man-made compounds are either synthesized or result as a byproduct of industrial processing. Many POPs have also been classified as endocrine disruptors [174, 175]. A recent study showed that metabolically abnormal obese individuals have higher levels of POPs in plasma compared to metabolically healthy obese individuals [176]. Obesity is primarily classified based on body mass index (BMI) or waist to hip circumference ratio. However, many individuals that surpass the obesity threshold set for these parameters do not exhibit the altered metabolic state linked to increased risk for obesity related diseases such as diabetes and CVD. Taken together with these new findings, it is possible that for some individuals “obesity” status alone may not pose the extended disease risk assumed and additional factors are required to result in disease-related metabolic state. New studies allow us to speculate that the propensity for abnormal metabolic state associated with obesity may be influenced by POPs. Thus evaluating the extent of environmental pollutant exposure may be of importance in linking obesity to further metabolic disruption. Further detailed in vivo studies need to be done to determine the involvement of epigenetic mechanisms influencing the role of POPs in obesity. Cell culture studies show that POPs such as 2,2’,4,4’-tetrabrominated diphenyl ether (BDE-47) and tributyltin (TBT) at concentrations of 2.5–25uM and ≥10nM, respectively, increase murine adipocyte differentiation compared to controls [160]. Furthermore, BDE-47 exposure but not TBT exposure was linked to decreased global methylation. A more recent study with similar results showed the increase in adipocyte differentiation caused by TBT exposure is linked to the PPARG signaling pathway for which we have discussed various pathways of epigenetic regulation [177].
3. Conclusions
In Table 1, we provide a compilation of epigenetic mechanisms acting within several major molecular pathways of obesity. Taken together, we show that many of the perturbed epigenetic mechanisms are consistent between model organisms with supportive human subject data; between nutrient and toxicant models; and are supported by genetic models of disrupted epigenetic regulators. Nevertheless, many of these findings are only correlative and we conclude that much more research is required to fully understand the role of such perturbations in obesity; to determine whether these changes are stable or change further with progression of obesity and its related diseases; and to determine whether different types of metabolic syndromes are associated with specific epigenetic profiles, pathway dysregulation or linked to a particular environmental stimulus (i.e. diet, toxicant etc.). Studies to determine whether such epigenetic defects are reversible and the agents that reverse them will likely identify potential targets for diagnosis and treatment of obesity and obesity-related disease.
Table 1.
Summary of epigenetically altered genes/pathways in obesity models.
| Altered genes/ pathways |
Influential tissues |
Obesogenic influences |
Epigenetic modifications |
Epigenetic regulators |
Model | Dietary influences |
Genetic influences |
Toxicant influences |
Refs |
|---|---|---|---|---|---|---|---|---|---|
| POMC | hypothalamus | homeostatic eating |
promoter DNA methylation |
N.D. | Human | caloric restriction | N.D. | N.D. | [71] |
| N.D. | Mouse | maternal CLAs supplementation |
N.D. | N.D. | [65] | ||||
| N.D. | Rat | neonatal overnutrition | N.D. | N.D. | [67] | ||||
| N.D. | Rat | maternal and neonatal folate supplementation |
N.D. | N.D. | [68] | ||||
| N.D. | Rat | maternal high fat diet | N.D. | N.D. | [69, 70] | ||||
| N.D. | Mouse | N.D. | Mecp2 knockout | N.D. | [72] | ||||
| Mecp2 | Mouse | N.D. | N.D. | maternal alcohol exposure |
[170] | ||||
|
Dnmtl, Mecp2 and active histone marks |
Rat | maternal choline supplementation |
N.D. | [168] | |||||
| N.D. | Rat | N.D. | N.D. | [157] | |||||
| N.D. | miR-103 | Mouse | N.D. | Dicer1 knockout | N.D. | [78] | |||
| ST8SIA4 | N.D. | promoter binding of histone acetyltransferase KAT8 |
Mouse | high fat diet | Kat8 silencing | N.D. | [76, 77] | ||
| leptin pathway |
adipose tissue and hypothalamus |
homeostatic eating |
promoter DNA methylation |
N.D. | Human | caloric restriction | N.D. | N.D. | [99] |
| N.D. | Mouse | maternal high fat diet or undernutrition |
N.D. | N.D. | [93,94] | ||||
| N.D. | Rat | high fat and sugar diet |
N.D. | N.D. | [97] | ||||
| promoter DNA methylation and histone acetylation |
promoter binding of DNMT1, MBD2 and RNA polymerase II |
Mouse | diet-induced obesity | N.D. | N.D. | [95] | |||
| N.D. | N.D. | Human | N.D. |
MECP2 loss of function mutation |
N.D. | [90] | |||
| N.D. | N.D. | Mouse | N.D. | Mecp2 knockout | N.D. | [91] | |||
| N.D. | N.D. | Mouse | N.D. | Dnmt3a knockout | N.D. | [92] | |||
| N.D. | N.D. | Mouse | high fat diet | N.D. | maternal BPA exposure |
[161] | |||
| N.D. | miR-200a, −200b, −429 | Mouse | N.D. | leptin nonsense mutation |
N.D. | [103] | |||
| N.D. | miR-200a | Rat | N.D. | N.D. | N.D. | [102] | |||
| N.D. | miR-132, −145 and −200a | Rat | high fat diet | N.D. | N.D. | [104] | |||
| N.D. | MiR-145 | Human | N.D. | N.D. | N.D. | [100] | |||
| N.D. | N.D. | Mouse | N.D. | Dicer1 knockout | N.D. | [78] | |||
| liver | N.D. | miR-143,−145 | Mouse | high fat diet | leptin receptor missense mutation |
N.D. | [101] | ||
| SLC6A3 | central reward circuitry and hypothalamus |
Hedonic and homeostatic eating |
promoter DNA methylation |
N.D. | Mouse | high fat diet | N.D. | N.D. | [107] |
| N.D. | Mouse | maternal high fat diet | N.D. | N.D. | [115] | ||||
| TH | N.D. | Mouse | high fat diet | N.D. | N.D. | [107] | |||
| N.D. | Mouse | high fat diet | Dnmt3a knockout | N.D. | [92] | ||||
| CDKN1C | N.D. | Mouse | maternal protein restriction |
N.D. | N.D. | [116] | |||
| C/EBP | adipose tissue |
adipogenesis | promoter DNA methylation |
N.D. | Chicken | folate depletion | N.D. | N.D. | [32] |
| PPARG | N.D. | histone methyltransferases | Mouse | diet-induced obesity | leptin nonsense mutation |
N.D. | [28] | ||
| N.D. | Mouse | vitamin A deficiency | N.D. | N.D. | [30] | ||||
| adipocyte differentiation |
N.D. | miR-27a, 27b | Human | N.D. | N.D. | N.D. | [100] | ||
| N.D. | miR-27a, 27b | Cell culture |
N.D. | N.D. | N.D. | [137, 138] | |||
| N.D. | miR-27a | Mouse/ Cell Culture |
high fat diet | N.D. | N.D. | [140] | |||
| decreased global DNA methylation |
N.D. | Cell culture |
N.D. | N.D. | TBT exposure |
[160, 177] | |||
| N.D. | N.D. | Cell culture |
N.D. | N.D. | BPA exposure |
[160] | |||
| N.D. | N.D. | Cell culture |
N.D. | N.D. | BDE-47 exposure |
[160] | |||
| FASN | Lipid accumulation |
promoter DNA methylation |
N.D. | Rat | high fat diet | N.D. | N.D. | [125] | |
| N.D. | Rat | high fat diet (isocaloric) |
N.D. | N.D. | [136] | ||||
| NNMT | adipose tissue and liver |
Energy expenditure |
global and local H3K4 methylation |
N.D. | Mouse | diet-induced obesity | N.D. | N.D. | [147] |
N.D.; not determined. Refs; references.
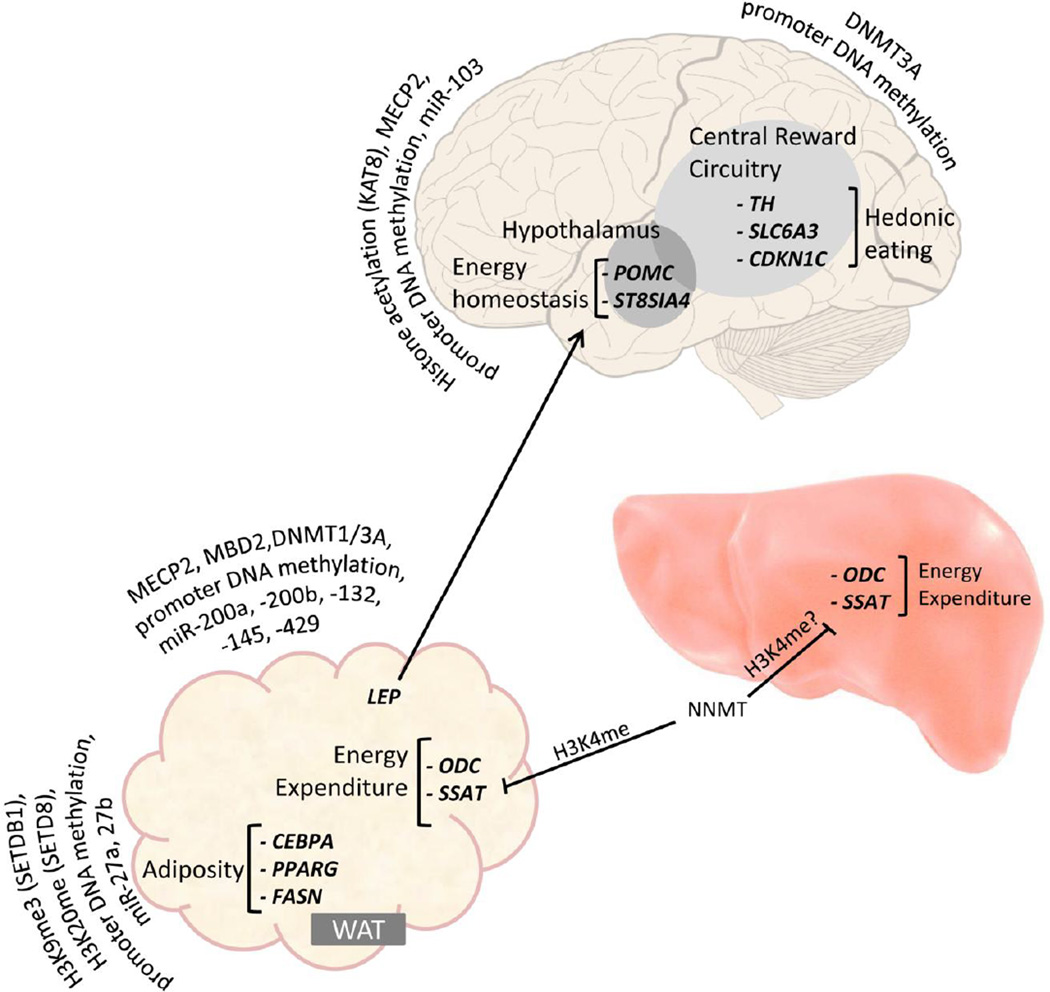
Figure 1 illustrates our current knowledge of some of the links between epigenetic mechanisms and molecular pathways of obesity including (1) homeostatic and hedonic eating behaviors controlled by brain and adipocyte-derived hormones, (2) adipocyte differentiation and fat accumulation, and (3) energy expenditure. This evidence of epigenetic dysregulation of molecular pathways of obesity supports the role on non-caloric mechanism of obesity, and further negates the paradigm that obesity is a disease caused merely by excess caloric intake. Epigenetic mechanisms can explain how both the source of fat and proportion of fat, even in an isocaloric diet can effect epigenetic mechanisms and lead to disruption of energy homeostatis via molecular pathways. For example, a recent study demonstrates that an isocaloric high fat diet in a rat model results in significantly increased body weight and fat mass, which is linked to hypomethylation upstream of the fatty acid synthase promoter (within a region from −700 to −1100 bp from the transcription start site) and decreased adipose fatty acid synthase mRNA levels and [136]. This suggests that in epigenetically-induced obesity, the proportion of fat in the diet may be more important in some contexts than the total amount of calories.
Figure 1.
Obesity-linked metabolic pathways susceptible to diet induced epigenetic perturbation. Hypothalamic proopiomelanocortin neurons control homeostatic eating by expressing appetite-inhibitory proopiomelanocortin. Obesogenic diet causes repression of proopiomelanocortin, which increases food intake. This is partially offset by acute proopiomelanocortin neuron plasticity change via upregulating ST8SIA4 as a feedback-loop. Adipose tissue secretes leptin to signal (indicated by pointed arrow) excessive energy storage in hypothalamus by upregulating proopiomelanocortin expression. Obesogenic diet causes blunted leptin response in proopiomelanocortin neurons and repressed leptin transcription in adipocyte. Meanwhile compulsive eating is promoted through diet-induced dysregulation of tyrosine hydroxylase, dopamine transporter and cyclin-dependent kinase inhibitor 1C in dopaminergic neurons in central reward circuitry, boosting desire for palatable food. Adipocyte differentiation as well as fat accumulation is perturbed by nutrition targeting CEBPA, PPARG and fatty acid synthase, causing excessive adiposity. Dietary components also influence energy expenditure, represented by regulating ornithine decarboxylase and spermidine-spermine N-acetyltransferase in fat and possibly liver through NNMT, which acts by inhibiting (indicated by flat-end arrow) the enrichment of H3K4 methylation at ornithine decarboxylase and spermidine-spermine N-acetyltransferase. WAT: white adipose tissue. Epigenetically regulated genes in each metabolic pathway are listed in bold and italics (POMC: proopiomelanocortin; ST8SIA4: ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4; TH: tyrosine hydroxylase; SLC6A3: dopamine transporter solute carrier family 6; CDKN1C: cyclin-dependent kinase inhibitor 1C; CEBPA: CCAAT/enhancer binding protein (C/EBP) alpha; PPARG: peroxisome proliferator-activated receptor gamma; LEP: leptin; FASN: fatty acid synthase; ODC: ornithine decarboxylase; SSAT: spermidine-spermine N-acetyltransferase). Epigenetic regulatory mechanisms are depicted for each pathway and include DNA methylation, histone marks and epigenetic regulatory genes (NNMT: Nicotinamide N-methyltransferase. SETDB1: SET domain bifurcated 1; SETD8: SET domain containing protein 8; MECP2: methyl CpG binding protein 2; MBD2: methyl-CpG binding domain protein 2; DNMT1/3A: DNA methyltransferase 1/3A; KAT8: lysine acetyltransferase 8).
Interestingly, our comparison of nutrient and toxicant exposure models indicates that similar epigenetic and molecular effects may have varied phenotypic outcomes. For example in the alcohol model discussed above there is no change in weight although DNA hypermethylation at the proopiomelanocortin promoter is detected as well as decreased proopiomelanocortin expression, a molecular effect linked to increased weight gain in the dietary models [170]. This finding indicates that although proopiomelanocortin expression plays a critical role in weight, either mRNA repression alone is not sufficient to result in weight gain or the levels induced by alcohol exposure in this model did not cross the threshold required to manifest the phenotype. Thus it is important to point out that changes in DNA methylation, even when coinciding with gene expression changes, may not result in altered phenotypic outcome. The implications are that use of epigenetic states as biomarkers of diseases such as obesity require careful determination of correlation with both gene expression and phenotypic outcome. Moreover, this brings to question whether gene dosage thresholds should be more carefully considered in determining the role of molecular changes in phenotypic outcomes.
Another potential factor contributing to discrepancies between exposure outcomes is the timing of exposure or the timing of molecular perturbation in the pathway of interest. These are not necessarily mutually inclusive. For example, when challenged by high fat diet, the length of BPA exposure during gestation resulted in differing phenotypic outcome. Adult male offspring from a maternal diet containing a maximum of 7.2 µg/kg/day BPA throughout gestation and lactation showed no change in caloric intake compared to unexposed controls [161], while those maternal BPA exposure at a dose of 10 µg/kg/day from gestational day 9 to 16 demonstrated greater caloric intake [178]. Timing of environmental exposure may target different windows of susceptibility. This is relevant for treatments performed at different stages of development (i.e. embryonic vs adult); different parental routes of exposure (maternal vs. paternal); and different timing of parental exposure (i.e. pre-gestation, gestation, lactation). In particular, in utero exposure effects will likely differ from adult exposure since fetal somatic tissues and germ cells are undergoing major epigenetic programming [179]. Importantly, in the case of fetal exposure, effects may not be observed until later life stages as in the case of developmental origins of adult disease. Another important aspect of timing of epigenetic changes related to obesity is the fact that it remains unclear whether obesity always precedes or follows the perturbed epigenetic state. Genetic models that act by knocking out/down the key metabolic or epigenetic regulator genes suggest that at least some observed initial epigenetic changes precede obesity-related phenotypes [72, 91, 92, 147, 170]. These may be potential targets of diagnoses, prevention or treatment.
Many cell culture models (including those discussed here) provide useful mechanistic information, however, without direct evidence in vivo, phenotypic and disease relevance cannot be definitively determined. Also, we cannot rule out other possible causes of disparities in cell culture and animal studies, such as uncontrolled exposure to other potentially confounding environmental stimuli. For example, epigenetic states at proopiomelanocortin are affected by diet, alcohol, POPs and even early life stress [65, 70, 168–170]. Ibrahim and colleagues [180] showed that POP exposure in mice fed contaminated whale meat did not lead to weight gain nor standard metabolic disruption in insulin signaling pathways. The authors suggest that the added nutritional components of the whale meat may have confounded the data since these mice actually weighed less compared to mice fed standard chow and lard based high fat diet.
Importantly, we highlight some overlapping mechanisms of obesity that are influenced by both nutrition and environmental toxicants. This area of research requires much more attention since clearly both combined play a major role in obesity either having separate or aggregate effects. For example, several studies including some discussed here show that the obesity-related effects of toxicant exposure are further exacerbated by dietary modulation, such as addition of high fat diet [161, 178, 181]. Interestingly, some studies suggest that increased adiposity caused by high fat diet may actually provide a protective effect during toxicant exposure. For example, exposure to polychlorinated biphenyls (PCB-77) in a mouse model has been shown to impair glucose homeostasis via a TNFα dependent pathway only when combined with weight loss [182]. This paradigm is supported by the lipophilic nature of many POPs allowing for sequestration in adipose tissue and thus limiting the potential adverse effects of circulation in the body. A separate study by the same group demonstrated rescue of PCB-induced impaired glucose homeostasis after treatment with resveratrol, a polyphenol found in many types of berries that has previously been shown to inhibit inflammatory cytokines such as TNFα [183]. Likewise is the case of rescue of alcohol-induced epigenetic changes by choline supplementation [157]. Thus characterization of overlapping mechanisms will clarify the potential for use of nutrition in the treatment of toxicant-induced obesity. Furthermore, by characterizing this complex phenotype through multiple approaches including nutrient, toxicant and genetic models, we will likely elucidate novel parts of the pathways involved while obtaining certainty of previously established mechanisms. However, it should be noted that in order to accurately compare the effects of diet vs toxicant or combined exposures there is a need for some level of standardized experimental design between projects and more controlled environment in order to allow for comparison.
A significant amount of progress has been made in determining the role of non-coding RNA in epigenetic mechanisms of obesity. In particular, as discussed above, miRNA have been repeatedly shown to be significantly associated with both leptin signaling and adipogenesis. However, it has remained a great challenge to identify the direct targets of such miRNA activity within obesity-related molecular pathways. This is in part due to the fact that miRNA-target binding patterns are highly variable and miRNA function is often tissue-specific [184]. In addition, a single miRNA can have hundreds of targets while multiple miRNAs can act coordinately on the same target [185]. It is also a challenge to assay differences in precursor species and cellular localization of the miRNA being measured. Because of these challenges, many studies rely on measuring the overall cellular levels of miRNA as a proxy for activity. Although this is not a direct measure for determining the impact on epigenetic activity of the miRNA at the target, it does provide correlative clues to potential interactions. Unfortunately, even when miRNA with perturbed expression levels are detected, co-regulation of multiple miRNAs due to close genomic proximity or shared regulators (i.e. transcription factors) make it difficult to determine which of the miRNA identified is causal in a particular pathway. Thus, a significant amount of research remains to be done in understanding the role of miRNA and other types of non-coding RNA in obesity-related phenotypes. We have not really even begun to dissect the role of nutrition and other environmental factors such as toxicants in perturbing non-coding RNA function with regards to obesity-related pathways.
Although there are ever increasing improvements in the types of technology available to assess epigenetic mechanisms, the interpretation of epigenetic findings in terms of biological relevance remains a significant challenge. This is in part due to the complexity of epigenetic mechanisms and the gaps in our understanding of how multiple epigenetic mechanisms work together to regulate gene expression. Epigenetic regulation of fatty acid synthase transcription is an excellent example of the complex interaction between epigenetic mechanisms and transcriptional machinery required for normal gene expression. In a recent study of a high fat diet rat model investigators observed substantial hypomethylation at −90 bp but also reported hypermethylation at −62 bp at the fatty acid synthase promoter correlated with increased fatty acid synthase mRNA level in adipose tissue [125]. To explain this differential methylation regulation, the −90bp locus is actually within the binding site of NF-Y and SP1 activation complex [133], while the −62bp locus is a binding site of upstream regulatory factor [134] and is essential for upstream regulatory factor and SREBF1-induced fatty acid synthase expression upon feeding [135]. Thus, even at a single gene promoter, contradicting DNA methylation states may be detected and only together explain dysregulation of transcription. Understanding which regulatory elements are present within epigenetically regulated regions is critical to understanding the epigenetic mechanisms of disruption and hence the biological relevance of changes.
In conclusion, this assessment only further highlights the fact that obesity is a highly complex and multifactorial disease. Therefore, we are only just beginning to fit the pieces together to define the pathways and modes of perturbation contributed by environmental exposure. One factor that likely contributes to mixed results from studies on human obese population is the recognition of metabolically normal and abnormal obesity. Approximately, half of the obese adults in the US are found to be metabolically normal as measured by several cardiometabolic parameters. On the other hand, up to one-fourth of non-obese adults are metabolically abnormal and at risk for many of the diseases associated with obesity [186]. Interestingly, data shows that metabolically normal obese people are less susceptible to certain weight-related phenotypes [187], and are less susceptible to the effects of POPs compared to metabolically abnormal counterparts [176]. Thus, inconsistencies and non-reproducibility of studies on obese population could be attributed to metabolic heterogeneity in obese subjects, which underlines the importance of differentiating obese subjects using parameters for various metabolic functions.
Acknowledgments
Grants, sponsors and funding sources: This work was supported by the National Institutes of health (NIH) Transition to Independent Environmental Health Research Career Development Award from NIEHS KES023849A to F.Y.I.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nilsson EE, Skinner MK. Environmentally induced epigenetic transgenerational inheritance of disease susceptibility. Transl Res. 2014;165(1) doi: 10.1016/j.trsl.2014.02.003. 12-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lutter M, Nestler EJ. Homeostatic and hedonic signals interact in the regulation of food intake. J Nutr. 2009;139(3) doi: 10.3945/jn.108.097618. 629-32-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang Y, Chen Y, Mao Y Group COW. The contribution of excess weight to prevalent diabetes in Canadian adults. Public health. 2008 Mar;122(3):271–276. doi: 10.1016/j.puhe.2007.06.002. PubMed PMID: 17931673. [DOI] [PubMed] [Google Scholar]
- 4.Power C, Pinto Pereira SM, Law C, Ki M. Obesity and risk factors for cardiovascular disease and type 2 diabetes: investigating the role of physical activity and sedentary behaviour in mid-life in the 1958 British cohort. Atherosclerosis. 2014 Apr;233(2):363–369. doi: 10.1016/j.atherosclerosis.2014.01.032. PubMed PMID: 24530764. [DOI] [PubMed] [Google Scholar]
- 5.van der Leeuw J, van der Graaf Y, Nathoe HM, de Borst GJ, Kappelle LJ, Visseren FL, et al. The separate and combined effects of adiposity and cardiometabolic dysfunction on the risk of recurrent cardiovascular events and mortality in patients with manifest vascular disease. Heart. 2014 Sep 15;100(18):1421–1429. doi: 10.1136/heartjnl-2014-305490. PubMed PMID: 24817441. [DOI] [PubMed] [Google Scholar]
- 6.McCrindle BW. Cardiovascular Consequences of Childhood Obesity. The Canadian journal of cardiology. 2015 Feb;31(2):124–130. doi: 10.1016/j.cjca.2014.08.017. PubMed PMID: 25661547. [DOI] [PubMed] [Google Scholar]
- 7.Ezenwaka CE, Okoye O, Esonwune C, Onuoha P, Dioka C, Osuji C, et al. High prevalence of abdominal obesity increases the risk of the metabolic syndrome in Nigerian type 2 diabetes patients: using the International Diabetes Federation worldwide definition. Metabolic syndrome and related disorders. 2014 Jun;12(5):277–282. doi: 10.1089/met.2013.0139. PubMed PMID: 24601861. [DOI] [PubMed] [Google Scholar]
- 8.Oberman B, Khaku A, Camacho F, Goldenberg D. Relationship between obesity, diabetes and the risk of thyroid cancer. American journal of otolaryngology. 2015 Mar 3; doi: 10.1016/j.amjoto.2015.02.015. PubMed PMID: 25794786. [DOI] [PubMed] [Google Scholar]
- 9.Benedetto C, Salvagno F, Canuto EM, Gennarelli G. Obesity and female malignancies. Best practice & research Clinical obstetrics & gynaecology. 2015 Feb 7; doi: 10.1016/j.bpobgyn.2015.01.003. PubMed PMID: 25779915. [DOI] [PubMed] [Google Scholar]
- 10.Agalliu I, Williams S, Adler B, Androga L, Siev M, Lin J, et al. The impact of obesity on prostate cancer recurrence observed after exclusion of diabetics. Cancer Causes Control. 2015 Mar 14; doi: 10.1007/s10552-015-0554-z. PubMed PMID:25771797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mo F, Morrison H, Neutel IC. Population attributable risk from obesity to arthritis in the Canadian Population Health Longitudinal Survey 1994–2006. International journal of rheumatic diseases. 2014 Jul;17(6):628–634. doi: 10.1111/1756-185X.12372. PubMed PMID: 24802703. [DOI] [PubMed] [Google Scholar]
- 12.Visser AW, loan-Facsinay A, de Mutsert R, Widya RL, Loef M, de Roos A, et al. Adiposity and hand osteoarthritis: the Netherlands Epidemiology of Obesity study. Arthritis research & therapy. 2014;16(1):R19. doi: 10.1186/ar4447. PubMed PMID: 24447395. Pubmed Central PMCID: 3978723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leisegang K, Bouic PJ, Menkveld R, Henkel RR. Obesity is associated with increased seminal insulin and leptin alongside reduced fertility parameters in a controlled male cohort. Reproductive biology and endocrinology: RB&E. 2014;12:34. doi: 10.1186/1477-7827-12-34. PubMed PMID: 24885899. Pubmed Central PMCID: 4019561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammoud AO, Meikle AW, Reis LO, Gibson M, Peterson CM, Carrell DT. Obesity and male infertility: a practical approach. Seminars in reproductive medicine. 2012 Dec;30(6):486–495. doi: 10.1055/s-0032-1328877. PubMed PMID: 23074007. [DOI] [PubMed] [Google Scholar]
- 15.Grundy A, Cotterchio M, Kirsh VA, Kreiger N. Associations between anxiety, depression, antidepressant medication, obesity and weight gain among Canadian women. PLoS ONE. 2014;9(6):e99780. doi: 10.1371/journal.pone.0099780. PubMed PMID: 24932472. Pubmed Central PMCID: 4059657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molyneaux E, Poston L, Ashurst-Williams S, Howard LM. Obesity and mental disorders during pregnancy and postpartum: a systematic review and meta-analysis. Obstetrics and gynecology. 2014 Apr;123(4):857–867. doi: 10.1097/AOG.0000000000000170. PubMed PMID: 24785615., Pubmed Central PMCID: 4254698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;(33 Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 18.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 19.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69(6):915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 20.Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem. 2002;278(6) doi: 10.1074/jbc.M210256200. 4035-40-40. [DOI] [PubMed] [Google Scholar]
- 21.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393(6683):386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 22.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nature reviews Genetics. 2009 May;10(5):295–304. doi: 10.1038/nrg2540. PubMed PMID: 19308066. [DOI] [PubMed] [Google Scholar]
- 23.Kuroishi T, Rios-Avila L, Pestinger V, Wijeratne SS, Zempleni J. Biotinylation is a natural, albeit rare, modification of human histones. Mol Genet Metab. 2011 Dec;104(4):537–545. doi: 10.1016/j.ymgme.2011.08.030. PubMed PMID: 21930408. Pubmed Central PMCID: 3224183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, et al. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007 Oct 18;449(7164):933–937. doi: 10.1038/nature06166. PubMed PMID: 17898714. [DOI] [PubMed] [Google Scholar]
- 25.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014 Mar 27;157(1):77–94. doi: 10.1016/j.cell.2014.03.008. PubMed PMID: 24679528. [DOI] [PubMed] [Google Scholar]
- 26.Delatte B, Deplus R, Fuks F. Playing TETris with DNA modifications. EMBO J. 2014 Jun 2;33(11):1198–1211. doi: 10.15252/embj.201488290. PubMed PMID: 24825349. Pubmed Central PMCID: 4198024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007 May 18;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. PubMed PMID: 17512414. [DOI] [PubMed] [Google Scholar]
- 28.Wakabayashi K-i, Okamura M, Tsutsumi S, Nishikawa NS, Tanaka T, Sakakibara I, et al. The peroxisome proliferator-activated receptor gamma/retinoid X receptor alpha heterodimer targets the histone modification enzyme PR-Set7/Setd8 gene and regulates adipogenesis through a positive feedback loop. Mol Cell Biol. 2009;29(13) doi: 10.1128/MCB.01856-08. 3544-55-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okamura M, Inagaki T, Tanaka T, Sakai J. Role of histone methylation and demethylation in adipogenesis and obesity. Organogenesis. 2010;6(1):24–32. doi: 10.4161/org.6.1.11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ribot J, Felipe F, Bonet ML, Palou A. Changes of adiposity in response to vitamin A status correlate with changes of PPARv2 expression. Obesity research. 2001 doi: 10.1038/oby.2001.65. [DOI] [PubMed] [Google Scholar]
- 31.Tian Y-J, Luo N, Chen N-N, Lun Y-Z, Gu X-Y, Li Z, et al. Maternal nicotinamide supplementation causes global DNA hypomethylation, uracil hypo-incorporation and gene expression changes in fetal rats. Br J Nutr. 2014;111(9) doi: 10.1017/S0007114513004054. 1594-601-601. [DOI] [PubMed] [Google Scholar]
- 32.Yu X, Liu R, Zhao G, Zheng M, Chen J, Wen J. Folate supplementation modifies CCAAT/enhancer-binding protein alpha methylation to mediate differentiation of preadipocytes in chickens. Poult Sci. 2014 Oct;93(10):2596–2603. doi: 10.3382/ps.2014-04027. PubMed PMID: 25037819. [DOI] [PubMed] [Google Scholar]
- 33.Mahaffey KR, Vanderveen JE. Nutrient-toxicant interactions: susceptible populations. Environ Health Perspect. 1979;29:81–87. doi: 10.1289/ehp.792981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho MR, Shin JY, Hwang JH, Jacobs DR, Jr, Kim SY, Lee DH. Associations of fat mass and lean mass with bone mineral density differ by levels of persistent organic pollutants: National Health and Nutrition Examination Survey 1999–2004. Chemosphere. 2011 Feb;82(9):1268–1276. doi: 10.1016/j.chemosphere.2010.12.031. PubMed PMID: 21196025. [DOI] [PubMed] [Google Scholar]
- 35.Kim M-J, Marchand P, Henegar C, Antignac J-P, Alli R, Poitou C, et al. Fate and complex pathogenic effects of dioxins and polychlorinated biphenyls in obese subjects before and after drastic weight loss. Environ Health Perspect. 2010;119(3):377–383. doi: 10.1289/ehp.1002848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong NS, Kim KS, Lee IK, Lind PM, Lind L, Jacobs DR, et al. The association between obesity and mortality in the elderly differs by serum concentrations of persistent organic pollutants: a possible explanation for the obesity paradox. Int J Obes (Lond) 2012 Sep;36(9):1170–1175. doi: 10.1038/ijo.2011.187. PubMed PMID: 21946706. [DOI] [PubMed] [Google Scholar]
- 37.Tutelyan VA, Kravchenko LV, Aksenov IV, Trusov NV, Guseva GV, Kodentsova VM, et al. Activity of xenobiotic-metabolizing enzymes in the liver of rats with multi-vitamin deficiency. Int J Vitam Nutr Res. 2013;83(1):5–13. doi: 10.1024/0300-9831/a000140. [DOI] [PubMed] [Google Scholar]
- 38.Singh M, Kumar D, Yusuf MA, Sardar M, Sarin NB. Effects of wild-type and a-tocopherol-enriched transgenic Brassica juncea on the components of xenobiotic metabolism, antioxidant status, and oxidative stress in the liver of mice. Transgenic Res. 2013;22(4) doi: 10.1007/s11248-013-9689-4. 813-22-22. [DOI] [PubMed] [Google Scholar]
- 39.Mustacich DJ, Gohil K, Bruno RS, Yan M, Leonard SW, Ho E, et al. Alpha-tocopherol modulates genes involved in hepatic xenobiotic pathways in mice. J Nutr Biochem. 2008;20(6) doi: 10.1016/j.jnutbio.2008.05.007. 469-76-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Canistro D, Pozzetti L, Sapone A, Broccoli M, Bonamassa B, Longo V, et al. Perturbation of rat hepatic metabolising enzymes by folic acid supplementation. Mutat Res. 2007;637(1–2):16–22. doi: 10.1016/j.mrfmmm.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 41.Jedrychowski W, Masters E, Choi H, Sochacka E, Flak E, Mroz E, et al. Pre-pregnancy dietary vitamin A intake may alleviate the adverse birth outcomes associated with prenatal pollutant exposure: epidemiologic cohort study in Poland. Int J Occup Environ Health. 2007;13(2):175–180. doi: 10.1179/oeh.2007.13.2.175. [DOI] [PubMed] [Google Scholar]
- 42.Kulkarni NN, Yi Z, Huehnken C, Agerberth B, Gudmundsson GH. Phenylbutyrate induces cathelicidin expression via the vitamin D receptor: Linkage to inflammatory and growth factor cytokines pathways. Mol Immunol. 2014;63(2):530–539. doi: 10.1016/j.molimm.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 43.Wahlang B, Falkner KC, Gregory B, Ansert D, Young D, Conklin DJ, et al. Polychlorinated biphenyl 153 is a diet-dependent obesogen that worsens nonalcoholic fatty liver disease in male C57BL6/J mice. J Nutr Biochem. 2013;24(9) doi: 10.1016/j.jnutbio.2013.01.009. 1587-95-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lammon CA, Hood RD. Effects of protein deficient diets on the developmental toxicity of inorganic arsenic in mice. Birth Defects Res B Dev Reprod Toxicol. 2004;71(3) doi: 10.1002/bdrb.20006. 124-34-34. [DOI] [PubMed] [Google Scholar]
- 45.Wu X, Barnhart C, Lein PJ, Lehmler HJ. Hepatic metabolism affects the atropselective disposition of 2,2’,3,3’,6,6’-hexachlorobiphenyl (PCB 136) in mice. Environmental science & technology. 2015 Jan 6;49(1):616–625. doi: 10.1021/es504766p. PubMed PMID: 25420130. Pubmed Central PMCID: 4291784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guzmán C, García R. Maternal protein restriction during pregnancy and/or lactation negatively affects follicular ovarian development and steroidogenesis in the prepubertal rat offspring. Archives of medical research. 2014 doi: 10.1016/j.arcmed.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 47.Sui S, He Jia Y, Li R, Cai D, Li X, et al. Maternal protein restriction during gestation and lactation programs offspring ovarian steroidogenesis and folliculogenesis in the prepubertal gilts. J Steroid Biochem Mol Biol. 2014;143:267–276. doi: 10.1016/j.jsbmb.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 48.Park EC, II, Kim S, Hong Y, Hwang JW, Cho G-S, Cha H-N, et al. Inhibition of CYP4A reduces hepatic endoplasmic reticulum stress and features of diabetes in mice. Gastroenterology. 2014;147(4) doi: 10.1053/j.gastro.2014.06.039. 860-9-9. [DOI] [PubMed] [Google Scholar]
- 49.Douard V, Patel C, Lee J, Tharabenjasin P, Williams E, Fritton JC, et al. Chronic high fructose intake reduces serum 1,25 (OH)2D3 levels in calcium-sufficient rodents. PLoS ONE. 2014;9(4):e93611. doi: 10.1371/journal.pone.0093611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Radhakrishnakartha H, Appu AP, Indira M. Ascorbic acid supplementation enhances recovery from ethanol induced inhibition of Leydig cell steroidogenesis than abstention in male guinea pigs. Eur J Pharmacol. 2013;723:73–79. doi: 10.1016/j.ejphar.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 51.Chang HJ, Park JS, Lee EK, Kim MH, Baek MK, Kim HR, et al. Ascorbic acid suppresses the 2,3,7,8- tetrachloridibenxo-p-dioxin (TCDD)-induced CYP1A1 expression in human HepG2 cells. Toxicol In Vitro. 2009;23(4):622–626. doi: 10.1016/j.tiv.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 52.Cai D, Jia Y, Lu J, Yuan M, Sui S, Song H, et al. Maternal dietary betaine supplementation modifies hepatic expression of cholesterol metabolic genes via epigenetic mechanisms in newborn piglets. Br J Nutr. 2014;112(9) doi: 10.1017/S0007114514002402. 1459-68-68. [DOI] [PubMed] [Google Scholar]
- 53.Erkekoglu P, Giray BK, Caglayan A, Hincal F. Selenium and/or iodine deficiency alters hepatic xenobiotic metabolizing enzyme activities in rats. Journal of trace elements in medicine and biology: organ of the Society for Minerals and Trace Elements. 2012 Jan;26(1):36–41. doi: 10.1016/j.jtemb.2011.12.002. PubMed PMID: 22366236. [DOI] [PubMed] [Google Scholar]
- 54.Yeo GS, Farooqi IS, Challis BG, Jackson RS, O’Rahilly S. The role of melanocortin signalling in the control of body weight: evidence from human and murine genetic models. QJM. 2000;93(1):7–14. doi: 10.1093/qjmed/93.1.7. [DOI] [PubMed] [Google Scholar]
- 55.Page-Wilson G, Freda PU, Jacobs TP, Khandji AG, Bruce JN, Foo ST, et al. Clinical utility of plasma POMC and AgRP measurements in the differential diagnosis of ACTH-dependent Cushing’s syndrome. J Clin Endocrinol Metab. 2014;99(10):E1838–E1845. doi: 10.1210/jc.2014-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ehrlich S, Weiss D, Burghardt R, Infante-Duarte C, Brockhaus S, Muschler MA, et al. Promoter specific DNA methylation and gene expression of POMC in acutely underweight and recovered patients with anorexia nervosa. J Psychiatr Res. 2010;44(13) doi: 10.1016/j.jpsychires.2010.01.011. 827-33-33. [DOI] [PubMed] [Google Scholar]
- 57.Zemel MB, Shi H. Pro-opiomelanocortin (POMC) deficiency and peripheral melanocortins in obesity. Nutr Rev. 2000;58(6):177–180. doi: 10.1111/j.1753-4887.2000.tb01857.x. [DOI] [PubMed] [Google Scholar]
- 58.Ozen S, Ozcan N, Uçar SK, Göksen D, Darcan S. Unexpected clinical features in a female patient with proopiomelanocortin (POMC) deficiency. J Pediatr Endocrinol Metab. 2014 doi: 10.1515/jpem-2014-0324. [DOI] [PubMed] [Google Scholar]
- 59.Sergeyev V, Broberger C, Hökfelt T. Effect of LPS administration on the expression of POMC, NPY, galanin, CART and MCH mRNAs in the rat hypothalamus. Brain Res Mol Brain Res. 2001;90(2):93–100. doi: 10.1016/s0169-328x(01)00088-2. [DOI] [PubMed] [Google Scholar]
- 60.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang F, Gelernter J, Kranzler HR, Zhang H. Identification of POMC exonic variants associated with substance dependence and body mass index. PLoS ONE. 2012;7(9):e45300. doi: 10.1371/journal.pone.0045300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baker M, Gaukrodger N, Mayosi BM, Imrie H, Farrall M, Watkins H, et al. Association between common polymorphisms of the proopiomelanocortin gene and body fat distribution: a family study. Diabetes. 2005;54(8) doi: 10.2337/diabetes.54.8.2492. 2492-6-6. [DOI] [PubMed] [Google Scholar]
- 63.Newell-Price J. Proopiomelanocortin gene expression and DNA methylation: implications for Cushing’s syndrome and beyond. J Endocrinol. 2003;177(3) doi: 10.1677/joe.0.1770365. 365-72-72. [DOI] [PubMed] [Google Scholar]
- 64.Yoo JY, Lee S, Lee HA, Park H, Park YJ, Ha EH, et al. Can proopiomelanocortin methylation be used as an early predictor of metabolic syndrome? Diabetes care. 2014 Mar;37(3):734–739. doi: 10.2337/dc13-1012. PubMed PMID: 24222450. [DOI] [PubMed] [Google Scholar]
- 65.Zhang X, Yang R, Jia Y, Cai D, Zhou B, Qu X, et al. Hypermethylation of Sp1 binding site suppresses hypothalamic POMC in neonates and may contribute to metabolic disorders in adults: impact of maternal dietary CLAs. Diabetes. 2014 May;63(5):1475–1487. doi: 10.2337/db13-1221. PubMed PMID: 24379351. [DOI] [PubMed] [Google Scholar]
- 66.Yang G, Lim C-Y, Li C, Xiao X, Radda GK, Li C, et al. FoxOI inhibits leptin regulation of proopiomelanocortin promoter activity by blocking STAT3 interaction with specificity protein 1. J Biol Chem. 2008;284(6) doi: 10.1074/jbc.M804965200. 3719-27-27. [DOI] [PubMed] [Google Scholar]
- 67.Plagemann A, Harder T, Brunn M, Harder A, Roepke K, Wittrock-Staar M, et al. Hypothalamic proopiomelanocortin promoter methylation becomes altered by early overfeeding: an epigenetic model of obesity and the metabolic syndrome. J Physiol (Lond) 2009;587(Pt 20) doi: 10.1113/jphysiol.2009.176156. 4963-76-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cho CE, Sanchez-Hernandez D, Reza-Lopez SA, Huot PS, Kim Yl, Anderson GH. High folate gestational and post-weaning diets alter hypothalamic feeding pathways by DNA methylation in Wistar rat offspring. Epigenetics. 2013 Jul;8(7):710–719. doi: 10.4161/epi.24948. PubMed PMID: 23803567. Pubmed Central PMCID: 3781190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marco A, Kisliouk T, Tabachnik T, Meiri N, Weller A. Overweight and CpG methylation of the Pomc promoter in offspring of high-fat-diet-fed dams are not “reprogrammed” by regular chow diet in rats. FASEB J. 2014 Jun 13; doi: 10.1096/fj.14-255620. PubMed PMID: 24928196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paternain L, Batlle MA, De la Garza AL, Milagro Fl, Martinez JA, Campion J. Transcriptomic and epigenetic changes in the hypothalamus are involved in an increased susceptibility to a high-fat-sucrose diet in prenatally stressed female rats. Neuroendocrinology. 2012;96(3):249–260. doi: 10.1159/000341684. PubMed PMID: 22986707. [DOI] [PubMed] [Google Scholar]
- 71.Crujeiras AB, Campion J, Diaz-Lagares A, Milagro Fl, Goyenechea E, Abete I, et al. Association of weight regain with specific methylation levels in the NPY and POMC promoters in leukocytes of obese men: a translational study. Regul Pept. 2013 Sep 10;186:1–6. doi: 10.1016/j.regpep.2013.06.012. PubMed PMID: 23831408. [DOI] [PubMed] [Google Scholar]
- 72.Wang X, Lacza Z, Sun YE, Han W. Leptin resistance and obesity in mice with deletion of methyl-CpG-binding protein 2 (MeCP2) in hypothalamic pro-opiomelanocortin (POMC) neurons. Diabetologia. 2013;57(1) doi: 10.1007/s00125-013-3072-0. 236-45-45. [DOI] [PubMed] [Google Scholar]
- 73.Kudo S. Methyl-CpG-binding protein MeCP2 represses Sp1-activated transcription of the human leukosialin gene when the promoter is methylated. Mol Cell Biol. 1998 Sep;18(9):5492–5499. doi: 10.1128/mcb.18.9.5492. PubMed PMID: 9710633. Pubmed Central PMCID: 109134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gouaze A, Brenachot X, Rigault C, Krezymon A, Rauch C, Nedelec E, et al. Cerebral cell renewal in adult mice controls the onset of obesity. PLoS ONE. 2013;8(8):e72029. doi: 10.1371/journal.pone.0072029. PubMed PMID: 23967273. Pubmed Central PMCID: 3742483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seifert A, Glanz D, Glaubitz N, Horstkorte R, Bork K. Polysialylation of the neural cell adhesion molecule: interfering with polysialylation and migration in neuroblastoma cells. Arch Biochem Biophys. 2012;524(1):56–63. doi: 10.1016/j.abb.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 76.Brenachot X, Rigault C, Nédélec E, Laderriére A, Khanam T, Gouaze A, et al. The histone acetyltransferase MOF activates hypothalamic polysialylation to prevent diet-induced obesity in mice. Mol Metab. 2014;3(6):619–629. doi: 10.1016/j.molmet.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Benani A, Hryhorczuk C, Gouaze A, Fioramonti X, Brenachot X, Guissard C, et al. Food intake adaptation to dietary fat involves PSA-dependent rewiring of the arcuate melanocortin system in mice. J Neurosci. 2012;32(35) doi: 10.1523/JNEUROSCI.0624-12.2012. 11970-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vinnikov IA, Hajdukiewicz K, Reymann J, Beneke J, Czajkowski R, Roth LC, et al. Hypothalamic miR-103 protects from hyperphagic obesity in mice. J Neurosci. 2014 Aug 6;34(32):10659–10674. doi: 10.1523/JNEUROSCI.4251-13.2014. PubMed PMID: 25100599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kowarsch A, Preusse M, Marr C, Theis FJ. miTALOS: analyzing the tissue-specific regulation of signaling pathways by human and mouse microRNAs. Rna. 2011 May;17(5):809–819. doi: 10.1261/rna.2474511. PubMed PMID: 21441347. Pubmed Central PMCID: 3078731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berthoud H-R, Morrison C. The brain, appetite, and obesity. Annu Rev Psychol. 2007;59:55–92. doi: 10.1146/annurev.psych.59.103006.093551. [DOI] [PubMed] [Google Scholar]
- 81.Cowley MA, Smart JL, Rubinstein M, Cerdán MG, Diano S, Horvath TL, et al. Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature. 2001;411(6836):480–484. doi: 10.1038/35078085. [DOI] [PubMed] [Google Scholar]
- 82.Ahima RS. Revisiting leptin’s role in obesity and weight loss. J Clin Invest. 2008;118(7) doi: 10.1172/JCI36284. 2380-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morton GJ, Cummings DE, Baskin DG, Barsh GS, Schwartz MW. Central nervous system control of food intake and body weight. Nature. 2006;443(7109):289–295. doi: 10.1038/nature05026. [DOI] [PubMed] [Google Scholar]
- 84.Farooqi IS, Matarese G, Lord GM, Keogh JM, Lawrence E, Agwu C, et al. Beneficial effects of leptin on obesity, T cell hyporesponsiveness, and neuroendocrine/metabolic dysfunction of human congenital leptin deficiency. J Clin Invest. 2002;110(8) doi: 10.1172/JCI15693. 1093-103-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pelleymounter MA, Cullen MJ, Baker MB, Hecht R, Winters D, Boone T, et al. Effects of the obese gene product on body weight regulation in ob/ob mice. Science. 1995;269(5223):540–543. doi: 10.1126/science.7624776. [DOI] [PubMed] [Google Scholar]
- 86.Chen Y, Wu R, Chen H-Z, Xiao Q, Wang W-J, He J-P, et al. Enhancement of hypothalamic STAT3 acetylation by nuclear receptor Nur77 dictates leptin sensitivity. Diabetes. 2015 doi: 10.2337/db14-1206. [DOI] [PubMed] [Google Scholar]
- 87.Al Rayyan N, Zhang J, Burnside AS, Good DJ. Leptin signaling regulates hypothalamic expression of nescient helix-loop-helix 2 (Nhlh2) through signal transducer and activator 3 (Stat3) Mol Cell Endocrinol. 2014;384(1–2):134–142. doi: 10.1016/j.mce.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.El-Haschimi K, Pierroz DD, Hileman SM, Bjørbaek C, Flier JS. Two defects contribute to hypothalamic leptin resistance in mice with diet-induced obesity. J Clin Invest. 2000;105(12) doi: 10.1172/JCI9842. 1827-32-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ernst MB, Wunderlich CM, Hess S, Paehler M, Mesaros A, Koralov SB, et al. Enhanced Stat3 activation in POMC neurons provokes negative feedback inhibition of leptin and insulin signaling in obesity. J Neurosci. 2009;29(37) doi: 10.1523/JNEUROSCI.5712-08.2009. 11582-93-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Blardi P, de Lalla A, D’Ambrogio T, Vonella G, Ceccatelli L, Auteri A, et al. Long-term plasma levels of leptin and adiponectin in Rett syndrome. Clin Endocrinol (Oxf) 2008;70(5) doi: 10.1111/j.1365-2265.2008.03386.x. 706-9-9. [DOI] [PubMed] [Google Scholar]
- 91.Fyffe SL, Neul JL, Samaco RC, Chao HT, Ben-Shachar S, Moretti P, et al. Deletion of Mecp2 in Sim1-expressing neurons reveals a critical role for MeCP2 in feeding behavior, aggression, and the response to stress. Neuron. 2008 Sep 25;59(6):947–958. doi: 10.1016/j.neuron.2008.07.030. PubMed PMID: 18817733., Pubmed Central PMCID: 2597031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kohno D, Lee S, Harper MJ, Kim KW, Sone H, Sasaki T, et al. Dnmt3a in sim1neurons is necessary for normal energy homeostasis. J Neurosci. 2014;34(46):15288–15296. doi: 10.1523/JNEUROSCI.1316-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Khalyfa A, Carreras A, Hakim F, Cunningham JM, Wang Y, Gozal D. Effects of late gestational high-fat diet on body weight, metabolic regulation and adipokine expression in offspring. Int J Obes (Lond) 2013;37(11):1481–1489. doi: 10.1038/ijo.2013.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jousse C, Parry L, Lambert-Langlais S, Maurin A-C, Averous J, Bruhat A, et al. Perinatal undernutrition affects the methylation and expression of the leptin gene in adults: implication for the understanding of metabolic syndrome. FASEB J. 2011;25(9) doi: 10.1096/fj.11-181792. 3271-8-8. [DOI] [PubMed] [Google Scholar]
- 95.Shen W, Wang C, Xia L, Fan C, Dong H, Deckelbaum RJ, et al. Epigenetic modification of the leptin promoter in diet-induced obese mice and the effects of N-3 polyunsaturated fatty acids. Sci Rep. 2014;4:e5282. doi: 10.1038/srep05282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Polson DA, Thompson MP. Macronutrient composition of the diet differentially affects leptin and adiponutrin mRNA expression in response to meal feeding. J Nutr Biochem. 2004;15(4) doi: 10.1016/j.jnutbio.2003.11.009. 242-6-6. [DOI] [PubMed] [Google Scholar]
- 97.Uriarte G, Paternain L, Milagro Fl, Martinez JA, Campió J. generating maturen Shifting to a control diet after a high-fat, high-sucrose diet intake induces epigenetic changes in retroperitoneal adipocytes of Wistar rats. J Physiol Biochem. 2013;69(3) doi: 10.1007/s13105-012-0231-6. 601-11-11. [DOI] [PubMed] [Google Scholar]
- 98.Bouchard L, Rabasa-Lhoret R, Faraj M, Lavoie M-E, Mill J, Pérusse L, et al. Differential epigenomic and transcriptomic responses in subcutaneous adipose tissue between low and high responders to caloric restriction. Am J Clin Nutr. 2009;91(2) doi: 10.3945/ajcn.2009.28085. 309-20-20. [DOI] [PubMed] [Google Scholar]
- 99.Cordero P, Campión J, Milagro Fl, Goyenechea E, Steemburgo T, Javierre BM, et al. Leptin and TNF-alpha promoter methylation levels measured by MSP could predict the response to a low-calorie diet. J Physiol Biochem. 2011;67(3) doi: 10.1007/s13105-011-0084-4. 463-70-70. [DOI] [PubMed] [Google Scholar]
- 100.Viesti ACR, Salgado W, Jr, Pretti da Cunha Tirapelli D, dos Santos JS. The expression of LEP, LEPR, IGF1 and IL10 in obesity and the relationship with microRNAs. PLoS ONE. 2014;9(4):e93512. doi: 10.1371/journal.pone.0093512. PubMed PMID: 24690978. Pubmed Central PMCID: 3972109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jordan SD, Kruger M, Willmes DM, Redemann N, Wunderlich FT, Bronneke HS, et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nature cell biology. 2011 Apr;13(4):434–446. doi: 10.1038/ncb2211. PubMed PMID: 21441927. [DOI] [PubMed] [Google Scholar]
- 102.Benoit C, Ould-Hamouda H, Crepin D, Gertler A, Amar L, Taouis M. Early leptin blockade predisposes fat-fed rats to overweight and modifies hypothalamic microRNAs. J Endocrinol. 2013 Jul;218(1):35–47. doi: 10.1530/JOE-12-0561. PubMed PMID: 23576026. [DOI] [PubMed] [Google Scholar]
- 103.Crepin D, Benomar Y, Riffault L, Amine H, Gertler A, Taouis M. The over-expression of miR-200a in the hypothalamus of ob/ob mice is linked to leptin and insulin signaling impairment. Mol Cell Endocrinol. 2014 Mar 25;384(1–2):1–11. doi: 10.1016/j.mce.2013.12.016. PubMed PMID: 24394757. [DOI] [PubMed] [Google Scholar]
- 104.Sangiao-Alvarellos S, Pena-Bello L, Manfredi-Lozano M, Tena-Sempere M, Cordido F. Perturbation of hypothalamic microRNA expression patterns in male rats after metabolic distress: impact of obesity and conditions of negative energy balance. Endocrinology. 2014 May;155(5):1838–1850. doi: 10.1210/en.2013-1770. PubMed PMID: 24517225. [DOI] [PubMed] [Google Scholar]
- 105.Shang J, Li J, Keller MP, Hohmeier HE, Wang Y, Feng Y, et al. Induction of miR-132 and miR-212 Expression by Glucagon-Like Peptide 1 (GLP-1) in Rodent and Human Pancreatic beta-Cells. Molecular endocrinology. 2015 Jul 28;:e20141335. doi: 10.1210/me.2014-1335. PubMed PMID: 26218441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Heneghan HM, Miller N, McAnena OJ, O’Brien T, Kerin MJ. Differential miRNA expression in omental adipose tissue and in the circulation of obese patients identifies novel metabolic biomarkers. The Journal of clinical endocrinology and metabolism. 2011 May;96(5):E846–E850. doi: 10.1210/jc.2010-2701. PubMed PMID: 21367929. [DOI] [PubMed] [Google Scholar]
- 107.Vucetic Z, Carlin JL, Totoki K, Reyes TM. Epigenetic dysregulation of the dopamine system in diet-induced obesity. J Neurochem. 2012;120(6) doi: 10.1111/j.1471-4159.2012.07649.x. 891-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Morton GJ, Meek TH, Schwartz MW. Neurobiology of food intake in health and disease. Nature reviews Neuroscience. 2014 Jun;15(6):367–378. doi: 10.1038/nrn3745. PubMed PMID: 24840801., Pubmed Central PMCID: 4076116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu S, Borgland SL. Regulation of the mesolimbic dopamine circuit by feeding peptides. Neuroscience. 2015;289C:119–142. doi: 10.1016/j.neuroscience.2014.12.046. [DOI] [PubMed] [Google Scholar]
- 110.Schwartz MW, Woods SC, Porte D, Seeley RJ, Baskin DG. Central nervous system control of food intake. Nature. 2000;404(6778):661–671. doi: 10.1038/35007534. [DOI] [PubMed] [Google Scholar]
- 111.Johnson PM, Kenny PJ. Dopamine D2 receptors in addiction-like reward dysfunction and compulsive eating in obese rats. Nat Neurosci. 2010;13(5):635–641. doi: 10.1038/nn.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Geiger BM, Behr GG, Frank LE, Caldera-Siu AD, Beinfeld MC, Kokkotou EG, et al. Evidence for defective mesolimbic dopamine exocytosis in obesity-prone rats. FASEB J. 2008;22(8) doi: 10.1096/fj.08-110759. 2740-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Daubner SC, Le T, Wang S. Tyrosine hydroxylase and regulation of dopamine synthesis. Arch Biochem Biophys. 2010;508(1):1–12. doi: 10.1016/j.abb.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Missale C, Nash SR, Robinson SW, Jaber M, Caron MG. Dopamine receptors: from structure to function. Physiol Rev. 1998;78(1):189–225. doi: 10.1152/physrev.1998.78.1.189. [DOI] [PubMed] [Google Scholar]
- 115.Vucetic Z, Kimmel J, Totoki K, Hollenbeck E, Reyes TM. Maternal high-fat diet alters methylation and gene expression of dopamine and opioid-related genes. Endocrinology. 2010;151(10):4756–4764. doi: 10.1210/en.2010-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vucetic Z, Totoki K, Schoch H, Whitaker KW, Hill-Smith T, Lucki I, et al. Early life protein restriction alters dopamine circuitry. Neuroscience. 2010;168(2):359–370. doi: 10.1016/j.neuroscience.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Alsió J, Olszewski PK, Norbäck AH, Gunnarsson ZEA, Levine AS, Pickering C, et al. Dopamine D1 receptor gene expression decreases in the nucleus accumbens upon long-term exposure to palatable food and differs depending on diet-induced obesity phenotype in rats. Neuroscience. 2010;171(3):779–787. doi: 10.1016/j.neuroscience.2010.09.046. [DOI] [PubMed] [Google Scholar]
- 118.Huang X-F, Yu Y, Zavitsanou K, Han M, Storlien L. Differential expression of dopamine D2 and D4 receptor and tyrosine hydroxylase mRNA in mice prone, or resistant, to chronic high-fat diet-induced obesity. Brain Res Mol Brain Res. 2005;135(1–2):150–161. doi: 10.1016/j.molbrainres.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 119.Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev. 2000 Jun 1;14(11):1293–1307. PubMed PMID: 10837022. [PubMed] [Google Scholar]
- 120.Berndt J, Kovacs P, Ruschke K, Klöting N, Fasshauer M, Schön MR, et al. Fatty acid synthase gene expression in human adipose tissue: association with obesity and type 2 diabetes. Diabetologia. 2007;50(7) doi: 10.1007/s00125-007-0689-x. 1472-80-80. [DOI] [PubMed] [Google Scholar]
- 121.Schweizer M, Roder K, Zhang L, Wolf SS. Transcription factors acting on the promoter of the rat fatty acid synthase gene. Biochem Soc Trans. 2002;30(Pt6):1070–1072. doi: 10.1042/bst0301070. [DOI] [PubMed] [Google Scholar]
- 122.Lodhi IJ, Yin L, Jensen-Urstad APL, Funai K, Coleman T, Baird JH, et al. Inhibiting adipose tissue lipogenesis reprograms thermogenesis and PPARv activation to decrease diet-induced obesity. Cell Metab. 2012;16(2):189–201. doi: 10.1016/j.cmet.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Narvaez CJ, Matthews D, Broun E, Chan M, Welsh J. Lean phenotype and resistance to diet-induced obesity in vitamin D receptor knockout mice correlates with induction of uncoupling protein-1 in white adipose tissue. Endocrinology. 2009 Feb;150(2):651–661. doi: 10.1210/en.2008-1118. PubMed PMID: 18845643. Pubmed Central PMCID: 2646525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bhat M, Noolu B, Qadri SSYH, Ismail A. Vitamin D deficiency decreases adiposity in rats and causes altered expression of uncoupling proteins and steroid receptor coactivator3. J Steroid Biochem Mol Biol. 2014;144(PtB):304–312. doi: 10.1016/j.jsbmb.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 125.Gracia A, Elcoroaristizabal X, Fernández-Quintela A, Miranda J, Bediaga NG, de Pancorbo MM, et al. Fatty acid synthase methylation levels in adipose tissue: effects of an obesogenic diet and phenol compounds. Genes Nutr. 2014;9(4):411–419. doi: 10.1007/s12263-014-0411-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lee J, Saha PK, Yang Q-H, Lee S, Park JY, Suh Y, et al. Targeted inactivation of MLL3 histone H3-Lys-4 methyltransferase activity in the mouse reveals vital roles for MLL3 in adipogenesis. Proc Natl Acad Sci USA. 2008;105(49) doi: 10.1073/pnas.0810100105. 19229-34-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cho Y-W, Hong S, Jin Q, Wang L, Lee J-E, Gavrilova O, et al. Histone methylation regulator PTIP is required for PPARgamma and C/EBPalpha expression and adipogenesis. Cell Metab. 2009;10(1):27–39. doi: 10.1016/j.cmet.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hemming S, Cakouros D, Isenmann S, Cooper L, Menicanin D, Zannettino A, et al. EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells. 2014 Mar;32(3):802–815. doi: 10.1002/stem.1573. PubMed PMID: 24123378. [DOI] [PubMed] [Google Scholar]
- 130.Lefterova Ml, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22(21) doi: 10.1101/gad.1709008. 2941-52-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang Z-C, Liu Y, Li S-F, Guo L, Zhao Y, Qian S-W, et al. Suv39h1 mediates AP-2a–dependent inhibition of C/EBPa expression during adipogenesis. Mol Cell Biol. 2014;34(12) doi: 10.1128/MCB.00070-14. 2330-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Payne VA, Au W-S, Lowe CE, Rahman SM, Friedman JE, O’Rahilly S, et al. C/EBP transcription factors regulate SREBPIc gene expression during adipogenesis. Biochem J. 2009;425(1):215–223. doi: 10.1042/BJ20091112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Roder K, Wolf SS, Beck KF, Schweizer M. Cooperative binding of NF-Y and Sp1 at the DNase I- hypersensitive site, fatty acid synthase insulin-responsive element 1, located at −500 in the rat fatty acid synthase promoter. The Journal of biological chemistry. 1997 Aug 22;272(34):21616–21624. doi: 10.1074/jbc.272.34.21616. PubMed PMID: 9261184. [DOI] [PubMed] [Google Scholar]
- 134.Yuyama M, Fujimori K. Suppression of adipogenesis by valproic acid through repression of USF1- activated fatty acid synthesis in adipocytes. Biochem J. 2014;459(3):489–503. doi: 10.1042/BJ20131476. [DOI] [PubMed] [Google Scholar]
- 135.Latasa MJ, Griffin MJ, Moon YS, Kang C, Sul HS. Occupancy and function of the −150 sterol regulatory element and −65 E-box in nutritional regulation of the fatty acid synthase gene in living animals. Mol Cell Biol. 2003 Aug;23(16):5896–5907. doi: 10.1128/MCB.23.16.5896-5907.2003. PubMed PMID: 12897158. Pubmed Central PMCID: 166350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lomba A, García-Díaz DF, Paternain L, Marti A. Weight gain induced by an isocaloric pair-fed high fat diet: a nutriepigenetic study on FASN and NDUFB6 gene promoters. Mol Genet Metab. 2010;101(2–3):273–278. doi: 10.1016/j.ymgme.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 137.Kang T, Lu W, Xu W, Anderson L, Bacanamwo M, Thompson W, et al. MicroRNA-27 (miR-27) targets prohibitin and impairs adipocyte differentiation and mitochondrial function in human adipose-derived stem cells. The Journal of biological chemistry. 2013 Nov 29;288(48):34394–34402. doi: 10.1074/jbc.M113.514372. PubMed PMID: 24133204. Pubmed Central PMCID: 3843054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Karbiener M, Fischer C, Nowitsch S, Opriessnig P, Papak C, Ailhaud G, et al. microRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochemical and biophysical research communications. 2009 Dec 11;390(2):247–251. doi: 10.1016/j.bbrc.2009.09.098. PubMed PMID: 19800867. [DOI] [PubMed] [Google Scholar]
- 139.Liu D, Lin Y, Kang T, Huang B, Xu W, Garcia-Barrio M, et al. Mitochondrial dysfunction and adipogenic reduction by prohibitin silencing in 3T3-L1 cells. PLoS ONE. 2012;7(3):e34315. doi: 10.1371/journal.pone.0034315. PubMed PMID: 22479600. Pubmed Central PMCID: 3316679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kim SY, Kim AY, Lee HW, Son YH, Lee GY, Lee JW, et al. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARgamma expression. Biochemical and biophysical research communications. 2010 Feb 12;392(3):323–328. doi: 10.1016/j.bbrc.2010.01.012. PubMed PMID: 20060380. [DOI] [PubMed] [Google Scholar]
- 141.Matsubara K, Aoyama K, Suno M, Awaya T. N-methylation underlying Parkinson’s disease. Neurotoxicol Teratol. 2002;24(5):593–598. doi: 10.1016/s0892-0362(02)00212-x. [DOI] [PubMed] [Google Scholar]
- 142.Riederer M, Erwa W, Zimmermann R, Frank S, Zechner R. Adipose tissue as a source of nicotinamide N- methyltransferase and homocysteine. Atherosclerosis. 2009 Jun;204(2):412–417. doi: 10.1016/j.atherosclerosis.2008.09.015. PubMed PMID: 18996527. [DOI] [PubMed] [Google Scholar]
- 143.Ulanovskaya OA, Zuhl AM, Cravatt BF. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation +sink. Nat Chem Biol. 2013;9(5):300–306. doi: 10.1038/nchembio.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bi HC, Pan YZ, Qiu JX, Krausz KW, Li F, Johnson CH, et al. N-methylnicotinamide and nicotinamide N- methyltransferase are associated with microRNA-1291-altered pancreatic carcinoma cell metabolome and suppressed tumorigenesis. Carcinogenesis. 2014 Oct;35(10):2264–2272. doi: 10.1093/carcin/bgu174. PubMed PMID: 25115443. Pubmed Central PMCID: 4178474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Watala C, Kazmierczak P, Dobaczewski M, Przygodzki T, Bartus M, Lomnicka M, et al. Anti-diabetic effects of 1-methylnicotinamide (MNA) in streptozocin-induced diabetes in rats. Pharmacol Rep. 2009 Jan-Feb;61(1):86–98. doi: 10.1016/s1734-1140(09)70010-6. PubMed PMID: 19307696. [DOI] [PubMed] [Google Scholar]
- 146.Kannt A, Pfenninger A, Teichert L, Tonjes A, Dietrich A, Schon MR, et al. Association of nicotinamide-N- methyltransferase mRNA expression in human adipose tissue and the plasma concentration of its product, 1- methylnicotinamide, with insulin resistance. Diabetologia. 2015 Apr;58(4):799–808. doi: 10.1007/s00125-014-3490-7. PubMed PMID: 25596852. Pubmed Central PMCID: 4351435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kraus D, Yang Q, Kong D, Banks AS, Zhang L, Rodgers JT, et al. Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature. 2014;508(7495):258–262. doi: 10.1038/nature13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Perez-Leal O, Merali S. Regulation of polyamine metabolism by translational control. Amino Acids. 2011;42(2–3) doi: 10.1007/s00726-011-1036-6. 611-7-7. [DOI] [PubMed] [Google Scholar]
- 149.Zafrir B, Jain M. Lipid-lowering therapies, glucose control and incident diabetes: evidence, mechanism and clinical implications. Cardiovasc Drugs Ther. 2014;28(4) doi: 10.1007/s10557-014-6534-9. 361-77-77. [DOI] [PubMed] [Google Scholar]
- 150.Blond E, Rieusset J, Alligier M, Lambert-Porcheron S, Bendridi N, Gabert L, et al. Nicotinic acid effects on insulin sensitivity and hepatic lipid metabolism: an in vivo to in vitro study. Horm Metab Res. 2014;46(6) doi: 10.1055/s-0034-1372600. 390-6-6. [DOI] [PubMed] [Google Scholar]
- 151.Romano ME, Webster GM, Vuong AM, Thomas Zoeller R, Chen A, Hoofnagle AN, et al. Gestational urinary bisphenol A and maternal and newborn thyroid hormone concentrations: The HOME Study. Environmental research. 2015 Mar 17;138:453–460. doi: 10.1016/j.envres.2015.03.003. PubMed PMID: 25794847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.McGuinn LA, Ghazarian AA, Joseph Su L, Ellison GL. Urinary bisphenol A and age at menarche among adolescent girls: evidence from NHANES 2003–2010. Environmental research. 2015 Jan;136:381–386. doi: 10.1016/j.envres.2014.10.037. PubMed PMID: 25460659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proceedings of the National Academy of Sciences of the United States of America. 2007 Aug 7;104(32):13056–13061. doi: 10.1073/pnas.0703739104. PubMed PMID: 17670942. Pubmed Central PMCID: 1941790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Medici V, Schroeder Dl, Woods R, LaSalle JM, Geng Y, Shibata NM, et al. Methylation and gene expression responses to ethanol feeding and betaine supplementation in the cystathionine beta synthase- deficient mouse. Alcohol Clin Exp Res. 2014;38(6) doi: 10.1111/acer.12405. 1540-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Cordero P, Milagro Fl, Campíon J, Martínez JA. Maternal methyl donors supplementation during lactation prevents the hyperhomocysteinemia induced by a high-fat-sucrose intake by dams. Int J Mol Sci. 2013;14(12) doi: 10.3390/ijms141224422. 24422-37-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Carlin J, George R, Reyes TM. Methyl donor supplementation blocks the adverse effects of maternal high fat diet on offspring physiology. PLoS ONE. 2013;8(5):e63549. doi: 10.1371/journal.pone.0063549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bekdash RA, Zhang C, Sarkar DK. Gestational choline supplementation normalized fetal alcohol-induced alterations in histone modifications, DNA methylation, and proopiomelanocortin (POMC) gene expression in β- endorphin-producing POMC neurons of the hypothalamus. Alcohol Clin Exp Res. 2013;37(7) doi: 10.1111/acer.12082. 1133-42-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bhandari R, Xiao J, Shankar A. Urinary bisphenol A and obesity in U.S children. American journal of epidemiology. 2013 Jun 1;177(11):1263–1270. doi: 10.1093/aje/kws391. PubMed PMID: 23558351. Pubmed Central PMCID: 3664337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Song Y, Hauser R, Hu FB, Franke AA, Liu S, Sun Q. Urinary concentrations of bisphenol A and phthalate metabolites and weight change: a prospective investigation in US women. Int J Obes (Lond) 2014;38(12):1532–1537. doi: 10.1038/ijo.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sales LB, Kamstra JH, Cenijn PH, van Rijt LS, Hamers T, Legler J. Effects of endocrine disrupting chemicals on in vitro global DNA methylation and adipocyte differentiation. Toxicol In Vitro. 2013;27(6):1634–1643. doi: 10.1016/j.tiv.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 161.Mackay H, Patterson ZR, Khazall R, Patel S, Tsirlin D, Abizaid A. Organizational effects of perinatal exposure to bisphenol-A and diethylstilbestrol on arcuate nucleus circuitry controlling food intake and energy expenditure in male and female CD-1 mice. Endocrinology. 2013;154(4):1465–1475. doi: 10.1210/en.2012-2044. [DOI] [PubMed] [Google Scholar]
- 162.Doshi T, Mehta SS, Dighe V, Balasinor N, Vanage G. Hypermethylation of estrogen receptor promoter region in adult testis of rats exposed neonatally to bisphenol A. Toxicology. 2011 Nov 18;289(2–3):74–82. doi: 10.1016/j.tox.2011.07.011. PubMed PMID: 21827818. [DOI] [PubMed] [Google Scholar]
- 163.Susiarjo M, Sasson I, Mesaros C, Bartolomei MS. Bisphenol a exposure disrupts genomic imprinting in the mouse. PLoS genetics. 2013 Apr;9(4):e1003401. doi: 10.1371/journal.pgen.1003401. PubMed PMID: 23593014. Pubmed Central PMCID: 3616904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Susiarjo M, Xin F, Bansal A, Stefaniak M, Li C, Simmons RA, et al. Bisphenol a exposure disrupts metabolic health across multiple generations in the mouse. Endocrinology. 2015 Jun;156(6):2049–2058. doi: 10.1210/en.2014-2027. PubMed PMID: 25807043. Pubmed Central PMCID: 4430620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Thorvaldsen JL, Mann MR, Nwoko O, Duran KL, Bartolomei MS. Analysis of sequence upstream of the endogenous H19 gene reveals elements both essential and dispensable for imprinting. Mol Cell Biol. 2002 Apr;22(8):2450–2462. doi: 10.1128/MCB.22.8.2450-2462.2002. PubMed PMID: 11909940. Pubmed Central PMCID: 133727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Werts RL, Van Calcar SC, Wargowski DS, Smith SM. Inappropriate feeding behaviors and dietary intakes in children with fetal alcohol spectrum disorder or probable prenatal alcohol exposure. Alcoholism, clinical and experimental research. 2014 Mar;38(3):871–878. doi: 10.1111/acer.12284. PubMed PMID: 24164456. Pubmed Central PMCID: 3959629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Fuglestad AJ, Boys CJ, Chang PN, Miller BS, Eckerle JK, Deling L, et al. Overweight and obesity among children and adolescents with fetal alcohol spectrum disorders. Alcoholism, clinical and experimental research. 2014 Sep;38(9):2502–2508. doi: 10.1111/acer.12516. PubMed PMID: 25159809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Govorko D, Bekdash RA, Zhang C, Sarkar DK. Male germline transmits fetal alcohol adverse effect on hypothalamic proopiomelanocortin gene across generations. Biological psychiatry. 2012 Sep 1;72(5):378–388. doi: 10.1016/j.biopsych.2012.04.006. PubMed PMID: 22622000. Pubmed Central PMCID: 3414692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wu Y, Patchev AV, Daniel G, Almeida OFX, Spengler D. Early-life stress reduces DNA methylation of the Pomc gene in male mice. Endocrinology. 2014;155(5):1751–1762. doi: 10.1210/en.2013-1868. [DOI] [PubMed] [Google Scholar]
- 170.Gangisetty O, Bekdash R, Maglakelidze G, Sarkar DK. Fetal alcohol exposure alters proopiomelanocortin gene expression and hypothalamic-pituitary-adrenal axis function via increasing MeCP2 expression in the hypothalamus. PLoS ONE. 2014;9(11):e113228. doi: 10.1371/journal.pone.0113228. PubMed PMID: 25409090. Pubmed Central PMCID:4237387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ricceri L, De Filippis B, Fuso A, Laviola G. Cholinergic hypofunction in MeCP2-308 mice: beneficial neurobehavioural effects of neonatal choline supplementation. Behav Brain Res. 2011;221(2):623–629. doi: 10.1016/j.bbr.2011.03.051. [DOI] [PubMed] [Google Scholar]
- 172.Ward BC, Kolodny NH, Nag N, Berger-Sweeney JE. Neurochemical changes in a mouse model of Rett syndrome: changes over time and in response to perinatal choline nutritional supplementation. J Neurochem. 2008;108(2) doi: 10.1111/j.1471-4159.2008.05768.x. 361-71-71. [DOI] [PubMed] [Google Scholar]
- 173.Nag N, Berger-Sweeney JE. Postnatal dietary choline supplementation alters behavior in a mouse model of Rett syndrome. Neurobiol Dis. 2007;26(2):473–480. doi: 10.1016/j.nbd.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 174.Kim S, Park J, Kim HJ, Lee JJ, Choi G, Choi S, et al. Association between several persistent organic pollutants and thyroid hormone levels in serum among the pregnant women of Korea. Environment international. 2013 Sep;59:442–448. doi: 10.1016/j.envint.2013.07.009. PubMed PMID: 23928038. [DOI] [PubMed] [Google Scholar]
- 175.Gonzalez-Jauregui M, Valdespino C, Salame-Mendez A, Aguirre-Leon G, Rendon-Vonosten J. Persistent organic contaminants and steroid hormones levels in Morelet’s crocodiles from the Southern Gulf of Mexico. Archives of environmental contamination and toxicology. 2012 Apr;62(3):445–454. doi: 10.1007/s00244-011-9716-5. PubMed PMID: 22002785. [DOI] [PubMed] [Google Scholar]
- 176.Gauthier M-S, Rabasa-Lhoret R, Prud’homme D, Karelis AD, Geng D, van Bavel B, et al. The metabolically healthy but obese phenotype is associated with lower plasma levels of persistent organic pollutants as compared to the metabolically abnormal obese phenotype. J Clin Endocrinol Metab. 2014;99(6):E1061–E1066. doi: 10.1210/jc.2013-3935. [DOI] [PubMed] [Google Scholar]
- 177.Biemann R, Fischer B, Blüher M, Santos AN. Tributyltin affects adipogenic cell fate commitment in mesenchymal stem cells by a PPARv independent mechanism. Chem Biol Interact. 2014;214:1–9. doi: 10.1016/j.cbi.2014.01.021. [DOI] [PubMed] [Google Scholar]
- 178.Garcia-Arevalo M, Alonso-Magdalena P, Rebelo Dos Santos J, Quesada I, Carneiro EM, Nadal A. Exposure to bisphenol-A during pregnancy partially mimics the effects of a high-fat diet altering glucose homeostasis and gene expression in adult male mice. PLoS ONE. 2014;9(6):e100214. doi: 10.1371/journal.pone.0100214. PubMed PMID: 24959901. Pubmed Central PMCID: 4069068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001 Aug 10;293(5532):1089–1093. doi: 10.1126/science.1063443. PubMed PMID: 11498579. [DOI] [PubMed] [Google Scholar]
- 180.Ibrahim MM, Fjaere E, Lock EJ, Froyland L, Jessen N, Lund S, et al. Metabolic impacts of high dietary exposure to persistent organic pollutants in mice. Toxicol Lett. 2012 Nov 23;215(1):8–15. doi: 10.1016/j.toxlet.2012.09.022. PubMed PMID: 23041606. [DOI] [PubMed] [Google Scholar]
- 181.Gardebjer EM, Anderson ST, Pantaleon M, Wlodek ME, Moritz KM. Maternal alcohol intake around the time of conception causes glucose intolerance and insulin insensitivity in rat offspring, which is exacerbated by a postnatal high-fat diet. FASEB J. 2015 Mar 2; doi: 10.1096/fj.14-268979. PubMed PMID: 25733565. [DOI] [PubMed] [Google Scholar]
- 182.Baker NA, Karounos M, English V, Fang J, Wei Y, Stromberg A, et al. Coplanar polychlorinated biphenyls impair glucose homeostasis in lean C57BL/6 mice and mitigate beneficial effects of weight loss on glucose homeostasis in obese mice. Environ Health Perspect. 2013 Jan;121(1):105–110. doi: 10.1289/ehp.1205421. PubMed PMID: 23099484. Pubmed Central PMCID: 3553436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Baker NA, English V, Sunkara M, Morris AJ, Pearson KJ, Cassis LA. Resveratrol protects against polychlorinated biphenyl-mediated impairment of glucose homeostasis in adipocytes. The Journal of nutritional biochemistry. 2013 Dec;24(12):2168–2174. doi: 10.1016/j.jnutbio.2013.08.009. PubMed PMID: 24231106. Pubmed Central PMCID: 4066417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Akbari Moqadam F, Pieters R, den Boer ML. The hunting of targets: challenge in miRNA research. Leukemia. 2013 Jan;27(1):16–23. doi: 10.1038/leu.2012.179. PubMed PMID: 22836911. [DOI] [PubMed] [Google Scholar]
- 185.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005 May;37(5):495–500. doi: 10.1038/ng1536. PubMed PMID: 15806104. [DOI] [PubMed] [Google Scholar]
- 186.Wildman RP, Muntner P, Reynolds K, McGinn AP, Rajpathak S, Wylie-Rosett J, et al. The obese without cardiometabolic risk factor clustering and the normal weight with cardiometabolic risk factor clustering: prevalence and correlates of 2 phenotypes among the US population (NHANES 1999–2004) Arch Intern Med. 2008;168(15) doi: 10.1001/archinte.168.15.1617. 1617-24-24. [DOI] [PubMed] [Google Scholar]
- 187.Fabbrini E, Yoshino J, Yoshino M, Magkos F, Tiemann Luecking C, Samovski D, et al. Metabolically normal obese people are protected from adverse effects following weight gain. J Clin Invest. 2015 Feb;125(2):787–795. doi: 10.1172/JCI78425. PubMed PMID: 25555214. [DOI] [PMC free article] [PubMed] [Google Scholar]