Abstract
Endogenous labeling with stable isotopes is used to study the metabolism of proteins in vivo. However, traditional detection methods such as GC/MS cannot measure tracer enrichment in multiple proteins simultaneously, and multiple reaction monitoring MS cannot measure precisely the low tracer enrichment in slowly turning-over proteins as in HDL. We exploited the versatility of the high-resolution/accurate mass (HR/AM) quadrupole Orbitrap for proteomic analysis of five HDL sizes. We identified 58 proteins in HDL that were shared among three humans and that were organized into five subproteomes according to HDL size. For seven of these proteins, apoA-I, apoA-II, apoA-IV, apoC-III, apoD, apoE, and apoM, we performed parallel reaction monitoring (PRM) to measure trideuterated leucine tracer enrichment between 0.03 to 1.0% in vivo, as required to study their metabolism. The results were suitable for multicompartmental modeling in all except apoD. These apolipoproteins in each HDL size mainly originated directly from the source compartment, presumably the liver and intestine. Flux of apolipoproteins from smaller to larger HDL or the reverse contributed only slightly to apolipoprotein metabolism. These novel findings on HDL apolipoprotein metabolism demonstrate the analytical breadth and scope of the HR/AM-PRM technology to perform metabolic research.
Keywords: coronary heart disease, size fractionation, mass spectrometry, compartmental modeling, metabolism
Stable isotope amino acid tracers are commonly used to study the kinetics of proteins in circulation (1). GC/MS usually determines isotope enrichment by measuring the derivatized forms of D0 and trideuterated leucine (D3-Leu) (2, 3), a method with high cost and low sensitivity and specificity. Recently, proteomics-based triple quadrupole multiple reaction monitoring (MRM) permitted a more practical and highly specific multipeptide approach to in vivo kinetic studies (4, 5). However, MRM relies on low-resolution readouts (unit mass resolution) that do not readily permit precise quantification of tracer enrichment that is lower than 1%, which is common in apolipoprotein kinetics (5, 6). Factors contributing to low precision include interference by not only the sister isotope 13C15N M3 ion but also background ions. In this study, we aim to extend further the scope of in vivo kinetics by exploiting the recently developed high-resolution/accurate mass parallel reaction monitoring (HR/AM-PRM) method performed on the quadrupole Orbitrap mass spectrometer (7, 8). The HR/AM fragment ion scan feature has the potential to measure D3-Leu enrichment between 0.03% and 1.0%, a low incorporation range that is a consequence of a bolus-administered tracer, useful in revealing tracer-tracee relationships.
Epidemiologic studies have consistently shown that low HDL, either measured by the levels of its cholesterol content (HDL-C) or its primary protein (apoA-I), is an independent risk factor for coronary heart disease (CHD) (9, 10). However, genetic variations associated with differences in HDL-C but not in LDL cholesterol (LDL-C) and TGs are not associated with CHD risk (11). Treatments that raise HDL-C or apoA-I have so far failed to show beneficial effects in CHD prevention (12–15). HDL is a complex population of lipoproteins with a diverse content of proteins and lipids. Targeting HDL simply by increasing its cholesterol and/or apoA-I concentration alone may not suffice to reduce risk (16).
Apolipoprotein metabolism has been extensively studied in VLDL and LDL (17–19), but to a lesser extent in HDL. HDL apolipoprotein metabolism is commonly studied by labeling apoA-I using radioisotopes or by stable isotope amino acids such as D3-Leu. The kinetic parameters, fractional catabolic rate (FCR) and production rate (PR), are typically determined for HDL using total plasma apoA-I, whereas only a handful of studies have looked into apoA-I HDL metabolism by size: large HDL2 and small HDL3 (20, 21); prebeta and α HDL (22); and very recently, prebeta, α3, α2, and α1 HDL (3). In this latter study, Mendivil et al. (3), who utilized GC/MS for tracer enrichment analysis, demonstrated that the canonical HDL size-expansion model (prebeta is converted to α3, then to α2 and then to α1 HDL), which is posited to play a role in reverse cholesterol transport (23), could not explain their apoA-I tracer enrichment data. In fact, the final best-fit model indicated that the majority of apoA-I on α HDL originated from the source compartment, presumably the liver mainly and intestine secondarily; size expansion pathways only provided a minor contribution to apoA-I secretion on α HDL (3). Besides apoA-I, the kinetics of other HDL apolipoproteins, such as apoA-II, apoE, apoC-III, and apoA-IV remain unexplored in the context of HDL size (24–26). The kinetics of others, such as apoM and apoD, have yet to be studied.
We present a comprehensive workflow for a mass spectrometric-dependent characterization of the HDL proteome. We investigated the distribution of HDL proteins in humans across five HDL sizes by both spectral counting and stable isotope dilution quantification methods. Also, we present the first ever report of HR/AM-PRM to measure D3-Leu tracer enrichment in the range of 0.03% to 1.0%, which is necessary to determine in vivo the kinetic parameters of HDL apolipoproteins, and applied the method to seven apolipoproteins across five HDL size fractions. The enrichment curves and plasma pool sizes were then used for multicompartmental modeling to identify the source, conversion, and removal pathways for each apolipoprotein across the HDL size fractions. These analyses corroborate the recent findings by Mendivil et al. that like apoA-I, the other apolipoproteins appear on HDL primarily from their respective source compartments.
The kinetics of the proteins that HDL contains may lead to an improved understanding of HDL function in humans and eventually the development of more reliable clinical markers of CHD risk and HDL-based targets for treatment.
MATERIALS AND METHODS
Subject screening and enrollment
We studied 3 participants, two female (250 and 225) and one male (243), who were ages 25, 33, and 49 years old; were overweight or obese with a BMI of 26, 31, and 31 kg/m2; and had low HDL-C levels of 48, 37, and 24 mg/dl, respectively (supplementary Table 1). Exclusion criteria included high LDL-C (>190 mg/dl); low HDL-C (<20 mg/dl); very high fasting TG (>500 mg/dl); apoE genotypes E2E2, E2E4, or E4E4; use of medications or therapies that can alter lipid levels (lipid-lowering medications, beta blockers, some psychiatric medicines, and hormone replacement); secondary hyperlipidemia (such as untreated hypothyroidism); any hepatic or renal complications (as identified by chemistry panel); diabetes mellitus; pregnancy; or individuals who refuse to eat the study diet and abstain from alcohol consumption.
Subjects who expressed interest in the study were initially screened for eligibility by phone call followed by a visit to the Center for Clinical Investigation (CCI) at Brigham and Women’s Hospital. During the CCI visit, a diet questionnaire and a blood sample were collected to assess eligibility. All subjects who were deemed eligible and wanted to participate in the study gave written informed consent. This study was approved by the Institutional Review Board of Brigham and Women’s Hospital and Harvard School of Public Health.
Dietary protocol
The three participants consumed a high-unsaturated-fat diet consisting of 37% calories from fat (8% saturated, 24% monounsaturated, and 5% polyunsaturated). The diet adhered to the Institute of Medicine Dietary Reference Intake guidelines for healthy nutrient intake (http://ods.od.nih.gov/Health_Information/Dietary_Reference_Intake.aspx) and was formulated by Brigham and Women’s Hospital CCI nutrition research unit. All food and beverages were provided for the duration of the study. Alcoholic beverages were not part of the study diet, and intake was not permitted. Participants visited the CCI every Monday, Wednesday, and Friday where they picked up food, completed a food diary, and had their body weight measured. Calories were adjusted to compensate for any complaints of hunger or satiety or changes in body weight.
Tracer infusion protocol
The three participants ate a controlled, high-unsaturated-fat diet for 32 days, 28 days prior to the kinetic study, and 4 days during the kinetic study. On the morning of day 28, participants were admitted to Brigham and Women’s Hospital CCI where they received an intravenous bolus injection of the stable isotope tracer D3-Leu at a concentration of 10 mg/kg over 10 min. Blood was sampled immediately before the bolus injection (time 0 h), and at 0.5, 1, 1.5, 2, 3, 4, 6, 8, 10, 12, 14, 16, 18, 22, 46, 70, and 94 h postinfusion. After the 22 h sample was collected, participants were discharged. Over the next 3 days, the 46, 70, and 94 h postinfusion blood samples were collected at the ambulatory CCI.
Total plasma leucine enrichment quantification
Total plasma leucine (D3-Leu labeled and endogenous) was isolated from 0.2 ml of plasma from time points 0, 1, 2, 3, 4, 6, 8, 10, 12, 14, 18, 22, 46, and 70 h postinfusion using an AG 50W-X8 cation exchange resin (Bio-Rad). The isolated amino acids were then dried under nitrogen, derivatized to heptafluorobutiric acid esters, and measured using GC/MS (Agilent 6890 GC, 5973 MS). The total plasma tracer (D3-Leu) enrichment was quantified by taking the area under the curve of the tracer divided by the area under the curve of total plasma leucine (D3-Leu tracer + Leu tracee).
HDL preparation and native gel electrophoresis
Immediately after each time point collection, plasma was isolated from blood by refrigerated centrifugation and then aliquotted and stored at −80°C. HDL was isolated from time points 0, 0.5, 1, 1.5, 2, 4, 6, 8, 10, 12, 14, 16, 18, 22, and 70 h for participant 243, and 0, 2, 4, 6, 12, 18, 22, and 70 h for participants 225 and 250. HDL was purified from 1 ml of plasma by overnight incubation with anti-apoA-I immunoglobulin (Academy Biomedical, Houston, TX) bound to sepharose 4B resin (Academy Biomedical). The unbound non-apoA-I-containing fraction was collected by gravity flow, and the bound apoA-I-containing fraction was eluted using 3 M NaSCN (Sigma-Aldrich, St. Louis, MO). To evaluate whether any apoB lipoproteins that contained apoA-I were affecting the HDL measurements, we removed apoB lipoproteins by dextran sulfate/magnesium chloride precipitation from participant 243 time points 0, 2, 4, 6, 12, 18, 22, and 70 h (27). HDL purification was then carried out as described above. HDL was separated by size using nondenaturing polyacrylamide gel electrophoresis (ND-PAGE) on a 4–30% gradient gel (Jule INC) run at 15 mA for 16 h. A molecular weight standard from the GE/Amersham calibration kit (catalog no. 17-0445-01) was run alongside the samples. After completion of the run, the gel was stained for 1–2 h in Coomassie Brilliant Blue (Invitrogen, Grand Island, NY) and destained in ddH2O until the gel background was mostly clear. Using the molecular weight standard as a guide, portions of the gel corresponding to each HDL size were excised: above 12.2 nm, α0; between 12.2 nm and 9.5 nm, α1; between 9.5 nm and 8.2 nm, α2; between 8.2 and 7.2 nm, α3; and band at 7.1 nm, prebeta (28) (supplementary Fig. 1A). Because the uppermost gel fraction has not been previously characterized, we have referred to this fraction as α0, adhering to the α and prebeta size fraction nomenclature. However, previous two-dimensional electrophoresis studies have indicated this slowest migrating apoA-I fraction may comprise a second, larger-sized prebeta subpopulation (prebeta-2) as well as very large α HDL (28, 29).
Human apoA-I protein/FLEXIQuant standard
A plasmid containing human apoA-I (clone HsCD00000689) was purchased from the Dana Farber/Harvard Cancer Center DNA Resource Core. The gene was subcloned into the FLEXIQuant vector containing the N-terminal 6XHistine tag (30) using the KpnI and NotI cloning sites. The KpnI-based primer, GGGGTACCATGAAAGCTGCGGTGCTG, and the NotI-based primer, GGGCGGCCGCTCACTGGGTGTTGAGCTTCTTAG, were purchased from Integrated DNA Technologies. The underlined nucleotides correspond to apoA-I sequences. ApoA-I in vitro transcription and translation was done using the CellFree Sciences Kit [WEPRO2240H; Cambridge Isotope Laboratories (CIL)]. L-Arg-13C8-15N2 (catalog no. CNLM-539-H-PK), L-Lys-13C6-15N2 (catalog no. CNLM-291-H-PK), and L-Leu-(5-5-5-2H3/D3-Leu) (catalog no. DLM-1259-1) were purchased from CIL. All 20 natural amino acids were purchased from Sigma. The final labeled FLEXIQuant standard is referred to herein as FLEX-apoA-I. The protein was purified by nickel sepharose (GE Healthcare) based affinity for the N-terminal 6XHIS tag (4). The 13C15N Arg/Lys-labeled FLEX-apoA-I used for absolute quantification studies was eluted from nickel sepharose and kept in solution; a small aliquot was used for ELISA assays and the rest for in-solution trypsin digestion. The 13C15N Arg/Lys-labeled FLEX-apoA-I was quantified using the in vitro synthesized reference light FLEX peptide (TENLYFQGDISR; New England Peptide) during MS analysis (and by ELISA). Light and D3-Leu-labeled apoA-I mixtures used for PRM standard curves were SDS-PAGE analyzed for in-gel trypsin digestion. The alkylation step for both in-solution and in-gel (FLEX-apoA-I standard and HDL proteins) trypsin reactions was omitted to increase throughput of samples. Trypsin reactions were done for 4 h (4). Peptide samples were resuspended in 5% acetonitrile, 0.5% formic acid prepared in HPLC-grade water.
Relative and absolute quantification of apolipoproteins
Spectral counting was used for the relative quantification of the apolipoproteins: a given protein’s peptide spectrum matches (PSMs, derived from the sum of three time points, i.e., 0, 4, and 12 h) for each HDL size fraction were normalized to the total PSMs for all size fractions for that protein (sum normalization) per participant (supplementary Fig. 1B, C). The PSMs were normalized this way in lieu of normalization to the entire HDL proteome because with the latter approach, apoA-I dominated the output, compressing the signals of the remaining apolipoproteins. In addition to the FLEX-apoA-I standard above, in vitro synthesized peptides for absolute quantification (AQUA peptides; supplementary Fig. 2A) were used to quantify six additional apolipoproteins, including apoA-II (SPELQAEA[K-label]), apoA-IV (TQVNTQAEQL[R-label]), apoC-III (DALSSVQESQVAQQA[R-label]), apoD (VLNQEL[R-label]), apoE (LGPLVEQG[R-label]), and apoM (FLLYN[R-label]), purchased from New England Peptides (NEP) and quantified by the absolute amino acid method (NEP), including the light FLEX peptide above. The peptide standards were chosen based on the following criteria: 1) fully cleaved, 2) devoid of methionines and cysteines, and 3) not reported to be posttranslationally modified (reference: Uniprot.org). We also ensured that our standard peptides were on the higher end of ionization signal intensity [as judged by the area under the curve of the extracted ion chromatogram (XIC)], when compared with other peptides from the same protein, which decreases the likelihood that the peptides are posttranslationally modified (30, 31). Arginines were labeled with 13C8,15N2; lysines, with 13C6,15N2. We established the appropriate spike-in amount for the peptides by determining the linear range of ionization (area under the curve of the M0 ion) for both the standard and sample-derived peptides (supplementary Fig. 2B, C). Quantification of the labeled apoA-I standard mixture was calculated by the unlabeled FLEX peptide tag (30, 32). The optimal peptide spiking mixture comprised FLEX-apoA-I tryptic peptides and the six AQUA peptides at a final on-column amount of 10 fmol FLEX peptide and 1 fmol each of the other apolipopeptides. The HDL sample peptides were diluted 1 in 40 (supplementary Fig. 2B, C).
LC-MS/MS
Peptide samples were analyzed with the Q Exactive mass spectrometer fronted with a Nanospray FLEX ion source, and coupled to an Easy-nLC1000 HPLC pump (Thermo Scientific, Bremen, Germany). The peptides were subjected to a dual-column setup: an Acclaim PepMap RSLC C18 trap column (75 μm × 20 mm) and an Acclaim PepMap RSLC C18 analytical column (75 μm × 150 mm; Thermo Scientific). The analytical gradient was run at 250 nl/min from 5% to 18% solvent B (acetonitrile/0.1% formic acid) for 10 min or 30 min, followed by 5 min of 95% solvent B. Solvent A was 0.1% formic acid. All reagents were HPLC grade. For standard data-dependent MS/MS, the instrument was set to 140 K resolution, and the top 10 precursor ions (within a scan range of m/z 380–1,500) were subjected to higher energy collision-induced dissociation (HCD), collision energy 25% (stepped CE ± 10%), isolation width m/z 3, and 17.5 K resolution. For HR/AM-PRM acquisitions, the DIA module was used by including an MS1 isolation list comprising a 2 min to 3 min retention time window for a given peptide. The peptide retention time overlaps were minimized in order to dedicate high-resolution acquisitions on a single peptide per scan (supplementary Fig. 3 and supplementary Table 2). Each peptide MS1 isolation window was centered on the average m/z of M0 and M3 peaks (or M6 for 2 leucines), ± m/z 5. The maximum ion cutoff was set to 5e6. The HCD collision energy was set to 25 ± 10%. The MS data were queried against the Human UniProt database (downloaded August 1, 2014) using the HT-SEQUEST search algorithm (33), via the Proteome Discoverer (PD) Package (version 1.4; Thermo Scientific), using a 10 ppm tolerance window in the MS1 search space and a 0.02 Da fragment tolerance window for HCD data. Methionine oxidation was set as a variable modification. Recombinant APOA-I (FLEX-apoA-I) was searched against a custom database, which comprised the Saccharomyces cerevisiae UniProt database (downloaded March 27, 2012) to which the FLEX-APOA-I sequence was added. The peptide false discovery rate was calculated using Percolator provided by PD.
Peptide enrichment quantification
Skyline (https://skyline.gs.washington.edu) was used as a visualization tool for analyzing MS/MS data and for candidate fragment ion filtering, as well as for MS1-based quantification. Manual quantification of the M0 and 2H M3 fragment peaks was done by calculating the XICs of the area under the peaks using the Xcalibur software (Thermo Scientific). If the peak valley was >10% of the peak height (i.e., for ions approximately greater than m/z 500), the extraction was taken from the valley point. The XIC values were recorded and plotted in Excel (Microsoft).
Calculation of apolipoprotein pool sizes
The pool size (total milligrams of protein in plasma) for apoA-I, apoA-II, apoA-IV, apoC-III, apoD, apoE, and apoM in the α0, α1, α2, α3, and prebeta HDL size fractions was determined by first converting the femtomoles on-column of each protein per size fraction to milligrams of protein per 1 ml of plasma. To determine the amount of sample loss during preparation, the mg/ml of apoA-I per size fraction were summed to get an estimated total apoA-I concentration. This estimated total apoA-I concentration was then compared with the total plasma apoA-I concentration, as determined by ELISA using anti-apoA-I antibodies (Academy Biomedical). The sample loss “correction factor” was calculated by dividing the ELISA total apoA-I concentration by the estimated total apoA-I concentration and was determined to be 119 (±48). Assuming that sample loss was similar for all size fractions, the milligrams per milliliter estimated protein concentrations for each size fraction were then multiplied by the correction factor to determine the milligrams per milliliter concentration of each protein in each HDL size fraction. The milligrams per milliliter protein concentrations per size were then multiplied by the total plasma volume to determine the protein pool size per HDL size fraction. Plasma volume for each subject was calculated by the following formula: plasma volume (dl) = ideal body weight (kg) × 0.44 + excess weight (kg) × 0.1 (34).
Multicompartmental modeling
Kinetic modeling was performed using SAAM II modeling software (SAAM Institute, Seattle, WA). A separate model was generated for apoA-I, apoA-II, apoE, apoM, apoC-III, and apoA-IV and describes the kinetic behaviors of each apolipoprotein on each size fraction. ApoD was excluded from model development due to the small number of data points that showed tracer enrichment. The apoA-I model used in this study was based on the model recently developed by Mendivil et al. (3).
Each model contains an input, a source, and plasma HDL compartments. The input compartment is the plasma amino acid precursor pool (D3-Leu tracer enrichment in plasma) expressed as a forcing function that drives the appearance of plasma D3-Leu tracer in the model. Each participant’s D3-Leu tracer enrichment curve was used for all six of the apolipoprotein models. The source compartment accounts for the time necessary for labeled protein to appear on the HDL size fractions in plasma. The appearance of labeled apoA-I, apoA-II, and apoE on plasma HDL was rapid and occurred by 30 min. In contrast, label first appeared in apoM, apoC-III, and apoA-IV after 30 min; therefore, a delay compartment was used as the source compartment. Two to five additional compartments were added to each model that represented the HDL size fractions. The protein pool size and enrichment, [tracer (2H M3) / tracer (2H M3) + tracee (M0)], were the data assigned to each compartment. The FCR, the fraction of a plasma protein pool turned over per day, was determined for each protein in each HDL size by taking the sum of the rate constants exiting that compartment. PR, the amount (mg) of protein produced or transferred to HDL per day per kg of body weight, was calculated using the following formula:
For all the proteins, a direct secretion pathway into each size fraction was required for satisfactory fitting. Multiple pathways among the sizes were tested, but only those pathways that had a nonzero flux and improved model fits for each participant were included in the final model. All size fractions with discernible enrichment curves for at least two participants were included in the final model for each apolipoprotein: apoA-I, α0, α1, α2, α3, prebeta; apoA-II, α1, α2, α3; apoE, α0, α1, α2, α3; apoC-III, α1, α2, α3; and apoA-IV, α3, prebeta. Only the α1 and α2 HDL sizes were included in the apoM model; α0 and α3 were excluded due to their small pool size (<4% total apoM on HDL in plasma) and variable enrichment curves across participants. Besides the apoA-I HDL size pools, two additional compartments were included in the apoA-I model: 1) a delay compartment connecting the source and prebeta pools. This may represent an extravascular delay (EVD) processing compartment that includes apoA-I prebeta that has been secreted but is outside the systemic circulation (3). 2) A compartment connecting the α3 and prebeta pools. This compartment may represent lipidated apoA-I (LA1) that has been released from α3 and is then used to generate prebeta (3). Steady-state kinetics were assumed for all apolipoproteins.
Data representation
Heat maps and hierarchical clustering were executed using the Qlucore software package (www.qlucore.com). All Excel plots and figures were compiled in PowerPoint (Microsoft) and Photoshop (Adobe). Results are presented as median or mean (SD) unless otherwise specified.
RESULTS
Study workflow
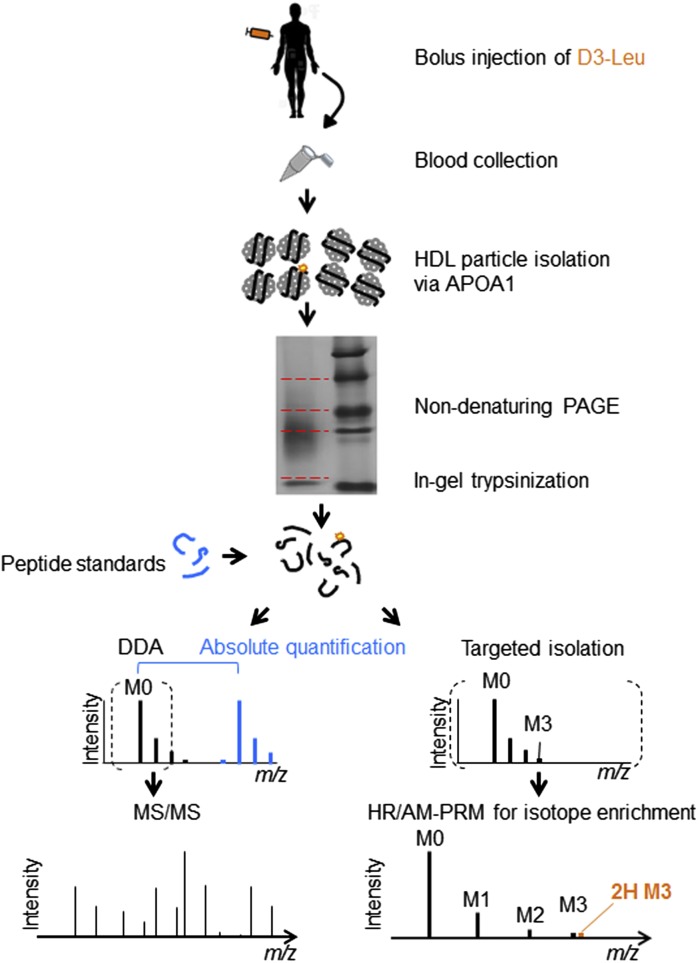
Three participants (supplementary Table 1) received a bolus injection of 10 mg/kg D3-Leu and provided blood samples up to 70 h postinjection. HDL particles were isolated from plasma by anti-apoA-I immunopurification and separated by size using ND-PAGE (Fig. 1). The upper- to lowermost gel fractions comprise the largest- to smallest-sized lipoprotein populations, respectively (28, 29) (supplementary Fig. 1A). The subsequent MS included DDA for protein identification, absolute quantification by spiking of standard peptides for apolipoprotein pool size determination, and HR/AM-PRM for D3-Leu detection (Fig. 1).
Fig. 1.
An overview of the in vivo HDL apolipoprotein study. Workflow for the comprehensive MS-based HDL proteome profiling and quantification of D3-Leu-labeled apolipoproteins isolated from three participants. Dashed red lines indicate segments for in-gel digestions. DDA, data-dependent acquisition.
The HDL size fraction subproteomes
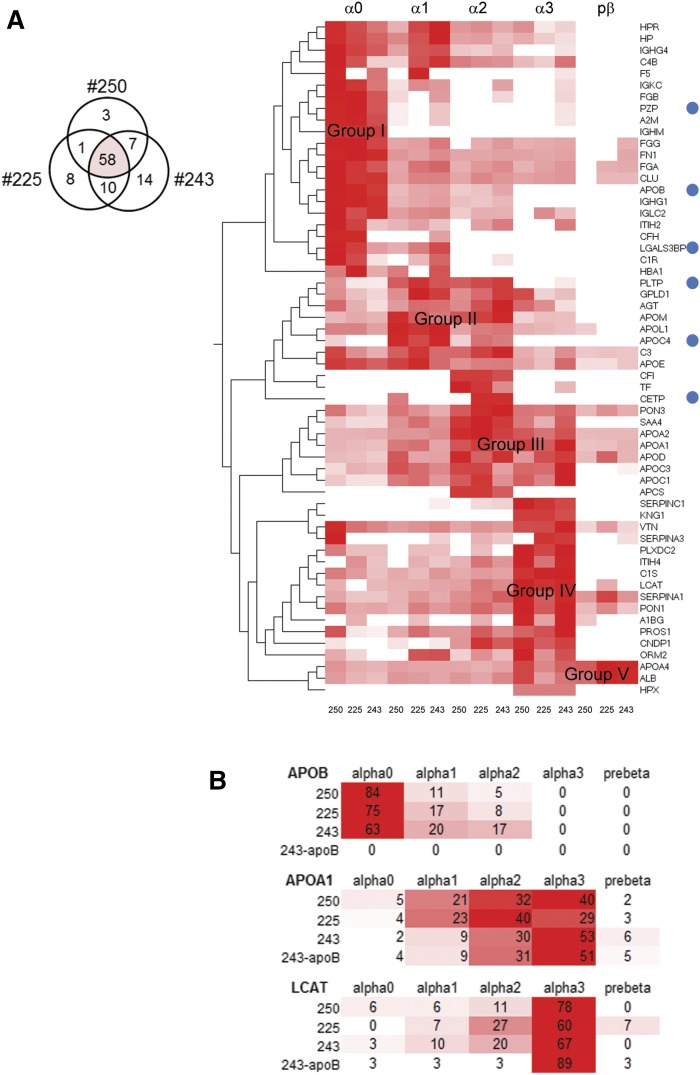
HDL from the three participants shared 58 proteins (Fig. 2A). Our analysis confirmed the presence of classical apolipoproteins, complement factors, fibrinogen, serpins, LCAT, and serum paraoxonase/arylesterase 1 (PON1), all of which were previously reported using other HDL isolation methods (35, 36). The protein-specific sum-normalized spectral counts (per participant) estimated the relative proportion of protein abundances across the five native gel size fractions. Hierarchical clustering then compared the three proteomes. The heat map reveals that five major protein groups (I to V) emerge when the cluster matrix is sorted by decreasing lipoprotein size (Fig. 2A).
Fig. 2.
Hierarchical cluster analysis of the HDL proteome. A: Heat map of the HDL proteome intersect (Venn diagram) for the three participants (250, 225, and 243). The sum-normalized spectral counts were clustered based on the similarity of the protein abundance profiles across the HDL size fractions and were grouped by participant. HDL sizes are indicated on the top. Blue circles indicate proteins that were not detected in the sample depleted in apoB by dextran sulfate and magnesium chloride (supplementary Fig. 1C). B: A heat map that compares the relative abundances of apoA-I, LCAT, and apoB across the HDL sizes to highlight differences among the participants and to highlight the effects of apoB depletion. The sum-normalized percent abundance values are provided. Scale: Red to white indicates highest to lowest (no signal). LGALS3BP, galectin-3-binding protein; PZP, pregnancy zone protein.
As expected, apoA-I is present on all HDL sizes; however, its signal predominates in α2 and α3 and forms part of the group III protein cluster (Fig. 2A). Group I represents the 22 proteins that predominate in α0 size, including apoB and fibrinogen (Fig. 2A). The presence of apoB perhaps reflects small LDL and small chylomicron remnants that contain low levels of apoA-I (36–38). Group II represents the proteins that predominate in α1 and α2 size HDL and contains 11 proteins including apoE, apoM, apoL-I, apoC-IV, cholesteryl ester transfer protein (CETP), and phospholipid transfer protein (PLTP). Small amounts of apoB in this group (Fig. 2A) also may indicate smaller-sized LDL particles (39) or small proteolytic fragments of apoB that adhere to HDL. In addition to apoA-I, group III contains eight proteins including apoA-II, apoD, apoC-I, apoC-III, PON3, and SAA4 that are present in α2 and α3 (Fig. 2A). Group IV contains 14 proteins located in α3 including PON1, SERPINC1, SERPINA1, SERPINA3, and the hallmark for smaller-sized HDL particles, LCAT (40, 41). Forming a fifth cluster, albumin and apoA-IV are main components of the prebeta HDL size, which also includes small amounts of additional proteins such as apoE, apoA-II, PON1, PON3, apoD, and SERPINA1 (Fig. 2A). These distinct groupings of HDL proteins, distributed heterogeneously to specific size fractions, are consistent in the three participants (Fig. 2A). This consistency raises the possibility that specific functions of HDL, whether related to reverse cholesterol transport or other processes, take place within specific HDL sizes.
LDL and small chylomicron remnants have low amounts of apoA-I (37, 38) and thus could explain the presence of apoB in apoA-I lipoproteins in the present study. It is also possible that a small LDL and its associated proteins are in the larger HDL size range (42, 43). Thus, we performed a second HDL isolation experiment using participant 243. We chose this participant because he had the highest apoB content in the larger HDL size range. We removed apoB-containing lipoproteins by dextran sulfate/magnesium chloride precipitation prior to the apoA-I immunoaffinity purification (27). When compared with the proteome profile obtained from the three non-apoB-depleted samples, apoB, apoC-IV, PLTP, CETP, LGALS3BP, and the protease inhibitor PZP were not present in the apoB-depleted sample (Fig. 2A; supplementary Fig. 1C). Despite the removal of apoB lipoproteins, many proteins remained in the α0 fraction including clusterin/apoJ and apoE (supplementary Fig. 1C), indicating that the slowest migrating fraction also contains very large HDL particles. The addition of the apoB-depleted data set into the hierarchical analysis did not disrupt the general architecture observed in the nondepleted data sets (supplementary Fig. 1C). More specifically, the relative distributions of apoA-I and LCAT signals were preserved (Fig. 2B), indicating that apoB depletion did not interfere with the overall integrity of the HDL proteome.
HDL apolipoprotein pool size
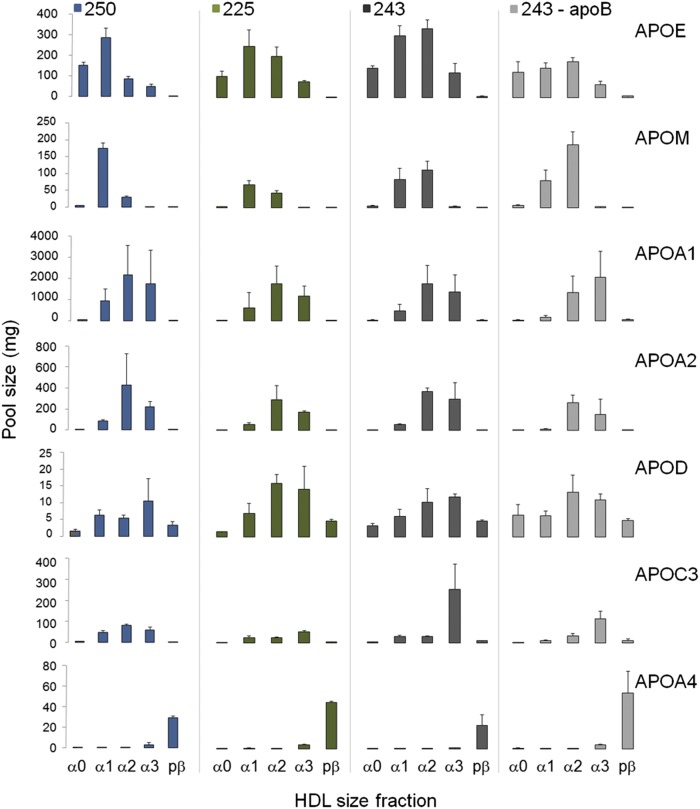
The calculated protein pool sizes for the apolipoproteins ranged from ∼15 mg of apoD to 2,000 mg of apoA-I (Fig. 3). Moreover, the relative distribution of protein pool sizes across the HDL sizes (Fig. 3) was consistent with the spectral counting data (Fig. 2A). These distributions are consistent among the three participants and after apoB depletion (Fig. 3). The total apoA-I pool size (sum of apoA-I pools in all HDL sizes) was 4,897, 3,521, and 3,630 mg for participants 250, 225, and 243 within the expected apoA-I pool size range for individuals with low HDL-C and who are overweight or obese (44). The total protein pool sizes for the other proteins were ∼10-fold (apoE, apoM, apoA-II, apoC-III) to 100-fold (apoD, apoA-IV) smaller than total apoA-I, as shown previously (43).
Fig. 3.
Absolute quantification of HDL apolipoproteins. Histogram of the calculated apolipoprotein pool sizes in plasma as determined by stable isotope-labeled peptide standards for the three participants, 250, 225, and 243. ApoB-depleted sample indicated as 243-apoB. Error bars indicate standard deviation of n = 2–4 replicates.
HR/AM-PRM for quantification of low enrichment of D3-Leu peptides
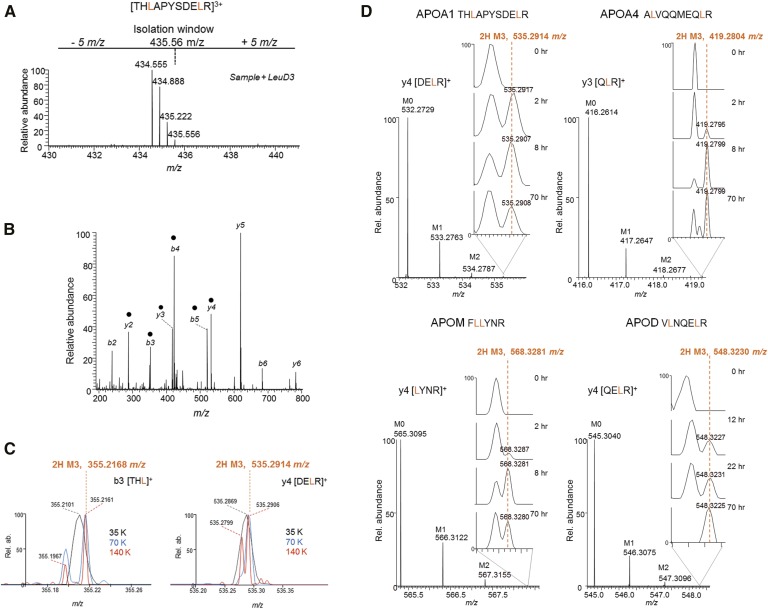
The quadrupole Orbitrap performs HR/AM full mass spectrometric scans of fragment ions, also known as HR/AM-PRM (8, 45). With a resolution setting of R = 140 K at m/z 200, a peptide with a leucine(s) within the first five amino acids from either terminus (Fig. 4A) is desirable because its fragment ions will fall within the lower m/z range (Fig. 4B) permitting deconvolution of the 2H M3 isotope from background ions (Fig. 4C). While an in-depth comparison between PRM and MRM has been described previously (8), we summarize the salient features here: PRM permits coisolation of the D0 native and D3-Leu-labeled peptides simultaneously, as demonstrated by a SIM scan that reflects the m/z 10 isolation window used in this study (Fig. 4A). Unlike MRM, all fragment ions are detected in a single scan in PRM; therefore, the benefits of increasing the scan resolution (e.g., from 35 K to 140 K) is readily evaluated for a wide m/z range at once (Fig. 4C). Two fragment ions of an apoA-I peptide demonstrate the advantage of HR/AM scan readouts on resolving the 2H M3 ion from background peaks (Fig. 4C), thereby permitting accurate relative peak quantification, [tracer (2H M3) / (tracer (2H M3) + tracee (M0)], for subsequent kinetic profiling. In this study, a setting of resolution = 140 K was the default setting for the HR/AM-PRM method.
Fig. 4.
HR/AM-PRM for quantification of low D3-Leu tracer enrichment in apolipoproteins. A: A selected ion monitoring (SIM) scan reflecting the isolation window of an APOA1 peptide whose D3-Leu enrichment is <1.0% and cannot be readily detected in the SIM scan. B: The PRM scan for the peptide in A. Black circles indicate ions used for quantification. C: The 2H M3 ion (theoretical m/z value is indicated in orange) can be resolved from surrounding background peaks when the scanning resolution (R = 35, 70 or 140K at 200 m/z) is increased. D: Example fragment ion peak profiles from the indicated apolipoprotein peptide. The appearance of the 2H M3 ion over time is highlighted in the zoomed in m/z range (inset).
Apolipoprotein peptides for HR/AM-PRM
We synthesized full-length FLEX-tagged apoA-I in either its native D0 or D3-Leu-labeled forms using a cell-free wheat germ extract-based system (32) and performed a series of serial spiking experiments aimed to evaluate the linearity of HR/AM-PRM-based quantification strategy for D3-Leu enrichment of <1%. A D0-Leu to D3-Leu mixing standard curve was prepared from 1:1 to 100:1 and analyzed by HR/AM-PRM, and the 2H M3 to M0 ratios of two to five fragment ions for each peptide were determined (supplementary Table 2; supplementary Fig. 3A). The calculated ratios varied slightly for each peptide as determined by the slope of their standard curves (supplementary Fig. 3A). However, when we calculated the z-scores for each mixing ratio and built a regression plot, the slope was zero (supplementary Fig. 3A), indicating that the technical variance observed within each peptide can be resolved by taking the median of all three. We then performed a second serial dilution experiment using the 100:1 (D0-Leu/D3-Leu) mixing ratio sample to determine the effects of absolute injection amounts on the observed ratios. Two of the three peptides demonstrated consistent ratio values across four orders of magnitude of absolute intensity, and one across the two orders of magnitude (supplementary Fig. 3B). The M0 signal intensities (area under the curves) for these case peptide fragments fall within the dynamic range of apolipoprotein peptide intensities (supplementary Fig. 2C); therefore, the linear responses observed here are expected to be applicable for the other apolipoprotein peptides.
For the remaining six apolipoproteins, we evaluated the candidacy of their respective leucine peptides for HR/AM-PRM based on the ability to detect the 2H M3 ion directly from HDL from participants 250 and 225. With the exception of apoD, two to three peptides with at least two fragment ions were monitored for each protein (supplementary Table 2). The peptide retention time overlaps were minimized in order to dedicate HR/AM-PRM on a single peptide at a time, resulting in two to three separate injection runs to minimizing the overlap. If more than one peptide per apolipoprotein yielded similar enrichment profiles as verified by both participant 250 and 225 data, we reduced the HR/AM-PRM acquisition method down to one peptide for most of the apolipoproteins, in order to increase the throughput of sample collection on the instrument (supplementary Table 2; supplementary Fig. 3C). The HR/AM-PRM scans for example fragment ions for apoA-I, apoA-IV, apoM, and apoD demonstrate the emergence and disappearance of the 2H M3 ion across the sampled time points (Fig. 4D). The HR/AM scans in combination with the contrasting absence of signal at the 0 h time point enabled confident identification of the 2H M3 ion, most notably for when it appeared as a shoulder peak of a nonspecific peak (i.e., apoM y4 ion 2 h; Fig. 4D).
To illustrate further the practicality of HR/AM-PRM for the detection of very low tracer enrichment (<1.0%), we aimed to quantify the tracer in participant 243’s α3 apoE and apoA-I using an MS1-dependent method (supplementary Fig. 4). The samples were analyzed using the Orbitrap Elite (R = 240 K at m/z 400, dynamic range = 10 K). Unlike our HR/AM-PRM strategy, which is limited to peptides with terminal leucines, the MS1-based quantification is conducive to any leucine containing peptide, irrespective of the leucine’s placement within the amino acid sequence. As shown with HR/AM-PRM, the lower the m/z value, the more likely the 2H M3 isotopes can be resolved from interfering peaks (Fig. 4C); however, this m/z constraint limits even the choice of peptides for MS1-based quantification to those on the lower end of the m/z range. The peptides for apoE and apoA-I were between m/z 380 and 700 (supplementary Fig. 4A). For apoE, the MS1 and HR/AM-PRM enrichment curves were similar, reaching nearly 6% enrichment by 2 h; however, the descending slope of the MS1-generated enrichment curve (R2 = 0.38) was not as defined as that from the HR/AM-PRM data (R2 = 0.87) (supplementary Fig. 4B). More drastically, no discernible curve was observed for apoA-I using the MS1-based method, despite the use of seven peptides for quantification. On the other hand, only three peptides (10 fragment ions) were required for the HR/AM-PRM to observe an enrichment curve (supplementary Fig. 4B). A closer look at example peptide spectra reinforces that signal interference in the MS1 reduces reliable detection of the 2H M3 ion (supplementary Fig. 4C). The candidate 2H M3 ion for the apoE peptide is discernible from surrounding peaks; however, that of the apoA-I peptide is not observed. The relatively high enrichment for apoE tracer (>1.0%) compensates for interference in MS1 scans; however, despite the >10-fold increase in absolute signal over apoE, apoA-I’s tracer is too low for detection. Although HR/AM MS1 is often used for stable isotope-based quantification, this approach is most practical when isotope pairs are within 1- to 100-fold difference in intensity.
The HDL apolipoprotein tracer enrichment curves
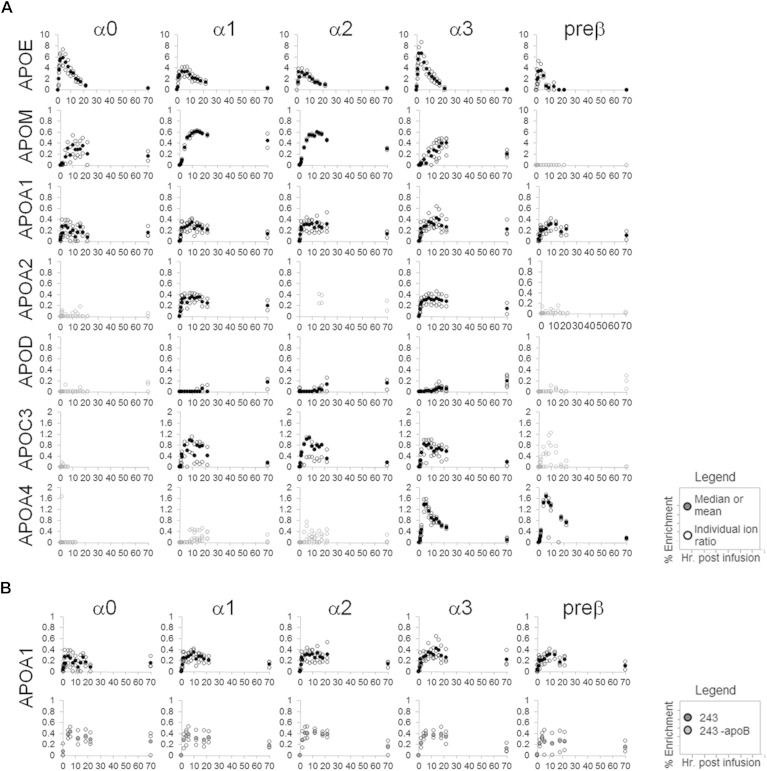
The HR/AM-PRM-enabled isotope enrichment profiling revealed a diverse array of apolipoprotein enrichment curves (Fig. 5A; supplementary Fig. 5). A comparison of the tracer enrichment curves for the apolipoproteins across the HDL size fractions demonstrates that no two apolipoproteins share similar curves. In contrast, the tracer enrichment curves for each apolipoprotein are similar across the three participants, although apoC-III is an exception (supplementary Fig. 5A–C). The most defining feature that distinguishes the various apolipoprotein tracer enrichment curves from each other is the hour at which the isotope enrichment is highest (Fig. 5A; supplementary Fig. 5A–C). ApoA-I and apoA-II peak between 8 h and 12 h with 0.2–0.4% peak tracer enrichment. On the other hand, apoE peaks the earliest, usually by 2 h to 4 h, and reaching up to 8%. ApoA-IV peaks between 4 h and 6 h (up to 2% enrichment), and apoM between 10 h and 14 h (0.6% enrichment). In stark contrast to the other apolipoproteins, apoD enrichment was not discernable until 12 h. The highest isotope enrichment point recorded for apoD was at the 70 h time point with 0.2% enrichment (Fig. 5A; supplementary Fig. 5A–C). ApoB depletion did not affect the enrichment curves of most apolipoproteins (supplementary Fig. 5C, D). For instance, the five HDL size curves for apoA-I are similar between the depleted and nondepleted samples: neither the time range of peak enrichment nor the enrichment maximum was altered after apoB depletion (Fig. 5B). Only a few minor changes were observed after apoB depletion, one being apoE, which exhibited a slightly higher peak enrichment of 6% in α1 and α2, when compared with no depletion (4%); however, the time of peak enrichment (2–4 h) and general apoE enrichment curve structure were maintained (supplementary Fig. 5C, D). ApoE on HDL has been shown to bind polyanions (46). Because apoB precipitation by dextran sulfate/magnesium chloride depends on polyanion binding, it is likely that the minor enrichment curve differences seen after apoB depletion are due to apoE on α1 and α2 HDL binding to and being removed by the precipitation reagent.
Fig. 5.
HDL apolipoprotein isotope enrichment curves. A: Isotope enrichment curves for participant 243. B: ApoA-I before and after apoB depletion. Open circles are individual fragment ion enrichment values; closed circles are the median values or mean (apoA-I, mean of three peptides).
Multicompartmental analysis
Our goal for multicompartmental analysis was to construct a comprehensive kinetic model that describes the source and removal pathways for each apolipoprotein among the HDL size fractions. The final models for each protein showed excellent fitting of the enrichment curves and pool sizes for all participants (supplementary Fig. 6). The kinetic parameters including the PR, FCR, and pool sizes are provided in Table 1 and supplementary Table 3.
TABLE 1.
Kinetic parameters of HDL apolipoproteins
| Apolipoprotein | HDL Size | FCR (pools/day) | PR (mg/kg/day) | PS (mg) |
| ApoE | ||||
| α0 | 4.05 (1.97) | 4.85 (1.30) | 116.85 (50.99) | |
| α1 | 2.26 (0.61) | 7.11 (1.20) | 270.83 (25.20) | |
| α2 | 3.59 (2.35) | 7.03 (4.18) | 190.61 (123.16) | |
| α3 | 7.14 (4.88) | 5.52 (2.92) | 72.34 (30.19) | |
| ApoM | ||||
| α1 | 0.61 (0.31) | 0.67 (0.16) | 107.00 (58.06) | |
| α2 | 0.60 (0.31) | 0.41 (0.25) | 60.84 (43.31) | |
| ApoA-I | ||||
| α0 | 0.48 (0.28) | 0.06 (0.03) | 14.83 (13.33) | |
| α1 | 0.67 (0.36) | 4.59 (1.45) | 671.10 (241.21) | |
| α2 | 0.76 (0.52) | 14.76 (8.95) | 1,801.94 (333.09) | |
| α3 | 0.50 (0.26) | 7.61 (2.39) | 1,421.63 (298.52) | |
| Prebeta | 6.06 (0.71) | 1.30 (1.09) | 17.85 (14.01) | |
| ApoA-II | ||||
| α1 | 0.42 (0.35) | 0.33 (0.16) | 73.16 (20.34) | |
| α2 | 0.70 (0.62) | 2.62 (1.53) | 359.52 (97.22) | |
| α3 | 0.36 (0.22) | 0.80 (0.27) | 194.69 (33.71) | |
| ApoC-III | ||||
| α1 | 1.79 (0.52) | 0.52 (0.09) | 26.57 (3.53) | |
| α2 | 1.81 (0.37) | 0.52 (0.09) | 26.01 (1.26) | |
| α3 | 1.76 (0.59) | 2.48 (1.85) | 151.90 (147.49) | |
| ApoA-IV | ||||
| α3 | 3.29 (0.50) | 0.10 (0.05) | 2.33 (1.07) | |
| Prebeta | 2.74 (0.82) | 1.11 (0.21) | 36.12 (11.78) |
PS, pool size. Parameter estimates are expressed as mean (SD) for participants 250, 225, and 243. Parameter estimates were made using the SAAM II software.
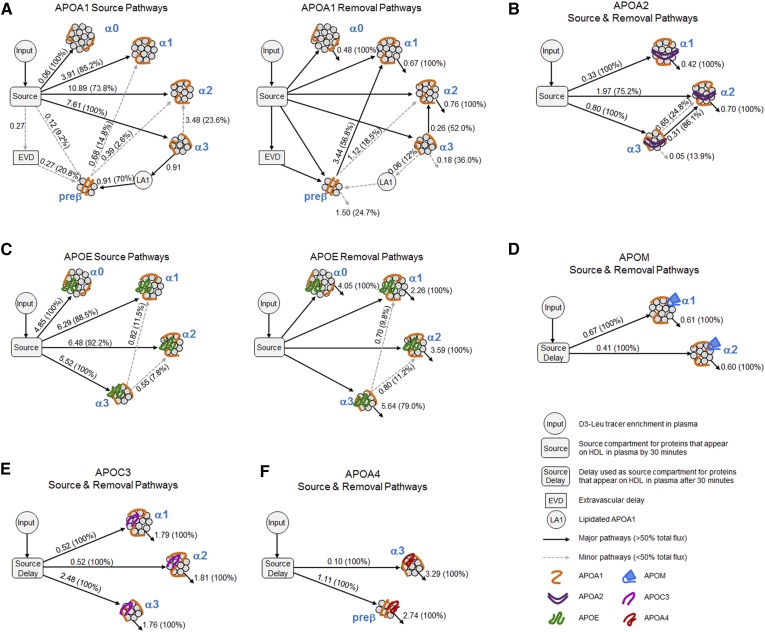
The majority apoA-I on α0 (100%), α1 (85.2%), α2 (73.8%), and α3 (100%) HDL came directly from the source compartment (Fig. 6A, source pathways, solid arrows). In contrast, apoA-I prebeta originated mostly from α3 (70.0%) with only minor amounts from the source (9.2%) and EVD (20.8%) compartments. Size expansion was only a minor contributing source of apoA-I α1 and α2 HDL (α1, 14.8% from prebeta; α2, 23.6% from α3 and 2.6% from prebeta) (Fig. 6A, source pathways, dashed arrows). Removal of apoA-I α0 (100%), α1 (100%), and α2 (100%) was due to direct removal from plasma, whereas transfer of apoA-I from α3 to α 2 (52.0%) and prebeta to α1 (56.8%) provided the major pathways of their respective apoA-I removal (Fig. 6A, removal pathways, solid arrows).
Fig. 6.
Kinetic models describing the in vivo metabolism of the HDL apolipoproteins on each HDL size fraction. SAAM II models highlighting the synthesis and removal pathways for each apolipoprotein across the HDL size fractions: ApoA-I (A), ApoA-II (B), ApoE (C), ApoM (D), ApoC-III (E) and ApoA-IV (F). Each model contains an input compartment, which represent the plasma amino acid precursor pool (D3-Leu tracer enrichment in plasma) expressed as a forcing function. The same D3-Leu tracer enrichment curve was used for all protein models per participant. The source compartment accounts for the time necessary for labeled protein to appear on each HDL size fraction in plasma. Source pathways: number next to arrow represents average flux (mg/kg/d) through each pathway for the three participants. Numbers in parentheses represent average percent source of each protein into each HDL size fraction. Removal pathways: numbers next to arrows are average rates (pools/day), and numbers in parentheses are average percent of protein removed by that pathway. EVD may represent a processing compartment that includes apoA-I on prebeta that has been secreted but is outside the systemic circulation. LA1 prebeta may be generated from α3 HDL through the release of LA1. ApoA-II α3 and α2 arrows: top dashed arrow is a source and bottom solid arrow is a removal pathway.
Similar to apoA-I on the α fractions, the source compartment accounted for the majority of apoA-II α1 (100%), α2 (75.2%), and α3 (100%) (Fig. 6B, solid arrows out of source compartment). Minor flux pathways from smaller to larger HDL sizes were also seen for apoA-II α2 (24.8% from α3) (Fig. 6B, α3 to α2 dashed arrow). Interestingly, the α3 to α2 transfer pathway detected for apoA-II was also the only transfer pathway detected among the α fractions for apoA-I (Fig. 6A). The major removal pathway for apoA-II α1 (100%) and α2 (100%) is clearance from circulation (Fig. 6B). On the other hand, the majority of apoA-II on α3 (86.1%) was transferred to α2 (Fig. 6B, α3 to α2 solid arrow).
Although the majority of apoE α0 (100%), α1 (88.5%), α2 (92.2%), and α3 (100%) originated directly from the source compartment, minor flux pathways from smaller to larger HDL sizes were also observed for apoE α1 (11.5% from α3) and α2 (7.8% from α3) (Fig. 6C, source pathways). The majority of apoE was removed directly from the HDL size fractions (α0, 100%; α1, 100%; α2, 100%; and α3, 79.0%) while small amounts of apoE on α3 HDL were removed and transferred to α1 (9.8%) and α2 (11.2%) HDL (Fig. 6C, removal pathways).
The apoM, apoC-III, and apoA-IV models showed similar source and removal pathways across the size fractions. All of apoM (α1 and α2), apoC-III (α1, α2, and α3), and apoA-IV (α3 and prebeta) originated from the source compartment (Fig. 6D–F, arrows out of source compartment) and were removed directly from HDL (Fig. 6D–F, arrows out of size fraction protein pools) with no evidence of flux pathways between the HDL sizes that were modeled.
DISCUSSION
We used a multi-mass spectrometric approach to characterize the distribution and metabolism of apolipoproteins in five HDL sizes. Spectral counting provided qualitative information regarding the distribution of the proteome across the HDL sizes. Hierarchical cluster analysis revealed that the HDL proteome is organized into five subproteomes, each associating with an HDL size range (Fig. 2). We then performed kinetic analysis on seven apolipoproteins by determining their pool sizes per HDL size fraction (Fig. 3) and by quantifying their tracer enrichment by HR/AM-PRM.
The apolipoprotein pool sizes were determined using stable isotope peptide standards under the assumption that peptide recovery from each HDL size/gel fraction was the same. Consequently, the sample loss correction factor determined for apoA-I was applied to the remaining six apolipoproteins. The total apoA-I pool size was 4,897, 3,521, and 3,630 mg for participants 250, 225, and 243, respectively, the expected apoA-I pool size range for individuals with low HDL-C and who are overweight or obese (44). Separating total apoA-I into the five HDL size fractions showed that the majority of apoA-I (>65%) resides in the α2 and α3 pools. This apoA-I distribution was maintained in all three participants. The total protein pool sizes for the other proteins were ∼10-fold (apoE, apoM, apoA-II, apoC-III) to 100-fold (apoD, apoA-IV) smaller than total apoA-I, similar to what has been shown previously (43).
HR/AM-PRM can detect trace signals with high confidence as demonstrated by the ability to reliably detect tracer enrichment of <1% for any given peptide in all three participants (Fig. 5; supplementary Fig. 5). HR/AM-PRM performed on a quadrupole Orbitrap first selects the peptide of interest and then fragments and scans all fragment ions simultaneously (7, 8, 45, 47). Because PRM itself is a relatively recent method, MRM [sometimes referred to as selected reaction monitoring (SRM)] is the more commonly used method to detect and quantify target peptides. MRM/SRM relies on scans of a single fragment ion at a time but usually monitors multiple fragment ions per protein in succession. Nonetheless, this approach requires optimization steps to determine which fragment ions are suitable for subsequent quantification studies (48, 49). A recent study by Ronsein et al. (50) determined that PRM and SRM are comparable in terms of sensitivity and accuracy for quantification of an apoA-I protein standard curve, which spanned a femtomole to picomole range. The same study (50) used a narrow peptide isolation window (±1 Da), and the PRM scans were set to the lowest resolution setting of 17,500 K (at m/z 200), making the method comparable to MRM. Our approach (Fig. 4) demonstrated further the versatility of PRM by quantifying differences in relative abundance up to ∼3,000-fold (in the attomole to femtomole range) within a single scan (140,000 K). As a consequence of this dynamic range capability (dynamic range limit is 5,000 for the Q Exactive), we were able to observe the markedly diverse kinetics of seven HDL apolipoproteins across the five HDL size fractions for the first time.
A very recent study by Mendivil et al. (3) on apoA-I metabolism, which used GC/MS for tracer enrichment analysis, reported similar apoA-I enrichment curves across the HDL sizes in 12 participants with high or low HDL-C. Mendivil et al. (3) tested several kinetic models exploring the potential apoA-I metabolic pathways across the HDL size fractions and demonstrated that the canonical HDL size-expansion model, in which prebeta is the nascent HDL particle appearing in plasma and undergoes stepwise enlargement to α3, 2, and 1, could not explain the apoA-I tracer enrichment data. The final best-fit model instead showed that the majority of apoA-I on α HDL originated from the source compartment; size expansion pathways only provided a minor contribution to apoA-I on α HDL (3). These same findings were replicated in our compartmental analysis (Fig. 6A); thus, this HR/AM-PRM apoA-I metabolism study has provided an independent confirmation of the new model for HDL metabolism recently described using GC/MS.
Because the HR/AM-PRM approach allowed us to measure multiple apolipoproteins simultaneously, we were also able to perform compartmental modeling for apoA-II, apoE, apoM, apoC-III, and apoA-IV across the HDL sizes for the first time. Similarly to apoA-I on the α sizes, the majority of the other apolipoproteins appear on HDL directly from the source compartment. This result is not surprising because the enrichment curves for a given apolipoprotein generally look similar across the HDL size fractions (Fig. 5).
Yet, subtle differences in the enrichment curves between the HDL sizes were discernible for most apolipoproteins, notably apoE in all participants. For the apolipoproteins and HDL sizes that were modeled, some of these differences proved to be meaningful; they represented flux between the HDL sizes (Fig. 6). For instance, we determined that apoA-II flux occurred from α3 to α2 HDL. Interestingly, this was also the only pathway detected between the α sizes observed for apoA-I. It is known that >60% of apoA-I particles contain apoA-II (51); therefore, it is likely that we are describing the kinetics of the same particles. For apoE, pathways between α3 and α2 and between α3 and α1 were identified. PLTP has been shown to remodel small spherical apoE-HDL into larger particles by promoting particle fusion (52). The α3 to α1 and α3 to α2 pathways may reflect this process. These results are also compatible with transfer of apoE protein itself from small to large HDL. ApoE has been shown to exchange between HDL and apoB-containing lipoproteins, so it is likely that transfer pathways between the HDL sizes also exist (53, 54).
In contrast to apoA-I, apoA-II, and apoE, no pathways were detected between the apoM, apoC-III, and apoA-IV size fractions. However, this may be due to several reasons: exchange may not be occurring, the time points monitored were not optimal for observing exchange, or exchange is occurring at the same rate in both directions between all HDL sizes so no difference in enrichment is observed. Moreover, the molecular weight size windows for the ND-PAGE are specific for apoA-I/HDL. Our study has demonstrated that apoA-I protein interactions are nonuniform across the HDL sizes (Fig. 2). Therefore, the potential to detect these additional transfer pathways may be in part dependent on a reoptimization of the molecular weight size cutoffs for a given HDL subproteome.
The overall findings of the compartmental modeling demonstrated that the majority of the apolipoproteins monitored appear on HDL directly from their respective source compartments. ApoA-I, apoA-II, apoE, apoM, and apoC-III are synthesized primarily in the liver while apoA-IV is synthesized in the small intestine. However, the appearance of a given apolipoprotein on HDL may not be due to direct section from the primary area of synthesis per se. We cannot rule out intermediate transfer to a compartment not measured in our system. For instance, the delayed appearance of apoM, apoC-III, and apoA-IV on HDL in plasma may reflect an intermediary step of transfer from an apoB-containing particle. ApoA-IV is secreted into the lymphatics on the surface of chylomicrons and then enters blood where it can be transferred to HDL (55). Evidence in the human intestinal CaCo-2 cell line and in rodents also suggests that apoA-IV α3 and prebeta HDL may be synthesized by the small intestine and secreted into circulation directly (56) or through the lympathics (57–60). One or both of these mechanisms may occur and account for the delayed appearance of apoA-IV on α3 and prebeta HDL in plasma. Additional causes for a delayed appearance onto HDL also include mechanisms controlling protein synthesis, processing, and secretion that may vary across the apolipoproteins.
It is interesting to note the delayed appearance (12 h to 22 h) and extremely low peak enrichment (∼0.2%) of apoD on HDL. The value of the HR/AM-PRM method is also highlighted by the ability to accurately and reliably measure the tracer enrichment range (0.03–0.2%) of apoD. Without previous knowledge of apoD metabolism, we did not anticipate the need to collect additional blood samples at later time points of the 3 day study. As a consequence, apoD compartmental modeling could not be performed due to insufficient measurements at time points beyond 12 h.
In closing, we have presented a novel application for the HR/AM-PRM technology to measure tracer enrichment between 0.03% and 1.0% for several HDL apolipoproteins, simultaneously. It is likely that the diversity in metabolism observed among the 7 apolipoproteins reflects the depth that exists in the full array of the remaining 50 or more HDL proteins described in our study. The power of HR/AM-PRM technology thus provides a foundation to design in vivo/clinical metabolism studies aimed to understand the metabolism of specific HDL apolipoproteins that have unique relations to risk of diseases such as diabetes and cardiovascular disease.
Supplementary Material
Acknowledgments
The authors thank Mr. Iwao Yamada and Mr. Katsutoshi Miyosawa for their technical assistance.
Footnotes
Abbreviations:
- CCI
- Center for Clinical Investigation
- CHD
- coronary heart disease
- D3-Leu
- trideuterated leucine
- EVD
- extravascular delay
- FCR
- fractional catabolic rate
- HCD
- higher energy collision-induced dissociation
- HDL-C
- HDL cholesterol
- HR/AM-PRM
- high-resolution/accurate mass parallel reaction monitoring
- LA1
- lipidated apoA-I
- MRM
- multiple reaction monitoring
- ND-PAGE
- nondenaturing polyacrylamide gel electrophoresis
- PLTP
- phospholipid transfer protein
- PON1
- paraoxonase/arylesterase 1
- PR
- production rate
- PSM
- peptide spectrum match
- SIM
- selected ion monitoring
- SRM
- selected reaction monitoring
- XIC
- extracted ion chromatogram
This work was supported by research grants from Kowa Company Ltd. (Nagoya, Japan; M.A.) and the National Institutes of Health [R01HL107550 (M.A.); UL1 RR 025758-01; and R01HL095964 (F.M.S.)].
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Bateman R. J., Munsell L. Y., Morris J. C., Swarm R., Yarasheski K. E., and Holtzman D. M.. 2006. Human amyloid-beta synthesis and clearance rates as measured in cerebrospinal fluid in vivo. Nat. Med. 12: 856–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan D. C., Barrett P. H., and Watts G. F.. 2004. Lipoprotein transport in the metabolic syndrome: methodological aspects of stable isotope kinetic studies. Clin. Sci. (Lond.). 107: 221–232. [DOI] [PubMed] [Google Scholar]
- 3.Mendivil C. O., Furtado J., Morton A. M., Wang L., and Sacks F. M.. 2016. Novel pathways of apolipoprotein A-I metabolism in high-density lipoprotein of different sizes in humans. Arterioscler. Thromb. Vasc. Biol. 36: 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lassman M. E., McAvoy T., Lee A. Y., Chappell D., Wong O., Zhou H., Reyes-Soffer G., Ginsberg H. N., Millar J. S., Rader D. J., et al. 2014. Practical immunoaffinity-enrichment LC-MS for measuring protein kinetics of low-abundance proteins. Clin. Chem. 60: 1217–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee A. Y., Yates N. A., Ichetovkin M., Deyanova E., Southwick K., Fisher T. S., Wang W., Loderstedt J., Walker N., Zhou H., et al. 2012. Measurement of fractional synthetic rates of multiple protein analytes by triple quadrupole mass spectrometry. Clin. Chem. 58: 619–627. [DOI] [PubMed] [Google Scholar]
- 6.Zheng C., Ikewaki K., Walsh B. W., and Sacks F. M.. 2006. Metabolism of apoB lipoproteins of intestinal and hepatic origin during constant feeding of small amounts of fat. J. Lipid Res. 47: 1771–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallien S., Duriez E., Crone C., Kellmann M., Moehring T., and Domon B.. 2012. Targeted proteomic quantification on quadrupole-orbitrap mass spectrometer. Mol. Cell. Proteomics. 11: 1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peterson A. C., Russell J. D., Bailey D. J., Westphall M. S., and Coon J. J.. 2012. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol . Cell. Proteomics. 11: 1475–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Angelantonio E., Sarwar N., Perry P., Kaptoge S., Ray K. K., Thompson A., Wood A. M., Lewington S., Sattar N., Packard C. J., et al. 2009. Major lipids, apolipoproteins, and risk of vascular disease. J. Am. Med. Assoc. 302: 1993–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson A., and Danesh J.. 2006. Associations between apolipoprotein B, apolipoprotein AI, the apolipoprotein B/AI ratio and coronary heart disease: a literature-based meta-analysis of prospective studies. J. Intern. Med. 259: 481–492. [DOI] [PubMed] [Google Scholar]
- 11.Voight B. F., Peloso G. M., Orho-Melander M., Frikke-Schmidt R., Barbalic M., Jensen M. K., Hindy G., Holm H., Ding E. L., Johnson T., et al. 2012. Plasma HDL cholesterol and risk of myocardial infarction: a Mendelian randomisation study. Lancet. 380: 572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boden W. E., Probstfield J. L., Anderson T., Chaitman B. R., Desvignes-Nickens P., Koprowicz K., McBride R., Teo K., and Weintraub W.. 2011. Niacin in patients with low HDL cholesterol levels receiving intensive statin therapy. N. Engl. J. Med. 365: 2255–2267. [DOI] [PubMed] [Google Scholar]
- 13.Barter P. J., Caulfield M., Eriksson M., Grundy S. M., Kastelein J. J., Komajda M., Lopez-Sendon J., Mosca L., Tardif J. C., Waters D. D., et al. 2007. Effects of torcetrapib in patients at high risk for coronary events. N. Engl. J. Med. 357: 2109–2122. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz G. G., Olsson A. G., Abt M., Ballantyne C. M., Barter P. J., Brumm J., Chaitman B. R., Holme I. M., Kallend D., Leiter L. A., et al. 2012. Effects of dalcetrapib in patients with a recent acute coronary syndrome. N. Engl. J. Med. 367: 2089–2099. [DOI] [PubMed] [Google Scholar]
- 15.Feig J. E., Hewing B., Smith J. D., Hazen S. L., and Fisher E. A.. 2014. High-density lipoprotein and atherosclerosis regression: evidence from preclinical and clinical studies. Circ. Res. 114: 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng C., and Aikawa M.. 2012. High-density lipoproteins: from function to therapy. J. Am. Coll. Cardiol. 60: 2380–2383. [DOI] [PubMed] [Google Scholar]
- 17.Grundy S. M., Vega G. L., and Kesaniemi Y. A.. 1985. Abnormalities in metabolism of low density lipoproteins associated with coronary heart disease. Acta Med. Scand. Suppl. 701: 23–37. [DOI] [PubMed] [Google Scholar]
- 18.Zheng C., Khoo C., Furtado J., and Sacks F. M.. 2010. Apolipoprotein C–III and the metabolic basis for hypertriglyceridemia and the dense low-density lipoprotein phenotype. Circulation. 121: 1722–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Packard C. J., Demant T., Stewart J. P., Bedford D., Caslake M. J., Schwertfeger G., Bedynek A., Shepherd J., and Seidel D.. 2000. Apolipoprotein B metabolism and the distribution of VLDL and LDL subfractions. J. Lipid Res. 41: 305–318. [PubMed] [Google Scholar]
- 20.Walsh B. W., Li H., and Sacks F. M.. 1994. Effects of postmenopausal hormone replacement with oral and transdermal estrogen on high density lipoprotein metabolism. J. Lipid Res. 35: 2083–2093. [PubMed] [Google Scholar]
- 21.Brinton E. A. 1996. Oral estrogen replacement therapy in postmenopausal women selectively raises levels and production rates of lipoprotein A-I and lowers hepatic lipase activity without lowering the fractional catabolic rate. Arterioscler. Thromb. Vasc. Biol. 16: 431–440. [DOI] [PubMed] [Google Scholar]
- 22.Li X., Stolinski M., and Umpleby A. M.. 2012. Development of a method to measure prebetaHDL and alphaHDL apoA-I enrichment for stable isotopic studies of HDL kinetics. Lipids. 47: 1011–1018. [DOI] [PubMed] [Google Scholar]
- 23.Cohen D. E., and Fisher E. A.. 2013. Lipoprotein metabolism, dyslipidemia, and nonalcoholic fatty liver disease. Semin. Liver Dis. 33: 380–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malmendier C. L., Lontie J. F., Lagrost L., Delcroix C., Dubois D. Y., and Gambert P.. 1991. In vivo metabolism of apolipoproteins A-IV and A-I associated with high density lipoprotein in normolipidemic subjects. J. Lipid Res. 32: 801–808. [PubMed] [Google Scholar]
- 25.Cohn J. S., Batal R., Tremblay M., Jacques H., Veilleux L., Rodriguez C., Mamer O., and Davignon J.. 2003. Plasma turnover of HDL apoC-I, apoC-III, and apoE in humans: in vivo evidence for a link between HDL apoC-III and apoA-I metabolism. J. Lipid Res. 44: 1976–1983. [DOI] [PubMed] [Google Scholar]
- 26.Ji J., Watts G. F., Johnson A. G., Chan D. C., Ooi E. M., Rye K. A., Serone A. P., and Barrett P. H.. 2006. High-density lipoprotein (HDL) transport in the metabolic syndrome: application of a new model for HDL particle kinetics. J. Clin. Endocrinol. Metab. 91: 973–979. [DOI] [PubMed] [Google Scholar]
- 27.Warnick G. R., Benderson J., and Albers J. J.. 1982. Dextran sulfate-Mg2+ precipitation procedure for quantitation of high-density-lipoprotein cholesterol. Clin. Chem. 28: 1379–1388. [PubMed] [Google Scholar]
- 28.Asztalos B. F., Sloop C. H., Wong L., and Roheim P. S.. 1993. Two-dimensional electrophoresis of plasma lipoproteins: recognition of new apo A-I-containing subpopulations. Biochim. Biophys. Acta. 1169: 291–300. [DOI] [PubMed] [Google Scholar]
- 29.Rosenson R. S., Brewer H. B. Jr., Chapman M. J., Fazio S., Hussain M. M., Kontush A., Krauss R. M., Otvos J. D., Remaley A. T., and Schaefer E. J.. 2011. HDL measures, particle heterogeneity, proposed nomenclature, and relation to atherosclerotic cardiovascular events. Clin. Chem. 57: 392–410. [DOI] [PubMed] [Google Scholar]
- 30.Singh S., Springer M., Steen J., Kirschner M. W., and Steen H.. 2009. FLEXIQuant: a novel tool for the absolute quantification of proteins, and the simultaneous identification and quantification of potentially modified peptides. J. Proteome Res. 8: 2201–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pratt J. M., Simpson D. M., Doherty M. K., Rivers J., Gaskell S. J., and Beynon R. J.. 2006. Multiplexed absolute quantification for proteomics using concatenated signature peptides encoded by QconCAT genes. Nat. Protoc. 1: 1029–1043. [DOI] [PubMed] [Google Scholar]
- 32.Singh S., Kirchner M., Steen J. A., and Steen H.. 2012. A practical guide to the FLEXIQuant method. Methods Mol. Biol. 893: 295–319. [DOI] [PubMed] [Google Scholar]
- 33.MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., and MacCoss M. J.. 2010. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 26: 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikkilä E. A., and Kekki M.. 1972. Plasma triglyceride metabolism in thyroid disease. J. Clin. Invest. 51: 2103–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gordon S. M., Deng J., Tomann A. B., Shah A. S., Lu L. J., and Davidson W. S.. 2013. Multi-dimensional co-separation analysis reveals protein-protein interactions defining plasma lipoprotein subspecies. Mol. Cell. Proteomics. 12: 3123–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Zychlinski A., Williams M., McCormick S., and Kleffmann T.. 2014. Absolute quantification of apolipoproteins and associated proteins on human plasma lipoproteins. J. Proteomics. 106: 181–190. [DOI] [PubMed] [Google Scholar]
- 37.Banfi C., Brioschi M., Barcella S., Wait R., Begum S., Galli S., Rizzi A., and Tremoli E.. 2009. Proteomic analysis of human low-density lipoprotein reveals the presence of prenylcysteine lyase, a hydrogen peroxide-generating enzyme. Proteomics. 9: 1344–1352. [DOI] [PubMed] [Google Scholar]
- 38.Alonzi T., Mancone C., Amicone L., and Tripodi M.. 2008. Elucidation of lipoprotein particles structure by proteomic analysis. Expert Rev. Proteomics. 5: 91–104. [DOI] [PubMed] [Google Scholar]
- 39.Windmueller H. G., Herbert P. N., and Levy R. I.. 1973. Biosynthesis of lymph and plasma lipoprotein apoproteins by isolated perfused rat liver and intestine. J. Lipid Res. 14: 215–223. [PubMed] [Google Scholar]
- 40.Glomset J. A., Norum K. R., and King W.. 1970. Plasma lipoproteins in familial lecithin: cholesterol acyltransferase deficiency: lipid composition and reactivity in vitro. J. Clin. Invest. 49: 1827–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marcel Y. L., and Vezina C.. 1973. Lecithin: cholesterol acyltransferase of human plasma. Role of chylomicrons, very low, and high density lipoproteins in the reaction. J. Biol. Chem. 248: 8254–8259. [PubMed] [Google Scholar]
- 42.Vaisar T., Pennathur S., Green P. S., Gharib S. A., Hoofnagle A. N., Cheung M. C., Byun J., Vuletic S., Kassim S., Singh P., et al. 2007. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Invest. 117: 746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davidson W. S., Silva R. A., Chantepie S., Lagor W. R., Chapman M. J., and Kontush A.. 2009. Proteomic analysis of defined HDL subpopulations reveals particle-specific protein clusters: relevance to antioxidative function. Arterioscler. Thromb. Vasc. Biol. 29: 870–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ooi E. M., Watts G. F., Farvid M. S., Chan D. C., Allen M. C., Zilko S. R., and Barrett P. H.. 2005. High-density lipoprotein apolipoprotein A-I kinetics in obesity. Obes. Res. 13: 1008–1016. [DOI] [PubMed] [Google Scholar]
- 45.Michalski A., Damoc E., Hauschild J. P., Lange O., Wieghaus A., Makarov A., Nagaraj N., Cox J., Mann M., and Horning S.. 2011. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol. Cell. Proteomics. 10: M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orsoni A., Saheb S., Levels J. H., Dallinga-Thie G., Atassi M., Bittar R., Robillard P., Bruckert E., Kontush A., Carrie A., et al. 2011. LDL-apheresis depletes apoE-HDL and pre-beta1-HDL in familial hypercholesterolemia: relevance to atheroprotection. J. Lipid Res. 52: 2304–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miyosawa K., Watanabe Y., Murakami K., Murakami T., Shibata H., Iwashita M., Yamazaki H., Yamazaki K., Ohgiya T., Shibuya K., et al. 2015. New CETP inhibitor K-312 reduces PCSK9 expression: a potential effect on LDL cholesterol metabolism. Am. J. Physiol. Endocrinol . M etab . 309: E177–E190. [DOI] [PubMed] [Google Scholar]
- 48.Singh S. A., Miyosawa K., and Aikawa M.. 2015. Mass spectrometry meets the challenge of understanding the complexity of the lipoproteome: recent findings regarding proteins involved in dyslipidemia and cardiovascular disease. Expert Rev. Proteomics. 12: 519–532. [DOI] [PubMed] [Google Scholar]
- 49.Gallien S., and Domon B.. 2015. Detection and quantification of proteins in clinical samples using high resolution mass spectrometry. Methods. 81: 15–23. [DOI] [PubMed] [Google Scholar]
- 50.Ronsein G. E., Pamir N., von Haller P. D., Kim D. S., Oda M. N., Jarvik G. P., Vaisar T., and Heinecke J. W.. 2015. Parallel reaction monitoring (PRM) and selected reaction monitoring (SRM) exhibit comparable linearity, dynamic range and precision for targeted quantitative HDL proteomics. J. Proteomics. 113: 388–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheung M. C., and Albers J. J.. 1984. Characterization of lipoprotein particles isolated by immunoaffinity chromatography. Particles containing A-I and A-II and particles containing A-I but no A-II. J. Biol. Chem. 259: 12201–12209. [PubMed] [Google Scholar]
- 52.Rye K. A., and Barter P. J.. 2014. Regulation of high-density lipoprotein metabolism. Circ. Res. 114: 143–156. [DOI] [PubMed] [Google Scholar]
- 53.Mahley R. W., Innerarity T. L., Pitas R. E., Weisgraber K. H., Brown J. H., and Gross E.. 1977. Inhibition of lipoprotein binding to cell surface receptors of fibroblasts following selective modification of arginyl residues in arginine-rich and B apoproteins. J. Biol. Chem. 252: 7279–7287. [PubMed] [Google Scholar]
- 54.Blum C. B. 1982. Dynamics of apolipoprotein E metabolism in humans. J. Lipid Res. 23: 1308–1316. [PubMed] [Google Scholar]
- 55.Ghiselli G., Krishnan S., Beigel Y., and Gotto A. M. Jr. 1986. Plasma metabolism of apolipoprotein A-IV in humans. J. Lipid Res. 27: 813–827. [PubMed] [Google Scholar]
- 56.Brunham L. R., Kruit J. K., Iqbal J., Fievet C., Timmins J. M., Pape T. D., Coburn B. A., Bissada N., Staels B., Groen A. K., et al. 2006. Intestinal ABCA1 directly contributes to HDL biogenesis in vivo. J. Clin. Invest. 116: 1052–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nazih H., Nazih-Sanderson F., Krempf M., Michel Huvelin J., Mercier S., and Marie Bard J.. 2001. Butyrate stimulates ApoA-IV-containing lipoprotein secretion in differentiated Caco-2 cells: role in cholesterol efflux. J. Cell. Biochem. 83: 230–238. [DOI] [PubMed] [Google Scholar]
- 58.Green P. H., Tall A. R., and Glickman R. M.. 1978. Rat intestine secretes discoid high density lipoprotein. J. Clin. Invest. 61: 528–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamaguchi S., Zhang B., Tomonaga T., Seino U., Kanagawa A., Segawa M., Nagasaka H., Suzuki A., Miida T., Yamada S., et al. 2014. Selective evaluation of high density lipoprotein from mouse small intestine by an in situ perfusion technique. J. Lipid Res. 55: 905–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dallinga-Thie G. M., van Tol A., van’t Hooft F. M., and Groot P. H.. 1986. Distribution of apolipoproteins A-I and A-IV among lipoprotein classes in rat mesenteric lymph, fractionated by molecular sieve chromatography. Biochim. Biophys. Acta. 876: 108–115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.