Abstract
Sec14-like phosphatidylinositol transfer proteins (PITPs) play important biological functions in integrating multiple aspects of intracellular lipid metabolism with phosphatidylinositol-4-phosphate signaling. As such, these proteins offer new opportunities for highly selective chemical interference with specific phosphoinositide pathways in cells. The first and best characterized small molecule inhibitors of the yeast PITP, Sec14, are nitrophenyl(4-(2-methoxyphenyl)piperazin-1-yl)methanones (NPPMs), and a hallmark feature of NPPMs is their exquisite targeting specificities for Sec14 relative to other closely related Sec14-like PITPs. Our present understanding of Sec14::NPPM binding interactions is based on computational docking and rational loss-of-function approaches. While those approaches have been informative, we still lack an adequate understanding of the basis for the high selectivity of NPPMs among closely related Sec14-like PITPs. Herein, we describe a Sec14 motif, which we term the VV signature, that contributes significantly to the NPPM sensitivity/resistance of Sec14-like phosphatidylinositol (PtdIns)/phosphatidylcholine (PtdCho) transfer proteins. The data not only reveal previously unappreciated determinants that govern Sec14-like PITP sensitivities to NPPMs, but enable predictions of which Sec14-like PtdIns/PtdCho transfer proteins are likely to be NPPM resistant or sensitive based on primary sequence considerations. Finally, the data provide independent evidence in support of previous studies highlighting the importance of Sec14 residue Ser173 in the mechanism by which NPPMs engage and inhibit Sec14-like PITPs.
Keywords: lipid signaling, lipid transfer proteins, phospholipids, phosphatidylinositol transfer protein, phosphatidylcholine, phosphoinositides
Phosphoinositides (PIPs) are phosphorylated derivatives of phosphatidylinositol (PtdIns), and the metabolism of these lipids constitutes a major membrane-associated signaling system in eukaryotic cells (1–5). The chemical heterogeneity that distinguishes individual PIP species forms one basis for functionally compartmentalizing signaling platform identities on membrane surfaces (6, 7). Yet while the chemical heterogeneity of PIP species is simple, it translates to an enormous diversity of biological outcomes that derive from PIP signaling. In that regard, recent studies demonstrate additional layers of functional specification for PIP signaling that are of such resolution that production of an individual PIP species by a specific PtdIns kinase yields multiple biological outcomes in the same cell (8). We now appreciate that PtdIns transfer proteins (PITPs) play critical roles in functional compartmentalization of PIP signaling reactions by channeling PtdIns to PtdIns kinases and, subsequently, to distinct sets of effector proteins (9–11). The Sec14-like PITPs are best studied in this regard, and the multiplicity of Sec14-like PITPs expressed in even simple unicellular eukaryotes highlights the high degree of functional specification for these proteins (12, 13).
Emerging evidence that PITPs instruct the biological outcomes of PtdIns kinase activities recommends these proteins as novel targets for chemical intervention with PIP signaling pathways in cells (11, 14). The advantages of targeting PITPs for this purpose are that such interventions can be imposed with selectivities superior to those possible by popular strategies that either target individual PtdIns-kinase isoforms or individual PIP species (15, 16). Proof of concept is exemplified by the identification and validation of several classes of small molecule inhibitors (SMIs) that target Sec14, the major PITP of yeast. Sec14 binds both PtdIns and phosphatidylcholine (PtdCho) in vitro, and it coordinates PtdIns-4-phosphate production with PtdCho metabolism so as to promote membrane trafficking through trans-Golgi network and endosomal compartments in vivo (17–19). Of the SMIs that target Sec14 activity, the nitrophenyl(4-(2-methoxyphenyl)piperazin-1-yl)methanones (NPPMs) are best understood (14, 20, 21). These SMIs inhibit Sec14 by loading into the phospholipid binding pocket of this PITP and preventing subsequent rounds of lipid exchange (14).
An attractive property of NPPMs is their exquisite selectivity for Sec14 as a target. Even though yeast expresses five other Sec14-like PITPs (13), none of these proteins are at all sensitive to inhibition by NPPMs. This selectivity is, in most cases, accounted for by the mechanism by which NPPMs are bound by Sec14. Available data project that NPPM invades the hydrophobic lipid-binding pocket of Sec14 during a lipid-exchange cycle where the inhibitor engages both residues within PtdIns- and PtdCho-acyl chain space and residues essential for coordination of the PtdCho headgroup. Computational docking simulations, coupled with rational mutagenesis experiments, indicate the latter binding interactions are crucial. The NPPM-activated aryl halide moiety is projected to engage residue Ser173, a key component of the PtdCho headgroup-coordinating substructure, via a halogen bond (14). Because four of the other five yeast Sec14-like PITPs (Sfh2–Sfh5) do not conserve this PtdCho-coordinating unit (9), the NPPM resistance of those proteins is readily explained by their lack of the structural elements essential for NPPM binding.
The Sec14 specificity of NPPM remains completely obscure in other cases. For example, the Sec14-like PITP, Sfh1, is highly homologous to Sec14. Sfh1 has both PtdIns- and PtdCho-binding/transfer activities, as it shares with Sec14 the same PtdIns-binding motif and, more strikingly, the same PtdCho-coordinating substructure that is critical for NPPM binding (9, 13, 22). Yet, Sfh1 lipid exchange activities are impervious to NPPM challenge (14). Moreover, Sec14-like PtdIns/PtdCho-transfer proteins from other fungal pathogens are even more similar to Sec14 than is Sfh1, but are similarly resistant to inhibition by NPPMs (see Results). Those exceptional cases indicate that, despite a detailed and experimentally well-supported description for how NPPMs dock into the Sec14 lipid-binding pocket (14), there remain substantial gaps in our knowledge regarding the mechanism of Sec14 inhibition by NPPMs. Herein, we exploit unbiased approaches to identify a new Sec14 motif, the VV signature, that contributes to NPPM sensitivity. The data empower prediction of which Sec14-like PtdIns/PtdCho-transfer proteins are likely to be NPPM resistant or sensitive by extending our understanding of the underlying mechanisms that govern the sensitivities of Sec14-like PITPs to NPPMs.
MATERIALS AND METHODS
Yeast media and methods
Yeast complex and minimal media and genetic methods followed standard procedures (23–25). Yeast strains used in this study included the wild-type strain CTY182 (MATa ura3-52 lys2-801 his3Δ-200) and its isogenic sec14-1ts and spo14Δ derivatives, CTY1-1A (MATa ura3-52 lys2-801 his3Δ-200 s14-1ts), and CTY1092 (MATa ura3-52 lys2-801 his3 his3Δ-200 spo14Δ), respectively (26, 27). An isogenic pdr5Δ derivative of strain CTY1079 was designated YKM03 (MATa ura3-52 lys2-801 his3Δ-200 spo14Δ pdr5Δ) (this study). The gene replacement cassette plasmids used are listed in supplementary Table 1.
Small molecule inhibitors
Small molecules of interest were purchased from ChemBridge Chemical Store (San Diego, CA.), dissolved in DMSO to a final stock concentration of 20 mM, and stored in the dark at −20°C.
Unbiased cell-based screen
Spontaneous NPPMR mutants were generated using yeast strain CTY1079 (MATa ura3-52 lys2-801 his3Δ-200 spo14Δ SEC14) or the isogenic pdr5Δ strain, YKM03, and seeded onto YPD plates individually supplemented with the appropriate NPPM at concentrations of 10 μM and 2 μM, respectively. Approximately 1 × 107 cells from 45 independent overnight cultures were each seeded onto a YPD agar plate, and the seeded plates were incubated at 30°C for 96 h. One colony was picked from each plate and purified by two rounds of dilution streaking for isolated colonies. The NPPMR phenotype of each mutant colony was verified on YPD agar supplemented with NPPM and the colonies were expanded to generate individual frozen stock cultures.
Amplification and DNA sequencing of SEC14 genes from isolated NPPMR yeast isolates
Genomic DNA from each independently isolated NPPMR mutant was prepared and the SEC14 gene amplified via PCR using the DKO98 and 99 primer pair (supplementary Table 2). The SEC14 nucleotide sequences were determined in both directions (Eton), and aligned using the multiple sequence alignment program, Clustal Omega, with wild-type SEC14 as query sequence (27, 28).
Protein expression construct generation
NPPMR missense mutations and VV bar code mutants were generated by site-directed mutagenesis using pET28b (His8-SEC14) as mutagenesis substrate. Site-directed mutations were generated using QuikChange II (Stratagene) and confirmed by nucleotide sequencing. SEC14CA was amplified from genomic DNA via two rounds of PCR using the DKO1,2 and DKO14,15 primer sets (supplementary Table 2) and subcloned into pET28b, taking advantage of plasmid NcoI and SacI restriction sites. SEC14KL was amplified using oligonucleotides KL100,101 and subcloned into the SacII and SphI restriction sites of plasmid pVB16. An internal NcoI restriction site in SEC14KL was destroyed by incorporating a sense mutation using the KL92,93 mutagenic primer pair followed by a second round of amplification using primers KL90,91 to incorporate an N-terminal His8 tag. That PCR product was subcloned as an NcoI-SacI restriction fragment into pET28b. The SEC14CG gene was similarly amplified using oligonucleotides CG86,87 as primers, subcloned into pVB16, and a natural NcoI site was removed by site-directed mutagenesis. The modified gene was amplified in a second round using CG89,90 as primer pair and the amplified gene was subcloned into pET28b as an NcoI-Sac1 restriction fragment. Primer sequences are listed in supplementary Table 2. Protein expression plasmids used in this study are listed in supplementary Table 3.
Protein purification
Recombinant proteins were purified essentially as described (9). Briefly, Escherichia coli BL21 (DE3) cells carrying the appropriate expression plasmids were incubated at 37°C with shaking until cultures reached the desired cell densities (A600 = 0.8). Recombinant protein expression was induced with 60 μM isopropyl β-D-1-thiogalactopyranoside and cultures were incubated for an additional 18 h at 16°C. Cells were pelleted, resuspended in 300 mM NaCl, 25 mM Na2HPO4, and 1 mM phenylmethanesulfonylfluoride (pH 7.8), and subsequently disrupted by two successive passages through a French press at 10,000 psi. Cell-free lysates were clarified by serial centrifugations at 2,800 g (20 min) and 27,000 g (60 min). Clarified lysates were incubated with Co2+ TALON metal affinity beads overnight at 4°C with agitation, and washed exhaustively with 300 mM NaCl, 25 mM Na2HPO4, 5 mM 2-mercapthoethanol, and 5 mM imidazole (pH 7.8). Bound proteins were eluted with a continuous 10–200 mM imidazole gradient in 300 mM NaCl, 25 mM Na2HPO4, and 5 mM 2-mercapthoethanol (pH 7.8). Peak fractions were pooled, dialyzed against 300 mM NaCl, 25 mM Na2HPO4 (pH 7.8), and 5 mM 2-mercaptoethanol, and concentrated by using Amicon Ultra filter centrifugation (EMD Millipore). Protein concentrations were estimated by SDS-PAGE and visual comparisons to BSA titration series, and by A280 measurements.
PtdIns transfer assays
Assays were performed by previously established methods (9, 14). Recombinant Sec14 proteins were preincubated with acceptor membranes in 300 mM NaCl and 25 mM Na2HPO4 (pH 7.5) and SMI or DMSO (vehicle control), as appropriate, for 30 min at 37°C. Donor membranes (rat-liver microsomes) were added to initiate the assay, and reactions were incubated for an additional 30 min at 37°C. [3H]PtdIns transfer activities in the presence of SMIs were normalized to mock DMSO controls.
Preparation of structural files for docking simulations
A homology model for the closed Sec14 conformer was generated based on structural templates for both the open Sec14 conformer [Protein Data Bank (PDB) identification, 1AUA] and the closed conformer of Sfh1 bound to PtdIns (PDB ID, 3B7N). Proteins were prepared for docking using the Protein Preparation Wizard panel in Schrödinger Suite and complete structure was optimized to relieve all atom and bond constraints after adding all side chains and missing atoms (29).
Docking simulations
Two independent docking platforms were used. Computational docking used the genetic algorithm-based ligand docking program, GOLD (version 5.2.1), which exhaustively explores ligand conformations and provides limited flexibility to protein side chains with -OH groups by reorienting the H-bond donor and acceptor moieties. The GOLD scoring function is based on favorable conformations documented in the Cambridge Structural Database, and on empirical results of weak chemical interactions. The active site was defined by a single solvent accessible point near the center of the protein active site, a radius of ∼10 Å, and the GOLD cavity detection algorithm. To maintain consistency in docking, Sfh1 crystal structure was superposed on Sec14 coordinates and the binding cavity was defined using the same centroid that was used to define the Sec14 binding pocket. This ensured that the binding site interaction grids generated before docking shared relatable spatial coordinates and allowed direct comparisons of Sfh1 and Sec14 6748-481 docking results. GOLD docking was unconstrained to obtain unbiased results and to explore all possible ligand binding modes. Ligand was docked in independent runs, the 10 best solutions were produced for each run, and early termination of ligand docking was switched off. All other parameters used default settings (30).
For Glide docking routines, proteins and ligands were prepared using Protein Preparation Wizard and the LigPrep module of Maestro 10.1 Interface of Schrödinger Suite (Schrödinger Suite 2015, Glide version 6.6). Receptor grids were generated without constraints and with standard settings. Docking was performed using XP scoring function. No similarity torsional and inter-molecular interaction (hydrogen bonding or hydrophobic) constraints were used. Ligand was docked flexibly with nitrogen inversions and ring sampling turned on with post-docking minimization (31).
Molecular mechanics with generalized born and surface area
Molecular mechanics with generalized born and surface area (MM-GBSA) solvation is a widely applied method to calculate the free energy of the binding of ligands to proteins. A MM-GBSA approach was applied to calculate ligand binding energies and ligand strain energies in wild-type and mutant protein-ligand complexes. The docked solutions generated from GOLD and Glide ligand docking were used for calculating ligand binding energies of complexes. Residues within a 10 Å radius from the ligand were included in the flexible region with implicit solvent. MM-GBSA is implemented in the Prime module of the Schrödinger software package (32). Structural coordinates of Sec14::NPPM complexes overlayed on model Sec14 structures from Candida albicans, Candida glabrata, and Kluyveromyces lactis in PDB format are available upon request.
RESULTS
Comparisons of NPPM 6748-481 docking poses in Sec14 versus Sfh1
We sought to capitalize on high resolution Sfh1 crystal structures and a detailed computational model for how Sec14 binds NPPM 6748-481 to produce a NPPM-sensitive Sfh1 for the purpose of direct structural analysis of an Sfh1::NPPM complex. This approach was recommended by the fact that the modeled NPPM-binding pose in the Sec14 hydrophobic pocket is experimentally supported by rational mutagenesis and chemical structure-activity relationship data (14), that the Sfh1 and Sec14 binding pockets share 79% primary sequence identity and 89% similarity in the residues that form the boundaries of their respective phospholipid binding cavities, and that those two PITPs share essentially identical phospholipid binding properties (9). The high degree of structural conservation between these two cavities is further emphasized by the 1.06 Å root mean square deviation (RMSD) calculated after superposition of the two cavity structures. In those comparative analyses, the binding pockets were defined by the binocular criteria of Sfh1 versus Sec14 structural alignments and residues of interest falling within 4.5 Å of bound 6748-481 (as represented by the experimentally favored Sec14::NPPM 6748-481 pose previously described) (14).
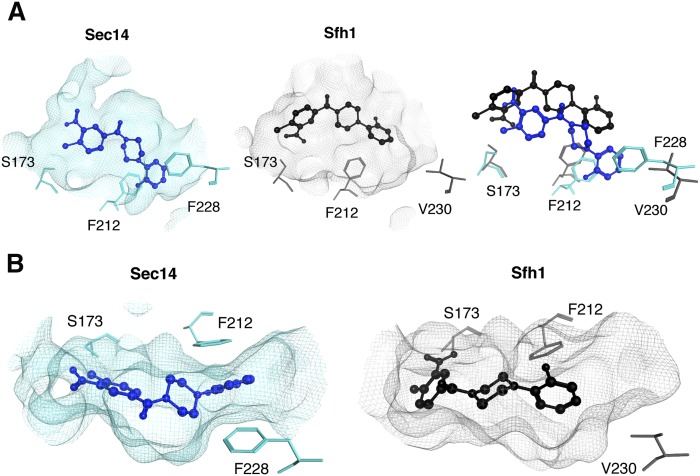
To query why Sfh1 is naturally resistant to inhibition by NPPMs, 6748-481 was docked into the phospholipid-binding cavity of Sfh1 (see Materials and Methods), and those results were compared with the model pose for 6748-481 bound in the Sec14 hydrophobic pocket. First, the Sfh1 crystal structure was superposed on Sec14 coordinates and the respective phospholipid-binding cavities were related by defining a reference centroid within the superposed cavity environments. As had previously been done with Sec14 (14), the Sfh1 pocket environment was defined as the sampling space for 6748-481 docking simulations. The reference centroid ensured that the Sec14 and Sfh1 interaction grids shared relatable spatial coordinates, thereby allowing direct comparisons of Sfh1 and Sec14 6748-481 docking results. Comparison of the highest-scoring 6748-481 dock pose indicated the Sfh1 solution shared the same coordinate space within the hydrophobic pocket, and a similar conformation as that calculated for Sec14. That is, the chloro-nitrophenyl group of 6748-481 was oriented toward the polar subregion of the pocket (Fig. 1A). However, unlike the case of the Sec14::6748-481 complex, where the chloro-nitrophenyl headgroup coordinates with the S173 -OH group, the Sfh1::6748-481 complex has the NPPM headgroup rotated 180° around the single phenyl-acyl rotatable bond that links the chloro-nitrophenyl headgroup moiety with the ketone functional group of the NPPM. This same rotamer was also represented in potential docking solutions for NPPM 6748-481 with Sec14, and it was initially considered a plausible binding mode pending experimental test (14). Short MDS runs (10 ns) showed the rotamer readily flips its headgroup orientation to adopt the 6748-481 pose in the Sec14 cavity supported by experiment (supplementary Movie 1). Nonetheless, the Sec14 and Sfh1 6748-481 docking poses remain significantly different (RMSD > 3 Å, Fig. 1B). Of particular note, the distal fluoro-benzyl moiety of 6748-481 is wedged in a deep hydrophobic cleft of the Sec14 lipid-binding cavity where it engages in stacking interactions with residues F228 and F212. In the Sfh1 case, those hydrophobic interactions are not evident (Fig. 1B), and this disparity reflects the fact that the Sec14 and Sfh1 pocket residues diverge most significantly along this subregion of their respective lipid-binding cavities.
Fig. 1.
Comparisons of NPPM 6748-481 docking poses in the hydrophobic pockets of Sec14 and Sfh1. A: Modeled NPPM 6748-481 poses within the hydrophobic pockets of Sec14 (left panel, blue) and Sfh1 (middle panel, black) are shown. Residues of interest are labeled. The right panel shows a superposition of the binding modes in the two distinct pocket environments and highlights the differences between the two binding modes. Sec14 residue F228 is projected to stabilize NPPM binding via stacking interactions, and these interactions are not available in the Sfh1 context. B: Top views of NPPM-481 docked to Sec14 (left panel) and Sfh1 (right panel) are shown. The protein pocket is rendered in surface mesh with residues of interest highlighted in ball and stick representation.
Rational structure-based engineering of the Sfh1 pocket is insufficient to produce an NPPM-sensitive Sfh1
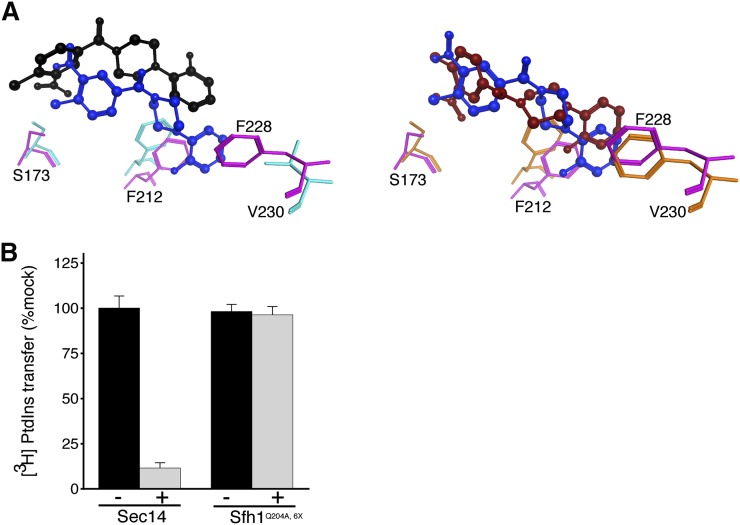
We sought to use the structural and 6748-481 docking information to design an Sfh1 with a Sec14-like pocket with the expectation that such an Sfh1 variant would now acquire NPPM sensitivity. To that end, structural and primary sequence alignments of Sfh1 and Sec14 identified six divergent residues in the cohort of amino acids that line the binding pocket and lie within 4.5 Å of the “bound” 6748-481. Except for F156 (as numbered in Sfh1) most of the nonconserved residues are positioned toward the hydrophobic subregion of the proposed NPPM binding site (Fig. 2A). Those six residues were converted en bloc to the cognate residues in Sec14 (F153V, L176M, I193V, V196A, A197S, and V227F) to generate what we term the Sfh16X mutant. Computational docking experiments were then run using the same parameters and settings as those employed in the Sfh1/Sec14 comparative docking experiments. Gratifyingly, the highest ranked docking solution was much more similar to the favored Sec14::6748-481 pose (RMSD = 1.0 Å), and the distal fluoro-benzyl group now intercalated into the hydrophobic cleft where it engaged in stacking interaction with the newly converted F230 (Fig. 2A). While, the chloro-nitrophenyl headgroup of 6748-481 still adopted the 180° rotamer conformation relative to the Sec14::6748-481 pose, we considered this a conformer that could rotate the activated aryl-halide functional group to a pose nearly identical to that which we predict describes bound 6748-481 in the Sec14 lipid-binding cavity (14).
Fig. 2.
A rational approach for engineering Sfh1 sensitivity to NPPM 6748-481. A: Superposition of the 6748-481 poses in Sec14 (blue) and Sfh1 (black) binding pockets is shown in ball and stick representation (left panel). This is to be compared with the improved superposition of 6748-481 poses in Sec14 (blue, ball and stick) versus Sfh16X (red, ball and stick) binding pockets (right panel). Key residues in the Sfh1 and Sfh16X binding pockets are represented in orange and magenta, respectively. B: Biochemical characterization of purified recombinant proteins is shown. Sec14/Sfh1Q204A,6X activity was measured in in vitro [3H]PtdIns transfer assays, in the absence (black bars) and presence (gray bars) of 6748-481. The proteins were preincubated with DMSO or 20 μM 6748-481 in the presence of PtdCho liposomes, as appropriate, prior to assay (see Materials and Methods). The values reported for assays with inhibitor are related to mock DMSO vehicle controls. Average values and standard deviations are given (n = 3). The intrinsic PtdIns-transfer activities measured for Sec14Q204,6X and other assay statistics are provided in supplementary Table 4.
To test whether the reengineered protein was sensitive to inhibition by NPPM, a Q204A missense substitution was incorporated into the Sfh16X protein. This was done because Q204A endows Sfh1 the desirable properties of stimulated lipid exchange activity in vitro and enhanced Sec14-like properties in vivo (14, 22). Those features made it an ideal experimental scaffold for manipulating, and subsequently monitoring, the NPPM-resistance/sensitivity properties of Sfh1 and its variants. Surprisingly, while purified recombinant Sfh1Q204A,6X exhibited robust [3H]PtdIns transfer activity in vitro, this activity remained indifferent to a high concentration of 6748-481 (20 μM, ∼100 times Sec14 IC50; Fig. 2B).
Genetic screen for Sec14 variants resistant to NPPM
The inadequacy of the working Sec14::NPPM 6748-481 docking solution in guiding successful engineering of an NPPM-sensitive version of Sfh1 indicated that our understanding of the inhibitor binding mechanism was, at best, incomplete or, at worst, incorrect. We therefore sought to gain additional insight regarding mechanisms of NPPM-binding by Sec14 (and NPPM-resistance for Sfh1) via an approach that did not rely on the necessarily targeted logic inherent to structure-based regimes. To that end, an unbiased genetic screen was performed that selected for mutant Sec14s ablated for NPPM sensitivity, while at the same time demanding maintenance of biologically sufficient levels of protein activity. One important consideration in designing the screen was limiting re-isolation of previously characterized “bypass Sec14” mutations that represented loss-of-function mutations in structural genes of the CDP-choline pathway for PtdCho biosynthesis (17), the SAC1 gene which encodes the major yeast PtdIns-4-P phosphatase (33–35) or the sterol- and PtdIns-4-P-binding protein, Kes1/Osh4 (19, 36–38). These bypass Sec14 mutations occur spontaneously at high frequencies [∼5 × 10−5 per cell per generation (17, 33)], would pass selection by conferring viability to yeast cells deficient in normally essential Sec14 activities (14, 21), and dominate the results of the screen. We therefore took advantage of the fact that all known bypass Sec14 mechanisms require activity of the normally nonessential phospholipase D enzyme encoded by the SPO14 gene to alleviate the cellular Sec14 requirement, and spo14Δ yeasts fail to yield spontaneously occurring bypass Sec14 mutants at all [<10−10 per cell per generation (26)]. Thus, the NPPMR screen was conducted using an otherwise wild-type spo14Δ yeast mutant as parental strain to preclude any background of known bypass Sec14 mutants.
Parental cells were seeded onto YPD agar plates individually supplemented with NPPMs, 6748-481, 4130-1276, or 67170-49, to final concentrations of 10 μM (supplementary Fig. 1A). While these NPPMs vary in their potencies as Sec14 inhibitors (6748-481 > 67170-49 > 4130-1276), all three NPPMs strongly inhibited yeast cell proliferation when incorporated in growth medium at this concentration. After incubation at 30°C for 96 h, the frequencies of emerging NPPMR colonies were scored. As expected, those frequencies were inversely proportional to NPPM potency as Sec14 inhibitor. Colonies resistant to 4130-1276 or 67179-49 intoxication arose at frequencies of ∼3 × 10−5 and 10−6 per cell per generation, respectively, and those resistant clones started appearing within 48 h of incubation. By contrast, colonies resistant to the most potent inhibitor, 6748-481, appeared only after approximately 96 h of incubation, and those colonies emerged at much lower frequencies (∼10−7 per cell per generation) (supplementary Fig. 1B). A parallel 6748-481 resistance selection was also performed in an isogenic pdr5Δ genetic background where the structural gene for the major yeast drug pump was deleted (39). In that version of the screen, the 6748-481 concentration for selection was lowered to 2 μM, as the pdr5Δ parental strain is approximately five times more sensitive to the inhibitor (unpublished observations). The frequencies of the 6748-481R colonies obtained in this sensitized selection regime were reduced even further (∼3 × 10−8 per cell per generation). As existing Sec14::NPPM docking poses were built using 6748-481 as ligand (14), subsequent analyses were restricted to yeast isolates resistant to NPPM 6748-481. A total of 45 independently isolated 6748-481-resistant clones were recovered from the two parallel screens, these clones were purified by at least two rounds of single colony isolation, and were analyzed in further detail.
Sec14 missense substitutions that confer NPPM resistance
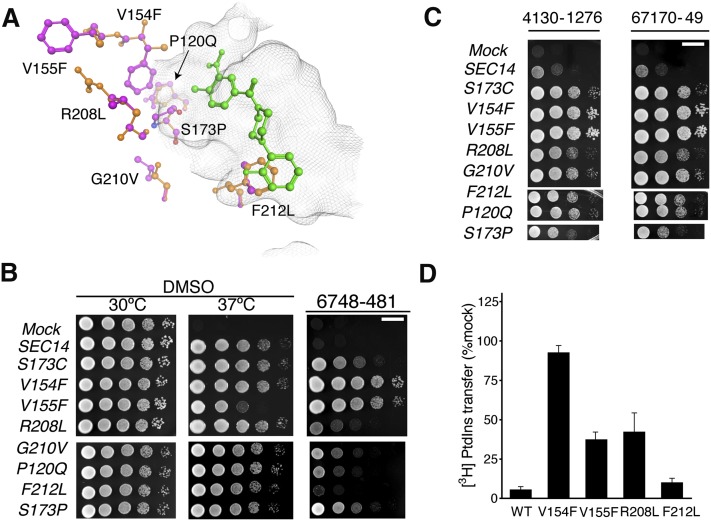
The SEC14 gene was amplified from each of the isolates by PCR using genomic DNA as template, and the nucleotide sequences of the recovered SEC14 genes were determined in their entirety. Of the genes so analyzed, 35 exhibited wild-type SEC14 sequence, while the remaining 10 carried single missense mutations in SEC14. Those 10 mutant SEC14 genes represented a total of seven unique SEC14 missense mutations that resulted in the following single amino acid changes to the Sec14 protein sequence: P120Q, V154F, V155F, S173P, R208L, G210V, and F212L. Superposition of the NPPMR missense substitutions onto high resolution Sec14 structural models demonstrated that, consistent with our previous in silico docking solution (14), all substitutions involved residues positioned in the immediate vicinity of the Sec14 lipid binding pocket (Fig. 3A). Indeed, we had previously identified S173 and F212 in Sec14::NPPM interaction fingerprint analyses where those two residues engaged the activated aryl halide and fluoro-benzyl moieties of NPPM 6748-481 via polar and hydrophobic/π-π stacking interactions, respectively (14). Recovery of Sec14S173P was particularly satisfying, as this variant was altered for the very residue identified by our previous docking simulations and rational mutagenesis studies as essential for the halogen-bonding mechanism that we propose governs Sec14::NPPM binding interactions (14). By contrast, the remaining five residues exhibited either weak scores in those analyses (V154 and R208) or were excluded from the interaction fingerprint list entirely (P120, V155, and G210).
Fig. 3.
Functional characterization of NPPM-resistant Sec14 proteins. A: The seven independently isolated Sec14 missense substitutions identified in NPPMR isolates are highlighted in ball and stick representation with mutant residues depicted in magenta and the corresponding wild-type residues in orange. The positions of the corresponding side chains are related to the NPPM 6748-481 pose depicted in green ball and stick. The surface of the Sec14 hydrophobic pocket is rendered as a gray wire-mesh. B: The indicated SEC14 genes were integrated into the LEU2 locus of a sec14-1ts strain and expressed under the control of the S. cerevisiae SEC14 promoter to generate strains exhibiting “physiological” levels of each Sec14 variant. The integrants were subsequently dilution spotted onto YPD plates supplemented with vehicle control DMSO or 20 μM NPPM, as indicated at top, and incubated for 48 h at the indicated temperatures. The mock condition documents the phenotype of an isogenic strain where a SEC14-less integration cassette was transplaced into the LEU2 locus. That expression of each Sec14 protein was sufficient to rescue sec14-1ts growth defects at the restrictive temperature of 37°C is demonstrated by comparison of the growth profiles in the left (30°C) and center (37°C) panels of the integrants relative to mock controls. The NPPMR phenotypes are displayed in the right panel. The plates were incubated for 48 h at the indicated temperatures before imaging. The NPPM481-resistance phenotypes were scored at 30°C. Note that R208L and F212L were weak mutants. Scale bar, 1 cm. C: Indicates that spontaneous mutants generated in response to 6748-481 confer pan-NPPM resistance to Sec14. The experiment is the same as above in (B) except that the plates were supplemented with NPPMs 4130-1276 and 67170-49, as indicated (20 μM final concentration). The mock condition documents the phenotype of an isogenic strain where a SEC14-less integration cassette was transplaced into the LEU2 locus. Scale bar, 1 cm. (D) NPPMR Sec14 proteins were expressed and purified as recombinant proteins and [3H]PtdIns transfer activities were measured in vitro in the presence of 20 μM NPPM. Protein concentration was clamped at 287 nM. Inhibition was normalized to the DMSO vehicle control. Values are representative of mean ± SEM; triplicate measurements from three independent experiments. The intrinsic PtdIns-transfer activities measured for each Sec14R and other assay statistics are provided in supplementary Table 5.
To verify that these seven missense substitutions were sufficient to confer NPPM 6748-481 resistance to Sec14, each corresponding mutation was individually incorporated into the genome of the naïve parental strain (SPO14+ in this case) and resistance to NPPM 6748-481 was subsequently queried as an unselected phenotype. Survey of NPPMR as an unselected phenotype circumvented the possibility of mistaking that phenotype for a bypass Sec14 phenotype. In every case, the mutation of interest was sufficient to render the recipient yeast strain resistant to the NPPM 6748-481 concentration used as selection filter in the NPPMR screen, irrespective of whether the naïve recipient was of a SPO14+ (Fig. 3B) or a spo14Δ (supplementary Fig. 2) genetic background. However, in both genetic backgrounds, we noted that the NPPMR phenotype of the SEC14F212L allele was very weak. Each SEC14R allele held the added feature of conferring an unselected NPPM 4130-1276 and 67170-49 resistance to naïve yeast cells and, when interrogated in the context of these NPPMs, the resistance phenotype associated with the SEC14F212L allele was clear (Fig. 3C). We inferred from the collective data that the mutant Sec14Rs were rendered insensitive to challenge with any of these three NPPMs.
SEC14R gene products exhibit NPPMR lipid exchange activities
As Sec14 is required for yeast cell viability (17, 27, 33), the NPPMR screen demanded that any Sec14R proteins passing through the screen must retain sufficient PtdIns- and PtdCho-exchange activities for biological function, but that those activities would be intrinsically resistant (at least to a substantial degree) to inhibition by NPPM. To examine those biochemical properties directly, representative Sec14Rs were produced as recombinant protein in bacteria, purified, and the NPPM-resistance properties tested in vitro. We were unable to analyze Sec14S173P or Sec14P120Q in this manner because those two mutant proteins exhibited insufficient activity for confident analysis when purified from the bacterial host (unpublished observations). As expected, the Sec14R proteins exhibited a range of basal PtdIns transfer activities (supplementary Table 5).
Given previous analyses of Sec14S173C and Sec14S173A substitutions whose design was guided by the Sec14::NPPM 6748-481 docking pose (14), the Sec14V154F was of particular interest as it was impervious to challenge with 20 μM NPPM 6748-481 in vitro (Fig. 3D). Indeed, it exhibited an essentially complete resistance to NPPM 6748-481 challenge at near saturating aqueous concentrations of inhibitor (100 μM; data not shown) even though V154 was not identified by our working Sec14::NPPM 6748-481 pose as a residue critical to the mechanism by which the NPPM is coordinated within the Sec14 lipid-binding cavity (14). As shown in Fig. 3D, the [3H]PtdIns transfer activities of Sec14V155F and Sec14R208L were also significantly more resistant to 6748-481 challenge compared with Sec14 in transfer assays spiked with 20 μM NPPM, but all those proteins nevertheless retained measurable sensitivities to the NPPM at this concentration [∼100 times IC50 Sec14 (14)]. In line with the weakness of the associated NPPMR phenotype, Sec14F212L was the least resistant to inhibition by NPPM481 at this concentration of inhibitor. The PtdIns transfer activity of this mutant exhibited an IC50 (642.7 ± 1.2 nM) that was only modestly increased relative to that of wild-type Sec14 protein (287.8 ± 1.1 nM).
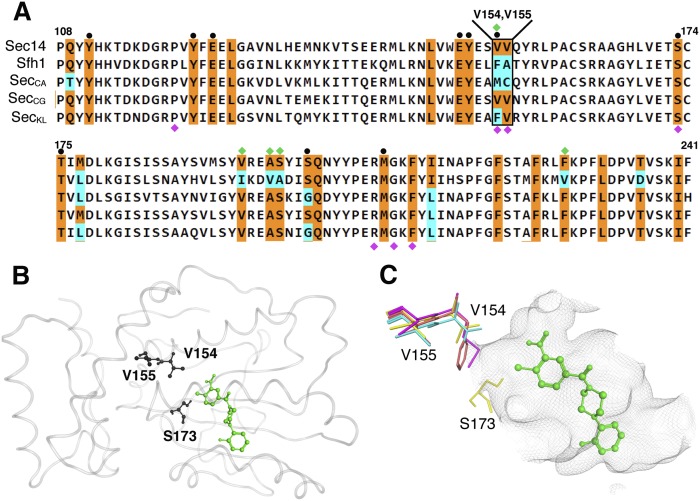
A VV signature correlated with NPPM sensitivity
Of the seven Sec14 residues identified in the unbiased NPPM-resistance screen, we noted that P120, S173, R208, G210, and F212 are precisely conserved between Sec14 and Sfh1. However, V154 and V155 are not conserved at all (Fig. 4A). The Sec14 V154V155 motif is diverged to F156A157 in Sfh1 and, strikingly, this very V154F substitution conferred upon Sec14 an essentially complete resistance to inhibition by NPPMs (see above). We therefore investigated the possibility that the Sec14 V154V155 motif is a signature of NPPM-sensitive Sec14-like PtdIns/PtdCho-transfer proteins. To that end, primary sequence alignments highlighting residues that line the Sec14 and Sfh1 phospholipid-binding pockets were generated. Those alignments were subsequently compared with the corresponding residues projected to line the phospholipid-binding cavities of Sec14 orthologs from C. albicans (Sec14CA), C. glabrata (Sec14CG), and K. lactis (Sec14KL). The essentially complete conservation, across this set of proteins, of the distinct substructure that coordinates PtdCho-binding emphasizes, yet again, the close similarities of the various pocket architectures (Fig. 4A). Even in this extended analysis, five out of seven residues identified in the NPPMR screen (P120, S173, R208, G210, and F212) are absolutely preserved across this cohort of Sec14-like PITPs. By contrast, while the Sec14 V154V155 motif is conserved between Saccharomyces cerevisiae and C. glabrata (V152V153 in the latter), K. lactis exhibits an F152V153 version, whereas the motif is diverged to M154C155 in C. albicans. (Fig. 4A). Although the Sec14 V154V155 motif is not predicted to interact directly with bound NPPM 6748-481 (Fig. 4B), we noted that the cognate V154 polymorphisms in K. lactis and C. albicans Sec14 proteins (F and M, respectively) had the effect of extending hydrophobic side chains deeper into the hydrophilic microenvironment that defines the PtdCho-headgroup coordination substructure of Sec14 (Fig. 4C).
Fig. 4.
The Sec14 VV motif is not conserved among closely related Sec14 PtdIns/PtdCho-transfer proteins. A: Shown is a partial primary sequence alignment of S. cerevisiae Sec14 with Sfh1 and Sec14 PITPs from C. albicans, C. glabrata, and K. lactis. The residues corresponding to the lipid-binding cavity are highlighted in color. Residues conserved between S. cerevisiae Sec14 and other Sec14-like proteins are colored in orange with nonconserved residues in cyan. At top, the Sec14 VV motif is identified. For reference, PtdCho-binding barcode residues (black circles) and residues altered to produce the Sfh16X mutant (green diamonds) are also highlighted at top. Labeled at bottom are residues where substitutions confer NPPM resistance to Sec14 (magenta diamonds). B: The S. cerevisiae Sec14::NPPM 6748-481 dock model is shown. Protein is rendered in gray transparent loop with residues V154, V155, and S173 shown in black ball and stick render. NPPM 6748-481 is highlighted in green ball and stick mode. C: A magnified view shows the spatial orientation of the Sec14 V154V155 side chains (in stick render) relative to the model NPPM 6748-481 pose (in green ball and stick mode), residue S173 is indicated, as is the Sec14 hydrophobic pocket surface (rendered as a gray wire-mesh surface). The cognate VV motif residues from S. cerevisiae (yellow), C. albicans (magenta), C. glabrata (cyan), and K. lactis (orange) are overlayed.
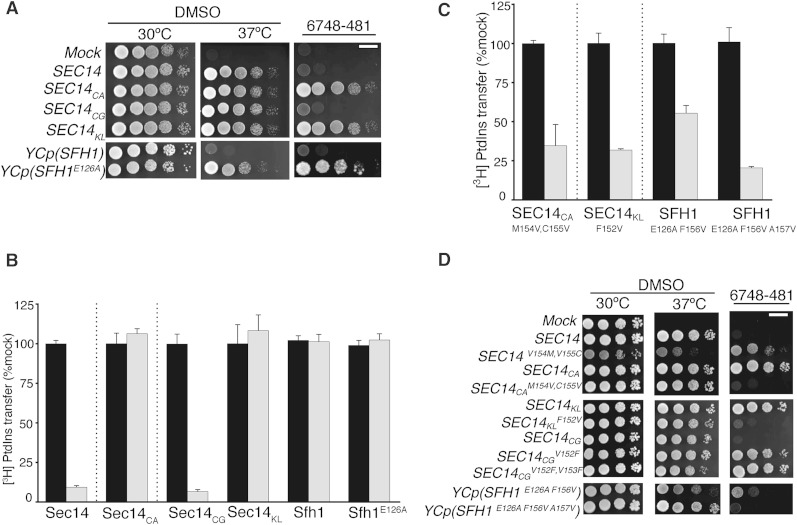
To test the predictive power of the VV signature for NPPM sensitivity of a Sec14 protein, the corresponding SEC14 genes were amplified from genomic DNAs of the corresponding fungal species, the amplified products were subcloned into the appropriate transplacement cassettes, and the genes of interest were individually integrated into the S. cerevisiae genome. All of those Sec14 orthologs scored as functional proteins when expressed in yeast, as judged by their abilities to rescue growth of sec14-1ts mutants on YPD agar at a restrictive temperature of 37°C (Fig. 5A). When these yeast strains were challenged with 20 μM NPPM 6748-481, the Sec14- and Sec14CG-expressing strains failed to proliferate, whereas yeast expressing Sec14KL or Sec14CA were indifferent to the chemical challenge. We inferred from those results that Sec14CG was sensitive to NPPM 6748-481, whereas Sec14KL and Sec14CA were not (Fig. 5A). As control, the Sfh1E126A protein was similarly analyzed, and proliferation of yeast expressing this protein was insensitive to inhibition by NPPM 6748-481 (Fig. 5A). The inferences derived from in vivo data were confirmed by in vitro [3H]PtdIns transfer assays that directly assessed the intrinsic sensitivities of these Sec14 proteins to NPPM 6748-481. Whereas Sec14CG activity was strongly inhibited by NPPM 6748-481 to an extent similar to that observed for S. cerevisiae Sec14, activities of the Sec14CA and Sfh1E126A proteins were completely impervious to challenge with this compound. Sec14KL, which has an incomplete VV signature (FV), was also resistant to inhibition by NPPM 6748-481 in vitro (Fig. 5B). These collective data report a strong correlation between the VV signature and NPPM sensitivity in closely related Sec14 proteins.
Fig. 5.
The VV motif is a signature of NPPM-sensitive yeast Sec14-like PtdIns/PtdCho-transfer proteins. A (top panels): SEC14 genes from the indicated fungal species were integrated into the LEU2 locus of a sec14-1ts strain and expressed under the control of the S. cerevisiae SEC14 promoter to generate strains exhibiting physiological levels of Sec14CA, Sec14CG, and Sec14KL. The integrants were subsequently dilution spotted onto YPD plates supplemented with vehicle control DMSO or NPPM, as indicated at top, and incubated for 48 h at the indicated temperatures. The mock condition documents the phenotype of an isogenic strain where a SEC14-less integration cassette was transplaced into the LEU2 locus. That expression of each heterologous Sec14 protein was sufficient to rescue sec14-1ts growth defects at the restrictive temperature of 37°C is demonstrated by comparison of the growth profiles in the left (30°C) and center (37°C) panels of the integrants relative to mock controls. A (right panel): NPPM 6748-481 resistance or sensitivity of the corresponding integrants was scored by spotting cells on YPD plates supplemented with 20 μM 6748-481 and incubating at 30°C for 48 h, at which time images were taken. The mock condition documents the phenotype of an isogenic strain where a SEC14-less integration cassette was transplaced into the LEU2 locus. The SFH1 control (little Sec14-like function in vivo) and the test SFH1E126A (enhanced Sec14-like function in vivo) were expressed from low-copy centromeric plasmids (YCp) as ectopic expression of this sort was required for visualization of the enhanced Sec14-like properties of Sfh1E126A. B: Characterization of the in vitro sensitivities of purified recombinant Sec14/Sec14-like proteins and the indicated mutant variants is shown. Protein concentrations were clamped at 287 nM throughout and NPPM 6748-481 concentrations were fixed at 20 μM. Values represent the mean ± SEM of triplicate measurements from three independent experiments. The intrinsic PtdIns-transfer activities measured for each Sec14 PITP and other assay statistics are provided in supplementary Table 6. C: Characterization of the in vitro sensitivities of purified recombinant Sec14/Sec14-like proteins with transplaced VV motifs is shown. Protein concentrations were clamped at 287 nM and NPPM 6748-481concentrations were fixed at 20 μM. Values represent the mean ± SEM of triplicate assay determinations from three independent experiments. The intrinsic PtdIns-transfer activities measured for each Sec14 PITP, the indicated variants, and other assay statistics are provided in supplementary Table 6. D: Figure organization and experimental conditions are as described for (A). The abilities of the indicated SEC14 and SFH1 variants to complement sec14-1ts growth defects at 37°C (middle panels) and to endow naïve yeast with resistance to NPPM challenge (right panels) were tested. Left panels represent vehicle growth controls under nonchallenge conditions (30°C), whereas the NPPM challenge plates were also incubated at 30°C. Images were taken after 48 h. The mock condition documents the phenotype of an isogenic strain where a SEC14-less integration cassette was transplaced into the LEU2 locus.
Reconstitution of a VV signature sensitizes Sec14-like PITPs to NPPM
The second approach for examining the relationship between the VV signature and NPPM sensitivity of Sec14-like PtdIns/PtdCho-transfer proteins interrogated whether transplacement of a VV signature into a naturally NPPMR Sec14-like PITP sensitizes the protein to this class of inhibitor. To that end, the divergent M154C155 and F156A157 motifs of Sec14CA and Sfh1E126A, respectively, were each converted to VV signatures. The NPPM-resistance properties of the mutant proteins were subsequently examined in vitro and in vivo. [3H]PtdIns transfer assays demonstrated that purified Sec14CAM154V,C155V had now acquired a significant sensitivity to NPPM 6748-481. While this sensitivity was not as marked as that of Sec14, it was dramatic, nonetheless, when gauged against the complete resistance of the parental Sec14CA to inhibitor. Similarly, whereas partial reconstitution of a VV motif in Sfh1E126A (by incorporation of the single F156V substitution) endowed modest NPPM sensitivity, full reconstitution of a VV signature further enhanced the sensitivity of Sfh1E126A,F156V,A157V to inhibition by NPPM 6748-481 (Fig. 5C). Consistent results were also obtained when the F152V substitution was transplaced into Sec14KL to reconstitute a VV signature in that protein. Although Sec14CGV152F,V153F also showed a reduced sensitivity to NPPM inhibition, as expected, those data came with the caveat that this mutant protein exhibited poor PtdIns- and PtdCho-transfer activity in vitro (unpublished observations). Thus, these substitutions come with a structural cost and, as described above, the mutant Sec14 variants exhibited a range of basal PtdIns-transfer activities in vitro (supplementary Table 6).
The biochemical results were supported by in vivo experiments where S. cerevisiae strains individually expressing Sec14CAM154V,C155V or Sec14KLF152V were tested for sensitivity to intoxication with NPPM 6748-481. Whereas proliferation of yeast cells expressing the wild-type versions of these Sec14 proteins was resistant to inclusion of this NPPM (20 μM) in the culture medium, the growth of cells expressing these same proteins with reconstituted VV signatures was arrested in the presence of inhibitor (Fig. 5D). The same result was observed in the case of Sfh1E126A, where full reconstitution of a VV signature rendered the Sfh1E126A,F156V,A157V sensitive to inhibition by NPPM 6748-481. Reciprocally, conversion of the Saccharomyces Sec14 VV motif to that of the Candida PITP (Sec14V154M and Sec14V154M,V155C) yielded mutant Sec14s that were resistant to NPPM 6748-481 (Fig. 5D). Taken together, the data indicate that the VV signature is not only a strong predictor of NPPM sensitivity in Sec14-like PITPs, but that transplacement of a VV signature into a naturally NPPM-resistant Sec14-like PtdIns/PtdCho-transfer protein is sufficient to confer NPPM sensitivity to that PITP.
Degrees of freedom in the Sec14 VV signature
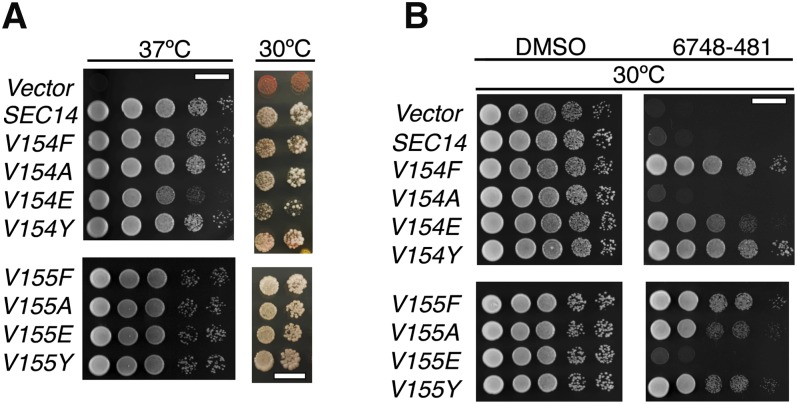
Given that divergence from the VV signature exerted significant effects of the sensitivity of Sec14-like PtdIns/PtdCho-transfer proteins to NPPM 6748-481, we wished to better assess the degrees of freedom permitted in the VV motif associated with NPPM sensitivity of the PITP. To that end, a series of single missense substitutions was incorporated into the Sec14 V154V155 motif, and the NPPM sensitivities of the individual mutants were tested by integrating the mutant SEC14 alleles into the genome of a naïve yeast strain and subjecting the transplacement mutants to an NPPM challenge. Yeasts expressing the Sec14V154F, Sec14V154E, Sec14 V154Y, Sec14V155F, Sec14V155A, or Sec14V155Y proteins were completely resistant to NPPM 6748-481, indicating that those polypeptides were functional PITPs with the property of being NPPMR (Fig. 6A). The former property was demonstrated in sec14-1ts growth rescue experiments at 37°C, and in plasmid shuffle experiments where rescue of the lethal sec14Δ allele was scored (Fig. 6B). By contrast, expression of the Sec14V154A and Sec14V155E mutants failed to confer an NPPMR phenotype to yeast cells (Fig. 6B), and this failure was not the trivial consequence of those proteins having been inactivated for PITP activity, as both proteins scored as active by the sec14-1ts complementation assay (Fig. 6B). These collective results indicate that Sec14 tolerates nonconservative changes in its VV motif from both a protein structure and protein function point of view, and with regard to its NPPM sensitivity properties. However, Sec14 does not tolerate incorporation of residues with side chains bulkier than Val, or of charged residues, at those positions from the standpoint of maintaining sensitivity to inhibition by NPPM.
Fig. 6.
Functional analysis of the VV motif in Sec14. SEC14 genes carrying the indicated for V154 and V155 missense substitutions (Phe, Ala, Glu, Tyr) were integrated into the LEU2 locus of a sec14-1ts strain. The mock condition documents the phenotype of an isogenic strain where a SEC14-less integration cassette was transplaced into the LEU2 locus. A: Integrants were interrogated for in vivo function by scoring rescue of sec14-1ts growth defects at the restrictive temperature of 37°C (left panel) or complementation of the normally lethal sec14Δ allele (right panel). Left panel: The YPD plates upon which the cells were spotted were incubated at 37°C for 48 h and images taken. Right panel: A colony color-based plasmid shuffle assay was used for determining whether the mutant Sec14 proteins could fulfill all biological functions of Sec14. Each mutant SEC14 gene was integrated into the MET17 locus of an ade2 ade3 sec14Δ strain carrying a Yep (SEC14, LEU2, ADE3) plasmid [strain CTY558 (9)]. The mock condition documents the phenotype of an isogenic strain where a SEC14-less integration cassette was transplaced into the MET17 locus. Plates were incubated at 30°C for 48 h and images were taken. The ability of mutant Sec14 expression to rescue lethality of the sec14Δ allele is recognized by loss of the Yep (SEC14, LEU2) plasmid upon relief of nutritional selection for LEU2 selection by spotting cells onto YPD agar. Plasmid loss is recognized by appearance of white colony segregants from the background of red plasmid-bearing colonies. All mutant proteins retained biological activity in this assay. B: Integrants expressing the indicated mutant Sec14 proteins in sec14-1ts genetic background were examined for NPPM 6748-481 sensitivity by dilution spotting on DMSO and drug-supplemented YPD agar. Plates were incubated at 30°C for 48 h and images taken. For both panels, the mock condition documents the phenotype of an isogenic strain where a SEC14-less integration cassette was transplaced into the LEU2 locus.
DISCUSSION
A sophisticated appreciation of how NPPMs bind and inhibit Sec14 activity is essential for further progress in several important areas, including rational design of SMIs with desirable properties, e.g., increased potency and a suitably broadened range of activity among closely related Sec14-like PITPs, etc. Our previous efforts to understand Sec14::NPPM binding interactions relied on rational loss-of-function approaches. That is, functional properties of Sec14 and of the NPPM were altered, using computational docking model-guided logic, with a view toward compromising protein::inhibitor interactions (14). Herein, we exploit an unbiased NPPM-resistance genetic screen to identify a new Sec14 motif, termed the VV signature, that contributes to NPPM sensitivity. We further demonstrate that suitable transplacement of a VV signature into naturally NPPM-resistant Sec14-like PtdIns/PtdCho-transfer proteins confers an NPPM-sensitivity onto those PITPs. The collective data both extend our appreciation of the determinants that govern Sec14-like PITP sensitivities to NPPMs and facilitate primary sequence predictions of which Sec14-like PtdIns/PtdCho-transfer proteins are likely to be NPPM resistant or sensitive. Moreover, these data further emphasize the importance of S173 in the mechanism by which NPPMs engage and inhibit Sec14-like PITPs.
A hallmark feature of NPPMs as PITP inhibitors is their exquisite specificity. Even very closely related Sec14-like PITPs exhibit differential sensitivities to these inhibitors, and the detailed structural pose for NPPM-binding to Sec14 does not offer a satisfactory structural basis for explaining those variable sensitivities. The Sec14-like Sfh1 makes a case in point. Unlike Sec14, this protein is completely resistant to NPPM challenge even though it preserves all obvious structural features required for NPPM-binding, as defined by the model Sec14::NPPM pose (14). As described herein, attempts to capitalize on the proposed Sec14::NPPM 6748-481 pose to engineer an NPPM-sensitive Sfh1 failed. In those analyses, we presumed that the NPPM pose closely approximated the stably bound configuration of the inhibitor within the Sec14 lipid-binding cavity.
There are several potential mechanisms by which a Sec14-like PITP can be intrinsically resistant to NPPM inhibition, however. First, the PITP may simply fail to bind the SMI. Such defects could be manifested anywhere along the NPPM binding trajectory from initial association of SMI with the protein’s surface to assumption of its resting pose within the PITP lipid-binding cavity. In such cases, the computed Sec14::NPPM pose would be of limited utility in deciphering mechanisms of resistance. Alternatively, NPPM binding might occur, but could be of sufficiently reduced affinity that the inhibitor becomes an exchangeable ligand (like PtdIns and PtdCho) so that SMI binding is readily reversible. In such circumstances, subtle differences in Sec14-like PITP pocket geometries could well determine NPPM sensitivity/resistance. Subtle differences of this nature might be difficult to recognize when examination of the problem relies strictly on a dock model in the absence of a high resolution Sec14::NPPM crystal structure.
Unbiased genetic screens cast a wider net in terms of what types of NPPM-resistant Sec14 variants one might recover, and these strategies powerfully complement structural approaches. Using such screens, we uncovered seven unique missense substitutions (P120Q, V154F, V155F, S173P, R208L, G210V, and F212L) that confer varying degrees of NPPM resistance to Sec14. This set is noteworthy on several counts. First, the S173P substitution involves the very Ser residue that is essential for the proposed halogen-bonding interaction required for stable binding of NPPM-481 (14). Second, five of the seven substitutions do not involve residues previously highlighted for rational mutagenesis on the basis of the proposed NPPM-481 binding pose (S173 and F212 are the exception). Third, while S173 is the only one of these seven residues projected to contact bound NPPM directly, six of the seven substitutions lie within 4.5 Å of NPPM-481, as it is configured in the proposed Sec14::NPPM binding pose (V155 is the exception). Interpretation of the NPPM resistance phenotypes associated with these missense substitutions provides detailed new insights into what properties determine NPPM resistance/sensitivity in Sec14-like PITPs.
Taken together, the results from the genetic screen, while highlighting new residues for consideration, nonetheless strongly support the proposed Sec14::NPPM docking pose (14). What is striking is that, contrary to the structure-based logic that drove engineering of the Sfh16X variant, all but one of the residues identified in the screen lie in the subregion of the Sec14 lipid-binding cavity projected to coordinate binding of the NPPM-481 activated aryl-halide headgroup. Only one (F212) lies in the hydrophobic region of the cavity proposed to coordinate the distal fluorobenzene moiety of bound NPPM-481, and the F212L substitution is the weakest of the substitutions recovered in terms of conferring NPPM resistance to Sec14. Residue F212 resides on the cavity floor and is projected to engage in π-π stacking interactions with the NPPM fluorobenzene moiety, as the NPPM intercalates into the narrow hydrophobic cleft bounded by residues F212, M177 (also on the cavity floor), and F228 (on the helical gate). We suggest the F212L substitution disorders that hydrophobic cleft and thereby weakens NPPM binding by interfering with its normal intercalation into this substructure. It is a plausible notion that this substitution results in NPPM becoming an exchangeable ligand and, therefore, a poor inhibitor of Sec14F212L.
Of the remaining six missense substitutions, five can be interpreted in the context of the previously proposed Sec14::NPPM docking pose that is anchored by a strong halogen bond interaction between the headgroup halide and residue S173 (14). Obviously, recovery of S173P in the unbiased genetic screen is particularly satisfying in that regard, and this mutant is essentially indifferent to NPPM challenge. However, the strong NPPM-resistance phenotypes associated with the R208L, G210V, and P120Q missense substitutions can be interpreted in terms of a highly conserved “S173 coordination envelope” that governs the essential nature of the S173 interaction with NPPM (Fig. 7). The R208 backbone engages in an H-bond with the backbone amide group of S173, whereas the G210 backbone amide coordinates with the S173 backbone carbonyl oxygen. Similarly, the S173 backbone amide is coordinated with the carbonyl oxygen of P120 (Fig. 7). Thus, these three residues not only reside within 4.5 Å of the NPPM binding site, but make direct contact with residue S173 and contribute to the spatial positioning of its side chain. Even subtle conformational perturbations in this S173 coordination envelope are expected to be of sufficient magnitude to upset the strict geometric requirements for formation of the critical S173-NPPM halogen bond and thereby compromise NPPM-binding, even though PtdCho-binding is not catastrophically affected given that these remain functional proteins in cells.
Fig. 7.
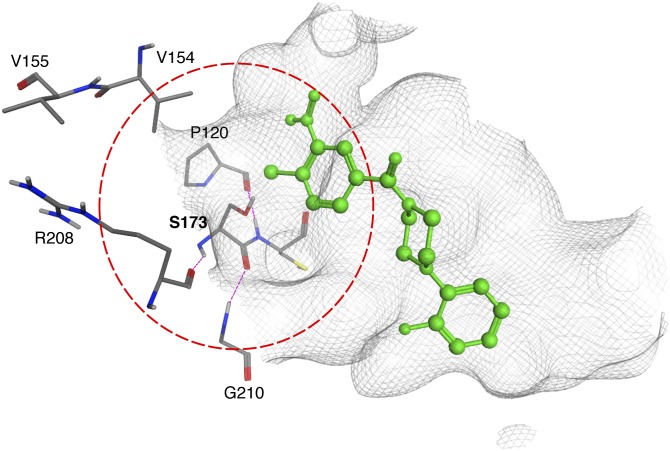
The Ser173 coordination envelope. The boundaries of the Sec14 S173 coordination envelope are defined by the dotted circle (red). Residues that coordinate S173 via H-bond interactions (magenta) are shown in stick render with carbon atoms in gray, oxygen in red and nitrogen in blue. The NPPM::6748-481 pose is shown in green and the binding pocket surface is rendered as a gray wire-mesh.
The remaining two Sec14 residues, V154 and V155, are of particular interest on several counts. First, these represent divergent residues among the Sec14-like PtdIns/PtdCho-transfer proteins, in contrast to the other five residues identified in the screen, which are highly conserved. Second, Sec14V154F resembles Sec14S173P in its complete indifference to NPPM-481, and Sec14V155F is also significantly resistant to NPPM challenge. Third, and most strikingly, NPPM sensitivity among Sec14-like PITPs tracks closely with this VV signature and transplacement of this signature into resistant Sec14-like PITPs confers significant NPPM sensitivity to those otherwise naturally NPPM-resistant PITPs. Residues V154 and V155 reside at the conserved kink in the long α-helix A8 that frames one side of the hydrophobic cavity of Sec14-like proteins (9, 40, 41). Both residues are positioned in proximity to S173, and V154 lies within the S173 coordination envelope. Although neither V154 nor V155 are projected to make any direct contacts with bound NPPM, one reasonable interpretation of NPPMR substitutions at these positions is that these perturb the S173 coordination envelope. In support of this, structural modeling experiments show the V154F, V154M, and V154E substitutions, associated with NPPM-resistance, intrude into the hydrophilic microenvironment of the lipid binding pocket that coordinates the S173::NPPM interaction (Fig. 7). Consistent with this idea, substitution of V154 with a small nonpolar residue (Ala) does not perturb the S173 coordination envelope in this manner and does not compromise NPPM binding by Sec14. Why bulky amino acid substitutions for V155 can endow NPPM resistance to Sec14 is less obvious, but indirect perturbation of the S173 coordination envelope is a tenable possibility given the close proximity of this residue to the coordination envelope boundary. Another possibility is that the VV motif directly binds NPPM during the binding trajectory of the SMI into its most stable configuration within the hydrophobic cavity. Polymorphisms within the VV motif may thereby sufficiently weaken Sec14 affinity for the NPPM to render it an easily exchangeable ligand and, therefore, an ineffectual inhibitor.
A detailed understanding of how NPPMs bind and inhibit Sec14 activity requires high resolution crystal structures of those complexes, and such information is essential for progress in the rational design of NPPMs with increased potencies and suitably broadened ranges of activity among closely related Sec14-like PITPs. The latter goal is desirable, as it would provide new avenues for crystallization of PITP::NPPM complexes. That is, the engineering of NPPM binding activity to mutant versions of naturally NPPM-resistant Sec14-like PITPs, which efficiently crystallize with bound ligands [e.g., Sfh1 (9)]. High resolution structures of such complexes are required for understanding how NPPMs inhibit Sec14-like PITPs, and directly determining whether the proposed S173-NPPM halogen-bonding interaction precisely describes the mechanism by which NPPMs inhibit Sec14-like PtdIns/PtdCho-transfer proteins.
Supplementary Material
Acknowledgments
The authors are grateful to the Texas A&M University High Performance Research Computing and Laboratory for Molecular Simulation for providing computational modeling support and computer time.
Footnotes
Abbreviations:
- MM-GBSA
- molecular mechanics with generalized born and surface area
- NPPM
- nitrophenyl(4-(2-methoxyphenyl)piperazin-1-yl)methanone
- PDB
- Protein Data Bank
- PIP
- phosphoinositide
- PITP
- phosphatidylinositol transfer protein
- PtdCho
- phosphatidylcholine
- PtdIns
- phosphatidylinositol
- SMI
- small molecule inhibitor
This work was supported by the National Institutes of Health grant GM44530 and from the Robert A. Welch Foundation grant BE-0017 to V.A.B. O.D. was supported by a Division of Undergraduate Education grant (NSF DBI-1358941) awarded to Texas A&M University AgriLife Research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Zhang X., Loijens J. C., Boronenkov I. V., Parker G. J., Norris F. A., Chen J., Thum O., Prestwich G. D., Majerus P. W., and Anderson R. A.. 1997. Phosphatidylinositol-4-phosphate 5-kinase isozymes catalyze the synthesis of 3-phosphate-containing phosphatidylinositol signaling molecules. J. Biol. Chem. 272: 17756–17761. [DOI] [PubMed] [Google Scholar]
- 2.Fruman D. A., Meyers R. E., and Cantley L. C.. 1998. Phosphoinositide kinases. Annu. Rev. Biochem. 67: 481–507. [DOI] [PubMed] [Google Scholar]
- 3.Ile K. E., Schaaf G., and Bankaitis V. A.. 2006. Phosphatidylinositol transfer proteins and cellular nanoreactors for lipid signaling. Nat. Chem. Biol. 2: 576–583. [DOI] [PubMed] [Google Scholar]
- 4.McLaughlin S., and Murray D.. 2005. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 438: 605–611. [DOI] [PubMed] [Google Scholar]
- 5.Strahl T., and Thorner J.. 2007. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1771: 353–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balla T. 2005. Inositol-lipid binding motifs: signal integrators through protein-lipid and protein-protein interactions. J. Cell Sci. 118: 2093–2104. [DOI] [PubMed] [Google Scholar]
- 7.Lemmon M. A. 2008. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9: 99–111. [DOI] [PubMed] [Google Scholar]
- 8.Bankaitis V. A., Mousley C. J., and Schaaf G.. 2010. The Sec14 superfamily and mechanisms for crosstalk between lipid metabolism and lipid signaling. Trends Biochem. Sci. 35: 150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaaf G., Ortlund E. A., Tyeryar K. R., Mousley C. J., Ile K. E., Garrett T. A., Ren J., Woolls M. J., Raetz C. R., Redinbo M. R., et al. 2008. Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily. Mol. Cell. 29: 191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nile A. H., Bankaitis V. A., and Grabon A.. 2010. Mammalian diseases of phosphatidylinositol transfer proteins and their homologs. Clin. Lipidol. 5: 867–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grabon A., Khan D., and Bankaitis V. A.. 2015. Phosphatidylinositol transfer proteins and instructive regulation of lipid kinase biology. Biochim. Biophys. Acta. 1851: 724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Routt S. M., Ryan M. M., Tyeryar K., Rizzieri K. E., Mousley C., Roumanie O., Brennwald P. J., and Bankaitis V. A.. 2005. Nonclassical PITPs activate PLD via the Stt4p PtdIns-4-kinase and modulate function of late stages of exocytosis in vegetative yeast. Traffic. 6: 1157–1172. [DOI] [PubMed] [Google Scholar]
- 13.Li X., Routt S. M., Xie Z., Cui X., Fang M., Kearns M. A., Bard M., Kirsch D. R., and Bankaitis V. A.. 2000. Identification of a novel family of nonclassic yeast phosphatidylinositol transfer proteins whose function modulates phospholipase D activity and Sec14p-independent cell growth. Mol. Biol. Cell. 11: 1989–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nile A. H., Tripathi A., Yuan P., Mousley C. J., Suresh S., Wallace I. M., Shah S. D., Pohlhaus D. T., Temple B., Nislow C., et al. 2014. PITPs as targets for selectively interfering with phosphoinositide signaling in cells. Nat. Chem. Biol. 10: 76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang-Ileto B., Frere S. G., and Di Paolo G.. 2012. Acute manipulation of phosphoinositide levels in cells. Methods Cell Biol. 108: 187–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keaney E. P., Connolly M., Dobler M., Karki R., Honda A., Sokup S., Karur S., Britt S., Patnaik A., Raman P., et al. 2014. 2-Alkyloxazoles as potent and selective PI4KIIIbeta inhibitors demonstrating inhibition of HCV replication. Bioorg. Med. Chem. Lett. 24: 3714–3718. [DOI] [PubMed] [Google Scholar]
- 17.Cleves A. E., McGee T. P., Whitters E. A., Champion K. M., Aitken J. R., Dowhan W., Goebl M., and Bankaitis V. A.. 1991. Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell. 64: 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bankaitis V. A., Aitken J. R., Cleves A. E., and Dowhan W.. 1990. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature. 347: 561–562. [DOI] [PubMed] [Google Scholar]
- 19.Li X., Rivas M. P., Fang M., Marchena J., Mehrotra B., Chaudhary A., Feng L., Prestwich G. D., and Bankaitis V. A.. 2002. Analysis of oxysterol binding protein homologue Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J. Cell Biol. 157: 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoon S., Smith A. M., Wallace I. M., Suresh S., Miranda M., Fung E., Proctor M., Shokat K. M., Zhang C., Davis R. W., et al. 2008. An integrated platform of genomic assays reveals small-molecule bioactivities. Nat. Chem. Biol. 4: 498–506. [Erratum. 2008. Nat. Chem. Biol. 4: 632.] [DOI] [PubMed] [Google Scholar]
- 21.Lee A. Y., St Onge R. P., Proctor M. J., Wallace I. M., Nile A. H., Spagnuolo P. A., Jitkova Y., Gronda M., Wu Y., Kim M. K., et al. 2014. Mapping the cellular response to small molecules using chemogenomic fitness signatures. Science. 344: 208–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaaf G., Dynowski M., Mousley C. J., Shah S. D., Yuan P., Winklbauer E. M., de Campos M. K., Trettin K., Quinones M. C., Smirnova T. I., et al. 2011. Resurrection of a functional phosphatidylinositol transfer protein from a pseudo-Sec14 scaffold by directed evolution. Mol. Biol. Cell. 22: 892–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito H., Fukuda Y., Murata K., and Kimura A.. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153: 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rothstein R. J. 1983. One-step gene disruption in yeast. Methods Enzymol. 101: 202–211. [DOI] [PubMed] [Google Scholar]
- 25.Sherman F., Fink G. R., and Hicks J. B.. 1983. Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor Press, Cold Spring Harbor, NY. [Google Scholar]
- 26.Xie Z., Fang M., Rivas M. P., Faulkner A. J., Sternweis P. C., Engebrecht J. A., and Bankaitis V. A.. 1998. Phospholipase D activity is required for suppression of yeast phosphatidylinositol transfer protein defects. Proc. Natl. Acad. Sci. USA. 95: 12346–12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bankaitis V. A., Malehorn D. E., Emr S. D., and Greene R.. 1989. The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J. Cell Biol. 108: 1271–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engel S. R., Dietrich F. S., Fisk D. G., Binkley G., Balakrishnan R., Costanzo M. C., Dwight S. S., Hitz B. C., Karra K., Nash R. S., et al. 2014. The reference genome sequence of Saccharomyces cerevisiae: then and now. G3 (Bethesda). 4: 389–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrödinger Release 2015-3: Schrödinger Suite 2015-3 Protein Preparation Wizard; Epik version 3.3, Schrödinger, LLC, New York, NY; Impact version 6.8, Schrödinger, LLC, New York, NY. [Google Scholar]
- 30.Jones G., Willett P., Glen R. C., Leach A. R., and Taylor R.. 1997. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 267: 727–748. [DOI] [PubMed] [Google Scholar]
- 31.Friesner R. A., Murphy R. B., Repasky M. P., Frye L. L., Greenwood J. R., Halgren T. A., Sanschagrin P. C., and Mainz D. T.. 2006. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 49: 6177–6196. [DOI] [PubMed] [Google Scholar]
- 32. Schrödinger Release 2015-3: Prime version 4.1, Schrödinger, LLC, New York, NY. [Google Scholar]
- 33.Cleves A. E., Novick P. J., and Bankaitis V. A.. 1989. Mutations in the SAC1 gene suppress defects in yeast Golgi and yeast actin function. J. Cell Biol. 109: 2939–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo S., Stolz L. E., Lemrow S. M., and York J. D.. 1999. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J. Biol. Chem. 274: 12990–12995. [DOI] [PubMed] [Google Scholar]
- 35.Rivas M. P., Kearns B. G., Xie Z., Guo S., Sekar M. C., Hosaka K., Kagiwada S., York J. D., and Bankaitis V. A.. 1999. Pleiotropic alterations in lipid metabolism in yeast sac1 mutants: relationship to “bypass Sec14p” and inositol auxotrophy. Mol. Biol. Cell. 10: 2235–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang M., Kearns B. G., Gedvilaite A., Kagiwada S., Kearns M., Fung M. K., and Bankaitis V. A.. 1996. Kes1p shares homology with human oxysterol binding protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. EMBO J. 15: 6447–6459. [PMC free article] [PubMed] [Google Scholar]
- 37.Im Y. J., Raychaudhuri S., Prinz W. A., and Hurley J. H.. 2005. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 437: 154–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Saint-Jean M., Delfosse V., Douguet D., Chicanne G., Payrastre B., Bourguet W., Antonny B., and Drin G.. 2011. Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J. Cell Biol. 195: 965–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katzmann D. J., Burnett P. E., Golin J., Mahe Y., and Moye-Rowley W. S.. 1994. Transcriptional control of the yeast PDR5 gene by the PDR3 gene product. Mol. Cell. Biol. 14: 4653–4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sha B., Phillips S. E., Bankaitis V. A., and Luo M.. 1998. Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein. Nature. 391: 506–510. [DOI] [PubMed] [Google Scholar]
- 41.Ren J., Pei-Chen Lin C., Pathak M. C., Temple B. R., Nile A. H., Mousley C. J., Duncan M. C., Eckert D. M., Leiker T. J., Ivanova P. T., et al. 2014. A phosphatidylinositol transfer protein integrates phosphoinositide signaling with lipid droplet metabolism to regulate a developmental program of nutrient stress-induced membrane biogenesis. Mol. Biol. Cell. 25: 712–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.