SUMMARY
Clinically acquired resistance to MAPK inhibitor (MAPKi) therapies for melanoma cannot be fully explained by genomic mechanisms and may be accompanied by co-evolution of intra-tumoral immunity. We sought to discover non-genomic mechanisms of acquired resistance and dynamic immune compositions by a comparative, transcriptomic-methylomic analysis of patient-matched melanoma tumors biopsied before therapy and during disease progression. Transcriptomic alterations across resistant tumors were highly recurrent, in contrast to mutations, and were frequently correlated with differential methylation of tumor cell-intrinsic CpG sites. We identified in the tumor cell compartment supra-physiologic c-MET up-expression, infra-physiologic LEF1 down-expression, and YAP1 signature enrichment as drivers of acquired resistance. Importantly, high intra-tumoral cytolytic T-cell inflammation prior to MAPKi therapy preceded CD8 T-cell deficiency/exhaustion and loss of antigen-presentation in half of disease-progressive melanomas, suggesting cross-resistance to salvage anti-PD-1/PD-L1 immunotherapy. Thus, melanoma acquires MAPKi-resistance with highly dynamic and recurrent non-genomic alterations and co-evolving intra-tumoral immunity.
INTRODUCTION
Understanding how melanomas acquire resistance to BRAF inhibitors (BRAFi) via genetic alterations shown to reactivate the MAPK pathway (Nazarian et al., 2010; Shi et al., 2014a; Shi et al., 2014b; Shi et al., 2012a; Shi et al., 2012b; Van Allen et al., 2014; Wagle et al., 2011) has guided the clinical development of BRAFi+MEKi combinatorial therapy. Despite superior clinical benefits, the double-drug approach commonly fails due to acquired resistance (Larkin et al., 2014; Long et al., 2014b) caused by a similar set of mutant genes responsible for acquired resistance to BRAFi monotherapy (Long et al., 2014a; Moriceau et al., 2015; Villanueva et al., 2013; Wagle et al., 2014). These shared mutations, which include V600EBRAF amplification and single nucleotide variants (SNVs) in NRAS, KRAS, MEK1/2, PTEN, CDKN2A and DUSP4, indicate that the reservoir of genomic diversity strongly limits the long-term efficacy of dual (i.e., BRAFi+MEKi) or likely higher-order (i.e., BRAFi+MEKi+ERKi) MAPKi therapy.
In addition to harboring heterogeneous genetic alterations in the MAPK and PI3K-PTEN-AKT core pathways, melanomas at distinct sites with acquired BRAFi resistance in any given patient display extensively branched evolution (Shi et al., 2014a; Shi et al., 2014b). Furthermore, many on-treatment tumors re-grow without any clear genetic mechanism (Rizos et al., 2014; Shi et al., 2014b). These observations suggested that a diverse array of melanoma sub-clones, sometimes concurrent intra-tumorally, evolve to circumvent the “bottleneck” of BRAFi therapy (Shi et al., 2014b) and that exome-scale dissection of acquired MAPKi resistance falls short of fully explaining clinical resistance.
Earlier (Johannessen et al., 2010; Nazarian et al., 2010) studies have pointed to transcriptome-based mechanisms of acquired BRAFi resistance. Given these leads, there is a clear need for comprehensive analyses of transcriptomic and epigenetic alterations underlying acquired MAPKi resistance in patient-derived melanoma samples. Identification of highly recurrent, non-genomic mechanisms may open the door to new combinatorial therapeutic strategies.
In the current therapeutic landscape, salvage therapies for patients with disease progression on MAPKi often involve immunotherapies, e.g., inhibitors of CTLA-4, PD-1 checkpoints or CSF-1R on tumor-associated macrophages. But it is not known whether MAPKi-resistant melanomas are distinct in their immuno-phenotypes and susceptibilities to anti-CTLA-4 or -PD-1 therapies. In fact, studies are emerging which support immune microenvironment modulation by BRAFi as a contributor to in vivo anti-tumor effects. Thus, immune evasion may contribute to acquired MAPKi-resistance (Ferrari de Andrade et al., 2014; Knight et al., 2013)
Hence, we sought a landscape perspective on the relative contributions of genomic and non-genomic mechanisms to acquired MAPKi resistance and co-evolutionary dynamics of the intra-tumoral immune microenvironment in patient-derived melanoma tissues.
RESULTS
Genetic Mechanisms of Acquired MAPKi Resistance in Melanoma
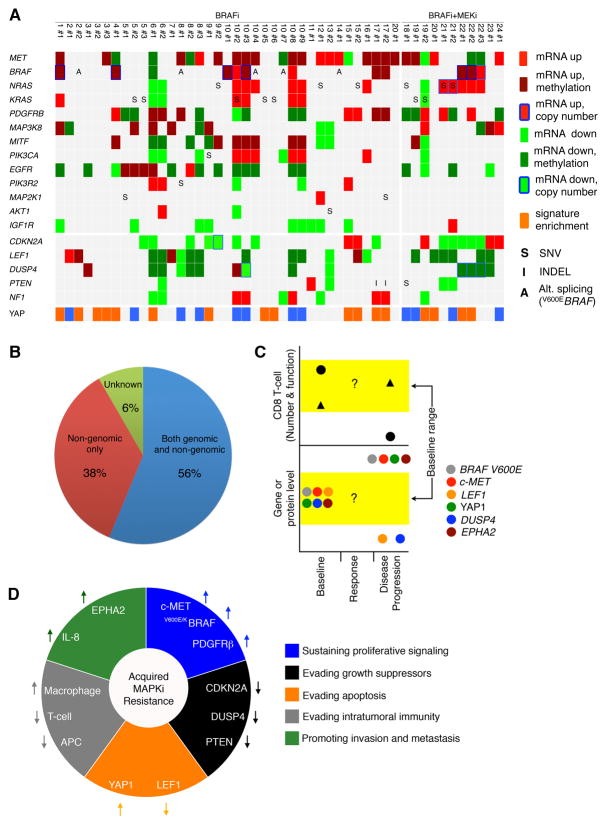
We analyzed whole-exome sequences (WES) (Table S1A) of serial tumor biopsies (baseline and acquired resistant tumors) and normal tissues from patients with advanced melanoma treated with MAPK inhibitor (MAPKi) regiments, which included single-drug (i.e., BRAFi) or double-drug (i.e., BRAFi+MEKi) therapies. When multiple disease-progressive or acquired MAPKi-resistant tumors were obtained from patients, they were compared to the same patient-matched baseline tumors. To assess the degree to which functionally validated genetic mechanisms account for clinically acquired MAPKi-resistance, we visualized the recurrence of these mutations specific to or highly enriched in single-drug and double-drug disease-progressive (DP and DD-DP, respectively) melanomas (n=67) relative to matched baseline tumors (Figure 1A). These functionally validated mutations (Moriceau et al., 2015; Shi et al., 2014b) included gain-of-function (GOF) events in V600E/KBRAF, NRAS, KRAS, MEK1 or MAP2K1, PIK3CA, AKT1, AKT3 and loss-of-function (LOF) events in PIK3R2, DUSP4, CDKN2A, PTEN. The most recurrent resistance mutations were detected almost mutually exclusively in V600E/KBRAF (copy number gains in 15 of 67 or 22%) or RAS (single-nucleotide variants with or without copy number gains in 17 of 67 or 25%). Less prevalent resistance mutations occurred at ≤9% (in PTEN, DUSP4) or as singleton events. Mutations in MITF, MEK2, RAC1 and NF1 were not specifically associated with resistant tumors. Importantly, 26 of 67 or 39% of resistant melanomas were not accounted for by any validated mutational mechanism.
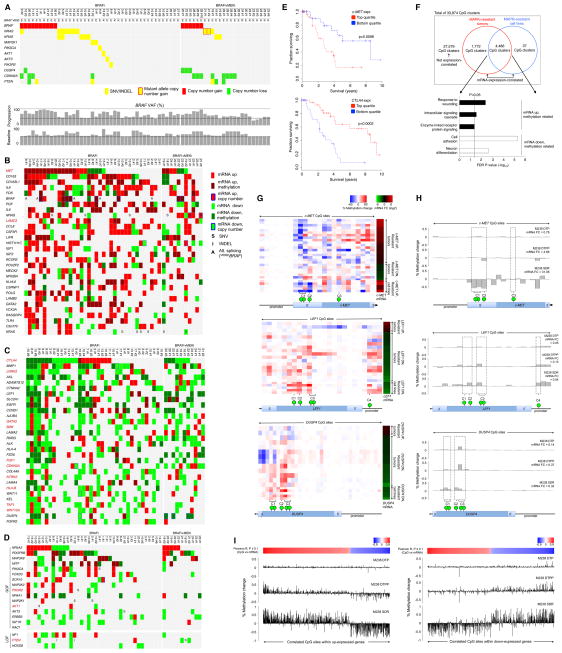
Figure 1. Landscape of Genomic, Transcriptomic and Methylomic Alterations in Melanoma with Acquired MAPKi Resistance.
(A) Matrix of disease progressive melanomas (n=67; indicated by patient and then tumor numbers) on BRAFi or BRAFi+MEKi therapies and of genes whose mutations cause acquired MAPKi resistance. Bottom, BRAF variant allelic frequencies or VAFs (resulting in V600E/K) adjusted by estimated tumor purities.
(B and C) Tiling of top 30 recurrent GOF (B) or LOF (C) gene-based events among cancer/melanoma/immune genes across 48 disease-progressive V600BRAF mutant melanoma samples relative to patient-matched baseline melanomas (left, BRAFi; right, BRAFi+MEKi). GOF or LOF events defined as GOF:LOF ratio ≥ 2 or LOF:GOF ratio ≥ 2, respectively. SNV, expressed non-synonymous single-nucleotide variants; INDELs, expressed small insertion-deletions. Darker colors, differential mRNA expression significantly correlated with differential CpG cluster methylation. Splice variants based on RT-PCR detection reported for BRAF only. Genes in red, expression levels correlated with survival in the TCGA Melanoma data.
(D) Resistance driver genes proposed in the literature and their genetic and non-genetic alterations in acquired MAPKi-resistant samples.
(E) Kaplan-Meier 10-year survival curves for c-MET and CTLA4 expression groups among TCGA patients with BRAF mutant melanoma (n=118) and whose follow-up durations were within 10 years. P-values, log rank test.
(F) Numbers of mRNA expression-correlated CpG clusters in resistant melanoma tissues and/or cell lines. Annotated genes from the overlapping CpG clusters grouped by up- or down-expression and Gene Ontology term enrichments.
(G) Expression-correlated CpG clusters on c-MET, LEF1, and DUSP4. Green bubble, a CpG site with anti-correlated differential mRNA expression vs. gDNA methylation. Heatmaps showing (left) % methylation change at all profiled CpG sites (red, hyper- and blue, hypo-methylation) across all resistant tumors and cell lines sorted by fold change (FC) of mRNA expression (right) of each gene (red, up- and green, down-expression).
(H) Percent methylation changes at profiled CpG sites (green bubbles, CpG sites nominated by aggregate tumor and cell line analysis as expression-correlated) and mRNA expression fold changes (FC). Both % methylation change and mRNA FC for each cell line sub-population were expressed relative to vehicle-treated M238.
(I) Percent methylation changes across all expression-correlated CpG sites (top heat scale, Pearson correlation R values) nominated by aggregate tumor and cell line analysis and located within up-(left) or down-(right) expression genes in the SDR sub-population (vs. vehicle-treated parental M238).
Landscape of Transcriptomic Alterations in Acquired MAPKi Resistance
We profiled the temporal transcriptomic alterations in 48 DP or DD-DP compared with patient-matched baseline melanoma tissues (Table S1A) and integrated analysis (Figure S1A) of temporal transcriptomic with expressed exomic alterations to assess the combined recurrences of GOF and LOF gene-based events. We rank-ordered recurrences of resistance-specific alterations based on the number of resistant samples and (in cases of ties) of patients. The gene list included 855 cancer-, melanoma-, and MAPKi resistance-, and immunotherapy-related genes (Table S1B). Importantly, among the top 30 GOF and LOF genes, transcriptomic alterations were generally more recurrent per gene and affected more genes than exomic alterations (Figures 1B, 1C and S1B; Table S1C and S1D). Notably, transcriptional up- or down-expression occurred recurrently and respectively in bona fide, mutated GOF (i.e., BRAF, NRAS, KRAS) and LOF (i.e., CDKN2A, DUSP4) resistance genes, with the transcriptomic events exceeding the mutational events in some cases (i.e., CDKN2A, DUSP4). Interestingly, V600E/KBRAF was subject to not only mutational alterations (copy number gain) (8 of 16) (Shi et al., 2012b) (Figure 1A and 1B), transcriptional up-expression (3 of 16) and alternative splicing (5 of 16) (Figure 1B) but also, in the absence of aforementioned mechanisms, mutant allele-selective expression (Figure S1B; Table S1E). In total, GOF events in BRAF occurred in 16 of 48 or 33% of resistant tumors. Similarly, NRAS and KRAS GOF events (26 of 48 or 54%) were a mixture of mRNA up-expression (16 of 26), mutational activation (11 of 26), and mutant allele-specific gene amplification (2 of 26) (Moriceau et al., 2015). In contrast to BRAF where both genetic and non-genetic alterations affected the mutant gene selectively, up-expression of WT NRAS or KRAS was commonly detected in acquired MAPKi-resistant melanoma tumors and was capable of conferring MAPKi resistance to sensitive melanoma lines (Lidsky et al., 2014; Shi et al., 2014b) (Figure S1C). Also, we observed statistically significant overlaps between genes that were recurrently mutated and recurrently differentially expressed (maximal recurrence capped at n=5 resistance tumors; Table S1F). This association was particularly strong with recurrent copy number alterations. Non-copy number genetic alterations tended to be differentially expressed, although the statistical significance was weaker. Of interest, four genes were recurrently mutated in a GOF manner (excluding CNVs) in at least five resistant tumors. These genes were recurrently up-expressed and included NRAS and KRAS.
Most highly recurrent GOF events (Figure 1B; Table S1C) were purely transcriptomic and could involve either tumor cell-intrinsic or stromal differential gene expression. c-MET and IL-8 (Sanchez-Laorden et al., 2014) were up-expressed respectively in 21 of 48 or 44% and 19 of 48 or 40% of resistant tumors. Furthermore, c-FOS and MEOX2 encode tumor cell-intrinsic transcriptional factors implicated in MAPKi resistance (Johannessen et al., 2013). Other purely transcriptomic, highly recurrent GOF events involved the macrophage markers (CD163 and CD163L1). Up-expression of other genes such as CCL8 (critical for chemotaxis of monocytes, lymphocytes, and granulocytes), CSF3R (critical for granulocyte function) and NFKBIA suggested inflammatory tumor infiltration. Thus, highly recurrent and GOF transcriptomic events may reflect evolution in both the tumor cell and immune compartments of acquired MAPKi-resistant melanoma tissues.
The majority of highly recurrent LOF gene-based events (Figure 1C; Table S1D) arose from transcriptomic down-expressions. These involved a gene most commonly mutated in Parkinson’s disease (LRRK2); an immune response modulation gene (CTLA4); antigen presentation genes (B2M, HLA-A, HLA-B and TAP1); Wnt signaling genes (LEF1, FZD6, WNT11, and WNT10A); and RTK genes (AXL, EGFR, ALK, NTRK2, and FGFR2). CTLA4 may be down-expressed in both the immune and melanoma compartments, since in three melanoma expression data sets CTLA4 expression, in contrast to PDCD1 (PD-1) expression, was less correlated with the expression of T cell genes CD3, CD4, or CD8 (Table S1G). Also, we have observed CTLA4 down-expression in several acquired MAPKi-resistant melanoma cell lines compared to their parental counterparts (data not shown). In the melanoma compartment, CTLA4 may be a direct therapeutic target of ipilimumab (Laurent et al., 2013). Moreover, the finding here of frequent AXL and EGFR down-expression in acquired MAPKi-resistant melanoma contrasted with previous cell line-based observations (Girotti et al., 2013; Muller et al., 2014). We therefore evaluated systematically the in vivo relevance of resistance mechanisms previously proposed based on functional studies in cell lines (Figure 1D). Despite the clear importance of CRAF as a convergent signaling node for various mechanisms of resistance (Moriceau et al., 2015; Nazarian et al., 2010; Shi et al., 2012b), CRAF (RAF1) itself was not subject to a single genetic or non-genetic alteration.
Beyond assessing recurrence as evidence of selection, we systematically gauged the impact of gene expression levels (top vs. bottom quartiles) on TCGA melanoma patients’ 10-year survival (significance cutoff, log-rank test p ≤ 0.05). Importantly, c-MET and CTLA4 were not only the most recurrently up- and down-expressed genes, respectively, among resistant melanomas (Figure 1B and 1D) but also genes whose expression levels portended survival significance (Figure 1E; Table S1H), even after adjusting for age, tumor ulceration and stage (c-MET: Cox HR=2.67 for the top quartile group, p-value=0.002, 95% CI=[1.4–5.0]; CTLA4: Cox HR=2.0 for the bottom quartile group, p-value=0.024, 95% CI=[1.1–3.8]). In addition to assessing the survival impacts of gene expression levels, we validated our recurrent transcriptomic GOF and LOF events using a published microarray study of an independent set of tissues (Long et al., 2014a; Rizos et al., 2014). Again, c-MET was recurrently up-expressed in 8 of 35 (23%) MAPKi-resistant tumors from 7 of 26 (27%) patients (Table S1I); CTLA4 was down-expressed in 9 of 35 (26%) MAPKi-resistant tumors from 7 of 26 (27%) patients (Table S1J).
A Common Methylomic Basis of Transcriptomic Alterations
An integrated transcriptome-methylome analysis (Figure 1B and 1C) revealed a subset of recurrent differential mRNA expression events, including those affecting c-MET, LEF1, and DUSP4, as highly correlated with differential genomic DNA (gDNA) CpG methylation (Table S1K). Methylation levels at 6,295 of all 33,874 (18.6%) CpG clusters were significantly correlated with differential mRNA expression (Figure 1F; Supplemental Experimental Procedures). To estimate the scope of tumor cell-intrinsic events, we calculated the numbers of expression-correlated CpG clusters across MAPKi-resistant tumors (n=43 pairs) and cell lines (n=5 pairs) and found that 4,486 of 6,295 (71.2%) expression-correlated CpG clusters were found in both (Figures 1F and Figure S1D). We then performed Gene Ontology (GO) enrichment analysis of genes annotated to overlapping expression-correlated CpG clusters; these genes included c-MET, LEF1, and DUSP4 (Figure 1G; Table S1K). Genes with methylation-correlated up-expression were enriched for wound healing and receptor-linked or intracellular signaling, whereas genes displaying methylation-correlated down-expression were enriched for cell adhesion and neuron differentiation (Figure 1F). In c-MET, three CpG clusters (C1-3) with differential methylation were negatively correlated with differential mRNA expression (Figure 1G). The extent and significance of these correlations compared favorably with those between mRNA levels of c-MET and its positive transcriptional factors (TFs) (Figure S1E). In nearly all (90%) pair wise comparisons of tumors and cell lines, differential c-MET mRNA expression could be accounted for by at least one differential CpG cluster methylation (Δβ ≥ 10% and FDR adjusted p ≤ 0.05). In contrast, only 48% displayed a concordant differential expression pattern of at least one c-MET TF (Figure S1F). Similarly for LEF1, DUSP4, and EPHA2, differential methylation at specific CpG clusters negatively correlated with differential mRNA expression (Figures 1G, S1G and S1H; Table S1K).
To corroborate expression-methylation correlations, we analyzed data from 335 TCGA melanoma samples. The Pearson correlation between the absolute methylation levels at C1 and normalized mRNA expression of c-MET was −0.48 (p < 2.2 e-16), while the correlation with MITF was 0.42 (p = 2.2e-15). We also compared the top and bottom quartiles of methylation levels (β values) in the c-MET CpG clusters and their mRNA expression levels. In particular, C1 CpG hypo-methylation associated strongly (Wilcoxon ranksum test p < 2.2 e-16) with high levels of c-MET mRNA expression (Figures S1I; C2 CpG cluster, not covered). Similar analysis for LEF1 and DUSP4 also supported expression-methylation correlations at specific CpG clusters (Figures S1J and S1K). Overall, among 7,769 individual expression-correlated CpG sites from 6,295 CpG clusters (Figure 1F), 4,086 CpG sites (53%) showed concordant differential mRNA expression and CpG site methylation (top vs. bottom quartiles) among the TCGA melanoma tumors. Furthermore, we examined our tissue-derived data for phenotypes inferred from expression patterns of methylation-regulated genes such as c-MET, LEF1, and DUSP4 (Table S1L). Genes whose differential expression positively correlated with that of c-MET were enriched in the GO term “pigmentation during development”; those negative correlated for “cell adhesion” and “positive regulation of cell proliferation”, which suggested a motile phenotype (Data S1A and S1B). On the other hand, differential expression of LEF1 and DUSP4 correlated strongly with enrichment of a mutant BRAF signature (LEF1, Pearson R=0.62 P=8.9e-6; DUSP4, Pearson R=0.60 P=1.8e-5) (Data S1C and S1D), which associated LEF1 and DUSP4 down-expression with reduced MAPK-addiction. Taken together, MAPK inhibition in BRAF mutant melanoma may lead to epigenetic transcriptomic alterations with functional consequences.
We then assessed whether BRAF inhibition in melanoma cell lines would lead to temporally incremental and correlated alterations between methylation and mRNA levels in c-MET, LEF1, DUSP4 and in general. Using two human V600EBRAF melanoma cell lines, we profiled the methylomes and transcriptomes of vemurafenib (BRAFi)-selected sub-populations over time, including drug-tolerant persisters (DTPs) (days of treatment), drug-tolerant proliferating persisters (DTPPs) (weeks), and single-drug resistant (SDR) sub-lines (months to years) as described (Shi et al., 2014a). We observed in time-dependent BRAFi-selected sub-populations (relative to vehicle-treated cells) methylation decreases at c-MET’s nominated CpG clusters along with mRNA increases. In contrast, methylation at nominated CpG clusters in LEF1 and DUSP4 increased while their mRNA levels decreased with BRAFi treatment duration (Figure 1H; Data S1E). We also examined the temporal methylation changes at all CpG sites within all genes displaying differential expression between vehicle-treated cells vs. their isogenic SDR sub-lines. Importantly, the magnitudes of methylation changes were time-dependent; the directions of methylation changes were concordant with the sites’ correlation scores independently derived from the aggregate analysis of the tissue and cell line pairs (Figure 1I; Data S1F and S1G). Thus, BRAFi treatments of cell lines led to progressive methylation-expression changes akin to observations across MAPKi-sensitive vs. resistant tumors.
Highly Recurrent c-MET Up-expression Mediates MAPKi Resistance
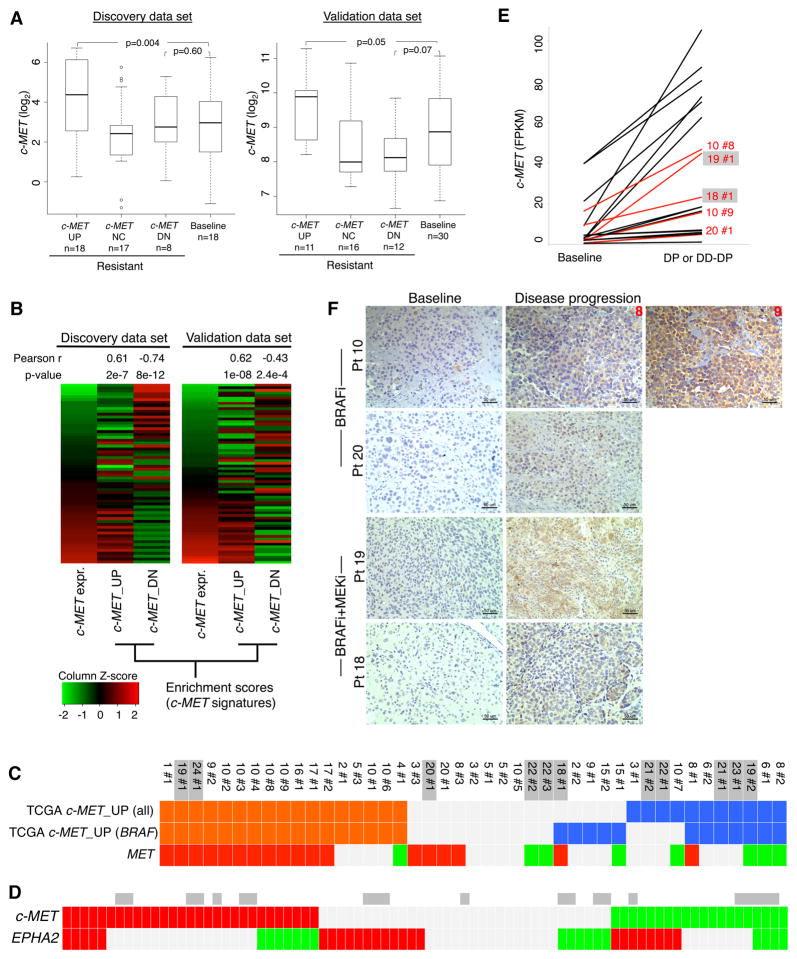
Because acquired MAPKi-resistant melanomas displayed highly recurrent c-MET up-expression (Figure 1B), we compared the absolute c-MET expression levels in three resistant and one sensitive melanoma sub-groups: (1) c-MET-up-expression (c-MET UP), (2) no c-MET differential expression, (3) c-MET down-expression, and (4) baseline (Figure 2A) and observed that c-MET UP resistant-melanomas in particular displayed supra-physiologic levels of c-MET transcripts compared to the sensitive, drug-naïve melanomas in both our and the validation cohorts. Additionally, the overall c-MET expression levels across the spectrum of sensitive- and resistant-melanomas correlated with single-sample enrichments of c-MET signatures generated from the TCGA Melanoma data set (Figure 2B and Table S2). Genes up-expressed (log2 FC≥2, FDR adjusted Wilcoxon p-value≤0.05) in the top quartile of c-MET expression (vs. the bottom quartile) defined the c-MET_UP signature, and genes up-expressed in the bottom quartile defined the c-MET_DOWN or DN signature. Importantly, c-MET up-expression in MAPKi-resistant melanomas generally concurred with positive enrichment of the c-MET_UP TCGA signatures (and down-expression with negative enrichment) (Figure 2C). By combining the transcriptomic analysis of RTK genes in both discovery and validation data sets (n=82 differential expressions), we found that another RTK EPHA2, which like c-MET has been nominated as a cancer metastasis gene, was up-expressed (n=25) in a largely mutually exclusive manner to c-MET up-expression (n=29) (one-side Fisher exact test p-value=0.031; odds ratio 0.34) (Figure 2D). EPHA2 up-expression among MAPKi-resistant melanomas also occurred in a supra-physiologic range (Figure S2A), and its top-quartile expression associated significantly with worse patient survival (Figure S2B). Using avaliable patient-matched FFPE samples (Figure 2E), we detected relative c-MET protein up-expression in disease progressive tissue sections (Figure 2F) using a validated antibody (Figure S2C) and found c-MET up-expression in MAPKi-resistance to be tumor cell-intrinsic.
Figure 2. Recurrent c-MET Up-expression in Disease Progressive Melanomas.
(A) c-MET mRNA expression levels in distinct subsets of disease progressive sample. P-values, Wilcoxon rank-sum test.
(B) Pearson correlation (P-values, t-test) of the c-MET mRNA expression levels in all baseline and disease progressive melanoma samples (discovery, n=59; validation, n=61) with GSVA enrichment scores of the TCGA melanoma-derived c-MET signatures.
(C, D) Tiling of c-MET signature enrichment (orange and blue, positive and negative enrichments) (C) and differential c-MET (C, D) and EPHA2 (D) expression (red and green, up- and down-expression) across disease progressive melanomas (DD-DP samples, grey) (both discovery and validation cohorts, D).
(E–F) Levels of c-MET mRNA (E) and protein (F) (for samples in red) in patient-matched pairs where resistance-associated c-MET mRNA up-expression was detected (ruler, 50 micron).
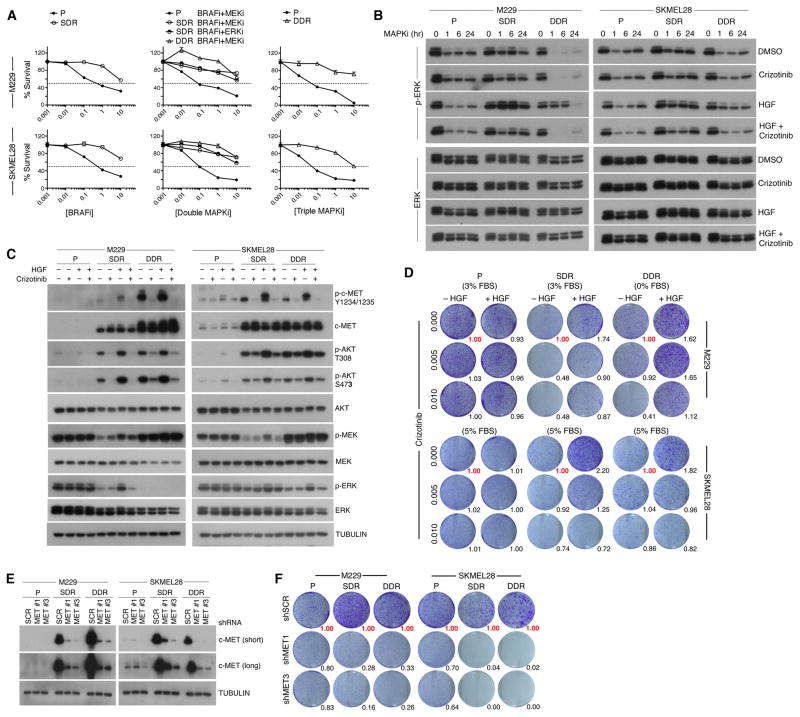
We then assessed the functional role of tumor cell-intrinsic, supra-physiologic c-MET up-expression using two triplets of isogenic cell lines where the drug-resistant sub-lines were (i) derived from the M229 and SKMEL28 human V600EBRAF melanoma cell lines by chronic BRAFi (vemurafenib) (single-drug resistance or SDR) or BRAFi (vemurafenib)+MEKi (selumetinib) (double-drug resistance or DDR) treatment (Figure 3A) and (ii) shown to display dramatic mRNA and protein up-expression of c-MET compared to their parental cell lines. In stark contrast to acquired MAPKi-resistant cell lines driven by genetic mechanisms such as NRAS mutations, V600EBRAF amplification, and/or MEK1 mutations, acquired SDR or DDR sub-lines up-expressing c-MET was highly refractory to downstream MAPK suppression (Figure 3A). Despite this, stimulation of cells by HGF addition during the course of BRAFi (for parental and SDR cell lines) or BRAFi+MEKi (for DDR cell lines) treatment accelerated p-ERK recovery or reactivation only in resistant cell lines and in a manner reversible by co-treatment with an inhibitor of c-MET, crizotinib (Figure 3B). c-MET up-expression was accompanied by enhanced activation-associated phosphorylation (Y1234/1235) and p-AKT (T308, S473) induction, which could be augmented by HGF stimulation and repressed by crizotinib treatment (Figure 3C). These studies indicate that supra-physiologic c-MET up-expression in SDR and DDR sub-lines mediates MAPK-redundant survival signaling.
Figure 3. c-MET Up-expression Drives Acquired MAPKi resistance.
(A) Three-day MTT survival assays of two isogenic melanoma triplets in response to single MAPKi (BRAFi), double MAPKi (as indicated) or triple MAPKi (BRAFi+MEKi+ERKi). BRAFi, vemurafenib; MEKi, selumetinib; ERKi, SCH772984 (all in μM). Error bars, SEM, n=5; normalized to DMSO vehicle as 100%.
(B) Western blot (WB) analysis of p-ERK recovery in both isogenic triplet cell lines in response to a single-dose of 1 μM BRAFi (P, SDR) or BRAFi+MEKi (DDR) under four conditions (DMSO/PBS, crizotinib (0.1 μM)/PBS, DMSO/HGF (20 ng/ml), and HGF/crizotinib). SDR and DDR sub-lines were first plated without MAPKi for 16 h. Loading controls, ERK.
(C) WB levels of activation-associated phosphorylation and total levels of indicated proteins in response to vehicle, crizotinib, and/or HGF treatments (1 h). TUBULIN, loading control.
(D) Long-term (10 d) clonogenic assay (P, no inhibitor; SDR, 1 μM BRAFi; DDR, 1 μM BRAFi+MEKi) −/+ HGF and crizotinib (μM). Growth quantifications relative to cultures without HGF and crizotinib (in red).
(E) WB analysis of c-MET knockdown (short and long exposures).
(F) Long-term (10 d) clonogenic assay −/+ c-MET knockdown. Growth quantifications relative to shScramble (shSCR) controls.
Importantly, HGF stimulation enhanced the clonogenic survival of M229 and SKMEL28 SDR and DDR sub-lines cultured with MAPK inhibitor(s) but not the parental cell lines (without MAPKi) (Figure 3D), indicating that the growth-promoting effect of HGF depended on c-MET up-expression. Also, treatments with low concentrations of crizotinib reduced the clonogenic growth of SDR and DDR sub-lines, with or without HGF stimulation, but not the parental melanoma cell lines. Furthermore, c-MET knockdown using two independent shRNAs (Figure 3E) preferentially reduced the clonogenic growth of resistant melanoma sub-lines (Figure 3F). Collectively, these data argue that melanoma cells can acquire MAPKi resistance via addiction to c-MET up-expression and hyper-activity.
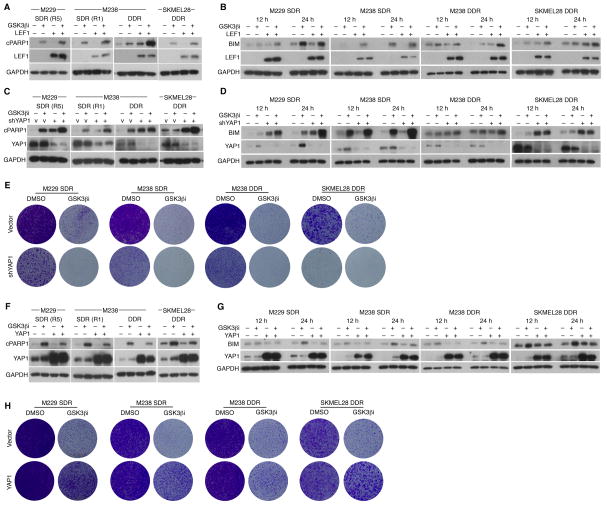
Altered β-catenin-LEF1 and YAP1 Signaling Reduces Apoptosis and Promotes MAPKi Resistance
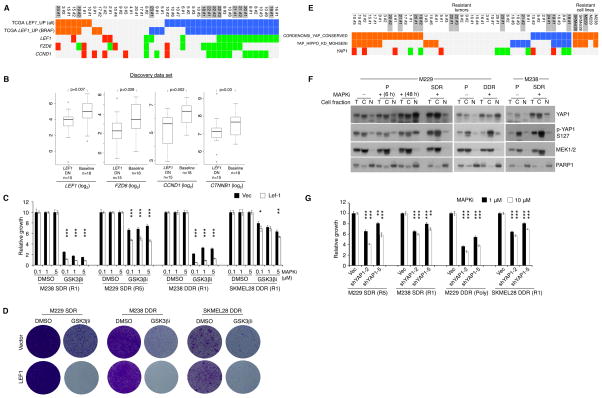
Whereas supra-physiologic c-MET up-expression in resistant tumors correlated strongly with its down-methylation, LEF1 down-expression correlated with its up-methylation (Figures 1C and 1G) and resulted in infra-physiologic expression levels (Figures 4A, 4B, and S3A). Moreover, LEF1 (and its related pathway genes, FZD6 and CCND1) down-expression concurred strongly with negative enrichment of TCGA melanoma LEF1_UP signatures (Figure 4A; Table S3), indicating down-regulation of β-catenin-LEF1 transcriptional activity. Consistently, we detected robust LEF1 protein down-expression in resistant tumors using a FFPE-validated antibody (Figures S3B and S3C). We also detected LEF1 down-expression in a panel of SDR or DDR cell lines (vs. parental cell lines) (Figure S3D). To understand whether β-catenin-LEF1 signal down-regulation promoted MAPKi resistance, we tested whether restoration of LEF1 expression and/or β-catenin-LEF1 signaling in these MAPKi-resistant melanoma cell lines would re-sensitize them to MAPKi. GSK3β inhibition using CHIR99021 strongly decreased p-β-catenin (Ser33/37 and Thr41) and increased total β-catenin levels in the parental cell lines (e.g., Figure S3E); these effects of GSK3β inhibition were weaker in the SDR and DDR cell lines (Figure S3F), consistent with a compromised β-catenin-LEF1 pathway. Importantly, GSK3βi strongly re-sensitized SDR and DDR melanoma cell lines to BRAFi and BRAFi+MEKi, respectively, and this re-sensitization to MAPKi was augmented by LEF1 re-expression in short-term (Figure 4C) and long-term (Figure 4D) survival assays. Hence, recurrent β-catenin-LEF1 down-regulation promotes MAPKi insensitivity.
Figure 4. β-catenin-LEF1 Up- and YAP1 Down-regulation Sensitize Resistant Melanoma to MAPKi.
(A) Tiling of LEF1_UP gene signature enrichment and differential expression of LEF1, FZD6 and CCND1 across disease progressive melanomas.
(B) Indicated mRNA expression levels in LEF1 down-expressed, MAPKi-resistant melanomas vs. baseline melanomas. P-values, Wilcoxon rank-sum test.
(C) Three-day survival assays of resistant melanoma cell lines at varying respective [BRAFi] and [BRAFi+MEKi], −/+ GSK3βi (10 μM CHIR99021) or LEF1 over-expression.
(D) Clonogenic assays of resistant cell lines (cultured with 1 μM MAPKi) −/+ LEF1 over-expression, with DMSO (9 d) or GSK3βi (10 μM,18 d).
(E) As in A except signature enrichment and differential expression indicated for YAP1.
(F) Western blot analysis of indicated parental (P) and isogenic resistant cell lines for levels of total and phospho-YAP1 and fraction marker proteins in total (T), cytoplasmic (C), and nuclear (N) fractions. MAPKi, 1 μM.
(G) Three-day survival assays of resistant melanoma cell lines at two indicated [BRAFi] or [BRAFi+MEKi], −/+ YAP1 knockdown.
*<0.01, **<0.001, ***<0.0001, Student t-test p-values, −/+ LEF1 over-expression.
Although analysis of recurrent, differential mRNA expression can uncover key transcriptome- and methylome-based resistance genes such as c-MET and LEF1, we asked whether gene signature-based analysis would identify resistance genes with post-transcriptional mechanism. One such candidate resistance gene, YAP1, was supported by recurrent signature enrichment without necessarily its mRNA up-expression in both MAPKi-resistant tumors and cell lines (Figure 4E). Importantly, Western blot analysis showed that these acquired MAPKi-resistant cell lines harbored increased total levels of YAP1 and phospho-YAP1, in the cytoplasmic, nuclear or both compartments, compared with levels in the untreated parental cell lines (Figure 4F). In fact, BRAFi treatment of the M229 parental line led to an accumulation of YAP1 protein but not its mRNA level. Consistently, we showed, using a validated anti-YAP1 antibody (Figure S3G), that YAP1 protein was up-expressed in disease progressive melanoma tissues (despite the lack of YAP1 mRNA up-expression) compared to their baseline tissues (Figure S3H). Notably, in SDR and DDR melanoma cell lines with positive enrichment of YAP1 signatures, YAP1 knockdown (Figure S3I) re-sensitized these resistant cell lines to BRAFi or BRAFi+MEKi (Figure 4G).
Given known intersections between the β-catenin-LEF1 and YAP1 signaling pathways in other biological contexts, we explored whether β-catenin-LEF1 down-regulation and YAP1 up-regulation may co-regulate resistance. Inhibition of GSK3β with over-expression of LEF1 in YAP1 signature-enriched, MAPKi-resistant cell lines strongly induced apoptosis, as measured by PARP1 cleavage (cPARP1) (Figure 5A), suggesting that β-catenin-LEF1 signaling promoted apoptotic sensitivity to MAPKi. Since BIM levels are known to modulate melanoma sensitivity to apoptotic induction, we tested whether GSK3βi treatment and restoration of LEF1 expression would promote the levels of the pro-apoptotic protein BIM. Indeed, this combination accelerated and/or augmented BIM accumulation in all acquired MAPKi-resistant cell lines tested (Figure 5B). Interestingly, while GSK3β inhibition and YAP1 knockdown each induced apoptosis, they together induced greater apoptosis (Figure 5C) in association with BIM accumulation (Figure 5D). Consistently, GSK3β inhibition with YAP1 knockdown resulted in the most extensive clonogenic growth suppression of acquired MAPKi-resistant melanoma cell lines (Figure 5E). Conversely, heterologous over-expression of YAP1, beyond the endogenously up-expressed levels in these resistant cell lines, reduced apoptosis and BIM induction by GSK3β inhibition (Figures 5F and 5G). Accordingly, exogenous YAP1 over-expression strongly rescued MAPKi-resistant melanoma cells from clonogenic growth suppression elicited by GSK3β inhibition (Figure 5H). Together, these results support the concept that β-catenin-LEF1 and YAP1 signaling antagonistically co-regulate the tumor cell-intrinsic, apoptotic threshold of melanoma to MAPKi.
Figure 5. β-catenin-LEF1 and YAP1 Co-regulate Apoptotic Sensitivity of Acquired MAPKi-resistant Melanoma Cells.
(A) Western blot (WB) analysis of indicated resistant melanoma cell lines (cultured with 1 μM MAPKi) for levels of cleaved PARP1 (cPARP1), LEF1, and GAPDH (loading control). Cells were treated with DMSO or GSK3βi (10 μM CHIR99021), −/+ LEF1 over-expression, for 1–2 days.
(B–D) As in A, except cell lysates collected at 12 and 24 h and probed for BIM levels (B, D) or harbored shVector (V) or shYAP1 (C, D).
(E) Clonogenic assays of indicated resistant cell lines (cultured with 1 μM MAPKi) treated with DMSO or GSK3βi (10 μM CHIR99021) for 14 d, −/+ YAP1 knockdown.
(F, G) As in A, except cells −/+ YAP1 over-expression and WBs were probed for YAP1 and cPARP1 (F) or BIM (G).
(H) As in E, except −/+ YAP1 over-expression; all cultures for 14 d except M238 DDR cultures with GSK3βi (22 d).
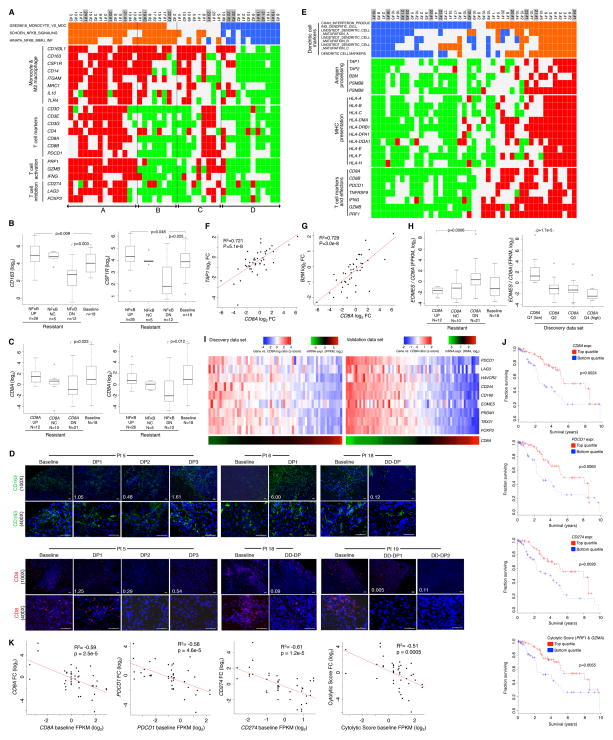
Acquiring MAPKi-resistance Can Deplete and Exhaust Intra-tumoral CD8 T-cells
Among the most recurrent gene-based transcriptomic alterations in acquired MAPKi resistant melanomas were those related to tumor-associated immune cells or inflammatory states (Figures 1B and 1C). By quantifying changes in gene set enrichment values between resistant vs. matched baseline tumors, we found that the most highly recurrent net positive enrichments were in signatures related to NFκB signaling or inflammation (from C6 oncogenic signatures, Broad Institute), monocyte functions (C7 immune signatures) and additional immune and inflammation signatures related to, for instance, T-cell function, serum response, and NFκB signaling (C2 chemical and genetic perturbation signatures) (Data S2A, S2B, and S2C). In assessing a potential relationship between monocyte function and intra-tumoral inflammation, we found that the enrichments (positive or negative) of NFκB/inflammation and of monocyte activation signatures aligned correspondingly with each other (Figure 6A). Additionally, positive enrichment of NFκB or inflammation signatures related strongly to the up-expression of a panel of M2 macrophage markers, including those among the top GOF genes such as CD163 and CD163L1 (Figure 1B). Thus, changes in tumor-associated macrophages likely contributed to distinct inflammatory states in melanomas with acquired MAPKi resistance.
Figure 6. A Pro-tumorigenic Immune Microenvironment Co-evolves with MAPKi Resistance.
(A) Tiling of differential gene signature enrichment (orange, positive; blue, negative) and expression (red, up; green, down) across disease progressive melanomas (DD-DP samples shaded grey).
(B, C) Boxplots of mRNA levels detected in baseline vs. subsets of MAPK-resistant melanomas categorized by enrichment status of the SCHOEN_NFKB_SIGNALING signature (B, C) or differential CD8A expression status (C).
(D) Anti-CD8 and -CD163 immunofluorescence of formalin-fixed, patient-matched melanoma tissues (ruler, 50 micron; white text, values of mRNA fold change).
(E) Refer to A.
(F–G) Correlations between mRNA levels of TAP1 (E) or B2M (F) vs. CD8A.
(H) Boxplots of the ratio of EOMES/CD8A mRNA levels in baseline vs. distinct subsets of MAPKi-resistant melanoma based on CD8A fold change (FC) status (left) and in each quartile of CD8A expression across all tumor samples (right).
(I) Heatmaps (left, discovery; right, validation) showing expression ratios of T-cell exhaustion genes to CD8A. Bottom, absolute mRNA levels of CD8A.
(J) Ten-year survival of TCGA melanoma patients in the top and bottom quartile expression groups of indicated genes (P-values, log rank test).
(K) Pearson correlations between mRNA levels at baseline vs. the FC from baseline to MAPKi-resistant melanomas.
P-values by t-test for Pearson correlations and by one-sided Wilcoxon rank-sum test for boxplots. FC cutoff at ≥1.5.
Given that tumor-associated M2 macrophages can antagonize the recruitment and effector functions of T-cells, we analyzed the relationship between macrophage-associated inflammation with expression markers of T-cell abundance/function. Interestingly, we found that a subset (group B, Figure 6A) among MAPKi-resistant melanomas with enhanced expression of macrophage-associated inflammation (groups A+B, Figure 6A) was strongly associated with reduced expression of T-cell marker/function. CSF1R and CD163 (M2 macrophage markers) expression levels displayed a highly positive correlation that was comparable to the correlation between CD8A and CD8B expression levels (Data S2D and S2E), indicating that the majority of CSF1R up-expression occurred in CD163-positive macrophage cells. Furthermore, negative enrichment of the NFκB signature in resistant tumors was significantly associated with expression loss of tumor-associated M2 macrophage markers, CD163 or CSF1R (Figure 6B). The putative deficiency of tumor-associated M2 macrophages and a pausi-inflammatory tumor microenvironment marked a second subset of intra-tumoral T-cell loss. Importantly, a similar pattern of immune re-composition was observed the validation data set (Data S2F). Thus, the states of macrophage and T-cell inflammation co-evolved with MAPKi resistance, suggesting the potential utility of CSF1R inhibitors.
We investigated further the dynamic loss of CD8 T-cells in a significant subset of acquired MAPKi-resistance. We observed: (1) a concurrent down-expression of CD8A, CD8B, PDCD1 (PD-1) and TNFRSF9 (4-1BB/CD137), where PDCD1 (PD-1) and TNFRSF9 (4-1BB/CD137) expression marks the melanoma tumor-reactive CD8 T-cell population (Gros et al., 2014) (Table S4), and (2) a suppression of CD8 T-cell numbers (reflected by the absolute CD8A expression values) to a level significantly below the baseline range, and (3) CD8 T-cell suppression in association with down-enrichment of the NFκB signature (Figure 6C; Data S2G). We could directly visualize suppression of CD8 T-cells using anti-CD8 (along with differential levels of macrophages using anti-CD163) immunofluorescence of fixed tissues from tumor sections adjacent to those subjected to RNASeq (Figure 6D). Given highly recurrent down-expression of antigen presentation genes (e.g., B2M, HLA-A, HLA-B and TAP1) (Figure 1C), we analyzed the potential relationship in the dynamic expression of genes related to antigen presentation, dendritic cells and CD8 T-cells (Figure 6E). This revealed a consistent concurrence between levels of intra-tumoral antigen presentation and CD8 T-cells/function in both the discovery and validation tissue cohorts, indicating that ~50% of all resistant melanoma displayed a relative loss of CD8 T-cells and their function (Data S2H). This was not surprising considering a general and tight correlation between CD8A vs. TAP1 or B2M expression levels among all melanomas in our cohort (Figures 6F and 6G), in the validation cohort (Figure S4A) and in the TCGA melanoma cohort (Figure S4B). Importantly, the subgroups of resistant melanomas with CD8A down-expression or of all melanomas with the lowest quartile of CD8A expression displayed a high ratio of EOMES/CD8A expression (where EOMES is a transcription factor related to T-cell exhaustion (Twyman-Saint Victor et al., 2015)) (Figures 6H and S4C). Reduced intra-tumoral CD8A expression was associated with increased ratios of expression in a panel of CD8 T-cell exhaustion genes over CD8A (Figure 6I), and the sub-group of resistant tumors with CD8A down-expression was strongly associated with T-cell exhaustion (Figure S4D). Moreover, expression levels of the CD8 T-cell marker (CD8A), its functional feedback (PDCD1/PD-1, CD274/PD-L1) driven by IFNγ, and its effector function (geometric average of PRF1-, GZMA-expressions defined as a cytolytic score (Rooney et al., 2015)) (Figure 6E) were found to be clinically important, as they impacted patient survival in the TCGA melanoma data (Figure 6J). Lastly, the dynamic nature of intra-tumoral CD8 T-cells before and during progression on MAPKi therapy was further underscored by an inverse relationship in the expression levels of these CD8 T-cell marker/function genes at baseline and during disease progression (Figures 6K and S4E). In short, MAPKi resistance and CD8 T-cell deficiency/exhaustion co-evolve frequently with down-regulation of the antigen presentation machinery.
DISCUSSION
Unraveling the complexities of cancer genomics has relied heavily on the recurrence of genetic events as a sine qua non evidence for functional selection. However, when the evolution of melanoma treated with MAPK inhibitor(s) was actually sampled by serial biopsies, the genetic variants positively selected by the inhibitors were not highly recurrent and together could not explain clinical relapse comprehensively. Here, we showed that gene-and signature-based transcriptomic alterations in acquired MAPKi-resistant melanoma were highly recurrent. Transcriptomic alterations, unlike mutations, could not be attributed to tumor or stromal/immune cells without specific validations through histologic analysis of tissues and functional analysis of cell line models of acquired resistance. We highlighted specific genes (c-MET, LEF1, YAP1) and pathways subject to recurrent differential regulation in resistant tumor cells. That c-MET up- and LEF1 down-expression cause acquired MAPKi-resistance is reminescent of findings that HGF stimulation and β-catenin activation can modulate innate BRAFi sensitivity (Biechele et al., 2012; Straussman et al., 2012). Thus, determinants of innate vs. acquired MAPKi resistance may converge on pathways. This is further exemplified in studies showing PI3K-AKT activation in both early and late resistance (Obenauf et al., 2015; Shi et al., 2014a; Shi et al., 2014b; Shi et al., 2011). As YAP1 signal activation in resistant melanoma appeared post-transcriptional, its altered post-translational regulation requires additional studies. Overall, genetic and epigenetic mechanisms can account broadly for disease progression on MAPKi therapies and contribute extensively to intra-tumor/patient and inter-patient tumor heterogeneity (Figures 7A and 7B).
Figure 7. Melanoma Evolution Driven by MAPK-targeted Therapies.
(A) Recurrence and heterogeneity of acquired resistance genes and mechanisms.
(B) Scope of genomic and/or non-genomic acquired resistance mechanisms.
(C) Dynamic gene expression alterations during the evolution of acquired resistance. Black circle and triangle, distinct tumor subsets.
(D) Contributions of non-genomic alterations to cancer phenotypes of acquire MAPKi-resistant melanoma.
For some of the gene targets of transcriptomic and functional alterations identified in MAPKi-resistance (e.g., c-MET, TAP1, B2M), their baseline expression ranges in the TCGA data were shown to impact patient survival. This point is of particular importance given the highly dynamic (i.e., out-of-range) up-expression (c-MET, YAP1, EPHA2 (Paraiso et al., 2014)) or down-expression (LEF1, TAP1, B2M, CD8A, DUSP4) events occurring with the evolution of MAPKi-resistant in melanoma (Figure 7C). It may not be surprising to find that the expression levels of genes reflective of CD8 T-cell and antigen presentation abundance and function were linked to patient survival given the relatively high mutation/neoantigen load and immunogenicity of melanoma and the clinical efficacy of PD-1 targeting in melanoma. We also presented evidence that differential CpG methylation likely underlay dynamic expression of the tumor cell-intrinsic transcriptome during the evolution of MAPKi-resistance. Broadly, the selection of distinct transcriptomic-methylomic state(s) imposed by MAPK-targeting likely impacts the panoply of melanoma phenotypes or “hallmarks” (Figure 7D).
Transcriptomic analysis of temporally paired tumor biopsies revealed highly recurrent evolutionary events in the immune compartment. That half of all melanoma with acquired MAPKi-resistance displayed a profound CD8 T-cell deficiency and exhaustion should bear on the selection of patients for salvage immunotherapies, specifically PD-1 inhibitors, and the clinical sequencing of immune checkpoint vs. MAPK inhibitors. We showed that the expression levels of PD-1, T-cell effector genes, and a marker of melanoma tumor-reactive CD8 T-cells, TNFRSF9 (4-1BB/CD137), tightly correlated with CD8A expression. Specifically, CD8A expression in disease progressive tumors can decrease with respect to not only the patient-matched baseline expression level but also the general baseline expression range (i.e., infra-physiologic). This distinctive expression pattern of CD8A, in both relative and absolute terms, denotes both CD8 T-cell depletion and exhaustion. Finally, since high intra-tumoral CD8 T-cell inflammation before therapy was correlated with a loss of intra-tumoral CD8 T-cell inflammation at disease progression, studies are warranted to examine the functional contributions of immune evasion to acquired MAPKi-resistance, as CD8 T-cells may contribute to the anti-tumor response of BRAF inhibition in vivo (Knight et al., 2013; Mok et al., 2015).
To anticipate cancer evolution, the iterative process of understanding acquired resistance and informing next-generation therapies should incorporate analysis of both genomic and non-genomic selection, including tumor and host-immune co-evolution. The extent of non-genomic and immune evolution in acquired MAPKi resistance documented here mandates a comprehensive analysis of early tumor responses to therapies in order to understand the true influence of targeted therapies on cancer evolution.
EXPERIMENTAL PROCEDURES
Analyses of Tumor Specimens
Patient-matched normal tissues and melanoma tumors (pre-treatment, during disease progression) were obtained with the approval of Institutional Review Boards (IRB) and patients’ consents. Ninety specimens were subjected to exome, transcriptome, and methylome profiling. WES and mRNA expression profiles were performed using pair-end sequencing with read length of 2×100 bps (Illumina HiSeq2000), except that microarray was used for tumors from patients #10–14 where data for patient #11–14 were taken from a published study (Long et al., 2014a; Rizos et al., 2014)). Paired methylome profiles were generated from the Illumina Infinium Methyl450K array.
Bioinformatic Analysis
We re-analyzed WES data from previous studies (Moriceau et al., 2015; Shi et al., 2014b) and defined differential gene expression (DGE) events based on the RNASeq (2-fold cutoff) as concordant DGE calls from at least two of three programs (Supplemental Experimental Procedures). Analysis of the microarray data (DGE cutoff at 1.5-fold) of patient #10 was performed using the Oligo R package and of the validation data set using the beadarray R package. For DGE of immune genes, we relaxed the cutoffs to 1.5-fold (RNASeq) and 1.25-fold (microarray), as the immune compartments were smaller than the tumor cell compartments. In addition to DGE, for each baseline and DP or DD-DP sample, we tabulated the normalized gene expression levels, which were expressed in FPKM. Expressed SNVs or INDELs were defined by FPKM values ≥ 0.1. CNV-related DGE events were defined as concurrent copy number gain and RNA up-expression (log2 FC ≥ 1 with q-value ≤ 0.05) or copy number loss and RNA down-expression (log2 FC ≤ −1, q-value ≤ 0.05). To analyze gene set enrichment of paired samples, we estimated the enrichment of a gene set based on the rank sum of fold changes of genes in the set compared to all fold changes in the sample (Wilcoxon rank-sum test; p-value cutoff ≤ 0.05; median of up- or down-expression across all genes in the gene set ≥ 10%). For single-sample gene set enrichments were derived using the GSVA program. GO enrichments were computed using DAVID.
For each CpG site in the array, the methylation change was measured by the percent methylation difference (Δβ) between baseline and resistant samples. The p-values for the change were corrected for multiple hypotheses testing with false discovery rates (FDR) q-values ≤ 0.05 defining differential methylation. CpG clusters were defined as a set of consecutive CpG sites whose methylation changes correlated with their nearest target gene’s mRNA expression changes across all resistant samples and cell lines with Those with Pearson R correlation coefficient (adjusted) P-value ≤ 0.1 were defined as expression-correlated, and their Pearson R coefficients were defined as correlation scores. For each sample, we identified all CpG clusters with significant differential methylation (q-value ≤ 0.05, |Δβ| ≥ 10%) and significant DGE (q-value ≤ 0.05, |log2| FC| ≥ 1) and assessed whether the direction of the changes of methylation and mRNA expression was consistent with the overall correlation between the CpG cluster and gene expression across all samples. Based on this, we nominated DGE events as driven by differential methylation at the expression-correlated CpG cluster(s).
Cell Culture, Inhibitors, and Constructs
Cell lines were maintained in DMEM with 10% heat-inactivated FBS, 2 mmol/L glutamine in a humidified 5% CO2 incubator. Stocks and dilutions of crizotinib (Selleck Chemicals), PLX4032 (Plexxikon, Berkeley, CA, USA), AZD6244 (Selleck Chemicals), and CHIR99021 (Tocris Bioscience) were made in DMSO. HGF (Life technologies) was suspended in PBS. MTT and clonogenic assays were performed and quantified as described (Moriceau et al., 2015). shc-METs were cloned using the pLL3.7-GFP vector (sequences available upon request); shYAP1s were purchased from GE Dharmacon (vector pLK0.1); and LEF1, YAP1 and NRAS WT or Q61R were constructed in pRRLsin-cPPT-CMV-IRES-GFP and pLVX-Tight-puro lentiviral vectors.
Protein Detection
Cell lysates for Western blots were made in RIPA buffer (Sigma) supplemented with protease (Roche) and phosphatase (Santa Cruz Biotechnology) inhibitor cocktails. We used the NE-PER® Nuclear and Cytoplasmic extraction reagents (Pierce Biotechnology) for cellular fractionation. For IHC, after deparaffinization and rehydration, tissue sections were antigen-retrieved at 95°C for 30 minutes. Immunostaining with anti-c-MET (Cell Signaling Technology) was performed using a streptavidin–biotin, horseradish peroxidase and DAB chromogen (Vector Labs). IHC with anti-YAP1 (Santa Cruz Biotechnology) and anti-LEF1 (Santa Cruz Biotechnology) was performed using alkaline phosphatase, vulcan fast red chromogen (Biocare Medical), and hematoxylin counterstain (Thermo Scientific). Immunostaning with anti-CD8 (Dako) and anti-CD163 (Abcam) was visualized by TRITC- and FITC-labeled secondary antibodies, respectively, and nuclei were counterstained by DAPI.
Supplementary Material
(A) Integrated Analysis of Exome, Transcriptome, and Methylome Alterations in Acquired MAPKi-resistant Melanoma. Genomic DNA (gDNA) and mRNA samples were extracted from the source tissues and subjected to profiling by the indicated omic platforms. Specific bioinformatic algorithms (text in grey) were used to compile lists of SNVs (single nucleotide variants), INDELs (small insertion and deletions), CNVs (copy number variations), differentially methylated CpG clusters, normalized gene expression levels, and DGE (differential gene expression). Integrated alterations specific to disease progressive melanoma samples (vs. patient-matched baseline samples) were binned into four broad classes. Cancer-related gene alterations were quantified based on recurrence and classified into gain-of-function (GOF) and loss-of-function (LOF) events based on the indicated criteria (text in grey). In parallel to single gene-based analyses, concurrent up- or down- expression of genes in oncogenic and immune-related expression signatures and self-curated TCGA Melanoma signatures were assessed by functional gene set enrichment and quantified based on recurrence.
(B) BRAF reference and variant RNASeq raw read counts are shown (C) to highlight relative expression levels of the wild type and variant alleles in the RNASeq data derived from progression tumor tissues.
(C) M229 cells were engineered to express the empty vector, low or high levels of indicated NRAS mRNA, as measured by Q-RT-PCR (top). In NRAS Q61R expressing lines, the ratio of mutant to WT expression was estimated by Sanger sequencing of the total cDNA (middle). (Bottom) three-day MTT survival assays of engineered M229 cell lines in response to BRAFi or BRAFi+MEKi treatment. BRAFi, vemurafenib; MEKi, selumetinib (all in μM). Error bars, SEM, n=5; normalized to DMSO-treatment as 100%.
(D) The relative proportions of expression-correlated vs. non-correlated CpG clusters in resistant melanoma tissues, cell lines, or both (in at least one resistant tumor and one cell line) across all chromosomes (chromosome Y excluded due to insufficient number of events).
(E) Correlation and significance values comparing differential c-MET mRNA expression vs. (1) differential gDNA methylation at CpG clusters and (2) differential mRNA expression of TF genes known to regulate c-MET expression in melanoma.
(F–H) Patterns of differential methylation on c-MET (F), LEF1 (G) or DUSP4 (H) CpG clusters across melanoma tumors and cell lines with acquired MAPKi-resistance which displayed differential c-MET, LEF1 or DUSP4 expression (BRAFi+MEKi resistant tumors or cell lines shaded grey). For F, differential mRNA expressions of c-MET TFs are also shown.
(I–K) c-MET (I), LEF1 (J) or DUSP4 (K) mRNA expression levels among samples in the top and bottom quartile methylation levels of proposed expression-correlated CpG clusters in the TCGA Melanoma data set. For I, c-MET mRNA expression levels are also shown for the top and bottom quartile mRNA expression groups of the c-MET TFs MITF and PAX3. P-values, one-sided Wilcoxon rank-sum test.
Table S1: Related to Figure 1. Provided as an Excel file.
(A) EPHA2 log2 mRNA expression levels in distinct subsets of disease progressive samples based on differential expression status of EPHA2 and in baseline melanomas (left, discovery RNASeq data set; right, validation microarray data set).
(B) Kaplan-Meier curves based on top and bottom quartiles of expression levels of EPHA2 in the TCGA overall (top) or BRAF mutant only (bottom) melanoma data sets.
(C) Anti-c-MET immunohistochemistry of M229 Parental (P, MAPKi sensitive, in DMSO) and Single Drug Resistant (SDR, BRAFi resistant, in 1 μM vemurafenib). Cell lines were pelleted, resuspended in Histogel, formalin-fixed and paraffin-embedded (FFPE). Ruler, 50 microns.
(A) LEF1, FZD6, CCDN1 and CTNNB1 mRNA expression levels (RMA in log2) in LEF1 down-expressed, MAPKi-resistant melanomas vs. baseline, MAPKi-sensitive melanomas (validation data set).
(B) Anti-LEF1 IHC using FFPE cell pellets of M229 P (DMSO) and M229 SDR (1 μM, vemurafenib).
(C) IHC using validated antibody in (B) showing LEF1 protein levels in patient-matched, paired melanoma samples where the disease progressive sample down-expressed LEF1 mRNA.
(D) Western blot (WB) analysis of relative LEF1 expression levels in isogenic melanoma cell lines (P, parental; SDR, single-drug resistant; DDR, double-drug resistant). TUBULIN, loading control.
(E, F) WB analysis of p-β-catenin and total β-catenin levels over a concentration range (E, parental cell line) or treatment time course (F, drug-resistant cell lines) of GSK3βi (CHIR99021). Experiments in F were performed using SDR and DDR cell lines in the absence or presence of FLAG-LEF1 over-expression.
(G) Anti-YAP1 IHC using FFPE cell pellets of M207 P (DMSO) and M207 P treated acutely with a MEKi (which down-regulated YAP1 on WB analysis).
(H) IHC using validated antibody in (G) showing YAP1 protein levels in patient-matched, paired melanoma samples where the disease progressive sample down-expressed YAP1 mRNA.
(I) WB analysis of YAP1 knockdown in indicated MAPKi-resistant cell lines.
(A) Pearson correlations between mRNA expression levels of CD8A vs. TAP1 or B2M in the validation data set. Expression levels of genes were expressed either as log2 fold change (FC) in MAPKi-resistant vs. pre-treatment, MAPKi-sensitive melanomas or as absolute normalized expression levels (RMA) of all samples, inclusive of sensitive and resistant melanomas.
(B) Pearson correlations between absolute, log2 normalized mRNA expression levels (FPKM) of CD8A vs. TAP1 or B2M in the TCGA Melanoma data set.
(C) Boxplots of the ratio of EOMES/CD8A log2 mRNA expression levels in each quartile of CD8A expression across all tumor samples (left, validation data set; right, TCGA melanoma).
(D) Heatmaps (left, discovery; right, validation) showing expression level ratios of T-cell exhaustion genes to CD8A within each of four sub-groups of tumors. Bottom, absolute expression levels of CD8A.
(E) Pearson correlations between mRNA expression levels of indicated genes at baseline vs. the log2 FC in the expression of the respective genes from baseline to MAPKi-resistant melanomas (validation data set).
Grey shade, BRAFi+MEKi resistant tumors. Orange, positive signature enrichment; blue, negative enrichment. Red, up-expression (FC≥1.25 fold); green, down-expression (FC≤1.25 fold). Pearson correlation analysis p-values, Student t-test. Expression level difference (boxplots) p-values, Wilcoxon rank-sum test.
(A–D) Heatmaps (A, C, D) of mRNA expression fold change (FC) of c-MET, LEF1, and DUSP4 and their top 20 positively and negative correlated genes and the respective relative enrichment values of a mutant BRAF signature. (B) GO terms enriched among genes whose differential expression (FC) was positively or negatively correlated with c-MET FC.
(E) Diagrams of indicated genes showing % methylation changes at all profiled CpG sites, including those sites (green bubbles) and clusters (e.g., C1) nominated by aggregate tumor and cell line analysis as expression-correlated, and mRNA expression fold changes (FC). For each melanoma cell line sub-population (PLX4032 2 days or PLX2D, DTP, DTPP, SDR) selected by increasing durations of BRAFi (vemurafenib, 1 μM) treatments, the changes in methylation (%) or mRNA expression (FC) were expressed relative to those levels in vehicle treated M229.
(F–G) Progressive % methylation changes in BRAFi (vemurafenib, 1 μM)-treated M229 or M238 cells (with increasing treatment durations) across all expression-correlated CpG sites (Pearson correlation R values shown on the top heat scale) nominated by aggregate tumor and cell line analysis and found within up- (left) or down-(right) expression genes in the SDR sub-population with respect to the vehicle-treated parental population. (F, Pearson R correlation values continuous; G, discontinuous).
(A, B, C) Top 20 enriched signatures in the discovery data set (A, C6 Oncogenic; B, C7 Immunologic; C, C2 Curated Chemical and Genetic Perturbations; from the Molecular Signature Database).
(D, E) Pearson correlations between mRNA expression levels of CSF1R vs. CD163 (D) or CD8B vs. CD8A (E). Expression levels of genes were expressed as log2 fold change (FC) in MAPKi-resistant vs. pre-treatment, MAPKi-sensitive melanomas (left) or as absolute normalized expression levels (FPKM) of all samples, inclusive of sensitive and resistant melanomas (right).
(F) Combined recurrence of NFκB and monocyte signature enrichments and up- or down-expression events of genes in the indicated panels (validation data set).
(G) Boxplots of CD8A log2 mRNA expression levels in baseline vs. distinct subsets of MAPKi-resistant melanoma (validation data set).
(H) Combined recurrence of dendritic cell signatures and up- or down-expression events of genes in the indicated panels (validation data set).
Table S2: Related to Figure 2. Provided as an Excel file.
Table S3: Related to Figure 4. Provided as an Excel file.
Table S4: Related to Figure 6. Provided as an Excel file.
Acknowledgments
We are grateful to G. Bollag (Plexxikon Inc.) for providing PLX4032 and Donald Hucks for tissue processing. This work was funded by Burroughs Wellcome Fund (to R.S.L), Stand Up To Cancer (to R.S.L.), Melanoma Research Alliance (to R.S.L.), the National Institutes of Health (1R01CA176111 to R.S.L; 1P01CA168585 to A.R. and R.S.L.; K12CA0906525 to D.B.J.), Wade F.B. Thompson/Cancer Research Institute CLIP Grant (to R.S.L), the SWOG/Hope Foundation (to R.S.L. and A.R.), the Ressler Family Foundation (to R.S.L. and A.R.), the Ian Copeland Melanoma Fund (to R.S.L.), the T.J. Martell Foundation and the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation (to K.B.D.).
Footnotes
Accession Number
The GEO accession number for the transcriptome and methylome data sets reported in this paper is GSE65186.
AUTHOR CONTRIBUTIONS
W.H., H.S., L.S., M.P., C.S., X.K., and R.S.L. designed and performed experiments and analyzed data. X.K., G.M. A.H., K.B.D., D.B.J., J.A.S. A.R. and R.S.L. provided reagents. All authors contributed to manuscript preparation. R.S.L. supervised the study. R.S.L. and W.H. wrote the paper. W.H., H.S., L.S., and M.P. are equal contributing authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Biechele TL, Kulikauskas RM, Toroni RA, Lucero OM, Swift RD, James RG, Robin NC, Dawson DW, Moon RT, Chien AJ. Wnt/beta-catenin signaling and AXIN1 regulate apoptosis triggered by inhibition of the mutant kinase BRAFV600E in human melanoma. Sci Signal. 2012;5:ra3. doi: 10.1126/scisignal.2002274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari de Andrade L, Ngiow SF, Stannard K, Rusakiewicz S, Kalimutho M, Khanna KK, Tey SK, Takeda K, Zitvogel L, Martinet L, et al. Natural Killer Cells Are Essential for the Ability of BRAF Inhibitors to Control BRAFV600E-Mutant Metastatic Melanoma. Cancer Res. 2014;74:7298–7308. doi: 10.1158/0008-5472.CAN-14-1339. [DOI] [PubMed] [Google Scholar]
- Girotti MR, Pedersen M, Sanchez-Laorden B, Viros A, Turajlic S, Niculescu-Duvaz D, Zambon A, Sinclair J, Hayes A, Gore M, et al. Inhibiting EGF receptor or SRC family kinase signaling overcomes BRAF inhibitor resistance in melanoma. Cancer Discov. 2013;3:158–167. doi: 10.1158/2159-8290.CD-12-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros A, Robbins PF, Yao X, Li YF, Turcotte S, Tran E, Wunderlich JR, Mixon A, Farid S, Dudley ME, et al. PD-1 identifies the patient-specific CD8(+) tumor-reactive repertoire infiltrating human tumors. J Clin Invest. 2014;124:2246–2259. doi: 10.1172/JCI73639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP, Barretina J, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen CM, Johnson LA, Piccioni F, Townes A, Frederick DT, Donahue MK, Narayan R, Flaherty KT, Wargo JA, Root DE, et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature. 2013;504:138–142. doi: 10.1038/nature12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight DA, Ngiow SF, Li M, Parmenter T, Mok S, Cass A, Haynes NM, Kinross K, Yagita H, Koya RC, et al. Host immunity contributes to the anti-melanoma activity of BRAF inhibitors. J Clin Invest. 2013;123:1371–1381. doi: 10.1172/JCI66236. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Larkin J, Ascierto PA, Dreno B, Atkinson V, Liszkay G, Maio M, Mandala M, Demidov L, Stroyakovskiy D, Thomas L, et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med. 2014;371:1867–1876. doi: 10.1056/NEJMoa1408868. [DOI] [PubMed] [Google Scholar]
- Laurent S, Queirolo P, Boero S, Salvi S, Piccioli P, Boccardo S, Minghelli S, Morabito A, Fontana V, Pietra G, et al. The engagement of CTLA-4 on primary melanoma cell lines induces antibody-dependent cellular cytotoxicity and TNF-alpha production. J Transl Med. 2013;11:108. doi: 10.1186/1479-5876-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidsky M, Antoun G, Speicher P, Adams B, Turley R, Augustine C, Tyler D, Ali-Osman F. Mitogen-activated protein kinase (MAPK) hyperactivation and enhanced NRAS expression drive acquired vemurafenib resistance in V600E BRAF melanoma cells. J Biol Chem. 2014;289:27714–27726. doi: 10.1074/jbc.M113.532432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long GV, Fung C, Menzies AM, Pupo GM, Carlino MS, Hyman J, Shahheydari H, Tembe V, Thompson JF, Saw RP, et al. Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nat Commun. 2014a;5:5694. doi: 10.1038/ncomms6694. [DOI] [PubMed] [Google Scholar]
- Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, Garbe C, Jouary T, Hauschild A, Grob JJ, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med. 2014b;371:1877–1888. doi: 10.1056/NEJMoa1406037. [DOI] [PubMed] [Google Scholar]
- Mok S, Tsoi J, Koya RC, Hu-Lieskovan S, West BL, Bollag G, Graeber TG, Ribas A. Inhibition of colony stimulating factor-1 receptor improves antitumor efficacy of BRAF inhibition. BMC Cancer. 2015;15:356. doi: 10.1186/s12885-015-1377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriceau G, Hugo W, Hong A, Shi H, Kong X, Yu CC, Koya RC, Samatar AA, Khanlou N, Braun J, et al. Tunable-Combinatorial Mechanisms of Acquired Resistance Limit the Efficacy of BRAF/MEK Cotargeting but Result in Melanoma Drug Addiction. Cancer Cell. 2015;27:240–256. doi: 10.1016/j.ccell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Krijgsman O, Tsoi J, Robert L, Hugo W, Song C, Kong X, Possik PA, Cornelissen-Steijger PD, Foppen MH, et al. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat Commun. 2014;5:5712. doi: 10.1038/ncomms6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obenauf AC, Zou Y, Ji AL, Vanharanta S, Shu W, Shi H, Kong X, Bosenberg MC, Wiesner T, Rosen N, et al. Therapy-induced tumour secretomes promote resistance and tumour progression. Nature. 2015;520:368–372. doi: 10.1038/nature14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraiso KH, Das Thakur M, Fang B, Koomen JM, Fedorenko IV, John JK, Tsao H, Flaherty KT, Sondak VK, Messina JL, et al. Ligand independent EphA2 signaling drives the adoption of a targeted therapy-mediated metastatic melanoma phenotype. Cancer Discov. 2014;5:264–273. doi: 10.1158/2159-8290.CD-14-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizos H, Menzies AM, Pupo GM, Carlino MS, Fung C, Hyman J, Haydu LE, Mijatov B, Becker TM, Boyd SC, et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: spectrum and clinical impact. Clin Cancer Res. 2014;20:1965–1977. doi: 10.1158/1078-0432.CCR-13-3122. [DOI] [PubMed] [Google Scholar]
- Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Laorden B, Viros A, Girotti MR, Pedersen M, Saturno G, Zambon A, Niculescu-Duvaz D, Turajlic S, Hayes A, Gore M, et al. BRAF inhibitors induce metastasis in RAS mutant or inhibitor-resistant melanoma cells by reactivating MEK and ERK signaling. Sci Signal. 2014;7:ra30. doi: 10.1126/scisignal.2004815. [DOI] [PubMed] [Google Scholar]
- Shi H, Hong A, Kong X, Koya RC, Song C, Moriceau G, Hugo W, Yu CC, Ng C, Chodon T, et al. A Novel AKT1 Mutant Amplifies an Adaptive Melanoma Response to BRAF Inhibition. Cancer Discov. 2014a;4:69–79. doi: 10.1158/2159-8290.CD-13-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G, Chodon T, Guo R, Johnson DB, Dahlman KB, et al. Acquired Resistance and Clonal Evolution in Melanoma during BRAF Inhibitor Therapy. Cancer Discov. 2014b;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Kong X, Ribas A, Lo RS. Combinatorial treatments that overcome PDGFRβ-driven resistance of melanoma cells to B-RAF(V600E) inhibition. Cancer Research. 2011;71:5067–5074. doi: 10.1158/0008-5472.CAN-11-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Moriceau G, Kong X, Koya RC, Nazarian R, Pupo GM, Bacchiocchi A, Dahlman KB, Chmielowski B, Sosman JA, et al. Preexisting MEK1 exon 3 mutations in V600E/KBRAF melanomas do not confer resistance to BRAF inhibitors. Cancer Discov. 2012a;2:414–424. doi: 10.1158/2159-8290.CD-12-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Moriceau G, Kong X, Lee MK, Lee H, Koya RC, Ng C, Chodon T, Scolyer RA, Dahlman KB, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat Commun. 2012b;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J, Frederick DT, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twyman-Saint Victor C, Rech AJ, Maity A, Rengan R, Pauken KE, Stelekati E, Benci JL, Xu B, Dada H, Odorizzi PM, et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature. 2015;520:373–377. doi: 10.1038/nature14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, Place CS, Taylor-Weiner A, Whittaker S, Kryukov GV, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva J, Infante JR, Krepler C, Reyes-Uribe P, Samanta M, Chen HY, Li B, Swoboda RK, Wilson M, Vultur A, et al. Concurrent MEK2 mutation and BRAF amplification confer resistance to BRAF and MEK inhibitors in melanoma. Cell Rep. 2013;4:1090–1099. doi: 10.1016/j.celrep.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, Kehoe SM, Johannessen CM, Macconaill LE, Hahn WC, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle N, Van Allen EM, Treacy DJ, Frederick DT, Cooper ZA, Taylor-Weiner A, Rosenberg M, Goetz EM, Sullivan RJ, Farlow DN, et al. MAP Kinase Pathway Alterations in BRAF-Mutant Melanoma Patients with Acquired Resistance to Combined RAF/MEK Inhibition. Cancer Discov. 2014;4:61–68. doi: 10.1158/2159-8290.CD-13-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Integrated Analysis of Exome, Transcriptome, and Methylome Alterations in Acquired MAPKi-resistant Melanoma. Genomic DNA (gDNA) and mRNA samples were extracted from the source tissues and subjected to profiling by the indicated omic platforms. Specific bioinformatic algorithms (text in grey) were used to compile lists of SNVs (single nucleotide variants), INDELs (small insertion and deletions), CNVs (copy number variations), differentially methylated CpG clusters, normalized gene expression levels, and DGE (differential gene expression). Integrated alterations specific to disease progressive melanoma samples (vs. patient-matched baseline samples) were binned into four broad classes. Cancer-related gene alterations were quantified based on recurrence and classified into gain-of-function (GOF) and loss-of-function (LOF) events based on the indicated criteria (text in grey). In parallel to single gene-based analyses, concurrent up- or down- expression of genes in oncogenic and immune-related expression signatures and self-curated TCGA Melanoma signatures were assessed by functional gene set enrichment and quantified based on recurrence.
(B) BRAF reference and variant RNASeq raw read counts are shown (C) to highlight relative expression levels of the wild type and variant alleles in the RNASeq data derived from progression tumor tissues.
(C) M229 cells were engineered to express the empty vector, low or high levels of indicated NRAS mRNA, as measured by Q-RT-PCR (top). In NRAS Q61R expressing lines, the ratio of mutant to WT expression was estimated by Sanger sequencing of the total cDNA (middle). (Bottom) three-day MTT survival assays of engineered M229 cell lines in response to BRAFi or BRAFi+MEKi treatment. BRAFi, vemurafenib; MEKi, selumetinib (all in μM). Error bars, SEM, n=5; normalized to DMSO-treatment as 100%.
(D) The relative proportions of expression-correlated vs. non-correlated CpG clusters in resistant melanoma tissues, cell lines, or both (in at least one resistant tumor and one cell line) across all chromosomes (chromosome Y excluded due to insufficient number of events).
(E) Correlation and significance values comparing differential c-MET mRNA expression vs. (1) differential gDNA methylation at CpG clusters and (2) differential mRNA expression of TF genes known to regulate c-MET expression in melanoma.
(F–H) Patterns of differential methylation on c-MET (F), LEF1 (G) or DUSP4 (H) CpG clusters across melanoma tumors and cell lines with acquired MAPKi-resistance which displayed differential c-MET, LEF1 or DUSP4 expression (BRAFi+MEKi resistant tumors or cell lines shaded grey). For F, differential mRNA expressions of c-MET TFs are also shown.
(I–K) c-MET (I), LEF1 (J) or DUSP4 (K) mRNA expression levels among samples in the top and bottom quartile methylation levels of proposed expression-correlated CpG clusters in the TCGA Melanoma data set. For I, c-MET mRNA expression levels are also shown for the top and bottom quartile mRNA expression groups of the c-MET TFs MITF and PAX3. P-values, one-sided Wilcoxon rank-sum test.
Table S1: Related to Figure 1. Provided as an Excel file.
(A) EPHA2 log2 mRNA expression levels in distinct subsets of disease progressive samples based on differential expression status of EPHA2 and in baseline melanomas (left, discovery RNASeq data set; right, validation microarray data set).
(B) Kaplan-Meier curves based on top and bottom quartiles of expression levels of EPHA2 in the TCGA overall (top) or BRAF mutant only (bottom) melanoma data sets.
(C) Anti-c-MET immunohistochemistry of M229 Parental (P, MAPKi sensitive, in DMSO) and Single Drug Resistant (SDR, BRAFi resistant, in 1 μM vemurafenib). Cell lines were pelleted, resuspended in Histogel, formalin-fixed and paraffin-embedded (FFPE). Ruler, 50 microns.
(A) LEF1, FZD6, CCDN1 and CTNNB1 mRNA expression levels (RMA in log2) in LEF1 down-expressed, MAPKi-resistant melanomas vs. baseline, MAPKi-sensitive melanomas (validation data set).
(B) Anti-LEF1 IHC using FFPE cell pellets of M229 P (DMSO) and M229 SDR (1 μM, vemurafenib).
(C) IHC using validated antibody in (B) showing LEF1 protein levels in patient-matched, paired melanoma samples where the disease progressive sample down-expressed LEF1 mRNA.
(D) Western blot (WB) analysis of relative LEF1 expression levels in isogenic melanoma cell lines (P, parental; SDR, single-drug resistant; DDR, double-drug resistant). TUBULIN, loading control.
(E, F) WB analysis of p-β-catenin and total β-catenin levels over a concentration range (E, parental cell line) or treatment time course (F, drug-resistant cell lines) of GSK3βi (CHIR99021). Experiments in F were performed using SDR and DDR cell lines in the absence or presence of FLAG-LEF1 over-expression.
(G) Anti-YAP1 IHC using FFPE cell pellets of M207 P (DMSO) and M207 P treated acutely with a MEKi (which down-regulated YAP1 on WB analysis).
(H) IHC using validated antibody in (G) showing YAP1 protein levels in patient-matched, paired melanoma samples where the disease progressive sample down-expressed YAP1 mRNA.
(I) WB analysis of YAP1 knockdown in indicated MAPKi-resistant cell lines.
(A) Pearson correlations between mRNA expression levels of CD8A vs. TAP1 or B2M in the validation data set. Expression levels of genes were expressed either as log2 fold change (FC) in MAPKi-resistant vs. pre-treatment, MAPKi-sensitive melanomas or as absolute normalized expression levels (RMA) of all samples, inclusive of sensitive and resistant melanomas.
(B) Pearson correlations between absolute, log2 normalized mRNA expression levels (FPKM) of CD8A vs. TAP1 or B2M in the TCGA Melanoma data set.
(C) Boxplots of the ratio of EOMES/CD8A log2 mRNA expression levels in each quartile of CD8A expression across all tumor samples (left, validation data set; right, TCGA melanoma).
(D) Heatmaps (left, discovery; right, validation) showing expression level ratios of T-cell exhaustion genes to CD8A within each of four sub-groups of tumors. Bottom, absolute expression levels of CD8A.
(E) Pearson correlations between mRNA expression levels of indicated genes at baseline vs. the log2 FC in the expression of the respective genes from baseline to MAPKi-resistant melanomas (validation data set).
Grey shade, BRAFi+MEKi resistant tumors. Orange, positive signature enrichment; blue, negative enrichment. Red, up-expression (FC≥1.25 fold); green, down-expression (FC≤1.25 fold). Pearson correlation analysis p-values, Student t-test. Expression level difference (boxplots) p-values, Wilcoxon rank-sum test.
(A–D) Heatmaps (A, C, D) of mRNA expression fold change (FC) of c-MET, LEF1, and DUSP4 and their top 20 positively and negative correlated genes and the respective relative enrichment values of a mutant BRAF signature. (B) GO terms enriched among genes whose differential expression (FC) was positively or negatively correlated with c-MET FC.
(E) Diagrams of indicated genes showing % methylation changes at all profiled CpG sites, including those sites (green bubbles) and clusters (e.g., C1) nominated by aggregate tumor and cell line analysis as expression-correlated, and mRNA expression fold changes (FC). For each melanoma cell line sub-population (PLX4032 2 days or PLX2D, DTP, DTPP, SDR) selected by increasing durations of BRAFi (vemurafenib, 1 μM) treatments, the changes in methylation (%) or mRNA expression (FC) were expressed relative to those levels in vehicle treated M229.
(F–G) Progressive % methylation changes in BRAFi (vemurafenib, 1 μM)-treated M229 or M238 cells (with increasing treatment durations) across all expression-correlated CpG sites (Pearson correlation R values shown on the top heat scale) nominated by aggregate tumor and cell line analysis and found within up- (left) or down-(right) expression genes in the SDR sub-population with respect to the vehicle-treated parental population. (F, Pearson R correlation values continuous; G, discontinuous).
(A, B, C) Top 20 enriched signatures in the discovery data set (A, C6 Oncogenic; B, C7 Immunologic; C, C2 Curated Chemical and Genetic Perturbations; from the Molecular Signature Database).
(D, E) Pearson correlations between mRNA expression levels of CSF1R vs. CD163 (D) or CD8B vs. CD8A (E). Expression levels of genes were expressed as log2 fold change (FC) in MAPKi-resistant vs. pre-treatment, MAPKi-sensitive melanomas (left) or as absolute normalized expression levels (FPKM) of all samples, inclusive of sensitive and resistant melanomas (right).
(F) Combined recurrence of NFκB and monocyte signature enrichments and up- or down-expression events of genes in the indicated panels (validation data set).
(G) Boxplots of CD8A log2 mRNA expression levels in baseline vs. distinct subsets of MAPKi-resistant melanoma (validation data set).
(H) Combined recurrence of dendritic cell signatures and up- or down-expression events of genes in the indicated panels (validation data set).
Table S2: Related to Figure 2. Provided as an Excel file.
Table S3: Related to Figure 4. Provided as an Excel file.
Table S4: Related to Figure 6. Provided as an Excel file.