Abstract
Interconnected molecular networks are at the heart of signaling pathways that mediate adaptive plasticity of eukaryotic cells. To gain deeper insights into the underlying molecular mechanisms, a comprehensive and representative analysis demands a deep and parallel coverage of a broad spectrum of molecular species. Therefore, we introduce a simultaneous metabolite, protein, lipid extraction (SIMPLEX) procedure, a novel strategy for the quantitative investigation of lipids, metabolites, and proteins. Compared with unimolecular workflows, SIMPLEX offers a fundamental turn in study design since multiple molecular classes can be accessed in parallel from one sample with equal efficiency and reproducibility. Application of this method in mass-spectrometry-based workflows allowed the simultaneous quantification of 360 lipids, 75 metabolites, and 3327 proteins from 106 cells. The versatility of this method is shown in a model system for adipogenesis— peroxisomal proliferator-activated receptor gamma (PPARG) signaling in mesenchymal stem cells—where we utilized SIMPLEX to explore cross-talk within and between all three molecular classes and identified novel potential molecular entry points for interventions, indicating that SIMPLEX provides a superior strategy compared with conventional workflows.
The cross-talk between lipid metabolism and protein-based signaling imposes interactions at various levels that are not well understood. Such interactions play a central role in the pathophysiology of many metabolic disorders such as insulin resistance, cancer, and obesity (1–3), which increases the demand for novel methodology to tackle this problem from a global and representative perspective (4). A major limitation in the study of such interactions is the known dependence of the regulations of interconnected systems, such as nuclear receptor signaling, on a multitude of factors. Important factors are activity, localization and abundance of proteins, the overall lipid distribution including the concentration of certain signaling lipids, and the accessibility of metabolites as building blocks. Evident examples of such consolidated, heterogeneous signaling systems are the ceramide and peroxisomal proliferator-activated receptor (PPAR) signaling pathways, which are both deeply intertwined with lipid metabolism (5, 6). In ceramide signaling, the formation of ceramides is regulated both by the sphingolipid metabolism and by signaling events such as ceramide-mediated activation of protein phosphatase 2A (PP2A), cathepsin D, or p38 MAPK and their downstream effects on apoptosis and proliferation (7–10). Fat cell differentiation, known as adipogenesis, is controlled by a complex interconnected system with PPARG as the master regulator of this system. During adipogenesis, high initial levels of glucocorticoids, cAMP and PPARG ligand trigger differentiation, leading to an increased PPARG and CCAAT/enhancer-binding protein alpha (CEBPA) protein expression level as well as to an elevated insulin sensitivity (11–13).
These and other coupled signaling systems illustrate an unmet need for a parallel analysis of lipid- and protein-based signaling in order to understand complex mechanisms at a systems biology level. An essential prerequisite is the availability of methods that enable the simultaneous, comprehensive, unbiased, and quantitative analysis of proteins, lipids, and metabolites from a single sample, rather than separate analysis with unimolecular strategies. While independent molecular extraction techniques may allow for the correlation of individual molecular classes, the additional experimental deviations, increased duration of the sample preparation, and the high sample consumption that would be needed for such parallel large-scale studies poses several limitations in study design and is a particular challenge for clinically derived tissues or cells.
The few current approaches that focus on individual molecular classes (14–16) to analyze e.g. lipid protein cross-talk cannot be used for multimolecular studies from the very same sample and thus neglect important and possibly direct interplay between metabolic and signaling events. Here, we demonstrate that lipids, metabolites, and proteins as well as protein post-translational modifications (PTMs) can be extracted and analyzed from the same sample in an unbiased and reproducible fashion, thus supporting parallel systems-wide quantification. We developed a biphasic organic extraction protocol for simultaneous metabolite, proteins lipid extraction (SIMPLEX), which is adapted to state-of-the-art lipidomics, metabolomics, and proteomics workflows. The performance, sensitivity, and reproducibility of SIMPLEX are comparable in quality to currently used, well-established unimolecular protocols. Finally, the applicability of SIMPLEX from a systems biology perspective was validated and confirmed by the study of the PPAR signaling network during the onset of adipogenesis.
EXPERIMENTAL PROCEDURES
Reagents and Standards
Methyl-tert-butyl-ether (MTBE)1, chloroform, iodacetamide, calcium chloride (CaCl2), ammonium bicarbonate, triethylammonium bicarbonate, ammonium acetate, and ammonium hydroxide were purchase from Sigma Aldrich (Steinheim, Germany). Water with 0.1% ammonium acetate (LC-MS grade) was purchased from Fluka (Buchs, Switzerland). Acetonitrile, 2-propanol, and methanol, with high chemical purity and high UV transmission (ULC/MS grade)-grade were obtained from Biosolve (Valkenswaard, The Netherlands). Sequencing grade Trypsin was obtained from Promega (Mannheim, Germany). Dithiothreitol (DTT) and benzonase were purchased from Roche Diagnostics (Mannheim, Germany). Tris(hydroxymethyl)-aminomethane (Tris) was purchased from Applichem (Darmstadt, Germany), sodium dodecyl sulfate (SDS) from Roth (Karlsruhe, Germany), and sodium chloride (NaCl) from Merck (Darmstadt, Germany). Bicinchoninic acid (BCA) assay was purchased from Thermo Scientific (Schwerte, Germany). The lipid standards: cholesteryl ester 22:0, triacylglycerol (TAG) 14:0/14:0/14:0, diacylglycerol 10:0/10:0, phosphatidic acid (PA) 17:0/14:1, phosphatidiylserine 12:0/13:0, phosphatidylethanolamine (PE) 21:0/22:6, lysophosphatidylcholine 19:0, phosphatidylcholine (PC) 21:0/22:6, phosphatidylglycerol (PG) 17:0/20:4, phosphatidylinositol (PI) 17:0/14:1, and ceramide/sphingoid mixture II were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). Metabolite standards (nonisotope-labeled) were serine, proline, riboflavin, succinic acid, tryptophan, reduced gluthathione, adenosine, and nicotinamide adenine dinucleotide from Sigma Aldrich and the corresponding isotope-labeled compounds: serine (2,3,3-D3, 98%), proline (2,3,3,4,4,5,5-D7, 97–98%), riboflavin (13C4, 99%; 15N2, 98%), succinic acid (D4, 98%), tryptophan (indole-D5, 98%), reduced glutathione (glycine-13C2, 98%; 15N, 96–99%), and adenosine (ribose-13C5, 98%) from Euriso-Top (Saarbrücken, Germany) and nicotinamide adenine dinucleotide (13C5, 96%) from Syncom (Groningen, The Netherlands).
Phosphate buffered saline (PBS, pH = 7.4), fetal bovine serum (FBS), minimum essential media (MEM-α), and penicillin-streptomycin-glutamine (100x) solution were obtained from Gibco while, goat anti-mouse antibody Alexa Fluor® 488 and BODIPY® 493/503 were purchased from Life Technologies (Darmstadt, Germany) and DAPI from AppliChem GmbH (Darmstadt, Germany). Rosiglitazone was obtained from Biomol (Hamburg, Germany) and the mouse PPARG antibody from Santa Cruze Biotechnologies (Germany).
Cell Culturing
OP9, a mesenchymal cell line from mouse bone marrow, was purchased from the Tokyo Metropolitan Institute of Medical Science. OP9 cells were grown in growth medium consisting of MEM-α, 2 mm l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin, plus 20% FBS, collected by centrifugation, washed once with PBS solution, and resuspended in 0.1% ammonium acetate. To study PPARG activation, for each condition, three biological replicates were prepared; the cells were grown as described above but with 10% FBS, and were treated for 48 h with different concentrations (1 nm to 1 μm) or fixed concentration (1 μm) of rosiglitazone, a PPARG activator. Thereby, a 0.01% DMSO solution served as a control.
Immunofluorescence Staining
OP9 cells were fixed with 5% formaldehyde in PBS for 30 min. Then the cells were gently washed three times with PBS and permeabilized with 0.05% saponin (Sigma 47036), blocked with 3% bovine serum albumin (Sigma 7906) and stained with DAPI (1:1000), anti-PPARG (1:500 Santa Cruz Biotechnology SC-7273) or BODIPY 493/503 (1 μg/ml, Molecular Probes d-3922) and with Alexa Fluor-488 as secondary antibody for the anti PPARG staining.
Experimental Design
For each extraction method, three replicates were carried out. Based on previous studies with this model system (12) also, three biological experiments were employed for the PPARG activation. A molecular species (lipid, metabolite, or protein) was confirmed as regulated if a fold change of 2 in the 95% confidence interval (Student's t test) was achieved.
SIMPLEX Protocol
For extraction, three aliquots, each containing 106 cells were used and treated with MeOH in tubes containing the internal lipid standard. 225 μl of cold MeOH were added to the cells, and they were vortexed for 20 s and incubated in liquid nitrogen for 1 min. Further, the samples were allowed to thaw at room temperature (RT) and were sonicated for 10 min at 4 °C. The cell lysis cycle and the sonication step were repeated three times. Then, 750 μl of cold MTBE were added, and the mixture was incubated for 1 h at 4 °C under agitation. Phase separation was induced by adding 188 μl water with 0.1% ammonium acetate. The extract was centrifuged at 10,000 g for 5 min, and the upper phase was collected, dried under nitrogen flow, and dissolved in 100 μl 2-Prop/MeOH/CHCl3 (4:2:1, v/v/v) containing 7.5 mm ammonium acetate for further analysis (lipid-containing fraction). For protein precipitation, MeOH was added to the remaining lower phase in a final ratio of 4:1, v/v MeOH/H2O, and the samples were incubated for 1 h at −20 °C, followed by 12 min centrifugation (13,000 g) at 4 °C. The resulting supernatant was removed (metabolite-containing fraction), and the remaining pellet dissolved in 1% SDS, 150 mm NaCl, 50 mm Tris (pH 7.8) for further proteomics experiments (protein-containing fraction). The dried extracts of metabolites were resuspended in 100 μl acetonitrile/water (ACN/H2O) (9:1, v/v), mixed, centrifuged for 12 min (13,000 g) at 4 °C to remove any insoluble debris, and then stored at −20 °C prior to LC/MS analysis.
Proteomics Sample Preparation
BCA assay was performed to determine protein concentrations. The disulfide bonds were reduced with 10 mm DTT for 30 min at 56 °C, and free sulfhydryl groups were alkylated using 30 mm iodacetamide for 30 min at room temperature in the dark. Samples were processed using filter-aided sample preparation (FASP) (17, 18), with minor changes. Briefly, samples were diluted to 10 mm SDS using freshly prepared 8 m urea, 100 mm Tris-HCl, pH 8.5, and loaded onto a spin filter by centrifugation for 25 min at 13,500 g. Afterward, 8 m urea/100 mm Tris-HCl at pH 8.5 was placed on the filter, followed by centrifugation for 15 min at 13,500 g; this step was repeated three times. Finally, the samples were washed three times with 50 mm triethylammonium bicarbonate, and the digestion was performed in 50 mm triethylammonium bicarbonate, 0.2 m GuHCl, 2 mm CaCl2, using a trypsin to protein ratio of 1:20 (w/w) for 12 h at 37 °C (19). Peptides were eluted by centrifugation, with 50 mm triethylammonium bicarbonate and acidified to a final concentration of 1% TFA. Digestion efficiency was controlled by monolithic reverse phase (RP) separation and the samples were stored at −40 °C (20).
Phosphopeptides Enrichment
Phosphopeptides were enriched using titanium dioxide, as previously described (21). Samples were dried under vacuum and resuspended in TiO2 loading buffer (80% ACN, 5% TFA, and 1 m glycolic acid). TiO2 beads were added in a bead-to-peptide ratio of 6:1. The suspension was incubated for 10 min at room temperature, and centrifuged to pellet the beads containing the phosphopeptides. This step was repeated twice using 1/2 and 1/4 of the amount of beads previously used for the three enrichment steps. Beads were pooled and washed with 80% ACN, 1% TFA, followed by a second washing step with 10% ACN, 0.1% TFA. Phosphopeptides were eluted with 1% ammonium hydroxide, pH 11.3. The eluted phosphopeptides were recovered and dried under vacuum, followed by a second enrichment step as previously described (19), however, using loading buffer 2 (70% ACN, 2% TFA). The eluates were acidified using formic acid and TFA (pH < 2), desalted using Oligo R3 microcolumns, and stored at −40 °C until analysis.
Recovery of Metabolite Standards
Aliquots of a mixture of unlabeled metabolite standards were pipetted into 2 ml Eppendorf tubes and dried under N2 flow. A second mixture containing stable isotope labeled versions of these metabolites was added to three out of nine aliquots (control), dried under N2 flow, resuspended in ACN/H2O (9:1, v/v), and stored at −20 °C prior to LC/MS analysis. The other aliquots were subjected to either SIMPLEX or MeOH extraction protocols as described above. Metabolite-containing pellets were redissolved in the isotopic-labeled mix, dried, and prepared for LC MS/MS analysis as the control samples. Metabolite analysis was performed as described above. The metabolite standard recovery was determined as the ratio of the peak areas of the labeled and the unlabeled standards.
Amino Acid Analysis
The absolute protein amount was determined before SIMPLEX and after SIMPLEX by amino acid analysis. Therefore, OP9 cells were lysed in 1% SDS buffer and divided in two sets of samples with three technical replicates, where each replicate represented ∼106 cells. Nine volumes of ice cold ethanol were added to the lysates and the samples were incubated overnight at −40 °C. Afterward, the supernatant was removed by centrifugation at 20,000 g for 30 min at 4 °C. From the resulting pellets, one set of samples (control) was stored at −80 °C for amino acid analysis, and the second set was subjected to the SIMPLEX workflow, as described above, prior to amino acid analysis. Next, protein pellets were hydrolyzed with acidic gas phase hydrolysis for 24 h at 110 °C using 6 N HCl and 0.2% phenol. Subsequently, amino acids were derivatized according to Cohen et al. (22) and norleucine was added as internal standard prior to this step. The derivatized amino acids were analyzed on an Ultimate 3000 system with a fluorescence detector (Thermo Scientific). As mobile phase, a modified buffer system as previously described (23) was used, and the amino acids were separated on a 130Å, 2.5 μm, 1 × 100 mm XBridge C18 column (Waters) using a flow rate of 100 μl/min in a 45 min gradient. Quantification of total protein amount was performed based on a five-point calibration curve (5–25 pmol/μl).
Lipid Analysis, Direct Infusion MS, and MS/MS analysis
The lipid extracts were diluted in 2-Prop/MeOH/CHCl3 (4:2:1, v/v/v) with 7.5 mm ammonium acetate in a 96-well plate (Eppendorf, Hamburg, Germany) and then infused via robotic nanoflow ion source TriVersa NanoMate (Advion BioSciences, Ithaca NY, USA) into an LTQ Orbitrap Velos instrument (Thermo Fisher Scientific, Bremen, Germany) using chips with spraying nozzles of 4.1 μm. The ion source was controlled by Chipsoft 8.3.1 software (Advion Biosciences). Ionization voltage was set to +1.25 kV in positive and –1.25 kV in negative mode; backpressure was set at 0.95 psi in both modes. The temperature of the ion transfer capillary was adjusted to 175 °C and the s-lens level was 63%. Polarity switch of the TriVersa NanoMate ion source was triggered by the mass spectrometer via contact closure signal as described previously (24).
For data-dependent experiments, full MS spectra were acquired under the target mass resolution Rm/z 400 of 100,000 (full width at half maximum at m/z 400) and background peaks of octadecyl (di-tert-buthylhydroxyphenyl) propionate ([M-H]−, m/z 529.4626), and decaethylene glycol ([M+NH4]+, m/z 476.3063) were used as lock masses. MS/MS experiments were carried out using higher energy collision dissociation in the higher energy collision dissociation collision cell. Precursors were selected within the m/z window of 1.4, and the fragment ions were detected in the Orbitrap mass analyzer using a target mass resolution Rm/z 400 of 7 500 full width at half maximum (FWHM). Data-dependent acquisition (DDA) experiments were repeated three times for each sample.
All spectra were imported by LipidXplorer software into a MasterScan database under the following settings: mass tolerance 5 ppm; range of m/z 400–1200; min occupation of 1; intensity threshold 1 × 104. Lipid identification was carried on as described by Herzog et al., (25, 26). For each glycerophospholipid class, the average number of carbon atoms and the average number of double bonds was calculated as previously described (27).
Metabolite Analysis, LC-MS/MS
The metabolite analyses were performed on an UltiMate 3000 system coupled to a QTRAP 6500 mass spectrometer (AB SCIEX). For separation, hydrophilic interaction liquid chromatography (HILIC) was carried out on a Luna-HILIC column (150 × 2 mm, 3-μm particle size, 200 Å pore size) from Phenomenex® (CA, USA) equipped with a HILIC guard column of 4 × 2.0 mm. The mobile phases were 95% acetonitrile (A) and 10 mm ammonium acetate and 10 mm ammonium hydroxide in 95% H2O (B). The gradient eluted isocratically with 95% ACN for 3 min followed by an increase to 50% over 6 min, held at 50% for 1 min, and increased to 65% over 5 min. Subsequent reconstitution of the starting conditions and re-equilibration with 95% A for 10 min resulted in a total analysis time of 25 min. The LC separation was carried out at 25 °C under a flow rate of 180 μl min-1, and 5 μl of sample were injected. After each sample injection, one blank run was performed to avoid any carry-over effects.
ESI Turbo V source parameters were set as follows: curtain gas, 30 arbitrary units; temperature, 350 °C; ion source gas 1, 40 arbitrary units; ion source gas 2, 65 arbitrary units; collision gas, medium; ion spray voltage, 5500 V/-4500 V (positive mode/negative mode). For the selected reaction monitoring (SRM) mode, Q1 and Q3 were set to unit resolution and the transitions for all compounds were adapted (28, 29). The SRM conditions can be found in the Table S4. For data acquisition and analysis Analyst (1.6.2) and MultiQuant 3.0 were used, respectively.
Protein Analysis, Nano-LC-MS/MS and SRM
Samples were separated on an Ultimate 3000 Rapid Separation Liquid Chromatography (RSLC) system (Dionex, Amsterdam, Netherlands) coupled to an Orbitrap Fusion for label-free analysis, a Q Exactive Plus for the PPARG end point study, or Orbitrap Elite mass spectrometer (Thermo Scientific) for phosphopeptide analysis, using data-dependent MS/MS acquisition. Peptides were preconcentrated on a 100 μm inner diameter trapping column (Acclaim C18 PepMap100, 100 μm × 2 cm, Thermo Scientific) followed by separation on a 75 μm inner diameter (ID) RP main column (Acclaim C18 PepMap100, 75 μm × 50 cm, Thermo Scientific) using a binary gradient (solvent A: 0.1% formic acid and solvent B: 0.1% formic acid in 84% ACN) at a flow rate of 250 nl/min. The gradient increased linearly from 5% to 45% B over 200 min and from 3% to 42% B over 108 min for phosphopeptide analysis. In order to minimize carry-over effects from previous samples, a dedicated LC-wash program with high organic content was used prior to each sample injection (20).
Survey scans were acquired in the Orbitrap with a resolution of 120 000 (Fusion) or 60 000 (Elite) FWHM. Data-dependent MS/MS of the 20 (Fusion) or 15 (Elite) most intense signals were acquired in the ion trap using collision-induced dissociation for Fusion and Elite. MS target values and maximum injection times were 4 × 105 ions and 50 ms for the Orbitrap Fusion, and 1 × 106 ions and 100 ms for the Orbitrap Elite. For MS/MS scans, the target values were set to 1 × 104 ions and 35 ms for Fusion and 1 × 104 ions and 50 ms for Elite. For the Q Exactive analysis, full MS scans were acquired at a resolution of 70 000 FWHM, followed by MS/MS of the 15 most abundant ions at 17,500 FWHM. Target value and maximum injection time were set to 3 × 106 ions and 120 ms for the full scan and 5 × 104 ions and 250 ms for MS/MS scans. Only precursors with charge states between +2 and +5 were selected for fragmentation.
Scheduled SRM experiments were conducted on a TSQ Vantage (Thermo Fisher Scientific) in the positive ion mode with the following tuning parameters: retention time window of 5 min, a cycle time of 1 s, and an average dwell time of 26 ms; the Q1 and Q3 were set to 0.70 u (FWHM), emitter voltage was set to 1500 V, and the temperature of the transfer capillary to 270 °C.
Label-Free Data Analysis
Label-free quantification was performed using the Progenesis LC-MS software version 4.1 from Nonlinear Dynamics (Newcastle upon Tyne, UK). MS raw data were aligned in automatic mode. Exported peak lists were searched using searchGUI 1.14.4 (30), Mascot 2.4 (Matrix Science), and X!Tandem Jackhammer (2013.06.15), using a concatenated target/decoy version of the mouse Uniprot database (downloaded on December 11, 2013, containing 20,273 target sequences). Data analysis was done using PeptideShaker 0.28.0 (http://code.google.com/p/peptide-shaker/) in order to maximize the number of identified peptides and proteins. The following search parameters were used: trypsin as protease with a maximum of two missed cleavages; carbamidomethylation of Cys as fixed and oxidation of Met as variable modification. MS and MS/MS tolerances were set to 10 ppm and 0.5 Da. In PeptideShaker (31), search results were combined and filtered at a false discovery rate of 1% on the protein level prior to export, and reimported into Progenesis. Peptide sequences containing oxidized Met and pyro-Glu (derived from X!Tandem second pass search) were omitted from further data analysis. Only proteins with at least two unique peptides were considered for quantification. For each protein, the average of the normalized abundances (obtained from three replicates processed with Progenesis) was calculated to determine the ratios between SIMPLEX and FASP and for the comparison of PPARG activated versus nonactivated cells, respectively. Finally, only proteins that were (i) commonly quantified in all six samples with (ii) at least two unique peptides, (iii) an analysis of variance p value of < 0.05 Progenesis, Anova, and (iv) an average ratio < 0.5 or > 2 (corresponding to twofold regulation) were considered as regulated. The transitions list of the investigated proteins can be found in the Supplemental material (Tab. S5). The mass spectrometry data may be downloaded from ProteomeXchange (32) (username: reviewer45796@ebi.ac.uk, password: ITWoxgTY).
Phosphoproteomics Data Analysis
Phosphopeptide data analysis was done using Proteome Discoverer version 1.4 (Thermo Scientific). Data were searched against the mouse Uniprot database (downloaded on December 11, 2013, containing 20,273 target sequences) using Mascot and Sequest. Search settings were as described above. Target decoy peptide spectrum matches validator was used to assess 1% false discovery rate (FDR) at the level of peptide spectrum matches. Finally, PhosphoRS 3.1 was used to determine phosphorylation site localization (33). Only phosphopeptide peptide spectrum matches with site localization ≥99% were considered. The ready-to-use Excel macro provided by Mechtler lab (http://ms.imp.ac.at/?goto = phosphors) was used to determine the confident phosphorylation sites for each peptide, as well as the position of the phosphorylation within the protein sequence. Further, only unique phopsho-sites were considered by using R-scripts. Peptides were considered as nonredundant if they differed either in their amino acid sequences in phosphorylation site position or number of phosphorylation sites. The mass spectrometry proteomics data were deposited to the ProteomeXchange (32) (username: reviewer05714@ebi.ac.uk, password: er1uFZAg).
Network Analysis
For pathway and network visualization upon PPARG activation, we used the browser -software String 9.1 (http://string-db.org/) and Cytoscape 3.2.0, including the plugin ClueGO for GO-Term analysis. In String 9.1, a confidence score with a cutoff at 0.5 was applied, correlating in this way coexpression, experimental, knowledge, and text-mining scores.
To visualize the dynamics upon PPARG activation, we color coded the nodes of the visualized network. Proteins showing a fold change ≥ 2 were considered up-regulated and marked in red, whereas for down-regulated proteins fold change ≤ 0.5 are indicated in blue. For the visualization of the different pathways that are built up from the regulated proteins, we performed a GO-Term analysis using the plugin ClueGO for Cytoscape. The ontologies were taken from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (updated in ClueGO on November 27, 2014). The network specificity was set between global and medium by showing only significant pathways with a p value of 0.05. The Kappa score for GO Term/Pathway network connectivity was set to 0.4.
RESULTS
The present study introduces SIMPLEX—a workflow for the simultaneous isolation of lipids, metabolites, and proteins to study interconnected networks at the systems scale. The protocol is based on the previously published lipid extraction protocol of Matyash et al. (34). In brief, the cells were incubated with cold MeOH and MTBE, and H2O was added to induce phase separation. Fractions containing lipids (top phase), metabolites (lower phase), and proteins (pellet) were then subjected to individual well-established omics workflows.
Extraction and Workflow Reliability
In order to investigate and analyze the cross talk between proteins, lipids, and metabolites, it is crucial that all chemical species are extracted and measured with high reproducibility and confidence. Therefore, three independent SIMPLEX extractions were conducted sequentially using OP9 cells, a mouse mesenchymal stem cell line with the ability to differentiate to adipocytes (35, 36). Cells were harvested and subsequently lipids, metabolites, and proteins were extracted by MTBE/MEOH, a lipid extraction procedure previously reported by Matyash et al. (34). The obtained methanol, ether, and protein fractions were subjected to the individual molecular-class-dependent omics workflow and analyzed by MS (Fig. 1A). For the proteomics pipeline, precipitated proteins were processed and analyzed after a quality control step using nano LC-MS on a high resolution MS instrument. Raw data were processed with multiple search engines for protein identification together with Progenesis (Nonlinear Dynamics) for label-free quantification.
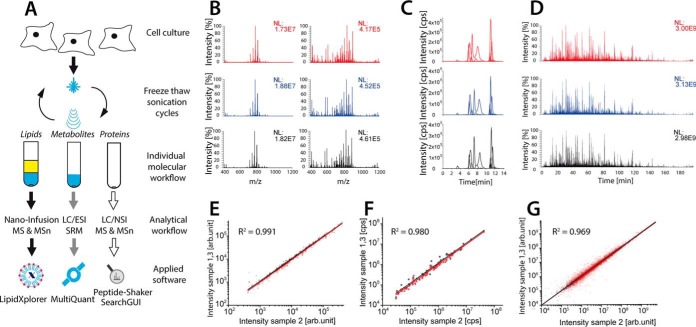
Fig. 1.
SIMPLEX a reproducible workflow to isolate and analyze lipids, metabolites and proteins. (A) For lipid extraction MTBE is added and phase separation is induced. The individual fractions containing lipids (top phase), metabolites (lower phase), and proteins (pellet) are then subjected to the individual omics workflows. Thereby, the lipidomics workflow employs shotgun lipidomics to achieve a quantitative picture of extracted lipids. LipidXplorer is then utilized for identification and quantification. For metabolomics the methanol fraction was subjected to a HILIC separation followed by a selected reaction monitoring (SRM) MS approach; the data were subsequently analyzed by MultiQuant. The remaining protein pellet was dissolved, reduced, alkylated, and trypsinated. Subsequently, the samples are separated by nano-RP liquid chromatography and analyzed by a top 20 MS/MS experiment. For data analysis in all proteomics experiments a combination of different search algorithms was used. (B) FT-MS spectra of the lipid fraction with a resolution of 100,000 (m/z = 400) obtained in positive (left) and negative ionization mode (right). (C) SRM extracted ion chromatograms of monitored metabolite transitions separated on a HILIC column and analyzed with fast polarity switching. (D) Total ion chromatogram obtained from nano-RP peptide separation. For all three extractions, the peak or chromatographic areas of the identified molecular features (lipidomics, metabolomics, and proteomics) were sorted by intensity and a linear fit was applied. The average coefficients of determination were calculated for the (E) lipidomics (F) metabolomics, and (G) proteomics data sets, respectively.
Lipid fractions were analyzed by individual direct infusion experiments, applying a combination of MS and MS/MS-based analysis. The obtained spectra were processed using LipidXplorer (25, 26), a de novo lipid sequencing platform for identification and quantification at the fatty acid scan species level. Extracted metabolites were identified and quantified using selected reaction monitoring (SRM) in HILIC mode, engaging fast polarity switching in order to ensure simultaneous analysis of negatively and positively charged metabolites.
Applying the SIMPLEX protocol and simple MS or LC-MS workflows, 360 lipids, 75 metabolites, and 3327 proteins (with at least two unique peptides per protein) were identified and quantified in a label free fashion from just one sample containing ∼1 million cells per replicate. For all three omics approaches, we achieved an excellent reproducibility, on both the spectral and chromatographic level, even when polarity switching was applied (Figs. 1B-1G). To illustrate the reliability of the workflow, the intensity of each identified molecular feature (protein, lipid, or metabolite) was plotted against the same feature of the two other extraction replicates (Figs. 1E–1G). All plots show coefficients of determination over 0.96, demonstrating the robustness of the applied protocol. An average relative standard deviation (RSD) of 5% for lipids, 15% for metabolites, and 18% for proteins, all over 3–4 orders of magnitude was achieved. Notably, our results are in line with previously published studies in lipidomics, metabolomics, and proteomics reporting similar errors and dynamic ranges for eukaryotic cell culture models (18, 37, 38).
Validation and Comparison with Standard Proteomics and Metabolomics Workflows
To validate the efficiency of our protocol, we directly compared SIMPLEX with state-of-the-art extractions procedures currently used in the fields of metabolomics and proteomics. To assess and verify the protein and metabolite extraction efficiency in comparison to standard proteomics and metabolomics workflows, SIMPLEX was evaluated against filter-aided sample preparation (17, 39) (FASP) for proteomics and MeOH extraction for metabolomics (40).
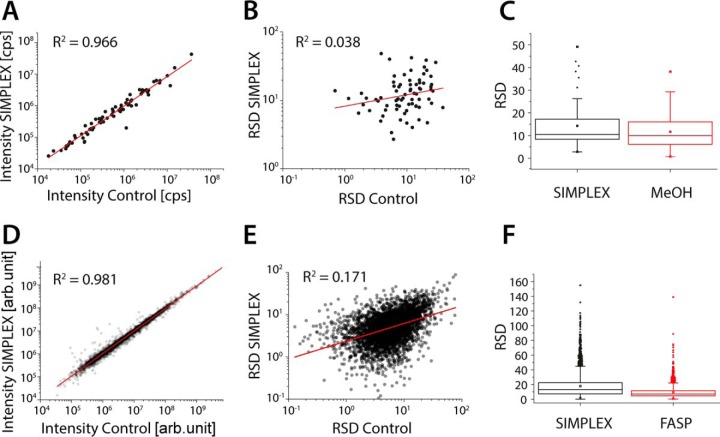
For metabolites, both the SIMPLEX and the standard metabolomics extraction method relied on MeOH/water; consequently similar metabolite intensities are expected, as reflected by the coefficient of determination of R2 = 0.97 (Fig. 2A, Table S1). For both protocols, the quantified metabolites spanned a dynamic range of three to four orders of magnitude and high abundant metabolites such as glutamate and glutathione (mm cellular concentration) could be analyzed next to low abundant metabolites such as adenosine and adenine (low μm cellular concentration) (28). Using stable isotope-labeled standards, we determined the absolute recoveries for eight metabolites representing polar amino acids (serine), aromatic amino acids (tryptophan), nonpolar amino acids (proline), organic acids (succinic acid), cofactors (nicotinamide adenine nucleotide, NAD+), vitamins (riboflavin), peptides (reduced glutathione), and nucleosides (adenosine) using either SIMPLEX or MeOH extraction in triplicate. Recoveries achieved with both methods were very similar for glutathione, succinic acid, serine, proline, and NAD+ with an average of 102 ± 2% for SIMPLEX and 101 ± 2% for MeOH (Fig. S1A). The notable exceptions for SIMPLEX were adenosine, tryptophan, and riboflavin with an average recovery of 64 ± 0.5%, probably due to the partitioning of the compounds, as compared with 90 ± 0.8% for MeOH. Further, we did not observe any bias regarding the error distribution (Fig. 2B) with average RSDs of 12%, and 15% for MeOH and SIMPLEX, respectively (Fig. 2C). For all detected metabolites, the average and median standard deviations were below 15% (Fig. 2C), which is within the range of the instrumentation variability and in a good agreement with previous metabolomics studies (41, 42).
Fig. 2.
SIMPLEX is as efficient as state-of-the-art protocols for metabolomics and proteomics. To compare SIMPLEX against other extraction protocols the proteins were extracted with and without MeOH precipitation prior FASP, followed by a standard proteomics workflow. For metabolites, a standard MeOH extraction versus the MTBE/MeOH extraction was used to demonstrate the comparability of the proposed strategy. Correlation plots and linear fits of the identified molecular features (A) metabolites (control versus SIMPLEX) and (D) proteins (control versus FASP). To investigate if the error between the samples is correlating, the relative standard deviation (RSD) was plotted for each identified feature against the S.D. of the corresponding feature from the control experiment for metabolites (B) and proteins (E). (C) and (F), displaying the average, median RSD for metabolites and proteins for the SIMPLEX and control experiment as well as their error distribution (n = 3 for all experiments).
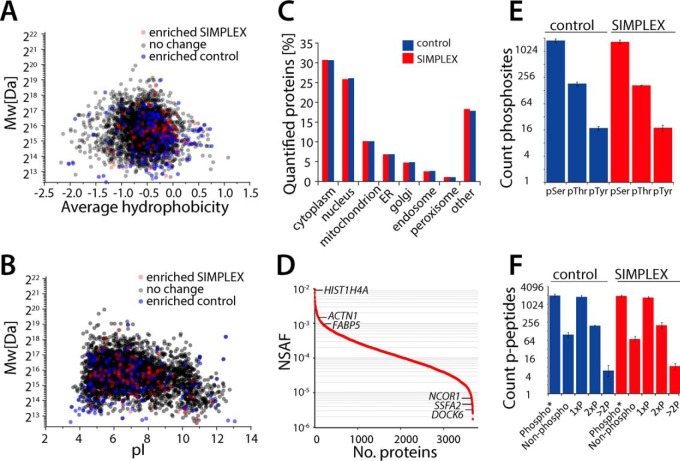
A broad variety of standardized protocols for sample preparation is available in proteomics. Here, we selected the FASP protocol (39) due to its high reproducibility, applicability, and its wide use within the field. Comparing SIMPLEX and the FASP protocol, we obtained a coefficient of determination R2 = 0.997 for all quantified proteins across three replicates (Fig. 2D, Table S2). For both methods, the dynamic range spanned over four orders of magnitude, and the RSDs were 18 and 10% for SIMPLEX and FASP, respectively (Fig. 2F). Importantly, we did not observe a bias concerning standard deviations (Fig. 2E) for particular protein groups or regarding molecular features such as molecular weight (MW), isoelectric point (pI), or hydrophobicity (Figs. 3A and 3B). Furthermore, we performed amino acid analysis to absolutely quantify the total protein content before and after SIMPLEX (biological triplicates) where a recovery of 115% ± 3% was achieved (Fig. S1B), confirming that there is no significant protein content loss. Notably, recoveries slightly above 100% are within the expected accuracy of the respective assays. Both the FASP and SIMPLEX protocol enables the simultaneous analysis of high abundant proteins (> 1,000,000 copies/cell) such as alpha-actinin-1 (ACTN1), fatty acid-binding protein, epidermal (FABP5), and histone H4 (HIST1H4a) as well as low abundant proteins (< 1000 copies/cell) such as nuclear receptor corepressor 1 (NCOR1), sperm-specific antigen 2 homolog (SSFA2), and dedicator of cytokinesis protein 6 (DOCK6) (43) (Fig. 3D), rendering SIMPLEX as an excellent and convenient protocol for label-free quantitative proteomics.
Fig. 3.
SIMPLEX displays no bias regarding protein properties, number of quantified proteins or identified phosphopeptides. (A) Hydrophobicity versus molecular weight plot of every quantified protein. All proteins in black are not significantly changed between the two protocols; the proteins in red show a significant change (> 2.5 or < 0.4) in the SIMPLEX extraction or in blue in the FASP control. (B) The isoelectric point is plotted against the molecular weight whereas the same criteria are applied as in A. (C) Relative protein distribution over different cellular compartments between the SIMPLEX and the control workflow, overall 3327 proteins were quantified. (D) Protein abundance as normalized spectral abundance factor, whereby the protein abundance is covering four orders of magnitude. (E) Number of identified Ser/Thr/Tyr phosphorylation sites between the SIMPLEX and the FASP protocol. (F) Distribution of the phosphorylation sites per peptide between the two protocols, in total 2018 phosphorylation sites were identified with both protocols (n = 3 technical replicates).
Next, we evaluated whether the SIMPLEX method could be employed beyond the application of global proteome analysis. A phosphoproteome analysis was performed by using titanium dioxide in order to enrich phosphopeptides from the digested samples. Using only 60 μg per sample and 2 h LC-MS analysis time, it was possible to identify, on average, between the three replicates, 1957 and 1846 phospho-sites for FASP and SIMPLEX (Fig. 3E, Table S3 a,b), where similar serine, threonine, and tyrosine site distributions and number of phosphorylation sites were obtained (Figs. 3E and 3F).
Moreover, the approach can be carried out in the normal time frame of a proteomics experiment with only an additional 3 h for the SIMPLEX extraction (3 h for the SIMPLEX extraction and 15 h for the proteomics sample preparation).
Together, these experiments provide evidence for the versatility and robustness of our method. Applying SIMPLEX to singular samples resulted in an excellent reproducibility, without any bias and limits from a restricted amount of cells.
Accessibility of the Different Molecular Classes
In order to decipher systematic correlative relationships across molecular layers, a deep coverage of the different molecular classes is mandatory. To ensure the suitability of our SIMPLEX method that can equally identify lipids, metabolites, and proteins with distinct chemical and functional properties, we investigated the class-specific coverage of SIMPLEX.
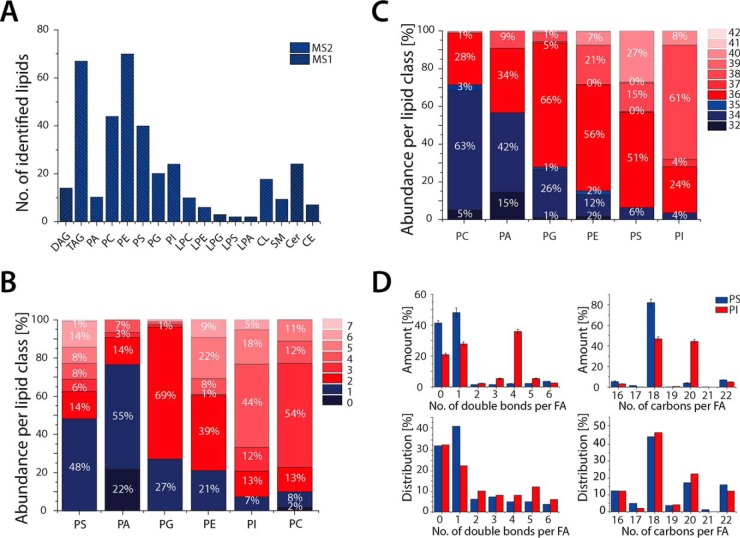
Notably, the SIMPLEX workflow does not interfere with lipid identification. Consequently, the analysis at both the species level (MS) and the fatty acid scan species level (MS/MS) of main glycerophospholipid classes (phosphatidylcholine, PE, phosphatidic acid, PI, phosphatidylglycerol, and phosphatidylserine), glycerolipids (diacylglycerol, TAG), steryl esters, and sphingolipids (ceramide and sphingomyelin) (Fig. 4A) can be carried out (24, 44), exploiting the same advantages of the extraction procedure as described by Matyash et al. (34), namely faster, cleaner lipid extraction and similar or better recoveries compared with Folch or Bligh and Dyer protocols (45, 46). Performing data analysis for glycerophospholipids (PL) at the species level for both fatty acid chains, total carbon counts ranging from 32 to 42 atoms were covered with a higher abundance of species containing 34 and 36 carbon atoms (Fig. 4C). Moreover, the detected glycerophospholipids displayed zero to seven double bonds per lipid species, with one to four double bonds per species being the most abundant at the individual class level (Fig. 4B). These findings are in agreement with other eukaryotic model systems such as Drosophila or human plasma where similar distributions were observed and numbers of species identified (27, 47).
Fig. 4.
Lipid composition of mesenchymal stem cells. (A) Identified lipid classes by direct infusion high resolution MS and MS/MS experiments. In total, 360 lipids were identified. (B) Sum of double bonds in fatty acids for all major phospholipid classes. (C) Total length analysis of glycerophospholipids, as sum of fatty acids, pointing out the most abundant species per class. (D) Investigation at the fatty acid scan species level applying MS/MS experiments elucidates the unsaturation level and chain length per fatty acid and is demonstrating that this distribution is distinct between different glycerophospholipid classes. Examples of phosphatidylserine (PS) and phosphatidylinositol (PI) are given in (D), the upper panel displays the abundance of double bonds and fatty acid chain length (number of carbons), and the lower panel the number of total identified fatty acids with a certain number of double bonds and chain length (n = 3 technical replicates for all performed experiments).
Considering that the lipid analysis was conducted on both MS and MS/MS levels, a further characterization at the fatty acid scan species level was feasible (48, 49), allowing the determination of individual chain length and number of double bonds per chain (Fig. 4D). This revealed that the length of fatty acid chains and number of double bonds differ significantly among various phospholipid classes. For instance, in the class of PI the most abundant fatty acids displayed chain length of 18 or 20 carbon atoms with zero, one, and four double bonds whereas phosphatidylserine's fatty acids have had a chain length of 18 carbon atoms and zero to one double bond per fatty acid. Here, in addition to even-numbered carbon fatty acid chains, less abundant odd chain fatty acids were also detected with chain length ranging from 17 to 21 carbon atoms, confirming the presence of odd chain fatty acid in mammalian cell systems (50).
To investigate correlative effects and interconnectivity between lipids and proteins, metabolites are important entities, as they occupy key positions in signaling and serve as building blocks for lipid and protein synthesis. Considering that probably only about 10% of all existing metabolites are known (51), here we utilized a targeted approach for metabolites involved into central metabolic functions such as amino acid, nucleotides, and nucleosides metabolism (Table S4). We were able to detect intracellular metabolites such as nucleosides, nucleotides and analogues (23), organic acids and derivatives (11), amino acids and derivatives (25), redox cofactors, and others (11), as well as carbohydrates and conjugates (5), thus providing access to metabolites at the systems level.
In addition to the lipidome and metabolome analysis, a significant coverage of the proteome was achieved: 3327 proteins with at least two unique peptides (1% false discovery rate) were identified and quantified without any fractionation steps and only by single-shot LC-MS analyses (Fig. 3C, Table S2). In this study, the identified proteins represent the protein content of the entire cell without bias and are of cytosolic (1016), nuclear (837), mitochondrial (340), endoplasmatic reticular (236), Golgi apparatus (161), endosomal (87), and lysosomal (59) origin, as assigned by GO-term analysis (Fig. 3C). Major metabolic pathways such as oxidative phosphorylation (62), purine metabolism (51), glycolysis/gluconeogenesis (30), citric acid cycle (20), and lipid metabolism (37) were detected in addition to signaling networks with metabolic impact such as insulin (42), PPAR (2522), indicating a representative proteomic coverage of the OP9 proteome. Finally, even with single-shot MS experiments, these findings match or outperform previous reports for other model systems where similar numbers of proteins (900–3287), metabolites (45–100), and lipids (250) were quantitatively accessed (27, 43, 52–54).
To the best of our knowledge, this is the first study quantifying and identifying 3762 molecular species, including lipids, metabolites, and proteins, from one sample. Thus, the application of SIMPLEX enabled us to generate the most exhaustive molecular study of mesenchymal stem cells currently available.
Analysis of a Molecular Network System via SIMPLEX
In order to determine whether this protocol is suitable to identify cross talk between metabolism and signaling, we employed SIMPLEX to study the network of PPAR signaling upon PPARG activation. PPARG activity was monitored by measuring the abundance of PPARG and fatty acid-binding protein (FABP4), a downstream target of PPARG with several PPARG-binding motifs upstream of its corresponding promoter region.
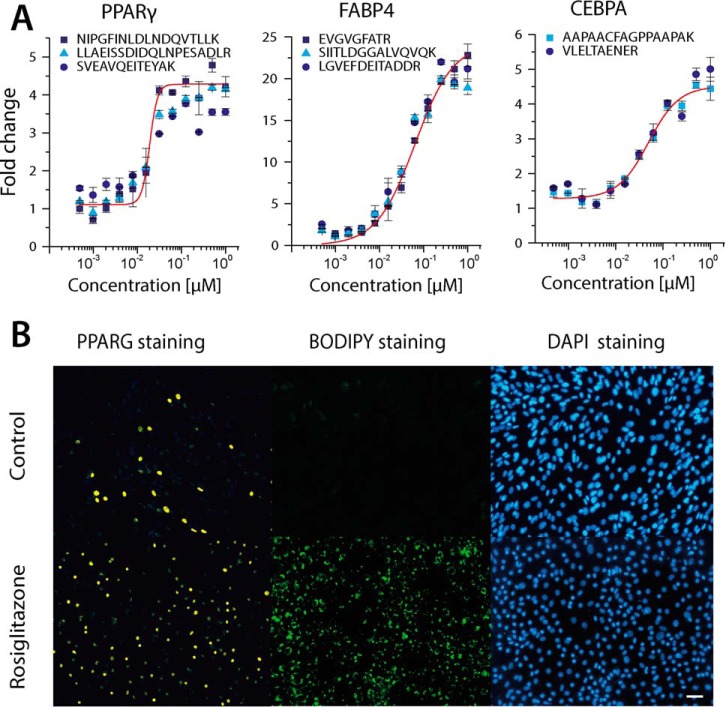
Over a time period of 48 h, PPARG was activated with an increasing agonist stimulus (rosiglitazone) and analyzed by SRM (Fig. 5, Table S5). The sigmoidal increase of PPARG and the induction of FABP4 and CEBPA were monitored as indicators of active PPAR signaling (12, 55). To study the effect of PPARG activation on the proteome, lipidome, and metabolome, we conducted a differential end-point study utilizing samples with the lowest and highest levels of PPARG expression (Fig. 5A). Overall, the abundance of 910 molecules was significantly regulated with a fold change ≥ 2 or ≤ 0.5 in the 95% confidence interval (Figs. 6A–6C). Among those we identified, 154 regulated lipids at the species level, 48 regulated metabolites, and 718 regulated proteins. Taken together, this renders the present work as the most comprehensive study describing the effect of PPARG activation on a cellular system to date. In addition to already known PPARG downstream targets such as platelet glycoprotein 4 (CD36), fatty acid-binding protein (FABP4), solute carrier family 2, facilitated glucose transporter member 4 (GLUT4), and phosphatidate phosphatase LPIN1 (LIPIN1) (11, 56), we identified several so far undescribed targets, including focal adhesion proteins alpha-actinin-1 (ACTN1), zyxin (ZYX), serine/threonine-protein phosphatase PP1-beta catalytic subunit (PPP1CB), myosin regulatory light polypeptide 9 (MYL9) and proteins involved in lipid synthesis (Table S6). Notably, the strongest regulations were observed for the following protein groups: (i) glycerolipid and glycerophospholipid metabolism, (ii) PPAR signaling, (iii) glycolysis/gluconeogeneses, (iv) pentosephosphate pathway, (v) insulin signaling, and (vi) focal adhesion. In total, 420 proteins were found to be up-regulated and 298 were found to be down-regulated after PPARG activation (Fig. 6E; Table S6).
Fig. 5.
PPARG activation induces downstream targets and lipid droplet formation. (A) To activate PPARG signaling confluent OP9 cells were treated over 48 h with different amounts of rosiglitazone ranging from 0.005 to 1 μm. Subsequently, the cells were harvested and subjected to a targeted proteomics workflow monitoring PPARG as well as its downstream targets FABP4 and CEBPA with at least two peptides. (B) To further validate obtained MS data the expression of adipogenesis master regulator PPARG and lipid accumulation after 48 h compared with the control (0.01% DMSO v/v) is illustrated protocols (n = 3 biological replicates). Immunohistochemistry staining of OP9 cells using specific antibodies to visualize PPARG (yellow), BODIPY 493/503 to visualize lipid droplets (green), and DAPI to visualize nuclei (blue) is presented. Scale bar, 100 μm.
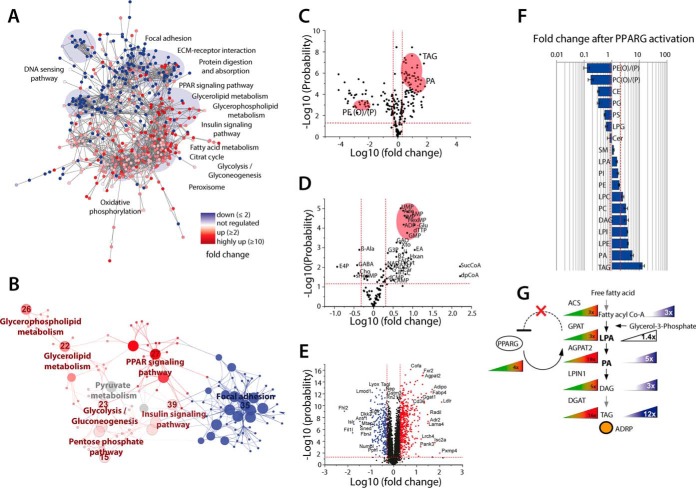
Fig. 6.
Activated PPAR signaling alters major metabolic pathways and lipid distribution. Mesenchymal OP9 mouse stem cells with the capability to differentiate to adipocytes were treated with 1 μm rosiglitazone for 48 h to induce PPARG signaling. After induction of PPARG, three induced and three unstimulated controls were subjected to the SIMPLEX workflow. (A) Cytoscape network analysis in which the nodes display the quantified proteins and the strings describe the relations between those at the confidence level. All proteins in red are at least twofold up-regulated whereas proteins in blue are at least twofold down-regulated. (B) The prominent regulated pathways at the protein level, here the same color code is applied. (C) Volcano plot of quantified lipids with 222 regulated species after the activation of PPARG signaling pathway, indicating a major remodeling of the entire lipidome. The most relevant molecules regarding PPARG signaling are indicated in red: TAG, PA, and plasmanyl or plasmenylphosphatidyletanolamine PE(O/P). (D) Volcano plot of the regulation of metabolites after PPARG activation. (E) Volcano plot of 3327 quantified proteins, 285 were classified as down-regulated and 435 proteins as up-regulated. (F) Bar plot of major regulated lipid classes. (G) Triacylglycerol synthesis pathway with all major enzymes and lipid abundance changes that are going along with activation of PPARG. For all experiments, the following criteria were applied n = 3 biological replicates and n = 2 for technical replicates.
The dynamic changes induced upon PPARG activation were as well reflected at the lipidome and metabolome level, as many lipid classes were significantly altered, with exception of ceramide, sphingomyelin, PI, and LPA (Fig. 6F, Fig. S3, Table S7). As expected from the global proteome analysis, the levels of lipids and metabolites involved in TAG synthesis were increased (Figs. 6C and 6F). Consequently, our data reflect the induction of adipogenesis and nascent lipid droplet formation, which is in agreement with lipid droplet staining (Fig. 5B). Next to the abundance changes at the lipid class level, the total number of detected lipid species was also increased with a gain in species that were strongly up-regulated at the class level including PE, phosphatidic acid, and TAG (Fig. 6F). Since lipid identification was conducted at the fatty acid scan species level, it was possible for the first time to observe changes in the chain length and double bond distribution that were induced upon PPARG activation. The abundance and number of short chain lipids were significantly increased while the level of unsaturated lipids was decreased (Fig. S2), most likely leading to the same membrane fluidity but an increased protection against oxidative stress (57) that is known to be elevated in adipogenesis (58).
In addition, all targeted metabolites were detectable in all samples generated prior and after PPARG induction, with the exception of succinyl-CoA and dephospho-CoA whose levels were not detected without stimulation (Table S8). Nonetheless, our analysis revealed a strong regulation of nucleosides and nucleotides (Fig. 6D) upon PPARG activation. These higher amounts of nucleotides and nucleosides reflects the final clonal expansion step that is required prior to the differentiation into mature adipocytes (55). As expected, for the formation of lipid droplet membranes, building blocks also required for phospholipid lipid biosynthesis, such as glycerol-3-phosphate and ethanolamine, were increased.
Notably, correlative effects between proteome and lipidome were most obvious for the TAG synthesis pathway for which the enzyme concentration levels seem to be tightly controlled by PPARG (Figs. 6F and 6G). The up-regulation of all proteins in this pathway is likely due to the presence of PPARG binding sites at the promoter regions of their corresponding genes (44), repetitively indicating that the entire pathway is controlled by PPARG. This protein regulation goes along with an up-regulation of lipids and metabolites of the TAG synthesis pathway, obviously needed for nascent lipid droplets, with the exception of LPA levels which remained constant (Figs. 6F and 6G). Therefore, we presume that the LPA level might be exactly controlled by PPARG since elevated LPA levels are known to blunt PPARG activation (59). Within the PPARG pathway, the concentration of LPA directly depends on the amount of glycerol-3-phosphate acyltransferase, the LPA synthesizing enzyme (GPAT), and on 1-acylglycerol-3- phosphate-O-acyltransferase (AGPAT2), which further processes LPA to phosphatidic acid (60). On average, all GPAT isoforms are modestly up-regulated (3x), whereas AGPAT2, the predominant isoform in preadipocytes, is strongly induced (19x) by PPARG (Fig. 6G). This strong induction is ensuring the synthesis of PA and results in a low, constant level of LPA, therefore avoiding a negative feedback to PPARG and favoring adipogenesis. Our finding points out that a tight control of LPA is needed for adipogenesis, which is further supported by the down-regulation of PPARG in AGPAT2 knockdown and knockout models (61) as well as the development of steatosis and insulin resistance in an AGPAT2-deficient mouse model (62).
DISCUSSION
The development of strategies for the description of relationships between gene expression, protein abundance, and metabolite concentrations including lipids is a considerable challenge due to the multiple layers of regulation among these different molecular classes. Many of the regulatory mechanisms involved, such as metabolic control and feedback between those molecular classes, are insufficiently understood on the systems scale. As an answer to these challenges, we are proposing SIMPLEX, an extraction strategy capable of bridging the currently existing gaps between these different molecular levels. The major advantages of SIMPLEX are (i) parallel access to different molecular classes, along with further advantages including, (ii) reduced amounts of required sample, (iii) straightforward handling of small sample amounts, (iv) prevention of contamination through distinct layers between molecular classes, and (v) low sample handling errors by a reduced number of extraction steps. The introduced workflow quantified over 3762 molecular species. This thorough characterization at the lipid layer provides valuable insights for future studies into physical properties of membranes such as membrane thickness, rigidity, transverse forces, and lateral pressure that are directly controlled by the length and saturation level of fatty acids, as well as by lipid composition (63). In addition to even chain fatty acids accounting for 95% of the total fatty acid content, we also observed odd chain fatty acids with up to 5% per class (Fig. 4). The origin and presence of these odd chain fatty acids in lipids is controversial since it is still unclear whether these occur as endogenously synthesized products or as artifacts derived from cell culture. In a case of preadipocyte culture, it is most likely that lipids with odd chain fatty acids derive from alpha-oxidation, which is increased during the transition process to mature adipocytes (64, 65).
Whereas the current study is a proof-of-concept, the SIMPLEX approach will have to be adapted to more complex samples such as tissues or body fluids. We are aware of the limitations of SIMPLEX, which can be mainly attributed to the large variety of molecular characteristics. One can expect that certain low abundant or very hydrophilic lipids will be hard to detect and might escape the hydrophobic phase. A similar behavior is likely for low abundant, lipidated proteins (66) or amphiphilic metabolites. In such cases, additional enrichment steps and re-extraction will be required, respectively. Furthermore, we are aware that amphiphilic metabolites might be distributed over more than one phase, and this could hamper their analysis if they are already low abundant or difficult to detect.
In addition, future experiments on bigger cohorts (e.g. dynamic processes or clinical samples) have to be carried out to further prove the robustness of the here developed approach. Nonetheless, we believe that SIMPLEX permits a fundamental turn in study design that will provide novel insights into the systems biology of multimolecular cellular processes and the etiopathology of metabolic disorders such insulin resistance and lipid storage diseases as well as obesity. This is emphasized by an analysis of the mesenchymal stem cell line OP9 (preadipocytes), which is able to differentiate into fat cells.
Here, the identification of a close collaboration between the PPARG signaling on the protein level and TAG metabolism on the metabolite level proves our initial motivation to investigate the system within the cellular context rather than a restricted single-sided way and allowed for the elucidation of a further level of feedback-based PPARG control derived from the lipid level.
In summary, we established a protocol that can deliver a representative picture of an entire biological system by extending the detection capabilities to more than one molecular class. As mass spectrometry instrumentation continuously advances regarding sensitivity and speed, we expect that this novel workflow will soon accommodate more quantitatively accessible molecular species and will deliver new exiting perspectives into metabolic disorders.
Supplementary Material
Acknowledgments
We thank SUPR-G team, together with members of the systems analysis lab, for thoughtful comments and discussions, Michelle Protzek for the cell culturing, and Susanne Krois for the amino acid analysis, as well as Stefan Loroch for data interpretation with R-scripts.
Footnotes
Author contributions: R.A. designed research; C.C., F.A.S., A.H., and R.A. performed the research; C.C., A.S., R.P.Z., and R.A. contributed new reagents or analytic tools; C.C., F.A.S., A.H., R.P.Z., and R.A. analyzed data; and C.C. and R.A. wrote the paper.
* This work is supported by the Ministry for Innovation, Science and Research of the Federal State of North Rhine-Westphalia, and by the BMBF SUPR-G e:Med program (Code 01ZX1401C).
 This article contains supplemental materials.
This article contains supplemental materials.
Conflict of Interest: We declare no conflict of interest.
1 The abbreviations used are:
- MTBE
- methyl-tert-buthyl-ether
- IAA
- iodacetamide
- ABC
- ammonium bicarbonate
- TEAB
- triethylammonium bicarbonate
- BCA
- bicinchoninic acid
- NL
- neutral loss
- Cer
- ceramide
- SM
- sphingomyelin
- PL
- glycerophospholipids
- PC
- phosphatidylcholine
- PG
- phosphatidylglycerol
- PS
- phosphatidylserine
- PI
- phosphatidylinositol
- PE
- phosphatidylethanolamine
- PA
- phosphatidic acid
- LPA
- lysophosphatidic acid
- LPC
- lysophosphatidylcholine
- LPS
- lysophosphatidylserine
- PE (O/P)
- plasmanyl or plasmenylphosphatidyletanolamine
- TAG
- triacylglycerol
- DAG
- diacylglycerol
- CE
- cholesteryl ester
- FDR
- false discovery rate
- LTQ
- linear ion trap
- HILIC
- hydrophilic interaction liquid chromatography
- FASP
- filter aided sample preparation
- AGC
- automatic gain control
- MS/MS
- tandem mass spectrometry
- SRM
- selected reaction monitoring
- LC-MS
- liquid chromatography coupled mass spectrometry.
REFERENCES
- 1.Chavez J. A., and Summers S. A. (2012) A ceramide-centric view of insulin resistance. Cell Metabolism 15, 585–594 [DOI] [PubMed] [Google Scholar]
- 2.Ouchi N., Parker J. L., Lugus J. J., and Walsh K. (2011) Adipokines in inflammation and metabolic disease. Nature Rev. Immunol. 11, 85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morad S. A., and Cabot M. C. (2013) Ceramide-orchestrated signalling in cancer cells. Nature Rev. Cancer 13, 51–65 [DOI] [PubMed] [Google Scholar]
- 4.Zechner R., Zimmermann R., Eichmann T. O., Kohlwein S. D., Haemmerle G., Lass A., and Madeo F. (2012) Fat signals—Lipases and lipolysis in lipid metabolism and signaling. Cell Metabolism 15, 279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haemmerle G., Moustafa T., Woelkart G., Büttner S., Schmidt A., van de Weijer T., Hesselink M., Jaeger D., Kienesberger P. C., Zierler K., Schreiber R., Eichmann T., Kolb D., Kotzbeck P., Schweiger M., Kumari M., Eder S., Schoiswohl G., Wongsiriroj N., Pollak N. M., Radner F. P., Preiss-Landl K., Kolbe T., Rulicke T., Pieske B., Trauner M., Lass A., Zimmermann R., Hoefler G., Cinti S., Kershaw E. E., Schrauwen P., Madeo F., Mayer B., and Zechner R. (2011) ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-alpha and PGC-1. Nature Med. 17, 1076–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitra P., Maceyka M., Payne S. G., Lamour N., Milstien S., Chalfant C. E., and Spiegel S. (2007) Ceramide kinase regulates growth and survival of A549 human lung adenocarcinoma cells. FEBS Lett. 581, 735–740 [DOI] [PubMed] [Google Scholar]
- 7.Dobrowsky R. T., Kamibayashi C., Mumby M. C., and Hannun Y. A. (1993) Ceramide activates heterotrimeric protein phosphatase 2A. J. Biol. Chem. 268, 15523–15530 [PubMed] [Google Scholar]
- 8.Heinrich M., Wickel M., Schneider-Brachert W., Sandberg C., Gahr J., Schwandner R., Weber T., Saftig P., Peters C., Brunner J., Krönke M., and Schütze S. (1999) Cathepsin D targeted by acid sphingomyelinase-derived ceramide. EMBO J. 18, 5252–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim H. J., Oh J. E., Kim S. W., Chun Y. J., and Kim M. Y. (2008) Ceramide induces p38 MAPK-dependent apoptosis and Bax translocation via inhibition of Akt in HL-60 cells. Cancer Lett. 260, 88–95 [DOI] [PubMed] [Google Scholar]
- 10.Mukhopadhyay A., Saddoughi S. A., Song P., Sultan I., Ponnusamy S., Senkal C. E., Snook C. F., Arnold H. K., Sears R. C., Hannun Y. A., and Ogretmen B. (2009) Direct interaction between the inhibitor 2 and ceramide via sphingolipid-protein binding is involved in the regulation of protein phosphatase 2A activity and signaling. FASEB J. 23, 751–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahrends R., Ota A., Kovary K. M., Kudo T., Park B. O., and Teruel M. N. (2014) Controlling low rates of cell differentiation through noise and ultrahigh feedback. Science 344, 1384–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park B. O., Ahrends R., and Teruel M. N. (2012) Consecutive positive feedback loops create a bistable switch that controls preadipocyte-to-adipocyte conversion. Cell Rep. 2, 976–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Z., Rosen E. D., Brun R., Hauser S., Adelmant G., Troy A. E., McKeon C., Darlington G. J., and Spiegelman B. M. (1999) Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol. Cell 3, 151–158 [DOI] [PubMed] [Google Scholar]
- 14.Buczynski M. W., Stephens D. L., Bowers-Gentry R. C., Grkovich A., Deems R. A., and Dennis E. A. (2007) TLR-4 and sustained calcium agonists synergistically produce eicosanoids independent of protein synthesis in RAW264.7 cells. J. Biol. Chem. 282, 22834–22847 [DOI] [PubMed] [Google Scholar]
- 15.Sabidó E., Quehenberger O., Shen Q., Chang C. Y., Shah I., Armando A. M., Andreyev A., Vitek O., Dennis E. A., and Aebersold R. (2012) Targeted proteomics of the eicosanoid biosynthetic pathway completes an integrated genomics-proteomics-metabolomics picture of cellular metabolism. Mol. Cell. Proteomics 11, M111.014746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meierhofer D., Weidner C., and Sauer S. (2014) Integrative analysis of transcriptomics, proteomics, and metabolomics data of white adipose and liver tissue of high-fat diet and rosiglitazone-treated insulin-resistant mice identified pathway alterations and molecular hubs. J. Proteome Res. 13, 5592–5602 [DOI] [PubMed] [Google Scholar]
- 17.Manza L. L., Stamer S. L., Ham A. J., Codreanu S. G., and Liebler D. C. (2005) Sample preparation and digestion for proteomic analyses using spin filters. Proteomics 5, 1742–1745 [DOI] [PubMed] [Google Scholar]
- 18.Hahne H., Pachl F., Ruprecht B., Maier S. K., Klaeger S., Helm D., Médard G., Wilm M., Lemeer S., and Kuster B. (2013) DMSO enhances electrospray response, boosting sensitivity of proteomic experiments. Nat. Methods 10, 989–991 [DOI] [PubMed] [Google Scholar]
- 19.Dickhut C., Radau S., and Zahedi R. P. (2014) Fast, efficient, and quality-controlled phosphopeptide enrichment from minute sample amounts using titanium dioxide. Meth. Mol. Biol. 1156, 417–430 [DOI] [PubMed] [Google Scholar]
- 20.Burkhart J. M., Schumbrutzki C., Wortelkamp S., Sickmann A., and Zahedi R. P. (2012) Systematic and quantitative comparison of digest efficiency and specificity reveals the impact of trypsin quality on MS-based proteomics. J. Proteomics 75, 1454–1462 22166745 [DOI] [PubMed] [Google Scholar]
- 21.Engholm-Keller K., Birck P., Størling J., Pociot F., Mandrup-Poulsen T., and Larsen M. R. (2012) TiSH–a robust and sensitive global phosphoproteomics strategy employing a combination of TiO2, SIMAC, and HILIC. J. Proteomics 75, 5749–5761 [DOI] [PubMed] [Google Scholar]
- 22.Cohen S. A., and Michaud D. P. (1993) Synthesis of a fluorescent derivatizing reagent, 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate, and its application for the analysis of hydrolysate amino acids via high-performance liquid chromatography. Anal. Biochem. 211, 279–287 [DOI] [PubMed] [Google Scholar]
- 23.Shindo N., Nojima S., Fujimura T., Taka H., Mineki R., and Murayama K. (1997) Separation of 18 6-aminoquinolyl-carbamyl-amino acids by ion-pair chromatography. Anal. Biochem. 249, 79–82 [DOI] [PubMed] [Google Scholar]
- 24.Schuhmann K., Almeida R., Baumert M., Herzog R., Bornstein S. R., and Shevchenko A. (2012) Shotgun lipidomics on a LTQ Orbitrap mass spectrometer by successive switching between acquisition polarity modes. J. Mass Spectrom. 47, 96–104 [DOI] [PubMed] [Google Scholar]
- 25.Herzog R., Schuhmann K., Schwudke D., Sampaio J. L., Bornstein S. R., Schroeder M., and Shevchenko A. (2012) LipidXplorer: A software for consensual cross-platform lipidomics. PloS One 7, e29851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herzog R., Schwudke D., Schuhmann K., Sampaio J. L., Bornstein S. R., Schroeder M., and Shevchenko A. (2011) A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol. 12, R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carvalho M., Sampaio J. L., Palm W., Brankatschk M., Eaton S., and Shevchenko A. (2012) Effects of diet and development on the Drosophila lipidome. Mol. Syst. Biol. 8, 600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennett B. D., Kimball E. H., Gao M., Osterhout R., Van Dien S. J., and Rabinowitz J. D. (2009) Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li K., Wang X., Pidatala V. R., Chang C. P., and Cao X. (2014) Novel quantitative metabolomic approach for the study of stress responses of plant root metabolism. J. Proteome Res. 13, 5879–5887 [DOI] [PubMed] [Google Scholar]
- 30.Vaudel M., Barsnes H., Berven F. S., Sickmann A., and Martens L. (2011) SearchGUI: An open-source graphical user interface for simultaneous OMSSA and X!Tandem searches. Proteomics 11, 996–999 [DOI] [PubMed] [Google Scholar]
- 31.Vaudel M., Burkhart J. M., Zahedi R. P., Oveland E., Berven F. S., Sickmann A., Martens L., and Barsnes H. (2015) PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nature Biotechnol. 33, 22–24 [DOI] [PubMed] [Google Scholar]
- 32.Vizcaíno J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolomé S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nature Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taus T., Köcher T., Pichler P., Paschke C., Schmidt A., Henrich C., and Mechtler K. (2011) Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 10, 5354–5362 [DOI] [PubMed] [Google Scholar]
- 34.Matyash V., Liebisch G., Kurzchalia T. V., Shevchenko A., and Schwudke D. (2008) Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 49, 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao J., Yan X. L., Li R., Liu Y., He W., Sun S., Zhang Y., Liu B., Xiong J., and Mao N. (2010) Characterization of OP9 as authentic mesenchymal stem cell line. J. Genet. Genom. 37, 475–482 [DOI] [PubMed] [Google Scholar]
- 36.Wolins N. E., Quaynor B. K., Skinner J. R., Tzekov A., Park C., Choi K., and Bickel P. E. (2006) OP9 mouse stromal cells rapidly differentiate into adipocytes: characterization of a useful new model of adipogenesis. J. Lipid Res. 47, 450–460 [DOI] [PubMed] [Google Scholar]
- 37.Fhaner C. J., Liu S., Ji H., Simpson R. J., and Reid G. E. (2012) Comprehensive lipidome profiling of isogenic primary and metastatic colon adenocarcinoma cell lines. Anal. Chem. 84, 8917–8926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan M., Breitkopf S. B., Yang X., and Asara J. M. (2012) A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nature Protocols 7, 872–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiśniewski J. R., Zougman A., Nagaraj N., and Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 40.Benton H. P., Wong D. M., Trauger S. A., and Siuzdak G. (2008) XCMS2: Processing tandem mass spectrometry data for metabolite identification and structural characterization. Anal. Chem. 80, 6382–6389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abdel Rahman A. M., Pawling J., Ryczko M., Caudy A. A., and Dennis J. W. (2014) Targeted metabolomics in cultured cells and tissues by mass spectrometry: method development and validation. Anal. Chim. Acta 845, 53–61 [DOI] [PubMed] [Google Scholar]
- 42.Wagner R., Li J., Kenar E., Kohlbacher O., Machicao F., Häring H. U., Fritsche A., Xu G., and Lehmann R. (2014) Clinical and non-targeted metabolomic profiling of homozygous carriers of transcription factor 7-like 2 variant rs7903146. Scientific Rep. 4, 5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., and Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 44.Schwudke D., Oegema J., Burton L., Entchev E., Hannich J. T., Ejsing C. S., Kurzchalia T., and Shevchenko A. (2006) Lipid profiling by multiple precursor and neutral loss scanning driven by the data-dependent acquisition. Anal. Chem. 78, 585–595 [DOI] [PubMed] [Google Scholar]
- 45.Bligh E. G., and Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 46.Folch J., Lees M., and Sloane Stanley G. H. (1957) A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 47.Stegemann C., Pechlaner R., Willeit P., Langley S. R., Mangino M., Mayr U., Menni C., Moayyeri A., Santer P., Rungger G., Spector T. D., Willeit J., Kiechl S., and Mayr M. (2014) Lipidomics profiling and risk of cardiovascular disease in the prospective population-based Bruneck study. Circulation 129, 1821–1831 [DOI] [PubMed] [Google Scholar]
- 48.Foster J. M., Moreno P., Fabregat A., Hermjakob H., Steinbeck C., Apweiler R., Wakelam M. J., and Vizcaíno J. A. (2013) LipidHome: A database of theoretical lipids optimized for high throughput mass spectrometry lipidomics. PloS One 8, e61951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liebisch G., Vizcaíno J. A., Kofeler H., Trötzmuller M., Griffiths W. J., Schmitz G., Spener F., and Wakelam M. J. (2013) Shorthand notation for lipid structures derived from mass spectrometry. J. Lipid Res. 54, 1523–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jenkins B., West J. A., and Koulman A. (2015) A review of odd-chain fatty acid metabolism and the role of pentadecanoic acid (c15:0) and heptadecanoic acid (c17:0) in health and disease. Molecules 20, 2425–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jansson J., Willing B., Lucio M., Fekete A., Dicksved J., Halfvarson J., Tysk C., and Schmitt-Kopplin P. (2009) Metabolomics reveals metabolic biomarkers of Crohn's disease. PloS One 4, e6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Onjiko R. M., Moody S. A., and Nemes P. (2015) Single-cell mass spectrometry reveals small molecules that affect cell fates in the 16-cell embryo. Proc. Natl. Acad. Sci. U.S.A. 112, 6545–6550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Molina H., Yang Y., Ruch T., Kim J. W., Mortensen P., Otto T., Nalli A., Tang Q. Q., Lane M. D., Chaerkady R., and Pandey A. (2009) Temporal profiling of the adipocyte proteome during differentiation using a five-plex SILAC based strategy. J. Proteome Res. 8, 48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adachi J., Kumar C., Zhang Y., and Mann M. (2007) In-depth analysis of the adipocyte proteome by mass spectrometry and bioinformatics. Mol. Cell. Proteomics 6, 1257–1273 [DOI] [PubMed] [Google Scholar]
- 55.Tang Q. Q., Zhang J. W., and Daniel Lane M. (2004) Sequential gene promoter interactions by C/EBPbeta, C/EBPalpha, and PPARgamma during adipogenesis. Biochem. Biophys. Res. Commun. 318, 213–218 [DOI] [PubMed] [Google Scholar]
- 56.Mikkelsen T. S., Xu Z., Zhang X., Wang L., Gimble J. M., Lander E. S., and Rosen E. D. (2010) Comparative epigenomic analysis of murine and human adipogenesis. Cell 143, 156–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pamplona R., Barja G., and Portero-Otín M. (2002) Membrane fatty acid unsaturation, protection against oxidative stress, and maximum life span: Ahomeoviscous-longevity adaptation? Ann. N.Y. Acad. Sci. 959, 475–490 [DOI] [PubMed] [Google Scholar]
- 58.Lee H., Lee Y. J., Choi H., Ko E. H., and Kim J. W. (2009) Reactive oxygen species facilitate adipocyte differentiation by accelerating mitotic clonal expansion. J. Biol. Chem. 284, 10601–10609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simon M. F., Daviaud D., Pradère J. P., Grès S., Guigné C., Wabitsch M., Chun J., Valet P., and Saulnier-Blache J. S. (2005) Lysophosphatidic acid inhibits adipocyte differentiation via lysophosphatidic acid 1 receptor-dependent down-regulation of peroxisome proliferator-activated receptor gamma2. J. Biol. Chem. 280, 14656–14662 [DOI] [PubMed] [Google Scholar]
- 60.Takeuchi K., and Reue K. (2009) Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis. Am. J. Physiol. Endocrinol. Metabolism 296, E1195–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gale S. E., Frolov A., Han X., Bickel P. E., Cao L., Bowcock A., Schaffer J. E., and Ory D. S. (2006) A regulatory role for 1-acylglycerol-3-phosphate-O-acyltransferase 2 in adipocyte differentiation. J. Biol. Chem. 281, 11082–11089 [DOI] [PubMed] [Google Scholar]
- 62.Cortés V. A., Curtis D. E., Sukumaran S., Shao X., Parameswara V., Rashid S., Smith A. R., Ren J., Esser V., Hammer R. E., Agarwal A. K., Horton J. D., and Garg A. (2009) Molecular mechanisms of hepatic steatosis and insulin resistance in the AGPAT2-deficient mouse model of congenital generalized lipodystrophy. Cell Metabolism 9, 165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Janmey P. A., and Kinnunen P. K. (2006) Biophysical properties of lipids and dynamic membranes. Trends Cell Biol. 16, 538–546 [DOI] [PubMed] [Google Scholar]
- 64.Roberts L. D., Virtue S., Vidal-Puig A., Nicholls A. W., and Griffin J. L. (2009) Metabolic phenotyping of a model of adipocyte differentiation. Physiolog. Genomics 39, 109–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Su X., Han X., Yang J., Mancuso D. J., Chen J., Bickel P. E., and Gross R. W. (2004) Sequential ordered fatty acid alpha oxidation and Delta9 desaturation are major determinants of lipid storage and utilization in differentiating adipocytes. Biochemistry 43, 5033–5044 [DOI] [PubMed] [Google Scholar]
- 66.Hentschel A., Zahedi R. P., and Ahrends R. (2015) Protein lipid modifications—More than just a greasy ballast. Proteomics. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.