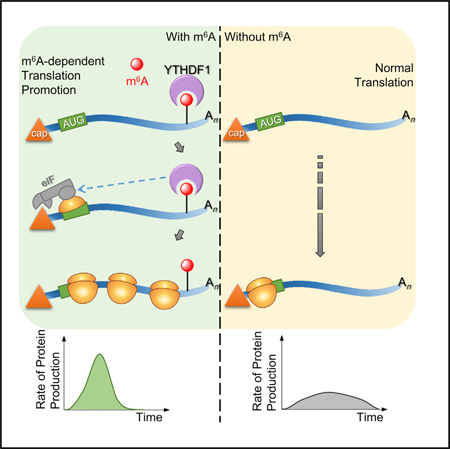
SUMMARY
N6-methyladenosine (m6A) is the most abundant internal modification in mammalian mRNA. This modification is reversible and non-stoichiometric and adds another layer to the dynamic control of mRNA metabolism. The stability of m6A-modified mRNA is regulated by an m6A reader protein, human YTHDF2, which recognizes m6A and reduces the stability of target transcripts. Looking at additional functional roles for the modification, we find that anotherm6A reader protein, human YTHDF1, actively promotes protein synthesis by interacting with translation machinery. In a unified mechanism of m6A-based regulation in the cytoplasm, YTHDF2-mediated degradation controls the lifetime of target transcripts, whereas YTHDF1-mediated translation promotion increases translation efficiency, ensuring effective protein production from dynamic transcripts that are marked by m6A. Therefore, the m6A modification in mRNA endows gene expression with fast responses and controllable protein production through these mechanisms.
Graphical abstract
INTRODUCTION
In eukaryotic cells, the control of mRNA translation and degradation is critical for managing the quantity and duration of gene expression. When compared to the transcriptional regulation of mRNA, regulatory pathways at the post-transcriptional level have distinct advantages, including the ability to promptly respond to stimuli, to fine-tune protein amounts, and to execute localized control (Moore, 2005). Global translation regulation is typically achieved by modulating both the activity of translation initiation factors and the availability of ribosomes (Hershey et al., 2012; Sonenberg and Hinnebusch, 2009). The regulation of functionally distinct gene groups also proves critical for the realization of complex phenotypic tasks in cell growth, division, and differentiation. Such processes are specified by cis-acting signals on RNAs (e.g., AU-rich element, iron-responsive element) and are mediated by cognate trans-acting factors, including RNA-binding proteins and small complementary RNAs (microRNAs and short interfering RNAs). However, as indicated by most recent evidence, dynamic modifications of mRNA, including 5-methylcytidine (Hussain et al., 2013; Squires et al., 2012), pseudouridine (Carlile et al., 2014; Schwartz et al., 2014), and N6-methyladenosinde (Fu et al., 2014; Lee et al., 2014; Meyer and Jaffrey, 2014; Nilsen, 2014; Wang and He, 2014a), have emerged as potential new mechanisms of post-transcriptional gene regulation.
N6-methyladenosine (m6A) is the most prevalent internal modification existing on eukaryotic mRNA (Desrosiers et al., 1974), and it impacts a variety of physiological events. On average, every mammalian mRNA contains over three sites of m6A within a G(m6A)C (70%) or A(m6A)C (30%) consensus sequence (Wei et al., 1976; Wei and Moss, 1977). The m6A modification on mRNA functions as a dynamic mark that is analogous to methylations on DNA and histone tails: m6A on mRNA is post-transcriptionally installed, erased, and recognized bym6A methyltransferases (“writer”; Bokar et al., 1997; Liu et al., 2014; Ping et al., 2014; Tuck, 1992; Wang et al., 2014b), demethylases (“eraser”; Jia et al., 2011; Zheng et al., 2013), and m6A-specific binding proteins (“reader”; Dominissini et al., 2012; Wang et al., 2014a), respectively. m6A methyltransferase is crucial for yeast meiosis (Shah and Clancy, 1992; Schwartz et al., 2013), the differentiation of mouse embryonic stem cells (Geula et al., 2015; Wang et al., 2014b), the development of fruit flies (Hongay and Orr-Weaver, 2011) and plants (Zhong et al., 2008), and the viability of human cells (Bokar, 2005). The two m6A demethylases (FTO and AlkBH5) have been associated with human body weight (Dina et al., 2007; Frayling et al., 2007; Do et al., 2008) and mouse fertility (Zheng et al., 2013), respectively.
In addition to the physiological importance, one of the fundamental cellular functions of m6A has been directly connected to the stability of mRNA. The m6A-modified transcripts inherently possess shorter half-lives than non-methylated ones in HeLa cells (Fu et al., 2014). The human YTH domain family protein 2 (YTHDF2) has been shown to affect the stability of m6A-modified RNA by localizing them to mRNA decay sites (Sheth and Parker, 2003; Wang et al., 2014a). The mRNA targets of YTHDF2 contain many transcription factors, indicating that the m6A-dependent mRNA turnover could serve to dynamically adjust the expression of regulatory genes (Wang and He, 2014b). This notion has been supported by recent studies of two yeast homologs of human YTH domain proteins that appear to affect mRNA stability (Harigaya et al., 2006; Hiriart et al., 2012; Kang et al., 2014) and the findings that m6A directs the expression of pluripotency regulators in mouse embryonic stem cells (Batista et al., 2014; Geula et al., 2015; Wang et al., 2014b).
Whereas mRNA is the direct output of gene transcription, proteins are the ultimate products of gene expression. Whetherm6A could affect protein production and how it would do so represent an unaddressed key question with regard tom6A-mediated gene regulation. In this work, we report that the human m6A reader protein YTHDF1 directly promotes the translation of methylated mRNAs. This function, together with YTHDF2-mediated mRNA decay (Wang et al., 2014a), depicts a dynamic and multi-dimensional mechanism of mRNA methylation in modulating gene expression.
RESULTS
YTHDF1 Binds m6A inside Cells
Out of five YTH domain family proteins encoded in the human genome, YTHDF1, YTHDF2, YTHDF3, and YTHDC1 have been confirmed for the selective binding of m6A in RNA (Wang et al., 2014a; Xu et al., 2014). Recent structural characterizations have revealed a hydrophobic pocket used by these proteins to preferentially bind the methyl group of m6A (Li et al., 2014; Luo and Tong, 2014; Theler et al., 2014; Xu et al., 2014). Residues that form this pocket are conserved from yeast to human, indicating that the function of selectivem6A binding is evolutionally preserved.
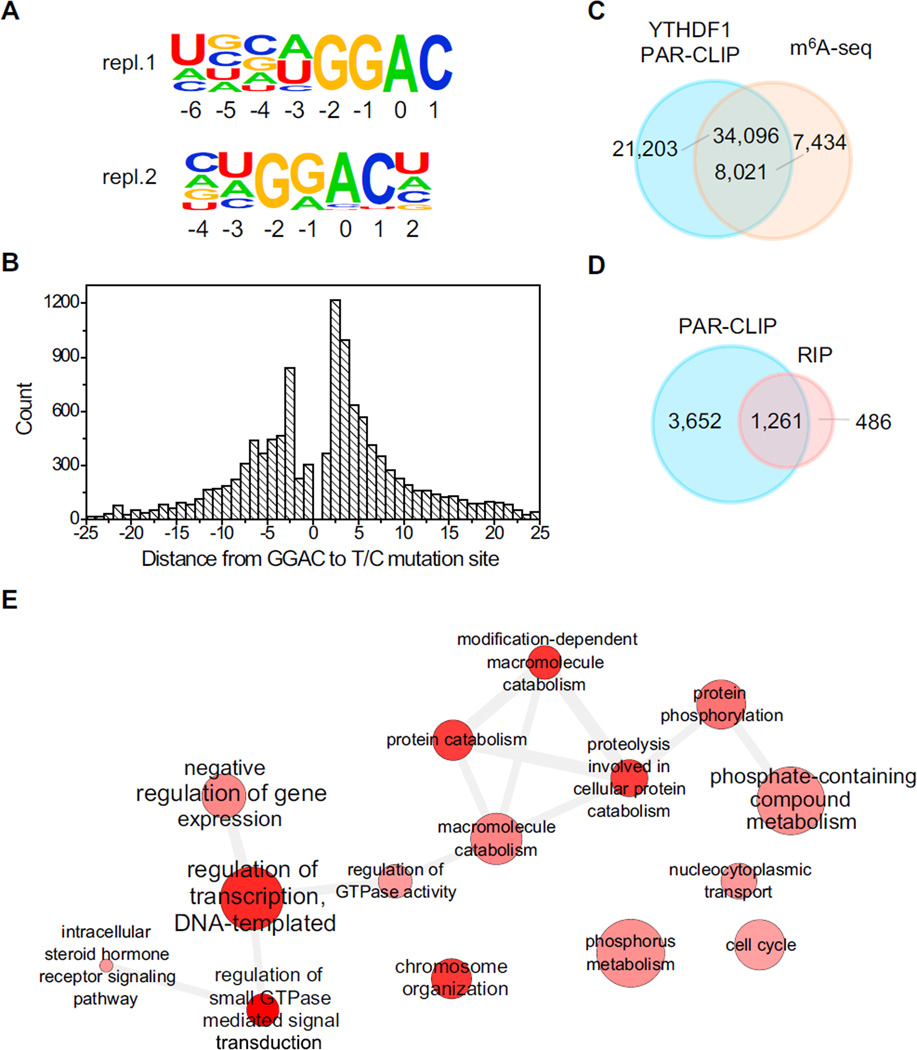
To characterize the binding sites of YTHDF1, we applied photoactivatable ribonucleoside crosslinking and immunoprecipitation (PAR-CLIP), which utilizes a photoreactive nucleoside such as 4-thiouridine (4SU) to crosslink RNA with protein and produce a detectable T-to-C mutation. This experiment identified 4,951 mRNA transcripts as potential targets of YTHDF1 (Figure S1A). A GRAC (R is G or A) motif, coinciding with the m6A consensus motif, was repeatedly identified from two biological replicates of PAR-CLIP samples (Figure 1A), covering over 50% of all PAR-CLIP clusters in each sample. It should be noted that the crosslinking of YTHDF1 to target RNA requires an appropriately spaced 4SU residue near the binding site. The distribution of the distances between GGAC and T-to-C mutation sites shows that the optimum crosslinking positions are +2, +3, and −3 (setting A as position 0, Figure 1B), suggesting that YTHDF1 binds at exact GRAC sites instead of other motifs that co-exist in the proximity of m6A sites. In addition, 62% of the PAR-CLIP peaks of YTHDF1 overlap with m6A peaks as determined by antibody-based m6A profiling (Figure 1C); YTHDF1-binding sites also cluster around the stop codon, resembling the distribution pattern of m6A sites on mRNA (Figure S1B). Taking these analyses together, we conclude that YTHDF1 recognizes m6A on RNA transcripts inside cells.
Figure 1. Transcriptome-wide Identification of YTHDF1 mRNA Targets.
(A) YTHDF1-binding motifs identified by HOMER from PAR-CLIP peaks of two biological replicates. Motif length was restricted to 6–8 nucleotides. The motif with the lowest p value of replicate 1 (repl.1) was found in 53.8% of 24,753 sites (p = 1 × 10−703), and that of replicate 2 (repl.2) was found in 54.5% of 58,549 sites (p = 1 × 10−1093).
(B) The distribution of the distance from GGAC to T-to-C mutation sites. The optimum crosslinking sites are at the −3, +2, and +3 positions.
(C) Overlap of YTHDF1 PAR-CLIP peaks and m6A-seq peaks in HeLa cells.
(D) Overlap of target genes identified by PAR-CLIP and RIP-seq for YTHDF1.
(E) Gene Ontology analysis of YTHDF1 mRNA targets.
See also Figure S1 and Table S1.
In addition, we sequenced RNA obtained from the immuno-purified ribonucleoprotein complex of YTHDF1 as a complementary method (RIP-seq) to reveal YTHDF1-bound RNA. Among 1,714 transcripts identified with enrichment greater than 2-fold (Figure S1C), 74% overlap with PAR-CLIP targets (Figure 1D). The 1,261 shared genes (CLIP+IP) were defined as high-confident targets of YTHDF1. Gene ontology (GO) analysis of these RNA revealed several distinct gene clusters (Figure 1E). Three hundred and seven (25%) of these genes are enriched under the GO term of regulation of transcription. Other notable pathways include phosphate metabolic process (105 genes), chromosome organization (70 genes), and regulation of small GTPase-mediated signal transduction (49 genes), further indicating that m6A tends to mark transcripts with regulatory functions.
YTHDF1 Promotes Ribosome Occupancy of Its Target mRNA
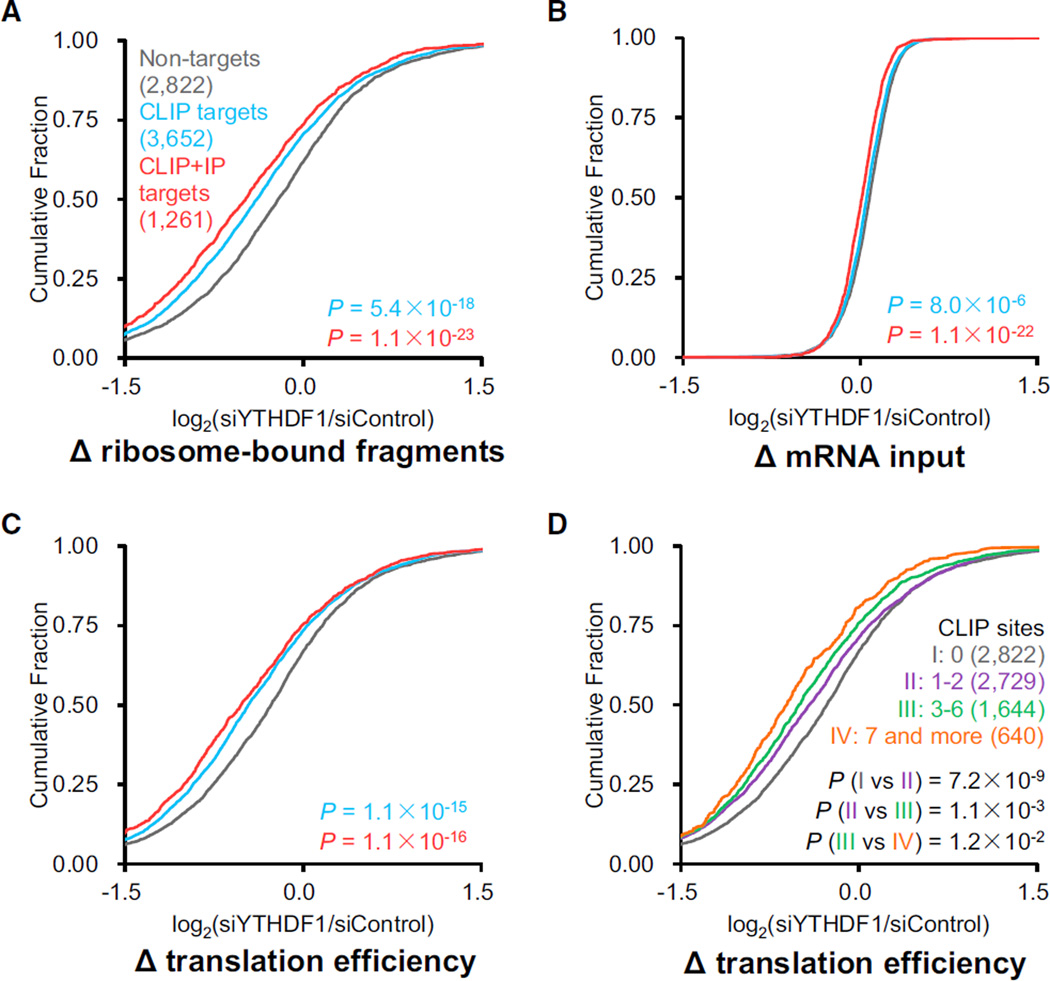
To explore whether YTHDF1 involves mRNA translation, we used ribosome profiling to assess ribosome density of each transcript mRNA with or without perturbation of YTHDF1. HeLa cells transfected with YTHDF1 or control siRNA were subjected to ribosome profiling and mRNA sequencing. Two biological replicates with the use of different YTHDF1 siRNA sequences were studied to avoid off-target effects. Transcripts presented (reads per kilobase per million reads [RPKM] > 1) in both ribosome profiling and mRNA sequencing samples were analyzed in parallel. These transcripts were then categorized as non-targets (absent from PAR-CLIP and RIP), PAR-CLIP only targets, and common targets of PAR-CLIP and RIP. A significant decrease in ribosome-bound mRNA reads for YTHDF1 targets was observed in the YTHDF1 knockdown samples compared to the controls (p < 0.001, Mann-Whitney U test) (Figure 2A). In contrast, the differences of mRNA inputs between groups in the knockdown and the control samples were small (Figure 2B). Overall, the knockdown of YTHDF1 led to reduced translation efficiency of its target transcripts (Figure 2C), suggesting a functional role of YTHDF1 in mRNA translation. The extent of the ribosome occupancy reduction caused by YTHDF1 knockdown correlates with the YTHDF1 knockdown efficiency (Figure S2) and the number of YTHDF1-binding sites on the transcripts (Figure 2D).
Figure 2. Knockdown of YTHDF1 Leads to Reduced Translation of Its mRNA Targets.
(A–C) Cumulative distribution log2-fold changes of ribosome-bound fragments (A), mRNA input (B), and translation efficiency (C, ratio of ribosome-bound fragments and mRNA input) between siYTHDF1 and siControl for non-targets (gray), PAR-CLIP-only targets (blue), and common targets of PAR-CLIP and RIP (red). p values were calculated using a two-sided Mann-Whitney test. (D) The mRNA lifetime log2-fold changes were further grouped and analyzed on the basis of the number of CLIP sites on each transcript. The extent of translation reduction caused by YTHDF1 knockdown correlates with the number of YTHDF1-binding sites for mRNA targets of YTHDF1. p values were calculated using a Kruskal-Wallis test.
See also Figure S2 and Table S1.
YTHDF1 Modulates the Translation Dynamics of m6A-Modified mRNA
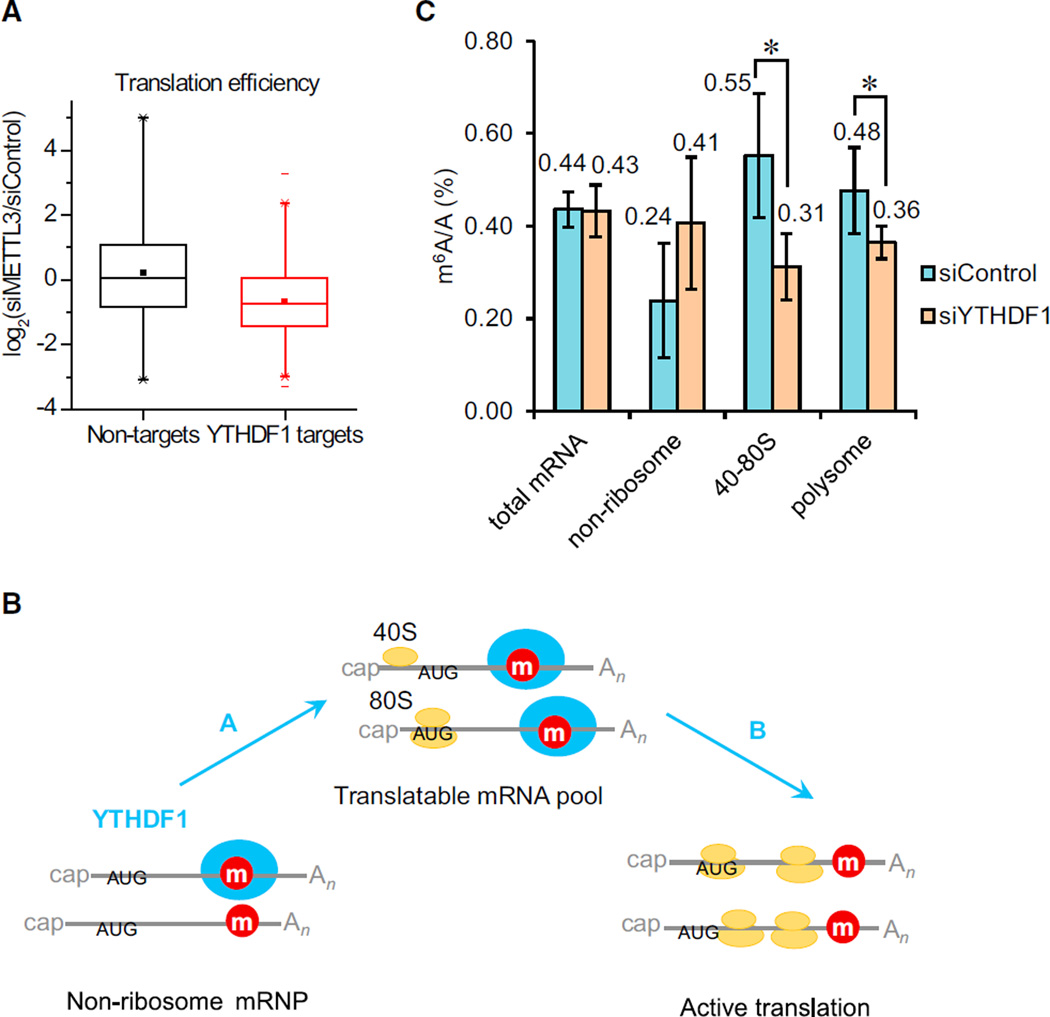
With the finding that YTHDF1 promotes ribosome loading of its target RNA, the next question to ask is whether this effect is m6A dependent. By knocking down m6A methyltransferase (METTL3) and performing ribosome profiling, we observed an overall decreased translation efficiency of YTHDF1 target transcripts with the reduction of m6A (Liu et al., 2014), compared to non-targets (Figure 3A). The results confirm that YTHDF1 promotes translation efficiency in an m6A-dependent manner.
Figure 3. YTHDF1 Enhances the Translation of m6A-Modified RNAs.
(A) Knockdown of the m6A methyltransferase (METTL3) reduced the translation efficiency of YTHDF1 target transcripts. Cumulative distribution log2-fold changes of the translation efficiency between siMETTL3 and siControl for non-targets (black) and YTHDF1 RNA targets (red). p = 0, two-sided Mann-Whitney test.
(B) A diagram illustrating that YTHDF1 plays two potential roles in the translation of m6A-modified RNAs: in Role A, YTHDF1 shuttles more mRNAs to translation machinery; in Role B, YTHDF1 accelerates the translation initiation rate of methylated mRNAs.
(C) Quantification of them6A/A ratio of total mRNA, the non-ribosome portion, 40S–80S, and polysome determined by LC-MS/MS for the YTHDF1 knockdown samples compared to controls after 48 hr. p values were determined using a two-sided Student’s t test for paired samples. Error bars represent mean ± SD. For total mRNA, n = 8 (four biological replicates × two technical replicates), p = 0.71. For the rest, n = 5 (two biological replicates, two technical replicates + three technical replicates), p = 0.083, 0.035, 0.049 for nonribosome, 40S–80S, and polysome fractions, respectively.
See also Figure S3.
The translation status of cytoplasmic mRNAs can be generally categorized into non-ribosome mRNPs (mRNA–protein particles), translatable mRNA pool (mRNPs associated with translation factors but not being actively translated), and actively translating polysome (Figure 3B). To further uncover the effects of YTHDF1 on m6A-modified RNA, sucrose gradient was utilized to fractionate cytoplasmic mRNAs into fractions with sedimentation coefficients of 20–35S that are exclusively non-ribosome mRNPs; 40–80S fractions containing the translatable mRNA pool and other large mRNPs; and the rest polysome fractions (Figure S3A). The knockdown of YTHDF1 does not change the m6A/A ratio of total mRNA (Figure 3C). Hence, YTHDF1 does not alter the overall methylated mRNA level but affects the association of its target mRNA with ribosome. If this is the case, upon the knockdown of YTHDF1, the amount of methylated mRNA should increase in mRNPs and decrease within the translating and translatable pools. Indeed, we observed an ~71% increase in the m6A/A ratio of mRNA isolated from non-ribosome mRNPs, an ~44% decrease from 40–80S fractions, and a 25% decrease from polysome fractions (Figure 3C). Thus, YTHDF1 modulates the subcellular distribution and translation status of the m6A-modified mRNA. It is worth noting that the role of YTHDF1 in mRNA stability cannot be completely excluded because an ~24% increase of m6A/A ratio in mRNA was observed after 24 hr overexpression of YTHDF1 (Figure S3B). Because mRNA translation and degradation often closely correlate, an elevated level of YTHDF1 might retain mRNA in translation and slow down decay as a secondary effect. These results do indicate that YTHDF1 is functionally distinct from YTHDF2 and may not be directly involved in mRNA decay.
To confirm the observed trend, two YTHDF1-targeted RNAs and a control were selected for validation: the SON mRNA (CDS methylated), the CREBBP mRNA (3′ UTR methylated), and a non-target RPL30 mRNA as a control. As detected by gene-specific PCR after reverse transcription (RT-PCR), both SON and CREBBP showed noticeable decreases in the polysome portion and increases in the non-ribosome mRNP portion after 48 hr YTHDF1 knockdown, whereas the control RPL30 mRNA did not show a decrease in the polysome portion (Figures S3C– S3E). These results confirm an active role of YTHDF1 in trafficking more transcripts to translation machinery (Role A, Figure 3B).
We next performed quantitative translation initiation sequencing (qTI-seq, Figures S3F and S3G), a method that measures the ribosome occupancy at translation initiation sites (TIS) (Gao et al., 2015), in order to probe initiation as a rate-limiting step in translation. We observed an increase of the TIS occupancy in YTHDF1 knockdown samples compared to control samples (Figure S3H). An increase of the ribosome occupancy at TIS could be caused by either increased mRNA population in the translation initiation step or slowed translation-initiation rate, or both. If YTHDF1 promotes mRNA translation solely by delivering more cellular mRNAs to translation machinery (Role A), a decrease of TIS occupancy is expected under the YTHDF1 knockdown conditions. The increased TIS occupancy observed upon YTHDF1 knockdown suggests that YTHDF1 also directly accelerates the translation initiation rate of the ribosome-bound mRNA (Role B, Figure 3B); this effect might be more dominant than recruiting mRNA to the translation machinery in this system.
YTHDF1 Binding Is Sufficient to Promote mRNA Translation
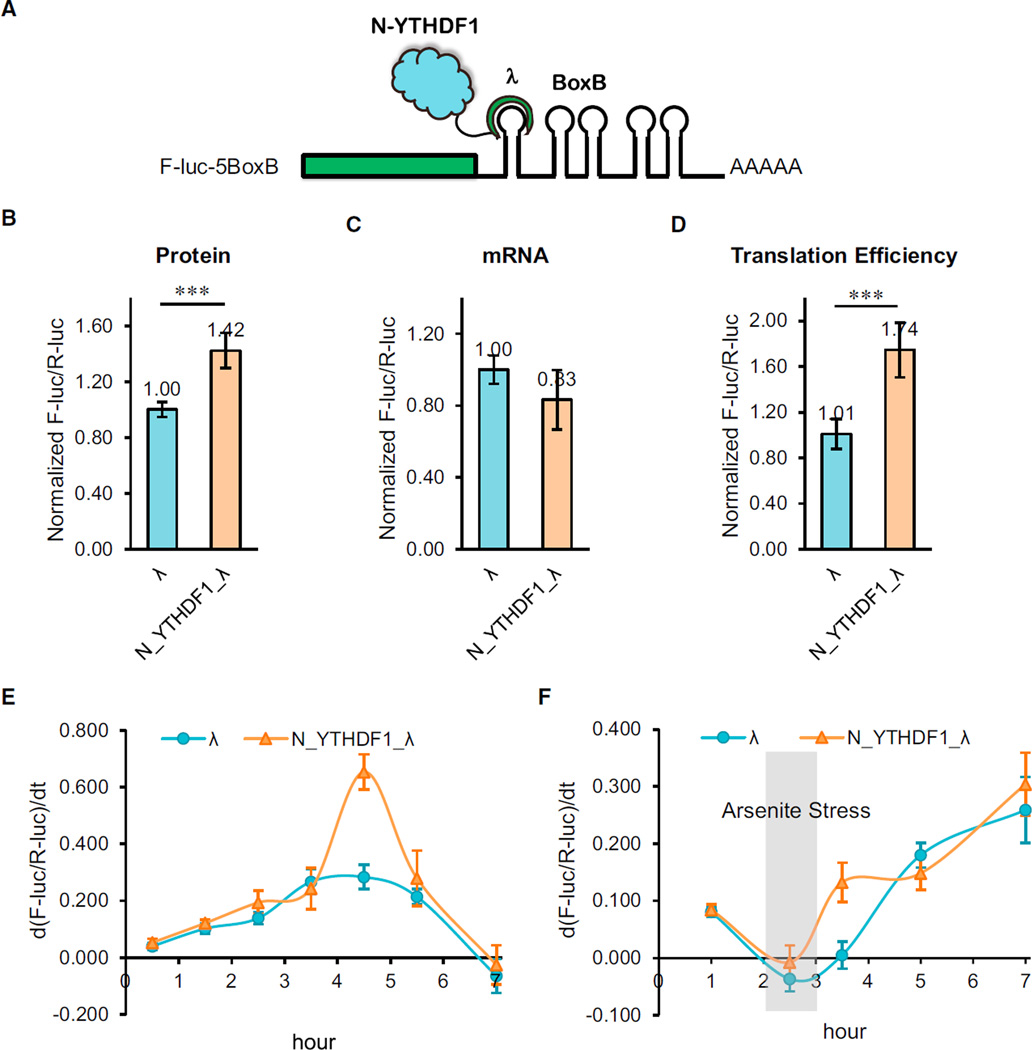
The increased ribosome loading mediated by YTHDF1 on its target transcripts could be attributed to either increased translation or potential hindered elongation. To probe the exact role of YTHDF1 on translation, we combined a switchable gene-expression system with a luciferase-based tethered reporter assay (Behm-Ansmant et al., 2006). As the C-terminal YTH domain of all YTH family proteins is primarily engaged in m6A binding (Wang et al., 2014a; Xu et al., 2014), we studied the function of the N-terminal domain of YTHDF1 (N_YTHDF1). N_YTHDF1 was fused with λ peptide (N_YTHDF1_λ), which specifically and tightly binds F-luc-5BoxB (five Box B sequence inserted into the 3′ UTR of the luciferase reporter). A Tet-Off inducible promoter was installed onto the F-luc-5BoxB construct, which blocks its transcription in the presence of doxycycline (DOX), thus enabling the evaluation of protein-expression dynamics upon DOX removal (Figure 4A). We first compared luciferase expression with and without the N_YTHDF1 tether after an 8 hr induction. The result showed an ~42% increase in translation with a slight decrease in the mRNA abundance when YTHDF1 was tethered to the luciferase mRNA (Figures 4B and 4C). The overall translation efficiency of the YTHDF1-tethered transcript had an ~72% increase over the control, showing a major effect of YTHDF1 in promoting translation efficiency (Figure 4D). In order to reveal how YTHDF1 dynamically promotes translation, we treated cells with a 2 hr pulse induction of transcription (removing DOX for 2 hr before adding back) and monitored the dynamics of the reporter luciferase protein expression. Compared to the control group, cells with the N_YTHDF1-tethered reporter showed a slightly elevated expression rate of luciferase at the beginning but a noticeably increased translation rate after the pulse induction period (Figure 4E). This observation confirms that YTHDF1 binding directly elevates mRNA translation efficiency.
Figure 4. The N-Terminal Domain of YTHDF1 Promotes Protein Production in a Tethering Assay.
(A) Construct of the tethering reporter assay. The mRNA reporter consists of an inducible promoter, firefly luciferase as the coding region, and five Box B sequence at 3′ UTR (F-luc-5BoxB). The N-terminal domain of YTHDF1 (N_YTHDF1) was fused with λ peptide (N_YTHDF1_λ), which recognizes Box B RNA with a high affinity. R-luc lacks the inducible promoter and was used as an internal control to normalize the F-luc signal.
(B) Under constant induction, the tethering of N_YTHDF1_λ to F-luc-5BoxB led to an on-average 42% increased translation in comparison with the control. The translation outcome was determined as a relative signal of F-luc divided by R-luc. Error bars, mean ± SD, p = 5.9 × 10−4 (two-sided Student’s t test for paired samples), n = 6 (three biological replicates × two technical replicates).
(C) Under constant induction, the mRNA abundance decreased slightly in the N_YTHDF1_λ-tethered group compared with the control. The mRNA abundance was determined by qRT-PCR of F-luc and R-luc. Error bars, mean × SD, p = 0.031, n = 6.
(D) The translation efficiency of the reporter mRNA increased by ~72% in the N_YTHDF1_λ-tethered group compared with the control. The translation efficiency is defined as the quotient of reporter protein production (F-luc/R-luc) divided by mRNA abundance. Error bars, mean ± SD, p = 3.2 × 10−5, n = 6.
(E) F-luc-5BoxB was induced with a pulse expression for 2 hr. The mRNA reporter showed higher translation when tethered with N_YTHDF1_λ compared with the control. y axis, d(F-luc/R-luc)/dt, indicating the changing rate of protein production. Error bars, mean ± SD, n = 4.
(F) After a 2 hr pulse expression and a 1 hr arsenite (1 mM) stress treatment, translation of the reporter protein was largely diminished. The translation recovery was assessed after the stress was released. The result showed that the N_YTHDF1_λ-tethered group exhibited faster translation recovery than the control group. Error bars, mean ± SD, n = 4.
We next investigated effects of YTHDF1 on the luciferase reporter expression during stress response. The tethered cells were given a 2 hr pulse induction followed by arsenite treatment for 1 hr, after which protein expression dynamics were monitored for another 5 hr. The result showed that although arsenite stress largely diminished protein expression, cells could gradually restore translation when the stress was lifted (Figure 4F). During the recovery stage, the N_YTHDF1-tethered transcript exhibited a faster restoration rate, indicating that YTHDF1 could facilitate the stress-recovery response of cells by promoting translation.
YTHDF1 Interacts with Initiation Factors to Promote Translation
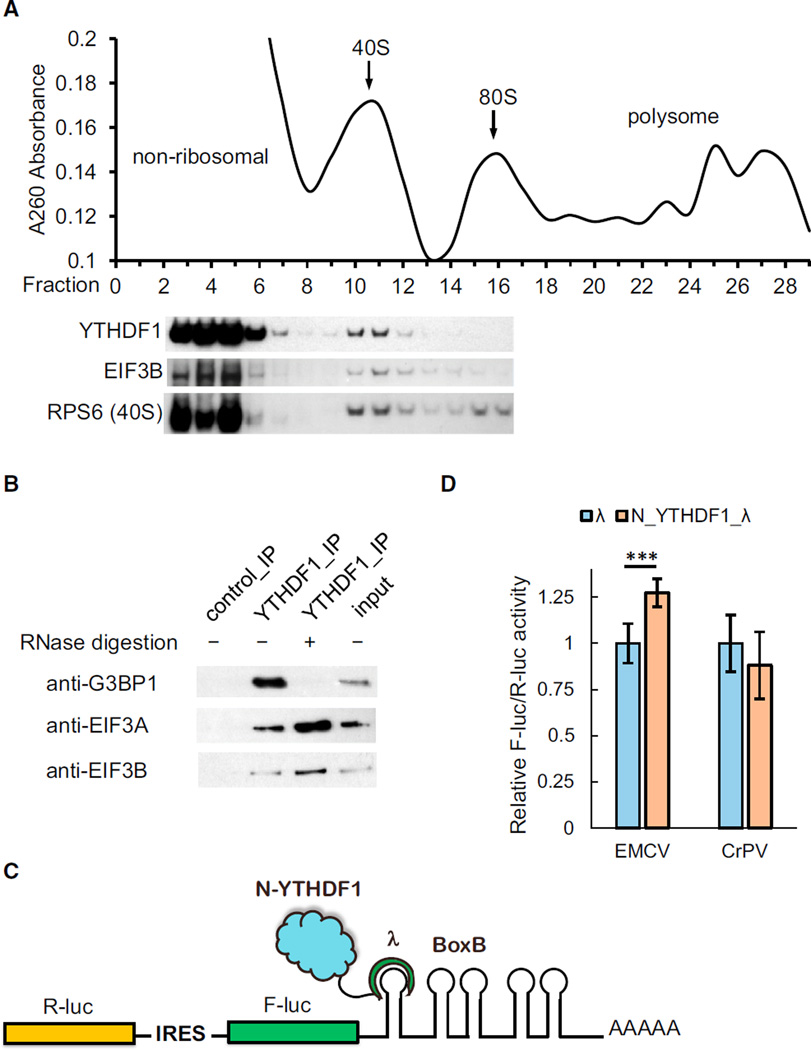
We constructed a HeLa cell line stably expressing an epitope-tagged YTHDF1 (N-terminal Flag and HA tags in tandem) near the endogenous level. By dissecting different ribosome fractions, we found an enrichment of YTHDF1 co-existing with ribosomal subunits and translation initiation factors in the 40S portion (Figure 5A). Under mild formaldehyde fixation, YTHDF1 was also observed in the 80S portion, but not 60S (Figure S4A). We next studied the composition of the YTHDF1-containing complex using tandem-affinity purification of the epitope-tagged YTHDF1 and protein mass spectrometry. A control sample stably expressing Flag-HA peptide without YTHDF1 was processed in parallel. The result revealed translation as the main theme of the protein interactome of YTHDF1 (Figure S4B). Compared to the control group, 119 unique proteins were co-purified with YTHDF1, 62 of which involve translation, including 27 out of 33 subunits of 40S, 23 out of 47 subunits of 60S, and 6 out of 13 subunits of translation initiation factor complex 3 (eIF3) (Table S2). The interaction between YTHDF1 and eIF3 was further confirmed by YTHDF1 immunopreciptitation and western blotting (Figure 5B).
Figure 5. YTHDF1 Associates with Translation Initiation Factors, Ribosome, and Stress Granule Marker.
(A) Western blotting of Flag-tagged YTHDF1 on each fraction of 10%–50% sucrose gradient showing that YTHDF1 associates with ribosome. The fractions were grouped to non-ribosomal mRNPs, 40S–80S, and polysome. RPS6 is a protein subunit of 40S ribosome, and eIF3B is a component of the translation initiation complex (each lane is aligned to the corresponding fraction on the upper plot).
(B) Western blotting showing that eIF3A, eIF3B, and G3BP1 co-immunoprecipitated with YTHDF1. The association of YTHDF1 with G3BP1 requires RNA, and those with eIF3A/eIF3B are independent of RNA.
(C) Construct of the bicistronic IRES reporter assay. As the first coding region, R-luc reports cap-dependent translation of the mRNA and serves as control. The second coding region encodes F-luc whose translation is controlled by different types of upstream IRES elements. Five Box B sequence was inserted at the 3′ UTR as the tethering site for the N-terminal domain of YTHDF1.
(D) Tethering N_YTHDF1_λ to the EMCV IRES reporter led to an on-average 27% increased translation of F-luc compared with R-luc, whereas N_YTHDF1_λ had no effect on the CrPV IRES reporter. Error bars, mean ± SD, p (EMCV) = 9.5 × 10−5, P (CrPV) = 0.050, two-sided Student’s t test for paired samples, n = 8 (biological replicates).
See also Figures S4 and S5 and Table S2.
Several RNA-binding proteins with known roles in translational control were also identified, e.g., YBX1, IGF2BP1, G3BP1, and PCBP2. These trans-acting factors may function collectively with YTHDF1 to affect translation of methylated mRNA. Interestingly, the interaction between YTHDF1 and stress granule marker G3BP1 (Kedersha and Anderson, 2007) is RNA dependent, whereas the interaction between YTHDF1 with eIF3 is not (Figure 5B). This observation suggests that the association of YTHDF1 with translation machinery could be a direct binding, whereas its involvement with stress granules is more contingent on the presence of bound mRNA.
The differential interaction of YTHDF1 with the aforementioned two classes of protein partners was verified with colocalization patterns under fluorescence immuno-staining (Figure S5). We treated cells with arsenite to induce stress and formation of stress granules. The co-localization of YTHDF1 with eIF3 is significant under both normal and stress conditions. The co-localization of YTHDF1 with G3BP1 increased by more than 75% when stimulated with arsenite stress, suggesting that the association between YTHDF1 and stress granules is conditional and likely driven by the dynamics of bound mRNA.
Next, we employed IRES reporters to further understand the mechanism of YTHDF1-dependent translation promotion. The translation of IRES reporters bypasses the requirement for the cap and cap-binding factor eIF4E (Figures 5C, 5D, S5C, and S5D) (Fraser and Doudna, 2007). The EMCV IRES directly binds the eIF4G subunit of the eIF4 complex. The HCV IRES bypasses the eIF4 complex and eIF4G-iduced loop formation by directly recruiting 40S and eIF3 (Hertz et al., 2013). The CrPV IRES recruits the ribosome completely independent of initiation factors (eIFs). We constructed reporters with the N-terminal domain of YTHDF1 (N_YTHDF1) tethered to the 3′ UTR of these IRES reporters (Figure 5C). Translation assays revealed that YTHDF1 can promote the translation of both cistron-encoding cap (Renilla luciferase) and IRES reporters (Firefly luciferase, Figure S5C), supporting the role of YTHDF1 in trafficking mRNA to active translation. The role of YTHDF1 in translation initiation could be evaluated by pairwise comparisons of IRES- versus cap-dependent translation. Tethering of N_YTHDF1 enhanced the translation of EMCV IRES reporter by ~27% in comparison to cap-dependent translation (Figure 5D). In contrast, the CrPV and HCV IRES reporters afforded 12%–26% less protein production increases compared with the corresponding cap-dependent translation in the presence of N_YTHDF1 tethering (Figures 5D and S5D). These results indicate that YTHDF1-dependent translation requires eIFs and likely relies on the eIF4G-dependent loop formation.
YTHDF1 and YTHDF2 in Methylated mRNA Translation and Metabolism
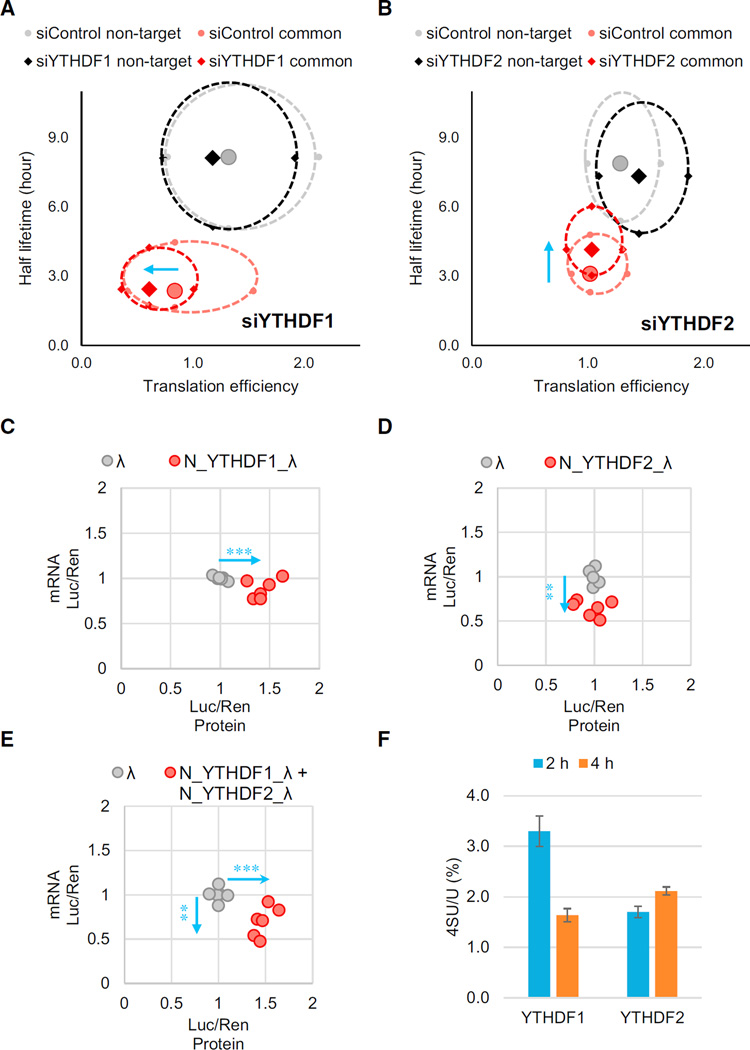
It has been previously reported that m6A reader protein YTHDF2 decreases the stability of its m6A-modified target mRNA (Wang et al., 2014a), which seems to contradict these findings about the translation-promotion role of m6A. YTHDF1 and YTHDF2 may regulate their own subsets of mRNA targets independently; however, they also share ~50% common target transcripts (Figure S6A). We investigated the effects of perturbing YTHDF1 and YTHDF2 on the translation and the stability of their common targets from ribosome profiling and mRNA lifetime profiling. The transcripts were categorized into non-targets and shared targets of YTHDF1 and YTHDF2 (other transcripts were omitted for clarity). We assessed both the lifetime and translation efficiency of mRNA and drew a two-dimensional plot (Figures 6A and 6B). The result showed that the shared targets behaved quite differently compared to non-targets. With YTHDF1 knockdown, the translation efficiency of shared targets decreased notably, whereas the average lifetime did not change. The knockdown of YTHDF2 resulted in a substantial increase in the lifetime of the shared targets but only a slight difference in the translation efficiency. These data verified the mRNA-destabilizing role of YTHDF2 and the translation-promotion role of YTHDF1. Whereas YTHDF2 controls the lifetime of the methylated transcripts, YTHDF1 ensures the efficient protein expression form these shared transcripts.
Figure 6. Translation Efficiency of the Common mRNA Targets of YTHDF1 and YTHDF2 is Affected by Both m6A Readers.
(A and B) Evaluations of translation efficiency and mRNA half-lifetimes of shared targets or non-targets of YTHDF1 and YTHDF2 with and without perturbation. Ribosome profiling and mRNA-seq data were collected under YTHDF1 knockdown (A) or YTHDF2 knockdown (B) conditions. Transcripts were categorized into common targets of YTHDF1 and YTHDF2 (red) or non-targets (black) under the knockdown conditions, and each compared with their corresponding control (pink and gray, respectively). The solid diamonds and circles represent median of the translation efficiency and half-lifetime. The four periphery dots surrounding each median are data quartiles (25% and 75% of each variance) and connected by dashed lines. The blue arrows denote the directions of changes compared to the control.
(C–E) Evaluations of both mRNA abundance and protein production of the reporter transcripts using the tethering assay. Cells transfected with inducible luciferase genes were tethered by YTHDF1 (C), YTHDF2 (D), or both (E). Protein production was calculated by normalized luciferase signal (F-luc/R-luc). mRNA abundance was quantified by qRTPCR with normalization. Data points representing the tethered group (red dots) were compared to those of the control group (tethered with control λ peptide, gray dots), and the directions of changes were shown by blue arrows.
(F) Quantification of the 4-thiouridine (4SU) /U ratio of YTHDF1- and YTHDF2-bound RNAs. Nascent transcribed mRNAs were labeled by 4SU for 1 hr. RNAs bound by YTHDF1 or YTHDF2 were isolated at 2 or 4 hr post-labeling, then analyzed by LC-MS/ MS. Error bars represent mean ± SD, n = 2. The results indicate that YTHDF1 binds nascent RNA before YTHDF2.
See also Figure S6.
Next, in order to provide direct evidence that YTHDF1 and YTHDF2 function by binding directly to their target mRNA, we used the same tethered luciferase reporter assay (Figure 5A) to guide N_YTHDF1_λ or N_YTHDF2_λ to the reporter transcript and assessed the impact on the cognate mRNA abundance and protein production (Figures 6C–6E). Similar to the ribosome profiling results, N_YTHDF1 tethering led to an increased protein production, N_YTHDF2 tethering gave a decreased mRNA abundance, and combining N_YTHDF1_λ and N_YTHDF2_λ resulted in both elevated protein expression and reduced mRNA abundance. These results validated that YTHDF1 and YTHDF2 possess distinct functions: YTHDF1 promotes mRNA translation, whereas YTHDF2 facilitates degradation. When these two proteins cooperate to regulate the shared target transcripts, a relatively shallow transcript signal could be converted to a sharp protein production profile.
Both YTHDF1 and YTHDF2 locate in the cytoplasm. To probe the temporal order of their binding to common RNA targets, we performed metabolic labeling of nascent mRNA in pulse-chase experiments and evaluated the association of nascent mRNA with YTHDF1 or YTHDF2, respectively (Figures 6F and S6B–S6D). The results showed that YTHDF1 binds to nascent mRNA transcripts earlier than YTHDF2, which is consistent with the expectation that translation of most of these mRNAs should occur before degradation under normal growth conditions.
DISCUSSION
We present herein a systematic study of mRNA recognition, protein interaction, and consequent translational control of the human mRNA by m6A reader protein YTHDF1. Transcriptome-wide characterization of YTHDF1-binding sites supports that YTHDF1 recognizes m6A-modified mRNA inside mammalian cells. High-throughput sequencing measurements uncovered a positive correlation between the ribosome loading of YTHDF1-targeted mRNAs and the number of YTHDF1-binding sites on the target mRNAs. As a consequence of YTHDF1 knockdown, the m6A-modified mRNAs were less associated with polysome. The causality between YTHDF1 binding and translation promotion was established by directly tethering YTHDF1 to a reporter transcript. We further showed that YTHDF1 interacts with eIFs and ribosomes. Integration of these results led us to propose a mechanism of m6A-dependent mRNA translation promotion through YTHDF1.
RNA m6A Methylation as a Mechanism of Translational Control
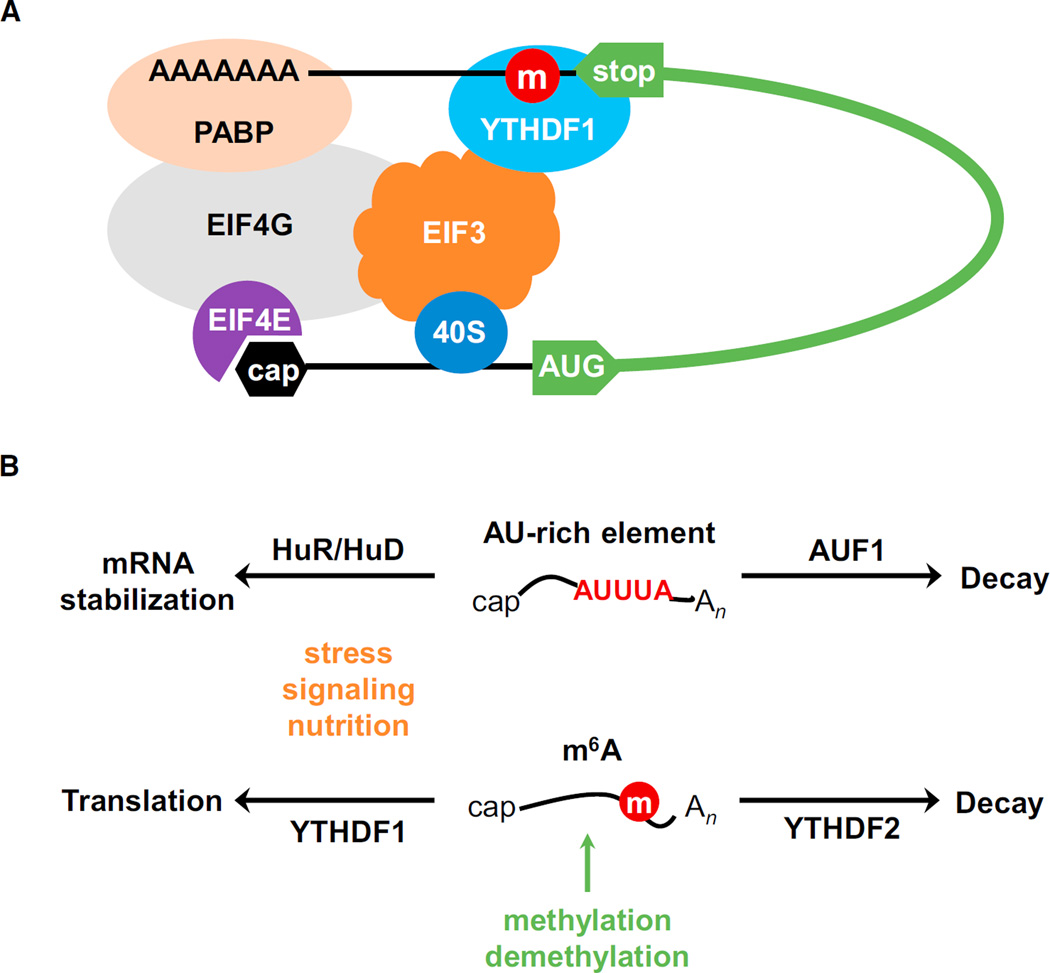
Our experimental evidence supports a dual role of YTHDF1 in delivering cellular mRNAs to translation machinery and directly facilitating translation initiation (Figure 3B). Initiation is typically the rate-limiting step of translation (Sonenberg and Hinnebusch, 2009), which includes several steps: (1) 40S ribosomal subunit, eIF3, and the eIF2–GTP–tRNAMet ternary complex assemble into the 43S pre-initiation complex; (2) mRNA is recruited to 43S through an association between the eIF4G and eIF3; (3) the resulting 48S initiation complex scans along the 5′ UTR to encounter the AUG start codon, which leads to the dissociation of eIFs and binding of the 60S subunit to form the 80S ribosome. The m6A methylation of mammalian mRNA occurs mostly at CDS and 3′ UTR but peaks around the stop codon, as do the YTHDF1-binding sites. But how do the protein-binding events at the 3′ end affect translation initiation happening at the 5′ end? There are different mechanistic possibilities. YTHDF1 could be spatially in proximity with translation initiation sites bridged by eIF4G (Figure 7A). During the canonical translation initiation, eIF4G binds both cap-binding protein eIF4E and poly(A)-binding protein to form a “closed loop.” Based on this model, if the eIF4G-mediated loop structure fails to form, the effects of YTHDF1 on mRNA translation should be weakened. The translation initiation step of HCV and CrPV IRES is independent of the eIF4G-mediated loop formation. Indeed, when tethered at the 3′ UTR, YTHDF1 does not improve the translation of HCV or CrPV IRES reporters as efficiently as that of the EMCV reporter (Figures 5D and S5D). In real biological systems, certain mRNA could have more complex 3′ UTRs and/orm6A sites at coding regions/ 5′ UTRs away from the 3′ end. The recruitment of YTHDF1 to the initiation site could be more complex compared to the tethered reporters tested here. We attributed the association of YTHDF1 with the translation initiation complex largely to the interaction between YTHDF1 and eIF3. This interaction could occur with YTHDF1 binding to m6A sites located at 3′ UTRs, coding regions, and 5′ UTRs of mRNA to promote translation. The detailed mechanisms and functional implications for the YTHDF1-promoted translation should be further studied in the future.
Figure 7. A Proposed Model of Translation Promotion by YTHDF1.
(A) YTHDF1 recruits m6A-modified transcripts to facilitate translation initiation. The association of YTHDF1 with translation initiation machinery may be dependent on the loop structure mediated by eIF4G and the interaction of YTHDF1 with eIF3.
(B) The m6A-based regulation through binding of YTHDF1 and YTHDF2 shares similarities with that of the AU-rich element which is regulated by HuR/ HuD and AUF1.
The looping model has been proposed for many RNA-binding proteins that affect translation from the 3′ end (Mazumder et al., 2003). This model, if true, also provides a reasonable explanation for the prompt translation recovery of the YTHDF1-tethered mRNA reporter after arsenite treatment, which induces the formation of stress granules (Kedersha and Anderson, 2007). Stress granules contain mRNAs stalled in translational initiation, usually a consequence of stress-induced phosphorylation of eIF2α (Buchan and Parker, 2009; Kedersha et al., 1999). It is possible that YTHDF1 mediates/stabilizes the formation of the stalled initiation complex. Therefore, once stress is released, the YTHDF1-bound mRNA can quickly resume translation initiation from its stalled position.
Unified Regulation through m6A in the Cytoplasm
Our results show that YTHDF1 promotes the translation of mRNA whereas YTHDF2 facilitates the degradation of mRNA in HeLa cells under normal growth conditions. Intuitively, translation and degradation are two opposite fates of mRNA, which suggest a more complex picture of the overall function of m6A on mRNA. YTHDF1 and YTHDF2 each have their own sets of target mRNAs that can be differentially regulated in response to different cellular signals. They also share a large set of common target mRNAs (Figure S6A). Regarding these shared mRNA targets, we observed that YTHDF1 binds RNA earlier during the mRNA life cycle than YTHDF2 does, suggesting that YTHDF1 and YTHDF2 function together to promote translation efficiency of transcripts that are dynamic and require transient controls. Their cooperation could provide potential functional benefits:
To achieve fast response with sufficient gene expression. Protein production is determined by mRNA abundance and translation efficiency. The m6A-modified RNAs tend to be dynamic and possess relatively shorter half-lives (Batista et al., 2014; Fu et al., 2014; Wang et al., 2014b). Unstable mRNAs approach new steady-state levels more quickly than stable transcripts following changes in the synthetic rates, thus decreasing the response time between stimulus and phenotypic output. For relatively short-lived mRNA to achieve sufficient protein production, translation efficiency needs to be elevated. YTHDF1 and YTHDF2 together shape a group of dynamically regulated genes. During cell differentiation or development events, YTHDF1 may activate the translation of methylated mRNAs to achieve sufficient protein productions, whereas YTHDF2 functions to limit the lifetimes of these mRNAs for proper cell development or differentiation.
To maintain stable protein quantity and reduce gene-expression noise. Transcription in eukaryotic cells occurs in bursts (Raj and van Oudenaarden, 2008). One task of post-transcriptional gene regulation is to reduce transcription noise (Siciliano et al., 2013). RNA methylation may serve as one of the mechanisms by which cells balance translation level despite varying transcription: more decay when transcripts are in excess; translation promotion when transcripts are in scarcity. This possibility will require further experimental validations from single-cell or single-molecule studies.
The m6A mark on mRNA shares similarities with the AU-rich element (ARE, Figure 7B) (Barreau et al., 2005). As a major function, they all mark unstable mRNA. AUF1 promotes fast decay of the ARE-containing transcripts (Gratacó s and Brewer, 2010), whereas YTHDF2 mediates degradation of methylated RNA under normal growth conditions (Wang et al., 2014a). In both cases, trans-acting factors also contribute to more complex gene-regulation modes. HuR stabilizes ARE mRNA, and such stabilization is more significant during nutrition shortage (Brennan and Steitz, 2001; Yaman et al., 2002). HuD also stabilizes ARE mRNA and is a key regulator of neuronal plasticity (Deschênes-Furry et al., 2006). Similarly, we show here that YTHDF1 elevates translation of the m6A-modified mRNA; such an effect is particularly obvious during stress response. Similar comparisons can be made between m6A and other RNA cis-elements that are encoded by primary sequence and play dual or multiple roles in mRNA metabolism. The m6A mark could work in combination with these other functional RNA elements, akin to DNA methylation and transcription factors. Over 7,000 human genes contain m6A sites, which is more widespread than other mRNA elements. The m6A-mediated translational and stability control is particularly unique in its reversibility, dynamics, and non-stoichiometric nature, which is different from elements encoded in the primary sequence. Because the methylation status of each site is not fixed, the landscape of m6A can shift according to cell cycle, differentiation status, and tissue type. The non-stoichiometry of m6A sites also enables fine-tuning of regulatory strength in addition to RNA-protein affinity.
In summary, the role of YTHDF1 in promoting mRNA translation efficiency fills a critical missing piece of the m6A-dependent gene regulation. This YTHDF1-dependent function could complement the mRNA decay pathway mediated through YTHDF2 to implement dynamic gene-expression regulation and offer an additional pathway for translation recovery from stress. We envision that the combination of functions of YTHDF2 and YTHDF1 allows for more precise and effective control of protein production during various biological transformations, such as stem cell differentiation, circadian rhythms, gametogenesis, and animal development.
EXPERIMENTAL PROCEDURES
High-Throughput RNA Sequencing
For PAR-CLIP, RIP-seq, and mRNA lifetime profiling, we followed previously reported procedures (Hafner et al., 2010; Peritz et al., 2006; Wang et al., 2014a). Ribosome profiling was conducted with the ARTseq Ribosome Profiling Kit (Mammalian, Epicentre). The sequencing data obtained from ribosome profiling were denoted as ribosome-bound fragments and those from RNA input as mRNA input. Translation efficiency was defined as the ratio of ribosome-protected fragments and mRNA input (Ingolia et al., 2009). The processed sequencing data were summarized in Table S1.
Polysome Profiling
HeLa cells were subjected to 48 hr knockdown and treated with cycloheximide (CHX) at 100 µg ml−1 for 2 min before collection. Cells were pelleted, lysed on ice, and centrifuged. The supernatant (~1.2 ml) was collected and loaded onto a 10/50%w/v sucrose gradient prepared in a lysis buffer without Triton X-100. The gradients were centrifuged at 4°C for 4 hr at 27,500 rpm (Beckman, rotor SW28). The sample was then fractioned and analyzed by Gradient Station (BioCamp) equipped with an ECONO UV monitor (BioRad) and fraction collector (FC203B, Gilson). The fractions were categorized and used for western blotting or pooled to isolate total RNA by TRIzol reagent for RT–PCR and mRNA for LC-MS/MS test.
Tether Assay and Stress Treatment
The reporter plasmid (pmirGlo-pTight-5BoxB) and the effecter plasmids (λ, N_YTHDF1_λ, N_YTHDF2_λ in pcDNA3.0) were used to transfect HeLa cells at 1:9 ratio under doxycycline (100 ng ml−1) inhibition. After 6 hr, transfection mixture was replaced with fresh media containing doxycycline. After 18 hr, cells were trypsin-digested, extensively washed with PBS, and re-seeded without doxycycline. Eight hours after re-seeding and induction, cells were either assayed by Dual-Glo Luciferase Assay Systems (Promega) to test protein production or processed to extract total RNA (DNase I digested), followed by RT–PCR quantification.
For pulse-induction: 2 hr after re-seeding, 500 ng ml−1 doxycycline was added into the wells, ending the 2 hr pulse induction of transcription. Then the translation dynamics was monitored by assaying cells each hour from re-seeding (set as t = 0) to 8 hr after re-seeding (except 7 hr).
For stress recovery: 2 hr after re-seeding, 500 ng ml−1 doxycycline was added into the wells, ending the 2 hr pulse induction of transcription. At the same time, 1 mM sodium arsenite was also added into the wells and removed at 3 hr, completing the 1 hr stress treatment. The translation dynamics was monitored by assaying cells each hour from re-seeding (set as t = 0) to 8 hr after re-seeding (except 7 hr).
For IRES tether reporter assay: 5 ng reporter plasmid (pRF-HCV-5BoxB, pRF-EMCV-5BoxB, pRF-CrPV-5BoxB), 45 ng effecter plasmid (pTight-GGS-λ, pTight-N_YTHDF1_λ), and 5 ng control plasmid (pJ7-LacZ) were used to transfect ~1.5 × 105 HeLa cells in each well of a 96-well plate. After 24 hr, half of cells were assayed to measure Firefly luciferase (F-luc) and Renilla luciferase (R-luc) activity, whereas the other half were assayed to measure the LacZ activity, which was then used to normalize both F-luc and R-luc signals. Finally, F-luc activity was normalized by R-luc to evaluate IRES-dependent expression compared to cap-dependent expression.
Supplementary Material
Highlights.
YTHDF1 selectively recognizes m6A-modified mRNAs
YTHDF1 promotes ribosome loading of m6A-modified mRNAs
YTHDF1 interacts with initiation factors to facilitate translation initiation
m6A modulates gene expression via control of mRNA translation and degradation
Acknowledgments
We thank Professor Tao Pan for insightful discussion and kindly providing the ribosome profiling instrument. We thank Mr. Wesley Clark for help in the ribosome profiling experiment. We thank Professor Anne E. Willis for providing the IRES reporter plasmids. We thank Institutes of Biomedical Sciences at Fudan University for protein mass spectrometry experiments. We thank Ms. Sarah Frank Reichard for editing the manuscript. C.H. is an investigator of the Howard Hughes Medical Institute (HHMI). B.S.Z. is supported by an HHMI International Student Fellowship. H.M. is supported by the Postdoctoral International Exchange Program of the China Postdoctoral Council (CPC). X. Weng is partially supported by the China Scholarship Council (CSC). The Mass Spectrometry Facility of the University of Chicago is funded by the National Science Foundation (CHE-1048528).
Footnotes
ACCESSION NUMBERS
Sequencing data have been deposited in NCBI’s GEO under accession number GSE63591.
SUPPLEMENTAL INFORMATION
Supplemental information includes Supplemental Experimental Procedures, six figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2015.05.014.
AUTHOR CONTRIBUTIONS
X. Wang and B.S.Z. contributed equally to this work. X. Wang, B.S.Z., and C.H. designed experiments. X. Wang and B.S.Z. performed experiments and analyzed data. I.A.R. assisted experiments and provided valuable discussion. X. Wang, Z.L., and D.H. processed the high-throughput RNA sequencing and proteomic data. H.M. helped the generation of the YTHDF1 stable line and protein mass spectroscopy experiment, and X. Weng, K.C., and H.S. assisted experiments and data analysis. X. Wang, B.S.Z., and C.H. wrote the manuscript.
REFERENCES
- Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:1–13. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar JA. The biosynthesis and functional roles of methylated nucleosides in eukaryotic mRNA. In: Grosjean H, editor. Fine-Tuning of RNA Functions by Modification and Editing. Springer; 2005. pp. 141–177. [Google Scholar]
- Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- Brennan CM, Steitz JA. HuR and mRNA stability. Cell. Mol. Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschênes-Furry J, Perrone-Bizzozero N, Jasmin BJ. The RNA-binding protein HuD: a regulator of neuronal differentiation, maintenance and plasticity. BioEssays. 2006;28:822–833. doi: 10.1002/bies.20449. [DOI] [PubMed] [Google Scholar]
- Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dina C, Meyre D, Gallina S, Durand E, Körner A, Jacobson P, Carlsson LMS, Kiess W, Vatin V, Lecoeur C, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- Do R, Bailey SD, Desbiens K, Belisle A, Montpetit A, Bouchard C, Pérusse L, Vohl MC, Engert JC. Genetic variants of FTO influence adiposity, insulin sensitivity, leptin levels, and resting metabolic rate in the Quebec Family Study. Diabetes. 2008;57:1147–1150. doi: 10.2337/db07-1267. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Fraser CS, Doudna JA. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat. Rev. Microbiol. 2007;5:29–38. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JRB, Elliott KS, Lango H, Rayner NW, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- Gao X, Wan J, Liu B, Ma M, Shen B, Qian S-B. Quantitative profiling of initiating ribosomes in vivo. Nat. Methods. 2015;12:147–153. doi: 10.1038/nmeth.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. http://dx.doi.org/10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- Gratacós FM, Brewer G. The role of AUF1 in regulated mRNA decay. Wiley Interdiscip Rev RNA. 2010;1:457–473. doi: 10.1002/wrna.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp A-C, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and micro-RNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature. 2006;442:45–50. doi: 10.1038/nature04881. [DOI] [PubMed] [Google Scholar]
- Hershey JW, Sonenberg N, Mathews MB. Principles of translational control: an overview. Cold Spring Harb. Perspect. Biol. 2012;4:a011528. doi: 10.1101/cshperspect.a011528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz MI, Landry DM, Willis AE, Luo G, Thompson SR. Ribosomal protein S25 dependency reveals a common mechanism for diverse internal ribosome entry sites and ribosome shunting. Mol. Cell. Biol. 2013;33:1016–1026. doi: 10.1128/MCB.00879-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiriart E, Vavasseur A, Touat-Todeschini L, Yamashita A, Gilquin B, Lambert E, Perot J, Shichino Y, Nazaret N, Boyault C, et al. Mmi1 RNA surveillance machinery directs RNAi complex RITS to specific meiotic genes in fission yeast. EMBO J. 2012;31:2296–2308. doi: 10.1038/emboj.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongay CF, Orr-Weaver TL. Drosophila Inducer of MEiosis 4 (IME4) is required for Notch signaling during oogenesis. Proc. Natl. Acad. Sci. USA. 2011;108:14855–14860. doi: 10.1073/pnas.1111577108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Aleksic J, Blanco S, Dietmann S, Frye M. Characterizing 5-methylcytosine in the mammalian epitranscriptome. Genome Biol. 2013;14:215. doi: 10.1186/gb4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Jeong SJ, Kim KN, Baek IJ, Chang M, Kang CM, Park YS, Yun CW. A novel protein, Pho92, has a conserved YTH domain and regulates phosphate metabolism by decreasing the mRNA stability of PHO4 in Saccharomyces cerevisiae. Biochem. J. 2014;457:391–400. doi: 10.1042/BJ20130862. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 α to the assembly of mammalian stress granules. J. Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Kim B, Kim VN. Emerging roles of RNA modification: m(6)A and U-tail. Cell. 2014;158:980–987. doi: 10.1016/j.cell.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Li F, Zhao D, Wu J, Shi Y. Structure of the YTH domain of human YTHDF2 in complex with an m6A mononucleotide reveals an aromatic cage for m6A recognition. Cell Res. 2014;24:1490–1492. doi: 10.1038/cr.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Tong L. Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc. Natl. Acad. Sci. USA. 2014;111:13834–13839. doi: 10.1073/pnas.1412742111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder B, Seshadri V, Fox PL. Translational control by the 3′-UTR: the ends specify the means. Trends Biochem. Sci. 2003;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014;15:313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Nilsen TW. Molecular biology. Internal mRNA methylation finally finds functions. Science. 2014;343:1207–1208. doi: 10.1126/science.1249340. [DOI] [PubMed] [Google Scholar]
- Peritz T, Zeng F, Kannanayakal TJ, Kilk K, Eiríksdóttir E, Langel U, Eberwine J. Immunoprecipitation of mRNA-protein complexes. Nat. Protoc. 2006;1:577–580. doi: 10.1038/nprot.2006.82. [DOI] [PubMed] [Google Scholar]
- Ping X-L, Sun B-F, Wang L, Xiao W, Yang X, Wang W-J, Adhikari S, Shi Y, Lv Y, Chen Y-S. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, Tabach Y, Mikkelsen TS, Satija R, Ruvkun G, et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell. 2013;155:1409–1421. doi: 10.1016/j.cell.2013.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, León-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah JC, Clancy MJ. IME4, a gene that mediates MAT and nutritional control of meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992;12:1078–1086. doi: 10.1128/mcb.12.3.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siciliano V, Garzilli I, Fracassi C, Criscuolo S, Ventre S, di Bernardo D. MiRNAs confer phenotypic robustness to gene networks by suppressing biological noise. Nat. Commun. 2013;4:2364. doi: 10.1038/ncomms3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theler D, Dominguez C, Blatter M, Boudet J, Allain FH-T. Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res. 2014;42:13911–13919. doi: 10.1093/nar/gku1116. http://dx.doi.org/10.1093/nar/gku1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck MT. Partial purification of a 6-methyladenine mRNA methyltransferase which modifies internal adenine residues. Biochem. J. 1992;288:233–240. doi: 10.1042/bj2880233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, He C. Dynamic RNA modifications in posttranscriptional regulation. Mol. Cell. 2014a;56:5–12. doi: 10.1016/j.molcel.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, He C. Reading RNA methylation codes through methyl-specific binding proteins. RNA Biol. 2014b;11:669–672. doi: 10.4161/rna.28829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014a;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014b;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CM, Moss B. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry. 1977;16:1672–1676. doi: 10.1021/bi00627a023. [DOI] [PubMed] [Google Scholar]
- Wei CM, Gershowitz A, Moss B. 5′-Terminal and internal methylated nucleotide sequences in HeLa cell mRNA. Biochemistry. 1976;15:397–401. doi: 10.1021/bi00647a024. [DOI] [PubMed] [Google Scholar]
- Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, Min J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 2014;10:927–929. doi: 10.1038/nchembio.1654. [DOI] [PubMed] [Google Scholar]
- Yaman I, Fernandez J, Sarkar B, Schneider RJ, Snider MD, Nagy LE, Hatzoglou M. Nutritional control of mRNA stability is mediated by a conserved AU-rich element that binds the cytoplasmic shuttling protein HuR. J. Biol. Chem. 2002;277:41539–41546. doi: 10.1074/jbc.M204850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang C-M, Li CJ, Vågbø CB, Shi Y, Wang W-L, Song S-H, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, Fray RG. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008;20:1278–1288. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.