Abstract
The transcription factor forkhead box M1 (FOXM1) is a validated oncoprotein in solid cancers, but its role in malignant plasma cell tumors such as multiple myeloma (MM) is unknown. We analyzed publicly available MM datasets and found that overexpression of FOXM1 prognosticates inferior outcome in a subset (~15%) of newly diagnosed cases, particularly patients with high-risk disease based on global gene expression changes. Follow-up studies using human myeloma cell lines (HMCLs) as the principal experimental model system demonstrated that enforced expression of FOXM1 increased growth, survival and clonogenicity of myeloma cells, whereas knockdown of FOXM1 abolished these features. In agreement with that, constitutive up-regulation of FOXM1 promoted HMCL xenografts in laboratory mice, whereas inducible knockdown of FOXM1 led to growth inhibition. Expression of cyclin dependent kinase 6 (CDK6) and NIMA-related kinase 2 (NEK2) was co-regulated with FOXM1 in both HMCLs and myeloma patient samples, suggesting interaction of these 3 genes in a genetic network that may lend itself to targeting with small-drug inhibitors for new approaches to myeloma therapy and prevention. These results establish FOXM1 as high-risk myeloma gene and provide support for the design and testing of FOXM1-targeted therapies specifically for the FOXM1High subset of myeloma.
Keywords: Plasma-cell neoplasm, cancer driver gene, targeted cancer therapy, small-drug inhibitor
Introduction
The prognosis of patients with multiple myeloma (MM), a difficult-to-cure blood cancer, depends in large measure on the genetic makeup of the myeloma cell. This is reflected in risk stratification models that consider cytogenetic features of myeloma, such as occurrence of oncogene-activating chromosomal translocations, and molecular features, such as gene expression changes1 measured with the assistance of the UAMS-702, EMC-923 or REL-174 gene test, to assign newly diagnosed cases to either standard-risk or high-risk groups. The distinction is clinically relevant because patients with high-risk myeloma have poor outcomes. Whereas overall survival (OS) for patients with standard-risk myeloma is 6–7 years, that for high-risk disease is no more than 2–3 years – despite the application of aggressive, risk-adapted therapies that include new myeloma drugs and, for eligible patients, high-dose therapy (HDT) followed by autologous stem-cell transplantation (ASCT)5. The unmet medical need of high-risk myeloma calls for dedicated efforts to elucidate the underlying genetic networks and develop approaches for their therapeutic targeting. This study demonstrates the involvement of the transcription factor forkhead box M1 (FOXM1) in a significant subset of high-risk myeloma (~15%) and suggests that FOXM1 provides a molecularly targeted treatment opportunity specifically for this group of patients.
In 2010, based on its enormous potential for the diagnosis and therapy of solid cancers, the AAAS journal Science bestowed upon FOXM1 the “Breakthrough of the Year” award. FOXM1, a member of the large forkhead box (FOX) family of proteins (n≈50), is a validated oncogene in carcinomas6 but has received little attention in myeloma and related plasma cell malignancies. Findings implicating FOXM1 in the maintenance and self-renewal of carcinoma stem cells7 have raised the question of whether it is similarly important for the putative myeloma stem cell8 – yet the elusiveness of this cell9 stands in the way of resolving the issue. Consistent with results that the expression of FOXM1 is tightly regulated in normal cells to ensure mitotic fidelity throughout the cell cycle10, deregulated expression of FOXM1 in carcinoma cells leads to centrosome amplification, mitotic catastrophe and other cytogenetic aberrations typically seen in cancer cells11. Whether FOXM1 governs genomic instability of MM12, a notorious but ill-explained feature of the neoplasia, is unclear. In diffuse large B-cell lymphoma, in which levels of the FOXM1 mRNA and the encoded protein are high, targeted inhibition of FOXM1 augments cell killing when combined with normally subtoxic doses of the proteasome inhibitor, bortezomib13. Whether this holds true for MM has not been established.
Our interest in FOXM1 began with a comparative gene expression analysis of B-lymphoma counterparts in humans and mice, implicating the transcription factor in an evolutionarily conserved pathway of neoplastic B-cell development14. Further encouraged by new evidence indicating that FOXM1 (1) drives tumor development and progression15–19 by virtue of a complex mechanism that includes enhanced cell proliferation, migration and invasion6, regulation of the DNA damage response20 and changes in the cancer epigenome21, (2) promotes cancer-cell resistance to ionizing radiation22 and cytotoxic drugs23, (3) governs, in part, the survival and tissue-regenerating capacity of both normal hematopoietic stem cells24 and malignant stem cell-like cells25, (4) links acquired resistance to cancer therapy with cancer stemness26 and (5) owing to the development of specific small-molecule inhibitors27, may soon be targeted more effectively than possible in the past28, we here decided to evaluate whether FOXM1 might play an important but heretofore overlooked role in plasma cell myeloma.
Material and Methods
FOXM1 expression and survival analysis in patients with myeloma
Levels of FOXM1 mRNA in myeloma cells were determined using Affymetrix U133Plus 2.0 microarrays (Santa Clara, CA) as previously described1. Results are available in the NIH Gene Expression Omnibus (GEO) under accession number GSE2658. Microarray data on MGUS and normal plasma cells are available at GSE5900. Statistical analysis of microarray results relied on GCOS1.1 software (Affymetrix, Santa Clara, CA), including log-rank tests for univariate association with disease-related survival.
Human myeloma cell lines (HMCLs), antibodies, and reagents
HMCLs, designated H929 or APR1, were chosen for studies on inducible knockdown (KD) of FOXM1. HMCLs, CAG or XG1, were used for studies on constitutive overexpression (OE) of FOXM1. All 4 cell lines had secretion of IgA29–32 and in vitro culture conditions (37 °C, 5% CO2) in common. Oncogene-activating chromosomal translocations took the form of t(4;14) in case of H929 cells33, t(11;14) in XG1 cells30 and t(14;16) in ARP1 and CAG cells33, 34. Gene expression spikes on microarrays corresponded to the translocation status: FGFR3 and WHSC1 (better known as MMSET) in case of H929 cells, CCND1 (cyclin D1) in XG1 cells and MAF (c-MAF) in ARP1 and CAG cells35. The status of the tumor suppressor p53 was wild-type in case of H929 cells36, mutated in XG1 cells37 and null in ARP1 and CAG cells34. Antibodies for Western blotting were purchased from Santa Cruz Biotechnology (FOXM1, catalog number sc-500; CDK6, sc-7961; β-actin, sc-47778) or Cell Signaling Technology (caspase-8, 4927; cleaved caspase-8, 9496; caspase-9, 9502; caspase-3, 9668; cleaved caspase-3, 9661; PARP, 9542). Doxycycline and thiostrepton (TS) were from Sigma.
Quantitative, reverse transcription (RT) polymerase chain reaction (qRT-PCR)
Total RNA was extracted using Quick-RNA MiniPrep (Zymo Research) and reverse transcribed using oligo dT primers and SuperScript III RT (Invitrogen). Data analysis relied on the ΔΔCt method. Primers were purchased from Integrated DNA Technologies (Coralville, Iowa). Sequences are available upon request.
Western blotting and co-immunoprecipitation (Co-IP) assays
Whole myeloma cell lysates were prepared using the Mammalian Cell Extraction kit (K269-500) from Biovision. Proteins (30 µg per sample) were fractionated on 4%–12% polyacrylamide gels blocked with 5% non-fat milk in Tris-buffered saline containing 0.05% Tween 20. Proteins were transferred to nitrocellulose membranes, incubated with primary antibody (dilution 10−3) overnight (4 °C), and visualized with HRP-conjugated secondary antibody using SuperSignal West Pico (Pierce). Blots were stripped, re-probed for β-actin, and evaluated by densitometry to estimate protein abundance. Co-IPs using the Pierce™ Direct Magnetic IP/Co-IP kit (Thermo Scientific) and antibodies to FOXM1 and CDK6 were performed as recently described38. IgG from Bethyl Laboratories was used as control.
Soft-agar clonogenicity assay
Clonogenic growth of myeloma was evaluated by seeding 1 × 104 cells in 0.5 mL RPMI 1640 (Invitrogen) supplemented with 0.33% agar and 10% FBS. Cells were grown in vitro (37 °C, 5% CO2) for 3 weeks, replenishing cell culture medium twice weekly. In some cases, cells were treated during weeks 2 and 3 with the FOXM1-inhibiting thiazole antibiotic, thiostrepton (TS). Cell clones, defined as tight aggregates of ≥40 myeloma cells, were enumerated using photographic images of soft-agar plates uploaded to Image J.
Cell proliferation and viability
Cell proliferation rate and viability were determined in 6-well plates using a hemacytometer and the trypan blue exclusion assay (0.4% dye in phosphate-buffered saline, pH 7.3).
Myeloma xenografts in immune compromised laboratory mice
To evaluate the impact of FOXM1 knockdown (KD) on myeloma growth, 2 × 106 H929 cells expressing either normal (FOXM1N) or reduced (FOXM1KD) levels of FOXM1 were injected subcutaneously (SC) into the right and left flank, respectively, of NSG mice (Jackson Laboratory, Bar Harbor, Maine). To induce FOXM1 knockdown (KD) in incipient tumors, doxycycline (2mg/mL) was added to the drinking water, beginning on day 10 following myeloma cell transfer.
Similarly, to assess the effect of FOXM1 overexpression (OE) on myeloma growth, paired samples of CAG cells (2 million in each flank) that expressed either elevated levels of FOXM1 (FOXM1OE) or normal levels (FOXM1N) were transferred SC to NSG mice (Jackson Laboratory). Ten days later, some mice were treated with intraperitoneal (IP) administrations of thiostrepton (30 mg/kg) twice weekly. In all cases, tumor growth was measured 2–3 times weekly, using a pair of calipers. Mice were sacrificed using CO2 asphyxiation when tumors reached 20 mm in diameter. All studies were approved under protocol 1301010 of the Institutional Animal Care and Use Committee of The University of Iowa.
Statistical analysis
Two-tailed Student’s t-test was used to compare two experimental groups. One-way analysis of variance (ANOVA) was used to evaluate more than two groups. The Kaplan-Meier method was used to determine myeloma patient survival in accordance with FOXM1 expression. In all cases, p ≤ 0.05 was considered significant.
Results
Heightened FOXM1 expression predicts poor survival in patients with myeloma
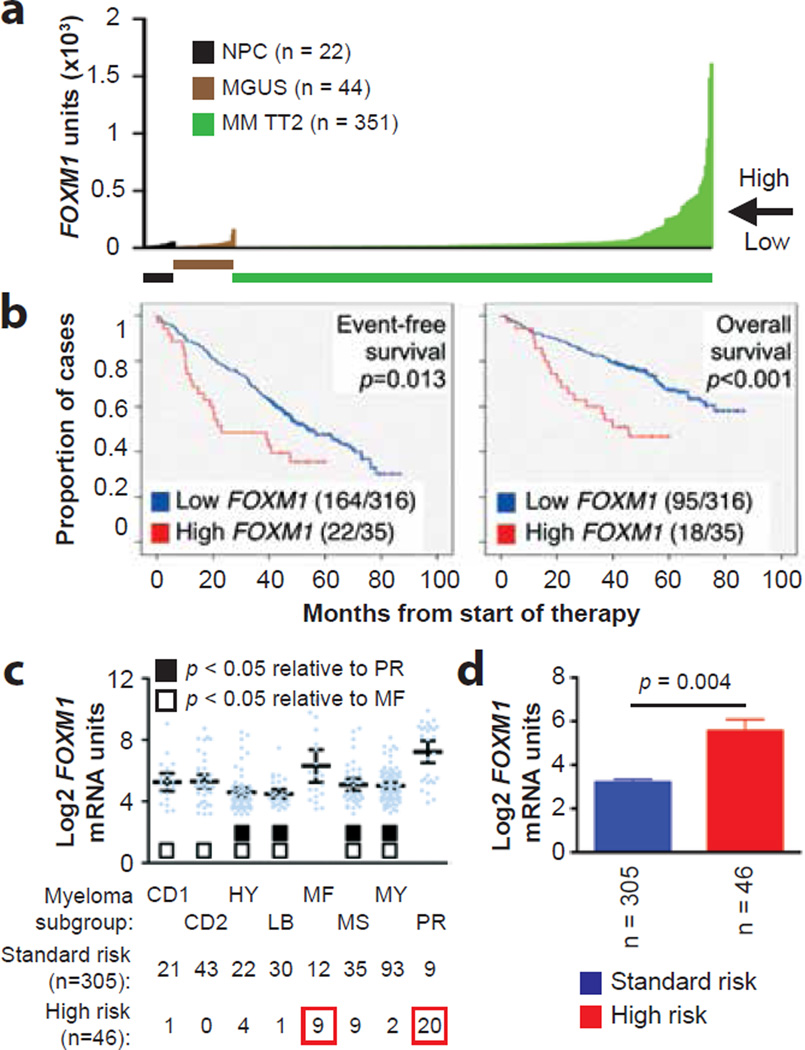
To evaluate the possibility that FOXM1 is important for myeloma, we analyzed a well-annotated, mature dataset, designated Total Therapy 2 (TT2), for which microarray-based gene expression and clinical outcome data were available. The levels of FOXM1 in 351 patients with newly diagnosed myeloma from that dataset are presented in Figure 1a, according to increasing gene expression. With 200 units of FOXM1 message used as a cutoff, the great majority of cases (316/351, 90%) exhibited the same expression levels seen in normal plasma cells (NPCs) from the bone marrow (BM; n = 22) or in BM plasma cells from individuals with monoclonal gammopathy of undetermined significance (MGUS; n = 44), a precursor of frank myeloma39. However, in ten percent of myelomas (35/351) FOXM1 levels were grossly elevated. The distinction between high and low FOXM1 was of prognostic significance, as both event-free survival (EFS) and overall survival (OS) were reduced in cases exhibiting high FOXM1 expression (Figure 1b). A very similar fraction of “FOXM1High” myelomas (18/149, 12.1%; Supplemental Figure 1a) associated with the same kind of survival disadvantage (Supplemental Figure 1b) was seen in the Total Therapy 3 (TT3) cohort, which comprised intensively treated patients (n = 149) that had received tandem ASCT and bortezomib/thalidomide-based induction and maintenance40. Likewise, elevated FOXM1 message predicted poor OS of patients with myeloma (n = 247) treated upfront using HDT/ASCT at the University of Heidelberg, Germany (Supplemental Figure 2)41. In agreement with the variable FOXM1 message levels seen in TT2/3 myelomas, a pilot immunolabeling study of archival myeloma-laden bone marrow sections pointed to variable amounts and distribution patterns of FOXM1 protein in myeloma cells (Supplemental Figure 3). These findings indicated that up-regulation of FOXM1 in a subset of myeloma (10–12%) leads to inferior outcome.
Figure 1. FOXM1 mRNA levels predict poor survival in a subset of patients with newly diagnosed myeloma.
(a) Line graph depicting the range of FOXM1 mRNA levels (gene probe ID 202580) in normal bone marrow (BM) plasma cells (NPC), “premalignant” BM plasma cells from individuals with monoclonal gammopathy of undetermined significance (MGUS) or malignant plasma cells from patients with newly diagnosed multiple myeloma (MM) from the University of Arkansas Total Therapy 2 (TT2) cohort. Specimens exhibiting less and more than 200 units of FOXM1 message were categorized as FOXM1Low and FOXM1High, respectively. This is indicated by the horizontal, labeled arrow pointing left.
(b) Reduced event-free survival (EFS) and overall survival (OS) in newly diagnosed TT2 patients harboring high FOXM1 levels. Of 351 myeloma cases, 316 (90%) had low FOXM1 levels (blue curve) and 35 (10%) had high FOXM1 levels (red curve). EFS and OS data were available from 186 (53%) and 113 (32%) patients, respectively.
(c) Mean values of FOXM1 levels in 8 molecular subgroups of MM: CD1, CCND1/CCND3 group 1; CD2, CCND1/CCND3 group 2; HY, hyperdiploid; LB, low bone disease; MF, MAF/MAFB; MS, MMSET; MY, myeloid; PR, proliferation2. FOXM1 is significantly elevated in MF myelomas as compared to 6 subgroups with low FOXM1 levels (open squares), and in the PR myelomas as compared to 4 such subgroups (closed squares), as assessed using the Bonferoni t test. The number of patients within each molecular subgroup who exhibit the standard-risk or high-risk UAMS-70-gene signature is indicated at the bottom. In total, 46 of 351 patients fell into the high-risk category with at least one case in each of the molecular subgroups except CD2.
(d) FOXM1 expression in high-risk MM, as defined by the UAMS-70-gene signature (n = 46), is elevated compared to that in low-risk MM (n = 305; Mann-Whitney test).
FOXM1 is a high-risk myeloma gene
We asked whether heightened FOXM1 expression in the TT2 cohort might be associated with a particular molecular subgroup of myeloma. Figure 1c presents the mean values of FOXM1 levels in 8 widely recognized subgroups, showing that elevated FOXM1 was particularly prevalent in 2 known to confer high risk in terms of clinical course and outcome: MAF/MAFB (MF) and proliferation (PR). A more recently developed molecular genetic approach to distinguishing high- and standard-risk disease; i.e., the UAMS 70-gene signature2, afforded another way of testing whether up-regulated FOXM1 might be a feature of high-risk myeloma. Statistical comparison of the mean FOXM1 levels in the 46 and 305 cases identified as high and standard risk according to this signature supported this contention (Figure 1d). In sync with that, not only was FOXM1High status statistically linked (χ2 contingency analysis) with a positive score in the 70-gene test (p < 10−4), almost two thirds of the UAMS-70 high-risk cases (29/46, 63%) fell into the PR (n=20) and MF (n=9) subgroups of myeloma (indicated by red squares in Figure 1c, bottom). The TT3 myelomas exhibited the same preponderance of FOXM1High in the MF/PR subgroups and the same association of FOXM1High status and positive 70-gene test score seen in the TT2 sample (Supplemental Figure 1c and d). The findings in more than 500 myelomas from the TT2/3 studies led us to conclude that FOXM1 is a bona fide high-risk myeloma gene.
Inducible downregulation of FOXM1 inhibits myeloma cells in vitro
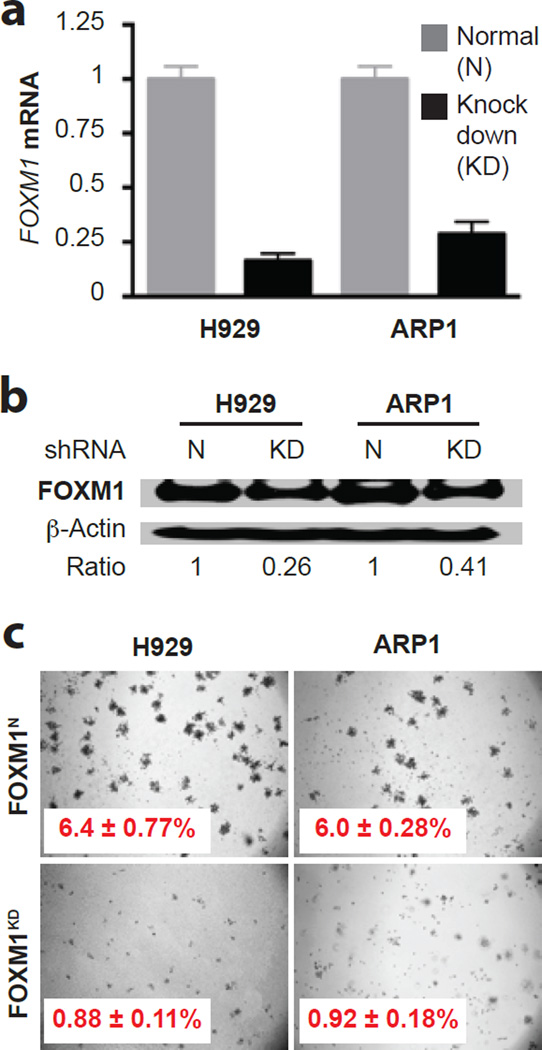
Following up on the clinical observations described above, we decided to elucidate the role of FOXM1 in myeloma biology using 2 independent human myeloma cell lines (HMCLs), designated H929 and ARP1, that harbored ample amounts of FOXM1 protein. To evaluate whether shRNA-mediated knockdown (KD) of FOXM1 message blunts the growth and survival of myeloma, we transduced H929 and ARP1 cells with lentivirus that encoded a FOXM1-targeted shRNA under control of a doxycycline (Dox)-inducible gene promoter: FOXM1KD cells. Cells transduced with a Dox-inducible “scrambled” shRNA, not targeting any expressed gene in mice and therefore leaving FOXM1 at normal levels (N), were used as control: FOXM1N cells. qRT-PCR and Western analyses demonstrated successful knockdown of FOXM1 in both cell lines at the mRNA (Figure 2a) and protein (Figure 2b) level. Significant reductions in FOXM1KD cell numbers, relative to FOXM1N controls, after 2, 3 and 4 days of Dox-dependent expression of shRNA indicated that partial loss of FOXM1 hampers myeloma growth in vitro (Supplemental Figure 4a). The diminished growth rate of FOXM1KD cells was attributed to both increased apoptotic cell death evidenced by increased proteolytic cleavage of poly (ADP-ribose) polymerase and caspases 3, 8 and 9 (Supplemental Figure 4b) and reduced cell cycle progression. The latter was revealed by flow cytometric analysis of DNA content that demonstrated an increase of cells in G1 and a concomitant decrease in S and G2/M (not shown). Soft agar clonogenicity assays demonstrated a striking (~7-fold) drop in size and number of FOXM1KD versus FOXM1N colonies: 0.88% vs. 6.4% in case of H929 cells and 0.92% vs. 6.0% in case of ARP1 cells (Figure 2c). These results suggested that FOXM1 regulates, in part, the growth and survival of myeloma cells.
Figure 2. Genetic knockdown of FOXM1 mitigates clonogenicity of myeloma in vitro.
(a) FOXM1 message levels in H929 and ARP1 myeloma cells, using qRT-PCR as measurement tool. Cells either under-expressed FOXM1 due to lentiviral transduction of a FOXM1–targeted shRNA “knockdown” construct (KD) or expressed FOXM1 at normal levels (N) following transduction of a non-targeted or “scrambled” shRNA used as control. Average loss of FOXM1 mRNA upon gene KD was ~80% and ~70% in H929 and ARP1 cells, respectively.
(b) Western analysis of samples included in panel a. Whole cell lysates were electrophoretically fractionated and immunoblotted using antibodies to FOXM1 and β-actin. Densitometry was employed to determine the FOXM1-to-β-actin ratio. Loss of FOXM1 protein in H929 and ARP1 KD cells amounted to 74% and 59%, respectively.
(c) Photographic images of representative soft-agar plates indicating the decreased clonogenic growth of FOXM1KD cells (bottom) compared to FOXM1N cells (top).
Genetic targeting of FOXM1 retards myeloma growth in mice
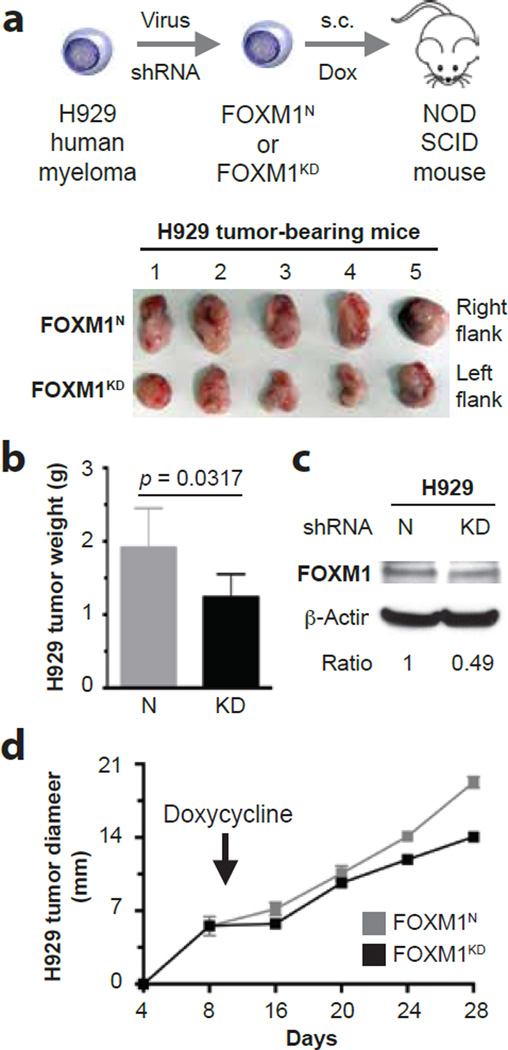
To determine whether inducible knockdown of FOXM1 inhibits myeloma in vivo, we xenografted FOXM1KD and FOXM1N H929 cells under the skin of the left and right abdominal flank of NSG mice (n = 5), respectively. Ten days after in vivo transfer of two million cells to each side, the mice were administered Dox in the drinking water to induce the expression of FOXM1-targeted or scrambled shRNA in the malignant plasma cells (Figure 3a, top). Tumor diameters were measured daily, using a pair of calipers, to compare the growth rate of the paired FOXM1KD and FOXM1N xenografts. In 5 of 5 hosts, the FOXM1KD tumors harvested on day 28 (endpoint of study) were visibly smaller than their FOXM1N counterparts (Figure 3a, bottom). The mean weight of FOXM1KD tumors (1.24 g) was 35% lower than that of FOXM1N tumors (1.91 g; Figure 3b). Compared to FOXM1N tumors, FOXM1KD tumors contained reduced amounts of FOXM1 protein (Figure 3c). Comparison of growth rates demonstrated that the FOXM1KD tumors lagged behind their FOXM1N counterparts (Figure 3d), indicating stable expression of the FOXM1-targeted shRNA during the 28-day growth period. These results demonstrated that genetic targeting of FOXM1 inhibits myeloma in vivo.
Figure 3. Inducible downregulation of FOXM1 inhibits myeloma xenografts in NSG mice.
(a) Shown at top is a scheme of the study design. FOXM1KD and FOXM1N H929 cells, generated using lentiviral shRNA transduction, were xenografted subcutaneously (s.c.) into the left and right flank of NSG hosts, respectively. Ten days later, mice received doxycycline in the drinking water to induce FOXM1-targeted shRNA in case of KD cells and scrambled shRNA in case of N cells. On day 28, xenografts were harvested and photographic images were taken (bottom).
(b) Mean weight of FOXM1KD and FOXM1N xenografts on day 28 post myeloma cell transfer.
(c) Western blot comparing FOXM1 protein levels in day-28 FOXM1KD and FOXM1N xenografts.
(d) Time course of tumor growth in NSG mice. Doxycycline treatment of mice began 10 days after myeloma cell transfer, as indicated by a vertical, labeled arrow pointing down. Tumor diameters were measured using a caliper, beginning on day 8 after xenografting. Mice were euthanized on day 28. Mean tumor diameters (squares) and standard deviations of the mean (short vertical lines with error bars) are plotted. Regression analysis of growth rates demonstrated that the FOXM1KD tumors (y = 0.665x + 0.0295; r2 = 0.948; p < 10−3) lagged behind their FOXM1N counterparts (y = 0.896x + 0.0280; r2 = 0.973; p < 10−3) by ~25%. The area under the curve of the FOXM1KD tumors (160) was ~15% smaller than that of the FOXM1N (188) tumors.
Enforced expression of FOXM1 promotes myeloma in vitro
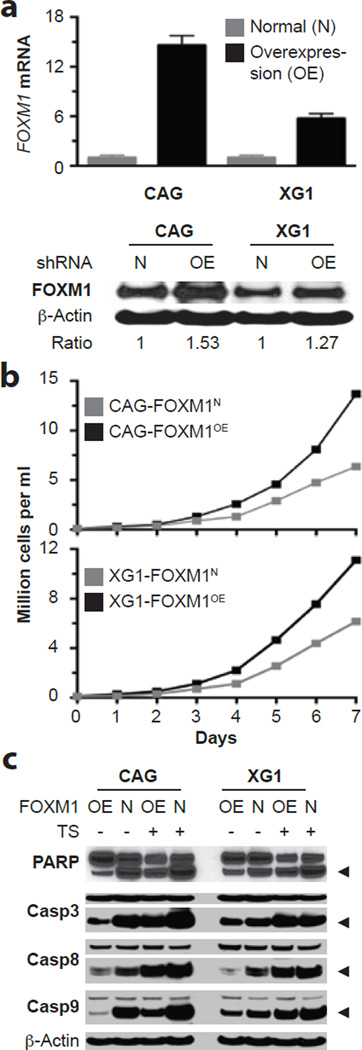
To complement the KD studies described above with the opposite experimental approach, we assessed whether enforced transgenic expression of FOXM1 might enhance myeloma growth. To that end, we transfected two independent HMCLs containing moderate amounts of FOXM1 protein, CAG and XG1, with lentivirus that encoded a FOXM1 cDNA gene under control of the EF1α promoter. Compared to cells transduced with non-coding “empty” virus that left FOXM1 levels unchanged (FOXM1N, used as control), cells over-expressing FOXM1 (FOXM1OE) contained elevated amounts of FOXM1 message (Figure 4a, top) and FOXM1 protein (Figure 4a, bottom). Up-regulation of FOXM1 promoted cell growth (Figure 4b), which was abolished by the FOXM1-inhibiting thiazole antibiotic, thiostrepton (TS)42 (Supplemental Figure 5a). Increased growth of FOXM1OE cells was attributed to enhanced cell cycle progression (not shown) and survival. The latter was reflected by less pronounced activation of PARP and caspases in FOXM1OE cells compared to FOXM1N cells (Figure 4c). Clonogenic growth in soft agar was moderately heightened (by ~30%) in untreated FOXM1OE vs. FOXM1N cells, but more significantly elevated (2–3 fold) in TS-treated FOXM1OE vs. FOXM1N cells (Supplemental Figure 5b and 5c). These results agreed with the FOXM1 knockdown studies and strengthened the contention that FOXM1 regulates growth and survival of myeloma cells in vitro.
Figure 4. Enforced expression of FOXM1 promotes growth and survival of myeloma cells in vitro.
(a) FOXM1 message levels measured by qRT-PCR (top) and FOXM1 protein levels determined by Western blotting (bottom) in CAG and XG1 myeloma cells that were either overexpressing FOXM1 constitutively due to lentiviral transduction of a FOXM1 cDNA gene (OE) or containing normal amounts of FOXM1 (N) due to transduction of an “empty” virus. The average increase in FOXM1 mRNA in OE cells was ~14-fold and ~6-fold in CAG and XG1 cells, respectively. The corresponding increase in FOXM1 protein was more modest, as indicated by the FOXM1-to-actin ratio below the Western blot.
(b) Line graphs depicting the growth of FOXM1OE and FOXM1N CAG (top) or XG1 (bottom) cells during one week in cell culture. OE cells grew faster than N cells, using two-way ANOVA for statistical comparison (p < 0.05).
(c) Western blots of whole-cell lysates of FOXM1OE and FOXM1N CAG (left) and XG1 (right) cells, using as detection tools specific antibodies to poly (ADP-ribose) polymerase (PARP) or three members of the apoptosis-related cysteine peptidase family of caspase proteins. Myeloma cells were either treated (indicated by “+” sign) using the FOXM1 inhibitor thiostrepton (TS) or left untreated (indicated by “-“ sign).
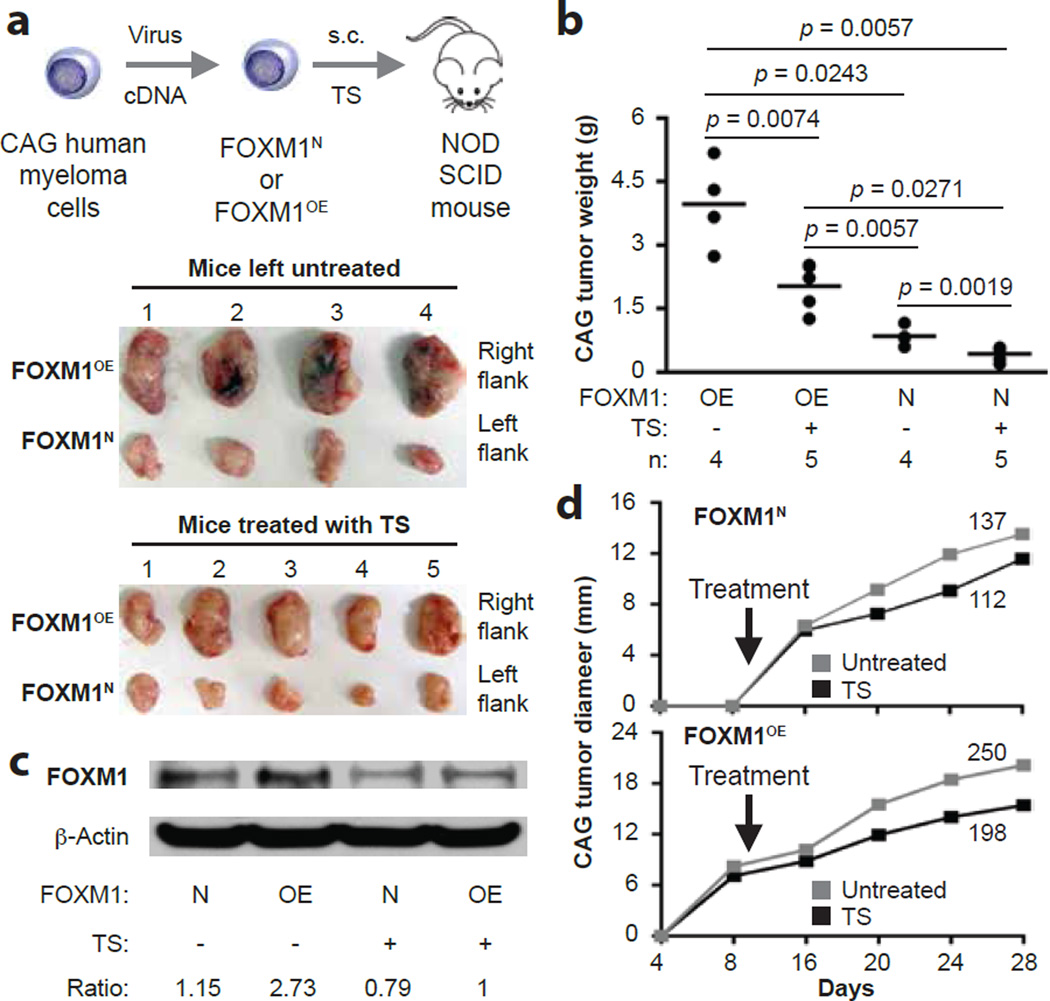
Overexpression of FOXM1 promotes myeloma xenografts in NSG mice
We transferred FOXM1OE and FOXM1N CAG cells to NSG mice treated with TS or left untreated (Figure 5a, top). In all cases, FOXM1OE tumors harvested at study endpoint (day 28) were larger than the FOXM1N tumors (Figure 5a, bottom). In untreated mice, the mean weight of FOXM1OE tumors (3.97 g) was significantly higher than that of FOXM1N tumors (0.848 g): a ratio of 4.7 (Figure 5b). The OE-to-N ratio (2.03 g / 0.432 g, 4.7) was the same in TS-treated mice (Figure 5b), which agreed with the expectation that TS inhibits FOXM1-expressing tumors, but did not support the possibility that FOXM1OE tumors were more sensitive to the drug than their FOXM1N counterparts. In both TS-treated and untreated mice, FOXM1OE tumors contained elevated levels of FOXM1 protein relative to FOXM1N controls (Figure 5c). Time course analyses of tumor size, analogous to those carried out in the KD studies, underlined the myeloma-promoting effect of FOXM1. Using the area under the curve as metric, growth rates of FOXM1OE tumors (198) were elevated by 77% in TS-treated mice compared to FOXM1N tumors (112; Figure 5d). Similarly, in untreated mice, growth rates of FOXM1OE tumors (250) were 83% higher than FOXM1N tumors (137; Figure 5d). These results added confidence to the contention that FOXM1 promotes myeloma in vivo, yet also indicated that TS may not be as active in mice as it is in vitro.
Figure 5. Treatment of NSG mice with thiostrepton inhibits FOXM1OE xenografts more effectively than FOXM1N xenografts.
(a) Scheme of experimental approach (top) and photographic images of myeloma xenografts harvested upon study termination on day 28 (bottom). FOXM1OE and FOXM1N CAG cells were generated using in vitro lentiviral gene transduction, followed by xenografting s.c. into the right and left flank of NSG hosts, respectively. One half of the study group was treated with TS (30 mg/kg IP twice weekly) beginning on day 10 post cell transfer, while the other half was left untreated.
(b) Mean tumor weights (indicated by horizontal lines) in the 4 experimental groups at end of study on day 28 post cell transfer. Tumor weights in TS-treated mice were smaller than in untreated mice (p values of Mann-Whitney tests are indicated) in case of both FOXM1OE and FOXM1N xenografts. The magnitude of TS-dependent tumor reduction was ~8 times higher in OE samples (4.2 g – 2.6 g = 1.6 g) compared to N samples (0.8 g – 0.6 g = 0.2 g).
(c) Representative Western blot of FOXM1 protein levels in FOXM1OE and FOXM1N xenografts collected from TS-treated (“+”) or untreated (“-“) hosts on day 28 post cell transfer. The ratios of FOXM1 to β-actin are indicated below the blot.
(d) Time course of tumor growth in NSG mice treated with TS or left untreated. Mean values (squares) are plotted. Areas under the curve, a metric of tumor growth that ranged from 112 to 250 in 4 experimental groups, are also indicated.
Coordinated expression of FOXM1 and CDK6 in myeloma
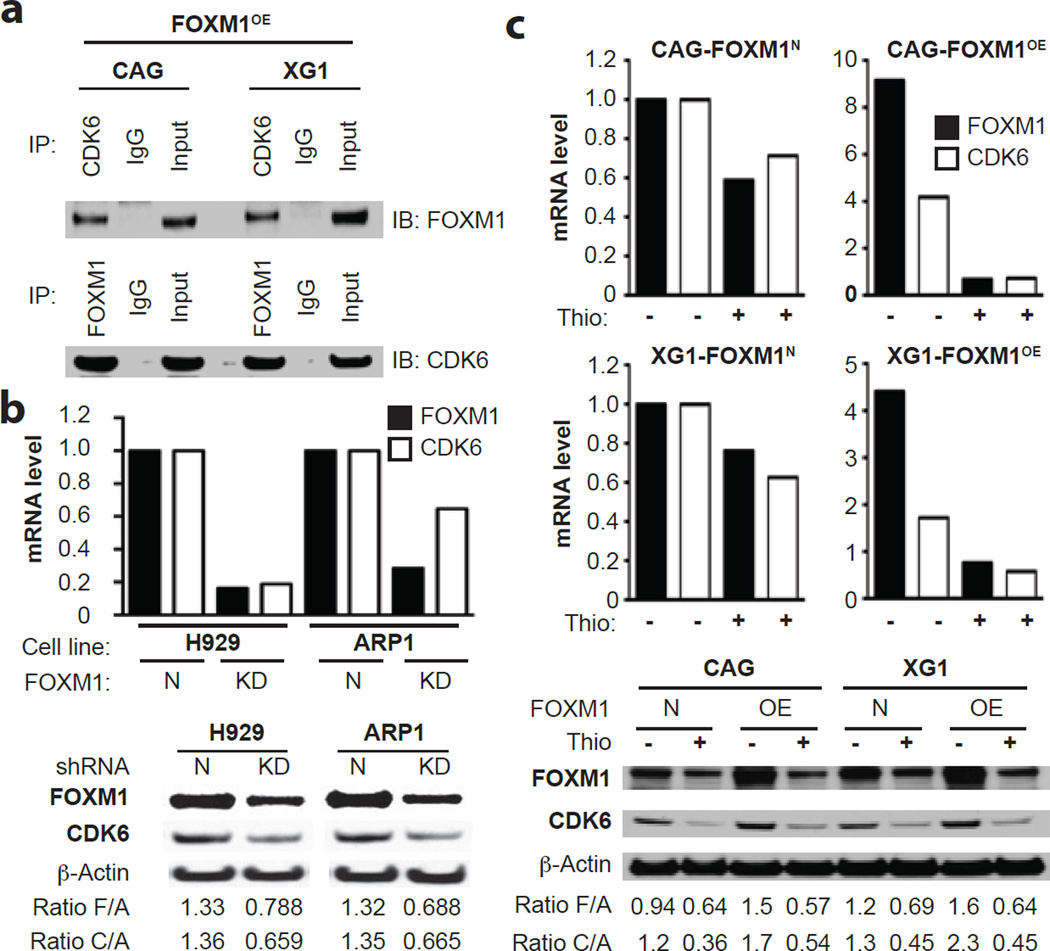
To elucidate the mechanism by which FOXM1 promotes myeloma, we followed up on publicly available UCSC ChIP-Seq results and published findings43, both indicating that CDK6 is a transcriptional target of FOXM1 in normal and malignant cells. There is also strong evidence that FOXM1 is a direct phosphorylation target of CDK6 in cancer, suggesting a positive auto-regulatory FOXM1-CDK6 feed-forward loop that supports the malignant state44. Co-IP analysis of FOXM1OE CAG and XG1 cells – which demonstrated that antibody to CDK6 (bait) pulled down FOXM1 (Figure 6a, top), whereas antibody to FOXM1 (bait) pulled down CDK6 (Figure 6a, bottom) – pointed to physical interaction of both proteins in myeloma cells. Co-detection of FOXM1 and CDK6 in FOXM1OE CAG cells by means of immunofluorescence microscopy supported this interpretation (not shown). qRT-PCR (Figure 6b, top) and Western blotting (Figure 6b, bottom) of FOXM1KD and FOXM1N H929 and ARP1 cells showed that FOXM1 and CDK6 expression may be co-regulated in myeloma. Consistent with that, pharmacological inhibition of FOXM1 using TS caused a coordinated drop of FOXM1 and CDK6 message in CAG and XG1 cells – more steeply in FOXM1OE than FOXM1N cells (Figure 6c, top). Corresponding changes in FOXM1 protein levels were also seen, but their magnitude was not as high as in case of mRNA (Figure 6c, bottom). In sync with the laboratory findings, FOXM1 and CDK6 expression were correlated in the TT2 and TT3 patient cohorts (Supplemental Figure 5b) and predictive of survival (Supplemental Figure 5c). Interestingly, prior to adopting the UAMS 70-gene signature for stratifying standard-risk and high-risk myeloma2, a 3-gene mini-signature had been developed for the same purpose. It relied on CDK6; one of its regulators, CKS1B, which encodes a member of the highly conserved cyclin kinase subunit 1 family of proteins45; and OPN3 (opsin 3), the role of which in myeloma is obscure (FZ, unpublished result).
Figure 6. Physical interaction and co-expression of FOXM1 and CDK6 in myeloma cells.
(a) Co-immunoprecipitation (Co-IP) result indicating physical interaction of FOXM1 and CDK6 in FOXM1-overexpressing (OE) CAG cells (left) and XG1 cells (right). Immunoblots using specific antibodies (Ab’s) to FOXM1 (after IP using Ab to CDK6) or CDK6 (after IP using Ab to FOXM1) are shown on top of each other. IgG isotype controls (labeled “IgG”) and samples of whole cell lysates not subjected to Co-IP (labeled “Input”) were included as controls.
(b) FOXM1 message (black bars) and CDK6 message (white bars) determined by qRT-PCR (top) and corresponding protein levels determined by immunoblotting (bottom) in FOXM1KD and FOXM1N H929 (left) and XG1 (right) myeloma cells. The ratios of target proteins to the house keeping protein, β-actin, are indicated below the Western blots. F/A and C/A denote the ratios including FOXM1 and CDK6, respectively.
(c) FOXM1 and CDK6 mRNA (top) and protein (bottom) levels in FOXM1OE and FOXM1N CAG or XG1 myeloma cells treated with TS (+) or left untreated (-). F/A and C/A ratios are as in panel b.
Co-expression of FOXM1 and NEK2 in myeloma
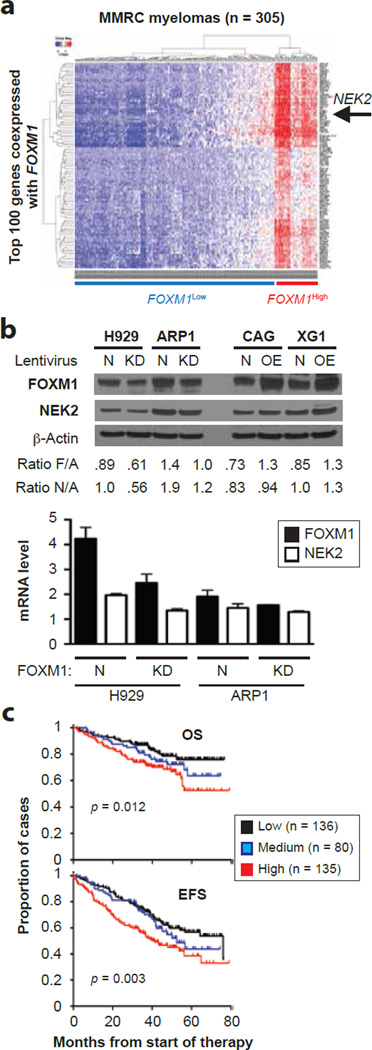
To identify additional network genes that may collaborate with FOXM1 in promoting myeloma, we interrogated the MMRC dataset available online for genes tightly co-expressed with FOXM1 (Figure 7a). Among the top 10 genes (r2 = 0.838) was NIMA-related kinase 2 or NEK2. Because NEK2 is not only a well-established transcriptional target of FOXM1 in various cell lineages46, 47 but also a driver of drug resistance in myeloma and other cancers48–50, we sought to confirm the co-expression of NEK2 and FOXM1 in independent datasets. This was the case in TT2 and TT3 (Supplemental Figure 6a, left) and also in MMRF’s CoMMpass℠ study (Supplemental Figure 6a, right), in which FOXM1High status conferred the same kind of survival disadvantage (Supplemental Figure 6b) seen in the TT2 (Figure 1b) and TT3 (Supplemental Figure 1) cohorts. In the CoMMpass℠ dataset, NEK2 was among the top 20 differentially expressed genes (p = 0.000331) in a 130-gene list (p < 0.05) that distinguished FOXM1High from FOXM1Low tumors (Supplemental Figure 6c). Gene set enrichment analysis (GSEA) using this gene list as input revealed a FOXM1-dependent network of transcription factors, DNA replication and cell division pathways in myeloma (Supplemental Figure 6d). These in silico findings prompted us to determine whether FOXM1 and NEK2 might be co-expressed in the 4 HMCLs used throughout this study. Western blotting showed that FOXM1 and NEK2 proteins shifted coordinately in accordance with FOXM1 status in 4 of 4 cell lines (Figure 7b, top). At the message level, results were less consistent; i.e., although NEK2 mRNA dropped upon knockdown of FOXM1 in H929 and ARP1 cells (Figure 7b, bottom), NEK2 did not rise upon overexpression of FOXM1 in CAG and XG1 cells (results not shown). Despite the latter finding, FOXM1 and NEK2 expression status was statistically linked with survival in the TT2 cohort (Figure 7c). These results provided evidence that FOXM1 and NEK2 are co-regulated in myeloma.
Figure 7. Coordinated expression of FOXM1 and NEK2 in myeloma cells.
(a) Co-expression of FOXM1 and NEK2 in the MMRC dataset (305 patients) publicly available at the Broad Institute’s Myeloma Genome Portal. The heat map contains the top 100 genes co-expressed with FOXM1. Each row and column represents one specific gene and patient, respectively. The position of NEK2, which is among the top 10 co-regulated genes, is indicated by a labeled arrow that points left.
(b) Shown at top is an immunoblot analysis of the FOXM1 and NEK2 protein levels in paired FOXM1KD/FOXM1N samples of H929 and ARP1 myeloma cells (left) or paired FOXM1OE/FOXM1N samples of CAG and XG1 myeloma cells (right). The ratios of target to house keeping protein (β-actin) are indicated below the Western blots: F/A for FOXM1 and N/A for NEK2. Presented at bottom is the result of a qRT-PCR analysis of FOXM1 and NEK2 expression in FOXM1KD and FOXM1N H929 (left) and ARP1 (right) cells, demonstrating that genetic down regulation of FOXM1 leads to a corresponding drop in NEK2 message.
(c) Expression levels of FOXM1 and NEK2 are associated with survival in TT2 myeloma patients. Cases were stratified as high or low expressers when both FOXM1 and NEK2 message were above (indicated in red) or below (black) the medium level in the TT2 dataset. All remaining cases (i.e., FOXM1High/NEK2Low or FOXM1Low/NEK2High) were designated as medium expressers (blue). Event-free survival (EFS) and overall survival (OS) in all 3 groups was plotted and statistically compared using log-rank analysis.
Discussion
The main finding of this study is clinical and experimental evidence for the involvement of FOXM1 in a relatively small (~15%) but aggressive subset of myeloma (chiefly subgroups MF and PR). FOXM1 expands the list of candidate genes uncovered by us35, 38, 48, 51 and others52, 53 that seem to render myeloma a high-risk disease by regulating pathways of tumor progression and stemness, acquisition of drug resistance and, ultimately, refractory relapse. A distinctive feature of FOXM1, compared to all other candidate genes identified thus far, is its discovery in a comprehensive gene expression analysis of mature B-cell lymphoma counterparts in humans and mice14. Cross-species analyses of this sort, which afford a unique and powerful approach to identifying genetic networks of neoplastic growth conserved over millions of years of evolution, have been successfully utilized in research on solid cancers but largely neglected in myeloma14. The results on FOXM1 reported here demonstrate the utility of this approach for myeloma and suggest that comparative oncogenomics of myeloma-like tumors in genetically engineered mouse models (GEMMs) may not only further our understanding of the natural history of myeloma, but also reveal new targets for treatment and prevention. The ongoing development of new GEMMs54–56 and continuous refinement of established ones57–59 are generating great promise along this line.
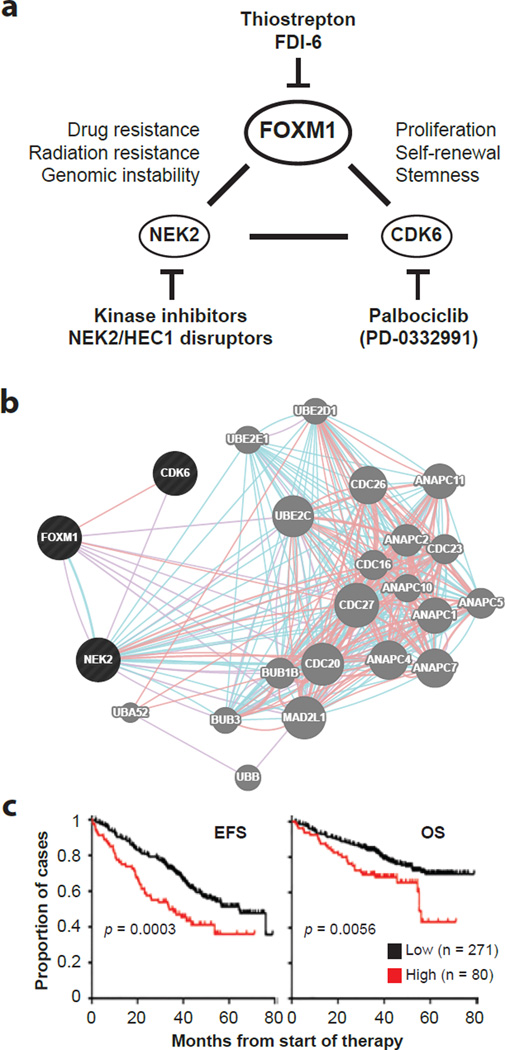
The results reported here may be summarized in a working model of FOXM1’s function in myeloma (Figure 8a), which considers (1) the gene’s co-regulation with CDK6 and NEK2, (2) the output of a network analysis that relied on the GeneMANIA online tool to predict genetic interactions of FOXM1 in myeloma cells (Figure 8b) and (3) the ability of a novel 3-gene mini-signature, comprised of FOXM1, CDK6 and NEK2, to prognosticate survival of patients with myeloma (Figure 8c). Although the underlying molecular genetics of the interaction depicted in Figure 8a needs to be elucidated in greater depth, the scheme may provide a useful blueprint for designing combination treatments for FOXM1High myelomas. This may take advantage of small-molecule inhibitors of (a) FOXM1, such as FDI-6, which binds directly to the transcription factor and displaces it from genomic targets in cancer cells27, (b) CDK6, such as palbociclib, which is not highly specific (it also targets CDK4) but has already demonstrated therapeutic activity (in conjunction with bortezomib and dexamethasone) in a phase 1/2 clinical trial of relapsed/refractory MM60 and (c) NEK2, such as aminopyridine scaffold compounds, which inhibit NEK2’s kinase activity61, or unrelated inhibitors, which trigger NEK2 degradation indirectly, using a mechanism that involves the disruption of NEK2 binding to kinetochore complex component, NDC80, better known as highly expressed in cancer 1 or HEC162. Despite broad cancer-suppressing activities63, the cyclic oligopeptide antibiotic, thiostrepton (TS), which has been used here as a FOXM1-inhibiting compound, is an unlikely candidate for further therapeutic development because its extensive use in veterinary medicine revealed severe toxicity issues.
Figure 8. FOXM1 may interact with CDK6 and NEK2 to shorten survival of patients with high-risk myeloma.
(a) Working model on the interaction of FOXM1, CDK6 and NEK2 in myeloma. Although FOXM1 is most firmly established as a proliferation-associated gene, new findings indicating that FOXM1 governs self-renewal and tumorigenicity of cancer stem cell-like cells in glioblastoma25, and that the FOXM1 target, CDK6, serves as a key regulator of leukemia stem cell activation64, raise the possibility that the interaction of FOXM1 and CDK6 in myeloma is also important for stemness. Additionally, FOXM1 may collaborate with NEK2 to drive resistance of myeloma cells to cancer therapy, given that NEK2 has been implicated in acquired drug resistance of many cancers48–50 and specifically shown to activate certain ABC drug transporters in myeloma48. Specific inhibitors of all 3 genes have been developed. Palbocicblib has already demonstrated activity in clinical trials on myeloma.
(b) Genetic interaction network of FOXM1, CDK6 and NEK2 (indicated in black to the left) generated with the help of the GeneMANIA online tool. Blue and pink lines denote pathways and physical interactions, respectively. Network genes are indicated by grey circles to the right that are labeled. The network’s apparent enrichment for ubiquination genes (not shown) points to the proteasome, suggesting in turn that the FOXM1-CDK6-NEK2 network core is involved in the response of myeloma cells to proteasome inhibition, a widely used treatment for myeloma.
(c) Kaplan-Meier plots of event-free survival (EFS, left) and overall survival (OS, right) of patients with myeloma from the TT2 cohort stratified according to high levels (red) or low levels (black) of FOXM1, CDK6 and NEK2 message upon microarray analysis. Myelomas containing higher than median amounts of mRNA of all three genes were designated as high (n = 80), whereas myelomas that did not meet this criterion were designated as low (n = 271). The differences in survival were significant using log-rank analysis (p < 0.05).
In conclusion, FOXM1 appears to be a bone fide high-risk myeloma gene that interacts with CDK6 and NEK2 to facilitate myeloma xenografts in mice and promote the growth, clonogenic self-renewal and survival of myeloma cells in vitro. Clinical studies are warranted to further validate FOXM1 as a potential therapeutic target in FOXM1High high-risk myeloma.
Supplementary Material
Acknowledgments
The expert technical assistance by Dr. Xuefang Jing and the provision of NSG mice by Dr. Weizhou Zhang (both Department of Pathology, University of Iowa) are gratefully acknowledged. This work was supported in part by NIH Training Grant T32-HL07734 and National Natural Science Foundation of China (NNSFC) Grant 81250110552 (both to VT); by NCI R01CA152105, Leukemia & Lymphoma Society Translational Research Program Awards 6246-11 and 6094-12, and NNSFC Award 81228016 (all to F.Z.); by institutional start-up funds from the Department of Internal Medicine, CCOM, UI (to FZ and GT); by NCI Core Grant P30CA086862 in support of The University of Iowa Holden Comprehensive Cancer Center; and by NCI R01CA151354 (to SJ).
Abbreviations
- CDK6
cyclin dependent kinase 6
- EFS
event-free survival
- FOXM1
forkhead box M1
- HMCL
human myeloma cell line
- KD
knockdown
- MM
multiple myeloma
- NEK2
NIMA (never in mitosis gene a)-related kinase 2
- OE
overexpression
- OS
overall survival
- TS
thiostrepton
Footnotes
Conflict-of-interest
The authors declare no competing financial interests.
Supplementary information
Supplementary information is available at Leukemia's website.
References
- 1.Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, et al. The molecular classification of multiple myeloma. Blood. 2006 Sep 15;108(6):2020–2028. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaughnessy JD, Jr, Zhan F, Burington BE, Huang Y, Colla S, Hanamura I, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007 Mar 15;109(6):2276–2284. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- 3.Kuiper R, Broyl A, de Knegt Y, van Vliet MH, van Beers EH, van der Holt B, et al. A gene expression signature for high-risk multiple myeloma. Leukemia. 2012 Nov;26(11):2406–2413. doi: 10.1038/leu.2012.127. [DOI] [PubMed] [Google Scholar]
- 4.Wu P, Walker BA, Broyl A, Kaiser M, Johnson DC, Kuiper R, et al. A gene expression based predictor for high risk myeloma treated with intensive therapy and autologous stem cell rescue. Leukemia & lymphoma. 2015 Mar;56(3):594–601. doi: 10.3109/10428194.2014.911863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajkumar SV. Multiple myeloma: 2012 update on diagnosis, risk-stratification, and management. Am J Hematol. 2012 Jan;87(1):78–88. doi: 10.1002/ajh.22237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koo CY, Muir KW, Lam EW. FOXM1: From cancer initiation to progression and treatment. Biochim Biophys Acta. 2012 Jan;1819(1):28–37. doi: 10.1016/j.bbagrm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Gong A, Huang S. FoxM1 and Wnt/beta-catenin signaling in glioma stem cells. Cancer research. 2012 Nov 15;72(22):5658–5662. doi: 10.1158/0008-5472.CAN-12-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsui W, Wang Q, Barber JP, Brennan S, Smith BD, Borrello I, et al. Clonogenic multiple myeloma progenitors, stem cell properties, and drug resistance. Cancer research. 2008 Jan 1;68(1):190–197. doi: 10.1158/0008-5472.CAN-07-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hajek R, Okubote SA, Svachova H. Myeloma stem cell concepts, heterogeneity and plasticity of multiple myeloma. British journal of haematology. 2013 Dec;163(5):551–564. doi: 10.1111/bjh.12563. [DOI] [PubMed] [Google Scholar]
- 10.Wonsey DR, Follettie MT. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer research. 2005 Jun 15;65(12):5181–5189. doi: 10.1158/0008-5472.CAN-04-4059. [DOI] [PubMed] [Google Scholar]
- 11.Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta. 2007 Jan;1775(1):92–102. doi: 10.1016/j.bbcan.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 12.Neri P, Bahlis NJ. Genomic instability in multiple myeloma: mechanisms and therapeutic implications. Expert Opin Biol Ther. 2013 Jun;13(Suppl 1):S69–S82. doi: 10.1517/14712598.2013.814637. [DOI] [PubMed] [Google Scholar]
- 13.Uddin S, Hussain AR, Ahmed M, Siddiqui K, Al-Dayel F, Bavi P, et al. Overexpression of FoxM1 offers a promising therapeutic target in diffuse large B-cell lymphoma. Haematologica. 2012 Jul;97(7):1092–1100. doi: 10.3324/haematol.2011.053421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tompkins VS, Han SS, Olivier A, Syrbu S, Bair T, Button A, et al. Identification of candidate B-lymphoma genes by cross-species gene expression profiling. PLoS One. 2013;8(10):e76889. doi: 10.1371/journal.pone.0076889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong X, Li L, Li Z, Le X, Huang C, Jia Z, et al. Dysregulated expression of FOXM1 isoforms drives progression of pancreatic cancer. Cancer research. 2013 Jul 1;73(13):3987–3996. doi: 10.1158/0008-5472.CAN-12-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai Y, Balli D, Ustiyan V, Fulford L, Hiller A, Misetic V, et al. Foxm1 expression in prostate epithelial cells is essential for prostate carcinogenesis. The Journal of biological chemistry. 2013 Aug 2;288(31):22527–22541. doi: 10.1074/jbc.M113.455089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Zheng Y, Park HJ, Li J, Carr JR, Chen YJ, et al. Targeting FoxM1 effectively retards p53-null lymphoma and sarcoma. Mol Cancer Ther. 2013 May;12(5):759–767. doi: 10.1158/1535-7163.MCT-12-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Motiwala T, Kutay H, Zanesi N, Frissora FW, Mo X, Muthusamy N, et al. PTPROt-mediated regulation of p53/Foxm1 suppresses leukemic phenotype in a CLL mouse model. Leukemia. 2015 Jun;29(6):1350–1359. doi: 10.1038/leu.2014.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kopanja D, Pandey A, Kiefer M, Wang Z, Chandan N, Carr JR, et al. Essential roles of FoxM1 in Ras-induced liver cancer progression and in cancer cells with stem cell features. J Hepatol. 2015 Mar 28; doi: 10.1016/j.jhep.2015.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zona S, Bella L, Burton MJ, Nestal de Moraes G, Lam EW. FOXM1: an emerging master regulator of DNA damage response and genotoxic agent resistance. Biochim Biophys Acta. 2014 Nov;1839(11):1316–1322. doi: 10.1016/j.bbagrm.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teh MT, Gemenetzidis E, Patel D, Tariq R, Nadir A, Bahta AW, et al. FOXM1 induces a global methylation signature that mimics the cancer epigenome in head and neck squamous cell carcinoma. PLoS One. 2012;7(3):e34329. doi: 10.1371/journal.pone.0034329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eckers JC, Kalen AL, Sarsour EH, Tompkins VS, Janz S, Son JM, et al. Forkhead box M1 regulates quiescence-associated radioresistance of human head and neck squamous carcinoma cells. Radiat Res. 2014 Oct;182(4):420–429. doi: 10.1667/RR13726.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karunarathna U, Kongsema M, Zona S, Gong C, Cabrera E, Gomes AR, et al. OTUB1 inhibits the ubiquitination and degradation of FOXM1 in breast cancer and epirubicin resistance. Oncogene. 2015 Jul 6; doi: 10.1038/onc.2015.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou Y, Li W, Sheng Y, Li L, Huang Y, Zhang Z, et al. The transcription factor Foxm1 is essential for the quiescence and maintenance of hematopoietic stem cells. Nat Immunol. 2015 Jun 29; doi: 10.1038/ni.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong AH, Wei P, Zhang S, Yao J, Yuan Y, Zhou AD, et al. FoxM1 Drives a Feed-Forward STAT3-Activation Signaling Loop That Promotes the Self-Renewal and Tumorigenicity of Glioblastoma Stem-like Cells. Cancer research. 2015 Jun 1;75(11):2337–2348. doi: 10.1158/0008-5472.CAN-14-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiu WT, Huang YF, Tsai HY, Chen CC, Chang CH, Huang SC, et al. FOXM1 confers to epithelial-mesenchymal transition, stemness and chemoresistance in epithelial ovarian carcinoma cells. Oncotarget. 2015 Feb 10;6(4):2349–2365. doi: 10.18632/oncotarget.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gormally MV, Dexheimer TS, Marsico G, Sanders DA, Lowe C, Matak-Vinkovic D, et al. Suppression of the FOXM1 transcriptional programme via novel small molecule inhibition. Nat Commun. 2014;5:5165. doi: 10.1038/ncomms6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halasi M, Gartel AL. Targeting FOXM1 in cancer. Biochem Pharmacol. 2013 Mar 1;85(5):644–652. doi: 10.1016/j.bcp.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 29.Gazdar AF, Oie HK, Kirsch IR, Hollis GF. Establishment and characterization of a human plasma cell myeloma culture having a rearranged cellular myc proto-oncogene. Blood. 1986 Jun;67(6):1542–1549. [PubMed] [Google Scholar]
- 30.Zhang XG, Gaillard JP, Robillard N, Lu ZY, Gu ZJ, Jourdan M, et al. Reproducible obtaining of human myeloma cell lines as a model for tumor stem cell study in human multiple myeloma. Blood. 1994 Jun 15;83(12):3654–3663. [PubMed] [Google Scholar]
- 31.Feinman R, Koury J, Thames M, Barlogie B, Epstein J, Siegel DS. Role of NF-kappaB in the rescue of multiple myeloma cells from glucocorticoid-induced apoptosis by bcl-2. Blood. 1999 May 1;93(9):3044–3052. [PubMed] [Google Scholar]
- 32.Baughn LB, Di Liberto M, Wu K, Toogood PL, Louie T, Gottschalk R, et al. A novel orally active small molecule potently induces G1 arrest in primary myeloma cells and prevents tumor growth by specific inhibition of cyclin-dependent kinase 4/6. Cancer research. 2006 Aug 1;66(15):7661–7667. doi: 10.1158/0008-5472.CAN-06-1098. [DOI] [PubMed] [Google Scholar]
- 33.Lombardi L, Poretti G, Mattioli M, Fabris S, Agnelli L, Bicciato S, et al. Molecular characterization of human multiple myeloma cell lines by integrative genomics: insights into the biology of the disease. Genes Chromosomes Cancer. 2007 Mar;46(3):226–238. doi: 10.1002/gcc.20404. [DOI] [PubMed] [Google Scholar]
- 34.Yellapantula V, Divya T, Dinu V, Scotch M. Informatics approaches for integrative analysis of disparate high-throughput genomic datasets in cander. Arizona State University (ASU) Electronic Dissertations and Theses Digital Repository. 2014 [Google Scholar]
- 35.Yang Y, Shi J, Tolomelli G, Xu H, Xia J, Wang H, et al. RARalpha2 expression confers myeloma stem cell features. Blood. 2013 Aug 22;122(8):1437–1447. doi: 10.1182/blood-2013-02-482919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hurt EM, Wiestner A, Rosenwald A, Shaffer AL, Campo E, Grogan T, et al. Overexpression of c-maf is a frequent oncogenic event in multiple myeloma that promotes proliferation and pathological interactions with bone marrow stroma. Cancer cell. 2004 Feb;5(2):191–199. doi: 10.1016/s1535-6108(04)00019-4. [DOI] [PubMed] [Google Scholar]
- 37.Romagnoli M, Trichet V, David C, Clement M, Moreau P, Bataille R, et al. Significant impact of survivin on myeloma cell growth. Leukemia. 2007 May;21(5):1070–1078. doi: 10.1038/sj.leu.2404602. [DOI] [PubMed] [Google Scholar]
- 38.Yang Y, Shi J, Gu Z, Salama ME, Das S, Wendlandt E, et al. Bruton tyrosine kinase is a therapeutic target in stem-like cells from multiple myeloma. Cancer research. 2015 Feb 1;75(3):594–604. doi: 10.1158/0008-5472.CAN-14-2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiss BM, Abadie J, Verma P, Howard RS, Kuehl WM. A monoclonal gammopathy precedes multiple myeloma in most patients. Blood. 2009 May 28;113(22):5418–5422. doi: 10.1182/blood-2008-12-195008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Rhee F, Szymonifka J, Anaissie E, Nair B, Waheed S, Alsayed Y, et al. Total Therapy 3 for multiple myeloma: prognostic implications of cumulative dosing and premature discontinuation of VTD maintenance components, bortezomib, thalidomide, and dexamethasone, relevant to all phases of therapy. Blood. 2010 Aug 26;116(8):1220–1227. doi: 10.1182/blood-2010-01-264333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seckinger A, Meissner T, Moreaux J, Depeweg D, Hillengass J, Hose K, et al. Clinical and prognostic role of annexin A2 in multiple myeloma. Blood. 2012 Aug 2;120(5):1087–1094. doi: 10.1182/blood-2012-03-415588. [DOI] [PubMed] [Google Scholar]
- 42.Hegde NS, Sanders DA, Rodriguez R, Balasubramanian S. The transcription factor FOXM1 is a cellular target of the natural product thiostrepton. Nature chemistry. 2011 Sep;3(9):725–731. doi: 10.1038/nchem.1114. [DOI] [PubMed] [Google Scholar]
- 43.Sanders DA, Gormally MV, Marsico G, Beraldi D, Tannahill D, Balasubramanian S. FOXM1 binds directly to non-consensus sequences in the human genome. Genome Biol. 2015;16(1):130. doi: 10.1186/s13059-015-0696-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anders L, Ke N, Hydbring P, Choi YJ, Widlund HR, Chick JM, et al. A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer cell. 2011 Nov 15;20(5):620–634. doi: 10.1016/j.ccr.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang H, Jiang N, Jiang H, Saha MN, Qi C, Xu W, et al. CKS1B nuclear expression is inversely correlated with p27Kip1 expression and is predictive of an adverse survival in patients with multiple myeloma. Haematologica. 2010 Sep;95(9):1542–1547. doi: 10.3324/haematol.2010.022210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nischalke HD, Schmitz V, Luda C, Aldenhoff K, Berger C, Feldmann G, et al. Detection of IGF2BP3, HOXB7, and NEK2 mRNA expression in brush cytology specimens as a new diagnostic tool in patients with biliary strictures. PLoS One. 2012;7(8):e42141. doi: 10.1371/journal.pone.0042141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calvisi DF, Pinna F, Ladu S, Pellegrino R, Simile MM, Frau M, et al. Forkhead box M1B is a determinant of rat susceptibility to hepatocarcinogenesis and sustains ERK activity in human HCC. Gut. 2009 May;58(5):679–687. doi: 10.1136/gut.2008.152652. [DOI] [PubMed] [Google Scholar]
- 48.Zhou W, Yang Y, Xia J, Wang H, Salama ME, Xiong W, et al. NEK2 Induces Drug Resistance Mainly through Activation of Efflux Drug Pumps and Is Associated with Poor Prognosis in Myeloma and Other Cancers. Cancer cell. 2013 Jan 14;23(1):48–62. doi: 10.1016/j.ccr.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X, Gao Y, Lu Y, Zhang J, Li L, Yin F. Upregulation of NEK2 is associated with drug resistance in ovarian cancer. Oncol Rep. 2014 Feb;31(2):745–754. doi: 10.3892/or.2013.2910. [DOI] [PubMed] [Google Scholar]
- 50.Marina M, Saavedra HI. Nek2 and Plk4: prognostic markers, drivers of breast tumorigenesis and drug resistance. Front Biosci (Landmark Ed) 2014;19:352–365. doi: 10.2741/4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi L, Wang S, Zangari M, Xu H, Cao TM, Xu C, et al. Over-expression of CKS1B activates both MEK/ERK and JAK/STAT3 signaling pathways and promotes myeloma cell drug-resistance. Oncotarget. 2010 May;1(1):22–33. doi: 10.18632/oncotarget.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanno T, Lim Y, Wang Q, Chesi M, Bergsagel PL, Matthews G, et al. Growth Differentiating Factor 15 enhances the tumor initiating and self-renewal potential of multiple myeloma cells. Blood. 2013 Dec 17; doi: 10.1182/blood-2013-08-524025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ziv E, Dean E, Hu D, Martino A, Serie D, Curtin K, et al. Genome-wide association study identifies variants at 16p13 associated with survival in multiple myeloma patients. Nat Commun. 2015;6:7539. doi: 10.1038/ncomms8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu Y, Zheng M, Gali R, Tian Z, Topal Gorgun G, Munshi NC, et al. A novel rapid-onset high-penetrance plasmacytoma mouse model driven by deregulation of cMYC cooperating with KRAS12V in BALB/c mice. Blood Cancer J. 2013;3:e156. doi: 10.1038/bcj.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dechow T, Steidle S, Gotze KS, Rudelius M, Behnke K, Pechloff K, et al. GP130 activation induces myeloma and collaborates with MYC. The Journal of clinical investigation. 2014 Dec;124(12):5263–5274. doi: 10.1172/JCI69094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tompkins VS, Rosean TR, Holman CJ, DeHoedt C, Olivier AK, Duncan KM, et al. Adoptive B-cell transfer mouse model of human myeloma. Leukemia. 2015 Jul 23; doi: 10.1038/leu.2015.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chesi M, Matthews GM, Garbitt VM, Palmer SE, Shortt J, Lefebure M, et al. Drug response in a genetically engineered mouse model of multiple myeloma is predictive of clinical efficacy. Blood. 2012 Mar 26; doi: 10.1182/blood-2012-02-412783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee EC, Fitzgerald M, Bannerman B, Donelan J, Bano K, Terkelsen J, et al. Antitumor activity of the investigational proteasome inhibitor MLN9708 in mouse models of B-cell and plasma cell malignancies. Clin Cancer Res. 2011 Dec 1;17(23):7313–7323. doi: 10.1158/1078-0432.CCR-11-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duncan K, Rosean TR, Tompkins VS, Olivier A, Sompallae R, Zhan F, et al. (18)F-FDG-PET/CT imaging in an IL-6- and MYC-driven mouse model of human multiple myeloma affords objective evaluation of plasma cell tumor progression and therapeutic response to the proteasome inhibitor ixazomib. Blood Cancer J. 2013;3:e165. doi: 10.1038/bcj.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Niesvizky R, Badros AZ, Costa LJ, Ely SA, Singhal SB, Stadtmauer EA, et al. Phase 1/2 study of cyclin-dependent kinase (CDK)4/6 inhibitor palbociclib (PD-0332991) with bortezomib and dexamethasone in relapsed/refractory multiple myeloma. Leukemia & lymphoma. 2015 May 15;:1–9. doi: 10.3109/10428194.2015.1030641. [DOI] [PubMed] [Google Scholar]
- 61.Innocenti P, Woodward H, O'Fee L, Hoelder S. Expanding the scope of fused pyrimidines as kinase inhibitor scaffolds: synthesis and modification of pyrido[3,4-d]pyrimidines. Organic & biomolecular chemistry. 2015 Jan 21;13(3):893–904. doi: 10.1039/c4ob02238f. [DOI] [PubMed] [Google Scholar]
- 62.Hu CM, Zhu J, Guo XE, Chen W, Qiu XL, Ngo B, et al. Novel small molecules disrupting Hec1/Nek2 interaction ablate tumor progression by triggering Nek2 degradation through a death-trap mechanism. Oncogene. 2015 Mar 5;34(10):1220–1230. doi: 10.1038/onc.2014.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bhat UG, Halasi M, Gartel AL. FoxM1 is a general target for proteasome inhibitors. PLoS One. 2009;4(8):e6593. doi: 10.1371/journal.pone.0006593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scheicher R, Hoelbl-Kovacic A, Bellutti F, Tigan AS, Prchal-Murphy M, Heller G, et al. CDK6 as a key regulator of hematopoietic and leukemic stem cell activation. Blood. 2015 Jan 1;125(1):90–101. doi: 10.1182/blood-2014-06-584417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.