Abstract
Genome architecture is shaped by gene-rich and repeat-rich regions also known as euchromatin and heterochromatin, respectively. Under normal conditions, the repeat-containing regions undergo little or no meiotic crossover (CO) recombination. COs within repeats are risky for the genome integrity. Indeed, they can promote non-allelic homologous recombination (NAHR) resulting in deleterious genomic rearrangements associated with diseases in humans. The assembly of heterochromatin is driven by the combinatorial action of many factors including histones, their modifications, and DNA methylation. In this review, we discuss current knowledge dealing with the epigenetic signatures of the major repeat regions where COs are suppressed. Then we describe mutants for epiregulators of heterochromatin in different organisms to find out how chromatin structure influences the CO rate and distribution.
Keywords: DNA methylation, Histone acetylation, Histone methylation, Heterochromatin, Euchromatin, Meiosis
Introduction
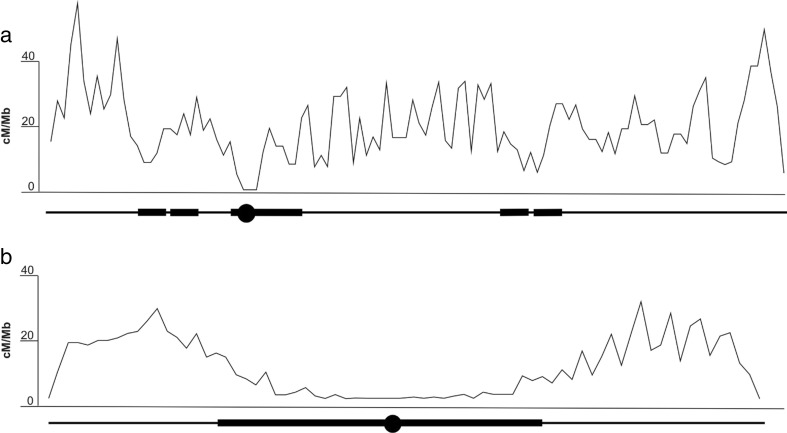
Genome architecture is shaped by gene-rich and repeat-rich regions also known as euchromatin and heterochromatin, respectively. Repeat sequences exist at a high number in the genomes of eukaryotes and are localized mostly in centromeres, telomeres, transposable elements, ribosomal (rRNA), and transfer RNA (tRNA) loci. Under normal conditions, the repeat-containing regions undergo little or no meiotic crossover recombination (Fig. 1) (Chen et al. 2008; Pan et al. 2011).
Fig. 1.
Recombination rate variation along human (a) and tomato (b) chromosome 12. Below each graph, chromosome structure is represented by euchromatin (thin line) and heterochromatin (thick line). Data have been extrapolated from Myers et al. (2005) (a) and The Tomato Genome Consortium (2012) (b)
Meiotic recombination is initiated by the SPO11 transesterase-dependent DNA double-strand breaks (DSBs) that are repaired as crossovers (COs), i.e., by reciprocal exchanges between the homologous chromosomes. In many organisms, more DSBs are formed than COs, thereby indicating alternative pathways of DSB repair including non-crossover (NCO), i.e., non-reciprocal exchange and inter-sister repair (IS) (Mercier et al. 2015; de Massy 2013). Multiple layers of CO control exist in most organisms. Firstly, recombination events are non-randomly distributed along chromosomes and they cluster in small regions called hot spots. Secondly, the phenomenon of CO interference leads nearby COs, so they occur further apart on a chromosome than would be expected by chance (Berchowitz and Copenhaver 2010). Finally, CO homeostasis maintains nearly constant CO number per meiosis despite variation in DSB numbers (Martini et al. 2006).
The mechanisms that are responsible of CO suppression in repeat regions can operate at different levels limiting the chromatin accessibility (Ben-Aroya et al. 2004), preventing DSBs, and repairing DSBs through NCO or IS. In budding yeast, it has been shown that open chromatin structure is required for SPO11 to access DNA for generating meiotic DSBs (Berchowitz et al. 2009; Pan et al. 2011). Vice versa, heterochromatic regions that have a closed conformation constrain SPO11 access (Pan et al. 2011). Notwithstanding the suppression, DSBs likely occur in repeats. In case that DSBs are repaired as COs within repeats, the genome integrity is at risk. Indeed, COs between repeats can promote non-allelic homologous recombination (NAHR) resulting in deleterious genomic rearrangements associated with human diseases (Shaw et al. 2002; Sasaki et al. 2010).
The assembly of heterochromatin is driven by the combinatorial action of many factors including histones, their modifications, and DNA methylation. Here, we discuss current knowledge concerning the epigenetic signatures of the major repeat regions where COs are suppressed. Then, we describe mutants for epiregulators of heterochromatin in different organisms to find out how chromatin structure influences the CO rate and distribution.
Where COs are suppressed
Centromeres
Centromeres are the chromosomal parts to which spindle microtubules become attached ensuring proper segregation during mitosis and meiosis. Centromeres are universally marked by the histone H3 variant CENH3, also known as CENP-A in mammals, which replaces canonical histone H3 in centromeric nucleosomes. In most eukaryotes, CENH3-containing chromatin is flanked by pericentromeric heterochromatin (Stimpson and Sullivan 2010). The pericentromeric domains are generally enriched in DNA methylation and histone modifications such as H3K9me2 (Simon et al. 2015). Except for budding yeast, centromeres are not characterized by specific DNA sequences. In budding yeast, the centromeric region is short (≈125 bp) and harbors a single nucleosome (Furuyama and Biggins 2007), whereas in most animals and plants, centromeric regions consist of several megabases (e.g., 1–2.5 Mb in human) (Steiner and Henikoff 2015). In the worm Caenorhabditis elegans, the holocentromere is formed by point centromeres distributed along the length of the chromosomes (Steiner and Henikoff 2014).
Unlike organisms with a localized centromere, during meiosis of C. elegans, the position of the centromere is dictated by the site of the CO, which is normally a single one per homolog pair (Schvarzstein et al. 2010). This exceptional behavior is likely due to the organization of holocentric chromosomes. Overall, the centromeric regions have a low recombination rate at several times below the genome average. In budding yeast, the nucleotide resolution map evidenced that DSBs are suppressed within 5-kb region around centromeres (Pan et al. 2011). Similarly, COs are almost depleted at centromeres in Arabidopsis, maize, and rice (Copenhaver et al. 1999; Drouaud et al. 2006; Shi et al. 2010; Si et al. 2015). However, NCOs were found in maize centromeric regions at a similar rate to the chromosome arms, thereby suggesting that DSBs are not prevented. In human, a drop of recombination rate estimated by CO maps was observed close to the centromeres (Myers et al. 2005; Lu et al. 2012; Hou et al. 2013). The mechanism by which centromeric recombination could interfere with chromosome function is not established. Presumably, COs too near the centromere are a constraint for kinetochore assembly (Ellermeier et al. 2010) with negative effects on chromosome segregation (Talbert and Henikoff 2010). For instance, centromeric COs can result in meiotic missegregation in fission yeast (Hall et al. 2003) and in humans where they can cause severe birth defects, such as trisomy 21 (Lamb et al. 1996; Ottolini et al. 2015).
Telomeres
Telomeres protect the ends of chromosomes. In a wide range of organisms, telomeres consist of short tandem repeats. In budding yeast, telomeres are short (∼300 bp of single repeats) and nucleosome-free while sub-telomeric repetitive elements, called TAS (telomere associated sequences), are likely organized in nucleosomes (Wellinger and Zakian 2012). On the other hand, telomeres of higher eukaryotes have extremely variable length (i.e., ∼2–15 kb of hexameric repeats in human somatic cells) and are organized in tightly packaged nucleosomes (Pisano et al. 2008). A conserved feature of telomeres is their enrichment in heterochromatic marks. For example, the mammalian telomeres are enriched in H3K9me3, H4K20me3, and hypoacetylated H3 and H4 histones. The function and structural integrity of telomeres depends on their repressive chromatin structure (reviewed in Peuscher and Jacobs 2012; Galati et al. 2013). Indeed, a decrease of above-mentioned heterochromatic marks in mice mutants result in aberrantly increased telomere length and chromosomal instability (García-Cao et al. 2004; Gonzalo et al. 2005; Palacios et al. 2010). Telomeres are suppressed for meiotic recombination. In budding yeast, 20-kb region adjacent to telomeres exhibit a significant lower meiotic DSB rate (Pan et al. 2011) consistent with previous observations about the reduction of COs (Barton et al. 2008; Chen et al. 2008). Interestingly, sub-telomeric regions show high levels of COs in the male meiosis of plants and mammals (Saintenac et al. 2009; Giraut et al. 2011; Paigen et al. 2008; de Boer et al. 2015). In the latter, sub-telomeric COs are promoted on both autosomes and sex chromosomes of the males. In particular, COs need to be formed in the sub-telomeric pseudoautosomal region of PAR, that is a very small region of homology between the X and Y chromosomes, to ensure their correct segregation (Kauppi et al. 2011; Hinch et al. 2014). In hermaphroditic plants lacking sex chromosomes, the sub-telomeric localization of recombination events in male meiosis could be associated with a selective advantage of the male gametophytes (Lenormand and Dutheil 2005). Conversely, the regions close to telomeres have a lower recombination in female meiosis than in male (Giraut et al. 2011; Paigen et al. 2008; de Boer et al. 2015). It is a widespread phenomenon termed “heterochiasmy” that is the difference in CO number and position between male and female meiosis (Lenormand 2003). In mice, the sex-specific differences in COs principally reflect differences in the recombination outcome, COs vs NCOs, between males and females (de Boer et al. 2015).
Telomeric COs may be detrimental to chromosome segregation (Ross et al. 1996; Su et al. 2000). In humans, the occurrence of a telomeric CO is associated with non-disjunction of chromosome 21 in female meiosis leading to Down syndrome (Oliver et al. 2012). The current model suggests that COs near telomeres have a negative effect on the cohesion between CO and the chromosome end, thereby threatening the bivalent maintenance in oocytes over the prolonged prophase I.
Ribosomal DNA
Ribosomal DNA (rDNA) has the essential role of encoding most of cellular RNA (Moss et al. 2007). It is composed of tandemly repeated genes and non-coding intergenic spacers (IGS), clustered in regions which are frequently sub-telomeric (McStay and Grummt 2008). In budding yeast, the rDNA cluster is organized into a single array of 100–200 units, whereas rDNA units can be repeated up to thousands of times in other organisms (Eickbush and Eickbush 2007). An rDNA array contains both transcriptionally active and transcriptionally silent repeats. For instance, ∼10 % of rDNA copies are usually active in Arabidopsis (Dvořáčková et al. 2015). The chromatin marks detected in a rDNA region are those typically associated to euchromatin or to heterochromatin depending on the activity of individual units (Bierhoff et al. 2014). In budding yeast, rDNA gene clusters are repressed for meiotic recombination (Gottlieb and Esposito 1989). Consistently, meiotic DSBs appeared almost absent from rDNA (Blitzblau et al. 2007) occurring 75-fold below genome average (Pan et al. 2011). Thus, the suppression of meiotic recombination at rDNA loci can mostly be attributed to the suppression of meiotic DSB formation. Additionally, some findings suggest that when DSBs are formed at rDNA loci they are repaired by IS, which also works to suppress NAHR (Petes 1980; Li et al. 2014).
Chromatin landscape of recombination suppression: histone variants and post-translational modifications
Histones and their modifications have a fundamental role in the assembly of chromatin domains (Kouzarides 2007). In the context of gene-rich regions, the prominent chromatin mark of meiotic recombination initiation is the histone H3 lysine 4 trimethylation (H3K4me3) in yeast (Borde et al. 2009; Sommermeyer et al. 2013; Acquaviva et al. 2013a) and mouse (Buard et al. 2009; Brick et al. 2012). Comprehensive literature reviewed the relationship between histone H3K4me3 and DSB formation (Borde and de Massy 2013; Acquaviva et al. 2013b). In Arabidopsis, in addition to H3K4me3, histone variant H2A.Z is found to be enriched at CO sites. In a mutant, which fails to deposit H2A.Z in the nucleosome, CO frequency is reduced (Choi et al. 2013).
Histone marks of repeat-associated chromatin differ between organisms. However, enrichment of histone H3 lysine 9 methylation (H3K9me) and wide histone de-acetylation are common features of heterochromatin (Table 1) (Nakayama et al. 2001; Suka et al. 2001). In budding yeast, the silent information regulator (Sir) genes are required for assembly of heterochromatin. In particular, Sir2 deacetylates H4K16 near the telomeres and within the rDNA cluster (Robyr et al. 2002). Loss of Sir2 alters the pattern of meiotic recombination (Gottlieb and Esposito 1989; Mieczkowski et al. 2007). Indeed, DSBs increased within 10 kb closest to telomeres and within the rDNA cluster, thereby avoiding the typical suppression. It was suggested that increased levels of acetylation in sir2 mutant open the chromatin structure by allowing recombination machinery easier access to DNA. Consistently, specific hyperacetylation at H4K16 has been demonstrated in vitro to directly influence higher-order chromatin structure by inhibiting the formation of compact 30-nm chromatin fiber (Shogren-Knaak et al. 2006). On the other hand, an increased suppression of DSBs occur in 20–120 kb near-telomeric regions in sir2 possibly determined by compensatory effects associated to interference. Although Sir2 prevents meiotic DSBs within rDNA, it has a DSB promoting effect on the rDNA borders, at the heterochromatin/euchromatin junctions. Since even DSBs adjacent to repetitive DNA can trigger NAHR, a border-specific protection system exists in budding yeast that counteracts DSB activities at rDNA borders. The buffer zones at the edges of the rDNA array are established by two interacting ATPases, Pch2 and Orc1, that prevent rDNA-proximal DSBs (Vader et al. 2011).
Table 1.
Main features associated to the different chromatin domains in higher eukaryotes
| Feature | Euchromatin | Heterochromatin |
|---|---|---|
| Structure | Loosely packed, open, accessible | Densely packed, closed, inaccessible |
| Composition | Mainly genes | Mainly repetitive elements |
| Activity | Expressed, active | Repressed, silent |
| DNA methylation | Hypomethylation | Hypermethylation |
| Histone post-translational modifications | Hyperacetylation of H3 and H4, H3K4me1, H3K4me2, H3K4me3, H3K27me3 | Hypoacetylation of H3 and H4 H3K9me2 |
| Histone variants | H2AX, H2A.Z | H2A.Z, CENH3 (CENP-A) |
H3K9me is enriched in domains of heterochromatin to maintain its silent and compact conformation (Lachner and Jenuwein, 2002; Mozzetta et al. 2015). A substantial amount of evidence suggests that H3K9me2 exerts a repressive role on meiotic recombination. In fission yeast, deletion mutants for components of Clr4–Rik1complex that methylates H3K9 in and near the centromeres had abundant centromeric recombination (Ellermeier et al. 2010). Acquisition of H3K9me2 at a targeted hotspot due to the increase in DNA methylation (see next section) is associated with recombination suppression in Arabidopsis (Yelina et al. 2015). The post-translational modifications of H3 at the same residue, lysine 9, i.e., methylation (H3K9me) or acetylation (H3K9ac), are antagonistic and have opposite effects on meiotic recombination. H3K9ac is found to mark regions that are proficient in meiotic recombination in fission yeast, Arabidopsis, and mice (Yamada et al. 2013; Perrella et al. 2010; Buard et al. 2009). Vice versa, in meiotic cells of mice, H3K9me3 was shown to be enriched in the recombinationally inactive allele relative to the active one at hotspot Psmb9 (Buard et al. 2009). In C. elegans, proper accumulation of H3K9me2 on meiotic prophase chromosomes is driven by HIM-17 which is essential for SPO11-induced DSBs (Reddy and Villeneuve 2004). However, timing of accumulation of cytologically detectable H3K9me2 does not correlate with DSB formation but instead with DSB repair. This evidence is consistent with an experiment in human mitotic cells showing a transient enrichment of H3K9me at DSBs. The authors suggested H3K9me to be critical for remodeling the damaged chromatin to allow efficient DNA repair (Ayrapetov et al. 2014).
Chromatin landscape of recombination suppression: DNA methylation
DNA methylation is a widespread epigenetic mark of repeat sequences associated with heterochromatin in eukaryote genomes of fungi, plants, and animals. DNA methylation is also found within bodies of active genes as a conserved feature between plants and animals (Zemach et al. 2010). Recently, DNA methylation was discovered in model species such as fruit fly (Takayama et al. 2014) and roundworm (Greer et al. 2015).
In different organisms, DNA methylation plays a repressive role on meiotic recombination. In the fungus Ascobolus immersus, the controlled addition of cytosine methylation (5mC) in a recombination interval encompassing a hotspot evidenced that the increase of DNA methylation has an inhibitory effect on COs by over a 100-fold (Maloisel and Rossignol 1998). In Arabidopsis and maize, genome-wide analysis of hotspots show low levels of DNA methylation (Choi et al. 2013; Rodgers-Melnick et al. 2015). In particular, the Arabidopsis region including hotspots 3a and 3b is characterized by a very low value of 5mC in all sequence contexts (CG, CHG, CHH where H = A, T, or C) compared with the genome average. De novo DNA methylation directed to this region by siRNAs causes significant suppression of recombination within the hotspots (Yelina et al. 2015). Mutants of Arabidopsis for METHYLTRANSFERASE1 (MET1), involved in the maintenance of 5mC methylation in CG context, and for DECREASED DNA METHYLATION 1 (DDM1), that encodes a SWI2/SNF2-like chromatin-remodeling protein necessary for DNA methylation, highlight that loss of DNA methylation increases meiotic recombination in the gene-rich chromosome arms (Mirouze et al. 2012; Melamed-Bessudo and Levy 2012). The mutants, however, do not show any increase of COs in heterochromatin, mainly pericentromeric in Arabidopsis (Colomé-Tatché et al. 2012; Mirouze et al. 2012; Melamed-Bessudo and Levy 2012; Yelina et al. 2012). DNA methylation is not, evidently, the only player in the suppression of COs in the plant repeat-containing regions. The occurrence of an epigenetic “double-lock” system was suggested by Melamed-Bessudo and Levy (2012). They hypothesize that, besides methylation-related condensation, recombination is inhibited by an additional factor controlling chromatin structure in heterochromatic regions.
A recent study revealed a histone-guided mechanism for the establishment of DNA methylation (Guo et al. 2015). In ddm1 heterochromatin, DNA methylation loss is connected to the decrease of H3K9me2 (Gendrel et al. 2002). Silenced hot spots, while increasing DNA methylation, gained H3K9me2 and nucleosome occupancy (Yelina et al. 2015). In Arabidopsis, the combination of changes for chromatin modifications including DNA methylation loss remodels the distribution of COs without affecting the total COs since the homeostatic effect is maintained. Particularly in met1, CO remodeling is suggested to be driven by loss of interfering COs from pericentromeric regions and increase in the euchromatic regions (Yelina et al. 2015).
In contrast with above-reported evidence, a positive correlation was found between germ-line DNA methylation and regional recombination rate in the human genome. In particular, DNA methylation increased in regions within recombination hot spots in male germ cells (Sigurdsson et al. 2009). This observation could be caused by the indirect effect of an increase in guanine and cytosine (GC) content at meiotic recombination hot spots due to the mechanism known as GC-biased gene conversion (Duret and Galtier 2009; Arbeithuber et al. 2015). Since CpG islands are the major sites of DNA methylation in mammals, this would result in higher methylation at recombination hot spots.
Conclusions
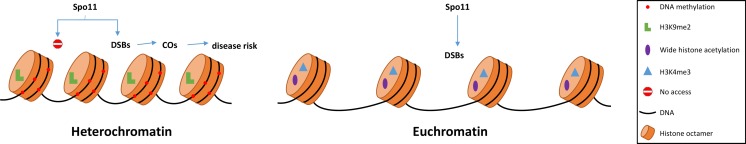
Direct and indirect evidence collected in the different organisms thus far highlight that epigenetic landscape of repeats is related to the suppression of DSBs/COs. Heterochromatin-enriched epigenetic marks likely operate at different levels to avoid meiotic recombination. In detail, they can influence the higher-order heterochromatin structure to make it more close than euchromatin creating a physical barrier to recombination machinery (Fig. 2). Furthermore, epigenetic marks can act as heterochromatin signatures to filter the access to the protein complexes of recombination machinery differentially. Finally, they exclude the epigenetic marks that are enriched in DSB/CO prone regions. The high-resolution mapping of DSB/CO formation in mutants for epiregulators of heterochromatin in various species will clarify the epigenetic framework of the meiotic recombination suppression. However, generation of high-resolution maps of DSBs/COs, their integration with epigenomic maps, and with chromosome conformation in meiotic cells are challenges in multicellular organisms. Maps of meiotic DSBs are not available in most species due to technical constraints. In addition, recombination maps generally do not include repeat-rich regions. To date, maps of DSBs at nucleotide resolution are available in budding and fission yeasts (Pan et al. 2011; Fowler et al. 2014). In mice and human genomes, repeat regions have been excluded in the high-resolution maps of male meiotic DSBs (Brick et al. 2012; Pratto et al. 2014).
Fig. 2.
Epigenetic landscape view of repeat-rich (heterochromatin) and gene-rich regions (euchromatin) associated with meiotic recombination suppression and proficiency, respectively. SPO11, an evolutionarily conserved protein, is the enzyme that promotes DNA DSBs and initiation of meiotic recombination
Acknowledgments
This study is funded by EUFP7/2009–2014 grant KBBE-2009-222883 (MeioSys) and Epigenomics Flagship Project (EPIGEN), MIUR-CNR. The authors thank Dr. Dianne Pickens for editing the manuscript.
Compliance with ethical standards
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
This article is part of a Special Issue on “Recent advances in meiotic chromosome structure, recombination and segregation” edited by Marco Barchi, Paula Cohen, and Scott Keeney.
References
- Acquaviva L, Székvölgyi L, Dichtl B, Dichtl BS, de La Roche Saint André C, Nicolas A, Géli V. The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science. 2013;339:215–218. doi: 10.1126/science.1225739. [DOI] [PubMed] [Google Scholar]
- Acquaviva L, Drogat J, Dehé PM, de La Roche S-AC, Géli V. Spp1 at the crossroads of H3K4me3 regulation and meiotic recombination. Epigenetics. 2013;8:355–360. doi: 10.4161/epi.24295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeithuber B, Betancourt AJ, Ebner T, Tiemann-Boege I. Crossovers are associated with mutation and biased gene conversion at recombination hotspots. Proc Natl Acad Sci U S A. 2015;112:2109–2114. doi: 10.1073/pnas.1416622112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, Xu Y, Price BD. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc Natl Acad Sci U S A. 2014;111:9169–9174. doi: 10.1073/pnas.1403565111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton AB, Pekosz MR, Kurvathi RS, Kaback DB. Meiotic recombination at the ends of chromosomes in Saccharomyces cerevisiae. Genetics. 2008;179:1221–1235. doi: 10.1534/genetics.107.083493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Aroya S, Mieczkowski PA, Petes TD, Kupiec M. The compact chromatin structure of a Ty repeated sequence suppresses recombination hot spot activity in Saccharomyces cerevisiae. Mol Cell. 2004;15:221–231. doi: 10.1016/j.molcel.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Berchowitz LE, Copenhaver GP. Genetic interference: don’t stand so close to me. Curr Genomics. 2010;11:91–102. doi: 10.2174/138920210790886835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz LE, Hanlon SE, Lieb JD, Copenhaver GP. A positive but complex association between meiotic double-strand break hotspots and open chromatin in Saccharomyces cerevisiae. Genome Res. 2009;19:2245–2257. doi: 10.1101/gr.096297.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierhoff H, Postepska-Igielska A, Grummt I. Noisy silence: non-coding RNA and heterochromatin formation at repetitive elements. Epigenetics. 2014;9:53–61. doi: 10.4161/epi.26485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitzblau HG, Bell GW, Rodriguez J, Bell SP, Hochwagen A. Mapping of meiotic single-stranded DNA reveals double-strand-break hotspots near centromeres and telomeres. Curr Biol. 2007;17:2003–2012. doi: 10.1016/j.cub.2007.10.066. [DOI] [PubMed] [Google Scholar]
- Borde V, de Massy B. Programmed induction of DNA double strand breaks during meiosis: setting up communication between DNA and the chromosome structure. Curr Opin Genet Dev. 2013;23:147–155. doi: 10.1016/j.gde.2012.12.002. [DOI] [PubMed] [Google Scholar]
- Borde V, Robine N, Lin W, Bonfils S, Géli V, Nicolas A. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 2009;28:99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642–645. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buard J, Barthès P, Grey C, de Massy B. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J. 2009;28:2616–2624. doi: 10.1038/emboj.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SY, Tsubouchi T, Rockmill B, Sandler JS, Richards DR, Vader G, Hochwagen A, Roeder GS, Fung JC. Global analysis of the meiotic crossover landscape. Dev Cell. 2008;15:401–415. doi: 10.1016/j.devcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Zhao X, Kelly KA, Venn O, Higgins JD, Yelina NE, Hardcastle TJ, Ziolkowski PA, Copenhaver GP, Franklin FC, McVean G, Henderson IR. Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat Genet. 2013;45:1327–1336. doi: 10.1038/ng.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomé-Tatché M, Cortijo S, Wardenaar R, Morgado L, Lahouze B, Sarazin A, Etcheverry M, Martin A, Feng S, Duvernois-Berthet E, Labadie K, Wincker P, Jacobsen SE, Jansen RC, Colot V, Johannes F. Features of the Arabidopsis recombination landscape resulting from the combined loss of sequence variation and DNA methylation. Proc Natl Acad Sci U S A. 2012;109:16240–16245. doi: 10.1073/pnas.1212955109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver GP, Nickel K, Kuromori T, Benito MI, Kaul S, Lin X, Bevan M, Murphy G, Harris B, Parnell LD, McCombie WR, Martienssen RA, Marra M, Preuss D. Genetic definition and sequence analysis of Arabidopsis centromeres. Science. 1999;286:2468–2474. doi: 10.1126/science.286.5449.2468. [DOI] [PubMed] [Google Scholar]
- de Boer E, Jasin M, Keeney S. Local and sex-specific biases in crossover vs. noncrossover outcomes at meiotic recombination hot spots in mice. Genes Dev. 2015;29:1721–1733. doi: 10.1101/gad.265561.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu Rev Genet. 2013;47:563–599. doi: 10.1146/annurev-genet-110711-155423. [DOI] [PubMed] [Google Scholar]
- Drouaud J, Camilleri C, Bourguignon PY, Canaguier A, Bérard A, Vezon D, Giancola S, Brunel D, Colot V, Prum B, Quesneville H, Mézard C. Variation in crossing-over rates across chromosome 4 of Arabidopsis thaliana reveals the presence of meiotic recombination “hot spots”. Genome Res. 2006;16:106–114. doi: 10.1101/gr.4319006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genomics Hum Genet. 2009;10:285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- Dvořáčková M, Fojtová M, Fajkus J. Chromatin dynamics of plant telomeres and ribosomal genes. Plant J. 2015;83:18–37. doi: 10.1111/tpj.12822. [DOI] [PubMed] [Google Scholar]
- Eickbush TH, Eickbush DG. Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics. 2007;175:477–485. doi: 10.1534/genetics.107.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier C, Higuchi EC, Phadnis N, Holm L, Geelhood JL, Thon G, Smith GR. RNAi and heterochromatin repress centromeric meiotic recombination. Proc Natl Acad Sci U S A. 2010;107:8701–8705. doi: 10.1073/pnas.0914160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler KR, Sasaki M, Milman N, Keeney S, Smith GR. Evolutionary diverse determinants of meiotic DNA break and recombination landscape across the genome. Genome Res. 2014;24:1650–1664. doi: 10.1101/gr.172122.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci U S A. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galati A, Micheli E, Cacchione S. Chromatin structure in telomere dynamics. Front Oncol. 2013;3:46. doi: 10.3389/fonc.2013.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Cao M, O’Sullivan R, Peters AH, Jenuwein T, Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39 h1 and Suv39 h2 histone methyltransferases. Nat Genet. 2004;36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science. 2002;297:1871–1873. doi: 10.1126/science.1074950. [DOI] [PubMed] [Google Scholar]
- Giraut L, Falque M, Drouaud J, Pereira L, Martin OC, Mézard C. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 2011;7:e1002354. doi: 10.1371/journal.pgen.1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S, García-Cao M, Fraga MF, Schotta G, Peters AH, Cotter SE, Eguía R, Dean DC, Esteller M, Jenuwein T, Blasco MA. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol. 2005;7:420–428. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- Gottlieb S, Esposito RE. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- Greer EL, Blanco MA, Gu L, Sendinc E, Liu J, Aristizábal-Corrales D, Hsu CH, Aravind L, He C, Shi Y. DNA methylation on N(6)-adenine in C. elegans. Cell. 2015;161:868–878. doi: 10.1016/j.cell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Wang L, Li J, Ding Z, Xiao J, Yin X, He S, Shi P, Dong L, Li G, Tian C, Wang J, Cong Y, Xu Y. Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature. 2015;517:640–644. doi: 10.1038/nature13899. [DOI] [PubMed] [Google Scholar]
- Hall IM, Noma K, Grewal SI. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc Natl Acad Sci U S A. 2003;100:193–198. doi: 10.1073/pnas.232688099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinch AG, Altemose N, Noor N, Donnelly P, Myers SR (2014) Recombination in the human pseudoautosomal region PAR1. PLoS Genet 10:e1004503. doi: 10.1371/journal.pgen.1004503 [DOI] [PMC free article] [PubMed]
- Hou Y, Fan W, Yan L, Li R, Lian Y, Huang J, Li J, Xu L, Tang F, Xie XS, Qiao J. Genome analyses of single human oocytes. Cell. 2013;155:1492–1506. doi: 10.1016/j.cell.2013.11.040. [DOI] [PubMed] [Google Scholar]
- Kauppi L, Barchi M, Baudat F, Romanienko PJ, Keeney S, Jasin M. Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science. 2011;331:916–920. doi: 10.1126/science.1195774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lachner M, Jenuwein T. The many faces of lysine methylation. Curr Opin Cell Biol. 2002;14:286–298. doi: 10.1016/S0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- Lamb NE, Freeman SB, Savage-Austin A, Pettay D, Taft L, Hersey J, Gu Y, Shen J, Saker D, May KM, Avramopoulos D, Petersen MB, Hallberg A, Mikkelsen M, Hassold TJ, Sherman SL. Susceptible chiasmate configurations of chromosome 21 predispose to non-disjunction in both maternal meiosis I and meiosis II. Nat Genet. 1996;14:400–405. doi: 10.1038/ng1296-400. [DOI] [PubMed] [Google Scholar]
- Lenormand T. The evolution of sex dimorphism in recombination. Genetics. 2003;163:811–822. doi: 10.1093/genetics/163.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand T, Dutheil J (2005) Recombination difference between sexes: a role for haploid selection. PLoS Biol 3(3):e63. doi:10.1371/journal.pbio.0030063 [DOI] [PMC free article] [PubMed]
- Li P, Jin H, Yu HG. Condensin suppresses recombination and regulates double-strand break processing at the repetitive ribosomal DNA array to ensure proper chromosome segregation during meiosis in budding yeast. Mol Biol Cell. 2014;25:2934–2947. doi: 10.1091/mbc.E14-05-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Zong C, Fan W, Yang M, Li J, Chapman AR, Zhu P, Hu X, Xu L, Yan L, Bai F, Qiao J, Tang F, Li R, Xie XS. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science. 2012;338:1627–1630. doi: 10.1126/science.1229112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloisel L, Rossignol JL. Suppression of crossing-over by DNA methylation in Ascobolus. Genes Dev. 1998;12:1381–1389. doi: 10.1101/gad.12.9.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini E, Diaz RL, Hunter N, Keeney S. Crossover homeostasis in yeast meiosis. Cell. 2006;126:285–295. doi: 10.1016/j.cell.2006.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- Melamed-Bessudo C, Levy AA. Deficiency in DNA methylation increases meiotic crossover rates in euchromatic but not in heterochromatic regions in Arabidopsis. Proc Natl Acad Sci U S A. 2012;109:E981–E988. doi: 10.1073/pnas.1120742109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Mézard C, Jenczewski E, Macaisne N, Grelon M. The molecular biology of meiosis in plants. Annu Rev Plant Biol. 2015;66:297–327. doi: 10.1146/annurev-arplant-050213-035923. [DOI] [PubMed] [Google Scholar]
- Mieczkowski PA, Dominska M, Buck MJ, Lieb JD, Petes TD. Loss of a histone deacetylase dramatically alters the genomic distribution of Spo11p-catalyzed DNA breaks in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2007;104:3955–3960. doi: 10.1073/pnas.0700412104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouze M, Lieberman-Lazarovich M, Aversano R, Bucher E, Nicolet J, Reinders J, Paszkowski J. Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc Natl Acad Sci U S A. 2012;109:5880–5885. doi: 10.1073/pnas.1120841109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T, Langlois F, Gagnon-Kugler T, Stefanovsky V. A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell Mol Life Sci. 2007;64:29–49. doi: 10.1007/s00018-006-6278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozzetta C, Boyarchuk E, Pontis J, Ait-Si-Ali S. Sound of silence: the properties and functions of repressive Lys methyltransferases. Nat Rev Mol Cell Biol. 2015;16:499–513. doi: 10.1038/nrm4029. [DOI] [PubMed] [Google Scholar]
- Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Oliver TR, Tinker SW, Allen EG, Hollis N, Locke AE, Bean LJ, Chowdhury R, Begum F, Marazita M, Cheung V, Feingold E, Sherman SL. Altered patterns of multiple recombinant events are associated with nondisjunction of chromosome 21. Hum Genet. 2012;131:1039–1046. doi: 10.1007/s00439-011-1121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottolini CS, Newnham LJ, Capalbo A, Natesan SA, Joshi HA, Cimadomo D, Griffin DK, Sage K, Summers MC, Thornhill AR, Housworth E, Herbert AD, Rienzi L, Ubaldi FM, Handyside AH, Hoffmann ER. Genome-wide maps of recombination and chromosome segregation in human oocytes and embryos show selection for maternal recombination rates. Nat Genet. 2015;47:727–735. doi: 10.1038/ng.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K, Szatkiewicz JP, Sawyer K, Leahy N, Parvanov ED, Ng SHS, Graber JH, Broman KW, Petkov PM. The recombinational anatomy of a mouse chromosome. PLoS Genet. 2008;4:e1000119. doi: 10.1371/journal.pgen.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios JA, Herranz D, De Bonis ML, Velasco S, Serrano M, Blasco MA. SIRT1 contributes to telomere maintenance and augments global homologous recombination. J Cell Biol. 2010;191:1299–1313. doi: 10.1083/jcb.201005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG, Tischfield SE, Zhu X, Neale MJ, Jasin M, Socci ND, Hochwagen A, Keeney S. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011;144:719–731. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrella G, Consiglio MF, Aiese-Cigliano R, Cremona G, Sanchez-Moran E, Barra L, Errico A, Bressan RA, Franklin FC, Conicella C. Histone hyperacetylation affects meiotic recombination and chromosome segregation in Arabidopsis. Plant J. 2010;62:796–806. doi: 10.1111/j.1365-313X.2010.04191.x. [DOI] [PubMed] [Google Scholar]
- Petes TD. Unequal meiotic recombination within tandem arrays of yeast ribosomal DNA genes. Cell. 1980;19:765–774. doi: 10.1016/S0092-8674(80)80052-3. [DOI] [PubMed] [Google Scholar]
- Peuscher MH, Jacobs JJ. Posttranslational control of telomere maintenance and the telomere damage response. Cell Cycle. 2012;11:1524–1534. doi: 10.4161/cc.19847. [DOI] [PubMed] [Google Scholar]
- Pisano S, Galati A, Cacchione S. Telomeric nucleosomes: forgotten players at chromosome ends. Cell Mol Life Sci. 2008;65:3553–3563. doi: 10.1007/s00018-008-8307-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratto F, Brick K, Khil P, Smagulova F, Petukhova GV, Camerini-Otero RD. DNA recombination. Recombination initiation maps of individual human genomes. Science. 2014;346:1256442. doi: 10.1126/science.1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KC, Villeneuve AM. C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell. 2004;118:439–452. doi: 10.1016/j.cell.2004.07.026. [DOI] [PubMed] [Google Scholar]
- Robyr D, Suka Y, Xenarios I, Kurdistani SK, Wang A, Suka N, Grunstein M. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell. 2002;109:437–446. doi: 10.1016/S0092-8674(02)00746-8. [DOI] [PubMed] [Google Scholar]
- Rodgers-Melnick E, Bradbury PJ, Elshire RJ, Glaubitz JC, Acharya CB, Mitchell SE, Li C, Li Y, Buckler ES. Recombination in diverse maize is stable, predictable, and associated with genetic load. Proc Natl Acad Sci U S A. 2015;112:3823–3828. doi: 10.1073/pnas.1413864112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross L, Maxfield R, Dawson D. Exchanges are not equally able to enhance meiotic chromosome segregation in yeast. Proc Natl Acad Sci U S A. 1996;93:4979–4983. doi: 10.1073/pnas.93.10.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintenac C, Falque M, Martin OC, Paux E, Feuillet C, Sourdille P. Detailed recombination studies along chromosome 3B provide new insights on crossover distribution in wheat (Triticum aestivum L.) Genetics. 2009;181:393–403. doi: 10.1534/genetics.108.097469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Lange J, Keeney S. Genome destabilization by homologous recombination in the germ line. Nat Rev Mol Cell Biol. 2010;11:182–195. doi: 10.1038/nrn2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvarzstein M, Wignall SM, Villeneuve AM. Coordinating cohesion, co-orientation, and congression during meiosis: lessons from holocentric chromosomes. Genes Dev. 2010;24:219–228. doi: 10.1101/gad.1863610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw CJ, Bi W, Lupski JR. Genetic proof of unequal meiotic crossovers in reciprocal deletion and duplication of 17p11.2. Am J Hum Genet. 2002;71:1072–1081. doi: 10.1086/344346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wolf SE, Burke JM, Presting GG, Ross-Ibarra J, Dawe RK. Widespread gene conversion in centromere cores. PLoS Biol. 2010;8(3):e1000327. doi: 10.1371/journal.pbio.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Si W, Yuan Y, Huang J, Zhang X, Zhang Y, Zhang Y, Tian D, Wang C, Yang Y, Yang S. Widely distributed hot and cold spots in meiotic recombination as shown by the sequencing of rice F2 plants. New Phytol. 2015;206:1491–1502. doi: 10.1111/nph.13319. [DOI] [PubMed] [Google Scholar]
- Sigurdsson MI, Smith AV, Bjornsson HT, Jonsson JJ. HapMap methylation-associated SNPs, markers of germline DNA methylation, positively correlate with regional levels of human meiotic recombination. Genome Res. 2009;19:581–589. doi: 10.1101/gr.086181.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon L, Voisin M, Tatout C, Probst AV. Structure and function of centromeric and pericentromeric heterochromatin in Arabidopsis thaliana. Front Plant Sci. 2015;6:1049. doi: 10.3389/fpls.2015.01049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommermeyer V, Béneut C, Chaplais E, Serrentino ME, Borde V. Spp1, a member of the Set1 complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol Cell. 2013;49:43–54. doi: 10.1016/j.molcel.2012.11.008. [DOI] [PubMed] [Google Scholar]
- Steiner FA, Henikoff S. Holocentromeres are dispersed point centromeres localized at transcription factor hotspots. Elife. 2014;3:e02025. doi: 10.7554/eLife.02025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner FA, Henikoff S. Diversity in the organization of centromeric chromatin. Curr Opin Genet Dev. 2015;31:28–35. doi: 10.1016/j.gde.2015.03.010. [DOI] [PubMed] [Google Scholar]
- Stimpson KM, Sullivan BA. Epigenomics of centromere assembly and function. Curr Opin Cell Biol. 2010;22:772–780. doi: 10.1016/j.ceb.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Su Y, Barton AB, Kaback DB. Decreased meiotic reciprocal recombination in subtelomeric regions in Saccharomyces cerevisiae. Chromosoma. 2000;109:467–475. doi: 10.1007/s004120000098. [DOI] [PubMed] [Google Scholar]
- Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell. 2001;8:473–479. doi: 10.1016/S1097-2765(01)00301-X. [DOI] [PubMed] [Google Scholar]
- Takayama S, Dhahbi J, Roberts A, Mao G, Heo SJ, Pachter L, Martin DI, Boffelli D. Genome methylation in D. melanogaster is found at specific short motifs and is independent of DNMT2 activity. Genome Res. 2014;24:821–830. doi: 10.1101/gr.162412.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. Centromeres convert but don’t cross. PLoS Biol. 2010;8(3):e1000326. doi: 10.1371/journal.pbio.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Tomato Genome Consortium Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader G, Blitzblau HG, Tame MA, Falk JE, Curtin L, Hochwagen A. Protection of repetitive DNA borders from self-induced meiotic instability. Nature. 2011;477:115–119. doi: 10.1038/nature10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger RJ, Zakian VA. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics. 2012;191:1073–1105. doi: 10.1534/genetics.111.137851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Ohta K, Yamada T. Acetylated Histone H3K9 is associated with meiotic recombination hotspots, and plays a role in recombination redundantly with other factors including the H3K4 methylase Set1 in fission yeast. Nucleic Acids Res. 2013;41:3504–3517. doi: 10.1093/nar/gkt049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelina NE, Choi K, Chelysheva L, Macaulay M, de Snoo B, Wijnker E, Miller N, Drouaud J, Grelon M, Copenhaver GP, Mezard C, Kelly KA, Henderson IR. Epigenetic remodeling of meiotic crossover frequency in Arabidopsis thaliana DNA methyltransferase mutants. PLoS Genet. 2012;8(8):e1002844. doi: 10.1371/journal.pgen.1002844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelina NE, Lambing C, Hardcastle TJ, Zhao X, Santos B, Henderson IR. DNA methylation epigenetically silences crossover hot spots and controls chromosomal domains of meiotic recombination in Arabidopsis. Genes Dev. 2015;29:2183–2202. doi: 10.1101/gad.270876.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]