Abstract
Background
Invasive lobular carcinoma (ILC) comprises approximately ~10–20% of breast cancers. In general, multifocal/multicentric (MF/MC) breast cancer has been associated with an increased rate of regional lymph node metastases. Tumor heterogeneity between foci represents a largely unstudied source of genomic variation in those rare patients with MF/MC ILC.
Methods
We characterized gene expression and copy number in 2 or more foci from 11 patients with MF/MC ILC (all ER+, HER2-) and adjacent normal tissue. RNA and DNA were extracted from 3x1.5mm cores from all foci. Gene expression (730 genes) and copy number (80 genes) were measured using Nanostring PanCancer and Cancer CNV panels. Linear mixed models were employed to compare expression in tumor versus normal samples from the same patient, and to assess heterogeneity (variability) in expression among multiple ILC within an individual.
Results
35 and 34 genes were upregulated (FC>2) and down-regulated (FC<0.5) respectively in ILC tumor relative to adjacent normal tissue, q<0.05. 9/34 down-regulated genes (FIGF, RELN, PROM1, SFRP1, MMP7, NTRK2, LAMB3, SPRY2, KIT) had changes larger than CDH1, a hallmark of ILC. Copy number changes in these patients were relatively few but consistent across foci within each patient. Amplification of three genes (CCND1, FADD, ORAOV1) at 11q13.3 was present in 2/11 patients in both foci. We observed significant evidence of within-patient between-foci variability (heterogeneity) in gene expression for 466 genes (p<0.05 with FDR 8%), including CDH1, FIGF, RELN, SFRP1, MMP7, NTRK2, LAMB3, SPRY2 and KIT.
Conclusions
There was substantial variation in gene expression between ILC foci within patients, including known markers of ILC, suggesting an additional level of complexity that should be addressed.
Introduction
Invasive lobular carcinoma (ILC) accounts for approximately 8% to 14% of all breast cancers with a predilection for multifocal (MF) and multicentric (MC) distribution and for bilaterality [1]. The largest study of multiple breast lesions (n = 8935, including lobular and ductal histology), reported the incidence of MF carcinoma, defined as the presence of multiple tumors within the same quadrant of the same breast, at 15.5% [2]. The same study reported the incidence of multicentric (MC) carcinoma, defined as multiple tumors in different quadrants of the same breast as 5.2%. Given the lower frequency of ILC compared to invasive ductal carcinoma (IDC) and the lower frequency of MF/MC compared to unifocal breast cancer, there is a dearth of data on molecular/genomic aspects of MF ILC specifically, with most studies focusing on either MF/MC versus unifocal carcinoma or lobular versus ductal histology. A study of 812 patients with ipsilateral invasive breast cancer reported 7.6% patients with lobular histology, 17.4% MF/MC and 2.1% (17/812) of patients with MF/MC ILC [3]. Additionally, the origin of MF breast cancer is unclear, plausible explanations include intramammary spread from a single primary tumor or alternatively, tumors arising from separate progenitor cells [4].
The characteristics and lack of molecular data in MF and ILC subtypes leads to a conundrum: The vast majority of ILCs are of lower histological grade, express estrogen receptor (ER), lack HER2 overexpression/gene amplification, and fall into the ‘luminal’ molecular subgroup, [1, 5, 6] characteristics associated with higher survival rates and relatively low recurrence rates [7–10]. Conversely, MF breast cancer is associated with an increased risk of regional lymph node metastases, increased risk of local relapse and worse outcome [11–14]. Stratification by intrinsic molecular subtyping in a study of 444 consecutive invasive breast cancer patients, showed that within the luminal A subtype (associated with higher survival rates), multifocal luminal A patients (n = 79) had significantly worse survival than unifocal luminal A (n = 212) patients and multifocal luminal B patients (n = 13) had significantly worse survival than unifocal luminal B patients (n = 29) [15]. The survival analyses were not further stratified for lobular and ductal histology within the luminal subgroups.
In this study we provide a high level molecular characterization of multiple foci from eleven patients with ER-positive, HER2-negative MF/MC ILC, herein referred to as MF ILC. Using a gene expression panel of 730 known cancer genes (606 genes from 13 canonical cancer pathways and 124 cancer associated driver genes), and a gene copy number panel of 80 known cancer genes, we examined the extent to which the genomic architecture varies between multiple foci in patients with ILC and between MF ILC and adjacent normal tissue at the level of individual genes and pathways. Our study design included multiple punches from each focus. This design allowed us to examine gene expression and gene copy number in this rare group of patients at the level of differences between ILC foci and adjacent normal tissue; intra-tumor heterogeneity; and the overall level of tumor-tumor variability (heterogeneity) within a patient relative to variability between tumors from different patients that is not attributable to intra-tumor variability.
Methods
Patient population
Genomic profiling was performed on eleven patients with invasive lobular MF breast cancer. Three x 1.5mm cores were punched from FFPE blocks of each primary focus by an experienced breast pathologist (XG), who selected areas that were >70% tumor cells in order to minimize the potential effect of differences in tumor versus stroma content. For patient 1, cores were punched from two lobular multifocal lesions in the right breast and one lobular unifocal lesion in the left breast; for all other patients the multiple lesions were in the same breast. 1.5mm cores were available from adjacent normal breast tissue in seven of the eleven multifocal patients and normal breast tissue from an additional six patients with other breast cancer subtypes.
All 11 patients and their foci were positive for ER and negative for HER2 by immunohistochemistry (IHC), while three patients were discrepant for PR status (Table 1). All tumors were confirmed by our pathologist (XG) as ILC pathology. All tumors with the exception of one were IHC negative for E-cadherin. One tumor (patient 7, tumor 1 was weakly positive for E-cadherin, detailed in S1 Fig). Patient material and clinical characteristics are summarized in Table 1.
Table 1. Patient material and clinical characteristics.
| Patient | Age/Sex/Race, Primary surgical treatment | Focus size/cm | Distance between foci/cm | ER | PR | HER2 | E-cad | Location | Histology/subtype | TNM stage | Final Stage | Grade | ALND, # pos LN | Genomic data | Adjacent normal available |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 44 / F / White | 4.0 | >4.0 | + | + | - | - | Right UOQ | Lobular/classic | T2 N3a M0 | IIIC | 1 | yes, right, 23/25 | yes | yes |
| R MRM and L total mastectomy | 1.8 | + | - | - | - | Right LOQ | Lobular/classic | T2 N3a M0 | 1 | yes | |||||
| 1.0 | + | + | - | Left UOQ | Lobular/classic | T1b N0 M0 | 1 | yes | |||||||
| 2 | 61 / F / White | 3.5 | 2.7 | + | - | - | - | Right UOQ | Lobular/pleomorphic | T2 N2a M0 | IIIC | 2 | yes, 7/18 | yes | yes |
| R mastectomy with ALND | 1.5 | + | + | - | - | Right UOQ | Lobular/classic | T2 N2a M0 | 1 | yes | |||||
| 1.0 | Right UOQ | Lobular/classic | T2 N2a M0 | 1 | no | ||||||||||
| 0.5 | Right UOQ | Lobular/classic | T2 N2a M0 | 1 | no | ||||||||||
| 3 | 66 / F / White | 5.0 | 1.4 | + | + | - | - | Right UOQ/midline | Lobular/pleomorphic | T2 N1a M0 | IIB | 2 | yes, 1/25 | yes | |
| R mastectomy with ALND | 1.4 | + | + | - | - | Right UIQ | Lobular/classic | T2 N1a M0 | 1 | yes | |||||
| 4 | 87 / F / White | 6.5 | >3.0 | + | + | - | - | Right UOQ | Lobular/alveolar | T3 N1a M0 | IIIA | 2 | yes, 1/23 | yes | yes |
| R mastectomy with ALND | 1.4 | + | + | - | - | Right LOQ | Lobular/classic | T3 N1a M0 | 1 | yes | |||||
| 5 | 66 / F / White | 2.5 | 3.5 | + | + | - | - | Right UOQ | Lobular/pleomorphic | T2 N0 M0 | IIA | 2 | no | yes | |
| R lumpectomy and SLNB | 2.2 | + | + | - | - | Right UOQ | Lobular/pleomorphic | T2 N0 M0 | 2 | yes | |||||
| 6 | 56 / F / White | 2.0 | 3.0 | + | + | - | - | Right UOQ | Lobular/classic | T1c N0 M0 | IA | 1 | no | yes | yes |
| R mastectomy and SLNB | 1.2 | + | + | - | - | Right UOQ | Lobular/classic | T1c N0 M0 | 1 | yes | |||||
| 7 | 65 / F / White | 1.2 | 11.0 | + | + | - | +* | Right UIQ | Lobular/trabecular | T1c N0 M0 | IA | 1 | no | yes | yes |
| bilateral mastectomy and SLNB | 0.8 | + | + | - | - | Right UOQ | Lobular/classic | T1c N0 M0 | 1 | yes | |||||
| 8 | 68 / F / White | 2.0 | 13.0 | + | - | - | - | Left LIQ | Lobular/classic | T1c N1a M0 | IIA | 1 | yes, 2/10 | yes | |
| L mastectomy with ALND | 1.5 | + | + | - | - | Left UOQ | Lobular/classic | T1c N1a M0 | 1 | yes | |||||
| 9 | 67 / F / White | 3.2 | 2.5 | + | + | - | - | Left UIQ | Lobular/pleomorphic | T2 N3a M0 | IIIC | 2 | yes, 11/33 | yes | |
| L mastectomy with ALND | 1.0 | + | + | - | - | Left UIQ | Lobular/classic | T2 N0 M0 | 1 | yes | |||||
| 10 | 60 / F / White | 6.0 | 3.0 | + | + | - | - | Right LIQ | Lobular/pleomorphic | T3 N3a M0 | IIIC | 2 | yes, 14/24 | yes | yes |
| R mastectomy with ALND | 0.7 | + | + | - | - | Right LOQ | Lobular/pleomorphic | T3 N3a M0 | 2 | yes | |||||
| 0.3 | Right UOQ | Lobular/pleomorphic | T3 N3a M0 | 2 | no | ||||||||||
| 11 | 73 / F / White | 4.5 | 1.5 | + | + | - | - | UI and UOQ | Lobular/pleomorphic | T2 N1a M0 | IIB | 2 | yes, 2/24 | yes | yes |
| R mastectomy with ALND | 0.6 | + | + | - | - | Right central | Lobular/pleomorphic | T2 N1a M0 | 2 | yes |
Abbreviations: MRM, modified radical mastectomy; UOQ, Upper outer quadrant; UIQ, upper inner quadrant; LOQ, left outer quadrant; ALND, Axillary lymph node dissection; SLNB, Sentinel lymph node biopsy.
* Weakly positive. This tumor was dominant classic lobular, with some mixed trabecular pattern.
Ethics Statement
All breast tumor samples were collected between 2012 and 2014 according to a protocol that was reviewed and approved by the Mayo Clinic Institutional Review Board under protocol 13–009696. The review board approved waiver of the requirement to obtain informed consent in accordance with 45 CFR 46.116. Patient records/information were anonymized and de-identified prior to analysis.
RNA and DNA extraction
RNA and DNA were extracted from each 1.5mm punch with the Qiagen AllPrep FFPE kit as per the manufacturer’s instructions, with the exception that proteinase K digestion times at 65°C were extended to overnight, as previously described [16].
Gene expression data
Gene expression on the NanoString® platform was assessed with the NanoString PanCancer Pathways panel of 730 genes designed to capture the activity of thirteen canonical hallmarks of cancer pathways (606 pathway genes: Notch, Wnt, Hedgehog, TGFB, MAPK, STAT, PI3K, RAS, Chromatin modification, transcriptional regulation, DNA damage control, cell cycle and apoptosis), 124 cancer associated driver genes and 40 reference genes. 100ng of each total RNA sample was prepared as per the manufacturer’s instructions under the high sensitivity protocol. Gene expression was quantified on the NanoString nCounterTM and raw counts were generated with nSolverTM. Raw counts were normalized against 36/40 reference genes, selected to have the least variance with the geNorm algorithm [17].
Normalized gene counts and raw data (RCC) files are available at GEO (accession number GSE79058).
Gene copy number data
Gene copy number on the NanoString platform was assessed with the NanoString Cancer CNV panel (version 1, average three probes/gene). 500ng of DNA were prepared as per the manufacturer’s instructions (Alu digest and high sensitivity protocols). Copy number was calculated within the nSolver software (NanoString). Briefly, the NanoString Cancer CNV panel contains 2–3 probes in each of 80 known cancer genes and 54 invariant control probes used for normalization. Copy number was estimated as twice the ratio of the average probe count per gene in each patient to the average probe count per gene in a set of 16 adjacent normal punches extracted with the same protocol as the multifocal tumors (seven of which were from the multifocal ILC patients marked in Table 1 and nine punches from adjacent normal waste tissue of patients with other types of breast cancer).
CCND1 amplification identified on the NanoString platform was validated with quantitative PCR (qPCR). Reactions were performed in duplicate with 20ng gDNA, TaqMan Universal PCR master mix, RNase P primer/ probe (4403328), and the CCND1 primer/probe set (Life Technologies). Amplification data were collected with an Applied Biosystems Viia7 sequence detector and analyzed with ViiA 7 RUO software. CT values were normalized to control RNase P, and abundance was calculated using the ΔΔCT method [18]. Copy number gains in individual tumor samples were calculated relative to copy number in adjacent normal tissue from the same patient.
Statistical analyses
Differential gene expression and heterogeneity analysis
Linear mixed models were used to assess between-tumor heterogeneity and difference in expression between foci and adjacent normal tissue. Normalized gene expression on the log (base 2) scale was the response variable. All models included both patient and tumor random effects. Tumor heterogeneity analyses included tumor grade (which correlated with lobular subtype: classic, pleomorphic, trabecular or alveolar) as an additional fixed effect. Fold change was estimated for the comparison of tumor versus adjacent normal analyses.
When conducting the heterogeneity analyses, the restricted maximum likelihood approach was used for model fitting to allow estimation of three standard deviation parameters for the three sources of variability: patient (σP), foci (σF), and punch (σIT). Using data from three punches for each tumor, we define the intra-tumor variability, as HET.IT = σIT2/ (σIT2+ σF2+ σP2) × 100%. We define the within-patient between-foci heterogeneity, as HET.F = σt2/ (σIT2+ σF2+ σP2) × 100%. This is the variation among foci from the same patient (σF2) expressed as a percentage of the total variation: the sum of the intra-tumor variability (σIT), the within-patient variability (σF2) and the patient to patient variability (σP2). Similarly, we define the between patient heterogeneity, as HET.P = σP2/ (σIT2+ σF2+ σP2) × 100%.
Likelihood ratio tests were used to test the null hypothesis, H0: HET.F = 0, or equivalently H0: σF = 0. Two approaches were used to account for multiple testing for all of these analyses: Holm adjusted p-values,[19] and false discovery rate (FDR) estimates, q-values, obtained via the Benjamini-Hochberg approach.[20] Statistical analyses were conducted with R version 3.0.2.
Differential gene pathway analysis
Pathway dysregulation was scored for each focus in thirteen canonical cancer pathways[21] within the NSolver software (NanoString) using Principal Component (PC) analysis with adjacent normal tissue as the baseline reference. Data for each pathway were scaled before taking the first PC by dividing each gene’s log2 expression values by the greater of either their standard deviation or 0.05. Using the pathway scores calculated in nSolver, we performed differential expression analysis using the same regression model as in the gene-level differential expression analysis. These regressions were used to calculate a p-value for the association of each pathway of tumor versus adjacent normal tissue. Global significance statistics were also calculated for each pathway by measuring the cumulative evidence for the differential expression of genes in a pathway. For MF ILC tumors, global significance of each pathway was calculated as the square root of the pathway’s average squared t-statistic. Global significance for each pathway was then plotted against linear association pathway scores.
Results
Differential Gene Expression between ILC tumors and adjacent normal tissue
Several studies report worse outcome of multifocal relative to unifocal breast cancer, [11–15] and some evidence (although limited) suggests that multifocal breast cancer results from intramammary spread from a single primary tumor [22–25]. Under this scenario, we hypothesized that potential prognostic markers of MF ILC would be common to both foci, in which case they could be identified by differential expression analyses between all tumors within our MF ILC sample set and adjacent normal tissue. Hence, using the Nanostring PanCancer pathways panel, we first compared gene expression of 730 known cancer genes in multiple ILC foci of 11 patients to adjacent normal tissue from 7 of those patients.
Amongst the 730 genes tested, (S1 Table) there was evidence of differential expression (unadjusted p<0.05) in tumor relative to adjacent normal tissue for 253 genes with an estimated false discovery rate (FDR) of 14%. The expression in ILC tissue was estimated to be two or more fold that of adjacent normal tissue for 73 of the 253 genes, and to be less than half (FC<0.5) that of expression in normal tissue for 60 of these genes. 34 genes were significantly down-regulated (FC<0.5) with q<0.05, nine of which (FIGF, RELN, PROM1, SFRP1, MMP7, NTRK2, LAMB3, SPRY2, KIT) showed larger fold changes (0.06 to 0.26) than CDH1 (0.28), which encodes for E-cadherin, the loss of which is a hallmark of ILC. COL11A1 and PKMYT1 showed the highest fold change of genes that were significantly up-regulated (with q<0.05) with FC of 25.0 and 11.7 respectively. Differentially expressed genes with FC>2.0 or 0.5 and q<0.05 are shown in Table 2. Results for all 730 genes are shown in S1 Table.
Table 2. Differential gene expression between MF ILC and adjacent normal tissue.
| Gene | FC | Unadjusted P-value | Adjusted P-value | qval (FDR) | Tumor expression |
|---|---|---|---|---|---|
| COL11A1 | 24.99 | 1.33E-03 | 8.97E-01 | 1.71E-02 | ↑ |
| PKMYT1 | 11.66 | 2.30E-05 | 1.67E-02 | 2.79E-03 | ↑ |
| COMP | 9.23 | 2.35E-03 | 1.00E+00 | 2.29E-02 | ↑ |
| COL1A1 | 8.40 | 2.96E-05 | 2.14E-02 | 3.08E-03 | ↑ |
| SIX1 | 7.93 | 2.15E-03 | 1.00E+00 | 2.20E-02 | ↑ |
| BMP8A | 6.94 | 2.01E-03 | 1.00E+00 | 2.16E-02 | ↑ |
| ZIC2 | 5.47 | 4.55E-03 | 1.00E+00 | 3.46E-02 | ↑ |
| CCNE2 | 5.05 | 6.68E-03 | 1.00E+00 | 4.47E-02 | ↑ |
| UBE2T | 5.02 | 2.04E-05 | 1.48E-02 | 2.79E-03 | ↑ |
| MCM2 | 4.56 | 2.74E-04 | 1.93E-01 | 7.40E-03 | ↑ |
| COL3A1 | 4.50 | 5.76E-05 | 4.16E-02 | 4.67E-03 | ↑ |
| FN1 | 4.36 | 2.36E-04 | 1.67E-01 | 7.11E-03 | ↑ |
| CREB3L1 | 4.30 | 2.43E-06 | 1.77E-03 | 8.87E-04 | ↑ |
| LEF1 | 4.19 | 9.45E-04 | 6.46E-01 | 1.47E-02 | ↑ |
| INHBA | 4.14 | 2.07E-03 | 1.00E+00 | 2.19E-02 | ↑ |
| CDKN2A | 4.08 | 4.68E-03 | 1.00E+00 | 3.52E-02 | ↑ |
| COL5A2 | 3.92 | 1.50E-05 | 1.09E-02 | 2.73E-03 | ↑ |
| E2F1 | 3.68 | 2.19E-04 | 1.56E-01 | 7.11E-03 | ↑ |
| COL1A2 | 3.58 | 4.26E-04 | 2.97E-01 | 9.26E-03 | ↑ |
| GATA3 | 3.50 | 2.25E-04 | 1.59E-01 | 7.11E-03 | ↑ |
| COL5A1 | 3.37 | 1.58E-04 | 1.13E-01 | 6.97E-03 | ↑ |
| CACNA1D | 3.05 | 2.58E-04 | 1.82E-01 | 7.23E-03 | ↑ |
| IL20RB | 3.00 | 6.78E-04 | 4.66E-01 | 1.15E-02 | ↑ |
| CDKN2B | 2.92 | 6.62E-05 | 4.78E-02 | 4.84E-03 | ↑ |
| CBLC | 2.91 | 3.58E-03 | 1.00E+00 | 2.90E-02 | ↑ |
| HIST1H3H | 2.76 | 1.62E-04 | 1.16E-01 | 6.97E-03 | ↑ |
| BRIP1 | 2.49 | 3.04E-03 | 1.00E+00 | 2.64E-02 | ↑ |
| PAX8 | 2.48 | 6.26E-03 | 1.00E+00 | 4.27E-02 | ↑ |
| POLE2 | 2.43 | 1.08E-05 | 7.90E-03 | 2.64E-03 | ↑ |
| TGFB3 | 2.41 | 1.01E-04 | 7.22E-02 | 5.00E-03 | ↑ |
| FEN1 | 2.38 | 1.33E-03 | 8.98E-01 | 1.71E-02 | ↑ |
| MAPT | 2.32 | 1.32E-03 | 8.90E-01 | 1.71E-02 | ↑ |
| CCND1 | 2.28 | 3.43E-04 | 2.40E-01 | 8.35E-03 | ↑ |
| EZH2 | 2.18 | 7.25E-04 | 4.98E-01 | 1.19E-02 | ↑ |
| CREB3L4 | 2.07 | 3.17E-03 | 1.00E+00 | 2.68E-02 | ↑ |
| FIGF | 0.06 | 1.50E-03 | 1.00E+00 | 1.80E-02 | ↓ |
| RELN | 0.12 | 4.05E-03 | 1.00E+00 | 3.11E-02 | ↓ |
| PROM1 | 0.13 | 1.40E-03 | 9.44E-01 | 1.74E-02 | ↓ |
| SFRP1 | 0.15 | 5.02E-03 | 1.00E+00 | 3.66E-02 | ↓ |
| MMP7 | 0.17 | 1.52E-03 | 1.00E+00 | 1.80E-02 | ↓ |
| NTRK2 | 0.22 | 3.62E-04 | 2.53E-01 | 8.52E-03 | ↓ |
| LAMB3 | 0.23 | 2.24E-03 | 1.00E+00 | 2.22E-02 | ↓ |
| SPRY2 | 0.24 | 2.13E-04 | 1.51E-01 | 7.11E-03 | ↓ |
| KIT | 0.26 | 2.17E-04 | 1.54E-01 | 7.11E-03 | ↓ |
| CDH1 | 0.28 | 4.57E-04 | 3.17E-01 | 9.26E-03 | ↓ |
| IL22RA1 | 0.29 | 4.86E-03 | 1.00E+00 | 3.59E-02 | ↓ |
| LIFR | 0.29 | 4.47E-04 | 3.11E-01 | 9.26E-03 | ↓ |
| MET | 0.30 | 5.16E-04 | 3.57E-01 | 9.90E-03 | ↓ |
| EGFR | 0.30 | 2.93E-04 | 2.06E-01 | 7.63E-03 | ↓ |
| FGF10 | 0.31 | 1.07E-03 | 7.31E-01 | 1.58E-02 | ↓ |
| ITGB4 | 0.31 | 8.07E-05 | 5.81E-02 | 5.00E-03 | ↓ |
| FLNC | 0.31 | 5.65E-03 | 1.00E+00 | 3.97E-02 | ↓ |
| PAK3 | 0.32 | 7.08E-03 | 1.00E+00 | 4.66E-02 | ↓ |
| ITGB3 | 0.35 | 1.66E-03 | 1.00E+00 | 1.90E-02 | ↓ |
| PDGFRA | 0.36 | 5.60E-04 | 3.87E-01 | 1.01E-02 | ↓ |
| CACNB2 | 0.37 | 1.03E-04 | 7.36E-02 | 5.00E-03 | ↓ |
| ITGB8 | 0.38 | 1.19E-03 | 8.05E-01 | 1.66E-02 | ↓ |
| ZBTB16 | 0.38 | 1.78E-03 | 1.00E+00 | 2.00E-02 | ↓ |
| FZD7 | 0.38 | 1.72E-04 | 1.23E-01 | 6.99E-03 | ↓ |
| CREB5 | 0.41 | 1.22E-03 | 8.27E-01 | 1.68E-02 | ↓ |
| KLF4 | 0.41 | 2.17E-03 | 1.00E+00 | 2.20E-02 | ↓ |
| TCF7L1 | 0.43 | 5.59E-04 | 3.87E-01 | 1.01E-02 | ↓ |
| MAML2 | 0.46 | 2.43E-04 | 1.72E-01 | 7.11E-03 | ↓ |
| MYC | 0.46 | 5.21E-03 | 1.00E+00 | 3.71E-02 | ↓ |
| PLD1 | 0.47 | 4.84E-04 | 3.36E-01 | 9.55E-03 | ↓ |
| GAS1 | 0.47 | 6.08E-03 | 1.00E+00 | 4.23E-02 | ↓ |
| ITGB6 | 0.48 | 3.36E-04 | 2.36E-01 | 8.35E-03 | ↓ |
| TGFBR2 | 0.48 | 6.75E-04 | 4.65E-01 | 1.15E-02 | ↓ |
| CDC14A | 0.50 | 3.32E-03 | 1.00E+00 | 2.75E-02 | ↓ |
Differential gene expression: ILC tumors versus adjacent normal tissue, FDR<0.05 and FC>2 or <0.5, ordered by direction and fold change. FC = fold change. FDR = false discovery rate.
Differential Pathway Expression between ILC and adjacent normal tissue
We used two different statistical approaches to assess significance of 13 canonical cancer pathways in ILC tumors relative to normal adjacent tissue (Fig 1A). Firstly, we scored each sample for pathway dysregulation and performed differential expression analysis of these scores to measure association of each pathway in ILC tumor (plotted as–log10 p-value. Secondly, we used global significance statistics as a measure of the cumulative evidence for differential expression of genes in each pathway.
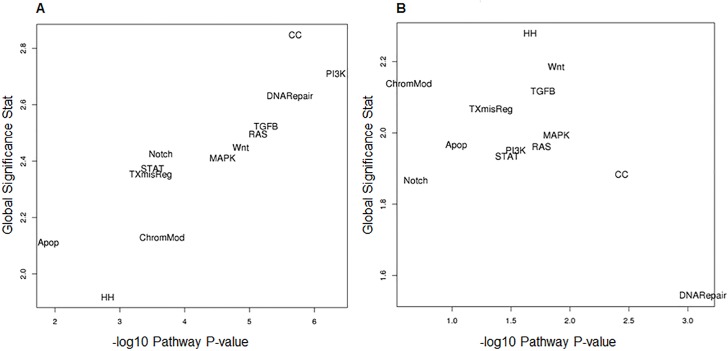
Fig 1. MF ILC pathway significance plots.
A. Differential pathway expression in ILC tumor relative to adjacent normal tissue and B. Differential pathway expression between patient foci. Using the pathway scores, we performed differential expression analysis using the same regression model as in the gene-level differential expression analysis. These regressions were used to calculate a p-value for the association of each pathway of focus versus adjacent normal tissue (A) and between patient foci (B). Global significance statistics were calculated for each pathway by measuring the cumulative evidence for the differential expression of genes in a pathway relative to adjacent normal tissue (A) and between patient foci (B). Global significance for each pathway was then plotted against linear association pathway scores. There is agreement on both scales with the greatest difference in the PI3K and cell cycle pathways in ILC foci relative to adjacent normal tissue (A) and little difference in any pathway as measured between foci within patients (B).
For the ILC tumors, the global significance score and pathway score p-value were in good agreement and all pathways were significantly different between ILC MF and adjacent normal tissue at p<0.05. The strongest associated pathways on both measures of association were PI3K and cell cycle, closely followed by DNA repair, TGFβ, RAS, Wnt and MAPK. Apoptosis and Hedgehog pathways were the least significant by both measures. The largest differences in absolute fold-change were PI3K (FC = 1098) and RAS (FC = 291) pathways (S2 Table).
Differential genomic architecture of multiple foci within ILC patients
Gene copy number in multiple foci in ILC patients
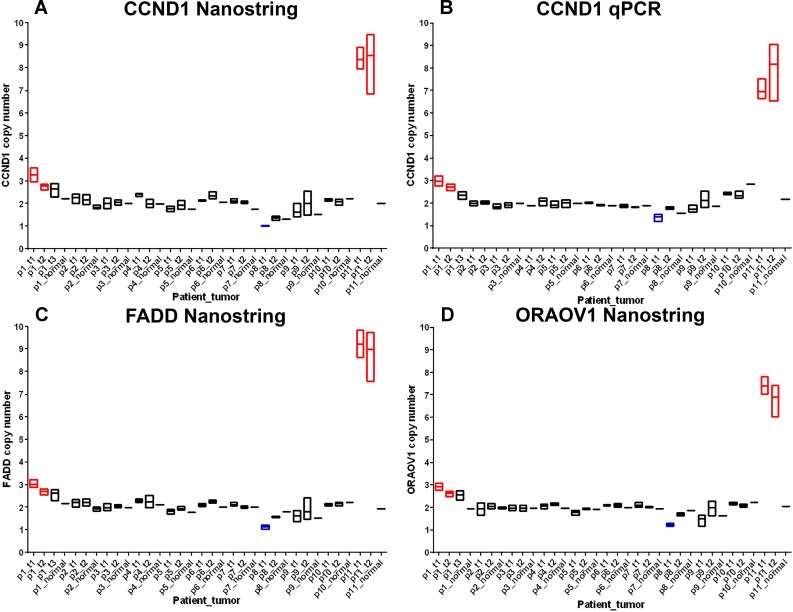
We assessed copy number in all foci in 80 known cancer genes. These data (with the limitation of 80 genes) were suggestive that multiple foci in the same patient are genetically homogeneous. Consistent copy number was observed across all three punches in each primary focus and between primary foci in each patient. We observed relatively few changes in gene copy number across MF ILC, the largest change being amplification of three genes (CCND1, FADD and ORAOV1) mapping to 11q13.3 in two of eleven patients (patients 1 and 11). Copy number data for this region are shown in Fig 2. 11q13.3 amplification has been previously described in breast cancer,[26, 27] specifically ILC, [28–30] and in oral squamous carcinoma where it was reported as prognostic of metastasis [31]. Within our dataset, amplifications were identified in CCND1, FADD and ORAOV1 in all three punches from both foci. Patient 11 showed higher copy number, ranging from 6.01 to 9.40 on the Nanostring platform. qPCR at the CCND1 locus confirmed the amplification in all three punches in both foci, copy number ranging 6.53–9.06. Amplification in patient 1 was more subtle, ranging 2.30–3.60 on the Nanostring platform and 2.17–3.2 with qPCR.
Fig 2. 11q13.3 gene copy number in 11 MF ILC patients.
Gene copy number was measured in three punches from each focus and a single punch from matched adjacent normal tissue where available. Ends of each box are minimum and maximum copy number and floating bar shows mean copy number. Copy number was measured by Nanostring and qPCR platforms. Patients are labelled p1-11 and foci are labelled t1, t2 and t3 in order of size, hence p1_t1 = patient 1, focus 1. Amplifications are highlighted in red and deletions in blue. A. CCND1 copy number by Nanostring; B. CCND1 copy number by qPCR; C. FADD copy number by Nanostring; D. ORAOV1 copy number by Nanostring.
A third patient (patient 8) showed some evidence of deletion at the CCND1, FADD and ORAOV1 genes in either one or both foci and consistent low level amplification across all three punches in each focus for AKT3 mapping to 1q43, copy number ranging 2.68–3.16 (mean 2.88, SD = 0.19) and MET mapping to 7q31.2, copy number ranging 2.52–3.93 (mean 3.24, SD = 0.53). In addition, patient 11 showed consistent low level amplification of ITGB4 at 17q25.1, copy number ranging 3.17–3.66 (mean 3.68, SD = 0.22) and MYC at 8q24.21, copy number ranging 2.96–3.51 (mean 3.32, SD = 0.22). All copy number changes are shown graphically across all punches for all 11 patients in S2 Fig.
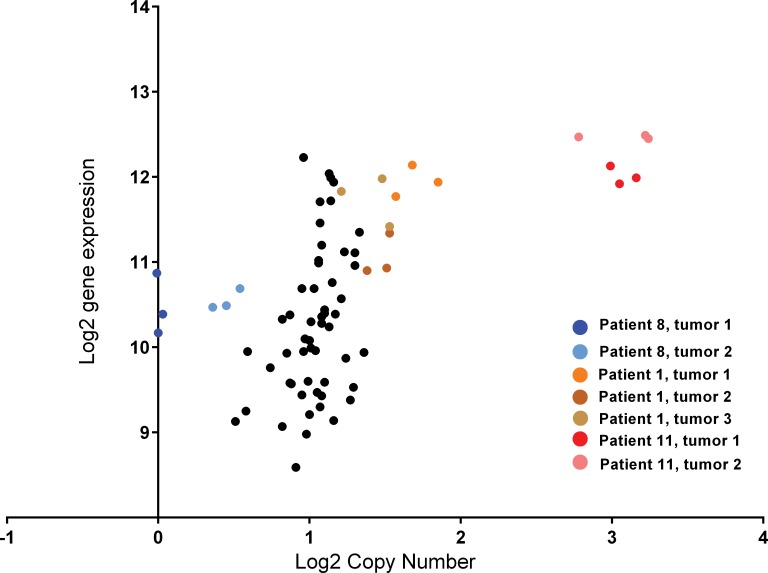
We then proceeded to correlate gene copy number with gene expression at 11q13.3 using CCND1 which is common to both NanoString PanCancer expression and cancer copy number panels (Fig 3). CCND1 gene expression was significantly correlated with copy number, Spearman r = 0.57, p<0.0001 when measured across the full sample of eleven patients. When considering just those patients with abnormal copy number: patient 8 (deletion) patient 1 (low level amplification) and patient 11 (high level amplification), correlation improves, Spearman r = 0.88.
Fig 3. CCND1 gene expression and gene copy number in 11 MF ILC patients.
Patient 8 (blue) deletion; Patient 1 (orange/brown) moderate copy number gain; Patient 11 (red/pink) high copy number gain.
Differential Pathway Expression in multiple foci within ILC patients
Using two independent methods to measure pathway significance, we did not observe any convincing evidence of differential pathway expression between patient foci across this patient group as a whole, (Fig 1B, S3 Table). However, visual inspection did suggest that the foci in at least 2/11 ILC patients (patients 4 and 7) were heterogeneous at the pathway level for Wnt, PI3K, RAS, Hedgehog, Transcription Misregulation, TGF-Beta, MAPK, STAT and Apoptosis pathways, S3 Fig. This may reflect a biological difference in that the foci in these patients are of different lobular subtypes. In patient 4, both foci are E-cadherin negative, but the larger focus is of the alveolar subtype and the smaller focus is classic lobular subtype. In patient 4, the larger focus is very weakly positive for E-cadherin and of the trabecular subtype and the smaller focus is E-cadherin negative and predominantly of the classic lobular subtype with some mixed trabecular subtype.
Alternatively, for all thirteen pathways in both patient 4 and patient 7, the pathway score of the smaller focus was always more similar to adjacent normal tissue than the larger focus, S4 Fig. These differences could also arise from cellularity in that the smaller focus was of higher normal tissue content (immune, stromal or epithelial). To examine cellularity we plotted copy number for each focus across 80 genes in the Nanostring Cancer CNV panel, S2 Fig. Very few copy number changes were observed in these patients, but where copy number did deviate slightly, the second focus (in both cases the classic lobular subtype focus) was closer to adjacent normal tissue.
Tumor heterogeneity
Our study design of three punches from every tumor allowed us to examine focal heterogeneity within MF ILC patients and tumor heterogeneity between MF ILC patients, for each gene, with tumor grade (lobular subtype) as a fixed effect. Variance, percentage heterogeneity of all three types (patient, foci, intra-tumor) and p-values for all 730 genes are shown in S4 Table.
There was strong evidence of heterogeneity between-foci within-patients. Of the 730 genes included in the analysis, 466 (64%) had unadjusted heterogeneity (Table 3, pval.F) p-values of <0.05 (with q-values<0.08), and 432 (59%) had a multiple-testing adjusted p-value of <0.05,suggesting that there is within-patient focus-to-focus heterogeneity for over half of the studied genes in addition to any patient-to-patient variability and variability in measures of gene expression among replicate punch samples from the same foci. There was also evidence of heterogeneity from patient to patient in addition to any variability between punches from the same foci and between foci within patients, although to a lesser extent than we observed for multiple foci within patients. Of the 730 genes included in this analysis, 292 (40%) had unadjusted heterogeneity (Table 3, pval.P) p-values of <0.05 (with q-values<0.12). These observations are very relevant when searching for markers of ILC relative to adjacent normal tissue or potential prognostic markers based on gene expression from a single specimen, and may be especially relevant to multifocal disease. This is illustrated in Table 3, where we show a breakdown of heterogeneity measures for CDH1, and the other nine genes identified with greater fold changes than CDH1 in our tumor versus adjacent normal analysis.
Table 3. Sources of heterogeneity in MF ILC differentially expressed genes.
| Gene | HET.P | HET.F | HET.IT | pval.P | pval.F |
|---|---|---|---|---|---|
| LAMB3 | 46.2 | 41.7 | 12.1 | 0.11 | 1.05E-09 |
| CDH1 | 0.0 | 73.8 | 26.2 | 1.00 | 3.16E-09 |
| SPRY2 | 0.0 | 75.1 | 24.9 | 1.00 | 3.42E-09 |
| NTRK2 | 0.0 | 65.5 | 34.5 | 1.00 | 1.19E-06 |
| FIGF | 1.5 | 62.0 | 36.5 | 1.00 | 4.56E-06 |
| SFRP1 | 5.5 | 54.9 | 39.6 | 1.00 | 4.82E-05 |
| KIT | 8.6 | 47.1 | 44.3 | 0.86 | 3.82E-04 |
| RELN | 15.1 | 42.4 | 42.6 | 0.69 | 7.53E-04 |
| MMP7 | 19.1 | 26.1 | 54.8 | 0.39 | 0.04 |
| PROM1 | 54.3 | 5.5 | 40.2 | 0.01 | 0.44 |
HET.P, variability between tumors in different patients, expressed as a percentage of total variability from: multiple punches in each tumor, variability between foci within patients and variability between tumors in different patients. pval.P, corresponding p-value for heterogeneity between tumors in different patients. HET.F, variability between foci within patients, expressed as a percentage of total variability. pval.F, corresponding p-value for heterogeneity between foci within patients. HET.IT, variability within tumors (intra-tumor heterogeneity) expressed as a percentage of total variability.
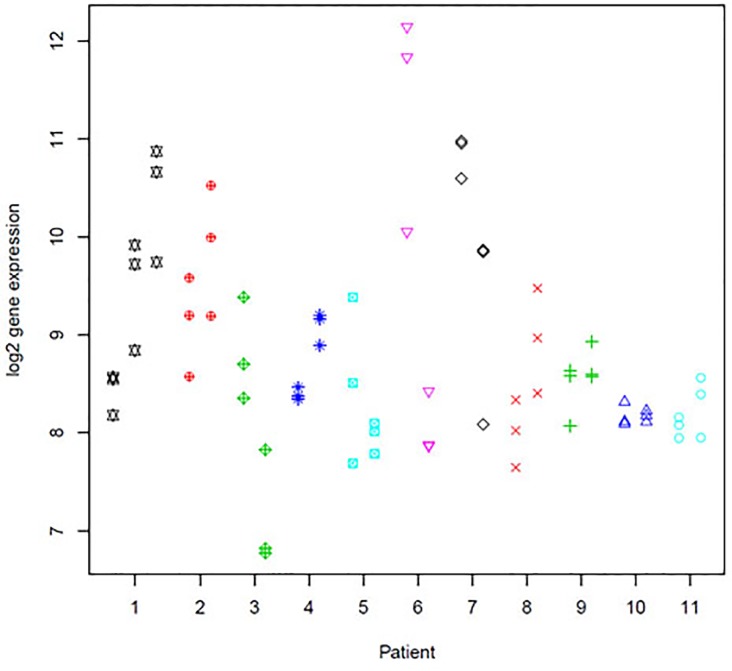
Gene expression measures for CDH1 shows very high evidence of within-patient between-tumor heterogeneity (73.8% heterogeneity, p = 3.16x10-9). Loss of CDH1 (E-cadherin) is an important diagnostic feature of ILC. Differential expression analyses of tumor versus adjacent normal tissue confirmed this finding with an absolute fold change of 0.28 in MF foci versus adjacent normal tissue (Table 2). In our heterogeneity analysis of CDH1 across multiple foci from patients (Fig 4), the median log2 gene expression is 8.56 (compared to 10.65 in adjacent normal tissue), but we see that in 5 of the 11 patients there is no overlap of CDH1 expression levels between foci, and in patient 6 there are ~3 orders of magnitude difference between foci. This observation is further illustrated in our breakdown of heterogeneity in Table 3 showing the greatest source of heterogeneity for this gene is between foci within patients (73.8%). This trend is also reflected for the nine other genes that were significantly down-regulated relative to adjacent normal tissue, with larger fold change than observed for CDH1.
Fig 4. CDH1 gene expression in 11 ILC patients with multiple foci.
Log2 gene expression is plotted for three punches from each foci in each patient. For each patient, foci are shown in order of size with punches from the largest focus always displayed to the left and punches from the smallest focus (for which tissue is available) to the right, in the same order as shown in Table 1.
Discussion
The molecular/genomic aspects of multifocal (MF) invasive lobular carcinoma (ILC) are poorly understood, despite the fact that these patients have significantly worse outcome than ILC patients with unifocal disease [11–14]. In this study we used detailed molecular characterization of multiple foci in a set of 11 MF ILC patients, all of whom were ER+ and HER2- in both foci.
Our first analyses of differential gene expression of tumor versus adjacent normal tissue were generally consistent with published studies of ILC [32–35]. We identified 253/730 genes from canonical cancer pathways that were significantly different, with estimated FDR = 14%. Loss of E-Cadherin (CDH1) is associated specifically with ILC and is an important diagnostic feature [32]. In this dataset of 11 MF ILC patients, CDH1 was significantly down-regulated, p = 4.6x10-4, q = 0.0093, absolute fold change 0.28.
Due to small sample size and different platforms used, the overlap of biomarkers (other than CDH1) between published studies of ILC genes is small. A meta-analysis of five gene expression studies identified THBS4 as a potential ILC biomarker.[34] THBS4 gene expression was also up-regulated in our ILC patients relative to adjacent normal tissue (absolute fold change 2.86, p = 0.027, q = 0.10). Korkola et al [33] defined 11 genes as capable of differentiating ILCs from ductal carcinoma, of which three (CDH1, SPRY1, THBS4) are present in the PanCancer pathways panel. All three genes were significantly differentially expressed relative to adjacent normal tissue in our sample set, p-values 4.6x10-4, 0.018 and 0.027 respectively, with estimated FDR associated with p<0.027 of 10%, (although we note that SPRY1 expression was lower in ILC relative to adjacent normal in our study and higher in ILC relative to IDC in the Korkola study). Turashvili et al [35] identified a number of genes as being significantly differentially expressed between lobular and ductal carcinoma relative to normal epithelial tissue, of which one, COL3A1 is also present on the PanCancer pathway panel. In agreement with this study, COL3A1 is significantly up-regulated in our ILC patients with a fold change of 4.5 and p = 5.76x10-5 that remained significant even after adjustment for multiple testing.
The PanCancer pathway panel includes multiple collagen genes and our data also showed significantly higher expression of COL11A1, fold change 24.99, p = 0.001, COL5A1, fold change 3.91, p = 2.0x10-8 and COL1A1, fold change 8.40, p = 5.65x10-8 in ILC tumor relative to adjacent normal tissue. Enhanced expression and deposition of collagens are associated with tumor development, progression [36–38] and specifically, breast cancer invasion and aggressiveness [39]. Collagens are the main structural extracellular matrix proteins and perhaps upregulation of these genes in multifocal ILC is related to the upregulation of THBS4, an extraceullalar glycoprotein. THBS4 plays an important role in interactions with the extracellular matrix and has also been shown to be expressed at higher levels in cancer associated stroma relative to normal stroma, with highest expression in tumors rich in stromal content, (ILC, ER positive low grade IDC; luminal A and normal-like subtypes) [34]. These findings suggest that increased THBS4 expression in breast cancer-associated extracellular matrix contributes to the activated stromal response exhibited during tumor progression and that this may facilitate invasion of tumor cells. Our pathway analysis of ILC tumor versus adjacent normal is supportive of these findings in that the collagen genes and THBS4 map to the PI3K pathway (the most dysregulated pathway in our tumor versus normal dataset). PI3K genes are expressed in both tumor and stromal cell types, allowing cross-talk between these cell types to modify the surrounding tumor microenvironment and promote tumorigenesis [40].
Our study identified FIGF, RELN, PROM1, SFRP1, MMP7, NTRK2, LAMB3, SPRY2 and KIT as being both significantly down-regulated in MF ILC (absolute fold changes 0.06, 0.12, 0.13, 0.15, 0.17, 0.22, 0.23, 0.24, 0.26, respectively) similar to CDH1 (absolute fold change 0.28) and significant with q<0.05. Given our study design of ILC tumor versus normal, rather than ILC versus IDC, it is possible that these genes are not specifically markers of ILC. We note that Turashvili et al [35] reported expression of SFRP1 to be significantly down-regulated in both ILC and IDC relative to matched normal lobular and ductal tissue respectively, and expression of MMP7 to be down-regulated in ductal carcinoma only. However, we also note that fold changes for all nine of these genes are in the same direction and of similar magnitude to CDH1, a hallmark of ILC. One possibility is that these decreases in expression are related to loss of CDH1 and are potential therapeutic targets in ILC. Four of these genes, (FIGF, RELN, LAMB3 and KIT), map to the PI3K pathway, two of which (FIGF and KIT) map to both PIK3 and RAS pathways. NTRK2 (Tyrosine kinase B neurotrophin receptor) functions in the MAP kinase pathway. Its kinase activity reportedly contributes to disease progression by inhibiting anoikis and promoting epithelial to mesenchymal transition, and PIK3 is an important downstream target of this cell survival pathway [41, 42]. SPRY2, an inhibitor of RAS/mitogen signaling, has been previously associated with prognosis of breast cancer [43]. We also examined differential pathway expression at the level of MF ILC versus adjacent normal tissue. All pathways were significant at p<0.05. We observed a high correlation between two independent measures of pathway significance allowing pathways to be ranked in order of significance. The most significant pathways were PI3K and cell cycle, although DNA repair, TGFβ, RAS, Wnt and MAP kinase were also highly significant, with PI3K and RAS showing the largest fold change. These data are also in agreement with current thinking that E-Cadherin-mediated adhesion inhibits tyrosine kinase receptor signaling; whereas loss of E-Cadherin, a salient feature of ILC, results in activation of receptor tyrosine kinase signaling pathways [44, 45].
We next sought to systematically examine differences between multiple foci within ILC patients. Firstly, we examined gene copy number across 80 known cancer genes. Copy number analysis showed a high level of consistency from multiple punches within each focus and between foci in the same patient. Two of eleven patients showed gain in copy number at 11q13.3. Amplification of three genes at this locus (CCND1, FADD and ORAOV1) was identified consistently in both foci of one patient (~8 copies), with a second patient showing small gain (3 copies) in both foci. Expression of CCND1 was also observed to be significantly up-regulated at the level of mRNA (q = 0.008) relative to adjacent normal tissue. CCND1 gain has been previously observed in both ILC and IDC [26, 28–30] with increased frequency in ILC relative to IDC [29]. The observation of this amplification consistently in both foci (from multiple sampling of three different punches in each foci), and the lack of other chromosomal aberrations is in line with previous studies of multifocal breast cancer, suggesting that the majority of multifocal lesions are clonally related [22–25]. We also observed some evidence for deletion of the same region in a single patient in either one or both foci, although given the difficulty in reliably estimating small copy number changes (particularly in samples from FFPE material), this could be due to artifact, and to our knowledge, deletion of CCND1 has not previously been reported.
Our copy number data suggest that multifocal ILC foci are, for the most part, very similar with respect to these genomic features. However, our unique study design of genomic data from three punches from each tumor in each patient allowed us to specifically separate intra and inter-tumor heterogeneity within patients and tumor heterogeneity between patients.
This analysis revealed strong evidence of within patient focus to focus heterogeneity for greater than half of the studied genes in addition to any between patient variability and variability in gene expression measures among punches from the same tumors after adjustment for tumor size. Put into the context of CDH1, a known hallmark of ILC, we observed significant loss of CDH1 expression across our sample of 11 MF ILC patients relative to adjacent normal tissue. We also observed, significant heterogeneity (73.8%, q = 9.60x10-8) of CDH1 gene expression between foci within the same patient, with one patient (patient 6) showing lower CDH1 expression by three orders of magnitude in the larger focus. Both foci in this patient were of the classic lobular subtype, and within the same quadrant (3cm apart). It is possible that this observation is an artifact due to different amounts of contaminating normal cells. However, our gene expression analyses showed relatively low level intra-tumor heterogeneity and our copy number analysis was uninformative: we did not observe any copy number changes <1.5 or ≥3.0 for this patient in either focus and our copy number panel of 80 genes did not include CDH1 or any other genes on chromosome 16.
Of the nine genes that were significantly down-regulated in tumor relative to adjacent normal at greater magnitude than CDH1, we also observed significant levels of within patient focus to focus heterogeneity for eight of nine genes. The same genes did not show significant heterogeneity between patients, which is likely why they we were able to detect significant difference in our tumor versus adjacent normal analysis. This opens the question, how many potential disease markers are missed due to patient heterogeneity in study designs of tumor versus normal, especially when using single specimens per tumor. Our study gives some estimation of this when ranking genes by the percentage of patient heterogeneity (S4 Table), which results in 13 genes: PPP2R2C, RAC1, IL20RB, MAPT, PPP2CB, MGMT, DDIT4, MAPK3, ZBTB16, FGFR3, LAMA1, RNF43 and IL23R, with >80% heterogeneity between patient tumors, and very little heterogeneity within tumors or between foci within the same patient.
Current literature suggests loss of CDH1 and other genes that potentially differentiate between ILC and IDC, promote epithelial to mesenchymal transition and activation of receptor tyrosine signaling pathways. Our data demonstrate an additional level of complexity of within patient foci heterogeneity and between patient tumors that could mask potential prognostic factors of MF ILC.
The main limitations of these observations are the limited sample size (eleven patients), and lack of point mutation analysis, as heterogeneity of driver mutations between foci would suggest a source of metastatic potential in MF ILC, that we were unable to address by gene expression analyses. Despite these limitations, this is the most detailed molecular study to date of MF ILC patients, all of whom were ER+ and HER2-. The importance of sequencing analyses of tumor heterogeneity and evolution was recently highlighted in two studies which included patients with MF ductal carcinoma (but not MF ILC). Desmedt et al [46] observed genomic heterogeneity between foci in 12/36 patients with MF IDC, despite similar pathological features. Yates et al [47] examined two to five foci from each of four patients with MF IDC, observing mutations in known driver genes that were private to one focus in three of four patients. They also found many private mutations with high variant allele fractions within individual foci, suggesting the occurrence of complete ‘clonal sweeps’ that replaced all other tumor cells within the focus. Our findings of genomic heterogeneity in MF ILC via gene expression analyses, multiregion sequencing studies of MF IDC [46, 47] and key questions established from an International meeting on the extent of tumor heterogeneity [48] suggest further exploitation of tumor heterogeneity in MF breast cancer will be crucial to our ability to design and select effective therapies and curtail treatment resistance.
Supporting Information
Patient 7, tumor 1: A) Hematoxylin and Eosin staining, trabecular subtype. B) E-cadherin staining, weakly positive. C) Cytoplasmic p120 staining. Patient 7, tumor 2: D) Hematoxylin and Eosin staining, dominant classic lobular with some mixed trabecular. E) E-cadherin staining, negative. F) Cytoplasmic p120 staining.
(TIF)
(PDF)
Boxes represent maximum, minimum and mean score observed from 3x1.5mm core punches from each tumor and a single punch for adjacent normal tissue where available. Patients are labelled p1-p11 and tumors are labelled T1 and T2. Patient 1 has three tumors labelled p1_T1, p1_T2 and p1_T3. Patients 4 and 7 with the most differences in pathway score between T1 and T2 are highlighted in red and blue respectively.
(PDF)
Each score from 3 core punches from each tumor and a single punch for adjacent normal tissue. Bars represent maximum, minimum and median score.
(TIF)
(XLSX)
(DOCX)
(DOCX)
This table shows the results of the heterogeneity linear mixed model analysis. The first column shows the gene name; columns B-D show the estimated standard deviations of the random effects for patient (σP), tumor foci (σF), intra-tumor punch sample (σIT) respectively. Columns E-G shows the estimated percent heterogeneity corresponding to each of these as described in the methods. Column H shows the p-value from the likelihood ratio test of the null hypothesis of no between patient variation, H0: σP = 0. Column I shows p-values adjusted for multiple testing using the Holm method, and column J shows q-values that are estimates of false discovery rate at each p-value cut-off. Columns K to M similarly show p-values and q-values corresponding to the test of the null hypothesis of no within-patient, between foci heterogeneity: H0: σF = 0.
(XLSX)
Acknowledgments
This study was funded by Mayo Clinic Cancer Focus Research Team, the 26.2 with Donna Foundation and the Serene M. and Frances C. Durling endowment.
Data Availability
All relevant data are within the paper and its Supporting Information files. Normalized gene counts and raw data files are available from the GEO database (GSE79058): (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE79058).
Funding Statement
This study was funded by Mayo Clinic Cancer Focus Research Team (AMA), the 26.2 with Donna Foundation (EAP) and the Serene M. and Frances C. Durling endowment (EAP).
References
- 1.Arpino G, Bardou VJ, Clark GM, Elledge RM. Infiltrating lobular carcinoma of the breast: tumor characteristics and clinical outcome. Breast Cancer Res. 2004;6(3):R149–56. Epub 2004/04/16. 10.1186/bcr767 bcr767 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolters R, Wockel A, Janni W, Novopashenny I, Ebner F, Kreienberg R, et al. Comparing the outcome between multicentric and multifocal breast cancer: what is the impact on survival, and is there a role for guideline-adherent adjuvant therapy? A retrospective multicenter cohort study of 8,935 patients. Breast Cancer Res Treat. 2013;142(3):579–90. Epub 2013/11/22. 10.1007/s10549-013-2772-y . [DOI] [PubMed] [Google Scholar]
- 3.Rezo A, Dahlstrom J, Shadbolt B, Rodins K, Zhang Y, Davis AJ. Tumor size and survival in multicentric and multifocal breast cancer. Breast. 2011;20(3):259–63. Epub 2011/02/18. 10.1016/j.breast.2011.01.005 S0960-9776(11)00008-7 [pii]. . [DOI] [PubMed] [Google Scholar]
- 4.Choi Y, Kim EJ, Seol H, Lee HE, Jang MJ, Kim SM, et al. The hormone receptor, human epidermal growth factor receptor 2, and molecular subtype status of individual tumor foci in multifocal/multicentric invasive ductal carcinoma of breast. Hum Pathol. 2012;43(1):48–55. Epub 2011/07/08. 10.1016/j.humpath.2010.08.026 S0046-8177(11)00156-0 [pii]. . [DOI] [PubMed] [Google Scholar]
- 5.Viale G, Rotmensz N, Maisonneuve P, Orvieto E, Maiorano E, Galimberti V, et al. Lack of prognostic significance of "classic" lobular breast carcinoma: a matched, single institution series. Breast Cancer Res Treat. 2009;117(1):211–4. Epub 2008/07/17. 10.1007/s10549-008-0112-4 . [DOI] [PubMed] [Google Scholar]
- 6.Weigelt B, Horlings HM, Kreike B, Hayes MM, Hauptmann M, Wessels LF, et al. Refinement of breast cancer classification by molecular characterization of histological special types. J Pathol. 2008;216(2):141–50. Epub 2008/08/23. 10.1002/path.2407 . [DOI] [PubMed] [Google Scholar]
- 7.Arvold ND, Taghian AG, Niemierko A, Abi Raad RF, Sreedhara M, Nguyen PL, et al. Age, breast cancer subtype approximation, and local recurrence after breast-conserving therapy. J Clin Oncol. 2011;29(29):3885–91. Epub 2011/09/09. 10.1200/JCO.2011.36.1105 JCO.2011.36.1105 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Metzger-Filho O, Sun Z, Viale G, Price KN, Crivellari D, Snyder RD, et al. Patterns of Recurrence and outcome according to breast cancer subtypes in lymph node-negative disease: results from international breast cancer study group trials VIII and IX. J Clin Oncol. 2013;31(25):3083–90. Epub 2013/07/31. 10.1200/JCO.2012.46.1574 JCO.2012.46.1574 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–26. Epub 2004/12/14. NEJMoa041588 [pii] 10.1056/NEJMoa041588 . [DOI] [PubMed] [Google Scholar]
- 10.Voduc KD, Cheang MC, Tyldesley S, Gelmon K, Nielsen TO, Kennecke H. Breast cancer subtypes and the risk of local and regional relapse. J Clin Oncol. 2010;28(10):1684–91. Epub 2010/03/03. 10.1200/JCO.2009.24.9284 JCO.2009.24.9284 [pii]. . [DOI] [PubMed] [Google Scholar]
- 11.Cabioglu N, Ozmen V, Kaya H, Tuzlali S, Igci A, Muslumanoglu M, et al. Increased lymph node positivity in multifocal and multicentric breast cancer. J Am Coll Surg. 2009;208(1):67–74. Epub 2009/02/21. 10.1016/j.jamcollsurg.2008.09.001 S1072-7515(08)01333-1 [pii]. . [DOI] [PubMed] [Google Scholar]
- 12.Chua B, Ung O, Taylor R, Boyages J. Frequency and predictors of axillary lymph node metastases in invasive breast cancer. ANZ J Surg. 2001;71(12):723–8. Epub 2002/03/22. 2266 [pii]. . [DOI] [PubMed] [Google Scholar]
- 13.Chung AP, Huynh K, Kidner T, Mirzadehgan P, Sim MS, Giuliano AE. Comparison of outcomes of breast conserving therapy in multifocal and unifocal invasive breast cancer. J Am Coll Surg. 2012;215(1):137–46; discussion 46–7. Epub 2012/05/23. 10.1016/j.jamcollsurg.2012.05.006 S1072-7515(12)00349-3 [pii]. . [DOI] [PubMed] [Google Scholar]
- 14.Coombs NJ, Boyages J. Multifocal and multicentric breast cancer: does each focus matter? J Clin Oncol. 2005;23(30):7497–502. Epub 2005/10/20. 23/30/7497 [pii] 10.1200/JCO.2005.02.1147 . [DOI] [PubMed] [Google Scholar]
- 15.Pekar G, Hofmeyer S, Tabar L, Tarjan M, Chen TH, Yen AM, et al. Multifocal breast cancer documented in large-format histology sections: long-term follow-up results by molecular phenotypes. Cancer. 2013;119(6):1132–9. Epub 2013/01/03. 10.1002/cncr.27877 . [DOI] [PubMed] [Google Scholar]
- 16.Norton N, Sun Z, Asmann YW, Serie DJ, Necela BM, Bhagwate A, et al. Gene expression, single nucleotide variant and fusion transcript discovery in archival material from breast tumors. PLoS One. 2013;8(11):e81925 Epub 2013/11/28. 10.1371/journal.pone.0081925 PONE-D-13-28059 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. Epub 2002/08/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. Epub 2002/02/16. 10.1006/meth.2001.1262 S1046-2023(01)91262-9 [pii]. . [DOI] [PubMed] [Google Scholar]
- 19.Holm S. A simple sequentially rejective multiple test procedure. Scandinavian Journal of Statistics. 1979;6:65–70. [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate—a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57(1):289–300. . [Google Scholar]
- 21.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–58. Epub 2013/03/30. 10.1126/science.1235122 339/6127/1546 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghazani AA, Arneson N, Warren K, Pintilie M, Bayani J, Squire JA, et al. Genomic alterations in sporadic synchronous primary breast cancer using array and metaphase comparative genomic hybridization. Neoplasia. 2007;9(6):511–20. Epub 2007/07/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noguchi S, Aihara T, Koyama H, Motomura K, Inaji H, Imaoka S. Discrimination between multicentric and multifocal carcinomas of the breast through clonal analysis. Cancer. 1994;74(3):872–7. Epub 1994/08/01. . [DOI] [PubMed] [Google Scholar]
- 24.Teixeira MR, Pandis N, Bardi G, Andersen JA, Bohler PJ, Qvist H, et al. Discrimination between multicentric and multifocal breast carcinoma by cytogenetic investigation of macroscopically distinct ipsilateral lesions. Genes Chromosomes Cancer. 1997;18(3):170–4. Epub 1997/03/01. [pii]. . [DOI] [PubMed] [Google Scholar]
- 25.Teixeira MR, Pandis N, Bardi G, Andersen JA, Mandahl N, Mitelman F, et al. Cytogenetic analysis of multifocal breast carcinomas: detection of karyotypically unrelated clones as well as clonal similarities between tumour foci. Br J Cancer. 1994;70(5):922–7. Epub 1994/11/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi EJ, Yun JA, Jabeen S, Jeon EK, Won HS, Ko YH, et al. Prognostic significance of TMEM16A, PPFIA1, and FADD expression in invasive ductal carcinoma of the breast. World J Surg Oncol. 2014;12:137 Epub 2014/06/03. 10.1186/1477-7819-12-137 1477-7819-12-137 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peters G, Fantl V, Smith R, Brookes S, Dickson C. Chromosome 11q13 markers and D-type cyclins in breast cancer. Breast Cancer Res Treat. 1995;33(2):125–35. Epub 1995/01/01. . [DOI] [PubMed] [Google Scholar]
- 28.Courjal F, Cuny M, Simony-Lafontaine J, Louason G, Speiser P, Zeillinger R, et al. Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: definition of phenotypic groups. Cancer Res. 1997;57(19):4360–7. Epub 1997/10/23. . [PubMed] [Google Scholar]
- 29.Gruel N, Lucchesi C, Raynal V, Rodrigues MJ, Pierron G, Goudefroye R, et al. Lobular invasive carcinoma of the breast is a molecular entity distinct from luminal invasive ductal carcinoma. Eur J Cancer. 2010;46(13):2399–407. Epub 2010/06/24. 10.1016/j.ejca.2010.05.013 S0959-8049(10)00385-0 [pii]. . [DOI] [PubMed] [Google Scholar]
- 30.Oyama T, Kashiwabara K, Yoshimoto K, Arnold A, Koerner F. Frequent overexpression of the cyclin D1 oncogene in invasive lobular carcinoma of the breast. Cancer Res. 1998;58(13):2876–80. Epub 1998/07/14. . [PubMed] [Google Scholar]
- 31.Noorlag R, van Kempen PM, Stegeman I, Koole R, van Es RJ, Willems SM. The diagnostic value of 11q13 amplification and protein expression in the detection of nodal metastasis from oral squamous cell carcinoma: a systematic review and meta-analysis. Virchows Arch. 2015;466(4):363–73. Epub 2015/02/11. 10.1007/s00428-015-1719-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dabbs DJ, Schnitt SJ, Geyer FC, Weigelt B, Baehner FL, Decker T, et al. Lobular neoplasia of the breast revisited with emphasis on the role of E-cadherin immunohistochemistry. Am J Surg Pathol. 2013;37(7):e1–11. Epub 2013/06/14. 10.1097/PAS.0b013e3182918a2b 00000478-201307000-00001 [pii]. . [DOI] [PubMed] [Google Scholar]
- 33.Korkola JE, DeVries S, Fridlyand J, Hwang ES, Estep AL, Chen YY, et al. Differentiation of lobular versus ductal breast carcinomas by expression microarray analysis. Cancer Res. 2003;63(21):7167–75. Epub 2003/11/13. . [PubMed] [Google Scholar]
- 34.McCart Reed AE, Song S, Kutasovic JR, Reid LE, Valle JM, Vargas AC, et al. Thrombospondin-4 expression is activated during the stromal response to invasive breast cancer. Virchows Arch. 2013;463(4):535–45. Epub 2013/08/15. 10.1007/s00428-013-1468-3 . [DOI] [PubMed] [Google Scholar]
- 35.Turashvili G, Bouchal J, Baumforth K, Wei W, Dziechciarkova M, Ehrmann J, et al. Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. BMC Cancer. 2007;7:55 Epub 2007/03/29. 1471-2407-7-55 [pii] 10.1186/1471-2407-7-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iyengar P, Espina V, Williams TW, Lin Y, Berry D, Jelicks LA, et al. Adipocyte-derived collagen VI affects early mammary tumor progression in vivo, demonstrating a critical interaction in the tumor/stroma microenvironment. The Journal of clinical investigation. 2005;115(5):1163–76. Epub 2005/04/21. 10.1172/JCI23424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Provenzano PP, Inman DR, Eliceiri KW, Knittel JG, Yan L, Rueden CT, et al. Collagen density promotes mammary tumor initiation and progression. BMC Med. 2008;6:11 Epub 2008/04/30. 10.1186/1741-7015-6-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shields MA, Dangi-Garimella S, Krantz SB, Bentrem DJ, Munshi HG. Pancreatic cancer cells respond to type I collagen by inducing snail expression to promote membrane type 1 matrix metalloproteinase-dependent collagen invasion. The Journal of biological chemistry. 2011;286(12):10495–504. Epub 2011/02/04. 10.1074/jbc.M110.195628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Acerbi I, Cassereau L, Dean I, Shi Q, Au A, Park C, et al. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr Biol (Camb). 2015;7(10):1120–34. Epub 2015/05/12. 10.1039/c5ib00040h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirsch E, Ciraolo E, Franco I, Ghigo A, Martini M. PI3K in cancer-stroma interactions: bad in seed and ugly in soil. Oncogene. 2014;33(24):3083–90. Epub 2013/07/31. 10.1038/onc.2013.265 . [DOI] [PubMed] [Google Scholar]
- 41.Douma S, Van Laar T, Zevenhoven J, Meuwissen R, Van Garderen E, Peeper DS. Suppression of anoikis and induction of metastasis by the neurotrophic receptor TrkB. Nature. 2004;430(7003):1034–9. Epub 2004/08/27. 10.1038/nature02765 nature02765 [pii]. . [DOI] [PubMed] [Google Scholar]
- 42.Geiger TR, Peeper DS. Critical role for TrkB kinase function in anoikis suppression, tumorigenesis, and metastasis. Cancer Res. 2007;67(13):6221–9. Epub 2007/07/10. 67/13/6221 [pii] 10.1158/0008-5472.CAN-07-0121 . [DOI] [PubMed] [Google Scholar]
- 43.Lo TL, Yusoff P, Fong CW, Guo K, McCaw BJ, Phillips WA, et al. The ras/mitogen-activated protein kinase pathway inhibitor and likely tumor suppressor proteins, sprouty 1 and sprouty 2 are deregulated in breast cancer. Cancer Res. 2004;64(17):6127–36. Epub 2004/09/03. 10.1158/0008-5472.CAN-04-1207 64/17/6127 [pii]. . [DOI] [PubMed] [Google Scholar]
- 44.Andl CD, Rustgi AK. No one-way street: cross-talk between e-cadherin and receptor tyrosine kinase (RTK) signaling: a mechanism to regulate RTK activity. Cancer Biol Ther. 2005;4(1):28–31. Epub 2005/01/22. 1431 [pii]. . [DOI] [PubMed] [Google Scholar]
- 45.Qian X, Karpova T, Sheppard AM, McNally J, Lowy DR. E-cadherin-mediated adhesion inhibits ligand-dependent activation of diverse receptor tyrosine kinases. EMBO J. 2004;23(8):1739–48. Epub 2004/04/02. 10.1038/sj.emboj.7600136 7600136 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Desmedt C, Fumagalli D, Pietri E, Zoppoli G, Brown D, Nik-Zainal S, et al. Uncovering the genomic heterogeneity of multifocal breast cancer. The Journal of pathology. 2015;236(4):457–66. Epub 2015/04/09. 10.1002/path.4540 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yates LR, Gerstung M, Knappskog S, Desmedt C, Gundem G, Van Loo P, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nature medicine. 2015;21(7):751–9. Epub 2015/06/23. 10.1038/nm.3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alizadeh AA, Aranda V, Bardelli A, Blanpain C, Bock C, Borowski C, et al. Toward understanding and exploiting tumor heterogeneity. Nature medicine. 2015;21(8):846–53. Epub 2015/08/08. 10.1038/nm.3915 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Patient 7, tumor 1: A) Hematoxylin and Eosin staining, trabecular subtype. B) E-cadherin staining, weakly positive. C) Cytoplasmic p120 staining. Patient 7, tumor 2: D) Hematoxylin and Eosin staining, dominant classic lobular with some mixed trabecular. E) E-cadherin staining, negative. F) Cytoplasmic p120 staining.
(TIF)
(PDF)
Boxes represent maximum, minimum and mean score observed from 3x1.5mm core punches from each tumor and a single punch for adjacent normal tissue where available. Patients are labelled p1-p11 and tumors are labelled T1 and T2. Patient 1 has three tumors labelled p1_T1, p1_T2 and p1_T3. Patients 4 and 7 with the most differences in pathway score between T1 and T2 are highlighted in red and blue respectively.
(PDF)
Each score from 3 core punches from each tumor and a single punch for adjacent normal tissue. Bars represent maximum, minimum and median score.
(TIF)
(XLSX)
(DOCX)
(DOCX)
This table shows the results of the heterogeneity linear mixed model analysis. The first column shows the gene name; columns B-D show the estimated standard deviations of the random effects for patient (σP), tumor foci (σF), intra-tumor punch sample (σIT) respectively. Columns E-G shows the estimated percent heterogeneity corresponding to each of these as described in the methods. Column H shows the p-value from the likelihood ratio test of the null hypothesis of no between patient variation, H0: σP = 0. Column I shows p-values adjusted for multiple testing using the Holm method, and column J shows q-values that are estimates of false discovery rate at each p-value cut-off. Columns K to M similarly show p-values and q-values corresponding to the test of the null hypothesis of no within-patient, between foci heterogeneity: H0: σF = 0.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. Normalized gene counts and raw data files are available from the GEO database (GSE79058): (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE79058).