Adeno-associated viral delivery of a microRNA targeting ATXN1 provides therapeutic benefit in mouse models of spinocerebellar ataxia type 1. Keiser et al. assess the translatability of this approach using adult rhesus macaques, and show that targeted delivery to deep cerebellar nuclei silences endogenous ATXN1 mRNA in multiple SCA1-affected sites.
Keywords: gene therapy, neurodegeneration, ataxia, spinocerebellar ataxia
Adeno-associated viral delivery of a microRNA targeting ATXN1 provides therapeutic benefit in mouse models of spinocerebellar ataxia type 1. Keiser et al. assess the translatability of this approach using adult rhesus macaques, and show that targeted delivery to deep cerebellar nuclei silences endogenous ATXN1 mRNA in multiple SCA1-affected sites.
Abstract
Spinocerebellar ataxia type 1 is one of nine polyglutamine expansion diseases and is characterized by cerebellar ataxia and neuronal degeneration in the cerebellum and brainstem. Currently, there are no effective therapies for this disease. Previously, we have shown that RNA interference mediated silencing of ATXN1 mRNA provides therapeutic benefit in mouse models of the disease. Adeno-associated viral delivery of an engineered microRNA targeting ATXN1 to the cerebella of well-established mouse models improved motor phenotypes, neuropathy, and transcriptional changes. Here, we test the translatability of this approach in adult rhesus cerebella. Nine adult male and three adult female rhesus macaque were unilaterally injected with our therapeutic vector, a recombinant adeno-associated virus type 1 (rAAV1) expressing our RNAi trigger (miS1) and co-expressing enhanced green fluorescent protein (rAAV1.miS1eGFP) into the deep cerebellar nuclei using magnetic resonance imaging guided techniques combined with a Stealth Navigation system (Medtronics Inc.). Transduction was evident in the deep cerebellar nuclei, cerebellar Purkinje cells, the brainstem and the ventral lateral thalamus. Reduction of endogenous ATXN1 messenger RNA levels were ≥30% in the deep cerebellar nuclei, the cerebellar cortex, inferior olive, and thalamus relative to the uninjected hemisphere. There were no clinical complications, and quantitative and qualitative analyses suggest that this therapeutic intervention strategy and subsequent reduction of ATXN1 is well tolerated. Collectively the data illustrate the biodistribution and tolerability of rAAV1.miS1eGFP administration to the adult rhesus cerebellum and are supportive of clinical application for spinocerebellar ataxia type 1.
Introduction
Spinocerebellar ataxia type 1 (SCA1), one of nine polyglutamine (polyQ) repeat diseases, is a dominantly inherited neurodegenerative disorder with no available treatment or cure. The incidence of SCA1 is ∼1–2 per 100 000 people, indicating that there are ∼3000 6000 patients in the USA alone (Submarony, 1998; Subramoni, 2014). SCA1 is caused by an unstable CAG expansion in the ATXN1 gene which encodes ataxin 1 (Orr et al., 1993; Banfi et al., 1994). Normally, a range of CAG repeats interspersed with 1–3 CATs are found in ATXN1. In SCA1 patients, the ATXN1 CAG repeat is >39, conferring a toxic gain-of-function to ataxin 1.
Although clinical onset of SCA1 may occur from childhood through adulthood, most patients present between 30–40 years of age with progressive wide-based gait, and difficulties with balance, speech, swallowing, coordination and spasticity. Extracerebellar dysfunction may also appear with increased deep tendon reflexes and oculomotor abnormalities. Mild cognitive impairment occurs in 10–20% of patients. Neuropathological studies of tissues from SCA1 patients show that the primary sites of degeneration are the dentate nucleus, the inferior olive and cerebellar Purkinje cells. There is more degeneration in the upper vermis, less in the lateral cerebellar cortex, and mild changes in the flocculonodular lobes. There is also involvement of brainstem nuclei and spinocerebellar tracts and variable reports of cerebral involvement (Robitaille et al., 1995, 1997; Donato et al., 2012; Rub et al., 2013). Ataxin 1 is ubiquitously expressed and is prevalent in cerebellar Purkinje cells (Servadio et al., 1995). Tissues from SCA1 patients also show ataxin 1-positive nuclear inclusions in Purkinje cells and brainstem neurons and in cerebrum (Duyckaerts et al., 1999).
Animal studies have been pivotal in defining the cellular and molecular mechanisms underlying SCA1 pathogenesis (Serra et al., 2004, 2006; Tsuda et al., 2005; Lam et al., 2006; Lambrechts and Carmeliet, 2006; Bowman et al., 2007; Cvetanovic et al., 2007; Lim et al., 2008; Morrison, 2009; Zoghbi and Orr, 2009; Lai et al., 2011; Orr, 2012; Rodriguez-Lebron et al., 2013). There is extensive evidence supporting the notion that the disease-causing mutation acts through a toxic gain-of-function mechanism, and that suppressing its expression would not only arrest disease progression, but may reverse disease phenotypes (Zu et al., 2004; Oz et al., 2011). Using a doxycycline-inducible transgenic mouse model for SCA1, Orr and colleagues (Zu et al., 2004) showed that repressing mutant protein expression 12 weeks after sustained expression significantly improved pathology and behavioural deficits. Thus, a window of opportunity for gene silencing strategies, initiated after disease onset, may exist.
Gene silencing approaches include RNA interference (RNAi) (Xia et al., 2002, 2004), antisense oligonucleotides (Kole et al., 2011), inhibitory antibodies, and more recently DNA editing approaches (Wood et al., 2011; Basu et al., 2015; Ousterout et al., 2015). We previously used recombinant adeno-associated viral vectors (rAAV) expressing short hairpin RNAs or artificial microRNAs (miRNAs) in mouse models of SCA1 to test the therapeutic utility of RNAi for SCA1 (Xia et al., 2004; Keiser et al., 2013, 2014). Those studies informed us about the neuroanatomical area of coverage and degree of silencing required for clinical efficacy in both transgenic and knock-in models of SCA1. These data showed that a single injection of rAAV1 expressing inhibitory RNAs to the deep cerebellar nuclei (DCN) afforded widespread transduction of Purkinje cells and brainstem nuclei. While promising, the scalability of this approach for SCA patient therapy is unknown.
In this work we devised and tested a surgical approach aimed at providing adequate coverage of brain regions affected in patients with SCA1. We show that directed delivery of rAAV1 to the DCN of the rhesus macaque brain results in broad transduction of Purkinje cells and brainstem neurons, achieving widespread gene silencing activity. These data also indicate the safety of ATXN1 silencing in primate brain.
Materials and methods
Animals and treatment groups
Nine adult male and three adult female rhesus monkeys (Macaca mulatta) were obtained from Abbott, Harlan, Oregon National Primate Research Center, and Tulane National Primate Research Center. Animals were pair-housed on a 12-h light/12-h dark cycle. Nine were adult male monkeys, 9.9 ± 0.8 years of age and weighed 15.7 ± 0.7 kg, and three were adult female monkeys, 7.3 ± 0.9 years old and weighed 8.3 ± 1.0 kg. All procedures were approved by the University of Illinois Chicago Institutional Animal Care and Use Committee and the Rush University Institutional Animal Care and Use Committee and accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. Animal care was supervised by veterinarians skilled in the care and maintenance of non-human primates.
Stereotaxic surgery
All surgical procedures were conducted under isoflurane anaesthesia (1–3% maintenance, inhalation) and sterile field conditions. Using MRI-guided techniques combined with a Stealth Navigation system (generously donated by Medtronics Inc.), a 100-µl Hamilton syringe fitted with a 30 G needle loaded with rAAV1.miS1eGFP or vehicle was lowered through small burr holes to the individual target deep cerebellar nuclei described in Table 1. Injection volumes shown in Table 1 were infused at 0.5–1.0 µl/min to minimize injectate reflux, inflammation or damage to the parenchyma. After the injection, the needle was left in place for 2 min, then slowly retracted. The following pre- and postoperative analgesics were given: meloxicam (0.1 mg/kg subcutaneously once daily ×3 days), hydromorphone (0.1 mg/kg intramuscular once preoperatively), and sustained release buprenorphine (0.2 mg/kg subcutaneously once postoperatively). Animals received cefazolin (25 mg/kg intravenously once preoperatively) and postoperatively (25 mg/kg intramuscular twice daily for 3 days).
Table 1.
Optimization of surgical sites and doses in non-human primates
| Study | Surgical target | Viral genomes | Volume | Total viral genomes | Total volume |
|---|---|---|---|---|---|
| 1 | Fastigial | 4.00 × 1010 | 50 µl | 8 × 1010 or 2.5 × 1011 | 100 µl |
| 1.25 × 1011 | 50 µl | ||||
| Interposed | 4.00 × 1010 | 50 µl | |||
| 1.25 × 1011 | 50 µl | ||||
| 2 | Fastigial | 5.20 × 1011 | 20 µl | 2.6 × 1012 | 100 µl |
| Interposed | 7.90 × 1011 | 30 µl | |||
| Dentate | 1.30 × 1012 | 50 µl |
General health assessment
Baseline body weights were obtained before surgery. Postoperatively, animals were monitored for activity, appetite, urine/faecal output, incision healing and general appearance daily ≥6 days by the University of Illinois Chicago veterinary staff for adverse reactions. None of the animals were reported to have any neurological deficits from the surgeries. All of the animals were reported being bright/alert/responsive within 2 days of surgery.
Necropsy and brain tissue processing
Six or 8 weeks post-surgery, animals were deeply sedated and euthanized by transcardial saline perfusion. The brain was removed from the calvarium and placed in a chilled coronal or sagittal block and 4-mm slabs were made. On ice, bilateral tissue punches from the cerebellar cortex, DCN, inferior olive, and thalamus were collected for molecular analyses and immediately frozen to preserve DNA, RNA and protein. Remaining tissue was post-fixed in 4% paraformaldehyde solution for 48 h then transferred to a 30% sucrose solution. Coronal or sagittal slices (40 µm) were sectioned on a microtome, and stored in cryoprotectant solution at −20°C until processed.
Semi-quantification of miRNA and ATXN1 messenger RNA expression
Total RNA was extracted from tissue punches using TRIzol® (Life Technologies). RNA was isolated from left and right cerebellar lobules, dentate nuclei, inferior olivary complexes from the medulla oblongata, and ventral lateral thalamic nuclei from each animal from both studies. Punches from uninjected hemispheres served as endogenous controls. MicroRNA (miRNA) specific complementary DNA was generated using miRNA stem-loop-specific primers and the High Capacity cDNA Reverse Transcription Kit (Life Technologies). Semi-quantitative PCR detected expression of S1 as previously described (Keiser et al., 2013) using the BIOLASE™ PCR Kit (Bioline). In addition, complementary DNA libraries were also generated. Endogenous mRNA levels of ATXN1 and rhesus GFAP were quantified by TaqMan® primer/probe sets (Applied Biosystems). Endogenous rhesus GAPDH was used to normalize expression across samples.
Histopathological examination
Free floating (40-µm thick) brain sections were processed by immunohistochemical visualization of enhanced GFP expression [Living Colors®A.V. Monoclonal Antibody (JL-8), 1:2000; Clontech], for activated microglia (Iba1, 1:1000; Wako), and for glial fibrillary acidic protein (GFAP, 1:1000; Dako) as previously described (McBride et al., 2011). Sagittal cerebellar sections (two to three from each animal) were immunostained for enhanced GFP and inspected using a Leica DM600B microscope. Enhanced GFP-positive Purkinje cells were visually scored as a percentage of the total number of Purkinje cells in lobules II–X.
Neutralizing antibody assay
Serum samples were heat inactivated at 56°C for 30 min. rAAV1CMVeGFP (multiplicity of infection: 5 × 103) was diluted in serum-free Dulbecco’s modified Eagle’s medium and incubated with 2-fold serial dilutions (initial dilution, 1:10) of heat-inactivated serum samples on Dulbecco’s modified Eagle’s medium for 1 h at 37°C. Subsequently, the serum-vector mixture was added to 96-well plates seeded with 4 × 104 HEK 293 cells/well. After 48 h, the intensity of enhanced GFP was measured with Spectra Max i3x Multi-Mode detection platform. The neutralizing antibody titre was reported as the highest serum dilution that inhibited rAAV1.CMVeGFP transduction (eGFP intensity) by 50% compared with the no serum control.
In vitro analysis
HEK293 cells were transfected using Lipofectamine® 2000 (Invitrogen™) with AAV shuttle plasmid expressing miS1 or miS1eGFP. Cells were collected 24 h later and RNA was extracted for quantitative real-time PCR analysis to assess silencing.
Statistical analysis
Statistical analyses were conducted using GraphPad Prism 6.0 software. For all analyses, P-values < 0.05 were considered statistically significant. Quantitative analyses between left and right hemispheres for each animal were performed using paired, two-tailed, t-tests.
Results
Verification of inhibitory RNA expression and ataxin 1 suppression in non-human primate
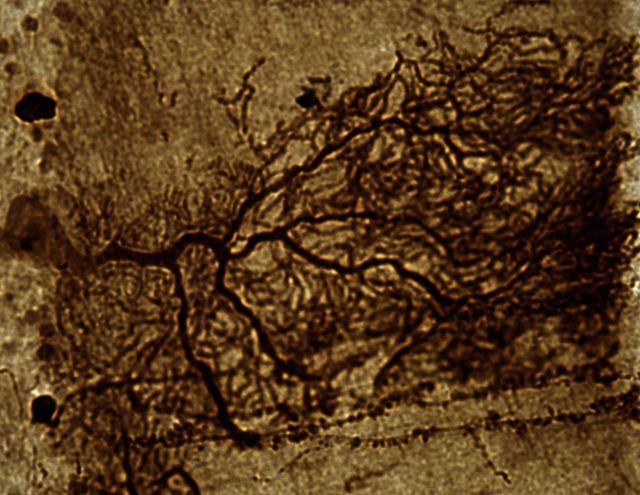
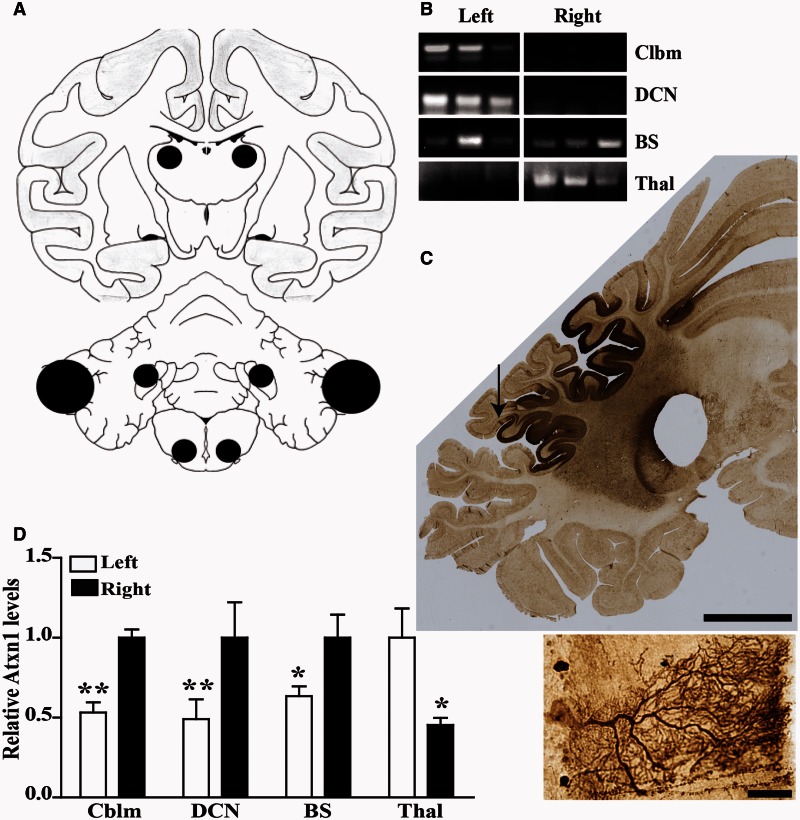
The study design is outlined in Fig. 1. First, we tested if rAAV1.miS1eGFP delivered to the DCN of non-human primates would transduce Purkinje cells as well as DCN neurons, and that expression of miS1 would reduce endogenous non-human primate ataxin 1. Tissue punches were taken from the site of injection (medial DCN), the cerebellar cortex, inferior olive (medulla), and the ventral lateral thalamic nuclei from both hemispheres for protein and RNA evaluation (Fig. 2A) and subjected to semi-quantitative PCR confirming miS1 expression in the left DCN and cerebellar cortex (Fig. 2B). miS1 was expressed in the left and right inferior olivary complexes due to contralateral and ipsilateral projections to the DCN. We also detected miS1 in the contralateral ventral lateral thalamic nuclei in axonal projections from the DCN. The remaining brain tissue was blocked in the coronal plane in 4-mm thick sections and post-fixed for histological analysis. Immunohistochemical analyses for enhanced GFP on coronal sections demonstrated that rAAV.miS1eGFP transduced neuronal cell bodies in the entire left DCN (fastigial, interposed and dentate) and 10–30% of the left cerebellar cortex (Fig. 2C). GFP-positive soma and dendritic arborization from a partially transduced lobule shows clear labelling of Purkinje cells (Fig 2C, inset). Quantitative real-time PCR data showed a significant reduction in rhesus ATXN1 mRNA levels in the cerebellar cortex, DCN, brainstem and contralateral thalamus (Fig. 2D). Biodistribution studies to assess if rAAV was present in peripheral tissues indicated that vector was not shed systemically. Vector genomes were undetectable in liver and testes as analysed by semi-quantitative PCR (Supplementary Fig. 1). These experiments demonstrate that delivery of rAAV to the DCN permits transduction of the cerebellar cortex and brainstem nuclei similar to what was found in mice, as well as cerebellar-thalami projections, a characteristic not previously noted in earlier mouse work.
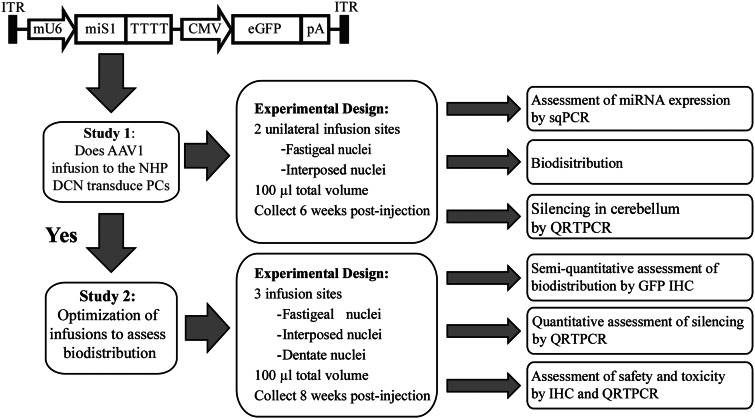
Figure 1.
rAAV1.miS1eGFP construct and experimental outline for Studies 1 and 2. sqPCR = semi-quantitative PCR; QRTPCR = quantitative real-time PCR; IHC = immunohistochemistry.
Figure 2.
Purkinje cell transduction and ATXN1 mRNA silencing. (A) Cartoon showing locations of tissue punches taken from the ventral lateral thalamic nuclei (top), the DCN, the cerebellar cortex, and the inferior olivary complex (bottom). (B) miRNA expression by semi-quantitative PCR from three representative animals from left and right cerebellum (Cblm), DCN, inferior olive from the brainstem (BS), and thalamus (Thal). (C) Representative stitched sagittal photomicrograph of the injected (left) hemisphere immunostained for enhanced GFP (dark brown). Scale bar = 1 mm. Arrow depicts region of inset (below), showing transduced Purkinje cell body and associated dendritic tree. Scale bar = 50 µm. (D) ATXN1 mRNA levels quantified from punches taken from left (white bars) and right (black bars) lobules (n = 6 punches per hemisphere per location) of the cerebellar cortex (Cblm: 0.52 ± 0.6), dentate nucleus (DCN: 0.49 ± 0.12), inferior olive (BS: 0.63 ± 0.06) and thalamus (Thal: 0.46 ± 0.04) expressed as a percentage of the average amount detected in the right hemisphere (Cblm: 1.00 ± 0.05; DCN: 1.00 ± 0.22; BS: 1.00 ± 0.14) or left hemisphere (Thal: 1.00 ± 0.18). *P < 0.05; **P < 0.01.
Improving biodistribution with delivery to the dentate nucleus
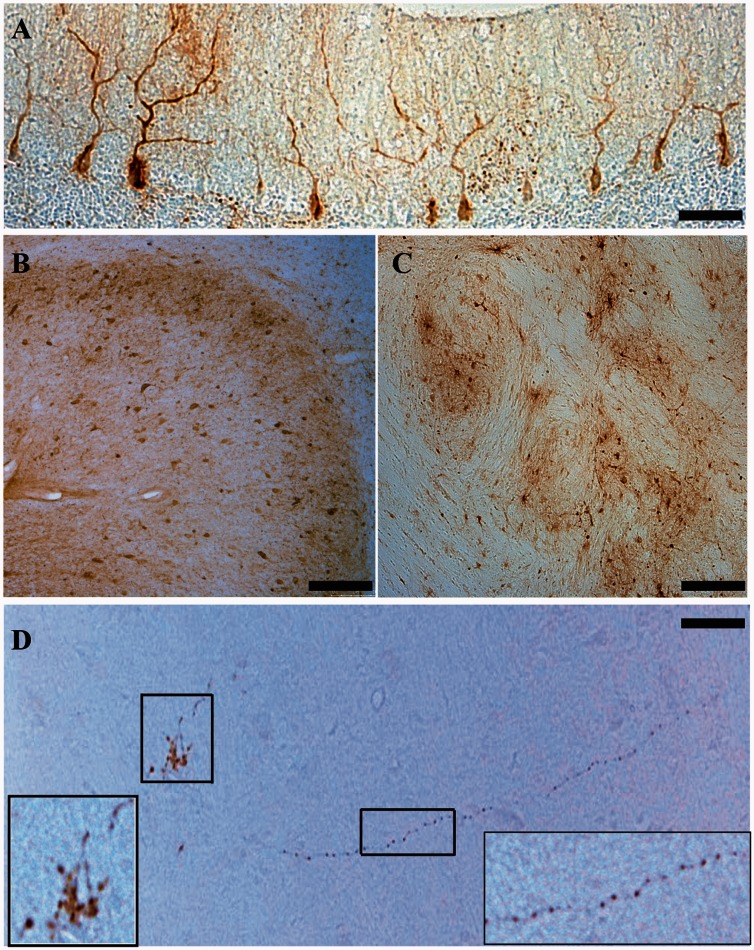
Because the dentate is one of the more severely affected structures affected in SCA1 (Robitaille et al., 1995; Donato et al., 2012), we next tested if an additional injection into the dentate was tolerated, and how the approach would impact transduction biodistribution. All animals (n = 6) received the same total volume of rAAV.miS1eGFP as the first cohort, but with a different infusion paradigm (Table 1) and a higher dose of vector genomes. This increase in vector genomes provides additional information on tolerability as we consider human dosing paradigms. As before, animals did not display overt neurological symptoms and were euthanized 8 weeks after injection. The cerebellum was removed, blocked in the sagittal plane in 4-mm thick slabs, and alternating slabs used for RNA extraction or post-fixed intact for immunohistological studies. GFP-positive Purkinje cells were found in most lobules of the cerebellar cortex (Fig. 3A). GFP-positive neurons were also notable in DCN and inferior olivary complex in the brainstem (Fig. 3B and C). GFP-positive axonal projections and presynaptic termini of DCN neurons were also detected in the contralateral thalamus, indicating anterograde (fibres) transport of rAAV1 and/or GFP (Fig. 3D).
Figure 3.
Enhanced GFP immunohistochemistry. (A) Representative sagittal section from the left hemisphere of an injected cerebellum. Enhanced GFP positive immunostaining in Purkinje cell somas and their dendritic arbors. Scale bar = 100 µm. (B) Coronal section of an injected cerebellum showing enhanced GFP positive neurons in the dentate nucleus. Scale bar = 400 µm (C) Enhanced GFP positive neurons within the left inferior olivary complex. Scale bar = 200 µm. (D) Enhanced GFP positive axonal projections from the contralateral DCN synapsing onto the ventral lateral nuclei of the right thalamus. Left inset shows enhanced GFP-positive presynaptic termini. Right inset shows enhanced GFP-positive magnified axon. Scale bar = 800 µm.
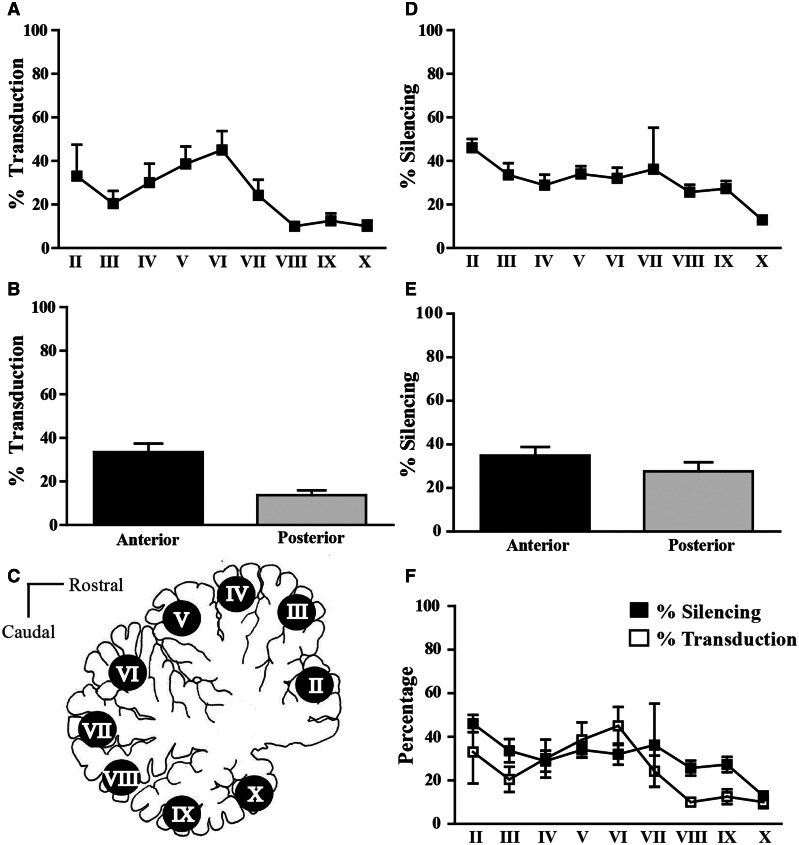
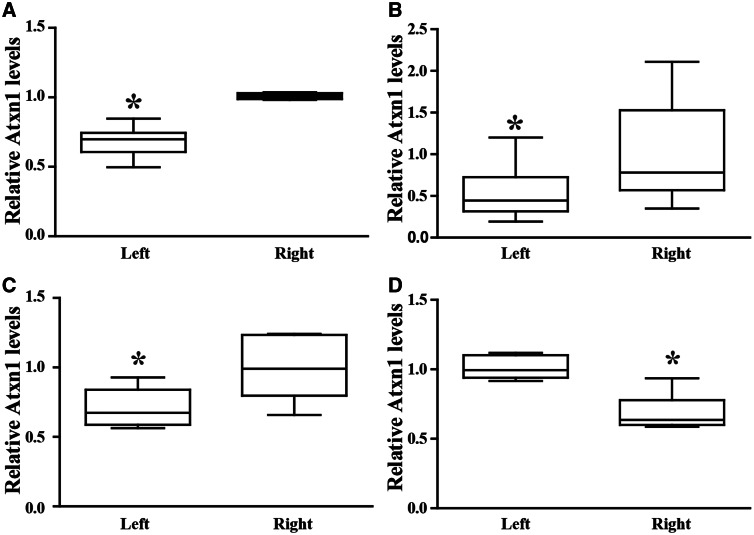
Representative sagittal sections from the cerebellum were scored for % Purkinje cells transduced (GFP immunohistochemistry) for each animal (Fig. 4A). We found greater coverage of the anterior lobe (paleocerebellum; 33.5% ±3.9; n = 49) compared to the posterior lobe (13.7% ± 2.3; n = 51) (Fig. 4B). Tissue punches taken from the cerebellar lobules and subjected to quantitative real-time PCR analysis showed significant suppression of endogenous ATXN1 distributed among the anterior (0.3497 ± 0.0397) and posterior lobes (0.28 ± 0.0413) relative to the uninjected cerebellar hemispheres (Fig. 4C–E). Cumulatively, punches taken from the left cerebellar hemisphere showed >30% lower levels of AXTN1 mRNA (0.68 ± 0.05) relative to the uninjected right hemisphere (1.01 ± 0.001) (*P < 0.05; Fig. 5A and Supplementary Fig. 2). Punches from the left DCN (0.54 ± 0.35), left inferior olive (0.71 ± 0.06) and right thalamus (0.69 ± 0.05) also showed significantly lower levels of ATXN1 mRNA compared to their uninjected hemisphere (paired t-test *P < 0.05; Fig. 5B–D). Notably, pre-existing neutralizing antibody titres did not impact knockdown efficacy (Supplementary Fig. 2). These data show that this delivery approach provides for widespread gene silencing in the primate cerebellum and associated structures.
Figure 4.
Transduction and reduction of ATXN1 mRNA levels among the cerebellar lobules. (A) Purkinje cells positively expressing enhanced GFP by immunohistochemistry were expressed as a percentage of the total number of Purkinje cells in each individual lobule (II–X) from 1–2 equivalent sagittal sections for all six animals. (B) Percentage of enhanced GFP-positive Purkinje cells per lobule represented as anterior and posterior lobes of the cerebellum. Lobules II–V averaged for the anterior lobe (black bar); lobules VI–X were averaged for the posterior lobe (grey bar). (C) Cartoon representation of tissue punches taken from the sagittal slab blocked 4–8 mm from the median. (D) Per cent ATXN1 mRNA silencing in tissue punches from each left (injected) cerebellar hemisphere lobule relative to the uninjected (right) side. (E) Per cent of ATXN1 mRNA silencing in the cerebellar lobes; lobules II–V were averaged for the anterior lobes (black bar); lobules VI–X averaged for the posterior lobes (grey bar). (F) Overlay of graphs A and D showing both % knockdown (black boxes) and % transduction (white boxes). Results are shown as mean ± SEM.
Figure 5.
ATXN1 mRNA levels. (A) Expression of ATXN1 mRNA levels from tissue punches taken from the left cerebellar cortex (0.68 ± 0.05) relative to the right (1.01 ± 0.001; n = 6; *P < 0.05). (B) Expression of ATXN1 mRNA levels from tissue punches taken from left dentate nuclei (0.54 ± 0.14) relative to the right (1.00 ± 0.26; n = 6; *P < 0.05). (C) Expression of ATXN1 mRNA levels from tissue punches taken from the left inferior olivary complex (0.71 ± 0.06) relative to the right (0.99 ± 0.09; n = 6; *P < 0.05). (D) Expression of ATXN1 mRNA levels from tissue punches taken from the right ventral lateral thalamic nuclei (0.69 ± 0.05) relative to the left (1.01 ± 0.03; n = 6; *P < 0.05). Results are shown as whisker plots.
rAAV1.miS1eGFP is well tolerated in non-human primate cerebellum
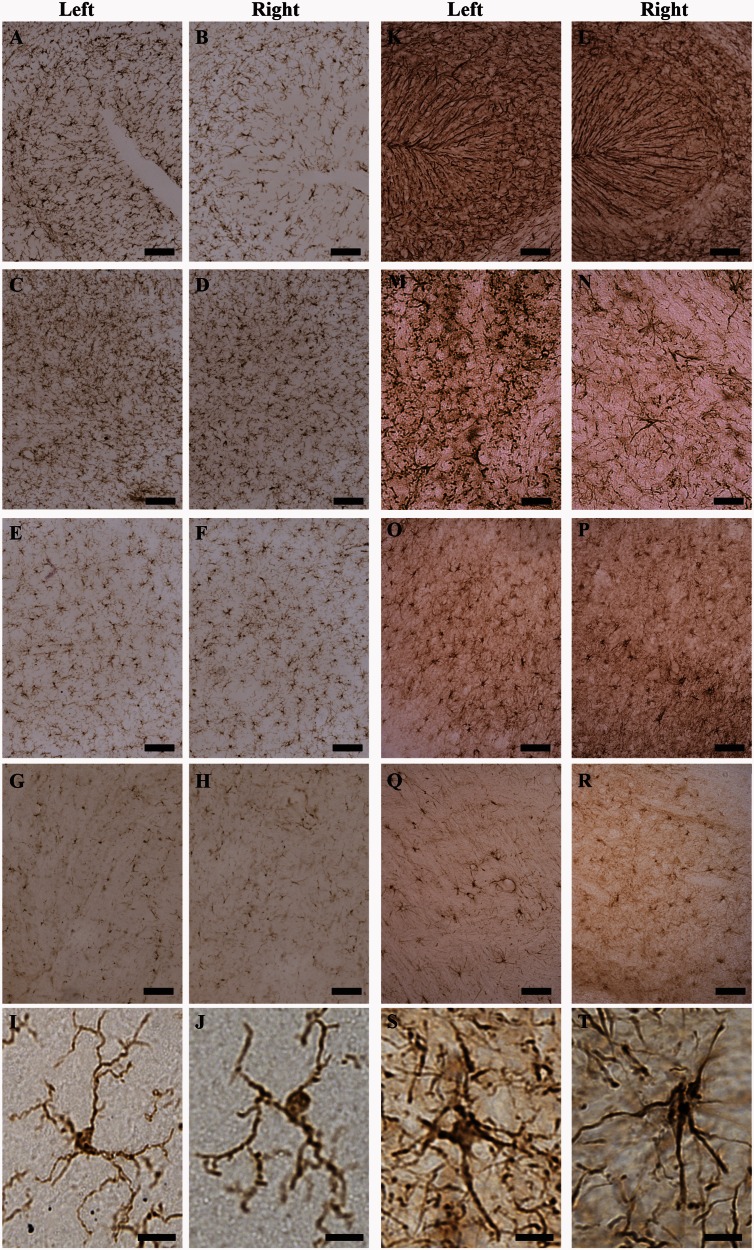
Inhibitory RNAs can cause toxicity in the brain (McBride et al., 2008) even when expressed in the context of an artificial miRNA (Monteys et al., 2014). Thus, we evaluated microglial and astroglial markers to gauge inflammatory responses after rAAV1.miS1eGFP delivery. Eight weeks post-injection, microglial activation, assessed by Iba1 immunohistochemistry, showed a slight enhancement in immunoreactivity in the left (Fig. 6A) versus the right (Fig. 6B) cerebellar cortex. There were no notable differences between the left and right hemispheres of the DCN (Fig. 6C and D), brainstem (Fig. 6E and F) or thalamus (Fig. 6G and H). Iba1-positive cells in DCNs from injected versus uninjected regions are shown for reference (Fig. 6I and J). Astrogliosis, assessed by GFAP immunohistochemistry, showed slight enhancement in immunoreactivity in the left (Fig. 6K) versus the right (Fig. 6L) cerebellar cortex and in the left (Fig. 6M) versus the right (Fig. 6N) DCN. There were no noticeable differences between the left and right hemisphere of the brainstem (Fig. 6O and P) or thalamus (Fig. 6Q and R). GFAP-positive cells from left and right DNCs (site of injection) are included for comparison (Fig. 6S and T). No significant differences in GFAP mRNA levels were found in punches harvested from these same areas (DCN, cerebellar cortex, inferior olive, and thalamus) as measured by quantitative real-time PCR, another sensitive indicator of glial activation (Supplementary Fig. 3).
Figure 6.
rAAV1.miS1eGFP is tolerated in non-human primate cerebellum. (A–J) IBA1 immunostaining in the left and right cerebellar cortex (A and B); left and right DCN (C and D); left and right inferior olivary complex (E and F); and left and right thalamus (G and H). Scale bars = 100 µm. Ramified microglia from left (I) and right (J) hemispheres. Scale bars = 50 µm. (K–T) GFAP immunostaining in the left and right cerebellar cortex (K and L); left and right DCN (M and N); left and right inferior olivary complex (O and P); and left and right thalamus (Q and R). Scale bars = 100 µm. Astroglia at higher magnification from left (S) and right (T) hemispheres. Scale bars = 50 µm.
Discussion
We previously demonstrated that injection of vectors expressing inhibitory RNAs targeted to Atxn1 mRNA into the DCN of SCA1 mice models improved behavioural and neuropathological phenotypes (Keiser et al., 2013, 2014). However, the technical feasibility and safety of this approach in primate brain was unknown, as was the biodistribution and extent of silencing that could be achieved. We show that rAAV1.miS1eGFP into the DCN of adult rhesus macaques results in broad distribution of transgene expression and reduction of ATXN1 mRNA in the cerebellum and extracerebellar areas affected in SCA1. rAAV1 was transported retrograde to Purkinje cells and neurons of the inferior olive, similar to what is seen in rodents after DCN injection (Keiser et al., 2014). rAAV1 vector is also transported anterograde allowing for expression in the presynaptic axonal projections from the DCN.
In earlier work, Bu and colleagues (2012) injected rAAV1 into the motor cortex, occipital cortex, striatum, thalamus, hippocampus, and the DCN to test an approach for overall CNS gene therapy. Our work is consistent with their findings from their DCN injections. We expanded on their approach by performing injections encompassing the medial, fastigial and dentate nuclei. As the extent of transduction from these approaches was not known, we quantified Purkinje cell transduction, and found 33% of Purkinje cells to be transduced on average, with the anterior lobules transduced more readily than posterior lobules. As the superior medial cerebellum (anterior lobe) is more affected than the lateral cortex in SCA1, these results are encouraging for translating this work to the clinic for patient treatment (Robitaille et al., 1995, 1997; Donato et al., 2012).
Prior work in mice models of SCA1 indicate that between 10–30% Purkinje cell transduction may be sufficient for clinical efficacy. We found significant benefit from transducing ∼10% of Purkinje cells after injection of rAAVs expressing a short hairpin RNA targeting human ATXN1 mRNA into the cerebellar cortex of a SCA1 transgenic mouse model (Xia et al., 2004). In later work in this same model, we found ∼30% reduction of mutant ATXN1 mRNA from directed delivery to the DCN, again with significant improvements in disease readouts. Finally, using a knock-in model but the same gene delivery approach, we found 27% reduction in murine Atxn1 mRNA; however, the tissue lysates in that work included transduced Purkinje cells and non-transduced cells in the granule and molecular layers. Similarly, we find ∼31% reduction in rhesus ATNX1 mRNA levels in cerebellar punches that include the molecular layer (Purkinje cell bodies, their dendrites and other cells within the molecular layer) and cells within the granule cell layer. These data indicate that infusions of rAAV into these deep cerebellar nuclei will provide sufficient coverage and knockdown of mutant ATXN1 mRNA levels in the cerebellar cortex for clinical benefit in patients.
In addition to the cerebellar cortex, the dentate and inferior olive are among the most severely affected regions (Robitaille et al., 1995; Donato et al., 2012), as well as the ventral lateral thalamic nuclei (Rub et al., 2012, 2013). In support of SCA1 therapy, we see ∼50% silencing in the DCN with transduction of most cells, and ∼30% silencing in the inferior olive and ventral lateral thalamus. Thus this delivery paradigm affords sufficient coverage of areas affected in SCA1, with reduction of ATXN1 mRNA (or mouse Atxn1 mRNA) to levels shown to be efficacious in mouse models (Xia et al., 2004; Keiser et al., 2013, 2014).
rAAV delivery is known to elicit a cellular immune response (Louis Jeune et al., 2013). This could be problematic if readministration was required. However, work by Bankiewicz and colleagues (Bankiewicz et al., 2006; Hadaczek et al., 2010) has shown that rAAV vector expression persists for up to 8 years in non-human primates and others have seen long lasting expression in dogs (Acland et al., 2005; Stieger et al., 2009), making readministration unlikely. Moreover, a recent publication demonstrated the safety and tolerability of readministering rAAVs to patients; Bennett et al. (2012) showed that readministration of rAAV to the retina contralateral to the one treated 1.7 to 3.3 years earlier was not only tolerated, but that there was no detrimental effect to the earlier beneficial effect from the eye treated first. In the case of delivery of rAAV to brains of primates with pre-existing immunities, McBride et al. (2011) have analysed the effects of neutralizing antibodies to rAAV1 in non-human primate on the volume of transduction in striatum. They saw no significant difference in the amount of striatal transduction in animals with higher levels of neutralizing antibody versus animals that had lower neutralizing antibody levels (McBride et al., 2011). Similarly, in this work, we saw no correlation between pre-existing neutralizing antibody titres and knockdown efficiency or biodistribution (Supplementary Fig. 2).
rAAV delivery for gene silencing in non-human primates has to date been reported for cerebral but not cerebellar targets. McBride et al. (2011) and Grondin et al. (2012) showed that 6 weeks or 6 months reduction of huntingtin in rhesus macaques was well tolerated. Other studies in rodents have shown that inhibitory RNAs, either in the form of short hairpin RNAs or miRNAs, can induce off-target silencing or cause toxicity through other mechanisms (McBride et al., 2008; Monteys et al., 2014), with associated glial activation and in some cases, cell loss (Gorbatyuk et al., 2010). In the present study, ATXN1 mRNA levels were reduced from 30–50% depending on the region examined, and both quantitative and qualitative observations using inflammatory markers for astroglia and microglia suggest that this reduction is well tolerated. Additionally, enhanced GFP can be toxic (Samaranch et al., 2014), which may underlie the modest qualitative increase in GFAP- and Iba1-immunoreactivity in the cerebellar cortex in the injected, versus the uninjected hemispheres. While there were no significant differences in GFAP mRNA levels, further studies with vectors lacking enhanced GFP will be important to fully assess the safety of our approach. Importantly, removing enhanced GFP from the expression cassette does not impact the potency of ATXN1 knockdown (Supplementary Fig. 4).
Overall, our data demonstrate the efficacy and tolerability of rAAV1.miS1eGFP administration to the DCN of non-human primate and the data are supportive of clinical application of this gene therapy in SCA1.
Supplementary Material
Acknowledgements
The authors would like to thank the University of Iowa Gene Transfer Vector Core and The Children’s Hospital of Philadelphia Research Vector Core for the viruses used in this work. The authors would also like to thank Scott Muller for his excellent technical assistance and Ryan Boudreau for his help in designing the miS1. We also acknowledge the support of Dr Harry Orr in raising funding for this study.
Glossary
Abbreviations
- DCN
deep cerebellar nuclei
- GFP
green fluorescent protein
- miRNA
microRNA
- rAAV
recombinant adeno-associated virus
- RNAi
RNA interference
- SCA1
spinocerebellar ataxia type 1
Funding
This work was funded by the National Institutes of Health, grant NS045667, the University of Iowa Roy J. Carver Trust, the National Ataxia Foundation, The Research Institute at The Children’s Hospital of Philadelphia and the University of Minnesota Foundation.
Supplementary material
Supplementary material is available at Brain online.
References
- Acland GM, Aguirre GD, Bennett J, Aleman TS, Cideciyan AQV, Bennicelli J, et al. Long-term restoration of rod and cone vision by single dose rAAV-mediated gene transfer to the retina in a canine model of childhood blindness. Mol Ther 2005; 12: 1072–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfi S, Servadio A, Chung MY, Kwiatkowski TJ, Jr, McCall AE, Duvick LA, et al. Identification and characterization of the gene causing type 1 spinocerebellar ataxia. Nat Genet 1994; 7: 513–20. [DOI] [PubMed] [Google Scholar]
- Bankiewicz KS, Forsayeth J, Eberling JL, Sanchez-Pernaute R, Pivirotto P, Bringas J, et al. Long-term clinical improvement in MPTP-lesioned primates after gene therapy with AAV-hAADC. Mol Ther 2006; 14: 564–70. [DOI] [PubMed] [Google Scholar]
- Basu S, Aryan A, Overcash JM, Samuel GH, Anderson MA, Dahlem TJ, et al. Silencing of end-joining repair for efficient site-specific gene insertion after TALEN/CRISPR mutagenesis in Aedes aegypti. Proc Natl Acad Sci USA 2015; 112: 4038–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett J, Ashtari M, Wellman J, Marshall KA, Cyckowski LL, Chung DC, et al. AAV2 gene therapy readministration in three adults with congenital blindness. Sci Transl Med 2012; 4: 120ra115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman AB, Lam YC, Jafar-Nejad P, Chen HK, Richman R, Samaco RC, et al. Duplication of Atxn1l suppresses SCA1 neuropathology by decreasing incorporation of polyglutamine-expanded ataxin-1 into native complexes. Nat Genet 2007; 39: 373–9. [DOI] [PubMed] [Google Scholar]
- Bu J, Ashe KM, Bringas J, Marshall J, Dodge JC, Cabrera-Salazar MA, et al. Merits of combination cortical, subcortical, and cerebellar injections for the treatment of Niemann-Pick disease type A. Mol Ther 2012; 20: 1893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetanovic M, Rooney RJ, Garcia JJ, Toporovskaya N, Zoghbi HY, Opal P. The role of LANP and ataxin 1 in E4F-mediated transcriptional repression. EMBO Rep 2007; 8: 671–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato SD, Mariotti C, Taroni F. Spinocerebellar ataxia type 1. Handb Clin Neurol 2012; 103: 399–421. [DOI] [PubMed] [Google Scholar]
- Duyckaerts C, Durr A, Cancel G, Brice A. Nuclear inclusions in spinocerebellar ataxia type 1. Acta Neuropathol 1999; 97: 201–7. [DOI] [PubMed] [Google Scholar]
- Gorbatyuk OS, Li S, Nash K, Gorbatyuk M, Lewin AS, Sullivan LF, et al. In vivo RNAi-mediated alpha-synuclein silencing induces nigrostriatal degeneration. Mol Ther 2010; 18: 1450–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondin R, Kaytor MD, Ai Y, Nelson PT, Thakker DR, Heisel J, et al. Six-month partial suppression of Huntingtin is well tolerated in the adult rhesus striatum. Brain 2012; 135(Pt 4): 1197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadaczek P, Eberling JL, Pivirotto P, Bringas J, Forsayeth J, Bankiewicz KS. Eight years of clinical improvement in MPTP-lesioned primates after gene therapy with AAV2-hAADC. Mol Ther 2010; 18: 1458–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiser MS, Boudreau RL, Davidson BL. Broad therapeutic benefit after RNAi expression vector delivery to deep cerebellar nuclei: implications for spinocerebellar ataxia type 1 therapy. Mol Ther 2014; 22: 588–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiser MS, Geoghegan JC, Boudreau RL, Lennox KA, Davidson BL. RNAi or overexpression: alternative therapies for Spinocerebellar Ataxia Type 1. Neurobiol Dis 2013; 56: 6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole R, Krainer AR, Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov 2011; 11: 125–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai S, O'Callaghan B, Zoghbi HY, Orr HT. 14‐3‐3 Binding to ataxin-1(ATXN1) regulates its dephosphorylation at Ser-776 and transport to the nucleus. J Biol Chem 2011; 286: 34606–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YC, Bowman AB, Jafar-Nejad P, Lim J, Richman R, Fryer JD, et al. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell 2006; 127: 1335–47. [DOI] [PubMed] [Google Scholar]
- Lambrechts D, Carmeliet P. VEGF at the neurovascular interface: therapeutic implications for motor neuron disease. Biochim Biophys Acta 2006; 1762: 1109–21. [DOI] [PubMed] [Google Scholar]
- Lim J, Crespo-Barreto J, Jafar-Nejad P, Bowman AB, Richman R, Hill DE, et al. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature 2008; 452: 713–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis Jeune V, Joergensen JA, Hajjar RJ, Weber T. Pre-existing anti-adeno-associated virus antibodies as a challenge in AAV gene therapy. Hum Gene Ther Methods 2013; 24: 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci USA 2008; 105: 5868–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride JL, Pitzer MR, Boudreau RL, Dufour B, Hobbs T, Ojeda SR, et al. Preclinical safety of RNAi-mediated HTT suppression in the rhesus macaque as a potential therapy for Huntington's disease. Mol Ther 2011; 19: 2152–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteys AM, Spengler RM, Dufour BD, Wilson MS, Oakley CK, Sowada MJ, et al. Single nucleotide seed modification restores in vivo tolerability of a toxic artificial miRNA sequence in the mouse brain. Nucleic Acids Res 2014; 42: 13315–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison DK. The 14‐3‐3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol 2009; 19: 16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HT. Polyglutamine neurodegeneration: expanded glutamines enhance native functions. Curr Opin Genet Dev 2012; 22: 251–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HT, Chung MY, Banfi S, Kwiatkowski TJ, Jr, Servadio A, Beaudet AL, et al. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat Genet 1993; 4: 221–6. [DOI] [PubMed] [Google Scholar]
- Ousterout DG, Kabadi AM, Thakore PI, Majoros WH, Reddy TE, Gersbach CA. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun 2015; 6: 6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oz G, Vollmers ML, Nelson CD, Shanley R, Eberly LE, Orr HT, et al. In vivo monitoring of recovery from neurodegeneration in conditional transgenic SCA1 mice. Exp Neurol 2011; 232: 290–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robitaille Y, Lopes-Cendes I, Becher M, Rouleau G, Clark AW. The neuropathology of CAG repeat diseases: review and update of genetic and molecular features. Brain Pathol 1997; 7: 901–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robitaille Y, Schut L, Kish SJ. Structural and immunocytochemical features of olivopontocerebellar atrophy caused by the spinocerebellar ataxia type 1 (SCA-1) mutation define a unique phenotype. Acta Neuropathol 1995; 90: 572–81. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lebron E, Liu G, Keiser M, Belhke MA, Davidson BL. Altered Purkinje cell miRNA expression and SCA1 pathogenesis. Neurobiol Dis 2013; 54: 456–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rub U, Burk K, Timmann D, den Dunnen W, Seidel K, Farrag K, et al. Spinocerebellar ataxia type 1 (SCA1): new pathoanatomical and clinico-pathological insights. Neuropathol Appl Neurobiol 2012; 38: 665–80. [DOI] [PubMed] [Google Scholar]
- Rub U, Schols L, Paulson H, Auburger G, Kermer P, Jen JC, et al. Clinical features, neurogenetics and neuropathology of the polyglutamine spinocerebellar ataxias type 1, 2, 3, 6 and 7. Prog Neurobiol 2013; 104: 38–66. [DOI] [PubMed] [Google Scholar]
- Samaranch L, San Sebastian W, Kells AP, Salegio EA, Heller G, Bringas JR, et al. AAV9-mediated expression of a non-self protein in nonhuman primate central nervous system triggers widespread neuroinflammation driven by antigen-presenting cell transduction. Mol Ther 2014; 22: 329–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra HG, Byam CE, Lande JD, Tousey SK, Zoghbi HY, Orr HT. Gene profiling links SCA1 pathophysiology to glutamate signaling in Purkinje cells of transgenic mice. Hum Mol Genet 2004; 13: 2535–43. [DOI] [PubMed] [Google Scholar]
- Serra HG, Duvick L, Zu T, Carlson K, Stevens S, Jorgensen N, et al. RORalpha-mediated Purkinje cell development determines disease severity in adult SCA1 mice. Cell 2006; 127: 697–708. [DOI] [PubMed] [Google Scholar]
- Servadio A, Koshy B, Armstrong D, Antalffy B, Orr HT, Zoghbi HY. Expression analysis of the ataxin-1 protein in tissues from normal and spinocerebellar ataxia type 1 individuals. Nat Genet 1995; 10: 94–8. [DOI] [PubMed] [Google Scholar]
- Stieger K, Schroeder J, Provost N, Mendes-Madeira A, Belbellaa B, Le Meur G, et al. Detection of intact rAAV particles up to 6 years after successful gene transfer in the retina of dogs and primates. Mol Ther 2009; 17: 516–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramony SH, Ashizawa T. Spinocerebellar Ataxia Type 1. Gene Rev [Internet] Seattle (WA); University of Washington, Seattle; 1993–2015. ISSN: 2372–0697. Submitted 1998 [Updated 2014 Jul 03]. [Google Scholar]
- Tsuda H, Jafar-Nejad H, Patel AJ, Sun Y, Chen HK, Rose MF, et al. The AXH domain of Ataxin-1 mediates neurodegeneration through its interaction with Gfi-1/Senseless proteins. Cell 2005; 122: 633–44. [DOI] [PubMed] [Google Scholar]
- Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, et al. Targeted genome editing across species using ZFNs and TALENs. Science 2011; 333: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H, Mao Q, Eliason SL, Harper SQ, Martins IH, Orr HT, et al. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat Med 2004; 10: 816–20. [DOI] [PubMed] [Google Scholar]
- Xia H, Mao Q, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol 2002; 20: 1006–10. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY, Orr HT. Pathogenic mechanisms of a polyglutamine-mediated neurodegenerative disease, spinocerebellar ataxia type 1. J Biol Chem 2009; 284: 7425–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Duvick LA, Kaytor MD, Berlinger MS, Zoghbi HY, Clark HB, et al. Recovery from polyglutamine-induced neurodegeneration in conditional SCA1 transgenic mice. J Neurosci 2004; 24: 8853–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.