Abstract
Pancreatic cancer is the fourth leading cause of cancer deaths in the US. Less than 6% of patients survive beyond the fifth year due to inadequate early diagnostics and ineffective treatment options. Our lab has developed an allogeneic, granulocyte-macrophage colony-stimulating factor (GM-CSF) secreting pancreatic cancer vaccine (GVAX) that has been tested in phase II clinical trials. Here, we employed a novel Serum Antibodies based SILAC-Immunoprecipitation (SASI) approach to identify proteins that elicit an antibody response after vaccination. The SASI approach utilizes immunoprecipitation with patient derived antibodies that is coupled to quantitative stable isotope–labeled amino acids in cell culture (SILAC). Using mass spectrometric analysis, we identified more than 150 different proteins that induce an antibody response after vaccination. The regulatory subunit 12A of protein phosphatase 1 (MYPT1 or PPP1R12A), regulatory subunit 8 of the 26S proteasome (PSMC5), and the transferrin receptor (TFRC) were shown to be pancreatic cancer associated antigens recognized by post-vaccination antibodies in the sera of patients with favorable disease-free survival after GVAX therapy. We further interrogated these proteins in over 80 GVAX-treated patients’ pancreases and uniformly found a significant increase in the expression of MYPT1, PSMC5, and TFRC in neoplastic compared to non-neoplastic pancreatic ductal epithelium. We show that the novel SASI approach can identify antibody targets specifically expressed in patients with improved disease-free survival after cancer vaccine therapy. These targets need further validation to be considered as possible pancreatic cancer biomarkers.
Keywords: Pancreatic cancer, cancer vaccine, proteomics, tumor immunology, tumor antigen
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDA) is notably the most aggressive and debilitating cancer with only 1% to 4% of patients having an overall survival of more than 5 years (1, 2). These low survival statistics are due to inadequate early diagnostics, and resistance to current chemoradiation therapies (3-5). Thus, alternative screening and treatment approaches are urgently needed for PDA.
We developed an allogeneic, granulocyte-macrophage colony-stimulating factor (GM-CSF)-secreting pancreatic cancer vaccine (GVAX), which has completed phase II clinical trials (6). A functional genomic approach identified a pancreatic cancer antigen, mesothelin, recognized by T cells (7). We reported the induction of mesothelin-specific T-cell responses only in patients with a disease-free survival (DFS) > 3 years, suggesting that the vaccine induces immunologically relevant T-cell responses (6). However, finding T cell antigens is limited by the need for patient-specific HLA reagents (8).
To circumvent this limitation, we developed a high-throughput, HLA-independent method to uncover serum antibodies induced by GVAX therapy in patients with extended disease-free survival. Antibodies may directly or indirectly remove malignant cells via opsonization, antigen presentation to T-cells, and by initiating natural killer cell or complement dependent cell toxicity (9). Examining antibody responses can also aid in the identification of T-cell antigens and T-cell responses that could be potentially useful as a predictive markers for survival or response to therapy. For example, melanocyte differentiation antigen, RAB38/NY-MEL-1, was initially identified by studying antibody responses in melanoma patients using the serological screening of cDNA expression library (SEREX) methodology. Spontaneous CD8+ T-cell responses are also detected to this antigen (10). Comparing pre- and post-vaccination western blots of PDA cell line lysates, separated by two-dimensional electrophoresis (2-DE) followed by mass-spectrometry analysis, led to the discovery of annexin A2 as a potential therapeutic target in PDA (11). This discovery has led to a deeper insight into PDA progression and metastasis and is rapidly being translated into annexin A2-based monoclonal antibody therapy for PDA. Thus, the serum of vaccinated patients with a favorable survival profile holds promise for identifying therapeutic targets in PDA.
The major drawbacks to current sera-based screening approaches are the inability to identify cell membrane proteins, and the low throughput and semi-quantitative readouts. We therefore developed a Serum Antibodies based SILAC-Immunoprecipitation (SASI) approach to identify proteins that elicit an antibody response after vaccination. This method takes advantage of stable isotope labeling of amino acids (SILAC) in PDA cell culture, immunoprecipitation with patient-derived antibodies and mass spectrometric analysis. The result is the subtraction of prevaccine sera from post-vaccine sera, providing a means to specifically study only vaccine-induced antibody responses.
This approach identified regulatory subunit 12A of protein phosphatase 1 (MYPT1 or PPP1R12A), regulatory subunit 8 of the 26S proteasome (PSMC5), and the transferrin receptor (TFRC) as targets of post-vaccination antibodies in the sera of patients that received GVAX and demonstrated a favorable disease-free survival. We further analyzed MYPT1, PSMC5, and TFRC expression in two independent sets of GVAX-treated patients’ normal and malignant pancreatic tumor specimens. We found significant expression of these proteins in malignant compared to normal duct epithelium. The antibody responses detected to these proteins in patients with an improved disease-free survival suggests that targeting of these proteins could have antitumor potential. Overall, our data demonstrates that this type of SASI approach can selectively identify new candidate biomarkers for screening and developing better targeted therapies.
Materials and Methods
Patients, serum, and tissue samples
Patients (N = 60)were enrolled in a phase II study of an allogeneic GM-CSF–secreting whole cell pancreatic cancer vaccine in compliance with the Johns Hopkins Medical Institution Institutional Review Board (IRB)-approved J9988 protocol (6). Blood samples were collected pre-vaccination, 14 days after 1st vaccination, and 28 days after each subsequent vaccination. Sera was collected by centrifugation, aliquoted, and stored at −80°C. Pancreatic tumor tissue samples were collect from patients at the time of pancreaticoduodenectomy and prior to vaccination. We also obtained tissue samples from a neoadjuvant study, J0810 for validation purposes (12).
Antibody purification
Antibodies were purified from pre- and post-vaccination sera using a protein G column (GE Healthcare) as per manufacturer's protocol. Quantification of purified antibodies was done with a NanoDrop spectrophotometer (Thermo Fisher Scientific).
SASI sample preparation
Panc 10.05 cells (American Type and Culture Colletion (ATCC) Manassas VA line CRL-2574) were developed (13) in Dr. E.M. Jaffee's lab and the cells were authenticated using short tandem repeat analysis in the Johns Hopkins Genetic Resource Core Facility at 6 month intervals. Panc 10.05 cells were grown in either light (12C6-Lys, 12C6-Arg) or heavy (13C6-Lys, 13C6-Arg) RPMI 1640 media containing 10% fetal bovine serum and antibiotics in a humidified incubator at 37°C with 5% CO2. Stable isotope containing amino acids, 13C6-arginine and 13C6-lysine, were purchased from Cambridge Isotope Laboratories. Arginine and lysine-free RPMI 1640 media, fetal bovine serum (FBS) and antibiotics (penicillin and streptomycin) were purchased from Invitrogen. The light and heavy cells were washed with phosphate buffered saline and harvested using M-PER buffer (Thermo Fisher Scientific) in the presence of cocktail protease inhibitors (Thermo Fisher Scientific). Protein was quantified using the Lowry method.
Immunoprecipitation for mass spectrometry
Equal amounts (10 mg) of light and heavy cell lysates were incubated with purified pre- and post-vaccination antibodies at 4°C overnight, respectively. On the following day, the two sets of lysate:antibody mixture were each incubated with protein G beads (Invitrogen) and washed using M-PER buffer. The immunoprecipitates were eluted by boiling in NuPAGE® LDS sample buffer (Invitrogen). The light and heavy eluted lysates were mixed 1:1. The mixture was concentrated and resolved by 10% SDS-PAGE. The gel was stained using a Coomassie dye staining kit (Invitrogen) prior to in-gel tryptic digestion for preparation of LC-MS/MS samples
Liquid chromatography tandem mass spectrometry and data analysis
In-gel digestion and LC-MS/MS analysis were done as described (14). The stained gel was excised into 18 bands and each band was destained in 40mM ammonium bicarbonate/40% acetonitrile solution. The samples were reduced with 5mM dithiothreitol/20% acetonitrile solution, alkylated with 10mM iodoacetamide and digested with trypsin. Sequencing grade modified porcine trypsin was purchased from Promega. The peptides were extracted, desalted, dried and reconstituted in 0.1% formic acid. The peptides were analyzed by reversed phase liquid chromatography tandem mass spectrometry (LC-MS/MS). Briefly, the peptides were separated using an on-line reverse phase nano high-performance liquid chromatography using a C18. Peptide samples related to patients 1 and 3 were analyzed on EASY-nLC (Thermo) coupled online to an LTQ-Orbitrap Elite mass spectrometer (Thermo) while peptide samples related to patient 6 was analyzed on the Eksigent Nano 2D high-performance liquid chromatography (HPLC) pumping system (Eksigent interfaced directly with an LTQ-Orbitrap XL mass spectrometer (Thermo Electron) Isolated proteins from each band were identified using an automated database search algorithm, MASCOT, within the Proteome Discoverer software platform (Thermo Electron) and processed Perseus software. Our data was searched at a mass tolerance of 10 ppm for precursor peptide ions and with carbamidomethylation of cysteine as a fixed modification and oxidation of methionine as a variable modification (14). In addition to SILAC labels (6 Da) on arginine and lysine as variable modifications. The proteolytic enzyme indicated was trypsin and we allowed up to two missed cleavage events.
Mass-spectrometry data validation
Panc 10.05 cells grown in light RPMI1640 media were lysed in M-PER buffer supplemented with protease inhibitor cocktail. The lysate was immunoprecipitated with either the pre- or post-vaccination purified antibodies using protein G beads. The immunoprecipitates were eluted by boiling in NUPAGE LDS sample buffer and resolved on a NuPAGE 4-12% Bis-Tris gel (Invitrogen). Proteins in the gel were transferred onto nitrocellulose membrane using a semi-dry apparatus (Invitrogen). The membrane was blocked in 5% bovine serum albumin (BSA, Invitrogen) in 0.1% Tween 20-PBS (PBS-T) buffer for 1 hour at room temperature and probed with the relevant primary antibody overnight at 4°C. Antibodies against galectin-3 (sc-19283), E3 ubiquitin protein ligase (sc-9561), Mesencephalic astrocyte-derived neurotrophic factor (sc-34560), Epidermal growth factor receptor kinase substrate 8-like protein 2 (sc-100722), Calpain-1 (sc-81171) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The membrane was incubated with the corresponding peroxidase conjugated secondary antibodies (Sigma) and then ECL Western Blotting Detection Reagents (GE Healthcare) was used for developing.
Western blot for detecting antibody responses in patients
Purified recombinant proteins, PSMC5 (TP301251), MYPT1 (TP323540) and TFRC (TP300980) expressed in human HEK293 cells were purchased from Origene. One microgram of purified protein was denatured by boiling in SDS-PAGE sample buffer and resolved on a NuPAGE 4-12% Bis-Tris gel (Invitrogen). Proteins in the gel were transferred onto nitrocellulose membrane using a semi-dry apparatus (Invitrogen). The membrane was cut into individual lanes and was blocked in 5% bovine serum albumin (BSA, Invitrogen) in 0.1% Tween 20-PBS (PBS-T) buffer for 1 hour at room temperature. After blocking, each individual lane was probed with either pre-vaccination or post-vaccination serum at 1:1000 dilution. A lane was used as a control and probed with mouse anti-FLAG antibody overnight at 4°C. The membrane was incubated with the peroxidase-conjugated secondary antibodies, goat anti-human IgG antibody (Sigma, A8419) for patient serum lanes or rabbit anti-mouse IgG (Sigma, A9044) for control lane. ECL Western Blotting Detection Reagents (GE Healthcare) was used for 1 minute at room temperature for developing. Chi squared analysis was used to test for statistical significance.
Immunohistochemistry
Staining protocols were optimized using the pancreatic cancer cell line Panc 10.05 as positive control and pancreatic tissue as negative control. Immunohistochemistry (IHC) was performed on formalin-fixed paraffin-embedded 5μm thick sections of the available pancreatic tumor tissue samples and the tissue microarrays. The tissue samples of the patients enrolled in the study were obtained from the Department of Pathology at Johns Hopkins Medical Institutions tissue archive. The tissue microarrays (TMA) were constructed from different types of malignant tumors and their companion normal tissues (15). The diagnoses were verified by evaluation of the histopathological and immunohistochemical stains by two reference pathologists (EDT and RAA).
A standard IHC protocol was applied using Bond-Leica autostainer (Leica Microsystems). Briefly, tissue sections were baked for 20 minutes at 65°C followed by deparaffinization and primary antibody incubation at optimal conditions. Bond polymer detection system was applied to develop the reaction. 3’,3’ diaminobenzidin (DAB) chromogen-substrate was utilized for visualization of reaction as per manufacturer's instructions (Leica Microsystems). All sections were then counterstained with hematoxylin, dehydrated and cover slipped. Antibody details: anti-PSMC5 from Rabbit 1:150 (Sigma), anti-PPP1R12A Rabbit (Sigma), anti-TFRC mouse clone H68.4) 1:2000 (Invitrogen)
Immunohistochemical stained pancreatic malignant and companion normal tissue were independently scored by two pathologists (E.D.T. and R.A.A.), blinded to study outcomes. Stained TMA slides were scored after conversion to digital images in a single-file pyramidal tiled TIFF format using Aperio® (Leica Biosystems) software and an acquisition magnification of 20X. For large tissue pancreatic cancer tissue sections five to ten representative 20x magnification fields were scored with a compound light microscope. Scoring consisted of the percentage of staining (distribution) in each cellular compartment (membrane, cytoplasm, and nucleus) of malignant and normal cells. The staining intensity was also graded as none (0), weak (1), and strong (2). In preparation for statistical analysis the staining intensity and distribution data for each tissue and TMA core studied were placed into two categories (negative and positive). Natural divisions in the staining data defined negative (<25% distribution with intensity of 0 or 1) and positive (>25% with intensity 1 or 2). All p-values were calculated using a JavaScript calculator using exact McNemar (PDA samples) or corrected Chi-squared or Fishers exact test (TMA).
RESULTS
Design and validation of quantitative proteomic approach
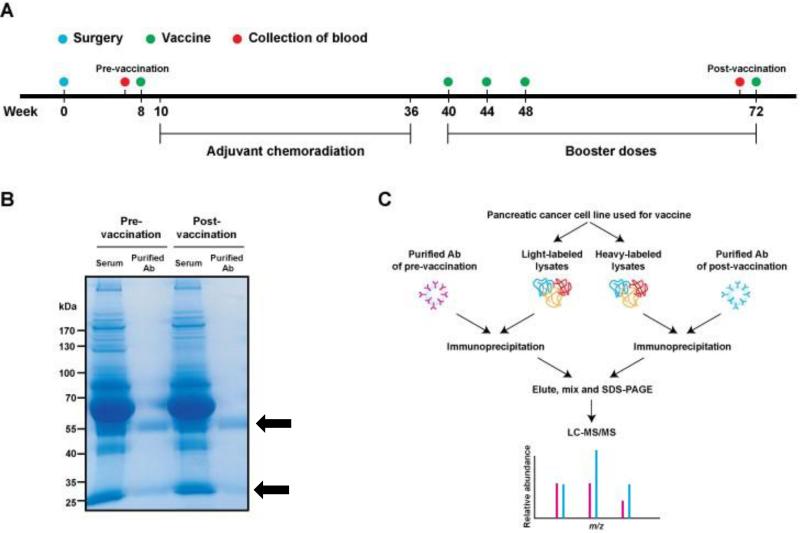
Serum samples used in this study were derived from a phase II, single institution study of 60 pancreatic cancer patients who underwent pancreaticoduodenectomy followed by adjuvant GVAX vaccinations integrated with 5-flourouracil based chemoradiation (6) (Fig. 1A). The patients were divided into 3 groups based on disease free survival (DFS) and the number of GVAX treatments. Group A = DFS > 3 years, five GVAX treatments (n = 12); group B = DFS < 3 years, 3–5 GVAX treatments (n = 21); group C = disease relapse before 2nd GVAX (n = 27). Total serum antibodies from 3 Group A patients (patients 1, 3 and 6) were purified from a protein G affinity column (Fig. 1B) and used to identify the targets of vaccine-induced antibodies.
Figure 1. Overview of the study design and SASI screen.
(A) Vaccination scheme based on Lutz et.al (6). The date of surgery was set as a start date for vaccination schedule (i.e. week 0). First vaccine was administered at week 8 prior to the adjuvant chemoradiation period (week 10 to week 36). Three consecutive booster injections of the vaccine were carried out at 40th, 44th and 48th weeks and then, last vaccine was administered at 72nd week. Blood samples collected before the first injection and before the last injection were used for the SASI approach. (B) Purification of serum antibodies. Serum antibodies from pre- and post-GVAX of patients with favorable DFS > 3 years were isolated with a protein G affinity column. Coomassie blue staining of the SDS-PAGE gel shows high purity of antibody fragments (black arrows) from serum samples and no significant difference was observed in the antibody amounts between pre- and post-vaccination serum samples. (C) Experimental scheme. One of the GVAX pancreatic cancer cell line was labeled by the SILAC method to produce light-labeled cell lysates and heavy-labeled cell lysates, each of which was then incubated with purified pre- and post-vaccination antibodies, respectively, for separate immunoprecipitations. Immunoprecipitated light- and heavy-labeled proteins were combined and separated on a SDS-PAGE gel. Peptides extracted from the gel were analyzed by LC-MS/MS.
The SASI screen was designed (Fig. 1C) to detect differentially produced antibodies that were present in patient serum before and after GVAX therapy. The proteome, from the Panc 10.05 cell line used in the GVAX vaccine, was differentially labeled using the SILAC technique with “light” (12C) and “heavy” (13C) lysines and arginines (16). Immunoprecipitation of the isotope-labeled proteins with purified patient antibodies, coupled with high resolution and high accuracy mass-spectrometry analysis, identified antibody targets (proteins) with fold-changes in post-versus pre-vaccination patient serum (Fig. 2, A-C). We then validated the SILAC data with western blot analysis. Three proteins, galectin-3, E3 ubiquitin-protein ligase UBR5, and mesencephalic astrocyte-derived neurotrophic factor had an increased antibody response post-vaccination by 15.3, 4.0, and 3.9-fold respectively, whereas two proteins showed a decreased antibody response, calpain-1 (0.4 fold) and epidermal growth factor receptor kinase substrate 8-like protein 2 (0.1 fold). By western blot analysis (Fig. 2D), galectin-3 protein increased more than 15-fold in the post-vaccination blot, whereas E3 ubiquitin-protein ligase UBR5 and mesencephalic astrocyte-derived neurotrophic factor increased by 4-fold. Conversely, calpain-1 expression decreased to 0.4-fold of pre-vaccination amounts after vaccination, and epidermal growth factor receptor kinase substrate 8 (EPS8)-like protein 2 decreased to 0.1-fold. These qualitative western blot results mirrored the trends we observed from the quantitative mass-spectrometry derived SILAC ratios.
Figure 2. Protein identification and validation using the SASI approach.
(A) Distribution of relative abundance of proteins precipitated with antibodies. Three biological experiments (denoted as Patients #1, #3 and #6) were performed, each quantitatively identifying about 800 to 900 proteins differentially precipitated between pre-vaccination and post-vaccination. Only a small fraction of proteins were observed to induce a highly differential antibody response after vaccination, indicating that most of antibodies may still target similar epitopes regardless of vaccine. (B) Pearson correlation. Pair-wise comparison of proteins observed in common showed higher similar pattern as evident by Pearson correlation (0.75, 0.85 and 0.86 for Patients #1 vs #6, #3 vs #6 and #1 vs #6, respectively). (C) A representative MS/MS spectrum. A peptide (VAVNDAHLLQYNHR) derived by trypsin protease from galectin-3 was identified and spectral assignment was shown. (D) Confirmation of the quantitative mass spectrometry (MS) results. Five molecules detected by the quantitative MS were used for the Western blot analysis. Pre- and post-vaccination antibodies of patient 6 (also used for SASI screen) with favorable outcome (DFS > 3 years) were used for immunoprecipitation. The precipitate was separated by SDS-PAGE followed by western blot using antibodies against the indicated proteins. The fold-change as detected by mass spectrometry in the SASI screen are shown to the right of each blot and correlates well with the qualitative results in the western blot, confirming that the MS data are all in keeping with the orthogonal method.
Identification of proteins by the SASI approach
The pre- and post-vaccination serum samples from three group A patients (GVAX responders) were used for the SASI screen. Patients 1, 3, and 6 had a prolonged DFS status (65, 61, and 43 months, respectively) and were alive at the end of the study. We identified 1280 unique proteins (Fig. 2, A-C) with a range of change from 16-fold increase to a 10-fold decrease in post-vaccination serum. Of the identified proteins 799 (62%) were found in at least two samples and 472 (37%) were found in all three samples (Supplemental Fig. S1). Annexin A2 protein, previously identified as biologically relevant PDA target (11), showed a median 1.3-fold increase and a maximum fold-change of 1.6. Thus, we narrowed our list (Supplemental Fig. S2) from 1280 proteins to 31 proteins (Supplemental Fig. S2) by employing annexin A2-based thresholds and other rigorous selection criteria. Of the 31 proteins, three proteins, galectin-3, annexin A2, and pyruvate kinase were identified previously by a 2-DE proteomic approach (8, 11) and are currently under investigation for their role in PDA pathogenesis and progression. Concordance of these two methods in identifying these three proteins (Table 1) further validated the capability of the SASI screen to uncover biologically relevant PDA targets. Another identified protein was HLA class I histocompatibility antigen (HLA) (Table 1). Antibody response to allogeneic non-self HLA expressed by the Panc 10.05 cell line used in the GVAX therapy is expected and provided another internal positive control, validating the SASI approach once again. The remaining 27 proteins provided us with a list of targets that warranted further study.
Table 1.
List of proteins of interest identified by the SASI approach.
| Protein | Gene symbol | Average fold change (post-vaccination/pre-vaccination) | Patients with increased antibody response (Total of 7 patients) |
|---|---|---|---|
| Galectin 3* | LGALS3 | 11.0 | N/A |
| MHC class I antigen A | HLA-A | 8.2 | N/A |
| 26S proteasome, regulatory subunit 8 | PSMC5 | 4.6 | 6/7 |
| HDGF-2 | HDGFRP2 | 3.2 | 5/7 |
| Prohibitin-2 | PHB2 | 2.4 | 4/7 |
| Retinol dehydrogenase 11 | RDH11 | 2.0 | 5/7 |
| MYPT1 | PPP1R12A | 1.7 | 6/7 |
| Transferrin receptor | TFRC | 1.7 | 6/7 |
| Pyruvate kinase* | PKM2 | 1.7 | N/A |
| Pyrroline-5-carboxylate reductase 1 | PYCR1 | 1.6 | 4/7 |
| 2,4-dienoyl-CoA reductase | DECR1 | 1.4 | 4/7 |
| Annexin A2* | ANXA2 | 1.4 | N/A |
The average fold change is determined by SILAC. The table also shows a summary of western blot analysis examining pre- and post-3rd vaccination antibody response of 7 (of the 12) Group A patients with favorable DFS for 8 proteins. PSMC5, MYPT1, TFRC, RDH11 and HDGFRP2 were selected for further evaluation because they had an increased antibody response in at least 5 out of the 7 patients tested. Asterisk indicates the proteins identified by previous approach. N/A: Not applicable.
PSMC5, MYPT1 and TFRC are antibody targets of the immune response against PDA
We pursued eight (Table 1) out of the 27 shortlisted proteins for validation partially based upon previous PDA serial analysis of gene expression (SAGE) data (17) and reagent availability. Using purified recombinant FLAG-tagged proteins expressed in HEK293 cells (obtained from Origene) for western blot analysis, we examined the pre- and post-third vaccination antibody responses of seven of 12 Group A patients with favorable DFS for eight proteins (Table 1). Anti-FLAG antibody served as a positive control to confirm the presence of the protein in the blot. Five proteins (PSMC5, MYPT1, TFRC, HDGFRP2 and RDH11) of the eight tested showed an increased antibody response post-vaccination in five or more patients studied. Therefore, these proteins were chosen for further analysis by western blot.
Next, we expanded our analysis and evaluated the prevalence of recognition of the 5 proteins by antibody responses pre- and post-vaccination from all 12 Group A patients who responded to the vaccine. We also compared sera from these patients with sera from 12 of the 21 Group B patients that did not respond to the vaccine. The selection of these 12 patients among 21 total in the group with DFS < 3 years was based on the number of vaccinations received. The 12 selected patients received at least three vaccinations, thereby allowing us to best compare the pre- and post-3rd vaccination antibody responses in both the responders and the non-responders.
When we examined the 12 group A patients, PSMC5 elicited an increased antibody response in 8 patients and a decrease in one (Table 2 and Supplemental Fig. S3A). When this was compared with 12 Group B patients (DFS < 3 years) where two patients had an increased antibody response and two patients had a decreased response, this difference was statistically significant (P < 0.05). For the 12 group A patients MYPT1, an increased antibody response was observed in 9 patients and a decrease in one (Table 2 and Supplemental Fig. S3B) compared with 12 Group B patients (DFS < 3 years) where five patients had an increased antibody response and four patients had a decreased antibody response,(P = 0.3). TFRC elicited an increased antibody response in eight of the 12 patients and a decrease in 2 patients (Table 2 and Supplemental Fig. S3C) compared with Group B patients (DFS < 3 years) where two of the 12 patients had an increased antibody response and two patients had a decreased response, this difference was statistically significant (p<0.05).
Table 2.
Correlation of increased antibody response to PSMC5, MYPT1 and TFRC post-vaccination with response to vaccine.
| Number of patients | |||||
|---|---|---|---|---|---|
| Total | Increase | Decrease | p-value* | ||
| PSMC5 | Responders | 12 | 8/12 (67%) | 1/12(8%) | < 0.05 |
| Non-responders | 12 | 2/12 (17%) | 2/12 (17%) | ||
| MYPT1 | Responders | 12 | 9/12 (75%) | 1/12(8%) | = 0.3 |
| Non-responders | 12 | 5/12 (42%) | 4/12 (33%) | ||
| TFRC | Responders | 12 | 8/12 (67%) | 1/12(8%) | < 0.05 |
| Non-responders | 12 | 2/12 (17%) | 2/12 (17%) | ||
Responders are patients with DFS > 3 years (Group A) whereas non-responders are patients with DFS < 3 years (Group B).
Fishers exact test
RDH11 and HDGFRP2 were two proteins identified by SASI that did not show any major difference in antibody response between the responders and non-responders. These proteins were therefore excluded from further analysis.
Increased PSMC5, MYPT1, TFRC tissue expression correlates with PDA development
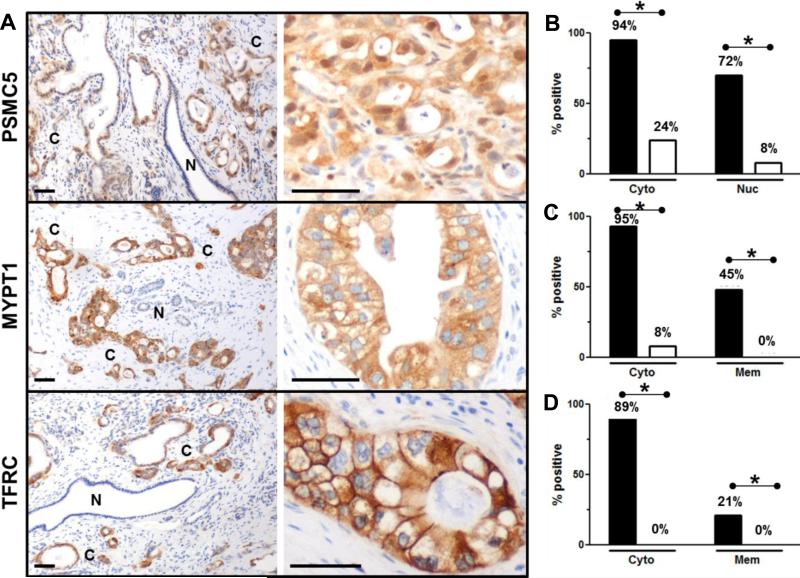
Antibody responses can be induced against oncoproteins due to changes in their expression levels, localization or post-translational modifications (18-21). Available serial analysis of gene expression (SAGE) data suggests that PSMC5, MYPT1, and TFRC may be overexpressed in PDA compared to normal pancreas (17). Therefore, we were interested in establishing the expression levels and tissue location of PSMC5 (Fig. 3, A and B), MYPT1 (Fig. 3, A and C) and TFRC (Fig. 3, A and D) in PDA samples. We analyzed paired PDA tissue samples from 45 of the 60 patients enrolled in the phase II study (J9988, all that was available) (6). The PSMC5 protein is a part of the 26S proteasome typically located in the cytoplasm. We found 94% of PDA express cytoplasmic PSMC5 in malignant compared to 24% non-malignant ductal epithelium (P < 0.01). There was significantly (P < 0.01) more nuclear PSMC5 in malignant (72%) cells compared to non-neoplastic ductal epithelium (8%), with an intermediate amount of expression in pre-neoplastic pancreatic intraepithelial neoplasia (PanIN) lesions. MYPT1, part of the Rho kinase pathway, showed significant expression in cytoplasm of the PDA (95%) compared to normal ductal epithelium (8%) (P < 0.01). MYPT1 expression was also observed in cancer that was invading pancreatic nerves and regional lymph nodes. Membranous MYPT1 (45%) was only observed in PDA and not in normal ductal epithelium (P < 0.01). Similarly, 89% of the PDA expressed cytoplasmic and 21% membranous TFRC (P < 0.01), whereas none of the normal ducts showed any cytoplasmic or membranous TFRC expression, consistent with previous reports (22). The fact that TFRC is a well-studied marker in PDA and other cancers further supports that the SASI approach is well suited in identifying biologically relevant oncoproteins.
Figure 3. Increased tissue expression of PSMC5, MYPT1 and TFRC parallels PDA development.
Representative tissue sections from the J9988 study stained for (A) PSMC5, MYPT1 and TFRC are shown. Normal non-neoplastic duct cells (indicated with an N) and cancer cells (indicated with a C). Scale bars = 50 μm. Staining summary for (B) PSMC5 (n = 45), (C) MYPT1 (n = 44) and (D) TFRC (n = 42). PDA glands (black bars) express PSMC5, MYPT1 and TFRC in both cytoplasm and nucleus/membrane. However, minimal staining is evident in non-neoplastic (white bars) pancreatic ducts. *P < 0.01 Fisher exact test.
To validate our staining results, we used PDA tissue from a neoadjuvant GVAX study (J0810), on which patients received a single GVAX vaccination 2 weeks prior to surgery. The expression of PSMC5 (Supplemental Fig. S4, A and B), MYPT1 (Supplemental Fig. S4, A and C) and TFRC (Supplemental Fig. S4, A and D) largely mirrored those we found in the samples from non-vaccinated patients (J9988 study). It is important to note that the expression data from the two studies differed in the amount of nuclear PSMC5 (J9988 study 72% vs J0810 study 27%) and membrane TFRC (J9988 study 21% vs J0810 study 45%) staining of the cancer cells. It is not clear if these differences represent variation in the patient populations in each of the GVAX study or performance of the antibody
Differential expression of nuclear PSMC5 and cytoplasmic and membrane MYPT1: a new diagnostic tool
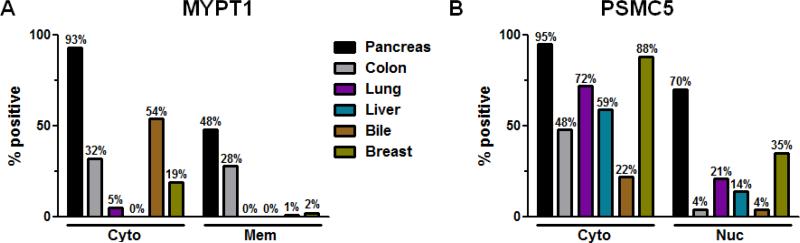
Since this is the first report that PSMC5 and MYPT1 are malignancy-associated markers, we further evaluated their prevalence of expression in a panel of cancers. Tissue microarrays of bile duct, lung, liver, colon, and breast cancers were evaluated for MYPT1 (Fig. 4A) and PSMC5 (Fig. 4B) expression. We found significantly greater cytoplasmic MYPT1 expression in pancreatic cancer (93% P < 0.01) compared to biliary (54%), colon (32%), breast (19%), lung (5%), and liver (0%) cancers. Membranous MYPT1 expression was also expessed significantly more in pancreatic cancers (28%) than all the other cancer types examined. Cytoplasmic PSMC5 expression was seen at relatively higher frequency in all cancer types examined except biliary (22%) cancers. Nuclear PSMC5 expression was high in pancreatic cancers (70%) with breast cancers (35%) the next most frequent cancer examined. It was interesting to see that there was significantly more cytoplasmic MYPT1 staining in ER+ (39%) compared to HER2+ (13%) and basal triple-negative (6%), breast cancer subtypes, whereas cytoplasmic PSMC5 was common in all types of breast cancer: basal (triple-negative, 94%), HER2+ (82%) and ER+ (88%). (Supplemental Fig. S5, A-D). Although these proteins are expressed in other cancer types, we found greater expression of cytoplasmic and membranous MYPT1 by pancreatic cancers/PDAs when compared to the other tumor types (P < 0.01). In addition, nuclear PSMC5 expression seems to be PDA-specific (P < 0.01). Thus, by utilizing differences in nuclear PSMC5, and cytoplasmic and membranous MYPT1 staining among various cancers, we can develop a differential diagnosis panel model for PDA that can be further validated in the clinic.
Figure 4. Evaluating MYPT1 and PSMC5 expression in biliary, lung, liver, colon and breast cancers using tissue microarrays.
(A) Comparing cytoplasmic and membranous MYPT1 staining in PDA (n = 44) versus biliary (n = 90), lung (n = 91), liver (n = 36), colon (n = 72) and breast (n = 54) cancers. Both cytoplasmic as well as membranous expression of MYPT1 is PDA-specific (P < 0.01). (B) Comparing cytoplasmic and nuclear PSMC5 staining in PDA (n = 45) versus biliary (n = 82), lung (n = 83), liver (n = 36), colon (n = 57) and breast cancers (n = 54). Nuclear expression of PSMC5 is PDA-specific (P < 0.01). However, cytoplasmic expression of PSMC5 is significant expressed in PDA compared to 72% lung, 59% liver and 48% colon but not 88% breast cancer.
DISCUSSION
We have developed a reliable quantitative proteomics approach we called “SASI”, to identify and categorize proteins that are potential therapeutic targets in pancreatic cancers. The SASI screen identified more than 2500 proteins, including those that are recognized by vaccine-induced differentially expressed antibodies found only in patients responding to therapy. This approach also identified new PDA-associated proteins that can be used to differentiate PDA from pre-malignant lesions, and from other non-PDA cancers.
We validated the design and application of this SASI approach in three distinct ways. First, SILAC ratios were mirrored by western blot analyses for the corresponding proteins. Second, SILAC identified post-vaccine–induced antibody responses to allogeneic HLA molecules, an expected finding (the vaccine comprises allogeneic whole tumor cells), allowing the allogeneic response to serve as a natural positive control. Third, three proteins (annexin a2, pyruvate kinase, and galectin 3) that were previously identified using a less specific 2-D approach were also identified by SASI (8, 11). All three proteins are being explored for biologic relevance in PDA. As an example, antibody responses to annexin A2 correlate with improved overall survival following GVAX therapy. In addition, we recently reported that annexin A2 induces epithelial mesenchymal transition (EMT) thereby facilitating metastases in a mouse model of PDA, and annexin A2 antibody therapy reduces the incidence of metastases (11).
Our results imply that the vaccine-induced antibody response to PSMC5, MYPT1, and TFRC may be a marker of clinical benefit. However, further studies with more patient samples are necessary to validate our findings. An increased antibody response post-vaccination correlates to a longer and favorable DFS, whereas a decreased response post-vaccination correlates to a shorter and unfavorable DFS. The data also suggests that these proteins are antigenic targets of vaccine-induced humoral responses in PDA patients. Most significantly, the antibody responses detected against these proteins in patients with DFS > 3 years suggests an anti-tumor potential of targeting these proteins.
We also noted a reduced antibody response to many proteins in patients that had vaccine-induced antibody responses. It is not clear if responders had serologic tolerance or if developing antibodies to a small number of proteins is critical to the response. These questions are under investigation.
It is interesting to note that the SASI approach identified PDA-associated proteins located in a number of sites within PDA cells. Our expression studies revealed that cytoplasmic and nuclear PSMC5, and cytoplasmic and membranous MYPT1 and TFRC, are preferentially expressed in PDA compared to normal pancreas tissue. Thus, this SASI approach is sensitive enough to identify differentially expressed proteins between malignant and nonmalignant cells. However, the mechanism by which antibody responses can be induced to nuclear and cytoplasmic proteins in this case is unclear. It is possible that GVAX-induced lymphoid aggregates (B-cells and T-cells observed to be infiltrating PDAs (12)), facilitate B cell responses to lysed tumor cells. Finally, TFRC is differentially expressed in PDA and other cancers (21), providing additional support for the SASI approach in identifying differentially expressed proteins.
TMA analyses revealed that expression of nuclear PSMC5, and cytoplasmic and membranous MYPT1, is highly specific for PDA compared to other cancers. The other cancer types analyzed that also stained positive for these markers are gastrointestinal-derived adenocarcinomas with similar morphologies. Colon cancer is one of the most common adenocarcinomas. Biliary cancer's presentation can overlap with PDA. Thus, by combining PSMC5 and MYPT1 staining, we can develop a differential diagnosis method. We have shown that PDA stains strongly for both PSMC5 and MYPT1. From the other tumors studied, only colon cancer stains highly for both markers. However, the nuclear staining seen in PDA is not as substantial in colon cancers. Thus, we can distinguish one cancer from the other using these two markers.
In summary, the SASI approach can be used to identify proteins that have a differential sera-antibody response among different patients or pre- and post-treatment from the same patient. Specifically, this approach can identify prognostic/predictive biomarkers of response to targeted therapies. In addition, SASI can identify protein targets of antibody responses associated with improved survival in patients with cancer. PDA-specific staining of cytoplasmic and membranous MYPT1 and nuclear PSMC5 can be coupled to the current PDA diagnosis protocol to improve specificity and sensitivity in successfully diagnosing PDA. Finally, these markers also hold potential to serve as novel therapeutic targets for PDA treatment. Future studies will include the testing of monoclonal antibody therapies targeting these proteins for treating PDA.
Supplementary Material
Acknowledgments
We gratefully acknowledge Drs. Christine Iacobuzio-Donahue, Edward Gabrielson and Peter Argani for providing the TMAs (all Johns Hopkins University School of Medicine). The authors acknowledge the joint participation by the Adrienne Helis Malvin Medical Research Foundation and the Diana Helis Henry Medical Research Foundation through its direct engagement in the continuous active conduct of medical research in conjunction with the Johns Hopkins Hospital, the Johns Hopkins University School of Medicine and the Foundation's Parkinson's Disease Programs.
FUNDING: This work was supported by the NIH K23 CA148964-01 (to L. Zheng), Johns Hopkins School of Medicine Clinical Scientist Award (to L. Zheng), an American Society of Clinical Oncology Young Investigator Award (to L. Zheng), Viragh Foundation and the Skip Viragh Pancreatic Cancer Center at Johns Hopkins (to D. Laheru, E.M. Jaffee, and L. Zheng), The National Pancreas Foundation (to L. Zheng), Lefkofsky Family Foundation (to L. Zheng and E. Jaffee), the NCI SPORE in Gastrointestinal Cancers P50 CA062924 (to E.M. Jaffee, L. Zheng, A. P. Klein and D Laheru), Lustgarten Foundation (to E.M. Jaffee and L. Zheng), and the Sol Goldman Pancreatic Cancer Center (L. Zheng). NIH 5K23 CA163672-02 (to D. T. Le) E.M. Jaffee is the first recipient of the Dana and Albert “Cubby” Broccoli Endowed Professorship. This study was also supported in part by Grant No. S10RR023025 from the High End Instrumentation Program of the NIH. NCI's Clinical Proteomic Tumor Analysis Consortium initiative (U24CA160036), a NIH grant (CA62924), a contract (HHSN268201000032C) from the National Heart, Lung and Blood Institute and the Sol Goldman Pancreatic Cancer Research Center.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST: Through a licensing agreement between Aduro Biotech and JHU, Dr. Jaffee could potentially receive royalties in the future on GVAX and Listeria vaccines for cancer. Dr. Jaffee receives research funding from ROCHE, Bristol-Meyers-Squibb and Aduro Biotech. R. Anders reports receiving commercial research support from FivePrime, Bristol Meyers Squibb, and is on the advisory board for Adaptive Biotechnologies.
Author contributions: D.T.J., M.S.K., L.H., R.S., L.Z., D.T.L., D.A.L., A.P., E.M.J., and R.A.A. conceived and designed the experiments. D.T.J., M.S.K., and R.S. performed the experiments. D.T.J., M.S.K., E.D.T., R.S., A.K., E.A.S., A.P., E.M.J., and R.A.A. analyzed the data. D.T.J., M.S.K., A.P., E.M.J., and R.A.A. wrote the manuscript.
Competing interests: Under a licensing agreement between Aduro BioTech, Inc. and the Johns Hopkins University, Dr. Elizabeth M. Jaffee and the University are entitled to milestone payments and royalty on sales of the vaccine product described in this manuscript. We do not have any other relevant conflicts of interest to be disclosed.
REFERENCES
- 1.C. Pierantoni C, Pagliacci A, Scartozzi M, Berardi R, Bianconi M, Cascinu S. Pancreatic cancer: progress in cancer therapy. Critical Reviews in Oncology/Hematology. 2008;67:27–38. doi: 10.1016/j.critrevonc.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 2.Press release “Survival Rate for Pancreatic Cancer Remains Unchanged While Other Leading Cancers See An Increase in Their Relative Survival Rates: January 5, 2012. The Association; Manhattan Beach, CA: c1999-2015. Pancreatic Cancer Action Network [Internet]. Available from: https://www.pancan.org/section-about/news-press-center/2012-press-releases/survival-rate-for-pancreatic-cancer-remains-unchanged-while-other-leading-cancers-see-an-increase-in-their-relative-survival-rates/ [Google Scholar]
- 3.Burris HA, 3rd, Moore MJ, Andersen J, Green MR, Rothenberg ML, Modiano MR, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. Journal of Clinical Oncology. 1997;15:2403–13. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 4.Conroy T, Desseigne F, Ychou M, Bouche O, Guimbaud R, Becouarn Y, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. The New England Journal of Medicine. 2011;364:1817–25. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 5.Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. The New England Journal of Medicine. 2013;369:1691–1703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lutz E, Yeo CJ, Lillemoe KD, Biedrzycki B, Kobrin B, Herman J, et al. A lethally irradiated allogeneic granulocyte-macrophage colony stimulating factor-secreting tumor vaccine for pancreatic adenocarcinoma. A Phase II trial of safety, efficacy, and immune activation. Annals of Surgery. 2011;253:328–35. doi: 10.1097/SLA.0b013e3181fd271c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas AM, Santarsiero LM, Lutz ER, Armstrong TD, Chen YC, Huang LQ, et al. Mesothelin-specific CD8(+) T cell responses provide evidence of in vivo cross-priming by antigen-presenting cells in vaccinated pancreatic cancer patients. The Journal of Experimental Medicine. 2004;200:297–306. doi: 10.1084/jem.20031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng L, Jaffee EM. Annexin A2 is a new antigenic target for pancreatic cancer immunotherapy. Oncoimmunology. 2012;1:112–4. doi: 10.4161/onci.1.1.18017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imai K, Takaoka A. Comparing antibody and small-molecule therapies for cancer. Nature Reviews Cancer. 2006;6:714–27. doi: 10.1038/nrc1913. [DOI] [PubMed] [Google Scholar]
- 10.Walton SM, Gerlinger M, de la Rosa O, Nuber N, Knights A, Gati A, et al. Spontaneous CD8 T cell responses against the melanocyte differentiation antigen RAB38/NY-MEL-1 in melanoma patients. Journal of Immunology. 2006;177:8212–18. doi: 10.4049/jimmunol.177.11.8212. [DOI] [PubMed] [Google Scholar]
- 11.Zheng L, Foley K, Huang L, Leubner A, Mo G, Olino K, et al. Tyrosine 23 phosphorylation-dependent cell-surface localization of annexin A2 is required for invasion and metastases of pancreatic cancer. PloS One. 2011;6:e19390. doi: 10.1371/journal.pone.0019390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lutz E, Wu A, Bigelow E, Sharma R, Mo G, Soares K, et al. Immunotherapy converts nonimmunogenic pancreatic tumors into immunogenic foci of immune regulation. Cancer Immunology Research. 2014;2:616–31. doi: 10.1158/2326-6066.CIR-14-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaffee EM, Schutte M, Gossett J, Morsberger LA, Adler AJ, Thomas M, et al. Development and characterization of a cytokine-secreting pancreatic adenocarcinoma vaccine from primary tumors for use in clinical trials. Cancer J. Sci. Am. 1998;4:194–203. [PubMed] [Google Scholar]
- 14.Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, et al. A draft map of the human proteome. Nature. 2014;509:575–81. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steinhardt AA, Gayyed MF, Klein AP, Dong J, Maitra A, Pan D, et al. Expression of Yes-associated protein in common solid tumors. Human pathology. 2008;39:1582–89. doi: 10.1016/j.humpath.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harsha HC, Molina H, Pandey A. Quantitative proteomics using stable isotope labeling with amino acids in cell culture. Nature Protocols. 2008;3:505–16. doi: 10.1038/nprot.2008.2. [DOI] [PubMed] [Google Scholar]
- 17.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kijanka G, Murphy D. Protein arrays as tools for serum autoantibody marker discovery in cancer. Journal of Proteomics. 2009;72:936–44. doi: 10.1016/j.jprot.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Desmetz C, Mange A, Maudelonde T, Solassol J. Autoantibody signatures: progress and perspectives for early cancer detection. Journal of Cellular and Molecular Medicine. 2011;15:2013–24. doi: 10.1111/j.1582-4934.2011.01355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casal JI, Barderas R. Identification of cancer autoantigens in serum: toward diagnostic/prognostic testing? Molecular Diagnosis & Therapy. 2010;14:149–54. doi: 10.1007/BF03256367. [DOI] [PubMed] [Google Scholar]
- 21.Tan HT, Low J, Lim SG, Chung MC. Serum autoantibodies as biomarkers for early cancer detection. The FEBS Journal. 2009;276:6880–904. doi: 10.1111/j.1742-4658.2009.07396.x. [DOI] [PubMed] [Google Scholar]
- 22.Ryschich E, Huszty G, Knaebel HP, Hartel M, Buchler MW, Schmidt J. Transferrin receptor is a marker of malignant phenotype in human pancreatic cancer and in neuroendocrine carcinoma of the pancreas. European Journal of Cancer. 2004;40:1418–22. doi: 10.1016/j.ejca.2004.01.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.